- 1Laboratory of Aquatic Animal Health Management, Department of Aquaculture, Faculty of Fisheries, Kasetsart University, Bangkok, Thailand
- 2Center of Excellence in Aquatic Animal Health Management, Faculty of Fisheries, Kasetsart University, Bangkok, Thailand
- 3Laboratory of Aquatic Animal Genetics, Department of Aquaculture, Faculty of Fisheries, Kasetsart University, Bangkok, Thailand
- 4Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
Two novel immunoglobulin heavy chain (Ighμ) transcripts encoding membrane-bound forms of IgM (mIgM) were discovered in bighead catfish, Clarias macrocephalus. The first transcript contains four constant and two transmembrane domains [Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2] that have never been reported in teleosts, and the second transcript is an unusual mIgM that has never been identified in any vertebrate [Cμ1-(Cδ2-Cδ3-Cδ4-Cδ5)-Cμ2-Cμ3-TM1-TM2]. Fluorescence in situ hybridization (FISH) in bighead catfish, North African catfish (C. gariepinus) and hybrid catfish revealed a single copy of Ighμ in individual parent catfish, while two gene copies were found in diploid hybrid catfish. Intensive sequence analysis demonstrated multiple distinct structural variabilities in the VH domain in Clarias, and hybrid catfish were defined and used to generate diversity with various mechanisms. Expression analysis of Ighμ in Aeromonas hydrophila infection of the head kidney, peripheral blood leukocytes and spleen revealed significantly higher levels in North African catfish and hybrid catfish than in bighead catfish.
Introduction
Vertebrate immunoglobulins (Igs) are the hallmark elements in adaptive immune responses to a particular antigen with high discrimination, specificity and long-term memory. Igs are generated by B cells and serve two purposes: 1) cell-surface receptors (membrane-bound forms; mIgs) for signaling and activation of cells and 2) soluble effector molecules (secreted forms; sIgs) for neutralization of microbes and toxins, opsonization (immunophagocytosis), antibody-dependent cell-mediated cytotoxicity (ADCC), and complement activation (immunolysis) (1).
The basic structure of Ig heavy chain (Igh) gene molecules consists of variable (VH) and constant (CH) regions (2). The gene sequence organizations of Igh vary depending on the species (3, 4). The immunologic effector functions of the Ig classes are determined by the different constant regions of the CH chain. In most mammals, five classes of Igs are categorized based on different gene sequences; these classes are μ, δ, γ, ϵ, and α, and they correspond to the five major isotypes of Igs, IgM, D, G, E, and A, respectively. In teleost fish, only three Igh isotypes have been identified, namely, IgM, D, and T/Z based on the gene sequences of the isotypes μ, δ, and τ/ζ, respectively (5–7). Additionally, distinct Igh classes have been identified in nonmammalian vertebrates, including IgNAR and IgW (in cartilaginous fish) (8), IgO (platypus), IgP (in Pleurodeles waltl) (9), IgX, IgF and IgY (in amphibians) (9, 10).
The evolution of Ig molecules in jawed vertebrates diverged from the “multicluster” type to the “translocon” type approximately 470 million years ago (11, 12). The Igh translocon configuration in jawed vertebrates consists of variable (VH), diversity (DH), joining (JH), and constant (CH) regions, although the organization of these regions varies depending on different species, such as mouse, VH-DH-JH-Cμ-Cδ-Cγ3-Cγ1-Cγ2b-Cγ2a-Cε-Cα; human, VH-DH-JH-Cμ-Cδ-Cγ3-Cγ1-ψε-Cα1Cγ2-Cγ4-Cε-Cα2; rabbit, VH-DH-JH-Cμ-Cγ-Cε-Cα-Cα (13 Cα genes were repeated); and cattle, VH-DH-JH-Cμ-Cδ-Cγ3-Cγ1-Cγ2-Cε-Cα4. In teleosts, gene organization appears to be an intermediate type between the multicluster and translocon types in its evolution from Chondrichthyes to tetrapods. The Igh genes possess a translocon configuration, VH-DH-JH-Cζ/τ-(VH)-DH-JH-Cμ-Cδ, similar to those of tetrapods, although there are differences in the Cζ/τ gene locations among teleost species and other vertebrate groups (13, 14). Interestingly, in catfish groups such as channel catfish (Ictalurus punctatus), the Cζ/τ genes are not found either in the 3’-region of the VH gene cluster or within the VH gene (15).
The Ighμ gene was the first Ig class identified in teleosts and has long been considered the most primitive and most prevalent Ig in fish plasma. It can be expressed as mIg or sIg. Secreted tetrameric IgM represents the main Ig in the serum of teleosts. In teleosts as well as a wide range of mammalian species, molecular characterization of the Ighμ gene has revealed that the secreted Ighμ transcripts consist of a rearranged VDJ region spliced to the Cμ1 domain, followed by Cμ2, Cμ3 or Cμ4 (VH-Cμ1-Cμ2-Cμ3- Cμ4). Interestingly, the unusual transcript patterns of the Ighμ gene indicate that the membrane Ighμ transcripts in teleost fish appear to be shorter than those in mammals since the first transmembrane (TM) exon is spliced directly to Cμ1, Cμ2 or Cμ3 thereby excluding the entire Cμ2 or/and Cμ3 or/and Cμ4 exons (VH-Cμ1-TM, or VH-Cμ1-Cμ2-TM, or VH-Cμ1-Cμ2-Cμ3-TM, respectively). These patterns have been identified and reported in many teleost species (3, 16–27).
In Thailand, the production of catfish is the second largest fish aquaculture industry, with approximately 159,314 million tons in 2004 (28). Two major species of catfish, bighead catfish (Clarias macrocephalus Gunther, 1864) and North African catfish (C. gariepinus), and their hybrid catfish (“pla-duk-big-uey” in Thai), are economically popular cultured species in southeast Asia, especially in Thailand, Vietnam, Malaysia and Indonesia. Notably, the hybrid catfish (C. macrocephalus × C. gariepinus) demonstrates many commercially desirable dominant characteristics from its parents, such as good quality of meat, rapid growth, better feed conversion, increased survival, resistance to many diseases and tolerance to many environmental conditions. Therefore, bighead catfish has become a remarkable supporter species in catfish production in many counties in southeast Asia.
Most of our current knowledge on the immune systems of catfish comes from other models and economically important fish species, such as zebrafish (Danio rerio) and medaka (Oryzias latipes) (14, 29–35). In addition, the structural, functional and genetic features of catfish Igs in the genus Clarias are still virtually absent.
The study of molecular Ig genes in catfish could provide a better understanding of their role in immunity, and these genes could be applied as tools for basic research, diagnosis, and therapy in catfish culture farming. Thus far, the study of Ig genes has focused on the molecular structure of Ighμ loci in catfish, particularly their splicing patterns, functions, diversification, expression and homogeneity or heterogeneity between species and their hybrids. In the present study, the organization of the Ighμ genes was analyzed at the chromosomal level. The obtained data provide new knowledge regarding catfish and may benefit applications in sustainable aquaculture industries.
Materials and Methods
Animals
Bighead catfish (Clarias macrocephalus Günther, 1864), North African catfish (Clarias gariepinus) and their hybrid catfish (C. macrocephalus × C. gariepinus), 90–120 g in body weight, were obtained from the Department of Aquaculture, Faculty of Fisheries, Kasetsart University, Thailand. The fish were acclimatized in a quarantine tank with aerated freshwater at temperatures between 28 and 31°C for 2 weeks before the start of the experiment. The experimental procedures performed with aquatic animals were carried out in accordance with the Ethical Principles and Guidelines for the Use of Animals National Research Council of Thailand for the care and use of animals for scientific purposes. The protocol was approved by the Animal Ethics Committee, Kasetsart University, Thailand (Ethics ID: ACKU61-FIS-004).
Isolation of Genomic DNA and RNA and cDNA Synthesis
Catfish genomic (g) DNA was isolated from whole blood tissues using a QIAamp DNA Blood and Tissue Mini Kit (QIAamp, CA, USA) according to the manufacturer’s protocols.
The PBLs of catfish are target tissues used to isolate total RNA and for cDNA synthesis. Total RNA was analyzed using NucleoZOL™ reagent (Clontech Laboratories, CA, USA) according to the manufacturer’s instructions. The obtained total RNA was then used as templates for first strand cDNA synthesis using the protocol described for the Thermo Scientific RevertAid Reverse Transcriptase Kit (Thermo Fisher Scientific, MA, USA). The products of the gDNAs and first-strand cDNA synthesis were stored at -80°C for further experiments.
Amplification of the Internal Constant Domain of cDNA Encoding Ighμ Genes
Degenerative primers were first designed to amplify the internal constant domain region of cDNA encoding Ighμ genes based on the highly conserved regions of Ighμ genes from the NCBI nucleotide database of closely related species of catfish, including Ictalurus punctatus (M27230), Hemibagrus macropterus (JF909893), Silurus meridionalis (KJ659069) and Pelteobagrus fulvidraco (JN202623). The primers were Cla_Ighμ_intr_f: 5′-GTYTMCMSYDTGGCARTGCGGCBC-3′ and Cla_Ighμ_intr_r: 5′-GARVYCTCTGGTGGAGSGAGCAMG-3′ with an approximate amplicon size of 950 bp. Reverse transcription PCRs (RT-PCRs) were carried out using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols. The PCR cycling conditions are one cycle of 95°C for 5 min, 30 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 90 sec, followed by a final extension at 72°C for 5 min. Then, PCR products encoding Ighμ genes were ligated to the pJET1.2/blunt cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols. Nucleotide sequencing of the recombinant plasmid was performed by the Macrogen sequencing service (Macrogen Inc., Seoul, South Korea) using pJET1.2 forward and reverse sequencing primers.
Recovery of 5′- and 3′-Constant Domains of cDNA Encoding Ighμ Genes
The 5′- and 3′- internal constant domain sequences of cDNA encoding Ighμ genes were recovered by rapid amplification of cDNA ends (RACE) PCR techniques using 5′- and 3′-SMARTer® (Clontech, Mountain View, CA, USA), according to the manufacturer’s protocol, with SMARTer® universal primers and gene-specific primers that were provided from the kits and designed from 5′- or 3′- internal constant domain sequences, respectively (Cla_Ighμ_5RACE_r: 5′-GTGCCGCTCGCATCCTTCCAAACG-3′ and Cla_Ighμ_3RACE_f: 5′-GGCTCAACTTCTCCAGTTAAGTG-3′). The 5′ and 3′ RACE-PCR products were cloned into the pJET1.2/blunt cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced using the Macrogen sequencing service (Macrogen, Inc., Seoul, South Korea) as described above.
Cloning and Characterization of Ighμ Genes of Catfish
To obtain and characterize the Ighμ genes of catfish, we used catfish gDNA to amplify the Ighμ genes of catfish using forward and reverse specific primers designed from the highly conserved region of the 5′ end of Cμ1 and the 3′UTR of full-length Ighμ cDNAs, respectively. The forward primer was Cla_Ighμ_Cμ1_f: 5′-GAACGTCGGTGACCGTAACTTCA-3′ for all Clarias catfish species. The reverse primers were Cla_mac_Ighμ_3UTR_r: 5′-GAACACACAAGCATCAGACAGACTG-3′ for big head catfish and hybrid catfish and Cla_gar_Ighμ_3UTR_r: 5′-CAAGAACACACAAGCATCAGACAG-3′ for North African catfish and hybrid catfish.
PCRs were carried out using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols. The PCR cycling conditions were one cycle of 95°C for 5 min, 30 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 5 min, followed by a final extension at 72°C for 10 min. The PCR products of Ighμ genes were cloned and sequenced as described above.
Sequence and Phylogenic Analyses of Full-Length cDNAs and Ighμ Gene
The obtained nucleotide sequences of Ighμ genes were characterized, analyzed, assembled, and aligned using bioinformatic software DNA sequences including GENETYX version 7.0, BLASTN (https://blast.ncbi.nlm.nih.gov/), BLASTX (https://blast.ncbi.nlm.nih.gov/), ExPASy (https://www.expasy.org/), DAS-Transmembrane Prediction server (www.sbc.su.se/~miklos/DAS/), MatGAT program version 2.0 (http://www.bitincka.com/ledion/matgat), and MEGA version 7.0 (http://www.megasoftware.net). Furthermore, the sequences were aligned with those of related Ighμ gene sequences in other vertebrate species using ClustalW. Phylogenetic trees were constructed using Molecular Evolutionary Genetics Analysis (MEGA) software, version 7.0 (Proprietary freeware, Japan) with neighbor-joining (NJ) algorithms with a bootstrap of 1000 replications.
Prediction of Protein Structures of Catfish Ighμ Genes
The protein structures of Ighμ molecules of catfish were determined to predict the possibility of constant domain structure using the structural bioinformatics web server SWISS-MODEL workspace integrates programs (https://swissmodel.expasy.org) according to the programs’ procedures (36). The amino acids in the constant domain of each Ighμ molecule were used to generate the target sequence for protein structure prediction.
