- 1State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 2Department of Gynecology and Obstetrics, Bethune International Peace Hospital, Shijiazhuang, China
- 3Institute of Military Cognition and Brain Sciences, Beijing, China
Specific therapeutics are not available for acute lung injury (ALI) induced by ricin toxin (RT). Inhibiting the host immune response in the course of pulmonary ricinosis is hypothesized to be of benefit and can be achieved by impairing granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling, thereby reducing the pro-inflammatory response to exogenous foreign body invasion. However, it is unknown whether mice with impaired GM-CSF signaling can survive after RT inhalation. To test this, colony stimulating factor 2 receptor alpha (Csf2ra) knockout (KO) mice that lack GM-CSF signaling and wild-type (WT) mice models of intratracheal exposure to a lethal dose (2× LD50) of RT were established. Survival was greater in Csf2ra KO mice 21 days after RT inhalation compared with WT mice. Highly co-expressed genes that probably attenuated the pro-inflammatory response in the lung of Csf2ra KO mice were identified. Bioinformatics analysis revealed that transcriptome changes involved mostly inflammation-related genes after RT exposure in both Csf2ra KO mice and WT mice. However, the activity levels of pro-inflammatory pathways, such as the TNF signaling pathway and NF-κB signaling pathway, in Csf2ra KO mice were significantly decreased and the degree of neutrophil chemotaxis and recruitment inhibited after RT-exposure relative to WT mice. RT-qPCR and flow cytometry validated results of RNA-Seq analysis. This work provides potential avenues for host-directed therapeutic applications that can mitigate the severity of ALI-induced by RT.
Introduction
Acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) are well characterized and readily recognized clinical disorders caused by clinical insults to the lung (1) that are the leading cause of acute respiratory failure, with a high mortality ratio (2). ALI/ARDS can result from direct injury caused by bacterial or viral pneumonia, as well as from toxin inhalation (3, 4).
Ricin toxin (RT) is a 60–65 kDa protein extracted from the seeds of the castor plant (Ricinus communis), a widespread plant in tropical regions that is readily available in the global market. RT belongs to the type 2 ribosome-inactivating proteins (RIP) family, is classified as a Category B agent by the U.S. Centers for Disease Control and Prevention (CDC) and is considered a potential bioterror agent due to its high availability and ease of preparation (5). The clinical manifestation of inhaled RT in animal models is closely related to ALI/ARDS, and involves pulmonary proinflammatory cytokine upregulation, massive neutrophil infiltration, and severe edema (5). Pulmonary RT intoxication may affect the transcriptome profile in lung tissues and lead to inflammatory responses that release inflammatory cytokines and activate pro-inflammatory signaling pathways. Currently, no specific therapeutics are available for the treatment of ALI/ARDS induced by RT (5). New therapeutics are urgently needed to effectively treat pulmonary RT toxicity. Emerging evidence suggests that the dramatic inflammatory response induced by RT exposure is the leading cause of mortality (6, 7). Pulmonary ricin toxicity is characterized by a lung pathology associated with a “cytokine storm” (8–10) that is directly caused by an imbalance between pro-inflammatory and anti-inflammatory cytokines (11). The cytokine storm results from a sudden acute increase in circulating levels of several pro-inflammatory cytokines, including IL-6, IL-1, TNF-α and interferon (12). Therefore, applying disease-modifying countermeasures, including inhibiting the host immune response, is a logical approach to treating the course of pulmonary ricinosis.
Granulocyte-macrophage colony-stimulating factor (GM-CSF) is recognized as a differentiation and growth factor for myeloid hematopoietic cells that regulates immune cell proliferation, growth, differentiation, survival, activation and function (13, 14). GM-CSF also bridges innate and adaptive immunity, playing important roles in modulating the immune response against foreign substances. The colony stimulating factor 2 receptor alpha (Csf2ra) gene encodes GM-CSF receptor α (CD116) in mice. Defective GM-CSF receptor signaling results in abnormal development and functionality of immune cells and impairment of the host immune response (14).
Our study aimed to look for effective immunotherapy targets for ALI induced by RT-exposure. We thus challenged Csf2ra knockout (KO) mice and wild-type (WT) mice with 2× LD50 of RT by aerosolized intratracheal inoculation and compared lung tissue pathology and survival in the groups. We also extracted mRNA at various time points post RT-exposure and applied bioinformatic techniques to characterize transcriptome changes. Our results indicate that Csf2ra KO mice have improved survival after RT-exposure and transcriptome analysis suggests this was a result of attenuated pro-inflammatory responses. This work provides direction for finding a therapeutic solution to mitigate severity of ALI-induced by RT.
Materials and methods
RT preparation
The RT used in this investigation was donated by the Immunology Laboratory of the Beijing Institute of Microbiology and Epidemiology. Extraction and purification methods for RT were done according to published literature (15). Purified RT was stored in a freezer at -80°C until use and all relevant experiments were conducted in a biosafety level-3 laboratory.
Laboratory animals
Female C57BL/6N WT mice (aged 8–10 weeks) and female Csf2ra-/- (Csf2ra KO) mice (aged 8–10 weeks) were obtained from Cyagen (Guangzhou, China). All experimental mice used were specific-pathogen-free (SPF). Mice were maintained on a 12:12 h light:dark cycle and adaptively raised for 1 week with free access to water and food before starting the experiment. This study was approved by the Institute of Animal Care and Use Committee (IACUC) at the Academy of Military Medical Sciences (AMMS), ethical approval number IACUC-AMMS-2017-031.
Mouse toxicity studies
Groups of Csf2ra KO mice (n = 10) and WT mice (n = 10) were inoculated intratracheally with 2× LD50 of RT (approximately 10 μg/kg body weight solution in PBS 50 μl per mouse). The method of intratracheal inoculation followed published literature (16). Briefly, mice were anesthetized with an intraperitoneal injection of 70 mg/kg body weight pentobarbital sodium. Mice were placed in a supine position, the mouth opened, and the tongue gently moved aside using forceps to better cannulate the trachea. The tracheal opening was visualized by inserting a laryngoscope (Huironghe Company, Beijing, China), then a micro sprayer (Huironghe Company) was inserted at the larynx (near the tracheal bifurcation) of the mouse and intratracheal inoculation of aerosol was administered. Mice were then allowed to recover from the anesthetic. The RT-challenged mice were housed in cages within a biosafety level 3 facility and monitored daily for survival.
Pathological analysis
Csf2ra KO and WT mice were sacrificed at 4 h (n = 3), 12 h (n = 5) and 72 h (n = 5) after intratracheal inoculation of RT. Lungs were dissected and hematoxylin-eosin staining (H&E) used to check for morphological changes in the lung tissue of Csf2ra KO and WT mice. The lungs of mice in all groups were immersed in a general-purpose tissue fixation solution for ≥48 h, embedded in paraffin, sectioned to a thickness of 4 µm, and then subjected to H&E and photographed. Pathological score was evaluated by two independent pathologists. A semi-quantitative assessment of lung injury was conducted by estimating the following parameters: alveolar septal thickening, perivasculitis, peribronchiolitis, neutrophil infiltration, lymphocytic infiltration, monocyte infiltration, vascular leakage, alveolar edema, hyaline membrane formation, bleeding, bronchial epithelial sloughing/necrosis, and endothelial injury. Each parameter was graded on a scale from 0 to 4, where 0 = normal; 1 = mild; 2 = moderate, 3 = severe, 4 = very severe injury. The total lung inflammation score was expressed as the summed parameter scores.
