- Department of Neuroscience, Physiology & Pharmacology, University College London, London, United Kingdom
The traditional view of the nuclear envelope (NE) was that it represented a relatively inert physical barrier within the cell, whose main purpose was to separate the nucleoplasm from the cytoplasm. However, recent research suggests that this is far from the case, with new and important cellular functions being attributed to this organelle. In this review we describe research suggesting an important contribution of the NE and its constituents in regulating the functions of cells of the innate and adaptive immune system. One of the standout properties of immune cells is their ability to migrate around the body, allowing them to carry out their physiological/pathophysiology cellular role at the appropriate location. This together with the physiological role of the tissue, changes in tissue matrix composition due to disease and aging, and the activation status of the immune cell, all result in immune cells being subjected to different mechanical forces. We report research which suggests that the NE may be an important sensor/transducer of these mechanical signals and propose that the NE is an integrator of both mechanical and chemical signals, allowing the cells of the innate immune system to precisely regulate gene transcription and functionality. By presenting this overview we hope to stimulate the interests of researchers into this often-overlooked organelle and propose it should join the ranks of mitochondria and phagosome, which are important organelles contributing to immune cell function.
Introduction
The nucleus, one of the defining features of all eukaryotic cells, houses most of the genetic information within the cell and is bounded by the nuclear envelope (NE). In the 1950s research began to elucidate the NE structure and its function (1–3). The prevailing view of this early research was that the NE was just a physical barrier separating nucleoplasm from cytoplasm; however evidence now indicates that this is a too simplistic view of this organelle and that the NE significantly contributes to cellular signalling and regulatory pathways (4–6).
The immune system is an integrated network of organs, cells and biochemical cascades which provides the host with protection against pathogens, noxious exogenous stimuli and trauma. The system also holds memory of pathogen components, allowing it to respond more efficiently when the pathogen is encountered again. While the immune system is required for the continued well-being of an individual, with increased susceptibility to microbial infections and cancers in individuals with supressed immune system; inappropriate activation of the immune system can lead to excessive inflammation, autoimmunity and host tissue destruction. To maintain a balance of appropriate activation, while guarding against self-induced tissue damage/pathology, the immune system employs numerous cellular sensors, regulatory pathways and extra- and intracellular signalling molecules to maintain a controlled appropriate response (7, 8).
Over the last 40 years various organelles, including lysosomes/phagosomes, mitochondria and endoplasmic reticulum (ER), have been shown to be important in regulating the activation and effector functions of both the innate and adaptive immune cells (9). Here, we review the current knowledge regarding the structure and function of NE in cells of the innate and adaptive immune systems and discuss the evidence which suggests the NE is an integrator of both mechanical and chemical signals encountered by immune cells during inflammation.
Structure and Function of NE
The structure of the NE (Figure 1) comprises two functionally distinct lipid membranes, the outer nuclear membrane (ONM) and the inner nuclear membrane (INM), which give rise to a functional compartment between them, known as the perinuclear space (1, 3). The ONM is continuous with the endoplasmic reticulum (ER) making the perinuclear space continuous with the ER lumen (4). The INM can directly interact with proteins within the nucleoplasm, with invaginations of INM reaching deep within the nucleoplasm, forming a nucleoplasmic reticulum (10, 11). An important interaction of the INM proteins is with type V intermediate filament proteins (nuclear lamins) to form the nuclear lamina at the nucleoplasmic surface of the INM (12). Divided into A/C-type and B-type based on their gene sequence, nuclear lamins are important for a number of nuclear functions (6, 13), including nuclear mechanical support and positioning (14, 15), chromatin organisation (16, 17), gene expression (18, 19), nuclear pore complex organisation (20, 21), NE breakdown/repair (22, 23), DNA synthesis (24), intracellular signalling (25–27), cell differentiation, polarisation and programmed cell death (28–30).
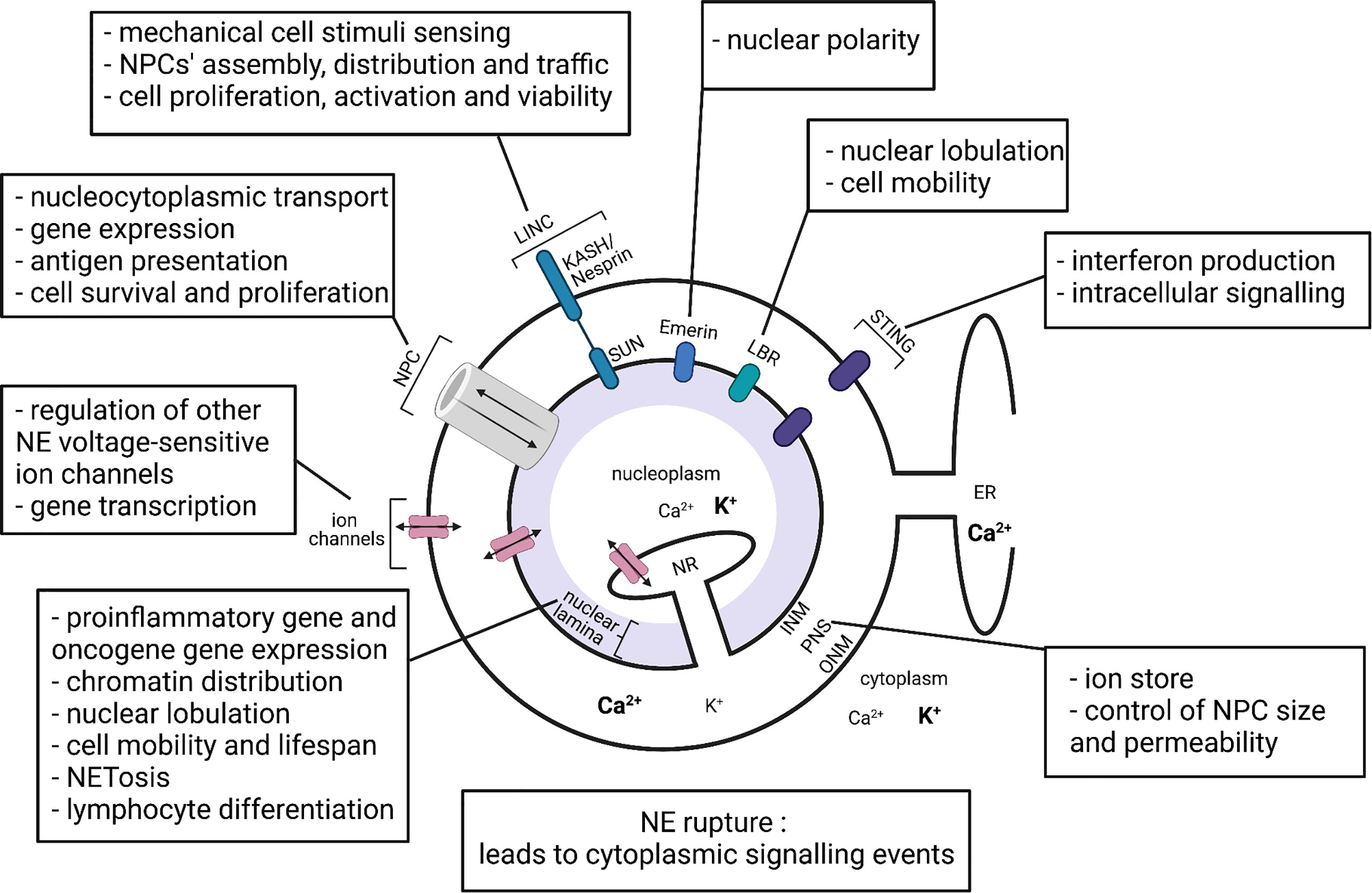
Figure 1 Representation of the nuclear envelope components and their proposed functions in immune cells. INM, inner nuclear membrane; ONM, outer nuclear membrane; PNS, perinuclear space; NR, nucleoplasmic reticulum; ER, endoplasmic reticulum; NPC, nuclear pore complex; LINC, linker of nucleoskeleton and cytoskeleton complex; LBR, lamin B receptor; STING, stimulator of interferon genes. Bold text represents higher ion concentration. Created with BioRender.com.
The INM and the ONM are only permeable to small non-polar molecules and sustain nucleoplasm membrane potential approximately -15mV with respect to the cytoplasm (31, 32). Polar molecules, ions and macromolecules are able to pass between nucleoplasm and cytoplasm through the nuclear pore complexes (NPCs), which are formed upon INM and ONM fusion (33).
Approximately 1% of human proteins have been experimentally demonstrated to be in the NE to date; and it is known that these proteins, and the complexes they form, add to the complexity of the NE structure and the signalling possibilities (34). The NE-associated proteins that have been the most studied in immune cells are the lamins.
Lamin Proteins
Lamins A and C, which are created via alternative splicing of the LMNA gene, are the major components of the nuclear lamina (Figure 1). Cellular functions of lamin A/C, include regulation of gene transcription, NPC formation, nuclear positioning and stability; and have been studied in a number of cell types (35). Several studies have demonstrated the important roles of NE lamins in various immune cell functions.
