- 1Medical School, Southeast University, Nanjing, China
- 2Department of Urology, Affiliated Zhongda Hospital of Southeast University, Nanjing, China
- 3Department of Urology, Nanjing Lishui District People’s Hospital, Zhongda Hospital, Southeast University, Nanjing, China
- 4Key Laboratory of Biomedical Information Engineering of Ministry of Education, Biomedical Informatics and Genomics Center, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an, China
The rapid development of multiple high-throughput sequencing technologies has made it possible to explore the critical roles and mechanisms of functional enhancers and enhancer RNAs (eRNAs). The inflammatory immune response, as a fundamental pathological process in infectious diseases, cancers and immune disorders, coordinates the balance between the internal and external environment of the organism. It has been shown that both active enhancers and intranuclear eRNAs are preferentially expressed over inflammation-related genes in response to inflammatory stimuli, suggesting that enhancer transcription events and their products influence the expression and function of inflammatory genes. Therefore, in this review, we summarize and discuss the relevant inflammatory roles and regulatory mechanisms of eRNAs in inflammatory immune cells, non-inflammatory immune cells, inflammatory immune diseases and tumors, and explore the potential therapeutic effects of enhancer inhibitors affecting eRNA production for diseases with inflammatory immune responses.
Highlights
This review summarized the relevant roles of eRNAs in inflammatory immune functions, mechanisms and therapies, and explored the research directions and target therapy prospects for inflammatory immune-related eRNAs.
Introduction
The rapid development of high-throughput sequencing technologies has made it possible to identify potential functional regulatory elements. To date, studies have identified numerous regulatory elements, such as enhancers, promoters and silencers (1).
Enhancers, which are distal regulatory DNA sequences, are approximately 500–2000 bp in length and function independently of orientation (2, 3). Since the discovery of the first enhancer SV40 by Banerji J et al. in 1981 (4), the specific function and mechanism of enhancers have been extensively explored. Numerous studies have revealed that pioneer transcription factors enhance nucleosome DNA to generate open chromatin (5), which promotes the recruitment of lineage-determining transcription factors (LDTFs) to maintain the activated state of enhancers (6). Subsequently, collaborative transcription factors (cTFs) and co-cofactors (CoFs), such as histone methyltransferases, are recruited, which promote histone 3 lysine 4 monomethylation (H3K4me1) and dimethylation (H3K4me2) (6–10). Following this, histone acetyltransferases (HAT) [e.g. CBP/p300 to promote H3K27 acetylation (ac)] (10, 11), general transcription factors (GTFs) (12) and RNA polymerase II (RNAPII) (3, 13) are further recruited to initiate enhancer-associated bidirectional transcription. Additionally, DNA demethylase (DME) is recruited during the initial phase of enhancer activation to regulate dynamic DNA methylation and p300 binding (14, 15) (Figure 1A). Consistent with previous studies, the above study suggests that enhancers positively regulate spatiotemporal gene expression (4, 16, 17).
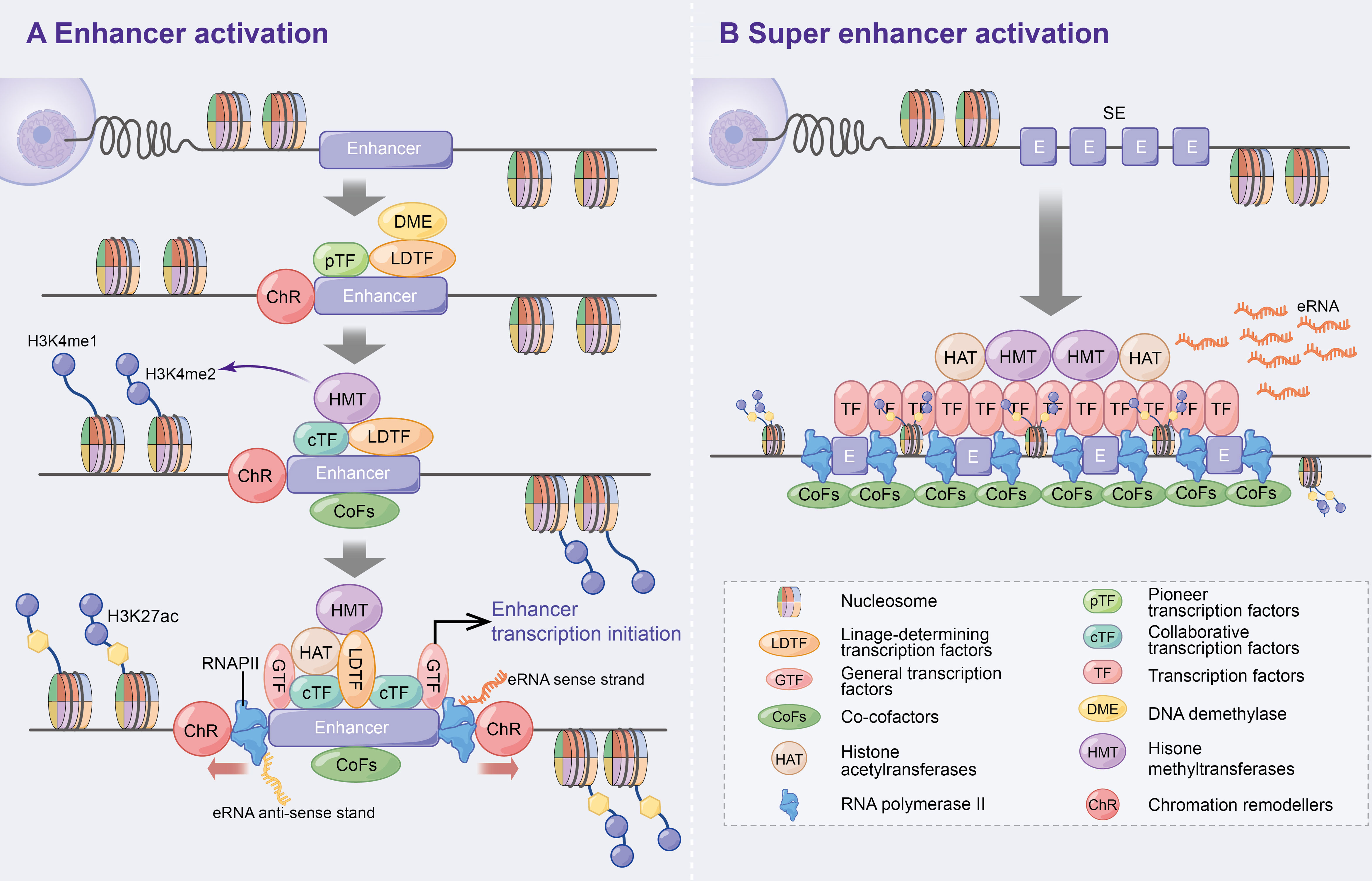
Figure 1 Activation mechanism of enhancers and super enhancer. (A) Enhancer activation. Multifactorial stimulation causes pTFs binding to nucleosomes leading to ChR and further recruitment of LDTFs, cTFs and CoFs that assist enhancer activation and histone methylation. On this basis, HAT, GTFs and RNAPII are recruited and ultimately contribute to enhancer transcription bi-directionally. In addition, DME are recruited to regulate DNA methylation and p300 status at the initial stage of enhancer activation. (B) Super enhancers are large clusters of enhancers with high levels of cell-specific transcription factors and cofactors that can ultimately express higher levels of eRNAs.
Super enhancers (SEs) were first found in mouse embryonic stem cells and tumour cell lines (18, 19). Compared to typical enhancers, SEs consist of large enhancer clusters with longer genomic regions, higher cell-specific transcription factor (TF) levels (e.g. Oct4, Sox2, Nanog, Eseeb and Klf4) (18, 20–22), CoFs (e.g. BRD4, Mediator and CDK9) (21), higher H3K4me1 expressions and H3K27ac modifications (23, 24). These characteristics allow SEs to highly express super enhancer derived super RNAs (seRNAs) (Figure 1B). Therefore, SEs have been speculated to be the key determinants of cell identity and fate (18, 23), and thereby lead to increased disease susceptibility when mutated (20, 23).
eRNA, as the enhancers transcription product, belongs to the class of non-coding RNA (ncRNA). Based on differences in function and size, ncRNAs are classified as long non coding RNA (lncRNA), promoter associated RNA (paRNA), enhancer RNA (eRNA), small RNA (sRNA), PIWI interacting RNA (piRNA), small nucleolar RNA (snoRNA), Small nuclear RNA (snRNA), ribosomal RNA (rRNA), micro RNA (miRNA) etc (25–28). Similarly, they are generated in different parts of the genome and can be obtained by unidirectional or bidirectional transcription, with variable stability and longevity (26–28). The section will briefly describe the differences and commonalities between eRNA and lncRNA. eRNAs are similar in length to lncRNAs, about 0.1-9 Kb, but are divided into two classes of ncRNAs by their associated histone profiles (29). In the 2010 study, Orom UA et al. (30) found that lncRNAs can regulate the expression of neighboring genes. Subsequently, this type of lncRNA was revealed to be an enhancer of some protein-coding genes, but its chromatin characteristics differ from typical enhancers and it is usually polyadenylated and does not have the characteristics of bidirectional transcription of eRNA. Further studies suggest that such lncRNAs carry intermediary complexes to neighboring genes via chromatin loops to influence their expression (30, 31). However, most eRNA transcripts are 5’ cap-shaped, unspliced, unpolyadenylated and have a short half-life, but are also dynamically regulated with signaling and correlate with increased expression of neighboring genes (32).
To date, numerous reviews have summarized the mechanisms and potential functions of enhancers, SEs and their transcription products (eRNAs) in tumours (33–36). Moreover, the expansion of the study dimension revealed that enhancers, SEs and their eRNAs were immediately altered in response to inflammatory stimuli, leading to abnormal inflammatory gene expression. This observation strongly suggests their involvement in biological processes, such as inflammation, immunity and neurodegeneration (24, 37–39). Yoshiki et al. (21) have summarized the potential role of SEs in inflammatory gene transcription.
However, to the best of our knowledge, the potential mechanisms and roles of eRNAs in inflammatory immune diseases are yet to be reviewed. This study aims to summarize the potential roles of eRNAs in inflammatory immune cells, non-inflammatory immune cells, inflammatory immune diseases and tumour inflammatory alterations, and thereby elucidate the relevance of eRNAs in inflammatory immunity (Figure 2). Additionally, the potential role of eRNAs as novel therapeutic targets and prognostic biomarkers for diseases with inflammatory immune alterations are explored.
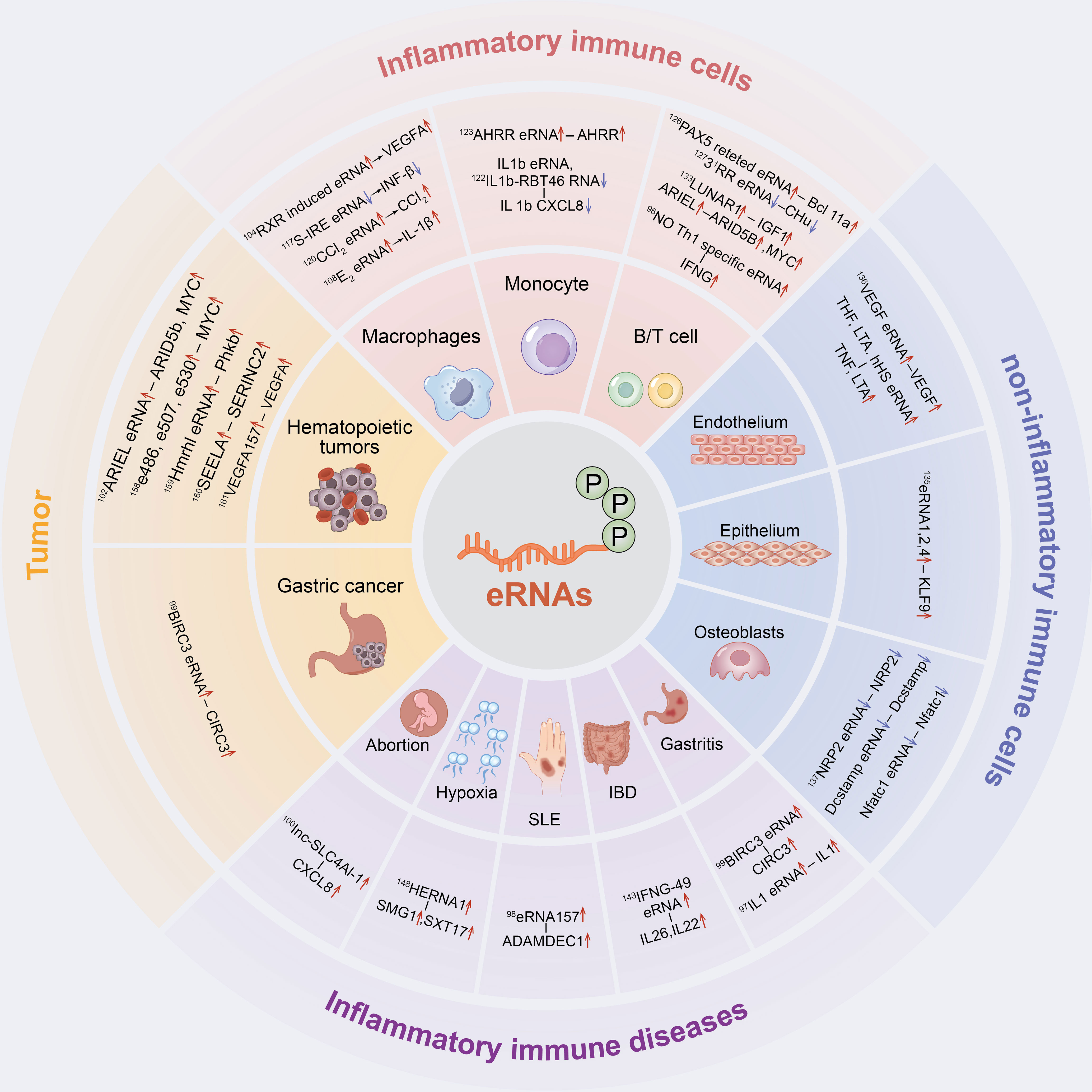
Figure 2 The inflammatory immune role of eRNAs in cells and diseases. eRNAs have a significant contribution in inflammatory immune cells, non-inflammatory immune cells, inflammatory immune diseases and tumor inflammatory alterations.
Results
Mechanism of Enhancer Transcription and eRNA in Regulating Target Genes
Effect of Enhancer Transcription on Their Products and Target Genes
The mechanism of enhancer activation has been discussed in the Introduction (Figure 1A); therefore, the regulation of downstream target genes by enhancer transcription (including initiation, elongation, termination and degradation) will be discussed in this section.
