- 1Infectious Diseases Clinic, IRCCS Policlinico San Martino Hospital, Genoa, Italy
- 2Department of Biomolecular Sciences, University of Urbino Carlo Bo, Urbino, Italy
- 3Biostatistics Unit, Department of Health Sciences, University of Genoa, Genoa, Italy
- 4Department of Informatics, Bioengineering, Robotics and System Engineering (DIBRIS), University of Genoa, Genoa, Italy
- 5Immunology Research Area, Bambino Gesù Children's Hospital IRCCS, Rome, Italy
- 6Department of Health Sciences, University of Genoa, Genoa, Italy
The quantification of proviral DNA is raising interest in view of clinical management and functional HIV eradication. Measures of all unintegrated HIV DNA (uDNA) forms in infected reservoir cells provides information on recent replication events that is not found from other proviral DNA assays. To evaluate its actual relevance in a cohort of perinatally-infected adult HIV patients (PHIV), we studied how peripheral blood mononuclear cell uDNA levels correlated with total HIV DNA (tDNA) and with overall replication or innate immune control parameters including NK cell activation/exhaustion and lymphoid turnover. Twenty-two PHIV were included, with successfully controlled HIV (HIV RNA <50 copies/mL) on combined antiretroviral therapy for mean of 8.7 ± 3.9 years. uDNA accounted for 16 [5.2-83.5] copies/µg and was strongly correlated with tDNA (ρ=0.700, p=0.001). Flow cytometric analysis of peripheral NK cells showed that CD69 expression was directly correlated uDNA (p=0.0412), but not with tDNA. Interestingly, CD56-CD16+NK cells which include newly described inflammatory precursors and terminally differentiated cells were directly correlated with uDNA levels (p<0.001), but not with tDNA, and an inverse association was observed between the proportion of NKG2D+ NK cells and uDNA (ρ=-0.548, p=0.015). In addition, CD34+DNAM-1brightCXCR4+ inflammatory precursor frequency correlated directly with uDNA levels (ρ=0.579, p=0.0075). The frequencies of CD56-CD16+ and CD34+DNAM-1brightCXCR4+ cells maintained association with uDNA levels in a multivariable analysis (p=0.045 and p=0.168, respectively). Thus, control of HIV-1 reservoir in aviremic patients on ART is an active process associated with continuous NK cell intervention and turnover, even after many years of treatment. Quantification of linear and circular uDNA provides relevant information on the requirement for ongoing innate immune control in addition to ART, on recent replication history and may help stratify patients for functional HIV eradication protocols with targeted options.
Introduction
A hallmark of lentivirus infection is the persistence of integrated or partly episomal DNA in long-lived cells—referred to as the reservoir—which guarantees lifelong infection (1). Accordingly, infection with human immunodeficiency virus type 1 (HIV-1) determines the establishment of the viral DNA reservoir of long-lived CD4+ cells (2). Indeed, lentiviral reservoirs are established very early in acute infections (3) and maintain replication competence even during antiretroviral treatment (ART), leading to a lifelong need for medication (4). The characterization of HIV reservoirs during ART over time and strategies for elimination or control represent a major scientific focus to achieve HIV eradication (5–7).
HIV DNA in CD4+ PBMC is a measure of total reservoir size (1, 5, 6) and rapidly declines after ART initiation, but it remains static afterward, tending to plateau after the first years of ART (8) and persisting even after 4-12 years of successful ART (9, 10). Within integrated nuclear HIV DNA sequences, partially modified or deleted HIV sequences may lead to attenuated disease in exceptional instances (11). About 3% of HIV reservoir sequences are fully replication competent in patients requiring ART to control HIV replication (12) and is in line with prompt viremic rebound on ART interruption (13) independent of more potent newer regimens (14). Low level viremic persistence indeed occurs in many patients on ART with suppressed plasma HIV RNA levels <50 copies/mL for a prolonged time, and a low level of viremia can still be detected by ultrasensitive assays (15, 16).
A fraction of HIV DNA is unintegrated (uDNA) and assays measuring these extrachromocosomal forms have been used to monitor ongoing intra-cellular viral replication events. Indeed, uDNA HIV in both linear and circular form represents a short-lived preintegration phase of HIV-1 latency and is considered a consequence of a residual viral replication in ART-treated patients (17, 18) Accordingly, uDNA assays are not used to estimate the viral reservoir because the majority of uDNA may degrade and contributes little to viral production (19).
Several studies showed that uDNA can still be measured during successful ART using an improved assay, even when plasma viremia is below the cut-off for common clinical tests, thus signaling recent replication events (17, 20, 21).
Reservoir persistence is long-lived and has been attributed to the inherent resistance of HIV reservoirs to HIV-specific CD8+ CTL (7, 22, 23). Indeed, in vitro suppression by CD8+ T cells of viral reservoir activation is largely independent of cytotoxic CD8+ T lymphocytes and relies on C-C chemokines and other soluble factors (24). Opposite to their potency in controlling virus replication CD8+ CTLs are apparently inefficient in controlling HIV reservoir. There are so far few or minimal indications that during HIV latency there is relevant antigen presentation to CD8+ CTLs. In addition using potent CD8+CTLs together with latency reversing agents and quantitative viral outgrowth assays, results in the elimination of a subset of CD4+ T cells cells harboring defective HIV proviruses, but not of those harboring infectious proviruses thus suggesting a relative inefficiency of CD8+CTLs in controlling viral reservoir (22). On the other hand, Natural Killer cells (NK) are instrumental in the control of HIV replication under specific conditions, such as in Elite- or HIV-controller patients where, in the absence of any antiretroviral treatment they associate with improved levels of HIV reservoir size and control (25–28). In this regard, NK cells appear to significantly contribute to shaping and containing the HIV-1 reservoir since their functional activity in both Elite Controller and ART-related suppressed patients inversely correlates with patients’ HIV reservoirs both in vivo and in vitro (29). Accordingly, their analysis could provide useful insights in the shaping of HIV reservoir under controlled conditions.
In perinatally HIV-infected children (PHIV) born to infected mothers, early ART shapes the HIV reservoir and the immune response (30–32). PHIV patients who were born in the early 90s and are now adolescents or young adults did not have the opportunity for early ART and were sometimes exposed to months or years of suboptimal antiretroviral regimens prior to combined antiretroviral therapy (cART) (33, 34). In this context, viremia copy years (VCY), a metric of cumulative HIV RNA burden calculated based on longitudinal viral load data, has been used to summarize in a single value the viral burden of years (35, 36). VCY is considered a reliable marker of past viral burden and has been associated with mortality and organ damage in people living with HIV and PHIV (36, 37).
Low-level viremic persistence in suppressed ART-treated patients contributes to sustain the reservoir size and may occur upon standard antigen-specific responses to daily non-HIV related antigenic stimulation of latently infected CD4+ T cells [e.g.: vaccination, transient infections (38, 39)]. These events occur in tissues where drug concentrations may be insufficient to fully suppress replication (40) thus leaving space for different immune control in different patients, and may be revealed by uDNA. These considerations together with the role played by Natural Killer (NK) cells in HIV reservoir control in Elite Controllers (29), raised the question whether circulating NK-cell phenotype and NK cell turnover could reveal their involvement in underlying HIV low-level replicative events in ART-treated patients. To this end we performed a combined evaluation of HIV DNA reservoirs and of VCY in a PHIV cohort that has survived two decades of infection with the aim of describing their relationship with NK cell activation/exhaustion markers and lymphoid turnover.
Materials and Methods
Study Cohort
Our center has been involved from the beginning of the HIV pandemic in the follow-up and description of the natural history of mother-to-child transmission (41). For this reason, we have continuously cared for patients surviving HIV infection since the early years when ART was unavailable. We here present data on the cohort of patients originally enrolled in the European Collaborative Study (42) who survived and were not lost to follow-up over the years.
The study was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments and in accordance with Italian national laws. All patients signed an informed consent form in which they agreed to the use of their clinical data in an anonymous form for scientific purposes. The use of the Ligurian HIV Network database (Medinfo) for scientific purposes was approved by the Ligurian Ethics Committee (date of approval: 28 August 2013). For the purpose of the present study, all patients were sampled at one timepoint in 2016 after providing informed consent for immune cell profiling to evaluate their innate immunity (IRB approval (IRCCS AOU San Martino-IST Genova n51/09, ALS 2 n°10/2011).
HIV RNA<50 copies/ml was a requirement for inclusion into the study. All demographic, laboratory, and therapeutic data were extrapolated from the electronic database Medinfo (43) that is automatically populated by an autonomous system according to international standard communication protocols based on HTTP (HyperText Transfer Protocol) and HSSP (Healthcare Services Specification Project). The Medinfo database allowed laboratory data analysis from 2010 to the present. Viremia copy-years (VCY), the principal exposure of interest, is a time-varying measure of cumulative plasma HIV burden that was calculated between 2010 and the time of study entry (2016). Patients had mean 2.96 HIV-RNA ( ± 0.69) evaluations per year (range 1-7 HIV-RNA evaluations/year) that were all included for VCY calculation during the study period. A threshold of <50 copies/mL was used to define undetectable HIV RNA across the study period. The trapezoidal rule was used to approximate the integral representing the area under each patient’s longitudinal HIV viral load (VL) curve. VL burden for each segment (time interval between two consecutive VL values) was calculated by multiplying the mean of the two VL values by the time interval (35). The copy years/mL for each segment of a patient’s VL curve were then summed to calculate viremia copy-years. Formally, VCY is the number of copies of HIV RNA per mL of plasma over time. 10.000 copy-years of viremia is equal to having a VL of 10.000 copies/mL each day for 1 year or a VL of 1.000 copies/mL for 10 years. Virological failure was defined as confirmed HIV RNA >200 copies/mL in two subsequent blood samples or as a single HIV RNA > 1.000 copies/mL (35, 44). We defined combined ART (cART) as the administration of at least 3 antiretroviral agents including either a ritonavir-boosted protease inhibitor (PI/r), a nonnucleoside reverse transcriptase inhibitor (NNRTI), or an integrase inhibitor (INSTI).
Sampling and Flow Cytometry
Peripheral blood (15 mL) was drawn by venipuncture and peripheral blood mononuclear cells (PBMC) were obtained by density gradient centrifugation (Ficoll-Hipaque) and cryopreserved until use. Cells were analyzed by multicolor flow cytometry(FACS Fortessa, BD, MountainView, CA, USA, as described previously (45). For analysis, cells were gated using forward and side light scatter parameters with acquisition of 10.000 events.
NK cells were defined as CD3-CD14-CD19-CD56+CD16+/-. Lin- cells were defined as CD3-CD14-CD19-. Inflammatory common lymphocyte precursors were defined by logical gates within Lin- subset as CD34+DNAM-1brightCXCR4+ cells and CD34-CD56-CD16+CD7- (45, 46). CD34-CD56-CD16+CD7- are recognized as a subset of CD56-CD16+ NK cells (46) previously described as “exhausted” NK cells (47).
