- 1Stem Cell Bank, National Institute of Hematology and Blood Transfusion, Pham Van Bach, Cau Giay, Hanoi, Vietnam
- 2Department of Hematology, Hanoi Medical University, 1 Ton That Tung, Dong Da, Hanoi, Vietnam
- 3Key Laboratory of Enzyme and Protein Technology, VNU University of Science, Vietnam National University-Hanoi, 334 Nguyen Trai, Thanh Xuan, Hanoi, Vietnam
- 4Faculty of Biology, VNU University of Science, Vietnam National University-Hanoi, 334 Nguyen Trai, Thanh Xuan, Hanoi, Vietnam
- 5Department of Probability and Statistics, Faculty of Mathematics–Mechanics–Informatics, VNU University of Science, Vietnam National University, 334 Nguyen Trai, Thanh Xuan, Hanoi, Vietnam
The frequencies and diversities of human leukocyte antigen (HLA) alleles and haplotypes are representative of ethnicities. Matching HLA alleles is essential for many clinical applications, including blood transfusion, stem cell transplantation, and tissue/organ transplantation. To date, the information about the frequencies and distributions of HLA alleles and haplotypes among the Kinh Vietnamese population is limited because of the small sample size. In this study, more than 3,750 cord blood units from individuals belonging to the Kinh Vietnamese population were genotyped using PCR sequence-specific oligonucleotide (PCR-SSO) for HLA testing. The results of the study demonstrated that the most frequently occurring HLA-A, -B, -C, and -DRB1 alleles were A*11:01 (25%), A*24:02 (12.3%), A*02:01 (11.2); A*03:03 (8.95%), A*02:03 (7.81%), A*29:01 (7.03%); B*15:02 (15.1%), B*46:01 (10.7%), B*58:01 (7.65%), B*38:02 (7.29%); C*08:01 (17.2), C*07:02 (16.2%), C*01:02 (15.2), C*03:02 (8.3%), C*15:05 (6.13); DRB1*12:02 (31.0%), DRB1*09:01 (10.47%), DRB1*15:02 (7.54%); DRB1*07:01 (6.68%), DRB1*10:01 (6.63%), respectively, with the highest allele diversity level observed in locus B (93 alleles). The most frequent haplotypes of two-locus combinations of HLA-A–B, HLA-A–C, HLA-A–DRB1, HLA-B–C, HLA-B–DRB1, and HLA-C–DRB1 haplotypes were A*11:01–B*15:02 (7.63%), A*11:01–C*08:01 (7.98%), A*11:01–DRB1*12:02 (10.56%), B*15:02–C*08:01 (14.0%), B*15:02–DRB1*12:02 (10.47%), and C*08:01–DRB1*12:02 (11.38%), respectively. In addition, the most frequent haplotypes of three- and four-locus sets of HLA-A–B–C, HLA-A–B–DRB1, HLA-A–C–DRB1, HLA-B–C–DRB1, and HLA-A–B–C–DRB1 were A*11:01–B*15:02–C*08:01 (7.57%), A*11:01–B*15:02–DRB1*12:02 (5.39%), A*11:01–C*08:01–DRB1*12:02 (5.54%), B*15:02–C*08:01–DRB1*12:02 (10.21%), and A*11:01–B*15:02–C*08:01–DRB1*12:02 (5.45%), respectively. This study provides critical information on the frequencies and distributions of HLA alleles and haplotypes in the Kinh Vietnamese population, accounting for more than 85% of Vietnamese citizens. It paves the way to establish an umbilical cord blood bank for cord blood transplantation programs in Vietnam.
Introduction
Haematopoietic stem cell transplantation is an effective treatment option for various blood diseases. Donors and recipients with blood relations are most suited for stem cell engraftment because of their high compatibility, ensuring the optimal grafting effect (1, 2). However, in many cases, patients do not have close relationships with the donors. Therefore, the patients could not have donors with high compatibility with the human leukocyte antigen (HLA) as finding a donor with suitable HLA for grafting is challenging. Consequently, cord blood banks have been developed (3–6). Cord blood, which is readily available and easy to collect, has an extremely low risk for donors and has been used in numerous clinics (5–8).
The HLA system includes two classes: HLA-A, -B, and -C, which correspond to major histocompatibility complex (MHC) class I, and HLA-DRB1, -DQA1, -DQB1, and -DPB1, which correspond to MHC class II. MHC class I is present on the surface of all cell types and is recognised byT-CD8+ cells (9, 10). Incompatibility in genotypes of both HLA classes I and II between the donor and recipient causes organ rejection and graft failure in haematopoietic stem cell transplantation (9). The primary requirement for cord blood grafting is HLA compatibility of at least four of the six loci from MHC classes I and II (10, 11).
Cord blood was previously regarded as clinical waste; however, it has recently been recognised as a promising and effective source for treating various diseases, particularly haematopoietic disorder-related diseases (5–8). When applying stem cells for transplantation, finding compatible donors among the relations is problematic. Therefore, cord blood samples stored in blood banks are an excellent alternative. Graft rejection is a serious complication that may occur after transplantation. Therefore, storing cord blood is beneficial for children and their relatives in the future. Because of its abundant sources, using cord blood in clinics will help reduce the treatment cost and time (3–6). Cord blood stem cells can differentiate into various cells, including red blood cells, white blood cells, and platelets. Thus, cord blood has become very promising for clinical applications in treating multiple aggressive pathologies, such as anaemia, lymphoma, and leukaemia (1, 5–8, 12).
Therefore, we investigated and reported information on the frequency, diversity, and distribution of alleles and haplotypes in four HLA loci, including HLA-A, HLA-B, HLA-C, and HLA-DRB1, in 3,750 cord blood units of the Kinh Vietnamese population, to establish an umbilical cord blood bank at the National Institute of Haematology and Blood Transfusion of Vietnam.
Materials And Methods
Statistical Analysis
Population statistical analyses and diversity of HLA genotypes were calculated using Arlequin 3.5 software. In particular, the individual allele frequencies of each locus and the common haplotype frequencies of two, three, and four loci were determined using the expectation–maximisation (EM) algorithm. An exact test using a Markov chain algorithm was performed to test for deviation from Hardy–Weinberg equilibrium (13, 14).
Population and Sample Collection
A cross-sectional study was conducted on a cohort of 3,776 Kinh Vietnamese healthy pregnant women for cord blood samples. The samples were collected between May 2014 and December 2019. Cord blood donors mostly came from every province in northern and north-central Vietnam. Cord blood samples of ≥80 mL in volume, without blood cluster or bizarre colours, collected within 24 h of birth, mean corpuscular volume (MCV) of ≥95 fl, no abnormal haemoglobin confirmed by high-performance liquid chromatography (HPLC) analysis, number of nucleated cells ≥109cells/mL, negative for infectious pathogens including hepatitis C virus (HCV), hepatitis B virus (HBV), human immunodeficiency virus (HIV), cytomegalovirus (CMV), and bacteria were collected and stored at −196°C in liquid nitrogen tanks at the Umbilical Cord Blood Bank, National Institute of Haematology and Blood Transfusion, Vietnam.
HLA Genotyping
All cord blood samples were used for DNA extraction using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), from which 40–120 ng of genomic DNA was required for each locus typing. Medium-to-high resolution (second field) HLA typing (MHR-2F) of four loci, including A, B, C, and DRB1, was conducted by PCR sequence-specific oligonucleotide (PCR-SSO) using the LIFECODES HLA SSO kit (Luminex, Austin, TX, USA) on a Luminex 200 system, and analysed with Xponent 3.1 software connected to the IMGT (International ImMunoGeneTics) database version 3.43 library to provide us with the well-documented allele (WDA) (15–17). Blind external controls supplied by UK External Quality Assessment Services (UK EQAS) were used to confirm the accuracy of the four HLA loci identified. The procedure has been standardized under ISO15189:2012 criteria and is routinely used at Stem Cell Bank, Vietnam National Institute of Haematology and Blood Transfusion. Heterogeneous or homogeneous forms of the HLA gene were found. HLA typing was performed from June 2015 to December 2020. A few samples with ambiguous results in the 4-digit assignment were excluded from further investigation in this study. HLA typing was performed from June 2015 to December 2020.
