- 1Curandis, New York, NY, United States
- 2Department of Gastroenterology, Affiliated Hospital of Guizhou Medical University, Guiyang, China
- 3Department of Pathology and Laboratory Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, United States
- 4Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY, United States
COVID-19 is accompanied by a myriad of both transient and long-lasting autoimmune responses. Dermatan sulfate (DS), a glycosaminoglycan crucial for wound healing, has unique affinity for autoantigens (autoAgs) from apoptotic cells. DS-autoAg complexes are capable of stimulating autoreactive B cells and autoantibody production. We used DS-affinity proteomics to define the autoantigen-ome of lung fibroblasts and bioinformatics analyses to study the relationship between autoantigenic proteins and COVID-induced alterations. Using DS-affinity, we identified an autoantigen-ome of 408 proteins from human HFL1 cells, at least 231 of which are known autoAgs. Comparing with available COVID data, 352 proteins of the autoantigen-ome have thus far been found to be altered at protein or RNA levels in SARS-CoV-2 infection, 210 of which are known autoAgs. The COVID-altered proteins are significantly associated with RNA metabolism, translation, vesicles and vesicle transport, cell death, supramolecular fibrils, cytoskeleton, extracellular matrix, and interleukin signaling. They offer clues to neurological problems, fibrosis, smooth muscle dysfunction, and thrombosis. In particular, 150 altered proteins are related to the nervous system, including axon, myelin sheath, neuron projection, neuronal cell body, and olfactory bulb. An association with the melanosome is also identified. The findings from our study illustrate a connection between COVID infection and autoimmunity. The vast number of COVID-altered proteins with high intrinsic propensity to become autoAgs offers an explanation for the diverse autoimmune complications in COVID patients. The variety of autoAgs related to mRNA metabolism, translation, and vesicles suggests a need for long-term monitoring of autoimmunity in COVID. The COVID autoantigen atlas we are establishing provides a detailed molecular map for further investigation of autoimmune sequelae of the pandemic, such as “long COVID” syndrome.
Summary Sentence: An autoantigen-ome by dermatan sulfate affinity from human lung HFL1 cells may explain neurological and autoimmune manifestations of COVID-19.
Introduction
The emergence of the novel coronavirus SARS-CoV-2 has dragged the world into a prolonged pandemic. Aside from the intensively studied ACE2, heparan sulfate is another crucial entry receptor for coronaviruses (1). Dermatan sulfate (DS), structurally and functionally similar to heparan sulfate and heparin, belongs to the glycosaminoglycan family. Many viruses, including Ebola, Vaccinia, Zika, Dengue, and Hepatitis C viruses, have been shown to interact with glycosaminoglycans (2–5). These polyanionic polysaccharides consist of disaccharide repeating units of amino sugars and uronic acids with varying degrees of sulfation. Glycosaminoglycans are major components of the extracellular matrix and basement membrane, act as a filler between cells and tissue fibers and have numerous biological functions.
DS is most abundant in the skin but is also found in lungs, blood vessels, heart valves, and tendons. DS plays important roles in cell death, wound healing, and tissue repair. In human wound fluid, DS is the most abundant glycosaminoglycan (6). Its biosynthesis is increased by fibroblasts, epithelial cells, and capillary endothelial cells in wounded skin, mucosal ulcers, and inflammation-associated angiogenesis (7–9). Its molecular size also changes during wound healing, with elongated DS polymers packing along thin collagen fibrils in wounded skin (10). After tissue injury, fibroblasts require DS to migrate from the stroma surrounding the injury into the fibrin-laden wound to facilitate granulation tissue formation and wound healing (11).
DS is also a key molecule in autoimmunity, as we have discovered (12–16). DS is the most potent among glycosaminoglycans in stimulating autoreactive B1 cells and autoantibody production (12, 13). DS has a peculiar affinity to apoptotic cells and their released autoantigens (autoAgs), and macromolecular autoAg-DS affinity complexes are capable of engaging autoBCRs in a dual signaling event to activate B1 cells (13, 14). Recently, we also found that DS may steer autoreactive B1 cell fate at the pre-B stage by regulating the immunoglobulin heavy chain of the precursor BCR (17). Our studies illustrate a unifying property of autoAgs, i.e., self-molecules with DS affinity have a high propensity to become autoAgs, which explains how seemingly unrelated self-molecules can all induce humoral autoimmunity via similar immunological signaling events. In support of our hypothesis and by using DS affinity, we have cataloged hundreds of classic and novel autoAgs (14–16, 18).
A diverse spectrum of autoimmune symptoms has been observed in COVID-19 patients, including autoimmune cytopenia, multisystem inflammatory syndrome in children, immune-mediated neurological syndromes, Guillain-Barré syndrome, connective tissue disease-associated interstitial lung disease, antiphospholipid syndrome, autoimmune hemolytic anemia, autoimmune encephalitis, systemic lupus erythematosus, optic neuritis and myelitis, and acquired hemophilia (19–26). Many autoantibodies have been identified in COVID patients, including ANA (antinuclear antibody), ENA (extractable nuclear antigen), ANCA (anti-neutrophil cytoplasmic antibody), lupus anticoagulant, antiphospholipid, anti-IFN, anti-myelin oligodendrocyte glycoprotein, and anti-heparin-PF4 complex (19–27).
To understand autoimmune sequelae of COVID, we aimed to establish a COVID autoantigen atlas that will serve as a molecular map to guide ongoing research into autoimmune sequelae of COVID (such as “long COVID” syndrome) and vaccine evaluation. Acute COVID leads to significant acute inflammatory lung injury and histologic remodeling that involves marked lung fibroblast activation and cell turnover (28). In this study, we identified an autoantigen-ome of 408 proteins from human fetal lung fibroblast HFL1 cells by DS-affinity fractionation and protein sequencing, with at least 231 being known autoAgs. We then compared these with currently available data from SARS-CoV-2-infected patients and cells (as of 12/14/2020 in Coronascape) (29–49). Remarkably, 352 (86.3%) of these proteins have been found to be altered (up- or down-regulated) at protein and/or RNA expression levels, and 210 of the COVID-altered proteins are known autoAgs in a great variety of autoimmune diseases and cancers. The COVID-altered proteins reveal intricate host responses to the viral infection and point to close associations with diverse disease manifestations of COVID-19.
Results and Discussion
An Autoantigen-Ome of 408 Proteins With DS-Affinity From HFL1 Cells
Proteins extracted from HFL1 cells were fractionated with DS-affinity resins. The DS-binding fraction eluting with 0.5 M NaCl yielded 306 proteins by mass spectrometry sequencing, corresponding to proteins with medium-to-strong DS affinity. The fraction eluting with 1.0 M NaCl yielded 121 proteins, corresponding to proteins with very strong DS affinity. After excluding redundancies, a total of 408 unique proteins were obtained (Table 1). To verify how many of these proteins are known autoAgs, we conducted an extensive literature search for autoantibodies specific for each protein. Remarkably, at least 231 (57%) of our DS-affinity proteins already have known associated specific autoantibodies in various diseases and are thus confirmed autoAgs, corresponding to 61% of proteins with very strong DS affinity and 54% of proteins with medium-to-strong DS affinity (see references in Table 1).
Of those not yet confirmed as autoAgs, a majority are similar to known autoAgs. As an example, we identified 18 ribosomal proteins, of which 9 have been individually identified as autoAgs (Table 1); however, anti-ribosomal autoantibodies are reported to react with a heterogeneous pool of many ribosomal proteins (206). Therefore, many of the ribosomal proteins we identified may be true but yet-to-be-confirmed autoAgs. As another example, autoantibodies against the 20S proteasome core are reported to be polyspecific and react with many subunits (250). Thus, although only 7 of 15 proteasome proteins we identified are thus far individually confirmed, the remainder may be true but yet-to-be-specified autoAgs. Similarly, some members of eukaryotic translation initiation and elongation factors are confirmed autoAgs, while others await confirmation. In summary, the putative autoantigen-ome from HFL1 cells provides at least 231 confirmed and 177 yet-to-confirm putative autoAgs (Table 1).
DS-Affinity Proteins Are Functionally Connected and Enriched
To find out whether DS-affinity-associated proteins are a random collection or biologically connected, we performed protein-protein interaction analyses with STRING (251). Of the 408 DS-associated proteins, 405 proteins recognized by STRING (ANP32C, ANXA2P2, HSP90AA2 excluded) have 7,582 interactions, whereas a random set of 405 proteins is expected to have only 3,060 interactions; hence, DS-affinity proteins represent a significantly connected network with PPI enrichment p-value <1.0E-6 (Figure 1). Based on cellular component classification, these proteins are highly concentrated in the nucleus (226 proteins), vesicles (111 proteins), ribonucleoprotein complexes (95 proteins), and the cytoskeleton (95 proteins).
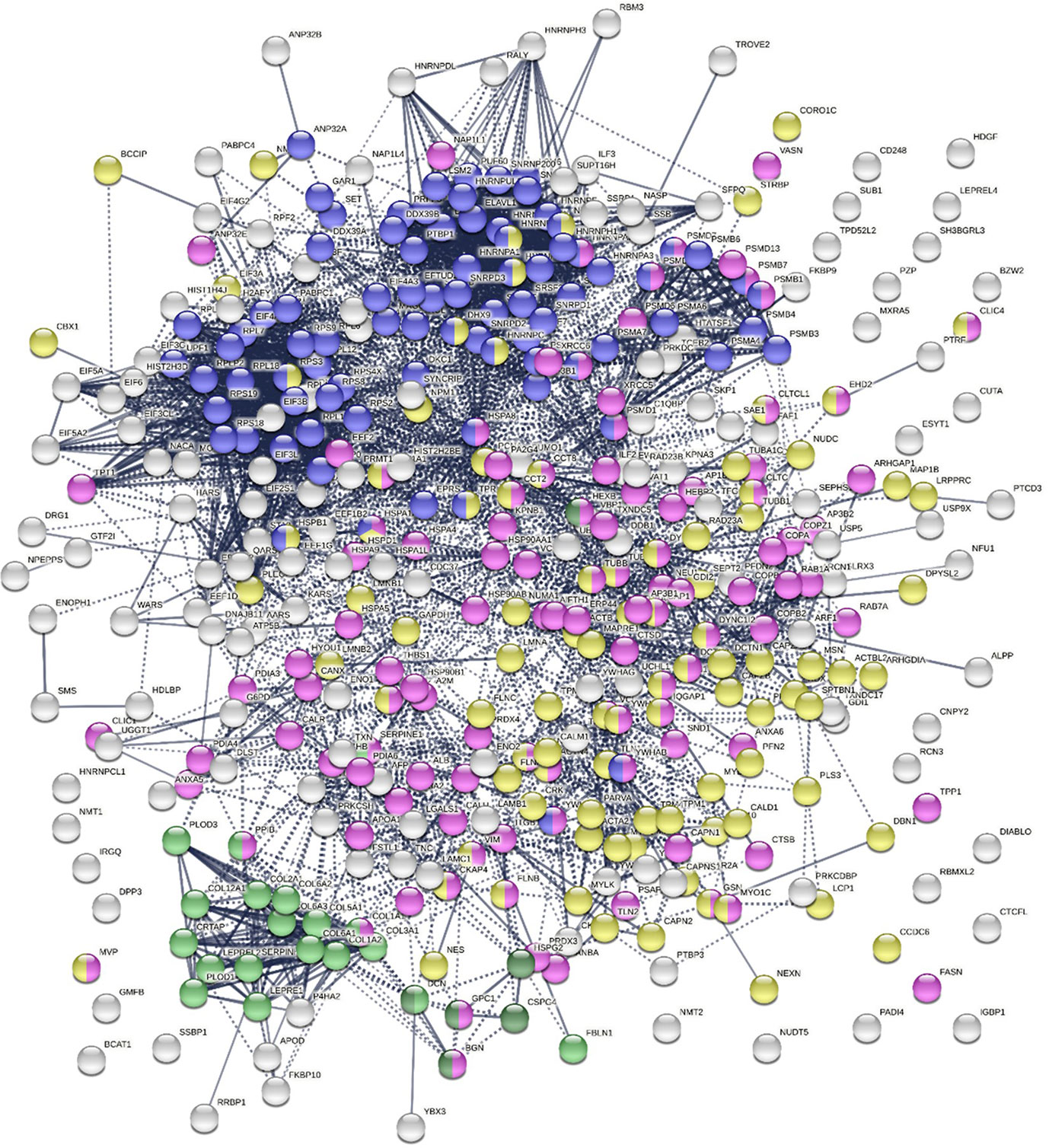
Figure 1 The 408-protein autoantigen-ome identified by DS-affinity from HFL1 cells forms a highly interacting network. Connecting lines represent interactions with high confidence (minimum interaction score of 0.7) as per STRING analysis. Colored proteins are involved in metabolism of RNA (blue), vesicles (pink), cytoskeleton (gold), collagen and elastic fibers (light green), and chondroitin sulfate/dermatan sulfate metabolism (dark green).
Pathway and process analyses by STRING and Metascape (29) revealed that the mRNA metabolic process is the most enriched GO Biological Process, and the top KEGG pathways are the spliceosome and protein processing in the endoplasmic reticulum. The top Reactome pathways are metabolism of RNA, metabolism of proteins, and axon guidance. The top local network clusters are GTP hydrolysis and joining of the 60S ribosomal subunits and mRNA splicing. The Molecular Complex Detection algorithm identified clusters related to eukaryotic translation elongation, cellular responses to stress, regulation of RNA stability, COPI-independent Golgi-to-ER retrograde traffic, and supramolecular fiber organization.
352 Known and Putative AutoAgs Are COVID-Altered Proteins
To find out which autoAgs may be involved in COVID-19, we compared the DS-affinity autoantigen-ome with proteins and genes that are up- or down-regulated in SARS-CoV-2 infection (Coronascape database comparison, Supplementary Table 1) (29–49). Remarkably, 352 (86.3%) of the 408 DS-affinity proteins have been found to be altered (up- and/or down-regulated at protein and/or mRNA levels) in COVID-19 patients or SARS-CoV-2 infected cells (Table 1). Of these, 260 are reported as up-regulated and 303 as down-regulated (including 211 that are both up- and down-regulated). The numbers are not conflicting, because the COVID data were generated by multiple proteomic and transcriptomic methods and different cells and tissues. A protein may not be overexpressed even when its mRNA is up-regulated, and a protein/gene may be up-regulated in one tissue or patient but down-regulated in another tissue or patient. A protein is considered altered if it is up- or down-regulated at the protein or RNA level and, in relation to SARS-CoV-2 infection, it is considered a COVID-altered protein.
Protein-interaction analysis revealed that 352 COVID-altered proteins form a highly connected network, exhibiting 6,286 interactions (vs. 2,451 expected; PPI enrichment p-value <1.0E-6) (Figure 2). Based on cellular component analysis, the altered proteins can be located to intracellular organelles (323 proteins), nucleus (199 proteins), endomembrane system (143 proteins), vesicles (99 proteins), ribonucleoprotein complex (87 proteins), cytoskeleton (84 proteins), ER (72 proteins), and cell projections (52 proteins). Organelles with significant numbers of component proteins identified include the melanosome (30/105 proteins in melanosome), proteasome (16/64), polysome (13/66), spliceosome (34/187), ficolin-1-rich granule lumen (22/125), azurophil granules (17/155), and myelin sheath (26/157).
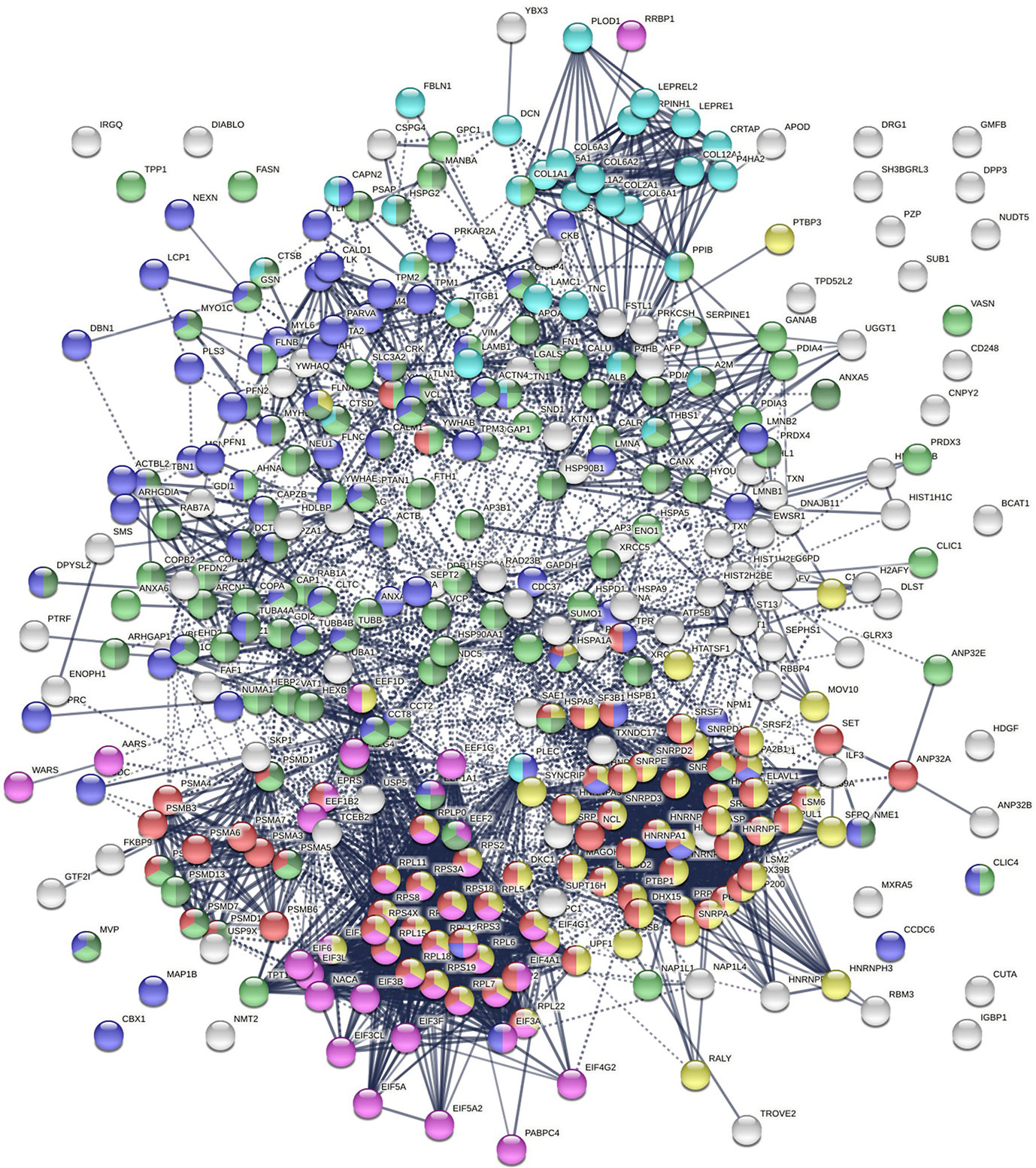
Figure 2 Network of 352 autoantigen-ome proteins that are altered in SARS-CoV-2 infected cells or patients. Connecting lines represent interactions with high confidence. Colored proteins are involved in metabolism of RNA (77 proteins, red), mRNA metabolic process (69 proteins, gold), translation (43 proteins, pink), vesicles (99 proteins, light green) and vesicle-mediated transport (84 proteins, dark green), cytoskeleton (84 proteins, blue), and extracellular matrix organization (32 proteins, aqua).
