- 1College of Basic Medical Science, Dalian Medical University, Dalian, China
- 2Division of Regulatory Glycobiology, Institute of Molecular Biomembrane and Glycobiology, Tohoku Pharmaceutical University, Sendai, Japan
- 3Research Institute for Microbial Diseases and World Premier International Immunology Frontier Research Center, Osaka University, Suita, Japan
Most of the membrane molecules involved in immune response are glycosylated. N-glycans linked to asparagine (Asn) of immune molecules contribute to the protein conformation, surface expression, stability, and antigenicity. Core fucosylation catalyzed by core fucosyltransferase (FUT8) is the most common post-translational modification. Core fucosylation is essential for evoking a proper immune response, which this review aims to communicate. First, FUT8 deficiency suppressed the interaction between μHC and λ5 during pre-BCR assembly is given. Second, we described the effects of core fucosylation in B cell signal transduction via BCR. Third, we investigated the role of core fucosylation in the interaction between helper T (TH) cells and B cells. Finally, we showed the role of FUT8 on the biological function of IgG. In this review, we discussed recent insights into the sites where core fucosylation is critical for humoral immune responses.
Introduction
Humoral immune response relies on intrinsic B cell development. The gradual stages of B lymphocyte differentiation are marked by the arrangement and expression of immunoglobulin (Ig) genes in the bone marrow (BM) (1). Early B lymphocytes are produced in the BM, in which there is a stepwise subgroup from hematopoietic stem cells (HSC) to immature B cells. The middle steps are composed of pro-B cells and pre-B cells. The V(D)J rearrangement of the μ-heavy chain (μHC) gene occurs in pro-B cells, and the gene rearrangement of light chains begins in the pre-B cell stage. Once a light chain gene is assembled and a completed IgM is expressed on the surface of immature B cells, the B cells differentiate further to become mature B cells expressing IgD and IgM. The mature B cells recirculate through secondary lymphoid tissues, where they may encounter foreign antigens (Ags). Ag-binding B cells are trapped in the T-cell zone of lymphoid tissue and activated by encounter with helper T (TH) cells, and differentiated into plasma cells, which secret large amounts of antibodies (Abs) to provide lasting protective immunity (2, 3).
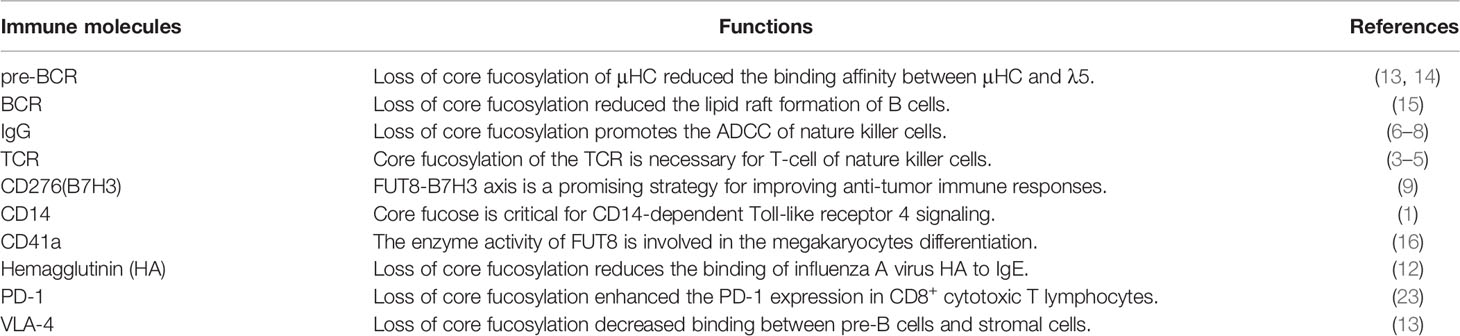
Almost all of the immunological molecules are glycosylated (4, 5). Glycosylation is one of the most common post-translational modifications of eukaryotic proteins. The glycans attached to these proteins can be classified into two major types: O-linked glycosylation, whose glycan chains are linked to the oxygen atom of serine or threonine residues in the Golgi, and N-linked glycosylation, whose glycan chains are covalently linked to the amide nitrogen of asparagine (Asn) residues of an Asn-X-Ser/Thr sequence (where X is not proline) by an N-glycosidic bond in the endoplasmic reticulum (ER). Despite their microheterogeneity, most N-glycans share a common N-glycan precursor, 14 monosaccharide residues (i.e., Glc3Man9GlcNAc2) that is pre-assembled on the ER membrane before it is transferred to protein. After the attachment of the N-glycan precursor to Asn-X-Ser/Thr in a protein, a series of processing reactions trim the N-glycan in the ER. After sequential trimming of the terminal monosaccharide residues (glucose and mannose), the glycoproteins exiting the ER en route to the Golgi carry N-glycans with a Man8GlcNAc2 isomer. Then, the glycoprotein undergoes further sugar chain elongation and “glycan maturation” (e.g., fucosylation, galactosylation, and terminal sialylation) by a suite of glycosyltransferases (6). The glycosyltransferases that assemble monosaccharide moieties of a simple nucleotide sugar donor substrate (e.g., UDP-Gal, GDP-Fuc, or CMP-Sia) into the acceptor substrate (linear and branched glycan chains). The ER-Golgi pathway harbors many glycan-modifying enzymes in eukaryotic cells. In the medial-Golgi, the N-glycan has two branches, which are initiated by the addition of two-terminA006C N-acetylglucosamine (GlcNAc) residues by N-acetylglucosaminyltransferase-I (GnT-I) and GnT-II. Additional branches can be initiated at C-4 of the core mannose α1,3 (by GnT-IV) and C-6 of the core mannose α1,6 (by GnT-V) to yield tri- and tetra-antennary N-glycans. N-glycans may carry a “bisecting” GlcNAc residue that is catalyzed by GnT-III. Further sugar additions, such as galactosylation by galactosyltransferases (GalT), fucosylation by fucosyltransferases (FUTs), and sialylation by sialyltransferases (STs), mostly occur in the trans-Golgi. FUTs consist of 13 members, including FUT1 to FUT11, protein O-fucosyltransferase 1 (POFUT1), and POFUT2. Core fucosylation is catalyzed by core fucosyltransferase (FUT8). Figure 1 depicts a bisecting N-GlcNAc on a tetra-antennary N-glycan, which may be present in all of the more highly branched structures. Glycosylation regulates several glycoprotein functions, including half-life on the membrane, sorting to specific subcellular sites, and folding in the ER (7).
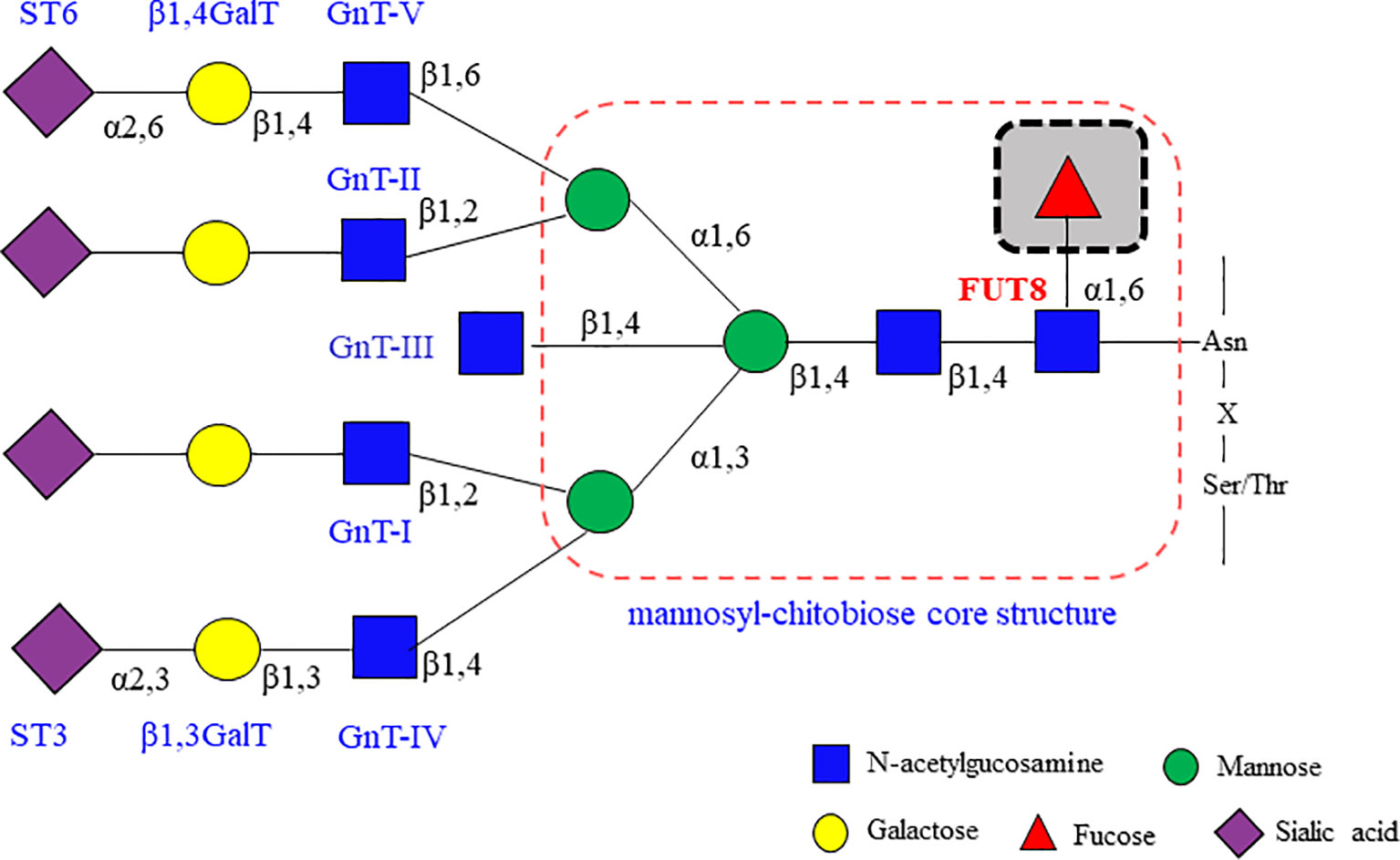
Figure 1 Glycosyltransferases involving in the biosynthesis of branching N-glycans. α2,6 sialyltransferases (ST6) and/or α2,3 sialyltransferases (ST3) that act on galactose, β1,4 galactosyltransferases (β1,4GalT) and/or β1,3 galactosyltransferases (β1,3GalT) that act on N-acetylglucosamine (GlcNAc), and the family of N-acetylglucosamine transferases (GnT-I, GnT-II, GnT-III, GnT-IV, GnT-V and so on) that participate in GlcNAc synthesis. Fucosyltransferases (FUT3, FUT4, FUT5, FUT6, FUT7, FUT8, FUT9) can attach fucose in either α1,3/α1,4 or α1,6 linkage. These enzymes act sequentially, so that the product of one enzyme yields a preferred acceptor substrate for the subsequent action of another. Different proteins may have different subsets of N-glycans, which is referred to as microheterogeneity. All N-glycans share a common core sugar sequence, Manα1–6(Manα1–3) Manβ1–4GlcNAcβ1–4GlcNAcβ1.
