- Shanghai Key Laboratory of Veterinary Biotechnology, Key Laboratory of Urban Agriculture (South), Ministry of Agriculture, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
Stimulator of the interferon gene (STING) is involved in mammalian antiviral innate immunity as an interferon (IFN) activator. However, there is still a lack of clarity regarding the molecular characterization of goose STING (GoSTING) and its role in the innate immune response. In the present study, we cloned GoSTING and performed a series of bioinformatics analyses. GoSTING was grouped into avian clades and showed the highest sequence similarity to duck STING. The in vitro experiments showed that the mRNA levels of GoSTING, IFNs, IFN-stimulated genes (ISGs), and proinflammatory cytokines were significantly upregulated in goose embryo fibroblast cells (GEFs) infected with Newcastle disease virus (NDV). Overexpression of GoSTING in DF-1 cells and GEFs strongly activated the IFN-β promoter as detected by a dual-luciferase reporter assay. Furthermore, overexpression of GoSTING induced the expression of other types of IFN, ISGs, and proinflammatory cytokines and inhibited green fluorescent protein (GFP)-tagged NDV (NDV-GFP) and GFP-tagged vesicular stomatitis virus (VSV) (VSV-GFP) replication in vitro. In conclusion, these data suggest that GoSTING is an important regulator of the type I IFN pathway and is critical in geese’s innate immune host defense against RNA viruses.
Introduction
The innate immune system is the host’s first line of defense against foreign pathogen invasion and endogenous damage, which initiates appropriate host defense mechanisms by detecting a series of pathogen-associated molecular patterns (PAMPs) and endogenous danger-associated molecular patterns (DAMPs) through pattern recognition receptors (PRRs) (1, 2). After PRRs are activated, downstream signaling pathways are triggered, resulting in the expression of chemokines, proinflammatory cytokines, and the synthesis of type I interferon (IFN) and type III IFN, which induces the expression of IFN-stimulated genes (ISGs) through IFN receptor and Janus kinase (JAK) - signal transducer and activator of transcription (STAT) signaling, to effectively inhibit the replication of pathogens, remove aliens, maintain the physiological balance of the body, and act as a driving force to influence subsequent adaptive immunity (3–6). Currently, the major families of PRRs include the Toll-like receptors (TLRs), the retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), and the nucleotide oligomerization domain-like receptors (NLRs, also called NACHT, LRR and PYD domain proteins) and cytosolic DNA sensors (7).
In addition to PRRs, several junction adaptor proteins are also critical for the induction of IFN, thereby ensuring the normal function of the innate immune system (8, 9). For example, all TLRs family members associate with corresponding adaptor proteins after sensing PAMPs or DAMPs, such as myeloid differentiation major response gene 88 (MyD88), MyD88-adaptor-like protein (MAL), TIR domain-containing adaptor inducing IFN-β (TRIF/TICAM1), TRIF-related adaptor molecule (TRAM/TICAM2), and ultimately activate downstream nuclear factor-κB (NF-κB), IFN regulatory factors (IRFs) and mitogen-activated protein kinase (MAPK) (10–12). Like MyD88, stimulator of the IFN gene (STING; also known as MITA, MYPS, ERIS, and TMEM173), a molecule in the endoplasmic reticulum (ER), is also a key adapter protein with a potent ability to induce type I IFNs, interleukins and other proinflammatory factors (9, 13, 14).
A large number of recent studies have confirmed that STING is the core molecule of innate immune response from pathogen cytosolic DNA and RNA (15). In the RNA-triggered pathway, STING is mainly involved in RIG-I rather than melanoma differentiation-associated gene 5 (MDA5) signaling and functions downstream of RIG-I and mitochondrial antiviral signaling protein (MAVS) and upstream of TANK-binding kinase 1 (TBK1) (9). Following the recognition of RNA ligands, RIG-I is activated by the virus and transmits the signal to MAVS located downstream of the mitochondria, where it interacts with STING. STING is then transported to the vesicle structure around the nucleus and acts as a reaction platform to recruit TBK1 to activate IFN regulator factor (IRF) 3, which forms a dimer and enters the nucleus to induce the synthesis of IFN (16). During the recognition of DNA viruses, the DNA sensors, such as DAI (17), IFI16 (18), DDX41 (19), and cGAS (20, 21), transmit the signal to STING after recognizing DNA, which then activates the IFN via the STING-TBK1-IRF3 pathway (8). IFI16 is mainly located in the nucleus and can recognize single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA) ligands in a length-dependent manner through its HIN domain (22). After binding to the ligand, IFI16 induces the expression of IFN-β through the activation of IRF3, and this expression is STING-dependent (23). DDX41 was identified as a cytoplasmic sensor capable of recognizing viral dsDNA. After sensing dsDNA through its DEADc domain, DDX41 binds to STING and initiates activation of the IFN pathway (19, 22). It can be seen that although the localization of various DNA sensors in cells, the nucleic acid forms recognized, and the site that binds to viral nucleic acids are different, the process involves STING as a key mediator. Overall, STING plays an important role in antiviral innate immunity as a key signaling molecule that regulates type I IFNs, which is essential for establishing antiviral status.
Most of the studies mentioned above on STING have focused on mammals, whereas STING signaling events in geese have not been studied. Geese, like ducks, belong to waterfowl and play a critical role in the transmission and dissemination of many important pathogens (14). In particular, Newcastle disease virus (NDV) and avian influenza virus (AIV) cause serious and economically significant diseases in almost all birds (24, 25). Chickens are susceptible to NDV and AIV due to a lack of RIG-I naturally. Instead, chickens express MDA5 or other as yet unidentified receptors which functionally compensate for the absence of RIG-I in the chicken genome (26–28). In addition, preliminary research in our laboratory demonstrated that chicken STING (chSTING) inhibited the replication of NDV and AIV and activated IRF-7 and NF-κB to induce the production of type I IFNs, possibly by participating in the MDA5-STING-IFN-β signaling pathway in chicken cells (29). Compared to chickens lacking RIG-I, ducks and geese encode RIG-I with a similar domain organization to mammals and are generally resistant to NDV and AIV (30). Overexpression of duck STING (DuSTING) has been shown to activate the type I IFN pathway and limit the replication of H9N2 AIV in our previous studies (31).
Interestingly, compared with duck RIG-I, goose RIG-I (GoRIG-I) exhibited a higher IFN-activating ability in DF-1 cells infected with or not infected with the influenza virus (30). Ding et al. have identified the key role of GoRIG-I in innate immunity against NDV infection, and goose MAVS was identified as a GoRIG-I interactive protein involved in the activation of type I IFN pathways goose cells (26, 32). However, whether goose STING (GoSTING), like its mammalian counterpart, also induces type I IFN signaling and exerts antiviral effects remains unclear.
In the present study, we cloned GoSTING and explored the function of GoSTING in innate immunity in geese. We investigated the function of GoSTING in RNA virus infection, and the effects of GoSTING on the inhibition of viral genomic RNA replication based on NDV infection were characterized in vitro. Furthermore, our results suggest that GoSTING is an important regulator of IFNs, proinflammatory cytokines, and ISGs in geese. These findings contribute to a more systematic understanding of the bird’s biological role of STING in the innate immune system and provide new insight into general and individual characteristics of the innate immune system in birds and mammals.
Materials and methods
Cells and viruses
DF-1 is a chicken embryonic fibroblast cell line from East Lansing strain eggs (33). Goose embryo fibroblast cells (GEFs) were prepared from 15-day-old goose embryos. The DF-1 cells and GEFs were maintained in high-glucose complete Dulbecco’s Modified Eagle’s Medium (DMEM; Corning, USA) supplemented with 10% fetal bovine serum (FBS; Nulen, Shanghai, China) and 1% penicillin-streptomycin (Gibco, USA). All cells were incubated at 37°C in a 5% CO2 incubator. The NDV strain NSD14 was isolated from chickens at a farm in Shandong Province, China. Green fluorescent protein (GFP) tagged NDV low virulent strain LaSota named NDV-GFP, and GFP tagged vesicular stomatitis virus (VSV) VSV-GFP were stored in our laboratory. These viruses were purified, propagated, and stored as described in our previous study (29).
