
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 19 October 2022
Sec. Mucosal Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.952994
This article is part of the Research Topic Interplay Between Diets, Microbiota, Bacterial Metabolites and Host for Intestinal Health and Disease View all 17 articles
 Seika Hashimoto-Hill
Seika Hashimoto-Hill Luisa Colapietro
Luisa Colapietro Vivienne Woo
Vivienne Woo Simona Antonacci
Simona Antonacci Jordan Whitt
Jordan Whitt Laura Engleman
Laura Engleman Theresa Alenghat*
Theresa Alenghat*Although diet has long been associated with susceptibility to infection, the dietary components that regulate host defense remain poorly understood. Here, we demonstrate that consuming rice bran decreases susceptibility to intestinal infection with Citrobacter rodentium, a murine pathogen that is similar to enteropathogenic E. coli infection in humans. Rice bran naturally contains high levels of the substance phytate. Interestingly, phytate supplementation also protected against intestinal infection, and enzymatic metabolism of phytate by commensal bacteria was necessary for phytate-induced host defense. Mechanistically, phytate consumption induced mammalian intestinal epithelial expression of STAT3-regulated antimicrobial pathways and increased phosphorylated STAT3, suggesting that dietary phytate promotes innate defense through epithelial STAT3 activation. Further, phytate regulation of epithelial STAT3 was mediated by the microbiota-sensitive enzyme histone deacetylase 3 (HDAC3). Collectively, these data demonstrate that metabolism of dietary phytate by microbiota decreases intestinal infection and suggests that consuming bran and other phytate-enriched foods may represent an effective dietary strategy for priming host immunity.
Intestinal infections pose a serious threat to public health worldwide, with reported cases exceeding two billion and over one million deaths each year (1). These infections are a leading cause of death among children, and surviving children can suffer from long-term health consequences such as delayed growth and vaccine failure (2, 3). Given that the intestinal mucosa is a primary site of exposure for multiple pathogens, deciphering the pathways that guide intestinal defense is critical for developing new approaches for treating and preventing infection.
Nutrition affects various aspects of physiology, including the immune system, as demonstrated by a strong correlation between malnutrition and infection morbidity (4, 5). Dietary components such as carbohydrates, lipids, amino acids, vitamins, and minerals have each been implicated in the regulation of host defense against pathogens (6, 7). For example, consumption of a high-fat diet increases disease in rodents following infection with C. difficile, C. rodentium, or Listeria monocytogenes (8–10). Trillions of commensal microbes reside in the mammalian intestine and are collectively referred to as the microbiota. Increasing evidence indicates that the metabolism of dietary nutrients by resident commensal microbes alters host physiology (11). Microbial metabolism results in the production of metabolites and small-molecule intermediates that regulate the symbiotic relationship between the microbiota and host (12). For example, microbiota-derived metabolites such as short-chain fatty acids (SCFAs) (13–15), indoles (16, 17), secondary bile acids (18, 19), and siderophores (20) have been described to protect during mouse models of infection. However, despite evidence linking nutrition and immunity, mechanistic insights needed to guide how diet can be modified to optimize host immunity are limited (11).
Intestinal epithelial cells (IECs) reside at the direct interface between the host and commensal microbes and, therefore, carry the potential to critically respond to signals from the diet, microbiota, and luminal metabolites (21–23). IECs provide the first line of defense against invading pathogens with constitutive expression of defense molecules, including antimicrobial peptides (AMPs), reactive oxygen species, and mucins (24, 25). Germ-free and microbiota depletion studies have shown that commensal microbial signals are required for basal expression of many AMPs (26, 27). IEC expression of the AMP regenerating islet-derived protein 3γ (Reg3γ) requires signaling through Toll-like receptors (TLRs) and other microbiota-sensitive pathways (28). In addition, nutritional regulation of AMP expression has also been suggested, and the timing of food intake can significantly alter IEC expression of AMPs (29, 30). However, the dietary factors that cooperate with the microbiota to prime IEC-mediated defense are not well known.
Phytate is in various foods, including bran, legumes, seeds, and nuts, and is enriched in diets such as vegetarian and Mediterranean (31–33). Mineral-chelating properties of phytate have been historically discussed in relation to rickets, which is caused by reduced calcium and phosphorus availability (34). Although phytate is no longer considered a primary pathogenic factor in rickets, absorption interference of iron, zinc, and other minerals with high doses of phytate could occur in the context of mineral deficiency, leading phytate to commonly be termed an anti-nutrient (35). However, diets containing phytate in combination with sufficient minerals do not present with disorders related to trace mineral absorption (33).
Rice bran is an abundant by-product generated during rice milling that has been gaining popularity as a food supplement over the past ten years (36, 37). Here, we discovered that ingestion of rice bran reduced infection burden and related pathology in a murine Citrobacter rodentium infection, the mouse model for enteropathogenic and enterohemorrhagic E. coli infection prevalent in the human population. Rice bran contains high amounts of phytate. Interestingly, phytate supplementation similarly protected against intestinal infection. This diet-induced protection was dependent on microbial digestion of phytate and the production of phytate metabolites. Mechanistically phytate induced STAT3 activation and the downstream defense pathway in the IECs. Phytate-induced STAT3 activation was mediated by the metabolite-sensitive enzyme HDAC3 in an IEC-intrinsic manner. Collectively, these findings reveal new diet-microbiota interactions that promote innate intestinal immunity and mammalian defense against infection.
Rice bran has been proposed to provide broad health benefits ranging from weight loss, cancer prevention, and protection from infection (37–40), provoking the hypothesis that rice bran may alter susceptibility to pathogens like E. coli. To test this, littermate mice were exclusively fed a custom diet containing 20% rice bran or a matched control diet for four weeks, and then infected with Citrobacter rodentium (Figure 1A). C. rodentium is a murine intestinal pathogen with similar pathogenesis to enteropathogenic and enterohemorrhagic E. coli, two leading causes of food-borne illnesses in humans (41–44). Interestingly, mice ingesting rice bran exhibited significantly reduced C. rodentium in the intestinal lumen (Figure 1B) and colonic tissue (Figure 1C) post-infection compared to control diet-fed mice (Figures 1B, C). Differences in pathogen burden were observed by day 3 post-infection, suggesting that bran diet decreased initial C. rodentium colonization and replication in the large intestinal mucosa. Consistent with decreased infection, C. rodentium-induced changes in stool consistency were less severe in the rice bran-fed mice compared to control diet-fed mice (Figure 1D). Pathological features of C. rodentium, such as colonic epithelial hyperplasia and leukocyte infiltration, were observed in control diet-fed mice (Figure 1E). However, these histologic features of C. rodentium infection were diminished in bran-fed mice (Figure 1E). Taken together, these findings suggest that a component of rice bran may decrease susceptibility to bacterial infection.