We have been devoted to the study that core fucosylation is an essential segment in the early B cell differentiation (8, 9), B cell activation (10, 11), Ab production (11, 12), systemic lupus erythematosus (SLE) development (11), infection (12, 13) and antitumor activity (14). The critical physiological function of the fucosylation is characterized by leukocyte adhesion deficiency II, which emphasizes the physiological significance of the fucosylation due to immunodeficiency caused by fucosylation deficiency (15). This review focuses on the unique roles of core fucosylation in adaptive humoral immune responses.
Core Fucosylation and Its Function
In mammalian cells, the core fucosylation catalyzed by FUT8 is a common post-translational modification (Figure 2) (16, 17). FUT8 is the only glycosyltransferase that catalyzes the transfer of fucose from GDP-fucose to the inner GlcNAc of N-glycans through α1,6-linkage (core fucosylation) in the Golgi apparatus. There are two different pathways for producing GDP-fucose, namely the salvage and de novo pathways. The salvage pathway utilizes free fucose derived from dietary sources. The de novo pathway (the major way) synthesizes GDP-fucose from GDP-mannose through oxidation, epimerization, and reduction catalyzed by two enzymes, including GDP-mannose 4,6-dehydratase (GMD) and GDP-4-keto-6-deoxy-mannose-3,5-epimerase-4-reductase (FX). Then, the GDP-fucose transporter (GFT) transfers the GDP-fucose from the cytosol into the Golgi apparatus.
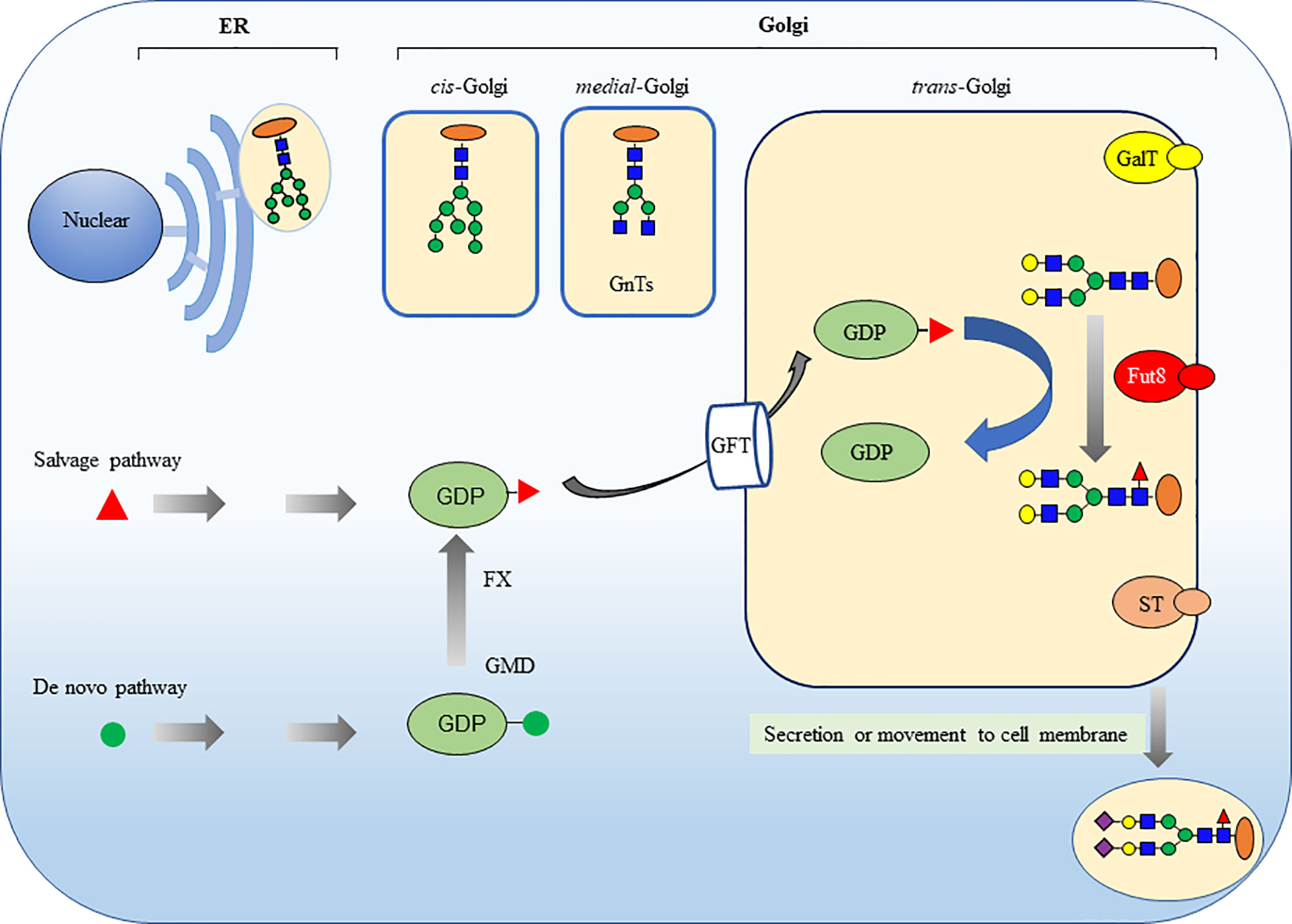
Figure 2 Core fucosylation modification in mammalian N-glycans is catalyzed by FUT8. The ER-Golgi pathway harbors many glycan-modifying enzymes in eukaryotic cells. The glycosyltransferases that operate in the ER are woven into the ER membrane. In contrast, the glycosyltransferases in Golgi compartments are generally type II membrane proteins with a small cytoplasmic amino-terminal domain, a single transmembrane domain, and a large lumenal domain that has an elongated stem region and a globular catalytic domain. FUT8 is a typical type II membrane protein and a Golgi apparatus–resident glycosyltransferase to catalyze the transfer of a fucose from GDP-fucose to the inner GlcNAc of N-glycans through α1,6-linkage (core fucosylation). There are two different pathways for producing GDP-fucose, namely salvage pathway and de novo pathway. The GDP-fucose is transferred from the cytosol into the Golgi apparatus by GDP-fucose transporter (GFT). The sugar additions, such as galactosylation by GalT, fucosylation by FUTs and sialylation by STs, mostly occurring in the trans-Golgi.
Previous studies have observed relationships between protein structures and the N-glycan repertoires (18). The underlying pentasaccharide (mannosyl-chitobiose: Man3GlcNAc2) core structure of all eukaryotic N-glycans serves as scaffolding. The flexible glycosidic linkages are often found in the pentasaccharide core structure of the N-glycans. The antenna flexibility could play a specific role in directing precise recognition in carbohydrate-protein interactions. N-linked glycosylation does not induce any secondary structure of proteins but that it does alter their conformational preferences in the vicinity of the glycosylation site, increasing the probability of more compact conformations (19). These effects seem to involve only the monosaccharide units of the N-glycan and are probably mediated by steric and hydrophobic or hydrophilic interactions between the N-glycan and the neighboring amino acid sidechains (20). Most glycoproteins are expressed on the cell surface. Each interaction between such proteins involved in cell-cell recognition events are frequently weak. Strong cell-cell interactions are obtained from many of these interactions occurring simultaneously on cell surface molecules. The N-glycans on these molecules influence their orientation and their packing of glycoproteins on the cell surface.
It has been reported that the core fucose influences the conformational flexibility of the core-fucosylated biantenna of N-glycans (21). The core fucose also plays a crucial role in the binding of an N-glycan to a plant lectin (22). As shown in the Table 1, core fucosylation regulates the conformation, stability and expression of immune molecules, such as pre-BCR, BCR, IgG, T cell receptor (TCR), CD276(B7H3), CD14, CD41a, hemagglutinin (HA), PD-1 and VLA-4. The association between an N-glycan and its Asn sidechain is relatively rigid and planar, with a tendency to extend the core sugar structure (24). Conversely, core fucosylation is strongly influenced by the structure of the proteins. The glycosylation sites of the proteins and their subcellular location determine whether the conjugated N-glycans will be modified by core fucosylation. It is reasonable to propose that core fucosylation could affect the glycosidic linkage flexibility and conformation of proteins, resulting in modification of protein interactions or assembly. Core fucosylation is ubiquitously expressed in mammalian tissues and participate in the regulation of numerous biological events of physiological and pathological conditions (25), including cell growth (8, 26), cell signal transduction (9, 10), protein-protein interaction (9, 10), cell-cell interaction (11, 12) and tumorigenesis (23, 27, 28). FUT8 knockout (FUT8-/-) mice exhibit early postnatal death (29), retardant growth (26, 30), emphysema-like changes (29, 31), schizophrenia-like phenotype (32) and so on.