RNA extraction, library construction and sequencing
In this experiment, Csf2ra KO and WT mice were divided randomly into four groups. The first was a control group that received an intratracheal inoculation of PBS without ricin and was then immediately sacrificed (0 h) via CO2 anesthetization followed by cervical dislocation and dissection. The remaining three experimental groups received intratracheal inoculation of aerosolized ricin and mice in each group were sacrificed via CO2 anesthetization followed by cervical dislocation and dissection at 4, 12 and 72 h after inoculation. During dissection, the lungs of mice were removed and lung lobes ground for RNA extraction and transcriptome sequencing.
Total RNA was extracted from the lung tissue using RNAprep Pure Tissue Kit (Tiangen, DP431), according to product instructions. The purity of the sample was determined by NanoPhotometer (IMPLEN, CA, USA). The concentration and integrity of RNA samples were tested using an Agilent 2100 RNA nano 6000 assay kit (Agilent Technologies, CA, USA). All samples were sent to Easyresearch Technology Company in China for sequencing. All cDNA libraries were sequenced using a paired-end strategy on the NovaSeq 6000 S4 platform, using NovaSeq 6000 S4 Reagent kit V1.5.
Pre-processing of RNA-Seq data
Sequencing data were filtered using a Perl script that: (1) removed reads containing the sequencing adapter; (2) removed reads with >15% low-quality base ratio (base quality ≤5); and (3) removed reads whose unknown base (‘N’ base) ratio was >5%. The resulting clean reads were stored in FASTQ format. Clean reads were mapped to the reference genome using HISAT2 (v2.1.0). Bowtie2 (v2.2.3) was used to align the clean reads to the reference coding gene set.
Statistical analysis of RNA-Seq data
Statistical analysis and plotting of RNA-Seq data were done using R (R version 4.1.0). Differentially expressed genes (DEGs) were identified using the EdgeR package (17, 18). Selection criteria for identifying DEGs were an adjusted P-value <0.05 and a |log2FoldChange| >1. The Bioconductor package clusterProfiler (19) for Gene Ontology (GO) and pathway-enrichment analysis (Kyoto Encyclopedia of Genes and Genomes, KEGG) was used to analyze DEGs and gene sets.
WGCNA
Weighted gene co-expression network analysis (WGCNA) was performed using the online website ImageGP (http://www.ehbio.com/Cloud_Platform/front/#/) with default values. Total expression variance in the top 5000 most variable genes in our study was used as input data to reduce amount of calculation required.
Time series analysis
The Bioconductor package maSigPro uses a two-step regression strategy for time series analysis. First, significant DEGs among different treatment groups are identified. Second, genes with similar expression patterns are grouped for cluster analysis and visualization. We used the maSigPro package (version 1.62.0) (20, 21) to analyze differences in gene expression patterns in Csf2ra KO and WT mice at 0, 4, 12 and 72 h post-inhalation.
Estimating the abundance of immune cells
The abundance of 10 types of immune cells was estimated using ImmuCellAI (22, 23) (http://bioinfo.life.hust.edu.cn/ImmuCellAI). This tool uses a gene set signature-based method with RNA-Seq data.
Flow cytometry analysis
Csf2ra KO mice and WT mice were sacrificed at 0 h (intratracheal inoculation of PBS) and 12 h (intratracheal inoculation of RT), and the lungs of mice were separated and immersed in tissue digestion solution (1.5 mg/ml collagenase A + 0.4 mg/ml DNase I + 1.5 U/ml dispase II in Hank’s Balanced Salt Solution (HBSS) containing 5% FBS and 10 mM HEPES) for 30 min. Digested tissues were ground, filtered and centrifuged at 3500 rpm for 10 min at 4°C. Red blood cells were lysed in 1 ml of RBC lysing buffer (Hao Yang Biological Products Technology Co., Ltd., Tianjin, China) and then single cell suspensions were filtered through a 40 µm nylon cell filter. Cells were counted by a Countess II FL automatic counter (Thermo Fisher Scientific, Waltham, MA, USA). For flow cytometry analysis, 2 × 106 cells were removed per tube and FcγR blocked for 20 min at 4°C using CD16/CD32 antibody (catalog no. 553141, BD Biosciences, San Jose, CA, USA). Cell suspensions were stained with fluorochrome-conjugated antibodies for 30 min and subjected to flow cytometry analysis using a BD FACSymphony A5 flow cytometer (BD Biosciences, Becton Dickinson, USA). The following fluorophore-conjugated antibodies were used: Fixable Viability Stain 510 (mouse, BD Biosciences, 564406); anti-CD45 (mouse, BD Biosciences, 564279); and anti-Ly6G (mouse, BioLegend, 127641). Data were analyzed with Flowjo version 10 (Ashland, OR, USA).
Flow cytometry data was analyzed using two-way ANOVAs followed by Sidak’s multiple comparisons tests. Statistical significance was defined as P <0.05. All data are expressed as mean ± standard deviation (SD) from three biological repeats. Statistical analyses and data plots for these data were done with Prism8 (GraphPad Software).
RT-qPCR confirmation
To validate RNA-Seq analysis, seven genes from the RNA-Seq analysis were selected for RT-qPCR, ensuring genes chosen were detected at 0, 4 and 12 h after inhalation. RNA was isolated from the tissue using RNAprep Pure Tissue (KitTiangen, DP431) according to the manufacturer’s instructions. RNA (0.8 µg) was used to synthesize cDNA using ReverTra Ace qPCR RT kit (TOYOBO, FSQ-301). The qPCR primers designed for the detection of mouse cDNA are given in Table 1. SYBR Green qPCR Master Mix (Servicebio, G3321-15) was used following the manufacturer’s instructions with 7500Fast DX Real-time PCR (Life Technologies Holdings Pte Ltd, T-1000-BYZB/SIN 5375-2013). All primers were verified for the production of a single specific PCR product with a melting curve program.
SDS–PAGE and western blotting
The lung of mice was lysed in RIPA lysis buffer (Solarbio, protease inhibitors and phosphatase inhibitors). Equivalent protein quantities were subjected to SDS–PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% non-fat milk or 5% BSA for 1 h at room temperature, and then probed with the indicated primary antibodies, followed by the appropriate HRP-conjugated anti-mouse/rabbit secondary antibodies (KPL). Immunoreactive bands were visualized with a chemiluminescence kit (Pierce). Antibodies against the following were used: p65 (1:1,000,CST), p-p65 (1:1,000, CST), IκB (1:1,000, CST), actin (1:5,000, proteintech), p-IκB (1:1,000, CST), TNF (1:1000, Abcam), GAPDH (1:5,000 proteintech).
