Corrigendum: A feedback regulatory loop involving dTrbd/dTak1 in controlling IMD signaling in Drosophila Melanogaster
- 1Centre for Developmental Biology, School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2School of Preclinical Medicine, Wannan Medical College, Wuhu, China
Negative regulators of the inflammatory responses are essential for the maintenance of immune homeostasis and organismal fitness. In Drosophila, the deubiquitinase (Dub) dTrbd selectively restricts the K63-linked ubiquitination modification of dTak1, a pivotal kinase of the IMD signaling pathway, to regulate the IMD innate immune response. However, which domain and how it functions to enable dTrbd’s activity remain unexplored. Here, we provide compelling evidence showing that the NZF domain of dTrbd is essential for its association with dTak1. Meanwhile, the Linker region of dTrbd is involved in modulating its condensation, a functional state representing the Dub enzymatical activity of dTrbd. Of interest, the activated IMD signals following bacterial stimuli enhance the dTrbd/dTak1 interaction, as well as the condensate assembly and Dub enzymatical activity of dTrbd. Collectively, our studies shed light on the dual mechanisms by which the IMD signaling-mediated feedback loop of dTrbd/dTak1 precisely regulates the innate immune response in Drosophila.
Introduction
As a rapid and critical defensive manner, innate immunity represents the first line of the host immune system (1–3). It is triggered by the pattern recognition receptors (PRRs), which are able to sense both extra-cellular germs and intra-cellular infections or damages within hours (1, 4). This triggering further initiates a non-specific inflammatory response, produces pleiotropic effective molecules without immune memory, and amplifies the protective reaction to other cells and organs to dominate adaptive immune responses (5).
Drosophila melanogaster has been one of the most well-known animal models for deciphering the fundamental molecular mechanisms underlying the precise regulation of host inflammatory reaction (2, 6). It harbors three main advantages: 1) as an insect whose defensive reaction only relies on innate immunity, Drosophila is an ideal model for investigating innate immune response without sophisticated interfering or cross-talk of adaptive immunity; 2) the great evolutionary congruence between Drosophila and mammals makes the pioneering discoveries regarding innate immunity of fruit flies be a potentially pivotal theoretic basis for studying mammalian systems (7); 3) performing genetic studies on Drosophila is highly convenient thanks to the powerful genetic manipulation tools and availability of various Drosophila mutants and transgenes.
In Drosophila, the innate immune response is mainly controlled by two signaling pathways, the Toll and the immune deficiency (IMD) pathways (2, 8, 9), which share substantial similarities with the mammalian MyD88-dependent Toll-like receptor (TLR) and the tumor necrosis factor receptor (TNFR) pathways, respectively (8, 10). The PRRs of both pathways sense the environmental pathogen-associated molecular patterns (PAMPs) and further activate a series of downstream caspase reactions, leading to the translocation of the transcription factor nuclear factor-kappa B (NF-κB) into the nucleus and the expression of a set of anti-microbial peptides (AMPs) (2, 6, 11, 12). Undoubtedly, a timely and robust innate immune response is pivotal for flies surviving in the battles against pathogenic invaders. Excessive activation of the innate immune response, however, is also deleterious, leading to detrimental consequences in various tissues/organs and even organismal mortality (8, 13–17). Thus, turning down the activated innate immune signaling pathways by negative regulators is fundamental for the maintenance of immune homeostasis and fitness (6, 8, 9, 16). It has been suggested that the IMD signaling pathway is precisely regulated by multiple negative modulators (8, 16). For instance, several proteins belonging to the peptidoglycan recognition protein (PGRP) family have been shown to be involved in the degradation of DAP- and Lys-type peptidoglycans (18–20), thereby antagonizing IMD signaling. Inside the cells, passive players such as Caspar (21), Cyld (22), Dnr1 (23, 24), dTrbd (13), Pirk (25, 26), Scny (27), and SkpA (28) have been demonstrated to play essential roles in down-regulating the IMD pathway through various regulatory manners. Among them, we noticed a deubiquitinase (Dub) dTrbd, which selectively modulates the K63-linked ubiquitination of dTak1 to attenuate IMD signaling. However, which domain is required for its biological function remains poorly understood.
In this study, we report a feedback regulatory loop involving the dTak1/dTrbd axis to control the IMD signaling pathway in Drosophila. We show that the N-terminal NZF domain of dTrbd is both required and sufficient for its binding to dTak1 for deubiquitination, whereas the Linker region mediates dTrbd condensation, which represents a functional state for executing its Dub enzymatical activity. Moreover, we find that bacterial stimuli-induced IMD activation not only reinforces the physical association of dTrbd/dTak1, but also positively facilitates dTrbd condensation to enhance the Dub enzymatical activity of dTrbd, thus negatively contributing to IMD signaling. Collectively, our results uncover dual mechanisms involving dTrbd/dTak1 to regulate the IMD signaling-mediated inflammatory reaction in Drosophila.
Results
Minor Alterations in dTrbd Expression During IMD Signaling
To explore whether dTrbd expression tightly parallels IMD signaling, we challenged Drosophila S2 cells with heat-killed Gram-negative bacterium Escherichia coli (E. coli) as previously described (29) and examined the mRNA profiles of dTrbd at various time points (0, 6, and 12 hours) via reverse transcription plus quantitative polymerase chain reaction (RT-qPCR) assays. As illustrated in Figure S1A, dTrbd transcript levels remained relatively similar along with the activation of the IMD signals, which were monitored by looking at the mRNA levels of the downstream AMPs genes attacin A (AttA) and cecropin A1 (CecA1) (Figures S1B, C) (30). Consistent results were obtained when we examined the protein levels of endogenous dTrbd in S2 cells by Western blot assays (Figures S1D, E) utilizing newly-generated anti-dTrbd antibodies (see Materials and Methods). To further validate our findings in vivo, we challenged male w1118 adults with freshly cultured E. coli. At various time points post bacterial infection (0, 6, and 12 hours), we collected fat body tissues from these flies to examine the dynamic expression profiles of dTrbd. As shown in Figures S1F-H, dTrbd was barely altered at the mRNA and protein levels in response to bacterial infection. Collectively, these results imply that dTrbd is not induced by pathogenic challenging both in cultured S2 cells and in Drosophila. Of note, the mRNA occurrences of dTak1, whose translational products are the regulatory targets of dTrbd in controlling the IMD signals (13), also displayed relatively potty amplitude fluctuations during IMD signaling both in vitro and in vivo (Figures S1I, J). These results were consistent with the previous findings in a microarray assay (25).
dTrbd Physically Associates With dTak1 via the N-terminal NZF Domain
We next sought to investigate the functional domain of dTrbd essential for its interaction with dTak1. The region between the N-terminal Npl4 Zinc-Finger (NZF) domain and the C-terminal Ovarian Tumor (OTU) domain was referred to as the Linker region (LR, Figure 1A). We constructed various truncated forms of dTrbd expressing plasmids (Figure 1A), namely dTrbdFL, dTrbdNZF, dTrbdLR, dTrbdOTU, dTrbdΔNZF, dTrbdΔLR, and dTrbdΔOTU, and transfected them together with plasmids expressing dTak1 into Drosophila S2 cells for co-immunoprecipitation (co-IP) experiments (GFP expressing or empty plasmids were utilized as the controls). As illustrated in Figure 1B, the N-terminal conserved NZF domain is both required and sufficient for the physical association between dTrbd and dTak1.