Core Fucosylation Regulates Pre-BCR Assembly
During early B cell differentiation, the formation of pre-BCR complex allows progression from the pro-B cell stage to pre-B cell stage (33). The assembly of the pre-BCR on the cell membrane is a critical checkpoint for B cell growth and differentiation in humans and mice (34–38). Suppression of pre-BCR complex formation inhibits the transition of B lymphocyte development from pro-B cell stage to pre-B cell stage (39, 40). The functional pre-BCR complex consists of immunoglobulin (Ig) μHC, surrogate light chain (SLC), and Igα/Igβ (CD79a/CD79b) (41). The μHCs assembled with SLC are classically obligatory for pre-BCR trafficking to the surface (42). The μHC (GI:90956) comprises the variable region of Ig μHC (VH) and the constant portion of the Ig μHC (CH). The λ5 (GI:54887631) and Vpre-B are non-covalently associated to form SLC. In a pre-BCR complex, λ5 binds to the CH1 domain of μHC, while Vpre-B interacted with the VH domain. Vpre-B is stabilized by a salt bridge between Glu106 residue of Vpre-B and Arg59 residue of VH (33). The N-glycans on μHC are of the high-mannose type (43). Ubelhart et al. (44) also discovered that μHC of mouse and human Igs contains the N-linked glycosylation site N46 and that a conserved N46-glycan at the CH1 domain of μHC was necessary for the interaction of μHC and λ5, followed by the formation of pre-BCR. There are no N-glycosylated sites on λ5, and it is covalently coupled to the CH1 domain of μHC via a carboxyl-terminal cysteine (42). Only pre-B cells that express the pre-BCR complex undergo the following clonal expansion.
In FUT8-/- BM, we found a profound and selective suppression in pre-B cell generations without a concomitant change in the pro-B cell population (8). To address the impairment of the pre-B cell population in FUT8-/- mice, we investigated the role of FUT8 in pre-BCR assembly. Indeed, μHC is a core fucosylated protein, and loss of core fucosylation of μHC reduces the binding affinity between μHC and λ5. Consistent with this result, the formation of pre-BCR is down-regulated in the FUT8 knockdown pre-B cell line (70Z/3-KD cells) and has been recovered in 70Z/3-KD-re cells (9). Indeed, the subpopulation of μHC+λ5+ cells is lower in FUT8-/- BM than in FUT8+/+ BM. Early pre-B cells expressing both μHC and SLC are in the BM and fetal liver; one produces SLC-pairing μHC, and the other produces SLC-nonpairing μHC (36). Only half of the in-frame rearranges μHCs pair correctly with SLCs (45). Subsequently, μHCs that bind poorly to SLCs are subsequently poorly represented among μHC families (46). The μHC undergoes glycosylation modification and is transported to the cell surface. The newly synthesized membrane form of μHC is Endo H-resistant, which reaches the cell surface, while the secretory form is Endo H-sensitive (47). Since FUT8 expression is significantly increased during the transition from pro-B to pre-B cell development (8), and since the core fucosylation is required for pre-BCR complex assembly (9), it is conceivable that FUT8 controls the core fucosylation of μHC, followed by pre-BCR complex assembly. There are no N-glycosylation sites in λ5, Vpre-B, and VH domains of μHC. When the binding of VH of μHC to Vpre-B is a protein-protein interaction by ionic bonding between Glu106 of Vpre-B and Arg59 residue of VH region, and the binding of CH1 of μHC to λ5 is a protein-glycoprotein interaction. It is reasonable to propose that the FUT8 regulated the interaction between CH1 of μHC and λ5, but did not influence the interaction between Vpre-B and μHC during pre-BCR assembly (Figure 3). The pre-BCR tunes pre-B cell repertoire by driving the preferential differentiation and expansion of cells with a higher quality of μHC (37). Thus, we propose a mechanism in which core fucosylated μHC has a higher potential to assemble with SLC and amplify the pre-B cell repertoire.
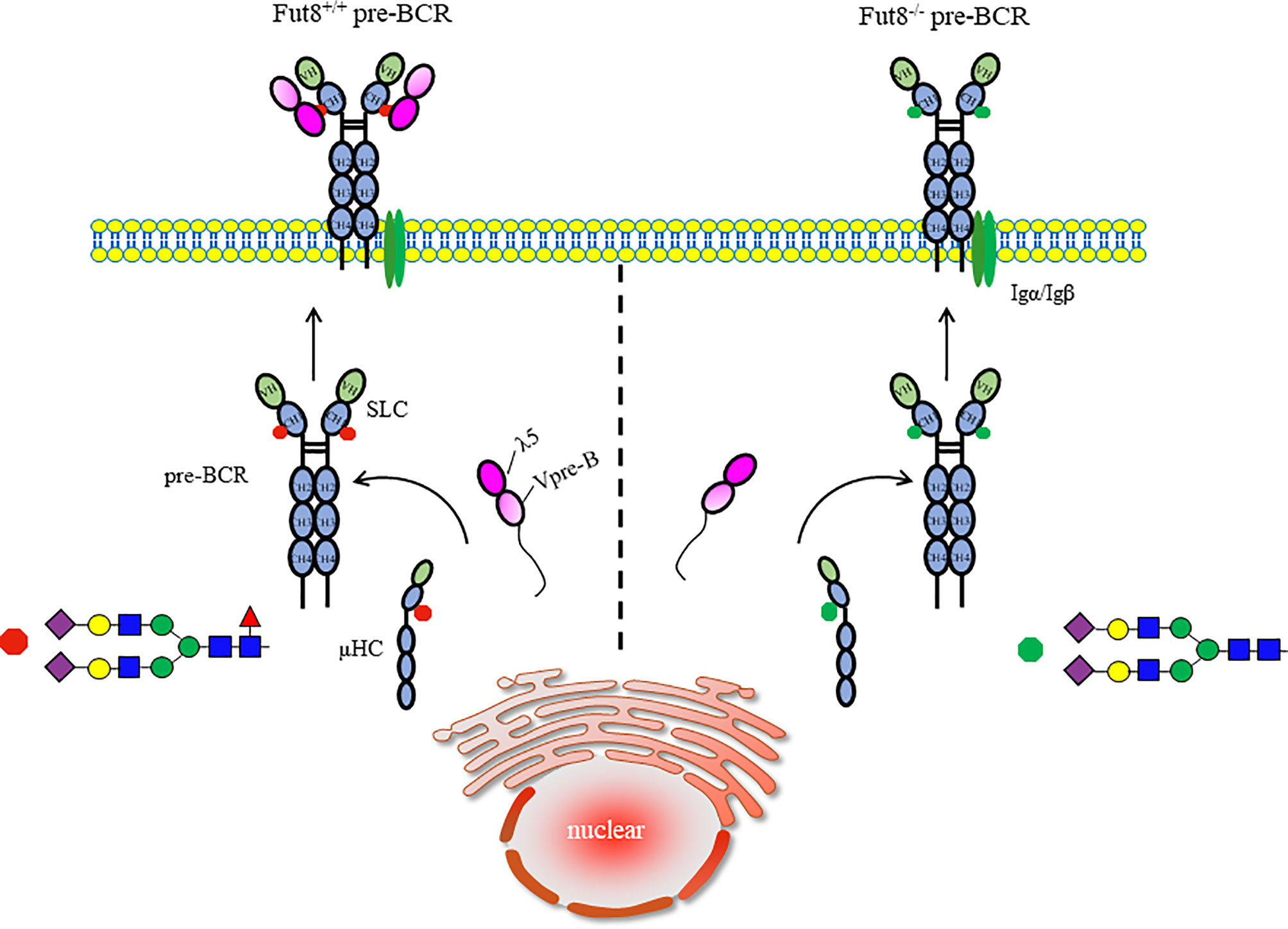
Figure 3 Core fucosylation regulates the assembly of pre-BCR. The early B cells growth depends on the assembly of pre-BCR. The conserved N46-glycosylation site on CH1 domain of μHC played a critical role in modulation of the interaction of μHC and λ5 before the formation of pre-BCR. Removal of core fucose of μHC impaired the interaction between μHC and λ5, and pre-BCR assembly, which is necessary and sufficient for signal transduction via pre-BCR and early B cell proliferation.
Pre-BCR expression is sufficient and required for constitutive cell signaling (48). Mutations of genes associated with the pre-BCR signal pathway culminate in agammaglobulinemia (35–38, 49). Core fucosylation is required for the appropriate binding of μHC to λ5, as well as pre-BCR assembly (9). Except for the signal transduction via pre-BCR, integrins bind to vascular adhesion molecule-1 (VCAM-1), thereby facilitating signaling with stimulation of IL-7 from stromal cells, which plays a significant role in the generation of B cell precursors (50). Integrins on pre-B cells, together with pre-BCR and galectin-1, could form a homogeneous lattice at the contact area between pre-B cells and stromal cells (51). FUT8 ablation reduces binding affinity between very late antigen 4 (VLA-4, α4β1 integrin) on pre-B cells and VCAM-1 on stromal cells, which impaired the colony expansion of pre-B cells in FUT8-/- BM (8).
Core Fucosylation Regulates the Signal Transduction via BCR
Upon B cell activation, B cells recognize both soluble and membrane-associated Ags by B-cell receptor (BCR), and respond to them (3). After the recognition of Ags, BCRs on B cells trigger signaling that eventually induced B cell activation and Abs production (52). First identified in 1970 (53), the BCR and Ab consisted of two heavy (H) chains and two light (L) chains, which are linked by disulfide bonds. Each H chain is composed of an N-terminal variable domain (VH) and three constant domains (CH1, CH2, CH3); the L chains consist of an N-terminal variable domain (VL) and one constant domain (CL). The ‘‘fragment antigen-binding’’ (Fab) domains, which mediates antigen recognition, contain the complementarity determining regions (CDRs), located in the N-terminal region of CH and CL, which define the antigen-specificity. The “fragment crystallizable’’ (Fc) of the C-terminal regions is composed of the two CH (CH2, CH3), which bind to the immune effector molecules, including Fc receptors. The BCR complex contains a membrane-bound immunoglobulin (mIg) with a small cytoplasmic domain and a disulfide-linked heterodimer Igα–Igβ (CD79a-CD79b), which contains the immunoreceptor tyrosine-based activation motif, ITAMs (54, 55).