Cytogenetic Mapping of Ighμ Genes in Catfish
Metaphase chromosomes of catfish were obtained from the head kidney of each catfish species treated with colchicine according to Karami et al. (2015) (37). Prepared tissues were stored in fixative solution at -20°C until use. The chromosomal spreads were analyzed by staining with 15% Giemsa for 45 min and examined under a light microscope (Olympus, MA, USA). The Ighμ gene probes for fluorescence in situ hybridization (FISH) used full-length Ighμ genes containing the constant domain Cμ1 to the 3′UTR of the Ighμ genes of each catfish species. DNA probes were performed according to standard procedures of FISH Tag detection kits (Thermo Fisher Scientific, MA, USA). Metaphase chromosome hybridization was carried out following a previously described method (38). Chromosomes and specific probes were performed with DAPI and Alexa Fluor 488 or 594 dyes following the manufacturer’s instructions (Thermo Fisher Scientific, MA, USA). Digital micrographs were recorded and processed using a Nikon/DigitaL Eclipse C2Si confocal microscope (Nikon Instruments Inc., NY, USA).
Diversity Analysis of the Variable Domain of Ighμ Genes of Catfish
The 5′ VH region of first-strand cDNAs encoding Ighμ genes was synthesized from total RNA of PBLs of each catfish species using a 5′ RACE-Ready cDNA synthesis kit (Clontech, Mountain View, CA, USA), as previously described. PCR was amplified using Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, USA) with the 5′ universal primer mix (UPM) and gene-specific primers (GSPs) from the kit and nucleotide sequences located in the 5′ region of the Cμ1 constant domain of Ighμ genes (Cla_Ighμ_Cμ1_r: 5′-GTGCCGCTCGCATCCTTCCAAACG-3′). The PCR cycling conditions were one cycle of 95°C for 5 min, 25 cycles of 95°C for 30 sec, 68°C for 30 sec, and 72°C for 120 sec, followed by a final extension at 72°C for 5 min. The 5′ RACE VH PCR products of Ighμ cDNA were ligated to the pJET1.2/blunt cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. A different hundred recombinant clones were randomly selected for sequencing by the Macrogen sequencing service (Macrogen Inc., Seoul, South Korea) as previously described.
Analysis of VH, DH and JH Segments
The 5′ VH nucleotide sequences of Ighμ genes were trimmed to three separated regions encoding the VH, DH and JH genes using an Ig variable domain sequence analysis tool (NCBI IgBLAST, https://www.ncbi.nlm.nih.gov/igblast/). The nucleotide sequences of each gene region were classified at the family level based on 80% similarity of nucleotide sequences using the matrix global alignment tool (MATGAT 2.0).
Analysis of Complementarity-Determining Regions (CDRs)
CDR nucleotide regions of each variable domain nucleotide sequence were first characterized using IgBLAST (https://www.ncbi.nlm.nih.gov/igblast/). The DH segments were predicted using the VDJsolver 1.0 server (https://services.healthtech.dtu.dk). The CDRs were analyzed for their characterization, including CDR length distribution and amino acid composition in each position, using the High V-QUEST IMGT tool (39).
Analysis of the Variability Plot of the Variable Domains of IgM Molecules
The variability plots of variable domain nucleotide sequences were analyzed using the Shannon (1948) (40) and Kabat and Wu (1971) (41) methods. The variability plots were illustrated using the online bioinformatics software protein variability server (PVS, https://imed.med.ucm.es/PVS/pvs-help.html).
Expression of Ighμ Genes in Catfish
The tissue distribution of Ighμ genes of catfish was addressed in both normal fish and fish that underwent Aeromonas hydrophila challenge. Fifty acclimatized catfish were randomly transferred into tanks containing 500 L of water. Catfish were used to determine the expression of the Ighμ gene in normal fish and infectious fish. The infectious catfish were intraperitoneally injected with 0.1 mL of virulent A. hydrophila AQH0018 bacterium (1 × 106 CFU/mL). A virulent A. hydrophila AQH0018 bacterium was grown in TSB medium at 32°C for 18 hr, and cell pellets were harvested by centrifugation at 2,500 rpm for 10 mins, washed and resuspended in 0.85% NaCl prior to injection. The effective doses were preliminarily determined to validate the optimum dose throughout the median lethal dose (LD50) assay (42). Sixteen organs, brain (BR), dendrite (DR), gall bladder (GB), gills (GIL), head kidney (HK), heart (HR), intestine (IN), liver (LI), muscle (MC), ovary (OV), peripheral blood lymphocytes (PBL), skin (SKN), spleen (SPL), stomach (STO), testes (TES) and trunk kidney (TK), were collected from three fish of each catfish before injection (normal fish) and post injection (infectious fish) at 0, 12, 24, 36, 48, 60, 72, 84, 96, 108, 120, 132, 144, 156 and 168 hr.
Total RNA and first-strand cDNA synthesis from sixteen organs of catfish were isolated using NucleoZOL™ reagent (Clontech Laboratories, CA, USA) and a Thermo Scientific RevertAid Reverse Transcriptase kit (Thermo Fisher Scientific, MA, USA) following the manufacturer’s instructions.
Quantification of Ighμ gene expression was performed using quantitative reverse transcriptase PCR (qRT-PCR) assays. qRT-PCR assays were performed with Brilliant III Ultra-Fast SYBR® Green (Agilent, CA, USA) in Mx3005P QPCR Systems (Agilent, CA, USA). qPCRs of the Ighμ gene and β-actin genes of each fish species were conducted using the specific primers Cla_Ighμ_qpcr_f: 5′-TGGACTGAGCTACGTTTGGAAGGA-3′ and Cla_ Ighμ_ qpcr_r: 5′-CGCCTGACTCACTGAGGAGTACTT-3′ with an amplicon size of 167 bp and Cla_b-actin_qpcr_f: 5′-GTCCGTGACATCAAGGAGAAGCTC-3′ and Cla_b-actin_qpcr_r: 5′-GGACTCCATACCCAGGAAAGATGG-3′ with an amplicon size of 189 bp for the Ighμ and β-actin genes, respectively. In addition, the expression distribution of two novel transcripts including Cm_mIgM2 and Cm_mIgM3 were performed in sixteen organs of healthy bighead catfish. The specific primers are Cm_mIgM2_f: 5′-GCACAGAGTTCACCTGCAATG-3′ and Cm_mIgM2_r: 5′-AAATGTTAAGGCATACAGACC-3′ with an amplicon size of 154 bp, and Cm_mIgM3_f: 5′-CCACGGATACGTCTGGAGAA-3′ and Cm_mIgM3_r: 5′-ACTCGCCTTGACTCTCACTG-3′ with an amplicon size of 163 bp.
qRT-PCR cycling conditions were one cycle of 95°C for 5 min, 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 90 sec, followed by a final extension at 72°C for 10 min. The expression of the β-actin gene was used as the housekeeping gene to standardize the results by eliminating variation in mRNA and cDNA quantity and quality. The relative expression of the Ighμ gene in catfish tissues was calculated using 2-ΔΔCT analysis according to the protocol by Schmittgen and Livak (2001) (43). The brain RT was used as calibrator. All reactions were done in triplicate.
Statistical Analysis
The relative expression levels of Ighμ genes were calculated using β-actin as a reference. All quantitative data are presented as the mean ± standard deviation (SD). Statistical analysis at each time course was performed using SPSS statistics 24.0 software using one-way analysis of variance (ANOVA) and Duncan’s new multiple range test (DMRT). The level of statistical significance between species in different organs is indicated as * (P<0.05), ** (P<0.01) and *** (P<0.001) using Student’s t-test.
Results
Characterization and Sequence Analysis of cDNA Transcripts Encoding a Constant Region of the Ighμ Gene
Through analysis of the cDNA transcripts encoding constant regions of the Ighμ genes in bighead catfish, North African catfish and their hybrid catfish, one hundred positive clones of each catfish were first cloned and sequenced to obtain the internal nucleotide sequences of the constant regions of the Ighμ genes. A single band of PCR fragment was shown for each catfish species. Sequence analysis indicated no variation in the nucleotide sequences of individual bighead catfish, North African catfish and their hybrid catfish. There were four exons encoding Cμs [Cμ1(partial)-Cμ2-Cμ3-Cμ4(partial)] of 954 and 949 bp for bighead catfish and North African catfish, respectively (Figure 1A), compared to the published Ighμ sequence of channel catfish (Ictalurus punctatus). In the hybrid, two different nucleotide sequences were observed; each sequence exhibited a 100% match with the Ighμ sequence of one of the parents. Among a hundred clones from hybrid catfish, the Ighμ nucleotide sequences shared a distribution of 28% and 72% to bighead catfish and North African catfish, respectively (Figure 1A).
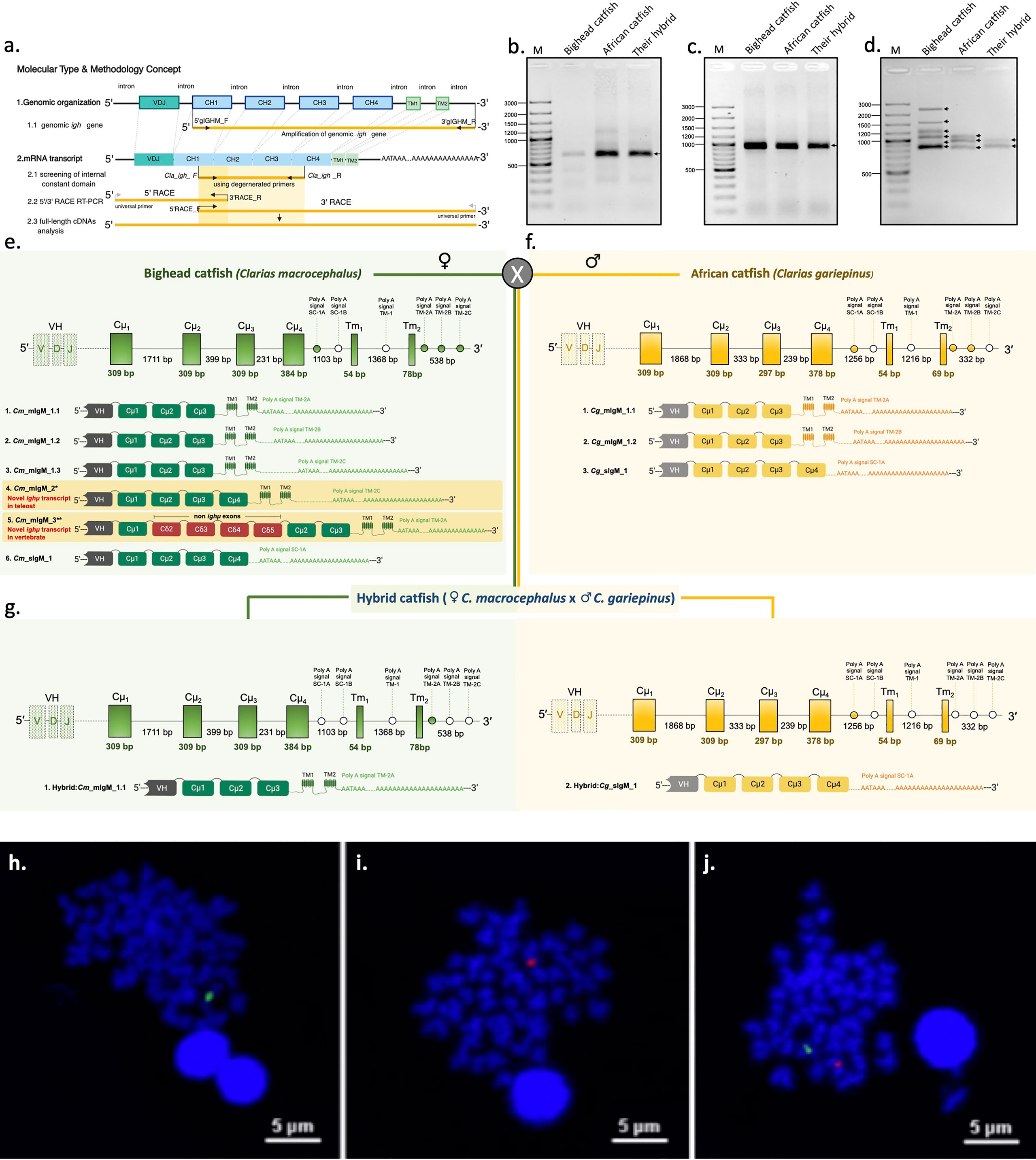
Figure 1 Representation of the genomic organization and splicing patterns of Ighμ in teleosts and catfish. Ighμ generally encodes a VH domain, four constant (Cμ1 to Cμ4) domains and two transmembrane domains. For full-length cDNA Ighμ transcripts in bighead catfish, North African catfish and their hybrid catfish, three major molecular techniques were performed (A). Patterns of PCR amplification products, 5’ RACE PCR (B), internal constant domain (C), and 3’ RACE PCR (D) of Ighμ of bighead catfish, North African catfish and their hybrid catfish. A single gene copy (NCBI accession no. MZ559374) and six different Ighμ transcripts were identified in bighead catfish, and five and one were expressed as membrane (NCBI accession nos. MN934742, MN934743, MN934744, MN934745 and MN934746) and secreted forms (NCBI accession no. MN934747), respectively. Moreover, two membrane forms of Ighμ transcripts had novel splicing patterns in teleosts and vertebrates (highlighted in yellow) (NCBI accession nos. MN934745 and MN934746) (E). A single gene copy (NCBI accession no. MZ559375) and three different Ighμ transcripts were identified in North African catfish; two and one were expressed as membrane (NCBI accession no. MN934748 and MN934749) and secreted forms (NCBI accession no. MN934750), respectively (F). Two gene copies (NCBI accession nos. MZ559376 and MZ559377) and two Ighμ transcripts were identified in the hybrid catfish (NCBI accession nos. MN934751 and MN934752) (G). Comparisons of cytogenetic localization of Ighμ gene loci in bighead catfish (H), North African catfish (I) and their hybrid catfish (J). All nucleotide sequences are illustrated in supplementary files and the NCBI database corresponding to the provided NCBI accession numbers.
