
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 30 June 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.872896
This article is part of the Research TopicEffects of Host–Microbiota Cross-Talk on the Immunity of Aquatic AnimalsView all 16 articles
 Huijie Chen1,2
Huijie Chen1,2 Yuanli Zhao1
Yuanli Zhao1 Kuangxin Chen1,3
Kuangxin Chen1,3 Yulai Wei1,3
Yulai Wei1,3 Hongrui Luo1
Hongrui Luo1 Yongming Li1
Yongming Li1 Fei Liu1,3
Fei Liu1,3 Zuoyan Zhu1
Zuoyan Zhu1 Wei Hu1
Wei Hu1 Daji Luo1,2,3*
Daji Luo1,2,3*Various bacterial diseases have caused great economic losses to the high-density and intensive aquaculture industry; however, the pathogenic mechanism underlying the large-scale challenged to caused by many bacteria remain unclear, making the prevention and treatment of these diseases difficult. In the present study, we isolated a bacterial strain from Cyprinus carpio having a typical bacterial disease and named it Cc2021. Through subsequent morphological observations, a regression challenge, biochemical identification, and 16S rRNA gene sequence analysis, we determined Cc2021 to be Plesiomonas shigelloides. Subsequently, we comprehensively investigated the pathogenicity of P. shigelloides in C. carpio through a regression challenge and assessed the underlying the pathogenic mechanism. Mortality results revealed that P. shigelloides is highly pathogenic and infects various tissues throughout the body, resulting in edema of the liver, spleen, and body and head kidneys. Histopathological analysis revealed obvious inflammation, bleeding, and necrosis in the intestine, spleen, and head kidney. The body’s immune tissues actively produce complement C3, superoxide dismutase, and lysozyme after a challenge to resist bacterial invasion. With regard to the underlying pathogenesis of P. shigelloides, comparative transcriptome analysis revealed 876 upregulated genes and 828 downregulated genes in the intestine of C. carpio after the challenge. Analysis of differentially expressed unigenes revealed the involvement of major immune pathways, particularly the TNF signaling pathway, interleukin (IL)-17 signaling pathway, and Toll-like receptor signaling pathway. The present study provides new valuable information on the immune system and defense mechanisms of P. shigelloides.
1. P. shigelloides was isolated and identified from C. carpio.
2. P. shigelloides challenge led to tissue edema, necrosis, and death in C. carpio.
3. The TNF signaling pathway in the intestinal tract of C. carpio was induced by P. shigelloides challenge.
Cyprinus carpio is a valuable and unique fish resource that belongs to the order Cypriniformes and family Cyprinidae (1). As a dominant cyprinid species, C. carpio has been reported to account for almost 10% of the annual freshwater aquaculture production (1, 2). However, with the deterioration of the natural environment and the increase in breeding density, the breeding environment of C. carpio has been destroyed. In particular, the increasing number of bacterial diseases has had a significant impact on the breeding industry of C. carpio and caused huge economic losses (3, 4). The prevention and control of bacterial diseases are areas that need to be urgently addressed for the healthy development of the C. carpio industry (5). Plesiomonas shigelloides, a member of the Vibrionaceae family, is found in the intestinal tracts of fishes, mammals, and humans (6–8). P. shigelloides is widely distributed in the aquatic environment and is considered a new pathogen that causes digestive tract inflammation in many aquatic animals (8). P. shigelloides can infect Trionyx sinensis, Takifugu obscurus, and other aquatic animals alone or in combination; this has resulted in great economic losses to the aquatic aquaculture industry and posed a certain safety hazard to consumers (9, 10). In recent years, P. shigelloides has been isolated and identified from several aquatic animals, such as Percocypris pingi, Lota, Mylopharyngodon piceus, and Carassius auratus (11–13); however, the pathogenicity of P. shigelloides in C. carpio remains unknown.
In humans, P. shigelloides has been associated with secondary challenges in immunocompromised states, including malignancy, blood disorders, and hepatobiliary diseases (14, 15). In addition to infecting humans, P. shigelloides can be pathogenic to aquatic animals, such as Percocypris pingi (Tchang), Huso huso♀ × Acipenser ruthenus♂, Takifugu obscurus, Ctenopharyngodon idella, and Pelteobagrus fulvidraco; it has been reported to be infect these species and even cause death (16, 17). The survival of aquatic animals as natural hosts are not affected under normal circumstances (18). The pathogenic bacteria obtained from Eriocheir sinensis H. milne Edwards were not found to be the cause of disease in a previous study (19). Moreover, symptoms were found to differ between cold-water fish and warm-water fish (20). Therefore, it can be said that P. shigelloides is an opportunistic pathogen that acts once the environment is suitable and has a high fatality rate after onset. After Ictalurus punctatus challenge, the mortality rate was found to be up to 60%–100% in a previous study (21). The median lethal dose for Nile tilapia was 1. 425 × 108 CFU/mL, while that for sturgeon was 1.0 × 105.8 CFU/mL (21, 22). P. shigelloides is increasingly causing damages to aquaculture; however, its pathogenic mechanism remains unknown. At present, there are many studies on the immunity of carp after a bacterial challenge; the commonly used effective methods are transcriptome sequencing and gene function study. However, few studies have assessed the similarities and differences in immune responses in C. carpio after a bacterial challenge based on transcriptome analysis.
Comparative transcriptome analysis has been widely used in immunological studies of different fishes, providing reliable data on immune mechanisms (23). The potential mechanism of antibacterial immunity and the candidate genes has been identified using comparative transcriptome analysis in Epinephelus lanceolatus (24). Transcriptome analysis of intestinal epithelial cells of I. punctatus has revealed that the key to the invasion of Edwardsiella ictaluri is the actin bone in the intestinal barrier (25). In addition to adult tissues and organs, the transcriptome has been used to assess the immune response of fish embryos and primary cells. The innate host immune response of zebrafish embryos infected with Salmonella Typhi has been studied by RNA-seq and marker-based sequencing (26). RNA-seq has been used to study the expression changes in the transcriptome before and after Poly (I:C) stimulation of rainbow trout embryo cells (26). Several differentially expressed genes (DEGs) have been found to be involved in multiple physiological systems, including endocrinal, reproductive, and immune systems. In a previous study, after the intraperitoneal injection of P. shigelloides solution, transcriptome sequencing was performed using the intestinal tissue samples of sturgeon. The results revealed that DEGs in the intestinal tissues were significantly enriched in biological processes, including the immune system process, immune effect process, and stimulus response (13). In the present study, a bacterium referred to as Cc2021 was isolated and purified as the key pathogenic bacterium from infected C. carpio. The pathogenic bacterium causing septicemia was identified through morphological observation, a regression challenge, biochemical identification, and 16S rRNA and gyrB gene sequence analysis. The mortality rate and effects of Cc2021 in C. carpio were analyzed on the basis of the tissue load, tissue edema, and histopathological findings. Comparative transcriptome analysis was performed to assess the immune response mechanism following Cc2021 challenge in both normal intestinal tissues and challenged intestinal tissues. The present study provides reference data regarding the pathogenic mechanism of P. shigelloides in C. carpio.
All experiments were conducted in accordance with the guidelines and regulations outlined by the Management and Use of Laboratory Animals of Hubei Province and complied with China’s existing laws and regulations for biological research. The present study did not involve any endangered or protected species.
The experimental fish C. carpio (mean weight, 50 – 100 g) was obtained from the Guanqiao Experimental Station (Wuhan City, Hubei Province, China) and acclimatized at 25 ± 1°C. All the fish were fed twice daily with commercial pellets for 2 weeks before the experiments.
Naturally infected C. carpio was obtained from the temperature-controlled Fish Culture Center of the Institute of Hydrobiology, Chinese Academy of Sciences (Wuhan City, Hubei Province, China). The body surface of C. carpio was disinfected with 70% alcohol and placed in a biosafety container for dissection. A sterilized inoculum ring was inserted deep into the lesion of the fish, the needle was gently rotated for sampling, and streaked inoculation was performed on the surface of sterile brain heart infusion (BHI) broth, followed by incubation at a constant temperature of 28°C for 48 h. A single dominant colony in the plate was selected and inoculated on another BHI plate thrice, and the purified pathogenic strain was obtained after pure culture. Finally, one part of the purified strain was used for subsequent experiments and the other part was stored in 50% glycerol at −80°C for future use.
Bacteria were analyzed using bacterial biochemical microidentification tubes and test papers (Hangzhou Microbial Reagent. Co., Ltd. Hangzhou, China). A single colony was selected, inoculated in a microbiological reaction tube, and cultured at 28°C for 48 h. Following this, the biochemical indicators were tested (Table 1). The bacterial strains were preliminarily identified according to Bergey’s Manual of Systematic Bacteriology and Common Bacterial System Identification Manual (27).
The 16S rRNA of the pathogenic bacterial strain was amplified using the 16S rRNA primers 27F (AGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT). The PCR cycling conditions were as follows: initial denaturation at 95°C for 5 min; followed by 35 cycles at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. The two primers used for PCR amplification of the gyrB gene sequence were as follows: UP1 (forward): 5′-GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYGA-3′ and UP2r (reverse): 5′-AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT-3′. The PCR reaction conditions were as follows: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, renaturation at 60°C for 1 min, and extension at 72°C for 1 min. After 30 cycles, incubation was performed at 72°C for 7 min. The reaction process was ended at 16°C, followed by preservation at 4°C. Amplification products were placed in 1% agarose gel for electrophoresis. Positive amplification products were recovered and sequenced by TSINGKE Biotechnology Co., Ltd (Wuhan, Hubei, China). The obtained 16S rRNA and gyrB gene sequences were analyzed by the NCBI BLAST retrieval system for sequence homology, and multiple alignments were made with the sequences of strains with high sequence similarity obtained from the GenBank database using ClustalX software. The neighbor-joining method was used to construct phylogenetic trees using Molecular Evolutionary Genetics Analysis (MEGA) 7.0 software. A bootstrap test was performed with 1000 repetitions.
The pathogenic bacterial strain was inoculated in BHI broth and cultured at 28°C under 200 rpm for 48 h. To determine the 50% lethal concentration of the pathogenic bacterial strain, four groups of C. carpio (n = 30) were intraperitoneally injected 100 μL of the bacterial strain (diluted in physiological saline) at concentrations of 1.0 × 107 CFU/mL, 5.0 × 107 CFU/mL, 1.0 × 108 CFU/mL, and 5.0 ×108 CFU/mL, while the control group was injected with sterile physiological saline (0.65% NaCl). Following this, mortality was monitored for the next 7 days.
After infecting the experimental fish (n = 100) with the measured half lethal concentration, the surviving fish were sampled from 0 to 7 days after the challenge. The blood, liver, head kidney, spleen, trunk kidney, and intestine (hindgut) of the challenged fish were randomly collected for the subsequent experiment (n = 5). Fish tissues injected with the same dose of saline were used as controls.
In total, 0.5 g of ground viscera (liver, head kidney, spleen, and intestine) was weighed and homogenized in BHI medium under sterile conditions using a Dounce tissue grinder (Sigma, USA). Serial dilutions (10−4, 10−5, and 10−6) were performed, 100 μL of the diluted sample was plated on BHI plates and incubated at 28°C for 48 h. Finally, the colony morphology was identified when the colonies were counted. CFU/g (log10) was calculated taking the dilution and total weight of the fish into consideration.
