- 1Sino-French Hoffmann Institute, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou, China
- 2State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 3The State Key Laboratory of Respiratory Disease, Guangzhou Medical University, Guangzhou, China
Animals adjust their lipid metabolism states in response to pathogens infection. However, the underlying molecular mechanisms for how lipid metabolism responds to infection remain to be elusive. In this study, we assessed the temporal changes of lipid metabolism profiles during infection by an integrated transcriptomics and lipidomics analysis. Ergosterol is identified to be required for proper host defense to pathogens. Notably, ergosterol level is increased in the hemolymph upon bacterial infection. We show that the increase of ergosterol level by food supplement or genetic depletion of Acsl, a long-chain fatty acid-CoA synthetase, promotes host survival against bacterial challenges. Together, our results suggest a critical role of lipid metabolism adaption in the process of host defense against invading pathogens.
Introduction
Animals evolve to form a set of sophisticated defense systems to cope with a variety of environmental stresses, such as pathogens infection. Innate immunity is the first line and most ancient host defense system against invading pathogens, including bacteria, fungi and viruses (1). The striking conservation of genetic regulation between flies and mammals together with the well-established genetic resources have made Drosophila an attractive model organism to decipher the principles of innate immune response (2, 3). In Drosophila, immune response comprises cellular and humoral immunity. In cellular immunity, hemocytes phagocytose or encapsulate, and trigger melanization, to destroy the invading pathogens (4). Humoral immunity, on the other hand, is responsible for the production of antimicrobial peptides (AMPs) mainly from the hemocytes and fat body, through the classical Toll and Imd signaling pathways (5). Although remarkable progress has been achieved during the past three decades in elucidating the underlying mechanisms for innate immune response and regulations, how the immune response activity is regulated physiologically remains largely unknown.
Innate immune system is critical for host survival, yet energetically expensive for a full protection during immune challenges, requiring proper re-distribution of energy (6). Upon pathogen invasion, Drosophila hemocytes sense and initiate a metabolic switch to aerobic glycolysis for boosting the immune response (7, 8). Furthermore, the activation of the Imd pathway and/or the Toll pathway in fat body disrupts insulin signaling, results in decreased triglyceride storage and impaired animal growth (9–11). More recently, dSTING, the conserved antiviral signaling pathway in both flies and humans (12, 13), is also reported to regulate lipid metabolism by modulating fatty acid synthesis (14, 15). The infection-induced metabolic adaption and reallocation of energy is indispensable for immune responses, because blocking the metabolic adjustment has been reported to impair host defense against pathogens (16).
Emerging evidence has established a linkage between lipid metabolism and innate immune response, however, the molecular mechanisms by which infection leads to lipid metabolism adaption and how lipid metabolites are involved in immune response are not well understood. In order to systemically assess the molecular connection between these two indispensable processes, here, we performed a time-course study for lipid metabolism genes and metabolites profile dynamics during bacterial infection by an integrated transcriptomics and lipidomics analysis, and identified ergosterol as a novel lipid metabolite involved in proper host defense to bacterial infection.
Materials and Methods
Fly Strains and Husbandry
The following fly stocks were used: The wild-type w1118 (#3605) flies were obtained from Bloomington Drosophila Stock Center. AcslTHU2816 and Acsl3222 RNAi lines were obtained from TsingHua Fly Center (THFC) and Vienna Drosophila Resource Center (VDRC). The key1, MyD88c03881 and w; pUbi-Gal4; pTub-Gal80ts lines were obtained from Dominique Ferrandon’s lab in Institut de Biologie Moléculaire et Cellulaire (IBMC). Unless specifically noted, all flies were kept on standard cornmeal food with yeast (refer to BDSC Cornmeal Food recipe) at 25°C. For yeast-free (YF) food, yeasts were not added to the standard cornmeal food. For ergosterol food, ergosterol (Macklin) was added to the yeast-free cornmeal food with ergosterol (100 mM). For bacterial infection, the newly eclosed flies (within 2 hour) were collected and transferred to YF, YF+ergosterol and normal food (NF), respectively and kept at 25°C for 3 days before bacterial infection. For Acsl RNAi experiment, Ubi-Gal4, UAS-Acsl RNAi, Tub-Gal80ts and control flies were kept at 29°C for 5 days before bacterial infection.
Bacterial Strains
Gram-negative bacteria Erwinia carotovora carotovora-15 (Ecc15, renamed as P. carotovorum) strain was obtained from Lei Pan’s lab in Institut Pasteur of Shanghai, Chinese Academy of Sciences (17). Serratia marcescens (S.m) (# 1.2818) and Enterococcus faecalis (E.fa) (#1.2135) were obtained from the China General Microbiological Culture Collection Centre (CGMCC).
Bacterial Infection
Ecc15, S.m and E.fa were cultured in standard LB medium overnight, with Ecc15 and S.m incubated at 30°C and E.fa at 37°C. Before infection, bacteria were washed by 1xPBS for 3 times and diluted to the indicated optical density (OD600) of 50, 2 and 0.1, respectively. For infection experiments, flies were anesthetized on CO2 pad before bacteria was injected quantitatively into the fly thorax using a microinjector (Nanoject III, Drummond Scientific Company). The injection volume of Ecc15, S.m and E.fa were 15, 2.3 and 9.2 nL per fly, respectively. Of note, the injection volume of Ecc15 in Figure 4A was 23 nL per fly.
Bacterial Load
To determine the CFU of Ecc15, S.m and E.fa-infected flies, individual fly was first homogenized gently in 200 μL 1xPBS at the indicated time points. In brief, a sterile zirconia bead (φ=3.0 mm) was added into the sample with 200 μL pre-cooled 1xPBS, then the sample was crashed by vibrating the zirconia bead with frequency of 30Hz for 30s in a Retsch MM400 grinding mixer. Then, 10 μL of homogenates were serial diluted at 1:104. 10 μL of the diluted-homogenates were placed on standard LB agar plate at 37°C overnight before counting. Each diluted sample was performed in duplicate.
qPCR
To measure mRNA levels of indicated fly genes, ~7 flies were collected for homogenate preparation. For homogenate preparation, a sterile zirconia bead was added into the sample with 200 μL pre-cooled RNAiso, then the sample was crashed by vibrating the zirconia bead with frequency of 30Hz for 3 min in a Retsch MM400 grinding mixer. Homogenates of fly for each treatment were then extracted by RNAiso Plus kit (TAKARA). RNA (1 μg) was reverse transcribed using PrimeScript™ RT reagent kit (TAKARA). qPCR analyses were preformed using the TB Green premix Ex Taq™ II kit on a BIO-RAD C1000 Touch™ Thermal Cycle. And the expression levels of target genes were normalize to rp49 (data are presented as ΔCT, 2^(Ctrp49 - Cttarget genes)). Primers used were in Supplementary Table 2.
Hemolymph Extraction
The Ecc15-infected w1118 female flies (OD=50, 15 nL) and the corresponding PBS-injected (control) flies were poked by needle at thorax and then placed at a 500 μL Eppendorf tube with a hole in the bottom, which was inserted in a 1.5 mL tube. Flies were centrifuged at 5,500 rpm for 15 min. The yielded hemolymph was gently mixed with TBST (0.1% Tween-20) and stored at -80°C until use.
