- 1Department of Animal Nutrition and Feed Science, College of Animal Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2The Cooperative Innovation Center for Sustainable Pig Production, Wuhan, China
Early life is a vital period for mammals to be colonized with the microbiome, which profoundly influences the development of the intestinal immune function. For neonates to resist pathogen infection and avoid gastrointestinal illness, the intestinal innate immune system is critical. Thus, this review summarizes the development of the intestinal microbiome and the intestinal innate immune barrier, including the intestinal epithelium and immune cells from the fetal to the weaning period. Moreover, the impact of the intestinal microbiome on innate immune development and the two main way of early-life intervention including probiotics and fecal microbiota transplantation (FMT) also are discussed in this review. We hope to highlight the crosstalk between early microbial colonization and intestinal innate immunity development and offer some information for early intervention.
1 Introduction
Neonates are more susceptible to infections and other gastrointestinal diseases such as necrotizing enterocolitis (NEC) due to the immature immune system (1, 2). The ability to fight infections is dependent on innate immunity in early life (3); furthermore, the development of innate immunity is earlier than adaptive immunity and contributes to the maturation of adaptive immunity (4). The intestine is not only the digestive organ but also the body’s largest organ of immunity. More than half of the immune cells are stationed in the intestine, and 70% of immunoglobulin A (IgA) is produced in the intestine (5). Furthermore, as the closest part of the body to the outside environment, the intestine is the first line of defense. Thus, the development of innate immunity is important for neonates, while the specific mechanisms of immune modulation in early life also remain largely unknown.
Recently, emerging studies have unraveled that the development of the intestinal immune system is correlated with the abundance and diversity of the intestinal microbiome (6). After birth, the structure and diversity of the intestinal microbiome become more complex, and in parallel to the changes in the neonatal intestinal microbiome, immune cells in the intestine gradually mature. For example, higher production of cytokines at 36 months of age is closely related to colonization with Bifidobacterium at 1 week old (7).
The intestinal microbiome is easily influenced by dietary changes and the environment in early life (8); thus, neonatal intestinal microbial initial colonization is an important window of opportunity. Early intervention of intestinal microbiome colonization contributes to the development of the intestinal microbiome of neonates and has an impact on host immune development (9, 10). Probiotics and fecal microbiota transplantation (FMT) are the two effective ways to modulate intestinal microecology, promote the development of intestinal immunity, and maintain the intestine mucosa homeostasis, which has attracted a lot of interest in researchers as two main strategies of microbial intervention (11, 12).
Several reviews have summarized that microbial colonization shapes the immune system (13) and the effect of breast milk and solid food on the intestinal immunity (14), while this review provides an overview of the influence of microbial colonization on the development of intestinal innate immunity in early life especially focusing on the interaction between the specific bacteria and innate immune cells. Moreover, the effect of two main strategies, probiotics and FMT, of early intervention on intestinal innate immunity is also concluded. The goal of this review is to highlight the crosstalk between early microbial colonization and intestinal innate immunity development in early life and offer some references for early intervention by a microbial strategy.
2 Intestinal microbiome maturation in early life
2.1 Fetal Microbiome Composition
It has long been thought that the fetus is sterile and the establishment of the fetal microbiome begins after birth. The microbial community has been detected on human placental samples, cord blood, and meconium (15), while some researchers report that the experiment procedure, including sample collection and reservation, sequencing, and choosing of reagents are all different, and there is contamination during the whole trial. Furthermore, a lot of studies suggest that the human placenta has no microbiome by using different methods including 16S sequence, microbial cultivation, quantitative real-time PCR (qPCR), metagenomic next-generation sequencing (mNGS), and setting the negative control containing air and water (16–18). Whether the fetal microbiome exists remains a controversial issue. Here, we just present the microbiome found in the fetus.
Low diversity and limited bacteria have been detected in the mammal fetal intestine, skin, and lung by mNGS, 16S sequence, RNA-in situ hybridization (RNA-ISH), and scanning electron microscopy (SEM) (19, 20). A labeled E. faecium isolated from a healthy woman can be found in the amniotic fluid of the inoculated mice, which also indicated that the maternal microbiome can be transferred into the fetus (21). Phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes are enriched in the fetal intestine, especially Enterobacteriaceae, Micrococcaceae, and Lactobacillus (22–24). Additionally, bacterial metabolites such as short-chain fatty acids (SCFAs), deoxynojirimycin, mitomycin, and tobramycin exist in the fetal intestine, which also implies that there is a microbiome in the fetal intestine (23, 25).
2.2 Neonatal Microbiome Maturation
The microbiome of the newborn is similar to that of the mother and is closest to the vaginal microbiome. The intestinal microbiome in early life matures and evolves along with infant growth, and generally stabilizes and evolves adult-like after weaning, which has been proved via different ages of animal models, including mice (26) and pig (27), and infants (28).
Previous research supports the idea that oxygen is consumed by pioneer facultative anaerobic bacteria in the aerobic gut in early life, which then provides a niche for obligate anaerobes to colonize (29). On the first day after birth, facultative anaerobes from the Proteobacteria and Firmicutes (Escherichia coli, Streptococcus, Enterococcus faecalis, and Enterobacter sp.) are identified, as well as a low relative abundance of strict anaerobes from the Bacteroidetes and Firmicutes (Bacteroides vulgatus, Parabacteroides, and Clostridium) (30).
The neonates’ intestinal core microbiome is characterized by the predominance of Bifidobacterium, Enterococcus, Lactobacillus, Escherichia, Bacteroides, and Clostridium (31), and the variation after birth is shown in Figure 1. Firmicutes and Bifidobacterium dominate in the early developmental stages, which are characterized by an unstable community structure and low microbiome maturation. Along with infants growing up, Bacteroides and Prevotella gradually take the place of Firmicutes and Bifidobacterium, becoming high-level bacterial genera in infants (28). Normally, it is characterized by the predominance of such microorganisms as Bifidobacterium longum and Bifidobacterium infantis, Bacteroidetes, Lactobacillus sp., Prevotella, and Atopobium, which determine the direction of the immune response and the formation of tolerance in neonates (32).
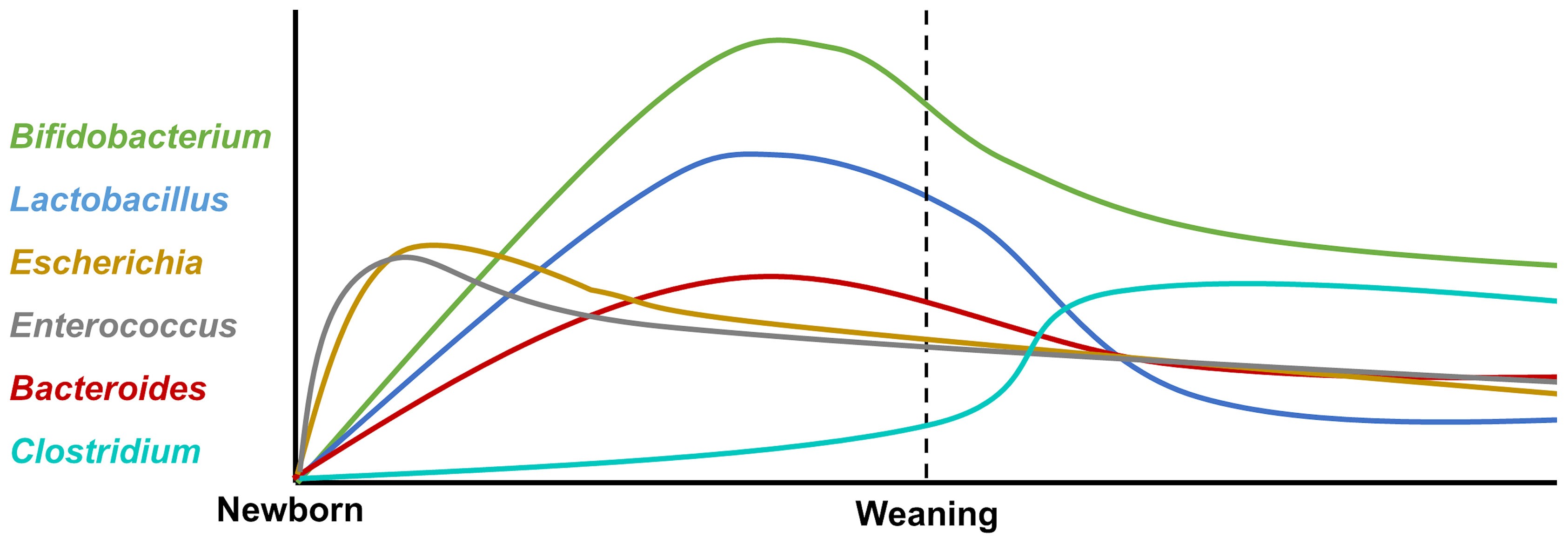
Figure 1 The variation of intestinal core microbiota after birth. The neonatal intestinal microbiota is mainly composed of Bifidobacterium, Enterococcus, Lactobacillus, Escherichia, Bacteroides, and Clostridium. After birth, facultative anaerobe bacteria Escherichia and Enterococcus increase rapidly and decrease gradually. The abundance of Bifidobacterium, Lactobacillus, and Bacteroides rises at the peak before weaning and then declines by degrees. Clostridium expands rapidly after weaning.
After weaning, dietary changes from breast milk to solid food fixed with carbohydrates and fibers lead to major structural changes in the intestinal flora and microbiome diversity increasing. Furthermore, the intestinal microbiome structure tends to be relatively mature and stable; Bifidobacterium, Lactobacillus, and Escherichia decrease after weaning; and the intestinal flora are dominated by Prevotella (33). The mature microbiome contributes to the development of the innate immune response; for instance, the levels of cytokines related to immune cell proliferation and maturation were increased in fecal water of offspring (33). Furthermore, enterocyte proliferation is induced by intestinal microbiome maturation during early human life via microbial metabolites (34).
3 Intestinal innate immune system development
The intestinal epithelium and innate immune cells are the two main functional cells of the mammalian intestine innate immunity system. The intestinal epithelium consists of epithelial cells, Paneth cells (PCs) and goblet cells (GCs), and innate immune cells including macrophages, dendritic cells (DCs), natural killer (NK) cells, and innate lymphoid cells (ILCs) (35).
3.1 The Intestinal Epithelium
3.1.1 Paneth Cells
PCs are housed in the crypt of the intestine and secrete antimicrobial peptides (AMPs), which are known to defend against pathogen infection (36, 37). Furthermore, PCs also play an essential role in intestinal homeostasis and microbiome colonization; for example, loss of PC function is implicated in microbiome dysbiosis and NEC in early life (38, 39). It has been found that Cdk5rap3 (a gene that serves a vital role in intestinal PC development and maintenance) knockout resulted in early embryonic lethality, which indicated that embryonic PC function may determine the embryo fate (40).
PCs are detected in the small intestine at 13.5 weeks’ gestational age in fetus (41), then significantly increase to quantities similar to adults after 29 weeks’ gestational age (42), whereas it is worth noticing that PCs do not develop until 7–10 days after birth in mice (43). AMP gene expression such as cryptdins increases in an age-dependent manner from 3 to 21 days in rat (44, 45). After weaning, the number of PCs and the mRNA expression levels of AMPs including RegIIIβ, RegIIIγ, and Ang4 in the intestine were increased in mice (46, 47).
It has been demonstrated that PC number and functions are regulated by the intestinal microbiome, and germ-free (GF) mice have fewer intestinal PC numbers and RegIIIγ secretion compared with specific pathogen-free (SPF) mice (48). Early-life microbiome participates in the function and maturation of PCs, and the gene expression of PC products including defensins, matrilysin, and phospholipase A2 was significantly downregulated after antibiotic treatment in neonates (49). Furthermore, the number of PCs in a humanized microbiota gnotobiotic mouse model is significantly decreased compared with SPF mice, and the transportation of normal infant intestinal microbiome can rescue the low number of PCs and increase the AMP (Cathelicidin-5 and Lysozyme-1) mRNA expression level in the humanized microbiota gnotobiotic mouse model (50). On the other hand, the loss of a protective mucus layer leads to increased epithelial–bacterial interactions in mice and then results in the upregulation of RegIIIβ and RegIIIγ expression (46). RegIIIγ mRNA expression increased in the small intestine of conventionally raised mice throughout the weaning period, but not in GF animals, indicating that the intestinal microbiome is crucial for PC function during the weaning period (51).
3.1.2 Goblet Cells
Intestinal GCs generate and secrete mucins, primarily mucin2 (MUC2), which are a major component of the mucus barrier, and serve as the first line of defense against pathogen infection and bacterial-derived product invasion (52, 53). Trefoil factor-2 (TFF2) deficiency in the neonatal small intestine allows the opportunistic neonatal pathogen E. coli K1 to permeate the weakened mucus layer in the distal small intestine and cause systemic infection (54). Intestinal GCs derive from intestinal stem cells (ISCs) that reside near the bottom of the intestinal crypt (55). It was recently discovered that GCs can form goblet cell-associated antigen passages (GAPs) and transfer luminal substances to antigen-presenting cells and DCs that underlie lamina propria (LP) and trigger adaptive immune responses (56).
In the human fetal small intestine, GCs first develop about 9–10 weeks of gestation, and both the stratified and simple columnar epithelium contains relatively undifferentiated oligomucous and mature GCs (57). Along with GCs development in the fetal intestine, MUC2 mRNA is detected as early as 9 weeks’ gestation, whereas MUC3 and MUC4, unlike MUC2, are expressed at a very early gestational age (as early as 6.5 weeks’ gestation) in humans, which expressed in the primitive gut endoderm before epithelial cytodifferentiation (58). In mice, shortly after villus emergence, around E16.5, the epithelium on the villi begins to undergo cytodifferentiation into GCs (59). After birth, the absolute number of GCs continues to expand, and the number of GCs in the intestines of 21-day-old piglets is almost double that of 1-day-old piglets (60).
The intestines of GF mice contain significantly fewer and smaller GCs compared with SPF mice (61). Furthermore, when GF mice are colonized with commensal bacteria, the size and number of GCs rapidly expand, and they release much Relm-β (62). Maturation of microbiome contributes to the function of GCs; functional maturation of GCs is more significant after exposure to adult microbiome than suckling microbiome, according to assessments of solute transporters and aquaporins (63). In a humanized microbiota gnotobiotic mouse model, the microbiome of an infant with poor weight gain has been shown to reduce the amount of GCs (50). The number of GCs was increased by transferring the microbiome from 15-day-old suckling conventional rats to the GF mice (63).
