- 1Department of Plant Protection, College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
- 2Department of Biotechnology, College of Bioscience and Biotechnology, Yangzhou University, Yangzhou, China
- 3Department of Entomology and Plant Pathology, Oklahoma State University, Stillwater, OK, United States
- 4Department of Entomology, College of Plant Protection, Northwest A&F University, Yangling, China
Nitric oxide (NO) at a high concentration is an effector to kill pathogens during insect immune responses, it also functions as a second messenger at a low concentration to regulate antimicrobial peptide (AMP) production in insects. Drosophila calcineurin subunit CanA1 is a ubiquitous serine/threonine protein phosphatase involved in NO-induced AMP production. However, it is unclear how NO regulates AMP expression. In this study, we used a lepidopteran pest Ostrinia furnacalis and Drosophila S2 cells to investigate how NO signaling affects the AMP production. Bacterial infections upregulated the transcription of nitric oxide synthase 1/2 (NOS1/2), CanA and AMP genes and increased NO concentration in larval hemolymph. Inhibition of NOS or CanA activity reduced the survival of bacteria-infected O. furnacalis. NO donor increased NO level in plasma and upregulated the production of CanA and certain AMPs. In S2 cells, killed Escherichia coli induced NOS transcription and boosted NO production, whereas knockdown of NOS blocked the NO level increase caused by E. coli. As in O. furnacalis larvae, supplementation of the NO donor increased NO level in the culture medium and AMP expression in S2 cells. Suppression of the key pathway genes showed that the IMD (but not Toll) pathway was involved in the upregulation of CecropinA1, Defensin, Diptericin, and Drosomycin by killed E. coli. Knockdown of NOS also reduced the expression of CanA1 and AMPs induced by E. coli, indicative of a role of NO in the AMP expression. Furthermore, CanA1 RNA interference and inhibition of its phosphatase activity significantly reduced NO-induced AMP expression, and knockdown of IMD suppressed NO-induced AMP expression. Together, these results suggest that NO-induced AMP production is mediated by CanA1 via the IMD pathway.
Introduction
Higher animals are armed with innate and adaptive immunity, but insects rely solely on less-specific innate immune responses to defend against invading pathogens in their habitats (1–3). In insects, bacterial and fungal pathogens trigger the host immune system via humoral and cellular components (4–6). Different immune challenges induce local and/or systemic responses (7, 8), suggesting that immune signaling pathways are extensively interlocked (9). Limited nutrients and short life spans for most insects require them to properly allocate energy between immune responses and other physiological processes such as development and reproduction (10–13). To maximize the reward of energy investment in immune responses, the interlocked immune signaling pathways must be regulated elaborately to avoid excessive immune responses. Reactive oxygen species (ROS) reaction and antimicrobial peptide (AMP) production are two primary humoral responses in the innate immune system of insects (4, 14). Insights into the cross-talk between them are important for understanding how different defense responses are coordinated to control infections.
ROS formation is a rapid, early response to pathogen invasion in insects. ROS can directly kill the invaders as effectors or function as signaling molecules to regulate the immune responses (15, 16). On the other hand, ROS can damage host cells as well (17). ROS include superoxide anion , H2O2, OH•, 1O2, and NO, each with a highly reactive oxygen atom (1, 18–20). They act as signaling compounds and/or toxic byproducts in cells (21). Among them, NO is a gaseous free radical functioning as a signal messenger for several physiological processes (22), including regulation of innate immunity (23–25). In mammals, NO is produced by nitric oxide synthase-2 (NOS2) in macrophages to control bacterial infection, which induces NOS2 transcription (26). In Drosophila melanogaster, NO is involved in the hemocyte encapsulation (27). In mosquitos, NO kills the Plasmodium parasites and increased NO to control the infection. Inhibition of NOS increases the rate of Plasmodium infection and that results in more deaths of infected mosquitos (28, 29). Blood meal taken by Anopheles stephensi catalyzes the conversion of NO to toxic metabolites, which kill the parasite in the gut (30).
AMP production is an effective immune response against microbial infection in insects (31, 32). They kill bacteria, fungi, and viruses sometimes (33–36). The Toll and IMD pathways actively participate in AMP production. Peptidoglycans (PGs) in the bacterial cell wall are recognized by the peptidoglycan recognition protein (PGRPs) to trigger the Toll and IMD pathways directly or indirectly. DAP-PGs from Gram- and some Gram+ bacteria are recognized by PGRP-LC/LE to induce the processing of IMD, FADD, Dredd, and Relish, and then the cleaved Relish enters the nucleus to activate AMP transcription (14, 36–40). Proteolytically activated Spätzle binds to the transmembrane receptor Toll to induce the intracellular signal transduction through MyD88, Tube/Pelle and Cactus. Finally, transcription factors such as Dorsal and Dif translocate into the nucleus to trigger AMP expression (14, 36, 38–40). Besides the classical Toll and IMD pathways, NO, eicosanoids, and calcineurin are also involved in the induced synthesis of AMPs in several model insects (25, 41).
Cross-talks among immune pathways keep the insect defense system running effectively and economically (15, 16, 42). Innate immunity is conserved at different levels in mammals, insects, and plants (43–45). In Drosophila larvae, NO activates the IMD pathway to produce Diptericin after infection by Gram-negative bacteria (46). The ROS stress upregulates NO production to enhance Diptericin synthesis in the adult gut (15). Furthermore, calcineurin subunit CanA1 is required for the NO regulation of AMP production in the fly (47). In Spodoptera exigua, injection of NOS inhibitor or knockdown of NOS reduced the AMP expression. In the absence of bacteria, an NO analog induced AMP expression (25). A cytokine named paralytic peptide induced NOS expression in the silkworm and triggered the AMP transcription in fat body (48). While AMP induction by NO is independent of the IMD or Toll pathway in Drosophila (41), this is dissimilar to the case in S. exigua (25), indicating that mechanism for NO regulation of AMP expression is unclear in insects. Comparative studies in different species are therefore needed to understand reasons for the discrepancy. Towards this goal, we used the Asia corn borer Ostrinia furnacalis as a model to investigate how NO signaling may communicate with the signaling pathways for AMP induction. NO strongly upregulated the AMP expression in O. furnacalis larvae, inhibition of NOS or CanA caused higher susceptibility of O. furnacalis to bacterial infection. In Drosophila S2 cells, IMD pathway connects NO signal to AMP production through CanA1.
Materials and Methods
Cell Culture and Insect Rearing
Drosophila S2 cells (Thermo Fisher, R69007) were maintained in Schneider’s Drosophila medium (Merck, S9895) containing 10% fetal bovine serum (FBS, Thermo Fisher, A3160802) (49). S2 cells were cultured in a 27°C incubator. All the S2 cells were plated in 12-well plates at 1×106 cells/well for different treatments (1 mL medium per well). Asian corn borers, O. furnacalis larvae were reared using an artificial diet at 25 ± 1°C, RH > 80%, and with a photoperiod of 16 h light and 8 h darkness (13, 50).
Bacterial Culture and Preparation of Dead Bacteria
Wild-type bacteria Escherichia coli, Pseudomonas aeruginosa and Micrococcus luteus (All the bacteria strains were kindly donated by Professor Zhiqiang Lu, Department of Entomology, College of Plant Protection, Northwest A & F University, China) picked from LB plates were grown overnight in Luria-Bertani (LB) medium at 37°C and 200 rpm. The 100 μL cultured bacteria were then inoculated into 10 mL fresh LB medium and cultured at 37°C until OD600 was close to 0.6. Finally, the bacteria were harvested by centrifugation at 8000×g for 10 min. After washing for 3 times, bacteria pellets were resuspended with phosphate buffered saline (PBS) for injection. To prepare the dead bacteria, E. coli and M. luteus cells from 100 mL LB medium were resuspended in 1 mL PBS and 40 mL 75% 2-propanol. After incubation for 1 h at 37°C and 200 rpm, dead bacteria were spun down and washed 3 times with PBS. Finally, the dead bacteria were resuspended in 1 mL PBS to treat S2 cells.
Survival Rate Assay of O. furnacalis Larvae after Infection
To determine the number of bacteria for injection, day 1, 4th instar O. furnacalis larvae were fed on artificial diet containing 5 μL (10 μg/μL) tetracycline that eliminates indigenous bacteria. The diet was replaced with fresh diet without antibiotic at 24 h post antibiotic treatment. Day 3, 4th instar larvae were injected with 1×103, 1×104, 1×105, or 1×106 live cells of P. aeruginosa or M. luteus. PBS was used as control. There were 20 larvae in each group. The survival in each group was recorded at 12 h intervals. All the data was analyzed by the log-rank test using Prism 5.0.
Treatment of S2 Cells and Infection of O. furnacalis Larvae
S2 cells cultured in 1 mL medium were incubated with 20 µL of killed bacteria at different amounts. At 24 h post bacterial exposure, the medium was collected for nitric oxide determination, and 500 µL Trizol (Invitrogen) was used to extract RNA from the S2 cells for qPCR analysis. Day 1, 4th instar larvae were fed on the diet containing 50 μg/μL tetracycline to eliminate indigenous bacteria before injection with bacteria as described previously (51). At 24 h after antibiotic feeding, larvae were transferred to fresh diet without antibiotic. Day 3, 4th instar larvae were injected with 1×104 of live P. aeruginosa and M. luteus or along with NOS inhibitor/CanA inhibitor (2 nmol each) for determination of survival curve or qPCR analysis. All the results generated in survival assay were recorded at 12 h intervals and the whole O. furnacalis larvae at certain times post infection were treated with Trizol regent for RNA extraction (Invitrogen). PBS was used as control. All the treatments were performed in triplicate.
RNA Interference
The dsRNA products were prepared as previously described (52). cDNA of Drosophila IMD, MyD88, NOS, and CanA1 and plasmid GFP- pEASY-T1 (TransGen) were used as templates for PCR amplification using gene-specific primers (Table S1). The conditioned medium (1 mL) from S2 cells cultured in 12 well plate was replaced with 0.5 mL of Schneider’s Drosophila medium containing 6 µg of dsRNA samples of IMD, MyD88, NOS, CanA1, or a mixture of dsIMD and dsMyD88 (6 µg each). After 1 h incubation, 0.5 mL of Schneider’s Drosophila medium containing 10% FBS was added to each well. Equal amount of GFP dsRNA was added as a control. RNAi efficiency was examined three days after dsRNA treatment using qPCR as described below. For the RNAi treatment combined with bacterial incubation, the killed bacteria were added to each well at 72 h post dsRNA treatment, and total RNA samples were prepared 24 h later.