Mean fluorescence intensity ratios (MFIr) were calculated using the formula MFI sample/MFI negative control—express mean cell molecule density. Data were analyzed using FlowJo (Tree Star, Inc.) (45, 46).
mAbs
The following panel of mouse anti-human mAbs was used: Anti-NKp44 (Z231, IgG1), (BAT221, IgG1), anti-DNAM-1 (F22, IgG1), anti-KIR2DL2/L3/S2 (CD158b1/b2,j), anti-KIR3DL1/S1 (CD158e1/e2), anti-KIR2DL1/S1 (CD158a/h), anti-NKG2A (Z270, IgG1; Z199, IgG2a), and anti-CD85j (F278, IgG1 kindly provided by Dr. D. Pende), all of which were produced in the laboratory (A. Moretta, Genova, Italy). Anti HLA-DR (D1-12, IgG2a) was kindly provided by Dr. R. S. Accolla (University of Insubria, Varese, Italy). Commercial Goat anti-mouse were used for indirect staining in some instances and complete commercial mAb list is given in the supplementary information (Supplementary Table 1).
IFN γ Production by NK Cells
PBMCs were stimulated using FcgR+ P815 target cells at a 10:1 E/T ratio in complete medium in the presence or absence of an anti-NKp30 and/or anti-NKp46 mAb mixture (0.1 μg/mL). Phorbol 12-myristate 13-acetate (25 ng/mL; Sigma-Aldrich, St Louis, Mo) + ionomycin (1 μg/mL; Sigma-Aldrich) was used for maximal production. GolgiPlug (BD Pharmingen) was added at 37°C from incubation start for 8 hours or after overnight incubation for 4 hours(o/n), as previously described (31). After incubation, cells were stained with mAbs followed by permeabilization/fixation (Citofix/Citoperm protocol; BD Pharmingen) and anti–IFN-γ+ in the presence of a permeabilizing solution. A total of 10,000 gated events were acquired.
Quantitative HIV DNA Analysis
The procedure for measuring HIV DNA levels has been described in detail previously (20, 48).
Briefly, we reported the workflow of experiments and any changes or improvements in the procedure. Cellular DNA was isolated from a pellet of 2x106 PBMCs by a DNA extraction kit following manufacturers’ instructions (QIAGEN QIAamp Blood Mini kit), following the manufacturers’ instructions. DNA amount was determined by a NanoVue Plus ND-1000 Spectrophotometer (GE Healthcare) and all purified DNAs had absorbance ratios A260/A280 of 1.7 (± 0.21). DNA recovery of 9.6 µg (± 2.5) was obtained for each sample.
Unintegrated HIV DNA (uDNA) (i.e., the ensemble of extrachromosomal viral cDNAs, including both linear cDNA and all the closed circular 1-LTR and 2-LTR and other rearranged forms) was obtained from cellular DNA by an optimized chromatographic procedure that separates high molecular weight DNA (HMW DNA) from low molecular weight DNA (LMW DNA). The uDNA was present in the eluate fraction.
Moreover the β-actin housekeeping gene was amplified in the eluate fraction and quantified on standard curve (ACTstd obtained with 10- and 2-fold serial dilutions of a reference genomic DNA (Promega) ranging from 1000 to 0.01 ng.) as control experiment on chromatographic separation demonstrating the feasibility of the procedure (49).
Total and unintegrated HIV DNA were simultaneously analyzed by a SYBR Green qPCR based method in a single run using a single set of specific primers selected in the 5’ LTR-Gag region of the HIV-1 genome, including the highly conserved primer-binding site (PBS); this method could detect all HIV-1 subtypes in the M group. PCR reactions were carried out in a 7500 real-time PCR system (Applied Biosystems, Thermo Fisher Scientific Inc) using the Hot-Rescue Real-Time PCR Kit Sybr Green (Diatheva srl). Each sample (cellular DNA or eluate fraction containing uDNA) was analyzed in six replicates, consisting of three wells containing 0.5 µg of DNA or the equivalent quantity of elution fraction and three wells containing 1.0 µg of DNA (4.5 µg, to ensure the detection of the target even in low copy numbers, i.e., near the quantification limit, QL). For samples with HIV DNA datum quantified near or detected below the QL, two additional 1 µg replicates were tested for a total of 6.5 µg of DNA (~106 PBMC). In the case of negative amplification, a PCR spike test was performed by adding two or ten copies of plasmid standard to the samples to exclude the presence of inhibitors.
The HIV DNA copy number was quantified by interpolating the experimentally determined threshold cycle (CT) based on standard curves generated using half-log serial dilutions from 105 to 2 copies (setting the quantification limits at 2 copies/PCR) and by adding up the copy number from the 0.5 and 1.0 µg replicates and expressed as copies/µg). The amount of integrated HIV DNA (iDNA) was obtained by subtracting the amount of uDNA from the amount of total HIV DNA (tDNA).
For the accurate enumeration of the analyzed cells in qPCR, the single copy housekeeping Rpp40 gene (part of the human RNAse P gene family) was amplified. This approach provided a control for cellular DNA degradation, presence of PCR inhibitors and input DNA normalization.
The standard curve (Rpp40std) for the quantification of a 100 bp fragment of the Rpp40 gene, (forward primer: 5’-CGTAAGCAAGTTTAGTGAATACCTGAA-3’ and the reverse primer: 5’-GCACAGCTTCCATCTTACTCAATC-3’) was made with 10- and 2-fold serial dilutions of a reference human genomic DNA (Promega) ranging from 100 to 0.01 ng [assuming 7.0 pg of DNA content per human diploid genome as conversion factor (Gregory TR. 2020. Animal Genome Size Database.
Statistical Analysis
Data were described using mean and standard deviation (SD) for normally distributed continuous variables, median and interquartile range [IQR] for not normally distributed continuous variables, and frequency (%) for categorical and ordinal variables. A logistic regression analysis using uDNA as dependent variable was performed. All covariates with P < 0.25 on their univariable association with detectable uDNA were triaged for inclusion into the multivariable model. A forward stepwise variable selection approach was used in this set of covariates to retain significant variables in the final multivariable logistic regression model.
In a sensitivity analysis, a multivariable LASSO regression model was performed to ensure that only the most relevant factors associated with presence of uDNA were identified.
This penalized regression method allows for the integration of a large number of possible correlated predictors into one model and to select amongst these despite a small sample size. A general linear model using tDNA as dependent variable was performed to assess its correlations with time from birth to ART and cART initiation, VCY, and Natural Killer (NK) cell molecules expression (CD69, CD56 CD16, NKp46, and NKp30 cells)
Spearman rank correlation coefficient (ρ) was used to assess correlations between variables and uDNA or tDNA. Significance tests were two-sided using 0.05 significance level (JMP 9.0.1, SAS Institute).
Results
Study Population
Overall, 22 PHIV patients (41% male, 91% Caucasian) born to HIV-transmitting mothers and subsequently followed up at Ospedale Policlinico San Martino-University of Genova were enrolled in this study. The mean (SD) age at the time of analysis was 22.6 (6.1) years and all had been followed up since birth. All had been on cART for mean of 8.7 (3.9) years. Relevant clinical data are indicated in Table 1. Since 18 patients were born before 1996, they had been administered either monotherapy or no treatment until cART became available. Accordingly, the mean (SD) time from birth to administration of the first antiretroviral drug was 3.264 (2.719) days and time to first cART was 5.330 (2.274) days. Study participants had mean CD4+T cell count 866.5 ( ± 405.9) cells/mmc, corresponding to an average CD4 of 28.8 ( ± 15.3) % at the time of uDNA evaluation. Of the 22 PHIV, 13 (59%) were successfully treated with ART from early childhood through to adult life, 1 (5%) was treated late after birth, although with very good adherence to ART over the years, and 8 (36%), whether treated early or late, had multiple virological failures due to low adherence to ART over the years. Figure 1 summarizes three patterns of viral replication, CD4+ cells and ART over a 9 year-period immediately before sampling for 3 representative patients. At the time of the study, shown by the red circle indicating HIV reservoir, participants had experienced different diseases courses with virological failures or viral blips over the years, as shown in Figure 1. Their mean VCY (from 2010 to 2016) was 20.802.6 (50.893,6) copy/years. Participants all had <50 copies/mL of HIV RNA when beginning the study. CD4+ T cell nadir was 442(236) cells/mmc, while CD4 frequency was 28.8% (15.3). Supplementary Figure 1 gives information on all the study participants.
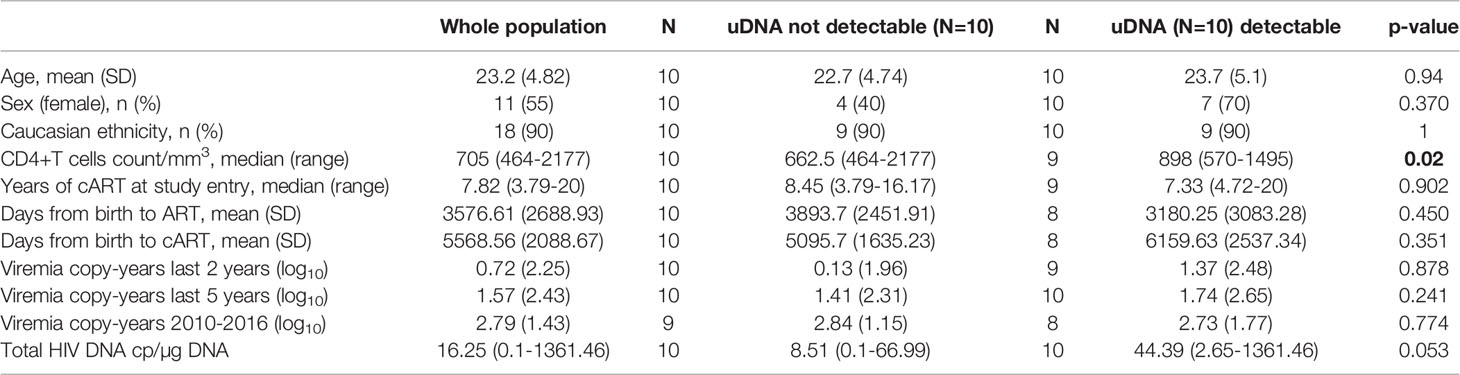
Table 1 Comparison of clinical and demographic and virological features of the study population, according to unintegrated DNA (uDNA) value, detectable vs. target not detected.
