
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 10 May 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.897754
Glioma is a brain tumor that arises in the central nervous system and is categorized according to histology and molecular genetic characteristics. Long non-coding RNAs (lncRNAs) are RNAs longer than 200 nucleotides in length. They have been reported to influence significant events such as carcinogenesis, progression, and increased treatment resistance on glioma cells. Long non-coding RNAs promote cell proliferation, migration, epithelial-to-mesenchymal transition and invasion in glioma cells. Various significant advancements in transcriptomic profiling studies have enabled the identification of immune-related long non-coding RNAs as immune cell-specific gene expression regulators that mediates both stimulatory and suppressive immune responses, implying lncRNAs as potential candidates for improving immunotherapy efficacy against tumors and due to the lack of different diagnostic and treatments for glioma, lncRNAs are potential candidates to be used as future diagnostic, prognostic biomarker and treatment tools for glioma. This review’s primary purpose is to concentrate on the role of long non-coding RNAs in early glioma identification, treatment, and immunotherapy.
Gliomas are the most often occurring malignant primary brain tumors in adults. They may occur anywhere in the central nervous system but are most often seen in the brain, developing in glial tissue (1). While these tumors are often malignant, some subtypes may not always behave malignantly. Moreover, they are the most frequently occurring primary intracranial tumor, accounting for 81% of malignant brain tumors (2). Although relatively rare, they cause significant mortality and morbidity (3). The United States’ incident rate is 3.20 per 100,000, and glioblastoma(GBM) occupies 60–70% of malignant glioma (4, 5). In addition, glioma is the 3rd most common cause of cancer mortality in people aged 15 to 34, accounting for 2.5 percent of cancer deaths globally; detailed data about mortality and incidences of glioma are present in Figure 1. Glioblastoma multiform accounts for 50% of gliomas, increasing frequency in persons over 65 (6, 7) and glioblastoma has a 5-year relative survival of ∼5% (8). Based on the world health organization, gliomas are classified based on the glial cells from which they originate, such as astrocytes, ependymal cells, and oligodendrocytes (9). Different risk factors have been identified to be related to glioma (10). Notably, they include an elevated risk associated with ionizing radiation exposure and lower risk associated with a history of allergies or atopic disorders (11).
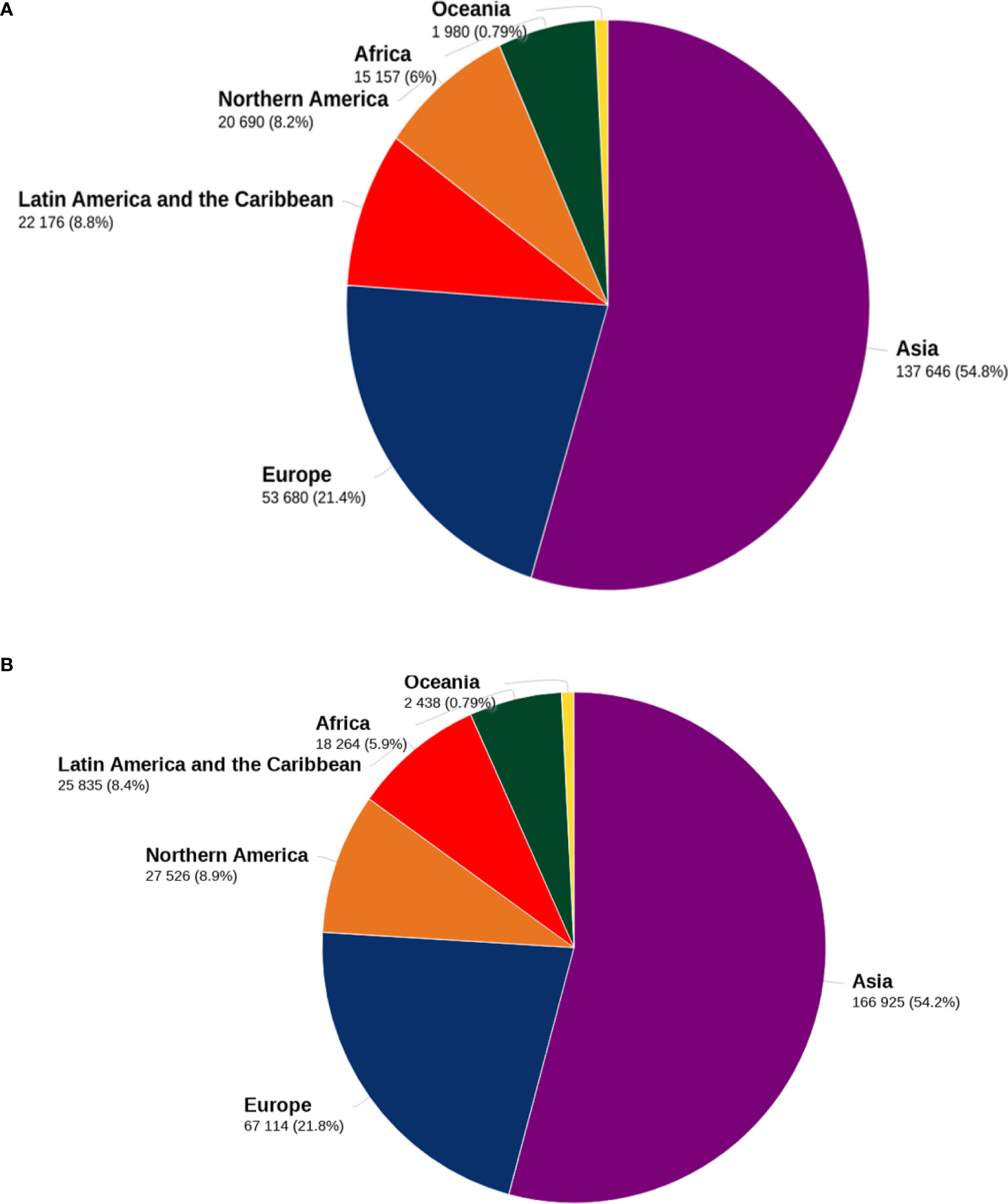
Figure 1 Death and incidence related to glioma. Figure 1 shows the distribution of mortality (A) and incidence (B) of brain tumors for both sexes and all ages by 2020 in major geographical areas. The data sources and methodologies used for each global region are available online from the global health data exchange and GLOBALCAN 2020 at the Global Cancer Observatory.
Currently, standard glioblastoma(GBM) treatments include maximal surgical resection and combined radio-chemotherapy (12). Apart from GBM’s rapid proliferation, extensive invasion, intra- and intertemporal genetic heterogeneity, and treatment resistance, the poor prognosis of GBM patients is also due to a lack of insights into the molecular pathogenesis and an absence of timely and sensitive diagnostic and therapeutic monitoring tools (13). Therefore, it is crucial to elucidate molecular mechanisms underlying glioma development and progression and further explore reliable biomarkers.
Approximately 98% of RNA transcribed from human DNA is not translated into protein and thus, is called non-coding RNA (ncRNA) (14), and some of them are classified as long non-coding RNAs which interfere with the transcription and translation of genes without altering DNA sequences (15, 16). Long non-coding RNAs(lncRNAs) are longer than 200 nucleotides; dysfunctions of several lncRNAs have often been reported in different tumors; and these dysfunctional lncRNAs are associated with the pathogenesis and regulation of glioma’s cell proliferation, cell motility, angiogenesis, drug resistance, and radiation resistance (16, 17). Originally, non-coding portions of the genome were referred to as junk DNA due to their inability to be translated into proteins. In addition, long non-coding RNAs, whose transcripts are longer than 200 nucleotides, have already been engaged in many biological activities (18). LncRNAs are hypothesized to influence gene expression and epigenetic, transcriptional, and post-transcriptional modification, and their expressions are connected with several biological characteristics, including cell survival (19, 20). Long non-coding RNA dysregulation is increasingly being related to various human disorders. Notably, long non-coding RNAs have unique expression patterns across multiple tumor types and function as tumor suppressors or promoters.
