
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 02 June 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.902941
Haematopoiesis in fish and mammals is a complex process, and many aspects regarding its model and the differentiation of haematopoietic stem cells (HSCs) still remain enigmatic despite advanced studies. The effects of microenvironmental factors or HSCs niche and signalling pathways on haematopoiesis are also unclear. This review presents Danio rerio as a model organism for studies on haematopoiesis in vertebrates and discusses the development of this process during the embryonic period and in adult fish. It describes the role of the microenvironment of the haematopoietic process in regulating the formation and function of HSCs/HSPCs (hematopoietic stem/progenitor cells) and highlights facts and research areas important for haematopoiesis in fish and mammals.
Haematopoiesis in vertebrates, despite many studies, remains to be fully characterised, and there are even controversies related to the model of this process and its details, particularly the differentiation of haematopoietic stem cells (HSCs) (1). As emphasized by Cheng et al. (1), these controversial observations might be related to differences in research techniques, especially methods for tracing the development/differentiation of HSCs and labelling efficiency of these cells. Haematopoiesis is still enigmatic, and perhaps this is the key issue, as a process strongly dependent on various factors of the microenvironment and signalling pathways that influence all stages of the formation and development of blood cells and their precursors (2). In the light of previous findings that confirmed the hierarchy of haematopoietic stem cells and hematopoietic stem/progenitor cells (HSPCs), as well as the differentiation models of HSCs (3–5), Cheng et al. (1), proposed new paradigms on haematopoietic stem cell differentiation, emphasizing at the same time the need for their further revision (6–10). In the proposed model (1) it is assumed that HSCs differentiate into multipotent progenitors (MPP), which include the subpopulation of short-term haematopoietic stem cells (MPP1/ST-HSC), giving rise to parallel subpopulations: MPP2, MPP3 and MPP4 (LMPP, lymphoid-primed multipotent progenitors). At the further stages of differentiation, the MPP2 subpopulation gives rise to pre-megakaryocyte-erythrocytes and ultimately the platelet lineage, with the indirect participation of the megakaryocyte progenitors and the erythrocyte lineage, mediated by the pre-colony forming unit-erythroid. The MPP3 subpopulation gives rise to granulocytes and monocytes that form from pre-granulocyte-macrophages and granulocyte-macrophage progenitors, and the MPP4 subpopulation (LMPP) differentiates, through common lymphoid progenitors mainly into lymphocytes. It should be stressed that the long-term research into details of haematopoiesis has provided many new data, especially on the differentiation of HSCs, giving reasons and foundations for the modification or even radical change of the model/paradigm of haematopoiesis (1). However, studies indicating that haematopoiesis may be characterised by the continuous acquisition of specific properties by hematopoietic stem/progenitor cells (HSCs/HSPCs) seem particularly interesting, since these cells are likely to have an epigenetic status allowing for the transformation towards a specific cell lineage or specific cell type (1, 11–15). Yokota (14) indicates the possible heterogeneity of HSCs and describes the process that corresponds to the “holacracy”. Xu et al. (16), by visualizing haematopoiesis as a stochastic process, showed that the formation of blood cells can be modelled as a dynamic process with a stochastic competition between the cell types. Haematopoiesis might also be a process based on deterministic events, as can be inferred from the study by Zhen et al. (17) in Danio rerio. Studies on zebrafish (D. rerio) also imply that haematopoiesis may be a continuous process of HSPCs’ differentiation associated with the simultaneous suppressive and/or stimulating transcriptional activity of genes that are responsible for the formation, proliferation and differentiation of cells specific for a particular haematopoietic lineage (18). Danio rerio (Kingdom – Animalia, Superphylum – Deuterostomia, Phylum – Chordata, Subphylum – Vertebrata, Class – Actinopterygii, Order – Cypriniformes Family – Cyprinidae, Subfamily – Danioninae, Genus – Danio, Species - D. rerio) due to its special biological characteristics is a model organism and a highly valuable and effective “tool” in studies on haematopoiesis and haematopoietic niches in vertebrates, including mammals and humans (19–22). Findings from studies on the zebrafish model have explained processes that influence the course of haematopoiesis and the development of HSCs, and have significant implications not only for general knowledge in the range of basic sciences, but most of all are important because of their potential applicability in regenerative medicine (2, 18, 19, 21–23).
Developmental processes and molecular mechanisms regulating haematopoiesis in embryos and larvae of D. rerio, as emphasized by Gore et al. (2), are conserved in evolutionarily younger organisms. It is also very important that the zebrafish has cells of all the haematopoietic lineages, available in each period of differentiation in the pronephros, which is the equivalent to mammalian bone marrow, and orthologs of many transcription factors (TFs), including TAL bHLH transcription factor 1 (TAL1, erythroid differentiation factor), GATA binding protein 2 (GATA2), RUNX family transcription factor 1 (RUNX1), MYB proto-oncogene, transcription factor (MYB, known as c-myb), and ETS transcription factor ERG (ERG), which play important roles in the process of haematopoiesis in mammals (24–30). The essential similarity between the haematopoiesis in zebrafish D. rerio and mammals also includes transcription mechanisms, more specifically the transcriptional status of cells, associated with the expression of genes coding regulatory factors, crucial for a specific cell lineage, as well as signalling pathways important for the regulation of haematopoiesis, including the Wnt signalling pathway and Notch signalling pathway (2, 18, 23, 31–33). It is noteworthy that Notch signaling targeting the transcription factor RUNX1 controls self-renewal of stem cells, and the Notch-Runx1 signaling pathway is essential for the fate of these cells (33). D. rerio is characterised by easy and fast reproduction (zebrafish is oviparous and fertilization is an external process); dynamic development (at 25-26°C); embryonic transparency, which enables observations and in vivo imaging of the development of embryos/larvae and haematopoiesis; ease of genetic testing and genetic modification to generate transgenic organisms using the Tol2 Transposase system (autonomous transposone identified in Japanese rice fish, Oryzias latipes, which is used to create the transgenic zebrafish) to obtain reporter lines suitable for specific labelling of certain types of cells with green fluorescent protein (GFP) (2, 18–20, 34–39). A model enabling the visualization of, inter alia, the hematopoietic process is Danio rerio double mutant - the nacre mutant and the spontaneous mutant the roy orbison (roy), known as the casper strain, which shows a complete lack of melanocytes and iridophores in embryogenesis and in adulthood. These fish retain the transparency of the outer shells throughout their lives and, very importantly, they are a tool with the expected sensitivity and resolution in imaging and analyzing the number and distribution of GFP-labeled stem cells in vivo (40–42). In zebrafish all events of blood cell formation and colonization of haematopoietic niches can be observed from the earliest stages of development (from a few hours and/or days after fertilization), at the single cell level. The effects of modified/downregulated expression of a specific gene (gene knockdown) or its removal or permanent deactivation (gene knockout) caused by the use of an antisense oligonucleotide (morpholino oligonucleotides) that binds to the coding gene or its mRNA, the CRISPR system and endonuclease Cas9 (CRISPR/Cas9) or transcription activator-like effector nucleases (TALEN) can also be analysed successfully in zebrafish (2, 19, 36). However, the analysis of the expression of a specific gene cannot ignore the fact that there are significant differences between phenotypes caused by genetic mutations and phenotypes caused by gene knockdown or knockout, also in D. rerio. It should also be borne in mind that harmful phenotype changes in mutants, but not in morphants, may be buffered by the activity of the mechanism underlying genetic compensation (43, 44). Therefore, conclusions reached from studies on haematopoiesis in zebrafish can be and are indeed used for modelling studies on haematopoiesis and its disorders in humans (20, 32). Nevertheless, it should be noted that despite the large research opportunities offered by model organisms of D. rerio, which complement the mammalian models, they are biased with certain limitations, which was indicated by Konantz et al. (20). For example, there is a limited availability of antibodies suitable for labelling cell surface markers or techniques for the simultaneous and selective expression of oncogenes in the tissues of adult D. rerio, which would offer more opportunities for the phenocopying of human disorders.