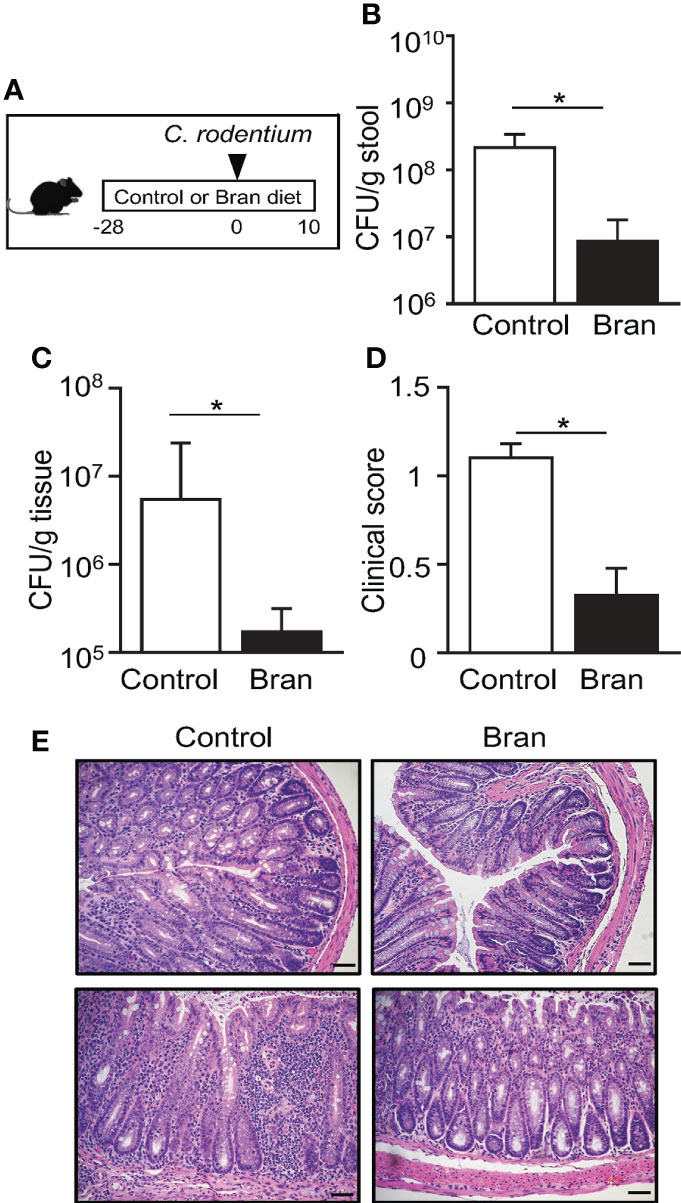
Figure 1 Consuming rice bran decreases susceptibility to C. rodentium infection. (A) Experimental approach. (B) Colony-forming units (CFUs) of C. rodentium in stool of infected control- or 20% rice bran diet-fed mice, normalized to sample weight, days 3 post-infection. (C) CFUs of C. rodentium in the colon tissues, day 10 post-infection. (D) Clinical scores representing severity of diarrhea, day 10 post-infection. (E) Histological staining of the colon tissues of infected mice, day 10 post-infection. Scale bars: 10 μm. Data are representative of 2-3 independent experiments. n = 4 per group. Results are mean ± SEM. *p < 0.05.
Rice bran is naturally enriched with phytate (31). Therefore, to test whether phytate itself alters C. rodentium infection, control mice and mice receiving 2% phytate were compared during infection (Figure 2A). Interestingly, similar to the outcome with rice bran ingestion, phytate consumption significantly decreased pathogen burden post-infection (Figure 2B). Consistent with decreased pathogen levels, clinical symptoms of infection were also less severe in phytate-treated mice relative to controls (Figure 2C). To examine pathology induced by C. rodentium infection, colonic tissue was examined following peak infection. As expected, C. rodentium induced colonic epithelial hyperplasia and leukocyte infiltration in control mice (Figure 2D). However, infection-associated pathology was reduced in mice fed phytate, recapitulating findings that occur with the rice bran diet (Figure 1). Collectively these data demonstrate that consuming phytate is sufficient to promote protection against intestinal bacterial infection.
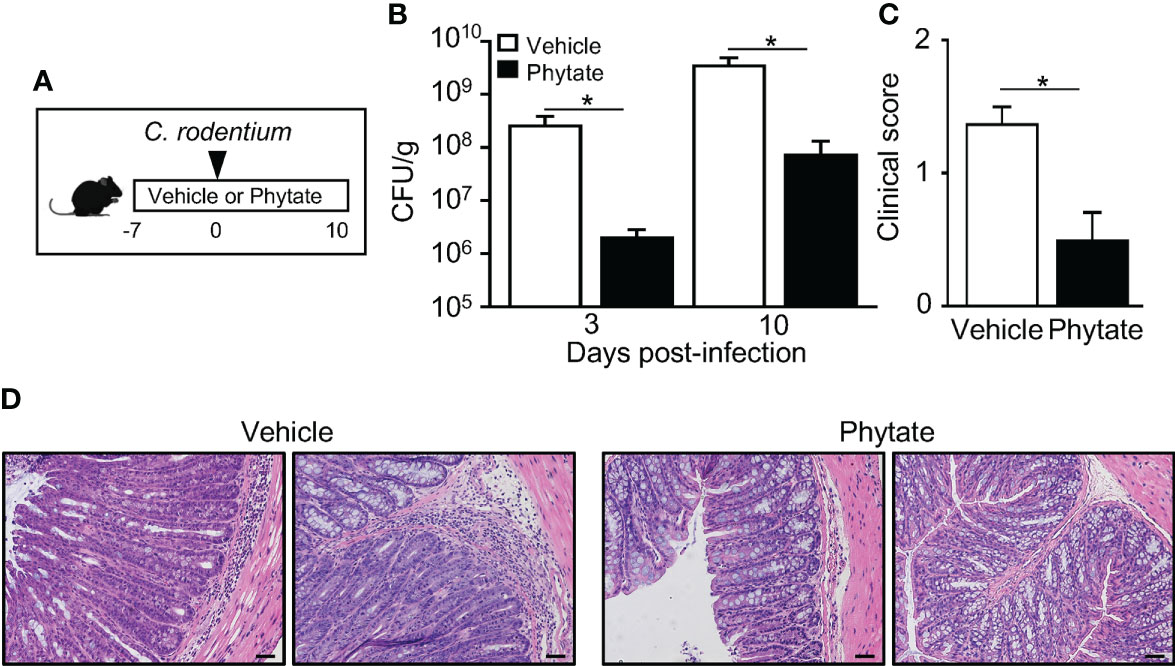
Figure 2 The natural component phytate in rice bran protects mice against C. rodentium. (A) Experimental approach. (B) CFUs of C. rodentium in stool of infected vehicle- or 2% phytate-treated mice, normalized to sample weight, days 3 and 10 post-infection. (C) Clinical scores representing severity of diarrhea, day 10 post-infection. (D) Histological staining of colon tissues of infected mice, day 10 post-infection. Scale bars: 10 μm. Data are representative of 2-3 independent experiments. n=4 per group. Results are mean ± SEM. *p < 0.05.
Monogastric mammals such as humans and mice do not produce the phytase enzyme that breaks down phytate in the intestinal lumen (45, 46). Instead, phytate digestion is dependent on phytase that is produced by bacteria residing in the intestine. Germ free (GF) animals exhibit lower concentrations of phytate metabolites in the gut lumen as microbial phytase catalyzes the removal of phosphorus from phytate to produce phosphorous and lower forms of inositol phosphates (47–49) (Figure 3A). Phytate supplementation to microbiota-replete, conventionally-raised (CNV) mice increased inositol trisphosphate (IP3) concentrations in intestinal contents (Figure 3B), confirming that commensal microbes in the mouse intestine break down phytate. To test whether microbiota are required for the phytate-mediated defense against C. rodentium infection, GF mice were treated with phytate prior to infection. (Figure 3C). Unlike CNV mice, GF mice exhibited comparable C. rodentium infection between vehicle and phytate-treated groups (Figure 3D). Given that phytate-induced protection was lost when mice lacked commensal microbes, the microbiota are required for regulation of host defense by phytate.