GTFs (e.g. TBP) and the serine 5-phosphorylated form of RNAPII (Ser5p was engaged in RNA capping mechanism) have been detected in the enhancer region in the transcription of lncRNAs or mRNAs (40). Moreover, global nuclear run-on sequencing (GRO-seq) and cap analysis gene expression (CAGE) results further suggest that eRNAs are capped (5′ end 7-methylguanosine (m7G) cap facilitates cap-binding complex (CBC) recruitment), with significant similarities in DNA sequences, core promoter elements and nucleosome spacing at enhancer and promoter transcription start sites (TSSs) (41–43), and promote bidirectional transcription (29, 32, 42, 44), thus indicating that the rules of transcription initiation apply to enhancers and promoters. Similar to promoters, the direction of enhancer transcription is also determined by the ratio of the relative density of polyA cleavage sites (PASs) and the U1 splice motif downstream of the TSSs, which affects the production of eRNAs initiated by RNAPII elongation (41, 42, 44–46) (Figure 3A).
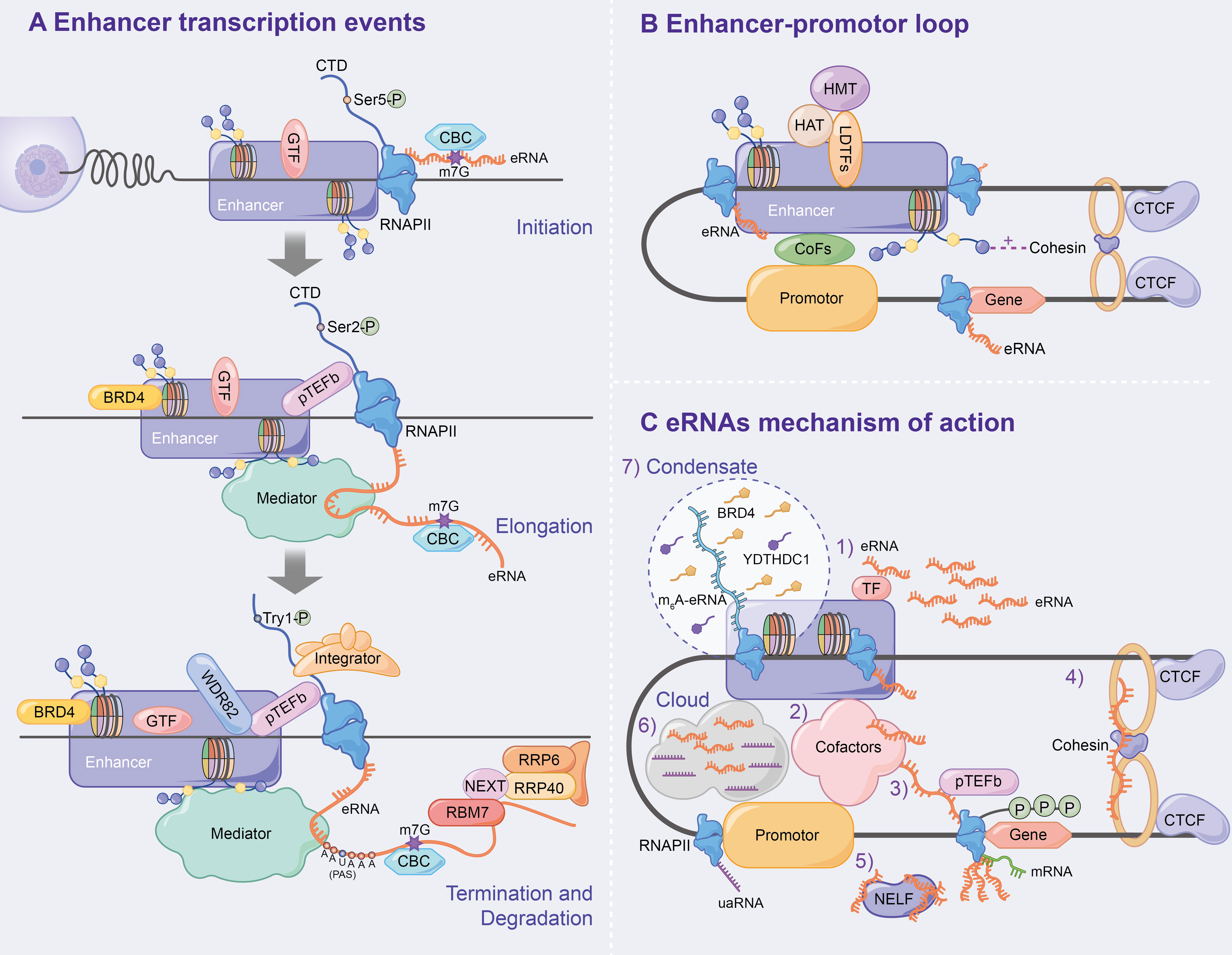
Figure 3 Mechanism of enhancer transcription and eRNAs to regulate target genes. (A) Following enhancer activation, GTFs and serine 5-phosphorylated form of RNAPII (Ser5p) are recruited, which leads CBC to binding to eRNA via m7G, ultimately initiating enhancer transcription. During the enhancer elongation, Ser2p is hypoenriched. pTEFb, BRD4 and the eRNAs being transcribed all affect transcription elongation. Subsequently, the nascent eRNAs interacts with RNAPII CTD to terminate transcription. In addition, the integrator, WDR82 and Tyr1p assist in correct enhancer transcription termination. Finally, the NEXT complex mediates the degradation of eRNAs. (B) Chromatin loops are generated under the influence of LDTFs, histone methylation or acetylation, cohesin-CTCF complex and interphase chromosome topologically associated domain, and ultimately activate target gene transcription. (C) Specific mechanisms have been demonstrated for eRNAs to affect enhancer activity, E-P loop formation, and transcription of downstream target genes.
The elongation, termination and RNA processing stages highlight transcriptional differences between enhancers and promoters. Ser2p, a form of RNAPII that is involved in elongation, is characterized by low enrichment, which results in low H3K36me3 levels and affects enhancer transcription elongation (40, 47, 48). The enhancer transcription elongation phase was found to be partially impacted by the overlapping mechanisms of the coding genes, including positive transcription elongation factor-b complex (pTEFb) and bromodomain-containing protein 4 (BRD4, which is recruited by H3K27ac) (10–12). Additionally, eRNAs undergoing transcription bind to the mediator complex and affect transcription elongation (18, 31, 49) (Figure 3A).
Numerous studies have found that PAS-mediated early termination regulates eRNA stability, and RNA exosomes degrade eRNAs. A study by Lai et al. confirmed that the integrator, which interacts with the RNAPII carboxy-terminal domain (RNAPII CTD), is an important regulator of eRNA termination (50) that functions after the nascent eRNAs have the PAS (AAUAAA) (42, 44, 50, 51). Integrator subunit deletion decreases eRNA levels and increases enhancer transcription activity, indicating the disruption of eRNA termination (50). Additionally, WD repeat-containing protein 82 (WDR82) acts as an adaptor protein that targets SET1 H3K4 methyltransferase, affecting enhancer transcription termination (52). Interestingly, Tyrosine 1-phosphorylated form of RNAPII (Tyr1p) was highly enriched in the active enhancer and PROMPT (upstream transcripts of the promoter) regions, whereas it was slightly enriched in the gene promoter region (53, 54). Moreover, Yeast Tyr1p was shown to be associated with eRNA termination (55, 56). In addition, under the mediation of the trimeric nuclear exosome targeting (NEXT) complex, wherein RBM7 directly binds to eRNAs, exosome component 10 (EXOSC10/RRP6) and EXOSC3 (RP40) were reported to be responsible for the final degradation of eRNAs (57). The recruitment of NEXT may also be facilitated by the CBC (57, 58) (Figure 3A).
The chromatin loop, including enhancer-promoter loop or E-P loop, as the optimal mediator of target gene expression, was found to play an important role in enhancer transcription and eRNA production via enhancer-promoter proximity facilitation (59–61) (Figure 3B). Since β-globin loci discovery, enhancer-promoter-mediated chromatin loops have been detected at multiple enhancer locus control regions (LCRs) interacting with target genes through chromosome conformation capture (3C) (62, 63). LDTFs [e.g. NF-E2 (64)] were found to directly anchor E-P loop regions to recruit cTFs, CoFs or histone-modifying enzymes, which affects E-P loop formation and enhancer activation (64–68). H3K4me1 improves interchromatin interactions between enhancers and promoters by promoting chromatin recruitment to the cohesin complex (69), whereas H3K27ac affects enhancer-promoter activity by destabilising nucleosomes or recruiting H3K27ac-binding proteins (70). CCCTC-binding factor (CTCF), a highly conserved zinc finger protein, is co-localized with cohesin and has an independent cohesion binding site (71, 72). The cohesin-CTCF complex creates repressor regions (blocking chromatin repressor region diffusions and blocking enhancer activity) and active regions (promoting enhancer to promoter proximity) to regulate chromatin homeostasis, which contributes to the formation and stabilization of long-term chromatin interactions, thus, affecting E-P loop formation and transcription progression (73–78). Using molecular dynamics models, Dusan et al. (79) demonstrated that transcription-induced superhelix in the interphase chromosome topologically associated domain (TAD) formation is the driving force for chromatin loop extrusion, which stimulated enhancer-promoter contacts and activates mRNA transcription in a given TAD.
Effect of eRNA on Target Genes
eRNAs, a transcription product of enhancers, affect enhancer activity, E-P loop formation and downstream target gene transcription. Numerous studies have found that eRNA has several mechanisms as follows (Figure 3C): 1) eRNA positively regulates enhancer transcription and stabilizes gene expression by binding to TFs (e.g. YY1) (12); 2) eRNA promotes active enhancer acetylation by recruiting Cofactors (e.g. CBP/300) (80) and interacts with various other Cofactors Complexes [e.g. RAD21 and heterogeneous nuclear ribonucleoprotein U (hnRNPU)] to stabilize the E-P loop and regulate target gene expression (81–83); 3) eRNAs directly induce E-P loop formation and histone modification by linking p300 (as Cofactors) and RNAP II (whose RNAP II are present on the enhancer and promoter, respectively) (84); 4) eRNA stabilizes the E-P loop structure by absorbing cohesin and subsequently regulating gene expression (85); 5) eRNA binds to the paused RNAPII by competing with mRNA, which allows the negative elongation factor (NELF) complex to separate from RNAPII and bind to eRNA, leading to RNAPII phosphorylation and the positive transcription elongation factor b (P-TEFb) recruitment and allowing RNAPII to enter the transcription elongation phase and produce mRNAs (86); 6) Cai et al. proposed for the first time that enhancers and promoters form a local molecular cloud during transcription, which consists of eRNA and uaRNA (eRNA promoter transcription analogues), bringing enhancers and promoters closer to facilitate E-P loop formation (87); 7) The recent studies have confirmed the prevalence of m6A modification in nascent eRNA, which recruits the nuclear m6A reader YTHDC1 to form a liquid-like condensate in a manner dependent on its C terminus intrinsically disordered region and arginine residues. Subsequently, the m6A-eRNA/YTHDC1 condensate co-mixes with and facilitates the formation of BRD4 coactivator condensate, ultimately activating the gene (88); 8) Aguilo et al. (89) demonstrated that NOP2/Sun RNA methyltransferase (NSUN7) can deposit 5-cytosine methylation on eRNA, which affects eRNA stability and regulates enhancer transcription; 9) eRNA regulates gene expression by modulating E-P interactions and higher chromatin structure topology (9).
Potential Regulatory Mechanisms of eRNA in Inflammatory Immunity
Acute and chronic inflammation is the adaptive response to the internal environment and external stimuli, and the underlying pathological events leading to atherosclerosis, cancer, infectious diseases and immune disorders. The production of active enhancers and eRNA production in the nucleus precedes inflammatory gene expression in response to lipopolysaccharide (LPS) stimuli (90–92). Comprehensive studies of eRNA inflammatory immune-related mechanisms have revealed that the binding of LDTF (e.g. PU.1/T-bet/AP-1 and C/EBP) to the enhancer regions under inflammatory mediator stimulation promotes nucleosome remodeling to create nucleosome-free regions and H3K4me1/2 modification and recruits RNAPII (13, 80, 93–96). Subsequently, inflammation-associated signal-dependent TF NFκB-p65/p50 binds to the enhancer region and leads to histone H3K27ac modification and eRNA transcriptional production (10, 13, 93, 97–100). The generated eRNAs assist the tight junctions of the mediator complexes (p300-BRD and pTEFb-MLL) using enhancer-promoter interactions to stabilize the E-P loop (13, 97, 99, 101, 102). Conversely, eRNAs coordinate cohesin-CTCF loop formation to promote chromosome cyclisation (13, 79, 103, 104). Therefore, eRNA regulates enhancers and downstream target gene transcription through the aforementioned methods, and the mRNA produced is translocated outside the nucleus and generates associated inflammatory factors (such as INF-β, IL-1β, TNF and CXCL8), which affect inflammatory immune cell response and related pathological responses.
In the following sections, we will summarize in detail the role of eRNAs in inflammatory immune cells, non-inflammatory immune cells, inflammatory immune diseases and tumors as well as their related mechanisms (Figure 2).
eRNA in Inflammatory Immune Cells
LPS, a major component of the outer wall of the gram-negative bacterial cell wall, activate mononuclear macrophages, lymphocyte, endothelial cells and epithelial cells through cellular signaling systems. Various cytokines and inflammatory mediators are synthesized and released in this process, which induces an inflammatory immune response (105–107). Nasun et al. (91) were the first to use toll-like receptor 4 signaling in macrophages as a model to clarify that SE-associated eRNA transcription is dynamically induced in most key genes driving innate immunity and inflammation using GRO-seq. Subsequently, CAGE data further verified that enhancer transcription preceded the target gene activation in monocytes under LPS stimulation (41, 90). Recently, Ma et al. (92) used simultaneous high-throughput ATAC and RNA expression with sequencing (SHARE-seq) to identify domains of regulatory chromatin (DORCs) that significantly overlap with SEs, which showed both chromatin accessibility and enhancer lineage-priming precede gene expression. This suggests that altered chromatin accessibility could be a pre-condition for cell lineage formation. Therefore, current studies have shown that both enhancer transcription and eRNA precede the transcriptional induction of inflammatory genes in the LPS response (90–92) and eRNA links enhancer activity to inflammatory gene expression through CBP-mediated H3K27 acetylation (80). Conversely, the plasticity of mature immune cells attracts us to explore the potential function of eRNAs in inflammatory immune cells (108, 109) (Table 1).
eRNA in Macrophages
Macrophages, the key cells that maintain the tissue’s internal environment and regulate the inflammatory immune response, perform tissue-specific functions and defend against infections (91, 120). Numerous studies have suggested the presence of PU.1 TFs in the active enhancer region of macrophages, which identify chromatin DNA recognition motifs and C/EBP, thereby creating nucleosome-free regions and undergoing histone tail H3K4me1/2 modifications (10, 13, 29, 54, 93, 95, 110–113, 121). Subsequently, similar signal-dependent TFs, p65 and NFκB, bind to the enhancer region under LPS stimulation, resulting in histone H3K27ac modifications and eRNA transcriptional generation (10, 93).
