
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 11 May 2022
Sec. Autoimmune and Autoinflammatory Disorders
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.893617
This article is part of the Research TopicNovel Therapeutic Targets in Liver DiseaseView all 6 articles
The prevalence and incidence of metabolic-associated fatty liver disease (MAFLD), a clinically heterogeneous disease whose primary clinical therapies include dietary control and exercise therapy, is increasing worldwide and constitutes a significant medical burden. Gut microbes influence the physiopathological processes of the liver through different mechanisms based on the gut-liver axis. Exosomes are essential carriers of intercellular communication. Most previous studies have focused on adipocyte- and hepatocyte-derived exosomes, while the critical role of microbial-derived exosomes and the molecular mechanisms behind them in MAFLD have received little attention. Therefore, we searched and screened the latest relevant studies in the PubMeb database to elucidate the link between microbial-derived exosomes and the pathogenesis of MAFLD, mainly in terms of insulin resistance, intestinal barrier, inflammatory response, lipid metabolism, and liver fibrosis. The aim was to provide a theoretical framework and support for clinical protocols and innovative drug development.
The global prevalence and medical burden of non-alcoholic fatty liver disease (NAFLD), a chronic liver disease that is mainly associated with environmental, dietary, genetic, intestinal, and immune factors, is increasing annually (1). Based on the consensus of expert panels in several countries, NAFLD was renamed as metabolic-associated fatty liver disease (MAFLD). MAFLD is associated with many diseases, including type 2 diabetes, hyperlipidaemia, and obesity; if left untreated, MAFLD can progress to steatohepatitis with liver fibrosis and even liver cancer (2). Its diagnostic criteria include the presence of one of the following three metrics based on hepatic steatosis: overweight/obesity, type 2 diabetes, or metabolic disorders (3–6). The intestine has the largest contact area with the external environment. Its intestinal barrier function is essential for maintaining homeostasis of the organism and includes the intestinal microbial barrier and mucus, gastrointestinal dynamics and secretion, epithelial barrier, immunity (innate and adaptive), and intestinal vascular and hepatic barriers (7). A trillion microorganisms are present in the human gastrointestinal tract. The number of these microorganisms dramatically exceeds the number of human cells. Intestinal microorganisms affect the physiological activities of the host, human metabolism, the immune system, and neurological diseases (8, 9). The intestine and liver may be linked through the portal vein and the transport of enteric-derived products to the liver, thereby affecting bile and associated antibodies in the intestine (10). Gut microbial-derived exosomes can be involved in the physiopathological conditions of the body by transporting a variety of substances. Therefore, in this review, we provide theoretical support for the development of clinical treatment protocols and drugs for MAFLD by discussing the relevance of gut microbial-derived exosomes in the pathogenesis of MAFLD.
Hundreds of millions of intestinal microorganisms live in the gut. These intestinal microbes encode millions of genes, 150 times the number of genes in the human genome, including a rich library of enzymes (11, 12). They provide uncoded enzymes that participate in human metabolism and maintain immune homeostasis through interactions with host cells (13). The gut microbiota is essential for host metabolism and immune homeostasis, and its components contribute significantly to shaping the host immune system (14). The quest to understand the relationship between the host and gut microbiota continues. Dysbiosis of the intestinal flora may lead to various diseases in the host, ultimately compromising health (8, 9). Dysbiosis of intestinal flora is an important pathogenic factor that induces the development and progression of MAFLD (15). The mechanisms by which bacteria influence the progression of metabolism-related diseases are largely unknown, although there is evidence that microbial-derived exosomes are associated with inflammation (16). Typically, bacterial cells communicate with their hosts and other bacteria through direct contact and secretion of soluble products such as metabolites (e.g., short-chain fatty acids), lipopolysaccharides, population-sensing peptides, nucleic acids, proteins, and extracellular vesicles (EVs) (17, 18). For example, butyrate from microbiota can upregulate miRNA-22 expression in hepatocytes, which can decrease Sirtuin1 expression and enhance reactive oxygen species (ROS) production to increase programmed cell death in hepatocytes (19).
Exosomes were initially thought to be a means of cellular waste disposal until further studies reported their role in mediating intercellular communication, thus attracting significant attention from researchers worldwide (20, 21). Exosomes are a class of EVs with diameters of approximately 30–150 nm (22, 23). Exosomes are derived from most cell types and are present in cell-conditioned media and different biological fluids such as serum, plasma, urine, saliva, ascites, cerebrospinal fluid, and amniotic fluid (24). Enteric bacteria (both pathogenic and commensal) derive EVs for communication with their hosts. These spherical membrane-encapsulated particles transmit some of the biological components of the parental bacteria to the extracellular environment (25). EVs perform different biological functions in the host through different synthetic pathways and mechanisms (26). EVs mainly consist of outer membrane vesicles (OMVs) released by gram-negative bacteria and membrane vesicles (MVs) released by gram-positive bacteria (25). Studies on gram-negative pathogens have shown that OMVs internalize in host cells and promote virulence by delivering cytotoxic factors as well as interfering with immune system mediators (27–29). Currently, microbiota vesicles are considered key players in the signaling process of the intestinal mucosa (25, 30). Gram-negative bacteria follow two major vesicle formation pathways. The first pathway of formation involves blistering of the outer membrane of the bacterial envelope to produce OMVs; the second pathway requires explosive cell lysis to form outer inner membrane vesicles (OIMVs) and explosive outer membrane vesicles (EOMVs) (18). Cytoplasmic membrane vesicles (CMVs) are produced by gram-positive bacteria through endolysin-triggered bulge cell death (26). OMVs are broadly defined as EVs. The blistering membrane process causes OMVs to disrupt the cross-linkage between the outer membrane and the underlying peptidoglycan cell wall layer. In the following sections, we collectively refer to all exosome subtypes as EVs. A variety of functions of exosomes have been characterized, including cell proliferation, differentiation, apoptosis, and immunomodulation (31, 32). Transcriptomic analysis revealed that EVs carry a variety of cargo, including many functional small coding miRNAs, messenger RNA (mRNA), and non-coding RNA (ncRNAs). Proteomic analysis has revealed that EVs also carry other cargo, including membrane-bound proteins, enzymes (e.g. autolysins), toxins, polysaccharides, and peptidoglycans (25, 33). EVs deliver these bioactive components to receptor cells to perform these functions (34).
Gut microbial release EVs that contain specific cargo molecules and have multiple functions (35). Various RNA species (e.g., mRNA, miRNA, tRNA) are among the biologically active components of EVs and may affect host gene expression when delivered to host cells (36–38). In a previous study, the role of OMV in signal transduction between the gut microbiota and the host was demonstrated. OMV produced by the symbiotic probiotic Escherichia coli induced the expression and secretion of several cytokines and chemokines in an in vitro model (39). EVs contain microRNAs (miRNAs), small coding RNA molecules that regulate gene expression after transcription. miRNAs remain biologically active after delivery to host cells (40). Transfection of Caco2-BBE cells derived from the parental Caco2 strain with mature miR-99b, miR-125a-5p, and miR-1269 decreased the cell growth rate and trans-epithelial resistance, indicating a shift toward the HT29-Cl.19A cell phenotype. EVs may support the transportation process for miRNA to affect cell function (41). Thus, intestinal bacteria use OMV as an essential strategy to communicate with and influence the host responses.