Neutrophils are usually the first cells of the innate immune system that migrate to sites of inflammation; and while they have potent anti-microbial actions, inappropriate accumulation of these active cells can contribute to immune paralysis and organ failure in severe inflammation (36). Low lamin A/C expression in neutrophils appears to contribute to a characteristic nuclear lobulation, flexibility and the ability of these cells to migrate through narrow spaces towards a site of infection (37–39). Interestingly, neutrophil nuclear morphology was reported to be of less significance than NE lamin content for neutrophil mobility (40). Mechanistically, a low lamin A/C NE is thought to reduce the prevalence of lamin-bound chromatin, hence altering chromatin distribution and increasing its mobility within the nucleus, facilitating chromatin flow in the direction of nuclear movement (16, 41, 42). Reduced lamin A/C expression was also suggested to facilitate NE rupture and nuclear protein transport during the formation of neutrophil extracellular traps (NETosis). There are also reports that low lamin A/C levels account for the short lifespan of neutrophils (43). How neutrophils control NE ruptures to facilitate NETosis requires further study.
With regards to the other major cell type of the innate immune system, monocytes/macrophages, it was reported that high levels of lamin A/C expression induced NF-κB nuclear translocation via the IKKβ/IκBα/NF-κB pathway and increased the expression of proinflammatory genes, such as Il6, Tnf, Ccl2, and Nos2, in adipose tissue macrophages (44). This is in line with the observation that lamin A/C deletion reduced the lipopolysaccharide-induced expression of these genes in macrophages. Importantly, the study established that macrophage lamin A/C levels are linked to adipose tissue inflammation and obesity-induced insulin resistance seen in type two diabetes (44). Lamins A/C levels were also suggested to affect tumour-associated macrophage activity. The c-Fos protein, which influences macrophage responses in cancer (45–47), was shown to interact with and be regulated by lamin A/C (25). Another study demonstrates that lamin A/C gene silencing reduces the number of Leishmania parasites per sampled macrophage and reduces the percentage of infected macrophages in Leishmaniasis (48). Generally, macrophages express higher levels of lamin A/C compared to some other immune cells, which may contribute to macrophages’ proinflammatory actions and comparably longer life span than other innate immune cell types (49). The level of lamin A/C expression in dendritic cells was reported to fall between that of macrophages and neutrophils (6, 50). The presence of functional lamin A/C in mature dendritic cells was demonstrated to limit the nuclear egress of viral capsids and the spread of infection in the case of herpes simplex virus type 1 (51). The role of lamins in dendritic cells, however, needs further investigation.
In the case of cells of the adaptive immune system, T lymphocytes transiently increase their lamin A/C expression upon antigen presentation, with none or weak lamin A/C expression being detected in resting human or mouse T and B lymphocytes (52–54). This demonstrates that NE protein composition can change depending on the cellular activation state. The T lymphocytes’ transient increase in lamin A/C expression was suggested to accelerate the immunological synapse formation between T lymphocytes and antigen presenting cells. Another possible explanation suggested was that transient increase in lamin A/C is related to the lymphocytes’ need to migrate out of lymph nodes and into target tissues (55). Interestingly, the role of lamin A/C may be more important in lymphocyte differentiation during infectious and autoimmune responses (56, 57). Lamin A/C absence reduces T helper 1 lymphocyte differentiation, but does not affect T helper 2 differentiation; this is consistent with significantly weakened lymphocyte responses to both viral and intracellular parasite infections seen in lamin A/C deficiency (58). In comparison, lamin A/C absence increased regulatory T lymphocyte differentiation, reduced T helper 1 differentiation and was protective in a model of inflammatory bowel disease (59, 60). However, exactly how T- lymphocytes regulate their lamin A/C expression is not clear with possible mechanisms including the AKT/protein kinase B signalling pathway (61, 62), microRNA (63) and/or retinoic acid (39, 64) being proposed.
In addition to lamin A/C, lamin B1 has been demonstrated to regulate lymphocyte somatic hypermutations and lymphoid malignancy progression (65). The loss of lamin B1 has also been suggested to contribute to increased inflammation seen with aging (66, 67), although the relative contribution of the innate immune cells in this observation requires further investigation. The Lamin B receptor (LBR) (Figure 1) appears critical for nuclear lobulation, rapid migration and normal function of neutrophils (68). Emerin, an INM protein (Figure 1), regulates nuclear polarity, a crucial feature of migratory cells (69). Finally, a pool of stimulator of interferon genes (STING), a DNA sensor and adaptor protein, was recently confirmed in the NE (70), which will be discussed later.
NPC and LINC Complexes
After the lamins the most well studied protein assemblies in the NE are the nuclear pore complexes (NPCs). The NPCs are multi-protein complexes, each comprising around 500 to 1000 individual protein molecules, of which the main structural proteins are termed nucleoporins (71, 72). There are around 40 different nucleoporins with their nomenclature being mostly based on their mass (73). The central channel of the NPC is located at sites of fused INM and ONM and is lined by nucleoporins. One of the most well-researched functions of NPCs is to control the movement of polar molecules and macromolecules across the NE. NPCs are also involved in chromatin organization, regulation of gene expression and DNA repair (33, 74).
Several studies have addressed the role of individual nucleoporins, and therefore by implication NPCs, in immune cells. Faria et al. (75) demonstrated that lower than normal levels of nucleoporin 96, a core element which is essential for the NPC assembly (71), resulted in altered immune responses, decreased interferon-mediated expression of major histocompatibility complexes, impaired antigen presentation by antigen presenting cells, which consequently reduced T lymphocyte proliferation and increased susceptibility to infection (75). Two other nucleoporins, nucleoporin 88 and nucleoporin 214, were shown to contribute to appropriate nuclear accumulation of some key immune transcription factors including NF-κB, thus regulating the relative strength and duration of immune responses at the level of the NPC in Drosophila and mammalian cells (76–78). Finally, nucleoporin 210 is critical for the survival of circulating T lymphocytes, regulates T cell receptor signalling and hence, adaptive immunity (79).
One important interaction of NPCs is with linker of nucleoskeleton and cytoskeleton (LINC) complexes. Evidence suggests (80–85) that this interaction couples the cytoskeleton and the nucleoskeleton, allowing cytoskeleton rearrangements activated by mechanical stimuli to regulate nucleocytoplasmic transport. LINC complexes comprise SUN proteins on the INM and KASH/Nesprin proteins on the ONM, which interact with each other across the perinuclear space (Figure 1). Mechanical stimuli have been demonstrated to propagate via a direct pathway from cell membrane integrins to SUN proteins on the NE, where SUN proteins transmit the signals via mechanical connections to NPCs, lamins and chromatin (80–83). The transmission of mechanical stimuli significantly affects the NPC conformation via the Nup153-SUN1 connection, the nucleocytoplasmic transport through NPCs and the state of chromatin packing (84). Importantly, different studies show that NPC and LINC complexes coexist on the NE and have a number of interdependent cellular functions. Firstly, LINC complexes regulate NPC assembly and distribution in the NE. SUN1 was shown to colocalize with NPCs and was suggested to reduce the spacing between the INM and the ONM, thus promoting membrane fusion at sites of NPC assembly (86–88). NPCs control the trafficking of SUN proteins across the NE for INM localization. For example, human SUN2 possesses a classical nuclear localization signal, that binds to the importin-alpha-importin-beta heterodimer and mediates transport through the NPC (89). There are also reports that LINC complexes can affect nucleocytoplasmic transport through NPCs, and a study by Li and Noegel (90) reported that SUN1, via interaction with nucleoporin 153, regulates efficient nuclear export of mRNA in mammalian cells (90).
While the significance of LINC complexes in immune cells is yet to be fully investigated, SUN2 regulates T lymphocyte proliferation, function and viability (91). In neutrophils, LINC expression along with lamin and LBR relative expression influence cell migration, which is critical for neutrophil function (92). With increasing recognition that mechanical stimuli can impact on immune cell function, see the following sections, the relationship between NPC-LINC and immune cell activation profiles merits further investigation.
NE Ion Channels
The vast majority of studies investigating ion channels have focused on ion channels in plasma membranes and intracellular double membrane systems of mitochondria and ER. In comparison, the double membranes of the NE have received relatively little attention. Nevertheless, the INM and ONM do contain a range of ion channels (Figure 1) (34, 93–95).
The lack of permeability of NE membranes to ions together with the presence of ion channels and transporters in the NE allow ionic gradients across INMs and ONMs to be generated and maintained irrespective of the NPCs’ activity. Various genetic, pharmacological and electrophysiological approaches have identified a number of ion channels present on INMs and ONMs of different cell types, from liver to immune cells. These include calcium ion channels which could contribute to the regulation of gene transcription, cell cycle control and other nuclear processes (34, 96), potassium ion channels which possibly regulate other NE voltage-sensitive channels by setting the membrane potential (32, 94, 97, 98) and chloride ion channels, potentially participating in osmotic volume regulation of the nucleus (32, 34, 98).
