- 1Eye Center, Renmin Hospital of Wuhan University, Wuhan, China
- 2School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Background: Behcet’s disease (BD) is a chronic immune disease that involves multiple systems. As the pathogenesis of BD is not clear, and new treatments are needed, we used bioinformatics to identify potential drugs and validated them in mouse models.
Methods: Behcet’s disease-related target genes and proteins were screened in the PubMed and UVEOGENE databases. The biological functions and pathways of the target genes were analyzed in detail by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. A protein-protein interaction (PPI) network was constructed by the STRING database, and hub genes were identified by the Cytoscape plug-in CytoHubba. Gene-drug interactions were identified from the DGIdb database. Experimental autoimmune uveitis (EAU) mice were used as an animal model for drug validation.
Results: A total of 249 target genes and proteins with significant differences in BD were screened, and the results of functional enrichment analysis suggested that these genes and proteins were more located on the cell membrane, involved in regulating the production of cytokines and affecting the activity of cytokines. They mainly regulated “Cytokine- Cytokine receptor interaction”, “Inflammatory bowel disease (IBD)” and “IL-17 signaling Pathway”. In addition, 10 hub genes were obtained through PPI network construction and CytoHubba analysis, among which the top 3 hub genes were closely related to BD. The DGIdb analysis enriched seven drugs acting together on the top 3 hub genes, four of which were confirmed for the treatment of BD or its complications. There is no evidence in the research to support the results in omeprazole, rabeprazole, and celastrol. However, animal experiments showed that rabeprazole and celastrol reduced anterior chamber inflammation and retinal inflammation in EAU mice.
Conclusions: The functional analysis of genes and proteins related to BD, identification of hub genes, and validation of potential drugs provide new insights into the disease mechanism and potential for the treatment of BD.
Introduction
Behcet’s disease (BD) is a chronic systemic disease involving multiple systems that generally presents with recurrent oral ulcers, genital ulcers, uveitis, vasculitis, skin lesions, and neurological and intestinal manifestations (1). BD is prevalent in Turkey, the Middle East, and East Asia (2, 3). However, the aetiology of BD is still unclear, and its diagnosis is complicated (4). Although occlusive vasculitis is a typical histopathological feature of BD, diagnosis still depends on typical clinical manifestations and clinical experience (5, 6).
Uveitis, one of the most common eye diseases in BD, is usually secondary to systemic manifestations, such as recurrent oral ulcers, after an average of 4 years (7). The typical characteristics of uveitis associated with BD are acute recurrent bilateral symmetrical or asymmetrical nongranulomatous panuveitis, accompanied by retinal vasculitis, and a tendency to self-heal (8). The treatment of BD mainly involves suppressing inflammation and reducing tissue damage. Although glucocorticoids and immunosuppressants are usually used for systemic anti-inflammatory treatment, treatment plans should be formulated based on clinical manifestations (9). Regarding the treatment of Behcet’s uveitis, in addition to glucocorticoids and immunosuppressants, other biological agents, such as infliximab (IFX) and adalimumab, have been proven to have certain therapeutic effects (10). Intravitreal injection of glucocorticoids is also an adjuvant treatment (11).
Currently, it is possible to control the progression of BD through the use of diverse therapeutic drugs and individualized treatments. However, it is also necessary to further understand the pathogenesis of this disease and introduce new, more effective drugs to control symptoms, improve prognosis, and even change the course of the disease (12). Here, bioinformatics was used to analyze genes or proteins most recently reported to be involved in BD to search for new potential drugs. Finally, we tested the possible impact of drug intervention on experimental autoimmune uveitis (EAU) mice, a model that has been widely used for human uveitis research (13). Overall, we aimed to provide new ideas for the treatment of BD.
Material and Methods
Identification of Behcet’s Disease-Related Target Genes and Proteins
Behcet’s disease-related target genes and proteins were obtained through the UVEOGENE database (http://www.uvogene.com) and PubMed (14) (15). We searched for papers related to BD from January 2016 to September 2021 in PubMed, and the relevant search terms included “Behcet’s disease”, “Neuro-Behcet’s disease”, “Bechet syndrome” and “Intestinal Behcet’s disease”. And detailed methods for screening target genes and proteins can be found in Section 1.1 of the Appendix.
Target Enrichment Analysis
GO and KEGG analyses of target genes or proteins were performed using the Metascape database (http://metascape.org/) and the UniProt database (https://www.uniprot.org/) Detailed methods are present in Section 1.2 of the Appendix.
Protein-Protein Interaction Network Analysis
A PPI network was constructed using the STRING database (https://string-db.org/) and Cytoscape software (version 3.8.2, California, USA). Detailed methods are present in Section 1.3 of the Appendix.
Acquisition of Hub Gene
The top ten hub genes in the PPI network were enriched by Cytoscape plug-in cytoHubba. Detailed methods are present in Section 1.4 of the Appendix.
Gene-Drug Interaction Analysis
Gene-drug interaction analysis can be achieved through the Drug Gene Interaction Database (DGIdb) version 4.2.0 (https://www.dgidb.org) (16). The drug-gene interaction network was constructed by the DGIdb database and Cytoscape Software. Detailed methods are present in Section 1.5 of the Appendix.
Construction of Mouse Models of Simulated Ocular Behcet’s Disease
The Jackson Lab (Bar Harbor, ME, USA) provided B10.RIII mouse parents. Mice were bred and raised under specific pathogen-free (SPF) conditions. The EAU model was constructed by subcutaneous injection of 200 µl of emulsifier per mouse. The emulsifier was prepared by mixing 50 µg IRBP with an equal volume of CFA containing 1.0 mg/ml of Mycobacterium tuberculosis strain (MTB) (17). Our study was approved by the Ethics Committee of The Renmin Hospital of Wuhan University (WDRM20210708A). We made every effort to minimize the harm to the animals.
Relative mRNA Expression of Hub Genes in EAU Models
TRIzol (Invitrogen, Carlsbad, CA, United States) was applied to extract total RNA from the retinas of EAU mice on the 14th day after injection. The extracted RNA was reverse transcribed into cDNA using the PrimeScript RT kit (Vazyme, China). SYBR premix (Vazyme, China) was used for final quantitative PCR detection. All the primers are listed in Table 1. The 2-△△Ct cycle threshold method was applied to calculate the relative mRNA expression.
Drug Intervention Corresponding to Hub Genes in EAU Models
Celastrol purchased from MedChemExpress (MCE, HY-13067) was dissolved in DMSO (Sigma) at a concentration of 74 mM (18). Celastrol was aliquoted and stored at -20°C. EAU mice were intraperitoneally injected with celastrol at a dose of 1 mg/kg/day from Day 7 to Day 14, and control EAU mice were intraperitoneally injected with PBS containing the same concentration of DMSO (18). Rabeprazole (MCE, HY-B0656A) was dissolved in sterile PBS and intraperitoneally injected into EAU mice at a dose of 60 mg/kg/day from Day 7 to Day 14 (19, 20). Control mice were given an intraperitoneal injection with an equal volume of PBS. From Day 7 to Day 14 of EAU progression, ocular inflammation was observed and recorded daily under a slit-lamp microscope. On Day 14 of EAU, the eyeballs or retinas of mice were removed for HE staining and quantitative PCR. The quantitative PCR method is as mentioned above. The clinical score and histological score followed the scoring criteria proposed by Caspi RR (21).
Statistical Analysis
All data were manifested as means ± standard error of the mean (Means ± SEM). Data were analyzed by GraphPad Prism 7.0(GraphPad Software, Inc, San Diego, CA, USA). The EAU score was processed by a Mann-Whitney U test. qRT-PCR experiments were performed by two-tailed Student’s t-test. P<0.05 was considered significant.
Results
Included Target Genes and Proteins Related to BD
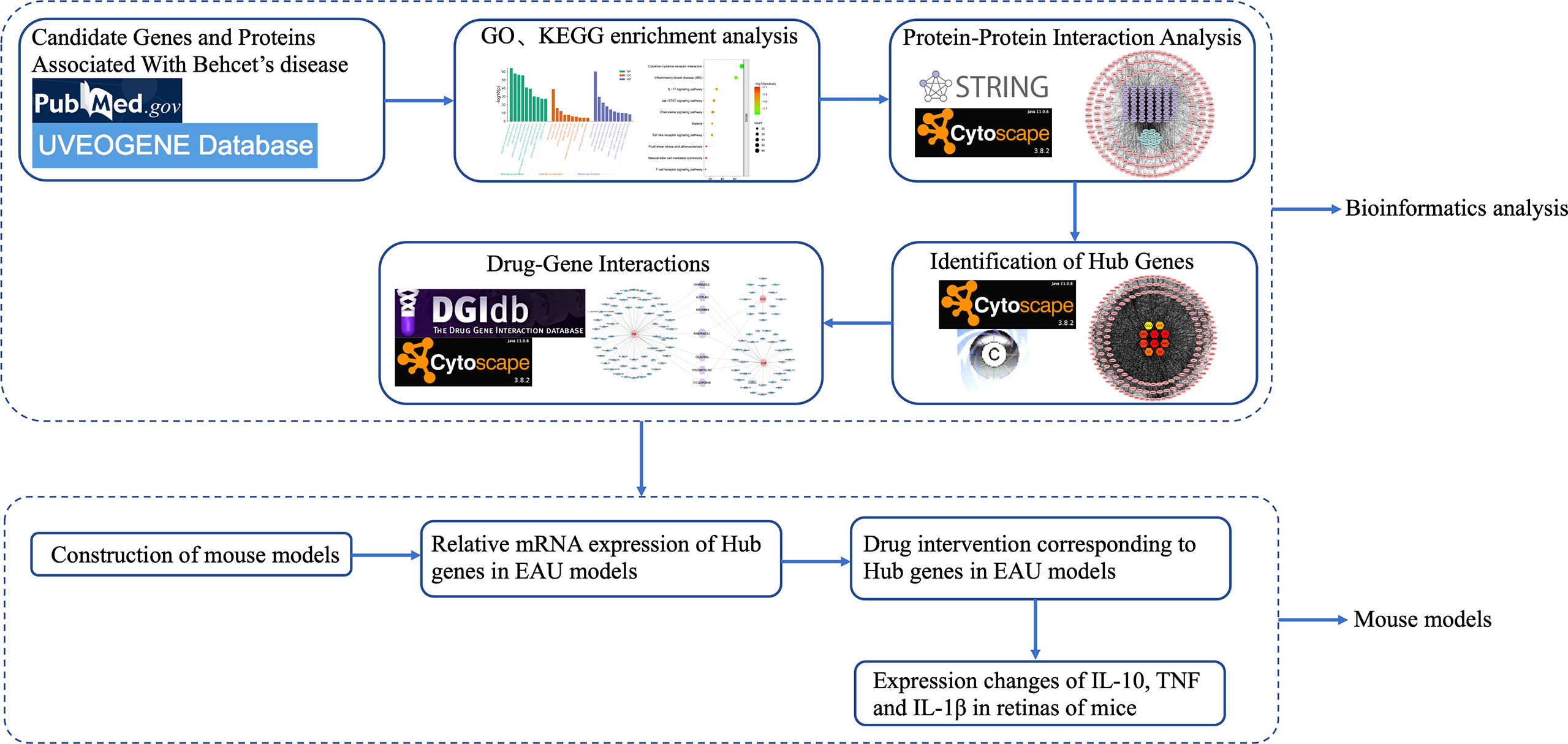
From January 2016 to September 2021, a total of 2654 articles were screened from PubMed, and 352 genes or proteins were selected according to the inclusion and exclusion criteria. After removing duplicates, a list of 249 differentially expressed genes or proteins was obtained that were significantly different from BD in the UVEOGENE database. Among them, 135 genes or proteins came from the UVEOGENE database and 114 from PubMed. The genes and proteins were normalized in the UniProt database, and the results are shown in Supplementary Table 1. The main research process of this study is shown in Figure 1.