Additionally, aberrant lncRNAs expression have been linked to the altered expression of genes involved in carcinogenesis, metastasis, and advancement of tumor stages in malignancies (21, 22). Rather than concentrating just on cancer cell proliferation and invasion, an emerging number of researches reveal the crucial function of the tumor microenvironment (TME) in the genesis and progression of gliomas. The TME is a complex structure composed of cancer cells and noncancerous cells such as endothelial cells, pericytes, fibroblasts, and immune cells (23). Microglia and macrophages make up to 30% to 50% of glioma cells, and tumor-associated microglia and macrophages (TAMs) in the brain are protumorigenic and increase proportionately to tumor grade (24, 25). Other immune cells, such as dendritic cells, have also been reported to play a critical role in cancer immunotherapy (26). As a result, it is essential to provide effective immunological predictors and prognostic markers to enhance glioma prognosis and guide specific treatment choices, including immunotherapy. This study aims to discuss the role of long non-coding RNAs in the early identification, treatment, and immunotherapy of glioma.
Long non-coding RNAs are non-coding RNAs longer than 200 nucleotides. During their biogenesisthey are systematically transcribed by RNA polymerase II, and long non-coding interfere with the transcription and translation of genes without altering DNA sequences. In addition, they are mostly polyadenylated and capped (27). Despite their increased nuclear distribution, more sparse structure, and lower evolutionary conservation, they have fewer exons than mRNAs (28). So, lncRNAs can be categorized into six categories based on their genomic positions: (a) sense, (b) antisense, (c) bidirectional, (d) intronic, (e)intergenic (Figure 2). Furthermore, lncRNA plays various roles in both the nucleus and cytoplasm. Their effects on the function and integrity of nuclear bodies play a role in chromatin remodeling, modifying chromosomal connections, transcription control, and post-transcriptional regulation of gene expression in the nucleus (29). Besides, they also regulate cellular mRNA turnover, translation, and post-translational modification within the cytoplasm. Micro-RNAs(mi-RNA) accessibility may be altered as competing endogenous RNAs (ceRNA) that act as a sponge for mi-RNAs.
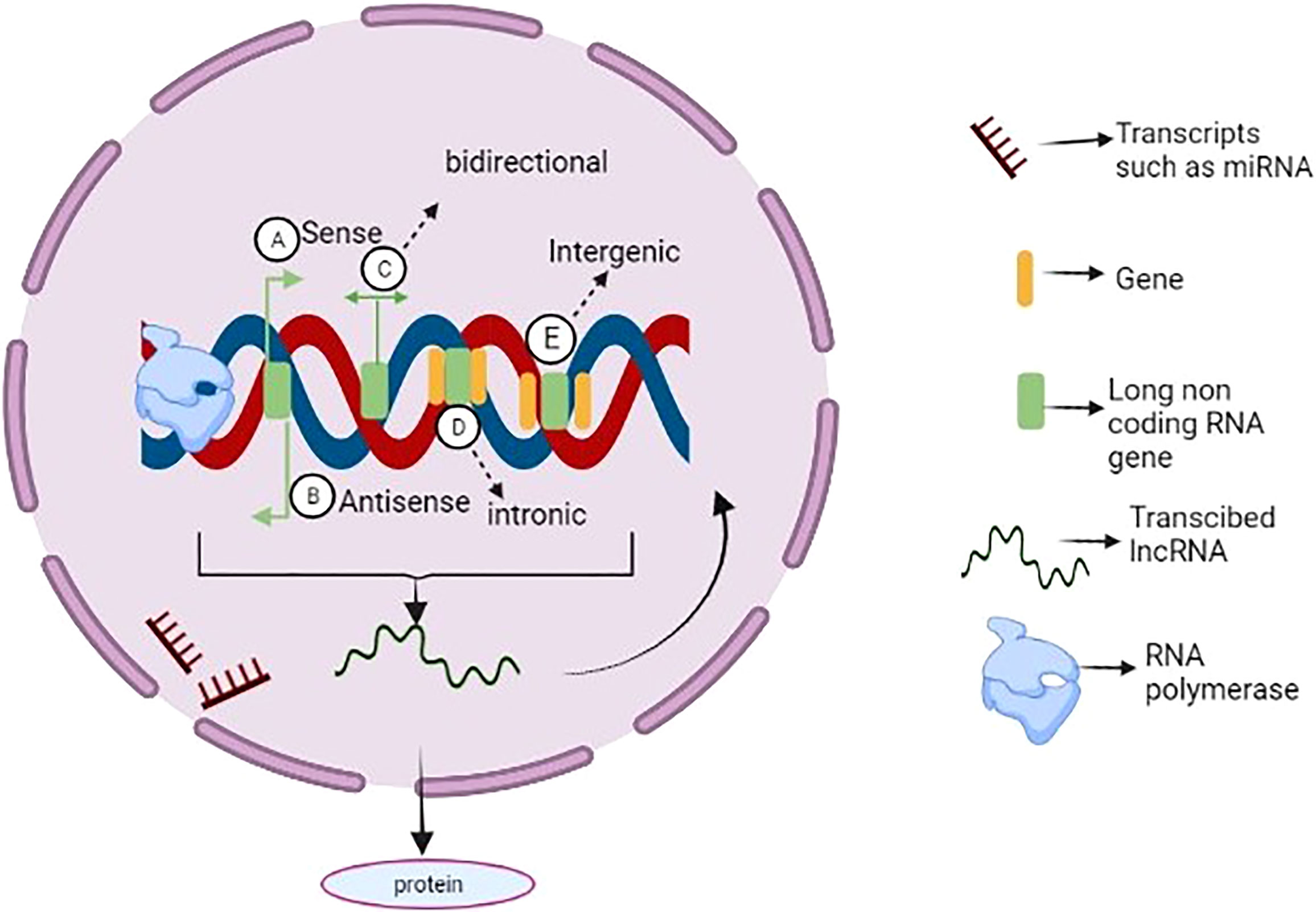
Figure 2 Long non coding RNA biogenesis and function. LncRNAs can be categorized into sense (A), antisense (B), bidirectional (C), intronic (D), and intergenic (E) lncRNAs based on their genomic location and Besides, some lncRNAs are reported to be transcribed from one of the strands of a DNA sequence. In addition to being transcribed by RNA polymerases, many of them are spliced, 5′ capped and polyadenylated. Although some lncRNAs can be translated into proteins, most of them serve as immediate regulators of genome function (e.g., microRNAs or very important binding partners of cellular components (e.g., proteins).
Moreover, lncRNAs act as decoys for RNA binding proteins involved in the mRNA degradation process or promote protein binding to lncRNAs; subsequently, by altering messenger RNA or interacting with ribosomes, lncRNAs significantly influence translation (30). At last, lncRNAs participate in post-transcriptional modifications, such as phosphorylation and ubiquitination (31).
Numerous studies have shown that long non-coding RNAs play critical roles in glioma formation by stimulating critical processes (Figure 3). For instance, a study conducted with the primary goal of identifying long non-coding RNA and clarifying its function and mode of action in glioma formation showed that LPP Antisense RNA 2(LPP-AS2) promotes glioma carcinogenesis through a miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop (32). Furthermore, a critical study discovered that clinical samples and cell lines had detectable quantities of FGD5 Antisense RNA 1(FGD5-AS1). By directly regulating the Wnt/-Catenin Pathway, the long non-coding RNA FGD5-AS1 promotes Glioma-related processes such as cell proliferation, migration, and invasion (33). Exosomes carrying long non-coding RNA have been reported to enhance glioma formation; for instance, recent research found that exosomes carrying Linc01060 increased glioma growth by effectively regulating the MZF1/c-Myc/HIF1 Axis (34); on the other hand, a study that examined the link between miR155HG and Annexin A2(ANXA2) to assess their malignancy in the formation of GBM discovered that the two, in combination with miR-185, enhanced glioma growth and development which proves the role of long non-coding RNA in glioma formation (35).
