- 1Cellular Immunology Unit, Department of Immunology, Transplantation and Infectious Diseases, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Ospedale San Raffaele, Milan, Italy
- 2Università Vita-Salute San Raffaele, Milan, Italy
The human microbiota is a unique set of microorganisms colonizing the human body and evolving within it from the very beginning. Acting as an insider, the microbiota provides nutrients, and mutualistically interacts with the host’s immune system, thus contributing to the generation of barriers against pathogens. While a strong link has been documented between intestinal dysbiosis (i.e., disruption to the microbiota homeostasis) and diseases, the mechanisms by which commensal bacteria impact a wide spectrum of mucosal and extramucosal human disorders have only partially been deciphered. This is particularly puzzling for multiple myeloma (MM), a treatable but incurable neoplasia of plasma cells that accumulate in the bone marrow and lead to end-organ damage. Here we revise the most recent literature on data from both the bench and the bedside that show how the gut microbiota modulates cancer immunity, potentially impacting the progression of asymptomatic monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM) to full blown MM. We also explore the effect of the gut microbiome on hematopoietic stem cell transplantation, chemotherapy, immunomodulating therapy and cancer immunotherapy in MM patients. Additionally, we identify the most cogent area of investigation that have the highest chance to delineate microbiota-related and pathobiology-based parameters for patient risk stratification. Lastly, we highlight microbiota-modulating strategies (i.e., diet, prebiotics, probiotics, fecal microbiota transplantation and postbiotics) that may reduce treatment-related toxicity in patients affected by MM as well as the rates of undertreatment of SMM patients.
Introduction
The very moment we open our eyes to the world, our body has already been colonized by symbiotic microorganisms that will increase in number and species and become established through the first years of life into our own microbiota (1). This also is the time in which the immune system gets forged to recognize and eliminate pathogens (2) while acquiring tolerance to the self (3) and the host microbiota that becomes an extended self (4). Of note, children share a stereotypic immune system development that is microbiota-driven (5). A strong link between gut microbiota and immune dynamics is also found in adults undergoing immune reconstitution after hematopoietic stem cell transplantation (HSCT) (6). The mechanisms by which the microbiota interacts with the host immune system, and eventually modulates and/or gets modulated by the immune system is under investigation (7–30). The modulatory effects of the microbiota on the immune system are not only local, but also systemic and affect distant organs (31). Therefore, disruption to the microbiota homeostasis (i.e., dysbiosis) associates with diseases that span from allergy (32) and other immune-mediated diseases (33) to obesity (34), psychiatric disorders (35) and cancer (36). The microbiota also directly impacts human pathologies. As few examples, selected species of Escherichia coli alkylate DNA on adenosine residues and induce double strand breaks, eventually favoring mutations in colorectal cancer (37); intestinal commensal bacteria lead to androgen biosynthesis, thus promoting endocrine resistance in prostate cancer (38). Additionally, the gut microbiota influences susceptibility of cancer patients to surgery (39), chemotherapy (40), radiotherapy (41) and immunotherapy (42). Indeed, fecal microbiota transplant (FMT) from donors who achieved complete response to anti-PD-1 monotherapy into anti-PD-1-refractory melanoma patients resulted safe, feasible and associated with clinical responses and improved cancer control by the immune system (43). Modulation of the gut microbiome has also been attempted in patients affected by hematologic malignancies because the microbiota is highly susceptible to most of the treatments proposed to these patients (44), and microbiota translocation into the bloodstream of patients with therapy-induced immunosuppression contributes to morbidity and mortality (45). In turn, treatment-induced dysbiosis can be corrected by probiotics, prebiotics and FMT (46).
We focused here on multiple myeloma (MM), a treatable but incurable neoplasia of plasma cells that mainly accumulate in the bone marrow (BM) causing anemia, hypercalcemia, renal insufficiency, and bone lesions (47). Full blown MM is often preceded by two potentially curable asymptomatic diseases: monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM) (48). Thus, identifying mechanisms by which MGUS and SMM patients progress to full-blown MM would represent a substantial clinical advancement. Microbiota-modulated immunity has been proposed as a mechanism of progression from SMM to MM (49). Additionally, several clinical and preclinical studies have highlighted the role of the gut microbiota in MM patients’ response to therapies (50, 51). Therefore, MM and its asymptomatic phases are examples of diseases in which alteration of the gut flora impacts disease progression, response to therapy and treatment-related toxicities. Information gathered in MM can be translated to other human diseases.
While we refer all interested readers to a more comprehensive review on this topic (52), in our short and more clinically-oriented paper we will start reporting data from both the bench and the bedside that show how the gut microbiota modulates MM. We will define the role of IL-17 in the crosstalk between MM and the intestinal microbiota. We will highlight how the gut microbiota is modified and can modify patients’ susceptibility to treatments. We will conclude with experimental and clinically strategies that can modulate the gut microbiota in patients affected by MGUS, SMM or MM.
Gut Microbiota and Multiple Myeloma
Diet can profoundly affect the gut microbiota. While a link between diet an MM has been investigated for decades (53), only recently a correlation between microbiome and progression of MM has been searched for. As for other human diseases (4), the gut microbiota from MM patients has a reduced richness in bacterial species (54). Bacteroides, Clostriudium leptum and Rothia are enriched in MM patients when compared to family members, who usually share the microbiome (55). Interestingly, the level of Clostridium leptum positively correlates with ISS stage in MM patients. Because Clostridium leptum and Rothia are butyric acid-producing bacteria, the authors hypothesized dysregulation of the sugar metabolism in the intestine of MM patients (54). Whether short-chain fatty acids (SCFAs) have direct effects on neoplastic plasma cells or anti-MM immunity (56) needs to be investigated.
Opportunistic nitrogen-recycling bacteria such as Klebsiella and Streptococcus are enriched in the gut microbiota of MM patients (57). Accordingly, MM patients showed increased urease and glutamine synthetase activity in their feces and more urea and less ammonia in their blood than healthy subjects. Mice treated with FMT from MM patients and challenged with 5TGM1 MM cells experienced accelerated tumor burden that associated with elevated L-glutamine levels in their blood (57). Thus, gut colonization by nitrogen-recycling bacteria accelerates MM by making available L-glutamine. Building on this knowledge, fluoroglutamine might work as PET tracer in MM (58).
Altogether, these findings suggest that metabolites produced by a dysbiotic microbiota impact MM progression. Indeed, genetic diversity in the microbiome provides a wide variety of enzymes that convert polysaccharides and oligosaccharides into SCFAs like acetate, propionate, and butyrate (59). In mice, acetate, butyrate and pentanoate exert partially overlapping effects on T cells, B cells and dendritic cells (60, 61). By activating receptors expressed on intestinal epithelial cells and hematopoietic cells, SCFAs reduce inflammation (62, 63). In the context of autologous HSCT, the presence of the butyrate-producing bacteria Eubacterium hallae or Faecalibacterium prausnitzii in the gut microbiota of MM patients positively correlates with increased rates of minimal residual disease negativity (64). In rats, butyrate administration ameliorates colitis by increasing numbers of regulatory T cell (Treg) and suppressing levels of the pro-inflammatory cytokine IL-17A in both plasma and colonic mucosa (65). Conversely, commensals favoring the expansion of intestinal T helper-17 (Th17) cells may accelerate MM progression in mice (66). Thus, a strong link exists among commensal bacteria, Treg/Th17 cell balance and MM.
IL-17 Swings the Balance Between Gut Microbiota and Multiple Myeloma
A different composition of the gut microbiota induces different immune responses locally and systemically (67). By producing IL-17, Th17 lymphocytes maintain a healthy intestinal mucosa and limit bacteria over-growth (68). Conversely, Treg cells allow tolerance to the extended self and limit excessive inflammation (69). Imbalance in the crosstalk between the host and the microbiota leads to excessive immune activation and expansion of Th17 cells.
