
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 03 May 2022
Sec. Vaccines and Molecular Therapeutics
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.884862
This article is part of the Research Topic Vaccine Delivery and Impact on Kinetics of Immune Responses View all 12 articles
The mRNA vaccines from Pfizer/BioNTech and Moderna were granted emergency approval in record time in the history of vaccinology and played an instrumental role in limiting the pandemic caused by SARS-CoV-2. The success of these vaccines resulted from over 3 decades of research from many scientists. However, the development of orally administrable mRNA vaccine development is surprisingly underexplored. Our group specializing in Salmonella-based vaccines explored the possibility of oral mRNA vaccine development. Oral delivery was made possible by the exploitation of the Semliki Forest viral replicon and Salmonella vehicle for transgene amplification and gene delivery, respectively. Herein we highlight the prospect of developing oral replicon-based mRNA vaccines against infectious diseases based on our recent primary studies on SARS-CoV-2. Further, we discuss the potential advantages and limitations of bacterial gene delivery.
Edward Jenner’s innovative contribution played a pivotal role in the ultimate eradication of smallpox and served as the harbinger of vaccination. This was followed by the works of Louis Pasteur, who spearheaded the development of live-attenuated cholera vaccine and inactivated anthrax vaccine in humans in 1897 and 1904, respectively. The field of vaccine research soon became popular, and vaccines were developed against a plethora of infectious diseases of medical and veterinary importance. First-generation traditional vaccines based on the use of live, live-attenuated, and inactivated organisms were instrumental in the control of measles, polio, rubella, mumps, classical swine fever, and many other diseases, and responsible for the eradication of smallpox in humans and rinderpest in cattle. Although live and live-attenuated vaccines are effective, they may pose significant health risks to vaccinated individuals, including the development of disease, transmission to healthy individuals, and reversion to a virulent form and particularly in individuals with compromised immune system (1–5). All this changed with the advent of molecular biology and recombinant DNA technology, which paved the way for the development of safer vaccines. However, DNA vaccines did not achieve their expected clinical success owing to limited or poor immunogenicity (6, 7). Technological refinements were made to improve DNA vaccine efficacy (8–15), but the risk of mutagenesis induced by exogenous DNA integration has limited their use in humans (16–19). This has led to a renewed interest in the use of RNA in vaccines and therapeutics.
Synthetic RNA vaccines fall into two main categories: non-replicating and self-amplifying mRNA vaccines. The non-replicating mRNA vaccine is a straightforward approach wherein administered mRNA is directly translated in the cytoplasm of transfected cells to produce immunogenic proteins. The extent of non-replicating mRNA vaccine-induced antigen expression is proportional to the number of transfected cells and thus, requires the injection of a large dose of mRNA. This can be overcome by the use of self-amplifying RNA replicons from alphaviruses, such as Sindbis virus (20), Semliki Forest virus (SFV) (21), and Venezuelan equine encephalitis virus (VEE) (22). Different vector systems, namely replication-competent viral particles, replication-deficient viral particles, and DNA-launched-mRNA vector approaches, have been exploited for transgene expression (reviewed in 23, 24). DNA-launched-mRNA vectors were engineered by deleting the structural genes from the genome and replacing them with the target genes (21, 25). The resulting vector backbone with non-structural proteins (nsp1–4) forms a replicase complex that drives efficient transgene expression by a self-amplifying mechanism (21, 24). The mRNA vaccines developed to combat SARS-CoV-2 constitute the first success story in the long history of mRNA vaccine development. Nonetheless, oral delivery of an mRNA vaccine has surprisingly not been exploited. In this article, we highlight a strategy for the development of oral replicon-based mRNA vaccines by taking cues from our recent publications and discussing the advantages of Salmonella-mediated oral gene delivery.
The vaccines developed against SARS-CoV-2 by Pfizer/BioNTech and Moderna constitute the first success stories in mRNA vaccine history. Although the delivery of mRNA wrapped in cationic liposomes was shown to produce proteins in human cells in 1989 (26), the potential of mRNA as a vaccine has yet to be exploited. During these past 3 decades, many scientists studied mRNA, and collective scientific advances enabled the production of the first successful mRNA vaccine in record time (Figure 1A). Some of the most important inventions to the adaptation of mRNA vaccination were the chemical modification of mRNA and lipid nanoparticles for delivery. Without lipid nanoparticle encapsulation, administered mRNA would be detected by the immune system and probably degraded by RNases. Of note, mRNA was shown to elicit TLR3-mediated immune activation of dendritic cells (DCs) (30), and bacterial RNA can prime DCs for higher IL-12 secretion (31). Replacing uridine with pseudouridine, the chemical modification that diminished immune recognition of administered mRNA, paved the way for mRNA treatments (32). The encapsulation of mRNA by lipid nanoparticles (LNPs) provided an effective and safe delivery platform (reviewed in 33). The discovery of increased protein expression and potent antibody responses to the SARS-CoV-2 spike protein in its stabilized prefusion conformation (34) is vital to the efficacy of mRNA vaccines. The developments and progress in mRNA vaccines against infectious diseases have been reviewed elsewhere (35, 36).
