- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu City, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu City, China
- 3Avian Disease Research Center, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu City, China
Viroporins are virally encoded transmembrane proteins that are essential for viral pathogenicity and can participate in various stages of the viral life cycle, thereby promoting viral proliferation. Viroporins have multifaceted effects on host cell biological functions, including altering cell membrane permeability, triggering inflammasome formation, inducing apoptosis and autophagy, and evading immune responses, thereby ensuring that the virus completes its life cycle. Viroporins are also virulence factors, and their complete or partial deletion often reduces virion release and reduces viral pathogenicity, highlighting the important role of these proteins in the viral life cycle. Thus, viroporins represent a common drug-protein target for inhibiting drugs and the development of antiviral therapies. This article reviews current studies on the functions of viroporins in the viral life cycle and their regulation of host cell responses, with the aim of improving the understanding of this growing family of viral proteins.
1 Introduction
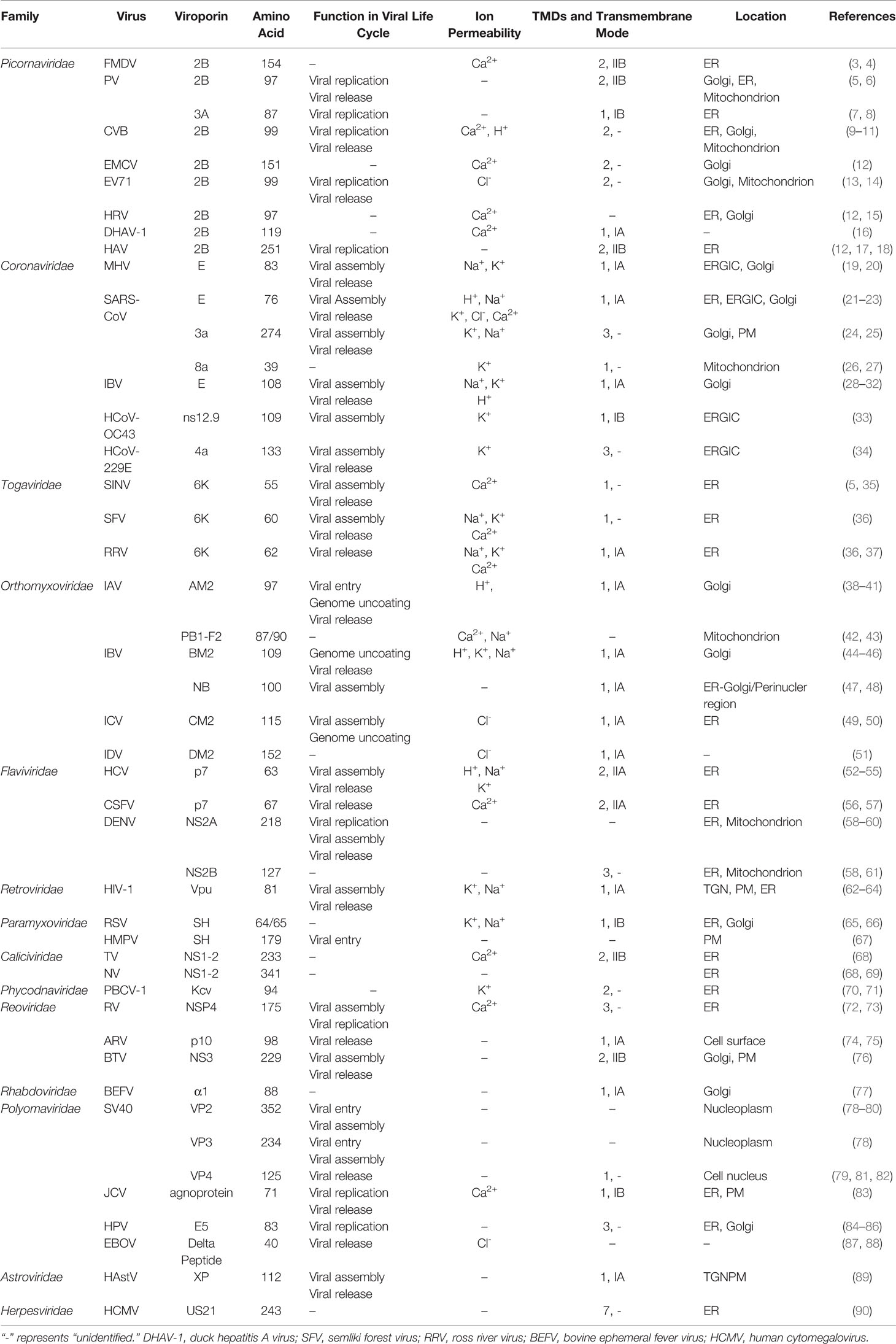
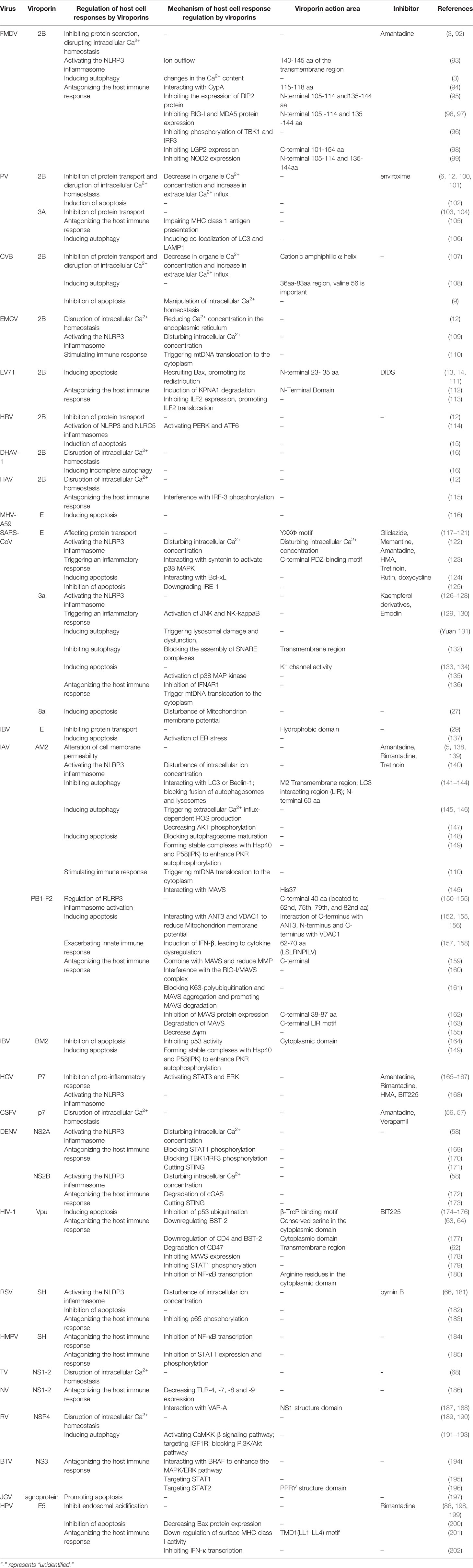
Viroporins are a class of small-molecule hydrophobic transmembrane proteins encoded by viruses, generally with 50-120 amino acid residues. A typical feature of viroporins is the presence of at least one transmembrane helix that anchors the protein into the membrane. Upon insertion into the membrane, their oligomerization produces hydrophilic channels or pores (1). Viroporins also have several characteristic structural motifs, including a set of basic residues (Lys or Arg) and an amphipathic α-helix, these basic amino acids are adjacent to the transmembrane domain and contribute to membrane binding, which controls the rhythm of viral reproduction for optimal spread by inducing membrane perforation at the correct cellular locations at different stages of the viral life cycle (2). Viroporins are essential for viral pathogenicity and replication and are involved in multiple processes including entry, uncoating, replication, assembly, and release in the viral life cycle (Table 1 and Figure 2). In addition, the ion channel activity of viroporins can cause the homeostasis of intracellular ions (e.g., Na+, K+, Ca2+, Cl-) (91). The functional activities of viroporins will affect the host cells and participate in defensive signaling pathways after virus infection of host cells, including autophagy (Figure 4), apoptosis (Figure 5), cellular immune responses (Figure 6), ensuring the completion of virus replication by disrupting the host cell physiology (Table 2). Viroporins were first identified in several RNA viruses, such as protein 2B (P2B) in picornaviruses (203) and matrix protein 2 (M2) in influenza A virus (IAV) (204), and subsequently more and more viroporins have been studied and reported. In addition to the well-known viroporins such as poliovirus 2B, alphavirus 6K, HIV Vpu, hepatitis C virus (HCV) p7 [reviewed in (1, 205)], human astrovirus (HAstV) XP (89), dengue virus (DENV) NS2A and NS2B (61), ebola virus (EBOV) delta peptide (88), Norwalk Viruses (NV) NS1-2 (68), classical swine fever virus (CSFV) p7 (56), and bluetongue virus (BTV) NS3 (76) have been reported to have viroporin-like activity and are proposed to be members of the viroporin family.
Depending on the internal nucleic acids of the viruses containing viroporins, they can be classified into viroporins encoded by DNA viruses (e.g., JC virus agnoprotein (83), human papillomavirus E5 (86), simian virus 40 VP4 (82) and viroporins encoded by RNA viruses (e.g., PV 2B (203, 206), IAV M2 (204), HCV p7 (54), BTV NS3 (76). According to the number of hydrophobic transmembrane domains (TMD) of viroporins, they are divided into two major classes: Class I and Class II, which can be further divided into subclasses A and B according to their different transmembrane modes (205) (Figure 1). Viroporins containing three hydrophobic transmembrane regions have also been identified in recent years, such as rotavirus (RV) NSP4 (189), the human papillomavirus(HPV) E5 protein (85), human coronavirus 229E (HCoV-229E) 4a (R. 34) and SARS-CoV 3a (24), but no further classification of such proteins has been performed.
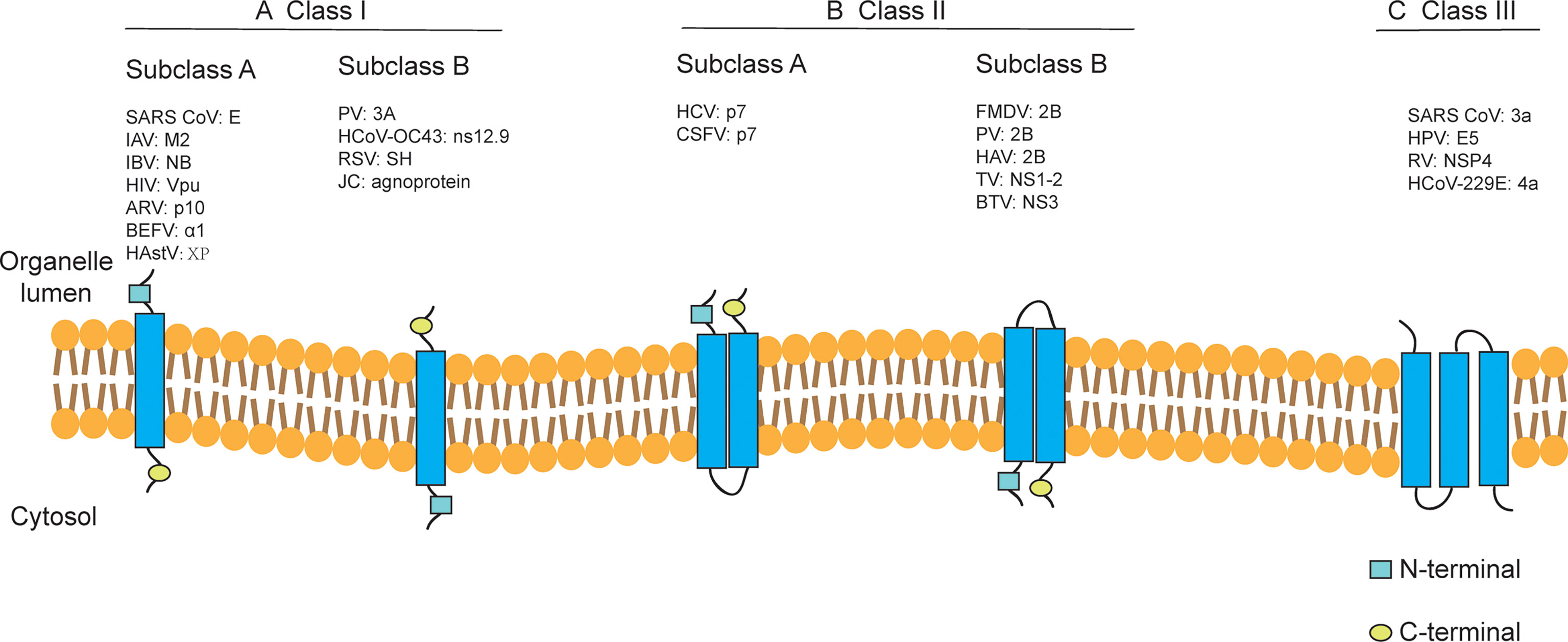
Figure 1 Classification of viroporins according to the number of transmembrane domains and the membrane topology of the constituent monomers. Class I and Class II viroporins have one and two TMD, respectively. (A) Class I A viroporins have their N-termini facing the lumenal side while Class I B have their N-termini in the cytosolic side. (B) Class II A viroporins have both the N- and C-termini in the lumenal side while Class II B have them facing the cytosol. (C) Class III viroporin with three TMDs. HCoV-OC43, human coronavirus OC43; TV, tulane virus. Figure adapted from (205).
Viroporins can form selective ion channels in the host cell membrane that mediate the transport of physiologically relevant ions (e.g., Na+, K+, Ca2+, Cl- or H+) (Table 1). For example, the IAV M2 protein can form a proton channel (41), while the Kcv protein encoded by paramecium bursaria chlorella virus 1(PBCV-1) is a K+ selective channel (71). However, most viroporins generally exhibit weak ion selectivity, and these channels generally do not show a preference for specific ions. For example, IAV PB1-F2 viroporin can generate conductance in the lipid bilayer without apparent selectivity and conducts both Ca2+ and Cl- (43). The SARS-CoV E protein is more selective for monovalent cations (Na+ and K+) than monovalent anions (Cl-) (207).
In addition, immunolocalization studies on virus-infected cells showed that most of the viroporins were localized on various intracellular organelles (e.g., Golgi, endoplasmic reticulum), while fewer are detected on the plasma membrane (Table 1). The names, amino acid sizes, roles in the viral cycle, and classification of the members of the viroporin family that have been proposed as a result of the studies are reported in Table 1. This article discusses the functions of currently reported viroporins in the viral life cycle and their regulation of host cell responses, emphasizing their potential as antiviral targets.
2 The Role of Viroporins in the Viral Life Cycle
Since viruses are obligate intracellular pathogens, they must depend on host cells for reproduction and metabolism. The life cycle of viruses varies greatly depending on the type and class of virus, but they follow the same basic stages of viral replication, i.e., adsorption, entry, uncoating, replication, assembly, and release. Although viroporins are involved in different stages of the viral life cycle (Table 1 and Figure 2), most viroporins are mainly involved in the later steps of the viral life cycle, such as assembly and release, as far as the current study has found. Viruses cannot complete proper assembly and release when function of viroporins is disrupted.
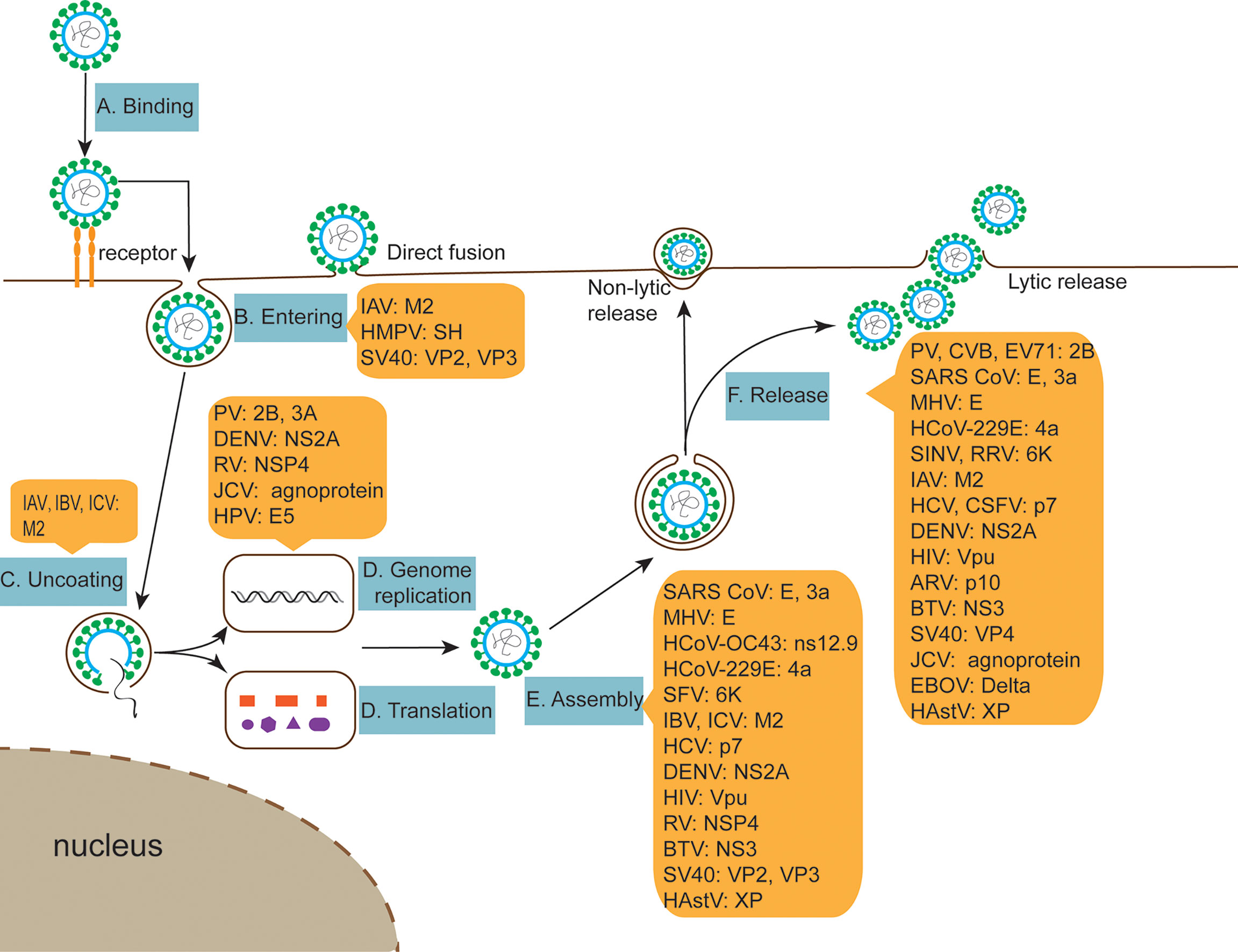
Figure 2 The role of viroporins in the viral life cycle. (A, B) Viroporins facilitate viral penetration of host plasma membrane into cells. (C) Viroporins trigger conformational changes in the virus, releasing the genome. (D) Viroporins-mediated viral replication. (E) Viroporins facilitate the assembly of new viral nucleic acids with protein capsids. (F) Viroporins promote virus release from host cells by budding or lysis.
2.1 Viral Entry and Uncoating
For infection to occur, the virus must first bind to and penetrate the host plasma membrane to deliver genetic material to the cytoplasm for replication. Enveloped and non-enveloped viruses employ different strategies to achieve the same goal. Viroporins, which are a structural component in enveloped viruses, actively facilitate the viral entry process, such as the influenza virus M2 protein. The influenza virus binds to the cell surface and is internalized into endosomes, and its encoded viroporin M2 is located in the viral lipid bilayer and is critical for infection. M2 acts as a proton-conducting channel in the viral envelope, supporting acidification of the viral interior in the endosome (208, 209). This process drives a series of conformational changes in the viral hemagglutinin (HA) protein, leading to the fusion of the viral envelope with the endosomal membrane and the delivery of nucleoprotein complexes into the cytoplasm (210). Acidification of M2 is also thought to promote the structural rearrangement of viral particles, which is required for the efficient uncoating of viral RNAs in the cytoplasm (45). Relevant studies have shown that the HMPV SH protein also has viroporin activity, which can regulate the fusion protein function during virus infection and may play a role in HMPV entering cells (67).