Th17 cells are pathogenic in MM (70–72). Alexandrakis et al. (73) originally observed higher IL-17A levels in the peripheral blood of MM patients of stage II and III compared to stage I and a positive correlation with VEGF and microvessel density in the BM, thus hypothesizing a proangiogenic role for IL-17A in MM. Th17 cells are enriched in the BM of MM patients (70), where they support local inflammation and favor bone disease by promoting osteoclast differentiation (71). IL-17A, whose levels are increased in the BM of MM patients, contributes to neoplastic plasma cell survival and proliferation through the autocrine release of IL-6 (74). Also mouse neoplastic plasma cells express functional 17RA/RC (66), and in vivo, MM plasma cells upregulate cell proliferation and cell-cycle progression pathways if stimulated with IL-17A (75); while genes related to antigen processing and presentation and leukocyte trafficking and activation are downregulated in response to IL-17A (75). Neoplastic plasma cells can produce IL-17A, and treatment with antibodies specific for human IL-17A delayed growth of human MM in immunodeficient mice (76). These findings spurred a clinical trial with anti-IL-17A antibodies in MM patients (NCT03111992). IL-17A also resulted detrimental in the context of allogeneic-HSCT (77, 78). These works also identified IL-17A as the main path linking microbiota to Graft-Versus-Host Disease (GVHD). Indeed, in the absence of donor IL-17A, HSCT was more effective in controlling mouse MM (75).
While IL-17A is also elevated in some MGUS patients (79), a direct link has been reported between gut microbiota, IL-17A and progression of asymptomatic MM to full-blown MM (66). In Vk*MYC mice developing de novo MM that invariably evolves from asymptomatic to symptomatic MM (80), Prevotella heparinolytica, a human commensal (81), induces expansion of Th17 cells in the intestinal mucosa. Gut-born Th17 cells migrate to the BM, where they promote neoplastic plasma cell proliferation and progression from asymptomatic to symptomatic MM. At odds, P. melaninogenica restrain MM progression by limiting expansion of Th17 cells. Similarly, in SMM patients, high levels of BM IL-17 predicted faster progression to active MM (66). Lack of IL-17A in MM mice, or treatment with antibiotics or antibodies blocking IL-17/IL-17R interactions delayed disease progression (66). Thus, targeting the microbiota-IL17A axis in SMM patients might block disease progression.
Role of the Gut Microbiota in Hematopoietic Stem Cell Transplantation
HSCT is a primary treatment for hematological malignancies and can be subdivided into autologous or allogeneic based on the use of self or donor-compatible HSCs, respectively (82). While the standard of care for MM patients is to receive high-dose chemotherapy followed by autologous-HSCT, allogeneic-HSCT can be proposed as part of a clinical trial and often associates with drawbacks like GVHD, a clinical condition in which the grafted immune system attacks tissues of the transplant recipient (83–85). The gut microbiota appears directly linked to allogeneic-HSCT success (86) and risk of GVHD (87). A large study including 111 MM patients across multiple clinical centers reported that lower diversity of intestinal microbiota associates with higher risk of transplant- and GHVD-related deaths (88).
Khan and colleagues highlighted interesting similarities in gut microbiota dysbiosis after both autologous- and allogeneic-HSCT in MM patients (89). Changes in the bacteriome and mycobiome also modulate early toxicity and the rate of neutrophil engraftment after autologous-HSCT (90). Generally, changes in bacterial abundances and species were linked to conditioning regimen or patient’s treatments (91, 92). Reduced bacterial diversity associates with increased immune activation, probability of relapse, GVHD and overall mortality (88, 91, 93–98). Enterobacteria and Proteobacteria abundance correlates with increasing probability of GVHD, pulmonary or gastro-intestinal complications and infection (92, 96, 98, 99). More in depth, enrichment in Clostridium difficile and Rothia associated with both autologous- and allogeneic-HSCT-related adverse events in MM patients (100), while colonization of species like Akkermansia muciniphila or Enterococcus faecium predispose to the dominance of other bacteria and further detrimental systemic consequences for patients (101). On the other hand, enrichment in Ruminococcaceae, Lachnospiraceae and Clostridiales correlates with higher transplantation efficiency and reduced GVHD (93, 102). Others identified Blautia, Actinomyces, Prevotella and Eubacterium limosum as commensals with protective effects (91, 98, 101).
Interestingly, a genus of bacteria may harbor species with opposing effects on the immune system (4). One example is Clostridium difficile that may increase risk of GVHD, disease relapse or mortality, contrary to other family members (6, 93, 99, 100). Different effects by different strains belonging to the same genus correlate to different metabolic activities. Butyrate and propionate can reduce GVHD, improve HSCT outcomes but also protect mice against radiation-induced injuries of the hematopoietic compartment (103). The microbiome also regulates energy uptake to improve allogeneic-HSCT outcomes in mice (97, 104), or lactose metabolism whose reduction, through diet or variation in microbial composition, limits pathological bacteria expansion and adverse effects in patients (105).
Impact of the Gut Microbiota in Therapies for MM Patients
Because the gut microbiota can influence response to therapy and toxicity across different treatments (40), also in MM patients a link between gut microbiota and response to therapies has been investigated.
Proteasome Inhibitors and Immunomodulating Drugs
MM patients benefit from combinations of proteasome inhibitors (PIs; bortezomib, carfilzomib, ixazomib) and immunomodulating drugs (IMiDs; thalidomide, lenalidomide, pomalidomide, dexamethasone) (106). PI treatment is burdened with gastrointestinal toxicity (107) that may also depend on gut microbiota (51). While the work by Pianko and colleagues did not investigate gastrointestinal toxicity in MM patients treated with PIs and/or IMiDs followed by autologous-HSCT, it showed an association between deep treatment response and enrichment in Eubacterium hallii and Faecalibacterium prausnitzii (64). F. prausnitzii usually associates with gut health, and both bacteria are SCFA producers (108), thus suggesting a potential link between SCFA producing bacteria a reduced gastrointestinal toxicity.
A higher prevalence of beneficial bacteria belonging to Bifidobacterium and Lactobacillus genus has been found in mice exposed to dexamethasone (109). Significantly decreased IL-17 levels in the intestinal mucosa and reduced colitis susceptibility were observed in mice receiving FMT from dexamethasone-conditioned donor mice (109). Thus, the immunosuppressive effect of dexamethasone may in part be supported by the induction of an anti-inflammatory microbiota (110).
Chemotherapies
Cyclophosphamide (CTX) is an alkylating agent that stimulates type I interferon response and Th1/Th17 lymphocyte polarization (111). Because of the dual role of Th17 cells in MM pathogenesis and gut homeostasis, the effect of CTX on intestinal microbiota is relevant to understand the therapeutic outcomes in MM patients. Bacteroidetes and Verrucomicrobia are significantly reduced in mice treated with CTX (112). CTX treatment also increases intestinal barrier permeability and translocation of Gram-positive commensals, which favor Th1 and Th17 cell differentiation and anti-tumor immunity (113, 114).
Similarly, melphalan administration in rats causes severe injury to the small intestine, weight loss and infections (115). These effects recapitulate the toxicity observed in patients and limit melphalan administration to elderly subjects (116). Indeed, melphalan induces dysbiosis, reduction of SCFA production and bacterial translocation (115). Thus, replenishing SCFA or normalization of microbiota composition might re-establish intestinal homeostasis and improve drug tolerability.
Cancer Immunotherapies
A strong correlation exists between gut microbiome and response to immune checkpoint inhibitors (ICIs) (43, 117–123). Interestingly, PD-L1 is expressed on malignant plasma cells, and PD-L1 predicts progression of SMM patients to MM (124). However, early-phase clinical trials with ICI as single agent showed modest activity in MM patients, whereas the combination with PIs ignited toxic events (125, 126). The latter might be linked to PI gastrointestinal toxicity (107). Andrews et al. associated defined microbiota signatures and abundance of Bacteroides intestinalis with upregulation of mucosal IL-1β and IL-17 in patients with ICI-related gastrointestinal adverse events (127). Administration of Bifidobacterium was sufficient to ameliorate ICI-related immunopathology in mice without dampening antitumor immunity (128). This strategy might also be adopted in MM patients.
The gut microbiota can promote the expansion and persistence of adoptively transferred cytotoxic T cells (CTLs) both in humans and mice (129). In mice, pentanoate and butyrate enhance anti-tumor activity of CTLs and chimeric antigen receptor (CAR)-T cells through metabolic and epigenetic reprogramming (130). A preliminary study on hematological patients receiving CAR-T cell therapy showed enrichment in Ruminococcacaeae, and Lachnospiraceae, which produce SCFA, in patients achieving complete remission (131). Because CAR-T cells are proposed to MM patients (132), it will be interesting to investigate how modulation of the gut microbiota can impact this treatment (133, 134).
