
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 24 March 2022
Sec. Autoimmune and Autoinflammatory Disorders
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.838884
This article is part of the Research Topic The Role of Omics Characteristics in the Diagnosis, Treatment, and Prognosis of Autoimmune Diseases View all 10 articles
 Cen Chang1,2†
Cen Chang1,2† Lingxia Xu1,2†
Lingxia Xu1,2† Runrun Zhang3
Runrun Zhang3 Yehua Jin1,2
Yehua Jin1,2 Ping Jiang1,2
Ping Jiang1,2 Kai Wei1,2
Kai Wei1,2 Linshuai Xu1,2
Linshuai Xu1,2 Yiming Shi1,2
Yiming Shi1,2 Jianan Zhao1,2
Jianan Zhao1,2 Momiao Xiong4
Momiao Xiong4 Shicheng Guo5,6*
Shicheng Guo5,6* Dongyi He1,2,7*
Dongyi He1,2,7*MicroRNAs (miRNAs) play crucial roles in regulating the transcriptome and development of rheumatoid arthritis (RA). Currently, a comprehensive map illustrating how miRNAs regulate transcripts, pathways, immune system differentiation, and their interactions with terminal cells such as fibroblast-like synoviocytes (FLS), immune-cells, osteoblasts, and osteoclasts are still laking. In this review, we summarize the roles of miRNAs in the susceptibility, pathogenesis, diagnosis, therapeutic intervention, and prognosis of RA. Numerous miRNAs are abnormally expressed in cells involved in RA and regulate target genes and pathways, including NF-κB, Fas-FasL, JAK-STAT, and mTOR pathways. We outline how functional genetic variants of miR-499 and miR-146a partly explain susceptibility to RA. By regulating gene expression, miRNAs affect T cell differentiation into diverse cell types, including Th17 and Treg cells, thus constituting promising gene therapy targets to modulate the immune system in RA. We summarize the diagnostic and prognostic potential of blood-circulating and cell-free miRNAs, highlighting the opportunity to combine these miRNAs with antibodies to cyclic citrullinated peptide (ACCP) to allow accurate diagnosis and prognosis, particularly for seronegative patients. Furthermore, we review the evidence implicating miRNAs as promising biomarkers of efficiency and response of, and resistance to, disease-modifying anti-rheumatic drugs and immunotherapy. Finally, we discuss the autotherapeutic effect of miRNA intervention as a step toward the development of miRNA-based anti-RA drugs. Collectively, the current evidence supports miRNAs as interesting targets to better understand the pathogenetic mechanisms of RA and design more efficient therapeutic interventions.
Rheumatoid arthritis (RA) is an autoimmune disease characterized by chronic joint inflammation and structural damage, accompanied by extra-articular manifestations such as rheumatoid nodules, interstitial pneumonia, vasculitis, and systemic complications. RA is typically progressive and insidious, with an incidence rate of 0.5–1% in Europe and North America (1). However, the precise mechanisms underlying the pathogenesis, disease activity, and severity of RA as well as the causes of different response to treatment are not fully understood. In view of current therapy strategies and treatment frames, early accurate diagnosis, effective and personalized treatment, and precision medicine have become increasingly urgent for patients with RA. A comprehensive understanding of RA is required from the perspectives of both genetics (2) [human leukocyte antigen (HLA) and non-HLA variants] and epigenetics [DNA methylation (3–5), microRNA (6), long non-coding RNA (7, 8), and histone modifications (9)].
MicroRNAs (miRNAs) are small endogenous non-coding RNAs with a length of around 22 nucleotides, and are involved in the post-transcriptional regulation of gene expression. In recent years, accumulating studies have demonstrated that miRNAs play a key role in various cancers (10–14) and autoimmune diseases, including RA, systemic lupus erythematosus (15, 16), Sjögren’s syndrome (17), and systemic sclerosis (18). In this review, we systematically summarize recent advances in understanding the role of miRNAs in RA. Emphasize the important role of miRNA in RA susceptibility, pathogenesis, and efficacy evaluation. Provide evidence supporting precision medicine research in RA.
Genome-wide association studies have identified >100 genetic factors for RA. However, these genetic variants only explain < 40% of the overall heritability of RA, and thus most of the heritability has not been explained, suggesting the need for more studies using different approaches and populations to identify the missing causes. Association studies of miRNA loci can reveal RA-associated functional or causal variants within different populations, such as Chinese (19, 20), Egyptian (21–23), Polish (24), Mexican (25), and Iranian (26) subjects. Gene expression and genetic polymorphisms of miR-146a and miR-499 showed diagnostic potential for RA (23). Consistently, the polymorphism rs3027898 in IRAK1, the target gene of miR-146a, is linked to RA in the Greek population (27). In contrast, miR-146a rs2431697 is associated with RA susceptibility in Chinese population (28). The rs3746444 (20q11.22, A>G) polymorphism of miR-499, which is encoded by the intron of MYH7B, is significantly linked to RA risk, disease activity, and methotrexate (MTX) toxicity. Interestingly, the AA genotype shows higher disease activity and MTX toxicity than the AG/GG genotypes (29). The AA and AG genotypes in the miRNA binding site rs3135500 of NOD2 are significantly associated with the risk of RA, with rs3135500 (A allele) showing a significant relationship with increased erythrocyte sedimentation rates (ESR) and C-reactive protein (CRP) concentrations (30). However, some studies showed inconsistent results in Polish (24), Mexican (25), and Chinese (19, 20, 31, 32) populations, suggesting that genetic polymorphisms of miR-146a and miR-499 are not significantly associated with RA susceptibility. For example, miR-499 rs3746444 A/G are not significantly associated with RA in Mexican people (25). miR-146a rs2910164 (20, 32) and miR-499 rs3746444 (20, 31) do not significantly correlate with RA in Chinese people. However, in addition to race, factors of individual heterogeneity and sample size should also be considered while evaluating inconsistent results. Moreover, the sample size of the above studies was not large. We recently demonstrated that meta-analysis could identify more significant single-nucleotide polymorphisms in a large sample size, and found that the interaction between HLA alleles and miRNA single-nucleotide polymorphisms (such as rs5997893 in miR-3928 and rs4947332 in HLA-DRB1) should be considered to explain susceptibility (33). In summary, genetic variations in miRNAs can help to explain the susceptibility to RA.
Fibroblast-like synoviocytes (FLS) and immune cells are the main cell types involved in the pathogenesis of RA. These cells can secrete exosomes and other substances to affect the occurrence and development of RA. Current researches have mainly focused on understanding miRNA-mediated transcriptional regulation of FAF1 (34), TNF-α (35), STAT1 (36), STAT3 (37), and mTOR (38, 39). miRNAs regulate inflammation, immune response, proliferation, and differentiation. Meanwhile, miRNA influence the micro-environment within synovial joints by targeting target genes and their related pathways, including Fas-FasL (34) and the NF-κB (40, 41) pathways. In this section, we summarize the regulatory roles of miRNAs in the main RA-associated cell entities, focusing on FLS, immune cells, and exosomes to highlight the importance of miRNAs in the pathogenesis of RA.
FLS in RA (RA-FLS) are key regulators of inflammation and bone destruction in RA. The aberrantly expression of miRNAs in RA-FLS play an important role in the pathogenesis of RA. For example, miR-625 is down-regulated in RA-FLS, which negatively impacts the expression of CTSC, KLF8, and EBF3. In contrast, miR-551b is up-regulated in RA-FLS, inhibiting the expression of ITGBL1 (42).
Dysregulation of miRNAs in RA-FLS affects biological functions such as cell proliferation, invasion, migration and apoptosis. Up-regulation of miR-145 affects all biological functions of RA-FLS by targeting SEMA3A (43). The expression of miR-29c-3p and miR-132-3p are decreased while miR-31-5p was increased in RA- FLS, and their dysregulation are associated with proliferation, invasion and migration of RA-FLS. Down-regulated miR-29c-3p promoted proliferation, invasion and migration of RA-FLS through up-regulation of COLA1 expression. Interestingly, up-regulated miR-31-5p and down-regulated miR-132-3p inhibited the proliferation, invasion and migration of RA-FLS by negatively regulating WASF3 and RB1, respectively, suggested that miR-31-5p and miR-132-3p are protective factors in RA (44). Down-regulation of miR-199a-3p (45), miR-449 (46), miR-431-5p (47), and up-regulation of miR-483-3p (48) can promote the proliferation and suppressed apoptosis by targeting RB1, HDAC1, XIAP, IGF-1, respectively. miR-124a is down-regulated and targets CDK2 and MCP-1 which only enhanced the proliferation of RA-FLS (49).