Enrichment Analysis of Target Genes and Proteins in BD
A total of 249 target genes and proteins were imported into the Metascape database for GO enrichment analysis and KEGG enrichment analysis. A total of 503 GO items were screened out in the Metascape database, including 337 BP items, 79 CC items, and 87 MF items, accounting for 67%, 15.7%, and 17.3%, respectively, and the top 10 items are shown in Figure 2. BP analysis showed that the included target genes and proteins primarily affected the “regulation of cytokine production”, “immune effector process”, “response to bacterium” and “cytokine-mediated signaling pathway”. In terms of cellular components (CC), these targets were mainly involved in the “side of membrane”, “endocytic vesicle” and “receptor complex”. In MF analysis, “cytokine activity”, “immune receptor activity” and “growth factor receptor binding” were affected. The KEGG enrichment analysis processed by the Metascape database screened 87 items (P<0.01) (Supplementary Table 2). The top 10 items are shown in Figure 3. The results suggested that these targets have a more significant impact on “Cytokine-cytokine receptor interaction”, “Inflammatory bowel disease (IBD) “and “ IL-17 signaling pathway” (Figure 3). In addition, 65 genes, 38 genes, and 25 genes were involved in these three pathways respectively. The diagram of the three signaling pathways is shown in Supplementary Figures 1–3.
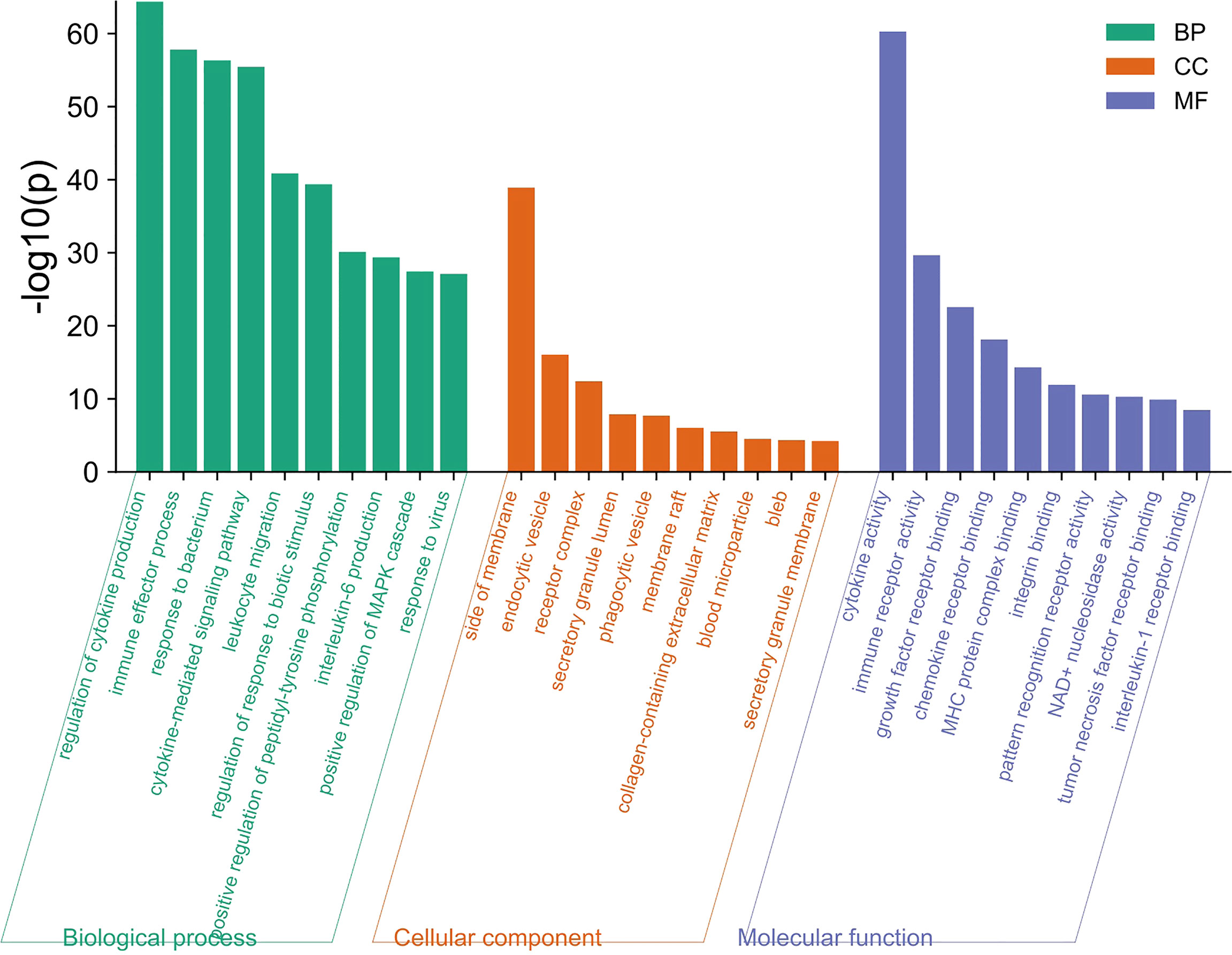
Figure 2 The top 10 items of GO enrichment analysis. The green, orange, and blue bars represent the analysis results of BP, CC, and MF, respectively. “Regulation of cytokine production”, “side of membrane”, “Cytokine activity” were the most significant difference in BP, CC, and MF respectively.
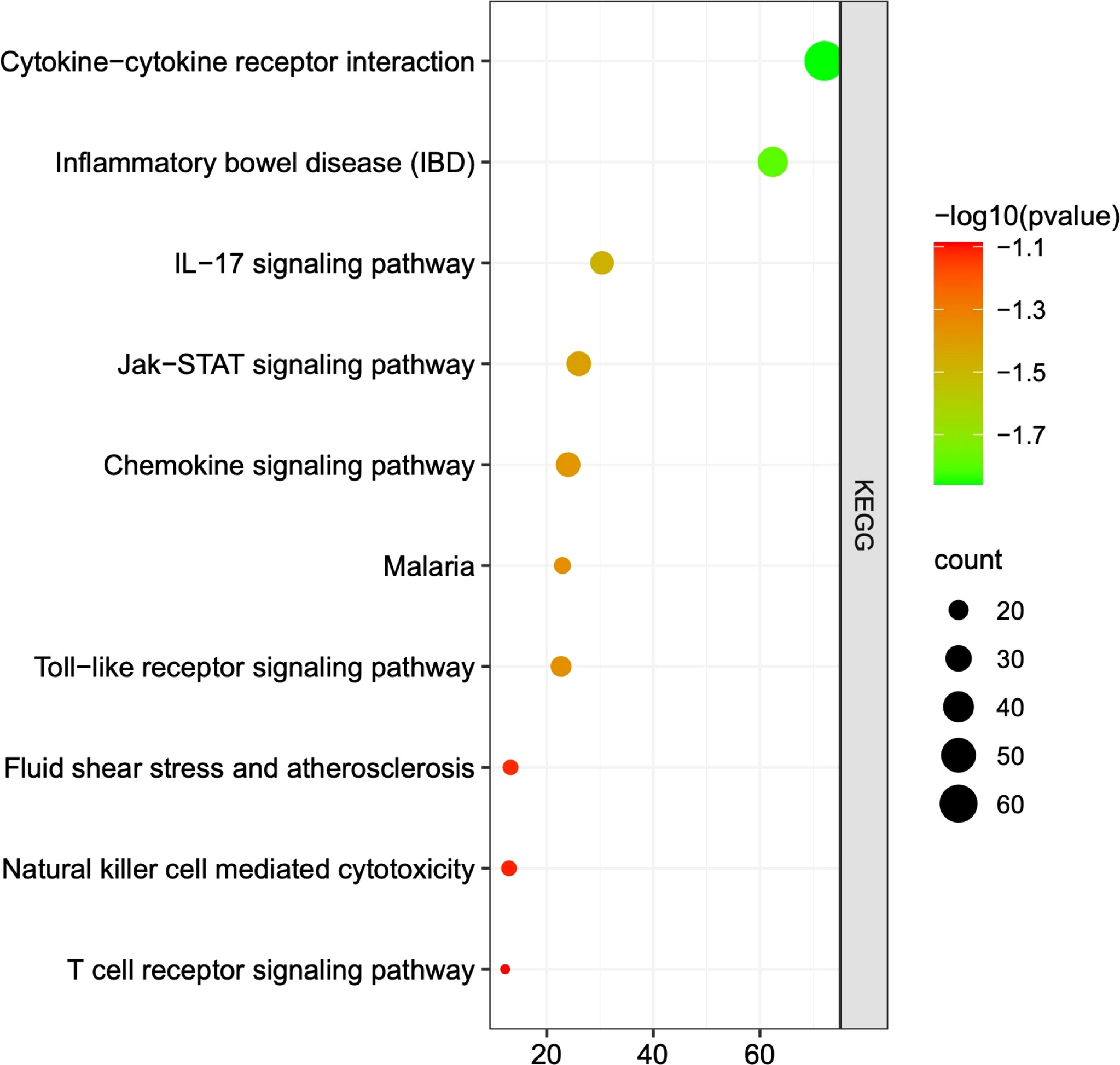
Figure 3 The top 10 pathways of KEGG analysis. Each dot represents a pathway, and the larger the dot is, the more genes it contains. The greener the dot is, the greater the difference. The three most significantly different pathways in KEGG analysis were “cytokine - cytokine receptor interaction”, “inflammatory bowel disease”, and “IL-17 signaling pathway”.