3.2 Immune Cells
3.2.1 Dendritic Cells
DCs are bone marrow-derived antigen-presenting cells that are at the front line in maintaining intestinal integrity, and they comprise two major subsets: conventional (or classical) DCs (cDCs) and plasmacytoid DCs (pDCs). Intestinal DCs are found within the intestinal LP (64), Peyer patches (PPs) (65), and mesenteric lymph nodes (MLNs) (66). CD103+ DCs are key players in the innate immune system, and the relative paucity of CD103+ DCs in the neonatal intestine contributes to the high susceptibility to intestinal infection (67).
DCs are a bridge to connect innate immunity with adaptive immunity, which have been shown to stimulate T helper (Th) cells’ [Th1, Th17, and regulatory T cells (Tregs)] response in the intestine (68) and induce the activation and differentiation of naive B cells to yield plasma cells that produce commensal-specific IgA in the LP (69). Neonatal DCs are fully capable of priming CD8+ T cells during RV infection, and mice can elicit a robust antigen-specific CD8+ T-cell response, including the production of interferon gamma (IFN-γ) in early life (70). ResDCs induce Foxp3+ Tregs to provide a robust feedback mechanism for the maintenance of intestinal tolerance for the antigen of food and commensal bacteria in early life (71).
Bacterial signals during DCs differentiation have a profound impact on DCs function. DCs do not develop until day 16 in GF mice (72). DCs are highly microbiome dependent, and are disturbed following broad-spectrum antibiotic treatment, but can be restored by FMT, commensal E. coli and Lactobacillus johnsonii strains (73, 74). Moreover, MLN DCs from GF mice were less stimulatory for T cells than their counterparts from SPF mice (75).
3.2.2 Innate Lymphoid Cells
ILCs are immune effector cells that contribute to host defense, metabolic homeostasis, and tissue repair (76). ILCs can be divided into three types: type 1 ILC includes IFN-γ producing ILC1, type 2 ILC includes interleukin-5 (IL-5) and IL-13 expressing ILC2, and type 3 ILC includes lymphoid tissue inducible cells (LTi) and IL-17 and IL-22 expressing ILC3 (77).
The early phase of ILC lineage commitment mainly occurred in the fetal liver and intestine. A considerable percentage of the lymphoid progenitor was found in the 8-week fetal intestine, which indicated that the intestine is an ILC-rich organ at fetal age, and ILC3 is a domain in the intestinal ILC family around week 12 (78). ILCs began working before the adaptive system has fully developed (79), early life is an important period for ILC maturation, and absolute ILC3 numbers increase dramatically from neonates to 14 days in mice (80). During the period of weaning, the proportion of phospho-signal transducer and activator of transcription 3 (pSTAT3)+ILC3 significantly increases, which indicates the activation of ILC3, and this phenomenon depends on IL-23 (79). After weaning, ILC3 activation is suppressed by the maturation of adaptive immune CD4+T cells, Treg, and Th17 cells (79).
It has been demonstrated that GF mice have been shown to have defective ILC3 development and lower production of IL-22, a key cytokine in epithelial responses to microorganisms in the small intestine (81, 82). Furthermore, prolonged transient neonatal antibiotic exposure suppresses IL-17A-producing ILC3 during the first 2 weeks of life, promoting bacterial translocation and increasing susceptibility to K. pneumoniae-induced sepsis, and recolonization with mature microbiome raises the level of IL-17A-producing ILC3 in mice, reducing their susceptibility to late-onset sepsis (LOS) (83).
3.2.3 Natural Killer Cells
NK cells are recognized as the only cytotoxic population of ILCs. NK cells take part in protecting against pathogen infection including Listeria monocytogenes, Salmonella, Citrobacter rodentium, and Yersinia enterocolitica infections (84–86).
NK cells are detected in the intestinal epithelium and LP, which can be seen in fetal intestines (87). After birth, the number of NK cells increased dramatically from day 1 to day 3 (88). Early in life, a large proportion of this population was discovered (40% of IE lymphocytes in 9-day-old rats), with a distinct age-decreasing pattern (89).
CD103+ NK cells are the main congenital lymphocyte group in the small intestine of infants. Compared with adult intestinal NK cells, infant intestinal NK cells showed a stronger effect, with higher expression of the disintermediating embryonic protein, perforin, and granase B, and stronger degranulation ability (90).
The lower presence of NK cells has been detected in the intestine of GF mice (91). Microbiome impacts the epigenome of intestinal epithelial cells (IECs) and modifies interaction between IECs and NK cells through altering methylation-modifying enzyme activity (91). However, whether the microbiome can directly interact with NK cells to regulate NK cell activity and function remains to be determined.
3.2.4 Macrophages
There is increasing evidence that macrophages play a crucial role in the maintenance of intestinal homeostasis and as key sentinels of the intestinal immune system. For example, macrophages enhance epithelial integrity by the production of prostaglandin E2 to stimulate ISC renewal (92). Moreover, macrophages maintain and expend Treg, Th17, and ILC3 through their secretion of cytokines such as IL-10, transforming growth factor beta (TGF-β), and IL-1β (93–95). Macrophages are first detected in the 11- to 12-week fetal intestine and the number rapidly increases from the 12-week to the 22-week period of gestation in human (96).
In comparison to conventionally housed controls, analysis of the colonic macrophage compartment in GF mice found markedly fewer monocyte–macrophage subsets in the colon (97). The microbiome also controls the homeostatic replenishment of monocyte-derived macrophages in the intestinal mucosa (97). In addition, genome-wide transcriptional profiling of macrophages from antibiotic-treated mice revealed a reduction in antiviral immunity genes (98). Furthermore, it has been demonstrated that early microbiome exposure stimulated tumor necrosis factor (TNF) production in spleen monocytes and macrophages of neonatal mice during the first 3 days of life (99) and the antibiotics caused an indirect or direct reduction in macrophage-like cells in the early life of chickens (100).
4 Microbiome and specific bacteria regulate the maturation of immunity in early life
In early life, the intestinal microbiome plays a crucial role in the development of the immune system. In detail, relative abundances of Bacteroides and Prevotella populations are highly correlated with responses to toll-like receptor-2 (TLR2) (101). Moreover, at 36 months of age, Bifidobacterium colonization is related to the greater production of IL-5, IL-6, IL-13, TNF, and IL-1. Gut colonization by Enterococcus, Staphylococcus aureus, or Clostridium in early childhood, on the other hand, was negatively associated with induced IL-13, IL-5, and TNF at 3 years of age (7). At 2 years of age, early S. aureus colonization was linked to a larger number of IL-4 and IL-10 generating cells (102).
Strong evidence has reported that early colonization of microbiome shapes the development of host immunity. Intestinal simulated epithelium exhibited a stronger epithelial barrier function and intestinal maturation when exposed to home-birth fecal supernatant, as well as a larger immune response (TLR4 route activation and pro-inflammatory cytokine release) (103). Furthermore, fungal colonization can lead to significant changes in intestinal bacterial composition in mice and independently affect systemic and intestinal innate and adaptive immunity in early life, and co-colonization of bacteria and fungi in the neonatal GF mice enhanced dextran sodium sulfate (DSS)-induced colonic inflammation in adulthood (104). As previously discussed, maternal microbial colonization during gestation increases ILC3 and mononuclear cell population size in offspring (105), further supporting the importance of the microbiome to the development of immunity in early life than in adults.
As mentioned before, the infant intestinal core microbiome is characterized by the predominance of Bifidobacterium, Enterococcus, Lactobacillus, Escherichia, Bacteroides, and Clostridium (31). Early colonization of the microbiome after birth is helping to shape the innate immune system (Figure 2).
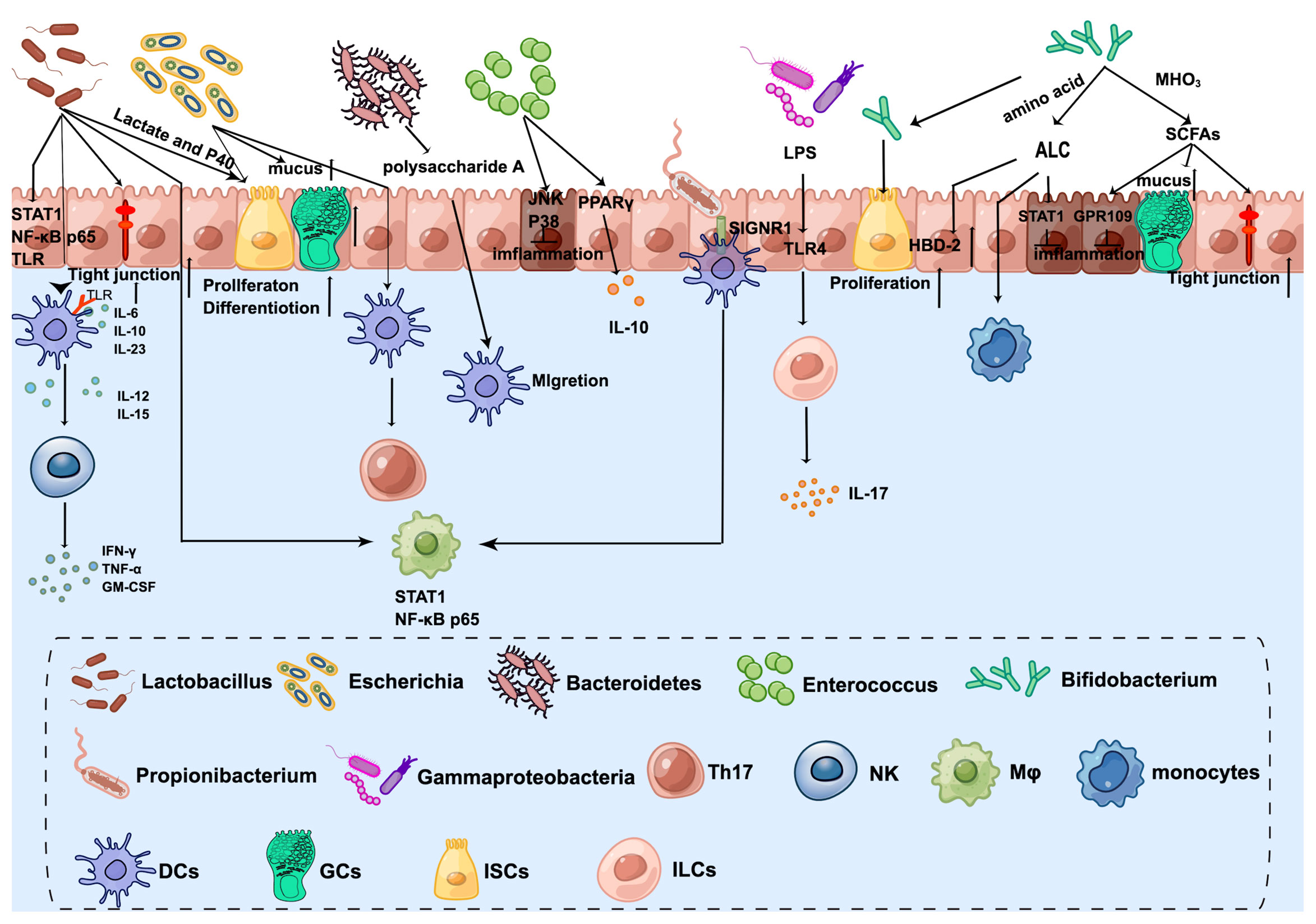
Figure 2 The crosstalk between microbiota and innate immune system in the neonatal intestine. Lactobacillus enhances DC differentiation and upregulates surface markers, which activates NK cells. Furthermore, STAT1 and NF-kB p65 nuclear translocation are influenced by Lactobacillus in both intestinal epithelial cells and underlying macrophages. Lactobacillus produced lactate and P40 can promote ISC-mediated epithelial development and tight junction formation. Bifidobacterium transfers amino acids into ALC, which activates monocytes and promotes the expression of beta-defensin2 of IECs. Additionally, ILA has anti-inflammatory effects on immature enterocytes through STAT1 pathways. Bifidobacterium releases SCFAs by metabolizing HMOs, which exert anti-inflammatory effects through GPR109A and increase tight junction and mucus gene expression. Furthermore, Bifidobacterium Tad pili stimulate the growth of the newborn mucosa. Escherichia increases the number of GCs and promotes mucus. The antigens of E. coli can be taken up by DCs and then present to CD4+ T cells to induce Th17. Bacteroides-produced Polysaccharide A mediates the migration of pDCs from the colon to the thymus to maintain PLZF+ lymphocyte homeostasis. Enterococcus attenuates proinflammatory cytokine secretions, especially IL-8 through JNK and p38 signaling pathways. Furthermore, Enterococcus promotes PPAR phosphorylation activity to activate the transcription of IL-10. Propionibacterium combines with SIGNR1 on the DCs to regulate macrophages. Gammaproteobacteria express cell-surface LPS, which is recognized by the host intestinal cells via TLR4 and then ILC3 produces the cytokine IL-17A. By Figdraw (www.figdraw.com).
4.1 Lactobacillus
Milk-promoted bacteria such as Lactobacillus are signs of normal breast milk, and neonates are quickly colonized by Lactobacillus by suckling, which affects immune development in early life (106). The abundance of Lactobacillus in newborn mice as well as treatment with specific Lactobacillus probiotics (L. murinus and L. johnsonii) were able to resist K. pneumoniae colonization and LOS induced by K. pneumoniae, in contrast to eradicating the transmission of Lactobacillus by maternal antibiotic exposure exacerbated disease (107). Moreover, the enrichment of Lactobacillus is associated with low NEC scores in the NEC rat model; in detail, L. murinus isolated from neonatal rats reduces molecular markers and the intestinal barrier damage of NEC (108).