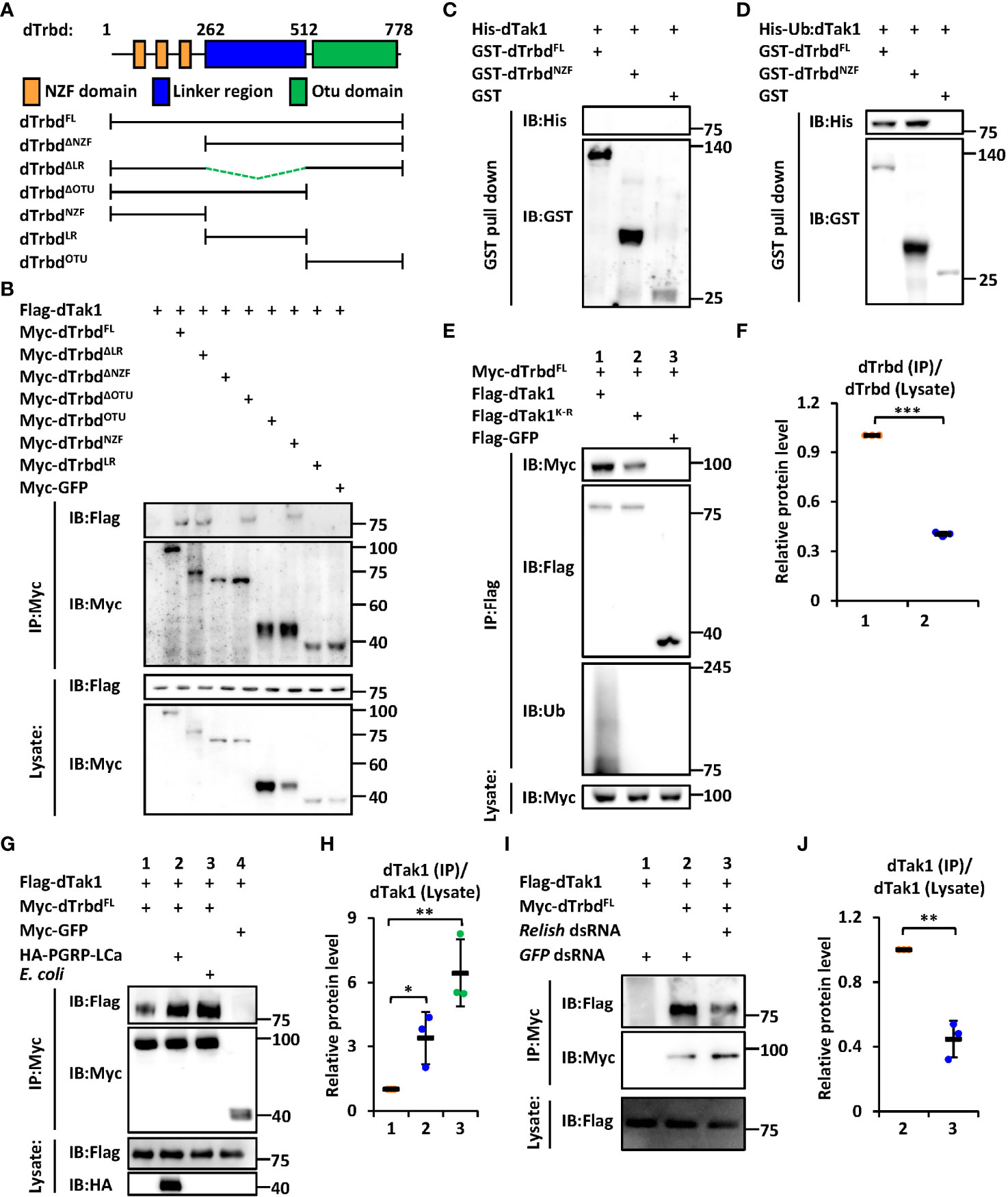
Figure 1 dTrbd associates with dTak1 via the N-terminal NZF domain. (A) Domain architecture of dTrbd. (B) Various combinations of expressing plasmids were transfected into S2 cells as indicated. 48 h post transfection, cells were lysed for immunoprecipitation assays using anti-Myc agarose beads, followed by Western blot assays. (C, D) Purified His-tagged dTak1 (C) or Ub-dTak1 (D) were incubated with GST-tagged dTrbdFL or dTrbdNZF (GST was used as the control). Samples were then subjected to GST pull-down and Western blot assays. (E, F) Indicated expressing plasmids were transfected into S2 cells for 48 h. Cell lysates were then prepared for immunoprecipitation and Western blot assays. Densitometry analysis to quantify the protein levels of Myc-dTrbd in the co-immunoprecipitant (E) is shown in (F). (G, H) S2 cells were transfected with various combinations of expressing plasmids for 36 h, followed by treatment of heat-killed E coli (Lane 4 in G) or PBS (Lanes 1-3 in G) for 12 h. Cells were then harvested for immunoprecipitation and Western blot assays as indicated (G). Densitometry analysis to quantify the protein levels of Flag-dTak1 in the co-immunoprecipitant (G) is shown in (H) (I, J) S2 cells were treated with different dsRNAs for 48 h and transfected with various expressing plasmids for 48 h as indicated. Cell lysates were then prepared for immunoprecipitation and Western blot assays (I). Densitometry analysis to quantify the protein levels of Flag-dTak1 in the co-immunoprecipitant (I) is shown in (J). In (F, H, J) data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
It has been suggested that the NZF domain functions mainly as a type of Ubiquitin-binding domains (31, 32). Based upon previous findings that dTak1 can undergo ubiquitination to regulate IMD signaling (13), we raised two explanations for the NZF domain binding to dTak1: 1) the NZF domain interacts with Ubiquitin, which behaves (directly or indirectly) as a “connective bridge” for the dTrbd/dTak1 association; 2) the NZF domain is also able to bind to other proteins/residues (dTak1 in our case) besides Ubiquitin. To further prove these hypotheses, we purified GST-tagged dTrbdFL and dTrbdNZF, as well as His-tagged dTak1 proteins from E. coli (Figure S2A), where it was not ubiquitinated (Figure S2B). As shown in the GST pull-down assays, we did not observe any detectable interactions of purified His-dTak1 with either GST-tagged dTrbdFL or dTrbdNZF (Figure 1C), suggesting dTrbd physically associates with dTak1 largely through Ubiquitin or indirectly. We next constructed a plasmid expressing dTak1 with a Ubiquitin at its N-terminus (referred to as Ub-dTak1) (Figure S2C). This fusion protein was further proven to be easily bound by both dTrbdFL and dTrbdNZF in the GST pull-down assays (Figure 1D), suggesting that the NZF domain of dTrbd is able to associate directly with the Ubiquitin-conjugated dTak1. To obtain more evidence, we selectively mutated all the lysine residues of dTak1 into arginines (referred to as dTak1K-R). Further co-IP experiments in Drosophila S2 cells displayed that dTak1K-R still readily co-immunoprecipitated with dTrbd when ubiquitination modifications were nearly fully abolished (Figures 1E, F). Notably, mutation of all the lysine residues in dTak1 somehow weakened its binding affinity to dTrbd (Figures 1E, F). Taken together, our results imply that dTrbd not only binds to the ubiquitinated dTak1, but also interacts with the non-ubiquitinated dTak1 through a(some) potential intermediator(s).
dTrbd Associates With dTak1 in an IMD Signal-Dependent Manner
To explore the involvement of IMD signaling in the binding of dTrbd to dTak1, we treated S2 cells with heat-killed E. coli or transfected them with plasmids expressing the constitutively active form of PGRP-LCa (33) to activate the IMD signaling pathway. These treatments indeed markedly induced downstream AMP expressions in S2 cells (Figures S3A, B). Our further co-IP approaches showed that the physical association between dTrbd and dTak1 was greatly strengthened when the IMD pathway was activated in S2 cells (Figures 1G, H). This observation was consistent with our in vivo examination of the dTrbd/dTak1 association (Figures S4A, B). In addition, silencing relish by the treatment of relish dsRNA in S2 cells largely prevented the interaction between dTrbd and dTak1 (Figures 1I, J), demonstrating that the IMD signaling pathway plays a critical role in impacting the dTrbd/dTak1 association.
The LR Regulates the Dub Enzymatical Activity of dTrbd
We next sought to identify the functional requirements of the domain(s) for dTrbd executing its Dub enzymatical activity. We transfected dTak1 expressing plasmids together with various truncated forms of dTrbd expressing plasmids into S2 cells and performed ubiquitination assays. As shown in Figures 2A, B and S5A, deletion of either the NZF or the OTU domain markedly prevented the functional roles of dTrbd in restricting the ubiquitination levels of dTak1 in S2 cells. It is not surprising for the authors to observe these results since the OTU domain is a necessarily catalytical center for dTrbd (13, 34), and the NZF domain is essential for dTrbd binding with its target (dTak1 in our case, Figure 1) for the catalytical reaction of deubiquitination.
