- 1Biotechnology Institute Thurgau (BITg) at the University of Konstanz, Kreuzlingen, Switzerland
- 2Graduate School for Cellular and Biomedical Sciences, University of Bern, Bern, Switzerland
- 3Faculty of Biology, University of Konstanz, Konstanz, Germany
- 4Theodor Kocher Institute, University of Bern, Bern, Switzerland
Chemokine-guided leukocyte migration is a hallmark of the immune system to cope with invading pathogens. Intruder confronted dendritic cells (DCs) induce the expression of the chemokine receptor CCR7, which enables them to sense and migrate along chemokine gradients to home to draining lymph nodes, where they launch an adaptive immune response. Chemokine-mediated DC migration is recapitulated and intensively studied in 3D matrix migration chambers. A major caveat in the field is that chemokine gradient formation and maintenance in such 3D environments is generally not assessed. Instead, fluorescent probes, mostly labelled dextran, are used as surrogate molecules, thereby neglecting important electrochemical properties of the chemokines. Here, we used site-specifically, fluorescently labelled CCL19 and CCL21 to study the establishment and shape of the chemokine gradients over time in the 3D collagen matrix. We demonstrate that CCL19 and particularly CCL21 establish stable, but short-distance spanning gradients with an exponential decay-like shape. By contrast, dextran with its neutral surface charge forms a nearly linear gradient across the entire matrix. We show that the charged C-terminal tail of CCL21, known to interact with extracellular matrix proteins, is determinant for shaping the chemokine gradient. Importantly, DCs sense differences in the shape of CCL19 and CCL21 gradients, resulting in distinct spatial migratory responses.
Introduction
Directed cell migration is a fundamental process that controls many physiological functions, including protective immunity (1). Immune cell migration is governed by the cell’s interaction with substrates of the environment and is orchestrated by extracellular directional guidance cues established primarily by members of the chemokine family (2, 3). As chemokines were originally described as secreted chemotactic cytokines, the paradigm of ‘chemotaxis’ emerged, in which cells migrate along a soluble, diffusion-based gradient towards the source of the chemoattractant (4). However, locally produced chemokines interact with extracellular matrix proteins of the surrounding environment (5), which prevents free diffusion and facilitate confined chemokine gradient formation (6–9). Importantly, negatively charged amino acid residues located typically at the C-terminal tail of chemokines control the interaction with extracellular substrates and likely gradient formation. However, information on chemokine distribution and quantitative assessments of chemokine gradients remain rare.
One of the best-studied chemokine gradient is that of CCL21 (8, 10). In the dermis, CCL21 is produced and secreted by lymphatic endothelial cells, kept locally through electrostatic interactions with the environment, and forms a steeply decaying gradient into the interstitial tissue (8). Pathogen experienced DCs are able to sense and migrate along this local CCL21 gradient in a CCR7 dependent manner to enter the lymph vessel on their way to the draining lymph node (8). Within the lymph node, CCL19, the second ligand for CCR7, seems to predominantly control DC migration and positioning (11). Notably, haptotactic DC migration along a substrate-bound CCL21 gradient and chemotactic, CCL19-mediated migration can be recapitulated in fabricated 3D collagen migration devices (12–14). However, which of the two CCR7 ligands is more potent in attracting DCs is controversial (13–16).
A major caveat in the field is that chemokine gradient formation and maintenance in 3D migration devices is generally not determined. Instead, fluorescent probes with comparable molecular weights to chemokines, mostly labelled dextran, are used as surrogate molecules (14, 17–19), thereby ignoring the electrostatic characteristics of chemokines. Importantly, the polar C terminal tail of CCL21 is well known to interact with extracellular matrix proteins, including collagen (20, 21). Hence, in the present study we set out to measure chemokine gradient formation and maintenance in time and space of a 3D collagen migration device. To do so, we used our recently developed site-specifically, fluorescently labelled CCL19 and CCL21 (22) in a side-by-site comparison with the commonly used surrogate molecule dextran.
Material and Methods
Production of Fluorescently Labelled Chemokines
Cloning and production of S6-tagged human CCL19 and CCL21 have been described previously (22). Human S6-tagged CCL21trunc, corresponding to amino acids 1-79 of the mature chemokine (23), was generated accordingly. Recombinant S6-tagged chemokines were site-specifically, enzymatically labelled using the phosphopantetheinyl-transferase Sfp with the fluorescent dye Dy549P1 (Dyomics, Jena, Germany) according to the established protocol and purified via C18 reverse phase HPLC (22).
Isolation of Human Monocyte-Derived Dendritic Cells (MoDCs)
Primary human monocytes were isolated from the blood of healthy donors, differentiated into MoDCs in GM-CSF/IL-4 and matured with a cytokine cocktail (TNFα/IL-6/IL-1β/PGE2) using standard protocols (24). Individual donors gave written consent and donations were approved by the local ethics committee.