Embryonic period haematopoiesis in D. rerio (Figure 1) has two stages, i.e. early-embryonic haematopoiesis (Figure 1.1A) and embryonic haematopoiesis (Figure 1.1B).
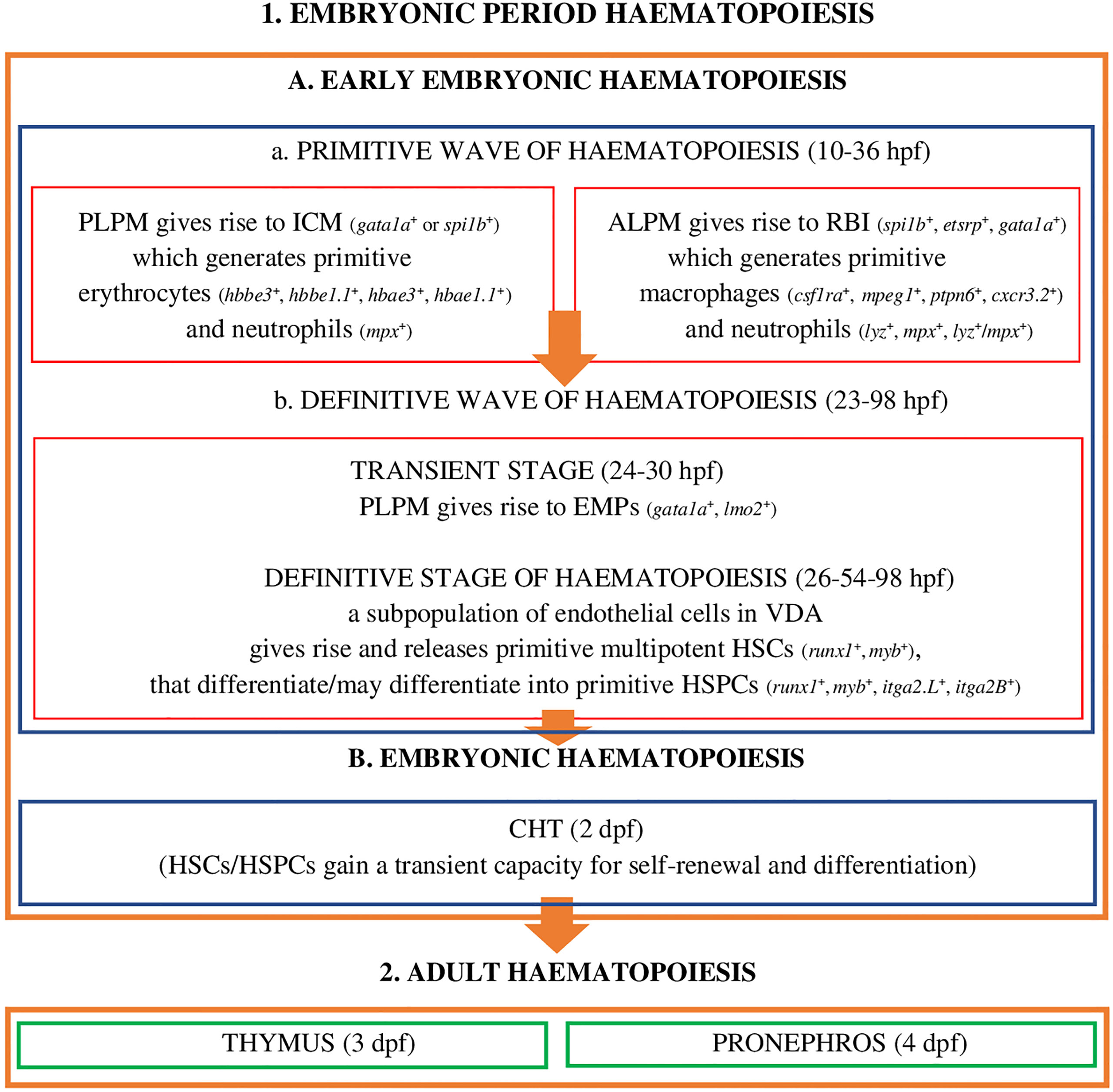
Figure 1 Embryonic period haematopoiesis and adult haematopoiesis in zebrafish (D. rerio) (references in the main text). Explanatory notes for part (A, B): PLPM, posterior lateral-plane mesoderm; ALPM, anterior lateral-plane mesoderm; ICM, intermediate cell mass; RBI, rostral blood islands; VDA, ventral wall of the dorsal aorta; CHT, caudal hematopoietic tissue; hpf, hours post fertilization; dpf, days after fertilization; spi1b, Spi-1 proto-oncogene b; etsrp, ETS1-related protein; gata1a, GATA binding protein 1a; hbbe3, haemoglobin beta embryonic-3, hbbe1.1, haemoglobin beta embryonic-1.1; hbae3, haemoglobin alpha embryonic-3, hbae1.1, haemoglobin alpha embryonic-1.1, lyz, lysozyme C, mpx, myeloid-specific peroxidase, lzm/mpx, double-positive neutrophils stain strongly with Sudan black; lmo2, LIM domain only 2; runx1, RUNX family transciption factor 1; myb, MYB proto-oncogene, transcription factor; itga2.L, integrin subunit alpha 2 L homeolog (itga2-A); itga2B, integrin subunit alpha 2 B (CD41).
At this stage of embryonic development, hematopoiesis in zebrafish (Figure 1.1A) is a process which, as in other vertebrates, develops in two successive waves: the primitive wave (Figure 1.1A.a) and the definitive wave (Figure 1.1A.b) (2, 20, 23, 45).
The primitive wave of the early-embryonic haematopoiesis (Figure 1.1A.a) starts in two different regions of the lateral mesoderm: in the posterior lateral-plate mesoderm (PLPM) and in the anterior lateral-plate mesoderm (ALPM). From PLPM, at the trunk midline of the fish embryo (2), the so-called intermediate cell mass (ICM) blood islands are formed (gata1a+ - GATA binding protein 1a or spi1b+ - Spi-1 proto-oncogene b, called pu.1) conceptually analogous to the extra - embryonic yolk sac blood islands of mammals and birds, which give rise to primitive erythroid cells - erythrocytes (E) (hbbe3+ - haemoglobin beta embryonic-3, hbbe1.1+ - haemoglobin beta embryonic-1.1, hbae3+ - haemoglobin alpha embryonic-3, hbae1.1+ - haemoglobin, alpha embryonic 1.1, known as hbae1), and 24 hours post fertilization (hpf) they enter the circulation as oval and nucleated cells and primitive myeloid cells - neutrophils (mpx+ - myeloid-specific peroxidase) (2, 19, 23, 28). Moreover, transcription factors Gata1a and Spi-1b exhibit a cross-inhibitory relationship to regulate the fates of primitive erythroid and myeloid cells (23). ALPM, which is the main myelopoietic site, gives rise to rostral blood islands (RBI) (etsrp+ - ETS1-related protein, spi1b+, gata1a+), where primitive macrophages are formed (csf1ra+ - colony stimulating factor 1 receptor a, mpeg1+ - macrophage expressed gene 1, ptpn6+ - protein tyrosine phosphatase non-receptor type 6, cxcr3.2+ - chemokine [C-X-C motif] receptor 3, tandem duplicate 2) as well as neutrophils (lyz+ - lysozyme C, mpx+, lyz+/mpx+ - double-positive neutrophils stain strongly with Sudan Black), which have a phagocytic capacity, and are involved in the formation of HSCs/HSPCs in the ventral wall of the dorsal aorta (VDA) and the migration of these cells to the vascular system/venous sinuses in the caudal region of the fish body, between the caudal artery and the vena cava, i.e. to the caudal haematopoietic tissue (CHT) (2, 19, 23, 46–53). As highlighted by Jagannathan-Bogdan and Zon (23), PLPM and ALPM in D. rerio co-expressing tal1, gata2, lmo2 (LIM domain only 2), fli1 (Fli-1 proto-oncogene, ETS transcription factor) and etsrp may give rise to angioblasts or HSCs, which confirms the presence of haemangioblasts - common precursors of endothelial cells and haematopoietic cells in zebrafish and humans.