PPI Network Analysis and Hub Gene Recognition
As shown in Figure 4, the PPI network was analyzed and constructed by the STRING database and Cytoscape software. In addition, 246 nodes and 3946 edges were involved in the PPI network. In addition, two clusters were obtained after processing and analysis by the MCODE plugin in Cytoscape software. Cluster 1containde 62 nodes and Cluster 2 contained 14 nodes (Figure 4). Moreover, the Cytohubba plugin was used to further analyze and process PPI network node signals. The MCC method was applied to select the top 10 hub genes with a score ≥5000 and node degree ≥10, including CCL2, IL-13, CSF2, IL-10, TNF, IL-17A, IL-1β, IFN-γ, IL-2, and IL-4 (Figure 5). The results calculated by the MCC method also suggested that TNF, IL-1β, and IL-10 were the top three hub genes respectively (Figure 5).
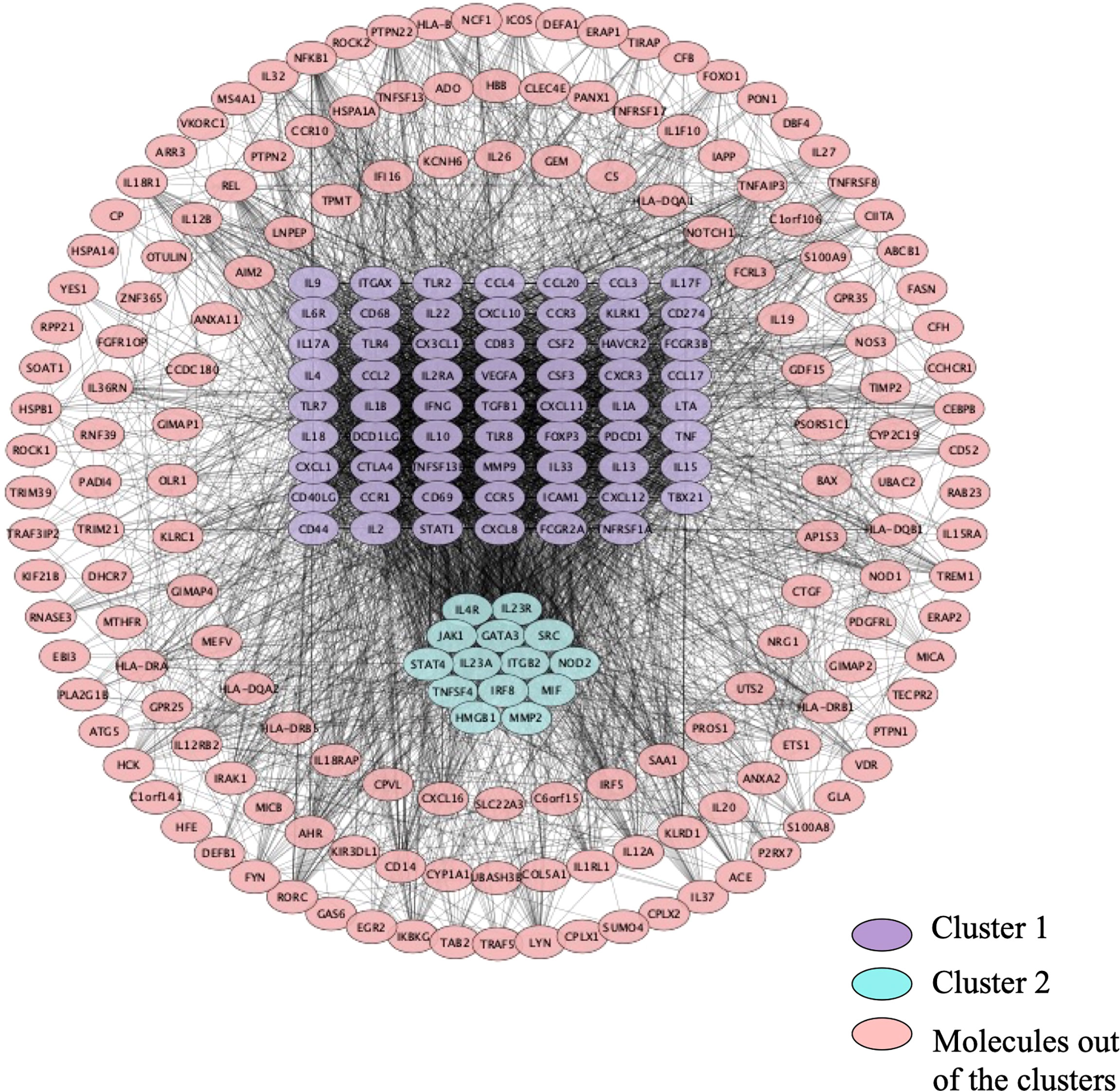
Figure 4 The PPI network. 246 nodes and 3946 edges were involved in the PPI network. Cluster 1 contains 62 nodes indicated in purple, and cluster 2 contains 14 nodes indicated in turquoise. Molecules in pink are not in any cluster.
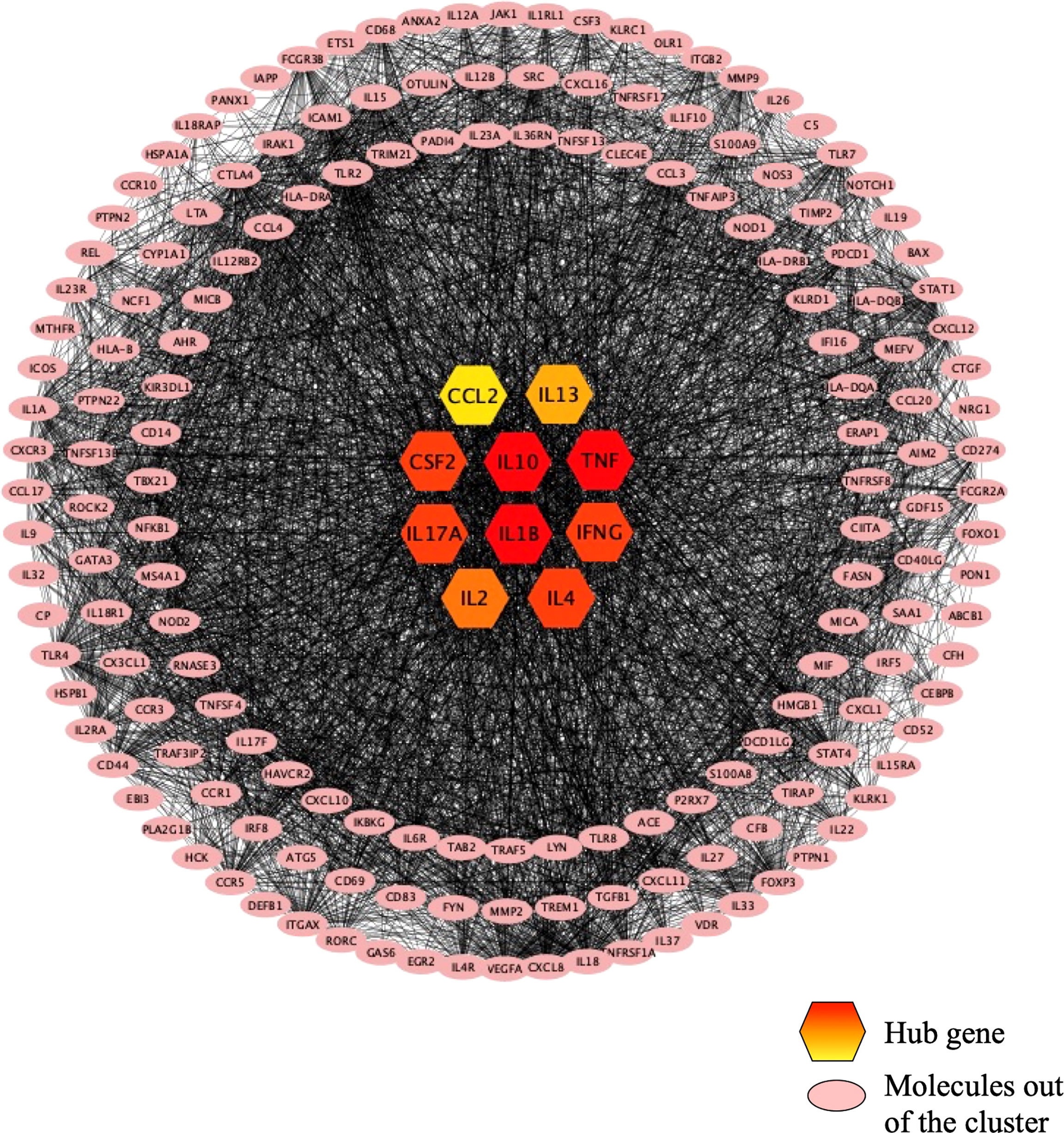
Figure 5 The top 10 hub genes recognized in the PPI network. The top 10 hub genes are in the center of the figure, and the color from red to yellow indicates the difference from large to small. TNF, IL1B, and IL10 are the top 3 hub genes. Molecules in pink are not in any cluster.
Drug-Gene Interaction
The DGIdb database was used to analyze the drugs interacting with the top three hub genes, and the results are shown in Figure 6. Among these drugs, 17 acted on IL-10, 37 acted on IL-1β, 68 acted on TNF, and 7 acted on any two or three genes simultaneously (Figure 6). All the results of the interaction with the first three hub genes are summarized in Supplementary Table 3, according to Interaction Type & Directionality, sources, Query Score, and Interaction Score. Among the seven drugs analyzed by the DGIdb database, infliximab, pentoxifylline, and cyclosporine have a clear therapeutic effect on BD. The usage, dosage, and side effects of these three drugs have been well studied (22). Alteplase, also known as recombinant human tissue plasminogen activator, rt-PA, is a thrombolytic agent primarily used for the treatment of Behcet’s disease-related thrombosis (23). However, the role of omeprazole, rabeprazole, and celastrol in BD has not been studied.