Dysregulation of miRNAs in RA-FLS can also affects the level of inflammation. Down-regulation of miR-126 (35), miR-23 (50) and up-regulation of miR-143 (43) can increase the release of inflammatory factors such as TNF-α, IL-1β, IL-6 through up-regulation of IL-23R, CXCL12 and down-regulation of IGFBP5 and thus affect the course of RA. What’s more, some miRNAs not only affects the level of inflammation, but also associated with biological functions of RA-FLS. For example, down-regulation of miR-137 (51) and miR-23a-5p (52) targeting LSD and TLR4 promotes proliferation, invasion, migration and inhibits apoptosis of RA-FLS and inhibits the release of inflammatory factors IL-1β and IL-6. Down-regulation of miR-29a (37) and miR-27a-3p (53) are associated with proliferation, apoptosis, and promoting secretion of TNF-α, IL-1, IL-6 and IL-8. Then, down-regulation of miR-22 (54), miR-124 (55) and miR-34a-5p (56) can enhanced proliferation of RA-FLS and the level of IL-6.
Additionally, the biological functions of RA-FLS and the level of inflammation are correlated with matrix metalloproteinases (MMPs) (57). Down-regulation of miR-203 (41, 42) and miR-147b-3p (58) increased the expression of MMP-1, MMP3 and MMP9, respectively, which in turn enhanced the expression of some inflammatory factors, such as IL-6 and TNF-α. Down-regulation of miR-27a (59) can contribute to MMPs gene expression by targeting the IL-17 pathways, thereby affecting the proliferation and invasion of RA-FLS. Conversely, up-regulated miR-155 could be a protective factor by inhibiting proliferation and invasion while attenuating the expression of MMP3 and IKBKE (60, 61).
In addition, dysregulation of miRNAs in RA-FLS affects joint bone erosion through the release of inflammatory cytokines, chemokines and MMPs, which may be an option for RA treatment (62, 63). For example, up-regulated miR-145-5p (40) and down-regulated miR-17-5p (64) affect bone and cartilage destruction through the release of IL-1β, IL-6, IL-8, MMP-1, MMP-3 and MMP-9. In vitro, overexpression of miR-221-3p inhibits osteoblast differentiation (65). Instead, miR-218 overexpression promotes osteogenic differentiation of RA-FLS by suppressing the Roundabout-1/Dickkopf-1 axis (66). miR-20a (67) and miR-21 (68) are targets of the TLR4/p38 and JAK/STAT3 signaling pathways respectively, affecting the proliferation and osteogenic differentiation of RA-FLS.
In summary, studies of miRNAs in RA-FLS have improved the understanding of the pathogenesis of RA. miRNAs are widely involved in the functions of FLS, and therefore are promising targets for drug development (Figure 1).
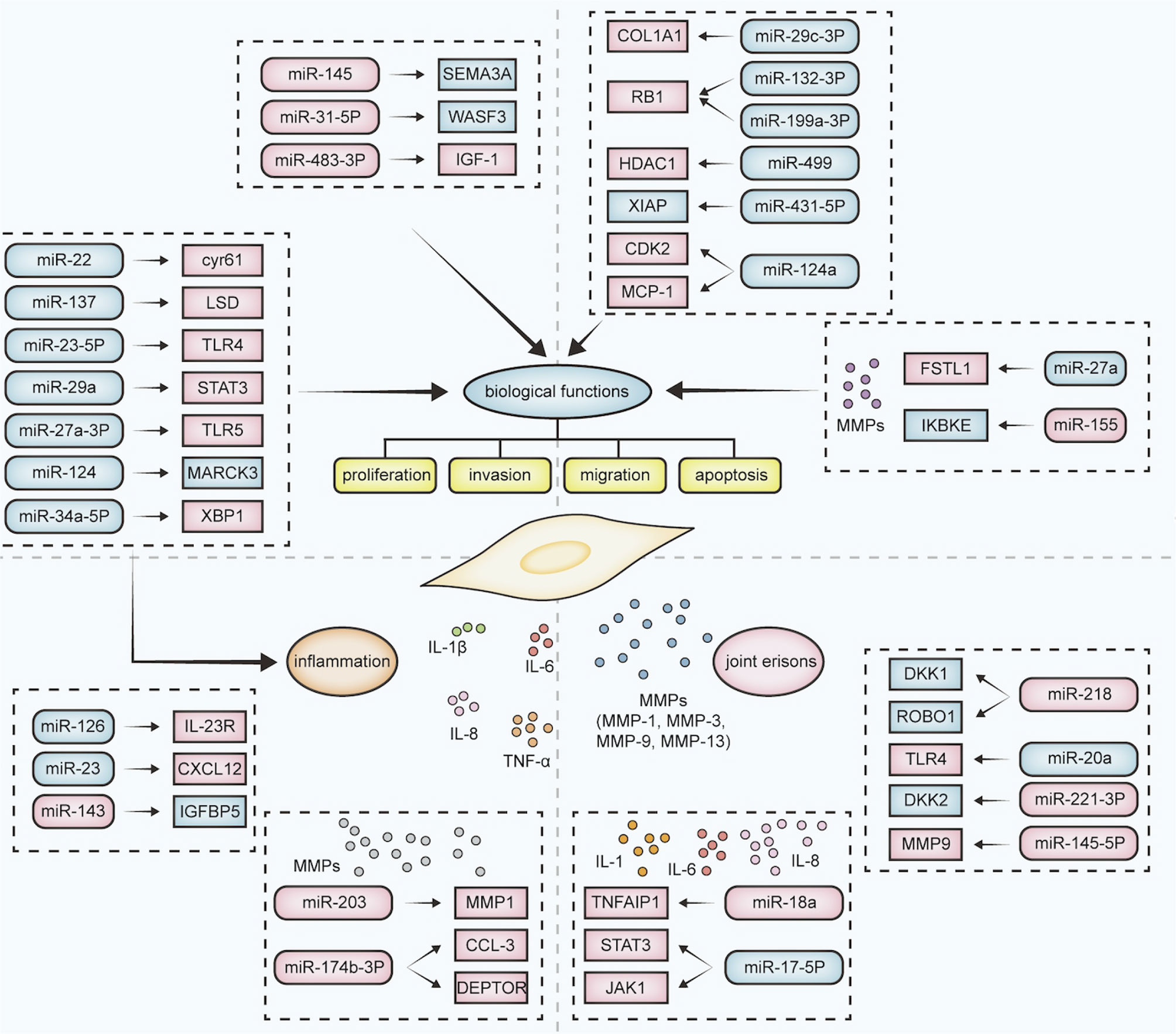
Figure 1 Effects of dysregulation of the miRNA-mRNA network on RA-FLS. The dysregulation of miRNAs and their target mRNAs in RA-FLS affects the biological function (such as proliferation, invasion, migration, and apoptosis), inflammatory levels, and joint bone destruction. Inflammatory levels are mainly related to the release of inflammatory factors such as IL-8, IL-6, and joint bone destruction is mainly related to the release of MMPs. Dysregulation of different miRNA-mRNA combinations affects different processes in RA-FLS. Rounded rectangles represent miRNAs; rectangles represent target mRNAs; pink represents upregulation; blue represents downregulation.
miRNAs have recently emerged as key regulators of the immune system, being involved in lymphocyte selection and proliferation, in T(reg) cells differentiation. In peripheral blood mononuclear cells (PBMCs), decreased expression of miR-671 and miR-7 may correlate with the expression of CDR1 and mTOR (38). And miR-29b enhances the anti-apoptotic effect by inhibiting the high-mobility group box-containing protein 1 (HBP1) (69).
In T cells sub-population derived from PBMCs, miR-99b-5p down-regulates mTOR and RASSF4, thereby inhibiting T cell apoptosis and promoting T cell proliferation and inflammatory response (39). Besides, miR-146a, miR-26, miR-let-7a, miR-146b, miR-150, miR-155 are increased and miR-363, miR-498 are decreased in the CD4+ T cell sub-type in PBMCs, Among these miRNAs, miR-146a may affect the apoptosis of T cell and RA progression by targeting IL-17 and Fas associated factor 1 (FAF1) (34, 70). Interestingly, miR-233 is highly expressed only in naive CD4+ lymphocytes but not in T(h)-17 cells, suggesting the importance to investigate the impact of miRNA on the pathogenesis of RA at the single-cell level (71).
There are numerous of miRNAs are also associated with other T cells sub-types, such as Treg cells and Th17 cells. The balance of Th17/Treg cells plays a crucial role in RA. IL-17 released by Th17 up-regulates the expression of RANKL on synovial fibroblasts stimulating the production of inflammatory cytokines such as TNF-α, IL-1, and IL-6 (72). Decreased levels of miR-20a (73) and miR-21 (74, 75) exacerbate the RA process by stimulating the NLRP3 inflammasome pathway and increasing STAT3 expression, respectively, while decreasing STAT5 expression, all of which are associated with the imbalance of Th17/Treg cells. Although miR-210-mediated negative regulation of HIF-1 also affects the dynamic equilibrium of Th17/Treg cells. Regrettably, the levels of miR-210 between RA and healthy controls have no significant difference (76). Interestingly, the expression of miR-146a is decreased in Treg cells during high RA activity, leading to a proinflammatory phenotype in these cells caused by concomitant up-regulation of its target STAT1 (36). For instance, miR-21 and miR-155 are related to the memory phenotype, and miR-92a relative to the naïve phenotype (77).