Monitoring of Chemokine Gradient Formation and MoDC Migration in 3D Matrix
Gradient formation of fluorescent chemokines and subsequent migration of matured human MoDCs within a 3D matrix was performed in Ibidi µ-slide chambers (Ibidi, Martinsried, Germany). Briefly, matured MoDCs were collected in RPMI 1640 medium supplemented with 10 % FCS and 1 % penicillin/streptomycin at a concentration of 107 cells/ml. For collagen polymerization, 20 μl 10× DMEM, 10 μL 7.5 % NaHCO3, and 150 μL PureCol collagen I (Advanced Biomatrix, Carlsbad, CA, USA) were carefully mixed with 90 μL cells. The cell-collagen mixture was mounted into µ-slides according to the manufacturer’s instructions. Tetramethylrhodamine conjugated dextran of 10 kDa (Invitrogen, ThermoFisher), or fluorescent chemokines, all at a concentration of 100 nM, were applied to the right reservoir, whereas DMEM without phenol red (Gibco, ThermoFisher) was added to the left reservoir of the Ibidi-chamber. DC migration and gradient formation was monitored and recorded by time-lapse bright field and fluorescence microscopy for 3 or 5 hours on a Axiovert 200M (Zeiss, Oberkochen, Germany) equipped with a Tokai Hit INU (Tokai, Shizuoka, Japan) incubation system acquiring images every 5 minutes.
Cellular 3D migration was evaluated using the ‘chemotaxis and migration tool’ (Ibidi) to determine the velocity and forward migration index. To assess gradient formation, images were analyzed with FIJI. Images were cropped to the region of interest represented by the collagen matrix to exclude the reservoirs. The distribution of mean fluorescent intensities was evaluated using the rectangular intensity histogram tool covering the entire matrix. Individual data sets were evaluated in Prism 9.0 (Graphpad, La Jolla, CA, USA) applying the one-phase decay model.
Results
Visualization and Quantification of Dextran, CCL19 and CCL21 Gradient Formation and Maintenance
Chemokine-guided leukocyte migration in a 3D environment is nowadays routinely monitored in standardized, commercially available devices. In such a device, e.g. the Ibidi µ-slide, cells are embedded in a defined gel matrix within an enclosed central chamber. The two connected adjacent reservoirs are filled with either a chemoattractant or medium that diffuse into the central gel following a source-to-sink model. Gel-embedded cells orient and migrate in 3D towards the higher concentration of the chemoattractant (Figure 1A). Due to the lack of tools to visualize chemokines in these 3D migration chambers, gradient formation has often been taken for granted or eventually estimated by the use of fluorescently labelled surrogate molecules. The most commonly used surrogate molecule is dextran with a molecular weight of 10 kDa to roughly match the size of a chemokine. We used tetramethylrhodamine-labelled dextran of 10 kDa (dextranTMR), applied it into one reservoir of the migration chamber and measured its diffusion into the 3D collagen I matrix over time by time-lapse fluorescence microscopy. Plotting the diffusion profiles of the dextranTMR intensity at the centered longitudinal axis of the gel chamber as a function of the distance to the reservoir for increasing time points revealed a stable, nearly linear gradient across the entire matrix chamber (which is 1000 microns) with only minor changes in the fluorescence distribution over the whole period of measurement, i.e. five hours (Figure 1B). Precise mapping of site-specifically labelled and fully functional CCL19-S6Dy549P1 (22) in the 3D collagen I matrix directly after its application to the reservoir revealed a high fluorescent signal near the reservoir edge, which rapidly decreased over the first hundred microns of the gel chamber (Figure 1C). After five hours of incubation, the distribution of CCL19-S6Dy549P1 within the 3D matrix converged to the one observed for dextranTMR. Notably, the CCL19-S6Dy549P1 chemokine gradient stretched across the entire matrix over the full period of measurement (Figure 1C). By contrast, fluorescently labelled CCL21-S6Dy549P1 (22) sparsely entered deep into the collagen matrix within the first 30 minutes and the fluorescence intensity derived from CCL21-S6Dy549P1 steeply decreased with the distance to the reservoir (Figure 1D). Notably and in marked contrast to dextranTMR and CCL19-S6Dy549P1, the high-slope exponential decay of the CCL21-S6Dy549P1 gradient was only measurable over a few hundred microns into the collagen matrix and remained undetectable at the opposite end of the chamber (Figure 1D). Such a steep and short-distance spanning CCL21 gradient shape has also been observed in the dermal interstitium (8). Although no in vivo data on a CCL19 gradient shape has been reported yet, our quantitative assessments of chemokine gradients in a 3D environment reveal distinct shapes for gradients with different chemokines, and importantly, uncovers the limitation of dextran as general surrogate marker for monitoring chemokine gradients.
