- 1Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison, WI, United States
- 2Wisconsin National Primate Research Center, University of Wisconsin-Madison, Madison, WI, United States
Knowledge of the MHC class I ligands of rhesus macaque killer-cell Ig-like receptors (KIRs) is fundamental to understanding the role of natural killer (NK) cells in this species as a nonhuman primate model for infectious diseases, transplantation and reproductive biology. We previously identified Mamu-AG as a ligand for KIR3DL05. Mamu-AG is a nonclassical MHC class I molecule that is expressed at the maternal-fetal interface of the placenta in rhesus macaques similar to HLA-G in humans. Although Mamu-AG and HLA-G share similar molecular features, including limited polymorphism and a short cytoplasmic tail, Mamu-AG is considerably more polymorphic. To determine which allotypes of Mamu-AG serve as ligands for KIR3DL05, we tested reporter cell lines expressing five different alleles of KIR3DL05 (KIR3DL05*001, KIR3DL05*004, KIR3DL05*005, KIR3DL05*008 and KIR3DL05*X) for responses to target cells expressing eight different alleles of Mamu-AG. All five allotypes of KIR3DL05 responded to Mamu-AG2*01:01, two exhibited dominant responses to Mamu-AG1*05:01, and three had low but detectable responses to Mamu-AG3*03:01, -AG3*03:02, -AG3*03:03 and -AG3*03:04. Since KIR3DL05*X is the product of recombination between KIR3DL05 and KIR3DS02, we also tested an allotype of KIR3DS02 (KIR3DS02*004) and found that this activating KIR also recognizes Mamu-AG2*01:01. Additional analysis of Mamu-AG variants with single amino acid substitutions identified residues in the α1-domain essential for recognition by KIR3DL05. These results reveal variation in KIR3DL05 and KIR3DS02 responses to Mamu-AG and define Mamu-AG polymorphisms that differentially affect KIR recognition.
Introduction
MHC class I molecules play a central role in both innate and adaptive immunity. In addition to presenting peptides derived from intracellular antigens on the cell surface for recognition by CD8+ T cells, MHC class I molecules serve as ligands for killer cell Ig-like receptors (KIRs) on natural killer (NK) cells (1, 2). Whereas the extensive polymorphism of classical MHC class I molecules ensures the presentation of a diverse repertoire of peptides derived from intracellular pathogens to CD8+ T cells, the limited polymorphism of nonclassical MHC class I molecules reflects their more specialized functions.
Macaques and other Old World monkeys express an expanded array of polymorphic MHC class I genes that correspond to the classical HLA-A and -B genes of humans (3–5). However, these species lack an ortholog of HLA-C as a consequence of the recent evolutionary origin of HLA-C as a duplication of an ancestral MHC-B gene that occurred after the divergence of apes and Old World monkeys (3, 6). Old World monkeys have accordingly expanded their lineage II KIR, which encode KIR3D receptors for MHC-A and -B, but lack lineage III KIR that encode KIR2D receptors for HLA-C in humans (7–13). Old World monkeys also have orthologs of each of the human nonclassical HLA-E, -F and -G genes. MHC-E and -F are well-conserved and broadly expressed in primates (14–16). However, the MHC-G genes of Old World monkeys have accumulated multiple stop codons and no longer encode functional proteins (17, 18). In the absence of a functional MHC-G locus, Old World monkeys have evolved another nonclassical MHC class I gene, designated MHC-AG, which appears to serve a similar function (19–22).
Macaques, vervets, and baboons express MHC-AG molecules with similar molecular features and tissue distribution as HLA-G (22, 23). MHC-AG sequences are more similar to MHC-A than to MHC-G as a consequence of their evolutionary origins as an MHC-A gene duplication (19–22). However like HLA-G, MHC-AG exhibits limited polymorphism, a truncated cytoplasmic tail, and is only expressed on placental trophoblasts that form the barrier between the mother and the developing fetus (19, 20, 23). Hence, Mamu-AG is believed to serve a similar function as HLA-G, namely contributing to pregnancy success through interactions with NK cells and myeloid cells that promote placental vascularization, tolerance to the haploidentical fetus, and protect the maternal-fetal interface from invading pathogens (23, 24).
The rhesus macaque (Macaca mulatta) is an important animal model for infectious diseases such as HIV/AIDS, CMV, Zika virus and SARS-CoV-2 (25–35), transplantation (36, 37), and reproductive biology (23, 38). We previously identified Mamu-AG as a ligand for KIR3DL05 (39). Here we show that KIR3DL05 recognizes certain allotypes of Mamu-AG, but not others, and define residues in the α1- and α2-domains of Mamu-AG essential for interactions with this receptor. We further identify Mamu-AG as a ligand for the activating receptor KIR3DS02. These observations provide an essential foundation for investigating the role of NK cells in the rhesus macaque as a nonhuman primate model for infectious diseases and reproductive biology. These findings also illustrate how polymorphisms in the α1-domain can lead to variation in KIR recognition of closely related MHC class I ligands.
Materials and Methods
MHC Class I-Transduced 721.221 Cells
Codon-optimized cDNA sequences encoding Mamu-AG molecules were synthesized by GenScript. These sequences were then cloned into the retroviral vector pQCXIP. Retroviral vectors were packaged into VSV-G-pseudotyped murine leukemia virus (MLV) particles by co-transfecting GP2-293 cells with pQCXIP-Mamu-AG constructs and pVSV-G (Clontech Laboratories). The cell culture supernatant was collected two days after transfection and cellular debris was removed by centrifugation followed by filtration through a 0.45 µm nylon membrane. HLA-null 721.221 cells were transduced by spinoculation for one hour with supernatant collected from the transfected GP2-293 cells. After two days, the transduced cells were placed under antibiotic selection with RPMI medium supplemented with 10% FBS, L-glutamine, penicillin, streptomycin (R10) and 0.4 µg/ml puromycin (Calbiochem). The concentration of puromycin in the medium was gradually increased to 1.0 µg/ml over 3-4 weeks to eliminate untransduced parental 721.221 cells. After 3-4 weeks of selection, cell lines were maintained in medium containing 0.4 µg/ml puromycin. Mamu-AG expression was confirmed by flow cytometry via surface staining with a PE-conjugated, pan-MHC class I-reactive monoclonal antibody (W6/32, Life Technologies). Mamu-AG-transduced 721.221 cell lines were maintained in R10 medium containing 0.4 µg/ml puromycin. The GenBank accession numbers for Mamu-AG alleles are as follows: Mamu-AG2*01:01 (U84783.1), -AG3*02:01:01:01 (U84785.1), -AG3*02:02:01 (U84786.1), -AG3*03:01:01:01 (U84787.1), -AG3*03:02 (U84789.1), -AG3*03:03 (KF855159.1), -AG3*03:04 (KF855160.1), and -AG1*05:01 (FJ409466). Only partial-length cDNA sequences were available for Mamu-AG3*03:03, -AG3*03:04, and -AG1*05:01. For constructs expressing these alleles, sequences encoding the α3-, transmembrane and cytoplasmic domains were therefore inferred from the Mamu-AG consensus sequence.
KIR-CD3ζ-Transduced Jurkat NFAT Luciferase Reporter Cells
Constructs for the expression of chimeric KIR-CD3ζ receptors with an N-terminal Flag-tag were engineered by cloning cDNA sequences into pQCXIP encoding the D0, D1, D2 and stem domains of KIR3DL05 downstream of sequences for the KIR3DL05*008 leader peptide and Flag-tag (DYKDDDDK) and upstream of sequences coding for the transmembrane and cytoplasmic domains of CD3ζ. Retroviral vectors were packaged into VSV-G-pseudotyped MLV particles as described above. Jurkat NFAT luciferase (JNL) cells (Signosis) were transduced by spinoculation for one hour with filtered supernatant from transfected GP2-293 cells. Two days after transduction, the JNL cells were placed under antibiotic selection in R10 medium containing 100 µg/ml hygromycin to maintain the luciferase reporter gene and 0.5 µg/ml puromycin to select for KIR-CD3ζ expression. The puromycin concentration in the medium was gradually increased to 1.0 µg/ml over 3-4 weeks to eliminate untransduced cells and then maintained at a concentration of 0.4 µg/ml. GenBank accession numbers for rhesus macaque KIR sequences are as follows: KIR3DL05*001/KIR3DLW36*001 (EU419045), KIR3DL05*004 (EU419066.1), KIR3DL05*005/KIR3DLW37*001 (EU419069), KIR3DL05*X (EU419067.1), KIR3DL05*008 (GU112291), and KIR3DS02*004 (EU419026).
Ligand Identification Assay
KIR-CD3ζ+ JNL cells (1x105) were co-cultured with Mamu-AG+ or parental 721.221 cells (1x105) overnight at 37°C and 5% CO2 in triplicate wells of white 96-well plates in 100 µl R10 medium without antibiotics. As a positive control, KIR-CD3ζ+ JNL cells were treated with 5 µg/ml of an anti-Flag-tag monoclonal antibody (GenScript) and 10 µg/ml of a goat anti-mouse secondary antibody (Poly4053, Biolegend). After 12-18 hours, 100 µl of BriteLite Plus luciferase substrate (PerkinElmer) was added to each well and luciferase activity in relative light units (RLU) was measured using a VICTOR X4 multiplate reader (PerkinElmer).