Results
Cohort Haematological Characteristics
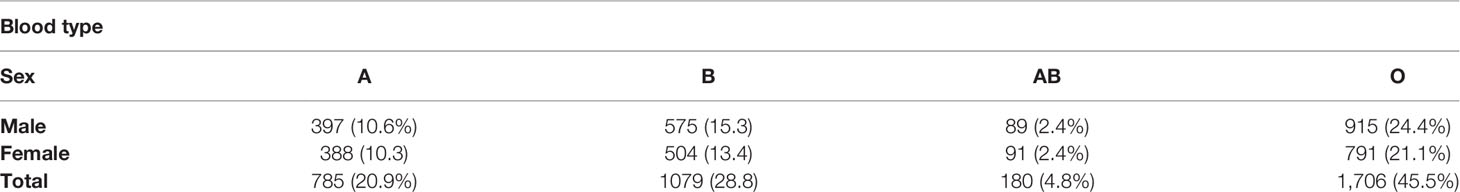
In this study, 3,776 cord blood samples were collected. The proportion of ambiguous results in the 4-digit assignment (“medium-to-high” resolution) was 0.68% (26/3,776) among the total 3,776 cord blood units. These 26 samples with ambiguous data were removed, and the remaining 3,750 samples were included in this research for further investigation. The characteristics of the study cohort are shown in Table 1. Among 3,750 donors, there were 1,976 men (52.7%) and 1,774 women (47.3%). The distribution of blood types, including A, B, AB, and O, was almost the same in men and women. The percentage of donors with O-type was the highest at 45.5%, the AB-type was the lowest at 4.8%, and the A-type and B-type contributed to 20.9% and 28.8%, respectively. In particular, the number of donors with the O-type was 2.2-, 1.6-, and 9.5-fold higher than the A-, B-, and AB-types, respectively (Table 1). This report agrees with previous reports on Kinh Vietnamese population (15, 18).
Individual Allele Frequencies
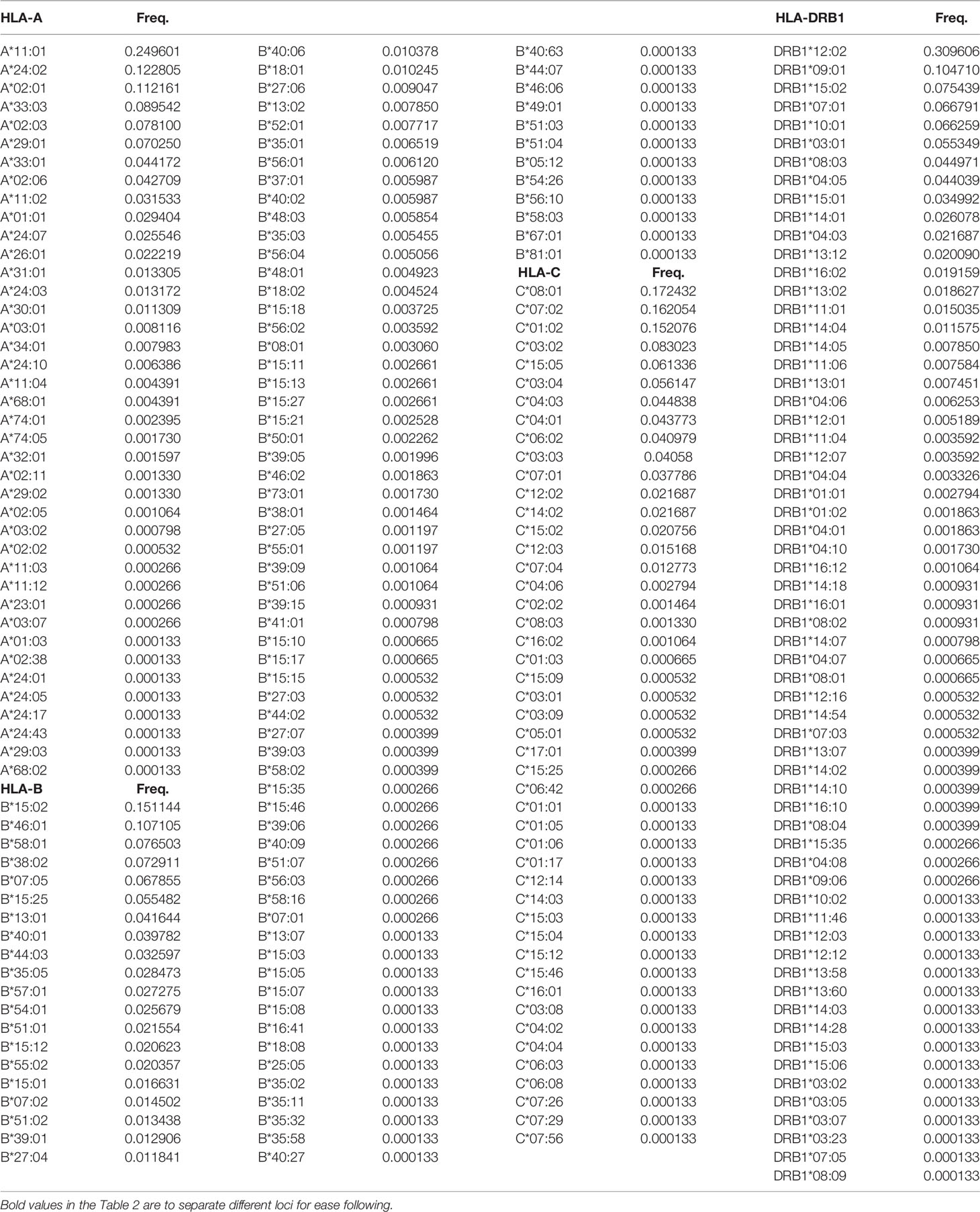
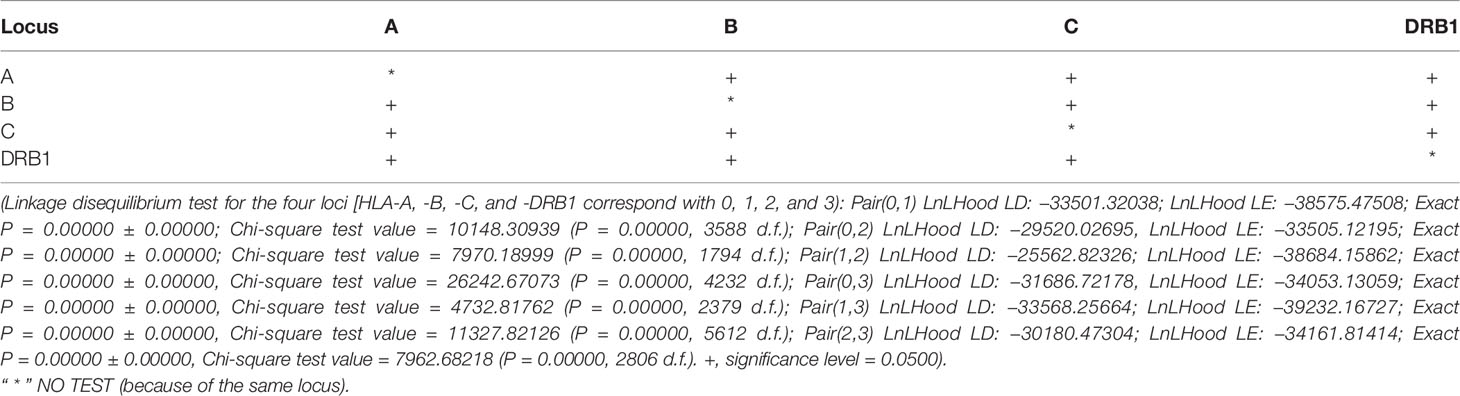
In this study, 40, 93, 47, and 62 individual alleles of HLA-A, -B, -C, and -DRB1, respectively, were found in the Kinh Vietnamese cohort (Table 2). This result indicated that locus B exhibited the highest polymorphism, with almost double the number of alleles than loci A, C, or DRB1. In the locus A (HLA-A), the most frequent alleles were A*11:01 (25%), A*24:02 (12.3%), A*02:01 (11.2), A*03:03 (8.95%), A*02:03 (7.81%), and A*29:01 (7.03%). In the locus B (HLA-B), the most frequent alleles were B*15:02 (15.1%), B*46:01 (10.7%), B*58:01 (7.65%), B*38:02 (7.29%), and B*15:25 (5.55%). In the locus C (HLA-C), the most frequent alleles were C*08:01 (17.2), C*07:02 (16.2%), C*01:02 (15.2), C*03:02 (8.3%), C*15:05 (6.13), and C*03:04 (5.61%). And, in the locus DRB1 (HLA-DRB1), the most frequent were DRB1*12:02 (31.0%), DRB1*09:01 (10.47%), DRB1*15:02 (7.54%); DRB1*07:01 (6.68%), DRB1*10:01 (6.63%), and DRB1*03:01 (5.53%). Of the 40 alleles at locus A, there were six alleles with frequencies of at least 5%, accounting for 72.3%, 15 alleles with frequencies of over 1%, accounting for 95.6, and 13 rare alleles with frequencies below 0.1%, accounting for 0.35%. Of the 93 alleles in locus B, only six alleles with frequencies of over 5% accounted for 53.1%, 21 alleles with frequencies greater than 1% accounted for 87.9%, and 42 rare alleles with frequencies below 0.1% accounted for 1.13%. In 47 alleles at locus C, there were six alleles with frequencies of over 5%, accounting for 68.7%, 16 alleles with frequencies of over 1%, accounting for 98.7%, and 26 rare alleles with frequencies below 0.1%, accounting for 0.63%. Among the 62 alleles of HLA-DRB1, there were six alleles with frequencies of over 5%, accounting for 67.8%, 16 alleles with frequencies greater than 1%, accounting for 93.4%, and 32 rare alleles with frequencies below 0.1%, accounting for 1.14% (Table 2). The allele HLA-DRB1*12:02 was the most common individual HLA allele present in four loci: HLA-A, -B, -C, and -DRB1, with a frequency of 31%. The Hardy–Weinberg equilibrium analysis results are presented in Table 3. The observed heterozygosity in all four loci of HLA-A, HLA-B, HLA-C, and HLA-DRB1 was significantly lower than expected by chance in this large cohort. With a small P value (<0.01) for each locus in this cohort, the observed heterozygosity was significantly less than expected. This suggests that the loci deviated from the Hardy–Weinberg equilibrium. An overview of the cumulative frequency at 50%, 75%, 90%, 95%, and 100% of the individual alleles of all four loci HLA-A, HLA-B, HLA-C, and HLA-DRB1 is presented in Figure 1, and their distributions are shown in Figure 2 (see also Supplemental Material 1). The polymorphic levels of HLA-A and HLA-C were almost the same and much lower than HLA-B, especially because of the number of frequent alleles (40 and 47 vs 93). The polymorphic HLA-DRB1 (62) was higher than HLA-A and HLA-C but lower than HLA-B. Other studies have also reported that HLA-A and HLA-C have a very similar amount of information, and HLA-B is the most polymorphic locus in not only three loci of HLA class I but in all six HLA loci for both HLA class I and HLA class II (19–23). Although the number of high-frequency (occurring frequency >5%) alleles in HLA-A, -B, -C, and -DRB1 were the same (six alleles for each), the number of rare alleles (occurring frequency <0.1%) in HLA-B was approximately 3-, 1.6-, and 1.3-fold higher than those in HLA-A, -C, and -DRB1, respectively. In addition, a test of linkage disequilibrium for all pairs of four loci (HLA-A, -B, -C, and -DRB1) was conducted, and the results are shown in Table 4. Significant linkage disequilibrium was observed for all pairs of the four loci (HLA-A, -B, -C, and -DRB1).
