
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 18 January 2023
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.947136
This article is part of the Research TopicTumor-Promoting Immune Cells: Cancer Immune Escape and BeyondView all 15 articles
 Jiawei Liu1,2†
Jiawei Liu1,2† Ning Fu2†
Ning Fu2† Zhenbang Yang3
Zhenbang Yang3 Ang Li3
Ang Li3 Hongjiao Wu3
Hongjiao Wu3 Ye Jin2
Ye Jin2 Qinqin Song1
Qinqin Song1 Shanshan Ji1
Shanshan Ji1 Hongxue Xu3
Hongxue Xu3 Zhi Zhang1*
Zhi Zhang1* Xuemei Zhang2,3*
Xuemei Zhang2,3*Background: CD55 plays an important role in the development of colon cancer. This study aims to evaluate the expression of CD55 in colon cancer and discover how it is regulated by transcriptional factors and miRNA.
Methods: The expression of CD55 was explored by TIMER2.0, UALCAN, and Human Protein Atlas (HPA) databases. TRANSFAC and Contra v3 were used to predict the potential binding sites of transcription factors in the CD55 promoter. TargetScan and starBase v2.0 were used to predict the potential binding ability of miRNAs to the 3′ untranslated region (3′UTR) of CD55. SurvivalMeth was used to explore the differentially methylated sites in the CD55 promoter. Western blotting was used to detect the expression of TFCP2 and CD55. Dual-luciferase reporter assay and chromatin immunoprecipitation (ChIP) assay were performed to determine the targeting relationship of TFCP2, NF-κB, or miR-27a-3p with CD55. CD55-related genes were explored by constructing a protein–protein interaction (PPI) network and performing pathway analysis by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG).
Results: CD55 was highly expressed in colon cancer tissues. The mRNA and protein expression levels of TFCP2 were reduced by si-TFCP2. NF-κB mRNA was obviously reduced by NF-κB inhibitor and increased by NF-κB activator. CD55 protein was also inhibited by miR-27a-3p. Dual-luciferase reporter assays showed that after knocking down TFCP2 or inhibiting NF-κB, the promoter activity of CD55 was decreased by 21% and 70%, respectively; after activating NF-κB, the promoter activity of CD55 increased by 2.3 times. As TFCP2 or NF-κB binding site was mutated, the transcriptional activity of CD55 was significantly decreased. ChIP assay showed that TFCP2 and NF-κB combined to the promoter of CD55. The luciferase activity of CD55 3′UTR decreased after being co-transfected with miR-27a-3p mimics and increased by miR-27a-3p antagomir. As the miR-27a-3p binding site was mutated, we did not find any significant effect of miR-27a-3p on reporter activity. PPI network assay revealed a set of CD55-related genes, which included CFP, CFB, C4A, and C4B. GO and KEGG analyses revealed that the target genes occur more frequently in immune-related pathways.
Conclusion: Our results indicated that CD55 is regulated by TFCP2, NF-κB, miR-27a-3p, and several immune-related genes, which in turn affects colon cancer.
According to GLOBOCAN 2020, colorectal cancer (CRC) ranks third in terms of morbidity and mortality in the world, presenting a serious threat to human health. Also, over the past decade, the rate of decline in CRC mortality has slowed (1). The incidence of CRC is known to be significantly influenced by early-life exposures (2). Thus, it is essential to find effective molecular targets for colon cancer treatment.
Gene expression is a highly regulated process. Given the complexity of gene regulation, it is not surprising that many human diseases, including various cancers, are caused by defective gene regulation. Transcription factors (TFs) are known to regulate chromatin and transcription by recognizing specific DNA sequences and subsequently forming a complex (3). MicroRNAs (miRNAs) are endogenous, and small non-coding RNAs are implicated in nearly all known physiological and pathological processes, such as cell proliferation, differentiation, and apoptosis. Importantly, miRNAs act as important regulators of several genes at the post-transcriptional level in almost all kinds of cancer cells. Multiple studies have reported that miRNAs exhibit aberrant expression in a variety of cancers.
DNA methylation is an epigenetic mechanism that tends to be crucial in the regulation of gene expression (4–6). Gene expression can be affected by the bonding of the methyl group to the cytosine nucleotide. DNA methylation in the promoter region of genes is often associated with the repression of transcription (7). The DNA methylation level in tumor tissues is mostly reported to be lower than that in normal adjacent tissues. During the development of cancer, the level of hypomethylation of genomic DNA increases as lesions progress from benign cell proliferation to aggressive cancer (8).
The complement system is a crucial element of innate immunity, and it also plays an essential role in the regulation of adaptive immunity (9). The activation of the complement system promotes cell proliferation in various cancers (10). CD55, also known as decay-accelerating factor (DAF), is one of the membrane-bound complement regulatory proteins (mCRPs) that play a key role in maintaining the homeostasis of the complement system. Dho and his colleagues found that CD55 was over-expressed in metastatic colon cancer tissues, and inhibition of CD55 could restrain colon cancer tumorigenesis and metastasis (11). The high expression of CD55 has a significantly decreased 7-year survival rate for colon cancer (12). Comprehensive bioinformatics analysis has also shown that CD55 was positively correlated with infiltration levels of CD8+ T cells, neutrophils, and dendritic cells in colon cancer, suggesting its potential role in tumor immune regulation (13).
Since CD55 might be used as a prognostic indicator and a therapeutic target in colon cancer, we investigated the possible mechanism of the regulation of CD55 in colon cancer and its impact on the molecular and immunological characteristics of colon cancer.
TIMER2.0 (http://timer.cistrome.org/) is a web server freely available to the research community. It consists of three main components: immunization, exploration, and estimation. The “Diff Exp” component allows users to compare gene expression differences between multiple tumor and normal tissues. In this study, we used TIMER2.0 to analyze the expression difference of CD55 between cancer tissues and normal tissues in a pan-cancer way. We analyzed the expression level of CD55 in colorectal cancer and its association with different histopathological types by UALCAN (http://ualcan.path.uab.edu), which contains cancer OMICS data. The protein expression of CD55 in human normal colon tissue and colon cancer tissue was verified by Human Protein Atlas (HPA; https://www.proteinatlas.org/). The HPA database, an antibody-based approach combined with transcriptomic data to outline global expression profiles, is the largest and most comprehensive public database of the spatial distribution of proteins in human tissues and cells (14). We downloaded the available miRNA data from the Gene Expression Omnibus (GEO) database (GSE48267 and GSE59856). We used the Limma program to analyze the expression of miR-27a in colorectal cancer.
From CD55, 2 kb of 5′ flanking region and 3′ untranslated region (3′UTR) was downloaded from the National Center for Biotechnology Information (NCBI) database. Transcription factor binding sites in the promoter of CD55 were predicted by using the TRANSFAC program (http://gene-regulation.com) and ConTra v3 online web tool (http://bioit2.irc.ugent.be/contra/v3/). “Vertebrates” was chosen as “Matrix groups”, and “cut-offs” was set to “minimize the sum of both error rates”. The potential miRNA binding sites in 3′UTR of CD55 were screened using TargetScanHuman 7.2 (https://www.targetscan.org/vert_72/) and starBase v2.0 (https://starbase.sysu.edu.cn/starbase2/index.php).
