
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 02 August 2022
Sec. Systems Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.940935
This article is part of the Research Topic Crosstalk between the microbiome, metabolome, and host immunity: a therapeutic perspective View all 4 articles
 Wiwat Chancharoenthana1,2*
Wiwat Chancharoenthana1,2* Nattawut Sutnu3,4
Nattawut Sutnu3,4 Peerapat Visitchanakun3,4
Peerapat Visitchanakun3,4 Vorthon Sawaswong5,6
Vorthon Sawaswong5,6 Suwalak Chitcharoen6
Suwalak Chitcharoen6 Sunchai Payungporn5,6
Sunchai Payungporn5,6 Alexandra Schuetz7,8,9
Alexandra Schuetz7,8,9 Marcus J. Schultz10,11,12
Marcus J. Schultz10,11,12 Asada Leelahavanichkul3,4*
Asada Leelahavanichkul3,4*Because studies on all fecal organisms (bacteria, fungi, and viruses) in sepsis are rare and bacteriophages during sepsis might have adapted against gut bacteria with possible pathogenicity, cecal ligation and puncture (CLP; a sepsis mouse model) was evaluated. In fecal bacteriome, sepsis increased Bacteroides and Proteobacteria but decreased Firmicutes, while fecal virome demonstrated increased Podoviridae when compared with sham feces. There was no difference in the fungal microbiome (predominant Ascomycota in both sham and CLP mice) and the abundance of all organisms between sepsis and control groups. Interestingly, the transfers of feces from CLP mice worsened sepsis severity when compared with sham fecal transplantation, as evaluated by mortality, renal injury (serum creatinine and histology), liver damage (liver enzyme and histology), spleen apoptosis, serum cytokines, endotoxemia, and bacteremia. In contrast, the transfers of fecal viral particles from sepsis mice, but not from sham mice, attenuated inflammation in CLP sepsis possibly through the decrease in several fecal pathogenic bacteria (such as Proteobacteria, Gammaproteobacteria, and Prevotellaceae) as evaluated by fecal microbiome analysis. Perhaps the isolation of favorable bacteriophages in sepsis feces and increased abundance ex vivo before oral treatment in a high concentration are beneficial.
Sepsis, an organ failure syndrome produced by a malfunction in the host’s immune system in response to a systemic infection, is one leading cause of death that has been recognized as a major global healthcare issue (1). The gut microbiome is closely associated with sepsis as gut barrier defect can enhance or cause sepsis referred to as ‘gut-origin sepsis’ (2, 3), and severe sepsis also induces intestinal integrity defect (sepsis-induced gut failure) through several mechanisms of intestinal injury (ischemic reperfusion injury, necrosis, and apoptosis) (4). Despite the fact that the pathophysiology of sepsis is complicated and incompletely understood, the disruption of the gut microbiota (gut dysbiosis) predisposes to sepsis may have a negative impact on sepsis outcomes. Indeed, the gut barrier consists of a single layer of epithelial cells, held together by epithelial tight junctions (TJs), with an approximate surface area of 20–30 m2, which serves as a selective physical barrier between the host and gastrointestinal (GI) contents, forming the intrinsic mucosal defense system (5). Gut barrier defect (leaky gut) allows the translocation of the microbial molecules (pathogen-associated molecular patterns (PAMPs)) or viable organisms from the gut into blood circulation or other sterile sites of the body (gut translocation) that facilitates systemic inflammation (2). In the gut, bacteria from the phyla Firmicutes (mostly Gram-positive anaerobes) and Bacteroidetes (obligate Gram-negative anaerobes) are the most predominant microbiota (6), followed by fungi, especially Candida albicans (in humans) and Candida non-albicans (in mice), which are the member of the phylum Ascomycota (7). Interestingly, the interactions between bacteria and fungi in the gut are important in sepsis, as fungal administration in mice enhances pathogenic bacteria (8) and antifungal drugs alter the bacterial community (9). Additionally, gut translocation of the major PAMPs of bacteria and fungi, including lipopolysaccharide (LPS) and (1→3)-β-D-glucan from Gram-negative bacteria and Candida spp., respectively, is partly a factor modifying sepsis severity, and control of the abundance of these organisms in the gut might attenuate sepsis (2).
Although the impact of bacteria and fungi in the gut on sepsis pathogenesis has been profoundly studied (10, 11), data on the whole gut microbiome, including the bacteriome, the virome, and mycobiome, in sepsis remains ill-defined. Bacteriophages (phages), the virus that infects and replicates within bacteria, have been proposed to be used against several antibiotic-resistant bacteria as adjunctive therapy (12, 13). Due to the specificity of phages against only limited bacterial strains, partly from the strain-specific molecules for the entry of viruses, the combination of phages is used to overcome this limitation (14). Because i) phages are a natural bacterial control in the ecosystem (15, 16), including the intestines (17, 18), and ii) sepsis (and systemic inflammation) alters gut microbiota (dysbiosis) and increases harmful microorganisms in the gut (19, 20), the increased bacteria with possible pathogenicity in the host with sepsis might naturally induce phage propagation, and these bacteria might partly be controlled by the induced phages. Thus, the adaptation of phages in the host might already be a combination of the necessary phages for the specific conditions, and the boost-up of these phage combinations might be beneficial as effective phages for individual patients.
Our goals were to investigate fecal organisms using a sepsis mouse model in order to test the following possibilities: i) sepsis may interfere with all gut organisms (bacteria, fungi, and viruses), ii) balanced homeostasis may exist in the sepsis intestine, and iii) gut microbial modification by the transfer of feces from normal mice (sham feces) or phages (viral particles) from sepsis mice may attenuate sepsis. Hence, the cecal ligation and puncture (CLP) sepsis mouse model was used for the experiments.
Male 8-week-old C57BL/6 mice were purchased from Siam Nomura (Samut Sakhon, Thailand) and had free access to water and chow before and after surgery. For sepsis induction, CLP was performed according to previous publications (21–24). Briefly, the cecum was ligated at 12 mm from the cecal tip, punctured twice with a 21-gauge needle, and gently pressed to express a small amount of fecal material before being placed into the abdominal cavity. In sham mice, the cecum was only identified through abdominal incision before suturing layer by layer with 6-0 nylon sutures. Then, 1 ml of prewarmed normal saline solution (NSS) with tramadol at 25 mg/kg/dose was subcutaneously administered after surgery, at 6 and 18 h post-CLP. Fecal samples for microbiome analysis were collected at 24 h of surgery (sham and CLP) after mice were sacrificed by cardiac puncture under isoflurane anesthesia.
Fecal samples from each mouse (0.25 g per mouse) from different cages in each experimental group were collected for microbiota analysis following a previous protocol (22, 25–28). In short, metagenomic DNA was extracted by DNeasy PowerSoil Kit (Qiagen, Maryland, USA) using the Universal prokaryotic 515F (forward, (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (reverse, 5′-GGACTACHVGGGTWTCTAAT-3′), with the Illumina adapter and Golay barcode sequences for 16S rRNA gene V4 library construction in Miseq300 platform (Illumina, San Diego, CA, USA). The raw sequences and operational taxonomic unit (OTU) were classified following Mothur’s standard operating platform. In parallel, to identify the taxonomic profiles of fungal microbiota (mycobiome) in feces, the DNA was extracted from fecal samples. The universal eukaryotic primers ITS3 (forward; 5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (reverse; 5′-TCCTCCGCTTATTGATATGC-3′) were used for identifying the gut mycobiota. The fungal DNA library was sequenced using the Miseq system (Illumina, San Diego, CA, USA) at Omics Sciences and Bioinformatics Center, Chulalongkorn University. Forward and reverse primers were removed from raw sequences using cutadapt version 1.18 and trimmomatic version 0.39 with the sliding window option to trim individual sequences where the average quality scores are less than 15 across four base pairs. To identify the composition of fungi in fecal samples, amplicon sequence variants (ASVs) were analyzed using the QIIME2 plugin DADA2 pipeline. Unclassified phylum or higher fungal classification was removed. To confirm non-fungal origin, the fungal classification was analyzed by a BLAST-based tool (https://blast.ncbi.nlm.nih.gov) (National Center for Biotechnology Information, Bethesda, MD, USA).
The procedure of virome sample processing was carried out following a previous study (29), with some modifications. Briefly, 50 mg of fecal sample was resuspended in 1 ml of phosphate-buffered saline (PBS) and centrifuged at 8,000 × g at 4°C for 5 min. The supernatant was filtered through a 0.45-µm syringe filter (Sartorius, Göttingen, Germany). The filtrate at 500 µl was then treated by a nuclease cocktail containing 30 U of RNase One (Promega, Madison, WI, USA), 3 U of Baseline-ZERO (Epicentre, Madison, WI, USA), 30 U of Benzonase (Novagen, Darmstadt, Germany), and 14 U of Turbo DNase (Ambion, Thermo Fisher Scientific, Waltham, MA, USA) in 1× Turbo DNase buffer. The sample was incubated at 37°C for 1.5 h and was extracted for viral nucleic acid using the MagMAX™ Viral RNA Isolation kit (Applied Biosystems, Thermo Fisher Scientific, USA). The cDNA was constructed based on the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA) with 100 pmol of Sol A random primer: 5′-GTT TCC CAC TGG AGG ATA NNN NNN NNN-3′ in 20 µl of reaction following the manufacturer’s protocol. After the first strand synthesis was done, 5 U of Klenow Fragment DNA polymerase (New England Biolabs, Ipswich, MA, USA) was added to the reaction and incubated at 37°C for 60 min and 75°C for 20 min. The random amplification of cDNA was performed based on a PCR comprising 2 µM of Sol B primer: 5′-GTT TCC CAC TGG AGG ATA-3′, 0.2 U of Phusion DNA polymerase (Thermo Fisher Scientific, USA), 0.25 of mM of dNTPs, 5 µl of cDNA template, and 1× Phusion HF buffer. The thermal profile was 95°C for 5 min, 5 cycles of 95°C for 30 s, 59°C for 60 s, and 72°C for 90 s, 35 cycles of 95°C for 30 s, 59°C for 30 s, and 72°C for 90 s (+2 s per cycle) followed by 72°C for 10 min and hold at 4°C. The random amplified products were then used for DNA library preparation using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, USA). Finally, the library was paired-end sequenced (2 × 250 cycles) using the Illumina MiSeq sequencing platform with 10% PhiX spike-in. The raw FASTQ reads were quality checked by FastQC. Then, the low-quality sequences (<Q20) and adaptor sequences were trimmed by Trimmomatic (version 0.36) (30). The host reads were filtered out by mapping the reads against the reference mouse genome (GRCm38) using Bowtie2 (31). The unmapped reads were de novo assembled by EnsembleAssembler (32). The contigs were BLASTx search against viral protein database (collected from ftp://ftp.ncbi.nih.gov/refseq/release/viral/) using with e−10 E−value cutoff. The taxonomically classified contigs were subsequently used as the reference sequence for mapping the viral reads to count the hits of each viral taxon. For quantitation of fecal viruses, the filtrated samples were diluted and stained with SYBR Gold (Thermo Fisher, CA) for DNA viruses and SYBR Green II (Solarbio, Beijing, China) for RNA viruses for 15 min before washing as previously described (33–35) and visualizing by LSM 800 Airyscan confocal laser scanning microscopy (CLSM; Carl Zeiss, Jena, Germany).