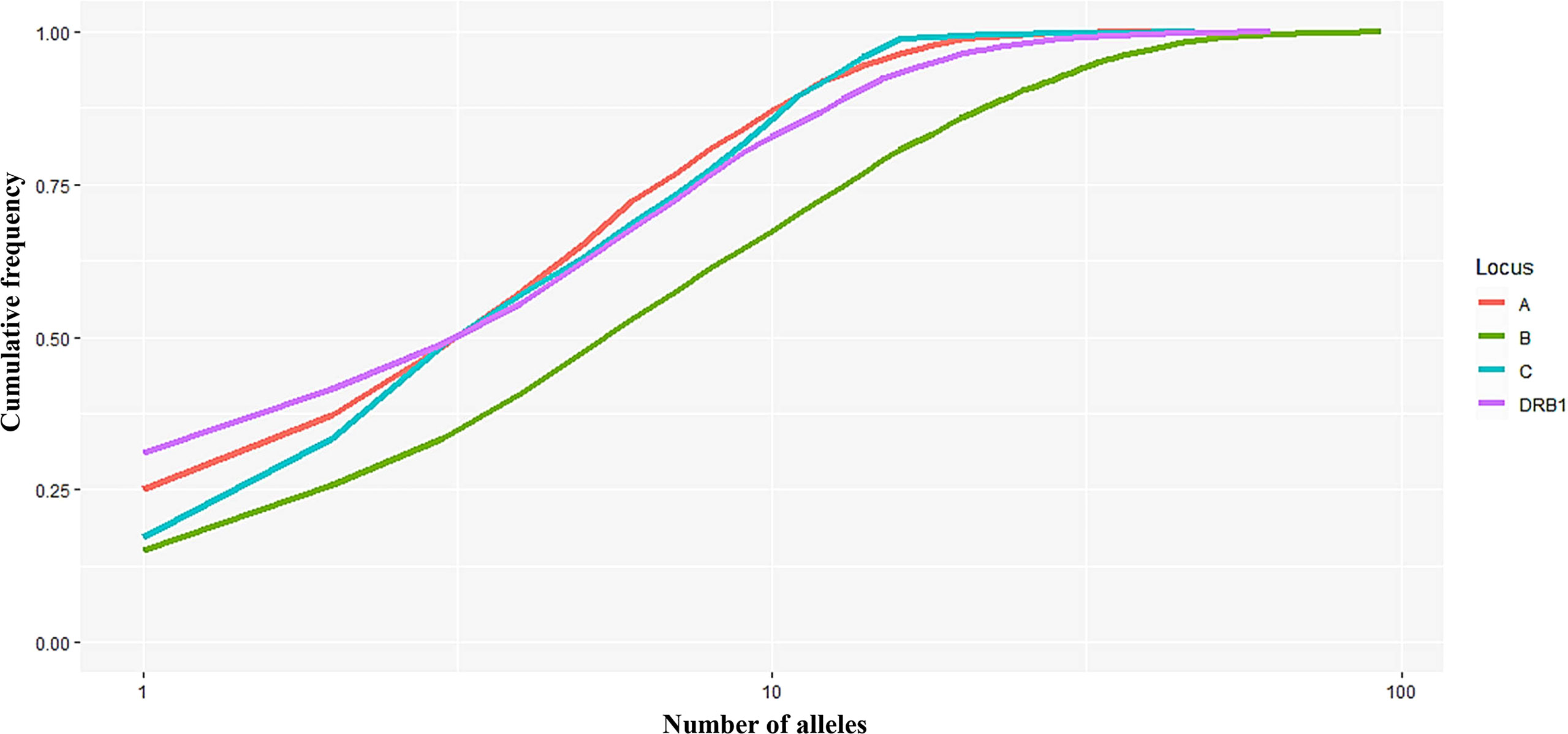
Figure 1 Curves showing the cumulative frequency of alleles for each HLA locus (A, B, C, and DRB1). The number of alleles (n) on the x-axis is expressed on a logarithmic scale.
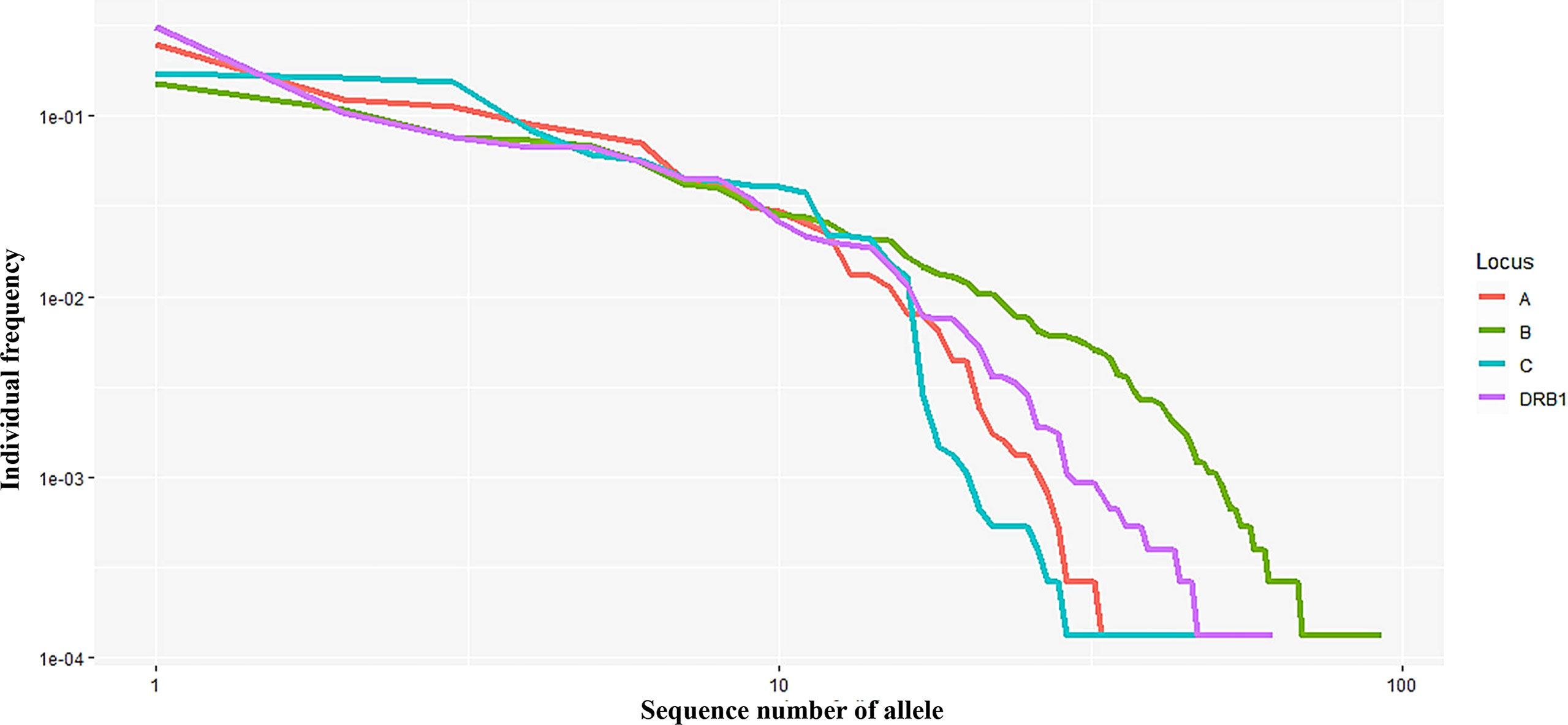
Figure 2 Curves showing the allele frequency of each HLA locus (A, B, C, and DRB1) in descending order.Both the x- and y-axes are expressed on a logarithmic scale.