Results
Csf2ra KO mice acquire protection from RT-induced mortality and morbidity
To investigate the role of Csf2ra on the survival time of mice after RT challenge, Csf2ra KO (n = 10) and WT (n = 10) mice were challenged with 2 × LD50 of RT by aerosolized intratracheal inoculation. We recorded death and survival events for the subsequent 21 days and survival curves are plotted in Figure 1A. The Csf2ra KO group began to die on day 10 post-RT exposure, while the WT group began to die on day 6 post-RT exposure. On day 21, the survival rates of mice in the Csf2ra KO and WT groups were 70% and 10%, respectively. These results indicate that Csf2ra deletion decreases mortality in mice after ALI induced by RT.
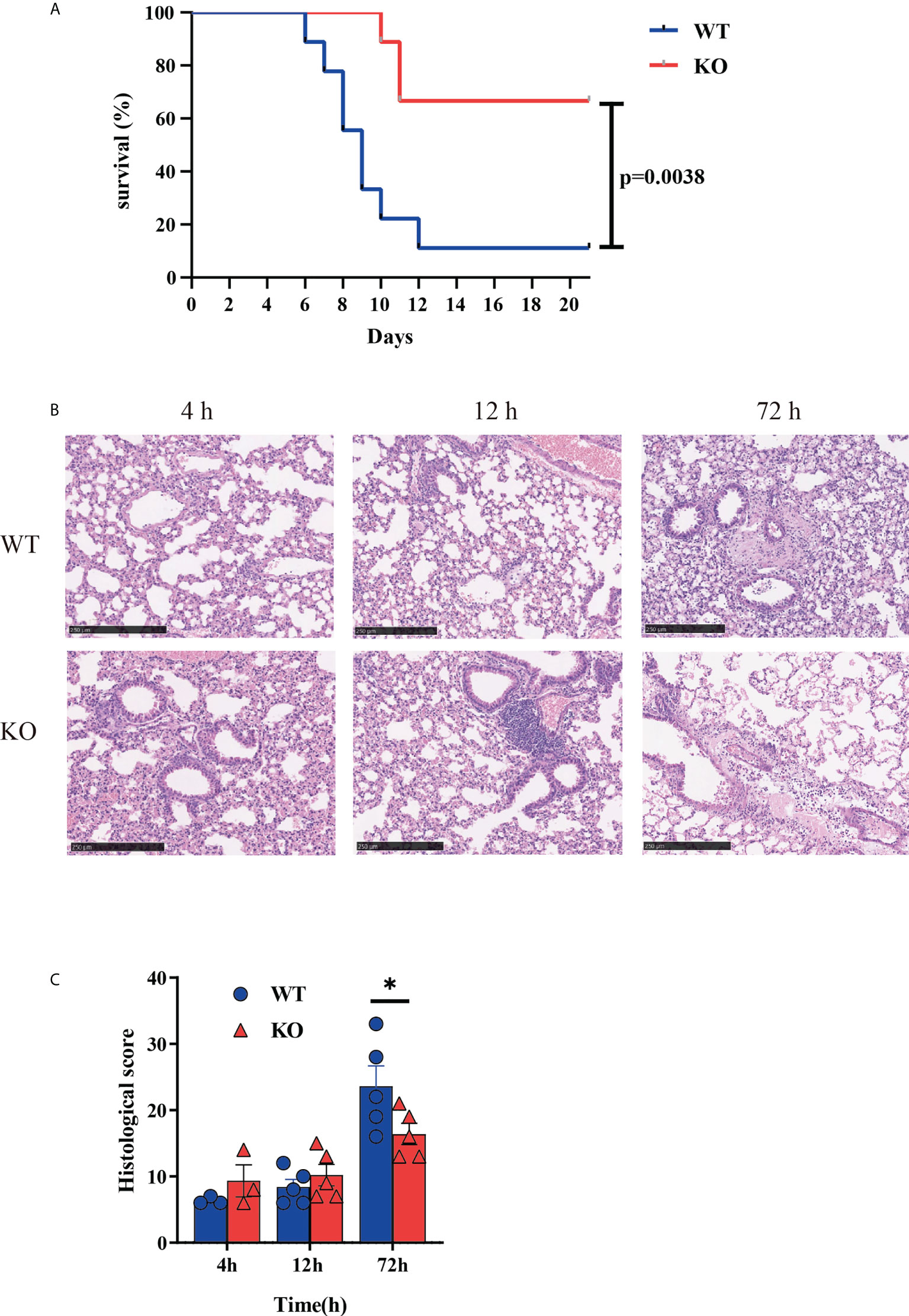
Figure 1 Csf2ra KO mice acquire protection from RT-induced mortality and morbidity. (A) Survival curves for Csf2ra KO mice (n = 10) and WT mice (n = 10); survival rate of the two groups is significantly different (P = 0.0038). (B) H&E stained lung tissue from each treatment group at three time points. Scale bar, 250 μm. (C) Pathological score for each group. *P <0.05.
Given the significantly reduced mortality in Csf2ra mice, we next examined whether Csf2ra deletion affected pathology of lungs in mice after RT exposure. Histopathological examination of the lung was performed for Csf2ra KO mice and WT mice at 0, 12 and 72 h post-exposure. At 72 h post-RT exposure, the lung of WT mice showed increased vascular leakage, the infiltration of inflammatory cells with edema, cellulose, necrosis and sloughing of bronchial epithelium (Figure 1B). Lung tissues from KO mice had significantly lower pathology scores than those in the WT group 72 h after RT exposure (Figure 1C).
WGCNA identified gene co-expression networks
A cluster analysis of cleaned RNA-Seq data identified 10 modules (Figure 2A). Analysis of the module–trait relationships revealed that module “black” with 48 genes was highly positively correlated with WT mice (r = 0.635, P < 0.001), while module “red” with 54 genes was highly positively correlated with Csf2ra KO mice (r = 0.684, P < 0.001). Subsequently, we analyzed the levels of expression of the modules associated with genes in the Csf2ra KO mice and WT mice at different time points (Supplementary Figures 1, 2). Module “black” included the genes Tnf, Cd14, Fas, Ccl4, Csf2rb and Tnfaip2. The expression levels of these genes in WT mice were higher than in Csf2ra KO mice. While the expression levels of genes in module “red” were lower in WT mice than in Csf2ra KO mice. To explore the biological processes in which these co-expressed genes participate, we conducted KEGG analysis and GO analysis. The KEGG analysis showed that the genes in module “black” were related to cytokine−cytokine receptor interaction, the toll−like receptor signaling pathway, the NF−κB signaling pathway and the chemokine signaling pathway (Figure 2B). Biological processes in the GO analysis showed that genes in the “red” module were related to immunoglobulin production, complement activation and B cell mediated immunity (Figure 2C).