After activation of the RXR signaling pathway, the RXR-induced eRNAs were detected on Vegfa and Tgm2 enhancers, with studies showing that the eRNAs maintained macrophage angiogenic activity via enhancer interactions (104). In 2018, a study by Bence et al. revealed that macrophages under multiple IL-4 stimulations showed increased IL-4-sensitive (Arg1 and Hbegf) and RSG/IL-4-sensitive (Tgm2) eRNA expressions, leading to active STAT6 recruitment, which induced macrophage phenotypic changes by affecting the nuclear receptor PPAR (122). Meanwhile, Zsolt et al. (111) revealed that eRNA expression levels in IL4-STAT6-mediated responses correlated with the enrichment of RNAPII-Ser5p and RNAPII-Ser2p and levels of the inhibitory and activating factor STAT6 locus H3K27ac, which reduced macrophage responsiveness to LPS and suppressed inflammatory responses, including inflammatory vesicle activation, IL-1b production and pyroptosis. Further studies have clarified that Kdm6a, a demethylase, not only promotes macrophage IL-6 expression through promoter H3K27me3 demethylation but also interacts with MLL4 to increase IFN-β-specific eRNA S-IRE1, thereby promoting IFN-β transcription in macrophages (114). Ha et al. (115) suggested that the PU.1-mediated E2 eRNA (E2 is a potential regulatory element of approximately 10 kbp that is located upstream of the TSS) in macrophages is essential for IL-1β mRNA transcription, which might influence the macrophage-assisted regulation of disease states, such as endotoxic shock, sepsis and infection. Additionally, Oishi et al. (116) found that RevErb expression regulated by Bmal1 was repressed in Arntl-/- macrophages, with further studies revealing that RevErbs repressed eRNA transcription by recruiting the NCoR- histone deacetylase3 (HDAC) repressor complex and increasing enhancer H3K27 acetylation, thereby regulating enhancer epigenetic state to control macrophage inflammatory response (43, 116). Huang et al. (123) used inflammatory macrophage activation models to demonstrate that the inflammation activation-associated corepressor (GPS2 and SMRT)-eRNA-CCL2 regulatory axis, in addition to finding that LNA-targeted Ccl2 enhancer E-transcribed eRNA in white adipose tissue macrophages of obese (ob/ob) mice, can partially reverse meta inflammation and insulin resistance.
Therefore, macrophages activate intranuclear enhancer transcription and eRNA production in response to inflammatory stimuli, which regulates inflammatory factors and chemokine release and affects macrophage polarisation. However, the specific mechanisms of inflammatory genotypic and phenotypic changes by eRNA remain unclear and require further experimental exploration.
eRNA in Monocytes
Circulating monocytes, innate immune response cells, prevent infection by rapidly removing invading pathogens. Heward (117) and IIott (118) reported in the same year that monocytes are differentially expressed with large amounts of eRNAs in response to LPS stimulation. Heward James et al. (117) identified the expression of six eRNAs induced by human monocyte THP1 cells after the LPS activation of the intrinsic immune response, including MARCKS-eRNA, ACSL1-eRNA, AZIN1-eRNA, TNFSF8-eRNA. SLC30A4-eRNA and SOCS3-eRNA. Moreover, the intracellular signaling pathways, such as NFκB and mitogen-activated protein kinase (MAPK), were demonstrated to regulate extracellular kinase 1/2 and p38, which could promote inflammation-associated eRNA expression (117). IIott Nicholas et al. (118) identified 76 differentially expressed eRNAs in primary human monocytes stimulated by LPS and found that the knockdown of the pro-inflammatory TFs, NFκB-mediated IL1b eRNA and IL1b-RBT46 eRNA, attenuated LPS-induced mRNA transcription and pro-inflammatory mediator release, including IL1b and CXCL8. Using reduced representation bisulfite sequencing (RRBS) technology, smoking-associated differentially methylated regions (SM-DMR) was found to up-regulate AHRR mRNA by activating the AHRR enhancer that expressed AHRR eRNA (124). Additionally, smoking-altered methylation and intragenic AHRR enhancer-produced eRNA were found to be necessary prerequisites for monocyte type-specific AHRR transcription (124). Moreover in THP-1 monocytes, hHS-8 was shown to target dCas9-KRAB at the IRF1 binding site to impair IFN-gamma expression on LPS-induced TNF genes and eRNA, thereby affecting monocyte inflammatory immune action (125). Current studies suggest that eRNAs regulate the direction of monocyte function; however, their functional mechanism remains unknown.
eRNA in Lymphocyte
As important cellular components of inflammatory immune response, lymphocytes are the main performers of the lymphatic system immune function. They are mainly responsible for fighting external infections and monitoring cellular mutations in the body, and are categorized into B lymphocytes, T lymphocytes and natural killer cells.
eRNA in B Lymphocytes
Studies have reported that eRNA transcription during B lymphocyte growth and development is closely associated with large-scale changes in DNA cytosine modifications (126). Moreover, Brazao et al. (127) identified 73 PAX5-dependent eRNAs near protein-coding genes in B-ALL cells, such as LNCGme00432, LNCGme0034 and LNCGme00345, that were the downstream genes of B-cell lymphoma 11a (Bcl11a), whose dysfunction may lead to the malignant development of B cells. Furthermore, Saintamand et al. (128) reported that LPS-induced stimulation in vitro, whereas 3RR eRNA deletion reduced transcription and disrupted downstream CH basal transcription, which affected the resting and active states of B cells.
Many eRNA studies have been conducted in the classical B lymphocyte line (GM12878); however, only a few studies have explored the direct effects of enhancer transcription and eRNAs on B lymphocytes. Kim et al. (119) confirmed that enhancers dynamically modulate the transcriptional activities of eRNAs and pre-mRNAs in B-lymphoblasts. Furthermore, the knockdown of TNFSF10-related eRNAs leads to selective regulation in interferon-induced apoptosis, indicating that eRNAs are necessary for target gene induction and can be potential target genes via transcriptional reprogramming (119). Katla et al. (129) identified quantitative trait loci associated with eRNA expression and direction-dependent enrichment at enhancer regions in human lymphoblastoid cell lines using capped-nascent-RNA sequencing. These loci were correlated with gene expression, defined central TF binding regions and flanking eRNA initiation cores, which are important indicators of non-coding regulatory variants. Therefore, B lymphocytes have important functionality in enhancer transcription and eRNAs, which affect the growth, differentiation and malignant progression of B lymphocytes; however, further extensive studies are required.
eRNA in T Lymphocytes
The exploration of acute T lymphocytic leukaemia leads to unrevealing the relationship between eRNAs and T lymphocytes, confirming the presence of a large number of eRNAs in T-ALL cells (130, 131). Ets1 is a sequence-specific transcription factor that plays an important role during hematopoiesis, and is essential for the transition of CD4−/CD8− double negative (DN) to CD4+/CD8+ double positive (DP) thymocytes. During early T cell differentiation, eRNA shows a DN to DP transition pattern and the Ets1 pattern in DP transition, similar to the RNAPII pattern, suggesting that eRNAs act as active regulatory elements that regulate Ets1 on nucleosome occupancy and enhancer activity to influence T cell differentiation (132). Trimarch et al. (133) identified the first functional eRNA (LUNAR1) in T-ALL cells in 2014, which enhances IGF1R mRNA expression and maintains the IGF1 signaling pathway via Notch signaling, thereby maintaining T-Acute lymphoblastic leukaemia (ALL) malignant progression. Subsequently, Tan et al. (102) identified the second functional eRNA (XLOC_005968) in T-ALL cells, namely ARIEL, which recruits intermediary proteins to the ARID5B enhancer, promotes enhancer-promoter interaction and activates ARID5B expression, and thereby positively regulating TAL1-induced transcription and MYC oncogene expression, to accelerate T cells malignant progression.
Recently, some studies have explored the effect of eRNAs on the generation and differentiation of T cells. Hertweck et al. (96) revealed that eRNAs transcribed by T-bet SE were mostly Th1-specific in T cells. In Th1 cells, eRNAs of IFNG upstream SEs were transcribed, while the downstream enhancer exhibited lower levels of P-TEFb occupancy and eRNA production, which affects Th cell differentiation. Luke et al. (125) found that activated CD4+ T cell increased hHS-8, TNF and LTA promoter H3K27 acetylation and nuclease sensitivity while synergistically inducing TNF, LTA and hHS-8 eRNA transcription to regulate TNF mRNA and LTA mRNA, affecting T cell pathology.
eRNA in Natural Killer Cells
To date, no studies have explored the effects of eRNAs on natural killer (NK) cells generation, development and differentiation. Only one study has shown that the H3K27me3 histone demethylase UTX controls specific gene expression programs during development of natural killer T cells through demethylase activity-dependent manner (134). We believe that enhancer transcription events and eRNAs have a potentially important role in NK cells as potential therapeutic targets and prognostic biomarkers in inflammatory immune diseases and tumors.
Inflammatory Function of eRNA in Non-Inflammatory Immune Cells
The inflammatory immune functions of eRNAs are not limited to inflammatory immune cells. The recent studies showed that non-immune inflammatory cells have inflammatory changes and altered cellular states under eRNA regulation (135–138). Isidore et al. (136) found that VEGFA-eRNA5 and VEGFC-eRNA3 in endothelial cells affect angiogenesis and lymphangiogenesis by regulating endogenous transcription and VEGFA and VEGFC expression. Additionally, eRNAs were found to be well correlated with VEGF expression across cell types and in response to hypoxic stimuli using GRO-Seq. Zhou et al. (138), in the same year, revealed that lncRNA-MAP3K affects inflammatory factor (e.g. ICAM-1, E-selectin and MCP-1) expression, reduces monocyte-endothelial cell adhesion and decreases TNF-α, IL-1β and COX2 expression in macrophages through the p38 MAPK signaling pathway and MAP3K4 cis-modulation, which ultimately regulates vascular inflammation. Despite that the overlapping features of lncRNA-MAP3K with 1D-eRNA, strong evidence supporting lncRNA-MAP3K4 as an eRNA that is transcribed from the MAP3K4 enhancer region is lacking (138). Conversely, glucocorticoids have been shown to induce KLF9 expression in lung epithelial cells, with the identification of three common glucocorticoid receptor binding sites that influenced KLF9 mRNA and protein expression levels by generating eRNAs, which ultimately affected glucocorticoid-induced anti-inflammatory effects (135). Additionally, Yukako et al. (137) showed that Nrp2-eRNA, Dcstamp-eRNA and Nfatc1-eRNAs could regulate the corresponding promoters to control gene expression, thereby positively regulating osteoclast differentiation to maintain bone resorption function. These studies, therefore, indicate the potential inflammatory role of eRNAs in non-inflammatory immune cells. However, further studies are required to explore their specific mechanisms and functions.
eRNA in Inflammatory Immune Diseases
NFκB is an important gene regulator involved in innate and adaptive immune responses as well as survival and proliferation of certain cell types (139), and eRNAs have been shown to be involved in the regulation of inflammatory transcriptional networks (10, 118, 140). Studies have shown that NFκB contributes to the synthesis of inflammatory gene-associated enhanced eRNAs, which further enhances transcription by looping enhancers and promoters or by recruiting RNA polymerase II to the promoter, creating a transcription-mediated multilevel cascade regulating transcription (10, 13, 81, 91). Therefore, this paper further summarizes the potential functions and mechanisms of eRNAs in various inflammatory immune-related diseases (Table 2).
eRNA in Gastritis
The chronic inflammation and apoptosis resistance associated with Helicobacter pylori infection contributes to gastric disease development, including gastritis and gastric cancer (142). Chen et al. (97) were the first to report that H. pylori stimulated the recruitment of RelA and Brd4 to inflammatory gene-related enhancers and promoters. Following this, IL1A and IL1B eRNA expressions were up-regulated to affect NFκB-dependent inflammatory gene expressions (e.g. IL1); however, JQ1 was found to attenuate the H. pylori-induced eRNA and mRNA synthesis of NFκB-dependent inflammatory gene subpopulations by inhibiting Brd4-related functions, which suppresses inflammatory immune cell proliferation in H. pylori-infected mice (97). Subsequently, Brd4 was shown to de-activate cIAP2 expression by activating BIRC3 eRNA synthesis in H. pylori infection, which in turn inhibited caspase-3 activation, and ultimately inhibiting cell apoptosis. Therefore, the novel role of BIRC3 eRNA in H. pylori-mediated apoptosis resistance was speculated (99).
eRNA in Inflammatory Bowel Disease
Inflammatory bowel disease (IBD), a chronic inflammatory intestinal immune disease, has an unknown molecular pathology. Studies have reported significant differences in eRNA expression levels among multiple chemokine gene-related enhancer regions (including CXCL1-3, CXCL5-6 and CXCL8), which are all up-regulated in IBD, Crohn’s disease (CD), ulcerative colitis (UC) and controls using CAGE and qPCR (143). Baillie et al. (90) used the human monocyte-derived macrophage as a model to explore the genetic aetiology of IBD. They found that transient eRNA transcripts at multiple loci precede promoter-associated transcripts under LPS induction, which affects the adaptation of monocytes to the gastrointestinal mucosal environment, thus leading to IBD. Additionally, Aune et al. (141) revealed a significant association between eRNA genomic location and disease-specific genetic polymorphisms in IBD. It also suggested that the transcription site of the IFNG-R-49 eRNA was more than 100 kb away from the IL26 and IL22 genes, which are consistent with the biological functions exhibited by the eRNA.
eRNA in Rheumatoid Arthritis
Rheumatoid arthritis (RA) is a systemic autoimmune disease characterized by chronic synovial inflammation. Various studies have identified the basic leucine zipper transcription factor 2 (BACH2) protein to be a key transcription factor of Treg cells for immune homeostasis. It regulates the expression of various cytokines, including INF-γ and cytokine receptors, whose mutations are associated with RA development and progression (144). BACH2 proteins have been reported to negatively regulate eRNA expression; however, eRNA types and potential mechanisms remain unclear (144). Many disease-associated variants in non-coding regions act by affecting gene transcription and are known as eQTL (145). Studies found that eQTLs were involved in eRNA transcriptional regulation and produced cell type-specific effects, such as STAT6 eQTL up-regulation in patients with RA upregulated inflammatory cytokine production (145–147). Unfortunately, there are no studies that specifically identify which eRNAs influence the occurrence and progression of RA by which mechanisms.
eRNA in Other Inflammatory Immune Diseases
Many studies have shown that eRNAs plays a critical role in various inflammatory diseases. The close correlation between ADAMDEC1 and ADAM28 in systemic lupus erythematosus (SLE) regulates the disease inflammatory process. Shi et al. (98) found that monocyte ADAMDEC1 over-expression in patients with SLE was induced by the stimulation of pro-inflammatory cytokines, moreover, under LPS stimulation, the binding of the p300-NFκB complex to enhancer 2 generates eRNA157 that promotes p300 activation, leading to an increase in H3K27ac at the enhancer and promoter region, thus, affecting the regulation of ADAMDEC1 mRNA and SLE-related inflammatory gene expression. Hertweck et al. (96) found that in a mouse model of autoimmune uveitis, T-bet allows mediator and P-TEFb recruitment in the form of SE by extending the SE-generating Ifng eRNA. Therefore, Th1 expression is activated, which triggers IFN-gamma-mediated CD4+ T cells to promote immunoretinol-like binding proteins in the retina. Additionally, flavanols and JQ1 can inhibit SE and its products to down-regulate related gene expression (such as Ifng, Tnf, Fasl, II18r1 and Ctla4), and ultimately decrease the severity of the disease. Furthermore, Huang et al. (100) first found that lnc-SLC4A1-1 was retained in the nucleus as an eRNA and facilitated TF NF-κB binding to CXCL8 promoter region, leading to an increase in H3K27ac in the CXCL8 promoter and subsequently elevated CXCL8 expression. CXCL8 activation is exacerbated by the induction of TNF-α and IL-1β inflammatory response in trophoblast cells, resulting in unexplained recurrent pregnancy loss. It was also found that hypoxia-inducible enhanced RNA 1 (HERNA1) is produced by direct hypoxia-inducible factor 1α binding to the hypoxia response element of histone h3-lysine27. Synaptotagmin XVII, membrane transport proteins, Ca2+ sensing protein and SMG1 are also encoded by phosphatidylinositol 3-kinase-related kinase, thereby regulating immune disease progression, metabolism and contraction (148). Spurlock et al. (149) found that whole blood eRNA expression data effectively classified and differentiated patients with multiple sclerosis from those with other inflammatory and non-inflammatory neurological diseases.