The 5′ and 3′ of the Ighμ gene constant regions in each catfish were consequently cloned and sequenced using 5′ and 3′ RACE techniques with specific primers designed from the 5′ end of the Cμ2 exon and 3′ end of Cμ1 for recovering the 5′ and 3′ nucleotide regions of Ighμ cDNAs, respectively. The obtained 5′ and 3′ nucleotide sequences of Ighμ genes corresponded to the Cμ1-Cμ2-Cμ3 and Cμ3-Cμ4-TM-polyA tails, respectively (Figure 1A). No variation difference in the nucleotide sequences was observed in the 5′ Ighμ constant region exon in bighead catfish and North African catfish. In addition, the hybrid catfish exhibited two different nucleotide sequences, one from each parent (Figures 1A, B). There was no variation in the internal nucleotide sequences of the constant regions of the Ighμ genes in all studied catfish. (Figure 1C). However, the sequences of the 3′ constant regions consisted of two major different Ig forms, (mIg and sIg), which are controlled by alternate mRNA processing events in certain species. Up to six nucleotide sequence patterns of 3′ region exons were observed in bighead catfish (Figure 1D): five patterns for mIg and one pattern for sIg. Among the five patterns of mIg, three corresponded to Cμ1-Cμ2-Cμ3-TM1-TM2 with differences in the polyadenylation signal region (polyA signal: TM-2A, TM-2B and TM-2C, respectively) of the untranslated 3′ sequence, one corresponded to Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2 containing the polyA signal TM-2C, and one corresponded to Cμ1-(non-Ighμ exons)-Cμ2-Cμ3-TM1-TM2 containing the polyA signal TM-2A. One sIg was encoded by exon Cμ1-Cμ2-Cμ3-Cμ4 and the polyA signal SC-1A (Figures 1D, E and Supplementary Figures 1–6).
Three nucleotide sequence patterns were observed in North African catfish, and they corresponded to two patterns for mIg and one pattern for sIg (Figure 1D). Two patterns of mIg corresponded to Cμ1-Cμ2-Cμ3-TM1-TM2 with differences in the polyadenylation signal region (polyA signal: TM-2A and TM-2B, respectively) of the untranslated 3′ sequence. Another pattern corresponded to sIg that contained four exons coding Cμ1-Cμ2-Cμ3-Cμ4 and the polyA signal SC-1A (Figures 1D, F and Supplementary Figures 7–9).
Two nucleotide sequence patterns of the 3′ region exon corresponding to mIg (Cμ1-Cμ2-Cμ3-TM1-TM2) and sIg (Cμ1-Cμ2-Cμ3-Cμ4) were observed in the hybrid. The mIg and sIg nucleotide sequence coding from Cμ1 through polyA tails were absolutely identical to their parent, bighead catfish and African catfish, respectively (Figures 1D, G and Supplementary Figures 1, 9).
Cytogenetic Mapping of Ighμ Genes in Catfish
In situ hybridization with specific probes to the Ighμ gene of individual catfish species, bighead catfish and North African catfish revealed that only a single copy of the Ighμ gene was found in bighead catfish (Figure 1H) and North African catfish (Figure 1I). In addition, a strong hybridization of both the Ighμ genes of bighead catfish and North African catfish was represented in their hybrid catfish (Figure 1J). The expression of Ighμ transcripts in the hybrid catfish from the two Ighμ genes received from the parents was confirmed.
Amino Acid Sequence Comparison of Ighμ Transcripts in Clarias Catfish
The amino acid sequences of Ighμ transcripts among bighead catfish, North African catfish and their hybrid catfish were aligned and compared to published complete IgH gene loci of channel catfish (Ictalurus punctatus) and zebrafish (Danio rerio). The aligned sequences are illustrated in Figure 2. The results showed several important features based on the overall relative size of the amino acid sequences of general Ighμ transcripts, with 436 and 430 residues for the constant (Cμ1-Cμ4) domain and 44 and 41 residues for the transmembrane domains of bighead catfish and North African catfish, respectively. The mIgM and sIgM forms of the hybrid catfish were similar in size to those of the parents, bighead catfish and North African catfish (Figure 2). The sequence similarity of the mIgM and sIgM forms of the hybrid catfish were absolutely conserved and similar to those of the parents. These results exhibited highly important significant differences between their phenotypic characterization and relationship to their parent.

Figure 2 Comparisons of translated amino acids among bighead catfish, North African catfish, their hybrid catfish, channel catfish and zebrafish in different Ighμ-coded exons found in this study: Cμ1 exon (A), non-Ighμ exon (B), Cμ2 exon (C), Cμ3 exon (D), Cμ4 exon (E) and Ighμ tail exon (F). *, fully conserved residue of amino acids that play the same role in the same column/position.
The translated amino acid sequences of the individual Cμ1, Cμ2, Cμ3, Cμ4 and transmembrane domains of bighead catfish, North African catfish and their hybrid catfish were used to define significant similarity among their relationships and evolutionary lineage. The amino acid sequences of the Cμ1 and Cμ2 domains of all catfish were similar in size, with 103 and 103 residues, respectively (Figure 2A). However, the Cμ1 and Cμ2 domains were slightly larger and smaller than the channel catfish Cμ1 and Cμ2 domains (102 and 104 residues, respectively) (Figure 2A). Additionally, bighead catfish shared amino acid similarity of Cμ1 and Cμ2 to African catfish, the hybrid catfish (mIgM), the hybrid catfish (sIgM), channel catfish and zebrafish with 82.4, 100.0, 82.4, 72.5 and 38.6% similarity for the Cμ1 domain and 77.2, 100.0, 727.2, 57.8 and 36.8% similarity for the Cμ2 domain, respectively (Supplementary Table 1). The amino acid similarity of the Cμ3 domains of bighead catfish shared 69.7, 100.0, 69.7, 45.9 and 32.9% and 74.4, 100.0, 74.4, 54.03 and 43.2% similarity in the Cμ4 domain to North African catfish, the hybrid catfish (mIgM), the hybrid catfish (sIgM), channel catfish and zebrafish, respectively. In addition, the amino acid similarity values of transmembrane domains of the Ighμ carboxyl-terminal region of bighead catfish were 97.5, 100.0, 84.0 and 76.0% for North African catfish, the hybrid catfish (mIgM), channel catfish and zebrafish, respectively. Overall, based on amino acid conservation analyses of five complete domains of Ighμ of bighead catfish, the Cμ1, Cμ2, Cμ3, Cμ4 and TM domains shared 75.8, 100.0, 75.8, 57.5 and 38.4% similarity with North African catfish, the hybrid catfish (mIgM), the hybrid catfish (sIgM), channel catfish and zebrafish, respectively (Supplementary Table 1).
The conservation of the Ighμ domain is evident in the catfish Cμ1, Cμ2, Cμ3, and Cμ4 of Ighμ structures. The cysteines that likely form the intradomain disulfide bridge (S-S) as well as the tryptophan located within the Cμ1, Cμ2, Cμ3, and Cμ4 regions were absolutely conserved among catfish, channel catfish and zebrafish (Figures 2A–E). The cysteine that likely participates in the disulfide linkage of the catfish chain to the light chain was also conserved in the Cμ1 regions (Figure 2A). Additionally, the predicted results of these protein tertiary structures of Ighμ transcripts in bighead catfish, North African catfish and their hybrid catfish are illustrated in Figure 3. The Ighμ molecules encoded by general coding exons found in teleost and catfish mIg (Cμ1-Cμ2-Cμ3-TM1-TM2) and sIg (Cμ1-Cμ2-Cμ3-Cμ4) exhibited similar predicted structures in all the studied catfish (Figures 3A–C). Additionally, the predicted results of these protein tertiary structures of Ighμ transcripts of unusual splicing patterns generated novel membrane forms of Ighμ transcripts in teleosts [Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2] and vertebrates [Cμ1-(Cδ2-Cδ3-Cδ4-Cδ5)-Cμ2-Cμ3-TM1-TM2] that were larger in size and rather complex compared to other general predicted structures (Figures 3D, E).
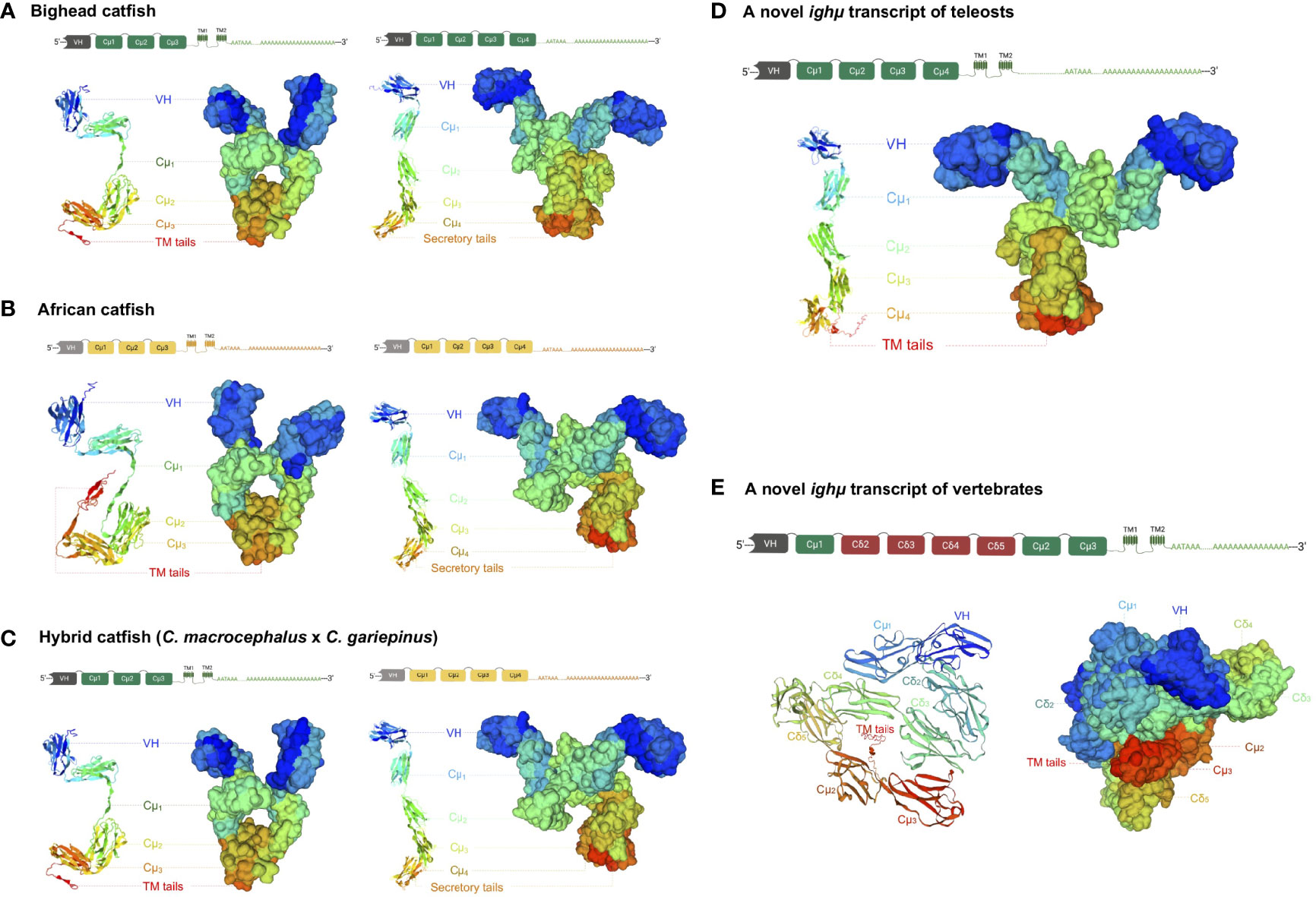
Figure 3 The predicted crystal protein structures of Ighμ transcripts expressed in the membrane and secreted forms in different catfish: bighead catfish (A), North African catfish (B), and their hybrid catfish (C). The more complex molecules of two novel Ighμ transcripts found in bighead catfish are illustrated in (D, E).
The evolutionary relationships of Ighμ constant regions among various vertebrate species are illustrated in Figure 4A, and the relationships of teleost Igh constant regions are illustrated in Figure 4B. In this study, the Clarias catfish Ighμ constant domains were cladded into the catfish group of vertebrate lineages and classified into the Ighμ group among the reported Igh in fish.
