
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 25 May 2022
Sec. Nutritional Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.881289
This article is part of the Research Topic Immune Pathogenesis of Intestinal Inflammatory Diseases View all 24 articles
Porcine epidemic diarrhea virus (PEDV) is an emerging coronavirus which causes acute diarrhea and destroys gastrointestinal barrier function in neonatal pigs. Trefoil factor 1 (TFF1) is a protective peptide for maintaining the integrity of gastrointestinal mucosa and reducing intestinal inflammation. However, its role in protecting intestinal epithelium against PEDV infection is still unclear. In this study, we discovered that TFF1 expression was activated in the jejunum of pigs with PEDV infection and TFF1 is required for the growth of porcine intestinal epithelial cells. For instance, inhibited cell proliferation and cell arrest were observed when TFF1 is genetically knocked-out using CRISPR-Cas9. Additionally, TFF1 depletion increased viral copy number and PEDV titer, along with the elevated genes involved in antiviral and inflammatory cytokines. The decreased TFF1 mRNA expression is in line with hypermethylation on the gene promoter. Notably, the strong interactions of protein-DNA complexes containing CCAAT motif significantly increased C/EBPα accessibility, whereas hypermethylation of mC-6 loci decreased C/EBPα binding occupancies in TFF1 promoter. Overall, our findings show that PEDV triggers the C/EBPα-mediated epigenetic regulation of TFF1 in intestine epithelium and facilitates host resistance to PEDV and other Coronavirus infections.
Porcine epidemic diarrhea is a highly contagious enteric disease caused by porcine epidemic diarrhea virus (PEDV) which is characterized by intestinal inflammation, watery diarrhea, vomiting, or dehydration. All ages and breeds of pigs are susceptible to PEDV, whereas it is most harmful to neonatal piglets (1). PEDV causes an 80-100% fatality rate in piglets, and more than 10% of the pig population is wiped out throughout the world (2). PEDV is a single-stranded positive-sense RNA virus which is classified into the genus α-Coronavirus of the Coronaviridae family. It contains four structural proteins: spike (S), envelope (E), matrix (M), and nucleocapsid (N) (3, 4). This virus infection is usually transmitted by the fecal-oral route or air from the nasal cavity to the intestinal mucosa and replicates principally in the intestinal epithelial cells (especially in the jejunum and ileum) (5, 6). Given that the functional integrity of intestinal epithelial barrier constitutes the main defense against pathogens invasion, its physiological homeostasis is critical for intestinal against PEDV infection.
The trefoil factor family (TFFs) is a group of protein polypeptides produced mainly by gastrointestinal mucus-secreting or mucin-secreting cells. TFFs are widely distributed in vertebrates and highly conserved in evolution, suggesting that they may be a class of proteins with important physiological functions (7, 8). TFF1 (also known as pS2), as a member of the TFFs, is a cysteine-rich secretory protein with the function of gastrointestinal protection (9, 10), mucosal defense, and injury healing (11–13). When co-expressed with the mucin MUC5AC, TFF1 works for mucosal protection by forming a stable mucus gel layer together with mucin. During the extracellular pathogen invasion, the anti-apoptotic activity and the epithelial cell migration are coordinately controlled by TFF1 to improve injury repair and immune response (14–16). Importantly, the TFF1 gene shows the tissue-specific expression pattern and is closely related to the ability of injury healing, activating the host’s resistance to pathogens infection (17, 18). Previously, we have demonstrated a 28.8-fold increase in TFF1 gene expression by transcriptome analysis in PEDV-infected jejunum (19). In this study, we aim to identify whether TFF1 is a potential target to benefit gut function in response to PEDV infection.
In recent years, the development of epigenetics provides new insights to solve the genetic mechanism of diseases. Among them, methylation of CpG residues is a key epigenetic modification of eukaryotic DNA which provides a stable gene silencing and is a major factor responsible for the suppression of gene expression (20). Importantly, heterochromatin domains are determined, in part, by methylation of cytosines at the 5th position of the pyrimidine ring. Indeed, previous reports revealed that the TFF1 gene expression is vulnerable to DNA methylation pattern (21, 22) and it has been documented that hypermethylation of TFF1 promoter triggers the gene downregulation in intestinal metaplasia and intestinal-type human cell lines (22, 23). So, this may be attributed to the impaired transcription factor like C/EBPα and/or HIF-1α modulation induced by methylation events (24, 25). Thus, this study aims to understand the TFF1 action of inflammatory inhibition in the PEDV-infected jejunum and the underlying mechanisms. Our findings exhibit that TFF1 is a resistant factor to a porcine coronavirus infection by the regulation of C/EBPα and DNA methylation.
The animal study proposal was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yangzhou University Animal Experiments Ethics Committee (permit number: SYXK (Su) IACUC 2012-0029). All experimental methods were conducted in accordance with the relevant guidelines for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China.
Four 7-day-old ternaries crossbred piglets (Duroc*Landrace*Yorkshire) were infected with PEDV featured with typical clinical symptoms of porcine epidemic diarrhea. In addition, four normal piglets were selected as the control. The herd of pigs was detected for PCR against PEDV, TGEV, PDCoV, and PoRV. All the animals were raised under the same conditions and humanely sacrificed by intravenous injection of pentobarbital sodium. After the sacrifice, duodenum, jejunum, and ileum samples were snap frozen immediately in liquid nitrogen for the subsequent experiments.
Jejunal contents were collected and diluted in 500 μL phosphate-buffered saline (PBS) buffer, then freeze-thawed repeatedly before the centrifugation to collect the supernatant containing viruses. The supernatants were added to the tubes containing TRIzol reagent (Takara Biotech, Dalian, China) and vortexed according to the manufacturer’s instruction. The cDNA was amplified with PrimeScript RT-PCR kit (Vazyme Biotech, Nanjing, China) to identify the infections of viruses, including PEDV, TGEV, PDCoV and PoRV. The primers are listed in Table 1.
Duodenum, jejunum, and ileum were fixed in 4% paraformaldehyde solution for 24 hours. Tissues were embedded in paraffin and cut into 5 μm pieces. For hematoxylin-eosin (HE) staining, the paraffin sections were conducted according to the routine procedure. The mounted slides were observed under Olympus iX53 (Tokyo, Japan) light microscope and the photographs were taken using Motic Image Advanced 3.2 Analysis System (Motic China group Co., Ltd., Xiamen, China).
Porcine intestinal epithelial cells (IPEC-J2) were kindly provided by the University of Pennsylvania. Vero kidney cell line (Vero CCL-81) were purchased from ATCC. IPEC-J2 and Vero cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, NY, USA) and 100 μg/mL penicillin/streptomycin (Solarbio, Beijing, China) in a humidified atmosphere containing 5% CO2 at 37°C. Cells were seeded in tissue culture flasks (Costar, Corning, MA, USA).
PEDV strains used in this study were the classic strain CV777, which was propagated and titrated in FBS-free DMEM medium supplemented with 8 μg/mL trypsin (Sigma-Aldrich, MO, USA) in Vero CCL-81 cells. Cells were infected with PEDV at MOI of 0.1 for 2 hours at 37°C (26). After 2 hours, the unattached virus inoculum was removed. Then, the cells were washed three times with cold PBS buffer and maintained in DMEM supplemented with 2% FBS. The infected cells and supernatants were collected after the indicated period and titrated according to TCID50 protocol. The viral titers were calculated using the Spearman-Karber equation. The PEDV genome copy number was calculated by the standard curve equation of CV777 well-established previously (26): y = −3.3354lg(x) + 37.832 (where y represents the number of Ct cycles, x represents the logarithm of the virus copy number based on the base 10).
To deplete TFF1 gene, three different single guide RNAs (sgRNAs) were designed using an CRISPR-Cas9 Design online software. Oligo nucleotides sequences of the sgRNAs were synthesized and cloned into the BbsI digested linearized pGK1.2 (EGFP + Puror) vector (Genloci Biotech, Nanjing, China) following the pGK1.2 oligo-cloning protocol. The primers are listed in Table 2. Colony PCR was performed to validate the sgRNA sequences using the VSP forward primer 5′-CATATGCTTACCGTAACTTGAAAG-3′ and three primer sets: primer set-1: (5’-GCCACAGGATTGAAGCACCA-3’), primer set-2: (5’-GTTGTCTGTGGGGTCATCAA-3’), primer set-3: (5’-GTGGGGTCATCAACGGCCAC-3’) as the reverse primers, respectively. The amplicons were performed by PCR-sequencing (Sangon Biotech, Shanghai, China) and the positive recombinant vectors were named as sgRNA1, sgRNA2 and sgRNA3, respectively. Subsequently, cells pool sequencing verified the efficiency of sgRNAs mixture. The primer used for cloning was as follows: TFF1-sgRNA forward primer (5’-CTTGGCCGGTACACTTTCAG-3’) and reverse primer (5’-CTTCACAGGTCCGTGGTTAGA-3’). IPEC-J2 were transfected with sgRNA1, sgRNA2 and sgRNA3 in 6-well tissue culture plates at a density of 2 × 105/mL. The culture medium with 3 μg/mL puromycin was added to the cells 72 hours later. For another 72 hours, positive monoclonal cells were harvested.
For TFF1 and C/EBPα overexpression, the full-length CDS of TFF1 and C/EBPα were amplified and cloned into a pcDNA3.1 vector (Invitrogen, CA, USA) with T4 DNA ligase for ligation by NheI and HindIII restriction sites, respectively. The recombinant overexpression vectors were defined as TFF1-oe and C/EBPα-oe, respectively. Subsequently, each of the overexpression vectors were transfected into IPEC-J2 using Lipofectamine™ 2000 (Invitrogen, CA, USA) following the manufacturer’s protocol. At 36 hours after transfection, the cells were passaged into the appropriate medium containing 400 µg/mL G418. The screening was conducted over 7 days and the stable cell lines constitute polyclonal pools of cells.
Small interfering RNAs (siRNAs) against C/EBPα mRNA sequences (si-C/EBPα-1, si-C/EBPα-2, and si-C/EBPα-3) of sus scrofa and negative control (si-C/EBPα-NC) were synthesized from GenePharma Corporation (Shanghai, China). The siRNAs and negative control were transfected into IPEC-J2 using Lipofectamine™ 2000 (Invitrogen, CA, USA) following the manufacturer’s instruction.
To assess cell viability, cells were seeded in 96-well plates with 5 × 103/well and incubated with PEDV at the indicated time. Then 10 μL cell counting kit-8 solution (CCK8, Dojindo Molecular Technologies Inc., Kumamoto, Japan) was added to each well followed by the incubation at 37°C for 2 hours. After the incubation, an absorbance at 450 nm was measured using a Multimode Microplate Reader (Spark™ 10M, Tecan GmbH, Austria). Caspase-3/7 activity was measured using a luminescence-based assay with the luminescent Caspase-Glo 3/7 assay kit (Promega Corporation, MA, USA) according to the manufacturer’s instruction. Each experiment was repeated three times and performed in triplicate.