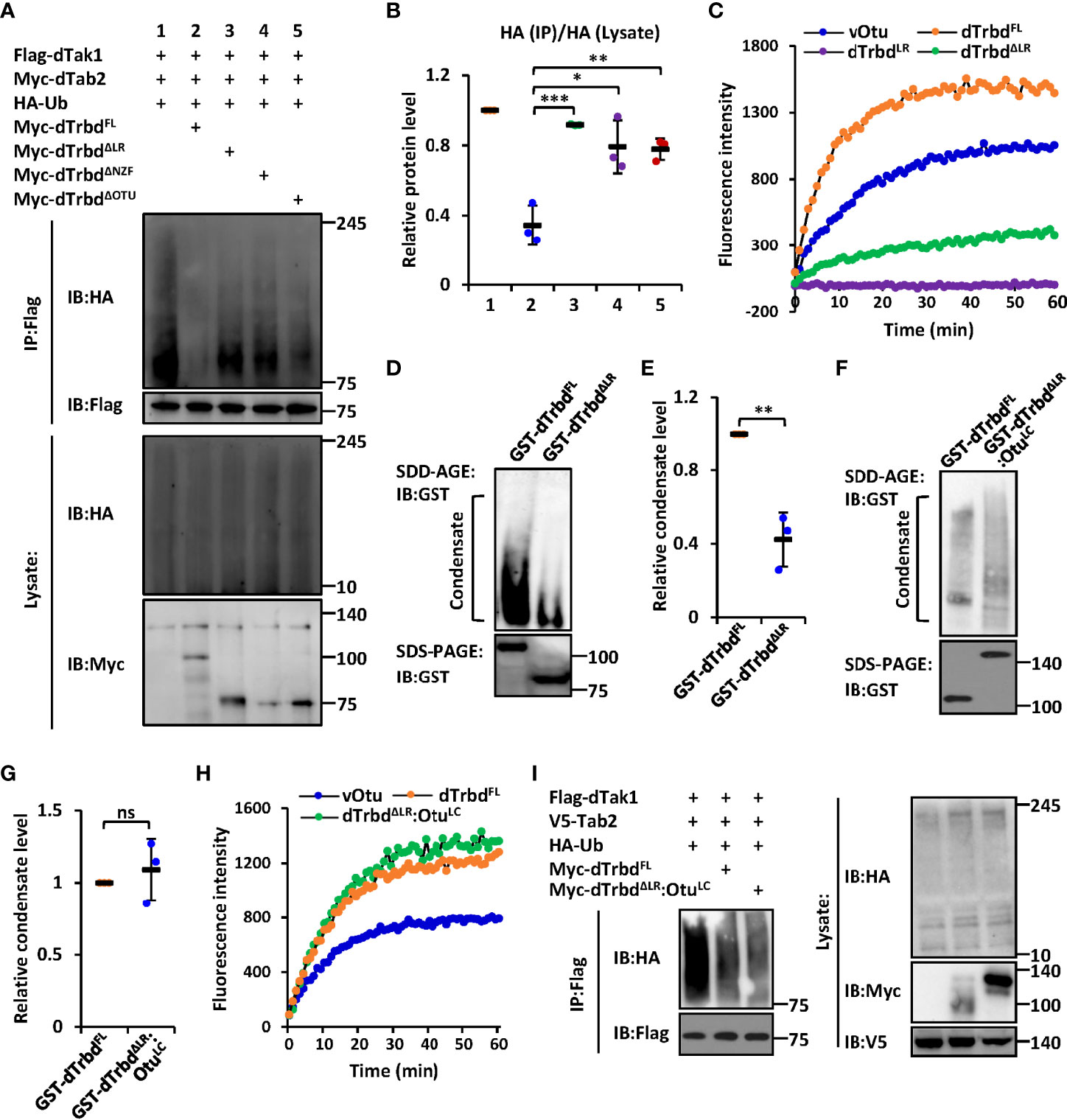
Figure 2 The LR-mediated condensation of dTrbd is essential for its Dub enzymatical activity. (A, B) S2 cells were transfected with various expressing plasmids for 48 h, followed by ubiquitination assays to examine the ubiquitination patterns of dTak1 as indicated (A). Densitometry analysis to quantify the ubiquitination levels of dTak1 (A) is shown in (B). (C) In vitro Dub assays displayed the enzymatical activity of purified proteins including dTrbdFL, dTrbdLR, and dTrbdΔLR. The viral Otu (vOtu) was used as the control. (D–G) Purified proteins [GST-tagged dTrbdFL and dTrbdΔLR in (D) as well as GST-tagged dTrbdFL and dTrbdΔLR : OtuLC in (F)] were subjected to SDD-AGE and SDS-PAGE assays. Densitometry analyses to quantify the condensate levels of indicated proteins (D, F) were shown in (E, G). (H) In vitro Dub assays showed that purified dTrbdΔLR : OtuLC enabled to cleave Ubiquitin from the substrate in the reaction. (I) Various combinations of expressing plasmids were transfected into S2 cells for 48 h, followed by ubiquitination assays as indicated. In (B, E, G) data are shown as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
The observation that over-expression of dTrbdΔLR largely lost the negative impact on the ubiquitination level of dTak1 in S2 cells (Figures 2A, B and S5A) prompted us to explore how the LR is involved in modulating the enzymatical ability of dTrbd. To do this, we first performed an in vitro Dub assay (35) using purified proteins namely dTrbdFL, dTrbdLR, and dTrbdΔLR to quantify the requirements of the LR for dTrbd’s Dub enzymatical activity (in this assay, the Ub-Rhodamine110 was used as the reaction substrate). As illustrated in Figure 2C, loss of the LR markedly down-regulated but not fully abolished the Dub enzymatical activity of dTrbd. Of note, dTrbdLR reasonably displayed no apparent Dub activity, since it is absent of the catalytical triad of the OTU domain (Figure 2C).
We then performed semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) assays to examine the condensation levels of purified dTrbdFL and other truncated forms of dTrbd proteins, as condensate assembly has recently been suggested to play essential roles in regulating protein enzymatical activities (33, 36–39). As shown in Figures 2D, E, signals representing aggregated forms of tested proteins could be easily detected in dTrbdFL but not in dTrbdΔLR, suggesting that the LR is required for dTrbd to form condensates. Of note, the LR itself is readily condensed in vitro (Figure S6A). The above observations are rather similar to our previous findings that the low-complexity (LC) domain-mediated condensation is essential for the Dub enzymatical activity of Drosophila Otu (33). However, the bioinformatic analysis suggested very low disordered property of dTrbdLR (Figure S6B). To our surprise, when we further replaced the LR of dTrbd with the LC domain of Otu, we found that the purified dTrbdΔLR : OtuLC protein easily formed condensate in our SDD-AGE assays (Figures 2F, G) and displayed comparable Dub enzymatical activity (Figure 2H), implying that dTrbd condensation is critical for its Dub enzymatical activity. Consistent results were also obtained when we further monitored the condensation patterns and Dub enzymatical activities of dTrbdFL and dTrbdΔLR : OtuLC in S2 cell cultures (Figures 2I, S6C).
dTrbd Undergoes Liquid-Liquid Phase Separation
According to our knowledge that a large amount of protein condensates can undergo phase separation to form membraneless organelles or granules, thus modulating enzymatical properties of proteins and the related biological processes (33, 36–39), we thus sought to identify whether the dTrbd condensates behave in a similar fashion. We first subjected purified dTrbdFL and dTrbdΔLR to ThT binding assays (33) to examine whether dTrbd could form amyloidal fibers in vitro. As shown in Figure 3A, dTrbdFL but not dTrbdΔLR easily bound to ThT. We then fused dTrbdFL and various truncated forms of dTrbd with GFP at their N-terminuses. Under confocal microscope, we observed that GFP-dTrbdFL underwent phase separation (Figures 3B-B’’’). In addition, both dTrbdΔNZF and dTrbdΔOTU also easily formed dense structures in a fairly short time except for dTrbdΔLR (Figures 3C-F). We last examined whether dTrbd forms dense structure in vivo. As illustrated in the immunostaining assays, dTrbd condensates can be easily observed in the fat body cells (Figures 3G-G’’).