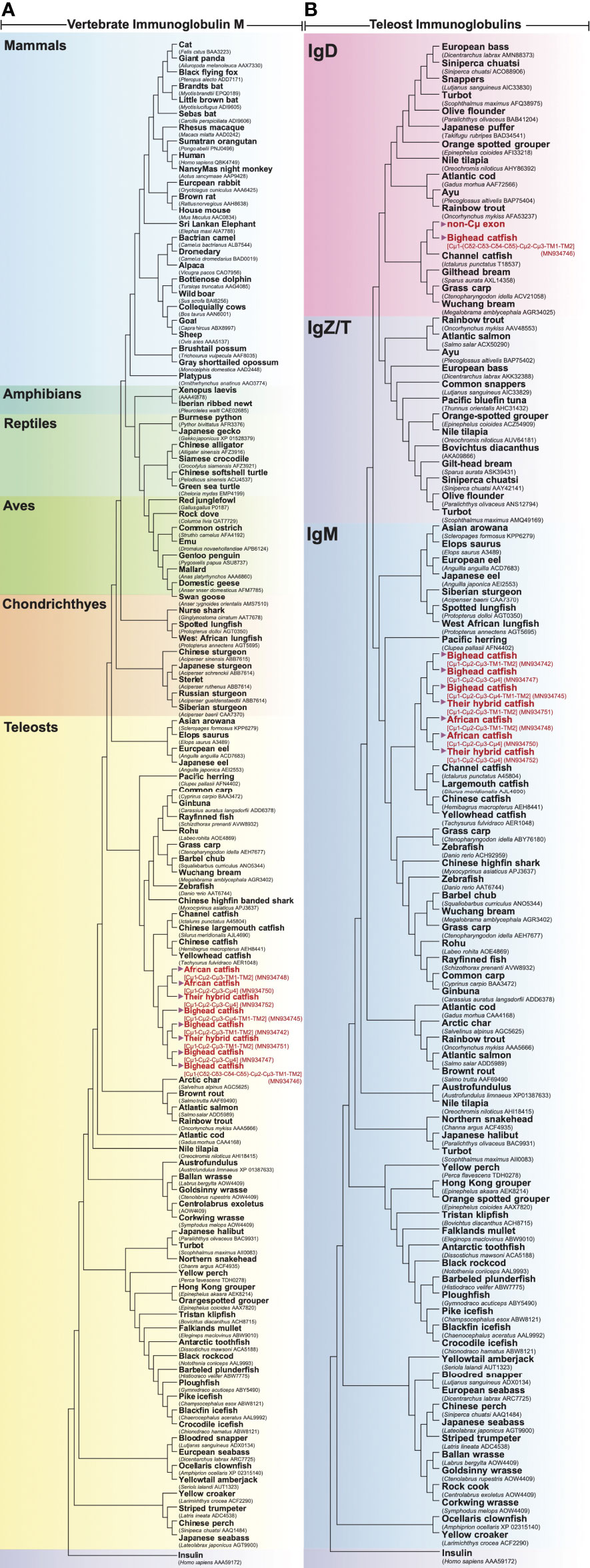
Figure 4 Evolutionary phylogenetic relationship of Ighμ between bighead catfish (NCBI accession nos. MN934742, MN934743, MN934744, MN934745, MN934746 and MN934747), North African catfish (NCBI accession nos. MN934748, MN934749 and MN934750), their hybrid catfish (NCBI accession nos. MN934751 and MN934752) and vertebrates, including mammals, amphibians, reptiles, aves, chondrichthyes and other teleosts (A). Phylogenetic relationship between immunoglobulin heavy chain, IgM, IgD and IgZ/T, expressed in reported teleost fish and bighead catfish, North African catfish and their hybrid catfish (B). Phylogenetic trees were constructed using neighbor-joining (NJ) algorithms with a bootstrap of 1000 replications.
Unusual Splicing Patterns Generate Novel Membrane Forms of Ighμ Transcripts in Teleosts and Vertebrates
The two novel and unusual splicing patterns of the Ighμ membrane form were found in only bighead catfish: one was associated with a new splicing pattern among teleosts, and the other among vertebrates. The constant region of the new Ighμ membrane form in teleosts is encoded by four Cμ4 exons and directly spliced into the first transmembrane exons (Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2), as reported in other vertebrates, such as tetrapods, birds, sharks, and mammals. However, the castratory event was observed by the lack of a cryptic donor splicing site at the 3′ region of the Cμ4 exon, which is important for splicing to the transmembrane domain to generate the membrane form as reported previously. The predicted crystal protein structure of this molecule is illustrated in Figure 3D. The predicted results of the protein tertiary structures are close to the actual crystal structures of reported Ighμ transcripts in bighead catfish (Figure 3A), North African catfish (Figure 3B) and their hybrid catfish (Figure 3C).
The other novel and unusual splicing pattern of the Ighμ transcript was notable for vertebrates. The largest molecule of the Ighμ membrane-bound form was defined together with the new Ighμ splicing pattern among vertebrate animals. The molecular size of the molecule was approximately 92.64 kDa by encoding two major constant IgH loci in teleosts: Cμ and non-Cμ gene loci. The molecule encoded three Cμs and non-Cμ constant exons. The first exon of the 5′N-terminal domain was the Cμ1 exon of the Ighμ gene, which was directly spliced to the non-Cμ exons (487 residues) (Figure 2B) and then spliced directly to link the Cμ2, Cμ3, TM1 and TM2 exons of the Ighμ gene of bighead catfish. The translated amino acid sequences of the Cμ exons, including Cμ1, Cμ2, Cμ3, TM1 and TM2, were identical (100%) to the previously described Ighμ gene of bighead catfish (Figures 1B, 2). In addition, the encoded non-Cμ constant region exons were studied via BLAST and compared to the published complete IgH gene loci of channel catfish and zebrafish. The translated amino acid sequences of the entire non-Cμ exons were 57.73 and 34.63% identical to the Ighδ gene locus of channel catfish and zebrafish, respectively (Supplementary Table 1). The evolutionary relationship analysis of non-Cμ exons of bighead catfish was confirmed; it was grouped into Ighδ among other teleosts and shared a branch with the Ighδ of the closely related teleost species channel catfish (Figure 4B). In addition, the nearest neighbors’ relationships of presumptive Ighδ constant exons were gilthead bream (Sparus aurata, AXL14358), grass carp (Ctenopharyngodon idella, ACV21058) and Wuchang bream (Megalobrama amblycephala, AGR34025). However, the presumptive non-Cμ exon sequences clustered within teleost Ighδ rather than Ighμ. The predicted protein tertiary structure of this molecule was larger in size and rather complex compared to other predicted structures (Figure 3E).
Genomic Organization of the Ighμ Gene Locus
To gain insight into the Ighμ locus of Clarias catfish and hybrid catfish, we also defined the complete genomic organization of the gene locus. The corresponding genomic sequences encoding the Ighμ constant domain were amplified, cloned, sequenced and compared to the published complete Igh gene locus in channel catfish. Consistent with this finding, only a single copy of the genomic Ighμ gene locus was found in bighead catfish and North African catfish (Figures 1E, F, H, I, and Supplementary Figures 10, 11). The complete constant region of the Ighμ locus, which encodes an Ig μ domain, contained approximately 6,793 kb (six exons, five introns and six polyadenylation signals) and 6,660 kb (six exons, five introns and six polyadenylation signals) of bighead catfish and North African catfish, respectively. However, the individual Ighμ loci of each bighead catfish and North African catfish were also found in the hybrid catfish, which had the transmembrane and secreted Ighμ transcripts, as mentioned before. The nucleotide identity between the two Ighμ gene loci in the hybrid catfish was identical to those of the parents, bighead catfish and African catfish (Figures 1G, J and Supplementary Figures 10, 11).
Diversity Analysis of the Variable Domain (VH) of Ighμ Genes
A total of 100 unique nonredundant clones of each catfish were characterized for VH sequences of the Ighμ gene. The functional regions of VH encoded by the VH, DH and JH segments were identified, and analysis of framework regions (FWs) and complementarity-determining regions (CDRs) was also performed (Figure 5A) according to the international immunogenetics (IMGT) information numbering system for Ig structural definition (59).
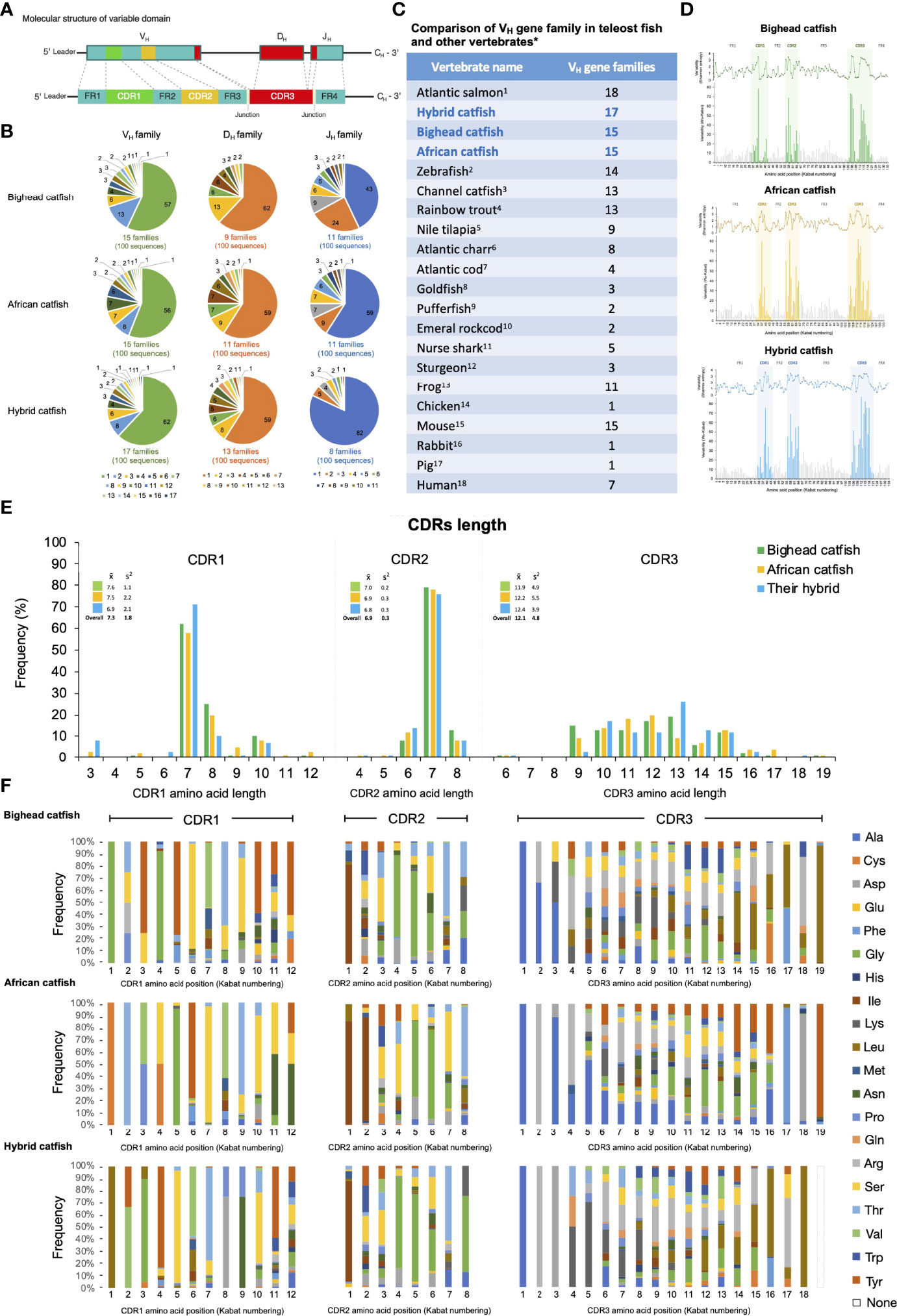
Figure 5 Representation of comparative diversity analyses of the VH domains of Ighμ bighead catfish, North African catfish and their hybrid catfish. General structures of the VH domain correspond to the VH, DH, and JH segments that consist of the FR1, CDR1, FR2, CDR2, FR3, CDR3 and FR4 regions found in all vertebrates (A). Comparative classification of the VH, DH, and JH gene segments in bighead catfish, North African catfish and their hybrid catfish (B). Comparison of VH gene family members among reported vertebrates (C). Degree of sequence variability of the VH repertoire in bighead catfish, North African catfish and their hybrid catfish, according to the Shannon (1948) and Kabat and Wu (1971) methods (D). Comparison of amino acid length in CDR1, CDR2, and CDR3 of bighead catfish, North African catfish and their hybrid catfish (E), x̅: average nucleotide length, S2: variance distribution. Percent frequency of amino acid variability and composition of CDR1 CDR2 and CDR3 regions of bighead catfish, North African catfish and their hybrid catfish (F). The NCBI accession numbers of VH nucleotide sequences are MN934442- MN934541, MN934542- MN934641 and MN934642- MN934741 for bighead catfish, North African catfish and their hybrid catfish, respectively.1Yasuike et al., 2010 (44); 2Danilova et al., 2005 (13); 3Yang et al., 2003 (45); 4Brown et al., 2006 (46); 5Phuyindee et al., 2015 (47); 6Andersson and Matsunaga, 1998 (48); 7Stenvik et al., 2000 (49); 8Wilson et al., 1991 (31); 9Peixoto and Brenner, 2000 (50); 10Coscia and Oreste, 2003 (22); 11Rumfelt et al., 2004 (51); 12Lundqvist et al., 1998 (52); 13Haire et al., 1990 (53);14Ota and Nei, 1995 (54); 15Mainville et al., 1996 (55); 16Mage et al., 1984 (56); 17Sun et al., 1994 (57) and 18Matsuda et al., 1998 (58).