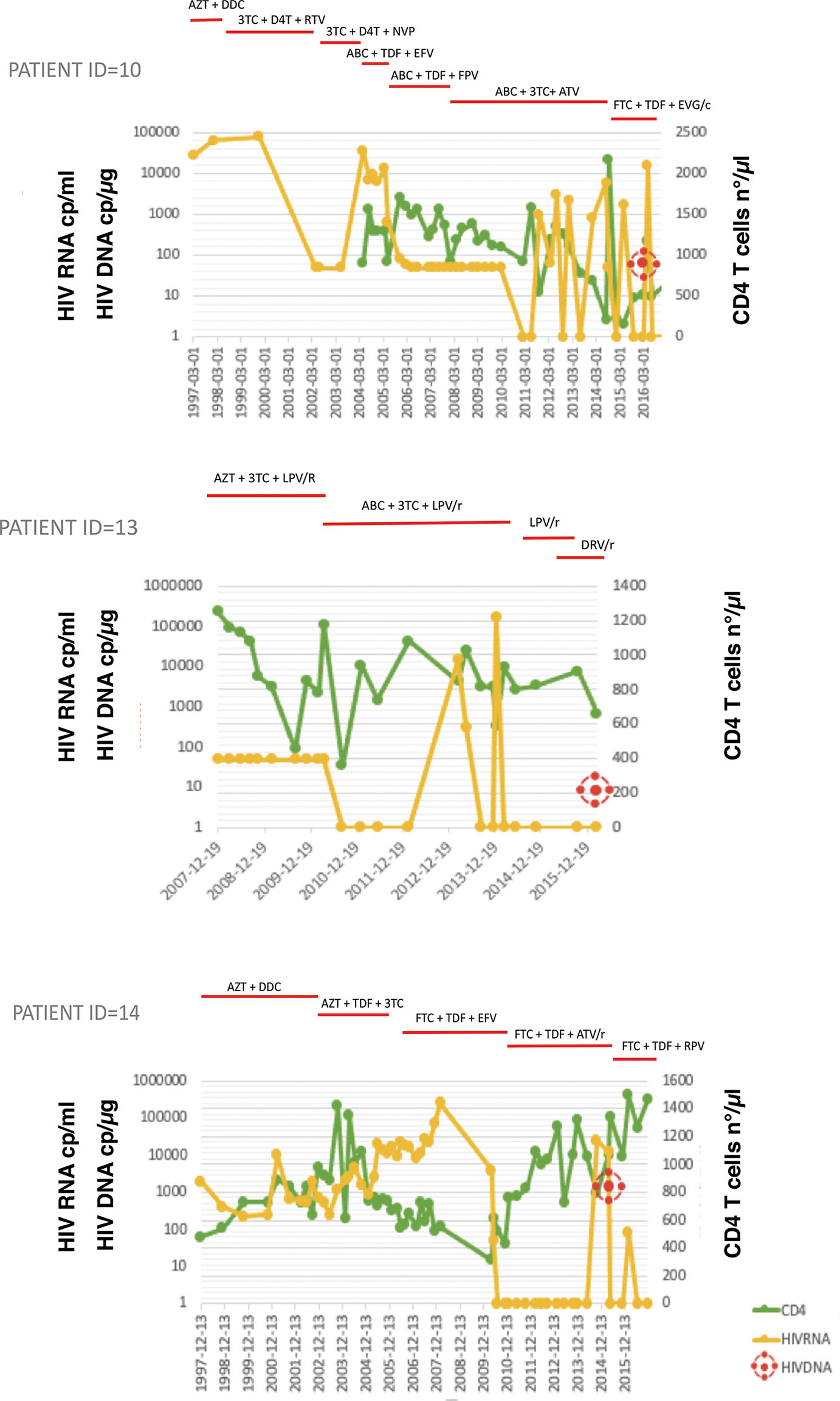
Figure 1 HIV-RNA and CD4+ T-cell count (CD4) trends over the years and total HIV DNA at study time in three representative patients. Treatments are shown on the top, bars indicate start and stop of relative antiretroviral medication. 3TC, lamivudine; ABC, abacavir; ATV, atazanavir; AZT, zidovudine; D4T, stavudine; DDC, zalcitabine; EFV, efavirenz; EVG/c, elvitegravir/cobicistat; FTC, emtricitabine; FPV, fosamprenavir; TDF, tenofovir disoproxil fumarate; NVP, nevirapine; RTV, ritonavir Top panel, Patient ID=10: multiple virological failures over the years due to low adherence to antiretroviral therapy (ART); Middle panel, patient ID=13: successfully treated from birth while maintaining good adherence to ART; Bottom panel, patient ID=14: treated late after birth, although with very good adherence to ART over the years.
uDNA Correlates With Total HIV DNA
Total and uDNA levels were measured for all patients, iDNA was obtained by subtraction of uDNA from tDNA. A negligible cross-contamination of HMW DNA (assayed by β-actin housekeeping gene amplification) which could harbor the iDNA was found in the LMW DNA fraction with no effect on the uDNA quantification (mean 5.1% (1.3%); Supplementary Table 2).
Individual measurements and mean values are shown in Figure 2A. Median [IQR] total (tDNA), unintegrated DNA (uDNA) and integrated (iDNA) HIV DNA, were 16 [5.2-83.5], 0.25 [0-4.2] and 14.5 [2.5-66.5] copies/μg, respectively (Figure 2B). We evaluated the relationship of uDNA with tDNA in each patient. Unintegrated DNA levels were strongly correlated with tDNA (ρ=0.700, p=0.001) and accounted for 7.3% (9.4%) of the tDNA. Interestingly, 10 patients did not have any level of uDNA with a measurable reservoir (tDNA) in 6 cases (Figure 2C).
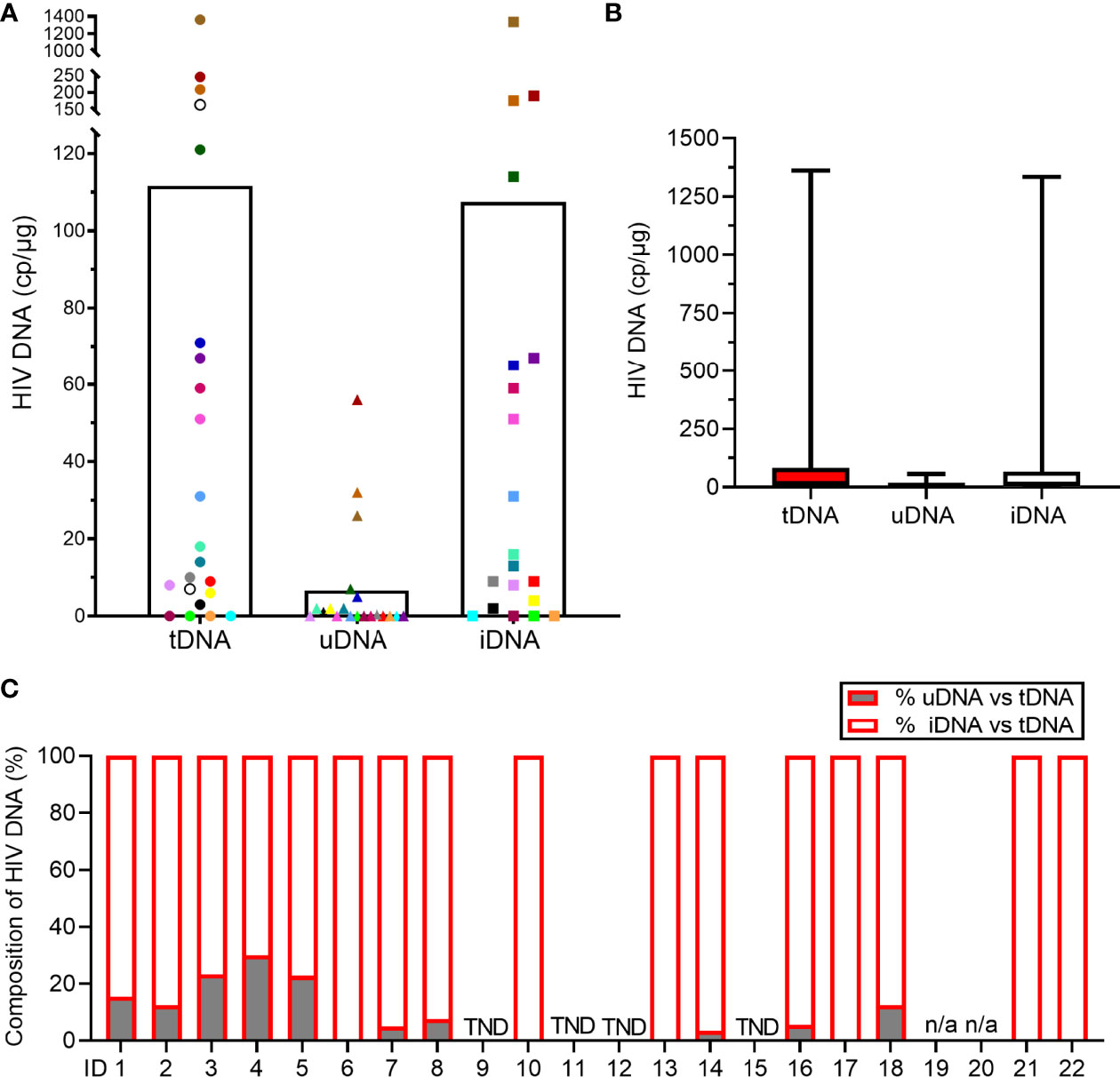
Figure 2 Measurements of HIV DNA reservoir in the study population. (A) Levels of total (tDNA), unintegrated (uDNA) and integrated (iDNA) HIV DNA. Each dot represents one patient sample and the white histogram represents the mean. Two patients’ samples did not yield sufficient cellular DNA for uDNA and iDNA analysis (open circles). (B) Levels of tDNA, uDNA and iDNA. Data represent the median [IQR]. (C) Percentages of uDNA and iDNA among tDNA in each study participant. When uDNA copy numbers were < QL of the assay, the % were reduced to 0% (ID 6, 10, 13, 17, 21, 22). When tDNA, uDNA and iDNA copy numbers were < QL, the sample is reported as TND (target not detected; ID 9, 11, 12, 15). Samples, ID 19 and ID 20, were not analyzed for uDNA and iDNA (n/a, not applicable).
In view of individual differences in initiation of ART treatment in the cohort, we controlled by correlation analysis whether differences in time to ART initiation played a role on HIV-reservoir. A trend towards longer intervals between birth and time at the start of combined ART (time to cART) was observed in PHIV patients with higher tDNA (Beta 0.394, 95%CI -2.05;+35.84, p=0.077), while no correlation was observed with uDNA levels using Spermans’ ρ for correlations regarding uDNA(ρ=0.35, p=0.154). As expected, no association was observed between HIV reservoir and the time to any ART, including monotherapy in early years (p=0.443 for tDNA and ρ =-0.079; p=0.8567 for uDNA). These findings suggest that shades of delay in ART initiation in this cohort associate with tDNA levels and are in line with previous reports in PHIV (50, 51). Interestingly, however, there was no relationship between uDNA levels and time to ART, suggesting that factors other than ART influence presence/absence of uDNA.