The whole target tissue was gathered and rinsed, and the immediate weight was recorded as the wet weight. Subsequently, the tissues were air dried for 2 days at 60°C, and their weight was recorded as the dry weight. The wet/dry weight ratio for each fish was calculated for evaluating tissue edema.
Tissue samples from the head kidney, spleen, liver, and intestine were dissected and immediately fixed in 10% neutral buffered formalin at 4°C, dehydrated in a graded ethanol series, leaned in xylene, embedded in paraffin, and cut into 4 μm sections using a rotary microtome (Leica, Germany). The sections were stained with HE staining for routine histological examination and observed using a light microscope (Olympus BX51, Japan).
Experimentally infected C. carpio was anesthetized with 3-aminobenzoic acid ethyl ester methanesulfonate (MS-222). Blood samples were collected from the caudal vein and placed for 1 h at room temperature. After centrifugation under 4500 rpm at 4°C for 15 min, the serum was collected and stored at −80°C. The serum biochemical indices of complement C3, superoxide dismutase (SOD), and lysozyme (LZM) were assessed using corresponding commercial kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The detection method refers to the kit instructions and previous reports (28).
Total RNA was extracted from six tissue samples (three from the control groups and three from the challenge groups, collected from the intestinal tissue after 3 days of P. shigelloides challenge) using TRIzol Reagent (Invitrogen, USA) and then treated with RNase-free DNase I (Thermo Scientific, USA) at 30°C for 30 min to remove genomic DNA contaminants. Subsequently, the purity, quantity, and integrity of the extracted total RNA samples were assessed using a NanoDropND-2000 spectrophotometer (Thermo, Waltham, USA), by 1.2% (w/v) agarose gel electrophoresis, and using an Agilent 2100 Bioanalyzer (Agilent Technologies, Richardson, USA), respectively. RNA samples with an RNA integrity number (RIN) > 8, 28S/18S > 0.7, and A260/280 of approximately 2.0 were used to construct the RNA-seq library. Poly (A) mRNA was isolated from total RNA using poly (dT) oligo-attached magnetic beads, and cDNA libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina, USA). In total, six cDNA libraries were sequenced using the Illumina Nova-Seq (BGI) sequencing platform to generate 150-bp paired-end reads.
Read quality of the collected RNA-seq data was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (29). The filtered clean data were subsequently mapped to the C. carpio genome (NCBI: ASM1834038v1) using HISAT2 v2.1.0 (30). All genes were annotated in the genome sequences available in six public databases, including NCBI’s non-redundant protein sequence (Nr) database, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG) (31), Gene Ontology (GO) (32), evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) (33), and Pfam (34).
DEGs between the control and challenge groups were identified using Baggerly’s test to calculate the fragments per kilobase of exon per million fragments mapped (FPKM) values. Benjamini–Hochberg correction was used to adjust the original P-values in Baggerly’s test in order to minimize the false discovery rate (FDR) (35). DEGs were identified if the associated PFDR was less than 0.05, and an absolute value of log2 (fold change) > 1 was regarded as the cut-off criterion. Clustering analysis was performed using the pheatmap package in R based on the FPKM values of DEGs. Subsequently, DEGs were enriched and subjected to GO term (36) and KEGG pathway analyses using clusterProfiler (37). A P-value < 0.05 indicated statistical significance, and the top 10 terms related to immunity were selected for visualization.
In total, six genes associated with immune-related gene testis development were randomly selected for qRT-PCR to validate the RNA-seq results. Reversed-transcribed cDNA obtained from total RNA used for transcriptome sequencing was synthesized using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Shanghai, China), according to the manufacturer’s protocol. All cDNA samples were diluted to 5 ng/µL and stored at −80°C until use. Specific primers were designed on the basis of NCBI Primer-BLAST (NCBI, USA), as listed in Table S1. All qRT-PCR reactions were performed in triplicate, and target specificity was determined on the basis of dissociation curve analysis. β-actin was selected as the internal control to normalize the expression level of each gene. The relative expression level of the target gene versus the β-actin gene was calculated using the 2−ΔΔCT method. The obtained data were statistically analyzed using GraphPad Prism 7.0 software.
The results have been reported as the means ± SEs after data preparation and statistical analysis using GraphPad Prism 7.0 software. Statistical significance of the findings in each experimental group relative to the control group was assessed using Student’s two-tailed t-test. Significance (P-value) has been indicated as follows: *(P < 0.05), **(P < 0.01), and ***(P < 0.001).
The symptoms of infected fish are shown in Figure 1; the symptoms mainly manifested as a mild floating head, outgroup swimming, slow swimming, and reduced feeding. In the middle stage of the disease, the fish stopped feeding completely, with slight eyeball protrusion, slight bleeding in the jaw and abdomen, and redness of the head (Figures 1A, B). In the later stage, death, abdominal bleeding, and swelling were noted (Figures 1C, D). After dissection, it was found that the dead fish had fluid accumulation (ascites) in the abdominal cavity. Moreover, the liver was swollen, with light yellow and irregular red spots distributed on it. Furthermore, the tissue felt slightly brittle, like bean paste (Figures 1E, F).
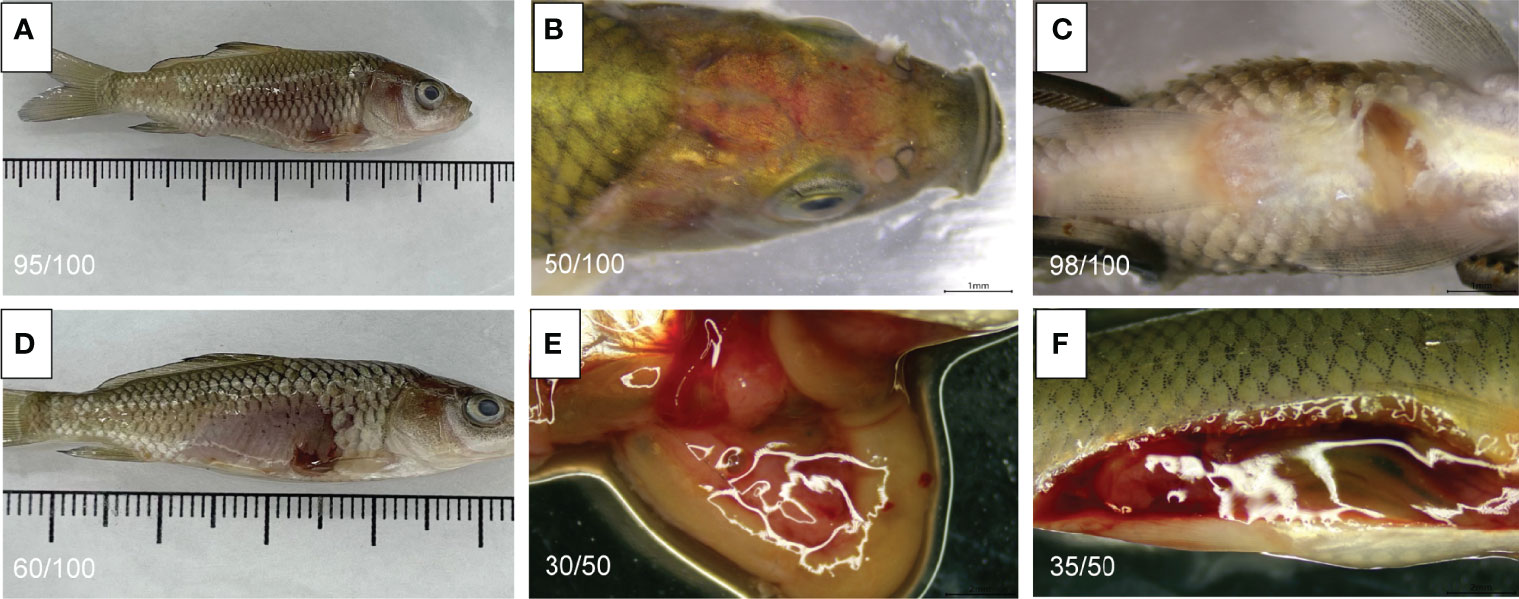
Figure 1 Symptoms of infected C carpio. (A) The abdominal scales of infected C carpio fell off, and the abdomen was red and swollen. (B) Red head of infected C carpio. (C) Swollen abdomen of infected C carpio. (D) The abdominal scales of infected C carpio fell off. The abdomen became red, with abscess-like fluid flowing out of it. (E) The visceral tissue was diffuse, and the liver was enlarged and pale. (F) The abdominal cavity of C carpio had a lot of yellowish fluid after dissection.
The dominant strain of pathogenic bacteria was isolated from the lesions of C. carpio (ulceration, ascites, hepatopancreas, and intestinal tract) and named Cc2021 (27). The bacterium grew well on BHI agar, and the colonies were round, beige opaque, smooth, moist, and slightly raised but with neat edges (Figure 2Aa). Gram staining results were microscopically observed. The bacterial cells stained red and appeared as short rods, which were arranged singly or in pairs, indicating that the strain was gram negative (Figure 2Ab). Negative stain electron microscopy revealed a straight rod with a terminal round shape, no spore or capsule, and two fascicular flagella at both ends. The size of the bacterium (short diameter × long diameter) was 0.8–1.0 μm × 3.0 μm (Figure 2Ac). These characteristics were consistent with the description provided in Bergey’s Manual of Systematic Bacteriology.
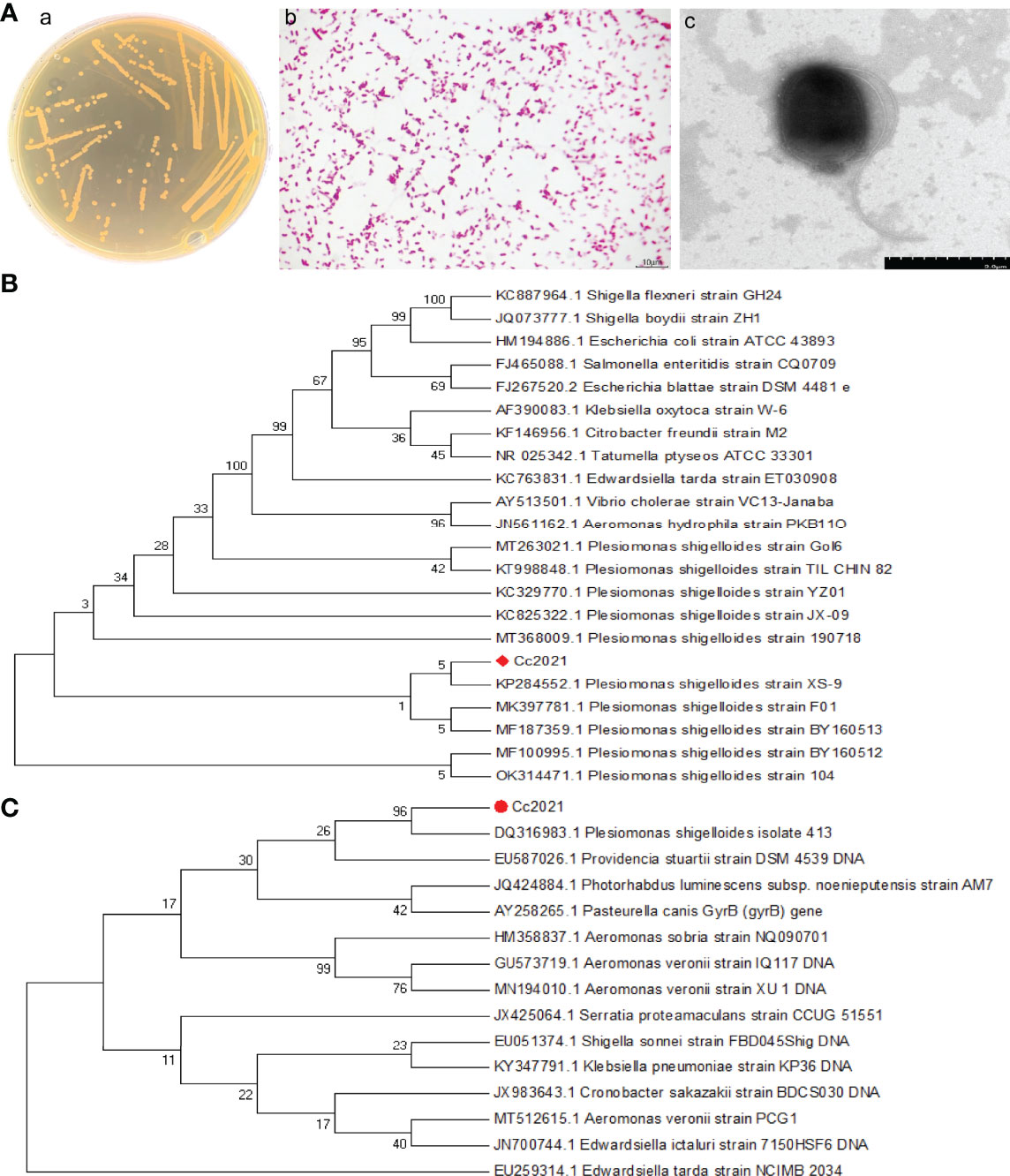
Figure 2 Identification of the pathogen’s morphological and molecular characteristics. (A) (a) Growth characteristics of isolates in BHI medium. (b) Gram staining results of isolates. (c) Negative staining results of isolates. (B) Phylogenetic tree based on the partial 16S rRNA gene sequences included in the expanded MicroSeq 500 library; the algorithm used to construct the tree was the unweighted pair group method using averages (UPGMA). (C) Phylogenetic tree based on the partial gyrB gene sequences included in the expanded MicroSeq 500 library; the algorithm used to construct the tree was UPGMA.
