
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 08 July 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.938895
This article is part of the Research Topic Host-Pathogen Interactions in Nontuberculous mycobacterial infections View all 7 articles
Non-tuberculous mycobacteria (NTM) are a heterogeneous group of originally environmental organi3sms, increasingly recognized as pathogens with rising prevalence worldwide. Knowledge of NTM’s mechanisms of virulence is lacking, as molecular research of these bacteria is challenging, sometimes more than that of M. tuberculosis (Mtb), and far less resources are allocated to their investigation. While some of the virulence mechanisms are common to several mycobacteria including Mtb, others NTM species-specific. Among NTMs, Mycobacterium abscessus (Mabs) causes some of the most severe and difficult to treat infections, especially chronic pulmonary infections. Mabs survives and proliferates intracellularly by circumventing host defenses, using multiple mechanisms, many of which remain poorly characterized. Some of these immune-evasion mechanisms are also found in Mtb, including phagosome pore formation, inhibition of phagosome maturation, cytokine response interference and apoptosis delay. While much is known of the role of Mtb-secreted effector molecules in mediating the manipulation of the host response, far less is known of the secreted effector molecules in Mabs. In this review, we briefly summarize the knowledge of secreted effectors in Mtb (such as ESX secretion, SecA2, TAT and others), and draw the parallel pathways in Mabs. We also describe pathways that are unique to Mabs, differentiating it from Mtb. This review will assist researchers interested in virulence-associated secretion in Mabs by providing the knowledge base and framework for their studies.
The non-tuberculous mycobacteria (NTM) family is comprised of over 150 species, some of which are increasingly recognized as emerging pathogens, causing infections in immunocompromised as well as immunocompetent patients (1, 2). While most NTM are of only little pathogenic potential to humans, some – like Mycobacterium marinum and Mycobacterium ulcerans, are important pathogens, whereas others, like Mycobacterium kansasii, Mycobacterium fortuitum and Mycobacterium abscessus (Mabs), are considered opportunistic pathogens, but nevertheless can cause severe, often deadly, pulmonary infections (3–5). Mabs, classified as a rapidly growing mycobacterium (RGM), causes skin, lung and soft tissues infections that are very difficult to eradicate (6). Mabs is many times – but not strictly – an intracellular pathogen, and is found within phagocytic immune cells (macrophages and dendritic cells) and within epithelial and endothelial cells (7). To allow intracellular survival, these bacteria require unique mechanisms of resistance to the mostly hostile intracellular niche.
Mycobacterium tuberculosis (Mtb) employs various strategies to evade the immune response inside the infected cell: phagosome perforation and maturation arrest, cytokine-response downregulation, inhibition of reactive oxygen species (ROS) secretion, apoptosis prevention and more (8–11). These are dependent on different effector proteins, transported via specialized secretion systems that interrupt host means of defense in versatile manners (12). Although secretion systems and their respective substrates have been under thorough investigation in Mtb, much less is known of their counterparts in Mabs. Progress in bioinformatic methods, secretome analysis and high-throughput sequencing, combined with improved tools for transposon mutagenesis and clues drawn from Mtb studies, allowed the discovery of new effectors in Mabs. Recently, a considerable amount of effort has been made to assess the contribution of secreted proteins to the virulence of Mabs. Most studies employed deletion mutants and evaluation of bacterial proliferation in ex-vivo and in-vivo models of infection. In this review, we follow the route of infection in Mtb, introducing the secreted effector proteins known to affect Mtb virulence, and then comparing them to their analogs in Mabs. This approach, although may miss completely novel effectors in Mabs (that are fundamentally different from those of Mtb), will provide the reader with a solid starting point in assessing these factors in Mabs. In addition, we outline the gaps of knowledge regarding factors yet to be identified in Mabs – gaps the filling of which will probably require more complicated analyses of the Mabs’s secretome and proteome, as well as in-depth bioinformatics look at the bacterial genome.
Secretion systems play an essential role in promoting virulence in mycobacteria (13–16). These sophisticated apparati enable the export of effector proteins across the thick membrane into the host cell, reducing immune response and promoting bacterial survival.
Three groups of secretion systems were identified in mycobacteria:
◼ Type VII secretion systems (T7SS): Known as ESX secretion systems, these were named after the 6kDa Early Secretory Antigenic Target (ESAT-6). Mtb’s ESAT-6 was established as a critical contributor to virulence, along with a second ESX-1 substrate CFP-10 (17). Partial deletion of the ESX-1 system accounts for the attenuation observed in BCG bacteria compared to their ancestor M. bovis (18, 19). The five different mycobacterial ESX systems (ESX-1-5) are encoded by paralogous loci that are widespread among the slow and rapidly growing species (20). In Mtb, ESX-1, ESX-3 and ESX-5 are critical components in virulence, while the roles of ESX-2 and ESX-4 remain obscure (21–24). Mabs, however, has only two identified ESX clusters: ESX-3, an essential secretion system involved in iron and zinc homeostasis; and ESX-4, recently shown to play a significant role in virulence, serving as an analogue for Mtb’s ESX-1[ (25, 26)].
◼ The Twin-Arginine Translocation (TAT) pathway: distinguished by the ability to transport polypeptides in their folded state. Substrates utilizing the TAT export system must hold a highly conserved twin-arginine leader motif (S/TRRXFLK) which is found in the N terminus of the protein (27, 28).
◼ The Sec pathway: The substrates exported across the cytoplasmic membrane by Sec systems are initially produced as precursor proteins, consisting of conserved amino- terminal signal sequences (29). During translocation, the signal peptide is cleaved to generate the mature exported protein. Two non-redundant Sec systems are identified in Mtb: SecA1 – an essential “housekeeping” system, and the accessory secretion factor - SecA2 (30, 31).
Generally, effector proteins secreted by mycobacteria each utilize a specific mechanism of export, depending on their individual traits.
Here, we will describe the main approaches Mtb uses to resist host cell’s antibacterial attempts, trying to unravel the analogous effectors in Mabs.
As mycobacteria are inhaled into the lungs and reach the alveolar cavity, they are phagocytized by alveolar macrophages (32). The bacilli are enclosed in a vesicular phagosome which is next subjected to fusion with the lysosome, forming a mature phagosome (e.g phagolysosome) (33). Permeabilizing the phagosomal membrane with eventual phagosomal escape is a pivotal step in immune resistance, allowing the pathogen to secrete effectors into the cytosol. A crucial effector involved in phagosome rupture is the ESX-1 substrate EsxA/ESAT-6 (Rv3875), that creates a heterodimer with another secreted protein, EsxB/CFP-10 (Rv3874) (17). Mtb mutants defective in secretion of EsxA and/or EsxB fail to translocate to the cytosol and are attenuated, as seen in the vaccine strain Mycobacterium bovis BCG and in the H37Ra strain (19, 34–37). The mechanism in which EsxA contributes to phagosome membrane rupture is still a matter of debate, but it appears to induce gross membrane disruption in a contact-dependent manner (38, 39). As previously mentioned, Mabs does not possess an ESX-1 secretion system. However, work conducted by Laencina et al. (26) presented evidence of Mabs EsxT (MAB_3753c) and EsxU (MAB_3754c) serving as functional analogues to EsxA and EsxB, respectively, as their deletion reduces phagosome-to-cytosol contact. Moreover, rupture of the phagosomal membrane occurs only in the presence of an intact eccB4 gene, a structural component of Mabs Esx-4 secretion system. Whether EsxT and EsxU directly damage the phagosome membrane is yet to be investigated.
After the phagosome internalizes the mycobacterium, a sequence of event initiates, which includes a decrease in pH and acquisition of antimicrobial properties. The late phagosome then fuses with the lysosome (Phagosome maturation), and the bacilli are processed into small particles, that are next presented to T-cells as antigens, initiating the adaptive immune response (40). Many mycobacterial secreted effectors target to disrupt this process.
