- 1Department of Clinical Laboratory, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
- 2State Key Laboratory of Complex, Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 3Department of Medical Research Center, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
Noncoding RNAs (ncRNAs) constitute more than 90% of the RNAs in the human genome. In the past decades, studies have changed our perception of ncRNAs from “junk” transcriptional products to functional regulatory molecules that mediate critical processes, including chromosomal modifications, mRNA splicing and stability, and translation, as well as key signaling pathways. Emerging evidence suggests that ncRNAs are abnormally expressed in not only cancer but also autoimmune diseases, such as systemic sclerosis (SSc), and may serve as novel biomarkers and therapeutic targets for the diagnosis and treatment of SSc. However, the functions and underlying mechanisms of ncRNAs in SSc remain incompletely understood. In this review, we discuss the current findings on the biogenetic processes and functions of ncRNAs, including microRNAs and long noncoding RNAs, as well as explore emerging ncRNA-based diagnostics and therapies for SSc.
Introduction
Systemic Sclerosis
Systemic sclerosis (SSc) is a rare, systemic autoimmune disease characterized by vascular damage, immune activation, and fibrosis of the skin and/or internal organs (1). The estimated prevalence of SSc was 88 per million in men and 514 per million in women, demonstrating sex-based differences (2). Patients with SSc are divided into different categories according to their skin condition: limited cutaneous SSc (lcSSc, skin fibrosis is restricted to the fingers, distal extremities, and face), diffuse cutaneous SSc (dcSSc, skin fibrosis is present in the trunk and proximal extremities), SSc sine scleroderma (no detectable skin involvement), and overlap syndrome (concomitant with another connective tissue disease) (3, 4). The definitive diagnosis of SSc is usually based on the 2013 European League Against Rheumatism (EULAR) and American College of Rheumatology (ACR) classification criteria (5, 6). Some key early clinical characteristics of SSc are associated with early internal organ involvement; however, there are no reliable criteria to identify patients with early symptoms. Thus, novel biomarkers and therapeutic targets of SSc are needed to improve its diagnosis and prognosis. Notably, numerous studies have reported that noncoding RNAs (ncRNAs), such as microRNAs (miRNAs) and long noncoding RNAs(lncRNAs), are implicated in the development of SSc.
ncRNAs
ncRNAs constitute more than 90% of the RNAs in the human genome. Scientists have discovered >50,000 ncRNAs in the past decade, but most remain unstudied (7). Unlike messenger RNAs (mRNAs), which transmit instructions encoded in the DNA for proteins production, ncRNAs are not involved in protein production (8). ncRNAs include intronic RNAs (9), miRNAs (10), lncRNAs (11), circular RNAs (12), extracellular RNAs (13), transfer RNA-derived small RNAs (14), and piwi-interacting RNAs (15). Although ncRNAs cannot encode proteins, they can modulate the expression of other genes through a variety of mechanisms at transcriptional and post-transcriptional levels, thereby affecting both normal cellular function and disease development. Studies on ncRNAs in SSc have focused on miRNAs and lncRNAs. This review summarizes the latest research on miRNAs and lncRNAs including their biosynthesis and functions, as well as their roles in SSc and its complications and as potential biomarkers.
Biosynthesis and Functions of miRNAs and lncRNAs
miRNA
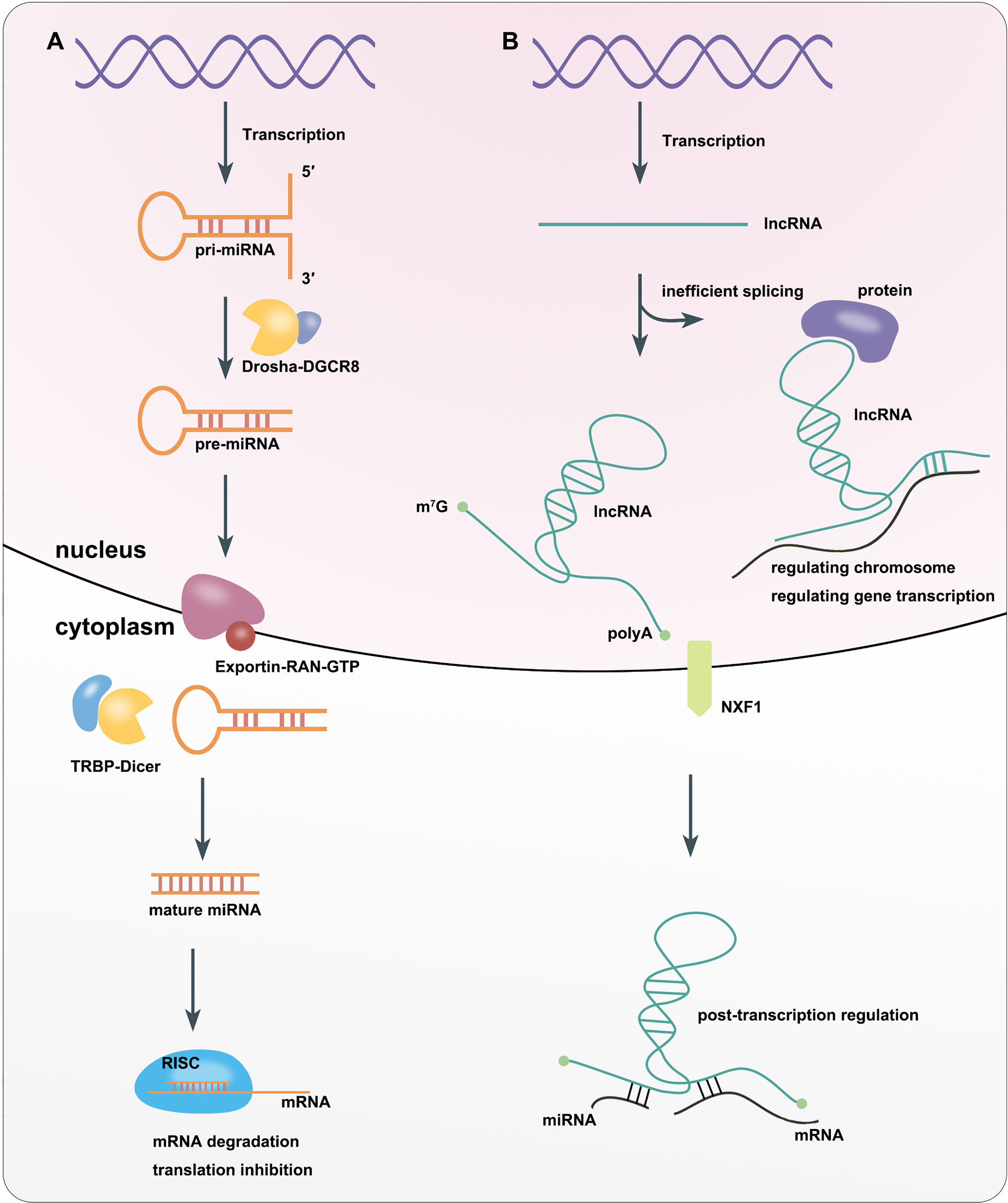
miRNAs are is a class of ncRNAs of that are 21–25 nucleotides (nt) in length. They were first discovered in Caenorhabditis elegans in 1993 (16). miRNAs can be divided into intracellular miRNAs, and extracellular miRNAs or circulating miRNAs (17). RNA polymerase II or III transcribes miRNA-related genes to primary miRNAs (pri-miRNAs) with thousands of nt in the nucleus. The core part of pri-miRNAs is a stem loop structure, whose double-stranded stem region contains functional miRNAs (18, 19). Then Drosha, a type III RNA cutting enzyme, forms a complex with DiGeorge syndrome critical region gene 8 (DGCR8), a double-stranded RNA-binding protein that recruits pri-miRNA substrates, which subsequently cleaves the pri-miRNA into pre-miRNA with only 60–70 nt. This leads to the formation of a hairpin structure (18, 20). Next, the Exportin5–RAN–GTP complex exports pre-miRNA from the nucleus into the cytoplasm (18, 21). In the cytoplasm, where the enzyme RNase Dicer enzymes, which binds to transactivation response element RNA-binding protein (TRBP), cleaves the pre-miRNA into mature-length miRNAs while the miRNA is still double-stranded (18, 22). One strand of the mature miRNA (the guide strand) interacts with Argonaute (AGO) proteins to enter the RNA-induced silencing complex (RISC). Finally, RISC induces target mRNA degradation or inhibiting gene translation by sequence complementary binding (23, 24) (Figure 1A).
