
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 16 May 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.855888
This article is part of the Research TopicInsect immunity and its interactions with microorganisms and parasitoidsView all 31 articles
 Saima Kausar1,2†
Saima Kausar1,2† Muhammad Nadeem Abbas1,2†
Muhammad Nadeem Abbas1,2† Isma Gul1,2†
Isma Gul1,2† Ruochen Liu1,2
Ruochen Liu1,2 Qianqian Li3
Qianqian Li3 Erhu Zhao1,2
Erhu Zhao1,2 Muhan Lv4*
Muhan Lv4* Hongjuan Cui1,2*
Hongjuan Cui1,2*Under different physiological conditions, such as microbial infection, epigenetic mechanisms regulate genes at the transcription level in living organisms. DNA methylation is a type of epigenetic mechanism in which DNA methyltransferases modify the expression of target genes. Here, we identified a full-length sequence of DNMT-1 and DNMT-2 from the Chinese oak silkworm, A. pernyi, which was highly similar to the homologous sequences of Bombyx mori. ApDNMT-1 and ApDNMT-2 have unique domain architectures of insect DNMTs, highlighting their conserved functions in A. pernyi. ApDNMT-1 and ApDNMT-2 were found to be widely expressed in various tissues, with the highest levels of expression in hemocytes, the ovary, testis, and fat bodies. To understand the biological role of these genes in microbial resistance, we challenged the fifth instar larvae of A. pernyi by administrating Gram-positive and Gram-negative bacteria and fungi. The results revealed that transcript levels of ApDNMT-1 and ApDNMT-2 were increased compared to the control group. The inhibition of these genes by a DNMTs inhibitor [5-azacytidine (5-AZA)] significantly reduced bacterial replication and larvae mortality. In addition, 5-AZA treatment modified the expression patterns of antimicrobial peptides (AMPs) in the A. pernyi larvae. Our results suggest that ApDNMT-1 and ApDNMT-2 seem to have a crucial role in innate immunity, mediating antimicrobial peptide responses against bacterial infection in A. pernyi.
The Chinese oak silkworm, A. pernyi, is broadly cultured for silk production in Asian countries, such as China, Korea, and India (1). This wild silk moth is also used as a model insect to investigate immunological response, development, metabolism regulation, and breeding technique. A. pernyi, like other insects, has strong and efficient innate immunity, including cellular and humoral components. Encapsulation, nodule formation, and phagocytosis are examples of cellular immune responses (2, 3). The humoral immune response includes the production of AMPs following a microbial infection (2, 4). In A. pernyi and other insects, complex sets of genes involved in the coordination of the innate immune system are related to the immune pathways such as Toll, Imd, and NF‐κB pathways (5). After pathogen detection, the activated cell surface receptors induce the downstream signaling cascade inside cells, stimulating the targeted immune response. Toll and IMD pathways produce AMPs in response to bacterial and fungal infection (5, 6).
DNA methylation is one of the critical epigenetic modifications that regulate gene transcription in eukaryotes and control various biological processes. DNA methyltransferases are responsible for DNA methylation (7, 8). Compared to mammals, studies on DNMTs and DNA methylation in insects remain relatively poor. However, some progress has been made in insects in recent years due to the development of innovative DNA methylation research methods. DNA methylation has been found in the genomes of insects such as silkworm, Nasonia Vitripennis, Tribolium castaneum, and Drosophila melanogaster, according to whole-genome bisulfite sequencing studies (9–12). However, the maintenance and regulatory mechanisms of DNA methylation in insects may vary significantly from those in other organisms (13–15). Insects have been shown to have different combinations of DNMT gene sets. The Apis mellifera genome contains DNMT-1, DNMT-2, and DNMT-3 genes (16). However, in silkworms and Tribolium castaneum, only two DNMT genes (DNMT-1 and DNMT-2) have been identified (16). In the living organisms lacking DNMT-3, including silkworms and beetles, DNMT-1 appears to be a de novo and maintenance DNMT. DNA methylation regulates various physiological processes in insects, such as immunity, memory, aging evolution, and others (17–21).
Insect immunological responses have not been extensively explored in terms of DNA methylation’s biological roles. It has been shown that Aedes aegypti genome wide-patterns of DNA methylation are disrupted by the infection by Wolbachia (22). Cytoplasmic polyhedrosis virus infection caused the differential expression of genes and affected the DNA methylation status in the fat bodies and midguts of silkworms (23). Subsequently, Wu et al. (15) observed the differential expression of DNMT genes in the Galleria mellonella larvae infected with three different strains of Metarhizium anisopliae, suggesting that DNA methylation in insects is likely involved in the immune response against microbial pathogens (15). In the present study, we identified the DNA methylation tool kit in the Chinese oak silkworm A. pernyi. Then we analyzed their spatial and temporal expression patterns in different tissues of A. pernyi larvae. Next, we treated A. pernyi larvae with 5-AZA, a DNA methylation inhibitor, and analyzed the effects of DNA methylation inhibition on the insect’s immune responses.
The model insect, A. pernyi larvae, was maintained at room temperature and fed on fresh oak leaves as described in previous reports (2, 4). Healthy fifth instar larvae were used in all of the following experiments.
To identify ApDNMT-1 and ApDNMT-2 sequences, we considered the genomic library of A. pernyi constructed in our laboratory. The retrieved sequence was further analyzed by the NCBI blastn tool for confirmation. Then the fragments of these genes were amplified by polymerase chain reaction using specific primers (Table 1). The ORF sequences of ApDNMT-1 and ApDNMT-2 were mapped into their respective sequences. The domain architecture was determined by using the blastp program. Multiple sequence alignments were performed with representative DNMT-1 and DNMT-2 sequences from other insect species retrieved from the GenBank database using Clustal software. The three-dimensional protein structure was analyzed by the SWISS-MODEL (http://Swiss-model.expasy.org). Phylogenetic analysis was carried out using Mega 6 software with 1000 bootstraps using the neighbor-joining method based on amino acid sequences.
To analyze tissue distribution patterns of DNMT genes, hemocytes, fat body, midgut, silk gland, testis, ovary, Malpighian tubules, and integument tissues were collected from the healthy A. pernyi larvae. Total RNA from each sample was extracted using TRIzol reagent (Invitrogen) according to the suppliers’ instructions, and 2 μg of total RNA was used to prepare cDNA. Oligonucleotide primers specific for the ApDNMT-1 and ApDNMT-2 sequences and the 18S rRNA sequence for internal control were designed using the Primer 3.0 online primer designing tool (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) (Table 1). We performed PCR and analyzed amplicons on the agarose gel before carrying out qRT-PCR to ensure the integrity and stability of 18S rRNA in the cDNA of all samples used in this study. The quantitative real-time polymerase chain reaction assay was performed with specific primers and synthesized cDNAs, and each reaction mixture contained 20 μL with 10 μL of SsoFast EvaGreen SuperMix (Bio-Rad, Hercules, California, USA), 1 μL of forward primer (200 mM), 1 μL of reverse primer (200 mM), 1 μL of diluted cDNA, and 7 μL of sterile water. The thermal cycling conditions were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Amplification was monitored on the iCycleriQ TM RT-PCR Detection System (Bio-Rad, Hercules, California, USA). The specificity of the SYBR-Green PCR signal was further confirmed by melting curve analysis. The experiments were repeated three times as independent biological replicates. The mRNA expression was quantified using the 2-ΔΔCTmethod (25).
For obtaining induction profiles of DNMT genes, fifth instar larvae of A. pernyi on the third day were divided into four groups with 54 larvae in each group. The A. pernyi larvae were injected with 5 μL each of E. coli (DH5α, 1 × 106 cells), B. cereus (1 × 106 cells), or B. bassiana (1 × 106 spores). The larvae injected with PBS were used as a control group. These microbial organisms were injected into the larval abdominal area using microliter syringes (Gaoge, Beijing, China), and injection sites were sealed with vaseline immediately after injection. The larvae were dissected, and immune tissues, including the fat bodies and hemolymph, were sampled at different time intervals (1.5, 3, 6, 12, 24, and 48 h) after injection. Three larvae were collected as one sample, and the biological sampling protocol was repeated three times. The transcript expression patterns of ApDNMT-1 and ApDNMT-2 genes were recorded using a qRT-PCR assay as described in the above section.
To determine the effect of 5-aza-2-deoxycytidine on the DNMT genes and, subsequently, on the immune responses against bacterial pathogens. 5-AZA can prevent de novo and maintenance of DNA methylation by binding covalently to DNMT genes (26). We purchased 5-AZA and diluted it in distilled water with 40 mM/1 μL. The fifth instar larvae of A. pernyi were injected with 1 μL 5-AZA into the abdominal area of the larvae, and the second injection was performed at 24 h intervals. Then we analyzed, the expression patterns of both the ApDNMT-1 and ApDNMT-2 genes using qRT-PCR. Bacterial pathogens (B. cereus or E. coli) were injected 24 h after the second 5-AZA injection. The larvae were used as control groups without any treatment, and the larvae were injected with distilled water.
To estimate the bacterial load, we first analyzed the bacterial clearance ability of larvae injected with 5-AZA and bacterial pathogens. After 6 h of bacterial injection, A. pernyi larvae legs were punctured to collect hemolymph from the 5-AZA treated group and control group (distilled water treated and untreated) and then immediately diluted. Fifty μL of each dilution was plated onto agar plates, incubated overnight at 37°C, and the colony-forming units were counted.
Furthermore, total genomic DNA was extracted from the hemolymph samples of treated (bacterial-injected and 5-AZA treated) and control A. pernyi larvae. The concentration of the extracted DNA was determined by a spectrophotometer, and 500 ng of total genomic DNA was used for each qRT-PCR reaction, and the analysis was performed as described in section 2.3. Specific primers for bacterial 16s rRNA were used in qRT-PCR, and the A. pernyi 18s rRNA gene was used as an internal control for normalization (24, 27). To better understand the insect-pathogen interaction, the mortality of A. pernyi was also analyzed using Graphpad Prism 6.0. Kaplan-Meier survival analysis.
A. pernyi larvae were treated with 5-AZA- bacteria (B. cereus or E. coli) or only bacteria or untreated larvae to determine the response of AMPs and their associated pathways. The larvae were dissected, and the fat bodies were used to isolate total RNA, which was then reverse transcribed to prepare cDNA. A quantitative RT-PCR assay was carried out (described in section 2.3) using AMPs (gloverin-like, cecropin, attacin, and lebocin) and their associated pathway genes by specific primers (Table 1).
In this study, all experiments were executed in triplicate and the obtained data represented the means ± S.E. The one-way analysis of variance (ANOVA) and Tukey’s multiple range tests were used to evaluate the difference between groups.