The definitive wave of early-embryonic haematopoiesis (Figure 1 - 1.Ab) includes the transient stage and the definitive stage of haematopoiesis. The transient stage takes place in PLPM, where multipotent erythromyeloid progenitors (24-30 hpf) are formed, also called erythroid-myeloid progenitors (gata1a+, lmo2+) (2, 19, 23, 54, 55). During the definitive stage of haematopoiesis the subpopulation of endothelial cells in VDA (the equivalent of the mammalian aorta-gonad-mesonephros [AGM] region), in the process of the so-called endothelial-hematopoietic transformation (EHT), also defined as the new type of cell behaviour, primitive multipotent HSCs (runx1+, myb+) are formed and released (26-54 hpf) that may differentiate into haematopoietic stem progenitor cells (HSPCs) expressing runx1, myb, itga2.L (integrin subunit alpha 2 L homeolog, known as itga2-A), itga2.B (integrin subunit alpha 2 B, known as CD41) (2, 4, 18, 19, 21, 47, 56, 57). According to Henninger et al. (58), about 30 HSCs or their clones are generated at this stage of embryonic development, and these cells are responsible for the formation of the haematopoietic system and life-long haematopoiesis in fish/vertebrates. Henninger et al. (58) also emphasized that HSCs in D. rerio in this period of embryonic development are the most productive. This event, i.e. the formation of HSCs/HSPCs, marks the beginning of the definitive haematopoiesis, in which blood cells are generated by self-renewal and differentiation of already existing HSCs/HSPCs but not those generated de novo (19, 58). HSCs/HSPCs are induced, for example, by Cxcl12b (chemokine [C-X-C motif] ligand 12b) produced by the specific population of endothelial precursors (endotome cells), and by TNF-α (tumour necrosis factor α), produced by primitive macrophages and neutrophils, and after moving into the vena cava they migrate to CHT (2 days post fertilization, dpf), which is the site of embryonic haematopoiesis (the equivalent of the foetal liver in mammals).
In CHT they are stimulated by cytokines: Kitlgb (kit ligand b), Osm (oncostatin M), Csf3a (colony stimulating factor 3 [granulocyte] a), Ccl25b (chemokine [C-C motif] ligand 25b), Cxcl8b (chemokine [C-X-C motif] ligand 8b) and Cxcl12a (chemokine [C-X-C motif] ligand 12a) as well as Klf6a (Krüppel-like factor 6a, transcription factor), and gain the capacity for self-renewal and differentiation. The development and expansion of these cells is a key property of CHT, supervised, inter alia, by the regulatory mechanism of non-hematopoietic CHT components, i.e. vascular endothelial cells, epithelial cells, fibroblasts, and nerve and muscle cells (59–61). It has been shown (59) that promoting the proliferation and differentiation of HSCs/HSPCs in the CHT niche, in addition to chemokines and cytokines, is also tightly controlled by various external and internal cellular factors, such as the cell cycle and transcriptomic features that are likely to affect cell heterogeneity in the parental and progenitor compartments. An example is the identified and characterized (59) vascular endothelial-specific factor, i.e. Gpr182 (G protein-coupled receptor 182), which plays a positive role in CHT remodeling favoring the expansion of HSPCs. It should be emphasized that the HSPCs population, in CHT D. rerio, includes four subpopulations, identified on the basis of different lineage-specific genes - HSPC1 cmyb+, a proliferative subpopulation not involved in the differentiation process and HSPC2 hemgn+, tmem14ca+, HSPC3 cebpb+, HSPC4 coro1a+, ccr9a+, rac2+, subpopulations capable of differentiating cells targeted to perform specialized functions (59). These subpopulations are also recognizable on the basis of the metabolic genes HSPC2 pcna+, cdk1+, slc11a2+, HSPC3 fosab+, HSPC4 actb1+, rac2+, litaf+, coro1a+ (59), suggesting their influence on HSPC heterogeneity (59). At this stage of development of hematopoiesis, HSCs/HSPCs give rise to embryonic macrophages, neutrophils and monocytes, they proliferate and migrate (19, 49, 56, 59–62) and finally colonize the developing thymus, where T lymphocytes are generated (3 dpf), along with the pronephros (4 dpf) (the equivalent to the bone marrow of mammals) (Figure 1. - 2) (2, 19, 32, 63, 64). In the pronephros, HSCs/HSPCs fulfil their life-long haematopoietic function (19).
The key sites in regulating the formation and function of stem and progenitor cells, HSCs/HSPCs, are specialized/specific anatomical regions called haematopoietic niches creating a microenvironment of the haematopoietic process. They have special anatomical and structural features, that is a specific subset of cells including vascular endothelial cells, mesenchymal stromal cells (MSC), macrophages and neutrophils, and regulatory agents that interact with stem cells and selectively orchestrate a development pathway for these cells (19, 36). Two haematopoietic niches have been identified in D. rerio: VDA, called the initiating haemopoietic niche, and CHT, defined as the primary tissue of embryonic haemopoiesis or the intermediate haematopoietic niche. In adult zebrafish, the haematopoietic niche is the pronephros, which accommodates self-renewing and differentiating HSCs/HSPCs, and generating all blood cells during the whole adult life of vertebrates (19, 28, 36).
Some vascular endothelial cells in VDA, called haemogenic endothelial cells (HE) in D. rerio give rise to HSCs (runx1+, myb+) and HSPCs (runx1+, myb+, itga2.L+, itga2B+), while others perform regulatory functions in the process of their formation. The formation of HSCs/HSPCs, as previously mentioned, is induced by the activity of a specific population of endothelial precursors, called “endotome cells” (at the primary stage of their formation in somite they are identified as meox1+, mesenchyme homeobox 1), which migrate and colonize VDA (65), and Cxcl12b produced by them. Further, due to the deactivation of meox1 (a gene coding a protein which may play a role in the molecular signalling network regulating the growth of somites) and increasing the number of “endotome cells”, the induction of HSCs/HSPCs is stronger and they are released to the circulation, with the involvement of cells other than endothelial cells from VDA, i.e. those from adjacent somites, as well as primitive macrophages and neutrophils by the release of cytokines and enzymes, such as TNFα or Mmp2 (matrix metalloproteinases 2 (19, 36, 47, 51, 58). Monteiro et al. (66) demonstrated that the formation of HSCs/HSPCs is also regulated by Tgf-β (transforming growth factor beta), an anti-inflammatory cytokine regulating the proliferation and differentiation of many types of cells. Tgf-β, by binding to a single type II serine-threonine kinase receptor (Tgf-βr2) in an autocrine mechanism activated by ligands, Tgf-β1a and Tgf-β1b, is involved in the determining/programming of endothelial cells towards the haemogenic endothelium (before EHT), and further, in a paracrine mechanism activated by Tgf-β3 (the source of the ligand is the notochord), it is involved in EHT and the generation of HSCs/HSPCs. Tgf-β, in both stages of its activity, also regulates the expression of Jag1a (jagged canonical Notch ligand 1a) activating the Notch1a receptor present on the surface of cells in VDA, which is a necessary process for the formation of haematopoietic stem cells (66, 67). The release of HSCs/HSPCs from VDA is regulated by Cbfβ (core binding factor subunit beta), coded by the cbfβ gene whose expression defines HE cells and determines EHT (68). A study by Bresciani et al. (68) revealed, however, the involvement and functional role of Cbfβ at different stages of HSCs/HSPCs development. Cbfβ is a subunit involved in the early stage of the definitive wave of haematopoiesis (Figure 1.1.A.b) by forming the Runx1/Cbfβ complex, which, as in mammals (mice), is a heterodimeric core-binding transcription factor) (68). In D. rerio both subunits, Cbfβ and Runx1, can be active independently of each other and participate in two different stages of the definitive haematopoietic wave (68). Runx1 is involved in the generation of HSCs (runx1+, myb+) in VDA, while Cbfβ is involved in the release of HSCs from VDA. The definitive wave of haematopoiesis in D. rerio is also regulated by two isoforms of the Scl transcription factor (stem cell leukaemia) - Sclβ and Sclα (17). These isoforms are involved in the defining and generation of HE – Sclβ (VDA scl-β+ cells, before EHT) and the generation, retention and release of HSCs/HSPCs - Sclα (generated HSCs scl-α+) (17). Of note is the fact that the expression of scl-β is the first molecular marker of haemogenic endothelial cells (17), and the process of HSCs generation is determined by a sequential activity of transcription factors: Scl-β, Runx1 (at the stage of EHT) and Scl-α. At this stage of haematopoiesis the differentiation of haematopoietic stem cells HSCs/HSPCs is also strongly regulated by Dnmt3bb.1 (DNA [cytosine-5-]-methyltransferase 3 beta, duplicate b.1), an enzyme that is one of six (dnmt3bb.1, dnmt3aa, dnmt3ab, dnmt3ba, dnmt3bb.2, dnmt3bb.3) homologs of DNMT3b in mammals (69, 70). The activity of Dnmt3bb.1 initiated in response to an increased expression of the runx1 gene (stimulated most likely by Scl-β (17),) and the Notch/Runx1 signalling pathway, sustains the expression of the myb gene and functional efficiency of HSCs/HSPCs (69, 70). Moreover, the vascular endothelial cells present in haematopoietic niches support the process of haematopoiesis regardless of their origin (VDA, CHT, pronephros) and different transcriptional characteristics (32, 71, 72). Importantly, the endothelial cells in the CHT are characterized by a specific alignment, for which HSCs/HSPCs migrating and released to the perivascular space must squeeze between them. Because of this, haematopoietic stem cells directly surrounding endothelial cells induce changes in their organization within the niche (a pocket is formed around HSCs/HSPCs) and probably also cause an increase in the concentration of cytokines and signalling molecules, which consequently influences the expansion and differentiation of HSCs/HSPCs and their effective interaction with stromal cells (19, 22, 36).