To determine the impact of fecal organisms on the CLP model, fecal transfers (fecal transplantation) were performed following a modified protocol (36). Briefly, fecal samples from cecum and colon in mice after 24 h post-CLP or post-sham were collected, diluted with PBS in a ratio of 1 g feces:0.5 ml PBS, and thoroughly mixed. Notably, colon fecal contents in sham mice need to be minced before being mixed with PBS, while CLP stools were usually liquified at 24 h post-surgery. After that, the solid fractions were separated by centrifugation at 13,000 × g for 10 min (at 4°C), and the fresh supernatant at 10 ml/kg was orally administered to the recipient mice once daily for 5 days. At 4 h after the last dose of fecal gavage, CLP was performed in the recipient mice as previously mentioned. Sham surgery was performed only in mice with fecal gavage by CLP feces, but not the gavage by sham feces, because all parameters of sham with fecal transplantation (sham feces or CLP feces) were not different from sham without fecal gavage (data not shown). Because the worsening of sepsis after transfers of sepsis feces might be due to fecal viruses, oral gavage of viral particles separated from sham and CLP mice after 24 h of surgery was also conducted. For the extraction of viruses, fecal samples from the cecum and colon were mixed with SM buffer (150 mM of NaCl, Tris-HCl pH 6.5, 10 mM of MgCl2, and 1 mM of CaCl2). The samples were centrifuged at 1,000 × g for 1 min to remove the tissue, followed by centrifugation at 5,000 × g for 30 min to separate the solid part.. The phage supernatants were collected and filtered through 0.45-µm pore-size syringe filters. All filtrated samples for viruses that were freshly extracted from the feces of one CLP mouse were transferred to one recipient mouse, and the process was repeated once daily for 5 days. Then, CLP or sham was performed for 4 h after the last dose of the phage administration.
Because the impact of crude fecal transplantation, similar to the administration of all fecal organisms (bacteria, virus, and fungi), could differ from the fecal transfer of viral particles (partly bacteriophages or phages) alone, viral particles freshly isolated from mice were orally administered thrice a day for 5 days prior to CLP (or sham) surgery. For the phage isolation, fecal samples from the cecum and colon were mixed with SM buffer (150 mM of NaCl, Tris-HCl pH 6.5, 10 mM of MgCl2, and 1 mM of CaCl2) before centrifugation at 1,000 × g for 1 min to remove the tissue, followed by a 5,000 × g centrifugation for 30 min to precipitate the bacterial cell. Then, the supernatants containing phages were collected and filtered through 0.45-µm syringe filters. Total feces per mouse after sham and CLP surgery with the approximate weight of 0.7 ± 0.3 and 0.4 ± 0.2 g, respectively, was diluted by 2 and 1.5 ml of SM buffer, respectively. The fecal samples of sham mice required a higher volume of dilution because of the well-formed consistency of the feces, while CLP fecal samples were liquefied for easier phage preparation. The abundance of viral particles in the samples was determined by staining with SYBR Gold (Thermo Fisher) and SYBR Green II (Solarbio, China) for 15 min before washing and visualizing by LSM 800 Airyscan CLSM (Carl Zeiss, Jena, Germany) as previously described (33–35). The total phage preparation from a mouse was isolated daily and divided into three times of oral administrations (keeping the preparations at 4°C between the doses). Then, the preparations at 0.5 × 109 viral particles per dose (approximately 10 ml/kg/dose) were orally administered every 4 h for one recipient mouse per day. After 5 days of administration, CLP surgery was performed the next day as described above.
At 24 h post-surgery, all mice were sacrificed through cardiac puncture under isoflurane anesthesia with sample collection (blood and organs). Kidney injury was determined by blood urea nitrogen (BUN) and serum creatinine using QuantiChrom (DIUR-500 and DICT-500, respectively) (BioAssay, Hayward, CA, USA). Liver damage and serum cytokines were determined by EnzyChrom Alanine Transaminase assay (EALT-100) (BioAssay) and enzyme-linked immunosorbent assays (ELISA) for mouse cytokines (Invitrogen, Carlsbad, CA, USA), respectively. In addition, blood leukocyte determination was performed by mixing blood with 3% acetic acid, a hemolytic solution, with a ratio of blood: acetic acid at 6:100 by volume before counting with a hemocytometer. In parallel, the Wright-stained blood smear was examined for the percentage of polymorphonuclear cells (PMNs) and lymphocytes, and the total cell numbers were calculated by the total count from the hemocytometer multiplied by the percentage of cells from the Wright-stained slide. Serum endotoxin (LPS) was measured by HEK-Blue LPS Detection (In vivoGen, San Diego, CA, USA), and the data were recorded as 0 when LPS values were less than 0.01 EU/ml because of the limited lower range of the standard curve. For bacterial burdens in blood, mouse blood samples in several dilutions were directly spread onto blood agar plates (Oxoid, Basingstoke, Hampshire, UK) and incubated at 37°C for 24 h before the enumeration of bacterial colonies. Gut permeability was determined by fluorescein isothiocyanate (FITC)-dextran assay (20, 37) using an oral administration of FITC-dextran, a non-absorbable molecule with 4.4-kDa molecular mass (Sigma-Aldrich, St. Louis, MO, USA), at 12.5 mg at 3 h before the detection in serum by fluorospectrometer (NanoDrop 3300; Thermo Fisher Scientific, Wilmington, DE, USA).
The semi-quantitative evaluation of renal histology on paraffin-embedded slides was performed after 10% neutral buffered formalin fixation, followed by hematoxylin and eosin (H&E) staining at ×200 magnification in 10 randomly selected fields for each mouse as previously described (26, 38). Briefly, the renal injury was defined as tubular epithelial swelling, loss of brush border, vacuolar degeneration, necrotic tubules, cast formation, and desquamation using the following scoring method: 0, area of damage <5%; 1, area of damage 5%–10%; 2, area of damage 10%–25%; 3, area of damage 25%–50%; and 4, area of damage >50%. In parallel, an anti-active caspase-3 antibody (Cell Signaling Technology, Beverly, MA, USA) was used for immunohistochemistry staining on 4-m paraffin sections, which were then evaluated in 10 randomly selected ×200 magnified fields as previously described (20). The spleen apoptosis score was expressed as positive cells per high-power field. However, the liver histological score was the sum of hepatocyte injury characteristics (cytoplasmic color fading, vacuolization, nuclear condensation, nuclear fragmentation, nuclear fading, and erythrocyte stasis) ranging from 0 to 5 multiplied by grades of damage of 0 = no, 1 = mild, 2 = moderate, and 3 = severe, following a publication (39).
Mean ± standard error (SE) was used for data presentation. The differences between groups were examined for statistical significance by one-way analysis of variance (ANOVA) followed by Tukey’s analysis or Student’s T-test for multiple and two-group comparisons, respectively. The time-point experiments and survival analysis were analyzed by the repeated-measures ANOVA and the log-rank test, respectively. All statistical analyses were performed with SPSS 11.5 software (SPSS, IL, USA) and GraphPad Prism version 9.3.1 software (La Jolla, CA, USA). A p-value of <0.05 was considered statistically significant.
Microbiome analysis from feces of mice after 24-h surgery demonstrated the prominent differences in bacteria and viruses, but not fungi, when compared between sham and CLP mice (Figures 1A–D). In fecal bacteriome, Firmicutes (the most prominent bacteria in a healthy host with possible beneficial effects) (40) was predominant in sham mice, while Proteobacteria (the Gram-negative aerobes and facultative anaerobes) (8) and Bacteroides (the most prominent Gram-negative anaerobes with possible pathogenesis in some conditions) (25) were predominant in CLP mice (Figure 1A, left, and Figure 2A). Despite no differences in the observed OTUs (the operational definition used to classify groups of closely related individuals) and the microbial diversity (Shannon and Chao-1), several Proteobacteria bacteria, including Enterobacter spp., Desulfovibrio spp., Oscillospira spp., and Halomonas spp., were identified in CLP mice (Figure 1A, middle and right, and Figure 1D). Notably, the similar OTUs between sham and CLP mice suggested a comparable number of bacterial species between these groups, which were supported by the representative microbial diversity score (Shannon and Chao-1) (41). For fecal mycobiome (fecal fungi), there was a subtle alteration in sham and CLP mice in all parameters (Figure 1B), partly due to the less abundance of fecal fungi in mice than in humans (19, 42, 43). Although Ascomycota (the phylum of C. albicans, the most prominent fungi in the human gut) was not different between sham and CLP mice, the fecal abundance of Myrothecium, a genus of fungi in the Ascomycota group with a limited report of infection (44), in CLP mice was less than that in sham mice (Figure 2B). In the fecal virome, the predominant viruses in sham mice were Siphoviridae (double-stranded DNA phages against some Lactobacilli spp.) (45) and Picobirnaviridae (the double-stranded RNA phages against several bacteria that could cause diarrhea) (46), and in CLP mice, the viruses were Siphoviridae and Podoviridae (the double-stranded DNA phages against some Salmonella spp.) (47) (Figures 1C, 2C). Meanwhile, Myoviridae (the double-stranded DNA phages against some Enterobacteria) (48) in sham mice was higher than in CLP mice (Figures 1C, 2D). Despite a limitation of random sequencing of virome on an analysis of the viral abundance in feces, direct fluorescent staining of viruses (DNA and RNA viruses) was not different between CLP and sham mice (Figure 2C).
