
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 25 May 2022
Sec. Microbial Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.842911
 Arno R. Bourgonje1†
Arno R. Bourgonje1† Geesje Roo-Brand2†
Geesje Roo-Brand2† Paola Lisotto2
Paola Lisotto2 Mehdi Sadaghian Sadabad2
Mehdi Sadaghian Sadabad2 Rosanne D. Reitsema3
Rosanne D. Reitsema3 Marcus C. de Goffau4,5
Marcus C. de Goffau4,5 Klaas Nico Faber1
Klaas Nico Faber1 Gerard Dijkstra1‡
Gerard Dijkstra1‡ Hermie J. M. Harmsen2*‡
Hermie J. M. Harmsen2*‡Introduction: Inflammatory bowel disease (IBD) is characterized by a disturbed gut microbiota composition. Patients with IBD have both elevated mucosal and serum levels of IgG-antibodies directed against bacterial antigens, including flagellins. In this study, we aimed to determine to which intestinal bacteria the humoral immune response is directed to in patients with IBD.
Methods: Fecal and serum samples were collected from patients with IBD (n=55) and age- and sex-matched healthy controls (n=55). Fecal samples were incubated with autologous serum and IgG-coated fractions were isolated by magnetic-activated cell sorting (MACS) and its efficiency was assessed by flow cytometry. The bacterial composition of both untreated and IgG-coated fecal samples was determined by 16S rRNA-gene Illumina sequencing.
Results: IgG-coated fecal samples were characterized by significantly lower microbial diversity compared to the fecal microbiome. Both in patients with IBD and controls, serum IgG responses were primarily directed to Streptococcus, Lactobacillus, Lactococcus, Enterococcus, Veillonella and Enterobacteriaceae, as well as against specific Lachnospiraceae bacteria, including Coprococcus and Dorea (all P<0.001), and to Ruminococcus gnavus-like bacteria (P<0.05). In contrast, serological IgG responses against typical commensal, anaerobic and colonic microbial species were rather low, e.g. to the Lachnospiraceae members Roseburia and Blautia, to Faecalibacterium, as well as to Bacteroides. Patients with IBD showed more IgG-coating of Streptococcus, Lactobacillus, and Lactococcus bacteria compared to healthy controls (all P<0.05). No differences in IgG-coated bacterial fractions were observed between Crohn’s disease and ulcerative colitis, between active or non-active disease, nor between different disease locations.
Conclusion: The IgG immune response is specifically targeted at distinct intestinal bacterial genera that are typically associated with the small intestinal microbiota, whereas responses against more colonic-type commensals are lower, which was particularly the case for patients with IBD. These findings may be indicative of a strong immunological exposure to potentially pathogenic intestinal bacteria in concordance with relative immune tolerance against commensal bacteria.
Inflammatory bowel diseases (IBD), encompassing Crohn’s disease (CD) and ulcerative colitis (UC), are chronic ulcerative inflammatory diseases of the gastrointestinal (GI) tract, which are characterized by an inappropriate and uncontrolled immune response against microbiota in genetically susceptible individuals (1). The etiology of IBD remains incompletely understood, although it is believed that a complex interplay between genetic and environmental factors, the gut microbiota and host immunity is driving disease pathogenesis (2, 3). Gut microbiota alterations are commonly observed in patients with IBD, which are characterized by decreased microbial diversity, increased proportions of potentially pathogenic bacteria, and decreased proportions of commensal, butyrate-producing bacteria (4–6). Gut bacteria closely interact with the intestinal barrier, where a prodigious passage of luminal antigens occurs, which are continuously sampled by the underlying mucosal immune system as an immune surveillance mechanism (7). A compromised mucosal barrier is a hallmark of IBD, which results in excessive translocation of luminal antigens, eliciting both mucosal and systemic immune responses, which may in turn trigger and/or aggravate intestinal inflammation (8, 9).
Antimicrobial antibody responses are frequently observed in patients with IBD, particularly in CD (10, 11). Circulating IgG may leak into the intestinal lumen when mucosal barrier integrity is compromised by, for example, ulcerative inflammation (12, 13). This may result in increased IgG-coating of fecal bacteria. Indeed, we previously observed that IgG-coated fecal bacteria were overrepresented in CD compared with healthy controls, which may also associate with disease activity (13–15). Notably, patients with CD demonstrated elevated binding of IgG derived from autologous serum to fecal bacteria, suggesting earlier exposure to these bacterial antigens, perhaps during disease flares when mucosal barrier function was impaired (13). However, the exact antigenic nature of these IgG immune responses is largely unknown, as well as the exact bacteria that are targeted by autologous serum IgG.
In this study, we aimed to investigate to which bacterial genera the humoral IgG response is directed to in patients with IBD and in healthy individuals. To do so, we leveraged magnetic-activated cell sorting (MACS) and flow cytometry (FC) procedures to quantify the fractions of different IgG-coated bacteria after incubating fecal samples from patients and healthy individuals with their own (autologous) serum. Furthermore, we characterized the fecal microbial composition of both untreated and autologous serum-incubated fecal samples using 16S rRNA gene sequencing.
Patients were included based on their participation in the String of Pearls initiative, a Dutch nation-wide project for which detailed phenotypic information and biological materials are collected from patients with IBD (16). Patients were recruited from the outpatient clinic of the department of gastroenterology in the University Medical Center Groningen (UMCG) and enrolled between 2010-2015. For this study, frozen fecal and serum samples were collected from patients with Crohn’s disease (CD) (n=34), ulcerative colitis (UC) (n=17) or inflammatory bowel disease unclassified (IBD-U) (n=4). For each patient, demographic and clinical information was collected, including age, sex, medication use (including biologicals), the Montreal disease classification, surgical history, and disease activity, all of which was assessed at time of sampling. Clinical disease activity was assessed using the Harvey-Bradshaw Index (HBI) for patients with CD and the Simple Clinical Colitis Activity Index (SCCAI) for patients with UC or IBD-U. The Montreal disease classification was recorded from their most recent visit to the outpatient clinic. Patients provided written informed consent for their participation in the study. This study was approved by the Institutional Review Board (IRB) of the UMCG (registered as no. 08/338) and was performed in accordance with the principles of the Declaration of Helsinki (2013). As controls, a cohort of 55 age- and gender-matched healthy individuals was included from the Northern Dutch Lifelines cohort study, from which only data on age and gender were recorded and who had self-proclaimed gastro-intestinal health. From these individuals, stool and serum samples were analyzed in a similar fashion as for those of patients with IBD.
Fecal samples were diluted 50-fold by adding 0.25 grams of fecal sample to 12.25 ml filtered phosphate-buffered saline (PBS). Homogenized samples were centrifuged at 700 xg for 5 min to remove large particulates. The supernatant was stored in 1 ml aliquots at -20°C until further analysis. For 16S rRNA gene sequencing, DNA was extracted from fecal samples using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and following the exact same procedure as described previously (17). In addition, a bead beating cell lysis step was performed using a Precellys 24 homogenizer (Bertin Technologies, Aix-en-Provence, France) and glass beads (0.1 mm) at 5.5 m/s in three rounds of 1 min at room temperature.
