- 1School of Medicine, Deakin University, Geelong, VIC, Australia
- 2Institute for Mental and Physical Health and Clinical Translation, Deakin University, Geelong, VIC, Australia
Cytokine receptor-like factor 3 (CRLF3) is an ancient protein conserved across metazoans that contains an archetypal cytokine receptor homology domain (CHD). This domain is found in cytokine receptors present in bilateria, including higher vertebrates, that play key roles in a variety of developmental and homeostatic processes, particularly relating to blood and immune cells. However, understanding of CRLF3 itself remains very limited. This study aimed to investigate this evolutionarily significant protein by studying its embryonic expression and function in early development, particularly of blood and immune cells, using zebrafish as a model. Expression of crlf3 was identified in mesoderm-derived tissues in early zebrafish embryos, including the somitic mesoderm and both anterior and posterior lateral plate mesoderm. Later expression was observed in the thymus, brain, retina and exocrine pancreas. Zebrafish crlf3 mutants generated by genome editing technology exhibited a significant reduction in primitive hematopoiesis and early definitive hematopoiesis, with decreased early progenitors impacting on multiple lineages. No other obvious phenotypes were observed in the crlf3 mutants.
Introduction
Cytokines and their specific receptors represent key components of cell-to-cell communication in multicellular organisms (1). Together they play significant roles in the production and function of blood and immune cells as well as other developmental and homeostatic processes (2). Cytokine receptors are complex proteins with a number of different domains, but all share a cytokine receptor homology domain (CHD) that consists of two fibronectin (FBN) type III folds bearing one or two paired cysteine residues with or without a WSXWS motif depending on whether it is a class I or class II receptor (1). Higher vertebrates possess a large family of cytokine receptors that contain one or more class I-type CHD, along with additional extracellular domains as well as a transmembrane and intracellular region, as well as a variety of receptors containing class II type CHDs which play important roles particularly in blood and immune cell development (3). Cytokine receptors arose in bilateria with the emergence of a protein related to the higher vertebrate GP130, typified by Domeless in fruit fly (Drosophila melanogaster) that acts as a classical cytokine receptor (3). All use the Janus kinase-Signal transducer and activator of transcription (JAK-STAT) pathway to mediate intracellular signaling (1).
Cytokine receptor like factor 3 (CRLF3) consists mostly of a class I CHD (3, 4). This ancient protein has been found across metazoan, suggesting it is the likely precursor of this important structural element of cytokine receptors (5), but is also retained in bilateria, including high vertebrate species (3). However, in stark contrast to cytokine receptors, little is known about CRLF3 and its function. Available data indicate broad expression in mice (6) and humans (7), including hematopoietic organs in both cases (6, 7). The limited functional studies have suggested potential neuronal functions in invertebrates (8, 9) and higher vertebrates (10). The invertebrate studies have also suggested that CRLF3 is activated by unknown cytokines (9) and utilizes the JAK-STAT pathway (8), although the details remain scant.
Vertebrate hematopoiesis occurs via a conserved program of development involving distinct waves (11). Primitive hematopoiesis in zebrafish commences in the anterior lateral mesoderm (ALM) and posterior lateral mesoderm (PLM) from precursors expressing scl that contribute to both blood and vascular development (12). Primitive myeloid cells are largely derived from spi1-expressing cells in the ALM whereas primitive erythrocytes develop from gata1-expressing cells in the PLM (13, 14), with a transient intermediate wave in the posterior blood island (PBI) contributing to both erythrocytes and granulocytes (15). Definitive hematopoiesis commences with the generation of multipotent hematopoietic stem cells (HSCs) expressing cmyb in the ventral wall of the dorsal aorta within the aorta-gonad-mesonephros region (16) and migrate first to the caudal hematopoietic tissue (CHT), equivalent to the mammalian fetal liver and then the kidney marrow, equivalent to the mammalian bone marrow, to generate multiple blood lineages (12, 17, 18). Lymphocyte progenitors expressing ikzf1 populate the zebrafish thymus early in embryogenesis with T lymphopoiesis well established by 5 dpf (19). Zebrafish B lymphocytes develop around 20 dpf (18), with NK-related cells evident at this time (20). Conserved cytokine receptor homologues have been shown to be expressed in and contribute to the development of blood and immune cells, including erythropoietin receptor (EPO-R) for erythrocytes (21), granulocyte colony-stimulating factor receptor (GCSF-R) for granulocytes (22) and interleukin 2 receptor (IL-2R) family members for T lymphocytes (23).
This study aimed to use zebrafish as a model to further investigate CRLF3. This involved analysis of its spatio-temporal expression pattern and employing genome editing to assess the impact of its ablation during early development, with an emphasis on hematopoiesis.
Materials and Methods
Zebrafish Husbandry
Wild-type and mutant zebrafish were maintained using standard husbandry practices (24). This included feeding thrice daily with a mixture of live feed (artemia and rotifers) and a dry granulated foodstuff (Otohime Hirame Japan). Fish were manually spawned and embryos maintained in a petri dish containing aquarium water with 0.00005% (w/v) methylene blue (Sigma-Aldrich) and raised in an incubator at 28.5°C with 0.03% (w/v) 1-phenyl-2-thio-urea (PTU) used from 8 h post fertilization (hpf) to inhibit pigmentation to enhance embryo transparency. All experiments were approved by the Deakin University Animal Welfare Committee.
Embryo Analysis
At appropriate time points embryos were collected and anesthetized with 0.4 mg/mL benzocaine and finally fixed with 4% (w/v) paraformaldehyde (PFA) made in phosphate-buffered saline (PBS). Fixed embryos were subjected to whole-mount in situ hybridization (WISH) with DIG-labeled probes as described (25). To make probes, 1-2 µg linearized DNA or 100-500 ng PCR products were incubated in DIG labelling mix (Roche) with 20 U RNase inhibitor and 40 U T7 or SP6 RNA polymerase. Following incubation at 37°C for 2 h, 20 U DNase I was added and the sample incubated at 37°C for a further 15 min before addition of EDTA (pH 8.0) to 20 mM, with the probe purified using G-50 gel filtration exclusion microcolumns (GE Health). Fixed embryos were rehydrated via a series of sequential 5 min washes in 75% (v/v) methanol, 50% (v/v) methanol, 25% (v/v) methanol and PBS before 4 × 5 min washes in PBS containing 0.1% Tween-20 (PBS-T). Embryos were then incubated at room temperature in PBS-T containing 10 µg/mL proteinase K for 1-30 min before fixing for 20 min in 4% (v/v) PFA/PBS followed by 5 × 5 min washes in PBS-T. Embryos were incubated in hybridization mix (HM) (50% formamide, 5×SSC, 0.1% Tween-20, 9.2 mM citric acid pH 6, 50 µg/mL heparin, 500 µg/mL tRNA) for 2-5 h at 70°C. Embryos were then bathed in probe solution (1/50–1/300 dilution of DIG-labelled RNA in HM+) and incubated at 70°C overnight. Probe solution was removed and embryos washed briefly in 100% HM– (50% formamide, 5×SSC, 0.1% Tween-20, 9.2 mM citric acid pH 6) and then sequentially for 15 min in 75% HM–/25% 2×SSC, 50% HM–/50% 2×SSC, 25% HM–/75% 2×SSC and 2×SSC, then twice in 0.2×SSC 30 min, then sequentially for 10 min in 75% 0.2×SSC/25% PBS-T, 50% 0.2×SSC/50% PBS-T, 25% 0.2×SSC/75% PBS-T and PBS-T. Embryos were then incubated in PBS-T containing 2% sheep serum and 2 mg/mL BSA at room temperature for 3 h then in antibody solution (1:5000 anti-DIG antibody in PBS-T, 2% sheep serum, 2 mg/mL BSA pre-absorbed against embryos) at 4°C overnight with agitation. Embryos were rinsed in PBS-T then 6 × 15 min washes in PBS-T at room temperature with gentle rocking and then 3 × 5 min washes in staining buffer (0.1 M Tris pH 9.5, 0.05 M MgCl2, 0.1 M NaCl, 0.1% (v/v) Tween 20) containing 25 µg/mL levamisol. Following this, embryos placed in staining buffer containing 0.225 mg/mL nitroblue tetrazolium (NBT) and 90.175 mg/mL 5-bromo,4-chloro,3-indolyl phosphate (BCIP) at room temperature in the dark for 4 h-3 d. Embryos were finally rinsed in PBS-T and fixed in 4% PFA/PBS when stained to an appropriate extent. In some experiments, embryos were injected at the 1-4 cell stage with standard control or lycat morpholinos as described (15), or bathed in 30 µM JAK3 inhibitor or DMSO as a control as published (26). Embryos were visualized using a MVX10 microscope (Olympus) with stage lighting provided using an LG-PS2 fibre optics light source (Olympus). Digital images were recorded using CellSens Dimension 1.6 software (Olympus) and DP72 camera (Olympus). Alternatively, cytospin preparations were prepared from embryonic blood, as described (27). These were fixed in 100% methanol for 1 min and stained with Giemsa for 20 min. Slides were viewed on a Leica DME stereomicroscope and differential counts performed, with images captured on a DFC290 digital camera (Leica).