Cells were seeded in 6-well plates and incubated with the indicated doses of PEDV. The apoptotic cells were detected by Annexin V-FITC/PI apoptosis analysis kit (Solarbio, Beijing, China) according to the manufacturer’s guide. The samples were analyzed by CytoFLEX flow cytometer (Beckman Coulter, CA, USA). For cell cycle detection, TFF1-depletion cells were seeded in tissue culture flask at a density of 2 × 106/mL. Cells were then washed with cold PBS and fixed overnight with cold 70% ethanol. The fixed cells were washed with PBS and stained with 1 mL PI staining reagent (50 mg/mL propidium iodide and 1 mg/mL RNase A in 1 mL of sodium citrate buffer) for 30 min at 37°C in a light-proof manner. Then the cell cycle profiles were analyzed by FACScan flow cytometer (Becton Dickinson, CA, USA). The percentage of cells at G0/G1, S and G2/M phases were calculated using Modifit software.
Cells were seeded on coverslips in 12-well tissue culture plates and infected with PEDV for 2 hours, rinsed, then incubated at 37°C. After 48 hours of PEDV infection, the cells were fixed in 4% paraformaldehyde for 15 min and permeabilized by 0.1% Triton X-100 in PBS for 10 min at RT. The cells were blocked with 5% bovine serum albumin (BSA) in PBS for 30 min at RT and then incubated with the anti-PEDV antibody PEDV N MAb (Medgene, SD-1-5, 1:500) overnight at 4°C. After washing thrice with PBS, the cells were incubated with a fluorochrome-conjugated secondary antibody (HuaBio, HA1015, 1:100) for 30 min at RT. The cell nuclei were stained with DAPI (1 μg/mL) for 5 min. The coverslips were mounted on microscope slides for the visualization under a fluorescence microscope (Olympus, Tokyo, Japan). Images were processed using ImageJ software.
Cells were treated with PEDV at the indicated time as above. RT-qPCR was performed to detect the mRNA expression of inflammatory cytokines (IL-2, IL-6, IL-8, IL-12, IL-α, IL-β, IFN-α, IFN-β, TNF-α) and antiviral genes (Mx1, Mx2, RIG). The primers are listed as shown in Table 3.
Total RNA was isolated from cells and intestinal samples using the TRIzol reagent (Takara Biotech, Dalian, China) following the manufacturer’s protocol. RT-qPCR amplification was performed using an ABI 7500 Fast Real-Time Quantitative PCR System (Applied Biosystems, Foster City, CA, USA) and relative gene expression was calculated using the 2−ΔΔCt method (27). The housekeeping gene GAPDH was used as an internal control for each experiment. The experiments were performed at least three times with data presented as means values ± SD. The primers are shown in Table 4.
Intestinal tissues and cells were lysed in radioimmunoprecipitation assay (RIPA) buffer that contains proteinase inhibitor cocktail on ice for 20 min. The supernatant was gathered, centrifuged at 12,000 g for 20 min, and the protein concentration was determined using the BCA Protein Assay Kit (CWBiotech, Beijing, China). Proteins were separated in SDS-PAGE and transferred to a PVDF membrane (Millipore, MA, USA). The membranes were blocked with 5% skimming milk in Tris-buffered saline-Tween (TBST) for 1 hour, then incubated at 4°C overnight with the primary antibodies [anti-TFF1 (LifeSpan BioScience, ls-c312842, 1:1000), anti-CDK4 (Proteintech Ltd, 11026-1-AP, 1:1000), anti-PCNA (Proteintech Ltd, 10205-2-AP, 1:1000), anti-Bax (HuaBio, ET1603-34, 1:1000), anti-Cleaved Caspase 3 (Cell Signaling Technology, #9664, 1:500), anti-PEDV-N (Youlong-Bio, DA0110,1:1000), anti-β-Actin (CWBIO, CW0096M, 1:1000) and anti-GAPDH (Proteintech Ltd, 10494-1-AP, 1:1000)], respectively. The membranes were rinsed and incubated with the HRP-conjugated secondary antibodies [goat anti-Rabbit IgG antibody (Abcam, ab205718, 1:20000) and anti-Mouse IgG antibody (HuaBio, HA1006, 1:10000)] for 1 hour at RT. The membranes were visualized with an Enhanced Chemiluminescent Detection kit (ThermoFisher Scientific, MA, USA) using the FluorChem FC3 Chemilumilescent system (ProteinSimple Ltd, CA, USA). The relative integrated density was measured and normalized against GAPDH or β-Actin expression. The experiments were performed at least three times with data presented as means values ± SD.
The genomic DNA was extracted from jejunum in diarrheic and normal piglets using TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China). The extracted DNA was bisulfite conversed using EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA). The conversion DNA was performed PCR amplification using ZymoTaq Premix, with the primers designed for amplification presented in Table 5. The amplification system contains 2 μL DNA, 12.5 μL ZYMO Taq Premix, 1 μL forward primer and 1 μL reverse primer (10 pmol/μL) and RNase-free H2O 8.5 μL. The following PCR reaction was performed to 95°C for 10 min, followed by 40 cycles with each cycle consisting of 95°C for 30 s, and 54°C for 30 s, extension at 72°C for 30 s, and a final 5 min extension at 72°C. The PCR products were purified by TIANquick Midi purification kit (Tiangen Biotech, Beijing, China) and ligated into pMD19-T vector (Takara Biotech, Dalian, China) at 16°C overnight and transformed into Escherichia coli DH-5α competent cells. More than 15 positive clones from each sample were randomly picked up for bisulfite-sequencing. The methylation status at each CpG site was aligned using QUMA online software.
The CpG islands in the promoter region of TFF1 were predicted by MethPrimer website. The parameters were set as follows: CpG islands length greater than 200 bp, GC content greater than 50%, and CpG o/e greater than 0.6. For the reporter-gene assays of methylation on TFF1 promoter, the CpG island fragment of TFF1 promoter (−145 to −478 bp) was ligated to the pGL3-basic vector. The vectors were treated with M.SssI (CpG) methyl transferase (New England Biolabs, Beijing, China) and transfected into IPEC-J2. Luciferase assays were performed 36 hours after transfection. The primer for CpG islands amplification is presented in Table 5. IPEC-J2 were treated with concentration of 0.1 μM 5’-Aza-2′-deoxycytidine (5’-Aza-2’-dC, Sigma-Aldrich, MO, USA) for 72 hours. Then the cells were harvested for mRNA expression analysis.
ChIP assay was performed using the Pierce Agarose ChIP Kit (ThermoFisher Scientific, MA, USA) following the manufacturer’s instructions. An amount of 80 mg sheared jejunum sample was minced and crosslinked in formaldehyde. After the chromatin digested with micrococcal nuclease, the crude chromatin fragments were immunoprecipitated with the specific anti-C/EBPα antibody (Santa Cruz, sc-365318, 1:500) overnight at 4°C and then incubated with protein-G magnetic beads. Next, the enriched chromatin was purified for ChIP-PCR amplification and the amplified products were examined with 2.0% agarose gel electrophoresis. The primer is presented as follows: forward primer (5’-CGCAGCATCTCTGCTGTGAA-3’) and reverse primer (5’-ACCCTCCCGCTAAGTCAACA-3’).
The probes for electrophoretic mobility shift assay (EMSA) were as follows: the wild-type probe (5′-ACCGCGCTGGCGCAGCATCT-3′) and the mutant-type probe (5′-ACCGCATCAATATGACATCT-3′). Briefly, an oligonucleotide containing the C/EBPα motif and its complementary sequences were synthesized and then annealed to double-stranded structures. The wild-type probe was treated by methyltransferase M.SssI (New England Biolabs, Beijing, China). Total nuclear proteins were extracted from trypsinized monolayers of IPEC-J2 by a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Nanjing, China). The double-stranded oligonucleotide was 5′ end-labeled with biotin-dUTP. For each binding reaction, 2 μg labeled probes, 2 μg nuclear extract, 4 μg poly (dI–dC), 10 μL binding buffer [20 mM Tris-HCl (pH 7.6), 50 mM KCl, 1 mM DTT, 0.5 mM EDTA, and 10% glycerol] were incubated at 25°C for 30 min. Then the protein–DNA complexes were separated on non-denaturing 6% polyacrylamide gels with precooled 0.5 × TBE buffer. For the super-shift assay, 1 μL anti-C/EBPα antibody (Santa Cruz, sc-365318, 1:500) was added to EMSA reaction mixtures and incubated for 60 min at 4°C prior to the labeled probe addition. Then, the protein–DNA complexes were transferred to nylon membrane at 60 v for 1 hour and autoradiography of the dried gel was detected immediately after UV cross-linking.
The TFF1 promoter sequences were analyzed by Alibaba2 online software to predict the potential transcription factor binding sites (TFBS). According to the core promoter prediction of BDGP online software, the PCR purified products were digested with KpnI and XhoI and ligated to the pGL3-basic luciferase reporter vector. Four plasmids with truncated promoters were constructed by inserting the DNA sequences of the TFF1 promoter ranging from −1200 to −1. The truncated recombinant vectors were defined as pGL3-control (−200 to −1), pGL3-TFF1-p1 (−360 to −1), pGL3-TFF1-p2 (−500 to −1) and pGL3-TFF1-p3 (−1200 to −1).
For the reporter-gene assays, pGL3-TFF1-wt was constructed by inserting the DNA fragments of the TFF1 promoter from −360 to −200 into pGL3-basic vector. The mutant form pGL3-TFF1-mut contains the sequences mutated from GCTGGCGCAG to ATCAATATGA. In addition, the sequences GCTGGCGCAG enrolled in pGL3-TFF1-wt was deleted and named as pGL3-TFF1-del. The IPEC-J2 were co-transfected with C/EBPα-oe and wild type, mutant or deletion forms of TFF1 promoter reporter constructs. The renilla was co-transfected for normalization. After 36 hours post-transfection, luciferase activity was analyzed with a Dual-Luciferase Assay system (Promega Corporation, Madison, USA) on a luminometer according to the manufacturer’s instruction. The relative fluorescence intensity was calculated as Firefly-Luc (Ff)/Renilla-Luc (Rn). All the transfections were performed in sextuplicate and each experiment was repeated at least three times.
Statistical analysis was performed by GraphPad Prism 8.0 software. All statistical details of experiments were included in the figure legends. All experiments were repeated at least three times and the results were shown as mean values ± SD.
The PCR products were amplified to detect PEDV, TGEV, PDCoV, PoRV in the small intestinal samples. The specific PCR products were only amplified in the infected samples with PEDV classic strain CV777 (Figure 1A). Our results show that PEDV is the unique infectious agent detected in diarrhea piglets (Supplementary Figure 1A). Histological analysis reveals that PEDV infection led to villi atrophy and partial shedding of villous epithelial cells (Figure 1B). The TFF1 mRNA expression was shown at higher levels in the porcine duodenum, jejunum, and ileum (Figure 1C). To further validate the relationship between TFF1 biological function and PEDV replication, we examined the mRNA expression of the TFF1 in vitro and in vivo, respectively. As shown in Figure 1D, the mRNA expression of TFF1 in jejunum and ileum was significantly up-regulated in the PEDV group compared to that of control (P < 0.01), and TFF1 protein expression was also significantly increased in jejunum (P < 0.01, Figure 1E; Supplementary Figure 1B). Furthermore, the mRNA expression of TFF1 was significantly up-regulated at 48 hours post infection of PEDV (P < 0.01, Figure 1F). These results provide evidence that TFF1 is associated with the host responses toward PEDV infection.