Infant’s feces-derived Lactobacillus appears to be a promising modulator of innate immune response, which can enhance DC differentiation including promoting the secretion of IL-6, IL-10, IL-12, IL-15, and IL-23 and upregulate surface markers (HLA-DR, CD86, CD83, and CCR7) in DCs (109), which activates NK cells and promotes the synthesis and secretion of INF-γ, TNF-α, and granulocyte-macrophage colony-stimulating factor (GM-CSF); the presence of Lactobacillus switches the immune system towards a Th1 response in early life (110, 111). Another type of Lactobacillus, isolated from exclusively breast-feeding infant feces, activates TLR signaling of DCs and reduces Salmonella-induced inflammation (112). Moreover, a co-culture model of the undeveloped small intestine and underlying macrophages showed that STAT1 and nuclear factor kappa B (NF-kB) p65 nuclear translocation are influenced by Lactobacillus in both IECs and underlying macrophages, which offers new insights into the potential routes by which Lactobacillus primes the early immune system to fight pathogens (113).
Though a lot of evidence shows that Lactobacillus modulates the innate immune system in early life, knowledge about the mechanisms underlying regulating innate immune development by Lactobacillus is limited. P40, a Lactobacillus rhamnosus GG-derived protein, enhances functional maturation of the intestine in early life including IEC proliferation, differentiation, and tight junction formation by activation of epidermal growth factor receptor (114). Furthermore, Lactobacillus’ SpaC pilus was found to adhere to the fetal gut and to modulate IECs’ innate immune gene TLR-related gene expression and attenuate pathogen-induced TNF-α mRNA expression (115). Lactate, mainly produced by Lactobacillus, is the most abundant metabolite in the intestine during the first 6 months of life (116), and is a key factor in promoting ISC-mediated epithelial development. In detail, lactate directly stimulates PCs, and intestinal stromal cells through Gpr81 activation of the Wnt3–β-catenin pathway and plays an important role in promoting the proliferation of Lgr5+ ISCs (117). Furthermore, the effect of lactate on the proliferation of ISCs in newborn mice was greater than that in adult mice (117).
4.2 Bifidobacterium
Bifidobacterium is among the earliest and most dominant colonizers of the neonatal gut and is closely related to intestinal development. Colonization of the infant intestine with Bifidobacterium is influenced by human milk oligosaccharides (HMOs) from breastmilk. HMOs stimulate the growth of Bifidobacterium in the intestine, which offers a nutrient source for the infant intestinal microbiome, especially Bifidobacterium species that encode HMO-utilizing genes (118). Lack of Bifidobacterium and its breast milk oligosaccharide utilization genes is associated with systemic inflammation and immune imbalance in early life (119).
Aromatic lactic acids (ALAs) include indolelactic acid (ILA), phenyllactic acid (PLA), and 4-hydroxyphenyllactic acid (4-OH-PLA), which is a metabolite produced from the aromatic amino acids tryptophan, phenylalanine, and tyrosine by Bifidobacterium utilizing aromatic lactate dehydrogenase. ALAs are associated with the activation of aryl hydrocarbon receptors (AhRs) and regulate intestinal homeostasis. It has been demonstrated that ILA acts as an agonist for both the AhR and the hydroxycarboxylic acid receptor 3 (HCA3) and affects ex vivo immunological responses of human CD4+ T cells and monocytes in a dose-dependent way (120). Furthermore, ILA activates AhR and attenuates damaging pro-inflammatory responses after IL-1β stimulus and increases the expression of pBD-2 in IECs (121, 122). Transcriptomic analysis was used to demonstrate that immature enterocytes are protected by ILA in a variety of ways, including anti-inflammatory and antiviral activities. STAT1 pathways are critical for IL-1β-induced inflammatory response, and STAT1 gene transcription by ILA is essential for maintaining healthy intestinal development (123).
SCFAs are released by Bifidobacterium species metabolism of HMOs using intracellular glycosyl hydrolases and are by-products that can be used by the host (124). The SCFA concentration in the intestine sharply increases after birth (125). SCFAs are known to have anti-inflammatory properties in mature enterocytes and immune cells. Their role in the innate immunity of the developing intestine is not well understood. A study suggested that an immature intestinal mechanism for reducing inflammation differs from that of the mature intestine. SCFAs activate G-protein-coupled receptor 109A (GPR109A) in immature enterocytes in response to IL-1 stimulation, then inhibit IL-1-induced activation of HDAC3 and HDAC5 genes to reduce inflammatory responses (126). Furthermore, when exposed to an IL-1β inflammatory stimulus, acetate, propionate, and butyrate were all anti-inflammatory. Butyrate induces an anti-IL-1 response by increasing tight junction and mucus gene expression (127). Sodium butyrate administration significantly reduced the expression of HMGB1 and pro-inflammatory cytokines and decreased intestinal inflammation in the NEC model (128).
Bifidobacterium Tad pili may play a role in the early maturation of the naive gut by producing a unique scaffold of extracellular protein structures that stimulate the growth of the newborn mucosa (129). Furthermore, in human embryonic colon epithelial cells, Bifidobacterium regulates intestinal development by stimulating cell proliferation and accelerating the maturation of the internal barrier via the Wnt/β-catenin and PI3K/Akt/mTOR signaling pathways (130).
4.3 Escherichia
Aerobic conditions initially help in the rapid colonization of facultative anaerobic species in the neonatal intestine, such as E. coli. Colonization in the early stages with E. coli causes epithelial remodeling in the colon, changing the epithelium’s structure as well as the mucus layer. For example, proliferative cell markers (Ki67, proliferating cell nuclear antigen, phospho-histone H3, and cyclin A) were higher in rats non-colonized with E. coli than in GF, and following E. coli inoculation, epithelial MUC2 and mucopolysaccharide levels decreased rapidly, implying that these components are released to join the mucus layer, which doubled in thickness after inoculation (131). Furthermore, the intestinal microbiome composition of newborns was closely related to that of their mothers. Intestine epithelial cell differentiation is regulated by transmitting E. coli from the mother to the neonates, which increases the numbers of GCs (132). It has been demonstrated that E. coli EC 25 isolated from neonatal rats protects neonatal rats from NEC (133).
During days 0–10 of life, immune system exposure to luminal bacteria is limited by growth factors. Then, bacterial antigens are delivered to the LP by GAPs and captured as well as processed by lamina propria DCs (134). The antigens of E. coli can be taken up by DCs and then present to CD4+ T cells to induce Th17 (135).
The oxygen-rich environment in the intestinal tract during the neonatal period is the main factor affecting the colonization of bacteria and driving the excessive growth of K. pneumoniae. The oxygen content in the intestinal tract decreases gradually with the development, forming mature bacteria dominated by anaerobic microorganisms that can resist K. pneumoniae (107). In addition, commensal Enterobacteriaceae contribute to colonization resistance by competing for oxygen with S. enteritidis, which is necessary for pathogen expansion (136).
4.4 Bacteroides
Bacteroides species are found in varying amounts in the baby’s intestinal microbiome, but by the first year of life, they are always present (137). Indeed, Polysaccharide A, produced by Bacteroides fragilis, promotes PLZF+ lymphocyte homeostasis in the thymus through TLR2 signaling, while the colony mediates the migration of pDCs from the colon to the thymus to maintain PLZF+ lymphocyte homeostasis, and intervening intestinal flora in early life by antibiotic affects the distribution of PLZF+ cells in the adult thymus and then increases susceptibility to DSS-induced colitis (124).
4.5 Enterococcus
E. faecalis dominates more than 60% of the total LAB in the stool of healthy newborns and represents a more effective competitor than the rest in initiating colonization of a newborn (138). In vivo and in vitro, E. faecalis attenuates inflammation by suppressing proinflammatory cytokine secretions, especially IL-8 (139). Furthermore, peroxisome proliferator-activated receptor (PPAR) activity can be altered by E. faecalis through phosphorylation, resulting in increased DNA binding and transcriptional activation of IL-10 (134).
4.6 Others
Human breast milk-fed preterm infants are colonized by commensal Propionibacterium with high abundance, and the P.UF1 bacterium combines with SIGNR1 on the DCs to regulate macrophage in the intestine, which contributes to protect against pathogen infection and mitigate induced NEC-like injury (140).
It has been demonstrated that there is a lower level of Akkermansia muciniphila in oversized infants, although there was no direct evidence showing the crosstalk between Akkermansia and intestinal immunity in newborns (141, 142). A study on enterocytes implies that A. muciniphila may alleviate obesity by strengthening the epithelial barrier in infants (143).
The bacteria that colonize a newborn’s gut in the first few days after birth are all from the class Gammaproteobacteria, Gram-negative bacteria that express cell-surface LPS, which is recognized by the host intestinal cells via TLR4. ILC3 produces the cytokine IL-17A as a result of this signaling sequence. Upregulation of IL-17A in the gut causes G-CSF to recruit neutrophils from the bone marrow into the bloodstream, increasing circulating neutrophils that effectively remove blood-borne infections like E. coli K1 (144).
5 Ways to early intervene
Early life is a critical period for the development of the intestinal innate immune system, and increased exposure to environmental microorganisms during this period may improve immune function by regulating the symbiotic microbiome. For example, microbial exposure early in life influences immune maturation; it may reduce the risk of certain conditions later in life, such as colitis, by enabling the intestinal microbiome to develop properly during the early stages of immune development (9, 10).
5.1 Probiotics
Probiotics are defined as live microorganisms that produce health benefits and have been investigated by humans for centuries. Strong evidence supports the contribution of probiotics to maintaining intestinal mucosa homeostasis and the development of immunity in early life. Furthermore, the ability to resist DSS- and C. rodentium-induced intestinal injury and intestinal tumor formation in adulthood can be enhanced by early-life intervention (114, 145, 146). Hence, the effect of probiotics on early life has attracted many researchers.
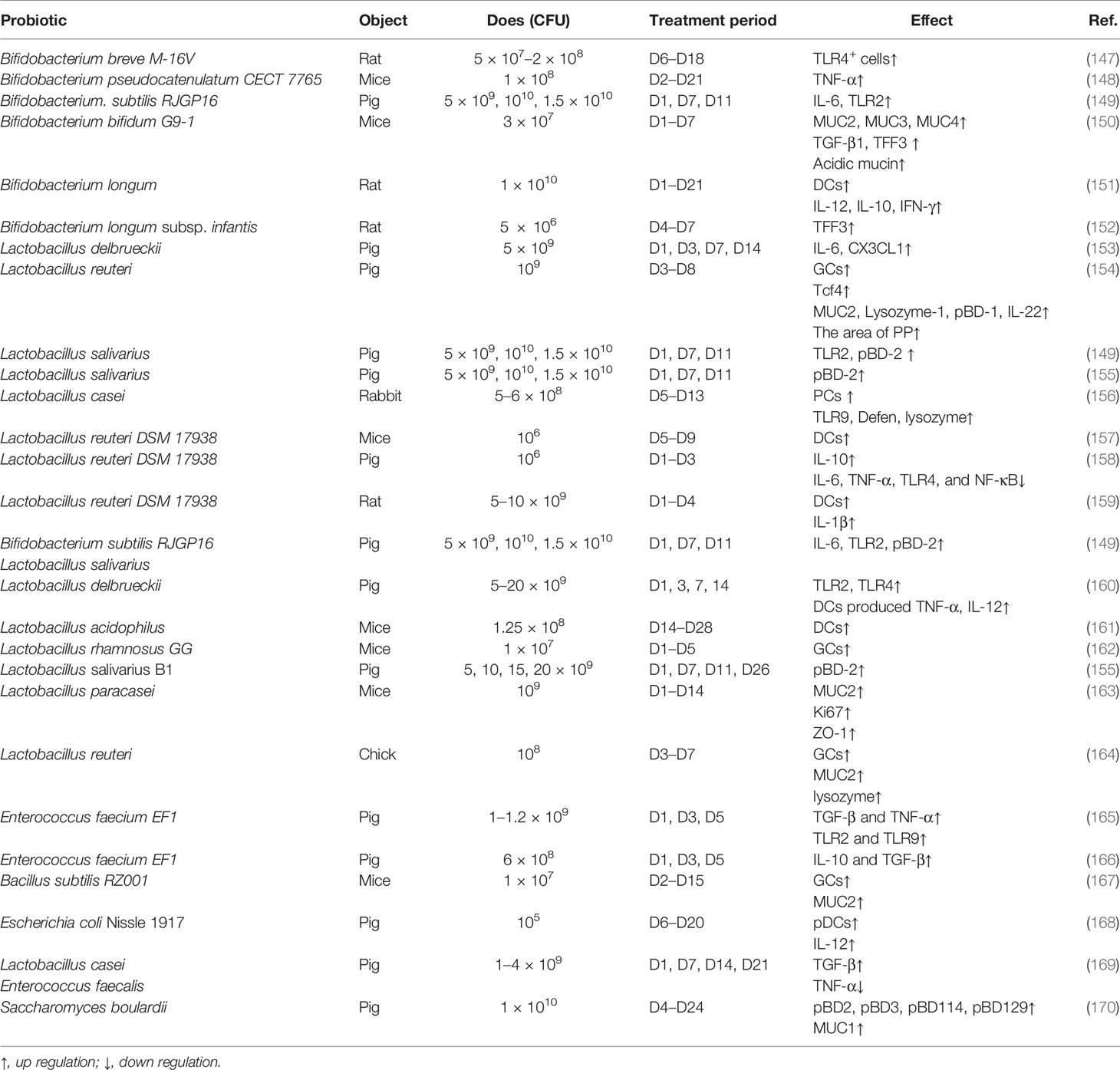
The impact of probiotics on intestinal innate immune development in early life is summarized in Table 1. As mentioned above, Lactobacillus and Bifidobacterium are milk-promoted bacteria in the neonatal intestine; thus, one of the early probiotics intervention strategies is to promote the colonization of these bacteria. Early administration of Bifidobacterium and Lactobacillus mainly enhanced the function of epithelial cells. The numbers and function of PCs and GCs are increased by supplementation with Bifidobacterium and Lactobacillus including the expression of TFF3, Lysozyme-1, β-defensin-1 (pBD-1), and pBD-2, and the secretion of MUC2, MUC3, and MUC4 (150, 154–156). The underlying mechanism may be through the bacteria-derived lactate (117). Furthermore, bacteria pilus was found to not only activate the TLR (115), and Bifidobacterium and Lactobacillus supplements in early life but also enhance the expression of TLR such as TLR2, TLR4, and TLR9. Moreover, early administration of Bifidobacterium enhances the secretion of TNF-α, IL-12, IL-10, IFN-γ, IL-6, CX3CL1, and IL-22. In addition, colonization of B. longum and Lactobacillus reuteri enhances the proliferation and the ability of antigen-presenting cells, which promote the development of adaptive immunity (151, 157, 159). It is worth noting that the co-supplementation of Bacillus subtilis and Lactobacillus salivarius stimulates a more significant mucosal immunity than the supplementation of each bacteria alone (149).
