- 1Translational Type 1 Diabetes Research, Department of Clinical Research, Steno Diabetes Center Copenhagen, Gentofte, Denmark
- 2Interventional Regenerative Medicine and Imaging Laboratory, Department of Radiology, Stanford University School of Medicine, Palo Alto, CA, United States
- 3Research Center for Molecular Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
- 4Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, Copenhagen, Denmark
- 5Center for Non-Coding RNA in Technology and Health, University of Copenhagen, Copenhagen, Denmark
- 6Department of Veterinary and Animal Sciences, University of Copenhagen, Copenhagen, Denmark
- 7Copenhagen Diabetes Research Center, Department of Pediatrics, Herlev University Hospital, Herlev, Denmark
- 8Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
Differential microRNA (miRNA or miR) regulation is linked to the development and progress of many diseases, including inflammatory bowel disease (IBD). It is well-established that miRNAs are involved in the differentiation, maturation, and functional control of immune cells. miRNAs modulate inflammatory cascades and affect the extracellular matrix, tight junctions, cellular hemostasis, and microbiota. This review summarizes current knowledge of differentially expressed miRNAs in mucosal tissues and peripheral blood of patients with ulcerative colitis and Crohn’s disease. We combined comprehensive literature curation with computational meta-analysis of publicly available high-throughput datasets to obtain a consensus set of miRNAs consistently differentially expressed in mucosal tissues. We further describe the role of the most relevant differentially expressed miRNAs in IBD, extract their potential targets involved in IBD, and highlight their diagnostic and therapeutic potential for future investigations.
Introduction
Inflammatory bowel disease (IBD) is an idiopathic, chronic inflammation that primarily affects the gastrointestinal tract. IBD patients experience frequent hospital admissions, many operations, and poor quality of life due to the disease complications (1, 2). Like many other immune-related diseases, the etiology of IBD is not well understood. However, it is generally believed to be a multifactorial disease where environmental factors, genetics, immune dysregulation, and microbiome dysbiosis trigger an inappropriate immune response in lamina-propria, which challenges mucosal homeostasis (3).
Ulcerative colitis (UC) and Crohn’s disease (CD) are the two major types of IBD. While CD shows a patchy transmural inflammatory pattern, UC is mainly limited to the innermost layers and rarely affects other layers of the intestine wall (1, 2). CD is associated with many pathophysiological complications, and its clinical symptoms vary according to the disease location (4). UC is more prevalent and mainly affects the colon (rectum) and generally has a milder course, with patients less prone to disease complications (5, 6). Genome-wide association studies (GWAS) identified 245 unique IBD loci. These susceptible loci are crucial in defining the disrupted intestinal immune system and disease pathways and constitute a solid genetic component of IBD (7–9).
Advances in IBD genetics, high-throughput sequencing technologies, and transcriptome studies provide new insights associated with noncoding RNAs, including long noncoding RNAs (lncRNAs) (10) and microRNAs (miRs or miRNAs) in various diseases (11, 12). Differentially expressed miRNAs are highly correlated with inflammatory and autoimmune disorders, including psoriasis (13), rheumatoid arthritis (14), multiple sclerosis (15), and IBD (16, 17). Mature miRNAs are short (~22 nt long) single-stranded noncoding RNAs derived from pre-miRNA hairpins (typically ~80 nt), and many of these are further processed from primary miRNA transcripts (pri-miRNA) of several hundred nucleotides when multiple pre-miRNAs are contained. The pri-miRNA can be intergenic or of intronic origin nucleotides and can be evolutionarily conserved. MiRNAs are involved in regulating gene expression post-transcriptionally (17–19), where the mature miRNA binds to its target typically with a seed sequence of 6 nucleotides from position 2-7 and with the remaining part binding often with a few nucleotide bulges.
Various studies indicate that differentially expressed miRNAs affect mRNA at several levels of regulation: transcriptional, post-transcriptional, chromatin modification, and genomic imprinting. miRNAs can affect biological processes through endogenous RNA competition, regulation of RNA transcription, protein sponges, and translation regulation. These regulations can cause decreased stability and translational repression that affects various biological functions, including proliferation, migration, cell signaling, autophagy, and apoptosis (3, 17, 20, 21). It is estimated that miRNAs regulate more than 60% of the mRNA through complementary pairing at 3′ untranslated regions (UTRs) (20). miRNAs are not only acting as local regulators within the cells; they also can be found in places far from their origin and are directly or indirectly involved in virtually all types of regulation of biological processes in living organisms (17, 21).
Furthermore, some miRNAs are stable in body fluids such as serum, plasma, urine, saliva, and other tissues (22–25). Many efforts are currently ongoing to identify differentially expressed miRNAs in IBD as biomarkers. Since the expression of differentially expressed miRNA in IBD and many other diseases seems to happen early in the disease, the evaluation of circulating miRNA or tissue-specific levels could be helpful for early diagnosis and successful treatments. Thus, it is highly important to study miRNA-expression profiles and their target genes as biomarkers for diagnosis, prognosis, progression, and treatment response.
This review presents an overview of current knowledge on differentially expressed miRNAs in IBD patients’ mucosal tissues and peripheral blood. To enrich the findings from the literature, we combined the literature curation with a meta-analysis of publicly available miRNA high-throughput datasets in mucosal tissues. We further discuss the importance of the most relevant miRNAs in the disease based on the available knowledge and suggest the miRNA participation role in developing chronic inflammation that characterizes pathogenesis. Finally, we discuss the relevance of miRNA differential expression for prediction/early diagnosis, disease progression and treatment responses, and the obstacles in the way.
Differentially Expressed miRNAs in IBD Patient’s Mucosal Tissues
Ulcerative Colitis
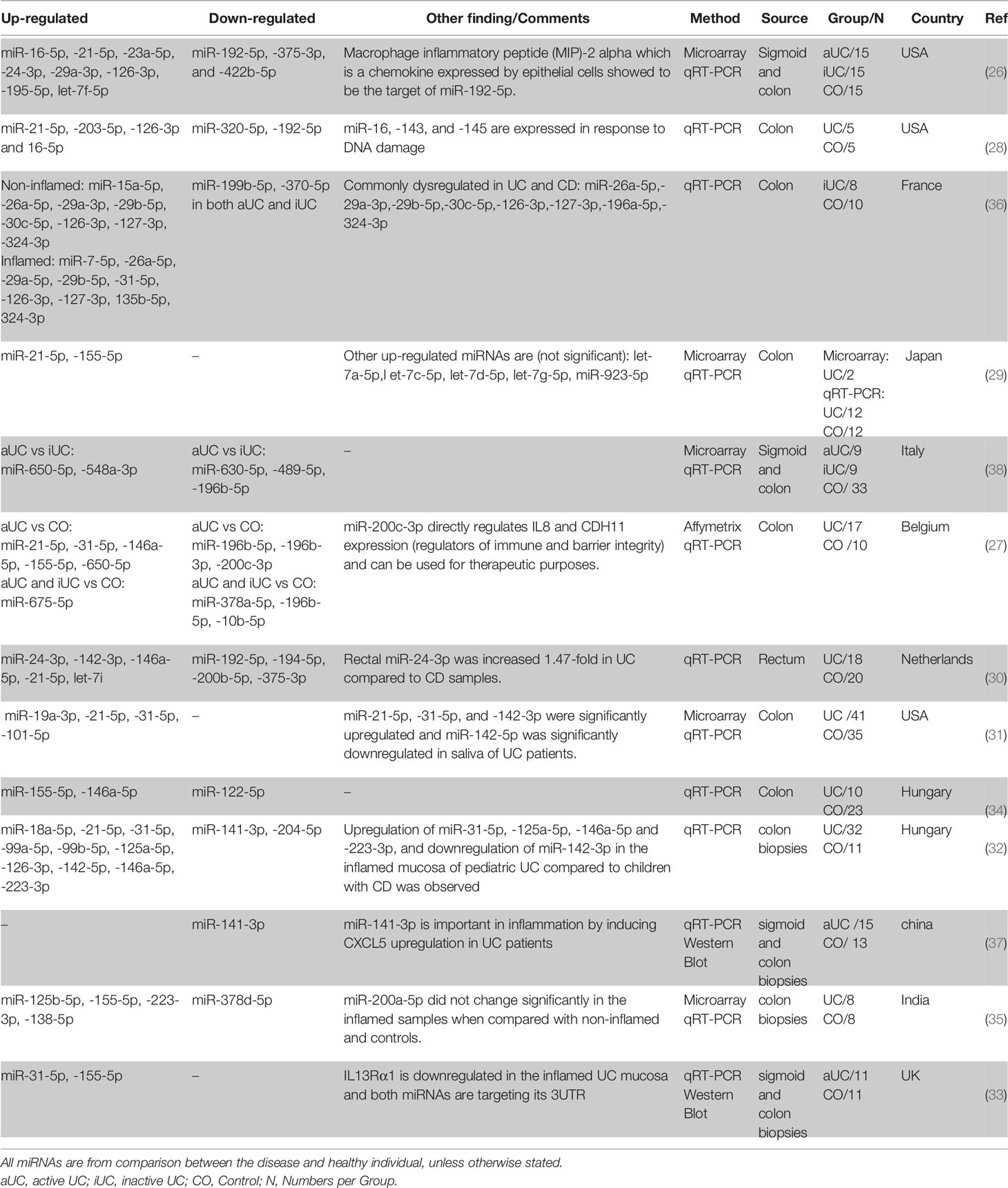
The first miRNA profiling study in IBD was performed in 2008 and compared biopsy samples from patients with active UC (aUC), inactive UC (iUC), chronic active CD (aCD), microscopic colitis, infectious colitis, and irritable bowel syndrome with healthy controls (26). Eleven miRNAs were differentially expressed in patients with aUC compared to the controls. miR-192-5p, miR-375-3p, and miR-422b-5p were significantly downregulated, and miR-16-5p, miR-21-5p, miR-23a-5p, miR-24-3p, miR-29a-3p, miR-126-3p, miR-195-5p, and let-7f-5p were significantly upregulated (26).
Following this pioneering observation, subsequent studies have identified many new miRNAs while reconfirming already identified ones. It is not surprising that the findings are not consistent as many variables differ between studies, including treatment, inflammatory status, disease duration, anatomical biopsy locations, different healthy control cohorts, and miRNAs profiling platforms. Regardless of these differences, several miRNAs are frequently reported as being differentially expressed. miR-21-5p (26–32), miR-155-5p (27, 29, 33–35), miR-31-5p (27, 31, 33, 36), miR-146a-5p (27, 30, 32, 34), miR-126-3p (26, 28, 32), miR-29a-3p (26, 36), miR-16-5p (26, 28), miR-223-3p (32, 35) and miR-24-3p (26, 30) showed to be constantly upregulated while miR-192-5p (26, 28, 30), miR-141-3p (32, 37), and miR-375-3p (26, 30) were downregulated in UC biopsies when compared to control biopsies (in at least two independent studies). Also, many miRNAs showed differential regulation when inactive UC is compared with active UC and control (Table 1).
Crohn’s Disease
In another pioneering study in 2010, Fasseu et al. identified 14 and 23 miRNAs differentially expressed (0.001< p <0.05) in iUC and inactive CD (iCD) patients, respectively (Tables 1, 2). Among them, 8 were commonly differentially expressed in iUC and iCD (miR-26a-5p, miR-29a-3p, miR-29b-5p, miR-30c-5p, miR-126-3p, miR-127-3p, miR-196a-5p, miR-324-3p). Further analysis showed that miR-26a-5p, miR-29b-5p, miR-126-3p, miR-127-3p, and miR-324-3p had coordinated differential regulation in the non-inflamed and inflamed colonic mucosa of IBD patients. On the other hand, miR-196b-5p, miR-199a-3p, miR-199b-5p, miR-320a-5p, miR-150-5p, and miR-223-3p demonstrated significant difference when non-inflamed UC and CD colonic biopsies were compared. Based on this screening, the authors suggested an important role of miRNAs in the inflammation at onset and/or relapse of IBD patients with quiescent mucosal tissues (36).
Succeeding studies have identified several miRNAs consistently shown to be differentially expressed between CD and control biopsies, including always upregulated miR-146a-5p (30–32, 34, 43), miR-21-5p (30–32, 39, 47), miR-31-5p (31, 32, 42, 45, 46), miR-223-3p (32, 39, 42, 48), miR-142-3p (30, 32, 45), let-7i-5p (30, 44), miR-23b-3p (39, 40), miR-106a-5p (39, 40) and constantly downregulated miR-192-5p (41, 44, 48), miR-194-5p (30, 48) and miR-375-3p (30, 31). There are also miRNAs with conflicting results including miR-150-5p (up in (32, 45), down in (42)), miR-19b-3p (up in (40), down in (39)), miR-215-5p (up in (42), down in (48)), and miR-629-5p (up in (40), down in (39)). Moreover, several miRNAs showed differential regulation when iCD compared with aCD and control (Table 2).