A 1 ml fecal sample aliquot (50-fold diluted) was centrifuged at 16,000 xg for 5 min, after which the bacterial pellet was dissolved in 1 ml PBS and centrifuged again. Subsequently, the bacterial pellet was resuspended in 100 μl autologous serum (100-fold diluted in PBS) and incubated on ice for 30 min. This suspension was added to 1 ml PBS/EDTA and centrifuged at 16,000 xg for 5 min. Next, the resulting bacterial pellet was resuspended in 500 μl Anti-Human IgG (γ-chain specific)-Biotin antibody (Sigma-Aldrich, St. Louis, MO, USA) which was 100-fold diluted in PBS/EDTA and incubated on ice for 30 min. Opsonized bacteria were sorted using a kit for magnetic-activated cell sorting (MACS) (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) as follows: 1 ml PBS/EDTA was added to the suspension and the mixture was subsequently centrifuged at 16,000 xg for 5 min. The bacterial pellet was suspended in 100 μl streptavidin-coated magnetic beads which was 10-fold diluted in PBS/EDTA and incubated on ice for 20 min, followed by another washing step with 1 ml PBS/EDTA and centrifugation at 16,000 xg for 5 min. Subsequently, the bacterial pellet was suspended in 500 μl PBS/EDTA. MS Columns were placed in the MACS Separator (magnetic plate) and were pre-rinsed with 500 μl PBS/EDTA. Next, 500 μl cell suspensions were applied onto a MS Column. Unlabeled cells were collected by washing the MS Columns three times with 500 μl PBS/EDTA. After this step, MS Columns were removed from the separator and placed on the new collection tube. In order to collect the magnetically labeled cells, 1.06 ml PBS/EDTA was applied onto the MS Column and immediately flushed out. The obtained liquids were stored at -20°C for DNA extraction and flow cytometry (FC) analysis.
A flow cytometry analysis was performed to confirm the efficiency of the MACS procedure. To do so, 50 μl of the magnetic bead-labeled fraction was resuspended in 100 μl Anti-Human IgG (γ-chain specific)-FITC antibody (Sigma-Aldrich, St. Louis, MO, USA) which was 100-fold diluted in PBS and incubated on ice in the dark for 30 min. This suspension was washed with 1 ml PBS and centrifuged at 16,000 xg for 5 min. The bacterial pellet was resuspended in 50 μl PBS while adding 10 μl propidium iodide (PI) (0.1 mg/ml) (Sigma-Aldrich, St. Louis, MO, USA). Fifty (50) μl of autologous serum-incubated stool supernatant suspension was labeled with Anti-Human IgG-FITC and another 50 μl was only incubated with PI as experimental controls. Samples were analyzed using the BD Accuri™ C6 flow cytometer (BD Biosciences, San Jose, CA, USA). Supernatant of three fecal samples with the largest bacterial pellet were washed with PBS and measured by flow cytometry to check the PI-induced background signal. The FL3-A signal above the threshold of these samples was measured in the analysis.
Fecal microbial composition was determined by Illumina MiSeq paired-end sequencing of the V3-V4 hypervariable region of the 16S rRNA gene (MiSeq Bentchtop Sequencer, Illumina Inc., San Diego, USA). Amplification of bacterial DNA was performed by PCR using modified 341F and 806R primers with a six-nucleotide barcode on the 806R primer for multiplexing (18, 19). Both primers contain an Illumina MiSeq adapter sequence, which is necessary for flow cell binding in the MiSeq machine. Primer sequences can be found in Supplementary Table S1. A detailed description of the PCR amplification procedure, DNA clean-up and MiSeq library preparation using a 2x300 cartridge can be found in the Supplementary Methods. Demultiplexing and clustering of sequencing reads was performed using Quantitative Insights In Microbial Ecology (QIIME) with UCLUST v.1.2.22q at 97% similarity (20, 21). Taxonomic profiling was performed using Paired-eND Assembler for DNA sequences (PANDAseq) (22). All sequences with quality scores <0.9 were discarded by PANDAseq to increase sequence read-out quality, and read numbers per sample were rarefied to 25,000 read/sample. QIIME was used to assign bacterial taxonomy down to family and genus level, and ARB was used to identify sequences further down to species level (23).
Demographic and clinical characteristics of the study population were presented as means ± standard deviations (SD), medians with interquartile ranges (IQR) or proportions n with corresponding percentages (%). Differences between groups were analyzed using Mann-Whitney U-tests, while paired differences were determined used Wilcoxon signed-rank tests while adjusting for multiple comparisons under a false discovery rate (FDR) of 5%. Bacterial genera having a mean relative abundance above 0.25 were considered eligible for analysis. Microbial richness and diversity of samples was estimated with the Shannon diversity index as a measure of microbial alpha-diversity using QIIME. The microbial dissimilarities matrix (Bray-Curtis distances) was obtained using vegdist from the vegan R package and used as a measure of beta-diversity (24). Principal coordinates were constructed using the cmdscale function. Permutational multivariate analysis of variance (PERMANOVA) using distance matrices (ADONIS) was performed to analyze the variance in the Bray-Curtis dissimilarity matrix that could be explained by sample origin (IBD vs HC) or sample treatment (MACS-sorted samples vs. unsorted samples). Statistical analyses were performed using R (v.3.5.2) and the Python programming language (v.3.8.5, Python Software Foundation, https://www.python.org), using the pandas (v.1.2.3), numpy (v.1.20.0) and scikit-bio (v.0.2.3) modules. Data visualization was performed using the seaborn (v.0.11.1) and matplotlib (v.3.4.1) packages in Python. Two-tailed P-values ≤ 0.05 were considered as statistically significant.
We included 55 patients with IBD (34 CD, 17 UC and 4 IBD-U) and 55 healthy controls (HC) (Table 1). Mean age was 44 (range: 20-64) years for patients with IBD and 46 (range: 20-64) years for HC. Among patients with IBD, 60% were female, which was comparable to 53% females among HC. Most patients with IBD were in clinical remission at the time of sampling (76%), whereas approximately one fourth of patients had active disease (24%).
Microbial richness and diversity of both untreated (full fecal microbiome) and IgG-coated fecal samples are presented in Figure 1. The median number of operational taxonomic units (OTUs) per sample, representing sample richness of the gut microbiota, was significantly lower in samples of patients with IBD compared with samples from HC (298 [interquartile range [IQR]: 200-467] vs. 625 [IQR: 504-717], P<0.001) (Figure 1A). No significant paired difference in OTU counts were observed between original and IgG-coated fecal samples of HC (635 [IQR:512-724] vs. 624 [IQR: 497-696], P>0.05), whereas IgG-coated fecal samples from patients with IBD showed a decreased OTU count (340 [206-530] vs. 287 [185-436], P<0.001) compared with untreated fecal samples. The median Shannon diversity index, which represents the alpha-diversity (richness and diversity) of the gut microbiota, was significantly lower in IBD samples compared with samples from HC (3.2 [IQR: 2.7-3.5] vs. 4.1 [IQR: 3.8-4.3], P<0.001) (Figure 1B). Similar to the OTU counts, no paired difference was observed between untreated and IgG-coated fecal samples from HC (4.2 [IQR: 3.8-4.4] vs. 4.0 [3.8-4.3], P>0.05), whereas there was a small difference in untreated vs. IgG-coated samples from patients with IBD (3.2 [IQR: 2.9-3.7] vs. 3.1 [2.7-3.4], P<0.001).