RNA-Seq and Bioinformatics Analyses
The total RNA of Ecc15-infected and PBS-injection control w1118 female flies were extracted at 12, 24, 48 and 72 hpi. The eukaryotic mRNA was enriched by Oligo (dT) beads, and rRNA was removed by Ribo-Zero™ Magnetic Kit (Epicentre Madison, WI, USA). Then, the RNA was quantified, reverse transcribed and sequenced by Illumina sequencing platform. These eukaryotic mRNA enrichment, rRNA elimination, cDNA library preparation and sequencing procedures were performed by Guangzhou Genedenovo Biotechnology Co., Ltd. The raw sequencing data were filtered by trim galore software (v0.6.4) to remove plausible remaining adapter sequences in reads and low quality (Q-value<=20) reads. The resulting clean data were aligned to fly’s genome dm6 using the STAR software (v2.7.2b) (18). Gene expression levels (gene counts and TPM- Transcripts Per Million) were quantified by rsem software (v1.3.1) (19). The following bioinformatics analyses were performed base on R software (v3.5.3). In detail, Weighted Correlation Network Analysis (WGCNA) was performed based on WGCNA package (v1.69) (20). In brief, the TPM gene expression matrix generated by rsem software was log2 transformed and was transposed; Second, soft thresholding powers were then calculated by WGCNA::pickSoftThreshold function; Third, automatic network construction and module detection were calculated by WGCNA::blockwiseModules function with soft thresholding powers calculated above (in this study, sft=14); Forth, the correlation value between module and time-course treatment (such as Ecc15 12 h, PBS 12 h et al.) were calculate by WGCNA::cor function; In the last step, the visualization of the correlation ship modules and time-course treatment by WGCNA:: labeledHeatmap function. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) analysis were performed using clusterProfiler package(v3.10.1) (21). In brief, for GO analysis, the indicated gene list were imported to the clusterProfiler::enrichGO function to preform biological process GO terms enrichment analysis. For GSEA analysis, the down-regulated genes of PBS vs Ecc15 12 h/24 h were imported to clusterProfiler::gseGO function to perform GSEA analysis. The visualizations of lipid metabolic process (GO:0006629) were plotted by clusterProfiler::gseaplot function. Transcription factor binding motifs enrichment analysis was performed by RcisTarget package (v1.2.1) (22). In brief, the lipid metabolism process related-genes identified by WGCNA analysis in the red module were imported to the RcisTarget::cisTarger function to identify DNA motifs that were significantly over-represented in the gene-set. In this step, the Hnf4 binding motif was identified (NES=9.72 and AUC =0.249) using dm6_motifRanking_mc8nr (https://resources-mirror.aertslab.org/cistarget); Second, to get the incident matrix of significant genes which were highly ranked for Hnf4 binding motif, RcisTarget::getSignificantGenes was used. In the last, the visualization of the incident matrix was plotted by Cytoscape software (v3.7.2)
Differential expression genes (DEGs) were identified by DEseq2 package (v1.22.2) (23). In brief, the gene counts expression matrix generated by rsem software were imported to the DESeq2::DESeq function to calculate differential expression analysis based on the Negative Binomial distribution, then the gene expression comparison results of each group (such as such as “PBS vs Ecc15 12 h”, “PBS vs Ecc15 24 h” et al.) were obtained by DESeq2::result function.
Lipid Extraction
Lipid were extracted from Drosophlia hemolymph using a modified version of the Bligh and Dyer’s method as described previously (24). In brief, add 750 µL of chloroform: methanol 1:2 (v/v) with 10% deionized water to samples. Then samples were incubated at 1,500 rpm for 1 h at 4°C. After the incubation, 350 µL of deionized water and 250 µL of chloroform were added to samples to induce phase separation. The samples were then centrifuged and the lower organic phase containing lipids was extracted and transferred into a clean tube. Lipid extraction was repeated once by adding 500 µL of chloroform to the remaining aqueous phase, and the lipid extracts were pooled into a single tube and dried in the SpeedVac under OH mode. Samples were stored at -80°C until further analysis.
Lipidomic Analyses
Polar lipids were analyzed using an Exion UPLC system coupled with a triple quadrupole/ion trap mass spectrometer (6500 Plus Qtrap; SCIEX) as described previously (25–27). Separation of individual lipid classes of polar lipids by normal phase (NP)-HPLC was conducted using a Phenomenex Luna 3µm-silica column (internal diameter 150 × 2.0 mm) with the following conditions: mobile phase A (chloroform: methanol: ammonium hydroxide, 89.5:10:0.5) and mobile phase B (chloroform: methanol: ammonium hydroxide: water, 55:39:0.5:5.5). MRM transitions were set up for comparative analysis of various polar lipids. Individual lipid species were quantified by referencing to spiked internal standards. PC-d31(16:0/18:1), PE-d31(16:0/18:1), PS-d31(16:0/18:1), PI-d31(16:0/18:1), PA(17:0/17:0), PG-d31(16:0/18:1), C17-Cer, C12-PECer were obtained from Avanti Polar Lipids. Dioctanoyl phosphatidylinositol (PI) (16:0-PI) was obtained from Echelon Biosciences, Inc. Glycerol lipids including diacylglycerols (DAGs) and triacylglycerols (TAGs) were quantified using a modified version of reverse phase HPLC/MRM. Separation of neutral lipids were achieved on a Phenomenex Kinetex-C18 2.6 µm column (i.d. 4.6x100 mm) using an isocratic mobile phase containing chloroform: methanol: 0.1 M ammonium acetate 100:100:4 (v/v/v) at a flow rate of 170 µL for 17 min. Levels of short-, medium-, and long-chain TAGs were calculated by referencing to spiked internal standards of TAG (16:0)3-d5 and TAG (18:0)3-d5 obtained from CDN isotopes, respectively. DAGs were quantified using d5-DAG16:0/16:0 as internal standards (Avanti Polar Lipids). Free cholesterols and cholesteryl esters were analyzed as described previously with d6-cholesterol and d6-cholesteryl ester (CE) (CDN isotopes) as internal standards.
Results
Time-Course Lipidomic Profiling of Drosophila Hemolymph After Ecc15 Infection Reveals a Dynamic Remodeling of Lipid Profiles
Immune challenges induce systemic adaptive changes of lipid metabolism to coordinate the proper mobilization of energy for host defense. Drosophila hemolymph, the open and circulating system, is the major platform for materials and energy exchange between tissues/organs (28). Thus, lipid metabolites profiling of the hemolymph during bacterial infection will provide important hints to assess the lipid metabolic status at the organismal level.
To this end, we collected the hemolymph from Ecc15 (renamed as P. carotovorum)-challenged and control (PBS) flies at different time points (12, 24, 48, 72 hpi), and performed a quantitative lipidomic analysis to assess their dynamic lipid metabolites profiles, including glycerides, phospholipids, glycerophosphatides, sphingolipids and sterols. Our lipidomic analysis revealed that several lipids, mainly phospholipids, sphingolipids and sterols, were significantly changed when flies were challenged with Ecc15 infection, such as diacylglycerols (DAGs), ceramides (Cer), hydroxyceramide (OH-Cer), phosphoinositides (PIs), and phosphatidylethanolamines (PEs). These results indicate that a systemic adaptive change of lipid metabolites occurs in Ecc15-challenged flies (Figure 1A, Supplementary Table 1). In particular, the level of PIs, commonly as membrane ingredients and intracellular signal transducing molecules, was down-regulated in both clean-wounded and Ecc15-infected flies (Figure 1B). The facts that supplementation of the diet with PIs, not other lipids, suppresses the proinflammatory cytokine levels in mammals (29), imply that the decrease of PIs in early stage of challenges is an active response of host to release the brakes for a full activation of immune response. Additionally, Cer and OH-Cer showed similar trend with PIs after infection (Figures 1C, D), which suggests that flies may increase the threshold to the oxidative stress damages through reducing the Cer and OH-Cer levels, as ceramides act as mediators of oxidative stress and inflammation in several human diseases (30–32). Infection-induced DAGs loss in the hemolymph (Figure 1E), likely either through renal purging of DAGs to prevent from oxidative stresses (33) or enhanced intake by surrounding tissues for energy supply (34).