Mesenchymal stromal cells (MSCs) in D. rerio originate from the ventral border of the caudal somites and are generated during the epithelial–mesenchymal transition (EMT). MSCs are present among the endothelial cells of vascular network/vascular sinuses forming CHT and may express Cxcl12a cytokines (19, 32, 73). MSCs come in contact with HSCs/HSPCs, anchor them and further orient their differentiation and division, which increases the population of stem cells. It is possible that this process is induced by the Cxcl12/CxcR4 signalling axis (36, 53). This is facilitated, as mentioned earlier, by the formation of ‘pockets’ around HSCs/HSPCs, increasing the local concentration of growth factors and signalling molecules, and creating the most productive conditions for the interaction between haematopoietic stem cells and stromal cells (36). Importantly, MSCs were previously described as fibroblastic reticular cells (FRCs), and later as stromal cells or stromal reticular cells (SRCs) also present in the pronephros in fish and, similar to CXCL12+ reticular cells, present in mammalian bone marrow (53).
Primitive macrophages and neutrophils, being the cells of the hematopoietic microenvironment, play an important regulatory role in the formation of HSCs/HSPCs in VDA and the migration of these cells to CHT and haematopoietic organs in adult individuals (pronephros, thymus) (2, 19, 23, 47–53). These cells, as mentioned earlier, develop at the early stage of embryonic haematopoiesis, i.e. during the primitive wave of haematopoiesis (16 hpf) (Figure 1.1.A.a) and the definitive wave of haematopoiesis (30 - 55 hpf) (Figure 1.1.A.b), when HSCs/HSPCs are generated (26-54 hpf). The presence of mpeg1+ macrophages was demonstrated in VDA and CHT (19, 46, 50). Travnickova et al. (50) reported that primitive macrophages, by releasing Mmp-9 (matrix metalloproteinases 9), induce the degradation of the extracellular matrix (ECM) and mobilize HSCs/HSPCs for migration. However, the actual role of these cells in niches of the early stage of embryonic haematopoiesis is still unclear (19, 37, 50, 67). Primitive neutrophils, like primitive macrophages, express the genes coding matrix metalloproteinases, Mmp-2 and Mmp-9, and these enzymes, apart from the degradation of extracellular matrix proteins, also stimulate the release of HSCs/HSPCs from VDA (Mmp-2) and CHT (Mmp-9) (19, 51). Theodore et al. (51) emphasized that Mmp-2 and Mmp-9 proteins are active at different stages of the formation, development and migration of HSCs/HSPCs and via discrete indirect or direct mechanisms, they are involved in processes associated with the signalling inflammatory process, extracellular matrix protein degradation, and the regulation of chemokines activity. In the formation of HSCs/HSPCs, the signalling pathway mediating sterile inflammation (in an environment without the inflammatory/damaging factor) triggered towards endothelial cells in the VDA also plays an important role. This process develops, for example, because of the activity of Tnf-α, a proinflammatory cytokine released mainly by primitive neutrophils (and to a lesser extent by the primitive macrophages), which activates the endothelial cells in VDA. In subsequent events of the process Tnf-α actives specific Tnfr2 (tumour necrosis factor receptor-2), and this increases the expression of Jag1a and activates Notch1a, which may be a receptor present on the surface of HSCs, and ultimately, through the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) active in the formed HSCs, transcription necessary for the release of HSCs/HSPCs from endothelial cells is triggered (67, 74). The formation, release and migration of HSCs/HSPCs is also controlled and promoted by the regulatory activity of Ifn-γ and Ifn type I – the equivalent of IFN-α in mammals (37). The regulatory properties of these cytokines, like Tnf-α, reflect their broad functional potential, which is very important, as it turns out, in the process of embryonic haematopoiesis that, of note, is not related to the inflammatory process/inflammation or protection against infection.
In adult fish, haematopoiesis develops in the pronephros. In this organ HSCs/HSPCs are self-renewing and differentiating, giving rise to all lineages of blood cells (19, 28, 36). However, there are still open questions about the site where HSCs are accommodated and differentiate, what the mechanisms regulating the formation of mature morphotic elements of blood in fish and other vertebrates are, and what is the most probable model/paradigm on haematopoiesis. The classical model of haematopoiesis in vertebrates, also in the light of recent studies on D. rerio (75), aided with state-of-the art research techniques and regarding “megakaryopoiesis” (18) or the molecular definition of cell populations/clusters in the pronephros and identification of marker genes for cells of specific haematopoietic lineages (75), is still being improved and is a subject of targeted verification (1, 11, 13–15, 18, 76). The proposed models of hematopoiesis and data on this process in vertebrates still raise further doubts and questions, for example about heterogeneity, the presence/localization of HSC/HSPC in the adult body or the definition of a hematopoietic niche, and thus whether the bone marrow in mammals or the anterior kidney in fish, they are the only/final sites of the blood formation process. Today we know that not (1, 14, 15).