The cDNA sequences of ApDNMT-1 and ApDNMT-2 were identified by using a genomic library of the Chinese oak silkworm A. pernyi, which was constructed in our laboratory. The full-length sequence of ApDNMT-1 comprised a total length of 5508 base pairs. The open reading frame (ORF) is 4344 bp and contains a 209 bp 5′ untranslated region (5′ UTR) and a 955 bp 3′ UTR (Supplementary Figure S1). The deduced ApDNMT-1 protein is comprised of 1447 amino acids with an approximate molecular weight of 163.182 kDa and isoelectric point of 6.53, respectively. The NCBI Conserved Domain Database was used to determine conserved domains of ApDNMT-1, which showed that this gene has four conserved domains: a Zinc finger domain, a Bromo adjacent homology domain, a DNA-cytosine methyltransferase domain, and a cysteine-rich ADD domain (Supplementary Figure S1; Figures 1, 3). Further, it was found that ApDNMT-1 has a comparable structure and domain architecture similar to that of silkworm in its tertiary structure (Figure 3A). Multiple sequence alignment analysis revealed that ApDNMT-1 has the highest similarity to B. mori (77.96%), followed by Athalia rosae (52.12%), Ceratina calcarata (49.74%), and so on. The full-length sequence of ApDNMT-2 contains an 1833 bp nucleotide sequence. The ORF of ApDNMT-2 was 888bp in length, with a 5′ UTR of 205 bp and a 3′ UTR of 740 bp. The deduced ApDNMT-2 protein contains 295 amino acid residues. The predicted molecular weight and isoelectric point are 33.89 kDa and 6.01, respectively. In a domain analysis of ApDNMT-2, we observed that this protein consisted of only one domain (the typical DNMT-2 cyt_C5_DNA methylase superfamily domain of DNMT-2 proteins), and its tertiary structure was highly comparable to that of the B. mori DNMT-2 protein (Supplementary Figure S2 and Figures 2, 3B). Additionally, multiple sequence alignment analysis revealed that ApDNMT-2 shares the maximum similarity with B. mori (66.78%), followed by Tribolium castaneum (48.41%), Drosophila melanogaster (43.73%), and other species. A phylogenetic analysis of amino acid sequences from diverse invertebrate and vertebrate species was carried out in order to better understand the evolutionary relationships of ApDNMT-1 and ApDNMT-2. The phylogenetic tree was constructed using the neighbor-joining method, which was performed using the Mega 6 software. The results revealed that ApDNMT-1 and ApDNMT-2 proteins are associated with the DNMT-1 and DNMT-2 proteins of animals, respectively (Figure 4).
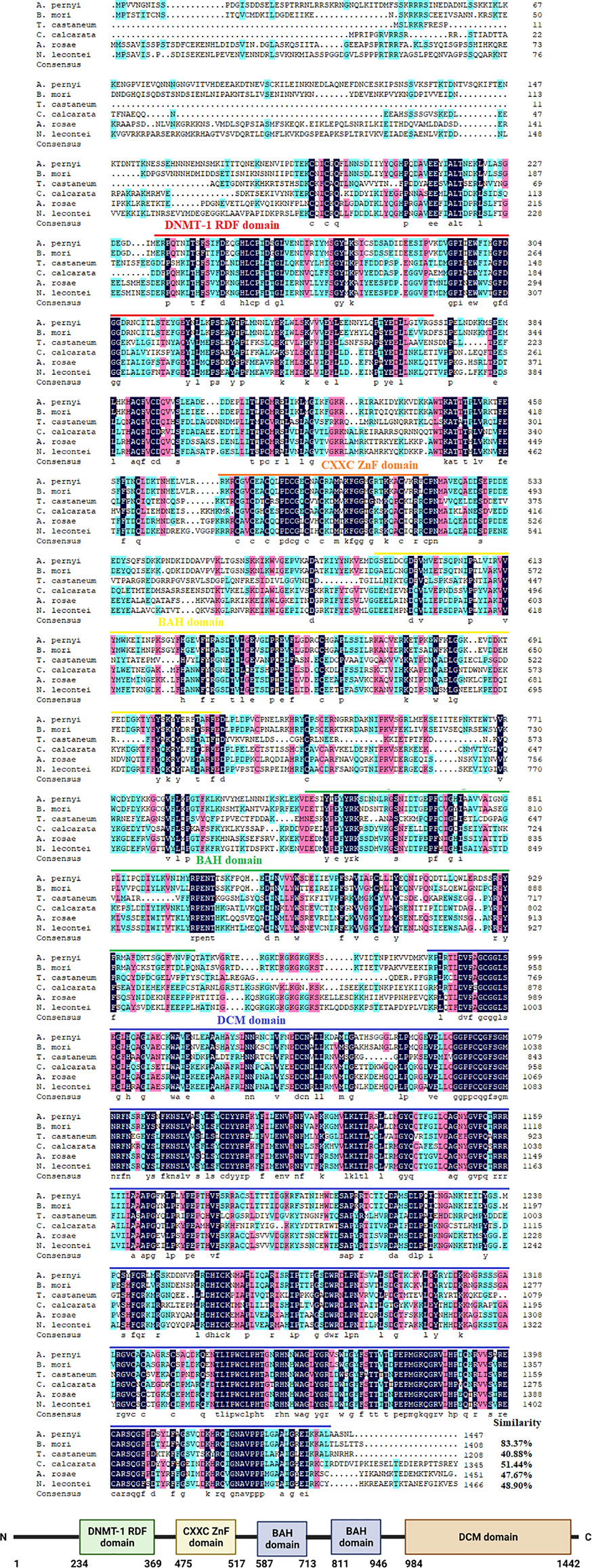
Figure 1 Alignment of the ApDNMT-1 protein with homologous proteins. The ApDNMT-1 deduced amino acid sequence was aligned with other DNMT-1 protein sequences. The sequences for alignment analysis are the following: accession no. NP_001036980.1 (Bombyx mori), accession no. ASA69505.1 (Tribolium castaneum), accession no. XP_026671099.1 (Ceratina calcarata), accession no. XP_012254091.1(Athalia rosae), and accession no. XP_015517160.1 (Neodiprion lecontei). Identified domains were labeled as DNMT1-RDF, DNA methyltransferase replication foci domain which is indicated by a red line; ZnF, Zinc finger domain is indicated by the orange line; BAH, Bromo adjacent homology domain is shown in yellow line; DCM, DNA-cytosine methyltransferases domain is shown in green line Cys-rich, cysteine-rich ADD domain is presented in the blue line.
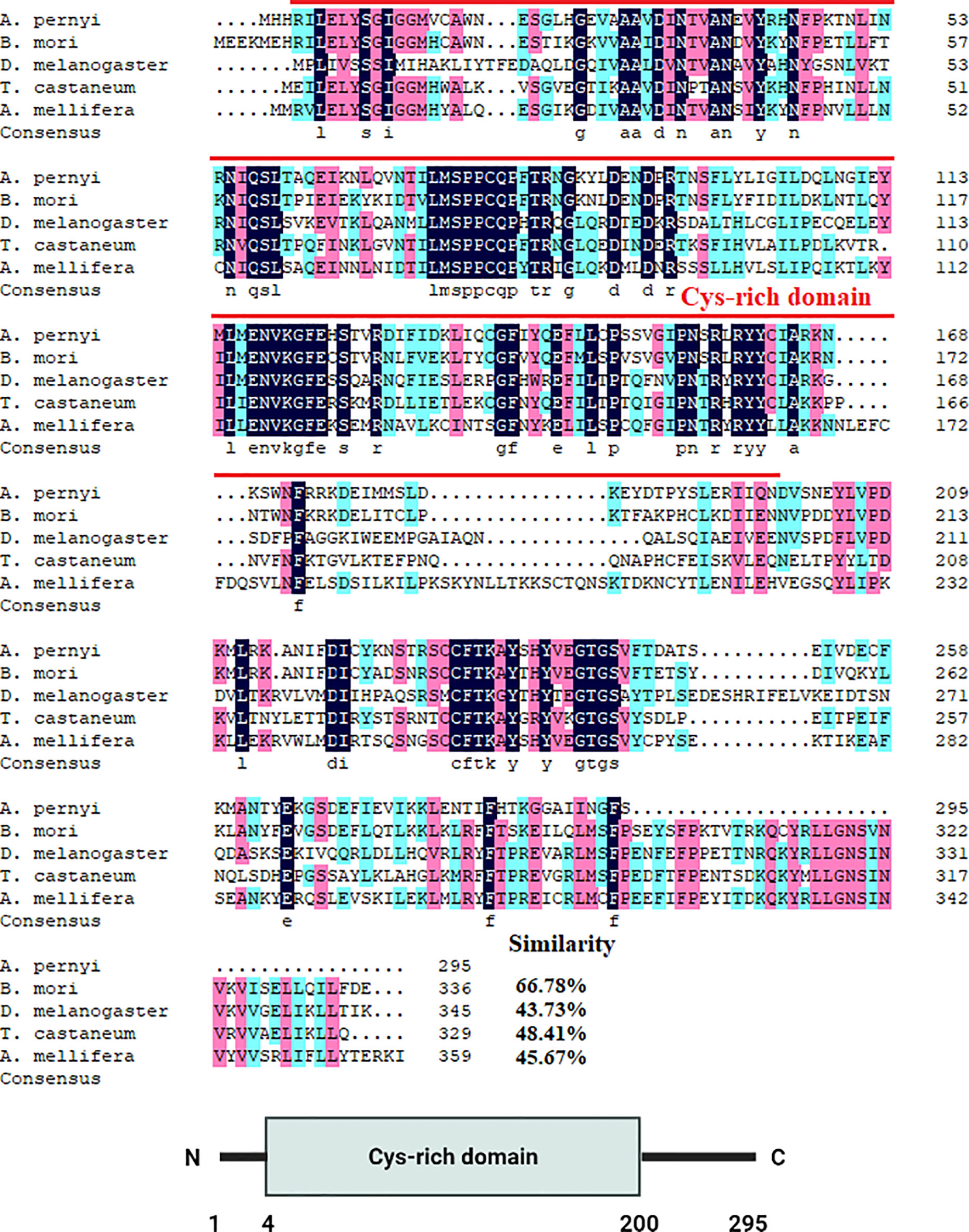
Figure 2 Alignment of the ApDNMT-2 protein with homologous proteins. The ApDNMT-2 deduced amino acid sequence was aligned with other DNMT-2 protein sequences. The sequences for alignment analysis are the following: accession no. NP_001036934.1 (Bombyx mori), accession no. AAF03835.1 (Drosophila melanogaster), accession no. (Tribolium castaneum), accession no. XP_006563008.1 (Apis mellifera). The putative DNMT2 cyt_C5_DNA methylase superfamily domain is indicated by red line.