The physiological and biochemical characteristics of the pathogen are provided in Table 1. According to Bergey’s Manual of Determinative Bacteriology (9th ed.), the bacterium could hydrolyze arginine, D-fructose, fucose, glucose, and myo-inositol but not adipic acid, citric acid, D-mannitol, phenylalanine aminotransferase, phenylacetic acid, sucrose, salicin, sorbitol, and urease. The results of methyl red and malic acid tests were also positive.
To further identify the strain, its 16S rRNA and gyrB genes were sequenced. NCBI BLAST was used for homology analysis based on the 16S rRNA and gyrB gene sequences of the strain; the results revealed that the strain was 99% similar to P. shigelloides. Phylogenetic analysis was performed after screening sequences with high similarity in terms of sequence alignment; the results (Figures 2B, C) revealed that both 16S rRNA and gyrB gene sequences of P. shigelloides formed a single cluster. Therefore, based on its morphological, physiological, biochemical, and molecular characteristics, the strain was identified to be P. shigelloides.
To assess the pathogenicity of P. shigelloides in C. carpio, one group of C. carpio was injected with physiological saline (65% NaCl) (control group), while four groups were injected with four different concentrations (1.0 × 107 CFU/mL, 5.0 × 107 CFU/mL, 1.0 × 108 CFU/mL, and 5.0 × 108 CFU/mL) of the pathogenic bacterial strain (experimental groups). Subsequently, deaths were recorded (Table 2). Both high and low concentrations of the bacterial suspension resulted in typical symptoms, similar to those observed in naturally infected fish. After the injection of a high concentration (5.0 × 108 CFU/mL) of the bacterial suspension, a large number of fish died within 2 days (73.3%), with the mortality reaching 100% within 7 days. The mortality rates of C. carpio were 80%, 53.3%, and 23.3% after the injection of 1.0 × 108 CFU/mL, 5.0 × 107 CFU/mL, and 1.0 × 107 CFU/mL concentrations, respectively. On the other hand, no death was noted in the control group. These mortality results served as the basis for the follow-up regression challenge experiment. Tissue edema and bacterial loads in C. carpio were assessed after P. shigelloides challenge. The dry/wet weight ratios of the liver, spleen, head kidney, and body kidney of C. carpio showed an increasing trend with an increase in the challenge time. After 2 days post-challenge, the dry/wet weight ratios of the liver and head kidney gradually increased, while those of the spleen and head kidney tended to decrease (Figures 3A–D). The bacterial load in the tissues from the intestine, hepatopancreas, head kidney, body kidney, and spleen of C. carpio infected with P. shigelloides and the blood bacterial concentrations increased with an increase in the challenge time. The bacterial load in the intestinal tissues was the highest, followed by that in the tissues from the hepatopancreas, head kidney, and spleen. The lowest bacterial load was noted in the blood and body kidney tissues (Figure 3E).
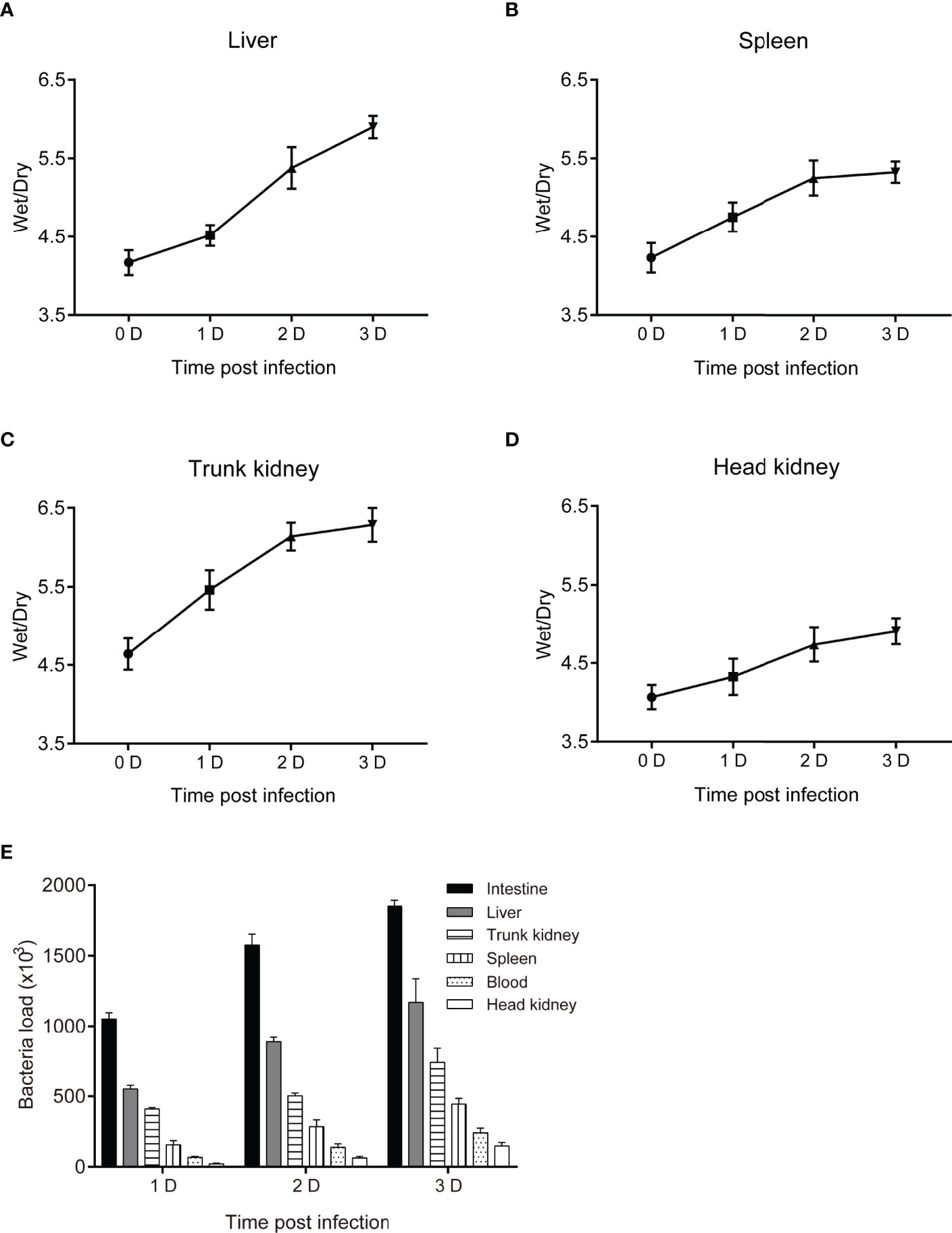
Figure 3 Tissue edema and bacterial load in C. carpio after P. shigelloides challenge. (A–D) The dry/wet ratio of the liver, spleen, body kidney, and head kidney of C. carpio 0–3 days after P. shigelloides challenge. (E) Bacterial load in C. carpio tissues after P. shigelloides challenge.
To observe the histopathological state of P. shigelloides-infected C. carpio, HE staining was performed using tissues from the liver, head kidney, spleen, and intestine at each period. The typical pathological states are shown in Figure 4. In the control group, the hepatic cells were rich in cytoplasm, had an obvious nucleus and cytoplasm, had a clear cell boundary, and were evenly dispersed (Figure 4Aa). After the artificial challenge, the inflammatory cells in the liver of infected C. carpio showed focal aggregation, with an unclear structure and a fuzzy boundary. The hepatic cells were denatured and necrotic, and the nucleus and cytoplasm were concentrated. Some hepatic cells were vacuolated and had dissolved nuclei. The tissue was loose and had edema, and degeneration and necrosis were noted around the sinuses. Moreover, the tissue structure was destroyed and several hepatocytes were vacuolated (Figure 4Ab). The renal structure of infected C. carpio was disorganized, with swelling and degeneration of cells, a narrow renal cavity, and a transparent tube type. The renal tissue was filled with lymphocytes, accompanied by multiple bleeding spots and an obvious inflammatory reaction (Figure 4Bb). Moreover, ferriflavin deposition in the splenic tissue, atrophy of the white pulp, and a reduced area occupied by splenic pulp hyperemia were noted (Figure 4Cb). The intestinal structure of C. carpio in the control group was intact; the inner wall of the tissue was not damaged, intestinal epithelial villi were arranged tightly and in an orderly manner, and basal epithelial tissues were obviously stratified (Figure 4Da). Villus epithelial cells in the intestinal tissues of infected C. carpio fell off, with the infiltration of local lymphocytes and the swelling of and increase in goblet cells in the mucosal epithelium being noted (Figure 4Db).