Moreover, other probiotics such as B. subtilis RZ001 and Enterococcus faecium EF1 also exert a regular function in innate immunity. Oral administration of B. subtilis RZ001 in suckling mice increases the number of PCs and the expression of MUC2 by promoting the proliferation of ISCs through Wnt signaling (167), while it is worth noticing that B. subtilis increased the relative abundance of lactate-producing bacteria, which may be the main reason for its effect. E. faecium EF1 supplementation in suckling pigs enhanced the innate immunity-related mRNA expression of toll-like receptors TLR2 and TLR9 and cytokines TGF-β and TNF-α (165).
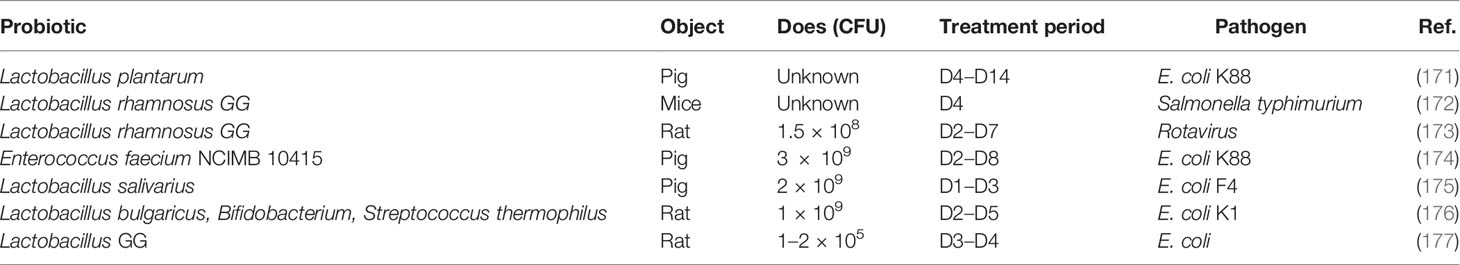
It has been demonstrated that early-life intervention by probiotics contributes to protecting against pathogen infection (Table 2). The defense against different kinds of invasive E. coli including K88, K1, and F4 was enhanced by supplanting probiotics by promoting the maturation of neonatal intestinal immune response and enhancing the mucosal barrier. For instance, co-administration of Lactobacillus bulgaricus, Bifidobacterium, and Streptococcus thermophilus promotes the production of mucin, ZO-1, and Ki67 in intestinal mucosa to prevent neonatal E. coli K1 translocation (176). Furthermore, LGG reverses the intestinal injury of neonatal mice against Salmonella challenge by increasing MUC2 and tight junction gene expression (172), and rotavirus infection can also be prevented by the early intervention of LGG (173).
A recent human intestinal mucosal transcriptome study assessing the effect of the administration of different probiotics has shown that changes in gene expression are more dependent on the individual than on the probiotic strain used (178); thus, the co-administration of probiotics is a good strategy to replace the single bacteria. Moreover, gene engineering strains are another useful strategy for early intervention. For example, a recombinant strain of L. reuteri secreting biologically active Lfampin (LR-LFCA) was developed; LR-LFCA enhances immune response and is beneficial for the intestinal health of neonatal piglets through improving intestinal barrier function and modulating the composition of gut (179).
Probiotics early intervention is increasingly being used in the clinic; a meta-analysis that included 10,812 infants demonstrated that early intervention with probiotics including Bifidobacterium spp., Lactobacillus spp., Saccharomyces spp., and Streptococcus spp. reduced the risk of NEC, late-onset invasive infection, and mortality in very-preterm or very-low-birth-weight infants (180). Moreover, the intervention that combined Bifidobacterium and Lacticaseibacillus accelerated microbial maturation such as α-diversity, stability, and interconnectivity and reduced the inflammatory response in extremely preterm infants (181). Probiotics are an effective strategy to prevent gastrointestinal disease in infants, and research showed that 14% of intensive care units administered probiotics to low-birth-weight premature babies between May and September 2015 in the USA (182). Clinical trials on probiotics aim to determine their safety and effectiveness, although their effect has been investigated in experimental animals such as mice, rats, and pigs.
5.2 FMT
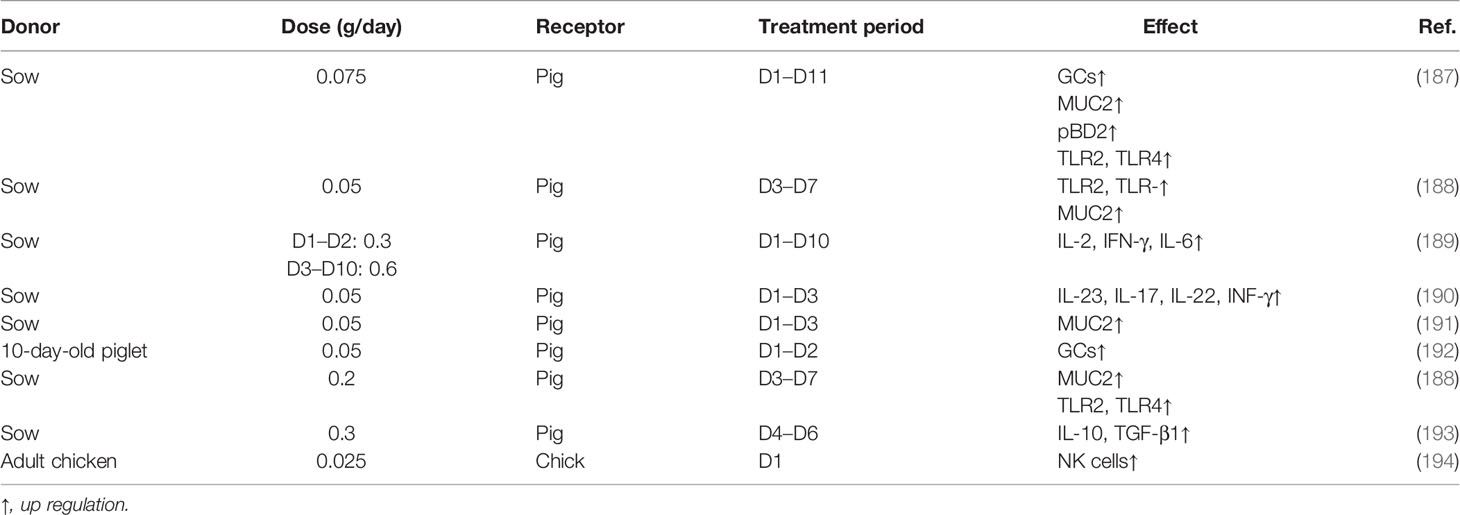
In recent years, as a strategy of intestinal microecological regulation apart from probiotics, FMT has become popular in recent years. FMT was mainly used to reverse the disorder gut microbiome ecosystem in Clostridioides difficile infection and inflammatory bowel disease (IBD) adulthood to achieve the therapeutic goal (183–185). In neonates, FMT not only promoted the diversity and composition of the intestine but also regulated the landscape of microbiota metabolites (186).
As shown in Table 3, there is little research on the effect of FMT in the development of intestinal innate immunity at present. FMT in the neonates promoted the development of GCs and the secretion of MUC2 (153, 187, 188, 192). Moreover, the expression of pBD-2, pBD-3, PMAP-23, and PAMP-37 by PCs was raised by FMT in the suckling piglet (187, 189). In addition, the expression of TLR such as TLR2 and TLR4, and the innate immune cytokines including IL-2, IL-6, IL-23, IL-17, IL-22, and INF-γ was enhanced after FMT (188–190), whereas the development and function of other types of innate immune cells including DCs, ILCs, NK, and macrophages were not been reported.
NEC is in part due to excessive inflammation in the immature intestine by colonizing bacteria because of an immature innate immune response (127). Evidence of a potential impact of FMT on reducing the risk of newborns with NEC has been shown. FMT before or after NEC induction can decrease the mucosa damage, inflammatory response (level of TNF-α, IL-1β, and IL-6), and oxidative damage [myeloperoxidase (MPO) activity] (195). Fecal filtrate transplantation (FFT) also prevented NEC in preterm neonates, and the orogastric administration was better than fecal filtrate transfer rectally (196).
Several studies have been reported on the FMT influence of pathogen infection. Compared with suckling pigs without FMT, FMT alleviated enterotoxigenic E. coli (ETEC) K88 infection caused by mucosa injury including the decrease in the number of GCs and the production of MUC2 (197). Lipopolysaccharide (LPS) is the major component of the outer membrane of Gram-negative bacteria, such as PAMPs, and LPS induces the damage of the mucosa integrity. FMT promoted the microbiome-derived indole acetic acid (IAA) from tryptophan, which maintains the intestinal barrier by the induction of IL-22, thus reducing susceptibility to LPS-induced destruction of epithelial integrity and severe inflammatory responses (186).
Of note, colonization of microbiome from different intestinal segments was region-specific, and compared with FMT, whole-intestinal microbiota transplantation (WIMT) is more conducive to the transplantation of the donor intestinal microbiome and its function into the recipient small intestine (198). However, it is worth noticing that not just any microbiome from any animal can promote immune system development and maturation in mice; the intestine immune system only depends on colonizing with a host-specific microbiome. For example, the number of GF mice’s T cells could only be reconstructed by mouse microbiome instead of human or rat microbiome (199).
6 Conclusion and perspectives
Intestinal innate immunity is one of the first defense lines of the host. The intestinal microbiome plays a crucial role in the development of the host’s immune system in early life. Here, we summarize how the intestine microbiome directs the development of the intestinal innate immune system composed of the epithelium and immune cells from early life. In neonates, the intestinal innate immunity is shaped by the milk-derived bacteria Lactobacillus and Bifidobacterium, and other commensal bacteria including Escherichia, Bacteroides, and Enterococcus. Early intervention by probiotics and FMT are two effective ways to modulate intestinal microecology, promote the development of intestinal immunity, and maintain intestinal mucosa homeostasis. Nevertheless, the impact of probiotics and FMT on other immune cells such as ILCs is not yet researched. Furthermore, a single probiotic or FMT may not be the best strategy for early intervention, and FMT carries the risk of facing an infectious disease because of the harmful strains (200). More focused efforts to uncover a combination of multiple bacteria can be developed to enhance neonatal intestinal health. For instance, the immune system was enhanced by colonizing the synthetic community (SYN) in the cecum, which is composed of 9 different commensal bacteria from chicken (201). Thus, the research on core bacteria and the combination of multiple bacteria is the trend for early intervention in the future. Moreover, clinical tools including sequence style and bioinformatics to detect and predict the intestinal microbiome in infants are worthy of further research. For example, bioinformatics accelerates the pharmaceutical industry (202). qPCR was used to detect the specific bacteria that correlated to disease or intestinal health in the clinic (203). Moreover, the occurrence of NGS and metagenomic sequencing of the microbiome helps us investigate the microbial composition deeply (204), and the different methods of analysis such as via a co-occurrence network, functional annotation, and random forest (181, 205) help us predict the maturation of the intestinal microbiome and exploit new probiotics for infants.