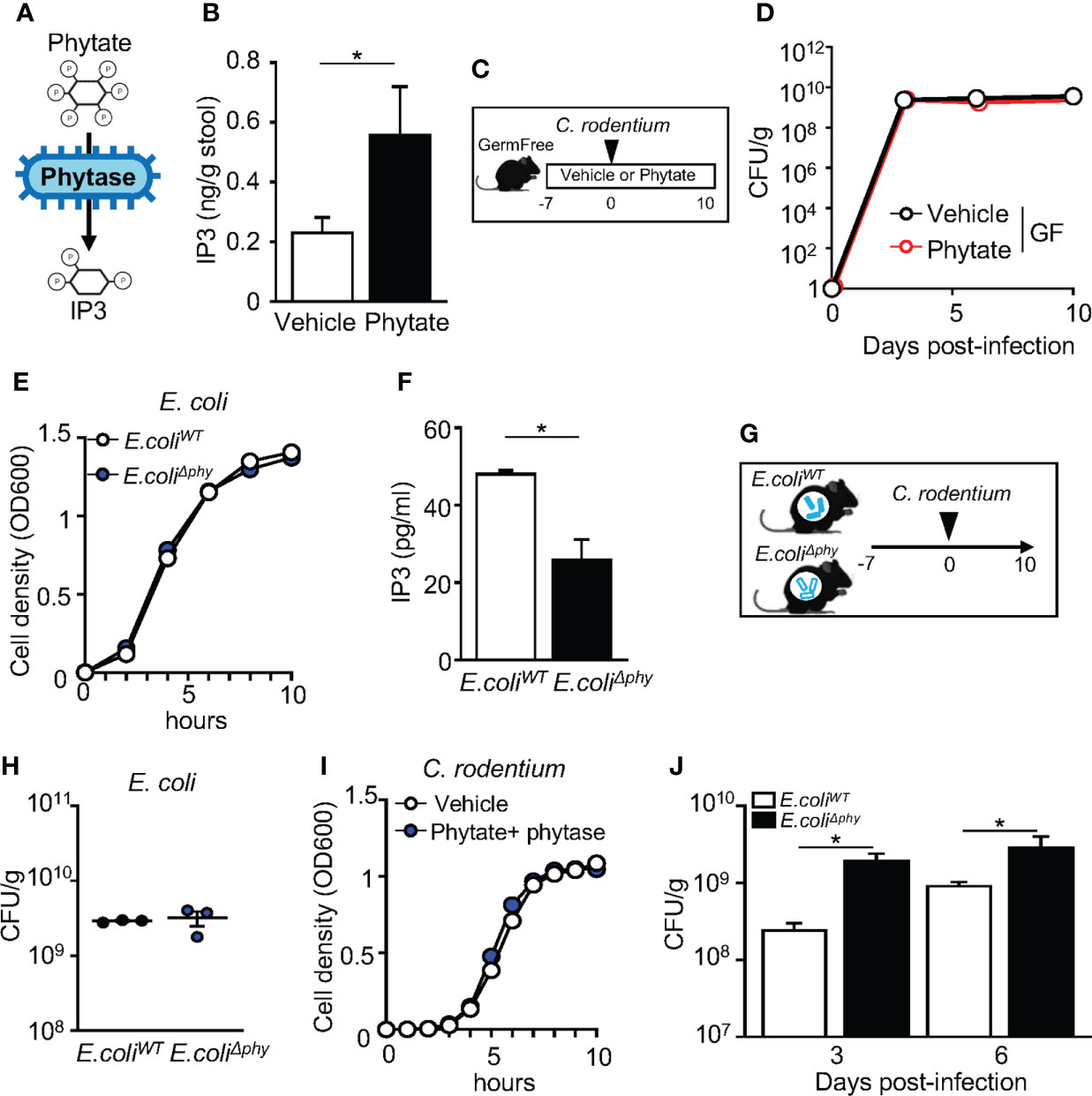
Figure 3 Metabolism of phytate by commensal bacteria mediates protection against infection. (A) Schematic of phytate metabolism by microbial phytase. (B) Inositol trisphosphate (IP3) concentration in fecal samples collected from vehicle- or 2% phytate-treated mice, normalized to sample weight. (C) Experimental approach. (D) CFUs of C. rodentium in stool of infected vehicle- or 2% phytate-treated GF mice, normalized to sample weight, days 3, 6 and 10 post-infection. (E) Bacterial cell density of E. coli. (F) IP3 concentration in media from E. coliWT and E. coliΔphy cultured with 1mM phytate, per 108 CFU of bacteria. (G) Experimental approach. (H) CFUs of E. coli in stool of monoassociated mice, normalized to sample weight, day 7 post-inoculation. (I) Bacterial cell density of C. rodentium. (J) CFUs of C. rodentium in stool of infected mice monoassociated with E. coliWT or E. coliΔphy, normalized to sample weight, days 3 and 6 post-infection. Data are representative of 2-3 independent experiments. n = 3-4 per group. Results are ± SEM. *p < 0.05.
These findings provoked the hypothesis that microbial-produced phytase is required for phytate-induced protection. To test this, we employed a commensal strain of E. coli that either expresses the wildtype phytase AppA gene (E. coliWT) or lacks the AppA gene (E. coliΔphy). To confirm phytase activity levels in these strains, E. coliWT and E. coliΔphy were cultured overnight in phytate-supplemented media, and the concentrations of IP3 in the supernatant were compared. E. coliWT and E. coliΔphy growth was similar (Figure 3E). However, significantly higher levels of IP3 were present in E. coliWT cultures compared to E. coliΔphy cultures, confirming impaired phytase activity in E. coliΔphy bacteria (Figure 3F). To next compare the role of bacterial phytase expression in vivo, GF mice were monoassociated with either E. coliWT or E. coliΔphy and fed phytate-containing chow (Figure 3G). Colonization was similar for both strains (Figure 3H), and phytate or phytase did not directly alter C. rodentium growth (Figure 3I). Remarkably, though, mice monoassociated with E. coliΔphy exhibited increased susceptibility to C. rodentium infection, relative to mice monoassociated with E. coliWT (Figure 3J), Therefore, phytase expressed by commensal bacteria enables dietary phytate to promote innate host defense against intestinal infection.
IECs provide initial defense against pathogenic infection in the intestine, in part through basal expression of antimicrobial molecules (50). Bran- and phytate-induced protection against C. rodentium occurred as early as day 3 post-infection, suggesting that phytate may regulate basal epithelial defense mechanisms. To test this hypothesis, global transcriptional profiles were compared in colonic IECs harvested from CNV mice treated with vehicle or phytate. These analyses identified numerous genes that were significantly upregulated or downregulated in IECs following phytate ingestion (Figure 4A). Interestingly, phytate significantly upregulated gene expression in host defense pathways. (Figures 4B, C). Phytate-induced defense genes included the antimicrobial peptide Reg3g and the bactericidal nitric oxide producer Nos2 (Figures 4C, D), both known mediators of early defense against C. rodentium (51, 52). Furthermore, IEC upregulation of Reg3g and Nos2 occurred in rice bran diet-fed mice compared to control mice (Figure 4E). Therefore, consuming rice bran or phytate induces basal epithelial antimicrobial defense mechanisms that protect against pathogenic bacterial infection in the intestine.