Flow Cytometry
KIR-CD3ζ expression was verified by surface staining parental and KIR-CD3ζ-transduced JNL cells with a PE-conjugated anti-Flag antibody (Miltenyi Biotec). MHC class I expression was confirmed by surface staining parental and Mamu-AG-transduced 721.221 cells with a PE-conjugated pan-MHC class I-specific monoclonal antibody (W6/32, eBioscience). JNL and 721.221 cells were also stained with live-dead near-IR fluorescent stain (Invitrogen) to exclude dead cells. Data was collected on a BD FACSymphony flow cytometer and analyzed using FlowJo software (TreeStar, Inc.). After gating on singlet, viable cells, the surface expression of KIR-CD3ζ and Mamu-AG on transduced cells was compared to parental JNL and 721.221 cells.
Statistical Analysis
The RLU data was analyzed by one-way ANOVA followed by Dunnett’s test for multiple comparisons of mean KIR-CD3ζ+ JNL cell responses to each of the Mamu-AG+ 721.221 cells with mean responses to the parental 721.221 cells as a negative control. For analysis of responses to the Mamu-AG2*02:01 variants, mean RLU values in response to 721.221 cells expressing each variant were compared with mean RLU values in response to 721.221 cells expressing wild-type Mamu-AG2*02:01 using an unpaired t test. Statistical analyses were performed using GraphPad Prism for Mac OS version 9.2.0.
Results
Allotypic Variation in KIR3DL05 Responses to Mamu-AG Ligands
Mamu-AG2*01:01 was previously identified as a ligand for KIR3DL05*008 (39). However, Mamu-AG and KIR3DL05 are both polymorphic, raising the possibility of allelic variation in ligand recognition. To assess the impact of polymorphisms in these molecules on receptor-ligand interactions, five allotypes of KIR3DL05 were tested for the recognition of eight allotypes of Mamu-AG. KIR3DL05*001, *004, *005, *X and *008 were selected from a previous study showing differences in KIR3DL05 binding to Mamu-A1*002-peptide complexes (40) (Figure 1) and Mamu-AG2*02:01, -AG3*02:01, -AG3*02:02, -AG3*03:01, -AG3*03:02, -AG3*03:03, -AG*03:04 and -AG1*05:01 were selected as representative allomorphs of predicted Mamu-AG loci (Figure 2A). Jurkat cells containing an NFAT-inducible luciferase reporter gene (JNL cells) were transduced with retroviral vectors expressing chimeric KIR-CD3ζ receptors consisting of the extracellular domains of KIR3DL05 (D0, D1, D2 and stem) fused to the transmembrane and cytoplasmic domains of human CD3ζ. To verify surface expression, a Flag-tag (DYKDDDDK) was appended to the N-terminus of the D0 domain of each of the KIR-CD3ζ chimeras (Figure 1B). 721.221 cells, which are deficient for the expression of endogenous HLA class I molecules (42), were in turn transduced with vectors expressing individual Mamu-AG alleles (Figure 2B).
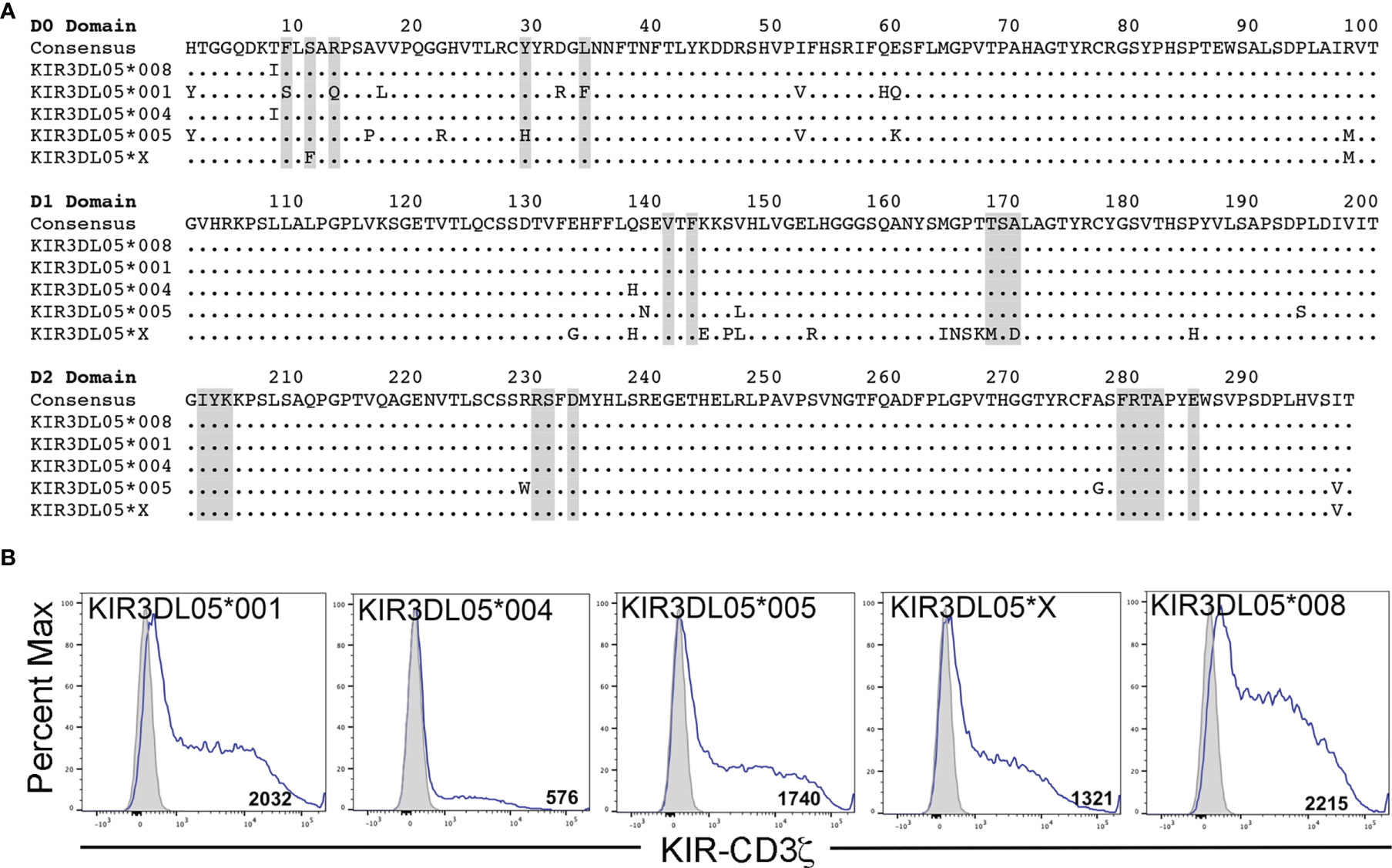
Figure 1 KIR3DL05 allotypes and surface expression of their KIR-CD3ζ chimeras on JNL cells. (A) Predicted amino acid sequence alignment for the D0, D1 and D2 domains of KIR3DL05*001, *004, *005, *X and *008. Regions shaded in gray indicate predicted MHC class I contact sites (41). Positions of amino acid identity are indicated by periods and amino acid differences are identified by their single-letter code. (B) KIR-CD3ζ+ JNL cells and parental JNL cells were stained with a PE-conjugated anti-Flag antibody and Near-IR LIVE/DEAD stain. After gating to exclude dead cells, the fluorescence intensity of KIR-CD3ζ (Flag-tag) staining on KIR-CD3ζ-transduced JNL cells (open) was compared to background staining on parental JNL cells (shaded). Flow cytometry data was analyzed using FlowJo 10.6 for Mac OSX. Values indicate geometric mean fluorescence intensity of staining.
