- 1Department of Obstetrics and Gynecology, Peking University Shenzhen Hospital, Shenzhen, China
- 2Institute of Obstetrics and Gynecology, Shenzhen Peking University-Hong Kong University of Science and Technology Medical Center (PKU-HKUST) Medical Center, Shenzhen, China
- 3Shenzhen Key Laboratory on Technology for Early Diagnosis of Major Gynecologic Diseases, Shenzhen, China
- 4Department of Biomedical Engineering, City University of Hong Kong, Hong Kong, Hong Kong SAR, China
Human papillomavirus (HPV) clearance is important in eliminating cervical cancer which contributes to high morbidity and mortality in women. Nevertheless, it remains largely unknown about key players in clearing pre-existing HPV infections. HPV antigens can be detected by the most important cervical antigen-presenting cells (Langerhans cells, LCs), of which the activities can be affected by cervicovaginal microbiota. In this review, we first introduce persistent HPV infections and then describe HPV-suppressed LCs activities, including but not limited to antigen uptake and presentation. Given specific transcriptional profiling of LCs in cervical epithelium, we also discuss the impact of cervicovaginal microbiota on LCs activation as well as the promise of exploring key microbial players in activating LCs and HPV-specific cellular immunity.
Introduction
Persistent high-risk human papillomavirus (hrHPV) infections causes the highest risk of invasive cervical cancer (ICC), one of the most common cancers threatening women’s health worldwide (1). Prophylactic HPV vaccines can not cover all HPV subtypes in ICC cases and thus provide limited benefits to eliminate pre-existing HPV infections (2, 3) which affect large populations in developing countries. Though screening is effective in preventing the progression of HPV infection and cervical dysplasia to ICC, it takes a long time to make it widely utilized, especially in low-income countries due to high financial and human resource burden. Clearance of HPV infection is encouraging alternative to eliminate ICC. Our and other prospective observational studies indicated that 85%, 50% and 5-10% of HPV infections could be cleared spontaneously in cervical intraepithelial neoplasia (CIN) 1, 2 and 3 respectively (4–6). Nevertheless, mechanisms of natural HPV clearance were largely unknown.
Langerhans cells in cervicovaginal mucosa represented key players in HPV antigen presentation and cellular immunity activation (7, 8). Emerging studies found LCs number or maturation levels reduced after HPV infections, and implicated the association of LCs with HPV clearance as well as CIN regression (9–14). In vitro experiments also indicated that LCs activation primed T cells and caused HPV-specific cellular immunity (15–18). Recent studies indicated unique transcriptional profiling of LCs in cervicovaginal microenvironments as compared to the skin and blood (19, 20), which may be caused by the impact of Lactobacillus and Candida on LCs immune functions (21, 22).
This review discusses how cervicovaginal HPV infections impair LCs immune functions, and what factors in cervicovaginal microenvironment hold the potential to activate LCs promoting HPV clearance, all of which will provide extensive insights into ICC prevention.
Carcinogenesis by persistent HPV infections
HPV infections require access of viral DNA, capsid protein L1 and L2 to the basal lamina binding to heparin sulfate proteoglycan on basal keratinocytes (23, 24). Once internalized, virions undergo endosomal transport and uncoating, with L2 protein-DNA complex (episome) ensuring correct nuclear entry of viral genomes while L1 protein retained in the endosome being degraded (25, 26). Before entry to the nucleus, E2 protein inhibits the expression of E6/E7 proteins which directly relate to the increasing severity of neoplasia by driving cell proliferation as well as inducing immune system dysfunction (27–29). If HPV episomes escape from host immune clearance, accumulation of E6/E7-induced genetic errors will eventually promote integration of viral episomes into the host cell chromosome (30–32) (Figure 1), together with overexpression of E6 and E7 led by dysregulation of E2 protein. Life cycle of HPV during persistent infections completes through the expression of minor coat protein L2 and major coat protein L1, as well as virus maturation in dying keratinocytes.
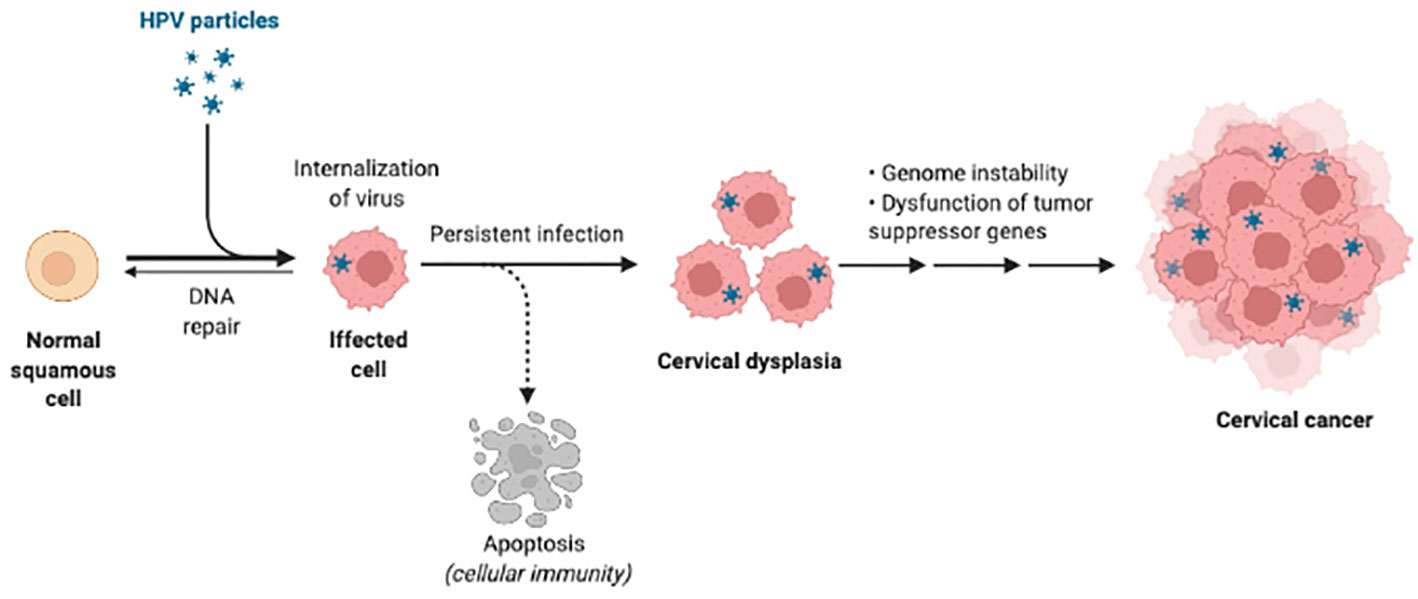
Figure 1 HPV infection and carcinogenesis. The HPV is internalized into the squamous cells first and persistent infection can cause DNA integration. Then infected cells are in dysfunction and develop to cancer. Created in BioRender.com.
Immune evasion plays key role in the above-mentioned life cycle of HPV. Keratinocytes function as immune sentinels via expressing pathogen recognition receptors (PRRs) to recognize pathogen-associated molecular patterns of HPV. Nevertheless, viral infection can suppress activities of PRRs such as toll-like receptors(TLRs) (33). For instance, E7 oncoprotein repressed TLR9 transcription by recruiting histone modifying enzyme EZH2 to the TLR9 promoter region (34). In consistence, E7 downregulates TLR9 expression through recruiting histone deacetylase HDAC1 and histone demethylase JARID1B to the regulatory region of the TLR9 promoter (35). Upon viral recognition, PRRs transduce intracellular signals to initiate the production of proinflammatory cytokines such as interferon(IFN)α and IFNβ. However, E6 and E7 proteins can block PRRs signal transduction cascades through two pathways: binding to interferon regulatory transcription factor (IRF) to inhibit its transcriptional activity and upregulating the deubiquitinating enzyme Ubiquitin C-Terminal Hydrolase L1 (UCHL1) to prevent TNF-receptor-associated factor 3 (TRAF3) activation (36–38).
The downregulation of NF-κB pathway is another critical strategy applied by HPV oncoproteins for immune evasion. NF-κB plays a key role in immune surveillance through promoting the expression of genes involved in antigen presentation and cytokine production (39). There is evidence supporting HPV16 E6 and E7 can inhibit NF-κB activity in keratinocytes which were cultured from the human cervical transformation zone (40). Moreover, high-risk HPV-infected keratinocytes upregulate UCHL1 to prevent the nuclear translocation of NF-κB (38). E6 and E7 can also bind to the coactivator of NF-κB [P300/CBP-associated factor (PCAF)] and then downregulate the NF-κB signaling pathway (41, 42).