Rab GTPases are molecular switches that coordinate the changes in phagosomal membrane upon internalizing the invader (41). Human Rab5 and Rab7 coordinate vesicle trafficking between the early phagosome to late endosome (Rab5) and from the late endosome to phagolysosome (Rab7) (42). Nucleoside diphosphate kinase A (NdkA, Rv2445c) is an Mtb GTPase secreted through the SecA2 pathway (30, 43, 44). NdkA binds Rab5 and Rab7 and facilitates the transition fromthe active GTP-bound state to the inactive GDP-bound state, interfering with the phagolysosome formation (45, 46). Also, NdkA inactivates Rac1 – a GTPase, required for activation of NADPH oxidase 2 (NOX2), thus blocking production of reactive oxygen species (ROS), and preventing bacterial killing (47). In Mabs the only annotated Ndk protein (MAB_1606) has an 86% similarity to Mtb’s NdkA, suggesting a similar role and function. Currently, no data on MAB_1606 deletion in Mabs has been published, leaving the role of Ndk in Mabs unexplored.
The next step in phagosome maturation that is manipulated by Mtb is the production of lipid regulator phosphatidylinositol 3-phosphate (PI3P) from phosphatidylinositol (PI). PI3P is a membrane tag that signals the macrophages to continue down the phagolysosome biogenesis pathway (48). Also exported by SecA2 pathway in Mtb (30), secreted acid phosphatase M (SapM, Rv3310) dephosphorylates PI3P, limiting its ability to recruit PI3P-binding proteins in the mycobacterium-containing vacuole (MCV) membrane, and blocking the maturation process (49–51). A deletion mutant of SapM in Mtb is attenuated in human macrophages and in-vivo in guinea pigs (50, 52, 53). However, although found in other slow-growing nontuberculous mycobacteria such as M. avium and M. marinum (50, 54, 55), a SapM analog has not yet been identified in M. abscessus.
The mature phagosome assembly is mediated by many RabGTPase proteins, each necessary for to complete the intricate process of phagolysosome fusion. Human Rab7L1, in its active GTP-bound form Rab7L1-GTP is recruited to bacilli-containing phagosomes, and signals other phago-lysosomal markers such as the previously mentioned RAB7 (56). In Mtb, secreted protein kinase G (PknG) interacts with Rab7L1, limiting the formation of the active GTP-bound state and interfering with the successive signaling process (57–59). A ΔpknG mutant in Mtb is attenuated in mice after intravenous injections, but not by aerosol delivery (60). Though PknG in Mabs is completely uninvestigated, we can speculate with confidence that it performs in the same manner as in Mtb. A putative protein encoded by MAB_4224 bears considerable similarity (82%) to Mtb’s PknG (Rv0410c). Furthermore, MAB_4244 lies between the gene encoding the probable glutamine binding protein H (glnH, MAB_4223) and the gene for acetate kinase A (ackA, MAB_4225c), exactly like it does in Mtb (glnH Rv0411c, ackA Rv0409). The close downstream proximity of pknG to glnH is of importance, since these two genes are suggested to be co-expressed in a conserved operon in Actinomycetes (61, 62). With the lack of a well-established mechanism, it is thought that GlnH, through protein-protein interactions, activates PknG via a probable transmembrane protein, GlnX (61). All these suggest Mabs PknG may play an important yet under-explored role in Mabs pathogenesis.
Another Mtb effector found to reduce the recruitment of EEA1, Rab5 and Rab7 and therefore inhibit phagosome maturation, is TlyA (Rv1694) (63). TlyA serves as a ribosomal RNA methyl transferase (rRNase) (64) and displays an additional role as a hemolysin, when purified from Mtb and expressed in M. smegmatis (Msme) (38, 65). Rahman et al. (66) showed that TlyA forms oligomers on phagosome membrane and red blood cells, finally leading to lysis. Mtb TlyA Knockout demonstrated reduced growth in ex-vivo infected macrophages and in mouse models, yet the lack of complementation experiments precluded from drawing a definitive conclusion regarding its role and effect (67). Mtb TlyA resides in close proximity to RecN (Rv1696). MAB_2359 is a putative rRNA methytransferase with 78% similarity to the Mtb’s TlyA, and is also located in proximity to Mabs recN, suggesting it is a TlyA analog, and that its role in pathogenesis should be explored further.
ESCRT (The host Endosomal Sorting Complexes Required for transport) pathway directs cargo destined for digestion in lysosomes, such as the Mtb-containing vacuole (MCV) (68). Moreover, ESCRT apparati facilitate antigen processing, therefore promoting T-cell activation during Mtb infection (69). The initial ESCRT machinery is assembled by several components, one of which is HRS (Hepatocyte growth factor-regulated tyrosine kinase substrate), a target for the Mtb Esx-3 secreted effector, EsxH (70). EsxH (Rv0288) forms a 1:1 heterodimer with another secreted molecule EsxG (Rv0287), together they serve as distinctly functional paralogues to the EsxA/EsxB complex in Mtb ESX-1 (71, 72). EsxH was demonstrated by co-immunoprecipitation assays to interact directly with HRS, diminishing the ESCRT assembly and inhibiting phago-lysosome fusion (70). Knock-down of HRS resulted in reduced MCV maturation, stressing the importance of this EsxH target. However, overexpression of Mtb’s EsxH in RAW cells had greater effect on phagosome maturation arrest than HRS depletion, suggesting that an additional target is aimed by EsxH (70). EsxH deletion caused considerable attenuation, with a 3-4 log decrease of CFU in the lungs of mice (69, 72) - however, some of this attenuation may be related to the role of ESX-3 in iron acquisition, rather than phagosome maturation arrest Among pathogenic mycobacteria, the Esx-3 cluster is highly conserved (73). Therefore, it is not unexpected to discover that Mabs EsxH (MAB_2228c) and EsxG (MAB_2229c) share relatively high similarity with their counterparts in Mtb (81% each). The contribution of Mabs ESX-3 secretion system to host responses was well-studied by Kim et el (25).. They created an Esx-3 deletion mutant (MAB_2224c-2234c, Δesx-3) to examine the effect on growth, inflammatory response and pathophysiology in ex- and in-vivo models. Although in-vitro growth of the mutant was not reduced, intracellular growth was significantly reduced in bone marrow derived macrophages (BMDM). Mice infected by the mutant exhibited lower bacillary loads 7 days post infection, but with no significant difference 14 days post infection. Also, mice infected by Δesx-3 showed less severe lung pathology and decreased granulomatous infiltrates. Serum levels of TNF-α and IL-6, as well as mRNA levels of proinflammatory cytokines were also decreased in Δesx-3 infected mice. A caveat of this study was the lack of complementation experiment, which make interpretation less straightforward. Additionally, since the ESX-3 secretion system is essential for maintaining homeostasis in an iron and zinc depleted environment, we cannot confidently attribute the consequences of its deletion to the absence of the EsxG/H complex and to it’s effect on phagosome maturation, from that on iron homeostasis. To do that would necessitate additional experiments which would attempt to separate the two functions of the ESX-3 system.