Human colon cancer cells HCT-116 and LOVO were provided by Procell (Wuhan, China). All plasmids (pGL3-Basic, pRL-SV40, and psiCHECK-2 plasmid) were purchased from Promega (Madison, WI, USA). The si-TFCP2, miR-27a-3p mimics, and miR-27a-3p antagomir were synthesized by GenePharma (Shanghai, China). The sequences of si-TFCP2 were 5′-GCU AAU CCA ACU CAA CUA ATT-3′ and 5′-UUA GUU GAG UUG GAU UAG CTT-3′. The sequences of siRNA control were 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and 5′-ACG UGA CAC GUU CGG AGA ATT-3′. The sequences of miR-27a-3p mimics were 5′-UUC ACA GUG GCU AAG UUC CGC-3′ and 5′-GGA ACU UAG CCA CUG UGA AUU-3′. The sequences of mimics control were 5′-UUC UCC GAA CGU GUC ACG UTT-3′ and 5′-ACG UGA CAC GUU CGG AGA ATT-3′. The sequence of miR-27a-3p antagomir was 5′-GCG GAA CUU AGC CAC UGU GAA-3′. The sequence of antagomir control was 5′-CAG UAC UUU UGU GUA CAA-3′. NF-κB inhibitor (B5556) and NF-κB activator (TNFα) (HZ-1014) were purchased from Sigma-Aldrich (New Jersey, USA) and ProteinTech (Chicago, USA), respectively.
The primer pair used to amplify the promoter of CD55 (1,963 bp) was CD55-PF (5′-GG GGTACC CCT CTC TAT GAA GGG CA-3′)/CD55-PR (5′-CCC AAGCTT GGG GAC GGC GGG AAC CAC GAC-3′) with protective bases and the recognition sites (underlined bases) of KpnI or HindIII in each primer. This PCR product was then cloned into a pGL3-Basic plasmid. The PCR primers to amplify the fragment of 3′UTR (1,237 bp) were CD553′UTR-PF (5′-CCG CTCGAG TGC CTT CAT TTA GGA TGC TTT CA-3′) and CD553′UTR-PR (5′-GTAA GCGGCCGC TTT ACA GTG AAA TGC CAT GAA CG-3′), of which protective bases and recognition sites (underlined) of XhoI and NotI sites were added. The 3′UTR PCR product was then cloned into a psiCHECK-2 plasmid. Successful plasmids were then designed wild-type plasmids as pGL3-CD55pro-Wt and psi-CD553′UTR-Wt. The CD55 promoter fragment containing TFCP2 or NF-κB binding sites was synthesized to generate mutant type (pGL3-CD55pro-TFCP2-Mut and pGL3-CD55pro-NF-κB-Mut) by Sangon Biotech (Shanghai, China). The CD55 3′UTR fragment containing miR-27a-3p binding sites was also synthesized to generate a mutant type (psi-CD553′UTR-Mut).
Human colon cancer cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, USA), 1% penicillin–streptomycin (100 U/ml of penicillin and 100 mg/ml of streptomycin, P&S; Thermo Fisher Scientific, USA) in an atmosphere with 5% CO2 at 37°C. Cells were transfected with Lipofectamine 2000 reagent (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. For TFCP2 binding analysis, 500 ng of pGL3-CD55pro/pGL3-CD55mut plasmid and 0.5 ng of pRL-SV40 plasmid were co-transfected with si-TFCP2 or negative control into HCT116 cells for 24 h. For NF-κB binding analysis, colon cancer cells were transfected with 500 ng of pGL3-CD55pro/pGL3-CD55mut and 0.5 ng of pRL-SV40 for 24 h and then treated with or without 20 μM of NF-κB inhibitor/50 ng of NF-κB activator for another 24 h. For miRNA binding analysis, 30 ng of psi-CD553′UTR-Wt/psi-CD553′UTR-Mut was co-transfected with 100 nM of miR-27a-3p mimics/control or 20 nM of miR-27a-3p antagomir/control into HCT116 cells for 24 h.
Total RNA was extracted from HCT116 cells using TRIzol reagent (Thermo Fisher Scientific, USA) and was then reversely transcribed into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). TFCP2 mRNA was detected using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA) in ABIPRISM® 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The amplification procedure was 50°C for 2 min and 95°C for 2 min, followed by 45 cycles of 95°C for 15 s and 60°C for 2 min. GAPDH was used as the reference gene. The primer pairs for the amplification of TFCP2, NF-κB, and GAPDH were TFCP2-PF/TFCP2-PR (5′-TCA CGT ATG TCA ATA ACT CCC CA-3′/5′-GTG TGG TTG GTA AGA GGT T-3′), NF-κB-PF/NF-κB-PR (5′-AAC AGA GAG GAT TTC GTT TCC G-3′/5′-TTT GAC CTG AGG GTA AGA CTT CT-3′), and GAPDH-PF/GAPDH-PR (5′-ACA ACT TTG GTA TCG TGG AAG G-3′/5′-GCC CAT CAC GCC ACA GTT TC-3′). Three repetitions were performed for each reaction. The relative mRNA expression was analyzed using the 2−ΔΔCt method.
Colon cancer cells were plated at 2 × 105 cells per well into 24-well plates. When cells reached 70%–80% confluence, each plasmid was then transfected into cells. After 24 h, cells were collected, and luciferase and Renilla reporter signals were detected using the Dual-luciferase Reporter Assay System (Promega, USA) by GloMax® 20/20 Luminometer (Promega, USA).
The colon cancer cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, USA). After being quantified and denatured, samples were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After being blocked for 1 h with 5% skimmed milk in TBST at room temperature, the membrane was incubated with the primary antibody overnight at 4°C and then washed and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. Specific protein was then developed by using enhanced chemiluminescence (ECL) luminescence reagents (Amersham, UK). The densitometry analysis was performed using ImageJ (National Institutes of Health, USA). β-Actin was applied as a reference control. The following primary antibodies were used: anti−CD55 (1:10,000 dilution; ab133684; Abcam, Cambridge, UK), anti−NF-κB p65 (1:5,000 dilution; ab32536; Abcam), and anti−TFCP2 (1:5,000 dilution; 15203-1-AP; ProteinTech).
LOVO cells (2 × 106) were plated in a 6-mm dish for the chromatin immunoprecipitation (ChIP) experiment by using a ChIP Assay kit (Thermo Fisher Scientific, USA) following the manufacturer’s protocol. To verify if NF-κB or TFCP2 binds to the promoter of CD55, a total of 5 μg of sheared DNA was used for chromatin immunoprecipitation using anti-NF-κB (1:250 dilution; ab32536; Abcam) or anti−TFCP2 antibody (1:250 dilution; 15203-1-AP; ProteinTech). The immunoprecipitated DNA was then amplified using specific primers to analyze the transcription factor binding site of CD55. The CD55 promoter-specific primers were described as follows: NF-κB site-PF/NF-κB site-PR (5′-CGT GTG GGG TGA GTA GGG-3′/5′-ATG CTG GTG AGC GGC GAG-3′) and TFCP2 site-PF/TFCP2 site-PR (5′-CGT CTT GTT TGT CCC ACC C-3′/5′-GCA GTA AGC AGA AGC CTC G-3′).