Microbiota-derived EVs are always localised in organs such as the gastrointestinal tract, which is in close contact with bacteria. The biodistribution of EVs derived from the human intestinal commensal bacterium Bacteroides thetaiotaomicron after gavage in mice with fluorescent labelling revealed that some EVs accumulated in the liver (42). In addition, by engineering Escherichia coli to express Cre recombinase (E. coli Cre) colonized into Rosa26.tdTomato-background mice, the Cre-LoxP system was used to report bacterial OMV transfer into intestinal epithelial cells and induce fluorescent reporter gene expression, including intestinal stem cells and mucosal immune cells such as macrophages. Outside the intestine, bacterial-derived Cre induces extended marker gene expression in various host tissues, including the heart, liver, kidney, spleen, and brain. E. coli OMVs are mainly concentrated in the hepatic confluent region (43). In contrast, endocytosis of endothelial cells is involved in the active transcellular migration of EVs across the epithelial monolayer (e.g., endocytosis, multivesicular body formation, and cytokinesis on the other side of the transcellular layer), which may be a pathway for EVs to cross the intestinal barrier and even the blood-brain barrier (44). Thus, EVs or OMVs released by microorganisms can act as biological shuttle systems for cross-border communication in the host, where bacteria transfer functional biomolecules via EVs to individual host cells and target organs, such as the liver. In summary, gut microbes may also be involved in the development of MAFLD through the release of multiple mediators, among which gut microbial-derived EVs may serve as important mediators of the association between gut microbes and MAFLD. Below, we describe the potential effects of gut microbe-derived EVs on various aspects of MAFLD, including insulin resistance, intestinal barrier function, inflammation, lipids, and liver fibrosis.
Gut microbial-derived EVs influence glucose metabolism by regulating insulin resistance. Insulin resistance is one of the core mechanisms of MAFLD (45). Thus, intestinal barrier dysfunction and increased intestinal permeability in MAFLD, which often implies absorption of host-microbial EVs and lipopolysaccharides (LPS), may lead to a state of insulin resistance (46). Similarly, a study characterising microbial-derived EVs from serum, urine, and stool samples from Korean T2DM subjects found that microbial-derived EVs were prevalent in other parts of the body, such as stool, serum, and urine, whereas gut microbial-derived EVs were less likely to cross the intestinal barrier in healthy individuals (47). It was found that fecal-derived EVs induced insulin resistance and poor glucose tolerance in high-fat diet (HFD)-fed mice compared to conventional diet-fed mice. Macrogenomic analysis revealed that Pseudomonas panacis (phylum Proteobacteria) EVs can pass through the intestinal barrier to insulin-responsive organs, such as the liver, skeletal muscle, and adipose tissue. They play multiple roles in HFD-fed mice, including blocking insulin signalling pathways in skeletal muscle and adipose tissue, inhibiting insulin-stimulated glucose uptake and GLUT4 translocation in myotubes, and suppressing pAKT expression levels in the skeletal muscle and adipose tissue of mice (48). The skeletal muscle is one of the major tissues involved in peripheral glucose uptake. When free fatty acids are elevated in the body, glucose utilisation by skeletal muscles is inhibited, resulting in insulin resistance (49).
As shown above, microbial-driven disruption of the intestinal epithelial and intestinal vascular barriers is a prerequisite for MAFLD, and disruption of the intestinal epithelial and vascular barriers is dependent on the Wnt/β-catenin signalling pathway to promote MAFLD (50). Gene (F11r) encoding junctional adhesion molecule A knockout mice with high fructose, high fat, and cholesterol induce defects in intestinal epithelial permeability, exhibiting more pronounced steatosis (51). The increased intestinal permeability allows the entry of multiple intestinal microbial-derived EVs and LPS, which may be a mechanism contributing to the development of MAFLD. Similar association studies have found increased levels of circulating bacterial EVs in patients with intestinal barrier dysfunction, such as untreated intestinal mucosal inflammation, and levels of bacterial EV-associated LPS correlated with plasma zonulin levels. This suggests that intestinal barrier dysfunction leads to the translocation of bacterial EVs (52). The microbiota affects miRNA expression in the caecum, and their target genes may control the synthesis of proteins related to immune system management and control of intestinal barrier function. Microbial-derived EVs may bridge these gaps and play an important role in maintaining intestinal barrier homeostasis. Microarray analysis of intestinal miRNAs from C57 mice raised under specific pathogen-free (SPF) and germ-free (GF) conditions revealed reduced intestinal miR-10a expression in SPF mice compared to that in GF mice, while GF mice showed decreased intestinal miR-10a expression after decolonization with field bacteria. Intestinal symbiotic bacteria maintain intestinal homeostasis by downregulating dendritic cell miR-10a expression through Toll-like receptor (TLR)-TLR legend (TLRL) interactions and the MyD88-dependent pathway, targeting IL-12/IL-23p40 expression (53). In addition, EVs are considered to have microbial-associated or pathogen-associated molecular patterns (MAMPs) (54). The content of MAMPs in EVs allows them to bind to host pattern recognition receptors (PRRs) in immune and non-immune cells to promote host pathology and immune tolerance, or confer protective immunity (54). Vibrio cholera was found to secrete OMV to induce the expression of pro-inflammatory cytokines (e.g., IL-8 and GM-CSF) and chemokines (e.g., CCL2, CCL20, and thymic stromal lymphopoietin) in intestinal epithelial cells in a NOD1-dependent manner through activation of the MAPK/NF-κB pathway. In vitro OMV-stimulated epithelial cells can promote the expression of high levels of costimulatory molecules and the release of pro-inflammatory cytokines and chemokines, such as IL-1, IL-6, TNF-α, CCL22, and CCL17, through the activation of dendritic cells (DCs). These cytokines activate CD4+ T cells and promote IL-4, IL-13, and IL-17 release (28). The human commensal bacterium Bacteroides fragilis produces a capsular polysaccharide (PSA)-dependent TLR-2 signaling pathway that promotes IL-10 production in dendritic cells (DCs) and prevents chemotactic colitis in mice (55). Thus, microbially derived EVs may be associated with inflammation through multiple immune cells and coordinate the release of multiple biological mediators, particularly MAFLD. For example, gram-negative EVs are characterized by internal phospholipid lobules and external lipopolysaccharide (LPS), a TLR-4 agonist (18). Escherichia coli releases OMV containing active pore components of LPS that drive inflammatory responses in human epithelial cells via Ca2+ signaling and activate TLR-4 (56). In the liver, activation of TLR-4 signaling in hepatocytes, accompanied by nuclear factor kappa B (NF-kB) activation and nuclear translocation, plays an important role in the initiation of MAFLD (57). TLR-4-deficient mice hepatocytes exhibit reduced insulin resistance and obesity-associated inflammation during high-quality diet feeding (58). In addition, systemic deletion of TLR-4 attenuates NASH development in MCD diet-fed mice (59). EVs can also deliver LPS into the cytosol of host cells and activate macrophages via TLR-4 receptors (60). In contrast, macrophage activation in MAFLD recruits inflammatory factors and is an essential driver of inflammation. Lipoteichoic acid (LTA) on the surface of gram-positive bacteria and their EVs can bind to TLR-2 to drive the immune response (26). Furthermore, peripheral blood mononuclear cells (PBMCs) stimulated of with Escherichia coli strain Nissle 1917 (EcN) or the commensal E. coli strain ECOR12 in vitro secreted OMV and LPS alone resulting in interleukin (IL)-10, macrophage inflammatory protein 1 alpha (MIP1a), tumor necrosis factor (TNF)-α, IL-6, and IL-8 secretion, implying intestinal inflammation and disruption of the intestinal barrier. Co-culture of intestinal epithelial Caco-2/PBMCs revealed that OMV was internalized by Caco2 cells, thereby preventing the overproduction of multiple pro-inflammatory factors and promoting the production of the anti-inflammatory cytokine IL-10 (39). EcN-derived OMV can promote IL-22 production, transforming growth factor (TGF)-β secretion, and Treg differentiation (39). IL-22 has been shown to enhance intestinal epithelial barrier function (61) and can cooperate with IL-6 to induce the transcription factor Runx1 to promote the differentiation of Th1 cells to Th17 cells, which promotes inflammation by secreting cytokines such as IL-17, while Treg cells can promote the secretion of the anti-inflammatory factor IL-10 to suppress inflammation (62, 63).