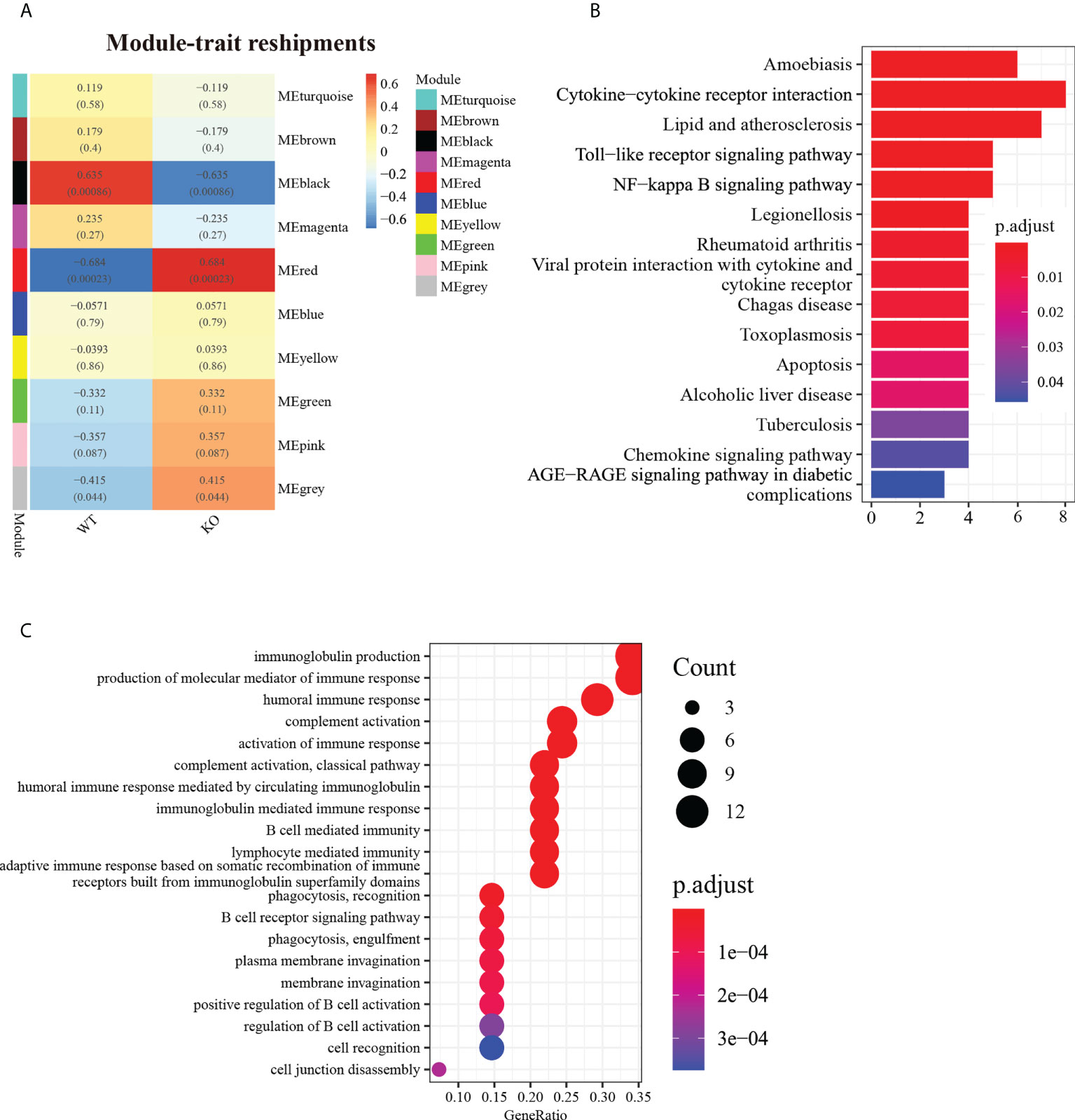
Figure 2 WGCNA identified gene co-expression networks in Csf2ra KO mice and WT mice. (A) Topological overlap matrix plot showing pairwise gene correlations within each module. Genes within different modules are labeled with different colors according to WGCNA’s conventions. (B) The top 15 KEGG entries for genes found in the “black” module. The x-axis represents gene count and the y-axis is KEGG terms. (C) The top 20 entries in GO analysis for biological processes in the “red” module.
DEGs analysis revealed that toxic effects induced by RT occurred in both WT mice and Csf2ra KO mice
In an effort to make sense of the host response in both the Csf2ra KO and WT groups, we compared Csf2ra KO mice and WT mice after RT inhalation to their respective steady-states for differential gene expression analysis. A total of 6343 genes were differentially expressed in the Csf2ra KO mice and WT mice at three time points (Supplementary Figure 3). The number of DEGs in the Csf2ra KO mice and WT mice gradually increased over time (Figure 3A). KEGG pathway analysis showed that the upregulated genes of each group were related to cytokine-cytokine receptor interaction, the TNF signaling pathway, the IL-17 signaling pathway and the NF-κB signaling pathway in both Csf2ra KO mice and WT mice at all time points after RT-exposure (Figure 3B). The DEG and KEGG analyses indicate that the lung tissue transcription profiles in the Csf2ra KO mice and WT mice were largely dominated by increases in the pro-inflammatory signaling pathway after RT inhalation.