Cell viability was analyzed by Cell Counting Kit 8 (CCK8; Dojindo, Kumamoto, Japan) according to the manufacturer’s protocol. Cells were seeded and cultured at a density of 5 × 103/well into 96-well microplates. After psi-CD553′UTR-Wt was co-transfected with miR-27a-3p mimics or mimics control for 24 h, 10 μl of CCK8 reagent was added and cultured for 30 mins. All experiments were performed in triplicate. The absorbance was measured at 450 nm using a microplate reader (Multiskan FC, Thermo Fisher Scientific, USA).
STRING consortium 2022 (https://string-db.org) aims to integrate all known and predicted associations between proteins, including both physical interactions and functional associations. We set the network type to “full STRING network”, the required score to “high confidence (0.700)”, and the size cutoff to “no more than 20 interactors” to obtain the CD55-related protein. We used the Cytoscape 3.9.1 platform to construct the protein interaction network (protein–protein interaction (PPI) network) and analyze the network characteristics.
We used the “ggplot2” R package and DAVID 6.8 (http://www.david.niaid.nih.gov) database to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of CD55-related proteins (p < 0.05). We performed the visualization of GO and KEGG enrichment by R version 3.6.1.
SurvivalMeth (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/) investigates the effect of DNA methylation-related functional elements on prognosis, which was developed by Harbin Medical University. We analyzed the methylation sites and their effect on the survival time of colon cancer patients of CD55 by using SurvivalMeth.
All analyses were performed with R version 3.6.1 and its appropriate packages. Statistical graphs were produced in GraphPad Prism 8. Statistical significance of the Wilcoxon rank sum test or t-test was used for two-group comparisons. All statistical tests were two-sided, and p < 0.05 was considered to be statistically significant.
Using the TIMER2.0 online program, we found that CD55 was highly expressed in a variety of solid cancers, including colorectal cancer (Figure 1A). There are two pathological types of colorectal cancer tissues, adenocarcinoma and mucinous adenocarcinoma. Using the UALCAN database, we evaluated the expression of CD55 in colorectal cancer tissues and its relationship with different pathological types and found that CD55 was highly expressed in either colorectal adenocarcinoma or mucinous adenocarcinoma when compared with that in adjacent normal tissues. Additionally, the average level of CD55 in mucinous adenocarcinoma tissue is significantly higher than that in adenocarcinoma tissue (Figures 1B–E). Immunohistochemical images from the HPA database showed that the expression CD55 was higher in colorectal cancer than in adjacent normal tissue (Figure 1F).
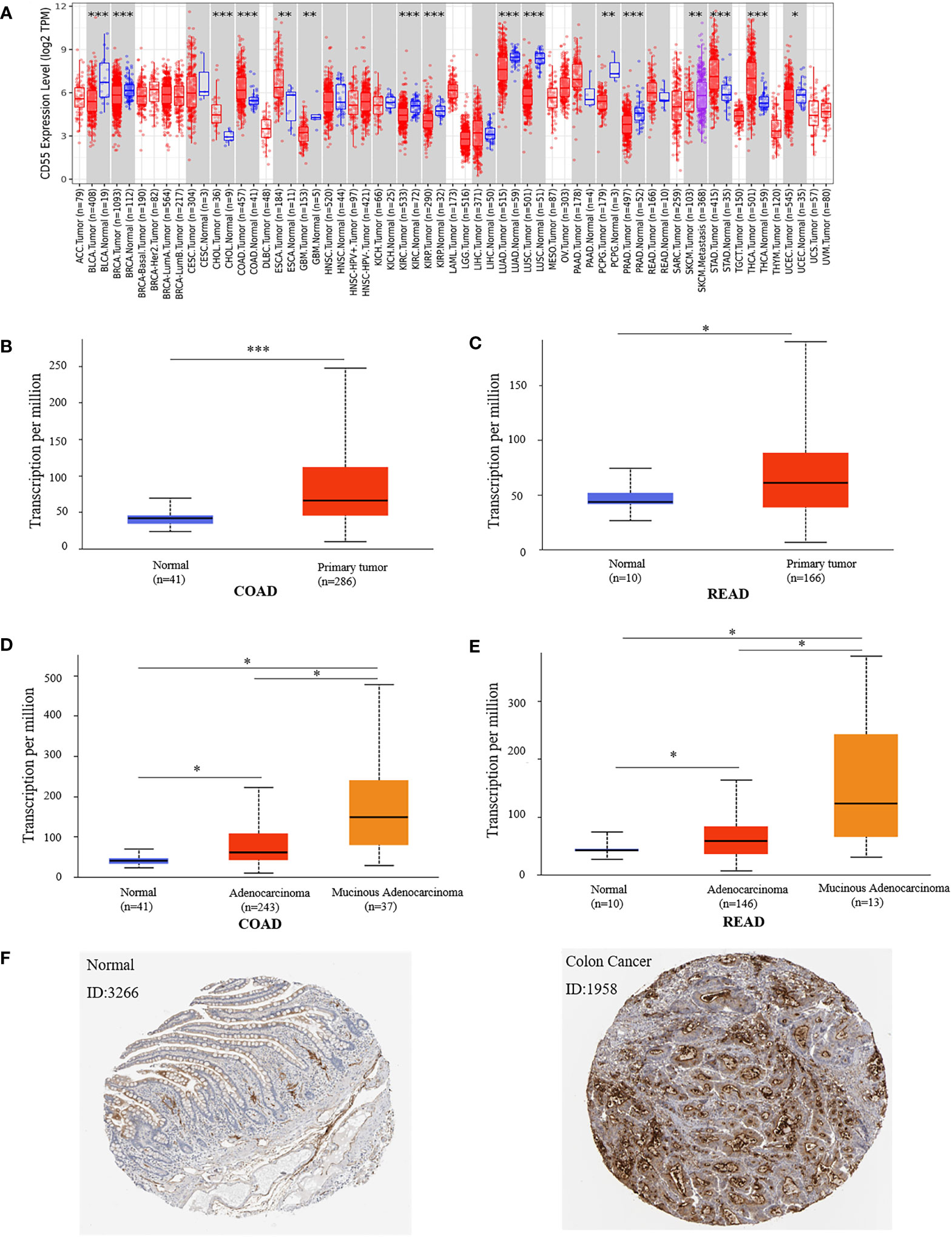
Figure 1 Expression of CD55 in colon cancer tissue and adjacent normal tissue. (A) CD55 expression levels in different tumor types from TCGA database were determined by TIMER2.0. (B, C) Quantification of CD55 expression levels in colorectal cancer (COAD, colon adenocarcinoma; READ, rectal adenocarcinoma) and paired adjacent normal tissues. (D, E) Quantification of CD55 expression in different pathological types of colorectal cancer. (F) Protein levels of CD55 in colorectal cancer and normal tissues by HPA database (*p < 0.05, **p < 0.01, ***p < 0.001). TCGA, The Cancer Genome Atlas.
To predict the potential TF binding sites of CD55, we used TRANSFAC online program, and it generated eight transcriptional factors, including COMP1, Hand1/E47, CDP CR1, Pax-4, TFCP2, Nkx2-5, c-Rel, and NF-κB (Table 1). According to the core binding characteristic of these TFs provided by the Contra v3 database, we found that TFCP2 and NF-κB were the most likely to bind to CD55 (Figures 2A–C).