Leaking bacterial products from the gut can also exacerbate the adipose tissue inflammatory response during the progression of MAFLD, leading to insulin resistance and having important effects on systemic metabolism. In addition to metabolites, microbial DNA accumulates in the peripheral circulation and adipose tissues of obese patients (64, 65). Obesity leads to a decrease in the number of complement receptors of the immunoglobulin superfamily (CRIg+) macrophages in human and mouse livers. This phenomenon diminishes the hepatic clearance of circulating bacterial EVs, leading to the spread of bacterial EVs to more distant adipose tissue and inducing inflammation and metabolic disturbances. The intervention of EVs in the intestinal flora of obese mice attenuated the reduction of insulin-stimulated AKT phosphorylation and activation of the cGAS/STING signalling pathway in adipocytes 3T3L-1 cells and enhanced the inflammatory response in adipose tissue (66). Bacterial EVs are enriched in β-cells of obese patients and promote islet inflammatory responses and impair insulin secretion from β-cells by activating the cGAS/STING signalling pathway (67). Similarly, EVs derived from adipocytes of obese patients can promote the release of inflammatory factors such as TNF-α and IL-6 from monocytes and upregulate hepatocyte TGF-β, PAI-1, MMP-7, and TIMP-1 expression to mediate hepatic steatosis and fibrosis (68).
Repeated inflammatory stimulation in MAFLD may lead to a poor healing response and, thus, a liver fibrotic response. EVs may be associated with liver fibrosis progression by regulating the expression of miRNAs and their target genes. By assessing miRNA expression profile changes through liver biopsies in patients with no fibrosis or severe liver fibrosis or cirrhosis, 30 upregulated and 45 downregulated miRNAs were found to be present in patients with liver fibrosis (69). In addition, serum levels of IL-6 and miR-233 in patients with NAFLD were correlated. Liver fibrosis was more severe after IL-6 bone marrow-specific knockdown in mice fed a high-fat diet. In vitro experiments suggested that the mechanism may involve IL-6 promoting the release of miR-233-containing exosomes from macrophages and reducing the pro-fibrotic gene expression of TAZ (70). Similarly, miR-21 regulates fibrosis progression in the liver (71). In addition to regulating miRNA expression, microbial-derived EVs can directly inhibit liver fibrosis by modulating inflammation and maintaining intestinal barrier homeostasis. Akkermansia muciniphila primarily inhabits the gastrointestinal mucus layer and is considered a probiotic strain for the treatment of obesity-related diseases (72). Previous studies have shown that heat-inactivated Akkermansia muciniphila can modulate the LPS-induced gene expression of liver fibrosis markers, including smooth muscle alpha-actin (α-SMA), tissue inhibitor of metalloproteinase (TIMP), collagen type 1 (Col1), TGF-β, TLR-4, and peroxisome proliferator-activated receptor gamma (PPARγ), in LX-2 cells and reverse the activation of hepatic stellate cell (HSC) in LPS-stimulated LX-2 cells. EVs (50 μg/ml) isolated from Akkermansia muciniphila inhibited TLR-2 and TLR-4 gene expression in LPS-stimulated LX-2 cells (73). They also reduced the release of serum cytokines TNF-α and IL-6, and increased the level of the anti-inflammatory factor IL-10, thereby suppressing inflammation (73). In addition, Akkermansia muciniphila and its EVs increased ZO-1 expression. They inhibited TNF-α and TLR-4 in the colonic tissues of HFD-fed mice, suggesting that they could improve intestinal barrier function and suppress inflammatory responses (73). EVs secreted by Akkermansia muciniphila also modulate intestinal integrity and restore disturbed intestinal flora by inhibiting the expression of liver fibrosis markers, including α-SMA, PDGF, TIMP, and Col1a1, and by suppressing inflammatory genes in experimental mice (73). In addition, MAFLD is associated with renal fibrosis. Live Akkermansia muciniphila bacteria and their EV attenuate HFD/CCL4-induced renal tissue injury and the associated gene expression of renal fibrosis, such as α-SMA, PDGF, Col1a1, and TGF-β (73).
Lipid deposition in MAFLD and lipotoxicity due to oxidative stress are essential drivers of inflammation, and microbial-derived EVs may affect hepatic lipid regulation via different mechanisms. Akkermansia muciniphila has also been shown to correlate with the lipid content. Changes in Akkermansia muciniphila are associated with lipid metabolism in adipose tissue and expression of inflammatory markers and glucose, insulin, triglyceride, and leptin levels in obese mouse models (74). A similar study found that post-pasteurization Akkermansia muciniphila can affect intestinal lipid absorption and metabolism, thus regulating body weight and fat gain by regulating perilipin2 lipid droplet-associated factors (75). In addition, gavage of A. muciniphila I (Amuc_GP01) in an HFD-induced experimental mouse model improved glucose tolerance, hyperlipidemia, and hepatic steatosis (76). Furthermore, in the HFD-induced intestinal microbiota of mice, triglyceride content in the liver was positively correlated with miR-21 expression, whereas hepatic miR-21 expression was positively correlated with Firmicutes and negatively correlated with Proteobacteria and Bacteroides acidifaciens. The ability of the bacterial antigens to regulate hepatic miRNAs was tested in vitro. The pro-inflammatory LPS of E. coli O55:B5 promoted miR-21 expression in primary wild-type mouse hepatocytes in a dose-dependent manner, suggesting that gut microbes, possibly through their antigens (e.g., LPS), regulate hepatic miRNA expression to influence the liver (77). The regulatory effects of miRNAs on gut bacteria and the liver need to be further validated by in vivo experiments. In aged DB/DB mice, long-term (18 weeks) inhibition of miR-21 reduces body weight and adipocyte size (78). In addition, it was found that intestinal flora disorders in MAFLD abnormally increased the Bacteroidetes ratio and Streptomyces spp., and that high mobility group box 1 (HMGB1) promoted hepatic steatosis through transfer from the intestine to the liver (79).