Genome Editing
The zebrafish crlf3 gene was targeted using genome editing with transcription activator-like effector nucleases (TALENs) and CRISPR/Cas9 (28). A guide RNA (gRNA) was designed to a region of exon 2 using the zifit protocol (29) with the primers 5’-TAGGCAGAGGCGTTGCTGCAGG and 5’-AAACCCTGCAGCAACGCCTCTG, while a pair of TALENs targeting a similar region were designed as described (30). Embryos were injected with 12.5 ng/μL gRNA and 300 ng/μL Cas9 mRNA or with 100 ng/μL mRNA encoding each TALEN and raised to adulthood. Founders were identified with high-resolution melt (HRM) analysis of PCR products with Precision Melt Suremix and Analysis Software (BioRad) (31) using primers spanning the targeted region (5’- CTATTTAGCAGCATGAGTTTACAGC and 5’-TAACAAGGGTTTCTGACTTCTATGC). These fish were outcrossed two times to wild-type fish to remove off-target mutations before in-crossing. Sequence analysis was performed at the Australian Genome Research Centre.
Statistics
Statistical analyses were performed using Graph Pad Prism (Version 8) software. To determine the statistical significance of various treatments, the unpaired independent student’s t test was employed, with Welch’s correction, where appropriate.
Bioinformatics
Protein sequences obtained from GenBank were aligned with Clustal Omega (version 1.2.4) (32), with a phylogenetic tree generated with NJ Plot (33).
Results
Expression of Zebrafish crlf3
Expression of crlf3 during zebrafish embryogenesis was examined by WISH with a full-length anti-sense probe in comparison to a control sense probe. The sense probe gave a small amount of diffuse background staining (Figures 1A, J). However, the anti-sense probe identified specific crlf3 expression, first evident from 10 hpf in adaxial cells in the presomitic mesoderm (Figure 1B) that continued until 18 hpf before waning (Figures 1D–G). Bilateral expression was observed from 12 hpf in the anterior lateral-plate mesoderm (ALP) (Figure 1C) and posterior lateral-plate mesoderm (PLM) (Figure 1D), which continued beyond 20 hpf when the PLM fuses to form the intermediate cell mass (ICM) (Figures 1F–H). To confirm this designation embryos were injected with a lycat antisense morpholino to ablate both hematopoietic and endothelial lineages in the ICM (34), which also greatly diminished crlf3 expression in this region (Figure 1I) compared to those injected with standard control morpholino (Figure 1H). By 4 dpf, crlf3 expression was observed in the thymus, dorsal midline of the midbrain and retina (Figures 1K–L), and from 7 dpf expression become stronger with additional expression observed in the exocrine pancreas and retina (Figures 1M–P). Embryos treated with a JAK3 inhibitor (35) that is able to ablate T lymphocytes in zebrafish (26) showed no crlf3 expression in the thymus (Figure 1R), while those treated with DMSO only exhibited robust expression (Figure 1Q).
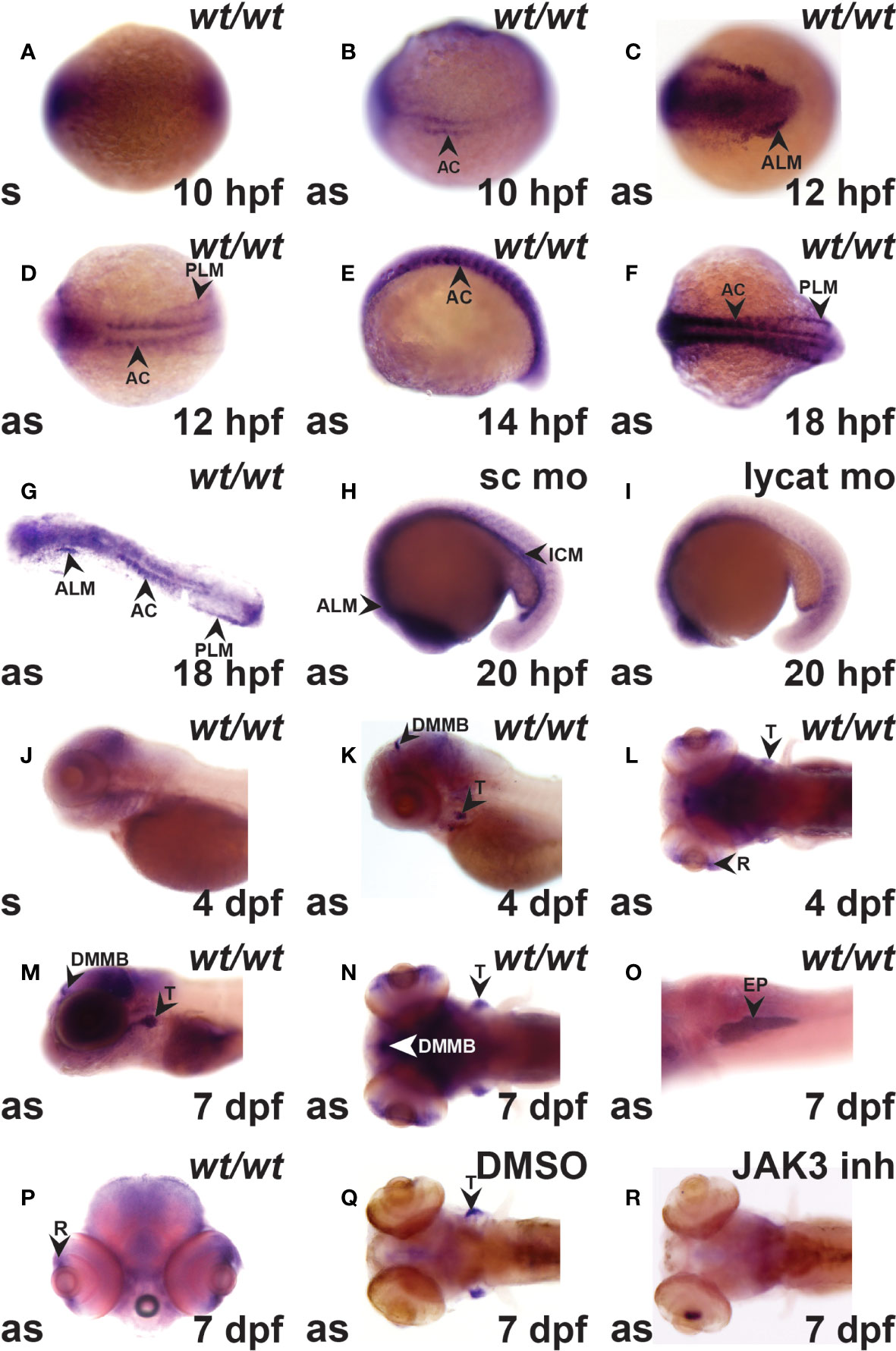
Figure 1 The crlf3 gene is expressed in hematopoietic and other tissues during zebrafish embryogenesis. (A–R). WISH with sense (s) or antisense (as) crlf3 probes at the indicated time points on wild-type embryos (wt/wt, A–G, J–P) or those injected with 1 mM standard control morpholino (sc mo, H) or lycat morpholino (lycat mo, I) or bathed from 56 hpf in DMSO vehicle control (DMSO, Q) or JAK3 inhibitor (JAK3 inh, R). Embryos are dorsal view with anterior to the left (A, B, D, F, L, N, Q, R); anterior view with dorsal to the left (C); lateral view with anterior to the left (E, J, K, M), except panel O that is ventral view with anterior to the left and P is anterior view with dorsal to the top. The indicated structures are: AC (adaxial cells), ALM (anterior lateral plate mesoderm), DMMB (dorsal midline of midbrain), EP (exocrine pancreas), ICM (intermediate cell mass), PLM (posterior lateral plate mesoderm), R (retina), and T (thymus).