Based on 80% nucleotide identity, the functional VH segments were classified into 15, 15 and 17 families for bighead catfish, North African catfish and their hybrid catfish, respectively. Between the most 3′ VH segment and the 5′ JH segment, 9, 11 and 13 functional DH segment families were defined in bighead catfish, North African catfish and their hybrid catfish, respectively. Altogether, the functional JH segments of catfish VH have been identified. There were 11, 11 and 8 families of bighead catfish, North African catfish and their hybrid catfish, respectively (Figure 5B). The comparisons of the VH gene family among catfish in this study and among reported vertebrates are illustrated in Figures 5B, C, respectively. The NCBI accession numbers of VH nucleotide sequences are MN934442-MN934541, MN934542-MN934641 and MN934642-MN934741 for bighead catfish, North African catfish and their hybrid catfish, respectively. In addition, adding P- and N-nucleotides was predominantly observed at the junction of the V and J segments for the generation of V(D)J junctional diversity in bighead catfish, North African catfish and their hybrid catfish (Supplementary Figures 12–14).
Analysis of Framework Regions (FWs) and Complementarity-Determining Regions (CDRs)
To determine the extent of diversification in the VH repertoire of the Ighμ gene, we calculated the length variation and percent deviations of amino acids in each position of the FWs and CDRs and compared them to those of germline VH genes from the international ImMunoGeneTics information system® (IMGT®, http://www.imgt.org) (60).
The FWs of bighead catfish, North African catfish and their hybrid catfish were similar in length and diversity of amino acids. The lengths of amino acids of FW1, FW2, FW3 and FW4 in all catfish were approximately 33, 14, 36 and 14 residues, respectively (Figure 5D). To examine the degree of sequence variability of the VH repertoire in Clarias catfish, plots of the variability according to the Shannon (1948) (40) and Kabat and Wu (1971) (41) methods were generated. The overall amino acid sequence variability of the VH region was mostly confined to the CDRs and particularly the CDR3s. In addition, the amino acid variability among CDRs (CDR1, CDR2 and CDR3) was highly similar to that of FR1 to FR4 of all studied catfish. Moreover, the comparisons of variability plots of VH sequences among the studied Clarias catfish were closely related in sequence patterns and variability level to each other based on both the Shannon and Kabat and Wu methods (Figure 5D). Shannon entropy plots of all catfish revealed increased diversity at amino acid positions 31 to 42 for CDR1, 58 to 65 for CDR2 and 102 to 120 for CDR3 (Figure 5D). They were generally related to the traditional definitions using the Kabat numbering convention (Figure 5D).
The amino acid length diagrams of CDRs of bighead catfish, North African catfish and their hybrid catfish in Figure 5E indicate that the amino acid length of CDR1 ranged from 3 to 12 residues with an average of 7.6, 7.5 and 6.9 residues, respectively. The most common length was seven for all catfish, with approximately 58 to 71% frequency.
Within nonredundant CDR1 (n = 100) sequences of each Clarias catfish, 6 major amino acids were observed in bighead catfish, which accounted for 79.7% of all residues: Ala (7.7%), Gly (16.5%), Ser (18.9%), Thr (9.1%), Val (4.1%) and Tyr (23.1%). For North African catfish, 8 major amino acids were observed, which accounted for 93.7% of all residues: Cys (11.7%), Gly (8.5%), Asn (8.0%), Pro (4.0%), Ser (30.2%), Thr (14.5%), Val (8.7%), and Tyr (7.9%). For their hybrid catfish, 7 major amino acids were observed, which accounted for 75.2% of all residues: Asp (6.8%), Gly (8.2%), Leu (10.2%), Ser (17.3%), Thr (8.1%), Val (9.7%), and Tyr (14.6%) (Figure 5F).
The amino acid sequences of the CDR2 were shorter overall than those of CDR1, which were 4 to 8 residues in bighead catfish, North African catfish and their hybrid catfish with average amino acid lengths of 7.0, 6.9 and 6.8 residues, respectively. Approximately 76 to 79% of nonredundant CDR2s were 7 amino acid residues in length. Shannon entropy plots of amino acid variation of nonredundant CDR1 and CDR2 were mostly similar in diversity to those of the related VH families within the species and among Clarias species. Within 8 residues in length of CDR2 revealed highly conserved residues at position one of the CDR1s (Figure 5F).
Six major amino acids were observed in nonredundant CDR2s (n = 100) sequences of bighead catfish, which accounted for 78.4% of all residues: Ala (5.4%), Asp (5.8%), Gly (25.3%), Ile (12.4%), Ser (11.7%) and Thr (17.5%). For North African catfish, 5 major amino acids were observed, which accounted for 78.2% of all residues: Asp (5.6%), Gly (21.0%), Ile (22.9%), Ser (17.4%) and Thr (11.1%). For their hybrid catfish, 5 major amino acids were also observed, which accounted for 73.2% of all residues: Asp (5.3%), Gly (31.8%), Ile (12.1%), Ser (11.3%) and Thr (12.4%) (Figure 5F).
The length distributions of the Clarias catfish CDR3 repertoire varied from 6 to 19 residues with averages of 11.9, 12.2 and 12.4 residues for bighead catfish, North African catfish and their hybrid catfish, respectively. The most common length was 10 to 15 residues for all catfish, with approximately 13 to 26% frequency. Analysis of amino acid frequencies for nonredundant CDR3s across all individual catfish revealed that the highest mutation rates in the VH repertoire were observed within CDR3 for all studied catfish. Highly conserved amino acid residues at positions one and two were observed in the CDR3 of all catfish. Seven major amino acids were observed in bighead catfish, which accounted for 66.3% of all residues: Ala (13.4%), Lys (7.1%), Leu (17.7%), Pro (3.6%), Gln (3.1%), Arg (17.1%) and Trp (4.0%). For North African catfish, 7 major amino acids were identified, which accounted for 80.3% of all residues: Ala (22.6%), Asp (7.5%), Gly (5.8%), His (9.5%), Ser (16.0%) Thr (4.3%) and Yyr (14.4%). For their hybrid catfish, 8 major amino acids were observed, which accounted for 77.4% of all residues: Ala (7.9%), Asp (5.9%), Gly (5.5%), Lys (10.7%), Leu (16.8%), Arg (21.0%), Ser (5.0%) and Thr (4.3%). Notably, three amino acids, Gly, Ser and Thr, were predominantly distributed in all CDR1s, CDR2s and CDR3s of all catfish in this study (Figure 5F).
Expression and Tissue Distribution of Ighμ Genes
To examine the distribution of Ighμ in Clarias catfish, we examined the potential functional coding segment of Ighμ, the Cμ2 region, in sixteen tissues of catfish. Ighμ was highly expressed in the head kidney, spleen and peripheral blood lymphocytes (PBLs) in all studied catfish (Figure 6A). The relative expression levels reached approximately 60-, 700- and 600-fold the accustomed expression in head kidney, an important fish immune response organ, of bighead catfish, North African catfish and their hybrid catfish, respectively. However, the lowest expression was also observed in various tissues, including the brain, dendrite, gallbladder, gills, heart, liver, muscle, ovary, skin, stomach, testes and trunk kidney, in all studied catfish (Figures 6A, B). Highly significant expression levels were observed in North African catfish and the hybrid catfish in several tissues, including the gills, head kidney, heart, intestine, liver, muscle, ovary, PBLs, skin, spleen, stomach, and testes, compared to those in bighead catfish (Figure 6B). No significant difference was observed in tissues of North African catfish and the hybrid catfish. However, the relative expression of Ighμ was not significantly different in dendrites, gall bladder or trunk kidney among all studied catfish (Figure 6B).
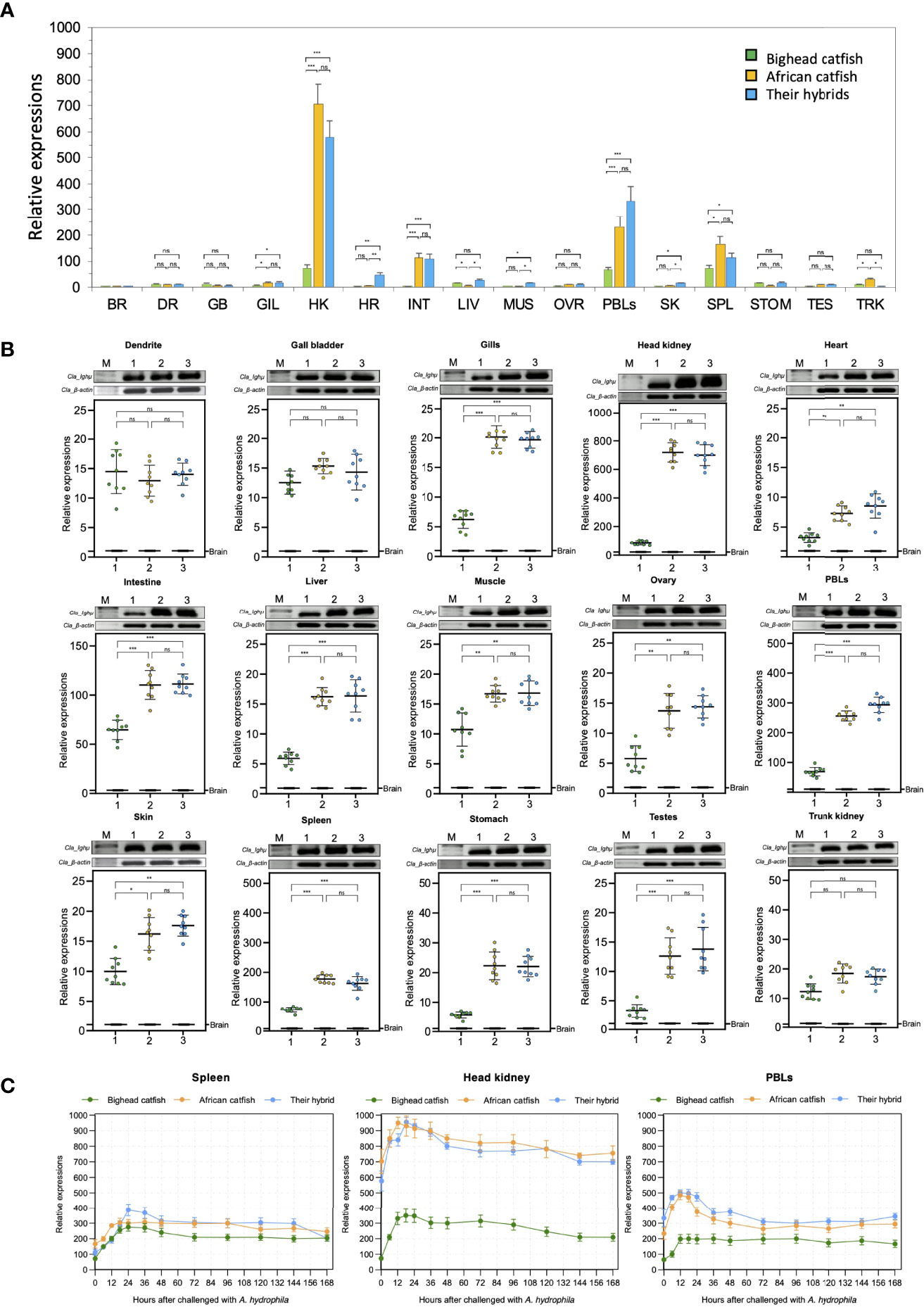
Figure 6 Representation of the overall relative expression and distribution of Ighμ in bighead catfish, North African catfish and their hybrid catfish in sixteen tissues (A). Comparison of the relative expression of Ighμ in sixteen different tissues (B). Expression responses of Ighμ in immune-related organs, including the spleen, head kidney and PBLs, after intraperitoneal injection of a virulent pathogen, Aeromonas hydrophila, of bighead catfish, North African catfish and their hybrid catfish (C). The relative expression levels of Ighμ genes were calculated using β-actin as a reference gene. All quantitative data are presented as the mean ± standard deviation (SD). The levels of statistical significance are indicated by *(P<0.05), **(P<0.01) or ***(P<0.001). The brain RT was used as the calibrator. All experiments are done in triplicate (n = 3). ns, not significant.
Intraperitoneal injection of virulent Aeromonas hydrophila clearly exhibited the pattern of immune responses of catfish, and its significance was defined among the three catfish. In the spleen, the expression of Ighμ was moderately increased approximately 200-400-fold after 12–168 hours (hr) of pathogen exposure for all catfish. For the head kidney, the expression of Ighμ was strongly increased approximately 200–300-, 850–950-, and 800–950-fold after 12–24 hr. of pathogen exposure for bighead catfish, North African catfish and their hybrid catfish, respectively. In the PBLs, Ighμ expression showed significant upregulation of approximately 200-, 450- and 500-fold after 12–24 hr. of exposure for bighead catfish, North African catfish and their hybrid catfish, respectively. In addition, an approximate upregulation of 200–300-fold was also observed in all catfish after 12–24 hr. of exposure (Figure 6C).
The relative expression of two novel transcripts, including Cm_mIgM2 and Cm_mIgM3 in bighead catfish tissues showed highly expression levels compared to brain, approximately 337- and 122-, 90- and 177-, 40- and 20-, 21- and 17-fold, in PBLs, head kidney, spleen, and liver of Cm_mIgM2 and Cm_mIgM3, respectively (Figures 7A, B).