Similarly, the group of 260 up-regulated proteins is highly connected (3,747 interactions vs. 1,424 expected) with significant enrichment in proteins associated with RNA and mRNA metabolism, translation, vesicles and vesicle-mediated transport, and regulation of cell death (Figure 3A). The group of 303 down-regulated proteins is also highly connected (4,860 interactions vs. 1,907 expected), and these proteins are significantly related to RNA metabolism, translation, vesicles, cytoskeleton, and extracellular matrix (Figure 3B).
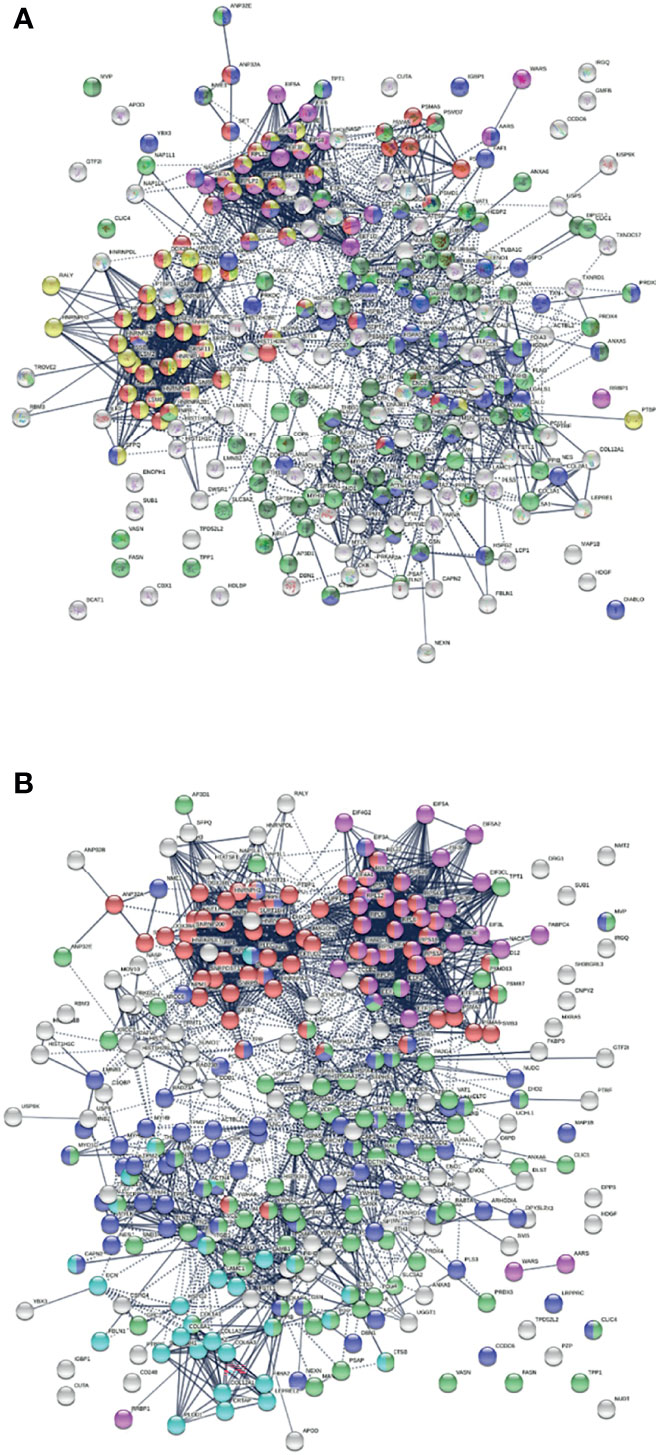
Figure 3 (A) Interaction network of 260 up-regulated proteins in SARS-CoV-2 infected cells or patients. Connecting lines represent interactions with high confidence (minimum interaction score of 0.7). Colored proteins are involved in metabolism of RNA (54 proteins, red), translation (28 proteins, pink), vesicles (82 proteins, light green) and vesicle-mediated transport (67 proteins, dark green), regulation of cell death (61 proteins, blue), and mRNA metabolic process (46 proteins, gold). (B) Interaction network of 303 down-regulated proteins in SARS-Cov-2 infected cells and patients. Connecting lines represent interactions with high confidence. Marked proteins are involved in RNA metabolism (64 proteins), translation (39 proteins, pink), vesicles (88 proteins, green), cytoskeleton (73 proteins, blue), and extracellular matrix organization (29 proteins, aqua).
Pathways and Processes Affected by COVID-Altered Proteins
Network enrichment analysis by Metascape revealed that the 352 COVID-altered proteins are most significantly enriched in RNA metabolism, axon guidance, and translation (Table 2). Many processes, e.g., regulated exocytosis, wound healing, supramolecular fiber organization, smooth muscle contraction, and platelet degranulation are significantly affected by COVID-altered proteins regardless of whether they are up- or down-regulated. The up-regulated proteins are more related to axon guidance and interleukin signaling, whereas down-regulated proteins are more related to cellular response to stress and apoptosis.
COVID-Altered AutoAgs Are Strongly Related to the Nervous System
COVID-19 patients frequently report neurological problems, such as loss of smell and taste, dizziness, headache, and stroke. While most symptoms are transient, some recovered patients are haunted by lingering neurological and psychological problems long after the viral infection. The underlying cause of transient and long-lasting neurological effects of COVID-19 has been puzzling. Analysis of COVID-altered proteins revealed a strong link to the nervous system. Of the 352 COVID-altered proteins, at least 150 are related to the nervous system (Figure 4A). More than 60 proteins are related to axon guidance based on ontology analyses (Table 2 and Figure 4A). In addition, there are 39 proteins related to neuron projection, 26 proteins related to myelin sheath, 25 proteins related to axon growth cone (252), 16 proteins related to neuronal cell body, 4 proteins related to cerebellar Purkinje cell layer, 3 proteins related to peripheral nervous system axon regeneration, and 2 proteins related to radial glial scaffolds. In particular, we found that 23 COVID-altered proteins are related to the olfactory bulb (253), which may explain the loss of smell in many COVID-19 patients.

Figure 4 (A) Nervous system-related proteins among COVID-altered proteins. Colored proteins are involved in axon guidance (62 proteins, aqua), axon growth cone (25 proteins, blue), myelin sheath (26 proteins, red), neuron projection (32 proteins, green) and neuron projection extension (7 proteins, dark green), neuronal cell body (16 proteins, gold), peripheral nervous system axon regeneration (3 proteins, brown), cerebellar Purkinje cell layer development (4 proteins, amber), and olfactory bulb (23 proteins, pink). (B) Neurological disease-related proteins among proteins altered in COVID. Colored are proteins found in neuronal infection with Japanese encephalitis virus (23 proteins, blue), neuroblastoma (21 proteins, red), glioblastoma (22 proteins, pink), neurodegeneration in Down syndrome (26 proteins, dark green), Alzheimer disease (22 proteins, aqua), schizophrenia (24 proteins, amber), cerebral ischemia induced neurodegenerative diseases (17 proteins, dark purple), Parkinson disease (17 proteins, brown), and neurodegeneration (21 proteins, green).
Most of these proteins are known autoAgs, e.g., ACTB, CANX, A2M, APOA1, CAPZA1, DPYSL2, FLNA, GDI2, LGALS1, MSN, PDIA3, PFN2, TNC, UCHL1, VCP, and VCL (see autoAg references in Table 1). Some yet-to-be-confirmed autoAgs with direct relation to the nerve system, e.g., NES (expressed mostly in nerve cells) and APOD (expressed by oligodendrocytes), warrant further investigation.
The COVID-altered proteins are also associated with a number of neurological diseases (Figure 4B). By comparing our data with published proteomes, 23 proteins were similarly found in neuronal infection by Japanese encephalitis virus (254), 21 proteins in neuroblastoma (255), 22 proteins in glioblastoma (256), 26 proteins in neurodegeneration in Down syndrome (257), 22 proteins in Alzheimer disease hippocampus (258), 24 proteins in schizophrenia (259), 17 proteins in cerebral ischemia (260), and 17 proteins in Parkinson disease (261).
Coronavirus-induced demyelination has been reported in a mouse model of multiple sclerosis (262), which may explain our identification of 26 altered proteins related to the myelin sheath in SARS-CoV-2 infection. In a mouse brain injury model, DS appears to play an important role in glial scar formation and regeneration of dopaminergic axons (263). Alterations of white matter DS and extracellular matrix are specific, dynamic, and widespread in multiple sclerosis patients (264). DS has recently been reported to promote neuronal differentiation in mouse and human neuronal stem cells (265). Given the various functional roles of DS, our identification of a large number of known and putative autoAgs with DS affinity related to the nervous system is a compelling finding.
COVID-Altered AutoAgs Are Related to Cell Death, Wound Healing, and Blood Coagulation
SARS-CoV-2 infection causes host cell death and leads to tissue injury. Wound healing, cellular response to stress, and apoptosis are among the most significant processes related to COVID-altered proteins (Table 2 and Figure 5A). For example, we identified 66 proteins related to regulation of cell death and 23 related to regulation of apoptotic signaling pathways. DS binds to apoptotic cells and autoAgs released from dying cells, which has led to our previous identification of hundreds of autoAgs (13–16, 18). Upon tissue injury, DS biosynthesis is ramped up by fibroblasts and epithelial and endothelial cells (7–9). After tissue injury, DS assists fibroblast migration into the wound to facilitate granulation tissue formation and wound healing (11). DS, similar to heparin, is also an important anticoagulant that inhibits clot formation via interaction with antithrombin and heparin cofactor II (266). Given these biological roles of DS, it is consistent that a large number of COVID-altered proteins related to cell death and tissue injury are identified by DS-affinity.
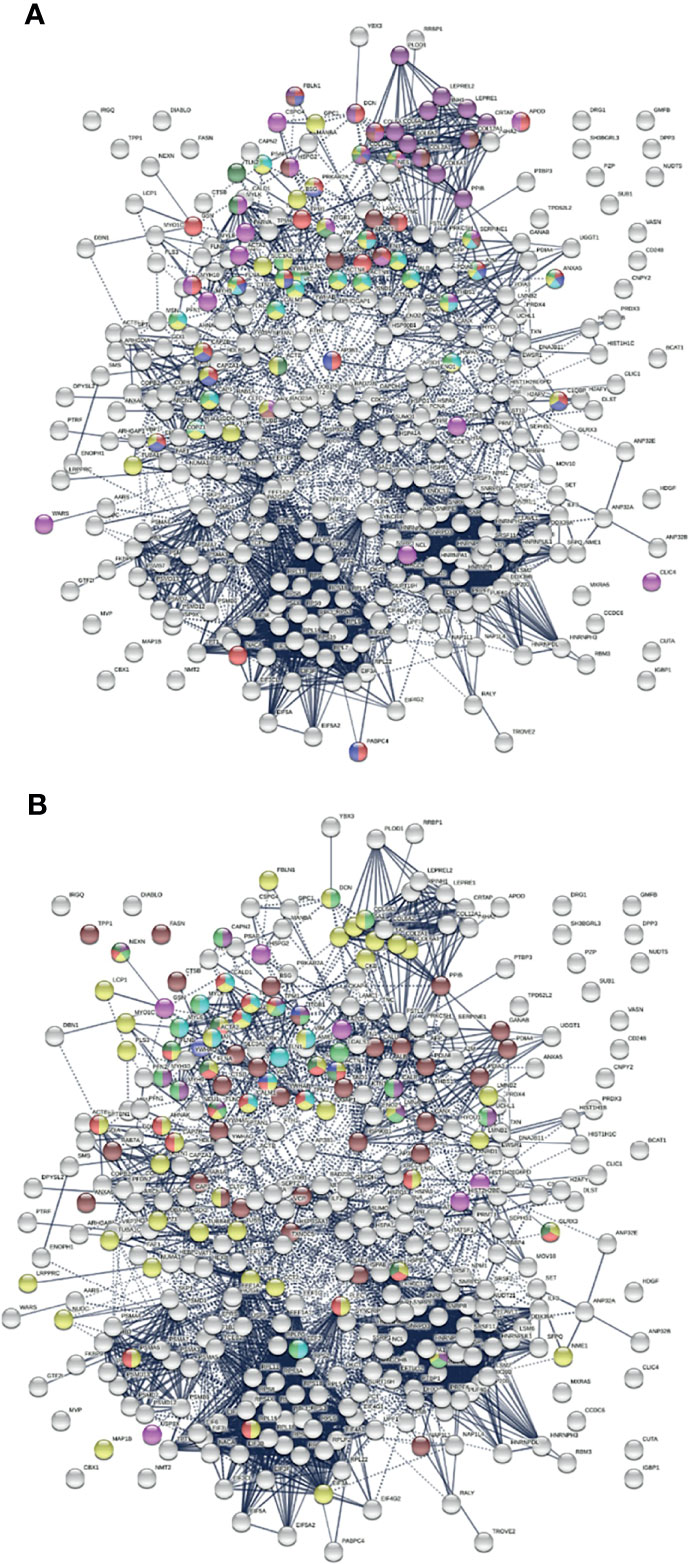
Figure 5 (A) Relation of COVID-altered proteins to wound healing and hemostasis. Response to wounding (25 proteins, red), blood vessel development (20 proteins, pink), blood coagulation (14 proteins, blue), collagen-containing extracellular matrix (13 proteins, brown), collagen biosynthesis and modifying enzymes (16 proteins, dark purple), platelet activation (3 proteins, dark green) and platelet activation signaling and aggregation (22 proteins, green), platelet degranulation (18 proteins, aqua), and hemostasis (35 proteins, gold). (B) Other significantly enriched groups among altered proteins. Supramolecular fiber (56 proteins, amber), melanosome (30 proteins, brown), striated muscle cell differentiation (11 proteins, purple), myofibril (23 proteins, red), muscle structure development (18 proteins, green), muscle contraction (13 proteins, aqua), Z disk (9 proteins, dark green), intercalated disk (4 proteins, blue), and amyloid fiber formation (6 proteins, pink).
Blood coagulation and thrombosis are frequent complications of COVID-19. Platelet degranulation is found to be significantly associated with at least 18 altered proteins (Table 2 and Figure 5A). COVID-altered proteins are related to blood coagulation, platelet activation, platelet alpha granules, fibrinogen binding, fibrinogen complex, platelet plug formation, von Willebrand factor A-like domain superfamily, and platelet-derived growth factor binding. Collagens, which support platelet adhesion and activation, and collagen biosynthesis and modifying enzymes are also among the COVID-altered proteins, e.g., collagen type VI trimer and type I trimer (Figure 5A). The majority of these altered proteins are known autoAgs, e.g., ALB, ANXA5, C1QBP, CALM1, CAPZB, COL1A1, COL1A2, COL6A1, FBLN1, FN1, PLEC, PPIB, THBS1, TLN1, TUBA4A, and YWHAZ (see autoAg references in Table 1). Some are unknown and await further investigation, e.g., AP3B1, CRK, CTSB, EHD2, PLOD1, PSAP, and PARKAR2A.
Supramolecular Fibril Alteration Offers Clues to Muscle Dysfunction and Fibrosis
Over 50 supramolecular filament proteins are identified by DS-affinity from HFL1 cells. Remarkably, nearly all (except for one) are found to be altered in SARS-CoV-2 infection, and the majority have already been reported as autoAgs (Table 1). They include various isoforms of actin, actinin, collagen, filamin, fibronectin, fibulin, dynactin, dynein, lamin, myosin, nestin, nexilin, profilin, plectin, plastin, proteoglycan, septin, spectrin, talin, tropomyosin, tubulin, vinculin, and vimentin (Table 1 and Figure 5B). These proteins are major components of the extracellular matrix, basement membrane, cell cytoskeleton, cytoskeletal motors, muscle filaments, and contractile motors of muscle cells.
A significant number of COVID-altered proteins are related. Emerin complex and smooth muscle contraction are among the top enriched biological processes of COVID-altered proteins (Table 2 and Figure 5B). Emerin is highly expressed in cardiac and skeletal muscle, and emerin mutations cause X-linked recessive Emery-Dreifuss muscular dystrophy, cardiac conduction abnormalities, and dilated cardiomyopathy. Smooth muscle resides primarily in the walls of hollow organs where it performs involuntary movements, e.g., respiratory tract, blood vessels, gastrointestinal tract, and renal glomeruli. In addition, we identified proteins with significant association to myofibrils (the contractile elements of skeletal and cardiac muscle; 23 proteins) (Figure 5B), stress fiber (a contractile actin filament bundle that consists of short actin filaments with alternating polarity: MYH9, MYLK, FLNB, TPM1, TPM2, TPM3, TPM4, ACTN1, ACTN4), muscle filament sliding (the sliding of actin thick filaments and myosin thick filaments past each other in muscle contraction), Z disk (plate-like region of a muscle sarcomere to which the plus ends of actin filaments are attached), intercalated disc (a cell-cell junction complex at which myofibrils terminate in cardiomyocytes, mediates mechanical and electrochemical integration between individual cardiomyocytes), and negative regulation of smooth muscle cell-matrix adhesion (2 proteins; SERPINE1, APOD).
Pulmonary fibrosis is prominent in COVID-19 and contributes to lethality in some cases (267, 268). Fibrosis, or fibrotic scarring, is pathological wound healing in which excessive extracellular matrix components are produced by fibroblasts and accumulate in the wounded area. Histopathological examination of COVID-19 patients found highly heterogenous injury patterns reminiscent of exacerbation of interstitial lung disease, including interstitial thickening, fibroblast activation, and deposition of collagen fibrils (22). We identified a significant number of COVID-altered proteins that are associated with collagen bundles and collagen biosynthesis and modifying enzymes (16 proteins), extracellular matrix organization (33 proteins), supramolecular fibers, and amyloid formation offering functional links to fibrosis (Figure 5B).
Potential AutoAgs in COVID-19 Patients and a Connection to the Melanosome
To find out how altered proteins may differ in patients, we compared our putative autoantigen-ome to published single-cell RNA sequencing data of 6 patients hospitalized for COVID-19 (29, 35) and identified 32-59 putative autoAgs per patient (Figure 6). Interestingly, while identified from different patients, the altered proteins/genes identified share involvement of leukocyte activation, vesicles and vesicle transport, protein processing in the ER (including antigen processing and presentation), regulation of cell death, translation, muscle contraction, myelin sheath, and curiously, the melanosome (Figure 6). The estrogen signaling pathway and thyroid hormone synthesis are found to be associated with altered proteins in some patients. Patient C2 has 5 altered proteins related to neuron differentiation regulation, and patient C4 has 6 altered proteins related to neuron death.