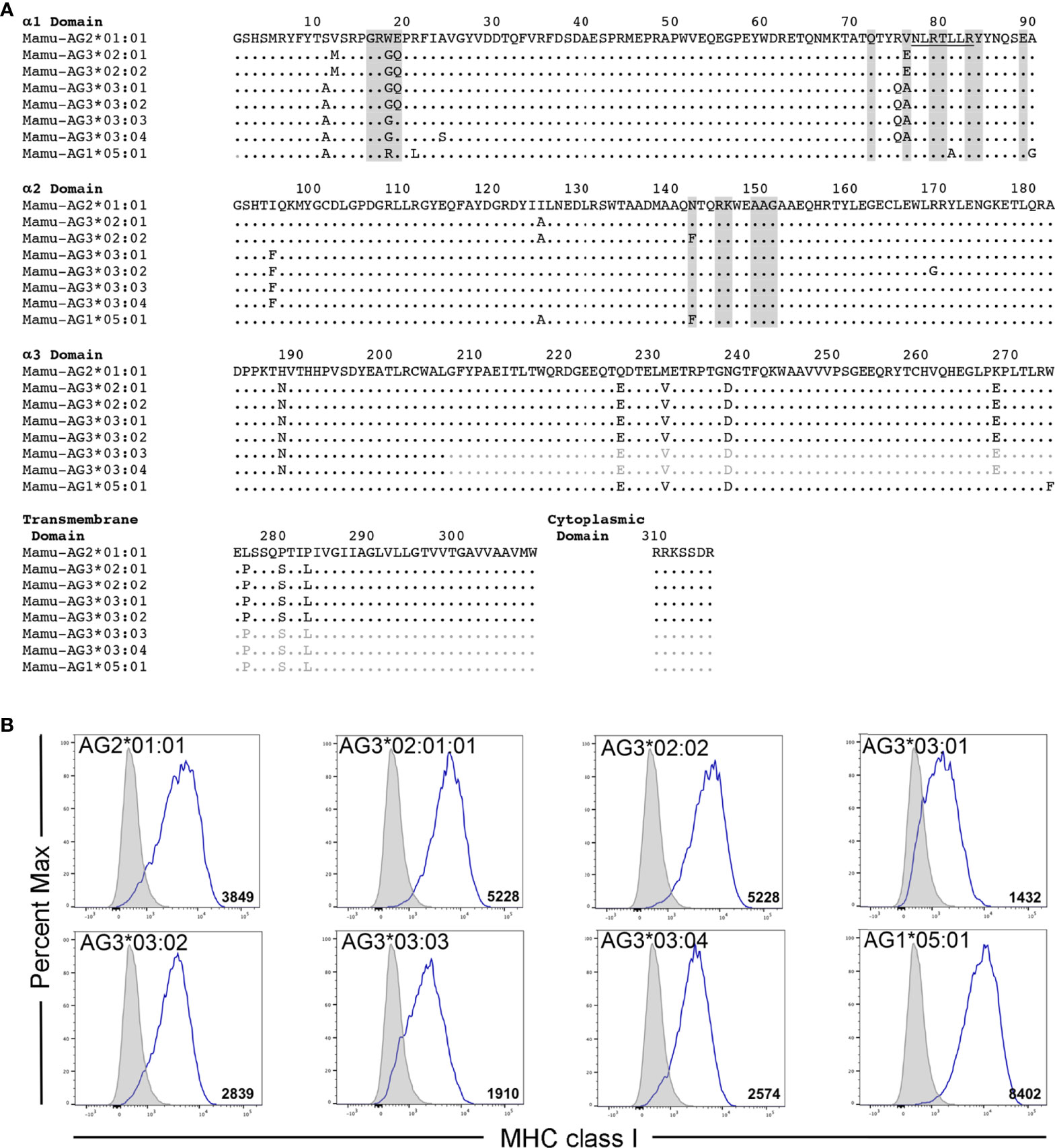
Figure 2 Mamu-AG allotypes and their surface expression on 721.221 cells. (A) An alignment showing the predicted amino acid sequences for Mamu-AG2*01:01, -AG3*02:01, -AG3*02:02, -AG3*03:01, -AG3*03:02, -AG3*03:03, -AG3*03:04, and -AG1*05:01. Regions shaded in gray indicate predicted KIR contact sites (41). Residues 77-83 that correspond to a Bw4 motif are underlined. Positions of amino acid identity are indicated by periods and amino acid differences are identified by their single-letter code. Sequences in the α3-, transmembrane and cytoplasmic domains of Mamu-AG3*03:03, -AG3*03:04, and -AG1*05:01 that were inferred from the consensus sequence are indicated in light gray. (B) Mamu-AG+ 721.221 cells and parental 721.221 cells were stained with a PE-conjugated pan-MHC class I-specific antibody (W6/32) and Near-IR LIVE/DEAD stain. After gating to exclude dead cells, the fluorescence intensity of MHC class I staining on Mamu-AG-transduced 721.221 cells (open) was compared to background staining of their respective parental cell lines (shaded). Flow cytometry data was analyzed using FlowJo 10.6 for Mac OSX. Values indicate geometric mean fluorescence intensity of staining.
Polymorphic differences were observed in KIR3DL05-mediated responses to different Mamu-AG molecules. Ligand recognition was detected by the MHC class I-dependent upregulation of luciferase by the KIR-CD3ζ+ JNL cells after an overnight incubation with each of the Mamu-AG+ 721.221 cells. Differences in luciferase induction were compared to parental 721.221 cells as a negative control and to KIR-CD3ζ+ JNL cells incubated with anti-Flag and anti-mouse IgG antibodies to cross-link receptors as a positive control for signaling. All five KIR3DL05 allotypes responded to Mamu-AG2*01:01 (Figure 3). Dominant responses to Mamu-AG1*05:01 were also observed for KIR3DL05*001 and KIR3DL05*X, and additional responses to Mamu-AG3*03:01, -AG3*03:02, -AG3*03:03 and -AG3*03:04 were detectable for KIR3DL05*004, KIR3DL05*X and KIR3DL05*008 (Figure 3). However, none of the KIR3DL05 allotypes responded to either Mamu-AG3*02:01 or -AG3*02:02 (Figure 3). These differences in ligand recognition did not correspond to differences in the surface expression of KIR-CD3ζ on JNL cells or Mamu-AG on 721.221 cells, but instead reflected polymorphisms in KIR3DL05 and Mamu-AG.
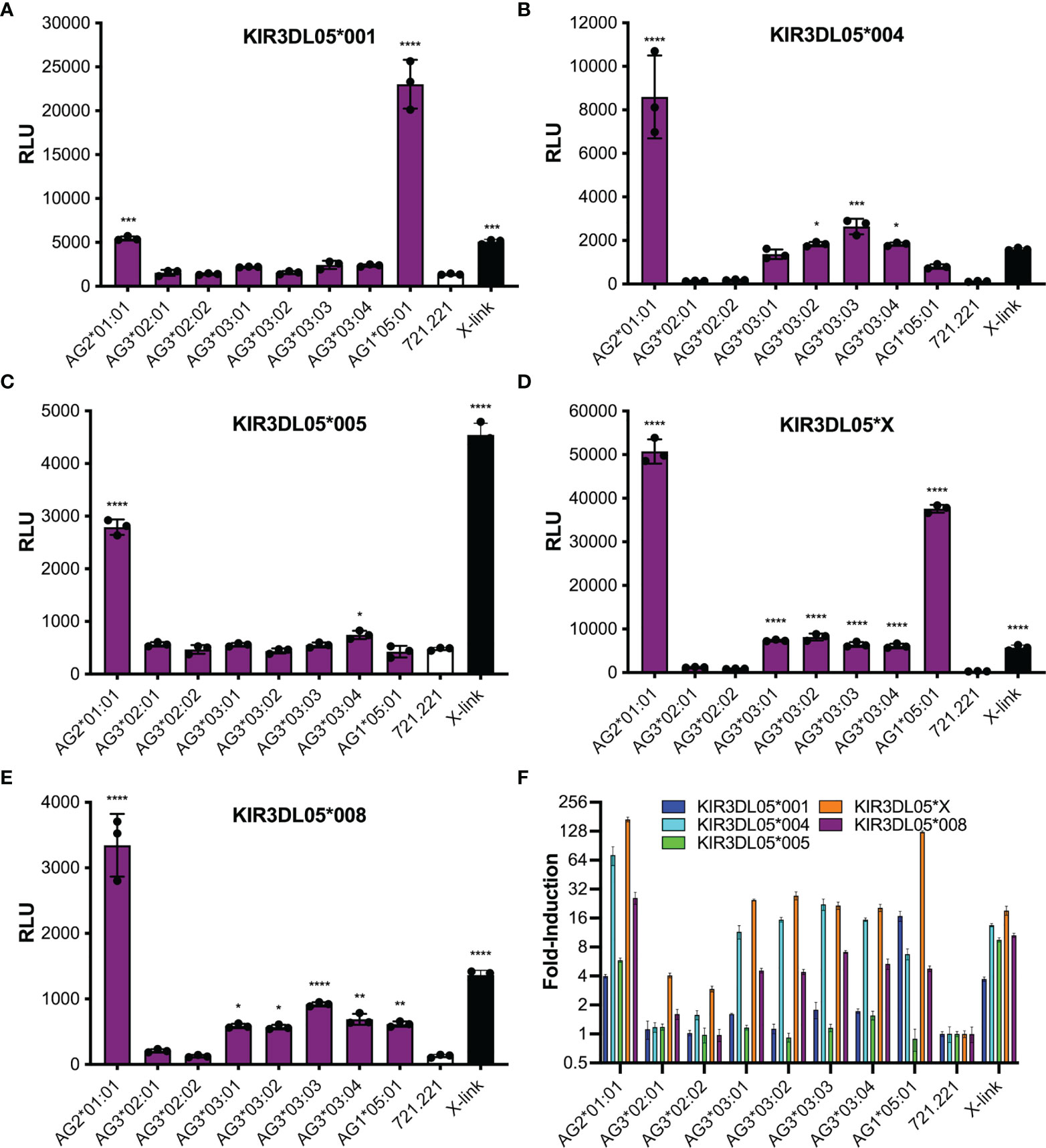
Figure 3 Mamu-AG recognition by five different allotypes of KIR3DL05. Jurkat NFAT luciferase (JNL) cells expressing the ectodomains of KIR3DL05*001 (A), KIR3DL05*004 (B), KIR3DL05*005 (C), KIR3DL05*X (D), or KIR3DL05*008 (E) fused to the transmembrane and cytoplasmic domains of CD3ζ were incubated at a 1:1 E:T ratio with 721.221 cells expressing eight different Mamu-AG allotypes. KIR-CD3ζ+ JNL cells were also incubated with parental 721.221 cells as a negative control and with a combination of anti-Flag and anti-mouse IgG antibodies as a positive control (X-link). Ligand recognition was detected by the upregulation of luciferase. (A–E) Bars indicate the mean relative light units (RLU) of luciferase activity in triplicate wells for each of the receptors plotted separately, or (F) the fold-induction of luciferase over background responses to parental 721.221 cells for each of the receptors plotted together. Error bars indicate SD of the mean and asterisks denote statistically significant differences (*p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001, one-way ANOVA with Dunnett’s test).