Cloning and bioinformatics analysis of GoSTING
Based on the predicted GoSTING sequence from the National Center for Biotechnology Information (NCBI), the primers GoSTING-F and GoSTING-R (Table 1), which were located outside of the GoSTING open reading frame (ORF), were designed to amplify potential GoSTING cDNA via RT-PCR on total RNA extracted from the GEF cells. The PCR product was ligated into a pTOPO-Blunt vector (Aidlab Biotech, Beijing, China) for sequencing, and the positive colonies were sent to the Beijing Genomics Institute (Beijing, China) for sequencing. The deduced amino acid sequence of GoSTING was analyzed using the SMART program. The amino acid sequence of GoSTING was aligned with the other animal STING proteins from ducks, chickens, humans, and pigs using Clustal W and edited with ESPript 3.0 (http://http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Sequence homology and phylogenetic analysis of amino acid sequences was constructed using DNASTAR software. A phylogenetic tree was constructed based on the STING from 13 different species, including mammals, birds, and fish. Homology modeling for GoSTING was conducted using the online protein-modeling server SWISS-MODEL (http://swissmodel.expasy.org/).
Construction of plasmids
The PCR primers are shown in Table 1. The expression construct pcDNA3.1-GoSTING-Flag was constructed by inserting full-length GoSTING into the Xho I and EcoR I sites of the pcDNA3.1-Flag expression vector via homologous recombination. The chicken IFN-β (ch-IFN-β) promoter-luciferase reporter plasmids pGL-IFN-β-Luc was constructed from chick embryo fibroblast genomic DNA using primers with Nhe I and Bgl II sites (IFN-β-P F and IFN-β-P R) to amplify -158 to +14 of the chicken IFN-β promoter motif, as described previously (29). The promoter fragment was inserted between Nhe I and Bgl II sites of the pGL3-basic luciferase reporter vector. The truncated plasmids of GoSTING, including d1-50 aa, d1-150 aa, d50-340 aa, d181-382 aa, d251-382 aa, d251-382 aa, d351-382 aa, d365-371 aa, d374-382 aa, d379-382 aa, and S369A were constructed using a modified homologous recombination method and the primers listed in Table 1.
Luciferase reporter assays
The DF-1 or GEF cells were plated in 24-well plates (NEST Biotechnology, Wuxi, China) and transiently transfected with the reporter plasmid pGL-IFN-β-Luc (0.12 μg/well) and internal control Renilla luciferase (PRL-TK, 0.06 μg/well) along with the indicated plasmids using Nulen PlusTrans™ Transfection Reagent (Nulen, Shanghai, China). According to the manufacturer’s instructions, the cells were lysed 24 hours after transfection, and luciferase activity was measured using a Dual-Luciferase Reporter Assay System kit (Promega, USA). Renilla luciferase activity was employed for normalization. All reporter assays were repeated at least three times.
Reverse transcription-quantitative real-time PCR
RNA was extracted from GEFs using an HP Total RNA kit (Omega, USA), and then the RNA was reverse-transcribed to cDNA using a cDNA synthesis kit (Vazyme). Reverse transcription-quantitative real-time PCR (qRT-PCR) tests were conducted according to the manufacturer’s instructions using a ChamQTM SYBR® qPCR Master Mix (Vazyme). The conditions and data processing method for the qRT-PCR test were previously described (29).
Virus infection and qRT-PCR analysis
For antiviral effect evaluation, GEF cells were transfected with pcDNA3.1-GoSTING-Flag plasmid or empty plasmid. After 24 hours, the GEF cells were washed twice with PBS (Gibco) and infected at 0.05 multiplicity of infection (MOI) with NSD14. The RNA from the cells, which were infected with the viruses at different times, was then collected for qRT-PCR to measure the mRNA level of GoSTING. The GoSTING-overexpressing and normal DF-1 cells were infected at 0.01 MOI with NDV-GFP or VSV-GFP, and fluorescence was measured 24h after infection using a fluorescence microscope.
Western blot analysis
The DF-1 cells were plated in 12-well plates at 1×106/mL and then transfected with a total of empty plasmid or GoSTING expression plasmid. At thirty-six hours post-transfection, cells were washed twice with phosphate buffer saline (PBS) (Gibco) and then lysed with a cell lysis buffer (Beyotime, Shanghai, China) containing an InStab™ protease cocktail (Yeasen, Shanghai, China) and phenylmethylsulfonyl fluoride (PMSF) (Yeasen). Lysates were centrifuged at 13,000 rpm for 15 minutes to obtain the supernatant and were eluted with a 5×SDS-PAGE loading buffer (Yeasen) and boiled for 10 min. Then the cell lysates were separated via SDS-PAGE and analyzed by Western blotting. Images were collected with the Tanon 5200 imaging system (Tanon, Shanghai, China), as described in our previous study (34).
Statistical analysis
Data were expressed as means ± standard deviations, with three biological replicates for each experiment. The two-tailed independent Student’s t-test was used to determine the significance. (*P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001).
Ethics statements
The studies involving goose embryos were conducted in the laboratory of Shanghai Veterinary Research Institute. The studies were reviewed and approved by the Animal Ethics Committee of Shanghai Veterinary Research Institute (20210521).
Results
Cloning and sequence analysis of GoSTING
Based on a predicted goose sequence (XM_013202032.1) from NCBI, primers GoSTING-F and GoSTING-R (Table 1), located outside of GoSTING ORF, were designed and used to amplify potential GoSTING cDNA using RT-PCR on total RNA extracted from the GEF cells.
Based on cDNA, the full-length GoSTING gene contains 1149 bp and encodes 382 amino acid (aa) residues (Figure 1A). Multiple sequence alignment showed that the amino acid sequences of GoSTING are 93.5, 44.0, and 61.8% identical to the STING gene in ducks (XP_027311055.1), humans (NP_938023.1), zebra finches (NP_001232785.1), respectively. The protein domains of GoSTING were predicted using the SMART program. The results show that GoSTING consists of a low compositional complexity region (31-42aa) and the TMEM173 (50-340aa) domain (Figure 1B).
