
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 06 May 2022
Sec. T Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.890056
This article is part of the Research Topic Molecular Switches of the Immune System: The E Protein/Id Axis in Hematopoietic Development and Function View all 11 articles
Immune responses are primarily mediated by adaptive and innate immune cells. Adaptive immune cells, such as T and B cells, evoke antigen-specific responses through the recognition of specific antigens. This antigen-specific recognition relies on the V(D)J recombination of immunoglobulin (Ig) and T cell receptor (TCR) genes mediated by recombination-activating gene (Rag)1 and Rag2 (Rag1/2). In addition, T and B cells employ cell type-specific developmental pathways during their activation processes, and the regulation of these processes is strictly regulated by the transcription factor network. Among these factors, members of the basic helix-loop-helix (bHLH) transcription factor mammalian E protein family, including E12, E47, E2-2, and HEB, orchestrate multiple adaptive immune cell development, while their antagonists, Id proteins (Id1-4), function as negative regulators. It is well established that a majority of T and B cell developmental trajectories are regulated by the transcriptional balance between E and Id proteins (the E-Id axis). E2A is critically required not only for B cell but also for T cell lineage commitment, whereas Id2 and Id3 enforce the maintenance of naïve T cells and naïve regulatory T (Treg) cells. Here, we review the current knowledge of E- and Id-protein function in T cell lineage commitment and Treg cell differentiation.
Innate immune cells and adaptive lymphocytes cooperatively evoke immune responses aimed at protecting our bodies from invasion of the pathogens. Innate immune cells, such as macrophages, neutrophils, and dendritic cells, are activated by pattern recognition receptors (PRRs) that recognize microbial components. On the other hand, adaptive lymphocyte T and B cells recognize specific antigens through diverse antigen receptors. This specific immune response relies on the V(D)J recombination of the immunoglobulin (Ig) and T cell receptor (TCR) genes mediated by the recombination-activating gene (Rag1/2). The assembly of the TCR and Ig genes from the arrays of variable (V), diversity (D), and joining (J) gene segments is initiated by a Rag1 and Rag2 protein complex, which recognizes and cleaves the recombination signal sequences (RSSs) flanking the V, D, and J segments of the Ig and TCR genes (1, 2). The expression of the Rag1/2 genes is stringently controlled. These genes are expressed only in T and B progenitor/precursor cells, meaning that Rag1/2 expression is a hallmark of the adaptive lymphocyte lineage.
Common lymphoid progenitors (CLPs) can give rise to T cells, B cells, innate lymphoid cells (ILCs) including natural killer (NK) cells, and dendritic cells (DCs). Once lymphoid progenitors from the fetal liver or bone marrow (BM) migrate into the thymus, they receive Notch1 receptor signaling through the interaction with Delta-like 4 (DL4)-expressing thymic epithelial cells and commit to the T cell lineage (3–6). After T cell lineage commitment, TCRβ and/or TCRγ/δ V(D)J gene recombination is initiated in immature CD4–CD8– (double negative; DN) cells. DN cells are divided into multiple distinct stages distinguished by surface expression of CD44 and CD25 (DN1-4). In DN1 cells, early T cell progenitors (ETPs) are defined by CD25–CD44+KIThi expression, and committed T progenitor (pro-T) cells start expressing CD25 (DN2) since CD25 is a direct target of Notch signaling. Following the success of productive TCRβ recombination in DN3 cells (CD44–CD25+), DN3 cells start proliferating and differentiating into DN4 cells and further into CD4+CD8+ (double positive; DP) cells (T precursor (pre-T) cells). Recombination of the TCRγ/δ gene occurs concurrently with TCRβ recombination in DN2-3 cells (7). Upon reaching the DP stage, thymocytes exit the cell cycle (resting DP cells) and start TCRα VJ recombination (8, 9). DP cells that succeed in the production of a functional TCRα/β undergo positive and negative selection, which permits the developmental progression of T cells that have acquired a TCR with moderate affinity for self-antigens associated with major histocompatibility complex (MHC) class I (for CD8 single-positive (CD8SP) cells) or class II (for CD4SP cells) (10). The population of CD4SP cells that react more strongly with self-antigens associated with the MHC in the thymus differentiates into distinct regulatory T cells (Tregs), which specifically express the transcription factor (TF) Foxp3 and play an indispensable role in suppressing autoimmunity and excessive immune responses (11). On the other hand, innate type of T cells also arise from DP cells, which are selected by CD1 for invariant natural killer T (iNKT) cells and by MHC-related protein MR1 for mucosal-associated invariant T (MAIT) cells (12, 13). In these processes, sequential expression of an ensemble of TFs specifies the lineage-specific gene expression program and function through the regulation of the enhancer repertoire and activities (14, 15). However, the precise molecular mechanisms of how lineage-specific TFs synergistically regulate enhancer activities and how these factors cooperatively orchestrate the changes in chromatin architecture for appropriate gene expression remain unclear.
E proteins are basic helix-loop-helix (bHLH) TFs involved in multiple hematopoietic developmental processes, and mammalian E proteins include E12, E47 (from the E2A;Tcf3 gene), E2-2 (Tcf4), and HEB (Tcf12). E proteins bind to the E-box motif (CANNTG) within the cis-regulatory element (CRE, enhancer region) of the target genes by forming homodimers or heterodimers. In contrast, Id proteins contain an HLH dimerization domain but lack the basic region that is required for DNA binding and form heterodimers with E proteins, antagonizing the DNA binding activity of E proteins and functioning as negative regulators of E proteins (16–18). Id proteins include Id1, Id2, Id3 and Id4, and hematopoietic cells primarily express Id2 and Id3. It is well established that the E and Id protein axis (the E-Id axis) regulates developmental trajectories of adaptive lymphocytes (19–21). The E2A gene encodes the E12 and E47 proteins, and E47 primarily regulates B cell lineage commitment, along with Ebf1, Pax5, and Foxo1 (22, 23). For T cell lineage commitment, E2A acts in pro-T cells along with HEB to establish a T cell-specific gene expression program and to suppress ILC development (24–28). HEB is also required for iNKT cell development from DP cells (29), and HEB and E2A play an important role in positive selection of DP cells (30). In contrast, Id3 is upregulated by pre-TCR and γδ TCR signaling through ERK-MAPK, Egr1, and NFAT and plays a central role in αβ/γδ T cell fate and maturation (31–33). Furthermore, a recent report revealed the importance of the Notch-E2A-Tcf1 axis in αβ versus γδT cell lineage bifurcation and γδT cell function (34). In addition, E2-2 is critically required for interferon-producing plasmacytoid DC (pDC) development, while Id2 regulates antigen-presenting classical DC (cDC) development by neutralizing E2-2 activity (35–37). Furthermore, Id2 is well known as a critical regulator of the development of all ILC subsets, including ILC1-3s, NK cells, and lymphoid tissue inducer (LTi) cells (38, 39).
Many reviews describing the role of the E-Id axis have focused on the lineage commitment of T and B cells and DCs and on development of conventional T cells, NK cells, γδT cells, and iNKT cells. In this review we focus on the roles of the E-Id axis in T cell lineage commitment, including adaptive versus innate lymphoid cells, and during Treg cell differentiation.
ILCs are a family of lymphocytes that do not have diversified antigen recognition receptors, such as Ig and TCR, and that primarily reside in various tissues and respond to infection, injury and damage (40). ILCs modulate immune responses and contribute to the maintenance of tissue homeostasis by sustaining appropriate immune responses at mucosal barriers and by enhancing immune responses through secretion of inflammatory cytokines. Functional similarities regulated by a common set of specific TFs may suggest that ILCs are the innate counterparts of T cells. ILCs can be segregated into distinct classes according to effector cytokine secretion and expression of specific TFs. ILC1s, including NK cells, are characterized by secretion of interferon-γ (IFN- γ) and expression of the specific TF T-bet. ILC2s express the TF Gata3 and Th2 cytokines (interleukin-4 (IL-4), IL-5, and IL-13). ILC3s, including LTi-like cells, express Rorγt and secrete IL-17/IL-22 and lymphotoxin (40, 41). Therefore, ILC1s, ILC2s, and ILC3s are counterparts of CD4 helper TH1, TH2, and TH17 cells, respectively, while NK cells mirror CD8 cytotoxic T cells. As well as adaptive T and B lymphocytes, ILCs develop from common lymphoid progenitors (CLPs), and lineage commitment into ILCs is regulated by sequential expression of an ensemble of TFs, including Nfil3, Tox, Id2, Tcf1, and Gata3 (42–48). In addition, PLZF in ILC precursors (ILCp), Bcl11b and Rorα in ILC2s, and Runx3 in ILC1s/3s are required for this process (49–52). In particular, it is well known that Gata3, Tcf1, and Bcl11b are also required for early T cell development (3, 53). These observations clearly show close similarities between ILC and T cell lineages not only in effector function but also in their development, and a combination of these shared TFs determines effector functions in each lineage of ILCs after passing the developmental bifurcation of adaptive and innate lymphoid lineage commitment. However, how these shared TFs play their distinct roles in early T cell and ILC development remains to be clarified. Therefore, it is important to understand what events result in the differences between T cells and ILCs during their development.