Since uDNA reflects early HIV entry events in susceptible cells, and its levels were not correlated with ART-free time after birth, we asked whether HIV uDNA might associate more adequately with replicative blips as expressed by VCY in the years 2010-2016. When testing this association, neither uDNA (ρ=-0.022; p=0.9332), nor tDNA (ρ=0.1997;p=0.4269), was correlated with VCY).
Taken together, these results confirm that tDNA, as a measure of the reservoir, is correlated with time to cART, and indicate that uDNA is not correlated with time to pharmacological control of HIV replication.
Increased uDNA Associates With NK Cell Activation and Exhaustion in cART Treated PHIV
In view of the role played by NK cells in host immune protection and control of HIV replication contributing to HIV reservoir size, we next focused on analysis of peripheral NK cells and their possible association with the viral reservoir size and its control in PHIV patients. Flow cytometric analysis was performed on CD3-CD14-CD19-CD56- PBMC (Figure 3A) to study receptor expression on CD56bright and CD56dim NK cells (Figure 3B). Gating strategy to study CD34+ and CD34- common lymphocyte precursors was performed on CD3-14-19-56- PBMC (Figure 3C). Accordingly, analysis of the whole cohort is shown as far as main circulating NK cells (Figure 3D), and expression of major activating NK cell receptors (Figure 3E), of inhibitory receptors (Figure 3F) and of activation surface antigens (Figure 3G). Frequencies of CD56bright and CD56dim NK cells and those for NK cells expressing major Natural Cytotoxicity Receptors and DNAM-1 and inhibitory receptors were in line with those observed in HIV adult patients (27, 52). Persistent expression of HLA-DR and CD69 activation markers on NK cells (Figure 3G) is higher compared to healthy uninfected donors and is in line with original reports showing persistent NK cell activation in the presence of successful ART with VL<50 copies/mL (52, 53). Since the aim of the study was not to compare PHIV with healthy donors but rather do provide in-depth analysis of immune markers with virological data, we next assessed correlations between HIV reservoir parameters from the direct assay and Natural Killer cell frequencies or surface molecule density using Spearman’s ρ test. Table 2 summarizes the associations observed between NK cells, circulating inflammatory precursor cells, and HIV-reservoir parameters (tDNA, uDNA).
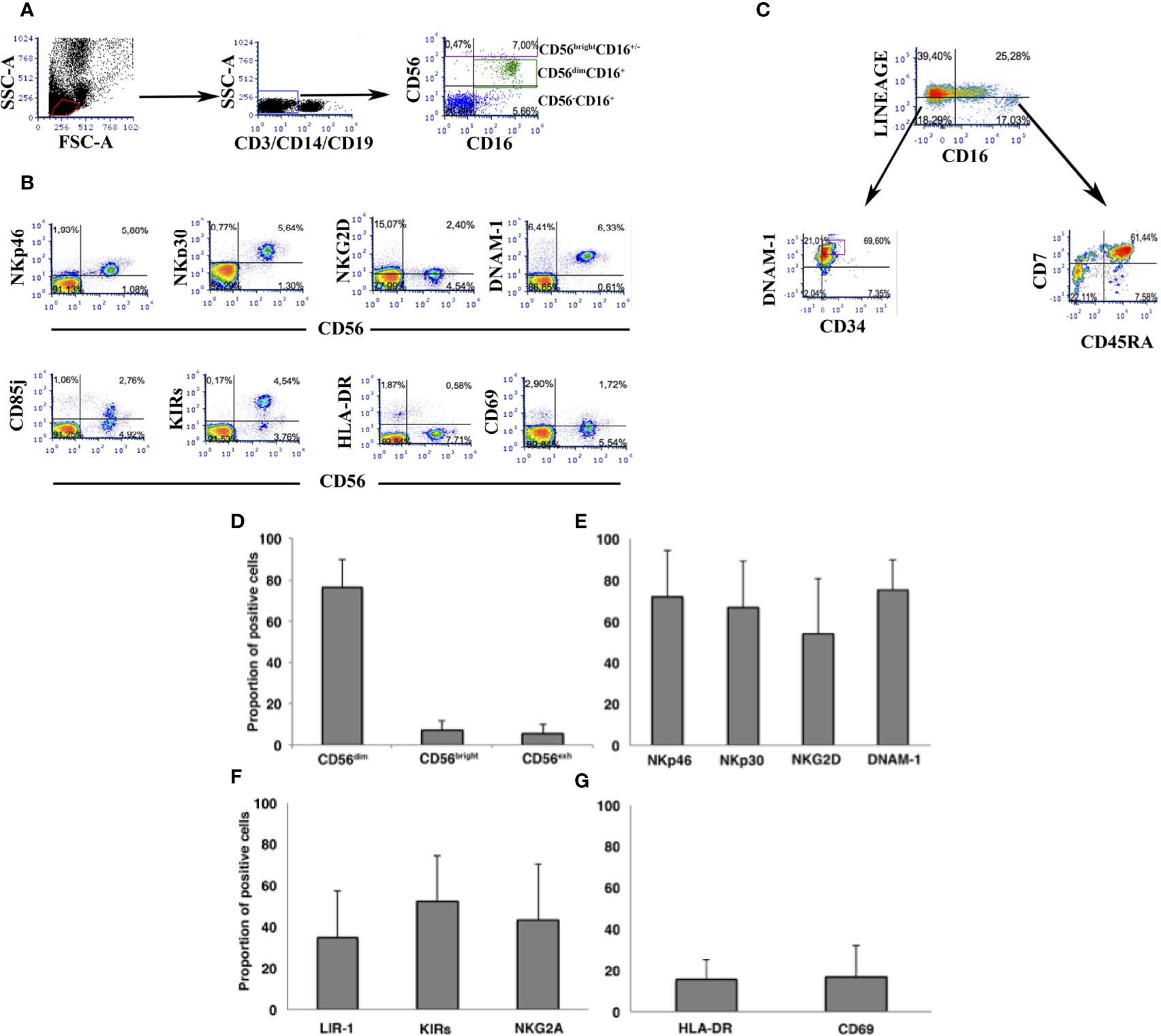
Figure 3 Flow cytometric analysis of peripheral NK cells in the cohort of PHIV. (A) Flowcytometric analysis of PBMC in a representative patient. Dot plots show gating strategy of PBMC to assess NK cell frequency. (B) Flowcytometric analysis of NK cells in a representative patient. CD3-14-19-56+ cells are stained for different NK cell surface antigens using directly labelled specific mAbs. (C) Flowcytometric analysis of a representative patient showing gating strategy and labelling to assess the frequency of (CD3-14-19-56-) and of CD34+CDNAM-1bright inflammatory cell precursors and of (CD3-14-19-56-). “Lineage” labels cells as lineage positive (CD3+14+19+56+) and lineage negative (CD3-14-19-56-)for gating and analysis of inflammatory precursors. (D-G) Histograms indicate mean ± SD of NK cells in PBMC of patients at the time of HIV reservoir determination. Frequencies are shown for the three major circulating NK cell subsets (D), for NK cells expressing activating receptors (E), inhibitory NK cell receptors (F), and activation molecules (G).
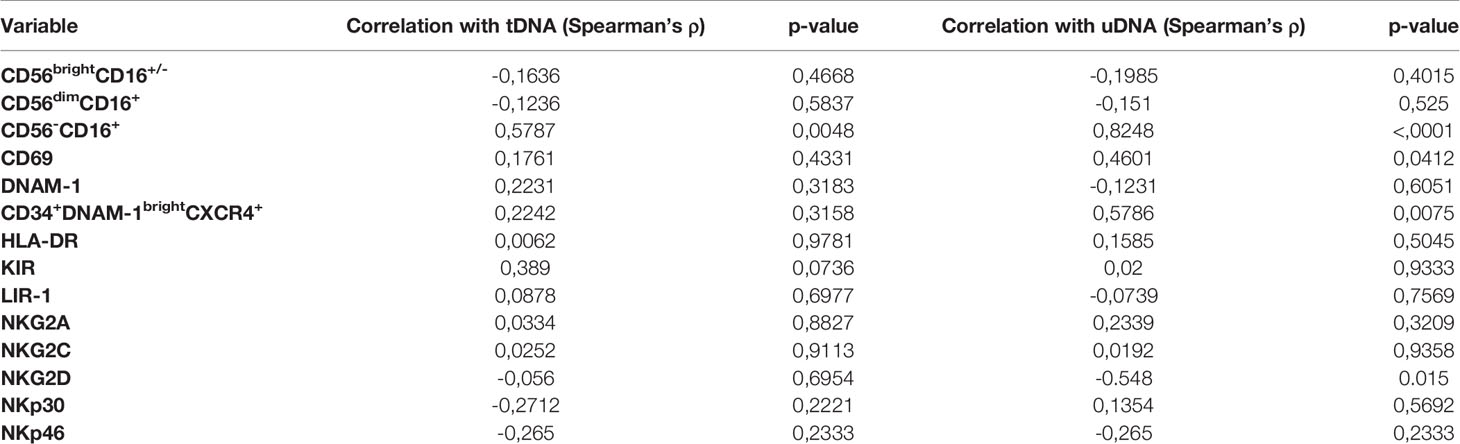
Table 2 Correlation analysis of Natural Killer cell subsets with total and unintegrated HIV HNA (tDNA and uDNA).
Molecule density of CD69 on NK cells (as determined by CD69 MFIr) and CD69 frequency were not correlated with tDNA and directly correlated with uDNA (ρ=0,457, p=0.0412; ρ=0.4601, p=0.0428 respectively)The frequency of CD56-CD16+NK cells which are composed for 50% by mature NK cells and in the rest by CD34-CD56-CD16+CD7- [Figure 3C and (54)], were also directly correlated with uDNA levels (ρ=0.8248, p<0.0001), but not with total or integrated HIV DNA. In addition, Spearman’s analysis revealed an inverse association between the proportion of NKG2D+ NK cells and uDNA (ρ=-0.548, p=0.015), but not with tDNA (ρ=-0.056, p= 0.6954). In contrast, the proportion of NK cells expressing the other activating NKp46 or NKp30 molecules was not correlated with any form of HIV reservoir (Table 2).
Circulating Inflammatory NK Cell Precursors (CD34+DNAM-1brightCXCR4+) Correlate With uDNA Viral Reservoir Forms
Following the present finding of an association between some NK cell subsets, including exhausted NK cells, and recent replication events or residual viral replication as determined by uDNA levels, we tested the hypothesis that continuous NK cell turnover may be observed in patients with higher uDNA levels. In the presence of increased peripheral turnover, there is increased bone marrow precursor activity (45, 46). To verify whether “inflammatory” NK cell precursors are released from the bone marrow in these patients, we used flow cytometry to assay the proportion of CD34+DNAM-1brightCXCR4+ precursors in PBMC. The proportion of these cells was directly correlated with uDNA levels (ρ=0.579, p=0.0075; Spearman’s test) thus supporting the hypothesis of increased lymphoid turnover in patients with higher uDNA levels. A direct correlation was also observed between CD34+DNAM-1brightCXCR4+ precursors and “exhausted” CD56-CD16+NK cells (ρ=0.520, p=0.013; Spearman’s test), thus further supporting the existence of chronic innate immune stimulation in patients with higher levels of uDNA (Table 2).