Differentially Expressed miRNAs in IBD Patient’s Peripheral Blood
Ulcerative Colitis
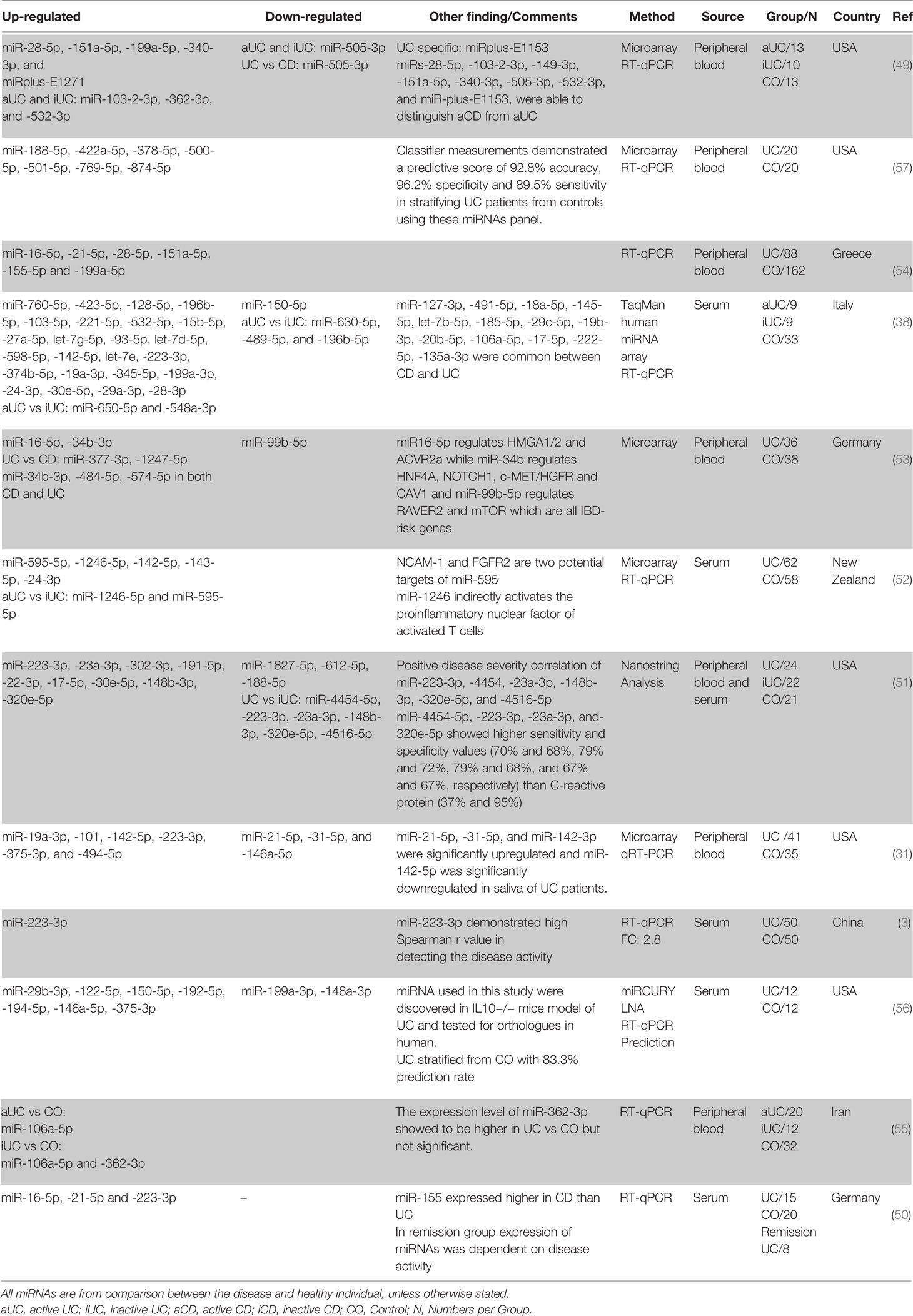
Similar to the findings in tissue biopsies, miRNAs are also differentially expressed in the peripheral blood of UC patients. In a first study, Wu et al. compared the circulating miRNA profile of whole blood of aUC and iUC patients and healthy individuals (49). Their microarray investigation showed a significant increase in the expression level of twelve miRNAs, while one, miR-505-3p, showed a significant decrease when comparing patients with aUC with healthy controls. miR-505-3p expression was decreased around 7-fold in active outpatient blood. In contrast, 3.1- and 5.2-fold expression increases were demonstrated in the blood of the active UC patients for miR-103-2-3p and miR-362-3p, respectively. Furthermore, a comparison between the circulating miRNA in the peripheral blood of UC patients with healthy individuals revealed a significant increase in the expression level of the miR-28-5p, miR-151a-5p, miR-199a-5p, miR-340-3p, and miRplus-E1271 in patients with aUC but not in iUC. Wu et al. further demonstrated that miRs-103-2-3p, miR-362-3p, and miR-532-3p are upregulated in both aUC and iUC. Following this initial study, in attempts to identify circulating miRNAs that contribute to UC development and to find proper biomarker candidates, many studies have been performed. From these studies miR-223-3p (3, 31, 38, 50, 51), miR-142-5p (31, 38, 52), miR-16-5p (50, 53, 54), miR-151a-5p (49, 54), miR-199a-5p (49, 54), miR-19a-3p (31, 38), miR-24-3p (38, 52), miR-28-5p (49, 54), miR-30e-5p (38, 51), miR-362-3p (49, 55) showed consistent upregulation in at least two independent studies, whereas none of the downregulated miRNAs had been validated in more than one study (possibly due to biases in which miRNAs are picked for validation). Moreover, miR-21-5p (up in (49, 50), down in (31)), miR-146a-5p (up in (56), down in (31)), miR-150-5p (up in (56), down in (38)), miR-188-5p (up in (57), down in (51)), miR-199a-3p (Up in (38), down in (56)) showed inconsistent differential regulation between different studies. miRNAs differential regulation was also detected when iUC was compared with aUC and control. miR-362-3p is the only miRNA that shows upregulation in two independent studies when iUC was compared with healthy control (49, 55) (Table 3).
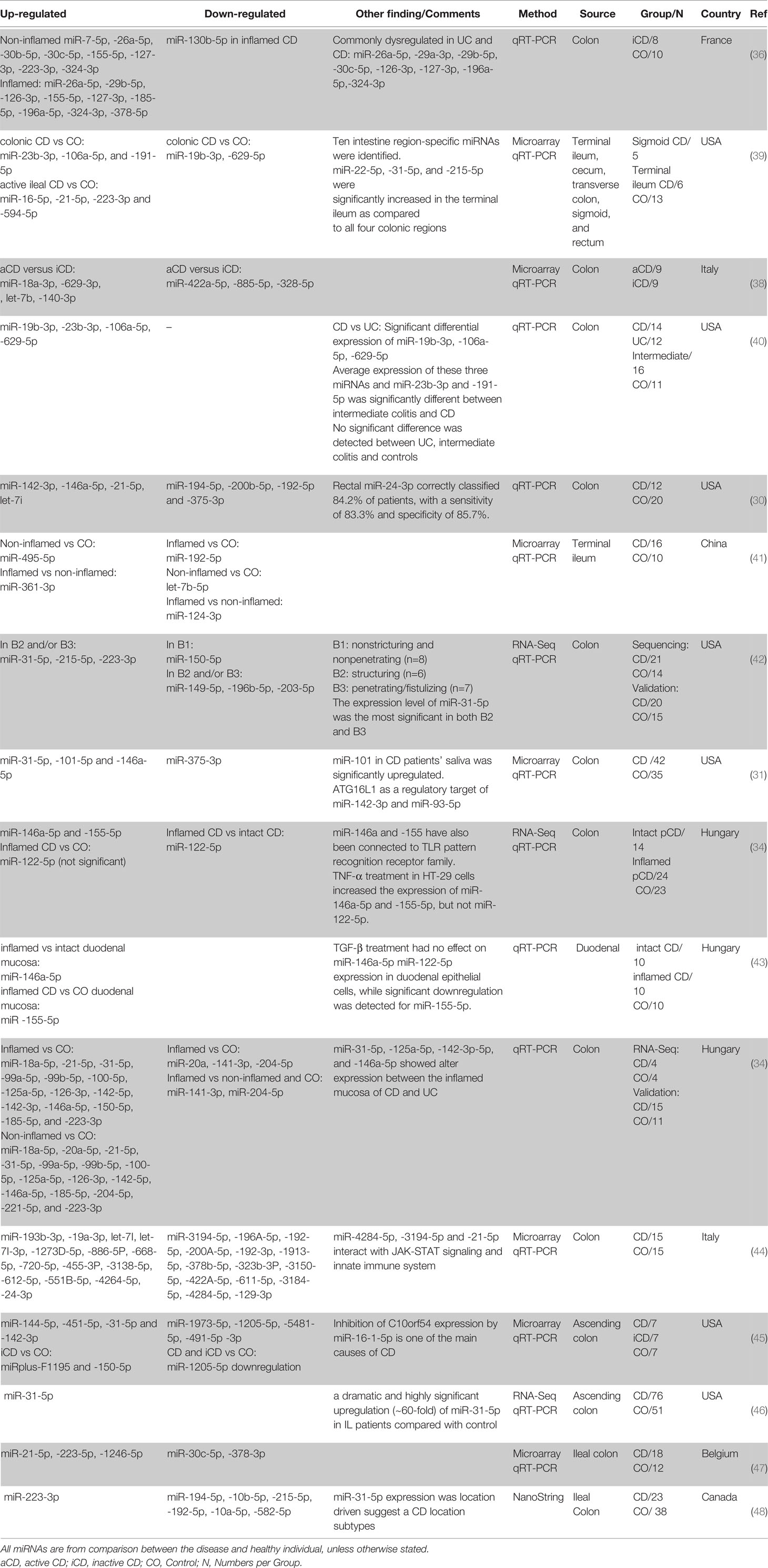
Crohn’s Disease
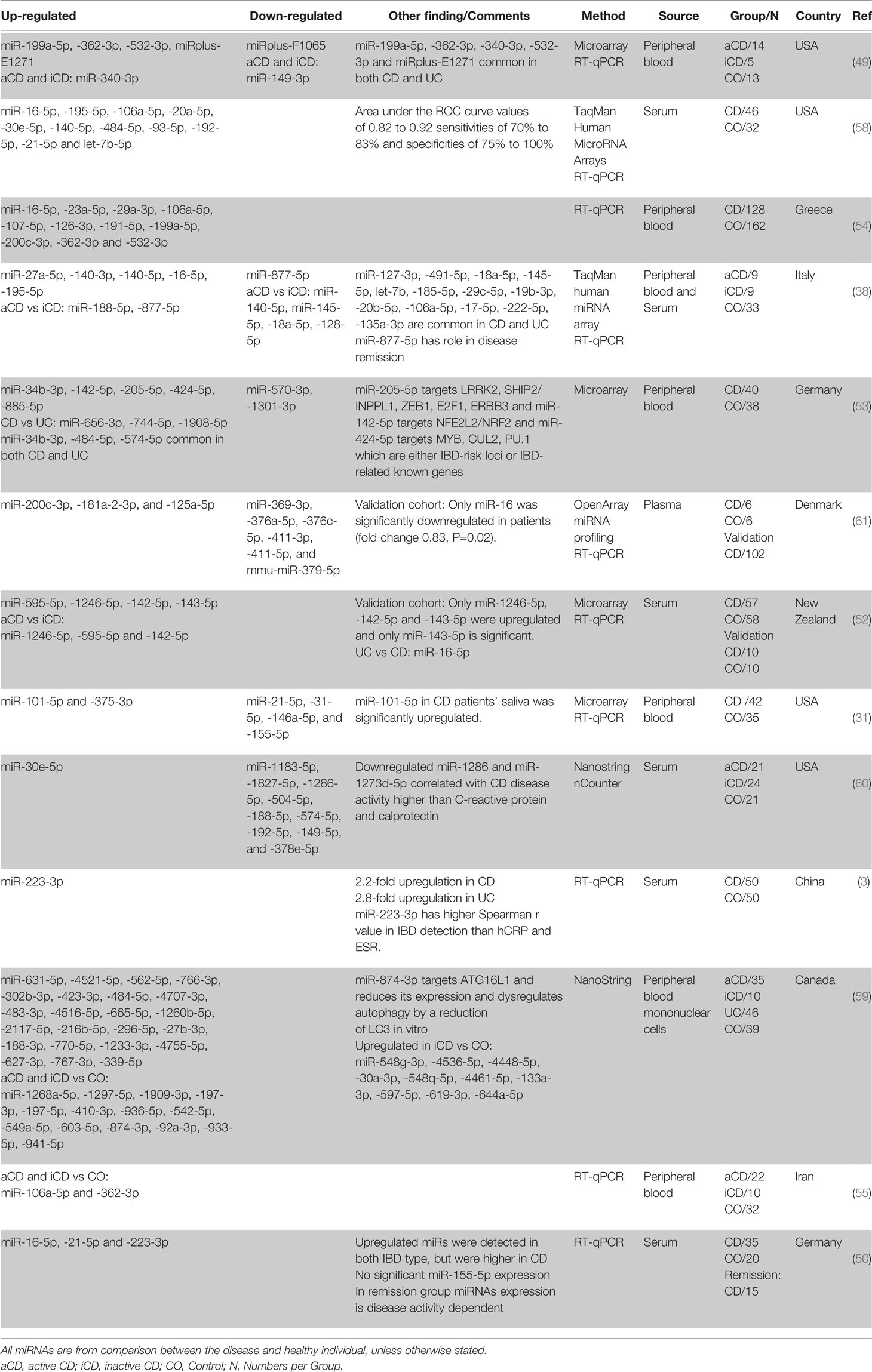
One of the first studies using whole blood for distinguishing CD patients from normal healthy individuals using miRNA profile was done by Wu et al. (49). Comparing the circulating miRNA of the aCD patients with healthy controls showed a significant increase in the expression of five miRNAs and a significant decrease in two others. Among them, miR-362-3p showed the most significant difference in expression of a 4.7-fold increase. Interestingly the expression of miR-340-3p showed a significant increase, and miR-149-3p showed a significant decrease in both active and inactive CD patients compared to the healthy controls.
Subsequent studies found miR-16-5p (38, 50, 54, 58), miR-484-5p (53, 58, 59), miR-362-3p (49, 54, 55), miR-106a-5p (54, 55, 58), miR-532-3p (49, 54), miR-30e-5p (58, 60), miR-223-3p (3, 50), miR-21-5p (50, 58), miR-200c-3p (54, 61), miR-199a-5p (49, 54), miR-195-5p (38, 58), miR-142-5p (52, 53), miR-140-5p (38, 58) to be consistently upregulated in CD patients in comparison with healthy controls (in at least two independent studies). However, similar to the UC studies, based on the lists manually curated from literature, no circulating miRNA is always downregulated when CD is compared to healthy controls (in more than one study). This could be because the main focus for blood-based biomarker discovery is on the upregulated miRNAs, not the downregulated ones. There are also miR-574-5p (up in (53), down in (60)) and miR-192-5p (up in (58), down in (60)) that were shown to be differentially expressed inconsistently between studies. Moreover, several circulating miRNAs showed differential regulation when iCD compared with aCD and control (Table 4).
Computational Meta-Analysis of Publicly Available High Throughput Studies
In addition to the literature curation, we also performed a meta-analysis of publicly available high throughput studies (microarray and RNA-Seq), including 3 UC (27, 62, 63) and 4 CD (42, 47, 62, 64) patient cohorts (Table 1). All included studies contained expression profiling at the level of the intestinal mucosa (colon or ileum). We combined the results of differential expression analysis between the UC or CD and the control group from each study as described in the supplementary section. The three UC datasets are consistent with each other, with most differentially expressed miRNAs being changed in the same direction, in contrast to the CD datasets, where many miRNAs are differentially expressed in opposite directions between the datasets (Supplementary Figure 1). The higher heterogeneity observed in the expression profiles from CD patients might be consistent with the more heterogeneous nature of CD compared to UC. There might also be other explanations, including different patients’ demographics, different sample handling, and data generation in different labs.
We obtained a final set of 158 miRNAs consistently differentially expressed between UC patients and controls and 69 miRNAs between CD patients and controls (p-value < 0.05 in at least two datasets and a global adjusted combined logit p-value < 0.05) and consistent in the direction of regulation across all datasets (Supplementary Files 1, 2 and Supplementary Figure 2).
The meta-analysis confirms most of the literature-curated miRNAs and at the same time provides dozens of other miRNAs not previously reported in UC or CD mucosa (e.g., miR-378a-3p, miR-191-5p, miR-92a-3p in UC; miR-30e-5p, miR-26b-5p, let-7f-5p, let-7g-5p, in CD; miR-146b-5p, miR-30d-5p, miR-148a-3p, miR-151a-5p in both UC and CD). In addition, few miRNAs showed different or no significant differential regulation compared to what was found in the literature, including miR-142-3p (30, 32, 45) in CD, which in literature curation showed to be constantly upregulated, while in the meta-analysis, it was constantly downregulated.
Moreover, miRNAs reported in the literature are predominantly upregulated (specifically for UC); however, the meta-analysis indicates an almost equal number of up- and downregulated miRNAs. This might be ascribed to the ease/bias of validation for the upregulated miRNAs for diagnostic purposes with available techniques. Furthermore, the downregulated miRNAs showed a higher average expression, possibly indicating a more substantial functional role of these miRNAs (65) (Supplementary Figure 3).
One of the studies (GSE89667) contained both UC and CD cohorts (62), and we used the UC versus CD comparison (adjusted p-value < 0.05), in conjunction with the results of the meta-analysis, to find a set of 18 miRNAs differentially expressed between UC and CD. Among these, e.g., miR-29a-3p, miR-155-5p, or miR-454-3p are upregulated in UC compared to CD, while miR-28-3p, miR-378a-5p or miR-422a are downregulated in UC compared to CD (Supplementary Data, Sheet 3).
Overlap of Colon and Blood miRNAs in UC and CD
There is great potential in identifying disease-specific miRNAs for diagnosis, progression, and therapeutic response. Consistently differentially expressed miRNAs in the colon and blood may have the highest clinical potential. From literature curation, 29 miRNAs were consistently differentially expressed in at least two studies in colon or blood of UC or CD (Figure 1).