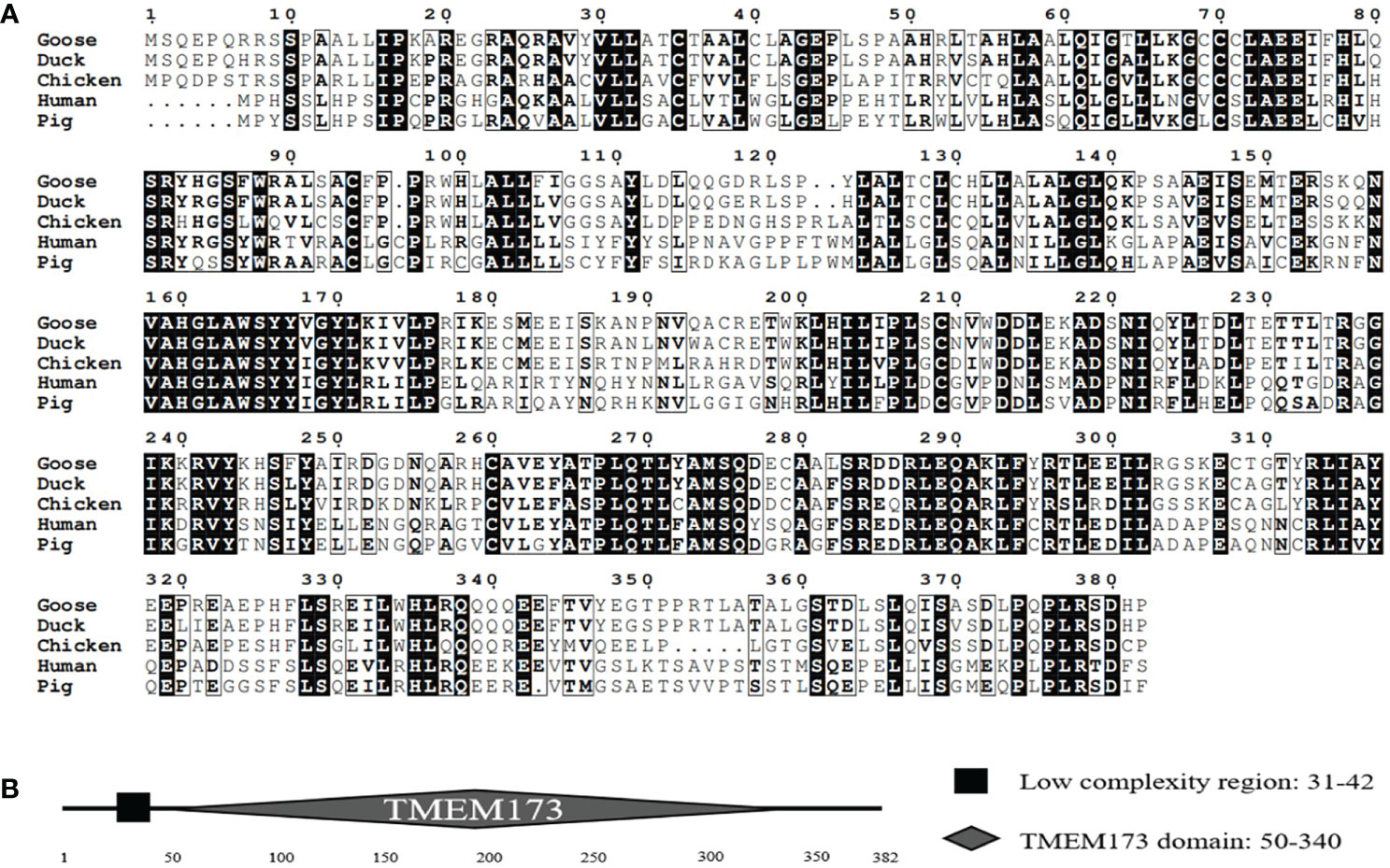
Figure 1 (A) The alignment of the deduced amino acid sequence of GoSTING with other animal STING proteins from the ducks, chickens, humans, and pigs was performed using the Clustal W program and edited with ESPript 3.0. The black shading indicates the identity of the amino acid, and the gray shading indicates similarity. (50% threshold). (B) The prediction of protein domains of GoSTING.
Phylogenetic tree analyses and the three-dimensional structure of GoSTING
The amino acid sequence homologies of different animals were conducted using MegAlign, and the results are shown in Figure 2A. A phylogenetic tree was developed based on multiple alignments of STING from various species, including fish, birds, and mammals. Phylogenetic analysis showed that the goose, duck, zebra finch, and chicken STING protein sequences were in the same subgroup. STING from mammals, including goats, cattle, pigs, cats, chimpanzees, humans, monkeys, and mice, was in another subgroup, and fish STING was in a third subgroup (Figure 2B). The predicted three-dimensional structures of GoSTING are shown in Figure 2C.
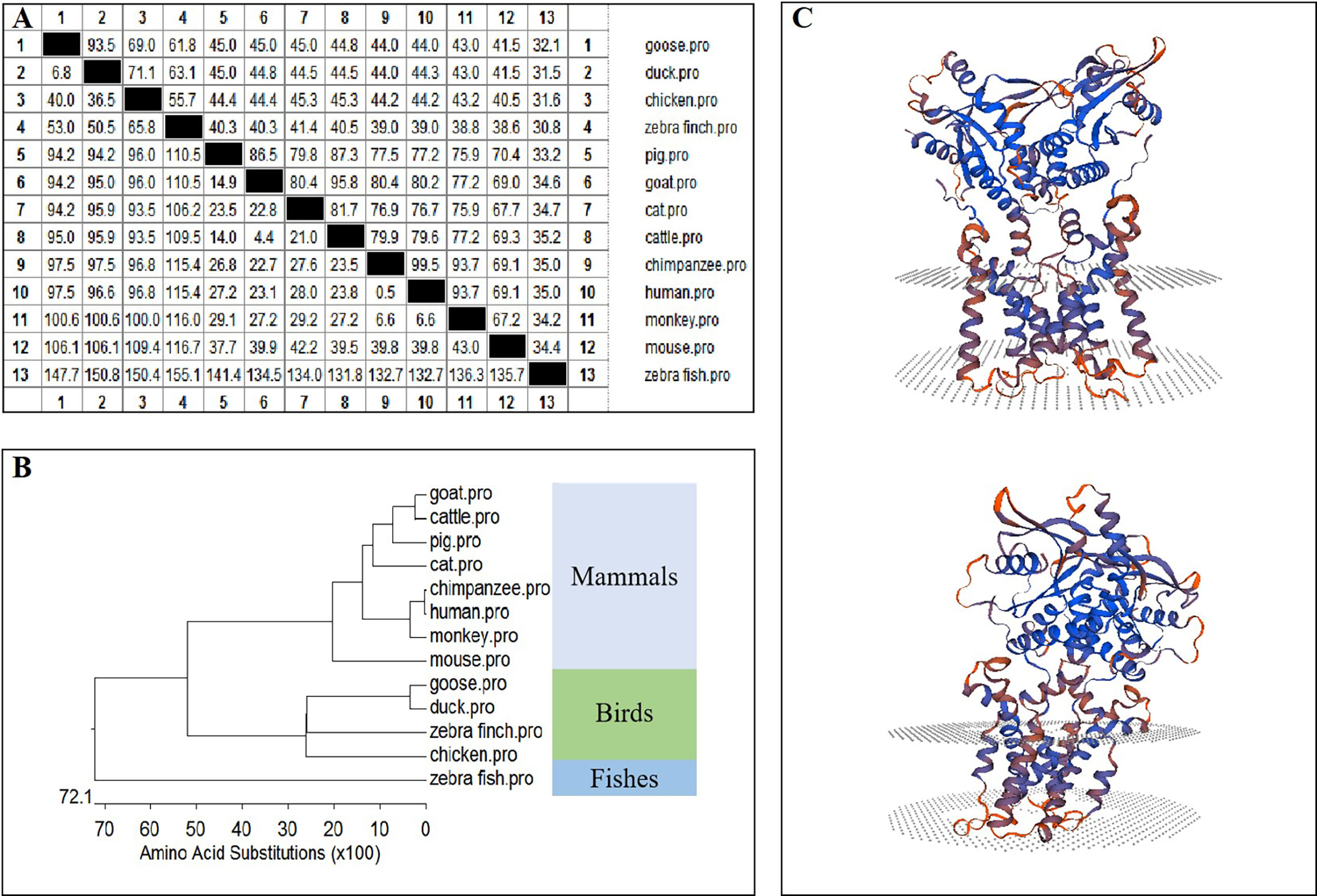
Figure 2 (A) The amino acid sequence homology of different animals. (B) Phylogenetic tree of the deduced amino acid sequence of GoSTING and other animal STING proteins. (C) Three-dimensional structure of GoSTING predicted by SWISS-MODEL.
Upregulation of GoSTING expression during viral infection
In mammals, STING is involved in the type I IFN-mediated antiviral innate immune response. However, the role of GoSTING in the antiviral response remains unclear. Upregulation of some immune-related genes is an important strategy for the host to fight infection. To determine whether GoSTING could respond to the RNA virus NDV, we analyzed the expression of GoSTING, some cytokines, and the ISGs in GEFs following infection with NDV using qRT-PCR. The results illustrated that the mRNA levels of GoSTING in the NDV-infected GEF cells were significantly upregulated during the early stages of infection (Figure 3A). The mRNA levels of IFNs (IFN-α, IFN-γ) (Figures 3B, C), IL-6 (Figure 3D), and ISGs (Mx-1 and PKR) (Figures 3E, F) were significantly upregulated as well. Cells resist the invasion of foreign viruses by upregulating the expression of these genes.