Acidification of the late phagosome is prompted by the presence of vacuolar ATPase (V-ATPase) proton pumps on the phagosomal membrane (74). One of the three phosphatases secreted by Mtb, protein tyrosine phosphatase A (PtpA, Rv2234), binds subunit H of V-ATPase (75, 76). However, the interaction between PtpA and subunit H was found to be through protein-protein interaction, suggesting that V-ATPase is not the catalytic substrate of PtpA (75). Rather, the interaction of PtpA with V-ATPase is a prerequisite for the main purpose of PtpA – dephosphorylating, and as a result inactivating, vacuolar protein sorting 33B (VPS33B) (75, 77). VPS33B, a member of ESCRT machinery, is involved in vesicle trafficking and responsible for the necessary alterations of membranes to promote phago-lysosome fusion. Bach et al. (77)showed direct binding and co-localization of PtpA and VPS33B in the cytosol and impaired recruitment of VPS33B to the lysosome in Mtb-infected macrophages. Mtb PtpA knockout (ΔptpA) fails to inhibit phagosome acidification and maturation in human THP-1 macrophages (75, 77). Interestingly, ΔptpA is attenuated for growth in ex-vivo infected macrophages, but not within in-vivo infected mouse model (78). However, this appears to be specific to the mouse model, and does not undermine the importance of PtpA in pathogenesis in humans. The putative protein encoded by MAB_1900c shares great similarity (81%) with PtpA in Mtb. The proximal gene MAB_1901c contains considerable similarity (75%) to Rv2232, the adjacent gene to ptpA in Mtb. Rv2232 encodes for protein tyrosine kinase (PtkA), which phosphorylates and activates PtpA (77). Deleting ptkA in Mtb also leads to growth reduction in infected macrophages (79). The presence of this cluster in Mabs indicates an opportunity for further investigation of the contribution of Mabs- PtpA to virulence.
Another factor playing a role in blocking phagosome maturation of Mtb is lipoamide dehydrogenase C (LpdC, Rv0462). LpdC participates in the metabolism of branched-chain amino acids and is also secreted via the SecA2 pathway (30, 80). Its role in phagosome maturation inhibition was found as it demonstrated a cholesterol-dependent interaction with coronin-1 found on macrophages infected with Mtb and BCG (81, 82). MAB_4127c has an 88% similarity, and although some databases automatically annotated the gene as lpdA, it is probably the closest analog to the lpdC (Rv0426) from Mtb, whereas the true analog of Mtb’s LpdA, is MAB_3656c.
Overall, phagosome maturation seems to be a target for manipulation by Mabs, much like Mtb. The effectors involved in inhibiting this process are understudied in Mabs, and their investigation promises to be both fruitful and interesting.
Autophagy is a conserved degradation process of the cell in which unnecessary components and dysfunctional organelles are discarded in a regulated manner, allowing elimination of some and recycling of other materials through lysosome digestion (83). Proper and regulated autophagy is important for immune control of mycobacterial infections (84). Mycobacteria have several effectors aimed at disrupting this process. One anti-autophagy effector secreted by Mtb is the Enhanced Intracellular Survival (Eis, Rv2416c) protein (85). Mtb Eis has an Nε-acetyltransferase activity (86). Upon infection Eis acetylates DUSP16/MKP-7, a JNK-specific phosphatase therefore preventing DUSP16/MKP=7 from activating beclin1 (BCLN1), a necessary protein in autophagy regulation (86, 87). Eis is also proposed to inhibit autophagy through IL-10 upregulation (88), and through a JNK-dependent mechanism, affecting ROS production (89). An Mtb Eis deletion (Δeis) causes increase in JNK activity, leading to elevated ROS and increased autophagy, proinflammatory response and host cell death. Nevertheless, Δeis does not show reduced virulence in mice (89, 90). Mabs harbors two Eis encoding genes, Eis1 (MAB_4124), and Eis2 (MAB_4532c). Eis1 seems to be the closest homologue to Mtb’s Eis by Bidirectional Best Hit (BBH) search (91). However, it was shown that Mabs’ Eis1 does not modify aminoglycosides, and the basis for this lack of activity was pinpointed to structural reasons, where the active site is too narrow to accommodate large substrates like these antibiotics (92). Curiously, Mabs Eis2 deletion mutant (Δeis2) was dramatically attenuated in murine macrophages, showing increased ROS levels. Moreover, Δeis2 was unable to penetrate the phagosomal membrane and induce phagosome-to-cytosol contact compared to WT Mabs (91).
As previously mentioned, ROS are produced within phagosomes of infected macrophage as an additional antimicrobial ammunition. NADPH oxidase 2 (NOX2), the enzyme producing ROS, is a multiprotein complex that promotes a distinct phagosome maturation and autophagy-related pathway called LC-3 associated phagocytosis (LAP) (93). In the process of LAP, one membrane engulfs a pathogen, or pathogenic residues, instead of a double membrane in autophagy. Mtb CpsA (Rv3484) interferes with the recruitment of NOX2 to the phagosome and inhibits LAP through an unknown target (94). However, no clear analog in Mabs was found, and it is unclear if Mabs manipulates this pathway in the macrophage.
Mycobacterial infection induces the secretion of a large number of cytokines, including interferon gamma (IFN-γ), interleukin-1 (IL-1), IL-2, IL-6, IL-10, IL12, IL-18 and tumor necrosis factor alpha (TNF-α) (11, 95). The immune response is provoked by some and downregulated by other cytokines to protect the host against unfavorable pathology. Mtb survival depends on its ability to subvert the immune response, in order to promote its proliferation and cell-to-cell spread. The following are mechanism of such immune-response and cytokine release modulation.
Enoyl coA hydratase A1 (EchA1, Rv0222) is secreted by Mtb through an unknown mechanism and reaches the host cell cytosol. Wang et al. found that EchA1 is subjected to ubiquitination by the host cell ubiquitin ligase ANAPC2 (96). Ubiquitinated EchA1 promotes the recruitment of SHP1 – a protein tyrosine phosphatase that interacts with TNF receptor-associated factor 6 (TRAF6) and inhibit its ubiquitination. Since ubiquitinated TRAF6 is a mediator in IL-1 signaling, its inhibition by Mtb EchA1 impairs the production of proinflammatory cytokines. Mice infected with Mtb EchA1 mutants produced much higher levels of IL-1b, IL-6 and IL-12 in lung tissue, and attenuation of ΔechA1 growth was demonstrated in mice following aerosol infection (96). We found a probable enoyl coA hydratase gene in Mabs genome (MAB_0606c) with 44% similarity to Rv0222. One should note, though, that a 44% similarity is quite low, and take this homology with caution. No other information regarding this protein in Mabs is currently known.
Disulfide bond forming DsbE (also known as Mpt53, Rv2878c) is an effector that was found to activate protective host responses through promoting proinflammatory cytokine production. Predicted to be secreted by SecA1/2 (43, 97), DsbE binds, hence increases phosphorylation of TGF-β-activated kinase 1 (TAK1), an important signaling molecule downstream to TLR/TRAF6/TAB2 or TAB3 signaling pathway (98). Phosphorylated Tak1 activates the NF-kb pathway and the kinases JNK and p38, leading to biosynthesis of proinflammatory cytokines TNF, IL-6 and IL-12. An Mtb ΔdsbE mutant induces less TNF and IL-6 production in ex-vivo macrophage models and in-vivo in mice lungs (98). Moreover, ΔdsbE was hypervirulent in mice, with 1-2 log increase in CFU at 21 days post infection (98). It is therefore tempting to speculate whether Mabs probable DsbE (99), with relatively high resemblance to Mtb protein (78% similarity), stimulates similar responses.
Apoptosis and necrosis are two variants of cell death which greatly differ in all aspects, such as energy requirements, regulation, organelles fate and causes (100). Mtb produces effectors that favor the development of necrosis and inhibit the cell’s processes promoting apoptosis (101). Since the final consequence of many augmented immune response processes is delayed cell death, many effectors manipulate apoptosis indirectly. For example, Mtb protein tyrosine phosphatase B (PtpB, Rv0153c) is a broad-spectrum phosphatase that was shown to inhibit the IFN-γ-mediated activation of ERK1/2 and p38 signaling pathway, hence inducing production of IL-6 and inhibiting host cell death (102). However, no clear orthologue proteins to PtpB were described in Mabs. The closest protein found is MAB_4591, carrying a low similarity of 42%, which should be taken very cautiously.