Generation of crlf3 Knockout Zebrafish
The zebrafish Crlf3 protein (Figure 2A) is encoded by a gene that consists of nine exons and eight introns (Figure 2B). To study the role of crlf3 both TALEN (Figure 2D) and CRISPR-Cas9 (Figure 2E) based genome editing approaches were designed to target adjacent sites in the coding region of exon 2 of the wild-type (wt) gene (Figure 2C). One-cell stage embryos were injected with either TALEN mRNA or Cas9 mRNA plus a guide RNA and raised to adulthood and crossed with wild-type embryos, with their progeny analyzed by high-resolution melt analysis and sequencing to identify potential heterozygote mutants. After a further round of out-crossing, heterozygote F2 mutants were in-crossed to yield homozygote F3 mutants. Sequencing identified two mutant alleles: a TALEN derived 1 bp deletion, designated mdu14 (Figure 2D), and a CRISPR-Cas9 derived 14 bp deletion, designated mdu15 (Figure 2E). Both of these mutations serve to severely truncate the encoded Crlf3 protein after just 8 or 7 residues, respectively, due to a frame-shift followed by an in-frame stop codon (Figure 2D, E), and so likely represent loss-of-function alleles.
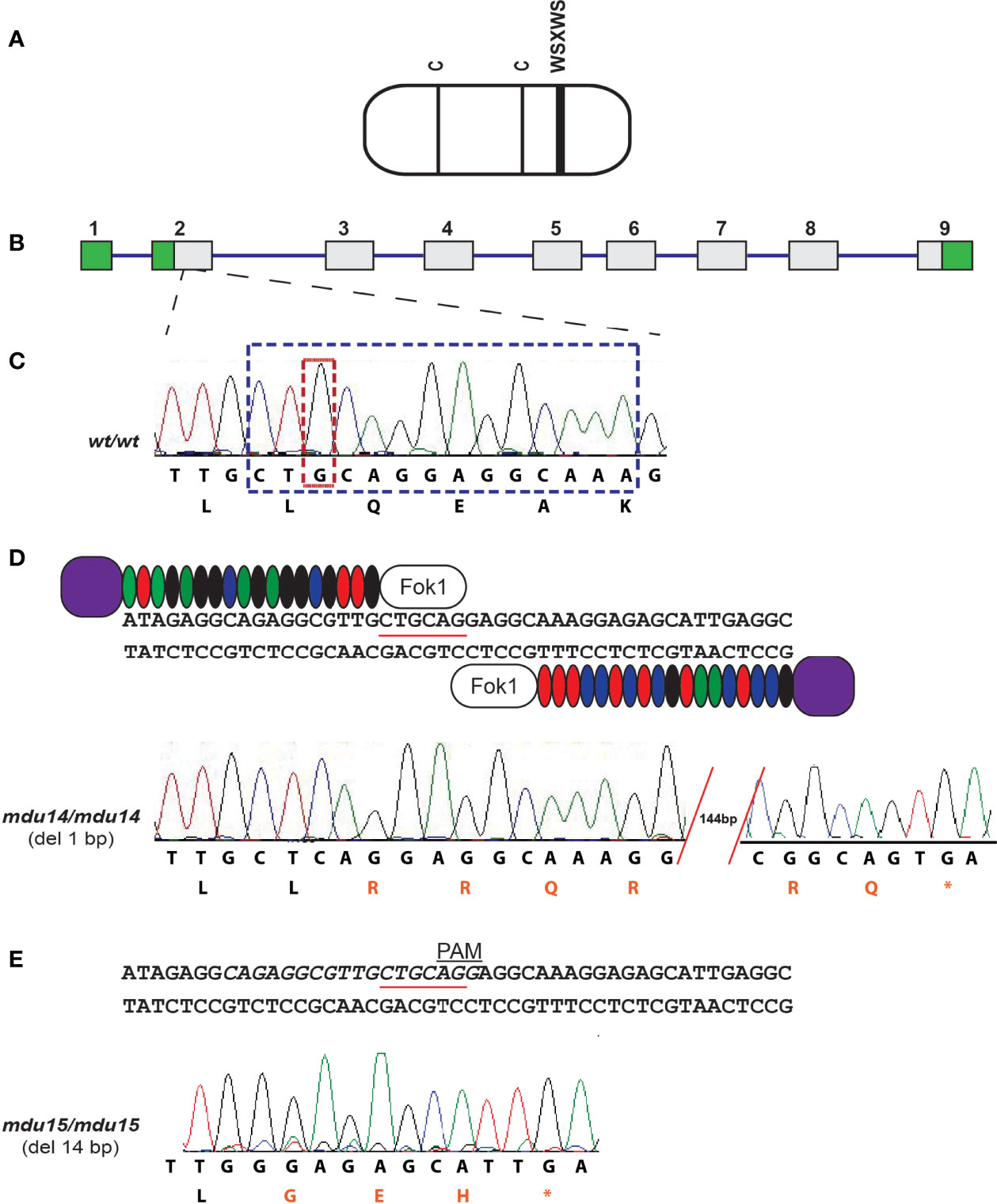
Figure 2 Generation of zebrafish crlf3 mutants using genome editing. (A) Schematic representation of the CRLF3 protein, consisting mostly of a cytokine receptor homology domain (rounded rectangle) containing two conserved cysteines (C, thin lines) and a WSXWS motif (thick line). (B) The intron/exon structure of the zebrafish crlf3 gene, with exons represented as numbered boxes, showing untranslated (green) and translated (gray) regions, and introns represented with intervening lines. (C) Sequence trace of homozygous wild-type crlf3wt/wt (wt/wt) and its corresponding nucleotide sequence and encoded amino acids shown below. Nucleotides deleted in mdu14 and mdu15 alleles are boxed in tan and blue, respectively. (D) Targeting of exon 2 with TALENs, with the PstI site used in RFLP analysis underlined, to generate the mdu14 allele, with the sequence of a homozygous crlf3mdu14/mdu14 (mdu14/mdu14) mutant shown. This represents a 1 bp deletion that causes a frameshift resulting in translation from an alternative reading frame (red) followed by a stop codon (*) that prematurely truncates the protein. (E) Targeting of exon 2 with CRISPR, with target site italicized and PAM site indicated, to generate the mdu15 allele, with the sequence of a homozygous crlf3mdu15/mdu15 (mdu15/mdu15) mutant shown. This 14 bp deletion also causes a frameshift and premature stop.