lncRNA
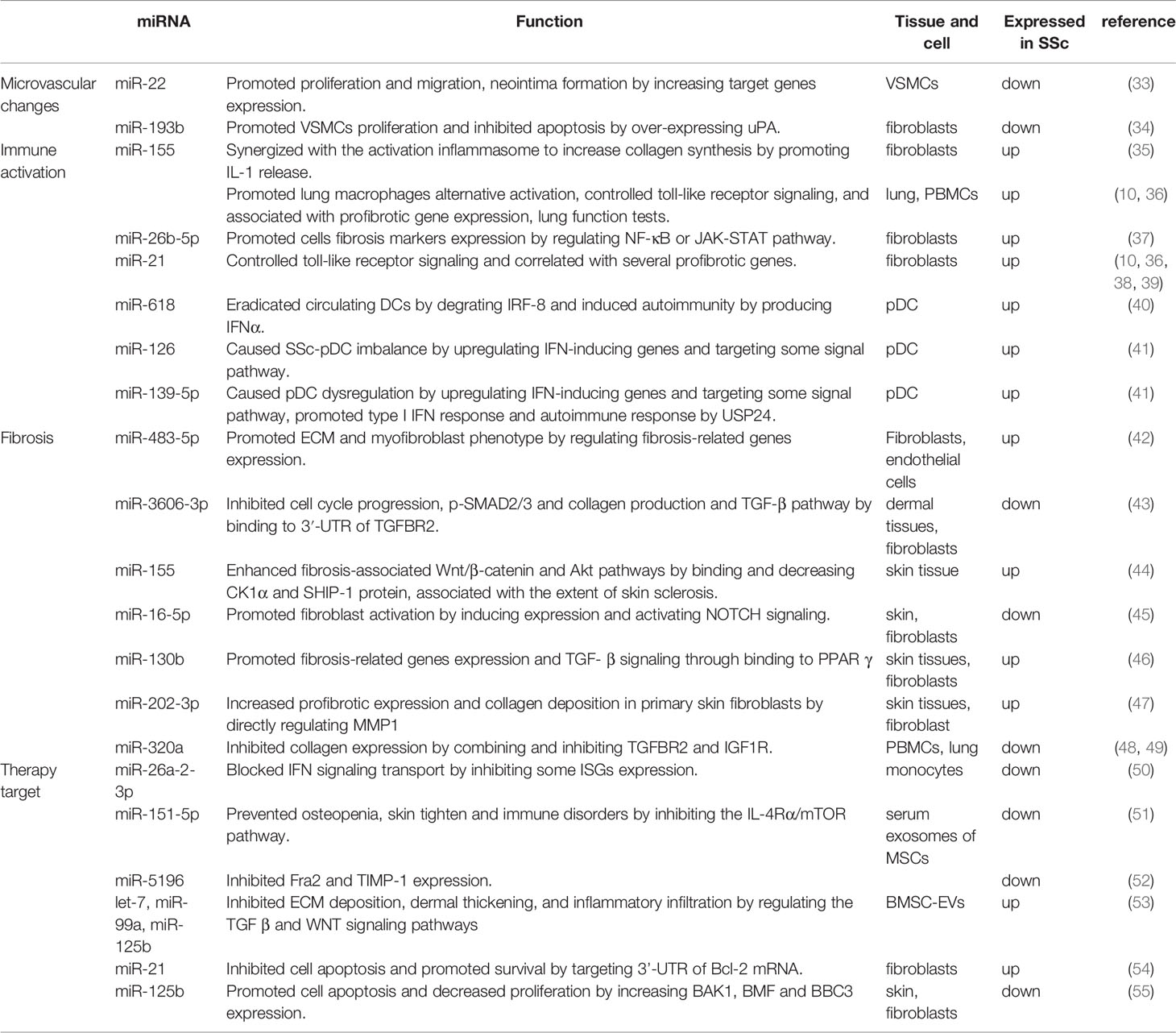
lncRNAs are defined as RNAs longer than 200 nt, which are not translated into functional proteins. lncRNAs are often capped by 7-methylguanosine (m7G) at their 5′ ends, polyadenylated at their 3′ ends, and spliced like mRNAs. But unlike mRNAs, many transcribed lncRNAs are retained in the nucleus due to inefficient processing by RNA polymerase II (Pol II), whereas others are spliced and exported to the cytoplasm by nucleus RNA export factor 1 (NXF1) (25–27). lncRNAs localized in the nucleus can interact with DNA, RNA, proteins, and other molecules to regulate the structure and function of chromosomes and cis–trans gene transcription as well as influence mRNA splicing, stability and translation. lncRNAs in the cytoplasm mostly trans-regulate gene expression at the post-transcriptional level, including regulating mRNA translation and degradation or intracellular signaling pathways. In addition, some lncRNAs can localize to organelles and regulate their function and metabolism, such as mitochondrial oxidation and homeostasis (28) (Figure 1B).
ncRNAs in SSc
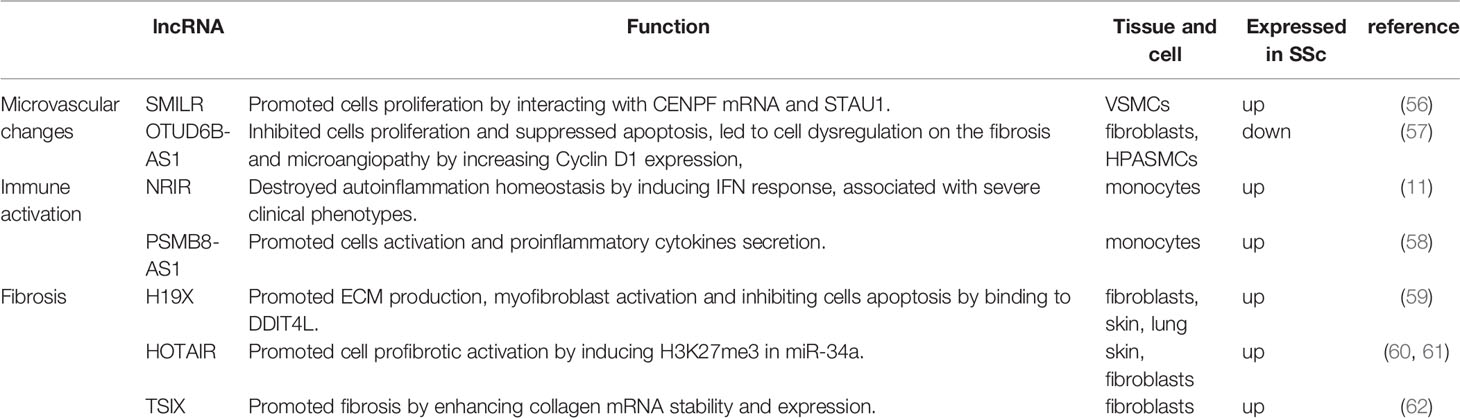
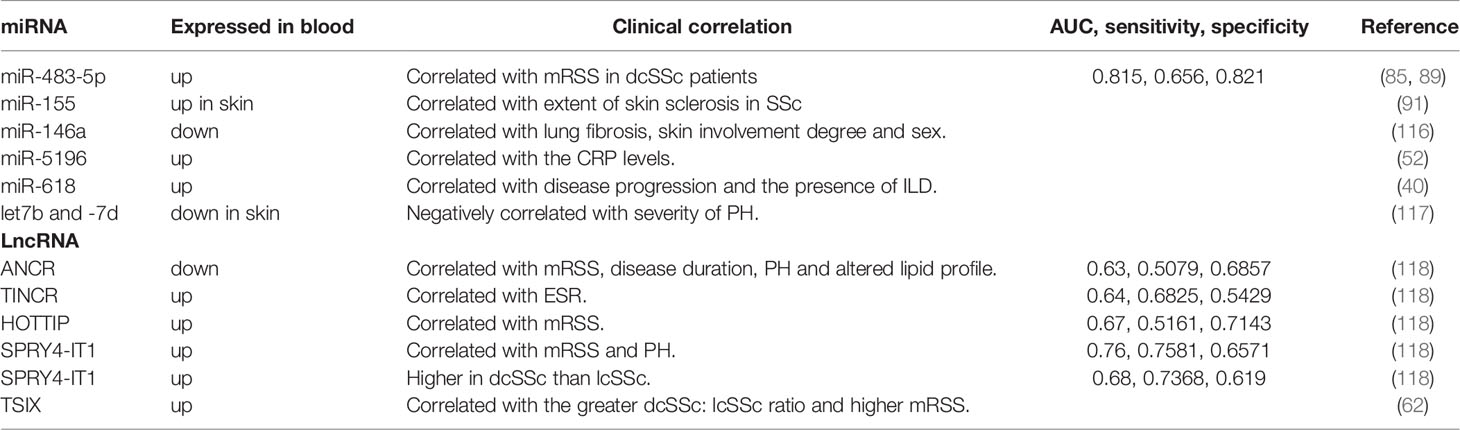
SSc is a chronic, progressive multifocal disease characterized by vascular alterations, autoimmunity, and fibrogenesis caused by environmental factors in genetically susceptible individuals (1). Its earliest stages feature microvascular injury and endothelial cell activation (29). Altered endothelial cells and infiltrating immune cells interact to secret proinflammatory cytokines and prefibrotic mediators, leading to myofibroblast transition and excessive extracellular matrix (ECM) deposition, culminating in fibrosis (30–32). Dysregulated miRNAs and lncRNAs in SSc tissues and blood act as inducers or suppressors, correlating with vascular changes, immune activation, and fibrosis. Herein, we summarize recent studies on the role of miRNAs (Table 1) and lncRNAs (Table 2) in SSc development.
Microvascular Changes
Microvascular injury and endothelial cell activation are the earliest and possibly primary events of SSc (5). Vascular injury leads to a decrease in the number of capillaries, proliferation of intimal and smooth muscle cells, thickening of the vascular wall, and narrowing of the lumen, consequently resulting in tissue hypoxia and oxidative stress (5). In addition, activated endothelial cells promote the expression of adhesion molecule, leading to the recruitment of inflammatory cells and secrete profibrotic factors [e.g., endothelin 1 and connective tissue growth factor (CTGF)] and profibrotic mediators [e.g., TGFβ, platelet-derived growth factor (PDGF), interleukin (IL)-1, and IL-6], which stimulate vascular smooth muscle cell (VSMC) proliferation and ECM production (5).
miRNA
miR-22 has been shown to modulate the expression and phenotype of VSMCs in human femoral arteries, and its overexpression reduces the proliferation and migration of VSMC, formation of neointima by binding to 3′ UTRs of target genes (MECP2, HDAC4, and EVI1) and degrating them (33). miR-193b-3p (miR-193b) was downregulated in fibroblasts and skin biopsy samples of patients with SSc but was unaffected by major profibrotic cytokines and hypoxia. Additionally, miR-193b inhibition resulted in the overexpression of its target, urokinase-type plasminogen activator (uPA), which was strongly expressed in VSMCs in SSc skin section and induced proliferation and inhibited the apoptosis of human pulmonary artery smooth muscle cells (HPSMCs) in an uPAR-independent manner. This led to the proliferative vasculopathy with intimal hyperplasia characteristic in SSc. Such vascular changes reduced blood flow and contributed to pulmonary arterial hypertension. Therefore, miR-193b, as a post-transcriptional regulator of uPA, could be a therapeutic target for SSc-related vasculopathy (34).
lncRNA
Smooth muscle enriched lncRNA (SMILR) is enriched in VSMCs; this recently discovered intergenic lncRNA specifically targets the late mitotic pathway in proliferating VSMCs by interacting with centromere protein F (CENPF) mRNA and STAU1 (56). CENPF controls mitosis, transcriptional regulation, and muscle cell differentiation (63). SMILR knockdown has been shown to silence CENPF, leading to the accumulation of binucleated cells and reduced proliferation (56). Other studies have indicated that STAU1 overexpression affects mitotic entry and impairs the proliferation of transformed cells (64). SMILR and STAU1 reportedly interact to inhibit STAU1 function during the cell cycle, thereby regulating the proliferation of VSMCs. Thus, SMILR could be a novel target for the treatment of aberrant growth of VSMCs (56). Antisense(AS) lncRNAs are transcribed from the opposite strand of protein-coding genes and overlap with one or more exons and introns of the gene’s sense. The expression of most AS lncRNAs is approximately 10-fold lower than that of their coding gene and is more tissue-specific (65–67). AS lncRNAs not only regulate sense gene expression but also act independently of the sense gene (7). lncOTUD6B-AS1 was significantly downregulated in the involved skin tissues compared with noninvolved skin tissues of patients with SSc. OTUD6B-AS1 knockdown significantly reduced proliferation in both dermal fibroblasts and HPASMCs by increasing target cyclin D1 expression at the mRNA and protein levels. It’s report that overexpression of cyclin D1 prevented DNA repair and prevented cells from entering in S phase (68). In addition, OTUD6B-AS1 knockdown favored cells apoptosis resistance, which can be regarded as a compensatory mechanism for inhibition of cells proliferation. Therefore, downregulated OTUD6B-AS1 resulted in cell dysregulation in SSc-related fibrosis and microangiopathy (57).