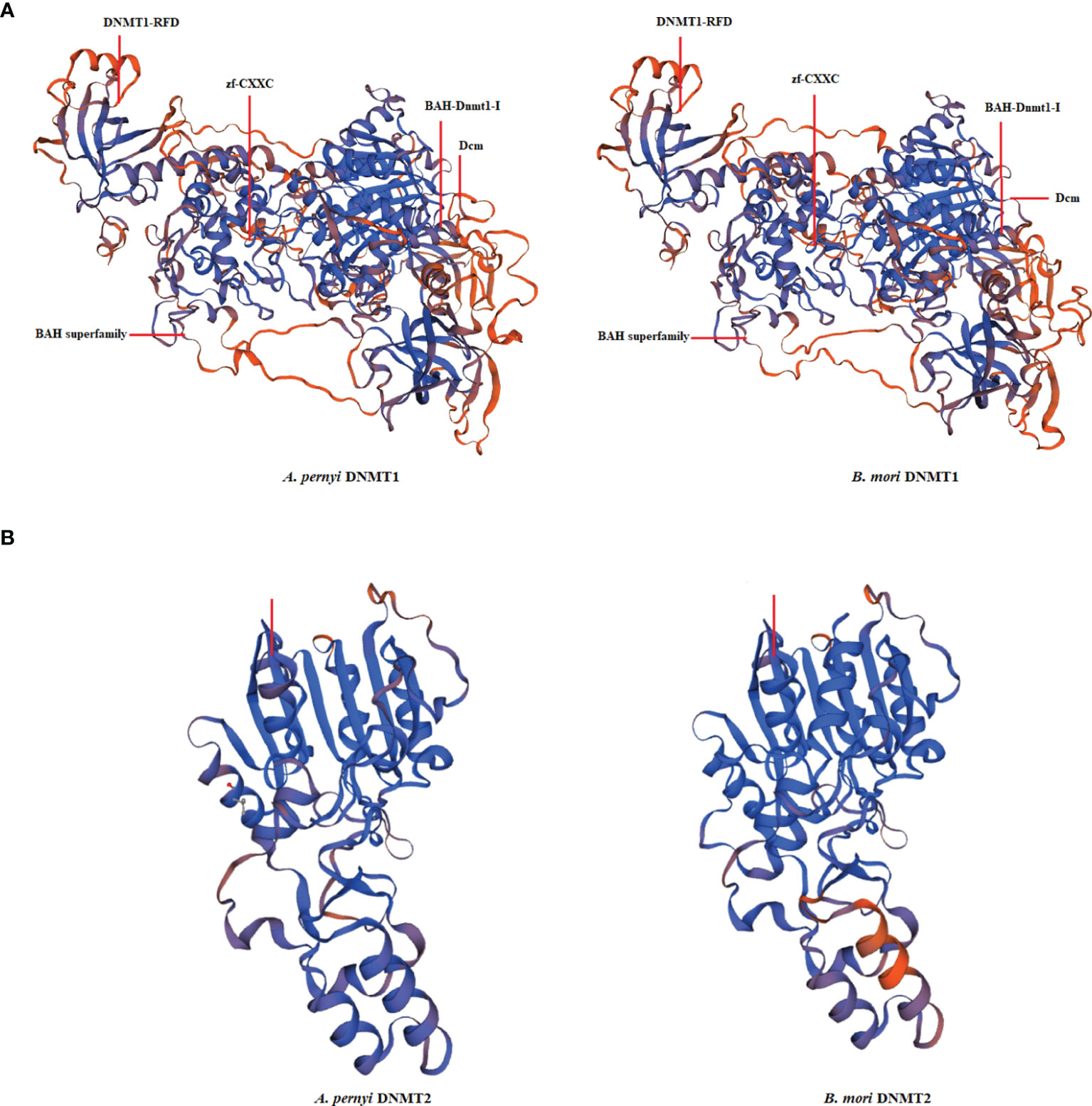
Figure 3 The tertiary structure of A. pernyi DNMT-1 (A) and DNMT-2 (B) proteins in comparison with Bombyx mori DNMT-1 and DNMT-2 proteins.
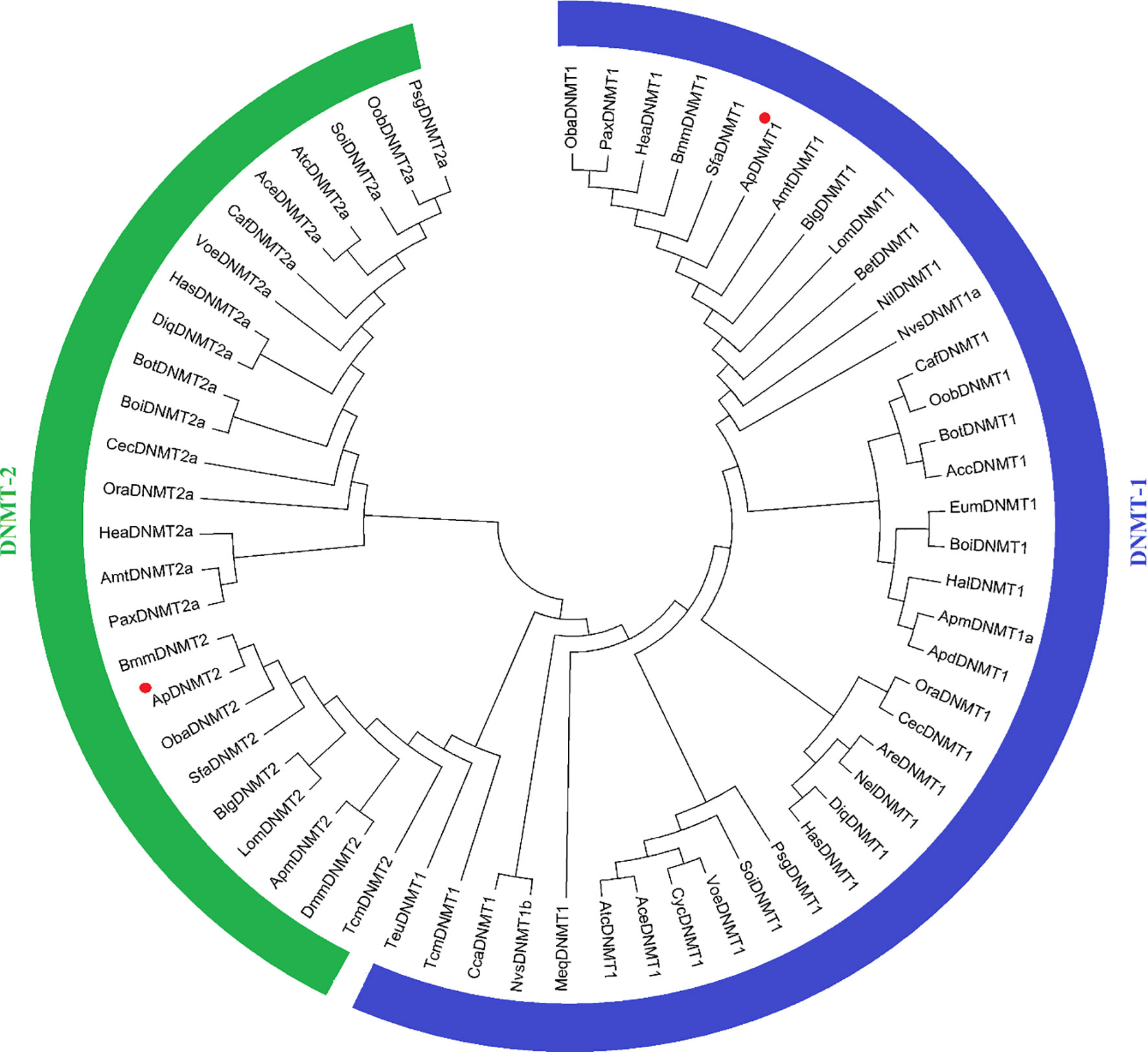
Figure 4 Phylogenetic analysis of A. pernyi ApDNMT-1 and ApDNMT-2 proteins with other DNMT-1 and DNMT-2 proteins from vertebrate and invertebrate species. The deduced amino acid sequences were aligned; later a phylogenetic tree was constructed using the neighbor-joining method.
To understand the biological role of ApDNMT-1 and ApDNMAT-2, we measured their transcription levels in different tissues (hemocyte, fat body, midgut, Malpighian tubules, integument, head, testis, ovary, and silk gland) of A. pernyi by performing qRT-PCR analysis. Using a tissue-specific mRNA expression analysis, it was found that ApDNMT-1 was transcribed in all of the tested tissues, with mRNA expression levels being notably high in the ovary, hemocytes, and fat bodies (Figure 5A). The ApDNMT-2 gene was observed in all of the tissues examined, although its mRNA expression levels varied in each tissue. ApDNMT-2 expression levels were shown to be higher in the ovary, hemocytes, and fat bodies, similar to ApDNMT-1 expression levels (Figure 5B). These findings show that these proteins have a broader range of biological roles in insects.
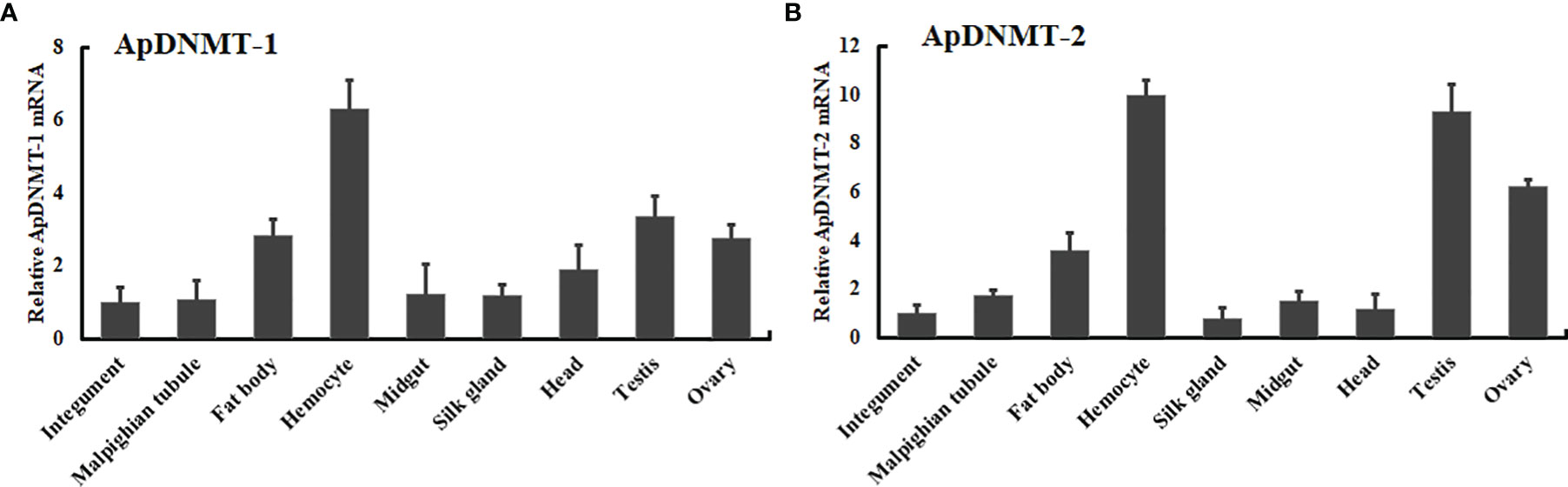
Figure 5 Tissue distribution profiles of ApDNMT-1 and ApDNMT-2 in various tissues of A. pernyi (A) Analysis of ApDNMT-1 transcript expression in various tissues of silkworm larvae by qRT-PCR, (B) the ApDNMT-2 transcript expression in various tissues of silkworm larvae by qRT-PCR. The mRNA level in the midgut was used as the calibrator. Bars exhibit the mean ± S.E (n = 3).