Univariate logistic regression included relevant associations with uDNA for CD56-CD16+, CD69MFIr, NKG2D, 2yrVCY, NKG2D, tDNA, CD34+DNAM-1brightCXCR4+ and female sex (p<0.25 in all cases). In the multivariable logistic model the frequency of CD56-CD16+, and CD34+DNAM-1brightCXCR4+ cell proportions, confirmed an association with uDNA levels (adjusted OR, aOR 1.61, 95%CI 1.01-2.58, p=0.045 and aOR 1.09, 95%CI 0.97-1.22, p=0.168, respectively) (Table 3).
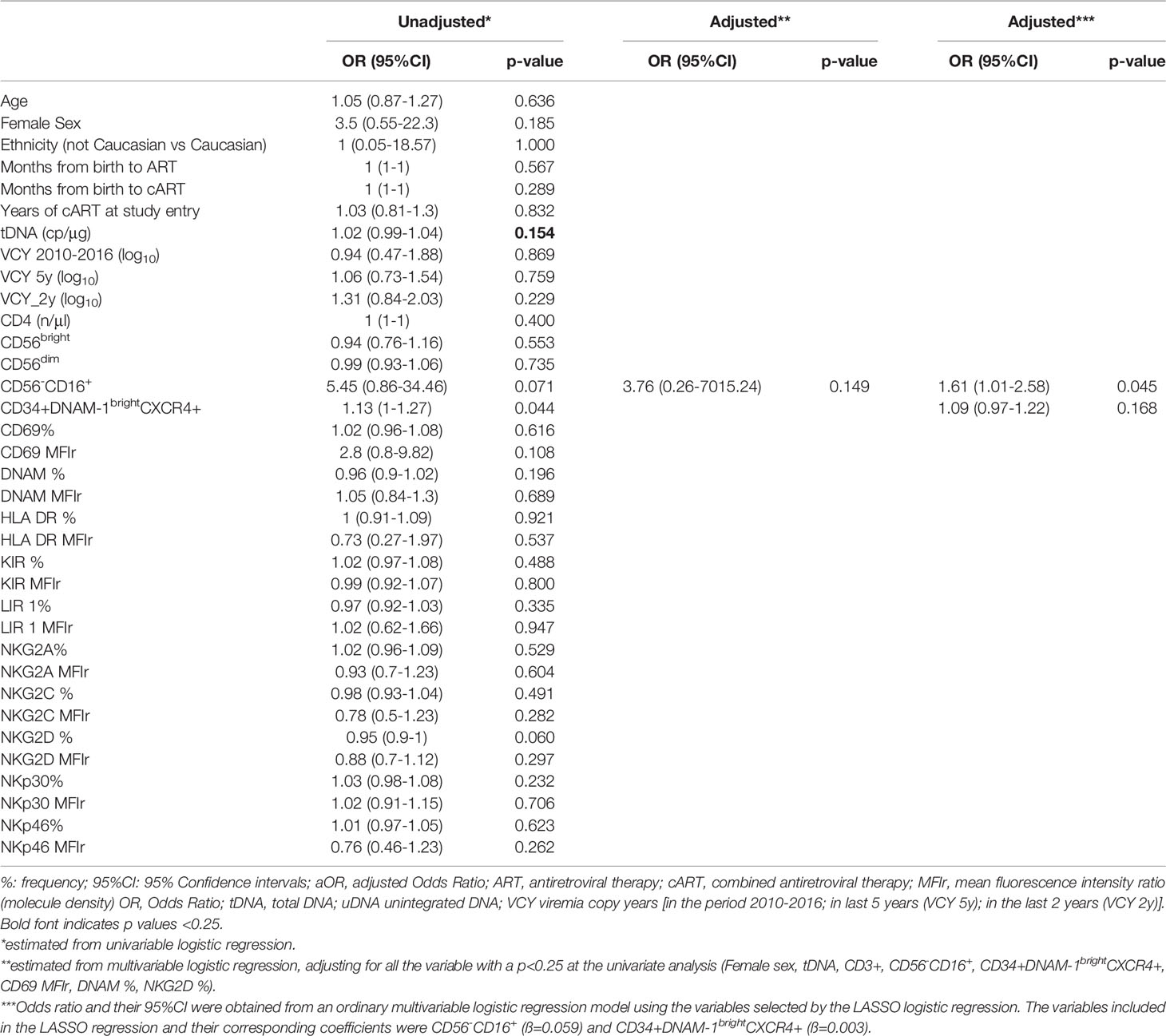
Table 3 Association between presence of unintegrated DNA (uDNA), clinical, laboratory data and Natural Killer (NK) cell molecules expression.
To provide further visual consistency to correlation analysis, we also show stratified patients according to presence (uDNA+) or absence (uDNA-) of linear and circular forms of uDNA in their CD4+ PBMC in Figure 4. Patients with detectable uDNA had significantly increased CD69 expression, increased circulation of inflammatory CD34+ precursors and of “exhausted”CD56-16+ cells (including CD34- inflammatory precursors) and had lower frequencies of NK cells expressing the activating NKG2D receptor.
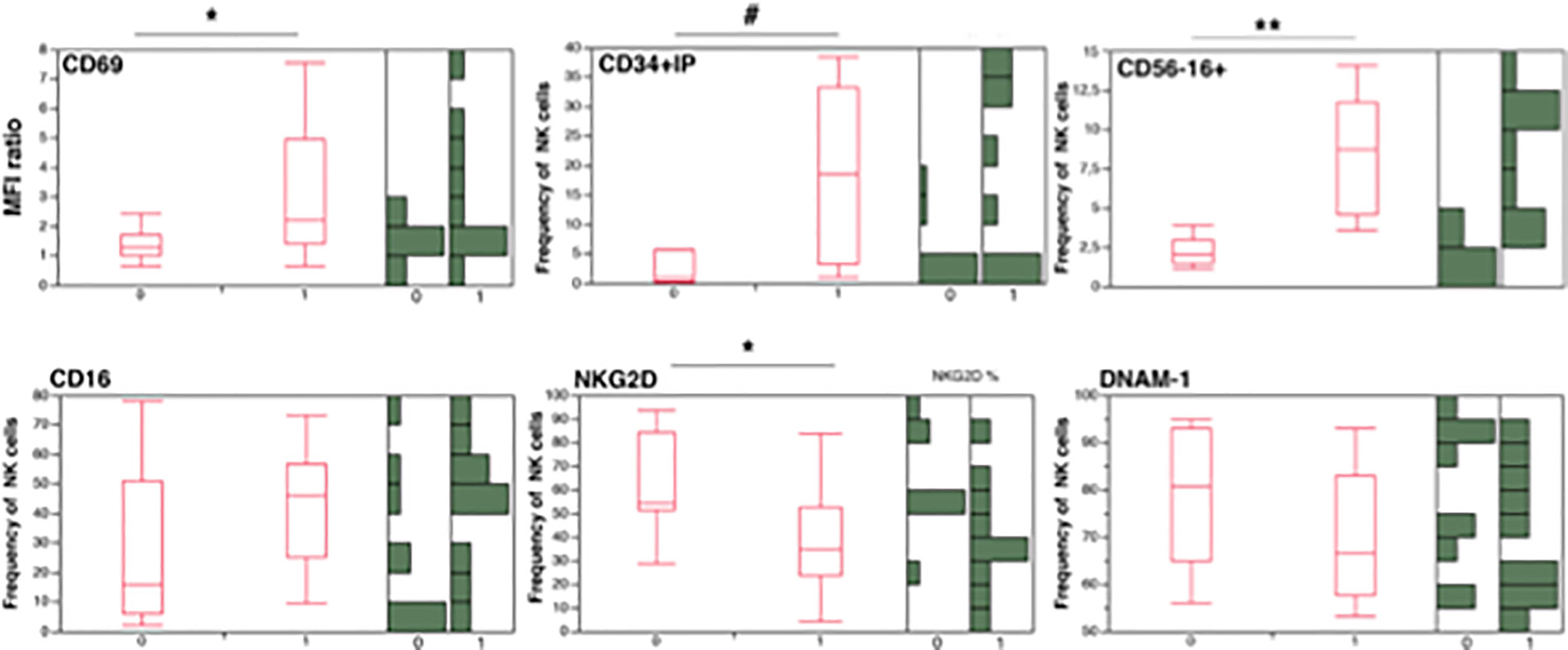
Figure 4 Natural killer and inflammatory precursor cell frequencies in PHIV patients after stratification for presence (1 = uDNA+) or absence (0 =uDNA-) of detectable uDNA. Lateral histograms (Green) show individual measurement frequencies *p<0.05; **p<0.01; #p<0.001; CD34+IP= CD34+ inflammatory precursors (CD34+DNAM-1brightCXCR4+).
These observations raised a relevant issue on whether increased NK cell activation and turnover in uDNA+ virologically suppressed cART-treated patients would be due to defective NK cell function, or rather whether it could reflect increased requirements of NK cell surveillance with conserved NK cell activity thus leading to increased peripheral turnover. To address this question, we therefore studied NK cell function in uDNA+ and uDNA- patients upon cytokine stimulation. NK cell exposure to rIL-12 and rIL-15 was used to reproduce in vitro NK cell interaction with mature DCs. As shown in Figure 5, in a representative uDNA+ patient, the specific IFNγ production (see methods) was 24.6 and 65.8% after 8 and 16 hours (o/n) stimulation while in uDNA- patients NK cell IFNγ production was 14% and 32.8%. Thus, we detected intact and possibly increased IFNγ production upon rIL-12+rIL-15 stimulation in patients with residual replication as shown by uDNA detection in viral reservoir.