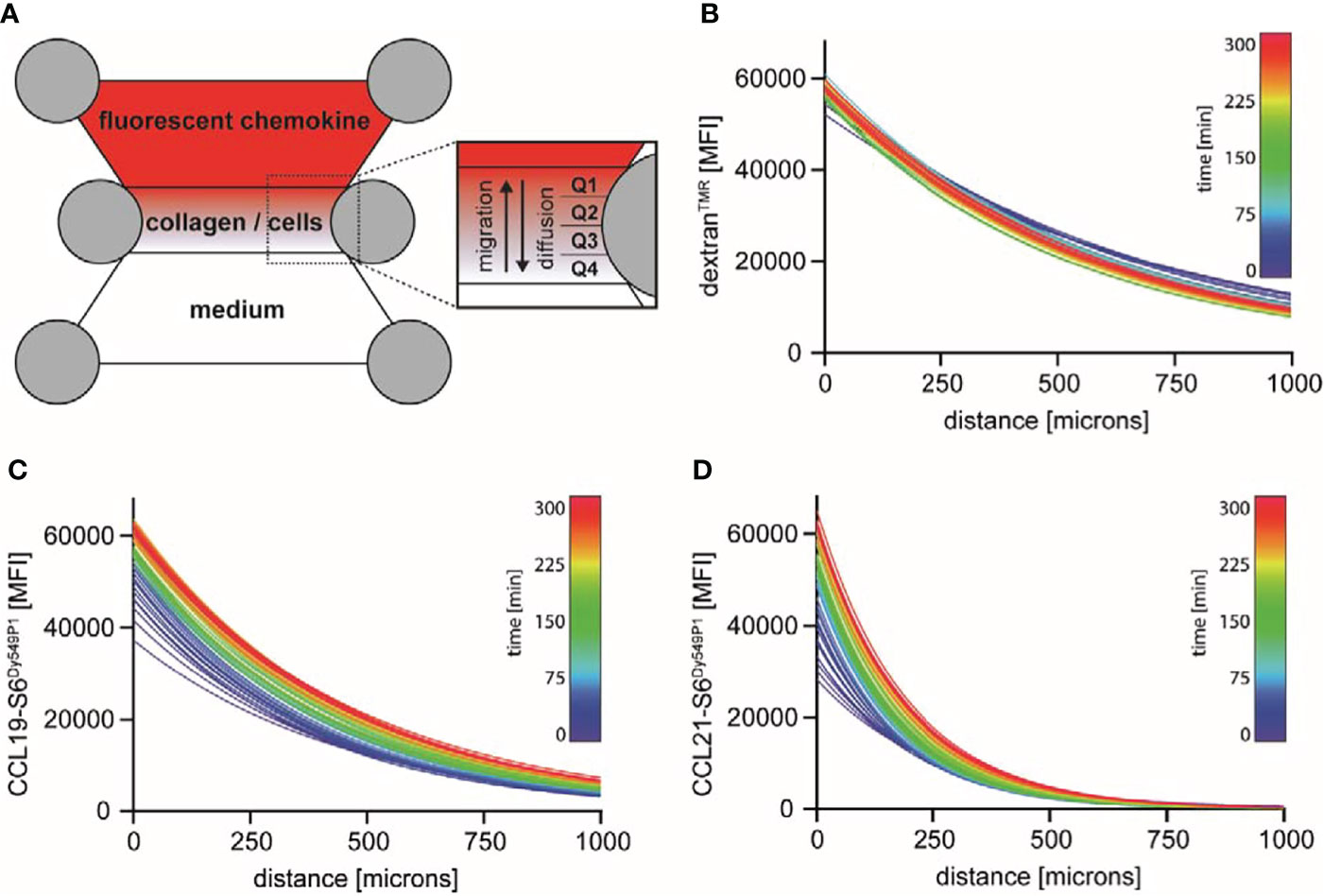
Figure 1 Visualization and characterization of fluorescently labelled dextran, CCL19 and CCL21 gradients in 3D migration chambers. (A) Schematic representation of a 3D migration chamber. Cells are embedded in a defined 3D collagen I matrix of an Ibidi μ-slide suitable for time-lapse fluorescence microscopy. Fluorescently labelled molecules and control medium are filled into the adjacent connected reservoirs, such that a gradient is established over time following a source-and-sink principle. The central chamber corresponds to the observation area, which we subdivided into four quarters (Q1-Q4) for certain analysis. (B-D) Distribution of fluorescently labelled molecules in the central observation chamber was measured by time-lapse fluorescent microscopy. One reservoir was filled with 100 nM dextranTMR (B), CCL19-S6Dy549P1 (C) or CCL21-S6Dy549P1 (D). Mean fluorescence intensity (MFI) of the labelled proteins within the 3D matrix was monitored over time for 5 hours, fitted with a one-phase decay model and color graded from blue (t = 0min) to red (t = 300min). One out of two independent experiments is shown.