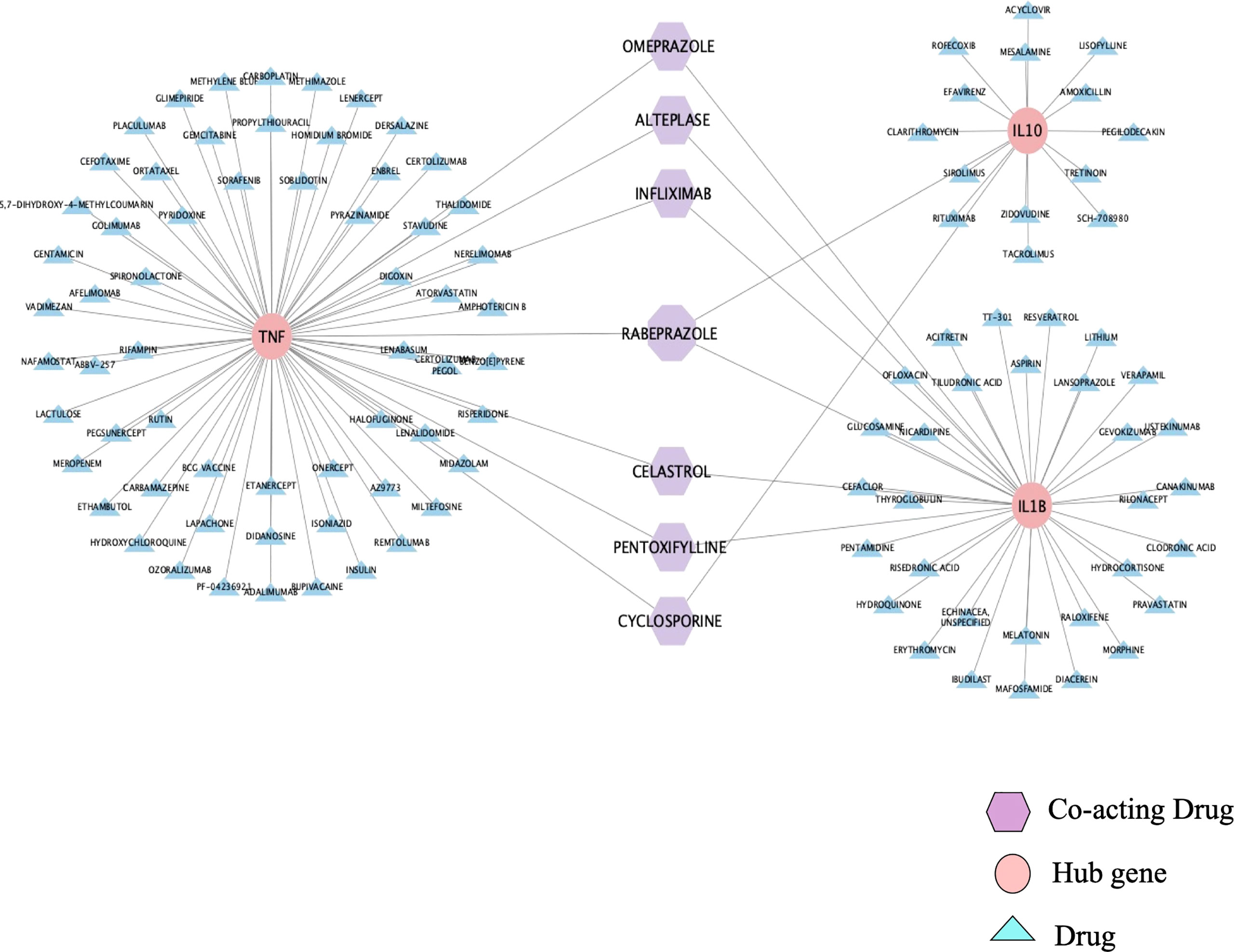
Figure 6 Drug-Gene Interaction. DGIdb database predicted six drugs acting on two hub genes and one drug acting on three hub genes. The pink represents the hub genes, the purple represents drugs that co-act on the hub genes, and the blue represents drugs that only act on one hub gene.
Expression of Hub Genes in EAU
Real-time PCR was applied to detect the expression of hub genes in the retinas of EAU mice. We compared the relative mRNA expression of hub genes in the retinas of naive and EAU mice on Day 14. The results showed that the relative mRNA expression of CCL2, CSF2, IL-2, IL-13, IL-4, IFN-γ, IL-1β, IL-17A, and TNF increased in the retina of EAU mice on Day 14, while IL-10 was decreased in the retina of EAU mice (Figure 7). CCL2, CSF2, IL-2, IL-13, IL-4, IFN-γ, IL-1β, IL-17A, TNF, and Il-10 showed statistically significant differences compared with naive mice (Figure 7).
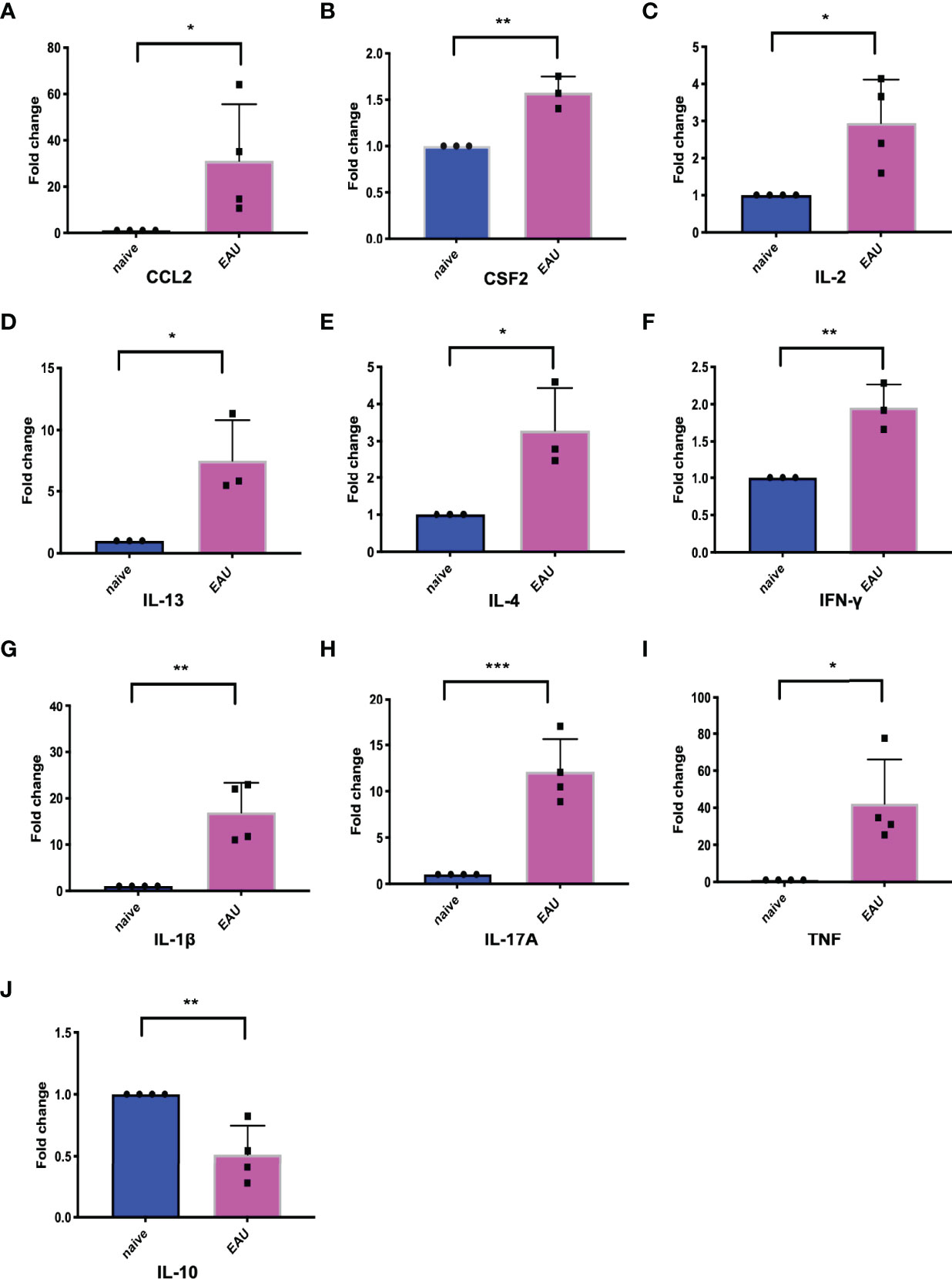
Figure 7 The expression of 10 hub genes in retinas of EAU mice and naive mice. The blue represents naive mice, and the red represents EAU mice. (A-J) represents the expression changes of CCL2, CSF2, IL-2, IL-13, IL-4, IFN-γ, IL-1β, IL-17A, TNF and IL-10, respectively. Except for the decreased expression of IL-10 in EAU, the other 9 hub genes were upregulated in EAU. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed student’s t-test). Graphs show mean ± SEM.
Celastrol Alleviated EAU
In EAU mice treated with celastrol, the inflammatory response of the anterior segment of the mouse was substantially reduced. Clear conjunctival hyperemia was still visible compared with naive mice (Figure 8A). A representative photograph of the anterior segment of the mouse was recorded on the 14th day, as shown in Figure 8A. Corneal edema and anterior chamber exudation were observed in the vehicle group, while other structures were not seen (Figure 8A). The clinical scores on Day 14 also showed that the scores of mice treated with celastrol were substantially lower than those of the vehicle group, and the difference was statistically significant (Figure 8C). In addition, the HE staining results also revealed that the EAU mice treated with celastrol had less structural damage to the retina on the 14th day and less inflammatory infiltration than the vehicle group, and there was a significant difference compared with the vehicle group (Figures 8B–D).