Immune Activation
Dysregulation of both innate and adaptive immunity may promote SSc. Signs of immune activation in SSc include the presence of inflammatory cells and inflammatory signatures in organs such as the skin and lungs, changes in the number and function of circulating immune cells, presence of type I interferon (IFN) signature in immune cells; and presence of serum autoantibodies. Polymorphisms of IFN regulatory factor 5 (IRF5) and signal transducer and activator of transcription 4 (STAT4), as well as several other immune pathway genes are linked to SSc (69).
miRNA
miR-155 regulates innate and adaptive immune responses (70, 71). Artlett et al. have showed that NLRP3 inflammasomes can enhance IL-1 transcription and promote autocrine signaling, further driving miR-155 expression. Then miR-155 synergizes with the inflammasome to induce a positive feed-forward signal that further promotes IL-1 release, leading to continual collagen expression (35). Bioinformatics analyses revealed that miR-26b-5p functions in fibroblasts and targets CXCL9 and CXCL13 (37). An miR-26b-5p inhibitor was previously reported to promote the activation of negative regulators of the NF-κB and JAK-STAT pathway, thereby hindering the expression of CXCL9 and CXCL13 and subsequently decreasing the mRNA levels of two known fibrosis markers, α-SMA and fibroblast activation protein (FAP), as well as the collagen-type I alpha 2 (Col1A2) and collagen-type IV alpha 1 (Col4A1), consequently impeding fibrosis (37, 72). In addition, Christmann et al. found dysregulated miRNAs in lung and in peripheral blood mononuclear cells (PBMCs) in SSc with interstitial lung disease (SSc-ILD) patients. For example, miR-155, which is highly expressed in SSc-IDL patients, is associated with profibrotic gene expression (SPP1 and POSTN) and lung function test findings, such as high-resolution computed tomography lung score, forced vital capacity, and diffusing capacity of the lung for carbon monoxide. However, miR-155 knockdown blocked the alternative activation of lung macrophages, leading to longer survival and less aggressive lung fibrosis in mice model (10). Increased miR-21 in SSc-IDL lung fibroblasts also correlates with the upregulation of several profibrotic genes, such as Col3a1 and POSTN (10). miR-21 along with miR-155 control toll-like receptor signaling (38), suggesting that they regulate immune activation in SSc-ILD.
In SSc, dysregulation of the immune system precedes effects on skin and lungs. Lung inflammation correlates with progressive lung fibrosis marked by TGFβ (73). miRNAs are a class of regulators that tightly control the overall inflammatory response. Rossato et al. reported the upregulation of miR-618 in plasmacytoid dendritic cells (pDCs) of patients with SSc, which led to the degradation of its target IFN regulatory factor 8 and consequently the eradication of circulating DCs (40), resulting in severe immunodeficiency in the affected patients (74, 75). Furthermore, miR-618 induced pDCs to produce a greater amount of IFNα in response to toll-like receptor 9 stimulation, contributing to the type I IFN signature observed in SSc patients. Such early molecular changes in the pDCs of patients with SSc suggested that miR-618 was an important epigenetic target to regulate immune system homeostasis and had important effects in the immune system and induce autoimmunity (40). Similarly, Chouri et al. showed that miR-126 and miR-139-5p were increased in pDC of patients with SSc in the earliest stage. Their expression upregulated IFN-inducing genes as well as IFIT3, IFI6, IFIT1, and CXCL10, which activate pDCs and trigger SSc via TLR9-mediated response and IFN signaling. Both miRNAs mainly target the PDGF, insulin-like growth factor, vascular endothelial growth factor, and IL signaling pathways, which also contribute to SSc–pDC imbalance. Further, miR-139-5p inhibited the expression of the deubiquitinating enzyme USP24 (41), which negatively regulates type I IFN response and autoimmune response (76). Altogether, the elevated levels of miR-126 and miR-139-5p might reflect the activation of circulating pDCs in SSc.
lncRNA
Monocytes of patients with lcSSc and noncutaneous SSc (ncSSc) showed higher expression of lncNRIR. As an IFN-dependent lncRNA, NRIR positively regulates LPS-induced IFN response in human monocytes. Silencing NRIR suppresses the IFN-stimulated genes CXCL10 and CXCL11 (11), which are linked to type I IFN signature, more severe clinical phenotypes, and lung and kidney involvement (77–79). Knocking down NRIR blocks IFN receptor signaling in SSc, maintaining autoinflammation homeostasis (11). LncPSMB8-AS1 is also upregulated in the blood monocytes of patients with early SSc, ncSSc and dsSSc and is implicated in immune cell activation and vesicle-related transport. PSMB8-AS1 knockdown resulted in decreased concentrations of IL-6 and TNFα in the cell-free supernatant (58), which are linked to fibrotic (80, 81), and ILD (82, 83). Thus, PSMB8-AS1 can modulate monocyte activation in SSc and secretion of proinflammatory cytokines (58).
Fibrosis
The accumulation of a fibrous matrix composed of collagen, elastin, glycosaminoglycan, and fibronectin is a hallmark of SSc. Activated stromal cells increasing synthesis, prolyl and lysyl oxidase and transglutaminase 2 enhancing assembly and crosslinking catalysed, as well as defective degradation together result in excessive accumulation of ECM during fibrosis. This process replaces normal tissue architecture with connective tissue, leading to permanent scarring (84, 85). α-SMA-positive and apoptosis-resistant myofibroblasts in fibrotic tissue secrete matrix molecules, TGFβ, and other profibrotic mediators, promoting ECM accumulation and remodeling (86).
miRNA
miR-483-5p is upregulated in localized scleroderma and SSc (from the early stages of the disease onwards) but not in other systemic autoimmune diseases, indicating that this miRNA could be linked to scleroderma-specific fibrosis. The overexpression of miR-483-5p in fibroblasts and endothelial cells regulates the expression of fibrosis-related genes, such as COL4A1 and COL4A2 encoding for collagen IV (42), a primary collagen in the basement membrane surrounding blood vessels and in the dermoepidermal junction in the skin that positively correlates with the modified Rodnan skin score (mRSS) (87). On the other hand, high miR-483-5p levels in endothelial cells enhance the transcriptional levels of α-SMA and SM22A (two myofibroblast differentiation markers), which promote the myofibroblast phenotype. In addition, miR-483-5p disrupts collagen homeostasis by downregulating Fli-1 and promoting ECM production. Serum exosome samples obtained from patients with SSc have high levels of miR-483-5p, which may contribute to the disease’s fibrotic phase (42). In a study by Shi, the levels of miR-3606-3p were significantly reduced in dermal tissues and primary fibroblasts. miR-3606-3p may function as a novel antifibrotic miRNA by targeting TGF-βreceptor types 2 (TGFBR2): overexpressed miR-3606-3p could significantly decrease the mRNA and protein levels of TGFBR2 by targeting its 3′-UTR, consequently reducing the production of p-SMAD2/3 and type I collagen as well as hindering cell cycle progression and TGF-βassociated signaling (43). The TGF-β pathway promotes fibrosis in SSc by modulating proliferation, activation, and accumulation of fibroblasts and stimulating ECM production (88). Upon stimulation, TGFBR2 first binds to TGF-β, autophosphorylates, and subsequently activates TGFBR1 to trigger a cascade response that regulates a series of fibrosis-related factors, such as collagens, α-SMA, CTGF, MMPs, tissue inhibitor of metalloproteinases (TIMPs), and fibronectin (89–92). Elevated miR-155 expression in skin tissue from SSc patients and mice model, which corresponded to the extent of skin sclerosis, and silencing miR-155 could inhibit fibrosis-associated Wnt/β-catenin and Akt pathways by upregulating the protein levels of CK1αand SHIP-1. In addition, antagomiR-155 treatment effectively reduced bleomycin-induced skin fibrosis in mice, suggesting that miR-155 is a potential therapeutic target for scleroderma (44). Another study reported reduced miR-16-5p expression in patients with SSc compared with healthy controls. miR-16-5p knockdown induced NOTCH2 mRNA expression and activated NOTCH signaling in human skin fibroblasts by hindering binding to NOTCH2 mRNA for degradation, which promoted the expression of α-SMA, Col 1α, and CTGF and inhibited the expression of collagenases MMP1 and MMP8. Indeed, miR-16-5p suppressed myofibroblast activation by decreasing NOTCH signaling and may prove promising in clinical treatments (45). miR-130b was upregulated in the dermis skin biopsy samples of patients with SSc, fibroblasts and skin tissues of fibrosis-mice model. miR-130b increased expression of fibrosis-related genes COL1A1, COL1A2, α-SMA and Fn, enhancing TGF- β signaling by negatively regulating PPARγ, an antifibrotic molecule (46). Similarly, miR-202-3p accumulated in skin tissues of patients with SSc and primary fibroblast compared with normal tissues and increased profibrotic gene expression as well as collagen deposition in primary skin fibroblasts by directly regulating MMP1 (47), which is involved in ECM degradation (93). miR-320a plays an antifibrotic role in SSc-ILD. PBMCs obtained from patients with SSc-ILD as well as lung tissues obtained from bleomycin-induced SSc-ILD mouse models showed reduced levels of miR-320a, which negatively regulates the expression of collagen genes by directly binding to and inhibiting TGFBR2 and IGF1R (48). TGFBR2 is a transmembrane serine/threonine kinase that forms a heterodimeric complex with TGFBR1, which can bind to TGF-β, which subsequently phosphorylates downstream proteins that promote fibrosis (94). IGF1R is the respective receptor of IGF1 signaling pathway, associating with pathological fibrosis states (95, 96).