In addition to the potential impact of microbial-derived EVs on lipid metabolism in MAFLD, most studies have found relevance of other source-derived exosome secretions to MAFLD, particularly lipid-regulated EVs release. For example, plasma exosome levels are significantly elevated in MAFLD patients. Fatty acid-induced damage-regulated autophagy modulator (DRAM) enhances hepatocyte-associated exosome release by promoting lysosomal localization of stomatin upon interaction with stomatin, which may serve as a potential biomarker of MAFLD (80). The presence of many membrane-bound vesicles generated by hepatocytes in experimental mice with steatohepatitis that respond to FFA-derived EVs correlates significantly with disease severity (81). Similarly, adipocytes can endocytose HepG2 cell-derived exosomes, thereby promoting their inflammatory phenotypic differentiation by activating phosphokinase and NF-kB signalling and recruiting inflammatory macrophages to co-promote the inflammatory milieu (82). The most relevant research advances have been reviewed widely (83–86). Without going into much detail, we note that the potential relevance of excessive lipid deposition in MAFLD and its regulation in microbially derived EVs deserves further investigation.
Many cited studies still use the term “NAFLD” and link to other metabolic diseases such as T2M. In view of the update of NAFLD terminology and diagnostic criteria to further standardise and advance clinical and research progress in the discipline of hepatology, we use the term “MAFLD” instead of “NAFLD” throughout the text. We believe that it is extremely important to standardise and update this terminology; however, the diagnostic distinction between them should be noted in the future. Sumida et al. summarised the current pharmacological treatments for MAFLD and the possible challenges (87). In summary, there are no FDA-approved treatment options for MAFLD, and diet control and exercise remain important lifestyle interventions. In addition, several drugs have potential therapeutic effects by targeting different mechanisms, such as lipid deposition (elafibranor), oxidative stress, inflammation, cell death (emricasan), intestinal microenvironment and metabolism (IMMe124), and antifibrotic drugs (cenicriviroc), some of which are already in clinical trials (87). As mentioned above, gut microbial-derived EVs might affect MAFLD through different mechanisms; therefore, the proposed increase in beneficial gut microbes (flora transplantation, genetic engineering) or reduction of harmful gut microbial populations (lifestyle changes, pharmacological interventions) are potentially important approaches. Probiotics, prebiotics (fermented products that improve the flora composition of patients) (88) and prebiotics (combination of probiotics and dietary or food components) can improve the extent of experimental animal models and MAFLD patients. For example, the intake of Lactobacillus acidophilus La5 and Bifidobacterium lactis Bb12 improved liver enzymes, total serum cholesterol, and LDL cholesterol levels in patients with MAFLD (89). Prebiotics can significantly reduce TNF-α, CRP, liver enzymes, and steatosis in patients (90). Most of the current studies on MAFLD and gut flora have focused on changes in the abnormal gut flora and the influence of the flora on a particular aspect of MAFLD and have not directly focused on the role of gut microbial-derived EVs. This is an important mechanism by which the intestinal flora exerts its physiopathological functions. Considering the preliminary studies on the potential benefits of probiotic transplants, prebiotics, and prebiotics for MAFLD treatment in combination with the role of EVs in MAFLD, we believe this will be a direction with great research potential in the future; however, further well-designed preclinical and clinical trials are still needed for in-depth studies.
Based on the gut-liver axis theory, gut microbes are an essential component of the human body, and their derived EVs, which contain abundant microbial DNA, proteins, and lipids, have been gradually recognized to contribute to the pathological mechanisms of various liver diseases, especially MAFLD (see Figure 1). In this article, we provide a theoretical basis for developing innovative clinical treatment options for MAFLD based on gut microbial-derived EVs by discussing their potential relationship with insulin resistance, the intestinal barrier, inflammation, lipid metabolism, and liver fibrosis and the improvement of MAFLD through probiotic colonization. There is great potential to elucidate the molecular composition of the cargoes carried by EVs and to explore how they interact with receptor cells and their mechanisms by combining a variety of modern molecular biology techniques and multi-omics applications.
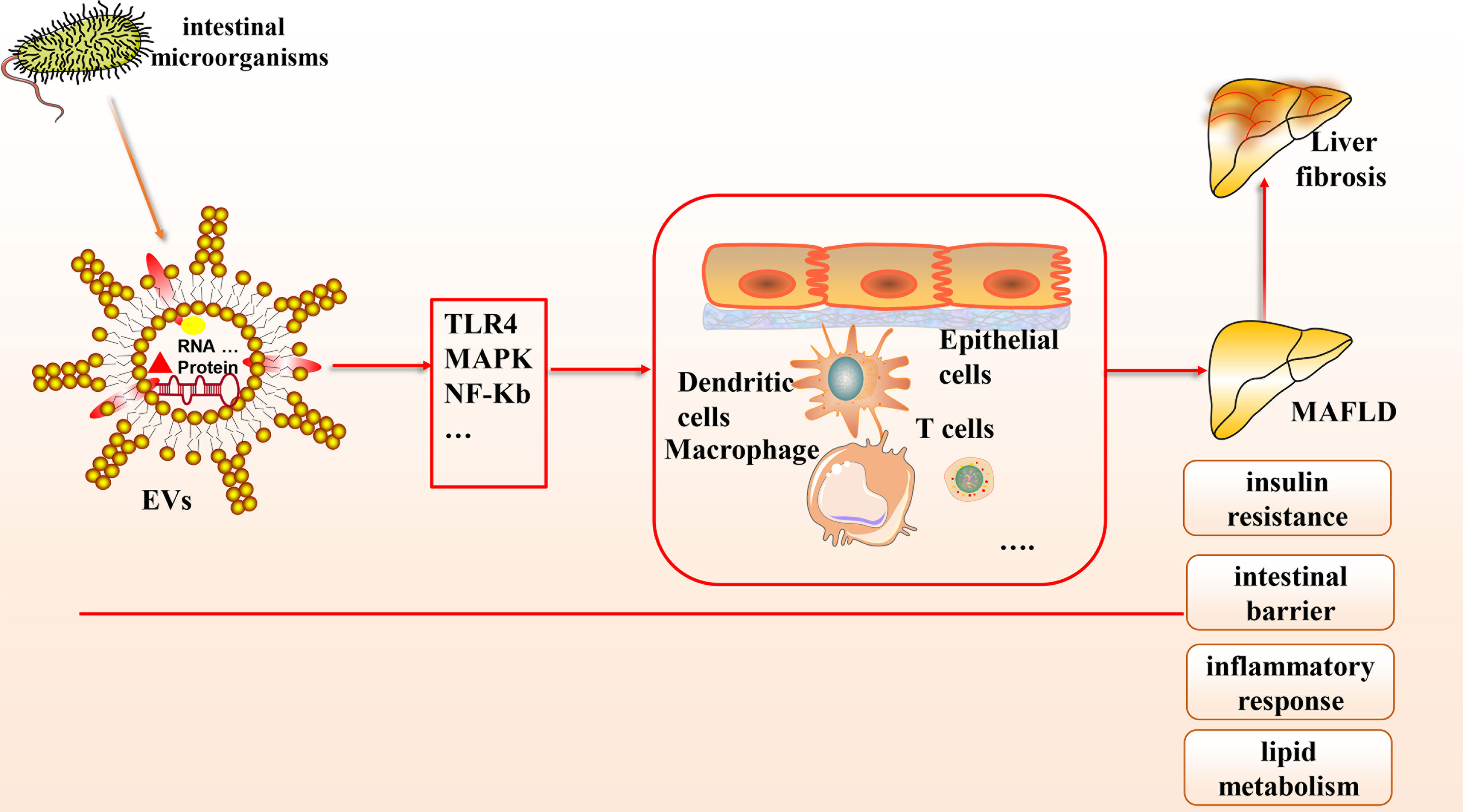
Figure 1 The connection between gut microbial-derived exosomes and MAFLD. Many microorganisms exist in the human body, which can secrete EVs, a bilayer lipid membrane structure that can include the outermost LPS, cytoplasmic and membrane-bound proteins, DNA, multiple protein components, etc. Based on the gut-liver axis theory, EVs can carry various biological mediators and affect multiple cells through many signaling pathways to influence pathological mechanisms in MAFLD, including intestinal barrier homeostasis, insulin resistance, lipid metabolism, inflammatory response, etc.