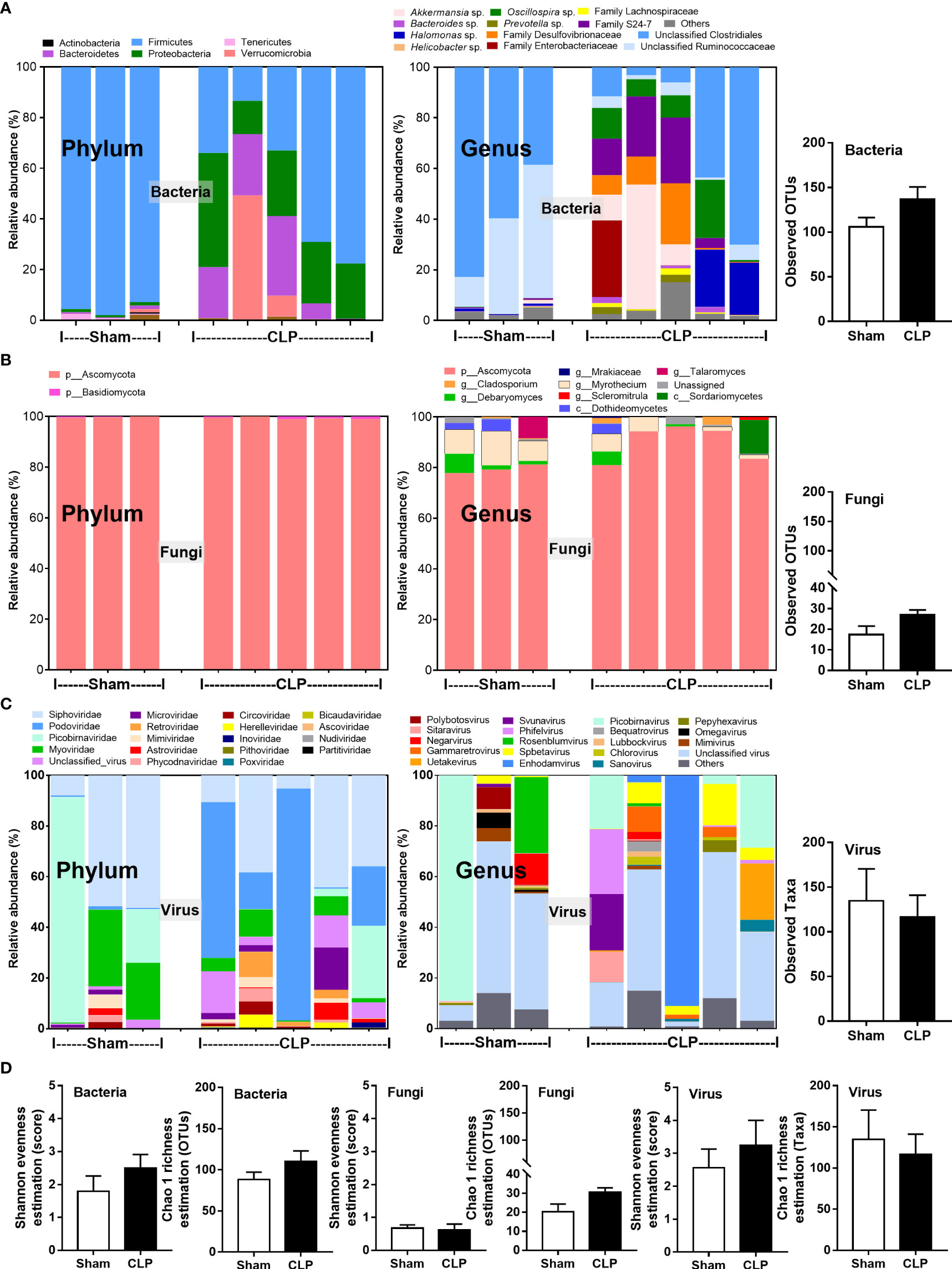
Figure 1 The fecal abundance of microbiome analysis on 16S rRNA (bacteria) (A) and nuclear ribosomal internal transcribed spacer (ITS) (fungi) (B) or random sequence (viruses) (C) from feces of mice after 24 h of sham or cecal ligation and puncture (CLP) surgery in phylum level and genus level together with the observed operational taxonomy units (OTUs) of bacterial and fungal analysis and observed taxa of virome analysis are demonstrated. Alpha diversity (Shannon and Chao-1 analysis) of bacteriome, mycobiome, and virome (D) are also indicated.

Figure 2 The fecal abundance of bacteria (Firmicutes, Bacteroides, and Proteobacteria) (A), fungi (Debaryomyces and Myrothecium) (B), and viruses (Siphoviridae, Podoviridae, and Myoviridae) (C) with quantitative analysis on DNA and RNA viruses by fluorescent staining (D) from feces of mice after 24 h of sham or cecal ligation and puncture (CLP) surgery are demonstrated (n = 3 in sham and 5 in CLP). *, p < 0.05 between the indicated group.
Although it has been previously mentioned that the transfers of feces from healthy and sepsis mice attenuate and worsen sepsis, respectively (49–51), the studies using the transfers of fresh feces are still very limited, and the conclusions are still inconsistent. Here, daily fecal gavage from CLP mice for 5 days before CLP surgery worsened sepsis severity as compared with CLP operation on sham fecal transfer recipients. Accordingly, CLP in mice with oral gavage by sepsis feces demonstrated a higher mortality rate, serum cytokines, endotoxin, bacteremia, gut leakage (FITC-dextran), and organ injury (liver and kidney) when compared with CLP with sham feces, despite the non-difference in spleen apoptosis (Figures 3A–L, 4A–D) possibly due to the pathogenic organisms from the orally administered sepsis feces. Accordingly, the kidney injury (mainly determined through renal tubular damage such as vacuolization, swelling, and desquamation) and liver damage (mainly evaluated by cell vacuolization) in CLP with sepsis feces were higher than in CLP with sham feces, while cell apoptosis (brown color stained in activated caspase 3 staining) in the spleen was not different between these groups (Figures 4A–D). Different from the fecal gavage of sepsis feces (36, 52, 53), we explore the impacts of the transfers of fecal viral particles from sepsis feces. As such, oral gavage of fecal viral particles that separated from feces of sepsis mice attenuated several sepsis parameters, including kidney and liver injuries (serum creatinine and alanine transaminase), bacteremia, endotoxemia, gut barrier defect (FITC-dextran assay), and serum cytokines (TNF-α, IL-6, and IL-10), but not survival analysis (Figures 5A–I).
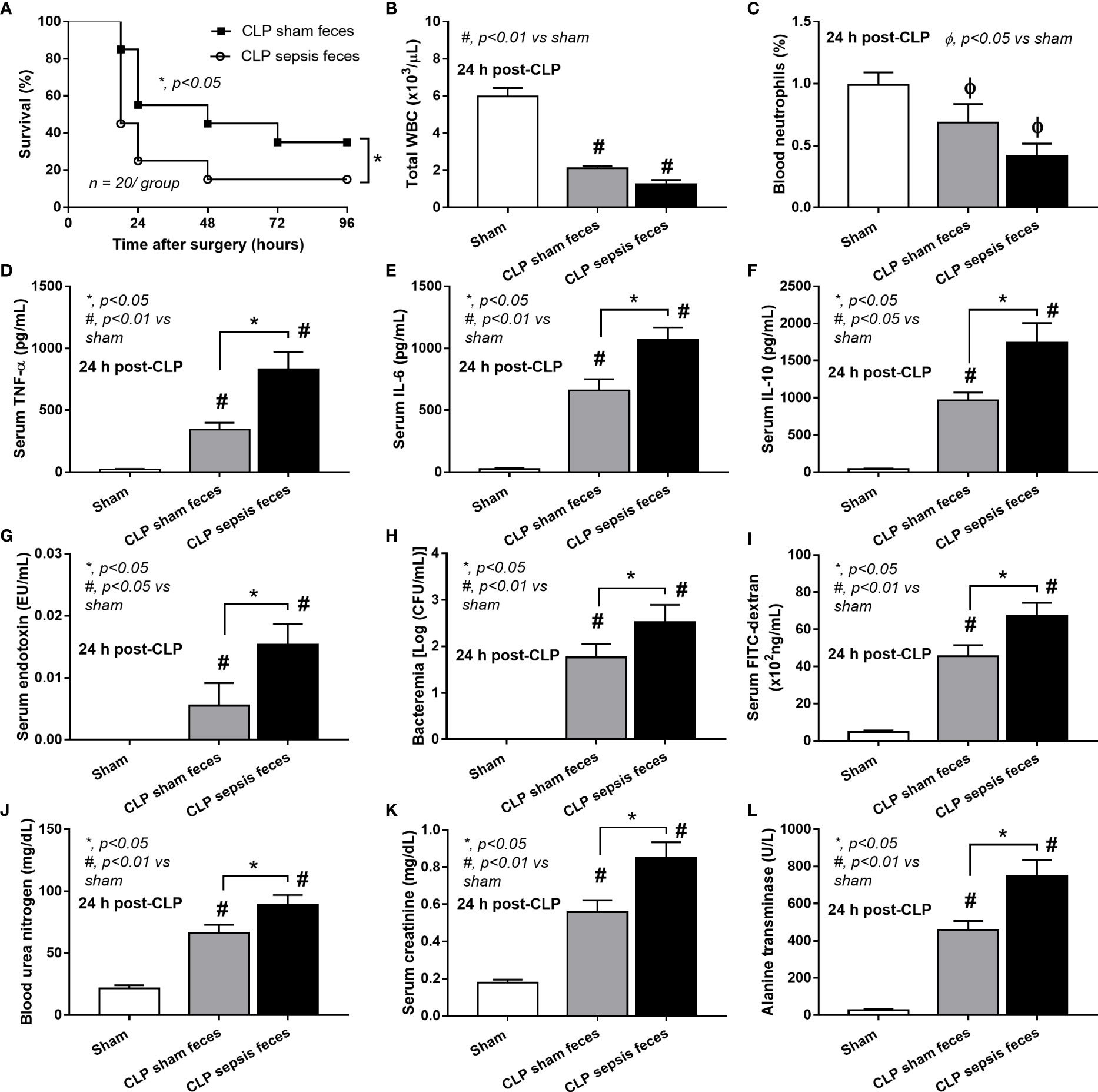
Figure 3 Characteristics of cecal ligation and puncture (CLP) mice with fecal transplantation of sham feces or CLP feces or sham mice with CLP feces (Sham) as evaluated by survival analysis (A), peripheral blood leukocytes and neutrophils (B, C), serum cytokines (TNF-α, IL-6, and IL-10) (D–F), serum endotoxin (G), bacteremia (H), gut leakage (FITC-dextran) (I), renal injury (blood urea nitrogen and serum creatinine) (J, K), and liver enzyme (serum alanine transaminase) (L) are demonstrated (n = 6–8 per group). *, p < 0.05 between the indicated group; #, p < 0.01 vs sham.