ILCs are derived from CLPs in the fetal liver (FL) and adult bone marrow (BM), and differentiate into functional mature ILCs in the resident tissues, while CD4 helper T cells and CD8 cytotoxic T cells mature in the thymus. The frequencies of ILCs, including mature Id2- and Gata3-expressing ILC2s and PLZF-expressing ILCps, are considerably low in the thymus of normal adult mice (54), because the majority of thymocytes in adult thymus are developing T cells. Consistent with a report that Rag1/2-mediated TCR recombination is dispensable for ILC development (55, 56), we and another group observed both the absence of D-J and V-DJ recombination of the TCRβ gene in ILC2s from wild-type lung tissue and aberrant ILC2s in the thymus from E2A/HEB-deficient mice (28, 57). According to these observations, the cell fate of the T versus ILC lineage must be principally determined by the thymic microenvironment. Notch signaling is one of the most likely external or environmental factors that distinguish T cells from the ILC lineage. In the absence of DL4 in thymic stromal cells, aberrant ILC2s are observed in the thymus, and constitutive Notch signaling completely blocks the ILC lineage in vivo. However, the proliferation of committed ILC precursors require mild to moderate Notch signaling, and short exposure to a Notch ligand combined with a high amount of IL-7 in CLPs leads to ILC2 generation in vitro (6, 58). Interestingly, recent studies have revealed an unexpectedly close relationship between T cells and ILCs (57, 59). Specifically, ILCps in BM express high levels of TCRβ constant region transcripts, and a proportion of tissue-resident ILC2s have undergone TCRγ gene recombination and express high levels of mRNAs of TCRβ and TCRγ4 constant regions (Cβ1/2 and Cγ4); however, the frequency of these TCRγ gene recombination is low, compared to that in γδT cells, and the recombination in these cells are nonfunctional (28, 57). Consistent with this observation, a high level of mRNA expression and broad chromatin accessibility in the TCRβ constant region with little or no expression of any TCR Vβ region in E2A/HEB-deficient ETPs, which tend toward an aberrant ILC lineage, were detected (28). According to these observations, T precursor cells that fail to properly undergo TCR recombination, especially TCRγ/δ recombination, may be able to convert their cell lineage into ILCs (56, 57). However, the numbers of mature ILC2s and PLZF-expressing ILCps in Rag2-deficient thymuses remain low; this phenomenon cannot explain why TCRγ/δ genes, but not TCRβ D-J gene, recombination are observed in ILCs, although TCRβ D-J and TCRγ/δ recombination occurs concurrently in the DN2 stage (28, 57). In contrast to these reports, the Sun group demonstrated that ILC2s in the thymus and lug from wild-type and E2A/HEB deletion (plck-Cre) mice, but not from Id1-transgenic (Id1-Tg) mice, exhibited TCRβ D-J and V-DJ gene recombination, which are detected by Southern blotting, and estimated that around 10% of ILC2s performed these recombination (60). In this report, even committed DN3 cells have a potential to differentiate into ILC2s in vitro, suggesting the lineage conversion of T cells to ILCs (60). Although these phenomena remain puzzling, T cells and ILCs are very close counterparts, and Rag1/2-mediated TCRβ recombination and its expression seem to be functional hallmarks of physiological T cell lineage commitment in vivo. A recent study provided an important clue regarding the mystery of the checkpoint for T cells and ILC2s in the thymus (61). During embryogenesis, functional ILC2s differentiate from ETPs in the fetal thymus, and these ILC2s preferentially migrate to mucosal tissues and reside for a long period. In this time-restricted thymic ILC2 development, specific TF RORα is the key factor that promotes ILC2 development and simultaneously suppresses the T cell lineage program by inducing Id2 expression, leading to E2A function antagonism (61, 62). This study demonstrated that ILC2 development in E2A/HEB-deficient mice does not represent simple aberrant ILC development and instead may be an implication of the physiological embryonic thymocyte development toward the ILC2 lineage. Although Id2 expression is a critical regulator of the ILC lineage, Id2 deletion in E2A/HEB deficiency leads to thymic ILC development as well as E2A/HEB deficiency, and transient Id2 expression induced by doxycycline can induce aberrant ILC2 development in adult thymus. Thus, T cell and ILC lineages may simply depend on the magnitude of E protein activity, and Id2 may function as a lineage switch for ILCs (28). Therefore, we conclude that after the enhancer repertoire associated with each lineage regulated by the E-Id axis is established, an ensemble of shared TFs, such as Tcf1, Bcl11b, and Gata3, instructs the lineage-specific gene expression programs in both T cells and ILCs (Figure 1). Indeed, Bcl11b binds to different sites in a lineage-specific manner associated with cell type-specific protein complexes (63). Interestingly, some members of these factors are dynamically recruited to the regulatory regions not only in a lineage-specific manner but also in a developmental stage-specific manner (64).
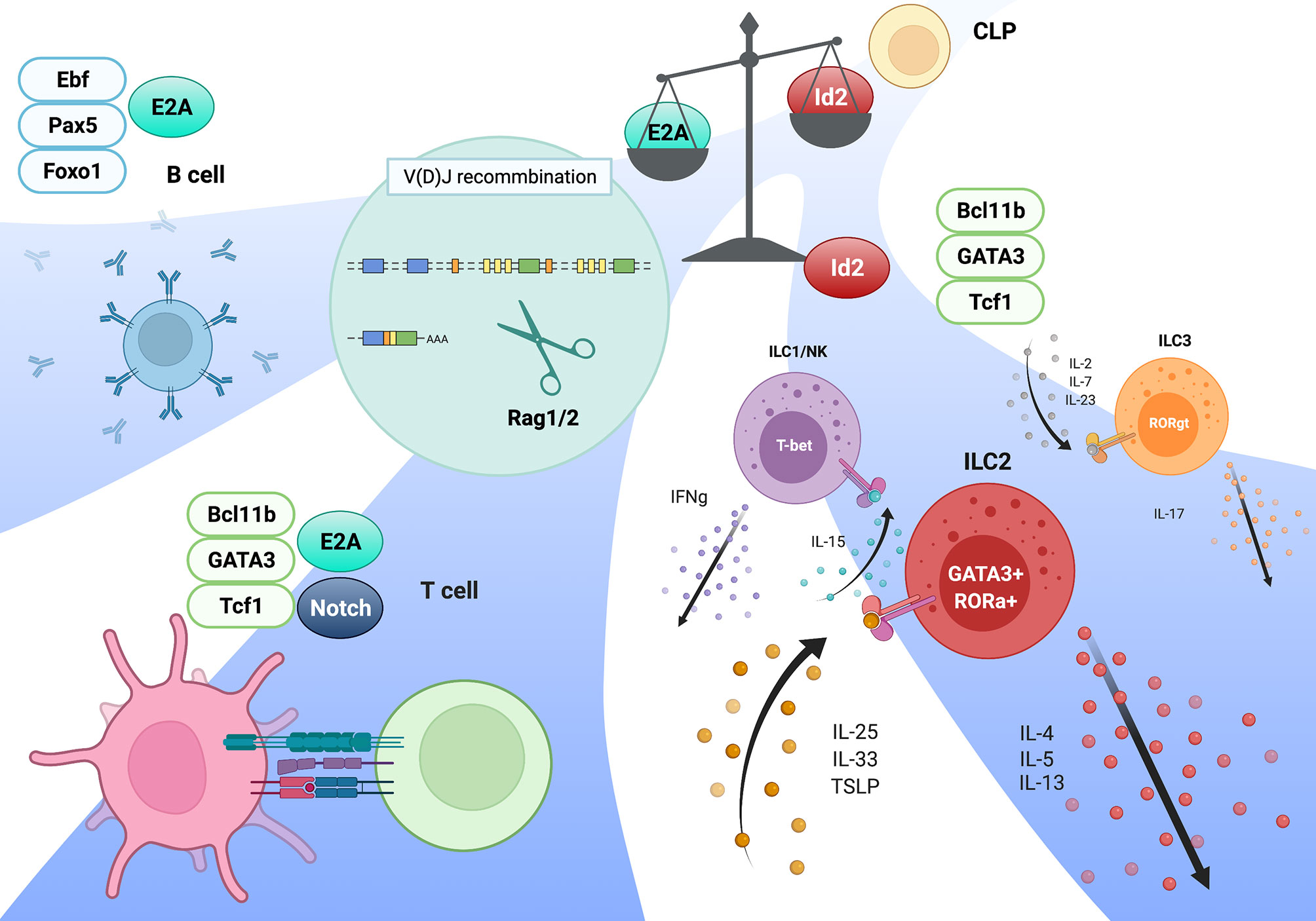
Figure 1 Model of adaptive and innate lymphocytes lineages mediated by the E-Id axis. The magnitude of E protein transcriptional activity determines the lineage commitments of adaptive versus innate lymphocytes. Following this process, an ensemble of TFs specific for each lineages validates lineage-specific gene expression program, along with E proteins in T and B cells. This figure was created with BioRender.com.
However, it remains unclear whether the loss of E protein activity in ETPs induces only ILC lineage commitment or also leads to the expansion of ILC precursors or mature ILCs. Since Id2 is continuously expressed at high levels after ILC lineage commitment, the magnitude of E protein activity may control not only the ILC versus T lineage commitment but also the expansion or activation of ILCs after the commitment, which is antagonized by Id2. Because E2A functions both as an initiator upon T cell lineage commitment and as a gatekeeper at β-selection (65), the loss of E protein activity in ILCs may play a role in the activation or expansion of ILCs.