Besides to impaired immune alarm functions of keratinocytes, above-mentioned inhibition of keratinocyte activities also modulates immune network to further facilitate persistent infection. For example, E5 protein reduces the expression of the major histocompatibility complex (MHC) I and CD1d to the cell surface, and then prevents cytotoxic T lymphocytes recognition of HPV antigens (43–45). In addition, E7 protein can interact with MHC I promoter, leading to repression of MHC I, LMP2, and TAP1 gene (46–48). Furthermore, HPV oncoproteins are capable of inhibiting the migration of antigen-presenting cells (APCs) to infected sites and repressing the APCs activities (49–53), further facilitating persistent infection.
Suppressed LCs activities caused by cervical HPV infections
Since LCs are the only APCs with which HPV will come into contact during HPV infections and initiate cell-mediated immune responses against HPV (7, 8, 19, 20), LCs are the most important and primary antigen-presenting cells (APCs) in the cervical epithelium. Suppression of LCs activities facilitated an immunosuppressive microenvironment that is permissive for HPV persistence (51–53), which was documented by reduced LCs number and maturation levels in cervical epithelium under HPV infections and squamous intraepithelial lesion (9–12).
Recruiting and remaining LCs in epidermis
Monocytes should be differentiated into competent and immature Langerhans cells which can detect pathogenic antigens. Nonetheless, HPV infection can block this differentiation and then inhibit the production of LCs (15). Immature LCs maintain the immune surveillance of the epithelium by presenting antigens and stimulating T lymphocytes following antigen uptake. After viral infections, immature LCs are recruited to the epidermis via chemokine (C-C-motif) receptor 6 (CCR6), which is the receptor for chemokine (c-c-motif) ligand 20 (CCL20) increasingly expressed by infected cervical keratinocytes (54, 55). Then LCs adhere to infected keratinocytes via LCs-expressed E-cadherin, as shown in Figure 2A. Nevertheless, HPV was reported to interfere with keratinocyte-derived CCL20 expression and HPV E6/E7 proteins promoted downregulation of CCL20, inhibiting LCs migration to the epidermis of inflammation (49, 50). Reduced E-cadherin levels caused by E7 protein also suggested the blockade of LCs adhesion with infected keratinocytes (56, 57), negatively affecting antigen uptake in epidermis after HPV infections. Moreover, the LC-keratinocyte crosstalk was dysregulated including decreased expression of interleukin-34 and depletion of immune-stimulatory LCs (58).
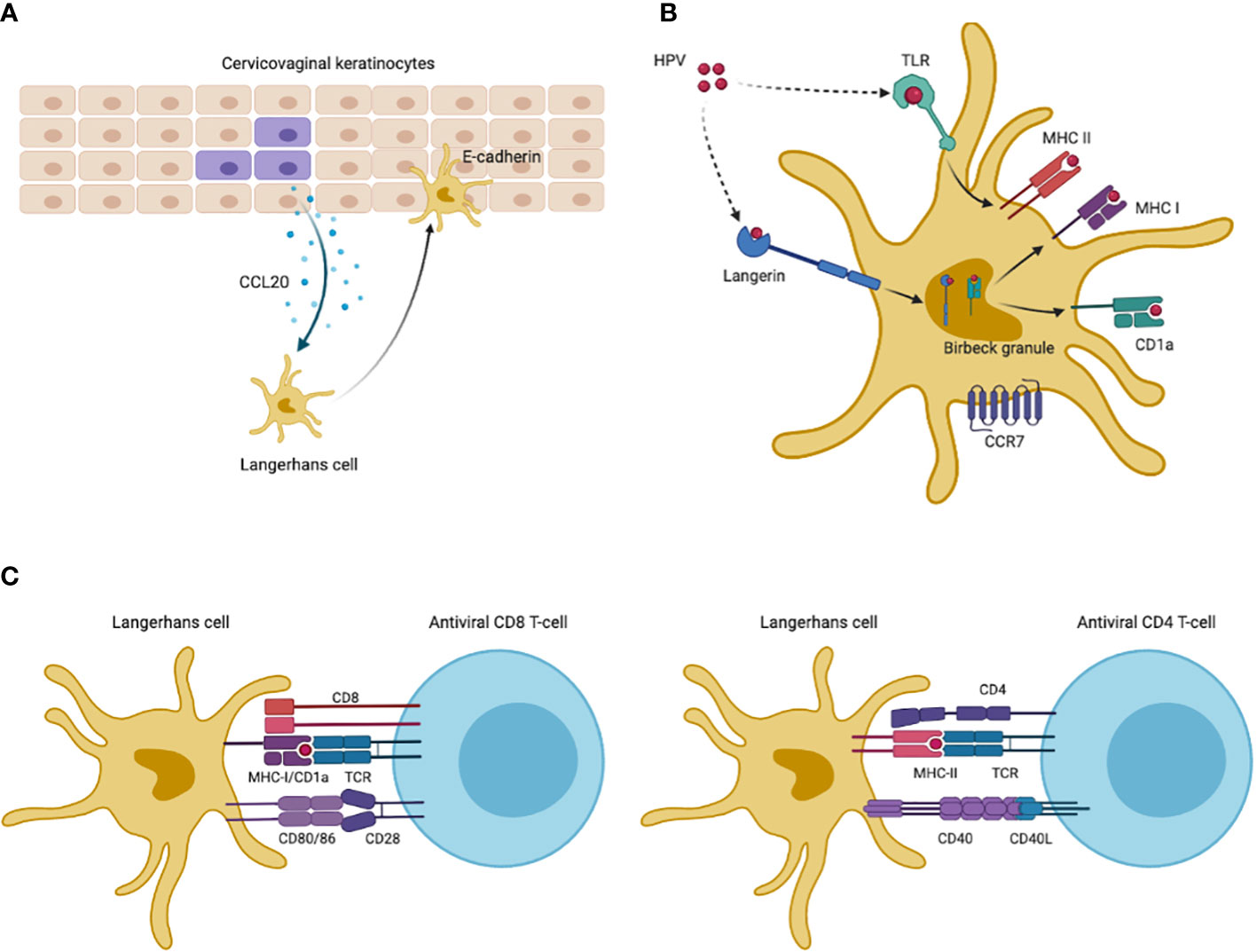
Figure 2 HPV antigen uptake and presentation of LCs. (A) Infected keratinocytes increase expression of CCL20 to attract Langerhans cells, which then migrate to inflammatory epidermis and adhere to keratinocytes via LCs-derived E-cadherin. (B) LCs maintained in inflammatory site capture HPV antigens via TLRs and langerin. Antigen uptake by TLRs are displayed on cell-membrane MHC-II molecules for presentation. Langerin-captured antigens are processed in intracellular Birbeck granules, being loaded on CD1a molecules in Birbeck granules or displayed in MHC-I molecules. CD1a molecules then cycle back to cell surfaces to present antigens. After antigen uptake and processing, LCs are migratory to lymph nodes via CCR7 which are receptors for T cell-derived CCL21. (C) Co-stimulatory molecules promote antigen presentation to CD4+ and CD8+ T cells. This Figureure was created in BioRender.com.
Capturing and processing antigens by LCs
When remained in cervical epidermis, immature LCs can capture HPV antigens via C-type lectin langerin (CD207) and toll-like receptors (TLRs) (59), as shown in Figure 2B. Cell-membrane langerin functions as endocytic receptor and internalizes antigens to LCs-specific intracellular Birbeck granules where antigens are degraded or delivered to antigen-presenting CD1a and major histocompatibility complex I (MHC-I) (60–64). Nevertheless, prior reports suggested that HPV significantly impaired langerin-CD1a antigen processing (10, 12). Besides to langerin, LCs also capture HPV antigens via TLRs which are common pathogen-recognition receptors (PRRs) on cell surface (59). However, LCs isolated from cervical tumors had decreased TLRs expression, such as TLR7/8/9, and were functionally anergic to TLR ligands, while a number of studies found that selective TLR agonists could promote the maturation of LCs (65–69). Additionally, HPV oncoproteins can activate the phosphoinositide 3-kinase (PI3-K) pathway to inhibit immune responses to HPV infection (16).