Treatment of S2 Cells With Compounds
Stock solution (250 mM) of diethylamine NONOate (Sigma D184, an NO releasing compound or NOC) was dissolved in water prior to use. To treat S2 cells, the NOC at 2.5 mM final concentration was used to increase NO level in the medium. S2 cell and medium samples were collected at 0, 6, 12, 24, and 48 h after NOC addition. Calcineurin A inhibitor FK506 (Sigma, F4679) was dissolved in DMSO to make a 100 mM stock. FK506 at 0, 10, 20, 30 and 50 mM along with 2.5 mM NOC was used to treat S2 cells and test influence of FK506 on the regulation of AMP production by NO. S2 cell and medium samples were collected at 24 h post NOC-FK506 treatment, PBS was used as control. Stock solution (400 mM) of Nω-nitro-L-arginine methyl ester (Sigma N5751, L-NAME, a NOS inhibitor) was dissolved in water and used at 200 μM along with dead bacteria in the medium as indicated to test the effect of L-NAME on AMP expression. S2 cells and medium samples were collected at 24 h post treatment. PBS and killed bacteria were used as negative and positive controls, respectively.
qPCR Analysis
S2 cells (1×106) or 3 whole larvae were collected from each biological treatment and replicate. Total RNA was extracted using 1 mL Trizol, RNA concentrations were determined on an Eppendorf BioPhotometer D30, and RNA integrity was examined by 1% agarose gel electrophoresis. cDNA templates were generated from 1 μg total RNA using HiScript III RT SuperMix for qPCR in the presence of genomic DNA wiper (Vazyme, Nanjing, China). Diluted cDNA (1:10, 1 μL) was used for qPCR analysis on a Bio-Rad CFX96 Real Time Detection System (Bio-Rad, CA, United States) in 20 μL reaction containing 1 μL of cDNA, 10 μL of AceQ Universal SYBR qPCR Master Mix (Vazyme), 1.0 μL each of forward and reverse primers (10 μM) and 7 μL ddH2O. The thermal cycling conditions were initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10 s and annealing-extension at 60°C for 30 s, with melting curve measured from 60 to 95°C. All the treatments were in triplicate. O. furnacalis reference gene ribosomal protein L8 (RPL8) gene (53) and D. melanogaster reference gene ribosomal protein 49 (RP49) (54)were used to calibrate the relative expressions of target genes. The mRNA level changes of interested genes were determined using the relative quantitative method (2−ΔΔCt) (55). qPCR data were plotted using GraphPad (Version 9.0.2) for statistical analysis. Student’s t-test results are shown as ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. Results of one-way ANOVA followed by Tukey’s test are marked similarly.
Determination of Nitric Oxide Concentration
To determinate NO concentrations in the media of S2 cells and hemolymph of O. furnacalis larvae, the samples were collected by centrifugation at 16,000×g for 30 s to remove cells, and the supernatants were used for measuring NO concentrations. The supernatants of medium samples and 1:100 diluted larval plasma (30 μL) were taken to measure NO levels using Total Nitric Oxide Assay Kit (Beyotime, Beijing, China) according to the manufacturer’s instructions (56).
Results
Inhibition of NOS and CanA Increased the Mortality of O. furnacalis After Bacterial Infection
To assess the immune stimulatory effect of P. aeruginosa and M. luteus on O. furnacalis, we injected larvae with different numbers of live bacteria and found that the larvae reached 50% mortality after injected with about 1×104 CFUs of P. aeruginosa or M. luteus (Figure S1). Thus, 1×104 CFUs of these two bacteria were used to challenge O. furnacalis larvae in later experiments. Injection of the NOS or CanA inhibitor caused higher mortality of larvae upon bacterial challenge (Figure 1), suggesting an involvement of NOS and CanA in the immune responses to bacterial infection in O. furnacalis.
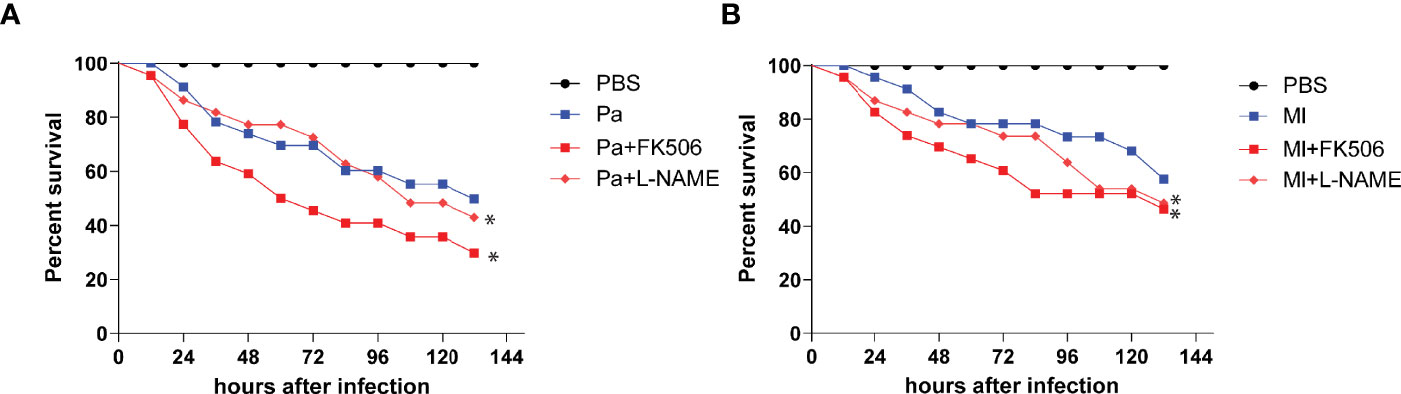
Figure 1 Inhibition of CanA and NOS caused more deaths of infected O. furnacalis larvae. After elimination of indigenous bacteria using 50 μg/μL tetracycline, day 3, 4th instar larvae were injected with 1× 104 cells of P. aeruginosa (A) or M. luteus (B) cells or along with 2 nmol FK506 or L-NAME, using PBS as control. All the data was analyzed using the log-rank test. *, p <0.05; FK506, CanA inhibitor; L-NAME, NOS inhibitor; Pa, P. aeruginosa; Ml, M. luteus.
Induction of NOS, CanA and AMPs by Bacterial Infection in O. furnacalis Larvae
NO and AMPs are effectors that eliminate invading bacteria in insects, and some research indicated cross-talks between ROS and AMP production (47). To investigate whether or not NOS and CanA are involved in the processes in O. furnacalis larvae, we first measured the transcript levels of NOS, CanA and AMPs under immune stress (Figure 2). NOS1 but not NOS2 mRNA level was strongly induced (Figures 2A, B), and the expression levels of NOS1 and NOS2 in different tissues showed that NOS1 was mainly expressed in hemocytes, while NOS2 were mainly expressed in fat body (Figure S5). NOS1 and CanA showed a similar expression pattern, which were mainly upregulated at 4 and 12 h post infection (Figures 2A, C). In addition, we found that CanA was also mainly expressed in hemocytes (Figure S5). The AMP effector genes were upregulated following the increase of NOS1 and CanA expression (Figures 2D−H). These data provided clues for us to explore the mechanism for NO-regulated AMP expression during immune responses.
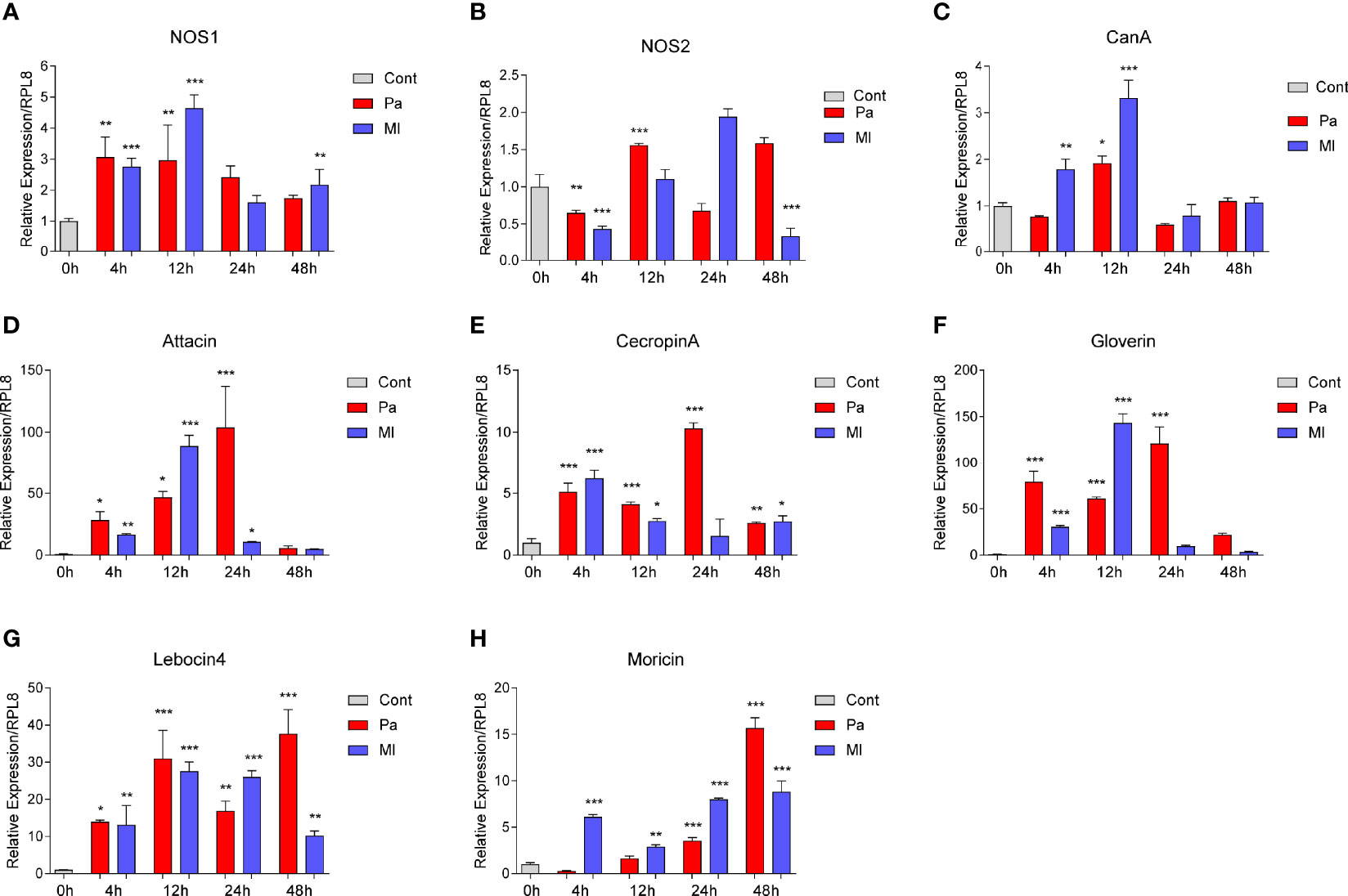
Figure 2 Expression changes of NOS, CanA and AMPs in O. furnacalis larvae after bacterial infection. mRNA level changes in NOS (A, B), CanA (C), Attacin (D), Cecropin A (E), Gloverin (F), Lebocin4 (G), and Moricin (H) in whole O. furnacalis larvae at certain times post 1×104 cells of P. aeruginosa or M. luteus infection. Cont, PBS as control; CanA, calcineurin A; NOS, nitric oxide synthase; Pa, P. aeruginosa; Ml, M. luteus. One-way ANOVA analysis followed by Tukey’s test was used to compare the control and infected groups. *p < 0.05; **p < 0.01; ***p < 0.001.