Haplotype Frequencies
Haplotype frequencies of two, three, and four loci combinations (A–B, A–C, A–DRB1, B–C, B–DRB1, C–DRB1, A-B-C, A–B–DRB1, A–C–DRB1, B–C–DRB1, and A-B-C–DRB1) are shown in Supplemental Material 2. The combinations of alleles in the two loci created 300, 341, 439, 471, 592, and 695 haplotypes of B–C, A–C, C–DRB1, A–DRB1, A–B, and B–DRB1, respectively. In the combinations of three loci, 695, 976, 1,071, 1,633, and 1,743 haplotypes of A–B–C, B–C–DRB1, A–C–DRB1, A–B–DRB1, and A–B–DRB1, respectively, were found. A total of 2,087 haplotypes of four HLA loci (A-B-C–DRB1) were recorded. The number of haplotypes with a frequency of over 5% of A–B, A–C, B–C, A–DRB1, B–DRB1, and C–DRB1 was only 3, 4, 5, 2, 2, and 2, respectively. The number of haplotypes with a frequency of over 1% of A–B, A–C, B–C, A–DRB1, B–DRB1, and C–DRB1 was 19, 24, 20, 20, 18, and 19, respectively (see Supplemental Material 2).
The highest haplotype frequency of HLA-A–B was A*11:01–B*15:02 (7.63%), followed by A*02:01–B*46:01 (5.97%), and A*29:01–B07:05 (5.34%). The highest haplotype frequency of HLA-A–C was A*11:01–C*08:01 (7.98%), followed by A*02:01–C*01:02 (6.51%), A*29:01–C*15:05 (5.37%), and A*11:01–C*07:02 (5.05%). The highest haplotype frequency of HLA-B–C was B*15:02–C*08:01 (14.0%), followed by B*46:01–C*01:02 (10.23%), B*58:01–C*03:02 (7.49%), B*38:02–C*07:02 (7.18%), and B*07:05–C*15:05 (5.83%). The highest haplotype frequency of HLA-A–DRB1 was A*11:01–DRB1*12:02 (10.56%), followed by A*24:02–DRB1*12:02 (5.04%). The highest haplotype frequency of HLA-B–DRB1 was B*15:02–DRB1*12:02 (10.47%), followed by B*46:01–DRB1*09:01 (5.20%). The highest haplotype frequency of HLA-C–DRB1 was C*08:01–DRB1*12:02 (11.38%), followed by HLA-C*01:02–DRB1*09:01 (5.49%).
In addition, the frequencies of haplotypes with combinations of three and four loci were also estimated and are shown in Supplemental Material 2. The highest haplotype frequency of HLA-A–B–C was HLA-A*11:01–B*15:02–C*08:01 (7.57%), followed by A*02:01–B*46:01–C*01:02 (5.89%), and A*29:01–B*07:05–C*15:05 (5.27%). The most frequent alleles of HLA-A–B–DRB1, HLA-A–C–DRB1, HLA-B–C–DRB1, and HLA-A–B–C–DRB1 were A*11:01–B*15:02–DRB1*12:02 (5.39%), A*11:01–C*08:01–DRB1*12:02 (5.54%), B*15:02–C*08:01–DRB1*12:02 (10.21%), and A*11:01–B*15:02–C*08:01–DRB1*12:02 (5.45%), respectively. In combinations of three or four loci, the haplotype distributions varied widely, with many haplotypes having frequencies below 0.1%. In particular, among 885 haplotypes of HLA-A–B–C, 1,743 haplotypes of HLA-A–B–DRB1, 1,633 haplotypes of HLA-A–C–DRB1, 1,071 haplotypes of HLA-B–C–DRB1, and 2,087 haplotypes of HLA-A–B–C–DRB1, 746 haplotypes (18.95%), 1,583 haplotypes (37.49%), 1,098 haplotypes (27.47%), 936 haplotypes (22.93%), and 1,939 haplotypes (41.53%), respectively, with rare frequencies of less than 0.1% (see Supplemental Material 2). The cumulative frequency is shown in Figures 3, 4. The numbers of haplotypes covering the corresponding 50%, 75%, 90%, 95%, and 100% alleles are shown in Table 5. A total of 976, 1,633, 1,071, 1,743, and 2,087 haplotypes of HLA-A–B–C, HLA-A–B–DRB1, HLA-A–C–DRB1, HLA-B–C–DRB1, and HLA-A–B–C–DRB1, respectively, were recorded.
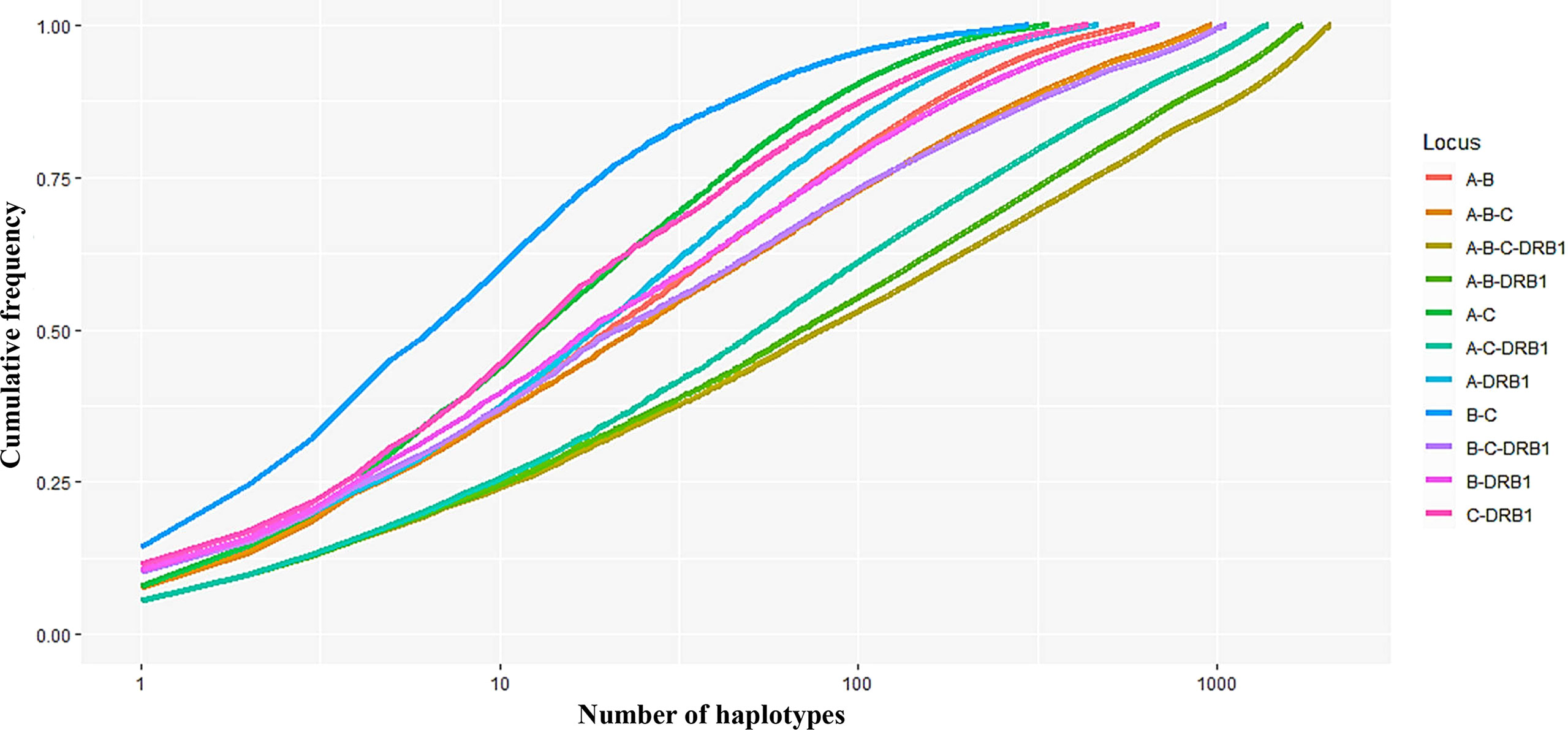
Figure 3 Curves showing the cumulative haplotype frequencies of each HLA locus-combination set (A–B, A–C, A–DRB1, B–C, B–DRB1, C–DRB1, A–B–C, A–B–DRB1, A–C–DRB1, B–C–DRB1, A–B–C–DRB1). The number of haplotypes (n) on the x-axis is expressed on a logarithmic scale.