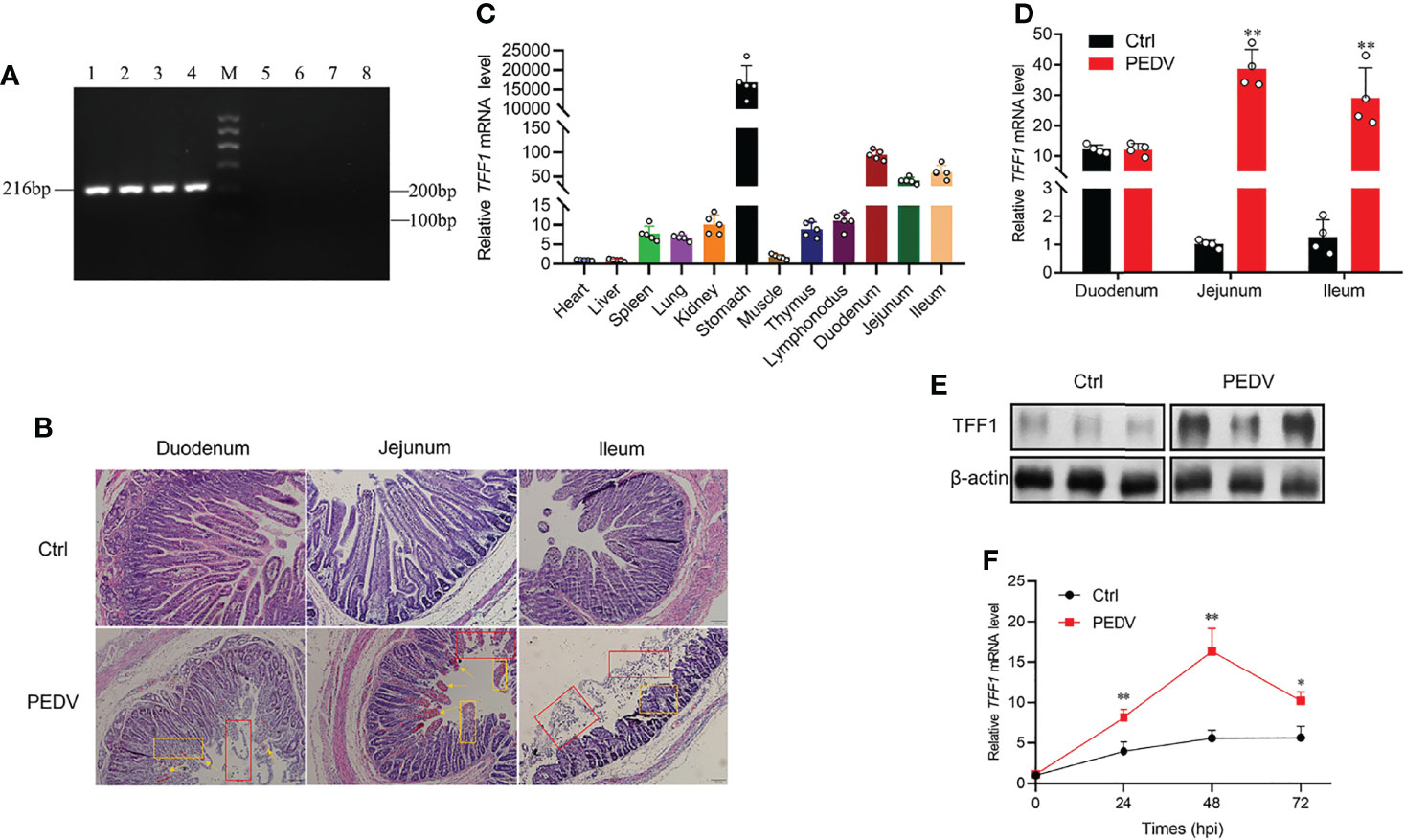
Figure 1 TFF1 is a key candidate responsible for PEDV infection. (A) RT-qPCR detection of M gene expression by 2.0% agarose gel electrophoresis. Lanes 1-4, intestinal contents of diarrhea piglets; Lane M, Marker I marker; Lanes 5-8, intestinal contents of control piglets. (B) Paraffin sections microscopy of duodenum, jejunum, and ileum tissues (100×). Different symbols in the pictures represented different pathological symptoms; yellow arrows represented congestion in the lamina propria, yellow boxes represented infiltration of inflammatory cells, and red boxes represented breakage and shedding of epithelial cells in the intestinal mucosa. (C) Porcine tissue expression distribution of TFF1 in 11 tissues. (D) Differential expression of TFF1 gene in duodenum, jejunum, and ileum. (E) Western blotting analysis of TFF1 protein level in jejunum. β-actin was used as internal reference proteins. (F) IPEC-J2 were infected with PEDV at MOI of 0.1. Cells were collected at 24, 48, and 72 hours post infection and detected mRNA expression of TFF1 by RT-qPCR. Data are presented as mean values ± SD, using two tailed Student’s t-test. *p < 0.05, **p < 0.01. Each treatment has triplicate biological replicates at least.
To verify the function of TFF1 during PEDV replication, we constructed TFF1 knockout and overexpression IPEC-J2 cell lines (Figures 2A–D; Supplementary Figures 2A–E). We found that knockout of TFF1 markedly up-regulated the mRNA expression of M gene, whereas TFF1 overexpression down-regulated its expression upon PEDV infection (Figure 2E). The knockout also resulted in an enhanced viral particle, while TFF1 overexpression led to a marked reduction of PEDV infectiousness (Figures 2F, G). Similarly, TFF1 knockout significantly increased PEDV genome copy number (P < 0.01, Figure 2H). In addition, the cell culture supernatants were titrated for TCID50 and it reveals that the infectious titers were significantly enhanced in TFF1-knockout cells (P < 0.01, Figure 2I). Overall, these results suggest that the loss of TFF1 expression exacerbates PEDV replication.
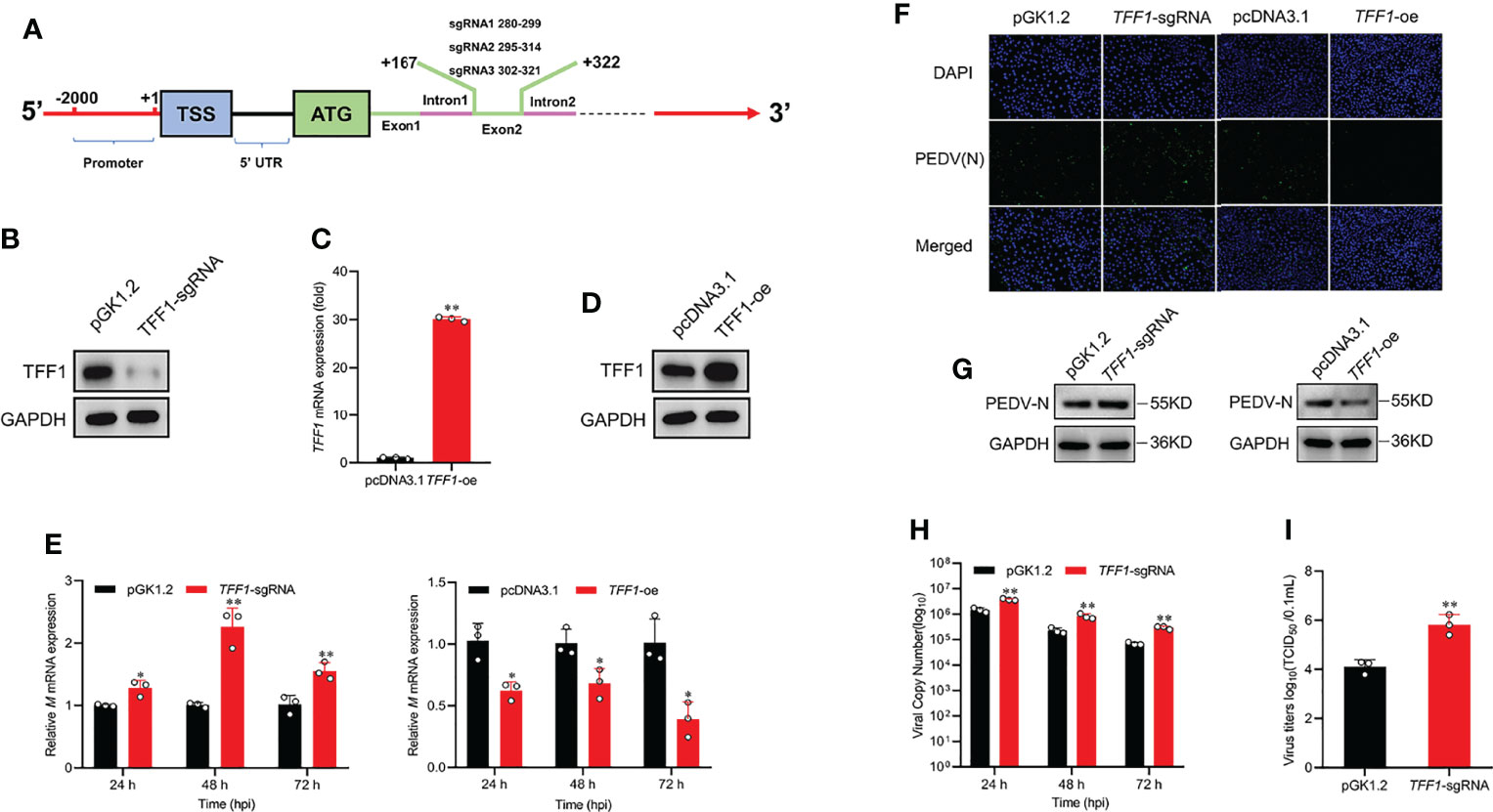
Figure 2 TFF1 inhibits PEDV replication. (A) The location of the three sgRNAs (sgRNA1, sgRNA2, sgRNA3) used for TFF1 knockout. (B) Western blot was performed to detect the knockdown efficiency of TFF1-sgRNA mixture pool cells compared with the pGK1.2 vector in IPEC-J2 cell lines. (C, D) Protein and mRNA levels of TFF1 were determined in IPEC-J2 transfected with TFF1-oe. (E) mRNA levels of M gene in IPEC-J2 transfected with TFF1-sgRNA and/or TFF1-oe after PEDV infection at MOI of 0.1 for 24, 48, and 72 hours. (F, G) The indirect immunofluorescence assay and western blotting were performed to detect PEDV (N) in PEDV-infected cells. TFF1-sgRNA and TFF1-oe cells were infected with PEDV at MOI of 0.1, respectively. (H) The PEDV genome copy number was measured by RT-qPCR. (I) Cell culture supernatants were collected at 24, 48, and 72 hours PEDV post infection and titrated for TCID50. Data are presented as mean values ± SD, using two tailed Student’s t-test. *p < 0.05, **p < 0.01. Each treatment has triplicate biological replicates at least.