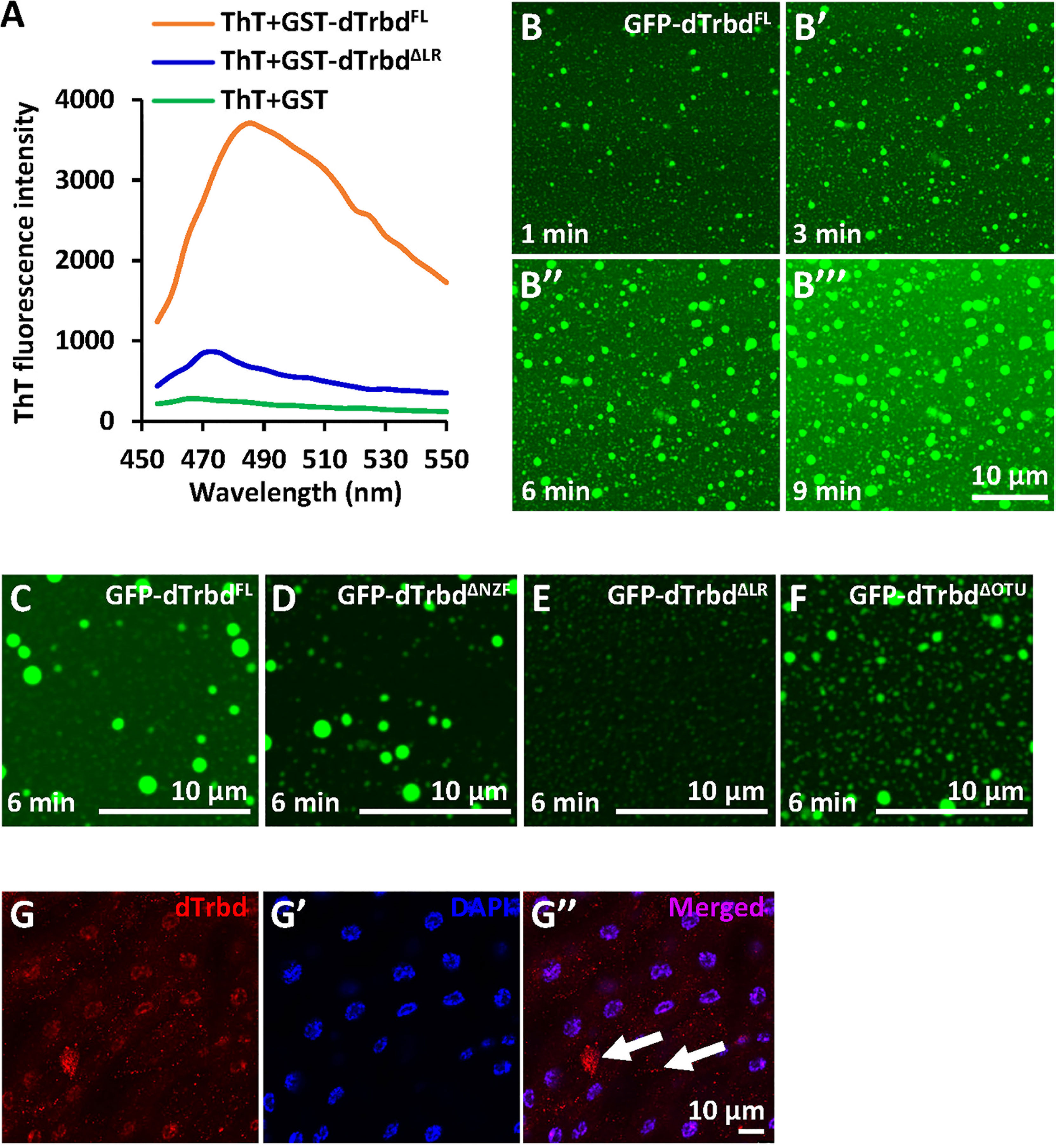
Figure 3 dTrbd undergoes phase separation depending on the Linker region. (A) dTrbdFL but not dTrbdΔLR binds to the amyloid binding dye Thioflavin T (ThT). (B-B’’’) Phase separation of dTrbdFL at various time points (1, 3, 6, and 9 min, respectively). Scale bars, 10 μm. (C-F) Different truncated forms of dTrbd (dTrbdFL, dTrbdΔNZF, and dTrbdΔOtu) can undergo phase separation except dTrbdΔLR. Scale bars, 10 μm. (G) Confocal imaging of w1118 larval fat bodies that were stained with anti-dTrbd (red) antibody. Scale bars, 10 μm.
To further explore the involvement of the LR in the condensate assembly of dTrbd in vivo, we constructed two types of transgenic flies including P{Uasp-GFP-dTrbdFL} and P{Uasp-GFP-dTrbdΔLR}, and crossed them with flies of P{c564-gal4}, which is a fat body specific driver (40). In the fat body cells of these progenies, we found that GFP-dTrbdFL other than GFP-dTrbdΔLR formed dense structures (Figures 4A-D’’).
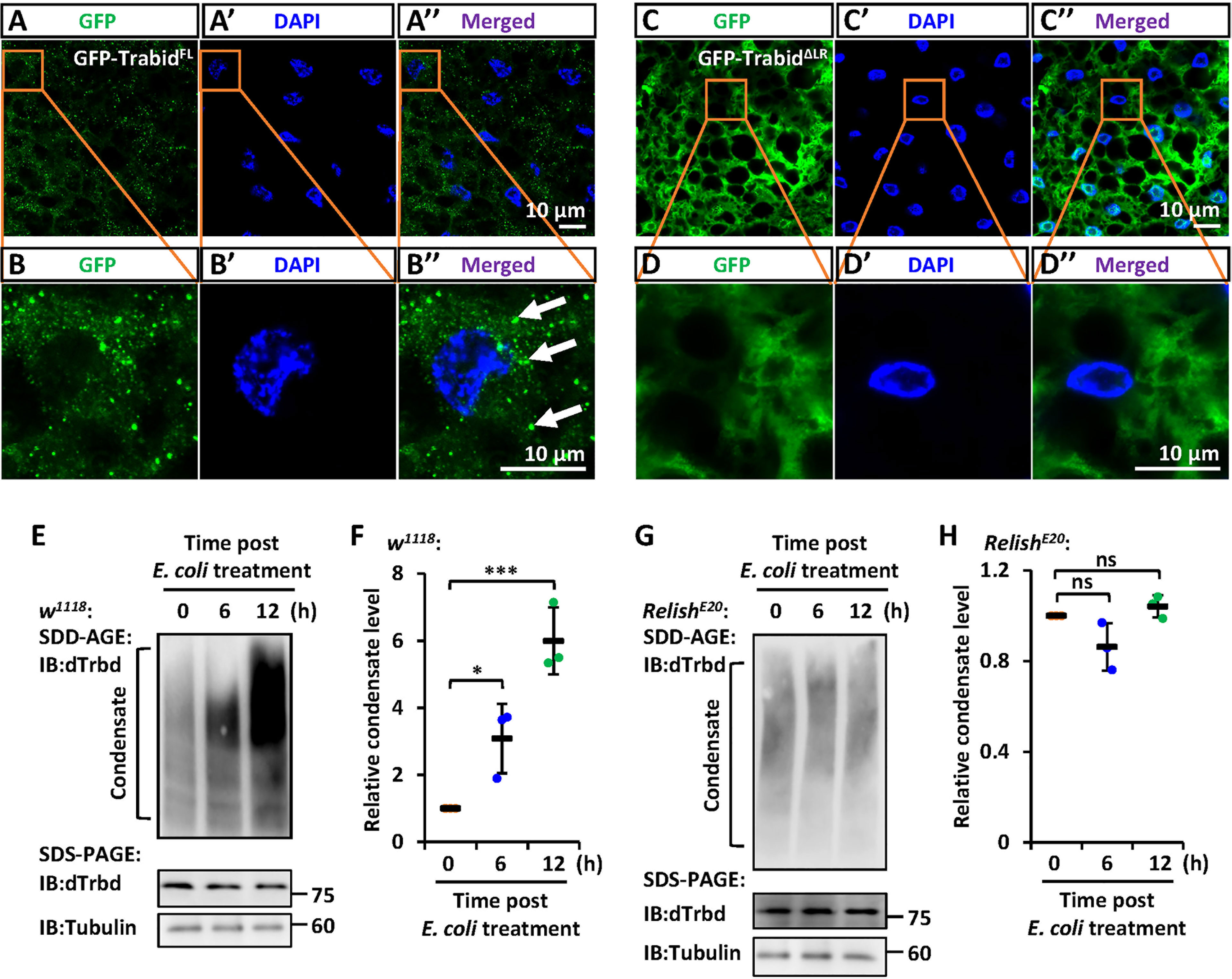
Figure 4 IMD signaling regulates the LR-mediated condensation of dTrbd. (A–D) Confocal imaging of fat bodies dissected from larvae including c564ts>GFP-dTrbdFL (A, B) and c564ts>GFP-dTrbdΔLR (C, D). Scale bars, 10 μm. (E, F) Male w1118 adults were infected with freshly-cultured E. coli. At various time points (0, 6, and 12 h) post infection, fat bodies were dissected and subjected to SDD-AGE (upper panel in E) or SDS-PAGE (lower panel in E) assays. Densitometry analysis to quantify the condensate level of samples in (E) is shown in (F). (G, H) Similar as in (E, F) except the RelishE20 mutants were used. In (F, H) data are shown as mean ± SD. *p < 0.05; ***p < 0.001; ns, not significant.