Besides, in macrophages, binding of miR-6089 and lncRNA-HIX003209 enhances the expression of TLR4 and exacerbates inflammation via the TLR4/NF-κB pathway (78). Up-regulation of miR-33 induces the expression of NLRP3 and 73 (79). Overall, miRNAs cooperate with other non-coding RNAs to alter the DNA methylation and/or expression of their targets, thus regulating innate and adaptive immune cells differentiation and apoptosis, ultimately influencing the inflammatory and autoimmune response in RA (Table 1).
Exosomes are secreted from cells and contain signal molecules such as miRNA, protein, and DNA, which have biological functions. Exosomal miRNA derived from bone marrow-derived mesenchymal stem cells has been shown to be closely related to the occurrence and development of RA. Among these exosomes, MSCs-drived miR-124a over-expression exosomes inhibit the proliferation and migration and promote the apoptosis of RA-FLS (80). Over-expression of miR-23b (83) and miR-34a (81) can inhibit the differentiation of Th17 cells, by reducing IL-17 secretion and targeting the cyclin I/ATM/ATR/p53 signaling pathway, respectively. Up-regulated of miR-21 (82) which targets TET1, reduce RA inflammation. Macrophage-derived exosomes miR-506-3p (84) and miR-103a (85) regulate the progression of RA by inhibiting the RANKL/NFATc1 signaling pathway and activating the JAK/STAT3 signaling pathway. miR-132 secreted by aryl hydrocarbon receptor activation Th17 in extracellular vesicles acts as a pro-inflammatory mediator to reduce the production of COX2, to increase the production of osteoclast (86). In addition, cell-derived small extracellular vesicles of miR-574-5p induces osteoclast differentiation by targeting TLR 7/8 (87), whereas miR-150-5p exosomes alleviates RA-FLS proliferation and angiogenesis and reduces RA joint destruction by targeting MMP14 and VEGF (88). Based on these results, miRNAs play an important role in the pathogenesis of RA and may represent promising outcome biomarkers and novel drug targets to decrease disease severity.
Emerging evidence indicates the potential of blood-circulating miRNAs associated with RA as biomarkers for early prevention. The levels of miR-371b, miR-483, and miR-642b are significantly up-regulated, whereas miR-25 and miR-378d are down-regulated in PBMCs in individuals who eventually develop RA from early undifferentiated arthritis (89). Additionally, miR-22 (90), miR-361-5p (91), and miR-223-3p (91) are significantly up-regulated in high-risk or CCP-positive populations. All these miRNAs may therefore be useful biomarkers for the early diagnosis of RA. Expression of miR-103a-3p is significantly increased in autoantibody-positive, symptomatic first-degree relatives and patients with RA, suggesting it as a potential biomarker for predicting imminent disease in individuals at risk for developing RA (92). Additionally, higher level of miR-99b-5p is found in the plasma of patients with early RA who progress to bone erosion after 12 months, indicating that miR-99b-5 can be monitored for bone erosion surveillance in RA patients (93).
In addition to playing a role in the early prevention of RA, the expression of some miRNAs can aid in improving the accuracy of RA diagnosis (94). The expression of miR-146a and miR-155 are significantly increased in RA PBMCs and whole blood (95). The levels of miR-24 and miR-125a are significantly higher in the serum of patients with RA regardless of the CCP status (96). Interestingly, analysis of miR-24-3p, miR-26a-5p, and miR-125a-5p levels in combination are a better diagnostic tool for RA, even though these miRNAs are not related to disease activity (97). Furthermore, miR-122-3p, miR-3925-3p, miR-342-3p, and miR-4764-5p show differential expression not only between healthy individuals and RA patients, but also between patients with RA and patients with osteoarthritis, systemic lupus erythematosus, or Graves, which show great potential as biomarkers to distinguish RA patients from HC or other diseases (98). The serum levels of miR-146a (99, 100), miR-22-3p (101), miR-5571-3p (102), and miR-135b-5p (102) are significantly higher in RA patients than in healthy controls and osteoarthritis patients. Other differentially expressed miRNAs in patients with RA serum include miR-4634, miR-181d, miR-3926, miR-9-5p, miR-219-2-3p6, miR-221, miR-222, miR-532, miR-106a, and miR-98 highlighting their potential as RA-specific diagnostic markers (98, 103). Nevertheless, the above miRNAs should be selected as biomarkers with caution. Their sensitivity and specificity need to be taken into consideration because they were only compared with patients with osteoarthritis (OA) or healthy control, and rarely analyzed with patients with other inflammatory autoimmune diseases, such as ankylosing spondylitis. In addition, it should be determined if the miRNA as diagnostic markers are expressed differently in patients before the onset of clinical symptoms. The above studies were conducted after patients were confirmed with RA diagnosis. Thus, more robust sample studies are needed to validate these markers in early RA.
The expression of miR-451 in T cells is significantly increased, which is positively correlated with the levels of disease activity score 28 (DAS-28), ESR, and serum IL-6 in RA (77). The level of miR-146a is positively correlated with the level of ESR and DAS-28 (99), whereas miR-5571-3p (102) correlates with the level of ESR and CRP, and miR-135b-5p only correlate with CRP (102). These miRNAs may therefore be suitable markers of disease activity in patients with RA. Increased serum miR-194-5p level is associated with disease recurrence (104). Concentration of circulating miR-23b, which positively correlates with ESR, CRP, and DAS-28, is significantly up-regulated after appropriate treatment, indicating that miR-23b is a dual marker for disease activity and prognosis (105). Similarly, miR-96-5p, miR-134-5p, miR-140-3p, miR-627-5p, miR-224, miR-760, miR-483-5p, miR-378, and miR-375 are not only diagnostic markers for RA, but also mirror disease activity (106, 107). However, these studies are still descriptive. Therefore, the underlying pathophysiology needs to be validated using other techniques.
Common and widely used anti-rheumatic drugs include cDMARDs (MTX, sulfasalazine, and hydroxychloroquine), bDMARDs (TNF-α inhibitors, rituximab, and tocilizumab), tsDMARDs (tofacitinib, barretinib, and filgotinib). Several studies have explored the relationship between serum miRNA levels and drug response. Evidence shows that high serum levels of miR-10 in patients with RA is correlated with good response to MTX (108). After 3 months of adalimumab/MTX combined treatment, the level of miR-27a-3p significantly decreased and clinical symptoms significantly improved (109). The reduced serum level of miR-5196 is positively correlated with the delta DAS28 after anti-TNF-α therapy (110). The level of miR-146a is increased in RA patients who respond well to anti-TNF therapy and, interestingly, can be considered as predictors of the response to anti-TNFα therapy together with CRP (24, 111, 112). In contrast, the serum levels of miR-23 and miR-223 are increased in patients with RA who respond well to anti-TNF-α/DMARD combination therapy, but correlate negatively with the response to anti-TNF drugs (111). High serum level of miR-125b is an indicator for good clinical response to rituximab therapy (113). Notably, miR-432-5p is significantly down-regulated in RA patients who are responsive to tofacitinib therapy but up-regulated in patients showing RA relapse (104). In RA, treatment with rituximab increases the levels of miR-16-5p and miR-23a-3p in the peripheral blood (114). The expression of miR-550b-2-5p, miR-4797-5p, miR-6509-5p, miR-378g, miR-4720-5p, miR-374b-5p, and miR-185-3p are different between individuals who show good vs poor responses to treatment with tripterygium glycosides (115, 116). Finally, the expression of miR-124a in FLS is increased following geniposide treatment; however, the relevance of this finding has not been assessed in clinical response studies (117).
In addition to DMARD treatment, alternative and complementary medicine preparations and mesenchymal stem cell treatments are also used in clinical practice. The auto-therapeutic effect of miRNAs has been demonstrated in mouse models of RA-FLS and autoimmune arthritis. For example, miR-449a mimics also inhibit the proliferation, migration, and IL-6 production of RA-FLS by regulating HMGB1 and YY1 expression (118). In the rat model with collagen-induced arthritis, miR-708-5p mimic improved pathological changes by inhibiting inflammatory cell infiltration, synovial hyperplasia, and cartilage destruction (119). An miR-126 agonist inhibits the expression of IL-23R, TNF-α, and IFN-γ in FLS (35). Furthermore, miR-26b-5p, miR-487b-3p, and miR-495-3p are significantly up-regulated in responders to adipose-derived mesenchymal stem cell treatment (120).
In summary, the changes of circulation miRNA in RA provide a promising opportunity for standard treatment, as well as indicate disease activity and predict RA outcomes.