Impact of crlf3 Ablation on Primitive Hematopoiesis
No evidence of overt developmental perturbation during embryogenesis was observed in crlf3 mutants by light microscopy. However, given the strong expression of crlf3 at the sites of embryonic blood and immune cell development, and the extensive involvement of cytokine receptors in these lineages, this was explored in more detail. The crlf3 mutants were first analyzed for primitive hematopoiesis using WISH. At 14 hpf, crlf3mdu14/mdu14 mutants and their wild-type siblings showed equivalent expression of scl, a marker of hemangioblasts (36), both caudally and rostrally (Figures 3A–D), and of fli1, a marker of vascular precursors (36), both caudally and rostrally (Figures 3E–H). In contrast, decreased expression for spi1, a marker of myeloid precursors (37, 38), was observed in crlf3mdu14/mdu14 embryos that reached significance for the rostral domain (Figures 3I–L), as well as for gata1, a marker erythroid precursors expressed solely in the caudal region of the embryo (39) (Figures 3M–O). Expression of ikzf1, which marks hematopoietic progenitors during primitive hematopoiesis (40), was also decreased at 20 hpf (Figures 3P–R), whereas expression of fli1 remained unchanged at 22 hpf (Figure 3S–U). However, crlf3mdu14/mdu14 embryos showed decreased numbers of cells expressing lcp1 (Figures 3V–X) and mpo (Figure 3Y–AA), markers of monocyte/macrophages (41) and neutrophils (13), respectively. In addition, a more subtle but still statistically significant decrease in expression was observed for hbbe (Figures 3AB–AD), a marker of mature erythroid cells (42). Analysis of crlf3mdu15/mdu15 mutants confirmed the results for ikzf1 (Supplementary Figures 1A–C), lcp1 (Supplementary Figure 1D), mpo (Supplementary Figure 1E) and hbbe (Supplementary Figure 1F).
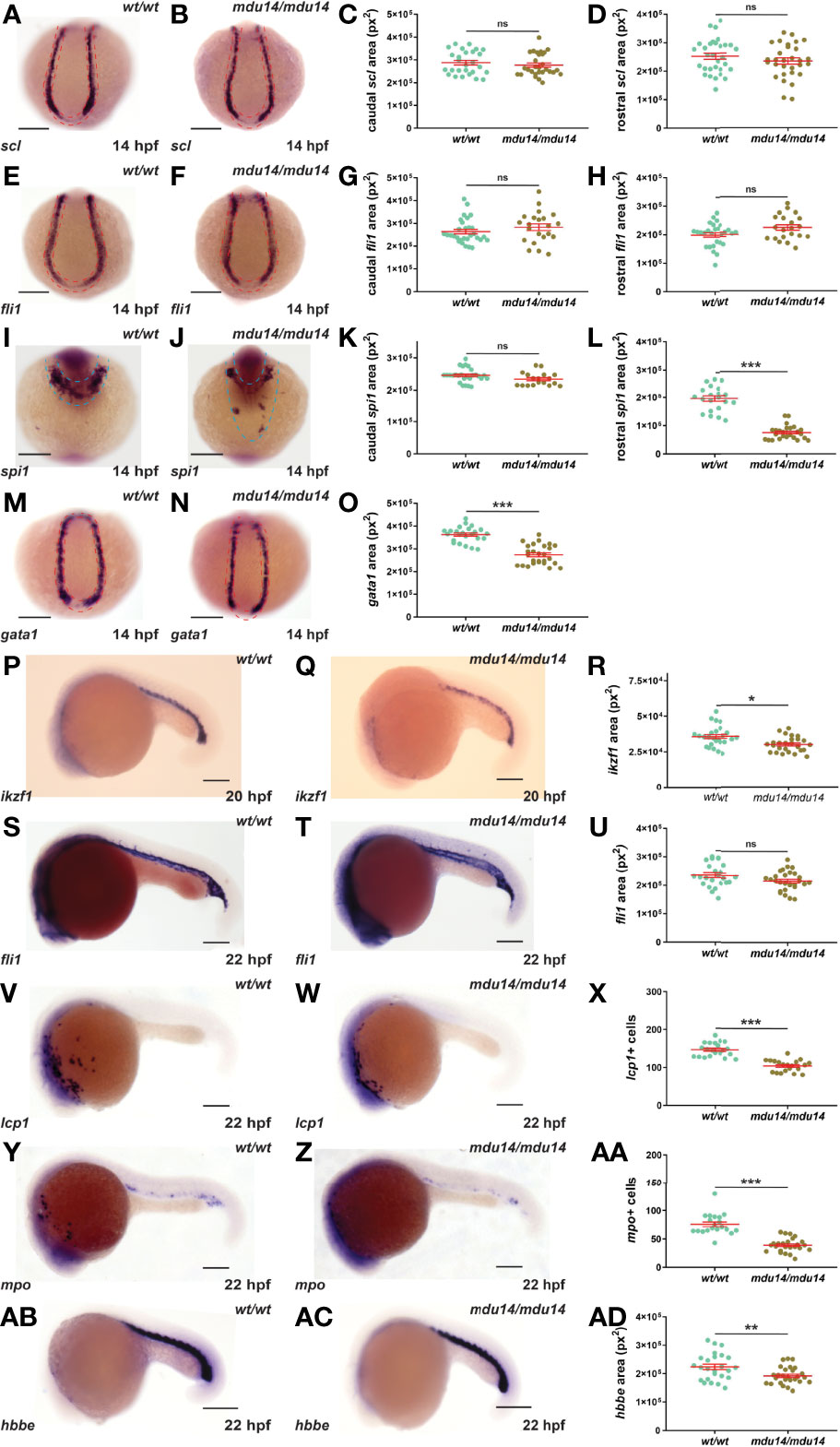
Figure 3 Mutation of crlf3 impacts primitive hematopoiesis. (A–AD). Homozygous crlf3wt/wt (wt/wt), and crlf3mdu14/mdu14 (mdu14/mdu14) embryos were subjected to WISH at 14 hpf with scl (A, B), fli1 (E, F), spi1 (I, J) and gata1 (M, N), at 20 hpf with ikzf1 (P, Q), and at 22 hpf with fli1 (S, T), lcp1 (V, W), mpo (Y, Z) and hbbe (AB, AC) (scale bar = 200 μm; red and blue dotted areas depict caudal and rostral expression domains, respectively). Individual embryos were assessed for area of staining or cell number at the indicated locations for scl (C, D), fli1 (G, H, U), spi1 (K, L), gata1 (O), ikzf1 (R), lcp1 (X), mpo (AA) and hbbe (AD), with the mean and SEM shown in red and level of statistical-significance indicated (***p < 0.001, **p < 0.01, *p < 0.05, ns, not significant). Welch’s correction was used for panel (R).
Impact of crlf3 Ablation on Early Definitive Hematopoiesis
The crlf3 mutants were also examined with respect to early definitive hematopoiesis, which generates both myeloid and lymphoid cells from around 2-3 dpf (43). Decreased expression of cmyb, a marker of hematopoietic stem cells in the caudal hematopoietic tissue (16), was observed in crlf3mdu14/mdu14 embryos at 4 dpf (Figures 4A–C). This correlated with a substantial reduction in the number of cells expressing lcp1 in (Figures 4D–F) and of those expressing mpo (Figures 4G–I) at 5 dpf. The crlf3mdu14/mdu14 embryos also showed decreased expression of hbbe (Figures 4J–L). Expression of ikzf1, a marker of lymphocyte precursors in the thymus (40), was not significantly different between crlf3mdu14/mdu14 embryos and their wild-type counterparts (p = 0.083) (Figures 4M–O). However, a small decrease was seen in the expression of two markers of mature lymphoid cells, rag1 (44) (p < 0.001) (Figures 4P–R) and tcra (45) (p < 0.001) (Figures 4S–U). Analysis of blood smears from 5 dpf embryos identified a decrease in circulating monocytes (p = 0.022) and neutrophils (p < 0.001) (Figures 4V–X), highlighting that these populations were particularly impacted. Analysis of crlf3mdu15/mdu15 mutants confirmed the results for cmyb (Supplementary Figures 1G–I), lcp1 (Supplementary Figure 1J), mpo (Supplementary Figure 1K) and hbbe (Supplementary Figure 1L).