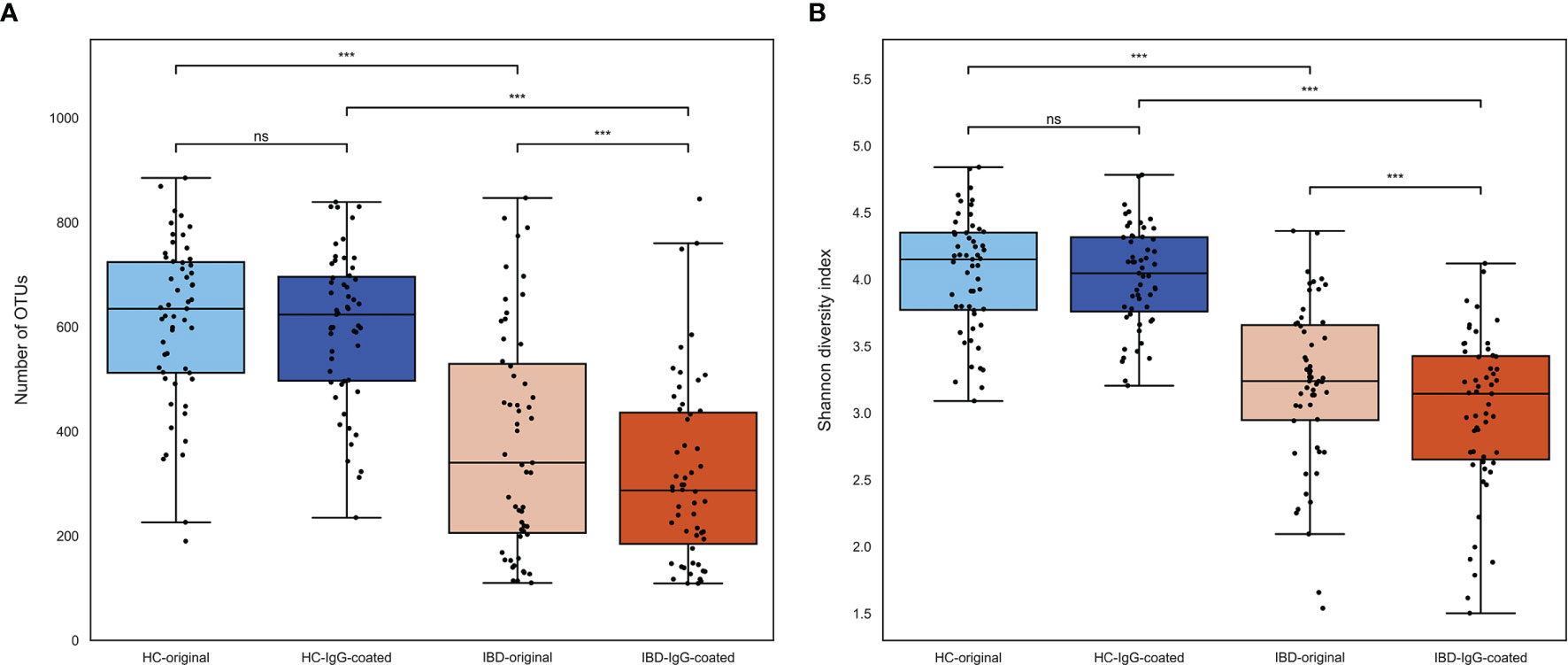
Figure 1 (A, B) Microbial richness and evenness of fecal samples from patients with IBD (orange colored) and HC (blue colored). (A) The number of OTUs was significantly lower in fecal samples from patients with IBD compared with samples from HC. OTUs were not significantly different after IgG-coating in samples from HC, whereas there was a small decrease in IgG-coated vs. untreated fecal samples from patients with IBD (paired analysis, P < 0.001). (B) The Shannon diversity index, representing the alpha-diversity of the gut microbiota, was significantly lower in patients with IBD vs. HC. Similar to the OTUs, IgG-coated vs. untreated samples showed no significant change in Shannon diversity index among HC, whereas there was a small decrease observed in IgG-coated samples from patients with IBD. HC, healthy controls; IBD, inflammatory bowel disease; MACS, magnetic-activated cell sorting; ns, non-significant; OTUs, operational taxonomic units. ***P < 0.001.
The differences in microbial abundances between different fecal samples were evaluated by quantifying the beta-diversity of the gut microbiota, which was performed by calculating Bray-Curtis dissimilarity distances. The beta-diversity was significantly different between IBD samples and samples from HC in the first two principal coordinates (P<0.001), as can be observed from the principal coordinate analysis (PCoA) plots shown in Figure 2. Permutational multivariate analysis of variance using distance matrices (PERMANOVA, using the ADONIS function) demonstrated that sample origin (IBD vs. HC) was able to explain 20.5% of the variation in the beta-diversity of the gut microbiota (P=0.01). In contrast, beta-diversity was not significantly changed after the MACS procedure in the first two principal coordinates (both PCoA1 and PCoA2: P>0.05), which was confirmed after ADONIS analysis which revealed that only 2.2% of the variation (P=0.02) could be explained by sample handling (IgG-coating procedure vs. no coating). No significant associations between shifts (before and after IgG-coating) in the first four PCoAs versus clinical characteristics were observed (Supplementary Figure S1).
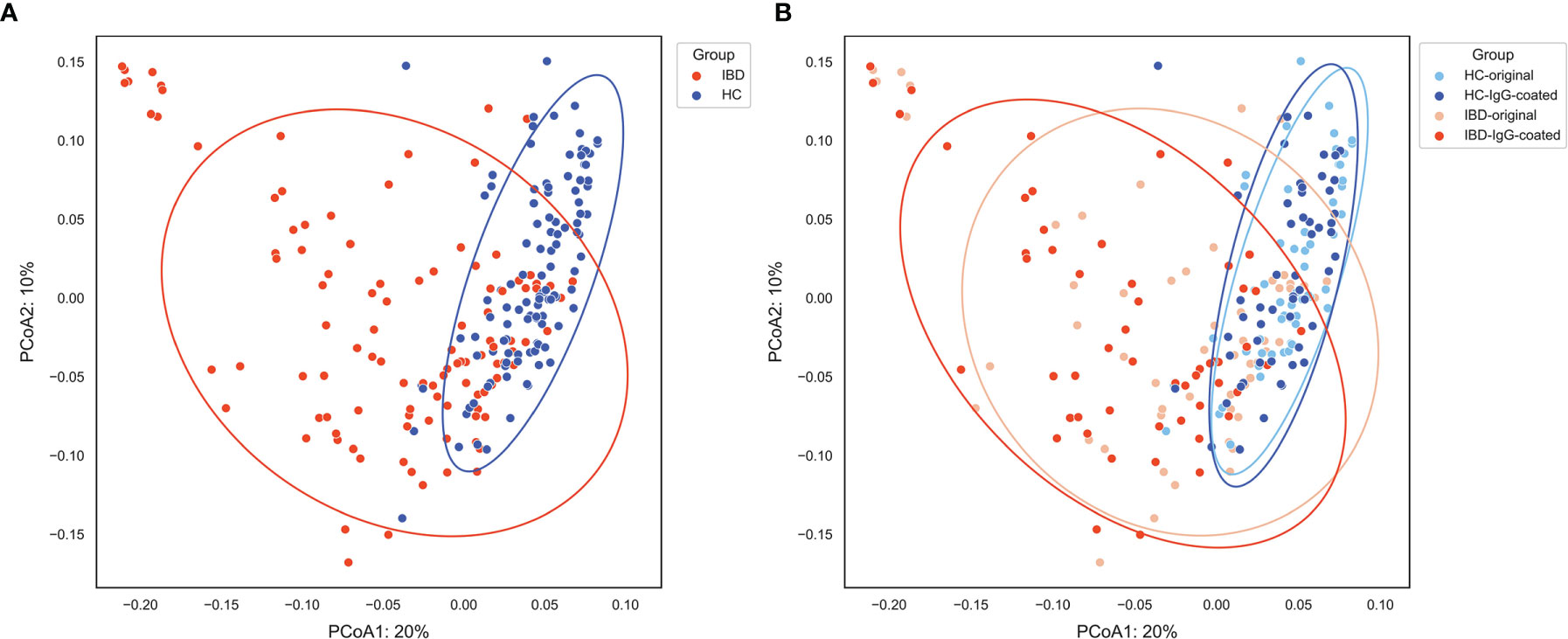
Figure 2 (A, B) Principal coordinate analysis (PCoA) of fecal samples from patients with IBD (orange) and HC (blue) visualizing the beta-diversity of the gut microbiota as represented by Bray-Curtis dissimilarities. (A) Sample origin (IBD vs. HC) shows an evident separation of the first two principal coordinates, which was statistically confirmed (PCoA1 and PCoA2: P < 0.001). (B) Paired analysis of IgG-coated vs. untreated fecal samples showed no significant change in the first two principal coordinates. Sample origin (IBD vs. HC) and sample handling (IgG-coated vs. untreated samples) significantly explained 20.5% and 2.2% of the variation in the gut microbiota, respectively. HC, healthy control; IBD, inflammatory bowel disease; MACS, magnetic-activated cell sorting; PCoA1, principal coordinate 1; PCoA2, principal coordinate 2.