Author Contributions
ZY designed and wrote the manuscript. XL completed the table. YW drew the figure. HW and JP designed the manuscript and provided funding and modified the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Hubei Province Science and Technology Innovation Major Project (2021BBA083) and China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gentile LF, Nacionales DC, Lopez MC, Vanzant E, Cuenca A, Cuenca AG, et al. Protective Immunity and Defects in the Neonatal and Elderly Immune Response to Sepsis. J Immunol (2014) 192:3156–65. doi: 10.4049/jimmunol.1301726
2. Wheeler DS, Wong HR, Zingarelli B. Pediatric Sepsis - Part I: "Children are Not Small Adults!". Open Inflamm J (2011) 4:4–15. doi: 10.2174/1875041901104010004
3. Wynn JL, Levy O. Role of Innate Host Defenses in Susceptibility to Early-Onset Neonatal Sepsis. Clin Perinatol (2010) 37:307–37. doi: 10.1016/j.clp.2010.04.001
4. Marshall JS, Warrington R, Watson W, Kim HL. An Introduction to Immunology and Immunopathology. Allergy Asthma CL IM (2018) 14:49. doi: 10.1186/s13223-018-0278-1
5. Chassaing B, Kumar M, Baker MT, Singh V, Vijay-Kumar M. Mammalian Gut Immunity. BioMed J (2014) 37:246–58. doi: 10.4103/2319-4170.130922
6. Lazar V, Ditu L-M, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol (2018) 9:1830. doi: 10.3389/fimmu.2018.01830
7. Rabe H, Lundell A-C, Sjoberg F, Ljung A, Strombeck A, Gio-Batta M, et al. Neonatal Gut Colonization by Bifidobacterium is Associated With Higher Childhood Cytokine Responses. Gut Microbes (2020) 12:1–14. doi: 10.1080/19490976.2020.1847628
8. Schmidt TSB, Raes J, Bork P. The Human Gut Microbiome: From Association to Modulation. Cell (2018) 172:1198–215. doi: 10.1016/j.cell.2018.02.044
9. Roslund MI, Puhakka R, Grönroos M, Nurminen N, Oikarinen S, Gazali AM, et al. Biodiversity Intervention Enhances Immune Regulation and Health-Associated Commensal Microbiota Among Daycare Children. Sci Adv (2020) 6:eaba2578. doi: 10.1126/sciadv.aba2578
10. Miyoshi J, Miyoshi S, Delmont TO, Cham C, Lee STM, Sakatani A, et al. Early-Life Microbial Restitution Reduces Colitis Risk Promoted by Antibiotic-Induced Gut Dysbiosis in Interleukin 10(-/-) Mice. Gastroenterology (2021) 161:940–52.e15. doi: 10.1053/j.gastro.2021.05.054
11. Guo FL, Cai DM, Li YW, Gu HT, Qu H, Zong QF, et al. How Early-Life Gut Microbiota Alteration Sets Trajectories for Health and Inflammatory Bowel Disease? Front Nutr (2021) 8:690073, 1–7. doi: 10.3389/fnut.2021.690073
12. Fuentes S, de Vos WM. How to Manipulate the Microbiota: Fecal Microbiota Transplantation. Microbiota Hum Body: Implications Health Dis (2016) 902:143–53. doi: 10.1080/19490976.2017.1290757
13. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How Colonization by Microbiota in Early Life Shapes the Immune System. Science (2016) 352:539–44. doi: 10.1126/science.aad9378
14. Parigi SM, Eldh M, Larssen P, Gabrielsson S, Villablanca EJ. Breast Milk and Solid Food Shaping Intestinal Immunity. Front Immunol (2015) 6:415. doi: 10.3389/fimmu.2015.00415
15. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med (2014) 6:237ra65–ra65. doi: 10.1126/scitranslmed.3008599
16. Leiby JS, McCormick K, Sherrill-Mix S, Clarke EL, Kessler LR, Taylor LJ, et al. Lack of Detection of a Human Placenta Microbiome in Samples From Preterm and Term Deliveries. Microbiome (2018) 6:1–11. doi: 10.1186/s40168-018-0575-4
17. Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, et al. Does the Human Placenta Delivered at Term Have a Microbiota? Results of Cultivation, Quantitative Real-Time PCR, 16s rRNA Gene Sequencing, and Metagenomics. Am J Obstet Gynecol (2019) 220:267. e1–. e39. doi: 10.1016/j.ajog.2018.10.018
18. Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of Placenta Samples With Contamination Controls Does Not Provide Evidence for a Distinct Placenta Microbiota. Microbiome (2016) 4:1–11. doi: 10.1186/s40168-016-0172-3
19. Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, et al. Microbial Exposure During Early Human Development Primes Fetal Immune Cells. Cell (2021) 184:3394–409. doi: 10.1016/j.cell.2021.04.039
20. Al Alam D, Danopoulos S, Grubbs B, Ali N, MacAogain M, Chotirmall SH, et al. Human Fetal Lungs Harbor a Microbiome Signature. Am J Respir (2020) 201:1002–6. doi: 10.1164/rccm.201911-2127LE
21. Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of Commensal Bacteria From Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr Microbiol (2005) 51:270–4. doi: 10.1007/s00284-005-0020-3
22. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable Bacterial Colonization is Highly Limited in the Human Intestine In Utero. Nat Med (2020) 26:599–607. doi: 10.1038/s41591-020-0761-3
23. Bi Y, Tu Y, Zhang N, Wang S, Zhang F, Suen G, et al. Multiomics Analysis Reveals the Presence of a Microbiome in the Gut of Fetal Lambs. Gut (2021) 70:853–64. doi: 10.1136/gutjnl-2020-320951
24. Husso A, Lietaer L, Pessa-Morikawa T, Gronthal T, Govaere J, Van Soom A, et al. The Composition of the Microbiota in the Full-Term Fetal Gut and Amniotic Fluid: A Bovine Cesarean Section Study. Front Microbiol (2021) 12:626421. doi: 10.3389/fmicb.2021.626421
25. Li Y, Toothaker JM, Ben-Simon S, Ozeri L, Schweitzer R, McCourt BT, et al. In Utero Human Intestine Harbors Unique Metabolome, Including Bacteria Metabolites. JCI Insight (2020) 5:e138751. doi: 10.1172/jci.insight.138751
26. Van Best N, Rolle-Kampczyk U, Schaap F, Basic M, Damink SO, Bleich A, et al. Bile Acids Drive the Newborn’s Gut Microbiota Maturation. Nat Commun (2020) 11:1–13. doi: 10.1038/s41467-020-17183-8
27. Yang Y, Liu Y, Liu J, Wang H, Guo Y, Du M, et al. Composition of the Fecal Microbiota of Piglets at Various Growth Stages. Front Vet Sci (2021) 8:661671. doi: 10.3389/fvets.2021.661671
28. Xiao L, Wang J, Zheng J, Li X, Zhao F. Deterministic Transition of Enterotypes Shapes the Infant Gut Microbiome at an Early Age. Genome Biol (2021) 22:1–21. doi: 10.1186/s13059-021-02463-3
29. Wang J, Dominguez-Bello MG. Microbial Colonization Alters Neonatal Gut Metabolome. Nat Microbiol (2020) 5:785–6. doi: 10.1038/s41564-020-0734-9
30. Bittinger K, Zhao C, Li Y, Ford E, Friedman ES, Ni J, et al. Bacterial Colonization Reprograms the Neonatal Gut Metabolome. Nat Microbiol (2020) 5:838–47. doi: 10.1038/s41564-020-0694-0
31. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol Mol Biol Rev (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
32. Kosenkova TV, Novikova VP, Boytsova EA, Zazerskaya IE, Lavrova OV, Gurina OP, et al. The Effect of the Course of Pregnancy on Vaginal Microbiocenosis and Development of Atopia in Children Born to Mothers With Bronchial Asthma. Nutrition (2020) 10:15–23. doi: 10.20953/2224-5448-2020-4-15-23
33. Khine WWT, Rahayu ES, See TY, Kuah S, Salminen S, Nakayama J, et al. Indonesian Children Fecal Microbiome From Birth Until Weaning was Different From Microbiomes of Their Mothers. Gut Microbes (2020) 12:1761240. doi: 10.1080/19490976.2020.1761240
34. Dougherty MW, Kudin O, Muhlbauer M, Neu J, Gharaibeh RZ, Jobin C. Gut Microbiota Maturation During Early Human Life Induces Enterocyte Proliferation via Microbial Metabolites. BMC Microbiol (2020) 20:1–14. doi: 10.1186/s12866-020-01892-7
35. Thaiss CA, Zmora N, Levy M, Elinav E. The Microbiome and Innate Immunity. Nature (2016) 535:65–74. doi: 10.1038/nature18847
36. Bevins CL, Salzman NH. Paneth Cells, Antimicrobial Peptides and Maintenance of Intestinal Homeostasis. Nat Rev Microbiol (2011) 9:356–68. doi: 10.1038/nrmicro2546
37. Martinez Rodriguez NR, Eloi MD, Huynh A, Dominguez T, Lam AHC, Carcamo-Molina D, et al. Expansion of Paneth Cell Population in Response to Enteric Salmonella Enterica Serovar Typhimurium Infection. Infect Immun (2012) 80:266–75. doi: 10.1128/IAI.05638-11
38. Kelleher SL, Alam S, Rivera OC, Barber-Zucker S, Zarivach R, Wagatsuma T, et al. Loss-Of-Function SLC30A2 Mutants are Associated With Gut Dysbiosis and Alterations in Intestinal Gene Expression in Preterm Infants. Gut Microbes (2022) 14:2014739. doi: 10.1080/19490976.2021.2014739
39. Lueschow SR, Stumphy J, Gong H, Kern SL, Elgin TG, Underwood MA, et al. Loss of Murine Paneth Cell Function Alters the Immature Intestinal Microbiome and Mimics Changes Seen in Neonatal Necrotizing Enterocolitis. PloS One (2018) 13:e0204967. doi: 10.1371/journal.pone.0204967
40. Quintero M, Liu S, Xia Y, Huang Y, Zou Y, Li G, et al. Cdk5rap3 is Essential for Intestinal Paneth Cell Development and Maintenance. Cell Death Dis (2021) 12:1–14. doi: 10.1038/s41419-021-03401-8
41. Rumbo M, Schiffrin EJ. Ontogeny of Intestinal Epithelium Immune Functions: Developmental and Environmental Regulation. Cell Mol Life Sci (2005) 62:1288–96. doi: 10.1007/s00018-005-5033-3
42. Heida FH, Beyduz G, Bulthuis MLC, Kooi EMW, Bos AF, Timmer A, et al. Paneth Cells in the Developing Gut: When do They Arise and When are They Immune Competent? Pediatr Res (2016) 80:306–10. doi: 10.1038/pr.2016.67
43. Bry L, Falk P, Huttner K, Ouellette A, Midtvedt T, Gordon JI. Paneth Cell Differentiation in the Developing Intestine of Normal and Transgenic Mice. P Natl Acad Sci USA (1994) 91:10335–9. doi: 10.1073/pnas.91.22.10335
44. Underwood MA, Kananurak A, Coursodon CF, Adkins-Reick CK, Chu H, Bennett SH, et al. Bifidobacterium Bifidum in a Rat Model of Necrotizing Enterocolitis: Antimicrobial Peptide and Protein Responses. Pediatr Res (2012) 71:546–51. doi: 10.1038/pr.2012.11
45. Inoue R, Tsuruta T, Nojima I, Nakayama K, Tsukahara T, Yajima T. Postnatal Changes in the Expression of Genes for Cryptdins 1-6 and the Role of Luminal Bacteria in Cryptdin Gene Expression in Mouse Small Intestine. FEMS Immunol Med Microbiol (2008) 52:407–16. doi: 10.1111/j.1574-695X.2008.00390.x
46. Burger-van Paassen N, Loonen LMP, Witte-Bouma J, Korteland-van Male AM, de Bruijn ACJM, van der Sluis M, et al. Mucin Muc2 Deficiency and Weaning Influences the Expression of the Innate Defense Genes Reg3 Beta, Reg3 Gamma and Angiogenin-4. PloS One (2012) 7:e38798. doi: 10.1371/journal.pone.0038798
47. Park CJ, Shaughnessy MP, Cowles RA. Mucosal Characteristics Vary Across Developmental Stages in the Small Intestine of C57BL/6J Mice. Life Sci (2020) 260:118428. doi: 10.1016/j.lfs.2020.118428
48. Schoenborn AA, von Furstenberg R, Valsaraj S, Hussain FS, Stein M, Shanahan M, et al. The Enteric Microbiota Regulates Paneth Cell Number and Function Without Affecting Intestinal Stem Cells. Gastroenterology (2017) 152:S13–S. doi: 10.1016/s0016-5085(17)30418-3
49. Schumann A, Nutten S, Donnicola D, Comelli EM, Mansourian R, Cherbut C, et al. Neonatal Antibiotic Treatment Alters Gastrointestinal Tract Developmental Gene Expression and Intestinal Barrier Transcriptome. Physiol Genomics (2005) 23:235–45. doi: 10.1152/physiolgenomics.00057.2005
50. Yu YY, Lu L, Sun J, Petrof EO, Claud EC. Preterm Infant Gut Microbiota Affects Intestinal Epithelial Development in a Humanized Microbiome Gnotobiotic Mouse Model. Am J Physiol Gastrointest (2016) 311:G521–G32. doi: 10.1152/ajpgi.00022.2016
51. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science (2006) 313:1126–30. doi: 10.1126/science.1127119
52. Pelaseyed T, Bergstrom JH, Gustafsson JK, Ermund A, Birchenough GMH, Schutte A, et al. The Mucus and Mucins of the Goblet Cells and Enterocytes Provide the First Defense Line of the Gastrointestinal Tract and Interact With the Immune System. Immunol Rev (2014) 260:8–20. doi: 10.1111/imr.12182
53. Johansson ME, Larsson JM, Hansson GC. The Two Mucus Layers of Colon are Organized by the MUC2 Mucin, Whereas the Outer Layer is a Legislator of Host-Microbial Interactions. Proc Natl Acad Sci USA (2011) 108 Suppl 1:4659–65. doi: 10.1073/pnas.1006451107
54. McCarthy AJ, Birchenough GMH, Taylor PW. Loss of Trefoil Factor 2 Sensitizes Rat Pups to Systemic Infection With the Neonatal Pathogen Escherichia Coli K1. Infect Immun (2019) 87:e00878–18. doi: 10.1128/iai.00878-18
55. van der Flier LG, Clevers H. Stem Cells, Self-Renewal, and Differentiation in the Intestinal Epithelium. Annu Rev Physiol (2009) 71:241–60. doi: 10.1146/annurev.physiol.010908.163145
56. Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial Sensing by Goblet Cells Controls Immune Surveillance of Luminal Antigens in the Colon. Mucosal Immunol (2015) 8:198–210. doi: 10.1038/mi.2014.58
57. Kim YS, Ho SB. Intestinal Goblet Cells and Mucins in Health and Disease: Recent Insights and Progress. Curr Gastroenterol Rep (2010) 12:319–30. doi: 10.1007/s11894-010-0131-2