Apoptosis is a redox-sensitive process. Mtb exploits this sensitivity and inhibits apoptosis by blocking ROS release by host cell into the phagosome. Superoxide dismutase A (SodA, Rv3846) is secreted by SecA2 and suspected to be involved in neutralization of superoxides produced by NOX2 in the MCV (31). Since SodA in an essential gene in Mtb, no deletion mutant could be established. However, a mutant with reduced SodA activity was constructed in Mtb and demonstrated in-vitro to be highly susceptible to killing by hydrogen superoxide (103). Moreover, mice infected by SodA-defective Mtb exhibited at least 10-fold more apoptotic cells in their lungs as compared to those infected by the WT strain (103). Mabs SodA (MAB_0118c) shares great similarity to its Mtb counterpart (89%). It will be intriguing to test whether Mabs mutants defective in SodA activity will have a phenotype consistent with the one in Mtb.
Mtb, as well as other pathogenic mycobacteria aspires to escape the intracellular niche in order to spread to other host cells. Encouraging necrosis is an effective approach to accomplish this goal (104). The outer membrane channel protein CpnT is required for efficient nutrient uptake in Mtb (105). While its N-terminal harbors a pore-forming capacity for this matter, its C-terminal domain, also called tuberculosis necrotizing toxin (TNT), can be released following proteolytic cleavage. TNT targets host cell coenzyme NAD+, thereby leading to its depletion (106). The necroptosis RIPK3/MLKL pathway is then activated, prompting the host cell death (107). Since deleting TNT does not attenuate bacilli growth in-vivo, it is suggested that Mtb has alternative pathways to promote necrosis (105). An analogous gene to Mtb CpnT/TNT analytic was not identified in the Mabs genome. However, a recent study found a cassette in a Mabs prophage with a polymorphic toxin (PT) that contains a C-terminal domain related to Mtb TNT (108). This finding offers a direction for further investigating CpnT/TNT analogues in Mabs, potentially involved in virulence.
Like most intracellular pathogens, mycobacteria, while in the macrophage, secrete a myriad of effector molecules into the phagolysosome as well as the cytosol in order to counter the many mechanisms at the macrophage’s disposal aimed at destroying this very pathogen. Knowledge of these secreted effectors and their mechanisms of action can be implemented into novel therapeutics, construction of attenuated mutants for vaccine purposes and predictions of disease severity. Whereas in M. tuberculosis there has been considerable progress in the characterization of the compendium of secreted effectors, this is not the case in most non-tuberculous mycobacteria, including M. abscessus. Most known M. abscessus-secreted effectors are at least structurally related to well-characterized secreted effectors in M. tuberculosis. Novel, yet undiscovered effectors probably exist, and may be shared by other rapid-growing mycobacteria (such as M. fortuitum) – but as data on this is still scarce, it is difficult to provide specific examples. Additional data and research specifically into the pathogenesis of M. abscessus is needed to characterize such effectors. In this review we attempted to provide the “basic footwork” for researchers interested in exploring the secreted effectors of M. abscessus – by summarizing the knowledge on those effectors that have been, at least partially, characterized, and by providing the basic links connecting characterized M. tuberculosis effectors with genes and proteins in M. abscessus, that may – or may not – play a homologous role in this pathogen. These were also summarized in Table 1. Figure 1 illustrates most of these effectors in a graphic manner. Obviously, this approach may miss completely novel and Mabs specific effectors that may very well exist - however their identification will necessitate more complicated experimental and bioinformatic analyses of the M. abscessus secretome, proteome and genome.
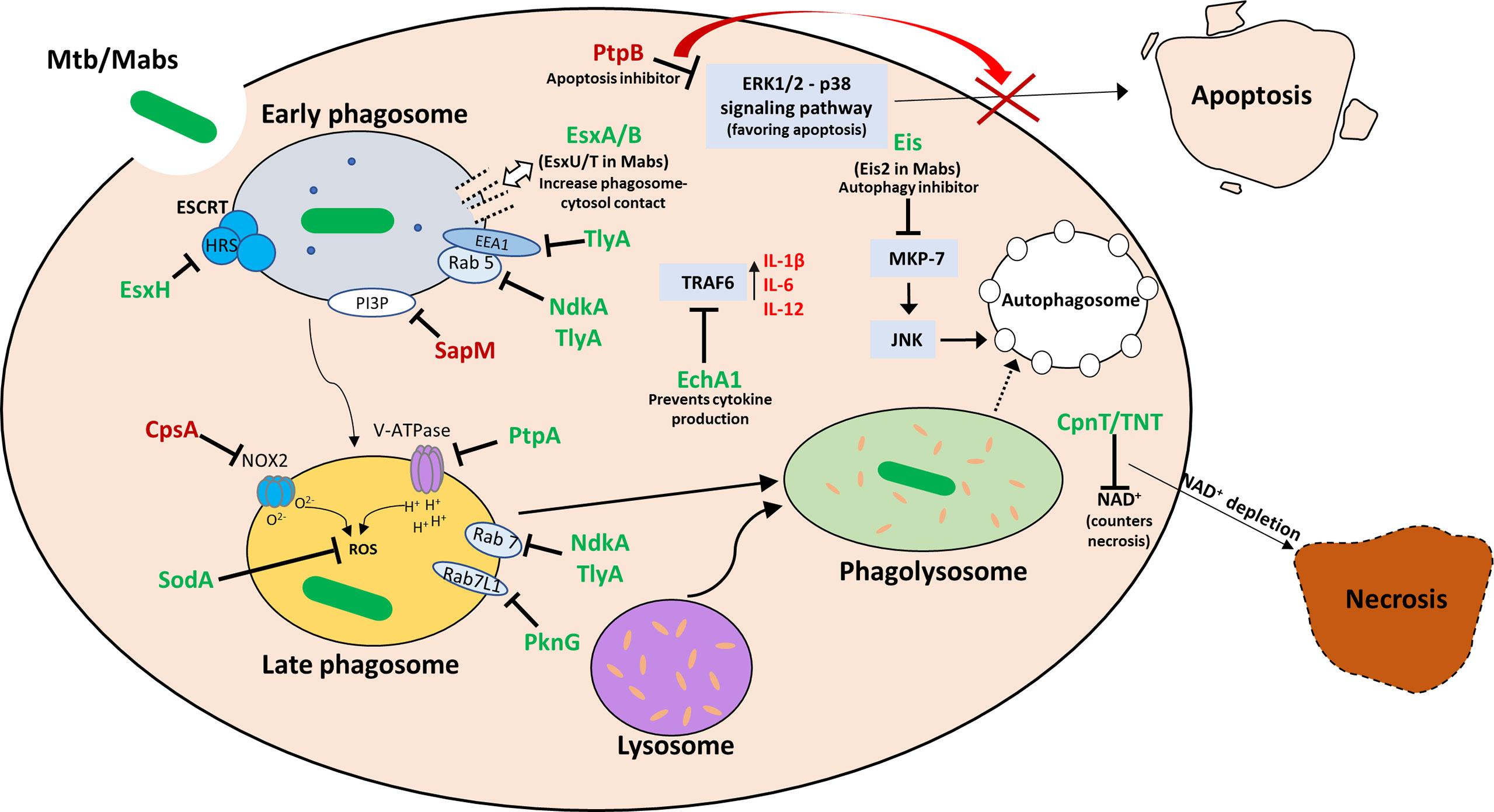
Figure 1 Visual summary of the secreted effectors in Mtb and Mabs and their intracellular targets. Once the bacilli are phagocytosed by the macrophage, it goes through a series of processing stages: early and late phagosome formation, phagosome acidification and phago-lysosome fusion. Necrosis is an undesirable outcome for the host, while it promotes cell-to-cell spread of the bacteria. Apoptosis, while leading to macrophage death, promotes effective immune response, and is therefore detrimental to the bacteria in the infection process. During each stage, the mycobacteria attempts to block phagosome maturation and acidification, prevent apoptosis and promote necrosis. Effectors are marked in green when a MABS analog of the Mtb protein is either identified or is presumed to exist, and in red when no MABS analog has been identified or suggested.