Non-envelope virions lack viroporins as structural components, and the involvement of such proteins in the process of virus entry into cells has been less well reported. A study found that antiretroviral (ARV) p10 is shown to play a key role in its viral fusion process, and the expression of p10 induces extensive cell-cell fusion in transfected cells. Thus, p10 is not only the first non-enveloped viral protein capable of promoting fusion from within, but also the first non-structural viral protein capable of inducing cell-cell fusion (75). Non-enveloped simian virus 40 (SV40) encodes three proteins with viroporin activity, VP2, VP3, and VP4, which play an important role in the life cycle of SV40. During infection, SV40 binds to the cell surface and is endocytosed into caveolin-coated vesicles (211) and subsequently translocated to the ER, where capsid reorganization and entry of viral particles into the cytoplasm occurs. VP2 and VP3 can integrate into the ER membrane and play an important role in the uncoating stage of the viral genome (78, 79). In addition to triggering viral particle infection, several viroporins encoded by influenza virus and SV40 play other roles in the viral life cycle, as will be discussed later.
2.2 Viral Replication and Assembly
After the release of the viral genome, the virus controls cellular proteins and organelles to achieve replication. Studies have shown that rearranged membranes are utilized during viral replication (104), and picornaviruses induce rearrangement of the host cell inner membrane to create structures that serve as functional scaffolds for genome replication (212). One of the most striking morphological changes that can be observed in enterovirus-infected cells is the massive accumulation of small ER and Golgi membrane vesicles in the cytoplasm (213). These vesicles are the site where viral RNA replication occurs. The 2BC protein has been implicated in the accumulation of these vesicles, where mutations that interfere with the pore formation capacity of 2B lead to defects in the early stages of viral RNA replication (213). Suhy et al. found that the poliovirus (PV) 2B protein alters cell membrane permeability and together with the 3A protein induces significant rearrangement of the intracellular membrane (104). PV 2B and 3A viroporins can also inhibit the cellular protein secretion pathway by disassembling the Golgi complex or blocking ER-Golgi transport, leading to the accumulation of membrane vesicles in the cytoplasm (101, 214). In addition, polioviruses harboring mutations in the 3A protein result in a marked reduction in positive-strand RNA synthesis (8). Picornavirus 2B protein also facilitates viral release by increasing the permeability of the plasma membrane, which will be discussed in the next section.
Throughout the replication cycle of coronaviruses, viruses use significant rearrangements of the host membrane for replication, protein expression, assembly, and release. The coronavirus E protein may also promote membrane rearrangement. When expressed alone in BHK-21 cells, mouse hepatitis virus (MHV) E can drive the intracellular formation of ERGIC-derived electron-dense membranes (19). The higher oligomers of infectious bronchitis virus (IBV) E are required for the production of virus-like particles (VLPs), suggesting that this form of protein is involved in the assembly of viral particles (215). Studies have shown that the MHV E protein is palmitoylated after translation, which contributes to the assembly of virions (216). The M, E, and N structural proteins of SARS-CoV are required for efficient assembly, transport, and release of virus-like particles (23). Recombinant CoVs (rCoVs) lacking the E gene (ΔE) exhibit abnormal morphology (217), suggesting that the E protein is involved in the assembly process. The function of the E protein is not to coordinate viral assembly but to induce bending of the viral envelope membrane so that the CoV particles acquire their characteristic spherical shape and morphology. Another important role of the coronavirus E protein is to regulate the pH within the lumen of intracellular organelles (e.g., the Golgi apparatus). For example, expression of the E protein of infectious bronchitis virus increases the pH within the organelle, induces neutralization of the Golgi pH, which results in a protective effect, allows isolation of the IBV spike-in protein from protein hydrolysis, and promotes viral assembly and release (32). Furthermore, the colocalization of ORF 3a with the M and E proteins essential for viral assembly suggests that ORF3a is important in the assembly or budding of SARS-CoV (25).
The rotavirus (RV) NSP4 protein also plays an important role in the viral assembly process. NSP4 manipulates the autophagic membrane trafficking process to take viral protein-containing membranes and transport them to viral replication sites for infectious particle assembly (191). The increase in cytoplasmic Ca2+ concentration mediated by NSP4 viroporin activates specific Ca2+-inducible signaling pathways to initiate autophagy, which allows the transport of the NSP4 and VP7 proteins to the virion for subsequent viral assembly (192). NSP4 binds to immature particles in the virion and triggers particle budding through these membranes to facilitate the assembly of capsid proteins on the particles to form mature infectious viral particles. Lopez et al. showed that small interfering RNAs that inhibit NSP4 expression in rotavirus-infected cells affect the distribution of other viral proteins, mRNA synthesis, and virion formation for viral RNA replication, suggesting a previously unrecognized function for NSP4 in RV replication (72).
The M2 protein is required for influenza virus assembly and budding. When viral budding begins, M2 is located at the edge of the budozone and induces a negative membrane curvature (218), thereby stabilizing the HA and M-induced positive membrane curvature in the budozone. Experimental results suggest that targeting M2 away from the apical plasma membrane may disrupt influenza virus assembly, budding, and replication (219). Phosphorylation of influenza virus CM2 promotes efficient virus replication (50). Deletion of CM2 results in impaired packaging and uncoating of virus-like particles (VLPs) and recombinant influenza viruses (49). The cytoplasmic structural domain of the BM2 protein performs the function of M1 binding to the viral ribonucleoprotein complex alone at the viral particle budding site (44, 220) and plays an important role in viral assembly. IAV M2 ubiquitination plays an important role in the production of infectious viruses by coordinating the efficient packaging of the viral genome into viral particles and the timing of virus-induced cell death (221). In addition, HCV p7 protein and HIV Vpu protein have also been shown to be involved in their viral assembly and release processes (52, 55, 62–64). The positively charged residue R84 in the dengue virus NS2A protein is essential for both viral RNA synthesis and intracellular viral particle assembly and maturation (60). Lulla et al. explored the role of the human astrovirus X protein in the viral life cycle by knocking it out and showed that the X protein may play a role in viral particle formation and viral release (89).
2.3 Viral Release
After assembly into infectious viral particles, viruses are released from host cells by budding (e.g., IAV, coronaviruses) or lysis (e.g., picornavirus, SV40). Alterations in plasma membrane permeability are important for cell lysis and the release of viral progeny (203). Enteroviruses are non-enveloped viruses that require the lysis of host cells to release newly formed virions. The expression of Enterovirus 2B protein disrupts intracellular Ca2+ homeostasis by progressively enhancing membrane permeability, leading to increased plasma membrane permeability and ultimately to membrane damage and release of the virus (6). For other viruses that are non-enveloped or are enveloped only by a protein coat, viral release often involves cell membrane perforation. For example, SV40 VP4 facilitates virus release by forming pores of approximately 3 nm in diameter in the host cell membrane to penetrate the membrane (82).
For enveloped viruses, budding and division are typically used to release virus from infected cells, and the mechanisms involved in the release process vary from virus to virus. During influenza virus release, the M2 protein is located at the neck of the budding virion, and the amphiphilic helix inserts into the membrane, inducing bending of the positive membrane, and finally pinching off the budding virion (218, 222). Mutations or deletions in the M2 cytoplasmic tail structural domain (CTD) significantly impair the assembly of viral proteins and genomic fragments into viral particles and viral particle release (218, 223, 224). A recent study found that the reduced hydrophobicity of the 91-94 motif of the IAV M2 protein significantly affected the budding ability of the M2 protein and compromised the bilayer membrane integrity of mutant viruses. It was suggested that the hydrophobic residues of the intracellular domain of the M2 protein play an important role in the release and membrane integrity of influenza virus (225). Several studies have shown that the coronavirus E protein is important for viral particle release. IBV E proteins have been shown to function in the secretory pathway, altering the luminal environment and rearranging secretory organelles, ultimately facilitating the efficient transport of viral particles (29–32). The E protein targets the Golgi apparatus and directs the release of virus-like particles (28). Mutations introduced into the HD of MHV or IBV affect the release of infectious particles from the cell (20, 29, 31). These findings suggest that IBV and MHV E viroporins may mediate viral release through their ion channel activity. In addition, Lu et al. found a significant reduction in viral release in cells transfected with ORF3a-specific siRNA (24), suggesting that the 3a protein may act as an ion channel promoting the viral release, but the mechanism by which the 3a protein affects virus release requires further study.
Unlike other viruses that mature in the lumen of the ER, RV is not released via the classical cytosolic pathway. Instead, recent data suggest that this virus may be transported from the ER directly through vesicles to the apical plasma membrane of polarized epithelial cells without passing through the Golgi complex. RV NSP4 interacts with immature particles to trigger outgrowth and synthesis into ER transmembrane proteins, and this rotavirus “budding” maturation process occurs through autophagy-hijacked COPII vesicle membranes (226). BTV NS3 protein of the same family as rotaviruses may function as the membrane protein of enveloped viruses, responsible for intracellular trafficking and budding of viral particles. Therefore, the NS3 protein is considered to act as a bridge between mature virions and cellular proteins during viral shedding (227). Recent studies have found that filovirus delta peptides function as viroporin to enhance the release of viral particles across host cell membranes (88). In addition, other viroporins such as alphavirus 6K protein, HCV p7 protein, HIV Vpu protein also play an important role in the virus release process (36, 55, 228) (Table 1 and Figure 2).
3 Regulation of Host Cell Responses by Viroporins
Viroporins encoded by viruses can affect a variety of host cell responses (Table 2), such as altering cell membrane permeability, activation of the NLRP3 inflammasome (Figure 3), regulating apoptosis (Figure 4) and autophagy (Figure 5), and affecting host immune responses (Figure 6). Viroporins promote their replication and proliferation by regulating these responses.
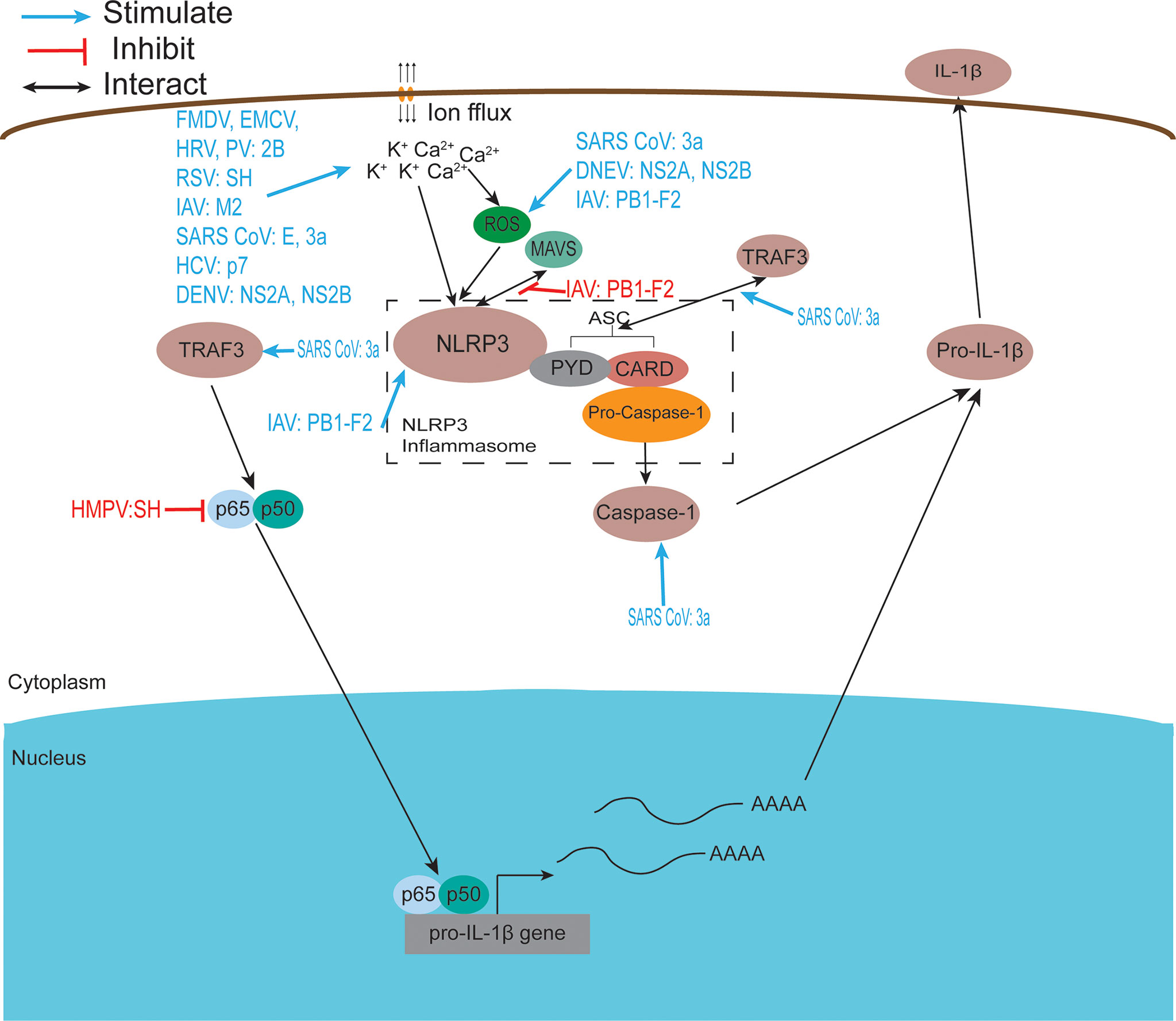
Figure 3 Viroporins regulate inflammasome activation. The NACHT, LRR, and PYD domain-containing protein 3 (NLRP3) inflammasome is an oligomeric complex composed of the NOD-like receptor NLRP3, the adaptor protein ASC, and Caspase-1 (229). Mitochondrial damage, protein aggregation, and abnormal ion concentrations caused by viral infection can activate the NLRP3 inflammasome leading to the secretion of IL-1β and IL-18. Most viroporins activate the NLRP3 inflammasome by disturbing intracellular ion concentrations. Some viroporins can activate NLRP3 through mitochondrial damage and increased ROS production.
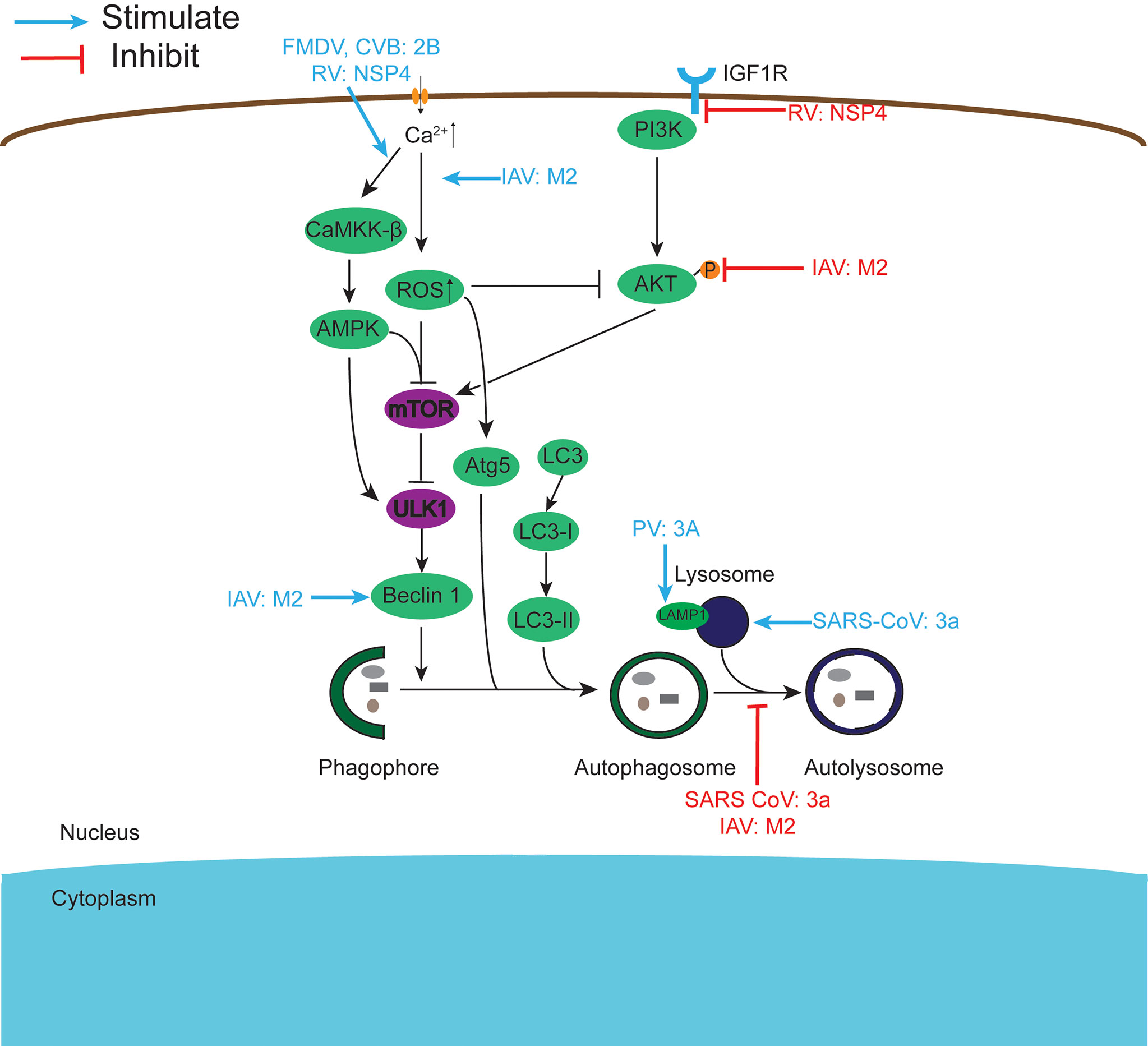
Figure 4 Viroporins regulate autophagy. Autophagy can be regulated by the PI3K-AKT-mTOR signaling pathway and the AMPK-TSC1/2-mTOR signaling pathway. Viroporins can regulate autophagy by regulating upstream signaling cascades, interfering with the formation of autophagosomes to activate, inhibit autophagy and fuse with lysosomes, and interact with key molecules of autophagy.
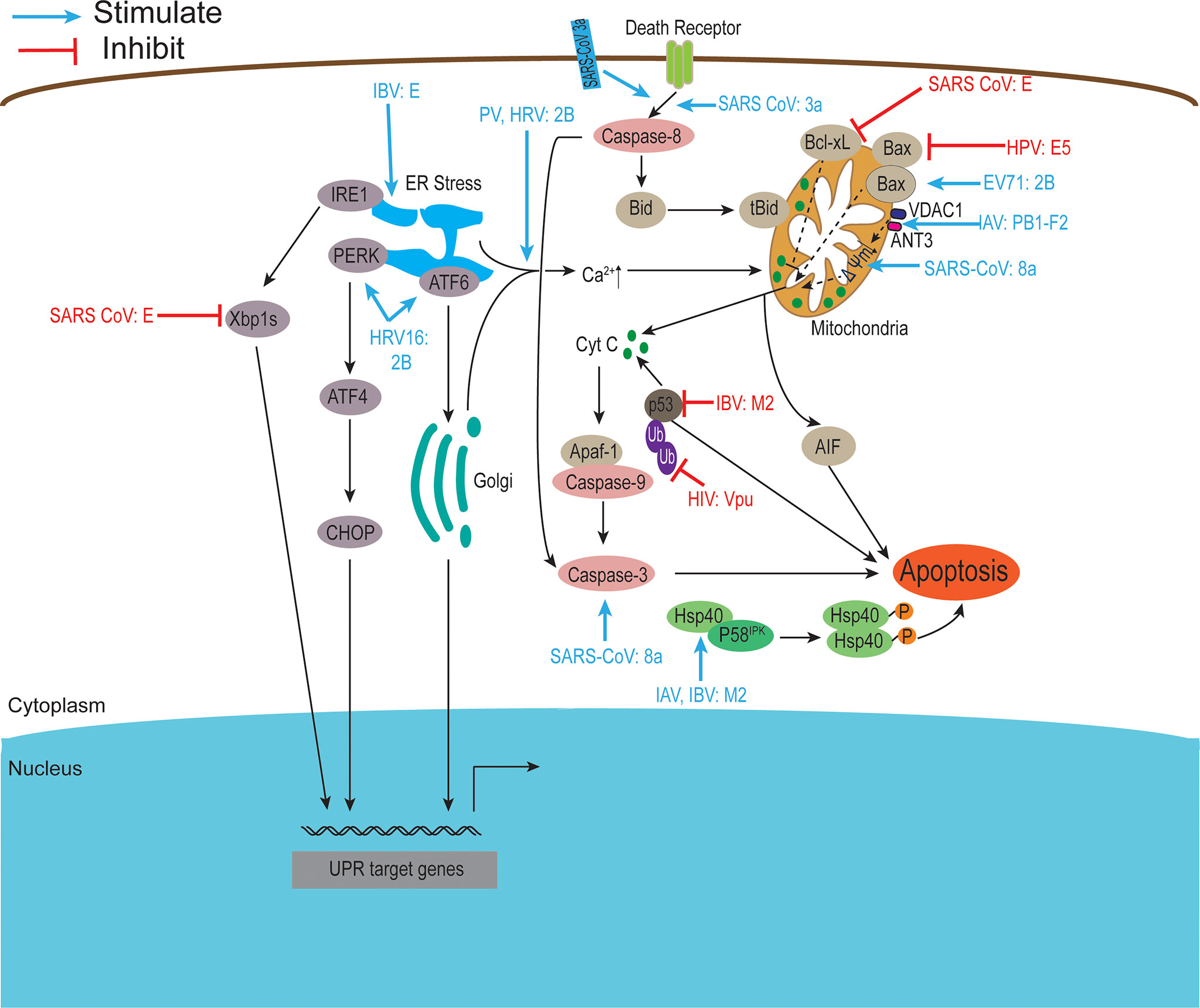
Figure 5 Viroporins regulate apoptosis. Apoptosis can be activated through two major signaling pathways, the death receptor-mediated pathway, and the mitochondrial pathway. Viroporins can induce apoptosis by changing calcium ion concentration, reducing mitochondrial membrane potential, recruiting apoptosis-related factors, and activating endoplasmic reticulum stress. Some viroporins can also inhibit apoptosis. Ub, Ubiquitin. Figure adapted from (230).