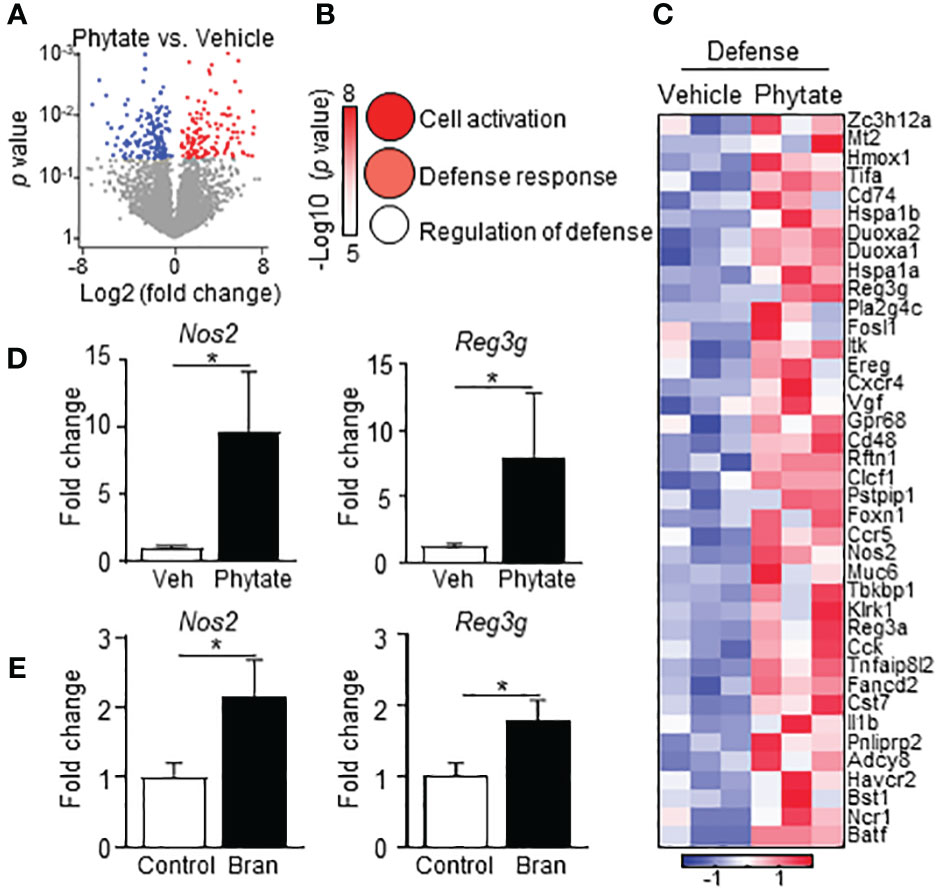
Figure 4 Phytate consumption primes elevated epithelial antimicrobial defense. (A) Volcano plot showing differentially expressed LI IEC genes between Vehicle- and 2% phytate-treated naïve mice identified by RNA-seq. Red: upregulated, blue: downregulated with phytate, p < 0.05. (B) Ontology of the genes upregulated with phytate in (A). Circle sizes correspond to gene numbers in pathways. (C) Heatmap of relative mRNA expression of IEC defense response genes upregulated in phytate. (D) mRNA expression levels in LI IEC, normalized to vehicle. (E) mRNA expression in large intestinal IECs, normalized to control. Data are representative of 2-3 independent experiments. n = 3-4 per group. Results are mean ± SEM. *p < 0.05.
To further dissect the mechanism of phytate-induced epithelial regulation, network analyses were conducted on the enriched phytate-induced defense genes. Interestingly, these analyses identified the transcription factor Signal Transducer And Activator Of Transcription 3 (STAT3) as the most central and essential factor for regulating this network (Figures 5A, B). Consistently, multiple known STAT3-controlled downstream genes were upregulated in IECs of phytate-fed mice (Figure 5C). This finding led to the hypothesis that phytate activates IEC-intrinsic STAT3. STAT3 activation is characterized by phosphorylation of its tyrosine 705 residue. Therefore, to test whether phytate alters STAT3 activation, pSTAT3 (Y705) in the large intestinal epithelium of control mice and mice receiving 2% phytate were compared. Interestingly, phytate feeding significantly increased phosphorylation of STAT3 in IECs relative to control mice (Figures 5D, E). Further, IECs harvested from rice bran-fed naïve mice also exhibited enhanced STAT3 activation (Figure 5F). Collectively, these data reveal that consuming phytate-enriched diets can increase epithelial STAT3 activation and therefore prime expression of antimicrobial targets that are regulated by STAT3.
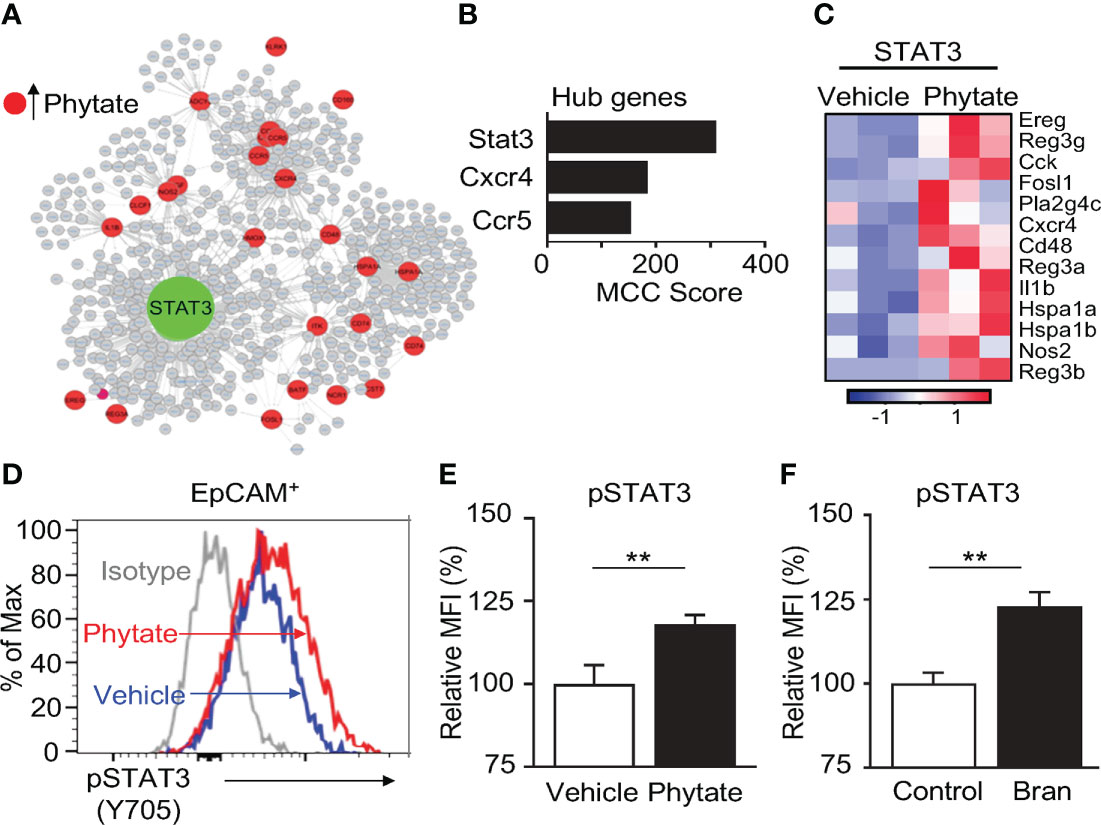
Figure 5 Epithelial STAT3 activation in the intestine is induced by phytate. (A) Interaction network of phytate-induced IEC defense genes. (B) Top hub genes in (A). (C) Heatmap of relative mRNA expression of IEC STAT3-target genes upregulated in phytate-treated. (D) Representative flow cytometry plots from IECs of vehicle- or 2% phytate-treated mice, gated on live EpCAM+ cells. (E, F) Mean Fluorescence Intensity (MFI) of pSTAT3 (Y705), normalized to vehicle (E) or control (F). Data are representative of 2-3 independent experiments. n=4 per group. Results are mean ± SEM. **p < 0.01.