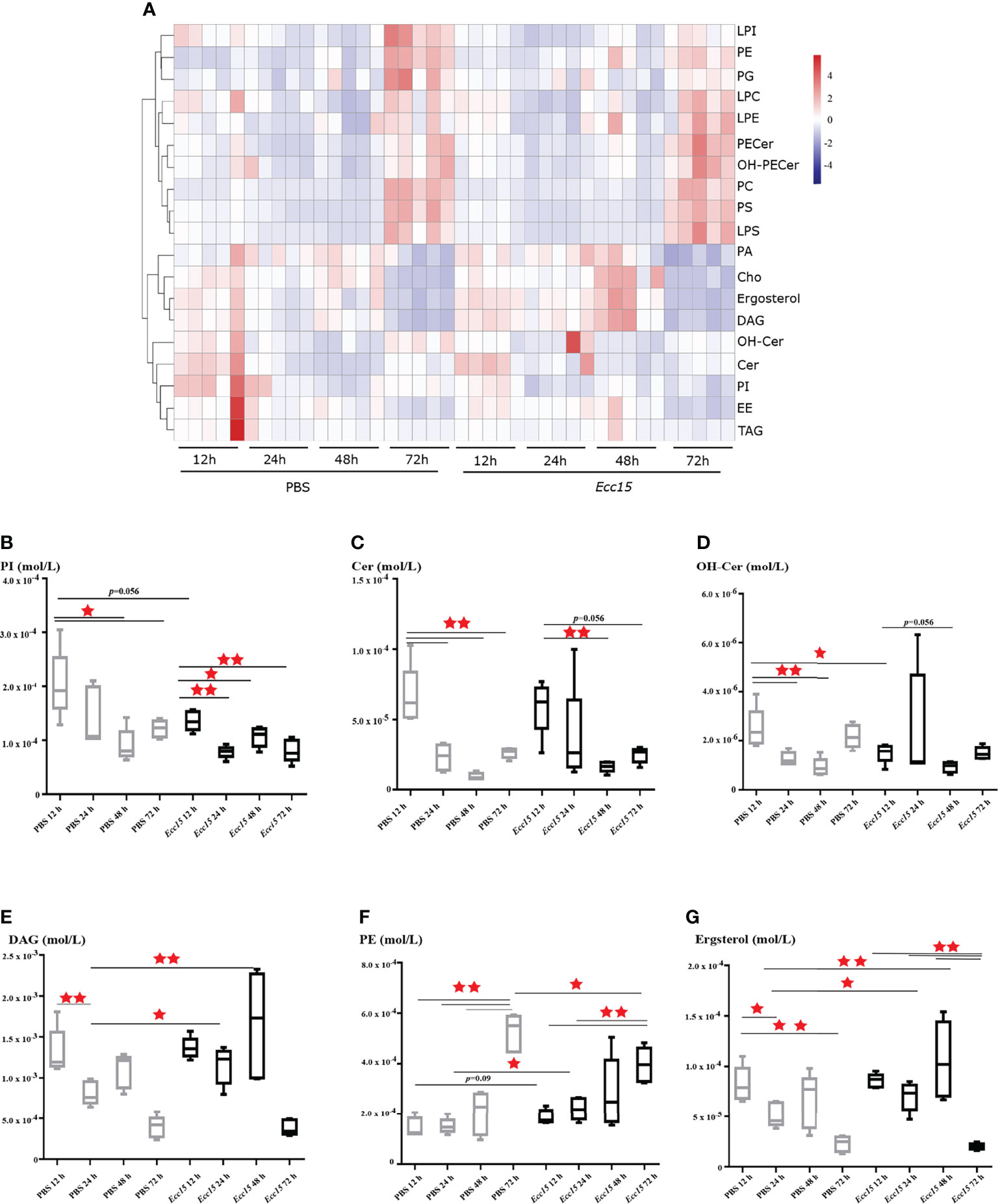
Figure 1 Time-course lipidomic profiling of Drosophila hemolymph after Ecc15 infection reveals a dynamic remodeling of lipid profiles. (A). Heatmap for selected lipid species profiles at indicated time points (12, 24, 48, 72 hpi) in control (PBS) and Ecc15-challenged (Ecc15) flies. 5 duplicates were performed for each group. (B–G). Box-plot of phosphatidylinositols (PI) (B), Ceramides (Cer) (C), Hydroxyceramide (OH-Cer) (D), Diacylglycerols (DAG) (E), Phosphatidylethanolamines (PE) (F), ergosterol (G) at indicated time points (12, 24, 48, 72 hpi) in control (PBS) and Ecc15-challenged (Ecc15) flies. *, p<0.05; **, p<0.001. (wilcox test).
Unlike PIs and Cer, the level of PEs, the second most abundant glycerophospholipid in flies, was significantly increased in Ecc15-infected flies compared to control (Figure 1F). Interestingly, we found that the Ecc15-infected flies contained high level of ergosterol, the major sterol in flies, in the hemolymph, compare with PBS-injected flies (Figure 1G).
Taken together, our temporal lipidomic analysis reveals that activation of immune response by bacterial stimuli results in the intense dynamic remodeling of lipid metabolism profiles, leading to the reallocation of large quantities of lipid metabolites during host defense.
Ergosterol Is Required for Host Defense of Bacterial Infection in Drosophila
Above results indicate that ergosterol level in hemolymph is significantly increased during Ecc15 infection, implying a potential role of ergosterol in host defense to bacterial infection. Unlike the well-characterized ergosterol synthesis pathway in fungi, there are no genes encoding the enzymes required for ergosterol de novo synthesis in Drosophila genome (35). Thus, ergosterol can only be ingested by flies from food. In standard fly food, yeast is the main source for ergosterol (35). Therefore, to investigate the potential role(s) of ergosterol in immune response, we reared the adult flies on yeast-free (YF) food and YF food with 100 mM ergosterol supplement (YF + ergosterol), respectively, and examined their susceptibility to Ecc15 infection. We found that flies reared on ergosterol food were more resistant to Ecc15 infection, as indicated by the extended survival curve (Figure 2A). Moreover, ergosterol supplement also rendered flies more resistant to S.m and E.fa infection (Figures 2B, C). Further, we wonder whether the increased host defense to infection was contributed by their enhanced immune response activity by ergosterol supplement. To address this question, we determined the transcriptional level of antimicrobial peptides Attactin A (AttA) and Diptericin (Dpt), which are indicators for the activation of Imd immune response pathway after Ecc15 infection. However, ergosterol feeding did not increase the AttA and Dpt mRNA level (Figures 2D, E). Besides, ergosterol feeding did not change the bacterial load (Figure 2H). Similar results were observed in S.m and E.fa-challenged flies (Figures 2F, G, I, J). Together, these results demonstrate that ergosterol, as a potent immune modulator, enhances host defense against bacterial infection, without changing the immune response strength.