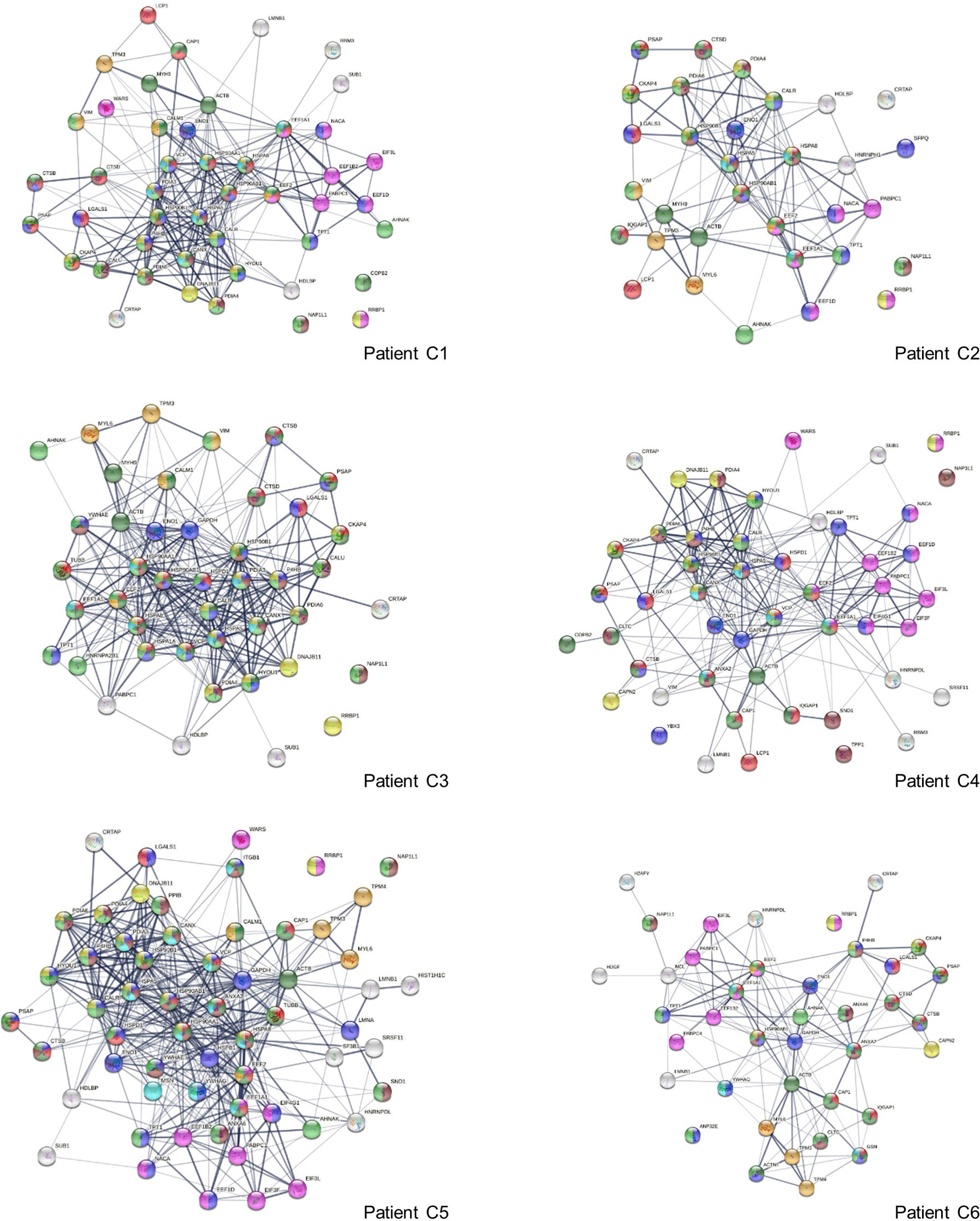
Figure 6 Interaction network of altered proteins in 6 COVID-19 patients. Colored proteins are associated with leukocyte activation involved in immune response (red), vesicles (light green) and vesicle-mediated transport (dark green), protein processing in the ER (yellow), regulation of cell death (blue), translation (pink), melanosome (brown), myelin sheath (aqua), and muscle contraction (amber).
Eleven altered proteins were identified in all 6 patients, including known autoAgs (ACTB, EEF1A1, EEF2, ENO1, LGALS1, PABPC1) and unknown ones (CRTAP, NAP1L1, PSAP, RRBP1, TPT1) (Table 1). AHNAK (neuroblast differentiation-associated protein, a known autoAg in lupus) was identified in 5 patients. Overall, a majority of the altered proteins identified from the 6 COVID patients are known autoAgs, e.g., CALM1, CALR, CALU, CANX, DNAJB11, HDGF, HSPA5 (BiP), IQGAP1, LCP1, LMNB1, MYH9, NACA, P4HB, SFPQ, PDIA3, TPM3, TUBB, VCP, VIM, WARS, and YB3 (Table 1). Unknown or putative autoAgs include CAP1, CTSB, HDLBP, HYOU1, SND1, and SUB1.
We initially identified 30 DS-affinity proteins from HFL1 cells related to the melanosome, and, intriguingly, all of these are also COVID-altered proteins (Figure 5B). Based on STRING GO analysis, the melanosome is the most significant cellular component related to altered proteins in all 6 patients (with false discovery rates ranging from 1.52E-8 to 1.11E-23). In HIV infection, melanosome production is stimulated in some patients and leads to an increase in pigmented lesions (269). However, melanosome involvement in COVID-19 is not known. Two Wuhan doctors in intensive care for COVID temporally turned dark, although the cause was thought to be a drug reaction. A COVID patient has been reported with acute flaccid tetraparesis and maculopapular pigmented plaques on the limbs (270). In mice, coronavirus induces an acute and long-lasting retinal disease, with initial retinal vasculitis followed by retinal degeneration that is associated with retinal autoantibodies and retinal pigment epithelium autoantibodies (271). Future research will be needed to investigate the interaction between COVID and melanosome activation.
Association Between Autoimmunity and Virus Infections
We identified COVID-altered proteins with DS-affinity that are involved in the host response to various aspects of viral infection and that possess a high propensity to become autoAgs. For example, viral RNA metabolism, translation, vesicles, and vesicle transport contribute a large number of known and putative autoAgs. In addition, viral processes, particularly symbiont processes and interspecies interactions between host and viruses, contribute significantly to altered proteins (Figure 7A). For example, among altered proteins related to response to viral processes, HSPA8, DDB1, RAD23A, PABPC1, PPIB, P4HB, LGALS1, GSN, and ILF3 are known autoAgs (Table 1).
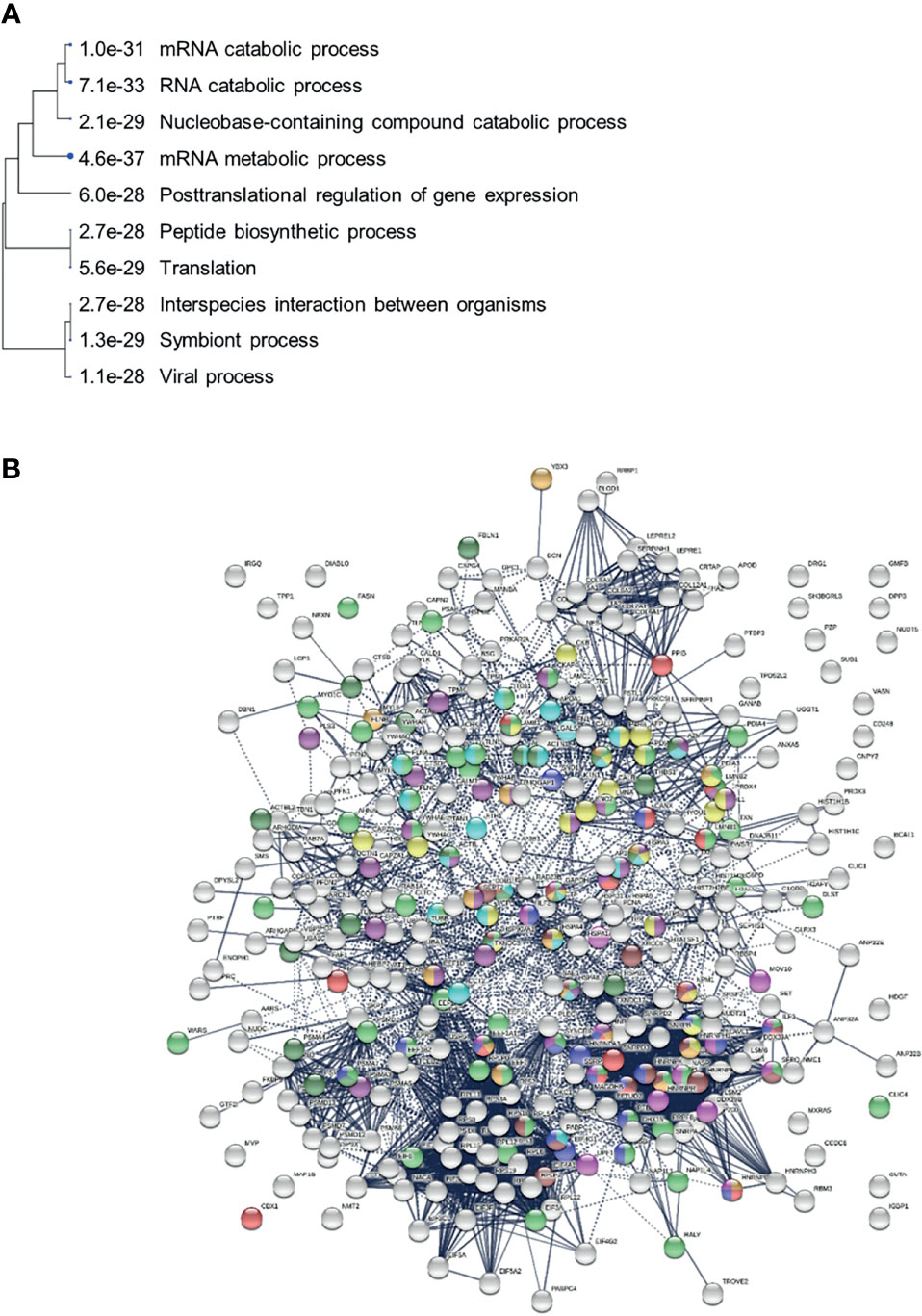
Figure 7 (A) Hierarchical clustering of top 10 pathways involving COVID-altered proteins. Analysis based on hypergeometric distribution followed by FDR correction. (B) COVID-altered host proteins with DS-affinity found in various viral infections. Porcine reproductive and respiratory syndrome (56 proteins, green), H5N1 avian influenza virus (27 proteins, dark purple), Japanese encephalitis virus (23 proteins, gold), Rift Valley fever virus (24 proteins, aqua), Hepatitis B virus (22 proteins, dark green), HIV (identified in different studies, 18 amber, 18 brown, 18 red and 17 pink), and shared among positive-sense RNA viruses (20 proteins, blue).
In particular, COVID-altered cytoskeletal filament proteins shed light on viral trafficking in host cells. SARS-CoV-2 infection induces profound remodeling of the cytoskeleton, and replicating viral vesicles are surrounded by a network of intermediate filaments (272). The cytoskeletal network appears to facilitate coronavirus transport and expulsion, with thickening actin filaments providing the bending force to extrude viral vesicles (273). We identified 84 altered proteins related to the cytoskeleton and 84 altered proteins related to vesicle-mediated transport (Figure 2). These altered proteins are implicated in various processes, including cytoskeleton-dependent intracellular transport, actin fiber-based movement, actin-mediated cell contraction, microtubule-dependent trafficking from the Golgi to the plasma membrane, and transport along microtubules.
Many positive-strand RNA viruses (including SARS-CoV-2, Enterovirus, Hepatitis C virus, Norovirus, and Poliovirus) hijack a common group of nuclear factors to support the biosynthetic functions required for viral replication and propagation (274). 20 of these hijacked nuclear proteins are identified by DS-affinity in our study (Figure 7). In addition, altered proteins are found in other viral infections, including porcine reproductive and respiratory syndrome virus (275), H5N1 avian influenza viruses (276, 277), Japanese encephalitis virus (254), Rift Valley fever virus (278), Hepatitis B virus (279), HIV (280–282), Herpes Simplex virus (283), and Epstein-Barr virus infection (Figure 7B and STRING ontology analysis). In some cases, viral infections may have both enhancing and protective effects on autoimmunity in type 1 diabetes (284).
Our study identified a large number of known and putative autoAgs that are related to mRNA metabolism, translation, vesicles, and vesicle trafficking (Figures 1, 2). This finding begs us to wonder whether mRNA vaccines may induce unintended autoimmune consequences in the long term. To induce protective immunity, mRNA vaccine vesicles will need to be transported into cells where they use the host cell machinery to produce a viral protein antigen, whereupon the antigen will be processed and presented by MHC molecules to induce B and T cell responses.
mRNA translation requires ribosomes, translation initiation factors, aminoacyl-tRNA synthetases, and elongation factors. We identified 18 ribosomal proteins by DS-affinity, all of which are altered in SARS-CoV-2 infection and 9 of which are known autoAgs (see references in Table 1). We also identified 15 eukaryotic translation initiation factor proteins, with 12 of them being COVID-altered and 4 being known autoAgs (Table 1). Six elongation factor proteins (5 subunits of EEF1 complex, EEF2) were identified by DS-affinity, of which all 6 are COVID-altered and 3 are known autoAgs (Table 1). Six tRNA synthetases were identified, with 5 being known autoAgs and 3 (AARS, EPRS, WARS) COVID-altered (Table 1). Autoantibodies to AARS are associated with interstitial lung disease and myositis (51, 285). EPRS appears to regulate pro-fibrotic protein synthesis during cardiac fibrosis (286). Gene mutations of WARS cause an autosomal dominant neurologic disorder characterized by slowly progressive distal muscle weakness and atrophy affecting both the lower and upper limbs (242, 287).
Once synthesized, the exogenous protein antigens are degraded by proteasomes, and the resulting peptides are transported into the ER where they are loaded onto MHC molecules by peptide loading complexes for presentation to T cells. In relation to these steps, 15 proteasome subunits were identified by DS-affinity, with 12 being COVID-altered and 7 being known autoAgs (Table 1). Nine proteins related to antigen processing and presentation are found to be altered in the 6 COVID-19 patients analyzed in this study, including HSPA1A, HSPA8, HSP90AA1, HSPAB1, HSPA5, PDIA3, CANX, CALR, and CTSB, with 7 being known autoAgs (Figure 5 and Table 1).
In addition, among the 352 COVID-altered proteins identified in this study, 69 proteins are associated with mRNA metabolism (Figure 2). Many of these proteins may be irrelevant to non-replicating mRNA molecules in mRNA vaccines, however, some are likely needed in processes such as 3’ end processing, deadenylation, and nonsense-mediated decay. For example, we identified poly(A) tail binding proteins PABPC1 and PABPC4 as COVID-altered proteins, both of which have been reported as autoAgs (Table 1).
Our study identified 99 altered proteins associated with vesicles and 84 proteins associated with vesicle-mediated transport (Figures 1, 2, 5). Although it is not clear which host molecules are involved in extra- and intracellular transport and uptake of mRNA vaccine vesicles, some of the vesicle-related proteins identified as DS-affinity proteins may be involved, e.g., proteins of receptor-mediated endocytosis (APOA1, CALR, CANX, CAP1, CLTC, HSP90AA1, HSP90B1, HSPG2, ITGB1, YWHAH) or phagocytosis (ACTB, CRK, GSN, HSP90AA1, HSP90AB1, MYH9, MYO1C, PDIA6, RAB7A, THBS1, TXNDC5).
Overall, a significant number of autoAgs related to different steps of mRNA vaccine action were identified in this study; however, our findings do not mean that these autoAgs will lead to aberrant autoimmune reactions as a result of mRNA vaccination. The development of autoimmune diseases or autoimmunity-related diseases entails a complex cascade of molecular and cellular interactions. Long-term monitoring of autoimmune adverse effects will be needed.
Conclusion
This study identifies an autoantigen-ome of 408 proteins from human fetal lung fibroblast HFL1 cells by DS-affinity and protein sequencing, of which at least 231 proteins are confirmed autoAgs. Of these, 352 (86.3%) are found to be altered in SARS-CoV-2 infection when compared to published data, with at least 210 COVID-altered proteins being known autoAgs. The altered proteins are significantly enriched in a number of pathways and processes and are closely connected to various disease manifestations of COVID-19, particularly neurological problems, fibrosis, muscle dysfunction, and thrombosis.
Viral infections cause significant perturbations of normal cellular and tissue component molecules in the host, leading to cell death and tissue injury. Autoantigens resulting from molecular alterations may result directly from the injury or indirectly from responses to the injury. As a stress response, DS biosynthesis may be ramped up to facilitate wound healing and dead cell clearance. DS associates with autoAgs and stimulates autoreactive B cells and autoantibody production. Specific autoantibodies that are initially induced in response to a certain injury site may circulate and attack secondary sites where the autoAgs are also expressed, leading to a complex array of local and systemic autoimmune diseases.
This study supports a connection between COVID and autoimmunity. We have shown in a series of papers on autoimmune disease that proteins with high affinity for DS possess intrinsic propensity to become recognized by the humoral immune system and serve as autoantigens (12–16, 18). We have shown in a prior paper that proteins that are, by themselves, not immunogenic can be turned into potent autoantigens and induce an autoantibody response if they are engineered to bind to DS and are exposed as DS-autoAg complexes to the immune system (14). The list of proteins enriched by DS-affinity in lung fibroblasts is, at first, only a putative catalogue of autoantigens. Intriguingly, when we performed a literature analysis of all DS-enriched proteins, we found that a very high proportion of them correspond to known autoantigens (this enrichment is much higher than would be expected by chance). Many of the COVID-induced autoantibodies described in a recent study correspond to autoantigens identified in our study (e.g., ribosomal P proteins, Ro/La, U1-snRNP, and chromatin histones) (288). While likely also autoantigens, we label proteins that have not been observed as autoantigens in the literature as “putative autoAgs.” We then show that among the DS-affinity proteins, there are many proteins that are also affected by COVID (many more than would be expected by statistical chance). Taking all these observations together, we hypothesize that our findings provide a rationale for why SARS-CoV-2 infection may induce autoimmune sequelae. Future serological studies will be needed to further confirm this hypothesis, but our dataset, together with the comprehensive list of possible autoAg targets, will be a valuable guide and map for these ongoing investigations. We believe that our dataset will be of great interest and value for research groups worldwide that are attempting to tackle the autoimmune aspects of COVID.
The COVID-19 autoantigen-ome provides a detailed molecular map for investigating the diverse spectrum of autoimmune sequelae caused by the pandemic. The COVID autoantigen atlas we are establishing will serve as a detailed molecular map and reference for ongoing research into COVID-induced autoimmunity and possible autoimmune causes of “long COVID” syndrome. It will thus serve as an important resource for the scientific community.
Materials and Methods
HFL1 Cell Culture
The HFL1 cell line was obtained from the ATCC (Manassas, VA, USA) and cultured in Eagle’s Minimum Essential Medium supplemented with 10% fetal bovine serum (Thermo Fisher) and a penicillin-streptomycin-glutamine mixture (Thermo Fisher) at 37°C.
Protein Extraction
About 100 million cells were harvested and suspended in 10 ml of 50 mM phosphate buffer (pH 7.4) containing the Roche Complete Mini protease inhibitor cocktail. Cells were homogenized on ice with a microprobe sonicator until the turbid mixture became nearly clear with no visible cells left. The homogenate was centrifuged at 10,000 g at 4°C for 20 min, and the supernatant was collected as the total protein extract. Protein concentration was measured with the RC DC protein assay (Bio-Rad).
