
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 21 July 2022
Sec. Alloimmunity and Transplantation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.938206
 Shingo Suzuki1
Shingo Suzuki1 Satoko Morishima2
Satoko Morishima2 Makoto Murata3
Makoto Murata3 Masafumi Tanaka1
Masafumi Tanaka1 Atsuko Shigenari1
Atsuko Shigenari1 Sayaka Ito1
Sayaka Ito1 Uma Kanga4
Uma Kanga4 Jerzy K. Kulski1,5
Jerzy K. Kulski1,5 Yasuo Morishima6,7
Yasuo Morishima6,7 Takashi Shiina1*
Takashi Shiina1*Acute graft-versus-host disease (aGVHD) is defined as a syndrome of an immunological response of graft to the host that occurs early after allogeneic hematopoietic stem cell transplantation (HCT). This disease is frequently observed even in HCT matched for human leukocyte antigen (HLA) alleles at multiple gene loci. Although the HLA region represents complex and diverse genomic characteristics, detailed association analysis is required for the identification of uncharacterized variants that are strongly associated with aGVHD. We genotyped three loci, OR2H2, HLA-F-AS1, and HLA-G, that are located in the 460 kb of HLA telomeric region and statistically analyzed the genotypes including HLA-DPB1 with clinical and transplantation outcomes using 338 unrelated bone marrow transplantation (UR-BMT) patient–donor pairs who were matched for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (HLA-10/10). Multivariate analyses demonstrated that HLA-F-AS1 and HLA-DPB1 mismatches were associated with grade II–IV aGVHD (hazard ratio (HR), 1.76; 95% CI, 1.07–2.88; p = 0.026; and HR, 1.59; CI, 1.02–2.49; p = 0.042, respectively). There was no confounding between HLA-F-AS1 and HLA-DPB1 (p = 0.512), suggesting that the HLA-F-AS1 mismatch has a strong effect on aGVHD independently of HLA-DPB1. Moreover, a stratified analysis suggested possible associations of HLA-F-AS1, HLA-DPB1, and/or HLA-G mismatches with grade II–IV aGVHD and the more severe grade III–IV aGVHD. These findings provide new insights into understanding the molecular mechanism of aGVHD caused by HLA-matched UR-BMT.
Unrelated donor hematopoietic stem cell transplantation (UR-HCT) is an established curative therapy for patients with hematologic malignancies and other hematologic and immunologic disorders in cases where no human leukocyte antigen (HLA)-matched relatives are found. An HLA mismatch between a donor’s cells or tissues (graft) transplanted into an unrelated patient (host) is well known to cause acute graft-versus-host disease (aGVHD) in many cases of UR-HCT (1, 2). The onset of aGVHD usually is defined as a syndrome characterized by rash, jaundice, and diarrhea, which occur early after allogeneic HCT with an immunological response of the graft to the host, in a graft-versus-host (GVH) direction, which may lead to graft rejection and death of the transplant recipient (3). The severity of aGVHD is graded between I and IV, with grade I the least severe (clinical manifestations limited to less than 50% of body skin involvement) and grade IV the most severe (skin, gastrointestinal tract, and/or liver involvement) stage prior to mortality (4).
Numerous association analyses between the HLA genotypes within the major histocompatibility complex (MHC) genomic region on chromosome 6 (6p21.3) and clinical and transplantation outcomes have reported that aGVHD strongly associates with patient–donor HLA mismatches (5–11). Therefore, the primary strategy to avoid aGVHD development is to match the HLA gene loci and alleles between donors and recipients (HLA-matched transplantation), so that, for example, HLA-A, HLA-B, HLA-C, and HLA-DRB1 (HLA-8/8) or HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (HLA-10/10) are matched between donors and recipients in a clinical setting (12–15). However, 11% of recipients with HLA-8/8-matched UR-HCT and 17% of recipients with HLA-10/10-matched-related HCT or UR-HCT develop grade III–IV aGVHD (2, 16). This means that there are uncharacterized variations involved in the development of aGVHD in addition to the classical HLA loci traditionally used for HLA-matched HCT. Since the MHC genomic region is composed of more than 400 genes together with the classical HLA class I and class II genes, it is possible that one or more of the unidentified neighboring genes were unmatched and might have contributed to aGVHD and the rejection of the graft.
In the HLA genomic region, HLA-DPB1 has been characterized as an aGVHD-related HLA locus, as follows: 1) HLA-DPB1 mismatch associates with severe aGVHD (15, 17), 2) T-cell epitope (TCE) algorism of DPB1 (18), 3) highly expressed HLA-DPB1 alleles associate with aGVHD (19), and 4) evolutionarily highly conserved gene structures in HLA-DPB1 associate with aGVHD, which are different from the TCE algorism (20). Hence, the importance of HLA-DPB1 for transplantation medicine is increasingly recognized with suggestions that HLA-DPB1 matched donors should be added to the traditional HLA-10/10 genotyping set. Also, the non-classical MHC gene, HLA-G, is known to associate with transplantation outcomes; for example, the insertion alleles of 14-bp insertion or deletion (indel) polymorphism (rs371194629) in the 3′-untranslated region (3′UTR) of HLA-G were reported to associate with aGVHD (21).
HLA-G and HLA-F genes are both located on the telomeric end of the MHC genomic region that represents a haplotypic segment with diverse genomic characteristics and that might be associated with aGVHD but has yet not been analyzed in detail (22). The HLA-G molecule binds to inhibitory receptors such as leukocyte Ig-like receptors B1 and B2 (LILRB1 and LILRB2), and the molecule has immunological functions such as inhibition of the cytotoxic activity of natural killer (NK) cells, induction of apoptosis and dendritic cells, and inhibition of CD4+ T-cell proliferation and antigen-presenting cells (23–27). Although the biological function of HLA-F has not been investigated in the same detail or as broadly as HLA-G, HLA-F alleles and HLA-F gene expression were identified to be involved with pathological and physiological processes such as viral infection, glioblastoma, alloimmunization in pregnancy, and autoimmune diseases like systemic lupus erythematosus (SLE) and breast cancer (28–32). The HLA-F molecule binds to inhibitory killer immunoglobulin-like receptors (KIRs) (KIR3DL2 and ILT2) and activated KIRs (KIR2DS4 and KIR3DS1) as open conformers (OCs) on NK cells (33–35). Also, receptors on NK cells are known to recognize peptides bound by HLA-F molecules or OC-type HLA-F molecules and function as immunoregulators (36). Moreover, a recent HCT study has indicated that soluble HLA-F might inhibit NK and T cells to favor allograft survival and that the upregulation of HLA-F might increase the severity of GVHD (37). Therefore, the HLA telomere genomic region that harbors HLA-F and HLA-G seems to be worth investigating in more detail to determine whether nucleotide sequence variants within the vicinity of the HLA-F and HLA-G gene loci are strongly associated with aGVHD.
In this study, to investigate whether novel MHC single-nucleotide polymorphisms (SNPs) might be associated with aGVHD, we performed the following: 1) analyzed a 460-kb haplotype block located within the telomeric region between MAS1L and HLA-A using SNP data of Japanese subjects in the International HapMap Project and the gene composition using gene mapping data from the National Center for Biotechnology Information (NCBI); 2) genotyped three loci, OR2H2 (olfactory receptor family 2 subfamily H member 2), HLA-F-AS1 (HLA-F antisense RNA 1), and HLA-G, that are located in the 460-kb haplotype block using 338 Japanese patient–donor pairs of unrelated bone marrow transplantation (UR-BMT) who were previously matched for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 (HLA-10/10); and 3) statistically analyzed the genotypes including HLA-DPB1 with clinical and transplantation outcome by univariate and multivariate analyses.
