
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 03 June 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.916383
This article is part of the Research TopicStructural Immunology of Molecular Innate ImmunityView all 14 articles
 Xuan Shi†
Xuan Shi† Sheng Wang†
Sheng Wang† Yutong Wu†
Yutong Wu† Quanfu Li
Quanfu Li Tong Zhang
Tong Zhang Keting Min
Keting Min Di Feng
Di Feng Meiyun Liu
Meiyun Liu Juan Wei
Juan Wei Lina Zhu
Lina Zhu Wei Mo
Wei Mo Zhuoran Xiao
Zhuoran Xiao Hao Yang
Hao Yang Yuanli Chen*
Yuanli Chen* Xin Lv*
Xin Lv*Background and aims: Cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) (cGAMP) synthase (cGAS) and stimulator of interferon genes (STING) are key components of the innate immune system. This study aims to evaluate the research of cGAS-STING pathway and predict the hotspots and developing trends in this field using bibliometric analysis.
Methods: We retrieved publications from Science Citation Index Expanded (SCI-expanded) of Web of Science Core Collection (WoSCC) in 1975-2021 on 16 March 2022. We examined the retrieved data by bibliometrix package in R software, VOSviewer and CiteSpace were used for visualizing the trends and hotspots of research on the cGAS-STING pathway.
Results: We identified 1047 original articles and reviews on the cGAS-STING pathway published between 1975 and 2021. Before 2016, the publication trend was increasing steadily, but there was a significant increase after 2016. The United States of America (USA) produced the highest number of papers (Np) and took the highest number of citations (Nc), followed by China and Germany. The University of Texas System and Frontiers in Immunology were the most prolific affiliation and journal respectively. In addition, collaboration network analysis showed that there were tight collaborations among the USA, China and some European countries, so the top 10 affiliations were all from these countries and regions. The paper published by Sun LJ in 2013 reached the highest local citation score (LCS). Keywords co-occurrence and co-citation cluster analysis revealed that inflammation, senescence, and tumor were popular terms related to the cGAS-STING pathway recently. Keywords burst detection suggested that STING-dependent innate immunity and NF-κB-dependent broad antiviral response were newly-emerged hotspots in this area.
Conclusions: This bibliometric analysis shows that publications related to the cGAS-STING pathway tend to increase continuously. The research focus has shifted from the mechanism how cGAS senses dsDNA and cGAMP binds to STING to the roles of the cGAS-STING pathway in different pathological state.
Over the past two decades, in mammalian cells, recognition of pathogens’ nucleic acids has been a key feature to sense microbial pathogens. In the field of sensing double-stranded DNA (dsDNA), cGAS is an important DNA-binding protein that represents the initiator of sensing dsDNA. Three strategies have been reported for cGAS to recognize pathogens efficiently. Firstly, cGAS is discovered in the cytoplasm, plasma membrane, and nucleus, it can rapidly recognize DNA and initiate the downstream immune response (1–4). Secondly, the recognition would be strengthened by high-mobility group box 1 protein (HMGB1), mitochondrial transcription factor A (TFAM) and modified by reactive oxygen species (ROS) (5, 6). Thirdly, the second messenger cyclic GMP-AMP (cGAMP) from these infected cells would show alarm to bystander cells to activate cGAS-STING pathway in these cells (7). When combined with dsDNA, the structure of cGAS would change and affect catalytic pockets. ATP and GTP in this pocket are catalyzed by cGAMP (8). As a second messenger, cGAMP is detected by STING, a cyclic-dinucleotide sensor (9, 10). Then STING is transported from the endoplasmic reticulum (ER) to Golgi through ER-Golgi intermediate compartment and sets off downstream signaling reaction (10, 11). STING is regarded as the central molecule of the downstream of I IFN (12, 13). STING is reported to enhance the activity of RIG-1-like receptors (RLR) signaling pathway (14) and the activity of interferons-β (IFN-β) which is dependent on interferon regulator factor 3 (IRF3) (15–17). In addition, the activation of STING can activate TANK-binding kinase 1 (TBK1) and IκB kinase (IKK). p-TBK1 phosphorylates interferon regulatory factor 3 (IRF3), and IRF3 translocated to the nucleus to transcript IFN-I (18). IKK is also recruited by STING, phosphorylates IκBα and induces NF-κB to translocate to nucleus. After that, lots of cytokines are transcribed to induce inflammatory and immune responses (19). In recent years, scholars have done lots of research about the cGAS-STING pathway. It is important to explore the hotspots and development trends of the cGAS-STING pathway in the past 10 years with CiteSpace and VOSviewer software.
Bibliometric analysis is a useful method by which scholars can evaluate the history, current, and future of publications and their quantity and quality (20). Bibliometrics can analyze publications (books, journals, and so on) by applying the literature system and metrology as objects. In addition, it can provide useful information to help to write the guideline, make decisions and treat diseases (21–23). In these years, many bibliometric analyses have been published in the biological field. However, bibliometric analysis on the cGAS-STING pathway remains a void. So the study aims to systematically analyze the research on the cGAS-STING pathway to digest the current state and the hotspots in this field.
The Science Citation Index Expanded (SCI-expanded) of Web of Science Core Collection (WoSCC) in 1975-2021 was systematically searched from 1 January 1975 to 31 December 2021 and was downloaded in a single day (2022.03.17) to avoid deviations. The search terms were set as follows: TS = (“stimulator of interferon genes” OR “transmembrane protein 173” OR STING OR ERIS OR MITA OR MPYS OR NET23 OR TMEM173) AND TS = (“Mab-21 domain containing 1” OR “E330016A19Rik” OR “cyclic guanosine monophosphate-adenosine monophosphate synthase” OR “cGAMP synthase” OR “cyclic GMP-AMP synthase” OR “MB21D1” OR “cGAS”). Two reviewers (XS and YW) independently identified these data search and then discussed the potential differences, the final agreement reached 0.90 (24). These two reviewers then sent these original articles and reviews into Endnote for further validation. Finally, 1047 original articles and reviews written in English were included. Figure 1 is the flowchart of literature selection.
Duplicate authors and misspelled elements were removed, and we used a thesaurus file to merge duplicates into one word, delete the useless words and correct the misspelled elements. Then, the clean data were imported to VOSviewer v.1.6.15.0, CiteSpace version 5.8.R3, and the “bibliometrix package 3.2.1” of R software (Version 4.1.3) for bibliometric analysis.
We used the numbers of papers and citations to represent the bibliographic material as previously reported (25). The productivities of papers were represented by the numbers of publications (Np), the impacts were represented by the numbers of citations (without self-citations) (Nc) and the numbers of average citations (Na) were Nc/Np, which represented the qualities of publications. These elements were regarded as three main perspectives to evaluate the levels of researches. In some cases, H-index was also developed to evaluate individual academic achievements, the publication output of a region or a nation, an institution, or a journal (26). What’s more, the impact factor (IF) from the latest version of Journal Citation Reports (JCR), and local citation score (LCS) also indicated the value of an article (27, 28).
VOSviewer, CiteSpace, and R (Version 4.1.3) are used for statistical computing and graphics. VOSviewer is a program to establish bibliometric maps by using the data collected from Web of Science Core Collection (29). It can provide a general comprehensive and detailed view of bibliometric maps based on collaborative data. CiteSpace is a program to analyze the potential knowledge contained in the scientific literature and visualize collected data (30). R software (Version 4.1.3) is the language and environment, which is wildly used for statistical computing and graphics (31). In this study, the bibliometrix package 3.2.1 in R was used to analyze data and perform a basic bibliometric analysis (32).
The number of original articles (785) and reviews (262) published was 1047, the total Nc for retrieved articles and reviews was 33357, the average Nc per article was 31.86. The H-index of all original articles and reviews was 102.
The annual Np related to the cGAS-STING pathway was shown in Figure 2A. The numbers of annual papers rose rapidly from 16 in 2013 to 332 in 2021 and the correlation coefficient R2 is 0.9863. The rapid increase indicated that more and more researchers were paying attention to this area.