Migration of mature LCs to lymph node
LCs downregulate their endocytic and antigen processing capacity after binding pathogens, and then become mature as well as migratory (70). To present captured antigens, LCs should migrate to the lymph nodes via CCR7 which is only expressed by mature LCs and functions as receptor for T cell-derived chemokine CCL21 (55, 71), as shown in Figure 2B. Though there lacked reports on CCR7 levels of LCs after HPV infections, cervical cancer cells suppressed the induction of CCR7 in phenotypically mature dendritic cells (DCs), while specific co-stimulatory molecules upregulated CCR7 expression on LCs surface (65, 68, 72, 73).
Presenting antigens via LCs to activate cell-mediated immunity
CD1a, which is a lipid-presenting molecule abundantly expressed on LCs, can bind antigens in Birbeck granules and recycle to the cell surface presenting lipid/glycolipid antigens to CD1a-restricted T cells (63). HPV infections reduced the density of CD1a+ LCs in cervical epithelium, and CD1a+ LCs number correlated positively with prognosis of HPV-infected cases (10, 12, 13). Cell-membrane MHC-I molecules cross-present pathogen-derived peptides which can be recognized by cytotoxic lymphocytes, while MHC-II molecules are mainly expressed on APCs surfaces presenting antigens to helper T cells. HPV E7 protein was reported to directly block the MHC-I heavy chain promoter and E5 protein retained human leukocyte antigen A/B (HLA-A/B) (44, 46, 74), which may explained the partial loss of MHC-I in cervical cancer (75). MHC-II levels on LCs surface also reduced in cervical biopsy collected from CIN patients (11), while TLR agonists activating LCs immune functions or bacterial vector vaccines could up-regulate expression of MHC molecules and elicit CD4+/CD8+ CMI (65, 68, 76, 77).
Unlike typical DCs, co-stimulatory molecules CD80/86 and CD40 on LCs, belonging to tumor necrosis factor (TNF) receptor family, are needed in presenting antigens to T cells and inducing CMI (78–81) (Figure 2C). CD80/86 binds T cell-derived CD28 to prevent the induction of T cell anergy, while the CD80/86 ligand (cytotoxic T lymphocyte associate protein-4, CTLA-4) plays an inhibitory role by inducing T cell anergy. CD40 is another type of TNF receptors on LCs and functions by binding T cell membrane CD40L to prevent anergic status of T cells. In vitro experiments found higher levels of CD80/86 and CD40 in monocytes-derived LCs exposed to HPV VLPs after introduction of TLR agonists (65, 68).
Modulation of LCs activities and clearance of HPV infections in cervicovaginal microenvironment
Given the key roles of mucosal LCs in presenting HPV antigens and inducing CMI in cervical microenvironment, activation of suppressed LCs holds the potential to promote HPV clearance, partially supported by prior reports suggesting that LCs number was a strong and independent prognostic factor for HPV-infected cases in cervical, head and neck, lung carcinoma (13, 14, 82). Dorotheíe Duluc et al. identified cervical mucosa-specific LCs transcriptional fingerprints compared to skin and blood (19, 20), suggesting the impact of cervicovaginal microenvironment on LCs activities.
The complexity of cervicovaginal microbiota
A number of studies found predominance of one or few Lactobacillus species in CVM in healthy lower reproductive tract, like Lactobacillus crispatus (community-state type I, CST I), Lactobacillus gasseri (CST II), Lactobacillus iners (CST III) and Lactobacillus jensenii (CST V) () (83–87). In contrast, depletion of Lactobacillus (CST IV) was frequently found in women with genital infections (88–95). Microbe-microbe and -host interactions determined the complexity of CVM. Vaginal Lactobacillus spp. can produce lactic acid via glycogen fermentation, maintaining acidic environment to inhibit the colonization of pathogens such as Chlamydia trachomatis, Neisseria gonorrhoeae and Gardnerella vaginalis. In addition, Lactobacillus-derived bacteriocins exhibited inhibitory effects on common pathogenic bacteria and certain fungi, like G. vaginalis and Candida (96–98). Biosurfactants excretion was also applied by Lactobacillus to alter surface tension and bacterial adhesion, thus then preventing overgrowth of pathogenic anaerobes, especially G. vaginalis (99–102). Additionally, CVM can modulate a finely-tuned immune response balancing reproductive tolerance with protection against genital infections (83, 103, 104).
The association of CVM with HPV infections and clearance
Imbalanced CVM was widely identified in women with persistent HPV infections and cervical intraepithelial neoplasia (CIN), including decreased bacterial diversity, depletion of Lactobacillus and accumulation of Gardnerella and Sneathia (88–93). Further analysis found significant differences of CVM and cervical immune microenvironment between HPV- or CIN-negative women and HPV-positive women with CIN or cervical cancer (105–108). For example, inhibitory immune checkpoint protein PD-L1 and LAG-3 were in negative correlation with Lactobacillus levels, whereas TLR2 correlated positively with Lactobacillus abundance. In contrast, PD-L1 and LAG-3 positively correlated with pathogen Gardnerella, Sneathia, Atopobium and Prevotella. Additionally, L. jensenii and L. crispatus were in negative relationship with PD-L1, and L. gasseri was negatively associated with LAG-3. A 12-month observational study also demonstrated the critical role of cervicovaginal bacteria in modulation of cervicovaginal immune responses and the host susceptibility to HIV (103). Additional studies showed that L. cripatus-dominated CVM provided higher protection against pathogenic infections, compared to L. iners-dominated and non-Lactobacillus-dominated CVM (109–112) (Figure 3). Longitudinal studies further suggested high proportion of HPV clearance or CIN regression for HPV-positive populations with dominant L. crispatus in CVM (111, 112). Besides to bacterial components in CVM, vaginal fungi were associated with persistence of HPV infections (112), though a retrospective study involving 100,605 women found that Candida was not in significant association with the risk of CIN (113).
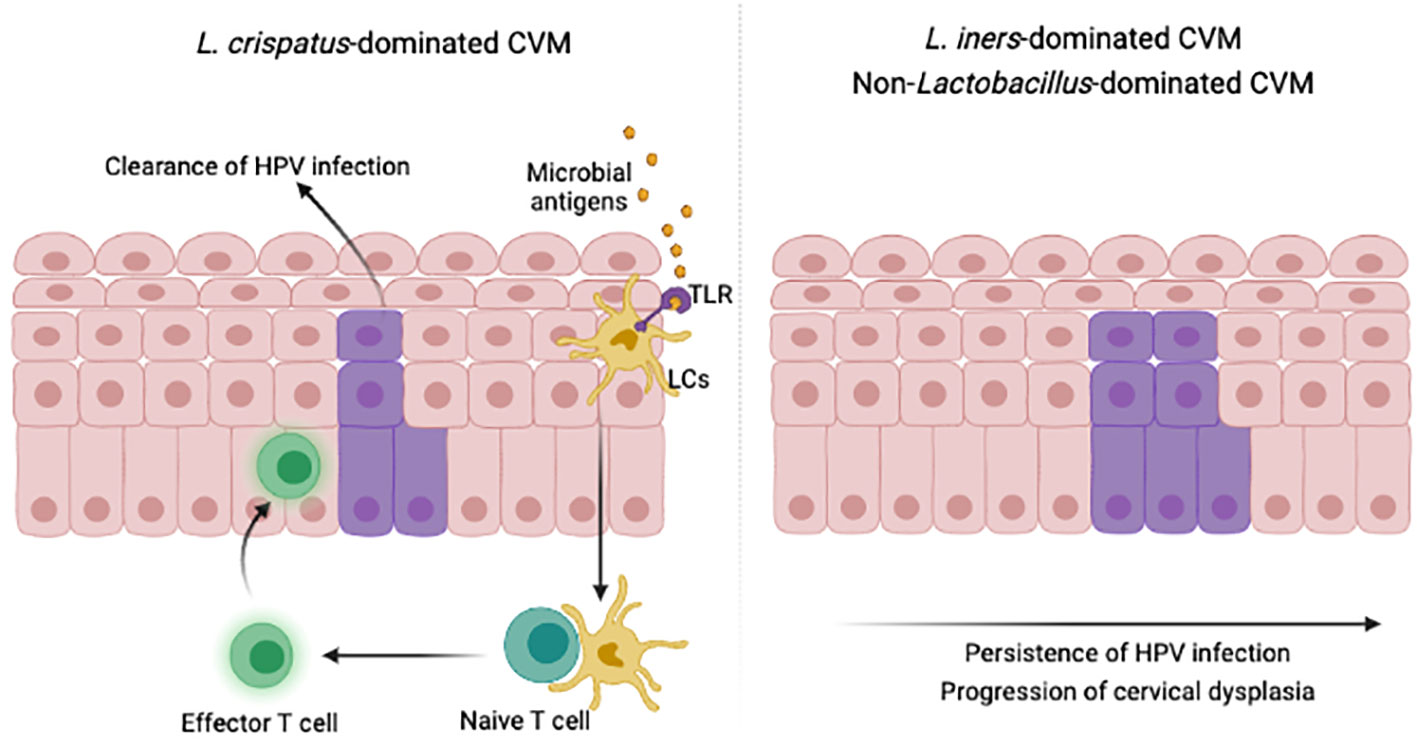
Figure 3 CVM modulate immunity to promote HPV clearance. Microbial key players in specific cervicovaginal environment activate suppressed LCs, migrating to lymph nodes, presenting antigens and attracting effector T cells to infected epidermis. This illustration was created in BioRender.com.