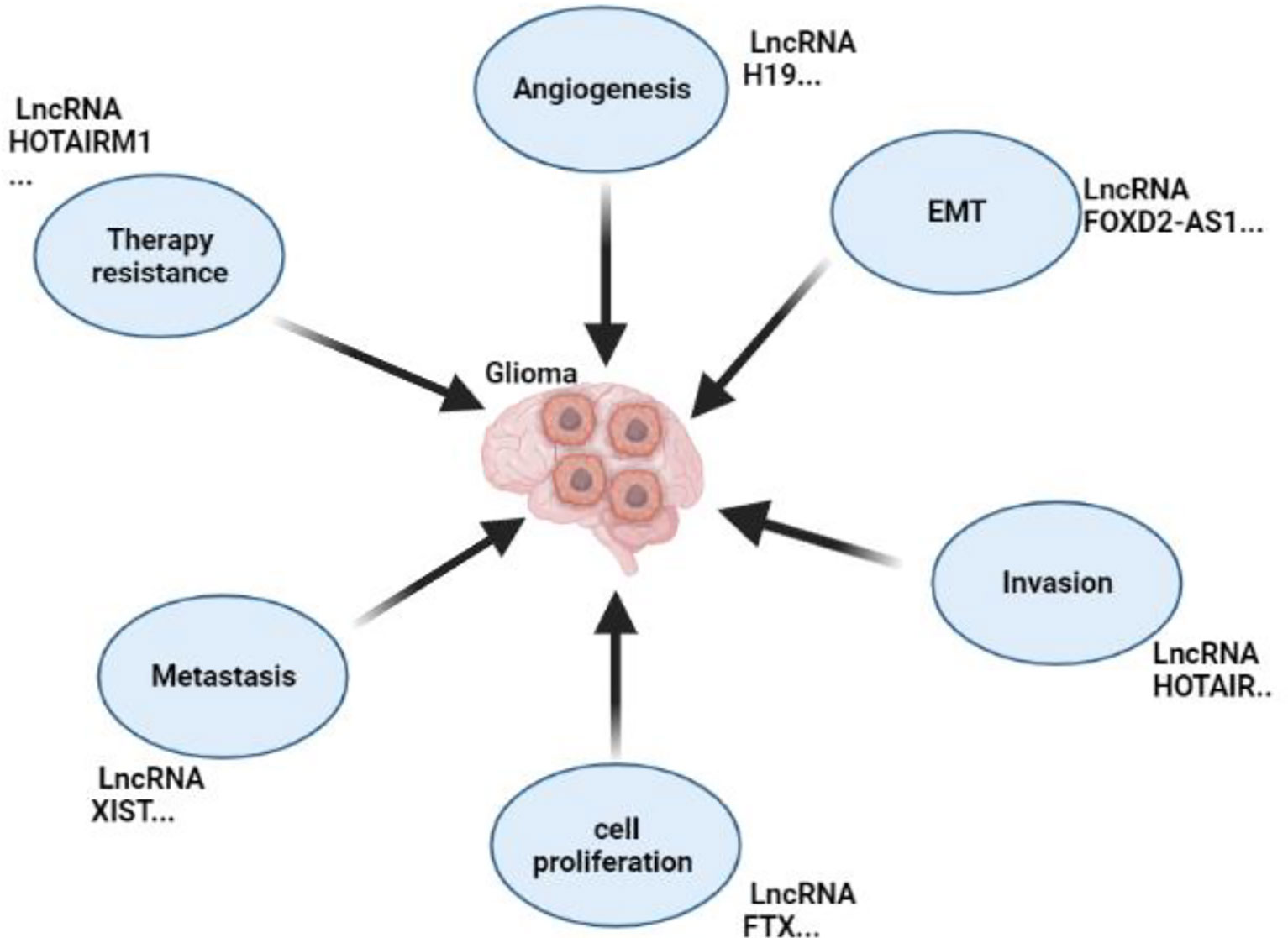
Figure 3 Roles of non-coding RNA in Glioma’s Development. This figure shows the roles of lncRNAs in glioma by promoting metastasis, EMT, invasion, proliferation, angiogenesis, and resistance to chemotherapy.
Furthermore, Zhang W et al. found that long non-coding RNA FTX was high in gliomas. By inhibiting miR-342-3p, this long non-coding RNA has been found to promote both cell proliferation and survival of glioma cells (36). Gliomas also undergo metastasis at this point in their development, and different researches have highlighted that long non-coding RNAs strongly influence and play a role in glioma metastasis. For example, Huixiao Dong et al. demonstrated that camp responsive element binding protein 1(CREB1) triggers long non-coding adjacent opposite strand RNA 1 (FOXD2-AS1), leading to increased cell proliferation and metastasis in gliomas via miR-185 sponging by targeting AKT Serine/Threonine Kinase 1(AKT1) (37). Besides, Chixing Luo and his colleagues found a correlation between miR-133a/SOX4 and lncRNA XIST promoting glioma proliferation and metastasis (38).
There are shreds of evidence showing that long non-coding RNAs may be involved in the promotion of glioma’s angiogenesis, which is a very ultimate factor that affects glioma’s development; recently, a study reported that lncRNA H19 promotes angiogenesis by activating the miR-138/HIF-1*/VEGF pathway, suggesting that targeting this pathway could be essential for successfully stopping angiogenesis (16), also, a study conducted by Zhihua Cheng et al., reported that long non-coding RNA XIST inhibited miR-429, led to glioma tumorigenesis and angiogenesis (39). Furthermore, it has also been reported that long non-coding RNAs promote EMT, an important event in cancer progression, including gliomas. For instance, research conducted by Juan Zhao showed that lncRNA FOXD2-AS1 speeds up numerous crucial events in gliomas, such as proliferation, metastasis, and EMT (40). It is evident that long non-coding RNAs play an essential role in the development of glioma by promoting different important glioma events. Additional information can be found in Table 1.
Long non-coding RNAs are adaptable molecules that interact with RNA, DNA, and proteins to influence the expression of protein-coding genes (47). Several immune cells contain lncRNAs that affect both innate and adaptive immunity. In recent years, researchers have become more aware of the importance of lncRNAs in controlling the tumor immune microenvironment (TME) (48, 49). Furthermore, tumor microenvironments contain immunosuppressive cells that inhibit antitumor immunity (50). Patients with malignant glioma present several immunological defects, including CD4 lymphopenia, elevated regulatory T cells (Tregs) in peripheral blood, immune-suppressive macrophage infiltration, and decreased Th1 cytokine production impaired cell-mediated immunity (51, 52). Besides, some studies have examined lncRNA expression patterns in glioblastoma and have discovered immune-related expression patterns (53, 54), with proven predictive value in glioma development and prognosis.
Moreover, lncRNAs control inflammation genes, which may affect the immune system. The lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1), for example, modulates the transcription of IL-8 and affects the activity of various immune cells (55); this is the same as lncRNA-Cox2, which has been found to inhibit the SWI/SNF complex’s chromatin remodeling activity in macrophages in addition to suppressing inflammatory genes in macrophages (56); and, another study found that hypoxia-related lncRNA in lower-grade gliomas correlates with prognosis and regulates the immune microenvironment (57). Understanding immune-related lncRNAs in gliomas may help understand the involvement of lncRNA in TME and the immune system which will significantly help to get more ways to deal with glioma aggressiveness and development.
It is essential to identify cancer as early as possible to fight it in its early stage. Identifying different types of cancer, including gliomas, requires biomarkers (58, 59). Upregulation and downregulation of long non-coding RNAs can reveal other cancer statuses, making them suitable biomarkers for specific forms of cancer, such as gliomas (60). Numerous studies demonstrate that lncRNAs have thus become a valuable tool for predicting gliomas. For example, researchers have developed a nomogram (based on m6A-LPS, age, and WHO grade) that is a good predictor of low-grade gliomas (LGGs) patients’ overall survival in both data sets, concluding that N6-Methylandenosine-Related lncRNAs are particularly useful biomarkers for predicting overall survival in lower-grade gliomas (61). In addition, a different study found that serum levels of lncRNA HOX antisense intergenic RNA (HOTAIR) were elevated in tumor samples compared with standard specimens. In addition, they found that long non-coding RNA HOTAIR was highly diagnostic, suggesting that it could serve as a potential biomarker to diagnose and predict gliomas (62).