Inflammatory Function of eRNA in Cancers
Cancer is currently a significant cause of death worldwide and it is a heterogeneous disease controlled by genetic and epigenetic alterations and transcriptional dysregulation (150). Numerous studies have been conducted to show that the abnormal expression levels of eRNAs, as an excellent marker of active enhancers and genes, are associated with dysregulation of enhancer transcription and gene expression in tumors (34, 151–153). Santanu Adhikary (33), Joo-Hyung Lee (34) and Zhao Zhang et al. (154) have each summarized in detail the potential functions, regulatory mechanisms and clinical therapeutic implications of eRNAs in cancer. Nevertheless, this study will focus on the potential functions and mechanisms of eRNAs in tumorigenesis development on its immune microenvironment and related inflammation (Table 3).
eRNA in Haematologic Malignancies
The relevant functions and mechanisms of eRNAs in inflammatory immune cells have been previously summarized, along with the exploration of the potential functions and mechanisms of eRNAs in immune hematopoietic system-related tumors. Almamun et al. (155) were the first to report that the aberrant methylation of enhancer regions was associated with the altered expression of neighboring genes involved in cell cycle processes, lymphocyte activation and apoptosis in pre-B ALL. Further studies have suggested an overall downregulation of eRNA transcripts in patients with pre-B ALL, which may affect the downregulation of target genes (such as ICOSLG, IRF4 and MSA1) in B-cell migration, proliferation and apoptosis (156). Further, Teppo et al. (157) used eRNA quantification to elucidate the aberrant transcriptional activity downstream of fusion TFs and demonstrated that the ETV6-RUNX1 axis regulates cell adhesion and transmembrane signaling pathways, which ultimately disrupts normal B lymphangiogenesis. Tan et al. (102) were the first to demonstrate the involvement of lncRNA in the regulation of TAL1-induced T-ALL oncogenic regulatory program. They also showed that XLOC_005968, the ARIEL eRNA, is oncogenic and positively regulated by ARID5B and MYC oncogene expression in T-ALL cells by recruiting mediator complexes and promoting ARID5B enhancer-promoter interactions (102). Additionally, Kaposi’s sarcoma-associated herpesvirus (KSHV), a human tumorigenic γ-2 herpesvirus, is the pathogen responsible for Kaposi’s sarcoma and primary effusion lymphoma. It is proposed that KSHV reactivation decreases MYC gene expression by downregulating MYC eRNA expression and enhancer activity, and shRNA-mediated and vIRF4-mediated cIRF4 suppression, which promotes lytic replication (158).
In leukaemia, a novel Hmrhl eRNA was shown to be highly upregulated in chronic granulocytic leukaemia (CML) cells and positively regulate its host gene phkb expression (159). Additionally, Fang et al. (160) found that eRNAs such as SEELA are widely activated in mixed-lineage leukaemia and demonstrated that SEELA directly binds to amino acid K31 of histone 4 and mediates the cis-activated transcription of the neighboring oncogene serine incorporate 2, which regulates oncogene transcription and tumour metabolism (sphingolipid synthesis) to influence leukaemia progression. The demethylation of vascular endothelial growth factor A (VEGFA) enhancer in CML promotes the overexpression of cancer signature genes. A study by Dahan showed that VEGFA+157 eRNA regulates its selective splicing, which affects CML cell proliferation by increasing RNAPII elongation via CCNT2 (161).
eRNA in Other Cancers
To date, eRNA inflammatory immune-related functions are only marginally studied in solid tumors. H. pylori infection is a major cause of gastric cancer, and its pathogenicity is associated with chronic inflammation induction and apoptosis resistance. The inflammatory immune role of eRNA in tumors was first reported by Chen et al. (99), demonstrating that H. pylori stimulate bromodomain-containing factor Brd4 recruitment to the BIRC3 enhancer, which promotes BIRC3 eRNA synthesis and cIAP2 expression, which inhibits caspase-3 activation and enhances apoptosis resistance in gastric epithelial cells. Additionally, some studies have suggested the presence of oncogenic SEs in colorectal cancer and confirmed their involvement in regulating oncogenic and immune pathways in colorectal cancer by modulating IL-20RA expression, affecting cell proliferation and immune evasion-related gene ecpression (162). Pancreatitis accelerates Kras mutation-driven tumorigenesis in mice, which is mostly found in pancreatic ductal adenocarcinoma (163). Li et al. (163) reported that under inflammatory stimulation, KrasG12D mutation targets a transient enhancer network driving proto-oncogene transcription and provides a sustained Kras-dependent oncogenic program to drive tumour tissue-specific progression. However, they did not explore the specific mechanism and types of enhancers and eRNAs.
The rapidly rising development of bioinformatic technologies provides novel means and directions to study the role of eRNA in inflammatory immunity in tumors. Various bioinformatics analyses have initially suggested that eRNA expression levels are significantly correlated with malignancy prognosis and can affect the tumour immune microenvironment (164–170). Furthermore, Xiao et al. (164) found that LINC02257 eRNA was significantly associated with cancer survival and immunotherapy-related indicators (e.g. tumour microenvironment, tumour mutational load and microsatellite instability). A study by Wang et al. (165) identified WAKMAR2 eRNA as a key candidate biomarker in invasive breast cancer, which may influence the tumour microenvironment by regulating the relevant immune-related genes, such as RAC2, IL27RA, IGLV1-51, IGHD, IGHA1 and FABP7. AC003092.1 eRNA and glioblastoma multiforme (GBM) overall survival were significantly correlated, and AC003092.1 eRNA was shown to be associated with the immunosuppressive microenvironment of GBM using single gene set enrichment analysis and CIBERSORTx system analysis (166). Using similar bioinformatic techniques, several studies have suggested that functional FOXO6-eRNA can regulate FOXO6 expression to influence EGFR and SOX2 expression and function in lung cancer progression (167); furthermore, the aberrant expression of LINC00987/A2M axis was closely associated with immune cell infiltration in lung adenocarcinoma (168). AC007255.1 in esophageal cancer was reported to be closely associated with tumour immune response and neutrophil activation (169). Additionally, AP001056.1 was shown to be enriched in the biological function analysis of head and neck squamous cell carcinoma, mainly in immune system processes (170). However, bioinformatic analyses can only tentatively suggest the potential role of relevant eRNAs in tumors. Therefore, further basic research and clinical trials are required to validate these results.
Clinical Perspectives of Enhancer Inhibitors Affecting eRNA Production
Many eRNAs are significantly differentially expressed in tumour tissues compared with paracancerous tissues (151, 154), which is consistent with the results of enhancer overactivation in cancer (171–174). Therefore, eRNAs can be considered as potential targets to overcome enhancer activation in cancer therapy. eRNAs have high specificity across tissues and tumours (41, 151, 154), with the antisense oligomer-based targeting of specific eRNAs effectively inhibiting target genes and tumour growth without theoretically affecting other unrelated tissues (49, 82, 102, 154, 175, 176). Therefore, eRNAs can be used as effective and highly precise therapeutic targets in cancer therapy. Regrettably, no relevant eRNA-targeted drugs are currently on the market or in clinical trials. In this section, we will therefore concentrate mainly on enhancer inhibitors affecting eRNA production to elucidate their potential therapeutic effects on inflammatory immune diseases and even cancer. The eRNA-related agents in cancer have been previously reviewed, including bromodomain and extra-terminal (BET) inhibitors, cyclin-dependent kinases (CDKs), HAT inhibitors and HDAC inhibitors (33, 34, 177). Since inhibitors that fully target eRNAs in inflammatory immune-related diseases have been rarely studied, the combined inhibitors of eRNAs and active enhancers, which produce eRNAs, have been summarized along with their therapeutic potential in inflammatory immune diseases in Figure 4.
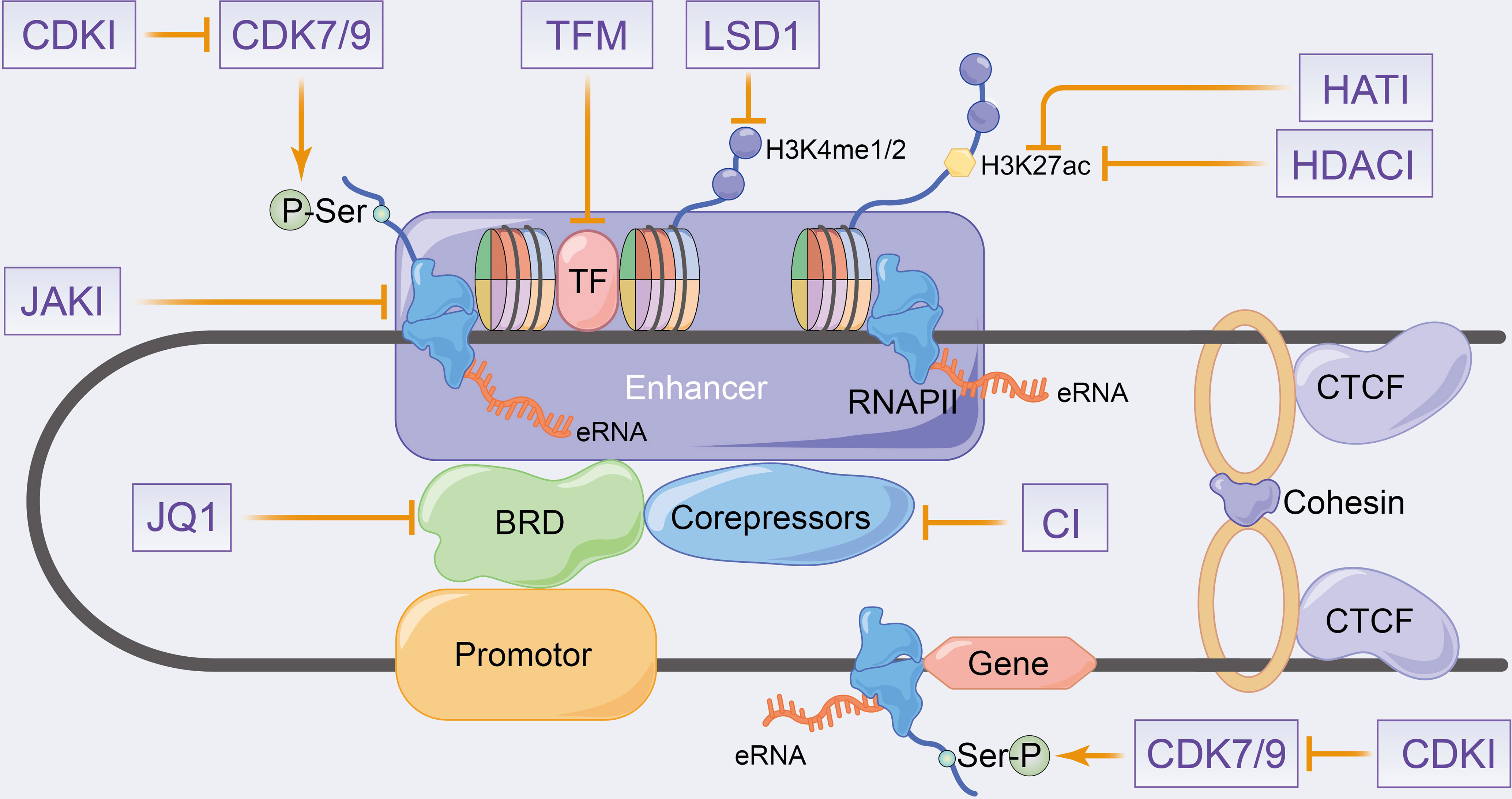
Figure 4 Schematic representation of putative therapeutic targets to enhancer transcription events and eRNA landscapes. CDKI, cyclin-dependent kinase inhibitors; HATI, histone acetyltransferase inhibitors; HDACI, histone deacetylase inhibitors; LSD1, lysine-specific demethylase 1; JAKI, janus kinase inhibitors; TFM, transcription factors inhibitors; CI, co-repressors inhibitors.
ET Inhibitors in eRNA
Estradiol cypionate (ET) inhibitors, such as JQ1 and I-BET762, can recognize acetylated histones by interfering with the BET family proteins BRD2, BRD3, BRD4 and BRDT (178). Thus, targeting this protein class can promote LPS-induced inflammatory gene expression in macrophages and significantly improve survival, as observed in an in vivo sepsis model (178, 179). JQ1 primarily targets BRD4 to inhibit TNF-α or IL-1β-induced inflammatory cytokines expression and reduces enhancer-mediated inflammatory responses and diseases (180–183) (Figure 4). H. pylori stimulates NFκB-dependent BRD4 to enhance inflammatory gene-related eRNA synthesis, similarly, JQ1 inhibits BRD4 to reduce eRNA synthesis and inhibits RNAPII recruitment that is induced by BRD4 interaction with RelA. Additionally, JQ1 has been reported to inhibit inflammatory gene expression and inflammatory immune cell proliferation in H. pylori-infected mice (97). Arnulf et al. (96) demonstrated that the treatment of Th1 cells with JQ1 and xanthinol resulted in the inhibition of P-TEFb, which produced a significant reduction in eRNA levels, including Ifng eRNA. This also promoted reduction in SE-related Th1 gene expression (e.g. Ifng, Tnf, Fasl, IL18 and Ctla4), which caused disease remission in autoimmune uveitis mice. Angela et al. (158) reported that the associated MYC eRNAs (including e486, e507 and e530) was significantly decreased after the JQ1 treatment of KSHV-infected primary effusion lymphomas, which inhibited MYC expression and KSHV cleavage gene expression induction. Additionally, a decrease in MYC mRNA was found on the knockdown of the corresponding eRNA (158). JQ1 was demonstrated to decrease the transcriptionally activated eRNAs of SEs, causing the down-regulation of IL-20RA expression and inhibition of growth, metastasis and immune escape in colorectal cancer (162). Therefore, JQ1 regulates enhancer transcription and eRNA synthesis by affecting the mediator complex, which regulates the downstream target genes to modulate the disease process.