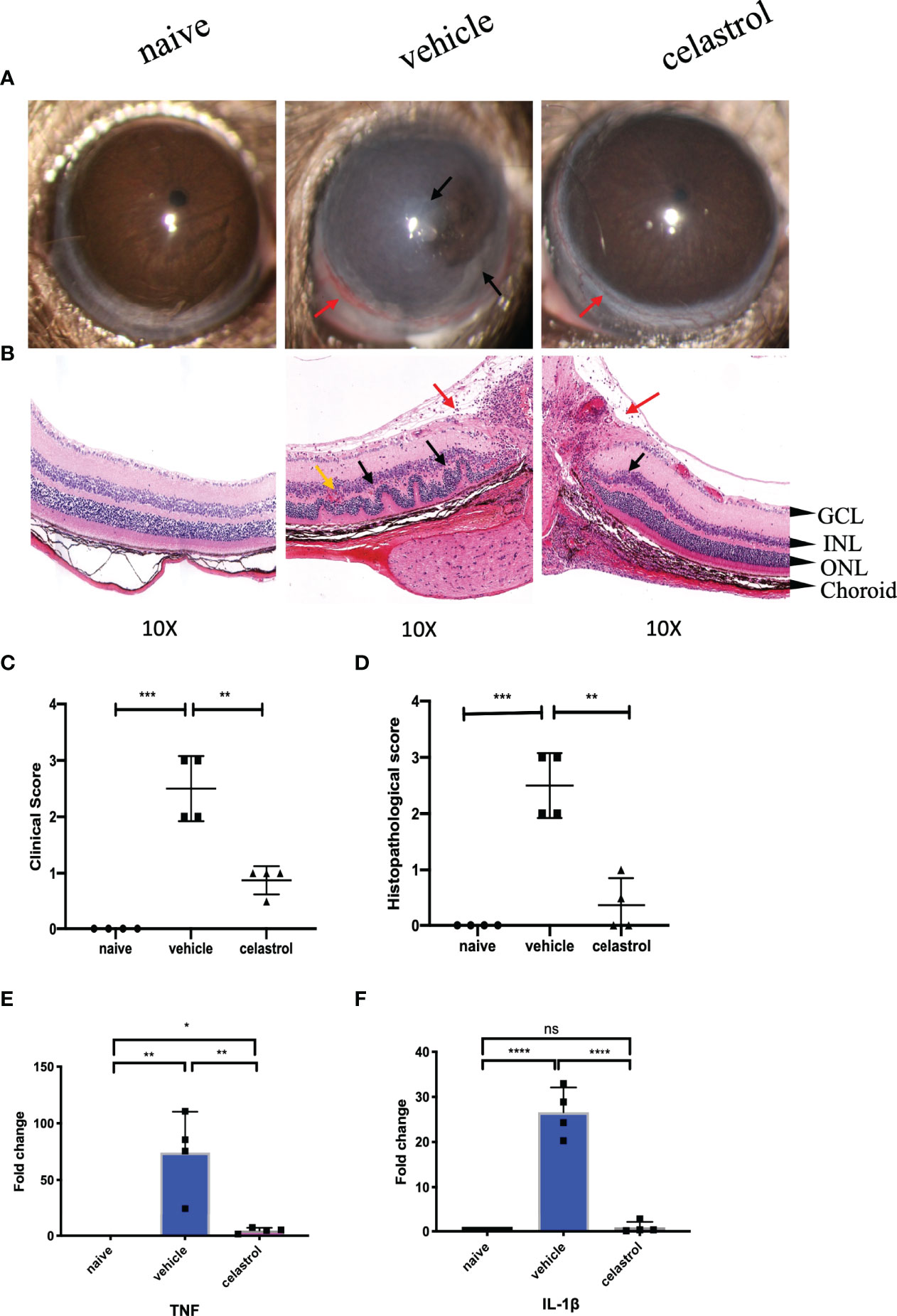
Figure 8 Celastrol alleviated EAU and inhibited the expression of TNF and IL-1β in the retinas of EAU. (A) Representative slit-lamp photographs of the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 4/group). Red arrow, conjunctival hyperemia. Black arrow, inflammatory exudation. (B) Representative staining images of HE sections in the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 4/group). Red arrow, vitreous inflammatory cell infiltration. Black arrow, retinal fold. Yellow arrow, neovascularization, and hemorrhage (GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer). (C) The clinical score of the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 4/group). (D) The histopathological score of the naive group, the vehicle intervention group, and the celastrol intervention group (n=4/group). (E) qRT-PCR analysis of TNF in retinas of the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 4/group). (F) qRT-PCR analysis of IL-1β in retinas of the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 4/group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-tailed student’s t-test). Graphs show mean ± SEM.
Celastrol Inhibited the Expression of TNF and IL-1β in EAU the Retina of EAU Mouse Retinas
After 7 days of celastrol intervention, real-time PCR was used to detect the expression changes of TNF and IL-1β mRNA in the retina. Our results showed that TNF and IL-1β were upregulated in the retinas of mice in the vehicle group compared with those in the naive group, with significant statistical differences. However, TNF and IL-1β were significantly downregulated in the retinas of mice treated with celastrol, with significant differences from the vehicle group. The difference was statistically significant compared with the naive group (Figures 8E, F).
Rabeprazole Alleviated EAU
In the rabeprazole group on Day 14, the anterior images showed moderate iridocyclitis with clear pupils and obvious conjunctival hyperemia, while in the vehicle group, corneal edema was more serious with unclear pupils and obvious conjunctival hyperemia (Figure 9A). Clinical scores showed significant differences between the two groups (Figure 9C). Meanwhile, the HE staining results on Day 14 indicated that the retinal structure of mice treated with rabeprazole was clear, the damage was lighter than that of the vehicle group, and there were fewer inflammatory cells found in the vitreous than were found in that of the vehicle group. Histological scores also showed significant differences between the two groups (Figures 9B–D).
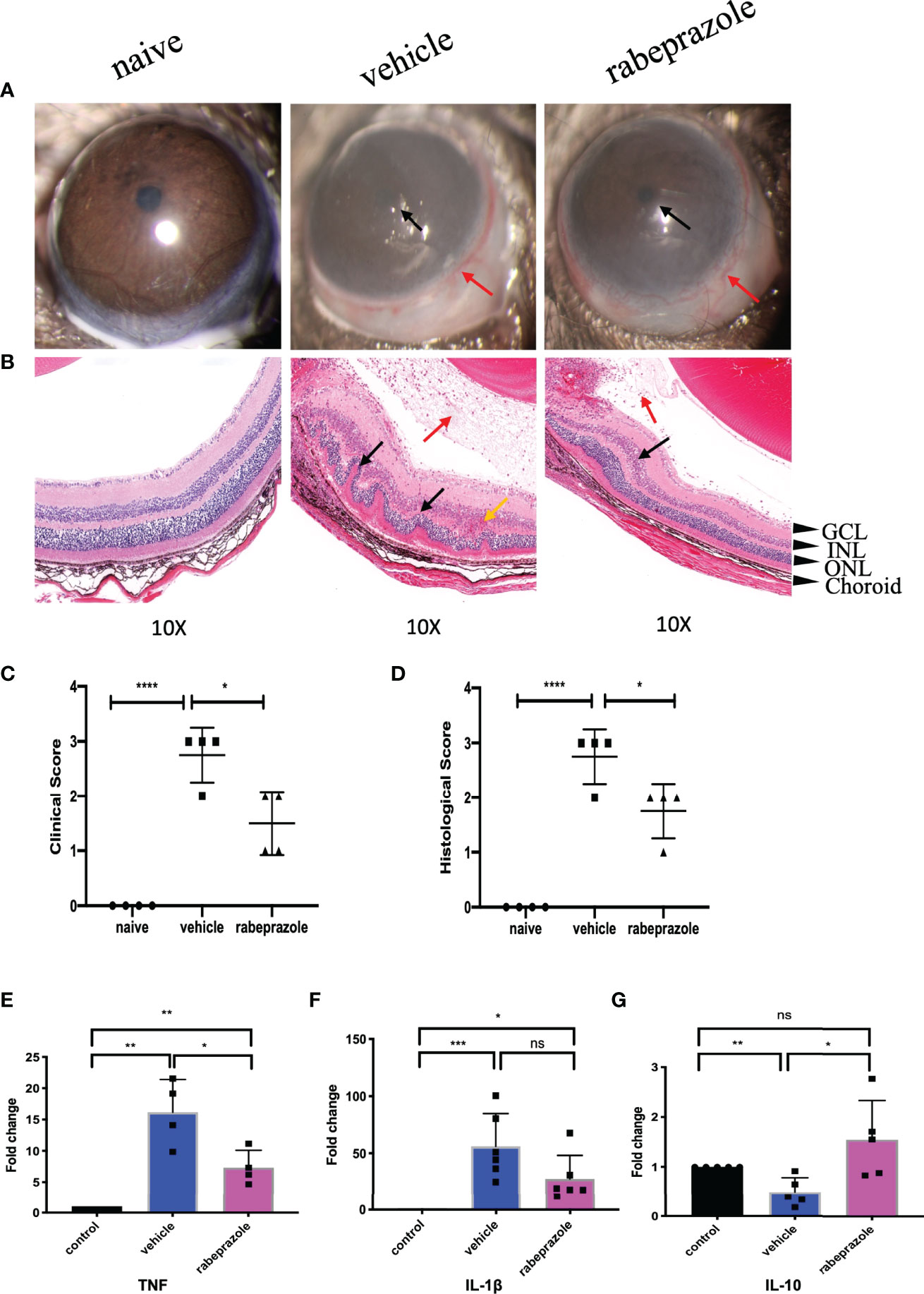
Figure 9 Rabeprazole alleviated EAU and inhibited TNF and IL-1β and promoted the expression of IL-10 in the retinas of EAU. (A) Representative slit-lamp photographs of the naive group, the vehicle intervention group, and the rabeprazole intervention group on day 14 (n = 4/group). Red arrow, conjunctival hyperemia. Black arrow, posterior synechiae, and inflammatory exudation. (B) Representative staining images of HE sections in the naive group, the vehicle intervention group, and the rabeprazole intervention group on day 14 (n = 4/group). Red arrow, vitreous inflammatory cell infiltration. Black arrow, retinal fold. Yellow arrow, neovascularization, and hemorrhage (GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer). (C) The clinical score of the naive group, the vehicle intervention group, and the rabeprazole intervention group on day 14 (n = 4/group). (D) The histopathological score of the naive group, the vehicle intervention group, and the rabeprazole intervention group (n = 4/group). (E) qRT-PCR analysis of TNF in retinas of the naive group, the vehicle intervention group, and the rabeprazole intervention group on day 14 (n = 4/group). (F) qRT-PCR analysis of IL-1β in retinas of the naive group, the vehicle intervention group, and the celastrol intervention group on day 14 (n = 6/group). (G) qRT-PCR analysis of IL-10 in retinas of the naive group, the vehicle intervention group and the rabeprazole intervention group on day 14 (n = 5/group) *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (two-tailed student’s t-test). Graphs show mean ± SEM.