To investigate the effects of TFF1 depletion on the growth and survival of IPEC-J2, we found that TFF1 knockout decreased the viability of IPEC-J2 and promoted the development of cell lesion in a time-dependent manner (Figures 3A, B). Similarly, the knockout of TFF1 also resulted in a poor survival of IPEC-J2 as measured by CCK8 and caused the pronounced apoptosis after PEDV infection, as reflected by the activation of caspase3/7 with flow cytometry analysis (P < 0.01, Figures 3C, D). Moreover, TFF1 knockout led to the significantly decreased - proteins which are important for cell proliferation, growth, and survival, along with an increased levels of Bax and Cleaved-Caspase 3 (Figure 3E). Additionally, we found that TFF1 knockout reduced the S-phase but increased the cell growth in the G0/G1 phase (Figure 3F). The expression of mRNA and protein for cell cycle genes were significantly down-regulated (Figures 3G, H). Furthermore, the mRNA expression of proinflammatory cytokines and antiviral genes were significantly up-regulated after TFF1 knockout (Figures 3I, J). These results indicate that TFF1 depletion alters the cell cycle and exacerbates PEDV-induced cell death and inflammatory responses.
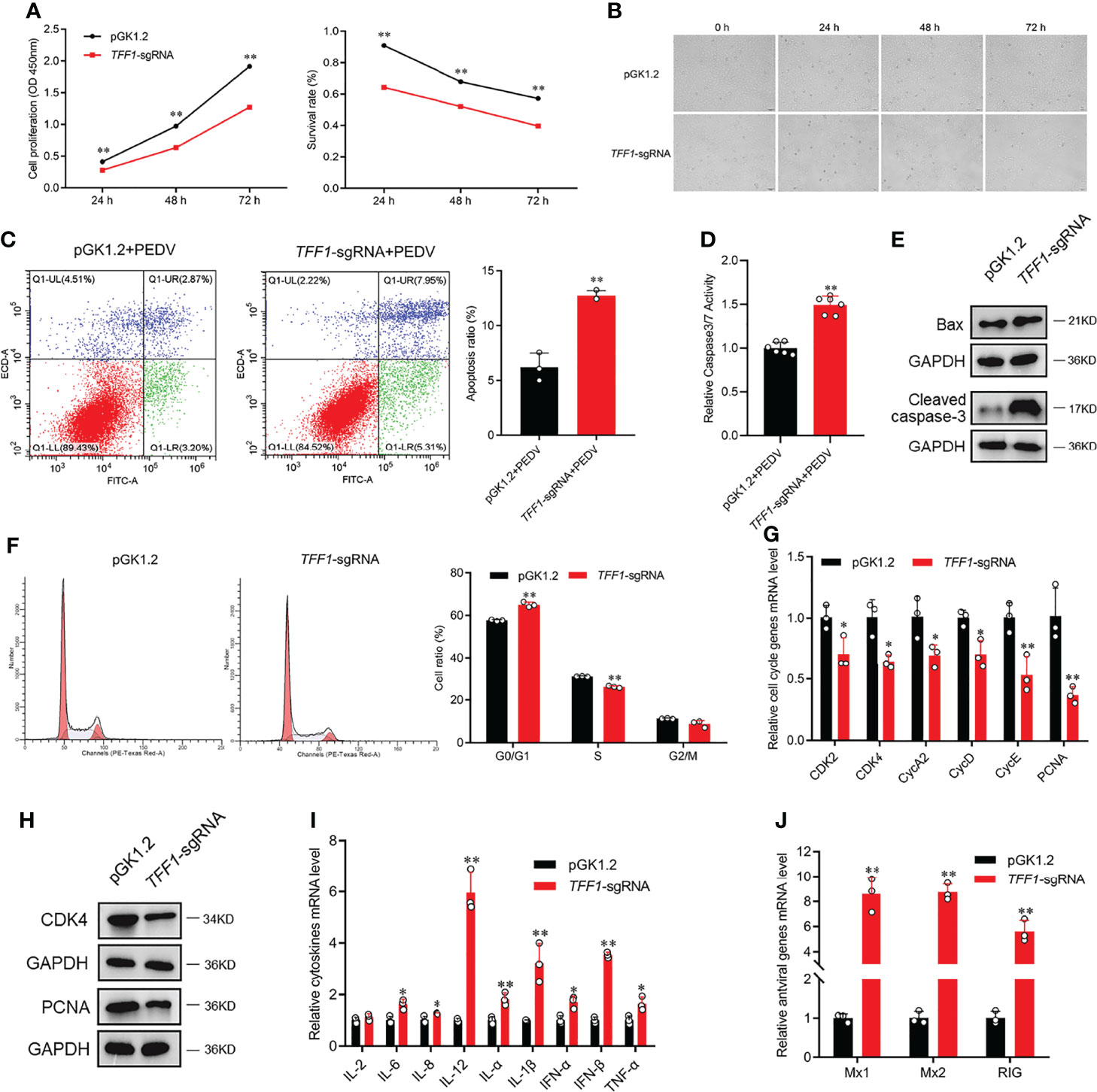
Figure 3 TFF1 reduces PEDV-induced intestinal inflammation. (A) The proliferation and cytotoxicity of TFF1-knockout were detected by CCK-8 assay in IPEC-J2. (B) Morphological changes in TFF1-knockout cells after PEDV infection for 0, 24, 48 and 72 hours. (C, D) Cell apoptotic was detected by caspase3/7 activity and flow cytometry. (E) Protein levels of cell proliferation regulators were detected by western blotting in response to PEDV infection at MOI of 0.1 in TFF1-knockout cells. (F) Cell cycle distribution was detected by flow cytometry. (G, H) mRNA and protein levels of cell cycle genes were determined in TFF1-sgRNA cell. (I, J) mRNA expression of inflammatory cytokines and antiviral genes were detected after PEDV infection at MOI of 0.1 in TFF1-sgRNA cells. Data are presented as mean values ± SD, using two tailed Student’s t-test. *p < 0.05, **p < 0.01. Each treatment has triplicate biological replicates at least.
The CpG islands (located at −477 to −299 bp) were predicted by MethPrimer (Figure 4A; Supplementary Figure 3A). As shown in Figure 4B, bisulfite-sequencing results reveal that there was different methylation status of 12 CpG sites. Methylation analysis demonstrates that mC-2 loci and mC-6 loci presented hypomethylation status compared to that of the control (P < 0.05, Figure 4C). Pearson correlation analysis shows that there was a significantly negative correlation between methylation status and TFF1 mRNA expression in the mC-6 loci (R =−0.78, r0.05 = 0.707, P =0.022, Figure 4D). According to the predicted core promoter sequences, we constructed four truncated recombinant vectors (Figure 4E; Supplementary Figure 3B). As shown in Figure 4F, luciferase reporter assay shows that the transcriptional activity of pGL3-TFF1-p1 was remarkedly higher than that of pGL3-control (P < 0.01), indicating that the core promoter region of TFF1 was located at −360 to −200 bp. To investigate the epigenetic mechanisms of TFF1 regulation, we first generated a methylated vector DNA substrate with the methyl transferase of M.SssI. Then, we transiently transfected the C/EBPα overexpression vector that failed to activate methylated reporter of TFF1 compared to the unmethylated reporter (Figure 4G). However, the mRNA expression of TFF1 was markedly upregulated after 5’-Aza-2’-dC treatment compared to that of control, indicating that DNA methylation inhibition strongly enhanced gene expression of TFF1 (Figure 4H). Furthermore, M.SssI or 5’-Aza-2’-dC treatment before PEDV infection considerably regulated PEDV entry in IPEC-J2. As shown in Figure 4I, PEDV invasion was significantly inhibited by 5’-Aza-2’-dC, while the expression of M gene was significantly upregulated with M.SssI treatment. These results suggest that the hypermethylation of TFF1 facilitates PEDV replication.
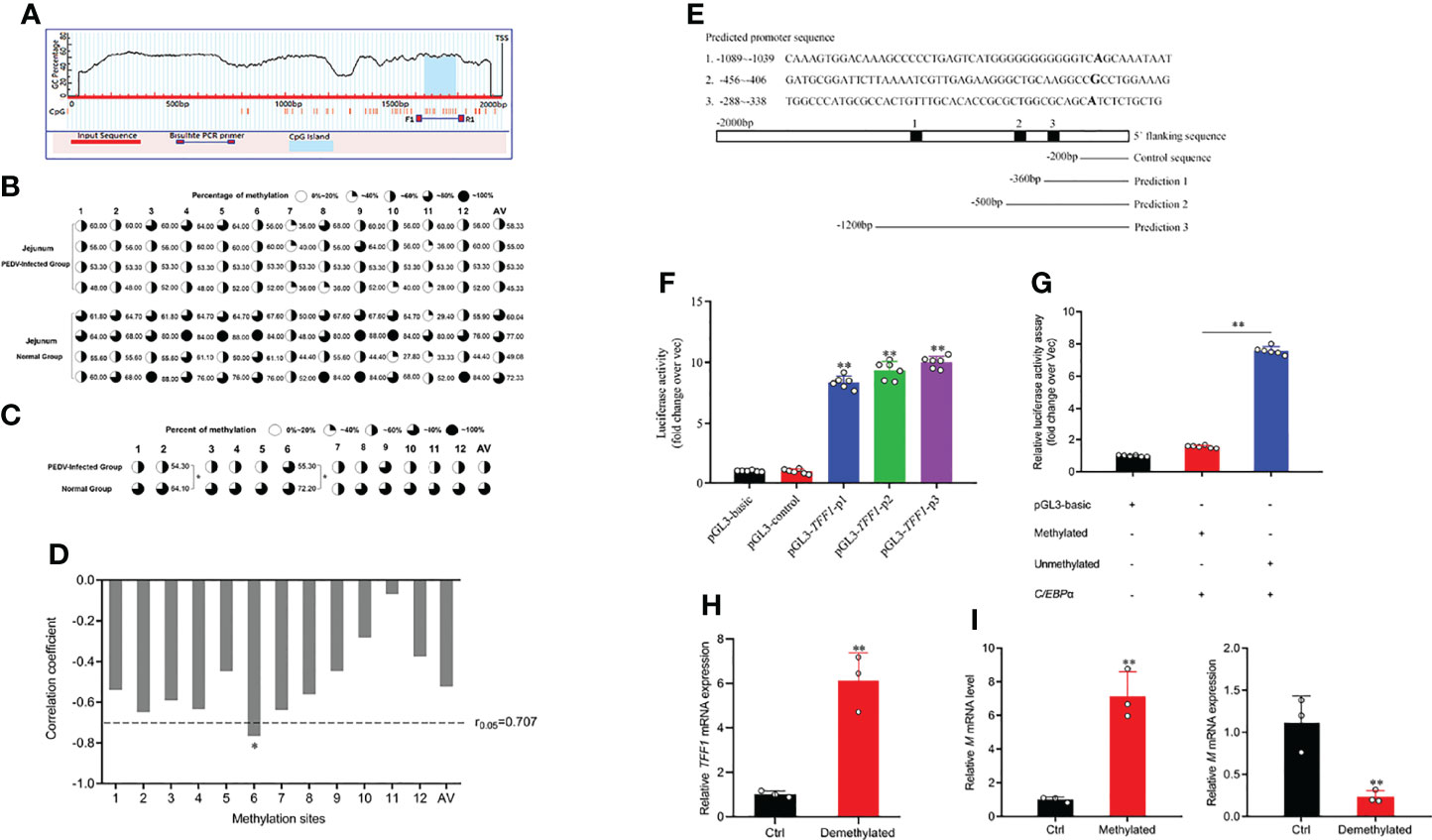
Figure 4 mC-6 methylation is involved in C/EBPα-mediated TFF1. (A) CpG island prediction of porcine TFF1 upstream 2000 bp promoter region. The long red bar represents the promoter sequence; dark blue line shows designed methylation primer; blue bar indicates the CpG island, TSS means transcriptional start site. (B, C) Bisulfite genomic sequencing analysis of methylation profile in 12 CpG sites and average methylation status in CpG island of the TFF1 promoter. AV, average methylation status. (D) The correlation analysis between methylation status of each CpG site and mRNA expression of TFF1 in jejunum. (E) Prediction of TFF1 core promoter region and amplification of truncated fragments. (F) Identification of core promoter region of TFF1 by four different truncated plasmids. (G) The relative fluorescence intensity of TFF1 was determined by luciferase reporter assay in vectors treated with or without methyl transferase M.SssI. IPEC-J2 were co-transfected with C/EBPα-oe and methylated or unmethylated vectors. (H) Relative mRNA expression of TFF1 was determined by RT-qPCR in IPEC-J2 treated with or without 0.1 μM 5’-Aza-2’-dC for 72 hours. (I) Relative mRNA expression of M gene was determined by RT-qPCR in IPEC-J2 treated with or without M.SssI after PEDV infection at MOI of 0.1. Data are presented as mean values ± SD, using two tailed Student’s t-test analysis. *p < 0.05, **p < 0.01. Each treatment has triplicate biological replicates at least.