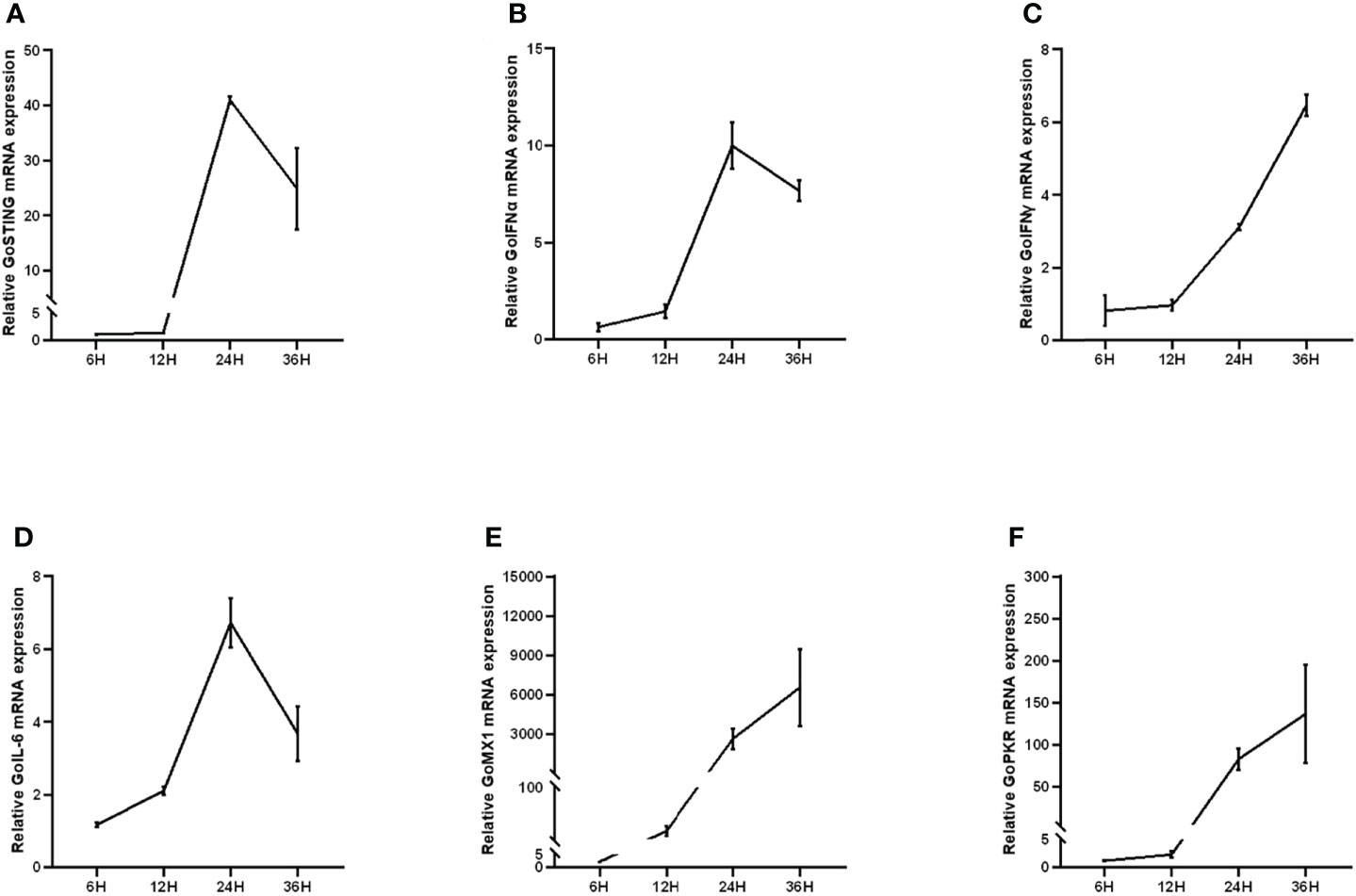
Figure 3 (A) Upregulation of GoSTING in GEF cells infected with NDV at 0.05 MOI. (B, C) Upregulation of IFNs (IFN-α and IFN-γ) in GEFs infected with NDV at 0.05 MOI. (D) Upregulation of IL-6 in GEFs infected with NDV at 0.05 MOI. (E, F) Upregulation of ISGs (Mx-1, PKR) in GEFs infected with NDV at 0.05 MOI. Error bars represent standard deviations.
GoSTING involved in the regulation of IFNs
STING has been a critical mediator of virus-triggered type I IFN signaling in chicken and duck cells through different pathways (14, 29, 31). To investigate whether GoSTING is also involved in the type I IFN signaling pathway, we transfected DF-1 cells with constructs expressing GoSTING and the empty vector, respectively, and examined the IFN-β activation with a luciferase reporter assay. The results showed that the overexpression of GoSTING resulted in a remarkable activation of the chIFN-β promoter in DF-1 cells (Figure 4A), and the activation of IFN-β exhibited a positive correlation with a dosage of the GoSTING plasmid (Figure 4B). To further confirm the ability of IFN activation of GoSTING, we prepared primary GEFs. Furthermore, luciferase assays were conducted with GEFs. Similarly, the overexpression of GoSTING in GEFs activated the IFN-β promoter (Figure 4C).
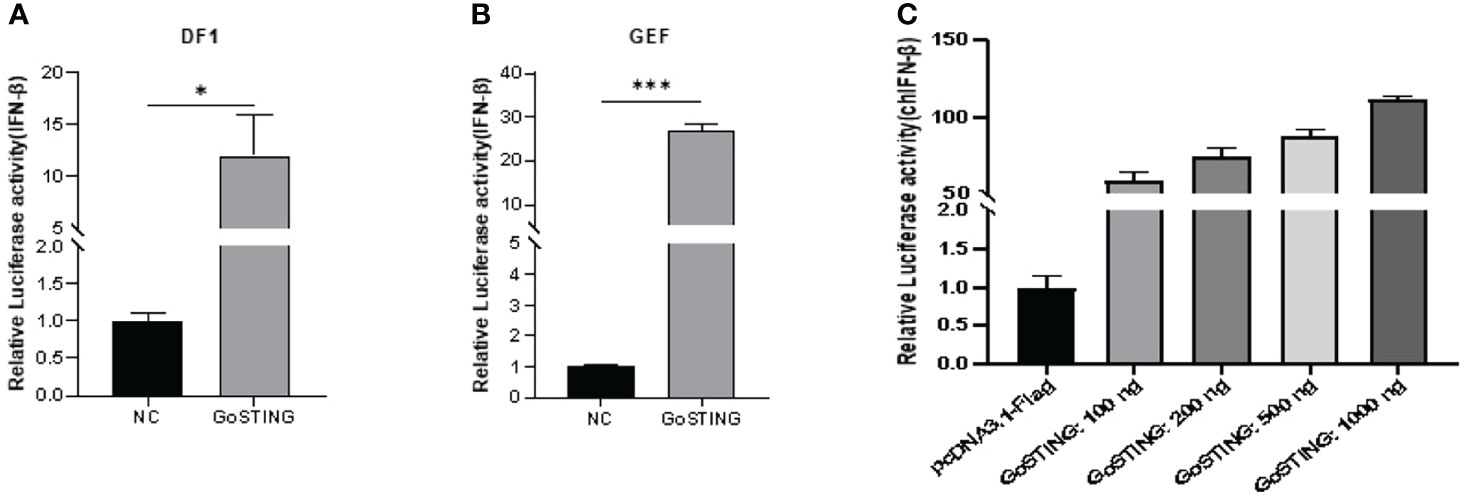
Figure 4 GoSTING is involved in regulating IFN-β. (A) DF-1 cells were cotransfected with luciferase reporter plasmids (pRL-TK and pGL-IFN-β-Luc) and pcDNA3.1-GoSTING-Flag or empty plasmid. Luciferase assays were performed after 24 hours of cotransfection. (B) GEFs were cotransfected with IFN-β luciferase reporter plasmids and with pcDNA3.1-GoSTING-Flag or pcDNA3.1-Flag. Luciferase assays were performed 24 hours after transfection. (C) GoSTING dose-independently induced IFN-β induction. The difference between the experimental and control groups was *p < 0.05 or ***p < 0.001.