According to a study, which also examined the expression of MAGI2 antisense RNA 3(MAGI2AS3) and its clinical significance in gliomas, it was found that the expression of the lncRNA was higher in tumors than in normal samples; in glioma patients, there was a direct correlation between the MAGI2-AS3 expression and cancer world health organization (WHO) grade and The Karnofsky Performance Scale (KPS) score. Kaplan-Meier analysis \showed that patients with low MAGI2 antisense RNA 3 (MAGI2-AS3) levels had a worse overall survival rate than those with high MAGI2-AS3 levels. Furthermore, the researchers discovered that the expression of MAGI2-AS3 in glioma tissues was an independent predictor of overall survival when analyzed by multivariate logistics (63).
Moreover, Jun-Chi Mei et al. revealed that the amount of long non-coding RNA ELF3 antisense RNA 1(ELF3-AS1) was significantly more tremendous in glioma specimens than in non- tumor samples adjacent to the tumor. Considering its prognostic and diagnostic importance, it could be regarded as a viable marker of glioma (64).On the other hand, F Shang and his Colleagues found that PXN-AS1-L was overexpressed in glioma. It can be used as a potential biomarker for glioma patients’ unfavorable outcomes (65). There’s been a link between metastasis-related lung adenocarcinoma transcript 1 (MALAT1) expression and greater chemoresistance to TMZ, suggesting it could be a prognostic marker for glioma (66). According to a recent analysis of LGG data from the cancer genome atlas (TCGA), 16 immune-related lncRNAs were significantly linked to patient outcomes (67).
Additionally, six lncRNA signatures associated with immunity within the TCGA offer excellent prognostic information for patients with GBM (68). Also, Wang S and his colleague revealed that the long non-coding RNA RPSAP52 could help predict postoperative survival for people with GBM since it is expressed in these cells (69). It was also reported that ADAMTS9-AS2 and HOXA11-AS gene expression increase with tumor grade. In addition, the CASC2 gene expression decreased dramatically with rising tumor grade, and the authors concluded that lncRNA ADAMTS9-AS2, HOXA11-AS, and CASC2 are potential biomarkers for glioma prognosis (70). Continuing to study lncRNAs will provide more effective and sensitive biomarkers to diagnose glioma early.
Due to the diversity of lncRNA activity, these transcripts are attractive as potential therapeutic targets due to their low abundance; broad and severe phenotypic effects (71). There is a poor prognosis for patients with GBM due to the heterogeneity and aggressive nature of glioma cells, an immune-privileged brain environment, the absence of effective treatments, and the inability to penetrate the BBB of most therapies (72). Another significant study developed a generalizable approach for rapidly identifying novel therapeutic targets using CRISPRi-based genome-wide screening. Additionally, this research discovered that antisense oligonucleotides targeting lncGRS-1 suppressed glioma tumor formation in 3D culture and increased the radiation sensitivity of glioma cells (73). According to Ke Zhou et al., nuclear enriched abundant transcript 1 (NEAT1) silencing inhibits motility and invasion of glioma cells by modifying SRY-Box Transcription Factor 2 (SOX2), which miR-132 targets. These findings establish NEAT1 as a potential therapeutic target (42). Earlier this year, it was discovered that inhibiting Wnt/-catenin signaling suppresses the growth of glioblastoma cells by blocking the processing of the lncRNA MIR22HG; for people with glioblastoma, targeting this RNA might represent a novel therapeutic strategy (74).
Additionally, Ning Guan and colleagues noticed that lncRNA Neuroblastoma Associated Transcript 1(NBAT1) was able to halt the evolution of glioma through the miR-21/SOX7 axis, demonstrating its therapeutic potential (75). Another challenge that physicians face while treating a patient with glioma is treatment resistance. LncRNAs have been implicated in improving resistance to several medicines. For instance, lncSBF2-AS1 SBF2-AS1 expression was elevated in TMZ-resistant glioblastoma (GBM) cells and tissues. That overexpression of SBF2-AS1 facilitated TMZ resistance but silenced SBF2-AS1 sensitized resistant GBM cells to TMZ (76). Furthermore, another research revealed that directly targeting miR-10a, the lncRNA TUSC7 decreased temozolomide resistance in glioblastoma (77). Yu L et al. demonstrated that silencing Long intergenic non-protein coding RNA 00475 could act as a tumor suppressor in gliomas via microRNA-449b-5p-dependent AGAP2 upregulation; this indicates the potential for its therapeutic use in the treatment of gliomas (78).
Moreover, another research discovered that lncRNA brain cytoplasmic RNA 1(BCYRN1) served as a miR-619-5p sponge to regulate the cue domain containing 2(CUEDC2) expression in the PTEN/AKT/p21 pathway; this combination reduced glioma development. This research indicated that the lncRNA brain cytoplasmic RNA 1(BCYRN1)acts as a tumor suppressor and may help diagnose and treat glioma (79). The studies discussed above provide critical evidence that lncRNAs may be used as therapeutic agents by suppressing glioma.
Determining therapy responses is critical throughout the cancer treatment process. LncRNAs are among the most effective techniques for monitoring glioma therapy. For example, metastasis-related lung adenocarcinoma transcript 1(MALAT1) has a role in tumor chemosensitivity and is overexpressed in temozolomide (TMZ)-resistant glioblastoma cells. Its silencing lowered TMZ resistance in vivo and in vitro (80). Similarly, TMZ-resistant tumors displayed reduced maternally expressed 3(MEG3) expression (41). Cisplatin sensitivity was boosted by increasing MEG3 expression. At the same time, chemoresistance was created by silencing MEG3 using si-RNA (81), and the authors discovered that MEG3 is a critical tool for tracking chemotherapy response in glioblastoma. Additionally, it was reported that SHG-44 cells resistant to paclitaxel expressed high levels of plasmacytoma variant translocation 1(PVT1) in vitro and the knocking down of this gene improved the chemo-response; the above effects resulted in this long coding RNA being able to track the response to paclitaxel in glioma patients.
The lncRNAs are crucial in many immune responses, including inflammation, cell differentiation, immune cell maturation, and infiltration (82, 83). The expression of immune-related lncRNAs can also be used to classify tumors since their expression is correlated with immunologic molecules such as cytokines and chemokines (84, 85). Due to these circumstances, it is vital to identify immune-related lncRNAs that affects glioma-associated immune cells, which can help develop glioma immunotherapies. For example, one study found a significant connection between immune-related lncRNA levels and immune cell infiltration in various tumors. This research comprehensively charted the landscape of lncRNA regulation in the immunome across 33 cancer types, showing that cancers with similar tissue origin are likely to share lncRNA immune regulators. Moreover, the immune-related lncRNAs are likely to show expression perturbation in cancer and lncRNAs are significantly correlated with immune cell infiltration. They found that immune-related lncRNAs can help prioritize cancer-related lncRNAs and further identify three molecular subtypes (proliferative, intermediate, and immunological, makinge them potential oncogenic biomarkers (86).
In response to toll-like receptors (TLRs) activation, NF-κB (Nuclear factor-κB) induces or suppresses the expression of lncRNAs in various immune cells, which influences immune responses (87). Specific lncRNAs have been linked to tumor growth by dysregulation of proliferation, apoptosis, metastasis, and angiogenesis. Furthermore, lncRNAs mainly target immunological checkpoints and cytokines, facilitating the establishment of an immunosuppressive milieu conducive to tumor proliferation and medication resistance. For instance, a study of the cancer genome atlas (TCGA) database indicated that lncRNA AC003092.1 is associated with an immunosuppressive microenvironment in glioblastoma (88). Through the research of the TCGA and the Chinese glioma genome atlas (CGGA) databases, researchers discovered that lncRNA SBF2-AS1 is linked with immunity in lower-grade glioma (89).