Having demonstrated the crucial role of TFF1 in IPEC-J2, we next examined whether C/EBPα participated in the transcriptional regulation of TFF1. As shown in Figure 5A, we further predicted the potential transcription factors in 12 CpG sites of TFF1 promoter, in which mC-6 loci was found to be within the binding domain of C/EBPα. We then performed the reporter-gene assays with the promoter of TFF1 and found that it was highly responsive to C/EBPα-mediated transactivation. The mutation or deletion of the putative C/EBPα binding site effectively diminished the C/EBPα-dependent activation (Figures 5B, C). Using transient co-transfection and reporter gene assays, we found that overexpression of C/EBPα resulted in transactivation of the TFF1 gene promoter, suggesting that C/EBPα can act as an activator of TFF1 (Figure 5D). Finally, a positive DNA fragment was detectable in the products amplified with chromatin fragments that were precipitated using specific anti-C/EBPα antibody (Figure 5E). Together, the data indicate that C/EBPα activates TFF1 target promoters via the putative C/EBPα binding site. To investigate the influence of DNA methylation on the nuclear protein binding properties, we performed EMSA using 5′-biotinylated probes corresponding to putative TFF1 sequences including the mC-6 loci. Specifically, two different double-stranded oligonucleotides (wild or mutant type) were treated with or without methyl transferase M.SssI and incubated with nuclear extracts from IPEC-J2. Thereafter, the wild-type probes resulted in significantly increased mobility shift of nuclear proteins compared with the mutant-type and methylated wild-type probes. The shift can be competed by an excess of unlabeled 5′-biotinylated probes which restrain C/EBPα binding to TFF1. Additionally, the specific binding of C/EBPα to the TFF1 was further confirmed using an anti-C/EBPα antibody in a super-shift assay (Figure 5F). These results show that methylation at mC-6 loci blocks the C/EBPα binding to TFF1 promoter region.
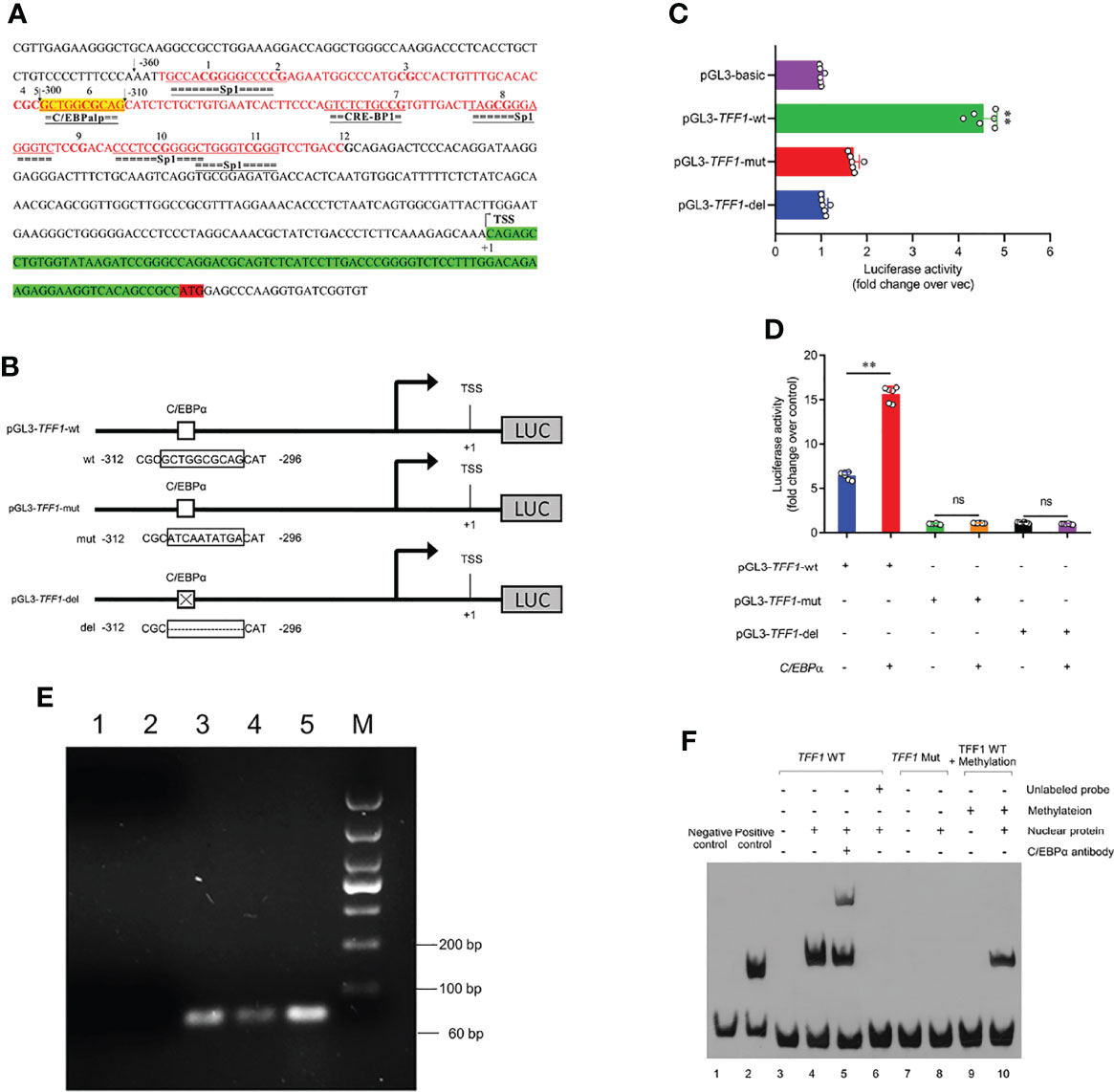
Figure 5 C/EBPα activates the TFF1 expression. (A) Schematic representation of the TFF1 promoter sequences. C/EBPα potential binding site is highlighted in yellow. CpG island is highlighted in red and CG sites are shown in bold font. TSS was the transcription start site, and defined as +1. (B, D) The pGL3-TFF1 recombinant vectors (with or without C/EBPα consensus site) were constructed and co-transfected with C/EBPα-oe into IPEC-J2. (C) The pGL3-TFF1 constructs (pGL3-TFF1-wt, pGL3-TFF1-mut and pGL3-TFF1-del) were transfected into IPEC-J2. Luciferase reporter assays were performed 36 hours post transfection. (E) Identification of C/EBPα binding to TFF1 promoter by ChIP-PCR. Lane 1, PCR amplification with RNase-free H2O (blank control); Lane 2, PCR amplification with Rabbit IgG antibody (negative control); Lane 3, PCR amplification with anti-C/EBPα antibody; Lane 4, PCR amplification with RNA Polymerase II antibody (positive control); Lane 5, PCR amplification with Input DNA; Lane M, DL1000 DNA marker. (F) EMSAs confirmed the binding of the C/EBPα protein to the TFF1 promoter region. All free biotin-labeled probes (TFF1 WT probes, Lane3-6, and Lane9-10; TFF1 Mut probes, Lane7-8) were examined by streptavidin-HRP conjugate. Lane 5, Super-shift mobility band was confirmed complex of biotin-labeled TFF1 WT probes and C/EBPα protein. Lane 6, unlabeled probes was a cold competitor probe. Lane 9-10, TFF1 WT probes were treated by methyl transferase M.SssI. Data are presented as mean values ± SD, using two tailed Student's t-test analysis. **P < 0.01, ns, not significant. Each treatment has triplicate biological replicates at least.
To examine the regulatory role of C/EBPα in PEDV infection, the cells with PEDV treatment resulted in a similar up-regulation of C/EBPα with that of TFF1 gene (Figure 6A). Therefore, siRNAs of C/EBPα were transfected into IPEC-J2 (Figure 6B and Supplementary Figure 4A). Interestingly, C/EBPα silencing played an important role in the viral replication, exhibiting that mRNA expression of M gene was significantly up-regulated (Figure 6C). Subsequently, C/EBPα overexpression vector was constructed (Supplementary Figures 4B, C) and co-transfected with promoter region of TFF1 significantly reduced the invasion rates of PEDV (Figure 6D), indicating that the C/EBPα plays an effective role in antiviral target activator on regulating TFF1 expression.