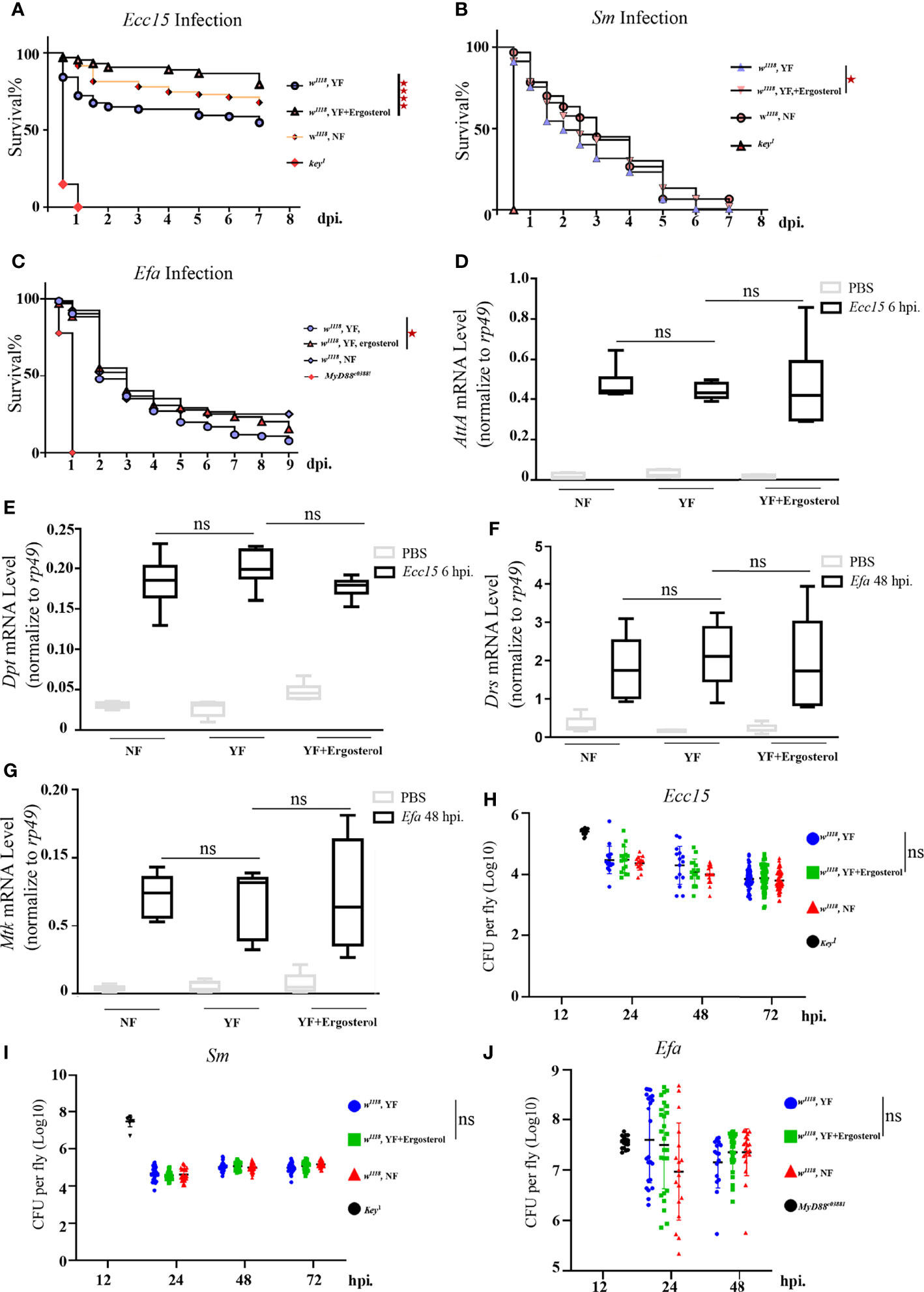
Figure 2 Ergosterol is required for host defense of bacterial infection in Drosophila. (A–C). Survival curves of w1118 and key1 or Myd88c03881 flies infected with Ecc15 (A), S.m (B), and E.fa (C), reared on different food. ****, p<0.0001 (Log-rank test). (D, E). Q-PCR plot of AttA and Dpt mRNA level in flies infected with Ecc15 at 6hpi, reared on different food. ns, not significant. (One-way ANOVA test with a Sidak test). (F, G). Q-PCR plot of Drs and Mtk mRNA level in flies infected with E.fa at 48hpi, reared on different food. ns, not significant. (One-way ANOVA test with a Sidak test). (H–J). CFU assay of w1118 and key1 or Myd88c03881 flies infected with Ecc15 (H), S.m (I), and E.fa (J), reared on different food. ns, not significant. (One-way ANOVA test with a Sidak test). NF, normal food; YF, yeast-free food; YF+ergosterol, yeast-free food containing ergosterol (100 mM).
Time-Course Transcriptomic Analysis Reveals That Changes of Lipid Metabolism Related Gene Expression Profiles Occur in Response to Ecc15 Infection
Next, we wondered how the ergosterol level in the hemolymph was regulated during infection. It is reported that bacterial infection decreases the feeding behavior by triggering food avoidance in Drosophila larvae (36) and pathogens infection also initiates a rapid renal purging of hemolymph lipids to reduce the lipid peroxidation level, lessening tissue damages (33). These studies suggest that the elevated ergosterol level in the hemolymph after infection is unlikely due to increased feeding or decreased defecating behaviors in response to pathogens invasion, raising the possibility that infection induced ergosterol level change is regulated intrinsically by host lipid metabolism machineries.
To investigate the regulatory mechanism for ergosterol level, we collected the whole flies at indicated time points (12, 24, 48 and 72 hpi) after Ecc15 infection, and performed a temporal transcriptomic analysis for their global RNA expression profiles (Supplementary Table 3). Large number of genes were upregulated after Ecc15 infection, compared with PBS-injected control flies (Figure 3A). Interestingly, along with the infection time, the numbers of upregulated genes increased (154, 170, 203 and 217) while the numbers of down-regulated decreased (146, 127, 6 and 4) (Figure 3A), indicating the different response profiles at different time after infection.
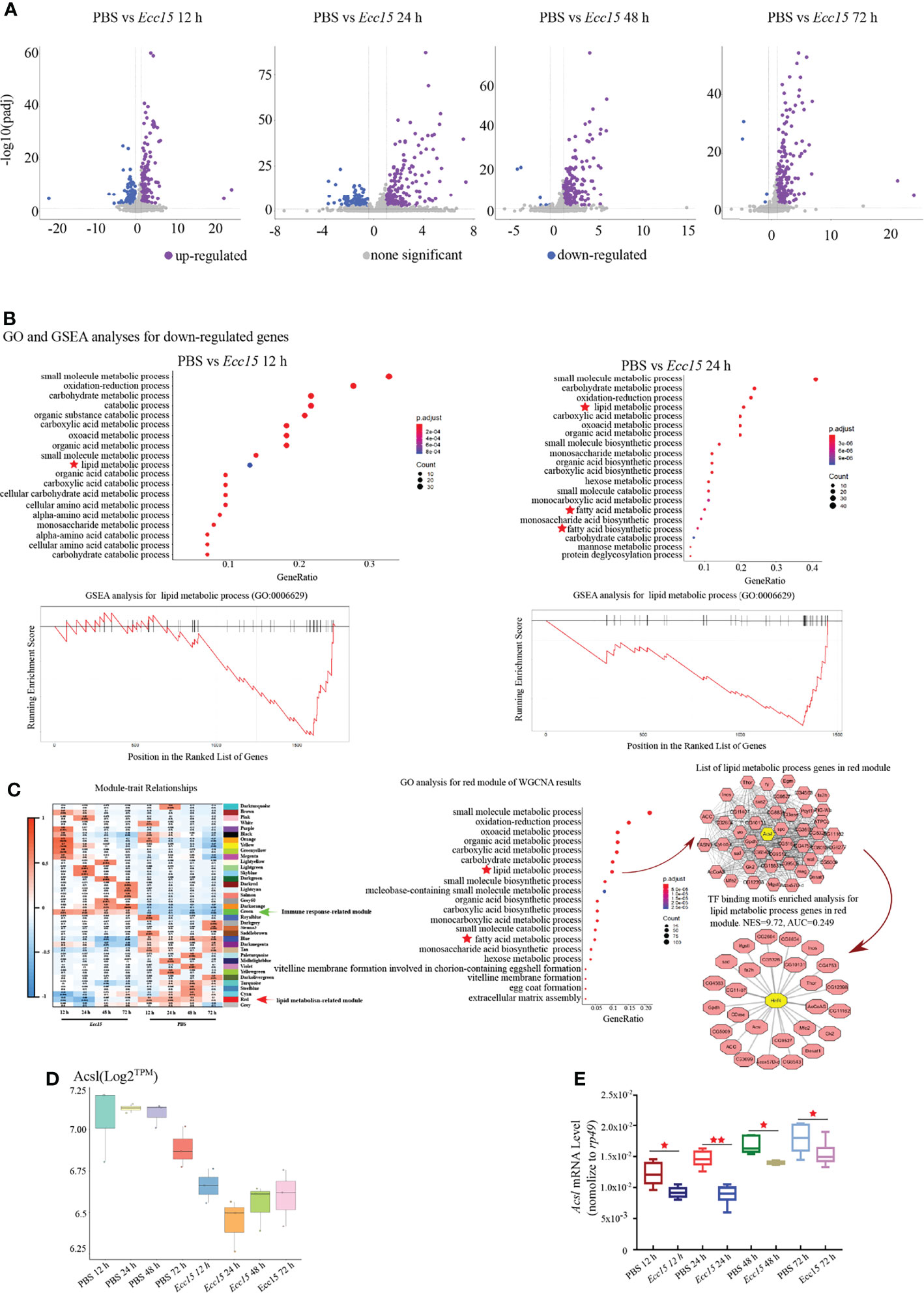
Figure 3 Time-course transcriptomic analysis reveals that changes of lipid metabolism related gene expression profiles occur in response to Ecc15 infection. (A). Volcano plot of differentially expressed genes (DEGs) with >2-fold up- or >0.7-fold down-regulation changes (p-adj<0.001) in flies infected with Ecc15 at indicated time points (12, 24, 48, 72 hpi), compared with control flies. (B). GO analysis (upper panel) for the down-regulated genes in (A) and GSEA analysis (lower panel) for genes with GO termed “lipid metabolic process” (GO:0006629) in down-regulated genes at indicated time points (12, 24 hpi). (C). Left panel, WGCNA analysis for time-course transcriptomic data. Middle panel, GO analysis for Module Red of WGCNA result. Right panel, hub-gene analysis (right-upper) and TF binding motifs enrichment analysis (right-lower) for genes with GO termed “lipid metabolic process” (GO:0006629) in Module Red. (D, E). Transcripts per million (TPM) (D) and Q-PCR (E) plot of Acsl mRNA level in control (PBS) and Ecc15-challenged (Ecc15) flies at indicated time points (12, 24, 48, 72 hpi). *, p < 0.05; **, p < 0.001. (One-way ANOVA test with a Sidak test).