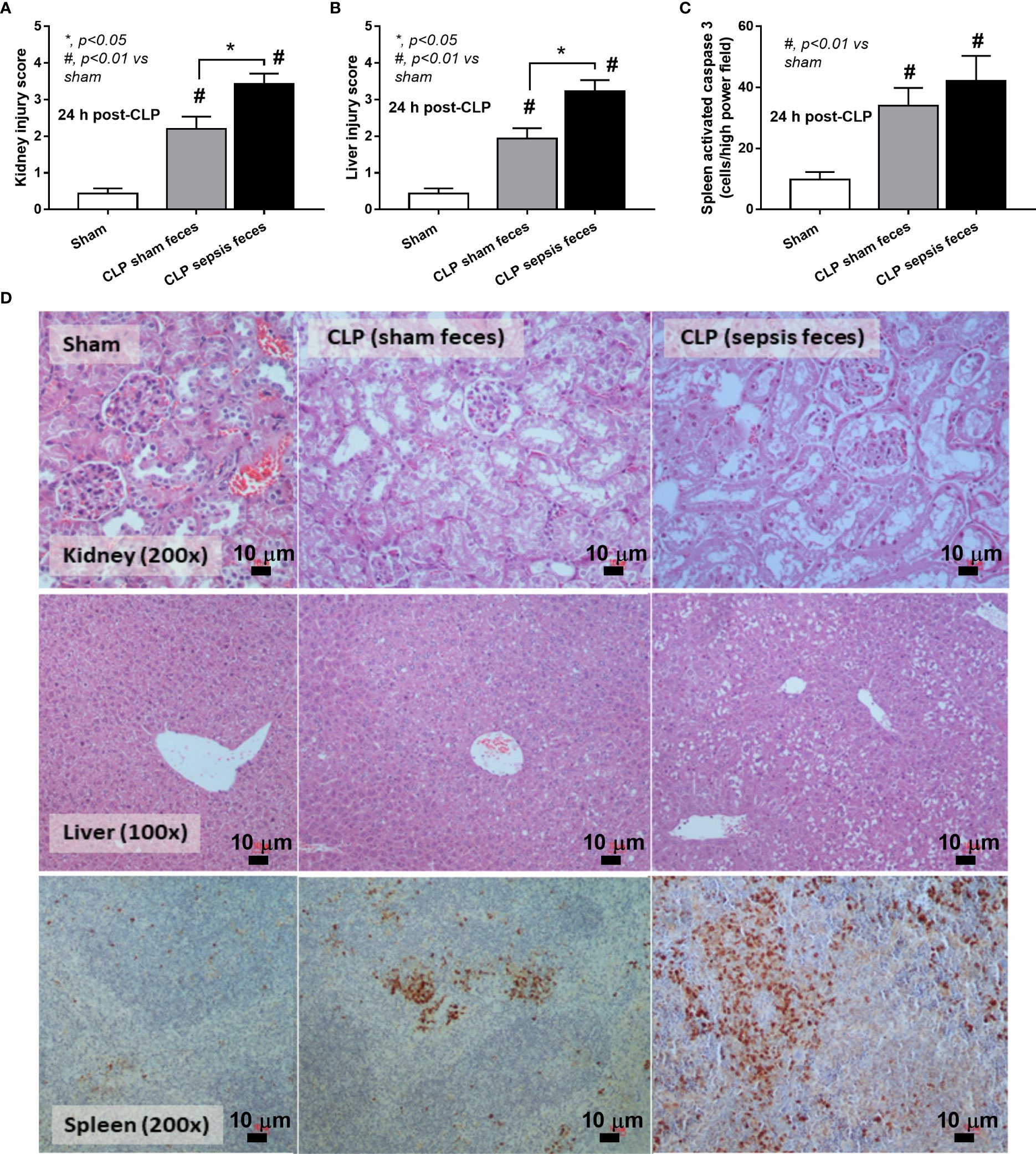
Figure 4 Characteristics of cecal ligation and puncture (CLP) mice with fecal transplantation of sham feces or CLP feces or sham mice with CLP feces (Sham) as evaluated by a histological score of kidney and liver (A, B) and spleen apoptosis (C) with the representative pictures (D) (original magnification ×200) of hematoxylin and eosin (H&E) staining (kidney and liver) and activated caspase 3-stained apoptosis from spleen are demonstrated (n = 6–8 per group). *, p < 0.05 between the indicated group; #, p < 0.01 vs sham.
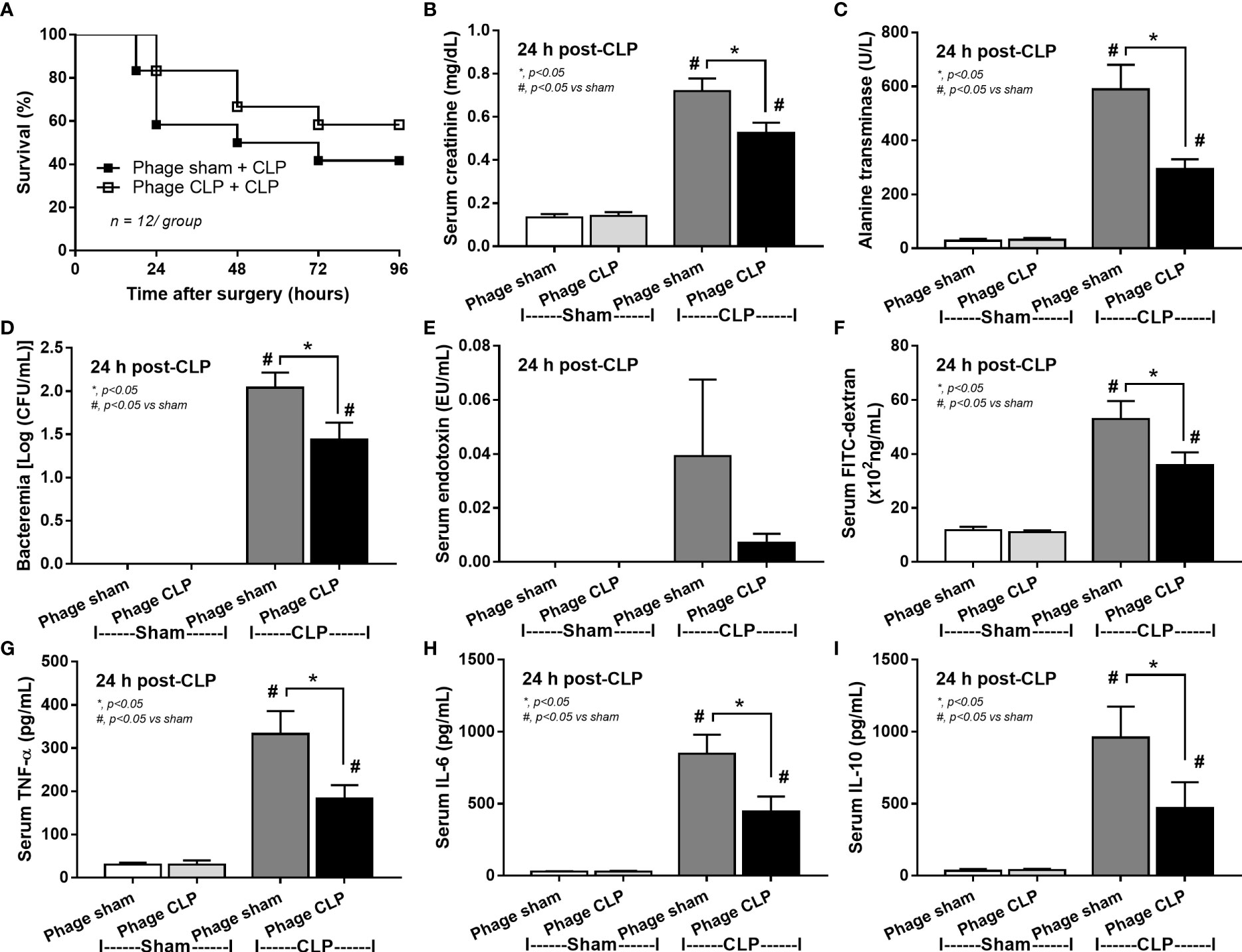
Figure 5 Characteristics of cecal ligation and puncture (CLP) mice with transfers of viral particles that were isolated from sham feces (phage sham) or CLP feces (phage CLP) before performing sham or CLP surgery as evaluated by survival analysis (A), serum creatinine (B), serum alanine transaminase (C), bacteremia (D), endotoxemia (E), gut barrier defect (FITC-dextran assay) (F), and serum cytokines (TNF-α, IL-6, and IL-10) (G–I) are demonstrated (n = 12 per group for A and 8–10/group for B–I). *, p < 0.05 between the indicated group; #, p < 0.05 vs sham.
Because of the possible different influences of fecal bacteriophages between sham and CLP mice, bacteriome analysis in mouse feces at 24 h post-CLP after the 5 days of gavage by viral particles from sham and CLP mice was conducted (Figures 6, 7, 8). Accordingly, viral particles from CLP reduced Proteobacteria (the pathogenic bacteria) and increased Firmicutes (the possible beneficial bacteria) (25), while they did not alter the abundance of Bacteroides (in phylum level of the analysis) and total fecal Gram-negative bacteria (Figure 8A). In the class and family levels of analysis, CLP viral particles decreased pathogenic bacteria, such as Gammaproteobacteria and Desulfovibrio (54, 55), and increased some beneficial bacteria, such as Bacilli (including probiotics Lactobacillus spp.) and Verrucomicrobia (including Akkermansia spp. with the possible enhanced gut integrity) (56, 57) (Figures 8B, C). However, we could not isolate phages from feces of both sham and sepsis mice using the culture with crude feces (aerobic and anaerobic processes) or the high abundance of bacteria that were isolated from feces (Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa) (data not shown). Perhaps, i) the beneficial phages are against several specific species of bacteria and ii) the combination of phages in the preparation of fecal viral particles, but not a single dominant phage is important for the sepsis attenuation effect..