Considering the NE of innate immune system cells, we have recently reported the expression of large conductance (~100-300pS) voltage and calcium activated potassium ion channels (BK channels; KCa1.1) (94). Importantly this study showed that nuclear BK channels regulate cAMP response element binding protein (CREB) phosphorylation in RAW264.7 macrophages. CREB is an important transcription factor with major roles in immune cells (99–101). Moreover, the study reported that nuclear calcium and calmodulin dependent kinases II and IV were involved in regulation of CREB by nuclear BK channels in macrophages (94). Similar findings were previously reported in hippocampal neurones (102). Based on knowledge from other cell types, one can hypothesise that blockade of nuclear BK channels in macrophages influences the activity of other nuclear ion channels or the Ca2+ filling state of the perinuclear space, possibly changing the NE membrane potential. This eventually could lead to increased nuclear calcium, activation of calcium-dependent kinases and, finally, activation of various transcription factors, including CREB. BK channels have also been identified on the NE of microglia, which are the resident macrophage-like cells of the central nervous system (103). In this study it was hypothesised that NE BK channels are involved in microglia response to stimuli, facilitating nitric oxide and cytokine production, potentially by regulating calcium and potassium fluxes in the nucleus (103).
To date only a few papers have addressed the role of NE ion channels in lymphocytes. Franco-Obregon et al. (95) reported that chloride channels (105pS) predominated on the ONM of T lymphocytes, while channels on the ONM of B lymphocytes were primarily cation selective (52pS) (95). Although only anion channels were detected in T lymphocyte ONM, both anion and cation channels were observed in B lymphocyte ONM (95). Generally, INMs are considered to have a different ion channel composition compared to ONMs (95, 104). On the INM of T-lymphocytes the presence of both large conductance anion selective channels (370pS) and cation selective (152pS) channels was reported (93). However further studies are required to fully characterise the ion channels present in the NE of lymphocytes. It will be interesting to see if NE ion channels in lymphocytes regulate aspects of T-lymphocyte physiology such as gene transcription, proliferation, differentiation and apoptosis.
Evidence is now beginning to accumulate that suggests NE ion channels are important in immune cell functions and activation. However, this is a complex system with ion channel type, location (INM or OMN), and subunit composition all impacting the functional significance of the NE ion channels.
Perinuclear Ion Store
As already mentioned, the two membranes of the NE form a lumen, referred to as the perinuclear space (Figure 1). The selective permeability of INM and ONM to ions contributes to ion concentration gradients across the two membranes and allows the perinuclear space to act as a cellular ion store. It is not unreasonable to assume that changes in ion concentration within the perinuclear space might have significant effects on nuclear/cellular functions. The idea that the nucleus possesses an autonomous calcium store and signalling system, which generates its own calcium transients independent of cytoplasmic calcium changes, has been debated for years. Supporting this idea several studies suggest that nuclear calcium is independent of cytoplasmic calcium and NPC-mediated calcium transport (102, 105–109). In liver cells, Leite et al. (105) showed that upon ATP stimulation nucleoplasmic calcium signalling preceded the cytoplasmic calcium signal, thus leading to the suggestion of an autonomous nuclear calcium signalling system (105). Similarly, in hippocampal neurones, Li et al. (102) reported that nuclear BK channel block induced nucleoplasmic calcium elevation without changing cytoplasmic calcium concentration (102). Other studies identified the nucleoplasmic reticulum, formed from INM invaginations, as a nuclear calcium store and regulator. The presence of calcium storage proteins, calreticulins (110), calcium releasing inositol-1,4,5-trisphosphate receptors (105) and calcium pumps (111, 112) in the NE and its invaginations further supports the idea of a fairly autonomous NE calcium store (107, 108). There are also calcium-mobilizing ryanodine receptors in lymphocyte nuclei, while in monocytes and neutrophils their localization is reported to be extranuclear (113). Perinuclear calcium, when released into the nucleoplasm, is thought to regulate nuclear calcium-sensitive gene expression through regulation of calcium-dependent enzymes and transcription factors (94, 102). In relation to cells of the immune system, nuclear calcium was found to control T lymphocyte fate decision between active proliferation and a hyporesponsive state (114). Moreover, it has been debated if the Ca2+ filling state of the perinuclear space influences NPC size and permeability. Several contradictory reports exist. Some show that NE calcium depletion significantly increases the proportion of NPCs blocked by a central plug and attenuates the nuclear influx of proteins, while others argue that depletion of calcium from the NE lumen does not significantly alter the movement of the green fluorescent protein across the NE (115–122).
Is the NE an Integrator of Mechanical and Chemical Signalling Pathways in Inflammation?
It is now apparent that along with the diverse array of extracellular chemical signals that immune cells respond to, such as pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), these cells can also respond to their physical environment. This mechanosensing may be of particular relevance during an inflammatory response where the leukocytes of the innate immune system need to essentially “travel and deform” to perform their function. During inflammation leukocytes can be subjected to changes in mechanical and compressive forces, such as those experienced during cell migration and cell extravasation; osmotic forces, which can occur during cell swelling and tissue damage; and forces associated with phagocytosis such as deformation of the leukocytes and engagement with targets of different shape and orientation. Tissue composition such as extracellular matrix stiffness, hydrostatic pressure and cell crowding can also provide mechanical signals to leukocytes. A complete map of the cellular pathways activated by mechanosensing in immune cells is yet to be fully elucidated but appears to involve plasma membrane ion channels such as PIEZO1 (123) and TRPV4 (124), adhesion molecules and cytoskeleton rearrangement, and the reader is pointed towards several recent reviews on the subject (125–127). The NE and its associated proteins are also involved in the signal transduction mechanisms activated by changes in mechanical forces through interactions between the cytoskeleton, LINC protein complexes and the lamin network which attaches the INM to lamin-associated-domains (LADs) within chromatin and thus influences chromatin dynamics (128, 129). Along with evidence that the NE represents a core component of mechanical signal transduction, a number of seminal studies have now also demonstrated that the NE itself is a sensor of mechanical forces (130–132), a function it may be well suited for (133). However, to date only a few reports have investigated the role of NE components in immune cells in response to mechanical forces (134).
As discussed above, the NE-associated proteins have effects on the cellular functions of immune cells and the NE is involved in the transduction pathways of many mechanical stimuli encountered by immune cells during an inflammatory response. In addition, the NE contains and interacts with a variety of structural proteins, ion channels, pore proteins and transporters, which suggests the NE may be important in cell signalling cascades which regulate nuclear functions such as heterochromatin formation, gene expression, nuclear morphology and nuclear repair. For example, ion channels within the NE could regulate Ca2+ levels in the perinuclear space which can regulate gene transcription (135), see the above section. But is there evidence that mechanosensing by NE can regulate cell signalling cascades and, therefore, immune cell function during an inflammatory event?
Yes-associated protein (YAP) is a transcription cofactor that regulates macrophage innate inflammatory pathways (136). Importantly, YAP activation and translocation into nucleus has been demonstrated to be regulated by NE flattening and nuclear pore opening caused by mechanical forces and extracellular matrix (ECM) stiffness (137, 138). The NE protein Emerin, which appears to be an important component in mechanotransduction pathways in the NE (139), binds and activates histone deacetylase 3 (HDAC3) (140), an important regulator of autoimmunity and inflammatory gene expression (141, 142). Finally, osmotic cell swelling, which causes increased NE tension, results in cPLA2 localisation to the INM of zebrafish leukocytes. This mechanism in the dying cells appeared to facilitate the mobilization of other leukocytes to clear necrotic debris, presumably by the release of eicosanoids (134). It will be interesting if these final observations can be extended to mammalian leukocytes and lymphocytes.
An important emerging principal is that the magnitude of NE perturbations will determine the cellular pathways activated (143), with the most severe mechanical stressors, or possibly a milder mechanical stressor but in a permissive cellular environment, leading to NE rupture. This does not necessarily lead to cell death as NE ruptures can be repaired. However, NE ruptures can cause the mixing of material from the nucleoplasm with the cytoplasm and this may be an important mechanism of leukocyte activation. The DNA sensing cGAS-STING and AIM-2 pathways are part of the innate immune response to viruses, whereby cells are activated by inappropriate cellular localisation of viral DNA which results in the production of type I interferons (144). However, these pathways can be activated by endogenous DNA spilling from the nucleus, and this may contribute to inflammatory diseases including Aicardi-Goutières Syndrome (145–148). NE rupture may also contribute to the low-grade chronic inflammation associated with old age, referred to as inflammaging. NE instability increases in senescent cells and is believed to contribute to the establishment of the senescence-associated secretory phenotype (SASP) (149, 150). SASP, which is also associated with ER stress, is proposed to be one of the main contributors to inflammaging (151). It will be important to investigate the effects of NE perturbations caused by mechanical stimuli in inflammatory environments. Taken together, it would appear that the NE is important in multiple immune functions of leukocytes.