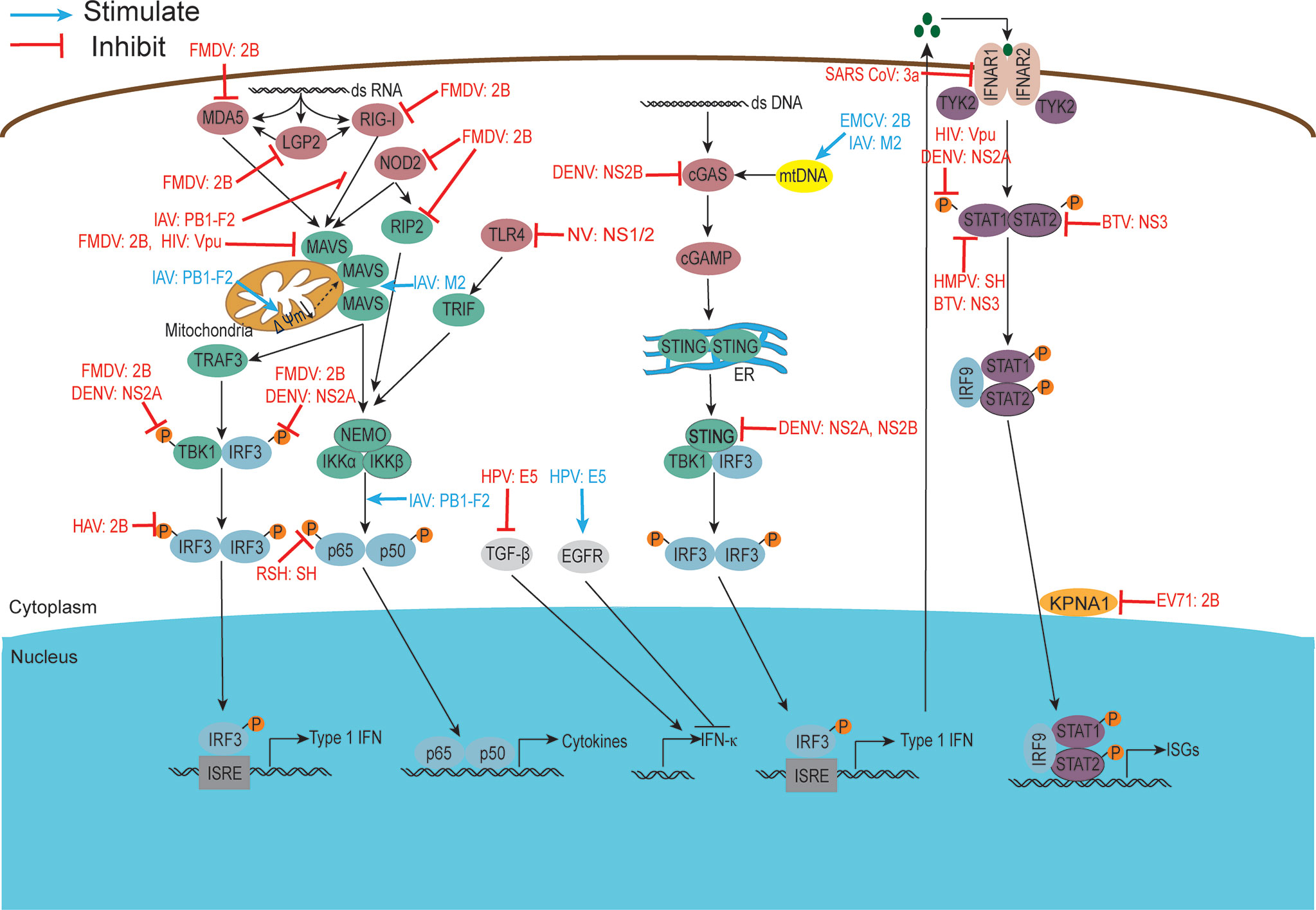
Figure 6 Viroporins regulate host immune responses. viroporins modulate host cell immune responses by interfering with PRRs recognition, interfering with bridging molecules, kinases, and downstream effectors in the innate immune signaling pathway, and interfering with IFN-mediated signaling. Figure adapted from (231).
3.1 Alteration of Host Cell Membrane Permeability and Disruption of Intracellular Ion Concentration Balance
Induction of changes in cell membrane permeability is a key function of viroporins (57, 83, 206, 207, 232). In addition to this, many virus-encoded viroporins can allow different ions (e.g., H+ and K+) across the cell membrane, affecting the concentration of ions inside and outside the cell as well as the membrane permeability to these ions. After infection of cells by picornavirus, increased membrane permeability leads to disruption of the extracellular ion gradient and thus disrupts the host intracellular ion concentration balance, in which 2B viroporin plays an irreplaceable role. 2B proteins such as foot-and-mouth disease virus (FMDV), PV, and Coxsackievirus B (CVB) can increase intracellular Ca2+ ion concentration, but by different mechanisms. The expression of CVB3 and PV 2B proteins leads to a decrease in Ca2+ concentration in the ER and Golgi complex as well as in Ca2+ uptake by mitochondria. At the same time, an increased influx of extracellular Ca2+ leads to increased cytoplasmic Ca2+ levels (9, 12). Similarly, the expression of human rhinovirus (HRV) 2B protein showed a decrease in Ca2+ concentration in the ER and Golgi apparatus, while encephalomyocarditis virus (EMCV) 2B protein only significantly decreased Ca2+ concentration in the ER (12). In contrast, other studies have shown that the expression of hepatitis A virus (HAV) and FMDV 2B proteins increased cytoplasmic Ca2+ levels but did not alter the levels of Ca2+ stored in organelles (3, 12). Specifically, the experimental results of Xie et al. suggest that the enterovirus (EV) 71 2B protein may mediate Cl- dependent currents in African Xenopus oocytes (14). The reason for this 2B-induced ion concentration variation among different small ribonucleic acid viruses may stem from differences in the proteins themselves and experimental settings and systems, and no studies are showing the permeability of other 2B proteins to Cl-. Therefore, further analysis of 2B protein-mediated ion permeability under viral infection and separate expression is required. Also significant for Ca2+ homeostasis is the RV NSP4 protein, whose effect on Ca2+ levels, along with that of the enterovirus 2B protein, has been characterized using fluorescent Ca2+ imaging (233). NSP4 is an ER transmembrane glycoprotein that activates ER calcium sensor matrix interaction molecule 1 (STIM1), leading to the plasma membrane (PM) Ca2+ inward flow (190). NSP4 also elevates cellular Ca2+ levels through a phospholipase C (PLC)-independent pathway, suggesting that it disrupts Ca2+ homeostasis and releases Ca2+ channels (189).
Experimental results suggest that viroporin E protein may play an important role in regulating ion homeostasis and the microenvironment of host cells (234, 235). Westerbeck et al. found that IBV E protein transient overexpression alters Golgi pH, providing the first evidence of a coronavirus-mediated alteration of the secretory pathway tubular microenvironment to facilitate infectious virus production (32). A recent study showed that the intracellular expression of SARS-CoV-2 E protein increases intra-Golgi PH (21). IAV M2 also downregulates the expression and function of two host ion channels, the amiloride-sensitive epithelial sodium channel (ENaC) (236) and the cystic fibrosis transmembrane conductance regulator (CFTR) (237) chloride channel, which promotes influenza pathogenesis.
In addition to altering the permeability of the invading cells themselves, cell permeabilization by PV 2B viroporin triggers bystander permeabilization in neighboring cells through a mechanism involving gap junctions, and proteins from MHV E, sindbis virus (SINV) 6K, and HCV NS4A are also able to penetrate neighboring cells to varying degrees (238). Some viroporins (e.g., PV 2B (5)and 3A (103, 104), IBV E (29)) are also able to target intracellular compartments that affect pH or Ca2+ homeostasis, thereby blocking protein secretion.
3.2 Regulation of the Activation of the Inflammasome
The inflammasome is a molecular platform activated upon cellular infection or stress, triggering the maturation of pro-inflammatory cytokines such as IL-1β, and participating in innate immune defense. The NACHT, LRR, and PYD domain-containing protein 3 (NLRP3) inflammasome is an oligomeric complex composed of the NOD-like receptor NLRP3, the adaptor protein ASC, and Caspase-1 (229). This complex is critical in the host antiviral immune response as it promotes IL-1β and IL-18 secretion and induces pyroptosis (239). Increasing evidence suggests that the effect of viroporins on membrane permeability and the subsequent disruption of ion homeostasis in cellular compartments may be the activation signal required to activate the NLRP3 inflammasome and produce IL-1β and IL-18 (Figure 3). This highlights the important role of ion concentration imbalance caused by the ion channel activity of viroporin in activating the NLRP3 inflammasome. Viroporins such as FMDV and HRV 2B protein (93, 114), respiratory syncytial virus (RSV) SH protein (66), IAV M2 protein (140), EMCV 2B protein (109), SARS-CoV E protein (122), SARS 3a protein (126), and HCV p7 protein (168) was reported to activate the NLRP3 inflammasome by disturbing intracellular ion concentrations (K+, Ca2+, Na+). The 2B protein from a variety of picornaviruses (including EMCV, PV, and EV 71), the DNEV NS2A protein and the SARS-Cov E protein were shown to induce NLRP3 cytoplasmic relocalization and inflammasome activation in an intracellular Ca2+-mediated manner (58, 109, 122). Other experimental results have shown that influenza virus M2 proton channel activates the NLRP3 inflammasome pathway by regulating intracellular K+, Na+, and Ca2+ concentrations (140).
In addition to disrupting intracellular ion concentrations, viroporins activate the inflammasome in other ways, resulting in an inflammatory response. DNEV NS2A and NS2B protein expression increase apoptosis-associated speck-like protein-containing caspase recruitment domain (ASC) oligomerization and secretion of IL-1β through caspase 1 activation (58). SARS 3a activates NLRP3 inflammasome by inducing disruption of intracellular ion concentration, mitochondrial damage, and TNF receptor-associated factor 3 (TRAF3)-mediated ubiquitination of ASC (126, 128, 131). Paradoxically, Siu et al. found that the oligomerization of ORF 3a was dispensable for the activation of NF-κB or NLRP3 inflammasome, indicating how the viral ion channel protein works in an ion channel-independent manner (128). In contrast, results from another related study showed that SARS-CoV 3a activates the NLRP3 inflammasome in an ion channel-dependent manner (126). This discrepancy may require further studies in the same experimental system to resolve. In addition to this, 3a upregulates the production of pro-inflammatory cytokines and chemokines by activating C-Jun N-terminal kinase (JNK) and the transcription factor nuclear factor-kappa B (NF-kappaB) (129). In contrast, human metapneumovirus (hMPV) SH has been reported to inhibit NF-κB transcriptional activity in airway epithelial cells (184). Other studies have shown that RSV SH viroporin accumulates in the Golgi apparatus within lipid raft structures and may form ion channels that trigger NLRP3 translocation from the cytoplasm to the Golgi apparatus and activate NLRP3 inflammasome (66). HCV-p7 induces the molecular mechanism of SOCS3 through STAT3 and ERK activation and has been shown to inhibit p7 in response to TNF-α pro-inflammatory response (165).
The IAV PB1-F2 protein has a special performance in regulating inflammatory response, exhibiting two distinct roles in promoting and inhibiting NLRP3 inflammasome. On the one hand, PB1-F2 can be integrated into the phagocytic lysosomal compartment triggering NLRP3 inflammasome activation, inducing the secretion of the IL-1β, which leads to severe pathophysiology (153). Several experimental results have shown that PB1-F2 activates NLRP3 inflammasomes and NLRP3-dependent cell recruitment (154), inducing lung inflammation (152). On the other hand, the PB1-F2 protein inhibits the activation of the RLRP3 inflammasome under certain conditions (155). The highly pathogenic H7N9 PB1-F2 protein selectively inhibits RNA-induced NLRP3 inflammasome activation by inhibiting MAVS-NLRP3 interactions in infected cells, but intracellular PB1-F2 does not affect extracellular PB1-F2-induced NLRP3 inflammasome maturation (151). H5N1 and H3N2 PB1-F2 expression reduced IL-1β levels secreted by infected macrophages (150). The intrinsic link between the activation and inhibition of the RLRP3 inflammasome by viroporin PB1-F2 is unclear, but ultimately it is for the better survival and reproduction of the virus in the host cell.
3.3 Regulation of Autophagy
Autophagy is a physiological catabolic process in which cells degrade internalized pathogens or worn organelles by forming membrane-enclosed autophagosomes. Although viruses must escape autophagic destruction, some viruses can also disrupt autophagy for their benefit. Studies have shown that viroporins can manipulate autophagy (Figure 4) and thus promote viral replication, and among the viroporins that have a strong effect on cellular autophagy are mainly the small ribonucleic acid virus 2B protein, coronavirus 3a protein, influenza virus M2 protein, and rotavirus NSP4 protein. FMDV and CVB3 2B proteins induce robust autophagy in host cells (3, 108). Simultaneous expression of PV 2BC and 3A proteins induces the co-localization of LC3 and LAMP1, which induces autophagy to promote viral replication (106). According to reports, NSP4 can induce autophagy by activating the Ca2+/calmodulin-dependent kinase kinase-β (CaMKK-β) signaling pathway or targeting IGF1R (191, 192), and further studies revealed that COPII vesicle transport plays an important role in this (226). Furthermore, early in viral infection, the miRNA encoded by the NSP4 gene targets IGF1R, blocking the PI3K/Akt pathway and triggering autophagy, but ultimately inhibiting autophagic maturation (193).
SARS-CoV 3a proteins can both induce autophagy by inserting into the lysosomal membrane, causing lysosomal damage and dysfunction (Yuan 131), and inhibit autophagosome degradation and thus cellular autophagy by preventing the assembly of the SNARE complex required for HOPS complex-mediated autolysosome formation (132). By comparison, it is found that 3a needs its viroporin activity to promote autophagy, while whether preventing autophagy also needs to be further studied. Similarly, the influenza virus M2 protein also exhibits both autophagy-promoting and autophagy-inhibiting aspects. IAV M2 inhibits autophagy by directly interacting with the autophagy proteins LC3 or Beclin-1, blocking the fusion of autophagosomes and lysosomes (141–143). In contrast, Wang et al. showed that M2 triggers extracellular Ca2+ influx-dependent ROS production, which subsequently leads to activation of ATG5 and inhibition of AKT/PKB and MTOR activities via the class I phosphatidylinositol 3-kinase (PI3K)-AKT-MTOR signaling pathway, ultimately triggering activation of autophagy (145). The inhibition of autophagy by 3a and M2 proteins may be a strategy employed by viruses during the pre-infection phase to survive in the host cell while releasing daughter viral particles from the cell by promoting autophagy after the maturation of their viral particles. At present, further studies on the regulation of host cell autophagy by viroporins other than the above-mentioned proteins are underway.
3.4 Regulation of Apoptosis
Apoptosis is a genetically programmed mechanism used by the host to eliminate damaged or unwanted cells by activating the caspases cascade and can be activated through two major signaling pathways: the extrinsic or death receptor-mediated pathway and the intrinsic or mitochondrial pathway. In addition, excessive Ca2+ loading in mitochondria can induce apoptosis by causing the opening of permeability transition pores, permeability swelling of mitochondria, and rupture of the outer mitochondrial membrane, which leads to the release of pro-apoptotic factors such as cytochrome C (230). Some virus-encoded viroporins can promote their propagation by affecting host cell apoptosis (Figure 5).
Accumulating evidence suggests that picornavirus 2B proteins can regulate host cell apoptotic responses in multiple ways. HRV16 2B protein activates PERK and ATF6 but not IRE1 to trigger ER stress (15). EV 71 2B protein induces apoptosis by recruiting and directly interacting with the pro-apoptotic protein Bax or by regulating the redistribution and activation of Bax (13, 111). Furthermore, PV 2B protein induces permeabilization of the plasma membrane, triggering apoptosis through the mitochondrial pathway (102). CVB 2B proteins play a major role in inhibiting apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis (9). Other viroporins also regulate host cell apoptosis in different ways. Studies have shown that overexpression of the E protein of the family Coronaviridae can induce apoptosis (124, 137) and the SARS-CoV E protein can interact with Bcl-xL, making the anti-apoptotic protein Bcl-xL unable to function normally (124). SARS-CoV E activates ER stress and induces pro-inflammatory cytokines to promote apoptosis during late infection (137). In contrast, DeDiego et al. used a microarray-based approach to show that SARS-CoV E may also have anti-apoptotic effects during infection (125). The 3a and 8a proteins of SARS-CoV also induce apoptosis (27, 134). The 8a protein is located in mitochondria, where it can perturb mitochondrial membrane potential and induce apoptosis through a caspase-3-dependent pathway (27).
In addition, the influenza virus M2 protein induces host cell apoptosis by blocking autophagosome maturation (148). It has been reported that Hsp40 acts as a regulator of PKR signaling by interacting with the PKR cytostatic p58IPK. Guan et al. found that M2 protein interacts with Hsp40 both in vitro and in vivo, speculating that it may enhance PKR autophosphorylation by forming a stable complex with Hsp40 and P58IPK, thereby inducing cell death (149). Another viroporin PB1-F2 encoded by the influenza virus interacts with two mitochondrial proteins, adenine nucleotide transporter (ANT3) and voltage-dependent anion channel 1 (VDAC1), which are present in the inner and outer mitochondrial membranes, respectively, leading to dissipation of mitochondrial membrane potential, inducing cell death (156). In addition, the PB1-F2 protein can also translocate to the inner mitochondrial membrane space via the TOMM40 channel, resulting in a decrease in mitochondrial membrane potential (MMP) and induction of apoptosis (155). In contrast, the BM2 protein interacts with p53 and inhibits its transcriptional and apoptotic activities (164). Some other viroporins (e.g., human respiratory syncytial virus SH protein, HPV E5 protein) have also been reported to have anti-apoptotic activity (174, 176, 182, 200).
3.5 Regulation of the Host Cellular Immune Response
The innate immune response is the first line of defense against viral infection, and to break through this line of defense and replicate effectively in vivo, many virus-encoded viroporins disrupt host immune defenses in a variety of ways (Figure 6). The regulation of host cell immune response by viroporins is mainly manifested as antagonism.