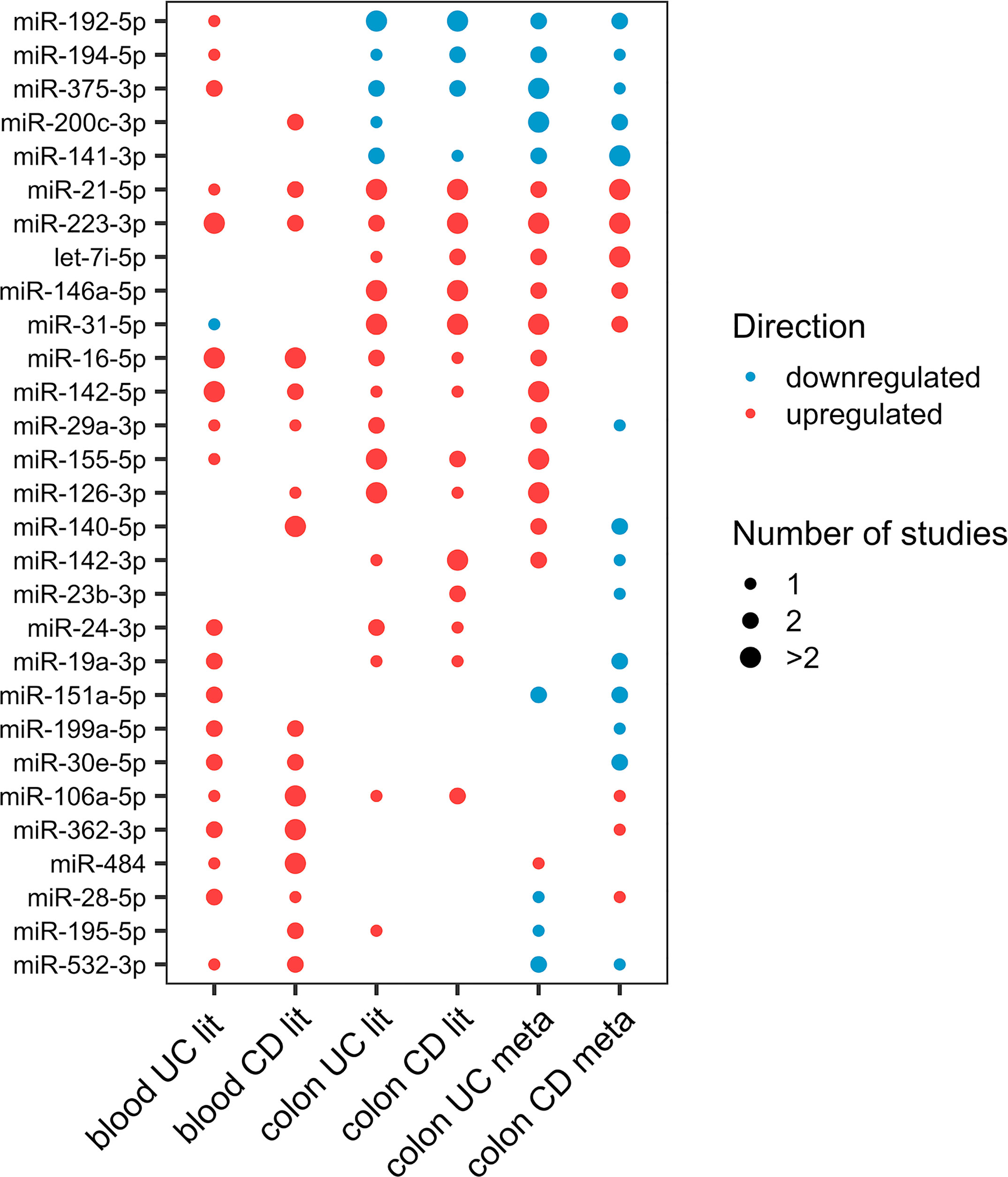
Figure 1 Dot-plot of the 29 differentially expressed miRNAs (at least two studies) in either colon or blood of UC or CD from literature. The node size represents the number of studies, and the node color corresponds to the expression statues, where red means upregulation and blue means downregulation.
Ulcerative Colitis
From the miRNAs with consistent differential regulation in at least two independent studies, miR-223-3p, miR-16-5p, and miR-24-3p showed upregulation in both mucosa and blood of UC patients compared with healthy individuals. miR-21-5p and miR-146a-5p were also shown to be differentially expressed in both tissues. However, the blood data for these two miRNAs were inconsistent. Considering only one study, 18 miRNAs were commonly differentially expressed in both tissues (Supplementary Data, Sheet 4).
Crohn’s Disease
From the miRNAs with consistent differential regulation in at least two independent studies in CD miR-223-3p and miR-21-5p showed upregulation in both mucosa and blood of patients compared with healthy individuals. miR-192-5p was also common and frequently downregulated in the mucosa; however, since the data for this miRNA in the blood is inconsistent, it was excluded. Finally, considering miRNAs differentially expressed in only one study, 13 miRNAs were shown to be commonly differentially expressed in both tissues (Supplementary Data, Sheet 5).
UC and CD miRNA Profile Similarities and Differences
Even the most experienced clinicians have problems in the initial diagnosis of IBD and stratifying its subtypes. Stratifying UC and CD has always been a challenge ascribed to their overlapping features. Although these IBD subtypes have common characteristics, significant genetic and clinical differences exist. Consequently, different transcriptome profiles, specifically distinct miRNAs signatures, might improve IBD subtype classification.
Colon
Many studies compared individuals with and without the diseases to stratify UC and CD based on mucosa biopsy miRNA signature (30–32, 34, 36, 38). Considering miRNAs validated to be differentially expressed in at least two studies in both UC and CD mucosa, miR-21-5p, miR-31-5p, miR-146a-5p, miR-223-3p showed to be commonly up- and miR-192-5p and miR-375-3p downregulated in both phenotypes.
Furthermore, considering miRNAs with consistent differential regulation in at least two independent studies, miR-155-5p, miR-126-3p, miR-29a-3p, miR-141-3p, miR-16-5p and miR-24-3p showed to be differentially expressed mainly in UC mucosa, while miR-142-3p, miR-150-5p, let-7i-5p, miR-23b-3p, miR-19b-3p, miR-215-5p, miR-629-5p, miR-194-5p and miR-106a-5p showed to be more frequently differentially expressed in CD mucosa.
To confirm the above observation, these miRNAs (from at least two studies) were more intersected against the literature miRNA lists, this time one study and more. The comparison showed that miR-29a-3p is only reported as significantly differentially expressed (SDE) in UC, and miR-23b-3p is only reported as SDE in CD. Moreover, the results for miR-150-5p and miR-215-5p were inconsistent.
Blood
Similar attempts to stratify UC and CD based on the blood miRNA profile of patients versus healthy individuals were made (3, 31, 38, 49, 50, 52–55). Considering frequently differentially expressed miRNAs in UC and CD blood, miR-223-3p, miR-142-5p, miR-16-5p, miR-199a-5p, miR-30e-5p, miR-362-3p were significantly differentially upregulated and were common between both phenotypes and thus could be considered as IBD biomarkers.
Furthermore, considering miRNAs with consistent differential regulation in at least two independent studies, miR-146a-5p, miR-150-5p, miR-151a-5p, miR-188-5p, miR-199a-3p, miR-19a-3p, miR-24-3p, miR-28-5p showed to be mainly differentially expressed in UC. miR-484-5p, miR-106a-5p, miR-574-5p, miR-532-3p, miR-200c-3p, miR-195-5p, miR-192-5p, miR-140-5p showed to be more frequently differentially expressed in CD blood.
To confirm this observation, these differentially expressed miRNAs (from at least two studies) in each phenotype were once more intersected against the literature miRNA lists, this time one study and more. The results showed miR-146a-5p, miR-150-5p, miR-151a-5p, miR-199a-3p, miR-19a-3p, miR-24-3p were only SDE in UC. The results for miR-150-5p and miR-199a-3p were inconsistent. Furthermore, miR-200c-3p, miR-195-5p, and miR-140-5p showed only SDE in CD.
Most Relevant Differentially Expressed miRNAs
To develop miRNA-based novel diagnostics and therapeutics for IBD, it is vital to understand the miRNAs expression changes in correlation with the disease phenotype, underlying mechanisms that regulate miRNAs, the target genes, and their interplay. Despite the heterogeneity of differentially expressed miRNAs in IBD, 66 miRNAs were identified from literature curation and meta-analysis as relevant candidates for diagnostic or therapeutic purposes that might also represent causative agents in disease development (Supplementary data, Sheet 6). For this set of miRNAs, we extracted “experimentally observed targets” from QIAGEN Ingenuity Pathway Analysis (IPA) software program v70750971 (66) and intersected these targets with genes related to IBD extracted from IPA and literature (Supplementary data, Sheet 7). This resulting list of 28 miRNAs with at least one IBD target was visualized in Cytoscape (67) (Figure 2). In the following, we discuss most of these miRNAs in more detail.
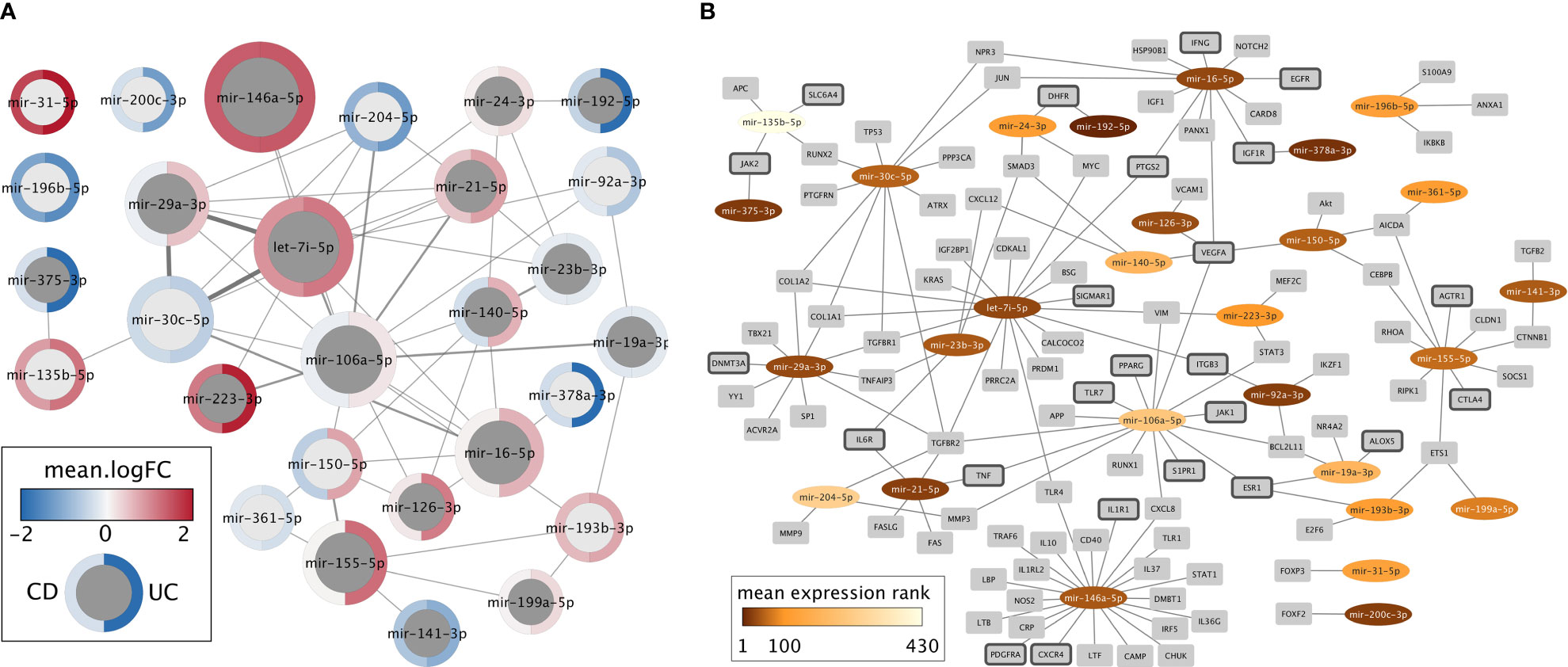
Figure 2 Network representations of the 28 miRNAs with at least one experimentally determined target known to be related to IBD. (A) Network of miRNAs only. Dark gray nodes represent miRNAs detected by literature curation, while light gray nodes were not identified in the literature, but only in the meta-analysis. The size of each miRNA node corresponds to the number of IBD targets this miRNA has, and the width of the edges represents the number of shared IBD targets. The mean logFC of each miRNA, according to the meta-analysis, is shown for CD (left) and UC (right) using a blue-white-red gradient on the node border. (B) Network of miRNAs (oval nodes) and their target genes (rectangle nodes). miRNAs are colored based on their mean expression rank. Target genes that code for proteins with a clinically approved drug according to the Pharos database are highlighted by dark gray node border color.
Let-7i-5p: Let-7i-5p is the regulator of TLR4, which is important in cytokine-mediated responses and a regulator of IL-6 (68). In THP-1 cells transfected with let-7i-5p mimics, both mRNA and protein levels of TLR4 showed downregulation (69). Let-7i-5p seems to assist cells in resetting their protein profile in response to external stimuli in allergic inflammation; the exact mechanism is not yet clear (70). Let-7i-5p regulates collagens, IL-6, TGF-βR1, IGF-1, and caspase-3 as primary regulators of inflammation, fibrosis, hypertrophy, and apoptosis (68).
miR-16-5p: miR-16-5p in the colonic UC mucosa partly regulates the inflammatory responses through negative regulation of A2aAR (NF-κB inhibitor) expression. miR-16-5p mimics transfection in colonic epithelial cells, demonstrated to increase nuclear translocation of NF-κB p65 protein and thus increase the expression of IFN-γ and IL-8 as important pro-inflammatory cytokines (71).
miR-19a-3p: Serum miRNA profiling of CD patients with and without strictures showed miR-19a-3p and miR-19b-3p as potential pathogenic markers (72). Low levels of miR-19a-3p and miR-19b-3p were strongly correlated with stricturing CD and independent of site, gender, age, disease duration, and activity (72). Moreover, it has been reported that miR-19a-3p decreases the SOCS3 expression, which consequently enhances IFN-α and IL-6 signal transduction (73).
miR-21-5p: miR-21-5p showed an essential role in colon epithelial cell hemostasis (74), adaptive immune responses (75), cytokine regulation (76), and IBD-related complications (77). It has been demonstrated that in response to epithelial damage, miR‐21-5p causes more intestinal permeability. Transfection of miR-21-5p mimics resulted in the loss of tight junction proteins, increased barrier permeability (74), and decreased CD3 and CD68 positive cells in the UC mouse model (78). The miR-21-5p knockout mice model also showed high resistance to dextran sulfate sodium (DSS) induced colitis, suggesting the pro-apoptotic effect of this miRNA. miR-21-5p also demonstrated an essential role in adaptive immune responses in T-cell function, with the highest detected expression in effector T cells, memory T cells, and the lowest in naive T cells (75). miR-21-5p has a regulatory role in innate immunity and is involved in TLR4 activation and monocyte differentiation. It is also induced by danger signals, such as activators of NF-kB in a negative feedback loop, to prevent damage (79). miR-21-5p is associated with disease activity in UC patients (80). Moreover, this miRNA regulates IL-12 release from dendritic cells and macrophages by targeting the IL-12p35 receptor (76). On the other hand, the association of this miRNA with irreversible IBD fibrosis and its increased level was observed in serum of humans with significant fibrosis (77) and development of dysplasia (81). It is noteworthy that several cellular injury models have shown to be TNF-α dependent with subsequent miR-21-5p induction (77, 82).
miR-23b-3p: miR-23b-3p represses autoimmune inflammation by suppressing (IL-17, TNF-α, IL-1β)-induced NF-κB activation, inflammatory cytokine expression by targeting TGF-β-activated kinase 1/MAP3K7 binding protein 2 (TAB2), TAB3 and inhibitor of NF-κB kinase subunit α. Conversely, IL-17 contributes to autoimmune pathogenesis by suppressing miR-23b-3p expression and promoting proinflammatory cytokine expression (83).