We selected 338 pairs (676 individuals) including patients with acute myeloid leukemia (AML) from 2,344 UR-BMT Japanese patient–donor pairs who previously were studied as part of the Japan Marrow Donor Program (JMDP) from 2006 to 2010 (17) for the present retrospective study using the following selection criteria: 1) patients transplanted from donors were HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 matched at the field-2 level (HLA-10/10) and were HLA-DPB1 matched or mismatched at the field-2 level in the GVH direction; 2) transplantation pairs were retyped for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 alleles at the field-2 level PCR-SSOP or PCR-SBT methods (17); 3) transplanted non-T cell-depleted marrow were without in vivo use of anti-thymocyte globulin (ATG) for GVHD prophylaxis; 4) first transplantation; 5) Japanese ethnicity; and 6) survival was for more than 7 days after transplantation. The patient and donor characteristics of the 338 pairs are shown in Table 1; 193 were HLA-DPB1 mismatched; 160 were diagnosed as grade 0, 81 as grade I, 67 as grade II, 26 as grade III, and 4 as grade IV.
Because an olfactory receptor gene cluster (ORGC) is located on the adjoining telomeric side of the HLA-F and HLA-G haplotype segment, we decided to include a portion of the ORGC genomic region from MAS1L to HCG4P11 (Figure 1) as part of our linkage disequilibrium (LD) analysis of inferred polymorphic haplotypes. In this regard, we also chose to genotype the polymorphic olfactory receptor gene, OR2H2, within the ORGC genomic region as a potential, a priori, negative control. To understand the inferred haplotype structure in the 460-kb HLA telomeric genomic region (Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13), chr6: 29486712 to 29946600) between MAS1L and HLA-A, we extracted all SNPs in this segment using the polymorphism data from 113 Japanese subjects analyzed by the International HapMap Project (hapmap3_r3, https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html) (38). The LD map was constructed by HaploView software Ver.4.2 using the extracted SNPs and was used to narrow down genomic positions for designing DNA polymorphism markers (39).
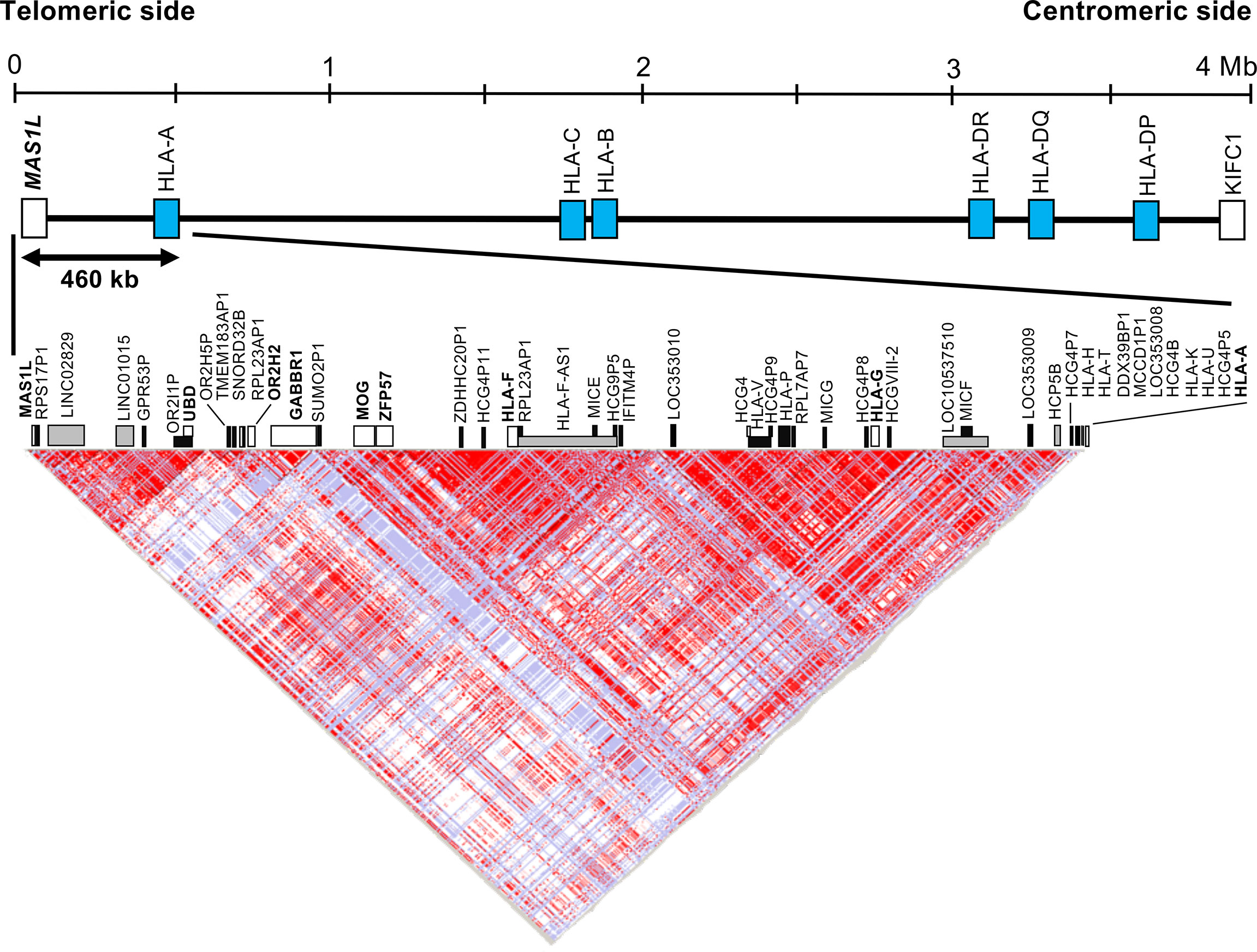
Figure 1 Haplotype structure of the human leukocyte antigen (HLA) telomeric genomic region between MAS1L and HLA-A. The linkage disequilibrium (LD) plot was generated with the program HaploView Ver.4.2. The top part shows the simplified gene composition of the 4-megabase (Mb) HLA region between MAS1L and KIFC. Blue box indicates representative classical HLA loci. The bottom part shows the LD map constructed by the HaploView software. A total of 603 single-nucleotide polymorphisms (SNPs) extracted from the HapMap data (hapmap3_r3) of 113 Japanese subjects in the 460-kb HLA telomeric genomic region between MAS1L and HLA-A (GRCh38.p13, chr6: 29486712 to 29946600) were used for this analysis. White, gray, and black boxes indicate coding genes, non-coding RNA (ncRNA), and pseudogenes, respectively. Each dot represents the value of D′/LOD, with the standard LD color scheme: white, LOD < 2 and D′ < 1; shade of pink/red, LOD > 2 and D′ < 1; blue, LOD < 2 and D′ = 1; bright red, LOD > 2 and D′ = 1.
Of the genomic DNA sequences that cover the entire HLA region derived from 95 HLA homozygous cell lines (40), we selected 43 haplotype sequences containing HLA-A alleles, which are frequently observed in the Japanese population (>0.2% allele frequency). After repeated sequences of the DNA sequences were masked by RepeatMasker software Ver. open-4.0.9 (http://www.repeatmasker.org), the unique DNA sequences between MAS1L and HLA-A were aligned by Sequencher Ver.5.0.1 DNA sequence assembly software (GENCODES, Ann Arbor, MI, USA). To perform effective genotyping by using a combination of PCR amplification and Sanger sequencing, we searched for genomic sites within the nucleotide alignments for a SNP density of more than five SNPs concentrated within less than 1 kb of sequence.