There are glycosylation sites at Asn297 of the Fc region but not in the VH of IgG-BCR. Microheterogeneity of N-glycans on IgG is detected by the linkage of galactose–sialic acid or galactose at one or both of the terminal GlcNAc or linkage of a third GlcNAc arm. The profile of N-glycans at Asn297 on BCR is of the complex type (51). Furthermore, BCR is a highly core fucosylated glycoprotein, and the core fucosylation regulates the function of BCR to distinguish Ag (10). Indeed, FUT8 ablation impaired the binding affinity of IgG-BCRs to their ligands. Since core fucosylation could modulate the conformation and ligand affinity of the membrane receptors (8–11, 26, 29) and could influence the flexibility of N-glycans antenna (21), it is suggested that core fucosylation could control the axial rotation of the Fab arms and the recognition of BCR to Ag. Core fucosylation of μHC impaired the interaction between μHC and λ5 of the SLC. However, FUT8 gene knockout/knockdown does not influence the BCR expression on the cell surface. It is conceivable that the difference of N-glycan in heavy chains is attributed to the distinct assembly between pre-BCR and BCR.
Lipid rafts are heterogeneous, dynamic and transient plasma membrane entities enriched in saturated phosho-, sphingo- and glycolipids, cholesterol, lipidated proteins and glycosylphosphatidylinositol (GPI)-anchored proteins (56). The BCR is found in detergent-soluble membrane fractions in resting cells, but is acquired in detergent-resistant membrane fractions (raft domains) after receptor activation (57–59). Fluorescence resonance energy transfer confocal microscopy revealed that the association of BCR with lipid raft occurred within seconds, resulting in the cross-linking of BCR (60). Upon Ag stimulation, core fucosylation influences the binding of BCR to Ag peptide at 30 seconds, the formation of lipid raft at 3 min, and signal transduction via BCR after 5 min (10). FUT8 ablation suppresses the together with BCR, Lyn, and GM1 or caveolin-1 in lipid raft domains. Except for their role in signaling molecules concentrating, lipid rafts play a significant role in protein internalization through the endocytic pathway (60). The FUT8 gene disruption significantly reduces the Ag endocytosis of B cells (10). Several mechanisms may contribute to the reduced lipid raft formation by FUT8 ablation. First, because of the conformation with an increasing affinity for lipid rafts caused by Ag-driven oligomerization of the BCR (58), the failed oligomerizations of BCR by afucosylation result in reduced lipid raft formation. Second, caveolin-1 is a scaffolding protein of cholesterol-rich caveolae lipid rafts (57), the inhibited recruitment of caveolin-1 by FUT8 ablation influences the efficient coalescing of the lipid raft in the plasma membrane. Third, the depletion of core fucose increased the hydrophilicity of immune molecules and impaired lipid raft formation (61) (Figure 4). Core fucosylation controls several parameters associated with B cell activation, including Ag recognition, BCR oligomerization, signal transduction via BCR, and lipid raft cluster formation.
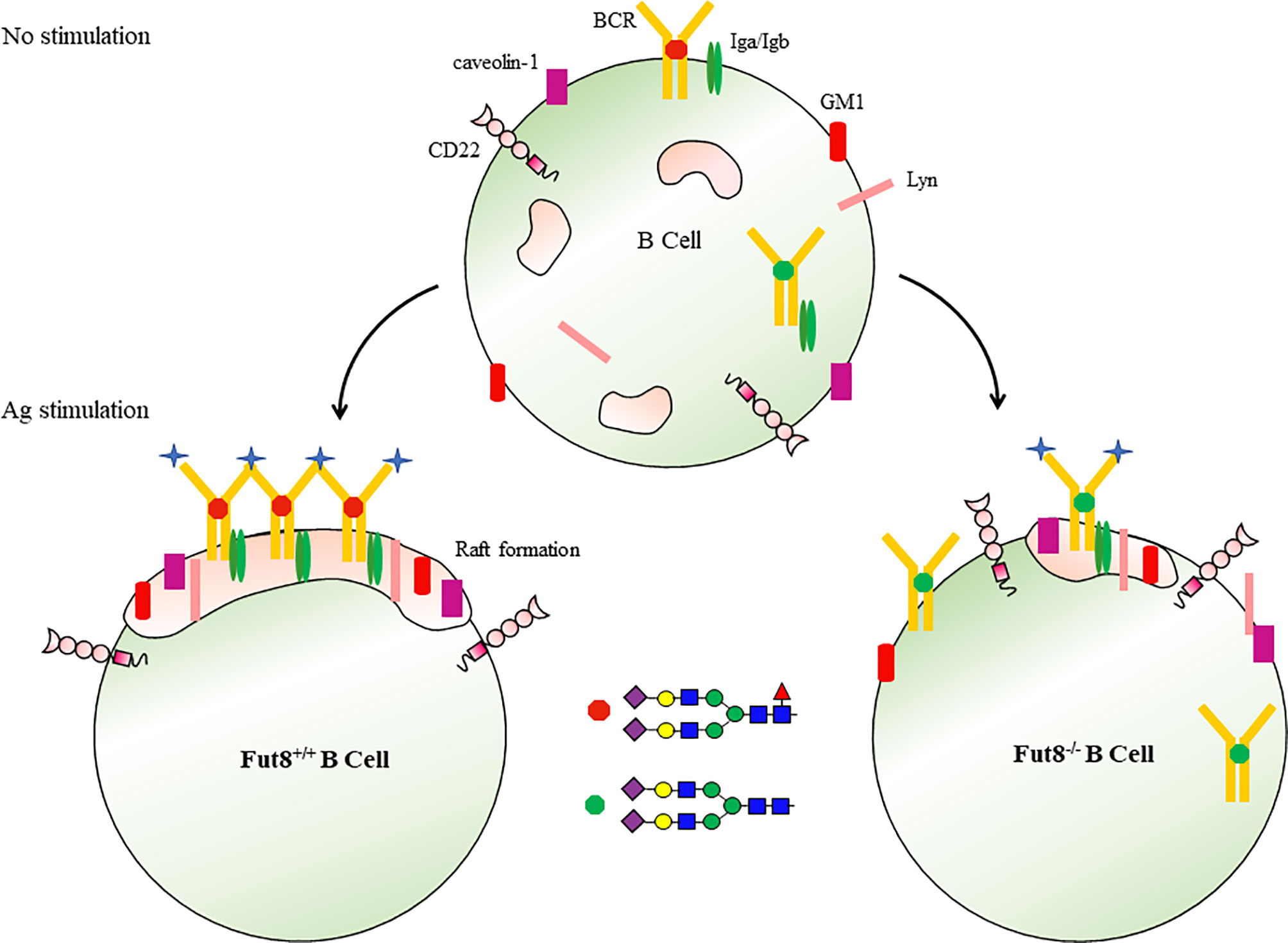
Figure 4 Core fucosylation regulates the signal transduction via BCR. Upon Ag engagement, BCRs on the surface of B cells associated with lipid rafts and triggers signal transductions required for the activation of B cells. Core fucosylation promoted B cells to form a highly efficient lipid raft domain, thereby regulating signal transduction via BCR.
Core Fucosylation Regulates the TH-B Cell Interaction
Upon Ag engagement by BCR, B cells process and present the Ag peptide in association with the major histocompatibility complex class II (MHC-II) molecules via BCR-mediated endocytosis (3). Then, the peptide-loaded MHC-II complex (pMHC-II) on B cells can be recognized by Ag-specific armed TH cells via T cell receptor (TCR) (62). During TH–B cell interaction, activated TH cells produce cytokines for the B lymphocytes clonal expansion and differentiate into Ab-secreting cells (58). Several studies revealed the contribution of glycosylation in the TH-B cell interaction (20). Loss of GlcNAcT-V glycosyltransferase (GnT-V), for example, could promote the recruitment of TCR to the synapse and enhance TCR internalization (63). FUT1 overexpressed T cells enhance the signaling pathway via TCR and apoptosis required for thymocyte maturation arrest (64). The α2,6-sialyltransferase (ST6Gal-1) is required for the development of humoral immunity (65, 66). The removal of N-glycan in T cells improves the functional avidity of TCR to Ag recognition (67). Notably, the most immune molecules participated in T and B cell activation are core-fucosylated glycoproteins. To assess the function of FUT8 in the T–B cell interactions, we obtained FUT8+/+OT-II and FUT8-/-OT-II mice by crossing FUT8+/- mice with OT-II mice (these transgenic mice express CD4+ TCR specific for chicken ovalbumin 323-339, OVA323–339). Most key proteins associated with Ag recognition and the orchestration including MHC-I and MHC-II, are glycosylated (68). Although I-Ad has a single N-glycan (69), core fucosylation cannot change the OVA323–339 presentation abilities of MHC-II (11, 12). However, compared to FUT8+/+OT-II cells, the communication of T–B cells (MHC-II+ TCRβ+ cells) is significantly decreased in FUT8-/-OT-II cells (11). Also, ZAP-70 and Syk phosphorylation was significantly reduced in FUT8-/-OT-II cells with interaction by OVA323–339-loaded B cells. Given that the pMHC on B cells induces a particular conformational change of TCR on T cells (70, 71), and the core fucose is expressed on the cell surface of T and B cells and could affect the flexibility and the conformation of proteins (21). It is reasonable to propose that core fucosylation would influence the conformational stability and geometry of any TCR–pMHC clusters in the TH–B cell interactions. Alternatively, a diverse range of glycan-binding proteins, such as galectins and siglecs, bind to the sugar chains on these glycoproteins, thereby modulating cell signaling and cell-cell interactions (21). Galectin-1-/- mice show abnormal thymocyte selection, causing alteration in T cell responses (72). CD22 selectin, which recognizes α2,6-linked sialylated glycans, is a B cell co-receptor that decreases the signaling via BCR (73). Fucose-specific lectins (74) enable to participate the events in the interactions between T and B cells.