Rabeprazole Inhibited TNF and IL-1β and Promoted the Expression of IL-10 in the Retinas of EAU Mice
To investigate whether rabeprazole affects the expression of the top 3 hub genes TNF, IL-1β, and IL-10 in the retina, we also extracted the retinas of EAU mice 7 days after rabeprazole intervention and measured the expression of these three genes by real-time PCR. They were compared with naive mice and vehicle mice. Our results showed that TNF expression increased in the retinas of mice on Day 14 after treatment with rabeprazole, which was significantly lower than that of the vehicle group, and there were significant differences between the naive group and vehicle group (Figure 9E). IL-1β expression was lower compared to that in the vehicle group, but there was no significant difference with the vehicle group (Figure 9F). Meanwhile, the expression of IL-10 in the retinas of the rabeprazole group was increased, and the difference was statistically significant compared with that in the vehicle group, but not statistically significant compared with that in the naive group (Figure 9).
Discussion
BD is a refractory disease that affects patients worldwide (24). The treatment of BD remains extremely challenging, although it is possible to select appropriate treatment strategies for patients with specific phenotypes (25, 26). Advances in bioinformatics research provide new approaches for us to process the biological information for some diseases and predict possible therapeutic drugs. In this study, we analyzed the function and pathways of BD-related target genes through bioinformatics, and enriched hub genes to further explore potential investigational drugs.
Combining the results of the searches of the PubMed and UVEOGENE databases, we found 249 genes and proteins that were significantly different in BD. Subsequently, we performed GO and KEGG enrichment analyses on the 249 genes and proteins using the Metascape database. The results of the GO enrichment analyses indicated that these genes and proteins were closely related to “regulation of cytokine production”, “immune effector process “, “response to bacterium” and “cytokine activity”. Indeed, multiple studies of BD have confirmed that a large number of inflammatory cytokines are involved in BD, such as TNF-α, IL-6, IL-17A, and IL-10 (27–29). In addition, a pathogenic theory for BD suggests that it may be caused by the activation of the innate or adaptive immune system due to bacterial or viral infection or autoantigen damage, thus inducing the production of a large number of inflammatory cytokines and chemokines that participate in the immune-inflammatory response (2).
KEGG enrichment analyses showed that the 249 genes and proteins were mainly enriched in the Cytokine - Cytokine receptor interaction, IBD, and IL-17 signaling pathways. Currently, multiple cytokine-cytokine receptors interactions, such as IL-23 and IL-23R, TNF-α and TNFR1 or TNFR2, and IL-17A or Il-17F and IL-17RA or IL-17RC, have been confirmed in BD (2, 30, 31). As a member of the IL-12 family, IL-23 is an important cytokine that promotes Th17-cell differentiation (32). The binding of IL-23 to IL-23R can activate Th17 cells, promote the release of inflammatory cytokines, such as IL-17, IL-6, and TNF-α, and enhance the inflammatory response (33). In addition, activation of the IL-23/IL-17 pathway is involved in IBD and BD (29). Furthermore, our results showed that 25 target genes were enriched in the IL-17 pathway, which also suggests that the IL-17 pathway is important in BD. Moreover, identification of the IBD pathway also explained that the occurrence of immune inflammation and the activation of Th1, Th17, and Th2 cells were closely related to the activation of multiple pathways such as the Nod-like receptor signaling pathway, Toll-like receptor pathway, and Cytokine - Cytokine pathway.
To explore the connections among target genes or proteins, we constructed a protein-protein interaction network that divided target genes or proteins into three categories according to the interaction scores: one was clustered with a purple background, one was clustered with a green background, and the rest were placed outside the clusters. Subsequently, the Cytoscape plugin was applied to enrich 10 hub genes (CCL2, IL-13, CSF2, IL-10, TNF, IL-17A, IL-1β, IFN-γ, IL-2, IL4). Among the 10 hub genes, CCL2 was also named MCP1, and CSF2 was also named GM-CSF; nine of the hub genes, excluding CSF2, came from UVEOGENE databases. Furthermore, the enrichment results for the hub genes also suggested that TNF, IL-1β, and IL-10 are the most critical hub genes. TNF is a key pathogenic factor of BD, and some studies have shown that TNF is related to disease activity in BD (34) (35). TNF is highly expressed in various tissues in BD patients, such as oral ulcer tissue, the aqueous humour, and intestinal lesions (36–38). At present, there are many kinds of TNF-α blockers available for the treatment of BD, such as IFX, alemtuzumab (ADA), and etanercept (ETC) (39). Moreover, IFX and ADA have been recommended as first-line treatments for severe posterior uveitis associated with BD (40). IL-1β, a proinflammatory factor, is mainly secreted by monocytes, macrophages, and dendritic cells (DCs) and can induce the release of various inflammatory chemokines and promote the production of immune inflammation (25) IL-1β is highly expressed in the serum of patients with BD, and targeted inhibition of IL-1β has also been shown to be effective in the treatment of BD (41). Currently, canakinumab, gevokizumab, and anakinra, which are all IL-1 blockers, have been proven to be effective in the treatment of BD (42–44). IL-10 is an anti-inflammatory cytokine secreted by Th2 cells that mainly antagonizes proinflammatory factors, and promotes the activation of B cells to produce antibodies (45, 46). Moreover, three BD risk alleles rs1518111A, rs1800872A, and rs1800871T were related to IL-10 (47, 48).
To explore potential drugs, we analyzed the association between genes or drugs and the top 3 hub genes through the DGIdb database. Our results were centered on seven drugs, including IFX, pentoxifylline, and cyclosporine, which are clinically appropriate for the treatment of BD, and alteplase can be used for the treatment of BD complications (22, 23). However, the remaining three drugs have not been proven to be effective in the treatment of BD. Celastrol is a bioactive ingredient of Tripterygium wilfordii, which has been proven to have anti-inflammatory, antitumor, and antineovascularization effects in some studies (49, 50). Several studies have shown that celastrol has a certain anti-inflammatory effect on IBD, experimental autoimmune encephalomyelitis (EAE), and psoriasis models (51–54). In recent years, Chinese doctors have paid more attention to clinical experiments evaluating the use of celastrol in treating rheumatoid arthritis, idiopathic membranous nephropathy, and IBD (NCT01613079, NCT01161459, NCT02044952). Omeprazole and Rabeprazole are proton pump inhibitors (PPIs) that are currently mainly used for the treatment of gastrointestinal diseases, including gastroesophageal reflux disease, gastric and duodenal ulcers, and Zollinger-Ellison syndrome (55). Omeprazole is a first-generation PPI approved for clinical treatment, and it was followed by lansoprazole, rabeprazole, and other PPIs (56). Although PPIs are primarily used to treat digestive diseases, studies have shown that they may have anti-inflammatory effects in addition to inhibiting acid production (57). In recent years, PPIs have played an important role in inhibiting antigen presentation by inhibiting the expression of TNF-α and IL-1β induced by different pathogen-associated molecular patterns (PAMPs). In addition, PPIs inhibit the release of a large number of inflammatory factors caused by Toll-like receptor signaling activation in monocytes (58). Both omeprazole and rabeprazole have been reported to exert anti-inflammatory effects related to the inhibition of NF- κB activation (59–61). Furthermore, a number of studies have confirmed that PPIs can not only reduce inflammation in the human gastrointestinal epithelium but also reduce inflammation in the human lung epithelium and psoriasis (60, 62, 63). However, the treatment of other systemic inflammatory diseases with PPIs has rarely been reported.
Next, to verify whether these drugs might inhibit the inflammatory response in BD, we constructed an EAU mouse model with B10RIII mice (64). We compared the expression of hub genes in the retinas of EAU mice between a pre-modeling timepoint and Day 14 after modeling. The results showed that IL-10 expression was reduced in the retinas of EAU mice, but the expression of the other 9 hub genes was increased; these 10 hub genes significantly differed between the model and control mice. The data suggested that these 10 hub genes are involved in EAU and that EAU is a good research model for BD uveitis. Subsequently, we selected celastrol and rabeprazole from among the three unproven drugs to treat EAU mice. Although our study results pointed to both omeprazole and rabeprazole, considering that both drugs are metabolized by the CYP450 enzyme, we chose rabeprazole for experimental verification and found that rabeprazole had little effect on the enzyme CYP450 (65). In the EAU mouse model, EAU usually starts between Days 7 and 9, peaks on approximately Day 14, and then gradually resolves (17). We chose to administer treatment from the onset of EAU to the peak of the disease. Our results showed that after treatment with celastrol for 7 days, anterior chamber inflammation was significantly reduced, the retinal structure was slightly intact, and the retinal pro-inflammatory factors TNF and IL-1β were significantly downregulated, suggesting that celastrol could effectively alleviate EAU. In addition, in mice treated with rabeprazole, the inflammation in the anterior part of the mice was slightly reduced compared with that in vehicle-treated mice, and the retinal damage was less severe. The expression levels of the proinflammatory factors TNF and IL-1β in the retinas were lower in rabeprazole-treated mice than in vehicle-treated mice, while the expression of the anti-inflammatory factor IL-10 was upregulated compared with that in vehicle mice. These results indicated that rabeprazole can alleviate EAU to a certain extent.
In a short, target functional analysis and potential drug mining for BD highlighted the effectiveness of network pharmacological approaches. However, uveitis represented by the EAU model is just one of the typical manifestations of BD, so these agents need to be validated in many models. We also demonstrated the anti-inflammatory effect of rabeprazole in EAU for the first time, but the mechanism and side effects remained unclear. These results are still worthy of further study, which may provide some inspiration for the treatment of BD. In addition, the bioinformatics analysis may be exploratory, our study focused on PPIs, which are not clinically used as anti-inflammatory agents, it is also worth noting that an increasing number of studies have shown the anti-inflammatory properties of PPIs. Furthermore, our bioinformatic analysis focused on the existing data and the addition of future information that could affect our results. Despite these limitations, this study is the first preliminary and basic experimental verification of BD target gene enrichment-related drugs and provides new insights into the disease mechanisms and therapeutic potential of BD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of the Renmin Hospital of Wuhan University (WDRM20210708A).