Mamu-AG Recognition by an Activating KIR
KIR3DL05*X is the product of a recombination event that resulted in a hybrid receptor with D0 and D1 domains corresponding to KIR3DS02 and a D2 domain typical of KIR3DL05. We therefore tested KIR3DS02*004, which shares identical D0 and D1 domains with KIR3DL05*X, to determine if this activating KIR could also recognize Mamu-AG (Figure 4A). JNL cells expressing a KIR3DS02*004-CD3ζ fusion responded to 721.221 cells expressing Mamu-AG2*01:01, but not to cells expressing other Mamu-AG alleles (Figures 4B, C). These results identify Mamu-AG2*01:01 as a ligand for KIR3DS02*004 and suggest that the more restricted pattern of responses to Mamu-AG compared to KIR3DL05*X reflects differences in the D2 domain.
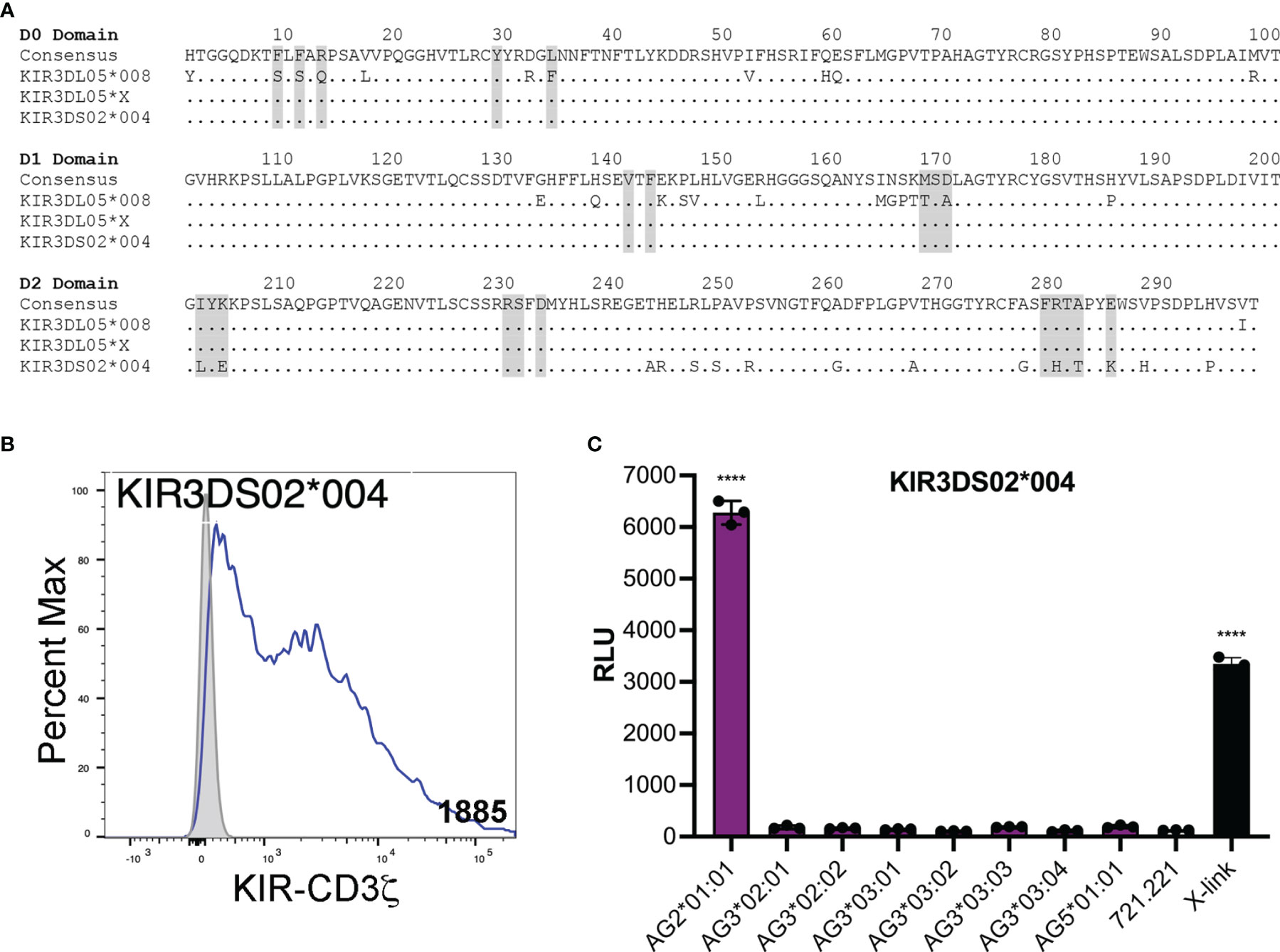
Figure 4 KIR3DS02 recognition of Mamu-AG. (A) Predicted amino acid sequence alignment for the D0-D2 domains of KIR3DL05*008, KIR3DL05*X and KIR3DS02*004. Regions shaded in gray indicate predicted MHC class I contact sites (41). Positions of amino acid identity are indicated by periods and amino acid differences are identified by their single-letter code. (B) KIR3DS02*004-CD3ζ+ JNL cells and parental JNL cells were stained with a PE-conjugated anti-Flag antibody and Near-IR LIVE/DEAD stain. After gating to exclude dead cells, the fluorescence intensity of KIR-CD3ζ (Flag-tag) staining on KIR-CD3ζ-transduced JNL cells (open) was compared to background staining on parental JNL cells (shaded). Geometric MFI values indicated. Flow cytometry data was analyzed using FlowJo 10.6 for Mac OSX. (C) KIR3DS02*004-CD3ζ+ JNL cells were incubated with 721.221 cells expressing eight different Mamu-AG allotypes at a 1:1 E:T ratio. KIR-CD3ζ+ JNL cells were also incubated with parental 721.221 cells as a negative control and with a combination of anti-Flag and anti-mouse IgG antibodies as a positive control (X-link). Ligand recognition was detected by the upregulation of luciferase. Bars indicate the mean relative light units (RLU) of luciferase activity in triplicate wells. Error bars indicate SD of the mean and asterisks denote statistically significant differences (****p < 0.0001, one-way ANOVA with Dunnett’s test).
Mamu-AG Determinants of Recognition by KIR3DL05
To define Mamu-AG polymorphisms that account for differences in KIR3DL05 recognition, amino acid substitutions were introduced into Mamu-AG2*01:01 at positions that differ from Mamu-AG3*02:01 and -AG3*03:01. 721.221 cells expressing these variants were then tested for recognition by KIR3DL05-CD3ζ+ JNL cells (Figure 5). The replacement of any one of three amino acids in the α1-domain of Mamu-AG2*01:01 with the corresponding residues of Mamu-AG3*02:01 (W18G, E19Q or V76E) partially or completely impaired KIR3DL05 recognition (Figure 5A). Likewise, individually exchanging three amino acids of Mamu-AG2*01:01 with the corresponding residues of Mamu-AG3*03:01 (S11A, R75Q or I95F) significantly impaired interactions with KIR3DL05 (Figure 5A). Conversely, a combination of three amino acid changes in the α1-domain of Mamu-AG3*02:01 (G18W, Q19E and E76V) was sufficient to restore interactions with KIR3DL05 to a similar level as Mamu-AG2*01:01 (Figure 5A). Thus, similar to the KIR recognition of classical MHC class I ligands, the specificity of KIR3DL05 is primarily determined by polymorphisms in the α1-domain of Mamu-AG.