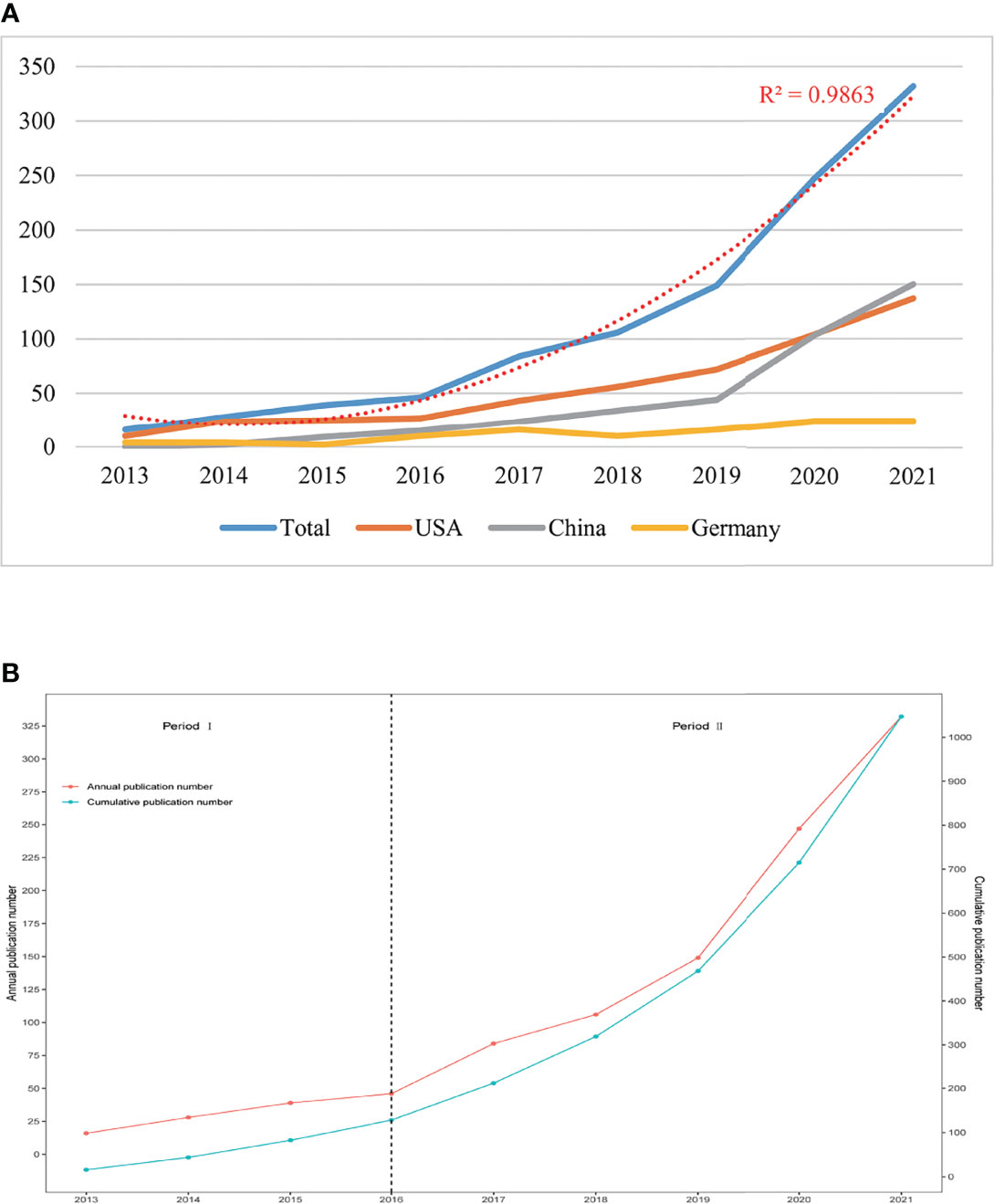
Figure 2 (A) The total numbers of publications and top three countries from 2013 to 2021. (B) The numbers of publications by year and accumulation from 2013 to 2021.
From 2013 to 2021, the Np in the USA had increased steadily. When it comes to China, before 2019, the Np was at a low level and was almost as half as that in the USA. However, the Np in China had reached the first place in 2021. This might be related to the increased investments of the Chinese government in scientific research.
In Figure 2B, it was interesting to note that the number of annual publications can be divided into two stages. With the model of research development (33), we found that from 2013 to 2016 (period I), publications outputs were at a low level. Theories in this area were not completed and the cGAS-STING pathway just began to come into focus. From 2016 to now (period II), a rapid increase occurred and the publication outputs had been over 1000 in 2021, which represented that more scholars were conducting research in this field and theories about the cGAS-STING pathway were booming. Since the cGAS-STING pathway has attracted more attention, a spurt would occur shortly.
A total of 1047 articles were published from 54 countries and regions. We ranked the top 10 output countries and regions of all authors according to the number of Np (Table 1). Because we used the bibliometrix package in R (Version 4.1.3) software to analyze all data, the data of England, Scotland, and Wales were merged automatically by the package in analysis of countries, and finally these were shown as UK. In Figure 3A, the Np in other countries was relatively at low levels and remained steady except the USA and China. China ranked first in Np in 2021 but the LCS of China was much lower than that of the USA (Figure 3B), representing that qualities of publications in China were still at a relative low level. The Np and LCS in the USA both increased rapidly, which means the publications about the cGAS-STING pathway in the USA were not only for quantity but also for quality. Figure 3C represents the distributions of publications in different countries and regions. Cooperation among different countries is an important driving force to promote the development of scientific research. To this point, close cooperation among different countries were shown in Figure 3D. The lines donated the cooperation between countries. The wider the lines, the closer the cooperation. However, most countries lacked lines, which means they lacked stable cooperation and communication.
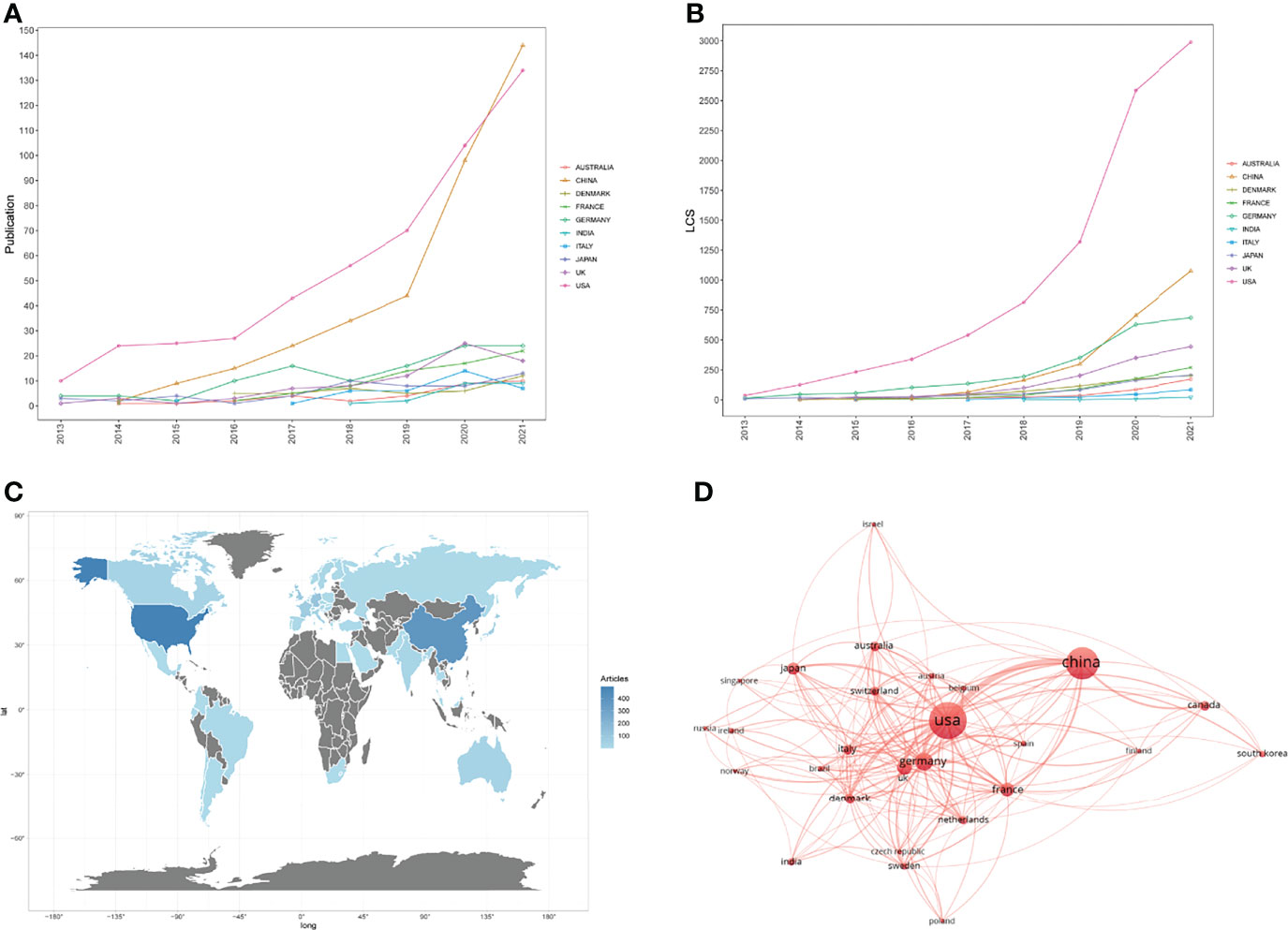
Figure 3 (A) Publications in the 10 most productive countries/regions. (B) The yearly number of local citation scores (LCS) of the 10 most productive countries/regions. (C) Distributions of countries/regions of publications. (D) The cooperation map of countries involved in the cGAS-STING pathway.