Dynamic Regulation of the dTrbd Condensate During IMD Signaling
We next sought to explore whether IMD signaling affects the condensate assembly of dTrbd. We infected male w1118 flies with freshly-cultured E. coli, and dissected the fat bodies at various time points. As shown in the SDD-AGE assays, bacterial challenging dramatically increased the dTrbd condensation (Figures 4E, F). These results imply that activation of the IMD signaling pathway positively contributes to dTrbd condensation, thereby enhancing its Dub enzymatical activity to turn down the IMD signaling. Consistently, in the Relish loss-of-function mutants (RelishE20), we failed to observe apparent alterations of dTrbd condensate (Figures 4G, H).
dTrbd Functions In Vivo via Multiple Domains
To further confirm the requirements of both the NZF domain and the LR for dTrbd functioning in vivo, we injected Erwinia carototovovora Asp 15 (Ecc15) into adult flies including 1) c564ts>+ (control), 2) c564ts>GFP-dTrbdFL, 3) c564ts>GFP-dTrbdΔNZF, and 4) c564ts>GFP-dTrbdΔLR. We observed much lower mRNA occurrences of AttA and CecA1 in dTrbdFL overexpressing (c564ts>GFP-dTrbdFL) flies compared to those of the control (Figures 5A, B). However, the mRNA levels of these AMP genes were relatively comparable between samples from c564ts>GFP-dTrbdΔNZF, c564ts>GFP-dTrbdΔLR, and the control flies (Figures 5A, B), suggesting both the NZF domain and the LR are required for dTrbd to negatively govern IMD signaling post microbial infection.
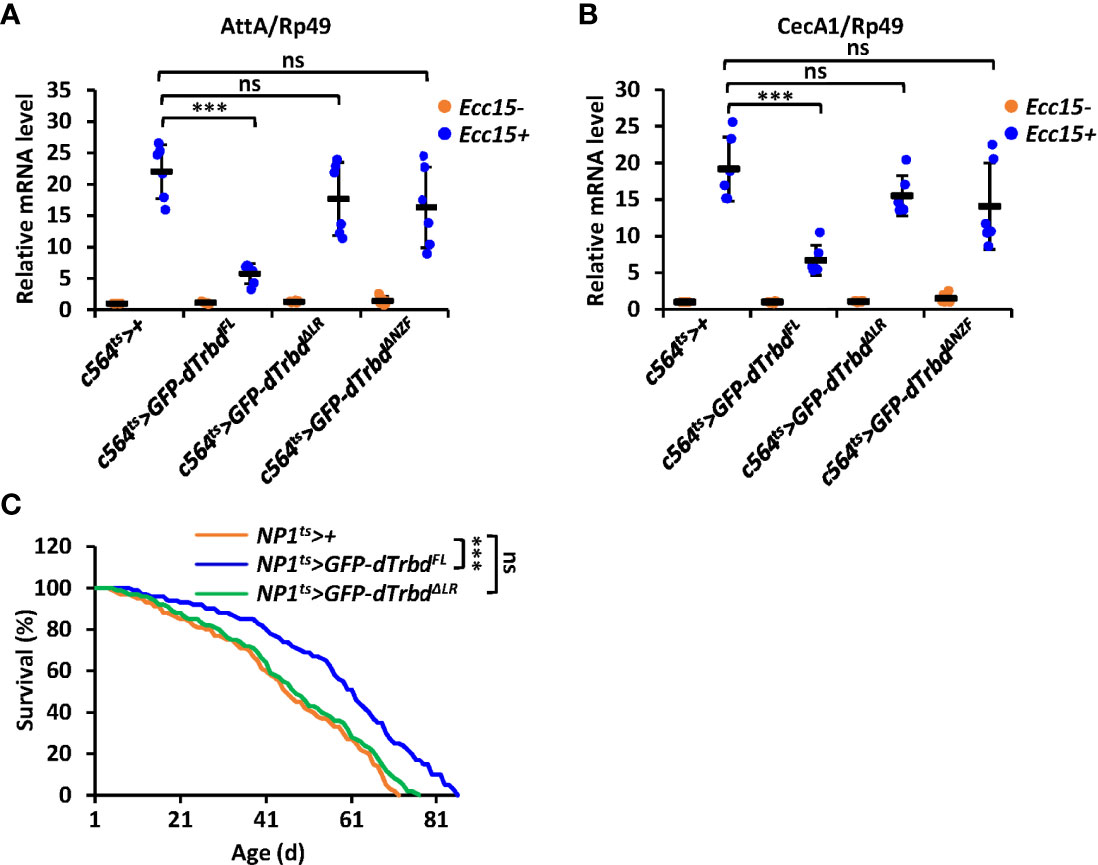
Figure 5 dTrbd functions in vivo via multiple domains. (A, B) Male adults including c564ts>+ (control), c564ts>GFP-dTrbdFL, c564ts>GFP-dTrbdΔNZF, c564ts>GFP-dTrbdΔLR, and c564ts>GFP-dTrbdΔLR were infected with freshly cultured Ecc15 (Ecc15 +) or not (Ecc15 -). 12 h later, flies were harvested and lysed for RT-qPCR assays to examine the mRNA levels of AttA (A) or CecA (B). (C) Male adults including NP1ts>+ (control), NP1ts>GFP-dTrbdFL, and NP1ts>GFP-dTrbdΔLR were subjected to lifespan assays. In (A, B) data are shown as mean ± SD. ***p < 0.001; ns, not significant.
Previous studies have shown that preventing the IMD signaling pathway in gut tissues positively contributes to the life longevity of Drosophila (15, 41–43). We thus sought to identify whether dTrbd modulates lifespan via multiple domains. As shown in Figure 5C, flies with ectopic expression of dTrbdFL in the guts displayed a prolonged lifespan. Additionally, overexpression of dTrbd without the LR was dispensable for impacting the fly lifespan (Figure 5C), strengthening the conclusion that dTrbd relies on multiple structural domains to execute its functional role in vivo.
Discussion
To date, various contributors have been thought to play passive roles in controlling the Drosophila IMD signaling pathway (44–46), providing potential theoretical bases for understanding how NF-κB dependent immune reactions are tightly regulated to avoid excessive inflammation. However, in the presence of so many negative modulators, how can IMD signaling be efficiently activated to clear invading microbes? One would naturally propose a possible working model in which the activities of these negative factors are precisely controlled during IMD signaling. In this study, we focus on a Dub dTrbd, which has recently been shown to restrict the K63-linked ubiquitination of dTak1, thereby attenuating IMD signaling in flies (13). We show that although dTrbd is not induced upon activation of the IMD signals by microbial stimuli, its binding ability to dTak1 and Dub enzymatical processivity are enhanced. Collectively, our studies uncover an IMD signaling-dependent feedback regulatory loop involving dTrbd/dTak1 to precisely control Drosophila innate immune response via diverse mechanisms.