58. Buisine MP, Devisme L, Savidge TC, Gespach C, Gosselin B, Porchet N, et al. Mucin Gene Expression in Human Embryonic and Fetal Intestine. Gut (1998) 43:519–24. doi: 10.1136/gut.43.4.519
59. Chin AM, Hill DR, Aurora M, Spence JR. Morphogenesis and Maturation of the Embryonic and Postnatal Intestine. Semin Cell Dev Biol (2017) 66:81–93. doi: 10.1016/j.semcdb.2017.01.011
60. Yuan C, Zhang P, Jin Y, Ullah Shah A, Zhang E, Yang Q. Single-Blinded Study Highlighting the Differences Between the Small Intestines of Neonatal and Weaned Piglets. Animals (2021) 11:271. doi: 10.3390/ani11020271
61. Kandori H, Hirayama K, Takeda M, Doi K. Histochemical, Lectin-Histochemical and Morphometrical Characteristics of Intestinal Goblet Cells of Germfree and Conventional Mice. Exp Anim TOKYO (1996) 45:155–60. doi: 10.1538/expanim.45.155
62. Allaire JM, Morampudi V, Crowley SM, Stahl M, Yu H, Bhullar K, et al. Frontline Defenders: Goblet Cell Mediators Dictate Host-Microbe Interactions in the Intestinal Tract During Health and Disease. Am J Physiol Gastrointest Liver Physiol (2018) 314:G360–g77. doi: 10.1152/ajpgi.00181.2017
63. Tomas J, Wrzosek L, Bouznad N, Bouet S, Mayeur C, Noordine M-L, et al. Primocolonization is Associated With Colonic Epithelial Maturation During Conventionalization. FASEB J (2013) 27:645–55. doi: 10.1096/fj.12-216861
64. Chirdo FG, Millington OR, Beacock-Sharp H, Mowat AM. Immunomodulatory Dendritic Cells in Intestinal Lamina Propria. Eur J Immunol (2005) 35:1831–40. doi: 10.1002/eji.200425882
65. Kelsall BL, Strober W. Distinct Populations of Dendritic Cells are Present in the Subepithelial Dome and T Cell Regions of the Murine Peyer's Patch. J Exp Med (1996) 183:237–47. doi: 10.1084/jem.183.1.237
66. Veenbergen S, van Berkel LA, du Pré MF, He J, Karrich JJ, Costes LM, et al. Colonic Tolerance Develops in the Iliac Lymph Nodes and can be Established Independent of CD103(+) Dendritic Cells. Mucosal Immunol (2016) 9:894–906. doi: 10.1038/mi.2015.118
67. Lantier L, Lacroix-Lamande S, Potiron L, Metton C, Drouet F, Guesdon W, et al. Intestinal CD103+Dendritic Cells Are Key Players in the Innate Immune Control of Cryptosporidium Parvum Infection in Neonatal Mice. PloS Pathog (2013) 9:e1003801. doi: 10.1371/journal.ppat.1003801
68. Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, et al. Comparative Transcriptional and Functional Profiling Defines Conserved Programs of Intestinal DC Differentiation in Humans and Mice. Nat Immunol (2014) 15:98–108. doi: 10.1038/ni.2768
69. Min YW, Rhee PL. The Role of Microbiota on the Gut Immunology. Clin Ther (2015) 37:968–75. doi: 10.1016/j.clinthera.2015.03.009
70. Sun T, Rojas OL, Li C, Ward LA, Philpott DJ, Gommerman JL. Intestinal Batf3-Dependent Dendritic Cells are Required for Optimal Antiviral T-Cell Responses in Adult and Neonatal Mice. Mucosal Immunol (2017) 10:775–88. doi: 10.1038/mi.2016.79
71. Pezoldt J, Pasztoi M, Zou M, Wiechers C, Beckstette M, Thierry GR, et al. Neonatally Imprinted Stromal Cell Subsets Induce Tolerogenic Dendritic Cells in Mesenteric Lymph Nodes. Nat Commun (2018) 9:1–14. doi: 10.1038/s41467-018-06423-7
72. Williams AM, Probert CSJ, Stepankova R, Tlaskalova-Hogenova H, Phillips A, Bland PW. Effects of Microflora on the Neonatal Development of Gut Mucosal T Cells and Myeloid Cells in the Mouse. Immunol (2006) 119:470–8. doi: 10.1111/j.1365-2567.2006.02458.x
73. Ekmekciu I, von Klitzing E, Neumann C, Bacher P, Scheffold A, Bereswill S, et al. Fecal Microbiota Transplantation, Commensal Escherichia Coli and Lactobacillus Johnsonii Strains Differentially Restore Intestinal and Systemic Adaptive Immune Cell Populations Following Broad-Spectrum Antibiotic Treatment. Front Microbiol (2017) 8:2430. doi: 10.3389/fmicb.2017.02430
74. Ekmekciu I, von Klitzing E, Fiebiger U, Escher U, Neumann C, Bacher P, et al. Immune Responses to Broad-Spectrum Antibiotic Treatment and Fecal Microbiota Transplantation in Mice. Front Immunol (2017) 8:397. doi: 10.3389/fimmu.2017.00397
75. Stagg AJ, Norin KE, Midtvedt T, Kamm MA, Knight SC, Bjoerksten B. Mesenteric Dendritic Cells From Germ-Free Mice Cause Less T-Cell Stimulation But Still Induce Alpha 4P7 Integrin. Microb Ecol Health Dis (2007) 19:171–83. doi: 10.1080/08910600701474263
76. Zook EC, Kee BL. Development of Innate Lymphoid Cells. Nat Immunol (2016) 17:775–82. doi: 10.1038/ni.3481
78. Liu C, Gong Y, Zhang H, Yang H, Zeng Y, Bian Z, et al. Delineating Spatiotemporal and Hierarchical Development of Human Fetal Innate Lymphoid Cells. Cell Res (2021) 31:1106–22. doi: 10.1038/s41422-021-00529-2
79. Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, et al. Innate and Adaptive Lymphocytes Sequentially Shape the Gut Microbiota and Lipid Metabolism. Nature (2018) 554:255–9. doi: 10.1038/nature25437
80. Qi X, Qiu J, Chang J, Ji Y, Yang Q, Cui G, et al. Brg1 Restrains the Pro-Inflammatory Properties of ILC3s and Modulates Intestinal Immunity. Mucosal Immunol (2021) 14:38–52. doi: 10.1038/s41385-020-0317-3
81. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and Commensal Microflora are Required for the Differentiation of Mucosal Interleukin 22-Producing NKp46+ Cells. Nat Immunol (2009) 10:83–91. doi: 10.1038/ni.1684
82. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial Flora Drives Interleukin 22 Production in Intestinal NKp46+ Cells That Provide Innate Mucosal Immune Defense. Immunity (2008) 29:958–70. doi: 10.1016/j.immuni.2008.11.001
83. Niu X, Daniel S, Kumar D, Ding EY, Savani RC, Koh AY, et al. Transient Neonatal Antibiotic Exposure Increases Susceptibility to Late-Onset Sepsis Driven by Microbiota-Dependent Suppression of Type 3 Innate Lymphoid Cells. Sci Rep (2020) 10:1–10. doi: 10.1038/s41598-020-69797-z
84. Naper C, Shegarfi H, Inngjerdingen M, Rolstad B. The Role of Natural Killer Cells in the Defense Against Listeria Monocytogenes Lessons From a Rat Model. J Innate Immun (2011) 3:289–97. doi: 10.1159/000324143
85. Ashkar AA, Reid S, Verdu EF, Zhang K, Coombes BK. Interleukin-15 and NK1.1+ Cells Provide Innate Protection Against Acute Salmonella Enterica Serovar Typhimurium Infection in the Gut and in Systemic Tissues. Infect Immun (2009) 77:214–22. doi: 10.1128/iai.01066-08
86. Hall LJ, Murphy CT, Hurley G, Quinlan A, Shanahan F, Nally K, et al. Natural Killer Cells Protect Against Mucosal and Systemic Infection With the Enteric Pathogen Citrobacter Rodentium. Infect Immun (2013) 81:460–9. doi: 10.1128/iai.00953-12
87. Li N, van Unen V, Höllt T, Thompson A, van Bergen J, Pezzotti N, et al. Mass Cytometry Reveals Innate Lymphoid Cell Differentiation Pathways in the Human Fetal Intestine. J Exp Med (2018) 215:1383–96. doi: 10.1084/jem.20171934
88. Pérez-Cano FJ, Castellote C, González-Castro AM, Pelegrí C, Castell M, Franch A. Developmental Changes in Intraepithelial T Lymphocytes and NK Cells in the Small Intestine of Neonatal Rats. Pediatr Res (2005) 58:885–91. doi: 10.1203/01.pdr.0000182187.88505.49
89. Marín-Gallén S, Pérez-Cano FJ, Castell M, Castellote C, Franch A. Intestinal Intraepithelial NK and NKT Cell Ontogeny in Lewis Rats. Dev Comp Immunol (2008) 32:1405–8. doi: 10.1016/j.dci.2008.06.011
90. Sagebiel AF, Steinert F, Lunemann S, Körner C, Schreurs RR, Altfeld M, et al. Tissue-Resident Eomes+ NK Cells are the Major Innate Lymphoid Cell Population in Human Infant Intestine. Nat Commun (2019) 10:1–13. doi: 10.1038/s41467-018-08267-7
91. Poupeau A, Garde C, Sulek K, Citirikkaya K, Treebak JT, Arumugam M, et al. Genes Controlling the Activation of Natural Killer Lymphocytes are Epigenetically Remodeled in Intestinal Cells From Germ-Free Mice. FASEB J (2019) 33:2719–31. doi: 10.1096/fj.201800787R
92. Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, Legrand F, et al. Inflammatory Monocytes Regulate Pathologic Responses to Commensals During Acute Gastrointestinal Infection. Nat Med (2013) 19:713–21. doi: 10.1038/nm.3189
93. Kim M, Galan C, Hill AA, Wu WJ, Fehlner-Peach H, Song HW, et al. Critical Role for the Microbiota in CX(3)CR1(+) Intestinal Mononuclear Phagocyte Regulation of Intestinal T Cell Responses. Immunity (2018) 49:151–63.e5. doi: 10.1016/j.immuni.2018.05.009
94. Shaw MH, Kamada N, Kim YG, Núñez G. Microbiota-Induced IL-1β, But Not IL-6, is Critical for the Development of Steady-State TH17 Cells in the Intestine. J Exp Med (2012) 209:251–8. doi: 10.1084/jem.20111703
95. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-Dependent Crosstalk Between Macrophages and ILC3 Promotes Intestinal Homeostasis. Science (2014) 343:1249288. doi: 10.1126/science.1249288
96. Maheshwari A, Kurundkar AR, Shaik SS, Kelly DR, Hartman Y, Zhang W, et al. Epithelial Cells in Fetal Intestine Produce Chemerin to Recruit Macrophages. Am J Physiol Gastrointest Liver Physiol (2009) 297:G1–g10. doi: 10.1152/ajpgi.90730.2008
97. Bain CC, Bravo-Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, et al. Constant Replenishment From Circulating Monocytes Maintains the Macrophage Pool in the Intestine of Adult Mice. Nat Immunol (2014) 15:929–37. doi: 10.1038/ni.2967
98. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity (2012) 37:158–70. doi: 10.1016/j.immuni.2012.04.011
99. Köhler A, Delbauve S, Smout J, Torres D, Flamand V. Very Early-Life Exposure to Microbiota-Induced TNF Drives the Maturation of Neonatal Pre-Cdc1. Gut (2021) 70:511–21. doi: 10.1136/gutjnl-2019-319700
100. Schokker D, Jansman AJM, Veninga G, de Bruin N, Vastenhouw SA, de Bree FM, et al. Perturbation of Microbiota in One-Day Old Broiler Chickens With Antibiotic for 24 Hours Negatively Affects Intestinal Immune Development. BMC Genom (2017) 18:1–14. doi: 10.1186/s12864-017-3625-6
101. Amenyogbe N, Dimitriu P, Smolen KK, Brown EM, Shannon CP, Tebbutt SJ, et al. Biogeography of the Relationship Between the Child Gut Microbiome and Innate Immune System. Mbio (2021) 12:e03079–20. doi: 10.1128/mBio.03079-20
102. Johansson MA, Saghafian-Hedengren S, Haileselassie Y, Roos S, Troye-Blomberg M, Nilsson C, et al. Early-Life Gut Bacteria Associate With IL-4-, IL-10-and IFN-Gamma Production at Two Years of Age. PloS One (2012) 7:e49315. doi: 10.1371/journal.pone.0049315
103. Selma-Royo M, Calatayud Arroyo M, Garcia-Mantrana I, Parra-Llorca A, Escuriet R, Martinez-Costa C, et al. Perinatal Environment Shapes Microbiota Colonization and Infant Growth: Impact on Host Response and Intestinal Function. Microbiome (2020) 8:1–19. doi: 10.1186/s40168-020-00940-8
104. van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin J-B, et al. Intestinal Fungi are Causally Implicated in Microbiome Assembly and Immune Development in Mice. Nat Commun (2020) 11:1–16. doi: 10.1038/s41467-020-16431-1
105. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The Maternal Microbiota Drives Early Postnatal Innate Immune Development. Science (2016) 351:1296–302. doi: 10.1126/science.aad2571
106. Witkowska-Zimny M, Kaminska-El-Hassan E. Cells of Human Breast Milk. Cell Mol Biol Lett (2017) 22:1–11. doi: 10.1186/s11658-017-0042-4
107. Singer JR, Blosser EG, Zindl CL, Silberger DJ, Conlan S, Laufer VA, et al. Preventing Dysbiosis of the Neonatal Mouse Intestinal Microbiome Protects Against Late-Onset Sepsis. Nat Med (2019) 25:1772–82. doi: 10.1038/s41591-019-0640-y
108. Isani M, Bell BA, Delaplain PT, Bowling JD, Golden JM, Elizee M, et al. Lactobacillus Murinus HF12 Colonizes Neonatal Gut and Protects Rats From Necrotizing Enterocolitis. PloS One (2018) 13:e0196710. doi: 10.1371/journal.pone.0196710
109. Haileselassie Y, Navis M, Vu N, Qazi KR, Rethi B, Sverremark-Ekstrom E. Lactobacillus Reuteri and Staphylococcus Aureus Differentially Influence the Generation of Monocyte-Derived Dendritic Cells and Subsequent Autologous T Cell Responses. Immun Inflammation Dis (2016) 4:315–26. doi: 10.1002/iid3.115
110. Fink LN, Zeuthen LH, Christensen HR, Morandi B, Frøkiaer H, Ferlazzo G. Distinct Gut-Derived Lactic Acid Bacteria Elicit Divergent Dendritic Cell-Mediated NK Cell Responses. Int Immunol (2007) 19:1319–27. doi: 10.1093/intimm/dxm103
111. Rizzello V, Bonaccorsi I, Dongarrà ML, Fink LN, Ferlazzo G. Role of Natural Killer and Dendritic Cell Crosstalk in Immunomodulation by Commensal Bacteria Probiotics. J BioMed Biotechnol (2011) 2011:473097. doi: 10.1155/2011/473097
112. Bermudez-Brito M, Munoz-Quezada S, Gomez-Llorente C, Matencio E, Bernal MJ, Romero F, et al. Human Intestinal Dendritic Cells Decrease Cytokine Release Against Salmonella Infection in the Presence of Lactobacillus Paracasei CNCM I-4034, a Novel Strain Isolated From Breast-Fed Newborns. Proc Nutr Soc (2013) 72:E59–E. doi: 10.1017/s002966511300061x