NO Increased mRNA Levels of CanA and Some AMPs in O. furnacalis Larvae
NOS catalyzes the production of NO from an endogenous substrate L-arginine. After infection with P. aeruginosa or M. luteus, NO concentrations in hemolymph increased significantly at 4, 12, 24 and 48 h (Figures 3A, B). Injection the diethylamine NONOate (NOC, an NO donor) also increased NO concentration to a similar level in hemolymph (Figure 3C). NO also induced the expression of CanA, and Defensin, Lebocin4 and Moricin (Figures 3D−G). However, NOC did not induce Attacin, CecropinA or Gloverin expression (Figure S2), suggesting NO has some specificity in inducing AMP production. Thus, we hypothesized that CanA may participate in bacteria-induced NO production to regulate the expression of certain AMPs.
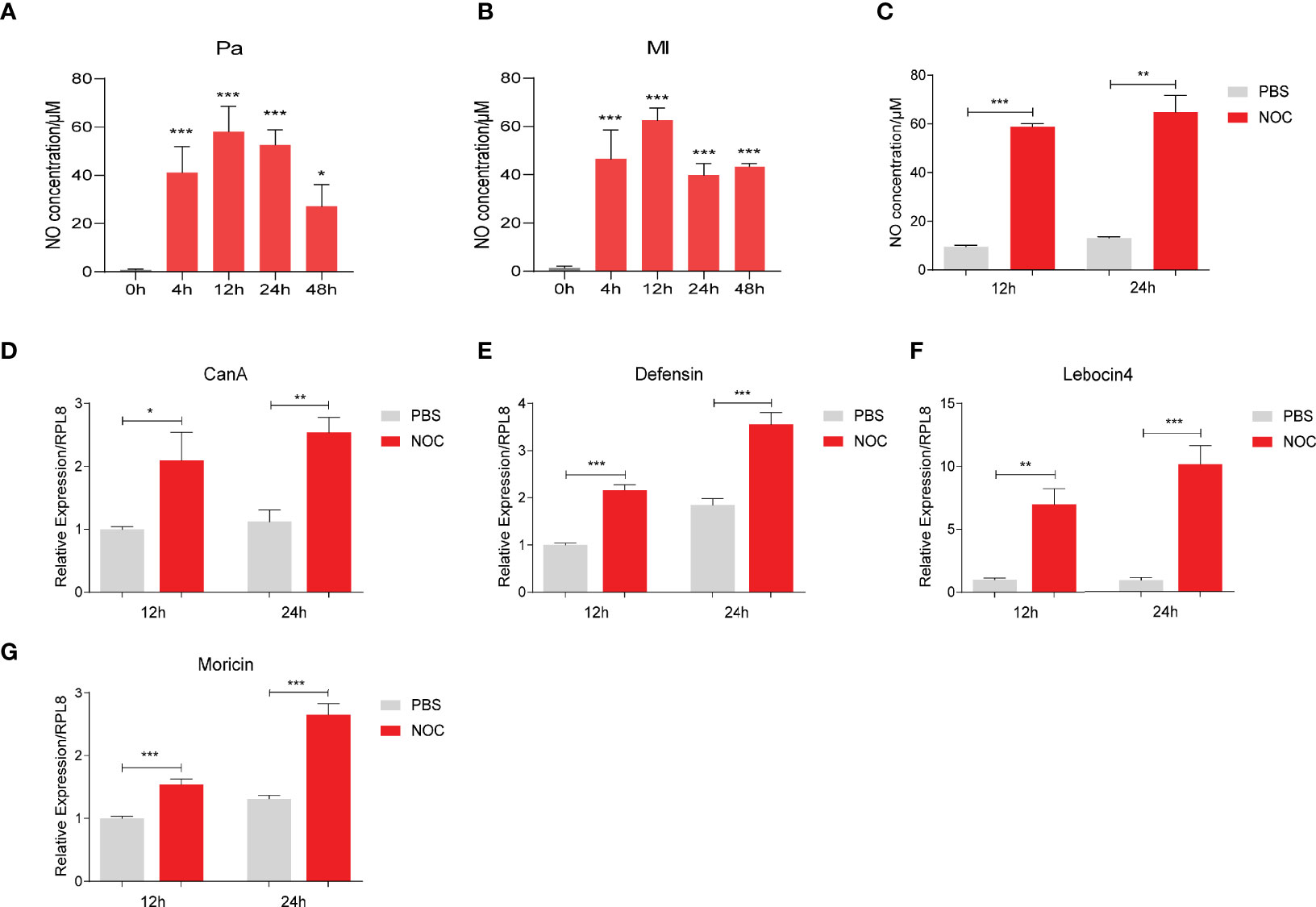
Figure 3 NO increased CanA and AMP expression in O. furnacalis larvae. NO concentrations in hemolymph after infection by 1×104 CFUs of P. aeruginosa (A) and M. luteus (B) at 4 to 48 h post injection. NO concentrations in larval hemolymph after injection of the NOC (C). Transcript levels of CanA (D), Defensin (E), Lebocin 4 (F) and Moricin (G) after NOC injection. One-way ANOVA followed by Tukey’s test was used to compare the control and infected groups (A, B). Student’s t-test was used to compare PBS- and NOC-treated groups (C−G). *p < 0.05; **p < 0.01; ***p < 0.001.
E. coli and NO Releasing Compound (NOC) Increased NO Concentration in the Medium of S2 Cells
To understand how NO may regulate AMP production, we used Drosophila S2 cells in further tests. Incubation with dead M. luteus and E. coli induced S2 cells to make AMPs (Figure S3). E. coli from 1 mL culture at OD600 = 1.0 led to a stronger AMP response than the Gram-positive bacteria. Thus, this amount of dead E. coli was chosen to treat S2 cells in the later experiments. We found that the NOS expression and NO production were strongly induced by E. coli, as in O. furnacalis larvae (Figures 4A, B, 2A). Knockdown of NOS reduced the NO level increased by E. coli (Figure 4C). After incubation with the NOC, NO concentration in the cell culture medium increased and lasted for two days at least (Figure 4D). Therefore, Drosophila S2 cells appear to be a good model for investigating the link between NO and AMP production.
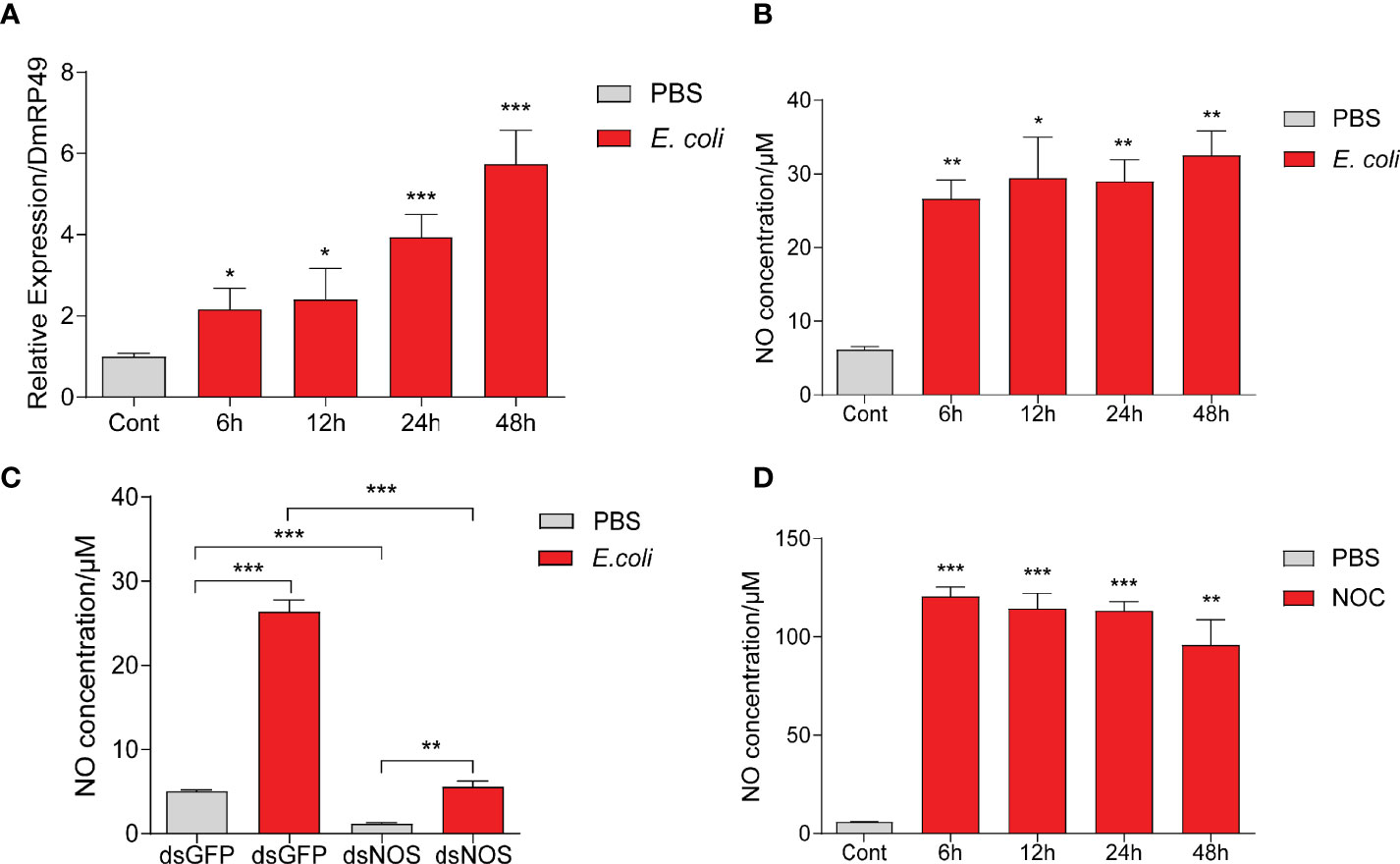
Figure 4 E coli and NOC treatments stimulated NO production in Drosophila S2 cells. NOS mRNA levels (A) and NO concentrations (B) after E coli infection. NO levels in the medium samples after NOS RNAi and treatment with killed E coli (C) or the NOC (D). Cont, PBS treatment at 0 h One-way ANOVA followed by Tukey’s test was used to compare control and treatment groups (A, B, D). Student’s t-test was used to analyze significance in (C). *p < 0.05; **p < 0.01; ***p < 0.001.