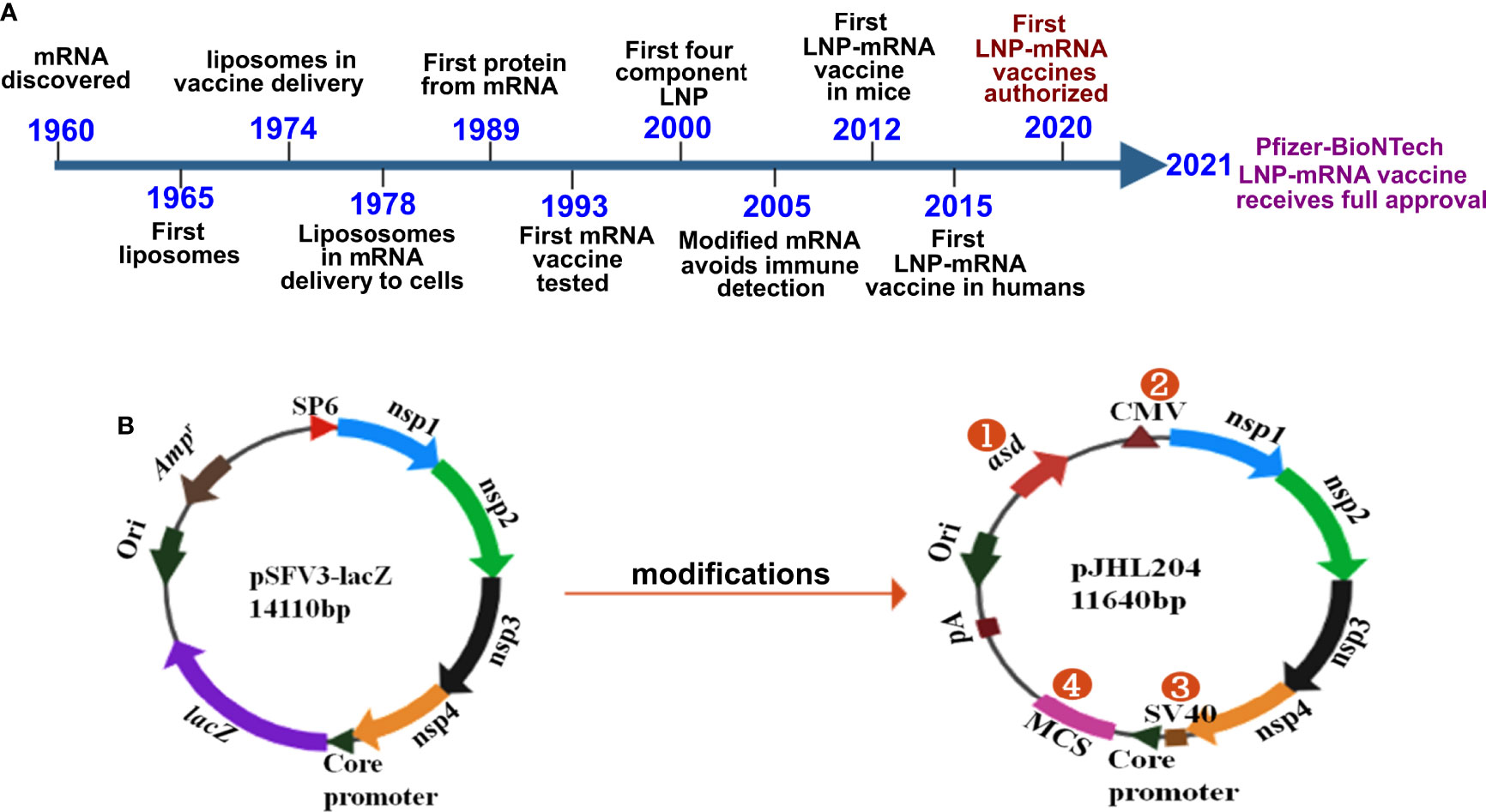
Figure 1 History and design of mRNA vaccines. (A) Timeline depicting some of the key milestones that contributed to the first successful mRNA vaccines developed against COVID-19. The timeline was adapted from Sahin et al., 2014 (ref. 27) Hou et al., 2021 (28); and Dolgin, 2021 (29). (B) The DNA-launched-mRNA vaccine design for bacterial delivery. pSFV3-lacZ, an SFV replicon-based vector, was used after making several modifications. 1- The ampicillin resistance marker was replaced with asd, an auxotrophic marker to enable antibiotic-free maintenance and delivery of the vector. 2- The SP6 promoter was replaced with the cytomegalovirus (CMV) promoter. 3- The SV40 promoter was placed just before the SFV sub-genomic core promoter to enable direct nuclear transcription of the vaccine constructs. 4- lacZ was replaced with multiple cloning site (MCS) sequences. nsp 1-4 from SFV constitute the replicon, which drives efficient transgene expression through RdRp. pA, polyadenylation signal; Ori, pBR origin of replication; RdRp, RNA dependent RNA polymerase; nsp, non-structural protein; SFV, Semliki Forest virus.
The poor uptake of mRNA by cells is associated with the rapid degradation of naked mRNA by extracellular RNAses (37–41). Developments of efficient mRNA delivery platforms have been fruitful in the last decade. From advancements in transfection reagents and liposomes to nanoparticles and nanoemulsions, in vivo antigen presentation and the immune response to mRNA-based vaccines have recently improved (42–51). The aforementioned mRNA complexing strategies have been shown to affect mRNA stability during storage (52). Thus, a continuous supply of raw materials is crucial for the uninterrupted production of mRNA vaccines. Such requirements can prove challenging at times when the demand is high (52–57). Additionally, substituting rare codons with frequently used synonymous codons and introducing modified nucleosides have been shown to enhance mRNA translation and stability in the context of vaccination (reviewed in 38). A major disadvantage associated with base modifications is that they may result in altered mRNA secondary structure, which may influence translation and protein folding. These alterations may, in turn, prove detrimental to efficacy (58–60). One of the major drawbacks with in vitro transcribed RNA is the presence of dsRNAs that trigger the innate immune response and reduces the vaccine efficacy. Advancement such as cellulose-based purification was shown to remove the dsRNA byproducts leading to the lower type I interferon response and improving the efficacy of a self-amplifying mRNA vaccine against Zika virus (61). Continuous efforts have been made to minimize the drawbacks associated with mRNA vaccines, enabling an array of these vaccines to enter phase IIb clinical trials (38, 62–67). Most of the current mRNA vaccines against infectious disease are administered using the conventional delivery routes, namely intramuscular, subcutaneous, intradermal, or intranodal routes. Most of these routes of administration require injection and specific conditions for storage and transport. Furthermore, the concerns associated with the stability of these vaccines and the addition of adjuvants to enhance immunogenicity increase the cost of production and pose toxicity threats (68, 69). Despite the success of mRNA vaccines in controlling infectious diseases, the limitations associated with their production and administration demonstrate the need to develop better and safer routes of administration for mRNA vaccines (70, 71).