Ab production is one of the major events in the adaptive humoral immune response. The titer of anti-OVA IgG is markedly decreased after the OVA immunization in FUT8-/- mice (11). Moreover, compared to FUT8+/+ SPLs, the number of IgG-producing cells is obviously decreased in FUT8-/- SPLs. During S. typhimurium infection, the production of IgG and sIgA specific for bacteria is also decreased in FUT8-/- mice with attenuated the TH–B cell interactions, but not in communication between T cells and DCs (12), indicating that core fucosylation is essential for the efficient Ab production. Ig class-switching recombination generates an isotype-switched Abs, which are essential for mediating effective humoral immunity (75). The isotype switching of a mature B cell via its BCR from one class to another depends upon the interaction of Ag-stimulated B cells with TH cells. To assess FUT8 function in Ig class-switching, IgGs of different subclasses (class-switched) and IgM (non-switched) are measured in the sera of FUT8-/- mice. The amounts of IgG subclasses, IgG1, IgG2a, IgG2b, and IgG3 are significantly reduced in FUT8-/- mice due to low levels of cytokines, such as IL-4, IL-5, IL-6, IFNγ, and TGF secreted by FUT8-/-TH cells, while amounts of IgM are relatively normal (11). Core fucosylation plays a crucial role in all steps required for Abs production. First, in the TH-B cell interactions, core fucosylation regulates the geometry and conformation of TCR-pMHC clusters in lipid rafts. Second, in cytokine production, core fucosylation controls the expression level of TH2-type cytokines (IL-4, IL-6) associated with the activation of TH and B cells (12). Third, in B cell generation, FUT8 affects the population of CD19+ B cells.
Several studies have been reported that aberrant cues of glycosylation contribute to autoimmune disease (AD) pathogenesis (76–78). For example, the low levels of sialylated IgG are detected in the sera of rheumatoid arthritis and Wegener’s granulomatosis patients, while the sialylated IgG is increased during remission (78, 79). Galactosylated IgG1 is significantly reduced in the sera of rheumatoid arthritis patients (77). GnT-V ablation induces the coclustering of TCRs at the cell surface, reducing the threshold for T cell activation and causing multiple sclerosis (63). SLE is a typical autoimmune disease whose characteristics are inflammatory disorders and autoantibodies production (80). The level of core fucosylated IgG is significantly increased in the sera of SLE patients (11, 76, 81). Indeed, TH cell activation in the peripheral blood of SLE patients plays a crucial role in SLE pathogenesis, and the hyperactivity of B cells is TH cell-dependent in SLE (82, 83). The remarkable differences of glycosylation on TH cells have been observed in active SLE patients (84). Core fucosylation is required for TCR activation in TH cells (61). Increased-core fucosylation induces TH cell hyper-activation and contributes to the SLE severity and pathogenesis.
Core Fucosylation Regulates the Function of IgGs
Glycosylation occurs normally in human IgGs, and plays a significant role in IgG function (85–87). Structural evidence proved that each IgG molecule has a highly conserved N-glycan at Asn297 in the CH2/CH3 domain of Fc regions, and plays a crucial role in sustaining the conformation of Fc domain of IgGs. Multiple sugar chain moieties extend from Fc domains toward each other into the interchain region of IgG, and then they stabilize the IgG framework (88). Differences in the sugar chains determine the variances in the orientation of the protein surface (88).
Several functions of IgGs are mediated through Fcγ receptors (FcγRs) on the effector cells. Because the N-glycan at Asn297 on the Fc domain is located next to the FcγRs binding interface, glycosylation controls the biological function of IgGs (88). The Fc domain sialylation triggers conformational changes in IgG1 that enable interactions with type II FcγRs, while core fucose alters type I FcγRs binding of IgG1 by modulating the Fc’s affinity for FcγRIIIa. Core fucosylated N-glycans attached to the Fc region is a critical determinant of antibody-dependent cell-mediated cytotoxicity (ADCC), as the deletion of core fucose from the Fc region enhances its binding affinity to the FcγRs and significantly improves ADCC (Figure 5) (89–92). The highly de-fucosylated (∼60%) IgG1 exhibits 100-fold ADCC compared to hyper fucosylated (∼10% defucosylated) IgG1, without any difference for Ag binding. Dengue virus infection increases the afucosylation level of IgG1, and the level of afucosylated IgG1 could predict the severity of dengue disease (93). Moreover, afucosylated IgG1 plays a critical role in immune responses to enveloped viruses, including COVID-19 (94). Also, afucosylated IgG efficiently induced FcγRs-dependent natural killer (NK) cell degranulation in malaria (95). As a result of these advances, Ab “glycoengineering” is currently gaining attention as an approach to enhance the effects of therapeutic Abs for tumor and virus infection (96).
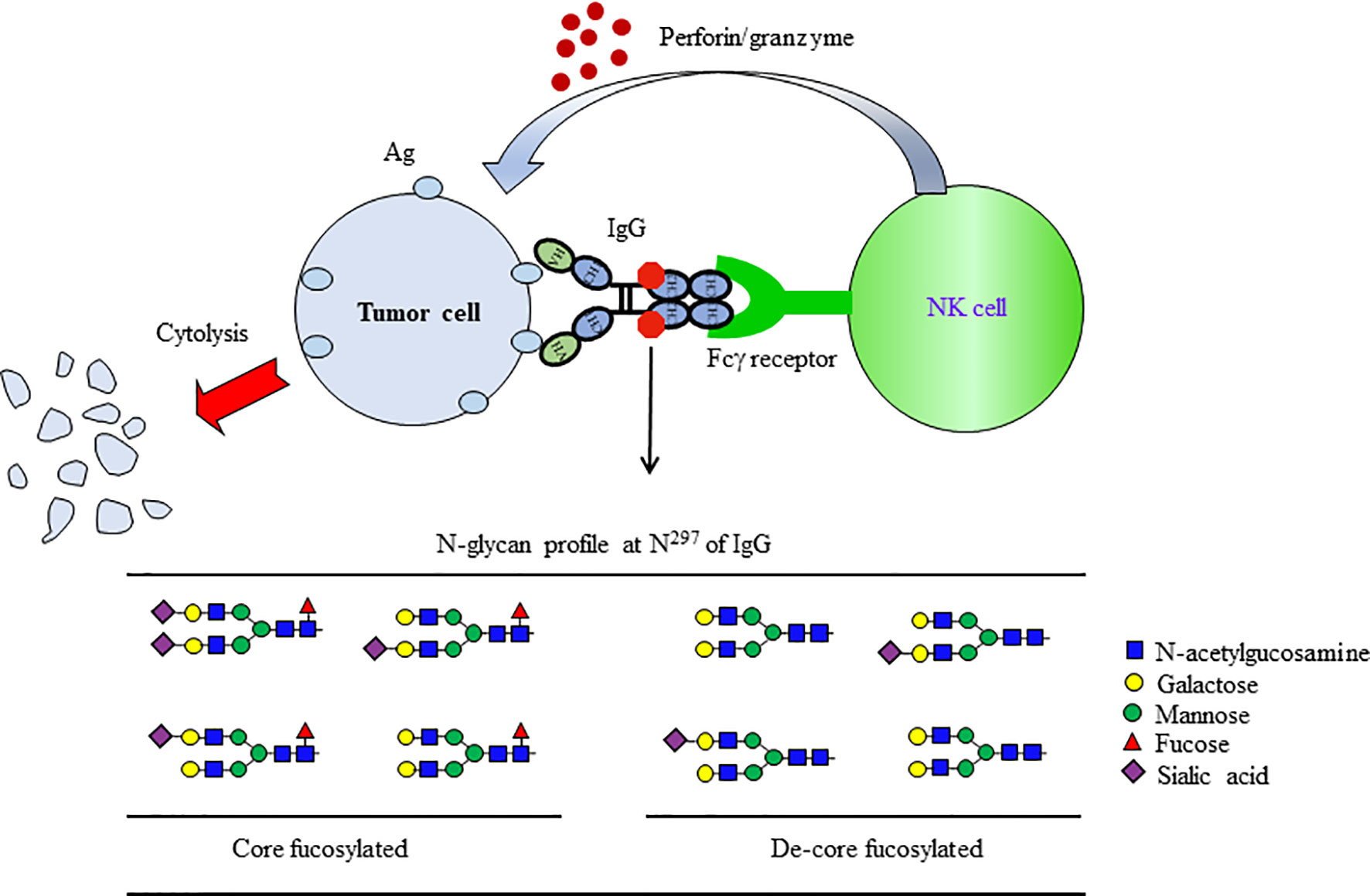
Figure 5 De-fucosylated IgG-Fc domain enhanced the induction of ADCC. ADCC is a specific effector mechanism of natural killer (NK) cells. ADCC refers to the killing of a target cell, which is coated with Abs by NK cells of the immune system. When the antibodies bind to the target Ag (tumor Ag), the IgG Fc is recognized by NK cells via FcγRs on their surfaces. Following recognition, the NK cells release chemicals, which then lyse and kill the target cell bound to the Ab. The removal of core fucosylation at Asn297 on IgG1 results in a 50~100-fold enhancement of ADCC.
In human serum, FUT8 is derived in about 95% from blood platelets (97). Platelets release the FUT8 with components of vesicles during blood coagulation (98). Serum IgG contains more than 30 different N-glycan profiles, and all of the major N-glycans were core fucosylated (99). Core fucosylated glycans is increased on the IgG of SLE patients (81). The hyper-core fucosylation frequently occurs in the sera of epithelial ovarian cancer patients with cisplatin resistance (23) and lung adenocarcinoma patients (14), but the core fucosylation level is down-regulated in the sera of patients with cervical cancer (100). Core fucosylation is also increased in intestinal tissues of patients with inflammatory bowel disease (61).
Conclusion and Perspectives
Appropriate B cell responses are critical for adaptive humoral immunity. Early B cell differentiation, activation and population of B cells, TH-B cell interaction and Ab production are processes carefully orchestrated by a complex network of immune molecules. During B cell development, approximately 90% of B cells are eliminated due to the greatly restricting mature immune repertoire of BCR available for Ag recognition (101). Hence, the mechanisms regulating B cell development are crucial in treating of ADs and improving vaccination strategies.