The most common bacterial family observed in fecal samples was Lachnospiraceae (IBD: 40%; HC: 38%), followed by Ruminococcaceae (IBD: 14%; HC: 26%) and Bifidobacteriaceae (IBD: 6.7%; HC: 6.8%). The bacterial family of Bacteroidaceae accounted for a lower fraction of the microbial composition (IBD: 0.8%; HC: 2.0%). More variation in fecal microbial composition was observed on the genus level while comparing mean relative abundances (%) between IgG-coated vs. untreated fecal samples from patients with IBD and from HC (Figure 3 and Table 2). Anti-IgG-based flow cytometry of fecal samples of patients with IBD revealed that between 13-77% (mean: 38%) of bacteria showed IgG-coating after incubation with autologous serum, which further increased to a range of 21-75% (mean: 45%) of enriched IgG-bound bacteria after MACS-sorting occurring in ~84% of samples (Supplementary Table S2). After the MACS procedure, several bacterial genera were significantly enriched in fecal samples from both patients with IBD and HC, including Streptococcus, Coprococcus, Dorea, Ruminococcus gnavus-like bacteria, Lactobacillus, Dialister, Veillonella, and Turicibacter (all P<0.05 in both groups). In total, 10 out of 21 (48%) bacterial genera demonstrated significant changes in abundance in IgG-coated vs. untreated fecal samples in both patients with IBD and healthy controls. The bacterial genera Clostridium, Enterococcus, and Collinsella were only significantly enriched in samples from HC, whereas Lactococcus was only significantly enriched in patients with IBD (Figure 4). In contrast to these enrichments, relative abundances of the bacterial genera Faecalibacterium, Roseburia, and Bacteroides were significantly decreased after IgG-coating in samples from both patients with IBD and HC (all P<0.05). In addition, the relative abundances of Blautia, Eubacterium and Enterobacteriaceae-like bacteria were solely decreased after IgG-coating in patients with IBD (P<0.01). Unpaired analysis between samples from patients with IBD and HC revealed that the bacterial genera Streptococcus, Lactobacillus and Lactococcus were more abundant in IBD (P<0.05).
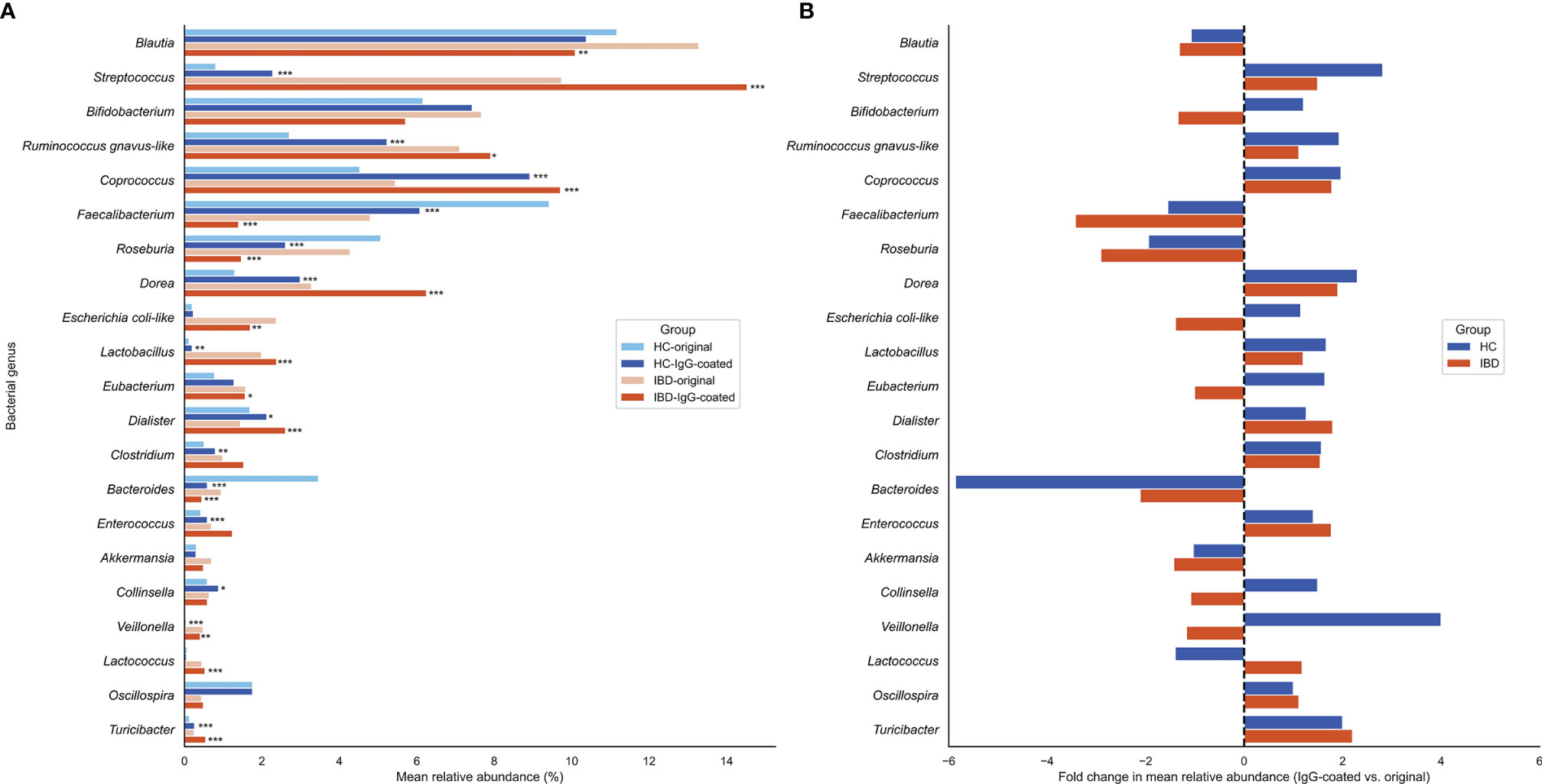
Figure 3 (A, B) Changes in relative abundances of fecal bacterial genera before and after IgG-coating in patients with IBD and HC. (A) Changes in mean relative abundances (%) after IgG-coating in fecal samples of patients with IBD (orange) and HC (blue). (B) Fold changes in mean relative abundances (%) before and after IgG-coating in IBD (orange) and HC (blue). Fold changes represent coating indices and were computed as follows: relative abundance (IgG-coated)/relative abundance (untreated). Relative increases in bacterial abundances are depicted on the right side of the plot, whereas decreases are displayed on the left side of the plot. Bacterial genera with relative abundances >0.25 were taken into statistical analysis. (*) indicate statistically significant increases or decreases in bacterial abundances: *P < 0.05; **P < 0.01; ***P < 0.001. HC, healthy control; IBD, inflammatory bowel disease.