Modulating LCs activities via cervicovaginal microbial components
Though the potential role of CVM in modulating immune responses is largely unknown, co-culturing of monocytes-induced LCs and vaginal Lactobacillus indicated that peptidoglycans (PGN) in cell wall of specific L. crispatus strain increased the levels of LCs surface marker (langerin) which is pivotal in antigen capture (21). Further analysis demonstrated that PGN induced higher levels of langerin via elevated expression of recognized receptors of L. crispatus, such as TLR2 and TLR6 (21). Other studies also suggested that lipid polysaccharides (LPS), common bacterial product and potent TLR4 agonist activated LCs being assessed by upregulated MHC-II and CD80/86 molecules (72).
Given effective role of TLR agonists in therapeutic vaccines against HPV infections (114–119), emerging reports suggested the promise of selective microbial components as “natural” adjuvant in therapeutic HPV vaccines (111–125), partly due to their potential in enhancing LCs functions via activating TLRs through wide range of pathogen-associated molecular pattern molecules and damage-associated molecular pattern molecules. For instance, mouse model indicated that Lactobacillus lactis and Lactobacillus casei immobilizing HPV E7 peptides were effective in inducing HPV-specific CMI (124, 125). Candin, produced by common vaginal fungi Candida, was also applied as effective adjuvant in therapeutic vaccine against HPV infections, inducing wart resolution and elevated T-helper type 1 cells which promoted viral clearance (111, 112).
Concluding remarks and future perspectives
Cervicovaginal LCs play an important role in recognizing HPV antigens and activating HPV-specific CMI, and prior reports found the association of natural clearance of HPV infections with increased LCs activities (13, 14, 82). Though some LCs activators, like TLR agonists and products of vaginal microbes can activate LCs (21, 22, 65, 68), associated adjuvants were not always effective in promoting HPV clearance and CIN regression (114–116, 118–125). This may be explained by inter-individual CVM differences which can impact LCs functions as well as natural clearance of pre-existing HPC infections. Nevertheless, it remains unclear for the mechanisms how CVM modulate LCs activities to promote HPV clearance. Though in vitro experiments indicated the positive impact of specific vaginal Lactobacillus strains on LCs activities (21), functional redundancy for microbiota (126–128) and Lactobacillus strain-specific functions (129–133) necessitated the exploration of “functionally key microbial products” in modulating LCs activities.
To determine the promise of activating cervicovaginal LCs as novel therapies to clear HPV infections, the following research strategies are recommended. Firstly, prospective cohort study on HPV-positive women without CIN or with low-grade CIN are conducted to analyze the association of CVM-host interactions with clinical outcomes of HPV infections in cervicovaginal microenvironment. Secondly, multi-omics technologies, such as meta-transcriptomics, should be applied to explore the candidates of microbial products which can increase the level of infected keratinocyte-derived CCL20, attracting migration of LCs to HPV-infected site. Additionally, we also need to analyze candidate microbial products and associated gene sequences which hold the potential to activate LCs as TLR agonists or co-stimulatory molecules. Thirdly, in vivo cellular and animal models should be applied to assess the role of aforementioned microbial products in modulating LCs activities and clearing HPV, including W12 cell lines which were isolated from women with HPV16 infection and low-grade CIN as well as dog, rabbit and mouse models with relative papillomavirus (134). Lastly, sub-clinical and even clinical trial will be applied to assess the efficacy of above-mentioned microbial products in activating LCs and promoting HPV clearance.
Though activating cervicovaginal LCs is promising in clearing HPV infections, more solid evidence will be needed to explore the association of LCs activities with HPV clearance. For instance, the NF-κB signaling pathway that can regulate the expression of CCL20 and a variety of cytokines was inhibited in HPV-associated pre-cancerous CIN but activated significantly in cervical cancer (38–42, 135), which can be explained by dramatically changed immune microenvironment for cervical cancer (39, 135–138). Given ever-updating findings on HPV-host interaction (138–140), immune microenvironments such as lymph nodes and LCs-related T cell responses should be considered when exploring the role of CVM-LCs interactions in HPV clearance.
Author contributions
WD, LG and RW made substantial contributions to the design and writing of this manuscript. HD and SL contributed to the discussion and conception of the work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Shenzhen High-level Hospital Construction Fund (YBH2019-260), Shenzhen Key Medical Discipline Construction Fund (No.SZXK027) and Sanming Project of Medicine in Shenzhen (No.SZSM202011016) and Scientific Research Foundation of PEKING UNIVERSITY SHENZHEN HOSPITAL (No. KYQD2021075).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Hildesheim A, Herrero R, Wacholder S, Rodriguez AC, Solomon D, Bratti MC, et al. Effect of human papillomavirus 16/18 L1 viruslike particle vaccine among young women with preexisting infection: a randomized trial. JAMA (2007) 298(7):743–53. doi: 10.1001/jama.298.7.743
3. Group FIS. Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N Engl J Med (2007) 356(19):1915–27. doi: 10.1056/NEJMoa061741
4. Song F, Du H, Xiao A, Wang C, Huang X, Liu Z, et al. Type-specific distribution of cervical hrHPV infection and the association with cytological and histological results in a Large population-based cervical cancer screening program: Baseline and 3-year longitudinal data. J Cancer (2020) 11(20):6157–67. doi: 10.7150/jca.48357
5. Plummer M, Schiffman M, Castle PE, Maucort-Boulch D, Wheeler CM, ALTS Group. A 2-year prospective study of human papillomavirus persistence among women with a cytological diagnosis of atypical squamous cells of undetermined significance or low-grade squamous intraepithelial lesion. J Infect Dis (2007) 195(11):1582–9. doi: 10.1086/516784
6. McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol (2008) 9(5):425–34. doi: 10.1016/S1470-2045(08)70103-7
7. Morris HH, Gatter KC, Stein H, Mason DY. Langerhans' cells in human cervical epithelium: an immunohistological study. Br J Obstet Gynaecol (1983) 90(5):400–11. doi: 10.1111/j.1471-0528.1983.tb08935.x
8. Strobl H, Krump C, Borek I. Micro-environmental signals directing human epidermal langerhans cell differentiation. Semin Cell Dev Biol (2019) 86:36–43. doi: 10.1016/j.semcdb.2018.02.016
9. Leong CM, Doorbar J, Nindl I, Yoon HS, Hibma MH. Loss of epidermal langerhans cells occurs in human papillomavirus alpha, gamma, and mu but not beta genus infections. J Invest Dermatol (2010) 130(2):472–80. doi: 10.1038/jid.2009.266
10. Hubert P, Caberg JH, Gilles C, Bousarghin L, Franzen-Detrooz E, Boniver J, et al. E-cadherin-dependent adhesion of dendritic and langerhans cells to keratinocytes is defective in cervical human papillomavirus-associated (pre)neoplastic lesions. J Pathol (2005) 206(3):346–55. doi: 10.1002/path.1771
11. Mota F, Rayment N, Chong S, Singer A, Chain B. The antigen-presenting environment in normal and human papillomavirus (HPV)-related premalignant cervical epithelium. Clin Exp Immunol (1999) 116(1):33–40. doi: 10.1046/j.1365-2249.1999.00826.x
12. Connor JP, Ferrer K, Kane JP, Goldberg JM. Evaluation of langerhans' cells in the cervical epithelium of women with cervical intraepithelial neoplasia. Gynecol Oncol (1999) 75(1):130–5. doi: 10.1006/gyno.1999.5559
13. Shannon B, Yi TJ, Perusini S, Gajer P, Ma B, Humphrys MS, et al. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol (2017) 10(5):1310–9. doi: 10.1038/mi.2016.129
14. Miyagi J, Kinjo T, Tsuhako K, Higa M, Iwamasa T, Kamada Y, et al. Extremely high langerhans cell infiltration contributes to the favourable prognosis of HPV-infected squamous cell carcinoma and adenocarcinoma of the lung. Histopathology (2001) 38(4):355–67. doi: 10.1046/j.1365-2559.2001.01067.x
15. Iijima N, Goodwin EC, Dimaio D, Iwasaki A. High-risk human papillomavirus E6 inhibits monocyte differentiation to langerhans cells. Virology (2013) 444(1-2):257–62. doi: 10.1016/j.virol.2013.06.020
16. Fausch SC, Fahey LM, Da Silva DM, Kast WM. Human papillomavirus can escape immune recognition through langerhans cell phosphoinositide 3-kinase activation. J Immunol (2005) 174(11):7172–8. doi: 10.4049/jimmunol.174.11.7172
17. Fausch SC, Da Silva DM, Kast WM. Differential uptake and cross-presentation of human papillomavirus virus-like particles by dendritic cells and langerhans cells. Cancer Res (2003) 63(13):3478–82.