miR-24-3p: miR-24-3p is reported to be involved in T cells proliferation, differentiation, and immune response (84). It is also reported that miR-24-3p targets Bcl-2 and PAK4 as prosurvival genes, thus, inducing cell death (85). Overexpression of PMS2L2 prompts miR-24-3p gene methylation, resulting in its inhibition. PMS2L2 overexpression, stimulated by LPS, is shown to promote Bcl-2 expression and to inhibit Bax, cleaved-caspase-3, and cleaved-caspase-9 expressions (86). Furthermore, miR-24-3p regulates the processing of latent TGF-β1 release by furin targeting (87). miR-24-3p is reported to downregulate not only TGF-β1, furin, and TNFAIP3 (88).
miR-28-5p: miR-28-5p are shown to be involved in cell proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT) (89). miR-28-5p can silence PD1 genes and regulate the PD1, Foxp3 positive and TIM3, Foxp3 positive, exhaustive Treg cells (90).
miR-29a-3p: miR-29a-3p has a seven-nucleotide wide binding site on the 3’UTR of the MCL-1 gene and could be involved in the UC pathogenesis through regulating this gene. Mcl-1 gene knockout is shown to cause apoptosis in the colonic epithelial HT29 cells (91). Increased expression of miR-29a-3p in the colon tissues of patients with irritable bowel syndrome increased intestinal membrane permeability, regulating the GLUL gene (92). Moreover, miR-29a-39 is reported to regulate pro-inflammatory cytokine secretion and scavenger receptor expression via LPL targeting in ox LDL-stimulated dendritic cells (93).
miR-30d-5p and miR-30c-5p: Oral administration of miR-30d-5p mimic ameliorates experimental autoimmune encephalomyelitis (EAE) through expansion of Tregs. In Akkermansia muciniphila, miR-30d-5p regulates lactase expression and increases Akkermansia abundance in the gut. Consequently, Akkermansia increases Tregs to suppress EAE symptoms (94). miR-30c-5p regulates ATG5 expression by targeting the 3’UTR (95). The inverse correlation between miR-30c-5p and ATG5 is not only observed in CD patients and intestinal epithelial T84 cells infected with the adherent-invasive Escherichia coli (AIEC) (95). The NF-κB pathway was shown to be activated in AIEC infected T84 cells, which induced the up-regulation of miR-30c-5p and consequently inhibited the ATG5 expression (95). It has further been reported that the autophagic activity inhibition by miR-30c-5p increased AIEC persistence within T84 cells and increased pro-inflammatory cytokines production (95). miR-30c-5p is also believed to regulate Th17 cells differentiation by targeting its negative regulators such as SMAD2, SMAD4, TGFβR2, SOCS3, FOXO3, and TSC1 (96). Thus, their differential regulation might cause an increase or decrease in Th17 cell numbers. ETS1, BCL6, and STAT1 are also among the important targets of miR-30c-5p (96).
miR-31-5p: miR-31-5p showed a gradual upregulation from normal to IBD conditions and seemed to target FIH-1, the inhibitor of HIF-1α protein (97). Also, in psoriasis, miR-31-5p inhibition in keratinocytes was shown to suppress NF-kB–driven promoter-luciferase activity and production of IL-1β, CXCL1, and CXCL5. miR-31-5p regulates these cytokine and chemokine expressions in endothelial cells and attracts leukocytes via STK40 as its primary target (98). miR-31-5p also targets Gprc5a, which is shown to be a critical regulator for peripherally derived regulatory T cells generation. miR-31-5p conditional deletion enhances induction of these regulatory T cells and decreases the severity of experimental autoimmune encephalomyelitis (99). IL-13 is a necessary type-2 T-helper cytokine, controlling epithelium function through the IL-13 receptor -A1. It has been shown that the transfection of miR-31-5p and miR-155-5p mimics reduces the expression of the IL-13 receptor, increases and blocks the phosphorylation of STAT6, and the expression of SOCS1 and CCL26 in the gut epithelium cell line, and therefore may contribute to disease aggravation (33). Furthermore, miR-31-5p is differentially expressed in post-ablation epithelium with increased barrier permeability (100).
miR-106a-5p: Serum level of miR-106a-5p in both CD and UC patients correlates with disease severity (55). Upon T cell activation, while most miRNAs are downregulated, miR-106a-5p is upregulated (101). In addition, in macrophages, miR-106a-5p can regulate SIRPα synthesis and, therefore, SIRPα-mediated inflammatory responses (102). miR-106a-5p deficiency showed to promote Treg induction IL-10 production and attenuate adoptive transfer colitis in T cell restricted deficiency (103). In non-colonic cell lines, miR-106a-5p regulates IL-10 expression (103). Moreover, in CD4+ T cells, miR-106a-5p miRNA family deletion also attenuated the inflammation in lymphopenic recipients. Global knock-out of miR-106a-5p was also shown to attenuate chronic murine ileitis (104). TGFβ appears to suppress miR-106a under physiological conditions to aid Treg induction. TNFα, on the other hand, appears to drive upregulation of miR-106a-5p under inflammatory conditions through NF-κB-dependent induction of the miR-106a-5p promoter, resulting in temporary suppression of normal immune regulation (104).
miR-126-3p: IκBα as the inhibitor of NF-κB was shown to be markedly decreased in active UC tissues (105). miR-126-3p and IκBα expression are inversely correlated in patients with active UC. miR-126-3p is shown to contribute to UC pathogenesis through binding to the 3′- UTR of IκBα and inhibiting the NF-κB signaling pathway (105). Anti-inflammatory activities of the red wine polyphenols were partly mediated through miR-126-3p induction (106). Polyphenolic red wine extract (WE) inhibited inflammation in LPS-stimulated human colon-derived CCD-18Co cells by inhibiting NF-κB and down-regulating pro-inflammatory agents, including TNF-α, IL-6, and CAMs. miR-126-3p was upregulated upon WE treatment in these cells, and NF-κB and VCAM-1 showed downregulation (107). VCAM-1 is one of the miR-126-3p targets (108). miR-126-3p knockdown is reported to up-regulate the PIK3R2 in CD8+ T cells (109) and alter the PI3K/Akt pathway activation responsible for regulatory T cells reduced induction and suppressive function (109). Moreover, IκB, an inhibitor of NFκB, is another target of miR-126-3p (109).
miR-140-5p: miR-140-5p is shown to downregulate TLR4 by being directly bound to its 3′UTR, which inhibits inflammatory cytokines secretion (110). Moreover, it has been demonstrated that miR-140-5p inhibited IL-6 and IL-8 secretion by regulating TLR4 expression (110).
miR-141-3p: miR-141-3p is aberrantly expressed in IBD and other autoimmune diseases, including lupus and psoriasis (111, 112). miR-141-3p targets CXCL12β (113), an epithelial cell-expressed chemokine whose inverse correlation with miR-141 is shown in the inflammation. Therefore, it is suggested that targeting CXCL12β by miR-141-3p might influence inflammatory cell trafficking into the inflamed sites. Thus, inhibiting colonic CXCL12β expression and blocking immune cell recruitment might be valuable for the CD treatment (113). miR-141-3p is also reported to suppress STAT4, thus, inhibiting inflammatory factors (114). miR-141-3p upregulation reduces the IL-1β, TNF-α, and IL-6 levels, consequently attenuating the chronic inflammatory pain severity (115). Furthermore, during Th17 cell induction, miR-141-3p expression is reported to be significantly upregulated (116). miR-141-3p can also exert protective effects on cell damage (114). It is also reported that miR-141-3p alleviates LPS-induced intestinal epithelial cell injury by inhibiting RIPK1-mediated necroptosis and inflammation (117).
miR-142-5p(-3p): In thymically derived Tregs, miR-142-5p is the predominant isoform. Tregs limit the development of autoimmunity by suppressing self-reactive peripheral T effector cell responses (118). miR-142-5p is shown to target SMAD3, CYR61 (119), and PD-L1 (120). Regulation of PD-L1 expression is through binding to its UTR and inversely correlated with miR-142-5p (121). TNF-α, IFN-γ, and IL-10, as prominent players in the immune response, are related to the PD-L1/PD-1 pathway. It has been shown that miR-142-5p overexpression results in TNF-α and IFN-γ upregulation and IL-10 downregulation (121). ATG16L1, as one of the most commonly detected genetic variations in CD patients, is predicted to be the target of miR-142-3p (122, 123). miR-142-3p negatively regulates ATG16L1 in CD colon epithelial cells. Upregulation of miR-142-3p reduced the autophagic activity of thymic-derived regulatory T cells by decreasing the expression of ATG16L1 (124). miR-142-3p binds directly to KDM6A (a lysine demethylase), resulting in the demethylation of H3K27me3, an epigenetic modification to the DNA packaging protein Histone H3. This, in turn, upregulates the expression of the anti-apoptotic protein Bcl-2. It has also been shown that antagomir-mediated knockdown of miR-142-3p can affect the induced regulatory T cells regulatory function, cytokine expression, and apoptosis through Foxp3 expression (125). Moreover, downregulation of miR-142-3p in macrophages of aged mice contributed to IL-6-associated aging disorders and consequently age-related inflammatory diseases (126).
miR-146a-5p and miR-146b-5p(-3p): miR-146a-5p has previously been shown to regulate the innate immune responses and TNF-α pathway in skin inflammation (127). miR-146a-5p deficient mice also develop immune disorders (128). In IBD, this miRNA regulates NOD2 derived gut inflammation and promotes proinflammatory cytokines released from activated macrophages (129). Moreover, upregulation of miR-146a-5p in monocytes in response to LPS resulted in the downregulation of TLR4 signaling pathway downstream genes (130). On the other hand, in mouse colitis, miR-146b-5p overexpression was shown to alleviate intestinal injury via NF-κB activation, epithelial barrier function improvement, and increased survival rate (131). miR-146b-5p seems to up-regulate NFκB via siah2 suppressing. Siah2 prompts TRAF proteins ubiquitination which is upstream of NFκB (131). miR-146b-3p, another member of the miR-146 family, is shown to negatively regulate lipid kinase PI3Kγ in (132), suppress proinflammatory ADA2, and block TNF-α secretion (133). Furthermore, miRNA-146b-3p expression is significantly downregulated by increased STAT3 activation (134).
miR-149-5p: Through targeting MyD88, miR-149-5p negatively regulates TLR triggered inflammatory cytokine production (135). MyD88 is involved in the TLR/NF-κB pathway. miR-149-5p is also associated with an increased IBD risk in the Chinese population (136).
miR-150-5p: miR-150-5p is proposed as one of the primary regulators of immune diseases (137), mainly through inhibiting inflammatory cytokines including IL-6, IL-1β, and TNF-α (138). It is also reported that the miR-150-5p upregulation in immune cells promotes the proliferation and maturation of myeloid cells and lymphocytes (139). c-Myb, a target of miR-150-5p, is reported to be significantly downregulated in UC patients’ colon and DSS-treated mice. miR-150-5p overexpression is reported to enhance apoptosis through targeting c-Myb, which damages the intestinal epithelial barrier (140).
miR-155-5p: miR-155-5p has shown a central regulatory role in innate and acquired immune systems. miR-155-5p is expressed in response to inflammatory mediators such as LPS, TLR ligands, and IFN-β and is induced in antigen-presenting cells, including plasmacytoid dendritic cells and macrophages. It has also been found that antigen-stimulated B and T cells induce miR-155-5p expression (141). Moreover, SOCS1, a negative regulator for activation of LPS-induced macrophage, JAK/STAT signal pathway, and antigen presentation by dendritic cells, is one of the main targets of miR-155-5p (142). In addition, Anti-miR-155-5p has been reported to suppress G-CSF, a regulator of granulopoiesis produced by macrophages during acute inflammation (143). Increasing expression of the level of this miRNA has also been shown in other inflammatory disorders, such as rheumatoid arthritis (144), atopic dermatitis (145), and multiple sclerosis (146). In addition, it has been reported that miR-155-5p is an oncogene (147).
miR-192-5p: miR-192-5p is shown to target MIP-2α (CXCL2), a CXC chemokine expressed by epithelial cells and essential in murine and human IBD. miR-192-5p is downregulated in inactive UC and demonstrated an inverse correlation with MIP2-α. miR-192-5p mimic was reported to inhibit MIP2-α induced MIP-2a expression (26). miR-192-5p is induced by TGF-β and TNF-α (26, 39) and regulates the collagen and chemokine expression, which are critical in inflammation and fibrosis (148). miR-192-5p is also identified as a tumor suppressor that can induce cell cycle arrest (149).
miR-193b-3p: miR-193b-3p differential regulation has been detected in several autoimmune diseases (150), mainly through inflammatory chemokines regulation (151). miR-193b-3p has been shown to target TGF-β2 and TGFBR3 3′-untranslated regions (152) and contribute to Th17 cells differentiation by inhibiting the negative regulators of Th17 differentiation expression and possibly through regulating TLR and Notch signaling pathways. Thus, suggesting the possible involvement of miR-193b-3p in the inflammatory response and Th17 function (153).
miR-194-5p: miR-194-5p is abundant in intestinal epithelial cells (39) and is shown to regulate the MAP4K4/c-Jun/MDM2 signaling pathway (154). Overexpression of miR-194-5p in the liver mesenchymal cells reduced the N-cadherin (155). In the Caco-2 intestinal epithelial cell model, HNF-1α induced miR-194-5p suggesting the influence on epithelial cell differentiation (156).
miR-195-5p: miR-195-5p is shown to correlate with IBD severity. An increase in miR-195-5p level can decrease c-Jun and p65 expression. Instead, miR-195-5p decreased expression increases Smad7 expression and consequently p65 and the AP-1 upregulation, which might explain the steroid resistance mechanism in some UC patients (157). miR-195-5p overexpression was shown to reduce M1-like macrophage polarization. miR-195-5p levels are reported as upregulated in M2c macrophages. LPS and IFN-γ stimulated THP-1 macrophages had reduced TLR2 levels following miR-195-5p overexpression. miR-195-5p also significantly decreased IL-1β, IL-6, and TNF-α levels in M1-stimulated macrophage supernatant cultures. In addition, levels of phosphorylated forms of p54 JNK, p46 JNK and p38 MAPK were shown to decrease by adding miR-195-5p in M1 macrophages upon stimulation. Altogether it seems like miR-195-5p is involved in macrophage polarization by inhibiting TLR2 inflammatory pathway mediators (158).
miR-199a-5p: miR-199a-5p showed significant upregulation in blood from UC patients compared with healthy controls (54). miR-199a-5p seems to suppress HIF-1α and SIRT1 and play a role in Treg cell differentiation by inhibiting genes involved in Th17 differentiation while activating others in Treg development (159, 160). RORγt is a lineage-specific transcription factor for Th17 differentiation. In multiple sclerosis, RORγt expression, a predicted target for miR-199a-5p (using miRWalk, miRTarBase, DIANA miRPath, UniGene), showed a significantly higher level in the relapsing phase versus remitting phase. This is consistent with the upregulation of miR-199a-5p, which correlates with lower Th17 cells and lower expression of RORγt in remitting phase (96). It has also been reported that miR-199-5p targets the activin A receptor type 1B gene that causes decreased CCAAT/enhancer-binding protein α expression and eventually monocyte/macrophage differentiation inhibition (161).
miR-200c-3p: miR-200c-3p plays a role in the FN1 post-transcriptional regulation; hence, EMT triggers by their downregulation (162, 163) most probably by regulating the E-cadherin transcriptional repressors ZEB1 and SIP1 (164). miR-200c-3p is reported to suppress the IL-6, CXCL9, and TNF-α expression (165). IL-6 intensifies inflammation through miR-200c-3p downregulation (166). In a macrophage-like human monocytic cell line exposed to the TLR4 ligand LPS, miR-200c-3p inhibits NF-κB activation in response to a TLR4 agonist. miR-200c-3p is known to regulate the TLR4 signaling efficiency through the MyD88-dependent pathway (167).