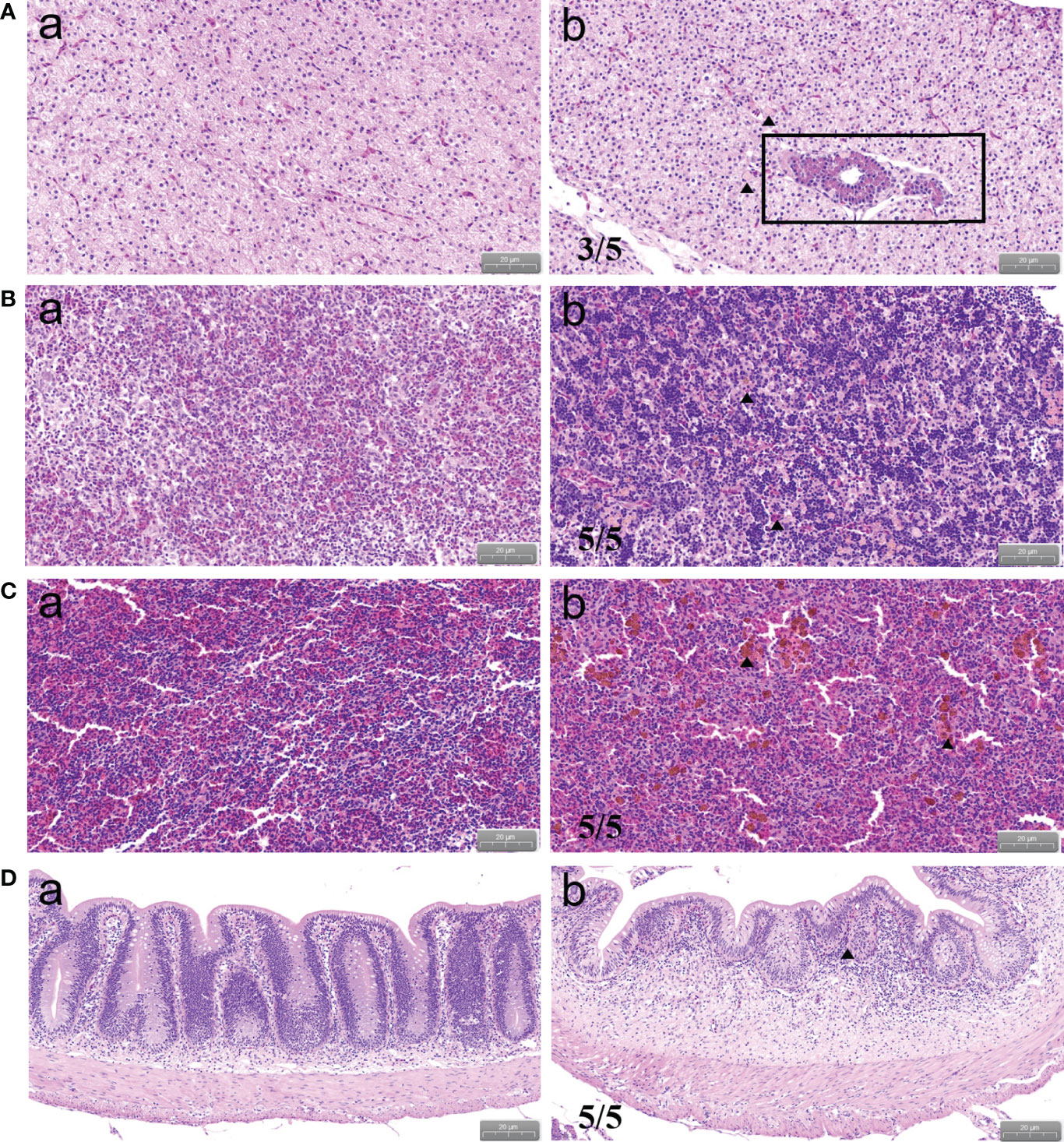
Figure 4 Histopathological changes in C carpio tissues after P. shigelloides challenge. (A) Infected liver. (B) Infected head kidney. (C) Infected spleen. (D) Infected intestine. Pictures of typical symptoms are shown. Scale bar of A, B, and C = 20 μm. Scale bar of D = 50 μm.
To assess the degree of activation of antibacterial immune responses in the key immune organs and bloodstream of C. carpio during P. shigelloides challenge, C3, SOD, and LZM activities were assessed at different stages after the challenge. The expression level of C3 in the serum increased from 1 to 7 days and reached its maximum on the 5th day post-challenge, becoming nearly three times the normal expression level (Figure 5A). The expression levels of LZM in all the tissues of healthy common carp were similar. The LZM levels in the blood, head kidney, liver, and intestine significantly increased after the challenge. Moreover, the LZM levels in all the tissues continued to increase from 1 to 7 days post-challenge. LZM activity was the highest in the intestine, followed by the liver and blood, while it was the lowest in the head kidney after the challenge (Figure 5B). The SOD levels in each tissue of C. carpio showed a trend of first increasing and then decreasing after the challenge; the increase was most obvious on 1 and 3 days, followed by a decrease (Figure 5C). The highest SOD activity was noted in the intestinal tissue after the challenge; this may be related to enteritis caused by the bacterium.
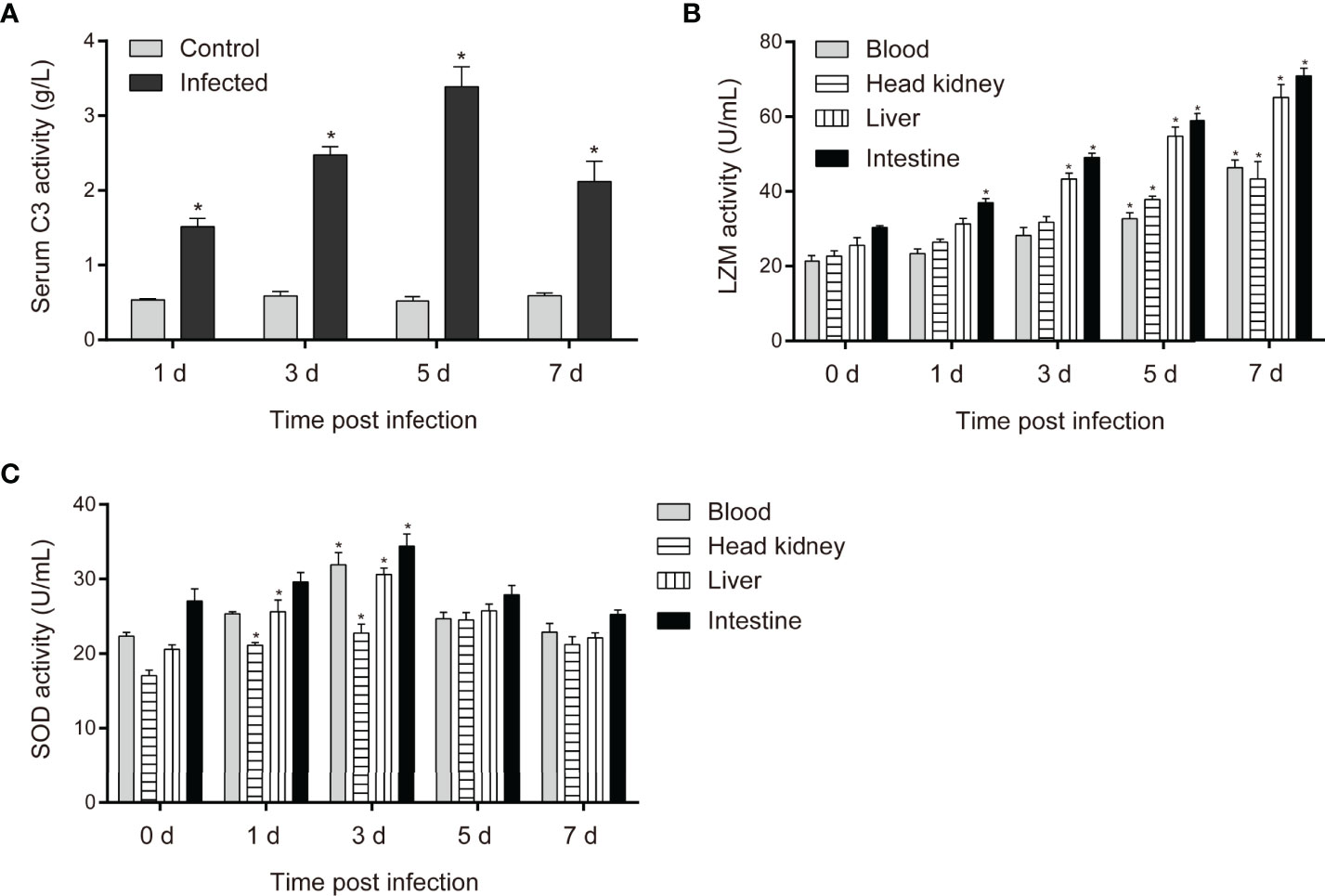
Figure 5 (A) Changes in complement C3 levels in the serum of C carpio with time after P. shigelloides challenge. (B) Changes in LZM activity in tissues of C carpio with time after P. shigelloides challenge. (C) Changes in SOD activity in tissues of C carpio with time after P. shigelloides challenge. "*" means P < 0.05.
All raw data generated in the present study have been uploaded to the NCBI Sequence Read Archive (SRA) under the registration number PRJNA793911. The original sequencing data included reads having low quality, joint contamination, and a high unknown base N content, which needed to be removed before data analysis in order to ensure the reliability of the results. After sequence filtration, a comparison of all genomes was performed, as shown in Figure S1A. Expression profiles of all the genes were obtained in both the control and challenge groups, with half reaching at least one FPKM (Figure S1B). Correlation analysis of the samples revealed that the correlation coefficients of Challenge 2 and Control 1 within the group were far less than 0.7; thus, these two samples were removed from subsequent analysis (Figure S1C). Analysis of FPKM expression levels for all the genes in the visualized heatmap revealed that the expression levels of Control 2 and Control 3 genes were consistent and those of Challenge 1 and Challenge 3 genes were consistent (Figure S1D).
To further assess the immune response mechanism of C. carpio during P. shigelloides challenge, comparative transcriptome analysis was performed for comparing the gene expression profiles in the intestinal tract of C. carpio between the control and challenge groups. For this analysis, three control group samples and three challenge group samples, collected on the 3rd day after the infection, were included. Based on sequence homology, GO enrichment analysis of DEGs in the intestinal tract was performed after P. shigelloides challenge, and the similarities and differences in these terms were analyzed. To further understand the functions of DEGs and the signaling pathways that they participate in, all DEGs were classified into three GO categories: biological process (BP; 20 subclasses), molecular function (MF; 20 subclasses), and cellular component (CC; 20 subclasses) (Figure S2A). KEGG pathway analysis of the intestinal tissue after P. shigelloides challenge revealed that there were 21 KEGG metabolic pathways involving different genes, mainly including the complement and coagulation cascade pathway, TNF signaling pathway, interleukin (IL)-17 signaling pathway, and toxoplasmosis and systemic lupus erythematosus pathway (Figure S2B).
The identification of DEGs can provide a deeper understanding of the changes in the immune mechanism of P. shigelloides-infected C. carpio. The heatmap in Figure 6A shows the expression of DEGs in each group. Intestinal gene expression was higher in the challenge group. Importantly, there was no significant difference in DEGs between the control and challenge groups. In total, 1704 genes were differentially expressed in the control and challenge groups, including 876 upregulated genes and 828 downregulated genes (Figure 6B). The upregulation level of genes was thus significantly higher than the downregulation level, which may be attributed to the cascade amplification of immune signal transmission.