BZ and JZ are responsible for the collection, collation and writing of the original manuscript. MJ, and DP are responsible for the collection of the original manuscript. XD, YS, and JS are responsible for the revision and review of the manuscript. All authors reviewed and accepted with the final version.
This work was funded by the National Natural Science Funds of China (81570524), Zhejiang Provincial Basic Public Welfare Research Project (GF20H030035), Major Projects of Hangzhou Medical and Health Science and Technology Program (0020191059).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Di Ciaula A, Baj J, Garruti G, Celano G, De Angelis M, Wang HH, et al. Liver Steatosis, Gut-Liver Axis, Microbiome and Environmental Factors. A Never-Ending Bidirectional Cross-Talk. J Clin Med (2020) 9(8):2648. doi: 10.3390/jcm9082648
2. Lindenmeyer CC, McCullough AJ. The Natural History of Nonalcoholic Fatty Liver Disease-An Evolving View. Clin Liver Dis (2018) 22(1):11–21. doi: 10.1016/j.cld.2017.08.003
3. Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, et al. A New Definition for Metabolic Dysfunction-Associated Fatty Liver Disease: An International Expert Consensus Statement. J Hepatol (2020) 73(1):202–9. doi: 10.1016/j.jhep.2020.03.039
4. Shiha G, Alswat K, Al Khatry M, Sharara AI, Örmeci N, Waked I, et al. Nomenclature and Definition of Metabolic-Associated Fatty Liver Disease: A Consensus From the Middle East and North Africa. Lancet Gastroenterol Hepatol (2021) 6(1):57–64. doi: 10.1016/S2468-1253(20)30213-2
5. Shiha G, Korenjak M, Eskridge W, Casanovas T, Velez-Moller P, Högström S, et al. Redefining Fatty Liver Disease: An International Patient Perspective. Lancet Gastroenterol Hepatol (2021) 6(1):73–9. doi: 10.1016/S2468-1253(20)30294-6
6. Mendez-Sanchez N, Arrese M, Gadano A, Oliveira CP, Fassio E, Arab JP, et al. The Latin American Association for the Study of the Liver (ALEH) Position Statement on the Redefinition of Fatty Liver Disease. Lancet Gastroenterol Hepatol (2021) 6(1):65–72. doi: 10.1016/S2468-1253(20)30340-X
7. Portincasa P, Bonfrate L, Khalil M, Angelis M, Calabrese FM, D'Amato M, et al. Intestinal Barrier and Permeability in Health, Obesity and NAFLD. Biomed (2021) 10(1):83. doi: 10.3390/biomedicines10010083
8. Li D, Wang P, Wang P, Hu X, Chen F. The Gut Microbiota: A Treasure for Human Health. Biotechnol Adv (2016) 34(7):1210–24. doi: 10.1016/j.biotechadv.2016.08.003
9. Chang CS, Kao CY. Current Understanding of the Gut Microbiota Shaping Mechanisms. J BioMed Sci (2019) 26(1):59. doi: 10.1186/s12929-019-0554-5
10. Yang X, Lu D, Zhuo J, Lin Z, Yang M, Xu X. The Gut-Liver Axis in Immune Remodeling: New Insight Into Liver Diseases. Int J Biol Sci (2020) 16(13):2357–66. doi: 10.7150/ijbs.46405
11. El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The Abundance and Variety of Carbohydrate-Active Enzymes in the Human Gut Microbiota. Nat Rev Microbiol (2013) 11(7):497–504. doi: 10.1038/nrmicro3050
12. Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nat (2019) 570(7762):462–7. doi: 10.1038/s41586-019-1291-3
13. Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, et al. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur J Nutr (2018) 57(1):1–24. doi: 10.1007/s00394-017-1445-8
14. Rooks MG, Garrett WS. Gut Microbiota, Metabolites and Host Immunity. Nat Rev Immunol (2016) 16(6):341–52. doi: 10.1038/nri.2016.42
15. Schnabl B, Brenner DA. Interactions Between the Intestinal Microbiome and Liver Diseases. Gastroenterol (2014) 146(6):1513–24. doi: 10.1053/j.gastro.2014.01.020
16. Yu YJ, Wang XH, Fan GC. Versatile Effects of Bacterium-Released Membrane Vesicles on Mammalian Cells and Infectious/Inflammatory Diseases. Acta Pharmacol Sin (2018) 39(4):514–33. doi: 10.1038/aps.2017.82
17. Hughes DT, Sperandio V. Inter-Kingdom Signalling: Communication Between Bacteria and Their Hosts. Nat Rev Microbiol (2008) 6(2):111–20. doi: 10.1038/nrmicro1836
18. Tulkens J, De Wever O, Hendrix A. Analyzing Bacterial Extracellular Vesicles in Human Body Fluids by Orthogonal Biophysical Separation and Biochemical Characterization. Nat Protoc (2020) 15(1):40–67. doi: 10.1038/s41596-019-0236-5
19. Pant K, Yadav AK, Gupta P, Islam R, Saraya A, Venugopal SK. Butyrate Induces ROS-Mediated Apoptosis by Modulating miR-22/SIRT-1 Pathway in Hepatic Cancer Cells. Redox Biol (2017) 12:340–9. doi: 10.1016/j.redox.2017.03.006
20. Li S, Zhan JK, Wang YJ, Lin X, Zhong JY, Wang Y, et al. Exosomes From Hyperglycemia-Stimulated Vascular Endothelial Cells Contain Versican That Regulate Calcification/Senescence in Vascular Smooth Muscle Cells. Cell Biosci (2019) 9:1. doi: 10.1186/s13578-018-0263-x
21. Lin X, Li S, Wang YJ, Wang Y, Zhong JY, He JY, et al. Exosomal Notch3 From High Glucose-Stimulated Endothelial Cells Regulates Vascular Smooth Muscle Cells Calcification/Aging. Life Sci (2019) 232:116582. doi: 10.1016/j.lfs.2019.116582
22. Lakshmi S, Hughes TA, Priya S. Exosomes and Exosomal RNAs in Breast Cancer: A Status Update. Eur J Cancer (2021) 144:252–68. doi: 10.1016/j.ejca.2020.11.033
23. Zhang L, Yu D. Exosomes in Cancer Development, Metastasis, and Immunity. Biochim Biophys Acta Rev Cancer (2019) 1871(2):455–68. doi: 10.1016/j.bbcan.2019.04.004