Deacetylation of STAT3 by the histone deacetylase HDAC3 promotes STAT3 activation in hepatocytes and lymphoma cells (53, 54). HDAC3 is an enzyme sensitive to environmental signals, and IEC expression of HDAC3 is critical in host defense against C. rodentium (55, 56). The phytate-derived metabolite inositol trisphosphate activates HDAC3 (47), and consistent with this, phytate-fed mice displayed increased mucosal HDAC activity relative to vehicle-fed mice (Figure 6A). Therefore, we first hypothesized that HDAC3 regulates STAT3 activation in IECs. To test this hypothesis, we utilized an IEC-specific HDAC3 knockout mouse model (57). Interestingly, IECs isolated from mice lacking IEC-intrinsic HDAC3 (HDAC3ΔIEC) displayed displayed significantly lower pSTAT3 levels relative to floxed littermate mice (HDAC3FF), indicating that STAT3 activation in IECs requires HDAC3 (Figures 6B, C). We next hypothesized that phytate-induced STAT3 activation was HDAC3-dependent. To test this, HDAC3ΔIEC mice and littermate HDAC3FF mice were treated with 2% phytate, and the levels of IEC pSTAT3 (Y705) were compared. The results exhibited that phytate ingestion increased STAT3 activation in IECs of HDAC3FF mice, but not IECs of HDAC3ΔIEC mice (Figure 6D). Taken together, these data demonstrate that dietary phytate mechanistically induces activation of epithelial STAT3 through regulation of HDAC3.
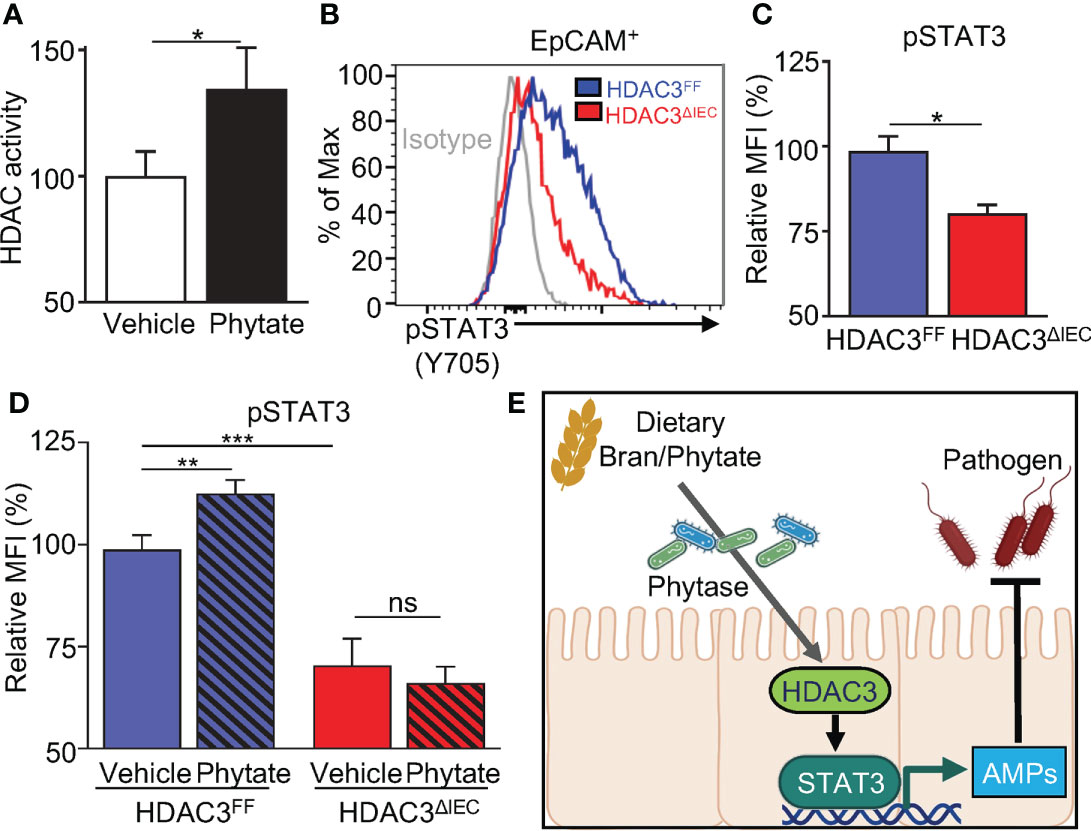
Figure 6 Phytate-mediated epithelial STAT3 activation requires HDAC3. (A) Intestinal epithelial HDAC activity of vehicle- or phytate-treated mice. (B) Representative flow cytometry plots from IECs of vehicle- or 2% phytate-treated mice, gated on live EpCAM+ cells. (C) Mean Fluorescence Intensity (MFI) of pSTAT3 (Y705), normalized to FF. (D) MFI of IEC pSTAT3 of vehicle- or 2% phytate-treated HDAC3FF or HDAC3ΔIEC mice, normalized to FF-vehicle. (E) Through phytate metabolism by microbial phytases, rice bran and its component phytate activate IEC STAT3 and downstream defense mechanisms against enteric infection. Data are representative of 2-3 independent experiments. n=4 per group. Results are mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
In this study, we identified that consumption of rice bran or phytate induces innate intestinal epithelial pathways that decrease susceptibility to bacterial infection (Figure 6E). Furthermore, this diet-induced protection is mediated by the commensal bacterial metabolism of phytate and subsequent activation of the HDAC3-STAT3 axis in epithelial cells (Figure 6E). Phytate-induced activation of IEC-intrinsic STAT3 required expression of HDAC3, as phytate failed to induce STAT3 activation in the absence of HDAC3 expression. Direct deacetylation of the lysine 685 residue of STAT3 by HDAC3 has been described to precede phosphorylation and nuclear translocation of STAT3 in hepatocytes (53), however, indirect mechanisms may also contribute. Similarly, inhibition of HDAC3 in lymphoma cells resulted in nuclear export of STAT3 to the cytoplasm, thus hindering STAT3 function as a transcriptional factor (54). Similar to these cells, our analyses demonstrate that loss of HDAC3 impairs STAT3 activation in IECs.
Although phytate induces HDAC3-mediated activation of STAT3 in IECs, involvement of other STAT3 activators, such as IL-22, IL-18, and IL-1β (58–60) cannot be excluded. IL-22, a well-characterized STAT3 activator, is produced by several different immune cells in the intestine in response to cytokines IL-6 and IL-23, and aryl hydrocarbon receptor (AhR) ligands (51, 61, 62). Further, vitamin A, tryptophan metabolites, and short-chain fatty acids have been shown to induce IL-22 production from intestinal immune cells (63–65). In addition, IECs can propagate the immune cell IL-22 response through secretion of chemoattractant resistin-like molecule-beta (RELM-β) that recruits IL-22 producing CD4+ T cells to the sites of infection (66). Thus, beyond epithelial-intrinsic regulation, future studies will be necessary to determine the contribution of phytate to local and systemic immune cell responses that promote host defense. This will include extending the analyses of phytate effects on immune follicles and mucin-producing goblet cells (66), as well as additional sites such as the small intestine and extra-intestinal tissue.