Table 2 showed the top 10 affiliations with the highest number of publications related to the cGAS-STING pathway. University of Texas System had the highest Np (88, 8.40%) among all affiliations, which were almost as twice as Chinese Academy of Sciences (51, 4.87%). The team of Zhijian James Chen, who discovered cGAS for the first time and explained its function, is from University of Texas System. The publications from this team contributed a lot for the first place of University of Texas System. The Nps of affiliations ranking three to five were the same, Howard Hughes Medical Institute (48, 4.58%), University of California System (48, 4.58%), and Harvard University (48, 4.58%). Among the top 10 affiliations, half of them belonged to the USA. This was related to its high investment and strong technical strength. In addition, University of Texas System got the highest H-index (38) followed by Howard Hughes Medical Institute (37), scholars in this area should focus on their high-quality research notably.
The 10 journals with the most research in cGAS-STING area, along with H-index and impact factor (IF) Eigenfactor Score as indicators of impact were listed in Table 3. These journals were more likely to accept articles on cGAS-STING pathway because they had produced the most publications on the related topics recently. Scholars in cGAS-STING area should focus on research published in these journals. The highest IF belonged to Nature (IF=49.926), followed by Immunity (IF=31.745), Cell Host & Microbe (IF=21.023), Nature Communication (IF=14.919) and Proceedings of the National Academy of Sciences of the United States of America (PNAS) (IF=11.205). The IF of these 5 journals were over 10 and they published over 1/10 papers in this area in the past, representing that cGAS-STING is a popular research orientation and it is not difficult for studies in cGAS-STING area to publish in top journals.
The LCS analysis provided detailed information for articles with high local citations. The numbers of LCS per year for the top 15 articles were presented in Figure 4A and S Table 1. Interestingly, 8 of them were from the team of Zhijian James Chen, the pioneer and founder of the cGAS-STING area. These research outputs of Chen’s lab were leading the trend and breakthrough in this area. The paper written by Sun LJ, the Ph.D. student in Chen’s lab, got the highest LCS score (555). In this paper, the authors firstly discovered an enzyme named cyclic GMP–AMP synthase (cGAS), which can detect DNA and active I IFN signaling pathway (1). Apart from two reviews (Chen Q, 2016; Harding SM, 2017) (8, 34), the other 13 high LCS studies were all from 2013 to 2015, when the area just emerged. After 2016, scholars around the world are conducted more and more research based on these classical research. Visualization of the top 50 LCS articles was shown in Figure 4B. In this network, each node represented a cited article, the size of each node was proportional to the frequency of this article by the other 49 articles.
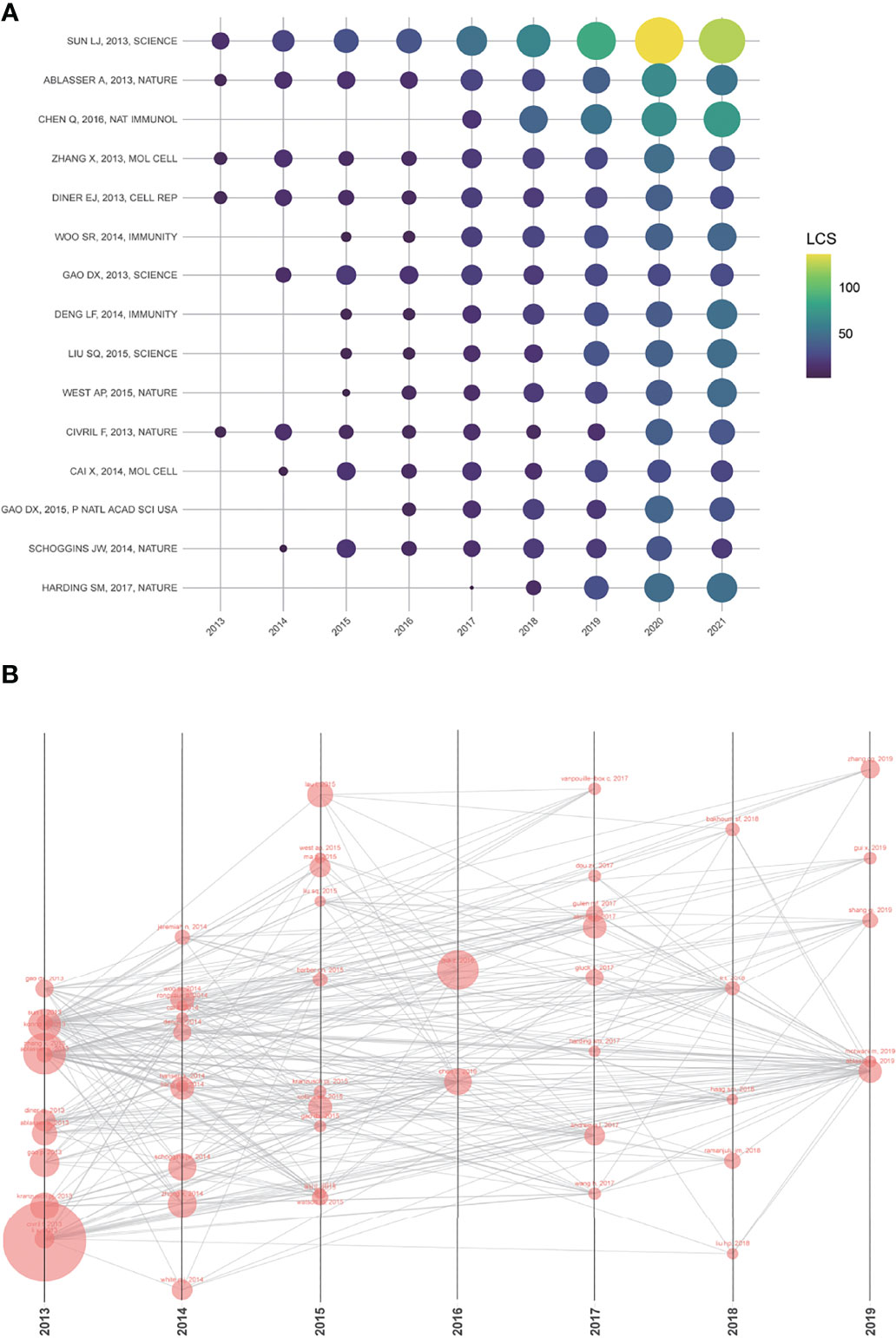
Figure 4 (A) The yearly number of local citations of papers with high local citation scores (LCS). The size and colors of the circle present the LCS of papers in that year. (B) One paper cited by the other papers with the top 50 LCS. The size of each node is proportional to the frequency of this article by the other 49 articles.