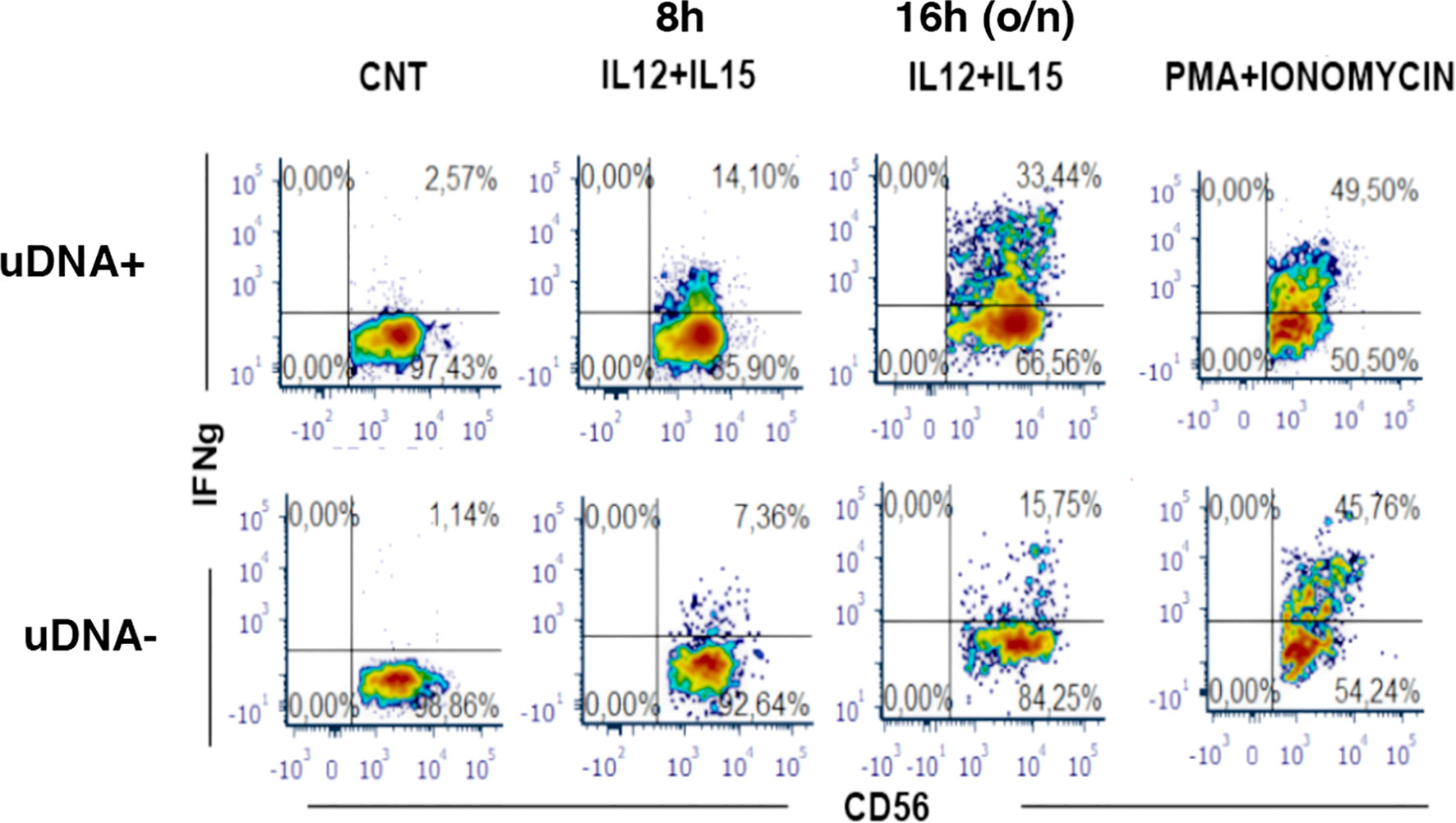
Figure 5 IFN-γ production is conserved in peripheral blood Natural Killer cells from cART-treated and plasma HIV-RNA suppressed patients with presence of uDNA. Flow cytometric analysis of IFN-γ production by CD56+ NK cells upon 8h- and 16h-stimulation in vitro with IL-12+IL-15. Medium alone as negative control (CNT) or maximal stimulus as positive control (PMA+IONO) are also shown. Upper row: uDNA positive patient (uDNA +), lower row: uDNA negative patient (uDNA -).
These data therefore indicate that in virologically suppressed patients with residual replication (uDNA+) NK cells are functional and are continuously recruited as shown by the increase and correlation with “inflammatory” precursors and signal increased peripheral turnover.
This increased NK cell turnover with conserved NK cell function may be poised to control residual replication events escaping cART.
Discussion
In the present work, we observed direct correlations between uDNA levels (which include both linear and circular HIV DNA) and tDNA, and overall activation and turnover of NK cells.
Patients in this cohort were infected perinatally at times with no or little access to immediate or early ART, which was unavailable at the time. Accordingly, they may be representative of PHIV patients currently born in areas where access to ART is still difficult (55). In PHIV, the HIV DNA reservoir set point is rapidly established after acute HIV Infection (56), which corresponds to neonatal age in vertically infected patients, and builds up through all stages of physical and immunological maturation. Early ART induces a sizeable containment of intact proviral sequences and affects adaptive and innate immune parameters (30), while delayed treatment initiation leads to the persistence of intact provirus for at least 7-9 years (57). Accordingly, reduced tDNA levels have been associated with younger age at ART initiation in PHIV patients (58), while higher levels are considered predictive of HIV disease progression (59).
We here observed a direct correlation between uDNA and tDNA, in line with previous reports (60, 61). The transcription and translation of uDNA prior to integration is thought to aid productive viral infection and to result in either preintegration latency or productive infection, and might reflect recent HIV entry in a cell and a viral reservoir, at least in some circumstances (62). Accordingly, these data suggest that uDNA could be used, even in patients with undetectable plasma HIV RNA, to estimate a recently established component of the HIV reservoir.
The present observation that uDNA—despite its lability—provided a reliable estimate of viral burden could be explained by higher levels of total reservoir (tDNA) in PHIV patients with ongoing viral replication that is below the threshold of detectability (25, 26). This point is supported by the observation that uDNA levels were directly associated with tDNA and only had a trend towards association with VCY over shorter periods of observation, thus suggesting that uDNA may reflect recent replication and preintegration events, in line with previous reports (63). VCY directly correlates with higher mortality and organ damage in people living with HIV, and is considered a reliable marker of viral burden over years (35, 36). Since its estimation is unfeasible in patients with incomplete past virological history, uDNA levels could be a more readily available useful surrogate marker for VCY, thus integrating in a single sampling point information on recent virological history of any patient. Accordingly, future work could usefully validate uDNA assays to provide a proxy for ongoing transient blips in replication in larger populations.
In view of the known role of NK cells in HIV-1 reservoir containment (26, 27, 29) and the positive effects of the successful control of viral replication on NK cell phenotypic landscape (30), we studied the relationship between DNA forms (uDNA, tDNA, iDNA) and NK cell phenotype and turnover by analyzing CD34+DNAM-1brightCXCR4+ inflammatory common lymphocyte precursors circulating in PBMC (45, 46). PHIV patients with higher uDNA levels had higher NK cell turnover, as shown by increased exhausted NK cells (CD56-CD16+) and increased CD34+DNAM-1brightCXCR4+ inflammatory precursors. Moreover, the expression of CD69, a marker of NK activation (27), was directly correlated with uDNA in PBMC, indicating higher activation in patients with recent replication events. Taken together, these data are in line with the known direct role of innate immunity in controlling HIV replication. In this context, the presence of residual HIV replication, as expressed by detectable uDNA levels, does not represent a lack of ART activity alone since all patients were successfully on treatment and had undetectable HIV RNA (<50 cp/ml) at the time of testing, but only some of them had undetectable uDNA. Accordingly, persistence of uDNA in only a fraction of them accompanied to NK cell parameters of activation and by increased output of “emergency” precursors to mature NK cells may be interpreted as an imbalance in immune control of HIV in these patients. This is supported by the notion that NK cell function and phenotype contribute to controlling both HIV replication (21, 28–30) and the HIV DNA reservoir (22). At the same time, the high frequency of NK CD34+DNAM-1brightCXCR4+ precursors in the peripheral blood of patients with higher proportions of exhausted CD56-CD16+NK cells could be interpreted as an attempt to rapidly reconstitute mature NK cells undergoing peripheral turnover in patients with detectable uDNA and incomplete immunological control (46). In addition, clonal expansion directed towards diverse antigens including CMV, flu, EBV rather than passive expansion could represent an important aspect in reservoir maintenance. Depending on efficiency of translocation, viral replication, innate immune mechanisms and CD8+CTL function, variable CD4 clonal expansion could be envisaged, with different levels of involvement of NK cells to control the renovating reservoir (64).
Measures of intact proviral copies provide useful information for cure-oriented attempts towards functional cure and do not provide data on recent replication events (12, 65, 66). In the present work, uDNA provides direct information on HIV reservoir dynamic status and correlates with residual “immune-fatigue” and peripheral turnover as shown by the present data on inflammatory precursor and NK cell phenotype/function. This may be regarded as information complementary to other HIV reservoir assays that may become clinically useful to support and monitor functional cure trials.
Some limitations apply to the present analysis. We had no control over ART adherence, possibly confounding study results. In addition, we evaluated HIV DNA in a single sample and could not assess its trend over time over repeated measures, which will be needed in future to transfer this knowledge into clinical usefulness. However, the study has the strengths of evaluating, in a very specific population of vertically infected youths, a new way to estimate recent HIV reservoir generation that cannot be evaluated by other assays and linking reservoir clearance with a study of innate immune response.
In conclusion, in aviremic PHIV patients even after prolonged suppression of viral replication on ART, HIV reservoir displays signs of turnover. Detection of HIV-1 uDNA associates with evidences of NK cell intervention and peripheral turnover without functional impairment. Measures of turnover in HIV reservoir and in immune responses with uDNA and inflammatory precursor assays may offer additional insight to targeted personalized choices for functional HIV eradication.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Ligurian Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LT: Data Curation, Investigation, Formal analysis, Validation, Visualization, writing. FeB: Data Curation, Investigation, Formal analysis, Validation, Visualization. AC Investigation, Formal analysis, Validation, Writing. CO: Data Curation, Investigation, Formal analysis, Validation. FrB: Statistical analysis, Validation. SM: Formal analysis, Validation. MG: Formal analysis, Validation. LM: Investigation, Funding acquisition, Writing. MM: Validation, Writing. AB: Conceptualization, Supervision, Data curation, Validation. AM: Conceptualization, Formal analysis, Funding acquisition, Validation, Writing, Supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants awarded by 5x1000 - Immunity in Cancer Spreading and Metastasis (ISM) Project Code: 21147; IG 5x1000 Molecular Clinical Oncology Extension Program - Project Code: 9962; AIRC INVESTIGATOR GRANT (LM) IG 2017 - Project Code: 19920; IG 2014 - Project Code: 15283; Istituto Superiore di Sanita` (I.S.S.): Programma nazionale di ricerca sull’AIDS, Accordi di collaborazione scientifica 45G.11 (A.D.M.), 40H69 (A.D.M.); MIUR: FISR2020IP_02937 (ADM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank all the boys and girls living with HIV, and their legal guardians, for participating in the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.847816/full#supplementary-material
Supplementary Figure 1 | Description of replication and immune parameters in the patient cohort. One panel for each patient. Each panel represents HIV RNA (cp/mL), HIV DNA (cp/106 PBMC), and CD4 T cell counts (cells/mmc) from 2000 to 2016 in the 22 PHIV enrolled in the study. Triangles indicate uDNA levels at the time of the assay in representative patients.