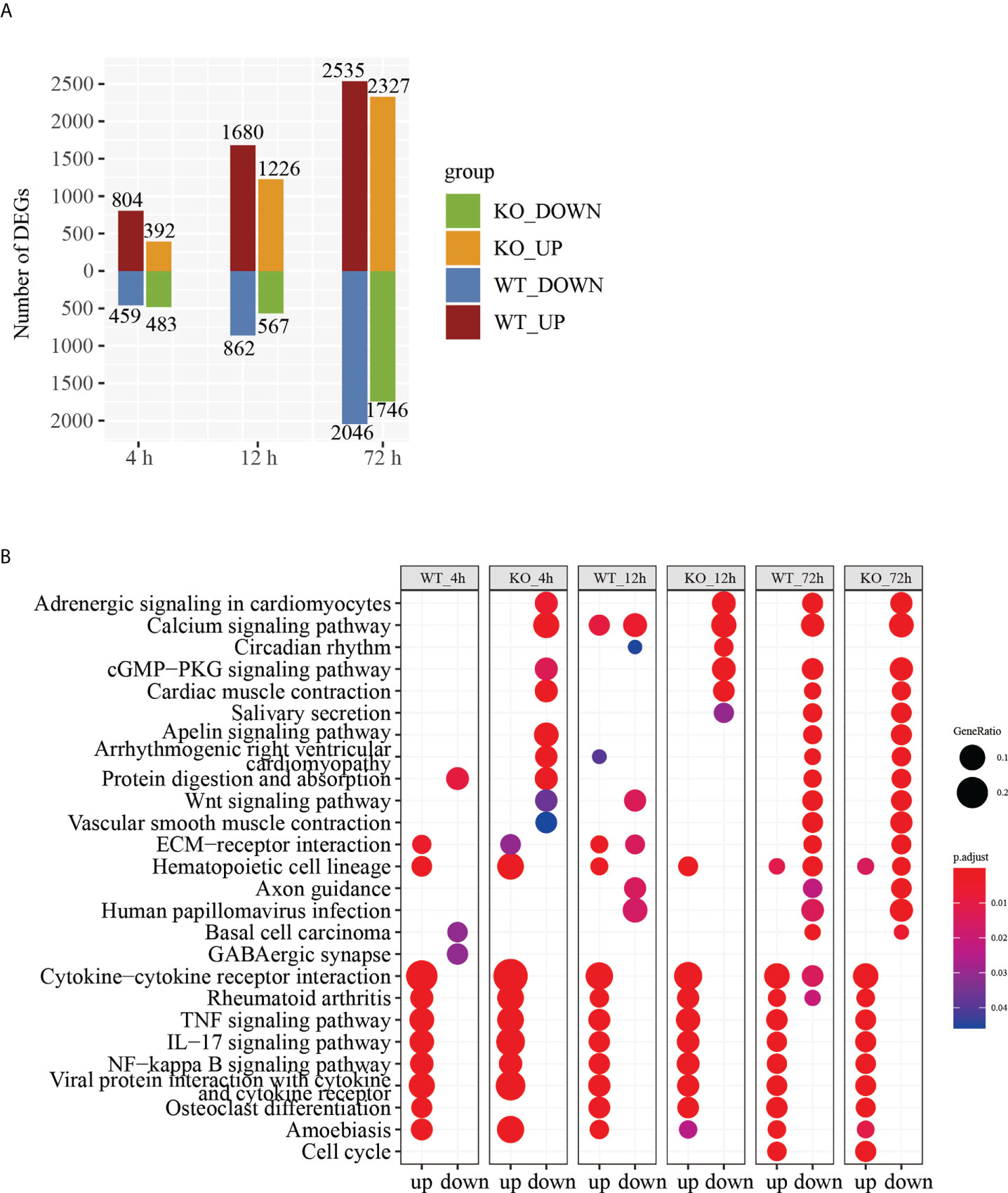
Figure 3 DEGs analysis of RNA-Seq in lung tissue of Csf2a KO mice and WT mice after RT-exposure compared to control. (A) Bar chart showing the number of DEGs under different experimental conditions. (B) The top 25 entries in the KEGG pathway with significant accumulation of up- and downregulated genes in each group under different experimental conditions.
DEGs analysis of TNF signaling pathway and NF-κB signaling pathway indicate delayed activation decreased in Csf2ra KO mice compared to WT mice
Within-group DEGs analysis revealed pro-inflammatory responses happened in both Csf2ra KO mice and WT mice. Next, we analyzed relative pro-inflammatory signal intensity for Csf2ra KO mice and WT mice at the same time points. KEGG pathway analysis of the DEGs in the Csf2ra KO mice and WT mice indicated that the TNF and NF-κB signaling pathways were significantly downregulated in Csf2ra KO mice relative to WT mice at 4, 12 and 72 h post-inhalation (Figure 4A).
Figure 4 DEGs analysis of RNA-Seq in lung tissue of Csf2ra KO mice compared to WT mice at 4, 12 and 72 h post RT-inhalation. (A) The top 17 entries in the KEGG pathway of upregulated genes in Csf2ra KO mice or WT mice under different experimental conditions. (B) The TNF signaling pathway was significantly downregulated in Csf2ra KO mice relative to WT mice at 4 h post-RT inhalation. (C) The NF-κB pathway was significantly downregulated in Csf2ra KO mice relative to WT mice at 4 h post-inhalation. (D) RT-qPCR verification of genes identified in transcriptome analysis. Two-way ANOVAs followed by Sidak’s multiple comparison tests were used; *P <0.05. (E) Western blot analysis for p-p65, p65, p-IκB, IκB and TNF protein expression levels.
Having found that DEGs are involved in activating the TNF and NF-κB signaling pathways, we next looked at which genes associated with these pathways were affected by Csf2ra deletion using the path-view package in R. Genes in the TNF (Figure 4B) and NF-κB (Figure 4C) signaling pathways were significantly downregulated in Csf2ra KO mice relative to WT mice at 4 and 12 h after RT exposure (Supplementary Figures 4, 5). We then chose three essential genes (Tnf, Il1b) for validation by RT-qPCR (Figure 4D). The protein levels of p-p65, p65, p-IκB, IκB and TNF were estimated using Western blot (Figure 4E). Csf2ra KO mice decreased p-p65, p-IκB and TNF expression at 4 h and 12 h after RT exposure relative to WT mice. In general, Csf2ra deletion in mice results in reduced levels of the TNF and NF-κB signaling pathways both at the transcript level and at the protein level after RT exposure.
Dynamic gene expression landscape of Csf2ra KO mice and WT mice
To understand transcriptome changes in the two groups, we used RNA sequencing (RNA-Seq) to document the global expression profiles of each group in a time-dependent manner (Figure 5A). For a holistic view, DEGs were clustered based on their temporal expression patterns. Based on the expression patterns of Csf2ra KO mice and WT groups, genes were divided into nine clusters; expression differences represent alterations in the transcription levels between groups. For example, cluster 5 had 117 genes and increased expression at 4, 12, and 72 h post-inhalation in both groups. The magnitude of increase in Csf2ra KO mice, however, was significantly lower than in WT mice. These genes were enriched for terms such as response to interleukin-1, eosinophil chemotaxis, and positive regulation of cytokine production (Figure 5B). Cluster 8 had 23 genes and increased in WT mice sharply at 4 and 12 h post-inhalation, but did not consistently increase in Csf2ra KO mice post-inhalation. GO analysis revealed that these gene are involved in leukocyte chemotaxis, neutrophil chemotaxis and the cytokine-mediated signaling pathway (Figure 5C). Next, we selected four genes (Csf2ra, F7, Atp6v0d2, Itgax) that consistently showed low expression in Csf2ra KO mice from Cluster 8 (Figure 5D) for validation by RT-QPCR (Figure 5E). In summary, Csf2ra deletion in mice reduced host response after RT-exposure.