Additionally, researchers have revealed that the expression of the lncRNA MIR155 host gene (MIR155HG) is elevated in GBM tissues in comparison to their equivalent standard counterparts and that its expression is associated with immune checkpoint inhibitors such as Programmed cell death one ligand (PD-L1) and T cell immunoglobulin and mucin domain 3 (TIM-3) in GBM and that its expression is associated with poor prognosis (90). It was also found that the lncRNA RP11-838N2.4 enhanced the cytotoxic effects of temozolomide in glioblastoma cells by inhibiting miR-10a functions (91). Furthermore, another study identified nine immune-associated lncRNAs predictive of low-grade glioma (AC009283.1, AC009227.1, AL121899.1, LINC00174, LINC02166, AC018647.1, AC061961.1, NRAV, and LINC00320). They concluded that these nine lncRNAs represent independent prognostic factors for developing low-grade gliomas and that these RNAs modulate immunological responses and cancer pathways to affect tumorigenesis and prognosis of patients with low-grade gliomas (92). Functional analysis of immune-related lncRNAs revealed that the differences in overall survival between groups with high and low RS might be explained by differences in cell differentiation, microtubule polymerization, and other processes. This study demonstrated that those immune-related lncRNAs contribute to glioma pathogenesis and clinical treatment, and It may be a potential therapeutic for glioma diagnosis (53).
Furthermore, another research discovered 11 immunological lncRNAs that may be used as prognostic indicators for individuals with LGG. The prognosis prediction performance of the nomogram produced according to these lncRNAs is quite effective. In addition, these 11 immune lncRNAs are associated with the infiltration of immune cell subtypes in tumor tissues, which have a possible role in the development of LGG via several routes (93). Furthermore, in glioblastomas, a research identified four prognostic lncRNAs (AK098425, AL833059, AK056155, and CR613436) that co-express protein-coding genes and are strongly associated with immune responses, including the inflammatory response, the innate immune response, and B cell-mediated immunity (68). The lncRNA HOTAIR myeloid-specific 1 (HOTAIRM1), which is overexpressed in gliomas, promotes malignancy by interacting with miR-129-5p as a ceRNA associated with immune activation, which enhances T cell-mediated immune responses (94).. Therefore, these lncRNAs are advantageous for developing immunotherapies specific to glioma. Considering the studies mentioned above, it is evident that studying in deep long non-coding RNAs will reveal new immunotherapeutic targets to develop various treatment options for cancer. This will include effective treatments for gliomas, as well.
Clinical trials assist scientists and physicians in evaluating diagnostic and therapeutic methods for different types of cancer, including gliomas. Numerous clinical studies have been conducted to demonstrate the use of lncRNA in treating and diagnosing a variety of malignancies, including hepatocellular carcinoma (NCT05088811), lung cancer, and prostate cancer (NCT03830619). Additionally, some other clinical trials on immunotherapies for glioma have been conducted, such as a phase I clinical trial evaluating cellular immunotherapy with intratumoral alloreactivity cytotoxic T lymphocytes and interleukin-2 for the treatment of recurrent malignant gliomas or meningioma one of such clinical studies (NCT01144247). However, no clinical studies investigated lncRNAs as diagnostic or therapeutic tools for glioma. Thus, there is a critical need for more clinical studies to demonstrate the functions of lncRNA in glioblastoma and establish them as standard therapy and biomarkers for glioma and other malignancies; details on the aforementioned clinical trials and others are in Table 2.
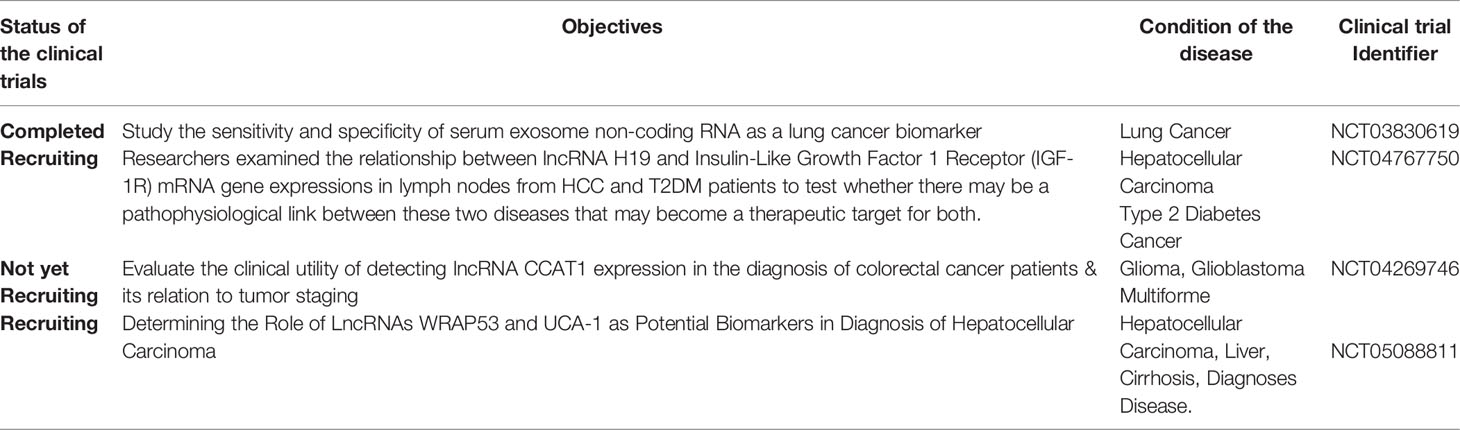
Table 2 Examples of clinical trials done on the roles of lncRNAs as biomarkers or in treatment agents.
Glioblastoma is one of the fatal types of brain cancer. While surgical and radiation advancements have been beneficial, the development of non-invasive treatments remains a necessity for genuinely outstanding treatment alternatives. Long non-coding RNAs have recently received renewed attention in glioma research and may be a viable tool for targeted therapy. It is well established that lncRNAs engage in various cellular regulatory mechanisms that influence the development of carcinogenesis. Numerous aberrant lncRNAs have been linked with glioma. They have been implicated in almost all of its characteristics, including cell proliferation, motility, angiogenesis, stemless, tumor recurrence, severity, and chemoresistance. Innovative biochemical and molecular techniques have aided in elucidating how lncRNAs govern and regulate many functionalities. While it is encouraging to discover potential diagnostics and therapies to increase glioma survival utilizing lncRNAs, most lncRNAs have different undefined properties. The distribution of medications into the brain across an intact BBB is an essential element to consider when developing helpful therapeutics. Due to their low stability and poor drug absorption, therapies based on lncRNAs are limited to in vivo usage. Thus, future research should focus on detecting the cell-to-cell heterogeneity of lncRNAs in gliomas, as this would aid in the creation of innovative RNA-based methodologies for treating this malignancy and instill fresh confidence in patients with glioma also, as recent research has shown that lncRNAs operate as epigenetic regulators that promote innate immune memory responses. Further investigation of immune-related lncRNA in terms of immune response prediction is necessary, as they may serve as candidates for particular immune cell-mediated tumor evasion and cellular death.
XW designed and drafted the manuscript. LY, JW, YH, CW, and ZL discussed and revised the manuscript. All authors read and approved the final manuscript.
This study was supported by the Clinical Medicine Science and Technology Innovation Project of JINAN (No. 202019020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
NEAT1, Nuclear enriched abundant transcript 1; Cox-2, Cyclooxygenase-2; FGD5-AS1, FGD5 antisense RNA 1; MZF1, Myeloid zinc finger 1; HIF1, Hypoxia-inducible factor 1-alpha; ANXA2, Annexin A2; CREB1, CAMP Responsive element-binding protein 1; FOXD2-AS1, Forkhead box D2 adjacent apposite strand RNA 1; XIST, X-inactive specific transcript;
VEGF, Vascular endothelial growth factor; EMT, Epithelial-to-Mesenchymal Transition;
HOTAIRM1, HOXA transcript antisense RNA, myeloid-Specific 1; TMPO-AS1, TMPO antisense RNA 1; TME, tumor microenvironment;
LGGs, Low-grade gliomas; MAGI2-AS3, MAGI2 antisense RNA 3; ELF3-AS1, ELF3 antisense RNA 1; MALAT1, Metastasis-associated lung adenocarcinoma transcript 1;
TMZ, Temozolomide; AGAP2, ArfGAP with GTPase domain, ankyrin repeat, and ph domain 2; BCYRN1, Brain cytoplasmic RNA 1; CUEDC2, CUE domain containing 2; BBB, Blood-brain barrier.
1. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The Epidemiology of Glioma in Adults: A "State of the Science" Review. Neuro Oncol (2014) 16(7):896–913. doi: 10.1093/neuonc/nou087
2. Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006-2010. Neuro Oncol (2013) 15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151
3. Kan LK, Drummond K, Hunn M, Williams D, Brien TJ, Monif M. Potential Biomarkers and Challenges in Glioma Diagnosis, Therapy, and Prognosis. BMJ Neurol Open (2020) 2(2):e000069. doi: 10.1136/bmjno-2020-000069
4. Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2010-2014. Neuro-Oncology (2017) 19:v1–88. doi: 10.1093/neuonc/nox158
5. Wen PY, Kesari S. Malignant Gliomas in Adults. N Engl J Med (2008) 359:492–507. doi: 10.1056/NEJMra0708126
6. Silantyev AS, Falzone L, Libra M, Gurina OI, Kardashova KS, Nikolouzakis TK, et al. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells (2019) 8:863. doi: 10.3390/cells8080863
7. Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis Through Clinical Presentation and Treatment. Asian Pacific J Cancer Prev (APJCP) (2017) 18(1):3. doi: 10.22034/APJCP.2017.18.1.3
8. Tykocki T, Eltayeb M. Ten-Year Survival in Glioblastoma. A Systematic Review. J Clin Neurosci (2018) 54:7–13. doi: 10.1016/j.jocn.2018.05.002
9. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathologica (2016) 131(6):803–20. doi: 10.1007/s00401-016-1545-1
10. Nørøxe DS, Yde CW, Østrup O, Michaelsen SR, Schmidt AY, Kinalis S, et al. Genomic Profiling of Newly Diagnosed Glioblastoma Patients and Its Potential for Clinical Utility – A Prospective, Translational Study. Mol Oncol (2020) 14(11):2727–43. doi: 10.1002/1878-0261.12790
11. Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of Gliomas. Cancer Treat Res (2015) 163:1–14. doi: 10.1007/978-3-319-12048-5_1
12. Minniti G, De Sanctis V, Muni R, Filippone F, Bozzao A, Valeriani M, et al. Radiotherapy Plus Concomitant and Adjuvant Temozolomide for Glioblastoma in Elderly Patients. J Neuro-Oncol (2008) 88(1):97–103. doi: 10.1007/s11060-008-9538-0
13. Alifieris C, Trafalis DT. Glioblastoma Multiforme: Pathogenesis and Treatment. Pharmacol Ther (2015) 152:63–82. doi: 10.1016/j.pharmthera.2015.05.005
14. Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends In Biochem Sci (2019) 44(1):33–52. doi: 10.1016/j.tibs.2018.09.012
15. Buruiană A, Florian ȘI, Florian AI, Timiș T-L, Mihu CM, Miclăuș M, et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int J Mol Sci (2020) 21(6):1950. doi: 10.3390/ijms21061950
16. Liu ZZ, Tian YF, Wu H, Ouyang SY, Kuang WL. LncRNA H19 Promotes Glioma Angiogenesis Through miR-138/HIF-1α/VEGF Axis. Neoplasma (2020) 67(1):111–8. doi: 10.4149/neo_2019_190121N61
17. Jia P, Cai H, Liu X, Chen J, Ma J, Wang P, et al. Long Non-Coding RNA H19 Regulates Glioma Angiogenesis and the Biological Behavior of Glioma-Associated Endothelial Cells by Inhibiting microRNA-29a. Cancer Lett (2016) 381(2):359–69. doi: 10.1016/j.canlet.2016.08.009
18. Kopp F, Mendell JTJC. Functional Classification and Experimental Dissection of Long Non-Coding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
19. Bali KK, Kuner RJ. Noncoding RNAs: Key Molecules in Understanding and Treating Pain. (2014) 20(8):437–48. doi: 10.1016/j.molmed.2014.05.006
20. Batista PJ, Chang HYJC. Long Non-Coding RNAs: Cellular Address Codes in Development and Disease. Cell (2013) 152(6):1298–307. doi: 10.1016/j.cell.2013.02.012
21. Kim S-H, Lim K-H, Yang S, Joo J-Y. Long Non-Coding RNAs in Brain Tumors: Roles and Potential as Therapeutic Targets. J Hematol Oncol (2021) 14(1):77. doi: 10.1186/s13045-021-01088-0
22. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for Glioma: Current Management and Future Application. Cancer Lett (2020) 476:1–12. doi: 10.1016/j.canlet.2020.02.002
23. Simon T, Jackson E, Giamas G. Breaking Through the Glioblastoma Micro-Environment via Extracellular Vesicles. Oncogene (2020) 39(23):4477–90. doi: 10.1038/s41388-020-1308-2
24. Geribaldi-Doldán N, Fernández-Ponce C, Quiroz RN, Sánchez-Gomar I, Escorcia LG, Velásquez EP, et al. The Role of Microglia in Glioblastoma. Front Oncol (2021) 10(2979):10:603495. doi: 10.3389/fonc.2020.603495
25. Chaudhary R, Morris RJ, Steinson E. The Multifactorial Roles of Microglia and Macrophages in the Maintenance and Progression of Glioblastoma. J Neuroimmunol (2021) 357:577633. doi: 10.1016/j.jneuroim.2021.577633
26. Zhou L, Zhu Y, Sun D, Zhang Q. Emerging Roles of Long Non-Coding RNAs in The Tumor Microenvironment. Int J Biol Sci (2020) 16(12):2094–103. doi: 10.7150/ijbs.44420
27. Cech TR, Steitz JA. The Non-Coding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell (2014) 157(1):77–94. doi: 10.1016/j.cell.2014.03.008
28. Yao R-W, Wang Y, Chen L-L. Cellular Functions of Long Non-Coding RNAs. Nat Cell Biol (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
29. Woolard E, Chorley BN. Chapter 3-1 - The Role of Noncoding RNAs in Gene Regulation. In: McCullough SD, Dolinoy DC, editors. Toxicoepigenetics. Cambridge, Massachusetts: Academic Press (2019). p. 217–35.