How is the E-Id axis regulated? While E2A and HEB mRNA expression levels are relatively consistent throughout the thymocyte development (ImmGen data; https://www.immgen.org/), the E2A protein level is high in ETPs and is the highest in DN2 cells; this level is downregulated in resting DP cells, as revealed by E2A-GFP fusion knock-in mouse analysis, indicating the presence of posttranslational regulation of the E2A protein (66–68). On the other hand, Id3 is upregulated by TCR signaling, including pre- and γδ-TCR, during thymocyte development and remains at a high level in peripheral naïve T and Treg cells (32, 67, 69). In peripheral T cells, TCR stimulation induces E2A protein expression, which is required for rapid memory-precursor formation of CD8 T cells, while Id2 and 3 function as regulators of CD8 T cell responses (70). Surprisingly, differential Id2 and Id3 expression in CD4 T cells during viral infection regulates TH1 or TFH cell development, respectively (71). During ILC lineage commitment, Id2 is initially upregulated in PLZF-expressing ILC precursors in which E2A protein is already downregulated, and this induction of Id2 expression is associated with the IL7R expression level, suggesting the involvement of cytokine signaling in Id2 expression (28). Consistently, the cis-regulatory element of the Id2 gene, which expresses the long noncoding RNA Rroid, controls ILC1 function by regulating Stat5 deposition at the Id2 promoter region; however, this locus is dispensable for Id2 expression in other ILCs (72). Therefore, Id2 expression in ILC lineages, which is probably mediated by cytokine signaling, is required not only for ILC lineage commitment but also for ILC maintenance.
As we discussed in the introduction, Rag1/2 gene expression discriminates between adaptive and innate lymphoid lineages. This indicates that TFs responsible for Rag1/2 expression are critical regulators of T and B lineage commitment (73). There are two waves of Rag1/2 expression during T and B cell development (74). The first wave of Rag1/2 expression is required for the assembly of IgH and TCRβ genes in pro-B and pro-T cells, respectively. After the selection of pre-TCR (TCRβ) or pre-BCR (IgH), Rag1 expression is transiently downregulated during the transition from the progenitors to precursors. In the precursor stage, Rag1/2 are re-expressed for IgL and TCRα gene recombination. Following the positive and negative selection of the TCR or BCR, the Rag1/2 genes are suppressed in mature naïve T and B cells and are never expressed for further recombination of the TCR and Ig genes. During these developmental processes, Rag1/2 gene expression is tightly regulated, and other types of immune cells never express the Rag1/2 genes. However, the molecular mechanisms of Rag1/2 gene expression remained to be determined. Both in vivo and in vitro studies have attempted to define the enhancer regions and TFs responsible for Rag1/2 expression (75). Both T and B progenitor/precursor cells express Rag1/2 and require distinct enhancers of these genes. The deletion of Erag (Enhancer of Rag), which is located at 23 kb upstream of the Rag2 promoter, resulted in a significant reduction in Rag1/2 expression and partial developmental defects during B cell development, without affecting thymocyte development (76). A study has reported that this Erag region is positively regulated by Foxo1 and negatively regulated by Gfi1b, Ebf1, and c-Myb (77–80). In contrast, an anti-silencer element (ASE), which is 8 kb in length and located 73 kb upstream of the Rag2 promoter, is required for Rag1/2 gene expression in DN3 and DP cells but not in developing B cells (81). In ChIP-seq data, most of T cell TFs includng E2A, Bcl111b, Tcf1, Gata3, Runx1, and Ikaros bound to ASE regions, while B cell TFs such as E2A, Pax5, and Irf4, but not Ebf1, bound to Erag region (82, 84).
The Krangel group demonstrated that the chromatin organizer mediates the interaction between ASE and Rag1/2 promoters to promote optimal expression of the Rag1/2 genes in DP cells and suggested that the ASE and Rag1 promoter regions function as a chromatin hub (82). Furthermore, this group proved that Gata3 and E2A regulate the ASE region, and Rag1 promoter activity relies on Runx1 and E2A binding in the VL3-3M2DP thymocyte cell line (83). A study also identified T or B cell-specific enhancer elements that drive Rag1/2 expression using the E2A ChIP-seq and ATAC-seq data from pro-T and pro-B cells to clarify the regulatory mechanisms of adaptive versus innate lineage choice. Two B cell-specific enhancers (Rag B cell enhancer 1 and 2; R1B (5 kb upstream of the Rag1 promoter) and R2B (partially overlapping with Erag)) and one T cell-specific enhancer (Rag-T cell enhancer (R-TEn)) were identified (84). A common E2A-binding region near the Rag1 promoter (R1pro), which is shared between T and B cells, was also identified. R1B/R2B and R-TEn uniquely bind to the Rag1/Rag2 promoter regions and form distinct chromatin structures in developing T and B cells, respectively. Deletion of both R1B and R2B in mice resulted in a severe developmental block at the pro-B stage, but not in T-cell development, resulting from drastic impairments in Rag-mediated IgH gene recombination, whereas single deletion of either R1B or R2B resulted in mild-to-moderate defects in B cell development that also occurred in Erag deletion mice (76, 84). This finding suggests enhancer redundancy in Rag1/2 expression in B cells. In contrast, R-TEn deletion resulted in severe developmental defects in β-selection of DN3 cells and positive selection of DP cells without affecting B cell development (84). These results raised the question of what TF regulates these Rag gene enhancer regions.
E2A is especially notable among TFs responsible for adaptive lymphocyte development because Rag1/2 gene expression was significantly reduced in E2A-deficient lymphoid-primed multipotent progenitors (LMPPs) and T progenitor cells (28, 85, 86). A mutation of the E-box motifs in the R-TEn (R-TEn-E-box-mutant), which blocks E-protein binding without affecting the recruitment of other TFs to this enhancer, directly proves that the E2A/E protein regulates this enhancer. R-TEn-E-box-mutant mice showed developmental defects in β-selection and positive selection, resulting from a severe reduction in Rag1/2 gene expression in DN3 and DP cells. Furthermore, genome structures, chromatin accessibility, histone H3 lysine K27 acetylation (H3K27ac), and cohesin recruitment were completely lost only at the Rag gene locus, indicating that the E2A/E protein binding to the enhancer region induces and promotes cell type-specific superenhancer (SE) formation (84). How does the E2A/E protein induce SE formation? bHLH TFs, such as E2A, were reported to interact with the histone acetyltransferase (HAT) CBP/P300 and SAGA proteins through the PECT motif within the activation domain 1 (AD1) of the E protein and recruit these coactivators to enhancer regions, thus inducing and promoting H3K27 acetylation (87–91). Active enhancers are accompanied by high levels of H3K27ac, CBP/P300, chromatin remodeler Brg1, and RNA polymerase II (PolII) to facilitate the recruitment of cohesin-loader NIPBL and the cohesin complex, which induce large-scale structural changes of the chromatin and may switch the locus from transcriptionally repressive (B) to permissive (A) compartments (92, 93). Simultaneously, E2A and other specific TFs also recruit the ten-eleven translocation (TET) family proteins to the enhancers to remove DNA methylation of the CpG islands in enhancers, which is associated with the SE function in developing and activated B cells (94, 95). SEs regulate certain genes that play characteristic roles in cell type-specific functions, thereby establishing cell identity (96, 97). Because the properties of SEs are based on highly cooperative interactions between cell type-specific TFs, transcriptional mediators, and RNA PolII and due to vulnerability to a perturbation of the key protein components (98), E2A functions in adaptive lymphocyte-specific enhancer regions as a pioneer and maintainer. Additionally, E2A plays an essential role in Rag1 expression in vivo through the regulation of the promoter activity. Surprisingly, E-box motif mutations in the Rag1-promoter region (R1pro-E-box-mutant) alone in mice are sufficient to inhibit the Rag1 gene expression, which leads to the developmental arrest at both the T and B cell progenitor stages, similar to those in Rag1-deficient mice. However, Rag2 expression and enhancer regions (R-TEn and R1B/R2B) are not affected in R1pro-E-box-mutant DN3 and pro-B cells (84). This result indicates that both cell type-specific enhancer and promoter regions independently rely on the recruitment of the E2A/E protein and that E protein-mediated interactions between enhancer and promoter regions determine adaptive lymphocyte-specific expression of the Rag gene. We summarised these regulatory regions in Table 1.
Overall, the binding of the E2A/E proteins to the E-box motifs in the cell type-specific cis-regulatory regions induces the recruitment of P300, other transcription mediators, the NIPBL/cohesin-complex, and chromatin organizers to orchestrate 3D structural changes of the genomes to initiate and maintain cell type-specific gene expression. In contrast, high expression levels of Id2 prevents Rag gene SE formation by antagonizing the E2A activity, and the Rag gene is sequestered in repressive chromatin (B) compartment (Figure 2). Curiously, sequence similarities of T and B cell-specific Rag gene enhanceres are conserved among mammals, birds and reptiles, but not in amphibians and fish. In addition, these conserved enhancer regions have been shown to harbor the E-box motifs conserved among these species (84). Thus, we propose that terrestrial animals evolutionarily acquired the gene regulatory mechanism mediated by the E proteins as enhancers to achieve higher Rag gene expression, which enables a diverse range of TCR and Ig gene recombination to protect against a wide range of the pathogens (99).