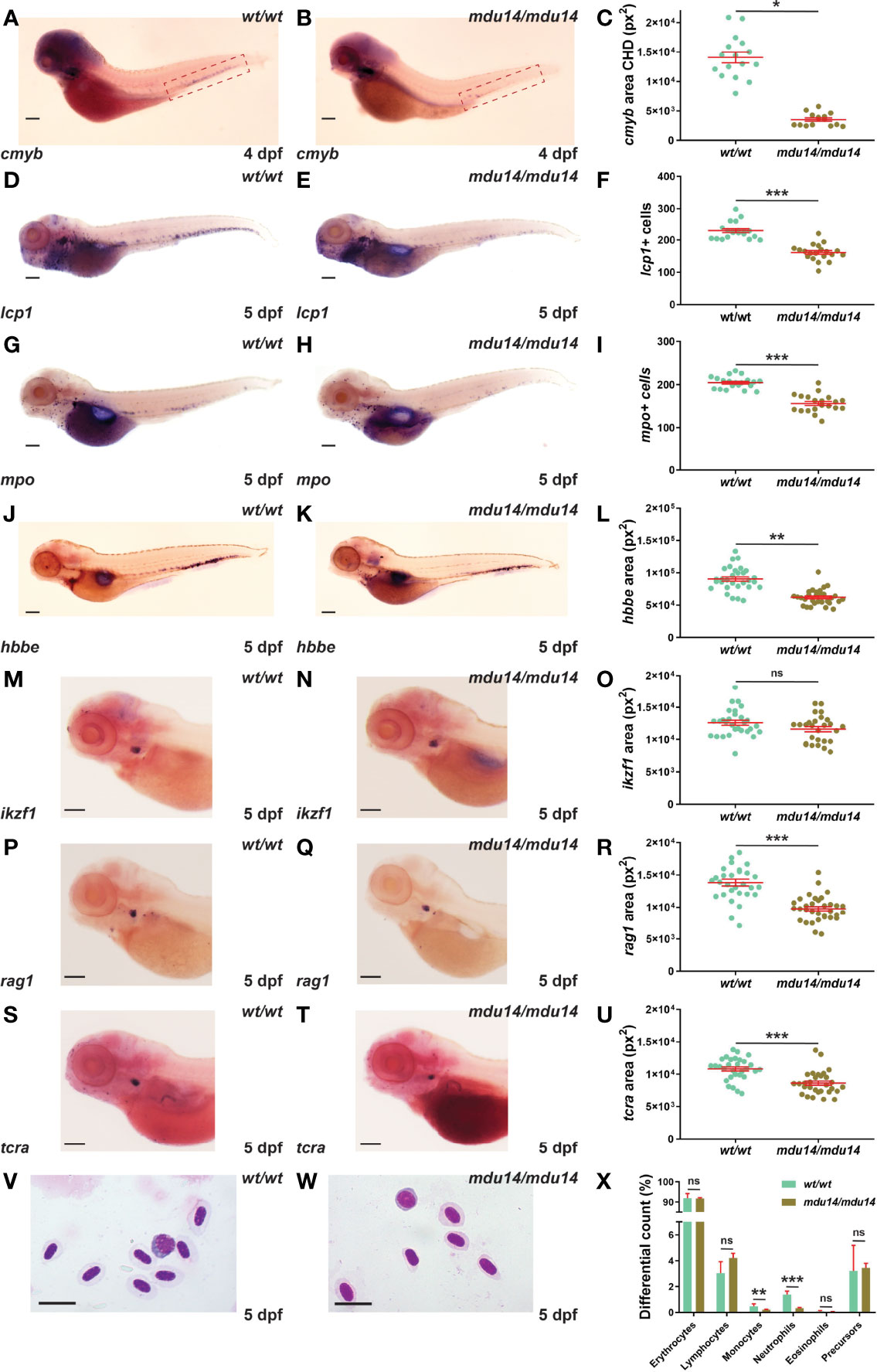
Figure 4 Mutation of crlf3 impacts early definitive hematopoiesis. (A–X). Homozygous wild-type crlf3wt/wt (wt/wt) and crlf3mdu14/mdu14 (mdu14/mdu14) embryos were subjected to WISH with cmyb (A, B) at 4 dpf, and lcp1 (D, E), mpo (G, H), hbbe (J, K), ikzf1 (M–N), rag1 (P, Q) and tcra (S, T) at 5 dpf (scale bar = 200 μm), or underwent blood analysis (V–W) at 5 dpf (scale bar = 10 μm). Individual embryos were assessed for the area of staining of cmyb CHD region (dotted boxes in panels A and B) (C), hbbe (L), ikzf1 (O), rag1 (R) and tcra (U), as well as the number of lcp1+ (F) and mpo+ (L) cells or for blood differential counts (X), with mean and SEM in red and statistical significance indicated (***p < 0.001, **p < 0.01, *p < 0.05, ns, not significant). Welch’s correction was used for panels (C, L).
Impact of crlf3 Ablation on Other Aspects of Embryogenesis
Given the embryonic expression pattern of crlf3, the expression of markers specific for somites, eye, brain and pancreas were also examined. No difference in expression between crlf3mdu14/mdu14 mutant and wild-type embryos was observed for myod, a marker of muscle lineages (46), at 14 hpf (Figures 5A, B) and 22 hpf (Figures 5C, D), pax6, a marker for retinal progenitor cells and ganglion cells in the eye and brain, respectively (47), at 5 dpf (Figures 5E, F), and trypsin, a marker of the exocrine pancreas (48) at 7 dpf (Figures 5G, H).
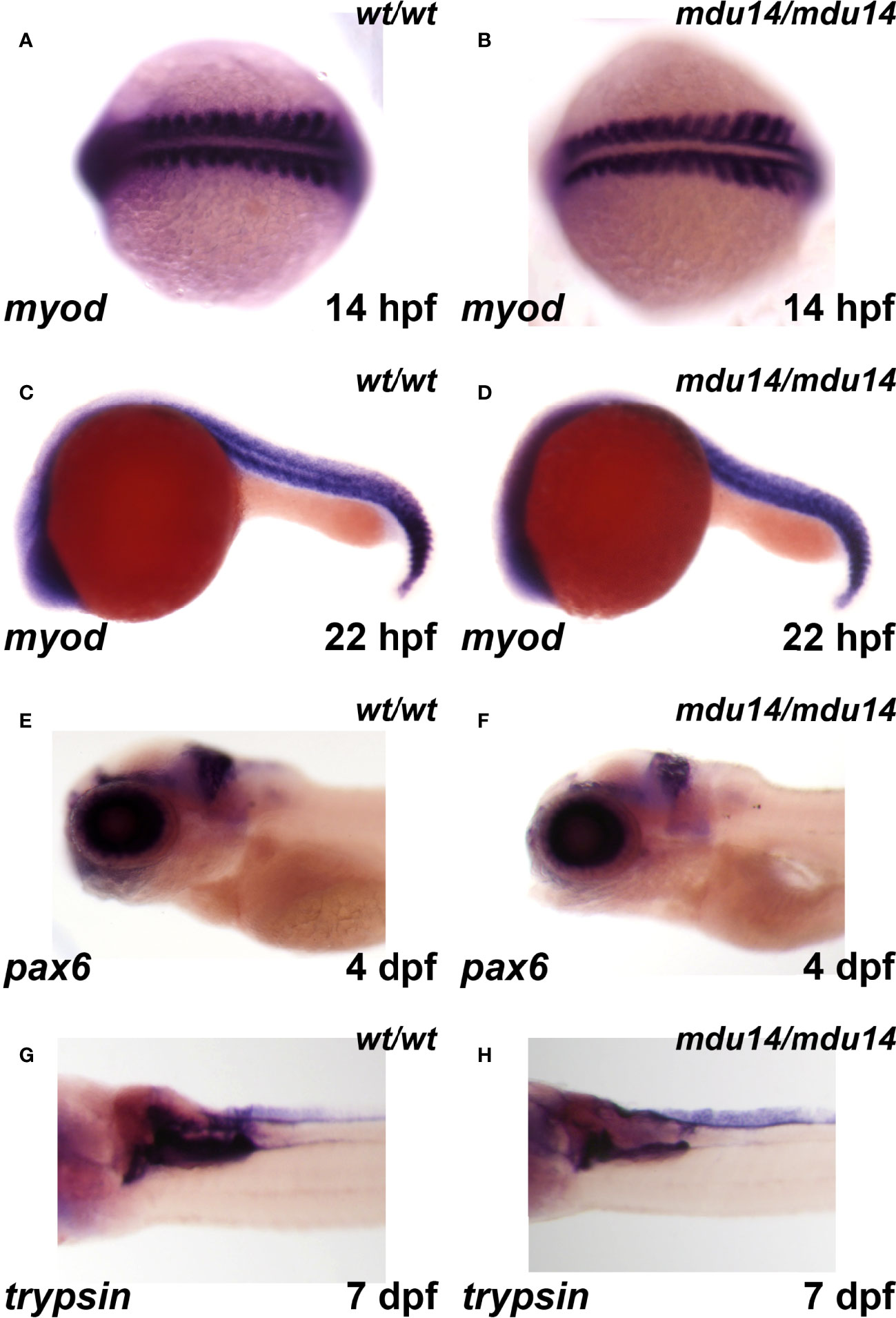
Figure 5 Mutation of crlf3 fails to affect other relevant embryonic tissues. (A–H) Homozygous wild-type crlf3wt/wt (wt/wt), and crlf3mdu14/mdu14 (mdu14/mdu14) embryos at the indicated ages were subjected to WISH with myod (A–D), pax6 (E, F) and trypsin (G, H), presented as dorsal view (A, B), lateral view (C–F) or ventral view (G, H) with anterior to the left in each case.