In conclusion, miRNAs play multiple roles in the development of RA, from susceptibility to pathogenesis. Blood and serum-circulating miRNAs have been explored as important biomarkers for early diagnosis, prognosis, and drug response prediction. Furthermore, miRNAs have been proposed for autotherapeutic approaches and as novel drug targets for the treatment of RA. Genetic variants in specific miRNAs can increase or decrease the risk and disease activity of RA in various racial. Meanwhile they are associated with methotrexate toxicity and responses to other treatments. Moreover, changes in miRNAs in various cells are related to the pathogenesis of RA, such as the proliferation and differentiation of immune cells, proliferation and apoptosis of synovial cells, and synovial inflammation and cartilage destruction. Research has remarkably progressed towards the development of miRNAs as biomarkers in the diagnosis, prognosis, disease activity, and response to therapeutic drugs with RA, providing a direction for early diagnosis and accurate treatment of RA, to achieve better treatment efficiency and precision medicine. Numerous miRNAs have been shown to act as therapeutic targets in RA-FLS and collagen-induced arthritis rat models. Furthermore, miRNAs show potential for identifying the subtypes of RA. For example, the levels of miR-7 and miR-214-5p are significantly increased in the serum of patients with RA associated-interstitial lung disease (121), and miR-9-5p targets the REST/miR-132 pathway to protect Schwann cells from inflammatory damage in RA-induced peripheral neuropathy (122). Although we have reached exciting milestones in the research on the multiple roles of miRNA in RA, further studies should be performed to translate this knowledge for clinical applications and resolve the current inconsistent results among different studies employing different methods or populations. For example, studies of miR-99, miR-143, and miR-197 as landmark miRNAs for predicting the response to anti-TNF-α therapy have failed to yield consistent results (123). Finally, future development of miRNA-based baseline RA polygenetic risk score models, particularly in conjunction with HLA, is needed. miRNA-based early diagnosis, prognosis, and drug response prediction models can be applied in the clinic. With the identification of additional miRNAs-based drug targets in clinical research, miRNA-based autotherapeutic treatments may show more promising results.
SG and DH conceived the content. CC and LXX wrote the manuscript. RZ, YJ, PJ, KW, and MX edited the manuscript. YS, JZ and LSX checked the manuscript information. LXX wrote the manuscript and LSX checked the manuscript information.
This work was funded by the National Natural Science Funds of China (82074234 and 82004166); Shanghai Chinese Medicine Development Office, National Administration of Traditional Chinese Medicine, Regional Chinese Medicine (Specialist) Diagnosis and Treatment Center Construction Project-Rheumatology; State Administration of Traditional Chinese Medicine, National TCM Evidence-Based Medicine Research and Construction Project, Basic TCM Evidence-Based Capacity Development Program; Shanghai Municipal Health Commission, East China Region based Chinese and Western Medicine Joint Disease Specialist Alliance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ibanez-Cabellos JS, Seco-Cervera M, Osca-Verdegal R, Pallardo FV, Garcia-Gimenez JL, et al. Epigenetic Regulation in the Pathogenesis of Sjogren Syndrome and Rheumatoid Arthritis. Front Genet (2019) 10:1104. doi: 10.3389/fgene.2019.01104
2. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery. Nature (2014) 506(7488):376–81. doi: 10.1038/nature12873
3. Guo S, Zhu Q, Jiang T, Wang R, Shen Y, Zhu X, et al. Genome-Wide DNA Methylation Patterns in CD4+ T Cells From Chinese Han Patients With Rheumatoid Arthritis. Mod Rheumatol (2017) 27(3):441–7. doi: 10.1080/14397595.2016.1218595
4. Chen S, Pu W, Guo S, Jin L, He D, Wang J. Genome-Wide DNA Methylation Profiles Reveal Common Epigenetic Patterns of Interferon-Related Genes in Multiple Autoimmune Diseases. Front Genet (2019) 10:223. doi: 10.3389/fgene.2019.00223
5. Guo S, Xu L, Chang C, Zhang R, Jin Y, He D. Epigenetic Regulation Mediated by Methylation in the Pathogenesis and Precision Medicine of Rheumatoid Arthritis. Preprints (2020) 11:811. doi: 10.3389/fgene.2020.00811
6. Furer V, Greenberg JD, Attur M, Abramson SB, Pillinger MH. The Role of microRNA in Rheumatoid Arthritis and Other Autoimmune Diseases. Clin Immunol (2010) 136(1):1–15. doi: 10.1016/j.clim.2010.02.005
7. Guo S, Liu J, Jiang T, Lee D, Wang R, Zhou X, et al. (5r)-5-Hydroxytriptolide (LLDT-8) Induces Substantial Epigenetic Mediated Immune Response Network Changes in Fibroblast-Like Synoviocytes From Rheumatoid Arthritis Patients. Sci Rep (2019) 9(1):11155. doi: 10.1038/s41598-019-47411-1
8. Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, et al. LncRNA PICSAR Promotes Cell Proliferation, Migration and Invasion of Fibroblast-Like Synoviocytes by Sponging miRNA-4701-5p in Rheumatoid Arthritis. EBioMedicine (2019) 50:408–20. doi: 10.1016/j.ebiom.2019.11.024
9. Angiolilli C, Kabala PA, Grabiec AM, Van Baarsen IM, Ferguson BS, Garcia S, et al. Histone Deacetylase 3 Regulates the Inflammatory Gene Expression Programme of Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Ann Rheum Dis (2017) 76(1):277–85. doi: 10.1136/annrheumdis-2015-209064
10. Ding H, Gao G, Zhang L, Shen G, Sun W, Gu Z, et al. The Protective Effects of Curculigoside A on Adjuvant-Induced Arthritis by Inhibiting NF-Small Ka, CyrillicB/NLRP3 Activation in Rats. Int Immunopharmacol (2016) 30:43–9. doi: 10.1016/j.intimp.2015.11.026
11. Fan L, Chen L, Ni X, Guo S, Zhou Y, Wang C, et al. Genetic Variant of miR-4293 Rs12220909 Is Associated With Susceptibility to Non-Small Cell Lung Cancer in a Chinese Han Population. PloS One (2017) 12(4):e0175666. doi: 10.1371/journal.pone.0175666
12. He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, et al. Hypomethylation of the hsa-miR-191 Locus Causes High Expression of Hsa-Mir-191 and Promotes the Epithelial-to-Mesenchymal Transition in Hepatocellular Carcinoma. Neoplasia (2011) 13(9):841–53. doi: 10.1593/neo.11698
13. Lin S, Pan L, Guo S, Wu J, Jin L, Wang JC, et al. Prognostic Role of microRNA-181a/B in Hematological Malignancies: A Meta-Analysis. PloS One (2013) 8(3):e59532. doi: 10.1371/journal.pone.0059532
14. Zhang P, Wang J, Lu T, Wang X, Zheng Y, Guo S, et al. miR-449b Rs10061133 and miR-4293 Rs12220909 Polymorphisms Are Associated With Decreased Esophageal Squamous Cell Carcinoma in a Chinese Population. Tumour Biol (2015) 36(11):8789–95. doi: 10.1007/s13277-015-3422-2
15. Xie L, Xu J. Role of MiR-98 and Its Underlying Mechanisms in Systemic Lupus Erythematosus. J Rheumatol (2018) 45(10):1397–405. doi: 10.3899/jrheum.171290
16. Senousy MA, Helmy HS, Fathy N, Shaker OG, Ayeldeen GM. Association of MTMR3 Rs12537 at miR-181a Binding Site With Rheumatoid Arthritis and Systemic Lupus Erythematosus Risk in Egyptian Patients. Sci Rep (2019) 9(1):12299. doi: 10.1038/s41598-019-48770-5