CDK Inhibitors in eRNA
CDK7 is a component of the transcription initiation factor IIH (TFIIH) in the GTF complex that regulates enhancer and target gene transcription by phosphorylating Ser5 and Ser7 on RNAPII (184). Additionally, CDK7 activates and phosphorylates the P-TEFb catalytic subunit of CDK9, which phosphorylates Ser2 in the RNAPII CTD to control transcriptional elongation and termination (184) (Figure 4). Currently, transcriptional CDKs are considered potent targets for cancer therapy (185–189), and recently, some studies have also evaluated the role of CDK inhibitors in inflammatory-immune diseases (190–192). The therapeutic role of CDK7 in haematologic malignancies has been widely reported that demonstrate the deletion of oncogenic transcription factors, such as RUNX1, by small-molecule CDK inhibitors in acute T-lymphoblastic leukaemia (189). Moreover, SY-1365, a highly selective CDK7 inhibitor, is currently used in clinical trials in patients with ovarian and breast cancer (187). These studies have confirmed that CDK7 inhibitors reduce enhancer-associated oncogene expression by modulating eRNA expression levels. CDK inhibitors such as THZ1, NVP-2 and THZ531 that inhibit CDK7, CDK9, CDK12 and CDK13, respectively, have been shown to downregulate SE-related oncogene expression and promote DNA damage response gene loss in chordoma and acute T-lymphoblastic leukaemia cells, respectively (185, 186, 188). Furthermore, the blocking of CDK7 has been reported to regulate the onset and intensity of immune-inflammatory responses by activating the tumour immune response and regulating granulocyte apoptosis and cytokine secretion (193, 194). Recently, transcriptional CDKs have been speculated to play an important role in pro-inflammatory gene expression (190), with Wei et al. to be the first to demonstrate in the cytokine release syndrome (CRS) that the CDK7 covalent inhibitor THZ1 downregulates inflammatory gene transcription in macrophages after preferential target inhibition associated with SEs, such as STAT1 and IL1, decreases cytokine release, alleviates the hyperinflammatory state and rescues lethal CRS mice (191).
Other Inhibitors in eRNA
Furthermore, enhancer transcription analyses and eRNA generation processes revealed that TF modulators, HAT inhibitors and HDAC inhibitors influence eRNA and mRNA production by modulating enhancer epigenetic characteristics (177) (Figure 4). HATs, such as p300-CBP, enable histone tail acetylation modifications (195), whereas polycomb repressive complex (PRC) mediates histone methylation. The drugs targeting PRC inhibit leukaemia-associated enhancer transcription, control pro-apoptotic B cell lymphoma-2 like 11 and mediate apoptosis in breast cancer cells (196, 197). HDACs mediate histone deacetylation, with numerous studies demonstrating the effect of HDAC inhibitors on enhancer landscape in various cancer types (197–199). Lysine-specific demethylase 1 (LSD1) was identified as a selective mediator of H3K4 demethylation, with LSD1 inhibitors affecting the progression of acute myeloid leukaemia by disrupting the enhancer with the SNAG structural domain transcriptional repressor GFI1 (200). Additionally, LSD1 inhibitors have been shown to affect enhancer activity in various tumors, such as androgen receptor function in prostate cancer (201, 202) and ERα activity in breast cancer (203). However, relevant studies exploring the potential therapeutic efficacy of HAT inhibitors and HDAC inhibitors in inflammatory immunity as along with their mechanisms and function on eRNA are lacking.
Notably, a study by Huang et al. demonstrated that corepressor recruitment (GPS2 and SMRT) is a genome-wide signature of inflammatory immune enhancers, which antagonizes eRNA transcription and CBP-mediated H3K27 acetylation. This reverses subinflammation and insulin resistance by providing targeted eRNA therapeutics for immunometabolic diseases (123). A Janus kinase inhibitor (tofacitinib) has been shown to block cytokine signaling in T cells, thereby affecting RA risk gene expressions and SE structure, which ultimately targets autoimmune diseases (144). Therefore, targeted small molecule drugs that affect enhancer transcription and eRNAs can be considered as potential therapeutic targets for inflammatory immune-related diseases and tumors. Although enhancer inhibitors have great potential in diseases with inflammation immune alterations, further clinical trials are needed to validate. In addition, small molecule inhibitors specifically targeting eRNA still have great potential for exploration and development.
Discussion
Rapid advances in sequencing and microscope technologies suggest the potential contribution of enhancer transcription and eRNAs in inflammatory immune-altered diseases. eRNAs have been considered to have induced a breakthrough in the field of targeted therapy, spurring various studies centered on transcriptional precision and complexity. The activation modes of enhancers and SEs and regulatory mechanisms of enhancer transcription and eRNAs on target genes have been extensively analysed in this review. However, owing to the limited understanding of enhancer transcription and eRNA biology, multiple questions remain to be addressed: 1) What are the basic features of enhancer-promoter communication, interdependence and base coexistence sequence? 2) How does enhancer transcription regulate eRNA and mRNA expression? 3) How does eRNA activate paired-promoter gene transcription? 4) How do specific structures (e.g. molecular clouds and condensates) formed by eRNA regulate the enhancer and promoter gene and affect related diseases? 5) How does the eRNA epigenetic modification affect the enhancer and promoter gene transcription?
Although studies on enhancer transcription and eRNAs are limited to the fields of cancer development and differentiation, recent studies demonstrate that both enhancer transcription activation and eRNA expression are preferentially expressed over inflammatory immune-related genes under LPS induction. This suggests a potential regulatory mechanism between eRNAs and inflammatory immune genes, which alters inflammatory immune responses in diseases. This review summarizes that eRNA expression levels in inflammatory and non-inflammatory immune cells are significantly correlated with inflammatory gene expression in response to inflammatory stimuli, leading to a rapid transition from a quiescent to an inflammatory transcriptional program, thus affecting the development and differentiation of associated immune and non-immune cells. Notably, the inflammation-associated NFκB signaling pathway contributes to inflammation-associated eRNA synthesis and positively regulates enhancer transcription to form a multi-cascade regulatory transcription that affects various inflammatory immune diseases, such as gastritis, SLE and inflammatory bowel disease. Currently, the inflammatory immunomodulatory role of eRNAs in tumors is limited to hematopoietic malignancies, while studies in substantive tumors are scarce. Consequently, the enhancer transcription processes and eRNAs have been speculated to not only affect the development and differentiation of inflammatory immune cells (monocyte-macrophage and lymphocytes predominantly) but also lead to the alteration in inflammatory-immune responses in various diseases, including tumors. However, the specific mechanisms or signaling pathways by which eRNAs affect cells and diseases remain unclear. However, bioinformatic analyses have made it possible to identify and screen functional eRNAs associated with inflammatory immunity and utilize them as a basis for extensive functional, mechanistic and therapeutic exploration.
Significant advances have been made in the treatment of inflammatory immune diseases and tumors using small-molecule drugs targeting enhancer transcription processes, which provide novel therapeutic directions and tools for diseases with inflammatory immune alterations and their drug resistance. However, further studies are required as the regulation of downstream target genes by eRNAs and the instability and dynamics of eRNA have been scarcely explored. Currently, no relevant studies have explored briefly the efficacy of small-molecule drugs that directly target eRNA on lesioned cells or diseases. However, some RNA-based inhibitors such as locked nucleic acid antisense oligonucleotides (LNA ASOs) have been to identified to have significant efficacy in targeting lncRNAs for silencing, which predicts that LNA ASO may be a potential targeting inhibitor in the eRNA field. Nevertheless, eRNA-associated protein chaperones can be identified using genomics and proteomics, and structure analyses can help in designing small-molecule regulators that specifically target eRNA protein chaperones. Additionally, further studies considering altered eRNA epigenetic modifications (e.g. 5-cytosine methylation, n6-adenosine methylation) and eRNA interactions in higher-order chromatin organization as potential eRNA targets are required. Therefore, this review aims to highlight the usefulness of eRNA as an effective potential therapeutic target and prognostic biomarker for inflammatory immune diseases and tumors with inflammatory immune alterations. However, further studies are required to confirm the functions and regulatory mechanisms of eRNAs in inflammatory-immune alterations and explore the potential therapeutic effects of relevant eRNA small molecule inhibitors in diseases with inflammatory-immune alterations.
Author Contributions
Overview concept: LW and BX. Literature data collection: LW and YH. Literature analysis: LW, YH, MC, and BX. Funding acquisition: YM, MC, and BX. Software and Visualization: LW. Writing-original draft: LW. Writing-review and editing: LW, WL, YH, MC, and BX.
Funding
National Natural Science Foundation of China (No. 81872089, 81370849, 81672551); Six talent peaks project in Jiangsu Province; Jiangsu Provincial Medical Innovation Team (CXTDA2017025); The National Key Research and Development Program of China (SQ2017YFSF090096); Innovative Team of Jiangsu Provincial (2017ZXKJQWO7).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Heintzman ND, Ren B. Finding Distal Regulatory Elements in the Human Genome. Curr Opin Genet Dev (2009) 19, 541–9. doi: 10.1016/j.gde.2009.09.006
2. Tippens ND, Vihervaara A, Lis JT. Enhancer Transcription: What, Where, When, and Why? Genes Dev (2018) 32:1–3. doi: 10.1101/gad.311605.118
3. Li W, Notani D, Rosenfeld MG. Enhancers as Non-Coding RNA Transcription Units: Recent Insights and Future Perspectives. Nat Rev Genet (2016) 17:207–23. doi: 10.1038/nrg.2016.4
4. Banerji J, Rusconi S, Schaffner W. Expression of a Beta-Globin Gene Is Enhanced by Remote SV40 DNA Sequences. Cell (1981) 27:299–308. doi: 10.1016/0092-8674(81)90413-x
5. Zaret KS, Carroll JS. Pioneer Transcription Factors: Establishing Competence for Gene Expression. Genes Dev (2011) 25:2227–41. doi: 10.1101/gad.176826.111
6. Spitz F, Furlong EE. Transcription Factors: From Enhancer Binding to Developmental Control. Nat Rev Genet (2012) 13:613–26. doi: 10.1038/nrg3207
7. Junion G, Spivakov M, Girardot C, Braun M, Gustafson E, Birney EH, et al. A Transcription Factor Collective Defines Cardiac Cell Fate and Reflects Lineage History. Cell (2012) 148:473–86. doi: 10.1016/j.cell.2012.01.030
8. Liu Z, Merkurjev D, Yang F, Li W, Oh S, Friedman M, et al. Enhancer Activation Requires Trans-Recruitment of a Mega Transcription Factor Complex. Cell (2014) 159:358–73. doi: 10.1016/j.cell.2014.08.027
9. Rothschild G, Basu U. Lingering Questions About Enhancer RNA and Enhancer Transcription-Coupled Genomic Instability. Trends Genet (2017) 33:143–54. doi: 10.1016/j.tig.2016.12.002
10. Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the Enhancer Landscape During Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell (2013) 51:310–25. doi: 10.1016/j.molcel.2013.07.010
11. Kanno T, Kanno Y, LeRoy G, Campos E, Sun H, Brooks SR, et al. BRD4 Assists Elongation of Both Coding and Enhancer RNAs by Interacting With Acetylated Histones. Nat Struct Mol Biol (2014) 21:1047–57. doi: 10.1038/nsmb.2912
12. Sigova AA, Abraham JB, Ji X, Molinie B, Hannett NM, Guo YE, et al. Transcription Factor Trapping by RNA in Gene Regulatory Elements. Science (2015) 350:978–81. doi: 10.1126/science.aad3346
13. Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and Regulated Transcriptional Programs. Trends Biochem Sci (2014) 39:170–82. doi: 10.1016/j.tibs.2014.02.007
14. Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung H, et al. Genome-Wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell (2013) 153:692–706. doi: 10.1016/j.cell.2013.04.002
15. Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, et al. Genome-Wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell (2013) 153:678–91. doi: 10.1016/j.cell.2013.04.001
16. Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal Interactions and Olfactory Receptor Choice. Cell (2006) 126:403–13. doi: 10.1016/j.cell.2006.06.035