Despite three decades of history supporting mRNA vaccine development and the successful rollout of mRNA vaccines during the COVID-19 pandemic, the possibility of oral delivery has surprisingly been underexplored (71). This could be attributed to the highly unstable nature of mRNA and the gut posing a significant barrier for mRNA delivery. However, some of the oral antigen delivery strategies such as yeast ghosts, microencapsulated antigens and microbial adhesions have been developed to overcome the harsh conditions in the gut (reviewed in 72). But they suffer from major limitation of poor intestinal epithelial barrier crossing and have not been explored to deliver mRNA (72). Further, lipid-based approaches such as liposomes, bilosomes and immunestimulating complexes (ISCOMs) also provide with a potential delivery vehicle for oral biologic delivery (reviewed in 71). The oral delivery of mRNA vaccines is possible due to the exploitation of an alphaviral replicon and Salmonella bactofection for mRNA amplification and gene delivery, respectively. Our group specialized in the development of Salmonella-based vaccines against diseases of veterinary and medical importance (73–79), exploited this platform for the development of an oral mRNA vaccine. Further, we exploited the Semliki Forest virus replicon for mRNA amplification (23, 24). We made several modifications to the original vector backbone (pSFV3) to enable transcription in host cells and plasmid maintenance in bacteria (Figure 1B) (25). The SP6 promoter was replaced with the Cytomegalovirus (CMV) promoter to enable transcription by mammalian RNA polymerase. The replacement of the ampicillin selection marker with the aspartate-semialdehyde dehydrogenase (asd) auxotrophic marker allows for antibiotic-free plasmid maintenance and delivery (80). The Salmonella strains used for gene delivery carry a deletion in the asd gene, creating balanced-lethal host-vector systems. Diaminopimelic acid (DAP), the product of asd, is a vital component of the bacterial cell wall, and asd mutants will not survive unless DAP is supplemented in growth media or the asd gene is complemented from a plasmid vector. Thus, asd serves as a powerful antibiotic-independent selection maker for bacterial delivery. This DNA-launched-mRNA vector design was exploited for the Salmonella-enabled oral delivery of a replicon-based mRNA vaccine against SARS-CoV-2 (25, 81, 82). The detailed mechanism of vector delivery, transgene amplification, and the generation of an immune response upon oral administration of Salmonella carrying the SFV replicon vector encoding vaccine immunogens is furnished in Figure 2. The findings demonstrate the possibility and potential of bacteria-mediated gene delivery for the development of oral replicon-based mRNA vaccines against infectious diseases.
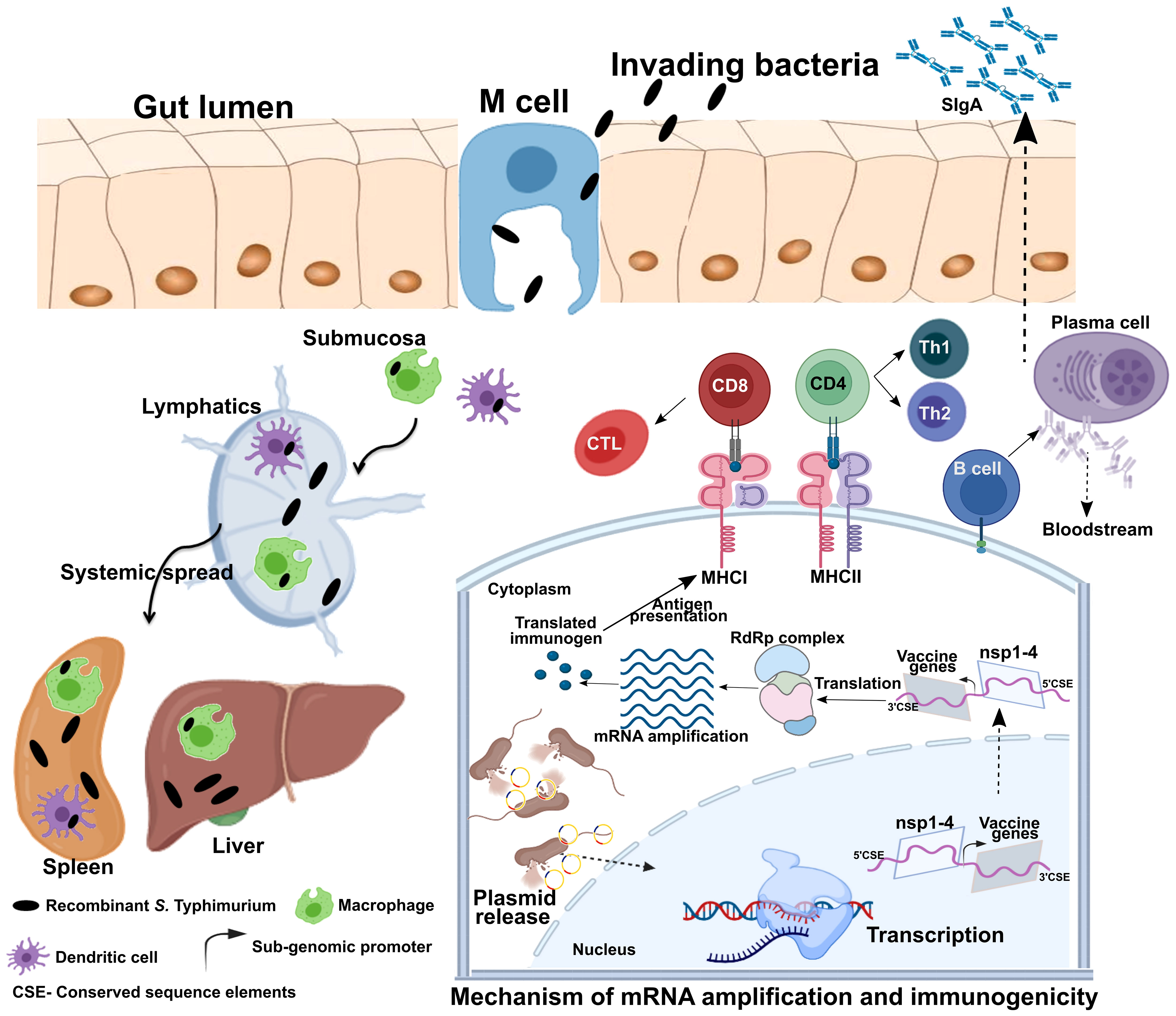
Figure 2 Mechanism of gene delivery, transgene expression, and induction of immune response. Upon oral administration, Salmonella Typhimurium is translocated from the luminal surface to submucosa by specialized M cells in the gut epithelium. Bacteria then invade antigen-presenting cells (APCs), such as macrophages and dendritic cells (DCs), and spread to different organs like the liver and spleen through lymphatics and the bloodstream. The vector encoding the Semliki Forest virus (SFV) replicon (nsp1-4) and SARS-CoV-2 immunogens is released within the host cell cytoplasm through bacterial lysis. Transcription of the delivered plasmid takes place in the cell nucleus, and, following in situ translation, the nsp1-4 proteins form an RNA-dependent RNA polymerase (RdRp) complex. The RdRp complex then recognizes the sub-genomic promoter and flanking conserved sequence elements (CSE), leading to enhanced mRNA amplification of vaccine genes. The resulting mRNAs translated to produce immunogenic proteins. The APCs process and present antigens to CD8+ and CD4+ T cells via MHC I and MHC II, respectively, leading to the elicitation of the T cell response. DCs can present antigens directly to B cells or follicular DCs (FDCs). FDC stores antigens for a longer time, periodically displaying them to cognate B cells. B cells then differentiate to specific antibody-secreting plasma cells and memory B cells. MHC, major histocompatibility complex; nsp, non-structural protein; CD, cluster of differentiation; CTL, cytotoxic T cell; Th, T helper cell; CSE- Conserved sequence elements. This figure was created with the help of the Biorender online tool (https://app.biorender.com/). The figure and description are reproduced with permission from Jawalagatti et al., 2022 (reference 82). ©The American Society of Gene and Cell Therapy.