By analyzing keywords, readers can easily summarize the topic of one study and explore the hotpots and directions in this area. Thus, co-occurrence analysis described of hot topics in this area (Figure 5A). There were five clusters: cGAS-STING pathway in inflammation and tumor immunology (red), cGAS-STING pathway sensing virus and its structure foundation (green), cGAS-STING pathway in innate immunity (purple), in ROS-induced inflammasome activation (yellow), and in autoimmune disease (blue). In overlay visualization, keywords were colored differently according to their average publication year (Figure 5B). For instance, ‘DNA sensor’ and ‘2nd-messenger’ were appeared at the beginning of the discovery of this area, whereas keywords ‘Inflammation’ and ‘Tumor’ were more recent. Senescence (cluster1, APY: 2020.143), Cancer therapy (cluster2, 2020), Neuroinflammation (cluster3, APY: 2020.539), and Neurodegeneration (cluster3, APY: 2020.3) are colored in yellow, which indicated that these fields had grown in popularity recently and would become hotspots soon (S Table 2).
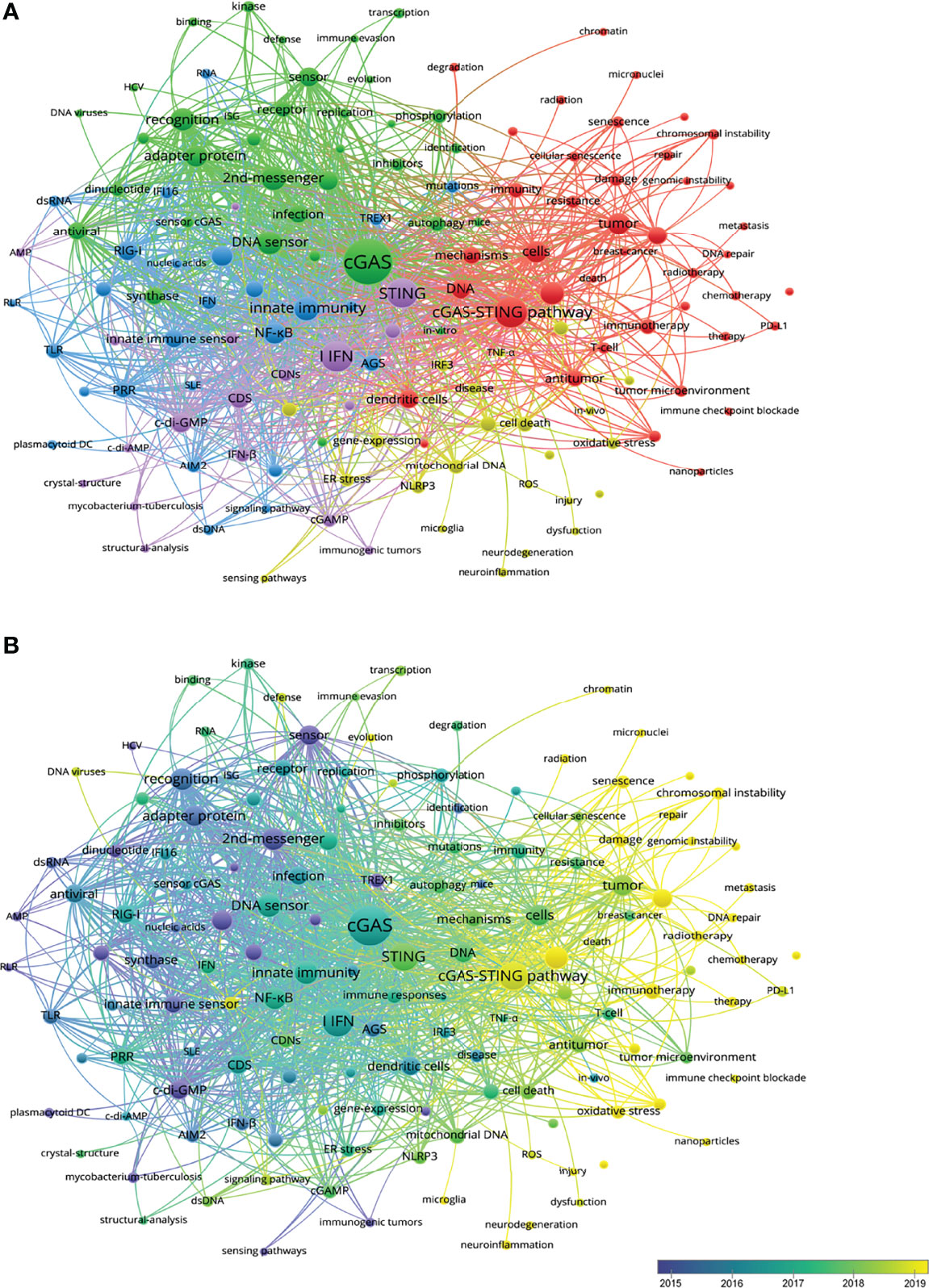
Figure 5 (A) CiteSpace visualization map of keywords clustering analysis related to the cGAS-STING pathway. The size of each nodes represents the frequency of occurrences. (B) Visualization of keywords according to the average publication year. Keywords in yellow appeared later than that in blue.
A co-citation network is a network of references co-cited by one or more papers at the same time. Conceptual clusters were created when a set of manuscripts were cited repeatedly together. Figure 6A showed the different clusters of these co-cited references, and 15 clusters were divided by CiteSpace: STING agonist, dsDNA-induced oligomerization, genomic instability, interferon response, acute kidney injury, NF-κB-dependent broad antiviral response, DNA damage response, human cytomegalovirus tegument protein, STING-dependent innate immunity, small molecule, cGAS-STING pathway, STING-dependent cytosolic DNA, early stage, LRRC8 volume-regulated anion channel, and other retroviruses. A timeline view of distinct clusters was presented in Figure 6B. It showed that cluster 1, dsDNA-induced oligomerization, had the most citation burst. Moreover, the hotspots were shifting from dsDNA-induced oligomerization to genomic instability, STING-dependent innate immunity, and NF-κB-dependent broad antiviral response.