We examined the transcription levels of the ApDNMT-1 and ApDNMT-2 genes in hemocytes and fat bodies from A. pernyi at 1.5, 3, 6, 12, 24, and 48 h after injection with B. bassiana, E. coli, and B. cereu to investigate the influence of fungal and bacterial infections on ApDNMT-1 and ApDNMT-2 genes. The qRT-PCR analysis revealed that both fungal and bacterial pathogens induced transcription patterns of the ApDNMT-1 and ApDNMT-2 genes in hemocytes and fat bodies that were similar to one another. During the early hours after infection, the expression levels of ApDNMT-1 and ApDNMT-2 genes were slightly altered. ApDNMT-1 gene expression was shown to be notably upregulated in the hemocytes and fat bodies at critical timepoints of infection (e.g., 6, 12, 24 h) (Figure 6). After the fungal and bacterial infections, the transcription patterns of ApDNMT-2 were found to be approximately similar to those of ApDNMT-1 (Figure 7). Following these timepoints, however, the transcription levels of these genes start to drop.
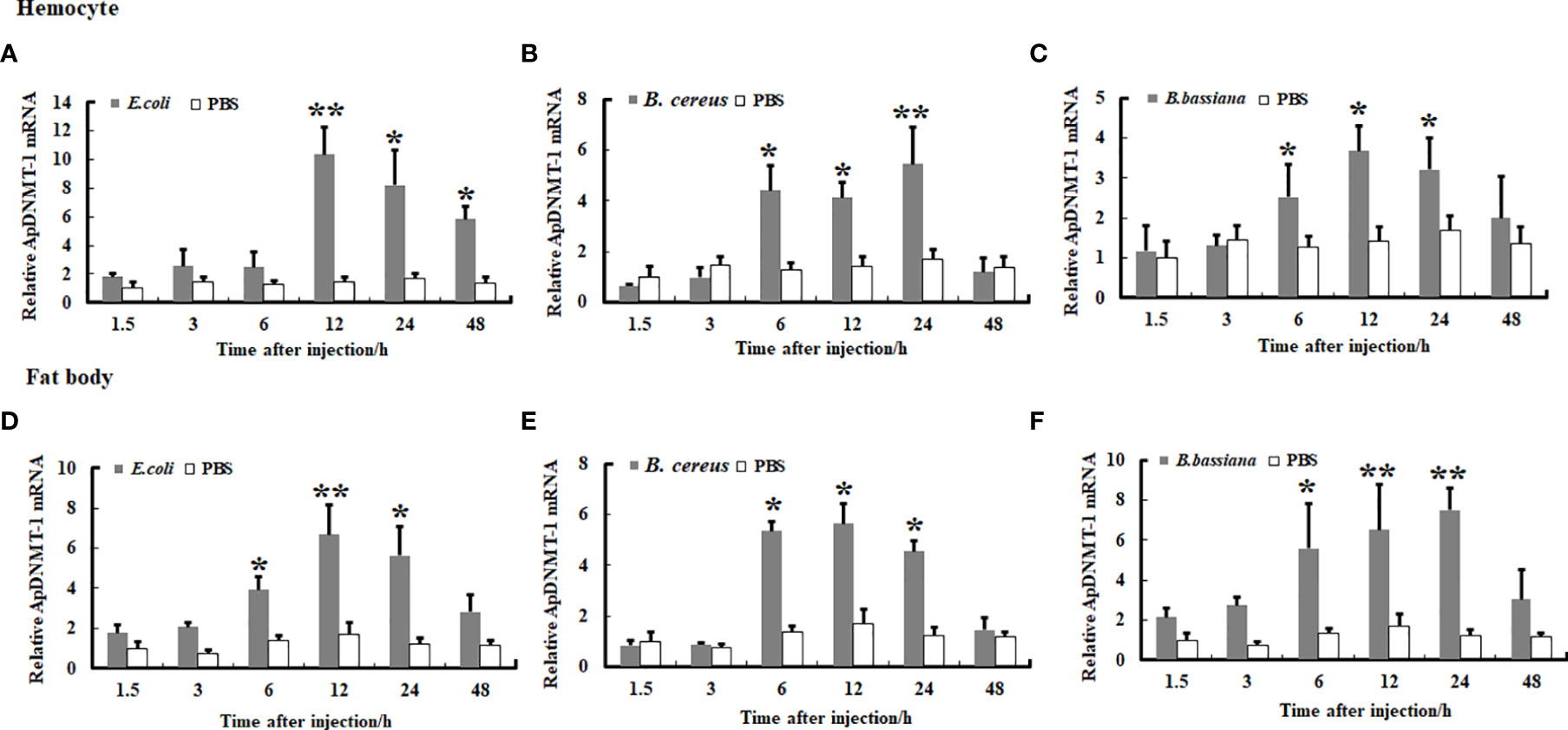
Figure 6 Expression profile of ApDNMT-1 in hemocyte and fat body following microbial challenge. (A) Expression profile following E. coli challenge in hemocyte, (B) Expression profile following B. cereus challenge in hemocyte, (C) Expression profile following B. bassiana challenge in hemocyte, (D) Expression profile following E. coli challenge in the fat body, (E) Expression profile following B. cereus challenge in the fat body, (F) Expression profile following B. bassiana challenge in the fat body. PBS was injected as the control. Bars show mean ± S.E. (n = 3), and asterisks indicate significant differences (*p < 0.05, **p < 0.01).
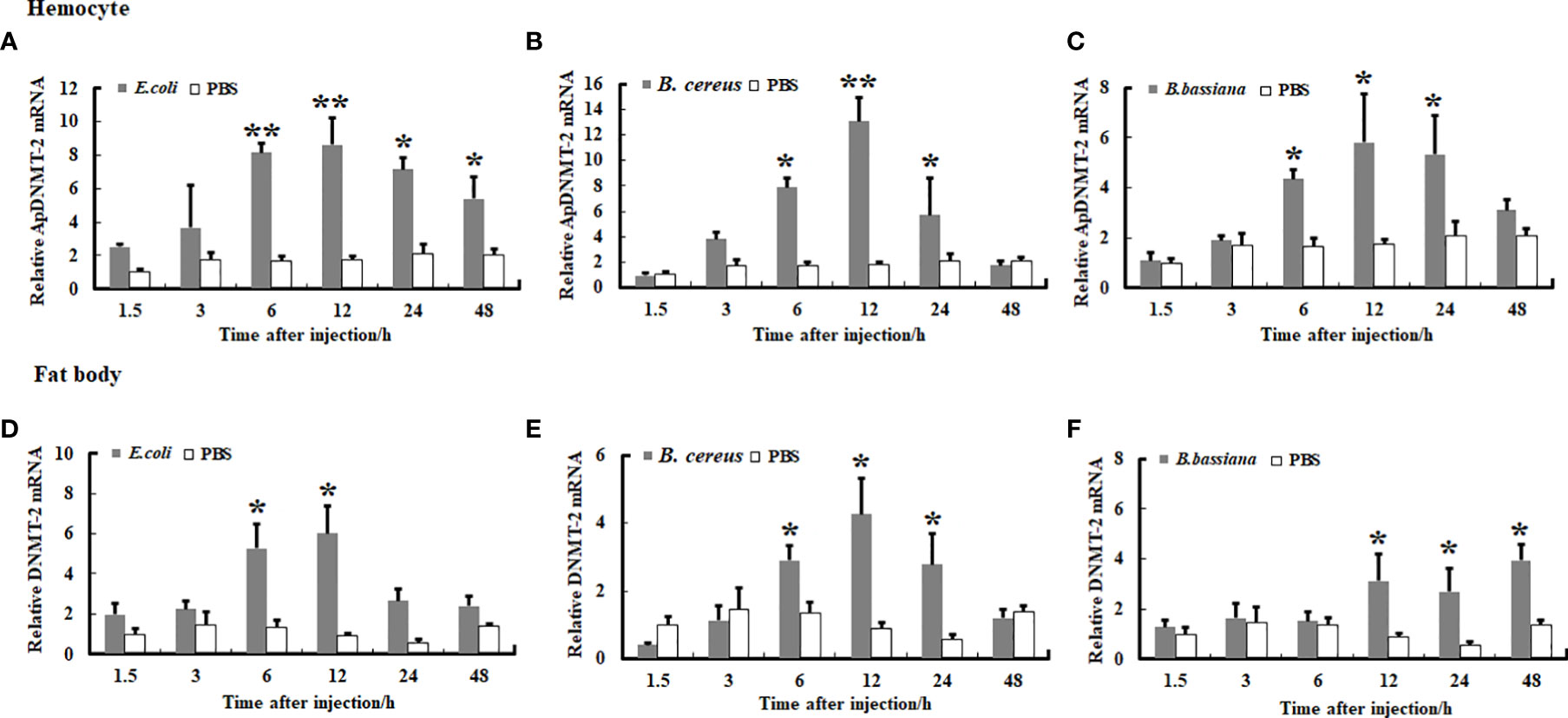
Figure 7 Expression profile of ApDNMT-2 in hemocyte and fat body following microbial challenge. (A) Expression profile following E. coli challenge in hemocyte, (B) Expression profile following B. cereus challenge in hemocyte, (C) Expression profile following B. bassiana challenge in hemocyte, (D) Expression profile following E. coli challenge in the fat body, (E) Expression profile following B. cereus challenge in fat body, (F) Expression profile following B. bassiana challenge in the fat body. PBS was injected as the control. Bars show mean ± S.E. (n = 3), and asterisks indicate significant differences (*p < 0.05, **p < 0.01).
Because of the remarkable variation in ApDNMT-1 and ApDNMT-2 gene transcription that was observed in A. pernyi larvae after fungal and bacterial infections, we next planned to investigate the importance of ApDNMT-1 and ApDNMT-2 involvement in innate immune responses. For this reason, we inhibited the DNMTs in the fifth instar larvae by injecting 5-AZA into the larvae, followed by B. cereus and E. coli injections. Before bacterial injection, we confirmed the suppression of ApDNMT-1 and ApDNMT-2 following the 5-AZA administration (Figure 8A). Total genomic DNA was extracted from the hemolymph samples of treated (bacterial-injected and 5-AZA treated) and control larvae. To perform qRT-PCR, we used 500 ng total genomic DNA for each qRT-PCR reaction, and the DNA concentrations were measured using a spectrophotometer. In qRT-PCR, specific primers for the 16s rRNA of the bacteria were used, and the insect 18s rRNA gene was used as an internal control for normalization. For each experiment, three biological replicates were replicated three times. Our results showed that 5-AZA treatment of larvae reduced replication of the pathogenic bacteria when compared to control larvae (Figures 8A, B). We also analyzed the mortality rate of bacteria-infected larvae and found that injection of 5-AZA attenuated larval mortality (Figure 8C).