The Charged C-Terminus Is Determinant for Shaping the CCL21 Gradient
Our data indicate a time-dependent formation and shaping of the chemokine gradients. This poses the question of how long it takes the chemokines to establish stable gradients within the 3D collagen matrix. Both fluorescent chemokines rapidly (in the time range of minutes) establish well-defined gradients within the 3D collagen matrix, although the steepness and width of the gradients adjust over time, until quasi-stable gradients are attained after 180 minutes that do not substantially change any longer (Figures 2A, B). The exponential decay, particularly of the CCL21-S6Dy549P1 gradient, clearly indicates that the chemokine cannot freely diffuse within the 3D collagen matrix like its surrogate dextranTMR. The steeper gradient of CCL21-S6Dy549P1 is likely to have its nature in the polar C-terminal tail, which is known to electrostatically interact with collagen and other extracellular matrix proteins (20, 21). To substantiate this, we fluorescently labelled a naturally occurring CCL21 variant [amino acids 1-79 of the mature CCL21 peptide (23)], referred to as CCL21trunc-S6Dy549P1 which lacks the charged C-terminal tail. Indeed, CCL21trunc-S6Dy549P1 showed a close to linear distribution within the 3D collagen matrix (Figure 2B).
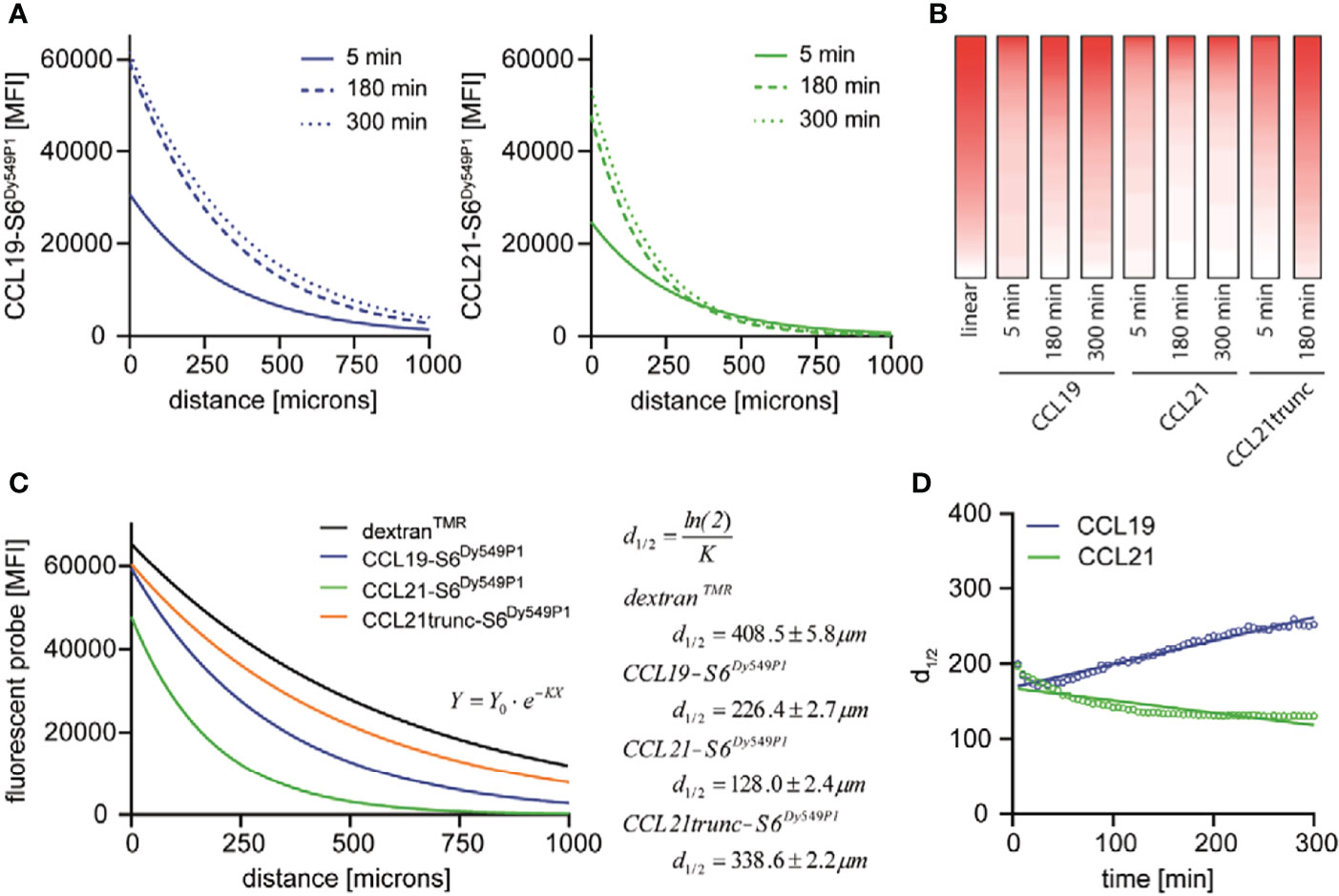
Figure 2 CCL19, CCL21 and CCL21trunc establish stable gradients of distinct shapes. (A) Mean fluorescence intensity (MFI) of CCL19-S6Dy549P1 and CCL21-S6Dy649P1 in relation to the distance to the reservoir measured after 5 min, 180 min and 300 min of incubation. (B) MFI values of CCL19-S6Dy549P1, CCL21-S6Dy649P1 [taken from (A)] and CCL21trunc-S6Dy549P1 were translated in a pseudocolor representation, and supplemented with an artificial linear gradient for comparison. (C) Applying a one-phase exponential decay function reveals different 50 % initial fluorescence values (d1/2) for dextranTMR, CCL19-S6Dy549P1, CCL21-S6Dy549P1 and CCL21trunc-S6Dy549P1 after 180 min. (D) Spatial distribution of d1/2 as the ‘mean chemokine diffusion’ over time increases for CCL19-S6Dy549P1, but not for CCL21-S6Dy549P1.