Considering the latest findings on haematopoiesis in vertebrates, including mammals and fish, the conventional concept of HSCs, including their presence, development and differentiation, is evolving into a completely different paradigm. HSCs/HSPCs form a flexible heterogeneous population of cells with different potential for self-renewal and differentiation (14, 18, 77, 78). In mammals these cells in the state of homeostasis circulate and generate peripheral haematopoiesis, respond to differentiating signals and the presence of antigens, and produce myeloid cells, including dendritic cells (DC), in the periphery (14, 79). HSCs and HSPCs have been detected in the lungs and the gut of mammals - humans and mice (14, 80), and it was demonstrated (79) that these cells can migrate to other organs/tissues and return from the periphery to the bone marrow. It should be emphasized that also in the periphery, the integrity of HSCs is protected by the haematopoietic microenvironment, i.e. niches that are heterogeneous by nature (14), because they are formed, for example, by osteoblasts, endothelial cells, mesenchymal stromal cells, nerve cells and megakaryocytes (ME) (14). Moreover, when exposed to microenvironmental factors, HSCs develop and change autonomously and heteronomously (14). Cheng et al. (1), in the presence of data obtained through the use of advanced single-cell ‘omics’ techniques, indicated a model of continuous differentiation of HSCs, which is characterized by the absence of a discernible hierarchy. HSCs develop and differentiate gradually, in many directions, continuously and without creating a distinct hierarchy of organized progenitor populations (Figure 2). Cheng et al. (1) assumed that because of the identified heterogeneity of HSCs, the ability of the development of these cells into specific lineages is already acquired before their differentiation. Studies on samples of human bone marrow (11) and in D. rerio (18) revealed that profiled cells with a restricted monolineage originate from the continuum of undifferentiated HSPCs, i.e. cells that have the potential of myeloid and lymphoid cells, either innate or adaptive (Figure 2). A separate haematopoietic lineage was identified among HSCs, ‘preventing’ the formation of megakaryocytes (the equivalent of thrombocytes in teleost fish) and platelets, which acquire properties specific just for that lineage (76). This fact is particularly interesting, also in consideration of the functional potential (haemostatic and immune properties) of these cells in vertebrates (platelets in mammals and thrombocytes in fish) at various stages of phylogeny (5). Brown and Ceredig (15) indicated that it is very likely that in mammals HSCs and their ‘progeny’ are pluripotent, because regardless of the predetermined fate of their development they may switch it to an alternative one, closely related, and during their development/differentiation they are sensitive to many cytokines (THPO - thrombopoietin, EPO - erythropoietin, CSF3 - colony stimulating factor 3 [granulocyte-colony stimulating factor - G-CSF], CSF1 - colony stimulating factor 1 [macrophage-colony stimulating factor - M-CSF], CSF2 - colony stimulating factor 2 [granulocyte-macrophage- colony stimulating factor - GM-CSF], FLT3L - FMS-like tyrosine kinase 3 ligand), which determine the profiling of HSCs and formation of a specific lineage of haemopoietic cells. The local increase in the concentration of certain cytokines and the autoregulated expression of the receptor specific for this cytokine most likely determine the fate of HSCs’ differentiation process, which depends on the effects of cytokine-receptor interaction (15).
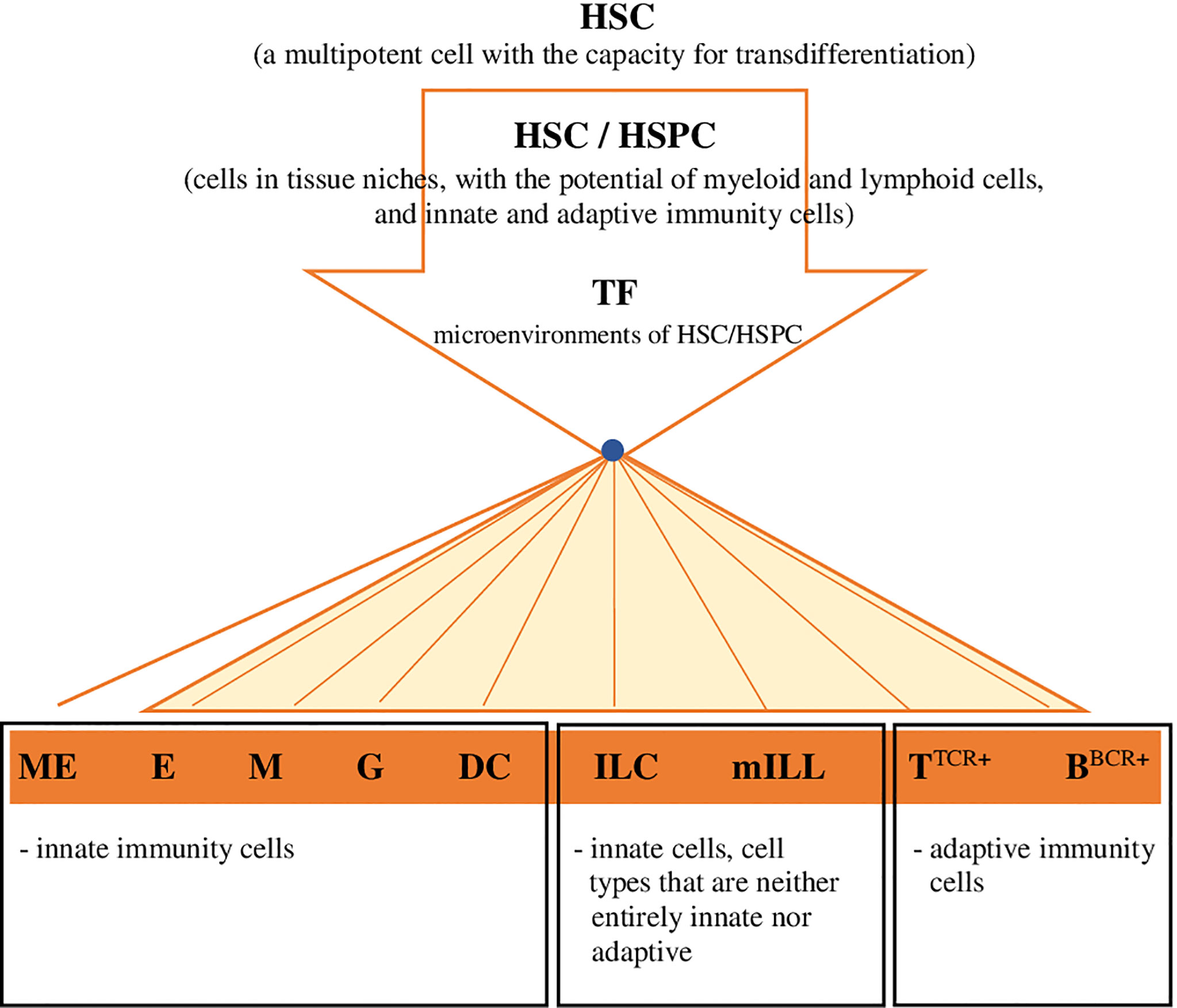
Figure 2 Hypothetical model for the development and differentiation of HSCs, in multiple directions specific for lineages, continuous and without creating a distinct hierarchy of organized progenitor populations (proposed by the authors). Explanatory noted: HSC, haematopoietic stem cell; HSC/HSPC, haematopoietic stem cell/hematopoietic progenitor cell (migratory/tissue,resident); TF, transcription factor (or sequence,specific DNA,binding factor) in the environment of HSC-HSPC cells; ME, megakaryocytes/source of platelets in mammals/thrombocytes in fish; E, erthcrocytes; M, monocytes/macrophages; G, granulocytes; DC, dendritic cells; ILC (ILC1; ILC2; ILC3/ NK; LTi), innate lymphoid cells; mILL (Tγδ lymohocytes [IEL, intraepithelial lymphocytes]; mucosal associated invariant T cells [MAIT]; NKT cells; B1 lymphocytes [unconventional BCD5+ lymphocytes]; marginal zone B cells), mammalian unconventional or innate-like lymphocytes; TTCR+, TTCR+ lymphocytes and their subpopulations; BBCR+, B BCR+ lymphocytes and their subpopulations.