As mentioned in the Introduction and above, the ubiquitinated dTak1 is targeted by the Dub dTrbd for deubiquitination to prevent Imd signaling (13). Nevertheless, which domain is essential for dTrbd to fulfill its biological role remains unexplored. To address this issue, we first explored whether dTrbd associates with dTak1 in a domain-dependent manner. In the domain mapping and the GST pulldown assays, we observed that dTrbd binds to dTak1 via its N-terminal conserved NZF domain. Interestingly, this domain has long been considered as a typical Ubiquitin binding domain (31, 32), which is consistent with our findings that dTrbdNZF can only bind physically to purified Ub-dTak1 but not dTak1. However, in cultured Drosophila S2 cells, our point mutation and co-IP experiments imply that dTrbd can interact with non-ubiquitinated dTak1. Put together, these data prompted us to hypothesize that dTrbd is capable of binding not only to the ubiquitinated dTak1 but also to the non-ubiquitinated dTak1 via an(some) intermediator(s). This hypothesis is consistent with our findings that dTrbd/dTak1 association is mediated by IMD signaling. A highly possible regulator of the IMD pathway, for instance, is the Drosophila Tab2 (dTab2), which has been shown to associate with both dTrbd (13) and dTak1 (47, 48). To our knowledge, there has been no evidence supporting the notion that dTab2 is able to bind to Ubiquitin, although the mammalian TAB2 homologs have been proven to harbor a relatively high Ubiquitin-binding affinity via the NZF domain (49). It will be worthwhile to identify the potential dTab2/Ubiquitin association and its involvement in dTrbd interacting with dTak1.
Recently, protein condensation and phase separation have been shown to participate in regulating the mammalian innate immune responses via impacting various cytosolic signaling pathways, namely the retinoic acid-inducible gene I protein (RIG-I) (50), the cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) (51), and the NF-κB (52) pathways. Unfortunately, very few studies have focused on the relationships between phase separation and inflammation in insects. We and colleagues recently found that In Drosophila, protein condensation and phase separation can mediate the Dub enzymatical activity of Otu to negatively contribute to the IMD innate immune pathway via the low-complexity (LC) domain (33). In this study, we demonstrate that the Dub enzymatical activity of dTrbd is also affected by the assembly of the functional condensate. However, dTrbd condensation is surprisingly dependent on the Linker region lacking LC properties, implying diverse regulatory mechanisms by which protein condensation mediates the Dub enzymatical processivity and the related biological processes. Notably, an outstanding study by Kleino et al. suggested that the RHIM motif in proteins such as Imd, PGRP-LC, and PGRP-LE is required for the assembly of functional amyloids (53). Therefore, it is thus probable that such motif exists in dTrbd and plays a role in governing its behavior.
Materials and methods
Drosophila Strain and Husbandry
All flies used in this study were raised in standard Drosophila culture medium (corn flour and agar) and cultured at 25°C. The w1118 strain was used as the control and the host for P-element-mediated transformation. The following Gal4 lines and transgenes were used in this study: 1) P{c564-Gal4}, P{NP1-Gal4}, and P{Tub-gal80ts} were obtained from Bloomington stock center; 2) P{UAS-GFP-dTrbdFL}, P{UAS-GFP-dTrbdΔNZF}, and P{UAS-GFP-dTrbdΔLR}, in which the full-length dTrbd, dTrbdΔNZF, and dTrbdΔLR were placed under the control of the UASp promoter, respectively.
Antibodies
The following primary antibodies were used for Western blot and immunostaining assays: Mouse anti-Flag (1:5000, Sigma-Aldrich, Cat#F1804); Rabbit anti-Myc (1:3000, Medical & Biological Laboratories, Cat#562); Mouse anti-dTrbd (1:1000), which was generated to an N-terminal fragment of dTrbd (amino acids 155-253) fused to His and purified from E. coli; Rabbit anti-dTak1 (1:1000, Abcam, Cat#239353); Mouse anti-β-Tubulin (1:3000, Cowin, Cat#CW0098M); Rabbit anti-HA (1:2500, Medical & Biological Laboratories, Cat#561); Mouse anti-GFP (1:2000, Sigma-Aldrich, Cat#G6539); Mouse anti-V5 (1:5000, Sigma-Aldrich, Cat#V8012).
The secondary antibodies for Western blot and immunostaining assays include Goat anti-Mouse IgG H&L (HRP); Goat anti-Rabbit IgG H&L (HRP); Goat anti-Mouse IgG H&L (Alexa Fluor 555) (1:1000, Abcam, Cat#ab150078).
RT-qPCR Assays
Total RNA was isolated with Trizol Reagent (Invitrogen), followed by cDNA synthesis using the first-strand cDNA synthesis kit (Transgen) according to the manufacturer’s instructions. RT-qPCR was performed in triplicate using SYBR Green Master Mix (Thermo) on a Light Cycler 480. Concentrations of specific targets were normalized to endogenous reference Rp49. Data shown are relative mRNA abundance compared to that of the control. Primers used in RT-qPCR assays were shown in Supplementary Table 1.
S2 Cell Transfection and Co-IP Assays
S2 cells were cultured in insect medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Hyclone) at 27°C. Lipofectamine 2000 (Invitrogen) was used for transfection of all cells in this study. For Co-IP assays, cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5% Triton X-100, 10 mg/ml aprotinin, 10 mg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride). Anti-Myc agarose beads (Abmart) or Anti-Flag agarose beads (Sigma) were used for indicated immunoprecipitation experiments. Immunoprecipitants were subjected to Western blot assays to detect the indicated protein levels.
Protein Purification and GST Pull-Down Assays
The GST-tagged or His-tagged proteins were expressed and purified from E. coli strain BL21. Briefly, protein expression was induced by adding isopropyl-beta-D-thiogalactopyranoside (final concentration of 1 mM) into BL21 bacterial medium when the OD600 of the culture reached 0.4-0.6, followed by overnight incubation at 18°C. Bacterium were pelleted and resuspended in 15 ml of lysis buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA). Cell integrity was disrupted by sonicating each sample for 30 min using an EpiShear™ Probe Sonicator (pulse 4 s on, 6 s off, 40% amplitude). Protein content was collected as the supernatant of the following centrifugation at 10,000 rpm, 4°C for 20 min. The Glutathione Sepharose 4B (Sigma, 17-0756-01) or BeyoGold™ His-tag Purification Resin (Beyotime, P2218) were used for purification of GST-tagged or His-tagged proteins, respectively. Each sample (1 μg) was finally resuspended in loading buffer and subjected to the Coomassie brilliant blue assay.
For GST pull-down assay, indicated proteins were incubated with the Glutathione Sepharose 4B (10 μl for each sample) at 4°C for 3 h. Samples were then washed with wash buffer (PBS with 1% TritonX-100) 3 times (1 h in total) and subjected to Western blot assays.
RNAi in S2 Cells by Treatment of dsRNA
All dsRNAs were synthesized using the in vitro T7 transcription Kit (Promega) according to the manufacture’s protocol. S2 cells were harvested and then diluted into fresh medium at a density of 1×106 cells per ml and treated with dsRNAs immediately. 1 h later, FBS (Hyclone) was added to the culture medium at a final concentration of 10%. Primers for dsRNA synthesis are shown in Supplementary Table 1.
SDD-AGE Assays
For samples of cell lysate, transfected S2 cells were harvested and lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, and 1mM phenylmethylsulfonyl fluoride) over ice for 30 min, followed by centrifugation (13000 rpm at 4°C) for 10 min. Supernatant was carefully collected and loaded with loading buffer (0.5 × TBE, 10% glycerol, 2% SDS, 0.0025% bromophenol blue) at room temperature for 15 min. For samples of purified proteins, indicated amounts of proteins were directly loaded with loading buffer at room temperature. Newly prepared 1.5% agarose gel with 0.1% SDS was pre-run by electrophoresis in running buffer (1×TBE and 0.1% SDS) for 1 h at 4°C. Loaded protein samples were then subjected to electrophoresis for 1 h and transferred to PVDF membrane (Millipore) for immunoblotting analysis.
In Vitro DUB Assays
The Ub rhodamine 110 (Boston Biochem) was used as the substrate for in vitro deubiquitination assays. In brief, proteins (100 ng for each sample) with Ub rhodamine 110 were incubated in the assay buffer (150 mM NaCl, 5 mM MgCl2, 50 mM Tris-HCl, pH 7.5). The mixture was added into 384-well low volume plate and dynamic fluorescence was monitored with excitation and emission wavelengths set at 485/20 and 535/20 nm, respectively, utilizing the MD SpectraMax M5 Microplate Reader. Fluorescence intensity for each condition was plotted as a function of time.