18. Fausch SC, Da Silva DM, Rudolf MP, Kast WM. Human papillomavirus virus-like particles do not activate langerhans cells: a possible immune escape mechanism used by human papillomaviruses. J Immunol (2002) 169(6):3242–9. doi: 10.4049/jimmunol.169.6.3242
19. Duluc D, Banchereau R, Gannevat J, Thompson-Snipes L, Blanck JP, Zurawski S, et al. Transcriptional fingerprints of antigen-presenting cell subsets in the human vaginal mucosa and skin reflect tissue-specific immune microenvironments. Genome Med (2014) 6(11):98. doi: 10.1186/s13073-014-0098-y
20. Duluc D, Gannevat J, Anguiano E, Zurawski S, Carley M, Boreham M, et al. Functional diversity of human vaginal APC subsets in directing T-cell responses. Mucosal Immunol (2013) 6(3):626–38. doi: 10.1038/mi.2012.104
21. Song J, Lang F, Zhao N, Guo Y, Zhang H. Vaginal lactobacilli induce differentiation of monocytic precursors toward langerhans-like cells: in vitro evidence. Front Immunol (2018) 9:2437. doi: 10.3389/fimmu.2018.02437
22. Nakagawa M, Coleman HN, Wang X, Daniels J, Sikes J, Nagarajan UM. IL-12 secretion by langerhans cells stimulated with Candida skin test reagent is mediated by dectin-1 in some healthy individuals. Cytokine (2014) 65(2):202–9. doi: 10.1016/j.cyto.2013.11.002
23. Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, et al. Arrangement of L2 within the papillomavirus capsid. J Virol (2008) 82(11):5190–7. doi: 10.1128/JVI.02726-07
24. Finnen RL, Erickson KD, Chen XS, Garcea RL. Interactions between papillomavirus L1 and L2 capsid proteins. J Virol (2003) 77(8):4818–26. doi: 10.1128/jvi.77.8.4818-4826.2003
25. Bergant Marusic M, Ozbun MA, Campos SK, Myers MP, Banks L. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic (2012) 13(3):455–67. doi: 10.1111/j.1600-0854.2011.01320.x
26. Schelhaas M, Shah B, Holzer M, Blattmann P, Kuhling L, Day PM, et al. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PloS Pathog (2012) 8(4):e1002657. doi: 10.1371/journal.ppat.1002657
27. Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol (2011) 19(1):33–9. doi: 10.1016/j.tim.2010.10.002
28. Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev (2009) 23(2):181–94. doi: 10.1101/gad.1735109
29. Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, et al. The biology and life-cycle of human papillomaviruses. Vaccine (2012) 30 Suppl 5:F55–70. doi: 10.1016/j.vaccine.2012.06.083
30. Lehoux M, D'Abramo CM, Archambault J. Molecular mechanisms of human papillomavirus-induced carcinogenesis. Public Health Genomics (2009) 12(5-6):268–80. doi: 10.1159/000214918
31. White EA, Kramer RE, Tan MJ, Hayes SD, Harper JW, Howley PM. Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity. J Virol (2012) 86(24):13174–86. doi: 10.1128/JVI.02172-12
32. Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc Natl Acad Sci U.S.A. (2006) 103(2):437–42. doi: 10.1073/pnas.0510012103
33. Miller LS. Toll-like receptors in skin. Adv Dermatol (2008) 24:71–87. doi: 10.1016/j.yadr.2008.09.004
34. Pacini L, Savini C, Ghittoni R, Saidj D, Lamartine J, Hasan UA, et al. Downregulation of toll-like receptor 9 expression by beta human papillomavirus 38 and implications for cell cycle control. J Virol (2015) 89:11396–405. doi: 10.1128/JVI.02151-15
35. Hasan UA, Zannetti C, Parroche P, Goutagny N, Malfroy M, Roblot G, et al. The human papillomavirus type 16 E7 oncoprotein induces a transcriptional repressor complex on the toll-like receptor 9 promoter. J Exp Med (2013) 210:1369–87. doi: 10.1084/jem.20122394
36. Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor- 3 and inhibits its transcriptional activity. Genes Dev (1998) 12:2061–72. doi: 10.1101/gad.12.13.2061
37. Park JS, Kim EJ, Kwon HJ, Hwang ES, Namkoong SE, Um SJ. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem (2000) 275:6764– 9. doi: 10.1074/jbc.275.10.6764
38. Karim R, Tummers B, Meyers C, Biryukov JL, Alam S, Backendorf C, et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PloS Pathog (2013) 9:e1003384. doi: 10.1371/journal.ppat.1003384
39. Tilborghs S, Corthouts J, Verhoeven Y, Arias D, Rolfo C, Trinh XB, et al. The role of nuclear factor-kappa b signaling in human cervical cancer. Crit Rev Oncol /Hematol (2017) 120:141–50. doi: 10.1016/j.critrevonc.2017.11.001
40. Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, et al. Human papillomavirus type 16 E6 and e 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF- kB promotes cell growth and immortalization. Virology (2012) 425:53–60. doi: 10.1016/j.virol.2011.12.023
41. Huang SM, McCance DJ. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol (2002) 76:8710– 21. doi: 10.1128/JVI.76.17.8710-8721.2002
42. Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J (1999) 18:5061–72. doi: 10.1093/emboj/18.18.5061
43. Cortese MS, Ashrafi GH, Campo MS. All 4 di-leucine motifs in the first hydrophobic domain of the E5 oncoprotein of human papillomavirus type 16 are essential for surface MHC class I downregulation activity and E5 endomembrane localization. Int J Cancer (2010) 126:1675– 82. doi: 10.1002/ijc.25004
44. Gruener M, Bravo IG, Momburg F, Alonso A, Tomakidi P. The E5 protein of the human papillomavirus type 16 down-regulates HLA-I surface expression in calnexin-expressing but not in calnexin-deficient cells. Virol J (2007) 4:116. doi: 10.1186/1743-422X-4-116
45. Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y, et al. CD1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (HPV) E5 protein: a possible mechanism for immune evasion by HPV. J Virol (2010) 84:11614– 23. doi: 10.1128/JVI.01053-10
46. Georgopoulos NT, Proffitt JL, Blair GE. Transcriptional regulation of the major histocompatibility complex (MHC) class I heavy chain, TAP1 and LMP2 genes by the human papillomavirus (HPV) type 6b, 16 and 18 E7 oncoproteins. Oncogene (2000) 19:4930–5. doi: 10.1038/sj.onc.1203860
47. Heller C, Weisser T, Mueller-Schickert A, Rufer E, Hoh A, Leonhardt RM, et al. Identification of key amino acid residues that determine the ability of high risk HPV16-E7 to dysregulate major histocompatibility complex class I expression. J Biol Chem (2011) 286:10983–97. doi: 10.1074/jbc.M110.199190
48. Li H, Ou X, Xiong J, Wang T. HPV16E7 mediates HADC chromatin repression and downregulation of MHC class I genes in HPV16 tumorigenic cells through interaction with an MHC class I promoter. Biochem Biophys Res Commun (2006) 349:1315–21. doi: 10.1016/j.bbrc.2006.08.182
49. Sperling T, Oldak M, Walch-Ruckheim B, Wickenhauser C, Doorbar J, Pfister H, et al. Human papillomavirus type 8 interferes with a novel C/EBPbeta-mediated mechanism of keratinocyte CCL20 chemokine expression and langerhans cell migration. PloS Pathog (2012) 8(7):e1002833. doi: 10.1371/journal.ppat.1002833