Induction of AMPs by NO in S2 Cells
To further uncover the role of NO in AMP production, we added NOC (NO donor) to the culture of S2 cells. At 24 and 48 h, the transcript levels of CecropinA1, Defensin, Diptericin and Drosomycin increased significantly (Figure 5). The effect was observed for CecropinA1 at 6 and 12 h, suggestive of a more sensitive response to NO for this gene. NO concentration elevations caused by E. coli or the NOC were detected at 6, 12, 24, and 48 h (Figures 4B, D). The major induction of AMPs at 24 and 48 h suggested that the NO involvement in AMP production may be indirect, relying on protein products of intermediate gene(s).
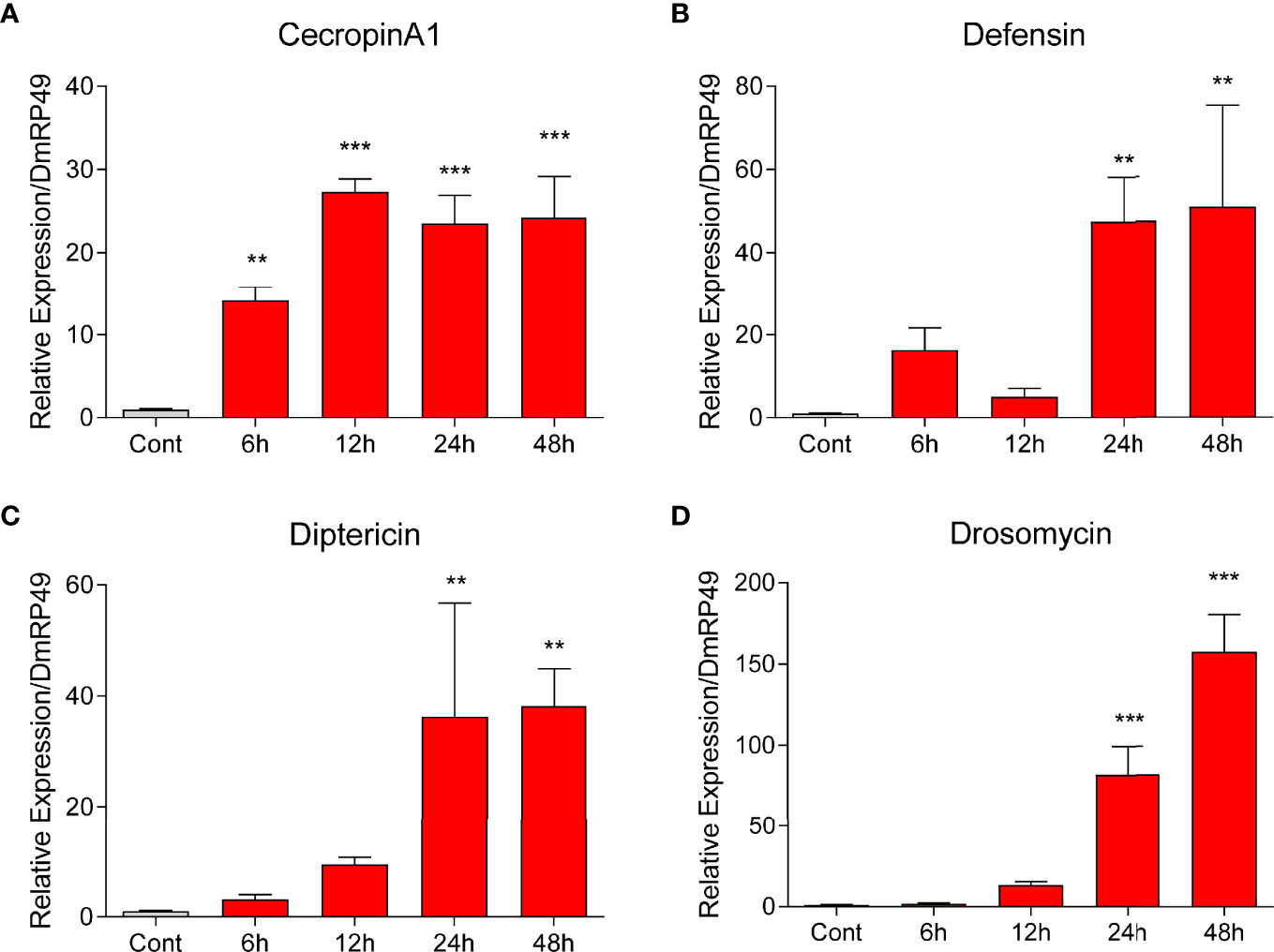
Figure 5 NOC induced AMP expression in Drosophila S2 cells. The NOC at a final concentration of 2.5 mM was used to treat S2 cells. Transcript levels of the four AMPs Cecropin A1 (A), Defensin (B), Diptericin (C), and Drosomycin (D) at different time points were measured by qPCR. One-way ANOVA followed by Tukey’s test was used to compare control and treated groups. **p < 0.01; ***p < 0.001.
IMD Pathway Connected the NO Signal to AMP Production in S2 Cells
Since AMP expression is known to be controlled by the Toll and IMD pathways (14, 37, 38, 57), how may NO-induced AMP production in S2 cells (Figure 5) and O. furnacalis larvae (Figure 3) be linked to the two classic pathways? To address this question, we employed RNA interference to knockdown the pathway components and determine whether the NO-induced AMP production is affected in S2 cells. We found the increases in mRNA levels of CecropinA1, Defensin, Diptericin and Drosomycin caused by dead E. coli were dramatically suppressed after IMD had been knocked down (Figures 6, S4). Treatment with dsRNA of MyD88 had a lesser effect. Therefore, NO-induced AMP production was regulated mainly by the IMD pathway but not much by Toll signaling. Similarly, the AMP transcription increases were partly suppressed by NOS dsRNA, suggesting the NOS may take part in the AMP induction upon E. coli treatment. Furthermore, knockdown of IMD in S2 cells significantly reduced CecropinA1, Defensin, Diptericin and Drosomycin expression, which induced by NOC treatment (Figures 6E–H). Together, these data suggested that NO is involved in the upregulation of CecropinA1, Defensin, Diptericin and Drosomycin transcription through the IMD pathway.
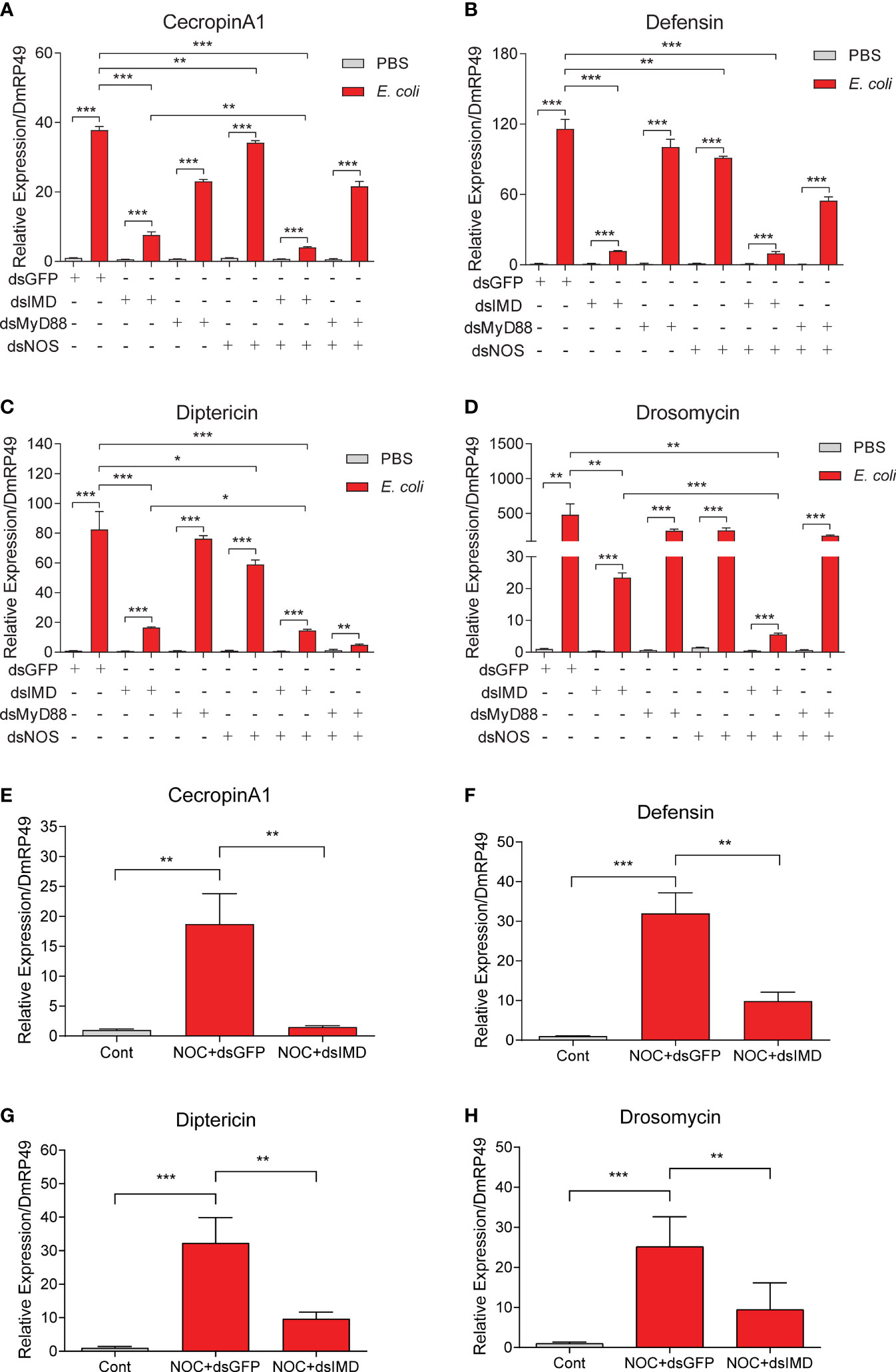
Figure 6 Effects of IMD, MyD88 or NOS knockdown on AMP expression in Drosophila S2 cells. To analyze possible roles of Toll/IMD pathway and NOS in NO-induced AMP expression, RNAi of IMD, MyD88 or NOS was performed in S2 cells for 48 h in advance of the treatment by killed E coli. Transcript levels of Cecropin A1 (A), Defensin (B), Diptericin (C) and Drosomycin (D) were analyzed by qPCR. The effects of IMD knockdown on induction of CecropinA1 (E), Defensin (F), Diptericin (G) and Drosomycin (H) by the NOC were detected. Student’s t-test was used to analyze significance, *p < 0.05; **p < 0.01; ***p < 0.001.