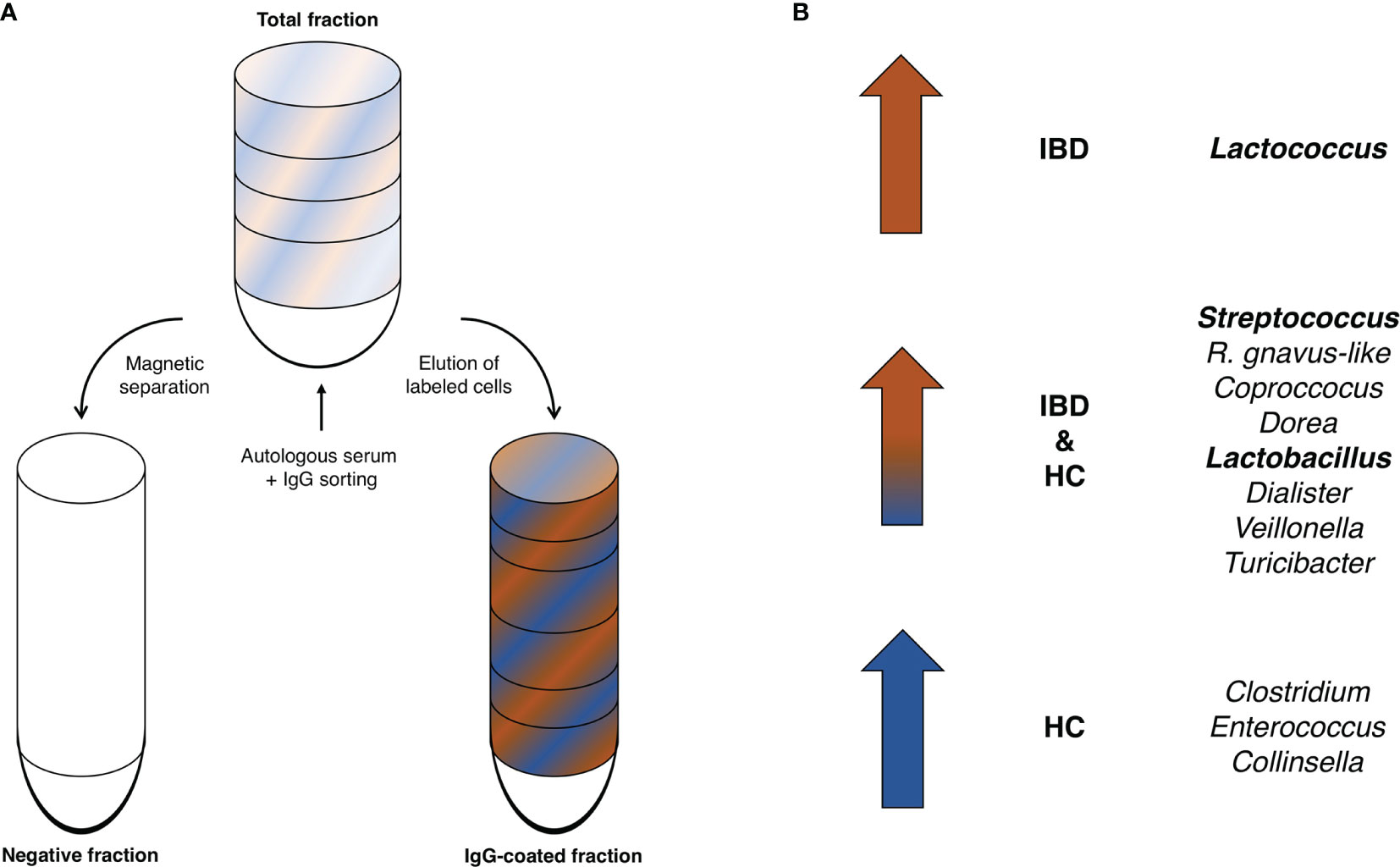
Figure 4 (A, B) Enriched IgG-coated fecal bacterial genera in patients with IBD and HC. (A) Schematic illustration of the IgG-based MACS-sorting procedure. The total fraction (top) represents the original, untreated sample, while the IgG-coated fraction represents the fraction left after magnetic separation of the non-IgG-coated bacterial fraction (negative fraction). (B) The bacterial genera Streptococcus, Coprococcus, Dorea, Lactobacillus, Dialister, Veillonella, and Turicibacter as well as R. gnavus-like bacteria were significantly enriched in IgG-coated samples in both patients with IBD and HC. Lactococcus bacteria were significantly enriched after IgG-coating solely in patients with IBD and Clostridium, Enterococcus and Collinsella solely in HC. Bold indicates bacterial genera that were significantly more enriched in IBD vs. HC.
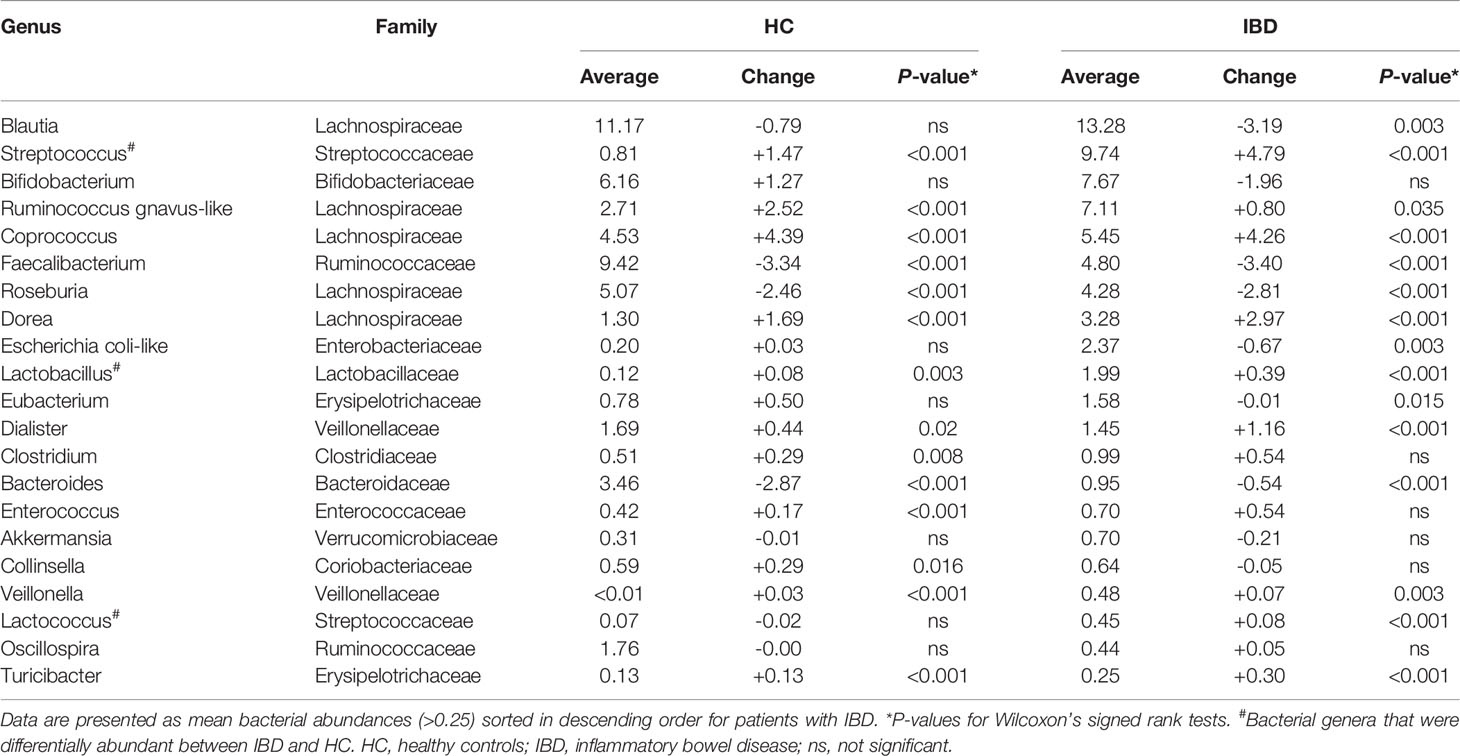
Table 2 Paired analysis of mean bacterial abundances (>0.25) in original, untreated fecal samples versus IgG-coated fecal samples.