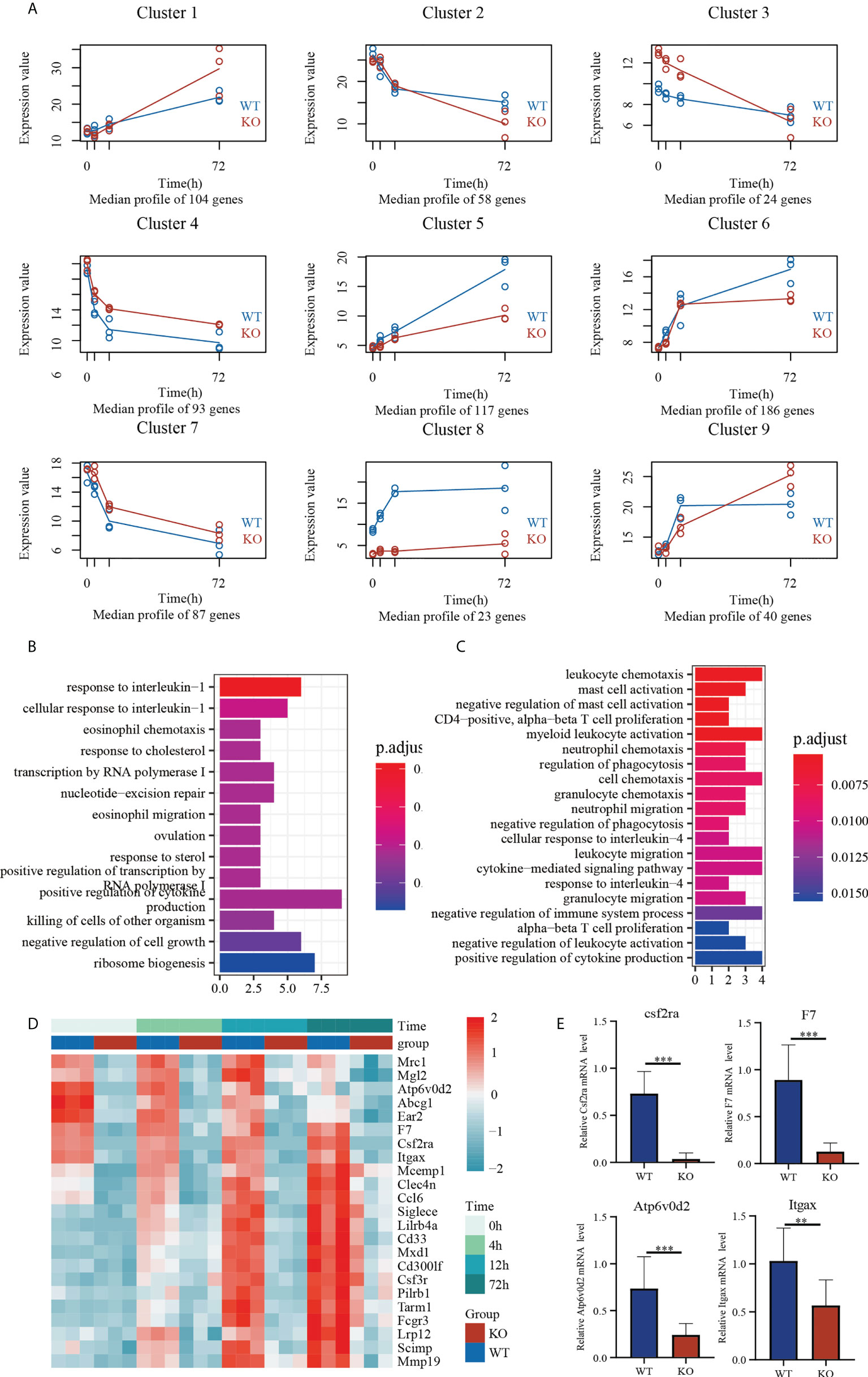
Figure 5 Temporal transcriptome analysis of lung tissues in Csf2ra KO mice and WT mice. (A) The nine clusters identified by the maSigPro algorithm for the selected genes. (B) Analysis of GO biological processes in Cluster 5. (C) Analysis of GO biological processes in Cluster 8. (D) Heat maps showing expression levels of genes in Cluster 8 in each group. (E) RT-qPCR verification of genes with consistently low expression in Csf2ra KO mice. Welch’s t test; **P <0.01, ***P <0.0001.
The degree of neutrophil chemotaxis and recruitment in Csf2ra KO mice was inhibited compared to WT mice at early time-points
Given the cytokines and pro-inflammatory signaling pathway alteration between Csf2ra KO mice and WT mice after RT exposure, we next investigated whether Csf2ra affects the accumulation of immune cells in lung tissue after RT exposure. The relative abundances of 11 types of infiltrating immune cells (TIICs) were quantified with ImmuCellAI in Csf2ra KO mice and WT mice (Figure 6A). The proportion of neutrophils differed significantly between Csf2ra KO mice and WT mice at 4 and 12 h after RT exposure. The proportion of neutrophils in WT mice at 0 h was almost undetectable, while neutrophils were present in Csf2ra KO mice at 0 h; this result is consistent with pathological observations in lung tissue. Importantly, the speed of neutrophil chemotaxis and recruitment toward lung tissue appeared inhibited in Csf2ra KO mice compared to WT mice.
Figure 6 The degree of neutrophil chemotaxis and recruitment in Csf2ra-/- mice was decreased relative to WT mice. (A) Immune cell abundance analyzed by ImmuCellAI at different timepoints for Csf2ra KO mice and WT mice. (B) Bar graph showing the percentage of neutrophils measured by flow cytometry. Two-way ANOVAs followed by Sidak’s multiple comparison tests; *P <0.05.
To test the validity of inferring that neutrophil chemotaxis and recruitment toward lung tissue was inhibited in the Csf2ra KO mice at early time-points, we used flow cytometry to identify changes in neutrophil populations during disease progression. The proportion of neutrophils in the lung tissues of WT mice was significantly higher than in Csf2ra KO mice at 12 h post-inhalation (Figure 6B), confirming the gene set signature‐based method for cell-type composition and that the degree of neutrophil chemotaxis and recruitment was inhibited in Csf2ra KO mice compared to WT mice at early time-points.
Discussion
The therapeutic window for intervention after RT exposure is narrow. The primary treatment strategy is typically supportive care (24, 25). Hence, finding new targets for the treatment of ALI induced by RT inhalation is of major clinical importance. Previous work from our laboratory has shown that expression levels of multiple pro-inflammatory cytokines and transcription factors increased significantly at the early stage of RT-induced ALI (4 h post-exposure) (6). Parajuli et al. (26) demonstrated that GM-CSF priming increased LPS-induced NF-κB nuclear translocation and production of IL-1β, IL-6, TNF-α. De Alessandris et al. (27) confirmed that neutrophilic activation via GM-CSFR can play an important role in neutrophilic lung inflammation. Here we show that, GM-CSF signaling deletion attenuated NF-κB and TNF pathway activity following RT challenge in mice. Meanwhile, neutrophil infiltration was significantly reduced in Csf2ra KO mice. We speculate blockage of cytokine induction is one of the most direct tactics to blunt the cytokine storm causing ALI/ARDS and lethality.
GM-CSF signaling deletion in mice increases risk for bacterial infection (28, 29). While the exact relationships between RT and GM-CSF signaling deletion remain unclear, the survival data suggest that GM-CSF signal deletion promotes survival in mice after RT exposure. WGCNA was used to identify a distinct co-expression network module associated with host immune response caused by Csf2ra KO. Genes in module “black” had the most negatively significant correlation with Csf2ra KO mice and genes in module “red” had the most positively significant correlation with Csf2ra KO mice. Enrichment analysis indicated that the pathways of genes in the “black” module were linked to cytokine-cytokine receptor interaction, the NF-κB signaling pathway and the chemokine signaling pathway. These genes decrease the likelihood of host immune responses post RT-challenge. Genes in the “red” module were linked to immunoglobulin production, B cell mediated immunity and complement activation. The expression levels of these genes were increased in Csf2ra KO mice compared to WT mice, especially in steady states (without RT-exposure). GM-CSF signaling also can act as an anti-inflammatory/regulatory cytokine (30). We speculate that abnormal activation of B cell and complement may partially explain inflammatory cell infiltration in Csf2ra KO mice at steady states.