Conclusions
The information gathered so far strongly suggest that interfering with the insider (i.e., modifying the intestinal microbiota) may limit MM progression and increase susceptibility to therapies. These strategies include dietary intervention, administration of prebiotics, probiotics and postbiotics, but also FMT (Figure 1). While the concept of nutritional support is well established in the clinical practice (135), a more precise modulation of the gut microbiota has been attempted only recently, thanks to the acquisition of technologies for microbiome identification (36). Interestingly, unique microbial signatures can also be found in the peripheral blood within and between several cancer types (136).
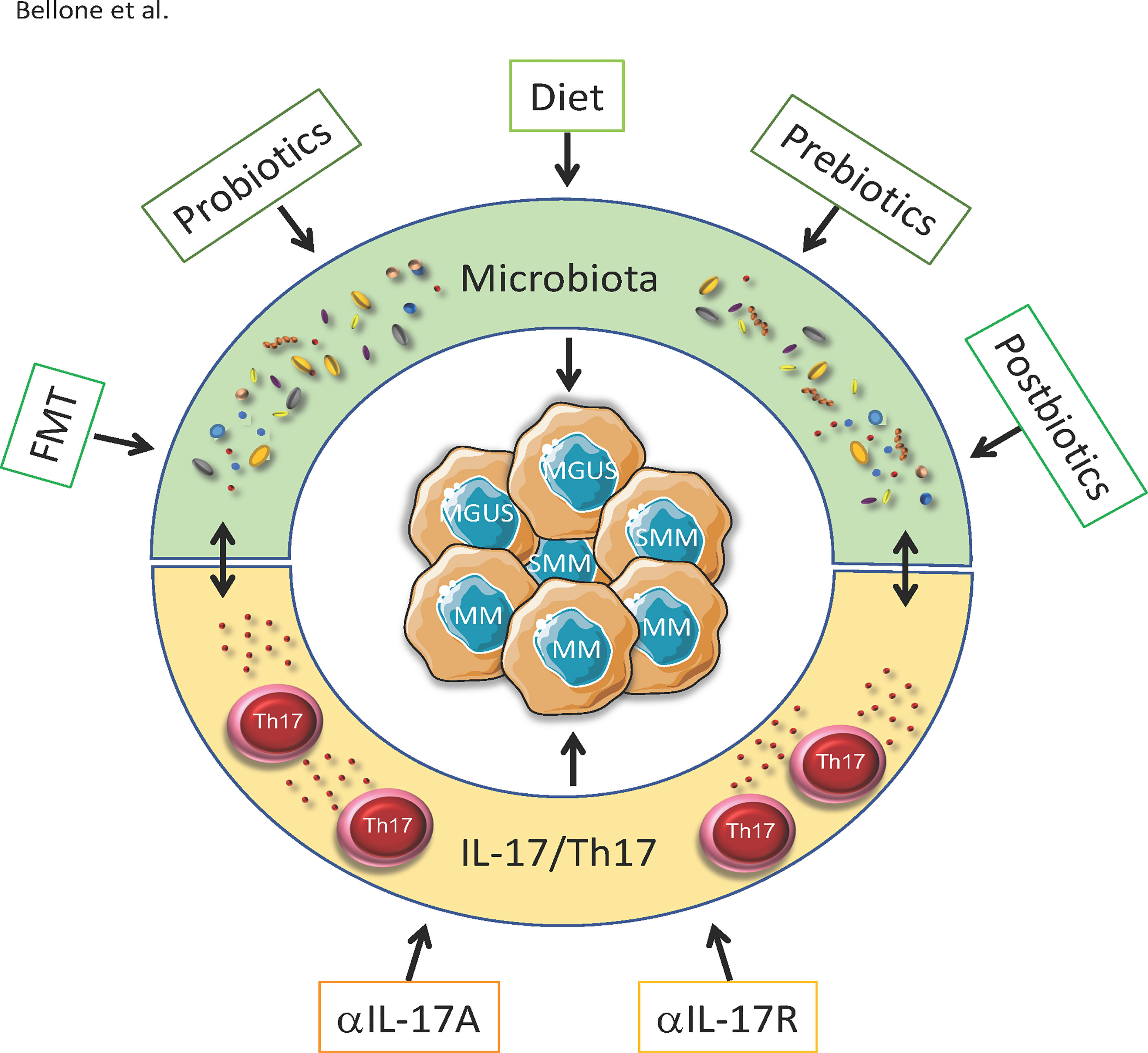
Figure 1 Strategies to impact MGUS, SMM and MM by targeting microbiota and IL-17/Th17. Progression from asymptomatic MGUS and SMM to symptomatic MM partially depends on the microbiota-Th17 axis. Peculiar composition of the gut microbiota locally induces the differentation of Th17 cells, which migrates in the BM and supports disease progression. Strategies to impact the disease by targeting the microbiota include diets enriched of specific nutrients(e.g.SCFA, vitamins, plant-specific foods etc.) autologous or heterologous FMT, prebiotics probiotic bacteria and postibiotics. All these strategies indirectly also reduce IL-17/Th17 accumulating in the gut and in the BM. Other than through the microbiota, monoclonal antibodies againstIL-17 and Il-17R interfere with its pathway and prevent the progression from SMM to MM in mice.
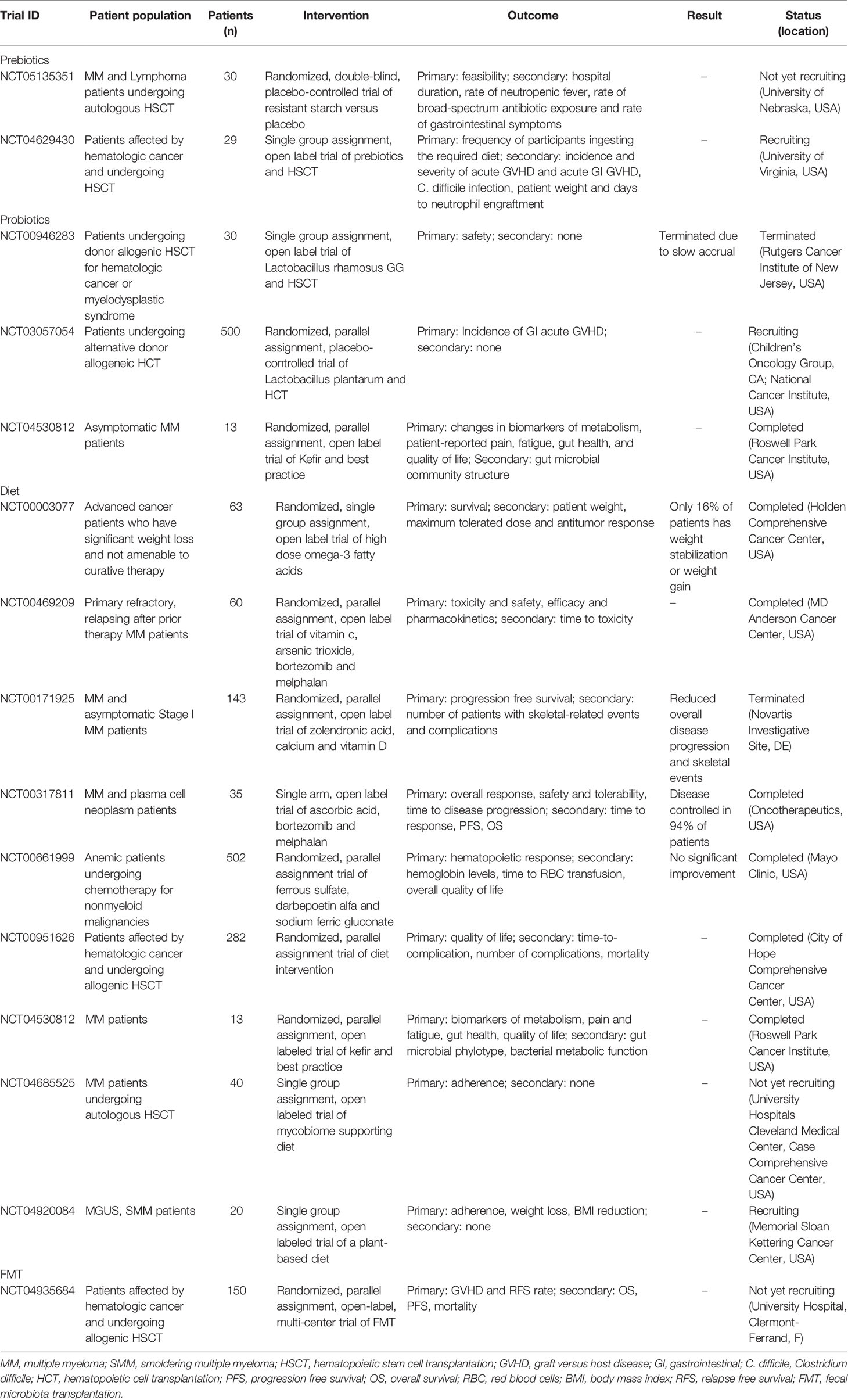
Clinical trials aimed at modulating the gut microbiome to improve therapeutic response in hematopoietic malignancies are ongoing (Table 1) (137). Many of these trials propose administration of 1-6 different commensals with or without dietary intervention, and several of them focus on FMT. Major outcomes for patients undergoing HSCT are safety and GVHD control. Recently, a randomized trial has been launched to assess FMT efficacy in preventing allogeneic-HSCT complications in MM patients (NCT04935684). In other ongoing clinical trials, the microbiota is investigated in correlation to taste function (NCT03276481); to the supplementation of probiotic fermented milk product (NCT04530812); and to combined therapies (i.e., Selinixor, carfizomib and daratumumab or pomalidomide; NCT04661137). Many more studies deal with dietary intervention. As few examples, NCT04920084 will investigate whether a plant-rich diet is feasible and prevent MM in overweight individuals with MGUS or SMM. Another trial will determine if a specific mycobiome supporting diet can reduce gut inflammation in patients undergoing autologous-HSCT (NCT04685525). Resistant starch versus maltodextrin will be compared in a randomized trial involving candidates to autologous-HSCT (NCT05135351).