NOS Was Required for the Upregulation of CanA1 in S2 Cells Induced by E. coli
In O. furnacalis larvae, bacterial infections increased the transcript levels of CanA and AMPs, whereas inhibition of CanA reduced resistance to the infections and resulted in more death (Figures 1, 2C), suggesting the involvement of CanA in the resistance to bacterial infection. To investigate whether CanA is involved in the resistance through regulating the expression of AMPs, S2 cells were used for further studies. We found that dead E. coli induced the expression of CanA1 at 12, 24, and 48 h in S2 cells (Figure 7A), and that addition of NO donor NOC also induced the expression of CanA1 at 12 and 24 h (Figure 7B), which indicates that bacterial infection and NO increased expression of CanA1. However, knockdown of NOS suppressed the induction of CanA1 by E. coli (Figure 7C), suggesting that CanA1 might be downstream of NO signal.
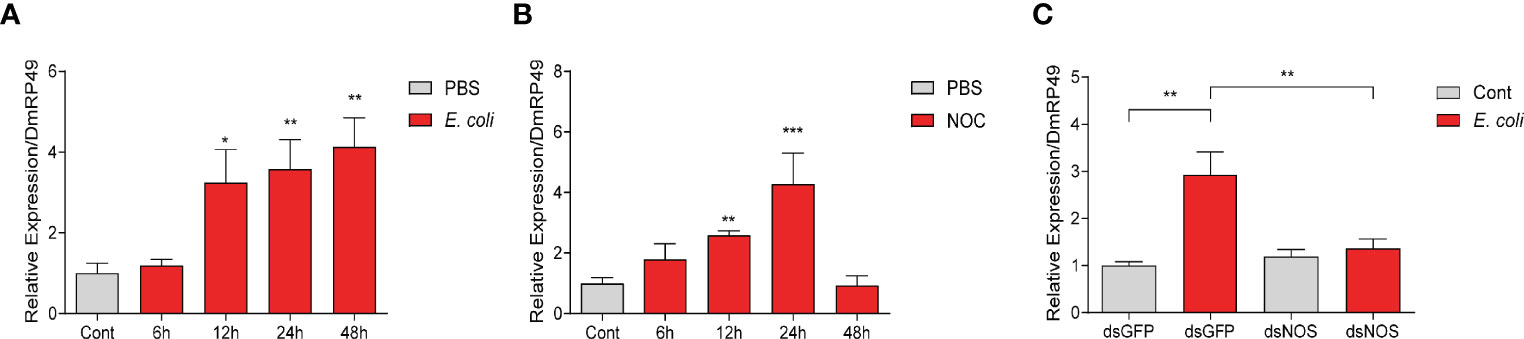
Figure 7 NO contributed to the upregulation of CanA1 induced by killed E coli in Drosophila S2 cells. The expression pattern of CanA1 in response to E coli treatment (A), NO donor (B), and NOS knockdown followed by E coli treatment (C) was determined by qPCR. Cont: PBS treatment; NOC: Nitric oxide donor. One-way ANOVA followed by Tukey’s test was used to compare control and treated groups (A, B). Student’s t-test was used to analyze significance (C), *p < 0.05; **p < 0.01; ***p < 0.001.
Knockdown and Inhibition of CanA1 Can Block the Upregulation of AMPs by NO in S2 Cells
To further confirm the relationship between CanA1 and NO on controlling AMPs expression in S2 cells, we used CanA1 inhibitor and knockdown to inhibit the activity of CanA1 and reduce the transcript level of CanA1, respectively. We found that CanA1 inhibitor could significantly block the expression of CecropinA1, Defensin, Diptericin and Drosomycin, which were induced by NO (Figures 8A–D). In addition, knockdown of CanA1 in S2 cells also decreased the expression of CanA1 induced by the NOC (Figure 8E), and significantly suppressed the upregulation of CecropinA1, Defensin, Diptericin and Drosomycin by NO (Figures 8F–I). These results directly indicated that the AMP expression induced by NO was mediated by CanA1.
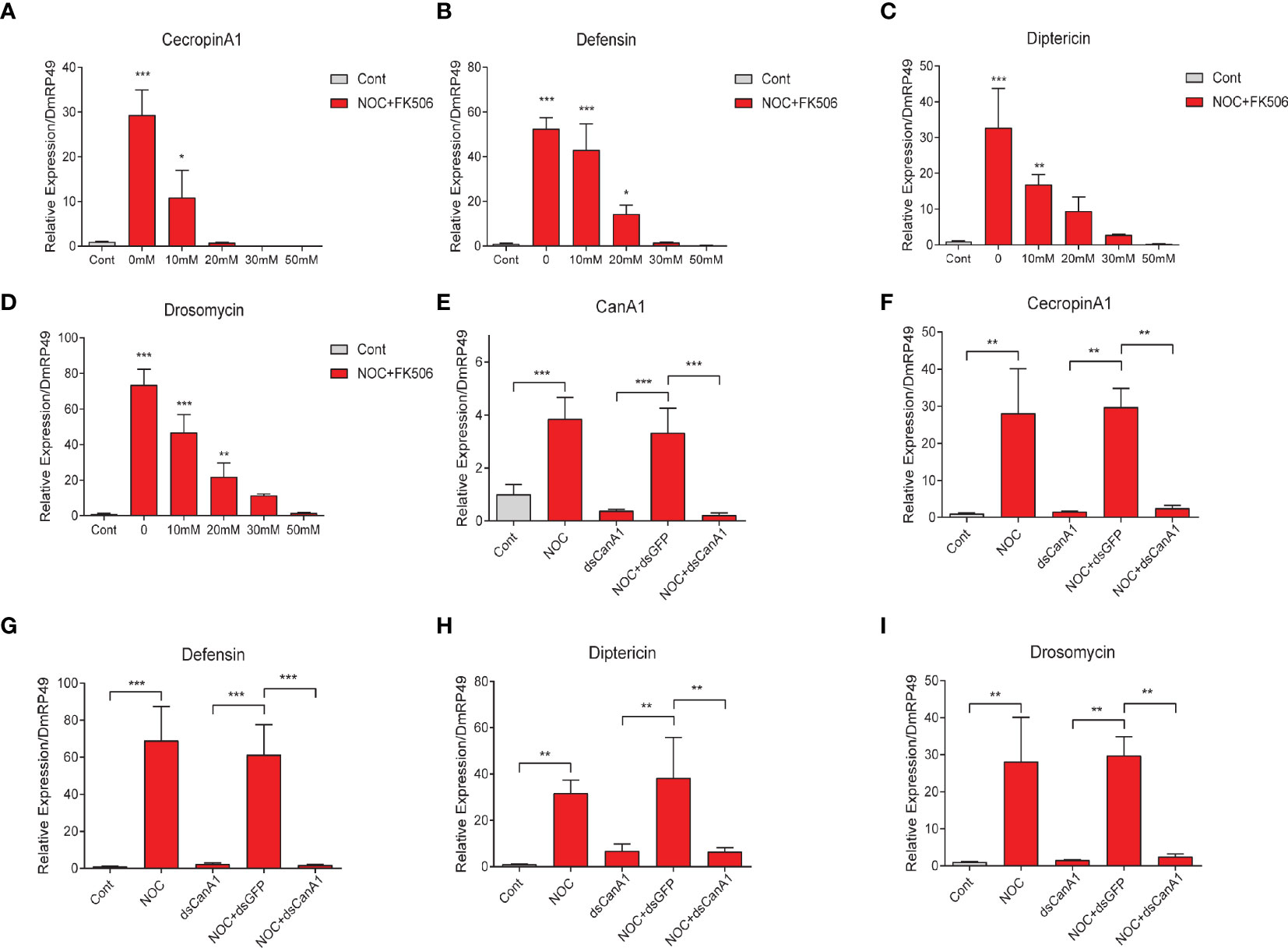
Figure 8 Effects of CanA1 knockdown and CanA1 inhibitor on NOC-induced AMP expression. Transcript level changes of Cecropin A (A), Defensin (B), Diptericin (C) and Drosomycin (D) in NOC treated S2 cells after different concentrations of CanA1 inhibitor treatment were determined by qPCR, The concentrations of FK506 were indicated. dsRNA of CanA1 was used to evaluate the effects of NOC on the expression of CanA1 (E), Cecropin A1 (F), Defensin (G), Diptericin (H) and Drosomycin (I) in response to NOC challenge. Cont, PBS used as control; NOC, nitric oxide donor; CanA1, CalcineurinA1. One-way ANOVA followed by Tukey’s test was used to compare control and treatment groups (A-D). Student’s t-test was used to analyze significance (E-I), *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
After insect innate immunity was discovered, the robust defense system has been well investigated in different species (8, 58). In particular, the immune signaling pathways such as ROS reaction, Toll and IMD pathways, and PPO cascade are well studied (14, 37, 38, 58–60). As a member of the ROS family, NO induced by pathogen infections participates in the regulation of AMP production, mainly regulated via classic Toll and IMD pathways. That means that there are cross-talks between the ROS reaction and AMP signaling pathway. In this study, we used a lepidopteran pest O. furnacalis to analyze the regulatory mechanism of NO on AMP production and found that bacterial infection can upregulate the expression of NOS, CanA and certain AMPs through NO production. NO and CanA are needed to fight against bacterial infection in O. furnacalis. Using S2 cells, we confirmed that CanA1 mediated the regulation of AMP productions between NO signal and IMD pathway. Our work suggests that NO signal might play as the messenger between rapid ROS reaction and AMP signaling pathway.