Ubiquitination Assays
Transfected S2 cells were harvested and lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 10% glycerol, and 1% SDS). Samples were then heated at 95°C for 5 min and combined with binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, and 10% glycerol) to adjust the SDS to a final concentration of 0.1%. Lysates were subjected to sonication and immunoprecipitated utilizing anti-Myc agarose beads (Abmart) or anti-Flag M2 beads (Sigma) for 4 h. The immune complexes were washed with wash buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.5% Nonidet P-40, and 10% glycerol) 3 times (1 h in total), and subjected to Western blot assays to detect the ubiquitination patterns of indicated proteins.
Phase Separation
Purified proteins (10 mg/ml) were incubated at room temperature with 50 mM Tris-HCl, pH 7.5, and 150 mM NaCl. Protein phase separation was examined using Zeiss LSM 710 Meta confocal microscope, and images were captured at indicated time points.
Immunostaining Assays
Fat bodies were dissected from larvae and fixed in fix buffer (4% formaldehyde, 0.25% Tween-20, and 75% n-heptane in 1×PBS) for 30 min at room temperature. Samples were then washed 3 times (total 1 h) and blocked (2 h) in PBTA buffer (0.3% Tween-20 and 1.5% BSA in 1×PBS). After incubation with Mouse anti-dTrbd (diluted in PBTA) antibody at 4°C overnight, samples were washed 3 times (total 1 h) at room temperature, and then incubated with Goat anti-Mouse (Alexa Fluor 555) antibody (diluted in PBTA) at room temperature for 2.5 h with DAPI (1:1000) to mark the cell nucleus. Finally, samples were washed 3 times (1 h in total), pressed into tablets, and observed under confocal microscope.
Pathogenic Infection of Flies and Larvae
Overnight bacterial culture was collected and diluted in sterile 1×PBS dilution at a concentration of OD600 = 1. Male adult flies were injected with 4.6 nl of the diluted bacterium or the same volume of PBS as controls. For larvae, they were washed three times with sterile H2O and placed in a small drop of water on a black rubber block. Injection was performed using a sharp capillary in the posterolateral body with 100 nl bacterial suspension (same volume of PBS was used for the control). Treated flies or larvae were transferred to a vial containing regular food and collected for further analysis as indicated.
Statistical Analyses
All statistical analyses were performed by using GraphPad Prism 8. Data were shown as mean and standard errors. Statistical significance was determined by using the two-tailed Student’s t or Mann-Whitney tests except for lifespan assays, in which the Log-Rank test (Kaplan-Meier method) was used for statistical analyses. The p value of less than 0.05 was considered statistically significant. * p < 0.05; ** p < 0.01; *** p < 0.001; ns, not significant.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
YHua, QC, and SJ conceived and designed the experiments. YHua, YHu, FK, RD, CZ, CCZ, SZ, YJ, YY, and QC performed the experiments. YHua, YZ, YHu, QC, and SJ analyzed the data. YHua, YZ, and YHu performed statistical analyses. YHua, QC, and SJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31871470 and 32100702) and the Anhui Provincial Natural Science Foundation (2008085J14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We deeply appreciate the critical discussion with Prof. Jules Hoffmann (University of Strasbourg) when we prepared this manuscript. We are indebted to Dr. Huanjie Song (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for generating the Mouse anti-dTrbd antibody. We thank Bloomington Drosophila Stock Center for the fly resource.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.932268/full#supplementary-material
References
1. Beutler B. Innate Immunity: An Overview. Mol Immunol (2004) 40(12):845–59. doi: 10.1016/j.molimm.2003.10.005
3. Fitzgerald KA, Kagan JC. Toll-Like Receptors and the Control of Immunity. Cell (2020) 180(6):1044–66. doi: 10.1016/j.cell.2020.02.041
4. Akira S. Innate Immunity to Pathogens: Diversity in Receptors for Microbial Recognition. Immunol Rev (2009) 227(1):5–8. doi: 10.1111/j.1600-065X.2008.00739.x
5. Hsu AP, Holland SM. Host Genetics of Innate Immune System in Infection. Curr Opin Immunol (2022) 74:140–9. doi: 10.1016/j.coi.2021.11.003
6. Lemaitre B, Hoffmann JA. The Host Defense of Drosophila Melanogaster. Annu Rev Immunol (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
7. Hoffmann JA, Reichhart JM. Drosophila Innate Immunity: An Evolutionary Perspective. Nat Immunol (2002) 3(2):121–6. doi: 10.1038/ni0202-121
8. Kleino A, Silverman N. The Drosophila IMD Pathway in the Activation of the Humoral Immune Response. Dev Comp Immunol (2014) 42(1):25–35. doi: 10.1016/j.dci.2013.05.014
9. Valanne S, Wang JH, Ramet M. The Drosophila Toll Signaling Pathway. J Immunol (2011) 186(2):649–56. doi: 10.4049/jimmunol.1002302
10. Nie L, Cai SY, Shao JZ, Chen J. Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Front Immunol (2018) 9:1523. doi: 10.3389/fimmu.2018.01523
11. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The Dorsoventral Regulatory Gene Cassette Spatzle/Toll/Cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell (1996) 86:973–83. doi: 10.1016/s0092-8674(00)80172-5
12. Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila Immune Deficiency (IMD) Is a Death Domain Protein That Activates Antibacterial Defense and can Promote Apoptosis. Dev Cell (2001) 1(4):503–14. doi: 10.1016/s1534-5807(01)00059-4
13. Fernando MDA, Kounatidis I, Ligoxygakis P. Loss of Trabid, a New Negative Regulator of the Drosophila Immune-Deficiency Pathway at the Level of Tak1, Reduces Life Span. PLoS Genet (2014) 10(2):e1004117. doi: 10.1371/journal.pgen.1004117
14. Paredes JC, Welchman DP, Poidevin M, Lemaitre B. Negative Regulation by Amidase PGRPs Shapes the Drosophila Antibacterial Response and Protects the Fly From Innocuous Infection. Immunity (2011) 35(5):770–9. doi: 10.1016/j.immuni.2011.09.018
15. Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 Promotes Gut Immune Homeostasis to Limit Commensal Dysbiosis and Extend Lifespan. Cell (2014) 156(1-2):109–22. doi: 10.1016/j.cell.2013.12.018
16. Zhai Z, Huang X, Yin Y. Beyond Immunity: The Imd Pathway as a Coordinator of Host Defense, Organismal Physiology and Behavior. Dev Comp Immunol (2018) 83:51–9. doi: 10.1016/j.dci.2017.11.008
17. Garschall K, Flatt T. The Interplay Between Immunity and Aging in Drosophila. F1000Research (2018) 7:160. doi: 10.12688/f1000research.13117.1
18. Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila Immune Response by Peptidoglycan-Recognition Proteins SC1 and SC2. PLoS Pathog (2006) 2:0139–47. doi: 10.1371/journal.ppat.0020014
19. Mellroth P, Karlsson J, Steiner H. A Scavenger Function for a Drosophila Peptidoglycan Recognition Protein. J Biol Chem (2003) 278(9):7059–64. doi: 10.1074/jbc.M208900200
20. Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila Amidase PGRP-LB Modulates the Immune Response to Bacterial Infection. Immunity (2006) 24(4):463–73. doi: 10.1016/j.immuni.2006.02.012
21. Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a Suppressor of Antibacterial Immunity in Drosophila. Proc Natl Acad Sci U S A (2006) 103(44):16358–63. doi: 10.1073/pnas.0603238103
22. Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, et al. A Drosophila Ortholog of the Human Cylindromatosis Tumor Suppressor Gene Regulates Triglyceride Content and Antibacterial Defense. Development (2007) 134(14):2605–14. doi: 10.1242/dev.02859
23. Foley E, O'Farrell PH. Functional Dissection of an Innate Immune Response by a Genome-Wide RNAi Screen. PLoS Biol (2004) 2(8):E203. doi: 10.1371/journal.pbio.0020203
24. Guntermann S, Primrose DA, Foley E. Dnr1-Dependent Regulation of the Drosophila Immune Deficiency Signaling Pathway. Dev Comp Immunol (2009) 33(1):127–34. doi: 10.1016/j.dci.2008.07.021
25. De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-Wide Analysis of the Drosophila Immune Response by Using Oligonucleotide Microarrays. Proc Natl Acad Sci U S A (2001) 98(22):12590–5. doi: 10.1073/pnas.221458698
26. Kleino A, Myllymaki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, et al. Pirk Is a Negative Regulator of the Drosophila Imd Pathway. J Immunol (2008) 180(8):5413–22. doi: 10.4049/jimmunol.180.8.5413
27. Thevenon D, Engel E, Avet-Rochex A, Gottar M, Bergeret E, Tricoire H, et al. The Drosophila Ubiquitin-Specific Protease Dusp36/Scny Targets IMD to Prevent Constitutive Immune Signaling. Cell Host Microbe (2009) 6(4):309–20. doi: 10.1016/j.chom.2009.09.007
28. Khush RS, Cornwell WD, Uram JN, Lemaitre B. A Ubiquitin-Proteasome Pathway Represses the Drosophila Immune Deficiency Signaling Cascade. Curr Biol (2002) 12(20):1728–37. doi: 10.1016/s0960-9822(02)01214-9
29. Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, et al. Inhibitor of Apoptosis 2 and TAK1-Binding Protein Are Components of the Drosophila Imd Pathway. EMBO J (2005) 24(19):3423–34. doi: 10.1038/sj.emboj.7600807
30. Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, et al. Cell-Surface Localization of Pellino Antagonizes Toll-Mediated Innate Immune Signalling by Controlling MyD88 Turnover in Drosophila. Nat Commun (2014) 5:3458. doi: 10.1038/ncomms4458
31. Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, et al. Ubiquitin Interactions of NZF Zinc Fingers. EMBO J (2004) 23(7):1411–21. doi: 10.1038/sj.emboj.7600114
32. Kristariyanto YA, Abdul Rehman SA, Campbell DG, Morrice NA, Johnson C, Toth R, et al. K29-Selective Ubiquitin Binding Domain Reveals Structural Basis of Specificity and Heterotypic Nature of K29 Polyubiquitin. Mol Cell (2015) 58(1):83–94. doi: 10.1016/j.molcel.2015.01.041
33. Ji S, Luo Y, Cai Q, Cao Z, Zhao Y, Mei J, et al. LC Domain-Mediated Coalescence Is Essential for Otu Enzymatic Activity to Extend Drosophila Lifespan. Mol Cell (2019) 74(2):363–77.e5. doi: 10.1016/j.molcel.2019.02.004
34. Licchesi JD, Mieszczanek J, Mevissen TE, Rutherford TJ, Akutsu M, Virdee S, et al. An Ankyrin-Repeat Ubiquitin-Binding Domain Determines TRABID's Specificity for Atypical Ubiquitin Chains. Nat Struct Mol Biol (2011) 19(1):62–71. doi: 10.1038/nsmb.2169
35. Ji S, Li C, Hu L, Liu K, Mei J, Luo Y, et al. Bam-Dependent Deubiquitinase Complex can Disrupt Germ-Line Stem Cell Maintenance by Targeting Cyclin a. Proc Natl Acad Sci U S A (2017) 114:6316–21. doi: 10.1073/pnas.1619188114
36. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-Free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers Within Hydrogels. Cell (2012) 149(4):753–67. doi: 10.1016/j.cell.2012.04.017
37. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell (2015) 163(1):123–33. doi: 10.1016/j.cell.2015.09.015
38. Hallegger M, Chakrabarti AM, Lee FCY, Lee BL, Amalietti AG, Odeh HM, et al. TDP-43 Condensation Properties Specify Its RNA-Binding and Regulatory Repertoire. Cell (2021) 184(18):4680–96.e22. doi: 10.1016/j.cell.2021.07.018
39. Shimobayashi SF, Ronceray P, Sanders DW, Haataja MP, Brangwynne CP. Nucleation Landscape of Biomolecular Condensates. Nature (2021) 599(7885):503–6. doi: 10.1038/s41586-021-03905-5
40. Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. Role for Sumoylation in Systemic Inflammation and Immune Homeostasis in Drosophila Larvae. PLoS Pathog (2010) 6(12):e1001234. doi: 10.1371/journal.ppat.1001234
41. Zhu Y, Cai Q, Zheng X, Liu L, Hua Y, Du B, et al. Aspirin Positively Contributes to Drosophila Intestinal Homeostasis and Delays Aging Through Targeting Imd. Aging Dis (2021) 12(7):1821–34. doi: 10.14336/AD.2020.1008
42. Cai Q, Ji S, Li M, Zheng S, Zhou X, Guo H, et al. Theaflavin-Regulated Imd Condensates Control Drosophila Intestinal Homeostasis and Aging. iScience (2021) 24(3):102150. doi: 10.1016/j.isci.2021.102150
43. Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, et al. Distinct Shifts in Microbiota Composition During Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell Rep (2015) 12(10):1656–67. doi: 10.1016/j.celrep.2015.08.004
44. Myllymaki H, Valanne S, Ramet M. The Drosophila Imd Signaling Pathway. J Immunol (2014) 192(8):3455–62. doi: 10.4049/jimmunol.1303309
45. Lee KZ, Ferrandon D. Negative Regulation of Immune Responses on the Fly. EMBO J (2011) 30(6):988–90. doi: 10.1038/emboj.2011.47
46. Imler JL. Overview of Drosophila Immunity: A Historical Perspective. Dev Comp Immunol (2014) 42(1):3–15. doi: 10.1016/j.dci.2013.08.018
47. Zhuang ZH, Sun L, Kong L, Hu JH, Yu MC, Reinach P, et al. Drosophila TAB2 Is Required for the Immune Activation of JNK and NF-Kappab. Cell Signal (2006) 18(7):964–70. doi: 10.1016/j.cellsig.2005.08.020
48. Geuking P, Narasimamurthy R, Basler K. A Genetic Screen Targeting the Tumor Necrosis Factor/Eiger Signaling Pathway: Identification of Drosophila TAB2 as a Functionally Conserved Component. Genetics (2005) 171(4):1683–94. doi: 10.1534/genetics.105.045534
49. Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-Sided Ubiquitin Binding Explains Specificity of the TAB2 NZF Domain. Nat Struct Mol Biol (2009) 16(12):1328–30. doi: 10.1038/nsmb.1731
50. Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS Forms Functional Prion-Like Aggregates to Activate and Propagate Antiviral Innate Immune Response. Cell (2011) 146(3):448–61. doi: 10.1016/j.cell.2011.06.041
51. Du MJ, Chen ZJ. DNA-Induced Liquid Phase Condensation of cGAS Activates Innate Immune Signaling. Science (2018) 361(6403):704–9. doi: 10.1126/science.aat1022
52. Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. Respiratory Syncytial Virus Sequesters NF-kappaB Subunit P65 to Cytoplasmic Inclusion Bodies to Inhibit Innate Immune Signaling. J Virol (2020) 94(22):e01380–20. doi: 10.1128/JVI.01380-20
Keywords: dTrbd/dTak1 association, dTrbd condensation and phase separation, dub enzymatical activity, IMD innate immune pathway, Drosophila melanogaster
Citation: Hua Y, Zhu Y, Hu Y, Kong F, Duan R, Zhang C, Zhang C, Zhang S, Jin Y, Ye Y, Cai Q and Ji S (2022) A Feedback Regulatory Loop Involving dTrbd/dTak1 in Controlling IMD Signaling in Drosophila Melanogaster. Front. Immunol. 13:932268. doi: 10.3389/fimmu.2022.932268
Received: 29 April 2022; Accepted: 23 June 2022;
Published: 14 July 2022.
Edited by:
Shoichiro Kurata, Tohoku University, JapanReviewed by:
Lele Zhu, University of Texas MD Anderson Cancer Center, United StatesJana Alonso, Spanish National Research Council (CSIC), Spain
Copyright © 2022 Hua, Zhu, Hu, Kong, Duan, Zhang, Zhang, Zhang, Jin, Ye, Cai and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingshuang Cai, 17610679696@163.com; Shanming Ji, jism@ahau.edu.cn
†These authors have contributed equally to this work
 Yongzhi Hua1†
Yongzhi Hua1† Shanming Ji
Shanming Ji