In this study, we demonstrate that a variety of IgG immune responses are directed against the majority of the main fecal bacterial genera in both patients with IBD and healthy individuals. The IgG-coated samples showed a great overlap in enrichment of specific IgG-coated bacterial genera between both groups. In particular, serum IgG was directed against the bacterial genera Streptococcus, Lactobacillus, Lactococcus, and Veillonella. In contrast, bacterial genera like Faecalibacterium, Roseburia, and Blautia were decreased after the IgG-coating procedure, which are members of the families Lachnospiraceae and Ruminococcaceae, representing two dominant colonic microbial groups (Figure 3). This may suggest that there is less IgG reactivity against commensal (butyrate-producing) bacteria, however other Lachnospiraceae members, e.g. Dorea and Coprococcus, were significantly enriched after IgG-coating. This indicates that there were IgG immune responses against several types of commensal anaerobic microbiota. Furthermore, we showed that IgG-coating had a small but statistically significant impact on sample microbial diversity, which was to be expected as several types of bacteria were separated out after the MACS procedure. No significant differences in IgG-coated bacteria were observed between patients with CD and UC, patients with quiescent or active disease or patients with different disease locations, although numbers were actually too small to allow for reliable subgroup analyses. Collectively, these findings emphasize that both patients with IBD and HC have IgG immune responses against the fecal microbiota, many of which are rather prominent in patients with IBD.
The observed IgG immune responses, particularly in patients with IBD, were preferentially directed against typical members of the small intestinal microbiota, including Streptococcus, Lactobacillus, Lactococcus, and Veillonella. These findings are in line with what is known on the composition of the small intestinal microbiota in the context of IBD. Bacteria that are typically enriched in the small intestine of patients with IBD as compared with population controls comprise, among others, Streptococcus, Veillonella, Clostridium, Lactobacillus, Klebsiella, Enterococcus, Lactococcus, and Actinomyces species (25–27). Moreover, some of these bacteria, especially Streptococcus and Lactobacillus, have also been found to be abundantly present within the mucosa-associated microbiota in inflamed biopsies from patients with IBD, residing within a thinner mucus layer compared to healthy controls (28). Considering these observations, one may speculate that a compromised small intestinal barrier integrity may lead to higher exposure of these bacteria to the mucosal immune system as they are in close proximity, resulting in increased specific IgG immune responses. However, this speculation was not sustained by our data as it can be anticipated that patients with CD, who often have small intestinal disease involvement, would exhibit increased IgG responses towards these bacteria compared to patients with UC, which was however not observed. In addition, our study was not sufficiently powered to reliably establish these potential differences between subtypes of IBD, and thus warrants further investigation. In keeping with this, the decreased IgG immune responses against commensal, anaerobic, butyrate-producing bacteria like Roseburia and Faecalibacterium may be explained by the fact that these bacteria confer barrier-protective and anti-inflammatory properties (29, 30). Butyrate is known for its anti-inflammatory and anti-carcinogenic effects, and it contributes to the preservation of the intestinal barrier by acting as an energy source for epithelial cells (30–32). Furthermore, these butyrate-producing bacteria are characterized by high oxygen sensitivity, which limits their passage of the intestinal barrier (as oxygen levels increase towards the intestinal mucosa), and, in turn, their potential exposure to the immune system (33). Of note, we observed a surprisingly low relative abundance and low IgG immune response against Bacteroides, whereas this genus is usually one of the most dominant bacterial groups detected in fecal samples (34). Therefore, the abundance of Bacteroides as observed in the present study (<1%) greatly underestimates its actual prevalence. One potential explanation for a relatively low IgG immune response could be that IgG responses to other bacterial genera were strongly enhanced, which could make the response to Bacteroides seem relatively lower since we have worked with relative bacterial quantifications. Another possible explanation could be that the specific primers that were used (341F/806R) may have failed to adequately amplify Bacteroides, but variations in sample storage conditions and DNA extraction procedures may also be responsible for such decreased abundance (35).
The observed enrichment after IgG-coating of several types of Lachnospiraceae (e.g. Dorea and Coprococcus) that belong to Clostridium cluster XIVa, but also that of many other bacteria like Clostridium, Lactococcus, Lactobacillus, and Enterococcus species could be explained by the fact that at least some species (e.g., Enterocloster bolteae and Enterocloster clostridioforme, or Ruminococcus gnavus belonging to the family of Lachnospiraceae) belonging to these bacterial genera are flagellated (10, 36, 37). Bacterial flagellae or flagellins are highly immunogenic proteins and dominant antigens in the context of IBD (36). Recently, a study demonstrated strong IgG immune responses against Lachnospiraceae flagellins in patients with CD, particularly those that were localized within the small intestine (11). This anti-flagellin antibody signature has been reported to be indicative of a more complicated disease course in CD, characterized by small intestinal disease involvement, frequent exacerbations and increased surgery rates (38, 39). Well-established flagellin antigens include CBir1, Fla-X, and A4-Fla2 (associated with Lachnospiraceae bacteria) (40, 41). Circulating anti-flagellin or antimicrobial antibodies are able to identify individuals who will develop CD years before the actual diagnosis, serving as serological predictors of the disease (42–45). A recent study showed that pre-existent anti-flagellin antibodies associated with future development of CD independently of subclinical inflammation, genetic susceptibility and intestinal barrier function (45). This suggests that the formation of adaptive immune responses against microbiota flagellins occurs very early in disease pathogenesis. Apart from the highly immunogenic properties of flagellins, bacterial flagella may facilitate transport across the epithelial mucus layer and enhance contact with a disrupted epithelial barrier, which may translate into an increased propensity to be exposed to the mucosal immune system (41, 46, 47).
An important observation of our study was that many of the enriched IgG-coated bacterial genera are consistently among those reported to be increased in abundance in patients with IBD (5, 6, 33). For example, this includes the previously mentioned (potentially pathogenic) small intestinal-type bacteria, but also Enterobacteriaceae, certain Lachnospiraceae (e.g., Ruminococcus gnavus), and Clostridium species. Similarly, patients with IBD consistently have lower numbers of Faecalibacterium, Roseburia, Blautia and Bifidobacterium, which were also not significantly enriched within the IgG-coated fractions in this study (48–50). Thus, bacterial groups that are commonly reported to be more abundant in patients with IBD compared with healthy individuals also largely overlap with those suggested to be more immunogenic based on findings from the present study.
Strengths of the present study include the fact that this study presents a unique characterization of antimicrobial immune responses against fecal bacteria in patients with IBD. Furthermore, the concurrent availability of an age- and sex-matched cohort of population controls enabled us to directly compare autologous IgG-coated fractions of fecal bacteria to determine their specificity for IBD. However, several limitations also warrant recognition. For instance, we could not confirm enrichment after IgG-coating for all fecal samples of patients with IBD (or we could not demonstrate this by flow cytometry), and the relative enrichment was also not that high when compared to earlier reports (51). One explanation to the limited enrichment observed in fecal samples of patients with IBD could be that anti-IgG-FITC is less effective when bacteria are already pre-treated with anti-IgG-biotin antibodies, which may in turn lead to underestimation of enrichment by anti-IgG-biotin/streptavidin-coated magnetic beads. In addition, the bacterial diversity in fecal samples from patients with IBD was significantly reduced after IgG-coating, suggesting that some degree of selection had occurred during the MACS-procedure, which might indicate the existence of selective immune responses towards specific bacteria in the context of IBD. Indeed, bacterial sequence results of the different sorted fractions demonstrated that there was a selection of specific bacterial groups. Another limitation to this study pertains to the relatively small sample size, which did not permit us to perform reliable subgroup analyses (e.g. a comparison between patients with CD and UC) as statistical power was very limited for this. In addition, we had to rely on clinical assessment of disease activity, as data on fecal calprotectin levels or endoscopic disease activity were not recorded at time of sampling. However, the majority of our cohort (~75%) was in disease remission and previous efforts already indicated that the degree of IgG-coating does not seem to be affected by disease activity (13).