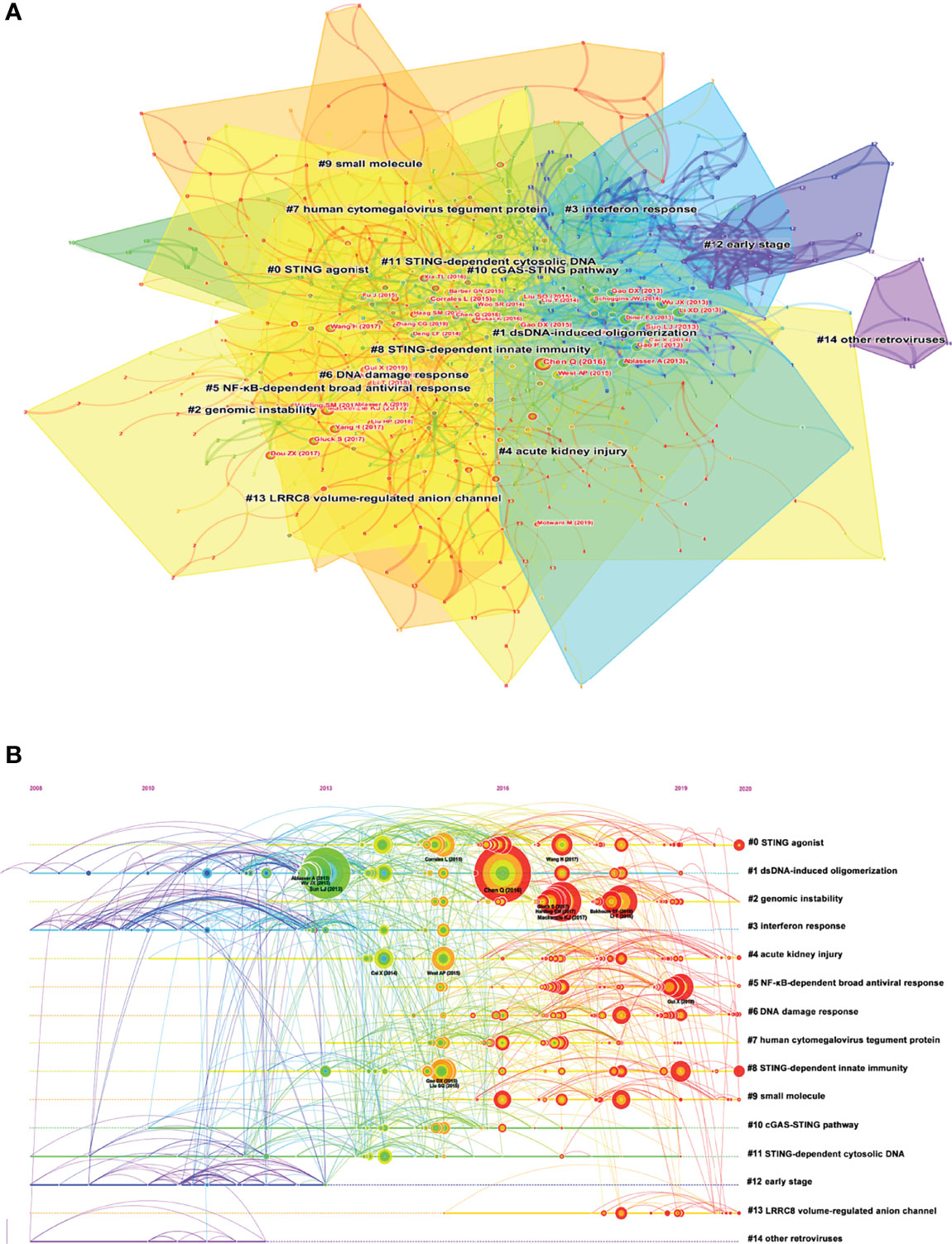
Figure 6 (A) The clustered network map of co-cited references on the cGAS-STING pathway. (B) The timeline view of co-citation clusters with their cluster-labels on the right.
Burst detection is used to reveal the hot references with an abrupt increase over time. In Figure 7, nodes represented articles, those nodes with red circles represented burst articles in this area. Figure 8 showed the most burst of co-cited references began in 2013, the year when the team of Zhijian James Chen discovered cGAS-STING pathway. 4 of 5 top strongest citation bursts were from the team of Zhijian James Chen, which also indicated his team’s great influence in this field.
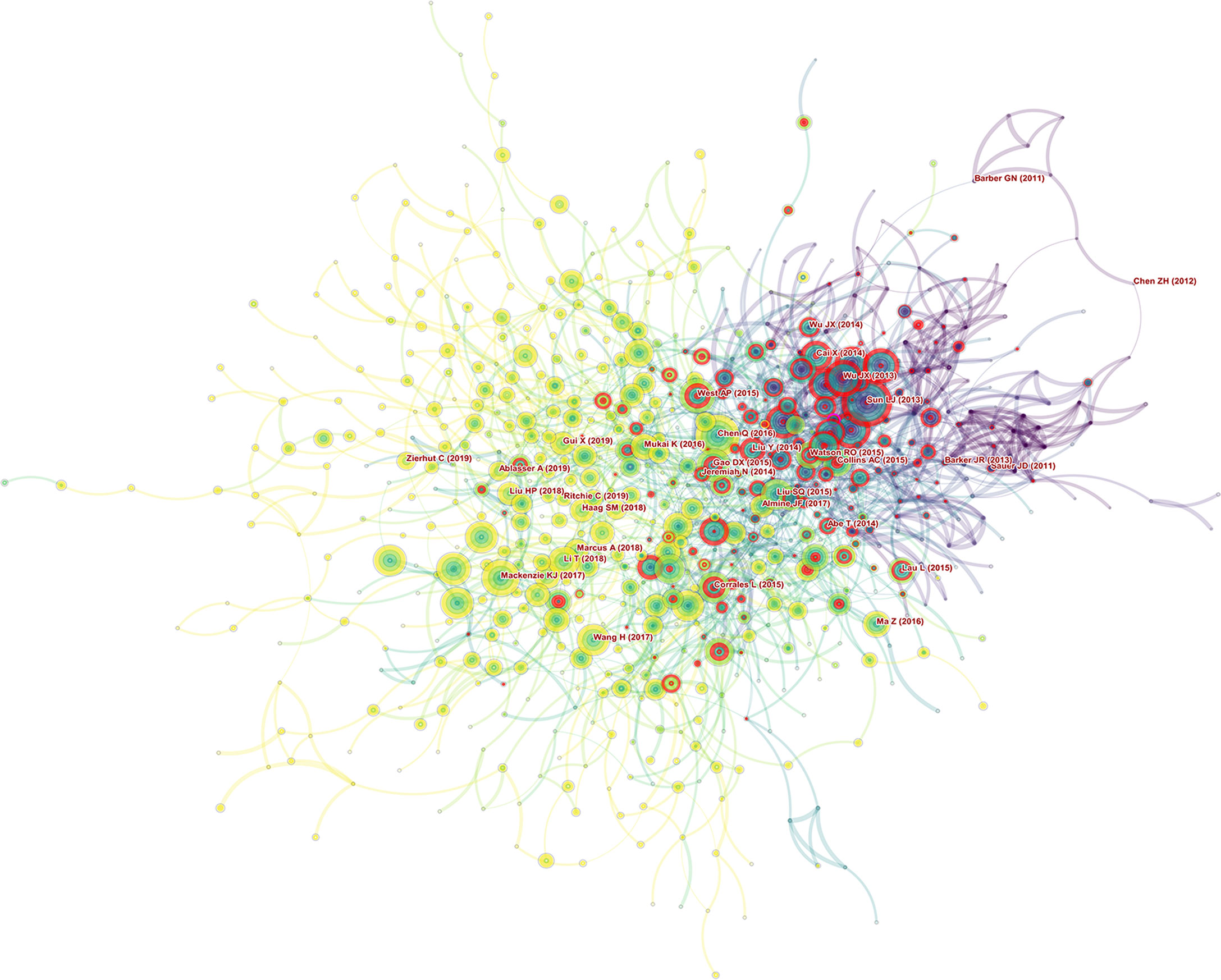
Figure 7 Papers with the strongest citation bursts in original articles on the cGAS-STING pathway between 2013 and 2021.