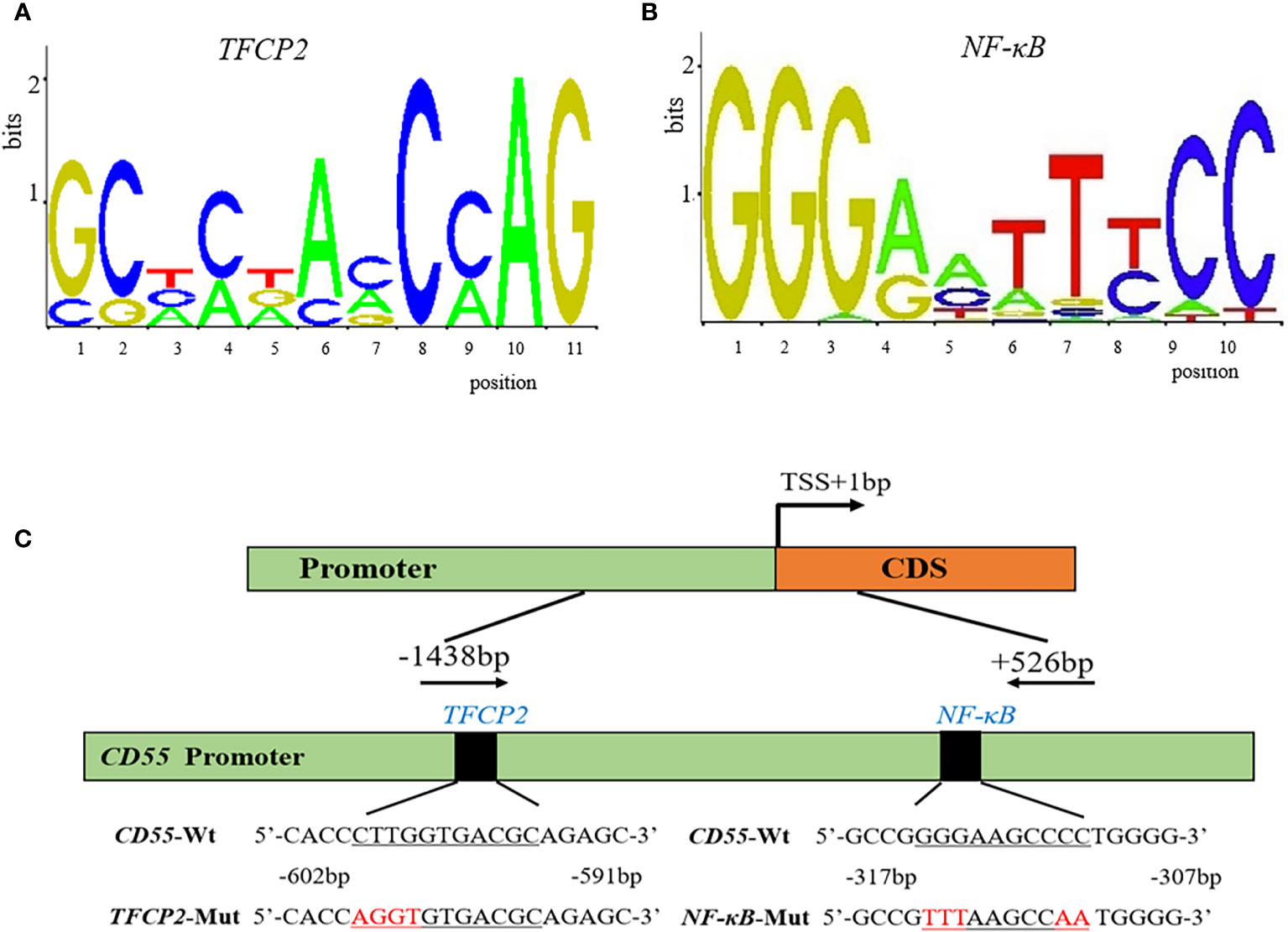
Figure 2 The potential binding sites and the diagram of the CD55 promoter. (A, B) The potential conserved sequence of TFCP2 and NF-κB binding site in the promoter of CD55 by Contra v3. (C) A diagram of the CD55 promoter region that was cloned in the luciferase reporter vector.
To confirm the regulation of TFCP2 to the expression of CD55, we used TFCP2 siRNA to treat HCT116 cells. We found that TFCP2 siRNA effectively reduced the expression of TFCP2 (Figures 3A, B). Dual-luciferase reporter assay showed that the reporter activity of pGL3-CD55pro-Wt with TFCP2 knockdown decreased by 21% compared with that without TFCP2 knockdown (p < 0.05) (Figure 3C). After the TFCP2 binding site was mutated, the promoter activity of CD55 was reduced by 66% (Figure 3G).

Figure 3 The regulation of transcription factors to the promoter of CD55. (A) The mRNA level of TFCP2 by RT-PCR after HCT116 cells transfected with si-TFCP2 or siRNA control for 24 h. (B) The protein expression of TFCP2 by Western blotting after HCT116 cells transfected with si-TFCP2 or siRNA control for 48 h. (C) Dual-luciferase assay of pGL3-CD55pro-Wt in HCT116 cells after co-transfected with si-TFCP2. (D) The mRNA level of NF-κB in HCT116 cells treated with NF-κB inhibitor and that in LOVO cells treated with NF-κB activator for 24 h. (E) The luciferase activity of pGL3-CD55pro-Wt in HCT116 cells treated with or without NF-κB inhibitor. (F) The luciferase activity of pGL3-CD55pro-Wt and pGL3-CD55pro-NF-κB-Mut in LOVO cells treated with or without NF-κB activator (TNFα). (G) CD55 promoter activity assay in pGL3-CD55pro-Wt, pGL3-CD55pro-NF-κB-Mut, and pGL3-CD55pro-TFCP2-Mut plasmids. Luciferase activity was normalized to Renilla luciferase activity (L/R). (H) The binding of TFCP2 and NF-κB to the promoter of CD55 by ChIP. IgG from rabbits served as a control (*p < 0.05, **p < 0.01, ***p < 0.001, ns, no significance). ChIP, chromatin immunoprecipitation.
We also evaluated the effect of NF-κB on the promoter activity of CD55. We found that the expression of NF-κB was significantly decreased by the NF-κB inhibitor and increased by the NF-κB activator (Figure 3D). The luciferase reporter analysis showed that the promoter activity of CD55 was decreased by 70% due to the NF-κB inhibitor (p < 0.01) (Figure 3E). The transcriptional activity of CD55 was activated by the NF-κB activator; however, after we mutated the binding site of NF-κB, we did not see the effect of the NF-κB activator on the luciferase reporter activity (Figure 3F). After the NF-κB binding site was mutated, the promoter activity of CD55 was reduced by 42% (Figure 3G).
ChIP assay presented that TFCP2 and NF-κB were combined to the promoter of CD55 (Figure 3H), which further supported the binding capability of TFCP2 and NF-κB to the promoter of CD55.