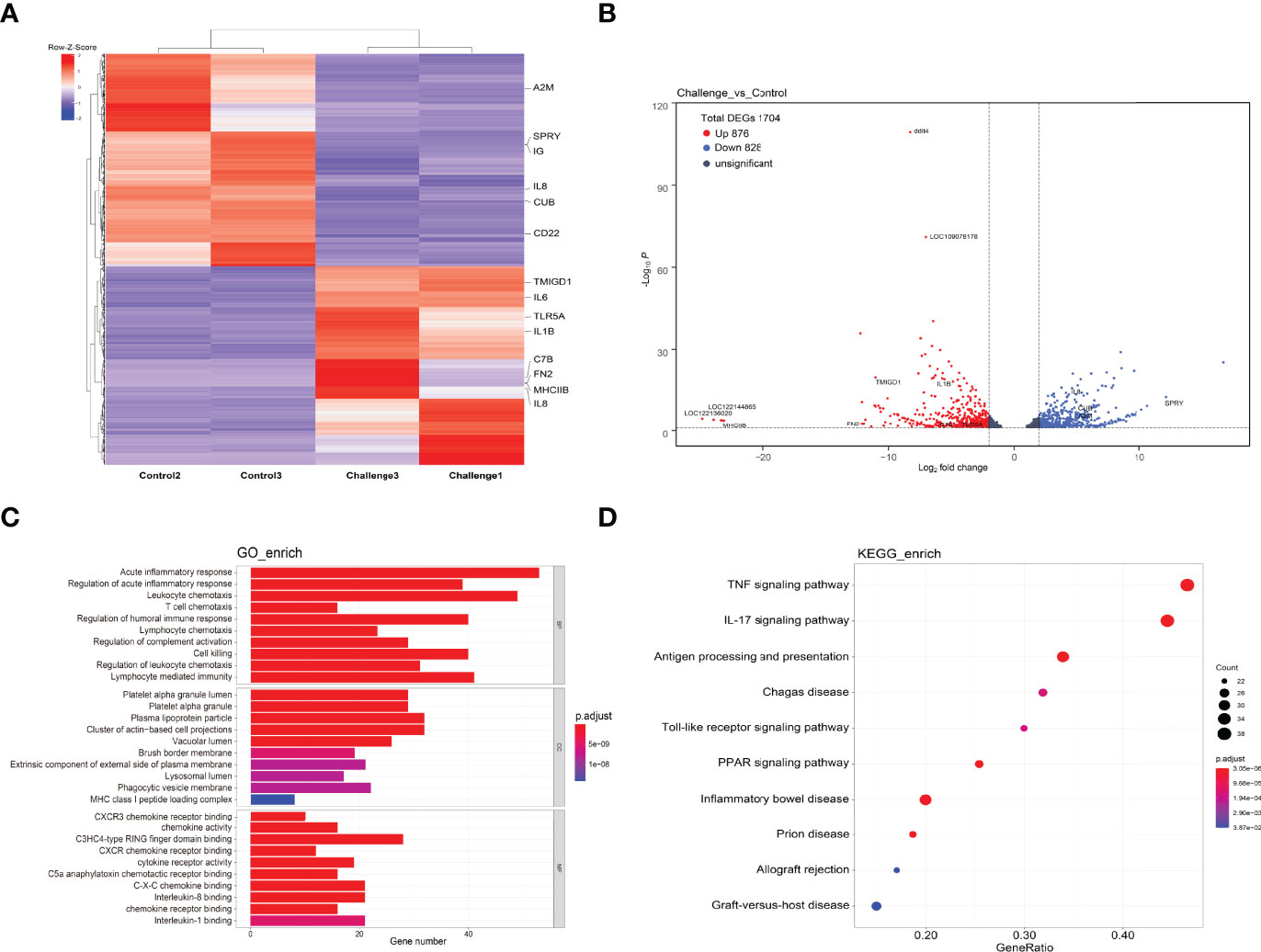
Figure 6 Differentially expressed genes. (A) Heatmap showing the expression of differentially expressed genes in each sample. (B) Volcano map showing the expression of differentially expressed genes in each group of samples. (C) GO enrichment analysis was performed for differentially expressed genes, and three categories of the top 10 immune-related pathways were selected for display. (D) KEGG enrichment analysis was performed for differentially expressed genes, and the top 10 immune-related pathways were selected for display.
To further assess the immune response after P. shigelloides challenge, we focused on immune-related DEGs classified into BP, MF, and CC categories, as shown in Figure 6C. The top 10 KEGG pathway terms are shown in Figure 6D. GO enrichment analysis of this module was dominated by functional categories related to immune system. In the BP category, acute inflammatory response, regulation of acute inflammatory response, and leukocyte chemotaxis were the top three terms. In the CC category, platelet alpha granule lumen, platelet alpha granule, and plasma lipoprotein particle were the top three GO terms. In the MF category, CXCR3 chemokine receptor binding, chemokine activity, and C3HC4-type RING finger domain binding were the top three terms. Furthermore, KEGG pathway enrichment of immune-related DEGs was analyzed. The immune-related signaling pathways of all enriched immune-related DEGs were selected as the top 10 ones, e.g., the TNF signaling pathway, IL-17 signaling pathway, antigen processing and presentation pathway, Chagas disease pathway, and Toll-like receptor (TLR) signaling pathway (Figure 6D). Some immune-related DEGs are shown in Table 3. These results suggested that both the innate and adaptive immune responses of C. carpio played vital roles in its response to P. shigelloides challenge. The discovery of the immune-related pathways and genes could provide a theoretical basis to understand the molecular mechanism of C. carpio infected with P. shigelloides or other bacteria.
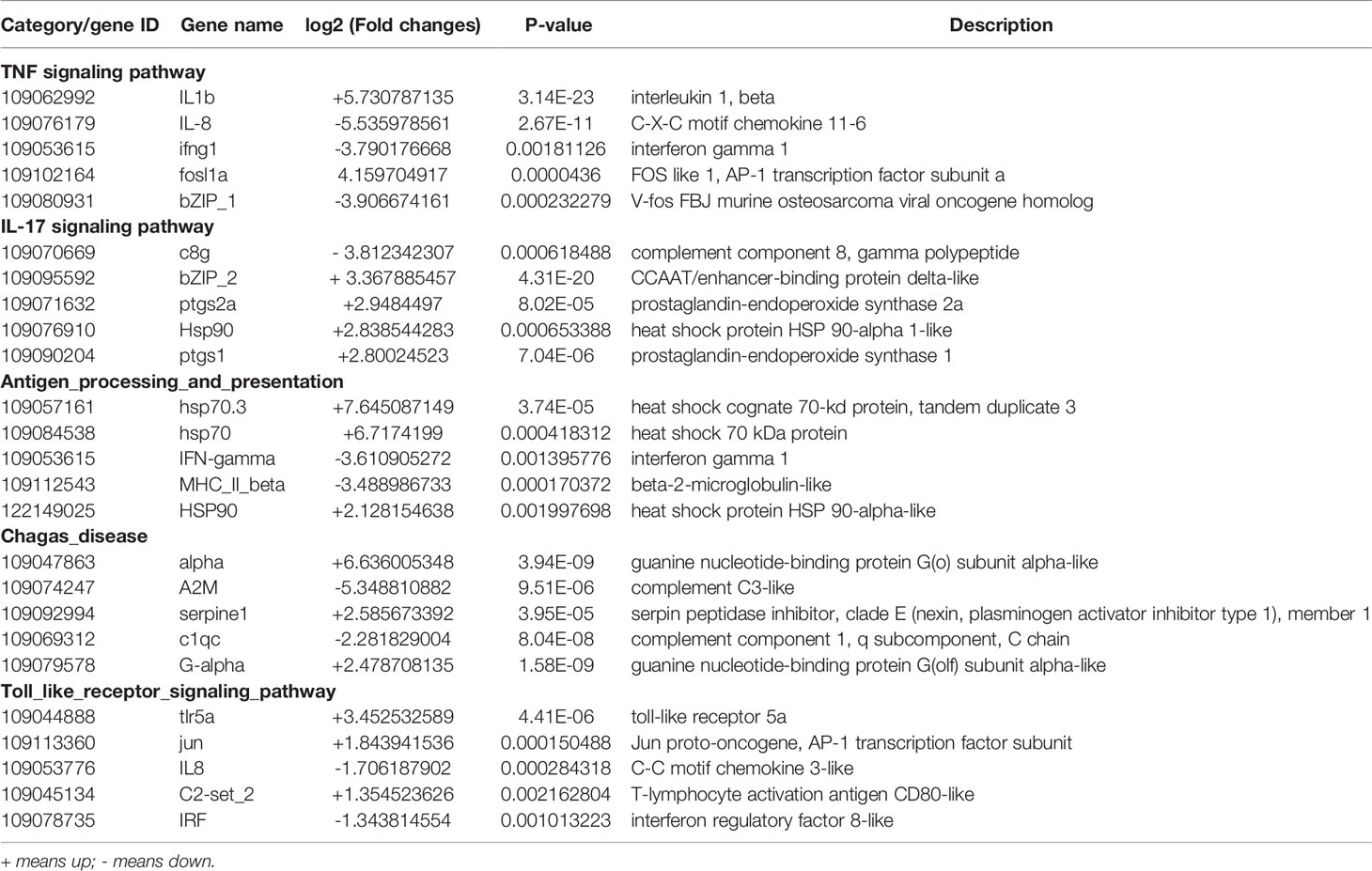
Table 3 Immune-related differentially expressed following P. shigelloides challenge based on the KEGG analysis.
To verify significant immune response candidates activated by P. shigelloides, qRT-PCR was performed on several candidates selected from the comparative transcriptome. The expression levels of IL-1β, Hsp70, TLR5a, MCH II, CD22, and IL-8 were verified in the blood, spleen, head kidney, and intestine at 0, 1, 3, 5, and 7 days after the challenge (Figure 7). The mRNA expression level of IL-1β dramatically increased and immediately reached the highest level on the 1st day after the challenge, following which it began to decrease gradually. The expression level of IL-1β was the highest in the intestine (Figure 7A). The mRNA expression level of Hsp70 gradually increased in all immune tissues from 1 to 7 days after the challenge; the expression level was higher in the head kidney and spleen (Figure 7B). The mRNA expression level of TLR5a was upregulated to a similar extent in all the tissues after the challenge, with the increase being the highest at 3 and 5 days (Figure 7C). Moreover, the mRNA expression level of MHC II was continuously upregulated after the challenge, with the most significant increase being noted in the intestine (Figure 7D). In contrast, the mRNA expression levels of CD22 and IL-8 were downregulated to varying degrees after the challenge in all the tissues (Figures 7E, F).