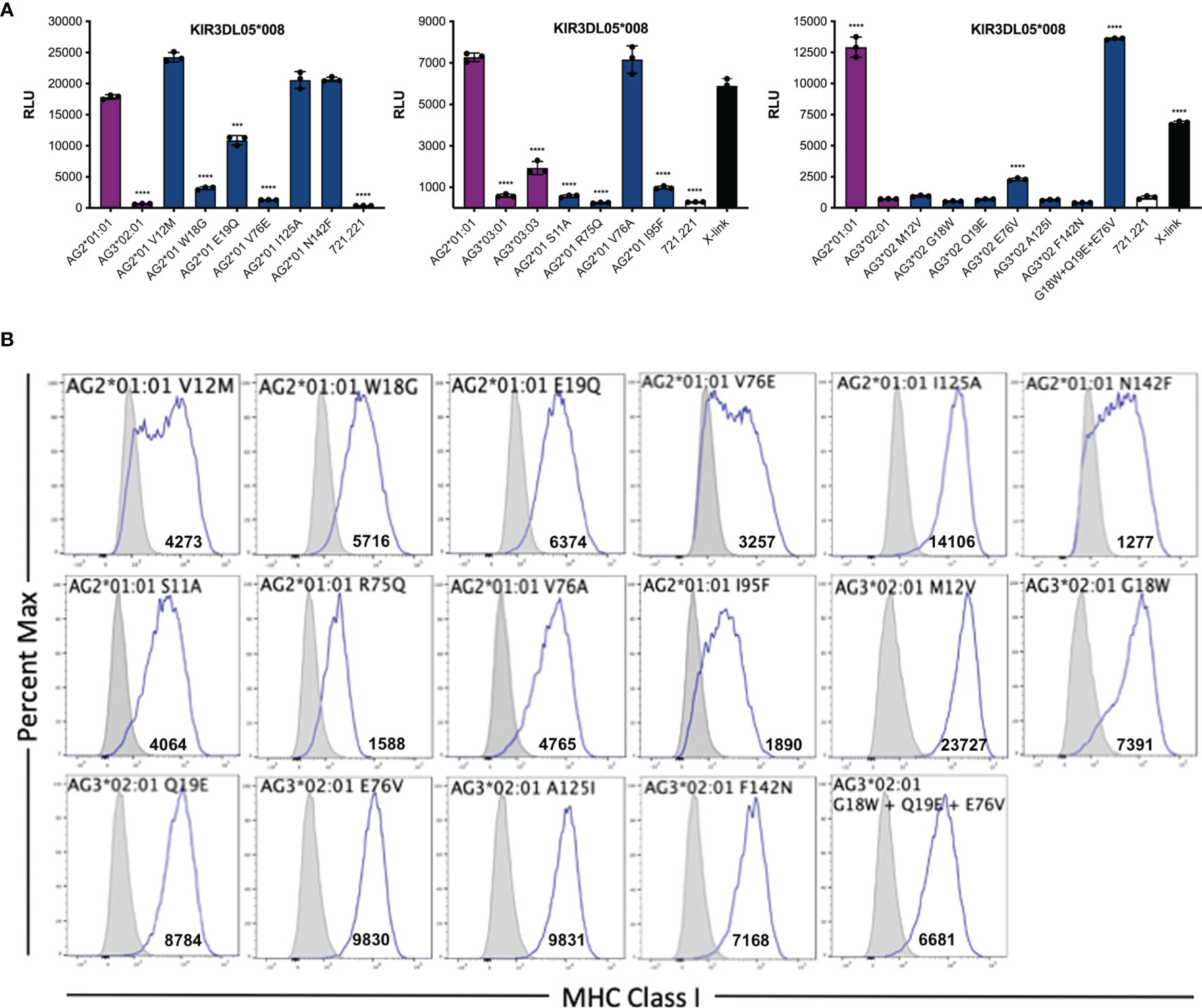
Figure 5 Identification of residues in the α1- and α2-domains of Mamu-AG that contribute to recognition by KIR3DL05. (A) 721.221 cells expressing Mamu-AG2*01:01 variants with single amino acid substitutions at positions that differ from Mamu-AG3*02:01 or -AG3*03:01 and Mamu-AG3*02:01 variants with substitutions at positions that differ from Mamu-AG2*01:01 were co-incubated with of KIR3DL05*008-CD3ζ+ JNL cells overnight at a 1:1 E:T ratio. KIR-CD3ζ+ JNL cells were also incubated with parental 721.221 cells as a negative control and with anti-Flag and anti-mouse IgG antibodies as positive control (X-link). Bars represent the mean luciferase activity (RLU) from triplicate wells. Error bars indicate SD of the mean and asterisks denote statistically significant differences (***p < 0.0005, ****p < 0.0001, one-way ANOVA with Dunnett’s test). (B) Mamu-AG+ 721.221 cells and parental 721.221 cells were stained with a PE-conjugated pan-MHC class I-specific antibody (W6/32) and Near-IR LIVE/DEAD stain. After excluding dead cells, the fluorescence intensity of MHC class I staining on the surface of Mamu-AG-transduced 721.221 cells (open) was compared to background staining on parental 721.221 cells (shaded). Flow cytometry data was analyzed using FlowJo 10.6 for Mac OSX. Values indicate geometric mean fluorescence intensity of staining.
The amino acid residues of Mamu-AG2*01:01 that participate in interactions with KIR3DL05 were modeled by in silico replacement of the corresponding residues in a three-dimensional structure of Mamu-A1*002. Mamu-A1*002 was selected for modeling due to its general sequence similarity with Mamu-AG and because it is also a ligand for KIR3DL05 (43). All of the polymorphic residues that affect KIR3DL05 recognition are in close proximity to the Bw4 motif (residues 77-83) at the C-terminal end of the α1-domain (Figure 6). Residues 75 and 76, for which the R75Q and V76E polymorphisms had the greatest impact on KIR3DL05 responses, are part of the α-helical region directly adjacent to the Bw4 motif (Figure 6). Residues 18 and 19 are located in an underlying loop that was previously identified as a KIR contact site based on the crystal structure of human KIR3DL1 in complex with HLA-B*5701 (Figure 6) (41). The side chains of these residues are surface exposed in a region where the W18G and E19Q polymorphisms would be expected to contribute to a patch of sequence diversity together with polymorphisms at positions 75 and 76. In contrast, residues 11 and 95 are located in the floor of the peptide binding pocket and are unlikely to be surface exposed. The partial effects of the S11A and I95F on KIR3DL05 responses may therefore reflect differences in peptide binding and/or conformational changes to the peptide-MHC class I complex. Overall, these results are consistent with the structure of KIR3DL1 in complex with HLA-B*5701 showing extensive contacts between the D0 and D1 domains of the KIR and the α1-domain of the MHC class I molecule (41). These findings also illustrate that while polymorphisms in residues 77-83 may determine broad patterns of KIR recognition (e.g. Bw4 specificity), polymorphisms in adjoining regions can contribute to variation in responses to closely related MHC class I allotypes.
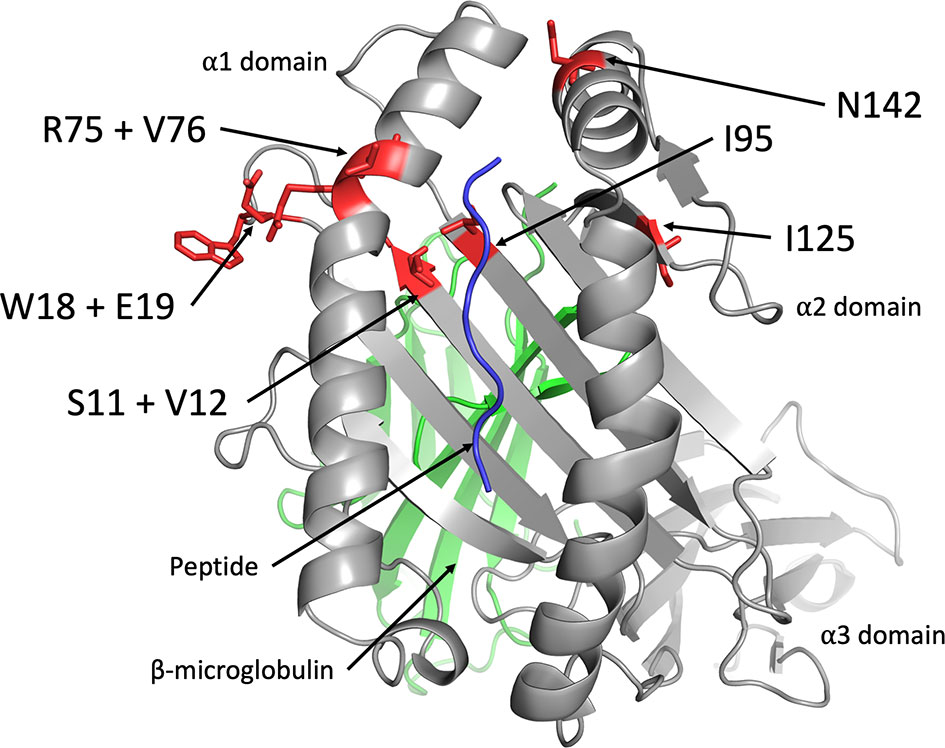
Figure 6 Location of amino acid polymorphisms in Mamu-AG that affect recognition by KIR3DL05. The side chains of Mamu-AG2*01:01 residues that affect recognition by KIR3DL05*008 (red) were modeled on a three-dimensional crystal structure of Mamu-A1*002 (grey) in complex with peptide (blue) and β2-microglobulin (green) (PDB 3jtt) using PyMol software.
Discussion
Definition of the MHC class I ligands of rhesus macaque KIRs is foundational for studying NK cell responses in this species as an animal model for infectious diseases, transplantation and reproductive biology. In a previous survey of KIR-MHC class I interactions in the rhesus macaque, Mamu-AG2*01:01 was identified as a ligand for KIR3DL05*008 (39). Although Mamu-AG does not have a direct human ortholog, it shares similar molecular features and tissue distribution as HLA-G (19, 20, 23). However, Mamu-AG is considerably more polymorphic than HLA-G owing to allelic variation at multiple loci (44). KIR3DL05 is also polymorphic with at least 33 different KIR3DL05 alleles identified so far (45). To gain a better understanding of the determinants of KIR3DL05 specificity for Mamu-AG, we investigated the impact of Mamu-AG polymorphisms on this receptor-ligand interaction.