The mechanism of how CanA regulates the AMPs expression is still unclear. As a member of the protein phosphatase 2B family, calcineurin is a Ca2+-dependent phosphatase, involved in many physiological processes such as regulation of Ca2+ homeostasis, transcription, and innate immunity (61–63). Calcineurin comprises a catalytic subunit A and a regulatory subunit B (41). In Drosophila, there are three catalytic subunits including calcineurin A1, protein phosphatase 2B-14D and calcineurin A-14F (64). Protein phosphatase 2B-14D and calcineurin A-14F can respond to Gram-positive bacterial infection and activate Dorsal to induce AMPs production (41). While calcineurin A1 has effects on regulation of AMPs production via Relish in response to Gram-negative bacterial infections or NO challenge (41, 46). Subunit calcineurin A1 can directly receive the signal of NO and act on Relish without the components of IMD pathway, subunits protein phosphatase 2B-14D and calcineurin A-14F directly activate Dorsal/Dif activity dependent on the calcium level altered by Gram-positive bacterial infections (41), suggesting that the regulation of AMPs production by calcineurin subunit A is directly mediated by NF-κB and independent of Toll/IMD pathways. In contrast, the NO-induced AMPs production is dependent on Toll/IMD pathway in S. exigua (25). In this study, our results indicate that IMD pathway is required for calcineurin A to mediate the regulation of AMPs by NO. These differences between non-NO and NO mediated AMPs production via the CanA1 regulation remain to be fully deciphered in the future. In B. mori and D. melanogaster, eicosanoids are involved in AMPs production (65, 66). Inhibition of phospholipase A2 (PLA2) activity can reduce the biosynthesis of eicosanoids, and finally decreases AMPs production in S. exigua (67). In addition, NO increased the activity of PLA2, and PLA2 was capable to upregulate the AMP production via eicosanoids in S. exigua (25, 68). Therefore, whether or not calcineurin A can regulate the PLA2 to alter AMPs production via eicosanoids need further investigation.
There is organ-to-organ communication during immune responses. In the Drosophila gut, the ROS reaction and IMD pathway producing AMPs play primary roles in the elimination of gut microbes (8). Enterobacteria Ecc15 oral infection can locally trigger the expression of AMPs and ROS reaction in adult Drosophila gut (4), although Ecc15 can’t cross through the gut and enter the hemolymph, the local infection in the gut also upregulates AMPs expression in fat body (69, 70). Furthermore, ROS stress induced by local Ecc15 oral infection in Drosophila gut upregulates the production of NO in gut, and then the NO signal as a messenger is relayed by hemocytes to trigger the expression of AMP Diptericin in the remote organ fat body (15). In this study, we found that NOS1 in hemocytes was the primary NOS in response to bacterial infections in O. furnacalis (Figures 2, S5), while AMPs were mainly expressed in gut and fat body (8, 25), we inferred that there might be also a link between hemocyte producing NO and fat body expressing AMPs in O. furnacalis. Based on the tissue expression analysis of CanA, NOS1 and NOS2, and the responses of these three genes to bacterial infections in O. furnacalis, we inferred NO production induced by bacteria in hemocytes increased the expression of CanA in hemocytes together with some unknown factors, which were released from hemocytes to induce AMPs expression in fat body via IMD pathway (Figure 9). To further confirm the organ-to-organ immune signals in O. furnacalis, Ex vivo assay using hemocytes and fat body from O. furnacalis larvae might need to be set up in the future. Using condition medium collected from dead bacteria-challenged hemocytes to stimulate the germ-free fat body may be an ideal approach to investigate the organ-to-organ communication and identify the unknown factors from hemocytes to fat body.
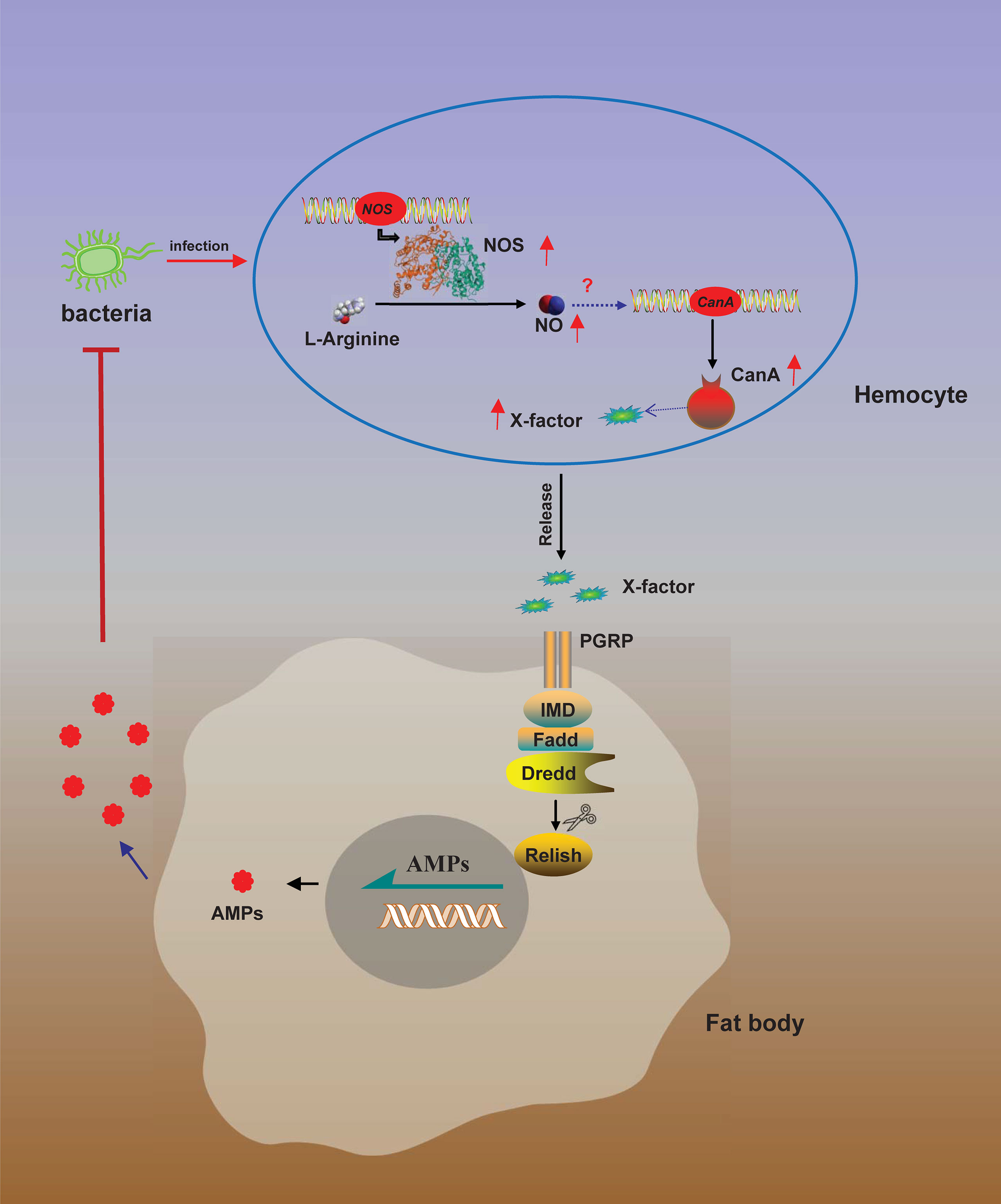
Figure 9 A model for NO-regulated AMP production. Bacterial infection induces NOS expression in hemocytes to convert L-arginine to NO by NOS. NO then induces CanA expression to produce and release of an unknown factor (X) from hemocytes. Released X activates AMP expression in fat body via IMD pathway to eliminate the invading bacteria.
NO plays a minor role in bacteria-induced AMP production. The classic pathways regulating AMP productions are Toll and IMD pathways in several model insects (71, 72). Toll pathway and IMD pathway are activated by Lys-peptidoglycans (from Gram-positive bacteria) and DAP-peptidoglycans (DAP: meso-diaminopimelic acid, mainly from Gram-negative bacteria), respectively (73). In this study, different amounts of killed Gram-positive and Gram-negative bacteria M. luteus and E. coli were used to treat S2 cells, even a small amount of E. coli showed stronger activity than M. luteus to induce AMPs expression in S2 cells. We found that knockdown of NOS did not totally block the induction of AMPs after bacterial infection, and NO only induced considerable AMPs, indicating that the AMPs production induced by NO takes part in the AMP production through IMD pathway in S2 cells.
So far, the mechanism of NO production after bacterial infection is unclear. In S. exigua, the expression of AMPs is under the regulation of Toll pathway and IMD pathway like that in other insects (14, 25, 74, 75). NO also can regulate the expression of AMPs in S. exigua, while knockdown of Toll or Relish decreases the expression of NOS, and reduces NO concentration in response to bacterial infection, suggesting that the NO signal is downstream of IMD and Toll pathway in S. exigua (25). In Drosophila, the NO-induced AMPs production is independent of Toll and IMD pathways (41). In this study, our result showed that NO upregulated the expression of CanA1, and then CanA1 activated the expression of AMPs via IMD pathway in S2 cells. Knockdown of both NOS and IMD could block expression of AMPs similar as knockdown of IMD only did, suggesting that the NO signaled IMD pathway to regulate the AMP expression.
Conclusion
In our study, we found that NO donor could induce the expression of Cecropin A, Defensin, Diptericin and Drosomycin in S2 cells (Figure 5), while in O. furnacalis larvae, NO donor significantly induced the expression of Defensin, Lebocin4 and Moricin but not Attacin, Cecropin A1 and Gloverin (Figures 3, S1). NO donor can upregulate the expression of Attacin1/2, Defensin and Gloverin in S. exigua (25). We also used B. mori to analyze the AMPs expression after bacterial infection or NO donor treatment (Figure S6A), and found that NO donor NOC had strong activity to induce the expression of AMPs Cecropin D, Cecropin E, Lebocin, Moricin and Defensin A. Moreover, inhibition of NOS using L-NAME increased the death of bacterial infected B. mori larvae (Figure S6B), which was consistent with the survival assay using O. furnacalis larvae. These data indicated that NO can specifically induce certain AMPs in different insects.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
CF, KC and ZL designed the study. XWa, JC and XWe performed the experiments. KC and JC performed statistical analysis, KC prepared the first draft. CF, HJ, ZL and YW revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (31901876, 31871952, and 31970467), Natural Science Foundation of Jiangsu Province (KB20190900), China Postdoctoral Science Foundation (2018M642343) and NIH grant GM58634.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.905419/full#supplementary-material
Supplementary Table 1 | Primers used in this study.