Through TargetScan and starBase v2.0 database, we found that there was one potential binding site of miR-27a-3p in the 3′UTR of CD55 (Figure 4A). Using the dataset from GEO, we found that miR-27a was downregulated in colorectal cancer (Figure 4B). We then conducted a CCK8 assay to see the effect of miR-27a on cell proliferation and found that miR-27a inhibited cell proliferation by 37.8%. We constructed luciferase reporter plasmid of CD55 3′UTR with or without mutated miR-27a-3p binding site. By co-transfecting with miR-27a-3p mimics or its mimics control into HCT116 or using an antagomir of miR-27a-3p or its antagomir control, we found that the luciferase reporter activity of CD55 3′UTR reduced by 56% by miR-27a-3p mimics and increased by 33% by miR-27a-3p antagomir (p < 0.001) (Figure 4C). Next, after transfecting miR-27a-3p mimics into HCT116, we found that CD55 was upregulated by miR-27a-3p (Figure 4D).
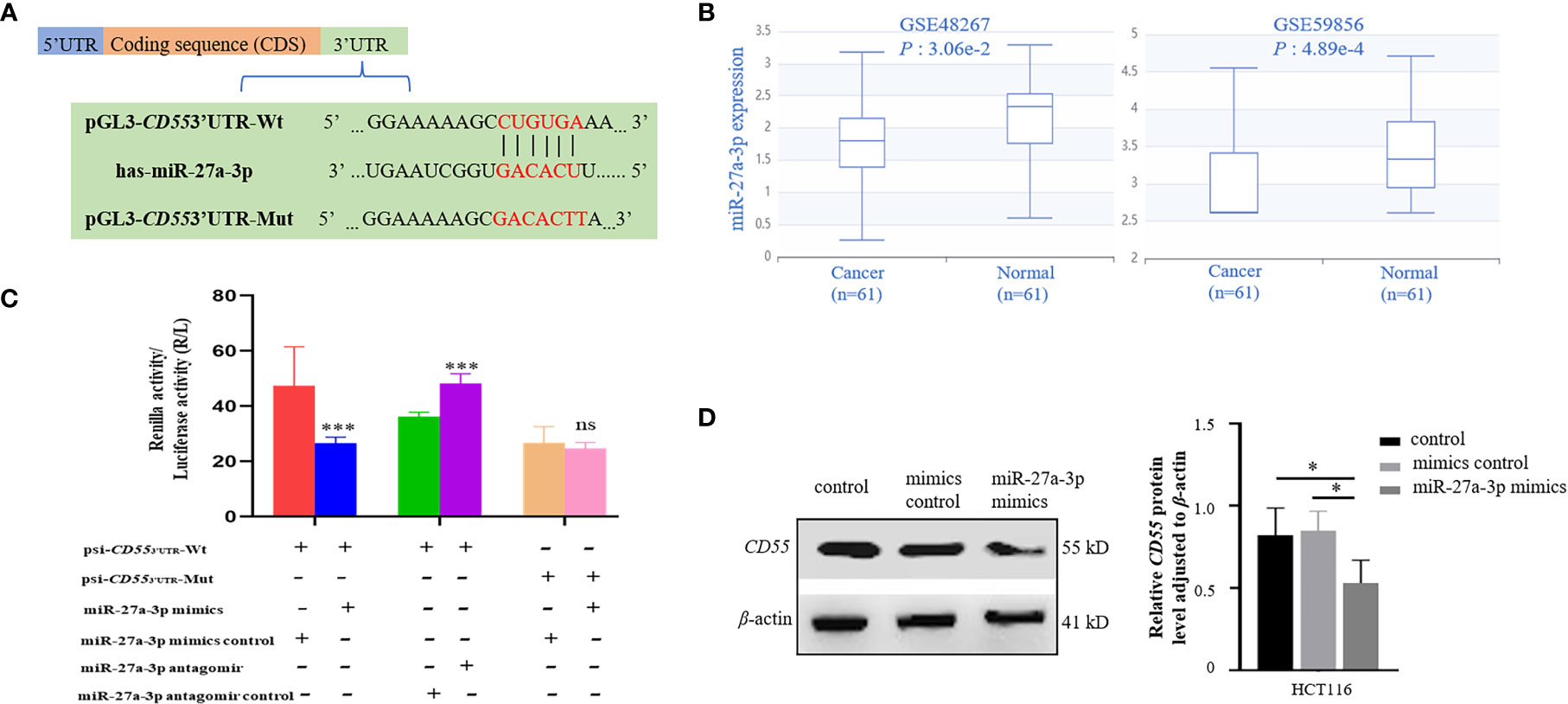
Figure 4 Binding and regulation of miRNA to CD55 3′UTR. (A) TargetScan and starBase v2.0 database predicted miR-27a-3p binding site in the 3′UTR of CD55. (B) The differential expression of miR-27a in two colon cancer GEO datasets (p < 0.05). (C) Verification of the potential binding of miR-27a-3p mimics or antagomir with CD55 using luciferase reporter assay. The luciferase reporter activity of CD55 3′UTR reduced by miR-27a-3p mimics and increased miR-27a-3p antagomir (p < 0.001). The relative luciferase activity was normalized to the Renilla luciferase activity. Each experiment was performed in triplicate. Data are presented as the mean value ± SD (*p < 0.05; **p < 0.01; ***p < 0.001, ns, no significance). (D) The protein expression of CD55 in HCT116 cells determined by Western blotting and densitometry analysis after cells were transfected with or without miR-27a-3p mimics (negative control) or with mimics control for 48 h (*p < 0.05). GEO, Gene Expression Omnibus.
SurvivalMeth presented that three methylation sites (cg00797651, cg22048546, and cg25771140) appeared in the promoter of CD55. Among these sites, cg00797651 has a lower DNA methylation level in tumor samples than in normal samples (p < 0.01) (Figure 5A). Then, we divided the samples into high- and low-risk groups by differentially methylated sites and compared the methylation level of CpGs between the two groups. We found that the high-risk group had lower methylation levels (Figures 5B, C).

Figure 5 Methylation modification of CD55 promoter in colon cancer. (A) The cg sites of CD55 promoter region in COAD by SurvivalMeth. (B) The methylation level of CpGs in high- and low-risk groups. (C) The heatmap of three cg sites and their methylation level (*p < 0.05, ***p < 0.001). COAD, colon adenocarcinoma.
We constructed a PPI regulatory network for CD55-related proteins using the STRING database. When setting the size cutoff to “no more than 20 interactors”, CFP, CFB, C4A, C4B, C5AR1, C3, C3AR1, C2, EGF, LCK, CD59, PIGA, PGAP1, ICAM1, EMR2, and CD97 proteins were incorporated (Figure 6). The top three GO enrichment involved in complement activation and apoptotic cell clearance at the biological process level was as follows: extracellular exosome, extracellular space, and plasma membrane at the cellular component level, and complement binding, endopeptidase inhibitor activity, and complement receptor activity at the molecular function level (Figure 7A; Table 2). KEGG pathway enrichment analysis showed that these genes were significantly enriched in complement and coagulation cascade pathways, Staphylococcus aureus infection pathways, coronavirus disease–COVID-19, alcoholic liver disease pathways, pertussis pathways, etc. (Figure 7B; Table 3).