lncRNA
LncRNA H19X is a key mediator of matrix remodeling in fibroblasts and SSc-related cell types during the fibrotic phase of SSc. Indeed, H19X is upregulated in the skin of patients with SSc when compared with that of HCs. Lung samples of patients with SSc-ILD also show H19X upregulation. This overexpression is not surprising considering that H19X is related to the TGF-β pathway, and TGF-β can promote H19X expression in a time- and dose-dependent manner in SSc-related cell types. The activated TGF-β pathway induces H19X expression, which promoted ECM production, myofibroblast activation and inhibited fibroblast apoptosis. As a pathogenic factor in SSc, H19X inhibits DNA damage-inducible transcript 4-like protein (DDIT4L) expression through the direct contact of H19X RNA with a DNA regulatory element upstream of the DDIT4L (59). DDIT4L inhibits ECM production under normal conditions and may be useful for predicting radiation-induced fibrosis (97). H19X acts as a key effector of TGF-β-induced ECM remodeling and fibrosis (59). Fibroblasts and skin tissues of patients with SSc showed high expression of lncHOTAIR, a potential factor involved in SSc development that drives the profibrotic activation of dermal fibroblasts via the enhancer of zeste 2 (EZH2)/miR-34a/NOTCH/collagen/α-SMA pathway. HOTAIR directs modulates EZH2 to induce H3K27me3 of miR-34a, thereby inhibiting miR-34a expression (60), which consequently suppresses NOTCH (98). This results in increased levels of collagen and α-SMA in vitro and in vivo (60). Other studies also showed that HOTAIR could activate dermal fibroblasts via the EZH2/miR-34a/NOTCH/GIL2 pathway. Upregulated HOTAIR caused NOTCH inhibition by EZH2/miR-34a in above mechanism, leading to GLI2 transcription, which promoted profibrotic markers (collagen, α-SMA, CTGF) expression (61). Dermal fibroblasts upregulate another downstream target of TGF-β, lncTSIX. TSIX knockdown was previously reported to reduce reduced the expression of type I collagen mRNA, which hindered SSc-related fibrosis (62). Messemaker et al. have also found that the expression levels of AS lncRNAs, including CTBP1-AS2, AGAP2-AS1, and OTUD6B-AS1, in SSc patients skin tissues showed variations: CTBP1-AS2 and AGAP2-AS1 increased, whereas OTUD6B-AS1 decreased. These three AS lncRNAs are related to their coding genes, indicating that they play functional roles in SSc pathogenesis (99).
Figure 2 is an illustration of the regulatory effects of various miRNAs and lncRNAs involved in SSc.
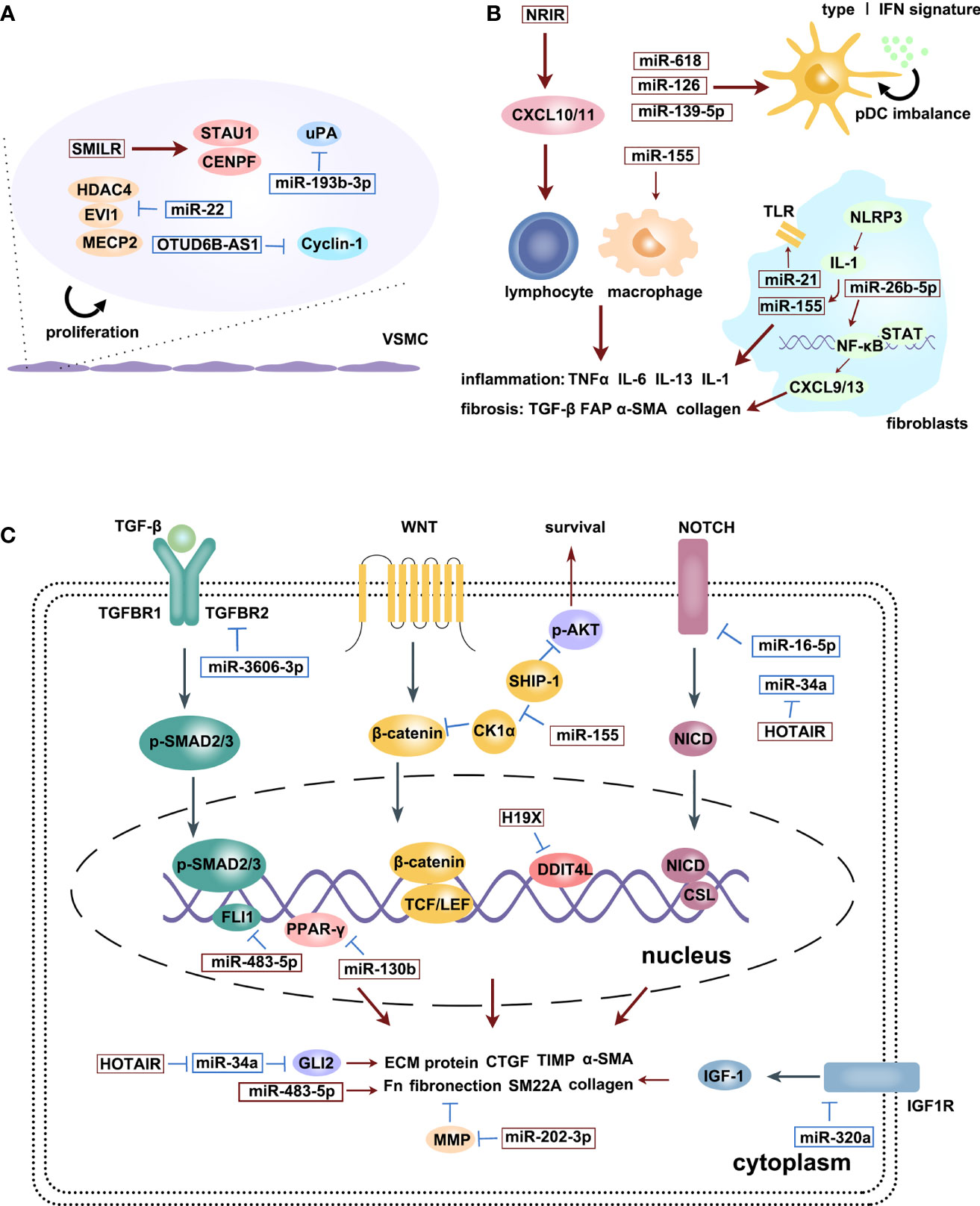
Figure 2 The regulatory effects of various miRNAs and lncRNAs involved in SSc process: microvascular changes (A), immune activation (B) and fibrosis (C). Upregulation (red squares) and downregulation (blue squares) of diverse ncRNAs participate in different signaling pathways, promoting SSc progression. Red arrows are promotion symbols, blue lines are inhibition symbols.
miRNAs as Therapeutic Targets in SSc
The ability to regulate multiple genes enables miRNAs to promote cancer and autoimmune diseases (36, 39). To treat such conditions, miRNAs could be used as therapeutics (miRNA mimics) or as targets of therapeutics (anti-miRs) (49), which rely on the natural RNAi mechanism of miRNAs in cells. miRNAs only need to be partially complementary to their target mRNAs to exert its post-transcriptional and translational inhibitory effects, thereby achieving the therapeutic silencing of multiple target genes (100). The expression of miR-26a-2-3p is lower in the monocytes of patients with SSc than in those of HCs. Exogenous delivery of miRNA-26a-2-3p to TLR-stimulated monocyte can inhibit some IFN-stimulated genes, such as IRF6, GBP2, IFI44L, Siglec1, and IFN-β, thereby blocking IFN signal transduction in SSc development. These findings support the use of miRNA therapeutics as a novel therapy for SSc (50). The serum of patients with SSc showed decreased levels of miR-151-5p. Deriving from exosomes of mesenchymal stem cells, miR-151-5p determined osteogenic versus adipogenic fate and rescued the osteopenia phenotype in Tsk/+ mice by inhibiting the IL-4Rα/mTOR pathway (51). Low levels of IL-4 or IL-4Rα can inhibit the fibrosis and osteopenia phenotypes (101, 102). Systemic delivery of miR-151-5p also prevented osteopenia, skin tightening, and reduced TH2 cell differentiation and T-cell infiltration in the hyperdermal area, indicating that miR-151-5p-based gene therapy could achieve therapeutic effects in multiple organs in Tsk/+mice. Considering that miR-151-5p present in mice and humans is 100% identical, it could be a suitable target for SSc treatment (51). The exogenous transfection of miR-5196 significantly reduced the expression of Fos-related antigen 2 (Fra2, AP-1 transcription factor) by binding to its five seed regions within the 3′-UTR and indirectly inhibited TIMP-1, indicating its potential suppression in SSc (52).
On the other hand, miRNAs in exosomes can also be an effective therapy method for SSc. Exosomes are microvesicles with lipid bilayers, containing miRNAs, proteins, metabolites, and other molecules, which can be secreted by cancer cells and a variety of stromal cells. After secretion, exosomes transduce endocrine, paracrine and autocrine signals and can mediate intercellular communication by releasing their contents into target cells through fusion (103). Remarkably, bone marrow mesenchymal stem cell-derived extracellular vesicles (BMSC-EVs), which contain several highly expressed miRNAs such as let-7 family, miR-99a, and miR-125b, were reported to reduce bleomycin-induced dermal thickening and fibrosis as well as abnormal deposition of collagen in a mouse model of SSc. BMSC-EVs treated SSc mice have decreased TGF-β1-positive cells,α-SMA-positive myofibroblasts, mast cells and infiltrating macrophages as well as lower mRNA levels of the inflammatory cytokines IL6, IL10, and TNF-α. BMSC-EVs might be useful for treating SSc using miRNAs, which could alleviate ECM deposition and inflammatory cell infiltration by regulating the TGFβ and Wnt signaling pathways (53). A previous study reported that there are 6 profibrotic miRNAs and 10 antifibrotic miRNAs in the serum exosomes of patients with SSc; further, the levels of 8 existing miRNAs were significantly different between the exosomes of patients with limited and diffuse SSc. The serum exosomes of patients with SSc patients stimulated normal fibroblast expressing of ECM components, such as Col 1A1, Col 3A1, FN1, and also induced the expression of three myofibroblast-associated genes COMP, α-SMA, and FN-EDA in dose-dependent manner. These findings suggest that exosomes containing miRNAs release their profibrotic contents upon fusion with target cells in their extracellular environment (104).