DS-Sepharose Resin Preparation
20 ml of EAH Sepharose 4B resins (GE Healthcare Life Sciences) were washed with distilled water three times and mixed with 100 mg of DS (Sigma-Aldrich) in 10 ml of 0.1 M MES buffer, pH 5.0. 500 mg of N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (Sigma-Aldrich) powder was added to the mixture. The reaction proceeded by end-over-end rotation at 25°C for 16 h. After coupling, resins were washed with water and equilibrated first with a low-pH buffer (0.1 M acetate, 0.5 M NaCl, pH 5.0) and then with a high-pH buffer (0.1 M Tris, 0.5 M NaCl, pH 8.0).
DS-Affinity Fractionation
The total proteins extracted from HFL1 cells were fractionated on DS-Sepharose columns with a BioLogic Duo-Flow system (Bio-Rad). About 40 mg of proteins in 40 ml of 10 mM phosphate buffer (pH 7.4; buffer A) were loaded onto the column at a rate of 1 ml/min. Unbound proteins were washed off with 60 ml of buffer A, and weakly bound proteins were eluted with 40 ml of 0.2 M NaCl in buffer A. DS-binding proteins were eluted with sequential 40-ml step gradients of 0.5 M and 1.0 M NaCl in buffer A. Fractions were desalted and concentrated to 0.5 ml with 5-kDa cut-off Vivaspin centrifugal filters (Sartorius). Fractionated proteins were separated by 1-D SDS-PAGE in 4-12% Bis-Tris gels, and the gel lanes corresponding to 1.0 M or 0.5 M NaCl elutions were divided into two or three sections for sequencing.
Mass Spectrometry Sequencing
Fractionated proteins with different affinity to DS were separated on 1D SDS PAGE in 4-12% NuPAGE Novex Bis-Tris gels (Invitrogen). Based on protein band intensity, the protein lanes containing proteins eluting at 0.5 M or 1.0 M NaCl were each cut into 2 sections, containing top and bottom bands, respectively. Gel sections were transferred into 1-mL tubes, cut into 1-mm3 pieces, dehydrated with acetonitrile, and dried in a speed-vac. Protein sequencing was performed at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. The gel pieces were rehydrated with 50 mM NH4HCO3 containing 12.5 µg/mL modified sequencing-grade trypsin (Promega) at 4°C for 45 min. Tryptic peptides were separated on a nano-scale C18 HPLC capillary column and analyzed after electrospray ionization in an LTQ linear ion-trap mass spectrometer (Thermo Fisher). The reference human proteome database was downloaded from UniProt (updated until March 2021). Peptide sequences and protein identities were assigned by matching the measured fragmentation patterns with protein or translated nucleotide databases using Sequest software. Peptides were required to be fully tryptic peptides with XCorr values of at least 1.5 for 1+ ions, 1.5 for 2+ ions, or 3.0 for 3+ ions. All data were manually inspected. Only proteins with ≥2 unique peptide matches were considered positively identified using a false discovery rate of <1% at peptide and protein levels (Supplemental Table 2).
COVID Data Comparison With Coronascape
DS-affinity proteins were compared with currently available proteomic and transcriptomic data from SARS-CoV-2 infection compiled in the Coronascape database (as of 12/14/2020) (29–49). These data had been obtained with proteomics, phosphoproteomics, interactome, ubiquitome, and RNA-seq techniques. Up- and down-regulated proteins or genes were identified by comparing COVID-19 patients vs. healthy controls and cells infected vs. uninfected by SARS-CoV-2. Similarity searches were conducted between our data and the Coronascape database to identify DS-affinity proteins (or their corresponding genes) that are up- and/or down-regulated in the viral infection.
Pathway and Process Enrichment Analysis
Pathways and processes enriched in the putative autoantigen-ome were analyzed with Metascape (29). The analysis was performed with various ontology sources, including KEGG Pathway, GO Biological Process, Reactome Gene Sets, Canonical Pathways, CORUM, TRRUST, and DiGenBase. All genes in the genome were used as the enrichment background. Terms with a p-value <0.01, a minimum count of 3, and an enrichment factor (ratio between the observed counts and the counts expected by chance) >1.5 were collected and grouped into clusters based on their membership similarities. The most statistically significant term within a cluster was chosen to represent the cluster. Pathway hierarchical clustering was obtained with ShinyGo (289).
Protein-Protein Interaction Network Analysis
Protein-protein interactions among collections of DS-affinity proteins were analyzed by STRING (251), including both direct physical interaction and indirect functional associations. Interactions are derived from genomic context predictions, high-throughput lab experiments, co-expression, automated text mining, and previous knowledge in databases. Each interaction is annotated with a confidence score from 0 to 1, with 1 being the highest, indicating the likelihood of an interaction to be true. Only interactions with high confidence (a minimum score of 0.7) are shown in the figures.
Literature Text Mining
Literature searches in Pubmed were performed for every DS-affinity protein identified in this study. Search keywords included the protein name, its gene symbol, alternative names and symbols, and the MeSH keyword “autoantibodies”. Only proteins with their specific autoantibodies reported in PubMed-listed journal articles were considered “confirmed” autoAgs in this study.
Data Availability Statement
The original contributions presented in the study are included in the article or the Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
JW directed the study, analyzed data, and wrote the manuscript. WZ performed some experiments and reviewed the manuscript. VR and MWR assisted in data analysis and manuscript preparation. MHR consulted on the study, analyzed data, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
MHR acknowledges grants from the NIH/NCI (R21 CA251992 and R21 CA263262), a Cycle for Survival Equinox Innovation Grant, an Investigator Grant from the Neuroendocrine Tumor Research Foundation (NETRF), and support from the Farmer Family Foundation. Parts of the study were supported by the MSKCC NCI Cancer Center Support Grant (P30 CA008748). The funding bodies were not involved in the design of the study or the collection, analysis, or interpretation of data.
Conflict of Interest
JW is the founder and Chief Scientific Officer of Curandis. MWR and VR are volunteers of Curandis. MHR is a member of the Scientific Advisory Boards of Trans-Hit Bio (Azenta Life Sciences), Proscia, and Universal DX.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jung-hyun Rho for technical assistance with experiments. We thank Ross Tomaino and the Taplin Biological Mass Spectrometry facility of Harvard Medical School for expert service with protein sequencing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.831849/full#supplementary-material
References
1. Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell (2020) 183(4):1043–57.e1015. doi: 10.1016/j.cell.2020.09.033
2. Tamhankar M, Gerhardt DM, Bennett RS, Murphy N, Jahrling PB, Patterson JL. Heparan Sulfate Is an Important Mediator of Ebola Virus Infection in Polarized Epithelial Cells. Virol J (2018) 15(1):135. doi: 10.1186/s12985-018-1045-0
3. Luteijn RD, Van Diemen F, Blomen VA, Boer IGJ, Manikam Sadasivam S, van Kuppevelt TH, et al. A Genome-Wide Haploid Genetic Screen Identifies Heparan Sulfate-Associated Genes and the Macropinocytosis Modulator TMED10 as Factors Supporting Vaccinia Virus Infection. J Virol (2019) 93(13):e02160–18. doi: 10.1128/JVI.02160-18
4. Gao H, Lin Y, He J, Zhou S, Liang M, Huang C, et al. Role of Heparan Sulfate in the Zika Virus Entry, Replication, and Cell Death. Virology (2019) 529:91–100. doi: 10.1016/j.virol.2019.01.019
5. Xu Y, Martinez P, Séron K, Luo G, Allain F, Dubuisson J, et al. Characterization of Hepatitis C Virus Interaction With Heparan Sulfate Proteoglycans. J Virol (2015) 89(7):3846–58. doi: 10.1128/JVI.03647-14
6. Penc SF, Pomahac B, Winkler T, Dorschner RA, Eriksson E, Herndon M, et al. Dermatan Sulfate Released After Injury Is a Potent Promoter of Fibroblast Growth Factor-2 Function. J Biol Chem (1998) 273(43):28116–21. doi: 10.1074/jbc.273.43.28116
7. Pohle T, Altenburger M, Shahin M, Konturek JW, Kresse H, Domschke W. Expression of Decorin and Biglycan in Rat Gastric Tissue: Effects of Ulceration and Basic Fibroblast Growth Factor. Scand J Gastroenterol (2001) 36(7):683–9. doi: 10.1080/003655201300191932
8. Nelimarkka L, Salminen H, Kuopio T, Nikkari S, Ekfors T, Laine J, et al. Decorin is Produced by Capillary Endothelial Cells in Inflammation-Associated Angiogenesis. Am J Pathol (2001) 158(2):345–53. doi: 10.1016/S0002-9440(10)63975-2
9. Lee PH, Trowbridge JM, Taylor KR, Morhenn VB, Gallo RL. Dermatan Sulfate Proteoglycan and Glycosaminoglycan Synthesis Is Induced in Fibroblasts by Transfer to a Three-Dimensional Extracellular Environment. J Biol Chem (2004) 279(47):48640–6. doi: 10.1074/jbc.M407241200
10. Kuwaba K, Kobayashi M, Nomura Y, Irie S, Koyama Y. Size Control of Decorin Dermatan Sulfate During Remodeling of Collagen Fibrils in Healing Skin. J Dermatol Sci (2002) 29(3):185–94. doi: 10.1016/S0923-1811(02)00023-3
11. Clark RA, Lin F, Greiling D, An J, Couchman JR. Fibroblast Invasive Migration Into Fibronectin/Fibrin Gels Requires a Previously Uncharacterized Dermatan Sulfate-CD44 Proteoglycan. J Invest Dermatol (2004) 122(2):266–77. doi: 10.1046/j.0022-202X.2004.22205.x
12. Wang JY, Roehrl MH. Glycosaminoglycans Are a Potential Cause of Rheumatoid Arthritis. Proc Natl Acad Sci USA (2002) 99(22):14362–7. doi: 10.1073/pnas.222536599
13. Wang JY, Lee J, Yan M, Rho JH, Roehrl MH. Dermatan Sulfate Interacts With Dead Cells and Regulates CD5(+) B-Cell Fate: Implications for a Key Role in Autoimmunity. Am J Pathol (2011) 178(5):2168–76. doi: 10.1016/j.ajpath.2011.01.028
14. Rho JH, Zhang W, Murali M, Roehrl MH, Wang JY. Human Proteins With Affinity for Dermatan Sulfate Have the Propensity to Become Autoantigens. Am J Pathol (2011) 178(5):2177–90. doi: 10.1016/j.ajpath.2011.01.031
15. Zhang W, Rho JH, Roehrl MH, Wang JY. A Comprehensive Autoantigen-Ome of Autoimmune Liver Diseases Identified From Dermatan Sulfate Affinity Enrichment of Liver Tissue Proteins. BMC Immunol (2019) 20(1):21. doi: 10.1186/s12865-019-0304-1
16. Zhang W, Rho JH, Roehrl MW, Roehrl MH. Wang JY. A Repertoire of 124 Potential Autoantigens for Autoimmune Kidney Diseases Identified by Dermatan Sulfate Affinity Enrichment of Kidney Tissue Proteins. PLoS One (2019) 14(6):e0219018. doi: 10.1371/journal.pone.0219018
17. Lee J, Rho J-H, Roehrl MH, Wang JY. Dermatan Sulfate Is a Potential Master Regulator of IgH via Interactions With Pre-BCR, GTF2I, and BiP ER Complex in Pre-B Lymphoblasts. bioRxiv Preprint Server Biol (2021) 12:680212. doi: 10.1101/2021.01.18.427153
18. Wang JY, Zhang W, Rho JH, Roehrl MW, Roehrl MH. A Proteomic Repertoire of Autoantigens Identified From the Classic Autoantibody Clinical Test Substrate HEp-2 Cells. Clin Proteomics (2020) 17:35. doi: 10.1186/s12014-020-09298-3
19. Zhou S, Jones-Lopez EC, Soneji DJ, Azevedo CJ, Patel VR. Myelin Oligodendrocyte Glycoprotein Antibody-Associated Optic Neuritis and Myelitis in COVID-19. J Neuro Ophthalmol (2020) 40(3):398–402. doi: 10.1097/WNO.0000000000001049
20. Bowles L, Platton S, Yartey N, Dave M, Lee K, Hart DP, et al. Lupus Anticoagulant and Abnormal Coagulation Tests in Patients With Covid-19. N Engl J Med (2020) 383(3):288–90. doi: 10.1056/NEJMc2013656
21. Gruber CN, Patel RS, Trachtman R, Lepow L, Amanat F, Krammer F, et al. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C). Cell (2020) 183(4):982–95.e914. doi: 10.1016/j.cell.2020.09.034
22. Gagiannis D, Steinestel J, Hackenbroch C, Schreiner B, Hannemann M, Bloch W, et al. Clinical, Serological, and Histopathological Similarities Between Severe COVID-19 and Acute Exacerbation of Connective Tissue Disease-Associated Interstitial Lung Disease (CTD-ILD). Front Immunol (2020) 11:587517. doi: 10.3389/fimmu.2020.587517
23. Lerma LA, Chaudhary A, Bryan A, Morishima C, Wener MH, Fink SL. Prevalence of Autoantibody Responses in Acute Coronavirus Disease 2019 (COVID-19). J Trans Autoimmun (2020) 3:100073. doi: 10.1016/j.jtauto.2020.100073
24. Fujii H, Tsuji T, Yuba T, Tanaka S, Suga Y, Matsuyama A, et al. High Levels of Anti-SSA/Ro Antibodies in COVID-19 Patients With Severe Respiratory Failure: A Case-Based Review : High Levels of Anti-SSA/Ro Antibodies in COVID-19. Clin Rheumatol (2020) 39(11):3171–5. doi: 10.1007/s10067-020-05359-y
25. Dragonetti D, Guarini G, Pizzuti M. Detection of Anti-Heparin-PF4 Complex Antibodies in COVID-19 Patients on Heparin Therapy. Blood Transfusion = Trasfusione del sangue (2020) 18(4):328. doi: 10.2450/2020.0164-20
26. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies Against Type I IFNs in Patients With Life-Threatening COVID-19. Science (New York NY) (2020) 370(6515):eabd4585. doi: 10.1126/science.abd4585
27. Kreye J, Reincke SM, Kornau HC, Sánchez-Sendin E, Corman VM, Liu H, et al. A Therapeutic Non-Self-Reactive SARS-CoV-2 Antibody Protects From Lung Pathology in a COVID-19 Hamster Model. Cell (2020) 183(4):1058–69.e1019. doi: 10.1016/j.cell.2020.09.049
28. Rendeiro AF, Ravichandran H, Bram Y, Chandar V, Kim J, Meydan C, et al. The Spatial Landscape of Lung Pathology During COVID-19 Progression. Nature (2021) 593(7860):564–9. doi: 10.1038/s41586-021-03475-6
29. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat Commun (2019) 10(1):1523. doi: 10.1038/s41467-019-09234-6
30. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-Cell Landscape of Immunological Responses in Patients With COVID-19. Nat Immunol (2020) 21(9):1107–18. doi: 10.1038/s41590-020-0762-x
31. Davies JP, Almasy KM, Mcdonald EF, Plate L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect Dis (2020) 6(12):3174–89. doi: 10.1021/acsinfecdis.0c00500
32. Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol Cell (2020) 80(1):164–74.e164. doi: 10.1016/j.molcel.2020.08.006
33. Sun J, Ye F, Wu A, Yang R, Pan M, Sheng J, et al. Comparative Transcriptome Analysis Reveals the Intensive Early Stage Responses of Host Cells to SARS-CoV-2 Infection. Front Microbiol (2020) 11:593857. doi: 10.3389/fmicb.2020.593857
34. Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. Proteomics of SARS-CoV-2-Infected Host Cells Reveals Therapy Targets. Nature (2020) 583(7816):469–72. doi: 10.1038/s41586-020-2332-7
35. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients With Severe COVID-19. Nat Med (2020) 26(7):1070–6. doi: 10.1038/s41591-020-0944-y
36. Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, et al. In Vivo Antiviral Host Transcriptional Response to SARS-CoV-2 by Viral Load, Sex, and Age. PLoS Biol (2020) 18(9):e3000849. doi: 10.1371/journal.pbio.3000849
37. Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 Antiviral Drugs Through Large-Scale Compound Repurposing. Nature (2020) 586(7827):113–9. doi: 10.1038/s41586-020-2577-1
38. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell (2020) 182(3):685–712.e619.
39. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell (2020) 181(5):1036–45.e1039. doi: 10.1016/j.cell.2020.04.026
40. Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell (2020) 182(1):59–72.e15. doi: 10.1016/j.cell.2020.05.032
41. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science (New York NY) (2020) 369(6499):50–4. doi: 10.1126/science.abc1669
42. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature (2020) 583(7816):459–68. doi: 10.1038/s41586-020-2286-9
43. Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic Characteristics of Bronchoalveolar Lavage Fluid and Peripheral Blood Mononuclear Cells in COVID-19 Patients. Emerg Microbes Infect (2020) 9(1):761–70. doi: 10.1080/22221751.2020.1747363
44. Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol (2020) 94(19):e00985–20. doi: 10.1128/JVI.00985-20
45. Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, et al. Dysregulation in Akt/mTOR/HIF-1 Signaling Identified by Proteo-Transcriptomics of SARS-CoV-2 Infected Cells. Emerg Microbes Infect (2020) 9(1):1748–60. doi: 10.1080/22221751.2020.1799723
46. Stukalov A, Girault V, Grass V, Bergant V, Karayel O, Urban C, et al. Multi-Level Proteomics Reveals Host-Perturbation Strategies of SARS-CoV-2 and SARS-CoV. bioRxiv Preprint Server Biol (2020) 594:246–52. doi: 10.1101/2020.06.17.156455
47. Wyler E, Mösbauer K, Franke V, Diag A, Gottula LT, Arsiè R, et al. Transcriptomic Profiling of SARS-CoV-2 Infected Human Cell Lines Identifies HSP90 as Target for COVID-19 Therapy. iScience (2021) 24(3):102151. doi: 10.1016/j.isci.2021.102151
48. Li Y, Wang Y, Liu H, Sun W, Ding B, Zhao Y, et al. Urine Proteome of COVID-19 Patients. Urine (Amst) (2020) 2:1-8. doi: 10.1016/j.urine.2021.02.001
49. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients With COVID-19. Nat Med (2020) 26(6):842–4. doi: 10.1038/s41591-020-0901-9
50. Saunders RD, Nakajima ST, Rai SN, Pan J, Gercel-Taylor C, Taylor DD. Alterations in Antibody Subclass Immune Reactivity to Trophoblast-Derived Fetal Fibronectin and α2-Macroglobulin in Women With Recurrent Pregnancy Loss. Am J Reprod Immunol (2012) 68(5):438–49. doi: 10.1111/j.1600-0897.2012.01182.x
51. Bunn CC, Bernstein RM, Mathews MB. Autoantibodies Against alanyl-tRNA Synthetase and Trnaala Coexist and are Associated With Myositis. J Exp Med (1986) 163(5):1281–91. doi: 10.1084/jem.163.5.1281
52. Mande PV, Parikh FR, Hinduja I, Zaveri K, Vaidya R, Gajbhiye R, et al. Identification and Validation of Candidate Biomarkers Involved in Human Ovarian Autoimmunity. Reprod BioMed Online (2011) 23(4):471–83. doi: 10.1016/j.rbmo.2011.06.013
53. Van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Bozic B, Pruijn GJ. The Rheumatoid Arthritis Synovial Fluid Citrullinome Reveals Novel Citrullinated Epitopes in Apolipoprotein E, Myeloid Nuclear Differentiation Antigen, and Beta-Actin. Arthritis Rheum (2013) 65(1):69–80. doi: 10.1002/art.37720
54. Hanrotel-Saliou C, Segalen I, Le Meur Y, Youinou P, Renaudineau Y. Glomerular Antibodies in Lupus Nephritis. Clin Rev Allergy Immunol (2011) 40(3):151–8. doi: 10.1007/s12016-010-8204-4
55. Wang T, Liu M, Zheng SJ, Bian DD, Zhang JY, Yao J, et al. Tumor-Associated Autoantibodies Are Useful Biomarkers in Immunodiagnosis of α-Fetoprotein-Negative Hepatocellular Carcinoma. World J Gastroenterol (2017) 23(19):3496–504. doi: 10.3748/wjg.v23.i19.3496
56. Sköldberg F, Rönnblom L, Thornemo M, Lindahl A, Bird PI, Rorsman F, et al. Identification of AHNAK as a Novel Autoantigen in Systemic Lupus Erythematosus. Biochem Biophys Res Commun (2002) 291(4):951–8. doi: 10.1006/bbrc.2002.6534
57. Nehring J, Schirmbeck LA, Friebus-Kardash J, Dubler D, Huynh-Do U, Chizzolini C, et al. Autoantibodies Against Albumin in Patients With Systemic Lupus Erythematosus. Front Immunol (2018) 9:2090. doi: 10.3389/fimmu.2018.02090
58. Lu D, Kuhn E, Bristow RE, Giuntoli RL, Kjær SK, Ie S, et al. Comparison of Candidate Serologic Markers for Type I and Type II Ovarian Cancer. Gynecol Oncol (2011) 122(3):560–6. doi: 10.1016/j.ygyno.2011.05.039
59. Caster DJ, Korte EA, Merchant ML, Klein JB, Wilkey DW, Rovin BH, et al. Autoantibodies Targeting Glomerular Annexin A2 Identify Patients With Proliferative Lupus Nephritis. Proteomics Clin Appl (2015) 9(11-12):1012–20. doi: 10.1002/prca.201400175
60. Hrycek A, Cieślik P. Annexin A5 and Anti-Annexin Antibodies in Patients With Systemic Lupus Erythematosus. Rheumatol Int (2012) 32(5):1335–42. doi: 10.1007/s00296-011-1793-2
61. Seko Y, Matsumoto A, Fukuda T, Imai Y, Fujimura T, Taka H, et al. A Case of Neonatal Lupus Erythematosus Presenting Delayed Dilated Cardiomyopathy With Circulating Autoantibody to Annexin A6. Int Heart J (2007) 48(3):407–15. doi: 10.1536/ihj.48.407
62. Jarius S, Wildemann B. ‘Medusa Head Ataxia’: The Expanding Spectrum of Purkinje Cell Antibodies in Autoimmune Cerebellar Ataxia. Part 3: Anti-Yo/CDR2, Anti-Nb/AP3B2, PCA-2, Anti-Tr/DNER, Other Antibodies, Diagnostic Pitfalls, Summary and Outlook. J Neuroinflamm (2015) 12:168. doi: 10.1186/s12974-015-0358-9
63. Vuilleumier N, Montecucco F, Hartley O. Autoantibodies to Apolipoprotein A-1 as a Biomarker of Cardiovascular Autoimmunity. World J Cardiol (2014) 6(5):314–26. doi: 10.4330/wjc.v6.i5.314
64. Creaney J, Dick IM, Yeoman D, Wong S, Robinson BW. Auto-Antibodies to β-F1-ATPase and Vimentin in Malignant Mesothelioma. PLoS One (2011) 6(10):e26515. doi: 10.1371/journal.pone.0026515
65. Polgár A, Falus A, Koó E, Ujfalussy I, Seszták M, Szuts I, et al. Elevated Levels of Synovial Fluid Antibodies Reactive With the Small Proteoglycans Biglycan and Decorin in Patients With Rheumatoid Arthritis or Other Joint Diseases. Rheumatol (Oxford England) (2003) 42(4):522–7. doi: 10.1093/rheumatology/keg168
66. Bhat NM, Adams CM, Chen Y, Bieber MM, Teng NN. Identification of Cell Surface Straight Chain Poly-N-Acetyl-Lactosamine Bearing Protein Ligands for VH4-34-Encoded Natural IgM Antibodies. J Immunol (Baltimore Md: 1950) (2015) 195(11):5178–88. doi: 10.4049/jimmunol.1501697
67. Beutgen VM, Schmelter C, Pfeiffer N, Grus FH. Autoantigens in the Trabecular Meshwork and Glaucoma-Specific Alterations in the Natural Autoantibody Repertoire. Clin Transl Immunol (2020) 9(3):e01101. doi: 10.1002/cti2.1101
68. Kishore U, Sontheimer RD, Sastry KN, Zappi EG, Hughes GR, Khamashta MA, et al. The Systemic Lupus Erythematosus (SLE) Disease Autoantigen-Calreticulin can Inhibit C1q Association With Immune Complexes. Clin Exp Immunol (1997) 108(2):181–90. doi: 10.1046/j.1365-2249.1997.3761273.x
69. Terrier B, Tamby MC, Camoin L, Guilpain P, Broussard C, Bussone G, et al. Identification of Target Antigens of Antifibroblast Antibodies in Pulmonary Arterial Hypertension. Am J Respir Crit Care Med (2008) 177(10):1128–34. doi: 10.1164/rccm.200707-1015OC
70. Weber CK, Haslbeck M, Englbrecht M, Sehnert B, Mielenz D, Graef D, et al. Antibodies to the Endoplasmic Reticulum-Resident Chaperones Calnexin, BiP and Grp94 in Patients With Rheumatoid Arthritis and Systemic Lupus Erythematosus. Rheumatol (Oxford England) (2010) 49(12):2255–63. doi: 10.1093/rheumatology/keq272
71. Matsuo K, Xiang Y, Nakamura H, Masuko K, Yudoh K, Noyori K, et al. Identification of Novel Citrullinated Autoantigens of Synovium in Rheumatoid Arthritis Using a Proteomic Approach. Arthritis Res Ther (2006) 8(6):R175. doi: 10.1186/ar2085
72. Li WH, Zhao J, Li HY, Liu H, Li AL, Wang HX, et al. Proteomics-Based Identification of Autoantibodies in the Sera of Healthy Chinese Individuals From Beijing. Proteomics (2006) 6(17):4781–9. doi: 10.1002/pmic.200500909
73. Furuta K, Hildebrandt B, Matsuoka S, Kiyosawa K, Reimer G, Luderschmidt C, et al. Immunological Characterization of Heterochromatin Protein P25beta Autoantibodies and Relationship With Centromere Autoantibodies and Pulmonary Fibrosis in Systemic Scleroderma. J Mol Med (Berl) (1998) 76(1):54–60. doi: 10.1007/s001090050190
74. Ohyama K, Baba M, Tamai M, Aibara N, Ichinose K, Kishikawa N, et al. Proteomic Profiling of Antigens in Circulating Immune Complexes Associated With Each of Seven Autoimmune Diseases. Clin Biochem (2015) 48(3):181–5. doi: 10.1016/j.clinbiochem.2014.11.008
75. Hirai K, Maeda H, Omori K, Yamamoto T, Kokeguchi S, Takashiba S. Serum Antibody Response to Group II Chaperonin From Methanobrevibacter Oralis and Human Chaperonin CCT. Pathog Dis (2013) 68(1):12–9. doi: 10.1111/2049-632X.12041
76. Ebrahimi M, Nylander E, Bäcklund B, Wahlin YB, Coates PJ, Nylander K. The Use of a Novel ELISA Method for Detection of Antibodies Against P63 in Sera From Patients Diagnosed With Oral and/or Genital and Skin Lichen Planus. J Oral Pathol Med (2010) 39(6):486–90. doi: 10.1111/j.1600-0714.2010.00890.x
77. Zhu L, Shen W, Zhu M, Coorey NJ, Nguyen AP, Barthelmes D, et al. Anti-Retinal Antibodies in Patients With Macular Telangiectasia Type 2. Invest Ophthalmol Vis Sci (2013) 54(8):5675–83. doi: 10.1167/iovs.13-12050
78. Koivula MK, Aman S, Karjalainen A, Hakala M, Risteli J. Are There Autoantibodies Reacting Against Citrullinated Peptides Derived From Type I and Type II Collagens in Patients With Rheumatoid Arthritis? Ann Rheum Dis (2005) 64(10):1443–50. doi: 10.1136/ard.2004.031211
79. Pardos-Gea J, Cortés-Hernández J, Castro-Marrero J, Balada E, Ordi-Ros J. Autoantibodies to Types I and IV Collagen and Heart Valve Disease in Systemic Lupus Erythematosus/Antiphospholipid Syndrome. Clin Rheumatol (2017) 36(6):1401–6. doi: 10.1007/s10067-017-3594-9
80. Araujo GR, Fonseca JE, Fujimura PT, Cunha-Junior JP, Silva CH, Mourão AF, et al. Anti-Type II Collagen Antibodies Detection and Avidity in Patients With Oligoarticular and Polyarticular Forms of Juvenile Idiopathic Arthritis. Immunol Lett (2015) 165(1):20–5. doi: 10.1016/j.imlet.2015.03.006
81. Nakos G, Adams A, Andriopoulos N. Antibodies to Collagen in Patients With Idiopathic Pulmonary Fibrosis. Chest (1993) 103(4):1051–8. doi: 10.1378/chest.103.4.1051
82. Hachem RR, Tiriveedhi V, Patterson GA, Aloush A, Trulock EP, Mohanakumar T. Antibodies to K-α 1 Tubulin and Collagen V Are Associated With Chronic Rejection After Lung Transplantation. Am J Transpl: Off J Am Soc Transplant Am Soc Transplant Surgeons (2012) 12(8):2164–71. doi: 10.1111/j.1600-6143.2012.04079.x
83. Nath DS, Basha HI, Mohanakumar T. Antihuman Leukocyte Antigen Antibody-Induced Autoimmunity: Role in Chronic Rejection. Curr Opin Organ Transplant (2010) 15(1):16–20. doi: 10.1097/MOT.0b013e3283342780
84. Vece TJ, Watkin LB, Nicholas S, Canter D, Braun MC, Guillerman RP, et al. Copa Syndrome: A Novel Autosomal Dominant Immune Dysregulatory Disease. J Clin Immunol (2016) 36(4):377–87. doi: 10.1007/s10875-016-0271-8
85. Yang Q, Qin J, Sun G, Qiu C, Jiang D, Ye H, et al. Discovery and Validation of Serum Autoantibodies Against Tumor-Associated Antigens as Biomarkers in Gastric Adenocarcinoma Based on the Focused Protein Arrays. Clin Transl Gastroenterol (2020) 12(1):e00284. doi: 10.14309/ctg.0000000000000284
86. Hong HS, Chung WH, Hung SI, Chen MJ, Lee SH, Yang LC. Clinical Association of Anti-Golgi Autoantibodies and Their Autoantigens. Scand J Immunol (2004) 59(1):79–87. doi: 10.1111/j.0300-9475.2004.01353.x
87. Dummer R, Mittelman A, Fanizzi FP, Lucchese G, Willers J, Kanduc D. Non-Self-Discrimination as a Driving Concept in the Identification of an Immunodominant HMW-MAA Epitopic Peptide Sequence by Autoantibodies From Melanoma Cancer Patients. Int J Cancer (2004) 111(5):720–6. doi: 10.1002/ijc.20310
88. Vetvicka V, Fusek M. Cathepsin D: Autoantibody Profiling as a Diagnostic Marker for Cancers. World J Clin Oncol (2013) 4(1):1–3. doi: 10.5306/wjco.v4.i1.1
89. Pitsch J, Kamalizade D, Braun A, Kuehn JC, Gulakova PE, Rüber T, et al. Drebrin Autoantibodies in Patients With Seizures and Suspected Encephalitis. Ann Neurol (2020) 87(6):869–84. doi: 10.1002/ana.25720
90. Brandsma CA, Kerstjens HA, Van Geffen WH, Geerlings M, Postma DS, Hylkema MN, et al. Differential Switching to IgG and IgA in Active Smoking COPD Patients and Healthy Controls. Eur Respir J (2012) 40(2):313–21. doi: 10.1183/09031936.00011211
91. Fritzler MJ, Hamel JC, Ochs RL, Chan EK. Molecular Characterization of Two Human Autoantigens: Unique cDNAs Encoding 95- and 160-kD Proteins of a Putative Family in the Golgi Complex. J Exp Med (1993) 178(1):49–62. doi: 10.1084/jem.178.1.49
92. Scofield RH. Do We Need New Autoantibodies in Lupus? Arthritis Res Ther (2010) 12(3):120. doi: 10.1186/ar2998
93. Fregeau DR, Prindiville T, Coppel RL, Kaplan M, Dickson ER, Gershwin ME. Inhibition of Alpha-Ketoglutarate Dehydrogenase Activity by a Distinct Population of Autoantibodies Recognizing Dihydrolipoamide Succinyltransferase in Primary Biliary Cirrhosis. Hepatology (1990) 11(6):975–81. doi: 10.1002/hep.1840110611
94. Oka M, Sato S, Soda H, Fukuda M, Kawabata S, Nakatomi K, et al. Autoantibody to Heat Shock Protein Hsp40 in Sera of Lung Cancer Patients. Japanese J Cancer Res: Gann (2001) 92(3):316–20. doi: 10.1111/j.1349-7006.2001.tb01097.x
95. Harper MM, Rudd D, Meyer KJ, Kanthasamy AG, Anantharam V, Pieper AA, et al. Identification of Chronic Brain Protein Changes and Protein Targets of Serum Auto-Antibodies After Blast-Mediated Traumatic Brain Injury. Heliyon (2020) 6(2):e03374. doi: 10.1016/j.heliyon.2020.e03374
96. Kim EG, Kwak SH, Hwang D, Yi EC, Park KS, Koo BK, et al. The Level of Autoantibodies Targeting Eukaryote Translation Elongation Factor 1 α1 and Ubiquitin-Conjugating Enzyme 2L3 in Nondiabetic Young Adults. Diabetes Metab J (2016) 40(2):154–60. doi: 10.4093/dmj.2016.40.2.154
97. Mooney CJ, Dunphy EJ, Stone B, Mcneel DG. Identification of Autoantibodies Elicited in a Patient With Prostate Cancer Presenting as Dermatomyositis. Int J Urol (2006) 13(3):211–7. doi: 10.1111/j.1442-2042.2006.01263.x
98. Fernández-Madrid F, Tang N, Alansari H, Granda JL, Tait L, Amirikia KC, et al. Autoantibodies to Annexin XI-A and Other Autoantigens in the Diagnosis of Breast Cancer. Cancer Res (2004) 64(15):5089–96. doi: 10.1158/0008-5472.CAN-03-0932
99. Bach M, Winkelmann G, Luhrmann R. 20S Small Nuclear Ribonucleoprotein U5 Shows a Surprisingly Complex Protein Composition. Proc Natl Acad Sci USA (1989) 86(16):6038–42. doi: 10.1073/pnas.86.16.6038
100. Waterman EA, Gawkrodger DJ, Watson PF, Weetman AP, Kemp EH. Autoantigens in Vitiligo Identified by the Serological Selection of a Phage-Displayed Melanocyte cDNA Expression Library. J Invest Dermatol (2010) 130(1):230–40. doi: 10.1038/jid.2009.207
101. Heo CK, Hwang HM, Lee HJ, Kwak SS, Yoo JS, Yu DY, et al. Serum Anti-EIF3A Autoantibody as a Potential Diagnostic Marker for Hepatocellular Carcinoma. Sci Rep (2019) 9(1):11059. doi: 10.1038/s41598-019-47365-4
102. Betteridge Z, Chinoy H, Vencovsky J, Winer J, Putchakayala K, Ho P, et al. Identification of a Novel Autoantigen Eukaryotic Initiation Factor 3 Associated With Polymyositis. Rheumatol (Oxford England) (2020) 59(5):1026–30. doi: 10.1093/rheumatology/kez406
103. Suwarnalata G, Tan AH, Isa H, Gudimella R, Anwar A, Loke MF, et al. Augmentation of Autoantibodies by Helicobacter Pylori in Parkinson’s Disease Patients May Be Linked to Greater Severity. PLoS One (2016) 11(4):e0153725. doi: 10.1371/journal.pone.0153725
104. Nabors LB, Furneaux HM, King PH. HuR, a Novel Target of Anti-Hu Antibodies, Is Expressed in Non-Neural Tissues. J Neuroimmunol (1998) 92(1-2):152–9. doi: 10.1016/S0165-5728(98)00196-9
105. Moscato S, Pratesi F, Sabbatini A, Chimenti D, Scavuzzo M, Passatino R, et al. Surface Expression of a Glycolytic Enzyme, Alpha-Enolase, Recognized by Autoantibodies in Connective Tissue Disorders. Eur J Immunol (2000) 30(12):3575–84. doi: 10.1002/1521-4141(200012)30:12<3575::AID-IMMU3575>3.0.CO;2-#
106. O’dwyer DT, Clifton V, Hall A, Smith R, Robinson PJ, Crock PA. Pituitary Autoantibodies in Lymphocytic Hypophysitis Target Both Gamma- and Alpha-Enolase - A Link With Pregnancy? Arch Physiol Biochem (2002) 110(1-2):94–8. doi: 10.1076/apab.110.1.94.897
107. Targoff IN, Trieu EP, Miller FW. Reaction of Anti-OJ Autoantibodies With Components of the Multi-Enzyme Complex of aminoacyl-tRNA Synthetases in Addition to isoleucyl-tRNA Synthetase. J Clin Invest (1993) 91(6):2556–64. doi: 10.1172/JCI116493
108. Garranzo-Asensio M, San Segundo-Acosta P, Povés C, Fernández-Aceñero MJ, Martínez-Useros J, Montero-Calle A, et al. Identification of Tumor-Associated Antigens With Diagnostic Ability of Colorectal Cancer by in-Depth Immunomic and Seroproteomic Analysis. J Proteomics (2020) 214:103635. doi: 10.1016/j.jprot.2020.103635
109. Leveque C, Hoshino T, David P, Shoji-Kasai Y, Leys K, Omori A, et al. The Synaptic Vesicle Protein Synaptotagmin Associates With Calcium Channels and is a Putative Lambert-Eaton Myasthenic Syndrome Antigen. Proc Natl Acad Sci USA (1992) 89(8):3625–9. doi: 10.1073/pnas.89.8.3625
110. Heo CK, Woo MK, Yu DY, Lee JY, Yoo JS, Yoo HS, et al. Identification of Autoantibody Against Fatty Acid Synthase in Hepatocellular Carcinoma Mouse Model and Its Application to Diagnosis of HCC. Int J Oncol (2010) 36(6):1453–9. doi: 10.3892/ijo_00000631
111. Forti S, Scanlan MJ, Invernizzi A, Castiglioni F, Pupa S, Agresti R, et al. Identification of Breast Cancer-Restricted Antigens by Antibody Screening of SKBR3 cDNA Library Using a Preselected Patient’s Serum. Breast Cancer Res Treat (2002) 73(3):245–56. doi: 10.1023/A:1015854415746
112. Kamhieh-Milz J, Sterzer V, Celik H, Khorramshahi O, Fadl Hassan Moftah R, Salama A. Identification of Novel Autoantigens via Mass Spectroscopy-Based Antibody-Mediated Identification of Autoantigens (MS-AMIDA) Using Immune Thrombocytopenic Purpura (ITP) as a Model Disease. J Proteomics (2017) 157:59–70. doi: 10.1016/j.jprot.2017.01.012
113. Adachi-Hayama M, Adachi A, Shinozaki N, Matsutani T, Hiwasa T, Takiguchi M, et al. Circulating Anti-Filamin C Autoantibody as a Potential Serum Biomarker for Low-Grade Gliomas. BMC Cancer (2014) 14:452. doi: 10.1186/1471-2407-14-452
114. Wang WY, Twu CW, Liu YC, Lin HH, Chen CJ, Lin JC. Fibronectin Promotes Nasopharyngeal Cancer Cell Motility and Proliferation. BioMed Pharmacother (2019) 109:1772–84. doi: 10.1016/j.biopha.2018.11.055
115. Dong X, Yang M, Sun H, Lü J, Zheng Z, Li Z, et al. Combined Measurement of CA 15-3 With Novel Autoantibodies Improves Diagnostic Accuracy for Breast Cancer. Onco Targets Ther (2013) 6:273–9. doi: 10.2147/OTT.S43122
116. Kit Y, Starykovych M, Vajrychova M, Lenco J, Zastavna D, Stoika R. Detection of Novel Auto-Antigens in Patients With Recurrent Miscarriage: Description of an Approach and Preliminary Findings. Croat Med J (2014) 55(3):259–64. doi: 10.3325/cmj.2014.55.259
117. Delunardo F, Soldati D, Bellisario V, Berry A, Camerini S, Crescenzi M, et al. Anti-GAPDH Autoantibodies as a Pathogenic Determinant and Potential Biomarker of Neuropsychiatric Diseases. Arthritis Rheumatol (2016) 68(11):2708–16. doi: 10.1002/art.39750
118. Kiyota A, Iwama S, Sugimura Y, Takeuchi S, Takagi H, Iwata N, et al. Identification of the Novel Autoantigen Candidate Rab GDP Dissociation Inhibitor Alpha in Isolated Adrenocorticotropin Deficiency. Endocr J (2015) 62(2):153–60. doi: 10.1507/endocrj.EJ14-0369
119. Massa O, Alessio M, Russo L, Nardo G, Bonetto V, Bertuzzi F, et al. Serological Proteome Analysis (SERPA) as a Tool for the Identification of New Candidate Autoantigens in Type 1 Diabetes. J Proteomics (2013) 82:263–73. doi: 10.1016/j.jprot.2013.02.030
120. Chung JM, Jung Y, Kim YP, Song J, Kim S, Kim JY, et al. Identification of the Thioredoxin-Like 2 Autoantibody as a Specific Biomarker for Triple-Negative Breast Cancer. J Breast Cancer (2018) 21(1):87–90. doi: 10.4048/jbc.2018.21.1.87
121. Biswas S, Sharma S, Saroha A, Bhakuni DS, Malhotra R, Zahur M, et al. Identification of Novel Autoantigen in the Synovial Fluid of Rheumatoid Arthritis Patients Using an Immunoproteomics Approach. PLoS One (2013) 8(2):e56246. doi: 10.1371/journal.pone.0056246
122. Rubin RL, Bell SA, Burlingame RW. Autoantibodies Associated With Lupus Induced by Diverse Drugs Target a Similar Epitope in the (H2A-H2B)-DNA Complex. J Clin Invest (1992) 90(1):165–73. doi: 10.1172/JCI115832
123. Burlingame RW, Boey ML, Starkebaum G, Rubin RL. The Central Role of Chromatin in Autoimmune Responses to Histones and DNA in Systemic Lupus Erythematosus. J Clin Invest (1994) 94(1):184–92. doi: 10.1172/JCI117305
124. Nahamura H, Yoshida K, Kishima Y, Enomoto H, Uyama H, Kuroda T, et al. Circulating Auto-Antibody Against Hepatoma-Derived Growth Factor (HDGF) in Patients With Ulcerative Colitis. Hepatogastroenterology (2004) 51(56):470–5.