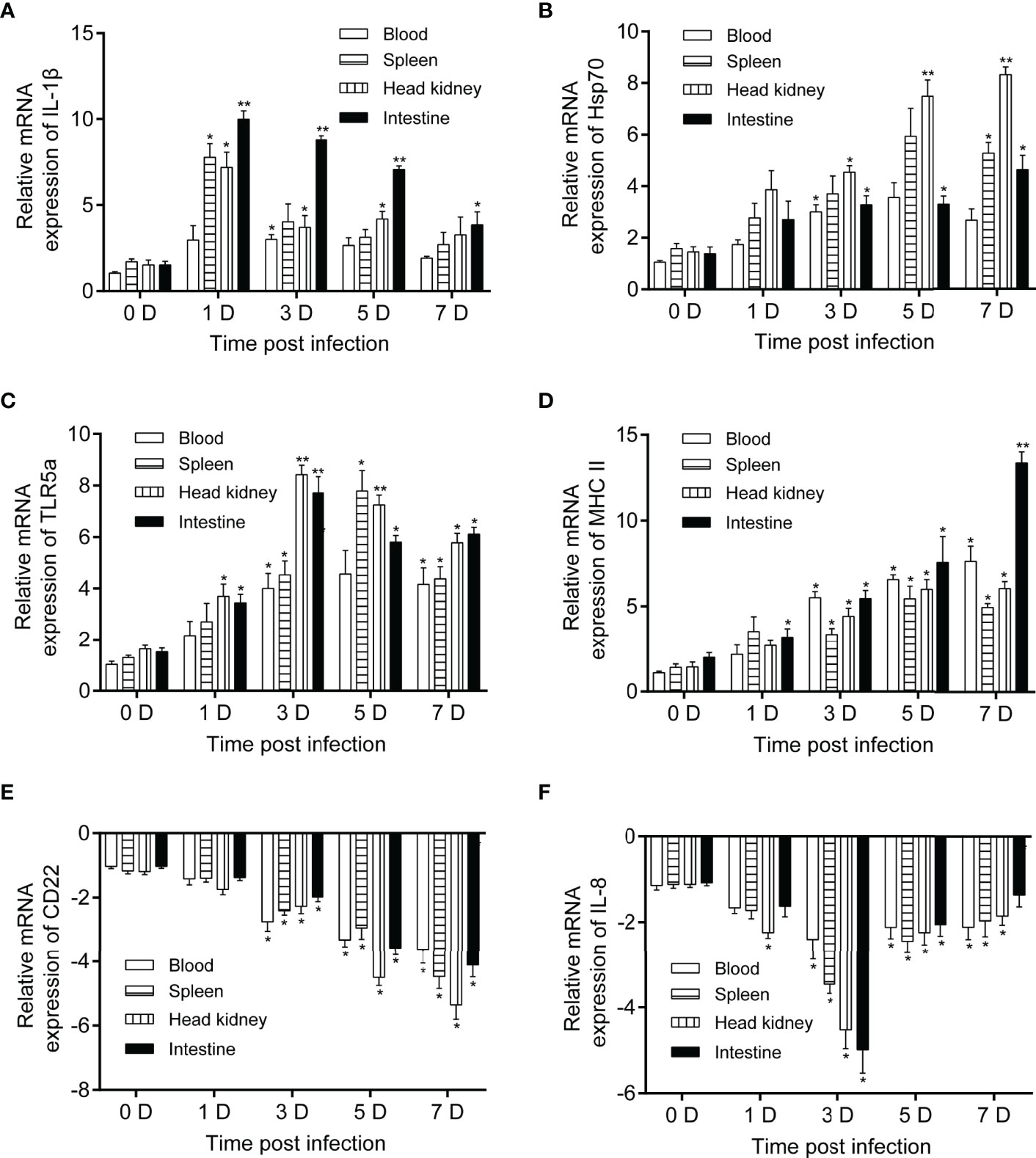
Figure 7 Relative mRNA expression levels of IL-1β, Hsp70, TLR5a, MCH II, CD22, and IL-8 in C carpio tissues after P. shigelloides challenge. mRNA expression levels of IL-1β (A), Hsp70 (B), TLR5a (C), MCH II (D), CD22 (E), and IL-8 (F) in the blood, spleen, head kidney, and intestine of C carpio at 0, 1, 3, 5, and 7 days after P. shigelloides challenge. The β-actin gene was used as a reference gene, and the data of day 0 were used for comparison with those of days 1, 3, 5, and 7 in order to analyze the differences. Data are presented as the means ± SEs (n = 4). Statistical analysis was performed using unpaired Student’s t-test (*P < 0.05 and **P < 0.01).
P. shigelloides, belonging to the order Eerobacterales, is widely distributed in nature. it is a kind of global distribution bacteria, its detection rate has obvious seasonal, high detection rate in summer, which may be related to the optimal growth temperature of the bacteria is higher. And P. shigelloides is a new pathogen of digestive tract inflammation in many aquatic animals, as well as a zoonotic pathogen, which is a pathogen of gastrointestinal infection outbreak transmitted by water and food (7). In recent years, there have been increasing reports on the pathogenicity of P. shigelloides in fishes. The pathogenic symptoms of P. shigelloides are slightly different in different fishes; this may be caused by the differences in fish species, culture environments, and strains (38, 39). However, few studies have been conducted on the pathogenic mechanism of P. shigelloides in fishes (40). Studies on the isolation and identification of bacterial species are essential for understanding the exact incidence of a disease in a given region and for planning epidemiological studies and control and eradication programs (41). Moreover, understanding how bacteria cause pathological symptoms can form the basis of preventing and treating bacterial diseases (42, 43). In the present study, we isolated a bacterium from C. carpio and identified it to be P. shigelloides by morphological, physiological, and biochemical methods; gene sequence analysis; and PCR. In addition, we used the isolated P. shigelloides strain to infect C. carpio and studied the changes in histopathology, tissue bacterial load, tissue edema, and immune gene expression to understand its pathogenic mechanism. Finally, and most importantly, through high-throughput sequencing technology, we explored the pathogenic mechanism of P. shigelloides and the immune response mechanism of C. carpio after P. shigelloides challenge. Our findings can serve as a reference for drug designing and vaccine adjuvant screening against pathogens.
P. shigelloides is widely distributed and found in different water bodies, animals, and humans (7). It can cause human acute gastroenteritis and other toxic symptoms. In recent years, it has been detected in different aquatic animals and has been found to be highly pathogenic to fishes. It can multiply in the intestines of fishes, damaging their organs and ultimately causing death (37). Grass carp mainly presents with redness and swelling of the body surface, liver, intestine, and anus (38, 39). Garra rufa infected with Streptococcus agalactiae presents with surface ulceration and intestinal and hepatic lesions (40). Pathological changes in the liver, stomach, intestine, kidney, throat, and other organs have been reported to be caused in Trionyx sinensis by Aeromonas hydrophila, with the findings in male and female gonads also being inconsistent (39). However, acute diarrhea and food poisoning have been found to be the main symptoms in humans after P. shigelloides challenge (41). In the present study, the disease in C. carpio was characterized by an abdominal cavity, the emergence of a few different levels of fish ascites, hepatomegaly, yellowish color, and an irregular distribution of red dots. Anatomic assessment revealed a crispy organization, light inflammation of the gills, erosion and rotting of the gills, bleeding of the gills, and significant intestinal inflammation. P. shigelloides is a pathogen that causes zoonotic diseases. It also causes a wide range of clinical symptoms in humans, such as acute gastroenteritis and diarrhea (7). Therefore, it is an important pathogen that can pose a risk to public health, especially to people who directly touch any infected fish.
Complement C3, SOD, and LZM in the serum are powerful indicators of the non-specific immune response of aquatic animals, which can identify pathogens, inactivate viruses, and reflect the degree of somatic cell damage to a certain extent (18). Moreover, they play an important role in the non-specific immunity of fishes (44). The complement system is an important component involved in the resistance of fishes to challenge and is involved in target cell lysis and conditioning reactions after activation. Complement C3 is the main component of the complement system (45). A previous study reported an increase in the complement C3 level of hybrid tilapia fed different doses of the commercial product DVAQUA (46). The serum complement C3 level of yellow catfish was found to significantly increase at 7, 14, 21, and 28 days after the injection of inactivated E. ictaluri vaccine, further providing that the vaccine could improve the non-specific immunity of yellow catfish (47). However, in the present study, the complement C3 level in the serum of P. shigelloides-infected C. carpio significantly increased, reached its maximum after 5 days post-challenge, and then significantly decreased. The results revealed that the non-specific immune mechanism was activated in C. carpio after P. shigelloides challenge and was maintained for a long time. SOD activity has been found to be closely related to the immune level of an organism. The SOD activity of Channa argus increased from 9 days after the injection of inactivated Nacardia seriolea, reaching its peak on the 12th day (48). In the present study, the SOD activity in the serum of C. carpio first increased after the injection of P. shigelloides and then decreased after reaching its maximum on day 3. This result indicated that C. carpio could produce a large number of free radicals, such as superoxide anions, at an early stage of the challenge and improve its antioxidant and immune capacity to resist the challenge. Moreover, LZM activity was found to increase in A. hydrophila-infected Chinese sturgeon (49). This result was consistent with that of the present study. In conclusion, the isolated bacterial strain could stimulate the body to produce an immune response after a challenge, which was manifested as increased C3, SOD, and LZM activities in the serum of C. carpio; this in turn stimulated the non-specific immunity of C. carpio to a certain extent.
Intestinal mucosal immunity is an important mechanism by which a host can resist a pathogen; it plays an important role in natural immunity (50). In previous studies, P. shigelloides challenge was found to destroy the intestinal tissue structure, reduce the number of mucosal immune-related cells, cause intestinal digestion and absorption dysfunction, and activate the mucosal immune response in hybrid sturgeon (12, 17). In the present study, the intestinal transcriptome of C. carpio was altered by P. shigelloides challenge. The results revealed that there were 1704 DEGs in the intestinal tissues of C. carpio after P. shigelloides challenge, including 876 upregulated DEGs and 828 downregulated DEGs. This may be related to extensive bacterial replication in C. carpio and the resistance of C. carpio to bacterial challenge. These results indicate that bacterial invasion into the digestive mucosa of C. carpio can induce a specific immune response through a non-specific immune function of the body, i.e., the phagocytosis of immune cells. Some pathogens, such as Legionella pneumophila, Listeria monocytogenes, and S. Typhi, recognize cytoplasmic pathogen-associated molecular patterns (PAMPs). The RIG-1-like receptor signaling pathway is activated to promote the expression of type 1 interferons and pro-inflammatory cytokines, thereby protecting against bacterial challenge (51, 52). Fish TLRs (such as TLR3) are responsible for recognizing not only nucleic acids produced by viruses but also bacterial components, thereby acting as antibacterial agents (53). Intracellular parasitic bacteria, such as L. monocytogenes and Mycobacterium tuberculosis, can activate the cellular solute DNA sensing pathway by recognizing and activating the cGAS-STING pathway. Following this, the TBK1/IRF3 signal cascade is activated to activate the type I IFN transcriptional response and exert antibacterial effects (54). In the present study, the main immune-related signaling pathways in the intestine of P. shigelloides-infected C. carpio were the TNF signaling pathway, IL-17 signaling pathway, antigen processing and presentation pathway, Chagas disease pathway, and TLR signaling pathway. These signaling pathways play an important role in immune cell activation, inflammatory response, DNA damage, lipid metabolism, and the inhibition of pathogenic bacteria.
To further understand the antibacterial mechanisms triggered by the innate immune response in C. carpio after P. shigelloides challenge, we analyzed the expression levels of six DEGs related to immunity in different tissues and at different times. The IL family is one of the largest cytokine families that plays an important role in the innate and acquired immune responses of the host (55). IL-1β was the first IL to be identified. It plays a critical role in the initiation and regulation of immune and inflammatory responses in many economically important teleost fishes (55). IL-1β in teleost fishes also plays an important role in host resistance to pathogens (56). Recent research has revealed that IL-1β plays an important role in the immune response of large yellow crocea against Vibrio alginolyticus (55). In the present study, after the challenge, the mRNA level of IL-1β rapidly increased and reached the highest level on day 1, following which it began to gradually decrease. The expression level in the intestine was the highest. IL-8, a CXC-type chemokine, plays a key role in acute inflammation by recruiting neutrophils in mammals. Moreover, fish IL-8 is a functional homolog of mammalian IL-8 (57). IL-8 is a well-known cytokine that plays a crucial role in inflammatory responses and is activated in response to various diseases (57). IL-8 was found to be expressed in various tissues of unchallenged olive flounder. In particular, its expression was the highest in the gills (58). The same results have been reported in rainbow trout, tongue sole (Cynoglossus semilaevis), and common carp (59). In the present study, the mRNA expression levels of IL-8 were downregulated to varying degrees after the challenge in all the tissues. These results suggest that IL-8 is closely related to inflammation and immune regulation in olive flounders and may be used as a basis for research on the immune systems of other fishes. HSP70 is an important member of the heat-shock protein superfamily; it appears in almost all species. HSP70 proteins also play a role in improving disease resistance (60). In the present study, the mRNA expression level of Hsp70 gradually increased in all the immune tissues 1–7 days after the challenge. This upregulation was most obvious in the head kidney and spleen. These results suggest that HSP70 is involved in the immune response against bacterial attacks and heat stress. TLR5 is associated with flagellin detection (61). Moreover, significant upregulation of TLR pathway genes has been reported after A. hydrophila challenge in other fish species (61). Increased expression levels of tlr5a and tlr5b genes were noted in turbot (Scophthalmus maximus L.) mucosal tissues (i.e., intestine and gills) in response to infection by the gram-positive, non-flagellated pathogen S. iniae (62). In the present study, the mRNA expression level of TLR5a was upregulated to a similar extent in all the tissues after the challenge, with the increase being the highest at 3 and 5 days. A similar upregulation of tlr5 in response to alive and formalin-killed R. salmoninarum has been reported (63, 64). Therefore, the role of TLR5 beyond the recognition of flagellin, particularly after exposure to non-flagellated bacteria in teleosts, warrants further investigation.