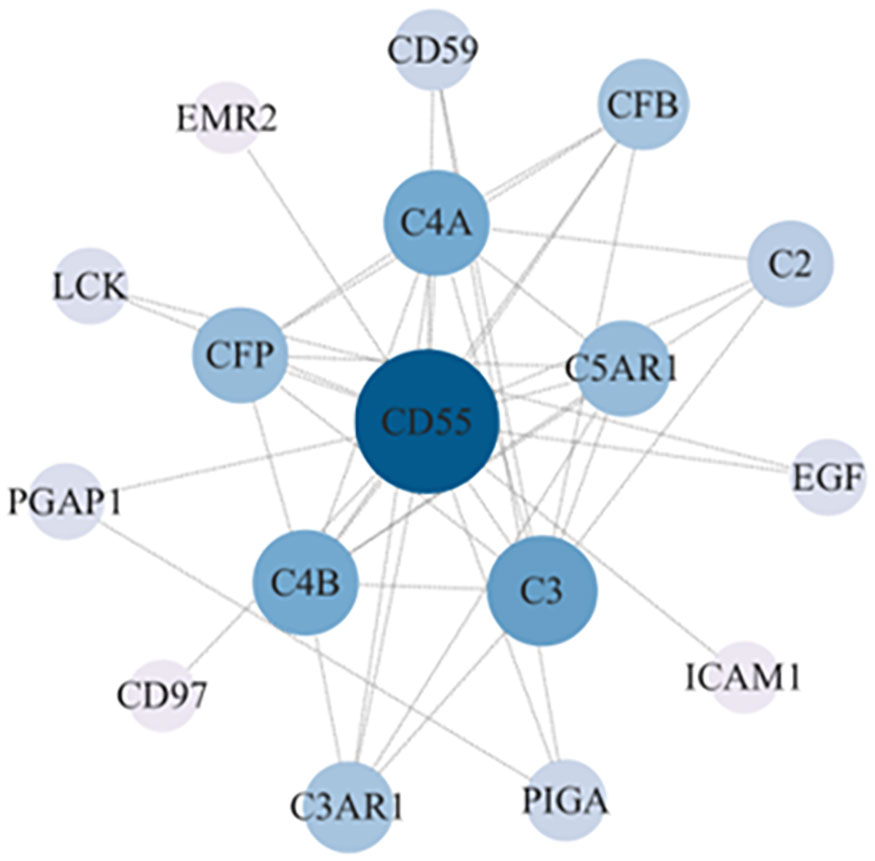
Figure 6 CD55 PPI network analysis. The PPI network of CD55 and related proteins were visualized by the STRING database. PPI, protein–protein interaction.
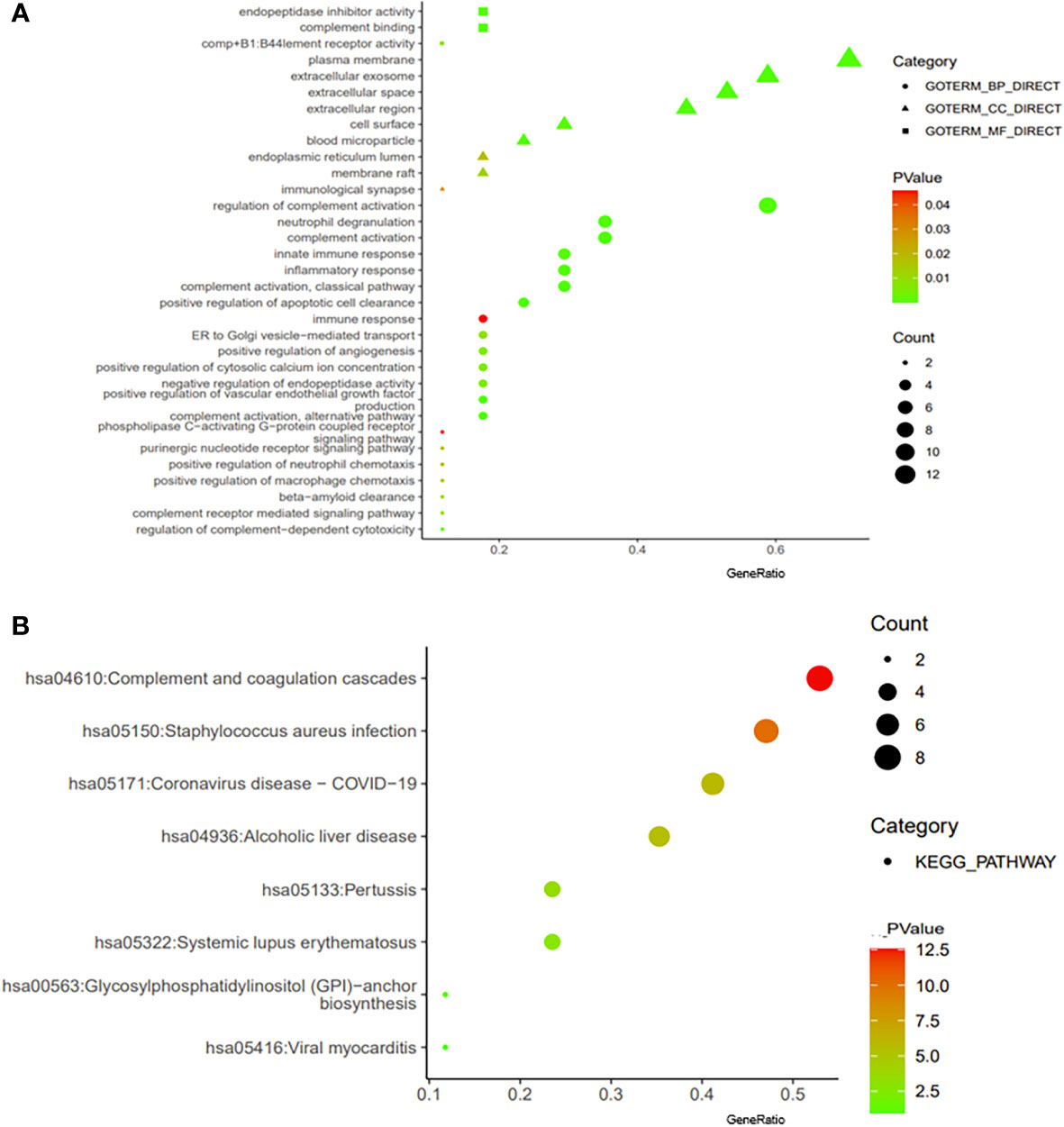
Figure 7 Enrichment analysis of CD55. The visualization of GO enrichment analysis (A) and KEGG enrichment analysis (B) of CD55 by DAVID 6.8. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The complement system in humans remains on standby and constantly on high alert for any potential intruders (15). The membrane complement regulatory proteins (CD35, CD46, CD55, and CD59) are important regulators of the complement system and are widely expressed on the surface of cells. In addition to the normal cells and tissues, the mCRPs also protect malignant cells from complement attack (16). For example, all hematopoietic cells, as well as endothelial and epithelial tissues, express CD55/DAF (17, 18). In comparison to the surrounding normal tissues, CD55/DAF is often expressed at substantially higher levels in a variety of malignant cells, including cells of CRC (19). The findings of the present study are consistent with those of the previous studies.