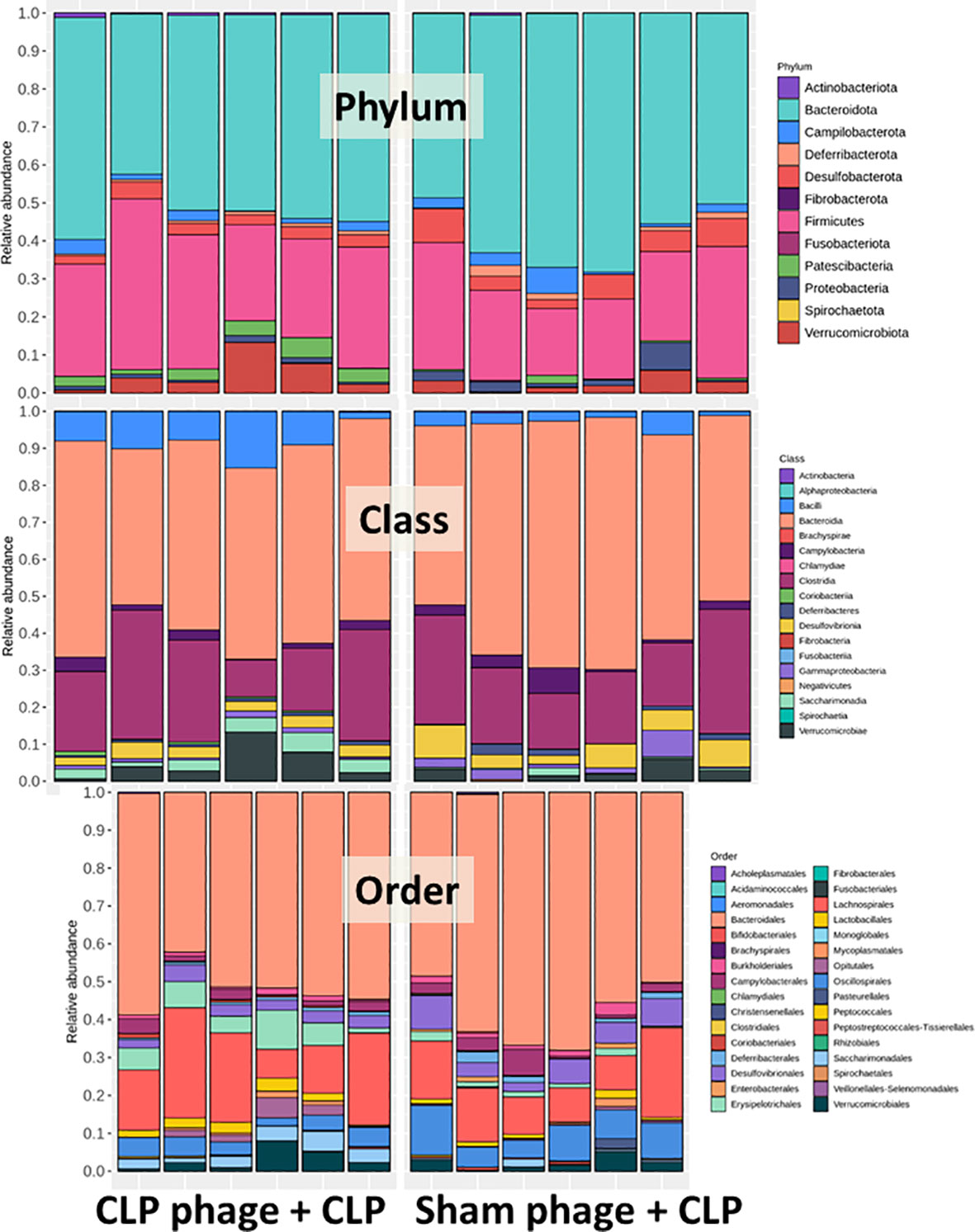
Figure 6 Fecal microbiota analysis from mice at 24 h after cecal ligation and puncture (CLP) surgery that was pre-conditioning by orally administration with the viral particles from sham (sham phage + CLP) or CLP mice (CLP phage + CLP) as determined by the relative abundance of bacterial diversity at phylum, class, and order levels of the analysis are demonstrated.
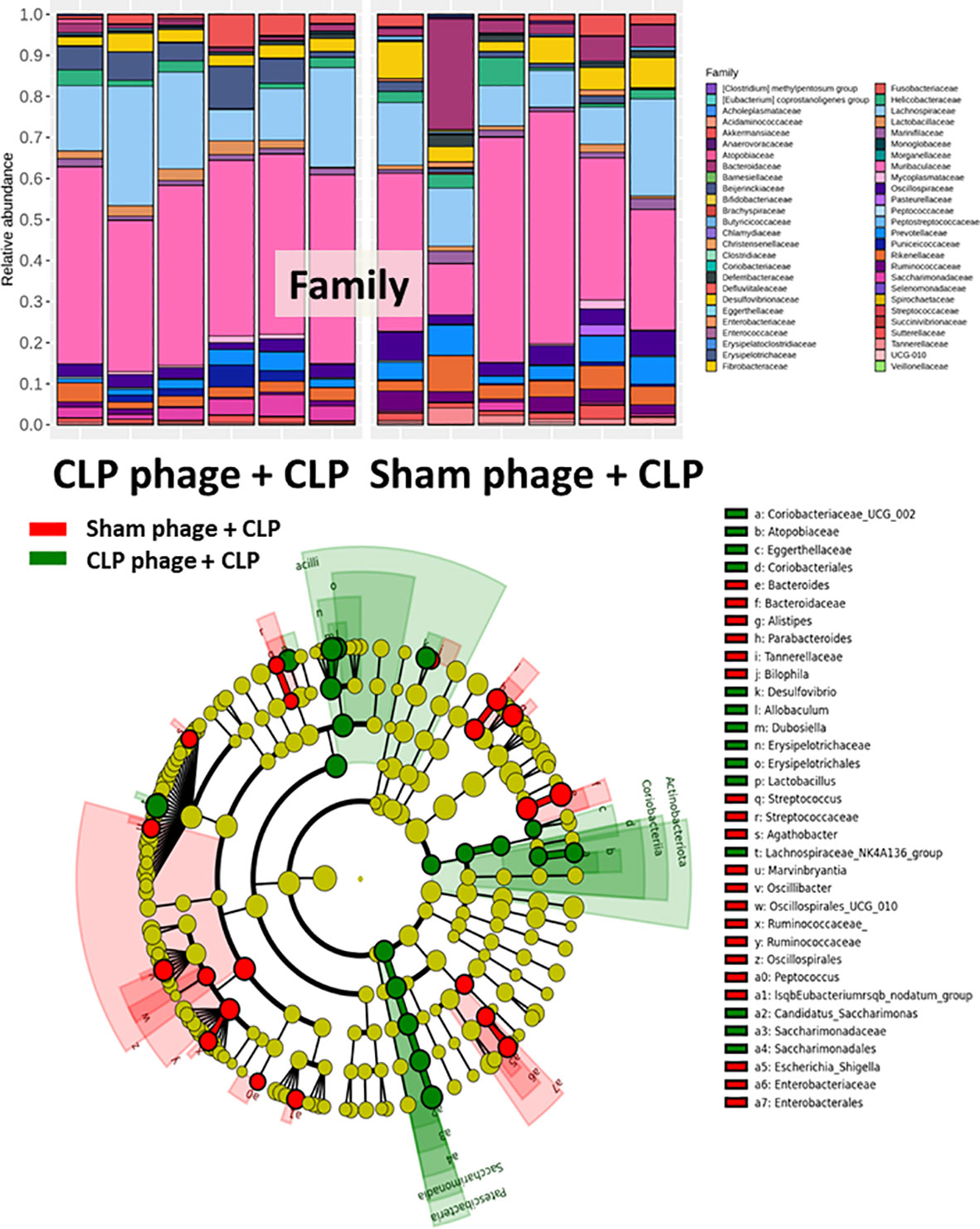
Figure 7 Fecal microbiota analysis from mice at 24 h after cecal ligation and puncture (CLP) surgery that were pre-conditioning by orally administration with the viral particles from sham (sham phage + CLP) or CLP mice (CLP phage + CLP) as determined by the relative abundance of bacterial diversity at the family level and cladogram plot are demonstrated.
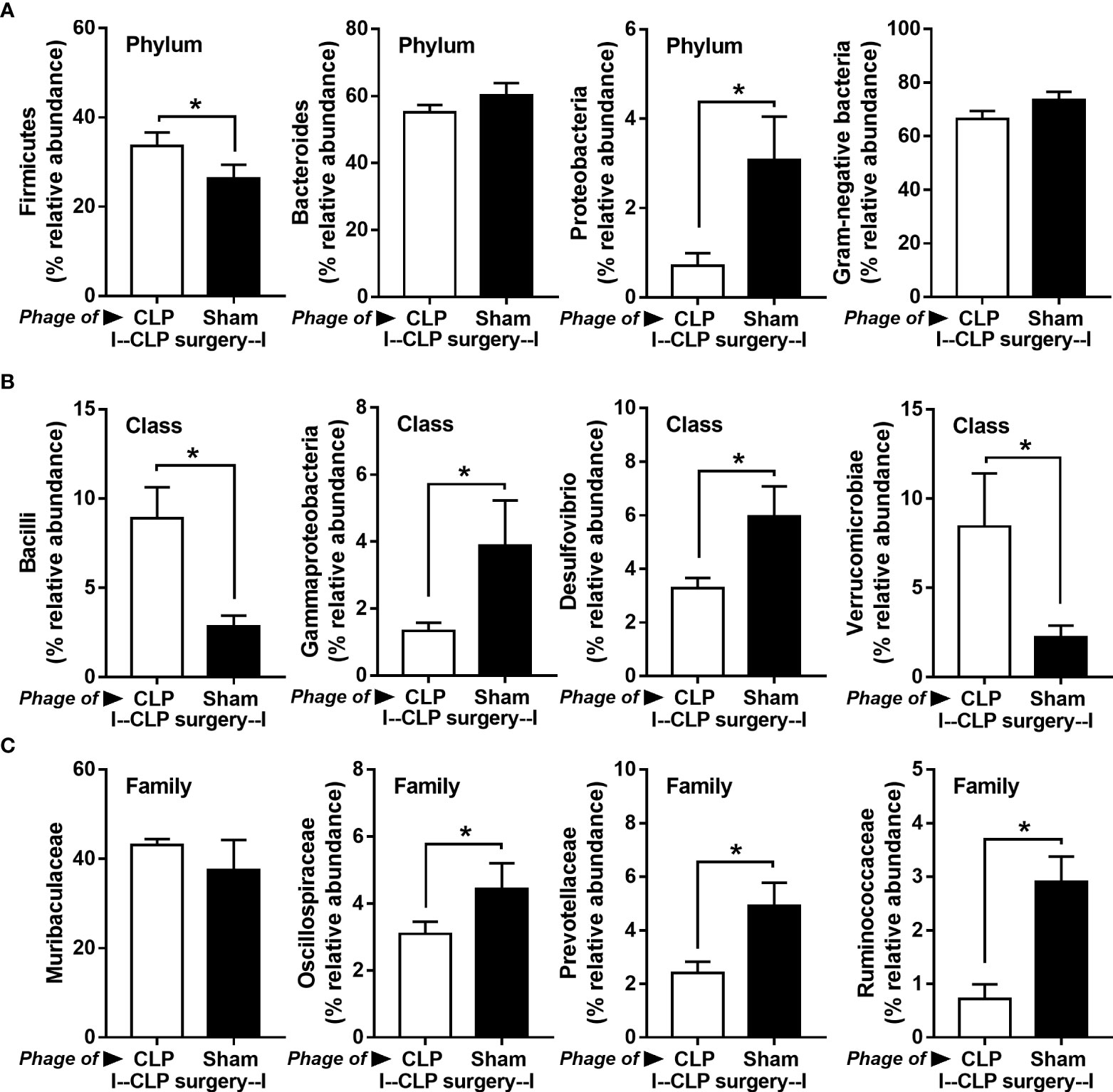
Figure 8 Fecal microbiota analysis from mice at 24 h after cecal ligation and puncture (CLP) surgery that were pre-conditioning by the oral administration with the viral particles from feces of sham (phage of sham) or CLP mice (phage of CLP) as indicated by graph presentation of the relative abundance of bacterial diversity for some important bacteria in phylum level (with total Gram-negative bacteria in feces) (A) and in class and family levels of the analysis (B, C) are demonstrated. *, p < 0.05 between the indicated group.