The OR2H2 and HLA-F-AS1 locus-specific primers are shown in Table S1. The primers were designed by Primer Express Ver.2.0 (Thermo Fisher Scientific, Waltham, MA, USA). The predicted amplicon sizes were 484 bp for OR2H2 and 1,036 bp for HLA-F-AS1. The 20 μl PCR mixture included 50 ng of genomic DNA, 0.5 units of PrimeSTAR GXL DNA Polymerase (1.25 U/μl, TaKaRa Bio, Otsu, Shiga, Japan), 4 μl of 5× PrimeSTAR GXL Buffer (5 mM of Mg2+), 1.6 μl of each dNTP (2.5 mM), and 1.0 μl (4 pmol/μl) of each primer. The cycling parameters were as follows: an initial denaturation with 94°C for 2 min followed by 30 cycles of 98°C/10 s, 60°C/15 s, and 68°C/1 min. PCRs were performed with the thermal cycler GeneAmp PCR system 9700 (Thermo Fisher Scientific, Foster City, CA, USA). Purified PCR products were sequenced directly with Big Dye Terminator Kit Ver.1.1 and ABI3130xl genetic analyzer (Thermo Fisher Scientific, Carlsbad, CA, USA). The DNA sequencing chromatogram data were analyzed by the Sequencher Ver.5.0.1 DNA sequence assembly software.
Since HLA-G is known to have low polymorphism over the entire gene region, the DNA sequences for full-length HLA-G alleles are difficult to determine by Sanger sequencing and short-read-based next-generation sequencing (NGS). Therefore, in-phase full-length DNA sequences were determined by the PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA, USA) using the long-ranged PCR products amplified by the previously designed HLA-G-specific primers (amplicon size, 5.6 kb) (41). We prepared pre-sequence samples according to the method for “preparing SMRTbell library with PacBio barcode universal primer for multiplexing amplicons” (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-SMRTbell-Libraries-using-PacBio-Barcoded-Universal-Primers-for-Multiplexing-Amplicons.pdf). The subsequent processes such as sequencing, clustering, phasing, consensus building, post-processing, and evaluation of the consensus sequences were performed according to our previous report (42). The HLA-G alleles were assigned by comparing the consensus sequences obtained by single-molecule real-time (SMRT) sequencing and the HLA-G allelic sequences released from the IPD-IMGT/HLA database (https://www.ebi.ac.uk/ipd/imgt/hla/) (43). The DNA sequencing chromatogram data were analyzed by the Sequencher ver.5.0.1 DNA sequence assembly software. Novel HLA-G allele candidates and HLA-G homozygote candidates were validated by Sanger sequencing using the ABI PRISM 3130xl Genetic Analyzer (Thermo Fisher Scientific, Foster City, CA, USA).
To evaluate associations of the mismatch information obtained from the four loci (HLA-G, HLA-F-AS1, HLA-DPB1, and OR2H2) and the impact on aGVHD, chronic GVHD (cGVHD), leukemia relapse, and transplant-related mortality, we performed the univariate and multivariable competing risk regression analyses using the Stata version 14 (StataCorp, Texas, USA) (44). The cumulative incidence of aGVHD was assessed by a method described previously (45). Confounders considered were patient age (linear), donor age (linear), gender (donor–recipient pair), risk of leukemia relapse (standard and high), GVHD prophylaxis (cyclosporine-based regimen, tacrolimus-based regimen, and other regimens without cyclosporine or tacrolimus), and conditioning regimen (reduced-intensity conditioning and myeloablative conditioning). Conditioning regimens were classified as myeloablative when total body irradiation was >8 Gy, oral busulfan was ≥9 mg/kg, intravenous busulfan was ≥7.2 mg/kg, or melphalan was >140 mg/m2, in accordance with Giralt et al. (46).
Expected and observed heterozygosity values were calculated by Excel function using the number of homozygotes for each allele. p-Values were calculated by the chi-square test using the expected and observed heterozygosity values.
Figure 1 and Table S2 show an LD map between two SNPs using the 603 SNPs, and 49 gene loci identified in the HLA telomeric region between MAS1L and HLA-A, respectively. In this region, a relatively large LD block was observed between SUMO2P1 and HLA-A, which was subdivided into two LD blocks: LOC353010–HLA-A block and SUMO2P1–IFITM4P block. HLA-G and HLA-F were included in the two different LD blocks: LOC353010–HLA-A block and SUMO2P1–IFITM4P block. Moreover, OR2H2 located on the telomeric side of HLA-F was included in another LD block independent of the other two blocks (Figure 1).
We identified two novel DNA polymorphic markers, HLA-F-AS1 and OR2H2, by comparative analysis among 43 genomic DNA sequences derived from HLA homozygous cell lines (40). HLA-F-AS1 is defined as a long non-coding RNA (lncRNA) located on the antisense side of HLA-F (Gene ID: 285830 in NCBI) (Figure S1), and seven known reference SNPs and seven haplotypes of HLA-F-AS1 were identified in the 43 genomic DNA sequences (Table 2). The OR2H2 SNP-dense site was composed of six known reference SNPs and six haplotypes (Table 2). Since HLA-G is included in the LD block adjoining HLA-F-AS1, we also selected this site as one of the three polymorphic DNA markers for genotyping UR-HCT cases with aGVHD. The physical distance between the three polymorphic markers is 140 kb between OR2H2 and HLA-F-AS1, 80 kb between HLA-F-AS1 and HLA-G, and 110 kb between HLA-G and HLA-A. A fourth polymorphic DNA marker, HLA-DPB1, in the HLA class II region (not shown) was included also for genotyping in this study.
From the polymorphism analyses of OR2H2 and HLA-F-AS1 markers using the 338 pairs (676 individuals), six and seven haplotypes were detected, respectively (GenBank Accession Nos. LC662707–LC662712 in OR2H2 and LC662713–LC662719 in HLA-F-AS1, Tables S3A, B). These were the same as the HLA-F-AS1 and OR2H2 haplotypes detected for the 43 genomic DNA sequences derived from HLA homozygous cell lines (Table 2). There was no statistical bias in the haplotype frequencies between donor and patient for both OR2H2 and HLA-F-AS1.
From the polymorphism analysis of HLA-G using the 338 pairs (676 individuals), 21 field-4 level (full-length level) allelic DNA sequences were identified (LC662720–LC662737 and LC662739–LC662741). By comparing these HLA-G allele sequences with those already within the IMGT/HLA database, we identified 12 novel alleles and 10 extended alleles (3,573–3,594 bp) that were in longer gene regions than in those that were previously determined (822–3,138 bp) (Table S4). The 14-bp indel polymorphism site (rs371194629) was included in all HLA-G allelic sequence analyses, and therefore the correspondence between the coding region’s polymorphisms and the 14-bp indel polymorphisms was clarified in all individuals. To investigate the association between HLA-G polymorphisms and aGVHD in detail, we created three datasets from the full-length level HLA-G sequences: the field-2 level alleles (HLA-G_Field-2), indel alleles in rs371194629 (HLA-G_ Indels), and haplotypes that combined both of them (HLA-G_Field-2+Indels). Seven, two, and eight alleles or haplotypes were detected in HLA-G_Field-2, HLA-G_Indels, and HLA-G_Field-2+Indels, respectively (Tables S3C–E). Furthermore, amino acid sequence polymorphisms deduced from the HLA-G alleles were not found in amino acid residues 100 and 103, which bind to KIR2DL4, and in residues 217–222 and 272, which bind to LILRB1/2 (47, 48).