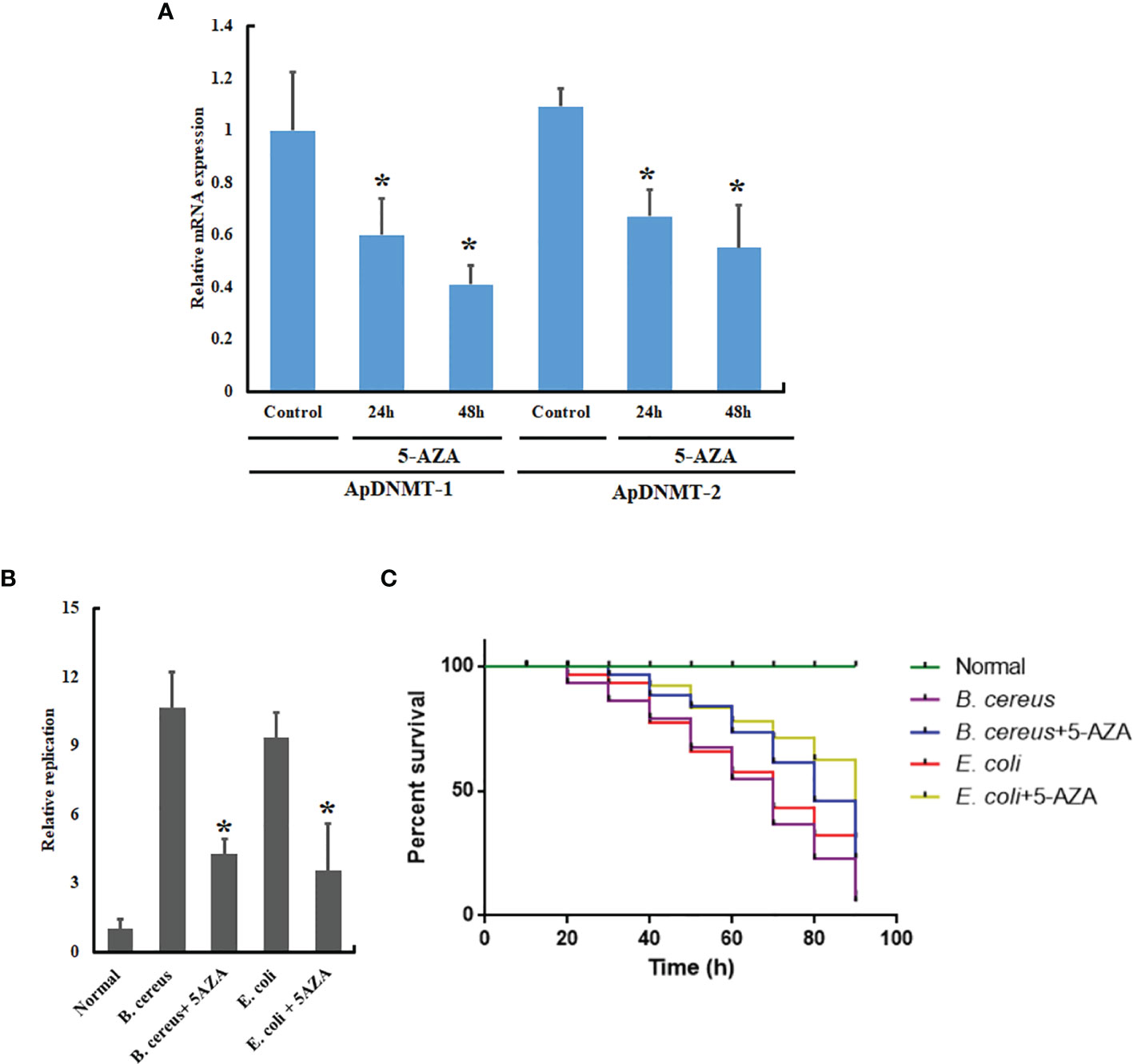
Figure 8 5-AZA administration reduced replication rates of the pathogenic bacteria and mortality rate of A. pernyi. (A) Relative mRNA expression levels of ApDNMT-1 and ApDNMT-2 in the 5-AZA-treated larvae and control group after 24 h and 48 h by qRT PCR. (B) qRT-PCR replication rate analysis showed that treatment of A. pernyi larvae with 5-AZA reduced replication rates of B. cereus and E. coli in A. pernyi. (C) 5-AZA application also decreased the mortality rate of A. pernyi following infection with the pathogenic bacteria. Bacterial DNA was quantified with universal 16S rRNA primers, and insect 18S rRNA was used as the reference control. Bars show mean ± S.E. (n = 3), and asterisks indicate significant differences (*p < 0.05).
In order to determine whether the presence of 5-AZA, a DNA methylation inhibitor, modulates the antimicrobial activity, we carried out a bacterial clearance assay using plasma collected from larvae that had been treated with 5-AZA and bacteria. The results revealed that plasma from A. pernyi larvae treated with 5-AZA had significantly greater antibacterial activity against gram-positive and gram-negative bacteria than in control groups. For example, the survival rates of bacteria (B. cereus and E. coli) in the 5-AZA treated larvae were much lower than in the distilled water treated larvae and the untreated control larvae (Figure 9).
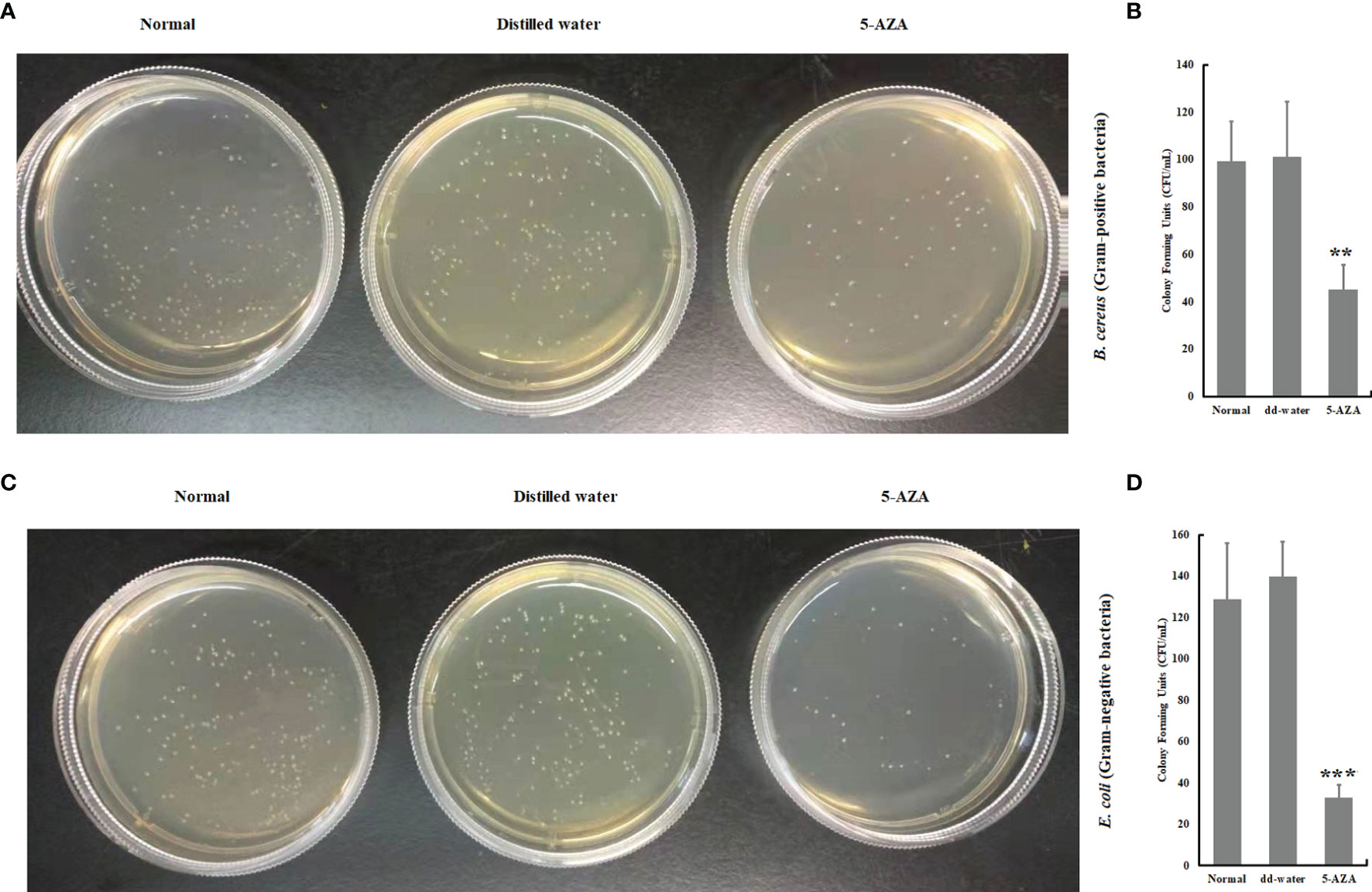
Figure 9 The plasma anti-bacterial activity was increased in the 5-AZA, and gram-positive bacteria (A, B) or 5-AZA and gram-negative bacteria (C, D) treated larval hemolymph. Plasma was sampled from A. pernyi treated with 5-AZA (n = 3) and distilled water. Equal volumes (10 μl) of plasma and bacterial suspension were incubated at 25°C for 1 h. Asterisks indicate significant differences (**p < 0.01, ***p < 0.001).
To evaluate the effects of 5-AZA on AMP gene expression, we quantified changes in the transcript levels of various AMPs in larvae that had been treated with 5-AZA and then challenged with bacteria. We injected A. pernyi larvae in the control group with distilled water or with bacteria only. We observed that among the four AMPs analyzed, the expression of attacin, cecropin with Gram-positive and gloverin-like, attacin, and lebocin with gram-positive bacteria was strongly increased in 5-AZA-injected larvae after challenge with B. cereus and E. coli as compared to the control group (Figure 10). To understand further how 5-AZA and bacterial treatments affect these AMP-associated pathways, we analyzed the Toll and IMD pathway-associated genes (Toll-like receptor, dorsal, and relish), and the results revealed that the expression of these genes was altered in the 5-AZA and bacterial treated larvae compared to control groups (Figure 11).