The essential domains of GoSTING in IFN activation
Based on the structural domains of GoSTING predicted by the SMART program, a series of truncated mutants lacking different function domains were constructed (Figure 5A). Their ability to activate the IFN-β promoter was assessed with dual-luciferase reporter assays. As shown in Figure 5B, the deletion of 50 (d1-50 aa) amino acids showed a significant decrease in the ability to activate IFN-β compared with the wild-type GoSTING. The further deletion of 290 residues in the GoSTING (d50-340aa) resulted in a remarkable decrease in promoter activity. In contrast, for the deletion mutant, GoSTING-d379-382aa, even with a deletion of only 4 aa at the C-terminal, led to such a strong decrease in IFN-β induction. The N-terminal deletion mutant (d1-150aa), the C-terminal deletion mutant (d181-382aa, d251-382aa, d351-382 aa, and d374-382aa) and the mutant deleted 365-371 amino acids (d365-371aa) failed to activate the IFN-β promoter. Moreover, the S369 (corresponds to the S366 in human STING) seems to play a decisive role in IFN activation since the S369A mutant failed to activate IFN-β completely.
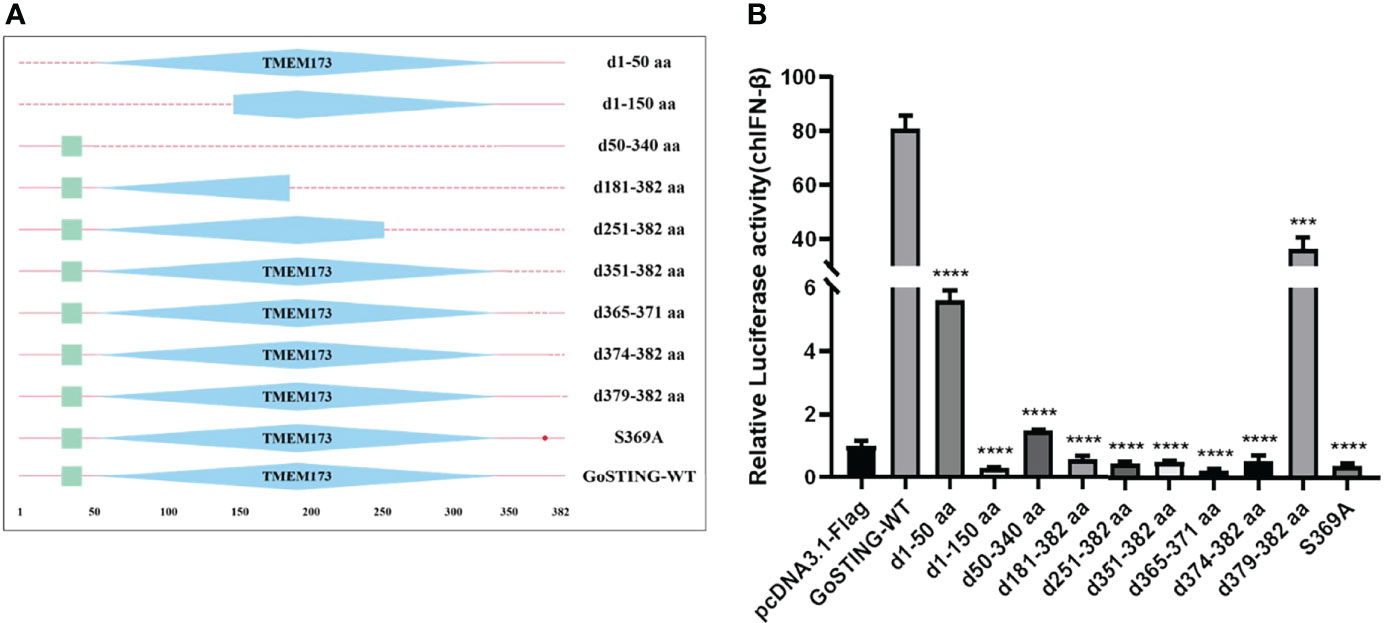
Figure 5 The Essential Domains of GoSTING in IFN Activation. (A) Schematic structure of GoSTING mutants. (B) The effects of GoSTING truncated mutants on IFN-β promoter activity. Cells were transfected with different expression plasmids of GoSTING and the reporter plasmids pGL-IFN-β-Luc and internal control Renilla luciferase (pRL-TK). Luciferase assays were performed 24h after transfection. All luciferase assays were repeated at least three times, and the difference between the experimental and control groups was ***p<0.001 or ****P<0.0001.
GoSTING plays an important role in anti-RNA viruses infection in vitro
To test the antiviral effects of GoSTING, the GoSTING-overexpressing and normal DF-1 cells were infected with NDV-GFP and VSV-GFP, respectively, and fluorescence was measured with a fluorescence microscope. The fluorescence intensities of both NDV-GFP and VSV-GFP in GoSTING overexpression cells were significantly lower than those in the control DF-1 cells at 14 and 24 h after viral infection (Figures 6A, B). To further investigate the GoSTING’s role during viral infection, the virus-infected cells were then lysed to detect the expression of NDV-GFP and VSV-GFP using Western blot. The protein band results showed that GoSTING could substantially reduce the expression of both NDV-GFP and VSV-GFP (Figures 6A, B). These results indicate that the overexpression of the GoSTING in DF-1 cells could inhibit NDV-GFP and VSV-GFP viral replication.