Future Prospective
When studying the role of the NE in immune cells a number of considerations need to be taken into account. As well as studying the NE in the setting of the whole cell it is also possible to carry out studies on isolated nuclei, NE preparations and reconstituted systems involving nuclei being placed is cytosolic preparations (94, 152, 153). The choice of experimental system employed will depend on the cellular process to be investigated and when using isolated nuclei or NE it is important to check for sample contamination with membrane material from other organelles.
Most modern cellular and biochemical techniques, for example immunofluorescence, Western blot analysis and electrophysiology, can be applied directly to isolated nuclei and NE, although determining if a protein is located within the inner or outer nuclear membrane still requires careful consideration. While nuclei isolation gives a direct access to the NE, it limits our understanding of how nuclei and the NE integrate within a functioning cell. Therefore, there is a need to develop methodologies which specifically target proteins in the nuclei and NE when investigating in whole cells. The reason for this is that many proteins in the nuclei and NE are not exclusive to these sites and can be found in other organelles and membranes within the cell. Most current pharmacological and genetic tools used to investigate protein function will not discriminate between the same protein found at different cellular locations, thus attributing specific cellular functions to NE located proteins is problematic. It will be important to develop pharmacological and genetic tools which specifically target the constituents of the nuclear membrane, both to facilitate basic research and hopefully future therapeutic development.
Considering the role of the NE in immune cells outstanding questions to be addressed are:
What are the differences and changes in NE composition between immune cell types and their various activation states?
Can selective targeting of NE components be achieved in a functioning immune cell and therefore help elucidate the role of these components in immune cells?
Can NE proteins influence gene transcription in immune cells, either by regulating transcription factor activation or by epigenetic mechanisms? And is this specific to certain genes?
Do NE components represent possible therapeutic targets for the treatment of autoimmune and inflammatory diseases?
Conclusion
The nucleus is sometimes referred to as “a cell within a cell”, and one would expect such an emotive phrase to be the focus of much research. It is surprising that the membrane which delineates this structure has received relatively little research attention. This may stem from the traditional view that the NE plays only a minor role in cellular physiology. One aim of this review is to increase the awareness of the roles that the NE may play in the cell and in particular in immune cell function.
The NE, through its various components such as lamins, NPC and LINC complexes, cation and anion channels has been shown to act as a regulator of immune cell function summarised in Figure 1. In the immune system the NE’s influences range from macrophage polarization and lymphocyte differentiation to cell migration and lifespan (Figure 1). In addition, the NE can also act as a sensor of intracellular and extracellular environments and physical stresses on the cell. However, the investigation of the NE in immune cells is still in its infancy. Today mitochondria and the ER are considered crucially important organelles in shaping the actions of cells of the immune system. Time will tell if the NE and its associated structures become the 3rd member of this trinity.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Callan HG, Tomlin SG. Experimental Studies on Amphibian Oocyte Nuclei. I. Investigation of the Structure of the Nuclear Membrane by Means of the Electron Microscope. R Soc (Great Britain) (1950) 137(888):367–78. doi: 10.1098/rspb.1950.0047
2. Heogeboom GH, Schneider WC. On the Nuclear Envelope. Science (1953) 118(3067):419. doi: 10.1126/science.118.3067.419
3. Watson ML. The Nuclear Envelope; its Structure and Relation to Cytoplasmic Membranes. J Biophys Biochem Cytol (1955) 1(3):257–70. doi: 10.1083/jcb.1.3.257
4. Hetzer MW, Walther TC, Mattaj IW. Pushing the Envelope: Structure, Function, and Dynamics of the Nuclear Periphery. Annu Rev (2005) 21(1):347–80. doi: 10.1146/annurev.cellbio.21.090704.151152
5. Hetzer MW, Wente SR. Border Control at the Nucleus: Biogenesis and Organization of the Nuclear Membrane and Pore Complexes. Dev Cell (2009) 17(5):606–16. doi: 10.1016/j.devcel.2009.10.007
6. Saez A, Herrero-Fernandez B, Gomez-Bris R, Somovilla-Crespo B, Rius C, Gonzalez-Granado JM. Lamin A/C and the Immune System: One Intermediate Filament, Many Faces. Int J Mol Sci (2020) 21:1–26. doi: 10.3390/ijms21176109
7. Sun L, Wang X, Saredy J, Yuan Z, Yang X, Wang H. Innate-Adaptive Immunity Interplay and Redox Regulation in Immune Response. Redox Biol (2020) 37:101759. doi: 10.1016/J.REDOX.2020.101759
8. Rawlings DJ, Metzler G, Wray-Dutra M, Jackson SW. Altered B Cell Signalling in Autoimmunity, Nature Reviews. Immunology (2017) 17(7):421. doi: 10.1038/NRI.2017.24
9. Harapas CR, Idiiatullina E, Al-Azab M, Hrovat-Schaale K, Reygaerts T, Steiner A, et al. Organellar Homeostasis and Innate Immune Sensing. Nat Rev Immunol (2022) 1–15. doi: 10.1038/s41577-022-00682-8
10. Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, et al. Subdiffraction Multicolor Imaging of the Nuclear Periphery With 3D Structured Illumination Microscopy. Science (2008) 320(5881):1332–6. doi: 10.1126/science.1156947
11. Malhas A, Goulbourne C, Vaux DJ. The Nucleoplasmic Reticulum: Form and Function. Trends Cell Biol (2011) 21(6):362–73. doi: 10.1016/j.tcb.2011.03.008
12. Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barkan R, et al. Nuclear Lamins: Key Regulators of Nuclear Structure and Activities. J Cell Mol Med (2009) 13(6):1059–85. doi: 10.1111/j.1582-4934.2008.00676.x
13. Patil S, Sengupta K. Role of A- and B-Type Lamins in Nuclear Structure–Function Relationships. Biol Cell (2021) 113(7):295–310. doi: 10.1111/boc.202000160
14. Nmezi B, Xu J, Fu R, Armiger TJ, Rodriguez-Bey G, Powell JS, et al. Concentric Organization of A- and B-Type Lamins Predicts Their Distinct Roles in the Spatial Organization and Stability of the Nuclear Lamina. Proc Natl Acad Sci USA (2019) 116(10):4307–15. doi: 10.1073/pnas.1810070116
15. Karoutas A, Szymanski W, Rausch T, Guhathakurta S, Rog-Zielinska EA, Peyronnet R, et al. The NSL Complex Maintains Nuclear Architecture Stability via Lamin A/C Acetylation. Nat Cell Biol (2019) 21(10):1248–60. doi: 10.1038/s41556-019-0397-z
16. Gesson K, Rescheneder P, Skoruppa MP, Von Haeseler A, Dechat T, Foisner R. A-Type Lamins Bind Both Hetero- and Euchromatin, the Latter Being Regulated by Lamina-Associated Polypeptide 2 Alpha. Genome Res (2016) 26(4):462–73. doi: 10.1101/gr.196220.115
17. Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear Lamins: Major Factors in the Structural Organization and Function of the Nucleus and Chromatin. Genes Dev (2008) 22(7)832–53. doi: 10.1101/gad.1652708
18. Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, et al. Altered Chromosomal Positioning, Compaction, and Gene Expression With a Lamin AC Gene Mutation. PLoS One (2010) 5(12):e14342. doi: 10.1371/journal.pone.0014342
19. Pradhan R, Nallappa MJ, Sengupta K. Lamin a/C Modulates Spatial Organization and Function of the Hsp70 Gene Locus via Nuclear Myosin I. J Cell Sci (2020) 133(4):jcs236265. doi: 10.1242/JCS.236265
20. Guo Y, Yixian Z. Lamins Position the Nuclear Pores and Centrosomes by Modulating Dynein. Mol Biol Cell (2015) 26(19):3379–89. doi: 10.1091/mbc.E15-07-0482
21. Kittisopikul M, Shimi T, Tatli M, Tran JR, Zheng Y, Medalia O, et al. Computational Analysis of Lamin Isoform Interactions With Nuclear Pore Complexes. bioRxiv (2020). doi: 10.1101/2020.04.03.022798
22. Ikegami K, Secchia S, Lieb J, Moskowitz I. Depolymerized Lamins Link Nuclear Envelope Breakdown to Mitotic Transcriptional Quiescence. bioRxiv (2018) 334110. doi: 10.1101/334110
23. Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear Envelope Breakdown Proceeds by Microtubule-Induced Tearing of the Lamina. Cell (2002) 108(1):83–96. doi: 10.1016/S0092-8674(01)00627-4
24. Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of Nuclear Lamin Organization Alters the Distribution of Replication Factors and Inhibits DNA Synthesis. J Cell Biol (1997) 136(6):1201–12. doi: 10.1083/jcb.136.6.1201
25. Ivorra C, Kubicek M, González JM, Sanz-González SM, Álvarez-Barrientos A, OConnor JE, et al. A Mechanism of AP-1 Suppression Through Interaction of C-Fos With Lamin A/C. Genes Dev (2006) 20(3):307–20. doi: 10.1101/gad.349506
26. Han X, Feng X, Rattner JB, Smith H, Bose P, Suzuki K, et al. Tethering by Lamin A Stabilizes and Targets the ING1 Tumour Suppressor. Nat Cell Biol (2008) 10(11):1333–40. doi: 10.1038/ncb1792
27. Wilson KL, Foisner R. Lamin-Binding Proteins. Cold Spring Harb Perspect Biol (2010) 2(4):a000554. doi: 10.1101/cshperspect.a000554
28. Maresca G, Natoli M, Nardella M, Arisi I, Trisciuoglio D, Desideri M, et al. LMNA Knock-Down Affects Differentiation and Progression of Human Neuroblastoma Cells. PLoS One (2012) 7(9):e45513. doi: 10.1371/journal.pone.0045513
29. Rao L, Perez D, White E. Lamin Proteolysis Facilitates Nuclear Events During Apoptosis. J Cell Biol (1996) 135(6):1441–55. doi: 10.1083/jcb.135.6.1441
30. Burke B. Lamins and Apoptosis: A Two-Way Street? J Cell Biol (2001) 153(3):F5–7. doi: 10.1083/jcb.153.3.f5
31. Loewenstein WR, Kanno Y. Some Electrical Properties of a Nuclear Membrane Examined With a Microelectrode. J Gen Physiol (1963) 46(6):1123–40. doi: 10.1085/jgp.46.6.1123
32. Mazzanti M, Bustamante JO, Oberleithner H. Electrical Dimension of the Nuclear Envelope, Physiological Reviews. Am Physiol Soc (2001) 81(1):1–19. doi: 10.1152/physrev.2001.81.1.1
33. Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Structure and Assembly of the Nuclear Pore Complex. Annu Rev Biophys (2019) 48(1):515–36. doi: 10.1146/annurev-biophys-052118-115308
34. Matzke AJM, Weiger TM, Matzke M. Ion Channels at the Nucleus: Electrophysiology Meets the Genome. Mol Plant (2010) 3(4):642–52. doi: 10.1093/mp/ssq013
35. Dubik N, Mai S. Lamin A/C: Function in Normal and Tumor Cells. Cancers (2020) 12(12):1–21. doi: 10.3390/CANCERS12123688
36. Leliefeld PHC, Wessels CM, Leenen LPH, Koenderman L, Pillay J. The Role of Neutrophils in Immune Dysfunction During Severe Inflammation. Crit Care (2016) 20:73. doi: 10.1186/s13054-016-1250-4
37. Olins AL, Zwerger M, Herrmann H, Zentgraf H, Simon AJ, Monestier M, et al. The Human Granulocyte Nucleus: Unusual Nuclear Envelope and Heterochromatin Composition. Eur J Cell Biol (2008) 87(5):279–90. doi: 10.1016/j.ejcb.2008.02.007
38. Olins AL, Ernst A, Zwerger M, Herrmann H, Olins DE. An In Vitro Model for Pelger-Huët Anomaly; Stable Knockdown of Lamin B Receptor in Hl-60 Cells. Nucleus (2010) 1(6):1–7. doi: 10.4161/nucl.1.6.13271
39. Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear Envelope and Chromatin Compositional Differences Comparing Undifferentiated and Retinoic Acid- and Phorbol Ester-Treated HL-60 Cells. Exp Cell Res (2001) 268(2):115–27. doi: 10.1006/excr.2001.5269
40. Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, et al. Nuclear Envelope Composition Determines the Ability of Neutrophil-Type Cells to Passage Through Micron-Scale Constrictions. J Biol Chem (2013) 288(12):8610–8. doi: 10.1074/jbc.M112.441535
41. Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ, King MC. The Tethering of Chromatin to the Nuclear Envelope Supports Nuclear Mechanics. Nat Commun (2015) 6(1):1–13. doi: 10.1038/ncomms8159
42. Manley HR, Keightley MC, Lieschke GJ. The Neutrophil Nucleus: An Important Influence on Neutrophil Migration and Function. Front Immunol (2018) 9:2867. doi: 10.3389/fimmu.2018.02867
43. Lahoz-Beneytez J, Elemans M, Zhang Y, Ahmed R, Salam A, Block M, et al. Human Neutrophil Kinetics: Modeling of Stable Isotope Labeling Data Supports Short Blood Neutrophil Half-Lives. Blood (2016) 127(26):3431–8. doi: 10.1182/blood-2016-03-700336
44. Kim Y, Bayona PW, Kim M, Chang J, Hong S, Park Y, et al. Macrophage Lamin A/C Regulates Inflammation and the Development of Obesity-Induced Insulin Resistance. Front Immunol (2018) 9:696. doi: 10.3389/fimmu.2018.00696
45. Hop HT, Arayan LT, Huy TXN, Reyes AWB, Vu SH, Min WG, et al. The Key Role of C-Fos for Immune Regulation and Bacterial Dissemination in Brucella Infected Macrophage. Front Cell Infect Microbiol (2018) 8:287. doi: 10.3389/fcimb.2018.00287
46. Ding Y, Hao K, Li Z, Ma R, Zhou Y, Zhou Z, et al. C-Fos Separation From Lamin A/C by GDF15 Promotes Colon Cancer Invasion and Metastasis in Inflammatory Microenvironment. J Cell Physiol (2020) 235(5):4407–21. doi: 10.1002/jcp.29317
47. Bottazzi B, Nobili N, Mantovani A. Expression of C-Fos Proto-Oncogene in Tumor-Associated Macrophages. J Immunol (1990) 144(12):4878–82.
48. Ovalle-Bracho C, Londonõ-Barbosa DA, Franco-Munõz C, Clavijo-Ramírez C. Functional Evaluation of Gene Silencing on Macrophages Derived From U937 Cells Using Interference RNA (shRNA) in a Model of Macrophages Infected With Leishmania (Viannia) Braziliensis. Parasitology (2015) 142(14):1682–92. doi: 10.1017/S0031182015001304
49. Röber RA, Gieseler RKH, Peters JH, Weber K, Osborn M. Induction of Nuclear Lamins A/C in Macrophages in In Vitro Cultures of Rat Bone Marrow Precursor Cells and Human Blood Monocytes, and in Macrophages Elicited In Vivo by Thioglycollate Stimulation. Exp Cell Res (1990) 190(2):185–94. doi: 10.1016/0014-4827(90)90184-C
50. Gieseler RKH, Xu H, Schlemminger R, Peters JH. Serum-Free Differentiation of Rat and Human Dendritic Cells, Accompanied by Acquisition of the Nuclear Lamins A/C as Differentiation Markers. Adv Exp Med Biol (1993) 329:287–91. doi: 10.1007/978-1-4615-2930-9_48
51. Turan A, Grosche L, Krawczyk A, Mühl-Zürbes P, Drassner C, Düthorn A., et al. Autophagic Degradation of Lamins Facilitates the Nuclear Egress of Herpes Simplex Virus Type 1. J Cell Biol (2019) 218:508–23. doi: 10.1083/jcb.201801151.