According to the UALCAN program, CRC tissues had higher levels of CD55 than normal tissue, which is supported by the data from the IHC and HPA databases (20). Notably, high expression of CD55 can also promote the dissemination of tumor cells in circulation. Blocking or downregulating CD55 may be an important step in advancing the efficacy of monoclonal antibody (mAb) immunotherapy for cancer (16).
Transcription factor CP2 (TFCP2) belongs to the TFCP2/Grainyhead family. The ubiquitous expression of TFCP2 suggests its involvement in comprehensive cellular functions and diseases such as cancer, Alzheimer’s disease, and AIDS (21). Previous studies have identified TFCP2 as a pro-cancer factor in hepatocellular carcinoma (22), pancreatic cancer (23), breast cancer (24), and CRC (25). Additionally, TFCP2 acts as a tumor suppressor, inhibiting the development of melanoma (26). Using the TRANSFAC online program, we found that TFCP2 potentially binds to the promoter of CD55. To verify this, we conducted a dual-luciferase reporter assay. After knocking down TFCP2 by siRNA, we found that the promoter activity of CD55 reduced by 21%; however, after the mutated TFCP2 binding site, it reduced by 66%. RNA interference (RNAi) is a natural process of target mRNA degradation. After we used si-TFCP2, the expression TFCP2 was decreased by more than 50%; however, TFCP2 could still bind to the promoter of CD55. After we mutated the binding site of TFCP2, TFCP2 hardly binds to the promoter of CD55 to regulate the expression of CD55. We then performed a ChIP assay and found that TFCP2 directly binds to the promoter of CD55 in colon cancer cells.
Nuclear factor-kappa B (NF-κB), belonging to the Rel/NF-κB family, regulates innate and adaptive immune responses (27). NF-κB is involved in tumorigenesis by regulating some important cell cycle-related genes, promoting cell proliferation, and inhibiting cell death (28). Using the TRANSFAC program, we found that NF-κB might bind to the promoter of CD55, which was subsequently confirmed by the dual-luciferase reporter assay. Colorectal cancer is a typical inflammation-dependent cancer (29, 30). NF-κB signaling is shown to link inflammation and the development of cancer (31). Moreover, ChIP provided clear evidence to support this finding.
MiRNAs are small non-coding regulatory RNAs (ncRNAs) that inhibit gene expression at a post-transcriptional level by binding to the 3′UTR of target mRNAs (32). Minor changes in miRNA have significant effects on gene expression (33). The dysregulation of miRNA was found in colon cancer tissues (34). Studies have shown that miR-27a-3p targets BTG1 to affect the biological phenotype of colorectal and ovarian cancer cells (35, 36). Additionally, in colon cancer cells, wild-type p53 downregulated the expression of miR-27a-3p (37). In the current study, through TargetScanHuman 7.2, we predicted that miR-27a-3p targeted the 3′UTR of CD55. Subsequently, CD55 was confirmed as a direct target of miR-27a-3p by a dual-luciferase reporter gene assay. It is reasonable to speculate that miR-27a-3p affects colon cancer cells by targeting CD55.
DNA methylation is a form of epigenetic modification. Aberrant DNA methylation has been identified as an important aspect of promoting CRC pathogenesis by silencing tumor suppressor genes. Studies have found that DNA hypermethylation could promote CRC metastasis by regulating CEBPB and TFCP2 (38). Using the SurvivalMeth program, we found that the promoter region of CD55 does have hypermethylation sites in colon cancer tissues. However, we did not find any overlap between hypomethylation sites in colorectal cancer cells and any of the putative binding sites identified on the CD55 promoter or impaired NF-κB binding to the CD55 promoter. Future research is still required to identify the mechanism.
The PPI network showed that CD55 and the key complement system components C3, C4A, and C4B are closely related to colon cancer. Studies have shown that C3 is also an immune-related core differential protein in colon adenocarcinoma (COAD) and is negatively correlated with the overall survival of COAD patients (39). Recent studies proposed that deficiency or pharmacological blockade of C5aR1 significantly impeded tumorigenesis of CRC, and the over-expression of C5aR1 contributed to the poor prognosis of CRC patients (40, 41). CFP plays a positive role in regulating the natural immune system in alternative pathways, and it is associated with immune infiltration in gastric cancer and lung cancer (42). We speculated that CFP and CD55 might be involved in the immune infiltration of tumor cells in colon cancer. This evidence indicated that CD55 promoted cancer by regulating complement activation either directly or indirectly (43).
In conclusion, the expression of CD55 in colon cancer was associated with the genetic regulation of TFCP2, NF-κB, epigenetic regulation of miR-27a-3p, and methylation modification. The genes associated with CD55 are probably involved in immune-related pathways in colon cancer. This study provides a theoretical basis and insight into the development of biomarkers for future research in the field of colon cancer.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
JL and NF: conceptualization, writing—original draft, and validation. ZY, AL, HW, YJ, HX, and QS: methodology and validation. AL and SJ: software and data curation. ZZ and XZ: supervision, writing—review and editing, and project administration. XZ: funding acquisition. All authors contributed to the article and approved the submitted version.
This study was supported by the Key Project of the Natural Science Foundation of Hebei province of China (grant number H2017209233).