24. Safdar A, Saleem A, Tarnopolsky MA. The Potential of Endurance Exercise-Derived Exosomes to Treat Metabolic Diseases. Nat Rev Endocrinol (2016) 12(9):504–17. doi: 10.1038/nrendo.2016.76
25. Kaparakis-Liaskos M, Ferrero RL. Immune Modulation by Bacterial Outer Membrane Vesicles. Nat Rev Immunol (2015) 15(6):375–87. doi: 10.1038/nri3837
26. Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the Wall: Extracellular Vesicles in Gram-Positive Bacteria, Mycobacteria and Fungi. Nat Rev Microbiol (2015) 13(10):620–30. doi: 10.1038/nrmicro3480
27. Ellis TN, Kuehn MJ. Virulence and Immunomodulatory Roles of Bacterial Outer Membrane Vesicles. Microbiol Mol Biol Rev (2010) 74(1):81–94. doi: 10.1128/MMBR.00031-09
28. Chatterjee D, Chaudhuri K. Vibrio Cholerae O395 Outer Membrane Vesicles Modulate Intestinal Epithelial Cells in a NOD1 Protein-Dependent Manner and Induce Dendritic Cell-Mediated Th2/Th17 Cell Responses. J Biol Chem (2013) 288(6):4299–309. doi: 10.1074/jbc.M112.408302
29. Schertzer JW, Whiteley M. Bacterial Outer Membrane Vesicles in Trafficking, Communication and the Host-Pathogen Interaction. J Mol Microbiol Biotechnol (2013) 23(1-2):118–30. doi: 10.1159/000346770
30. Olsen I, Amano A. Outer Membrane Vesicles - Offensive Weapons or Good Samaritans? J Oral Microbiol (2015) 7:27468. doi: 10.3402/jom.v7.27468
31. Couzin J. Cell Biology: The Ins and Outs of Exosomes. Sci (2005) 308(5730):1862–3. doi: 10.1126/science.308.5730.1862
32. Ni YQ, Lin X, Zhan JK, Liu YS. Roles and Functions of Exosomal Non-Coding RNAs in Vascular Aging. Aging Dis (2020) 11(1):164–78. doi: 10.14336/AD.2019.0402
33. Lee EY, Bang JY, Park GW, Choi DS, Kang JS, Kim HJ, et al. Global Proteomic Profiling of Native Outer Membrane Vesicles Derived From Escherichia Coli. Proteomics (2007) 7(17):3143–53. doi: 10.1002/pmic.200700196
34. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of Exosome Composition. Cell (2019) 177(2):428–45.e18. doi: 10.1016/j.cell.2019.02.029
35. Toyofuku M, Nomura N, Eberl L. Types and Origins of Bacterial Membrane Vesicles. Nat Rev Microbiol (2019) 17(1):13–24. doi: 10.1038/s41579-018-0112-2
36. Ahmadi Badi S, Moshiri A, Fateh A, Rahimi Jamnani F, Sarshar M, Vaziri F, et al. Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Front Microbiol (2017) 8:1610. doi: 10.3389/fmicb.2017.01610
37. Felli C, Baldassarre A, Masotti A. Intestinal and Circulating MicroRNAs in Coeliac Disease. Int J Mol Sci (2017) 18(9):1907. doi: 10.3390/ijms18091907
38. Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and Microbiota-Derived Extracellular Vesicles, Immune Function, and Disease Development. Int J Mol Sci (2019) 21(1):107. doi: 10.3390/ijms21010107
39. Fábrega MJ, Aguilera L, Giménez R, Varela E, Alexandra Cañas M, Antolin M, et al. Activation of Immune and Defense Responses in the Intestinal Mucosa by Outer Membrane Vesicles of Commensal and Probiotic Escherichia Coli Strains. Front Microbiol (2016) 7:705. doi: 10.3389/fmicb.2016.00705
40. Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles (2015) 4:27066. doi: 10.3402/jev.v4.27066
41. Dalmasso G, Nguyen HT, Yan Y, Laroui H, Srinivasan S, Sitaraman SV, et al. MicroRNAs Determine Human Intestinal Epithelial Cell Fate. Differentiation; Res Biol Diversity (2010) 80(2-3):147–54. doi: 10.1016/j.diff.2010.06.005
42. Jones EJ, Booth C, Fonseca S, Parker A, Cross K, Miquel-Clopés A, et al. The Uptake, Trafficking, and Biodistribution of Bacteroides Thetaiotaomicron Generated Outer Membrane Vesicles. Front Microbiol (2020) 11:57. doi: 10.3389/fmicb.2020.00057
43. Bittel M, Reichert P, Sarfati I, Dressel A, Leikam S, Uderhardt S, et al. Visualizing Transfer of Microbial Biomolecules by Outer Membrane Vesicles in Microbe-Host-Communication In Vivo. J Extracell Vesicles (2021) 10(12):e12159. doi: 10.1002/jev2.12159
44. Morad G, Carman CV, Hagedorn EJ, Perlin JR, Zon LI, Mustafaoglu N, et al. Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis. ACS Nano (2019) 13(12):13853–65. doi: 10.1021/acsnano.9b04397
45. Zhao J, Hu Y, Peng J. Targeting Programmed Cell Death in Metabolic Dysfunction-Associated Fatty Liver Disease (MAFLD): A Promising New Therapy. Cell Mol Biol lett (2021) 26(1):17. doi: 10.1186/s11658-021-00254-z
46. Erridge C, Attina T, Spickett CM, Webb DJ. A High-Fat Meal Induces Low-Grade Endotoxemia: Evidence of a Novel Mechanism of Postprandial Inflammation. Am J Clin Nutr (2007) 86(5):1286–92. doi: 10.1093/ajcn/86.5.1286
47. Nah G, Park SC, Kim K, Kim S, Park J, Lee S, et al. Type-2 Diabetics Reduces Spatial Variation of Microbiome Based on Extracellur Vesicles From Gut Microbes Across Human Body. Sci Rep (2019) 9(1):20136. doi: 10.1038/s41598-019-56662-x
48. Choi Y, Kwon Y, Kim DK, Jeon J, Jang SC, Wang T, et al. Gut Microbe-Derived Extracellular Vesicles Induce Insulin Resistance, Thereby Impairing Glucose Metabolism in Skeletal Muscle. Sci Rep (2015) 5:15878. doi: 10.1038/srep15878
49. Roden M. How Free Fatty Acids Inhibit Glucose Utilization in Human Skeletal Muscle. News Physiol Sci an Int J Physiol produced jointly by Int Union Physiol Sci Am Physiol Soc (2004) 19:92–6. doi: 10.1152/nips.01459.2003