17. Geyer PK, Green MM, Corces VG. Tissue-Specific Transcriptional Enhancers may Act in Trans on the Gene Located in the Homologous Chromosome: The Molecular Basis of Transvection in Drosophila. EMBO J (1990) 9:2247–56. doi: 10.1002/j.1460-2075.1990.tb07395.x
18. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell (2013) 153:307–19. doi: 10.1016/j.cell.2013.03.035
19. Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell (2013) 153:320–34. doi: 10.1016/j.cell.2013.03.036
20. Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin Stretch Enhancer States Drive Cell-Specific Gene Regulation and Harbor Human Disease Risk Variants. Proc Natl Acad Sci USA (2013) 110:17921–6. doi: 10.1073/pnas.1317023110
21. Higashijima Y, Kanki Y. Potential Roles of Super Enhancers in Inflammatory Gene Transcription. FEBS J (2021). doi: 10.1111/febs.16089
22. Siersbaek R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, et al. Transcription Factor Cooperativity in Early Adipogenic Hotspots and Super-Enhancers. Cell Rep (2014) 7:1443–55. doi: 10.1016/j.celrep.2014.04.042
23. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell (2013) 155:934–47. doi: 10.1016/j.cell.2013.09.053
25. Laham-Karam N, Laitinen P, Turunen TA, Yla-Herttuala S. Activating the Chromatin by Noncoding RNAs. Antioxid Redox Sign (2018) 29:813–31. doi: 10.1089/ars.2017.7248
26. Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating Gene Expression in Mammalian Cells With Promoter-Targeted Duplex RNAs. Nat Chem Biol (2007) 3:166–73. doi: 10.1038/nchembio860
27. Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
28. Sas-Chen A, Srivastava S, Yarden Y. The Short and the Long: Non-Coding RNAs and Growth Factors in Cancer Progression. Biochem Soc Trans (2017) 45:51–64. doi: 10.1042/BST20160131
29. De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A Large Fraction of Extragenic RNA Pol II Transcription Sites Overlap Enhancers. PloS Biol (2010) 8:e1000384. doi: 10.1371/journal.pbio.1000384
30. Orom UA, Derrien T, Beringer M, Gumireddy K, Gumireddy A, Bussotti G, et al. Long Noncoding RNAs With Enhancer-Like Function in Human Cells. Cell (2010) 143:46–58. doi: 10.1016/j.cell.2010.09.001
31. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs Associate With Mediator to Enhance Chromatin Architecture and Transcription. Nature (2013) 494:497–501. doi: 10.1038/nature11884
32. Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread Transcription at Neuronal Activity-Regulated Enhancers. Nature (2010) 465:182–7. doi: 10.1038/nature09033
33. Adhikary S, Roy S, Chacon J, Gadad SS, Das C. Implications of Enhancer Transcription and eRNAs in Cancer. Cancer Res (2021) 81:4174–82. doi: 10.1158/0008-5472.CAN-20-4010
34. Lee JH, Xiong F, Li W. Enhancer RNAs in Cancer: Regulation, Mechanisms and Therapeutic Potential. RNA Biol (2020) 17:1550–9. doi: 10.1080/15476286.2020.1712895
35. Thandapani P. Super-Enhancers in Cancer. Pharmacol Ther (2019) 199:129–38. doi: 10.1016/j.pharmthera.2019.02.014
36. Sengupta S, George RE. Super-Enhancer-Driven Transcriptional Dependencies in Cancer. Trends Cancer (2017) 3:269–81. doi: 10.1016/j.trecan.2017.03.006
37. Higashijima Y, Matsui Y, Shimamura T, Nakaki R, Nagai N, Tsutsumi S, et al. Coordinated Demethylation of H3K9 and H3K27 Is Required for Rapid Inflammatory Responses of Endothelial Cells. EMBO J (2020) 39:e103949. doi: 10.15252/embj.2019103949
38. Fanucchi S, Fok ET, Dalla E, Shibayama Y, Börner K, Chang EY, et al. Immune Genes Are Primed for Robust Transcription by Proximal Long Noncoding RNAs Located in Nuclear Compartments. Nat Genet (2019) 51:138–50. doi: 10.1038/s41588-018-0298-2
39. Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal Contact Permits Transcription Between Coregulated Genes. Cell (2013) 155:606–20. doi: 10.1016/j.cell.2013.09.051
40. Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, et al. Transcription Initiation Platforms and GTF Recruitment at Tissue-Specific Enhancers and Promoters. Nat Struct Mol Biol (2011) 18:956–63. doi: 10.1038/nsmb.2085
41. Andersson R, Gebhard C, Escalada IM, Hoof I, Bornholdt J, Boyd M, et al. An Atlas of Active Enhancers Across Human Cell Types and Tissues. Nature (2014) 507:455–61. doi: 10.1038/nature12787
42. Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT, et al. Analysis of Nascent RNA Identifies a Unified Architecture of Initiation Regions at Mammalian Promoters and Enhancers. Nat Genet (2014) 46:1311–20. doi: 10.1038/ng.3142
43. Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Heinz YT, et al. Rev-Erbs Repress Macrophage Gene Expression by Inhibiting Enhancer-Directed Transcription. Nature (2013) 498:511–5. doi: 10.1038/nature12209
44. Andersson R, Andersen PR, Valen E, Core LJ, Bornholdt J, Boyd M, et al. Nuclear Stability and Transcriptional Directionality Separate Functionally Distinct RNA Species. Nat Commun (2014) 5:5336. doi: 10.1038/ncomms6336
45. Ntini E, Järvelin AI, Bornholdt J, Chen Y, Boyd M, Jørgensen M, et al. Polyadenylation Site-Induced Decay of Upstream Transcripts Enforces Promoter Directionality. Nat Struct Mol Biol (2013) 20:923–8. doi: 10.1038/nsmb.2640
46. Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter Directionality Is Controlled by U1 snRNP and Polyadenylation Signals. Nature (2013) 499:360–3. doi: 10.1038/nature12349
47. Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, et al. Cotranscriptional Set2 Methylation of Histone H3 Lysine 36 Recruits a Repressive Rpd3 Complex. Cell (2005) 123:593–605. doi: 10.1016/j.cell.2005.10.025
48. Bonn S, Zinzen RP, Girardot C, Gustafson EH, Gonzalez AP, Delhomme N, et al. Tissue-Specific Analysis of Chromatin State Identifies Temporal Signatures of Enhancer Activity During Embryonic Development. Nat Genet (2012) 44:148–56. doi: 10.1038/ng.1064
49. Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, et al. Enhancer RNAs Participate in Androgen Receptor-Driven Looping That Selectively Enhances Gene Activation. Proc Natl Acad Sci USA (2014) 111:7319–24. doi: 10.1073/pnas.1324151111
50. Lai F, Gardini A, Zhang A, Shiekhattar R. Integrator Mediates the Biogenesis of Enhancer RNAs. Nature (2015) 525:399–403. doi: 10.1038/nature14906
51. Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R, et al. Integrator, a Multiprotein Mediator of Small Nuclear RNA Processing, Associates With the C-Terminal Repeat of RNA Polymerase II. Cell (2005) 123:265–76. doi: 10.1016/j.cell.2005.08.019
52. Austenaa LM, Barozzi I, Simonatto M, Masella S, Chiara GD, Ghisletti S, et al. Transcription of Mammalian Cis-Regulatory Elements Is Restrained by Actively Enforced Early Termination. Mol Cell (2015) 60:460–74. doi: 10.1016/j.molcel.2015.09.018
53. Hsin JP, Li W, Hoque M, Tian B, Manley JL. RNAP II CTD Tyrosine 1 Performs Diverse Functions in Vertebrate Cells. Elife (2014) 3:e02112. doi: 10.7554/eLife.02112
54. Descostes N, Heidemann M, Spinelli L, Schüller R, Maqbool MA, Fenouil R, et al. Tyrosine Phosphorylation of RNA Polymerase II CTD Is Associated With Antisense Promoter Transcription and Active Enhancers in Mammalian Cells. Elife (2014) 3:e02105. doi: 10.7554/eLife.02105
55. Bentley DL. Coupling mRNA Processing With Transcription in Time and Space. Nat Rev Genet (2014) 15:163–75. doi: 10.1038/nrg3662
56. Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, et al. CTD Tyrosine Phosphorylation Impairs Termination Factor Recruitment to RNA Polymerase II. Science (2012) 336:1723–5. doi: 10.1126/science.1219651
57. Lubas M, Andersen PR, Schein A, Dziembowski A, Kudla G, Jensen TH, et al. The Human Nuclear Exosome Targeting Complex Is Loaded Onto Newly Synthesized RNA to Direct Early Ribonucleolysis. Cell Rep (2015) 10:178–92. doi: 10.1016/j.celrep.2014.12.026
58. Muller-McNicoll M, Neugebauer KM. Good Cap/Bad Cap: How the Cap-Binding Complex Determines RNA Fate. Nat Struct Mol Biol (2014) 21:9–12. doi: 10.1038/nsmb.2751
59. Blackwood EM, Kadonaga JT. Going the Distance: A Current View of Enhancer Action. Science (1998) 281:60–3. doi: 10.1126/science.281.5373.60
60. Bulger M, Groudine M. Looping Versus Linking: Toward a Model for Long-Distance Gene Activation. Genes Dev (1999) 13:2465–77. doi: 10.1101/gad.13.19.2465
61. de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, et al. Three-Dimensional Organization of Gene Expression in Erythroid Cells. Curr Top Dev Biol (2008) 82:117–39. doi: 10.1016/S0070-2153(07)00005-1
62. Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-Range Chromatin Regulatory Interactions In Vivo. Nat Genet (2002) 32:623–6. doi: 10.1038/ng1051
63. Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction Between Hypersensitive Sites in the Active Beta-Globin Locus. Mol Cell (2002) 10:1453–65. doi: 10.1016/s1097-2765(02)00781-5
64. Kooren J, Palstra RJ, Klous P, Splinter E, Lindern MV, Grosveld F, et al. Beta-Globin Active Chromatin Hub Formation in Differentiating Erythroid Cells and in P45 NF-E2 Knock-Out Mice. J Biol Chem (2007) 282:16544–52. doi: 10.1074/jbc.M701159200
65. Kim SI, Bultman SJ, Kiefer CM, Dean A, Bresnick EH. BRG1 Requirement for Long-Range Interaction of a Locus Control Region With a Downstream Promoter. Proc Natl Acad Sci USA (2009) 106:2259–64. doi: 10.1073/pnas.0806420106
66. Kellner WA, Ramos E, Van Bortle K, Takenaka N, Corces VG. Genome-Wide Phosphoacetylation of Histone H3 at Drosophila Enhancers and Promoters. Genome Res (2012) 22:1081–8. doi: 10.1101/gr.136929.111
67. Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, et al. Proximity Among Distant Regulatory Elements at the Beta-Globin Locus Requires GATA-1 and FOG-1. Mol Cell (2005) 17:453–62. doi: 10.1016/j.molcel.2004.12.028
68. Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, et al. The Active Spatial Organization of the Beta-Globin Locus Requires the Transcription Factor EKLF. Genes Dev (2004) 18:2485–90. doi: 10.1101/gad.317004
69. Yan J, Chen SA, Local A, Liu T, Qiu Y, Dorighi KM, et al. Histone H3 Lysine 4 Monomethylation Modulates Long-Range Chromatin Interactions at Enhancers. Cell Res (2018) 28:204–20. doi: 10.1038/cr.2018.1
70. Catarino RR, Stark A. Assessing Sufficiency and Necessity of Enhancer Activities for Gene Expression and the Mechanisms of Transcription Activation. Genes Dev (2018) 32:202–23. doi: 10.1101/gad.310367.117
71. Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, et al. Cohesin Mediates Transcriptional Insulation by CCCTC-Binding Factor. Nature (2008) 451:796–801. doi: 10.1038/nature06634
72. Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins Functionally Associate With CTCF on Mammalian Chromosome Arms. Cell (2008) 132:422–33. doi: 10.1016/j.cell.2008.01.011
73. Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, et al. CTCF Mediates Long-Range Chromatin Looping and Local Histone Modification in the Beta-Globin Locus. Genes Dev (2006) 20:2349–54. doi: 10.1101/gad.399506
74. Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, et al. CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells. Nat Genet (2011) 43:630–8. doi: 10.1038/ng.857
75. Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and Inter-Chromosomal Interactions Correlate With CTCF Binding Genome Wide. Mol Syst Biol (2010) 6:426. doi: 10.1038/msb.2010.79
76. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature (2012) 485:376–80. doi: 10.1038/nature11082
77. Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, et al. Architectural Roles of Multiple Chromatin Insulators at the Human Apolipoprotein Gene Cluster. EMBO J (2009) 28:1234–45. doi: 10.1038/emboj.2009.81
78. Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, et al. Cohesin Regulates Tissue-Specific Expression by Stabilizing Highly Occupied Cis-Regulatory Modules. Genome Res (2012) 22:2163–75. doi: 10.1101/gr.136507.111
79. Racko D, Benedetti F, Dorier J, Stasiak A. Transcription-Induced Supercoiling as the Driving Force of Chromatin Loop Extrusion During Formation of TADs in Interphase Chromosomes. Nucleic Acids Res (2018) 46:1648–60. doi: 10.1093/nar/gkx1123
80. Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell (2017) 168:135–49.e122. doi: 10.1016/j.cell.2016.12.020
81. Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, et al. Functional Roles of Enhancer RNAs for Oestrogen-Dependent Transcriptional Activation. Nature (2013) 498:516–20. doi: 10.1038/nature12210
82. Jiao W, Chen Y, Song H, Li D, Mei H, Yang F, et al. HPSE Enhancer RNA Promotes Cancer Progression Through Driving Chromatin Looping and Regulating hnRNPU/P300/EGR1/HPSE Axis. Oncogene (2018) 37:2728–45. doi: 10.1038/s41388-018-0128-0
83. Pezone A, Zuchegna C, Tramontano A, Romano A, Russo G, Rosa M, et al. RNA Stabilizes Transcription-Dependent Chromatin Loops Induced By Nuclear Hormones. Sci Rep (2019) 9:3925. doi: 10.1038/s41598-019-40123-6
84. Yang Y, Su Z, Song X, Liang B, Zeng F, Chang X, et al. Enhancer RNA-Driven Looping Enhances the Transcription of the Long Noncoding RNA DHRS4-AS1, a Controller of the DHRS4 Gene Cluster. Sci Rep (2016) 6:20961. doi: 10.1038/srep20961