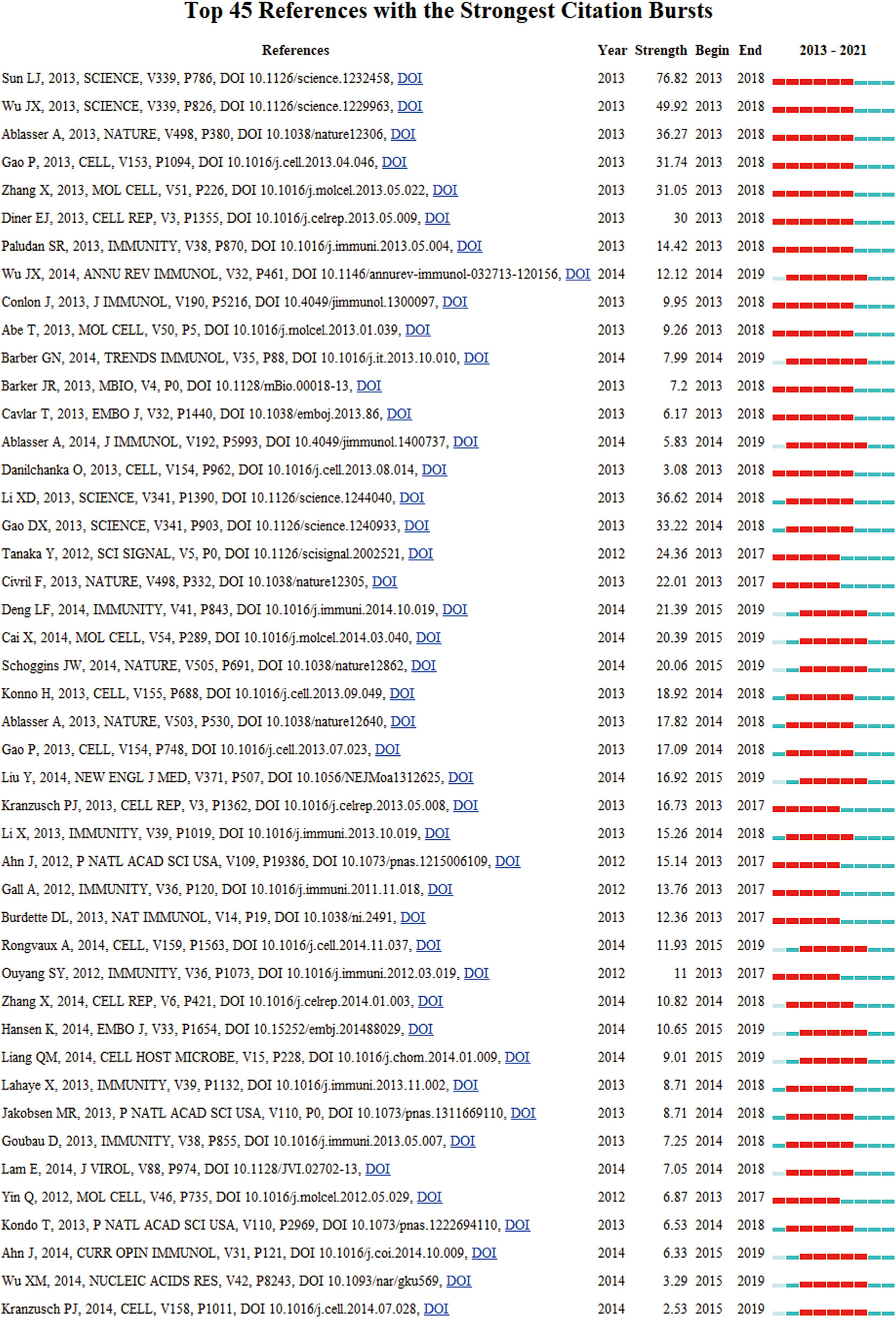
Figure 8 CiteSpace visualization map of the top 45 references with the strongest citation bursts involved in the cGAS-STING pathway.
In this study, we analyzed the development trends and hotspots of research on the cGAS-STING pathway by VOSviewer, CiteSpace and R (Version 4.1.3) software. We retrieved 1047 original articles and reviews published in 2013-2021. The annual numbers of publications showed an overall upward tread. Interestingly, the Np rocketed up after 2016. These publications with high LCS published before 2016 were the main reason for the rapid growth of the annual Np after 2016.
Among the top countries/regions, the USA ranked first in Np (493, 47.09%), followed by China (370, 35.34%), indicating that the USA and China are the leading countries in the cGAS-STING area. This was closely related to the large research expenditures of the USA and China in recent years. However, the Np, Na, and H-index in the USA were all higher than those in China. This may be because the cGAS-STING pathway was initially proposed by Zhijian James Chen (1), and then deep and extensive research were conducted by Chen’s team in the cGAS-STING area. What’s more, five of the top ten affiliations and seven of the top ten journals were from the USA. Because of these, the USA prevails in the cGAS-STING area. When it comes to affiliations, the H-index of Chinese Academy of Sciences (21) and Wuhan University (15) were similar to the other top 10 affiliations except for University of Texas System (38), but only one original study was included in the top 50 LCS (35), symbolizing that there were good studies in China which attracted the attention of international counterparts, but scholars and affiliations in China should make more efforts to promote the quality of their studies in this field. The other countries also made contributions to this field, although their influences of them were not as good as those in the USA and China.
As can be seen in the co-occurrence network, there were many lines from the USA and the line between the USA and China was the thickest, indicating that the collaboration between the USA and China was very close. Moreover, there were close collaborations between the USA and some countries in Europe, with 3 European affiliations listed in the top 10 affiliations. Therefore, institutions in the USA, China, and other countries should remove academic barriers, try to communicate to promote the development of the cGAS-STING pathway.
Notably, of the top 10 productive journals, the IFs of five were over 10, and the number of published papers in these journals accounted for 1/10 of that in the cGAS-STING area. This indicated that studies about the cGAS-STING pathway were of high quality. Frontiers in immunology (55, 5.33%) published the most articles in this area, followed by journal of immunology (35, 3.39%) and journal of virology (30, 2.91%), which reminded scholars to pay more attention to the roles of the cGAS-STING pathway in immunity and virus detection. In addition, the burst detection showed that STING-dependent innate immunity and NF-κB-dependent broad antiviral response were the hotspots recently. Scholars on this topic should pay more attention to these hotspots.
In the initial phase of one field, research is focused on the basic theories and mechanisms, which lay a solid foundation for further studies. Similarly, the hotspots of the cGAS-STING pathway have been changed from the mechanism to its roles in different diseases and translational medicine. In the first few years, scholars such as Zhijian James Chen and Veit Hornung discovered the role of cGAS in sensing dsDNA and activating the I IFN pathway. cGAS activates the second-messenger (36), which is essential for the STING activation (37–39). What’s more, scholars analyzed the structural mechanism how cGAS senses cytosolic DNA (40, 41).
In the next stage, scholars started to study the roles of the cGAS-STING pathway in different diseases and the influences of the cGAS-STING pathway in cell life activities. Immunity published two studies to demonstrate the roles of the cGAS-STING pathway in immunogenic tumors (42, 43), initiating studies of the cGAS-STING pathway in diseases. In these studies, the STING pathway was regarded as a key regulator of tumor immune responses. Researchers found that tumor-derived DNA was the ligand of STING pathway and was associated with phosphorylation of TBK1 and IRF3 and STING-dependent IFN-β. In STING-deficient mice, most of the therapeutic effects for the immune inhibitory factors were lost. In 2015, the relationships between the cGAS-STING pathway and apoptosis, autophagy, and inflammasome activation were studied by scholars (44–46). Based on these mechanistic investigations, the team of Zhijian James Chen and Blossom Damania reviewed the roles of the cGAS-STING pathway in autoimmune, inflammatory disease, and virus infection, respectively (8, 47). These two reviews concluded the studies between 2013 and 2016 and thus got high LCS.
In recent years, the keywords have focused on the roles of the cGAS-STING pathway in the treatments of diseases. In this period (period II in Figure 2B), publications increased rapidly based on previous studies. Article keywords demonstrated that scholars in the fields of cancer and neuroscience should pay more attention to the cGAS-STING pathway because these were hotspots in recent years. Shannon Grabosch’s study demonstrated that cisplatin activated the cGAS-STING pathway to modify tumor immunogenicity by increasing PD-L1, MHC I and calreticulin in tumor cells (48). In malignant tumors, scientists found that the expression of STING was positively correlated with immune cell infiltration (49). Inhibition of cGAS and STING in tomor cells can prevent tumor metastasis (50, 51). Scientists also found that cGAS-STING pathway promoted tumor progression in lewis lung cancer (LCC) (52), brain tumor (50), colon tumor (53), oral cancer (54), and tongue squamous cell carcinoma (55). In December 2017, Chukwuemika Aroh et al. firstly demonstrated that administration of cGAMP delivered by ultra-pH-sensitive nanoparticle can induce potent antiretroviral response against HIV-1 isolates (56). After that, more and more researchers paid attention to the nanoscience. Since the nanoparticle is a hotspot recently, with the development of interdisciplinary research, researchers should focus on the effects of the nanoparticle on diseases by interfering with the cGAS-STING pathway.