113. Trapecar M, Goropevsek A, Gorenjak M, Gradisnik L, Rupnik MS. A Co-Culture Model of the Developing Small Intestine Offers New Insight in the Early Immunomodulation of Enterocytes and Macrophages by Lactobacillus Spp. Through STAT1 and NF-kB P65 Translocation. PloS One (2014) 9:e86297. doi: 10.1371/journal.pone.0086297
114. Shen X, Liu LP, Peek RM, Acra SA, Moore DJ, Wilson KT, et al. Supplementation of P40, a Lactobacillus Rhamnosus GG-Derived Protein, in Early Life Promotes Epidermal Growth Factor Receptor-Dependent Intestinal Development and Long-Term Health Outcomes. Mucosal Immunol (2018) 11:1316–28. doi: 10.1038/s41385-018-0034-3
115. Ganguli K, Collado MC, Rautava J, Lu L, Satokari R, von Ossowski I, et al. Lactobacillus Rhamnosus GG and its SpaC Pilus Adhesin Modulate Inflammatory Responsiveness and TLR-Related Gene Expression in the Fetal Human Gut. Pediatr Res (2015) 77:528–35. doi: 10.1038/pr.2015.5
116. Tsukuda N, Yahagi K, Hara T, Watanabe Y, Matsumoto H, Mori H, et al. Key Bacterial Taxa and Metabolic Pathways Affecting Gut Short-Chain Fatty Acid Profiles in Early Life. Isme J (2021) 15:2574–90. doi: 10.1038/s41396-021-00937-7
117. Lee YS, Kim TY, Kim Y, Lee SH, Kim S, Kang SW, et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe (2018) 24:833–46.e6. doi: 10.1016/j.chom.2018.11.002
118. Thomson P, Medina DA, Garrido D. Human Milk Oligosaccharides and Infant Gut Bifidobacteria: Molecular Strategies for Their Utilization. Food Microbiol (2018) 75:37–46. doi: 10.1016/j.fm.2017.09.001
119. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-Mediated Immune System Imprinting Early in Life. Cell (2021) 184:3884–98. doi: 10.1016/j.cell.2021.05.030
120. Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, et al. Bifidobacterium Species Associated With Breastfeeding Produce Aromatic Lactic Acids in the Infant Gut. Nat Microbiol (2021) 6:1–16. doi: 10.1038/s41564-021-00970-4
121. Ehrlich AM, Pacheco AR, Henrick BM, Taft D, Xu G, Huda MN, et al. Indole-3-Lactic Acid Associated With Bifidobacterium-Dominated Microbiota Significantly Decreases Inflammation in Intestinal Epithelial Cells. BMC Microbiol (2020) 20:1–13. doi: 10.1186/s12866-020-02023-y
122. Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-Lactic Acid, a Metabolite of Tryptophan, Secreted by Bifidobacterium Longum Subspecies Infantis is Anti-Inflammatory in the Immature Intestine. Pediatr Res (2020) 88:209–17. doi: 10.1038/s41390-019-0740-x
123. Huang WY, Cho KY, Meng D, Walker WA. The Impact of Indole-3-Lactic Acid on Immature Intestinal Innate Immunity and Development: A Transcriptomic Analysis. Sci Rep (2021) 11:1–17. doi: 10.1038/s41598-021-87353-1
124. Stewart CJ. Breastfeeding Promotes Bifidobacterial Immunomodulatory Metabolites. Nat Microbiol (2021) 6:1335–6. doi: 10.1038/s41564-021-00975-z
125. Arnaud AP, Rome V, Richard M, Formal M, David-Le Gall S, Boudry G. Post-Natal Co-Development of the Microbiota and Gut Barrier Function Follows Different Paths in the Small and Large Intestine in Piglets. FASEB J (2020) 34:1430–46. doi: 10.1096/fj.201902514R
126. Nan Z, Yanan G, Weishu Z, Di M, Walker WA. Short Chain Fatty Acids Produced by Colonizing Intestinal Commensal Bacterial Interaction With Expressed Breast Milk are Anti-Inflammatory in Human Immature Enterocytes. PloS One (2020) 15:e0229283. doi: 10.1371/journal.pone.0229283
127. Gao YA, Davis B, Zhu WS, Zheng N, Meng D, Walker WA. Short-Chain Fatty Acid Butyrate, a Breast Milk Metabolite, Enhances Immature Intestinal Barrier Function Genes in Response to Inflammation In Vitro and In Vivo. Am J Physiol Gastrointest (2021) 320:G521–G30. doi: 10.1152/ajpgi.00279.2020
128. Sun Q, Ji YC, Wang ZL, She X, He Y, Ai Q, et al. Sodium Butyrate Alleviates Intestinal Inflammation in Mice With Necrotizing Enterocolitis. Mediators Inflammation (2021) 2021:6259381. doi: 10.1155/2021/6259381
129. Motherway MO, Houston A, O'Callaghan G, Reunanen J, O'Brien F, O'Driscoll T, et al. A Bifidobacterial Pilus-Associated Protein Promotes Colonic Epithelial Proliferation. Mol Microbiol (2019) 111:287–301. doi: 10.1111/mmi.14155
130. Jiaqi G, Fei L, Sijia Z, Evivie SE, Jialu S, Na L, et al. Effect of Bifidobacterium Longum Subsp. Longum on the Proliferative and Tight-Junction Activities of Human Fetal Colon Epithelial Cells. J Funct Foods (2021) 86:104715. doi: 10.1016/j.jff.2021.104715
131. Tomas J, Reygner J, Mayeur C, Ducroc R, Bouet S, Bridonneau C, et al. Early Colonizing Escherichia Coli Elicits Remodeling of Rat Colonic Epithelium Shifting Toward a New Homeostatic State. ISME J (2015) 9:46–58. doi: 10.1038/ismej.2014.111
132. Payros D, Secher T, Boury M, Brehin C, Ménard S, Salvador-Cartier C, et al. Maternally Acquired Genotoxic Escherichia Coli Alters Offspring's Intestinal Homeostasis. Gut Microbes (2014) 5:313–25. doi: 10.4161/gmic.28932
133. Thomas DM, Bell B, Papillon S, Delaplain P, Lim J, Golden J, et al. Colonization With Escherichia Coli EC 25 Protects Neonatal Rats From Necrotizing Enterocolitis. PloS One (2017) 12:e0188211. doi: 10.1371/journal.pone.0188211
134. Are A, Aronsson L, Wang SG, Greicius G, Lee YK, Gustafsson JA, et al. Enterococcus Faecalis From Newborn Babies Regulate Endogenous PPAR Gamma Activity and IL-10 Levels in Colonic Epithelial Cells. P Natl Acad Sci USA (2008) 105:1943–8. doi: 10.1073/pnas.0711734105
135. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu W-JH, Saldana-Morales FB, et al. Thymic Development of Gut-Microbiota-Specific T Cells. Nature (2021) 594:413–7. doi: 10.1038/s41586-021-03531-1
136. Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, et al. Commensal Enterobacteriaceae Protect Against Salmonella Colonization Through Oxygen Competition. Cell Host Microbe (2019) 25:128–39. doi: 10.1016/j.chom.2018.12.003
137. Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the Infant Gut Consume Milk Oligosaccharides via Mucus-Utilization Pathways. Cell Host Microbe (2011) 10:507–14. doi: 10.1016/j.chom.2011.10.007
138. Al-Balawi M, Morsy FM. Enterococcus faecalisIs a Better Competitor Than Other Lactic Acid Bacteria in the Initial Colonization of Colon of Healthy Newborn Babies at First Week of Their Life. Front Microbiol (2020) 11:2017. doi: 10.3389/fmicb.2020.02017
139. Wang SG, Hibberd ML, Pettersson S, Lee YK. Enterococcus Faecalis From Healthy Infants Modulates Inflammation Through MAPK Signaling Pathways. PloS One (2014) 9:e97523. doi: 10.1371/journal.pone.0097523
140. Colliou N, Ge Y, Sahay B, Gong MH, Zadeh M, Owen JL, et al. Commensal Propionibacterium Strain UF1 Mitigates Intestinal Inflammation via Th17 Cell Regulation. J Clin Investig (2017) 127:3970–86. doi: 10.1172/jci95376
141. Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The Microbiota of the Gut in Preschool Children With Normal and Excessive Body Weight. Obes (Silver Spring) (2012) 20:2257–61. doi: 10.1038/oby.2012.110
142. Corb Aron RA, Abid A, Vesa CM, Nechifor AC, Behl T, Ghitea TC, et al. Recognizing the Benefits of Pre-/Probiotics in Metabolic Syndrome and Type 2 Diabetes Mellitus Considering the Influence of Akkermansia Muciniphila as a Key Gut Bacterium. Microorganisms (2021) 9:618. doi: 10.3390/microorganisms9030618
143. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia Muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl Environ Microbiol (2015) 81:3655–62. doi: 10.1128/aem.04050-14
144. Thanabalasuriar A, Kubes P. Neonates, Antibiotics and the Microbiome. Nat Med (2014) 20:469–70. doi: 10.1038/nm.3558
145. Foye OT, Huang IF, Chiou CC, Walker WA, Shi HN. Early Administration of Probiotic Lactobacillus Acidophilus and/or Prebiotic Inulin Attenuates Pathogen-Mediated Intestinal Inflammation and Smad 7 Cell Signaling. FEMS Immunol Med Microbiol (2012) 65:467–80. doi: 10.1111/j.1574-695X.2012.00978.x
146. Liu X, Jin G, Tang Q, Huang S, Zhang Y, Sun Y, et al. Early Life Lactobacillus Rhamnosus GG Colonisation Inhibits Intestinal Tumour Formation. Br J Cancer (2022) 126:1–11. doi: 10.1038/s41416-021-01562-z
147. del Mar Rigo-Adrover M, Franch A, Castell M, Jose Perez-Cano F. Preclinical Immunomodulation by the Probiotic Bifidobacterium Breve M-16V in Early Life. PloS One (2016) 11:e0166082. doi: 10.1371/journal.pone.0166082
148. Moya-Perez A, Perez-Villalba A, Benitez-Paez A, Campillo I, Sanz Y. Bifidobacterium CECT 7765 Modulates Early Stress-Induced Immune, Neuroendocrine and Behavioral Alterations in Mice. Brain Behav Immun (2017) 65:43–56. doi: 10.1016/j.bbi.2017.05.011
149. Deng J, Li Y, Zhang J, Yang Q. Co-Administration of Bacillus Subtilis RJGP16 and Lactobacillus Salivarius B1 Strongly Enhances the Intestinal Mucosal Immunity of Piglets. Res Vet Sci (2013) 94:62–8. doi: 10.1016/j.rvsc.2012.07.025
150. Kawahara T, Makizaki Y, Oikawa Y, Tanaka Y, Maeda A, Shimakawa M, et al. Oral Administration of Bifidobacterium Bifidum G9-1 Alleviates Rotavirus Gastroenteritis Through Regulation of Intestinal Homeostasis by Inducing Mucosal Protective Factors. PloS One (2017) 12:e0173979. doi: 10.1371/journal.pone.0173979
151. Dong P, Yang Y, Wang W-p. The Role of Intestinal Bifidobacteria on Immune System Development in Young Rats. Early Hum Dev (2010) 86:51–8. doi: 10.1016/j.earlhumdev.2010.01.002
152. Underwood MA, Arriola J, Gerber CW, Kaveti A, Kalanetra KM, Kananurak A, et al. Bifidobacterium Longum Subsp. Infantis in Experimental Necrotizing Enterocolitis: Alterations in Inflammation, Innate Immune Response, and the Microbiota. Pediatr Res (2014) 76:326–33. doi: 10.1038/pr.2014.102
153. Li Y, Hou S, Chen J, Peng W, Wen W, Chen F, et al. Oral Administration of Lactobacillus Delbrueckii During the Suckling Period Improves Intestinal Integrity After Weaning in Piglets. J Funct Foods (2019) 63:103591. doi: 10.1016/j.jff.2019.103591
154. Wang M, Wu H, Lu L, Jiang L, Yu Q. Lactobacillus Reuteri Promotes Intestinal Development and Regulates Mucosal Immune Function in Newborn Piglets. Front Vet Sci (2020) 7:42. doi: 10.3389/fvets.2020.00042
155. Zhang J, Deng J, Li Y, Yang Q. The Effect of Lactobacillus on the Expression of Porcine Beta-Defensin-2 in the Digestive Tract of Piglets. Livest Sci (2011) 138:259–65. doi: 10.1016/j.livsci.2011.01.001
156. Shen XM, Cui HX, Xu XR. Orally Administered Lactobacillus Casei Exhibited Several Probiotic Properties in Artificially Suckling Rabbits. Asian-Australas J Anim Sci (2020) 33:1352–9. doi: 10.5713/ajas.18.0973
157. Hoang TK, He BK, Wang T, Tran DQ, Rhoads JM, Liu YY. Protective Effect of Lactobacillus Reuteri DSM 17938 Against Experimental Necrotizing Enterocolitis is Mediated by Toll-Like Receptor 2. Am J Physiol Gastrointest Liver Physiol (2018) 315:G231–G40. doi: 10.1152/ajpgi.00084.2017
158. Liu YY, Fatheree NY, Mangalat N, Rhoads JM. Lactobacillus Reuteri Strains Reduce Incidence and Severity of Experimental Necrotizing Enterocolitis via Modulation of TLR4 and NF-Kappa B Signaling in the Intestine. Am J Physiol Gastrointest Liver Physiol (2012) 302:G608–G17. doi: 10.1152/ajpgi.00266.2011
159. Hoang TK, Freeborn J, Wang T, Mai T, He B, Park S, et al. Human Breast Milk Promotes the Immunomodulatory Function of Probiotic Lactobacillus Reuteri DSM 17938 in the Neonatal Rat Intestine. J Probiotics Health (2019) 7:210. doi: 10.35248/2329-8901.19.7.210
160. Peng W, Li Y-H, Yang G, Duan J-L, Yang L-Y, Chen L-X, et al. Oral Administration of Lactobacillus Delbrueckii Enhances Intestinal Immunity Through Inducing Dendritic Cell Activation in Suckling Piglets. Food Funct (2022) 13:2570–80. doi: 10.1039/d1fo03864h
161. Chen CC, Chiu CH, Lin TY, Shi HN, Walker WA. Effect of Probiotics Lactobacillus Acidophilus on Citrobacter Rodentium Colitis: The Role of Dendritic Cells. Pediatr Res (2009) 65:169–75. doi: 10.1203/PDR.0b013e31818d5a06
162. Liu T, Song X, An Y, Wu X, Zhang W, Li J, et al. Lactobacillus Rhamnosus GG Colonization in Early Life Ameliorates Inflammaging of Offspring by Activating SIRT1/AMPK/PGC-1α Pathway. Oxid Med Cell Longev (2021) 2021:3328505. doi: 10.1155/2021/3328505
163. Wang S, Yan F, He F, Zhu H. Effects of Lactobacillus Paracasei N1115 on Intestinal Development in Neonatal Mice. Acta Nutrimenta Sin (2016) 38:71–4. doi: 10.1111/lam.13729
164. Xie S, Zhao SY, Jiang L, Lu LH, Yang Q, Yu QH. Lactobacillus Reuteri Stimulates Intestinal Epithelial Proliferation and Induces Differentiation Into Goblet Cells in Young Chickens. J Agric Food Chem (2019) 67:13758–66. doi: 10.1021/acs.jafc.9b06256
165. Li W-F, Huang Y, Li Y-L, Huang Q, Cui Z-w, Yu D-y, et al. Effect of Oral Administration of Enterococcus Faecium Ef1 on Innate Immunity of Sucking Piglets. PAK Vet J (2013) 33:9–13.