In conclusion, in the present study, a comprehensive and accurate method was used to identify pathogenic bacteria in C. carpio. Changes in the tissue bacterial load, tissue edema, histopathological parameters, innate immunity, and survival rates of P. shigelloides-infected C. carpio during the septicemia process were systemically analyzed. The results indicated that the host response to P. shigelloides challenge involved an inflammatory reaction. To further assess the pathogenesis, comparative transcriptome analysis was performed between the challenge and control groups at 3 days post-challenge. The analysis revealed 1704 genes that were differentially expressed. Further analysis revealed that about 10 KEGG pathways were involved in the host immune response to bacterial challenge; these included the TNF signaling pathway, IL-17 signaling pathway, antigen processing and presentation pathway, Chagas disease pathway, and TLR signaling pathway, among others. The present findings can serve as a reference for understanding the pathogenic mechanism of P. shigelloides in C. carpio and broaden the understanding of the systemic immune response of C. carpio.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA793911.
The animal study was reviewed and approved by Institute of Hydrobiology, Chinese Academy of Sciences. Written informed consent was obtained from the owners for the participation of their animals in this study.
DL conceived the project. DL and HC designed the study. HC and YZ performed most of the experiment and analyses. KC, YW, HL, and FL helped in experiment, data curation and analyses. YL helped in breeding and sampling. HC, YZ, DL, WH and ZZ prepared the draft and final version of the manuscript. All authors read and approved the final manuscript.
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31922085 to DL), the Strategic Priority Research Program of CAS (Grant No. XDA24010108to DL), Natural Science Foundation of of Hubei Province (Grant No. 2020CFA056 to DL), China Postdoctoral Science Foundation on the 70th finance (Grant No. 2021M703436 to YZ), and Postdoctoral innovation research positions of Hubei Province (YZ). The authors would like to thank Rui Li, Hairong Liu, Kaifeng Meng, Wentao Zhu and Chuang Xu for technical advice and assistance in experiments, Yuan Xiao and Zhenfei Xing at the Analysis and Testing Center of Institute of Hydrobiology for their assistance with providing Transmission Electron Microscope analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.872896/full#supplementary-material
1. Chen L, Xu J, Sun X, Xu P. Research Advances and Future Perspectives of Genomics and Genetic Improvement in Allotetraploid Common Carp. Rev Aquaculture (2021) 2(14):957–78. doi: 10.1111/raq.12636
2. Nakajima T, Hudson MJ, Uchiyama J, Makibayashi K, Zhang J. Common Carp Aquaculture in Neolithic China Dates Back 8,000 Years. Nat Ecol Evol (2019) 3(10):1415–8. doi: 10.1038/s41559-019-0974-3
3. Jung-Schroers V, Adamek M, Harris S, Syakuri H, Jung A, Irnazarow I, et al. Response of the Intestinal Mucosal Barrier of Carp (Cyprinus Carpio) to a Bacterial Challenge by Aeromonas Hydrophila Intubation After Feeding With Beta-1,3/1,6-Glucan. J Fish Dis (2018) 41(7):1077–92. doi: 10.1111/jfd.12799
4. Li H, Zhang F, Guo H, Zhu Y, Yuan J, Yang G, et al. Molecular Characterization of Hepcidin Gene in Common Carp (Cyprinus Carpio L.) and its Expression Pattern Responding to Bacterial Challenge. Fish Shellfish Immunol (2013) 35(3):1030–8. doi: 10.1016/j.fsi.2013.07.001
5. Dworaczek K, Kurzylewska M, Laban M, Pekala-Safinska A, Marczak M, Turska-Szewczuk A. Structure of the Disaccharide Repeating Unit of O-Specific Polysaccharide Isolated From Aeromonas Veronii Strain Bs8 Pathogenic to Common Carp (Cyprinus Carpio). Carbohydr Res (2021) 500:108210. doi: 10.1016/j.carres.2020.108210
6. Dou P, Wang L, Zhang H, Zheng Y. Molecular Identification and Drug Resistance Analysis of Plesiomonas Shigellode Isolated From Fish. Biotechnol Bull (2019) 35(11):118–23. doi: 10.1186/s12917-022-03208-5
7. Janda JM, Abbott SL, McIver CJ. Plesiomonas Shigelloides Revisited. Clin Microbiol Rev (2016) 29(2):349–74. doi: 10.1128/CMR.00103-15
8. Shinohara T, Okamoto K, Koyano S, Otani A, Yamashita M, Wakimoto Y, et al. Plesiomonas Shigelloides Septic Shock Following Ingestion of Dojo Nabe (Loach Hotpot). Open Forum Infect Dis (2021) 8(8):ofab401. doi: 10.1093/ofid/ofab401
9. Wu H-X. Identification and Inhibitory Effects of Drugs Against Pathogenic Bacteria of Soft-Shelled Turtle (Trionyx Sinensis) In Vitro. Fisheries Sci (Liaoning) (2004) 23(6):5–9. doi: 10.1007/s11769-002-0026-8
10. Huang Y, Li Y, Wang R, Xie M, Shi Y, Zhao Z. Calreticulin Functions in Antimicrobial Immunity of Obscure Puffer Takifugu Obscurus. Mol Immunol (2021) 140:77–86. doi: 10.1016/j.molimm.2021.10.008
11. Jahanbakhshi A, Pourmozaffar S, Adeshina I, Mahmoudi R, Erfanifar E, Ajdari A. Selenium Nanoparticle and Selenomethionine as Feed Additives: Effects on Growth Performance, Hepatic Enzymes' Activity, Mucosal Immune Parameters, Liver Histology, and Appetite-Related Gene Transcript in Goldfish (Carassius Auratus). Fish Physiol Biochem (2021) 47(2):639–52. doi: 10.1007/s10695-021-00937-6
12. Nadirah M, Ruhil HH, Jalal KC, Najiah M. Occurrence of Plesiomonas Shigelloides in Cultured Red Hybrid Tilapia (Oreochromis Niloticus) From Tropical Rivers, East Coast Malaysia. Pak J Biol Sci (2012) 15(12):600–3. doi: 10.3923/pjbs.2012.600.603
13. Dai YF, Shen YB, Wang ST, Zhang JH, Su YH, Bao SC, et al. RNA-Seq Transcriptome Analysis of the Liver and Brain of the Black Carp (Mylopharyngodon Piceus) During Fasting. Mar Biotechnol (NY) (2021) 23(3):389–401. doi: 10.1007/s10126-021-10032-9
14. Schneider F, Lang N, Reibke R, Michaely HJ, Hiddemann W, Ostermann H. Plesiomonas Shigelloides Pneumonia. Med Mal Infect (2009) 39(6):397–400. doi: 10.1016/j.medmal.2008.11.010
15. Ozdemir O, Sari S, Terzioglu S, Zenciroglu A. Plesiomonas Shigelloides Sepsis and Meningoencephalitis in a Surviving Neonate. J Microbiol Immunol Infect (2010) 43(4):344–6. doi: 10.1016/s1684-1182(10)60053-9
16. Herrera FC, Santos JA, Otero A, García-López ML. Occurrence of Plesiomonas Shigelloides in Displayed Portions of Saltwater Fish Determined by a PCR Assay Based on the hugA Gene. Int J Food Microbiol (2006) 108(2):233–8. doi: 10.1016/j.ijfoodmicro.2005.12.008
17. Cortes-Sanchez AJ, Espinosa-Chaurand LD, Diaz-Ramirez M, Torres-Ochoa E. Plesiomonas: A Review on Food Safety, Fish-Borne Diseases, and Tilapia. ScientificWorldJournal (2021) 2021:3119958. doi: 10.1155/2021/3119958
18. Zhu W, Zhang Y, Zhang J, Yuan G, Liu X, Ai T, et al. Astragalus Polysaccharides, Chitosan and Poly(I:C) Obviously Enhance Inactivated Edwardsiella Ictaluri Vaccine Potency in Yellow Catfish. Pelteobagrus fulvidraco Fish Shellfish Immunol (2019) 87:379–85. doi: 10.1016/j.fsi.2019.01.033
19. Pence MA. The Brief Case: Wound Infection With Plesiomonas Shigelloides Following a Freshwater Injury. J Clin Microbiol (2016) 54(5):1180–2. doi: 10.1128/JCM.02651-15
20. Mekasha S, Linke D. Secretion Systems in Gram-Negative Bacterial Fish Pathogens. Front Microbiol (2021) 12:782673. doi: 10.3389/fmicb.2021.782673
21. Pekala-Safinska A. Contemporary Threats of Bacterial Infections in Freshwater Fish. J Vet Res (2018) 62(3):261–7. doi: 10.2478/jvetres-2018-0037
22. Fernandes C, Flick G, Silva J, McCASKEY T. Comparison of Quality in Aquacultured Fresh Catfish Fillets II. Pathogens E. Coli O157:H7, Campylobacter, Vibrio, Plesiomonas, and Klebsiella. J Food Prot (1997) 60(10):1182–8. doi: 10.4315/0362-028x-60.10.1182
23. Wang YD, Wang YH, Hui CF, Chen JY. Transcriptome Analysis of the Effect of Vibrio Alginolyticus Infection on the Innate Immunity-Related TLR5-Mediated Induction of Cytokines in. Epinephelus lanceolatus Fish Shellfish Immunol (2016) 52:31–43. doi: 10.1016/j.fsi.2016.03.013
24. Sun F, Peatman E, Li C, Liu S, Jiang Y, Zhou Z, et al. Transcriptomic Signatures of Attachment, NF-kappaB Suppression and IFN Stimulation in the Catfish Gill Following Columnaris Bacterial Infection. Dev Comp Immunol (2012) 38(1):169–80. doi: 10.1016/j.dci.2012.05.006
25. Beck BH, Farmer BD, Straus DL, Li C, Peatman E. Putative Roles for a Rhamnose Binding Lectin in Flavobacterium Columnare Pathogenesis in Channel Catfish. Ictalurus punctatus Fish Shellfish Immunol (2012) 33(4):1008–15. doi: 10.1016/j.fsi.2012.08.018
26. Ordas A, Hegedus Z, Henkel C, Stockhammer O, Butler D, Jansen H, et al. Deep Sequencing of the Innate Immune Transcriptomic Response of Zebrafish Embryos to Salmonella Infection. Fish shellfish Immunol (2011) 31(5):716–24. doi: 10.1016/j.fsi.2010.08.022
27. Chen H, Yuan G, Su J, Liu X. Hematological and Immune Genes Responses in Yellow Catfish (Pelteobagrus Fulvidraco) With Septicemia Induced by. Edwardsiella ictaluri Fish Shellfish Immunol (2020) 97:531–9. doi: 10.1016/j.fsi.2019.11.071