In addition to OR2H2, HLA-F-AS1, and HLA-G, we performed an association analysis of HLA-DPB1 that included mismatched cases. Fourteen HLA-DPB1 alleles were detected from the previously genotyped HLA-DPB1 of the 338 pairs (Table S3F).
Heterozygosity values of OR2H2, HLA-F-AS1, HLA-G_Field-2, HLA-G_Indels, HLA-G_Field-2+Indels, and HLA-DPB1 were 0.73, 0.76, 0.49, 0.30, 0.57, and 0.85, respectively, and there was no statistically significant difference between the observed and expected values of heterozygotes (Table 3). Since these values suggest that allelic dropouts did not occur during PCR amplification, we proceeded with the following statistical analyses using genotype data of the four loci.
We initially evaluated mismatches between patient–donor pairs in the four loci of OR2H2, HLA-F-AS1, HLA-G, and HLA-DPB1 and the effects of transplant prognosis by the univariate analysis. From the evaluation of the association between the mismatches in GVH direction and aGVHD, the HLA-F-AS1 mismatch showed a high risk with grade III–IV and grade II–IV aGVHD (hazard ratio (HR), 2.89; 95% CI, 1.38–6.03; p = 0.005; and HR, 1.82; 95% CI, 1.16–2.85; p = 0.009, respectively). Similarly, the HLA-G (HLA-G_Field-2 and HLA-G_Field-2+Indels) and HLA-DPB1 mismatches showed high risk of grade III–IV (HR, 3.84; 95% CI, 1.52–9.75; p = 0.005; and HR, 3.12; 95% CI, 1.35–7.20; p = 0.008) and grade II–IV (HR, 1.72; 95% CI, 1.12–2.64; p = 0.012), respectively (Table 4). In contrast, OR2H2, which is located at the most genomic, telomeric end of the polymorphic markers, and the HLA-G_Indel mismatches were not associated with aGVHD. Therefore, of the HLA-G dataset, the HLA-G_Field-2, which is related at a much stronger level to aGVHD than the HLA-G_Field-2+Indels, was used for the following multivariate analyses.
Multivariate analysis of HLA-F-AS1, HLA-G, and HLA-DPB1 mismatches with clinical and transplantation results showed that the HLA-F-AS1 and HLA-DPB1 mismatches were strongly associated with grade II–IV aGVHD (HR, 1.76; 95% CI, 1.07–2.88; p = 0.026; and HR, 1.59; 95% CI, 1.02–2.49; p = 0.042) (Table 5). In contrast to the univariate analysis described above, the HLA-G mismatch did not show statistical significance in this multivariate analysis.
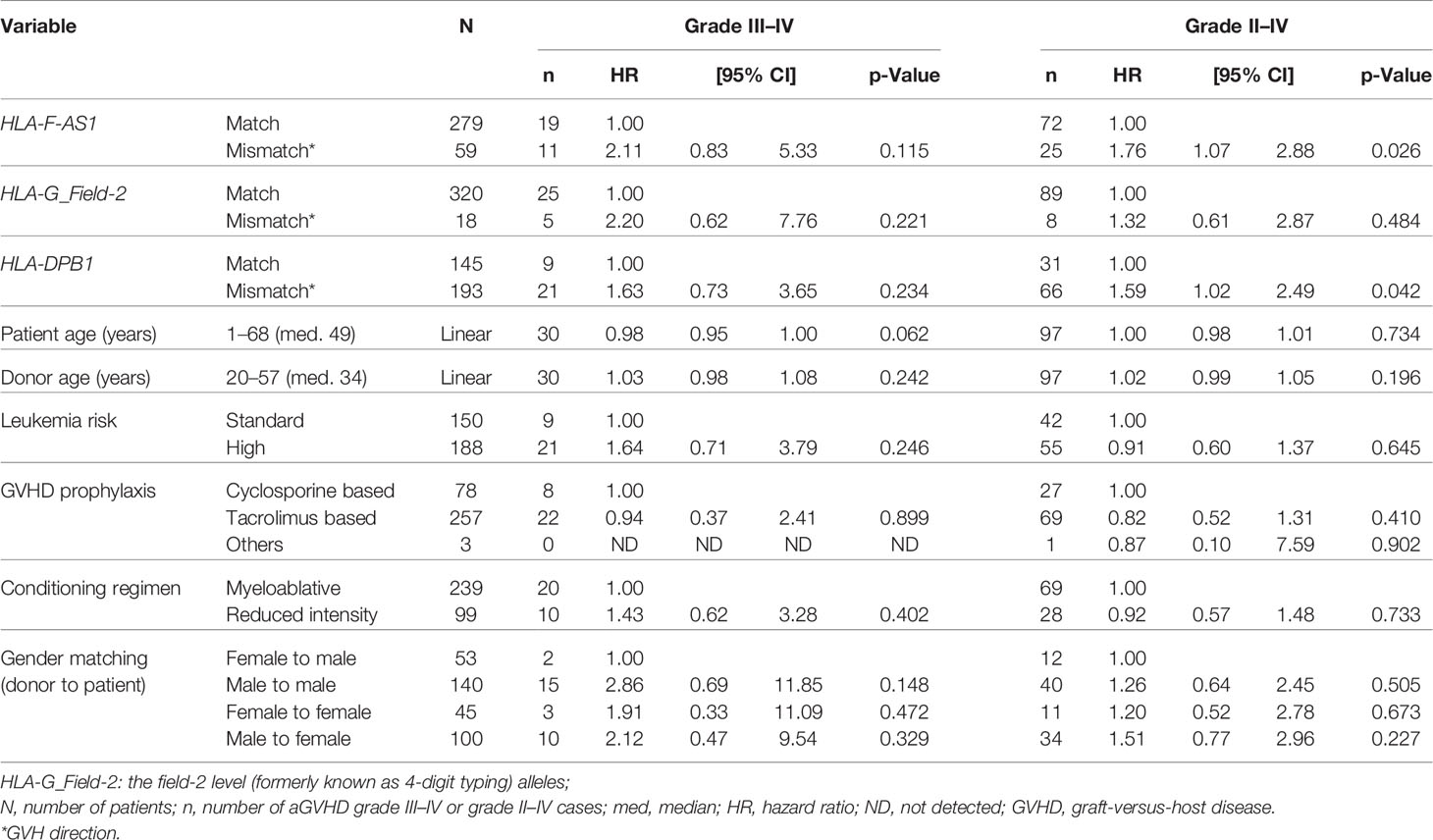
Table 5 Effect of HLA-F-AS1, HLA-G, and HLA-DPB1 mismatches on transplantation outcome by the multivariate analysis.
We performed a subgroup analysis of the risk by HLA-F-AS1, HLA-G, and/or HLA-DPB1 on aGVHD, which is shown in Table 6. The HLA-F-AS1 mismatch and HLA-F-AS1/HLA-DPB1 dual mismatches showed significant effects on grade II–IV aGVHD and/or grade III–IV aGVHD. These results suggest that the HLA-F-AS1 and HLA-DPB1 mismatches may be involved not only in the development of grade II–IV aGVHD but also in the development of the more severe grade III–IV aGVHD. Although the possibility of an HLA-G mismatch having an effect on grade II–IV and III–IV aGVHD is not conclusive because of the small number of mismatch samples in our study, it was nevertheless statistically significant (p < 0.05) in three of four mismatched categories (Table 6). The high HR values ranging between 8.11 and 13.41 for HLA-G alone or associated with HLA-F-AS1 and HLA-DPB1 mismatches and grade III–IV aGVHD suggest a possible haplotypic role for HLA-G with adverse clinical impacts.