The delivery of vaccines through the oral route can elicit a potent mucosal response considering the extensive presence of gut-associated lymphoid tissues (GALT). The bacterial species, Salmonella has the ability to interact with immune cells in Payer’s patch, leading to efficient induction of the mucosal response (83, 84). Mucosal vaccines play a pivotal role in limiting infections caused by digestive and respiratory pathogens. Moreover, gut bacteria can influence SIgA production in the lungs through CD103+ DCs (85). In agreement, we and others have documented the elicitation of mucosal response in respiratory sites by oral Salmonella-based vaccine administration (82, 86). Further, Salmonella can translocate through M cells in the intestine and reach organs such as the liver and spleen, eliciting a systemic response as well (87–89). One of the most important advantages of Salmonella is its innate ability to invade and proliferate in professional antigen-presenting cells (APCs), such as dendritic cells (DCs) (90) and macrophages (91), during which it directly delivers the DNA cargo to these cells. As antigens must be formed within the APC or cross-presented to an APC to elicit a cellular response (92), gene delivery and antigen expression within APCs result in robust cellular immunity along with the induction of a potent humoral response. Moreover, vaccine production can be easily scaled up, and a high number of doses can be prepared rapidly at an inexpensive rate. Importantly, bacteria-mediated vaccine delivery does not require additional adjuvants or delivery systems, which further cuts down the cost of manufacturing and limits the frequency of vaccine-associated adverse events (68, 69). Most important of all, the availability of licensed oral Salmonella Typhi vaccines provides the possibility of direct translation to humans. Further, the availability of a licensed live-attenuated Vibrio cholerae vaccine (Vaxchora; https://www.fda.gov/media/98688/download) provides with additional bacterial vector to develop vaccines against diseases of medical importance. The fact that Salmonella can be lyophilized permits a thermostable way to dispatch the vaccines and represents progress towards needle-free mass oral immunizations. Collectively, the data suggest the highly prospective nature of exploiting bacteria to develop oral mRNA vaccines with the ability to elicit potent systemic and mucosal immune responses. The intranasal delivery could also be exploited to develop potent mucosal mRNA vaccines. However, as the vaccine uses live-attenuated bacterium poses a significant safety and regulatory hurdle. The intranasal route is more suitable and safer for delivery of mRNA through polymeric delivery systems. The advantage of oral vaccine over an intranasal vaccine would be superior patient compliance and easy mass administration. Therefore, bacteria-mediated delivery of mRNA vaccines for mucosal vaccine development would be feasible when administered orally rather than intranasally.
One of the major limitations of live-attenuated bacteria is safety. However, the availability of tested and proven licensed vaccines provides safer delivery options. Furthermore, well-established tools to modify the bacterial genome provide an opportunity to create safer mutants (93). Another major limitation of using live-attenuated organisms for gene delivery is a hindrance from pre-existing immunity that can seriously affect vaccine efficacy (94, 95). Both SIgA and IgG could contribute to the pre-existing immunity against Salmonella. Nevertheless, this limitation could be overcome by deleting the O-antigen ligase (rfaL) or any other gene(s) from the bacterial genome that mask the bacteria from detection by the immune system (77). However, several studies have shown the positive influence of pre-existing immunity and recorded stronger immune responses against the delivered antigen by Salmonella vectors (reviewed in 96). Thus, the effect of pre-existing immunity on heterologous antigen delivery is likely negligible or less variable. Of note, the effect of pre-existing immunity on viral vectors is more pronounced than on bacterial vectors (96).
Our proof-of-principle studies using SARS-CoV-2 (25, 81, 82) provide evidence for the development of oral replicon-based mRNA vaccines against infectious diseases. Salmonella is an ideal bacterial vector owing to its unique ability to target GALT upon oral administration, resulting in both systemic and mucosal immune responses in vaccinated individuals. The possibility of oral delivery was partly enabled by creating a DNA-launched-mRNA design of the SFV replicon that essentially drives gene expression by a self-amplifying mRNA mechanism (25). Although the research on RNA vaccines and therapeutics spans over 3 decades, the possibility of oral mRNA vaccine delivery was yet to be explored. To the best of our knowledge, our studies are the first to demonstrate oral replicon-based mRNA vaccine delivery. To this end, we designed a multivalent SFV replicon-based vaccine targeting receptor-binding domain (RBD), heptad repeat domain (HR), membrane glycoprotein (M), and epitopes of nsp13 and employed Salmonella Typhimurium for gene delivery (25). The administration of the vaccine was highly safe in mice and hamsters inoculated both orally and intramuscularly (25, 82). The vaccine elicited potent Th1-dominated humoral and cellular immune responses in mice against all the target antigens, suggesting efficient antigen production and presentation (25, 82). Furthermore, RBD expressed after Salmonella delivery was confirmed to be antigenically intact in macrophage-like cells (82). We recorded the difference in mucosal immune response induction between oral and intramuscular routes of vaccine administration, highlighting the feasibility of exploiting oral administration for mucosal vaccine development (82). Most importantly, the vaccine protected hamsters against live SARS-CoV-2, and complete protection was elicited by oral immunization against viral replication and lung disease (82). Moreover, a robust cross-protection against the B.1.617.2 delta variant was evidenced following oral immunization in hamsters (82) and mice (81). The fact that an intranasal vaccine durably protected against SARS-CoV-2 variants (97, 98) and dimeric IgA had superior neutralizing activity (99) underscore the efficacy of the mucosal response exerted by oral vaccines in protection against rapidly replicating variants.
Our proof-of-principle studies have unraveled a novel method for the development of oral mRNA vaccines. The availability of some licensed live-attenuated bacterial vaccines increases the prospects of adopting such vaccines in the clinic. However, more studies using relevant bacterial species in suitable preclinical models are necessary to prove the hypothesis. Moreover, the possibility of other bacterial species, such as Shigella, could also be tested to optimize the choice of a bacterial vector.