28. Huo X, Fan C, Ai T, Su J. The Combination of Molecular Adjuvant CCL35.2 and DNA Vaccine Significantly Enhances the Immune Protection of Carassius Auratus Gibelio Against CyHV-2 Infection. Vaccines (Basel) (2020) 8(4):2076–393X. doi: 10.3390/vaccines8040567
29. Chen S, Zhou Y, Chen Y, Gu J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics (2018) 34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
30. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-Based Genome Alignment and Genotyping With HISAT2 and HISAT-Genotype. Nat Biotechnol (2019) 37(8):907–15. doi: 10.1038/s41587-019-0201-4
31. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG Resource for Deciphering the Genome. Nucleic Acids Res (2004) 32(Database issue):D277–80. doi: 10.1093/nar/gkh063
32. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: Tool for the Unification of Biology. Gene Ontology Consortium Nat Genet (2000) 25(1):25–9. doi: 10.1038/75556
33. Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, et al. eggNOG V4.0: Nested Orthology Inference Across 3686 Organisms. Nucleic Acids Res (2014) 42(Database issue):D231–9. doi: 10.1093/nar/gkt1253
34. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res (2021) 49(D1):D412–D9. doi: 10.1093/nar/gkaa913
35. BYaH Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc B (1995) 57(1):289–300. doi: 10.2307/2346101
36. Young M, Wakefield M, Smyth G, Oshlack A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol (2010) 11(2):R14. doi: 10.1186/gb-2010-11-2-r14
37. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res (2011) 39:W316–22. doi: 10.1093/nar/gkr483
38. Ekundayo T, Okoh A. Molecular Characterization, Intra-Species Diversity and Abundance of Freshwater Plesiomonas Shigelloides Isolates. Microorganisms (2020) 8(7):2076–607. doi: 10.3390/microorganisms8071081
39. González-Rey C, Svenson SB, Bravo L, Rosinsky J, Ciznar I, Krovacek K. Specific Detection of Plesiomonas Shigelloides Isolated From Aquatic Environments, Animals and Human Diarrhoeal Cases by PCR Based on 23S rRNA Gene. FEMS Immunol Med Microbiol (2000) 29(2):107–13. doi: 10.1111/j.1574-695X.2000.tb01512.x
40. East AK, Allaway D, Collins MD. Analysis of DNA Encoding 23S rRNA and 16S-23S rRNA Intergenic Spacer Regions From. Plesiomonas shigelloides FEMS Microbiol Lett (1992) 74(1):57–62. doi: 10.1016/0378-1097(92)90736-8
41. Gürtler V, Stanisich VA. New Approaches to Typing and Identification of Bacteria Using the 16s-23s rDNA Spacer Region. Microbiol (Reading) (1996) 50(6):3–16. doi: 10.1099/13500872-142-1-3
42. Edwards MS, McLaughlin RW, Li J, Wan X, Liu Y, Xie H, et al. Putative Virulence Factors of Plesiomonas Shigelloides. Antonie Van Leeuwenhoek (2019) 112(12):1815–26. doi: 10.1007/s10482-019-01303-6
43. Maciejewska A, Bednarczyk B, Lugowski C, Lukasiewicz J. Structural Studies of the Lipopolysaccharide Isolated From Plesiomonas Shigelloides O22:H3 (CNCTC 90/89). Int J Mol Sci (2020) 21(18):1422–0067. doi: 10.3390/ijms21186788
44. Chen XQ, Zhao W, Xie SW, Xie JJ, Zhang ZH, Tian LX, et al. Effects of Dietary Hydrolyzed Yeast (Rhodotorula Mucilaginosa) on Growth Performance, Immune Response, Antioxidant Capacity and Histomorphology of Juvenile Nile Tilapia (Oreochromis Niloticus). Fish Shellfish Immunol (2019) 90:30–9. doi: 10.1016/j.fsi.2019.03.068
45. Zhang X, Sun Z, Wang Y, Cao Y, Wang G, Cao F. Enhancement of Growth, Antioxidative Status, Nonspecific Immunity, and Disease Resistance in Gibel Carp (Carassius Auratus) in Response to Dietary Flos Populi Extract. Fish Physiol Biochem (2022) 48(1):67–83. doi: 10.1007/s10695-021-00992-z
46. He S, Zhou Z, Liu Y, Shi P, Yao B, Ringø E, et al. Effects of Dietary Saccharomyces Cerevisiae Fermentation Product (DVAQUA®) on Growth Performance, Intestinal Autochthonous Bacterial Community and non-Specific Immunity of Hybrid Tilapia (Oreochromis Niloticus ♀×O. Aureus ♂) Cultured in Cages. Aquaculture (2009) 294(1-2):99–107. doi: 10.1016/j.aquaculture.2009.04.043
47. Kui L, Liu Y. Effects of Inactivated Edwardsiella Ictaluri Vaccine on Expression of Immune-Related Genes and Activities of Catalase, Superoxide Dismutase and Complement C3 in Yellow Catfish. Fisheries Sci (2015) 34:150–4. doi: 10.16378/j.cnki.1003-1111.2015.03.004
48. Lin SM, Li FJ, Yuangsoi B, Doolgindachbaporn S. Effect of Dietary Phospholipid Levels on Growth, Lipid Metabolism, and Antioxidative Status of Juvenile Hybrid Snakehead (Channa Argusx Channa Maculata). Fish Physiol Biochem (2018) 44(1):401–10. doi: 10.1007/s10695-017-0443-3
49. Zhang HG, Jiang N, Zhang YL, Zhang YL, Ni Q, Fan YD. Effects of Tank Diameter and Water Depth on Growth Performance, Antioxidant Capacity and Immune Response of Chinese Sturgeon Acipenser Sinensis in Recirculating Aquaculture System. J Appl Ichthyology (2022) 38:84–92. doi: 10.1111/jai.14285
50. Perez-Lopez A, Behnsen J, Nuccio SP, Raffatellu M. Mucosal Immunity to Pathogenic Intestinal Bacteria. Nat Rev Immunol (2016) 16(3):135–48. doi: 10.1038/nri.2015.17
51. Ashley B, Yousef AK. Evolution of the Arsenal of Legionella Pneumophila Effectors To Modulate Protist Hosts. mBio (2018) 9(5):01313–8. doi: 10.1128/mBio.01313-18
52. Bermudez-Brito M, Munoz-Quezada S, Gomez-Llorente C, Matencio E, Bernal MJ, Romero F, et al. Cell-Free Culture Supernatant of Bifidobacterium Breve CNCM I-4035 Decreases Pro-Inflammatory Cytokines in Human Dendritic Cells Challenged With Salmonella Typhi Through TLR Activation. PloS One (2013) 8(3):e59370. doi: 10.1371/journal.pone.0059370
53. Sahoo BR. Structure of Fish Toll-Like Receptors (TLR) and NOD-Like Receptors (NLR). Int J Biol Macromol (2020) 161:1602–17. doi: 10.1016/j.ijbiomac.2020.07.293
54. Chai Q, Wang L, Liu CH, Ge B. New Insights Into the Evasion of Host Innate Immunity by. Mycobacterium tuberculosis Cell Mol Immunol (2020) 17(9):901–13. doi: 10.1038/s41423-020-0502-z
55. Dixon B, Barreda D, Sunyer J. Perspective on the Development and Validation of Ab Reagents to Fish Immune Proteins for the Correct Assessment of Immune Function. Front Immunol (2018) 9:2957. doi: 10.3389/fimmu.2018.02957
56. Khalil S, Reda R, Awad A. Efficacy of Spirulina Platensis Diet Supplements on Disease Resistance and Immune-Related Gene Expression in Cyprinus Carpio L. Exposed to Herbicide Atrazine Fish Shellfish Immunol (2017) 67:119–28. doi: 10.1016/j.fsi.2017.05.065
57. Wang GL, Wang MC, Zhang XW, Chang MX, Xie HX, Nie P. Molecular Cloning, Biological Effect, and Tissue Distribution of Interleukin-8 Protein in Mandarin Fish (Siniperca Chuasti) Upon Flavobacterium Columnare Infection. Fish Shellfish Immunol (2017) 66:112–9. doi: 10.1016/j.fsi.2017.05.016
58. Kim KH, Kim HC, Park CJ, Park JW, Lee YM, Kim WJ. Interleukin-8 (IL-8) Expression in the Olive Flounder (Paralichthys Olivaceus) Against Viral Hemorrhagic Septicemia Virus (VHSV) Challenge. Dev Reprod (2019) 23(3):231–8. doi: 10.12717/DR.2019.23.3.231
59. Laing K, Zou J, Wang T, Bols N, Hirono I, Aoki T, et al. Identification and Analysis of an Interleukin 8-Like Molecule in Rainbow Trout. Oncorhynchus mykiss Dev Comp Immunol (2002) 26(5):433–44. doi: 10.1016/s0145-305x(01)00092-1
60. Zhang C, Lu K, Wang J, Qian Q, Yuan X, Pu C. Molecular Cloning, Expression HSP70 and its Response to Bacterial Challenge and Heat Stress in. Microptenus salmoides Fish Physiol Biochem (2020) 46(6):2389–402. doi: 10.1007/s10695-020-00883-9
61. Pietretti D, Wiegertjes G. Ligand Specificities of Toll-Like Receptors in Fish: Indications From Infection Studies. Dev Comp Immunol (2014) 43(2):205–22. doi: 10.1016/j.dci.2013.08.010
62. Liu F, Su B, Fu Q, Shang M, Gao C, Tan F, et al. Identification, Characterization and Expression Analysis of TLR5 in the Mucosal Tissues of Turbot (Scophthalmus Maximus L.) Following Bacterial Challenge. Fish shellfish Immunol (2017) 68:272–9. doi: 10.1016/j.fsi.2017.07.021
63. Eslamloo K, Caballero-Solares A, Inkpen S, Emam M, Kumar S, Bouniot C, et al. Renibacterium Salmoninarumtranscriptomic Profiling of the Adaptive and Innate Immune Responses of Atlantic Salmon to Infection. Front Immunol (2020) 11:567838. doi: 10.3389/fimmu.2020.567838
Keywords: natural infection, bacteria identification, pathogenicity, histopathological analysis, tissue bacterial load, comparative transcriptome
Citation: Chen H, Zhao Y, Chen K, Wei Y, Luo H, Li Y, Liu F, Zhu Z, Hu W and Luo D (2022) Isolation, Identification, and Investigation of Pathogenic Bacteria From Common Carp (Cyprinus carpio) Naturally Infected With Plesiomonas shigelloides. Front. Immunol. 13:872896. doi: 10.3389/fimmu.2022.872896
Received: 10 February 2022; Accepted: 01 June 2022;
Published: 30 June 2022.
Edited by:
Yihong Chen, South China Normal University, ChinaReviewed by:
Eliška Zusková, University of South Bohemia, CzechiaCopyright © 2022 Chen, Zhao, Chen, Wei, Luo, Li, Liu, Zhu, Hu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daji Luo, bHVvZGFqaUBpaGIuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.