FUT8 can modify multiple proteins, and core fucosylation of N-glycans of the immune molecules could significantly alter their functions (14). Based on our previous study, core fucosylation is not only associated with pre-BCR assembly and Ag recognition of BCR but also plays a crucial role in effective lipid raft formation, TH–B cell interaction, and Ab production in guiding B lymphopoiesis to shape humoral immunity. It is worth noting that the loss of FUT8 inhibits effective Ab production, while hyper-activation of these cells results in SLE pathogenesis. There are 2937 single-nucleotide polymorphisms (SNPs) in the FUT8 gene region, and the FUT8 gene rs35949016 SNP could affect FUT8 expression (51, 102). Given that the balance between specific and degenerate immune responses holds a vital illumination for protective immunity versus autoimmunity, the relevance of FUT8 SNPs with immune regulatory activity will be considered. Several core fucosylated immune molecules have been identified thus far, and their significance in immune responses is becoming clear. However, the following points seem to be limitations of the research on core fucosylation. First, because FUT8 could regulate multiple glycoproteins, the widespread changes in core fucosylated proteins in FUT8-/- cells make it difficult to discern the role of core fucosylation in individual core fucosylated proteins. Second, very few core fucosylated glycoproteins crystallize for the entire N-glycan due to glycan microheterogeneity and flexibility. Third, due to the potential of flexibility in glycosidic linkages, the conformational requirements for efficient binding of core fucosylated N-glycans to proteins are difficult to analyze. The future ability to determine the sugar chains of immune receptors combined with the ability to selectively modulate cellular glycomes is expected to offer exciting chances to regulate adaptive immune responses. With a better understanding of how FUT8 regulates the adaptive immune system, its use in therapy for autoimmunity proves to be a helpful intervention from glycoimmunological aspects.
Author Contributions
YS and XL were mainly responsible to write the review and complete the Figures. TW collected and checked the References. WL designed the research and modified the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
WL is supported by grants from the National Nature Science Foundation of China (32171279, 31870797, 31570797, 31270864, 30972675) and Liaoning Provincial Program for Top Discipline of Basic Medical Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Azagra A, Marina-Zárate E, Ramiro AR, Javierre BM, Parra M. From Loops to Looks: Transcription Factors and Chromatin Organization Shaping Terminal B Cell Differentiation. Trends Immunol (2020) 41:46–60. doi: 10.1016/j.it.2019.11.006
2. Hardy RR, Hayakawa K. B Cell Development Pathways. Annu Rev Immunol (2001) 19:595–621. doi: 10.1146/annurev.immunol.19.1.595
3. Batista FD, Harwood NE. The Who, How and Where of Antigen Presentation to B Cells. Nat Rev Immunol (2009) 9:15–27. doi: 10.1038/nri2454
4. Baum LG, Cobb BA. The Direct and Indirect Effects of Glycans on Immune Function. Glycobiology (2017) 27:619–24. doi: 10.1093/glycob/cwx036
5. Wolfert MA, Boons GJ. Adaptive Immune Activation: Glycosylation Does Matter. Nat Chem Biol (2013) 9:776–84. doi: 10.1038/nchembio.1403
6. Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell (2006) 126:855–67. doi: 10.1016/j.cell.2006.08.019
8. Li W, Ishihara K, Yokota T, Nakagawa T, Koyama N, Jin J, et al. Reduced Alpha4beta1 Integrin/VCAM-1 Interactions Lead to Impaired Pre-B Cell Repopulation in Alpha 1,6-Fucosyltransferase Deficient Mice. Glycobiology (2008) 18:114–24. doi: 10.1093/glycob/cwm107
9. Li W, Liu Q, Pang Y, Jin J, Wang H, Cao H, et al. Core Fucosylation of μ Heavy Chains Regulates Assembly and Intracellular Signaling of Precursor B Cell Receptors. J Biol Chem (2012) 287:2500–8. doi: 10.1074/jbc.M111.303123
10. Li W, Yu R, Ma B, Yang Y, Jiao X, Liu Y, et al. Core Fucosylation of IgG B Cell Receptor Is Required for Antigen Recognition and Antibody Production. J Immunol (Baltimore Md (2015) 1950) 194:2596–606. doi: 10.4049/jimmunol.1402678
11. Liang W, Mao S, Sun S, Li M, Li Z, Yu R, et al. Core Fucosylation of the T Cell Receptor Is Required for T Cell Activation. Front Immunol (2018) 9:78. doi: 10.3389/fimmu.2018.00078
12. Zahid D, Zhang N, Fang H, Gu J, Li M, Li W. Loss of Core Fucosylation Suppressed the Humoral Immune Response in Salmonella Typhimurium Infected Mice. J Microbiol Immunol Infect = Wei mian yu gan ran za zhi (2020) 606–15. doi: 10.1016/j.jmii.2020.02.006
13. Hao S, Fan Q, Bai Y, Fang H, Zhou J, Fukuda T, et al. Core Fucosylation of Intestinal Epithelial Cells Protects Against Salmonella Typhi Infection via Up-Regulating the Biological Antagonism of Intestinal Microbiota. Front Microbiol (2020) 11:1097. doi: 10.3389/fmicb.2020.01097
14. Zhang N, Li M, Xu X, Zhang Y, Liu Y, Zhao M, et al. Loss of Core Fucosylation Enhances the Anticancer Activity of Cytotoxic T Lymphocytes by Increasing PD-1 Degradation. Eur J Immunol (2020) 1820–33. doi: 10.1002/eji.202048543
15. Sturla L, Fruscione F, Noda K, Miyoshi E, Taniguchi N, Contini P, et al. Core Fucosylation of N-Linked Glycans in Leukocyte Adhesion Deficiency/Congenital Disorder of Glycosylation IIc Fibroblasts. Glycobiology (2005) 15:924–34. doi: 10.1093/glycob/cwi081
16. Cai D, Xun C, Tang F, Tian X, Yang L, Ding K, et al. Glycoconjugate Probes Containing a Core-Fucosylated N-Glycan Trisaccharide for Fucose Lectin Identification and Purification. Carbohydr Res (2017) 449:143–52. doi: 10.1016/j.carres.2017.07.011
17. Ihara H, Ikeda Y, Toma S, Wang X, Suzuki T, Gu J, et al. Crystal Structure of Mammalian Alpha1,6-Fucosyltransferase, FUT8. Glycobiology (2007) 17:455–66. doi: 10.1093/glycob/cwl079
18. Thaysen-Andersen M, Packer NH. Site-Specific Glycoproteomics Confirms That Protein Structure Dictates Formation of N-Glycan Type, Core Fucosylation and Branching. Glycobiology (2012) 22:1440–52. doi: 10.1093/glycob/cws110
19. De Boeck H, Loontiens FG, Lis H, Sharon N. Binding of Simple Carbohydrates and Some N-Acetyllactosamine-Containing Oligosaccharides to Erythrina Cristagalli Agglutinin as Followed With a Fluorescent Indicator Ligand. Arch Biochem Biophys (1984) 234:297–304. doi: 10.1016/0003-9861(84)90352-7
20. Wormald MR, Dwek RA. Glycoproteins: Glycan Presentation and Protein-Fold Stability. Struct (London England: 1993) (1999) 7:R155–60. doi: 10.1016/S0969-2126(99)80095-1
21. Stubbs HJ, Lih JJ, Gustafson TL, Rice KG. Influence of Core Fucosylation on the Flexibility of a Biantennary N-Linked Oligosaccharide. Biochemistry (1996) 35:937–47. doi: 10.1021/bi9513719
22. Bourne Y, Mazurier J, Legrand D, Rougé P, Montreuil J, Spik G, et al. Structures of a Legume Lectin Complexed With the Human Lactotransferrin N2 Fragment, and With an Isolated Biantennary Glycopeptide: Role of the Fucose Moiety. Struct (London Engl (1994) 1993) 2:209–19. doi: 10.1016/S0969-2126(00)00022-8
23. Lv X, Song J, Xue K, Li Z, Li M, Zahid D, et al. Core Fucosylation of Copper Transporter 1 Plays a Crucial Role in Cisplatin-Resistance of Epithelial Ovarian Cancer by Regulating Drug Uptake. Mol Carcinog (2019) 58:794–807. doi: 10.1002/mc.22971
24. Wormald MR, Wooten EW, Bazzo R, Edge CJ, Feinstein A, Rademacher TW, et al. The Conformational Effects of N-Glycosylation on the Tailpiece From Serum IgM. Eur J Biochem (1991) 198:131–9. doi: 10.1111/j.1432-1033.1991.tb15995.x
25. Schneider M, Al-Shareffi E, Haltiwanger RS. Biological Functions of Fucose in Mammals. Glycobiology (2017) 27:601–18. doi: 10.1093/glycob/cwx034
26. Li W, Nakagawa T, Koyama N, Wang X, Jin J, Mizuno-Horikawa Y, et al. Down-Regulation of Trypsinogen Expression Is Associated With Growth Retardation in Alpha1,6-Fucosyltransferase-Deficient Mice: Attenuation of Proteinase-Activated Receptor 2 Activity. Glycobiology (2006) 16:1007–19. doi: 10.1093/glycob/cwl023
27. Osumi D, Takahashi M, Miyoshi E, Yokoe S, Lee SH, Noda K, et al. Core Fucosylation of E-Cadherin Enhances Cell-Cell Adhesion in Human Colon Carcinoma WiDr Cells. Cancer Sci (2009) 100:888–95. doi: 10.1111/j.1349-7006.2009.01125.x
28. Ito Y, Miyauchi A, Yoshida H, Uruno T, Nakano K, Takamura Y, et al. Expression of Alpha1,6-Fucosyltransferase (FUT8) in Papillary Carcinoma of the Thyroid: Its Linkage to Biological Aggressiveness and Anaplastic Transformation. Cancer Lett (2003) 200:167–72. doi: 10.1016/S0304-3835(03)00383-5
29. Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, et al. Dysregulation of TGF-Beta1 Receptor Activation Leads to Abnormal Lung Development and Emphysema-Like Phenotype in Core Fucose-Deficient Mice. Proc Natl Acad Sci USA (2005) 102:15791–6. doi: 10.1073/pnas.0507375102
30. Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. Core Fucosylation Regulates Epidermal Growth Factor Receptor-Mediated Intracellular Signaling. J Biol Chem (2006) 281:2572–7. doi: 10.1074/jbc.M510893200
31. Gao C, Maeno T, Ota F, Ueno M, Korekane H, Takamatsu S, et al. Sensitivity of Heterozygous α1,6-Fucosyltransferase Knock-Out Mice to Cigarette Smoke-Induced Emphysema: Implication of Aberrant Transforming Growth Factor-β Signaling and Matrix Metalloproteinase Gene Expression. J Biol Chem (2012) 287:16699–708. doi: 10.1074/jbc.M111.315333
32. Fukuda T, Hashimoto H, Okayasu N, Kameyama A, Onogi H, Nakagawasai O, et al. Alpha1,6-Fucosyltransferase-Deficient Mice Exhibit Multiple Behavioral Abnormalities Associated With a Schizophrenia-Like Phenotype: Importance of the Balance Between the Dopamine and Serotonin Systems. J Biol Chem (2011) 286:18434–43. doi: 10.1074/jbc.M110.172536
33. Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural Insight Into Pre-B Cell Receptor Function. Science (New York NY) (2007) 316:291–4. doi: 10.1126/science.1139412
34. Wasserman R, Li YS, Shinton SA, Carmack CE, Manser T, Wiest DL, et al. A Novel Mechanism for B Cell Repertoire Maturation Based on Response by B Cell Precursors to Pre-B Receptor Assembly. J Exp Med (1998) 187:259–64. doi: 10.1084/jem.187.2.259
35. Hess J, Werner A, Wirth T, Melchers F, Jäck HM, Winkler TH. Induction of Pre-B Cell Proliferation After De Novo Synthesis of the Pre-B Cell Receptor. Proc Natl Acad Sci USA (2001) 98:1745–50. doi: 10.1073/pnas.98.4.1745
36. Mårtensson IL, Almqvist N, Grimsholm O, Bernardi AI. The Pre-B Cell Receptor Checkpoint. FEBS Lett (2010) 584:2572–9. doi: 10.1016/j.febslet.2010.04.057
37. Kawano Y, Yoshikawa S, Minegishi Y, Karasuyama H. Selection of Stereotyped VH81X-{Micro}H Chains via Pre-B Cell Receptor Early in Ontogeny and Their Conservation in Adults by Marginal Zone B Cells. Int Immunol (2005) 17:857–67. doi: 10.1093/intimm/dxh265
38. Kawano Y, Yoshikawa S, Minegishi Y, Karasuyama H. Pre-B Cell Receptor Assesses the Quality of IgH Chains and Tunes the Pre-B Cell Repertoire by Delivering Differential Signals. J Immunol (Baltimore Md (2006) 1950) 177:2242–9. doi: 10.4049/jimmunol.177.4.2242
39. Zhang M, Srivastava G, Lu L. The Pre-B Cell Receptor and Its Function During B Cell Development. Cell Mol Immunol (2004) 1:89–94.