Discussion
Cytokines and cytokine receptors play a vital role in cell-to-cell communication important for hematopoiesis, immunity and other homeostatic processes (1). A defining characteristic of cytokine receptors is the presence of a conserved cytokine receptor homology domain (CHD) in the extracellular region. CRLF3 contains a CHD (3) that likely represents the archetypal CHD from which those found in all cytokine receptors were derived (5), with homologues of CRLF3 identified in a wide range of metazoan species (3). However, the function of this evolutionarily important protein remains poorly defined. This study used zebrafish as model to add to our understanding of CRLF3 in early development.
Zebrafish crlf3 was found to be expressed at several hematopoietic sites during embryogenesis. This included the ALM, PLM and ICM, sites of early myelopoiesis, as well the thymus, location of T lymphocyte development, the latter consistent with data from mouse embryos (6). Crlf3 has also been found to be expressed in the adult Xenopus thymus (49), adult mouse thymus and spleen (6) and adult human thymus, spleen and bone marrow (7). The zebrafish crlf3 gene was additionally expressed in a range of other embryonic tissues, including adaxial cells within the pre-somitic mesoderm, regions of the developing eye and brain, as well as the pancreas. Broad expression has also been observed in mouse embryos, with Crlf3 expressed in the mid-brain, sensory organs, ovary, testis, liver and other visceral organs (6). In adult Xenopus, crlf3 was expressed in the central nervous system and testis (49), with mammals also showing expression in the central nervous system, liver and a variety of other sites (6, 7). This collectively suggests potential roles throughout the life-course in blood and immune cells, where cytokine receptors play vital roles (2), as well as more broadly across other tissues.
Genome editing was used to generate two mutants of zebrafish crlf3. These most likely represent loss-of-function alleles since they only encoded 7 or 8 residues of the 444 amino acids comprising the CRLF3 protein (<2% of the total). Moreover, heterozygotes of either mutant produced no discernible phenotype (data not shown). Therefore, homozygous crlf3 mutants were analyzed with respect to early hematopoiesis in comparison to wild-type individuals. During the primitive wave of hematopoiesis, the crlf3 mutants showed no alterations in the expression of markers for bipotential hemangioblast cells or vascular precursors, but those for hematopoietic, myeloid and erythroid progenitors were reduced, as were markers for mature myeloid and erythrocytic cells. Collectively, these results indicate crlf3 likely impacts primitive hematopoiesis at the level of hematopoietic progenitors that affects downstream lineages. During definitive hematopoiesis, the markers for HSCs, mature myeloid and erythroid cells remained reduced, as did those for mature T lymphocytes – although not lymphocyte precursors. This suggests a broad role for crlf3 in definitive hematopoiesis, affecting HSCs and their downstream lineages as well as the production of mature lymphocytes. In each case, however, the impacts were relatively modest indicating it is not an essential regulator.
There is some evidence from other studies that CRLF3 contributes to blood and immune cell development and/or function throughout the life-course. Human CRLF3 variants have been associated with lymphocyte percentage in the blood (50) and risk of cutaneous leishmaniasis (51), while variants in the corresponding chicken gene are associated with an altered antibody response (52). It also forms part of a rare UTP6-CRLF3 fusion observed in human acute myeloid leukemia (53). Ablation in mice caused reduced platelet numbers due to a defect in platelet maturation, and consistent with an association between human CRLF3 variants and platelet distribution (50). Crlf3−/− mice also exhibited altered red blood cell parameters but no significant differences in immune cell populations or overall survival (54, 55). These observations are collectively consistent with a subtle role for CRLF3 in the hematopoietic system.
The broad embryonic expression of crlf3 promoted the analysis of other tissues. However, the expression of key markers of skeletal muscle, retinal progenitor cells and ganglion and exocrine pancreas were unaltered. Other studies have suggested roles in neuronal differentiation and function (10, 56), with a CRLF3 variant associated with the extent of autism in NF1 patients (56). On-line data on the Crlf3−/− mouse have also suggested reduced body weight and abnormal behaviors and tremors (55). However, no obvious differences in behavior or growth were observed in the crlf3 mutants (data not shown).
The mechanism of action for CRLF3 remains vague, although there is high conservation of the protein across bilaterians, including vertebrates and insects (Supplementary Figure 2). This is particular true in the CHD that incorporates the archetypal WSXWS domain, but also extends to residues in the C-terminus, suggesting both are important for function. Indeed, evidence from insect systems suggest that CRLF3 is activated by cytokines, with both exogenous mammalian erythropoietin and an as yet unidentified endogenous cytokine in the hemolymph able to elicit CRLF3-dependent responses (57), suggesting the CHD is functional in cytokine binding. The insect CRLF3 has also been shown to lie upstream of the JAK-STAT pathway (8), while mammalian studies have also identified STAT3 as a downstream mediator (58), actions presumably mediated by the C-terminus of the protein. In the latter context CRLF3 has been implicated in cell growth as a result of concomitant elevation of cyclin D proteins and NF-κb (58). This is consistent with the upregulation of CRLF3 observed during the early cancerous stage of actinic keratosis, cutaneous squamous cell carcinoma and non-melanoma skin cancer (59), and its involvement in the UTP6-CRLF3 fusion in acute myeloid leukemia (53). However, a distinct role is also emerging in the regulation of cell and organelle morphogenesis, with its ablation impacting neurite outgrowth (56), synaptic vesicle biogenesis (10) and the maturation of platelets from large pre-platelet precursors (50), while its enforced expression perturbed cell morphology and motility (58). These effects have been associated with altered microtubule stability (50) and disrupted RhoA signaling (56, 58), with the Hippo pathway also implicated (50). It is hoped that the crlf3 mutant will allow the further exploration of CRLF3’s mode of action.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Deakin University Animal Ethics Committee.
Author Contributions
TT, KP and CL performed experiments; TT, KP, AW and CL analyzed the results and prepared figures; YG, AW and CL designed the research; TT, KP, AW and CL wrote the paper, which was read and approved by all authors.