30. Yan Q, Zhu C, Guang S, Feng X. The Functions of Noncoding RNAs in rRNA Regulation. Front Genet (2019) 10(290):290. doi: 10.3389/fgene.2019.00290
31. Kaikkonen MU, Lam MTY, Glass CK. Non-Coding RNAs as Regulators of Gene Expression and Epigenetics. Cardiovasc Res (2011) 90(3):430–40. doi: 10.1093/cvr/cvr097
32. Zhang X, Niu W, Mu M, Hu S, Niu C. Long Non-Coding RNA LPP-AS2 Promotes Glioma Tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC Feedback Loop. J Exp Clin Cancer Res (2020) 39(1):196. doi: 10.1186/s13046-020-01695-8
33. Zhao JB, Xue JF, Zhang WZ, Ren YL, Yan DM. Long Noncoding RNA FGD5-AS1 Promotes Glioma Cell Proliferation, Migration and Invasion by Regulating Wnt/β-Catenin Pathway. Cancer Manage Res (2020) 12:6187–93. doi: 10.2147/CMAR.S250284
34. Li J, Liao T, Liu H, Yuan H, Ouyang T, Wang J, et al. Hypoxic Glioma Stem Cell-Derived Exosomes Containing Linc01060 Promote Progression of Glioma by Regulating the MZF1/c-Myc/Hif1α Axis. Cancer Res (2021) 81(1):114–28. doi: 10.1158/0008-5472.CAN-20-2270
35. Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The Mir155hg/miR-185/ANXA2 Loop Contributes to Glioblastoma Growth and Progression. J Of Exp Clin Cancer Res (2019) 38(1):133–3. doi: 10.1186/s13046-019-1132-0
36. Zhang W, Bi Y, Li J, Peng F, Li H, Li C, et al. Long Non-Coding RNA FTX is Upregulated in Gliomas and Promotes Proliferation and Invasion of Glioma Cells by Negatively Regulating miR-342-3p. Lab Investig A J Tech Methods Pathol (2017) 97(4):447–57. doi: 10.1038/labinvest.2016.152
37. Dong H, Cao W, Xue J. Long Non-Coding FOXD2-AS1 is Activated by CREB1 and Promotes Cell Proliferation and Metastasis in Glioma by Sponging miR-185 Through Targeting AKT1. Biochem Biophys Res Commun (2019) 508(4):1074–81. doi: 10.1016/j.bbrc.2018.12.050
38. Luo C, Quan Z, Zhong B, Zhang M, Zhou B, Wang S, et al. lncRNA XIST Promotes Glioma Proliferation and Metastasis Through miR-133a/SOX4. Exp Ther Med (2020) 19(3):1641–8. doi: 10.3892/etm.2020.8426
39. Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, et al. Long Non-Coding RNA XIST Promotes Glioma Tumorigenicity and Angiogenesis by Acting as a Molecular Sponge of miR-429. J Cancer (2017) 8(19):4106–16. doi: 10.7150/jca.21024
40. Zhao J, Zeng XB, Zhang HY, Xiang JW, Liu YS. Long Non-Coding RNA FOXD2-AS1 Promotes Cell Proliferation, Metastasis and EMT in Glioma by Sponging miR-506-5p. Open Med (Warsaw Poland) (2020) 15(1):921–31. doi: 10.1515/med-2020-0175
41. Momtazmanesh S, Rezaei N. Long Non-Coding RNAs in Diagnosis, Treatment, Prognosis, and Progression of Glioma: A State-Of-the-Art Review. Front. Oncol. (2021) 11(2698):712786. doi: 10.3389/fonc.2021.712786
42. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y, et al. Knockdown of Long Non-Coding RNA NEAT1 Inhibits Glioma Cell Migration and Invasion via Modulation of SOX2 Targeted by miR-132. Mol Cancer (2018) 17(1):105. doi: 10.1186/s12943-018-0849-2
43. Zhang D, Jiang H, Ye J, Gao M, Wang X, Lu E, et al. A Novel lncRNA, RPL34-AS1, Promotes Proliferation and Angiogenesis in Glioma by Regulating VEGFA. J Cancer (2021) 12(20):6189–97. doi: 10.7150/jca.59337
44. Liu G, Yang H, Cao L, Han K, Li G. LncRNA TMPO-AS1 Promotes Proliferation and Invasion by Sponging miR-383-5p in Glioma Cells. Cancer Manage Res (2020) 12:12001–9. doi: 10.2147/CMAR.S282539
45. Guo E, Liang C, He X, Song G, Liu H, Lv Z, et al. Long Noncoding RNA LINC00958 Accelerates Gliomagenesis Through Regulating miR-203/Cdk2. DNA Cell Biol (2018) 37(5):465–72. doi: 10.1089/dna.2018.4163
46. Sun WL, Kang T, Wang YY, Sun JP, Li C, Liu HJ, et al. Long Non-Coding RNA OIP5-AS1 Targets Wnt-7b to Affect Glioma Progression via Modulation of miR-410. Biosci Rep (2019) 39(1). doi: 10.1042/BSR20180395
47. Pi Y-N, Qi W-C, Xia B-R, Lou G, Jin W-L. Long Non-Coding RNAs in the Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Front Immunol (2021) 12(2719):697083. doi: 10.3389/fimmu.2021.697083
48. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune Cells Within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Lett (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
49. Jin MZ, Jin WL. The Updated Landscape of Tumor Microenvironment and Drug Repurposing. Signal Transduct Target Ther (2020) 5(1):166. doi: 10.1038/s41392-020-00280-x
50. Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front Immunol (2020) 11:940. doi: 10.3389/fimmu.2020.00940
51. DeCordova S, Shastri A, Tsolaki AG, Yasmin H, Klein L, Singh SK, et al. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front Immunol (2020) 11:1402. doi: 10.3389/fimmu.2020.01402
52. Vega EA, Graner MW, Sampson JH. Combating Immunosuppression in Glioma. Future Oncol (2008) 4(3):433–42. doi: 10.2217/14796694.4.3.433
53. Xia P, Li Q, Wu G, Huang Y. An Immune-Related lncRNA Signature to Predict Survival In Glioma Patients. Cell Mol Neurobiol (2021) 41(2):365–75. doi: 10.1007/s10571-020-00857-8
54. Wang X, Gao M, Ye J, Jiang Q, Yang Q, Zhang C, et al. An Immune Gene-Related Five-lncRNA Signature for to Predict Glioma Prognosis. Front Genet (2020) 11:612037. doi: 10.3389/fgene.2020.612037
55. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 Promotes Activation of Inflammasomes in Macrophages. Nat Commun (2019) 10(1):1495. doi: 10.1038/s41467-019-09482-6
56. Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages Through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol (Baltimore Md 1950) (2016) 196(6):2799–808. doi: 10.4049/jimmunol.1502146
57. Xu S, Tang L, Liu Z, Luo C, Cheng Q. Hypoxia-Related lncRNA Correlates With Prognosis and Immune Microenvironment in Lower-Grade Glioma. Front Immunol (2021) 12:731048. doi: 10.3389/fimmu.2021.731048
58. Yang X, Xie Z, Lei X, Gan R. Long Non-Coding RNA GAS5 in Human Cancer. Oncol Lett (2020) 20(3):2587–94. doi: 10.3892/ol.2020.11809
59. Al-Rugeebah A, Alanazi M, Parine NR. MEG3: An Oncogenic Long Non-Coding RNA in Different Cancers. Pathol Oncol Res (2019) 25(3):859–74. doi: 10.1007/s12253-019-00614-3
60. Chandra Gupta S, Nandan Tripathi Y. Potential of Long Non-Coding RNAs in Cancer Patients: From Biomarkers to Therapeutic Targets. Int J Cancer (2017) 140(9):1955–67. doi: 10.1002/ijc.30546
61. Tu Z, Wu L, Wang P, Hu Q, Tao C, Li K, et al. N6-Methylandenosine-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Lower-Grade Glioma Patients. Front Cell Dev Biol (2020) 8:642–2. doi: 10.3389/fcell.2020.00642
62. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum Long Non-Coding RNA HOTAIR as a Novel Diagnostic and Prognostic Biomarker in Glioblastoma Multiforme. Mol Cancer (2018) 17(1):74–4. doi: 10.1186/s12943-018-0822-0
63. Chen XD, Zhu MX, Wang SJ. Expression of Long Non-Coding RNA MAGI2−AS3 in Human Gliomas and Its Prognostic Significance. Eur Rev Med Pharmacol Sci (2019) 23(8):3455–60. doi: 10.1186/s12943-018-0822-0
64. Mei JC, Yan G, Mei SQ. Diagnostic and Prognostic Potentials of Long Noncoding RNA ELF3-AS1 in Glioma Patients. Dis Markers (2020) 2020:8871746. doi: 10.1155/2020/8871746