In conclusion, this study demonstrates that patients with IBD exhibit distinct IgG immune responses directed towards a smaller group of specific types of bacteria compared with healthy controls. More specifically, this IgG immune response seems to be directed against Streptococcus, Lactobacillus, and Lactococcus, among others, as well as bacteria belonging to the family of Lachnospiraceae and other Clostridium cluster XIVa bacteria (e.g. Coprococcus, Dorea, and Ruminococcus gnavus-group). In contrast, there was some degree of IgG reactivity against typical colonic microbiota including several members of Lachnospiraceae (e.g., Roseburia and Blautia), Ruminococcaceae (e.g., Faecalibacterium), and Bacteroides, which was not clearly different between patients with IBD and HC. In addition, no significant differences in IgG reactivity were observed between CD and UC, quiescent and active disease or different disease locations. Future research is warranted to further unravel the pathophysiological role of these IgG-bound bacterial groups within the context of IBD. Further investigation into the exact antigenic nature of these specific bacteria is paramount to better characterize host-microbiota interactions that are relevant to IBD.
The raw sequencing data used for this study are publicly available via the NCBI BioProject repository under accession number PRJNA816096.
The studies involving human participants were reviewed and approved by the Institutional Review Board (IRB) of the University Medical Center Groningen (full name in Dutch: “Medisch Ethische Toetsingscommissie”, METc). The patients/participants provided their written informed consent to participate in this study.
HH and GD were involved in conceptualization and study design. HH and GD were responsible for funding acquisition and resources. GR-B, MS, HM, and GD collected all study data. AB, GR-B, PL, MS, RR, MG, and HH performed data curation, data analysis, and visualization. AB, GR-B, and HH wrote the first draft of the manuscript. All authors contributed to results interpretation, critically reviewed the manuscript, contributed to manuscript revision, and read and approved the final version of the manuscript.
The research position of AB was supported by a JSM M.D.-Ph.D. trajectory grant from the Junior Scientific Masterclass (JSM) of the University of Groningen, the Netherlands (grant number: 17-57). The funders had no role in the design of the study, collection, analysis, or interpretation of data, nor in writing of the manuscript.
GD and HH received an unrestricted research grant from Royal DSM. GD received speaker fees from Pfizer, Abbvie and Janssen Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to express their gratitude towards all patients and healthy volunteers who participated in this study. The authors would also like to thank Carien Bus-Spoor, Joy Volkerink, Rudi Tonk, Tim Stobernack, and Solomon Mekonnen for their contributions to this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.842911/full#supplementary-material
1. Chang JT. Pathophysiology of Inflammatory Bowel Diseases. N Engl J Med (2020) 383(27):2652–64. doi: 10.1056/NEJMra2002697
2. Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-Wide Meta-Analysis Increases to 71 the Number of Confirmed Crohn’s Disease Susceptibility Loci. Nat Genet (2010) 42(12):1118–25. doi: 10.1038/ng.717
3. de Souza HSP, Fiocchi C, Iliopoulos D. The IBD Interactome: An Integrated View of Aetiology, Pathogenesis and Therapy. Nat Rev Gastroenterol Hepatol (2017) 14(12):739–49. doi: 10.1038/nrgastro.2017.110
4. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat Microbiol (2019) 4(2):293–305. doi: 10.1038/s41564-018-0306-4
5. Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut Microbiota Composition and Functional Changes in Inflammatory Bowel Disease and Irritable Bowel Syndrome. Sci Transl Med (2018) 10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914
6. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc Natl Acad Sci USA (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
7. Li H, Limenitakis JP, Greiff V, Yilmaz B, Schären O, Urbaniak C, et al. Mucosal or Systemic Microbiota Exposures Shape the B Cell Repertoire. Nature (2020) 584(7820):274–8. doi: 10.1038/s41586-020-2564-6
8. Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, et al. Intestinal Permeability—A New Target for Disease Prevention and Therapy. BMC Gastroenterol (2014) 14:189. doi: 10.1186/s12876-014-0189-7
9. Salim SY, Söderholm JD. Importance of Disrupted Intestinal Barrier in Inflammatory Bowel Diseases. Inflamm Bowel Dis (2011) 17(1):362–81. doi: 10.1002/ibd.21403
10. Christmann BS, Abrahamsson TR, Bernstein CN, Duck LW, Mannon PJ, Berg G, et al. Human Seroreactivity to Gut Microbiota Antigens. J Allergy Clin Immunol (2015) 136(5):1378–86.e1-5. doi: 10.1016/j.jaci.2015.03.036
11. Alexander KT, Zhao Q, Reif M, Rosenberg AF, Mannon PJ, Duck LW, et al. Human Microbiota Flagellins Drive Adaptive Immune Responses in Crohn’s Disease. Gastroenterology (2021) 161(2):522–35.e6. doi: 10.1053/j.gastro.2021.03.064
12. Monteleone G, Peluso I, Fina D, Caruso R, Andrei F, Tosti C, et al. Bacteria and Mucosal Immunity. Dig Liver Dis (2006) 38 (Suppl 2):S256–60. doi: 10.1016/S1590-8658(07)60005-X
13. Harmsen HJM, Pouwels SD, Funke A, Bos NA, Dijkstra G. Crohn’s Disease Patients Have More IgG-Binding Fecal Bacteria Than Controls. Clin Vaccine Immunol (2012) 19(4):515–21. doi: 10.1128/CVI.05517-11
14. van der Waaij LA, Kroese FGM, Visser A, Nelis GF, Westerveld BD, Jansen PLM, et al. Immunoglobulin Coating of Faecal Bacteria in Inflammatory Bowel Disease. Eur J Gastroenterol Hepatol (2004) 16(7):669–74. doi: 10.1097/01.meg.0000108346.41221.19
15. Masu Y, Kanazawa Y, Kakuta Y, Shimoyama Y, Onodera M, Naito T, et al. Immunoglobulin Subtype-Coated Bacteria are Correlated With the Disease Activity of Inflammatory Bowel Disease. Sci Rep (2021) 11(1):16672. doi: 10.1038/s41598-021-96289-5
16. Spekhorst LM, Imhann F, Festen EAM, van Bodegraven AA, de Boer NKH, Bouma G, et al. Cohort Profile: Design and First Results of the Dutch IBD Biobank: A Prospective, Nationwide Biobank of Patients With Inflammatory Bowel Disease. BMJ Open (2017) 7(11):e016695. doi: 10.1136/bmjopen-2017-016695
17. Heida FH, van Zoonen AGJF, Hulscher JBF, Te Kiefte BJC, Wessels R, Kooi EMW, et al. A Necrotizing Enterocolitis-Associated Gut Microbiota Is Present in the Meconium: Results of a Prospective Study. Clin Infect Dis (2016) 62(7):863–70. doi: 10.1093/cid/ciw016
18. Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of Multimillion-Sequence 16s rRNA Gene Libraries From Complex Microbial Communities by Assembling Paired-End Illumine Reads. Appl Environ Microbiol (2011) 77(11):3846–52. doi: 10.1128/AEM.02772-10
19. Bokulich NA, Joseph CM, Allen G, Benson AK, Mills DA. Next-Generation Sequencing Reveals Significant Bacterial Diversity of Botrytized Wine. PloS One (2012) 7(5):e36357. doi: 10.1371/journal.pone.0036357
20. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat Methods (2010) 7(5):335–6. doi: 10.1038/nmeth.f.303
21. Edgar RC. Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics (2010) 26(19):2460–1. doi: 10.1093/bioinformatics/btq461
22. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: Paired-End Assembler for Illumina Sequences. Bioinformatics (2012) 13:31. doi: 10.1186/1471-2105-13-31
23. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: A Software Environment for Sequence Data. Nucleic Acids Res (2004) 32(4):1363–71. doi: 10.1093/nar/gkh293
24. Oksanen JR, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package (2016). Available at: https://cran.r-project.org/web/packages/vegan/index.html.