3.5.1 Identification of Interfering PRRs
Recognition of pathogens is mainly mediated by pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), nucleotide oligomerization domain-like receptors (NLRs), cyclic GMP-AMP synthase (cGAS), and retinoic acid-inducible gene-1-like receptors (RLRs), which recognize pathogenic microbial infections and form the corresponding signal transduction to generate an immune response. RIG-I-like receptors (RLRs), including RIG-I, melanoma differentiation-associated protein (MDA)-5, and LGP2, are a series of cytoplasmic RNA unwinds that detect multiple viral RNAs accumulated during viral infection or replication. Activation of RIG-I is responsible for the induction of type I interferons (IFNs) and the expression of many cytokines and chemokines (240). To survive and multiply in host cells, viroporins can interfere with their viral recognition by PRRs.
It was shown that FMDV 2B protein interacts with and inhibits the expression of RIG-I and MDA5 to antagonize their mediated antiviral effects (97). In 2019, Liu H et al. found that FMDV infection triggered NOD2 transcription and reduced NOD2 protein expression. Further experimental results showed that 2B protein was one of the reasons for this phenomenon (95). Similarly, the IAV PB1-F2 protein exhibited type I IFN antagonism by interfering with the RIG-I/MAVS complex (160). The work of Lateef et al. showed that Toll-like receptors were targets of norovirus (NV) NS1/2, which decreased the expression of TLR-4, -7, -8, and -9 and increased the expression of several pro-inflammatory cytokines/chemokines (186). To avoid detection of mitochondrial DNA during infection, the dengue virus NS2B protein targets the DNA sensor cyclic GMP-AMP synthase (cGAS) for lysosomal degradation and results in the inhibition of type I interferon production in infected cells (172). Whether other viroporins are also involved in regulating host cell immune responses by interfering with PRRs recognition is currently under further investigation.
3.5.2 Interference With Adaptor Molecules, Kinases, and Downstream Effectors in Innate Immune Signaling Pathways
Virus-encoded viroporins have developed multiple mechanisms to disrupt the recruitment process or degrade linker molecules, as well as related kinases critical for signal transduction, to block subsequent signal transduction and antagonize the host antiviral response. It has been reported that phosphorylation of RIP2 was identified as a marker for activation of the NOD2-mediated NF-κB pathway (241). In 2021, Liu H et al. found that FMDV infection triggered the transcription of RIP2, and its 2B protein could reduce the expression of RIP2 protein (95). Both FMDV 2B and DENV NS2A inhibit the phosphorylation of TBK1 and IRF3 in the RLR signaling pathway (96, 170). Furthermore, DENV NS2A and NS2B proteins inhibit the production of type I IFN by cleaving STING (171, 173). According to literature reports, influenza virus-encoded viroporin PB1-F2 is a particularly potent inhibitor of antiviral signaling, inhibiting the expression of mitochondrial antiviral signaling protein (MAVS) and it is signaling through various means. PB1-F2 can degrade MAVS by inducing mitophagy and lead to the inhibition of type I interferon production (162, 163). Cheung et al. found that PB1-F2 protein interacts with TRIM31-MAVS by forming protein aggregates on mitochondria, thereby preventing K63-polyubiquitination and MAVS aggregation and promoting MAVS degradation, ultimately inhibiting host antiviral defense (161). In addition to this, an increasing number of studies have shown that the reduction of mitochondrial inner membrane potential is also an important way in which PB1-F2 protein inhibits MAVS signaling (155, 159). Several other studies have reported that HIV-1 Vpu effectively inhibits NF-κB-induced antiviral immune responses at the transcriptional level (180). HAV 2B is able to interfere with IRF-3 phosphorylation and inhibit IFNβ gene transcription (115).
In addition, some other viroporins (e.g., BTV NS3, RSV SH) resist the host defense response by interacting with BRAF or inhibiting p65 phosphorylation (183, 194). In particular, the influenza virus M2 protein antagonizes autophagy and interacts with MAVS, thereby increasing MAVS aggregation, positively regulating MAVS-mediated antiviral innate immune responses (145), however, overactivation of MAVS signaling could lead to detrimental levels of inflammation or other immunopathological consequences that are harmful to the host.
3.5.3 Interference With IFN-Mediated Signaling
Interferons play a crucial role in regulating and activating the host innate immune response to viral infection and limiting viral replication and spread and are also important targets for viroporins. Experimental results have shown that HMPV SH proteins downregulate type I IFN pathway signaling by affecting STAT1 expression and phosphorylation (185). Similarly, DENV NS2A, NS4A, and NS4B proteins complex together to block STAT1 phosphorylation and inhibit ISG production (169). BTV NS3 and NS4 synergistically antagonize type I interferon signaling by targeting STAT1 (195). In addition, BTV NS3 blocks IFN signaling by binding STAT2 to induce its degradation through an autophagy-dependent mechanism (196). Karyopherins (KPNAs) are cytoplasmic proteins essential for nuclear transport of p-STAT1/2 and translocation of ISGF complexes, and enterovirus A71 2B inhibits interferon-activated JAK/STAT signaling by inducing Caspase-3-dependent KPNA1 degradation (112). Whether other viroporins can also antagonize the host antiviral response by blocking the JAK/STAT pathway is unknown and requires further experimental investigation.
3.5.4 Other Ways to Resist the Host Immune Response
Major histocompatibility complex (MHC) class 1 is also an important player in the antiviral response. Studies have shown that PV 3A proteins can impair MHC class I antigen presentation (105). CVB3, which encodes 2B and 2BC proteins in the same family as PV, upregulates the endocytosis of MHC class I by focusing endocytic vesicles on the Golgi complex and rapidly removes proteins from the cell surface. This may render CD8+ T cells unrecognizable to CVB3-infected cells and therefore inaccessible to many antiviral effector molecules, which is important for immune evasion by CVB3 (242). Similarly, PB1-F2 induces IFNβ through the NF-κB pathway independently of the AP-1 and IRF3 pathways, and overexpression of the MHC-I gene is involved in suppressing antiviral immunity (157, 158). The interaction between NV NS1/2 and the vesicle-associated membrane protein VAP-A suggests that calicivirus manipulate intracellular trafficking, thereby inhibiting or blocking key innate immune proteins (TLR, IFN, MHC, etc.) to the cell surface (187, 188).
Vpu downregulates bone stromal cell antigen 2 (BST-2) protein from the cell surface, which protects HIV-infected CD4+ T cells from antibody-mediated cell lysis in addition to promoting self-release (63, 64), it also protects HIV-infected CD4+ T cells from antibody-mediated cell lysis (177). It should be noted that mutants that impair Vpu ion channels do not affect the downregulation of BST-2, suggesting that the downregulation of BST-2 is independent of the ion channel activity of Vpu (243). In addition, Vpu promotes phagocytosis of infected CD4+ T cells by macrophages through downregulation of CD47 to facilitate viral dissemination and promote immune evasion (62).
Cytoplasmic mitochondrial DNA (mtDNA) activates cGAS-mediated antiviral immune responses, and the viroporin activity of influenza virus M2 or EMCV 2B proteins triggers mtDNA translocation into the cytoplasm in a MAVS-dependent manner. Although influenza virus-induced cytoplasmic mtDNA stimulates cGAS and DDX41-dependent innate immune responses, influenza virus nonstructural protein 1 (NS1) binds to mtDNA to evade STING-dependent antiviral immunity (110). A recent study demonstrated that Cyclophilin A (CypA) plays a role in promoting RIG-I-mediated antiviral immune responses by controlling the ubiquitination of RIG-I and mitochondrial antiviral signaling protein (MAVS) (244). FMDV 2B attenuates CypA during FMDV infection by interacting with CypA mediated antiviral effects during FMDV infection, and further studies have shown that this interaction is specific to FMDV 2B in picornavirus viroporins (94). In addition to this, some other viroporins evade the host antiviral response by some other means (Table 2).
4 Viroporins Interact With Other Host or Viral Proteins to Promote Their Own Proliferation
Viruses lack the necessary machinery for self-replication and therefore rely on host cell machinery for reproduction. Many viruses utilize the host cell replication machinery to establish infection through host-virus protein-protein interactions (PPIs) (245). Some viruses encode viroporins that interact with host proteins to ensure the successful completion of their life cycle (Table 3).
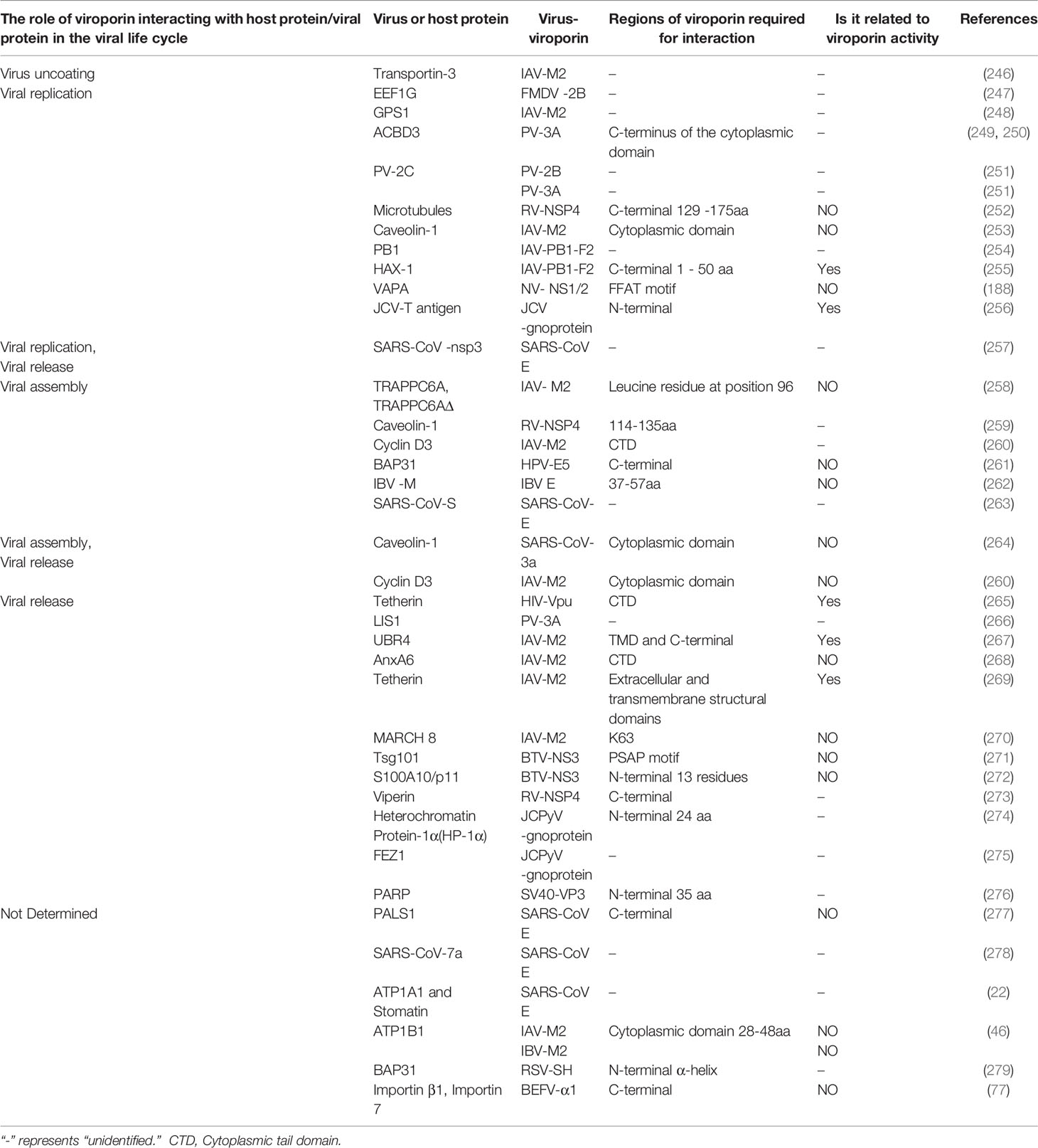
Table 3 The role of viroporins interacting with other host or viral proteins in the viral life cycle.
Current research data suggest that viroporins interact with host proteins and are mainly involved in regulating the later stages of the viral life cycle. Relevant studies have shown that viroporins such as Picornavirus 2B and 3A proteins, IAV M2 and PB1-F2 proteins, and coronavirus E protein can regulate the replication stage of the virus by interacting with different host proteins, of which influenza virus AM2 and PB1-F2 proteins are good examples. The M2 protein has been reported to interact with the G protein pathway repressor protein 1 (GPS1), which is involved in the transcription and replication of influenza virus genomic RNA by activating the NF-κB signaling pathway (248). Mazur et al. found that the PB1-F2 protein co-localizes and directly interacts with the viral PB1 polymerase protein, thereby determining the localization of the PB1 protein and enhancing viral polymerase activity, which ultimately leads to enhanced accumulation of viral RNA (254). Recent experimental results identified HCLS1-associated protein X1 (HAX-1) as a functional restriction factor of IAV polymerase that binds to PA subunits, and PB1-F2 protein promotes its efficient replication by binding to HAX-1 and relieving HAX-1-mediated restriction of avian viral polymerase PA (255). In addition, FMDV 2B and PV 3A proteins interact with eukaryotic translation elongation factor 1γ (eEF1G) and acyl-CoA-binding domain protein 3 (ACBD3), respectively, to promote self-replication. Further studies revealed that eEF1G may play a potential role in assisting protein 2B to produce virus-induced vesicles and induce cell lysis (247). ACBD3 is a Golgi-resident protein involved in maintaining Golgi structure and regulating intracellular trafficking between the ER and the Golgi (280). PV 3A interacts with it and counteracts the inhibitory effect of ACBD3 on viral replication (250). Several other studies have shown that 3A proteins can utilize ACBD3 to recruit PI4KIIIβ to viral replication sites to facilitate self-replication (249, 281). Furthermore, NV NS1/2 interacts with the host protein VAMP-associated protein A (VAPA) to promote self-replication (188).
After virus replication is completed and enters the assembly stage, viroporins such as influenza virus M2 protein and RV NSP4 protein play an important regulatory role by binding to host factors. Cyclin D3 is a key cell cycle regulator in the G0/G1 phase, and the M2 protein binds to it, inducing its redistribution from the nucleus to the cytoplasm and subsequent degradation by the proteasome, a process that facilitates the correct assembly of progeny virions (260). Caveolae are known to serve as a site for signal transduction, viral entry into cells, and viral assembly. Experimental results found that NSP4 interacts directly with caveolin-1 to facilitate the intracellular transport of NSP4 from the ER to the cell surface (259), and Sapin et al. suggested that this interaction may contribute to the final step in RV morphogenesis (282). Similarly, the coronavirus 3a protein plays an important role in assembly and release by interacting with the caveolin-1 protein (264). In addition, the HPV E5 protein can interact with BAP31 and can mediate post-transcriptional effects or promote viral particle assembly (261).
After the virus matures, its encoded viroporins can release the mature particles extracellularly by binding to host proteins. Currently, influenza virus M2 protein, BTV NS3 protein and JCV gnoprotein protein are more studied in this regard. It has been found that the influenza virus M2 protein binds to the ubiquitin-protein ligase E3 component N-recognition protein 4 (UBR4) or Tetherin (BST-2) protein and facilitates the release of the virus outside the cell by promoting apical translocation of viral proteins or by downregulating Tetherin through the proteasomal pathway (267, 269). BTV NS3 assists in the viral release by recruiting the ESCRT-I protein Tsg101 or interacting with the S100A10/p11 protein (271, 272). JCV gnoprotein promotes the release of progeny viruses by interacting with heterochromatin protein-1α (HP-1α) or fiber bundle and elongation protein zeta1 (FEZ1) (275). Furthermore, studies have reported that Vpu counteracts the viral release-limiting effect of tetherin by interacting with tetherin and downregulating tetherin in the plasma membrane (265). In contrast, some studies have found that host proteins inhibit viral release by binding to viroporins. The human protein annexin A6 (AnxA6) interacts with M2 and negatively regulates influenza virus infection by affecting viral budding (268). The membrane-associated RING-CH8 (MARCH 8) protein inhibits IAV release by redirecting the viral M2 protein from the plasma membrane to the lysosome for degradation (270). Similarly, the IFN-stimulating protein Viperin delays rotavirus release by inhibiting NSP4-induced intrinsic apoptosis (273). This negative regulation of viral release mediated by host proteins through interaction with viroporins may be a strategy for the host to combat viral infection, but the detailed molecular mechanisms need to be further investigated.
In addition to interacting with host proteins, viroporins have also been reported to bind to their other viral proteins to promote the completion of the viral life cycle. For example, both mammalian and yeast two-hybrid systems have shown that PV 3A multimerizes and interacts with 2B and 2C ATPases to promote viral replication (251). IBV E and M proteins interact through their cytoplasmic tails leading to the assembly of coronavirus-like particles (262). Recent studies have shown that the SARS-CoV E protein induces the retention of the S protein in the ERGIC by regulating the cellular secretory pathway, preventing the formation of syncytia, and promoting the assembly of SARS-CoV viral particles (263). The current growing family of viroporin and consequently the increasing number of host proteins interacting with them have been reported, and such interactions play an important role in regulating the viral life cycle and promoting viral proliferation, but the specific stages of the viral life cycle at which they act, and the mechanisms remain to be further investigated.
5 Conclusions
Viroporins are potential targets for antiviral therapy because of their significant impact on the viral life cycle and host and have become targets for inhibitory drug development and antiviral therapy. AM2, the first viroporin discovered, has a well-established biological role in viral pathogenesis and is a proven drug target (283). Amantadine is the first drug to inhibit influenza virus replication by inhibiting the ion channel activity of M2, which can mediate the conversion of hemagglutinin (HA) to its low pH conformation by interacting with the AM2 protein, thereby preventing proton conduction and inhibits viral entry (284). Amantadine has also been reported to have a targeted inhibitory effect on viroporins such as FMDV 2B, SARS-CoV E, HCV p7 (3, 54, 121), however, drug resistance limits its clinical application. The identification and application of tauroursodeoxycholic acid (TUDCA) as an M2 proton channel inhibitor will expand our understanding of IAV biology and complement the current anti-IAV arsenal (285). Subsequent experimental results showed that drugs such as hexamethylene amiloride (HMA) (286), gliclazide (120), memantine (120), and emodin (127) have inhibitory effects on the channel activity of some viroporins (Table 2). Recent studies have demonstrated synergistic antiviral effects achieved by the combination of adamantanes and novel compounds (287), which provides directions for the development of viroporins inhibitory drugs. Currently, some other pharmacological inhibitors against viroporins (e.g., pyrin B, BIT225, DIDS, and divalent copper complexes) have been reported (14, 166, 181, 288). However, none of these drugs has an effective inhibitory effect against most viroporin activity at this stage, and therefore, new anti-viroporins drugs are urgently needed to be developed. In addition to antiviral drug research, attenuated viruses lacking viroporins are increasingly being suggested as vaccine candidates (289–291).
Although viroporins are increasingly studied, their protein families are expanding, and their functions are better understood, many aspects remain unclear, and future studies on this area should continue to elucidate the structure, function, and mechanism of such viral proteins to develop a library of antiviral targets across multiple virus families.