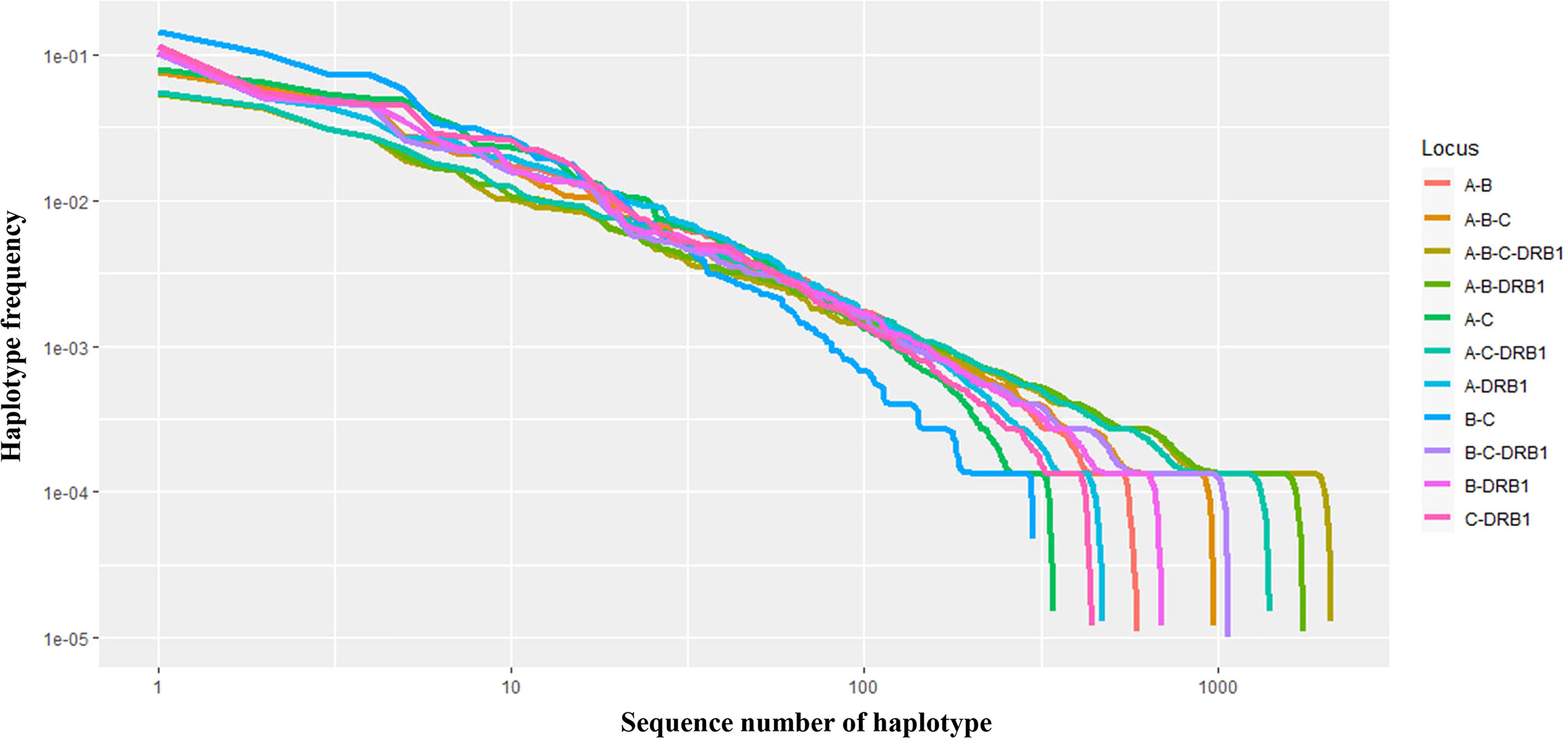
Figure 4 Curves showing the haplotype frequency of each set of loci (A–B, A–C, A–DRB1, B–C, B–DRB1, C–DRB1, A–B–C, A–B–DRB1, A–C–DRB1, B–C–DRB1, A–B–C–DRB1) in descending order. Both the x- and y-axes are expressed on a logarithmic scale.
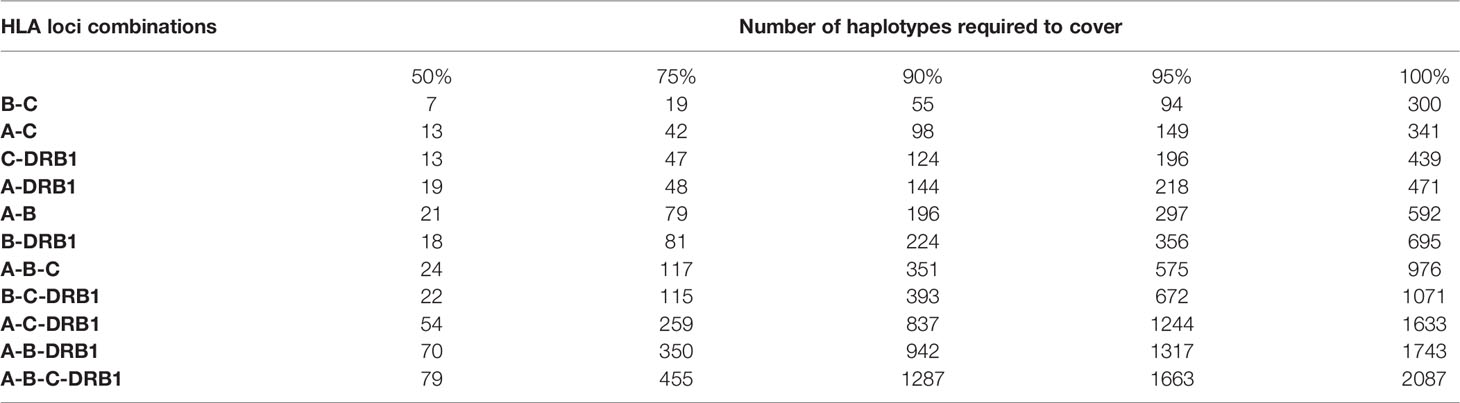
Table 5 Number of the most frequent haplotypes required to cover 50%, 75%, 90%, 95%, and 100% of the genes in each locus-combination set.
Discussion
Transplantation rejection is a common issue. Six HLA loci correspond to MHC class I and MHC class II, with three loci of HLA-A, -B, and -C relating to class I and HLA-DRB1, -DQA1, -DQB1, and -DPB1 related to class II. The level of HLA compatibility required varies depending on the transplantation technique used. In organ or stem cell transplantation, the compatibility of five or even six loci between the donor and the recipient is necessary; however, compatibility of at least four of the six loci is acceptable in the case of cord blood transplantation. Approximately 500 cord blood banks have been established in 97 countries worldwide (3–5).
The Kinh people were the most predominant ethnic community in Vietnam and accounted for more than 85% of the Vietnamese population (15, 18). However, information on the frequency and distribution of HLA alleles in the Kinh Vietnamese cord blood is yet to be thoroughly investigated. Several studies have examined HLA alleles and haplotypes in the Kinh Vietnamese, but the sample size was quite limited, with only around 100 donors (14, 15). The National Institute of Haematology and Blood Transfusion in Vietnam is one of the largest and most important storage facilities for umbilical cord blood in Vietnam. For future transplantation use, HLA genotypes of every cord blood sample should be tested.
In this study, cord blood units from the Kinh Vietnamese cohort were collected and stored in the cord blood bank at the National Institute of Haematology and Blood Transfusion, and PCR-SSO with Luminex Technology, a routine method in the institute, was used for HLA genotyping of these samples. Actually, there are several studies recorded that Luminex Technology for HLA-typing may cause a few percentage of ambiguous results as the Reviewer 2 have mentioned. However, many laboratories have used this method to identify HLA genotypes at 4-digit assignment (described as “medium-to-high” resolution) with good results (21, 24). Typically, Park et al. conducted genotyping of HLA-A, HLA-B and HLA-DRB1 of more than ten thousand (10,918) Koreans by using Luminex Technology and they confirmed that this platform was possible for genotyping most of the samples at the level of 4-digit assignment, there were only few cases with ambiguous results that must be reconfirmed by Sequence-Based Typing (SBT) from Abbot Molecular, USA (21). In this study, because the procedure has been standardized under ISO15189:2012 criteria and is routinely used at Stem Cell Bank, Vietnam National Institute of Haematology and Blood Transfusion, the proportion of ambiguous results in the 4-digit assignment (“medium-to-high” resolution) was very low, only 0.68% (26/3,776) among the total 3,776 cord blood units. As we concentrated on big data analysiswith a large sample size, therefore, for convenience, we did not include these 26 samples in this report, and the remaining 3,750 samples were analysed.