The natural tropism of bacteria for tumors have been exploited to design bacterial therapy for cancer (138). Commensals can be engineered to deliver drugs into tumors (36), and strategies have been devised to implement bacterial lysis with release of genetically encoded cargo within tumors only when a predefined population density of bacteria is reached (139–141).
Microbiota-derived metabolites (142) and microorganism-associated molecular patterns (143) can protect the BM from ionizing radiation toxicity. Additionally, recovery of lymphocytes and neutrophils after irradiation largely depends on gut microbiota, which also supports nutrients and caloric uptake (97). Because disruption of the intestinal microbiota occurs frequently in HSCT recipients as consequence of the conditioning regimens and wide-spectrum antibiotic use, peri-transplant treatment with simple nutrients, commensal-derived metabolites, selected bacterial species, or even FMT (144) should support optimal immune reconstitution and protection from GVHD and transplant-associated nutritional alterations (135). Of note, FMT in allogeneic-HCT recipients induced protection from intestinal GVHD that associated with increased abundance of butyrate-producing bacteria (144). Butyrate and propionate levels also associate with protection from chronic GVHD in patients affected by MM (145). Administration of resistant starch and prebiotics to allogeneic-HSCT recipients reduced the incidence of acute GVHD that associated with preservation of butyrate-producing commensals (146).
Modulation of the gut microbiota and its derivatives should be time and context dependent. While propionate can provide radioprotection (142), butyrate might limit the immunostimulatory activity of radiotherapy by decreasing dendritic cell antigen presentation (147). In the same vein, melanoma patients resistant to anti-CTLA-4 blockade showed high blood propionate and butyrate levels and higher proportion of Tregs (56). Thus, an excess of SCFAs, while protective in some contexts (60–63, 65, 93, 108), may be detrimental for the induction of efficient anti-cancer CTL responses (56, 147, 148). The situation is even more complex in MM patients, in which modulation of the gut microbiota and its metabolic derivatives should aim at restraining Th17 cell numbers in favor of optimal CTL responses (4). It will be interesting to investigate in animal models of MM if resistant fiber or SCFA supplementation in combination with immune checkpoint blockade limit the expansion of Th17 cells in favor of a potent anti-tumor immunity.
Manipulation of the gut microbiota is not without risks (149). Probiotic strains (e.g., Lactobacilli) can cause bacteriemia although the mechanism of transmission from probiotic to blood is unclear (150). Probiotics may also impair microbiota reconstitution after antibiotic-induced dysbiosis (151). Thus, further investigation is warranted to better understand the mechanistic links between prokaryotic and eukaryotic cells sharing our body space.
Author Contributions
MB designed the review. MB, AB, LC, ML, and BM performed the literature review and wrote the manuscript. AB designed and created the table and the figure. All authors reviewed and edited the manuscript. All authors also approved the final version of the manuscript.
Funding
The research leading to these results has received funding from Associazione Italiana per la Ricerca sul Cancro (AIRC) under IG2018- ID. 21808 to MB and was supported by a grant from the Leukemia & Lymphoma Society (# 6618-21) to MB. AB was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro/AIRC (Grants #22316 to AB).
Conflict of Interest
Matteo Bellone and Arianna Brevi are co-owner of the following patent: WO2020/109620A2. bacterial strains for medical uses.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize to any investigators whose work has not been underlined in this review due to oversight or space limitations. LC conducted this study in partial fulfillment of her PhD in Molecular Medicine, Program in Molecular Oncology and Immunology, Università Vita-Salute San Raffaele, Milan, Italy.
References
1. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current Understanding of the Human Microbiome. Nat Med (2018) 24:392–400. doi: 10.1038/nm.4517
2. Caballero S, Pamer EG. Microbiota-Mediated Inflammation and Antimicrobial Defense in the Intestine. Annu Rev Immunol (2015) 33:227–56. doi: 10.1146/annurev-immunol-032713-120238
3. Bellone M. Autoimmune Disease: Pathogenesis. In: eLS Encyclopedia of Life Science. John Wiley & Sons, Ltd (Ed.). (2015). doi: 10.1002/9780470015902.a0001276.pub4
4. Bellone M, Brevi A, Huber S. Microbiota-Propelled T Helper 17 Cells in Inflammatory Diseases and Cancer. Microbiol Mol Biol Rev (2020) 84(2):e00064–19. doi: 10.1128/MMBR.00064-19
5. Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J, et al. Stereotypic Immune System Development in Newborn Children. Cell (2018) 174:1277–92 e14. doi: 10.1016/j.cell.2018.06.045
6. Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, et al. The Gut Microbiota is Associated With Immune Cell Dynamics in Humans. Nature (2020) 588:303–7. doi: 10.1038/s41586-020-2971-8
7. Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, et al. Thymus-Derived Regulatory T Cells Contribute to Tolerance to Commensal Microbiota. Nature (2013) 497:258–62. doi: 10.1038/nature12079
8. Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, et al. Control of TH17 Cells Occurs in the Small Intestine. Nature (2011) 475:514–8. doi: 10.1038/nature10228
9. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral Education of the Immune System by Colonic Commensal Microbiota. Nature (2011) 478:250–4. doi: 10.1038/nature10434
10. Nakajima A, Negishi N, Tsurui H, Kadowaki-Ohtsuji N, Maeda K, Nanno M, et al. Commensal Bacteria Regulate Thymic Aire Expression. PloS One (2014) 9:e105904. doi: 10.1371/journal.pone.0105904
11. Nutsch K, Chai JN, Ai TL, Russler-Germain E, Feehley T, Nagler CR, et al. Rapid and Efficient Generation of Regulatory T Cells to Commensal Antigens in the Periphery. Cell Rep (2016) 17:206–20. doi: 10.1016/j.celrep.2016.08.092
12. Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, et al. Individual Intestinal Symbionts Induce a Distinct Population of RORgamma(+) Regulatory T Cells. Science (2015) 349:993–7. doi: 10.1126/science.aaa9420
13. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A Defined Commensal Consortium Elicits CD8 T Cells and Anti-Cancer Immunity. Nature (2019) 565:600–5. doi: 10.1038/s41586-019-0878-z
14. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell (2017) 168:928–43 e11. doi: 10.1016/j.cell.2017.01.022
15. Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, et al. Maturation of the Gut Microbiome During the First Year of Life Contributes to the Protective Farm Effect on Childhood Asthma. Nat Med (2020) 26:1766–75. doi: 10.1038/s41591-020-1095-x
16. Nicholas DA, Proctor EA, Agrawal M, Belkina AC, Van Nostrand SC, Panneerseelan-Bharath L, et al. Fatty Acid Metabolites Combine With Reduced Beta Oxidation to Activate Th17 Inflammation in Human Type 2 Diabetes. Cell Metab (2019) 30:447–61.e5. doi: 10.1016/j.cmet.2019.07.004