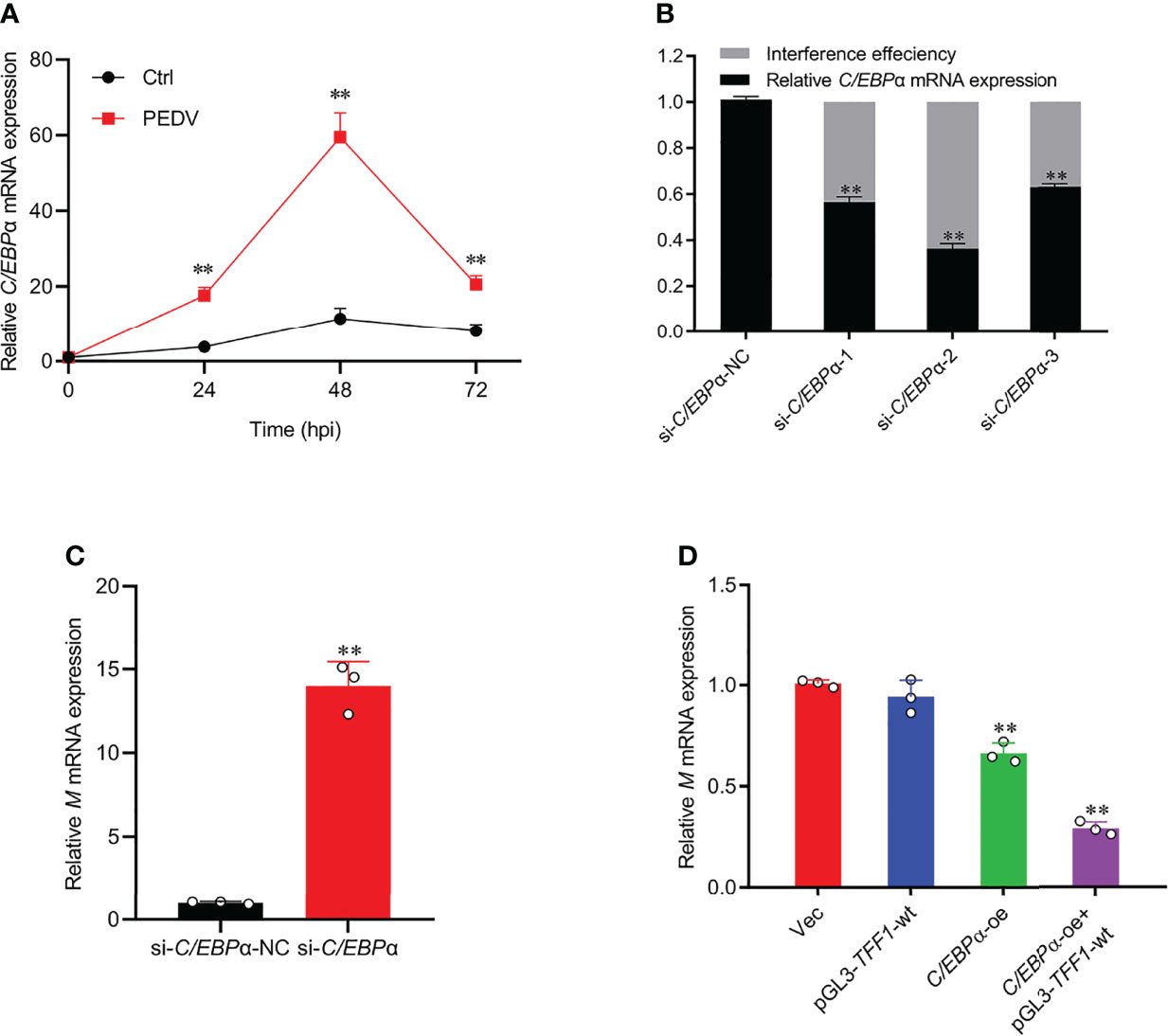
Figure 6 C/EBPα exhibited antiviral activity. (A) IPEC-J2 were infected with PEDV at MOI of 0.1. Cells were collected at 24, 48, and 72 hours post infection and detected mRNA expression of C/EBPα by RT-qPCR. (B) Knockdown efficiency of si-C/EBPα-1, si-C/EBPα-2, and si-C/EBPα-3 was detected by RT-qPCR. (C) Relative mRNA expression of M gene was determined by RT-qPCR in IPEC-J2 transfected with si-C/EBPα and si-C/EBPα-NC. (D) Relative mRNA expression of M gene was determined by RT-qPCR in IPEC-J2 co-transfected with pGL3-TFF1-wt and C/EBPα-oe after PEDV infection. Data are presented as means ± SD, using two tailed Student’s t-test analysis. **p < 0.01. Each treatment has triplicate biological replicates at least.
The intestinal barrier integrity is essential for the maintenance of homeostasis, while its dysfunction causes significant consequences and thereby gastrointestinal diseases, such as viral and bacterial infection. PEDV strains have been characterized by rapid proliferation in and destruction of intestinal epithelial cells (IECs) among suckling piglets (28) but milder in adult pigs (29, 30). A variety of viruses was considered a leading cause of severe gastroenteritis and diarrhea (31, 32), determined by the process of intestinal development in young animals. There were vacuolated small intestinal epithelial cells in most 2-week-old pigs but hardly detected after 3-4 weeks from the newborn (33). The turnover of the small intestinal epithelium in the newborns (7-10 days) was slower than that of the weaned piglets (2-4 days) (34). The blunted recovery may result from the susceptibility to various infections like PEDV at the newborn stage. Thus, it is vital to determine the PEDV and host mucosa interactions and the onset of the disease. Our results and others have demonstrated that TFF1 functions to restore and maintain the gastrointestinal mucosal barrier integrity (35, 36). While TFF1 abnormal expression, together with the characteristics of diarrhea population is subjected to intestinal villous atrophy, deepening crypts depth in the PEDV-infected suckling piglets. The specific responses of TFF1 hyper-expression have been revealed by Ren (37) and Soutto (38), who found pathologically increased TFF1 expression in gastroenteritis and gastrointestinal ulcers. In previous studies, a significant up-regulation of TFF1 expression after PEDV infection has been proved (39). Herein, we addressed that TFF1 was highly expressed in the jejunum in response to PEDV and revealed it as the primary loci, where TFF1 started to defend further PEDV entry by the enhanced transcription. Indeed, TFF1 knockout mouse has been generated previously and exhibited pro-inflammatory responses in the small intestine with an abnormal mucosal epithelium proliferation and an obvious impaired barrier function (40). Being consistent with the actions of TFF1 deletion, the knockout of the TFF1 gene in IPEC-J2 significantly suppresses cell proliferation and induces apoptosis. In our model, there is also an enhanced viral particle, copy number, and virus titer of PEDV. In this regard, as we assumed that TFF1 is one of the key factors to resist the PEDV infection and the ectopic expression of TFF1 strongly inhibited PEDV replication. Notably, TFF1 functions enrolled in cell growth and early differentiation contribute to its protective role probably through proliferation-associated β-catenin signaling pathway (41). Thus, we provided evidence for supporting TFF1-controlled cell cycle and arrest. Our evidence exhibited that a significantly increased cell proportion at the G1 phase, with a drastic reduction at S phase, was caused by TFF1 knockout. According to these clues of cell proliferation by TFF1 depletion during PEDV invasion, we suggest that TFF1 possibly re-establishes the epithelium community via promoting cell proliferation. It is well-known that the viral infection-triggers severe inflammatory responses as one of the main events of pathogenesis (42). Importantly, IECs integrate signals to modulate inflammatory responses and facilitate intestinal homeostasis as reported (43). It is reasonable that couples of inflammatory cytokines were secreted which helps to prevent viral entry of the gut barrier (44). These responses have also been displayed in PEDV infection. Herein, we showed the altered RIG-I signaling pathway, RIG-I-like receptors, and Mx1 and Mx2 in infected intestine. The selections are critical for anti-viral actions in IECs (45, 46), inducing IFN-I (IFN-α/IFN-β) production and also working for the host innate immune system when confronted with the viral invasion (47). Given the fact that the pro-inflammatory cytokines, such as TNF-α and interleukins, are activated by the innate immune system, we found that the cytokines associated with antiviral genes were all elevated. This suggests that innate immune responses functioned through cytokines and are involved in the PEDV-infection for the defense responses.
The transcription factor CCAAT enhancer binding protein α (C/EBPα) is essential for normal cell development and evidence suggests that impaired function resulting from silencing C/EBPα contributes directly to disease development (48). In agreement with a previous study, we also verified that C/EBPα knockdown was significantly associated with enhanced PEDV virulence. Furthermore, C/EBPα is involved in various innate immune functions such as inflammatory cytokines production and immune response against HIV infection by TLR3-mediated (49). Therefore, up-regulated C/EBPα expressions may prime the cells for generating a robust immune response against PEDV infection. C/EBPβ would prime cell differentiation and promote the expression of TFF1 by inducing C/EBPα expression in vivo. In parallel, TFF1 expression is upregulated in C/EBPα-transfected cells in vitro (24). Activating TFF1 production, an important event in PEDV infection suppression, could follow the overexpression of C/EBPα and/or promoter hypomethylation. Abnormal alterations of methylation status are always associated with the transcription silencing and contribute to disease development (50). By preventing the binding of transcription factors to their motifs, DNA methylation can therefore block transcription activation of gene promoters (51). In previous studies, the epigenetic regulation modulated TFF1 expression has been proved (52, 53), whereas the epigenetic modifications of TFF1 function under PEDV infection is still unknown. As we assumed, PEDV changed the DNA methylation profiles in which hypomethylation of mC-6 loci was observed to in association with TFF1 expression. Being consistent with the actions of mC-6 loci methylated, TFF1 gene transcriptional activity was reduced. Notably, the mC-6 loci containing the C/EBPα binding domain was also located in the TFF1 core promoter region. It is worth mentioning that TFF1 transcription was highly responsive to C/EBPα-mediated transactivation and, therefore, plays a crucial role in the activation of gene transcription, cell proliferation, and differentiation (54). In agreement with the results of Gonçalo Regalo et al. (24), we verified that C/EBPα can bind to the putative motif in porcine TFF1 promoter and perform its transactivated action. This is associated with the finding of C/EBPα-medicated TFF1 by inhibiting the Ras/MAPK pathway due to their co-localization (24). Indeed, Ras/MAPK signaling is heavily involved in C/EBPα-dependent regulation of specific antivirus genes, which may be an alternative way to modify TFF1 expression during PEDV infection and need to be further validated.
A graphic illustration of the C/EBPα-mediated epigenetic regulation of TFF1 in PEDV infection is shown in Figure 7. This study indicated that TFF1 is highly expressed in the inflamed jejunum epithelium during PEDV-infection. The TFF1 expression improves the intestinal functions through down-regulating pro-inflammatory cytokines gene expression like IFNs and TNF-α, and facilitate the inhibition of the virus replication by increasing antivirus genes likes RIG, Mx1 and Mx2. This process further benefits the porcine intestinal epithelial cell proliferation and growth by inhibiting apoptotic kinases such as Bax and Caspase 3. Therefore, the repairment helps the host to fight against the porcine coronavirus infection. Mechanistically, this virus affects the DNA methylation profile, especially the mC-6 loci methylation, including the key factor C/EBPα binding sites. The transformation alteration of C/EBPα is detrimental to the specific bind to TFF1. This process drives the TFF1 action of jejunum inflammatory responses when suffering from PEDV invasion.