The findings in this study regarding frequencies and distributions of alleles and haplotypes in the Kinh Vietnamese cohort were similar to those in previous reports (15, 18), but there were a few differences. Hoa et al. (15) and Do et al. (18) found that the three most frequent alleles for HLA-A were A∗11:01, A∗24:02, and A∗33:03, with frequencies of 22.77%, 12.87%, 10.89%, and 22.9%, and 13.8% and 11.5%, respectively; however, in this study, the three most frequent alleles in HLA-A were A*11:01, A*24:02, and A*02:01 with frequencies of 25%, 12.3%, and 11.2%, respectively. This indicated that in the small sample size, A*02:01 was not listed as a dominant allele with a frequency of less than 5%, but in the larger sample size, A*02:01 was listed in the top three among the most frequent alleles. Similarly, regarding the allele frequencies at locus B, although the B∗40:01 allele was classified as a dominant allele with a frequency of more than 6% in Hoa et al. (6.2%) (15) and Do et al. (7.92%) (18), it only occurred with a frequency of 3.98% in this study. Furthermore, although the three most frequent alleles at locus C were the same in the studies by Hoa et al. (15), Do et al. (18), and the current study, the ranked positions were not. In particular, C∗07:02 (21.8%), C∗01:02 (13.4%), and C∗08:01 (12.9%) were listed as dominant HLA-C alleles in Do et al. (15); C*01:02 (16.5%), C*08:01 (15.6%), and C*07:02 (14.7%) in Hoa et al. (15); and C*08:01 (17.1%), C*07:02 (16.2%), and C*01:02 (15.2%) in the current study. In the HLA-DRB1 locus, the three most frequent alleles in Hoa et al. (15), Do et al. (18), and this study were the same, with the top three alleles being DRB1*12:02, DRB1∗09:01, and DRB1*15:02.
The diversity in individual allele frequencies and distributions resulted in differences in frequencies and distributions of HLA haplotypes of combination sets of two, three, or four loci. For instance, the most frequent haplotypes of the HLA-A–B set found in our study were A*11:01–B*15:02 (7.63%) and A*02:01–B*46:01 (5.97%), whereas those sets in Do et al. (18) were A∗29:01–B∗07:05 (6.93%), A∗33:03–B∗58:01 (6.43%), and A∗11:01–B∗15:02 (5.87%); and in Hoa et al. (15) were A*1101–B*15:02 (8.3%) and A*33:03–B*58:01 (5.6%). The existence of A*02:01–B*46:01 in the current study indicates a significant difference. The most frequent haplotypes in the three-locus HLA-A-B-C set by Do et al. (18) were A∗29:01–C∗15:05–B∗07:05, whereas in Hoa et al. (15) and our study were A*11:01–B*15:02–C*08:01. The most frequent haplotype in the three-locus HLA-A–B–DRB1 in Do et al. (18) was A*33:03–B∗58:01–DRB1∗03:01, in Hoa et al. (15) was A*29:01–B*07:05–DRB1*10:01, and in this work was A*11:01–B*15:02–DRB1*12:02.
Many previous studies have reported frequencies of HLA alleles in various populations (19–23). In Korean population (16–18), the most frequent alleles of HLA-A, -B, and -DRB1 were A*02:01, A*02:06, A*11:01, A*24:02, A*31:01, A*33:03 (HLA-A); B*15:01, B*44:03, B*51:01, B*54:01, B*58:01 (HLA-B); and DRB1*01:01, DRB1*04:05, DRB1*07:01, DRB1*08:03, DRB1*09:01, DRB1*13:02, DRB1*15:01 (HLA-DRB1). In the Saudi population, the most common alleles of HLA-A, -B, -C, and -DRB1 are HLA-A∗02:01, A∗24:02 (HLA-A); B∗51:01, B∗50:01 (HLA-B), C∗06:02, C∗07:02 (HLA-C), and DRB1∗07:01, DRB1∗03:01 (HLA-DRB1) (23). Nevertheless, A*24:02, A*33:03, A*02:03 (HLA-A); B*40:01, B*46:01, B*58:01, B*13:01, B*15:02, B*38:02 (HLA-B); C*07:02, C*01:02, C*08:01, C*03:04 (HLA-C); and DRB1*09:01, DRB1*12:02, DRB1*15:01 (HLA-DRB1) were the most common alleles among Han Chinese (22). The most frequent alleles of HLA-A, HLA-B, HLA-C, and HLA-DRB1 in the European American population were A*02:01, A*01:01, and A*03:01 (HLA-A); B*07:02, B*08:01 (HLA-B), C*07:01, and C*07:02 (HLA-C); and DRB1*15:01, DRB1*07:01, and DRB1*03:01 (HLA-DRB1) (25). Furthermore, the most frequent alleles of HLA-A, HLA-B, HLA-C, and HLA-DRB1 in Kinh Vietnamese population found in this study were A*11:01, A*24:02, A*02:01, A*03:03, A*02:03, A*29:01 (HLA-A); B*15:02, B*46:01, B*58:01, B*38:02, B*15:25 (HLA-B); C*08:01, C*07:02, C*01:02, C*03:02, C*15:05, C*03:04 (HLA-C); and DRB1*12:02, DRB1*09:01, DRB1*15:02; DRB1*07:01, DRB1*10:01, DRB1*03:01 (HLA-DRB1).