17. Jang SI, Tandon M, Teos L, Zheng C, Warner BM, Alevizos I. Dual Function of miR-1248 Links Interferon Induction and Calcium Signaling Defects in Sjogren’s Syndrome. EBioMedicine (2019) 48:526–38. doi: 10.1016/j.ebiom.2019.09.010
18. Iwamoto N, Vettori S, Maurer B, Brock M, Pachera E, Jungel A, et al. Downregulation of miR-193b in Systemic Sclerosis Regulates the Proliferative Vasculopathy by Urokinase-Type Plasminogen Activator Expression. Ann Rheum Dis (2016) 75(1):303–10. doi: 10.1136/annrheumdis-2014-205326
19. Yang B, Chen J, Li Y, Zhang J, Li D, Huang Z, et al. Association of Polymorphisms in pre-miRNA With Inflammatory Biomarkers in Rheumatoid Arthritis in the Chinese Han Population. Hum Immunol (2012) 73(1):101–6. doi: 10.1016/j.humimm.2011.10.005
20. Yang B, Zhang JL, Shi YY, Li DD, Chen J, Huang ZC, et al. Association Study of Single Nucleotide Polymorphisms in Pre-miRNA and Rheumatoid Arthritis in a Han Chinese Population. Mol Biol Rep (2011) 38(8):4913–9. doi: 10.1007/s11033-010-0633-x
21. El-Shal AS, Aly NM, Galil SM, Moustafa MA, Kandel WA. Association of microRNAs Genes Polymorphisms With Rheumatoid Arthritis in Egyptian Female Patients. Joint Bone Spine (2013) 80(6):626–31. doi: 10.1016/j.jbspin.2013.03.005
22. Shaker OG, El Boghdady NA, El Sayed AE. Association of MiRNA-146a, MiRNA-499, IRAK1 and PADI4 Polymorphisms With Rheumatoid Arthritis in Egyptian Population. Cell Physiol Biochem (2018) 46(6):2239–49. doi: 10.1159/000489592
23. Ayeldeen G, Nassar Y, Ahmed H, Shaker O, Gheita T. Possible Use of miRNAs-146a and -499 Expression and Their Polymorphisms as Diagnostic Markers for Rheumatoid Arthritis. Mol Cell Biochem (2018) 449(1-2):145–56. doi: 10.1007/s11010-018-3351-7
24. Bogunia-Kubik K, Wysoczańska B, Piątek D, Iwaszko M, Ciechomska M, Świerkot J. Significance of Polymorphism and Expression of miR-146a and NFkB1 Genetic Variants in Patients With Rheumatoid Arthritis. Arch Immunol Ther Exp (Warsz) (2016) 64(Suppl 1):131–6. doi: 10.1007/s00005-016-0443-5
25. Aleman-Avila I, Jimenez-Morales M, Beltran-Ramirez O, Barbosa-Cobos RE, Jimenez-Morales S, Sanchez-Munoz F, et al. Functional Polymorphisms in Pre-Mir146a and Pre-Mir499 Are Associated With Systemic Lupus Erythematosus But Not With Rheumatoid Arthritis or Graves’ Disease in Mexican Patients. Oncotarget (2017) 8(54):91876–86. doi: 10.18632/oncotarget.19621
26. Hashemi M, Eskandari-Nasab E, Zakeri Z, Atabaki M, Bahari G, Jahantigh M, et al. Association of Pre-miRNA-146a Rs2910164 and premiRNA-499 Rs3746444 Polymorphisms and Susceptibility to Rheumatoid Arthritis. Mol Med Rep (2013) 7(1):287–91. doi: 10.3892/mmr.2012.1176
27. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. A Polymorphism in the 3’-UTR of Interleukin-1 Receptor-Associated Kinase (IRAK1), a Target Gene of miR-146a, Is Associated With Rheumatoid Arthritis Susceptibility. Joint Bone Spine (2010) 77(5):411–3. doi: 10.1016/j.jbspin.2010.05.013
28. Zhang LL, Wu XX, Wang XF, Di DS, Huang Q, Liu RS, et al. Genetic Variant in microRNA-146a Gene Is Associated With Risk of Rheumatoid Arthritis. Ann Med (2021) 53(1):824–9. doi: 10.1080/07853890.2021.1933163
29. Toraih EA, Ismail NM, Toraih AA, Hussein MH, Fawzy MS. Precursor miR-499a Variant But Not miR-196a2 Is Associated With Rheumatoid Arthritis Susceptibility in an Egyptian Population. Mol Diagn Ther (2016) 20(3):279–95. doi: 10.1007/s40291-016-0194-3
30. Ehtesham N, Alani B, Mortazavi D, Azhdari S, Kenarangi T, Esmaeilzadeh E, et al. Association of Rs3135500 and Rs3135499 Polymorphisms in the MicroRNA-Binding Site of Nucleotide-Binding Oligomerization Domain 2 (NOD2) Gene With Susceptibility to Rheumatoid Arthritis. Iran J Allergy Asthma Immunol (2021) 20(2):178–87. doi: 10.18502/ijaai.v20i2.6051
31. Yang XK, Li P, Zhang C, Leng RX, Li S, Liu J, et al. Association Between IRAK1 Rs3027898 and miRNA-499 Rs3746444 Polymorphisms and Rheumatoid Arthritis: A Case Control Study and Meta-Analysis. Z Rheumatol (2017) 76(7):622–9. doi: 10.1007/s00393-016-0169-0
32. Zhou X, Zhu J, Zhang H, Zhou G, Huang Y, Liu R. Is the microRNA-146a (Rs2910164) Polymorphism Associated With Rheumatoid Arthritis? Association of microRNA-146a (Rs2910164) Polymorphism and Rheumatoid Arthritis Could Depend on Gender. Joint Bone Spine (2015) 82(3):166–71. doi: 10.1016/j.jbspin.2014.12.009
33. Guo S, Jin Y, Zhou J, Zhu Q, Jiang T, Bian Y, et al. MicroRNA Variants and HLA-miRNA Interactions Are Novel Rheumatoid Arthritis Susceptibility Factors. Front Genet (2099) 2021:12. doi: 10.3389/fgene.2021.747274
34. Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, et al. Altered microRNA Expression Profile With miR-146a Upregulation in CD4+ T Cells From Patients With Rheumatoid Arthritis. Arthritis Res Ther (2010) 12(3):R81. doi: 10.1186/ar3006
35. Gao J, Kong R, Zhou X, Ji L, Zhang J, Zhao D. MiRNA-126 Expression Inhibits IL-23R Mediated TNF-Alpha or IFN-Gamma Production in Fibroblast-Like Synoviocytes in a Mice Model of Collagen-Induced Rheumatoid Arthritis. Apoptosis (2018) 23(11-12):607–15. doi: 10.1007/s10495-018-1474-7
36. Zhou Q, Haupt S, Kreuzer JT, Hammitzsch A, Proft F, Neumann C, et al. Decreased Expression of miR-146a and miR-155 Contributes to an Abnormal Treg Phenotype in Patients With Rheumatoid Arthritis. Ann Rheum Dis (2015) 74(6):1265–74. doi: 10.1136/annrheumdis-2013-204377
37. Liu J, Fei D, Xing J, Du J, et al. MicroRNA-29a Inhibits Proliferation and Induces Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Repressing STAT3. BioMed Pharmacother (2017) 96:173–81. doi: 10.1016/j.biopha.2017.09.120
38. Tang X, Wang J, Xia X, Tian J, Rui K, Xu H, et al. Elevated Expression of ciRS-7 in Peripheral Blood Mononuclear Cells From Rheumatoid Arthritis Patients. Diagn Pathol (2019) 14(1):11. doi: 10.1186/s13000-019-0783-7
39. Zhu X, Wu L, Mo X, Xia W, Guo Y, Wang M, et al. Identification of PBMC-Expressed miRNAs for Rheumatoid Arthritis. Epigenetics (2020) 15(4):386–97. doi: 10.1080/15592294.2019.1676613
40. Wang X, Tang K, Wang Y, Chen Y, Yang M, Gu C, et al. Elevated Microrna1455p Increases Matrix Metalloproteinase9 by Activating the Nuclear factorkappaB Pathway in Rheumatoid Arthritis. Mol Med Rep (2019) 20(3):2703–11. doi: 10.3892/mmr.2019.10499
41. Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, et al. Altered Expression of microRNA-203 in Rheumatoid Arthritis Synovial Fibroblasts and its Role in Fibroblast Activation. Arthritis Rheum (2011) 63(2):373–81. doi: 10.1002/art.30115
42. de la Rica L, Urquiza JM, Gomez-Cabrero D, Islam AB, Lopez-Bigas N, Tegner J, et al. Identification of Novel Markers in Rheumatoid Arthritis Through Integrated Analysis of DNA Methylation and microRNA Expression. J Autoimmun (2013) 41:6–16. doi: 10.1016/j.jaut.2012.12.005
43. Hong BK, You S, Yoo SA, Park D, Hwang D, Cho CS, et al. MicroRNA-143 and -145 Modulate the Phenotype of Synovial Fibroblasts in Rheumatoid Arthritis. Exp Mol Med (2017) 49(8):e363. doi: 10.1038/emm.2017.108