Apoptosis refers to the endogenous programmed cell death that maintains homeostasis (105). The inhibition of apoptosis is a common hallmark of several diseases, such as cancer (106), fibrosis (107), and autoimmune diseases (108). As is known, fibroblasts are the central mediators of fibrotic manifestations in SSc. In SSc, fibroblasts in lesional areas, which are mostly myofibroblasts, secrete ECM, resulting in excessive scarring of the skin (109). The decreased susceptibility of fibroblasts to apoptosis drives fibrosis in SSc and represents a potential therapeutic target (107). Farsangi et al. showed that miR-21 upregulation enhanced Bcl-2 expression in both SSc and normal fibroblasts (54) by directly targeting Bcl-2 mRNA at the 3′-UTR region (110), thereby preventing apoptosis and promoting cell survival in SSc fibroblasts (54). Another study showed that miR-125b is downregulated in the skin and primary dermal fibroblasts of patients with SSc. Further, miR-125b knockdown decreased the number of α-SMA-positive cells, increased apoptosis, and decreased the proliferation of dermal fibroblasts by increasing the expression of BAK1, BMF, and BBC3, which are involved in the mitochondrial apoptotic pathway. Thus, miR-125b may act as a disease suppressor, exerting compensatory protective effects directed against excessive skin fibrosis (55).
As biomarkers in SSc
Early detection and treatment are key to improving SSc prognosis. As miRNAs and lncRNAs mainly function upstream of various signaling pathways, making them promising diagnostic tools. miRNAs can be released into the extracellular environment by binding to RNA-binding proteins or by secretion from cell-derived plasma exosomes (111–113). Extracellular miRNA expression levels can reflect one’s physiological state, as indicated by the profiles of dysregulated miRNAs in plasma and serum (112, 114). Recent developments in next-generation sequencing have made profiling miRNA levels in biological fluids a viable approach to discover biomarkers (115). Additionally, miRNAs in peripheral blood are stable and resistant to degradation by ribonucleases in the body fluids (112). Thus, miRNAs in blood represent promising new complementary biomarkers for SSc. However, currently, there are few studies on lncRNAs as biomarkers of autoimmune diseases, with only one study on SSc. Herein, we summarized the recent findings on miRNAs and lncRNAs as biomarkers present in the blood of patients with SSc (Table 3).
miRNA
The circulating serum miR-483-5p and snRNA-U6 show elevated expression levels in all SSc disease subsets (early SSc, ncSSc, dcSSc, and lcSSc). Their expression levels are particularly correlated with mRSS in patients with dcSSc. Receiver operating characteristic curve (ROC) analysis revealed the high predictive ability of miR-483-5p as a biomarker of SSc, with area under the curve, sensitivity, and specificity of 0.815, 0.656, and 0.821, respectively. More than 60% of SSc patients have miR-483-5p levels exceeding the potential optimal cut-off of normal levels (89). Vreca et al. showed that miR-146a relates to the predisposition and phenotypic heterogeneity of SSc, namely the higher prevalence of miR-146a rs2910164 C > G genotype in patients with SSc having lung fibrosis and an active form of the disease. In addition, miR-146a was downregulated in PBMCs of patients with SSc compared with those of HCs, and the levels of miR-146a were lower in women than in men. Patients with SSc showing mild skin involvement (mRSS score ≤ 10) expressed 36% less miR-146a than those with severe skin involvement. Altogether, reduced miR-146a expression represents a risk factor for the development and progression of SSc (116). Another study reported elevated circulating miR-5196 in the serum and monocytes of patients with SSc, which were positively correlated with C-reactive protein levels. This suggests miR-5196 can be used as a biomarker of inflammation in SSc (52). Rossato’s study showed that miR-618 is incrementally upregulated in pDCs of patients with early SSc (fold change, 1.35), SSc patients without fibrosis (fold change, 1.41), and patients with overt fibrosis (fold change, 3.85), including those with dcSSc with a disease duration of ≤2 years (fold change, 3.68). Higher levels of miR-618 were also associated with ILD (40). Rusek et al. found that miR-4484 was also upregulated up to 18-folds in the serum of patients with SSc compared with that of HCs (119).
Pulmonary hypertension (PH) is one of the most serious complications in patients with SSc. A meta-analysis reported that the prevalence of PH is 5%–14% among patients with SSc (120). Identifying biomarkers that can predict the presence and progression of PH is crucial because it worsens the prognosis of patients with SSc (121). Izumiya et al. found that five let-7 family members (let-7a, -7d, -7e, -7f,and -7g) were downregulated in six PH skin tissues obtained from patients with SSc; let-7b and -7d expression levels were significantly and negatively correlated with the severity of PH in patients with SSc, indicating that skin miRNA levels could be a useful marker for predicting the presence and severity of PH in patients with SSc (117).
lncRNA
Four lncRNAs associated with skin biology (122)—ANCR, TINCR, HOTTIP, and SPRY4-IT1 —have been identified as novel candidate biomarkers for SSc. Recent evidence suggests that TINCR, HOTTIP, and SPRY4-IT1 are upregulated, whereas ANCR is downregulated in plasma of patients with SSc compared with HCs. SPRY4-IT1 was significantly overexpressed in patients with dcSSC than in those with lcSSc, indicating its potential in the diagnosis of subtypes. SPRY-IT1, HOTTIP, and ANCR were also correlated with mRSS: ANCR expression was related to disease duration, the presence of PH, and altered lipid profile in patients with SSc, TINCR was positively correlated with ESR; and SPRY4-IT1 was associated with the presence of PH (118). Wang et al. found upregulated levels of lncTSIX in the serum of patients with SSc and fibroblast-conditioned medium, suggesting that dermal fibroblasts release and increase TSIX levels in patients with SSc. Elevated serum TSIX levels are a potential SSc biomarker as they are related to a significantly greater dcSSc:lcSSc ratio and higher mRSS (62).
Conclusion
The high mortality and difficult diagnosis of SSc burden patients and society. Currently, next-generation sequencing technologies and other high-throughput methods can be used to readily detect the different expression levels of miRNAs and lncRNAs in healthy and disease conditions. Normally, the levels of miRNAs and lncRNAs are stable owing to the physiological balance between synthesis and degradation. However, dysregulated miRNA or lncRNA expression changes their functions and may promote the onset of SSc. In this review, we elaborated on the propathogenesis and antipathogenesis mechanisms of ncRNAs (miRNAs and lncRNAs) in the different stages of SSc, including microvascular changes, inflammation, and fibrosis. Several databases such as miRBase and starBase can be used to predict the functions of miRNAs and lncRNAs and their interactions with targeted proteins. Research on miRNAs and lncRNAs in SSc has focused on the transcriptional and post-transcriptional levels rather than post-translation level. Hence, further research on post-translational modifications is warranted to better understand the pathogenesis of autoimmune diseases. Recent studies have shown that some miRNAs and lncRNAs are highly expressed in the blood, indicating their potential value as noninvasive biomarkers for the diagnosis and prognosis of SSc. Compared with traditional proteins markers, ncRNAs appear more precise and sensitive, although their practicality is yet to be tested. Moreover, miRNAs may represent therapeutic targets to inhibit key pathogenic molecules and switch phenotypic changes involved in the development of SSc via siRNA, AS oligonucleotides, or CRISPR–Cas9-mediated genome editing. However, few applications of miRNAs for SSc-targeted therapy exist. Advances described in this review may stimulate additional research necessary to better understand the basic biology of miRNAs and lncRNAs and explore their potential as tools for clinical application in cases of SSc and other autoimmune diseases.
Author Contributions
YZL and YML provided direction and guidance throughout the preparation of this manuscript. HZ, HL, XL, and YH collected and prepared the related literature. YML drafted the manuscript. YZL, LC, HZ, and HL reviewed and made significant revisions to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFE0207300), the National Natural Science Foundation of China (81871302), Beijing Municipal Science & Technology Commission (Z211100002521021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Shangqing Yang (engineer) for providing guidance on figure preparation.
Abbreviations
AGO, Argonaute; BMSC-EVs, bone marrow mesenchymal stem cell -derived extracellular vesicles; circRNAs, circular RNAs; CENPE, centromere protein F; Col1A2, collagen-typeIalpha 2; Col4A1, collagen-typeIValpha 1; CTGF, connective tissue growth factor; DDIT4L, DNA damage-inducible transcript 4-like protein; ECM, excessive extracellular matrix; EZH2, enhancer of zeste 2; FAP, fibroblast activation protein; HPASMCs, human pulmonary artery smooth muscle cells; IFN, interferon; IRF5, IFN regulatory factor 5; mRNA, message RNA; miRNAs, microRNAs; mRSS, modified Rodnan skin score; m7G, 7-methyl guanosine; lncRNAs, long non-coding RNAs; ncRNAs, non-coding RNAs; ncSSc, non-cutaneous SSc; NXF1, nucleus RNA export factor 1; PBMCs, peripheral blood mononuclear cells; pDCs, plasmacytoid dendritic cells; PDGF, platelet-derived growth factor; piRNAs, piwi-interacting RNAs; pri-miRNA, primary miRNA; RISC, RNA-induced silencing complex; STAT4, signal transducer and activator of transcription 4; SMILR, Smooth muscle enriched long non-coding RNA; SSc, systemic sclerosis; lcSSc, limited cutaneous SSc; dcSSc, diffuse cutaneous SSc; TGFBR2, TGF-β receptor types 2; TIMP-1, tissue inhibitor of metalloproteinases 1; tsRNAs, transfer RNA-derived small RNAs; TRBP, transactivation response element RNA-binding protein; TLR-9, toll-like receptor 9; UTR, untranslated region; uPA, urokinase-type plasminogen activator; VSMC, vascular smooth muscle cell.