Next, we performed a Gene Ontology (GO) analysis for the up-regulated genes in Ecc15-challenged flies. The GO analysis revealed that genes related to host defense were enriched (Supplementary Figures S1A, B), as expected. Notably, genes involving lipid metabolism was significantly enriched in the downregulated genes (designated as “lipid metabolic process”, “fatty acid metabolic process” and “fatty acid biosynthetic process”) (Figure 3B; Supplementary Table 4), suggesting that lipid metabolism was extensively regulated during infection. Next, we performed Gene Set Enrichment Analysis (GSEA) analysis for the down-regulated genes after Ecc15 infection, genes involving lipid metabolic process were highly enriched (Figure 3B). These data indicated a systemic adaption in host lipid metabolism during bacterial infection.
To further identify the genes or gene sets whose expression profiles were most tightly correlated with host defense in the bacterial infection processes, a weighted correlation network analysis (WGCNA) was performed. We identified 37 signature modules with distinctive correlation features to Ecc15 infection (Figure 3C). Module Green was significantly positively correlated to infection at 12 and 24 hpi, in which the immune related genes (49 genes in GO:0006955) were significantly enriched, suggesting that flies initiated an extensive immune response against bacterial stress at a relatively early stage (Figure 3C, Supplementary Figure 2 and Supplementary Table 5). Interestingly, Module Red, which exhibited significantly negative correlation with Ecc15 infection, was identified (Figure 3C). In this module, a bunch of lipid metabolism-related genes (47 genes in GO:0006629, designated as “lipid metabolic process” and “fatty acid metabolic process”) was enriched (Figure 3C and Supplementary Table 6). Further network analysis identified Acsl, a long-chain fatty acid-CoA synthetase, as a hub gene in the Module Red. Both RNA-Seq and qPCR results confirmed that Acsl were down-regulated in Ecc15-infected flies (Figures 3D, E).
To find out the key regulator(s) responsible to these down-regulated lipid metabolic genes in this module, we performed a transcription factor (TF) binding motifs enrichment analysis, and the result supported a pivotal role of HNF4, a master regulator for lipid mobilization and fatty acid beta-oxidation, as a core transcription factor for lipid metabolism change during Ecc15 infection (Figure 3C). Altogether, these data demonstrate that host coordinates multiple biological machineries, especially lipid metabolism processes, to efficiently fight with pathogen invasion.
Flies With Acsl Depletion Are More Resistant to Bacterial Infection Than the Wild Type
Our previous results have established the pro-survival role of ergosterol in flies challenged with bacteria. Thus, we wonder whether Acsl, the hub gene in infection-induced lipid metabolism profile, also regulates ergosterol level and plays a role in host defense by regulating ergosterol level. Systemic knockdown of Acsl lead to flies lethal, thus, we utilized temperature-sensitive (with Tub-Gal80ts) Gal4/UAS system to spare the normal functions of Acsl during development. Taking this system, we ubiquitously knockdown Acsl mRNA level in adult flies (Supplementary Figure 3), and examined their susceptibility to bacterial infection. Indeed, the survival rates of Acsl RNAi flies were significantly higher than wild type flies against systemic Ecc15 infection (Figure 4A). Notably, Acsl knockdown also rendered flies more resistant when challenged with E.fa, a Gram positive bacteria (Figure 4B). Next, we collected the fly hemolymph and determined the ergosterol level by LC/MS in Acsl RNAi flies. Ergosterol level in the hemolymph was significantly increased when flies were ubiquitously depletion of Acsl (Figure 4C). These results suggest that modulation of ergosterol level through Acsl manipulation regulates the sensitivity of flies to bacterial infection, representing a previously uncharacterized role of Acsl-ergosterol metabolism pathway in host defense to pathogen invasion.
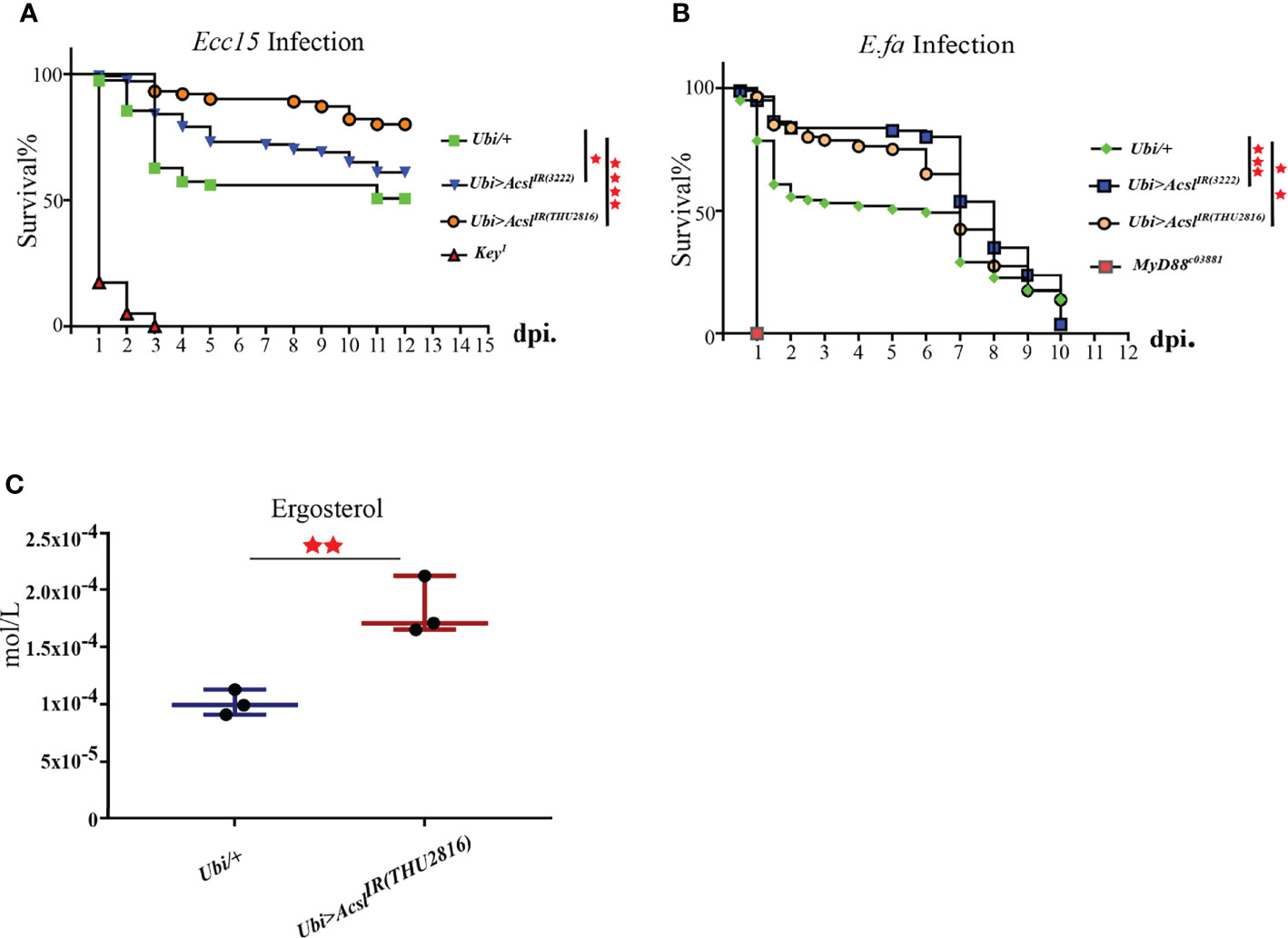
Figure 4 Flies with Acsl depletion are more resistant to bacterial infection than the wild type. (A). Survival curves of Acsl RNAi and key1 flies infected with Ecc15. *, p<0.05; ****, p<0.001. (Gehan-Breslow-Wilcoxon test). (B). Survival curves of Acsl RNAi and Myd88c03881 flies infected with E.fa. **, p<0.01; ***, p<0.005. (Gehan-Breslow-Wilcoxon test). (C). Box plot of ergosterol in control and Acsl RNAi flies. **, p<0.01 (wilcox test).