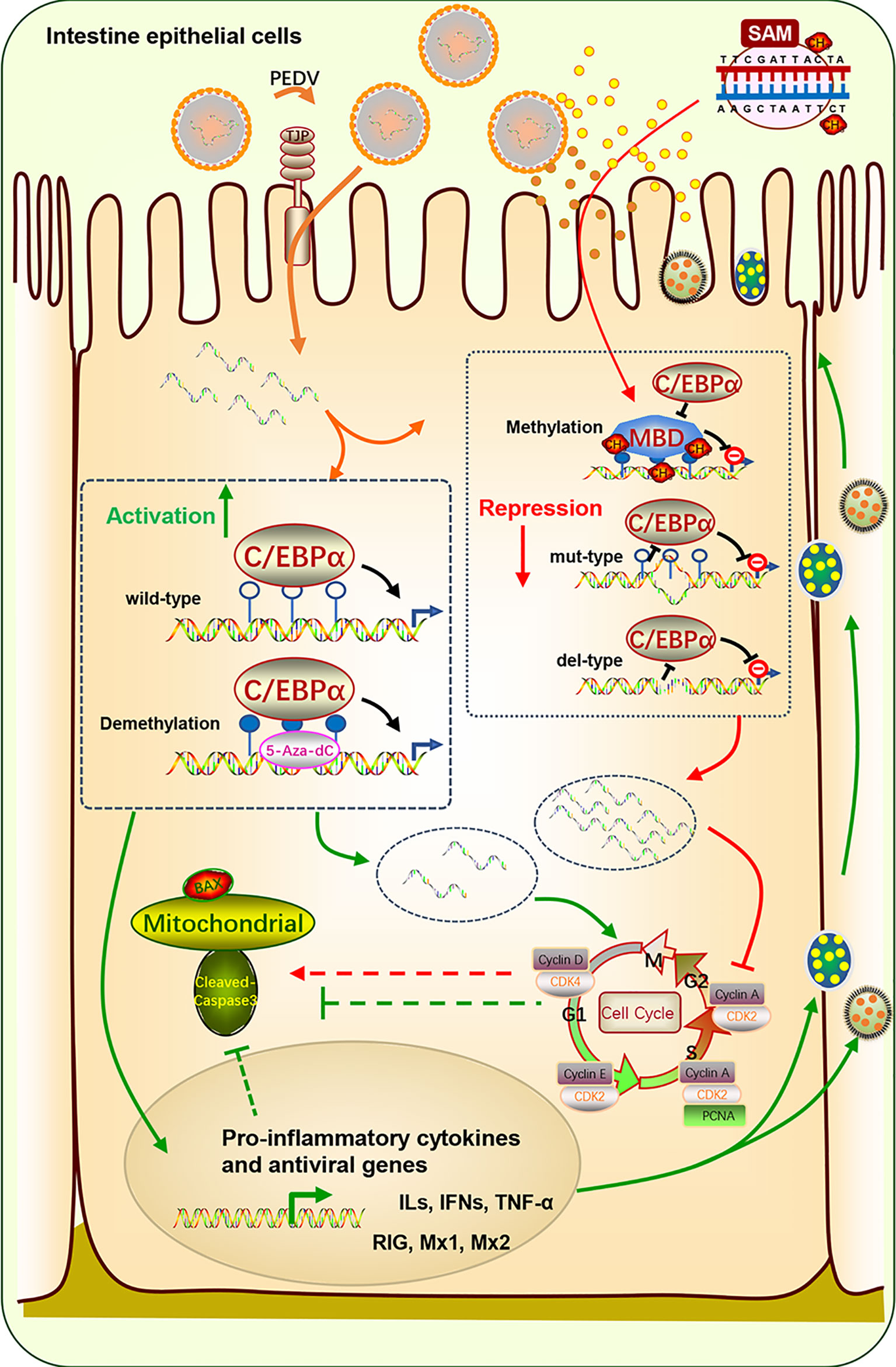
Figure 7 Schematic diagram of C/EBPα-modulated TFF1 via mC-6 methylation in the PEDV-induced jejunum inflammation. TFF1 is highly expressed in intestinal epithelium during PEDV-infected jejunum inflammation. It improves the intestinal functions through reducing pro-inflammatory cytokines and facilitating the inhibition of the virus replication by increasing antivirus genes. This process further benefits the porcine intestinal epithelium cell proliferation and growth by inhibiting apoptotic kinases. The mucous repairment is therefore help the host resistant to the porcine coronavirus infection. Mechanistically, this virus affects DNA methylation profile especially the mC-6 loci methylation including the key factor C/EBPα binding sites. The transformation alteration of C/EBPα is detrimental for the specific bind to TFF1. This process determinates the TFF1 action of jejunum inflammatory responses suffer from PEDV invasion.
In this study, we confirmed that high expression of TFF1 contributes to enhancing the host resistance to PEDV invasion. Additional, upregulated of C/EBPα expression also partially inhibited PEDV replication. Activating TFF1 production could follow the overexpression of C/EBPα and/or promoter hypomethylation, which contributes to enhancing piglets’ resistance to PEDV. Collectively, our results feature a novel connection between TFF1 and regulator C/EBPα for PEDV resistance in porcine intestinal epithelium. Our discoveries provide unique insights into the potential modulation of DNA methylation to mediate the C/EBPα-dependent TFF1. Our findings contribute to the field of intestinal protection, anti-inflammation, and anti-coronavirus in porcine and other species. Challenges remain as to address complex mechanisms of coronaviruses and TFF1 can be regarded as a potential biomarker candidate and therapy target for further studies.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The animal study proposal was approved by the Institutional Animal Care and Use Committee (IACUC) of the Yangzhou University Animal Experiments Ethics Committee (permit number: SYXK (Su) IACUC 2012-0029, Approval Date: 09-12-2012). All experimental methods were conducted in accordance with the relevant guidelines and regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China.
HQ and QZ performed most of the experiments; HW and SW participated in the experiments; WB and DC conceived the study, HQ, QZ, and DC participated in its design and coordination, and wrote the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (No. 31972535), the Jiangsu Agricultural Science And Technology Innovation Fund [CX(21)2014], the Science and Technology Support Program of Jiangsu Province (BE2019344) and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.881289/full#supplementary-material
1. Song D, Moon H, Kang B. Porcine Epidemic Diarrhea: A Review of Current Epidemiology and Available Vaccines. Clin Exp Vaccine Res (2015) 4:166–76. doi: 10.7774/cevr.2015.4.2.166
2. Liu C, Ma Y, Yang Y, Zheng Y, Shang J, Zhou Y, et al. Cell Entry of Porcine Epidemic Diarrhea Coronavirus Is Activated by Lysosomal Proteases. J Biol Chem (2016) 291:24779–86. doi: 10.1074/jbc.M116.740746
3. Song D, Park B. Porcine Epidemic Diarrhoea Virus: A Comprehensive Review of Molecular Epidemiology, Diagnosis, and Vaccines. Virus Genes (2012) 44:167–75. doi: 10.1007/s11262-012-0713-1
4. Jung K, Saif LJ. Porcine Epidemic Diarrhea Virus Infection: Etiology, Epidemiology, Pathogenesis and Immunoprophylaxis. Vet J (2015) 204:134–43. doi: 10.1016/j.tvjl.2015.02.017
5. Suzuki T, Shibahara T, Yamaguchi R, Nakade K, Yamamoto T, Miyazaki A, et al. Pig Epidemic Diarrhoea Virus S Gene Variant With a Large Deletion non-Lethal to Colostrum-Deprived Newborn Piglets. J Gen Virol (2016) 97:1823–8. doi: 10.1099/jgv.0.000513
6. Li Y, Wu Q, Huang L, Yuan C, Wang J, Yang Q. An Alternative Pathway of Enteric PEDV Dissemination From Nasal Cavity to Intestinal Mucosa in Swine. Nat Commun (2018) 9:3811. doi: 10.1038/s41467-018-06056-w
7. Hoffmann W, Hauser F. The P-domain or Trefoil Motif: A Role in Renewal and Pathology of Mucous Epithelia. Trends Biochem Sci (1993) 18:239–43. doi: 10.1016/0968-0004(93)90170-R
8. Rio MC, Lepage P, Diemunsch P, Roitsch C, Chambon P. Primary Structure of Human Protein pS2. C R Acad Sci III (1988) 307:825–31.
9. Thim L. Trefoil Peptides: From Structure to Function. Cell Mol Life Sci (1997) 53:888–903. doi: 10.1007/s000180050108
10. Ribieras S, Tomasetto C, Rio M. The pS2/TFF1 Trefoil Factor, From Basic Research to Clinical Applications. Biochim Et Biophys Acta (1998) 1378:F61–77. doi: 10.1016/s0304-419x(98)00016-x
11. Huynh E, Li J. Generation of Lactococcus Lactis Capable of Coexpressing Epidermal Growth Factor and Trefoil Factor to Enhance In Vitro Wound Healing. Appl Microbiol Biotechnol (2015) 99:4667–77. doi: 10.1007/s00253-015-6542-0
12. Vandenbroucke K, Hans W, Van Huysse J, Neirynck S, Demetter P, Remaut E, et al. Active Delivery of Trefoil Factors by Genetically Modified Lactococcus Lactis Prevents and Heals Acute Colitis in Mice. Gastroenterology (2004) 127:502–13. doi: 10.1053/j.gastro.2004.05.020
13. Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, Vanhoenacker P, Corveleyn S, et al. AG013, a Mouth Rinse Formulation of Lactococcus Lactis Secreting Human Trefoil Factor 1, Provides a Safe and Efficacious Therapeutic Tool for Treating Oral Mucositis. Oral Oncol (2010) 46:564–70. doi: 10.1016/j.oraloncology.2010.04.008
14. Lalani EN, Williams R, Jayaram Y, Gilbert C, Stamp G. Trefoil Factor-2, Human Spasmolytic Polypeptide, Promotes Branching Morphogenesis in MCF-7 Cells. Lab Investiga J Tech Methods Pathol (1999) 79:537–46.