Supplementary Figure 1 | Survival of O. furnacalis larvae after bacterial infection. Survival curves of the larvae after an exposure to live P. aeruginosa (A) and M. luteus (B) at different dosages (1×103 to 1×106 CFUs/larva)
Supplementary Figure 2 | NOC did not induce Attacin, Cecropin A or Gloverin synthesis in O. furnacalis larvae.
Supplementary Figure 3 | Induced transcription of Cecropin A1 (A, B), Diptericin (C, D), Defensin (E, F), and Drosomycin (G, H) in Drosophila S2 cells by different amounts of killed M. luteus (A, C, E, G) and E. coli (B, D, F, H).
Supplementary Figure 4 | The efficiency of RNAi against the target genes in Drosophila S2 cells.
Supplementary Figure 5 | Expression of CanA, NOS1 and NOS2 genes in different tissues of O. furnacalis larvae. Different letters above a given treatment indicate means significantly differ. One-way ANOVA followed by Tukey’s test was used to compare each other.
Supplementary Figure 6 | Effects of NO donor and bacterial infections on AMP expression (A) in and survival curves (B) of B. mori larvae. Day 3, 5th instar B. mori larvae were injected with NO donor NOC or S-nitroso-N-acetylpenicillamine (SNAP) (20 nmol/larva), 1×107 cells of P. aeruginosa or S. aureus, or 1×107 cells of P. aeruginosa or S. aureus together with NOS inhibitor L-NAME (2 nmol/larva). Fat body was collected at 6 h post infection, and transcript level change of Cecropin A, Cecropin B, Cecropin D, Cecropin E, Gloverin 4, Lebocin, Moricin and Defensin A were determined by PCR (A). The survival curve was also plotted after bacterial infection with or without NOS inhibitor L-NAME (B).
References
1. Gillespie JP, Kanost MR, Trenczek T. Biological Mediators of Insect Immunity. Annu Rev Entomol (1997) 42:611–43. doi: 10.1146/annurev.ento.42.1.611
2. Bulmer MS, Bachelet I, Raman R, Rosengaus RB, Sasisekharan R. Targeting an Antimicrobial Effector Function in Insect Immunity as a Pest Control Strategy. Proc Natl Acad Sci USA (2009) 106(31):12652–7. doi: 10.1073/pnas.0904063106
3. Welchman DP, Aksoy S, Jiggins F, Lemaitre B. Insect Immunity: From Pattern Recognition to Symbiont-Mediated Host Defense. Cell Host Microbe (2009) 6(2):107–14. doi: 10.1016/j.chom.2009.07.008
4. Lemaitre B, Hoffmann J. The Host Defense of Drosophila Melanogaster. Annu Rev Immunol (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
5. Strand MR. The Insect Cellular Immune Response. Insect Sci (2008) 15(1):1–14. doi: 10.1111/j.1744-7917.2008.00183.x
6. Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic Perspectives in Innate Immunity. Science (1999) 284(5418):1313–8. doi: 10.1126/science.284.5418.1313
7. Eleftherianos I, Heryanto C, Bassal T, Zhang W, Tettamanti G, Mohamed A. Haemocyte-Mediated Immunity in Insects: Cells, Processes and Associated Components in the Fight Against Pathogens and Parasites. Immunology (2021) 164(3):401–32. doi: 10.1111/imm.13390
8. Buchon N, Silverman N, Cherry S. Immunity in Drosophila Melanogaster- From Microbial Recognition to Whole-Organism Physiology. Nat Rev Immunol (2014) 14(12):796–810. doi: 10.1038/nri3763
9. Holmes VR, Johnston JS. The Innate Immune Response of Eusocial Hymenopterans to Viral Pathogen Challenge. Ann Entomol Soc Am (2021) 115(2):141–7. doi: 10.1093/aesa/saab047
10. Krams I, Daukste J, Kivleniece I, Kaasik A, Krama T, Freeberg TM, et al. Trade-Off Between Cellular Immunity and Life Span in Mealworm Beetles Tenebrio Molitor. Curr Zool (2013) 59(3):340–6. doi: 10.1093/czoolo/59.3.340
11. Bedick JC, Tunaz H, Aliza ARN, Putnam SM, Ellis MD, Stanley DW. Eicosanoids Act in Nodulation Reactions to Bacterial Infections in Newly Emerged Adult Honey Bees, Apis Mellifera, But Not in Older Foragers. Comp Biochem Physiol C (2001) 130(1):107–17. doi: 10.1016/S1532-0456(01)00226-5
12. Fellowes MDE, Kraaijeveld AR, Godfray HCJ. Cross-Resistance Following Artificial Selection for Increased Defense Against Parasitoids in Drosophila Melanogaster. Evolution (1999) 53(3):966–72. doi: 10.2307/2640737
13. Chen KK, Song JH, Song QS, Dou XY, Wang Y, Wei YH, et al. Transcriptomic Analysis Provides Insights Into the Immune Responses and Nutrition in Ostrinia Furnacalis Larvae Parasitized by Macrocentrus Cingulum. Arch Insect Biochem Physiol (2022) 109(3):e21863. doi: 10.1002/arch.21863
14. Chen KK, Lu ZQ. Immune Responses to Bacterial and Fungal Infections in the Silkworm, Bombyx Mori. Dev Comp Immunol (2018) 83:3–11. doi: 10.1016/j.dci.2017.12.024
15. Wu SC, Liao CW, Pan RL, Juang JL. Infection-Induced Intestinal Oxidative Stress Triggers Organ-to-Organ Immunological Communication in Drosophila. Cell Host Microbe (2012) 11(4):410–7. doi: 10.1016/j.chom.2012.03.004
16. Amcheslavsky A, Ip YT. Be a Good Neighbor: Organ-To-Organ Communication During the Innate Immune Response. Cell Host Microbe (2012) 11(4):323–4. doi: 10.1016/j.chom.2012.04.003
17. Kohchi C, Inagawa H, Nishizawa T, Soma G. ROS and Innate Immunity. Anticancer Res (2009) 29(3):817–21.
18. Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen Peroxide: A Metabolic By-Product or a Common Mediator of Ageing Signals? Nat Rev Mol Cell Biol (2007) 8(9):722–8. doi: 10.1038/nrm2240
19. Liochev SI. Reactive Oxygen Species and the Free Radical Theory of Aging. Free Radic Biol Med (2013) 60:1–4. doi: 10.1016/j.freeradbiomed.2013.02.011
20. Rhee SG, signaling. H2O2 C. A Necessary Evil for Cell Signaling. Science (2006) 312(5782):1882–3. doi: 10.1126/science.1130481
22. Miranda KM, Espey MG, Yamada K, Krishna M, Ludwick N, Kim S, et al. Unique Oxidative Mechanisms for the Reactive Nitrogen Oxide Species, Nitroxyl Anion. J Bio Chem (2001) 276(3):1720–7. doi: 10.1074/jbc.M006174200
23. Bredt DS, Snyder SH. Nitric Oxide: A Physiologic Messenger Molecule. Annu Rev Biochem (1994) 63:175–95. doi: 10.1146/annurev.bi.63.070194.001135
24. Kim Y, Ahmed S, Stanley D, An CJ. Eicosanoid-Mediated Immunity in Insects. Dev Comp Immunol (2018) 83:130–43. doi: 10.1016/j.dci.2017.12.005
25. Sadekuzzaman M, Kim Y. Nitric Oxide Mediates Antimicrobial Peptide Gene Expression by Activating Eicosanoid Signaling. PloS One (2018) 13(2):e0193282. doi: 10.1371/journal.pone.0193282
26. MacMicking J, Xie QW, Nathan C. Nitric Oxide and Macrophage Function. Annu Rev Immunol (1997) 15:323–50. doi: 10.1146/annurev.immunol.15.1.323
27. Nappi AJ, Vass E, Frey F, Carton Y. Nitric Oxide Involvement in Drosophila Immunity. Nitric Oxide (2000) 4(4):423–30. doi: 10.1006/niox.2000.0294
28. Rivero A. Nitric Oxide: An Antiparasitic Molecule of Invertebrates. Trends Parasitol (2006) 22(8):352–2. doi: 10.1016/j.pt.2006.02.014
29. Hillyer JF, Estevez-Lao TY. Nitric Oxide is an Essential Component of the Hemocyte-Mediated Mosquito Immune Response Against Bacteria. Dev Comp Immunol (2010) 34(2):141–9. doi: 10.1016/j.dci.2009.08.014
30. Peterson TML, Gow AJ, Luckhart S. Nitric Oxide Metabolites Induced in Anopheles Stephensi Control Malaria Parasite Infection. Free Radic Bio Med (2007) 42(1):132–42. doi: 10.1016/j.freeradbiomed.2006.10.037
31. Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect Antimicrobial Peptides and Their Applications. Appl Microbiol Biotechnol (2014) 98(13):5807–22. doi: 10.1007/s00253-014-5792-6
32. Wu Q, Patocka J, Kuca K. Insect Antimicrobial Peptides, a Mini Review. Toxin (Basel) (2018) 10(11):461. doi: 10.3390/toxins10110461
33. Koczulla AR, Bals R. Antimicrobial Peptides: Current Status and Therapeutic Potential. Drugs (2003) 63(4):389–406. doi: 10.2165/00003495-200363040-00005
34. Zhao PC, Li JJ, Wang Y, Jiang HB. Broad-Spectrum Antimicrobial Activity of the Reactive Compounds Generated In Vitro by Manduca Sexta Phenoloxidase. Insect Biochem Mol Biol (2007) 37(9):952–9. doi: 10.1016/j.ibmb.2007.05.001
35. Feng M, Fei SG, Xia JM, Labropoulou V, Swevers L, Sun JC. Antimicrobial Peptides as Potential Antiviral Factors in Insect Antiviral Immune Response. Front Immunol (2020) 11:2030. doi: 10.3389/fimmu.2020.02030
36. Jiang HB, Vilcinskas A, Kanost MR. Immunity in Lepidopteran Insects. Adv Exp Med Biol (2010) . 708:181–204. doi: 10.1007/978-1-4419-8059-5_10
37. Myllymaki H, Valanne S, Ramet M. The Drosophila Imd Signaling Pathway. J Immunol (2014) 192(8):3455–62. doi: 10.4049/jimmunol.1303309
38. Valanne S, Wang JH, Ramet M. The Drosophila Toll Signaling Pathway. J Immunol (2011) 186(2):649–56. doi: 10.4049/jimmunol.1002302
39. De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd Pathways are the Major Regulators of the Immune Response in Drosophila. EMBO J (2002) 21(11):2568–79. doi: 10.1093/emboj/21.11.2568