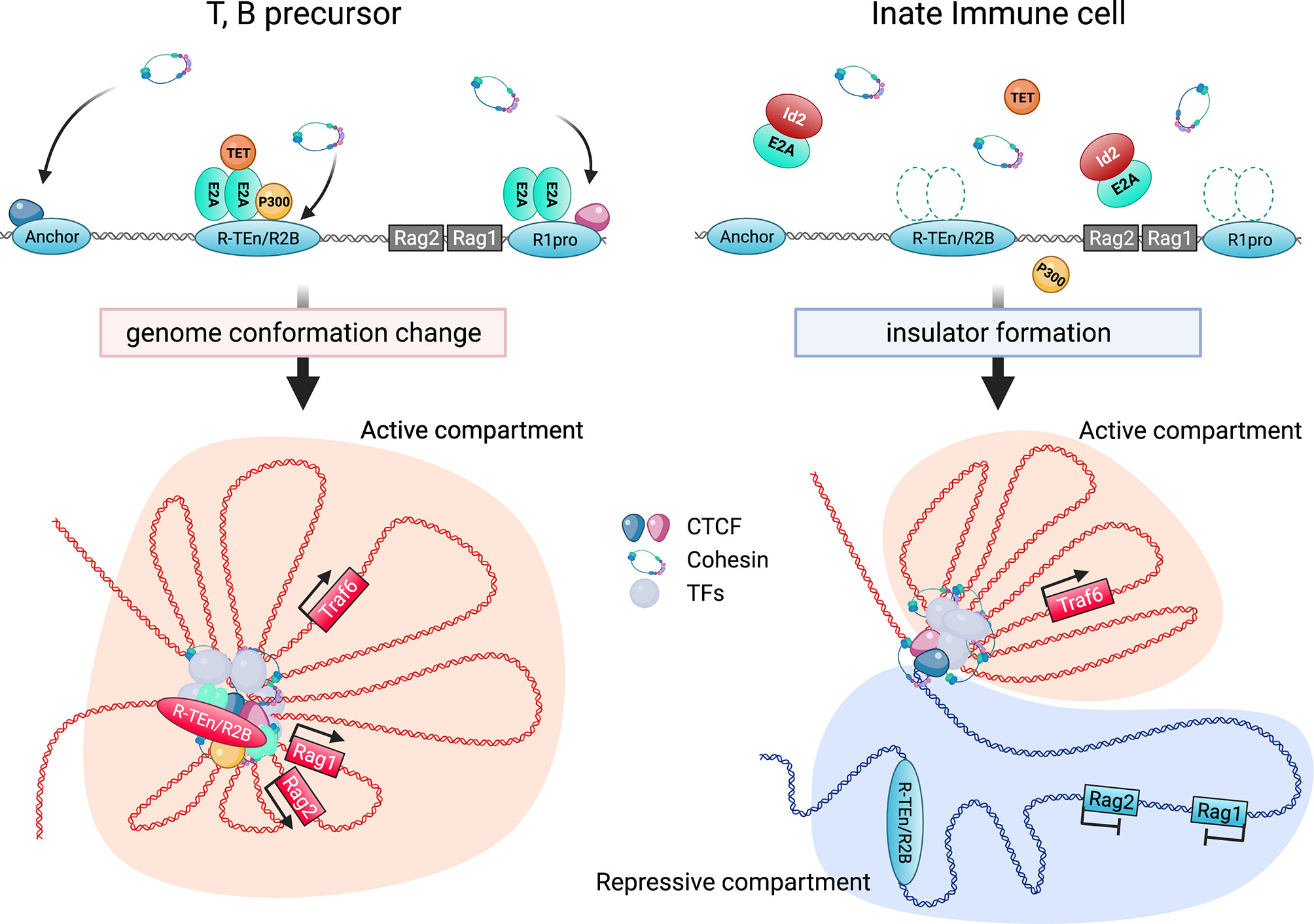
Figure 2 Regulation of Rag gene locus by E2A and cis-regulatory elements. E2A binding to the specific enhancer (R-TEn and R2B) and R1pro regions induces the genome conformation changes to form adaptive lymphocyte-specific SE through the recruitment of P300, TET, and NIPBL-cohesin complex (left; developing T and B cells). In contrast, Id2 prevents E2A/E proteins from binding to these regulatory regions, leading to the insulator formation to sequester the Rag genes in repressive chromatin compartment in innate immune cells (right; macrophage etc). This figure was created with BioRender.com.
E and Id proteins play a central role in effector/memory and tissue-resident cytotoxic CD8 T cell differentiation and the activation of helper CD4 T cells, including TH1 and follicular helper T (TFH) cells (67, 71, 100–105). However, to our knowledge, no review papers have addressed the role of the E-Id axis in Treg cells. In this section, we focus on the roles of Id and E proteins in Treg cells. Treg cells play a central role in the maintenance of immune homeostasis by suppressing autoimmunity and excessive inflammatory responses and by tissue repair after inflammation. Naturally occurring Treg cells differentiate in the thymus (natural Treg (nTreg) or thymic Treg (tTreg) cells), which constitutively express TF Foxp3, while a population of Foxp3-expressing Treg cells develops from naïve CD4 T cells in the periphery (peripheral Treg (pTreg) cells) (106). In addition, naïve CD4 T cells can develop into Foxp3-expressing Treg cells in vitro by TCR stimulation in the presence of TGF-β plus IL-2 (induced Treg (iTreg) cells) (107). Treg cells show functional heterogeneity to regulate a variety of immune responses, and each subset of Treg cells has a specialized gene expression program. As well as conventional CD4 T cells, Treg cells differentiate into effector subsets, named effector Treg (eTreg) cells, accompanied by Blimp1 and Irf4 TFs, and express unique migratory chemokine receptors to home to the site of inflammation and higher suppressive molecules such as IL-10 and CTLA-4 to control tissue inflammation (108–110). For instance, TH1-Treg cells express CXCR3, which is mediated by T-bet, to migrate into TH1 inflammatory sites (111). In addition, follicular regulatory T (TFR) cells, a specialized subset of Treg cells, regulate TFH cell function and germinal center B-cell responses for the humoral immunity (112–114). More recently, specialized subsets of Treg cells in nonlymphoid tissues, such as adipose tissue, muscle tissue, lung tissue, and the central nervous system, have been shown to play an important role in tissue homeostasis and regenerative functions, and amphiregulin and Notch ligand Jagged1 from Treg cells contribute to tissue regeneration (115–118). This subset of Treg cells is often referred to as tissue-resident Treg (TR-Treg) cells. They are derived from effector Treg cells, which in turn are instructed by TF Batf (119) (Figure 3).
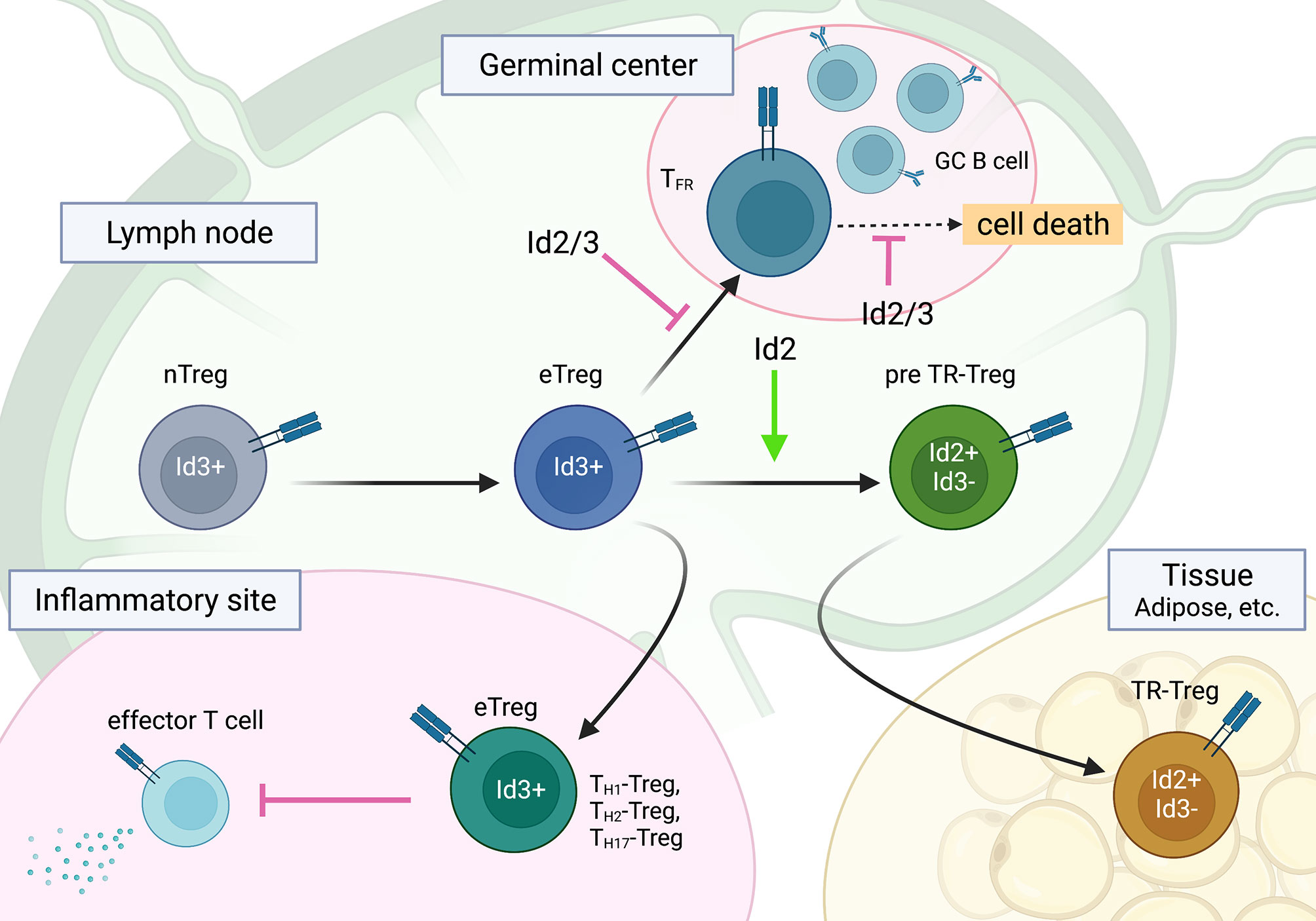
Figure 3 The roles of Id2 and Id3 in Treg cell differentiation into subsets of effector Treg cells. Id2 and Id3 enforce the naïve state of Treg cells, especially in TFR cells. A regulatory switch of Id3 to Id2 plays a role in TR-Treg cell differentiation and function. This figure was created with BioRender.com.