50. Guess JC, McCance DJ. Decreased migration of langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type 16 E6/E7 is related to reduced macrophage inflammatory protein-3alpha production. J Virol (2005) 79(23):14852–62. doi: 10.1128/JVI.79.23.14852-14862.2005
51. Denny LA, Franceschi S, de Sanjose S, Heard I, Moscicki AB, Palefsky J. Human papillomavirus, human immunodeficiency virus and immunosuppression. Vaccine (2012) 30 Suppl 5:F168–74. doi: 10.1016/j.vaccine.2012.06.045
52. Tang A, Dadaglio G, Oberkampf M, Di Carlo S, Peduto L, Laubreton D, et al. B cells promote tumor progression in a mouse model of HPV-mediated cervical cancer. Int J Cancer (2016) 139(6):1358–71. doi: 10.1002/ijc.30169
53. Bergot AS, Ford N, Leggatt GR, Wells JW, Frazer IH, Grimbaldeston MA. HPV16-E7 expression in squamous epithelium creates a local immune suppressive environment via CCL2- and CCL5- mediated recruitment of mast cells. PloS Pathog (2014) 10(10):e1004466. doi: 10.1371/journal.ppat.1004466
54. Le Borgne M, Etchart N, Goubier A, Lira SA, Sirard JC, van Rooijen N, et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity (2006) 24(2):191–201. doi: 10.1016/j.immuni.2006.01.005
55. Caux C, Vanbervliet B, Massacrier C, Ait-Yahia S, Vaure C, Chemin K, et al. Regulation of dendritic cell recruitment by chemokines. Transplantation (2002) 73(1 Suppl):S7–11. doi: 10.1097/00007890-200201151-00005
56. Brand A, Diener N, Zahner SP, Tripp C, Backer RA, Karram K, et al. E-cadherin is dispensable to maintain langerhans cells in the epidermis. J Invest Dermatol (2020) 140(1):132–42.e3. doi: 10.1016/j.jid.2019.06.132
57. Laurson J, Khan S, Chung R, Cross K, Raj K. Epigenetic repression of e-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis (2010) 31(5):918–26. doi: 10.1093/carcin/bgq027
58. Tuong ZK, Lukowski SW, Nguyen QH, Chandra J, Zhou C, Gillinder K, et al. A model of impaired langerhans cell maturation associated with HPV induced epithelial hyperplasia. iScience (2021) 24(11):103326. doi: 10.1016/j.isci.2021.103326
59. Ingber A. Langerhans cell receptors. Dermatol Clin (2007) 25(4):559–62. doi: 10.1016/j.det.2007.06.019
60. Fehres CM, Duinkerken S, Bruijns SC, Kalay H, van Vliet SJ, Ambrosini M, et al. Langerin-mediated internalization of a modified peptide routes antigens to early endosomes and enhances cross-presentation by human langerhans cells. Cell Mol Immunol (2017) 14(4):360–70. doi: 10.1038/cmi.2015.87
61. Valladeau J, Dezutter-Dambuyant C, Saeland S. Langerin/CD207 sheds light on formation of birbeck granules and their possible function in langerhans cells. Immunol Res (2003) 28(2):93–107. doi: 10.1385/IR:28:2:93
62. Valladeau J, Ravel O, Dezutter-Dambuyant C, Moore K, Kleijmeer M, Liu Y, et al. Langerin, a novel c-type lectin specific to langerhans cells, is an endocytic receptor that induces the formation of birbeck granules. Immunity (2000) 12(1):71–81. doi: 10.1016/s1074-7613(00)80160-0
63. Hunger RE, Sieling PA, Ochoa MT, Sugaya M, Burdick AE, Rea TH, et al. Langerhans cells utilize CD1a and langerin to efficiently present nonpeptide antigens to T cells. J Clin Invest (2004) 113(5):701–8. doi: 10.1172/JCI19655
64. Barral DC, Cavallari M, McCormick PJ, Garg S, Magee AI, Bonifacino JS, et al. CD1a and MHC class I follow a similar endocytic recycling pathway. Traffic (2008) 9(9):1446–57. doi: 10.1111/j.1600-0854.2008.00781.x
65. Fahey LM, Raff AB, Da Silva DM, Kast WM. Reversal of human papillomavirus-specific T cell immune suppression through TLR agonist treatment of langerhans cells exposed to human papillomavirus type 16. J Immunol (2009) 182(5):2919–28. doi: 10.4049/jimmunol.0803645
66. Le Poole IC, ElMasri WM, Denman CJ, Kroll TM, Bommiasamy H, Lyons Eiben G, et al. Langerhans cells and dendritic cells are cytotoxic towards HPV16 E6 and E7 expressing target cells. Cancer Immunol Immunother (2008) 57(6):789–97. doi: 10.1007/s00262-007-0415-z
67. Aliahmadi E, Gramlich R, Grutzkau A, Hitzler M, Kruger M, Baumgrass R, et al. TLR2-activated human langerhans cells promote Th17 polarization via IL-1beta, TGF-beta and IL-23. Eur J Immunol (2009) 39(5):1221–30. doi: 10.1002/eji.200838742
68. Da Silva DM, Woodham AW, Skeate JG, Rijkee LK, Taylor JR, Brand HE, et al. Langerhans cells from women with cervical precancerous lesions become functionally responsive against human papillomavirus after activation with stabilized poly-I:C. Clin Immunol (2015) 161(2):197–208. doi: 10.1016/j.clim.2015.09.003
69. Kumar MM, Adurthi S, Ramachandran S, Mukherjee G, Joy O, Krishnamurthy H, et al. Toll-like receptors 7, 8, and 9 expression and function in primary human cervical cancer langerhans cells: evidence of anergy. Int J Gynecol Cancer (2013) 23(1):184–92. doi: 10.1097/IGC.0b013e31827a2003
70. Kaplan DH, Kissenpfennig A, Clausen BE. Insights into langerhans cell function from langerhans cell ablation models. Eur J Immunol (2008) 38(9):2369–76. doi: 10.1002/eji.200838397
71. Dieu MC, Vanbervliet B, Vicari A, Bridon JM, Oldham E, Ait-Yahia S, et al. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J Exp Med (1998) 188(2):373–86. doi: 10.1084/jem.188.2.373
72. Da Silva DM, Movius CA, Raff AB, Brand HE, Skeate JG, Wong MK, et al. Suppression of langerhans cell activation is conserved amongst human papillomavirus alpha and beta genotypes, but not a micro genotype. Virology (2014) 452-453:279–86. doi: 10.1016/j.virol.2014.01.031
73. Pahne-Zeppenfeld J, Schroer N, Walch-Ruckheim B, Oldak M, Gorter A, Hegde S, et al. Cervical cancer cell-derived interleukin-6 impairs CCR7-dependent migration of MMP-9-expressing dendritic cells. Int J Cancer (2014) 134(9):2061–73. doi: 10.1002/ijc.28549
74. Ashrafi GH, Haghshenas MR, Marchetti B, O'Brien PM, Campo MS. E5 protein of human papillomavirus type 16 selectively downregulates surface HLA class I. Int J Cancer (2005) 113(2):276–83. doi: 10.1002/ijc.20558
75. Hasim A, Abudula M, Aimiduo R, Ma JQ, Jiao Z, Akula G, et al. Post-transcriptional and epigenetic regulation of antigen processing machinery (APM) components and HLA-I in cervical cancers from uighur women. PloS One (2012) 7(9):e44952. doi: 10.1371/journal.pone.0044952
76. Schnupf P, Portnoy DA. Listeriolysin O: a phagosome-specific lysin. Microbes Infect (2007) 9(10):1176–87. doi: 10.1016/j.micinf.2007.05.005
77. Watts C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat Immunol (2004) 5(7):685–92. doi: 10.1038/ni1088
78. Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev (2009) 229(1):152–72. doi: 10.1111/j.1600-065X.2009.00782.x
79. Goronzy JJ, Weyand CM. T-Cell co-stimulatory pathways in autoimmunity. Arthritis Res Ther (2008) 10 Suppl 1:S3. doi: 10.1186/ar2414
80. Kumamoto Y, Iwasaki A. Unique features of antiviral immune system of the vaginal mucosa. Curr Opin Immunol (2012) 24(4):411–6. doi: 10.1016/j.coi.2012.05.006
81. Chopin M, Nutt SL. Establishing and maintaining the langerhans cell network. Semin Cell Dev Biol (2015) 41:23–9. doi: 10.1016/j.semcdb.2014.02.001