15. Taupin DR, Kinoshita K, Podolsky DK. Intestinal Trefoil Factor Confers Colonic Epithelial Resistance to Apoptosis. Proc Natl Acad Sci (2000) 97:799–804. doi: 10.1073/pnas.97.2.799
16. Bossenmeyer-Pourieí C, Kannan R, Ribieras S, Wendling C, Stoll I, Thim L, et al. The Trefoil Factor 1 Participates in Gastrointestinal Cell Differentiation by Delaying G1-S Phase Transition and Reducing Apoptosis. J Cell Biol (2002) 157:761–70. doi: 10.1083/jcb200108056
17. Lugrin J, Rosenblatt-Velin N, Parapanov R, Liaudet L. The Role of Oxidative Stress During Inflammatory Processes. Biol Chem (2014) 395:203–30. doi: 10.1515/hsz-2013-0241
18. Park WS, Oh RR, Park JY, Lee JH, Shin MS, Kim HS, et al. Somatic Mutations of the Trefoil Factor Family 1 Gene in Gastric Cancer. Gastroenterology (2000) 119:691–8. doi: 10.1053/gast.2000.16483
19. Wang H, Yang L, Qu H, Feng H, Wu S, Bao W. Global Mapping of H3K4 Trimethylation (H3k4me3) and Transcriptome Analysis Reveal Genes Involved in the Response to Epidemic Diarrhea Virus Infections in Pigs. Animals (2019) 9:523. doi: 10.3390/ani9080523
20. Sharma S, Kelly TK, Jones PA. Epigenetics in Cancer. Carcinogenesis (2010) 31:27–36. doi: 10.1093/carcin/bgp220
21. Gonzaga IM, Soares Lima SC, Nicolau MC, Nicolau-Neto P, Da Costa NM, de Almeida Simão T, et al. TFF1 Hypermethylation and Decreased Expression in Esophageal Squamous Cell Carcinoma and Histologically Normal Tumor Surrounding Esophageal Cells. Clin Epigenet (2017) 9:130. doi: 10.1186/s13148-017-0429-0
22. Song M. DNA Methylation of Trefoil Factor 1 (TFF1) is Associated With the Tumorigenesis of Gastric Carcinoma. Mol Med Rep (2014) 9:109–17. doi: 10.3892/mmr.2013.1772
23. Carvalho R, Kayademir T, Soares P, Canedo P, Sousa S, Oliveira C, et al. Loss of Heterozygosity and Promoter Methylation, But Not Mutation, may Underlie Loss of TFF1 in Gastric Carcinoma. Lab Invest (2002) 82:1319–26. doi: 10.1097/01.LAB.0000029205.76632.A8
24. Regalo G, Resende C, Wen X, Gomes B, Durães C, Seruca R, et al. C/EBP Alpha Expression is Associated With Homeostasis of the Gastric Epithelium and With Gastric Carcinogenesis. Lab Invest (2010) 90(8):1132–9. doi: 10.1038/labinvest.2010.79
25. Romano E, Vllahu M, Bizzarro V, Belvedere R, Esposito R, Petrella A, et al. TFF1 Promotes EMT-like Changes Through an Auto-Induction Mechanism. Int J Mol Sci (2018) 19:2018. doi: 10.3390/ijms19072018
26. Wang S, Xu C, Shi J, Wang H, Bao W. Regulatory Effect and Mechanism of APN Gene on Porcine Epidemic Diarrhea Virus Resistance. Gene (2021) 775:145448. doi: 10.1016/j.gene.2021.145448
27. Livak KJ, Schmittgen TDL. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-DDCt Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
28. Jung K, Annamalai T, Lu Z, Saif LJ. Comparative Pathogenesis of US Porcine Epidemic Diarrhea Virus (PEDV) Strain PC21A in Conventional 9-Day-Old Nursing Piglets vs. 26-Day-Old Weaned Pigs. Vet Microbiol (2015) 178:31–40. doi: 10.1016/j.vetmic.2015.04.022
29. Shibata I, Tsuda T, Mori M, Ono M, Uruno K. Isolation of Porcine Epidemic Diarrhea Virus in Porcine Cell Cultures and Experimental Infection of Pigs of Different Ages. Vet Microbiol (2000) 72:173–82. doi: 10.1016/S0378-1135(99)00199-6
30. Thomas JT, Chen Q, Gauger PC, Giménez-Lirola LG, Sinha A, Harmon KM, et al. Effect of Porcine Epidemic Diarrhea Virus Infectious Doses on Infection Outcomes in Naïve Conventional Neonatal and Weaned Pigs. PloS One (2015) 10:e0139266. doi: 10.1371/journal.pone.0139266
31. Miyabe FM, Dall Agnol AM, Leme RA, Oliveira TES, Headley SA, Fernandes T, et al. Porcine Rotavirus B as Primary Causative Agent of Diarrhea Outbreaks in Newborn Piglets. Sci Rep (2020) 10:22002. doi: 10.1038/s41598-020-78797-y
32. Tizard IR. Vaccination Against Coronaviruses in Domestic Animals. Vaccine (2020) 38:5123–30. doi: 10.1016/j.vaccine.2020.06.026
33. Moon HW. Vacuolated Villous Epithelium of the Small Intestine of Young Pigs. Vet Pathol (1972) 9:3–21. doi: 10.1177/030098587200900102
34. Moon HW. Epithelial Cell Migration in the Alimentary Mucosa of the Suckling Pig. Proc Soc Exp Biol Med (1971) 137:151–4. doi: 10.3181/00379727-137-35533
35. Longman RJ, Douthwaite J, Sylvester PA, Poulsom R, Corfield AP, Thomas MG, et al. Coordinated Localisation of Mucins and Trefoil Peptides in the Ulcer Associated Cell Lineage and the Gastrointestinal Mucosa. Gut (2000) 47:792–800. doi: 10.1136/gut.47.6.792
36. Ruchaud-Sparagano MH, Westley BR, May FEB. The Trefoil Protein TFF1 is Bound to MUC5AC in Humangastric Mucosa. Cell Mol Life Sci (2004) 61:1946–54. doi: 10.1007/s00018-004-4124-x
37. Ren JL, Luo JY, Lu YP, Wang L, Shi HX. Relationship Between Trefoil Factor 1 Expression and Gastric Mucosa Injuries and Gastric Cancer. World J Gastroenterol (2005) 11:2674–7. doi: 10.3748/wjg.v11.i17.2674
38. Soutto M, Chen Z, Saleh MA, Katsha A, Zhu S, Zaika A, et al. TFF1 Activates p53 Through Down-Regulation of miR-504 in Gastric Cancer. Oncotarget (2014) 5:5663–73. doi: 10.18632/oncotarget.2156
39. Qi X, Xu Y, Qin W, Wang H, Wu S, Bao W. Preliminary Screening of Intestinal Barrier Genes Associated With Porcine Epidemic Diarrhea Virus Infection in Pigs. Rev Bras Zootecnia (2020) 49:e20200064. doi: 10.37496/rbz4920200064
40. Lefebvre O, Chenard MP, Masson R, Linares J, Dierich A, LeMeur M, et al. Gastric Mucosa Abnormalities and Tumorigenesis in Mice Lacking the pS2 Trefoil Protein. Science (1996) 274:259–62. doi: 10.1126/science.274.5285.259
41. Soutto M, Peng D, Katsha A, Chen Z, Piazuelo MB, Washington MK, et al. Activation of β-Catenin Signalling by TFF1 Loss Promotes Cell Proliferation and Gastric Tumorigenesis. Gut (2015) 64:1028–39. doi: 10.1136/gutjnl-2014-307191
42. Li S, Zhang Y, Guan Z, Li H, Ye M, Chen X. Sars-CoV-2 Triggers Inflammatory Responses and Cell Death Through Caspase-8 Activation. Signal Transduct Targeted Ther (2020) 5:1–10. doi: 10.1038/s41392-019-0089-y
43. Peterson LW, Artis D. Intestinal Epithelial Cells: Regulators of Barrier Function and Immune Homeostasis. Nat Rev Immunol (2014) 14:141–53. doi: 10.1038/nri3608
44. Zhang H, Liu Q, Su W, Wang J, Sun Y, Zhang J, et al. Genome-Wide Analysis of Differentially Expressed Genes and the Modulation of PEDV Infection in Vero E6 Cells. Microb Pathog (2018) 117:247–54. doi: 10.1016/j.micpath.2018.02.004
45. Hirata Y, Broquet AH, Menchén L, Kagnoff MF. Activation of Innate Immune Defense Mechanisms by Signaling Through RIG-I/IPS-1 in Intestinal Epithelial Cells. J Immunol (2007) 179:5425–32. doi: 10.4049/jimmunol.179.8.5425
46. Sheikh IA, Koley H, Chakrabarti MK, Hoque KM. The Epac1 Signaling Pathway Regulates Cl- Secretion Via Modulation of Apical KCNN4c Channels in Diarrhea. J Biol Chem (2013) 288:20404–15. doi: 10.1074/jbc.M113.467860
47. Wang S, Wu J, Wang F, Wang H, Wu Z, Wu S, et al. Expression Pattern Analysis of Antiviral Genes and Inflammatory Cytokines in PEDV-infected Porcine Intestinal Epithelial Cells. Front Vet Sci (2020) 7:75. doi: 10.3389/fvets.2020.00075
48. Pabst T, Mueller BU. Complexity of CEBPA Dysregulation in Human Acute Myeloid Leukemia. Clin Cancer Res (2009) 15(17):5303–7. doi: 10.1158/1078-0432.CCR-08-2941
49. Bhargavan B, Woollard SM, Kanmogne GD. Toll-Like Receptor-3 Mediates HIV-1 Transactivation Via Nfκb and JNK Pathways and Histone Acetylation, But Prolonged Activation Suppresses Tat and HIV-1 Replication. Cell Signal (2016) 28(2):7–22. doi: 10.1016/j.cellsig.2015.11.005
50. Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA Methylation Correlates With Differential Expression of Angiogenic Factors in Human Heart Failure. PloS One (2010) 5:e8564. doi: 10.1371/journal.pone.0008564
51. Greenberg M, Bourc'His D. The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat Rev Mol Cell Biol (2019) 20:590–607. doi: 10.1038/s41580-019-0159-6
52. Babyatsky MW, DeBeaumont M, Thim L, Podolsky DK. Oral Trefoil Peptides Protect Against Ethanol- and Indomethacin-Induced Gastric Injury in Rats. Gastroenterology (1996) 110:489–97. doi: 10.1053/gast.1996.v110.pm8566596
53. Chatagnon A, Ballestar E, Esteller M, Dante R. A Role for methyl-CpG Binding Domain Protein 2 in the Modulation of the Estrogen Response of pS2/TFF1 Gene. PloS One (2010) 5:e9665. doi: 10.1371/journal.pone.0009665
Keywords: TFF1, DNA methylation, C/EBPα, porcine epidemic diarrhea virus, intestinal epithelial barrier, coronavirus
Citation: Qu H, Zong Q, Wang H, Wu S, Cai D and Bao W (2022) C/EBPα Epigenetically Modulates TFF1 Expression via mC-6 Methylation in the Jejunum Inflammation Induced by a Porcine Coronavirus. Front. Immunol. 13:881289. doi: 10.3389/fimmu.2022.881289
Received: 23 March 2022; Accepted: 26 April 2022;
Published: 25 May 2022.
Edited by:
Wenkai Ren, South China Agricultural University, ChinaReviewed by:
Shu Zhu, Zhejiang University, ChinaCopyright © 2022 Qu, Zong, Wang, Wu, Cai and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbin Bao, d2JiYW9AeXp1LmVkdS5jbg==; Demin Cai, ZGVtaW5jYWlAeXp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.