Constituents of the microbiota can alter susceptibility to pathogenic C. rodentium infection through direct inhibition via nutritional competition or production of bacteriostatic molecules such as butyrate and sulfide (13, 67, 68). Phytate supplementation did not result in a significant deviation of microbiota composition or luminal SCFAs concentrations compared to vehicle-supplemented mice in dextran sulfate sodium (DSS)-treated mice (47). On the other hand, increased Lactobacillus spp. has been reported with phytate supplementation in high sucrose-fed rats (69). Increased levels of Lactobacillus spp. have also been reported with 4-week consumption of 10% bran diet in rodents (70). Microbial phytases encompass multiple classes of enzymes and are expressed by a variety of commensal bacterial and fungal species in the intestinal lumen (71). Our gnotobiotic model employing a phytase-deficient commensal strain demonstrated that microbial phytase was essential in protection against infection, verifying the importance of phytate and its metabolites in the regulation of host epithelial defense. Phytase deficiency in commensal E. coli did not result in growth disadvantage in the monocolonization model in our study. However, it is possible that commensal bacterial phytase-derived products also directly alter C. rodentium virulence and therefore not only promote protection via regulation of host immunity. Elucidating the association between phytase abundance and individuals’ responsiveness to dietary phytate may be essential for guiding the use of phytate for nutritional intervention strategies.
The data presented in this manuscript reveal a new mechanism of dietary regulation of mucosal immunity in which metabolism of phytate-rich foods by commensal bacteria primes epithelial defense against infection via an HDAC3-STAT3 pathway. Evidence also suggests that rice bran decreases susceptibility to other enteric pathogens, such as Salmonella and rotavirus (40, 72). Therefore, consuming bran and other phytate-enriched foods may represent an effective dietary strategy for broadly boosting innate mucosal defense against multiple intestinal pathogens. Discovery of this diet-microbiota-host regulatory mechanism provides fundamental insights that can guide practical and personalized nutritional interventions designed to enhance mucosal immunity.
All murine experiments were performed according to guidelines of the Institutional Animal Care and Use Committee. Animals were housed up to 4 per cage in a ventilated cage with 12 h light/dark cycle and free access to chow and water. Animals were provided with appropriate care by a licensed veterinarian. Floxed Hdac3 mice were bred to C57BL/6 mice expressing Cre-recombinase under the control of villin promoter to generate HDAC3ΔIEC mice (57). Gnotobiotic mice were maintained in sterile isolators (Class Biologically Clean) in the CCHMC Gnotobiotic Mouse Facility, fed autoclaved food and water, and routinely monitored to ensure the absence of microbial contamination. To establish E.coli and phytase-KO E. coli monoassociated mice, GF mice received 1 x109 CFU bacteria in PBS via oral gavage. Monoassociated mice were housed on a sealed positive pressure IVC rack (Allentown). Phytate (phytic acid sodium salt hydrate, Sigma) was dissolved in water to make 2% phytate and filtered (0.22μm) prior to providing to the mice as drinking water. Control diet (TD.160791) or 20% rice bran (NOW stabilized rice bran) diet (TD.210517) were custom made by Envigo with matched macronutrients, calories, minerals, vitamins, and fiber. Mice were infected with 1x109 CFU GFP-C. rodentium (DBS100) via oral gavage. Stool and colon tissues were homogenized in sterile PBS using a Tissue Lyser II, serially diluted and plated on MacConkey agar. CFUs were counted and normalized to stool weight after 16 hr. For histologic analyses, sections of colon were fixed in 10% neutralized formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin.
IECs were isolated from the large intestine by shaking tissue in 1mM EDTA/1mM DTT 5% FBS PBS at 37°C for 10 min as described previously (57). RNA was extracted from cells using the RNeasy Kit (Qiagen) according to manufacturer’s instructions. For RT-qPCR, RNA was reverse-transcribed with Verso reverse transcriptase (Invitrogen) and expression was compared using SYBR (Applied Biosystems) and analyzed in the linear range of amplification. Target gene expression was normalized to an unaffected control gene. For global expression analyses, 3 biological replicates of IECs from vehicle- and phytate-treated mice were compared. Reads were bar codes trimmed and mapped to mouse genome (GRCm38) using Bowtie2. The reads aligning to known transcripts were quantified using Seqmonk (V1.47.1) and visualized using Genepattern Multiplot Studio. Differential expression analysis was performed using EdgeR within Seqmonk (p<0.05, fold change >1.5). For pathway and ontological analyses, gene lists were submitted to the Toppgene database (toppgene.cchmc.org), which amasses ontological data from over 30 individual repositories. Network construction and identification of hub genes using Cytohubba were done on Cytoscape v3.9.1.
IECs were isolated as described above. Cells were stained using the following fluorescence-conjugated monoclonal antibodies diluted in FACS buffer (2% FBS, 0.01% sodium azide, PBS): Brilliant Violent 711 anti-CD326 (EpCAM) (Clone: G8.8, BD Biosciences), PE anti-pSTAT3 (Tyr 705) (Clone: LUVNKLA, Invitrogen), PE anti-Mouse IgG2bκ isotype (Clone: eBMG2b, Invitrogen). Dead cells were excluded with the Violet dead Cell Stain Kit (Invitrogen). For pSTAT3 staining, the cells were fixed in 4% PFA following dead cell staining, then permeabilized with methanol prior to surface and pSTAT3 or isotype staining. Samples were acquired on the Canto III and analyzed with FlowJo™ v10.8 Software (BD Life Sciences).
Fecal pellets were homogenized in cold PBS and extract was collected after centrifugation at 4°C. IP3 ELISA was performed on fecal extract or bacterial culture supernatant according to manufacturer instructions (MyBiosource). Briefly, samples were incubated with 50 µl of biotinylated detection antibody for 45 minutes at 37°C, washed 3 times, and incubated with HRP conjugate at 37°C for 30 minutes. The plate was rinsed with wash buffer followed by substrate incubation for 15 minutes at 37°C. The reaction was stopped, and the optical density of each well was measured using a micro-plate reader (Biotek Synergy 2) set to 450 nm.
Wild type commensal E. coli K-12 (ATCC 700926) and phytase-KO E. coli CU-1867 (ATCC 47092) colonies were grown on LB agar plate and inoculated in 10 ml LB broth for overnight culture prior to administration to GF mice in mono-association studies. For culture media IP3 quantification study, E. coli were cultured with 1 mM phytate. Mono-associated mice were monitored for contamination by quantitative PCR. Fecal samples were collected in 2ml pre-weighed sterile microcentrifuge tubes. Fecal bacterial DNA was isolated using QIAamp® Fast DNA Stool Mini Kit (Qiagen) following the kit protocol. Bacterial DNA was assessed by quantitative PCR (QuantStudio3; Applied Biosystems) using 16S-rRNA and bacterial-specific primer pairs (Invitrogen, MilliporeSigma). PCR of E. coli phytase gene (AppA) was performed to confirm lack of AppA in phytase-KO E. coli (47). To determine CFUs in mono-associated mice, stool was homogenized, serially diluted, and grown on Lennox LB plates in aerobic condition overnight. CFUs were normalized to stool weights.
LI mucosa was lysed in RIPA buffer and HDAC activity was assayed using a fluorometric assay (Active Motif). Briefly, 10 μg of cell lysate was incubated with 100 μM of HDAC substrate at 37°C for 1 hour. 50 μl of developer solution containing 2 μM of trichostatin A (TSA) was added at room temperature to stop the reaction. Fluorescence was measured using a fluorescent plate reader (Biotek Synergy 2) with an excitation wavelength of 340 nm and an emission wavelength of 460 nm.
All statistical analyses were performed using GraphPad Prism 8.0. Statistical significance was determined by students t-test or ANOVA. All data meet the assumptions of the statistical tests used. Results are shown as mean ± SEM and considered significant at p<0.05 (*); p<0.01 (**); p<0.001 (***).
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Cincinnati Children’s Institutional Animal Care and Use Committee.
TA and SH-H designed the studies and analyzed the data. SH-H, LC, VW, LE, SA, and JW carried out experiments. TA and SH-H wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research is supported by the National Institutes of Health (DK114123, DK116868 to TA). TA holds a Kenneth Rainin Foundation Award and Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. This project is supported in part by PHS grant P30 DK078392.