125. Wesierska-Gadek J, Penner E, Lindner H, Hitchman E, Sauermann G. Autoantibodies Against Different Histone H1 Subtypes in Systemic Lupus Erythematosus Sera. Arthritis Rheum (1990) 33(8):1273–8. doi: 10.1002/art.1780330830
126. Baranova SV, Dmitrienok PS, Ivanisenko NV, Buneva VN, Nevinsky GA. Antibodies to H2a and H2b Histones From the Sera of HIV-Infected Patients Catalyze Site-Specific Degradation of These Histones. Mol Biosyst (2017) 13(6):1090–101. doi: 10.1039/C7MB00042A
127. Bruschi M, Galetti M, Sinico RA, Moroni G, Bonanni A, Radice A, et al. Glomerular Autoimmune Multicomponents of Human Lupus Nephritis In Vivo (2): Planted Antigens. J Am Soc Nephrol (2015) 26(8):1905–24. doi: 10.1681/ASN.2014050493
128. Van Bavel CC, Dieker J, Muller S, Briand JP, Monestier M, Berden JH, et al. Apoptosis-Associated Acetylation on Histone H2B Is an Epitope for Lupus Autoantibodies. Mol Immunol (2009) 47(2-3):511–6. doi: 10.1016/j.molimm.2009.08.009
129. Baranova SV, Dmitrenok PS, Zubkova AD, Ivanisenko NV, Odintsova ES, Buneva VN, et al. Antibodies Against H3 and H4 Histones From the Sera of HIV-Infected Patients Catalyze Site-Specific Degradation of These Histones. J Mol Recognition: JMR (2018) 31(7):e2703. doi: 10.1002/jmr.2703
130. Barnay-Verdier S, Fattoum L, Borde C, Kaveri S, Gibot S, Maréchal V. Emergence of Autoantibodies to HMGB1 Is Associated With Survival in Patients With Septic Shock. Intensive Care Med (2011) 37(6):957–62. doi: 10.1007/s00134-011-2192-6
131. Guarneri C, Aguennouz M, Guarneri F, Polito F, Benvenga S, Cannavo SP. Autoimmunity to Heterogeneous Nuclear Ribonucleoprotein A1 in Psoriatic Patients and Correlation With Disease Severity. J Dtsch Dermatol Ges (2018) 16(9):1103–7. doi: 10.1111/ddg.13631
132. Konig MF, Giles JT, Nigrovic PA, Andrade F. Antibodies to Native and Citrullinated RA33 (hnRNP A2/B1) Challenge Citrullination as the Inciting Principle Underlying Loss of Tolerance in Rheumatoid Arthritis. Ann Rheum Dis (2016) 75(11):2022–8. doi: 10.1136/annrheumdis-2015-208529
133. Siapka S, Patrinou-Georgoula M, Vlachoyiannopoulos PG, Guialis A. Multiple Specificities of Autoantibodies Against hnRNP a/B Proteins in Systemic Rheumatic Diseases and hnRNP L as an Associated Novel Autoantigen. Autoimmunity (2007) 40(3):223–33. doi: 10.1080/08916930701352357
134. Heegaard NH, Larsen MR, Muncrief T, Wiik A, Roepstorff P. Heterogeneous Nuclear Ribonucleoproteins C1/C2 Identified as Autoantigens by Biochemical and Mass Spectrometric Methods. Arthritis Res (2000) 2(5):407–14. doi: 10.1186/ar119
135. Skriner K, Hueber W, Süleymanoglu E, Höfler E, Krenn V, Smolen J, et al. AUF1, the Regulator of Tumor Necrosis Factor Alpha Messenger RNA Decay, Is Targeted by Autoantibodies of Patients With Systemic Rheumatic Diseases. Arthritis Rheum (2008) 58(2):511–20. doi: 10.1002/art.23306
136. Zhang Y, Zhao H, Liu B, Li L, Zhang L, Bao M, et al. Low Level Antibodies Against Alpha-Tropomyosin Are Associated With Increased Risk of Coronary Heart Disease. Front Pharmacol (2020) 11:195. doi: 10.3389/fphar.2020.00195
137. Op De Beéck K, Maes L, Van Den Bergh K, Derua R, Waelkens E, Van Steen K, et al. Heterogeneous Nuclear RNPs as Targets of Autoantibodies in Systemic Rheumatic Diseases. Arthritis Rheum (2012) 64(1):213–21. doi: 10.1002/art.33327
138. Yang L, Fujimoto M, Murota H, Serada S, Fujimoto M, Honda H, et al. Proteomic Identification of Heterogeneous Nuclear Ribonucleoprotein K as a Novel Cold-Associated Autoantigen in Patients With Secondary Raynaud’s Phenomenon. Rheumatol (Oxford England) (2015) 54(2):349–58. doi: 10.1093/rheumatology/keu325
139. Hassfeld W, Chan EK, Mathison DA, Portman D, Dreyfuss G, Steiner G, et al. Molecular Definition of Heterogeneous Nuclear Ribonucleoprotein R (hnRNP R) Using Autoimmune Antibody: Immunological Relationship With hnRNP P. Nucleic Acids Res (1998) 26(2):439–45. doi: 10.1093/nar/26.2.439
140. Harlow L, Rosas IO, Gochuico BR, Mikuls TR, Dellaripa PF, Oddis CV, et al. Identification of Citrullinated Hsp90 Isoforms as Novel Autoantigens in Rheumatoid Arthritis-Associated Interstitial Lung Disease. Arthritis Rheum (2013) 65(4):869–79. doi: 10.1002/art.37881
141. Qin HY, Mahon JL, Atkinson MA, Chaturvedi P, Lee-Chan E, Singh B. Type 1 Diabetes Alters Anti-Hsp90 Autoantibody Isotype. J Autoimmun (2003) 20(3):237–45. doi: 10.1016/S0896-8411(03)00035-0
142. Cid C, Regidor I, Alcazar A. Anti-Heat Shock Protein 90beta Antibodies Are Detected in Patients With Multiple Sclerosis During Remission. J Neuroimmunol (2007) 184(1-2):223–6. doi: 10.1016/j.jneuroim.2006.11.001
143. Suzuki S, Utsugisawa K, Iwasa K, Satoh T, Nagane Y, Yoshikawa H, et al. Autoimmunity to Endoplasmic Reticulum Chaperone GRP94 in Myasthenia Gravis. J Neuroimmunol (2011) 237(1-2):87–92. doi: 10.1016/j.jneuroim.2011.06.011
144. Chen M, Aosai F, Mun HS, Norose K, Hata H, Yano A. Anti-HSP70 Autoantibody Formation by B-1 Cells in Toxoplasma Gondii-Infected Mice. Infect Immun (2000) 68(9):4893–9. doi: 10.1128/IAI.68.9.4893-4899.2000
145. Shimizu F, Schaller KL, Owens GP, Cotleur AC, Kellner D, Takeshita Y, et al. Glucose-Regulated Protein 78 Autoantibody Associates With Blood-Brain Barrier Disruption in Neuromyelitis Optica. Sci Transl Med (2017) 9(397):eaai9111. doi: 10.1126/scitranslmed.aai9111
146. Iannaccone A, Giorgianni F, New DD, Hollingsworth TJ, Umfress A, Alhatem AH, et al. Circulating Autoantibodies in Age-Related Macular Degeneration Recognize Human Macular Tissue Antigens Implicated in Autophagy, Immunomodulation, and Protection From Oxidative Stress and Apoptosis. PLoS One (2015) 10(12):e0145323. doi: 10.1371/journal.pone.0145323
147. Korneeva I, Bongiovanni AM, Girotra M, Caputo TA, Witkin SS. Serum Antibodies to the 27-Kd Heat Shock Protein in Women With Gynecologic Cancers. Am J Obstet Gynecol (2000) 183(1):18–21. doi: 10.1016/S0002-9378(00)72431-8
148. Fillit H, Shibata S, Sasaki T, Spiera H, Kerr LD, Blake M. Autoantibodies to the Protein Core of Vascular Basement Membrane Heparan Sulfate Proteoglycan in Systemic Lupus Erythematosus. Autoimmunity (1993) 14(3):243–9. doi: 10.3109/08916939309077372
149. Bremer HD, Landegren N, Sjoberg R, Hallgren A, Renneker S, Lattwein E, et al. ILF2 and ILF3 Are Autoantigens in Canine Systemic Autoimmune Disease. Sci Rep (2018) 8(1):4852. doi: 10.1038/s41598-018-23034-w
150. Presslauer S, Hinterhuber G, Cauza K, Horvat R, Rappersberger K, Wolff K, et al. RasGAP-Like Protein IQGAP1 Is Expressed by Human Keratinocytes and Recognized by Autoantibodies in Association With Bullous Skin Disease. J Invest Dermatol (2003) 120(3):365–71. doi: 10.1046/j.1523-1747.2003.12070.x
151. Ola TO, Biro PA, Hawa MI, Ludvigsson J, Locatelli M, Puglisi MA, et al. Importin Beta: A Novel Autoantigen in Human Autoimmunity Identified by Screening Random Peptide Libraries on Phage. J Autoimmun (2006) 26(3):197–207. doi: 10.1016/j.jaut.2006.01.003
152. Lu Y, Ye P, Chen SL, Tan EM, Chan EK. Identification of Kinectin as a Novel Behçet’s Disease Autoantigen. Arthritis Res Ther (2005) 7(5):R1133–1139. doi: 10.1186/ar1798
153. Inagaki J, Kondo A, Lopez LR, Shoenfeld Y, Matsuura E. Pregnancy Loss and Endometriosis: Pathogenic Role of Anti-Laminin-1 Autoantibodies. Ann New York Acad Sci (2005) 1051:174–84. doi: 10.1196/annals.1361.059
154. Peutz-Kootstra CJ, Hansen K, De Heer E, Abrass CK, Bruijn JA. Differential Expression of Laminin Chains and Anti-Laminin Autoantibodies in Experimental Lupus Nephritis. J Pathol (2000) 192(3):404–12. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH707>3.0.CO;2-L
155. Ueda K, Nakanishi T, Shimizu A, Takubo T, Matsuura N. Identification of L-Plastin Autoantibody in Plasma of Patients With Non-Hodgkin’s Lymphoma Using a Proteomics-Based Analysis. Ann Clin Biochem (2008) 45(Pt 1):65–9. doi: 10.1258/acb.2007.006230
156. Lutomski D, Joubert-Caron R, Lefebure C, Salama J, Belin C, Bladier D, et al. Anti-Galectin-1 Autoantibodies in Serum of Patients With Neurological Diseases. Clin Chim Acta (1997) 262(1-2):131–8. doi: 10.1016/S0009-8981(97)06544-3
157. Konstantinov KN, Galcheva-Gargova Z, Hoier-Madsen M, Wiik A, Ullman S, Halberg P, et al. Autoantibodies to Lamins A and C in Sera of Patients Showing Peripheral Fluorescent Antinuclear Antibody Pattern on HEP-2 Cells. J Invest Dermatol (1990) 95(3):304–8. doi: 10.1111/1523-1747.ep12485010
158. Von Mikecz A, Konstantinov K, Buchwald DS, Gerace L, Tan EM. High Frequency of Autoantibodies to Insoluble Cellular Antigens in Patients With Chronic Fatigue Syndrome. Arthritis Rheum (1997) 40(2):295–305. doi: 10.1002/art.1780400215
159. Brito J, Biamonti G, Caporali R, Montecucco C. Autoantibodies to Human Nuclear Lamin B2 Protein. Epitope Specificity in Different Autoimmune Diseases. J Immunol (Baltimore Md: 1950) (1994) 153(5):2268–77.
160. Tanaka M, Kishimura M, Ozaki S, Osakada F, Hashimoto H, Okubo M, et al. Cloning of Novel Soluble Gp130 and Detection of Its Neutralizing Autoantibodies in Rheumatoid Arthritis. J Clin Invest (2000) 106(1):137–44. doi: 10.1172/JCI7479
161. Gadoth A, Kryzer TJ, Fryer J, Mckeon A, Lennon VA, Pittock SJ. Microtubule-Associated Protein 1B: Novel Paraneoplastic Biomarker. Ann Neurol (2017) 81(2):266–77. doi: 10.1002/ana.24872
162. Suzuki K, Nagao T, Itabashi M, Hamano Y, Sugamata R, Yamazaki Y, et al. A Novel Autoantibody Against Moesin in the Serum of Patients With MPO-ANCA-Associated Vasculitis. Nephrol Dial Transplant (2014) 29(6):1168–77. doi: 10.1093/ndt/gft469
163. Marinou D, Katsifis G, Barouta G, Liaskos C, Sakkas LI, Tsakris A, et al. Major Vault Protein/Lung Resistance Related Protein: A Novel Biomarker for Rheumatoid Arthritis. Clin Exp Rheumatol (2020) 39(5):1033-42.
164. Von Muhlen CA, Chan EK, Peebles CL, Imai H, Kiyosawa K, Tan EM. Non-Muscle Myosin as Target Antigen for Human Autoantibodies in Patients With Hepatitis C Virus-Associated Chronic Liver Diseases. Clin Exp Immunol (1995) 100(1):67–74. doi: 10.1111/j.1365-2249.1995.tb03605.x
165. Zasońska BA, Hlídková H, Petrovský E, Myronovskij S, Nehrych T, Negrych N, et al. Monodisperse Magnetic Poly(Glycidyl Methacrylate) Microspheres for Isolation of Autoantibodies With Affinity for the 46 kDa Form of Unconventional Myo1C Present in Autoimmune Patients. Mikrochimica Acta (2018) 185(5):262. doi: 10.1007/s00604-018-2807-5
166. Mossabeb R, Seiberler S, Mittermann I, Reininger R, Spitzauer S, Natter S. Characterization of a Novel Isoform of Alpha-Nascent Polypeptide-Associated Complex as IgE-Defined Autoantigen. J Invest Dermatol (2002) 119(4):820–9. doi: 10.1046/j.1523-1747.2002.00518.x
167. Batova IN, Richardson RT, Widgren EE, O’rand MG. Analysis of the Autoimmune Epitopes on Human Testicular NASP Using Recombinant and Synthetic Peptides. Clin Exp Immunol (2000) 121(2):201–9. doi: 10.1046/j.1365-2249.2000.01303.x
168. Qin Z, Lavingia B, Zou Y, Stastny P. Antibodies Against Nucleolin in Recipients of Organ Transplants. Transplantation (2011) 92(7):829–35. doi: 10.1097/TP.0b013e31822d0977
169. Cortés-Sarabia K, Rodríguez-Nava C, Medina-Flores Y, Mata-Ruíz O, López-Meza JE, Gómez-Cervantes MD, et al. Production and Characterization of a Monoclonal Antibody Against the Sialidase of Gardnerella Vaginalis Using a Synthetic Peptide in a MAP8 Format. Appl Microbiol Biotechnol (2020) 104(14):6173–83. doi: 10.1007/s00253-020-10691-z
170. Le Naour F, Brichory F, Misek DE, Brechot C, Hanash SM, Beretta L. A Distinct Repertoire of Autoantibodies in Hepatocellular Carcinoma Identified by Proteomic Analysis. Mol Cell proteomics: MCP (2002) 1(3):197–203. doi: 10.1074/mcp.M100029-MCP200
171. Underwood JR, Csar XF, Veitch BA, Hearn MT. Characterization of the Specificity of a Naturally-Occurring Monoclonal Anti-Thymocyte Autoantibody Derived From an Unimmunized, Neonatal Balb/c Mouse. Thymus (1993) 21(4):199–219.