References
1. Katsumoto TR, Whitfield ML, Connolly MK. The Pathogenesis of Systemic Sclerosis. Annu Rev Pathol (2011) 6:509–37. doi: 10.1146/annurev-pathol-011110-130312
2. Barnes J, Mayes MD. Epidemiology of Systemic Sclerosis: Incidence, Prevalence, Survival, Risk Factors, Malignancy, and Environmental Triggers. Curr Opin Rheumatol (2012) 24(2):165–70. doi: 10.1097/BOR.0b013e32834ff2e8
3. Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, et al. Systemic Sclerosis. Nat Rev Dis Primers (2015) 1:15002. doi: 10.1038/nrdp.2015.2
4. Denton CP, Khanna D. Systemic Sclerosis. Lancet (2017) 390(10103):1685–99. doi: 10.1016/S0140-6736(17)30933-9
5. van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. Classification Criteria for Systemic Sclerosis: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann Rheum Dis (2013) 72(11):1747–55. doi: 10.1136/annrheumdis-2013-204424
6. Jordan S, Maurer B, Toniolo M, Michel B, Distler O. Performance of the New ACR/EULAR Classification Criteria for Systemic Sclerosis in Clinical Practice. Rheumatol (Oxf) (2015) 54(8):1454–8. doi: 10.1093/rheumatology/keu530
7. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
8. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting Noncoding RNAs in Disease. J Clin Invest (2017) 127(3):761–71. doi: 10.1172/JCI84424
9. Neil CR, Fairbrother WG. Intronic RNA: Ad'junk' Mediator of Post-Transcriptional Gene Regulation. Biochim Biophys Acta Gene Regul Mech (2019) 1862(11-12):194439. doi: 10.1016/j.bbagrm.2019.194439
10. Christmann RB, Wooten A, Sampaio-Barros P, Borges CL, Carvalho CR, Kairalla RA, et al. miR-155 in the Progression of Lung Fibrosis in Systemic Sclerosis. Arthritis Res Ther (2016) 18(1):155. doi: 10.1186/s13075-016-1054-6
11. Mariotti B, Servaas NH, Rossato M, Tamassia N, Cassatella MA, Cossu M, et al. The Long Non-Coding RNA NRIR Drives IFN-Response in Monocytes: Implication for Systemic Sclerosis. Front Immunol (2019) 10:100. doi: 10.3389/fimmu.2019.00100
12. Kristensen LS, Andersen MS, Stagsted L, Ebbesen KK, Hansen TB, Kjems J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat Rev Genet (2019) 20(11):675–91. doi: 10.1038/s41576-019-0158-7
13. Das S, Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, et al. The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell (2019) 177(2):231–42. doi: 10.1016/j.cell.2019.03.023
14. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, et al. A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature (2017) 552(7683):57–62. doi: 10.1038/nature25005
15. Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, et al. The Emerging Role of the piRNA/Piwi Complex in Cancer. Mol Cancer (2019) 18(1):123. doi: 10.1186/s12943-019-1052-9
16. Lee RC, Feinbaum RL, Ambros V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell (1993) 75(5):843–54.
17. Liu Q, Zhu Y, Zhu W, Zhang G, Yang YP, Zhao C. The Role of MicroRNAs in Tendon Injury, Repair, and Related Tissue Engineering. Biomaterials (2021) 277:121083. doi: 10.1016/j.biomaterials.2021.121083
18. He L, Hannon GJ. MicroRNAs: Small RNAs With a Big Role in Gene Regulation. Nat Rev Genet (2004) 5(7):522–31. doi: 10.1038/nrg1379
19. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA Maturation: Stepwise Processing and Subcellular Localization. EMBO J (2002) 21(17):4663–70. doi: 10.1093/emboj/cdf476
20. Burke JM, Kuny CV, Kincaid RP, Sullivan CS. Identification, Validation, and Characterization of Noncanonical miRNAs. Methods (2015) 91:57–68. doi: 10.1016/j.ymeth.2015.07.013
21. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear Export of microRNA Precursors. Science (2004) 303(5654):95–8. doi: 10.1126/science.1090599
22. Treiber T, Treiber N, Meister G. Regulation of microRNA Biogenesis and its Crosstalk With Other Cellular Pathways. Nat Rev Mol Cell Biol (2019) 20(1):5–20. doi: 10.1038/s41580-018-0059-1
23. Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the Assembly of the RNAi Enzyme Complex. Cell (2003) 115(2):199–208. doi: 10.1016/S0092-8674(03)00759-1
24. Hutvagner G. Small RNA Asymmetry in RNAi: Function in RISC Assembly and Gene Regulation. FEBS Lett (2005) 579(26):5850–7. doi: 10.1016/j.febslet.2005.08.071
25. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a Complete Map of the Human Long Non-Coding RNA Transcriptome. Nat Rev Genet (2018) 19(9):535–48. doi: 10.1038/s41576-018-0017-y
26. Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, et al. NONCODEV5: A Comprehensive Annotation Database for Long Non-Coding RNAs. Nucleic Acids Res (2018) 46(D1):D308–14. doi: 10.1093/nar/gkx1107
27. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet (2017) 33(8):540–52. doi: 10.1016/j.tig.2017.05.004
28. Gil N, Ulitsky I. Regulation of Gene Expression by Cis-Acting Long non-Coding RNAs. Nat Rev Genet (2020) 21(2):102–17. doi: 10.1038/s41576-019-0184-5
29. Gabrielli A, Avvedimento EV, Krieg T. Scleroderma. N Engl J Med (2009) 360(19):1989–2003. doi: 10.1056/NEJMra0806188
30. Abraham DJ, Varga J. Scleroderma: From Cell and Molecular Mechanisms to Disease Models. Trends Immunol (2005) 26(11):587–95. doi: 10.1016/j.it.2005.09.004
31. Varga J, Abraham D. Systemic Sclerosis: A Prototypic Multisystem Fibrotic Disorder. J Clin Invest (2007) 117(3):557–67. doi: 10.1172/JCI31139
32. Chizzolini C, Brembilla NC, Montanari E, Truchetet ME. Fibrosis and Immune Dysregulation in Systemic Sclerosis. Autoimmun Rev (2011) 10(5):276–81. doi: 10.1016/j.autrev.2010.09.016
33. Yang F, Chen Q, He S, Yang M, Maguire EM, An W, et al. miR-22 Is a Novel Mediator of Vascular Smooth Muscle Cell Phenotypic Modulation and Neointima Formation. Circulation (2018) 137(17):1824–41. doi: 10.1161/CIRCULATIONAHA.117.027799
34. Iwamoto N, Vettori S, Maurer B, Brock M, Pachera E, Jungel A, et al. Downregulation of miR-193b in Systemic Sclerosis Regulates the Proliferative Vasculopathy by Urokinase-Type Plasminogen Activator Expression. Ann Rheum Dis (2016) 75(1):303–10. doi: 10.1136/annrheumdis-2014-205326
35. Artlett CM, Sassi-Gaha S, Hope JL, Feghali-Bostwick CA, Katsikis PD. Mir-155 is Overexpressed in Systemic Sclerosis Fibroblasts and is Required for NLRP3 Inflammasome-Mediated Collagen Synthesis During Fibrosis. Arthritis Res Ther (2017) 19(1):144. doi: 10.1186/s13075-017-1331-z
36. Shenouda SK, Alahari SK. MicroRNA Function in Cancer: Oncogene or a Tumor Suppressor? Cancer Metastasis Rev (2009) 28(3-4):369–78. doi: 10.1007/s10555-009-9188-5
37. Sun YH, Xie M, Wu SD, Zhang J, Huang CZ. Identification and Interaction Analysis of Key Genes and MicroRNAs in Systemic Sclerosis by Bioinformatics Approaches. Curr Med Sci (2019) 39(4):645–52. doi: 10.1007/s11596-019-2086-3
38. Quinn SR, O'Neill LA. A Trio of microRNAs That Control Toll-Like Receptor Signalling. Int Immunol (2011) 23(7):421–5. doi: 10.1093/intimm/dxr034
39. Liu H, Chen Y, Yin G, Xie Q. Therapeutic Prospects of MicroRNAs Carried by Mesenchymal Stem Cells-Derived Extracellular Vesicles in Autoimmune Diseases. Life Sci (2021) 277:119458. doi: 10.1016/j.lfs.2021.119458
40. Rossato M, Affandi AJ, Thordardottir S, Wichers C, Cossu M, Broen J, et al. Association of MicroRNA-618 Expression With Altered Frequency and Activation of Plasmacytoid Dendritic Cells in Patients With Systemic Sclerosis. Arthritis Rheumatol (2017) 69(9):1891–902. doi: 10.1002/art.40163
41. Chouri E, Wang M, Hillen MR, Angiolilli C, Silva-Cardoso SC, Wichers C, et al. Implication of miR-126 and miR-139-5p in Plasmacytoid Dendritic Cell Dysregulation in Systemic Sclerosis. J Clin Med (2021) 10(3):491. doi: 10.3390/jcm10030491
42. Chouri E, Servaas NH, Bekker C, Affandi AJ, Cossu M, Hillen MR, et al. Serum microRNA Screening and Functional Studies Reveal miR-483-5p as a Potential Driver of Fibrosis in Systemic Sclerosis. J Autoimmun (2018) 89:162–70. doi: 10.1016/j.jaut.2017.12.015
43. Shi X, Liu Q, Li N, Tu W, Luo R, Mei X, et al. MiR-3606-3p Inhibits Systemic Sclerosis Through Targeting TGF-Beta Type II Receptor. Cell Cycle (2018) 17(16):1967–78. doi: 10.1080/15384101.2018.1509621
44. Yan Q, Chen J, Li W, Bao C, Fu Q. Targeting miR-155 to Treat Experimental Scleroderma. Sci Rep (2016) 6:20314. doi: 10.1038/srep20314
45. Yao Q, Xing Y, Wang Z, Liang J, Lin Q, Huang M, et al. MiR-16-5p Suppresses Myofibroblast Activation in Systemic Sclerosis by Inhibiting NOTCH Signaling. Aging (Albany NY) (2020) 13(2):2640–54.