Discussion
Pathogen invasion triggers an immune response in the host. Flies respond to immune challenges by activating a conserved Toll-Dif or/and Imd-Relish pathway that stimulates synthesis and secretion of substantial antimicrobial peptides (AMPs) into hemolymph (37). Usually, activation of innate immune signaling is energetically demanding, requiring the coordinating of nutritional supply system and host defense system, to achieve efficient pathogens killing. Lipid metabolism has been shown to be involved in providing energy and signal transduction processes during immune responses. In both flies and humans, excessive immune activation leads to metabolic dysregulation, while loss of metabolic homeostasis usually results in the weakening of the immune system, demonstrating an intimate link between lipid metabolism and immune response (38). The coordination of immune and metabolic pathways and functional conservation of these pathways between flies and mammals make Drosophila an ideal model for the study of immune-metabolism crosstalk at organismal level.
Metabolic adaption occurs in response to bacterial infection. However, how bacterial infection leads to lipid metabolism changes during infection and what are the main lipid metabolites that involving host defense are still largely unknown. We performed a temporal lipidomic and transcriptomic analysis to investigate the dynamic profiles of lipid metabolic genes and lipid metabolites in the hemolymph of flies challenged with Ecc15 infection. Systemic metabolic change was observed in Ecc15-infected flies, indicating a complicated role of specific lipid metabolites in respond to pathogen invasion.
Lipidomic analysis revealed profound changes of several components of phospholipids, sphingolipids and sterols, such as PIs, PEs, Cer/OH-Cer, DAGs, and ergosterol, implying their potential roles in host defense. Rather than cholesterol, ergosterol is the main sterol in flies (35). We wonder whether the increased ergosterol induced by Ecc15 infection is functional or not in immune system. Notably, artificially increase of ergosterol level in the hemolymph, by oral feeding or genetic manipulation, significantly enhanced host defense to bacterial infection by promoting host survival. Consistent with our finding, Adrien Franchet et al. found that feeding ergosterol promoted host survival against the microsporidium Tubulinosema ratisbonensis through an unknown mechanism (39).
Then, as the main sterol in flies, how does ergosterol function in immune response? We proposed that ergosterol may function as cholesterol substitute in immune response, based on the following: 1) Ergosterol and cholesterol share highly similar molecular structure. 2) They can substitute each other for supporting fly growth and survival. Based on these, it is plausible that ergosterol has the ability to be converted into hormones, such as ecdysone, which plays an important role in promoting innate immune response, by increasing phagocytic activities on blood cells or promoting pattern recognition receptors (i.e. PGRP-LC and PGRP-LE) expression (40). However, regardless of the structural similarity to cholesterol (41), ergosterol cannot be converted into ecdysone, instead, into 24-epi-MaA, dh-methylE and dhMaA, which can also support the whole life cycle of fruit fly (42). Our data showed that ergosterol did not alter AMPs mRNA level and bacterial load, indicating that ergosterol did not promote immune response signaling activation, even though ergosterol or its metabolites showed overlapped functions with ecdysone in supporting animal growth.
Ergosterol is the main sterols in the plasm membrane (PM) in flies, and sterols are required for special microdomain formation, for example, lipid rafts and immune-synapses, on the PM. We wonder whether ergosterol is required for host defense by boosting immune signaling activation through clustering immune components for pathogens recognition and sequential signal transduction. However, our findings are not supportive to this hypothesis due to the observations that ergosterol feeding did not change AMPs expression level. Interestingly, increased ergosterol level caused by Acsl depletion in Huang et al.’s study was associated with impaired Dpp signaling (43), implying a context (e.g., tissue, pathway)-dependent action of ergosterol in flies.
Alternatively, ergosterol severs as an integrate component of membrane and promotes membrane fusion (44, 45), raising the possibility that ergosterol tends to participate in maintaining the immune homeostasis and tissue repair (resilience), instead of killing pathogens or activating immune response (resistance). This possibility is supported by the findings that in yeast the ergosterol can sustain membrane integrity by regulating membrane fluidity and permeability (46) through interacting with phospholipids and membrane-anchored proteins (47). Additionally, the tolerance capacity of yeast to environmental stresses is tightly correlated to its intracellular ergosterol levels (48). Further studies are needed to decipher whether ergosterol functions in membrane repair and integrity.
Our further study identified Acsl as the key regulator for lipid metabolism adaption in respond to pathogen infection. It is reported that knockdown of Acsl increased the ergosterol level in fly brains (43). Consistently, our results showed ergosterol level in the hemolymph was increased in Acsl knockdown flies. So, how does Acsl affects ergosterol level in vivo? As a key player in fatty acid synthesis with its palmitoyl-CoA ligase activity, Acsl is localized on ER (43, 49), promotes de novo formation of lipid droplets (49), which are identified as the central hub integrating and coordinating cellular metabolism and the immune system (50), to cope with intrinsic and extrinsic stresses. Lipid droplets are primarily composed of polar lipids and residual proteins on the monolayer membrane surface with neutral lipids in the core content, including TAG and sterols (51). Interestingly, it is reported that in yeast the ergosterol can be converted into the form of steryl-esters (SEs) and stored in lipid droplets, serving as a sterol pool to maintain the balance of intracellular sterols (52, 53). Although lacking of direct evidence, it is possible that Acsl reduces the free ergosterol level by promoting the ergosterol packing into lipid droplets, while knockdown of Acsl increases ergosterol by breaking down the lipid droplets, releasing ergosterol into the hemolymph. Recently, Huang et al. reported that knockdown of Acsl in the fly brain resulted in significant changes of several lipids, such as ergosterol, phosphoethanolamine ceramide (CerPE), and mannosyl glucosylceramide (MacCer), by an unknown mechanism, impairing signaling transduction (43). In line with our observations, it is suggested that lipid metabolites may work synergistically to deal with intrinsic and extrinsic stresses.
Taken together, our study identified a previously unidentified role of Acsl-ergosterol metabolism axis in immune response, providing a potential alternative way for modulating host defense for infection diseases treatment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Genome Sequence Archive in BIG Data Center (https://bigd.big.ac.cn), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number: CRA006720.
Author Contributions
ZD, RJ and CW conceived the study, designed the experiments, discussed the results, and wrote the manuscript. ZD, YY, JZL, and BZ performed the experiments and analysis. ZD generated the figures. GS provided technical assistance in lipidomic assay. JYL discussed the results. All authors read and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study has been financially supported by the National Key R&D Program of China (2021YFA0805800 and 2020YFA0803202 to RJ), the National Natural Science Foundation of China (31970538 to RJ, 32100703 to CW, 32000574), Guangzhou Medical University Discipline Construction Funds (Basic Medicine) (JCXKJS2022A02 to RJ), the 111 Project (D18010 to RJ), the Local Innovative and Research Teams Project of Guangdong Perl River Talents Program (2017BT01S155 to RJ), the Special Innovation Projects of Universities in Guangdong Province (2018KTSCX182 to JL), the Medical Scientific Research Foundation of Guangdong Province (A2019292 to JL), the Natural Science Foundation of Guangdong, China (2017A030310403 to ZD), and the grant of the State Key Laboratory of Respiratory Disease, Guangdong-Hong Kong-Macao Joint Laboratory of Respiratory Infectious Disease (GHMJLRID-Z-202106 to CW).