Author Contributions
The design and conception of the manuscript were completed by YX and ZC. Data collection and collation were performed by QX and CL. Bioinformatics analysis was finished by CL and FL. The EAU model constructed by QX, BP, and XG. Drug validation-related experiments were performed by QX, FL, and BP. Data collation and mapping were finished by CL, QX, and HR. Manuscript writing and revision were performed by QX, CL, FL, YX, and ZC. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Hubei health and family planning commission supporting projects WJ2017Z004 and WJ2021M156.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.895869/full#supplementary-material
References
1. Mendes D, Correia M, Barbedo M, Vaio T, Mota M, Goncalves O, et al. Behcet's Disease–A Contemporary Review. J Autoimmun (2009) 32(3-4):178–88. doi: 10.1016/j.jaut.2009.02.011
2. Tong B, Liu X, Xiao J, Su G. Immunopathogenesis of Behcet's Disease. Front Immunol (2019) 10:665. doi: 10.3389/fimmu.2019.00665
3. Pineton de Chambrun M, Wechsler B, Geri G, Cacoub P, Saadoun D. New Insights Into the Pathogenesis of Behcet's Disease. Autoimmun Rev (2012) 11(10):687–98. doi: 10.1016/j.autrev.2011.11.026
4. Verity DH, Wallace GR, Vaughan RW, Stanford MR. Behcet's Disease: From Hippocrates to the Third Millennium. Br J Ophthalmol (2003) 87(9):1175–83. doi: 10.1136/bjo.87.9.1175
5. Marshall SE. Behcet's Disease. Best Pract Res Clin Rheumatol (2004) 18(3):291–311. doi: 10.1016/j.berh.2004.02.008
6. International Study Group for Behcet's Disease. Criteria for Diagnosis of Behcet's Disease. Lancet (1990) 335(8697):1078–80. doi: 10.1016/0140-6736(90)92643-V
7. Deuter CM, Kotter I, Wallace GR, Murray PI, Stubiger N, Zierhut M. Behcet's Disease: Ocular Effects and Treatment. Prog Retin Eye Res (2008) 27(1):111–36. doi: 10.1016/j.preteyeres.2007.09.002
8. Ksiaa I, Abroug N, Kechida M, Zina S, Jelliti B, Khochtali S, et al. Eye and Behcet's Disease. J Fr Ophtalmol (2019) 42(4):e133–e46. doi: 10.1016/j.jfo.2019.02.002
9. Kural-Seyahi E, Fresko I, Seyahi N, Ozyazgan Y, Mat C, Hamuryudan V, et al. The Long-Term Mortality and Morbidity of Behcet Syndrome: A 2-Decade Outcome Survey of 387 Patients Followed at a Dedicated Center. Med (Baltimore) (2003) 82(1):60–76. doi: 10.1097/00005792-200301000-00006
10. Jaffe GJ, Dick AD, Brezin AP, Nguyen QD, Thorne JE, Kestelyn P, et al. Adalimumab in Patients With Active Noninfectious Uveitis. N Engl J Med (2016) 375(10):932–43. doi: 10.1056/NEJMoa1509852
11. Ozguler Y, Leccese P, Christensen R, Esatoglu SN, Bang D, Bodaghi B, et al. Management of Major Organ Involvement of Behcet's Syndrome: A Systematic Review for Update of the EULAR Recommendations. Rheumatol (Oxford) (2018) 57(12):2200–12. doi: 10.1093/rheumatology/key242
12. Alpsoy E. Behcet's Disease: A Comprehensive Review With a Focus on Epidemiology, Etiology and Clinical Features, and Management of Mucocutaneous Lesions. J Dermatol (2016) 43(6):620–32. doi: 10.1111/1346-8138.13381
13. Caspi RR, Roberge FG, Chan CC, Wiggert B, Chader GJ, Rozenszajn LA, et al. A New Model of Autoimmune Disease. Experimental Autoimmune Uveoretinitis Induced in Mice With Two Different Retinal Antigens. J Immunol (1988) 140(5):1490–5.
14. Wang Q, Su G, Tan X, Deng J, Du L, Huang X, et al. UVEOGENE: An SNP Database for Investigations on Genetic Factors Associated With Uveitis and Their Relationship With Other Systemic Autoimmune Diseases. Hum Mutat (2019) 40(3):258–66. doi: 10.1002/humu.23702
15. Chen Z, Zhong Z, Zhang W, Su G, Yang P. Integrated Analysis of Key Pathways and Drug Targets Associated With Vogt-Koyanagi-Harada Disease. Front Immunol (2020) 11:587443. doi: 10.3389/fimmu.2020.587443
16. Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: A Redesign and Expansion of the Drug-Gene Interaction Database. Nucleic Acids Res (2018) 46(D1):D1068–D73. doi: 10.1093/nar/gkx1143
17. Yang H, Zheng S, Qiu Y, Yang Y, Wang C, Yang P, et al. Activation of Liver X Receptor Alleviates Ocular Inflammation in Experimental Autoimmune Uveitis. Invest Ophthalmol Vis Sci (2014) 55(4):2795–804. doi: 10.1167/iovs.13-13323
18. Venkatesha SH, Moudgil KD. Celastrol Suppresses Experimental Autoimmune Encephalomyelitis via MAPK/SGK1-Regulated Mediators of Autoimmune Pathology. Inflammation Res (2019) 68(4):285–96. doi: 10.1007/s00011-019-01219-x
19. Watanabe T, Higuchi K, Hamaguchi M, Shiba M, Tominaga K, Fujiwara Y, et al. Monocyte Chemotactic Protein-1 Regulates Leukocyte Recruitment During Gastric Ulcer Recurrence Induced by Tumor Necrosis Factor-Alpha. Am J Physiol Gastrointest Liver Physiol (2004) 287(4):G919–28. doi: 10.1152/ajpgi.00372.2003
20. Hamaguchi M, Fujiwara Y, Takashima T, Hayakawa T, Sasaki E, Shiba M, et al. Increased Expression of Cytokines and Adhesion Molecules in Rat Chronic Esophagitis. Digestion (2003) 68(4):189–97. doi: 10.1159/000075698
21. Caspi RR. Experimental Autoimmune Uveoretinitis in the Rat and Mouse. Curr Protoc Immunol (2003) 15:15.6. doi: 10.1002/0471142735.im1506s53
22. Karadag O, Bolek EC. Management of Behcet's Syndrome. Rheumatol (Oxford) (2020) 59(Suppl 3):iii108–iii17. doi: 10.1093/rheumatology/keaa086
23. Arif S, Arif S, Liaqat J, Slehria AU, Palwa AR. Transvenous Catheter-Based Thrombolysis With Continuous Tissue Plasminogen Activator Infusion for Refractory Thrombosis in a Patient With Behcet's Disease. Cureus (2020) 12(8):e10049. doi: 10.7759/cureus.10049
24. Sugimura N, Mizoshita T, Sugiyama T, Togawa S, Miyaki T, Suzuki T, et al. Real-World Efficacy of Adalimumab and Infliximab for Refractory Intestinal Behcet's Disease. Dig Liver Dis (2019) 51(7):967–71. doi: 10.1016/j.dld.2018.10.024
25. Dinarello CA, van der Meer JW. Treating Inflammation by Blocking Interleukin-1 in Humans. Semin Immunol (2013) 25(6):469–84. doi: 10.1016/j.smim.2013.10.008
26. Rotondo C, Lopalco G, Iannone F, Vitale A, Talarico R, Galeazzi M, et al. Mucocutaneous Involvement in Behcet's Disease: How Systemic Treatment Has Changed in the Last Decades and Future Perspectives. Mediators Inflamm (2015) 2015:451675. doi: 10.1155/2015/451675
27. Wakefield D, Lloyd A. The Role of Cytokines in the Pathogenesis of Inflammatory Eye Disease. Cytokine (1992) 4(1):1–5. doi: 10.1016/1043-4666(92)90028-p
28. Akman-Demir G, Tuzun E, Icoz S, Yesilot N, Yentur SP, Kurtuncu M, et al. Interleukin-6 in Neuro-Behcet's Disease: Association With Disease Subsets and Long-Term Outcome. Cytokine (2008) 44(3):373–6. doi: 10.1016/j.cyto.2008.10.007
29. Chi W, Zhu X, Yang P, Liu X, Lin X, Zhou H, et al. Upregulated IL-23 and IL-17 in Behcet Patients With Active Uveitis. Invest Ophthalmol Vis Sci (2008) 49(7):3058–64. doi: 10.1167/iovs.07-1390
30. Saadoun D, Bodaghi B, Bienvenu B, Wechsler B, Sene D, Trad S, et al. Biotherapies in Inflammatory Ocular Disorders: Interferons, Immunoglobulins, Monoclonal Antibodies. Autoimmun Rev (2013) 12(7):774–83. doi: 10.1016/j.autrev.2013.02.002
31. Sonmez C, Yucel AA, Yesil TH, Kucuk H, Sezgin B, Mercan R, et al. Correlation Between IL-17a/F, IL-23, IL-35 and IL-12/-23 (P40) Levels in Peripheral Blood Lymphocyte Cultures and Disease Activity in Behcet's Patients. Clin Rheumatol (2018) 37(10):2797–804. doi: 10.1007/s10067-018-4049-7
32. Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, et al. Divergent Pro- and Antiinflammatory Roles for IL-23 and IL-12 in Joint Autoimmune Inflammation. J Exp Med (2003) 198(12):1951–7. doi: 10.1084/jem.20030896
33. Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, et al. IL-23 Drives a Pathogenic T Cell Population That Induces Autoimmune Inflammation. J Exp Med (2005) 201(2):233–40. doi: 10.1084/jem.20041257
34. Gholijani N, Ataollahi MR, Samiei A, Aflaki E, Shenavandeh S, Kamali-Sarvestani E. An Elevated Pro-Inflammatory Cytokines Profile in Behcet's Disease: A Multiplex Analysis. Immunol Lett (2017) 186:46–51. doi: 10.1016/j.imlet.2016.12.001
35. Mesquida M, Molins B, Llorenc V, Sainz de la Maza M, Hernandez MV, Espinosa G, et al. Proinflammatory Cytokines and C-Reactive Protein in Uveitis Associated With Behcet's Disease. Mediators Inflamm (2014) 2014):396204. doi: 10.1155/2014/396204
36. Dalghous AM, Freysdottir J, Fortune F. Expression of Cytokines, Chemokines, and Chemokine Receptors in Oral Ulcers of Patients With Behcet's Disease (BD) and Recurrent Aphthous Stomatitis is Th1-Associated, Although Th2-Association is Also Observed in Patients With Bd. Scand J Rheumatol (2006) 35(6):472–5. doi: 10.1080/03009740600905380
37. El-Asrar AM, Struyf S, Kangave D, Al-Obeidan SS, Opdenakker G, Geboes K, et al. Cytokine Profiles in Aqueous Humor of Patients With Different Clinical Entities of Endogenous Uveitis. Clin Immunol (2011) 139(2):177–84. doi: 10.1016/j.clim.2011.01.014
38. Ferrante A, Ciccia F, Principato A, Giardina AR, Impastato R, Peralta S, et al. A Th1 But Not a Th17 Response is Present in the Gastrointestinal Involvement of Behcet's Disease. Clin Exp Rheumatol (2010) 28(4 Suppl 60):S27–30.