In D. rerio HSCs/HSPCs migrate from CHT and further colonize the thymus, where T lymphocytes are formed (3 dpf), and the pronephros, where erythroid, myeloid and lymphoid – B lymphocytes cells are formed, except T lymphocytes (4 dpf) (2, 19, 32, 56, 62–64, 81). As emphasized by Macaulay et al. (18), zebrafish have cells of all haematopoietic lineages and orthologs of transcription factors and other genes that are also involved in the process of haematopoiesis in mammals, namely tal1, lmo2, lyl1, gata2, runx1, meis1, myb and erg specific for HSCs, fli1, gfi1b, gata1, cd61, cd42b specific for megakaryocyte/erythroid lineages, and gfi1, spi1b specific for myeloid cell lineage. It should be added that D. rerio has two GATA2 orthologs, ie gata2a and gata2b. The dominant and required for maintenance of HSCs is gata2b, expressed on HSCs and HSPCs, while gata2a dominates in the vascular system, including hemogenic endothelial cells (30). The high degree of comparability/functional similarity between fish and mammals also relates to signalling pathways and transcription mechanisms that are active during haematopoiesis in these two taxonomic groups of vertebrates (18, 23). According to Macaulay et al. (18), adult haematopoiesis in D. rerio is, as in mammals, continuous and asynchronous/flexible, and the pronephros in fish, like the bone marrow in mammals, accommodates all types of haematopoietic cells at different stages of differentiation. This was shown in studies on the origin and development of thrombocytes (18), based on single cell RNA sequencing (scRNA-seq), which allows for the analysis of differential expression (DE), grouping and classification of cells, but also the reconstruction of cell differentiation trajectory. This study (18) revealed that the differentiation of cells (acquisition of specific phenotypes) progresses along a one-dimensional, non-branching path. It was also found (18) that this process is correlated/consistent with the transcriptional programme, which is reflected in the stimulated or suppressed expression of genes specific for the programme of development, differentiation and functional determination of cells from a particular lineage, or restricting the proliferation of cells and their translational capacity. Macaulay et al. (18) emphasized that as this process continues, the number of expressed genes and mRNA content in the cell is reduced and limited to those which define a specific cell lineage. In addition to these data, information on defined sets of genes specific for HSCs, HSPCs, megakaryocytes/thrombocytes runx1,cd41 (kidney marrow [pronephros]); neutrophils mpx (kidney marrow [pronephros]); NK cells lck/rag1-/- (kidney marrow [pronephros]); B cells rag2 (kidney marrow [pronephros]); mature T cells lck (thymus) is also important (18). The heterogeneity of cells in the pronephros in adult D. rerio was also demonstrated by Tang et al. (75), based on the massively parallel transcriptomic method using indexing droplets (InDrops), single-cell RNA sequencing and t-distributed stochastic neighbour embedding (tSNE). In these studies, major haematopoietic cells in the pronephros of zebrafish were defined, i.e. neutrophils, progenitors, erythroid cells, HSCs/thrombocytes, B cells, T/NK cells, myeloid cells, macrophages, as well as seven unique kidney stromal-cell types. Novel genes of specific haematopoietic cell lineages were also identified (HSCs/HSPCsrunx1, thrombocytescd41, neutrophilsmpx, T/NKlck cells, NKlck/rag1-/- cells, Brag2 cells), and a new population of NKLmpeg1.1,ccl33.3,nkl.3,nkl.4,prf1.2,prf1.7 cells (NK-like), which conserve the genes typical for cells with cytotoxic and lytic capacity despite the lack of their association with Tlck lymphocytes. The analysis of findings (75) suggests that classically defined HSPCs involved in the lineage of erythroids and thrombocytes may also include progenitor cells with closely related transcriptional programmes.
The presented information shows that haematopoiesis in D. rerio, as in mammals, is a complex process, both at the embryonic stage and in adult individuals. Findings on the development of haematopoiesis and the role of the microenvironment/niche in the regulation of the formation and function of HSCs/HSPCs shed a new light on this process, especially in the context of details associated with the differentiation of HSCs and the resulting need to modify the paradigm on haematopoiesis. Despite the discoveries made to date, the process of haematopoiesis in D. rerio and other vertebrates requires further studies to explain, for example, the epigenetic characteristics of cells from specific haematopoietic lineages, taking into account different stages of development and a continuum of differentiation, or the presence of HSCs outside the bone marrow/pronephros niche, and define the ‘local’ capacity of these cells for self-renewal and differentiation into mature forms of certain haematopoietic lineages.
All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cheng H, Zheng Z, Cheng T. New Paradigms on Hematopoietic Stem Cell Differentiation. Protein Cell (2020) 11:34–44. doi: 10.1007/s13238-019-0633-0
2. Gore AV, Pillay LM, Galanternik MV, Weinstein BM. The Zebrafish: A Fintastic Model for Hematopoietic Development and Disease. Wiley Interdiscip Rev Dev Biol (2018) 7:e312. doi: 10.1002/wdev.312
3. Kobayashi I, Kondo M, Yamamori S, Kobayashi-Sun J, Taniguchi M, Kanemaru K, et al. Enrichment of Hematopoietic Stem/Progenitor Cells in the Zebrafish Kidney. Sci Rep (2019) 9:14205. doi: 10.1038/s41598-019-50672-5
4. Svoboda O, Stachura DL, Machoňová O, Pajer P, Brynda J, Zon LI, et al. Dissection of Vertebrate Hematopoiesis Using Zebrafish Thrombopoietin. Blood (2014) 124:220–8. doi: 10.1182/blood-2014-03-564682
5. Stosik M, Tokarz-Deptuła B, Deptuła W. Characterisation of Thrombocytes in Osteichthyes. J Vet Res (2019) 63:123–31. doi: 10.2478/jvetres-2019-0017
6. Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-Cell Hi-C Reveals Cell-to-Cell Variability in Chromosome Structure. Nature (2013) 502:59–64. doi: 10.1038/nature12593
7. Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee M, et al. Integrated Single-Cell Analysis Maps the Continuous Regulatory Landscape of Human Hematopoietic Differentiation. Cell (2018) 173:1535–48.e16. doi: 10.1016/j.cell.2018.03.074
8. Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, et al. Joint Profiling of Chromatin Accessibility and Gene Expression in Thousands of Single Cells. Science (2018) 361:1380–5. doi: 10.1126/science.aau0730
9. Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, et al. A Single Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell (2018) 174:1309–24.e18. doi: 10.1016/j.cell.2018.06.052
10. Satpathy AT, Saligrama N, Buenrostro JD, Wei Y, Wu B, Rubin AJ, et al. Transcript-Indexed ATAC-Seq for Precision Immune Profiling. Nat Med (2018) 24:580–90. doi: 10.1038/s41591-018-0008-8
11. Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, et al. Human Haematopoietic Stem Cell Lineage Commitment is a Continuous Process. Nat Cell Biol (2017) 19:271–81. doi: 10.1038/ncb3493