Based on VOSviewer, CiteSpace and R (Version 4.1.3) software, we analyzed and made the visualization of the literature, and revealed the development trends and the hotspots in this field. At the same time, we used LCS to find the important literature, which led to the development of the cGAS-STING area and scholars in this field should pay close attention to these literature. Moreover, this study provided a better insight into the evolving research foci and trends when compared with traditional reviews. However, there are still some limitations. Firstly, only English articles and reviews from SCI-expanded were included. Secondly, because VOSviewer could not analyze the full texts of the publications, it may omit some information. Lastly, the publications included were from 2013 to 2021, the influential studies published in 2022 with low Nc were excluded, but this limitation would not change the results in this study. Therefore, future work should expand the research base to include non-English studies and the latest outstanding publications.
This bibliometric analysis revealed that the research on the innate immune DNA sensing cGAS-STING pathway were developing rapidly at present. The USA and China were the leading countries, and the USA has made many outstanding breakthroughs in this field. About 10% studies were published in high-quality journals. From 2013 to 2021, the foci of research on the cGAS-STING pathway has changed from the basic mechanism to treatments of diseases via the cGAS-STING pathway, especially cancer and nanoparticle, these would be hotspots of research recently and in the near future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
XS and SW conceived the study. XS, YW, QL, TZ, KM, DF, ML, and JW were involved in the data collection and analysis. LZ, WM, ZX, and HY re-examined the data. XS and SW drafted the manuscript. YC and XL revised the manuscript. All authors have provided final approval of the version to be submitted. XS, SW, and YW contributed equally to this work.
This work was supported by grants from the National Natural Science Foundation of China (No.81871601, No.82100090, and No.82000085), grants from the Program of Shanghai Academic Research Leader (No.21XD1402800), grants from the Shanghai “Rising Stars of Medical Talent” Youth Development Program: Outstanding Youth Medical Talents, grants from the Shanghai Natural Science Foundation (22ZR1452200), grants from the Shanghai Sailing Program (21YF1438400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer, DY, declared a shared affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.916383/full#supplementary-material
1. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science (2013) 339(6121):786–91. doi: 10.1126/science.1232458
2. Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction Between Self- and Viral DNA. Cell (2019) 176(6):1432–46.e1411. doi: 10.1016/j.cell.2019.01.049
3. Gentili M, Lahaye X, Nadalin F, Nader GPF, Puig Lombardi E, Herve S, et al. The N-Terminal Domain of cGAS Determines Preferential Association With Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Rep (2019) 26(9):2377–93.e2313. doi: 10.1016/j.celrep.2019.01.105. Erratum in: Cell Rep. 2019 Mar 26;26(13):3798.
4. Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS Surveillance of Micronuclei Links Genome Instability to Innate Immunity. Nature (2017) 548(7668):461–5. doi: 10.1038/nature23449
5. Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, et al. cGAS Senses Long and HMGB/TFAM-Bound U-Turn DNA by Forming Protein-DNA Ladders. Nature (2017) 549(7672):394–8. doi: 10.1038/nature23890
6. Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, et al. Oxidative Damage of DNA Confers Resistance to Cytosolic Nuclease TREX1 Degradation and Potentiates STING-Dependent Immune Sensing. Immunity (2013) 39(3):482–95. doi: 10.1016/j.immuni.2013.08.004
7. Zhou C, Chen X, Planells-Cases R, Chu J, Wang L, Cao L, et al. Transfer of cGAMP Into Bystander Cells via LRRC8 Volume-Regulated Anion Channels Augments STING-Mediated Interferon Responses and Anti-Viral Immunity. Immunity (2020) 52(5):767–81.e766. doi: 10.1016/j.immuni.2020.03.016
8. Chen Q, Sun L, Chen Z. Regulation and Function of the cGAS-STING Pathway of Cytosolic DNA Sensing. Nat Immunol (2016) 17(10):1142–9. doi: 10.1038/ni.3558
9. Zhong B, Yang Y, Li S, Wang Y, Li Y, Diao F, et al. The Adaptor Protein MITA Links Virus-Sensing Receptors to IRF3 Transcription Factor Activation. Immunity (2008) 29(4):538–50. doi: 10.1016/j.immuni.2008.09.003
10. Ishikawa H, Ma Z, Barber G. STING Regulates Intracellular DNA-Mediated, Type I Interferon-Dependent Innate Immunity. Nature (2009) 461(7265):788–92. doi: 10.1038/nature08476
11. Abe T, Barber G. Cytosolic-DNA-Mediated, STING-Dependent Proinflammatory Gene Induction Necessitates Canonical NF-κb Activation Through TBK1. J Virol (2014) 88(10):5328–41. doi: 10.1128/JVI.00037-14
12. Paludan SR, Bowie AG. Immune Sensing of DNA. Immunity (2013) 38(5):870–80. doi: 10.1016/j.immuni.2013.05.004
13. West AP, Shadel GS, Ghosh S. Mitochondria in Innate Immune Responses. Nat Rev Immunol (2011) 11(6):389–402. doi: 10.1038/nri2975
14. Ishikawa H, Barber GN. STING is an Endoplasmic Reticulum Adaptor That Facilitates Innate Immune Signalling. Nature (2008) 455(7213):674–8. doi: 10.1038/nature07317
15. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an Adaptor Triggering RIG-I- and Mda5-Mediated Type I Interferon Induction. Nat Immunol (2005) 6(10):981–8. doi: 10.1038/ni1243
16. Bhatia R, Shaffer TH, Hossain J, Fisher AO, Horner LM, Rodriguez ME, et al. Surfactant Administration Prior to One Lung Ventilation: Physiological and Inflammatory Correlates in a Piglet Model. Pediatr Pulmonol (2011) 46(11):1069–78. doi: 10.1002/ppul.21485
17. Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M Jr. The Mitochondrial Targeting Chaperone 14-3-3ϵ Regulates a RIG-I Translocon That Mediates Membrane Association and Innate Antiviral Immunity. Cell Host Microbe (2012) 11(5):528–37. doi: 10.1016/j.chom.2012.04.006
18. Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. Structural Basis of STING Binding With and Phosphorylation by TBK1. Nature (2019) 567(7748):394–8. doi: 10.1038/s41586-019-1000-2
19. Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are Essential Components of the IRF3 Signaling Pathway. Nat Immunol (2003) 4(5):491–6. doi: 10.1038/ni921
20. Cancino CA, Merigo JM, Coronado F, Dessouky Y, Dessouky M. Forty Years of Computers & Industrial Engineering: A Bibliometric Analysis. Comput Ind Eng (2017) 113:614–29. doi: 10.1016/j.cie.2017.08.033
21. Narotsky D, Green P, Lebwohl B. Temporal and Geographic Trends in Celiac Disease Publications: A Bibliometric Analysis. Eur J Gastroenterol Hepatol (2012) 24(9):1071–7. doi: 10.1097/MEG.0b013e328355a4ab
22. Seriwala H, Khan M, Shuaib W, Shah S. Bibliometric Analysis of the Top 50 Cited Respiratory Articles. Expert Rev Respir Med (2015) 9(6):817–24. doi: 10.1586/17476348.2015.1103649
23. Khan M, Ullah W, Riaz I, Bhulani N, Manning W, Tridandapani S, et al. Top 100 Cited Articles in Cardiovascular Magnetic Resonance: A Bibliometric Analysis. J Cardiovasc Magn Reson: Off J Soc Cardiovasc Magn Reson (2016) 18(1):87. doi: 10.1186/s12968-016-0303-9