VJ and PK: prepared the figures and wrote the article. JL: acquired funding and commented on the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A1A03033084).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Minor PD. Live Attenuated Vaccines: Historical Successes and Current Challenges. Virol (2015) 479–480:379–92. doi: 10.1016/j.virol.2015.03.032
2. Minor PD. The Polio-Eradication Programme and Issues of the End Game. J Gen Virol (2012) 93:457–74. doi: 10.1099/vir.0.036988-0
3. Marsden SA, Boulger LR, Magrath DI, Reeve P, Schild GC, Taffs LF. Monkey Neurovirulence of Live, Attenuated (Sabin) Type I and Type II Poliovirus Vaccines. J Biol Stand (1980) 8:303–9. doi: 10.1016/S0092-1157(80)80008-4
4. Shaghaghi M, Parvaneh N, Ostad-Rahimi P, Fathi SM, Shahmahmoodi S, Abolhassani H, et al. Combined Immunodeficiency Presenting With Vaccine-Associated Paralytic Poliomyelitis: A Case Report and Narrative Review of Literature. Immunol Invest (2014) 43:292–8. doi: 10.3109/08820139.2013.859156
5. Rubin LG, Levin MJ, Ljungman P, Davies EG, Avery R, Tomblyn M, et al. IDSA Clinical Practice Guideline for Vaccination of the Immunocompromised Host. Clin Infect Dis (2014) 58:e44–e100. doi: 10.1093/cid/cit684
6. Kutzler MA, Weiner DB. DNA Vaccines: Ready for Prime Time? Nat Rev Genet (2008) 9:776–88. doi: 10.1038/nrg2432
7. MacGregor RR, Boyer JD, Ugen KE, Lacy KE, Gluckman SJ, Bagarazzi ML, et al. First Human Trial of a DNA-Based Vaccine for Treatment of Human Immunodeficiency Virus Type 1 Infection: Safety and Host Response. J Infect Dis (1998) 178:92–100. doi: 10.1086/515613
8. Cheung Y-K, Cheng SC-S, Sin FW-Y, Xie Y. Plasmid Encoding Papillomavirus Type 16 (HPV16) DNA Constructed With Codon Optimization Improved the Immunogenicity Against HPV Infection. Vaccine (2004) 23:629–38. doi: 10.1016/j.vaccine.2004.07.010
9. Narum DL, Kumar S, Rogers WO, Fuhrmann SR, Liang H, Oakley M, et al. Codon Optimization of Gene Fragments Encoding Plasmodium Falciparum Merzoite Proteins Enhances DNA Vaccine Protein Expression and Immunogenicity in Mice. Infect Immun (2001) 69:7250–3. doi: 10.1128/IAI.69.12.7250-7253.2001
10. Yan J, Yoon H, Kumar S, Ramanathan MP, Corbitt N, Kutzler M, et al. Enhanced Cellular Immune Responses Elicited by an Engineered HIV-1 Subtype B Consensus-Based Envelope DNA Vaccine. Mol Ther (2007) 15:411–21. doi: 10.1038/sj.mt.6300036
11. Xu Z-L, Mizuguchi H, Ishii-Watabe A, Uchida E, Mayumi T, Hayakawa T. Optimization of Transcriptional Regulatory Elements for Constructing Plasmid Vectors. Gene (2001) 272:149–56. doi: 10.1016/S0378-1119(01)00550-9
12. Kaur R, Rauthan M, Vrati S. Immunogenicity in Mice of a Cationic Microparticle-Adsorbed Plasmid DNA Encoding Japanese Encephalitis Virus Envelope Protein. Vaccine (2004) 22:2776–82. doi: 10.1016/j.vaccine.2004.01.040
13. Pai Kasturi S, Qin H, Thomson KS, El-Bereir S, Cha S, Neelapu S, et al. Prophylactic Anti-Tumor Effects in a B Cell Lymphoma Model With DNA Vaccines Delivered on Polyethylenimine (PEI) Functionalized PLGA Microparticles. J Control Release (2006) 113:261–70. doi: 10.1016/j.jconrel.2006.04.006
14. Jilek S, Merkle H, Walter E. DNA-Loaded Biodegradable Microparticles as Vaccine Delivery Systems and Their Interaction With Dendritic Cells. Adv Drug Delivery Rev (2005) 57:377–90. doi: 10.1016/j.addr.2004.09.010
15. Vandervoort J. Microneedles for Transdermal Drug Delivery: A Minireview. Front Biosci (2008) 13:1711. doi: 10.2741/2794
16. Nichols WW, Ledwith BJ, Manam SV, Troilo PJ. Potential DNA Vaccine Integration Into Host Cell Genome. Ann N Y Acad Sci (1995) 772:30–9. doi: 10.1111/j.1749-6632.1995.tb44729.x
17. Martin T, Parker SE, Hedstrom R, Le T, Hoffman SL, Norman J, et al. Plasmid DNA Malaria Vaccine: The Potential for Genomic Integration After Intramuscular Injection. Hum Gene Ther (1999) 10:759–68. doi: 10.1089/10430349950018517
18. Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, et al. Detection of Integration of Plasmid DNA Into Host Genomic DNA Following Intramuscular Injection and Electroporation. Gene Ther (2004) 11:711–21. doi: 10.1038/sj.gt.3302213
19. Liu MA. DNA Vaccines: An Historical Perspective and View to the Future. Immunol Rev (2011) 239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x
20. Xiong C, Levis R, Shen P, Schlesinger S, Rice CM, Huang HV. Sindbis Virus: An Efficient, Broad Host Range Vector for Gene Expression in Animal Cells. Science (80-) (1989) 243:1188–91. doi: 10.1126/science.2922607
21. Liljeström P, Garoff H. A New Generation of Animal Cell Expression Vectors Based on the Semliki Forest Virus Replicon. Bio/Technology (1991) 9:1356–61. doi: 10.1038/nbt1291-1356
22. Davis NL, Willis LV, Smitht JF, Johnston RE. In Vitro Synthesis of Infectious Venezuelan Equine Encephalitis Virus RNA From a cDNA Clone: Analysis of a Viable Deletion Mutant. Virology (1989) 171:189–204. doi: 10.1016/0042-6822(89)90526-6
23. Lundstrom K. Biology and Application of Alphaviruses in Gene Therapy. Gene Ther (2005) 12:S92–7. doi: 10.1038/sj.gt.3302620
25. Jawalagatti V, Kirthika P, Park J-Y, Hewawaduge C, Lee JH. Highly Feasible Immunoprotective Multicistronic SARS-CoV-2 Vaccine Candidate Blending Novel Eukaryotic Expression and Salmonella Bactofection. J Adv Res (2022) 36:211–22. doi: 10.1016/j.jare.2021.07.007
26. Malone RW, Felgner PL, Verma IM. Cationic Liposome-Mediated RNA Transfection. Proc Natl Acad Sci (1989) 86:6077–81. doi: 10.1073/pnas.86.16.6077