Our findings reveal three general patterns of Mamu-AG recognition among the KIR3DL05 allotypes selected for this study. The most common pattern exhibited by KIR3DL05*004, *005 and *008 includes dominant responses to Mamu-AG2*01:01 and lower, more variable responses to Mamu-AG3*03:01, -AG3*03:02, -AG3*03:03 and -AG3*03:04. The nearly identical responses of KIR3DL05*004 and *008 reflect the overall similarity of their extracellular domains, which only differ by a single amino acid in the D1 domain (Q139H). KIR3DL05*005 differs from each of these KIRs by seven residues in D0 domain and two in the D1 domain. However, only one of these polymorphisms (Y29H) is located in a predicted MHC class I contact site. In contrast to the other allotypes, KIR3DL05*001 responded strongly to Mamu-AG1*05:01, weakly to Mamu-AG2*01:01, and not at all to Mamu-AG3. Since KIR3DL05*001 and *008 share identical D1 and D2 domains, but differ by nine amino acids in the D0 domain, including three residues in predicted MHC class I contact sites, this shift in Mamu-AG recognition is a function of polymorphisms in the D0 domain. KIR3DL05*X exhibited a more complex pattern that included dominant responses to both Mamu-AG2*01:01 and -AG1*05:01, and detectable responses to Mamu-AG3*03:01, -AG3*03:02, -AG3*03:03 and -AG3*03:04. Whereas KIR3DL05*X differs from KIR3DL05*008 by only two amino acids in the D0 domain, there are thirteen differences in the D1 domain. The more extensive differences in D1 reflect the hybrid origins of this KIR, which is comprised of the leader peptide, D0 and D1 domains of KIR3DS02 (exons 1-4) and the D2, stem, transmembrane and cytoplasmic domains of KIR3DL05 (exons 5-9) (40). Overall, these observations reveal differences in KIR3DL05 recognition of Mamu-AG molecules that are determined by polymorphisms in their D0 and D1 domains.
We also tested an allotype of KIR3DS02 with identical D0 and D1 domains to KIR3DL05*X. KIR3DS02*004 recognized Mamu-AG2*01:01, but did not respond to other Mamu-AG allotypes. These results identify Mamu-AG2*01:01 as a ligand for this activating KIR and indicate that differences in the D2 domains of KIR3DS02*004 and KIR3DL05*X account for differences in their responses to other Mamu-AG molecules.
An analysis of single amino acid variants identified Mamu-AG polymorphisms that affect KIR3DL05 recognition. These include differences in the α-helical region of the α1 domain (R75Q & V76E), an N-terminal loop of the α1 domain (W18G & E19Q), and the β-sheet that forms the floor of the peptide binding pocket (S11A & I95F). Residues 18, 19, 75 and 76 are located on the same face of the α1 domain and coincide with predicted KIR contact sites (41). Thus, the effects of polymorphisms at these positions may be attributed to interactions with KIR3DL05. On the other hand, residues 11 and 95 are not predicted to contact the KIR, but may alter the conformation of the bound peptide or adjacent residues in the α1 domain in a manner that indirectly affects interactions with KIR3DL05. These observations illustrate how a few amino acid differences in surfaces of the α1 domain can result in differential recognition of closely related MHC class I molecules and are consistent with the co-evolution of the D0 and D1 domains of KIRs in concert with the α1 domain of MHC class I ligands.
In humans, HLA-G is expressed by fetal trophoblasts that invade the uterine endometrium during early stages of pregnancy and on extravillous trophoblasts that remodel spiral arteries of the developing placenta (46). HLA-G is a ligand for KIR2DL4 (47, 48), which is an unusual 2 domain KIR with both activating and inhibitory features. KIR2DL4 has a positively charged arginine residue in the transmembrane domain for pairing with FcεRIγ to transduce activating signals and a long cytoplasmic tail with an immunotyrosine-based inhibitory motif (ITIM) (49). This receptor interacts with a soluble isoform of HLA-G in endosomes to stimulate the release of pro-inflammatory and pro-angiogenic factors by decidual NK cells that promote placental vascularization (50–52). Although the presence of HLA-E on extravillous trophoblasts is thought to play a dominant role in maintaining fetal tolerance via inhibitory interactions with CD94/NKG2A on decidual NK cells, HLA-G may facilitate tolerance through additional interactions with inhibitory receptors and by serving as a source of leader peptides bound by HLA-E (52, 53).
The ortholog of HLA-G (Mamu-G) in rhesus macaques is a pseudogene (17). However, macaques express another nonclassical MHC class I molecule with similar tissue distribution (19). Like HLA-G, Mamu-AG is expressed on fetal cytotrophoblasts that invade the endometrium and on syncytiotrophoblasts of the placental chorionic villi (23). While it has been difficult to experimentally verify the function of HLA-G in humans, passive transfer of a Mamu-AG-specific monoclonal antibody to pregnant macaques was shown to disrupt leukocyte responses to implantation, vascularization, and placental development (38). Thus, Mamu-AG appears to serve a similar function as HLA-G.
The identification of Mamu-AG as a ligand for KIR3DL05 and KIR3DS02 reveals that certain macaque KIRs are capable of mediating functional interactions with this nonclassical molecule. While macaques express a recognizable ortholog of KIR2DL4 (KIR2DL04) (7, 54), there is no reason to believe that this receptor interacts with Mamu-AG, which bears no homology to HLA-G. Nevertheless, it is likely that there are other receptors for Mamu-AG that are yet to be identified. These may include additional KIRs or other types of receptors that react more broadly with MHC class I molecules, such as the Ig-like transcripts 2 and 4 (ILT-2 and ILT-4) that are known to serve as receptors for HLA-G in humans (55).
A complex and precarious balance of NK cell activating and inhibitory receptor interactions is essential for normal placental development, fetal tolerance, and immune surveillance of the maternal-fetal interface. Precisely how KIR3DL05 and KIR3DS02 interactions with Mamu-AG contribute to this balance in macaques is presently unclear. The ligation of activating receptors on decidual NK cells may stimulate the release of pro-inflammatory and pro-angiogenic factors that promote placental vascularization and increase blood supply to the developing fetus, similar to the interactions of KIR2DL4 with soluble HLA-G or lineage III KIRs (KIR2Ds) with HLA-C in humans (52, 56). In the absence of HLA-G or HLA-C, it is conceivable that macaques depend on Mamu-AG recognition by activating KIRs such as KIR3DS02 to stimulate placental vascularization. Mamu-AG may also facilitate fetal tolerance through interactions with inhibitory KIRs such as KIR3DL05 or by providing leader peptides that stabilize Mamu-E on the surface of trophoblasts for inhibitory signaling through CD94/NKG2A (57).
Recent studies have demonstrated a role for decidual NK cells in protecting the maternal-fetal interface. Extravillous trophoblasts are susceptible to a number of pathogens that are transmitted in utero, including Zika virus (ZiKV), cytomegalovirus (CMV), and Toxoplasma gondii (58–63). In the case of ZiKV, infection of extravillous trophoblasts causes ER stress that results in the downregulation of HLA-C and -G, which renders these cells susceptible to killing by decidual NK cells (60). Similar antimicrobial functions of decidual NK cells have been reported for CMV and Listeria monocytogenes (64–66). Thus, disruption or alteration of Mamu-AG expression on macaque trophoblasts may trigger decidual NK cell responses through inhibitory or activating KIRs. KIR3DL05 and KIR3DS02 may thereby contribute to pregnancy success by protecting the maternal-fetal interface from invading microorganisms. Furthermore, polymorphic differences in these KIRs and Mamu-AG could account for individual variation in resistance to placental transmission of viral and bacterial pathogens.
The rhesus macaque has become an increasingly valuable animal model for studying viral pathogens such as CMV and Zika virus that are frequently transmitted to the developing fetus during pregnancy (67–70). Hence, the molecular interactions between macaque NK cell receptors and their MHC class I ligands that occur at the maternal-fetal interface may be especially important for understanding viral transmission and pathogenesis in this context. The present study defines many of the Mamu-AG ligands recognized by diverse allotypes of KIR3DL05 and identifies KIR3DS02*004 as an activating receptor for Mamu-AG2*01:01. These findings afford greater insight into NK cell recognition of a nonclassical molecule implicated in pregnancy and provide a broader foundation for investigating NK cell biology in the rhesus macaque as a nonhuman primate model for infectious diseases and reproductive biology.
Data Availability Statement
The GenBank accession numbers for the sequences presented in this study are provided in the Materials and Methods. All of the data presented in this study are included in the article. Further inquires can be directed to the corresponding author.
Author Contributions
RN, JA, WS, MW, and PB performed the experiments and analyzed the data. KS and SJ designed the experiments. DE conceived the project and wrote the manuscript. All of the authors contributed to the manuscript and approved the submitted version.