We thank the Way, Qualls, and Deshmukh labs for useful discussions and members of the Alenghat lab for critical reading of the manuscript. We thank CCHMC Veterinary Services, Research Flow Cytometry, and Pathology Research Core for services and technical assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Troeger C, Blacker BF, Khalil IA, Rao PC, Cao S, Zimsen SRM, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis [Internet]. (2018) 18(11):1211–28. doi: 10.1016/S1473-3099(18)30362-1
2. Troeger C, Colombara DV, Rao PC, Khalil IA, Brown A, Brewer TG, et al. Global disability-adjusted life-year estimates of long-term health burden and undernutrition attributable to diarrhoeal diseases in children younger than 5 years. Lancet Glob Heal [Internet]. (2018) 6(3):e255–69. doi: 10.1016/S2214-109X(18)30045-7
3. Naylor C, Lu M, Haque R, Mondal D, Buonomo E, Nayak U, et al. Environmental Enteropathy, Oral Vaccine Failure and Growth Faltering in Infants in Bangladesh. EBioMedicine [Internet]. (2015) 2(11):1759–66. doi: 10.1016/j.ebiom.2015.09.036
4. Ibrahim MK, Zambruni M, Melby CL, Melby PC. Impact of childhood malnutrition on host defense and infection. Clin Microbiol Rev (2017) 30(4):919–71. doi: 10.1128/CMR.00119-16
5. Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nat [Internet]. (2011) 474(7351):327–36. doi: 10.1038/nature10213
6. Cao Y, Liu J, Zhu W, Qin N, Ren X, Zhu B, et al. Impact of dietary components on enteric infectious disease. Crit Rev Food Sci Nutr (2021) 62, 1–26. doi: 10.1080/10408398.2021.1871587
7. Nobs SP, Zmora N, Elinav E. Nutrition regulates innate immunity in health and disease. Annu Rev Nutr (2020) 40:189–219. doi: 10.1146/annurev-nutr-120919-094440
8. An J, Zhao X, Wang Y, Noriega J, Gewirtz AT, Zou J. Western-style diet impedes colonization and clearance of Citrobacter rodentium. PloS Pathog [Internet]. (2021) 17(4):e1009497. doi: 10.1371/journal.ppat.1009497
9. Moore JH, Pinheiro CCD, Zaenker EI, Bolick DT, Kolling GL, van Opstal E, et al. Defined nutrient diets alter susceptibility to Clostridium difficile associated disease in a murine model. PloS One (2015) 10(7):e0131829. doi: 10.1371/journal.pone.0131829
10. Las Heras V, Clooney AG, Ryan FJ, Cabrera-Rubio R, Casey PG, Hueston CM, et al. Short-term consumption of a high-fat diet increases host susceptibility to Listeria monocytogenes infection. Microbiome (2019) 7(1):1–12. doi: 10.1186/s40168-019-0621-x
11. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol (2019) 16(1):35–56. doi: 10.1038/s41575-018-0061-2
12. McCarville JL, Chen GY, Cuevas VD, Troha K, Ayres JS. Microbiota Metabolites in Health and Disease. Annu Rev Immunol (2020) 38(1):147–70. doi: 10.1146/annurev-immunol-071219-125715
13. Osbelt L, Thiemann S, Smit N, Lesker TR, Schröter M, Gálvez EJC, et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PloS Pathog (2020) 16(3):e1008448. doi: 10.1371/journal.ppat.1008448
14. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe (2016) 20(2):202–14. doi: 10.1016/j.chom.2016.07.001
15. Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe (2018) 24(2):296–307.e7. doi: 10.1016/j.chom.2018.07.002
16. Metidji A, Omenetti S, Crotta S, Li Y, Nye E, Ross E, et al. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity (2018) 49(2):353–362.e5. doi: 10.1016/j.immuni.2018.07.010
17. Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, et al. Feedback control of AHR signalling regulates intestinal immunity. Nature (2017) 542(7640):242–245J. doi: 10.1038/nature21080
18. Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. MSphere (2016) 1(1):e00045–15. doi: 10.1128/mSphere.00045-15
19. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature (2015) 517(7533):205–8. doi: 10.1038/nature13828
20. Sassone-Corsi M, Chairatana P, Zheng T, Perez-Lopez A, Edwards RA, George MD, et al. Siderophore-based immunization strategy to inhibit growth of enteric pathogens. Proc Natl Acad Sci U.S.A. (2016) 113(47):13462–7. doi: 10.1073/pnas.1606290113
21. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol (2014) 14(3):141–53. doi: 10.1038/nri3608
22. Ramanan D, Cadwell K. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe (2016) 19(4):434–41. doi: 10.1016/j.chom.2016.03.003
23. Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, et al. A Map of Toll-like Receptor Expression in the Intestinal Epithelium Reveals Distinct Spatial, Cell Type-Specific, and Temporal Patterns. Immunity (2018) 49(3):560–575.e6. doi: 10.1016/j.immuni.2018.07.016
24. Allaire JM, Crowley SM, Law HT, Chang S-Y, Ko H-J, Vallance BA. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol (2018) 39(9):677–96. doi: 10.1016/j.it.2018.04.002
25. Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol (2012) 12(7):503–16. doi: 10.1038/nri3228
26. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science (2006) 313(5790):1126–30. doi: 10.1126/science.1127119
27. Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol [Internet]. (2003) 4(3):269–73. doi: 10.1038/ni888
28. Shipra V LBC, Lars E V. HL. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci [Internet]. (2008) 105(52):20858–63. doi: 10.1073/pnas.0808723105
29. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell (2017) 168(5):928–43. doi: 10.1016/j.cell.2017.01.022
30. Brooks JF II, Behrendt CL, Ruhn KA, Lee S, Raj P, Takahashi JS, et al. The microbiota coordinates diurnal rhythms in innate immunity with the circadian clock. Cell (2021) 184(16):4154–67. doi: 10.1016/j.cell.2021.07.001
31. Schlemmer U, Frølich W, Prieto RM, Grases F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol Nutr Food Res (2009) 53(S2):S330–75. doi: 10.1002/mnfr.200900099
32. Prieto RM, Fiol M, Perello J, Estruch R, Ros E, Sanchis P, et al. Effects of Mediterranean diets with low and high proportions of phytate-rich foods on the urinary phytate excretion. Eur J Nutr (2010) 49(6):321–6. doi: 10.1007/s00394-009-0087-x
33. Saunders AV, Craig WJ, Baines SK. Zinc and vegetarian diets. Med J Aust (2013) 199(4):S17–21. doi: 10.5694/mja11.11493
34. Bruce HM, Callow RK. Cereals and rickets. The role of inositolhexaphosphoric acid. Biochem J (1934) 28(2):517. doi: 10.1042/bj0280517
35. Nissar J, Ahad T, Naik HR, Hussain SZ. A review phytic acid: As antinutrient or nutraceutical. J Pharmacogn Phytochem. (2017) 6(6):1554–60.