Author Contributions
XX and AC contributed to the design of the manuscript. XO, DS, SM, JH, QY, YW, SC, SZ, and DZ, provided ideas contributing to the conception of this manuscript. RJ, ML, X-XZ, QG, and BT helped to create the figures. MW modified the manuscript. All the authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the China Agriculture Research System of MOF and MARA, and the Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gonzalez ME, Carrasco L. Viroporins. FEBS Lett (2003) 552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4
2. Giorda KM, Hebert DN. Viroporins Customize Host Cells for Efficient Viral Propagation. DNA Cell Biol (2013) 32(10):557–64. doi: 10.1089/dna.2013.2159
3. Ao D, Guo HC, Sun SQ, Sun DH, Fung TS, Wei YQ, et al. Viroporin Activity of the Foot-And-Mouth Disease Virus Non-Structural 2b Protein. PloS One (2015) 10(5):e0125828. doi: 10.1371/journal.pone.0125828
4. Gladue DP, Largo E, de la Arada I, Aguilella VM, Alcaraz A, Arrondo JLR, et al. Molecular Characterization of the Viroporin Function of Foot-And-Mouth Disease Virus Nonstructural Protein 2b. J Virol (2018) 92(23):e01360–18. doi: 10.1128/jvi.01360-18
5. Madan V, Castelló A, Carrasco L. Viroporins From RNA Viruses Induce Caspase-Dependent Apoptosis. Cell Microbiol (2008) 10(2):437–51. doi: 10.1111/j.1462-5822.2007.01057.x
6. Martínez-Gil L, Bañó-Polo M, Redondo N, Sánchez-Martínez S, Nieva JL, Carrasco L, et al. Membrane Integration of Poliovirus 2B Viroporin. J Virol (2011) 85(21):11315–24. doi: 10.1128/jvi.05421-11
7. Lama J, Carrasco L. Expression of Poliovirus Nonstructural Proteins in Escherichia Coli Cells. Modification of Membrane Permeability Induced by 2B and 3A. J Biol Chem (1992) 267(22):15932–7. doi: 10.1016/S0021-9258(19)49623-3
8. Teterina NL, Rinaudo MS, Ehrenfeld E. Strand-Specific RNA Synthesis Defects in a Poliovirus With a Mutation in Protein 3A. J Virol (2003) 77(23):12679–91. doi: 10.1128/jvi.77.23.12679-12691.2003
9. Campanella M, de Jong AS, Lanke KW, Melchers WJ, Willems PH, Pinton P, et al. The Coxsackievirus 2B Protein Suppresses Apoptotic Host Cell Responses by Manipulating Intracellular Ca2+ Homeostasis. J Biol Chem (2004) 279(18):18440–50. doi: 10.1074/jbc.M309494200
10. van Kuppeveld FJ, Galama JM, Zoll J, van den Hurk PJ, Melchers WJ. Coxsackie B3 Virus Protein 2B Contains Cationic Amphipathic Helix That is Required for Viral RNA Replication. J Virol (1996) 70(6):3876–86. doi: 10.1128/jvi.70.6.3876-3886.1996
11. van Kuppeveld FJ, Hoenderop JG, Smeets RL, Willems PH, Dijkman HB, Galama JM, et al. Coxsackievirus Protein 2B Modifies Endoplasmic Reticulum Membrane and Plasma Membrane Permeability and Facilitates Virus Release. EMBO J (1997) 16(12):3519–32. doi: 10.1093/emboj/16.12.3519
12. de Jong AS, de Mattia F, Van Dommelen MM, Lanke K, Melchers WJ, Willems PH, et al. Functional Analysis of Picornavirus 2B Proteins: Effects on Calcium Homeostasis and Intracellular Protein Trafficking. J Virol (2008) 82(7):3782–90. doi: 10.1128/jvi.02076-07
13. Cong H, Du N, Yang Y, Song L, Zhang W, Tien P. Enterovirus 71 2b Induces Cell Apoptosis by Directly Inducing the Conformational Activation of the Proapoptotic Protein Bax. J Virol (2016) 90(21):9862–77. doi: 10.1128/jvi.01499-16
14. Xie S, Wang K, Yu W, Lu W, Xu K, Wang J, et al. DIDS Blocks a Chloride-Dependent Current That is Mediated by the 2B Protein of Enterovirus 71. Cell Res (2011) 21(8):1271–5. doi: 10.1038/cr.2011.112
15. Song J, Chi M, Luo X, Song Q, Xia D, Shi B, et al. Non-Structural Protein 2B of Human Rhinovirus 16 Activates Both PERK and ATF6 Rather Than IRE1 to Trigger ER Stress. Viruses (2019) 11(2):133. doi: 10.3390/v11020133
16. Liu Z, Ye Q, Cheng A, Ou X, Mao S, Sun D, et al. A Viroporin-Like 2B Protein of Duck Hepatitis A Virus 1 That Induces Incomplete Autophagy in DEF Cells. Poult Sci (2021) 100(10):101331. doi: 10.1016/j.psj.2021.101331
17. Shukla A, Dey D, Banerjee K, Nain A, Banerjee M. The C-Terminal Region of the non-Structural Protein 2B From Hepatitis A Virus Demonstrates Lipid-Specific Viroporin-Like Activity. Sci Rep (2015) 5:15884. doi: 10.1038/srep15884
18. Vives-Adrián L, Garriga D, Buxaderas M, Fraga J, Pereira PJ, Macedo-Ribeiro S, et al. Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus. J Virol (2015) 89(7):3648–58. doi: 10.1128/jvi.02881-14
19. Raamsman MJ, Locker JK, de Hooge A, de Vries AA, Griffiths G, Vennema H, et al. Characterization of the Coronavirus Mouse Hepatitis Virus Strain A59 Small Membrane Protein E. J Virol (2000) 74(5):2333–42. doi: 10.1128/jvi.74.5.2333-2342.2000
20. Ye Y, Hogue BG. Role of the Coronavirus E Viroporin Protein Transmembrane Domain in Virus Assembly. J Virol (2007) 81(7):3597–607. doi: 10.1128/jvi.01472-06
21. Cabrera-Garcia D, Bekdash R, Abbott GW, Yazawa M, Harrison NL. The Envelope Protein of SARS-CoV-2 Increases Intra-Golgi pH and Forms a Cation Channel That is Regulated by pH. J Physiol (2021) 599(11):2851–68. doi: 10.1113/jp281037
22. Nieto-Torres JL, Dediego ML, Alvarez E, Jiménez-Guardeño JM, Regla-Nava JA, Llorente M, et al. Subcellular Location and Topology of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology (2011) 415(2):69–82. doi: 10.1016/j.virol.2011.03.029
23. Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, et al. The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles. J Virol (2008) 82(22):11318–30. doi: 10.1128/jvi.01052-08
24. Lu W, Zheng BJ, Xu K, Schwarz W, Du L, Wong CK, et al. Severe Acute Respiratory Syndrome-Associated Coronavirus 3a Protein Forms an Ion Channel and Modulates Virus Release. Proc Natl Acad Sci U.S.A. (2006) 103(33):12540–5. doi: 10.1073/pnas.0605402103
25. Yuan X, Li J, Shan Y, Yang Z, Zhao Z, Chen B, et al. Subcellular Localization and Membrane Association of SARS-CoV 3a Protein. Virus Res (2005) 109(2):191–202. doi: 10.1016/j.virusres.2005.01.001
26. Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio (2018) 9(3):e02325–17. doi: 10.1128/mBio.02325-17
27. Chen C-Y, Ping Y-H, Lee H-C, Chen K-H, Lee Y-M, Chan Y-J, et al. Open Reading Frame 8a of the Human Severe Acute Respiratory Syndrome Coronavirus Not Only Promotes Viral Replication But Also Induces Apoptosis. J Infect Dis (2007) 196(3):405–15. doi: 10.1086/519166
28. Corse E, Machamer CE. Infectious Bronchitis Virus E Protein is Targeted to the Golgi Complex and Directs Release of Virus-Like Particles. J Virol (2000) 74(9):4319–26. doi: 10.1128/jvi.74.9.4319-4326.2000
29. Ruch TR, Machamer CE. The Hydrophobic Domain of Infectious Bronchitis Virus E Protein Alters the Host Secretory Pathway and is Important for Release of Infectious Virus. J Virol (2011) 85(2):675–85. doi: 10.1128/jvi.01570-10
30. Ruch TR, Machamer CE. The Coronavirus E Protein: Assembly and Beyond. Viruses (2012) 4(3):363–82. doi: 10.3390/v4030363
31. Ruch TR, Machamer CE. A Single Polar Residue and Distinct Membrane Topologies Impact the Function of the Infectious Bronchitis Coronavirus E Protein. PloS Pathog (2012) 8(5):e1002674. doi: 10.1371/journal.ppat.1002674
32. Westerbeck JW, Machamer CE. The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus. J Virol (2019) 93(11):e00015–19. doi: 10.1128/jvi.00015-19
33. Zhang R, Wang K, Ping X, Yu W, Qian Z, Xiong S, et al. The Ns12.9 Accessory Protein of Human Coronavirus OC43 Is a Viroporin Involved in Virion Morphogenesis and Pathogenesis. J Virol (2015) 89(22):11383–95. doi: 10.1128/jvi.01986-15
34. Zhang R, Wang K, Lv W, Yu W, Xie S, Xu K, et al. The ORF4a Protein of Human Coronavirus 229E Functions as a Viroporin That Regulates Viral Production. Biochim Biophys Acta (2014) 1838(4):1088–95. doi: 10.1016/j.bbamem.2013.07.025
35. Sanz MA, Madan V, Carrasco L, Nieva JL. Interfacial Domains in Sindbis Virus 6K Protein. Detection and Functional Characterization. J Biol Chem (2003) 278(3):2051–7. doi: 10.1074/jbc.M206611200
36. Melton JV, Ewart GD, Weir RC, Board PG, Lee E, Gage PW. Alphavirus 6K Proteins Form Ion Channels. J Biol Chem (2002) 277(49):46923–31. doi: 10.1074/jbc.M207847200
37. Taylor A, Melton JV, Herrero LJ, Thaa B, Karo-Astover L, Gage PW, et al. Effects of an In-Frame Deletion of the 6k Gene Locus From the Genome of Ross River Virus. J Virol (2016) 90(8):4150–9. doi: 10.1128/jvi.03192-15
38. Madsen JJ, Grime JMA, Rossman JS, Voth GA. Entropic Forces Drive Clustering and Spatial Localization of Influenza A M2 During Viral Budding. Proc Natl Acad Sci U.S.A. (2018) 115(37):E8595–e8603. doi: 10.1073/pnas.1805443115
39. Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. Influenza Virus A M2 Protein Generates Negative Gaussian Membrane Curvature Necessary for Budding and Scission. J Am Chem Soc (2013) 135(37):13710–9. doi: 10.1021/ja400146z
40. Torabifard H, Panahi A, Brooks CL 3rd. M2 Amphipathic Helices Facilitate pH-Dependent Conformational Transition in Influenza A Virus. Proc Natl Acad Sci U.S.A. (2020) 117(7):3583–91. doi: 10.1073/pnas.1913385117
41. Vijayvergiya V, Wilson R, Chorak A, Gao PF, Cross TA, Busath DD. Proton Conductance of Influenza Virus M2 Protein in Planar Lipid Bilayers. Biophys J (2004) 87(3):1697–704. doi: 10.1529/biophysj.104.043018
42. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, et al. A Novel Influenza A Virus Mitochondrial Protein That Induces Cell Death. Nat Med (2001) 7(12):1306–12. doi: 10.1038/nm1201-1306
43. Henkel M, Mitzner D, Henklein P, Meyer-Almes FJ, Moroni A, Difrancesco ML, et al. The Proapoptotic Influenza A Virus Protein PB1-F2 Forms a Nonselective Ion Channel. PloS One (2010) 5(6):e11112. doi: 10.1371/journal.pone.0011112
44. Imai M, Watanabe S, Ninomiya A, Obuchi M, Odagiri T. Influenza B Virus BM2 Protein is a Crucial Component for Incorporation of Viral Ribonucleoprotein Complex Into Virions During Virus Assembly. J Virol (2004) 78(20):11007–15. doi: 10.1128/jvi.78.20.11007-11015.2004
45. Mandala VS, Loftis AR, Shcherbakov AA, Pentelute BL, Hong M. Atomic Structures of Closed and Open Influenza B M2 Proton Channel Reveal the Conduction Mechanism. Nat Struct Mol Biol (2020) 27(2):160–7. doi: 10.1038/s41594-019-0371-2
46. Mi S, Li Y, Yan J, Gao GF. Na(+)/K (+)-ATPase β1 Subunit Interacts With M2 Proteins of Influenza A and B Viruses and Affects the Virus Replication. Sci China Life Sci (2010) 53(9):1098–105. doi: 10.1007/s11427-010-4048-7
47. Demers A, Ran Z, Deng Q, Wang D, Edman B, Lu W, et al. Palmitoylation is Required for Intracellular Trafficking of Influenza B Virus NB Protein and Efficient Influenza B Virus Growth In Vitro. J Gen Virol (2014) 95(Pt 6):1211–20. doi: 10.1099/vir.0.063511-0
48. Sunstrom NA, Premkumar LS, Premkumar A, Ewart G, Cox GB, Gage PW. Ion Channels Formed by NB, an Influenza B Virus Protein. J Membr Biol (1996) 150(2):127–32. doi: 10.1007/s002329900037
49. Furukawa T, Muraki Y, Noda T, Takashita E, Sho R, Sugawara K, et al. Role of the CM2 Protein in the Influenza C Virus Replication Cycle. J Virol (2011) 85(3):1322–9. doi: 10.1128/jvi.01367-10
50. Goto T, Shimotai Y, Matsuzaki Y, Muraki Y, Sho R, Sugawara K, et al. Effect of Phosphorylation of CM2 Protein on Influenza C Virus Replication. J Virol (2017) 91(22):e00773–17. doi: 10.1128/jvi.00773-17
51. Kesinger E, Liu J, Jensen A, Chia CP, Demers A, Moriyama H. Influenza D Virus M2 Protein Exhibits Ion Channel Activity in Xenopus Laevis Oocytes. PloS One (2018) 13(6):e0199227. doi: 10.1371/journal.pone.0199227
52. Brohm C, Steinmann E, Friesland M, Lorenz IC, Patel A, Penin F, et al. Characterization of Determinants Important for Hepatitis C Virus P7 Function in Morphogenesis by Using Trans-Complementation. J Virol (2009) 83(22):11682–93. doi: 10.1128/jvi.00691-09
53. Carrère-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. Subcellular Localization and Topology of the P7 Polypeptide of Hepatitis C Virus. J Virol (2002) 76(8):3720–30. doi: 10.1128/jvi.76.8.3720-3730.2002
54. Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, et al. The P7 Protein of Hepatitis C Virus Forms an Ion Channel That is Blocked by the Antiviral Drug, Amantadine. FEBS Lett (2003) 535(1-3):34–8. doi: 10.1016/s0014-5793(02)03851-6
55. Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. Hepatitis C Virus P7 Protein is Crucial for Assembly and Release of Infectious Virions. PloS Pathog (2007) 3(7):e103. doi: 10.1371/journal.ppat.0030103
56. Gladue DP, Holinka LG, Largo E, Fernandez Sainz I, Carrillo C, O’Donnell V, et al. Classical Swine Fever Virus P7 Protein is a Viroporin Involved in Virulence in Swine. J Virol (2012) 86(12):6778–91. doi: 10.1128/jvi.00560-12
57. Guo HC, Sun SQ, Sun DH, Wei YQ, Xu J, Huang M, et al. Viroporin Activity and Membrane Topology of Classic Swine Fever Virus p7 Protein. Int J Biochem Cell Biol (2013) 45(7):1186–94. doi: 10.1016/j.biocel.2013.03.021
58. Shrivastava G, Visoso-Carvajal G, Garcia-Cordero J, Leon-Juarez M, Chavez-Munguia B, Lopez T, et al. Dengue Virus Serotype 2 and Its Non-Structural Proteins 2A and 2B Activate NLRP3 Inflammasome. Front Immunol (2020) 11:352. doi: 10.3389/fimmu.2020.00352
59. Wu RH, Tsai MH, Chao DY, Yueh A. Scanning Mutagenesis Studies Reveal a Potential Intramolecular Interaction Within the C-Terminal Half of Dengue Virus NS2A Involved in Viral RNA Replication and Virus Assembly and Secretion. J Virol (2015) 89(8):4281–95. doi: 10.1128/jvi.03011-14
60. Xie X, Gayen S, Kang C, Yuan Z, Shi PY. Membrane Topology and Function of Dengue Virus NS2A Protein. J Virol (2013) 87(8):4609–22. doi: 10.1128/jvi.02424-12
61. León-Juárez M, Martínez-Castillo M, Shrivastava G, García-Cordero J, Villegas-Sepulveda N, Mondragón-Castelán M, et al. Recombinant Dengue Virus Protein NS2B Alters Membrane Permeability in Different Membrane Models. Virol J (2016) 13:1. doi: 10.1186/s12985-015-0456-4
62. Cong L, Sugden SM, Leclair P, Lim CJ, Pham TNQ, Cohen É A. HIV-1 Vpu Promotes Phagocytosis of Infected CD4(+) T Cells by Macrophages Through Downregulation of CD47. mBio (2021) 12(4):e0192021. doi: 10.1128/mBio.01920-21
63. Neil SJ, Zang T, Bieniasz PD. Tetherin Inhibits Retrovirus Release and is Antagonized by HIV-1 Vpu. Nature (2008) 451(7177):425–30. doi: 10.1038/nature06553
64. Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, et al. The Interferon-Induced Protein BST-2 Restricts HIV-1 Release and is Downregulated From the Cell Surface by the Viral Vpu Protein. Cell Host Microbe (2008) 3(4):245–52. doi: 10.1016/j.chom.2008.03.001
65. Techaarpornkul S, Barretto N, Peeples ME. Functional Analysis of Recombinant Respiratory Syncytial Virus Deletion Mutants Lacking the Small Hydrophobic and/or Attachment Glycoprotein Gene. J Virol (2001) 75(15):6825–34. doi: 10.1128/jvi.75.15.6825-6834.2001
66. Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. Human Respiratory Syncytial Virus Viroporin SH: A Viral Recognition Pathway Used by the Host to Signal Inflammasome Activation. Thorax (2013) 68(1):66–75. doi: 10.1136/thoraxjnl-2012-202182
67. Masante C, El Najjar F, Chang A, Jones A, Moncman CL, Dutch RE. The Human Metapneumovirus Small Hydrophobic Protein has Properties Consistent With Those of a Viroporin and can Modulate Viral Fusogenic Activity. J Virol (2014) 88(11):6423–33. doi: 10.1128/jvi.02848-13
68. Strtak AC, Perry JL, Sharp MN, Chang-Graham AL, Farkas T, Hyser JM. Recovirus NS1-2 Has Viroporin Activity That Induces Aberrant Cellular Calcium Signaling To Facilitate Virus Replication. mSphere (2019) 4(5):e00506–19. doi: 10.1128/mSphere.00506-19
69. Hyde JL, Mackenzie JM. Subcellular Localization of the MNV-1 ORF1 Proteins and Their Potential Roles in the Formation of the MNV-1 Replication Complex. Virology (2010) 406(1):138–48. doi: 10.1016/j.virol.2010.06.047
70. Kang M, Moroni A, Gazzarrini S, DiFrancesco D, Thiel G, Severino M, et al. Small Potassium Ion Channel Proteins Encoded by Chlorella Viruses. Proc Natl Acad Sci U.S.A. (2004) 101(15):5318–24. doi: 10.1073/pnas.0307824100
71. Plugge B, Gazzarrini S, Nelson M, Cerana R, Van Etten JL, Derst C, et al. A Potassium Channel Protein Encoded by Chlorella Virus PBCV-1. Science (2000) 287(5458):1641–4. doi: 10.1126/science.287.5458.1641
72. López T, Camacho M, Zayas M, Nájera R, Sánchez R, Arias CF, et al. Silencing the Morphogenesis of Rotavirus. J Virol (2005) 79(1):184–92. doi: 10.1128/jvi.79.1.184-192.2005
73. Pham T, Perry JL, Dosey TL, Delcour AH, Hyser JM. The Rotavirus NSP4 Viroporin Domain is a Calcium-Conducting Ion Channel. Sci Rep (2017) 7:43487. doi: 10.1038/srep43487