Funding
This work was supported by Public Health Service Grants AI095098, AI161816, AI121135, AI148379 and AI098485 (to DE) and OD011106 (to the WNPRC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Roger Wiseman at the Wisconsin National Primate Research Center for his assistance with changes to the nonhuman primate KIR and MHC class I nomenclature. DE is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
References
1. Parham P. MHC Class I Molecules and KIRs in Human History, Health and Survival. Nat Rev Immunol (2005) 5:201–14. doi: 10.1038/nri1570
2. Lanier LL. NK Cell Recognition. Annu Rev Immunol (2005) 23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526
3. Boyson JE, Schufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Huges AL, et al. The MHC Class I Genes of the Rhesus Monkey: Different Evolutionary Histories of the MHC Class I and II Genes in Primates. J Immunol (1996) 156:4656–65. doi: 10.1016/0198-8859(96)85380-8
4. Otting N, Heijmans CMC, Noort RC, Groot N, Doxiadis GGM, vanRood JJ, et al. Unparalleled Complexity of the MHC Class I Region in Rhesus Macaques. Proc Natl Acad Sci USA (2005) 102:1626–31. doi: 10.1073/pnas.0409084102
5. Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, O’Connor DH. Haplessly Hoping: Macaque Major Histocompatibility Complex Made Easy. ILAR J Nat Res Council Inst Lab Anim Resour (2013) 54:196–210. doi: 10.1093/ilar/ilt036
6. Adams EJ, Parham P. Species-Specific Evolution of MHC Class I Genes in the Higher Primates. Immunol Rev (2001) 183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x
7. Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, et al. Single Haplotype Analysis Demonstrates Rapid Evolution of the Killer Immunoglobulin-Like Receptor (KIR) Loci in Primates. Genome Res (2005) 15:25–35. doi: 10.1101/gr.2381205
8. Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the Killer Cell Ig-Like Receptors of Rhesus Monkeys. J Immunol (2001) 166:4380–90. doi: 10.4049/jimmunol.166.7.4380
9. Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O’Connor DH. Complete Characterization of Killer Ig-Like Receptor (KIR) Haplotypes in Mauritian Cynomolgus Macaques: Novel Insights Into Nonhuman Primate KIR Gene Content and Organization. J Immunol (2008) 181:6301–8. doi: 10.4049/jimmunol.181.9.6301
10. Kruse PH, Rosner C, Walter L. Characterization of Rhesus Macaque KIR Genotypes and Haplotypes. Immunogenetics (2010) 62:281–93. doi: 10.1007/s00251-010-0433-4
11. Blokhuis JH, Wiel M, Doxiadis GG, Bontrop RE. The Mosaic of KIR Haplotypes in Rhesus Macaques. Immunogenetics (2010) 62:295–306. doi: 10.1007/s00251-010-0434-3
12. Palacios C, Cuervo LC, Cadavid LF. Evolutionary Patterns of Killer Cell Ig-Like Receptor Genes in Old World Monkeys. Gene (2011) 474:39–51. doi: 10.1016/j.gene.2010.12.006
13. Bimber BN, Evans DT. The Killer-Cell Immunoglobulin-Like Receptors of Macaques. Immunol Rev (2015) 267:246–58. doi: 10.1111/imr.12329
14. Boyson JE, McAdam SN, Gallimore A, Golos TG, Liu X, Gotch FM, et al. The MHC E Locus in Macaques Is Polymorphic and Is Conserved Between Macaques and Humans. Immunogenetics (1995) 41:59–68. doi: 10.1007/BF00182314
15. Knapp L, Cadavid LF, Watkins DI. The MHC-E Locus Is the Most Well Conserved of All Known Primate Class I Histocompatibility Genes. J Immunol (1998) 160:189–96.
16. Otting N, Bontrop RE. Characterization of the Rhesus Macaque (Macaca Mulatta) Equivalent of HLA-F. Immunogenetics (1993) 38:141–5. doi: 10.1007/BF00190901
17. Boyson JE, Iwanaga KK, Golos TG, Watkins DI. Identifiction of the Rhesus Monkey HLA-G Ortholog. Mamu-G Is a Pseudogene. J Immunol (1996) 157:5428–37.
18. Castro MJ, Morales P, Fernandez-Soria V, Suarez B, Recio MJ, Alvarez M, et al. Allelic Diversity at the Primate Mhc-G Locus: Exon 3 Bears Stop Codons in All Cercopithecinae Sequences. Immunogenetics (1996) 43:327–36. doi: 10.1007/BF02199801
19. Boyson JE, Iwanaga KK, Golos TG, Watkins DI. Identification of a Novel MHC Class I Gene, Mamu-AG, Expressed in the Placenta of a Primate With an Inactivated G Locus. J Immunol (1997) 159:3311–21.
20. Boyson JE, Iwanaga KK, Urvater JA, Hughes AL, Golos TG, Watkins DI. Evolution of a New Nonclassical MHC Class I Locus in Two Old World Primate Species. Immunogenetics (1999) 49:86–98. doi: 10.1007/s002510050467
21. Langat DK, Morales PJ, Fazleabas AT, Mwenda JM, Hunt JS. Baboon Placentas Express Soluble and Membrane-Bound Paan-AG Proteins Encoded by Alternatively Spliced Transcripts of the Class Ib Major Histocompatibility Complex Gene, Paan-AG. Immunogenetics (2002) 54:164–73. doi: 10.1007/s00251-002-0454-8
22. Bondarenko GI, Dambaeva SV, Grendell RL, Hughes AL, Durning M, Garthwaite MA, et al. Characterization of Cynomolgus and Vervet Monkey Placental MHC Class I Expression: Diversity of the Nonhuman Primate AG Locus. Immunogenetics (2009) 61:431–42. doi: 10.1007/s00251-009-0376-9
23. Slukvin II, Lunn DP, Watkins DI, Golos TG. Placental Expression of the Nonclassical MHC Class I Molecule Mamu-AG at Implantation in the Rhesus Monkey. Proc Natl Acad Sci USA (2000) 97:9104–9. doi: 10.1073/pnas.97.16.9104
24. Arnaiz-Villena A, Juarez I, Suarez-Trujillo F, Lopez-Nares A, Vaquero C, Palacio-Gruber J, et al. HLA-G: Function, Polymorphisms and Pathology. Int J Immunogenet (2020) 48(2):172–92. doi: 10.1111/iji.12513
25. Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, et al. Therapeutic Efficacy of Potent Neutralizing HIV-1-Specific Monoclonal Antibodies in SHIV-Infected Rhesus Monkeys. Nature (2013) 503:224–8. doi: 10.1038/nature12744
26. Esquivel RN, Patel A, Kudchodkar SB, Park DH, Stettler K, Beltramello M, et al. In Vivo Delivery of a DNA-Encoded Monoclonal Antibody Protects Non-Human Primates Against Zika Virus. Mol Ther (2019) 27:974–85. doi: 10.1016/j.ymthe.2019.03.005
27. Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, et al. Fc Receptor But Not Complement Binding Is Important in Antibody Protection Against HIV. Nature (2007) 449:101–4. doi: 10.1038/nature06106
28. Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, et al. Effective, Low-Titer Antibody Protection Against Low-Dose Repeated Mucosal SHIV Challenge in Macaques. Nat Med (2009) 15:951–4. doi: 10.1038/nm.1974
29. Magnani DM, Rogers TF, Maness NJ, Grubaugh ND, Beutler N, Bailey VK, et al. Fetal Demise and Failed Antibody Therapy During Zika Virus Infection of Pregnant Macaques. Nat Commun (2018) 9:1624. doi: 10.1038/s41467-018-04056-4
30. Martinez-Navio JM, Fuchs SP, Pantry SN, Lauer WA, Duggan NN, Keele BF, et al. Adeno-Associated Virus Delivery of Anti-HIV Monoclonal Antibodies Can Drive Long-Term Virologic Suppression. Immunity (2019) 50:567–75.e5. doi: 10.1016/j.immuni.2019.02.005
31. Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, et al. Vaccine-Induced Protection From Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity (2019) 50:241–52.e6. doi: 10.1016/j.immuni.2018.11.011
32. Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, et al. Passive Transfer of Modest Titers of Potent and Broadly Neutralizing Anti-HIV Monoclonal Antibodies Block SHIV Infection in Macaques. J Exp Med (2014) 211:2061–74. doi: 10.1084/jem.20132494
33. Welles HC, Jennewein MF, Mason RD, Narpala S, Wang L, Cheng C, et al. Vectored Delivery of Anti-SIV Envelope Targeting mAb via AAV8 Protects Rhesus Macaques From Repeated Limiting Dose Intrarectal Swarm SIVsmE660 Challenge. PLoS Pathog (2018) 14:e1007395. doi: 10.1371/journal.ppat.1007395
34. Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, et al. Comparison of Nonhuman Primates Identified the Suitable Model for COVID-19. Sig Transduct Target Ther (2020) 5:157. doi: 10.1038/s41392-020-00269-6
35. Roark HK, Jenks JA, Permar SR, Schleiss MR. Animal Models of Congenital Cytomegalovirus Transmission: Implications for Vaccine Development. J Infect Dis (2020) 221:S60–73. doi: 10.1093/infdis/jiz484
36. Torrealba JR, Katayama M, Fechner JH Jr., Jankowska-Gan E, Kusaka S, Xu Q, et al. Metastable Tolerance to Rhesus Monkey Renal Transplants Is Correlated With Allograft TGF-Beta 1+CD4+ T Regulatory Cell Infiltrates. J Immunol (2004) 172:5753–64. doi: 10.4049/jimmunol.172.9.5753
37. Forrest L, Fechner J, Post J, Van Asselt N, Kvasnica K, Haynes LD, et al. Tomotherapy Applied Total Lymphoid Irradiation and Allogeneic Hematopoietic Cell Transplantation Generates Mixed Chimerism in the Rhesus Macaque Model. Radiat Res (2021) 196(6):623–32. doi: 10.1667/RADE-20-00246.1