27. Sahin U, Karikó K, Türeci Ö. mRNA-Based Therapeutics — Developing a New Class of Drugs. Nat Rev Drug Discov (2014) 13:759–80. doi: 10.1038/nrd4278
28. Hou X, Zaks T, Langer R, Dong Y. Lipid Nanoparticles for mRNA Delivery. Nat Rev Mater (2021) 6:1078–94. doi: 10.1038/s41578-021-00358-0
29. Dolgin E. The Tangled History of mRNA Vaccines. Nature (2021) 597:318–24. doi: 10.1038/d41586-021-02483-w
30. Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA Is an Endogenous Ligand for Toll-Like Receptor 3. J Biol Chem (2004) 279:12542–50. doi: 10.1074/jbc.M310175200
31. Koski GK, Karikó K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. Cutting Edge: Innate Immune System Discriminates Between RNA Containing Bacterial Versus Eukaryotic Structural Features That Prime for High-Level IL-12 Secretion by Dendritic Cells. J Immunol (2004) 172:3989–93. doi: 10.4049/jimmunol.172.7.3989
32. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA Recognition by Toll-Like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
33. Buschmann MD, Carrasco MJ, Alishetty S, Paige M, Alameh MG, Weissman D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines (2021) 9:65. doi: 10.3390/vaccines9010065
34. Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, et al. Immunogenicity and Structures of a Rationally Designed Prefusion MERS-CoV Spike Antigen. Proc Natl Acad Sci (2017) 114:E7348–57. doi: 10.1073/pnas.1707304114
35. Bloom K, van den Berg F, Arbuthnot P. Self-Amplifying RNA Vaccines for Infectious Diseases. Gene Ther (2021) 28:117–29. doi: 10.1038/s41434-020-00204-y
36. Zhang C, Maruggi G, Shan H, Li J. Advances in mRNA Vaccines for Infectious Diseases. Front Immunol (2019) 10:594. doi: 10.3389/fimmu.2019.00594
37. Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol Ther (2019) 27:710–28. doi: 10.1016/j.ymthe.2019.02.012
38. Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA Vaccines — A New Era in Vaccinology. Nat Rev Drug Discov (2018) 17:261–79. doi: 10.1038/nrd.2017.243
39. Tsui NBY, Ng EKO, Lo YMD. Stability of Endogenous and Added RNA in Blood Specimens, Serum, and Plasma. Clin Chem (2002) 48:1647–53. doi: 10.1093/clinchem/48.10.1647
40. Kauffman KJ, Webber MJ, Anderson DG. Materials for Non-Viral Intracellular Delivery of Messenger RNA Therapeutics. J Control Release (2016) 240:227–34. doi: 10.1016/j.jconrel.2015.12.032
41. Guan S, Rosenecker J. Nanotechnologies in Delivery of mRNA Therapeutics Using Nonviral Vector-Based Delivery Systems. Gene Ther (2017) 24:133–43. doi: 10.1038/gt.2017.5
42. Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines Against H10N8 and H7N9 Influenza Viruses. Mol Ther (2017) 25:1316–27. doi: 10.1016/j.ymthe.2017.03.035
43. Pollard C, Rejman J, De Haes W, Verrier B, Van Gulck E, Naessens T, et al. Type I IFN Counteracts the Induction of Antigen-Specific Immune Responses by Lipid-Based Delivery of mRNA Vaccines. Mol Ther (2013) 21:251–9. doi: 10.1038/mt.2012.202
44. Uchida S, Kinoh H, Ishii T, Matsui A, Tockary TA, Takeda KM, et al. Systemic Delivery of Messenger RNA for the Treatment of Pancreatic Cancer Using Polyplex Nanomicelles With a Cholesterol Moiety. Biomaterials (2016) 82:221–8. doi: 10.1016/j.biomaterials.2015.12.031
45. Mockey M, Bourseau E, Chandrashekhar V, Chaudhuri A, Lafosse S, Le Cam E, et al. mRNA-Based Cancer Vaccine: Prevention of B16 Melanoma Progression and Metastasis by Systemic Injection of MART1 mRNA Histidylated Lipopolyplexes. Cancer Gene Ther (2007) 14:802–14. doi: 10.1038/sj.cgt.7701072
46. Perche F, Benvegnu T, Berchel M, Lebegue L, Pichon C, Jaffrès P-A, et al. Enhancement of Dendritic Cells Transfection In Vivo and of Vaccination Against B16F10 Melanoma With Mannosylated Histidylated Lipopolyplexes Loaded With Tumor Antigen Messenger RNA. Nanomed Nanotechnol Biol Med (2011) 7:445–53. doi: 10.1016/j.nano.2010.12.010
47. Démoulins T, Milona P, Englezou PC, Ebensen T, Schulze K, Suter R, et al. Polyethylenimine-Based Polyplex Delivery of Self-Replicating RNA Vaccines. Nanomed Nanotechnol Biol Med (2016) 12:711–22. doi: 10.1016/j.nano.2015.11.001
48. Maruggi G, Chiarot E, Giovani C, Buccato S, Bonacci S, Frigimelica E, et al. Immunogenicity and Protective Efficacy Induced by Self-Amplifying mRNA Vaccines Encoding Bacterial Antigens. Vaccine (2017) 35:361–8. doi: 10.1016/j.vaccine.2016.11.040
49. Brazzoli M, Magini D, Bonci A, Buccato S, Giovani C, Kratzer R, et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-Amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J Virol (2016) 90:332–44. doi: 10.1128/JVI.01786-15
50. Brito LA, Chan M, Shaw CA, Hekele A, Carsillo T, Schaefer M, et al. A Cationic Nanoemulsion for the Delivery of Next-Generation RNA Vaccines. Mol Ther (2014) 22:2118–29. doi: 10.1038/mt.2014.133
51. McCullough KC, Bassi I, Milona P, Suter R, Thomann-Harwood L, Englezou P, et al. Self-Replicating Replicon-RNA Delivery to Dendritic Cells by Chitosan-Nanoparticles for Translation In Vitro and In Vivo. Mol Ther - Nucleic Acids (2014) 3:e173. doi: 10.1038/mtna.2014.24
52. Crommelin DJA, Anchordoquy TJ, Volkin DB, Jiskoot W, Mastrobattista E. Addressing the Cold Reality of mRNA Vaccine Stability. J Pharm Sci (2021) 110:997–1001. doi: 10.1016/j.xphs.2020.12.006
53. Reichmuth AM, Oberli MA, Jaklenec A, Langer R, Blankschtein D. mRNA Vaccine Delivery Using Lipid Nanoparticles. Ther Deliv (2016) 7:319–34. doi: 10.4155/tde-2016-0006