12. Karamitros D, Stoilova B, Aboukhalil Z, Hamey F, Reinisch A, Samitsch M, et al. Single-Cell Analysis Reveals the Continuum of Human Lympho-Myeloid Progenitor Cells. Nat Immunol (2018) 19:85–97. doi: 10.1038/s41590-017-0001-2
13. Laurenti E, Göttgens B. From Haematopoietic Stem Cells to Complex Differentiation Landscapes. Nature (2018) 553:418–26. doi: 10.1038/nature25022
14. Yokota T. “Hierarchy” and “Holacracy”; A Paradigm of the Hematopoietic System. Cells (2019) 8:1138. doi: 10.3390/cells8101138
15. Brown G, Ceredig R. Modeling the Hematopoietic Landscape. Front Cell Dev Biol (2019) 7:104. doi: 10.3389/fcell.2019.00104
16. Xu J, Wang Y, Guttorp P, Abkowitz JL. Visualizing Hematopoiesis as a Stochastic Process. Blood Adv (2018) 2:2637–45. doi: 10.1182/bloodadvances.2018023705
17. Zhen F, Lan Y, Yan B, Zhang W, Wen Z. Hemogenic Endothelium Specification and Hematopoietic Stem Cell Maintenance Employ Distinct Scl Isoforms. Development (2013) 140:3977–85. doi: 10.1242/dev.097071
18. Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, et al. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep (2016) 14:966–77. doi: 10.1016/j.celrep.2015.12.082
19. Wattrus SJ, Zon LI. Stem Cell Safe Harbor: The Hematopoietic Stem Cell Niche in Zebrafish. Blood Adv (2018) 2:3063–9. doi: 10.1182/bloodadvances.2018021725
20. Konantz M, Schürch C, Hanns P, Müller JS, Sauteur L, Lengerke C. Modeling Hematopoietic Disorders in Zebrafish. Dis Model Mech (2019) 12(9):dmm040360. doi: 10.1242/dmm.040360
21. Ma D, Zhang J, Lin H, Italiano J, Handin RI. The Identification and Characterization of Zebrafish Hematopoietic Stem Cells. Blood (2011) 118:289–97. doi: 10.1182/blood-2010-12-327403
22. Avagyan S, Zon LI. Fish to Learn: Insights Into Blood Development and Blood Disorders From Zebrafish Hematopoiesis. Hum Gene Ther (2016) 27:287–94. doi: 10.1089/hum.2016.024
23. Jagannathan-Bogdan M, Zon LI. Hematopoiesis. Development (2013) 140:2463–7. doi: 10.1242/dev.083147
24. Hsia N, Zon LI. Transcriptional Regulation of Hematopoietic Stem Cell Development in Zebrafish. Exp Hematol (2005) 33:1007–14. doi: 10.1016/j.exphem.2005.06.013
25. Capron C, Lécluse Y, Kaushik AL, Foudi A, Lacout C, Sekkai D, et al. The SCL Relative LYL-1 is Required for Fetal and Adult Hematopoietic Stem Cell Function and B-Cell Differentiation. Blood (2006) 107:4678–86. doi: 10.1182/blood-2005-08-3145
26. Greig KT, Carotta S, Nutt SL. Critical Roles for C-Myb in Hematopoietic Progenitor Cells. Semin Immunol (2008) 20:247–56. doi: 10.1016/j.smim.2008.05.003
27. Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, et al. The Transcription Factor Erg is Essential for Definitive Hematopoiesis and the Function of Adult Hematopoietic Stem Cells. Nat Immunol (2008) 9:810–9. doi: 10.1038/ni.1617
28. Orkin SH, Zon LI. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell (2008) 132:631–44. doi: 10.1016/j.cell.2008.01.025
29. Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential Expression of Hox, Meis1, and Pbx1 Genes in Primitive Cells Throughout Murine Hematopoietic Ontogeny. Exp Hematol (2002) 30:49–57. doi: 10.1016/S0301-472X(01)00757-3
30. Gioacchino E, Koyunlar C, Zink J, de Looper H, de Jong M, Dobrzycki T, et al. Essential Role for Gata2 in Modulating Lineage Output From Hematopoietic Stem Cells in Zebrafish. Blood Adv (2021) 5:2687–700. doi: 10.1182/bloodadvances.2020002993
31. Clements WK, Traver D. Signalling Pathways That Control Vertebrate Haematopoietic Stem Cell Specification. Nat Rev Immunol (2013) 13:336–48. doi: 10.1038/nri3443
32. Wolf A, Aggio J, Campbell C, Wright F, Marquez G, Traver D, et al. Zebrafish Caudal Haematopoietic Embryonic Stromal Tissue (CHEST) Cells Support Haematopoiesis. Sci Rep (2017) 7:44644. doi: 10.1038/srep44644
33. Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic Stem Cell Fate is Established by the Notch–Runx Pathway. Genes Dev (2005) 19:2331–42. doi: 10.1101/gad.1337005
34. Kawakami K. Tol2: A Versatile Gene Transfer Vector in Vertebrates. Genome Biol (2007) 8:S7. doi: 10.1186/gb-2007-8-s1-s7
35. Carradice D, Lieschke GJ. Zebrafish in Hematology: Sushi or Science? Blood (2008) 111:3331–42. doi: 10.1182/blood-2007-10-052761
36. Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, et al. Hematopoietic Stem Cell Arrival Triggers Dynamic Remodeling of the Perivascular Niche. Cell (2015) 160:241–52. doi: 10.1016/j.cell.2014.12.032
37. Li P, Lahvic JL, Binder V, Pugach EK, Riley EB, Tamplin OJ, et al. Epoxyeicosatrienoic Acids Enhance Embryonic Haematopoiesis and Adult Marrow Engraftment. Nature (2015) 523:468–71. doi: 10.1038/nature14569
38. Bertrand JY, Traver D. Hematopoietic Cell Development in the Zebrafish Embryo. Curr Opin Hematol (2009) 16:243–8. doi: 10.1097/MOH.0b013e32832c05e4
39. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: A Multisite Gateway-Based Construction Kit for Tol2 Transposon Transgenesis Constructs. Dev Dyn (2007) 236:3088–99. doi: 10.1002/dvdy.21343
40. White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell (2008) 2:183–9. doi: 10.1016/j.stem.2007.11.002
41. D’Agati G, Beltre R, Sessa A, Burger A, Zhou Y, Mosimann C, et al. A Defect in the Mitochondrial Protein Mpv17 Underlies the Transparent Casper Zebrafish. Dev Biol (2017) 430:11–7. doi: 10.1016/j.ydbio.2017.07.017
42. Rajpurohit SK, Gopal A, Mon MY, Patel NG, Arora V. Development of Tg(UAS:SEC-Hsa.ANXA5-YFP,myl7:RFP); Casper(roy–/–,nacre–/–) Transparent Transgenic In Vivo Zebrafish Model to Study the Cardiomyocyte Function. Cells (2021) 10:1963. doi: 10.3390/cells10081963
43. Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, et al. Genetic Compensation Induced by Deleterious Mutations But Not Gene Knockdowns. Nature (2015) 524:230–3. doi: 10.1038/nature14580
44. Rouf MA, Wen L, Mahendra Y, Wang J, Zhang K, Liang S, et al. The Recent Advances and Future Perspectives of Genetic Compensation Studies in the Zebrafish Model. Genes Dis (2022). doi: 10.1016/j.gendis.2021.12.003
45. Paik EJ, Zon LI. Hematopoietic Development in the Zebrafish. Int J Dev Biol (2010) 54:1127–37. doi: 10.1387/ijdb.093042ep
46. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. Mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood (2011) 117:e49–56. doi: 10.1182/blood-2010-10-314120
47. Kissa K, Herbomel P. Blood Stem Cells Emerge From Aortic Endothelium by a Novel Type of Cell Transition. Nature (2010) 464:112–5. doi: 10.1038/nature08761
48. Mahony CB, Fish RJ, Pasche C, Bertrand JY. Tfec Controls the Hematopoietic Stem Cell Vascular Niche During Zebrafish Embryogenesis. Blood (2016) 128:1336–45. doi: 10.1182/blood-2016-04-710137
49. Mahony CB, Pasche C, Bertrand JY. Oncostatin M and Kit-Ligand Control Hematopoietic Stem Cell Fate During Zebrafish Embryogenesis. Stem Cell Rep (2018) 10:1920–34. doi: 10.1016/j.stemcr.2018.04.016
50. Travnickova J, Tran Chau V, Julien E, Mateos-Langerak J, Gonzalez C, Lelièvre E, et al. Primitive Macrophages Control HSPC Mobilization and Definitive Haematopoiesis. Nat Commun (2015) 6:6227. doi: 10.1038/ncomms7227