MBO wrote the manuscript. MM helped in editing the manuscript. DB edited and conceived the manuscript.
MM has a research grant from the Israeli Science Foundation (ISF). DB is supported by NIH R21 grant 1R21AI156415-01A1.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with the author (DB).
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. To K, Cao R, Yegiazaryan A, Owens J, Venketaraman V. General Overview of Nontuberculous Mycobacteria Opportunistic Pathogens: Mycobacterium Avium and Mycobacterium Abscessus. J Clin Med (2020) 9:1–24. doi: 10.3390/jcm9082541
2. Guglielmetti L, Mougari F, Lopes A, Raskine L, Cambau E. Human Infections Due to Nontuberculous Mycobacteria: The Infectious Diseases and Clinical Microbiology Specialists’ Point of View. Future Microbiol (2015) 10:1467–83.
3. Donohue MJ, Wymer L. Increasing Prevalence Rate of Nontuberculous Mycobacteria Infections in Five States, 2008-2013. Ann Am Thorac Soc (2016) 13:2143–50. doi: 10.1513/AnnalsATS.201605-353OC
4. Olivier KN, Weber DJ, Wallace RJ, Faiz AR, Lee JH, Zhang Y, et al. Nontuberculous Mycobacteria: I: Multicenter Prevalence Study in Cystic Fibrosis. Am J Respir Crit Care Med (2003) 167:828–34. doi: 10.1164/rccm.200207-678OC
5. Prevots DR, Marras TK. Epidemiology of Human Pulmonary Infection With Nontuberculous Mycobacteria a Review. Clinics Chest Med (2015) 36:13–34. doi: 10.1016/j.ccm.2014.10.002
6. Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. Mycobacterium Abscessus: A New Antibiotic Nightmare. J Antimicrobial Chemother (2012) 67:810–8. doi: 10.1093/jac/dkr578
7. Zhang C, Asif H, Holt GE, Griswold AJ, Campos M, Bejarano P, et al. Mycobacterium Abscessus—Bronchial Epithelial Cells Cross-Talk Through Type I Interferon Signaling. Front Immunol (2019) 10:2888. doi: 10.3389/fimmu.2019.02888
8. Queval CJ, Brosch R, Simeone R. The Macrophage: A Disputed Fortress in the Battle Against Mycobacterium Tuberculosis. Front Microbiol (2017) 8:2284. doi: 10.3389/fmicb.2017.02284
9. Liu CH, Liu H, Ge B. Innate Immunity in Tuberculosis: Host Defense vs Pathogen Evasion. Cell Mol Immunol (2017) 14:963–75. doi: 10.1038/cmi.2017.88
10. Upadhyay S, Mittal E, Philips JA. Tuberculosis and the Art of Macrophage Manipulation. Pathog Dis (2018) 76. doi: 10.1093/femspd/fty037
11. Sia JK, Rengarajan J. Immunology of Mycobacterium Tuberculosis Infections. Microbiol Spectr (2019) 7. doi: 10.1128/microbiolspec.gpp3-0022-2018
12. Augenstreich J, Briken V. Host Cell Targets of Released Lipid and Secreted Protein Effectors of Mycobacterium Tuberculosis. Front Cell Infect Microbiol (2020) 10:595029. doi: 10.3389/fcimb.2020.595029
13. Abdallah AM, Bestebroer J, Savage NDL, de Punder K, van Zon M, Wilson L, et al. Mycobacterial Secretion Systems ESX-1 and ESX-5 Play Distinct Roles in Host Cell Death and Inflammasome Activation. J Immunol (2011) 187:4744–53. doi: 10.4049/jimmunol.1101457
14. Gröschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX Secretion Systems: Mycobacterial Evolution to Counter Host Immunity. Nat Rev Microbiol (2016) 14:677–91. doi: 10.1038/nrmicro.2016.131
15. Kurtz S, McKinnon KP, Runge MS, Ting JP-Y, Braunstein M. The SecA2 Secretion Factor of Mycobacterium Tuberculosis Promotes Growth in Macrophages and Inhibits the Host Immune Response. Infect Immun (2006) 74:6855–64.
16. Vaziri F, Brosch R. ESX/Type VII Secretion Systems-An Important Way Out for Mycobacterial Proteins. Microbiol Spectr (2019) 7(4). doi: 10.1128/microbiolspec.PSIB
17. Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, et al. Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors CFP-10 and ESAT-6. EMBO J (2005) 24:2491–8. doi: 10.1038/sj.emboj.7600732
18. Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 Contributed to the Attenuation of the Live Tuberculosis Vaccines Mycobacterium Bovis BCG and Mycobacterium Microti. Tel (2002) 46(3):709–17. doi: 10.1046/j.1365-2958.2002.03237.x
19. Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, et al. The Primary Mechanism of Attenuation of Bacillus Calmette-Gué Rin is a Loss of Secreted Lytic Function Required for Invasion of Lung Interstitial Tissue. Proc Natl Acad Sci U S A (2003) 100(21):12420–5. doi: 10.1073/pnas.1635213100
20. Lagune M, Petit C, Sotomayor FV, Johansen MD, Beckham KSH, Ritter C, et al. Conserved and Specialized Functions of Type Vii Secretion Systems in non-Tuberculous Mycobacteria. Microbiol (United Kingdom) (2021) 167. doi: 10.1099/MIC.0.001054
21. Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJP, Garcia-Vallejo JJ, et al. The ESX-5 System of Pathogenic Mycobacteria Is Involved In Capsule Integrity and Virulence Through Its Substrate Ppe10. PLoS Pathog (2016) 12. doi: 10.1371/journal.ppat.1005696
22. Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, et al. Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) Protein Domains Target LipY Lipases of Pathogenic Mycobacteria to the Cell Surface via the ESX-5 Pathway. J Biol Chem (2011) 286:19024–34. doi: 10.1074/jbc.M110.204966
23. Serafini A, Pisu D, Palù G, Rodriguez GM, Manganelli R. The ESX-3 Secretion System Is Necessary for Iron and Zinc Homeostasis in Mycobacterium Tuberculosis. PLoS One (2013) 8. doi: 10.1371/journal.pone.0078351
24. Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey Van Pittius NC. The Plasmid-Mediated Evolution of the Mycobacterial ESX (Type VII) Secretion Systems. BMC Evolutionary Biol (2016) 16. doi: 10.1186/s12862-016-0631-2
25. Kim YS, Yang CS, Nguyen LT, Kim JK, Jin HS, Choe J, et al. Mycobacterium Abscessus ESX-3 Plays an Important Role in Host Inflammatory and Pathological Responses During Infection. Microbes Infect (2017) 19:5–17. doi: 10.1016/j.micinf.2016.09.001
26. Laencina L, Dubois V, le Moigne V, Viljoen A, Majlessi L, Pritchard J, et al. Identification of Genes Required for Mycobacterium Abscessus Growth In Vivo With a Prominent Role of the ESX-4 Locus. Proc Natl Acad Sci (2018), 201713195. doi: 10.1073/pnas.1713195115
27. Berks BC, Sargent F, Palmer T. The Tat Protein Export Pathway. Mol Microbiol (2000) 35(2):260–74. doi: 10.1046/j.1365-2958.2000.01719.x
28. Saint-Joanis B, Demangel C, Jackson M, Brodin P, Marsollier L, Boshoff H, et al. Inactivation of Rv2525c, a Substrate of the Twin Arginine Translocation (Tat) System of Mycobacterium Tuberculosis, Increases β-Lactam Susceptibility and Virulence. J Bacteriol (2006) 188:6669–79. doi: 10.1128/JB.00631-06
29. Tsirigotaki A, de Geyter J, Šoštarić N, Economou A, Karamanou S. Protein Export Through the Bacterial Sec Pathway. Nat Rev Microbiol (2017) 15:21–36. doi: 10.1038/nrmicro.2016.161
30. Zulauf KE, Sullivan JT, Braunstein M. The SecA2 Pathway of Mycobacterium Tuberculosis Exports Effectors That Work in Concert to Arrest Phagosome and Autophagosome Maturation. PLoS Pathog (2018) 14. doi: 10.1371/journal.ppat.1007011
31. Braunstein M, Espinosa BJ, Chan J, Belisle JT, R. Jacobs W. SecA2 Functions in the Secretion of Superoxide Dismutase A and in the Virulence of Mycobacterium Tuberculosis. Mol Microbiol (2003) 48:453–64.
32. Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, et al. Alveolar Macrophages Provide an Early Mycobacterium Tuberculosis Niche and Initiate Dissemination. Cell Host Microbe (2018) 24:439–46.e4. doi: 10.1016/j.chom.2018.08.001
33. Flannagan RS, Cosío G, Grinstein S. Antimicrobial Mechanisms of Phagocytes and Bacterial Evasion Strategies. Nat Rev Microbiol (2009) 7:355–66. doi: 10.1038/nrmicro2128
34. Gao L-Y, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A Mycobacterial Virulence Gene Cluster Extending RD1 is Required for Cytolysis, Bacterial Spreading and ESAT-6 Secretion. Mol Microbiol (2004) 53:1677–93. doi: 10.1111/j.1365-2958.2004.04261.x
35. Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, et al. Phagosomal Rupture by Mycobacterium Tuberculosis Results in Toxicity and Host Cell Death. PLoS Pathog (2012) 8:e1002507. doi: 10.1371/journal.ppat.1002507
36. van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. Tuberculosis and M. Leprae Translocate From the Phagolysosome to the Cytosol in Myeloid Cells. Cell (2007) 129:1287–98. doi: 10.1016/j.cell.2007.05.059
37. Zhang Q, Wang D, Jiang G, Liu W, Deng Q, Li X, et al. EsxA Membrane-Permeabilizing Activity Plays a Key Role in Mycobacterial Cytosolic Translocation and Virulence: Effects of Single-Residue Mutations at Glutamine 5. Sci Rep (2016) 6:32618. doi: 10.1038/srep32618
38. King CH, Mundayoor S, Crawford JT, Shinnick TM. Expression of Contact-Dependent Cytolytic Activity by Mycobacterium Tuberculosis and Isolation of the Genomic Locus That Encodes the Activity. Infect Immun (1993) 61:2708–12. doi: 10.1128/iai.61.6.2708-2712.1993
39. Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, et al. Mycobacterial ESX-1 Secretion System Mediates Host Cell Lysis Through Bacterium Contact-Dependent Gross Membrane Disruptions. Proc Natl Acad Sci U.S.A. (2017) 114:1371–6. doi: 10.1073/pnas.1620133114
40. Flannagan RS, Jaumouillé V, Grinstein S. The Cell Biology of Phagocytosis. Annu Rev Pathol (2012) 7:61–98. doi: 10.1146/annurev-pathol-011811-132445
41. Prashar A, Schnettger L, Bernard EM, Gutierrez MG. Rab GTPases in Immunity and Inflammation. Front Cell Infect Microbiol (2017) 7:435. doi: 10.3389/fcimb.2017.00435
42. Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab Conversion as a Mechanism of Progression From Early to Late Endosomes. Cell (2005) 122:735–49. doi: 10.1016/j.cell.2005.06.043
43. Målen H, Berven FS, Fladmark KE, Wiker HG. Comprehensive Analysis of Exported Proteins From Mycobacterium Tuberculosis H37Rv. Proteomics (2007) 7:1702–18. doi: 10.1002/pmic.200600853
44. Chopra P, Singh A, Koul A, Ramachandran S, Drlica K, Tyagi AK, et al. Cytotoxic Activity of Nucleoside Diphosphate Kinase Secreted From Mycobacterium Tuberculosis. Eur J Biochem (2003) 270:625–34. doi: 10.1046/j.1432-1033.2003.03402.x
45. Chopra P, Koduri H, Singh R, Koul A, Ghildiyal M, Sharma K, et al. Nucleoside Diphosphate Kinase of Mycobacterium Tuberculosis Acts as GTPase-Activating Protein for Rho-GTPases. FEBS Lett (2004) 571:212–6. doi: 10.1016/j.febslet.2004.06.073
46. Sun J, Wang X, Lau A, Liao T-YA, Bucci C, Hmama Z. Mycobacterial Nucleoside Diphosphate Kinase Blocks Phagosome Maturation in Murine RAW 264. 7 Macrophages PLoS One (2010) 5:e8769. doi: 10.1371/journal.pone.0008769
47. Sun J, Singh V, Lau A, Stokes RW, Obregón-Henao A, Orme IM, et al. Mycobacterium Tuberculosis Nucleoside Diphosphate Kinase Inactivates Small GTPases Leading to Evasion of Innate Immunity. PLoS Pathog (2013) 9:e1003499. doi: 10.1371/journal.ppat.1003499
48. Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of Phosphatidylinositol 3-Kinase and Rab5 Effectors in Phagosomal Biogenesis and Mycobacterial Phagosome Maturation Arrest. J Cell Biol (2001) 154:631–44. doi: 10.1083/jcb.200106049
49. Saleh MT, Belisle JT. Secretion of an Acid Phosphatase (SapM) by Mycobacterium Tuberculosis That is Similar to Eukaryotic Acid Phosphatases. J Bacteriol (2000) 182:6850–3. doi: 10.1128/JB.182.23.6850-6853.2000
50. Koliwer-Brandl H, Knobloch P, Barisch C, Welin A, Hanna N, Soldati T, et al. Distinct Mycobacterium Marinum Phosphatases Determine Pathogen Vacuole Phosphoinositide Pattern, Phagosome Maturation, and Escape to the Cytosol. Cell Microbiol (2019) 21:e13008. doi: 10.1111/cmi.13008
51. Vergne I, Chua J, Lee H-H, Lucas M, Belisle J, Deretic V. Mechanism of Phagolysosome Biogenesis Block by Viable Mycobacterium Tuberculosis. Proc Natl Acad Sci U.S.A. (2005) 102:4033–8. doi: 10.1073/pnas.0409716102
52. Puri RV, Reddy PV, Tyagi AK. Secreted Acid Phosphatase (SapM) of Mycobacterium Tuberculosis is Indispensable for Arresting Phagosomal Maturation and Growth of the Pathogen in Guinea Pig Tissues. PLoS One (2013) 8:e70514. doi: 10.1371/journal.pone.0070514
53. Saikolappan S, Estrella J, Sasindran SJ, Khan A, Armitige LY, Jagannath C, et al. The Fbpa/sapM Double Knock Out Strain of Mycobacterium Tuberculosis is Highly Attenuated and Immunogenic in Macrophages. PLoS One (2012) 7:e36198. doi: 10.1371/journal.pone.0036198
54. Arsenault RJ, Maattanen P, Daigle J, Potter A, Griebel P, Napper S. From Mouth to Macrophage: Mechanisms of Innate Immune Subversion by Mycobacterium Avium Subsp. Paratuberculosis Vet Res (2014) 45:54. doi: 10.1186/1297-9716-45-54
55. Festjens N, Bogaert P, Batni A, Houthuys E, Plets E, Vanderschaeghe D, et al. Disruption of the SapM Locus in Mycobacterium Bovis BCG Improves its Protective Efficacy as a Vaccine Against M. Tuberculosis. EMBO Mol Med (2011) 3:222–34. doi: 10.1002/emmm.201000125
56. MacLeod DA, Rhinn H, Kuwahara T, Zolin A, di Paolo G, McCabe BD, et al. RAB7L1 Interacts With LRRK2 to Modify Intraneuronal Protein Sorting and Parkinson’s Disease Risk. Neuron (2013) 77:425–39. doi: 10.1016/j.neuron.2012.11.033
57. Pradhan G, Shrivastva R, Mukhopadhyay S. Mycobacterial PknG Targets the Rab7l1 Signaling Pathway To Inhibit Phagosome-Lysosome Fusion. J Immunol (2018) 201:1421–33. doi: 10.4049/jimmunol.1800530
58. Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, et al. Protein Kinase G From Pathogenic Mycobacteria Promotes Survival Within Macrophages. Science (2004) 304:1800–4. doi: 10.1126/science.1099384
59. Prisic S, Husson RN. Mycobacterium Tuberculosis Serine/Threonine Protein Kinases. Microbiol Spectr (2014) 2. doi: 10.1128/microbiolspec.MGM2-0006-2013
60. Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, et al. The Mycobacterium Tuberculosis Protein Serine/Threonine Kinase PknG is Linked to Cellular Glutamate/Glutamine Levels and is Important for Growth In Vivo. Mol Microbiol (2004) 52:1691–702. doi: 10.1111/j.1365-2958.2004.04085.x
61. Bhattacharyya N, Nkumama IN, Newland-Smith Z, Lin L-Y, Yin W, Cullen RE, et al. An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria. mBio (2018) 9. doi: 10.1128/mBio.00931-18
62. Nguyen L, Walburger A, Houben E, Koul A, Muller S, Morbitzer M, et al. Role of Protein Kinase G in Growth and Glutamine Metabolism of Mycobacterium Bovis BCG. J Bacteriol (2005) 187:5852–6. doi: 10.1128/JB.187.16.5852-5856.2005
63. Mittal E, Kumar S, Rahman A, Krishnasastry M v. Modulation of Phagolysosome Maturation by Bacterial tlyA Gene Product. J Biosci (2014) 39:821–34. doi: 10.1007/s12038-014-9472-6
64. Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin Binds Across the Ribosomal Subunit Interface Using tlyA-Encoded 2’-O-Methylations in 16S and 23S rRNAs. Mol Cell (2006) 23:173–82. doi: 10.1016/j.molcel.2006.05.044
65. Wren BW, Stabler RA, Das SS, Butcher PD, Mangan JA, Clarke JD, et al. Characterization of a Haemolysin From Mycobacterium Tuberculosis With Homology to a Virulence Factor of Serpulina Hyodysenteriae. Microbiol (Reading) (1998) 144(Pt 5):1205–11. doi: 10.1099/00221287-144-5-1205
66. Rahman A, Srivastava SS, Sneh A, Ahmed N, Krishnasastry M v. Molecular Characterization of tlyA Gene Product, Rv1694 of Mycobacterium Tuberculosis: A non-Conventional Hemolysin and a Ribosomal RNA Methyl Transferase. BMC Biochem (2010) 11:35. doi: 10.1186/1471-2091-11-35
67. Rahman MA, Sobia P, Dwivedi VP, Bhawsar A, Singh DK, Sharma P, et al. Mycobacterium Tuberculosis TlyA Protein Negatively Regulates T Helper (Th) 1 and Th17 Differentiation and Promotes Tuberculosis Pathogenesis. J Biol Chem (2015) 290:14407–17. doi: 10.1074/jbc.M115.653600
68. Philips JA, Porto MC, Wang H, Rubin EJ, Perrimon N. ESCRT Factors Restrict Mycobacterial Growth. Proc Natl Acad Sci U.S.A. (2008) 105:3070–5. doi: 10.1073/pnas.0707206105
69. Portal-Celhay C, Tufariello JM, Srivastava S, Zahra A, Klevorn T, Grace PS, et al. Mycobacterium Tuberculosis EsxH Inhibits ESCRT-Dependent CD4+ T-Cell Activation. Nat Microbiol (2016) 2:16232. doi: 10.1038/nmicrobiol.2016.232
70. Mehra A, Zahra A, Thompson V, Sirisaengtaksin N, Wells A, Porto M, et al. Mycobacterium Tuberculosis Type VII Secreted Effector EsxH Targets Host ESCRT to Impair Trafficking. PLoS Pathog (2013) 9:e1003734. doi: 10.1371/journal.ppat.1003734
71. Ilghari D, Lightbody KL, Veverka V, Waters LC, Muskett FW, Renshaw PS, et al. Solution Structure of the Mycobacterium Tuberculosis EsxG•EsxH Complex: Functional Implications and Comparisons With Other M. Tuberculosis Esx Family Complexes. J Biol Chem (2011) 286:29993–30002. doi: 10.1074/jbc.M111.248732
72. Tufariello JM, Chapman JR, Kerantzas CA, Wong K-W, Vilchèze C, Jones CM, et al. Separable Roles for Mycobacterium Tuberculosis ESX-3 Effectors in Iron Acquisition and Virulence. Proc Natl Acad Sci U.S.A. (2016) 113:E348–57. doi: 10.1073/pnas.1523321113
73. Nath Y, Ray SK, Buragohain AK. Essential Role of the ESX-3 Associated Eccd3 Locus in Maintaining the Cell Wall Integrity of Mycobacterium Smegmatis. Int J Med Microbiol (2018) 308:784–95. doi: 10.1016/j.ijmm.2018.06.010
74. Cotter K, Stransky L, McGuire C, Forgac M. Recent Insights Into the Structure, Regulation, and Function of the V-ATPases. Trends Biochem Sci (2015) 40:611–22. doi: 10.1016/j.tibs.2015.08.005
75. Wong D, Bach H, Sun J, Hmama Z, Av-Gay Y. Mycobacterium Tuberculosis Protein Tyrosine Phosphatase (PtpA) Excludes Host Vacuolar-H+-ATPase to Inhibit Phagosome Acidification. Proc Natl Acad Sci U.S.A. (2011) 108:19371–6. doi: 10.1073/pnas.1109201108
76. Koul A, Choidas A, Treder M, Tyagi AK, Drlica K, Singh Y, et al. Cloning and Characterization of Secretory Tyrosine Phosphatases of Mycobacterium Tuberculosis. J Bacteriol (2000) 182:5425–32. doi: 10.1128/JB.182.19.5425-5432.2000
77. Bach H, Papavinasasundaram KG, Wong D, Hmama Z, Av-Gay Y. Mycobacterium Tuberculosis Virulence is Mediated by PtpA Dephosphorylation of Human Vacuolar Protein Sorting 33B. Cell Host Microbe (2008) 3:316–22. doi: 10.1016/j.chom.2008.03.008
78. Grundner C, Cox JS, Alber T. Protein Tyrosine Phosphatase PtpA is Not Required for Mycobacterium Tuberculosis Growth in Mice. FEMS Microbiol Lett (2008) 287:181–4. doi: 10.1111/j.1574-6968.2008.01309.x
79. Wong D, Li W, Chao JD, Zhou P, Narula G, Tsui C, et al. Protein Tyrosine Kinase, PtkA, is Required for Mycobacterium Tuberculosis Growth in Macrophages. Sci Rep (2018) 8:155. doi: 10.1038/s41598-017-18547-9
80. Venugopal A, Bryk R, Shi S, Rhee K, Rath P, Schnappinger D, et al. Virulence of Mycobacterium Tuberculosis Depends on Lipoamide Dehydrogenase, a Member of Three Multienzyme Complexes. Cell Host Microbe (2011) 9:21–31. doi: 10.1016/j.chom.2010.12.004
81. Deghmane A-E, Soualhine H, Soulhine H, Bach H, Sendide K, Itoh S, et al. Lipoamide Dehydrogenase Mediates Retention of Coronin-1 on BCG Vacuoles, Leading to Arrest in Phagosome Maturation. J Cell Sci (2007) 120:2796–806. doi: 10.1242/jcs.006221
82. Gatfield J, Pieters J. Essential Role for Cholesterol in Entry of Mycobacteria Into Macrophages. Science (2000) 288:1647–50. doi: 10.1126/science.288.5471.1647
83. Glick D, Barth S, Macleod KF. Autophagy: Cellular and Molecular Mechanisms. J Pathol (2010) 221:3–12. doi: 10.1002/path.2697