52. Contu F, Rangel-Pozzo A, Trokajlo P, Wark L, Klewes L, Johnson NA, et al. Distinct 3d Structural Patterns of Lamin a/C Expression in Hodgkin and Reed-Sternberg Cells. Cancers (2018) 10(9):E286. doi: 10.3390/cancers10090286
53. Guilly MN, Bensussan A, Bourge JF, Bornens M, Courvalin JC. A Human T Lymphoblastic Cell Line Lacks Lamins A and C. EMBO J (1987) 6(12):3795–9. doi: 10.1002/j.1460-2075.1987.tb02715.x
54. Rober RA, Sauter H, Weber K, Osborn M. Cells of the Cellular Immune and Hemopoietic System of the Mouse Lack Lamins A/C: Distinction Versus Other Somatic Cells. J Cell Sci (1990) 95(4): 587–98. doi: 10.1242/jcs.95.4.587
55. González-Granado JM, Silvestre-Roig C, Rocha-Perugini V, Trigueros-Motos L, Cibrián D, Morlino G, et al. Nuclear Envelope Lamin-A Couples Actin Dynamics With Immunological Synapse Architecture and T Cell Activation. Sci Signal (2014) 7(322):ra37. doi: 10.1126/scisignal.2004872
56. Rønningen T, Shah A, Oldenburg AR, Vekterud K, Delbarre E, Moskaug JO, et al. Prepatterning of Differentiation-Driven Nuclear Lamin a/C-Associated Chromatin Domains by GlcNAcylated Histone H2B. Genome Res (2015) 25(12):1825–35. doi: 10.1101/gr.193748.115
57. Lund E, Collas P. Nuclear Lamins: Making Contacts With Promoters. Nucleus (United States) (2013) 4(6):424–30. doi: 10.4161/nucl.26865
58. Toribio-Fernández R, Zorita V, Rocha-Perugini V, Iborra S, Martínez Del Hoyo G, Chevre R, et al. Lamin A/C Augments Th1 Differentiation and Response Against Vaccinia Virus and Leishmania Major Article. Cell Death Dis (2018) 9(1):1–15. doi: 10.1038/s41419-017-0007-6
59. Toribio-Fernández R, Herrero-Fernandez B, Zorita V, López JA, Vázquez J, Criado G, et al. Lamin A/C Deficiency in CD4 + T-Cells Enhances Regulatory T-Cells and Prevents Inflammatory Bowel Disease. J Pathol (2019) 249(4):509–22. doi: 10.1002/path.5332
60. Izcue A, Coombes JL, Powrie F. Regulatory T Cells Suppress Systemic and Mucosal Immune Activation to Control Intestinal Inflammation. Immunol Rev (2006) 212:256–71. doi: 10.1111/j.0105-2896.2006.00423.x
61. Bauer B, Baier G. Protein Kinase C and AKT/protein Kinase B in CD4+ T-Lymphocytes: New Partners in TCR/CD28 Signal Integration. Mol Immunol (2002) 38(15):1087–99. doi: 10.1016/S0161-5890(02)00011-1
62. Bertacchini J, Beretti F, Cenni V, Guida M, Gibellini F, Mediani L, et al. The Protein Kinase Akt/PKB Regulates Both Prelamin A Degradation and Lmna Gene Expression. FASEB J (2013) 27(6):2145–55. doi: 10.1096/fj.12-218214
63. Zuo B, Yang J, Wang F, Wang L, Yin Y, Dan J, et al. Influences of Lamin A Levels on Induction of Pluripotent Stem Cells. Biol Open (2012) 1(11):1118–27. doi: 10.1242/bio.20121586
64. Pino-Lagos K, Benson MJ, Noelle RJ. Retinoic Acid in the Immune System. Ann New York Acad Sci (2008) 1143:170–87. doi: 10.1196/annals.1443.017
65. Klymenko T, Bloehdorn J, Bahlo J, Robrecht S, Akylzhanova G, Cox K, et al. Lamin B1 Regulates Somatic Mutations and Progression of B-Cell Malignancies. Leukemia (2018) 32(2):364–75. doi: 10.1038/leu.2017.255
66. Chen H, Zheng X, Zheng Y. Age-Associated Loss of Lamin-B Leads to Systemic Inflammation and Gut Hyperplasia. Cell Cell (2014) 159(4):829–43. doi: 10.1016/J.CELL.2014.10.028
67. Tran JR, Chen H, Zheng X, Zheng Y. Lamin in Inflammation and Aging. Curr Opin Cell Biol (2016) 40:124–30. doi: 10.1016/J.CEB.2016.03.004
68. Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, et al. Mouse Neutrophils Lacking Lamin B-Receptor Expression Exhibit Aberrant Development and Lack Critical Functional Responses. Exp Hematol (2008) 36(8):965–76. doi: 10.1016/j.exphem.2008.04.006
69. Nastały P, Purushothaman D, Marchesi S, Poli A, Lendenmann T, Kidiyoor GR, et al. Role of the Nuclear Membrane Protein Emerin in Front-Rear Polarity of the Nucleus. Nat Commun (2020) 11(1):1–12. doi: 10.1038/s41467-020-15910-9
70. Dixon CR, Malik P, de las Heras JI, Saiz-Ros N, de Lima Alves F, Tingey M, et al. STING Nuclear Partners Contribute to Innate Immune Signalling Responses, Biorxiv. iScience (2020) 24(9):103055. doi: 10.1101/2020.12.21.423744
71. Krull S, Thyberg J, Björkroth B, Rackwitz H-R, Cordes VC. Nucleoporins as Components of the Nuclear Pore Complex Core Structure and Tpr as the Architectural Element of the Nuclear Basket. Mol Biol Cell (2004) 15(9):4261. doi: 10.1091/MBC.E04-03-0165
72. Sabinina VJ, Hossain MJ, Hériché JK, Hoess P, Nijmeijer B, Mosalaganti S, et al. Three-Dimensional Superresolution Fluorescence Microscopy Maps the Variable Molecular Architecture of the Nuclear Pore Complex. Mol Biol Cell (2021) 32(17):1523–33. doi: 10.1091/MBC.E20-11-0728
73. Lin DH, Hoelz A. The Structure of the Nuclear Pore Complex (An Update). Annu Rev Biochem (2019) 88:725–83. doi: 10.1146/annurev-biochem-062917-011901
74. Beck M, Hurt E. The Nuclear Pore Complex: Understanding its Function Through Structural Insight. Nat Rev Mol Cell Biol (2017) 18(2):73–89. doi: 10.1038/nrm.2016.147
75. Faria AMC, Levay A, Wang Y, Kamphorst AO, Rosa MLP, Nussenzveig DR, et al. The Nucleoporin Nup96 Is Required for Proper Expression of Interferon-Regulated Proteins and Functions. Immun Immun (2006) 24(3):295–304. doi: 10.1016/j.immuni.2006.01.014
76. Xylourgidis N, Roth P, Sabri N, Tsarouhas V, Samakovlis C. The Nucleoporin Nup214 Sequesters CRM1 at the Nuclear Rim and Modulates Nfκb Activation in Drosophila. J Cell Sci (2006) 119(21):4409–19. doi: 10.1242/jcs.03201
77. Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, Samakovlis C. The Drosophila Nucleoporin DNup88 Localizes DNup214 and CRM1 on the Nuclear Envelope and Attenuates NES-Mediated Nuclear Export. J Cell Biol (2003) 163(4):701–6. doi: 10.1083/jcb.200304046
78. Takahashi N, van Kilsdonk JWJ, Ostendorf B, Smeets R, Bruggeman SWM, Alonso A, et al. Tumor Marker Nucleoporin 88 kDa Regulates Nucleocytoplasmic Transport of NF-κb. Biochem Biophys Res Commun (2008) 374(3):424–30. doi: 10.1016/j.bbrc.2008.06.128
79. Borlido J, Sakuma S, Raices M, Carrette F, Tinoco R, Bradley LM, et al. Nuclear Pore Complex-Mediated Modulation of TCR Signaling is Required for Naïve CD4 + T Cell Homeostasis. Nat Immunol (2018) 19(6):594–605. doi: 10.1038/s41590-018-0103-5
80. Jean RP, Gray DS, Spector AA, Chen CS. Characterization of the Nuclear Deformation Caused by Changes in Endothelial Cell Shape. J Biomech Eng (2004) 126(5):552–8. doi: 10.1115/1.1800559
81. Wang N, Tytell JD, Ingber DE. Mechanotransduction at a Distance: Mechanically Coupling the Extracellular Matrix With the Nucleus. Nat Rev Mol Cell Biol (2009) 10(1):75–82. doi: 10.1038/NRM2594
82. Jahed Z, Shams H, Mehrbod M, Mofrad MRK. Mechanotransduction Pathways Linking the Extracellular Matrix to the Nucleus. Int Rev Cell Mol Biol (2014) 310:171–220. doi: 10.1016/B978-0-12-800180-6.00005-0
83. Kirby TJ, Lammerding J. Emerging Views of the Nucleus as a Cellular Mechanosensor. Nat Cell Biol (2018) 20:4. doi: 10.1038/s41556-018-0038-y
84. Donnaloja F, Jacchetti E, Soncini M, Raimondi MT. Mechanosensing at the Nuclear Envelope by Nuclear Pore Complex Stretch Activation and Its Effect in Physiology and Pathology. Front Physiol (2019) 10:896. doi: 10.3389/FPHYS.2019.00896
85. Jahed Z, Soheilypour M, Peyro M, Mofrad MRK. The LINC and NPC Relationship – its Complicated! J Cell Sci (2016) 129(17):3219–29. doi: 10.1242/jcs.184184
86. Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, et al. Functional Association of Sun1 With Nuclear Pore Complexes. J Cell Biol (2007) 178(5):785–98. doi: 10.1083/jcb.200704108
87. Lu W, Gotzmann J, Sironi L, Jaeger V-M, Schneider M, Lüke Y, et al. Sun1 Forms Immobile Macromolecular Assemblies at the Nuclear Envelope. Biochim Biophys Acta (BBA) (2008) 1783(12):2415–26. doi: 10.1016/j.bbamcr.2008.09.001
88. Talamas JA, Hetzer MW. POM121 and Sun1 Play a Role in Early Steps of Interphase NPC Assembly. J Cell Biol (2011) 194(1):27–37. doi: 10.1083/jcb.201012154
89. Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, et al. A Classical NLS and the SUN Domain Contribute to the Targeting of SUN2 to the Inner Nuclear Membrane. EMBO J (2010) 29(14):2262–75. doi: 10.1038/emboj.2010.119
90. Li P, Noegel AA. Inner Nuclear Envelope Protein SUN1 Plays a Prominent Role in Mammalian mRNA Export. Nucleic Acids Res (2015) 43(20):9874–88. doi: 10.1093/nar/gkv1058
91. Donahue DA, Porrot F, Couespel N, Schwartz O. SUN2 Silencing Impairs CD4 T Cell Proliferation and Alters Sensitivity to HIV-1 Infection Independently of Cyclophilin A. J Virol (2017) 91(6):2303–19. doi: 10.1128/jvi.02303-16
92. Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, et al. The LINC-Less Granulocyte Nucleus. Eur J Cell Biol (2009) 88(4):203–14. doi: 10.1016/j.ejcb.2008.10.001
93. Fedorenko OA, Volkova TM, Marchenko SM. New Cation Channel of the T-Lymphocyte Nuclear Membrane. Fiziol Zh (2006) 52(1):17–21.