36. Bodie AR, Micciche AC, Atungulu GG, Rothrock MJ, Ricke SC. Current Trends of Rice Milling Byproducts for Agricultural Applications and Alternative Food Production Systems. Front Sustain Food Syst (2019) 3:47. doi: 10.3389/fsufs.2019.00047
37. Gul K, Yousuf B, Singh AK, Singh P, Wani AA. Rice bran: Nutritional values and its emerging potential for development of functional food—A review. Bioact Carbohydrates Diet Fibre (2015) 6(1):24–30. doi: 10.1016/j.bcdf.2015.06.002
38. Yu Y, Zhang J, Wang J, Sun B. The anti-cancer activity and potential clinical application of rice bran extracts and fermentation products. RSC Adv (2019) 9(31):18060–9. doi: 10.1039/C9RA02439E
39. Yang S-C, Huang W-C, Ng XE, Lee M-C, Hsu Y-J, Huang C-C, et al. Rice Bran Reduces Weight Gain and Modulates Lipid Metabolism in Rats with High-Energy-Diet-Induced Obesity. Nutrients (2019) 11. doi: 10.3390/nu11092033
40. Kumar A, Henderson A, Forster GM, Goodyear AW, Weir TL, Leach JE, et al. Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC Microbiol (2012) 12(1):1–9. doi: 10.1186/1471-2180-12-71
41. Collins JW, Keeney KM, Crepin VF, Rathinam VAK, Fitzgerald KA, Finlay BB, et al. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol (2014) 12(9):612–23. doi: 10.1038/nrmicro3315
42. Bouladoux N, Harrison OJ, Belkaid Y. The Mouse Model of Infection with Citrobacter rodentium. Curr Protoc Immunol (2017) 119(1):19. doi: 10.1002/cpim.34
43. Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PloS Med (2015) 12(12):e1001921–e1001921. doi: 10.1371/journal.pmed.1001921
44. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M, Roy SL, et al. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg Infect Dis. (2011) 17(1):7-15. doi: 10.3201/eid1701.p11101
45. Iqbal TH, Lewis KO, Cooper BT. Phytase activity in the human and rat small intestine. Gut (1994) 35(9):1233 LP – 1236. doi: 10.1136/gut.35.9.1233
46. Wise A, Gilburt DJ. Phytate hydrolysis by germfree and conventional rats. Appl Environ Microbiol (1982) 43(4):753–6. doi: 10.1128/aem.43.4.753-756.1982
47. Wu S, Hashimoto-Hill S, Woo V, Eshleman EM, Whitt J, Engleman L, et al. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nat [Internet]. (2020) 586(7827):108–12. doi: 10.1038/s41586-020-2604-2
48. Schlemmer U, Jany K -D., Berk A, Schulz E, Rechkemmer G. Degradation of phytate in the gut of pigs - pathway of gastrointestinal inositol phosphate hydrolysis and enzymes involved. Arch für Tierernaehrung (2001) 55(4):255–80. doi: 10.1080/17450390109386197
49. Kumar V, Sinha AK, Makkar HPS, Becker K. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem [Internet]. (2010) 120(4):945–59. doi: 10.1016/j.foodchem.2009.11.052
50. Mukherjee S, Hooper LV. Antimicrobial Defense of the Intestine. Immunity (2015) 42(1):28–39. doi: 10.1016/j.immuni.2014.12.028
51. Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med (2008) 14(3):282–9. doi: 10.1038/nm1720
52. Vallance BA, Deng W, De Grado M, Chan C, Jacobson K, Finlay BB. Modulation of inducible nitric oxide synthase expression by the attaching and effacing bacterial pathogen Citrobacter rodentium in infected mice. Infect Immun (2002) 70(11):6424–35. doi: 10.1128/IAI.70.11.6424-6435.2002
53. Lu X-F, Cao X-Y, Zhu Y-J, Wu Z-R, Zhuang X, Shao M-Y, et al. Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell Death Dis (2018) 9(3):1–14. doi: 10.1038/s41419-018-0428-x
54. Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia (2012) 26(6):1356–64. doi: 10.1038/leu.2011.340
55. Woo V, Eshleman EM, Rice T, Whitt J, Vallance BA, Alenghat T. Microbiota inhibit epithelial pathogen adherence by epigenetically regulating C-type lectin expression. Front Immunol (2019) 10(MAY):1–10. doi: 10.3389/fimmu.2019.00928
56. Navabi N, Whitt J, Wu S, Woo V, Moncivaiz J, Jordan MB, et al. Epithelial Histone Deacetylase 3 Instructs Intestinal Immunity by Coordinating Local Lymphocyte Activation. Cell Rep (2017) 19(6):1165–75. doi: 10.1016/j.celrep.2017.04.046
57. Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CGK, Mullican SE, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature (2013) 504:153. doi: 10.1038/nature12687
58. Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med (2009) 206(7):1465–72. doi: 10.1084/jem.20082683
59. Ratsimandresy RA, Indramohan M, Dorfleutner A, Stehlik C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol (2017) 14(1):127–42. doi: 10.1038/cmi.2016.35
60. Overcast G, Eshleman E, Alenghat T, Pasare C. Epithelial cell intrinsic IL-1R signaling regulates intestinal homeostasis and inflammation. Am Assoc Immnol (2020) 204(1):Supplement 235.6.
61. Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, et al. Th22 Cells Are an Important Source of IL-22 for Host Protection against Enteropathogenic Bacteria. Immunity (2012) 37(6):1061–75. doi: 10.1016/j.immuni.2012.08.024
62. Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, et al. Aryl Hydrocarbon Receptor-Induced Signals Up-regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterol [Internet]. (2011) 141(1):237–248.e1. doi: 10.1053/j.gastro.2011.04.007
63. Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, Groom JR, et al. Retinoic acid expression associates with enhanced IL-22 production by T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med (2013) 210(6):1117–24. doi: 10.1084/jem.20121588
64. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun [Internet]. (2020) 11(1):4457. doi: 10.1038/s41467-020-18262-6
65. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immun [Internet]. (2013) 39(2):372–85. doi: 10.1016/j.immuni.2013.08.003
66. Bergstrom KSB, Morampudi V, Chan JM, Bhinder G, Lau J, Yang H, et al. Goblet Cell Derived RELM-β Recruits CD4+ T Cells during Infectious Colitis to Promote Protective Intestinal Epithelial Cell Proliferation. PloS Pathog (2015) 11(8):e1005108. doi: 10.1371/journal.ppat.1005108
67. Kamada N, Kim YG, Sham SH, Puente JL, Martens EC, Núñez G, et al. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science (2012) 336(6086):1325–9. doi: 10.1126/science.1222195
68. Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell (2021) 184(3):615–627.e17. doi: 10.1016/j.cell.2020.12.011
69. Okazaki Y, Katayama T. Phytic acid actions on hepatic lipids and gut microbiota in rats fed a diet high in sucrose is influenced by dietary fat level. Nutr Res (2020) 74:45–51. doi: 10.1016/j.nutres.2019.11.010
70. Henderson AJ, Kumar A, Barnett B, Dow SW, Ryan EP. Consumption of rice bran increases mucosal immunoglobulin A concentrations and numbers of intestinal Lactobacillus spp. J Med Food (2012) 15(5):469–75. doi: 10.1089/jmf.2011.0213
71. Balaban NP, Suleimanova AD, Shakirov EV, Sharipova MR. Histidine acid phytases of microbial origin. Microbiology (2018) 87(6):745–56. doi: 10.1134/S0026261718060024
Keywords: diet, microbiota, intestine, innate immunity, metabolite
Citation: Hashimoto-Hill S, Colapietro L, Woo V, Antonacci S, Whitt J, Engleman L and Alenghat T (2022) Dietary phytate primes epithelial antibacterial immunity in the intestine. Front. Immunol. 13:952994. doi: 10.3389/fimmu.2022.952994
Received: 25 May 2022; Accepted: 23 August 2022;
Published: 19 October 2022.
Edited by:
Silvia Melgar, University College Cork, IrelandReviewed by:
John MacSharry, University College Cork, IrelandCopyright © 2022 Hashimoto-Hill, Colapietro, Woo, Antonacci, Whitt, Engleman and Alenghat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Theresa Alenghat, dGhlcmVzYS5hbGVuZ2hhdEBjY2htYy5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.