There are many previous studies about the role of the E-Id axis in Treg cell development and activation. The expression of Id3 is high in naïve Treg cells and low in ICOS+ effector Treg cells, and TCR stimulation in Treg cells downregulates Id3. In contrast, Id2 levels are low in naïve Treg cells, and TCR stimulation induces the upregulation of Id2 in vitro (120, 121). It has been reported that E2A/HEB and Id3 are involved in the development of tTreg cell and iTreg cells; drastically increased tTreg cells were observed in a study of E2A/HEB-deficient thymus, while decreased tTreg cells were detected in a study of Id3–/– thymus (122, 123). In addition, blocking the E protein by Id1 overexpression in mice resulted in an increased frequency and number of tTreg cells due to the expansion of thymic Treg cells, while Foxp3 mRNA induced by TCR stimulation was significantly lower in naïve Id1-Tg CD4 T cells (124). However, the deletion of E2A and HEB in early stages blocks T cell lineage commitment, and their deletion in DP cells bypasses the TCR-mediated positive selection of DP cells, leading to the CD8SP stage accompanied by severe impairment of the CD4SP lineage (28, 125). In addition, Id3 is required for MHC-restricted positive selection of DP cells (126). The combined loss of Id2 and Id3 results in blockage of the transition from CD69+TCRβlo or – DP to fully TCR-selected CD69+TCRβhi DP cells at a young age; however, PLZF-expressing innate TFH cells expand with limited TCR repertoires and occupy the CD4SP population in adults, suggesting that in the absence of Id2 and Id3, conventional CD4 T cell development is severely affected (102). Therefore, it remains unclear whether changes in tTreg populations in these gene-deficient mice are reflected by the severely impaired CD4SP population and reduced strength of TCR signaling or whether E2A/HEB and Id3 are actually involved in the induction of Foxp3 expression or tTreg cell development. Furthermore, since Id3 enforces naïve T cell fate by antagonizing E2A activity and Id3-deficient CD4SP or CD8SP cells readily differentiate into IFN-γ-producing effector T cells,TFH cells (CD4SP), or innate-like CD8 T cells in the thymus (67, 127), attenuated iTreg cell development in Id3–/– mice is more likely the result of fewer naïve CD4 T cells in the periphery. However, from the result that the deletion of E2A/HEB led to increased iTreg development in vitro, E protein activity is thought to be involved in iTreg cell development (123). It was reported that E47 indirectly regulates Foxp3 expression through the regulation of Spi-B and SOCS3 in Id3-deficient Treg cells and that Foxp3 mRNA in Id2/Id3-deficient Treg cells is comparable to that in control Treg cells, indicating that E2A does not regulate Foxp3 gene expression (120, 128). In line with this, E2A occupancy around the Foxp3 gene locus, by ChIP-seq analysis, was not detected in Id2/Id3-deficient DP cells (129).
Although the role of Id and E proteins in tTreg development is unclear, the E-Id axis plays an important role in Treg cell function. Indeed, Treg-specific deletion of Id2 and Id3 using Foxp3-Cre in mice leads to fatal inflammatory disease, which is characterized by spontaneous TH2 inflammation in the lung, skin, and esophagus, similar to human atopic diseases such as bronchial asthma, atopic dermatitis, and eosinophilic esophagitis (120). Id2/Id3 depletion in Treg cells induces CXCR5, which is a direct target of the E2A-Id3 axis in TFR and TFH cell development and preferentially migrates to B-cell follicles. However, Id2/Id3-deficiency in Treg cells has been shown to result in compromised maintenance of Treg cells mediated by TCR stimulation in vitro. This result suggests that Id proteins function as gatekeepers for eTreg and TFR cells as well as CD4 T cells and control the maintenance of Treg cells. Although Id2 and Id3 compensate for each other in single KO Treg cells, Id2 and Id3 have distinct roles in Treg cell function. According to Id3 expression with CD62L and CD44, the Campbell group demonstrated stepwise developmental stages toward TR-Treg cells; Id3 was highly expressed in central naïve Treg cells and effector Treg cells, whereas ICOShi Id3lo TR-Treg precursor cells expressed Id2, suggesting a regulatory switch from Id3 to Id2 in Treg cells (121, 130). This seems to be similar to tissue resident effector/memory CD8 T cells (100, 105). Interestingly, consistent with the Id switch in Treg cells, a loss of Id2 expression in Treg cells results in decreased expression of TR-Treg cell-related functional molecules and leads to increased cell death of Treg cells, suggesting an Id2-dependent TR-Treg cell-specific program (131). Curiously, Treg cells lacking E2A and HEB exhibit effector phenotypes and increased stability, suggesting the linkage of E protein and TCR signaling in the gene signature of effector Treg cell development (132). In contrast, ectopic Id2 expression in Treg cells in mice enhance Treg cell plasticity and lead to a reduction in Treg cells (133). Taken together, although the underlying molecular mechanism remains to be determined, it now seems apparent that the E-Id axis orchestrates Treg cell differentiation toward the fate of TFR, eTreg and TR-Treg cells and dictates function and plasticity in lymphoid and nonlymphoid tissues (Figure 3).
The E-Id transcriptional axis plays an important role in T/B cell lineage commitment, discrimination between T cells and ILCs, including Rag gene expression, and T/Treg cell function. However, it remains to be investigated how the E-Id axis orchestrates cell type-specific enhancer activities in conjunction with other TFs associated with T cell activation and TCR signaling. Future experiments are warranted to explore the role of the E-Id axis in T and B cell activation under the inflammatory conditions. These findings may have implications for health and immunological disorders.
RH, KM and MM wrote the manuscript and figures. All authors contributed to the article and approved the submitted version.
This work was funded by the KAKENHI (Grants-in-Aid for Scientific Research) from the MEXT of Japan (19H03487 for MM), the Mochida Memorial Foundation, the Takeda Science Foundation, the FUJIWARA Memorial Foundation (MM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank C. Murre for kind suggestions and for reviewing the manuscript.
1. Schatz DG, Oettinger MA, Baltimore D. The V(D)J Recombination Activating Gene, RAG-1. Cell (1989) 59(6):1035–48. doi: 10.1016/0092-8674(89)90760-5
2. Schatz DG, Ji Y. Recombination Centres and the Orchestration of V(D)J Recombination. Nat Rev Immunol (2011) 11(4):251–63. doi: 10.1038/nri2941
3. Rothenberg EV. Programming for T-Lymphocyte Fates: Modularity and Mechanisms. Genes Dev (2019) 33(17-18):1117–35. doi: 10.1101/gad.327163.119
4. Koch U, Fiorini E, Benedito R, Besseyrias V, Schuster-Gossler K, Pierres M, et al. Delta-Like 4 is the Essential, Nonredundant Ligand for Notch1 During Thymic T Cell Lineage Commitment. J Exp Med (2008) 205(11):2515–23. doi: 10.1084/jem.20080829
5. Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, et al. Delta-Like 4 is Indispensable in Thymic Environment Specific for T Cell Development. J Exp Med (2008) 205(11):2507–13. doi: 10.1084/jem.20080134
6. Chea S, Schmutz S, Berthault C, Perchet T, Petit M, Burlen-Defranoux O, et al. Single-Cell Gene Expression Analyses Reveal Heterogeneous Responsiveness of Fetal Innate Lymphoid Progenitors to Notch Signaling. Cell Rep (2016) 14(6):1500–16. doi: 10.1016/j.celrep.2016.01.015
7. Capone M, Hockett RD Jr., Zlotnik A. Kinetics of T Cell Receptor Beta, Gamma, and Delta Rearrangements During Adult Thymic Development: T Cell Receptor Rearrangements are Present in CD44(+)CD25(+) Pro-T Thymocytes. Proc Natl Acad Sci USA (1998) 95(21):12522–7. doi: 10.1073/pnas.95.21.12522
8. Kreslavsky T, Gleimer M, Von Boehmer H. Alphabeta Versus Gammadelta Lineage Choice at the First TCR-Controlled Checkpoint. Curr Opin Immunol (2010) 22(2):185–92. doi: 10.1016/j.coi.2009.12.006
9. Krangel MS. Mechanics of T Cell Receptor Gene Rearrangement. Curr Opin Immunol (2009) 21(2):133–9. doi: 10.1016/j.coi.2009.03.009
10. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and Negative Selection of the T Cell Repertoire: What Thymocytes See (and Don't See). Nat Rev Immunol (2014) 14(6):377–91. doi: 10.1038/nri3667
11. Hori S, Nomura T, Sakaguchi S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science (2003) 299(5609):1057–61. doi: 10.1126/science.1079490