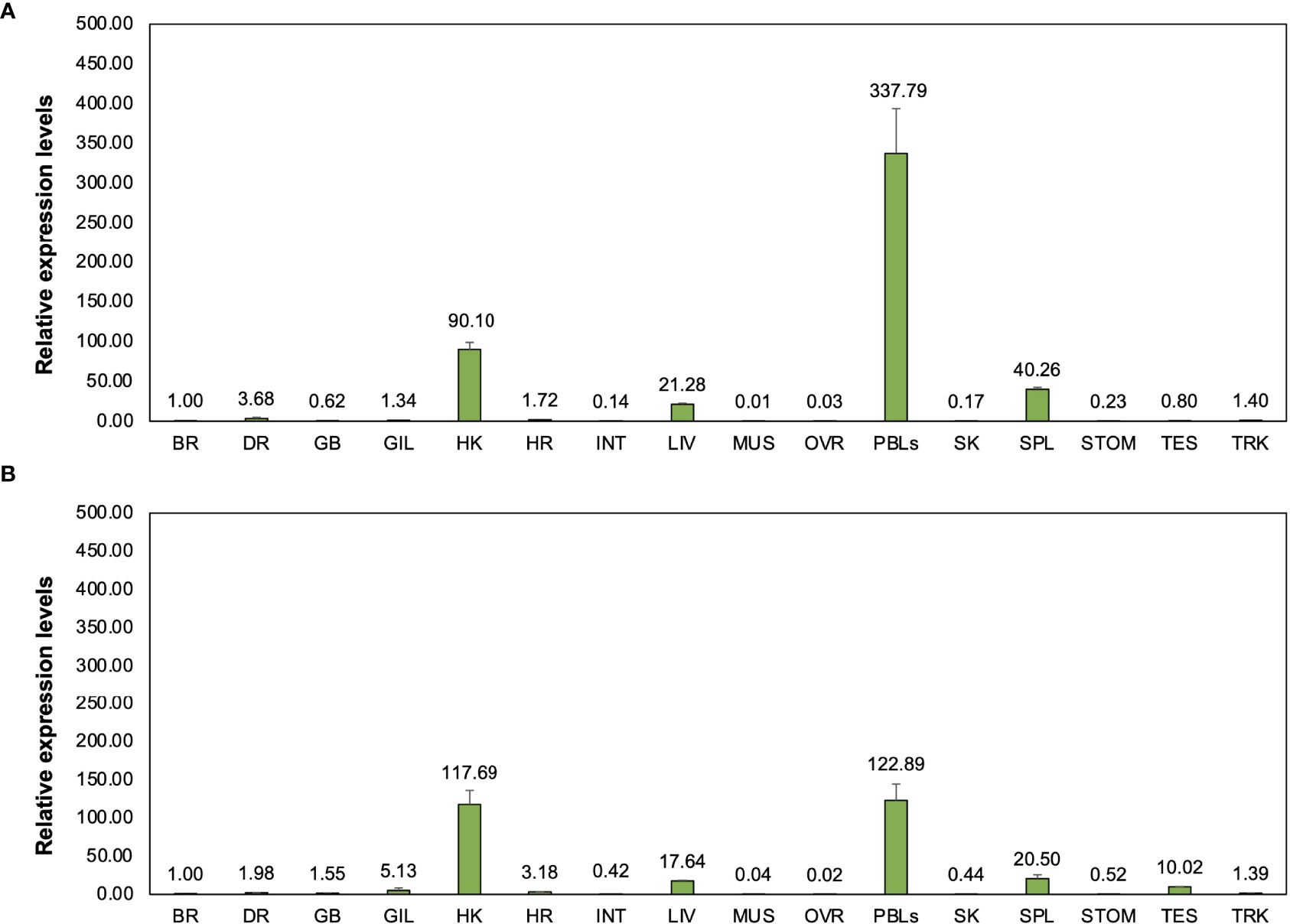
Figure 7 Representation of relative expression levels and distribution of two novel Ighμ molecules in bighead catfish including Cm_mIgM2 (A) and Cm_mIgM3 (B) in sixteen tissues. All experiments are done in triplicate (n=3).
Discussion
The specific features of Ighμ constant region transcripts found in bighead catfish, North African catfish and their hybrid catfish conform to the patterns of organization reported previously in other vertebrate and teleost Ighμ genes. The Ighμ constant regions of the secreted form of Ighμ are a typically spliced pattern, as found in other teleosts, which are encoded by four Cμ protein domains, Cμ1, Cμ2, Cμ3 and Cμ4, without a transmembrane protein domain at the carboxyl-terminal region (7, 61). In addition, the general mRNA splicing patterns to generate the membrane form of Ighμ were found in all studied catfish by direct splicing from Cμ3 to the first transmembrane exon with the lack of a cryptic donor splicing site (T/C/G↓GGTAAA), as reported in the 3′ region of the Cμ4 exon in mammalian, shark, and amphibian mIg molecules. In this study, two domains of transmembrane proteins of Ighμ were found in all studied catfish as well as in channel catfish and other teleosts (5, 14, 24, 29).
Two unexpected and unusual Ighμ constant region transcripts were found in bighead catfish. One is a novel Ighμ transcript for teleosts, and the other is a novel transcript for vertebrates. The form is unusual in that the transmembrane domains are directly spliced to exon Cμ4 to create a membrane-bound form of Ighμ that did not seem to conform to any patterns of the membrane form of the Ighμ structure previously reported in teleosts. The molecule corresponded with Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2. Although this molecular pattern feature is typical of most mammals and vertebrates examined previously (62, 63), the cryptic donor splicing site at the 3′ region of the Cμ4 exon, the acceptor site of the first transmembrane (TM1) exon (62, 64), was not found in the case of this study. In this regard, the presence of the Cμ4 exon between Cμ3 and the first transmembrane domain of the Ighμ membrane-bound form produced a unique characteristic of the mIg form as a novel membrane receptor feature of B cells in bighead catfish and teleosts.
Another important Ighμ transcript was discovered in bighead catfish, and it is interesting in that the membrane-bound form of Ighμ was encoded by two major exon domains, Cμ and non-Cμ exon domains. The molecular pattern was obviously characterized as Cμ1-(non-Cμ)-Cμ2-Cμ3-TM1-TM2. The presence of 487 amino acid residues of the non-Cμ exon between the Cμ1 and Cμ2 regions of the typical membrane-bound form of Ighμ was spliced directly to the end of the Cμ1 exon without any additional or lacking amino acids. Thus far, the non-Cμ exon was defined compared to the most closely and distinctly related published Igh locus, channel catfish (Ictalurus punctatus) (63, 65) and zebrafish (Danio rerio) (13, 66). The amino acid similarity of the non-Cμ exon was 57.73% and 34.63% to the Ighδ gene loci of channel catfish and zebrafish, respectively. It is also characterized as four Ighδ constant exons based on the Ighδ gene locus of channel catfish, Cδ2-Cδ3-Cδ4-Cδ5. There were 79, 220, 94 and 94 amino acid residues, respectively (63, 65). In this regard, it is interesting that the 3′ end of non-Cμ, Cδ5, was directly spliced to the Cμ2-Cμ3-TM1-TM2 exons with the identical polyadenylation signal at the C-terminus of the described typically membrane-bound form of the Ighμ molecule in bighead catfish. The evolutionary relationship analysis of this novel membrane-bound form seemed to confirm the identification by grouping into the Ighδ clade among other teleosts and sharing a branch to the Ighδ of the closely related teleost species, channel catfish (Figure 4). However, the presumptive non-Cμ exon sequences clustered within the teleost Ighδ rather than Ighμ gene domains. The new pattern of mRNA splicing of the Ighμ membrane formed by two major Ighμ and Ighδ constant exons could be defined as Cμ1-Cδ2-Cδ3-Cδ4-Cδ5-Cμ2-Cμ3-TM1 and TM2. The presence of encoding Ighδ constant exons inside the Ighμ molecule suggests that this was a novel splicing pattern among vertebrate animals that has never been present in any class of vertebrates thus far (5, 9, 25, 29, 66). However, the unusual splicing of the two patterns was not found in either North African catfish or the hybrid catfish. Therefore, it is possible that bighead catfish express Ighμ membrane receptors with at least three different mRNA splicing patterns.
Inside the gene organizations of constant Ighμ regions, the lengths of the five introns separating these six exons seem to be similar to those of the closely related species channel catfish42 and quite different from those of other vertebrates. The intron separating Cμ1 and Cμ2 was approximately 1.7 kb for bighead catfish, 1.8 kb for North African catfish and 1.7 kb for channel catfish (63). Xenopus also exhibited a long intron between Cμ1 and Cμ2 of approximately 1.6 kb (67, 68). Smaller introns separating Cμ2 to Cμ3 and Cμ3 to Cμ4 of approximately 0.2–0.4 kb and 0.2–0.3 kb were found in bighead catfish and North African catfish, respectively, and were also identified in channel catfish (63). However, a longer intron (approximately 1.4 kb) between exons Cμ2 and Cμ3 was found in the horn shark Heterodontus francisci (69). An extensive distance between Cμ4 and the first transmembrane domain exons of approximately 1–3 kb seems to be observed in bighead catfish, North African catfish and all reported vertebrates, such as mouse, horned shark and channel catfish (63, 67–69) . The introns separating Cμ1, Cμ2, Cμ3, and Cμ4 in mouse are all less than 0.5 kb (70, 71). In addition, a large intron separating TM1 and TM2 exons was also approximately 1.3 kb and 1.2 kb for bighead catfish and North African catfish, respectively, as found in Xenopus (approximately 1.1 kb) and channel catfish (approximately 2.3 kb) (63, 67, 69) . On the basis of the obtained data, this Ighμ gene organization was characteristically generalized to all teleost fish.
The gene localization of the Ighμ gene region of the IgH constant region locus of bighead catfish, North African catfish and their hybrid catfish confirms that bighead catfish and North African catfish possess only a single copy of this gene, whereas their hybrid catfish possesses two copies of this gene (Figure 1J). The finding that the Clarias catfish Ighμ gene is most likely found in a single copy in the genome suggests that it may be common to all teleost fish, e.g., in channel catfish (72) and several other teleosts (26, 73, 74). The obtained Ighμ constant gene organization of the hybrid catfish clearly showed that the sequences were identical to those of the parents. The presence of two copies of the Ighμ gene in the hybrid catfish may be normal in diploid hybrid animals. In this regard, it is interesting that the Ighμ transcripts of the hybrid catfish are processed from different gene loci, and the membrane-bound and secretory forms were made up of the Ighμ gene of bighead catfish and North African catfish, respectively. The degree similarity of the amino acid was also identical to those of the parents. This available evidence would seem thus far to support the disease resistance characteristics of North African catfish and the hybrid catfish in several research reports. This suggests that the soluble effector molecules secreted from Ighμ of North African catfish and the hybrid catfish may be strongly involved in specific immune responses, such as toxin or microbe neutralization, opsonization (immunophagocytosis), antibody-dependent cell-mediated cytotoxicity (ADCC), and complement activation (immunolysis) (1, 75).
Furthermore, the ability of Ig to interact and bind antigen was involved in the variable domains of Ighμ molecules. Specific pathogens or antigens are recognized via the fragment antigen-binding (Fab) variable region (49). Additional analysis of the VH domain provided extensive insight. In our findings, the overall structures of bighead catfish, North African catfish and their hybrid catfish VH region corresponded to the VH, DH, and JH segments that consist of FR1, CDR1, FR2, CDR2, FR3, CDR3 and FR4, as reported in all vertebrates examined previously (49, 76).
Analysis of the VH, DH and JH family classification based on their 80% similarities of amino acids interestingly demonstrated that a large number of VH gene families were defined in all studied catfish, of approximately 15–17 family members from one hundred nonredundant clones. There are similarities in some teleosts and some mammals, e.g., Atlantic salmon, zebrafish, and mice (14–18 families). However, there were also higher levels in the VH gene family than in vertebrates (Figure 5C) (13, 44, 58). Much less is known and reported about the DH and JH segments of teleosts than about the VH of teleosts. Bighead catfish, North African catfish and their hybrid catfish process approximately 9, 11, and 13 families and 11, 11, and 8 families for a hundred nonredundant clones of DH and JH segments, respectively, compared to 9 in channel catfish, 4 in mouse, 6 in human, 6 in O. mykiss, and 2 in G. morhua for the JH segment (73). To date, there have been few reports of DH region diversity in vertebrates and teleosts, and significant variability in DH segments can be inferred from the generation of variability in CDR3 defined in the different cDNA clone sequences (49, 73). This available evidence indicates that the mechanisms of VH diversity generalization of Ighμ are made up of several gene family members that are necessary for generating several distinct VH repertoires in all studied catfish. In addition, junctional diversity occurs at the junction of the VH-DH and DH-JH boundaries. These regions code for CDR3. Diversity is increased by the addition of P-nucleotides and N-nucleotides (49). P-nucleotides are often made-up palindromic sequences and added to the ends of the asymmetrical DNA strand. Moreover, the random nontemplate-encoded addition of 2 to 20 base pairs or N nucleotides changes the amino acid sequence in the hypervariable CDR3. P- and N-nucleotides are principally found in the VH-DH and DH-JH junctions of the assembled heavy chain gene (49, 76). Furthermore, it was suggested that inversion (D-D joining), nucleotide deletion, heavy-light chain pairing and somatic hypermutation contribute to diversity in VH antigenic recognition after naive B cells are released to the periphery and encounter antigens (49, 73). Based on the findings in the present study, these mechanisms may be used to generate diverse VH repertoires in bighead catfish, North African catfish, and their hybrid catfish.