44. Tseng CC, Wu LY, Tsai WC, Ou TC, Wu CC, Sung WY, et al. Differential Expression Profiles of the Transcriptome and miRNA Interactome in Synovial Fibroblasts of Rheumatoid Arthritis Revealed by Next Generation Sequencing. Diagn (Basel) (2019) 9(3). doi: 10.3390/diagnostics9030098
45. Wangyang Y, Yi L, Wang T, Feng Y, Liu G, Li D, et al. MiR-199a-3p Inhibits Proliferation and Induces Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes via Suppressing Retinoblastoma 1. Biosci Rep (2018) 38(6). doi: 10.1042/BSR20180982
46. Guo J, Cao X, Zhao W, Zhu H, Ma X, Hao C, et al. MicroRNA-449 Targets Histone Deacetylase 1 to Regulate the Proliferation, Invasion, and Apoptosis of Synovial Fibroblasts in Rheumatoid Arthritis. Ann Palliat Med (2021) 10(7):7960–9. doi: 10.21037/apm-21-1383
47. Wang Y, Zhang K, Yuan X, Xu N, Zhao S, Hou L, et al. miR-431-5p Regulates Cell Proliferation and Apoptosis in Fibroblast-Like Synoviocytes in Rheumatoid Arthritis by Targeting XIAP. Arthritis Res Ther (2020) 22(1):231. doi: 10.1186/s13075-020-02328-3
48. Wang Y, Hou L, Yuan X, Xu N, Zhao S, Yang L, et al. miR-483-3p Promotes Cell Proliferation and Suppresses Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Targeting IGF-1. BioMed Pharmacother (2020) 130:110519. doi: 10.1016/j.biopha.2020.110519
49. Nakamachi Y, Kawano S, Takenokuchi M, Nishimura K, Sakai Y, Chin T, et al. MicroRNA-124a Is a Key Regulator of Proliferation and Monocyte Chemoattractant Protein 1 Secretion in Fibroblast-Like Synoviocytes From Patients With Rheumatoid Arthritis. Arthritis Rheum (2009) 60(5):1294–304. doi: 10.1002/art.24475
50. Gao B, Sun G, Wang Y, Geng Y, Zhou L, Chen X, et al. microRNA-23 Inhibits Inflammation to Alleviate Rheumatoid Arthritis via Regulating CXCL12. Exp Ther Med (2021) 21(5):459. doi: 10.3892/etm.2021.9890
51. Sun W, Zhang Y, Wang G. MicroRNA-137-Mediated Inhibition of Lysine-Specific Demethylase-1 Prevents Against Rheumatoid Arthritis in an Association With the REST/mTOR Axis. Mol Pain (2021) 17:17448069211041847. doi: 10.1177/17448069211041847
52. Bao X, Ma L, He C. MicroRNA-23a-5p Regulates Cell Proliferation, Migration and Inflammation of TNF-α-Stimulated Human Fibroblast-Like MH7A Synoviocytes by Targeting TLR4 in Rheumatoid Arthritis. Exp Ther Med (2021) 21(5):479. doi: 10.3892/etm.2021.9910
53. Chen L, Lu Q, Chen J, Feng R, Yang C. Upregulating miR-27a-3p Inhibits Cell Proliferation and Inflammation of Rheumatoid Arthritis Synovial Fibroblasts Through Targeting Toll-Like Receptor 5. Exp Ther Med (2021) 22(5):1227. doi: 10.3892/etm.2021.10661
54. Lin J, Huo R, Xiao L, Zhu X, Xie J, Sun S, et al. A Novel P53/microRNA-22/Cyr61 Axis in Synovial Cells Regulates Inflammation in Rheumatoid Arthritis. Arthritis Rheumatol (2014) 66(1):49–59. doi: 10.1002/art.38142
55. Meng Q, Pan B, Sheng P. Histone Deacetylase 1 Is Increased in Rheumatoid Arthritis Synovium and Promotes Synovial Cell Hyperplasia and Synovial Inflammation in the Collagen-Induced Arthritis Mouse Model via the microRNA-124-Dependent MARCKS-JAK/STAT Axis. Clin Exp Rheumatol (2021) 39(5):970–81.
56. Song AF, Kang L, Wang YF, Wang M. MiR-34a-5p Inhibits Fibroblast−Like Synoviocytes Proliferation via XBP1. Eur Rev Med Pharmacol Sci (2020) 24(22):11675–82. doi: 10.26355/eurrev_202011_23812
57. Tolboom TC, Pieterman E, van der Laan WH, Toes RE, Huidekoper AL, Nelissen RG, et al. Invasive Properties of Fibroblast-Like Synoviocytes: Correlation With Growth Characteristics and Expression of MMP-1, MMP-3, and MMP-10. Ann Rheum Dis (2002) 61(11):975–80. doi: 10.1136/ard.61.11.975
58. Jiang C, Wu X, Li X, Li M, Zhang W, Tao P, et al. Loss of microRNA-147 Function Alleviates Synovial Inflammation Through ZNF148 in Rheumatoid and Experimental Arthritis. Eur J Immunol (2021) 51(8):2062–73. doi: 10.1002/eji.202048850
59. Shi DL, Shi GR, Xie J, Du XZ, Yang H. MicroRNA-27a Inhibits Cell Migration and Invasion of Fibroblast-Like Synoviocytes by Targeting Follistatin-Like Protein 1 in Rheumatoid Arthritis. Mol Cells (2016) 39(8):611–8. doi: 10.14348/molcells.2016.0103
60. Stanczyk J, Pedrioli DM, Brentano F, Sanchez-Pernaute O, Kolling C, Gay RE, et al. Altered Expression of MicroRNA in Synovial Fibroblasts and Synovial Tissue in Rheumatoid Arthritis. Arthritis Rheum (2008) 58(4):1001–9. doi: 10.1002/art.23386
61. Long L, Yu P, Liu Y, Wang S, Li R, Shi J, et al. Upregulated microRNA-155 Expression in Peripheral Blood Mononuclear Cells and Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. Clin Dev Immunol (2013) 2013p:296139. doi: 10.1155/2013/296139
62. Okamoto K, Nakashima T, Shinohara M, Negishi-Koga T, Komatsu N, Terashima A, et al. Osteoimmunology: The Conceptual Framework Unifying the Immune and Skeletal Systems. Physiol Rev (2017) 97(4):1295–349. doi: 10.1152/physrev.00036.2016
63. Rana AK, Li Y, Dang Q, Yang F, et al. Monocytes in Rheumatoid Arthritis: Circulating Precursors of Macrophages and Osteoclasts and, Their Heterogeneity and Plasticity Role in RA Pathogenesis. Int Immunopharmacol (2018) 65:348–59. doi: 10.1016/j.intimp.2018.10.016
64. Najm A, Masson FM, Preuss P, Georges S, Ory B, Quillard T, et al. MicroRNA-17-5p Reduces Inflammation and Bone Erosions in Mice With Collagen-Induced Arthritis and Directly Targets the JAK/STAT Pathway in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Arthritis Rheumatol (2020) 72(12):2030–9. doi: 10.1002/art.41441
65. Maeda Y, Farina NH, Matzelle MM, Fanning PJ, Lian JB, Gravallese EM. Synovium-Derived MicroRNAs Regulate Bone Pathways in Rheumatoid Arthritis. J Bone Miner Res (2017) 32(3):461–72. doi: 10.1002/jbmr.3005
66. Iwamoto N, Fukui S, Takatani A, Shimizu T, Umeda M, Nishino A, et al. Osteogenic Differentiation of Fibroblast-Like Synovial Cells in Rheumatoid Arthritis Is Induced by microRNA-218 Through a ROBO/Slit Pathway. Arthritis Res Ther (2018) 20(1):189. doi: 10.1186/s13075-018-1703-z
67. Kong XH, Shi SF, Hu HJ, Wang JX. MicroRNA-20a Suppresses RANKL-Modulated Osteoclastogenesis and Prevents Bone Erosion in Mice With Rheumatoid Arthritis Through the TLR4/p38 Pathway. J Biol Regul Homeost Agents (2021) 35(3):921–31. doi: 10.23812/20-604-a
68. Li HW, Zeng HS. Regulation of JAK/STAT Signal Pathway by miR-21 in the Pathogenesis of Juvenile Idiopathic Arthritis. World J Pediatr (2020) 16(5):502–13. doi: 10.1007/s12519-019-00268-w
69. Ren B, Liu J, Wu K, Zhang J, Lv Y, Wang S, et al. TNF-Alpha-Elicited miR-29b Potentiates Resistance to Apoptosis in Peripheral Blood Monocytes From Patients With Rheumatoid Arthritis. Apoptosis (2019) 24(11-12):892–904. doi: 10.1007/s10495-019-01567-3
70. Niimoto T, Nakasa T, Ishikawa M, Okuhara A, Izumi B, Deie M, et al. MicroRNA-146a Expresses in Interleukin-17 Producing T Cells in Rheumatoid Arthritis Patients. BMC Musculoskelet Disord (2010) 11:209. doi: 10.1186/1471-2474-11-209
71. Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 Is Overexpressed in T-Lymphocytes of Patients Affected by Rheumatoid Arthritis. Hum Immunol (2010) 71(2):206–11. doi: 10.1016/j.humimm.2009.11.008