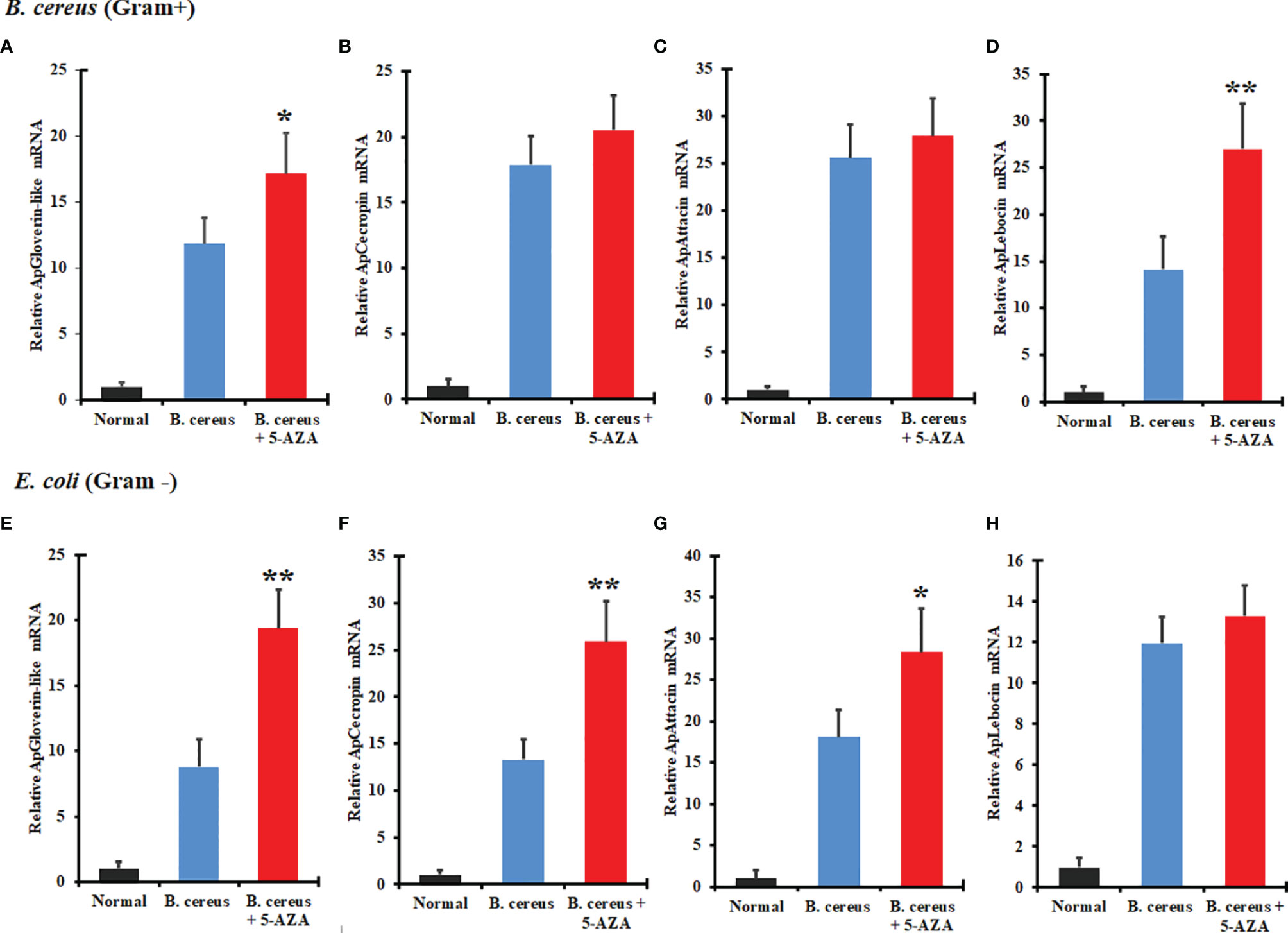
Figure 10 Treatment with 5-AZA modulated the expression of antimicrobial peptides (AMPs) in A. pernyi following bacterial infections. RT-qPCR analysis of gloverin-like (A, E), cecropin (B, F), attacin (C, G), and lebocin (D, H) confirmed that 5-AZA administration altered the expression levels of AMPs in response to B cereus (A–D) and E. coli (E–H). 18S was used as the reference gene in RT-qPCR. Reactions from three biological replicates were repeated three times. Bars show mean ± S.E. (n = 3), and asterisks indicate significant differences (*p < 0.05, **p < 0.01).
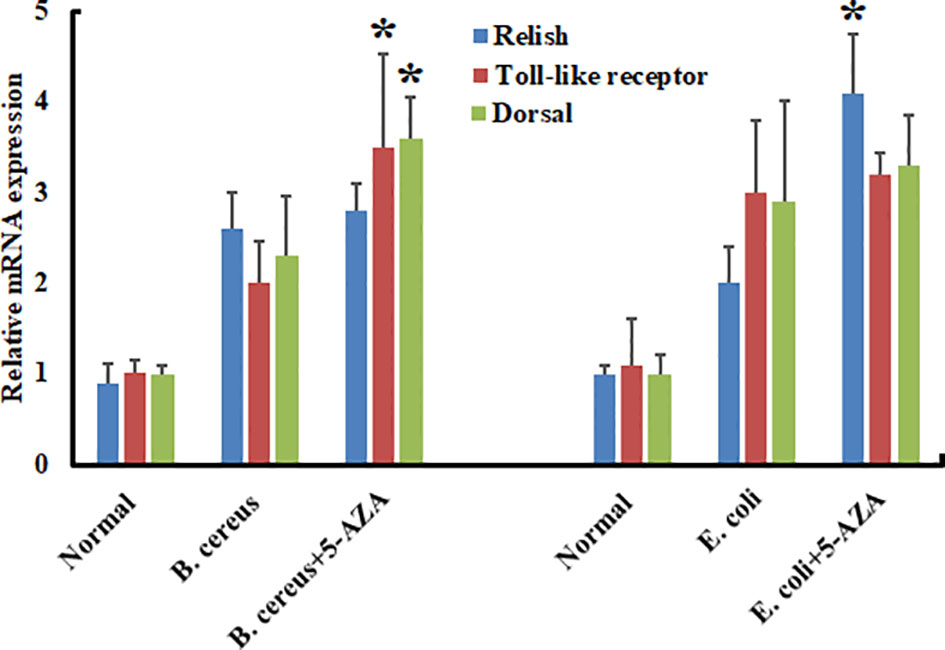
Figure 11 Treatment with 5-AZA modulated the expression of Toll and IMP pathway-associated genes in A. pernyi following bacterial infections. RT-qPCR analysis of relish, Toll-like receptor, and dorsal confirmed that 5-AZA administration altered the expression levels of these pathways-associated genes in response to B cereus and E. coli. 18S was used as the reference gene in RT-qPCR. Reactions from three biological replicates were repeated three times. Bars show mean ± S.E. (n = 3), and asterisks indicate significant differences (*p < 0.05).
In the present study, we identified two members of the DNA methyltransferase family, including DNMT-1 and DNMT-2, from the Chinese oak silkworm A. pernyi. These genes share high similarities with the DNMT-1 and DNMT-2 of other lepidopterans, particularly with B. mori. Previous studies have also reported the presence of only DNMT-1 and DNMT-2 genes in lepidopteran species (21, 28). These studies support the hypothesis of the loss of the DNMT-3 gene from lepidopteran species (29). Conserved domain analysis showed that ApDNMT-1 and ApDNMT-2 proteins have the typical domain composition of DNMT families. ApDNMT-1 has four conserved domains, while ApDNMT-2 has only 1 domain, suggesting that these proteins have different functions. The phylogenetic analysis of DNMTs also exhibited high similarities of ApDNMT-1 and ApDNMT-2 proteins to the homologous proteins in other lepidopterans, particularly B. mori. In the phylogenetic tree, we observed that DNMT-1 and DNMT-2 clades clustered separately, further strengthening the idea that these proteins have different biological roles in insects.
Furthermore, we analyzed the tissue distribution of these genes in A. pernyi larvae. Our tissue mRNA expression analysis showed that ApDNMT-1 and ApDNMT-2 expression was detectable in all A. pernyi larval tissues examined, but there were notable mRNA expression levels in the hemocytes, testis, and fat bodies. We presume that these variations in ApDNMT-1 and ApDNMT-2 transcription levels between tissues might have to do with different functions in insects. We suggest this possibility since many previous reports on insects have shown that DNA methylation regulates various physiological processes by modulating a wide range of gene expressions (21, 30). Kausar et al. (21) reviewed that in the insect’s DNA methylation levels of embryos, larvae, and adults, genes associated with innate immunity, development, growth, etc., respond to the changing levels of DNA methylation. In a previous study, the authors suggested constitutive expression of DNMTs in the insect tissues through their life cycle. This observation might be elucidated by the fact that insects undergo various physical and physiological changes during the life cycle, requiring a precise mechanism for the surveillance of these processes (21, 30). It is likely that we are seeing a similar factor affecting the expression of DNMTs. Because of the reasons above, it is not surprising that DNMTs are expressed constitutively in various tissues of A. pernyi larvae.
Moreover, mRNA expression analysis revealed that both ApDNMT-1 and ApDNMT-2 gene transcription levels were increased in the Chinese oak silkworm, A. pernyi, after microbial injections, particularly at critical timepoints of infection. The upregulation of DNMT genes transcription seems to be a general response of insects against microbial pathogens. Baradaran and his coworkers (30) reported the induction of DNMT-1 and DNMT-2 genes following bacterial infection. Besides, it has also been shown that E. coli infection induced DNMT-1 and DNMT-2 transcription levels and caused differential DNA methylation status of the genome in Manduca sexta (31) and Galleria mellonella (32). In addition, in Aedes aegypti and Drosophila melanogaster, increased DNMT-2 gene expression and hypermethylation have been observed following the Wolbachia infection (33–35). Although DNMT-3, a de novo DNA methyltransferase, is absent in lepidopteran insects, including A. pernyi, recent research has shown that in insects, particularly lepidopteran insects, DNMT-1 is responsible for DNA methylation rather than the DNMT-3 gene (28). In addition, this gene has also been reported to be involved in the maintenance of DNA methylation and also in de novo DNA methylation (36). In contrast, the presence of DNMT-2 in dipteran insects (e.g., A. aegypti and D. melanogaster) is responsible for changes in genome methylation (35, 37). The molecular mechanism by which pathogens modify DNA methylation is not clear in insects; however, this mechanism has been well explored in vertebrates (21). A previous study suggested that modulation of DNA methylation is caused by natural killer cell activity via IFN (interferon)-gamma (38). The superfamily of tripartite motif-containing proteins induces innate immunity and pathogen defense against a variety of pathogen infections in response to IFNs (39). Lim et al. found that the HBx-mediated reduction of TRIM22 may evade the antiviral effect of IFNs in a host, implying that HBx is one of the factors controlled by the pathogens to regulate DNA methylation. Subsequently, reducing IFN regulatory factor-1 binding affinity thereby decreases the IFN-stimulated induction of TRIM22h, which impedes IFN regulatory factor-1, which subsequently seems to influence DNA methyltransferase tool kit and DNA methylation (40). This process may help pathogens evade host immunity. Given our observations and these studies, it seems that the upregulation of ApDNMT-1 and ApDNMT-2 genes in A. pernyi after microbial infections can affect the DNA methylation status in this insect species, thereby modulating gene expression and affecting different physiological responses, including immunity.
Based on the above observations, we predict DNMT genes can affect the immune responses of the insect. Data are limited on the immunological functions of DNA methylation in insects, and we were interested in seeing whether DNA methylation might be implemented in the modulation of innate immune response in A. pernyi. For this purpose, we administrated 5-AZA to inhibit the activity of DNMT genes in A. pernyi. This DNMT inhibitor is commonly used to suppress DNA methylation in the genomes of insects (40–43). Our results revealed that bacterial replication and mortality rates were lower following 5-AZA treatment compared to the control group. Additionally, colony-forming units were also lower in the 5-AZA treated larvae, indicating that the DNMTs are essential for the optimal growth of bacteria in insects. To the best of our knowledge, there is only one report on the necessity of DNMTs for bacterial replication in insects (30). However, in humans, the tick-born bacterial pathogen Anaplasma phagocytophilum greatly triggers hypermethylation in the neutrophil genome after 24 h post-infection, followed by differential expression of genes. In addition, the number of bacteria (A. phagocytophilum) growing in the presence of 5-AZA was approximately 25% less than in the control group (44).