85. Tsai PF, Orso SD, Rodriguez J, Vivanco KO, Ko KD, Jiang K, et al. A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In Trans. Mol Cell (2018) 71:129–41.e128. doi: 10.1016/j.molcel.2018.06.008
86. Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK, et al. Enhancer RNA Facilitates NELF Release From Immediate Early Genes. Mol Cell (2014) 56:29–42. doi: 10.1016/j.molcel.2014.08.023
87. Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, et al. RIC-Seq for Global In Situ Profiling of RNA-RNA Spatial Interactions. Nature (2020) 582:432–7. doi: 10.1038/s41586-020-2249-1
88. Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, et al. Enhancer RNA M6a Methylation Facilitates Transcriptional Condensate Formation and Gene Activation. Mol Cell (2021) 81:3368–85.e3369. doi: 10.1016/j.molcel.2021.07.024
89. Aguilo F, Li S, Balasubramaniyan N, Sancho A, Benko S, Zhang F, et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1alpha. Cell Rep (2016) 14:479–92. doi: 10.1016/j.celrep.2015.12.043
90. Baillie JK, Arner E, Daub C, Hoon MD, Itoh M, Kawaji H, et al. Analysis of the Human Monocyte-Derived Macrophage Transcriptome and Response to Lipopolysaccharide Provides New Insights Into Genetic Aetiology of Inflammatory Bowel Disease. PloS Genet (2017) 13:e1006641. doi: 10.1371/journal.pgen.1006641
91. Hah N, Benner C, Chong LW, Yu RT, Downes M, Evans RM, et al. Inflammation-Sensitive Super Enhancers Form Domains of Coordinately Regulated Enhancer RNAs. Proc Natl Acad Sci USA (2015) 112:E297–302. doi: 10.1073/pnas.1424028112
92. Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, et al. Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell (2020) 183:1103–16.e1120. doi: 10.1016/j.cell.2020.09.056
93. Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, et al. Effect of Natural Genetic Variation on Enhancer Selection and Function. Nature (2013) 503:487–92. doi: 10.1038/nature12615
94. Ghisletti S, Barozzi I, Mietton F, Polletti S, Santa FD, Venturini E, et al. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity (2010) 32:317–28. doi: 10.1016/j.immuni.2010.02.008
95. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004
96. Hertweck A, Evans CM, Eskandarpour M, Lau JCH, Oleinika K, Jackson I, et al. T-Bet Activates Th1 Genes Through Mediator and the Super Elongation Complex. Cell Rep (2016) 15:2756–70. doi: 10.1016/j.celrep.2016.05.054
97. Chen J, Wang Z, Hu X, Chen R, Romero-Gallo J, Peek RM Jr, et al. BET Inhibition Attenuates Helicobacter Pylori-Induced Inflammatory Response by Suppressing Inflammatory Gene Transcription and Enhancer Activation. J Immunol (2016) 196:4132–42. doi: 10.4049/jimmunol.1502261
98. Shi L, Li S, Maurer K, Zhang Z, Petri M, Sullivan K. Enhancer RNA and NFkappaB-Dependent P300 Regulation of ADAMDEC1. Mol Immunol (2018) 103:312–21. doi: 10.1016/j.molimm.2018.09.019
99. Chen Y, Sheppard D, Dong X, Hu X, Chen M, Chen R, et al. H. Pylori Infection Confers Resistance to Apoptosis via Brd4-Dependent BIRC3 eRNA Synthesis. Cell Death Dis (2020) 11:667. doi: 10.1038/s41419-020-02894-z
100. Huang Z, Du G, Huang X, Han L, Han X, Xu B, et al. The Enhancer RNA lnc-SLC4A1-1 Epigenetically Regulates Unexplained Recurrent Pregnancy Loss (URPL) by Activating CXCL8 and Nfκb Pathway. EBioMedicine (2018) 38:162–70. doi: 10.1016/j.ebiom.2018.11.015
101. van Arensbergen J, van Steensel B, Bussemaker HJ. In Search of the Determinants of Enhancer-Promoter Interaction Specificity. Trends Cell Biol (2014) 24:695–702. doi: 10.1016/j.tcb.2014.07.004
102. Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MB, et al. The Enhancer RNA ARIEL Activates the Oncogenic Transcriptional Program in T-Cell Acute Lymphoblastic Leukemia. Blood (2019) 134:239–51. doi: 10.1182/blood.2018874503
103. Zhang X, Pang P, Jiang M, Cao Q, Li H, Xu Y, et al. eRNAs and Superenhancer lncRNAs Are Functional in Human Prostate Cancer. Dis Markers (2020) 2020:8847986. doi: 10.1155/2020/8847986
104. Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S, et al. The Active Enhancer Network Operated by Liganded RXR Supports Angiogenic Activity in Macrophages. Genes Dev (2014) 28:1562–77. doi: 10.1101/gad.242685.114
105. Bierhaus A, Chen J, Liliensiek B, Nawroth PP. LPS and Cytokine-Activated Endothelium. Semin Thromb Hemost (2000) 26:571–87. doi: 10.1055/s-2000-13214
106. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front Immunol (2019) 10:1084. doi: 10.3389/fimmu.2019.01084
107. Rathinam VAK, Zhao Y, Shao F. Innate Immunity to Intracellular LPS. Nat Immunol (2019) 20:527–33. doi: 10.1038/s41590-019-0368-3
108. Cobaleda C, Busslinger M. Developmental Plasticity of Lymphocytes. Curr Opin Immunol (2008) 20:139–48. doi: 10.1016/j.coi.2008.03.017
109. Kondilis-Mangum HD, Wade PA. Epigenetics and the Adaptive Immune Response. Mol Aspects Med (2013) 34:813–25. doi: 10.1016/j.mam.2012.06.008
110. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac Separates Active From Poised Enhancers and Predicts Developmental State. Proc Natl Acad Sci USA (2010) 107:21931–6. doi: 10.1073/pnas.1016071107
111. Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, et al. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity (2018) 48:75–90.e76. doi: 10.1016/j.immuni.2017.12.010
112. Hsu E, Zemke NR, Berk AJ. Promoter-Specific Changes in Initiation, Elongation, and Homeostasis of Histone H3 Acetylation During CBP/p300 Inhibition. Elife (2021) 10:e63512. doi: 10.7554/eLife.63512
113. Narita T, Ito S, Higashijima Y, Chu WK, Neumann K, Walter J, et al. Enhancers Are Activated by P300/CBP Activity-Dependent PIC Assembly, RNAPII Recruitment, and Pause Release. Mol Cell (2021) 81:2166–82.e2166. doi: 10.1016/j.molcel.2021.03.008
114. Li X, Zhang Q, Shi Q, Liu Y, Zhao K, Shen Q, et al. Demethylase Kdm6a Epigenetically Promotes IL-6 and IFN-Beta Production in Macrophages. J Autoimmun (2017) 80:85–94. doi: 10.1016/j.jaut.2017.02.007
115. Ha SD, Cho W, DeKoter RP, Kim SO. The Transcription Factor PU.1 Mediates Enhancer-Promoter Looping That Is Required for IL-1beta eRNA and mRNA Transcription in Mouse Melanoma and Macrophage Cell Lines. J Biol Chem (2019) 294:17487–500. doi: 10.1074/jbc.RA119.010149
116. Oishi Y, Hayashi S, Isagawa T, Oshima M, Iwama A, Shimba S, et al. Bmal1 Regulates Inflammatory Responses in Macrophages by Modulating Enhancer RNA Transcription. Sci Rep (2017) 7:7086. doi: 10.1038/s41598-017-07100-3
117. Heward JA, Roux BT, Lindsay MA. Divergent Signalling Pathways Regulate Lipopolysaccharide-Induced eRNA Expression in Human Monocytic THP1 Cells. FEBS Lett (2015) 589:396–406. doi: 10.1016/j.febslet.2014.12.026
118. NE II, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, et al. Long Non-Coding RNAs and Enhancer RNAs Regulate the Lipopolysaccharide-Induced Inflammatory Response in Human Monocytes. Nat Commun (2014) 5:3979. doi: 10.1038/ncomms4979
119. Kim YJ, Xie P, Cao L, Zhang MQ, Kim TH. Global Transcriptional Activity Dynamics Reveal Functional Enhancer RNAs. Genome Res (2018) 28:1799–811. doi: 10.1101/gr.233486.117
120. Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity (2016) 44:439–49. doi: 10.1016/j.immuni.2016.02.024
121. Link VM, Gosselin D, Glass CK. Mechanisms Underlying the Selection and Function of Macrophage-Specific Enhancers. Cold Spring Harb Symp Quant Biol (2015) 80:213–21. doi: 10.1101/sqb.2015.80.027367
122. Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Monroy IC, et al. The Nuclear Receptor PPARgamma Controls Progressive Macrophage Polarization as a Ligand-Insensitive Epigenomic Ratchet of Transcriptional Memory. Immunity (2018) 49:615–26.e616. doi: 10.1016/j.immuni.2018.09.005
123. Huang Z, Liang N, Goñi S, Damdimopoulos A, Wang C, Ballaire R, et al. The Corepressors GPS2 and SMRT Control Enhancer and Silencer Remodeling via eRNA Transcription During Inflammatory Activation of Macrophages. Mol Cell (2021) 81:953–68.e959. doi: 10.1016/j.molcel.2020.12.040
124. Wan M, Bennett BD, Pittman GS, Campbell MR, Reynolds LM, Porter DK, et al. Identification of Smoking-Associated Differentially Methylated Regions Using Reduced Representation Bisulfite Sequencing and Cell Type-Specific Enhancer Activation and Gene Expression. Environ Health Perspect (2018) 126:047015. doi: 10.1289/EHP2395
125. Jasenosky LD, Nambu A, Tsytsykova AV, Ranjbar S, Haridas V, Kruidenier L, et al. Identification of a Distal Locus Enhancer Element That Controls Cell Type-Specific TNF and LTA Gene Expression in Human T Cells. J Immunol (2020) 205:2479–88. doi: 10.4049/jimmunol.1901311
126. Benner C, Isoda T, Murre C. New Roles for DNA Cytosine Modification, eRNA, Anchors, and Superanchors in Developing B Cell Progenitors. Proc Natl Acad Sci USA (2015) 112:12776–81. doi: 10.1073/pnas.1512995112
127. Brazao TF, Johnson JS, Müller J, Heger A, Ponting CP, Tybulewicz VLJ, et al. Long Noncoding RNAs in B-Cell Development and Activation. Blood (2016) 128:e10–19. doi: 10.1182/blood-2015-11-680843
128. Saintamand A, Vincent-Fabert C, Marquet M, Ghazzaui N, Magnone V, Pinaud E, et al. Emu and 3’rr IgH Enhancers Show Hierarchic Unilateral Dependence in Mature B-Cells. Sci Rep (2017) 7:442. doi: 10.1038/s41598-017-00575-0
129. Kristjansdottir K, Dziubek A, Kang HM, Kwak H. Population-Scale Study of eRNA Transcription Reveals Bipartite Functional Enhancer Architecture. Nat Commun (2020) 11:5963. doi: 10.1038/s41467-020-19829-z
130. Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z, Yeoh AEJ, et al. Identification of Novel lncRNAs Regulated by the TAL1 Complex in T-Cell Acute Lymphoblastic Leukemia. Leukemia (2018) 32:2138–51. doi: 10.1038/s41375-018-0110-4
131. Wallaert A, Durinck K, Loocke WV, Walle IV, Matthijssens F, Volders PJ, et al. Long Noncoding RNA Signatures Define Oncogenic Subtypes in T-Cell Acute Lymphoblastic Leukemia. Leukemia (2016) 30:1927–30. doi: 10.1038/leu.2016.82
132. Cauchy P, Maqbool MA, Cabeza JZ, Vanhille L, Koch F, Fenouil R, et al. Dynamic Recruitment of Ets1 to Both Nucleosome-Occupied and -Depleted Enhancer Regions Mediates a Transcriptional Program Switch During Early T-Cell Differentiation. Nucleic Acids Res (2016) 44:3567–85. doi: 10.1093/nar/gkv1475
133. Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Favera RD, Tsirigos A, et al. Genome-Wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia. Cell (2014) 158:593–606. doi: 10.1016/j.cell.2014.05.049
134. Beyaz S, Kim JH, Pinello L, Xifaras ME, Hu Y, Huang J, et al. The Histone Demethylase UTX Regulates the Lineage-Specific Epigenetic Program of Invariant Natural Killer T Cells. Nat Immunol (2017) 18:184–95. doi: 10.1038/ni.3644
135. Mostafa MM, Bansa A, Michi AN, Sasse SK, Proud D, Gerber AN, et al. Genomic Determinants Implicated in the Glucocorticoid-Mediated Induction of KLF9 in Pulmonary Epithelial Cells. J Biol Chem (2021) 296:100065. doi: 10.1074/jbc.RA120.015755
136. Mushimiyimana I, Bosch VT, Niskanen H, Downes NL, Moreau PR, Hartigan K, et al. Genomic Landscapes of Noncoding RNAs Regulating VEGFA and VEGFC Expression in Endothelial Cells. Mol Cell Biol (2021) 41:e0059420. doi: 10.1128/MCB.00594-20
137. Sakaguchi Y, Nishikawa K, Seno S, Matsuda H, Takayanagi H, Ishii M, et al. Roles of Enhancer RNAs in RANKL-Induced Osteoclast Differentiation Identified by Genome-Wide Cap-Analysis of Gene Expression Using CRISPR/Cas9. Sci Rep (2018) 8:7504. doi: 10.1038/s41598-018-25748-3
138. Zhou H, Simion V, Pierce JB, Haemmig S, Chen AF, Feinberg MW, et al. LncRNA-MAP3K4 Regulates Vascular Inflammation Through the P38 MAPK Signaling Pathway and Cis-Modulation of MAP3K4. FASEB J (2021) 35:e21133. doi: 10.1096/fj.202001654RR
139. Chen LF, Greene WC. Shaping the Nuclear Action of NF-Kappab. Nat Rev Mol Cell Biol (2004) 5:392–401. doi: 10.1038/nrm1368
140. Lee TI, Young RA. Transcriptional Regulation and Its Misregulation in Disease. Cell (2013) 152:1237–51. doi: 10.1016/j.cell.2013.02.014
141. Aune TM, Crooke PS 3rd, Patrick AE, Tossberg JT, Olsen NJ, Spurlock CF 3rd, et al. Expression of Long Non-Coding RNAs in Autoimmunity and Linkage to Enhancer Function and Autoimmune Disease Risk Genetic Variants. J Autoimmun (2017) 81:99–109. doi: 10.1016/j.jaut.2017.03.014
142. Israel DA, Peek RM. Pathogenesis of Helicobacter Pylori-Induced Gastric Inflammation. Aliment Pharmacol Ther (2001) 15:1271–90. doi: 10.1046/j.1365-2036.2001.01052.x
143. Boyd M, Thodberg M, Vitezic M, Bornholdt J, Seerup KV, Chen Y, et al. Characterization of the Enhancer and Promoter Landscape of Inflammatory Bowel Disease From Human Colon Biopsies. Nat Commun (2018) 9:1661. doi: 10.1038/s41467-018-03766-z
144. Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, et al. Super-Enhancers Delineate Disease-Associated Regulatory Nodes in T Cells. Nature (2015) 520:558–62. doi: 10.1038/nature14154
145. Suzuki A, Terao C, Yamamoto K. Linking of Genetic Risk Variants to Disease-Specific Gene Expression via Multi-Omics Studies in Rheumatoid Arthritis. Semin Arthritis Rheum (2019) 49:S49–53. doi: 10.1016/j.semarthrit.2019.09.007
146. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery. Nature (2014) 506:376–81. doi: 10.1038/nature12873
147. Okada Y, Eyre S, Suzuki A, Kochi Y, Yamamoto K. Genetics of Rheumatoid Arthritis: 2018 Status. Ann Rheum Dis (2019) 78:446–53. doi: 10.1136/annrheumdis-2018-213678