166. Huang Y, Li YL, Huang Q, Cui ZW, Yu DY, Rajput IR, et al. Effect of Orally Administered Enterococcus Faecium EF1 on Intestinal Cytokines and Chemokines Production of Suckling Piglets. PAK Vet J (2012) 32:81–4.
167. Guo C, Li Y, Geng M, Gai S, Zhang T, Qi W, et al. Effects of Bacillus Subtilis RZ001 on Intestinal Development, Intestinal Microbiota and Expression of Wnt Signaling Pathway Related Genes of Suckling Mice. Chin J Anim Nutr (2021) 33:506–18. doi: 1006-267X(2021)33:1<506:KCYBGJ>2.0.TX;2-R
168. Vlasova AN, Shao L, Kandasamy S, Fischer DD, Rauf A, Langel SN, et al. Escherichia Coli Nissle 1917 Protects Gnotobiotic Pigs Against Human Rotavirus by Modulating pDC and NK-Cell Responses. Eur J Immunol (2016) 46:2426–37. doi: 10.1002/eji.201646498
169. Liu CQ, Zhu Q, Chang J, Yin QQ, Song AD, Li ZT, et al. Effects of Lactobacillus Casei and Enterococcus Faecalis on Growth Performance, Immune Function and Gut Microbiota of Suckling Piglets. Arch Anim Nutr (2017) 71:120–33. doi: 10.1080/1745039x.2017.1283824
170. Liu HB, Hou CL, Wang G, Jia HM, Yu HT, Zeng XF, et al. Lactobacillus Reuteri I5007 Modulates Intestinal Host Defense Peptide Expression in the Model of IPEC-J2 Cells and Neonatal Piglets. Nutrients (2017) 9:559. doi: 10.3390/nu9060559
171. Yang KM, Jiang ZY, Zheng CT, Wang L, Yang XF. Effect of Lactobacillus Plantarum on Diarrhea and Intestinal Barrier Function of Young Piglets Challenged With Enterotoxigenic Escherichia Coli K88. J Anim Sci (2014) 92:1496–503. doi: 10.2527/jas.2013-6619
172. Naik AK, Pandey U, Mukherjee R, Mukhopadhyay S, Chakraborty S, Ghosh A, et al. Lactobacillus Rhamnosus GG Reverses Mortality of Neonatal Mice Against Salmonella Challenge. Toxicol Res (Camb) (2019) 8:361–72. doi: 10.1039/c9tx00006b
173. Ventola H, Lehtoranta L, Madetoja M, Simonen-Tikka ML, Maunula L, Roivainen M, et al. Effects of the Viability of Lactobacillus Rhamnosus GG on Rotavirus Infection in Neonatal Rats. World J Gastroenterol (2012) 18:5925–31. doi: 10.3748/wjg.v18.i41.5925
174. Peng X, Wang R, Hu L, Zhou Q, Liu Y, Yang M, et al. Enterococcus Faecium NCIMB 10415 Administration Improves the Intestinal Health and Immunity in Neonatal Piglets Infected by Enterotoxigenic Escherichia Coli K88. J Anim Sci Biotechnol (2019) 10:1–15. doi: 10.1186/s40104-019-0376-z
175. Sayan H, Assavacheep P, Angkanaporn K, Assavacheep A. Effect of Lactobacillus Salivarius on Growth Performance, Diarrhea Incidence, Fecal Bacterial Population and Intestinal Morphology of Suckling Pigs Challenged With F4(+) Enterotoxigenic Escherichia Coli. Asian-Australas J Anim Sci (2018) 31:1308–14. doi: 10.5713/ajas.17.0746
176. Zeng Q, He XL, Puthiyakunnon S, Xiao HS, Gong ZL, Boddu S, et al. Probiotic Mixture Golden Bifido Prevents Neonatal Escherichia Coli K1 Translocation via Enhancing Intestinal Defense. Front Microbiol (2017) 8:1798. doi: 10.3389/fmicb.2017.01798
177. Sherman MP, Bennett SH, Hwang FFY, Yu C. Neonatal Small Bowel Epithelia: Enhancing Anti-Bacterial Defense With Lactoferrin and Lactobacillus Gg. Biometals (2004) 17:285–9. doi: 10.1023/b:Biom.0000027706.51112.62
178. van Baarlen P, Troost F, van der Meer C, Hooiveld G, Boekschoten M, Brummer RJ, et al. Human Mucosal In Vivo Transcriptome Responses to Three Lactobacilli Indicate How Probiotics may Modulate Human Cellular Pathways. Proc Natl Acad Sci U.S.A. (2011) 108 Suppl 1:4562–9. doi: 10.1073/pnas.1000079107
179. Xie WC, Song LY, Wang XY, Xu YG, Liu ZS, Zhao DF, et al. A Bovine Lactoferricin-Lactoferrampin-Encoding Lactobacillus Reuteri CO21 Regulates the Intestinal Mucosal Immunity and Enhances the Protection of Piglets Against Enterotoxigenic Escherichia Coli K88 Challenge. Gut Microbes (2021) 13:1956281. doi: 10.1080/19490976.2021.1956281
180. Sharif S, Meader N, Oddie SJ, Rojas-Reyes MX, McGuire W. Probiotics to Prevent Necrotising Enterocolitis in Very Preterm or Very Low Birth Weight Infants. Cochrane Database Syst Rev (2020) 10:Cd005496. doi: 10.1002/14651858.CD005496.pub5
181. Samara J, Moossavi S, Alshaikh B, Ortega VA, Pettersen VK, Ferdous T, et al. Supplementation With a Probiotic Mixture Accelerates Gut Microbiome Maturation and Reduces Intestinal Inflammation in Extremely Preterm Infants. Cell Host Microbe (2022) 30:696–711.e5. doi: 10.1016/j.chom.2022.04.005
182. Viswanathan S, Lau C, Akbari H, Hoyen C, Walsh MC. Survey and Evidence Based Review of Probiotics Used in Very Low Birth Weight Preterm Infants Within the United States. J Perinatol (2016) 36:1106–11. doi: 10.1038/jp.2016.144
183. Lai CY, Sung J, Cheng F, Tang W, Wong SH, Chan PKS, et al. Systematic Review With Meta-Analysis: Review of Donor Features, Procedures and Outcomes in 168 Clinical Studies of Faecal Microbiota Transplantation. Aliment Pharmacol Ther (2019) 49:354–63. doi: 10.1111/apt.15116
184. Pavel FM, Vesa CM, Gheorghe G, Diaconu CC, Stoicescu M, Munteanu MA, et al. Highlighting the Relevance of Gut Microbiota Manipulation in Inflammatory Bowel Disease. Diagnost (Basel) (2021) 11:1090. doi: 10.3390/diagnostics11061090
185. Negrut N, Bungau S, Behl T, Khan SA, Vesa CM, Bustea C, et al. Risk Factors Associated With Recurrent Clostridioides Difficile Infection. Healthc (Basel) (2020) 8:352. doi: 10.3390/healthcare8030352
186. Geng S, Cheng S, Li Y, Wen Z, Ma X, Jiang X, et al. Faecal Microbiota Transplantation Reduces Susceptibility to Epithelial Injury and Modulates Tryptophan Metabolism of the Microbial Community in a Piglet Model. J Crohns Colitis (2018) 12:1359–74. doi: 10.1093/ecco-jcc/jjy103
187. Hu L, Geng S, Li Y, Cheng S, Fu X, Yue X, et al. Exogenous Fecal Microbiota Transplantation From Local Adult Pigs to Crossbred Newborn Piglets. Front Microbiol (2018) 8:2663. doi: 10.3389/fmicb.2017.02663
188. Qi R, Zhang Z, Wang J, Qiu X, Wang Q, Yang F, et al. Introduction of Colonic and Fecal Microbiota From an Adult Pig Differently Affects the Growth, Gut Health, Intestinal Microbiota and Blood Metabolome of Newborn Piglets. Front Microbiol (2021) 12:623673. doi: 10.3389/fmicb.2021.623673
189. Teng T, Gao F, He W, Fu HY, Guo J, Bai GD, et al. An Early Fecal Microbiota Transfer Improves the Intestinal Conditions on Microflora and Immunoglobulin and Antimicrobial Peptides in Piglets. J Agric Food Chem (2020) 68:4830–43. doi: 10.1021/acs.jafc.0c00545
190. Xiang Q, Wu X, Pan Y, Wang L, Cui C, Guo Y, et al. Early-Life Intervention Using Fecal Microbiota Combined With Probiotics Promotes Gut Microbiota Maturation, Regulates Immune System Development, and Alleviates Weaning Stress in Piglets. Int J Mol Sci (2020) 21:503. doi: 10.3390/ijms21020503
191. Xiang Q, Wu X, Pan Y, Wang L, Guo Y, Cui C, et al. Early Intervention Using Fecal Microbiota Transplantation Combined With Probiotics Influence the Growth Performance, Diarrhea, and Intestinal Barrier Function of Piglets. Appl SCI BASEL (2020) 10:568. doi: 10.3390/app10020568
192. Brunse A, Martin L, Rasmussen TS, Christensen L, Skovsted Cilieborg M, Wiese M, et al. Effect of Fecal Microbiota Transplantation Route of Administration on Gut Colonization and Host Response in Preterm Pigs. Isme J (2019) 13:720–33. doi: 10.1038/s41396-018-0301-z
193. Ma X, Xu T, Qian M, Zhang Y, Yang Z, Han X. Faecal Microbiota Transplantation Alleviates Early-Life Antibiotic-Induced Gut Microbiota Dysbiosis and Mucosa Injuries in a Neonatal Piglet Model. Microbiol Res (2021) 255:126942. doi: 10.1016/j.micres.2021.126942
194. Meijerink N, Kers JG, Velkers FC, van Haarlem DA, Lamot DM, de Oliveira JE, et al. Early Life Inoculation With Adult-Derived Microbiota Accelerates Maturation of Intestinal Microbiota and Enhances NK Cell Activation in Broiler Chickens. Front Vet Sci (2020) 7:584561. doi: 10.3389/fvets.2020.584561
195. Prado C, Michels M, Ávila P, Burger H, Milioli MVM, Dal-Pizzol F. The Protective Effects of Fecal Microbiota Transplantation in an Experimental Model of Necrotizing Enterocolitis. J Pediatr Surg (2019) 54:1578–83. doi: 10.1016/j.jpedsurg.2018.10.045
196. Brunse A, Deng L, Pan X, Hui Y, Castro-Mejía JL, Kot W, et al. Fecal Filtrate Transplantation Protects Against Necrotizing Enterocolitis. Isme J (2021) 6:686–94. doi: 10.1038/s41396-021-01107-5
197. Cheng S, Ma X, Geng S, Jiang X, Li Y, Hu L, et al. Fecal Microbiota Transplantation Beneficially Regulates Intestinal Mucosal Autophagy and Alleviates Gut Barrier Injury. mSystems (2018) 3:e00137–18. doi: 10.1128/mSystems.00137-18
198. Li N, Zuo B, Huang S, Zeng B, Han D, Li T, et al. Spatial Heterogeneity of Bacterial Colonization Across Different Gut Segments Following Inter-Species Microbiota Transplantation. Microbiome (2020) 8:1–24. doi: 10.1186/s40168-020-00917-7
199. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut Immune Maturation Depends on Colonization With a Host-Specific Microbiota. Cell (2012) 149:1578–93. doi: 10.1016/j.cell.2012.04.037
200. Marrs T, Walter J. Pros and Cons: Is Faecal Microbiota Transplantation a Safe and Efficient Treatment Option for Gut Dysbiosis? Allergy (2021) 76:2312–7. doi: 10.1111/all.14750
201. Zenner C, Hitch TCA, Riedel T, Wortmann E, Tiede S, Buhl EM, et al. Early-Life Immune System Maturation in Chickens Using a Synthetic Community of Cultured Gut Bacteria. mSystems (2021) 6:e01300–20. doi: 10.1128/mSystems.01300-20
202. Behl T, Kaur I, Sehgal A, Singh S, Bhatia S, Al-Harrasi A, et al. Bioinformatics Accelerates the Major Tetrad: A Real Boost for the Pharmaceutical Industry. Int J Mol Sci (2021) 22:6184. doi: 10.3390/ijms22126184
203. Sagheddu V, Patrone V, Miragoli F, Morelli L. Abundance and Diversity of Hydrogenotrophic Microorganisms in the Infant Gut Before the Weaning Period Assessed by Denaturing Gradient Gel Electrophoresis and Quantitative PCR. Front Nutr (2017) 4:29. doi: 10.3389/fnut.2017.00029
204. Gu W, Miller S, Chiu CY. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu Rev Pathol (2019) 14:319–38. doi: 10.1146/annurev-pathmechdis-012418-012751
Keywords: early life, microbiome colonization, intestine, innate immunity, probiotics, FMT
Citation: Yang Z, Liu X, Wu Y, Peng J and Wei H (2022) Effect of the Microbiome on Intestinal Innate Immune Development in Early Life and the Potential Strategy of Early Intervention. Front. Immunol. 13:936300. doi: 10.3389/fimmu.2022.936300
Received: 05 May 2022; Accepted: 23 June 2022;
Published: 19 July 2022.
Edited by:
Qiushui He, University of Turku, FinlandReviewed by:
Simona Gabriela Bungau, University of Oradea, RomaniaCosmin Mihai Vesa, University of Oradea, Romania
Copyright © 2022 Yang, Liu, Wu, Peng and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Peng, cGVuZ2ppYW5AbWFpbC5oemF1LmVkdS5jbg==; Hongkui Wei, d2VpaG9uZ2t1aUBtYWlsLmh6YXUuZWR1LmNu
 Zhipeng Yang
Zhipeng Yang Xiangchen Liu1
Xiangchen Liu1 Hongkui Wei
Hongkui Wei