40. Conley ME, Rohrer J, Rapalus L, Boylin EC, Minegishi Y. Defects in Early B-Cell Development: Comparing the Consequences of Abnormalities in Pre-BCR Signaling in the Human and the Mouse. Immunol Rev (2000) 178:75–90. doi: 10.1034/j.1600-065X.2000.17809.x
41. Espeli M, Rossi B, Mancini SJ, Roche P, Gauthier L, Schiff C. Initiation of Pre-B Cell Receptor Signaling: Common and Distinctive Features in Human and Mouse. Semin Immunol (2006) 18:56–66. doi: 10.1016/j.smim.2005.11.002
42. Corcos D, Dunda O, Butor C, Cesbron JY, Lorès P, Bucchini D, et al. Pre-B-Cell Development in the Absence of Lambda 5 in Transgenic Mice Expressing a Heavy-Chain Disease Protein. Curr Biol: CB (1995) 5:1140–8. doi: 10.1016/S0960-9822(95)00230-2
43. Haimovich J, Ben Moshe N, Raviv Y, Hollander N. All Oligosaccharide Moieties of the μ Chains in the Pre-BCR Are of the High-Mannose Type. Mol Immunol (2010) 48:351–5. doi: 10.1016/j.molimm.2010.07.005
44. Ubelhart R, Bach MP, Eschbach C, Wossning T, Reth M, Jumaa H. N-Linked Glycosylation Selectively Regulates Autonomous Precursor BCR Function. Nat Immunol (2010) 11:759–65. doi: 10.1038/ni.1903
45. ten Boekel E, Melchers F, Rolink AG. Changes in the V(H) Gene Repertoire of Developing Precursor B Lymphocytes in Mouse Bone Marrow Mediated by the Pre-B Cell Receptor. Immunity (1997) 7:357–68. doi: 10.1016/S1074-7613(00)80357-X
46. Marshall AJ, Wu GE, Paige GJ. Frequency of VH81x Usage During B Cell Development: Initial Decline in Usage Is Independent of Ig Heavy Chain Cell Surface Expression. J Immunol (Baltimore Md (1996) 1950) 156:2077–84.
47. Rabinovich E, Bar-Nun S, Amitay R, Shachar I, Gur B, Taya M, et al. Different Assembly Species of IgM Are Directed to Distinct Degradation Sites Along the Secretory Pathway. J Biol Chem (1993) 268:24145–8. doi: 10.1016/S0021-9258(20)80503-1
48. Gauthier L, Rossi B, Roux F, Termine E, Schiff C. Galectin-1 Is a Stromal Cell Ligand of the Pre-B Cell Receptor (BCR) Implicated in Synapse Formation Between Pre-B and Stromal Cells and in Pre-BCR Triggering. Proc Natl Acad Sci USA (2002) 99:13014–9. doi: 10.1073/pnas.202323999
49. Lopez Granados E, Porpiglia AS, Hogan MB, Matamoros N, Krasovec S, Pignata C, et al. Clinical and Molecular Analysis of Patients With Defects in Micro Heavy Chain Gene. J Clin Invest (2002) 110:1029–35. doi: 10.1172/JCI0215658
50. Hynes RO. Integrins: Versatility, Modulation, and Signaling in Cell Adhesion. Cell (1992) 69:11–25. doi: 10.1016/0092-8674(92)90115-S
51. Rossi B, Espeli M, Schiff C, Gauthier L. Clustering of Pre-B Cell Integrins Induces Galectin-1-Dependent Pre-B Cell Receptor Relocalization and Activation. J Immunol (Baltimore Md (2006) 1950) 177:796–803. doi: 10.4049/jimmunol.177.2.796
52. Gauld SB, Dal Porto JM, Cambier JC. B Cell Antigen Receptor Signaling: Roles in Cell Development and Disease. Science (New York NY) (2002) 296:1641–2. doi: 10.1126/science.1071546
53. Raff MC, Sternberg M, Taylor RB. Immunoglobulin Determinants on the Surface of Mouse Lymphoid Cells. Nature (1970) 225:553–4. doi: 10.1038/225553a0
54. Gomes de Castro MA, Wildhagen H, Sograte-Idrissi S, Hitzing C, Binder M, Trepel M, et al. Differential Organization of Tonic and Chronic B Cell Antigen Receptors in the Plasma Membrane. Nat Commun (2019) 10:820. doi: 10.1038/s41467-019-08677-1
55. Hagman J. Conveying the Message: Identification of Ig-Alpha and Ig-Beta as Components of the B Cell Receptor Complex. J Immunol (Baltimore Md: 1950) (2009) 183:1503–4. doi: 10.4049/jimmunol.0990055
56. Sezgin E, Levental I, Mayor S, Eggeling C. The Mystery of Membrane Organization: Composition, Regulation and Roles of Lipid Rafts. Nat Rev Mol Cell Biol (2017) 18:361–74. doi: 10.1038/nrm.2017.16
57. Cheng PC, Dykstra ML, Mitchell RN, Pierce SK. A Role for Lipid Rafts in B Cell Antigen Receptor Signaling and Antigen Targeting. J Exp Med (1999) 190:1549–60. doi: 10.1084/jem.190.11.1549
58. Pierce SK. Lipid Rafts and B-Cell Activation. Nat Rev Immunol (2002) 2:96–105. doi: 10.1038/nri726
59. Gupta N, DeFranco AL. Visualizing Lipid Raft Dynamics and Early Signaling Events During Antigen Receptor-Mediated B-Lymphocyte Activation. Mol Biol Cell (2003) 14:432–44. doi: 10.1091/mbc.02-05-0078
60. Sohn HW, Tolar P, Jin T, Pierce SK. Fluorescence Resonance Energy Transfer in Living Cells Reveals Dynamic Membrane Changes in the Initiation of B Cell Signaling. Proc Natl Acad Sci USA (2006) 103:8143–8. doi: 10.1073/pnas.0509858103
61. Fujii H, Shinzaki S, Iijima H, Wakamatsu K, Iwamoto C, Sobajima T, et al. Core Fucosylation on T Cells, Required for Activation of T-Cell Receptor Signaling and Induction of Colitis in Mice, Is Increased in Patients With Inflammatory Bowel Disease. Gastroenterology (2016) 150:1620–32. doi: 10.1053/j.gastro.2016.03.002
62. Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, et al. The Crystal Structure of a T Cell Receptor in Complex With Peptide and MHC Class II. Science (New York NY) (1999) 286:1913–21. doi: 10.1126/science.286.5446.1913
63. Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative Regulation of T-Cell Activation and Autoimmunity by Mgat5 N-Glycosylation. Nature (2001) 409:733–9. doi: 10.1038/35055582
64. Moore GT, Brown SJ, Winterhalter AC, Lust M, Salvaris EJ, Selan C, et al. Glycosylation Changes in Hfut1 Transgenic Mice Increase TCR Signaling and Apoptosis Resulting in Thymocyte Maturation Arrest. Mol Immunol (2008) 45:2401–10. doi: 10.1016/j.molimm.2007.11.006
65. Hennet T, Chui D, Paulson JC, Marth JD. Immune Regulation by the ST6Gal Sialyltransferase. Proc Natl Acad Sci USA (1998) 95:4504–9. doi: 10.1073/pnas.95.8.4504
66. Irons EE, Lau JTY. Systemic ST6Gal-1 Is a Pro-Survival Factor for Murine Transitional B Cells. Front Immunol (2018) 9:2150. doi: 10.3389/fimmu.2018.02150
67. Kuball J, Hauptrock B, Malina V, Antunes E, Voss RH, Wolfl M, et al. Increasing Functional Avidity of TCR-Redirected T Cells by Removing Defined N-Glycosylation Sites in the TCR Constant Domain. J Exp Med (2009) 206:463–75. doi: 10.1084/jem.20082487
68. Johnson JL, Jones MB, Ryan SO, Cobb BA. The Regulatory Power of Glycans and Their Binding Partners in Immunity. Trends Immunol (2013) 34:290–8. doi: 10.1016/j.it.2013.01.006
69. Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal Structures of Two I-Ad-Peptide Complexes Reveal That High Affinity can be Achieved Without Large Anchor Residues. Immunity (1998) 8:319–29. doi: 10.1016/S1074-7613(00)80537-3
70. Alam SM, Davies GM, Lin CM, Zal T, Nasholds W, Jameson SC, et al. Qualitative and Quantitative Differences in T Cell Receptor Binding of Agonist and Antagonist Ligands. Immunity (1999) 10:227–37. doi: 10.1016/S1074-7613(00)80023-0
71. Schamel WW, Arechaga I, Risueño RM, van Santen HM, Cabezas P, Risco C, et al. Coexistence of Multivalent and Monovalent TCRs Explains High Sensitivity and Wide Range of Response. J Exp Med (2005) 202:493–503. doi: 10.1084/jem.20042155
72. Perillo NL, Pace KE, Seilhamer JJ, Baum LG. Apoptosis of T Cells Mediated by Galectin-1. Nature (1995) 378:736–9. doi: 10.1038/378736a0
73. Poe JC, Tedder TF. CD22 and Siglec-G in B Cell Function and Tolerance. Trends Immunol (2012) 33:413–20. doi: 10.1016/j.it.2012.04.010