Analysis of the CDR3s provided strong evidence that the CDR3 of all studied catfish is longer than those of Atlantic cod, trout, frog and mouse but shorter than those of rabbit, human and cattle (13, 44, 49, 58). In addition, overall analysis of the VH domain of Ighμ indicated that the amino acid deviation in the CDRs was higher than that in the FWs in all catfish. Notably, these findings generally indicated predominant conservation in the VH repertoire (49, 73). However, the calculated amino acid variability and composition of all CDRs in VH interestingly demonstrated that high variability plots and variability in amino acid composition were observed in all CDR1s, CDR2s and CDR3s of all studied catfish compared to those of reported vertebrates (49, 77, 78). Based on our findings, the potential for increasing the structural variability of the VH domain in Clarias catfish and their hybrids seems to be higher than that of teleosts, particularly given the number of VH and JH family members, long length of CDR3, and high variability in all CDR1s, CDR2s and CDR3s. This may be a general feature of Clarias catfish, which has never been reported thus far.
The functional expression of Ighμ in bighead catfish, North African catfish and their hybrid catfish was examined in different tissues. The Ighμ gene of all studied catfish and two novel Ighμ molecules of bighead catfish were typically expressed in any organ and predominantly expressed in immune-related organs, especially the head kidney, PBLs, spleen and liver tissues, which is in accordance with previous studies (13, 79). High expression of the two novel transcripts in immune-involved organs may indicate specific unknown functions that need further investigation.
In our study, significant responses of Ighμ expression among Clarias catfish and the hybrid catfish were observed after bacterial exposure. North African catfish and the hybrid catfish exhibited approximately 900–950-fold relative expression levels in head kidney and 400–500-fold in PBLs; these levels are much higher than those of bighead catfish by at least 2–2.5-fold changes. The head kidney in teleosts is considered a primary lymphoid organ similar to the mammalian bone marrow, a major hemopoietic organ and site of Ig and other immune cell production in teleosts (15). A. hydrophila is a significant pathogen of Clarias catfish in Thailand. It is highly pathogenic due to the presence of hemolysin and aerolysin virulence genes, which demonstrate high virulence in Clarias catfish acute septicemia. A virulent strain could cause death within 24 hrs. and resulted in abdominal effusion and varying degrees of internal organ hemorrhage. The precise mechanism by which specific IgM immune responses rapidly increased after 12-24 hrs. post-exposure is still uncertain in the A. hydrophila challenge experiment. It could be suggested in two ways. First, as a characteristic of acute septicemia, the virulent bacteria evenly divide speedily in the blood and are rapidly circulated to the major immune-related tissues such as the head and kidney, activating the chemokine signaling pathway and triggering an inflammatory response, as well as upregulating IgM in the host. Second, prior to conducting the experiment, all studied catfish were exposed to A. hydrophila in their surrounding environment, resulting in a strongly rapid upregulation of IgM specific to A. hydrophila 12-24 hrs. post-exposure.
Unexpectedly, the pattern of Ighμ expression in hybrid catfish is consistent with their fatherhood of North African catfish. This evidence may imply a long mysterious question regarding the well-known characteristics of the bacterial resistance of North African catfish and the hybrid catfish, which normally are much higher than those of bighead catfish.
These results indicate that the Ighμ transcripts in bighead catfish, North African catfish and the hybrid catfish are typically produced by splicing patterns of mRNA reported in all teleosts. Notably, in bighead catfish, two more unusual spicing Ighμ transcripts of membrane-bound forms, 1) Cμ1-Cμ2-Cμ3-Cμ4-TM1-TM2 and 2) Cμ1-(Cδ2-Cδ3-Cδ4-Cδ5)-Cμ2-Cμ3-TM1-TM2, represent novel patterns of mRNA splicing of the Ighμ membrane form for teleosts and vertebrates, respectively. However, the reasons for the addition of Ighδ to Ighμ molecules are still unknown. All Ighμ transcripts were produced from a single gene copy in individual bighead catfish and North African catfish. In addition, the different Ighμ transcripts, membrane and secreted forms, of diploid hybrid catfish consisted of two copies of the Ighμ gene from the parents, bighead catfish and North African catfish. Extensive insight into the VH domain could generate a number of distinct structural variabilities in the VH domain in Clarias catfish and the hybrid catfish. The overall functional significance of Ighμ in hybrid catfish is mostly similar to that in North African catfish.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by The Animal Ethics Committee, Kasetsart University, Thailand (Ethics ID: ACKU61-FIS-004). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
AB designed and performed all the experiments and wrote the original manuscript. UN-N and PS revised the manuscript. PS designed the experiments and revised the manuscript. All authors listed have read and approved the manuscript for publication.
Funding
This work was financially supported by the Thailand Research Fund (TRF) and the Betagro Science Center, Thailand. This study is part of the project entitled “Genetics and Biotechnology for Improvement of Aquatic Animal Production” (contract number DPG5980003) awarded to Professor Uthairat Na-Nakorn under the “Distinguished Research Professor 2016 Award.”
Conflict of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Associate Professor Dr. Kornsorn Srikulnath and his laboratory members, Department of Genetics, Faculty of Science, Kasetsart University, Bangkok Thailand, for training in chromosome preparation. In addition, we thank the Graduate School at Kasetsart University, Thailand, for the partial financial support of the student who is the first author of this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.884434/full#supplementary-material
References
1. Burton DR, Woof JM. Human Antibody Effector Function. Adv Immunol (1992) 51:1–84. doi: 10.1016/S0065-2776(08)60486-1
2. Warr GW. The Immunoglobulin Genes of Fish. Dev Comp Immunol (1995) 19:1–12. doi: 10.1016/0145-305X(94)00052-H
3. Coscia MR, Varriale S, De Santi C, Giacomelli S, Oreste U. Evolution of the Antarctic Teleost Immunoglobulin Heavy Chain Gene. Mol Phylogenet Evol (2010) 55:226–33. doi: 10.1016/j.ympev.2009.09.033
4. Stavnezer J, Amemiya CT. Evolution of Isotype Switching. Semin Immunol (2004) 16:257–75. doi: 10.1016/j.smim.2004.08.005
5. Hikima J, Jung TS, Aoki T. Immunoglobulin Genes and Their Transcriptional Control in Teleosts. Dev Comp Immunol (2011) 35:924–36. doi: 10.1016/j.dci.2010.10.011
6. Ohno S. Immunoglobulins Vol. 197-204. Litman GW, Good RA, editors. New York, NY, USA: Springer USA (1978).
7. Wilson MR, van Ravenstein E, Miller NW, Clem LW, Middleton DL, Warr GW. cDNA Sequences and Organization of IgM Heavy Chain Genes in Two Holostean Fish. Dev Comp Immunol (1995) 19:153–64. doi: 10.1016/0145-305X(94)00063-L
8. Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, et al. Structural Analysis of the Nurse Shark (New) Antigen Receptor (NAR): Molecular Convergence of NAR and Unusual Mammalian Immunoglobulins. Proc Natl Acad Sci USA (1998) 95:11804–9. doi: 10.1073/pnas.95.20.11804
9. Schaerlinger B, Bascove M, Frippiat J-P. A New Isotype of Immunoglobulin Heavy Chain in the Urodele Amphibian Pleurodeles Waltl Predominantly Expressed in Larvae. Mol Immunol (2008) 45:776–86. doi: 10.1016/j.molimm.2007.06.356
10. Golub R, Charlemagne J. Structure, Diversity, and Repertoire of VH Families in the Mexican Axolotl. J Immunol (1998) 160:1233–9.
11. Hsu E, Criscitiello MF. Diverse Immunoglobulin Light Chain Organizations in Fish Retain Potential to Revise B Cell Receptor Specificities. J Immunol (2006) 177:2452–62. doi: 10.4049/jimmunol.177.4.2452
12. Litman GW, Anderson MK, Rast JP. Evolution of Antigen Binding Receptors. Annu Rev Immunol (1999) 17:109–47. doi: 10.1146/annurev.immunol.17.1.109
13. Danilova N, Bussmann J, Jekosch K, Steiner LA. The Immunoglobulin Heavy-Chain Locus in Zebrafish: Identification and Expression of a Previously Unknown Isotype, Immunoglobulin Z. Nat Immunol (2005) 6:295–302. doi: 10.1038/ni1166
14. Savan R, Aman A, Nakao M, Watanuki H, Sakai M. Discovery of a Novel Immunoglobulin Heavy Chain Gene Chimera From Common Carp (Cyprinus Carpio L.). Immunogenetics (2005) 57:458–63. doi: 10.1007/s00251-005-0015-z
15. Bengtén E, Quiniou S, Hikima J, Waldbieser G, Warr GW, Miller NW, et al. Structure of the Catfish IGH Locus: Analysis of the Region Including the Single Functional IGHM Gene. Immunogenetics (2006) 58:831–44. doi: 10.1007/s00251-006-0139-9
16. Saha NR, Suetake H, Suzuki Y. Analysis and Characterization of the Expression of the Secretory and Membrane Forms of IgM Heavy Chains in the Pufferfish. Takifugu rubripes. Mol Immunol (2005) 42:113–24. doi: 10.1016/j.molimm.2004.06.034
17. Oreste U, Coscia M. Specific Features of Immunoglobulin VH Genes of the Antarctic Teleost. Trematomus bernacchii. Gene (2002) 295:199–204. doi: 10.1016/S0378-1119(02)00686-8
18. Merlino A, Varriale S, Coscia MR, Mazzarella L, Oreste U. Structure and Dimerization of the Teleost Transmembrane Immunoglobulin Region. J Mol Graph Model (2008) 27:401–7. doi: 10.1016/j.jmgm.2008.07.001
19. Magor BG, Wilson MR, Miller NW, Clem LW, Middleton DL, Warr GW. An Ig Heavy Chain Enhancer of the Channel Catfish Ictalurus Punctatus: Evolutionary Conservation of Function But Not Structure. J Immunol (1994) 153:5556–63.
20. Hordvik I, Voie AM, Glette J, Male R, Endresen C. Cloning and Sequence Analysis of Two Isotypic IgM Heavy Chain Genes From Atlantic Salmon, Salmo Salar L. Eur J Immunol (1992) 22:2957–62. doi: 10.1002/eji.1830221130
21. Ghaffari SH, Lobb CJ. Cloning and Sequence Analysis of Channel Catfish Heavy Chain cDNA Indicate Phylogenetic Diversity Within the IgM Immunoglobulin Family. J Immunol (1989) 142:1356–65.
22. Coscia MR, Oreste U. Limited Diversity of the Immunoglobulin Heavy Chain Variable Domain of the Emerald Rock Cod Trematomus Bernacchii. Fish Shellfish Immunol (2003) 14:71–92. doi: 10.1006/fsim.2002.0418
23. Cheng CA, John JA, Wu MS, Lee CY, Lin CH, Lin CH, et al. Characterization of Serum Immunoglobulin M of Grouper and cDNA Cloning of Its Heavy Chain. Vet Immunol Immunopathol (2006) 109:255–65. doi: 10.1016/j.vetimm.2005.08.029
24. Bengten E, Leanderson T, Pilstrom L. Immunoglobulin Heavy Chain cDNA From the Teleost Atlantic Cod (Gadus Morhua L.): Nucleotide Sequences of Secretory and Membrane Form Show an Unusual Splicing Pattern. Eur J Immunol (1991) 21:3027–33. doi: 10.1002/eji.1830211219
25. Andersson E, Peixoto B, Tormanen V, Matsunaga T. Evolution of the Immunoglobulin M Constant Region Genes of Salmonid Fish, Rainbow Trout (Oncorhynchus Mykiss) and Arctic Charr (Salvelinus Alpinus): Implications Concerning Divergence Time of Species. Immunogenetics (1995) 41:312–5. doi: 10.1007/BF00172156
26. Amemiya CT, Litman GW. Complete Nucleotide Sequence of an Immunoglobulin Heavy-Chain Gene and Analysis of Immunoglobulin Gene Organization in a Primitive Teleost Species. Proc Natl Acad Sci USA (1990) 87:811–5.