54. Hassett KJ, Benenato KE, Jacquinet E, Lee A, Woods A, Yuzhakov O, et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol Ther - Nucleic Acids (2019) 15:1–11. doi: 10.1016/j.omtn.2019.01.013
55. Lutz J, Lazzaro S, Habbeddine M, Schmidt KE, Baumhof P, Mui BL, et al. Unmodified mRNA in LNPs Constitutes a Competitive Technology for Prophylactic Vaccines. NPJ Vaccines (2017) 2:29. doi: 10.1038/s41541-017-0032-6
56. Sedic M, Senn JJ, Lynn A, Laska M, Smith M, Platz SJ, et al. Safety Evaluation of Lipid Nanoparticle–Formulated Modified mRNA in the Sprague-Dawley Rat and Cynomolgus Monkey. Vet Pathol (2018) 55:341–54. doi: 10.1177/0300985817738095
57. Golan MS, Mahoney E, Trump B, Linkov I. Resilience and Efficiency for the Nanotechnology Supply Chains Underpinning COVID-19 Vaccine Development. Curr Opin Chem Eng (2021) 34:100759. doi: 10.1016/j.coche.2021.100759
58. Mauro VP, Chappell SA. A Critical Analysis of Codon Optimization in Human Therapeutics. Trends Mol Med (2014) 20:604–13. doi: 10.1016/j.molmed.2014.09.003
59. Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, et al. Synonymous Codons Direct Cotranslational Folding Toward Different Protein Conformations. Mol Cell (2016) 61:341–51. doi: 10.1016/j.molcel.2016.01.008
60. Yu C-H, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, et al. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-Translational Protein Folding. Mol Cell (2015) 59:744–54. doi: 10.1016/j.molcel.2015.07.018
61. Zhong Z, McCafferty S, Opsomer L, Wang H, Huysmans H, De Temmerman J, et al. Corticosteroids and Cellulose Purification Improve, Respectively, the In Vivo Translation and Vaccination Efficacy of sa-mRNAs. Mol Ther (2021) 29:1370–81. doi: 10.1016/j.ymthe.2021.01.023
62. Jacobson JM, Routy J-P, Welles S, DeBenedette M, Tcherepanova I, Angel JB, et al. Dendritic Cell Immunotherapy for HIV-1 Infection Using Autologous HIV-1 RNA. JAIDS J Acquir Immune Defic Syndr (2016) 72:31–8. doi: 10.1097/QAI.0000000000000926
63. Schnee M, Vogel AB, Voss D, Petsch B, Baumhof P, Kramps T, et al. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection Against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PloS Negl Trop Dis (2016) 10:e0004746. doi: 10.1371/journal.pntd.0004746
64. Alberer M, Gnad-Vogt U, Hong HS, Mehr KT, Backert L, Finak G, et al. Safety and Immunogenicity of a mRNA Rabies Vaccine in Healthy Adults: An Open-Label, Non-Randomised, Prospective, First-in-Human Phase 1 Clinical Trial. Lancet (2017) 390:1511–20. doi: 10.1016/S0140-6736(17)31665-3
65. Gandhi RT, Kwon DS, Macklin EA, Shopis JR, McLean AP, McBrine N, et al. Immunization of HIV-1-Infected Persons With Autologous Dendritic Cells Transfected With mRNA Encoding HIV-1 Gag and Nef. JAIDS J Acquir Immune Defic Syndr (2016) 71:246–53. doi: 10.1097/QAI.0000000000000852
66. Routy J-P, Boulassel M-R, Yassine-Diab B, Nicolette C, Healey D, Jain R, et al. Immunologic Activity and Safety of Autologous HIV RNA-Electroporated Dendritic Cells in HIV-1 Infected Patients Receiving Antiretroviral Therapy. Clin Immunol (2010) 134:140–7. doi: 10.1016/j.clim.2009.09.009
67. Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, et al. Modified mRNA Vaccines Protect Against Zika Virus Infection. Cell (2017) 168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017
68. Ruiz JT, Luján L, Blank M, Shoenfeld Y. Adjuvants- and Vaccines-Induced Autoimmunity: Animal Models. Immunol Res (2017) 65:55–65. doi: 10.1007/s12026-016-8819-5
69. Nicholls EF, Madera L, Hancock REW. Immunomodulators as Adjuvants for Vaccines and Antimicrobial Therapy. Ann N Y Acad Sci (2010) 1213:46–61. doi: 10.1111/j.1749-6632.2010.05787.x
70. D’Amico C, Fontana F, Cheng R, Santos HA. Development of Vaccine Formulations: Past, Present, and Future. Drug Deliv Transl Res (2021) 11:353–72. doi: 10.1007/s13346-021-00924-7
71. Coffey JW, Das Gaiha G, Traverso G. Oral Biologic Delivery: Advances Toward Oral Subunit, DNA, and mRNA Vaccines and the Potential for Mass Vaccination During Pandemics. Annu Rev Pharmacol Toxicol (2021) 61:517–40. doi: 10.1146/annurev-pharmtox-030320-092348
72. Devriendt B, De Geest BG, Goddeeris BM, Cox E. Crossing the Barrier: Targeting Epithelial Receptors for Enhanced Oral Vaccine Delivery. J Control Release (2012) 160:431–9. doi: 10.1016/j.jconrel.2012.02.006
73. Jawale CV, Lee JH. Salmonella Enterica Serovar Enteritidis Ghosts Carrying the Escherichia Coli Heat-Labile Enterotoxin B Subunit Are Capable of Inducing Enhanced Protective Immune Responses. Clin Vaccine Immunol (2014) 21:799–807. doi: 10.1128/CVI.00016-14
74. Jawale CV, Chaudhari AA, Jeon BW, Nandre RM, Lee JH. Characterization of a Novel Inactivated Salmonella Enterica Serovar Enteritidis Vaccine Candidate Generated Using a Modified Ci857/λ P R/Gene E Expression System. Infect Immun (2012) 80:1502–9. doi: 10.1128/IAI.06264-11
75. Hyoung KJ, Hajam IA, Lee JH. A Consensus-Hemagglutinin-Based Vaccine Delivered by an Attenuated Salmonella Mutant Protects Chickens Against Heterologous H7N1 Influenza Virus. Oncotarget (2017) 8:38780–92. doi: 10.18632/oncotarget.16353
76. Kim JH, Hajam IA, Lee JH. Oral Immunization With a Novel Attenuated Salmonella Typhimurium Encoding Influenza HA, M2e and NA Antigens Protects Chickens Against H7N9 Infection. Vet Res (2018) 49:12. doi: 10.1186/s13567-018-0509-y