The shape of the chemokine gradients could best be fitted using an exponential one-phase decay function of the type Y = Y0 · e–KX after subtracting the background fluorescence by setting the plateau equal to zero. Here, Y0 describes the initial fluorescence, whereas K is the rate constant expressed in reciprocal of the X-axis units. Based on the calculation of K, the chemokine half-life corresponds to 50 % of the initial fluorescence derived by , resulting in d1/2. From this fit, d1/2 of CCL21-S6Dy549P1 was the shortest after three hours of gradient formation (d1/2 = 128.0 ± 2.4 μm) and remained relatively stable over time (Figure 2C). However, CCL19-S6Dy549P1 showed a more distant drop in fluorescence intensity (d1/2 = 226.4 ± 2.7 μm) (Figure 2C), whereas CCL21trunc-S6Dy549P1 and dextranTMR showed a comparable distribution among the gradient with comparable 50 % fluorescence intensities (d1/2 = 338.6 ± 2.2 μm for CCL21trunc-S6Dy549P1 and d1/2 = 408.5 ± 5.8 μm for dextranTMR, respectively) (Figure 2C). Moreover, calculating the chemokine specific diffusion d1/2 revealed a slight temporal decrease for CCL21-S6Dy549P1 as the initial fluorescence within the first chamber quarter raises over time and subsequently lowers the spatial localization of d1/2 (Figure 2D). By contrast, the spatiotemporal distribution of d1/2 for CCL19-S6Dy549P1 follows an ascendant line based on the forward shift of the 50 % fluorescence intensity caused by free chemokine diffusion underlining chemotactic gradient formation (Figure 2D).
DCs Sense Differences in the CCL19 and CCL21 Gradients Resulting in Distinct Migratory Responses
To investigate functional consequences for guiding DCs by the different shapes of the chemokine gradients, we embedded human matured MoDCs into the central 3D collagen matrix and let them migrate along the CCL19-S6Dy549P1 or CCL21-S6Dy549P1 gradient. We subdivided the migration chamber into four consecutive quarters, Q1 to Q4, of which Q1 is the one closest to the chemokine reservoir (Figure 3A). Cells in each quarter that moved more than a cell diameter were tracked to determine the velocity and the forward migration index (yFMI) of the migrating MoDCs. Notably, MoDCs that started in Q1 migrated with a comparable, statistically indistinguishable velocity and yFMI in either of the two chemokine gradients (Figure 3A). Moreover, all MoDCs sensed and migrated towards the chemokine source (Figure 3B). Intriguingly, a similar velocity of migrating MoDCs was observed in all four quarters for the two CCR7 ligands (Figure 3A). By contrast, MoDCs starting in Q2 of the CCL21-S6Dy549P1 gradient, although still motile, show a significantly reduced directional migration behavior as manifested with a significantly lower yFMI (Figure 3A). Notably, MoDCs from all quarters were able to sense and follow the CCL19-S6Dy549P1 gradient, whereas MoDCs in Q4 were motile but failed to sense and migrate along the CCL21-S6Dy549P1 gradient (Figure 3B).