94. Selezneva A, Yoshida M, Gibb A, Willis D. Nuclear BK Channels Regulate CREB Phosphorylation in RAW264.7 Macrophages. Pharmacol Rep (2021) 73:881–90. doi: 10.1007/s43440-021-00229-z
95. Franco-Obregón A, Wang HW, Clapham DE. Distinct Ion Channel Classes are Expressed on the Outer Nuclear Envelope of T- and B-Lymphocyte Cell Lines. Biophys J (2000) 79(1):202–14. doi: 10.1016/S0006-3495(00)76284-9
96. Bootman MD, Fearnley C, Smyrnias I, MacDonald F, Roderick HL. An Update on Nuclear Calcium Signalling. J Cell Sci J Cell Sci (2009) 122(14):2337–50. doi: 10.1242/jcs.028100
97. Mazzanti M, DeFelice LJ, Cohen J, Malter H. Ion Channels in the Nuclear Envelope. Nature (1990) 343(6260):764–7. doi: 10.1038/343764a0
98. Guihard G, Proteau S, Payet MD, Escande D, Rousseau E. Patch-Clamp Study of Liver Nuclear Ionic Channels Reconstituted Into Giant Proteoliposomes. FEBS Lett (2000) 476(3):234–9. doi: 10.1016/S0014-5793(00)01752-X
99. Park JM, Greten FR, Wong A, Westrick RJ, Arthur JSC, Otsu K, et al. Signaling Pathways and Genes That Inhibit Pathogen-Induced Macrophage Apoptosis - CREB and NF-κb as Key Regulators. Immunity (2005) 23(3):319–29. doi: 10.1016/j.immuni.2005.08.010
100. Roach SK, Lee SB, Schorey JS. Differential Activation of the Transcription Factor Cyclic AMP Response Element Binding Protein (CREB) in Macrophages Following Infection With Pathogenic and Nonpathogenic Mycobacteria and Role for CREB in Tumor Necrosis Factor Alpha Production. Infect Immun (2005) 73(1):514–22. doi: 10.1128/IAI.73.1.514-522.2005
101. Wen AY, Sakamoto KM, Miller LS. The Role of the Transcription Factor CREB in Immune Function. J Immunol Am Assoc Immunol (2010) 185(11):6413–9. doi: 10.4049/jimmunol.1001829
102. Li B, Jie W, Huang L, Wei P, Li S, Luo Z, et al. Nuclear BK Channels Regulate Gene Expression via the Control of Nuclear Calcium Signaling. Nat Neurosci (2014) 17(8):1055–63. doi: 10.1038/nn.3744
103. Yang X, Wang G, Cao T, Zhang L, Ma Y, Jiang S, et al. Large-Conductance Calcium-Activated Potassium Channels Mediate Lipopolysaccharide-Induced Activation of Murine Microglia. J Biol Chem (2019) 294(35):12921–32. doi: 10.1074/jbc.RA118.006425
104. Rousseau E, Michaud C, Lefebvre D, Proteau S, Decrouy A. Reconstitution of Ionic Channels From Inner and Outer Membranes of Mammalian Cardiac Nuclei. Biophys J (1996) 70:703–14. doi: 10.1016/S0006-3495(96)79610-8
105. Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, et al. Nuclear and Cytosolic Calcium are Regulated Independently. Proc Natl Acad Sci USA (2003) 100(5):2975–80. doi: 10.1073/pnas.0536590100
106. Gerasimenko OV, Gerasimenko JV, Tepikin AV, Petersen OH. ATP-Dependent Accumulation and Inositol Trisphosphate- or Cyclic ADP-Ribose-Mediated Release of Ca2+ From the Nuclear Envelope. Cell (1995) 80(3):439–44. doi: 10.1016/0092-8674(95)90494-8
107. Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of Calcium Signals in the Nucleus by a Nucleoplasmic Reticulum. Nat Cell Biol (2003) 5(5):440–6. doi: 10.1038/ncb980
108. Marius P, Guerra MT, Nathanson MH, Ehrlich BE, Leite MF. Calcium Release From Ryanodine Receptors in the Nucleoplasmic Reticulum. Cell Calcium (2006) 39(1):65–73. doi: 10.1016/j.ceca.2005.09.010
109. Higazi DR, Fearnley CJ, Drawnel FM, Talasila A, Corps EM, Ritter O, et al. Endothelin-1-Stimulated InsP3-Induced Ca2+ Release Is a Nexus for Hypertrophic Signaling in Cardiac Myocytes. Mol Cell (2009) 33(4):472–82. doi: 10.1016/j.molcel.2009.02.005
110. Balakier H, Dziak E, Sojecki A, Librach C, Michalak M, Opas M. Calcium-Binding Proteins and Calcium-Release Channels in Human Maturing Oocytes, Pronuclear Zygotes and Early Preimplantation Embryos. Hum Reproduct Oxford Univ Press (2002) 17(11):2938–47. doi: 10.1093/humrep/17.11.2938
111. Avedanian L, Jacques D, Bkaily G. Presence of Tubular and Reticular Structures in the Nucleus of Human Vascular Smooth Muscle Cells. J Mol Cell Cardiol (2011) 50(1):175–86. doi: 10.1016/j.yjmcc.2010.10.005
112. Collado-Hilly M, Shirvani H, Jaillard D, Mauger JP. Differential Redistribution of Ca2+-Handling Proteins During Polarisation of MDCK Cells: Effects on Ca2+ Signalling. Cell Calcium (2010) 48(4):215–24. doi: 10.1016/j.ceca.2010.09.003
113. Clark A, Petty H. Differential Intracellular Distributions of Inositol Trisphosphate and Ryanodine Receptors Within and Among Hematopoietic Cells. J Histochem Cytochem (2005) 53(7):913–6. doi: 10.1369/JHC.4B6610.2005
114. Monaco S, Jahraus B, Samstag Y, Bading H. Nuclear Calcium is Required for Human T Cell Activation. J Cell Biol (2016) 215(2):231–43. doi: 10.1083/jcb.201602001
115. Greber UF, Gerace L. Depletion of Calcium From the Lumen of Endoplasmic Reticulum Reversibly Inhibits Passive Diffusion and Signal-Mediated Transport Into the Nucleus. J Cell Biol (1995) 128(1–2):5–14. doi: 10.1083/jcb.128.1.5
116. Perez-Terzic C, Gacy AM, Bortolon R, Dzeja PP, Puceat M, Jaconi M, et al. Structural Plasticity of the Cardiac Nuclear Pore Complex in Response to Regulators of Nuclear Import. Circ Res (1999) 84(11):1292–301. doi: 10.1161/01.RES.84.11.1292
117. Wei X, Henke VG, Strübing C, Brown EB, Clapham DE. Real-Time Imaging of Nuclear Permeation by EGFP in Single Intact Cells. Biophys J (2003) 84(2 I):1317–27. doi: 10.1016/S0006-3495(03)74947-9
118. OBrien EM, Gomes DA, Sehgal S, Nathanson MH. Hormonal Regulation of Nuclear Permeability. J Biol Chem (2007) 282(6):4210–7. doi: 10.1074/jbc.M606300200
119. Stoffler D, Goldie KN, Feja B, Aebi U. Calcium-Mediated Structural Changes of Native Nuclear Pore Complexes Monitored by Time-Lapse Atomic Force Microscopy. J Mol Biol (1999) 287(4):741–52. doi: 10.1006/jmbi.1999.2637
120. Wang H, Clapham DE. Conformational Changes of the in Situ Nuclear Pore Complex. Biophys J (1999) 77(1):241–7. doi: 10.1016/S0006-3495(99)76885-2
121. Erickson E, Mooren O, Moore-Nichols D, Dunn R. Activation of Ryanodine Receptors in the Nuclear Envelope Alters the Conformation of the Nuclear Pore Complex. Biophys Chem (2004) 112(1):1–7. doi: 10.1016/J.BPC.2004.06.010
122. Erickson ES, Mooren OL, Moore D, Krogmeier JR, Dunn RC. The Role of Nuclear Envelope Calcium in Modifying Nuclear Pore Complex Structure. Can J Physiol Pharmacol (2006) 84(3-4):309–18. doi: 10.1139/Y05-109
123. Atcha H, Jairaman A, Holt J, Meli V, Nagalla R, Veerasubramanian P, et al. Mechanically Activated Ion Channel Piezo1 Modulates Macrophage Polarization and Stiffness Sensing. Nat Commun (2021) 12(1):3256. doi: 10.1038/S41467-021-23482-5