Funding
The authors recognize the support of funding from IMPACT at Deakin University. TT was supported by a Deakin University International Postgraduate Research Award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the Deakin University Animal House staff for exceptional assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.910428/full#supplementary-material
References
1. Liongue C, Sertori R, Ward AC. Evolution of Cytokine Receptor Signaling. J Immunol (2016) 197:11–8. doi: 10.4049/jimmunol.1600372
2. Robb L. Cytokine Receptors and Hematopoietic Differentiation. Oncogene (2007) 26:6715–23. doi: 10.1038/sj.onc.1210756
3. Liongue C, Ward AC. Evolution of Class I Cytokine Receptors. BMC Evol Biol (2007) 7:120. doi: 10.1186/1471-2148-7-120
4. Boulay JL, O'Shea JJ, Paul WE. Molecular Phylogeny Within Type I Cytokines and Their Cognate Receptors. Immunity (2003) 19:159–63. doi: 10.1016/S1074-7613(03)00211-5
5. Liongue C, Taznin T, Ward AC. Signaling via the CytoR/JAK/STAT/SOCS Pathway: Emergence During Evolution. Mol Immunol (2016) 71:166–75. doi: 10.1016/j.molimm.2016.02.002
6. Smith CM, Hayamizu TF, Finger JH, Bello SM, McCright IJ, Xu J, et al. The Mouse Gene Expression Database (GXD): 2019 Update. Nucleic Acids Res (2019) 47:D774–9. doi: 10.1093/nar/gky922
7. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-Based Map of the Human Proteome. Science (2015) 347:1260419. doi: 10.1126/science.1260419
8. Miljus N, Heibeck S, Jarrar M, Micke M, Ostrowski D, Ehrenreich H, et al. Erythropoietin-Mediated Protection of Insect Brain Neurons Involves JAK and STAT But Not PI3K Transduction Pathways. Neuroscience (2014) 258:218–27. doi: 10.1016/j.neuroscience.2013.11.020
9. Hahn N, Knorr DY, Liebig J, Wustefeld L, Peters K, Buscher M, et al. The Insect Ortholog of the Human Orphan Cytokine Receptor CRLF3 Is a Neuroprotective Erythropoietin Receptor. Front Mol Neurosci (2017) 10:223. doi: 10.3389/fnmol.2017.00223
10. Hashimoto Y, Muramatsu K, Kunii M, Yoshimura S, Yamada M, Sato T, et al. Uncovering Genes Required for Neuronal Morphology by Morphology-Based Gene Trap Screening With a Revertible Retrovirus Vector. FASEB J (2012) 26:4662–74. doi: 10.1096/fj.12-207530
11. Galloway JL, Zon LI. Ontogeny of Hematopoiesis: Examining the Emergence of Hematopoietic Cells in the Vertebrate Embryo. Curr Top Dev Biol (2003) 53:139–58. doi: 10.1016/S0070-2153(03)53004-6
12. Chen AT, Zon LI. Zebrafish Blood Stem Cells. J Cell Biochem (2009) 108:35–42. doi: 10.1002/jcb.22251
13. Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and Functional Characterization of Granulocytes and Macrophages in Embryonic and Adult Zebrafish. Blood (2001) 98:3087–96. doi: 10.1182/blood.V98.10.3087
14. Davidson AJ, Zon LI. The 'Definitive' (and 'Primitive') Guide to Zebrafish Hematopoiesis. Oncogene (2004) 23:7233–46. doi: 10.1038/sj.onc.1207943
15. Rasighaemi P, Basheer F, Liongue C, Ward AC. Zebrafish as a Model for Leukemia and Other Hematopoietic Disorders. J Hematol Oncol (2015) 8:29. doi: 10.1186/s13045-015-0126-4
16. Bertrand JY, Kim AD, Teng S, Traver D. CD41+ Cmyb+ Precursors Colonize the Zebrafish Pronephros by a Novel Migration Route to Initiate Adult Hematopoiesis. Development (2008) 135:1853–62. doi: 10.1242/dev.015297
17. Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, et al. Origins and Unconventional Behavior of Neutrophils in Developing Zebrafish. Blood (2008) 111:132–41. doi: 10.1182/blood-2007-06-095398
18. Page DM, Wittamer V, Bertrand JY, Lewis KL, Pratt DN, Delgado N, et al. An Evolutionarily Conserved Program of B-Cell Development and Activation in Zebrafish. Blood (2013) 122:e1–e11. doi: 10.1182/blood-2012-12-471029
19. Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, et al. In Vivo Tracking of T Cell Development, Ablation, and Engraftment in Transgenic Zebrafish. Proc Natl Acad Sci USA (2004) 101:7369–74. doi: 10.1073/pnas.0402248101
20. Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC, et al. Single-Cell Transcriptional Analysis of Normal, Aberrant, and Malignant Hematopoiesis in Zebrafish. J Exp Med (2016) 213:979–92. doi: 10.1084/jem.20152013
21. Paffett-Lugassy N, Hsia N, Fraenkel PG, Paw B, Leshinsky I, Barut B, et al. Functional Conservation of Erythropoietin Signaling in Zebrafish. Blood (2007) 110:2718–26. doi: 10.1182/blood-2006-04-016535
22. Basheer F, Rasighaemi P, Liongue C, Ward AC. Zebrafish Granulocyte Colony-Stimulating Factor Receptor Maintains Neutrophil Number and Function Throughout the Life Span. Infect Immun (2019) 87:e00793–18. doi: 10.1128/IAI.00793-18
23. Sertori R, Jones R, Basheer F, Rivera L, Dawson S, Loke S, et al. Generation and Characterization of a Zebrafish IL-2RγC SCID Model. Int J Mol Sci (2022) 23:2385. doi: 10.3390/ijms23042385
24. Lawrence C. The Husbandry of Zebrafish (Danio Rerio): A Review. Aquaculture (2007) 269:1–20. doi: 10.1016/j.aquaculture.2007.04.077
25. Thisse C, Thisse B. High-Resolution in Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat Protoc (2008) 3:59–69. doi: 10.1038/nprot.2007.514
26. Sertori R, Liongue C, Basheer F, Lewis KL, Rasighaemi P, de Coninck D, et al. Conserved IL-2RγC Signaling Mediates Lymphopoiesis in Zebrafish. J Immunol (2016) 196:135–43. doi: 10.4049/jimmunol.1403060
27. Lewis RS, Stephenson SEM, Ward AC. Constitutive Activation of Zebrafish Stat5 Expands Hematopoietic Cell Populations In Vivo. Exp Hematol (2006) 34:179–87. doi: 10.1016/j.exphem.2005.11.003
28. Sertori R, Trengove M, Basheer F, Ward AC, Liongue C. Genome Editing in Zebrafish: A Practical Overview. Brief Funct Genomics (2016) 15:322–30. doi: 10.1093/bfgp/elv051
29. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat Biotech (2013) 31:227–9. doi: 10.1038/nbt.2501
30. Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, et al. Simple Methods for Generating and Detecting Locus-Specific Mutations Induced With TALENs in the Zebrafish Genome. PLoS Genet (2012) 8:e1002861. doi: 10.1371/journal.pgen.1002861
31. Garritano S, Gemignani F, Voegele C, Nguyen-Dumont T, Le Calvez-Kelm F, De Silva D, et al. Determining the Effectiveness of High Resolution Melting Analysis for SNP Genotyping and Mutation Scanning at the TP53 Locus. BMC Genet (2009) 10:5. doi: 10.1186/1471-2156-10-5
32. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol Syst Biol (2011) 7:539. doi: 10.1038/msb.2011.75
33. Perriere G, Gouy M. WWW-Query: An on-Line Retrieval System for Biological Sequence Banks. Biochimie (1996) 78:364–9. doi: 10.1016/0300-9084(96)84768-7