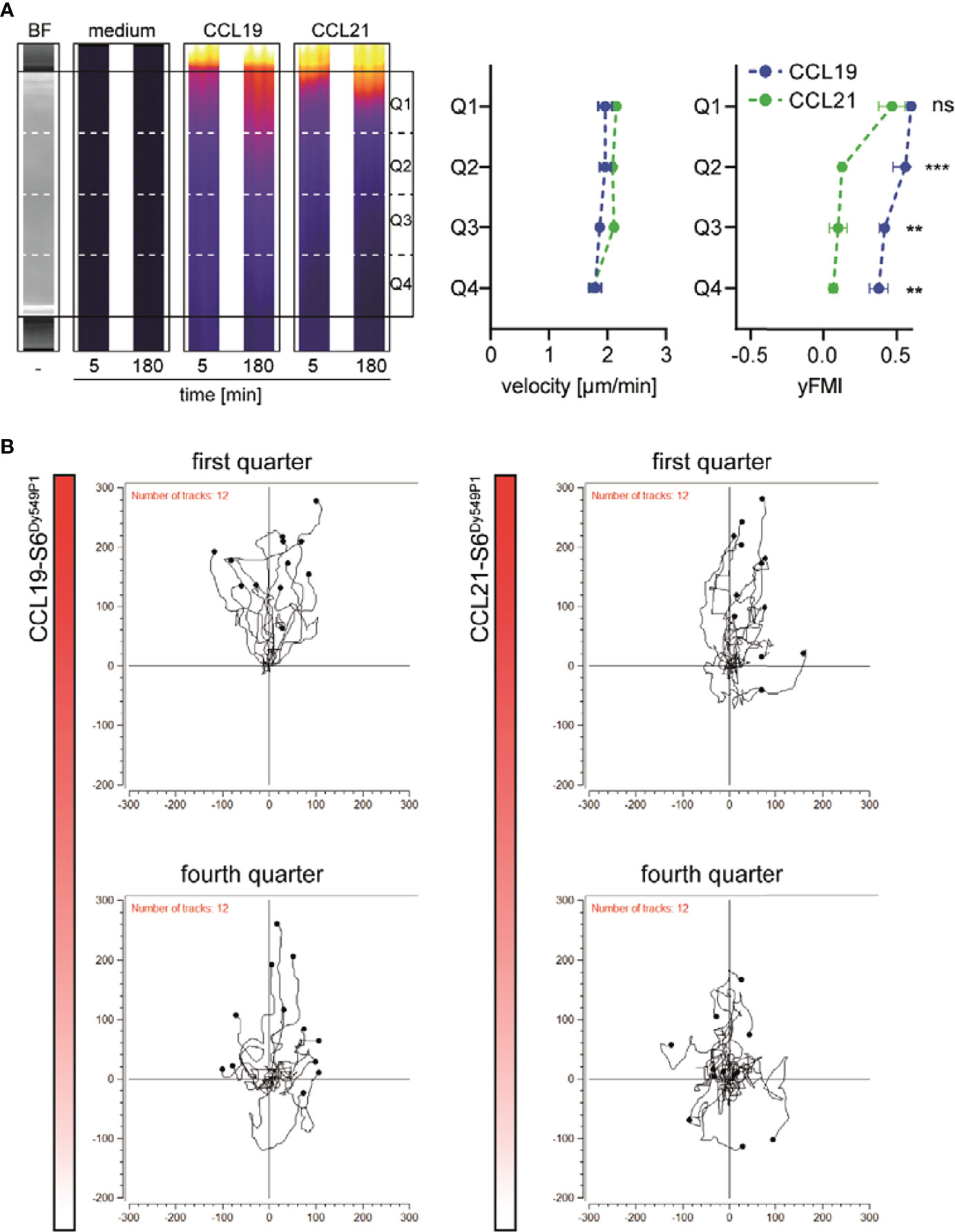
Figure 3 Distinct shapes of CCL19 and CCL21 gradients translate into different migratory responses of human DCs. (A) Representative cropped microscopic images of the central 3D migration chamber. Bright field (BF) as well as pseudocolored fluorescence images of medium, CCL19-S6Dy549P1 and CCL21-S6Dy549P1 at indicated time points derived from one out of three independent experiments are shown (left panel). Velocity and forward migration index in the y-axis (yFMI) of human matured MoDCs migrating along the chemokine gradients in each quarter are shown in the right panel. Mean values ± SD for n = 3, statistical analysis: two-way ANOVA with Sidak post-test. (B) Spider plots of individual tracks of migrating MoDCs in quarters Q1 and Q4.
Discussion
Cell migration along chemokine gradients is a fundamental process not only during development but also in orchestrating protective immune responses. Major advances have been achieved in the design and fabrication of sophisticated devices to study cell migration dynamics in 3D environments in real-time. By contrast, how chemokines distribute in such devices and how gradients are formed and maintained is sparsely investigated. The main reason for the poor information is the lack of appropriate tools, i.e. the availability of fluorescently labelled chemokines. Fluorescent surrogate molecules like fluorescently labelled dextran of the molecular weight of chemokines have been used instead (14, 17–19). Therefore, we recently took on this problem and have designed and produced a first and a second generation of fluorescently labelled chemokines (22, 25).
In the present study, we now show the limitations of such surrogates in a defined 3D matrix. We demonstrate that the surrogate dextran, which owns non-charged surface characteristics, essentially follows a near free, linear diffusion over time within the 3D matrix. In marked contrast, chemokines possess clusters of charged amino acids exposed at their surface, and hence establish exponential-like gradients in the 3D collagen migration chamber. Indeed, CCL21 with its extended, highly polar C-terminus formed the steepest gradient. This goes along with its profound interaction with glycosaminoglycan structures and other extracellular substrate components that is abrogated in CCL21trunc (20, 21). Importantly, electrostatic interactions between the chemokine and the extracellular matrix substrate restrain the free diffusion of the chemokine, such that chemokine gradients are of much shorter distance than surrogate dextran gradients. Consequently, these particular chemokine properties must be taken into consideration when cell migration along chemokine gradients is investigated. In fact, the distance from the chemokine source and the shape of the chemokine gradient is decisive whether a cell is able to sense and follow the guidance cue or not. Consistent with a previous study on murine chemokines, we found that a stable gradient was established after about two to three hours for CCL19-S6Dy549P1 and CCL21-S6Dy549P1, whereas gradient formation of dextranFITC was not temporally affected (13). The steep gradient shape of CCL21-S6Dy549P1 in our 3D matrix mimics the fast decay of the murine CCL21 gradient away from the lymphatic vessels reported in the dermis (8, 26). We further show that gradients formed by two ligands for CCR7 are of a different shape and point to a limited meaningfulness of using dextran as surrogate molecule to characterize chemokine gradients.