The cumulative incidence curves of grade II–IV aGVHD by the number of HLA-F-AS1 and HLA-DPB1 locus mismatches showed clear risk differences compared to the matched cases (Figure 2).
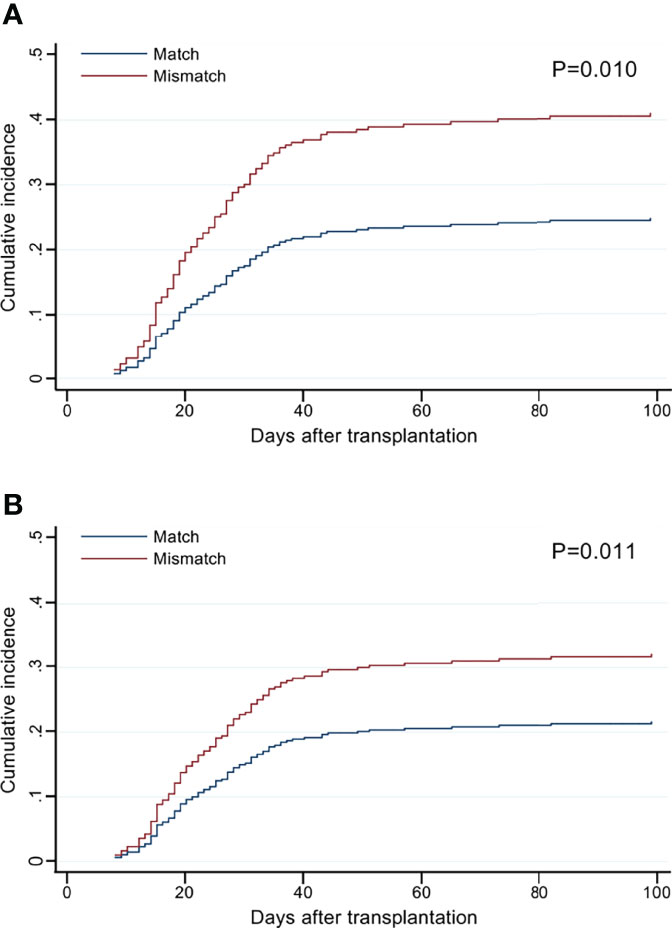
Figure 2 Cumulative incidence of grade II–IV acute graft-versus-host disease (aGVHD) by the mismatch number of HLA-F-AS1 and HLA-DPB1 at the allele level in the GVH direction. (A, B) Cumulative incidence curves of HLA-F-AS1 and HLA-DPB1, respectively. Cumulative incidences at 100 days were HLA-F-AS1 match, 26% (95% CI, 21%–31%); HLA-F-AS1 mismatch, 42% (30%–55%); HLA-DPB1 match, 21% (15%–28%); and HLA-DPB1 mismatch, 34% (28%–41%).
The multivariate analysis of HLA-F-AS1, HLA-G, and HLA-DPB1 mismatches with cGVHD, relapse, and mortality showed that only HLA-DPB1 tended to associate with cGVHD (HR, 1.58; 95% CI, 1.00–2.50; p = 0.050) but not with relapse and mortality (Table S5).
Association analysis of HLA-G_Indels showed a risk of grade II–IV aGVHD in patients with the deletion/insertion (Del/Ins) genotype (HR, 1.63; 95% CI, 1.07–2.49; p = 0.024) (Table 7). In contrast, there were no significant differences between the insertion allele and aGVHD in donors.
In this case–control study of the HLA telomeric region using the 338 UR-BMT pairs, we discovered that the HLA-F-AS1 mismatch showed a relatively strong association with aGVHD. The HLA-F-AS1 mismatch showed a similar aGVHD risk (HR, 1.76 vs. 1.59) as HLA-DPB1 (Table 5). However, no confounding was observed between the two polymorphic DNA markers (p = 0.512), which suggests that their effects on aGVHD are independent of different genetic factors (Table S6). Therefore, a novel aGVHD-related polymorphism appears to be located on the genomic segment of HLA-F-AS1 or its neighborhood. Also, the mismatches of HLA-F-AS1 and/or HLA-DPB1 together may have an effect on the development of both grade II–IV and grade III–IV aGVHD.
We chose to genotype the polymorphic olfactory receptor gene, OR2H2, within the ORGC genomic region telomeric of HLA-F-AS1 (Figure 1) as a potential, a priori, negative control. In this regard, the genotyped OR2H2 marker located within the ORGC on the most telomeric side of HLA-F-AS1 did not show an association with aGVHD, confirming that the significant SNP associations with aGVHD are located within a 330-kb region on the centromeric side of OR2H2.
In the HLA genomic region, duplication units, including HLA class I genes, HLA-F-AS1, and repetitive sequences such as retrotransposons, formed by segmental duplication events are densely distributed in the telomere side of HLA-A (22, 49). From our genomic sequencing of the primate MHC regions, we previously suggested that the MHC diversity is not limited to antigen/T-cell receptor (TCR) interacting sites of the classical HLA class I molecules but spreads to the surrounding genomic segments as “hitchhiking diversity” owing to the accumulated effect of overdominant selection acting on the classical HLA loci (41). Harmless hitchhiking mutations generated during primate evolution may have been changed pathologically in modern humans by structural and physiological changes and/or life–environmental changes. In fact, several disease-related genes, such as diffuse panbronchiolitis, psoriasis vulgaris, rheumatoid arthritis, and sarcoidosis, were identified in the hitchhiking areas (50). HLA-F-AS1 is included in the hitchhiking area, along with MICA (51, 52), HLA-G (21), and the rs887464 SNP (53) associated with transplantation outcome. Also, some HLA-F-AS1 variant(s) might be associated together as a haplotype with the HLA-F gene variant and Alu-HF and SVA-HF insertion variants that are located within close proximity (49). Therefore, uncharacterized aGVHD-related polymorphisms may still be present in the hitchhiking areas of the HLA genomic region.
Previously, SNP-based association studies in the HLA telomeric region were performed using the Affymetrix Human Mapping 500K array set and the 1,228 SNPs located in the HLA genomic region of the Illumina FastTrack Genotyping Service (Illumina, Inc.) (7, 53). However, the HLA-F-AS1 polymorphism was not identified because the SNP set used for genotyping did not contain the seven SNPs composed of HLA-F-AS1 markers. Association analyses of transplant recipients with aGVHD using microsatellite markers have also been reported, but the HLA-F-AS1 genomic region has not been analyzed so far (54). Therefore, the identification of aGVHD-related polymorphisms may have been overlooked previously due to the extraordinarily complex genomic structure of the region.
Because HLA-F-AS1 is defined as an lncRNA (55), its involvement with aGVHD appears to be different from the traditional notion of an antigen presentation system mediated by HLA molecules and their associated antigenic peptides. For example, HLA-F-AS1 expression has been shown to influence the production and migration of macrophages via intermediates such as microRNA (miRNA) and profilin 1 in colorectal cancer (55, 56). Moreover, macrophage infiltration of skin lesions after HCT was suggested to be an important predictor of aGVHD as well as a negative prognostic factor of overall survival (OS) (57). Therefore, HLA-F-AS1 may be associated with transplantation outcomes via macrophage production, migration, and infiltration and the regulation of HLA expression levels. However, since there is currently no direct evidence that HLA-F-AS1 is a true aGVHD-related gene, detailed polymorphism analysis of the LD block containing the HLA-F-AS1 marker is still required to identify true aGVHD-related polymorphisms with stronger genetic effects. Also, validation of the association between HLA-F-AS1 and aGVHD is necessary for another cohort because of the relatively small cohort (338 patient–donor pairs) that we used in this study. Furthermore, there may be ethnic differences that have a stronger effect on HLA-F-AS1 than what was detected in the Japanese in the present study.