40. Kaneko T, Silverman N. Bacterial Recognition and Signalling by the Drosophila IMD Pathway. Cell Microbiol (2005) 7(4):461–9. doi: 10.1111/j.1462-5822.2005.00504.x
41. Li YX, Dijkers PF. Specific Calcineurin Isoforms are Involved in Drosophila Toll Immune Signaling. J Immunol (2015) 194(1):168–76. doi: 10.4049/jimmunol.1401080
42. Dionne MS, Schneider DS. Models of Infectious Diseases in the Fruit Fly Drosophila Melanogaster. Dis Models Mech (2008) 1(1):43–9. doi: 10.1242/dmm.000307
43. Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate Immunity in Plants and Animals: Striking Similarities and Obvious Differences. Immunol Rev (2004) 198:249–66. doi: 10.1111/j.0105-2896.2004.0119.x
44. Hoffmann JA, Reichhart JM. Drosophila Innate Immunity: An Evolutionary Perspective. Nat Immunol (2002) 3(2):121–6. doi: 10.1038/ni0202-121
45. Buchmann K. Evolution of Innate Immunity: Clues From Invertebrates via Fish to Mammals. Front Immunol (2014) 5:459. doi: 10.3389/fimmu.2014.00459
46. Foley E, O'Farrell PH. Nitric Oxide Contributes to Induction of Innate Immune Responses to Gram-Negative Bacteria in Drosophila. Genes Dev (2003) 17(1):115–25. doi: 10.1101/gad.1018503
47. Dijkers PF, O'Farrell PH. Drosophila Calcineurin Promotes Induction of Innate Immune Responses. Curr Bio (2007) 17(23):2087–93. doi: 10.1016/j.cub.2007.11.001
48. Ishii K, Adachi T, Hamamoto H, Oonishi T, Kamimura M, Imamura K, et al. Insect Cytokine Paralytic Peptide Activates Innate Immunity via Nitric Oxide Production in the Silkworm Bombyx Mori. Dev Comp Immunol (2013) 39(3):147–53. doi: 10.1016/j.dci.2012.10.014
49. Zhou Y, Guo J, Wang XY, Cheng Y, Guan JW, Barman P, et al. FKBP39 Controls Nutrient Dependent Nprl3 Expression and TORC1 Activity in Drosophila. Cell Death Dis (2021) 12(6):571. doi: 10.1038/s41419-021-03860-z
50. Chen KK, Tang T, Song QS, Wang ZY, He KL, Liu X, et al. Transcription Analysis of the Stress and Immune Response Genes to Temperature Stress in Ostrinia Furnacalis. Front Physiol (2019) 10:1289. doi: 10.3389/fphys.2019.01289
51. Chen KK, Liu C, He Y, Jiang HB, Lu ZQ. A Short-Type Peptidoglycan Recognition Protein From the Silkworm: Expression, Characterization and Involvement in the Prophenoloxidase Activation Pathway. Dev Comp Immunol (2014) 45(1):1–9. doi: 10.1016/j.dci.2014.01.017
52. Feng CJ, Zhao Y, Chen KK, Zhai HF, Wang ZY, Jiang HB, et al. Clip Domain Prophenoloxidase Activating Protease is Required for Ostrinia Furnacalis Guenee to Defend Against Bacterial Infection. Dev Comp Immunol (2018) 87:204–15. doi: 10.1016/j.dci.2018.06.014
53. Wang WX, Wang YP, Deng XJ, Dang XL, Tian JH, Yi HY, et al. Molecular and Functional Characterization of a C-Type Lysozyme From the Asian Corn Borer, Ostrinia Furnacalis. J Insect Sci (2009) 9(1):17. doi: 10.1673/031.009.1701
54. Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of Potential Reference Genes for Reverse Transcription-qPCR Studies of Physiological Responses in Drosophila Melanogaster. J Insect Physiol (2011) 57:840–50. doi: 10.1016/j.jinsphys.2011.03.014
55. Schmittgen TD, Livak KJ. Analyzing Real-Time PCR Data by the Comparative C(T) Method. Nat Protoc (2008) 3(6):1101–8. doi: 10.1038/nprot.2008.73
56. Wang RJ, Chen KK, Xing LS, Lin Z, Zou Z, Lu ZQ. Reactive Oxygen Species and Antimicrobial Peptides are Sequentially Produced in Silkworm Midgut in Response to Bacterial Infection. Dev Comp Immunol (2020) 110:103720. doi: 10.1016/j.dci.2020.103720
57. Horng T, Medzhitov R. Drosophila MyD88 is an Adapter in the Toll Signaling Pathway. Proc Natl Acad Sci USA (2001) 98(22):12654–8. doi: 10.1073/pnas.231471798
58. Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-Kappa B/Rel Proteins and the Humoral Immune Responses of Drosophila Melanogaster. Nf-Kb Health Dis (2011) 349:25–60. doi: 10.1007/82_2010_107
59. Han M, Qin S, Song XJ, Li YF, Jin P, Chen LM, et al. Evolutionary Rate Patterns of Genes Involved in the Drosophila Toll and Imd Signaling Pathway. BMC Evol Biol (2013) 13:245. doi: 10.1186/1471-2148-13-245
60. Kellenberger C, Leone P, Coquet L, Jouenne T, Reichhart JM, Roussel A. Structure-Function Analysis of Grass Clip Serine Protease Involved in Drosophila Toll Pathway Activation. J Biol Chem (2011) 286(14):35087–95. doi: 10.1074/jbc.M110.182741
61. Furman JL, Norris CM. Calcineurin and Glial Signaling: Neuroinflammation and Beyond. J Neuroinflamm (2014) 11:158. doi: 10.1186/s12974-014-0158-7
62. Saraf J, Bhattacharya P, Kalia K, Borah A, Sarmah D, Kaur H, et al. A Friend or Foe: Calcineurin Across the Gamut of Neurological Disorders. ACS Cent Sci (2018) 4(7):805–19. doi: 10.1021/acscentsci.8b00230
63. Kang YJ, Kusler B, Otsuka M, Hughes M, Suzuki N, Suzuki S, et al. Calcineurin Negatively Regulates TLR-Mediated Activation Pathways. J Immunol (2007) 179(7):4598–607. doi: 10.4049/jimmunol.179.7.4598
64. Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, et al. A Genome-Wide Drosophila RNAi Screen Identifies DYRK-Family Kinases as Regulators of NFAT. Nature (2006) 441(7093):646–50. doi: 10.1038/nature04631
65. Morishima I, Yamano Y, Inoue K, Matsuo N. Eicosanoids Mediate Induction of Immune Genes in the Fat Body of the Silkworm, Bombyx Mori. FEBS Lett (1997) 419(1):83–6. doi: 10.1016/S0014-5793(97)01418-X
66. Yajima M, Takada M, Takahashi N, Kikuchi H, Natori S, Oshima Y, et al. A Newly Established In Vitro Culture Using Transgenic Drosophila Reveals Functional Coupling Between the Phospholipase A(2)-Generated Fatty Acid Aascade and Lipopolysaccharide-Dependent Activation of the Immune Deficiency (Imd) Pathway in Insect Immunity. Biochem J (2003) 371:205–10. doi: 10.1042/Bj20021603
67. Hwang J, Park Y, Kim Y, Hwang J, Lee D. An Entomopathogenic Bacterium, Xenorhabdus Nematophila, Suppresses Expression of Antimicrobial Peptides Controlled by Toll and Imd Pathways by Blocking Eicosanoid Biosynthesis. Arch Insect Biochem Physiol (2013) 83(3):151–69. doi: 10.1002/arch.21103
68. Ji JY, Yin ZH, Zhang SS, Shen DX, An CJ. PLA(2) Mediates the Innate Immune Response in Asian Corn Borer, Ostrinia Furnacalis. Insect Sci (2022) 29(1):245–58. doi: 10.1111/1744-7917.12932
69. Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, et al. The Phytopathogenic Bacteria Erwinia Carotovora Infects Drosophila and Activates an Immune Response. Proc Natl Acad Sci USA (2000) 97(7):3376–81. doi: 10.1073/pnas.070357597
70. Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, et al. Tissue-Specific Inducible Expression of Antimicrobial Peptide Genes in Drosophila Surface Epithelia. Immunity (2000) 13(5):737–48. doi: 10.1016/S1074-7613(00)00072-8
71. Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, et al. A Genome-Wide Analysis of Immune Responses in Drosophila. Proc Natl Acad Sci USA (2001) 98(26):15119–24. doi: 10.1073/pnas.261573998
72. Zou Z, Evans JD, Lu ZQ, Zhao PC, Williams M, Sumathipala N, et al. Comparative Genomic Analysis of the Tribolium Immune System. Genome Biol (2007) 8(8):R177. doi: 10.1186/gb-2007-8-8-r177
73. Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial Detection by Drosophila Peptidoglycan Recognition Proteins. Microbes Infect (2009) 11(6-7):631–6. doi: 10.1016/j.micinf.2009.03.004
74. Cao XL, He Y, Hu Y, Wang Y, Chen YR, Bryant B, et al. The Immune Signaling Pathways of Manduca Sexta. Insect Biochem Mol Biol (2015) 62:64–74. doi: 10.1016/j.ibmb.2015.03.006
Keywords: insect immunity, reactive oxygen species, Ostrinia furnacalis, nitric oxide synthase, signal transduction
Citation: Chen K, Wang X, Wei X, Chen J, Wei Y, Jiang H, Lu Z and Feng C (2022) Nitric Oxide-Induced Calcineurin A Mediates Antimicrobial Peptide Production Through the IMD Pathway. Front. Immunol. 13:905419. doi: 10.3389/fimmu.2022.905419
Received: 27 March 2022; Accepted: 20 April 2022;
Published: 18 May 2022.
Edited by:
Zhen Zou, Institute of Zoology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Chunju An, China Agricultural University, ChinaPaulina Schmitt, Pontificia Universidad Católica de Valparaíso, Chile
Copyright © 2022 Chen, Wang, Wei, Chen, Wei, Jiang, Lu and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Congjing Feng, ZmVuZ2NqQHl6dS5lZHUuY24=; Zhiqiang Lu, emhpcWlhbmcubHVAbndhZnUuZWR1LmNu
 Kangkang Chen
Kangkang Chen Xinyan Wang
Xinyan Wang Xiangyi Wei
Xiangyi Wei Jiaqian Chen
Jiaqian Chen Youheng Wei
Youheng Wei Haobo Jiang
Haobo Jiang Zhiqiang Lu
Zhiqiang Lu Congjing Feng
Congjing Feng