Data Availability Statement
Datasets for this study are deposited on Zenodo and are publicly available under a Creative Commons Attribution 4.0 International license, doi: 10.5281/zenodo.6362275. The data can be found at https://zenodo.org/record/6362275#.Yp81SKjMIuU.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of the University of Konstanz. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization, MA and DL. Methodology, MA, OG, and VP. Investigation, MA, OG, and VP. Visualization, MA, and DL. Writing - original draft, MA and DL. Writing - review and editing, all authors. Supervision, DL. Project administration, DL. Funding acquisition, DL. All authors contributed to the article and approved the submitted version.
Funding
This research was supported in parts by research funding from the Swiss National Science Foundation, grant number 310030_189144, the Thurgauische Stiftung für Wissenschaft und Forschung, and the State Secretariat for Education, Research and Innovation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. SenGupta S, Parent CA, Bear JE. The Principles of Directed Cell Migration. Nat Rev Mol Cell Biol (2021) 22:529–47. doi: 10.1038/s41580-021-00366-6
2. Rot A, von Andrian UH. Chemokines in Innate and Adaptive Host Defense: Basic Chemokinese Grammar for Immune Cells. Annu Rev Immunol (2004) 22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543
3. Legler DF, Thelen M. Chemokines: Chemistry, Biochemistry and Biological Function. Chimia. (2016) 70:856–9. doi: 10.2533/chimia.2016.856
4. Insall RH. Understanding Eukaryotic Chemotaxis: A Pseudopod-Centred View. Nat Rev Mol Cell Biol (2010) 11:453–8. doi: 10.1038/nrm2905
5. Proudfoot AEI, Johnson Z, Bonvin P, Handel TM. Glycosaminoglycan Interactions With Chemokines Add Complexity to a Complex System. Pharmaceuticals. (2017) 10:E70. doi: 10.3390/ph10030070
6. Bao X, Moseman EA, Saito H, Petryniak B, Thiriot A, Hatakeyama S, et al. Endothelial Heparan Sulfate Controls Chemokine Presentation in Recruitment of Lymphocytes and Dendritic Cells to Lymph Nodes. Immunity. (2010) 33:817–29. doi: 10.1016/j.immuni.2010.10.018
7. Sarris M, Masson JB, Maurin D, van der Aa LM, Boudinot P, Lortat-Jacob H, et al. Inflammatory Chemokines Direct and Restrict Leukocyte Migration Within Live Tissues as Glycan-Bound Gradients. Curr Biol (2012) 22:2375–82. doi: 10.1016/j.cub.2012.11.018
8. Weber M, Hauschild R, Schwarz J, Moussion C, de Vries I, Legler DF, et al. Interstitial Dendritic Cell Guidance by Haptotactic Chemokine Gradients. Science. (2013) 339:328–32. doi: 10.1126/science.1228456
9. Graham GJ, Handel TM, Proudfoot AEI. Leukocyte Adhesion: Reconceptualizing Chemokine Presentation by Glycosaminoglycans. Trends Immunol (2019) 40:472–81. doi: 10.1016/j.it.2019.03.009
10. Ulvmar MH, Werth K, Braun A, Kelay P, Hub E, Eller K, et al. The Atypical Chemokine Receptor CCRL1 Shapes Functional CCL21 Gradients in Lymph Nodes. Nat Immunol (2014) 15:623–30. doi: 10.1038/ni.2889
11. Luther SA, Bidgol A, Hargreaves DC, Schmidt A, Xu Y, Paniyadi J, et al. Differing Activities of Homeostatic Chemokines CCL19, CCL21, and CXCL12 in Lymphocyte and Dendritic Cell Recruitment and Lymphoid Neogenesis. J Immunol (2002) 169:424–33. doi: 10.4049/jimmunol.169.1.424
12. Schumann K, Lammermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, et al. Immobilized Chemokine Fields and Soluble Chemokine Gradients Cooperatively Shape Migration Patterns of Dendritic Cells. Immunity. (2010) 32:703–13. doi: 10.1016/j.immuni.2010.04.017
13. Haessler U, Pisano M, Wu M, Swartz MA. Dendritic Cell Chemotaxis in 3D Under Defined Chemokine Gradients Reveals Differential Response to Ligands CCL21 and CCL19. Proc Natl Acad Sci USA (2011) 108:5614–9. doi: 10.1073/pnas.1014920108