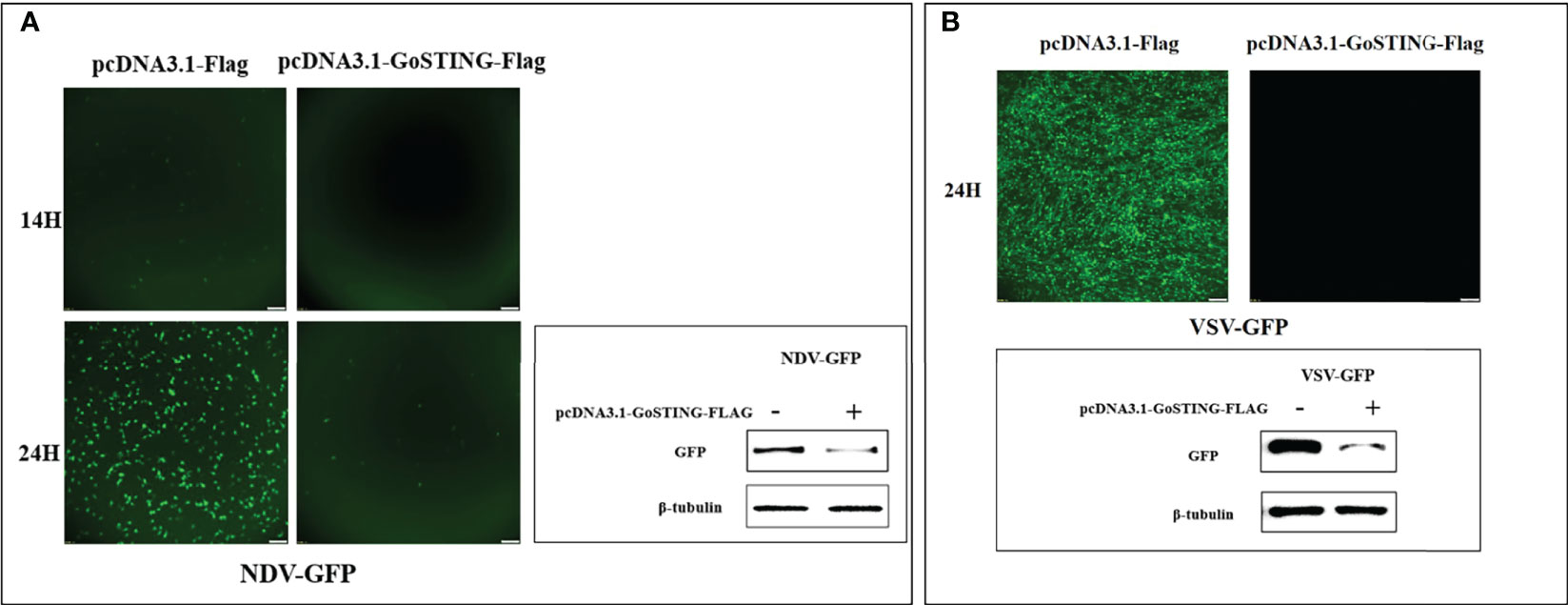
Figure 6 GoSTING inhibits viral yield. (A) Viral fluorescence in DF-1 cells transfected with pcDNA3.1-Flag or pcDNA3.1-GoSTING-Flag and infected with NDV-GFP at 0.01 MOI. Error bars represent standard deviations and Western blots for the expression of the NDV-GFP. (B) Viral fluorescence in DF-1 cells transfected with pcDNA3.1-Flag or pcDNA3.1-GoSTING-Flag and infected with VSV-GFP at 0.01 MOI. Error bars represent standard deviations and Western blots for the expression of the VSV-GFP.
Discussion
STING is a key signaling molecule that regulates innate immune signaling processes. Previous studies in our laboratory found that overexpression of chSTING and DuSTING in their respective cells could activate the IFN-β promoter and exert antiviral effects. Compared with chickens and ducks, the ability of geese to resist NDV and AIV showed a more significant advantage (35, 36). STING as a key IFN regulator may be one of the reasons for the difference in antiviral ability. A better understanding of the functions of GoSTING may help explain these differences. Currently, the functional characterization of GoSTING is pending and controversial. Therefore, it is necessary to carry out functional research on GoSTING.
In this study, GoSTING was identified with an open reading frame of 1149 bp, encoding 382 amino acid residues (Figure 1A). According to the prediction of the SMART website, GoSTING contains a TMEM173 (50–340 aa) domain (Figure 1B), which is highly conserved in GoSTING and other mammalian STINGs, indicating its important function in the host. Using MegAlign software alignment, the amino acid sequence of GoSTING was 93.5% similar to that of DuSTING, far exceeding that of other species (range from 32.1% to 69%), even to its closest relative birds, chickens, and zebra finches, the amino acid similarities were only 69% and 61.8% (Figure 2A). Similar results can be obtained by phylogenetic tree analysis (Figure 2B). The STING protein sequences of geese, ducks, zebra finches and chickens belong to one subgroup. The STING of mammals, including goats, cattle, pigs, cats, chimpanzees, humans, monkeys, and mice, belong to another subgroup. STING sequences from zebrafish belong to a third subgroup. The above results reflect that GoSTING has a closer genetic relationship with poultry, especially ducks. The predicted three-dimensional structures of GoSTING are shown in Figure 2C.
For the host, the production of IFN to induce the expression of ISGs, which is a powerful viral restriction factor in establishing an antiviral state, is a common strategy to resist viral infection (3). GEF cells were transfected with GoSTING or empty vector and then infected with NDV. As expected, the qRT-PCR test showed that the virus could significantly upregulate the mRNA levels of IFNs (IFN-α and IFN-γ) (Figures 3B, C) and downstream ISGs, including Mx-1 and PKR (Figures 3E, F), which have been shown to play an important role in the antiviral innate immune defense of IFN (37, 38). The proinflammatory factor IL-6 showed the same expression trend (Figure 3C). Many studies have shown that mammalian STING can act as an IFN-activated gene. To elucidate whether GoSTING has the same function, we overexpressed GoSTING in GEF or DF-1 cells and examined the activity of the IFN-β promoter by a dual-luciferase reporter assay. The results showed that overexpression of GoSTING could strongly activate the IFN-β promoter, and this induction was positively correlated with the dose of transfected GoSTING (Figures 4A–C). Based on the above findings, we infer that GoSTING inhibits NDV replication and exerts immunomodulatory effects by activating IFN pathway disorders and inducing some ISGs in the early stage of viral infection.
To identify the GoSTING domains important for IFN induction, a series of truncated forms of GoSTING mutants were generated, and their relative induction of IFN-β promoter activity was measured (Figure 5). The results showed that GoSTING mutants’ ability to miss the entire TMEM173 domain to activate the IFN-β promoter was significantly reduced, indicating that the TMEM173 domain was necessary for GoSTING to activate IFN-β, which was consistent with the previous TMEM173 domain of mammalian STING. Subtle changes in the amino acid sequence of TMEM173 affect STING-dependent innate immune signaling by reducing the ability to activate type I IFNs (13). Chen et al. determined that the carboxy-terminal region of STING is required and sufficient for activation of TBK1 and stimulation of IRF3 phosphorylation (39). Deletion of GoSTING C-terminal amino acid fragments of different sizes (d181-382aa, d251-382aa, d351-382aa, d374-382aa, and d379-382aa) resulted in a marked reduction in their ability to activate the IFN-β promoter. GoSTING-d1-50aa and GoSTING-d1-150aa, lacking 50 and 150 amino acids at the N-terminus of GoSTING, respectively, also had significantly reduced activation ability compared with wild-type GoSTING, which we speculate may be due to a barrier in its localization to the organelle. The underlying mechanism of STING regulation is phosphorylation at some sites in response to stimulation of cytoplasmic DNA (39). Our results show that the S369A point mutant abolished its IFN-β activation, suggesting that serine 369 may be an important serine site for STING activation.
Recently, the role of STING in inhibiting RNA viruses has attracted increasing research interest. RNA virus can activate STING and upregulate its expression after invading the host (40, 41). Deletion of STING renders murine embryonic fibroblasts (STING-/-MEFs) highly susceptible to infection by minus-strand viruses, including vesicular stomatitis virus (VSV) (42). ChSTING exhibits antiviral function against RNA viruses NDV and VSV (29, 43). We performed a series of experiments to clarify whether GoSTING also has antiviral activity. By monitoring the GoSTING mRNA level, we found that NDV can regulate GoSTING at the transcriptional level and upregulate the expression of GoSTING, and this phenomenon is particularly evident in the early and middle stages of virus infection, which indicates that GoSTING may play an important role in NDV infection. We thus explored the effect of GoSTING on viral replication. The results showed that GoSTING overexpression in DF-1 cells significantly inhibited the viral replication of NDV-GFP and VSV-GFP (Figure 6). In fact, research on STING for confinement of DNA viruses has already started (44). STING has been reported to be involved in innate immune defenses triggered by adenovirus, herpes simplex virus, and papilloma virus (45–47). Studies on chickens and ducks have also shown that in addition to RNA viruses, both chSTING and DuSTING show resistance to DNA viruses (14, 48, 49). Based on the conservation of amino acids in the STING protein that are critical for the recognition of various exogenous nucleic acid moieties (30) and the close kinship of geese to two other avian species, we speculate that GoSTING is required for host responses to both DNA and RNA viruses. This paper demonstrates that GoSTING plays an important role in RNA virus infection, but the role of GoSTING in DNA virus needs further study.