Conflict of Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Fly stocks were obtained from Bloomington Drosophila Stock Center (USA), Vienna Drosophila Resource Center (Austria) and TsingHua Fly Center (China). We are grateful to Profs Lei Pan and Dominique Ferrandon for their kind share of bacterial and fly strains used in this study. We thank all the Jiao Lab members for constructive discussion and assistance in this project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.933137/full#supplementary-material
Supplementary Figure 1 | GO analysis and Heatmap for up-regulated genes after Ecc15 infection. (A1-A4) GO analysis for the up-regulated genes at indicated time points (12, 24, 48, 72 hpi). (B) Heatmap plot for AMPs genes of Imd pathway at indicated time points (12, 24, 48, 72 hpi).
Supplementary Figure 2 | GO analysis for genes in Module Green in WGCNA result.
Supplementary Figure 3 | Knockdown efficiency of Acsl RNAi lines. Q-PCR plot of Acsl mRNA level in control and Acsl RNAi flies. Ubi/w1118, w; pUbi-Gal4/+; pTub-Gal80ts/+. Ubi>AcslIR(3222), w; pUbi-Gal4/UAS-AcslIR(3222); pTub-Gal80ts/+. Ubi> AcslIR(THU2816), w; pUbi-Gal4/+; pTub-Gal80ts/UAS- AcslIR(THU2816). ****, p<0.001. (One-way ANOVA test with a Sidak test).
Supplementary Table 1 | List of lipid metabolites detected in lipidomic analysis.
Supplementary Table 2 | Primers used in this study.
Supplementary Table 3 | Gene expression matrix (TPM and Counts) in Ecc15 challenged flies at indicated time points.
Supplementary Table 4 | Top 50 GO list in down-regulated DEGs gene after Ecc15 infection.
Supplementary Table 5 | Top 50 GO list in WGCNA Green Module.
Supplementary Table 6 | Top 50 GO list in WGCNA Red Module.
References
1. Hsu AP, Holland SM. Host Genetics of Innate Immune System in Infection. Curr Opin Immunol (2022) 74:140–9. doi: 10.1016/j.coi.2021.11.003
2. Buchon N, Silverman N, Cherry S. Immunity in Drosophila Melanogaster - From Microbial Recognition to Whole-Organism Physiology. Nat Rev Immunol (2014) 14:796–810. doi: 10.1038/nri3763
3. Kounatidis I, Ligoxygakis P. Drosophila as a Model System to Unravel the Layers of Innate Immunity to Infection. Open Biol (2012) 2:120075. doi: 10.1098/rsob.120075
4. Parsons B, Foley E. Cellular Immune Defenses of Drosophila Melanogaster. Dev Comp Immunol (2016) 58:95–101. doi: 10.1016/j.dci.2015.12.019
5. Vesala L, Hultmark D, Valanne S. Recent Advances in Drosophila Cellular and Humoral Innate Immunity. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.598618
6. Hotamisligil G. Inflammation, metaflammation and immunometabolic disorders. Nature (2017) 542:177–85. doi: 10.1038/nature21363
7. Krejcova G, Danielova A, Nedbalova P, Kazek M, Strych L, Chawla G, et al. Drosophila Macrophages Switch to Aerobic Glycolysis to Mount Effective Antibacterial Defense. Elife (2019) 8:e50414. doi: 10.7554/eLife.50414
8. Martinez BA, Hoyle RG, Yeudall S, Granade ME, Harris TE, Castle JD, et al. Innate Immune Signaling in Drosophila Shifts Anabolic Lipid Metabolism From Triglyceride Storage to Phospholipid Synthesis to Support Immune Function. PloS Genet (2020) 16(11):e1009192. doi: 10.1101/2020.03.01.972208
9. DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The Immune Response Attenuates Growth and Nutrient Storage in Drosophila by Reducing Insulin Signaling. Proc Natl Acad Sci United States America (2009) 106:20853–8. doi: 10.1073/pnas.0906749106
10. Molaei M, Vandehoef C, Karpac J. NF-Kappa B Shapes Metabolic Adaptation by Attenuating Foxo-Mediated Lipolysis in Drosophila. Dev Cell (2019) 49(5):802. doi: 10.1016/j.devcel.2019.04.009
11. Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, Watnick PI. The Drosophila Immune Deficiency Pathway Modulates Enteroendocrine Function and Host Metabolism. Cell Metab (2018) 28:449–+. doi: 10.1016/j.cmet.2018.05.026
12. Goto A, Okado K, Martins N, Cai H, Barbier V, Lamiable O, et al. The Kinase IKK Beta Regulates a STING- and NF-Kappa B-Dependent Antiviral Response Pathway in Drosophila. Immunity (2018) 49:225–+. doi: 10.1016/j.immuni.2018.07.013
13. Martin M, Hiroyasu A, Guzman RM, Roberts SA, Goodman AG. Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep (2018) 23:3537–+. doi: 10.1016/j.celrep.2018.05.029
14. Akhmetova K, Balasov M, Chesnokov I. Drosophila STING Protein has a Role in Lipid Metabolism. Elife (2021) 10:e67358. doi: 10.7554/eLife.67358
15. Vila IK, Chamma H, Steer A, Saccas M, Taffoni C, Turtoi E, et al. STING Orchestrates the Crosstalk Between Polyunsaturated Fatty Acid Metabolism and Inflammatory Responses. Cell Metab (2022) 34:125–+. doi: 10.1016/j.cmet.2021.12.007
16. Koyama T, Texada MJ, Halberg KA, Rewitz K. Metabolism and Growth Adaptation to Environmental Conditions in Drosophila. Cell Mol Life Sci (2020) 77:4523–51. doi: 10.1007/s00018-020-03547-2
17. He X, Yu J, Wang M, Cheng Y, Han Y, Yang S, et al. Bap180/Baf180 is Required to Maintain Homeostasis of Intestinal Innate Immune Response in Drosophila and Mice. Nat Microbiol (2017) 2:17056. doi: 10.1038/nmicrobiol.2017.56
18. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
19. Li B, Dewey CN. RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf (2011) 12:323. doi: 10.1186/1471-2105-12-323
20. Langfelder P, Horvath S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
21. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS (2012) 16:284–7. doi: 10.1089/omi.2011.0118
22. Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-Cell Regulatory Network Inference and Clustering. Nat Methods (2017) 14:1083–6. doi: 10.1038/nmeth.4463
23. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data With Deseq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
24. Lam SM, Zhang C, Wang Z, Ni Z, Zhang S, Yang S, et al. A Multi-Omics Investigation of the Composition and Function of Extracellular Vesicles Along the Temporal Trajectory of COVID-19. Nat Metab (2021) 3:909–22. doi: 10.1038/s42255-021-00425-4
25. Lam SM, Wang R, Miao H, Li B, Shui G. An Integrated Method for Direct Interrogation of Sphingolipid Homeostasis in the Heart and Brain Tissues of Mice Through Postnatal Development Up to Reproductive Senescence. Anal Chim Acta (2018) 1037:152–8. doi: 10.1016/j.aca.2018.01.015
26. Lu J, Lam SM, Wan Q, Shi L, Huo Y, Chen L, et al. High-Coverage Targeted Lipidomics Reveals Novel Serum Lipid Predictors and Lipid Pathway Dysregulation Antecedent to Type 2 Diabetes Onset in Normoglycemic Chinese Adults. Diabetes Care (2019) 42:2117–26. doi: 10.2337/dc19-0100