74. Manabe Y, Marchetti R, Takakura Y, Nagasaki M, Nihei W, Takebe T, et al. The Core Fucose on an IgG Antibody Is an Endogenous Ligand of Dectin-1. Angewandte Chemie (International Ed English) (2019) 58:18697–702. doi: 10.1002/anie.201911875
75. Chen Z, Wang JH. Signaling Control of Antibody Isotype Switching. Adv Immunol (2019) 141:105–64. doi: 10.1016/bs.ai.2019.01.001
76. Vučković F, Krištić J, Gudelj I, Teruel M, Keser T, Pezer M, et al. Association of Systemic Lupus Erythematosus With Decreased Immunosuppressive Potential of the IgG Glycome. Arthritis Rheumatol (Hoboken NJ) (2015) 67:2978–89. doi: 10.1002/art.39273
77. Ercan A, Cui J, Chatterton DE, Deane KD, Hazen MM, Brintnell W, et al. Aberrant IgG Galactosylation Precedes Disease Onset, Correlates With Disease Activity, and Is Prevalent in Autoantibodies in Rheumatoid Arthritis. Arthritis Rheum (2010) 62:2239–48. doi: 10.1002/art.27533
78. Matsumoto A, Shikata K, Takeuchi F, Kojima N, Mizuochi T. Autoantibody Activity of IgG Rheumatoid Factor Increases With Decreasing Levels of Galactosylation and Sialylation. J Biochem (2000) 128:621–8. doi: 10.1093/oxfordjournals.jbchem.a022794
79. Espy C, Morelle W, Kavian N, Grange P, Goulvestre C, Viallon V, et al. Sialylation Levels of Anti-Proteinase 3 Antibodies Are Associated With the Activity of Granulomatosis With Polyangiitis (Wegener’s). Arthritis Rheum (2011) 63:2105–15. doi: 10.1002/art.30362
80. Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, et al. Normalization of CD4+ T Cell Metabolism Reverses Lupus. Sci Trans Med (2015) 7:274ra18. doi: 10.1126/scitranslmed.aaa0835
81. Sun Y, Li Z, Liang W, Zhang Y, Song W, Song J, et al. A Novel Immunochromatographic Strips Assay for Rapid and Simple Detection of Systemic Lupus Erythematosus. Sci Rep (2020) 10:14178. doi: 10.1038/s41598-020-71137-0
82. Rupanagudi KV, Kulkarni OP, Lichtnekert J, Darisipudi MN, Mulay SR, Schott B, et al. Cathepsin S Inhibition Suppresses Systemic Lupus Erythematosus and Lupus Nephritis Because Cathepsin S Is Essential for MHC Class II-Mediated CD4 T Cell and B Cell Priming. Ann Rheuma Dis (2015) 74:452–63. doi: 10.1136/annrheumdis-2013-203717
83. Mao L, Hou H, Wu S, Zhou Y, Wang J, Yu J, et al. TIGIT Signalling Pathway Negatively Regulates CD4(+) T-Cell Responses in Systemic Lupus Erythematosus. Immunology (2017) 151:280–90. doi: 10.1111/imm.12715
84. Ramos-Martínez E, Lascurain R, Tenorio EP, Sánchez-González A, Chávez-Rueda K, Chávez-Sánchez L, et al. Differential Expression of O-Glycans in CD4(+) T Lymphocytes From Patients With Systemic Lupus Erythematosus. Tohoku J Exp Med (2016) 240:79–89. doi: 10.1620/tjem.240.79
85. Donadel G, Calabro A, Sigounas G, Hascall VC, Notkins AL, Harindranath N. Human Polyreactive and Monoreactive Antibodies: Effect of Glycosylation on Antigen Binding. Glycobiology (1994) 4:491–6. doi: 10.1093/glycob/4.4.491
86. Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural Analysis of Human IgG-Fc Glycoforms Reveals a Correlation Between Glycosylation and Structural Integrity. J Mol Biol (2003) 325:979–89. doi: 10.1016/S0022-2836(02)01250-0
87. Burton DR, Dwek RA. Immunology. Sugar Determines Antibody Activity. Science (New York NY) (2006) 313:627–8. doi: 10.1126/science.1131712
88. Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The Structure of a Human Type III Fcgamma Receptor in Complex With Fc. J Biol Chem (2001) 276:16469–77. doi: 10.1074/jbc.M100350200
89. Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, et al. The Absence of Fucose But Not the Presence of Galactose or Bisecting N-Acetylglucosamine of Human IgG1 Complex-Type Oligosaccharides Shows the Critical Role of Enhancing Antibody-Dependent Cellular Cytotoxicity. J Biol Chem (2003) 278:3466–73. doi: 10.1074/jbc.M210665200
90. Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, et al. Unique Carbohydrate-Carbohydrate Interactions Are Required for High Affinity Binding Between FcgammaRIII and Antibodies Lacking Core Fucose. Proc Natl Acad Sci USA (2011) 108:12669–74. doi: 10.1073/pnas.1108455108
91. Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, et al. IgG Subclass-Independent Improvement of Antibody-Dependent Cellular Cytotoxicity by Fucose Removal From Asn297-Linked Oligosaccharides. J Immunol Methods (2005) 306:151–60. doi: 10.1016/j.jim.2005.08.009
92. Lu J, Chu J, Zou Z, Hamacher NB, Rixon MW, Sun PD. Structure of Fcγri in Complex With Fc Reveals the Importance of Glycan Recognition for High-Affinity IgG Binding. Proc Natl Acad Sci USA (2015) 112:833–8. doi: 10.1073/pnas.1418812112
93. Bournazos S, Vo HTM, Duong V, Auerswald H, Ly S, Sakuntabhai A, et al. Antibody Fucosylation Predicts Disease Severity in Secondary Dengue Infection. Science (New York NY) (2021) 372:1102–5. doi: 10.1126/science.abc7303
94. Larsen MD, de Graaf EL, Sonneveld ME, Plomp HR, Nouta J, Hoepel W, et al. Afucosylated IgG Characterizes Enveloped Viral Responses and Correlates With COVID-19 Severity. Science (New York NY) (2021) 371:eabc8378. doi: 10.1126/science.abc8378
95. Larsen MD, Lopez-Perez M, Dickson EK, Ampomah P, Tuikue Ndam N, Nouta J, et al. Afucosylated Plasmodium Falciparum-Specific IgG Is Induced by Infection But Not by Subunit Vaccination. Nat Commun (2021) 12:5838. doi: 10.1038/s41467-021-26118-w
96. Le NP, Bowden TA, Struwe WB, Crispin M. Immune Recruitment or Suppression by Glycan Engineering of Endogenous and Therapeutic Antibodies. Biochim Biophys Acta (2016) 1860:1655–68. doi: 10.1016/j.bbagen.2016.04.016
97. Kościelak J, Pacuszka T, Miller-Podraza H, Zdziechowska H. Activities of Fucosyltransferases in Sera of Leukaemic Patients: Platelet Origin of Serum Alpha-6-L-Fucosyltransferase. Biochem Soc Trans (1987) 15:603–6. doi: 10.1042/bst0150603
98. Antoniewicz J, Bykowska K, Zdebska E, Kościelak J. Human Platelets Release Alpha-6-L-Fucosyltransferase Upon Activation. FEBS Lett (1989) 244:388–90. doi: 10.1016/0014-5793(89)80569-1
99. Lu G, Holland LA. Profiling the N-Glycan Composition of IgG With Lectins and Capillary Nanogel Electrophoresis. Anal Chem (2019) 91:1375–83. doi: 10.1021/acs.analchem.8b03725
100. Fan Q, Wu Y, Li M, An F, Yao L, Wang M, et al. Lactobacillus spp. Create a Protective Micro-Ecological Environment Through Regulating the Core Fucosylation of Vaginal Epithelial Cells Against Cervical Cancer. Cell Death Dis (2021) 12:1094. doi: 10.1038/s41419-021-04388-y
101. Chung JB, Silverman M, Monroe JG. Transitional B Cells: Step by Step Towards Immune Competence. Trends Immunol (2003) 24:343–9. doi: 10.1016/S1471-4906(03)00119-4
Keywords: core fucosylation, pre-B cell, BCR, IgG, humoral immune response
Citation: Sun Y, Li X, Wang T and Li W (2022) Core Fucosylation Regulates the Function of Pre-BCR, BCR and IgG in Humoral Immunity. Front. Immunol. 13:844427. doi: 10.3389/fimmu.2022.844427
Received: 28 December 2021; Accepted: 25 February 2022;
Published: 25 March 2022.
Edited by:
Irena Trbojević-Akmačić, Genos Glycoscience Research Laboratory, CroatiaReviewed by:
József Prechl, Diagnosticum Zrt., HungaryAnja Lux, University of Erlangen Nuremberg, Germany
Copyright © 2022 Sun, Li, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhe Li, bGl3ZW56aGVAZG11LmVkdS5jbg==
†These authors have contributed equally to this work
 Yuhan Sun1,2†
Yuhan Sun1,2† Xueying Li
Xueying Li Wenzhe Li
Wenzhe Li