Nowadays some progress has been made in the research of STING in the innate immunity of birds. In this study, the amino acid sequence alignment showed that the homology of GoSTING to DuSTNG and chSTING was 93.5% and 69.0%, respectively (Figure 2A). It can be concluded that GoSTING has high homology with STING of birds, especially ducks. In addition, the results of protein domain prediction showed that the TMEM173 domain is conserved in birds, suggesting that birds may be similar in the activation of STING and the recognition of PAMPs. Previous studies have shown that overexpression of chSTING in DF-1 cells can significantly inhibit the replication of AIV and NDV, accompanied by an increase in pro-inflammatory cytokines such as IFN-β, IL-1β, and IL-2 (29). The effect of DuSTING on IFN activation and anti-RNA virus has also been elucidated by multiple investigators (31, 50). This study determined the functional characterization of GoSTING and found that GoSTING also has similar functions. We therefore conclude that STING is an essential IFN mediator that plays a role in avian innate immunity against RNA viruses. It is worth mentioning that both chSTING and DuSTING show resistance to DNA viruses, but whether GoSTING has the ability to resist DNA viruses remains to be studied.
However, the IFN signaling mechanisms of STING in chickens, ducks, and geese may be different, although they belong to the same bird species. At present, a relatively comprehensive study of the innate immune signaling pathway in chickens has been carried out. The biggest difference between the RLR pathway of chickens, ducks and geese is that chickens lack RIG-I (51), a key receptor for sensing many RNA viruses in birds, including AIV and NDV (32), making chickens more susceptible to some viruses, especially RNA viruses that require RIG-I for recognition. Previous studies in our laboratory showed that chSTING senses AIV virus by using MDA5 to compensate for RIG-I, and conducts signal transduction through MDA5-STING-IFN pathway (29). Nonetheless, chicken MDA5 is not sufficient against AIV, and AIV often causes lethal death in chickens (30). In contrast, ducks and geese tend to be natural hosts for many asymptomatic AIV subtypes (30), which are related to the molecular basis of their RIG-I. DuSTING was identified as an important receptor that responds to AIV infection and induces IFN-β production, but how RIG-I works for the function of DuSTING in RIG-I present ducks is unclear (31). Our results suggest that GoSTING is an important regulator of IFN, pro inflammatory cytokines and ISGs, and plays a role in antiviral innate immunity in geese. However, the current research on the RLR pathway of waterfowl, especially geese, is still relatively fragmented. Although both GoRIG-I (32) and GoMDA5 (52) have been shown to play a role in the anti-RNA virus innate immunity of geese, whether pathogen-associated RNA triggers STING signaling through RIG-I or MDA5 or whether RIG-I and MDA5 share the downstream STING signaling pathway is unclear. Future experiments based on RIG-I or MDA5 knockout duck cell lines may be required for further validation.
To sum up, our findings suggest that GoSTING is an important innate immune modulatory molecule involved in antiviral innate immunity in geese through its involvement in the type I IFN signaling pathway. The overexpression of GoSTING can upregulate several important pivotal ISGs and proinflammatory factors and combat NDV infection. Our study complements the functional characteristics of GoSTING, enriches the overall understanding of avian STING, and contributes to a more comprehensive and systematic understanding of the anti-RNA virus innate immune signaling pathway of avian STING.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving goose embryos were conducted in the laboratory of Shanghai Veterinary Research Institute. The studies were reviewed and approved by the Animal Ethics Committee of Shanghai Veterinary Research Institute (20210521). The animal study was reviewed and approved by the Animal Ethics Committee of Shanghai Veterinary Research Institute.
Author contributions
YC and JS designed the experiment. FF and ZL performed the majority of the experiments. JW, PL, YanL and YawL helped with the experiments. FF, ZL and YC wrote the paper. ZW, JM, and YY helped analyze the experimental results. All authors contributed to the article and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Natural Science foundation of Shanghai (20ZR1425100), the National Natural Science Foundation of China (32072865, and 32072864), Science and Technology Commission of Shanghai Municipality (21N41900100), the Interdisciplinary Program of Shanghai Jiao Tong University (YG2021QN108), and State Key Laboratory of Veterinary Biotechnology Foundation Grant (SKLVBF202107).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Aoshi T, Koyama S, Kobiyama K, Akira S, Ishii KJ. Innate and adaptive immune responses to viral infection and vaccination. Curr Opin Virol (2011) 1:226–32. doi: 10.1016/j.coviro.2011.07.002
2. Fischer S. Pattern recognition receptors and control of innate immunity: Role of nucleic acids. Curr Pharm Biotechnol (2018) 19:1203–9. doi: 10.2174/138920112804583087
3. Carty M, Guy C, Bowie AG. Detection of viral infections by innate immunity. Biochem Pharmacol (2021) 183:114316. doi: 10.1016/j.bcp.2020.114316
4. Damania B, Blackbourn DJ. Innate barriers to viral infection. Future Microbiol (2012) 7:815–22. doi: 10.2217/fmb.12.52
5. Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science (2010) 327:291–5. doi: 10.1126/science.1183021
6. Lin R, Xing J, Zheng C. Editorial: Sensing DNA in antiviral innate immunity. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.644310
7. Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses (2011) 3:920–40. doi: 10.3390/v3060920
8. Cheng Y, Ma J, Liu Y, Gao Q, Yan Y, Wang H, et al. Chicken TBK1 interacts with STING and is involved in IFN-β signaling regulation. Dev Comp Immunol (2017) 77:200–9. doi: 10.1016/j.dci.2017.08.011
9. Chen H, Jiang Z. The essential adaptors of innate immune signaling. Protein Cell (2013) 4:27–39. doi: 10.1007/s13238-012-2063-0
10. Gupta S, Tsoporis JN, Jia SH, Dos Santos CC, Parker TG, Marshall JC. Toll-like receptors, associated biochemical signaling networks, and S100 ligands. Shock (2021) 56:167–77. doi: 10.1097/SHK.0000000000001704
11. Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell (2020) 180:1044–66. doi: 10.1016/j.cell.2020.02.041
12. Asami J, Shimizu T. Structural and functional understanding of the toll-like receptors. Protein Sci (2021) 30:761–72. doi: 10.1002/pro.4043
13. Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol (2015) 15:760–70. doi: 10.1038/nri3921
14. Chen S, Wu Z, Zhang J, Wang M, Jia R, Zhu D, et al. Duck stimulator of interferon genes plays an important role in host anti-duck plague virus infection through an IFN-dependent signalling pathway. Cytokine (2018) 102:191–9. doi: 10.1016/j.cyto.2017.09.008
15. Liu Y, Goulet M-L, Sze A, Hadj SB, Belgnaoui SM, Lababidi RR, et al. RIG-I-Mediated STING upregulation restricts herpes simplex virus 1 infection. J Virol (2016) 90:9406–19. doi: 10.1128/JVI.00748-16