65. Shang F, Du SW, Ma XL. Up−regulation of lncRNA PXN-AS1-L is Associated With Unfavorable Prognosis in Patients Suffering From Glioma. Eur Rev Med Pharmacol Sci (2019) 23(20):8950–5. doi: 10.26355/eurrev_201910_19293
66. Chen W, Xu X-K, Li J-L, Kong K-K, Li H, Chen C, et al. MALAT1 is a Prognostic Factor in Glioblastoma Multiforme and Induces Chemoresistance to Temozolomide Through Suppressing miR-203 and Promoting Thymidylate Synthase Expression. Oncotarget (2017) 8(14):22783–99. doi: 10.18632/oncotarget.15199
67. Li X, Meng Y. Survival Analysis of Immune-Related lncRNA in Low-Grade Glioma. BMC Cancer (2019) 19(1):813. doi: 10.1186/s12885-019-6032-3
68. Zhou M, Zhang Z, Zhao H, Bao S, Cheng L, Sun J. An Immune-Related Six-lncRNA Signature to Improve Prognosis Prediction of Glioblastoma Multiforme. Mol Neurobiol (2018) 55(5):3684–97. doi: 10.1007/s12035-017-0572-9
69. Wang S, Guo X, Lv W, Li Y, Zhang L, Dong C, et al. LncRNA RPSAP52 Upregulates TGF-β1 to Increase Cancer Cell Stemness and Predict Postoperative Survival in Glioblastoma. Cancer Manage Res (2020) 12:2541–7. doi: 10.2147/CMAR.S227496
70. Ebrahimi AA, Ashoori H, Vahidian F, Mosleh IS, Kamian S. Long Non-Coding RNA Panel as a Molecular Biomarker in Glioma. J Egyptian Natl Cancer Inst (2021) 33(1):31. doi: 10.1186/s43046-021-00090-4
71. Stackhouse CT, Gillespie GY, Willey CD. Exploring the Roles of lncRNAs in GBM Pathophysiology and Their Therapeutic Potential. Cells (2020) 9(11):2369. doi: 10.3390/cells9112369
72. Zeng T, Li L, Zhou Y, Gao L. Exploring Long Non-Coding RNAs in Glioblastoma: Regulatory Mechanisms and Clinical Potentials. Int J Genomics (2018) 2018:2895958. doi: 10.1155/2018/2895958
73. Liu SJ, Malatesta M, Lien BV, Saha P, Thombare SS, Hong SJ, et al. CRISPRi-Based Radiation Modifier Screen Identifies Long Non-Coding RNA Therapeutic Targets in Glioma. Genome Biol (2020) 21(1):83. doi: 10.1186/s13059-020-01995-4
74. Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, et al. Interfering With Long Non-Coding RNA MIR22HG Processing Inhibits Glioblastoma Progression Through Suppression of Wnt/β-Catenin Signalling. Brain (2020) 143(2):512–30. doi: 10.1093/brain/awz406
75. Guan N, Wang R, Feng X, Li C, Guo W. Long Non-Coding RNA NBAT1 Inhibits the Progression of Glioma Through the miR-21/SOX7 Axis. Oncol Lett (2020) 20(3):3024–34. doi: 10.3892/ol.2020.11847
76. Zhang Z, Yin J, Lu C, Wei Y, Zeng A, You Y. Exosomal Transfer of Long Non-Coding RNA SBF2-AS1 Enhances Chemoresistance to Temozolomide in Glioblastoma. J Exp Clin Cancer Res (2019) 38(1):166. doi: 10.1186/s13046-019-1139-6
77. Shang C, Tang W, Pan C, Hu X, Hong Y. Long Non-Coding RNA TUSC7 Inhibits Temozolomide Resistance by Targeting miR-10a in Glioblastoma. Cancer Chemother Pharmacol (2018) 81(4):671–8. doi: 10.1007/s00280-018-3522-y
78. Yu L, Gui S, Liu Y, Qiu X, Qiu B, Zhang X, et al. Long Intergenic Non-Protein Coding RNA 00475 Silencing Acts as a Tumor Suppressor in Glioma Under Hypoxic Condition by Impairing microRNA-449b-5p-Dependent AGAP2 Up-Regulation. Ther Adv Med Oncol (2020) 12:1758835920940936. doi: 10.1177/1758835920940936
79. Mu M, Niu W, Zhang X, Hu S, Niu C. LncRNA BCYRN1 Inhibits Glioma Tumorigenesis by Competitively Binding With miR-619-5p to Regulate CUEDC2 Expression and the PTEN/AKT/p21 Pathway. Oncogene (2020) 39(45):6879–92. doi: 10.1038/s41388-020-01466-x
80. Kim SS, Harford JB, Moghe M, Rait A, Pirollo KF, Chang EH. Targeted Nanocomplex Carrying siRNA Against MALAT1 Sensitizes Glioblastoma to Temozolomide. Nucleic Acids Res (2018) 46(3):1424–40. doi: 10.1093/nar/gkx1221
81. Ma B, Gao Z, Lou J, Zhang H, Yuan Z, Wu Q, et al. Long Non-Coding RNA MEG3 Contributes to Cisplatin−Induced Apoptosis via Inhibition of Autophagy in Human Glioma Cells. Mol Med Rep (2017) 16(3):2946–52. doi: 10.3892/mmr.2017.6897
82. Chen YG, Satpathy AT, Chang HY. Gene Regulation in the Immune System by Long Non-Coding RNAs. Nat Immunol (2017) 18(9):962–72. doi: 10.1038/ni.3771
83. Geng H, Tan X-D. Functional Diversity of Long Non-Coding RNAs in Immune Regulation. Genes Dis (2016) 3(1):72–81. doi: 10.1016/j.gendis.2016.01.004
84. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front Immunol (2019) 10:379. doi: 10.3389/fimmu.2019.00379
85. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell (2018) 33(4):690–705.e699. doi: 10.1016/j.ccell.2018.03.014
86. Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Et Al: Pan-Cancer Characterization of Immune-Related lncRNAs Identifies Potential Oncogenic Biomarkers. Nat Commun (2020) 11(1):1000. doi: 10.1038/s41467-020-14802-2
87. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of Long Noncoding RNAs. Annu Rev Immunol (2017) 35(1):177–98. doi: 10.1146/annurev-immunol-041015-055459
88. Guo XY, Zhong S, Wang ZN, Xie T, Duan H, Zhang JY, et al. Immunogenomic Profiling Demonstrate AC003092.1 as an Immune-Related eRNA in Glioblastoma Multiforme. Front Genet (2021) 12:633812. doi: 10.3389/fgene.2021.633812
89. Zhang Q, Liu XJ, Li Y, Ying XW, Chen L. Prognostic Value of Immune-Related lncRNA SBF2-AS1 in Diffuse Lower-Grade Glioma. Technol Cancer Res Treat (2021) 20:15330338211011966. doi: 10.1177/15330338211011966
90. Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a Prognostic Biomarker and Associated With Immune Infiltration and Immune Checkpoint Molecules Expression in Multiple Cancers. Cancer Med (2019) 8(17):7161–73. doi: 10.1002/cam4.2583
91. Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang Z, et al. Long Non-Coding RNA RP11-838N2.4 Enhances the Cytotoxic Effects of Temozolomide by Inhibiting the Functions of miR-10a in Glioblastoma Cell Lines. Oncotarget (2016) 7(28):43835–51. doi: 10.18632/oncotarget.9699
92. Maimaiti A, Jiang L, Wang X, Shi X, Pei Y, Hao Y, et al. Identification and Validation of an Individualized Prognostic Signature of Lower-Grade Glioma Based on Nine Immune Related Long Non-Coding RNA. Clin Neurol Neurosurg (2021) 201:106464. doi: 10.1016/j.clineuro.2020.106464
93. Wen J, Wang Y, Luo L, Peng L, Chen C, Guo J, et al. Identification and Verification on Prognostic Index of Lower-Grade Glioma Immune-Related LncRNAs. Front Oncol (2020) 10:578809–9. doi: 10.3389/fonc.2020.578809
Keywords: glioma, biomarkers, long non-coding RNA, prognosis, immunotherapy
Citation: Wu X, Yang L, Wang J, Hao Y, Wang C and Lu Z (2022) The Involvement of Long Non-Coding RNAs in Glioma: From Early Detection to Immunotherapy. Front. Immunol. 13:897754. doi: 10.3389/fimmu.2022.897754
Received: 16 March 2022; Accepted: 19 April 2022;
Published: 10 May 2022.
Edited by:
Mohd Wajid Ali Khan, University of Hail, Saudi ArabiaReviewed by:
Ipek Erdogan, Izmir Institute of Technology, TurkeyCopyright © 2022 Wu, Yang, Wang, Hao, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiming Lu, bHV6aGltaW5nc2R1QDE2My5jb20=; Changyin Wang, MTc5MzE1MjkwM0BxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.