Finally, we analyzed whether the mRNA expression of A. pernyi AMPs could be stimulated after an E. coli challenge and whether DNA methylation inhibition by 5-AZA had any effect on the transcription levels of these antimicrobial peptides. We observed that inhibition of DNMT inhibitor regulated AMPs following the bacterial infection. These results show that DNA methylation has a potential effect on the expression patterns of these peptides under bacterial infection, leading us to suggest that in A. pernyi, DNA methylation influences transcription of AMPs in response to Gram-negative E. coli. In humoral immunity, AMPs have a crucial role in suppressing microbial infection in insects (45, 46). Thus, the variation in the transcription levels of AMPs can shape the insect-bacterial pathogen interactions. Considering the increase of some AMPs transcription and the reduced colony-forming units and bacterial replication in A. pernyi following 5-AZA treatment, it also suggests that changes in the production of AMPs are one of the key molecular mechanisms that attenuate the replication of bacteria. In human cells, it has been shown that 5-AZA has an immunomodulatory function (47). In addition to insects, it has also been demonstrated in human that DNA methylation can be changed in a short period of time in response to microbial infection and that these changes in DNA methylation can influence immune cell responsiveness (48), suggesting that microbes, particularly bacteria, use DNA methylation strategies to evade host immune responses (49–51). A recent study analyzed the global transcriptional patterns of gene expression after M. tuberculosis, and found that the majority of differentially expressed genes were hypermethylated, which could play a biological role in the activation of innate and adaptive immune cells (51). Helicobacter (H.) pylori are also one of the most studied pathogens in terms of DNA methylation in humans. H. pylori causes changes in DNA methylation, and this abnormal DNA methylation in H. pylori-infected gastric mucosae has been linked to a higher risk of gastric cancer (52). DNA hypermethylation caused by H. pylori infection can be partially reversible once the bacterium is eliminated or 5-AZA is administered, resulting in a lower risk of gastric cancers triggered by H. pylori infection (53, 54). Besides, A non-catalytic biological role has also been described for the DNMT-1 gene since it can control expression levels without any variation in DNA methylation patterns, probably via interactions with other enzymatic proteins associated with epigenetic gene modulation (55, 56). Overall, whether the changes in the AMPs transcription patterns are due to the methylation status of these genes, or some other molecular mechanism, is still unexplored; however, the molecular mechanism could also be via variations in the gene transcriptional machinery triggered by the lack of DNMTs’ activity.
Furthermore, the upregulation of AMPs found in this study was mainly controlled by the Toll and IMD signaling pathways in insects, implying that DNA methylation may influence the activity of these pathways. Our analysis also confirmed that the 5-AZA and bacterial treatments influenced the expression of Toll and IMD pathway-associated genes. Our findings are consistent with previous studies; for example, evidence indicates that despite changes in AMPs transcription caused by DNA methylation (57–59), the Toll and IMD pathway-associated genes are modulated by epigenetic mechanisms, particularly by DNA methylation (60, 61). Thus, we conclude that DNA methylation influences the expression patterns of AMPs directly or indirectly, which helps a host organism to attenuate microbial infections.
In summary, our study is the first to identify and clone DNA methyltransferases (DNMT-1 and DNMT-2 genes) from the Chinese oak silkworm A. pernyi. Phylogenetic and structural analysis revealed a high similarity of these proteins with their respective homologous enzymes in other insects. ApDNMT-1 and ApDNMT-2 were observed in all tissues examined, and their transcription patterns changed in response to microbial infection. As a DNMTs inhibitor, 5-AZA treatment reduced the replication rate, colony-forming units, and mortality of larvae compared to the control group. Additionally, after 5-AZA treatment, transcription patterns of AMPs and their associated pathway-related genes were altered in response to bacterial infection. Altogether, our data uncovered a novel immune regulatory function of DNMTs that shapes the insect-pathogen interaction in the Chinese oak silkworm A. pernyi.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
SK, MA, and IG: study design, data acquisition, results analysis, and prepared manuscript. RL, EZ, and QL: analysis of results and modification of the manuscript. HC and ML critical review, modification of manuscript and fund acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the China Postdoctoral Science Foundation (2021M692676), Chongqing -Postdoctoral Science Foundation (cstc2021jcyj-bsh0128), the National Natural Science Foundation of China (31802142), the Doctorial Start-up Fund of Southwest University (SWU020023), the Fundamental Research Funds for the Central Universities (XDJK2019C089), and the Foundation of State Key Laboratory of Silkworm Genome Biology (SKLSGB-ORP202003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.855888/full#supplementary-material
Supplementary Figure 1 | ApDNMT-1 nucleotide and deduced amino acid sequence of A. pernyi. The ApDNMT-1 deduced amino acid sequence is presented below the nucleotide sequence of cDNA, and the one-letter codes are aligned with the second nucleotide of each codon. The nucleotide and amino acid sequences are numbered at the left. Identified domains were labeled as DNMT1-RDF: DNA methyltransferase replication foci domain which is indicated by a red box, ZnF: Zinc finger domain is indicated by an orange colour box, BAH: Bromo adjacent homology domain is shown in yellow colour box, DCM: DNA-cytosine methyltransferases domain is shown in green box Cys-rich: cysteine-rich ADD domain is presented in a blue box.
Supplementary Figure 2 | ApDNMT-2 nucleotide and deduced amino acid sequence of A. pernyi. The ApDNMT-2 deduced amino acid sequence is presented below the nucleotide sequence of cDNA, and the one-letter codes are aligned with the second nucleotide of each codon. The nucleotide and amino acid sequences are numbered at the left. The putative DNMT2 cyt_C5_DNA methylase superfamily domain is shown in the red box.
1. Li W, Zhang Z, Lin L, Terenius O. Antheraea pernyi (Lepidoptera: Saturniidae) and Its Importance in Sericulture, Food Consumption, and Traditional Chinese Medicine. J Econ Entomol (2017) 110(4):1404–11. doi: 10.1093/jee/tox140
2. Kausar S, Abbas MN, Qian C, Zhu BJ, Sun Y, Sun YX, et al. Serpin-14 Negatively Regulates Prophenoloxidase Activation and Expression of Antimicrobial Peptides in Chinese Oak Silkworm Antheraea pernyi. Dev Comp Immunol (2017) 76:45–55. doi: 10.1016/j.dci.2017.05.017
3. Wang G, Na S, Qin L. Uncovering the Cellular and Humoral Immune Responses of Antheraea pernyi Hemolymph to Antheraea pernyi Nucleopolyhedrovirus Infection by Transcriptome Analysis. J Invert Pathol (2019) 166:107205. doi: 10.1016/j.jip.2019.107205
4. Kausar S, Abbas MN, Qian C, Zhu BJ, Gao J, Sun Y, et al. Role of Antheraea pernyi Serpin 12 in Prophenoloxidase Activation and Immune Responses. Arch Insect Biochem Physiol (2018) 97:e21435. doi: 10.1002/arch.21435
5. Hillyer JF. Insect Immunology and Hematopoiesis. Dev Comp Immunol (2016) 58:102–18. doi: 10.1016/j.dci.2015.12.006
6. Kausar S, Abbas MN, Zhao Y, Cui H. Immune Strategies of Silkworm, Bombyx mori Against Microbial Infections. Invert Surviv J (2019) 16:130–40. doi: 10.25431/1824-307X/isj.v0i0.130-140
7. Bestor TH. The DNA Methyltransferases of Mammals. Hum Mol Genet (2000) 9:2395–402. doi: 10.1093/hmg/9.16.2395
8. Li E. Chromatin Modification and Epigenetic Reprogramming in Mammalian Development. Nat Rev Genet (2002) 3:662–73. doi: 10.1038/nrg887
9. Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, et al. Single Base-Resolution Methylome of the Silkworm Reveals a Sparse Epigenomic Map. Nat Biotechnol (2010) 28:516–20. doi: 10.1038/nbt.1626
10. Feliciello I, Parazajder J, Akrap I, Ugarkovic D. First Evidence of DNA Methylation in Insect Tribolium Castaneum: Environmental Regulation of DNA Methylation Within Heterochromatin. Epigenetics (2013) 8:534–41. doi: 10.4161/epi.24507
11. Beeler SM, Wong GT, Zheng JM, Bush EC, Remnant EJ, Oldroyd BP, et al. Whole-Genome DNA Methylation Profile of the Jewel Wasp (Nasonia Vitripennis). G3 (Bethesda) (2014) 4:383–8. doi: 10.1534/g3.113.008953
12. Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA Methylation is Found in Drosophila Melanogaster But Absent in Saccharomyces Cerevisiae, Schizosaccharomyces Pombe, and Other Yeast Species. Anal Chem (2014) 86:3697–702. doi: 10.1021/ac500447w
13. Glastad KM, Hunt BG, Yi SV, Goodisman MA. DNA Methylation in Insects: on the Brink of the Epigenomic Era. Insect Mol Biol (2011) 20:553–65. doi: 10.1111/j.1365-2583.2011.01092.x
14. Standage DS, Berens AJ, Glastad KM, Severin AJ, Brendel VP, Toth AL. Genome, Transcriptome and Methylome Sequencing of a Primitively Eusocial Wasp Reveal a Greatly Reduced DNA Methylation System in a Social Insect. Mol Ecol (2016) 25:1769–84. doi: 10.1111/mec.13578
15. Wu P, Jie W, Shang Q, Annan E, Jiang X, Hou C, et al. DNA Methylation in Silkworm Genome may Provide Insights Into Epigenetic Regulation of Response to Bombyx mori Cypovirus Infection. Sci Rep (2017) 7:16013. doi: 10.1038/s41598-017-16357-7
16. Lyko F, Maleszka R. Insects as Innovative Models for Functional Studies of DNA Methylation. Trends Genet (2011) 27:127–31. doi: 10.1016/j.tig.2011.01.003
17. Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, et al. Genome-Wide and Caste-Specific DNA Methylomes of the Ants Camponotus Floridanus and Harpegnathos Saltator. Curr Biol (2012) 22:1755–64. doi: 10.1016/j.cub.2012.07.042
18. Yan H, Bonasio R, Simola DF, Liebig J, Berger SL, Reinberg D. DNA Methylation in Social Insects: How Epigenetics can Control Behavior and Longevity. Annu Rev Entomol (2015) 60:435–52. doi: 10.1146/annurev-ento-010814-020803
19. Biergans SD, Giovanni GC, Judith R, Charles C. Dnmts and Tettarget Memory-Associated Genes After Appetitive Olfactory Training in Honey Bees. Sci Rep (2015) 5:21656. doi: 10.1038/srep16223
20. Herb BR, Wolschin F, Hansen KD, Aryee MJ, Langmead B, Irizarry R, et al. Reversible Switching Between Epigenetic States in Honeybee Behavioral Subcastes. Nat Neurosci (2012) 15:1371–3. doi: 10.1038/nn.3218
21. Kausar S, Abbas MN, Cui H. A Review on the DNA Methyltransferase Family of Insects: Aspect and Prospects. Int J Biol Macromol (2021) 186:289–302. doi: 10.1016/j.ijbiomac.2021.06.205
22. Ye YH, Woolfit M, Huttley GA, Rances E, Caragata EP, Popovici J, et al. Infection With a Virulent Strain of Wolbachia Disrupts Genome Wide-Patterns of Cytosine Methylation in the Mosquito Aedes Aegypti. PloS One (2013) 8:e66482. doi: 10.1371/journal.pone.0066482
23. Vilcinkas A. The Role of Epigenetics in Host–Parasite Coevolution: Lessons From the Model Host Insects Galleria Mellonella and Tribolium Castaneum. Zoology (2016) 119:273–80. doi: 10.1016/j.zool.2016.05.004
24. Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. Improvement of Phylum- and Class-Specific Primers for Real-Time PCR Quantification of Bacterial Taxa. J Microbiol Methods (2011) 86:351–6. doi: 10.1016/j.mimet.2011.06.010
25. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta CT) Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
26. Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as Inhibitors of DNA Methylation: Mechanistic Studies and Their Implications for Cancer Therapy. Oncogene (2002) 21:5483–95. doi: 10.1038/sj.onc.1205699
27. Abbas MN, Kausar S, Gul I, Ke XX, Dong Z, Lu X, et al. Suppressor of Cytokine Signalling 6 is a Potential Regulator of Antimicrobial Peptides in the Chinese Oak Silkworm, Antheraea pernyi. Mole Immunol (2021) 140:12–21. doi: 10.1016/j.molimm.2021.10.001
28. Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. Evolution of DNA Methylation Across Insects. Mol Biol Evol (2017) 34:654–65. doi: 10.1093/molbev/msw264
29. Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics Resolves the Timing and Pattern of Insect Evolution. Science (2014) 346:763–7. doi: 10.1126/science.1257570
30. Baradaran E, Moharramipour S, Asgari S, Mehrabadi M. Induction of DNA Methyltransferase Genes in Helicoverpa Armigera Following Injection of Pathogenic Bacteria Modulates Expression of Antimicrobial Peptides and Affects Bacterial Proliferation. J Insect Physiol (2019) 118:103939. doi: 10.1016/j.jinsphys.2019.103939
31. Heitmueller M, Billion A, Dobrindt U, Vilcinskas A, Mukherjee K. Epigenetic Mechanisms Regulate Innate Immunity Against Uropathogenic and Commensal-Like Escherichia Coli in the Surrogate Insect Model Galleria Mellonella. Infect Immun (2017) 85:e00336–00317. doi: 10.1128/IAI.00336-17
32. Gegner J, Baudach A, Mukherjee K, Halitschke R, Vogel H, Vilcinskas A. Epigenetic Mechanisms are Involved in Sex-Specific Trans-Generational Immune Priming in the Lepidopteran Model Host Manduca Sexta. Front Physiol (2019) 10:137. doi: 10.3389/fphys.2019.00137
33. Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia Uses a Host microRNA to Regulate Transcripts of a Methyltransferase, Contributing to Dengue Virus Inhibition in Aedes Aegypti. Proc Natl Acad Sci USA (2013) 110:10276–81. doi: 10.1073/pnas.1303603110
34. LePage DP, Jernigan KK, Bordenstein SR. The Relative Importance of DNA Methylation and Dnmt2-mediated Epigenetic Regulation on Wolbachia Densities and Cytoplasmic Incompatibility. PeerJ (2014) 2:e678.