References
1. Maldarelli F. The Role of HIV Integration in Viral Persistence: No More Whistling Past the Proviral Graveyard. J Clin Invest (2016) 126(2):438–47. doi: 10.1172/JCI80564
2. Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science (1997) 278(5341):1295–300. doi: 10.1126/science.278.5341.1295
3. Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, et al. Rapid Seeding of the Viral Reservoir Prior to SIV Viraemia in Rhesus Monkeys. Nature (2014) 512(7512):74–7. doi: 10.1038/nature13594
4. Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, et al. Presence of an Inducible HIV-1 Latent Reservoir During Highly Active Antiretroviral Therapy. Proc Natl Acad Sci (1997) 94(24):13193–7. doi: 10.1073/pnas.94.24.13193
5. Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, et al. Recommendations for Measuring HIV Reservoir Size in Cure-Directed Clinical Trials. Nat Med (2020) 26(9):1339–50. doi: 10.1038/s41591-020-1022-1
6. Fromentin R, Chomont N. HIV Persistence in Subsets of CD4+ T Cells: 50 Shades of Reservoirs. Semin Immunol (2020) 51:101438. doi: 10.1016/j.smim.2020.101438
7. Sadowski I, Hashemi FB. Strategies to Eradicate HIV From Infected Patients: Elimination of Latent Provirus Reservoirs. Cell Mol Life Sci (2019) 76(18):3583–600. doi: 10.1007/s00018-019-03156-8
8. Hocqueloux L, Avettand-Fènoël V, Jacquot S, Prazuck T, Legac E, Mélard A, et al. Long-term Antiretroviral Therapy Initiated During Primary HIV-1 Infection Is Key to Achieving Both Low HIV Reservoirs and Normal T Cell Counts. J Antimicrobial Chemother (2013) 68(5):1169–78. doi: 10.1093/jac/dks533
9. Viard JP, Burgard M, Hubert JB, Aaron L, Rabian C, Pertuiset N, et al. Impact of 5 Years of Maximally Successful Highly Active Antiretroviral Therapy on CD4 Cell Count and HIV-1 DNA Level. Aids (2004) 18(1):45–9. doi: 10.1097/00002030-200401020-00005
10. Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, et al. HIV-1 DNA Decay Dynamics in Blood During More Than a Decade of Suppressive Antiretroviral Therapy. Clin Infect Dis (2014) 59(9):1312–21. doi: 10.1093/cid/ciu585
11. Learmont J, Tindall B, Evans L, Cunningham A, Cunningham P, Wells J, et al. Long-Term Symptomless HIV-1 Infection in Recipients of Blood Products From a Single Donor. Lancet (1992) 340(8824):863–7. doi: 10.1016/0140-6736(92)93281-q
12. Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, et al. A Quantitative Approach for Measuring the Reservoir of Latent HIV-1 Proviruses. Nature (2019) 566(7742):120–5. doi: 10.1038/s41586-019-0898-8
13. Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, et al. HIV-1 and T Cell Dynamics After Interruption of Highly Active Antiretroviral Therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA (1999) 96(26):15109–14. doi: 10.1073/pnas.96.26.15109
14. Sneller MC, Huiting ED, Clarridge KE, Seamon C, Blazkova J, Justement JS, et al. Kinetics of Plasma HIV Rebound in the Era of Modern Antiretroviral Therapy. J Infect Dis (2020) 222(10):1655–9. doi: 10.1093/infdis/jiaa270
15. Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, et al. Low-Level Viremia Persists For At Least 7 Years in Patients on Suppressive Antiretroviral Therapy. Proc Natl Acad Sci USA (2008) 105(10):3879–84. doi: 10.1073/pnas.0800050105
16. Frenkel LM, Wang Y, Learn GH, McKernan JL, Ellis GM, Mohan KM, et al. Multiple Viral Genetic Analyses Detect Low-Level Human Immunodeficiency Virus Type 1 Replication During Effective Highly Active Antiretroviral Therapy. J Virol (2003) 77(10):5721–30. doi: 10.1128/jvi.77.10.5721-5730.2003
17. Petitjean G, Al Tabaa Y, Tuaillon E, Mettling C, Baillat V, Reynes J, et al. Unintegrated HIV-1 Provides an Inducible and Functional Reservoir in Untreated and Highly Active Antiretroviral Therapy-Treated Patients. Retrovirology (2007) 4(60):1–12. doi: 10.1186/1742-4690-4-60
18. Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, et al. The Decay of the Latent Reservoir of Replication-Competent HIV-1 Is Inversely Correlated With the Extent of Residual Viral Replication During Prolonged anti-retroviral therapy. Nat Med (2000) 6(1):82–5. doi: 10.1038/71577
19. Zack JA, Haislip AM, Krogstad P, Chen IS. Incompletely Reverse-Transcribed Human Immunodeficiency Virus Type 1 Genomes in Quiescent Cells Can Function as Intermediates in the Retroviral Life Cycle. J Virol (1992) 66(3):1717–25. doi: 10.1128/jvi.66.3.1717-1725.1992
20. Orlandi C, Canovari B, Bozzano F, Marras F, Pasquini Z, Barchiesi F, et al. A Comparative Analysis of Unintegrated HIV-1 DNA Measurement as a Potential Biomarker of the Cellular Reservoir in the Blood of Patients Controlling and Non-Controlling Viral Replication. J Transl Med (2020) 18(1):204. doi: 10.1186/s12967-020-02368-y
21. Pierson TC, Zhou Y, Kieffer TL, Ruff CT, Buck C, Siliciano RF. Molecular Characterization of Preintegration Latency in Human Immunodeficiency Virus Type 1 Infection. J Virol (2002) 76(17):8518–31. doi: 10.1128/jvi.76.17.8518-8513.2002
22. Huang S-H, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, et al. Latent HIV Reservoirs Exhibit Inherent Resistance to Elimination By CD8+ T Cells. J Clin Invest (2018) 128(2):876–89. doi: 10.1172/JCI97555
23. Stevenson EM, Ward AR, Truong R, Thomas AS, Huang SH, Dilling TR, et al. HIV-Specific T Cell Responses Reflect Substantive In Vivo Interactions With Antigen Despite Long-Term Therapy. JCI Insight (2021) 6(3):1–14. doi: 10.1172/jci.insight.142640
24. Chun TW, Justement JS, Moir S, Hallahan CW, Ehler LA, Liu S, et al. Suppression of HIV Replication in the Resting CD4+ T Cell Reservoir By Autologous CD8+ T Cells: Implications for the Development of Therapeutic Strategies. Proc Natl Acad Sci USA (2001) 98(1):253–8. doi: 10.1073/pnas.98.1.253
25. Malnati MS, Ugolotti E, Monti MC, Battista D, Vanni I, Bordo D, et al. Activating Killer Immunoglobulin Receptors and HLA-C: A Successful Combination Providing HIV-1 Control. Sci Rep (2017) 7(42470):1–10. doi: 10.1038/srep42470
26. Tomescu C, Duh FM, Hoh R, Viviani A, Harvill K, Martin MP, et al. Impact of Protective Killer Inhibitory Receptor/Human Leukocyte Antigen Genotypes on Natural Killer Cell and T-Cell Function in HIV-1-Infected Controllers. Aids (2012) 26(15):1869–78. doi: 10.1097/QAD.0b013e32835861b0
27. Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, et al. Natural Killer Cells in HIV Controller Patients Express an Activated Effector Phenotype and Do Not Up-Regulate NKp44 on IL-2 Stimulation. Proc Natl Acad Sci USA (2013) 110(29):11970–5. doi: 10.1073/pnas.1302090110
28. Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential Natural Killer Cell-Mediated Inhibition of HIV-1 Replication Based on Distinct KIR/HLA Subtypes. J Exp Med (2007) 204(12):3027–36. doi: 10.1084/jem.20070695
29. Marras F, Casabianca A, Bozzano F, Ascierto ML, Orlandi C, Di Biagio A, et al. Control of the HIV-1 DNA Reservoir Is Associated In Vivo and In Vitro With NKp46/NKp30 (CD335 CD337) Inducibility and Interferon Gamma Production by Transcriptionally Unique NK Cells. J Virol (2017) 91(23):1–19. doi: 10.1128/jvi.00647-17
30. Garcia-Broncano P, Maddali S, Einkauf KB, Jiang C, Gao C, Chevalier J, et al. Early Antiretroviral Therapy in Neonates With HIV-1 Infection Restricts Viral Reservoir Size and Induces a Distinct Innate Immune Profile. Sci Transl Med (2019) 11(520):1–27. doi: 10.1126/scitranslmed.aax7350
31. Uprety P, Chadwick EG, Rainwater-Lovett K, Ziemniak C, Luzuriaga K, Capparelli EV, et al. Cell-Associated HIV-1 DNA and RNA Decay Dynamics During Early Combination Antiretroviral Therapy in HIV-1-Infected Infants. Clin Infect Dis (2015) 61(12):1862–70. doi: 10.1093/cid/civ688
32. Avettand-Fenoel V, Blanche S, Le Chenadec J, Scott-Algara D, Dollfus C, Viard JP, et al. Relationships Between HIV Disease History and Blood HIV-1 DNA Load in Perinatally Infected Adolescents and Young Adults: The ANRS-EP38-IMMIP Study. J Infect Dis (2012) 205(10):1520–8. doi: 10.1093/infdis/jis233
33. Ungaro R, Taramasso L, Bruzzone B, Vicenti I, Galli L, Borghi V, et al. Prevalence of Acquired Resistance Mutations in a Large Cohort of Perinatally Infected HIV-1 Patients. Clin Microbiol Infect (2019) 25(11):1443–6. doi: 10.1016/j.cmi.2019.07.004
34. Contreras GA, Bell CS, Del Bianco G, Pérez N, Benjamins L, Kleinosky MT, et al. Incidence and Predictors of Antiretroviral Resistance in Perinatally HIV-1 Infected Children and Adolescents. J Infect (2016) 72(3):353–61. doi: 10.1016/j.jinf.2015.12.005
35. Mugavero MJ, Napravnik S, Cole SR, Eron JJ, Lau B, Crane HM, et al. Viremia Copy-Years Predicts Mortality Among Treatment-Naive HIV-Infected Patients Initiating Antiretroviral Therapy. Clin Infect Dis (2011) 53(9):927–35. doi: 10.1093/cid/cir526
36. Wang R, Haberlen SA, Palella FJ Jr, Mugavero MJ, Margolick JB, Macatangay BJC, et al. Viremia Copy-Years and Mortality Among Combination Antiretroviral Therapy-Initiating HIV-Positive Individuals: How Much Viral Load History Is Enough? AIDS (2018) 32(17):2547–56. doi: 10.1097/QAD.0000000000001986
37. Sarteschi G, Di Biagio A, Focà E, Taramasso L, Bovis F, Celotti A, et al. Viremia Copy-Years and Risk of Estimated Glomerular Filtration Rate Reduction in Adults Living With Perinatal HIV Infection. PloS One (2020) 15(10):e0240550. doi: 10.1371/journal.pone.0240550
38. Jones RB, Kovacs C, Chun TW, Ostrowski MA. Short communication: HIV Type 1 Accumulates in Influenza-Specific T Cells in Subjects Receiving Seasonal Vaccination in the Context of Effective Antiretroviral Therapy. AIDS Res Hum Retroviruses (2012) 28(12):1687–92. doi: 10.1089/aid.2012.0115
39. Miller RL, Ponte R, Jones BR, Kinloch NN, Omondi FH, Jenabian M-A, et al. HIV Diversity and Genetic Compartmentalization in Blood and Testes During Suppressive Antiretroviral Therapy. J Virol (2019) 93(17):e00755–19. doi: 10.1128/JVI.00755-19
40. Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, et al. Persistent HIV-1 Replication Maintains the Tissue Reservoir During Therapy. Nature (2016) 530(7588):51–6. doi: 10.1038/nature16933
41. Newell ML, Dunn D, Peckham C, De Maria A, Ferrazin A, De Rossi A, et al. Detection of Virus in Vertically Exposed HIV-Antibody-Negative Children. Lancet (1996) 347(8996):213–5. doi: 10.1016/S0140-6736(96)90401-8
42. European Collaborative Study. Children Born to Women With HIV-1 Infection: Natural History and Risk of Transmission. Lancet (1991) 337(8736):253–60. doi: 10.1016/0140-6736(91)90866-N
43. Gazzarata R, Giannini B, Giacomini M. A SOA-Based Platform to Support Clinical Data Sharing. J Healthcare Eng (2017) 2017:2190679. doi: 10.1155/2017/2190679
44. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. In: Department of Health and Human Services (2021). Available at: https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/AdultandAdolescentGL.pdf.