27. Lam SM, Wang Z, Li J, Huang X, Shui G. Sequestration of Polyunsaturated Fatty Acids in Membrane Phospholipids of Caenorhabditis Elegans Dauer Larva Attenuates Eicosanoid Biosynthesis for Prolonged Survival. Redox Biol (2017) 12:967–77. doi: 10.1016/j.redox.2017.05.002
28. Yang HR, Hultmark D. Tissue Communication in a Systemic Immune Response of Drosophila. Fly (2016) 10:115–22. doi: 10.1080/19336934.2016.1182269
29. Vessichelli M, Mariggio S, Varone A, Zizza P, Di Santo A, Amore C, et al. The Natural Phosphoinositide Derivative Glycerophosphoinositol Inhibits the Lipopolysaccharide-Induced Inflammatory and Thrombotic Responses. J Biol Chem (2017) 292:12828–41. doi: 10.1074/jbc.M116.773861
30. Gaggini M, Ndreu R, Michelucci E, Rocchiccioli S, Vassalle C. Ceramides as Mediators of Oxidative Stress and Inflammation in Cardiometabolic Disease. Int J Mol Sci (2022) 23:2719. doi: 10.3390/ijms23052719
31. Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, et al. Involvement of Oxidative Stress-Induced Abnormalities in Ceramide and Cholesterol Metabolism in Brain Aging and Alzheimer’s Disease. Proc Natl Acad Sci (2004) 101:2070–5. doi: 10.1073/pnas.0305799101
32. Kota V, Hama H. 2′-Hydroxy Ceramide in Membrane Homeostasis and Cell Signaling. Adv Biol Regul (2014) 54:223–30. doi: 10.1016/j.jbior.2013.09.012
33. Li XX, Rommelaere S, Kondo S, Lemaitre B. Renal Purge of Hemolymphatic Lipids Prevents the Accumulation of ROS-Induced Inflammatory Oxidized Lipids and Protects Drosophila From Tissue Damage. Immunity (2020) 52:374–+. doi: 10.1016/j.immuni.2020.01.008
34. Heier C, Kuhnlein RP. Triacylglycerol Metabolism in Drosophila Melanogaster. Genetics (2018) 210:1163–84. doi: 10.1534/genetics.118.301583
35. Knittelfelder O, Prince E, Sales S, Fritzsche E, Wöhner T, Brankatschk M, et al. Sterols as Dietary Markers for Drosophila Melanogaster. Biochim Biophys Acta (BBA)-Mol Cell Biol Lipids (2020) 1865:158683. doi: 10.1016/j.bbalip.2020.158683
36. Surendran S, Huckesfeld S, Waschle B, Pankratz MJ. Pathogen-Induced Food Evasion Behavior in Drosophila Larvae. J Exp Biol (2017) 220:1774–80. doi: 10.1242/jeb.153395
37. Imler JL. Overview of Drosophila Immunity: A Historical Perspective. Dev Comp Immunol (2014) 42:3–15. doi: 10.1016/j.dci.2013.08.018
38. Dionne MS. Immune-Metabolic Interaction in Drosophila. Fly (2014) 8:75–9. doi: 10.4161/fly.28113
39. Franchet A, Niehus S, Caravelle G, Ferrandon D. Phosphatidic Acid as a Limiting Host Metabolite for the Proliferation of the Microsporidium Tubulinosema Ratisbonensis in Drosophila Flies. Nat Microbiol (2019) 4:645–55. doi: 10.1038/s41564-018-0344-y
40. Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, et al. Ecdysone Triggered PGRP-LC Expression Controls Drosophila Innate Immunity. EMBO J (2013) 32:1626–38. doi: 10.1038/emboj.2013.100
41. Silva L, Coutinho A, Fedorov A, Prieto M. Competitive Binding of Cholesterol and Ergosterol to the Polyene Antibiotic Nystatin A Fluorescence Study. Biophys J (2006) 90:3625–31. doi: 10.1529/biophysj.105.075408
42. Lavrynenko O, Rodenfels J, Carvalho M, Dye NA, Lafont R, Eaton S, et al. The Ecdysteroidome of Drosophila: Influence of Diet and Development. Development (2015) 142:3758–68. doi: 10.1242/dev.124982
43. Huang Y, Huang S, Lam SM, Liu ZH, Shui GH, Zhang YQ. Acsl, the Drosophila Ortholog of Intellectual-Disability-Related ACSL4, Inhibits Synaptic Growth by Altered Lipids. J Cell Sci (2016) 129:4034–45. doi: 10.1242/jcs.195032
44. Hurst LR, Fratti RA. Lipid Rafts, Sphingolipids, and Ergosterol in Yeast Vacuole Fusion and Maturation. Front Cell Dev Biol (2020) 8. doi: 10.3389/fcell.2020.00539
45. Jin H, McCaffery JM, Grote E. Ergosterol Promotes Pheromone Signaling and Plasma Membrane Fusion in Mating Yeast. J Cell Biol (2008) 180:813–26. doi: 10.1083/jcb.200705076
46. Dupont S, Lemetais G, Ferreira T, Cayot P, Gervais P, Beney L. Ergosterol Biosynthesis: A Fungal Pathway for Life on Land? Evol: Int J Organic Evol (2012) 66:2961–8. doi: 10.1111/j.1558-5646.2012.01667.x
47. Krumpe K, Frumkin I, Herzig Y, Rimon N, Özbalci C, Brügger B, et al. Ergosterol Content Specifies Targeting of Tail-Anchored Proteins to Mitochondrial Outer Membranes. Mol Biol Cell (2012) 23:3927–35. doi: 10.1091/mbc.e11-12-0994
48. Aguilera F, Peinado R, Millan C, Ortega J, Mauricio J. Relationship Between Ethanol Tolerance, H+-ATPase Activity and the Lipid Composition of the Plasma Membrane in Different Wine Yeast Strains. Int J Food Microbiol (2006) 110:34–42. doi: 10.1016/j.ijfoodmicro.2006.02.002
49. Zhang Y, Chen D, Wang Z. Analyses of Mental Dysfunction-Related ACSl4 in Drosophila Reveal its Requirement for Dpp/BMP Production and Visual Wiring in the Brain. Hum Mol Genet (2009) 18:3894–905. doi: 10.1093/hmg/ddp332
50. Bosch M, Sanchez-Alvarez M, Fajardo A, Kapetanovic R, Steiner B, Dutra F, et al. Mammalian Lipid Droplets are Innate Immune Hubs Integrating Cell Metabolism and Host Defense. Science (2020) 370:309–+. doi: 10.1126/science.aay8085
51. Walther TC, Farese RV. Lipid Droplets and Cellular Lipid Metabolism. Annu Rev Biochem (2012) 81:687–714. doi: 10.1146/annurev-biochem-061009-102430
52. Choudhary V, Schneiter R. Pathogen-Related Yeast (PRY) Proteins and Members of the CAP Superfamily are Secreted Sterol-Binding Proteins. Proc Natl Acad Sci (2012) 109:16882–7. doi: 10.1073/pnas.1209086109
Keywords: Drosophila, innate immunity, lipid metabolism, lipidomics, transcriptomics, ergosterol, ACSL
Citation: Deng Z, Yang Y, Luo J, Zhang B, Liu J, Shui G, Jiao R and Wei C (2022) An Integrated Transcriptomics and Lipidomics Analysis Reveals That Ergosterol Is Required for Host Defense Against Bacterial Infection in Drosophila. Front. Immunol. 13:933137. doi: 10.3389/fimmu.2022.933137
Received: 30 April 2022; Accepted: 15 June 2022;
Published: 07 July 2022.
Edited by:
Akira Goto, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Likui Feng, The Rockefeller University, United StatesKwang-Zin Lee, Fraunhofer Society (FHG), Germany
Copyright © 2022 Deng, Yang, Luo, Zhang, Liu, Shui, Jiao and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Renjie Jiao, cmppYW9AZ3pobXUuZWR1LmNu; Chuanxian Wei, d2VpY2h1YW54aWFuQGd6aG11LmVkdS5jbg==
 Zihao Deng1
Zihao Deng1 Jiazhen Luo
Jiazhen Luo Renjie Jiao
Renjie Jiao Chuanxian Wei
Chuanxian Wei