51. Theodore LN, Hagedorn EJ, Cortes M, Natsuhara K, Liu SY, Perlin JR, et al. Distinct Roles for Matrix Metalloproteinases 2 and 9 in Embryonic Hematopoietic Stem Cell Emergence, Migration, and Niche Colonization. Stem Cell Rep (2017) 8:1226–41. doi: 10.1016/j.stemcr.2017.03.016
52. Davidson AJ, Zon LI. The ‘Definitive’ (and ‘Primitive’) Guide to Zebrafish Hematopoiesis. Oncogene (2004) 23:7233–46. doi: 10.1038/sj.onc.1207943
53. Murayama E, Sarris M, Redd M, Le Guyader D, Vivier C, Horsley W, et al. NACA Deficiency Reveals the Crucial Role of Somite-Derived Stromal Cells in Haematopoietic Niche Formation. Nat Commun (2015) 6:8375. doi: 10.1038/ncomms9375
54. Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive Hematopoiesis Initiates Through a Committed Erythromyeloid Progenitor in the Zebrafish Embryo. Development (2007) 134:4147–56. doi: 10.1242/dev.012385
55. Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch Signaling Distinguishes 2 Waves of Definitive Hematopoiesis in the Zebrafish Embryo. Blood (2010) 115:2777–83. doi: 10.1182/blood-2009-09-244590
56. Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D. Haematopoietic Stem Cells Derive Directly From Aortic Endothelium During Development. Nature (2010) 464:108–11. doi: 10.1038/nature08738
57. Ciau-Uitz A, Monteiro R, Kirmizitas A, Patient R. Developmental Hematopoiesis: Ontogeny, Genetic Programming and Conservation. Exp Hematol (2014) 42:669–83. doi: 10.1016/j.exphem.2014.06.001
58. Henninger J, Santoso B, Hans S, Durand E, Moore J, Mosimann C, et al. Clonal Fate Mapping Quantifies the Number of Haematopoietic Stem Cells That Arise During Development. Nat Cell Biol (2017) 19:17–27. doi: 10.1038/ncb3444
59. Xia J, Kang Z, Xue Y, Ding Y, Gao S, Zhang Y, et al. A Single-Cell Resolution Developmental Atlas of Hematopoietic Stem and Progenitor Cell Expansion in Zebrafish. Proc Natl Acad Sci (2021) 118:e2015748118. doi: 10.1073/pnas.2015748118
60. Xue Y, Lv J, Zhang C, Wang L, Ma D, Liu F. The Vascular Niche Regulates Hematopoietic Stem and Progenitor Cell Lodgment and Expansion via Klf6a-Ccl25b. Dev Cell (2017) 42:349–62.e4. doi: 10.1016/j.devcel.2017.07.012
61. Li D, Xue W, Li M, Dong M, Wang J, Wang X, et al. VCAM-1+ Macrophages Guide the Homing of HSPCs to a Vascular Niche. Nature (2018) 564:119–24. doi: 10.1038/s41586-018-0709-7
62. Boisset J-C, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In Vivo Imaging of Haematopoietic Cells Emerging From the Mouse Aortic Endothelium. Nature (2010) 464:116–20. doi: 10.1038/nature08764
63. Chen AT, Zon LI. Zebrafish Blood Stem Cells. J Cell Biochem (2009) 108:35–42. doi: 10.1002/jcb.22251
64. Jin H, Xu J, Wen Z. Migratory Path of Definitive Hematopoietic Stem/Progenitor Cells During Zebrafish Development. Blood (2007) 109:5208–14. doi: 10.1182/blood-2007-01-069005
65. Nguyen PD, Hollway GE, Sonntag C, Miles LB, Hall TE, Berger S, et al. Haematopoietic Stem Cell Induction by Somite-Derived Endothelial Cells Controlled by Meox1. Nature (2014) 512:314–8. doi: 10.1038/nature13678
66. Monteiro R, Pinheiro P, Joseph N, Peterkin T, Koth J, Repapi E, et al. Transforming Growth Factor β Drives Hemogenic Endothelium Programming and the Transition to Hematopoietic Stem Cells. Dev Cell (2016) 38:358–70. doi: 10.1016/j.devcel.2016.06.024
67. Espín-Palazón R, Stachura DL, Campbell CA, García-Moreno D, Cid ND, Kim AD, et al. Proinflammatory Signaling Regulates Hematopoietic Stem Cell Emergence. Cell (2014) 159:1070–85. doi: 10.1016/j.cell.2014.10.031
68. Bresciani E, Carrington B, Wincovitch S, Jones M, Gore AV, Weinstein BM, et al. Cbfβ and RUNX1 Are Required at 2 Different Steps During the Development of Hematopoietic Stem Cells in Zebrafish. Blood (2014) 124:70–8. doi: 10.1182/blood-2013-10-531988
69. Gore AV, Weinstein BM. DNA Methylation in Hematopoietic Development and Disease. Exp Hematol (2016) 44:783–90. doi: 10.1016/j.exphem.2016.04.013
70. Gore AV, Athans B, Iben JR, Johnson K, Russanova V, Castranova D, et al. Epigenetic Regulation of Hematopoiesis by DNA Methylation. eLife (2016) 5:e11813. doi: 10.7554/eLife.11813
71. Campbell C, Su T, Lau RP, Shah A, Laurie PC, Avalos B, et al. Zebrafish Embryonic Stromal Trunk (ZEST) Cells Support Hematopoietic Stem and Progenitor Cell (HSPC) Proliferation, Survival, and Differentiation. Exp Hematol (2015) 43:1047–61. doi: 10.1016/j.exphem.2015.09.001
72. Stachura DL, Reyes JR, Bartunek P, Paw BH, Zon LI, Traver D. Zebrafish Kidney Stromal Cell Lines Support Multilineage Hematopoiesis. Blood (2009) 114:279–89. doi: 10.1182/blood-2009-02-203638
73. Glass TJ, Lund TC, Patrinostro X, Tolar J, Bowman TV, Zon LI, et al. Stromal Cell–Derived Factor-1 and Hematopoietic Cell Homing in an Adult Zebrafish Model of Hematopoietic Cell Transplantation. Blood (2011) 118:766–74. doi: 10.1182/blood-2011-01-328476
74. Quillien A, Moore JC, Shin M, Siekmann AF, Smith T, Pan L, et al. Distinct Notch Signaling Outputs Pattern the Developing Arterial System. Development (2014) 141:1544–52. doi: 10.1242/dev.099986
75. Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, et al. Dissecting Hematopoietic and Renal Cell Heterogeneity in Adult Zebrafish at Single-Cell Resolution Using RNA Sequencing. J Exp Med (2017) 214:2875–87. doi: 10.1084/jem.20170976
76. Carrelha J, Meng Y, Kettyle LM, Luis TC, Norfo R, Alcolea V, et al. Hierarchically Related Lineage-Restricted Fates of Multipotent Haematopoietic Stem Cells. Nature (2018) 554:106–11. doi: 10.1038/nature25455
77. Quesenberry PJ, Goldberg LR, Dooner MS. A Stem Cell Apostasy: A Tale of 4 H Words. Stem Cells (2015) 33:15–20. doi: 10.1002/stem.1829
78. Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, et al. Clonal Analysis Unveils Self-Renewing Lineage-Restricted Progenitors Generated Directly From Hematopoietic Stem Cells. Cell (2013) 154:1112–26. doi: 10.1016/j.cell.2013.08.007
79. Massberg S, Schaerli P, Knezevic-Maramica I, Köllnberger M, Tubo N, Moseman EA, et al. Immunosurveillance by Hematopoietic Progenitor Cells Trafficking Through Blood, Lymph, and Peripheral Tissues. Cell (2007) 131:994–1008. doi: 10.1016/j.cell.2007.09.047
80. Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, et al. The Lung is a Site of Platelet Biogenesis and a Reservoir for Hematopoietic Progenitors. Nature (2017) 544:105–9. doi: 10.1038/nature21706
Keywords: haematopoietic stem cells, embryonic period haematopoiesis, post-embryonic haematopoiesis, haematopoietic niches, Danio rerio (zebrafish)
Citation: Stosik M, Tokarz-Deptuła B and Deptuła W (2022) Haematopoiesis in Zebrafish (Danio Rerio). Front. Immunol. 13:902941. doi: 10.3389/fimmu.2022.902941
Received: 23 March 2022; Accepted: 05 May 2022;
Published: 02 June 2022.
Edited by:
Geert Wiegertjes, Wageningen University and Research, NetherlandsReviewed by:
Jian Huang, Coriell Institute For Medical Research, United StatesCopyright © 2022 Stosik, Tokarz-Deptuła and Deptuła. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beata Tokarz-Deptuła, YmVhdGEudG9rYXJ6LWRlcHR1bGFAdXN6LmVkdS5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.