38. Bondarenko GI, Burleigh DW, Durning M, Breburda EE, Grendell RL, Golos TG. Passive Immunization Against the MHC Class I Molecule Mamu-AG Disrupts Rhesus Placental Development and Endometrial Responses. J Immunol (2007) 179:8042–50. doi: 10.4049/jimmunol.179.12.8042
39. Banerjee P, Ries M, Janaka SK, Grandea AG 3rd, Wiseman R, O’Connor DH, et al. Diversification of Bw4 Specificity and Recognition of a Nonclassical MHC Class I Molecule Implicated in Maternal-Fetal Tolerance by Killer Cell Ig-Like Receptors of the Rhesus Macaque. J Immunol (2018) 201:2776–86. doi: 10.4049/jimmunol.1800494
40. Colantonio AD, Bimber BN, Neidermyer WJ, Reeves RK, Alter G, Altfeld M, et al. KIR Polymorphisms Modulate Peptide-Dependent Binding to an MHC Class I Ligand With a Bw6 Motif. PLoS Pathog (2011) 7:e1001316. doi: 10.1371/journal.ppat.1001316
41. Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, et al. Killer Cell Immunoglobulin-Like Receptor 3DL1-Mediated Recognition of Human Leukocyte Antigen B. Nature (2011) 479:401–5. doi: 10.1038/nature10517
42. Shimizu Y, Koller B, Geraghty D, Orr H, Shaw S, Kavathas P, et al. Transfer of Cloned Human Class I Major Histocompatibility Complex Genes Into HLA Mutant Human Lymphoblastoid Cells. Mol Cell Biol (1986) 6:1074–87. doi: 10.1128/mcb.6.4.1074-1087.1986
43. Feng Y, Qi J, Zhang H, Wang J, Liu J, Jiang F, et al. X-Ray Crystallographic Characterization of Rhesus Macaque MHC Mamu-A*02 Complexed With an Immunodominant SIV-Gag Nonapeptide. Acta Crystallogr Sect F Struct Biol Cryst Commun (2006) 62:13–5. doi: 10.1107/S1744309105038704
44. Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic Divergence of the Rhesus Macaque Major Histocompatibility Complex. Genome Res (2004) 14:1501–15. doi: 10.1101/gr.2134504
45. Bruijnesteijn J, de Groot NG, Otting N, Maccari G, Guethlein LA, Robinson J, et al. Nomenclature Report for Killer-Cell Immunoglobulin-Like Receptors (KIR) in Macaque Species: New Genes/Alleles, Renaming Recombinant Entities and IPD-NHKIR Updates. Immunogenetics (2020) 72:37–47. doi: 10.1007/s00251-019-01135-8
46. Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: At the Interface of Maternal-Fetal Tolerance. Trends Immunol (2017) 38:272–86. doi: 10.1016/j.it.2017.01.009
47. Allan DS, Colonna M, Lanier LL, Churakova TD, Abrams JS, Ellis SA, et al. Tetrameric Complexes of Human Histocompatibility Leukocyte Antigen (HLA)-G Bind to Peripheral Blood Myelomonocytic Cells. J Exp Med (1999) 189:1149–56. doi: 10.1084/jem.189.7.1149
48. Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G-Specific Receptor Expressed on All Natural Killer Cells. J Exp Med (1999) 189:1093–100. doi: 10.1084/jem.189.7.1093
49. Faure M, Long EO. KIR2DL4 (CD158d), an NK Cell-Activating Receptor With Inhibitory Potential. J Immunol (2002) 168:6208–14. doi: 10.4049/jimmunol.168.12.6208
50. Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, et al. Activation of NK Cells by an Endocytosed Receptor for Soluble HLA-G. PLoS Biol (2006) 4:e9. doi: 10.1371/journal.pbio.0040009
51. Rajagopalan S. Endosomal Signaling and a Novel Pathway Defined by the Natural Killer Receptor KIR2DL4 (CD158d). Traffic (2010) 11:1381–90. doi: 10.1111/j.1600-0854.2010.01112.x
52. Rajagopalan S. HLA-G-Mediated NK Cell Senescence Promotes Vascular Remodeling: Implications for Reproduction. Cell Mol Immunol (2014) 11:460–6. doi: 10.1038/cmi.2014.53
53. Ishitani A, Sageshima N, Lee N, Dorofeeva N, Hatake K, Marquardt H, et al. Protein Expression and Peptide Binding Suggest Unique and Interacting Functional Roles for HLA-E, F, and G in Maternal-Placental Immune Recognition. J Immunol (2003) 171:1376–84. doi: 10.4049/jimmunol.171.3.1376
54. Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. Evidence for Balancing Selection Acting on KIR2DL4 Genotypes in Rhesus Macaques of Indian Origin. Immunogenetics (2009) 61:503–12. doi: 10.1007/s00251-009-0379-6
55. Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human Inhibitory Receptors Ig-Like Transcript 2 (ILT2) and ILT4 Compete With CD8 for MHC Class I Binding and Bind Preferentially to HLA-G. Proc Natl Acad Sci USA (2003) 100:8856–61. doi: 10.1073/pnas.1431057100
56. Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, et al. Maternal Activating KIRs Protect Against Human Reproductive Failure Mediated by Fetal HLA-C2. J Clin Invest (2010) 120:4102–10. doi: 10.1172/JCI43998
57. Dambaeva SV, Bondarenko GI, Grendell RL, Kravitz RH, Durning M, Golos TG. Non-Classical MHC-E (Mamu-E) Expression in the Rhesus Monkey Placenta. Placenta (2008) 29:58–70. doi: 10.1016/j.placenta.2007.10.001
58. Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, et al. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe (2016) 20:155–66. doi: 10.1016/j.chom.2016.07.002
59. Robbins JR, Bakardjiev AI. Pathogens and the Placental Fortress. Curr Opin Microbiol (2012) 15:36–43. doi: 10.1016/j.mib.2011.11.006
60. Sen Santara S, Crespo AC, Mulik S, Ovies C, Boulenouar S, Strominger JL, et al. Decidual NK Cells Kill Zika Virus-Infected Trophoblasts. Proc Natl Acad Sci USA (2021) 118(47):e2115410118. doi: 10.1073/pnas.2115410118
61. Vigliani MB, Bakardjiev AI. Intracellular Organisms as Placental Invaders. Fetal Matern Med Rev (2014) 25:332–8. doi: 10.1017/S0965539515000066
62. Cao B, Mysorekar IU. Intracellular Bacteria in Placental Basal Plate Localize to Extravillous Trophoblasts. Placenta (2014) 35:139–42. doi: 10.1016/j.placenta.2013.12.007
63. Weisblum Y, Panet A, Zakay-Rones Z, Haimov-Kochman R, Goldman-Wohl D, Ariel I, et al. Modeling of Human Cytomegalovirus Maternal-Fetal Transmission in a Novel Decidual Organ Culture. J Virol (2011) 85:13204–13. doi: 10.1128/JVI.05749-11
64. Crespo AC, Strominger JL, Tilburgs T. Expression of KIR2DS1 by Decidual Natural Killer Cells Increases Their Ability to Control Placental HCMV Infection. Proc Natl Acad Sci USA (2016) 113:15072–7. doi: 10.1073/pnas.1617927114
65. Siewiera J, El Costa H, Tabiasco J, Berrebi A, Cartron G, Le Bouteiller P, et al. Human Cytomegalovirus Infection Elicits New Decidual Natural Killer Cell Effector Functions. PLoS Pathog (2013) 9:e1003257. doi: 10.1371/journal.ppat.1003257
66. Crespo AC, Mulik S, Dotiwala F, Ansara JA, Sen Santara S, Ingersoll K, et al. Decidual NK Cells Transfer Granulysin to Selectively Kill Bacteria in Trophoblasts. Cell (2020) 182:1125–39.e18. doi: 10.1016/j.cell.2020.07.019
67. Itell HL, Kaur A, Deere JD, Barry PA, Permar SR. Rhesus Monkeys for a Nonhuman Primate Model of Cytomegalovirus Infections. Curr Opin Virol (2017) 25:126–33. doi: 10.1016/j.coviro.2017.08.005
68. Vera Cruz D, Nelson CS, Tran D, Barry PA, Kaur A, Koelle K, et al. Intrahost Cytomegalovirus Population Genetics Following Antibody Pretreatment in a Monkey Model of Congenital Transmission. PLoS Pathog (2020) 16:e1007968. doi: 10.1371/journal.ppat.1007968
69. Mostrom MJ, Scheef EA, Sprehe LM, Szeltner D, Tran D, Hennebold JD, et al. Immune Profile of the Normal Maternal-Fetal Interface in Rhesus Macaques and Its Alteration Following Zika Virus Infection. Front Immunol (2021) 12:719810. doi: 10.3389/fimmu.2021.719810
Keywords: KIR, MHC, Mamu-AG, macaque, HLA-G, placenta, pregnancy
Citation: Nicholas RE, Sandstrom K, Anderson JL, Smith WR, Wetzel M, Banerjee P, Janaka SK and Evans DT (2022) KIR3DL05 and KIR3DS02 Recognition of a Nonclassical MHC Class I Molecule in the Rhesus Macaque Implicated in Pregnancy Success. Front. Immunol. 13:841136. doi: 10.3389/fimmu.2022.841136
Received: 22 December 2021; Accepted: 01 March 2022;
Published: 23 March 2022.
Edited by:
Ronald Bontrop, Biomedical Primate Research Centre (BPRC), NetherlandsReviewed by:
Lutz Walter, Leibniz Institute for Primate Research, GermanyJesse Bruijnesteijn, Biomedical Primate Research Centre (BPRC), Netherlands
Copyright © 2022 Nicholas, Sandstrom, Anderson, Smith, Wetzel, Banerjee, Janaka and Evans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David T. Evans, dtevans2@wisc.edu
 Rachel E. Nicholas
Rachel E. Nicholas Kjell Sandstrom
Kjell Sandstrom Jennifer L. Anderson1
Jennifer L. Anderson1 Molly Wetzel
Molly Wetzel Priyankana Banerjee
Priyankana Banerjee Sanath Kumar Janaka
Sanath Kumar Janaka David T. Evans
David T. Evans