82. Kindt N, Descamps G, Seminerio I, Bellier J, Lechien JR, Pottier C, et al. Langerhans cell number is a strong and independent prognostic factor for head and neck squamous cell carcinomas. Oral Oncol (2016) 62:1–10. doi: 10.1016/j.oraloncology.2016.08.016
83. Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun (2017) 8(1):875. doi: 10.1038/s41467-017-00901-0
84. Ma B, France MT, Crabtree J, Holm JB, Humphrys MS, Brotman RM, et al. A comprehensive non-redundant gene catalog reveals extensive within-community intraspecies diversity in the human vagina. Nat Commun (2020) 11(1):940. doi: 10.1038/s41467-020-14677-3
85. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U.S.A. (2011) 108 Suppl 1:4680–7. doi: 10.1073/pnas.1002611107
86. Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med (2012) 4(132):132ra52. doi: 10.1126/scitranslmed.3003605
87. Chaban B, Links MG, Jayaprakash TP, Wagner EC, Bourque DK, Lohn Z, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome (2014) 2:23. doi: 10.1186/2049-2618-2-23
88. Cheng L, Norenhag J, Hu YOO, Brusselaers N, Fransson E, Ahrlund-Richter A, et al. Vaginal microbiota and human papillomavirus infection among young Swedish women. NPJ Biofilms Microbiomes (2020) 6(1):39. doi: 10.1038/s41522-020-00146-8
89. Zhou Y, Wang L, Pei F, Ji M, Zhang F, Sun Y, et al. Patients with LR-HPV infection have a distinct vaginal microbiota in comparison with healthy controls. Front Cell Infect Microbiol (2019) 9:294. doi: 10.3389/fcimb.2019.00294
90. Oh HY, Kim BS, Seo SS, Kong JS, Lee JK, Park SY, et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin Microbiol Infect (2015) 21(7):674.e1–9. doi: 10.1016/j.cmi.2015.02.026
91. Mitra A, MacIntyre DA, Lee YS, Smith A, Marchesi JR, Lehne B, et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci Rep (2015) 5:16865. doi: 10.1038/srep16865
92. Borgogna JC, Shardell MD, Santori EK, Nelson TM, Rath JM, Glover ED, et al. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG (2020) 127(2):182–92. doi: 10.1111/1471-0528.15981
93. Lee JE, Lee S, Lee H, Song YM, Lee K, Han MJ, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PloS One (2013) 8(5):e63514. doi: 10.1371/journal.pone.0063514
94. Chao XP, Sun TT, Wang S, Fan QB, Shi HH, Zhu L, et al. Correlation between the diversity of vaginal microbiota and the risk of high-risk human papillomavirus infection. Int J Gynecol Cancer (2019) 29(1):28–34. doi: 10.1136/ijgc-2018-000032
95. Gao W, Weng J, Gao Y, Chen X. Comparison of the vaginal microbiota diversity of women with and without human papillomavirus infection: a cross-sectional study. BMC Infect Dis (2013) 13:271. doi: 10.1186/1471-2334-13-271
96. Kovachev S. Defence factors of vaginal lactobacilli. Crit Rev Microbiol (2018) 44(1):31–9. doi: 10.1080/1040841X.2017.1306688
97. Stoyancheva G, Marzotto M, Dellaglio F, Torriani S. Bacteriocin production and gene sequencing analysis from vaginal lactobacillus strains. Arch Microbiol (2014) 196(9):645–53. doi: 10.1007/s00203-014-1003-1
98. Dover SE, Aroutcheva AA, Faro S, Chikindas ML. Natural antimicrobials and their role in vaginal health: A short review. Int J Probiotics Prebiotics (2008) 3(4):219–30.
99. Ojala T, Kankainen M, Castro J, Cerca N, Edelman S, Westerlund-Wikstrom B, et al. Comparative genomics of lactobacillus crispatus suggests novel mechanisms for the competitive exclusion of gardnerella vaginalis. BMC Genomics (2014) 15:1070. doi: 10.1186/1471-2164-15-1070
100. Reid G, Heinemann C, Velraeds M, van der Mei HC, Busscher HJ. Biosurfactants produced by lactobacillus. Methods Enzymol (1999) 310:426–33. doi: 10.1016/s0076-6879(99)10033-8
101. Zarate G, Nader-Macias ME. Influence of probiotic vaginal lactobacilli on in vitro adhesion of urogenital pathogens to vaginal epithelial cells. Lett Appl Microbiol (2006) 43(2):174–80. doi: 10.1111/j.1472-765X.2006.01934.x
102. Strus M, Brzychczy-Wloch M, Gosiewski T, Kochan P, Heczko PB. The in vitro effect of hydrogen peroxide on vaginal microbial communities. FEMS Immunol Med Microbiol (2006) 48(1):56–63. doi: 10.1111/j.1574-695X.2006.00120.x
103. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity (2015) 42(5):965–76. doi: 10.1016/j.immuni.2015.04.019
104. Laniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol (2020) 17(4):232–50. doi: 10.1038/s41585-020-0286-z
105. Laniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-Hispanic and Hispanic women. Sci Rep (2018) 8(1):7593. doi: 10.1038/s41598-018-25879-7
106. Ilhan ZE, Laniewski P, Thomas N, Roe DJ, Chase DM, Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine (2019) 44:675–90. doi: 10.1016/j.ebiom.2019.04.028
107. Laniewski P, Cui H, Roe DJ, Barnes D, Goulder A, Monk BJ, et al. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci Rep (2019) 9(1):7333. doi: 10.1038/s41598-019-43849-5
108. Laniewski P, Cui H, Roe DJ, Chase DM, Herbst-Kralovetz MM. Vaginal microbiota, genital inflammation, and neoplasia impact immune checkpoint protein profiles in the cervicovaginal microenvironment. NPJ Precis Oncol (2020) 4:22. doi: 10.1038/s41698-020-0126-x
109. Wang H, Ma Y, Li R, Chen X, Wan L, Zhao W. Associations of cervicovaginal lactobacilli with high-risk human papillomavirus infection, cervical intraepithelial neoplasia, and cancer: A systematic review and meta-analysis. J Infect Dis (2019) 220(8):1243–54. doi: 10.1093/infdis/jiz325
110. Di Paola M, Sani C, Clemente AM, Iossa A, Perissi E, Castronovo G, et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk human papillomavirus infection. Sci Rep (2017) 7(1):10200. doi: 10.1038/s41598-017-09842-6
111. Mitra A, MacIntyre DA, Ntritsos G, Smith A, Tsilidis KK, Marchesi JR, et al. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat Commun (2020) 11(1):1999. doi: 10.1038/s41467-020-15856-y
112. Usyk M, Zolnik CP, Castle PE, Porras C, Herrero R, Gradissimo A, et al. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PloS Pathog (2020) 16(3):e1008376. doi: 10.1371/journal.ppat.1008376
113. Engberts MK, Verbruggen BS, Boon ME, van Haaften M, Heintz AP. Candida and dysbacteriosis: a cytologic, population-based study of 100,605 asymptomatic women concerning cervical carcinogenesis. Cancer (2007) 111(5):269–74. doi: 10.1002/cncr.22947
114. Da Silva DM, Skeate JG, Chavez-Juan E, Luhen KP, Wu JM, Wu CM, et al. Therapeutic efficacy of a human papillomavirus type 16 E7 bacterial exotoxin fusion protein adjuvanted with CpG or GPI-0100 in a preclinical mouse model for HPV-associated disease. Vaccine (2019) 37(22):2915–24. doi: 10.1016/j.vaccine.2019.04.043
115. Maynard SK, Marshall JD, MacGill RS, Yu L, Cann JA, Cheng LI, et al. Vaccination with synthetic long peptide formulated with CpG in an oil-in-water emulsion induces robust E7-specific CD8 T cell responses and TC-1 tumor eradication. BMC Cancer (2019) 19(1):540. doi: 10.1186/s12885-019-5725-y