The TNF and the NF-κB signaling pathway were downregulated in Csf2ra KO mice compared to WT mice after RT exposure. Tumor necrosis factor (TNF), as a critical cytokine, can induce a wide range of intracellular signal pathways, including apoptosis and cell survival, as well as inflammation and immunity. TNFR1 signaling induces activation of numerous genes that are primarily controlled by two distinct pathways: the NF-κB pathway and the MAPK cascade, or apoptosis and necroptosis. TNFR2 signaling activates the NF-κB pathway, including the PI3K-dependent NF-κB pathway and the JNK pathway promoting cell survival. Different from bacterial or viral infection, the pro-inflammation associated genes in ALI/ARDS induced by toxins have mainly harmful effects. Therefore, the delayed cytokine expression level of genes associated with pro-inflammatory cytokines in Csf2ra KO mice after RT-exposure may reduce incidence of ARDS, leading to increased survival.
Another characteristic of pulmonary ricin toxicity is rapid neutrophil infiltration, which ultimately leads to respiratory failure and death. Neutrophils are considered a major hallmark of ALI/ARDS, whether caused by ricin (8–10) or not (31–33). Aggressive or prolonged neutrophil responses result in deleterious inflammatory conditions and tissue destruction. Decreased pulmonary neutrophil infiltration is associated with attenuation of injury severity. In our investigation, the degree of neutrophil infiltration into lung tissue in Csf2ra KO mice was much lower than in WT mice. GM-CSF signaling deletion attenuated inflammation and imbalance of the immune system and alleviated severity of ALI induced by RT.
The mitigating effect of Csf2ra knockout in mice on RT-mediated lethality may involve multiple aspects. First, the maturation and function of alveolar macrophages (AMs) requires GM-CSF signaling (34). Given their physiological location in lungs, AMs may be the first immune cell to respond to inhaled RT exposure. Moreover, as the time since induction of the RT challenge increased, the number of AMs gradually decreased (35). Thus, AMs may play a central role in the mechanism of ricin toxicity. Korcheva etal. (36) investigated the immunologic properties of AMs in vitro and discovered AMs may be the upstream regulators of inflammatory cascades that occur with RT exposure. The in vivo results of our study also suggest this. Second, blocking GM-CSF signaling may effectively protect mice from death after RT inhalation by alleviating inflammation. In inflammation, GM-CSF serves as a communication conduit between tissue-invading lymphocytes and myeloid cells (14). A positive feedback loop caused by GM-CSF leads to fast inflammatory cell infiltration and increasing pro-inflammatory cytokine secretion (37, 38). In our research, the lack of GM-CSF signaling in Csf2ra KO mice eliminates this pro-inflammatory response, reducing the severity of ALI/ARDS induced by RT. Our study does have limitations. We cannot distinguish between dysfunction of AMs and lack of GM-CSF signaling as the main factor alleviating ALI/ARDS induced by RT. Future studies using conditional knockout mice are needed to explore this question.
In summary, our data revealed that a Csf2ra deficiency protects mice from mortality and morbidity induced by RT. The majority of the protective mechanism of ALI/ARDS is via reduction in cytokine secretion, decreased activation of pro-inflammatory signaling pathways and decreased neutrophils infiltration. The present study provides a treatment perspective for suppressing the host immune response to protect the host from mortality. This work provides potential therapeutic targets for mitigating the severity of ALI induced by RT.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: NCBI Gene Expression Omnibus; GSE199606.
Ethics statement
The animal study was reviewed and approved by Institute of Animal Care and Use Committee.
Author contributions
FZ: Methodology, Formal analysis, Investigation, Data curation, Writing - Original Draft. sl: Conceptualization, Methodology, Formal analysis, Investigation, Validation. NX: Methodology, Validation. MD: Methodology, Validation. ZZ: Methodology, Validation. YW: Methodology, Formal analysis. DS: Methodology, Formal analysis. BG: Methodology, Formal analysis, Investigation, Resources. DZ: Methodology, Formal analysis, Investigation, Resources. LH: Conceptualization, Investigation, Resources, Data curation, Writing - Review & Editing, Supervision. HY: Conceptualization, Investigation, Resources, Data curation, Writing - Review & Editing, Supervision. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.900755/full#supplementary-material
References
1. Wheeler AP, Bernard GR. Acute lung injury and the acute respiratory distress syndrome: a clinical review. Lancet (2007) 369:1553–64. doi: 10.1016/S0140-6736(07)60604-7
2. Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, et al. Incidence and outcomes of acute lung injury. New Engl J Med (2005) 353:1685–93. doi: 10.1056/NEJMoa050333
3. Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med (2000) 342:1334–49. doi: 10.1056/NEJM200005043421806
4. Matthay MA, Ware LB, Zimmerman GA. The acute respiratory distress syndrome. J Clin Invest (2012) 122:2731–40. doi: 10.1172/JCI60331
5. Gal Y, Mazor O, Falach R, Sapoznikov A, Kronman C, Sabo T. Treatments for pulmonary ricin intoxication: Current aspects and future prospects. Toxins (Basel) (2017) 9(10):311. doi: 10.3390/toxins9100311
6. Jiao Z, Li S, Su D, Gan C, Zhao X, Gao B, et al. Time-course transcriptome analysis of lungs from mice exposed to ricin by intratracheal inoculation. Toxicol Lett (2021) 337:57–67. doi: 10.1016/j.toxlet.2020.11.012
7. Pillar N, Haguel D, Grad M, Shapira G, Yoffe L, Shomron N. Characterization of MicroRNA and gene expression profiles following ricin intoxication. Toxins (Basel) (2019) 11(5):250. doi: 10.3390/toxins11050250
8. Gal Y, Mazor O, Alcalay R, Seliger N, Aftalion M, Sapoznikov A, et al. Antibody/doxycycline combined therapy for pulmonary ricinosis: Attenuation of inflammation improves survival of ricin-intoxicated mice. Toxicol Rep (2014) 1:496–504. doi: 10.1016/j.toxrep.2014.07.013
9. Gal Y, Sapoznikov A, Falach R, Ehrlich S, Aftalion M, Sabo T, et al. Potent antiedematous and protective effects of ciprofloxacin in pulmonary ricinosis. Antimicrob Agents Chemother (2016) 60:7153–8. doi: 10.1128/AAC.01696-16