72. van Hamburg JP, Tas SW. Molecular Mechanisms Underpinning T Helper 17 Cell Heterogeneity and Functions in Rheumatoid Arthritis. J Autoimmun (2018) 87:69–81. doi: 10.1016/j.jaut.2017.12.006
73. Jin S, Sun S, Ling H, Ma J, Zhang X, Xie Z, et al. Protectin DX Restores Treg/T(h)17 Cell Balance in Rheumatoid Arthritis by Inhibiting NLRP3 Inflammasome via miR-20a. Cell Death Dis (2021) 12(3):280. doi: 10.1038/s41419-021-03562-6
74. Dong L, Wang X, Tan J, Li H, Qian W, Chen J, et al. Decreased Expression of microRNA-21 Correlates With the Imbalance of Th17 and Treg Cells in Patients With Rheumatoid Arthritis. J Cell Mol Med (2014) 18(11):2213–24. doi: 10.1111/jcmm.12353
75. Guggino G, Orlando V, Saieva L, Ruscitti P, Cipriani P, La Manna MP, et al. Downregulation of Mirna17-92 Cluster Marks Vgamma9Vdelta2 T Cells From Patients With Rheumatoid Arthritis. Arthritis Res Ther (2018) 20(1):236. doi: 10.1186/s13075-018-1740-7
76. Huang Q, Chen SS, Li J, Tao SS, Wang M, Leng RX, et al. miR-210 Expression in PBMCs From Patients With Systemic Lupus Erythematosus and Rheumatoid Arthritis. Ir J Med Sci (2018) 187(1):243–9. doi: 10.1007/s11845-017-1634-8
77. Smigielska-Czepiel K, van den Berg A, Jellema P, van der Lei RJ, Bijzet J, Kluiver J, et al. Comprehensive Analysis of miRNA Expression in T-Cell Subsets of Rheumatoid Arthritis Patients Reveals Defined Signatures of Naive and Memory Tregs. Genes Immun (2014) 15(2):115–25. doi: 10.1038/gene.2013.69
78. Yan S, Wang P, Wang J, Yang J, Lu H, Jin C, et al. Long Non-Coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-kappaB Signaling Pathway in Rheumatoid Arthritis. Front Immunol (2019) 10:2218. doi: 10.3389/fimmu.2019.02218
79. Xie Q, Wei M, Zhang B, Kang X, Liu D, Zheng W, et al. MicroRNA33 Regulates the NLRP3 Inflammasome Signaling Pathway in Macrophages. Mol Med Rep (2018) 17(2):3318–27. doi: 10.3892/mmr.2017.8224
80. Meng HY, Chen LQ, Chen LH. The Inhibition by Human MSCs-Derived miRNA-124a Overexpression Exosomes in the Proliferation and Migration of Rheumatoid Arthritis-Related Fibroblast-Like Synoviocyte Cell. BMC Musculoskelet Disord (2020) 21(1):150. doi: 10.1186/s12891-020-3159-y
81. Wu H, Zhou X, Wang X, Cheng W, Hu X, Wang Y, et al. miR-34a in Extracellular Vesicles From Bone Marrow Mesenchymal Stem Cells Reduces Rheumatoid Arthritis Inflammation via the Cyclin I/ATM/ATR/p53 Axis. J Cell Mol Med (2021) 25(4):1896–910. doi: 10.1111/jcmm.15857
82. Li GQ, Fang YX, Liu Y, Meng FR, Wu X, Zhang CW, et al. MicroRNA-21 From Bone Marrow Mesenchymal Stem Cell-Derived Extracellular Vesicles Targets TET1 to Suppress KLF4 and Alleviate Rheumatoid Arthritis. Ther Adv Chronic Dis (2021) 12:20406223211007369. doi: 10.1177/20406223211007369
83. Hu R, Lv W, Zhang S, Liu Y, Sun B, Meng Y, et al. Combining miR-23b Exposure With Mesenchymal Stem Cell Transplantation Enhances Therapeutic Effects on EAE. Immunol Lett (2021) 229:18–26. doi: 10.1016/j.imlet.2020.11.007
84. Dinesh P, Kalaiselvan S, Sujitha S, Rasool M. miR-506-3p Alleviates Uncontrolled Osteoclastogenesis via Repression of RANKL/NFATc1 Signaling Pathway. J Cell Physiol (2020) 235(12):9497–509. doi: 10.1002/jcp.29757
85. Chen M, Li MH, Zhang N, Sun WW, Wang H, Wang YA, et al. Pro-Angiogenic Effect of Exosomal microRNA-103a in Mice With Rheumatoid Arthritis via the Downregulation of Hepatocyte Nuclear Factor 4 Alpha and Activation of the JAK/STAT3 Signaling Pathway. J Biol Regul Homeost Agents (2021) 35(2):629–40. doi: 10.23812/20-537-a
86. Donate PB, Alves de Lima K, Peres RS, Almeida F, Fukada SY, Silva TA, et al. Cigarette Smoke Induces miR-132 in Th17 Cells That Enhance Osteoclastogenesis in Inflammatory Arthritis. Proc Natl Acad Sci USA (2021) 118(1). doi: 10.1073/pnas.2017120118
87. Hegewald AB, Breitwieser K, Ottinger SM, Mobarrez F, Korotkova M, Rethi B, et al. Extracellular miR-574-5p Induces Osteoclast Differentiation via TLR 7/8 in Rheumatoid Arthritis. Front Immunol (2020) 11:585282. doi: 10.3389/fimmu.2020.585282
88. Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic Potential of Mesenchymal Cell-Derived miRNA-150-5p-Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF. J Immunol (2018) 201(8):2472–82. doi: 10.4049/jimmunol.1800304
89. Kurowska W, Kuca-Warnawin E, Radzikowska A, Jakubaszek M, Maślińska M, Kwiatkowska B, et al. Monocyte-Related Biomarkers of Rheumatoid Arthritis Development in Undifferentiated Arthritis Patients - A Pilot Study. Reumatologia (2018) 56(1):10–6. doi: 10.5114/reum.2018.74742
90. Ouboussad L, Hunt L, Hensor EMA, Nam JL, Barnes NA, Emery P, et al. Profiling microRNAs in Individuals at Risk of Progression to Rheumatoid Arthritis. Arthritis Res Ther (2017) 19(1):288. doi: 10.1186/s13075-017-1492-9
91. Romo-García MF, Bastian Y, Zapata-Zuñiga M, Macías-Segura N, Castillo-Ortiz JD, Lara-Ramírez EE, et al. Identification of Putative miRNA Biomarkers in Early Rheumatoid Arthritis by Genome-Wide Microarray Profiling: A Pilot Study. Gene (2019) 720:144081. doi: 10.1016/j.gene.2019.144081
92. Anaparti V, Smolik I, Meng X, Spicer V, Mookherjee N, El-Gabalawy H, et al. Whole Blood microRNA Expression Pattern Differentiates Patients With Rheumatoid Arthritis, Their Seropositive First-Degree Relatives, and Healthy Unrelated Control Subjects. Arthritis Res Ther (2017) 19(1):249. doi: 10.1186/s13075-017-1459-x
93. Yue J, Lau TCK, Griffith JF, Xu J, Xiao F, Shi L, et al. Circulating miR-99b-5p as a Novel Predictor of Erosion Progression on High-Resolution Peripheral Quantitative Computed Tomography in Early Rheumatoid Arthritis: A Prospective Cohort Study. Int J Rheum Dis (2019) 22(9):1724–33. doi: 10.1111/1756-185X.13644
94. Evangelatos G, Fragoulis GE, Koulouri V, Lambrou GI. MicroRNAs in Rheumatoid Arthritis: From Pathogenesis to Clinical Impact. Autoimmun Rev (2019) 18(11):102391. doi: 10.1016/j.autrev.2019.102391
95. Mookherjee N, El-Gabalawy HS. High Degree of Correlation Between Whole Blood and PBMC Expression Levels of miR-155 and miR-146a in Healthy Controls and Rheumatoid Arthritis Patients. J Immunol Methods (2013) 400-401:106–10. doi: 10.1016/j.jim.2013.10.001
96. Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M, et al. Comprehensive microRNA Analysis Identifies miR-24 and miR-125a-5p as Plasma Biomarkers for Rheumatoid Arthritis. PloS One (2013) 8(7):e69118. doi: 10.1371/journal.pone.0069118
97. Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM, et al. Utility of Select Plasma MicroRNA for Disease and Cardiovascular Risk Assessment in Patients With Rheumatoid Arthritis. J Rheumatol (2015) 42(10):1746–51. doi: 10.3899/jrheum.150232
98. Wang W, Zhang Y, Zhu B, Duan T, Xu Q, Wang R, et al. Plasma microRNA Expression Profiles in Chinese Patients With Rheumatoid Arthritis. Oncotarget (2015) 6(40):42557–68. doi: 10.18632/oncotarget.6449
99. Abou-Zeid A, Saad M, Soliman E. MicroRNA 146a Expression in Rheumatoid Arthritis: Association With Tumor Necrosis Factor-Alpha and Disease Activity. Genet Test Mol Biomarkers (2011) 15(11):807–12. doi: 10.1089/gtmb.2011.0026