46. Luo H, Zhu H, Zhou B, Xiao X, Zuo X. MicroRNA-130b Regulates Scleroderma Fibrosis by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Mod Rheumatol (2015) 25(4):595–602. doi: 10.3109/14397595.2014.1001311
47. Zhou B, Zhu H, Luo H, Gao S, Dai X, Li Y, et al. MicroRNA-202-3p Regulates Scleroderma Fibrosis by Targeting Matrix Metalloproteinase 1. BioMed Pharmacother (2017) 87:412–8. doi: 10.1016/j.biopha.2016.12.080
48. Li Y, Huang J, Hu C, Zhou J, Xu D, Hou Y, et al. MicroRNA-320a: An Important Regulator in the Fibrotic Process in Interstitial Lung Disease of Systemic Sclerosis. Arthritis Res Ther (2021) 23(1):21. doi: 10.1186/s13075-020-02411-9
49. Petrovic N, Ergun S. miRNAs as Potential Treatment Targets and Treatment Options in Cancer. Mol Diagn Ther (2018) 22(2):157–68. doi: 10.1007/s40291-017-0314-8
50. Ciechomska M, Wojtas B, Swacha M, Olesinska M, Benes V, Maslinski W. Global miRNA and mRNA Expression Profiles Identify miRNA-26a-2-3p-Dependent Repression of IFN Signature in Systemic Sclerosis Human Monocytes. Eur J Immunol (2020) 50(7):1057–66. doi: 10.1002/eji.201948428
51. Chen C, Wang D, Moshaverinia A, Liu D, Kou X, Yu W, et al. Mesenchymal Stem Cell Transplantation in Tight-Skin Mice Identifies miR-151-5p as a Therapeutic Target for Systemic Sclerosis. Cell Res (2017) 27(4):559–77. doi: 10.1038/cr.2017.11
52. Ciechomska M, Zarecki P, Merdas M, Swierkot J, Morgiel E, Wiland P, et al. The Role of microRNA-5196 in the Pathogenesis of Systemic Sclerosis. Eur J Clin Invest (2017) 47(8):555–64. doi: 10.1111/eci.12776
53. Jin J, Ou Q, Wang Z, Tian H, Xu JY, Gao F, et al. BMSC-Derived Extracellular Vesicles Intervened the Pathogenic Changes of Scleroderma in Mice Through miRNAs. Stem Cell Res Ther (2021) 12(1):327. doi: 10.1186/s13287-021-02400-y
54. Jafarinejad-Farsangi S, Farazmand A, Gharibdoost F, Karimizadeh E, Noorbakhsh F, Faridani H, et al. Inhibition of MicroRNA-21 Induces Apoptosis in Dermal Fibroblasts of Patients With Systemic Sclerosis. Int J Dermatol (2016) 55(11):1259–67. doi: 10.1111/ijd.13308
55. Kozlova A, Pachera E, Maurer B, Jungel A, Distler J, Kania G, et al. Regulation of Fibroblast Apoptosis and Proliferation by MicroRNA-125b in Systemic Sclerosis. Arthritis Rheumatol (2019) 71(12):2068–80. doi: 10.1002/art.41041
56. Mahmoud AD, Ballantyne MD, Miscianinov V, Pinel K, Hung J, Scanlon JP, et al. The Human-Specific and Smooth Muscle Cell-Enriched LncRNA SMILR Promotes Proliferation by Regulating Mitotic CENPF mRNA and Drives Cell-Cycle Progression Which Can Be Targeted to Limit Vascular Remodeling. Circ Res (2019) 125(5):535–51. doi: 10.1161/CIRCRESAHA.119.314876
57. Takata M, Pachera E, Frank-Bertoncelj M, Kozlova A, Jungel A, Whitfield ML, et al. OTUD6B-AS1 Might Be a Novel Regulator of Apoptosis in Systemic Sclerosis. Front Immunol (2019) 10:1100. doi: 10.3389/fimmu.2019.01100
58. Servaas NH, Mariotti B, van der Kroef M, Wichers C, Pandit A, Bazzoni F, et al. Characterization of Long Non-Coding RNAs in Systemic Sclerosis Monocytes: A Potential Role for PSMB8-AS1 in Altered Cytokine Secretion. Int J Mol Sci (2021) 22(9):4365. doi: 10.3390/ijms22094365
59. Pachera E, Assassi S, Salazar GA, Stellato M, Renoux F, Wunderlin A, et al. Long Noncoding RNA H19X is a Key Mediator of TGF-Beta-Driven Fibrosis. J Clin Invest (2020) 130(9):4888–905. doi: 10.1172/JCI135439
60. Wasson CW, Abignano G, Hermes H, Malaab M, Ross RL, Jimenez SA, et al. Long non-Coding RNA HOTAIR Drives EZH2-Dependent Myofibroblast Activation in Systemic Sclerosis Through miRNA 34a-Dependent Activation of NOTCH. Ann Rheum Dis (2020) 79(4):507–17. doi: 10.1136/annrheumdis-2019-216542
61. Wasson CW, Ross RL, Wells R, Corinaldesi C, Georgiou IC, Riobo-Del GN, et al. Long non-Coding RNA HOTAIR Induces GLI2 Expression Through Notch Signalling in Systemic Sclerosis Dermal Fibroblasts. Arthritis Res Ther (2020) 22(1):286. doi: 10.1186/s13075-020-02376-9
62. Wang Z, Jinnin M, Nakamura K, Harada M, Kudo H, Nakayama W, et al. Long Non-Coding RNA TSIX is Upregulated in Scleroderma Dermal Fibroblasts and Controls Collagen mRNA Stabilization. Exp Dermatol (2016) 25(2):131–6. doi: 10.1111/exd.12900
63. Ma L, Zhao X, Zhu X. Mitosin/CENP-F in Mitosis, Transcriptional Control, and Differentiation. J BioMed Sci (2006) 13(2):205–13. doi: 10.1007/s11373-005-9057-3
64. Boulay K, Ghram M, Viranaicken W, Trepanier V, Mollet S, Frechina C, et al. Cell Cycle-Dependent Regulation of the RNA-Binding Protein Staufen1. Nucleic Acids Res (2014) 42(12):7867–83. doi: 10.1093/nar/gku506
65. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
66. Fatica A, Bozzoni I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat Rev Genet (2014) 15(1):7–21. doi: 10.1038/nrg3606
67. Clark BS, Blackshaw S. Long Non-Coding RNA-Dependent Transcriptional Regulation in Neuronal Development and Disease. Front Genet (2014) 5:164. doi: 10.3389/fgene.2014.00164
68. Fukami-Kobayashi J, Mitsui Y. Cyclin D1 Inhibits Cell Proliferation Through Binding to PCNA and Cdk2. Exp Cell Res (1999) 246(2):338–47. doi: 10.1006/excr.1998.4306
69. Mayes MD, Bossini-Castillo L, Gorlova O, Martin JE, Zhou X, Chen WV, et al. Immunochip Analysis Identifies Multiple Susceptibility Loci for Systemic Sclerosis. Am J Hum Genet (2014) 94(1):47–61. doi: 10.1016/j.ajhg.2013.12.002
70. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, et al. Regulation of the Germinal Center Response by microRNA-155. Science (2007) 316(5824):604–8. doi: 10.1126/science.1141229
71. Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, et al. The microRNA miR-155 Controls CD8(+) T Cell Responses by Regulating Interferon Signaling. Nat Immunol (2013) 14(6):593–602. doi: 10.1038/ni.2576
72. Richmond A. Nf-Kappa B, Chemokine Gene Transcription and Tumour Growth. Nat Rev Immunol (2002) 2(9):664–74. doi: 10.1038/nri887
73. Christmann RB, Sampaio-Barros P, Stifano G, Borges CL, de Carvalho CR, Kairalla R, et al. Association of Interferon- and Transforming Growth Factor Beta-Regulated Genes and Macrophage Activation With Systemic Sclerosis-Related Progressive Lung Fibrosis. Arthritis Rheumatol (2014) 66(3):714–25. doi: 10.1002/art.38288
74. Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu Rev Immunol (2008) 26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400
75. Gottenberg JE, Cagnard N, Lucchesi C, Letourneur F, Mistou S, Lazure T, et al. Activation of IFN Pathways and Plasmacytoid Dendritic Cell Recruitment in Target Organs of Primary Sjogren's Syndrome. Proc Natl Acad Sci USA (2006) 103(8):2770–5. doi: 10.1073/pnas.0510837103
76. Chiang CY, Engel A, Opaluch AM, Ramos I, Maestre AM, Secundino I, et al. Cofactors Required for TLR7- and TLR9-Dependent Innate Immune Responses. Cell Host Microbe (2012) 11(3):306–18. doi: 10.1016/j.chom.2012.02.002
77. Eloranta ML, Franck-Larsson K, Lovgren T, Kalamajski S, Ronnblom A, Rubin K, et al. Type I Interferon System Activation and Association With Disease Manifestations in Systemic Sclerosis. Ann Rheum Dis (2010) 69(7):1396–402. doi: 10.1136/ard.2009.121400
78. Farina G, Lafyatis D, Lemaire R, Lafyatis R. A Four-Gene Biomarker Predicts Skin Disease in Patients With Diffuse Cutaneous Systemic Sclerosis. Arthritis Rheum (2010) 62(2):580–8. doi: 10.1002/art.27220
79. Antonelli A, Ferri C, Fallahi P, Ferrari SM, Giuggioli D, Colaci M, et al. CXCL10 (Alpha) and CCL2 (Beta) Chemokines in Systemic Sclerosis–a Longitudinal Study. Rheumatol (Oxf) (2008) 47(1):45–9. doi: 10.1093/rheumatology/kem313
80. Gourh P, Arnett FC, Assassi S, Tan FK, Huang M, Diekman L, et al. Plasma Cytokine Profiles in Systemic Sclerosis: Associations With Autoantibody Subsets and Clinical Manifestations. Arthritis Res Ther (2009) 11(5):R147. doi: 10.1186/ar2821
81. Scala E, Pallotta S, Frezzolini A, Abeni D, Barbieri C, Sampogna F, et al. Cytokine and Chemokine Levels in Systemic Sclerosis: Relationship With Cutaneous and Internal Organ Involvement. Clin Exp Immunol (2004) 138(3):540–6. doi: 10.1111/j.1365-2249.2004.02642.x
82. De Lauretis A, Sestini P, Pantelidis P, Hoyles R, Hansell DM, Goh NS, et al. Serum Interleukin 6 Is Predictive of Early Functional Decline and Mortality in Interstitial Lung Disease Associated With Systemic Sclerosis. J Rheumatol (2013) 40(4):435–46. doi: 10.3899/jrheum.120725
83. Schmidt K, Martinez-Gamboa L, Meier S, Witt C, Meisel C, Hanitsch LG, et al. Bronchoalveoloar Lavage Fluid Cytokines and Chemokines as Markers and Predictors for the Outcome of Interstitial Lung Disease in Systemic Sclerosis Patients. Arthritis Res Ther (2009) 11(4):R111. doi: 10.1186/ar2766
84. Bhattacharyya S, Wei J, Varga J. Understanding Fibrosis in Systemic Sclerosis: Shifting Paradigms, Emerging Opportunities. Nat Rev Rheumatol (2011) 8(1):42–54.