17. Sullivan ZA, Khoury-Hanold W, Lim J, Smillie C, Biton M, Reis BS, et al. Gammadelta T Cells Regulate the Intestinal Response to Nutrient Sensing. Science (2021) 371(6535):eaba8310. doi: 10.1126/science.aba8310
18. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut Microbiota Metabolism of Dietary Fiber Influences Allergic Airway Disease and Hematopoiesis. Nat Med (2014) 20:159–66. doi: 10.1038/nm.3444
19. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal Microbiota-Derived Short-Chain Fatty Acids Regulation of Immune Cell IL-22 Production and Gut Immunity. Nat Commun (2020) 11:4457. doi: 10.1038/s41467-020-18262-6
20. Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B Cell-Intrinsic Epigenetic Modulation of Antibody Responses by Dietary Fiber-Derived Short-Chain Fatty Acids. Nat Commun (2020) 11:60. doi: 10.1038/s41467-019-13603-6
21. Garidou L, Pomie C, Klopp P, Waget A, Charpentier J, Aloulou M, et al. The Gut Microbiota Regulates Intestinal CD4 T Cells Expressing RORgammat and Controls Metabolic Disease. Cell Metab (2015) 22:100–12. doi: 10.1016/j.cmet.2015.06.001
22. Marino E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al. Gut Microbial Metabolites Limit the Frequency of Autoimmune T Cells and Protect Against Type 1 Diabetes. Nat Immunol (2017) 18:552–62. doi: 10.1038/ni.3713
23. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity (2019) 50:432–45.e7. doi: 10.1016/j.immuni.2018.12.018
24. Vogl T, Klompus S, Leviatan S, Kalka IN, Weinberger A, Wijmenga C, et al. Population-Wide Diversity and Stability of Serum Antibody Epitope Repertoires Against Human Microbiota. Nat Med (2021) 27:1442–50. doi: 10.1038/s41591-021-01409-3
25. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal Microbiota and Myelin Autoantigen Cooperate to Trigger Autoimmune Demyelination. Nature (2011) 479:538–41. doi: 10.1038/nature10554
26. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
27. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity (2016) 44:875–88. doi: 10.1016/j.immuni.2016.03.013
28. Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-Classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell (2018) 172:784–96 e18. doi: 10.1016/j.cell.2017.12.033
29. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, et al. Focused Specificity of Intestinal TH17 Cells Towards Commensal Bacterial Antigens. Nature (2014) 510:152–6. doi: 10.1038/nature13279
30. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WH, Saldana-Morales FB, et al. Thymic Development of Gut-Microbiota-Specific T Cells. Nature (2021) 594:413–7. doi: 10.1038/s41586-021-03531-1
31. Brown EM, Kenny DJ, Xavier RJ. Gut Microbiota Regulation of T Cells During Inflammation and Autoimmunity. Annu Rev Immunol (2019) 37:599–624. doi: 10.1146/annurev-immunol-042718-041841
32. Renz H, Skevaki C. Early Life Microbial Exposures and Allergy Risks: Opportunities for Prevention. Nat Rev Immunol (2021) 21:177–91. doi: 10.1038/s41577-020-00420-y
33. Ruff WE, Greiling TM, Kriegel MA. Host-Microbiota Interactions in Immune-Mediated Diseases. Nat Rev Microbiol (2020) 18:521–38. doi: 10.1038/s41579-020-0367-2
34. Canfora EE, Meex RCR, Venema K, Blaak EE. Gut Microbial Metabolites in Obesity, NAFLD and T2DM. Nat Rev Endocrinol (2019) 15:261–73. doi: 10.1038/s41574-019-0156-z
35. Nikolova VL, Smith MRB, Hall LJ, Cleare AJ, Stone JM, Young AH. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-Analysis. JAMA Psychiatry (2021) 78(12):1343–54. doi: 10.1001/jamapsychiatry.2021.2573
36. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The Microbiome and Human Cancer. Science (2021) 371. doi: 10.1126/science.abc4552
37. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational Signature in Colorectal Cancer Caused by Genotoxic Pks(+) E. Coli Nature (2020) 580:269–73. doi: 10.1038/s41586-020-2080-8
38. Pernigoni N, Zagato E, Calcinotto A, Troiani M, Mestre RP, Cali B, et al. Commensal Bacteria Promote Endocrine Resistance in Prostate Cancer Through Androgen Biosynthesis. Science (2021) 374:216–24. doi: 10.1126/science.abf8403
39. Guyton K, Alverdy JC. The Gut Microbiota and Gastrointestinal Surgery. Nat Rev Gastroenterol Hepatol (2017) 14:43–54. doi: 10.1038/nrgastro.2016.139
40. Roy S, Trinchieri G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat Rev Cancer (2017) 17:271–85. doi: 10.1038/nrc.2017.13
41. Tonneau M, Elkrief A, Pasquier D, Paz Del Socorro T, Chamaillard M, Bahig H, et al. The Role of the Gut Microbiome on Radiation Therapy Efficacy and Gastrointestinal Complications: A Systematic Review. Radiother Oncol (2021) 156:1–9. doi: 10.1016/j.radonc.2020.10.033
42. Derosa L, Routy B, Desilets A, Daillere R, Terrisse S, Kroemer G, et al. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov (2021) 11:2396–412. doi: 10.1158/2159-8290.CD-21-0236
43. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal Microbiota Transplant Promotes Response in Immunotherapy-Refractory Melanoma Patients. Science (2021) 371:602–9. doi: 10.1126/science.abb5920
44. D’Angelo CR, Sudakaran S, Callander NS. Clinical Effects and Applications of the Gut Microbiome in Hematologic Malignancies. Cancer (2021) 127:679–87. doi: 10.1002/cncr.33400
45. Song Y, Himmel B, Ohrmalm L, Gyarmati P. The Microbiota in Hematologic Malignancies. Curr Treat Options Oncol (2020) 21:2. doi: 10.1007/s11864-019-0693-7
46. Severyn CJ, Brewster R, Andermann TM. Microbiota Modification in Hematology: Still at the Bench or Ready for the Bedside? Blood Advances (2019) 3:3461–72. doi: 10.1182/bloodadvances.2019000365
47. Palumbo A, Anderson K. Multiple Myeloma. N Engl J Med (2011) 364:1046–60. doi: 10.1056/NEJMra1011442
48. Kyle RA, Larson DR, Therneau TM, Dispenzieri A, Kumar S, Cerhan JR, et al. Long-Term Follow-Up of Monoclonal Gammopathy of Undetermined Significance. N Engl J Med (2018) 378:241–9. doi: 10.1056/NEJMoa1709974
49. Brevi A, Cogrossi LL, Grazia G, Masciovecchio D, Impellizzieri D, Lacanfora L, et al. Much More Than IL-17a: Cytokines of the IL-17 Family Between Microbiota and Cancer. Front Immunol (2020) 11:565470. doi: 10.3389/fimmu.2020.565470
50. Ahmed N, Ghannoum M, Gallogly M, de Lima M, Malek E. Influence of Gut Microbiome on Multiple Myeloma: Friend or Foe? J Immunother Cancer (2020) 8(1):e000576. doi: 10.1136/jitc-2020-000576
51. Alkharabsheh O, Sidiqi MH, Aljama MA, Gertz MA, Frankel AE. The Human Microbiota in Multiple Myeloma and Proteasome Inhibitors. Acta Haematol (2020) 143:118–23. doi: 10.1159/000500976
52. Jasinski M, Bilinski J, Basak GW. The Role of the Gut Microbiome in Pathogenesis, Biology, and Treatment of Plasma Cell Dyscrasias. Front Oncol (2021) 11:741376. doi: 10.3389/fonc.2021.741376
53. Shapiro YN, Peppercorn JM, Yee AJ, Branagan AR, Raje NS, Donnell EKO. Lifestyle Considerations in Multiple Myeloma. Blood Cancer J (2021) 11:172. doi: 10.1038/s41408-021-00560-x
54. Zhang B, Gu J, Liu J, Huang B, Li J. Fecal Microbiota Taxonomic Shifts in Chinese Multiple Myeloma Patients Analyzed by Quantitative Polimerase Chain Reaction (QPCR) and 16S rRNA High-Throughput Sequencing. Med Sci Monit (2019) 25:8269–80. doi: 10.12659/MSM.919988
55. Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal Analysis of Microbial Interaction Between Humans and the Indoor Environment. Science (2014) 345:1048–52. doi: 10.1126/science.1254529
56. Coutzac C, Jouniaux JM, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts With Cancer. Nat Commun (2020) 11:2168. doi: 10.1038/s41467-020-16079-x
57. Jian X, Zhu Y, Ouyang J, Wang Y, Lei Q, Xia J, et al. Alterations of Gut Microbiome Accelerate Multiple Myeloma Progression by Increasing the Relative Abundances of Nitrogen-Recycling Bacteria. Microbiome (2020) 8:74. doi: 10.1186/s40168-020-00854-5
58. Valtorta S, Toscani D, Chiu M, Sartori A, Coliva A, Brevi A, et al. [(18)F](2S,4R)-4-Fluoroglutamine as a New Positron Emission Tomography Tracer in Myeloma. Front Oncol (2021) 11:760732. doi: 10.3389/fonc.2021.760732
59. Honda K, Littman DR. The Microbiome in Infectious Disease and Inflammation. Annu Rev Immunol (2012) 30:759–95. doi: 10.1146/annurev-immunol-020711-074937
60. Brevi A, Bellone M. Fatty is Not That Bad: Feeding Short-Chain Fatty Acids to Restrain Autoimmunity. Cell Mol Immunol (2017) 14(11):878–80. doi: 10.1038/cmi.2017.52
61. Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The Short-Chain Fatty Acid Pentanoate Suppresses Autoimmunity by Modulating the Metabolic-Epigenetic Crosstalk in Lymphocytes. Nat Commun (2019) 10:760. doi: 10.1038/s41467-019-08711-2
62. Maslowski KM, Mackay CR. Diet, Gut Microbiota and Immune Responses. Nat Immunol (2011) 12:5–9. doi: 10.1038/ni0111-5
63. Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, et al. Metabolite-Sensing Receptors GPR43 and GPR109A Facilitate Dietary Fibre-Induced Gut Homeostasis Through Regulation of the Inflammasome. Nat Commun (2015) 6:6734. doi: 10.1038/ncomms7734
64. Pianko MJ, Devlin SM, Littmann ER, Chansakul A, Mastey D, Salcedo M, et al. Minimal Residual Disease Negativity in Multiple Myeloma Is Associated With Intestinal Microbiota Composition. Blood Advances (2019) 3:2040–4. doi: 10.1182/bloodadvances.2019032276
65. Zhang M, Zhou Q, Dorfman RG, Huang X, Fan T, Zhang H, et al. Butyrate Inhibits Interleukin-17 and Generates Tregs to Ameliorate Colorectal Colitis in Rats. BMC Gastroenterol (2016) 16:84. doi: 10.1186/s12876-016-0500-x
66. Calcinotto A, Brevi A, Chesi M, Ferrarese R, Garcia Perez L, Grioni M, et al. Microbiota-Driven Interleukin-17-Producing Cells and Eosinophils Synergize to Accelerate Multiple Myeloma Progression. Nat Commun (2018) 9:4832. doi: 10.1038/s41467-018-07305-8
67. Kundu P, Blacher E, Elinav E, Pettersson S. Our Gut Microbiome: The Evolving Inner Self. Cell (2017) 171:1481–93. doi: 10.1016/j.cell.2017.11.024
68. Macpherson AJ, Uhr T. Induction of Protective IgA by Intestinal Dendritic Cells Carrying Commensal Bacteria. Science (2004) 303:1662–5. doi: 10.1126/science.1091334
69. Tanoue T, Atarashi K, Honda K. Development and Maintenance of Intestinal Regulatory T Cells. Nat Rev Immunol (2016) 16:295–309. doi: 10.1038/nri.2016.36
70. Dhodapkar KM, Barbuto S, Matthews P, Kukreja A, Mazumder A, Vesole D, et al. Dendritic Cells Mediate the Induction of Polyfunctional Human IL17-Producing Cells (Th17-1 Cells) Enriched in the Bone Marrow of Patients With Myeloma. Blood (2008) 112:2878–85. doi: 10.1182/blood-2008-03-143222
71. Noonan K, Marchionni L, Anderson J, Pardoll D, Roodman GD, Borrello I. A Novel Role of IL-17-Producing Lymphocytes in Mediating Lytic Bone Disease in Multiple Myeloma. Blood (2010) 116:3554–63. doi: 10.1182/blood-2010-05-283895
72. Bryant C, Suen H, Brown R, Yang S, Favaloro J, Aklilu E, et al. Long-Term Survival in Multiple Myeloma is Associated With a Distinct Immunological Profile, Which Includes Proliferative Cytotoxic T-Cell Clones and a Favourable Treg/Th17 Balance. Blood Cancer J (2013) 3:e148. doi: 10.1038/bcj.2013.34
73. Alexandrakis MG, Pappa CA, Miyakis S, Sfiridaki A, Kafousi M, Alegakis A, et al. Serum Interleukin-17 and its Relationship to Angiogenic Factors in Multiple Myeloma. Eur J Intern Med (2006) 17:412–6. doi: 10.1016/j.ejim.2006.02.012
74. Prabhala RH, Pelluru D, Fulciniti M, Prabhala HK, Nanjappa P, Song W, et al. Elevated IL-17 Produced by TH17 Cells Promotes Myeloma Cell Growth and Inhibits Immune Function in Multiple Myeloma. Blood (2010) 115:5385–92. doi: 10.1182/blood-2009-10-246660
75. Vuckovic S, Minnie SA, Smith D, Gartlan KH, Watkins TS, Markey KA, et al. Bone Marrow Transplantation Generates T Cell-Dependent Control of Myeloma in Mice. J Clin Invest (2019) 129:106–21. doi: 10.1172/JCI98888
76. Prabhala RH, Fulciniti M, Pelluru D, Rashid N, Nigroiu A, Nanjappa P, et al. Targeting IL-17A in Multiple Myeloma: A Potential Novel Therapeutic Approach in Myeloma. Leukemia (2016) 30:379–89. doi: 10.1038/leu.2015.228
77. Kappel LW, Goldberg GL, King CG, Suh DY, Smith OM, Ligh C, et al. IL-17 Contributes to CD4-Mediated Graft-Versus-Host Disease. Blood (2009) 113:945–52. doi: 10.1182/blood-2008-08-172155
78. Varelias A, Ormerod KL, Bunting MD, Koyama M, Gartlan KH, Kuns RD, et al. Acute Graft-Versus-Host Disease is Regulated by an IL-17-Sensitive Microbiome. Blood (2017) 129:2172–85. doi: 10.1182/blood-2016-08-732628
79. Bosseboeuf A, Allain-Maillet S, Mennesson N, Tallet A, Rossi C, Garderet L, et al. Pro-Inflammatory State in Monoclonal Gammopathy of Undetermined Significance and in Multiple Myeloma Is Characterized by Low Sialylation of Pathogen-Specific and Other Monoclonal Immunoglobulins. Front Immunol (2017) 8:1347. doi: 10.3389/fimmu.2017.01347
80. Calcinotto A, Grioni M, Jachetti E, Curnis F, Mondino A, Parmiani G, et al. Targeting TNF-Alpha to Neoangiogenic Vessels Enhances Lymphocyte Infiltration in Tumors and Increases the Therapeutic Potential of Immunotherapy. J Immunol (2012) 188:2687–94. doi: 10.4049/jimmunol.1101877
81. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the Human Gut Microbiome. Nature (2011) 473:174–80. doi: 10.1038/nature09944
82. Blazar BR, Hill GR, Murphy WJ. Dissecting the Biology of Allogeneic HSCT to Enhance the GvT Effect Whilst Minimizing GvHD. Nat Rev Clin Oncol (2020) 17:475–92. doi: 10.1038/s41571-020-0356-4
83. Bjorkstrand BB, Ljungman P, Svensson H, Hermans J, Alegre A, Apperley J, et al. Allogeneic Bone Marrow Transplantation Versus Autologous Stem Cell Transplantation in Multiple Myeloma: A Retrospective Case-Matched Study From the European Group for Blood and Marrow Transplantation. Blood (1996) 88:4711–8. doi: 10.1182/blood.V88.12.4711.bloodjournal88124711
84. Bruno B, Rotta M, Patriarca F, Mordini N, Allione B, Carnevale-Schianca F, et al. A Comparison of Allografting With Autografting for Newly Diagnosed Myeloma. N Engl J Med (2007) 356:1110–20. doi: 10.1056/NEJMoa065464
85. Giralt S, Costa LJ, Maloney D, Krishnan A, Fei M, Antin JH, et al. Tandem Autologous-Autologous Versus Autologous-Allogeneic Hematopoietic Stem Cell Transplant for Patients With Multiple Myeloma: Long-Term Follow-Up Results From the Blood and Marrow Transplant Clinical Trials Network 0102 Trial. Biol Blood Marrow Transplant (2020) 26:798–804. doi: 10.1016/j.bbmt.2019.11.018