77. Lalsiamthara J, Kim JH, Lee JH. Engineering of a Rough Auxotrophic Mutant Salmonella Typhimurium for Effective Delivery. Oncotarget (2018) 9:25441–57. doi: 10.18632/oncotarget.25192
78. Kim B, Won G, Lee JH. Construction of an Inactivated Typhoid Vaccine Candidate Expressing Escherichia Coli Heat-Labile Enterotoxin B Subunit and Evaluation of Its Immunogenicity in a Murine Model. J Med Microbiol (2017) 66:1235–43. doi: 10.1099/jmm.0.000543
79. Lalsiamthara J, Lee JH. Brucella Lipopolysaccharide Reinforced Salmonella Delivering Brucella Immunogens Protects Mice Against Virulent Challenge. Vet Microbiol (2017) 205:84–91. doi: 10.1016/j.vetmic.2017.05.012
80. Nakayama K, Kelly SM, Curtiss R. Construction of an ASD+ Expression-Cloning Vector: Stable Maintenance and High Level Expression of Cloned Genes in a Salmonella Vaccine Strain. Nat Biotechnol (1988) 6:693–7. doi: 10.1038/nbt0688-693
81. Jawalagatti V, Kirthika P, Hewawaduge C, Park J-Y, Yang M, Oh B, et al. A Simplified SARS-CoV-2 Mouse Model Demonstrates Protection by an Oral Replicon-Based mRNA Vaccine. Front Immunol (2022) 13:811802. doi: 10.3389/fimmu.2022.811802
82. Jawalagatti V, Kirthika P, Hewawaduge C, Yang M, Park J-Y, Oh B, et al. Bacteria-Enabled Oral Delivery of a Replicon-Based mRNA Vaccine Candidate Protects Against Ancestral and Delta Variant SARS-CoV-2. Mol Ther (2022) 30. doi: 10.1016/j.ymthe.2022.01.042
83. Martinoli C, Chiavelli A, Rescigno M. Entry Route of Salmonella Typhimurium Directs the Type of Induced Immune Response. Immunity (2007) 27:975–84. doi: 10.1016/j.immuni.2007.10.011
84. Sato A, Hashiguchi M, Toda E, Iwasaki A, Hachimura S, Kaminogawa S. CD11b + Peyer’s Patch Dendritic Cells Secrete IL-6 and Induce IgA Secretion From Naive B Cells. J Immunol (2003) 171:3684–90. doi: 10.4049/jimmunol.171.7.3684
85. Lycke NY, Bemark M. The Regulation of Gut Mucosal IgA B-Cell Responses: Recent Developments. Mucosal Immunol (2017) 10:1361–74. doi: 10.1038/mi.2017.62
86. Hopkins S, Kraehenbuhl JP, Schödel F, Potts A, Peterson D, de Grandi P, et al. A Recombinant Salmonella Typhimurium Vaccine Induces Local Immunity by Four Different Routes of Immunization. Infect Immun (1995) 63:3279–86. doi: 10.1128/iai.63.9.3279-3286.1995
87. Jang MH, Kweon M-N, Iwatani K, Yamamoto M, Terahara K, Sasakawa C, et al. Intestinal Villous M Cells: An Antigen Entry Site in the Mucosal Epithelium. Proc Natl Acad Sci (2004) 101:6110–5. doi: 10.1073/pnas.0400969101
88. Jones BD, Ghori N, Falkow S. Salmonella Typhimurium Initiates Murine Infection by Penetrating and Destroying the Specialized Epithelial M Cells of the Peyer’s Patches. J Exp Med (1994) 180:15–23. doi: 10.1084/jem.180.1.15
89. Penheiter KL, Mathur N, Giles D, Fahlen T, Jones BD. Non-Invasive Salmonella Typhimurium Mutants Are Avirulent Because of an Inability to Enter and Destroy M Cells of Ileal Peyer’s Patches. Mol Microbiol (1997) 24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x
90. Wick MJ. The Role of Dendritic Cells During Salmonella Infection. Curr Opin Immunol (2002) 14:437–43. doi: 10.1016/S0952-7915(02)00364-3
91. Gog JR, Murcia A, Osterman N, Restif O, McKinley TJ, Sheppard M, et al. Dynamics of Salmonella Infection of Macrophages at the Single Cell Level. J R Soc Interface (2012) 9:2696–707. doi: 10.1098/rsif.2012.0163
92. Liu MA. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines (2019) 7:37. doi: 10.3390/vaccines7020037
93. Datsenko KA, Wanner BL. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc Natl Acad Sci (2000) 97(12):6640–5. doi: 10.1073/pnas.120163297
94. Roberts M, Bacon A, Li J, Chatfield S. Prior Immunity to Homologous and Heterologous Salmonella Serotypes Suppresses Local and Systemic Anti-Fragment C Antibody Responses and Protection From Tetanus Toxin in Mice Immunized With Salmonella Strains Expressing Fragment C. Infect Immun (1999) 67:3810–5. doi: 10.1128/IAI.67.8.3810-3815.1999
95. Mok DZL, Chan KR. The Effects of Pre-Existing Antibodies on Live-Attenuated Viral Vaccines. Viruses (2020) 12:520. doi: 10.3390/v12050520
96. Saxena M, Van TTH, Baird FJ, Coloe PJ, Smooker PM. Pre-Existing Immunity Against Vaccine Vectors – Friend or Foe? Microbiology (2013) 159:1–11. doi: 10.1099/mic.0.049601-0
97. Hassan AO, Shrihari S, Gorman MJ, Ying B, Yaun D, Raju S, et al. An Intranasal Vaccine Durably Protects Against SARS-CoV-2 Variants in Mice. Cell Rep (2021) 36:109452. doi: 10.1016/j.celrep.2021.109452
98. Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts Against SARS-CoV-2. Cell (2020) 183:169–184.e13. doi: 10.1016/j.cell.2020.08.026
Keywords: bacterial delivery, alphaviral replicon, mRNA vaccine, oral, mucosal vaccine, SARS-CoV-2, infectious diseases
Citation: Jawalagatti V, Kirthika P and Lee JH (2022) Oral mRNA Vaccines Against Infectious Diseases- A Bacterial Perspective [Invited]. Front. Immunol. 13:884862. doi: 10.3389/fimmu.2022.884862
Received: 27 February 2022; Accepted: 11 April 2022;
Published: 03 May 2022.
Edited by:
Gabriel Pedersen, Statens Serum Institut (SSI), DenmarkReviewed by:
Irina V. Kiseleva, Institute of Experimental Medicine (RAS), RussiaCopyright © 2022 Jawalagatti, Kirthika and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Hwa Lee, am9obmhsZWVAamJudS5hYy5rcg==
†Present addresses: Vijayakumar Jawalagatti, Urology Department, Mayo Clinic, Rochester, MN, United States
Perumalraja Kirthika, Biochemistry and Molecular Biology Department, Mayo Clinic, Rochester, MN, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.