116. Gandhapudi SK, Ward M, Bush JPC, Bedu-Addo F, Conn G, Woodward JG. Antigen priming with enantiospecific cationic lipid nanoparticles induces potent antitumor CTL responses through novel induction of a type I IFN response. J Immunol (2019) 202(12):3524–36. doi: 10.4049/jimmunol.1801634
117. Vasievich EA, Chen W, Huang L. Enantiospecific adjuvant activity of cationic lipid DOTAP in cancer vaccine. Cancer Immunol Immunother (2011) 60(5):629–38. doi: 10.1007/s00262-011-0970-1
118. Daayana S, Elkord E, Winters U, Pawlita M, Roden R, Stern PL, et al. Phase II trial of imiquimod and HPV therapeutic vaccination in patients with vulval intraepithelial neoplasia. Br J Cancer (2010) 102(7):1129–36. doi: 10.1038/sj.bjc.6605611
119. Zom GG, Willems M, Khan S, van der Sluis TC, Kleinovink JW, Camps MGM, et al. Novel TLR2-binding adjuvant induces enhanced T cell responses and tumor eradication. J Immunother Cancer (2018) 6(1):146. doi: 10.1186/s40425-018-0455-2
120. Coleman HN, Greenfield WW, Stratton SL, Vaughn R, Kieber A, Moerman-Herzog AM, et al. Human papillomavirus type 16 viral load is decreased following a therapeutic vaccination. Cancer Immunol Immunother (2016) 65(5):563–73. doi: 10.1007/s00262-016-1821-x
121. Greenfield WW, Stratton SL, Myrick RS, Vaughn R, Donnalley LM, Coleman HN, et al. A phase I dose-escalation clinical trial of a peptide-based human papillomavirus therapeutic vaccine with candida skin test reagent as a novel vaccine adjuvant for treating women with biopsy-proven cervical intraepithelial neoplasia 2/3. Oncoimmunology (2015) 4(10):e1031439. doi: 10.1080/2162402X.2015.1031439
122. Kim KH, Horn TD, Pharis J, Kincannon J, Jones R, O'Bryan K, et al. Phase 1 clinical trial of intralesional injection of candida antigen for the treatment of warts. Arch Dermatol (2010) 146(12):1431–3. doi: 10.1001/archdermatol.2010.350
123. Kawana K, Adachi K, Kojima S, Taguchi A, Tomio K, Yamashita A, et al. Oral vaccination against HPV E7 for treatment of cervical intraepithelial neoplasia grade 3 (CIN3) elicits E7-specific mucosal immunity in the cervix of CIN3 patients. Vaccine (2014) 32(47):6233–9. doi: 10.1016/j.vaccine.2014.09.020
124. Ribelles P, Benbouziane B, Langella P, Suarez JE, Bermudez-Humaran LG. Protection against human papillomavirus type 16-induced tumors in mice using non-genetically modified lactic acid bacteria displaying E7 antigen at its surface. Appl Microbiol Biotechnol (2013) 97(3):1231–9. doi: 10.1007/s00253-012-4575-1
125. Lee TY, Kim YH, Lee KS, Kim JK, Lee IH, Yang JM, et al. Human papillomavirus type 16 E6-specific antitumor immunity is induced by oral administration of HPV16 E6-expressing lactobacillus casei in C57BL/6 mice. Cancer Immunol Immunother (2010) 59(11):1727–37. doi: 10.1007/s00262-010-0903-4
126. Tian L, Wang XW, Wu AK, Fan YH, Friedman J, Dahlin A, et al. Deciphering functional redundancy in the human microbiome. Nat Commun (2020) 11(1):6217. doi: 10.1038/s41467-020-19940-1
127. Ferrer M, Ruiz A, Lanza F, Haange SB, Oberbach A, Till H, et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol (2013) 15(1):211–26. doi: 10.1111/j.1462-2920.2012.02845.x
128. Morrison DJ, Preston T. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Gut Microbes (2016) 7(3):189–200. doi: 10.1080/19490976.2015.1134082
129. Liu JS, Hu DG, Chen YQ, Huang HX, Zhang H, Zhao JX, et al. Strain-specific properties of lactobacillus plantarum for prevention of salmonella infection. Food Funct (2018) 9(7):3673–82. doi: 10.1039/c8fo00365c
130. Atassi F, Ahn DLPV, Moal VLL. Diverse expression of antimicrobial activities against bacterial vaginosis and urinary tract infection pathogens by cervicovaginal microbiota strains of Lactobacillus gasseri and Lactobacillus crispatus. Front Microbiol (2019) 10:2900. doi: 10.3389/fmicb.2019.02900
131. Schmid M, Muri J, Melidis D, Varadarajan AR, Somerville V, Wicki A, et al. Comparative genomics of completely sequenced Lactobacillus helveticus genomes provides insights into strain-specific genes and resolves metagenomics data down to the strain level. Front Microbiol (2018) 9:63. doi: 10.3389/fmicb.2018.00063
132. Lee IC, Caggianiello G, Swam II, Taverne N, Meijerink M, Bron PA, et al. Strain-specific features of extracellular polysaccharides and their impact on Lactobacillus plantarum-host interactions. Appl Environ Microbiol (2016) 82(13):3959–70. doi: 10.1128/AEM.00306-16
133. Sadi RA, Nighot P, Nighot M, Haque M, Rawat M, Ma TY. Lactobacillus acidophilus induces a strain-specific and toll-like receptor 2-dependent enhancement of intestinal epithelial tight junction barrier and protection against intestinal inflammation. Am J Pathol (2021) 191(5):872–84. doi: 10.1016/j.ajpath.2021.02.003
134. Doorbar J. Model systems of human papillomavirus-associated disease. J Pathol (2016) 238(2):166–79. doi: 10.1002/path.4656
135. DAC RM, Bastos MM, Medeiros R, Oliveira PA. The NFkappaB signaling pathway in papillomavirus-induced lesions: friend or foe? Anticancer Res (2016) 36:2073–83.
136. Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases–elimination, equilibrium and escape. Curr Opin Immunol (2014) 27:16–25. doi: 10.1016/j.coi.2014.01.004
137. Rotman J, Otter LAS, Bleeker MCG, Samuels SS, Heeren AM, Roemer MGM, et al. PD-L1 and PD-L2 expression in cervical cancer- regulation and biomarker potential. Front Immunol (2020) 11:596825. doi: 10.3389/fimmu.2020.596825
138. Patel D, Huang SM, Baglia LA, McCance DJ. The role of semiquantitative evaluation of lympho-vascular space invasion in early stage cervical cancer patients. Gynecol Oncol (2021) 162(2):299–307. doi: 10.1016/j.ygyno.2021.06.002
139. Jin BY, Campbell TE, Draper LM, Stevanovic S, Weissbrich B, Yu ZY, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight (2018) 3(8):e99488. doi: 10.1172/jci.insight.99488
Keywords: human papillomavirus clearance, Langerhans cells, cervicovaginal microbiota, cellular immunity, human papillomavirus infection
Citation: Dai W, Gui L, Du H, Li S and Wu R (2022) The association of cervicovaginal Langerhans cells with clearance of human papillomavirus. Front. Immunol. 13:918190. doi: 10.3389/fimmu.2022.918190
Received: 12 April 2022; Accepted: 27 September 2022;
Published: 12 October 2022.
Edited by:
Fanghui Zhao, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaReviewed by:
Mayumi Nakagawa, University of Arkansas for Medical Sciences, United StatesMarcela Lizano, National Institute of Cancerology (INCAN), Mexico
Copyright © 2022 Dai, Gui, Du, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruifang Wu, d3VyZnB1c2hAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Wenkui Dai
Wenkui Dai Liming Gui
Liming Gui Hui Du1,2,3†
Hui Du1,2,3† Shuaicheng Li
Shuaicheng Li Ruifang Wu
Ruifang Wu