27. Nakao M, Moritomo T, Tomana M, Fujiki K, Yano T. Isolation of cDNA Encoding the Constant Region of the Immuoglobulin Heavy-Chain From Common Carp (Cyprinus Carpio L.). Fish Shellfish Immunol (1998) 8:425–34. doi: 10.1006/fsim.1998.0149
29. Srisapoome P, Ohira T, Hirono I, Aoki T. Genes of the Constant Regions of Functional Immunoglobulin Heavy Chain of Japanese Flounder. Paralichthys olivaceus. Immunogen (2004) 56:292–300. doi: 10.1007/s00251-004-0689-7
30. Picchietti S, Abelli L, Buonocore F, Randelli E, Fausto AM, Scapigliati G., et al. Immunoglobulin Protein and Gene Transcripts in Sea Bream (Sparus Aurata L.) Oocytes. Fish Shellfish Immunol (2006) 20:398–404. doi: 10.1016/j.fsi.2005.06.001
31. Wilson MR, Middleton D, Warr GW. Immunoglobulin VH Genes of the Goldfish, Carassius Auratus: A Re-Examination. Mol Immunol (1991) 28:449–57. doi: 10.1016/0161-5890(91)90158-G
32. Buonocore F, Stocchi V, Nunez-Ortiz N, Randelli E, Gerdol M, Pallavicini A, et al. Immunoglobulin T From Sea Bass (Dicentrarchus Labrax L.): Molecular Characterization, Tissue Localization and Expression After Nodavirus Infection. BMC Mol Biol (2017) 18:8. doi: 10.1186/s12867-017-0085-0
33. Bengten E, Stromberg S, Pilstrom L. Immunoglobulin VH Regions in Atlantic Cod (Gadus Morhua L.): Their Diversity and Relationship to VH Families From Other Species. Dev Comp Immunol (1994) 18:109–22. doi: 10.1016/0145-305X(94)90239-9
34. Magadan-Mompo S, Sanchez-Espinel C, Gambon-Deza F. Immunoglobulin Heavy Chains in Medaka (Oryzias Latipes). BMC Evol Biol (2011) 11:165. doi: 10.1186/1471-2148-11-165
35. Hu YL, Zhu LY, Xiang LX, Shao JZ. Discovery of an Unusual Alternative Splicing Pathway of the Immunoglobulin Heavy Chain in a Teleost Fish, Danio Rerio. Dev Comp Immunol (2011) 35:253–7. doi: 10.1016/j.dci.2010.10.009
36. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res (2018) 46:W296–303. doi: 10.1093/nar/gky427
37. Karami A, Eghtesadi Araghi P, Syed MA, Wilson SP. Chromosome Preparation in Fish: Effects of Fish Species and Larval Age. Int Aquat Res (2015) 7:201–10. doi: 10.1007/s40071-015-0104-z
38. Knoll JH, Lichter P. In Situ Hybridization to Metaphase Chromosomes and Interphase Nuclei. Curr Protoc Hum Genet (2005) 45:4.3.1–4.3.31. doi: 10.1002/0471142905.hg0403s45
39. Li S, Lefranc MP, Miles JJ, Alamyar E, Giudicelli V, Duroux P, et al. IMGT/HighV QUEST Paradigm for T Cell Receptor IMGT Clonotype Diversity and Next Generation Repertoire Immunoprofiling. Nat Commun (2013) 4:2333. doi: 10.1038/ncomms3333
40. Shannon CE. A Mathematical Theory of Communication. Bell Syst Tech (1948) 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
41. Kabat EA, Wu TT, Bilofsky H. Unusual Distributions of Amino Acids in Complementarity-Determining (Hypervariable) Segments of Heavy and Light Chains of Immunoglobulins and Their Possible Roles in Specificity of Antibody-Combining Sites. J Biol Chem (1977) 252:6609–16. doi: 10.1016/S0021-9258(17)39891-5
42. Chinedu E, Arome D, Ameh FS. A New Method for Determining Acute Toxicity in Animal Models. Int J Toxicol (2013) 20:224–6. doi: 10.4103/0971-6580.121674
43. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)). Method Methods (San Diego Calif.) (2001) 25:402–8. doi: 10.1006/meth.2001.1262
44. Yasuike M, de Boer J, von Schalburg KR, Cooper GA, McKinnel L, Messmer A, et al. Evolution of Duplicated IgH Loci in Atlantic Salmon. Salmo salar. BMC Genomics (2010) 11:486. doi: 10.1186/1471-2164-11-486
45. Yang F, Ventura-Holman T, Waldbieser GC, Lobb CJ. Structure, Genomic Organization, and Phylogenetic Implications of Six New VH Families in the Channel Catfish. Mol Immunol (2003) 40:247–60. doi: 10.1016/S0161-5890(03)00143-3
46. Brown GD, Kaattari IM, Kaattari SL. Two New Ig VH Gene Families in. Oncorhynchus mykiss. Immunogen (2006) 58:933–6. doi: 10.1007/s00251-006-0149-7
47. Phuyindee C, Unajak S, Srisapoome P. Diversity Analysis of the Immunoglobulin M Heavy Chain Gene in Nile Tilapia, Oreochromis Niloticus (Linnaeus). Afr J Biotechnol (2015) 14:2282–99. doi: 10.5897/AJB2014.14001
48. Andersson E, Matsunaga T. Evolutionary Stability of the Immunoglobulin Heavy Chain Variable Region Gene Families in Teleost. Immunogenetics (1998) 47:272–7. doi: 10.1007/s002510050357
49. Stenvik J, Lundbäck AS, Jørgensen TO, Pilström L. Variable Region Diversity of the Atlantic Cod (Gadus Morhua L.) Immunoglobulin Heavy Chain. Immunogenetics (2000) 51:670–80. doi: 10.1007/s002510000188
50. Peixoto BR, Brenner S. Characterization of Approximately 50 Kb of the Immunoglobulin VH Locus of the Japanese Pufferfish. Fugu rubripes. Immunogen (2000) 51:443–51. doi: 10.1007/s002510050643
51. Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and Repertoire of IgW and IgM VH Families in the Newborn Nurse Shark. BMC Immunol (2004) 5:8. doi: 10.1186/1471-2172-5-8
52. Lundqvist ML, Strömberg S, Pilström L. Ig Heavy Chain of the Sturgeon Acipenser Baeri: cDNA Sequence and Diversity. Immunogenetics (1998) 48:372–82. doi: 10.1007/s002510050448
53. Haire RN, Amemiya CT, Suzuki D, Litman GW. Eleven Distinct VH Gene Families and Additional Patterns of Sequence Variation Suggest a High Degree of Immunoglobulin Gene Complexity in a Lower Vertebrate. Xenopus laevis. J Exp Med (1990) 171:1721–37. doi: 10.1084/jem.171.5.1721
54. Ota T, Nei M. Evolution of Immunoglobulin VH Pseudogenes in Chickens. Mol Biol Evol (1995) 12:94–102. doi: 10.1093/oxfordjournals.molbev.a040194
55. Mainville CA, Sheehan KM, Klaman LD, Giorgetti CA, Press JL, Brodeur PH. Deletional Mapping of Fifteen Mouse VH Gene Families Reveals a Common Organization for Three IgH Haplotypes. J Immunol (1996) 156:1038–46.
56. Pinheiro A, Lanning D, Alves PC, Mage RG, Knight KL, van der Loo W, et al. Molecular Bases of Genetic Diversity and Evolution of the Immunoglobulin Heavy Chain Variable Region (IGHV) Gene Locus in Leporids. Immunogenetics (2011) 63:397–408. doi: 10.1007/s00251-011-0533-9
57. Sun J, Kacskovics I, Brown WR, Butler JE. Expressed Swine VH Genes Belong to a Small VH Gene Family Homologous to Human VHIII. J Immunol (1994) 153:5618–27.
58. Matsuda F, Ishii K, Bourvagnet P, Kuma KiHayashida H, Miyata T, et al. The Complete Nucleotide Sequence of the Human Immunoglobulin Heavy Chain Variable Region Locus. J Exp Med (1998) 188:2151–62. doi: 10.1084/jem.188.11.2151
59. Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, et al. IMGT, the International ImMunoGeneTics Information System. Nucleic Acids Res (2005) 33:D593–7. doi: 10.1093/nar/gki065
60. Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Müller W, Bontrop R, et al. IMGT, the International ImMunoGeneTics Database. Nucleic Acids Res (1999) 27:209–12. doi: 10.1093/nar/27.1.209
61. Hansen JD, Landis ED, Phillips RB. Discovery of a Unique Ig Heavy-Chain Isotype (IgT) in Rainbow Trout: Implications for a Distinctive B Cell Developmental Pathway in Teleost Fish. Proc Natl Acad Sci USA (2005) 102:6919–24. doi: 10.1073/pnas.0500027102
62. Fillatreau S, Six A, Magadan S, Castro R, Sunyer JO, Boudinot P. The Astonishing Diversity of Ig Classes and B Cell Repertoires in Teleost Fish. Front Immunol (2013) 4. doi: 10.3389/fimmu.2013.00028
63. Bengtén E, Quiniou SM, Stuge TB, Katagiri T, Miller NW, Clem LW, et al. The IgH Locus of the Channel Catfish, Ictalurus Punctatus, Contains Multiple Constant Region Gene Dequences: Different Genes Encode Heavy Chains of Membrane and Secreted IgD. J Immunol (2002) 169:2488. doi: 10.4049/jimmunol.169.5.2488
64. Bruce SR, Dingle RWC. Peterson Ml. B-cell Plasma-Cell Splicing Differences: A Potential Role Regulated Immunoglobulin RNA Processing. RNA (2003) 9:1264–73. doi: 10.1261/rna.5820103
65. Ventura-Holman T, Lobb CJ. Structural Organization of the Immunoglobulin Heavy Chain Locus in the Channel Catfish: The IgH Locus Represents a Composite of Two Gene Clusters. Mol Immunol (2002) 38:557–64. doi: 10.1016/S0161-5890(01)00075-X
66. Danilova N, Saunders HL, Ellestad KK, Magor BG. The Zebrafish IgH Locus Contains Multiple Transcriptional Regulatory Regions. Dev Comp Immunol (2011) 35:352–9. doi: 10.1016/j.dci.2010.10.010
67. Schwager J, Mikoryak CA, Steiner LA. Amino Acid Sequence of Heavy Chain From Xenopus Laevis IgM Deduced From cDNA Sequence: Implications for Evolution of Immunoglobulin Domains. Proc Natl Acad Sci USA (1988) 85:2245–9. doi: 10.1073/pnas.85.7.2245
68. Schwager J, Grossberger D, Du Pasquier L. Organization and Rearrangement of Immunoglobulin M Genes in the Amphibian Xenopus. EMBO J 19887(8):2409–15. doi: 10.1002/j.1460-2075.1988.tb03086.x
69. Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, et al. The Evolution of Multiple Isotypic IgM Heavy Chain Genes in the Shark. J Immunol (2008) 180:7461–70. doi: 10.4049/jimmunol.180.11.7461
70. McGuire KL, Duncan WR, Tucker PW. Phylogenetic Conservation of Immunoglobulin Heavy Chains: Direct Comparison of Hamster and Mouse Cμ Genes. Nucleic Acids Res (1985) 13:5611–28. doi: 10.1093/nar/13.15.5611
71. Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, et al. Two mRNAs Can Be Produced From a Single Immunoglobulin μ Gene by Alternative RNA Processing Pathways. Cell (1980) 20:313–9. doi: 10.1016/0092-8674(80)90617-0
72. Wilson MR, Marcuz A, van Ginkel F, Miller NW, Clem LW, Middleton D, et al. The Immunoglobulin M Heavy Chain Constant Region Gene of the Channel Catfish, Ictalurus Punctatus: An Unusual mRNA Splice Pattern Produces the Membrane Form of the Molecule. Nucleic Acids Res (1990) 18:5227–33. doi: 10.1093/nar/18.17.5227
73. Ghaffari SH, Lobb CJ. Nucleotide Sequence of Channel Catfish Heavy Chain cDNA and Genomic Blot Analyses. Implications Phylogeny Ig Heavy Chains. J Immunol (1989) 143:2730–9.
74. Lee MA, Bengtén E, Daggfeldt A, Rytting A-S, Pilström L. Characterisation of Rainbow Trout cDNAs Encoding a Secreted and Membrane-Bound Ig Heavy Chain and the Genomic Intron Upstream of the First Constant Exon. Mol Immunol (1993) 30:641–8. doi: 10.1016/0161-5890(93)90075-M
75. Harris LJ, Larson SB, McPherson A. Comparison of Intact Antibody Structures and the Implications for Effector Function. Adv Immunol (1999) 72:191–208. doi: 10.1016/S0065-2776(08)60021-8
76. Andersson E, Matsunaga T. Evolution of Immunoglobulin Heavy Chain Variable Region Genes: A VH Family Can Last for 150-200 Million Years or Longer. Immunogenetics (1995) 41:18–28. doi: 10.1007/BF00188428
77. Roman T, De Guerra A, Charlemagne J. Evolution of Specific Antigen Recognition: Size Reduction and Restricted Length Distribution of the CDRH3 Regions in the Rainbow Trout. Eur J Immunol (1995) 25:269–73. doi: 10.1002/eji.1830250144
78. Brodeur PH, Riblet R. The Immunoglobulin Heavy Chain Variable Region (Igh-V) Locus in the Mouse. I. One Hundred Igh-V Genes Comprise Seven Families Homologous Genes. Eur J Immunol (1984) 14:922–30. doi: 10.1002/eji.1830141012
Keywords: Clarias catfish, IgM, novel transcript, characterization, structural analysis
Citation: Bunnoy A, Na-Nakorn U and Srisapoome P (2022) Mystifying Molecular Structure, Expression and Repertoire Diversity of IgM Heavy Chain Genes (Ighμ) in Clarias Catfish and Hybrids: Two Novel Transcripts in Vertebrates. Front. Immunol. 13:884434. doi: 10.3389/fimmu.2022.884434
Received: 26 February 2022; Accepted: 12 May 2022;
Published: 17 June 2022.
Edited by:
Masahiro Sakai, University of Miyazaki, JapanReviewed by:
Yo Okamura, University of Washington, United StatesMadhubanti Basu, University of Alabama at Birmingham, United States
Copyright © 2022 Bunnoy, Na-Nakorn and Srisapoome. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prapansak Srisapoome, ZmZpc3Bzc3BAa3UuYWMudGg=
 Anurak Bunnoy
Anurak Bunnoy Uthairat Na-Nakorn
Uthairat Na-Nakorn Prapansak Srisapoome
Prapansak Srisapoome