86. Shono Y, van den Brink MRM. Gut Microbiota Injury in Allogeneic Haematopoietic Stem Cell Transplantation. Nat Rev Cancer (2018) 18:283–95. doi: 10.1038/nrc.2018.10
87. Fredricks DN. The Gut Microbiota and Graft-Versus-Host Disease. J Clin Invest (2019) 129:1808–17. doi: 10.1172/JCI125797
88. Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med (2020) 382:822–34. doi: 10.1056/NEJMoa1900623
89. Khan N, Lindner S, Gomes ALC, Devlin SM, Shah GL, Sung AD, et al. Fecal Microbiota Diversity Disruption and Clinical Outcomes After Auto-HCT: A Multicenter Observational Study. Blood (2021) 137:1527–37. doi: 10.1182/blood.2020006923
90. El Jurdi N, Filali-Mouhim A, Salem I, Retuerto M, Dambrosio NM, Baer L, et al. Gastrointestinal Microbiome and Mycobiome Changes During Autologous Transplantation for Multiple Myeloma: Results of a Prospective Pilot Study. Biol Blood Marrow Transplant (2019) 25:1511–9. doi: 10.1016/j.bbmt.2019.04.007
91. Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol (2017) 35:1650–9. doi: 10.1200/JCO.2016.70.3348
92. Rafei H, Jenq RR. Microbiome-Intestine Cross Talk During Acute Graft-Versus-Host Disease. Blood (2020) 136:401–9. doi: 10.1182/blood.2019000950
93. Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. Regulation of Intestinal Inflammation by Microbiota Following Allogeneic Bone Marrow Transplantation. J Exp Med (2012) 209:903–11. doi: 10.1084/jem.20112408
94. Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The Effects of Intestinal Tract Bacterial Diversity on Mortality Following Allogeneic Hematopoietic Stem Cell Transplantation. Blood (2014) 124:1174–82. doi: 10.1182/blood-2014-02-554725
95. Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, et al. Gut Microbiota Predict Pulmonary Infiltrates After Allogeneic Hematopoietic Cell Transplantation. Am J Respir Crit Care Med (2016) 194:450–63. doi: 10.1164/rccm.201507-1491OC
96. Mancini N, Greco R, Pasciuta R, Barbanti MC, Pini G, Morrow OB, et al. Enteric Microbiome Markers as Early Predictors of Clinical Outcome in Allogeneic Hematopoietic Stem Cell Transplant: Results of a Prospective Study in Adult Patients. Open Forum Infect Dis (2017) 4:ofx215. doi: 10.1093/ofid/ofx215
97. Staffas A, Burgos da Silva M, Slingerland AE, Lazrak A, Bare CJ, Holman CD, et al. Nutritional Support From the Intestinal Microbiota Improves Hematopoietic Reconstitution After Bone Marrow Transplantation in Mice. Cell Host Microbe (2018) 23:447–57 e4. doi: 10.1016/j.chom.2018.03.002
98. Greco R, Nitti R, Mancini N, Pasciuta R, Lorentino F, Lupo-Stanghellini MT, et al. Microbiome Markers are Early Predictors of Acute GVHD in Allogeneic Hematopoietic Stem Cell Transplant Recipients. Blood (2021) 137:1556–9. doi: 10.1182/blood.2020007158
99. Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic Analysis of the Stool Microbiome in Patients Receiving Allogeneic Stem Cell Transplantation: Loss of Diversity is Associated With Use of Systemic Antibiotics and More Pronounced in Gastrointestinal Graft-Versus-Host Disease. Biol Blood Marrow Transplant (2014) 20:640–5. doi: 10.1016/j.bbmt.2014.01.030
100. Alonso CD, Treadway SB, Hanna DB, Huff CA, Neofytos D, Carroll KC, et al. Epidemiology and Outcomes of Clostridium Difficile Infections in Hematopoietic Stem Cell Transplant Recipients. Clin Infect Dis (2012) 54:1053–63. doi: 10.1093/cid/cir1035
101. Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin Infect Dis (2012) 55:905–14. doi: 10.1093/cid/cis580
102. Weber D, Oefner PJ, Hiergeist A, Koestler J, Gessner A, Weber M, et al. Low Urinary Indoxyl Sulfate Levels Early After Transplantation Reflect a Disrupted Microbiome and are Associated With Poor Outcome. Blood (2015) 126:1723–8. doi: 10.1182/blood-2015-04-638858
103. Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, et al. Gut Microbiome-Derived Metabolites Modulate Intestinal Epithelial Cell Damage and Mitigate Graft-Versus-Host Disease. Nat Immunol (2016) 17:505–13. doi: 10.1038/ni.3400
104. Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc Natl Acad Sci USA (2004) 101:15718–23. doi: 10.1073/pnas.0407076101
105. Stein-Thoeringer CK, Nichols KB, Lazrak A, Slingerland AE, Slingerland JB, et al. Lactose Drives Enterococcus Expansion to Promote Graft-Versus-Host Disease. Science (2019) 366:1143–9. doi: 10.1126/science.aax3760
106. Rajkumar SV, Kumar S. Multiple Myeloma Current Treatment Algorithms. Blood Cancer J (2020) 10:94. doi: 10.1038/s41408-020-00359-2
107. Stansborough RL, Gibson RJ. Proteasome Inhibitor-Induced Gastrointestinal Toxicity. Curr Opin Support Palliat Care (2017) 11:133–7. doi: 10.1097/SPC.0000000000000266
108. Ferreira-Halder CV, Faria AVS, Andrade SS. Action and Function of Faecalibacterium Prausnitzii in Health and Disease. Best Pract Res Clin Gastroenterol (2017) 31:643–8. doi: 10.1016/j.bpg.2017.09.011
109. Huang EY, Inoue T, Leone VA, Dalal S, Touw K, Wang Y, et al. Using Corticosteroids to Reshape the Gut Microbiome: Implications for Inflammatory Bowel Diseases. Inflammation Bowel Dis (2015) 21:963–72. doi: 10.1097/MIB.0000000000000332
110. Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, et al. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl Environ Microbiol (2018) 84(4):e02288–17. doi: 10.1128/AEM.02288-17
111. Sistigu A, Viaud S, Chaput N, Bracci L, Proietti E, Zitvogel L. Immunomodulatory Effects of Cyclophosphamide and Implementations for Vaccine Design. Semin Immunopathol (2011) 33:369–83. doi: 10.1007/s00281-011-0245-0
112. Xu X, Zhang X. Effects of Cyclophosphamide on Immune System and Gut Microbiota in Mice. Microbiol Res (2015) 171:97–106. doi: 10.1016/j.micres.2014.11.002
113. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science (2013) 342:971–6. doi: 10.1126/science.1240537
114. Yang J, Liu KX, Qu JM, Wang XD. The Changes Induced by Cyclophosphamide in Intestinal Barrier and Microflora in Mice. Eur J Pharmacol (2013) 714:120–4. doi: 10.1016/j.ejphar.2013.06.006
115. Wardill HR, de Mooij CEM, da Silva Ferreira AR, van de Peppel IP, Havinga R, Harmsen HJM, et al. Translational Model of Melphalan-Induced Gut Toxicity Reveals Drug-Host-Microbe Interactions That Drive Tissue Injury and Fever. Cancer Chemother Pharmacol (2021) 88:173–88. doi: 10.1007/s00280-021-04273-7
116. van der Velden WJ, Herbers AH, Feuth T, Schaap NP, Donnelly JP, Blijlevens NM. Intestinal Damage Determines the Inflammatory Response and Early Complications in Patients Receiving Conditioning for a Stem Cell Transplantation. PloS One (2010) 5:e15156. doi: 10.1371/journal.pone.0015156
117. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science (2015) 350:1079–84. doi: 10.1126/science.aad1329
118. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Science (2015) 350:1084–9. doi: 10.1126/science.aac4255
119. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science (2018) 359:97–103. doi: 10.1126/science.aan4236
120. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The Commensal Microbiome is Associated With Anti-PD-1 Efficacy in Metastatic Melanoma Patients. Science (2018) 359:104–8. doi: 10.1126/science.aao3290
121. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Science (2018) 359:91–7. doi: 10.1126/science.aan3706
122. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-Derived Inosine Modulates Response to Checkpoint Inhibitor Immunotherapy. Science (2020) 369:1481–9. doi: 10.1126/science.abc3421
123. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal Microbiota Transplant Overcomes Resistance to Anti-PD-1 Therapy in Melanoma Patients. Science (2021) 371:595–602. doi: 10.1126/science.abf3363
124. Gorgun G, Samur MK, Cowens KB, Paula S, Bianchi G, Anderson JE, et al. Lenalidomide Enhances Immune Checkpoint Blockade-Induced Immune Response in Multiple Myeloma. Clin Cancer Res (2015) 21:4607–18. doi: 10.1158/1078-0432.CCR-15-0200
125. Jelinek T, Paiva B, Hajek R. Update on PD-1/PD-L1 Inhibitors in Multiple Myeloma. Front Immunol (2018) 9:2431. doi: 10.3389/fimmu.2018.02431
126. Lesokhin AM, Bal S, Badros AZ. Lessons Learned From Checkpoint Blockade Targeting PD-1 in Multiple Myeloma. Cancer Immunol Res (2019) 7:1224–9. doi: 10.1158/2326-6066.CIR-19-0148
127. Andrews MC, Duong CPM, Gopalakrishnan V, Iebba V, Chen WS, Derosa L, et al. Gut Microbiota Signatures are Associated With Toxicity to Combined CTLA-4 and PD-1 Blockade. Nat Med (2021) 27:1432–41. doi: 10.1038/s41591-021-01406-6
128. Wang F, Yin Q, Chen L, Davis MM. Bifidobacterium can Mitigate Intestinal Immunopathology in the Context of CTLA-4 Blockade. Proc Natl Acad Sci USA (2018) 115:157–61. doi: 10.1073/pnas.1712901115
129. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut Microbial Metabolites Facilitate Anticancer Therapy Efficacy by Modulating Cytotoxic CD8(+) T Cell Immunity. Cell Metab (2021) 33:988–1000 e7. doi: 10.1016/j.cmet.2021.03.002
130. Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A, et al. Microbial Short-Chain Fatty Acids Modulate CD8(+) T Cell Responses and Improve Adoptive Immunotherapy for Cancer. Nat Commun (2021) 12:4077. doi: 10.1038/s41467-021-24331-1
131. Smith M, Zakrzewski J, James S, Sadelain M. Posttransplant Chimeric Antigen Receptor Therapy. Blood (2018) 131:1045–52. doi: 10.1182/blood-2017-08-752121
132. van de Donk N, Usmani SZ, Yong K. CAR T-Cell Therapy for Multiple Myeloma: State of the Art and Prospects. Lancet Haematol (2021) 8:e446–e61. doi: 10.1016/S2352-3026(21)00057-0
133. Schubert ML, Rohrbach R, Schmitt M, Stein-Thoeringer CK. The Potential Role of the Intestinal Micromilieu and Individual Microbes in the Immunobiology of Chimeric Antigen Receptor T-Cell Therapy. Front Immunol (2021) 12:670286. doi: 10.3389/fimmu.2021.670286
134. Di Nicola M, Apetoh L, Bellone M, Colombo MP, Dotti G, Ferrone S, et al. Innovative Therapy, Monoclonal Antibodies and Beyond. Cytokine Growth Factor Rev (2017) 38:1–9. doi: 10.1016/j.cytogfr.2017.10.002
135. Baumgartner A, Bargetzi A, Zueger N, Bargetzi M, Medinger M, Bounoure L, et al, et al. Revisiting Nutritional Support for Allogeneic Hematologic Stem Cell Transplantation-a Systematic Review. Bone Marrow Transplant (2017) 52:506–13. doi: 10.1038/bmt.2016.310
136. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome Analyses of Blood and Tissues Suggest Cancer Diagnostic Approach. Nature (2020) 579:567–74. doi: 10.1038/s41586-020-2095-1
137. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the Microbiome to Improve Therapeutic Response in Cancer. Lancet Oncol (2019) 20:e77–91. doi: 10.1016/S1470-2045(18)30952-5
138. Zheng JH, Nguyen VH, Jiang SN, Park SH, Tan W, Hong SH, et al. Two-Step Enhanced Cancer Immunotherapy With Engineered Salmonella Typhimurium Secreting Heterologous Flagellin. Sci Transl Med (2017) 9(376):eaak9537. doi: 10.1126/scitranslmed.aak9537
139. Swofford CA, Van Dessel N, Forbes NS. Quorum-Sensing Salmonella Selectively Trigger Protein Expression Within Tumors. Proc Natl Acad Sci USA (2015) 112:3457–62. doi: 10.1073/pnas.1414558112
140. Din MO, Danino T, Prindle A, Skalak M, Selimkhanov J, Allen K, et al. Synchronized Cycles of Bacterial Lysis for In Vivo Delivery. Nature (2016) 536:81–5. doi: 10.1038/nature18930
141. Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable Bacteria Induce Durable Tumor Regression and Systemic Antitumor Immunity. Nat Med (2019) 25:1057–63. doi: 10.1038/s41591-019-0498-z
142. Guo H, Chou WC, Lai Y, Liang K, Tam JW, Brickey WJ, et al. Multi-Omics Analyses of Radiation Survivors Identify Radioprotective Microbes and Metabolites. Science (2020) 370(6516):eaay9097. doi: 10.1126/science.aay9097
143. Burdelya LG, Krivokrysenko VI, Tallant TC, Strom E, Gleiberman AS, Gupta D, et al. An Agonist of Toll-Like Receptor 5 has Radioprotective Activity in Mouse and Primate Models. Science (2008) 320:226–30. doi: 10.1126/science.1154986
144. van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor Fecal Microbiota Transplantation Ameliorates Intestinal Graft-Versus-Host Disease in Allogeneic Hematopoietic Cell Transplant Recipients. Sci Transl Med (2020) 12(556):aaz8926. doi: 10.1126/scitranslmed.aaz8926
145. Markey KA, Schluter J, Gomes ALC, Littmann ER, Pickard AJ, Taylor BP, et al. The Microbe-Derived Short-Chain Fatty Acids Butyrate and Propionate are Associated With Protection From Chronic GVHD. Blood (2020) 136:130–6. doi: 10.1182/blood.2019003369
146. Yoshifuji K, Inamoto K, Kiridoshi Y, Takeshita K, Sasajima S, Shiraishi Y, et al. Prebiotics Protect Against Acute Graft-Versus-Host Disease and Preserve the Gut Microbiota in Stem Cell Transplantation. Blood Advances (2020) 4:4607–17. doi: 10.1182/bloodadvances.2020002604
147. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gomez L, Verginadis I, Bittinger K, et al. Gut Microbiota Modulate Dendritic Cell Antigen Presentation and Radiotherapy-Induced Antitumor Immune Response. J Clin Invest (2020) 130:466–79. doi: 10.1172/JCI124332
148. Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, et al. Gut Microbiota Modulates Adoptive Cell Therapy via CD8alpha Dendritic Cells and IL-12. JCI Insight (2018) 3(4):e94952. doi: 10.1172/jci.insight.94952
149. Suez J, Zmora N, Segal E, Elinav E. The Pros, Cons, and Many Unknowns of Probiotics. Nat Med (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
150. Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, et al. Genomic and Epidemiological Evidence of Bacterial Transmission From Probiotic Capsule to Blood in ICU Patients. Nat Med (2019) 25:1728–32. doi: 10.1038/s41591-019-0626-9
Keywords: microbiota, multiple myeloma, monoclonal gammopathy of undetermined significance, smoldering multiple myeloma, prevotella, T helper 17, interleukin 17, gut micobiome
Citation: Brevi A, Cogrossi LL, Lorenzoni M, Mattorre B and Bellone M (2022) The Insider: Impact of the Gut Microbiota on Cancer Immunity and Response to Therapies in Multiple Myeloma. Front. Immunol. 13:845422. doi: 10.3389/fimmu.2022.845422
Received: 29 December 2021; Accepted: 25 February 2022;
Published: 17 March 2022.
Edited by:
Khashayarsha Khazaie, Mayo Clinic, United StatesReviewed by:
Nelson Chao, Duke University, United StatesCopyright © 2022 Brevi, Cogrossi, Lorenzoni, Mattorre and Bellone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matteo Bellone, YmVsbG9uZS5tYXR0ZW9AaHNyLml0
 Arianna Brevi
Arianna Brevi Laura Lucia Cogrossi1,2
Laura Lucia Cogrossi1,2 Matteo Bellone
Matteo Bellone