miR-223-3p: miR-223-3p is shown to be involved in the activation of granulocytes and is overexpressed in naive CD4+ T-lymphocytes (168). Furthermore, the downregulation of miR-223-3p in primary macrophages increased TLR4 and STAT3 basal expression and LPS-stimulated TLR4, STAT3, and NOS2 expression. On the contrary, miR-223-3p mimics treatment in primary macrophages has decreased TLR4 expression while negatively regulating FBXW7 expression, a well-known suppressor of TLR4 signaling. Based on these outcomes, it is concluded that miR-223-3p abundance in macrophages can change macrophage activation and modulate the response to stimuli via effects on the TLR4/FBXW7 axis (169). It has also been shown that miR-223-3p mediates the cross-talk between the intestinal barrier and the IL-23 pathway by targeting CLDN8, a claudin protein that constitutes the backbone of the intestinal barrier (170). miR-223-3p has also been used as a biomarker in IBD (3). Thus, the evidence suggests its proinflammatory role and highlights its potential as a RNA biomarker that seems to be conserved between different species. miR-223-3p is also produced by neutrophils and monocytes and acts as a controller of NLRP3 inflammasome activity, regulating the intestine inflammatory process by affecting IL-1β production (171).
miR-375-3p: miR-375-3p is reported to be downregulated in the intestinal mucosa of UC and CD patients. TLR4 is one of the main targets of miR-375-3p with inverse correlation. miR-375-3p mediated upregulation of TLR4 induces NF-κB activation, which leads to an increase in pro-inflammatory factors (172). Intestines show a high level of miR-375-3p expression. Cell death, including apoptosis and/or necrosis, results in the miR-375-3p leak from cellular to extracellular space, eventually ending in the blood. Therefore, it is suggested that elevated miR-375-3p in serum may be a predictor of tissue damage (173).
miR-378a-3p: miR-378a-3p expression is reported to be inversely correlated with IL-33 expression; IL-33 is a predicted target of miR-378a-3p (174). miR-378a-3p is highly conserved between species, but not IL-33 (175). The miR-378a-3p is located in intron 1 of the PPARGC1B gene that is differentially regulated in UC patients’ intestinal mucosa 26. PPARGC1B protein is highly expressed in the intestinal epithelium (176) and is involved in the control of mitogenesis and mitochondrial metabolism (177), energy production, and biogenesis (178). Therefore, it can be concluded that in inflamed mucosa, the miR-378a-3p decrease might reflect a metabolic shift, possibly related to the increment of energy expenditure and ROS overproduction (179).
miR-424-5p: miR-424-5p is shown to control monocyte/macrophage differentiation. miR-424-5p expression upregulation is regulated by transcription factor PU.1. When upregulated, miR-424-5p induces monocyte differentiation via NFI-A inhibition (180) as its main target.
miR-532-3p: miR-532-3p acts as an antagonist for LPS/TNF-α stimulated macrophages by targeting the ASK1/p38 MAPK signaling pathway, thus suppressing the inflammation, which is mediated through this pathway. Thus, it has been suggested as a potential target for treating autoimmune inflammatory diseases (181).
Concluding Remarks
Early diagnosis and treatment are vital in IBD, as induction of early remission and maintenance can prevent long-term complications and eliminate the need for surgery. However, due to insufficient clinical sensitivity and specificity of current biomarkers and a large population of patients with functional bowel disorders, there is often a delay in the confident diagnosis of IBD and its sub-classification into either UC or CD (182). At the same time, the primary way to overcome IBD is to induce and maintain early remission. Most current IBD diagnostic tests reflect generalized inflammation and do not discriminate between IBD subtypes (182).
Since their discovery, thousands of miRNAs have been identified. Accumulating evidence suggests that specific miRNA expression signatures contribute to the IBD development and progression. Most studies reveal correlations between IBD and differentially expressed miRNAs instead of causal relationships. As discussed above, only a few studies investigate the underlying molecular mechanisms of the disease; thus, the precise function of most miRNAs in IBD has yet to be clarified. Furthermore, there has been a lack of reproducibility between studies, partly ascribed to a lack of standardized study designs and different approaches.
Moreover, many variables differ between studies, including age, sex, various treatment regimens, disease activity level and duration, having different control groups, sampling from different anatomic locations, sampling method, preservation and processing of the samples, and the different criteria for measuring expression fold change and significances (e.g., different FC, log FC, p-value and p-adj criteria). Thus, it is essential to understand the conditions under which a differentially expressed miRNA was discovered. For instance, epigenetic regulations are among the primary factor stimulated by the environment. Stimuli such as diet, lifestyle, work condition, and stress are elements as important as the clinical and technical manifestations of signs of disease. Regardless of these differences, while being aware of them, in this review, we attempted to identify and give an overview of the most frequently differentially expressed miRNAs in colon and blood of both UC and CD across multiple studies from literature and meta-analysis and further described the roles of selected miRNAs in the disease pathogenesis and their connection to IBD.
For biomarker studies, circulating miRNAs (of saliva, serum, urine, plasma, and other body fluids) attracted great interest as non- or semi-invasive clinical biomarkers mainly due to ease of access, stability, conserved structure, and ease of detection by quantitative approaches like real-time PCR. The need for endoscopic examination and invasive sampling of biopsies limit the use of colonic miRNAs as biomarkers. Thus, if a miRNA demonstrates a similar consistent differential regulation in colonic biopsies and blood of the IBD patients compared with healthy control, it can be used as a proper disease biomarker signature. miR-223-3p, in this case, might be an excellent example of such miRNAs. This miRNA is significantly differentially expressed in both UC and CD in blood and tissue biopsies and thus can be considered a reliable IBD biomarker candidate.
Anti-cytokines therapies have been relatively successful; however, not all patients respond to these treatments. As important post-transcriptional gene regulators, miRNAs were shown to contribute to disease aggravation through immune responses, inflammation, mucus barrier, and epithelium function dysregulation; thus, miRNA-based therapy might be developed as a potential therapeutic approach. In this case, miRNAs complementary antisense oligonucleotides or miRNA mimics can be potential therapeutics that abolish or mimic miRNA’s function and, therefore, block inflammatory progression, modulate cytokines or chemokine hemostasis and increase the treatment sensitivity of conventional therapies. As such, miRNAs are used for modulating hypoxia (183, 184) and the inflammatory response by targeting major inflammatory pathways (185–189) and essential molecules, including tight junction proteins that maintain the integrity of the membrane (74, 190, 191).
Future Perspectives
Although progress has been made towards understanding the role of miRNAs in IBD pathophysiology, many conditions and many more miRNAs remain insufficiently characterized for diagnostic and therapeutic applications, partly as it is still a relatively young field. Also, as a chronic disease with flare-ups and remission, besides comparing disease versus control, it is relevant to look at disease subgroups, e.g., the differences between active/inactive and inflamed/not inflamed intestinal regions. While some studies grouped patients into active UC, inactive UC, inflamed UC, and non-inflamed UC, still further studies are needed to improve our understanding. In addition, it remains to be determined how associations with IBD risk loci might affect miRNA’s expression and the disease phenotype. Moreover, although it has been less focused on, the disease activity index can also be assessed by profiling miRNA specifically at different disease stages while maintaining that miRNA expression is often tissue or pathology specific.
Due to the IBD complexity and the lack of consistency between miRNA signatures, it is difficult to diagnose the disease, identify the subtypes, and monitor the disease status or location using a single or even a panel of miRNAs. Although there is an imperious need for faster ways to validate miRNAs as biomarkers, the sensitivity and specificity of miRNA candidates should be checked in large-scale studies to avoid false positive or false negative diagnosis.
Differentially expressed miRNAs profiling can be a valuable indication of phenotypic changes in IBD, showing an obvious correlation with disease evolution. However, differential expression per se does not indicate the ultimate role of the identified miRNAs in disease pathophysiology, as there are complex networks of interaction between miRNAs and their targets that also depend on the cell type, location, and tissue condition. It is noteworthy that many miRNAs might have the same target. Thus, when it comes to the therapeutic interventions using the miRNAs, the main issue is the side effects of miRNA-based drugs that need to be considered in extensive validation studies before miRNAs can enter the market and be incorporated into clinical practice. Also, miRNA expression as measured on high-throughput platforms, e.g., RNA-sequencing, has limitations. For example, if a highly expressed target is downregulated, the expression of the miRNA will appear as increased despite the miRNA being processed at the same rate, i.e., miRNA itself is not directly regulated. Extending miRNA analysis to be “target context-aware” rather than looking at miRNA solely from small RNA-sequencing will likely shed more nuances on to cause and effect of regulated miRNAs and thereby pave the way for considering miRNAs in diseases. Despite the present limitations, we anticipate that miRNAs application and targeting will become routine diagnostic and therapeutic approaches in clinical settings as current techniques evolve rapidly.
Author Contributions
All authors have made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Independent Danish Research Foundation, Technology, and Production, grants 4005-00443 and 8020-00300B, the Novo Nordisk Foundation, grant NNF14CC0001, Lundbeck Foundation, grant R303-2018-3148, and the Sehested Hansen foundation. The funders had no role in study design, data collection, analysis, or manuscript preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.865777/full#supplementary-material
References
1. Kalla R, Ventham NT, Satsangi J, Arnott IDR. Crohn’s Disease. BMJ (2014) 349:g6670. doi: 10.1136/bmj.g6670
2. Ford AC, Moayyedi P, Hanauer SB. Ulcerative Colitis. BMJ (2013) 346(feb05 2):f432–f. doi: 10.1136/bmj.f432
3. Wang H, Zhang S, Yu Q, Yang G, Guo J, Li M, et al. Circulating MicroRNA223 Is a New Biomarker for Inflammatory Bowel Disease. Medicine (2016) 95(5):e2703. doi: 10.1097/MD.0000000000002703
4. Baumgart DC, Sandborn WJ. Crohn’s Disease. Lancet (2012) 380(9853):1590–605. doi: 10.1016/S0140-6736(12)60026-9
5. Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, Grand RJ, et al. The Prevalence and Geographic Distribution of Crohn’s Disease and Ulcerative Colitis in the United States. Clin Gastroenterol Hepatol (2007) 5(12):1424–9. doi: 10.1016/j.cgh.2007.07.012
6. Danese S, Fiocchi C. Ulcerative Colitis. N Engl J Med (2011) 365(18):1713–25. doi: 10.1056/NEJMra1102942
7. de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, et al. Genome-Wide Association Study Implicates Immune Activation of Multiple Integrin Genes in Inflammatory Bowel Disease. Nat Genet (2017) 49(2):256–61. doi: 10.1038/ng.3760
8. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-Microbe Interactions Have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature (2012) 491(7422):119–24. doi: 10.1038/nature11582
9. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res (2019) 47(D1):D1005–D12. doi: 10.1093/nar/gky1120
10. Yarani R, Mirza AH, Kaur S, Pociot F. The Emerging Role of lncRNAs in Inflammatory Bowel Disease. Exp Mol Med (2018) 50(12):161. doi: 10.1038/s12276-018-0188-9
11. Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol (2010) 2(4):161–79.
12. Zhang L, Wu H, Zhao M, Chang C, Lu Q. Clinical Significance of miRNAs in Autoimmunity. J Autoimmun (2020) 109:102438. doi: 10.1016/j.jaut.2020.102438
13. Hawkes JE, Nguyen GH, Fujita M, Florell SR, Callis Duffin K, Krueger GG, et al. microRNAs in Psoriasis. J Invest Dermatol (2016) 136(2):365–71. doi: 10.1038/JID.2015.409
14. Chen X-M, Huang Q-C, Yang S-L, Chu Y-L, Yan Y-H, Han L, et al. Role of Micro RNAs in the Pathogenesis of Rheumatoid Arthritis: Novel Perspectives Based on Review of the Literature. Medicine (2015) 94(31):e1326. doi: 10.1097/MD.0000000000001326
15. Wu T, Chen G. miRNAs Participate in MS Pathological Processes and Its Therapeutic Response. Mediat Inflamm (2016) 2016:4578230. doi: 10.1155/2016/4578230
16. Chapman CG, Pekow J. The Emerging Role of miRNAs in Inflammatory Bowel Disease: A Review. Ther Adv Gastroenterol (2015) 8(1):4–22. doi: 10.1177/1756283X14547360
17. Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, et al. MicroRNAs: New Players in IBD. Gut (2015) 64(3):504–17. doi: 10.1136/gutjnl-2014-307891
18. Lin S-L, Miller JD, Ying S-Y. Intronic microRNA (miRNA). J BioMed Biotechnol (2006) 2006(4):26818. doi: 10.1155/JBB/2006/26818
19. MacFarlane L-A R, Murphy P. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics (2010) 11(7):537–61. doi: 10.2174/138920210793175895
20. Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most Mammalian mRNAs are Conserved Targets of microRNAs. Genome Res (2009) 19(1):92–105. doi: 10.1101/gr.082701.108
21. Kim DH, Saetrom P, Snøve O Jr., Rossi JJ. MicroRNA-Directed Transcriptional Gene Silencing in Mammalian Cells. Proc Natl Acad Sci USA (2008) 105(42):16230–5. doi: 10.1073/pnas.0808830105
22. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in Body Fluids–the Mix of Hormones and Biomarkers. Nat Rev Clin Oncol (2011) 8(8):467–77. doi: 10.1038/nrclinonc.2011.76
23. Silva SS, Lopes C, Teixeira AL, Carneiro de Sousa MJ, Medeiros R. Forensic miRNA: Potential Biomarker for Body Fluids? Forensic Sci Int Genet (2015) 14:1–10. doi: 10.1016/j.fsigen.2014.09.002
24. Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, et al. Robust microRNA Stability in Degraded RNA Preparations From Human Tissue and Cell Samples. Clin Chem (2010) 56(6):998–1006. doi: 10.1373/clinchem.2009.141580
25. Peiró-Chova L, Peña-Chilet M, López-Guerrero JA, García-Giménez JL, Alonso-Yuste E, Burgues O, et al. High Stability of microRNAs in Tissue Samples of Compromised Quality. Virchows Arch (2013) 463(6):765–74. doi: 10.1007/s00428-013-1485-2
26. Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are Differentially Expressed in Ulcerative Colitis and Alter Expression of Macrophage Inflammatory Peptide-2 Alpha. Gastroenterology (2008) 135(5):1624–35.e24. doi: 10.1053/j.gastro.2008.07.068
27. Van der Goten J, Vanhove W, Lemaire K, Van Lommel L, Machiels K, Wollants W-J, et al. Integrated miRNA and mRNA Expression Profiling in Inflamed Colon of Patients With Ulcerative Colitis. PloS One (2014) 9(12):e116117. doi: 10.1371/journal.pone.0116117
28. Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, et al. Diagnostic microRNA Markers for Screening Sporadic Human Colon Cancer and Active Ulcerative Colitis in Stool and Tissue. Cancer Genomics Proteom (2009) 6(5):281–95.
29. Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, et al. Increased Expression of microRNA in the Inflamed Colonic Mucosa of Patients With Active Ulcerative Colitis. J Gastroenterol Hepatol (2010) 25 Suppl 1:S129–33. doi: 10.1111/j.1440-1746.2009.06216.x
30. Zahm AM, Hand NJ, Tsoucas DM, Le Guen CL, Baldassano RN, Friedman JR. Rectal microRNAs are Perturbed in Pediatric Inflammatory Bowel Disease of the Colon. J Crohns Colitis (2014) 8(9):1108–17. doi: 10.1016/j.crohns.2014.02.012
31. Schaefer JS, Attumi T, Opekun AR, Abraham B, Hou J, Shelby H, et al. MicroRNA Signatures Differentiate Crohn’s Disease From Ulcerative Colitis. BMC Immunol (2015) 16(1). doi: 10.1186/s12865-015-0069-0
32. Béres NJ, Kiss Z, Sztupinszki Z, Lendvai G, Arató A, Sziksz E, et al. Altered Mucosal Expression of microRNAs in Pediatric Patients With Inflammatory Bowel Disease. Dig Liver Dis (2017) 49(4):378–87. doi: 10.1016/j.dld.2016.12.022
33. Gwiggner M, Martinez-Nunez RT, Whiteoak SR, Bondanese VP, Claridge A, Collins JE, et al. MicroRNA-31 and MicroRNA-155 Are Overexpressed in Ulcerative Colitis and Regulate IL-13 Signaling by Targeting Interleukin 13 Receptor α-1. Genes (2018) 9(2). doi: 10.3390/genes9020085
34. Béres NJ, Szabó D, Kocsis D, Szűcs D, Kiss Z, Müller KE, et al. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis (2016) 22(2):327–35. doi: 10.1097/MIB.0000000000000687
35. Valmiki S, Ahuja V, Paul J. MicroRNA Exhibit Altered Expression in the Inflamed Colonic Mucosa of Ulcerative Colitis Patients. World J Gastroenterol (2017) 23(29):5324–32. doi: 10.3748/wjg.v23.i29.5324
36. Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, et al. Identification of Restricted Subsets of Mature microRNA Abnormally Expressed in Inactive Colonic Mucosa of Patients With Inflammatory Bowel Disease. PloS One (2010) 5(10). doi: 10.1371/journal.pone.0013160
37. Cai M, Chen S, Hu W. MicroRNA-141 Is Involved in Ulcerative Colitis Pathogenesis via Aiming at CXCL5. J Interferon Cytokine Res (2017) 37(9):415–20. doi: 10.1089/jir.2017.0019
38. Iborra M, Bernuzzi F, Correale C, Vetrano S, Fiorino G, Beltrán B, et al. Identification of Serum and Tissue Micro-RNA Expression Profiles in Different Stages of Inflammatory Bowel Disease. Clin Exp Immunol (2013) 173(2):250–8. doi: 10.1111/cei.12104
39. Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, et al. Identification of microRNAs Associated With Ileal and Colonic Crohn’s Disease†. Inflamm Bowel Dis (2010) 16(10):1729–38. doi: 10.1002/ibd.21267
40. Lin J, Cao Q, Zhang J, Li Y, Shen B, Zhao Z, et al. MicroRNA Expression Patterns in Indeterminate Inflammatory Bowel Disease. Mod Pathol (2013) 26(1):148–54. doi: 10.1038/modpathol.2012.131
41. Guo Z, Wu R, Gong J, Zhu W, Li Y, Wang Z, et al. Altered microRNA Expression in Inflamed and non-Inflamed Terminal Ileal Mucosa of Adult Patients With Active Crohn’s Disease. J Gastroenterol Hepatol (2015) 30(1):109–16. doi: 10.1111/jgh.12644
42. Peck BCE, Weiser M, Lee SE, Gipson GR, Iyer VB, Sartor RB, et al. MicroRNAs Classify Different Disease Behavior Phenotypes of Crohn’s Disease and May Have Prognostic Utility. Inflamm Bowel Dis (2015) 21(9):2178–87. doi: 10.1097/MIB.0000000000000478
43. Szűcs D, Béres NJ, Rokonay R, Boros K, Borka K, Kiss Z, et al. Increased Duodenal Expression of miR-146a and -155 in Pediatric Crohn’s Disease. World J Gastroenterol (2016) 22(26):6027. doi: 10.3748/wjg.v22.i26.6027
44. Palmieri O, Creanza TM, Bossa F, Latiano T, Corritore G, Palumbo O, et al. Functional Implications of MicroRNAs in Crohn’s Disease Revealed by Integrating MicroRNA and Messenger RNA Expression Profiling. Int J Mol Sci (2017) 18(7). doi: 10.3390/ijms18071580
45. Wu LY, Ma XP, Shi Y, Bao CH, Jin XM, Lu Y, et al. Alterations in microRNA Expression Profiles in Inflamed and Noninflamed Ascending Colon Mucosae of Patients With Active Crohn’s Disease. J Gastroenterol Hepatol (2017) 32(10):1706–15. doi: 10.1111/jgh.13778
46. Keith BP, Barrow JB, Toyonaga T, Kazgan N, O’Connor MH, Shah ND, et al. Colonic Epithelial miR-31 Associates With the Development of Crohn’s Phenotypes. JCI Insight (2018) 3(19). doi: 10.1172/jci.insight.122788
47. Verstockt S, De Hertogh G, van der Goten J, Verstockt B, Vancamelbeke M, Machiels K, et al. Gene and Mirna Regulatory Networks During Different Stages of Crohn’s Disease. J Crohns Colitis (2019) 13(7):916–30. doi: 10.1093/ecco-jcc/jjz007
48. Mohammadi A, Kelly OB, Smith MI, Kabakchiev B, Silverberg MS. Differential miRNA Expression in Ileal and Colonic Tissues Reveals an Altered Immunoregulatory Molecular Profile in Individuals With Crohn’s Disease Versus Healthy Subjects. J Crohns Colitis (2019) 13(11):1459–69. doi: 10.1093/ecco-jcc/jjz076
49. Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, et al. Peripheral Blood microRNAs Distinguish Active Ulcerative Colitis and Crohn’s Disease. Inflamm Bowel Dis (2011) 17(1):241–50. doi: 10.1002/ibd.21450
50. Schönauen K, Le N, von Arnim U, Schulz C, Malfertheiner P, Link A. Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm Bowel Dis (2018) 24(7):1547–57. doi: 10.1093/ibd/izy046
51. Polytarchou C, Oikonomopoulos A, Mahurkar S, Touroutoglou A, Koukos G, Hommes DW, et al. Assessment of Circulating MicroRNAs for the Diagnosis and Disease Activity Evaluation in Patients With Ulcerative Colitis by Using the Nanostring Technology. Inflamm Bowel Dis (2015) 21(11):2533–9. doi: 10.1097/MIB.0000000000000547
52. Krissansen GW, Yang Y, McQueen FM, Leung E, Peek D, Chan YC, et al. Overexpression of miR-595 and miR-1246 in the Sera of Patients With Active Forms of Inflammatory Bowel Disease. Inflamm Bowel Dis (2015) 21(3):520–30. doi: 10.1097/MIB.0000000000000285
53. Hübenthal M, Hemmrich-Stanisak G, Degenhardt F, Szymczak S, Du Z, Elsharawy A, et al. Sparse Modeling Reveals miRNA Signatures for Diagnostics of Inflammatory Bowel Disease. PloS One (2015) 10(10):e0140155. doi: 10.1371/journal.pone.0140155
54. Paraskevi A, Theodoropoulos G, Papaconstantinou I, Mantzaris G, Nikiteas N, Gazouli M. Circulating MicroRNA in Inflammatory Bowel Disease. J Crohn’s Colitis (2012) 6(9):900–4. doi: 10.1016/j.crohns.2012.02.006
55. Omidbakhsh A, Saeedi M, Khoshnia M, Marjani A, Hakimi S. Micro-RNAs -106a and -362-3p in Peripheral Blood of Inflammatory Bowel Disease Patients. Open Biochem J (2018) 12:78–86. doi: 10.2174/1874091X01812010078
56. Viennois E, Zhao Y, Han MK, Xiao B, Zhang M, Prasad M, et al. Serum miRNA Signature Diagnoses and Discriminates Murine Colitis Subtypes and Predicts Ulcerative Colitis in Humans. Sci Rep (2017) 7(1):2520. doi: 10.1038/s41598-017-02782-1
57. Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, et al. Genome-Wide Maps of Circulating miRNA Biomarkers for Ulcerative Colitis. PloS One (2012) 7(2):e31241. doi: 10.1371/journal.pone.0031241
58. Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating microRNA is a Biomarker of Pediatric Crohn Disease. J Pediatr Gastroenterol Nutr (2011) 53(1):26–33. doi: 10.1097/MPG.0b013e31822200cc
59. Mohammadi A, Kelly OB, Filice M, Kabakchiev B, Smith MI, Silverberg MS. Differential Expression of microRNAs in Peripheral Blood Mononuclear Cells Identifies Autophagy and TGF-Beta-Related Signatures Aberrantly Expressed in Inflammatory Bowel Disease. J Crohns Colitis (2018) 12(5):568–81. doi: 10.1093/ecco-jcc/jjy010
60. Oikonomopoulos A, Polytarchou C, Joshi S, Hommes DW, Iliopoulos D. Identification of Circulating MicroRNA Signatures in Crohn’s Disease Using the Nanostring Ncounter Technology. Inflamm Bowel Dis (2016) 22(9):2063–9. doi: 10.1097/MIB.0000000000000883
61. Jensen MD, Andersen RF, Christensen H, Nathan T, Kjeldsen J, Madsen JS. Circulating microRNAs as Biomarkers of Adult Crohn’s Disease. Eur J Gastroenterol Hepatol (2015) 27(9):1038–44. doi: 10.1097/MEG.0000000000000430
62. Lin J, Zhang X, Zhao Z, Welker NC, Li Y, Liu Y, et al. Novel MicroRNA Signature to Differentiate Ulcerative Colitis From Crohn Disease: A Genome-Wide Study Using Next Generation Sequencing. Microrna (2016) 5(3):222–9. doi: 10.2174/2211536605666161117113031
63. Lorén V, Garcia-Jaraquemada A, Naves JE, Carmona X, Mañosa M, Aransay AM, et al. ANP32E, a Protein Involved in Steroid-Refractoriness in Ulcerative Colitis, Identified by a Systems Biology Approach. J Crohns Colitis (2019) 13(3):351–61. doi: 10.1093/ecco-jcc/jjy171
64. Ben-Shachar S, Yanai H, Sherman Horev H, Elad H, Baram L, Issakov O, et al. MicroRNAs Expression in the Ileal Pouch of Patients With Ulcerative Colitis Is Robustly Up-Regulated and Correlates With Disease Phenotypes. PloS One (2016) 11(8):e0159956. doi: 10.1371/journal.pone.0159956
65. Nielsen MM, Pedersen JS. miRNA Activity Inferred From Single Cell mRNA Expression. Sci Rep (2021) 11(1):9170. doi: 10.1038/s41598-021-88480-5
66. Krämer A, Green J, Pollard J Jr., Tugendreich S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics (2014) 30(4):523–30. doi: 10.1093/bioinformatics/btt703
67. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13(11):2498–504. doi: 10.1101/gr.1239303
68. Wang X, Wang H-X, Li Y-L, Zhang C-C, Zhou C-Y, Wang L, et al. MicroRNA Let-7i Negatively Regulates Cardiac Inflammation and Fibrosis. Hypertension (2015) 66(4):776–85. doi: 10.1161/HYPERTENSIONAHA.115.05548
69. Satoh M, Tabuchi T, Minami Y, Takahashi Y, Itoh T, Nakamura M. Expression of Let-7i is Associated With Toll-Like Receptor 4 Signal in Coronary Artery Disease: Effect of Statins on Let-7i and Toll-Like Receptor 4 Signal. Immunobiology (2012) 217(5):533–9. doi: 10.1016/j.imbio.2011.08.005
70. Zhai Y, Zhong Z, Chen C-YA, Xia Z, Song L, Blackburn MR, et al. Coordinated Changes in mRNA Turnover, Translation, and RNA Processing Bodies in Bronchial Epithelial Cells Following Inflammatory Stimulation. Mol Cell Biol (2008) 28(24):7414–26. doi: 10.1128/MCB.01237-08
71. Tian T, Zhou Y, Feng X, Ye S, Wang H, Wu W, et al. MicroRNA-16 is Putatively Involved in the NF-κb Pathway Regulation in Ulcerative Colitis Through Adenosine A2a Receptor (A2aAR) mRNA Targeting. Sci Rep (2016) 6:30824. doi: 10.1038/srep30824
72. Lewis A, Mehta S, Hanna LN, Rogalski LA, Jeffery R, Nijhuis A, et al. Low Serum Levels of MicroRNA-19 Are Associated With a Stricturing Crohn’s Disease Phenotype. Inflamm Bowel Dis (2015) 21(8):1926–34. doi: 10.1097/MIB.0000000000000443
73. Collins AS, McCoy CE, Lloyd AT, O’Farrelly C, Stevenson NJ. miR-19a: An Effective Regulator of SOCS3 and Enhancer of JAK-STAT Signalling. PloS One (2013) 8(7):e69090. doi: 10.1371/journal.pone.0069090
74. Yang Y, Ma Y, Shi C, Chen H, Zhang H, Chen N, et al. Overexpression of miR-21 in Patients With Ulcerative Colitis Impairs Intestinal Epithelial Barrier Function Through Targeting the Rho GTPase RhoB. Biochem Biophys Res Commun (2013) 434(4):746–52. doi: 10.1016/j.bbrc.2013.03.122
75. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA Profiling of Naïve, Effector and Memory CD8 T Cells. PloS One (2007) 2(10):e1020. doi: 10.1371/journal.pone.0001020
76. Sandborn WJ, Gasink C, Gao L-L, Blank MA, Johanns J, Guzzo C, et al. Ustekinumab Induction and Maintenance Therapy in Refractory Crohn’s Disease. N Engl J Med (2012) 367(16):1519–28. doi: 10.1056/NEJMoa1203572
77. Yang G, Yang L, Wang W, Wang J, Wang J, Xu Z. Discovery and Validation of Extracellular/Circulating microRNAs During Idiopathic Pulmonary Fibrosis Disease Progression. Gene (2015) 562(1):138–44. doi: 10.1016/j.gene.2015.02.065
78. Shi C, Liang Y, Yang J, Xia Y, Chen H, Han H, et al. MicroRNA-21 Knockout Improve the Survival Rate in DSS Induced Fatal Colitis Through Protecting Against Inflammation and Tissue Injury. PloS One (2013) 8(6):e66814. doi: 10.1371/journal.pone.0066814
79. Momen-Heravi F, Bala S. miRNA Regulation of Innate Immunity. J Leukocyte Biol (2018) 103(6):1205–17. doi: 10.1002/JLB.3MIR1117-459R
80. Yan H, Zhang X, Xu Y. Aberrant Expression of miR-21 in Patients With Inflammatory Bowel Disease: A Protocol for Systematic Review and Meta Analysis. Medicine (2020) 99(17):e19693. doi: 10.1097/MD.0000000000019693
81. Zhao J, Tang N, Wu K, Dai W, Ye C, Shi J, et al. MiR-21 Simultaneously Regulates ERK1 Signaling in HSC Activation and Hepatocyte EMT in Hepatic Fibrosis. PloS One (2014) 9(10):e108005. doi: 10.1371/journal.pone.0108005
82. Zarjou A, Yang S, Abraham E, Agarwal A, Liu G. Identification of a microRNA Signature in Renal Fibrosis: Role of miR-21. Am J Physiol Renal Physiol (2011) 301(4):F793–801. doi: 10.1152/ajprenal.00273.2011
83. Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, et al. The microRNA miR-23b Suppresses IL-17-Associated Autoimmune Inflammation by Targeting TAB2, TAB3 and IKK-α. Nat Med (2012) 18(7):1077–86. doi: 10.1038/nm.2815
84. Ye S-B, Zhang H, Cai T-T, Liu Y-N, Ni J-J, He J, et al. Exosomal miR-24-3p Impedes T-Cell Function by Targeting FGF11 and Serves as a Potential Prognostic Biomarker for Nasopharyngeal Carcinoma. J Pathol (2016) 240(3):329–40. doi: 10.1002/path.4781
85. Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, et al. MicroRNA-24 Regulates Vascularity After Myocardial Infarction. Circulation (2011) 124(6):720–30. doi: 10.1161/CIRCULATIONAHA.111.039008
86. Yu T, Meng F, Xie M, Liu H, Zhang L, Chen X. Long Noncoding RNA PMS2L2 Downregulates miR-24 Through Methylation to Suppress Cell Apoptosis in Ulcerative Colitis. Dig Dis (2021) 39(5):467–76. doi: 10.1159/000513330
87. Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. MicroRNA-24 Regulates the Processing of Latent Tgfβ1 During Cyclic Mechanical Stress in Human Trabecular Meshwork Cells Through Direct Targeting of FURIN. J Cell Physiol (2011) 226(5):1407–14. doi: 10.1002/jcp.22476
88. Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, et al. Comprehensive microRNA Analysis Identifies miR-24 and miR-125a-5p as Plasma Biomarkers for Rheumatoid Arthritis. PloS One (2013) 8(7):e69118. doi: 10.1371/journal.pone.0069118
89. Lv Y, Yang H, Ma X, Wu G. Strand-Specific miR-28-3p and miR-28-5p Have Differential Effects on Nasopharyngeal Cancer Cells Proliferation, Apoptosis, Migration and Invasion. Cancer Cell Int (2019) 19:187. doi: 10.1186/s12935-019-0915-x
90. Li Q, Johnston N, Zheng X, Wang H, Zhang X, Gao D, et al. miR-28 Modulates Exhaustive Differentiation of T Cells Through Silencing Programmed Cell Death-1 and Regulating Cytokine Secretion. Oncotarget (2016) 7(33):53735–50. doi: 10.18632/oncotarget.10731
91. Lv B, Liu Z, Wang S, Liu F, Yang X, Hou J, et al. MiR-29a Promotes Intestinal Epithelial Apoptosis in Ulcerative Colitis by Down-Regulating Mcl-1. Int J Clin Exp Pathol (2014) 7(12):8542–52.
92. Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a Regulates Intestinal Membrane Permeability in Patients With Irritable Bowel Syndrome. Gut (2010) 59(6):775–84. doi: 10.1136/gut.2009.181834
93. Chen T, Li Z, Tu J, Zhu W, Ge J, Zheng X, et al. MicroRNA-29a Regulates Pro-Inflammatory Cytokine Secretion and Scavenger Receptor Expression by Targeting LPL in oxLDL-Stimulated Dendritic Cells. FEBS Lett (2011) 585(4):657–63. doi: 10.1016/j.febslet.2011.01.027
94. Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, et al. Oral Administration of miR-30d From Feces of MS Patients Suppresses MS-Like Symptoms in Mice by Expanding Akkermansia Muciniphila. Cell Host Microbe (2019) 26(6):779–94.e8. doi: 10.1016/j.chom.2019.10.008
95. Nguyen HTT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s Disease-Associated Adherent Invasive Escherichia Coli Modulate Levels of microRNAs in Intestinal Epithelial Cells to Reduce Autophagy. Gastroenterology (2014) 146(2):508–19. doi: 10.1053/j.gastro.2013.10.021
96. Ghadiri N, Emamnia N, Ganjalikhani-Hakemi M, Ghaedi K, Etemadifar M, Salehi M, et al. Analysis of the Expression of Mir-34a, Mir-199a, Mir-30c and Mir-19a in Peripheral Blood CD4+T Lymphocytes of Relapsing-Remitting Multiple Sclerosis Patients. Gene (2018) 659:109–17. doi: 10.1016/j.gene.2018.03.035
97. Olaru AV, Selaru FM, Mori Y, Vazquez C, David S, Paun B, et al. Dynamic Changes in the Expression of MicroRNA-31 During Inflammatory Bowel Disease-Associated Neoplastic Transformation. Inflamm Bowel Dis (2011) 17(1):221–31. doi: 10.1002/ibd.21359
98. Xu N, Meisgen F, Butler LM, Han G, Wang X-J, Söderberg-Nauclér C, et al. MicroRNA-31 is Overexpressed in Psoriasis and Modulates Inflammatory Cytokine and Chemokine Production in Keratinocytes via Targeting Serine/Threonine Kinase 40. J Immunol (2013) 190(2):678–88. doi: 10.4049/jimmunol.1202695
99. Zhang L, Ke F, Liu Z, Bai J, Liu J, Yan S, et al. MicroRNA-31 Negatively Regulates Peripherally Derived Regulatory T-Cell Generation by Repressing Retinoic Acid-Inducible Protein 3. Nat Commun (2015) 6:7639. doi: 10.1038/ncomms8639
100. Jovov B, Shaheen NJ, Orlando GS, Djukic Z, Orlando RC. Defective Barrier Function in Neosquamous Epithelium. Am J Gastroenterol (2013) 108(3):386–91. doi: 10.1038/ajg.2012.440
101. Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, et al. T Cell Activation Induces Proteasomal Degradation of Argonaute and Rapid Remodeling of the microRNA Repertoire. J Exp Med (2013) 210(2):417–32. doi: 10.1084/jem.20111717
102. Zhu D, Pan C, Li L, Bian Z, Lv Z, Shi L, et al. MicroRNA-17/20a/106a Modulate Macrophage Inflammatory Responses Through Targeting Signal-Regulatory Protein α. J Allergy Clin Immunol (2013) 132(2):426–36.e8. doi: 10.1016/j.jaci.2013.02.005
103. Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, et al. Posttranscriptional Regulation of Interleukin-10 Expression by hsa-miR-106a. Proc Natl Acad Sci USA (2009) 106(14):5761–6. doi: 10.1073/pnas.0808743106
104. Sanctuary MR, Huang RH, Jones AA, Luck ME, Aherne CM, Jedlicka P, et al. miR-106a Deficiency Attenuates Inflammation in Murine IBD Models. Mucosal Immunol (2019) 12(1):200–11. doi: 10.1038/s41385-018-0091-7
105. Soukhtanloo M, Mohtashami E, Maghrouni A, Mollazadeh H, Mousavi SH, Roshan MK, et al. Natural Products as Promising Targets in Glioblastoma Multiforme: A Focus on NF-κb Signaling Pathway. Pharmacol Rep (2020) 72(2):285–95. doi: 10.1007/s43440-020-00081-7
106. Feng X, Wang H, Ye S, Guan J, Tan W, Cheng S, et al. Up-Regulation of microRNA-126 may Contribute to Pathogenesis of Ulcerative Colitis via Regulating NF-kappaB Inhibitor Iκbα. PloS One (2012) 7(12):e52782. doi: 10.1371/journal.pone.0052782
107. Angel-Morales G, Noratto G, Mertens-Talcott S. Red Wine Polyphenolics Reduce the Expression of Inflammation Markers in Human Colon-Derived CCD-18Co Myofibroblast Cells: Potential Role of microRNA-126. Food Funct (2012) 3(7):745–52. doi: 10.1039/c2fo10271d
108. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 Regulates Endothelial Expression of Vascular Cell Adhesion Molecule 1. Proc Natl Acad Sci USA (2008) 105(5):1516–21. doi: 10.1073/pnas.0707493105
109. Kim EH, Suresh M. Role of PI3K/Akt Signaling in Memory CD8 T Cell Differentiation. Front Immunol (2013) 4:20. doi: 10.3389/fimmu.2013.00020
110. Li H, Guan S-B, Lu Y, Wang F. MiR-140-5p Inhibits Synovial Fibroblasts Proliferation and Inflammatory Cytokines Secretion Through Targeting TLR4. BioMed Pharmacother (2017) 96:208–14. doi: 10.1016/j.biopha.2017.09.079
111. Chan EKL, Satoh M, Pauley KM. Contrast in Aberrant microRNA Expression in Systemic Lupus Erythematosus and Rheumatoid Arthritis: Is microRNA-146 All We Need? Arthritis Rheum (2009) 60(4):912–5. doi: 10.1002/art.24421
112. Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, et al. Deep Sequencing of Small RNAs From Human Skin Reveals Major Alterations in the Psoriasis Mirnaome. Hum Mol Genet (2011) 20(20):4025–40. doi: 10.1093/hmg/ddr331
113. Huang Z, Shi T, Zhou Q, Shi S, Zhao R, Shi H, et al. miR-141 Regulates Colonic Leukocytic Trafficking by Targeting CXCL12β During Murine Colitis and Human Crohn’s Disease. Gut (2014) 63(8):1247–57. doi: 10.1136/gutjnl-2012-304213
114. Pan A, Tan Y, Wang Z, Xu G. STAT4 Silencing Underlies a Novel Inhibitory Role of microRNA-141-3p in Inflammation Response of Mice With Experimental Autoimmune Myocarditis. Am J Physiol Heart Circ Physiol (2019) 317(3):H531–H40. doi: 10.1152/ajpheart.00048.2019
115. Shen WS, Xu XQ, Zhai NN, Zhou ZS, Shao J, Yu YH. Potential Mechanisms of microRNA-141-3p to Alleviate Chronic Inflammatory Pain by Downregulation of Downstream Target Gene HMGB1: In Vitro and In Vivo Studies. Gene Ther (2017) 24(6):353–60. doi: 10.1038/gt.2017.28
116. Bahmani L, Baghi M, Peymani M, Javeri A, Ghaedi K. MiR-141-3p and miR-200a-3p are Involved in Th17 Cell Differentiation by Negatively Regulating RARB Expression. Hum Cell (2021) 34(5):1375–87. doi: 10.1007/s13577-021-00558-4
117. Li X, Wang Y, Wang Y, He X. MiR-141-3p Ameliorates RIPK1-Mediated Necroptosis of Intestinal Epithelial Cells in Necrotizing Enterocolitis. Aging (2020) 12(18):18073–83. doi: 10.18632/aging.103608
118. Anandagoda N, Willis JCD, Hertweck A, Roberts LB, Jackson I, Refik Gökmen M, et al. microRNA-142–Mediated Repression of Phosphodiesterase 3B Critically Regulates Peripheral Immune Tolerance. J Clin Invest (2019) 129(3):1257–71. doi: 10.1172/JCI124725
119. Ashrafizadeh M, Rafiei H, Mohammadinejad R, Farkhondeh T, Samarghandian S. Wnt-Regulating microRNAs Role in Gastric Cancer Malignancy. Life Sci (2020) 250:117547. doi: 10.1016/j.lfs.2020.117547
120. Amato G, Vita F, Quattrocchi P, Minciullo PL, Pioggia G, Gangemi S. Involvement of miR-142 and miR-155 in Non-Infectious Complications of CVID. Molecules (2020) 25(20). doi: 10.3390/molecules25204760
121. Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, et al. miR-142-5p Regulates Tumor Cell PD-L1 Expression and Enhances Anti-Tumor Immunity. Biochem Biophys Res Commun (2017) 488(2):425–31. doi: 10.1016/j.bbrc.2017.05.074
122. Zhang H-F, Qiu L-X, Chen Y, Zhu W-L, Mao C, Zhu L-G, et al. ATG16L1 T300A Polymorphism and Crohn’s Disease Susceptibility: Evidence From 13,022 Cases and 17,532 Controls. Hum Genet (2009) 125(5-6):627–31. doi: 10.1007/s00439-009-0660-7
123. Lu C, Chen J, Xu H-G, Zhou X, He Q, Li Y-L, et al. MIR106B and MIR93 Prevent Removal of Bacteria From Epithelial Cells by Disrupting ATG16L1-Mediated Autophagy. Gastroenterology (2014) 146(1):188–99. doi: 10.1053/j.gastro.2013.09.006
124. Lu Y, Gao J, Zhang S, Gu J, Lu H, Xia Y, et al. miR-142-3p Regulates Autophagy by Targeting ATG16L1 in Thymic-Derived Regulatory T Cell (Ttreg). Cell Death Dis (2018) 9(3):290. doi: 10.1038/s41419-018-0298-2
125. Gao J, Gu J, Pan X, Gan X, Ju Z, Zhang S, et al. Blockade of miR-142-3p Promotes Anti-Apoptotic and Suppressive Function by Inducing KDM6A-Mediated H3K27me3 Demethylation in Induced Regulatory T Cells. Cell Death Dis (2019) 10(5):332. doi: 10.1038/s41419-019-1565-6
126. Liu Y, Song X, Meng S, Jiang M. Downregulated Expression of miR-142-3p in Macrophages Contributes to Increased IL-6 Levels in Aged Mice. Mol Immunol (2016) 80:11–6. doi: 10.1016/j.molimm.2016.10.009
127. Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs: Novel Regulators in Skin Inflammation. Clin Exp Dermatol (2008) 33(3):312–5. doi: 10.1111/j.1365-2230.2008.02804.x
128. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. miR-146a Is a Significant Brake on Autoimmunity, Myeloproliferation, and Cancer in Mice. J Exp Med (2011) 208(6):1189–201. doi: 10.1084/jem.20101823
129. Ghorpade DS, Sinha AY, Holla S, Singh V, Balaji KN. NOD2-Nitric Oxide-Responsive microRNA-146a Activates Sonic Hedgehog Signaling to Orchestrate Inflammatory Responses in Murine Model of Inflammatory Bowel Disease. J Biol Chem (2013) 288(46):33037–48. doi: 10.1074/jbc.M113.492496
130. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and Pathological Roles for microRNAs in the Immune System. Nat Rev Immunol (2010) 10(2):111–22. doi: 10.1038/nri2708
131. Nata T, Fujiya M, Ueno N, Moriichi K, Konishi H, Tanabe H, et al. MicroRNA-146b Improves Intestinal Injury in Mouse Colitis by Activating Nuclear Factor-κb and Improving Epithelial Barrier Function. J Gene Med (2013) 15(6-7):249–60. doi: 10.1002/jgm.2717
132. Liu Y, Zhu J-Q, Jin X-H, Dong M-P, Zheng J-F. Up-Regulation of miR-146b-3p Protects Septic Mice With Acute Respiratory Distress Syndrome by Inhibiting PI3K/AKT Signaling Pathway. J Bioenerg Biomembr (2020) 52(4):229–36. doi: 10.1007/s10863-020-09839-3
133. Fulzele S, El-Sherbini A, Ahmad S, Sangani R, Matragoon S, El-Remessy A, et al. MicroRNA-146b-3p Regulates Retinal Inflammation by Suppressing Adenosine Deaminase-2 in Diabetes. BioMed Res Int (2015) 2015:846501. doi: 10.1155/2015/846501
134. Cai F, Wu F, Cao J, Chen X. MicroRNA-146b-3p Regulates the Development and Progression of Cerebral Infarction With Diabetes Through RAF1/P38MAPK/COX-2 Signaling Pathway. Am J Transl Res (2018) 10(2):618–28.