39. Tracey D, Klareskog L, Sasso EH, Salfeld JG, Tak PP. Tumor Necrosis Factor Antagonist Mechanisms of Action: A Comprehensive Review. Pharmacol Ther (2008) 117(2):244–79. doi: 10.1016/j.pharmthera.2007.10.001
40. Desbois AC, Vallet H, Domont F, Comarmond C, Cacoub P, Saadoun D. Management of Severe Complications in Behcet's Disease With TNF Inhibitors. Expert Opin Biol Ther (2017) 17(7):853–9. doi: 10.1080/14712598.2017.1328496
41. Pay S, Erdem H, Pekel A, Simsek I, Musabak U, Sengul A, et al. Synovial Proinflammatory Cytokines and Their Correlation With Matrix Metalloproteinase-3 Expression in Behcet's Disease. Does Interleukin-1beta Play a Major Role in Behcet's Synovitis? Rheumatol Int (2006) 26(7):608–13. doi: 10.1007/s00296-005-0040-0
42. Vitale A, Rigante D, Caso F, Brizi MG, Galeazzi M, Costa L, et al. Inhibition of Interleukin-1 by Canakinumab as a Successful Mono-Drug Strategy for the Treatment of Refractory Behcet's Disease: A Case Series. Dermatology (2014) 228(3):211–4. doi: 10.1159/000358125
43. Gul A, Tugal-Tutkun I, Dinarello CA, Reznikov L, Esen BA, Mirza A, et al. Interleukin-1beta-Regulating Antibody XOMA 052 (Gevokizumab) in the Treatment of Acute Exacerbations of Resistant Uveitis of Behcet's Disease: An Open-Label Pilot Study. Ann Rheum Dis (2012) 71(4):563–6. doi: 10.1136/annrheumdis-2011-155143
44. Cantarini L, Vitale A, Scalini P, Dinarello CA, Rigante D, Franceschini R, et al. Anakinra Treatment in Drug-Resistant Behcet's Disease: A Case Series. Clin Rheumatol (2015) 34(7):1293–301. doi: 10.1007/s10067-013-2443-8
45. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and Functions of the IL-10 Family of Cytokines in Inflammation and Disease. Annu Rev Immunol (2011) 29:71–109. doi: 10.1146/annurev-immunol-031210-101312
46. Zhou ZY, Chen SL, Shen N, Lu Y. Cytokines and Behcet's Disease. Autoimmun Rev (2012) 11(10):699–704. doi: 10.1016/j.autrev.2011.12.005
47. Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, et al. Genome-Wide Association Study Identifies Variants in the MHC Class I, IL10, and IL23R-IL12RB2 Regions Associated With Behcet's Disease. Nat Genet (2010) 42(8):698–702. doi: 10.1038/ng.625
48. Mizuki N, Meguro A, Ota M, Ohno S, Shiota T, Kawagoe T, et al. Genome-Wide Association Studies Identify IL23R-IL12RB2 and IL10 as Behcet's Disease Susceptibility Loci. Nat Genet (2010) 42(8):703–6. doi: 10.1038/ng.624
49. Cascao R, Vidal B, Raquel H, Neves-Costa A, Figueiredo N, Gupta V, et al. Effective Treatment of Rat Adjuvant-Induced Arthritis by Celastrol. Autoimmun Rev (2012) 11(12):856–62. doi: 10.1016/j.autrev.2012.02.022
50. Li Z, Yao L, Li J, Zhang W, Wu X, Liu Y, et al. Celastrol Nanoparticles Inhibit Corneal Neovascularization Induced by Suturing in Rats. Int J Nanomedicine (2012) 7:1163–73. doi: 10.2147/IJN.S27860
51. Wang R, Gu X, Dai W, Ye J, Lu F, Chai Y, et al. A Lipidomics Investigation Into the Intervention of Celastrol in Experimental Colitis. Mol Biosyst (2016) 12(5):1436–44. doi: 10.1039/c5mb00864f
52. Xian J, Zhong X, Gu H, Wang X, Li J, Li J, et al. Colonic Delivery of Celastrol-Loaded Layer-By-Layer Liposomes With Pectin/Trimethylated Chitosan Coating to Enhance Its Anti-Ulcerative Colitis Effects. Pharmaceutics (2021) 13(12):2005. doi: 10.3390/pharmaceutics13122005
53. Yang H, Liu C, Jiang J, Wang Y, Zhang X. Celastrol Attenuates Multiple Sclerosis and Optic Neuritis in an Experimental Autoimmune Encephalomyelitis Model. Front Pharmacol (2017) 8:44. doi: 10.3389/fphar.2017.00044
54. Qiu F, Xi L, Chen S, Zhao Y, Wang Z, Zheng Y. Celastrol Niosome Hydrogel Has Anti-Inflammatory Effect on Skin Keratinocytes and Circulation Without Systemic Drug Exposure in Psoriasis Mice. Int J Nanomedicine (2021) 16:6171–82. doi: 10.2147/IJN.S323208
55. Hu ZH, Shi AM, Hu DM, Bao JJ. Efficacy of Proton Pump Inhibitors for Patients With Duodenal Ulcers: A Pairwise and Network Meta-Analysis of Randomized Controlled Trials. Saudi J Gastroenterol (2017) 23(1):11–9. doi: 10.4103/1319-3767.199117
56. Fontana RJ. Pathogenesis of Idiosyncratic Drug-Induced Liver Injury and Clinical Perspectives. Gastroenterology (2014) 146(4):914–28. doi: 10.1053/j.gastro.2013.12.032
57. Miyashita T, Shah FA, Harmon JW, Marti GP, Matsui D, Okamoto K, et al. Do Proton Pump Inhibitors Protect Against Cancer Progression in GERD? Surg Today (2013) 43(8):831–7. doi: 10.1007/s00595-012-0395-2
58. Balza E, Piccioli P, Carta S, Lavieri R, Gattorno M, Semino C, et al. Proton Pump Inhibitors Protect Mice From Acute Systemic Inflammation and Induce Long-Term Cross-Tolerance. Cell Death Dis (2016) 7:e2304. doi: 10.1038/cddis.2016.218
59. Handa O, Yoshida N, Fujita N, Tanaka Y, Ueda M, Takagi T, et al. Molecular Mechanisms Involved in Anti-Inflammatory Effects of Proton Pump Inhibitors. Inflammation Res (2006) 55(11):476–80. doi: 10.1007/s00011-006-6056-4
60. Xie J, Fan L, Xiong L, Chen P, Wang H, Chen H, et al. Rabeprazole Inhibits Inflammatory Reaction by Inhibition of Cell Pyroptosis in Gastric Epithelial Cells. BMC Pharmacol Toxicol (2021) 22(1):44. doi: 10.1186/s40360-021-00509-7
61. Kotsuka M, Hashimoto Y, Nakatake R, Okuyama T, Hatta M, Yoshida T, et al. Omeprazole Increases Survival Through the Inhibition of Inflammatory Mediaters in Two Rat Sepsis Models. Shock (2022) 57(3):444–56. doi: 10.1097/SHK.0000000000001897
62. Ebrahimpour A, Wang M, Li L, Jegga AG, Bonnen MD, Eissa NT, et al. Esomeprazole Attenuates Inflammatory and Fibrotic Response in Lung Cells Through the MAPK/Nrf2/HO1 Pathway. J Inflammation (Lond) (2021) 18(1):17. doi: 10.1186/s12950-021-00284-6
63. Bafutto M, Oliveira EC, Zaterka S. Evaluation of Psoriasis Treatment With Esomeprazole - A Pilot Study. Arq Gastroenterol (2019) 56(3):261–3. doi: 10.1590/S0004-2803.201900000-49
64. Ye Z, Zhang N, Wu C, Zhang X, Wang Q, Huang X, et al. A Metagenomic Study of the Gut Microbiome in Behcet's Disease. Microbiome (2018) 6(1):135. doi: 10.1186/s40168-018-0520-6
Keywords: Behcet’s disease, potential drugs, EAU, rabeprazole, celastrol
Citation: Xia Q, Lyu C, Li F, Pang B, Guo X, Ren H, Xing Y and Chen Z (2022) Candidate Drugs Screening for Behcet’s Disease Based on Bioinformatics Analysis and Mouse Experiments. Front. Immunol. 13:895869. doi: 10.3389/fimmu.2022.895869
Received: 14 March 2022; Accepted: 23 May 2022;
Published: 21 June 2022.
Edited by:
Raphaela Goldbach-Mansky, National Institutes of Health (NIH), United StatesReviewed by:
Haner Direskeneli, Marmara University, TurkeyRosaria Talarico, University of Pisa, Italy
Copyright © 2022 Xia, Lyu, Li, Pang, Guo, Ren, Xing and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiqiao Xing, WWlxaWFvX3hpbmc1N0B3aHUuZWR1LmNu; Zhen Chen, aGNoZW56aGVuQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Qinyun Xia1†
Qinyun Xia1† Xiaoyu Guo
Xiaoyu Guo Yiqiao Xing
Yiqiao Xing