Sepsis altered all organisms in the gut. The transfers of crude sepsis feces worsen sepsis, while the crude sham feces did not alter sepsis. However, the viral particles from sepsis feces, but not from sham feces, attenuated sepsis.
The enhanced bacteria with possible pathogenicity, especially Proteobacteria, in the gut after sepsis is well-known as a result of either the local intestinal inflammation (such as Clostridium infection) or systemic responses (bacteremia with septicemia) (58). While some intestinal gut microorganisms directly decrease the beneficial bacteria (partly through the competition for nutrients), systemic infection indirectly alters the gut microbiome through several mechanisms, for example, an intestinal oxygen alteration (vasodilatation-induced hyperoxygenation or low blood pressure-induced hypoxia) and the select growth of some organisms from phospholipids (derived from the dying enterocytes) or the antibiotic peptides (59). Unfortunately, most of the bacteria with possible pathogenicity usually have more protecting factors against harsh microenvironments than beneficial bacteria (59). Regarding the gut fungi, Ascomycota in the phylum-level analysis was similarly predominant in the feces of both sham and sepsis mice. However, the abundance of Myrothecium in the genus-level analysis in sepsis feces was lower than in the sham group, which might partly initiate a selective environment for some microorganisms with possible pathogenicity. Because sepsis significantly reduced Myrothecium, which produces some molecules against several harmful factors to the host, including some organisms (60) and toxic substances from enterocytes (61, 62), the reduced Myrothecium might be another factor associated with sepsis-induced gut dysbiosis.
With gut virome analysis, there were more prominent Myoviridae (in sham) and Podoviridae (in CLP), the main components of several phage cocktails in other studies (48), without the significant differences in other viral families between sham and sepsis. Both Myoviridae and Podoviridae are phages against several Staphylococci spp., while only Myoviridae can attack coagulase-negative staphylococci (63), the gut bacteria causing neonatal sepsis (64). Because Proteobacteria was enhanced by sepsis and Podoviridae eliminated some Proteobacteria (such as Citrobacter freundii) (47), the enhanced Podoviridae in sepsis might be the phage against sepsis-induced gut bacteria with possible pathogenicity. Although these sepsis-induced viruses might have a potential for ameliorating septic inflammation or pathology, Podoviridae is a family of very short-non-contractile tail viruses in the order Caudovirales with 130 species in this family, which possibly attenuate several important sepsis-associated bacteria (P. aeruginosa, E. coli, Salmonella spp., and Staphylococcus spp.) (47, 48, 65, 66). The subsequent tests of the Podoviridae cocktail in sepsis are of interest. Nevertheless, there are still limited data on the non-bacterial gut organisms in sepsis compared to the larger volume of literature on sepsis-induced bacterial dysbiosis. Here, we hypothesize that CLP sepsis induces the harsh microenvironment leading to the overgrowth of Proteobacteria with the reduced beneficial organisms resulting in an increase in phages against Proteobacteria as a natural organismal control. Unfortunately, during sepsis, the improved beneficial phages are unable to keep up with the bacteria with possible pathogenicity, leading to increased gut dysbiosis and sepsis disease progression. It would be interesting to see further research on the interkingdom association of gut microbes in sepsis.
The fecal transplantation of sepsis feces worsen sepsis, partly due to increased pathogenic bacteria and worsening of gut dysbiosis, supporting previous publications (36). Surprisingly, administration of viral particles from sepsis feces, but not from sham feces, attenuated sepsis as indicated by several parameters (kidney injury, liver damage, gut barrier defect, and serum cytokines). Despite the possibility of the transfer of other beneficial factors (such as short-chain fatty acids) from sepsis feces, the washing step during the preparation of viral particles (see Materials and methods) should decrease this contamination, and the experimental impacts might mainly be due to the viral particles. These data implied that there were higher beneficial phages in sepsis feces than in sham feces, possibly for natural control of the overgrowth of harmful microorganisms during sepsis. Although administration of viral particles from sepsis feces slightly decreased Firmicutes (the possible beneficial bacteria) (67), pathogenic Proteobacteria (68) were prominently reduced. Because phages infect only some specific bacteria that express similar molecules for viral entry (69, 70), the effects of viral particles on several bacteria imply the presence of multiple phages as the natural cocktails against pathogenic bacteria in sepsis feces. Although these beneficial phages might be the phages against both aerobes and anaerobes, the separation and identification of these phages from mouse feces are difficult. Indeed, we could not identify phages using either mixed or isolated bacteria (both aerobes and anaerobes) that were extracted from mouse feces (both sham and sepsis groups) (data not shown). Despite these limitations, isolation of viral particles against pathogenic bacteria from the infected host before the ex vivo phage propagation and re-administration of these natural phage cocktails into the host is an interesting hypothesis to test. With this strategy, the extensive preliminary preparation of phage cocktails and the pre-form test for phage bactericidal activity might not be necessary because the proper combination of phages is directly isolated from the host during the natural control of harmful microorganisms. More studies to test this hypothesis are warranted.
Because only a sepsis model induced by intra-abdominal infection using CLP surgery was used, the results might not refer to sepsis with other sources of infection (such as pneumonia or renal sepsis). Although there are several intra-abdominal sepsis models, including CLP, colon ascendens stent peritonitis (CASP), and cecal slurry injection (intraperitoneal injection of cecal contents from a donor rodent) (71), the CLP model was used here due to the possible less interference on the gut microbiota. While the foreign material of stent in CASP and the manipulation of feces from the rodent donor might interfere with the growth of some organisms, CLP seems to have less influence on gut microbiota.
Due to the specificity of the individual type of phages that can infect only a limited type of bacterial species, the preparation of a combination of a large variety of phages against a single bacterial species as ‘phage cocktails’ is necessary (72). For example, anti-Pseudomonas phages that separated from a patient with P. aeruginosa infection might not be effective in another patient with P. aeruginosa infection (the same bacteria with a subtle difference), and then the anti-Pseudomonas phage cocktails extracted from several patients will be necessary for clinical use. Hence, a huge collection of phages, possibly with a very high cost and long duration of preparation, will be necessary for the ready-to-use phage cocktails for clinical situations. From our data, i) there were phages against endogenous bacteria in the gut, ii) there were alterations of endogenous phages along with the change in gut bacteria (toward pathogenic bacteria in sepsis), and iii) there was not enough quantity of the endogenous phages for the effective inhibition of pathogenic bacteria during sepsis (Figure 9). Hence, for the patients infected with the endogenous bacteria, the extraction of the naturally developed endogenous phages, before administration back to the host, might overcome the necessity for the preparation of phage cocktails that will reduce the cost of phage therapy. The rapid processes of extraction and propagation of the endogenous phages from the patients themselves might be interesting for the adjuvant therapy in sepsis. More studies are warranted.
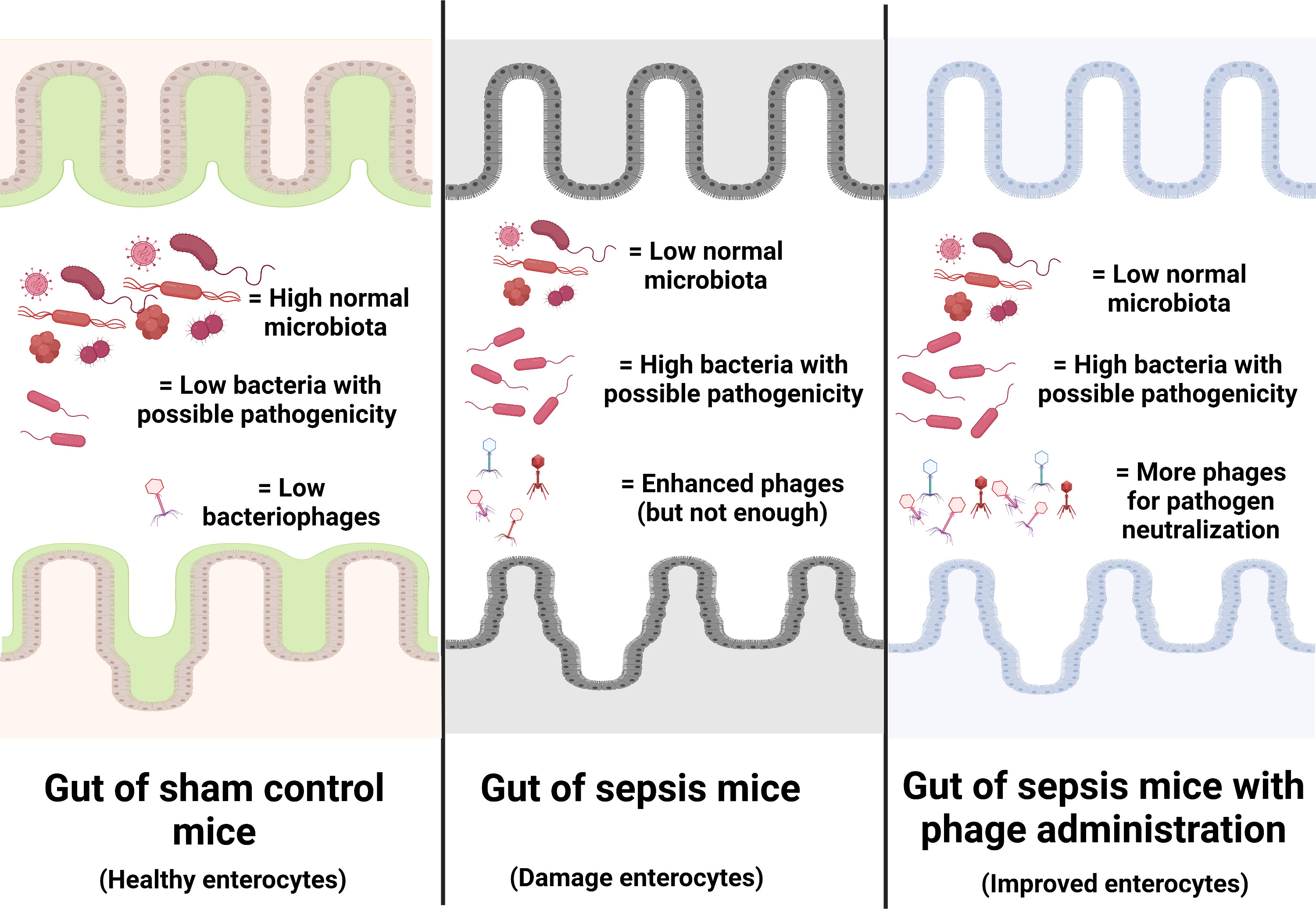
Figure 9 Illustration of the working hypothesis indicating gut dysbiosis (increase in bacteria with possible pathogenicity with decreased normal microbiota with possible enhanced bacteriophages) in the gut of sepsis mice when compared with sham mice. The administration of phages, isolated from other sepsis mice, increased phages to the level that is enough for the more effective neutralization of pathogenic bacteria. Phage isolation from the sepsis host with a rapid process of phage propagation before re-administration into the sepsis host is of interest for further test.