The manuscript has previously appeared in a preprint: (44).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
2. Siegel RL, Torre LA, Soerjomataram I, Hayes RB, Bray F, Weber TK, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut (2019) 68(12):2179–85. doi: 10.1136/gutjnl-2019-319511
3. Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell (2018) 172(4):650–65. doi: 10.1016/j.cell.2018.01.029
4. Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature (1983) 301(5895):89–92. doi: 10.1038/301089a0
5. Razin A, Riggs AD. DNA Methylation and gene function. Sci (New York NY) (1980) 210(4470):604–10. doi: 10.1126/science.6254144
6. Felsenfeld G, McGhee J. Methylation and gene control. Nature (1982) 296(5858):602–3. doi: 10.1038/296602a0
7. Matharu N, Ahituv N. Modulating gene regulation to treat genetic disorders. Nat Rev Drug Discovery (2020) 19(11):757–75. doi: 10.1038/s41573-020-0083-7
8. Esteller M. Epigenetics in cancer. New Engl J Med (2008) 358(11):1148–59. doi: 10.1056/NEJMra072067
9. Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res (2010) 20(1):34–50. doi: 10.1038/cr.2009.139
10. Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest (2017) 127(3):780–9. doi: 10.1172/jci90962
11. Dho SH, Cho EH, Lee JY, Lee SY, Jung SH, Kim LK, et al. A novel therapeutic anti−CD55 monoclonal antibody inhibits the proliferation and metastasis of colorectal cancer cells. Oncol Rep (2019) 42(6):2686–93. doi: 10.3892/or.2019.7337
12. Durrant LG, Chapman MA, Buckley DJ, Spendlove I, Robins RA, Armitage NC. Enhanced expression of the complement regulatory protein CD55 predicts a poor prognosis in colorectal cancer patients. Cancer immunol immunother CII (2003) 52(10):638–42. doi: 10.1007/s00262-003-0402-y
13. Bao D, Zhang C, Li L, Wang H, Li Q, Ni L, et al. Integrative analysis of complement system to prognosis and immune infiltrating in colon cancer and gastric cancer. Front Oncol (2020) 10:553297. doi: 10.3389/fonc.2020.553297
14. Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci Publ Protein Soc (2018) 27(1):233–44. doi: 10.1002/pro.3307
15. Liszewski MK, Farries TC, Lublin DM, Rooney IA, Atkinson JP. Control of the complement system. Adv Immunol (1996) 61:201–83. doi: 10.1016/s0065-2776(08)60868-8
16. Gorter A, Meri S. Immune evasion of tumor cells using membrane-bound complement regulatory proteins. Immunol Today (1999) 20(12):576–82. doi: 10.1016/s0167-5699(99)01537-6
17. Medof ME, Walter EI, Rutgers JL, Knowles DM, Nussenzweig V. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J Exp Med (1987) 165(3):848–64. doi: 10.1084/jem.165.3.848
18. Asch AS, Kinoshita T, Jaffe EA, Nussenzweig V. Decay-accelerating factor is present on cultured human umbilical vein endothelial cells. J Exp Med (1986) 163(1):221–6. doi: 10.1084/jem.163.1.221
19. Li L, Spendlove I, Morgan J, Durrant LG. CD55 is over-expressed in the tumour environment. Br J Cancer (2001) 84(1):80–6. doi: 10.1054/bjoc.2000.1570
20. Shang Y, Chai N, Gu Y, Ding L, Yang Y, Zhou J, et al. Systematic immunohistochemical analysis of the expression of CD46, CD55, and CD59 in colon cancer. Arch Pathol Lab Med (2014) 138(7):910–9. doi: 10.5858/arpa.2013-0064-OA
21. Veljkovic J, Hansen U. Lineage-specific and ubiquitous biological roles of the mammalian transcription factor LSF. Gene (2004) 343(1):23–40. doi: 10.1016/j.gene.2004.08.010
22. Santhekadur PK, Rajasekaran D, Siddiq A, Gredler R, Chen D, Schaus SE, et al. The transcription factor LSF: a novel oncogene for hepatocellular carcinoma. Am J Cancer Res (2012) 2(3):269–85.
23. Yuedi D, Yuankun C, Jiaying Z, Han L, Yueqi W, Houbao L, et al. TFCP2 activates beta-catenin/TCF signaling in the progression of pancreatic cancer. Oncotarget (2017) 8(41):70538–49. doi: 10.18632/oncotarget.19741
24. Zhao Y, Kaushik N, Kang JH, Kaushik NK, Son SH, Uddin N, et al. A feedback loop comprising EGF/TGFα sustains TFCP2-mediated breast cancer progression. Cancer Res (2020) 80(11):2217–29. doi: 10.1158/0008-5472.Can-19-2908
25. Shao K, Pu W, Zhang J, Guo S, Qian F, Glurich I, et al. DNA Hypermethylation contributes to colorectal cancer metastasis by regulating the binding of CEBPB and TFCP2 to the CPEB1 promoter. Clin Epigenet (2021) 13(1):89. doi: 10.1186/s13148-021-01071-z
26. Kotarba G, Krzywinska E, Grabowska AI, Taracha A, Wilanowski T. TFCP2/TFCP2L1/UBP1 transcription factors in cancer. Cancer Lett (2018) 420:72–9. doi: 10.1016/j.canlet.2018.01.078
27. Liang Y, Zhou Y, Shen P. NF-kappaB and its regulation on the immune system. Cell Mol Immunol (2004) 1(5):343–50.
28. Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev (2008) 18(1):19–26. doi: 10.1016/j.gde.2008.01.020
29. Coussens LM, Werb Z. Inflammation and cancer. Nature (2002) 420(6917):860–7. doi: 10.1038/nature01322
30. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell (2010) 140(6):883–99. doi: 10.1016/j.cell.2010.01.025
31. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol (2018) 18(5):309–24. doi: 10.1038/nri.2017.142
32. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol (2009) 4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222
33. Cao Y, Hu J, Fang Y, Chen Q, Li H. Association between a functional variant in microRNA-27a and susceptibility to colorectal cancer in a Chinese han population. Genet Mol Res GMR (2014) 13(3):7420–7. doi: 10.4238/2014.September.12.8
34. Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, et al. Prognostic values of microRNAs in colorectal cancer. biomark Insights (2006) 2:113–21. doi: 10.1177/117727190600100009
35. Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci (2014) 21(1):95. doi: 10.1186/s12929-014-0095-x
36. Su C, Huang DP, Liu JW, Liu WY, Cao YO. miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol Lett (2019) 18(3):2825–34. doi: 10.3892/ol.2019.10629
37. Li E, Han K, Zhou X. microRNA-27a-3p down-regulation inhibits malignant biological behaviors of ovarian cancer by targeting BTG1. Open Med (Wars) (2019) 14:577–85. doi: 10.1515/med-2019-0065
38. Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2006) 12(7 Pt 1):2014–24. doi: 10.1158/1078-0432.Ccr-05-1853
39. Liang JZ, Liang XL, Zhong LY, Wu CT, Zhang J, Wang Y. Comparative proteome identifies complement component 3-mediated immune response as key difference of colon adenocarcinoma and rectal adenocarcinoma. Front Oncol (2020) 10:617890. doi: 10.3389/fonc.2020.617890
40. Ding P, Li L, Li L, Lv X, Zhou D, Wang Q, et al. C5aR1 is a master regulator in colorectal tumorigenesis via immune modulation. Theranostics (2020) 10(19):8619–32. doi: 10.7150/thno.45058
41. Ding P, Xu Y, Li L, Lv X, Li L, Chen J, et al. Intracellular complement C5a/C5aR1 stabilizes β-catenin to promote colorectal tumorigenesis. Cell Rep (2022) 39(9):110851. doi: 10.1016/j.celrep.2022.110851
42. Cui G, Geng L, Zhu L, Lin Z, Liu X, Miao Z, et al. CFP is a prognostic biomarker and correlated with immune infiltrates in gastric cancer and lung cancer. J Cancer (2021) 12(11):3378–90. doi: 10.7150/jca.50832
43. Geller A, Yan J. The role of membrane bound complement regulatory proteins in tumor development and cancer immunotherapy. Front Immunol (2019) 10:1074. doi: 10.3389/fimmu.2019.01074
Keywords: complement, CD55, colon cancer, NF-κB, MiR-27a-3p
Citation: Liu J, Fu N, Yang Z, Li A, Wu H, Jin Y, Song Q, Ji S, Xu H, Zhang Z and Zhang X (2023) The genetic and epigenetic regulation of CD55 and its pathway analysis in colon cancer. Front. Immunol. 13:947136. doi: 10.3389/fimmu.2022.947136
Received: 18 May 2022; Accepted: 28 December 2022;
Published: 18 January 2023.
Edited by:
Nicolas Larmonier, Université de Bordeaux, FranceReviewed by:
Gustavo Pedraza-Alva, Institute of Biotechnology, National Autonomous University of Mexico, MexicoCopyright © 2023 Liu, Fu, Yang, Li, Wu, Jin, Song, Ji, Xu, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi Zhang, emhpMTk2OUAxNjMuY29t; Xuemei Zhang, emhhbmd4dWVtZWlAbmNzdC5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.