34. Xiong JW, Yu Q, Zhang J, Mably JD. An Acyltransferase Controls the Generation of Hematopoietic and Endothelial Lineages in Zebrafish. Circ Res (2008) 102:1057–64. doi: 10.1161/CIRCRESAHA.107.163907
35. Changelian PS, Flanagan ME, Ball DJ, Kent CR, Magnuson KS, Martin WH, et al. Prevention of Organ Allograft Rejection by a Specific Janus Kinase 3 Inhibitor. Science (2003) 302:875–8. doi: 10.1126/science.1087061
36. Dooley KA, Davidson AJ, Zon LI. Zebrafish Scl Functions Independently in Hematopoietic and Endothelial Development. Dev Biol (2005) 277:522–36. doi: 10.1016/j.ydbio.2004.09.004
37. Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, et al. Myelopoiesis in the Zebrafish, Danio Rerio. Blood (2001) 98:643–51. doi: 10.1182/blood.V98.3.643
38. Ward AC, McPhee DO, Condron MM, Varma S, Cody SH, Onnebo SMN, et al. The Zebrafish Spi1 Promoter Drives Myeloid-Specific Expression in Stable Transgenic Fish. Blood (2003) 102:3238–40. doi: 10.1182/blood-2003-03-0966
39. Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive Hematopoiesis Initiates Through a Committed Erythromyeloid Progenitor in the Zebrafish Embryo. Development (2007) 134:4147–56. doi: 10.1242/dev.012385
40. Willett CE, Kawasaki H, Amemiya CT, Lin S, Steiner LA. Ikaros Expression as a Marker for Lymphoid Progenitors During Zebrafish Development. Dev Dyn (2001) 222:694–8. doi: 10.1002/dvdy.1223
41. Berman JN, Kanki JP, Look AT. Zebrafish as a Model for Myelopoiesis During Embryogenesis. Exp Hematol (2005) 33:997–1006. doi: 10.1016/j.exphem.2005.06.010
42. Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, et al. Characterization of Embryonic Globin Genes of the Zebrafish. Dev Biol (2003) 255:48–61. doi: 10.1016/S0012-1606(02)00041-6
43. Bertrand JY, Traver D. Hematopoietic Cell Development in the Zebrafish Embryo. Curr Opin Hematol (2009) 16:243–8. doi: 10.1097/MOH.0b013e32832c05e4
44. Willett CE, Zapata A, Hopkins N, Steiner LA. Expression of Zebrafish Rag Genes During Early Development Identifies the Thymus. Dev Biol (1997) 182:331–41. doi: 10.1006/dbio.1996.8446
45. Danilova N, Hohman VS, Sacher F, Ota T, Willett CE, Steiner LA. T Cells and the Thymus in Developing Zebrafish. Dev Comp Immunol (2004) 28:755–67. doi: 10.1016/j.dci.2003.12.003
46. Schnapp E, Pistocchi AS, Karampetsou E, Foglia E, Lamia CL, Cotelli F, et al. Induced Early Expression of Mrf4 But Not Myog Rescues Myogenesis in the Myod/Myf5 Double-Morphant Zebrafish Embryo. J Cell Sci (2009) 122:481–8. doi: 10.1242/jcs.038356
47. Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, et al. Identification of Pax6-Dependent Gene Regulatory Networks in the Mouse Lens. PLoS One (2009) 4:e4159. doi: 10.1371/journal.pone.0004159
48. Wendik B, Maier E, Meyer D. Zebrafish Mnx Genes in Endocrine and Exocrine Pancreas Formation. Dev Biol (2004) 268:372–83. doi: 10.1016/j.ydbio.2003.12.026
49. Karimi K, Fortriede JD, Lotay VS, Burns KA, Wang DZ, Fisher ME, et al. Xenbase: A Genomic, Epigenomic and Transcriptomic Model Organism Database. Nucleic Acids Res (2017) 46:D861–8. doi: 10.1093/nar/gkx936
50. Bennett C, Lawrence M, Guerrero JA, Stritt S, Waller AK, Yan Y, et al. CRLF3 Plays a Key Role in the Final Stage of Platelet Genesis and is a Potential Therapeutic Target for Thrombocythaemia. Blood (2022) 139:2227–39. doi: 10.1182/blood.2021013113
51. Castellucci LC, Almeida L, Cherlin S, Fakiola M, Francis RW, Carvalho EM, et al. A Genome-Wide Association Study Identifies SERPINB10, CRLF3, STX7, LAMP3, IFNG-AS1, and KRT80 as Risk Loci Contributing to Cutaneous Leishmaniasis in Brazil. Clin Infect Dis (2021) 72:e515–25. doi: 10.1093/cid/ciaa1230
52. Polewko-Klim A, Lesinski W, Golinska AK, Mnich K, Siwek M, Rudnicki WR. Sensitivity Analysis Based on the Random Forest Machine Learning Algorithm Identifies Candidate Genes for Regulation of Innate and Adaptive Immune Response of Chicken. Poult Sci (2020) 99:6341–54. doi: 10.1016/j.psj.2020.08.059
53. Padella A, Simonetti G, Paciello G, Giotopoulos G, Baldazzi C, Righi S, et al. Novel and Rare Fusion Transcripts Involving Transcription Factors and Tumor Suppressor Genes in Acute Myeloid Leukemia. Cancers (2019) 11:1951. doi: 10.3390/cancers11121951
54. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-Throughput Discovery of Novel Developmental Phenotypes. Nature (2016) 537:508–14. doi: 10.1038/nature19356
55. Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Mouse Genome Database G. Mouse Genome Database (MGD) 2019. Nucleic Acids Res (2019) 47:D801–6. doi: 10.1093/nar/gky1056
56. Wegscheid ML, Anastasaki C, Hartigan KA, Cobb OM, Papke JB, Traber JN, et al. Patient-Derived iPSC-Cerebral Organoid Modeling of the 17q11.2 Microdeletion Syndrome Establishes CRLF3 as a Critical Regulator of Neurogenesis. Cell Rep (2021) 36:109315. doi: 10.1016/j.celrep.2021.109315
57. Knorr DY, Hartung D, Schneider K, Hintz L, Pies HS, Heinrich R. Locust Hemolymph Conveys Erythropoietin-Like Cytoprotection via Activation of the Cytokine Receptor CRLF3. Front Physiol (2021) 12:648245. doi: 10.3389/fphys.2021.648245
58. Yang F, Xu YP, Li J, Duan SS, Fu YJ, Zhang Y, et al. Cloning and Characterization of a Novel Intracellular Protein P48.2 That Negatively Regulates Cell Cycle Progression. Int J Biochem Cell Biol (2009) 41:2240–50. doi: 10.1016/j.biocel.2009.04.022
Keywords: CRLF3, hematopoiesis, cytokine receptor, zebrafish, embryogenesis
Citation: Taznin T, Perera K, Gibert Y, Ward AC and Liongue C (2022) Cytokine Receptor-Like Factor 3 (CRLF3) Contributes to Early Zebrafish Hematopoiesis. Front. Immunol. 13:910428. doi: 10.3389/fimmu.2022.910428
Received: 01 April 2022; Accepted: 24 May 2022;
Published: 20 June 2022.
Edited by:
Gregory Maniero, Stonehill College, United StatesReviewed by:
Valerie Kouskoff, The University of Manchester, United KingdomSherri L. Christian, Memorial University of Newfoundland, Canada
Christopher Mahony, University of Birmingham, United Kingdom
Copyright © 2022 Taznin, Perera, Gibert, Ward and Liongue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clifford Liongue, Yy5saW9uZ3VlQGRlYWtpbi5lZHUuYXU=
†Present address: Yann Gibert, Department of Cell and Molecular Biology, University of Mississippi Medical Centre, Jackson, MS, United States
‡These authors have contributed equally to this work
 Tarannum Taznin1
Tarannum Taznin1 Kaushalya Perera
Kaushalya Perera Yann Gibert
Yann Gibert Alister C. Ward
Alister C. Ward Clifford Liongue
Clifford Liongue