74. Bodelón G, Labrada L, Martínez-Costas J, Benavente J. Modification of Late Membrane Permeability in Avian Reovirus-Infected Cells: Viroporin Activity of the S1-Encoded Nonstructural P10 Protein. J Biol Chem (2002) 277(20):17789–96. doi: 10.1074/jbc.M202018200
75. Shmulevitz M, Duncan R. A New Class of Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by the non-Enveloped Fusogenic Reoviruses. EMBO J (2000) 19(5):902–12. doi: 10.1093/emboj/19.5.902
76. Han Z, Harty RN. The NS3 Protein of Bluetongue Virus Exhibits Viroporin-Like Properties. J Biol Chem (2004) 279(41):43092–7. doi: 10.1074/jbc.M403663200
77. Joubert DA, Blasdell KR, Audsley MD, Trinidad L, Monaghan P, Dave KA, et al. Bovine Ephemeral Fever Rhabdovirus α1 Protein has Viroporin-Like Properties and Binds Importin β1 and Importin 7. J Virol (2014) 88(3):1591–603. doi: 10.1128/jvi.01812-13
78. Daniels R, Rusan NM, Wadsworth P, Hebert DN. SV40 VP2 and VP3 Insertion Into ER Membranes is Controlled by the Capsid Protein VP1: Implications for DNA Translocation Out of the ER. Mol Cell (2006) 24(6):955–66. doi: 10.1016/j.molcel.2006.11.001
79. Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, et al. BAP31 and BiP are Essential for Dislocation of SV40 From the Endoplasmic Reticulum to the Cytosol. Nat Cell Biol (2011) 13(11):1305–14. doi: 10.1038/ncb2339
80. Giorda KM, Raghava S, Zhang MW, Hebert DN. The Viroporin Activity of the Minor Structural Proteins VP2 and VP3 is Required for SV40 Propagation. J Biol Chem (2013) 288(4):2510–20. doi: 10.1074/jbc.M112.428425
81. Daniels R, Sadowicz D, Hebert DN. A Very Late Viral Protein Triggers the Lytic Release of SV40. PloS Pathog (2007) 3(7):e98. doi: 10.1371/journal.ppat.0030098
82. Raghava S, Giorda KM, Romano FB, Heuck AP, Hebert DN. The SV40 Late Protein VP4 is a Viroporin That Forms Pores to Disrupt Membranes for Viral Release. PloS Pathog (2011) 7(6):e1002116. doi: 10.1371/journal.ppat.1002116
83. Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, et al. The Human Polyoma JC Virus Agnoprotein Acts as a Viroporin. PloS Pathog (2010) 6(3):e1000801. doi: 10.1371/journal.ppat.1000801
84. Conrad M, Bubb VJ, Schlegel R. The Human Papillomavirus Type 6 and 16 E5 Proteins are Membrane-Associated Proteins Which Associate With the 16-Kilodalton Pore-Forming Protein. J Virol (1993) 67(10):6170–8. doi: 10.1128/jvi.67.10.6170-6178.1993
85. Krawczyk E, Suprynowicz FA, Sudarshan SR, Schlegel R. Membrane Orientation of the Human Papillomavirus Type 16 E5 Oncoprotein. J Virol (2010) 84(4):1696–703. doi: 10.1128/jvi.01968-09
86. Wetherill LF, Holmes KK, Verow M, Müller M, Howell G, Harris M, et al. High-Risk Human Papillomavirus E5 Oncoprotein Displays Channel-Forming Activity Sensitive to Small-Molecule Inhibitors. J Virol (2012) 86(9):5341–51. doi: 10.1128/jvi.06243-11
87. He J, Melnik LI, Komin A, Wiedman G, Fuselier T, Morris CF, et al. Ebola Virus Delta Peptide Is a Viroporin. J Virol (2017) 91(16)e00438–17. doi: 10.1128/jvi.00438-17
88. Pokhrel R, Pavadai E, Gerstman BS, Chapagain PP. Membrane Pore Formation and Ion Selectivity of the Ebola Virus Delta Peptide. Phys Chem Chem Phys (2019) 21(10):5578–85. doi: 10.1039/c8cp07323f
89. Lulla V, Firth AE. A Hidden Gene in Astroviruses Encodes a Viroporin. Nat Commun (2020) 11(1):4070. doi: 10.1038/s41467-020-17906-x
90. Luganini A, Di Nardo G, Munaron L, Gilardi G, Fiorio Pla A, Gribaudo G. Human Cytomegalovirus US21 Protein is a Viroporin That Modulates Calcium Homeostasis and Protects Cells Against Apoptosis. Proc Natl Acad Sci U.S.A. (2018) 115(52):E12370–e12377. doi: 10.1073/pnas.1813183115
91. Nieto-Torres JL, Verdiá-Báguena C, Castaño-Rodriguez C, Aguilella VM, Enjuanes L. Relevance of Viroporin Ion Channel Activity on Viral Replication and Pathogenesis. Viruses (2015) 7(7):3552–73. doi: 10.3390/v7072786
92. Moffat K, Knox C, Howell G, Clark SJ, Yang H, Belsham GJ, et al. Inhibition of the Secretory Pathway by Foot-and-Mouth Disease Virus 2BC Protein is Reproduced by Coexpression of 2B With 2C, and the Site of Inhibition is Determined by the Subcellular Location of 2C. J Virol (2007) 81(3):1129–39. doi: 10.1128/jvi.00393-06
93. Zhi X, Zhang Y, Sun S, Zhang Z, Dong H, Luo X, et al. NLRP3 Inflammasome Activation by Foot-And-Mouth Disease Virus Infection Mainly Induced by Viral RNA and non-Structural Protein 2B. RNA Biol (2020) 17(3):335–49. doi: 10.1080/15476286.2019.1700058
94. Liu H, Xue Q, Cao W, Yang F, Ma L, Liu W, et al. Foot-And-Mouth Disease Virus Nonstructural Protein 2B Interacts With Cyclophilin A, Modulating Virus Replication. FASEB J (2018) 32(12):fj201701351. doi: 10.1096/fj.201701351
95. Liu H, Xue Q, Zhu Z, Yang F, Cao W, Liu X, et al. Foot-And-Mouth Disease Virus Inhibits RIP2 Protein Expression to Promote Viral Replication. Virol Sin (2021) 36(4):608–22. doi: 10.1007/s12250-020-00322-2
96. Li M, Xin T, Gao X, Wu J, Wang X, Fang L, et al. Foot-And-Mouth Disease Virus non-Structural Protein 2B Negatively Regulates the RLR-Mediated IFN-β Induction. Biochem Biophys Res Commun (2018) 504(1):238–44. doi: 10.1016/j.bbrc.2018.08.161
97. Zhu Z, Wang G, Yang F, Cao W, Mao R, Du X, et al. Foot-And-Mouth Disease Virus Viroporin 2b Antagonizes RIG-I-Mediated Antiviral Effects by Inhibition of Its Protein Expression. J Virol (2016) 90(24):11106–21. doi: 10.1128/jvi.01310-16
98. Zhu Z, Li C, Du X, Wang G, Cao W, Yang F, et al. Foot-And-Mouth Disease Virus Infection Inhibits LGP2 Protein Expression to Exaggerate Inflammatory Response and Promote Viral Replication. Cell Death Dis (2017) 8(4):e2747. doi: 10.1038/cddis.2017.170
99. Liu H, Zhu Z, Xue Q, Yang F, Cao W, Zhang K, et al. Foot-And-Mouth Disease Virus Antagonizes NOD2-Mediated Antiviral Effects by Inhibiting NOD2 Protein Expression. J Virol (2019) 93(11):e00124–19. doi: 10.1128/jvi.00124-19
100. Heinz BA, Vance LM. The Antiviral Compound Enviroxime Targets the 3A Coding Region of Rhinovirus and Poliovirus. J Virol (1995) 69(7):4189–97. doi: 10.1128/jvi.69.7.4189-4197.1995
101. Sandoval IV, Carrasco L. Poliovirus Infection and Expression of the Poliovirus Protein 2B Provoke the Disassembly of the Golgi Complex, the Organelle Target for the Antipoliovirus Drug Ro-090179. J Virol (1997) 71(6):4679–93. doi: 10.1128/jvi.71.6.4679-4693.1997
102. Madan V, Sánchez-Martínez S, Carrasco L, Nieva JL. A Peptide Based on the Pore-Forming Domain of Pro-Apoptotic Poliovirus 2B Viroporin Targets Mitochondria. Biochim Biophys Acta (2010) 1798(1):52–8. doi: 10.1016/j.bbamem.2009.10.013
103. Choe SS, Dodd DA, Kirkegaard K. Inhibition of Cellular Protein Secretion by Picornaviral 3A Proteins. Virology (2005) 337(1):18–29. doi: 10.1016/j.virol.2005.03.036
104. Suhy DA, Giddings TH Jr., Kirkegaard K. Remodeling the Endoplasmic Reticulum by Poliovirus Infection and by Individual Viral Proteins: An Autophagy-Like Origin for Virus-Induced Vesicles. J Virol (2000) 74(19):8953–65. doi: 10.1128/jvi.74.19.8953-8965.2000
105. Deitz SB, Dodd DA, Cooper S, Parham P, Kirkegaard K. MHC I-Dependent Antigen Presentation is Inhibited by Poliovirus Protein 3A. Proc Natl Acad Sci U.S.A. (2000) 97(25):13790–5. doi: 10.1073/pnas.250483097
106. Jackson WT, Giddings TH Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of Cellular Autophagosomal Machinery by RNA Viruses. PloS Biol (2005) 3(5):e156. doi: 10.1371/journal.pbio.0030156
107. de Jong AS, Visch HJ, de Mattia F, van Dommelen MM, Swarts HG, Luyten T, et al. The Coxsackievirus 2B Protein Increases Efflux of Ions From the Endoplasmic Reticulum and Golgi, Thereby Inhibiting Protein Trafficking Through the Golgi. J Biol Chem (2006) 281(20):14144–50. doi: 10.1074/jbc.M511766200
108. Wu H, Zhai X, Chen Y, Wang R, Lin L, Chen S, et al. Protein 2B of Coxsackievirus B3 Induces Autophagy Relying on Its Transmembrane Hydrophobic Sequences. Viruses (2016) 8(5):131. doi: 10.3390/v8050131
109. Ito M, Yanagi Y, Ichinohe T. Encephalomyocarditis Virus Viroporin 2B Activates NLRP3 Inflammasome. PloS Pathog (2012) 8(8):e1002857. doi: 10.1371/journal.ppat.1002857
110. Moriyama M, Koshiba T, Ichinohe T. Influenza A Virus M2 Protein Triggers Mitochondrial DNA-Mediated Antiviral Immune Responses. Nat Commun (2019) 10(1):4624. doi: 10.1038/s41467-019-12632-5
111. Han X, Cong H. Enterovirus 71 Induces Apoptosis by Directly Modulating the Conformational Activation of Pro-Apoptotic Protein Bax. J Gen Virol (2017) 98(3):422–34. doi: 10.1099/jgv.0.000705
112. Sun M, Lin Q, Wang C, Xing J, Yan K, Liu Z, et al. Enterovirus A71 2b Inhibits Interferon-Activated JAK/STAT Signaling by Inducing Caspase-3-Dependent Karyopherin-α1 Degradation. Front Microbiol (2021) 12:762869. doi: 10.3389/fmicb.2021.762869
113. Jin J, Wang W, Ai S, Liu W, Song Y, Luo Z, et al. Enterovirus 71 Represses Interleukin Enhancer-Binding Factor 2 Production and Nucleus Translocation to Antagonize ILF2 Antiviral Effects. Viruses (2019) 12(1):22. doi: 10.3390/v12010022
114. Triantafilou K, Kar S, van Kuppeveld FJ, Triantafilou M. Rhinovirus-Induced Calcium Flux Triggers NLRP3 and NLRC5 Activation in Bronchial Cells. Am J Respir Cell Mol Biol (2013) 49(6):923–34. doi: 10.1165/rcmb.2013-0032OC
115. Paulmann D, Magulski T, Schwarz R, Heitmann L, Flehmig B, Vallbracht A, et al. Hepatitis A Virus Protein 2B Suppresses Beta Interferon (IFN) Gene Transcription by Interfering With IFN Regulatory Factor 3 Activation. J Gen Virol (2008) 89(Pt 7):1593–604. doi: 10.1099/vir.0.83521-0
116. An S, Chen CJ, Yu X, Leibowitz JL, Makino S. Induction of Apoptosis in Murine Coronavirus-Infected Cultured Cells and Demonstration of E Protein as an Apoptosis Inducer. J Virol (1999) 73(9):7853–9. doi: 10.1128/jvi.73.9.7853-7859.1999
117. Bonzano C, Borroni D, Lancia A, Bonzano E. Doxycycline: From Ocular Rosacea to COVID-19 Anosmia. New Insight Into the Coronavirus Outbreak. Front Med (Lausanne) (2020) 7:200. doi: 10.3389/fmed.2020.00200
118. Dey D, Borkotoky S, Banerjee M. In Silico Identification of Tretinoin as a SARS-CoV-2 Envelope (E) Protein Ion Channel Inhibitor. Comput Biol Med (2020) 127:104063. doi: 10.1016/j.compbiomed.2020.104063
119. Minakshi R, Padhan K. The YxxΦ Motif Within the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3a Protein is Crucial for its Intracellular Transport. Virol J (2014) 11:75. doi: 10.1186/1743-422X-11-75
120. Singh Tomar PP, Arkin IT. SARS-CoV-2 E Protein is a Potential Ion Channel That can be Inhibited by Gliclazide and Memantine. Biochem Biophys Res Commun (2020) 530(1):10–4. doi: 10.1016/j.bbrc.2020.05.206
121. Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX, Gong X. Conductance and Amantadine Binding of a Pore Formed by a Lysine-Flanked Transmembrane Domain of SARS Coronavirus Envelope Protein. Protein Sci (2007) 16(9):2065–71. doi: 10.1110/ps.062730007
122. Nieto-Torres JL, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Castaño-Rodriguez C, Fernandez-Delgado R, et al. Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology (2015) 485:330–9. doi: 10.1016/j.virol.2015.08.010
123. Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, et al. The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein is a Determinant of Viral Pathogenesis. PloS Pathog (2014) 10(8):e1004320. doi: 10.1371/journal.ppat.1004320
124. Yang Y, Xiong Z, Zhang S, Yan Y, Nguyen J, Ng B, et al. Bcl-xL Inhibits T-Cell Apoptosis Induced by Expression of SARS Coronavirus E Protein in the Absence of Growth Factors. Biochem J (2005) 392(Pt 1):135–43. doi: 10.1042/bj20050698
125. DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Regla-Nava JA, Alvarez E, Oliveros JC, et al. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis. PloS Pathog (2011) 7(10):e1002315. doi: 10.1371/journal.ppat.1002315
126. Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front Microbiol 10 (2019) 50:50. doi: 10.3389/fmicb.2019.00050
127. Schwarz S, Wang K, Yu W, Sun B, Schwarz W. Emodin Inhibits Current Through SARS-Associated Coronavirus 3a Protein. Antiviral Res (2011) 90(1):64–9. doi: 10.1016/j.antiviral.2011.02.008
128. Siu KL, Yuen KS, Castaño-Rodriguez C, Ye ZW, Yeung ML, Fung SY, et al. Severe Acute Respiratory Syndrome Coronavirus ORF3a Protein Activates the NLRP3 Inflammasome by Promoting TRAF3-Dependent Ubiquitination of ASC. FASEB J (2019) 33(8):8865–77. doi: 10.1096/fj.201802418R
129. Kanzawa N, Nishigaki K, Hayashi T, Ishii Y, Furukawa S, Niiro A, et al. Augmentation of Chemokine Production by Severe Acute Respiratory Syndrome Coronavirus 3a/X1 and 7a/X4 Proteins Through NF-kappaB Activation. FEBS Lett (2006) 580(30):6807–12. doi: 10.1016/j.febslet.2006.11.046
130. Narayanan K, Huang C, Makino S. SARS Coronavirus Accessory Proteins. Virus Res (2008) 133(1):113–21. doi: 10.1016/j.virusres.2007.10.009
131. Yue Y, Nabar NR, Shi C-S, Kamenyeva O, Xiao X, Hwang I-Y, et al. SARS-Coronavirus Open Reading Frame-3a Drives Multimodal Necrotic Cell Death. Cell Death Dis (2018) 9(9):904. doi: 10.1038/s41419-018-0917-y
132. Miao G, Zhao H, Li Y, Ji M, Chen Y, Shi Y, et al. ORF3a of the COVID-19 Virus SARS-CoV-2 Blocks HOPS Complex-Mediated Assembly of the SNARE Complex Required for Autolysosome Formation. Dev Cell (2021) 56(4):427–442.e425. doi: 10.1016/j.devcel.2020.12.010
133. Chan CM, Tsoi H, Chan WM, Zhai S, Wong CO, Yao X, et al. The Ion Channel Activity of the SARS-Coronavirus 3a Protein is Linked to its Pro-Apoptotic Function. Int J Biochem Cell Biol (2009) 41(11):2232–9. doi: 10.1016/j.biocel.2009.04.019
134. Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, et al. The ORF3a Protein of SARS-CoV-2 Induces Apoptosis in Cells. Cell Mol Immunol (2020) 17(8):881–3. doi: 10.1038/s41423-020-0485-9
135. Padhan K, Minakshi R, Towheed MAB, Jameel S. Severe Acute Respiratory Syndrome Coronavirus 3a Protein Activates the Mitochondrial Death Pathway Through P38 MAP Kinase Activation. J Gen Virol 89(Pt (2008) 8):1960–9. doi: 10.1099/vir.0.83665-0
136. Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PloS One (2009) 4(12):e8342. doi: 10.1371/journal.pone.0008342
137. Li S, Yuan L, Dai G, Chen RA, Liu DX, Fung TS. Regulation of the ER Stress Response by the Ion Channel Activity of the Infectious Bronchitis Coronavirus Envelope Protein Modulates Virion Release, Apoptosis, Viral Fitness, and Pathogenesis. Front Microbiol (2019) 10:3022. doi: 10.3389/fmicb.2019.03022
138. Leonov H, Astrahan P, Krugliak M, Arkin IT. How do Aminoadamantanes Block the Influenza M2 Channel, and How Does Resistance Develop? J Am Chem Soc (2011) 133(25):9903–11. doi: 10.1021/ja202288m
139. Patel H, Kukol A. Prediction of Ligands to Universally Conserved Binding Sites of the Influenza a Virus Nuclear Export Protein. Virol (2019), 537:97–103. doi: 10.1016/j.virol.2019.08.013
140. Ichinohe T, Pang IK, Iwasaki A. Influenza Virus Activates Inflammasomes via its Intracellular M2 Ion Channel. Nat Immunol (2010) 11(5):404–10. doi: 10.1038/ni.1861
141. Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-Interacting Motif in the Influenza A Virus M2 Protein is Required to Subvert Autophagy and Maintain Virion Stability. Cell Host Microbe (2014) 15(2):239–47. doi: 10.1016/j.chom.2014.01.006
142. Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, et al. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion With Lysosomes. Cell Host Microbe (2009) 6(4):367–80. doi: 10.1016/j.chom.2009.09.005
143. Gannagé M, Rämer PC, Münz C. Targeting Beclin 1 for Viral Subversion of Macroautophagy. Autophagy (2010) 6(1):166–7. doi: 10.4161/auto.6.1.10624
144. Ren Y, Li C, Feng L, Pan W, Li L, Wang Q, et al. Proton Channel Activity of Influenza A Virus Matrix Protein 2 Contributes to Autophagy Arrest. J Virol (2016) 90(1):591–8. doi: 10.1128/jvi.00576-15
145. Wang R, Zhu Y, Lin X, Ren C, Zhao J, Wang F, et al. Influenza M2 Protein Regulates MAVS-Mediated Signaling Pathway Through Interacting With MAVS and Increasing ROS Production. Autophagy (2019) 15(7):1163–81. doi: 10.1080/15548627.2019.1580089
146. Zhirnov OP, Klenk HD. Influenza A Virus Proteins NS1 and Hemagglutinin Along With M2 are Involved in Stimulation of Autophagy in Infected Cells. J Virol (2013) 87(24):13107–14. doi: 10.1128/jvi.02148-13
147. Wang R, Zhu Y, Zhao J, Ren C, Li P, Chen H, et al. Autophagy Promotes Replication of Influenza A Virus In Vitro. J Virol (2019) 93(4):e01984–18. doi: 10.1128/jvi.01984-18
148. Rossman JS, Lamb RA. Autophagy, Apoptosis, and the Influenza Virus M2 Protein. Cell Host Microbe (2009) 6(4):299–300. doi: 10.1016/j.chom.2009.09.009
149. Guan Z, Liu D, Mi S, Zhang J, Ye Q, Wang M, et al. Interaction of Hsp40 With Influenza Virus M2 Protein: Implications for PKR Signaling Pathway. Protein Cell (2010) 1(10):944–55. doi: 10.1007/s13238-010-0115-x