14. Schwarz J, Bierbaum V, Merrin J, Frank T, Hauschild R, Bollenbach T, et al. A Microfluidic Device for Measuring Cell Migration Towards Substrate-Bound and Soluble Chemokine Gradients. Sci Rep (2016) 6:36440. doi: 10.1038/srep36440
15. Ricart BG, John B, Lee D, Hunter CA, Hammer DA. Dendritic Cells Distinguish Individual Chemokine Signals Through CCR7 and CXCR4. J Immunol (2010) 186:53–61. doi: 10.4049/jimmunol.1002358
16. Hjorto GM, Larsen O, Steen A, Daugvilaite V, Berg C, Fares S, et al. Differential CCR7 Targeting in Dendritic Cells by Three Naturally Occurring CC-Chemokines. Front Immunol (2016) 7:568. doi: 10.3389/fimmu.2016.00568
17. Raman D, Neel NF, Sai J, Mernaugh RL, Ham AJ, Richmond AJ. Characterization of Chemokine Receptor CXCR2 Interacting Proteins Using a Proteomics Approach to Define the CXCR2 “Chemosynapse”. Methods Enzymol. (2009) 460:315–30. doi: 10.1016/S0076-6879(09)05215-X
18. Petrie Aronin CE, Zhao YM, Yoon JS, Morgan NY, Prustel T, Germain RN, et al. Migrating Myeloid Cells Sense Temporal Dynamics of Chemoattractant Concentrations. Immunity. (2017) 47:862–74. doi: 10.1016/j.immuni.2017.10.020
19. Frick C, Dettinger P, Renkawitz J, Jauch A, Berger CT, Recher M, et al. Nano-Scale Microfluidics to Study 3D Chemotaxis at the Single Cell Level. PloS One (2018) 13:e0198330. doi: 10.1371/journal.pone.0198330
20. Patel DD, Koopmann W, Imai T, Whichard LP, Yoshie O, Krangel MS. Chemokines Have Diverse Abilities to Form Solid Phase Gradients. Clin Immunol (2001) 99:43–52. doi: 10.1006/clim.2000.4997
21. Hirose J, Kawashima H, Swope Willis M, Springer TA, Hasegawa H, Yoshie O, et al. Chondroitin Sulfate B Exerts its Inhibitory Effect on Secondary Lymphoid Tissue Chemokine (SLC) by Binding to the C-Terminus of SLC. Biochim Biophys Acta (2002) 1571:219–24. doi: 10.1016/S0304-4165(02)00232-5
22. Artinger M, Matti C, Gerken OJ, Veldkamp CT, Legler DF. A Versatile Toolkit for Semi-Automated Production of Fluorescent Chemokines to Study CCR7 Expression and Functions. Int J Mol Sci (2021) 22:4158. doi: 10.3390/ijms22084158
23. Barmore AJ, Castex SM, Gouletas BA, Griffith AJ, Metz SW, Muelder NG, et al. Transferring the C-Terminus of the Chemokine CCL21 to CCL19 Confers Enhanced Heparin Binding. Biochem Biophys Res Commun (2016) 477:602–6. doi: 10.1016/j.bbrc.2016.06.098
24. Hauser MA, Schaeuble K, Kindinger I, Impellizzieri D, Krueger WA, Hauck CR, et al. Inflammation-Induced CCR7 Oligomers Form Scaffolds to Integrate Distinct Signaling Pathways for Efficient Cell Migration. Immunity. (2016) 44:59–72. doi: 10.1016/j.immuni.2015.12.010
25. Purvanov V, Matti C, Samson GPB, Kindinger I, Legler DF. Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. Int J Mol Sci (2018) 19:E3876. doi: 10.3390/ijms19123876
Keywords: chemokine gradient formation and maintenance, CCL19, CCL21, CCR7, dendritic cell migration, fluorescent chemokines
Citation: Artinger M, Gerken OJ, Purvanov V and Legler DF (2022) Distinct Fates of Chemokine and Surrogate Molecule Gradients: Consequences for CCR7-Guided Dendritic Cell Migration. Front. Immunol. 13:913366. doi: 10.3389/fimmu.2022.913366
Received: 05 April 2022; Accepted: 09 May 2022;
Published: 13 June 2022.
Edited by:
Silvano Sozzani, Sapienza University of Rome, ItalyReviewed by:
Andy Chevigné, Luxembourg Institute of Health, Luxembourg, LuxembourgUrsula Norman, Monash University, Australia
Copyright © 2022 Artinger, Gerken, Purvanov and Legler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel F. Legler, ZGFuaWVsLmxlZ2xlckBiaXRnLmNo
 Marc Artinger
Marc Artinger Oliver J. Gerken1,2
Oliver J. Gerken1,2 Daniel F. Legler
Daniel F. Legler