50. Mouries J, Brescia P, Silvestri A, Spadoni I, Sorribas M, Wiest R, et al. Microbiota-Driven Gut Vascular Barrier Disruption is a Prerequisite for non-Alcoholic Steatohepatitis Development. J Hepatol (2019) 71(6):1216–28. doi: 10.1016/j.jhep.2019.08.005
51. Rahman K, Desai C, Iyer SS, Thorn NE, Kumar P, Liu Y, et al. Loss of Junctional Adhesion Molecule A Promotes Severe Steatohepatitis in Mice on a Diet High in Saturated Fat, Fructose, and Cholesterol. Gastroenterol (2016) 151(4):733–46.e12. doi: 10.1053/j.gastro.2016.06.022
52. Tulkens J, Vergauwen G, Van Deun J, Geeurickx E, Dhondt B, Lippens L, et al. Increased Levels of Systemic LPS-Positive Bacterial Extracellular Vesicles in Patients With Intestinal Barrier Dysfunction. Gut (2020) 69(1):191–3. doi: 10.1136/gutjnl-2018-317726
53. Singh N, Shirdel EA, Waldron L, Zhang RH, Jurisica I, Comelli EM. The Murine Caecal microRNA Signature Depends on the Presence of the Endogenous Microbiota. Int J Biol Sci (2012) 8(2):171–86. doi: 10.7150/ijbs.8.171
54. Riley DR, Sieber KB, Robinson KM, White JR, Ganesan A, Nourbakhsh S, et al. Bacteria-Human Somatic Cell Lateral Gene Transfer Is Enriched in Cancer Samples. PloS Comput Biol (2013) 9(6):e1003107. doi: 10.1371/journal.pcbi.1003107
55. Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer Membrane Vesicles of a Human Commensal Mediate Immune Regulation and Disease Protection. Cell Host Microbe (2012) 12(4):509–20. doi: 10.1016/j.chom.2012.08.004
56. Soderblom T, Oxhamre C, Wai SN, Uhlen P, Aperia A, Uhlin BE, et al. Effects of the Escherichia Coli Toxin Cytolysin A on Mucosal Immunostimulation via Epithelial Ca2+ Signalling and Toll-Like Receptor 4. Cell Microbiol (2005) 7(6):779–88. doi: 10.1111/j.1462-5822.2005.00510.x
57. Li L, Chen L, Hu L, Liu Y, Sun HY, Tang J, et al. Nuclear Factor High-Mobility Group Box1 Mediating the Activation of Toll-Like Receptor 4 Signaling in Hepatocytes in the Early Stage of Nonalcoholic Fatty Liver Disease in Mice. Hepatol (Baltimore Md) (2011) 54(5):1620–30. doi: 10.1002/hep.24552
58. Jia L, Vianna CR, Fukuda M, Berglund ED, Liu C, Tao C, et al. Hepatocyte Toll-Like Receptor 4 Regulates Obesity-Induced Inflammation and Insulin Resistance. Nat Commun (2014) 5:3878. doi: 10.1038/ncomms4878
59. Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-Like Receptor-4 Signaling and Kupffer Cells Play Pivotal Roles in the Pathogenesis of non-Alcoholic Steatohepatitis. J Hepatol (2007) 47(4):571–9. doi: 10.1016/j.jhep.2007.04.019
60. Gu L, Meng R, Tang Y, Zhao K, Liang F, Zhang R, et al. Toll-Like Receptor 4 Signaling Licenses the Cytosolic Transport of Lipopolysaccharide From Bacterial Outer Membrane Vesicles. Shock (2019) 51(2):256–65. doi: 10.1097/SHK.0000000000001129
61. Nikoopour E, Bellemore SM, Singh B. IL-22, Cell Regeneration and Autoimmunity. Cytokine (2015) 74(1):35–42. doi: 10.1016/j.cyto.2014.09.007
62. Sanjabi S, Zenewicz LA, Kamanaka M, Flavell RA. Anti-Inflammatory and Pro-Inflammatory Roles of TGF-Beta, IL-10, and IL-22 in Immunity and Autoimmunity. Curr Opin Pharmacol (2009) 9(4):447–53. doi: 10.1016/j.coph.2009.04.008
63. Liu HP, Cao AT, Feng T, Li Q, Zhang W, Yao S, et al. TGF-β Converts Th1 Cells Into Th17 Cells Through Stimulation of Runx1 Expression. Eur J Immunol (2015) 45(4):1010–8. doi: 10.1002/eji.201444726
64. Ortiz S, Zapater P, Estrada JL, Enriquez P, Rey M, Abad A, et al. Bacterial DNA Translocation Holds Increased Insulin Resistance and Systemic Inflammatory Levels in Morbid Obese Patients. J Clin Endocrinol Metab (2014) 99(7):2575–83. doi: 10.1210/jc.2013-4483
65. Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, et al. Intestinal Mucosal Adherence and Translocation of Commensal Bacteria at the Early Onset of Type 2 Diabetes: Molecular Mechanisms and Probiotic Treatment. EMBO Mol Med (2011) 3(9):559–72. doi: 10.1002/emmm.201100159
66. Luo Z, Ji Y, Gao H, Gomes Dos Reis FC, Bandyopadhyay G, Jin Z, et al. CRIg(+) Macrophages Prevent Gut Microbial DNA-Containing Extracellular Vesicle-Induced Tissue Inflammation and Insulin Resistance. Gastroenterol (2021) 160(3):863–74. doi: 10.1053/j.gastro.2020.10.042
67. Gao H, Luo Z, Ji Y, Tang K, Jin Z, Ly C, et al. Accumulation of Microbial DNAs Promotes to Islet Inflammation and β Cell Abnormalities in Obesity in Mice. Nat Commun (2022) 13(1):565. doi: 10.1038/s41467-022-28239-2
68. Li CJ, Fang QH, Liu ML, Lin JN. Current Understanding of the Role of Adipose-Derived Extracellular Vesicles in Metabolic Homeostasis and Diseases: Communication From the Distance Between Cells/Tissues. Theranostics (2020) 10(16):7422–35. doi: 10.7150/thno.42167
69. Leti F, Malenica I, Doshi M, Courtright A, Van Keuren-Jensen K, Legendre C, et al. High-Throughput Sequencing Reveals Altered Expression of Hepatic microRNAs in Nonalcoholic Fatty Liver Disease-Related Fibrosis. Transl Res (2015) 166(3):304–14. doi: 10.1016/j.trsl.2015.04.014
70. Hou X, Yin S, Ren R, Liu S, Yong L, Liu Y, et al. Myeloid-Cell-Specific IL-6 Signaling Promotes MicroRNA-223-Enriched Exosome Production to Attenuate NAFLD-Associated Fibrosis. Hepatology (Baltimore Md) (2021) 74(1):116–32. doi: 10.1002/hep.31658
71. Zhang J, Jiao J, Cermelli S, Muir K, Jung KH, Zou R, et al. miR-21 Inhibition Reduces Liver Fibrosis and Prevents Tumor Development by Inducing Apoptosis of CD24+ Progenitor Cells. Cancer Res (2015) 75(9):1859–67. doi: 10.1158/0008-5472.CAN-14-1254
72. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation With Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat Med (2019) 25(7):1096–103. doi: 10.1038/s41591-019-0495-2
73. Keshavarz Azizi Raftar S, Ashrafian F, Yadegar A, Lari A, Moradi HR, Shahriary A, et al. The Protective Effects of Live and Pasteurized Akkermansia Muciniphila and Its Extracellular Vesicles Against HFD/CCl4-Induced Liver Injury. Microbiol Spectr (2021) 9(2):e0048421. doi: 10.1128/Spectrum.00484-21
74. Schneeberger M, Everard A, Gómez-Valadés AG, Matamoros S, Ramírez S, Delzenne NM, et al. Akkermansia Muciniphila Inversely Correlates With the Onset of Inflammation, Altered Adipose Tissue Metabolism and Metabolic Disorders During Obesity in Mice. Sci Rep (2015) 5:16643. doi: 10.1038/srep16643
75. Depommier C, Van Hul M, Everard A, Delzenne NM, De Vos WM, Cani PD. Pasteurized Akkermansia Muciniphila Increases Whole-Body Energy Expenditure and Fecal Energy Excretion in Diet-Induced Obese Mice. Gut Microbes (2020) 11(5):1231–45. doi: 10.1080/19490976.2020.1737307
76. Deng L, Ou Z, Huang D, Li C, Lu Z, Liu W, et al. Diverse Effects of Different Akkermansia Muciniphila Genotypes on Brown Adipose Tissue Inflammation and Whitening in a High-Fat-Diet Murine Model. Microbial Pathogen (2020) 147:104353. doi: 10.1016/j.micpath.2020.104353
77. Blasco-Baque V, Coupe B, Fabre A, Handgraaf S, Gourdy P, Arnal JF, et al. Associations Between Hepatic miRNA Expression, Liver Triacylglycerols and Gut Microbiota During Metabolic Adaptation to High-Fat Diet in Mice. Diabetologia (2017) 60(4):690–700. doi: 10.1007/s00125-017-4209-3
78. Seeger T, Fischer A, Muhly-Reinholz M, Zeiher AM, Dimmeler S. Long-Term Inhibition of miR-21 Leads to Reduction of Obesity in Db/Db Mice. Obesity (Silver Spring) (2014) 22(11):2352–60. doi: 10.1002/oby.20852
79. Chen Y, Sun H, Bai Y, Zhi F. Gut Dysbiosis-Derived Exosomes Trigger Hepatic Steatosis by Transiting HMGB1 From Intestinal to Liver in Mice. Biochem Biophys Res Commun (2019) 509(3):767–72. doi: 10.1016/j.bbrc.2018.12.180
80. Zhang J, Tan J, Wang M, Wang Y, Dong M, Ma X, et al. Lipid-Induced DRAM Recruits STOM to Lysosomes and Induces LMP to Promote Exosome Release From Hepatocytes in NAFLD. Sci Adv (2021) 7(45):eabh1541. doi: 10.1126/sciadv.abh1541
81. Povero D, Eguchi A, Niesman IR, Andronikou N, de Mollerat du Jeu X, Mulya A, et al. Lipid-Induced Toxicity Stimulates Hepatocytes to Release Angiogenic Microparticles That Require Vanin-1 for Uptake by Endothelial Cells. Sci Signaling (2013) 6(296):ra88. doi: 10.1126/scisignal.2004512
82. Wang S, Xu M, Li X, Su X, Xiao X, Keating A, et al. Exosomes Released by Hepatocarcinoma Cells Endow Adipocytes With Tumor-Promoting Properties. J Hematol Oncol (2018) 11(1):82. doi: 10.1186/s13045-018-0625-1
83. Devhare PB, Ray RB. Extracellular Vesicles: Novel Mediator for Cell to Cell Communications in Liver Pathogenesis. Mol Aspects Med (2018) 60:115–22. doi: 10.1016/j.mam.2017.11.001
84. Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, et al. Extracellular Vesicles in Liver Pathobiology: Small Particles With Big Impact. Hepatology (Baltimore Md) (2016) 64(6):2219–33. doi: 10.1002/hep.28814
85. Wang W, Zhu N, Yan T, Shi YN, Chen J, Zhang CJ, et al. The Crosstalk: Exosomes and Lipid Metabolism. Cell Commun Signaling CCS (2020) 18(1):119. doi: 10.1186/s12964-020-00581-2
86. Dorairaj V, Sulaiman SA, Abu N, Abdul Murad NA. Extracellular Vesicles in the Development of the Non-Alcoholic Fatty Liver Disease: An Update. Biomolecules (2020) 10(11):1494. doi: 10.3390/biom10111494
87. Sumida Y, Yoneda M. Current and Future Pharmacological Therapies for NAFLD/NASH. J Gastroenterol (2018) 53(3):362–76. doi: 10.1007/s00535-017-1415-1
88. Davani-Davari D, Negahdaripour M, Karimzadeh I, Seifan M, Mohkam M, Masoumi SJ, et al. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods (2019) 8(3):92. doi: 10.3390/foods8030092
89. Nabavi S, Rafraf M, Somi MH, Homayouni-Rad A, Asghari-Jafarabadi M. Effects of Probiotic Yogurt Consumption on Metabolic Factors in Individuals With Nonalcoholic Fatty Liver Disease. J Dairy Sci (2014) 97(12):7386–93. doi: 10.3168/jds.2014-8500
Keywords: metabolic-associated fatty liver disease, gut microbial-derived exosomes, therapeutic approaches, insulin resistance, intestinal barrier, inflammatory response, lipid metabolism, liver fibrosis
Citation: Zhang B, Zhao J, Jiang M, Peng D, Dou X, Song Y and Shi J (2022) The Potential Role of Gut Microbial-Derived Exosomes in Metabolic-Associated Fatty Liver Disease: Implications for Treatment. Front. Immunol. 13:893617. doi: 10.3389/fimmu.2022.893617
Received: 10 March 2022; Accepted: 08 April 2022;
Published: 11 May 2022.
Edited by:
Maria Serena Longhi, Harvard Medical School, United StatesReviewed by:
Prashanth Thevkar Nagesh, Beth Israel Deaconess Medical Center and Harvard Medical School, United StatesCopyright © 2022 Zhang, Zhao, Jiang, Peng, Dou, Song and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junping Shi, MjAxMzEwMDRAaHpudS5lZHUuY24=; Yu Song, c29uZ3l1QHpjbXUuZWR1LmNu; Xiaobing Dou, eGJkb3U3N0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.