16. Ran JS, Jin J, Zhang XX, Wang Y, Ren P, Li JJ, et al. Molecular characterization, expression and functional analysis of chicken STING. Int J Mol Sci (2018) 19(12):3706. doi: 10.3390/ijms19123706
17. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature (2007) 448:501–5. doi: 10.1038/nature06013
18. Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA (2012) 109:E3008–17. doi: 10.1073/pnas.1211302109
19. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol (2011) 12:959–65. doi: 10.1038/ni.2091
20. Lee JH, Chiang C, Gack MU. Endogenous nucleic acid recognition by RIG-I-Like receptors and cGAS. J Interferon Cytokine Res (2019) 39:450–8. doi: 10.1089/jir.2019.0015
21. Wang J, Ba G, Han YQ, Ming SL, Wang MD, Fu PF, et al. Cyclic GMP-AMP synthase is essential for cytosolic double-stranded DNA and fowl adenovirus serotype 4 triggered innate immune responses in chickens. Int J Biol Macromol (2020) 146:497–507. doi: 10.1016/j.ijbiomac.2020.01.015
22. Yu H, Bruneau RC, Brennan G, Rothenburg S. Battle royale: Innate recognition of poxviruses and viral immune evasion. Biomedicines (2021) 9:765. doi: 10.3390/biomedicines9070765
23. Li D, Wu R, Guo W, Xie L, Qiao Z, Chen S, et al. STING-mediated IFI16 degradation negatively controls type I interferon production. Cell Rep (2019) 29:1249–1260.e4. doi: 10.1016/j.celrep.2019.09.069
24. Ganar K, Das M, Sinha S, Kumar S. Newcastle Disease virus: current status and our understanding. Virus Res (2014) 184:71–81. doi: 10.1016/j.virusres.2014.02.016
25. Mishra A, Vijayakumar P, Raut AA. Emerging avian influenza infections: Current understanding of innate immune response and molecular pathogenesis. Int Rev Immunol (2017) 36:89–107. doi: 10.1080/08830185.2017.1291640
26. Sun Y, Mao X, Zheng H, Wu W, Rehman ZU, Liao Y, et al. Goose MAVS functions in RIG-i-mediated IFN-β signaling activation. Dev Comp Immunol (2019) 93:58–65. doi: 10.1016/j.dci.2018.12.006
27. Karpala AJ, Stewart C, McKay J, Lowenthal JW, Bean AG. Characterization of chicken Mda5 activity: regulation of IFN-β in the absence of RIG-I functionality. J Immunol (2011) 186:5397–405. doi: 10.4049/jimmunol.1003712
28. Chen S, Cheng A, Wang M. Innate sensing of viruses by pattern recognition receptors in birds. Vet Res (2013) 44:82. doi: 10.1186/1297-9716-44-82
29. Cheng Y, Sun Y, Wang H, Yan Y, Ding C, Sun J. Chicken STING mediates activation of the IFN gene independently of the RIG-I gene. J Immunol (2015) 195:3922–36. doi: 10.4049/jimmunol.1500638
30. Neerukonda SN, Katneni U. Avian pattern recognition receptor sensing and signaling. Vet Sci (2020) 7(1):14. doi: 10.3390/vetsci7010014
31. Cheng Y, Liu Y, Shi S, Niu Q, Zhu W, Wang Z, et al. Functional characterization of duck STING in IFN-β induction and anti-H9N2 avian influenza viruses infections. Front Immunol (2019) 10:2224. doi: 10.3389/fimmu.2019.02224
32. Sun Y, Ding N, Ding SS, Yu S, Meng C, Chen H, et al. Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol Immunol (2013) 53:321–7. doi: 10.1016/j.molimm.2012.08.022
33. Vaheri A, Ruoslahti E, Hovi T, Nordling S. Stimulation of density-inhibited cell cultures by insulin. J Cell Physiol (1973) 81:355–63. doi: 10.1002/jcp.1040810308
34. Niu Q, Cheng Y, Wang H, Yan Y, Sun J. Chicken DDX3X activates IFN-β via the chSTING-chIRF7-IFN-β signaling axis. Front Immunol (2019) 10:822. doi: 10.3389/fimmu.2019.00822
35. Li ZJ, Li Y, Chang S, Ding Z, Mu LZ, Cong YL. Antigenic variation between Newcastle disease viruses of goose and chicken origin. Arch Virol (2010) 155:499–505. doi: 10.1007/s00705-010-0610-7
36. Pantin-Jackwood MJ, Swayne DE. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev Sci Tech (2009) 28:113–36. doi: 10.20506/rst.28.1.1869
37. Chen S, Zhang W, Wu Z, Zhang J, Wang M, Jia R, et al. Goose mx and OASL play vital roles in the antiviral effects of type I, II, and III interferon against newly emerging avian flavivirus. Front Immunol (2017) 8:1006. doi: 10.3389/fimmu.2017.01006
38. Liu WJ, Yang YT, Huang YM, Zhou DR, Xu DN, Cao N, et al. Identification of goose PKR gene: Structure, expression profiling, and antiviral activity against Newcastle disease virus. J Interferon Cytokine Res (2018) 38:333–40. doi: 10.1089/jir.2018.0025
39. Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal (2012) 5:ra20. doi: 10.1126/scisignal.2002521
40. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
41. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA (2009) 106:8653–8. doi: 10.1073/pnas.0900850106
42. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature (2008) 455:674–8. doi: 10.1038/nature07317
43. Li S, Yang J, Zhu Y, Ji X, Wang K, Jiang S, et al. Chicken DNA sensing cGAS-STING signal pathway mediates broad spectrum antiviral functions. Vaccines (Basel) (2020) 8(3):369. doi: 10.3390/vaccines8030369
44. Ni G, Ma Z, Damania B. cGAS and STING: At the intersection of DNA and RNA virus-sensing networks. PloS Pathog (2018) 14:e1007148. doi: 10.1371/journal.ppat.1007148
45. Lam E, Stein S, Falck-Pedersen E. Adenovirus detection by the cGAS/STING/TBK1 DNA sensing cascade. J Virol (2014) 88:974–81. doi: 10.1128/JVI.02702-13
46. Lau L, Gray EE, Brunette RL, Stetson DB. DNA Tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science (2015) 350:568–71. doi: 10.1126/science.aab3291
47. Ge R, Zhou Y, Peng R, Wang R, Li M, Zhang Y, et al. Conservation of the STING-mediated cytosolic DNA sensing pathway in zebrafish. J Virol (2015) 89:7696–706. doi: 10.1128/JVI.01049-15
48. Oliveira M, Rodrigues DR, Guillory V, Kut E, Giotis ES, Skinner MA, et al. Chicken cGAS senses fowlpox virus infection and regulates macrophage effector functions. Front Immunol (2021) 11:613079–9. doi: 10.3389/fimmu.2020.613079
49. Liu Y, Lin R, Olagnier D. RIGulation of STING expression: at the crossroads of viral RNA and DNA sensing pathways. Inflammation Cell Signal (2017) 4:e1491. doi: 10.14800/ics.1491
50. Zhang W, Jiang B, Zeng M, Duan Y, Wu Z, Wu Y, et al. Binding of duck tembusu virus nonstructural protein 2A to duck STING disrupts induction of its signal transduction cascade to inhibit beta interferon induction. J Virol (2020) 94(9):e01850–19. doi: 10.1128/JVI.01850-19
51. Magor KE, Miranzo Navarro D, Barber MRW, Petkau K, Fleming-Canepa X, Blyth GAD, et al. Defense genes missing from the flight division. Dev Comp Immunol (2013) 41:377–88. doi: 10.1016/j.dci.2013.04.010
Keywords: goose, STING, IFN, innate immunity, RNA virus
Citation: Fu F, Lin Z, Li Y, Wang J, Li Y, Liu P, Wang Z, Ma J, Yan Y, Sun J and Cheng Y (2022) Goose STING mediates IFN signaling activation against RNA viruses. Front. Immunol. 13:921800. doi: 10.3389/fimmu.2022.921800
Received: 16 April 2022; Accepted: 28 June 2022;
Published: 26 July 2022.
Edited by:
Rongtuan Lin, McGill University, CanadaReviewed by:
Shaopo Zu, Henan Agricultural University, ChinaHuibin Yu, Yale University, United States
Copyright © 2022 Fu, Lin, Li, Wang, Li, Liu, Wang, Ma, Yan, Sun and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqiang Cheng, d3l5Y3lxQHNqdHUuZWR1LmNu; Jianhe Sun, c3VuamhlQHNqdHUuZWR1LmNu
 Feiyu Fu
Feiyu Fu Zhenyu Lin
Zhenyu Lin Yanlin Li
Yanlin Li Jie Wang
Jie Wang Zhaofei Wang
Zhaofei Wang Yaxian Yan
Yaxian Yan Jianhe Sun
Jianhe Sun Yuqiang Cheng
Yuqiang Cheng