35. Bhattacharya T, Newton ILG, Hardy RW. Wolbachia Elevates Host Methyltransferase Expression to Block an RNA Virus Early During Infection. PloS Pathog (2017) 13:e1006427. doi: 10.1371/journal.ppat.1006427
36. Yarychkivska O, Tavana O, Gu W, Bestor TH. Independent Functions of DNMT1 and USP7 at Replication Foci. Epigenet Chromatin (2018) 11:9. doi: 10.1186/s13072-018-0179-z
37. Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like Protein Mediates DNA Methylation in Drosophila. Development (2003) 130:5083–90. doi: 10.1242/dev.00716
38. Okamoto Y, Shinjo K, Shimizu Y, Sano T, Yamao K, Gao W, et al. Hepatitis Virus Infection Affects DNA Methylation in Mice With Humanized Livers. Gastroenterology (2014) 146:562–72. doi: 10.1053/j.gastro.2013.10.056
39. Ozato K, Shin DM, Chang TH, Morse HC. TRIM Family Proteins and Their Emerging Roles in Innate Immunity. Nat Rev Immunol (2008) 8:849–60. doi: 10.1038/nri2413
40. Lim KH, Park ES, Kim DH, Cho KC, Kim KP, Park YK, et al. Suppression of Interferon-Mediated anti-HBV Response by Single CpG Methylation in the 5’-UTR of TRIM22. Gut (2018) 67:166–78. doi: 10.1136/gutjnl-2016-312742
41. Amarasinghe HE, Clayton CI, Mallon EB. Methylation and Worker Reproduction in the Bumble-Bee (Bombus Terrestris). Proc R Soc Lond B Biol Sci (2014) 281:20132502. doi: 10.1098/rspb.2013.2502
42. Kumar S, Kim Y. An Endoparasitoid Wasp Influences Host DNA Methylation. Sci Rep (2017) 7:43287. doi: 10.1038/srep43287
43. Xu G, Zhang J, Lyu H, Song Q, Feng Q, Xiang H, et al. DNA Methylation Mediates BmDeaf1-regulated Tissue-and Stage-Specific Expression of BmCHSA-2b in the Silkworm, Bombyx mori. Epigenet Chromatin (2018) 11:32. doi: 10.1186/s13072-018-0202-4
44. Sinclair SHG, Yegnasubramanian S, Dumler JS. Global DNA Methylation Changes and Differential Gene Expression in Anaplasma Phagocytophilum-Infected Human Neutrophils. Clin Epigenet (2015) 7:77. doi: 10.1186/s13148-015-0105-1
45. Abbas MN, Zhu BJ, Kausar S, Dai LS, Sun YX, Tian JW, et al. Suppressor of Cytokine Signaling 2-12 Regulates Antimicrobial Peptides and Ecdysteroid Signaling Pathways in Bombyx mori (Dazao). J Insect Physiol (2017) 103:47–56. doi: 10.1016/j.jinsphys.2017.10.004
46. Zhang K, Hu X, Zhao Y, Pan G, Li C, Ji H, et al. Scavenger Receptor B8 Improves Survivability by Mediating Innate Immunity in Silkworm, Bombyx mori. Dev Comp Immunol (2021) 116:103917. doi: 10.1016/j.dci.2020.103917
47. Sanchez-Abarca LI, Gutierrez-Cosio S, Santamaria C, Caballero-Velazquez T, Blanco B, Herreo-Sanchez C, et al. Immunomodulatory Effect of 5-Azacytidine (5-azaC): Potential Role in the Transplantation Setting. Blood (2010) 115:107–21. doi: 10.1182/blood-2009-03-210393
48. Pacis A, Tailleux L, morin AM, Lambourne J, MacIsaac JL, Yotova V, et al. Bacterial Infection Remodels the DNA Methylation Landscape of Human Dendritic Cells. Genome Res (2015) 25(12):1801–11. doi: 10.1101/gr.192005.115
49. Das K, Saikolappan S, Dhandayuthapani S. Differential Expression of miRNAs by Macrophages Infected With Virulent and Avirulent Mycobacterium Tuberculosis. Tuberculosis (Edinb) (2013) 93 Suppl:S47–50. doi: 10.1016/S1472-9792(13)70010-6
50. Das K, Garnica O, Dhandayuthapani S. Modulation of Host miRNAs by Intracellular Bacterial Pathogens. Front Cell Infect Microbiol (2016) 6:79. doi: 10.3389/fcimb.2016.00079
51. Looney M, Lorenc R. Halushka MK and Karakousis Pc. Key Macrophage Responses to Infection With Mycobacterium Tuberculosis are Co-Regulated by microRNAs and DNA Methylation. Front Immunol (2021) 12: 685237. doi: 10.3389/fimmu.2021.685237
52. Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, et al. High Levels of Aberrant DNA Methylation in Helicobacter Pylori-Infected Gastric Mucosae and its Possible Association With Gastric Cancer Risk. Clin Cancer Res (2006) 12:989–95. doi: 10.1158/1078-0432.CCR-05-2096
53. Sepulveda AR, Yao Y, Yan W, Park DI, Kim JJ, Gooding W, et al. Cpg Methylation and Reduced Expression of O6-methylguanine DNA Methyltransferase is Associated With Helicobacter Pylori Infection. Gastroenterology (2010) 138:1836–44. doi: 10.1053/j.gastro.2009.12.042
54. Niwa T, Toyoda T, Tsukamoto T, mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter Pylori-Induced Gastric Cancers in Gerbils by a DNA Demethylating Agent. Cancer Prev Res (Phila) (2013) 6:263–70. doi: 10.1158/1940-6207.CAPR-12-0369
55. Clements N, Hannigan M, Miller SL, Peel JL, Milford JB. Comparisons of Urban and Rural PM10–2.5 and PM2.5 Mass Concentrations and Semi-Volatile Fractions in Northeastern Colorado. Atmos Chem Phys (2016) 16:7469–84. doi: 10.5194/acp-16-7469-2016
56. Schulz JM, Knoflach F, Hernandez MC, Bischofberger J. Dendrite-Targeting Interneurons Control Synaptic NMDA-Receptor Activation Via Nonlinear A5-GABAA Receptors. Nat Commun (2018) 9:3576. doi: 10.1038/s41467-018-06004-8
57. Noh YH, Lee J, Seo SJ. Myung SCh. Promoter DNA Methylation Contributes to Human β-Defensin-1 Deficiency in Atopic Dermatitis. Anim Cells Sys (2018) 22:172–7. doi: 10.1080/19768354.2018.1458652
58. Chen X, Qi G, Qin M, Zou Y, Zhong K, Tang Y, et al. DNA Methylation Directly Downregulates Human Cathelicidin Antimicrobial Peptide Gene (CAMP) Promoter Activity. Oncotarget (2017) 8:27943–52. doi: 10.18632/oncotarget.15847
59. Kweh MF, Merriman KE, Nelson CD. Inhibition of DNA Methyltransferase and Histone Deacetylase Increases β-Defensin Expression But Not the Effects of Lipopolysaccharide or 1,25-Dihydroxyvitamin D3 in Bovine Mammary Epithelial Cells. J Dairy Sci (2018) 102:5706–12. doi: 10.3168/jds.2018-16141
60. Goto A, Fukuyama H, Imler JL, Hoffmann JA. The Chromatin Regulator DMAP1 Modulates Activity of the Nuclear Factor _B (Nf-_B) Transcription Factor Relish in the Drosophila Innate Immune Response. J Biol Chem (2014) 289:20470–6. doi: 10.1074/jbc.C114.553719
Keywords: silkworm, epigenetics, DNA methylation, pathogens, antimicrobial peptides
Citation: Kausar S, Abbas MN, Gul I, Liu R, Li Q, Zhao E, Lv M and Cui H (2022) Molecular Identification of Two DNA Methyltransferase Genes and Their Functional Characterization in the Anti-Bacterial Immunity of Antheraea pernyi. Front. Immunol. 13:855888. doi: 10.3389/fimmu.2022.855888
Received: 16 January 2022; Accepted: 11 April 2022;
Published: 16 May 2022.
Edited by:
Yonggyun Kim, Andong National University, South KoreaReviewed by:
Luc Swevers, National Centre of Scientific Research Demokritos, GreeceCopyright © 2022 Kausar, Abbas, Gul, Liu, Li, Zhao, Lv and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhan Lv, NTI3MjE3NTlAcXEuY29t; Hongjuan Cui, aGN1aUBzd3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.