135. Xu G, Zhang Z, Xing Y, Wei J, Ge Z, Liu X, et al. MicroRNA-149 Negatively Regulates TLR-Triggered Inflammatory Response in Macrophages by Targeting Myd88. J Cell Biochem (2014) 115(5):919–27. doi: 10.1002/jcb.24734
136. Zhu M, Li D, Jin M, Li M. Association Between microRNA Polymorphisms and the Risk of Inflammatory Bowel Disease. Mol Med Rep (2016) 13(6):5297–308. doi: 10.3892/mmr.2016.5157
137. Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic Potential of Mesenchymal Cell-Derived miRNA-150-5p-Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF. J Immunol (2018) 201(8):2472–82. doi: 10.4049/jimmunol.1800304
138. Tibaldi J, Pistorio A, Aldera E, Puzone L, El Miedany Y, Pal P, et al. Development and Initial Validation of a Composite Disease Activity Score for Systemic Juvenile Idiopathic Arthritis. Rheumatology (2020) 59(11):3505–14. doi: 10.1093/rheumatology/keaa240
139. Tsitsiou E, Lindsay MA. microRNAs and the Immune Response. Curr Opin Pharmacol (2009) 9(4):514–20. doi: 10.1016/j.coph.2009.05.003
140. Chen W-X, Ren L-H, Shi R-H. Implication of miRNAs for Inflammatory Bowel Disease Treatment: Systematic Review. World J Gastrointest Pathophysiol (2014) 5(2):63–70. doi: 10.4291/wjgp.v5.i2.63
141. Plank M, Maltby S, Mattes J, Foster PS. Targeting Translational Control as a Novel Way to Treat Inflammatory Disease: The Emerging Role of microRNAs. Clin Exp Allergy (2013) 43(9):981–99. doi: 10.1111/cea.12135
142. Evel-Kabler K, Song X-T, Aldrich M, Huang XF, Chen S-Y. SOCS1 Restricts Dendritic Cells’ Ability to Break Self Tolerance and Induce Antitumor Immunity by Regulating IL-12 Production and Signaling. J Clin Invest (2006) 116(1):90–100. doi: 10.1172/JCI26169
143. Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmén J, et al. Silencing of microRNA-155 in Mice During Acute Inflammatory Response Leads to Derepression of C/Ebp Beta and Down-Regulation of G-CSF. Nucleic Acids Res (2009) 37(17):5784–92. doi: 10.1093/nar/gkp577
144. Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, et al. Altered Expression of microRNA-203 in Rheumatoid Arthritis Synovial Fibroblasts and its Role in Fibroblast Activation. Arthritis Rheumatol (2011) 63(2):373–81. doi: 10.1002/art.30115
145. Sonkoly E, Janson P, Majuri M-L, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 Is Overexpressed in Patients With Atopic Dermatitis and Modulates T-Cell Proliferative Responses by Targeting Cytotoxic T Lymphocyte-Associated Antigen 4. J Allergy Clin Immunol (2010) 126(3):581–9.e1-20. doi: 10.1016/j.jaci.2010.05.045
146. Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 Ameliorates Experimental Autoimmune Encephalomyelitis. J Immunol (2011) 187(5):2213–21. doi: 10.4049/jimmunol.1003952
147. Soriano A, Jubierre L, Almazán-Moga A, Molist C, Roma J, de Toledo JS, et al. microRNAs as Pharmacological Targets in Cancer. Pharmacol Res (2013) 75:3–14. doi: 10.1016/j.phrs.2013.03.006
148. Archanioti P, Gazouli M, Theodoropoulos G, Vaiopoulou A, Nikiteas N. Micro-RNAs as Regulators and Possible Diagnostic Bio-Markers in Inflammatory Bowel Disease. J Crohns Colitis (2011) 5(6):520–4. doi: 10.1016/j.crohns.2011.05.007
149. Georges SA, Biery MC, Kim S-Y, Schelter JM, Guo J, Chang AN, et al. Coordinated Regulation of Cell Cycle Transcripts by P53-Inducible microRNAs, miR-192 and miR-215. Cancer Res (2008) 68(24):10105–12. doi: 10.1158/0008-5472.CAN-08-1846
150. Trevisani F, Ghidini M, Larcher A, Lampis A, Lote H, Manunta P, et al. MicroRNA 193b-3p as a Predictive Biomarker of Chronic Kidney Disease in Patients Undergoing Radical Nephrectomy for Renal Cell Carcinoma. Br J Cancer (2016) 115(11):1343–50. doi: 10.1038/bjc.2016.329
151. Arner E, Mejhert N, Kulyté A, Balwierz PJ, Pachkov M, Cormont M, et al. Adipose Tissue microRNAs as Regulators of CCL2 Production in Human Obesity. Diabetes (2012) 61(8):1986–93. doi: 10.2337/db11-1508
152. Zhou X, Li Q, Xu J, Zhang X, Zhang H, Xiang Y, et al. The Aberrantly Expressed miR-193b-3p Contributes to Preeclampsia Through Regulating Transforming Growth Factor-β Signaling. Sci Rep (2016) 6:19910. doi: 10.1038/srep19910
153. Shirani F, Baghi M, Rostamian Delavar M, Shoaraye Nejati A, Eshaghiyan A, Nasr-Esfahani MH, et al. Upregulation of miR-9 and miR-193b Over Human Th17 Cell Differentiation. Mol Genet Genom Med (2020) 8(12):e1538. doi: 10.1002/mgg3.1538
154. Wang B, Shen Z-L, Gao Z-D, Zhao G, Wang C-Y, Yang Y, et al. MiR-194, Commonly Repressed in Colorectal Cancer, Suppresses Tumor Growth by Regulating the MAP4K4/c-Jun/MDM2 Signaling Pathway. Cell Cycle (2015) 14(7):1046–58. doi: 10.1080/15384101.2015.1007767
155. Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, et al. miR-194 is a Marker of Hepatic Epithelial Cells and Suppresses Metastasis of Liver Cancer Cells in Mice. Hepatology (2010) 52(6):2148–57. doi: 10.1002/hep.23915
156. Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, et al. Inducible Expression of microRNA-194 is Regulated by HNF-1alpha During Intestinal Epithelial Cell Differentiation. RNA (2008) 14(7):1433–42. doi: 10.1261/rna.810208
157. Chen G, Cao S, Liu F, Liu Y. miR-195 Plays a Role in Steroid Resistance of Ulcerative Colitis by Targeting Smad7. Biochem J (2015) 471(3):357–67. doi: 10.1042/BJ20150095
158. Bras JP, Silva AM, Calin GA, Barbosa MA, Santos SG, Almeida MI. miR-195 Inhibits Macrophages Pro-Inflammatory Profile and Impacts the Crosstalk With Smooth Muscle Cells. PloS One (2017) 12(11):e0188530. doi: 10.1371/journal.pone.0188530
159. Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, et al. Downregulation of miR-199a Derepresses Hypoxia-Inducible Factor-1alpha and Sirtuin 1 and Recapitulates Hypoxia Preconditioning in Cardiac Myocytes. Circ Res (2009) 104(7):879–86. doi: 10.1161/CIRCRESAHA.108.193102
160. Rane S, He M, Sayed D, Yan L, Vatner D, Abdellatif M. An Antagonism Between the AKT and Beta-Adrenergic Signaling Pathways Mediated Through Their Reciprocal Effects on miR-199a-5p. Cell Signal (2010) 22(7):1054–62. doi: 10.1016/j.cellsig.2010.02.008
161. Lin H-S, Gong J-N, Su R, Chen M-T, Song L, Shen C, et al. miR-199a-5p Inhibits Monocyte/Macrophage Differentiation by Targeting the Activin A Type 1B Receptor Gene and Finally Reducing C/Ebpα Expression. J Leukoc Biol (2014) 96(6):1023–35. doi: 10.1189/jlb.1A0514-240R
162. Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c Binding to FN1 Suppresses the Proliferation, Migration and Invasion of Gastric Cancer Cells. BioMed Pharmacother (2017) 88:285–92. doi: 10.1016/j.biopha.2017.01.023
163. Tang O, Chen X-M, Shen S, Hahn M, Pollock CA. MiRNA-200b Represses Transforming Growth Factor-β1-Induced EMT and Fibronectin Expression in Kidney Proximal Tubular Cells. Am J Physiol-Renal Physiol (2013) 304(10):F1266–F73. doi: 10.1152/ajprenal.00302.2012
164. Chen Y, Xiao Y, Ge W, Zhou K, Wen J, Yan W, et al. miR-200b Inhibits TGF-β1-Induced Epithelial-Mesenchymal Transition and Promotes Growth of Intestinal Epithelial Cells. Cell Death Dis (2013) 4:e541. doi: 10.1038/cddis.2013.22
165. Zhang W, Fu Z, Yin H, Han Q, Fan W, Wang F, et al. Macrophage Polarization Modulated by Porcine Circovirus Type 2 Facilitates Bacterial Coinfection. Front Immunol (2021) 12:688294. doi: 10.3389/fimmu.2021.688294
166. Rokavec M, Wu W, Luo J-L. IL6-Mediated Suppression of miR-200c Directs Constitutive Activation of Inflammatory Signaling Circuit Driving Transformation and Tumorigenesis. Mol Cell (2012) 45(6):777–89. doi: 10.1016/j.molcel.2012.01.015
167. Wendlandt EB, Graff JW, Gioannini TL, McCaffrey AP, Wilson ME. The Role of microRNAs miR-200b and miR-200c in TLR4 Signaling and NF-κb Activation. Innate Immun (2012) 18(6):846–55. doi: 10.1177/1753425912443903
168. Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 is Overexpressed in T-Lymphocytes of Patients Affected by Rheumatoid Arthritis. Hum Immunol (2010) 71(2):206–11. doi: 10.1016/j.humimm.2009.11.008
169. Deiuliis JA, Syed R, Duggineni D, Rutsky J, Rengasamy P, Zhang J, et al. Visceral Adipose MicroRNA 223 Is Upregulated in Human and Murine Obesity and Modulates the Inflammatory Phenotype of Macrophages. PloS One (2016) 11(11):e0165962. doi: 10.1371/journal.pone.0165962
170. Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, et al. Pro-Inflammatory miR-223 Mediates the Cross-Talk Between the IL23 Pathway and the Intestinal Barrier in Inflammatory Bowel Disease. Genome Biol (2016) 17:58. doi: 10.1186/s13059-016-0901-8
171. Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, et al. Myeloid-Derived miR-223 Regulates Intestinal Inflammation via Repression of the NLRP3 Inflammasome. J Exp Med (2017) 214(6):1737–52. doi: 10.1084/jem.20160462
172. Wu CP, Bi YJ, Liu DM, Wang LY. Hsa-miR-375 Promotes the Progression of Inflammatory Bowel Disease by Upregulating TLR4. Eur Rev Med Pharmacol Sci (2019) 23(17):7543–9. doi: 10.26355/eurrev_201909_18871
173. Chiba M, Monzen S, Iwaya C, Kashiwagi Y, Yamada S, Hosokawa Y, et al. Serum miR-375-3p Increase in Mice Exposed to a High Dose of Ionizing Radiation. Sci Rep (2018) 8(1):1302. doi: 10.1038/s41598-018-19763-7
174. Dubois-Camacho K, Diaz-Jimenez D, de la Fuente M, Quera R, Simian D, Martínez M, et al. Inhibition of miR-378a-3p by Inflammation Enhances IL-33 Levels: A Novel Mechanism of Alarmin Modulation in Ulcerative Colitis. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.02449
175. Nozawa M, Fujimi M, Iwamoto C, Onizuka K, Fukuda N, Ikeo K, et al. Evolutionary Transitions of MicroRNA-Target Pairs. Genome Biol Evol (2016) 8(5):1621–33. doi: 10.1093/gbe/evw092
176. Bellafante E, Morgano A, Salvatore L, Murzilli S, Di Tullio G, D’Orazio A, et al. PGC-1β Promotes Enterocyte Lifespan and Tumorigenesis in the Intestine. Proc Natl Acad Sci USA (2014) 111(42):E4523–31. doi: 10.1073/pnas.1415279111
177. Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, et al. Control of Mitochondrial Metabolism and Systemic Energy Homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci USA (2012) 109(38):15330–5. doi: 10.1073/pnas.1207605109
178. Liu C, Lin JD. PGC-1 Coactivators in the Control of Energy Metabolism. Acta Biochim Biophys Sin (2011) 43(4):248–57. doi: 10.1093/abbs/gmr007
179. Sifroni KG, Damiani CR, Stoffel C, Cardoso MR, Ferreira GK, Jeremias IC, et al. Mitochondrial Respiratory Chain in the Colonic Mucosal of Patients With Ulcerative Colitis. Mol Cell Biochem (2010) 342(1-2):111–5. doi: 10.1007/s11010-010-0474-x
180. Rosa A, Ballarino M, Sorrentino A, Sthandier O, De Angelis FG, Marchioni M, et al. The Interplay Between the Master Transcription Factor PU.1 and miR-424 Regulates Human Monocyte/Macrophage Differentiation. Proc Natl Acad Sci USA (2007) 104(50):19849–54. doi: 10.1073/pnas.0706963104
181. Dinesh P, Kalaiselvan S, Sujitha S, Rasool M. MicroRNA-532-3p Regulates Pro-Inflammatory Human THP-1 Macrophages by Targeting ASK1/p38 MAPK Pathway. Inflammation (2021) 44(1):229–42. doi: 10.1007/s10753-020-01325-7
182. Soubières AA, Poullis A. Emerging Role of Novel Biomarkers in the Diagnosis of Inflammatory Bowel Disease. World J Gastrointest Pharmacol Ther (2016) 7(1):41–50. doi: 10.4292/wjgpt.v7.i1.41
183. Lou C, Li Y. Functional Role of microRNA-135a in Colitis. J Inflamm (2018) 15:7. doi: 10.1186/s12950-018-0181-z
184. Chen Y, Shan T, Qu H, Chen Y, Wang N, Xia J. Inhibition of miR-16 Ameliorates Inflammatory Bowel Disease by Modulating Bcl-2 in Mouse Models. J Surg Res (2020) 253:185–92. doi: 10.1016/j.jss.2020.03.037
185. Garo LP, Ajay AK, Fujiwara M, Gabriely G, Raheja R, Kuhn C, et al. MicroRNA-146a Limits Tumorigenic Inflammation in Colorectal Cancer. Nat Commun (2021) 12(1):2419. doi: 10.1038/s41467-021-22641-y
186. Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, et al. MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology (2015) 149(4):981–92 e11. doi: 10.1053/j.gastro.2015.05.057
187. Tian Y, Xu J, Li Y, Zhao R, Du S, Lv C, et al. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology (2019) 156(8):2281–96.e6. doi: 10.1053/j.gastro.2019.02.023
188. Peng L, Zhang H, Hao Y, Xu F, Yang J, Zhang R, et al. Reprogramming Macrophage Orientation by microRNA 146b Targeting Transcription Factor IRF5. EBioMedicine (2016) 14:83–96. doi: 10.1016/j.ebiom.2016.10.041
189. Peng Y, Wang Q, Yang W, Yang Q, Pei Y, Zhang W. MiR-98-5p Expression Inhibits Polarization of Macrophages to an M2 Phenotype by Targeting Trib1 in Inflammatory Bowel Disease. Acta Biochim Pol (2020) 67(2):157–63. doi: 10.18388/abp.2020_5152
190. Ye D, Guo S, Al-Sadi R, Ma TY. MicroRNA Regulation of Intestinal Epithelial Tight Junction Permeability. Gastroenterology (2011) 141(4):1323–33. doi: 10.1053/j.gastro.2011.07.005
Keywords: miRNA, ulcerative colitis, Crohn’s disease, inflammatory bowel disase, Transcriptomics
Citation: Yarani R, Shojaeian A, Palasca O, Doncheva NT, Jensen LJ, Gorodkin J and Pociot F (2022) Differentially Expressed miRNAs in Ulcerative Colitis and Crohn’s Disease. Front. Immunol. 13:865777. doi: 10.3389/fimmu.2022.865777
Received: 30 January 2022; Accepted: 13 April 2022;
Published: 06 June 2022.
Edited by:
Bertrand Kaeffer, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Amada Torres, Instituto de Biotecnología - UNAM, MexicoSwapna Mahurkar-Joshi, University of California, Los Angeles, United States
Copyright © 2022 Yarani, Shojaeian, Palasca, Doncheva, Jensen, Gorodkin and Pociot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Yarani, cmV6YS55YXJhbmkuMDFAcmVnaW9uaC5kaw==; Flemming Pociot, ZmxlbW1pbmcucG9jaW90QHJlZ2lvbmguZGs=
 Reza Yarani
Reza Yarani Ali Shojaeian
Ali Shojaeian Oana Palasca4,5,6
Oana Palasca4,5,6 Nadezhda T. Doncheva
Nadezhda T. Doncheva Lars Juhl Jensen
Lars Juhl Jensen Jan Gorodkin
Jan Gorodkin Flemming Pociot
Flemming Pociot