We also investigated the relationship between HLA-G_Field-2 mismatch and aGVHD, and found a significant association of p = 0.005 for grade III–IV aGVHD but not grade II–IV by the univariate analysis (Table 4). This suggests a possible effect of the coding exons of HLA-G on severe aGVHD. However, there was no significant effect of an HLA-G allelic mismatch on the outcome of the transplantation (Table 5). However, the Del/Ins genotype of the 14-bp indels (rs371194629) in 3′UTR of the HLA-G was suggested to associate with the risk of grade II–IV aGVHD (Table 7). Here, we were unable to evaluate the effect of the Ins/Ins genotype on aGVHD because the number of Ins/Ins genotypes was inadequate in both the patient and donor groups. However, Boukouaci et al. (21) suggested that the HLA-G low expressor 14-bp insertion allele constitutes a risk factor for the incidence of severe aGVHD in patients who received bone marrow as a stem cell source (21). The rs371194629 is involved in the stability of HLA-G mRNAs (58), and the 14-bp indel polymorphism regulates the binding of multiple miRNAs such as miR148 and miR152, along with neighborhood SNPs such as rs106332044 (59). Since these descriptions support our data, the insertion allele and Del/Ins genotype are considered a risk for aGVHD.
It seems that haplotype matching at all the gene loci of the MHC genomic region for unrelated HCT would be preferred to matching at only five HLA loci in order to reduce the risks of aGVHD (14, 15). MHC haplotype matching is not technically or financially feasible for most tissue and cell typing laboratories so at this time, matching at 5-HLA loci or haplo-identical (half-matched) remains the method of choice (60). Also, GVHD is still a significant and potentially life-threatening complication even when the HLA is matched at five or more loci for unrelated HCT (14, 15). To better understand the role of the HLA-F-AS1, HLA-F, and HLA-G independently or as a haplotypic segment in the development of aGVHD, we propose that future studies sequence this MHC genomic region by the PACBIO long-range sequencing method. A better understanding and definition of the haplotypes of the HLA-F/HLA-G genomic segment might be important in helping to reduce the risks of unrelated HCT and lead to more successful transplantation outcomes.
This study revealed that HLA-F-AS1 mismatches between sequence variants are significantly associated with the risk of developing grade II–IV aGVHD. The significant effect of the HLA-F-AS1 mismatch on aGVHD was independent of the classical HLA and HLA-G polymorphisms. Thus, the results of this research provide useful new information for the development of future BMT donor selection algorithms and the coordination of patients and donors. We expect that our findings will lead to further new insights into a better understanding of the molecular mechanism of aGVHD caused by HLA-matched UR-BMT.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Institutional Review Board for Clinical Research, Tokai University Ethics committee, JMDP. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
SS, MT, UK, MM, YM, and TS participated in the design of the study. SS, AS, and SI performed nucleotide sequencing and genotyping. SS, SM, and YM performed statistical data analyses. SS, MT, JK, and TS performed the analysis and wrote the paper. All authors reviewed and approved the final version of the paper.
This work was supported by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) (16H06502 and 22K08465) and a Practical Research Project for Allergic Diseases and Immunology from the Japan Agency for Medical Research and Development (JP22ek0510032).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank JMDP for providing peripheral blood mononuclear cell (PBMC) samples and clinical data and Dr. Takashi Hirano for the technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.938206/full#supplementary-material
1. Kernan NA, Bartsch G, Ash RC, Beatty PG, Champlin R, Filipovich A, et al. Analysis of 462 Transplantations From Unrelated Donors Facilitated by the National Marrow Donor Program. N Engl J Med (1993) 328(9):593–602. doi: 10.1056/NEJM199303043280901
2. Sasazuki T, Juji T, Morishima Y, Kinukawa N, Kashiwabara H, Inoko H, et al. Effect of Matching of Class I HLA Alleles on Clinical Outcome After Transplantation of Hematopoietic Stem Cells From an Unrelated Donor. Japan Marrow Donor Program. N Engl J Med (1998) 339(17):1177–85. doi: 10.1056/NEJM199810223391701
3. Jacobsohn DA, Vogelsang GB. Acute Graft Versus Host Disease. Orphanet J Rare Dis (2007) 2:35. doi: 10.1186/1750-1172-2-35
4. Przepiorka D, Weisdorf D, Martin P, Klingemann HG, Beatty P, Hows J, et al. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant (1995) 15(6):825–8.
5. Ratanatharathorn V, Nash RA, Przepiorka D, Devine SM, Klein JL, Weisdorf D, et al. Phase III Study Comparing Methotrexate and Tacrolimus (Prograf, FK506) With Methotrexate and Cyclosporine for Graft-Versus-Host Disease Prophylaxis After HLA-Identical Sibling Bone Marrow Transplantation. Blood (1998) 92(7):2303–14.
6. Loiseau P, Busson M, Balere ML, Dormoy A, Bignon JD, Gagne K, et al. HLA Association With Hematopoietic Stem Cell Transplantation Outcome: The Number of Mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 Is Strongly Associated With Overall Survival. Biol Blood Marrow Transplant (2007) 13(8):965–74. doi: 10.1016/j.bbmt.2007.04.010
7. Sato-Otsubo A, Nannya Y, Kashiwase K, Onizuka M, Azuma F, Akatsuka Y, et al. Genome-Wide Surveillance of Mismatched Alleles for Graft-Versus-Host Disease in Stem Cell Transplantation. Blood (2015) 126(25):2752–63. doi: 10.1182/blood-2015-03-630707
8. Morishima Y, Sasazuki T, Inoko H, Juji T, Akaza T, Yamamoto K, et al. The Clinical Significance of Human Leukocyte Antigen (HLA) Allele Compatibility in Patients Receiving a Marrow Transplant From Serologically HLA-A, HLA-B, and HLA-DR Matched Unrelated Donors. Blood (2002) 99(11):4200–6. doi: 10.1182/blood.v99.11.4200
9. Shaw BE, Gooley TA, Malkki M, Madrigal JA, Begovich AB, Horowitz MM, et al. The Importance of HLA-DPB1 in Unrelated Donor Hematopoietic Cell Transplantation. Blood (2007) 110(13):4560–6. doi: 10.1182/blood-2007-06-095265
10. Petersdorf EW, Malkki M, Hsu K, Bardy P, Cesbron A, Dickinson A, et al. 16th IHIW: International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Int J Immunogenet (2013) 40(1):2–10. doi: 10.1111/iji.12022
11. Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, et al. High-Resolution Donor-Recipient HLA Matching Contributes to the Success of Unrelated Donor Marrow Transplantation. Blood (2007) 110(13):4576–83. doi: 10.1182/blood-2007-06-097386
12. Furst D, Muller C, Vucinic V, Bunjes D, Herr W, Gramatzki M, et al. High-Resolution HLA Matching in Hematopoietic Stem Cell Transplantation: A Retrospective Collaborative Analysis. Blood (2013) 122(18):3220–9. doi: 10.1182/blood-2013-02-482547
13. Flomenberg N, Baxter-Lowe LA, Confer D, Fernandez-Vina M, Filipovich A, Horowitz M, et al. Impact of HLA Class I and Class II High-Resolution Matching on Outcomes of Unrelated Donor Bone Marrow Transplantation: HLA-C Mismatching Is Associated With a Strong Adverse Effect on Transplantation Outcome. Blood (2004) 104(7):1923–30. doi: 10.1182/blood-2004-03-0803
14. Morishima S, Ogawa S, Matsubara A, Kawase T, Nannya Y, Kashiwase K, et al. Impact of Highly Conserved HLA Haplotype on Acute Graft-Versus-Host Disease. Blood (2010) 115(23):4664–70. doi: 10.1182/blood-2009-10-251157
15. Petersdorf EW, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC Haplotype Matching for Unrelated Hematopoietic Cell Transplantation. PloS Med (2007) 4(1):e8. doi: 10.1371/journal.pmed.0040008
16. Granier C, Biard L, Masson E, Porcher R, Peffault de Latour R, Robin M, et al. Impact of the Source of Hematopoietic Stem Cell in Unrelated Transplants: Comparison Between 10/10, 9/10-HLA Matched Donors and Cord Blood. Am J Hematol (2015) 90(10):897–903. doi: 10.1002/ajh.24112
17. Morishima Y, Kashiwase K, Matsuo K, Azuma F, Morishima S, Onizuka M, et al. Biological Significance of HLA Locus Matching in Unrelated Donor Bone Marrow Transplantation. Blood (2015) 125(7):1189–97. doi: 10.1182/blood-2014-10-604785
18. Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon J-D, et al. Effect of T-Cell-Epitope Matching at HLA-DPB1 in Recipients of Unrelated-Donor Haemopoietic-Cell Transplantation: A Retrospective Study. Lancet Oncol (2012) 13(4):366–74. doi: 10.1016/s1470-2045(12)70004-9
19. Petersdorf EW, Malkki M, O'HUigin C, Carrington M, Gooley T, Haagenson MD, et al. High HLA-DP Expression and Graft-Versus-Host Disease. N Engl J Med (2015) 373(7):599–609. doi: 10.1056/NEJMoa1500140
20. Morishima S, Shiina T, Suzuki S, Ogawa S, Sato-Otsubo A, Kashiwase K, et al. Evolutionary Basis of HLA-DPB1 Alleles Affects Acute GVHD in Unrelated Donor Stem Cell Transplantation. Blood (2018) 131(7):808–17. doi: 10.1182/blood-2017-08-801449
21. Boukouaci W, Busson M, Fortier C, Amokrane K, de Latour RP, Robin M, et al. Association of HLA-G Low Expressor Genotype With Severe Acute Graft-Versus-Host Disease After Sibling Bone Marrow Transplantation. Front Immunol (2011) 2:74. doi: 10.3389/fimmu.2011.00074
22. Shiina T, Tamiya G, Oka A, Takishima N, Yamagata T, Kikkawa E, et al. Molecular Dynamics of MHC Genesis Unraveled by Sequence Analysis of the 1,796,938-Bp HLA Class I Region. Proc Natl Acad Sci USA (1999) 96(23):13282–7. doi: 10.1073/pnas.96.23.13282
23. Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human Inhibitory Receptors Ig-Like Transcript 2 (ILT2) and ILT4 Compete With CD8 for MHC Class I Binding and Bind Preferentially to HLA-G. Proc Natl Acad Sci U.S.A. (2003) 100(15):8856–61. doi: 10.1073/pnas.1431057100
24. Rouas-Freiss N, Marchal RE, Kirszenbaum M, Dausset J, Carosella ED. The Alpha1 Domain of HLA-G1 and HLA-G2 Inhibits Cytotoxicity Induced by Natural Killer Cells: Is HLA-G the Public Ligand for Natural Killer Cell Inhibitory Receptors? Proc Natl Acad Sci USA (1997) 94(10):5249–54. doi: 10.1073/pnas.94.10.5249
25. Fournel S, Aguerre-Girr M, Huc X, Lenfant F, Alam A, Toubert A, et al. Cutting Edge: Soluble HLA-G1 Triggers CD95/CD95 Ligand-Mediated Apoptosis in Activated CD8+ Cells by Interacting With CD8. J Immunol (2000) 164(12):6100–4. doi: 10.4049/jimmunol.164.12.6100
26. Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella ED. Soluble Hla-G Protein Secreted by Allo-Specific CD4+ T Cells Suppresses the Allo-Proliferative Response: A CD4+ T Cell Regulatory Mechanism. Proc Natl Acad Sci U.S.A. (2001) 98(21):12150–5. doi: 10.1073/pnas.201407398
27. LeMaoult J, Caumartin J, Daouya M, Favier B, Le Rond S, Gonzalez A, et al. Immune Regulation by Pretenders: Cell-To-Cell Transfers of HLA-G Make Effector T Cells Act as Regulatory Cells. Blood (2007) 109(5):2040–8. doi: 10.1182/blood-2006-05-024547
28. Lin A, Yan WH. The Emerging Roles of Human Leukocyte Antigen-F in Immune Modulation and Viral Infection. Front Immunol (2019) 10:964. doi: 10.3389/fimmu.2019.00964
29. Hrbac T, Kopkova A, Siegl F, Vecera M, Ruckova M, Kazda T, et al. HLA-E and HLA-F Are Overexpressed in Glioblastoma and HLA-E Increased After Exposure to Ionizing Radiation. Cancer Genomics Proteomics (2022) 19(2):151–62. doi: 10.21873/cgp.20311
30. Persson G, Picard C, Marin G, Isgaard C, Stæhr CS, Molinari N, et al. Maternal HLA Ib Polymorphisms in Pregnancy Allo-Immunization. Front Immunol (2021) 12:657217. doi: 10.3389/fimmu.2021.657217
31. Jucaud V, Ravindranath MH, Terasaki PI, Morales-Buenrostro LE, Hiepe F, Rose T, et al. Serum Antibodies to Human Leucocyte Antigen (HLA)-E, HLA-F and HLA-G in Patients With Systemic Lupus Erythematosus (SLE) During Disease Flares: Clinical Relevance of HLA-F Autoantibodies. Clin Exp Immunol (2016) 183(3):326–40. doi: 10.1111/cei.12724
32. Harada A, Ishigami S, Kijima Y, Nakajo A, Arigami T, Kurahara H, et al. Clinical Implication of Human Leukocyte Antigen (HLA)-F Expression in Breast Cancer. Pathol Int (2015) 65:569–74. doi: 10.1111/pin.12343
33. Garcia-Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR, et al. Open Conformers of HLA-F Are High-Affinity Ligands of the Activating NK-Cell Receptor Kir3ds1. Nat Immunol (2016) 17(9):1067–74. doi: 10.1038/ni.3513
34. Lepin EJM, Bastin JM, Allan DSJ, Roncador G, Braud VM, Mason DY, et al. Functional Characterization of HLA-F and Binding of HLA-F Tetramers to ILT2 and ILT4 Receptors. Eur J Immunol (2000) 30(12):3552–61. doi: 10.1002/1521-4141(200012)30:12<3552::Aid-immu3552>3.0.Co;2-l
35. Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK, et al. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor Kir3ds1. PloS One (2016) 11(9):e0163297. doi: 10.1371/journal.pone.0163297
36. Dulberger CL, McMurtrey CP, Holzemer A, Neu KE, Liu V, Steinbach AM, et al. Human Leukocyte Antigen F Presents Peptides and Regulates Immunity Through Interactions With NK Cell Receptors. Immunity (2017) 46(6):1018–29 e7. doi: 10.1016/j.immuni.2017.06.002