148. Mirtschink P, Bischof C, Pham MC, Sharma R, Khadayate S, Rossi G, et al. Inhibition of the Hypoxia-Inducible Factor 1alpha-Induced Cardiospecific HERNA1 Enhance-Templated RNA Protects From Heart Disease. Circulation (2019) 139:2778–92. doi: 10.1161/CIRCULATIONAHA.118.036769
149. Spurlock CF, Tossberg JT, Shaginurova G, Sriram S, Wingstrom J, Crooke PS, et al. A Molecular-Based Approach Using Long, Non-Coding RNA and Enhancer-Associated lncRNA Gene Expression Signatures to Classify Multiple Sclerosis Using Peripheral Whole Blood. Mult Scler J (2017) 23:94–4. doi: 10.1093/jmcb/mjz047
150. Suzuki A, Makinoshima H, Wakaguri H, Esumi H, Sugano S, Kohno T, et al. Aberrant Transcriptional Regulations in Cancers: Genome, Transcriptome and Epigenome Analysis of Lung Adenocarcinoma Cell Lines. Nucleic Acids Res (2014) 42:13557–72. doi: 10.1093/nar/gku885
151. Chen H, Li C, Peng X, Zhou Z, Weinstein JN, Liang H, et al. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell (2018) 173:386–99.e312. doi: 10.1016/j.cell.2018.03.027
152. Murakawa Y, Yoshihara M, Kawaji H, Nishikawa M, Zayed H, Suzuki H, et al. Enhanced Identification of Transcriptional Enhancers Provides Mechanistic Insights Into Diseases. Trends Genet (2016) 32:76–88. doi: 10.1016/j.tig.2015.11.004
153. Schmitt AD, Hu M, Ren B. Genome-Wide Mapping and Analysis of Chromosome Architecture. Nat Rev Mol Cell Biol (2016) 17:743–55. doi: 10.1038/nrm.2016.104
154. Zhang Z, Lee JH, Ruan H, Ye Y, Krakowiak J, Hu Q, et al. Transcriptional Landscape and Clinical Utility of Enhancer RNAs for eRNA-Targeted Therapy in Cancer. Nat Commun (2019) 10:4562. doi: 10.1038/s41467-019-12543-5
155. Almamun M, Levinson BT, Swaay AC, Johnson NT, McKay SD, Arthur GL, et al. Integrated Methylome and Transcriptome Analysis Reveals Novel Regulatory Elements in Pediatric Acute Lymphoblastic Leukemia. Epigenetics (2015) 10:882–90. doi: 10.1080/15592294.2015.1078050
156. Almamun M, Kholod O, Stuckel AJ, Levinson BT, Johnson NT, Johnson GL, et al. Inferring a Role for Methylation of Intergenic DNA in the Regulation of Genes Aberrantly Expressed in Precursor B-Cell Acute Lymphoblastic Leukemia. Leuk Lymphoma (2017) 58:1–12. doi: 10.1080/10428194.2016.1272683
157. Teppo S, Laukkanen S, Liuksiala T, Nordlund J, Oittinen M, Teittinen K, et al. Genome-Wide Repression of eRNA and Target Gene Loci by the ETV6-RUNX1 Fusion in Acute Leukemia. Genome Res (2016) 26:1468–77. doi: 10.1101/gr.193649.115
158. Park A, Oh S, Jung KL, Choi UY, Lee HR, Rosenfeld MG, et al. Global Epigenomic Analysis of KSHV-Infected Primary Effusion Lymphoma Identifies Functional MYC Superenhancers and Enhancer RNAs. Proc Natl Acad Sci USA (2020) 117:21618–27. doi: 10.1073/pnas.1922216117
159. Fatima R, Choudhury SR, Divya TR, Bhaduri U, Rao MRS. A Novel Enhancer RNA, Hmrhl, Positively Regulates Its Host Gene, Phkb, in Chronic Myelogenous Leukemia. Noncoding RNA Res (2019) 4:96–108. doi: 10.1016/j.ncrna.2019.08.001
160. Fang K, Huang W, Sun YM, Chen TQ, Zeng ZC, Yang QQ, et al. Cis-Acting lnc-eRNA SEELA Directly Binds Histone H4 to Promote Histone Recognition and Leukemia Progression. Genome Biol (2020) 21:269. doi: 10.1186/s13059-020-02186-x
161. Dahan S, Sharma A, Cohen K, Baker M, Taqatqa N, Bentata M, et al. Vegfa’s Distal Enhancer Regulates Its Alternative Splicing in CML. NAR Cancer (2021) 3:zcab029. doi: 10.1093/narcan/zcab029
162. Yu D, Yang X, Lin J, Cao Z, Lu C, Yang Z, et al. Super-Enhancer Induced IL-20ra Promotes Proliferation/Metastasis and Immune Evasion in Colorectal Cancer. Front Oncol (2021) 11:724655. doi: 10.3389/fonc.2021.724655
163. Li Y, He Y, Peng J, Su Z, Li Z, Zhang B, et al. Mutant Kras Co-Opts a Proto-Oncogenic Enhancer Network in Inflammation-Induced Metaplastic Progenitor Cells to Initiate Pancreatic Cancer. Nat Cancer (2021) 2:49–. doi: 10.1038/s43018-020-00134-z
164. Xiao J, Liu Y, Yi J, Liu X. LINC02257, an Enhancer RNA of Prognostic Value in Colon Adenocarcinoma, Correlates With Multi-Omics Immunotherapy-Related Analysis in 33 Cancers. Front Mol Biosci (2021) 8:646786. doi: 10.3389/fmolb.2021.646786
165. Wang L, Liu J, Tai J, Zhou N, Huang T, Xue Y, et al. A Prospective Study Revealing the Role of an Immune-Related eRNA, WAKMAR2, in Breast Cancer. Sci Rep (2021) 11:15328. doi: 10.1038/s41598-021-94784-3
166. Guo XY, Zhong S, Wang ZN, Xie T, Duan H, Zhang JY, et al. Immunogenomic Profiling Demonstrate AC003092.1 as an Immune-Related eRNA in Glioblastoma Multiforme. Front Genet (2021) 12:633812. doi: 10.3389/fgene.2021.633812
167. Qin N, Ma Z, Wang C, Zhang E, Li Y, Huang M, et al. Comprehensive Characterization of Functional eRNAs in Lung Adenocarcinoma Reveals Novel Regulators and a Prognosis-Related Molecular Subtype. Theranostics (2020) 10:11264–77. doi: 10.7150/thno.47039
168. Ma J, Lin X, Wang X, Min Q, Wang T, Tang C, et al. Reconstruction and Analysis of the Immune-Related LINC00987/A2M Axis in Lung Adenocarcinoma. Front Mol Biosci (2021) 8:644557. doi: 10.3389/fmolb.2021.644557
169. Wang Q, Yu X, Yang N, Xu L, Zhou Y. LncRNA AC007255.1, an Immune-Related Prognostic Enhancer RNA in Esophageal Cancer. PeerJ (2021) 9:e11698. doi: 10.7717/peerj.11698
170. Gu X, Wang L, Boldrup L, Coates PJ, Fahraeus R, Sgaramella N, et al. AP001056.1, A Prognosis-Related Enhancer RNA in Squamous Cell Carcinoma of the Head and Neck. Cancers (Basel) (2019) 11(3):347. doi: 10.3390/cancers11030347
171. Bahr C, Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, et al. A Myc Enhancer Cluster Regulates Normal and Leukaemic Haematopoietic Stem Cell Hierarchies. Nature (2018) 553:515–20. doi: 10.1038/nature25193
172. Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene Regulation. An Oncogenic Super-Enhancer Formed Through Somatic Mutation of a Noncoding Intergenic Element. Science (2014) 346:1373–7. doi: 10.1126/science.1259037
173. Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of Focally Amplified Lineage-Specific Super-Enhancers in Human Epithelial Cancers. Nat Genet (2016) 48:176–82. doi: 10.1038/ng.3470
174. Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The Chromatin Accessibility Landscape of Primary Human Cancers. Science (2018) 362(6413):eaav1898. doi: 10.1126/science.aav1898
175. Ding M, Zhan H, Liao X, Li A, Zhong Y, Gao Q, et al. Enhancer RNA - P2RY2e Induced by Estrogen Promotes Malignant Behaviors of Bladder Cancer. Int J Biol Sci (2018) 14:1268–76. doi: 10.7150/ijbs.27151
176. Liang J, Zhou H, Gerdt C, Tan M, Colson T, Kaye KM, et al. Epstein-Barr Virus Super-Enhancer eRNAs Are Essential for MYC Oncogene Expression and Lymphoblast Proliferation. Proc Natl Acad Sci USA (2016) 113:14121–6. doi: 10.1073/pnas.1616697113
177. Hamdan FH, Johnsen SA. Perturbing Enhancer Activity in Cancer Therapy. Cancers (Basel) (2019) 11 (5):634. doi: 10.3390/cancers11050634
178. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective Inhibition of BET Bromodomains. Nature (2010) 468:1067–73. doi: 10.1038/nature09504
179. Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of Inflammation by a Synthetic Histone Mimic. Nature (2010) 468:1119–23. doi: 10.1038/nature09589
180. Belkina AC, Nikolajczyk BS, Denis GV. BET Protein Function Is Required for Inflammation: Brd2 Genetic Disruption and BET Inhibitor JQ1 Impair Mouse Macrophage Inflammatory Responses. J Immunol (2013) 190:3670–8. doi: 10.4049/jimmunol.1202838
181. Khan YM, Kirkham P, Barnes PJ, Adcock IM. Brd4 Is Essential for IL-1beta-Induced Inflammation in Human Airway Epithelial Cells. PloS One (2014) 9:e95051. doi: 10.1371/journal.pone.0095051
182. Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, et al. Brd4 Maintains Constitutively Active NF-kappaB in Cancer Cells by Binding to Acetylated RelA. Oncogene (2014) 33:2395–404. doi: 10.1038/onc.2013.179
183. Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, et al. NF-KappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol Cell (2014) 56:219–31. doi: 10.1016/j.molcel.2014.08.024
184. Parua PK, Fisher RP. Dissecting the Pol II Transcription Cycle and Derailing Cancer With CDK Inhibitors. Nat Chem Biol (2020) 16:716–24. doi: 10.1038/s41589-020-0563-4
185. Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 Inhibition Suppresses Super-Enhancer-Linked Oncogenic Transcription in MYCN-Driven Cancer. Cell (2014) 159:1126–39. doi: 10.1016/j.cell.2014.10.024
186. Zhang T, Kwiatkowski N, Olson CM, Clarke SED, Abraham BJ, Greifenberg AK, et al. Covalent Targeting of Remote Cysteine Residues to Develop CDK12 and CDK13 Inhibitors. Nat Chem Biol (2016) 12:876–84. doi: 10.1038/nchembio.2166
187. Hu S, Marineau JJ, Rajagopal N, Hamman KB, Choi YJ, Schmidt DR, et al. Discovery and Characterization of SY-1365, a Selective, Covalent Inhibitor of CDK7. Cancer Res (2019) 79:3479–91. doi: 10.1158/0008-5472.CAN-19-0119
188. Sharifnia T, Wawer MJ, Chen T, Huang QY, Weir BA, Ann Sizemore A, et al. Small-Molecule Targeting of Brachyury Transcription Factor Addiction in Chordoma. Nat Med (2019) 25:292–300. doi: 10.1038/s41591-018-0312-3
189. Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, et al. Targeting Transcription Regulation in Cancer With a Covalent CDK7 Inhibitor. Nature (2014) 511:616–20. doi: 10.1038/nature13393
190. Schmitz ML, Kracht M. Cyclin-Dependent Kinases as Coregulators of Inflammatory Gene Expression. Trends Pharmacol Sci (2016) 37:101–13. doi: 10.1016/j.tips.2015.10.004
191. Wei Y, Li C, Bian H, Qian W, Jin K, Xu T, et al. Targeting CDK7 Suppresses Super Enhancer-Linked Inflammatory Genes and Alleviates CAR T Cell-Induced Cytokine Release Syndrome. Mol Cancer (2021) 20:5. doi: 10.1186/s12943-020-01301-7
192. Siebert S, Pratt AG, Stocken DD, Morton M, Cranston A, Cole M, et al. Targeting the Rheumatoid Arthritis Synovial Fibroblast via Cyclin Dependent Kinase Inhibition: An Early Phase Trial. Med (Baltimore) (2020) 99:e20458. doi: 10.1097/MD.0000000000020458
193. Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, et al. CDK7 Inhibition Potentiates Genome Instability Triggering Anti-Tumor Immunity in Small Cell Lung Cancer. Cancer Cell (2020) 37:37–54.e39. doi: 10.1016/j.ccell.2019.11.003
194. Cartwright JA, Lucas CD, Rossi AG. Inflammation Resolution and the Induction of Granulocyte Apoptosis by Cyclin-Dependent Kinase Inhibitor Drugs. Front Pharmacol (2019) 10:55. doi: 10.3389/fphar.2019.00055
195. Xu YM, Du JY, Lau AT. Posttranslational Modifications of Human Histone H3: An Update. Proteomics (2014) 14:2047–60. doi: 10.1002/pmic.201300435
196. Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, et al. Selective Inhibition of EZH2 and EZH1 Enzymatic Activity by a Small Molecule Suppresses MLL-Rearranged Leukemia. Blood (2015) 125:346–57. doi: 10.1182/blood-2014-06-581082
197. Huang JP, Ling K. EZH2 and Histone Deacetylase Inhibitors Induce Apoptosis in Triple Negative Breast Cancer Cells by Differentially Increasing H3 Lys(27) Acetylation in the BIM Gene Promoter and Enhancers. Oncol Lett (2017) 14:5735–42. doi: 10.3892/ol.2017.6912
198. Sanchez GJ, Richmond PA, Bunker EN, Karman SS, Azofeifa J, Garnett AT, et al. Genome-Wide Dose-Dependent Inhibition of Histone Deacetylases Studies Reveal Their Roles in Enhancer Remodeling and Suppression of Oncogenic Super-Enhancers. Nucleic Acids Res (2018) 46:1756–76. doi: 10.1093/nar/gkx1225
199. Mishra VK, Wegwitz F, Kosinsky BL, Sen M, Baumgartner R, Wulff T, et al. Histone Deacetylase Class-I Inhibition Promotes Epithelial Gene Expression in Pancreatic Cancer Cells in a BRD4- and MYC-Dependent Manner. Nucleic Acids Res (2017) 45:6334–49. doi: 10.1093/nar/gkx212
200. Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 From GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep (2018) 22:3641–59. doi: 10.1016/j.celrep.2018.03.012
201. Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, et al. LSD1 Demethylates Repressive Histone Marks to Promote Androgen-Receptor-Dependent Transcription. Nature (2005) 437:436–9. doi: 10.1038/nature04020
202. Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, et al. Phosphorylation of Histone H3T6 by PKCbeta(I) Controls Demethylation at Histone H3K4. Nature (2010) 464:792–6. doi: 10.1038/nature08839
Keywords: eRNA, immune inflammatory, enhancer transcription events, cancers, therapeutic prospects
Citation: Wan L, Li W, Meng Y, Hou Y, Chen M and Xu B (2022) Inflammatory Immune-Associated eRNA: Mechanisms, Functions and Therapeutic Prospects. Front. Immunol. 13:849451. doi: 10.3389/fimmu.2022.849451
Received: 06 January 2022; Accepted: 21 March 2022;
Published: 19 April 2022.
Edited by:
Andrea Baragetti, University of Milan, ItalyReviewed by:
Masaki Miyazaki, Kyoto University, JapanChangchang Cao, Institute of Biophysics (CAS), China
Copyright © 2022 Wan, Li, Meng, Hou, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Xu, bmp4YnNldUBzZXUuZWR1LmNu; Yue Hou, aG91eXVlQHhqdHUuZWR1LmNu; Ming Chen, bWluZ2NoZW5zZXVAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Lilin Wan
Lilin Wan Wenchao Li2†
Wenchao Li2† Ming Chen
Ming Chen Bin Xu
Bin Xu