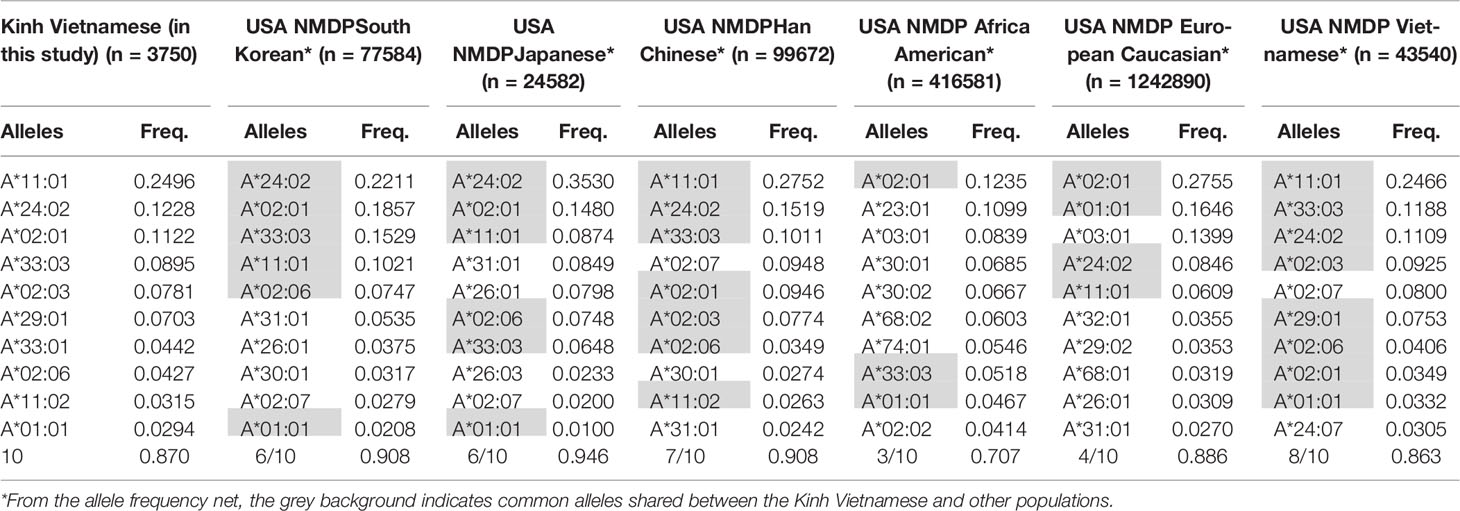
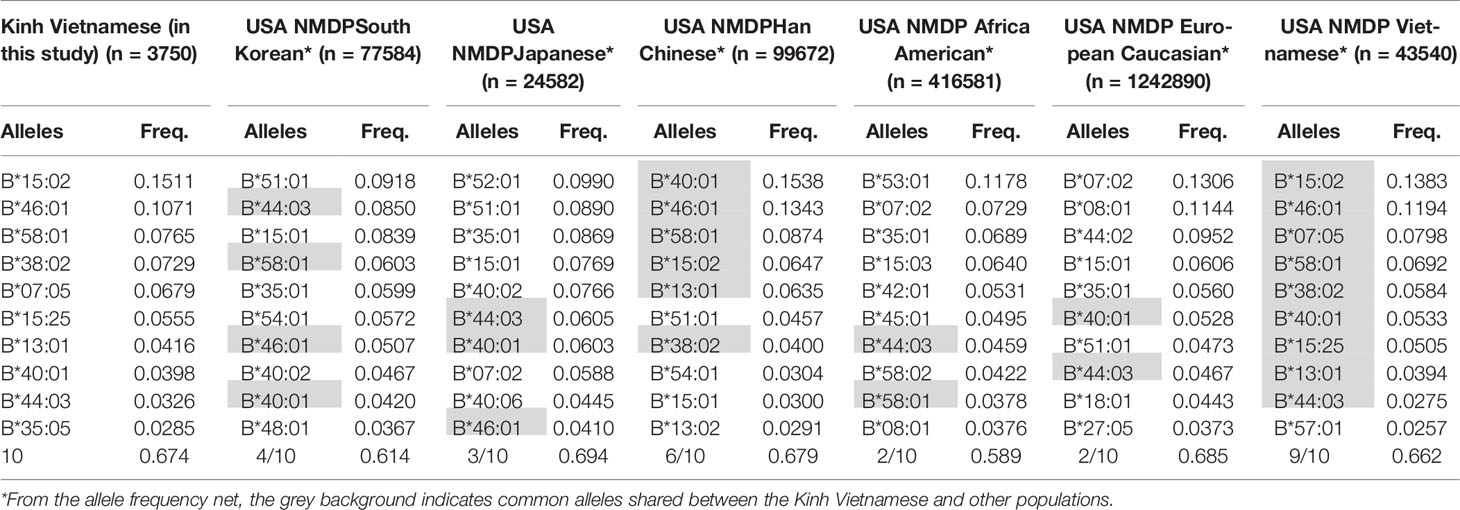
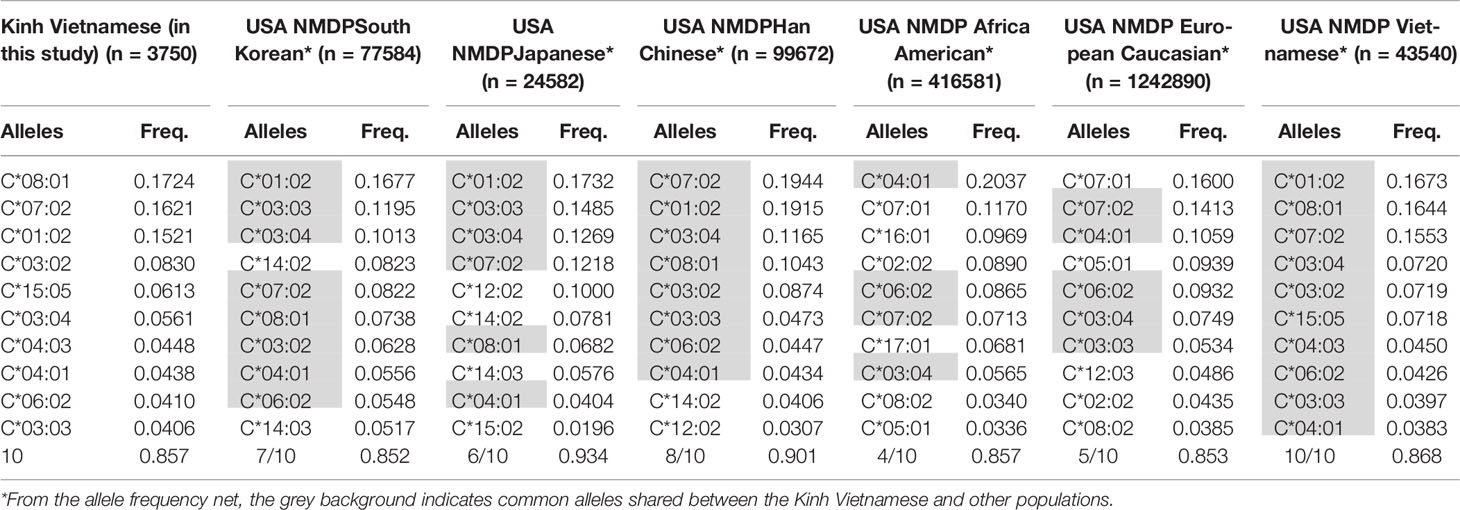
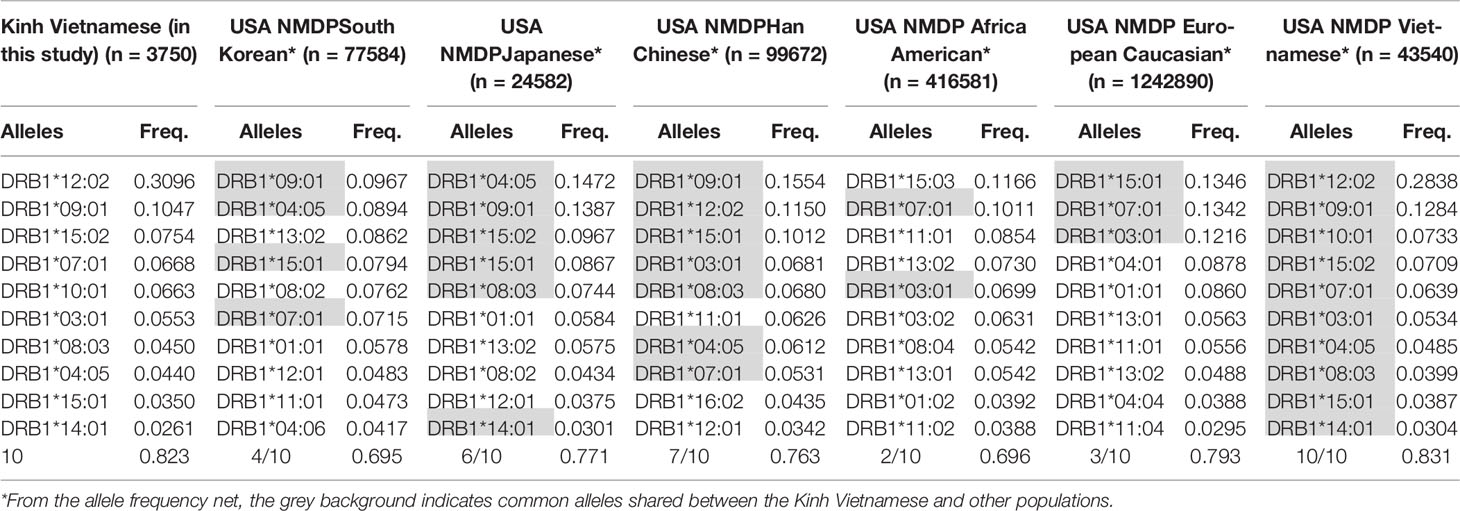
These data suggest that the distributions and the most frequent HLA alleles in Asian populations, such as Kinh Vietnamese, Han Chinese, and Koreans, were comparable; however, the level of similarity varied from locus to locus. HLA data were collected from a worldwide selection of populations in the Allelefriquencies.net database (http://www.allelefrequencies.net) (26) and used for comparisons. In particular, the distributions of the 10 most frequent alleles of HLA-A, -B, -C, and -DRB1 in Kinh Vietnamese in this study (n = 3,750), USA National Marrow Donor Program (NMDP) Vietnamese (n = 43,540), South Korean (n = 77,584), Japanese (n = 24,582), Han Chinese (n = 99,672), African American (n = 416,581), and European Caucasian (n = 1,242,890) were compared and are presented in Tables 6–9, respectively (26, 27). Although the ranking positions of the 10 most frequent alleles are somewhat different, the distributions of these alleles in the Kinh Vietnamese (observed in this study) and the USA NMDP Vietnamese recorded by ABC are very similar (Tables 6–9). In particular, eight of 10 HLA-A alleles, nine of 10 HLA-B alleles, 10 of 10 HLA-C alleles, and 10 of 10 HLA-DRB1 alleles among the 10 most frequent alleles in the USA NMDP Vietnamese were the same as those in Kinh Vietnamese. Furthermore, six of 10 HLA-A alleles, four of 10 HLA-B alleles, seven of 10 HLA-C alleles, and four of 10 HLA-DRB1 alleles in the 10 most frequent alleles in USA NMDP South Korea; six of 10 HLA-A alleles, three of 10 HLA-B alleles, six of 10 HLA-C alleles, and six of 10 HLA-DRB1 alleles in the 10 most frequent alleles in USA NMDP Japanese; seven of 10 HLA-A alleles, six of 10 HLA-B alleles, eight of 10 HLA-C alleles, and seven of 10 HLA-DRB1 alleles in the 10 most frequent alleles in USA NMDP Han Chinese; three of 10 HLA-A alleles, two of 10 HLA-B alleles, four of 10 HLA-C alleles, and two of 10 HLA-DRB1 alleles in the 10 most frequent alleles in USA NMDP Africa American; and four of 10 HLA-A alleles, two of 10 HLA-B alleles, five of 10 HLA-C alleles, and three of 10 HLA-DRB1 alleles in 10 highest frequent alleles in USA NMDP European Caucasian are the same as those in Kinh Vietnamese (Tables 5–8). These data demonstrated that, except for the USA NMDP Vietnamese, among these populations, the similarity of high-frequency HLA alleles in Kinh Vietnamese and Han Chinese was the highest, followed by South Korean, Japanese, European Caucasian, and African American. These data also indicated that the distributions and the most frequent HLA alleles in the Kinh Vietnamese population differed from those in European American and African American populations. These results suggest that Asian populations have different HLA allele frequencies than the American and European populations. Moreover, the diversity in the frequencies of individual alleles of HLA loci creates large differences in frequencies and distributions of two-, three-, or four-loci haplotypes among various populations (19–27). This suggests that transplantations between different populations are quite challenging, and therefore, establishing a bank to store cord blood samples for transplantation programs is important.
Institutional Review Board Statement
All participants provided informed consent for inclusion before participating in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Hanoi Medical University, No. 78/HDDDDHYHN (for DDNCYSH, approval date: 30 May 2017).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further enquiries can be directed to the corresponding author.