10. Lindauer ML, Wong J, Iwakura Y., Magun B. E. Pulmonary inflammation triggered by ricin toxin requires macrophages and IL-1 signaling. J Immunol (2009) 183:1419–26. doi: 10.4049/jimmunol.0901119
11. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev (2012) 76:16–32. doi: 10.1128/MMBR.05015-11
12. Ragab D, Salah EH, Taeimah M, Khattab R, Salem R. The COVID-19 cytokine storm; what we know so far. Front Immunol (2020) 11:1446. doi: 10.3389/fimmu.2020.01446
13. Broughton SE, Dhagat U, Hercus TR, Nero TL, Grimbaldeston MA, Bonder CS, et al. The GM-CSF/IL-3/IL-5 cytokine receptor family: from ligand recognition to initiation of signaling. Immunol Rev (2012) 250:277–302. doi: 10.1111/j.1600-065X.2012.01164.x
14. Becher B, Tugues S, Greter M. GM-CSF: From growth factor to central mediator of tissue inflammation. Immunity (2016) 45:963–73. doi: 10.1016/j.immuni.2016.10.026
15. Griffiths GD, Rice P, Allenby SC, Upshall DG. Inhalation toxicology and histopathology of ricin and abrin toxins. Inhal Toxicol (1995) 7 (2):269–88. doi: 10.3109/08958379509029098
16. Feng J, Hu X, Fu M, Dai L, Yu Y, Luo W, et al. Enhanced protection against q fever in BALB/c mice elicited by immunization of chloroform-methanol residue of coxiella burnetii via intratracheal inoculation. Vaccine (2019) 37:6076–84. doi: 10.1016/j.vaccine.2019.08.041
17. Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
18. Mccarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Resesrch (2012) 40:4288–97. doi: 10.1093/nar/gks042
19. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an r package for comparing biological themes among gene clusters. Omics (2012) 16:284–7. doi: 10.1089/omi.2011.0118
20. Conesa A, Nueda MJ, Ferrer A, Talón M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics (2006) 22:1096–102. doi: 10.1093/bioinformatics/btl056
21. Nueda MJ, Tarazona S, Conesa A. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics (2014) 30:2598–602. doi: 10.1093/bioinformatics/btu333
22. Miao YR, Zhang Q, Lei Q, Luo M, Xie GY, Wang H, et al. ImmuCellAI: A unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) (2020) 7:1902880. doi: 10.1002/advs.201902880
23. Miao Y-R, Xia M, Luo M, Luo T, Yang M, Guo A-Y. ImmuCellAI-mouse: a tool for comprehensive prediction of mouse immune cell abundance and immune microenvironment depiction. Bioinformatics (2022) 38:785–91. doi: 10.1093/bioinformatics/btab711
24. Respaud R, Marchand D, Pelat T, Tchou-Wong KM, Roy CJ, Parent C, et al. Development of a drug delivery system for efficient alveolar delivery of a neutralizing monoclonal antibody to treat pulmonary intoxication to ricin. J Control Release (2016) 234:21–32. doi: 10.1016/j.jconrel.2016.05.018
25. Feldberg L, Elhanany E, Laskar O, Schuster O. Rapid, sensitive and reliable ricin identification in serum samples using LC–MS/MS. Toxins (2021) 13:79. doi: 10.3390/toxins13020079
26. Parajuli B, Sonobe Y, Kawanokuchi J, Doi Y, Noda M, Takeuchi H, et al. GM-CSF increases LPS-induced production of proinflammatory mediators via upregulation of TLR4 and CD14 in murine microglia. J Neuroinflamm (2012) 9:268. doi: 10.1186/1742-2094-9-268
27. De Alessandris S, Ferguson GJ, Dodd AJ, Juss JK, Devaprasad A, Piper S, et al. Neutrophil GM-CSF receptor dynamics in acute lung injury. J Leukoc Biol (2019) 105:1183–94. doi: 10.1002/JLB.3MA0918-347R
28. Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun (2017) 8:1512. doi: 10.1038/s41467-017-01803-x
29. De Groote MA, Johnson L, Podell B, Brooks E, Basaraba R, Gonzalez-juarrero M. GM-CSF knockout mice for preclinical testing of agents with antimicrobial activity against mycobacterium abscessus. J Antimicrob Chemother (2014) 69:1057–64. doi: 10.1093/jac/dkt451
30. Bhattacharya P, Budnick I, Singh M, Thiruppathi M, Alharshawi K, Elshabrawy H, et al. Dual role of GM-CSF as a pro-inflammatory and a regulatory cytokine: Implications for immune therapy. J Interferon Cytokine Res (2015) 35:585–99. doi: 10.1089/jir.2014.0149
31. Till GO, Johnson KJ, KunkeL R, Ward PA. Intravascular activation of complement and acute lung injury. dependency on neutrophils and toxic oxygen metabolites. J Clin Invest (1982) 69:1126–35. doi: 10.1172/JCI110548
32. Yan B, Chen F, Xu L, Xing J, Wang X. HMGB1-TLR4-IL23-IL17A axis promotes paraquat-induced acute lung injury by mediating neutrophil infiltration in mice. Sci Rep (2017) 7:597. doi: 10.1038/s41598-017-00721-8
33. Koma T, Yoshimatsu K, Nagata N, Sato Y, Shimizu K, Yasuda SP, et al. Neutrophil depletion suppresses pulmonary vascular hyperpermeability and occurrence of pulmonary edema caused by hantavirus infection in C.B-17 SCID mice. J Virol (2014) 88:7178–88. doi: 10.1128/JVI.00254-14
34. Shima K, Arumugam P, Sallese A, Horia Y, Ma Y, Trapnell C, et al. A murine model of hereditary pulmonary alveolar proteinosis caused by homozygous Csf2ra gene disruption. Am J Physiol Lung Cell Mol Physiol (2022) 322(3):L438–48. doi: 10.1152/ajplung.00175.2021
35. Roy CJ, Ehrbar DJ, Bohorova N, Bohorov O, Kim D, Pauly M, et al. Rescue of rhesus macaques from the lethality of aerosolized ricin toxin. JCI Insight (2019) 4 (1):e124771. doi: 10.1172/jci.insight.124771
36. Korcheva V, Wong J, Lindauer M, Jacoby DB, Iordanov MS, Magun B. Role of apoptotic signaling pathways in regulation of inflammatory responses to ricin in primary murine macrophages. Mol Immunol (2007) 44:2761–71. doi: 10.1016/j.molimm.2006.10.025
37. Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell (2014) 25:605–20. doi: 10.1016/j.ccr.2014.03.021
Keywords: RT, Csf2ra, ALI, RNA-Seq, GM-CSF receptor, inflammation response
Citation: Zong F, Li S, Wang Y, Xiao N, Deng M, Zhang Z, Su D, Gao B, Zhou D, Hu L and Yang H (2022) Csf2ra deletion attenuates acute lung injuries induced by intratracheal inoculation of aerosolized ricin in mice. Front. Immunol. 13:900755. doi: 10.3389/fimmu.2022.900755
Received: 21 March 2022; Accepted: 06 September 2022;
Published: 20 September 2022.
Edited by:
Lisardo Bosca, Autonomous University of Madrid, SpainReviewed by:
Alfonso Luque, Instituto de Salud Carlos III (ISCIII), SpainBeatriz De Las Heras, Department of Pharmacology, (UCM), Spain
Copyright © 2022 Zong, Li, Wang, Xiao, Deng, Zhang, Su, Gao, Zhou, Hu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiying Yang, eWh5ODAzMjRAMTYzLmNvbQ==; Lingfei Hu, aHVsaW5nMDNAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Fuliang Zong
Fuliang Zong Sha Li1†
Sha Li1† Dongsheng Zhou
Dongsheng Zhou