In conclusion, CLP sepsis alters all organisms (bacteria, fungi, and viruses) in the gut, and the natural cocktails of phages that adapted against the altered bacteria might be beneficial for sepsis attenuation. The isolation of phages from the host with rapid ex vivo propagation before re-administration to the host is a proposed new strategy of sepsis adjuvant therapy.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: NCBI Sequence Read Archive; PRJNA838435
The animal care and use protocol (SST 010/2562) was certified by the Institutional Animal Care and Use Committee of Chulalongkorn University’s Faculty of Medicine in Bangkok, Thailand, in compliance with the US National Institutes of Health criteria.
WC and AL conceptualized, designed, and managed the study. WC, NS, PV, VS, SC, SP, and AL performed the experiments. WC and AL performed the data analysis. WC and AL provided the key reagents. WC and AL wrote the manuscript. WC received the funding. AL, AS, and MS performed supervision. All authors made substantial contributions to subsequent versions of the manuscript and approved the final version.
This research paper is supported by Specific League Funds from Mahidol University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the global burden of disease study. Lancet (2020) 395:200–11. doi: 10.1016/S0140-6736(19)32989-7
2. Amornphimoltham P, Yuen PST, Star RA, Leelahavanichkul A. Gut leakage of fungal-derived inflammatory mediators: Part of a gut-Liver-Kidney axis in bacterial sepsis. Dig Dis Sci (2019) 64:2416–28. doi: 10.1007/s10620-019-05581-y
3. Assimakopoulos SF, Triantos C, Thomopoulos K, Fligou F, Maroulis I, Marangos M, et al. Gut-origin sepsis in the critically ill patient: Pathophysiology and treatment. Infection (2018) 46:751–60. doi: 10.1007/s15010-018-1178-5
4. Haussner F, Chakraborty S, Halbgebauer R, Huber-Lang M. Challenge to the intestinal mucosa during sepsis. Front Immunol (2019) 10:891. doi: 10.3389/fimmu.2019.00891
5. Helander HF, Fändriks L. Surface area of the digestive tract - revisited. Scand J Gastroenterol (2014) 49:681–9. doi: 10.3109/00365521.2014.898326
6. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms (2019) 7:14. doi: 10.3390/microorganisms7010014
7. Pérez JC. Fungi of the human gut microbiota: Roles and significance. Int J Med Microbiol (2021) 311:151490. doi: 10.1016/j.ijmm.2021.151490
8. Hiengrach P, Panpetch W, Worasilchai N, Chindamporn A, Tumwasorn S, Jaroonwitchawan T, et al. Administration of candida albicans to dextran sulfate solution treated mice causes intestinal dysbiosis, emergence and dissemination of intestinal pseudomonas aeruginosa and lethal sepsis. Shock (2020) 53:189–98. doi: 10.1097/SHK.0000000000001339
9. Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe (2016) 19:865–73. doi: 10.1016/j.chom.2016.05.003
10. Adelman MW, Woodworth MH, Langelier C, Busch LM, Kempker JA, Kraft CS, et al. The gut microbiome’s role in the development, maintenance, and outcomes of sepsis. Crit Care (2020) 24:278. doi: 10.1186/s13054-020-02989-1
11. Saithong S, Saisorn W, Visitchanakun P, Sae-Khow K, Chiewchengchol D, Leelahavanichkul A. A synergy between endotoxin and (1→3)-Beta-D-Glucan enhanced neutrophil extracellular traps in candida administered dextran sulfate solution induced colitis in FcGRIIB-/- lupus mice, an impact of intestinal fungi in lupus. J Inflammation Res (2021) 14:2333–52. doi: 10.2147/JIR.S305225
12. Petrovic Fabijan A, Lin RCY, Ho J, Maddocks S, Ben Zakour NL, Iredell JR. Safety of bacteriophage therapy in severe staphylococcus aureus infection. Nat Microbiol (2020) 5:465–72. doi: 10.1038/s41564-019-0634-z
13. Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant mycobacterium abscessus. Nat Med (2019) 25:730–3. doi: 10.1038/s41591-019-0437-z
14. Lin DM, Koskella B, Lin HC. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther (2017) 8:162–73. doi: 10.4292/wjgpt.v8.i3.162
15. Maslov S, Sneppen K. Regime shifts in a phage-bacterium ecosystem and strategies for its control. mSystems (2019) 4:e00470-19. doi: 10.1128/mSystems.00470-19
16. Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev (2014) 38:916–31. doi: 10.1111/1574-6976.12072
17. Sutton TDS, Hill C. Gut bacteriophage: Current understanding and challenges. Front Endocrinol (Lausanne) (2019) 10:784. doi: 10.3389/fendo.2019.00784
18. Guerin E, Hill C. Shining light on human gut bacteriophages. Front Cell Infect Microbiol (2020) 10:481. doi: 10.3389/fcimb.2020.00481
19. Panpetch W, Hiengrach P, Nilgate S, Tumwasorn S, Somboonna N, Wilantho A, et al. Additional candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by lactobacillus rhamnosus L34. Gut Microbes (2020) 11:465–80. doi: 10.1080/19490976.2019.1662712
20. Visitchanakun P, Saisorn W, Wongphoom J, Chatthanathon P, Somboonna N, Svasti S, et al. Gut leakage enhances sepsis susceptibility in iron-overloaded β-thalassemia mice through macrophage hyperinflammatory responses. Am J Physiol Gastrointest Liver Physiol (2020) 318:G966–79. doi: 10.1152/ajpgi.00337.2019
21. Sae-Khow K, Charoensappakit A, Visitchanakun P, Saisorn W, Svasti S, Fucharoen S, et al. Pathogen-associated molecules from gut translocation enhance severity of cecal ligation and puncture sepsis in iron-overload β-thalassemia mice. J Inflammation Res (2020) 13:719–35. doi: 10.2147/JIR.S273329
22. Panpetch W, Sawaswong V, Chanchaem P, Ondee T, Dang CP, Payungporn S, et al. Candida administration worsens cecal ligation and puncture-induced sepsis in obese mice through gut dysbiosis enhanced systemic inflammation, impact of pathogen-associated molecules from gut translocation and saturated fatty acid. Front Immunol (2020) 11:561652. doi: 10.3389/fimmu.2020.561652
23. Issara-Amphorn J, Chancharoenthana W, Visitchanakun P, Leelahavanichkul A. Syk inhibitor attenuates polymicrobial sepsis in FcgRIIb-deficient lupus mouse model, the impact of lupus characteristics in sepsis. J Innate Immun (2020) 12:461–79. doi: 10.1159/000509111
24. Visitchanakun P, Tangtanatakul P, Trithiphen O, Soonthornchai W, Wongphoom J, Tachaboon S, et al. Plasma miR-370-3P as a biomarker of sepsis-associated encephalopathy, the transcriptomic profiling analysis of microrna-arrays from mouse brains. Shock (2020) 54:347–57. doi: 10.1097/SHK.0000000000001473
25. Ondee T, Pongpirul K, Visitchanakun P, Saisorn W, Kanacharoen S, Wongsaroj L, et al. Lactobacillus acidophilus LA5 improves saturated fat-induced obesity mouse model through the enhanced intestinal akkermansia muciniphila. Sci Rep (2021) 11:6367. doi: 10.1038/s41598-021-85449-2
26. Panpetch W, Kullapanich C, Dang CP, Visitchanakun P, Saisorn W, Wongphoom J, et al. Candida administration worsens uremia-induced gut leakage in bilateral nephrectomy mice, an impact of gut fungi and organismal molecules in uremia. mSystems (2021) 6:e01187-20. doi: 10.1128/mSystems.01187-20
27. Visitchanakun P, Panpetch W, Saisorn W, Chatthanathon P, Wannigama DL, Thim-Uam A, et al. Increased susceptibility to dextran sulfate-induced mucositis of iron-overload β-thalassemia mice, another endogenous cause of septicemia in thalassemia. Clin Sci (Lond) (2021) 135:1467–86. doi: 10.1042/CS20210328
28. Issara-Amphorn J, Dang CP, Saisorn W, Limbutara K, Leelahavanichkul A. Candida administration in bilateral nephrectomy mice elevates serum (1→3)-β-D-glucan that enhances systemic inflammation through energy augmentation in macrophages. Int J Mol Sci (2021) 22:5031. doi: 10.3390/ijms22095031
29. Sawaswong V, Fahsbender E, Altan E, Kemthong T, Deng X, Malaivijitnond S, et al. High diversity and novel enteric viruses in fecal viromes of healthy wild and captive Thai cynomolgus macaques (Macaca fascicularis). Viruses (2019) 11:971. doi: 10.3390/v11100971
30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
31. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods (2012) 9:357–9. doi: 10.1038/nmeth.1923
32. Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, et al. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res (2015) 43:e46. doi: 10.1093/nar/gkv002
33. Bao HD, Pang MD, Olaniran A, Zhang XH, Zhang H, Zhou Y, et al. Alterations in the diversity and composition of mice gut microbiota by lytic or temperate gut phage treatment. Appl Microbiol Biotechnol (2018) 102:10219–30. doi: 10.1007/s00253-018-9378-6