24. Landis J, Koch G. The Measurement of Observer Agreement for Categorical Data. Biometrics (1977) 33(1):159–74. doi: 10.2307/2529310
25. Wang S, Zhou H, Zheng L, Zhu W, Zhu L, Feng D, et al. Global Trends in Research of Macrophages Associated With Acute Lung Injury Over Past 10 Years: A Bibliometric Analysis. Front Immunol (2021) 12:669539. doi: 10.3389/fimmu.2021.669539
26. Hirsch J. An Index to Quantify an Individual’s Scientific Research Output. Proc Natl Acad Sci USA (2005) 102(46):16569–72. doi: 10.1073/pnas.0507655102
27. Jones T, Huggett S, Kamalski J. Finding a Way Through the Scientific Literature: Indexes and Measures. World Neurosurg (2011) 76:36–8. doi: 10.1016/j.wneu.2011.01.015
28. Roldan-Valadez E, Salazar-Ruiz S, Ibarra-Contreras R, Rios C. Current Concepts on Bibliometrics: A Brief Review About Impact Factor, Eigenfactor Score, CiteScore, SCImago Journal Rank, Source-Normalised Impact Per Paper, H-Index, and Alternative Metrics. Irish J Med Sci (2019) 188(3):939–51. doi: 10.1007/s11845-018-1936-5
29. van Eck N, Waltman L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics (2010) 84(2):523–38. doi: 10.1007/s11192-009-0146-3
30. Chen C. Searching for Intellectual Turning Points: Progressive Knowledge Domain Visualization. Proc Natl Acad Sci USA (2004) 101(Suppl 1):5303–10. doi: 10.1073/pnas.0307513100
31. Aria M, Cuccurullo C. Bibliometrix: An R-Tool for Comprehensive Science Mapping Analysis. J Informetr (2017) 11(4):959–75. doi: 10.1016/j.joi.2017.08.007
32. Hao X, Liu Y, Li X, Zheng J. Visualizing the History and Perspectives of Disaster Medicine: A Bibliometric Analysis. Disaster Med Public Health Preparedness (2019) 13:966–73. doi: 10.1017/dmp.2019.31
33. Frame JD, Baum JJ, Card M. Information Approach to Examining Developments in an Energy Technology - Coal-Gasification. J Am Soc Inf Sci (1979) 30(4):193–201. doi: 10.1002/asi.4630300404
34. Harding S, Benci J, Irianto J, Discher D, Minn A, Greenberg R. Mitotic Progression Following DNA Damage Enables Pattern Recognition Within Micronuclei. Nature (2017) 548(7668):466–70. doi: 10.1038/nature23470
35. Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS Suppresses DNA Repair and Promotes Tumorigenesis. Nature (2018) 563(7729):131–6. doi: 10.1038/s41586-018-0629-6
36. Kranzusch P, Lee A, Berger J, Doudna J. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Rep (2013) 3(5):1362–8. doi: 10.1016/j.celrep.2013.05.008
37. Ablasser A, Schmid-Burgk J, Hemmerling I, Horvath G, Schmidt T, Latz E, et al. Cell Intrinsic Immunity Spreads to Bystander Cells via the Intercellular Transfer of cGAMP. Nature (2013) 503(7477):530–4. doi: 10.1038/nature12640
38. Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov A, et al. Structure-Function Analysis of STING Activation by C[G(2’,5’)Pa(3’,5’)P] and Targeting by Antiviral DMXAA. Cell (2013) 154(4):748–62. doi: 10.1016/j.cell.2013.07.023
39. Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, et al. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages is an Endogenous High-Affinity Ligand for STING. Mol Cell (2013) 51(2):226–35. doi: 10.1016/j.molcel.2013.05.022
40. Civril F, Deimling T, de Oliveira Mann C, Ablasser A, Moldt M, Witte G, et al. Structural Mechanism of Cytosolic DNA Sensing by cGAS. Nature (2013) 498(7454):332–7. doi: 10.1038/nature12305
41. Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, et al. The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex With DNA and Undergoes Switch-Like Conformational Changes in the Activation Loop. Cell Rep (2014) 6(3):421–30. doi: 10.1016/j.celrep.2014.01.003
42. Woo S, Fuertes M, Corrales L, Spranger S, Furdyna M, Leung M, et al. STING-Dependent Cytosolic DNA Sensing Mediates Innate Immune Recognition of Immunogenic Tumors. Immunity (2014) 41(5):830–42. doi: 10.1016/j.immuni.2014.10.017
43. Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity (2014) 41(5):843–52. doi: 10.1016/j.immuni.2014.10.019
44. White M, McArthur K, Metcalf D, Lane R, Cambier J, Herold M, et al. Apoptotic Caspases Suppress mtDNA-Induced STING-Mediated Type I IFN Production. Cell (2014) 159(7):1549–62. doi: 10.1016/j.cell.2014.11.036
45. Watson R, Bell S, MacDuff D, Kimmey J, Diner E, Olivas J, et al. The Cytosolic Sensor cGAS Detects Mycobacterium Tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe (2015) 17(6):811–9. doi: 10.1016/j.chom.2015.05.004
46. Man S, Karki R, Malireddi R, Neale G, Vogel P, Yamamoto M, et al. The Transcription Factor IRF1 and Guanylate-Binding Proteins Target Activation of the AIM2 Inflammasome by Francisella Infection. Nat Immunol (2015) 16(5):467–75. doi: 10.1038/ni.3118
47. Ma Z, Damania B. The cGAS-STING Defense Pathway and Its Counteraction by Viruses. Cell Host Microbe (2016) 19(2):150–8. doi: 10.1016/j.chom.2016.01.010
48. Grabosch S, Bulatovic M, Zeng F, Ma T, Zhang L, Ross M, et al. Cisplatin-Induced Immune Modulation in Ovarian Cancer Mouse Models With Distinct Inflammation Profiles. Oncogene (2019) 38(13):2380–93. doi: 10.1038/s41388-018-0581-9
49. An X, Zhu Y, Zheng T, Wang G, Zhang M, Li J, et al. An Analysis of the Expression and Association With Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol Ther Nucleic Acids (2019) 14:80–9. doi: 10.1016/j.omtn.2018.11.003
50. Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-Astrocyte Gap Junctions Promote Brain Metastasis by cGAMP Transfer. Nature (2016) 533(7604):493–8. doi: 10.1038/nature18268
51. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal Instability Drives Metastasis Through a Cytosolic DNA Response. Nature (2018) 553(7689):467–72. doi: 10.1038/nature25432
52. Lemos H, Mohamed E, Huang L, Ou R, Pacholczyk G, Arbab AS, et al. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res (2016) 76(8):2076–81. doi: 10.1158/0008-5472.CAN-15-1456
53. Liang H, Deng L, Hou Y, Meng X, Huang X, Rao E, et al. Host STING-Dependent MDSC Mobilization Drives Extrinsic Radiation Resistance. Nat Commun (2017) 8(1):1736. doi: 10.1038/s41467-017-01566-5
54. Cheng AN, Cheng LC, Kuo CL, Lo YK, Chou HY, Chen CH, et al. Mitochondrial Lon-Induced mtDNA Leakage Contributes to PD-L1-Mediated Immunoescape via STING-IFN Signaling and Extracellular Vesicles. J Immunother Cancer (2020) 8(2):e001372. doi: 10.1136/jitc-2020-001372
55. Liang D, Xiao-Feng H, Guan-Jun D, Er-Ling H, Sheng C, Ting-Ting W, et al. Activated STING Enhances Tregs Infiltration in the HPV-Related Carcinogenesis of Tongue Squamous Cells via the C-Jun/CCL22 Signal. Biochim Biophys Acta (2015) 1852(11):2494–503. doi: 10.1016/j.bbadis.2015.08.011
Keywords: innate immunity, cGAS-STING pathway, bibliometrics, VOSviewer, CiteSpace
Citation: Shi X, Wang S, Wu Y, Li Q, Zhang T, Min K, Feng D, Liu M, Wei J, Zhu L, Mo W, Xiao Z, Yang H, Chen Y and Lv X (2022) A Bibliometric Analysis of the Innate Immune DNA Sensing cGAS-STING Pathway from 2013 to 2021. Front. Immunol. 13:916383. doi: 10.3389/fimmu.2022.916383
Received: 09 April 2022; Accepted: 09 May 2022;
Published: 03 June 2022.
Edited by:
Chaofeng Han, Second Military Medical University, ChinaReviewed by:
Degang Yang, Tongji University, ChinaCopyright © 2022 Shi, Wang, Wu, Li, Zhang, Min, Feng, Liu, Wei, Zhu, Mo, Xiao, Yang, Chen and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Lv, eGlubHZnQDEyNi5jb20=; Yuanli Chen, Y2hlbnl1YW5saTkwQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.