172. Brankin B, Skaar TC, Brotzman M, Trock B, Clarke R. Autoantibodies to the Nuclear Phosphoprotein Nucleophosmin in Breast Cancer Patients. Cancer Epidemiol Biomarkers Prev (1998) 7(12):1109–15.
173. Andrade LE, Chan EK, Peebles CL, Tan EM. Two Major Autoantigen-Antibody Systems of the Mitotic Spindle Apparatus. Arthritis Rheum (1996) 39(10):1643–53. doi: 10.1002/art.1780391006
174. Ochs RL, Stein TW Jr., Chan EK, Ruutu M, Tan EM. cDNA Cloning and Characterization of a Novel Nucleolar Protein. Mol Biol Cell (1996) 7(7):1015–24. doi: 10.1091/mbc.7.7.1015
175. Nagayama S, Yokoi T, Tanaka H, Kawaguchi Y, Shirasaka T, Kamataki T. Occurrence of Autoantibody to Protein Disulfide Isomerase in Patients With Hepatic Disorder. J Toxicol Sci (1994) 19(3):163–9. doi: 10.2131/jts.19.3_163
176. Becker A, Ludwig N, Keller A, Tackenberg B, Eienbroker C, Oertel WH, et al. Myasthenia Gravis: Analysis of Serum Autoantibody Reactivities to 1827 Potential Human Autoantigens by Protein Macroarrays. PLoS One (2013) 8(3):e58095. doi: 10.1371/journal.pone.0058095
177. Houng AK, Maggini L, Clement CY, Reed GL. Identification and Structure of Activated-Platelet Protein-1, a Protein With RNA-Binding Domain Motifs That Is Expressed by Activated Platelets. Eur J Biochem (1997) 243(1-2):209–18. doi: 10.1111/j.1432-1033.1997.0209a.x
178. Kaneda K, Takasaki Y, Takeuchi K, Yamada H, Nawata M, Matsushita M, et al. Autoimmune Response to Proteins of Proliferating Cell Nuclear Antigen Multiprotein Complexes in Patients With Connective Tissue Diseases. J Rheumatol (2004) 31(11):2142–50.
179. Caorsi C, Niccolai E, Capello M, Vallone R, Chattaragada MS, Alushi B, et al. Protein Disulfide Isomerase A3-Specific Th1 Effector Cells Infiltrate Colon Cancer Tissue of Patients With Circulating Anti-Protein Disulfide Isomerase A3 Autoantibodies. Transl Res (2016) 171:17–28.e11-12. doi: 10.1016/j.trsl.2015.12.013
180. Chang DC, Piaggi P, Hanson RL, Knowler WC, Bogardus C, Krakoff J. Autoantibodies Against PFDN2 are Associated With an Increased Risk of Type 2 Diabetes: A Case-Control Study. Diabetes Metab Res Rev (2017) 33(8):10.1002/dmrr.2922. doi: 10.1002/dmrr.2922
181. Orchard PJ, Nascene DR, Gupta A, Taisto ME, Higgins L, Markowski TW, et al. Cerebral Adrenoleukodystrophy is Associated With Loss of Tolerance to Profilin. Eur J Immunol (2019) 49(6):947–53. doi: 10.1002/eji.201848043
182. Frampton G, Moriya S, Pearson JD, Isenberg DA, Ward FJ, Smith TA, et al. Identification of Candidate Endothelial Cell Autoantigens in Systemic Lupus Erythematosus Using a Molecular Cloning Strategy: A Role for Ribosomal P Protein P0 as an Endothelial Cell Autoantigen. Rheumatol (Oxford England) (2000) 39(10):1114–20. doi: 10.1093/rheumatology/39.10.1114
183. Wieczorek M, Czernik A. Paraneoplastic Pemphigus: A Short Review. Clin Cosmet Investig Dermatol (2016) 9:291–5. doi: 10.2147/CCID.S100802
184. Kratz A, Harding MW, Craft J, Mackworth-Young CG, Handschumacher RE. Autoantibodies Against Cyclophilin in Systemic Lupus Erythematosus and Lyme Disease. Clin Exp Immunol (1992) 90(3):422–7. doi: 10.1111/j.1365-2249.1992.tb05862.x
185. Lin LH, Xu YW, Huang LS, Hong CQ, Zhai TT, Liao LD, et al. Serum Proteomic-Based Analysis Identifying Autoantibodies Against PRDX2 and PRDX3 as Potential Diagnostic Biomarkers in Nasopharyngeal Carcinoma. Clin Proteomics (2017) 14:6. doi: 10.1186/s12014-017-9141-5
186. Kobayashi S, Hiwasa T, Arasawa T, Kagaya A, Ishii S, Shimada H, et al. Identification of Specific and Common Diagnostic Antibody Markers for Gastrointestinal Cancers by SEREX Screening Using Testis cDNA Phage Library. Oncotarget (2018) 9(26):18559–69. doi: 10.18632/oncotarget.24963
187. Schild-Poulter C, Su A, Shih A, Kelly OP, Fritzler MJ, Goldstein R, et al. Association of Autoantibodies With Ku and DNA Repair Proteins in Connective Tissue Diseases. Rheumatol (Oxford England) (2008) 47(2):165–71. doi: 10.1093/rheumatology/kem338
188. Feist E, Kuckelkorn U, Dörner T, Donitz H, Scheffler S, Hiepe F, et al. Autoantibodies in Primary Sjögren’s Syndrome are Directed Against Proteasomal Subunits of the Alpha and Beta Type. Arthritis Rheum (1999) 42(4):697–702. doi: 10.1002/1529-0131(199904)42:4<697::AID-ANR12>3.0.CO;2-H
189. Feist E, Dorner T, Kuckelkorn U, Schmidtke G, Micheel B, Hiepe F, et al. Proteasome Alpha-Type Subunit C9 is a Primary Target of Autoantibodies in Sera of Patients With Myositis and Systemic Lupus Erythematosus. J Exp Med (1996) 184(4):1313–8. doi: 10.1084/jem.184.4.1313
190. Bohring C, Krause W. Characterization of Spermatozoa Surface Antigens by Antisperm Antibodies and its Influence on Acrosomal Exocytosis. Am J Reprod Immunol (2003) 50(5):411–9. doi: 10.1034/j.1600-0897.2003.00103.x
191. Sugimoto K, Hiwasa T, Shibuya K, Hirano S, Beppu M, Isose S, et al. Novel Autoantibodies Against the Proteasome Subunit PSMA7 in Amyotrophic Lateral Sclerosis. J Neuroimmunol (2018) 325:54–60. doi: 10.1016/j.jneuroim.2018.09.013
192. Scheffler S, Kuckelkorn U, Egerer K, Dörner T, Reiter K, Soza A, et al. Autoimmune Reactivity Against the 20S-Proteasome Includes Immunosubunits LMP2 (Beta1i), MECL1 (Beta2i) and LMP7 (Beta5i). Rheumatol (Oxford England) (2008) 47(5):622–6. doi: 10.1093/rheumatology/ken042
193. Mojtahedi Z, Safaei A, Yousefi Z, Ghaderi A. Immunoproteomics of HER2-Positive and HER2-Negative Breast Cancer Patients With Positive Lymph Nodes. OMICS (2011) 15(6):409–18. doi: 10.1089/omi.2010.0131
194. Montecucco C, Caporali R, Cobianchi F, Biamonti G. Identification of Autoantibodies to the I Protein of the Heterogeneous Nuclear Ribonucleoprotein Complex in Patients With Systemic Sclerosis. Arthritis Rheum (1996) 39(10):1669–76. doi: 10.1002/art.1780391009
195. Fiorentino DF, Presby M, Baer AN, Petri M, Rieger KE, Soloski M, et al. PUF60: A Prominent New Target of the Autoimmune Response in Dermatomyositis and Sjögren’s Syndrome. Ann Rheum Dis (2016) 75(6):1145–51. doi: 10.1136/annrheumdis-2015-207509
196. Schepens I, Jaunin F, Begre N, Läderach U, Marcus K, Hashimoto T, et al. The Protease Inhibitor Alpha-2-Macroglobulin-Like-1 is the P170 Antigen Recognized by Paraneoplastic Pemphigus Autoantibodies in Human. PLoS One (2010) 5(8):e12250. doi: 10.1371/journal.pone.0012250
197. Thébault S, Gilbert D, Hubert M, Drouot L, Machour N, Lange C, et al. Orderly Pattern of Development of the Autoantibody Response in (New Zealand White X BXSB)F1 Lupus Mice: Characterization of Target Antigens and Antigen Spreading by Two-Dimensional Gel Electrophoresis and Mass Spectrometry. J Immunol (Baltimore Md: 1950) (2002) 169(7):4046–53. doi: 10.4049/jimmunol.169.7.4046
198. Vaughan JH, Valbracht JR, Nguyen MD, Handley HH, Smith RS, Patrick K, et al. Epstein-Barr Virus-Induced Autoimmune Responses. I. Immunoglobulin M Autoantibodies to Proteins Mimicking and Not Mimicking Epstein-Barr Virus Nuclear Antigen-1. J Clin Invest (1995) 95(3):1306–15. doi: 10.1172/JCI117781
199. Doe K, Nozawa K, Hiruma K, Yamada Y, Matsuki Y, Nakano S, et al. Antibody Against Chromatin Assembly Factor-1 Is a Novel Autoantibody Specifically Recognized in Systemic Lupus Erythematosus. Lupus (2014) 23(10):1031–41. doi: 10.1177/0961203314536245
200. Wagatsuma M, Kimura M, Suzuki R, Takeuchi F, Matsuta K, Watanabe H. Ezrin, Radixin and Moesin Are Possible Auto-Immune Antigens in Rheumatoid Arthritis. Mol Immunol (1996) 33(15):1171–6. doi: 10.1016/S0161-5890(96)00083-1
201. Sato T, Uchiumi T, Kominami R, Arakawa M. Autoantibodies Specific for the 20-KDal Ribosomal Large Subunit Protein L12. Biochem Biophys Res Commun (1990) 172(2):496–502. doi: 10.1016/0006-291X(90)90700-W
202. Guialis A, Patrinou-Georgoula M, Tsifetaki N, Aidinis V, Sekeris CE, Moutsopoulos HM. Anti-5s RNA/protein (RNP) Antibody Levels Correlate With Disease Activity in a Patient With Systemic Lupus Erythematosus (SLE) Nephritis. Clin Exp Immunol (1994) 95(3):385–9. doi: 10.1111/j.1365-2249.1994.tb07008.x
203. Neu E, Von Mikecz AH, Hemmerich PH, Peter HH, Fricke M, Deicher H, et al. Autoantibodies Against Eukaryotic Protein L7 in Patients Suffering From Systemic Lupus Erythematosus and Progressive Systemic Sclerosis: Frequency and Correlation With Clinical, Serological and Genetic Parameters. The SLE Study Group. Clin Exp Immunol (1995) 100(2):198–204. doi: 10.1111/j.1365-2249.1995.tb03653.x
204. Elkon K, Bonfa E, Llovet R, Danho W, Weissbach H, Brot N. Properties of the Ribosomal P2 Protein Autoantigen Are Similar to Those of Foreign Protein Antigens. Proc Natl Acad Sci USA (1988) 85(14):5186–9. doi: 10.1073/pnas.85.14.5186
205. Luo LY, Herrera I, Soosaipillai A, Diamandis EP. Identification of Heat Shock Protein 90 and Other Proteins as Tumour Antigens by Serological Screening of an Ovarian Carcinoma Expression Library. Br J Cancer (2002) 87(3):339–43. doi: 10.1038/sj.bjc.6600439
206. Absi M, La Vergne JP, Marzouki A, Giraud F, Rigal D, Reboud AM, et al. Heterogeneity of Ribosomal Autoantibodies From Human, Murine and Canine Connective Tissue Diseases. Immunol Lett (1989) 23(1):35–41. doi: 10.1016/0165-2478(89)90152-1
207. Tycowski KT, Shu MD, Steitz JA. A Small Nucleolar RNA is Processed From an Intron of the Human Gene Encoding Ribosomal Protein S3. Genes Dev (1993) 7(7a):1176–90. doi: 10.1101/gad.7.7a.1176
208. Betteridge Z, Gunawardena H, North J, Slinn J, Mchugh N. Identification of a Novel Autoantibody Directed Against Small Ubiquitin-Like Modifier Activating Enzyme in Dermatomyositis. Arthritis Rheum (2007) 56(9):3132–7. doi: 10.1002/art.22862
209. Abreu-Velez AM, Howard MS, Hashimoto K, Hashimoto T. Autoantibodies to Sweat Glands Detected by Different Methods in Serum and in Tissue From Patients Affected by a New Variant of Endemic Pemphigus Foliaceus. Arch Dermatol Res (2009) 301(10):711–8. doi: 10.1007/s00403-009-0972-4
210. Margutti P, Sorice M, Conti F, Delunardo F, Racaniello M, Alessandri C, et al. Screening of an Endothelial cDNA Library Identifies the C-Terminal Region of Nedd5 as a Novel Autoantigen in Systemic Lupus Erythematosus With Psychiatric Manifestations. Arthritis Res Ther (2005) 7(4):R896–903. doi: 10.1186/ar1759
211. Bates RL, Payne SJ, Drury SL, Nelson PN, Isenberg DA, Murphy JJ, et al. The Prevalence and Clinical Significance of Autoantibodies to Plasminogen Activator Inhibitor 1 in Systemic Lupus Erythematosus. Lupus (2003) 12(8):617–22. doi: 10.1191/0961203303lu436oa
212. Yokota S, Kubota H, Matsuoka Y, Naitoh M, Hirata D, Minota S, et al. Prevalence of HSP47 Antigen and Autoantibodies to HSP47 in the Sera of Patients With Mixed Connective Tissue Disease. Biochem Biophys Res Commun (2003) 303(2):413–8. doi: 10.1016/S0006-291X(03)00352-8
213. Hwang HM, Heo CK, Lee HJ, Kwak SS, Lim WH, Yoo JS, et al. Identification of Anti-SF3B1 Autoantibody as a Diagnostic Marker in Patients With Hepatocellular Carcinoma. J Transl Med (2018) 16(1):177. doi: 10.1186/s12967-018-1546-z
214. Hosono Y, Nakashima R, Serada S, Murakami K, Imura Y, Yoshifuji H, et al. Splicing Factor Proline/Glutamine-Rich Is a Novel Autoantigen of Dermatomyositis and Associated With Anti-Melanoma Differentiation-Associated Gene 5 Antibody. J Autoimmun (2017) 77:116–22. doi: 10.1016/j.jaut.2016.11.006
215. Overzet K, Gensler TJ, Kim SJ, Geiger ME, van Venrooij WJ, Pollard KM, et al. Small Nucleolar RNP Scleroderma Autoantigens Associate With Phosphorylated Serine/Arginine Splicing Factors During Apoptosis. Arthritis Rheum (2000) 43(6):1327–36. doi: 10.1002/1529-0131(200006)43:6<1327::AID-ANR15>3.0.CO;2-S
216. Yamamoto AM, Amoura Z, Johannet C, Jeronimo AL, Campos H, Koutouzov S, et al. Quantitative Radioligand Assays Using De Novo-Synthesized Recombinant Autoantigens in Connective Tissue Diseases: New Tools to Approach the Pathogenic Significance of Anti-RNP Antibodies in Rheumatic Diseases. Arthritis Rheum (2000) 43(3):689–98. doi: 10.1002/1529-0131(200003)43:3<689::AID-ANR27>3.0.CO;2-U
217. Huntriss JD, Latchman DS, Williams DG. Lupus Autoantibodies Discriminate Between the Highly Homologous Sm Polypeptides B/B’ and SmN by Binding an Epitope Restricted to B/B’. Clin Exp Immunol (1993) 92(2):263–7. doi: 10.1111/j.1365-2249.1993.tb03390.x
218. Brahms H, Raymackers J, Union A, De Keyser F, Meheus L, Luhrmann R. The C-Terminal RG Dipeptide Repeats of the Spliceosomal Sm Proteins D1 and D3 Contain Symmetrical Dimethylarginines, Which Form a Major B-Cell Epitope for Anti-Sm Autoantibodies. J Biol Chem (2000) 275(22):17122–9. doi: 10.1074/jbc.M000300200
219. Mcclain MT, Ramsland PA, Kaufman KM, James JA. Anti-Sm Autoantibodies in Systemic Lupus Target Highly Basic Surface Structures of Complexed Spliceosomal Autoantigens. J Immunol (Baltimore Md: 1950) (2002) 168(4):2054–62. doi: 10.4049/jimmunol.168.4.2054
220. Satoh M, Chan JY, Ross SJ, Ceribelli A, Cavazzana I, Franceschini F, et al. Autoantibodies to Survival of Motor Neuron Complex in Patients With Polymyositis: Immunoprecipitation of D, E, F, and G Proteins Without Other Components of Small Nuclear Ribonucleoproteins. Arthritis Rheum (2011) 63(7):1972–8. doi: 10.1002/art.30349
221. Garbarz M, Dhermy D, Bournier O, Bezeaud A, Boivin P. Anti-Spectrin in Sera Containing Smooth Muscle Autoantibodies From Patients With Chronic Active Hepatitis. Clin Exp Immunol (1981) 43(1):87–93.
222. Zaninoni A, Vercellati C, Imperiali FG, Marcello AP, Fattizzo B, Fermo E, et al. Detection of Red Blood Cell Antibodies in Mitogen-Stimulated Cultures From Patients With Hereditary Spherocytosis. Transfusion (2015) 55(12):2930–8. doi: 10.1111/trf.13257
223. Santoro P, De Andrea M, Migliaretti G, Trapani C, Landolfo S, Gariglio M. High Prevalence of Autoantibodies Against the Nuclear High Mobility Group (HMG) Protein SSRP1 in Sera From Patients With Systemic Lupus Erythematosus, But Not Other Rheumatic Diseases. J Rheumatol (2002) 29(1):90–3.