150. Boal-Carvalho I, Mazel-Sanchez B, Silva F, Garnier L, Yildiz S, Bonifacio JP, et al. Influenza A Viruses Limit NLRP3-NEK7-Complex Formation and Pyroptosis in Human Macrophages. EMBO Rep (2020) 21(12):e50421. doi: 10.15252/embr.202050421
151. Cheung PH, Ye ZW, Lee TT, Chen H, Chan CP, Jin DY. PB1-F2 Protein of Highly Pathogenic Influenza A (H7N9) Virus Selectively Suppresses RNA-Induced NLRP3 Inflammasome Activation Through Inhibition of MAVS-NLRP3 Interaction. J Leukoc Biol (2020) 108(5):1655–63. doi: 10.1002/jlb.4ab0420-694r
152. Le Goffic R, Leymarie O, Chevalier C, Rebours E, Da Costa B, Vidic J, et al. Transcriptomic Analysis of Host Immune and Cell Death Responses Associated With the Influenza A Virus PB1-F2 Protein. PloS Pathog (2011) 7(8):e1002202. doi: 10.1371/journal.ppat.1002202
153. McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, et al. Activation of the NLRP3 Inflammasome by IAV Virulence Protein PB1-F2 Contributes to Severe Pathophysiology and Disease. PloS Pathog (2013) 9(5):e1003392. doi: 10.1371/journal.ppat.1003392
154. Pinar A, Dowling JK, Bitto NJ, Robertson AA, Latz E, Stewart CR, et al. PB1-F2 Peptide Derived From Avian Influenza A Virus H7N9 Induces Inflammation via Activation of the NLRP3 Inflammasome. J Biol Chem (2017) 292(3):826–36. doi: 10.1074/jbc.M116.756379
155. Yoshizumi T, Ichinohe T, Sasaki O, Otera H, Kawabata S, Mihara K, et al. Influenza A Virus Protein PB1-F2 Translocates Into Mitochondria via Tom40 Channels and Impairs Innate Immunity. Nat Commun (2014) 5:4713. doi: 10.1038/ncomms5713
156. Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P. Influenza Virus PB1-F2 Protein Induces Cell Death Through Mitochondrial ANT3 and VDAC1. PloS Pathog (2005) 1(1):e4. doi: 10.1371/journal.ppat.0010004
157. La Gruta NL, Thomas PG, Webb AI, Dunstone MA, Cukalac T, Doherty PC, et al. Epitope-Specific TCRbeta Repertoire Diversity Imparts No Functional Advantage on the CD8+ T Cell Response to Cognate Viral Peptides. Proc Natl Acad Sci U.S.A. (2008) 105(6):2034–9. doi: 10.1073/pnas.0711682102
158. Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, Leymarie O, et al. Influenza A Virus Protein PB1-F2 Exacerbates IFN-Beta Expression of Human Respiratory Epithelial Cells. J Immunol (2010) 185(8):4812–23. doi: 10.4049/jimmunol.0903952
159. Varga ZT, Grant A, Manicassamy B, Palese P. Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon by Binding to MAVS and Decreasing Mitochondrial Membrane Potential. J Virol (2012) 86(16):8359–66. doi: 10.1128/jvi.01122-12
160. Dudek SE, Wixler L, Nordhoff C, Nordmann A, Anhlan D, Wixler V, et al. The Influenza Virus PB1-F2 Protein has Interferon Antagonistic Activity. Biol Chem (2011) 392(12):1135–44. doi: 10.1515/bc.2011.174
161. Cheung PH, Lee TT, Kew C, Chen H, Yuen KY, Chan CP, et al. Virus Subtype-Specific Suppression of MAVS Aggregation and Activation by PB1-F2 Protein of Influenza A (H7N9) Virus. PloS Pathog (2020) 16(6):e1008611. doi: 10.1371/journal.ppat.1008611
162. Varga ZT, Ramos I, Hai R, Schmolke M, García-Sastre A, Fernandez-Sesma A, et al. The Influenza Virus Protein PB1-F2 Inhibits the Induction of Type I Interferon at the Level of the MAVS Adaptor Protein. PloS Pathog (2011) 7(6):e1002067. doi: 10.1371/journal.ppat.1002067
163. Wang R, Zhu Y, Ren C, Yang S, Tian S, Chen H, et al. Influenza A Virus Protein PB1-F2 Impairs Innate Immunity by Inducing Mitophagy. Autophagy (2021) 17(2):496–511. doi: 10.1080/15548627.2020.1725375
164. Zhang H, Yu H, Wang J, Zhang M, Wang X, Ahmad W, et al. The BM2 Protein of Influenza B Virus Interacts With P53 and Inhibits its Transcriptional and Apoptotic Activities. Mol Cell Biochem (2015) 403(1-2):187–97. doi: 10.1007/s11010-015-2349-7
165. Convery O, Gargan S, Kickham M, Schroder M, O’Farrelly C, Stevenson NJ. The Hepatitis C Virus (HCV) Protein, P7, Suppresses Inflammatory Responses to Tumor Necrosis Factor (TNF)-α via Signal Transducer and Activator of Transcription (STAT)3 and Extracellular Signal-Regulated Kinase (ERK)-Mediated Induction of Suppressor of Cytokine Signaling (SOCS)3. FASEB J (2019) 33(8):8732–44. doi: 10.1096/fj.201800629RR
166. Luscombe CA, Huang Z, Murray MG, Miller M, Wilkinson J, Ewart GD. A Novel Hepatitis C Virus P7 Ion Channel Inhibitor, BIT225, Inhibits Bovine Viral Diarrhea Virus In Vitro and Shows Synergism With Recombinant Interferon-Alpha-2b and Nucleoside Analogues. Antiviral Res (2010) 86(2):144–53. doi: 10.1016/j.antiviral.2010.02.312
167. Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM, et al. Intracellular Proton Conductance of the Hepatitis C Virus P7 Protein and its Contribution to Infectious Virus Production. PloS Pathog (2010) 6(9):e1001087. doi: 10.1371/journal.ppat.1001087
168. Farag NS, Breitinger U, El-Azizi M, Breitinger HG. The P7 Viroporin of the Hepatitis C Virus Contributes to Liver Inflammation by Stimulating Production of Interleukin-1β. Biochim Biophys Acta Mol Basis Dis (2017) 1863(3):712–20. doi: 10.1016/j.bbadis.2016.12.006
169. Green AM, Beatty PR, Hadjilaou A, Harris E. Innate Immunity to Dengue Virus Infection and Subversion of Antiviral Responses. J Mol Biol (2014) 426(6):1148–60. doi: 10.1016/j.jmb.2013.11.023
170. Dalrymple NA, Cimica V, Mackow ER. Dengue Virus NS Proteins Inhibit RIG-I/MAVS Signaling by Blocking TBK1/IRF3 Phosphorylation: Dengue Virus Serotype 1 NS4A Is a Unique Interferon-Regulating Virulence Determinant. mBio (2015) 6(3):e00553–00515. doi: 10.1128/mBio.00553-15
171. Aguirre S, Maestre AM, Pagni S, Patel JR, Savage T, Gutman D, et al. DENV Inhibits Type I IFN Production in Infected Cells by Cleaving Human STING. PloS Pathog (2012) 8(10):e1002934. doi: 10.1371/journal.ppat.1002934
172. Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, et al. Dengue Virus NS2B Protein Targets cGAS for Degradation and Prevents Mitochondrial DNA Sensing During Infection. Nat Microbiol (2017) 2:17037. doi: 10.1038/nmicrobiol.2017.37
173. Yu CY, Chang TH, Liang JJ, Chiang RL, Lee YL, Liao CL, et al. Dengue Virus Targets the Adaptor Protein MITA to Subvert Host Innate Immunity. PloS Pathog (2012) 8(6):e1002780. doi: 10.1371/journal.ppat.1002780
174. Akari H, Bour S, Kao S, Adachi A, Strebel K. The Human Immunodeficiency Virus Type 1 Accessory Protein Vpu Induces Apoptosis by Suppressing the Nuclear Factor kappaB-Dependent Expression of Antiapoptotic Factors. J Exp Med (2001) 194(9):1299–311. doi: 10.1084/jem.194.9.1299
175. Khoury G, Ewart G, Luscombe C, Miller M, Wilkinson J. Antiviral Efficacy of the Novel Compound BIT225 Against HIV-1 Release From Human Macrophages. Antimicrob Agents Chemother (2010) 54(2):835–45. doi: 10.1128/aac.01308-09
176. Verma S, Ali A, Arora S, Banerjea AC. Inhibition of β-TrcP-Dependent Ubiquitination of P53 by HIV-1 Vpu Promotes P53-Mediated Apoptosis in Human T Cells. Blood (2011) 117(24):6600–7. doi: 10.1182/blood-2011-01-333427
177. Pham TN, Lukhele S, Hajjar F, Routy JP, Cohen É A. HIV Nef and Vpu Protect HIV-Infected CD4+ T Cells From Antibody-Mediated Cell Lysis Through Down-Modulation of CD4 and BST2. Retrovirology (2014) 11:15. doi: 10.1186/1742-4690-11-15
178. Sanchez DJ, Miranda D Jr., Marsden MD, Dizon TM, Bontemps JR, Davila SJ, et al. Disruption of Type I Interferon Induction by HIV Infection of T Cells. PloS One (2015) 10(9):e0137951. doi: 10.1371/journal.pone.0137951
179. Nguyen NV, Tran JT, Sanchez DJ. HIV Blocks Type I IFN Signaling Through Disruption of STAT1 Phosphorylation. Innate Immun (2018) 24(8):490–500. doi: 10.1177/1753425918803674
180. Langer S, Hammer C, Hopfensperger K, Klein L, Hotter D, De Jesus PD, et al. HIV-1 Vpu is a Potent Transcriptional Suppressor of NF-κb-Elicited Antiviral Immune Responses. Elife (2019) 8:e41930. doi: 10.7554/eLife.41930
181. Li Y, To J, Verdià-Baguena C, Dossena S, Surya W, Huang M, et al. Inhibition of the Human Respiratory Syncytial Virus Small Hydrophobic Protein and Structural Variations in a Bicelle Environment. J Virol (2014) 88(20):11899–914. doi: 10.1128/jvi.00839-14
182. Fuentes S, Tran KC, Luthra P, Teng MN, He B. Function of the Respiratory Syncytial Virus Small Hydrophobic Protein. J Virol (2007) 81(15):8361–6. doi: 10.1128/jvi.02717-06
183. Pollock N, Taylor G, Jobe F, Guzman E. Modulation of the Transcription Factor NF-κb in Antigen-Presenting Cells by Bovine Respiratory Syncytial Virus Small Hydrophobic Protein. J Gen Virol (2017) 98(7):1587–99. doi: 10.1099/jgv.0.000855
184. Bao X, Kolli D, Liu T, Shan Y, Garofalo RP, Casola A. Human Metapneumovirus Small Hydrophobic Protein Inhibits NF-kappaB Transcriptional Activity. J Virol (2008) 82(16):8224–9. doi: 10.1128/jvi.02584-07
185. Hastings AK, Amato KR, Wen SC, Peterson LS, Williams JV. Human Metapneumovirus Small Hydrophobic (SH) Protein Downregulates Type I IFN Pathway Signaling by Affecting STAT1 Expression and Phosphorylation. Virology (2016) 494:248–56. doi: 10.1016/j.virol.2016.04.022
186. Lateef Z, Gimenez G, Baker ES, Ward VK. Transcriptomic Analysis of Human Norovirus NS1-2 Protein Highlights a Multifunctional Role in Murine Monocytes. BMC Genomics (2017) 18(1):39. doi: 10.1186/s12864-016-3417-4
187. Ettayebi K, Hardy ME. Norwalk Virus Nonstructural Protein P48 Forms a Complex With the SNARE Regulator VAP-A and Prevents Cell Surface Expression of Vesicular Stomatitis Virus G Protein. J Virol (2003) 77(21):11790–7. doi: 10.1128/jvi.77.21.11790-11797.2003
188. McCune BT, Tang W, Lu J, Eaglesham JB, Thorne L, Mayer AE, et al. Noroviruses Co-Opt the Function of Host Proteins VAPA and VAPB for Replication via a Phenylalanine-Phenylalanine-Acidic-Tract-Motif Mimic in Nonstructural Viral Protein Ns1/2. mBio (2017) 8(4):e00668-17. doi: 10.1128/mBio.00668-17
189. Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. Rotavirus Disrupts Calcium Homeostasis by NSP4 Viroporin Activity. mBio (2010) 1(5)e00265–10. doi: 10.1128/mBio.00265-10
190. Hyser JM, Utama B, Crawford SE, Broughman JR, Estes MK. Activation of the Endoplasmic Reticulum Calcium Sensor STIM1 and Store-Operated Calcium Entry by Rotavirus Requires NSP4 Viroporin Activity. J Virol (2013) 87(24):13579–88. doi: 10.1128/jvi.02629-13
191. Crawford SE, Estes MK. Viroporin-Mediated Calcium-Activated Autophagy. Autophagy (2013) 9(5):797–8. doi: 10.4161/auto.23959
192. Crawford SE, Hyser JM, Utama B, Estes MK. Autophagy Hijacked Through Viroporin-Activated Calcium/Calmodulin-Dependent Kinase Kinase-β Signaling is Required for Rotavirus Replication. Proc Natl Acad Sci U.S.A. (2012) 109(50):E3405–3413. doi: 10.1073/pnas.1216539109
193. Zhou Y, Geng P, Liu Y, Wu J, Qiao H, Xie Y, et al. Rotavirus-Encoded Virus-Like Small RNA Triggers Autophagy by Targeting IGF1R via the PI3K/Akt/mTOR Pathway. Biochim Biophys Acta Mol Basis Dis (2018) 1864(1):60–8. doi: 10.1016/j.bbadis.2017.09.028
194. Kundlacz C, Pourcelot M, Fablet A, Amaral Da Silva Moraes R, Léger T, Morlet B, et al. Novel Function of Bluetongue Virus NS3 Protein in Regulation of the MAPK/ERK Signaling Pathway. J Virol (2019) 93(16)e00336-19. doi: 10.1128/jvi.00336-19
195. Li Z, Lu D, Yang H, Li Z, Zhu P, Xie J, et al. Bluetongue Virus non-Structural Protein 3 (NS3) and NS4 Coordinatively Antagonize Type I Interferon Signaling by Targeting STAT1. Vet Microbiol (2021) 254:108986. doi: 10.1016/j.vetmic.2021.108986
196. Avia M, Rojas JM, Miorin L, Pascual E, Van Rijn PA, Martín V, et al. Virus-Induced Autophagic Degradation of STAT2 as a Mechanism for Interferon Signaling Blockade. EMBO Rep (2019) 20(11):e48766. doi: 10.15252/embr.201948766
197. Merabova N, Kaniowska D, Kaminski R, Deshmane SL, White MK, Amini S, et al. JC Virus Agnoprotein Inhibits In Vitro Differentiation of Oligodendrocytes and Promotes Apoptosis. J Virol (2008) 82(3):1558–69. doi: 10.1128/jvi.01680-07
198. Disbrow GL, Hanover JA, Schlegel R. Endoplasmic Reticulum-Localized Human Papillomavirus Type 16 E5 Protein Alters Endosomal pH But Not Trans-Golgi pH. J Virol (2005) 79(9):5839–46. doi: 10.1128/jvi.79.9.5839-5846.2005
199. Straight SW, Herman B, McCance DJ. The E5 Oncoprotein of Human Papillomavirus Type 16 Inhibits the Acidification of Endosomes in Human Keratinocytes. J Virol (1995) 69(5):3185–92. doi: 10.1128/jvi.69.5.3185-3192.1995
200. Oh JM, Kim SH, Cho EA, Song YS, Kim WH, Juhnn YS. Human Papillomavirus Type 16 E5 Protein Inhibits Hydrogen-Peroxide-Induced Apoptosis by Stimulating Ubiquitin-Proteasome-Mediated Degradation of Bax in Human Cervical Cancer Cells. Carcinogenesis (2010) 31(3):402–10. doi: 10.1093/carcin/bgp318
201. Cortese MS, Ashrafi GH, Campo MS. All 4 Di-Leucine Motifs in the First Hydrophobic Domain of the E5 Oncoprotein of Human Papillomavirus Type 16 are Essential for Surface MHC Class I Downregulation Activity and E5 Endomembrane Localization. Int J Cancer (2010) 126(7):1675–82. doi: 10.1002/ijc.25004
202. Scott ML, Woodby BL, Ulicny J, Raikhy G, Orr AW, Songock WK, et al. Human Papillomavirus 16 E5 Inhibits Interferon Signaling and Supports Episomal Viral Maintenance. J Virol (2020) 94(2):e01582–19. doi: 10.1128/jvi.01582-19
203. Aldabe R, Barco A, Carrasco L. Membrane Permeabilization by Poliovirus Proteins 2B and 2BC. J Biol Chem (1996) 271(38):23134–7. doi: 10.1074/jbc.271.38.23134
204. Pinto LH, Holsinger LJ, Lamb RA. Influenza Virus M2 Protein has Ion Channel Activity. Cell (1992) 69(3):517–28. doi: 10.1016/0092-8674(92)90452-i
205. Nieva JL, Madan V, Carrasco L. Viroporins: Structure and Biological Functions. Nat Rev Microbiol (2012) 10(8):563–74. doi: 10.1038/nrmicro2820
206. Agirre A, Barco A, Carrasco L, Nieva JL. Viroporin-Mediated Membrane Permeabilization. Pore Formation by Nonstructural Poliovirus 2B Protein. J Biol Chem (2002) 277(43):40434–41. doi: 10.1074/jbc.M205393200
207. Wilson L, McKinlay C, Gage P, Ewart G. SARS Coronavirus E Protein Forms Cation-Selective Ion Channels. Virology (2004) 330(1):322–31. doi: 10.1016/j.virol.2004.09.033
208. Wang K, Xie S, Sun B. Viral Proteins Function as Ion Channels. Biochim Biophys Acta (2011) 1808(2):510–5. doi: 10.1016/j.bbamem.2010.05.006
209. Watkins LC, DeGrado WF, Voth GA. Multiscale Simulation of an Influenza A M2 Channel Mutant Reveals Key Features of Its Markedly Different Proton Transport Behavior. J Am Chem Soc (2022) 144(2):769–76. doi: 10.1021/jacs.1c09281
210. Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing Infection of Individual Influenza Viruses. Proc Natl Acad Sci U.S.A. (2003) 100(16):9280–5. doi: 10.1073/pnas.0832269100
211. Pelkmans L, Kartenbeck J, Helenius A. Caveolar Endocytosis of Simian Virus 40 Reveals a New Two-Step Vesicular-Transport Pathway to the ER. Nat Cell Biol (2001) 3(5):473–83. doi: 10.1038/35074539
212. Guinea R, Carrasco L. Phospholipid Biosynthesis and Poliovirus Genome Replication, Two Coupled Phenomena. EMBO J (1990) 9(6):2011–6. doi: 10.1002/j.1460-2075.1990.tb08329.x
213. Bienz K, Egger D, Pfister T. Characteristics of the Poliovirus Replication Complex. Arch Virol Suppl (1994) 9:147–57. doi: 10.1007/978-3-7091-9326-6_15
214. Doedens JR, Kirkegaard K. Inhibition of Cellular Protein Secretion by Poliovirus Proteins 2B and 3A. EMBO J (1995) 14(5):894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x
215. Westerbeck JW, Machamer CE. A Coronavirus E Protein Is Present in Two Distinct Pools With Different Effects on Assembly and the Secretory Pathway. J Virol (2015) 89(18):9313–23. doi: 10.1128/jvi.01237-15
216. Boscarino JA, Logan HL, Lacny JJ, Gallagher TM. Envelope Protein Palmitoylations are Crucial for Murine Coronavirus Assembly. J Virol (2008) 82(6):2989–99. doi: 10.1128/jvi.01906-07
217. DeDiego ML, Alvarez E, Almazán F, Rejas MT, Lamirande E, Roberts A, et al. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene is Attenuated In Vitro and In Vivo. J Virol (2007) 81(4):1701–13. doi: 10.1128/jvi.01467-06
218. Rossman JS, Jing X, Leser GP, Lamb RA. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell (2010) 142(6):902–13. doi: 10.1016/j.cell.2010.08.029
219. Wohlgemuth N, Lane AP, Pekosz A. Influenza A Virus M2 Protein Apical Targeting Is Required for Efficient Virus Replication. J Virol (2018) 92(22):e01425–18. doi: 10.1128/jvi.01425-18
220. Imai M, Kawasaki K, Odagiri T. Cytoplasmic Domain of Influenza B Virus BM2 Protein Plays Critical Roles in Production of Infectious Virus. J Virol (2008) 82(2):728–39. doi: 10.1128/jvi.01752-07
221. Su WC, Yu WY, Huang SH, Lai MMC. Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles. J Virol (2018) 92(4):e01972–17. doi: 10.1128/jvi.01972-17