12. Constantinides MG, Bendelac A. Transcriptional Regulation of the NKT Cell Lineage. Curr Opin Immunol (2013) 25(2):161–7. doi: 10.1016/j.coi.2013.01.003
13. Napier RJ, Adams EJ, Gold MC, Lewinsohn DM. The Role of Mucosal Associated Invariant T Cells in Antimicrobial Immunity. Front Immunol (2015) 6:344. doi: 10.3389/fimmu.2015.00344
14. Hosokawa H, Rothenberg EV. How Transcription Factors Drive Choice of the T Cell Fate. Nat Rev Immunol (2021) 21(3):162–76. doi: 10.1038/s41577-020-00426-6
15. Taniuchi I. CD4 Helper and CD8 Cytotoxic T Cell Differentiation. Annu Rev Immunol (2018) 36:579–601. doi: 10.1146/annurev-immunol-042617-053411
16. Murre C, Mccaw PS, Baltimore D. A New DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, MyoD, and Myc Proteins. Cell (1989) 56(5):777–83. doi: 10.1016/0092-8674(89)90682-x
17. Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The Protein Id: A Negative Regulator of Helix-Loop-Helix DNA Binding Proteins. Cell (1990) 61(1):49–59. doi: 10.1016/0092-8674(90)90214-y
18. Murre C. Helix-Loop-Helix Proteins and the Advent of Cellular Diversity: 30 Years of Discovery. Genes Dev (2019) 33(1-2):6–25. doi: 10.1101/gad.320663.118
20. Belle I, Zhuang Y. E Proteins in Lymphocyte Development and Lymphoid Diseases. Curr Top Dev Biol (2014) 110:153–87. doi: 10.1016/b978-0-12-405943-6.00004-x
21. Miyazaki K, Miyazaki M, Murre C. The Establishment of B Versus T Cell Identity. Trends Immunol (2014) 35(5):205–10. doi: 10.1016/j.it.2014.02.009
22. Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A Global Network of Transcription Factors, Involving E2A, EBF1 and Foxo1, That Orchestrates B Cell Fate. Nat Immunol (2010) 11(7):635–43. doi: 10.1038/ni.1891
23. Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, et al. Global Changes in the Nuclear Positioning of Genes and Intra- and Interdomain Genomic Interactions That Orchestrate B Cell Fate. Nat Immunol (2012) 13(12):1196–204. doi: 10.1038/ni.2432
24. Wang HC, Qian L, Zhao Y, Mengarelli J, Adrianto I, Montgomery CG, et al. Downregulation of E Protein Activity Augments an ILC2 Differentiation Program in the Thymus. J Immunol (2017) 198(8):3149–56. doi: 10.4049/jimmunol.1602009
25. Ikawa T, Kawamoto H, Goldrath AW, Murre C. E Proteins and Notch Signaling Cooperate to Promote T Cell Lineage Specification and Commitment. J Exp Med (2006) 203(5):1329–42. doi: 10.1084/jem.20060268
26. Braunstein M, Anderson MK. HEB-Deficient T-Cell Precursors Lose T-Cell Potential and Adopt an Alternative Pathway of Differentiation. Mol Cell Biol (2011) 31(5):971–82. doi: 10.1128/MCB.01034-10
27. Braunstein M, Anderson MK. HEB in the Spotlight: Transcriptional Regulation of T-Cell Specification, Commitment, and Developmental Plasticity. Clin Dev Immunol (2012) 2012:678705. doi: 10.1155/2012/678705
28. Miyazaki M, Miyazaki K, Chen K, Jin Y, Turner J, Moore AJ, et al. The E-Id Protein Axis Specifies Adaptive Lymphoid Cell Identity and Suppresses Thymic Innate Lymphoid Cell Development. Immunity (2017) 46(5):818–34:e4. doi: 10.1016/j.immuni.2017.04.022
29. D'cruz LM, Knell J, Fujimoto JK, Goldrath AW. An Essential Role for the Transcription Factor HEB in Thymocyte Survival, Tcra Rearrangement and the Development of Natural Killer T Cells. Nat Immunol (2010) 11(3):240–9. doi: 10.1038/ni.1845
30. Jones ME, Zhuang Y. Acquisition of a Functional T Cell Receptor During T Lymphocyte Development is Enforced by HEB and E2A Transcription Factors. Immunity (2007) 27(6):860–70. doi: 10.1016/j.immuni.2007.10.014
31. Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the Helix-Loop-Helix Proteins, E2A and Id3, by the Ras-ERK MAPK Cascade. Nat Immunol (2001) 2(2):165–71. doi: 10.1038/84273
32. Koltsova EK, Ciofani M, Benezra R, Miyazaki T, Clipstone N, Zúñiga-Pflücker JC, et al. Early Growth Response 1 and NF-ATc1 Act in Concert to Promote Thymocyte Development Beyond the Beta-Selection Checkpoint. J Immunol (2007) 179(7):4694–703. doi: 10.4049/jimmunol.179.7.4694
33. Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, et al. Marked Induction of the Helix-Loop-Helix Protein Id3 Promotes the Gammadelta T Cell Fate and Renders Their Functional Maturation Notch Independent. Immunity (2009) 31(4):565–75. doi: 10.1016/j.immuni.2009.07.010
34. Fahl SP, Contreras AV, Verma A, Qiu X, Harly C, Radtke F, et al. The E Protein-TCF1 Axis Controls γδ T Cell Development and Effector Fate. Cell Rep (2021) 34(5):108716. doi: 10.1016/j.celrep.2021.108716
35. Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, et al. Transcription Factor E2-2 is an Essential and Specific Regulator of Plasmacytoid Dendritic Cell Development. Cell (2008) 135(1):37–48. doi: 10.1016/j.cell.2008.09.016
36. Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous Expression of the Transcription Factor E2-2 Maintains the Cell Fate of Mature Plasmacytoid Dendritic Cells. Immunity (2010) 33(6):905–16. doi: 10.1016/j.immuni.2010.11.023
37. Grajkowska LT, Ceribelli M, Lau CM, Warren ME, Tiniakou I, Nakandakari Higa S, et al. Isoform-Specific Expression and Feedback Regulation of E Protein TCF4 Control Dendritic Cell Lineage Specification. Immunity (2017) 46(1):65–77. doi: 10.1016/j.immuni.2016.11.006
38. Verykokakis M, Zook EC, Kee BL. ID'ing Innate and Innate-Like Lymphoid Cells. Immunol Rev (2014) 261(1):177–97. doi: 10.1111/imr.12203
39. Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional Regulation of Innate Lymphoid Cell Fate. Nat Rev Immunol (2015) 15(7):415–28. doi: 10.1038/nri3855
40. Cherrier DE, Serafini N, Di Santo JP. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity (2018) 48(6):1091–103. doi: 10.1016/j.immuni.2018.05.010
41. Klose CS, Artis D. Innate Lymphoid Cells as Regulators of Immunity, Inflammation and Tissue Homeostasis. Nat Immunol (2016) 17(7):765–74. doi: 10.1038/ni.3489
42. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 Upregulation Identifies Early Innate Lymphoid Progenitors in the Bone Marrow. Nat Immunol (2015) 16(10):1044–50. doi: 10.1038/ni.3248
43. Seehus CR, Aliahmad P, de la Torre B, Iliev ID, Spurka L, Funari VA, et al. The Development of Innate Lymphoid Cells Requires TOX-Dependent Generation of a Common Innate Lymphoid Cell Progenitor. Nat Immunol (2015) 16(6):599–608. doi: 10.1038/ni.3168
44. Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, et al. NFIL3 Orchestrates the Emergence of Common Helper Innate Lymphoid Cell Precursors. Cell Rep (2015) 10(12):2043–54. doi: 10.1016/j.celrep.2015.02.057
45. Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of Type 1 ILCs From a Common Progenitor to All Helper-Like Innate Lymphoid Cell Lineages. Cell (2014) 157(2):340–56. doi: 10.1016/j.cell.2014.03.030
46. Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, et al. Nfil3 is Required for the Development of All Innate Lymphoid Cell Subsets. J Exp Med (2014) 211(9):1733–40. doi: 10.1084/jem.20140145
47. Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The Transcription Factor GATA-3 Controls Cell Fate and Maintenance of Type 2 Innate Lymphoid Cells. Immunity (2012) 37(4):634–48. doi: 10.1016/j.immuni.2012.06.020
48. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The Transcription Factor GATA3 is Critical for the Development of All IL-7rα-Expressing Innate Lymphoid Cells. Immunity (2014) 40(3):378–88. doi: 10.1016/j.immuni.2014.01.012
49. Constantinides MG, Mcdonald BD, Verhoef PA, Bendelac A. A Committed Precursor to Innate Lymphoid Cells. Nature (2014) 508(7496):397–401. doi: 10.1038/nature13047
50. Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 Specifies Lineage Commitment of Innate Lymphoid Cells. Nat Immunol (2015) 16(11):1124–33. doi: 10.1038/ni.3272
51. Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A, et al. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity (2015) 43(2):354–68. doi: 10.1016/j.immuni.2015.07.005
52. Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, et al. Transcription Factor Rorα is Critical for Nuocyte Development. Nat Immunol (2012) 13(3):229–36. doi: 10.1038/ni.2208
53. Hosokawa H, Romero-Wolf M, Yui MA, Ungerbäck J, Quiloan MLG, Matsumoto M, et al. Bcl11b Sets Pro-T Cell Fate by Site-Specific Cofactor Recruitment and by Repressing Id2 and Zbtb16. Nat Immunol (2018) 19(12):1427–40. doi: 10.1038/s41590-018-0238-4
54. Jones R, Cosway EJ, Willis C, White AJ, Jenkinson WE, Fehling HJ, et al. Dynamic Changes in Intrathymic ILC Populations During Murine Neonatal Development. Eur J Immunol (2018) 48(9):1481–91. doi: 10.1002/eji.201847511
55. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate Production of T(H)2 Cytokines by Adipose Tissue-Associated C-Kit(+)Sca-1(+) Lymphoid Cells. Nature (2010) 463(7280):540–4. doi: 10.1038/nature08636
56. Shin SB, Mcnagny KM. ILC-You in the Thymus: A Fresh Look at Innate Lymphoid Cell Development. Front Immunol (2021) 12:681110. doi: 10.3389/fimmu.2021.681110
57. Shin SB, Lo BC, Ghaedi M, Scott RW, Li Y, Messing M, et al. Abortive γδtcr Rearrangements Suggest ILC2s are Derived From T-Cell Precursors. Blood Adv (2020) 4(21):5362–72. doi: 10.1182/bloodadvances.2020002758
58. Koga S, Hozumi K, Hirano KI, Yazawa M, Terooatea T, Minoda A, et al. Peripheral Pdgfrα(+)Gp38(+) Mesenchymal Cells Support the Differentiation of Fetal Liver-Derived ILC2. J Exp Med (2018) 215(6):1609–26. doi: 10.1084/jem.20172310
59. Harly C, Cam M, Kaye J, Bhandoola A. Development and Differentiation of Early Innate Lymphoid Progenitors. J Exp Med (2018) 215(1):249–62. doi: 10.1084/jem.20170832
60. Qian L, Bajana S, Georgescu C, Peng V, Wang HC, Adrianto I, et al. Suppression of ILC2 Differentiation From Committed T Cell Precursors by E Protein Transcription Factors. J Exp Med (2019) 216(4):884–99. doi: 10.1084/jem.20182100
61. Ferreira ACF, Szeto ACH, Heycock MWD, Clark PA, Walker JA, Crisp A, et al. Rorα is a Critical Checkpoint for T Cell and ILC2 Commitment in the Embryonic Thymus. Nat Immunol (2021) 22(2):166–78. doi: 10.1038/s41590-020-00833-w
62. Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R. A Single-Cell Transcriptomic Atlas of Thymus Organogenesis Resolves Cell Types and Developmental Maturation. Immunity (2018) 48(6):1258–70.e6. doi: 10.1016/j.immuni.2018.04.015
63. Hosokawa H, Romero-Wolf M, Yang Q, Motomura Y, Levanon D, Groner Y, et al. Cell Type-Specific Actions of Bcl11b in Early T-Lineage and Group 2 Innate Lymphoid Cells. J Exp Med (2020) 217(1). doi: 10.1084/jem.20190972
64. Hosokawa H, Masuhara K, Koizumi M. Transcription Factors Regulate Early T Cell Development via Redeployment of Other Factors: Functional Dynamics of Constitutively Required Factors in Cell Fate Decisions. Bioessays (2021) 43(5):e2000345. doi: 10.1002/bies.202000345
65. Engel I, Johns C, Bain G, Rivera RR, Murre C. Early Thymocyte Development is Regulated by Modulation of E2A Protein Activity. J Exp Med (2001) 194(6):733–45. doi: 10.1084/jem.194.6.733
66. Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An Analysis of T Cell Intrinsic Roles of E2A by Conditional Gene Disruption in the Thymus. J Immunol (2002) 168(8):3923–32. doi: 10.4049/jimmunol.168.8.3923
67. Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The Opposing Roles of the Transcription Factor E2A and its Antagonist Id3 That Orchestrate and Enforce the Naive Fate of T Cells. Nat Immunol (2011) 12(10):992–1001. doi: 10.1038/ni.2086
68. Teachenor R, Beck K, Wright LY, Shen Z, Briggs SP, Murre C. Biochemical and Phosphoproteomic Analysis of the Helix-Loop-Helix Protein E47. Mol Cell Biol (2012) 32(9):1671–82. doi: 10.1128/mcb.06452-11
69. Engel I, Murre C. The Function of E- and Id Proteins in Lymphocyte Development. Nat Rev Immunol (2001) 1(3):193–9. doi: 10.1038/35105060
70. D'cruz LM, Lind KC, Wu BB, Fujimoto JK, Goldrath AW. Loss of E Protein Transcription Factors E2A and HEB Delays Memory-Precursor Formation During the CD8+ T-Cell Immune Response. Eur J Immunol (2012) 42(8):2031–41. doi: 10.1002/eji.201242497
71. Shaw LA, Bélanger S, Omilusik KD, Cho S, Scott-Browne JP, Nance JP, et al. Id2 Reinforces TH1 Differentiation and Inhibits E2A to Repress TFH Differentiation. Nat Immunol (2016) 17(7):834–43. doi: 10.1038/ni.3461
72. Mowel WK, Mccright SJ, Kotzin JJ, Collet MA, Uyar A, Chen X, et al. Group 1 Innate Lymphoid Cell Lineage Identity Is Determined by a Cis-Regulatory Element Marked by a Long Non-Coding RNA. Immunity (2017) 47(3):435–49.e8. doi: 10.1016/j.immuni.2017.08.012
73. Miyazaki K, Miyazaki M. The Interplay Between Chromatin Architecture and Lineage-Specific Transcription Factors and the Regulation of Rag Gene Expression. Front Immunol (2021) 12:659761. doi: 10.3389/fimmu.2021.659761
74. Wilson A, Held W, Macdonald HR. Two Waves of Recombinase Gene Expression in Developing Thymocytes. J Exp Med (1994) 179(4):1355–60. doi: 10.1084/jem.179.4.1355
75. Kuo TC, Schlissel MS. Mechanisms Controlling Expression of the RAG Locus During Lymphocyte Development. Curr Opin Immunol (2009) 21(2):173–8. doi: 10.1016/j.coi.2009.03.008
76. Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, et al. A Conserved Transcriptional Enhancer Regulates RAG Gene Expression in Developing B Cells. Immunity (2003) 19(1):105–17. doi: 10.1016/s1074-7613(03)00181-x
77. Amin RH, Schlissel MS. Foxo1 Directly Regulates the Transcription of Recombination-Activating Genes During B Cell Development. Nat Immunol (2008) 9(6):613–22. doi: 10.1038/ni.1612
78. Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, et al. Distinct Functions for the Transcription Factor Foxo1 at Various Stages of B Cell Differentiation. Nat Immunol (2008) 9(12):1388–98. doi: 10.1038/ni.1667
79. Schulz D, Vassen L, Chow KT, Mcwhirter SM, Amin RH, Möröy T, et al. Gfi1b Negatively Regulates Rag Expression Directly and via the Repression of Foxo1. J Exp Med (2012) 209(1):187–99. doi: 10.1084/jem.20110645
80. Timblin GA, Xie L, Tjian R, Schlissel MS. Dual Mechanism of Rag Gene Repression by C-Myb During Pre-B Cell Proliferation. Mol Cell Biol (2017)37(12). doi: 10.1128/mcb.00437-16
81. Yannoutsos N, Barreto V, Misulovin Z, Gazumyan A, Yu W, Rajewsky N, et al. A Cis Element in the Recombination Activating Gene Locus Regulates Gene Expression by Counteracting a Distant Silencer. Nat Immunol (2004) 5(4):443–50. doi: 10.1038/ni1053
82. Hao B, Naik AK, Watanabe A, Tanaka H, Chen L, Richards HW, et al. An Anti-Silencer- and SATB1-Dependent Chromatin Hub Regulates Rag1 and Rag2 Gene Expression During Thymocyte Development. J Exp Med (2015) 212(5):809–24. doi: 10.1084/jem.20142207
83. Naik AK, Byrd AT, Lucander ACK, Krangel MS. Hierarchical Assembly and Disassembly of a Transcriptionally Active RAG Locus in CD4(+)CD8(+) Thymocytes. J Exp Med (2019) 216(1):231–43. doi: 10.1084/jem.20181402
84. Miyazaki K, Watanabe H, Yoshikawa G, Chen K, Hidaka R, Aitani Y, et al. The Transcription Factor E2A Activates Multiple Enhancers That Drive Rag Expression in Developing T and B Cells. Sci Immunol (2020) 5(51). doi: 10.1126/sciimmunol.abb1455
85. Dias S, Månsson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity (2008) 29(2):217–27. doi: 10.1016/j.immuni.2008.05.015
86. Xu W, Carr T, Ramirez K, Mcgregor S, Sigvardsson M, Kee BL. E2A Transcription Factors Limit Expression of Gata3 to Facilitate T Lymphocyte Lineage Commitment. Blood (2013) 121(9):1534–42. doi: 10.1182/blood-2012-08-449447
87. Bradney C, Hjelmeland M, Komatsu Y, Yoshida M, Yao TP, Zhuang Y. Regulation of E2A Activities by Histone Acetyltransferases in B Lymphocyte Development. J Biol Chem (2003) 278(4):2370–6. doi: 10.1074/jbc.M211464200
88. Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and Functional Collaboration of P300/CBP and bHLH Proteins in Muscle and B-Cell Differentiation. Genes Dev (1996) 10(19):2478–90. doi: 10.1101/gad.10.19.2478
89. Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A Conserved Motif Present in a Class of Helix-Loop-Helix Proteins Activates Transcription by Direct Recruitment of the SAGA Complex. Mol Cell (1999) 4(1):63–73. doi: 10.1016/s1097-2765(00)80188-4
90. Qiu Y, Sharma A, Stein R. P300 Mediates Transcriptional Stimulation by the Basic Helix-Loop-Helix Activators of the Insulin Gene. Mol Cell Biol (1998) 18(5):2957–64. doi: 10.1128/mcb.18.5.2957
91. Liang JJ, Peng H, Wang JJ, Liu XH, Ma L, Ni YR, et al. Relationship Between the Structure and Function of the Transcriptional Regulator E2A. J Biol Res (Thessalon) (2021) 28(1):15. doi: 10.1186/s40709-021-00146-5
92. Rowley MJ, Corces VG. Organizational Principles of 3D Genome Architecture. Nat Rev Genet (2018) 19(12):789–800. doi: 10.1038/s41576-018-0060-8
93. Zhu Y, Denholtz M, Lu H, Murre C. Calcium Signaling Instructs NIPBL Recruitment at Active Enhancers and Promoters via Distinct Mechanisms to Reconstruct Genome Compartmentalization. Genes Dev (2021) 35(1-2):65–81. doi: 10.1101/gad.343475.120
94. Lio CW, Zhang J, González-Avalos E, Hogan PG, Chang X, Rao A. Tet2 and Tet3 Cooperate With B-Lineage Transcription Factors to Regulate DNA Modification and Chromatin Accessibility. Elife (2016) 5. doi: 10.7554/eLife.18290