100. Ciechomska M, Wojtas B, Bonek K, Roszkowski L, Gluszko P, Benes V, et al. Comprehensive microRNA and Transcriptomic Profiling of Rheumatoid Arthritis Monocytes: Role of microRNA-146b in Proinflammatory Progression. Rheumatol (Oxford) (2021). doi: 10.1093/rheumatology/keab407
101. Ormseth MJ, Solus JF, Sheng Q, Ye F, Song H, Wu Q, et al. The Endogenous Plasma Small RNAome of Rheumatoid Arthritis. ACR Open Rheumatol (2020) 2(2):97–105. doi: 10.1002/acr2.11098
102. Liu C, Pan A, Chen X, Tu J, Xia X, Sun L, et al. MiR-5571-3p and miR-135b-5p, Derived From Analyses of microRNA Profile Sequencing, Correlate With Increased Disease Risk and Activity of Rheumatoid Arthritis. Clin Rheumatol (2019) 38(6):1753–65. doi: 10.1007/s10067-018-04417-w
103. Khalifa O, Pers YM, Ferreira R, Senechal A, Jorgensen C, Apparailly F, et al. X-Linked miRNAs Associated With Gender Differences in Rheumatoid Arthritis. Int J Mol Sci (2016) 17(11). doi: 10.3390/ijms17111852
104. Fernández-Ruiz JC, Ramos-Remus C, Sánchez-Corona J, Castillo-Ortiz JD, Castañeda-Sánchez JJ, Bastian Y, et al. Analysis of miRNA Expression in Patients With Rheumatoid Arthritis During Remission and Relapse After a 5-Year Trial of Tofacitinib Treatment. Int Immunopharmacol (2018) 63:35–42. doi: 10.1016/j.intimp.2018.07.028
105. Liu X, Ni S, Li C, Xu N, Chen W, Wu M, et al. Circulating microRNA-23b as a New Biomarker for Rheumatoid Arthritis. Gene (2019) 712:143911. doi: 10.1016/j.gene.2019.06.001
106. Ormseth MJ, Solus JF, Sheng Q, Ye F, Wu Q, Guo Y, et al. Development and Validation of a MicroRNA Panel to Differentiate Between Patients With Rheumatoid Arthritis or Systemic Lupus Erythematosus and Controls. J Rheumatol (2020) 47(2):188–96. doi: 10.3899/jrheum.181029
107. Abdelaleem OO, Fouad NA, Shaker OG, Ahmed TI, Abdelghaffar NK, Eid HM, et al. Serum miR-224, miR-760, miR-483-5p, miR-378 and miR-375 as Potential Novel Biomarkers in Rheumatoid Arthritis. Int J Clin Pract (2021) p:e14651. doi: 10.1111/ijcp.14651
108. Hong H, Yang H, Xia Y. Circulating miR-10a as Predictor of Therapy Response in Rheumatoid Arthritis Patients Treated With Methotrexate. Curr Pharm Biotechnol (2018) 19(1):79–86. doi: 10.2174/1389201019666180417155140
109. Sode J, Krintel SB, Carlsen AL, Hetland ML, Johansen JS, Hørslev-Petersen K, et al. Plasma MicroRNA Profiles in Patients With Early Rheumatoid Arthritis Responding to Adalimumab Plus Methotrexate vs Methotrexate Alone: A Placebo-Controlled Clinical Trial. J Rheumatol (2018) 45(1):53–61. doi: 10.3899/jrheum.170266
110. Ciechomska M, Bonek K, Merdas M, Zarecki P, Swierkot J, Gluszko P, et al. Changes in MiRNA-5196 Expression as a Potential Biomarker of Anti-TNF-Alpha Therapy in Rheumatoid Arthritis and Ankylosing Spondylitis Patients. Arch Immunol Ther Exp (Warsz) (2018) 66(5):389–97. doi: 10.1007/s00005-018-0513-y
111. Castro-Villegas C, Perez-Sanchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limon P, et al. Circulating miRNAs as Potential Biomarkers of Therapy Effectiveness in Rheumatoid Arthritis Patients Treated With Anti-TNFalpha. Arthritis Res Ther (2015) 17:49. doi: 10.1186/s13075-015-0555-z
112. Liu Y, Han Y, Qu H, Fang J, Ye M, Yin W, et al. Correlation of microRNA Expression Profile With Clinical Response to Tumor Necrosis Factor Inhibitor in Treating Rheumatoid Arthritis Patients: A Prospective Cohort Study. J Clin Lab Anal (2019) 33(7):e22953. doi: 10.1002/jcla.22953
113. Duroux-Richard I, Pers YM, Fabre S, Ammari M, Baeten D, Cartron G, et al. Circulating miRNA-125b Is a Potential Biomarker Predicting Response to Rituximab in Rheumatoid Arthritis. Mediators Inflamm 2014 (2014) p:342524. doi: 10.1155/2014/342524
114. Perez-Sanchez C, Cecchi I, Barbarroja N, Patino-Trives AM, Luque-Tevar M, Perez-Sanchez L, et al. Early Restoration of Immune and Vascular Phenotypes in Systemic Lupus Erythematosus and Rheumatoid Arthritis Patients After B Cell Depletion. J Cell Mol Med (2019) 23(9):6308–18. doi: 10.1111/jcmm.14517
115. Zhang Y, Wang H, Mao X, Guo Q, Li W, Wang X, et al. A Novel Circulating miRNA-Based Model Predicts the Response to Tripterysium Glycosides Tablets: Moving Toward Model-Based Precision Medicine in Rheumatoid Arthritis. Front Pharmacol (2018) 9:378. doi: 10.3389/fphar.2018.00378
116. Wang XY, Wang HL, Mao X, Li GY, Guo QY, Li WJ, et al. [Identification of Biomarkers to Response of Tripterygium Glycosides Tablets Acting on Rheumatoid Arthritis by Integrating Transcriptional Data Mining and Biomolecular Network Analysis]. Zhongguo Zhong Yao Za Zhi (2019) 44(16):3415–22. doi: 10.19540/j.cnki.cjcmm.20181031.001
117. Wang Y, Dai L, Wu H, Zhang ZR, Wang WY, Fu J, et al. Novel Anti-Inflammatory Target of Geniposide: Inhibiting Itgbeta1/Ras-Erk1/2 Signal Pathway via the miRNA-124a in Rheumatoid Arthritis Synovial Fibroblasts. Int Immunopharmacol (2018) 65:284–94. doi: 10.1016/j.intimp.2018.09.049
118. Cai Y, Jiang C, Zhu J, Xu K, Ren X, Xu L, et al. miR-449a Inhibits Cell Proliferation, Migration, and Inflammation by Regulating High-Mobility Group Box Protein 1 and Forms a Mutual Inhibition Loop With Yin Yang 1 in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Arthritis Res Ther (2019) 21(1):134. doi: 10.1186/s13075-019-1920-0
119. Wu J, Fan W, Ma L, Geng X. miR-708-5p Promotes Fibroblast-Like Synoviocytes’ Cell Apoptosis and Ameliorates Rheumatoid Arthritis by the Inhibition of Wnt3a/beta-Catenin Pathway. Drug Des Devel Ther (2018) 12:3439–47. doi: 10.2147/DDDT.S177128
120. Mallinson DJ, Dunbar DR, Ridha S, Sutton ER, De la Rosa O, Dalemans W, et al. Identification of Potential Plasma microRNA Stratification Biomarkers for Response to Allogeneic Adipose-Derived Mesenchymal Stem Cells in Rheumatoid Arthritis. Stem Cells Transl Med (2017) 6(4):1202–6. doi: 10.1002/sctm.16-0356
121. Oka S, Furukawa H, Shimada K, Hashimoto A, Komiya A, Fukui N, et al. Plasma miRNA Expression Profiles in Rheumatoid Arthritis Associated Interstitial Lung Disease. BMC Musculoskelet Disord (2017) 18(1):21. doi: 10.1186/s12891-017-1389-4
122. Li Z, Li Y, Li Q, Zhang Z, Jiang L, Li X, et al. Role of miR-9-5p in Preventing Peripheral Neuropathy in Patients With Rheumatoid Arthritis by Targeting REST/miR-132 Pathway. In Vitro Cell Dev Biol Anim (2019) 55(1):52–61. doi: 10.1007/s11626-018-0310-2
Keywords: rheumatoid arthritis, microRNA, susceptibility, pathogenesis, epigenetics
Citation: Chang C, Xu L, Zhang R, Jin Y, Jiang P, Wei K, Xu L, Shi Y, Zhao J, Xiong M, Guo S and He D (2022) MicroRNA-Mediated Epigenetic Regulation of Rheumatoid Arthritis Susceptibility and Pathogenesis. Front. Immunol. 13:838884. doi: 10.3389/fimmu.2022.838884
Received: 18 December 2021; Accepted: 02 March 2022;
Published: 24 March 2022.
Edited by:
Zhangran Chen, Xiamen University, ChinaReviewed by:
Zhigang Lu, Nanjing University of Chinese Medicine, ChinaCopyright © 2022 Chang, Xu, Zhang, Jin, Jiang, Wei, Xu, Shi, Zhao, Xiong, Guo and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shicheng Guo, c2hpY2hlbmcuZ3VvQHdpc2MuZWR1; Dongyi He, aGVkb25neWkxOTY3QHNodXRjbS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.