34. Khan Mirzaei M, Khan MAA, Ghosh P, Taranu ZE, Taguer M, Ru J, et al. Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe (2020) 27:199–212.e195. doi: 10.1016/j.chom.2020.01.004
35. Noble RT, Fuhrman JA. Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl Environ Microbiol (2000) 66:3790–7. doi: 10.1128/AEM.66.9.3790-3797.2000
36. Liu Z, Li N, Fang H, Chen X, Guo Y, Gong S, et al. Enteric dysbiosis is associated with sepsis in patients. FASEB J (2019) 33:12299–310. doi: 10.1096/fj.201900398RR
37. Thim-Uam A, Surawut S, Issara-Amphorn J, Jaroonwitchawan T, Hiengrach P, Chatthanathon P, et al. Leaky-gut enhanced lupus progression in the fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci Rep (2020) 10:777. doi: 10.1038/s41598-019-57275-0
38. Panpetch W, Somboonna N, Bulan DE, Issara-Amphorn J, Worasilchai N, Finkelman M, et al. Gastrointestinal colonization of candida albicans increases serum (1→3)-β-D-Glucan, without candidemia, and worsens cecal ligation and puncture sepsis in murine model. Shock (2018) 49:62–70. doi: 10.1097/SHK.0000000000000896
39. Yang J, Wu R, Qiang X, Zhou M, Dong W, Ji Y, et al. Human adrenomedullin and its binding protein attenuate organ injury and reduce mortality after hepatic ischemia-reperfusion. Ann Surg (2009) 249:310–7. doi: 10.1097/SLA.0b013e3181961d43
40. Issara-Amphorn J, Somboonna N, Pisitkun P, Hirankarn N, Leelahavanichkul A. Syk inhibitor attenuates inflammation in lupus mice from FcgRIIb deficiency but not in pristane induction: The influence of lupus pathogenesis on the therapeutic effect. Lupus (2020) 29:1248–62. doi: 10.1177/0961203320941106
41. Nguyen NP, Warnow T, Pop M, White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes. (2016) 2:16004. doi: 10.1038/npjbiofilms.2016.4
42. Panpetch W, Somboonna N, Palasuk M, Hiengrach P, Finkelman M, Tumwasorn S, et al. Oral candida administration in a clostridium difficile mouse model worsens disease severity but is attenuated by bifidobacterium. PLoS One (2019) 14:e0210798. doi: 10.1371/journal.pone.0210798
43. Panpetch W, Somboonna N, Bulan DE, Issara-Amphorn J, Finkelman M, Worasilchai N, et al. Oral administration of live- or heat-killed candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-β-D-glucan. PLoS One (2017) 12:e0181439. doi: 10.1371/journal.pone.0181439
44. Rameshkumar G, Sikha M, Ponlakshmi M, Selva Pandiyan A, Lalitha P. A rare case of myrothecium species causing mycotic keratitis: Diagnosis and management. Med Mycol Case Rep (2019) 25:53–5. doi: 10.1016/j.mmcr.2019.07.010
45. Bebeacua C, Lai L, Vegge CS, Brøndsted L, van Heel M, Veesler D, et al. Visualizing a complete siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol (2013) 87:1061–8. doi: 10.1128/JVI.02836-12
46. Ghosh S, Malik YS. The true host/s of picobirnaviruses. Front Vet Sci (2020) 7:615293. doi: 10.3389/fvets.2020.615293
47. Hamdi S, Rousseau GM, Labrie SJ, Kourda RS, Tremblay DM, Moineau S, et al. Characterization of five podoviridae phages infecting citrobacter freundii. Front Microbiol (2016) 7:1023. doi: 10.3389/fmicb.2016.01023
48. Kornienko M, Kuptsov N, Gorodnichev R, Bespiatykh D, Guliaev A, Letarova M, et al. Contribution of podoviridae and myoviridae bacteriophages to the effectiveness of anti-staphylococcal therapeutic cocktails. Sci Rep (2020) 10:18612. doi: 10.1038/s41598-020-75637-x
49. Gaines S, Alverdy JC. Fecal micobiota transplantation to treat sepsis of unclear etiology. Crit Care Med (2017) 45:1106–7. doi: 10.1097/CCM.0000000000002382
50. Wei Y, Yang J, Wang J, Yang Y, Huang J, Gong H, et al. Successful treatment with fecal microbiota transplantation in patients with multiple organ dysfunction syndrome and diarrhea following severe sepsis. Crit Care (2016) 20:332. doi: 10.1186/s13054-016-1491-2
51. Kim SM, DeFazio JR, Hyoju SK, Sangani K, Keskey R, Krezalek MA, et al. Fecal microbiota transplant rescues mice from human pathogen mediated sepsis by restoring systemic immunity. Nat Commun (2020) 11:2354. doi: 10.1038/s41467-020-15545-w
52. Keskey R, Cone JT, DeFazio JR, Alverdy JC. The use of fecal microbiota transplant in sepsis. Transl Res (2020) 226:12–25. doi: 10.1016/j.trsl.2020.07.002
53. Gai X, Wang H, Li Y, Zhao H, He C, Wang Z, et al. Fecal microbiota transplantation protects the intestinal mucosal barrier by reconstructing the gut microbiota in a murine model of sepsis. Front Cell Infect Microbiol (2021) 11:736204. doi: 10.3389/fcimb.2021.736204
54. Scales BS, Dickson RP, Huffnagle GB. A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J Leukoc Biol (2016) 100:943–50. doi: 10.1189/jlb.3MR0316-106R
55. Goldstein EJ, Citron DM, Peraino VA, Cross SA. Desulfovibrio desulfuricans bacteremia and review of human desulfovibrio infections. J Clin Microbiol (2003) 41:2752–4. doi: 10.1128/JCM.41.6.2752-2754.2003
56. Reid G. The scientific basis for probiotic strains of lactobacillus. Appl Environ Microbiol (1999) 65:3763–6. doi: 10.1128/AEM.65.9.3763-3766.1999
57. Fujio-Vejar S, Vasquez Y, Morales P, Magne F, Vera-Wolf P, Ugalde JA, et al. The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum verrucomicrobia. Front Microbiol (2017) 8:1221. doi: 10.3389/fmicb.2017.01221
58. Bachta KER, Allen JP, Cheung BH, Chiu CH, Hauser AR. Systemic infection facilitates transmission of pseudomonas aeruginosa in mice. Nat Commun (2020) 11:543. doi: 10.1038/s41467-020-14363-4
59. Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol (2017) 10:18–26. doi: 10.1038/mi.2016.75
60. Brown R, Priest E, Naglik JR, Richardson JP. Fungal toxins and host immune responses. Front Microbiol (2021) 12:643639. doi: 10.3389/fmicb.2021.643639
61. Hsu YH, Nakagawa M, Hirota A, Shima S, Nakayama M. Structure of myrocin b, a new diterpene antibiotic produced by. Myrothecium verrucaria. Agr Biol Chem (1988) 52:1305–7. doi: 10.1271/bbb1961.52.1305
62. Elkhateeb WA, Daba GM. Myrothecium as promising model for biotechnological applications, potentials and challenges. BioMed J Sci Tech Res (2019) 16:12126–31. doi: 10.26717/BJSTR.2019.16.002869
63. Azam AH, Tanji Y. Peculiarities of staphylococcus aureus phages and their possible application in phage therapy. Appl Microbiol Biotechnol (2019) 103:4279–89. doi: 10.1007/s00253-019-09810-2
64. Golińska E, Strus M, Tomusiak-Plebanek A, Więcek G, Kozień Ł, Lauterbach R, et al. Coagulase-negative staphylococci contained in gut microbiota as a primary source of sepsis in low- and very low birth weight neonates. J Clin Med (2020) 9:2517. doi: 10.3390/jcm9082517
65. Górski A, Jończyk-Matysiak E, Łusiak-Szelachowska M, Międzybrodzki R, Weber-Dąbrowska B, Borysowski J. The potential of phage therapy in sepsis. Front Immunol (2017) 8:1783. doi: 10.3389/fimmu.2017.01783
66. Alvi IA, Asif M, Tabassum R, Aslam R, Abbas Z, Rehman SU. RLP, a bacteriophage of the family podoviridae, rescues mice from bacteremia caused by multi-drug-resistant pseudomonas aeruginosa. Arch Virol (2020) 165:1289–97. doi: 10.1007/s00705-020-04601-x
67. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
68. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: A common factor in human diseases. BioMed Res Int (2017) 2017:9351507. doi: 10.1155/2017/9351507
69. Principi N, Silvestri E, Esposito S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front Pharmacol (2019) 10:513. doi: 10.3389/fphar.2019.00513
70. Salmond GP, Fineran PC. A century of the phage: past, present and future. Nat Rev Microbiol (2015) 13:777–86. doi: 10.1038/nrmicro3564
71. Lewis AJ, Seymour CW, Rosengart MR. Current murine models of sepsis. Surg Infect (Larchmt) (2016) 17:385–93. doi: 10.1089/sur.2016.021
Keywords: sepsis, CLP, microbiome, mycobiome, virome, fecal transplantation
Citation: Chancharoenthana W, Sutnu N, Visitchanakun P, Sawaswong V, Chitcharoen S, Payungporn S, Schuetz A, Schultz MJ and Leelahavanichkul A (2022) Critical roles of sepsis-reshaped fecal virota in attenuating sepsis severity. Front. Immunol. 13:940935. doi: 10.3389/fimmu.2022.940935
Received: 10 May 2022; Accepted: 11 July 2022;
Published: 02 August 2022.
Edited by:
Haitao Wen, The Ohio State University United StatesReviewed by:
Eun Jeong Park, Mie University, JapanCopyright © 2022 Chancharoenthana, Sutnu, Visitchanakun, Sawaswong, Chitcharoen, Payungporn, Schuetz, Schultz and Leelahavanichkul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wiwat Chancharoenthana, d2l3YXQuY2hhQG1haGlkb2wuYWMudGg=; Asada Leelahavanichkul, YWxlZWxhaGF2YW5pdEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.