95. Lio CJ, Shukla V, Samaniego-Castruita D, González-Avalos E, Chakraborty A, Yue X, et al. TET Enzymes Augment Activation-Induced Deaminase (AID) Expression via 5-Hydroxymethylcytosine Modifications at the Aicda Superenhancer. Sci Immunol (2019) 4(34). doi: 10.1126/sciimmunol.aau7523
96. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell (2013) 155(4):934–47. doi: 10.1016/j.cell.2013.09.053
97. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell (2013) 153(2):307–19. doi: 10.1016/j.cell.2013.03.035
98. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell (2017) 169(1):13–23. doi: 10.1016/j.cell.2017.02.007
99. Yoshikawa G, Miyazaki K, Ogata H, Miyazaki M. The Evolution of Rag Gene Enhancers and Transcription Factor E and Id Proteins in the Adaptive Immune System. Int J Mol Sci (2021) 22(11). doi: 10.3390/ijms22115888
100. Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, et al. The Transcriptional Regulators Id2 and Id3 Control the Formation of Distinct Memory CD8+ T Cell Subsets. Nat Immunol (2011) 12(12):1221–9. doi: 10.1038/ni.2158
101. Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, et al. Transcription Factor Achaete-Scute Homologue 2 Initiates Follicular T-Helper-Cell Development. Nature (2014) 507(7493):513–8. doi: 10.1038/nature12910
102. Miyazaki M, Miyazaki K, Chen S, Chandra V, Wagatsuma K, Agata Y, et al. The E–Id Protein Axis Modulates the Activities of the PI3K–AKT–mTORC1–Hif1a and C-Myc/p19Arf Pathways to Suppress Innate Variant TFH Cell Development, Thymocyte Expansion, and Lymphomagenesis. Genes Dev (2015) 29(4):409–25. doi: 10.1101/gad.255331.114
103. He R, Hou S, Liu C, Zhang A, Bai Q, Han M, et al. Follicular CXCR5- Expressing CD8(+) T Cells Curtail Chronic Viral Infection. Nature (2016) 537(7620):412–28. doi: 10.1038/nature19317
104. Omilusik KD, Nadjsombati MS, Shaw LA, Yu B, Milner JJ, Goldrath AW. Sustained Id2 Regulation of E Proteins is Required for Terminal Differentiation of Effector CD8(+) T Cells. J Exp Med (2018) 215(3):773–83. doi: 10.1084/jem.20171584
105. Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, Mcdonald B, et al. Heterogenous Populations of Tissue-Resident CD8(+) T Cells Are Generated in Response to Infection and Malignancy. Immunity (2020) 52(5):808–24.e7. doi: 10.1016/j.immuni.2020.04.007
106. Ohkura N, Sakaguchi S. Transcriptional and Epigenetic Basis of Treg Cell Development and Function: Its Genetic Anomalies or Variations in Autoimmune Diseases. Cell Res (2020) 30(6):465–74. doi: 10.1038/s41422-020-0324-7
107. Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of Peripheral CD4+CD25- Naive T Cells to CD4+CD25+ Regulatory T Cells by TGF-Beta Induction of Transcription Factor Foxp3. J Exp Med (2003) 198(12):1875–86. doi: 10.1084/jem.20030152
108. Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, et al. The Transcription Factors Blimp-1 and IRF4 Jointly Control the Differentiation and Function of Effector Regulatory T Cells. Nat Immunol (2011) 12(4):304–11. doi: 10.1038/ni.2006
109. Cretney E, Kallies A, Nutt SL. Differentiation and Function of Foxp3(+) Effector Regulatory T Cells. Trends Immunol (2013) 34(2):74–80. doi: 10.1016/j.it.2012.11.002
110. Wing JB, Sakaguchi S. Multiple Treg Suppressive Modules and Their Adaptability. Front Immunol (2012) 3:178. doi: 10.3389/fimmu.2012.00178
111. Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The Transcription Factor T-Bet Controls Regulatory T Cell Homeostasis and Function During Type 1 Inflammation. Nat Immunol (2009) 10(6):595–602. doi: 10.1038/ni.1731
112. Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, et al. Follicular Regulatory T Cells Expressing Foxp3 and Bcl-6 Suppress Germinal Center Reactions. Nat Med (2011) 17(8):983–8. doi: 10.1038/nm.2426
113. Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, et al. Foxp3+ Follicular Regulatory T Cells Control the Germinal Center Response. Nat Med (2011) 17(8):975–82. doi: 10.1038/nm.2425
114. Wing JB, Tekgüç M, Sakaguchi S. Control of Germinal Center Responses by T-Follicular Regulatory Cells. Front Immunol (2018) 9:1910. doi: 10.3389/fimmu.2018.01910
115. Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, et al. PPAR-γ is a Major Driver of the Accumulation and Phenotype of Adipose Tissue Treg Cells. Nature (2012) 486(7404):549–53. doi: 10.1038/nature11132
116. Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell (2015) 162(5):1078–89. doi: 10.1016/j.cell.2015.08.021
117. Ali N, Zirak B, Rodriguez RS, Pauli ML, Truong HA, Lai K, et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell (2017) 169(6):1119–29.e11. doi: 10.1016/j.cell.2017.05.002
118. Ito M, Komai K, Mise-Omata S, Iizuka-Koga M, Noguchi Y, Kondo T, et al. Brain Regulatory T Cells Suppress Astrogliosis and Potentiate Neurological Recovery. Nature (2019) 565(7738):246–50. doi: 10.1038/s41586-018-0824-5
119. Delacher M, Imbusch CD, Hotz-Wagenblatt A, Mallm JP, Bauer K, Simon M, et al. Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity (2020) 52(2):295–312.e11. doi: 10.1016/j.immuni.2019.12.002
120. Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu L-F, et al. Id2 and Id3 Maintain the Regulatory T Cell Pool to Suppress Inflammatory Disease. Nat Immunol (2014) 15(8):767–76. doi: 10.1038/ni.2928
121. Sullivan JM, Höllbacher B, Campbell DJ. Cutting Edge: Dynamic Expression of Id3 Defines the Stepwise Differentiation of Tissue-Resident Regulatory T Cells. J Immunol (2019) 202(1):31–6. doi: 10.4049/jimmunol.1800917
122. Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, et al. Control of the Differentiation of Regulatory T Cells and T(H)17 Cells by the DNA-Binding Inhibitor Id3. Nat Immunol (2011) 12(1):86–95. doi: 10.1038/ni.1965
123. Gao P, Han X, Zhang Q, Yang Z, Fuss IJ, Myers TG, et al. Dynamic Changes in E-Protein Activity Regulate T Reg Cell Development. J Exp Med (2014) 211(13):2651–68. doi: 10.1084/jem.20132681
124. Liu C, Wang HC, Yu S, Jin R, Tang H, Liu YF, et al. Id1 Expression Promotes T Regulatory Cell Differentiation by Facilitating TCR Costimulation. J Immunol (2014) 193(2):663–72. doi: 10.4049/jimmunol.1302554
125. Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are Required to Block Thymocyte Proliferation Prior to Pre-TCR Expression. J Immunol (2007) 178(9):5717–26. doi: 10.4049/jimmunol.178.9.5717
126. Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte Selection is Regulated by the Helix-Loop-Helix Inhibitor Protein, Id3. Immunity (2000) 12(1):17–26. doi: 10.1016/s1074-7613(00)80155-7
127. Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP Protein-Dependent Natural Killer T-Like Cells Regulate the Development of CD8(+) T Cells With Innate Lymphocyte Characteristics. Immunity (2010) 33(2):203–15. doi: 10.1016/j.immuni.2010.07.013
128. Rauch KS, Hils M, Lupar E, Minguet S, Sigvardsson M, Rottenberg ME, et al. Id3 Maintains Foxp3 Expression in Regulatory T Cells by Controlling a Transcriptional Network of E47, Spi-B, and SOCS3. Cell Rep (2016) 17(11):2827–36. doi: 10.1016/j.celrep.2016.11.045
129. Roy S, Moore AJ, Love C, Reddy A, Rajagopalan D, Dave SS, et al. Id Proteins Suppress E2A-Driven Invariant Natural Killer T Cell Development Prior to TCR Selection. Front Immunol (2018) 9:42. doi: 10.3389/fimmu.2018.00042
130. Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, et al. CCR7 Provides Localized Access to IL-2 and Defines Homeostatically Distinct Regulatory T Cell Subsets. J Exp Med (2014) 211(1):121–36. doi: 10.1084/jem.20131142
131. Frias AB Jr., Hyzny EJ, Buechel HM, Beppu LY, Xie B, Jurczak MJ, et al. The Transcriptional Regulator Id2 Is Critical for Adipose-Resident Regulatory T Cell Differentiation, Survival, and Function. J Immunol (2019) 203(3):658–64. doi: 10.4049/jimmunol.1900358
132. Han X, Huang H, Gao P, Zhang Q, Liu X, Jia B, et al. E-Protein Regulatory Network Links TCR Signaling to Effector Treg Cell Differentiation. Proc Natl Acad Sci USA (2019) 116(10):4471–80. doi: 10.1073/pnas.1800494116
Keywords: T cell versus ILCs, Rag gene expression, E-Id axis, T-lineage commitment, Treg differentiation
Citation: Hidaka R, Miyazaki K and Miyazaki M (2022) The E-Id Axis Instructs Adaptive Versus Innate Lineage Cell Fate Choice and Instructs Regulatory T Cell Differentiation. Front. Immunol. 13:890056. doi: 10.3389/fimmu.2022.890056
Received: 05 March 2022; Accepted: 12 April 2022;
Published: 06 May 2022.
Edited by:
Barbara L. Kee, The University of Chicago, United StatesReviewed by:
Hiroyuki Hosokawa, Tokai University, JapanCopyright © 2022 Hidaka, Miyazaki and Miyazaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaki Miyazaki, bW1peWF6YWtpQGluZnJvbnQua3lvdG8tdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.