25. Ruigrok RAAA, Collij V, Sureda P, Klaassen MAY, Bolte LA, Jansen BH, et al. The Composition and Metabolic Potential of the Human Small Intestinal Microbiota Within the Context of Inflammatory Bowel Disease. J Crohns Colitis (2021) 15(8):1326–38. doi: 10.1093/ecco-jcc/jjab020
26. Kastl AJ Jr, Terry NA, Wu GD, Albenberg LG. The Structure and Function of the Human Small Intestinal Microbiota: Current Understanding and Future Directions. Cell Mol Gastroenterol Hepatol (2020) 9(1):33–45. doi: 10.1016/j.jcmgh.2019.07.006
27. Villmones HC, Haug ES, Ulvestad E, Grude N, Stenstad T, Halland A, et al. Species Level Description of the Human Ileal Bacterial Microbiota. Sci Rep (2018) 8(1):4736. doi: 10.1038/s41598-018-23198-5
28. Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, et al. Mucosal Bacterial Microflora and Mucus Layer Thickness in Adolescents With Inflammatory Bowel Disease. World J Gastroenterol (2009) 15(42):5287–94. doi: 10.3748/wjg.15.5287
29. Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J Nutr (2009) 139(9):1619–25. doi: 10.3945/jn.109.104638
30. Wang HB, Wang PY, Wang X, Wan YL, Liu YC. Butyrate Enhances Intestinal Epithelial Barrier Function via Up-Regulation of Tight Junction Protein Claudin-1 Transcription. Dig Dis Sci (2012) 57(12):3126–35. doi: 10.1007/s10620-012-2259-4
31. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
32. Louis P, Flint HJ. Diversity, Metabolism and Microbial Ecology of Butyrate-Producing Bacteria From the Human Large Intestine. FEMS Microbiol Lett (2009) 294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x
33. von Martels JZH, Sadaghian Sadabad M, Bourgonje AR, Blokzijl T, Dijkstra G, Faber KN, et al. The Role of Gut Microbiota in Health and Disease: In Vitro Modeling of Host-Microbe Interactions at the Aerobe-Anaerobe Interphase of the Human Gut. Anaerobe (2017) 44:3–12. doi: 10.1016/j.anaerobe.2017.01.001
34. Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature (2012) 486(7402):207–14. doi: 10.1038/nature11234
35. Maukonen J, Simões C, Saarela M. The Currently Used Commercial DNA-Extraction Methods Give Different Results of Clostridial and Actinobacterial Populations Derived From Human Fecal Samples. FEMS Microbiol Ecol (2012) 79(3):697–708. doi: 10.1111/j.1574-6941.2011.01257.x
36. Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, et al. Bacterial Flagellin Is a Dominant Antigen in Crohn Disease. J Clin Invest (2004) 113(9):1296–306. doi: 10.1172/JCI20295
37. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature (2019) 569(7758):655–62. doi: 10.1038/s41586-019-1237-9
38. Targan SR, Landers CJ, Yang H, Lodes MJ, Cong Y, Papadakis KA, et al. Antibodies to CBir1 Flagellin Define a Unique Response That Is Associated Independently With Complicated Crohn’s Disease. Gastroenterology (2005) 128(7):2020–8. doi: 10.1053/j.gastro.2005.03.046
39. Papadakis KA, Yang H, Ippoliti A, Mei L, Elson CO, Hershberg RM, et al. Anti-Flagellin (CBir1) Phenotypic and Genetic Crohn’s Disease Associations. Inflamm Bowel Dis (2007) 13(5):524–30. doi: 10.1002/ibd.20106
40. Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, et al. Isolation of Flagellated Bacteria Implicated in Crohn’s Disease. Inflamm Bowel Dis (2007) 13(10):1191–201. doi: 10.1002/ibd.20237
41. Arnott ID, Landers CJ, Nimmo EJ, Drummond HE, Smith Bk, Targan SR, et al. Sero-Reactivity to Microbial Components in Crohn’s Disease Is Associated With Disease Severity and Progression, But Not NOD2/CARD15 Genotype. Am J Gastroenterol (2004) 99(12):2376–84. doi: 10.1111/j.1572-0241.2004.40417.x
42. Choung RS, Princen F, Stockfisch TP, Torres J, Maue AC, Porter CK, et al. Serologic Microbial Associated Markers can Predict Crohn’s Disease Behaviour Years Before Disease Diagnosis. Aliment Pharmacol Ther (2016) 43(12):1300–10. doi: 10.1111/apt.13641
43. van Schaik FD, Oldenburg B, Hart AR, Siersema PD, Lindgren S, Grip O, et al. Serological Markers Predict Inflammatory Bowel Disease Years Before the Diagnosis. Gut (2013) 62(5):683–8. doi: 10.1136/gutjnl-2012-302717
44. Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, et al. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology (2020) 159(1):96–104. doi: 10.1053/j.gastro.2020.03.007
45. Lee SH, Turpin W, Espin-Garcia O, Raygoza Garay JA, Smith MI, Leibovitzh H, et al. Anti-Microbial Antibody Response Is Associated With Future Onset of Crohn’s Disease Independent of Biomarkers of Altered Gut Barrier Function, Subclinical Inflammation, and Genetic Risk. Gastroenterology (2021) S0016-5085(21):03243–1. doi: 10.1053/j.gastro.2021.07.009
46. Gacesa R, Kurilshikov A, Vich Vila A, Sinha T, Klaassen MAY, Bolte LA, et al. Environmental Factors Shaping the Gut Microbiome in a Dutch Population. Nature (2022)604:732–9. doi: 10.1038/s41586-022-04567-7
47. Bourgonje AR, Andreu-Sánchez S, Vogl T, Hu S, Vich Vila A, Gacesa R, et al. In-Depth Characterization of the Serum Antibody Epitope Repertoire in Inflammatory Bowel Disease Using Phage-Displayed Immunoprecipitation Sequencing. bioRxiv (2021). doi: 10.1101/2021.12.07.471581
48. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium Prausnitzii is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc Natl Acad Sci USA (2008) 105(43):16731–6. doi: 10.1073/pnas.0804812105
49. Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A Pyrosequencing Study in Twins Shows That Gastrointestinal Microbial Profiles Vary With Inflammatory Bowel Disease Phenotypes. Gastroenterology (2010) 139(6):1844–54.e1. doi: 10.1053/j.gastro.2010.08.049
50. Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the Dominant Faecal Bacterial Groups in Patients With Crohn’s Disease of the Colon. Gut (2003) 52(2):237–42. doi: 10.1136/gut.52.2.237
Keywords: inflammatory bowel disease, immune system, immunoglobulin G (IgG), microbiota, humoral immunity
Citation: Bourgonje AR, Roo-Brand G, Lisotto P, Sadaghian Sadabad M, Reitsema RD, de Goffau MC, Faber KN, Dijkstra G and Harmsen HJM (2022) Patients With Inflammatory Bowel Disease Show IgG Immune Responses Towards Specific Intestinal Bacterial Genera. Front. Immunol. 13:842911. doi: 10.3389/fimmu.2022.842911
Received: 24 December 2021; Accepted: 25 April 2022;
Published: 25 May 2022.
Edited by:
Jochen Mattner, University of Erlangen Nuremberg, GermanyReviewed by:
Jessica Crothers, University of Vermont, United StatesCopyright © 2022 Bourgonje, Roo-Brand, Lisotto, Sadaghian Sadabad, Reitsema, de Goffau, Faber, Dijkstra and Harmsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hermie J. M. Harmsen, aC5qLm0uaGFybXNlbkB1bWNnLm5s
†These authors have contributed equally and share first authorship
‡These authors have contributed equally and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.