37. Neuchel C, Fürst D, Tsamadou C, Schrezenmeier H, Mytilineos J. Extended Loci Histocompatibility Matching in HSCT-Going Beyond Classical HLA. Int J Immunogenet (2021) 48(4):299–316. doi: 10.1111/iji.12545
38. International HapMap C, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating Common and Rare Genetic Variation in Diverse Human Populations. Nature (2010) 467(7311):52–8. doi: 10.1038/nature09298
39. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics (2005) 21(2):263–5. doi: 10.1093/bioinformatics/bth457
40. Norman PJ, Norberg SJ, Guethlein LA, Nemat-Gorgani N, Royce T, Wroblewski EE, et al. Sequences of 95 Human MHC Haplotypes Reveal Extreme Coding Variation in Genes Other Than Highly Polymorphic HLA Class I and II. Genome Res (2017) 27(5):813–23. doi: 10.1101/gr.213538.116
41. Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, et al. Rapid Evolution of Major Histocompatibility Complex Class I Genes in Primates Generates New Disease Alleles in Humans Via Hitchhiking Diversity. Genetics (2006) 173(3):1555–70. doi: 10.1534/genetics.106.057034
42. Suzuki S, Ranade S, Osaki K, Ito S, Shigenari A, Ohnuki Y, et al. Reference Grade Characterization of Polymorphisms in Full-Length HLA Class I and II Genes With Short-Read Sequencing on the ION PGM System and Long-Reads Generated by Single Molecule, Real-Time Sequencing on the Pacbio Platform. Front Immunol (2018) 9:2294. doi: 10.3389/fimmu.2018.02294
43. Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA Database. Nucleic Acids Res (2020) 48(D1):D948–D55. doi: 10.1093/nar/gkz950
44. Gooley TA, Leisenring W, Crowley J, Storer BE. Estimation of Failure Probabilities in the Presence of Competing Risks: New Representations of Old Estimators. Stat Med (1999) 18(6):695–706. doi: 10.1002/(sici)1097-0258(19990330)18:6<695::aid-sim60>3.0.co;2-o
45. Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. J Am Stat Assoc (1999) 94(446):496–509. doi: 10.2307/2670170
46. Giralt S, Ballen K, Rizzo D, Bacigalupo A, Horowitz M, Pasquini M, et al. Reduced-Intensity Conditioning Regimen Workshop: Defining the Dose Spectrum. Report of a Workshop Convened by the Center for International Blood and Marrow Transplant Research. Biol Blood Marrow Transplant (2009) 15(3):367–9. doi: 10.1016/j.bbmt.2008.12.497
47. Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G Alpha1 Domain Involve in KIR2DL4 Recognition. Cell Res (2005) 15(3):176–82. doi: 10.1038/sj.cr.7290283
48. Clements CS, Kjer-Nielsen L, Kostenko L, Hoare HL, Dunstone MA, Moses E, et al. Crystal Structure of HLA-G: A Nonclassical MHC Class I Molecule Expressed at the Fetal-Maternal Interface. Proc Natl Acad Sci U.S.A. (2005) 102(9):3360–5. doi: 10.1073/pnas.0409676102
49. Kulski JK, Suzuki S, Shiina T. SNP-Density Crossover Maps of Polymorphic Transposable Elements and HLA Genes Within MHC Class I Haplotype Blocks and Junction. Front Genet (2020) 11:594318. doi: 10.3389/fgene.2020.594318
50. Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA Genomic Loci Map: Expression, Interaction, Diversity and Disease. J Hum Genet (2009) 54(1):15–39. doi: 10.1038/jhg.2008.5
51. Carapito R, Jung N, Kwemou M, Untrau M, Michel S, Pichot A, et al. Matching for the Nonconventional MHC-I MICA Gene Significantly Reduces the Incidence of Acute and Chronic GVHD. Blood (2016) 128(15):1979–86. doi: 10.1182/blood-2016-05-719070
52. Fuerst D, Neuchel C, Niederwieser D, Bunjes D, Gramatzki M, Wagner E, et al. Matching for the MICA-129 Polymorphism Is Beneficial in Unrelated Hematopoietic Stem Cell Transplantation. Blood (2016) 128(26):3169–76. doi: 10.1182/blood-2016-05-716357
53. Petersdorf EW, Malkki M, Gooley TA, Spellman SR, Haagenson MD, Horowitz MM, et al. MHC-Resident Variation Affects Risks After Unrelated Donor Hematopoietic Cell Transplantation. Sci Transl Med (2012) 4(144):144ra01. doi: 10.1126/scitranslmed.3003974
54. Li S, Kawata H, Katsuyama Y, Ota M, Morishima Y, Mano S, et al. Association of Polymorphic MHC Microsatellites With GVHD, Survival, and Leukemia Relapse in Unrelated Hematopoietic Stem Cell Transplant Donor/Recipient Pairs Matched at Five HLA Loci. Tissue Antigens (2004) 63(4):362–8. doi: 10.1111/j.0001-2815.2004.00200.x
55. Huang Y, Sun H, Ma X, Zeng Y, Pan Y, Yu D, et al. HLA-F-AS1/miR-330-3p/PFN1 Axis Promotes Colorectal Cancer Progression. Life Sci (2020) 254:117180. doi: 10.1016/j.lfs.2019.117180
56. Zhang J, Li S, Zhang X, Li C, Zhang J, Zhou W. Lncrna HLA-F-AS1 Promotes Colorectal Cancer Metastasis by Inducing PFN1 in Colorectal Cancer-Derived Extracellular Vesicles and Mediating Macrophage Polarization. Cancer Gene Ther (2021) 28(12):1269–84. doi: 10.1038/s41417-020-00276-3
57. Nishiwaki S, Terakura S, Ito M, Goto T, Seto A, Watanabe K, et al. Impact of Macrophage Infiltration of Skin Lesions on Survival After Allogeneic Stem Cell Transplantation: A Clue to Refractory Graft-Versus-Host Disease. Blood (2009) 114(14):3113–6. doi: 10.1182/blood-2009-03-209635
58. Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P. The 14 Bp Deletion-Insertion Polymorphism in the 3' UT Region of the HLA-G Gene Influences HLA-G mRNA Stability. Hum Immunol (2003) 64(11):1005–10. doi: 10.1016/j.humimm.2003.08.347
59. Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-Specific Targeting of MicroRNAs to HLA-G and Risk of Asthma. Am J Hum Genet (2007) 81(4):829–34. doi: 10.1086/521200
Keywords: human leukocyte antigen, graft-versus-host disease, bone marrow transplantation, haplotype, hitchhiking diversity, genotyping
Citation: Suzuki S, Morishima S, Murata M, Tanaka M, Shigenari A, Ito S, Kanga U, Kulski JK, Morishima Y and Shiina T (2022) Sequence Variations Within HLA-G and HLA-F Genomic Segments at the Human Leukocyte Antigen Telomeric End Associated With Acute Graft-Versus-Host Disease in Unrelated Bone Marrow Transplantation. Front. Immunol. 13:938206. doi: 10.3389/fimmu.2022.938206
Received: 07 May 2022; Accepted: 20 June 2022;
Published: 21 July 2022.
Edited by:
Jukka Partanen, Finnish Red Cross Blood Service, FinlandReviewed by:
Katarzyna Bogunia-Kubik, Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences, PolandCopyright © 2022 Suzuki, Morishima, Murata, Tanaka, Shigenari, Ito, Kanga, Kulski, Morishima and Shiina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Shiina, dHNoaWluYUBpcy5pY2MudS10b2thaS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.