45. Bozzano F, Marras F, Ascierto ML, Cantoni C, Cenderello G, Dentone C, et al. ‘Emergency Exit’ of Bone-Marrow-Resident CD34(+)DNAM-1(bright)CXCR4(+)-Committed Lymphoid Precursors During Chronic Infection and Inflammation. Nat Commun (2015) 6:8109. doi: 10.1038/ncomms9109
46. Bozzano F, Della Chiesa M, Pelosi A, Antonini F, Ascierto ML, Del Zotto G, et al. HCMV-Controlling NKG2C(+) NK Cells Originate From Novel Circulating Inflammatory Precursors. J Allergy Clin Immunol (2021) 147(6):2343–57. doi: 10.1016/j.jaci.2020.12.648
47. Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, et al. Characterization of CD56-/CD16+ Natural Killer (NK) Cells: A Highly Dysfunctional NK Subset Expanded in HIV-Infected Viremic Individuals. Proc Natl Acad Sci USA (2005) 102(8):2886–91. doi: 10.1073/pnas.0409872102
48. Surdo M, Cortese MF, Orlandi C, Di Santo F, Aquaro S, Magnani M, et al. Different Kinetics of Viral Replication and DNA Integration in the Main HIV-1 Cellular Reservoirs in the Presence and Absence of Integrase Inhibitors. Antiviral Res (2018) 160:165–74. doi: 10.1016/j.antiviral.2018.10.017
49. Casabianca A, Orlandi C, Canovari B, Scotti M, Acetoso M, Valentini M, et al. A Real Time PCR Platform for the Simultaneous Quantification of Total and Extrachromosomal HIV DNA Forms in Blood of HIV-1 Infected Patients. PloS One (2014) 9(11):e111919. doi: 10.1371/journal.pone.0111919
50. Payne H, Chan MK, Watters SA, Otwombe K, Hsiao NY, Babiker A, et al. Early ART-Initiation and Longer ART Duration Reduces HIV-1 Proviral DNA Levels in Children From the CHER Trial. AIDS Res Ther (2021) 18(1):63. doi: 10.1186/s12981-021-00389-1
51. Avettand-Fenoel V, Lechenadec J, Diallo MS, Fillion M, Melard A, Samri A, et al. Initiating Antiretroviral Treatment Early in Infancy Has Long-term Benefits on the Human Immunodeficiency Virus Reservoir in Late Childhood and Adolescence. Clin Infect Dis (2021) 73(11):e4214–22. doi: 10.1093/cid/ciaa1931
52. Bisio F, Bozzano F, Marras F, Di Biagio A, Moretta L, De Maria A. Successfully Treated HIV-Infected Patients Have Differential Expression of NK Cell Receptors (NKp46 and NKp30) According to AIDS Status at Presentation. Immunol Lett (2013) 152(1):16–24. doi: 10.1016/j.imlet.2013.03.003
53. Lichtfuss GF, Cheng WJ, Farsakoglu Y, Paukovics G, Rajasuriar R, Velayudham P, et al. Virologically Suppressed HIV Patients Show Activation of NK Cells and Persistent Innate Immune Activation. J Immunol (2012) 189(3):1491–9. doi: 10.4049/jimmunol.1200458
54. Bozzano F, Della Chiesa M, Pelosi A, Antonini F, Ascierto ML, Del Zotto G, et al. HCMV-Controlling NKG2C(+) NK Cells Originate From Novel Circulating Inflammatory Precursors. J Allergy Clin Immunol (2021) 147(6):2343–57. doi: 10.1016/j.jaci.2020.12.648
55. O. World Health. Access to Antiretroviral Drugs in Low- and Middle-Income Countries: Technical Report July 2014. World Health Org Geneva (2014). Available at: https://apps.who.int/iris/bitstream/handle/10665/128150/9789241507547_eng.pdf;jsessionid=73267D3D606AF638C95AEEC0242E38B4?sequence=1.
56. Ananworanich J, Chomont N, Eller LA, Kroon E, Tovanabutra S, Bose M, et al. HIV DNA Set Point is Rapidly Established in Acute HIV Infection and Dramatically Reduced by Early ART. EBioMedicine (2016) 11:68–72. doi: 10.1016/j.ebiom.2016.07.024
57. Katusiime MG, Halvas EK, Wright I, Joseph K, Bale MJ, Kirby-McCullough B, et al. Intact HIV Proviruses Persist in Children Seven to Nine Years after Initiation of Antiretroviral Therapy in the First Year of Life. J Virol (2020) 94(4):e01519–19. doi: 10.1128/JVI.01519-19
58. Foster C, Domínguez-Rodríguez S, Tagarro A, Gkouleli T, Heaney J, Watters S, et al. The CARMA Study: Early Infant Antiretroviral Therapy—Timing Impacts on Total HIV-1 DNA Quantitation 12 Years Later. J Pediatr Infect Dis Soc (2020) 10(3):295–301. doi: 10.1093/jpids/piaa071
59. Tsiara CG, Nikolopoulos GK, Bagos PG, Goujard C, Katzenstein TL, Minga AK, et al. Impact of HIV Type 1 DNA Levels on Spontaneous Disease Progression: A Meta-Analysis. AIDS Res Hum Retroviruses (2011) 28(4):366–73. doi: 10.1089/aid.2011.0032
60. Orlandi C, Canovari B, Bozzano F, Marras F, Pasquini Z, Barchiesi F, et al. A Comparative Analysis of Unintegrated HIV-1 DNA Measurement as a Potential Biomarker of the Cellular Reservoir in the Blood of Patients Controlling and Non-Controlling Viral Replication. J Trans Med (2020) 18(1):204. doi: 10.1186/s12967-020-02368-y
61. Trémeaux P, Lenfant T, Boufassa F, Essat A, Mélard A, Gousset M, et al. Increasing Contribution of Integrated Forms to Total HIV DNA in Blood During HIV Disease Progression From Primary Infection. EBioMedicine (2019) 41:455–64. doi: 10.1016/j.ebiom.2019.02.016
62. Sloan RD, Wainberg MA. The Role of Unintegrated DNA in HIV Infection. Retrovirology (2011) 8(1):52. doi: 10.1186/1742-4690-8-52
63. Sharkey M, Triques K, Kuritzkes DR, Stevenson M. In Vivo Evidence for Instability of Episomal Human Immunodeficiency Virus Type 1 cDNA. J Virol (2005) 79(8):5203–10. doi: 10.1128/jvi.79.8.5203-5210.2005
64. Lau CY, Adan MA, Maldarelli F. Why the HIV Reservoir Never Runs Dry: Clonal Expansion and the Characteristics of HIV-Infected Cells Challenge Strategies to Cure and Control HIV Infection. Viruses (2021) 13(12):1–43. doi: 10.3390/v13122512
65. Gandhi RT, Cyktor JC, Bosch RJ, Mar H, Laird GM, Martin A, et al. Selective Decay of Intact HIV-1 Proviral DNA on Antiretroviral Therapy. J Infect Dis (2021) 223(2):225–33. doi: 10.1093/infdis/jiaa532
66. Papasavvas E, Azzoni L, Ross BN, Fair M, Yuan Z, Gyampoh K, et al. Intact Human Immunodeficiency Virus (HIV) Reservoir Estimated by the Intact Proviral DNA Assay Correlates With Levels of Total and Integrated DNA in the Blood During Suppressive Antiretroviral Therapy. Clin Infect Dis (2021) 72(3):495–8. doi: 10.1093/cid/ciaa809
Keywords: HIV, reservoir, natural killer, residual replication, inflammatory precursor, CD34, ART, unintegrated HIV-DNA
Citation: Taramasso L, Bozzano F, Casabianca A, Orlandi C, Bovis F, Mora S, Giacomini M, Moretta L, Magnani M, Di Biagio A and De Maria A (2022) Persistence of Unintegrated HIV DNA Associates With Ongoing NK Cell Activation and CD34+DNAM-1brightCXCR4+ Precursor Turnover in Vertically Infected Patients Despite Successful Antiretroviral Treatment. Front. Immunol. 13:847816. doi: 10.3389/fimmu.2022.847816
Received: 03 January 2022; Accepted: 21 March 2022;
Published: 26 April 2022.
Edited by:
Remi Cheynier, U1016 Institut Cochin (INSERM), FranceCopyright © 2022 Taramasso, Bozzano, Casabianca, Orlandi, Bovis, Mora, Giacomini, Moretta, Magnani, Di Biagio and De Maria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea De Maria, NTUyMjVAdW5pZ2UuaXQ=
†Present address: Federica Bozzano, Policlinico San Martino Hospital, Genoa, Italy
‡These authors have contributed equally to this work
 Lucia Taramasso
Lucia Taramasso Federica Bozzano
Federica Bozzano Anna Casabianca
Anna Casabianca Chiara Orlandi
Chiara Orlandi Francesca Bovis3
Francesca Bovis3 Mauro Giacomini
Mauro Giacomini Lorenzo Moretta
Lorenzo Moretta Mauro Magnani
Mauro Magnani Antonio Di Biagio
Antonio Di Biagio