222. Hu B, Siche S, Möller L, Veit M. Amphipathic Helices of Cellular Proteins Can Replace the Helix in M2 of Influenza A Virus With Only Small Effects on Virus Replication. J Virol (2020) 94(3):e01605–19. doi: 10.1128/jvi.01605-19
223. Chen BJ, Leser GP, Jackson D, Lamb RA. The Influenza Virus M2 Protein Cytoplasmic Tail Interacts With the M1 Protein and Influences Virus Assembly at the Site of Virus Budding. J Virol (2008) 82(20):10059–70. doi: 10.1128/jvi.01184-08
224. Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. Influenza Virus M2 Ion Channel Protein is Necessary for Filamentous Virion Formation. J Virol (2010) 84(10):5078–88. doi: 10.1128/jvi.00119-10
225. Bao D, Lu C, Ma T, Xu G, Mao Y, Xin L, et al. Hydrophobic Residues at the Intracellular Domain of the M2 Protein Play an Important Role in Budding and Membrane Integrity of Influenza Virus. J Virol (2022) 96(9):e0037322. doi: 10.1128/jvi.00373-22
226. Crawford SE, Criglar JM, Liu Z, Broughman JR, Estes MK. COPII Vesicle Transport Is Required for Rotavirus NSP4 Interaction With the Autophagy Protein LC3 II and Trafficking to Viroplasms. J Virol (2019) 94(1):e01341–19. doi: 10.1128/jvi.01341-19
227. Celma CC, Roy P. A Viral Nonstructural Protein Regulates Bluetongue Virus Trafficking and Release. J Virol (2009) 83(13):6806–16. doi: 10.1128/jvi.00263-09
228. Hsu K, Han J, Shinlapawittayatorn K, Deschenes I, Marbán E. Membrane Potential Depolarization as a Triggering Mechanism for Vpu-Mediated HIV-1 Release. Biophys J (2010) 99(6):1718–25. doi: 10.1016/j.bpj.2010.07.027
229. Broz P, Dixit VM. Inflammasomes: Mechanism of Assembly, Regulation and Signalling. Nat Rev Immunol (2016) 16(7):407–20. doi: 10.1038/nri.2016.58
230. Orrenius S, Zhivotovsky B, Nicotera P. Regulation of Cell Death: The Calcium-Apoptosis Link. Nat Rev Mol Cell Biol (2003) 4(7):552–65. doi: 10.1038/nrm1150
231. Zhang X, Paget M, Wang C, Zhu Z, Zheng H. Innate Immune Evasion by Picornaviruses. Eur J Immunol (2020) 50(9):1268–82. doi: 10.1002/eji.202048785
232. Lee HR, Lee GY, You DG, Kim HK, Yoo YD. Hepatitis C Virus P7 Induces Membrane Permeabilization by Interacting With Phosphatidylserine. Int J Mol Sci (2020) 21(3):897. doi: 10.3390/ijms21030897
233. Hyser JM, Estes MK. Pathophysiological Consequences of Calcium-Conducting Viroporins. Annu Rev Virol (2015) 2(1):473–96. doi: 10.1146/annurev-virology-100114-054846
234. Cao Y, Yang R, Wang W, Lee I, Zhang R, Zhang W, et al. Computational Study of the Ion and Water Permeation and Transport Mechanisms of the SARS-CoV-2 Pentameric E Protein Channel. Front Mol Biosci (2020) 7:565797. doi: 10.3389/fmolb.2020.565797
235. Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, et al. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. PloS Pathog (2014) 10(5):e1004077. doi: 10.1371/journal.ppat.1004077
236. Lazrak A, Iles KE, Liu G, Noah DL, Noah JW, Matalon S. Influenza Virus M2 Protein Inhibits Epithelial Sodium Channels by Increasing Reactive Oxygen Species. FASEB J (2009) 23(11):3829–42. doi: 10.1096/fj.09-135590
237. Londino JD, Lazrak A, Jurkuvenaite A, Collawn JF, Noah JW, Matalon S. Influenza Matrix Protein 2 Alters CFTR Expression and Function Through its Ion Channel Activity. Am J Physiol Lung Cell Mol Physiol (2013) 304(9):L582–592. doi: 10.1152/ajplung.00314.2012
238. Madan V, Redondo N, Carrasco L. Cell Permeabilization by Poliovirus 2B Viroporin Triggers Bystander Permeabilization in Neighbouring Cells Through a Mechanism Involving Gap Junctions. Cell Microbiol (2010) 12(8):1144–57. doi: 10.1111/j.1462-5822.2010.01460.x
239. Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 Inflammasome in Inflammatory Diseases. Nat Rev Drug Discovery (2018) 17(8):588–606. doi: 10.1038/nrd.2018.97
240. Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell (2011) 147(2):423–35. doi: 10.1016/j.cell.2011.09.039
241. Jing H, Fang L, Wang D, Ding Z, Luo R, Chen H, et al. Porcine Reproductive and Respiratory Syndrome Virus Infection Activates NOD2-RIP2 Signal Pathway in MARC-145 Cells. Virology (2014) 458-459:162–71. doi: 10.1016/j.virol.2014.04.031
242. Cornell CT, Kiosses WB, Harkins S, Whitton JL. Coxsackievirus B3 Proteins Directionally Complement Each Other to Downregulate Surface Major Histocompatibility Complex Class I. J Virol (2007) 81(13):6785–97. doi: 10.1128/jvi.00198-07
243. Mehnert T, Routh A, Judge PJ, Lam YH, Fischer D, Watts A, et al. Biophysical Characterization of Vpu From HIV-1 Suggests a Channel-Pore Dualism. Proteins (2008) 70(4):1488–97. doi: 10.1002/prot.21642
244. Liu W, Li J, Zheng W, Shang Y, Zhao Z, Wang S, et al. Cyclophilin A-Regulated Ubiquitination is Critical for RIG-I-Mediated Antiviral Immune Responses. Elife (2017) 6:e24425. doi: 10.7554/eLife.24425
245. Javier RT, Rice AP. Emerging Theme: Cellular PDZ Proteins as Common Targets of Pathogenic Viruses. J Virol (2011) 85(22):11544–56. doi: 10.1128/jvi.05410-11
246. Zou J, Yu L, Zhu Y, Yang S, Zhao J, Zhao Y, et al. Transportin-3 Facilitates Uncoating of Influenza A Virus. Int J Mol Sci (2022) 23(8):4128. doi: 10.3390/ijms23084128
247. Zhang Z, Pan L, Ding Y, Lv J, Zhou P, Fang Y, et al. Eef1g Interaction With Foot-and-Mouth Disease Virus Nonstructural Protein 2B: Identification by Yeast Two-Hybrid System. Microb Pathog (2017) 112:111–6. doi: 10.1016/j.micpath.2017.09.039
248. Kuwahara T, Yamayoshi S, Noda T, Kawaoka Y. G Protein Pathway Suppressor 1 Promotes Influenza Virus Polymerase Activity by Activating the NF-κb Signaling Pathway. mBio (2019) 10(6):e02867–19. doi: 10.1128/mBio.02867-19
249. Horova V, Lyoo H, Różycki B, Chalupska D, Smola M, Humpolickova J, et al. Convergent Evolution in the Mechanisms of ACBD3 Recruitment to Picornavirus Replication Sites. PloS Pathog (2019) 15(8):e1007962. doi: 10.1371/journal.ppat.1007962
250. Téoulé F, Brisac C, Pelletier I, Vidalain PO, Jégouic S, Mirabelli C, et al. The Golgi Protein ACBD3, an Interactor for Poliovirus Protein 3A, Modulates Poliovirus Replication. J Virol (2013) 87(20):11031–46. doi: 10.1128/jvi.00304-13
251. Teterina NL, Levenson E, Rinaudo MS, Egger D, Bienz K, Gorbalenya AE, et al. Evidence for Functional Protein Interactions Required for Poliovirus RNA Replication. J Virol (2006) 80(11):5327–37. doi: 10.1128/jvi.02684-05
252. Xu A, Bellamy AR, Taylor JA. Immobilization of the Early Secretory Pathway by a Virus Glycoprotein That Binds to Microtubules. EMBO J (2000) 19(23):6465–74. doi: 10.1093/emboj/19.23.6465
253. Sun L, Hemgård GV, Susanto SA, Wirth M. Caveolin-1 Influences Human Influenza A Virus (H1N1) Multiplication in Cell Culture. Virol J (2010) 7:108. doi: 10.1186/1743-422x-7-108
254. Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, Ludwig S. The Proapoptotic Influenza A Virus Protein PB1-F2 Regulates Viral Polymerase Activity by Interaction With the PB1 Protein. Cell Microbiol (2008) 10(5):1140–52. doi: 10.1111/j.1462-5822.2008.01116.x
255. Mazel-Sanchez B, Boal-Carvalho I, Silva F, Dijkman R, Schmolke M. H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-Mediated Restriction of Avian Virus Polymerase PA in Human Lung Cells. J Virol (2018) 92(11):e00425–18. doi: 10.1128/jvi.00425-18
256. Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC Virus Agno Protein With T Antigen Modulates Transcription and Replication of the Viral Genome in Glial Cells. J Virol (2001) 75(3):1476–86. doi: 10.1128/jvi.75.3.1476-1486.2001
257. Alvarez E, DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, Marcos-Villar L, Enjuanes L. The Envelope Protein of Severe Acute Respiratory Syndrome Coronavirus Interacts With the non-Structural Protein 3 and is Ubiquitinated. Virology (2010) 402(2):281–91. doi: 10.1016/j.virol.2010.03.015
258. Zhu P, Liang L, Shao X, Luo W, Jiang S, Zhao Q, et al. Host Cellular Protein Trappc6aΔ Interacts With Influenza A Virus M2 Protein and Regulates Viral Propagation by Modulating M2 Trafficking. J Virol (2017) 91(1):e01757–16. doi: 10.1128/jvi.01757-16
259. Parr RD, Storey SM, Mitchell DM, McIntosh AL, Zhou M, Mir KD, et al. The Rotavirus Enterotoxin NSP4 Directly Interacts With the Caveolar Structural Protein Caveolin-1. J Virol (2006) 80(6):2842–54. doi: 10.1128/jvi.80.6.2842-2854.2006
260. Fan Y, Mok CK, Chan MC, Zhang Y, Nal B, Kien F, et al. Cell Cycle-Independent Role of Cyclin D3 in Host Restriction of Influenza Virus Infection. J Biol Chem (2017) 292(12):5070–88. doi: 10.1074/jbc.M117.776112
261. Regan JA, Laimins LA. Bap31 is a Novel Target of the Human Papillomavirus E5 Protein. J Virol (2008) 82(20):10042–51. doi: 10.1128/jvi.01240-08
262. Lim KP, Liu DX. The Missing Link in Coronavirus Assembly. Retention of the Avian Coronavirus Infectious Bronchitis Virus Envelope Protein in the Pre-Golgi Compartments and Physical Interaction Between the Envelope and Membrane Proteins. J Biol Chem (2001) 276(20):17515–23. doi: 10.1074/jbc.M009731200
263. Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset FL, et al. The SARS-CoV-2 Envelope and Membrane Proteins Modulate Maturation and Retention of the Spike Protein, Allowing Assembly of Virus-Like Particles. J Biol Chem (2021) 296:100111. doi: 10.1074/jbc.RA120.016175
264. Padhan K, Tanwar C, Hussain A, Hui PY, Lee MY, Cheung CY, et al. Severe Acute Respiratory Syndrome Coronavirus Orf3a Protein Interacts With Caveolin. J Gen Virol 88(Pt (2007) 11):3067–77. doi: 10.1099/vir.0.82856-0
265. McNatt MW, Zang T, Bieniasz PD. Vpu Binds Directly to Tetherin and Displaces it From Nascent Virions. PloS Pathog (2013) 9(4):e1003299. doi: 10.1371/journal.ppat.1003299
266. Kondratova AA, Neznanov N, Kondratov RV, Gudkov AV. Poliovirus Protein 3A Binds and Inactivates LIS1, Causing Block of Membrane Protein Trafficking and Deregulation of Cell Division. Cell Cycle (2005) 4(10):1403–10. doi: 10.4161/cc.4.10.2041
267. Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe (2015) 18(6):723–35. doi: 10.1016/j.chom.2015.11.002
268. Ma H, Kien F, Manière M, Zhang Y, Lagarde N, Tse KS, et al. Human Annexin A6 Interacts With Influenza a Virus Protein M2 and Negatively Modulates Infection. J Virol (2012) 86(3):1789–801. doi: 10.1128/jvi.06003-11
269. Hu S, Yin L, Mei S, Li J, Xu F, Sun H, et al. BST-2 Restricts IAV Release and is Countered by the Viral M2 Protein. Biochem J (2017) 474(5):715–30. doi: 10.1042/bcj20160861
270. Liu X, Xu F, Ren L, Zhao F, Huang Y, Wei L, et al. MARCH8 Inhibits Influenza A Virus Infection by Targeting Viral M2 Protein for Ubiquitination-Dependent Degradation in Lysosomes. Nat Commun (2021) 12(1):4427. doi: 10.1038/s41467-021-24724-2
271. Wirblich C, Bhattacharya B, Roy P. Nonstructural Protein 3 of Bluetongue Virus Assists Virus Release by Recruiting ESCRT-I Protein Tsg101. J Virol (2006) 80(1):460–73. doi: 10.1128/jvi.80.1.460-473.2006
272. Celma CC, Roy P. Interaction of Calpactin Light Chain (S100A10/p11) and a Viral NS Protein is Essential for Intracellular Trafficking of Nonenveloped Bluetongue Virus. J Virol (2011) 85(10):4783–91. doi: 10.1128/jvi.02352-10
273. Sarkar R, Nandi S, Lo M, Gope A, Chawla-Sarkar M. Viperin, an IFN-Stimulated Protein, Delays Rotavirus Release by Inhibiting Non-Structural Protein 4 (NSP4)-Induced Intrinsic Apoptosis. Viruses (2021) 13(7):1324. doi: 10.3390/v13071324
274. Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, et al. Dissociation of Heterochromatin Protein 1 From Lamin B Receptor Induced by Human Polyomavirus Agnoprotein: Role in Nuclear Egress of Viral Particles. EMBO Rep (2005) 6(5):452–7. doi: 10.1038/sj.embor.7400406
275. Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, et al. Identification of FEZ1 as a Protein That Interacts With JC Virus Agnoprotein and Microtubules: Role of Agnoprotein-Induced Dissociation of FEZ1 From Microtubules in Viral Propagation. J Biol Chem (2005) 280(26):24948–56. doi: 10.1074/jbc.M411499200
276. Gordon-Shaag A, Yosef Y, Abd El-Latif M, Oppenheim A. The Abundant Nuclear Enzyme PARP Participates in the Life Cycle of Simian Virus 40 and is Stimulated by Minor Capsid Protein VP3. J Virol (2003) 77(7):4273–82. doi: 10.1128/jvi.77.7.4273-4282.2003
277. Teoh KT, Siu YL, Chan WL, Schlüter MA, Liu CJ, Peiris JS, et al. The SARS Coronavirus E Protein Interacts With PALS1 and Alters Tight Junction Formation and Epithelial Morphogenesis. Mol Biol Cell (2010) 21(22):3838–52. doi: 10.1091/mbc.E10-04-0338
278. Fielding BC, Gunalan V, Tan TH, Chou CF, Shen S, Khan S, et al. Severe Acute Respiratory Syndrome Coronavirus Protein 7a Interacts With hSGT. Biochem Biophys Res Commun (2006) 343(4):1201–8. doi: 10.1016/j.bbrc.2006.03.091
279. Li Y, Jain N, Limpanawat S, To J, Quistgaard EM, Nordlund P, et al. Interaction Between Human BAP31 and Respiratory Syncytial Virus Small Hydrophobic (SH) Protein. Virology (2015) 482:105–10. doi: 10.1016/j.virol.2015.03.034
280. Liao J, Guan Y, Chen W, Shi C, Yao D, Wang F, et al. ACBD3 is Required for FAPP2 Transferring Glucosylceramide Through Maintaining the Golgi Integrity. J Mol Cell Biol (2019) 11(2):107–17. doi: 10.1093/jmcb/mjy030
281. Greninger AL, Knudsen GM, Betegon M, Burlingame AL, Derisi JL. The 3A Protein From Multiple Picornaviruses Utilizes the Golgi Adaptor Protein ACBD3 to Recruit PI4KIIIβ. J Virol (2012) 86(7):3605–16. doi: 10.1128/jvi.06778-11
282. Sapin C, Colard O, Delmas O, Tessier C, Breton M, Enouf V, et al. Rafts Promote Assembly and Atypical Targeting of a Nonenveloped Virus, Rotavirus, in Caco-2 Cells. J Virol (2002) 76(9):4591–602. doi: 10.1128/jvi.76.9.4591-4602.2002
283. Pinto LH, Lamb RA. The M2 Proton Channels of Influenza A and B Viruses. J Biol Chem (2006) 281(14):8997–9000. doi: 10.1074/jbc.R500020200
284. Sugrue RJ, Bahadur G, Zambon MC, Hall-Smith M, Douglas AR, Hay AJ. Specific Structural Alteration of the Influenza Haemagglutinin by Amantadine. EMBO J (1990) 9(11):3469–76. doi: 10.1002/j.1460-2075.1990.tb07555.x
285. Li N, Zhang Y, Wu S, Xu R, Li Z, Zhu J, et al. Tauroursodeoxycholic Acid (TUDCA) Inhibits Influenza A Viral Infection by Disrupting Viral Proton Channel M2. Sci Bull (Beijing) (2019) 64(3):180–8. doi: 10.1016/j.scib.2018.08.013
286. Wilson L, Gage P, Ewart G. Hexamethylene Amiloride Blocks E Protein Ion Channels and Inhibits Coronavirus Replication. Virology (2006) 353(2):294–306. doi: 10.1016/j.virol.2006.05.028
287. Scott C, Kankanala J, Foster TL, Goldhill DH, Bao P, Simmons K, et al. Site-Directed M2 Proton Channel Inhibitors Enable Synergistic Combination Therapy for Rimantadine-Resistant Pandemic Influenza. PloS Pathog (2020) 16(8):e1008716. doi: 10.1371/journal.ppat.1008716
288. Gordon NA, McGuire KL, Wallentine SK, Mohl GA, Lynch JD, Harrison RG, et al. Divalent Copper Complexes as Influenza A M2 Inhibitors. Antiviral Res (2017) 147:100–6. doi: 10.1016/j.antiviral.2017.10.009
289. Fett C, DeDiego ML, Regla-Nava JA, Enjuanes L, Perlman S. Complete Protection Against Severe Acute Respiratory Syndrome Coronavirus-Mediated Lethal Respiratory Disease in Aged Mice by Immunization With a Mouse-Adapted Virus Lacking E Protein. J Virol (2013) 87(12):6551–9. doi: 10.1128/jvi.00087-13
290. Lu S, Xie XX, Zhao L, Wang B, Zhu J, Yang TR, et al. The Immunodominant and Neutralization Linear Epitopes for SARS-CoV-2. Cell Rep (2021) 34(4):108666. doi: 10.1016/j.celrep.2020.108666
Keywords: viroporins, function, viral life cycle, host cell response, interactions, inhibitors
Citation: Xia X, Cheng A, Wang M, Ou X, Sun D, Mao S, Huang J, Yang Q, Wu Y, Chen S, Zhang S, Zhu D, Jia R, Liu M, Zhao X-X, Gao Q and Tian B (2022) Functions of Viroporins in the Viral Life Cycle and Their Regulation of Host Cell Responses. Front. Immunol. 13:890549. doi: 10.3389/fimmu.2022.890549
Received: 06 March 2022; Accepted: 10 May 2022;
Published: 02 June 2022.
Edited by:
Pablo Alberto González, Pontificia Universidad Católica de Chile, ChileReviewed by:
Joseph Hyser, Baylor College of Medicine, United StatesHans-Georg Breitinger, German University in Cairo, Egypt
Copyright © 2022 Xia, Cheng, Wang, Ou, Sun, Mao, Huang, Yang, Wu, Chen, Zhang, Zhu, Jia, Liu, Zhao, Gao and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingshu Wang, bXNod2FuZ0AxNjMuY29t
†These authors have contributed equally to this work
 Xiaoyan Xia
Xiaoyan Xia Anchun Cheng
Anchun Cheng Mingshu Wang
Mingshu Wang Xumin Ou
Xumin Ou Di Sun1,2,3
Di Sun1,2,3 Sai Mao
Sai Mao Juan Huang
Juan Huang Qiao Yang
Qiao Yang Shun Chen
Shun Chen Shaqiu Zhang
Shaqiu Zhang Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Qun Gao
Qun Gao Bin Tian
Bin Tian