85. Castelino FV, Varga J. Emerging Cellular and Molecular Targets in Fibrosis: Implications for Scleroderma Pathogenesis and Targeted Therapy. Curr Opin Rheumatol (2014) 26(6):607–14. doi: 10.1097/BOR.0000000000000110
86. Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmouliere A, Varga J, et al. Recent Developments in Myofibroblast Biology: Paradigms for Connective Tissue Remodeling. Am J Pathol (2012) 180(4):1340–55. doi: 10.1016/j.ajpath.2012.02.004
87. Motegi SI, Sekiguchi A, Fujiwara C, Toki S, Ishikawa O. Possible Association of Elevated Serum Collagen Type IV Level With Skin Sclerosis in Systemic Sclerosis. J Dermatol (2017) 44(2):167–72. doi: 10.1111/1346-8138.13564
88. Pattanaik D, Brown M, Postlethwaite BC, Postlethwaite AE. Pathogenesis of Systemic Sclerosis. Front Immunol (2015) 6:272. doi: 10.3389/fimmu.2015.00272
89. Chu H, Jiang S, Liu Q, Ma Y, Zhu X, Liang M, et al. Sirtuin1 Protects Against Systemic Sclerosis-Related Pulmonary Fibrosis by Decreasing Proinflammatory and Profibrotic Processes. Am J Respir Cell Mol Biol (2018) 58(1):28–39. doi: 10.1165/rcmb.2016-0192OC
90. Wang JC, Lai S, Guo X, Zhang X, de Crombrugghe B, Sonnylal S, et al. Attenuation of Fibrosis In Vitro and In Vivo With SPARC siRNA. Arthritis Res Ther (2010) 12(2):R60. doi: 10.1186/ar2973
91. Chu H, Shi Y, Jiang S, Zhong Q, Zhao Y, Liu Q, et al. Treatment Effects of the Traditional Chinese Medicine Shenks in Bleomycin-Induced Lung Fibrosis Through Regulation of TGF-Beta/Smad3 Signaling and Oxidative Stress. Sci Rep (2017) 7(1):2252. doi: 10.1038/s41598-017-02293-z
92. Liu Q, Chu H, Ma Y, Wu T, Qian F, Ren X, et al. Salvianolic Acid B Attenuates Experimental Pulmonary Fibrosis Through Inhibition of the TGF-Beta Signaling Pathway. Sci Rep (2016) 6:27610. doi: 10.1038/srep27610
93. Gross J, Lapiere CM. Collagenolytic Activity in Amphibian Tissues: A Tissue Culture Assay. Proc Natl Acad Sci USA (1962) 48:1014–22.
94. Penttinen RP, Kobayashi S, Bornstein P. Transforming Growth Factor Beta Increases mRNA for Matrix Proteins Both in the Presence and in the Absence of Changes in mRNA Stability. Proc Natl Acad Sci USA (1988) 85(4):1105–8. doi: 10.1073/pnas.85.4.1105
95. Li J, Choi E, Yu H, Bai XC. Structural Basis of the Activation of Type 1 Insulin-Like Growth Factor Receptor. Nat Commun (2019) 10(1):4567. doi: 10.1038/s41467-019-12564-0
96. Uh ST, Inoue Y, King TJ, Chan ED, Newman LS, Riches DW. Morphometric Analysis of Insulin-Like Growth Factor-I Localization in Lung Tissues of Patients With Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med (1998) 158(5 Pt 1):1626–35. doi: 10.1164/ajrccm.158.5.9804025
97. Forrester HB, Li J, Leong T, McKay MJ, Sprung CN. Identification of a Radiation Sensitivity Gene Expression Profile in Primary Fibroblasts Derived From Patients Who Developed Radiotherapy-Induced Fibrosis. Radiother Oncol (2014) 111(2):186–93. doi: 10.1016/j.radonc.2014.03.007
98. Andersson ER, Sandberg R, Lendahl U. Notch Signaling: Simplicity in Design, Versatility in Function. Development (2011) 138(17):3593–612. doi: 10.1242/dev.063610
99. Messemaker TC, Chadli L, Cai G, Goelela VS, Boonstra M, Dorjee AL, et al. Antisense Long Non-Coding RNAs Are Deregulated in Skin Tissue of Patients With Systemic Sclerosis. J Invest Dermatol (2018) 138(4):826–35. doi: 10.1016/j.jid.2017.09.053
100. Labatut AE, Mattheolabakis G. Non-Viral Based miR Delivery and Recent Developments. Eur J Pharm Biopharm (2018) 128:82–90. doi: 10.1016/j.ejpb.2018.04.018
101. Kodera T, McGaha TL, Phelps R, Paul WE, Bona CA. Disrupting the IL-4 Gene Rescues Mice Homozygous for the Tight-Skin Mutation From Embryonic Death and Diminishes TGF-Beta Production by Fibroblasts. Proc Natl Acad Sci USA (2002) 99(6):3800–5. doi: 10.1073/pnas.052709999
102. Chen C, Akiyama K, Wang D, Xu X, Li B, Moshaverinia A, et al. mTOR Inhibition Rescues Osteopenia in Mice With Systemic Sclerosis. J Exp Med (2015) 212(1):73–91. doi: 10.1084/jem.20140643
103. Milman N, Ginini L, Gil Z. Exosomes and Their Role in Tumorigenesis and Anticancer Drug Resistance. Drug Resist Update (2019) 45:1–12. doi: 10.1016/j.drup.2019.07.003
104. Wermuth PJ, Piera-Velazquez S, Jimenez SA. Exosomes Isolated From Serum of Systemic Sclerosis Patients Display Alterations in Their Content of Profibrotic and Antifibrotic microRNA and Induce a Profibrotic Phenotype in Cultured Normal Dermal Fibroblasts. Clin Exp Rheumatol (2017) 35 Suppl 106(4):21–30.
105. Fadeel B, Orrenius S. Apoptosis: A Basic Biological Phenomenon With Wide-Ranging Implications in Human Disease. J Intern Med (2005) 258(6):479–517. doi: 10.1111/j.1365-2796.2005.01570.x
106. Cotter TG. Apoptosis and Cancer: The Genesis of a Research Field. Nat Rev Cancer (2009) 9(7):501–7. doi: 10.1038/nrc2663
107. Kissin E, Korn JH. Apoptosis and Myofibroblasts in the Pathogenesis of Systemic Sclerosis. Curr Rheumatol Rep (2002) 4(2):129–35. doi: 10.1007/s11926-002-0008-y
108. Eguchi K. Apoptosis in Autoimmune Diseases. Intern Med (2001) 40(4):275–84. doi: 10.2169/internalmedicine.40.275
109. Eckes B, Moinzadeh P, Sengle G, Hunzelmann N, Krieg T. Molecular and Cellular Basis of Scleroderma. J Mol Med (Berl) (2014) 92(9):913–24. doi: 10.1007/s00109-014-1190-x
110. Dong J, Zhao YP, Zhou L, Zhang TP, Chen G. Bcl-2 Upregulation Induced by miR-21 via a Direct Interaction is Associated With Apoptosis and Chemoresistance in MIA PaCa-2 Pancreatic Cancer Cells. Arch Med Res (2011) 42(1):8–14. doi: 10.1016/j.arcmed.2011.01.006
111. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange Between Cells. Nat Cell Biol (2007) 9(6):654–9. doi: 10.1038/ncb1596
112. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as Stable Blood-Based Markers for Cancer Detection. Proc Natl Acad Sci USA (2008) 105(30):10513–8. doi: 10.1073/pnas.0804549105
113. Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: Vehicles for the Transfer of Toxic Proteins Associated With Neurodegenerative Diseases? Front Physiol (2012) 3:124. doi: 10.3389/fphys.2012.00124
114. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat Cell Biol (2008) 10(12):1470–6. doi: 10.1038/ncb1800
115. Cheng L, Quek CY, Sun X, Bellingham SA, Hill AF. The Detection of microRNA Associated With Alzheimer's Disease in Biological Fluids Using Next-Generation Sequencing Technologies. Front Genet (2013) 4:150. doi: 10.3389/fgene.2013.00150
116. Vreca M, Andjelkovic M, Tosic N, Zekovic A, Damjanov N, Pavlovic S, et al. Impact of Alterations in X-Linked IRAK1gene and miR-146a on Susceptibility and Clinical Manifestations in Patients With Systemic Sclerosis. Immunol Lett (2018) 204:1–8. doi: 10.1016/j.imlet.2018.10.002
117. Izumiya Y, Jinnn M, Kimura Y, Wang Z, Onoue Y, Hanatani S, et al. Expression of Let-7 Family microRNAs in Skin Correlates Negatively With Severity of Pulmonary Hypertension in Patients With Systemic Scleroderma. Int J Cardiol Heart Vasc (2015) 8:98–102.
118. Abd-Elmawla MA, Hassan M, Elsabagh YA, Alnaggar A, Senousy MA. Deregulation of Long Noncoding RNAs ANCR, TINCR, HOTTIP and SPRY4-IT1 in Plasma of Systemic Sclerosis Patients: SPRY4-IT1 as a Novel Biomarker of Scleroderma and its Subtypes. Cytokine (2020) 133:155124. doi: 10.1016/j.cyto.2020.155124
119. Rusek M, Michalska-Jakubus M, Kowal M, Beltowski J, Krasowska D. A Novel miRNA-4484 is Up-Regulated on Microarray and Associated With Increased MMP-21 Expression in Serum of Systemic Sclerosis Patients. Sci Rep (2019) 9(1):14264. doi: 10.1038/s41598-019-50695-y
120. Avouac J, Airo P, Meune C, Beretta L, Dieude P, Caramaschi P, et al. Prevalence of Pulmonary Hypertension in Systemic Sclerosis in European Caucasians and Metaanalysis of 5 Studies. J Rheumatol (2010) 37(11):2290–8. doi: 10.3899/jrheum.100245
121. Koh ET, Lee P, Gladman DD, Abu-Shakra M. Pulmonary Hypertension in Systemic Sclerosis: An Analysis of 17 Patients. Br J Rheumatol (1996) 35(10):989–93. doi: 10.1093/rheumatology/35.10.989
Keywords: ncRNA, miRNA, lncRNA, systemic sclerosis, function, biomarker
Citation: Liu Y, Cheng L, Zhan H, Li H, Li X, Huang Y and Li Y (2022) The Roles of Noncoding RNAs in Systemic Sclerosis. Front. Immunol. 13:856036. doi: 10.3389/fimmu.2022.856036
Received: 16 January 2022; Accepted: 17 March 2022;
Published: 08 April 2022.
Edited by:
Sebastien Sanges, INSERM U1286 - INFINITE, FranceReviewed by:
Marco Matucci Cerinic, University of Florence, ItalyXiaoxiang Chen, Shanghai JiaoTong University, China
Copyright © 2022 Liu, Cheng, Zhan, Li, Li, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhe Li, eW9uZ3poZWxpcHVtY2hAMTI2LmNvbQ==
 Yongmei Liu
Yongmei Liu Linlin Cheng1,2
Linlin Cheng1,2 Haoting Zhan
Haoting Zhan Haolong Li
Haolong Li Xiaomeng Li
Xiaomeng Li Yuan Huang
Yuan Huang Yongzhe Li
Yongzhe Li