84. Bento CF, Empadinhas N, Mendes V. Autophagy in the Fight Against Tuberculosis. DMA Cell Biol (2015) 34(4):228–42.
85. Dahl JL, Wei J, Moulder JW, Laal S, Friedman RL. Subcellular Localization of the Iitracellular Survival-Enhancing Eis Protein of Mycobacterium Tuberculosis. Infect Immun (2001) 69:4295–302. doi: 10.1128/IAI.69.7.4295-4302.2001
86. Kim KH, An DR, Song J, Yoon JY, Kim HS, Yoon HJ, et al. Mycobacterium Tuberculosis Eis Protein Initiates Suppression of Host Immune Responses by Acetylation of DUSP16/MKP-7. Proc Natl Acad Sci U.S.A. (2012) 109:7729–34. doi: 10.1073/pnas.1120251109
87. Menon MB, Dhamija S. Beclin 1 Phosphorylation - at the Center of Autophagy Regulation. Front Cell Dev Biol (2018) 6:137. doi: 10.3389/fcell.2018.00137
88. Duan L, Yi M, Chen J, Li S, Chen W. Mycobacterium Tuberculosis EIS Gene Inhibits Macrophage Autophagy Through Up-Regulation of IL-10 by Increasing the Acetylation of Histone H3. Biochem Biophys Res Commun (2016) 473:1229–34. doi: 10.1016/j.bbrc.2016.04.045
89. Shin D-M, Jeon B-Y, Lee H-M, Jin HS, Yuk J-M, Song C-H, et al. Mycobacterium Tuberculosis Eis Regulates Autophagy, Inflammation, and Cell Death Through Redox-Dependent Signaling. PLoS Pathog (2010) 6:e1001230. doi: 10.1371/journal.ppat.1001230
90. Samuel LP, Song C-H, Wei J, Roberts EA, Dahl JL, Barry CE, et al. Expression, Production and Release of the Eis Protein by Mycobacterium Tuberculosis During Infection of Macrophages and its Effect on Cytokine Secretion. Microbiol (Reading) (2007) 153:529–40. doi: 10.1099/mic.0.2006/002642-0
91. Dubois V, Pawlik A, Bories A, le Moigne V, Sismeiro O, Legendre R, et al. Mycobacterium Abscessus Virulence Traits Unraveled by Transcriptomic Profiling in Amoeba and Macrophages. PLoS Pathog (2019) 15:e1008069. doi: 10.1371/journal.ppat.1008069
92. Ung KL, Kremer L, Blaise M. Structural Analysis of the N-Acetyltransferase Eis1 From Mycobacterium Abscessus Reveals the Molecular Determinants of its Incapacity to Modify Aminoglycosides. Proteins (2021) 89(1):94–106.
93. Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular Characterization of LC3-Associated Phagocytosis Reveals Distinct Roles for Rubicon, NOX2 and Autophagy Proteins. Nat Cell Biol (2015) 17:893–906. doi: 10.1038/ncb3192
94. Köster S, Upadhyay S, Chandra P, Papavinasasundaram K, Yang G, Hassan A, et al. Mycobacterium Tuberculosis is Protected From NADPH Oxidase and LC3-Associated Phagocytosis by the LCP Protein CpsA. Proc Natl Acad Sci U.S.A. (2017) 114:E8711–20. doi: 10.1073/pnas.1707792114
95. Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An Essential Role for Interferon Gamma in Resistance to Mycobacterium Tuberculosis Infection. J Exp Med (1993) 178:2249–54. doi: 10.1084/jem.178.6.2249
96. Wang L, Wu J, Li J, Yang H, Tang T, Liang H, et al. Host-Mediated Ubiquitination of a Mycobacterial Protein Suppresses Immunity. Nature (2020) 577:682–8. doi: 10.1038/s41586-019-1915-7
97. Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat Biotechnol (2019) 37:420–3. doi: 10.1038/s41587-019-0036-z
98. Wang L, Liu Z, Wang J, Liu H, Wu J, Tang T, et al. Oxidization of Tgfβ-Activated Kinase by MPT53 is Required for Immunity to Mycobacterium Tuberculosis. Nat Microbiol (2019) 4:1378–88. doi: 10.1038/s41564-019-0436-3
99. Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, et al. Non Mycobacterial Virulence Genes in the Genome of the Emerging Pathogen Mycobacterium Abscessus. PLoS One (2009) 4:e5660. doi: 10.1371/journal.pone.0005660
100. D’Arcy MS. Cell Death: A Review of the Major Forms of Apoptosis, Necrosis and Autophagy. Cell Biol Int (2019) 43:582–92. doi: 10.1002/cbin.11137
101. Moraco AH, Kornfeld H. Cell Death and Autophagy in Tuberculosis. Semin Immunol (2014) 26:497–511. doi: 10.1016/j.smim.2014.10.001
102. Zhou B, He Y, Zhang X, Xu J, Luo Y, Wang Y, et al. Targeting Mycobacterium Protein Tyrosine Phosphatase B for Antituberculosis Agents. Proc Natl Acad Sci U.S.A. (2010) 107:4573–8. doi: 10.1073/pnas.0909133107
103. Edwards KM, Cynamon MH, Voladri RK, Hager CC, DeStefano MS, Tham KT, et al. Iron-Cofactored Superoxide Dismutase Inhibits Host Responses to Mycobacterium Tuberculosis. Am J Respir Crit Care Med (2001) 164:2213–9. doi: 10.1164/ajrccm.164.12.2106093
104. Divangahi M, Behar SM, Remold H. Dying to Live: How the Death Modality of the Infected Macrophage Affects Immunity to Tuberculosis. Adv Exp Med Biol (2013) 783:103–20. doi: 10.1007/978-1-4614-6111-1_6
105. Danilchanka O, Sun J, Pavlenok M, Maueröder C, Speer A, Siroy A, et al. An Outer Membrane Channel Protein of Mycobacterium Tuberculosis With Exotoxin Activity. Proc Natl Acad Sci U.S.A. (2014) 111:6750–5. doi: 10.1073/pnas.1400136111
106. Sun J, Siroy A, Lokareddy RK, Speer A, Doornbos KS, Cingolani G, et al. The Tuberculosis Necrotizing Toxin Kills Macrophages by Hydrolyzing NAD. Nat Struct Mol Biol (2015) 22:672–8. doi: 10.1038/nsmb.3064
107. Pajuelo D, Gonzalez-Juarbe N, Tak U, Sun J, Orihuela CJ, Niederweis M. NAD+ Depletion Triggers Macrophage Necroptosis, a Cell Death Pathway Exploited by MycobacteYrium Tuberculosis. Cell Rep (2018) 24:429–40. doi: 10.1016/j.celrep.2018.06.042
Keywords: mycobacteria, abscessus, secretion, virulence, macrophage
Citation: Bar-Oz M, Meir M and Barkan D (2022) Virulence-Associated Secretion in Mycobacterium abscessus. Front. Immunol. 13:938895. doi: 10.3389/fimmu.2022.938895
Received: 08 May 2022; Accepted: 20 June 2022;
Published: 08 July 2022.
Edited by:
Nicola Ivan Lorè, Division of Immunology, Transplantation and Infectious Diseases, IRCCS San Raffaele Scientific Institute, ItalyReviewed by:
Vitor Mendes, University of Cambridge, United KingdomCopyright © 2022 Bar-Oz, Meir and Barkan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Barkan, ZGFuaWVsLmJhcmthbkBtYWlsLmh1amkuYWMuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.