224. Cortini A, Bembich S, Marson L, Cocco E, Edomi P. Identification of Novel non-Myelin Biomarkers in Multiple Sclerosis Using an Improved Phage-Display Approach. PLoS One (2019) 14(12):e0226162. doi: 10.1371/journal.pone.0226162
225. Meng Y, Zhang M, Zhao X, Cheng Y, Jia R, Wang Y, et al. Decreased Serum Thrombospondin-1 and Elevation of Its Autoantibody Are Associated With Multiple Exacerbated Clinical Manifestations in Systemic Lupus Erythematosus. Clin Rheumatol (2018) 37(10):2707–14. doi: 10.1007/s10067-018-4188-x
226. Muto M, Mori M, Hiwasa T, Takiguchi M, Iwadate Y, Uzawa A, et al. Novel Serum Autoantibodies Against Talin1 in Multiple Sclerosis: Possible Pathogenetic Roles of the Antibodies. J Neuroimmunol (2015) 284:30–6. doi: 10.1016/j.jneuroim.2015.05.005
227. Schwenzer A, Jiang X, Mikuls TR, Payne JB, Sayles HR, Quirke AM, et al. Identification of an Immunodominant Peptide From Citrullinated Tenascin-C as a Major Target for Autoantibodies in Rheumatoid Arthritis. Ann Rheum Dis (2016) 75(10):1876–83. doi: 10.1136/annrheumdis-2015-208495
228. Geng X, Biancone L, Dai HH, Lin JJ, Yoshizaki N, Dasgupta A, et al. Tropomyosin Isoforms in Intestinal Mucosa: Production of Autoantibodies to Tropomyosin Isoforms in Ulcerative Colitis. Gastroenterology (1998) 114(5):912–22. doi: 10.1016/S0016-5085(98)70310-5
229. Gajbhiye R, Sonawani A, Khan S, Suryawanshi A, Kadam S, Warty N, et al. Identification and Validation of Novel Serum Markers for Early Diagnosis of Endometriosis. Hum Reprod (2012) 27(2):408–17. doi: 10.1093/humrep/der410
230. Kimura A, Sakurai T, Yamada M, Koumura A, Hayashi Y, Tanaka Y, et al. Anti-Endothelial Cell Antibodies in Patients With Cerebral Small Vessel Disease. Curr Neurovasc Res (2012) 9(4):296–301. doi: 10.2174/156720212803530726
231. Enarson P, Rattner JB, Ou Y, Miyachi K, Horigome T, Fritzler MJ. Autoantigens of the Nuclear Pore Complex. J Mol Med (Berl) (2004) 82(7):423–33. doi: 10.1007/s00109-004-0554-z
232. Zhao X, Cheng Y, Gan Y, Jia R, Zhu L, Sun X. Anti-Tubulin-Alpha-1C Autoantibody in Systemic Lupus Erythematosus: A Novel Indicator of Disease Activity and Vasculitis Manifestations. Clin Rheumatol (2018) 37(5):1229–37. doi: 10.1007/s10067-018-4024-3
233. Matthes T, Wolff A, Soubiran P, Gros F, Dighiero G, Natural autoantibodies II. And Induced Antibodies Recognize Different Epitopes on the Tubulin Molecule. J Immunol (Baltimore Md: 1950) (1988) 141(9):3135–41.
234. Kimura A, Yoshikura N, Koumura A, Hayashi Y, Kobayashi Y, Kobayashi I, et al. Identification of Target Antigens of Naturally Occurring Autoantibodies in Cerebrospinal Fluid. J Proteomics (2015) 128:450–7. doi: 10.1016/j.jprot.2015.05.005
235. Prasannan L, Misek DE, Hinderer R, Michon J, Geiger JD, Hanash SM. Identification of Beta-Tubulin Isoforms as Tumor Antigens in Neuroblastoma. Clin Cancer Res (2000) 6(10):3949–56.
236. Muro Y, Ogawa Y, Kato Y, Hagiwara M. Autoantibody to Thioredoxin Reductase in an Ovarian Cancer Patient. Biochem Biophys Res Commun (1998) 242(2):267–71. doi: 10.1006/bbrc.1997.7914
237. Betteridge ZE, Gunawardena H, Chinoy H, North J, Ollier WE, Cooper RG, et al. Clinical and Human Leucocyte Antigen Class II Haplotype Associations of Autoantibodies to Small Ubiquitin-Like Modifier Enzyme, a Dermatomyositis-Specific Autoantigen Target, in UK Caucasian Adult-Onset Myositis. Ann Rheum Dis (2009) 68(10):1621–5. doi: 10.1136/ard.2008.097162
238. Li X, Sun J, Mu R, Gan Y, Wang G, He J, et al. The Clinical Significance of Ubiquitin Carboxyl Hydrolase L1 and its Autoantibody in Neuropsychiatric Systemic Lupus Erythematosus. Clin Exp Rheumatol (2019) 37(3):474–80.
239. Zhou Y, Cui J, Du H. Autoantibody-Targeted TAAs in Pancreatic Cancer: A Comprehensive Analysis. Pancreatology (2019) 19(5):760–8. doi: 10.1016/j.pan.2019.06.009
240. Miyachi K, Hosaka H, Nakamura N, Miyakawa H, Mimori T, Shibata M, et al. Anti-P97/VCP Antibodies: An Autoantibody Marker for a Subset of Primary Biliary Cirrhosis Patients With Milder Disease? Scand J Immunol (2006) 63(5):376–82. doi: 10.1111/j.1365-3083.2006.01747.x
241. Li FJ, Surolia R, Li H, Wang Z, Kulkarni T, Liu G, et al. Autoimmunity to Vimentin Is Associated With Outcomes of Patients With Idiopathic Pulmonary Fibrosis. J Immunol (Baltimore Md: 1950) (2017) 199(5):1596–605. doi: 10.4049/jimmunol.1700473
242. Paley EL, Alexandrova N, Smelansky L. Tryptophanyl-tRNA Synthetase as a Human Autoantigen. Immunol Lett (1995) 48(3):201–7. doi: 10.1016/0165-2478(95)02469-7
243. Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, et al. Isolation and Characterization of cDNA Encoding the 80-kDa Subunit Protein of the Human Autoantigen Ku (P70/P80) Recognized by Autoantibodies From Patients With Scleroderma-Polymyositis Overlap Syndrome. Proc Natl Acad Sci USA (1990) 87(5):1777–81. doi: 10.1073/pnas.87.5.1777
244. Hoa S, Hudson M, Troyanov Y, Proudman S, Walker J, Stevens W, et al. Single-Specificity Anti-Ku Antibodies in an International Cohort of 2140 Systemic Sclerosis Subjects: Clinical Associations. Med (Baltimore) (2016) 95(35):e4713. doi: 10.1097/MD.0000000000004713
245. Braunschweig D, Krakowiak P, Duncanson P, Boyce R, Hansen RL, Ashwood P, et al. Autism-Specific Maternal Autoantibodies Recognize Critical Proteins in Developing Brain. Transl Psychiatry (2013) 3:e277. doi: 10.1038/tp.2013.50
246. Kistner A, Bigler MB, Glatz K, Egli SB, Baldin FS, Marquardsen FA, et al. cir Association With Clinical Features in Newly Diagnosed Giant Cell Arteritis. Rheumatol (Oxford England) (2017) 56(5):829–34. doi: 10.1093/rheumatology/kew469
247. Van Beers-Tas MH, Marotta A, Boers M, Maksymowych WP, Van Schaardenburg D. A Prospective Cohort Study of 14-3-3eta in ACPA and/or RF-Positive Patients With Arthralgia. Arthritis Res Ther (2016) 18:76. doi: 10.1186/s13075-016-0975-4
248. Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, et al. Occurrence of Autoantibodies to Annexin I, 14-3-3 Theta and LAMR1 in Prediagnostic Lung Cancer Sera. J Clin Oncol (2008) 26(31):5060–6. doi: 10.1200/JCO.2008.16.2388
249. Chakravarti R, Gupta K, Swain M, Willard B, Scholtz J, Svensson LG, et al. 14-3-3 in Thoracic Aortic Aneurysms: Identification of a Novel Autoantigen in Large Vessel Vasculitis. Arthritis Rheumatol (2015) 67(7):1913–21. doi: 10.1002/art.39130
250. Feist E, Kuckelkorn U, Dorner T, Donitz H, Scheffler S, Hiepe F, et al. Autoantibodies in Primary Sjogren's Syndrome are Directed Against Proteasomal Subunits of the Alpha and Beta Type. Arthritis Rheum (1999) 42(4):697–702. doi: 10.1002/1529-0131(199904)42:4<697::AID-ANR12>3.0.CO;2-H
251. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–d613. doi: 10.1093/nar/gky1131
252. Estrada-Bernal A, Sanford SD, Sosa LJ, Simon GC, Hansen KC, Pfenninger KH. Functional Complexity of the Axonal Growth Cone: A Proteomic Analysis. PLoS One (2012) 7(2):e31858. doi: 10.1371/journal.pone.0031858
253. Wille M, Schümann A, Kreutzer M, Glocker MO, Wree A, Mutzbauer G, et al. The Proteome Profiles of the Olfactory Bulb of Juvenile, Adult and Aged Rats - an Ontogenetic Study. Proteome Sci (2015) 13:8. doi: 10.1186/s12953-014-0058-x
254. Sengupta N, Ghosh S, Vasaikar SV, Gomes J, Basu A. Modulation of Neuronal Proteome Profile in Response to Japanese Encephalitis Virus Infection. PLoS One (2014) 9(3):e90211. doi: 10.1371/journal.pone.0090211
255. Rozek W, Kwasnik M, Debski J, Zmudzinski JF. Mass Spectrometry Identification of Granins and Other Proteins Secreted by Neuroblastoma Cells. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2013) 34(3):1773–81. doi: 10.1007/s13277-013-0716-0
256. Mallawaaratchy DM, Hallal S, Russell B, Ly L, Ebrahimkhani S, Wei H, et al. Comprehensive Proteome Profiling of Glioblastoma-Derived Extracellular Vesicles Identifies Markers for More Aggressive Disease. J Neuro Oncol (2017) 131(2):233–44. doi: 10.1007/s11060-016-2298-3
257. Perluigi M, Di Domenico F, Buttterfield DA. Unraveling the Complexity of Neurodegeneration in Brains of Subjects With Down Syndrome: Insights From Proteomics. Proteomics Clin Appl (2014) 8(1-2):73–85. doi: 10.1002/prca.201300066
258. Ayyadevara S, Balasubramaniam M, Parcon PA, Barger SW, Griffin WS, Alla R, et al. Proteins That Mediate Protein Aggregation and Cytotoxicity Distinguish Alzheimer's Hippocampus From Normal Controls. Aging Cell (2016) 15(5):924–39. doi: 10.1111/acel.12501
259. Davalieva K, Maleva Kostovska I, Dwork AJ. Proteomics Research in Schizophrenia. Front Cell Neurosci (2016) 10:18. doi: 10.3389/fncel.2016.00018
260. Kahl A, Blanco I, Jackman K, Baskar J, Milaganur Mohan H, Rodney-Sandy R, et al. Cerebral Ischemia Induces the Aggregation of Proteins Linked to Neurodegenerative Diseases. Sci Rep (2018) 8(1):2701. doi: 10.1038/s41598-018-21063-z
261. Repici M, Hassanjani M, Maddison DC, Garção P, Cimini S, Patel B, et al. The Parkinson's Disease-Linked Protein DJ-1 Associates With Cytoplasmic mRNP Granules During Stress and Neurodegeneration. Mol Neurobiol (2019) 56(1):61–77. doi: 10.1007/s12035-018-1084-y
262. Mcmillan MT, Pan XQ, Smith AL, Newman DK, Weiss SR, Ruggieri M.R. C.OMMAS.R.X.X.X., et al. Coc Bladder Overactivity in a Mouse Model of Multiple Sclerosis. Am J Physiol Renal Physiol (2014) 307(5):F612–622. doi: 10.1152/ajprenal.00151.2014
263. Li HP, Komuta Y, Kimura-Kuroda J, Van Kuppevelt TH, Kawano H. Roles of Chondroitin Sulfate and Dermatan Sulfate in the Formation of a Lesion Scar and Axonal Regeneration After Traumatic Injury of the Mouse Brain. J Neurotrauma (2013) 30(5):413–25. doi: 10.1089/neu.2012.2513
264. Sobel RA, Ahmed AS. White Matter Extracellular Matrix Chondroitin Sulfate/Dermatan Sulfate Proteoglycans in Multiple Sclerosis. J Neuropathol Exp Neurol (2001) 60(12):1198–207. doi: 10.1093/jnen/60.12.1198
265. Ogura C, Hirano K, Mizumoto S, Yamada S, Nishihara S. Dermatan Sulfate Promotes Neuronal Differentiation in Mouse and Human Stem Cells. J Biochem (2021) 169(1):55–64. doi: 10.1093/jb/mvaa087
266. Sobczak AIS, Pitt SJ, Stewart AJ. Glycosaminoglycan Neutralization in Coagulation Control. Arteriosc Thromb Vasc Biol (2018) 38(6):1258–70. doi: 10.1161/ATVBAHA.118.311102
267. Li Y, Wu J, Wang S, Li X, Zhou J, Huang B, et al. Progression to Fibrosing Diffuse Alveolar Damage in a Series of 30 Minimally Invasive Autopsies With COVID-19 Pneumonia in Wuhan, China. Histopathology (2021) 78(4):542-55. doi: 10.1111/his.14249
268. Wigén J, Löfdahl A, Bjermer L, Elowsson-Rendin L, Westergren-Thorsson G. Converging Pathways in Pulmonary Fibrosis and Covid-19 - The Fibrotic Link to Disease Severity. Respir Med: X (2020) 2:100023. doi: 10.1016/j.yrmex.2020.100023
269. Smith KJ, Skelton HG, Heimer W, Baxter D, Angritt P, Frisman D, et al. Melanocytic Activation in HIV-1 Disease: HMB-45 Staining in Common Acquired Nevi. Military Medical Consortium for the Advancement of Retroviral Research. J Am Acad Dermatol (1993) 29(4):539–44. doi: 10.1016/0190-9622(93)70218-I
270. Boostani R, Talab FR, Meibodi NT, Zemorshidi F. COVID-19 Associated With Sensorimotor Polyradiculoneuropathy and Skin Lesions: A Case Report. J Neuroimmunol (2020) 350:577434. doi: 10.1016/j.jneuroim.2020.577434
271. Hooks JJ, Percopo C, Wang Y, Detrick B. Retina and Retinal Pigment Epithelial Cell Autoantibodies Are Produced During Murine Coronavirus Retinopathy. J Immunol (1993) 151(6):3381–9.
272. Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S, et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe (2020) 28(6):853–66.e855. doi: 10.1016/j.chom.2020.11.003
273. Ng ML, Lee JW, Leong ML, Ling AE, Tan HC, Ooi EE. Topographic Changes in SARS Coronavirus-Infected Cells at Late Stages of Infection. Emerg Infect Dis (2004) 10(11):1907–14. doi: 10.3201/eid1011.040195
274. Lloyd RE. Nuclear Proteins Hijacked by Mammalian Cytoplasmic Plus Strand RNA Viruses. Virology (2015) 479-480:457–74. doi: 10.1016/j.virol.2015.03.001
275. Chen Z, Liu S, Zhang S, Zhang Y, Yu J, Sun W, et al. Porcine Reproductive and Respiratory Syndrome Virus Strains With Higher Virulence Cause Marked Protein Profile Changes in MARC-145 Cells. Sci Rep (2018) 8(1):15000. doi: 10.1038/s41598-018-32984-0
276. Li Y, Ming F, Huang H, Guo K, Chen H, Jin M, et al. Proteome Response of Chicken Embryo Fibroblast Cells to Recombinant H5N1 Avian Influenza Viruses With Different Neuraminidase Stalk Lengths. Sci Rep (2017) 7:40698. doi: 10.1038/srep40698
277. Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, et al. Host- and Strain-Specific Regulation of Influenza Virus Polymerase Activity by Interacting Cellular Proteins. mBio (2011) 2(4):e00151-11. doi: 10.1128/mBio.00151-11
278. Nuss JE, Kehn-Hall K, Benedict A, Costantino J, Ward M, Peyser BD, et al. Multi-Faceted Proteomic Characterization of Host Protein Complement of Rift Valley Fever Virus Virions and Identification of Specific Heat Shock Proteins, Including HSP90, as Important Viral Host Factors. PLoS One (2014) 9(5):e93483. doi: 10.1371/journal.pone.0093483
279. Yang J, Yang L, Li B, Zhou W, Zhong S, Zhuang Z, et al. iTRAQ-Based Proteomics Identification of Serum Biomarkers of Two Chronic Hepatitis B Subtypes Diagnosed by Traditional Chinese Medicine. BioMed Res Int (2016) 2016:3290260. doi: 10.1155/2016/3290260
280. Csősz É, Tóth F, Mahdi M, Tsaprailis G, Emri M, Tőzsér J. Analysis of Networks of Host Proteins in the Early Time Points Following HIV Transduction. BMC Bioinf (2019) 20(1):398. doi: 10.1186/s12859-019-2990-3
281. Monette A, Panté N, Mouland AJ. HIV-1 Remodels the Nuclear Pore Complex. J Cell Biol (2011) 193(4):619–31. doi: 10.1083/jcb.201008064
282. Stake M, Singh D, Singh G, Marcela Hernandez J, Kaddis Maldonado R, Parent LJ, et al. HIV-1 and Two Avian Retroviral 5' Untranslated Regions Bind Orthologous Human and Chicken RNA Binding Proteins. Virology (2015) 486:307–20. doi: 10.1016/j.virol.2015.06.001
283. Tada S, Hamada M, Yura Y. Proteomic Analysis of Secretomes of Oncolytic Herpes Simplex Virus-Infected Squamous Cell Carcinoma Cells. Cancers (Basel) (2018) 10(2):28. doi: 10.3390/cancers10020028
284. Christen U, Von Herrath MG. Do Viral Infections Protect From or Enhance Type 1 Diabetes and How can We Tell the Difference? Cell Mol Immunol (2011) 8(3):193–8. doi: 10.1038/cmi.2010.71
285. Wilkes MR, Sereika SM, Fertig N, Lucas MR, Oddis CV. Treatment of Antisynthetase-Associated Interstitial Lung Disease With Tacrolimus. Arthritis Rheum (2005) 52(8):2439–46. doi: 10.1002/art.21240
286. Wu J, Subbaiah KCV, Xie LH, Jiang F, Khor ES, Mickelsen D, et al. Glutamyl-Prolyl-tRNA Synthetase Regulates Proline-Rich Pro-Fibrotic Protein Synthesis During Cardiac Fibrosis. Circ Res (2020) 127(6):827–46. doi: 10.1161/CIRCRESAHA.119.315999
287. Tsai PC, Soong BW, Mademan I, Huang YH, Liu CR, Hsiao CT, et al. A Recurrent WARS Mutation Is a Novel Cause of Autosomal Dominant Distal Hereditary Motor Neuropathy. Brain J Neurol (2017) 140(5):1252–66. doi: 10.1093/brain/awx058
288. Chang SE, Feng A, Meng W, Apostolidis SA, Mack E, Artandi M, et al. New-Onset IgG Autoantibodies in Hospitalized Patients With COVID-19. Nat Commun (2021) 12(1):5417. doi: 10.1038/s41467-021-25509-3
Keywords: COVID-19, SARS-CoV-2, autoantigens, autoantibodies, dermatan sulfate, autoimmunity
Citation: Wang JY, Zhang W, Roehrl VB, Roehrl MW and Roehrl MH (2022) An Autoantigen Atlas From Human Lung HFL1 Cells Offers Clues to Neurological and Diverse Autoimmune Manifestations of COVID-19. Front. Immunol. 13:831849. doi: 10.3389/fimmu.2022.831849
Received: 09 December 2021; Accepted: 21 February 2022;
Published: 24 March 2022.
Edited by:
Lazaros Ignatios Sakkas, University of Thessaly, GreeceCopyright © 2022 Wang, Zhang, Roehrl, Roehrl and Roehrl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Y. Wang, anVsaWFAY3VyYW5kaXMuY29t; Michael H. Roehrl, cm9laHJsbUBtc2tjYy5vcmc=
 Julia Y. Wang
Julia Y. Wang Wei Zhang2
Wei Zhang2 Michael H. Roehrl
Michael H. Roehrl