124. Dutta B, Goswami R, Rahaman S. TRPV4 Plays a Role in Matrix Stiffness-Induced Macrophage Polarization. Front Immunol (2020) 11:570195. doi: 10.3389/FIMMU.2020.570195
125. Orsini EM, Perelas A, Southern BD, Grove LM, Olman MA, Scheraga RG. Stretching the Function of Innate Immune Cells. Front Immunol (2021) 12:767319/BIBTEX. doi: 10.3389/FIMMU.2021.767319/BIBTEX
126. Santoni G, Amantini C, Santoni M, Maggi F, Morelli MB, Santoni A. Mechanosensation and Mechanotransduction in Natural Killer Cells. Front Immunol (2021) 12:688918/BIBTEX. doi: 10.3389/FIMMU.2021.688918/BIBTEX
127. Zhang X, Kim TH, Thauland TJ, Li H, Majedi FS, Ly C, et al. Unraveling the Mechanobiology of Immune Cells. Curr Opin Biotechnol (2020) 66:236. doi: 10.1016/J.COPBIO.2020.09.004
128. Goelzer M, Goelzer J, Ferguson ML, Neu CP, Uzer G. Nuclear Envelope Mechanobiology: Linking the Nuclear Structure and Function. Nucleus (2021) 12(1):90. doi: 10.1080/19491034.2021.1962610
129. Maurer M, Lammerding J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu Rev Biomed Eng (2019) 21:443–68. doi: 10.1146/ANNUREV-BIOENG-060418-052139
130. Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, et al. Isolated Nuclei Adapt to Force and Reveal a Mechanotransduction Pathway in the Nucleus. Nat Cell Biol (2014) 16:4. doi: 10.1038/ncb2927
131. Lomakin AJ, Cattin CJ, Cuvelier D, Alraies Z, Molina M, Nader GPF, et al. The Nucleus Acts as a Ruler Tailoring Cell Responses to Spatial Constraints. Sci (New York NY) (2020) 370(6514):eaba2894. doi: 10.1126/SCIENCE.ABA2894
132. Venturini V, Pezzano F, Castro FC, Häkkinen HM, Jiménez-Delgado S, Colomer-Rosell M, et al. The Nucleus Measures Shape Changes for Cellular Proprioception to Control Dynamic Cell Behavior. Sci (New York NY) (2020) 370(6514):2644. doi: 10.1126/SCIENCE.ABA2644
133. Enyedi B, Niethammer P. Nuclear Membrane Stretch and its Role in Mechanotransduction. Nucleus (2017) 8(2):156–61. doi: 10.1080/19491034.2016.1263411
134. Enyedi B, Jelcic M, Niethammer P. The Cell Nucleus Serves as a Mechanotransducer of Tissue Damage-Induced Inflammation. Cell (2016) 165(5):1160–70. doi: 10.1016/J.CELL.2016.04.016
135. Oliveira AG, Guimarães ES, Andrade LM, Menezes GB, Leite MF. Decoding Calcium Signaling Across the Nucleus. Am Physiol Soc Bethesda MD (2014) 29(5):361–8. doi: 10.1152/PHYSIOL.00056.2013
136. Wang S, Zhou L, Ling L, Meng X, Chu F, Zhang S, et al. The Crosstalk Between Hippo-YAP Pathway and Innate Immunity. Front Media S.A (2020) 11:323/BIBTEX. doi: 10.3389/FIMMU.2020.00323/BIBTEX
137. Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, et al. Force Triggers YAP Nuclear Entry by Regulating Transport Across Nuclear Pores. Cell (2017) 171(6):1397–410.e14. doi: 10.1016/J.CELL.2017.10.008
138. Meli VS, Atcha H, Veerasubramanian PK, Nagalla RR, Luu TU, Chen EY, et al. YAP-Mediated Mechanotransduction Tunes the Macrophage Inflammatory Response. Sci Adv (2020) 6(49):8471. doi: 10.1126/SCIADV.ABB8471
139. Szczesny SE, Mauck RL. The Nuclear Option: Evidence Implicating the Cell Nucleus in Mechanotransduction. J Biomechan Eng (2017) 139(2):0210061. doi: 10.1115/1.4035350
140. Demmerle J, Koch AJ, Holaska JM. The Nuclear Envelope Protein Emerin Binds Directly to Histone Deacetylase 3 (HDAC3) and Activates HDAC3 Activity. J Biol Chem (2012) 287(26):22080–8. doi: 10.1074/JBC.M111.325308
141. Emmett MJ, Lazar MA. Integrative Regulation of Physiology by Histone Deacetylase 3. Nat Rev Mol Cell Biol (2018) 20(2):102–15. doi: 10.1038/s41580-018-0076-0
142. Nguyen HCB, Adlanmerini M, Hauck AK, Lazar MA. Dichotomous Engagement of HDAC3 Activity Governs Inflammatory Responses. Nature (2020) 584:7820. doi: 10.1038/s41586-020-2576-2
143. Nader GP, de F, Williart A, Piel M. Nuclear Deformations, From Signaling to Perturbation and Damage. Curr Opin Cell Biol (2021) 72:137–45. doi: 10.1016/J.CEB.2021.07.008
144. Briard B, Place D, Kanneganti T. DNA Sensing in the Innate Immune Response. Physiol (Bethesda Md.) Physiol (Bethesda) (2020) 35(2):112–24. doi: 10.1152/PHYSIOL.00022.2019
145. Maciejowski J, Hatch EM. Nuclear Membrane Rupture and Its Consequences. Annu Rev (2020) 36:85–114. doi: 10.1146/ANNUREV-CELLBIO-020520-120627
146. Ablasser A, Chen ZJ. cGAS in Action: Expanding Roles in Immunity and Inflammation. Science (2019) 363(6431):eaat8657. doi: 10.1126/SCIENCE.AAT8657
147. Kumar V. A STING to Inflammation and Autoimmunity. J Leukocyte Biol (2019) 106(1):171–85. doi: 10.1002/JLB.4MIR1018-397RR
148. Kumari P, Russo AJ, Shivcharan S, Rathinam VA. AIM2 in Health and Disease: Inflammasome and Beyond. Immunol Rev (2020) 297(1):83–95. doi: 10.1111/IMR.12903
149. Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, et al. Cytoplasmic Chromatin Triggers Inflammation in Senescence and Cancer. Nature (2017) 550:7676. doi: 10.1038/nature24050
150. Glück S, Guey B, Gulen MF, Wolter K, Kang T-W, Schmacke NA, et al. Innate Immune Sensing of Cytosolic Chromatin Fragments Through cGAS Promotes Senescence. Nat Cell Biol (2017) 19:9. doi: 10.1038/ncb3586
151. Lopes-Paciencia S, Saint-Germain E, Rowell MC, Ruiz AF, Kalegari P, Ferbeyre G. The Senescence-Associated Secretory Phenotype and its Regulation. Cytokine (2019) 117:15–22. doi: 10.1016/J.CYTO.2019.01.013
152. Reddy SAG, Chaturvedi MM, Darnay BG, Chan H, Higuchi M, Aggarwal BB. Reconstitution of Nuclear Factor Kappa B Activation Induced by Tumor Necrosis Factor Requires Membrane-Associated Components. Comparison With Pathway Activated by Ceramide. J Biol Chem (1994) 269(41):25369–72. doi: 10.1016/S0021-9258(18)47258-4
Keywords: nuclear envelope, nucleus, immune cells, inflammation, lamins, ion channels
Citation: Selezneva A, Gibb AJ and Willis D (2022) The Nuclear Envelope as a Regulator of Immune Cell Function. Front. Immunol. 13:840069. doi: 10.3389/fimmu.2022.840069
Received: 20 December 2021; Accepted: 10 May 2022;
Published: 10 June 2022.
Edited by:
Claudia Milena Trujillo-Vargas, University of Antioquia, ColombiaReviewed by:
Jörg Renkawitz, Ludwig Maximilian University of Munich, GermanyYangnan Gu, University of California, Berkeley, United States
Copyright © 2022 Selezneva, Gibb and Willis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dean Willis, ZGVhbi53aWxsaXNAdWNsLmFjLnVr
 Anna Selezneva
Anna Selezneva Alasdair J. Gibb
Alasdair J. Gibb Dean Willis
Dean Willis