Author Contributions
TNQ and NDT: hypotheses and research questions. NDT, TNQ,NBK, BQK, and NTVA: research proposal. PDT, NBK, CVS, and TTTA: data analysis. NDT, TNQ, PDT, CVS, and NTVA: Data discussion. NDT, TNQ, TTTA, NTVA, and PDT: manuscript writing. NDT, PDT: Providing figures. NDT, NTVA, BQK,NBK, PDT: final paper review.
Funding
This study was supported by the National Institute of Haematology and Blood Transfusion in Vietnam and funded by National Research Grant code DTDL.CN-63/19 from the Ministry of Science and Technology, Vietnam.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.875283/full#supplementary-material
References
1. Berglund S, Magalhaes I, Gaballa A, Vanherberghen B, Uhlin M. Advances in Umbilical Cord Blood Cell Therapy: The Present and the Future. Expert Opin Biol Ther (2017) 17(6):691–9. doi: 10.1080/14712598.2017.1316713
2. Ballen KK, Gluckman E, Broxmeyer HE. Umbilical Cord Blood Transplantation: The First 25 Years and Beyond. Blood (2013) 122(4):491–8. doi: 10.1182/blood-2013-02-453175
3. Shearer WT, Lubin BH, Cairo MS, Notarangelo LD. Section on Hematology/Oncology; Section on Allergy and Immunology. Cord Blood Banking for Potential Future Transplantation. Pediatrics (2017) 140(5):e20172695. doi: 10.1542/peds.2017-2695
4. Fasouliotis SJ, Schenker JG. Human Umbilical Cord Blood Banking and Transplantation: A State of the Art. Eur J Obstet Gynecol Reprod Biol (2000) 90(1):13–25. doi: 10.1016/S0301-2115(99)00214-6
5. Mayani H, Wagner JE, Broxmeyer HE. Cord Blood Research, Banking, and Transplantation: Achievements, Challenges, and Perspectives. Bone Marrow Transplant (2020) 55(1):48–61. doi: 10.1038/s41409-019-0546-9
6. Ballen KK, Lazarus H. Cord Blood Transplant for Acute Myeloid Leukaemia. Br J Haematol (2016) 173(1):25–36. doi: 10.1111/bjh.13926
7. Kekre N, Antin JH. Cord Blood Versus Haploidentical Stem Cell Transplantation for Hematological Malignancies. Semin Hematol (2016) 53(2):98–102. doi: 10.1053/j.seminhematol.2016.01.007
8. Horwitz ME, Stiff PJ, Cutler C, Brunstein C, Hanna R, Maziarz RT, et al. Omidubicel vs Standard Myeloablative Umbilical Cord Blood Transplantation: Results of a Phase 3 Randomized Study. Blood (2021) 138(16):1429–40. doi: 10.1182/blood.2021011719
9. Dehn J, Spellman S, Hurley CK, Shaw BE, Barker JN, Burns LJ, et al. Selection of Unrelated Donors and Cord Blood Units for Hematopoietic Cell Transplantation: Guidelines From the NMDP/CIBMTR. Blood (2019) 134(12):924–34. doi: 10.1182/blood.2019001212
10. Fan WL, Shiao MS, Hui RC, Su SC, Wang CW, Chang YC, et al. HLA Association With Drug-Induced Adverse Reactions. J Immunol Res (2017) 2017:3186328. doi: 10.1155/2017/3186328
11. Wieczorek M, Abualrous ET, Sticht J, Álvaro-Benito M, Stolzenberg S, Noé F, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol (2017) 8:292. doi: 10.3389/fimmu.2017.00292
12. Sun JM, Kurtzberg J. Cord Blood for Brain Injury. Cytotherapy (2015) 17(6):775–85. doi: 10.1016/j.jcyt.2015.03.004
13. Excoffier L, Laval G, Schneider S. Arlequin (Version 3.0): An Integrated Software Package for Population Genetics Data Analysis. Evol Bioinform Online (2007) 1:47–50. doi: 10.1177/117693430500100003
14. Niu T. Algorithms for Inferring Haplotypes. Genet Epidemiol (2004) 27:334–47. doi: 10.1002/gepi.20024
15. Hoa BK, Hang NT, Kashiwase K, Ohashi J, Lien LT, Horie T, et al. HLA-A, -B, -C, -DRB1 and -DQB1 Alleles and Haplotypes in the Kinh Population in Vietnam. Tissue Antigens (2008) 71(2):127–34. doi: 10.1111/j.1399-0039.2007.00982.x
16. Testi M, Andreani M. Luminex-Based Methods in High-Resolution HLA Typing. Methods Mol Biol (2015) 1310:231–45. doi: 10.1007/978-1-4939-2690-9_19
17. Guarene M, Badulli C, Cremaschi AL, Sbarsi I, Cacciatore R, Tinelli C, et al. Luminex® xMAP® Technology Is an Effective Strategy for High-Definition Human Leukocyte Antigen Typing of Cord Blood Units Prior to Listing. Int J Artif Organs (2018) 41(5):284–8. doi: 10.1177/0391398818762356
18. Do MD, Le LGH, Nguyen VT, Dang TN, Nguyen NH, Vu HA, et al. High-Resolution HLA Typing of HLA-A, -B, -C, -DRB1, and -DQB1 in Kinh Vietnamese by Using Next-Generation Sequencing. Front Genet (2020) 11:383. doi: 10.3389/fgene.2020.00383
19. Huh JY, Yi DY, Eo SH, Cho H, Park MH, Kang MS. HLA-A, -B and -DRB1 Polymorphism in Koreans Defined by Sequence-Based Typing of 4128 Cord Blood Units. Int J Immunogenet (2013) 40(6):515–23. doi: 10.1111/iji.12067
20. Baek IC, Choi EJ, Shin DH, Kim HJ, Choi H, Kim TG. Allele and Haplotype Frequencies of Human Leukocyte Antigen-A, -B, -C, -DRB1, -DRB3/4/5, -DQA1, -DQB1, -DPA1, and -DPB1 by Next Generation Sequencing-Based Typing in Koreans in South Korea. PLos One (2021) 16(6):e0253619. doi: 10.1371/journal.pone.0253619
21. Park H, Lee YJ, Song EY, Park MH. HLA-A, HLA-B and HLA-DRB1 Allele and Haplotype Frequencies of 10 918 Koreans From Bone Marrow Donor Registry in Korea. Int J Immunogenet (2016) 43(5):287–96. doi: 10.1111/iji.12288
22. Trachtenberg E, Vinson M, Hayes E, Hsu YM, Houtchens K, Erlich H, et al. HLA Class I (A, B, C) and Class II (DRB1, DQA1, DQB1, DPB1) Alleles and Haplotypes in the Han From Southern China. Tissue Antigens (2007) 70(6):455–63. doi: 10.1111/j.1399-0039.2007.00932.x
23. Jawdat D, Uyar FA, Alaskar A, Müller CR, Hajeer A. HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 Allele and Haplotype Frequencies of 28,927 Saudi Stem Cell Donors Typed by Next-Generation Sequencing. Front Immunol (2020) 11:544768. doi: 10.3389/fimmu.2020.544768
24. Roh EY, In JW, Shin S, Yoon JH, Park KU, Song EY. Performance of LIFECODES HLA-DQB1 Typing Kit Using Luminex Platform in Koreans. Ann Lab Med (2015) 35(1):123–7. doi: 10.3343/alm.2015.35.1.123
25. Creary LE, Gangavarapu S, Mallempati KC, Montero-Martín G, Caillier SJ, Santaniello A, et al. Next-Generation Sequencing Reveals New Information About HLA Allele and Haplotype Diversity in a Large European American Population. Hum Immunol (2019) 80(10):807–22. doi: 10.1016/j.humimm.2019.07.275
26. Available at: http://allelefrequencies.net/hla6006a.asp?hla_population=3220.
Keywords: human leukocyte antigen (HLA), allele, haplotype, cord blood, Kinh Vietnamese population
Citation: Que TN, Khanh NB, Khanh BQ, Van Son C, Van Anh NT, Anh TTT, Tung PD and Thang ND (2022) Allele and Haplotype Frequencies of HLA-A, -B, -C, and -DRB1 Genes in 3,750 Cord Blood Units From a Kinh Vietnamese Population. Front. Immunol. 13:875283. doi: 10.3389/fimmu.2022.875283
Received: 15 February 2022; Accepted: 30 May 2022;
Published: 29 June 2022.
Edited by:
Pierre-Antoine GOURRAUD, Université de Nantes, FranceReviewed by:
Abeer Madbouly, National Marrow Donor Program, United StatesMartin Maiers, National Marrow Donor Program, United States
Copyright © 2022 Que, Khanh, Khanh, Van Son, Van Anh, Anh, Tung and Thang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nguyen Dinh Thang, bmR0aGFuZ0BodXMuZWR1LnZu
 Tran Ngoc Que
Tran Ngoc Que Nguyen Ba Khanh
Nguyen Ba Khanh Bach Quoc Khanh1,2
Bach Quoc Khanh1,2 Chu Van Son
Chu Van Son Nguyen Dinh Thang
Nguyen Dinh Thang