- 1Immuno-Oncology Laboratory, Department of Pathology, School of Basic Medicine, Central South University, Changsha, China
- 2Department of Immunobiology, Yale University School of Medicine, New Haven, CT, United States
Natural killer (NK) cells are an important component of the innate immune system due to their strong ability to kill virally infected or transformed cells without prior exposure to the antigen (Ag). However, the biology of human NK (hNK) cells has largely remained elusive. Recent advances have characterized several novel hNK subsets. Among them, adaptive NK cells demonstrate an intriguing specialized antibody (Ab)-dependent response and several adaptive immune features. Most adaptive NK cells express a higher level of NKG2C but lack an intracellular signaling adaptor, FcϵRIγ (hereafter abbreviated as FcRγ). The specific expression pattern of these genes, with other signature genes, is the result of a specific epigenetic modification. The expansion of adaptive NK cells in vivo has been documented in various viral infections, while the frequency of adaptive NK cells among peripheral blood mononuclear cells correlates with improved prognosis of monoclonal Ab treatment against leukemia. This review summarizes the discovery and signature phenotype of adaptive NK cells. We also discuss the reported association between adaptive NK cells and pathological conditions. Finally, we briefly highlight the application of adaptive NK cells in adoptive cell therapy against cancer.
1 Introduction
As a crucial member of the innate lymphoid cell (ILC) family, natural killer (NK) cells specialize in killing damaged, infected, or transformed cells and releasing cytokines as immune modulators (1, 2). Unlike adaptive immune cells, NK cells do not need to be exposed to the antigen (Ag) in advance to mature effector functions. Therefore, NK cells are considered at the forefront of the host immune system. The important role of NK cells in restricting viral infection or tumor metastasis has been observed in vivo in both murine and human studies (3, 4). However, although a series of stochastically expressed activating and inhibitory receptors have been described, the recognition of NK and target cells is far less clear (5). The final activity of NK cells depends on the balance between activating and inhibitory signals transduced by these receptors.
Accumulating studies have shown that NK cells demonstrate adaptive immune features, such as clonal expansion and immune memory, resulting in a diversely functional repertoire and stronger responses to previously encountered stimuli (6). For example, murine Ly49H-dependent NK cells are capable of responding specifically to murine cytomegalovirus (MCMV) and mount stronger reactions to secondary challenges (7, 8). Moreover, murine hepatic CXCR6+ NK cells mediate memory response to a secondary challenge to hapten or viruses in the absence of T and B cells (9–11).
The adaptive features of human NK (hNK) cells have been extensively studied during the last decade. Human cytomegalovirus (HCMV), an enveloped member of the Herpesviridae family, has been shown to extensively alter the phenotype and function of hNK cells, as reflected by a strong association with an NK subset expressing NKG2C, a receptor for HLA class I histocompatibility antigen, alpha chain E (HLA-E) (12, 13). Of interest, these NK cells specifically respond to HCMV peptides presented by HLA-E (14). Subsequent studies have also suggested that HCMV infection is positively associated with a group of peripheral blood (PB) NK cells, g-NK, characterized by the absence of FcRγ, a signaling adaptor protein. NKG2C+ NK and g-NK subsets are largely overlapping but not identical, and g-NK cells also demonstrate adaptive features, such as clonal expansion, augmented effector function, and extended lifespan (15–17) (Figure 1). In 2015, hNK cells lacking FcRγ, spleen tyrosine kinase (SYK), and EWS/FLI1-activated transcript 2 (EAT-2), along with silencing of promyelocytic leukemia zinc finger (PLZF) expression, were termed “adaptive NK cells” by Schlums et al. (18). A subset of NKG2C+ adaptive NK cells can specifically recognize HCMV strains encoding variable UL40 peptides and can further expand and differentiate in response to stimulation by pro-inflammatory factors (14). However, available evidence suggests that the above-mentioned recognition of NKG2C with HLA-E plus peptides is not observed in NKG2C- adaptive NK cells (19). Immunological memory is defined as the enhanced response to the re-challenge of the same Ag, and immunological memory cells can persist for years, even for a lifetime (20). Thus far, there are no data indicating that adaptive NK cells can recognize various Ags and mount an enhanced response. Rather, adaptive NK cells achieve a broader spectrum of Ag specificity through antibody (Ab)-dependent function. NK cells have long been considered short-lived innate effector cells. However, several lines of evidence indicate that adaptive NK cells have an unexpectedly long lifespan compared to conventional NK (c-NK) cells. Resembling adaptive memory cells, adaptive NK cells can live for months to years (16). Additionally, adaptive NK cells have been found to expand in response to other viral infections, such as hepatitis B virus (HBV), hepatitis C virus (HCV), human immunodeficiency virus (HIV), herpes simplex virus (HSV), and influenza virus (21–25). This review summarizes the research progress of adaptive NK cells, with a special focus on molecular hallmarks, epigenetic regulation, differentiation, and tissue distribution. Additionally, we focus on the correlation between adaptive NK and multiple clinical diseases, such as viral infection and cancer, and put forward prospects that target them to improve immunotherapy.
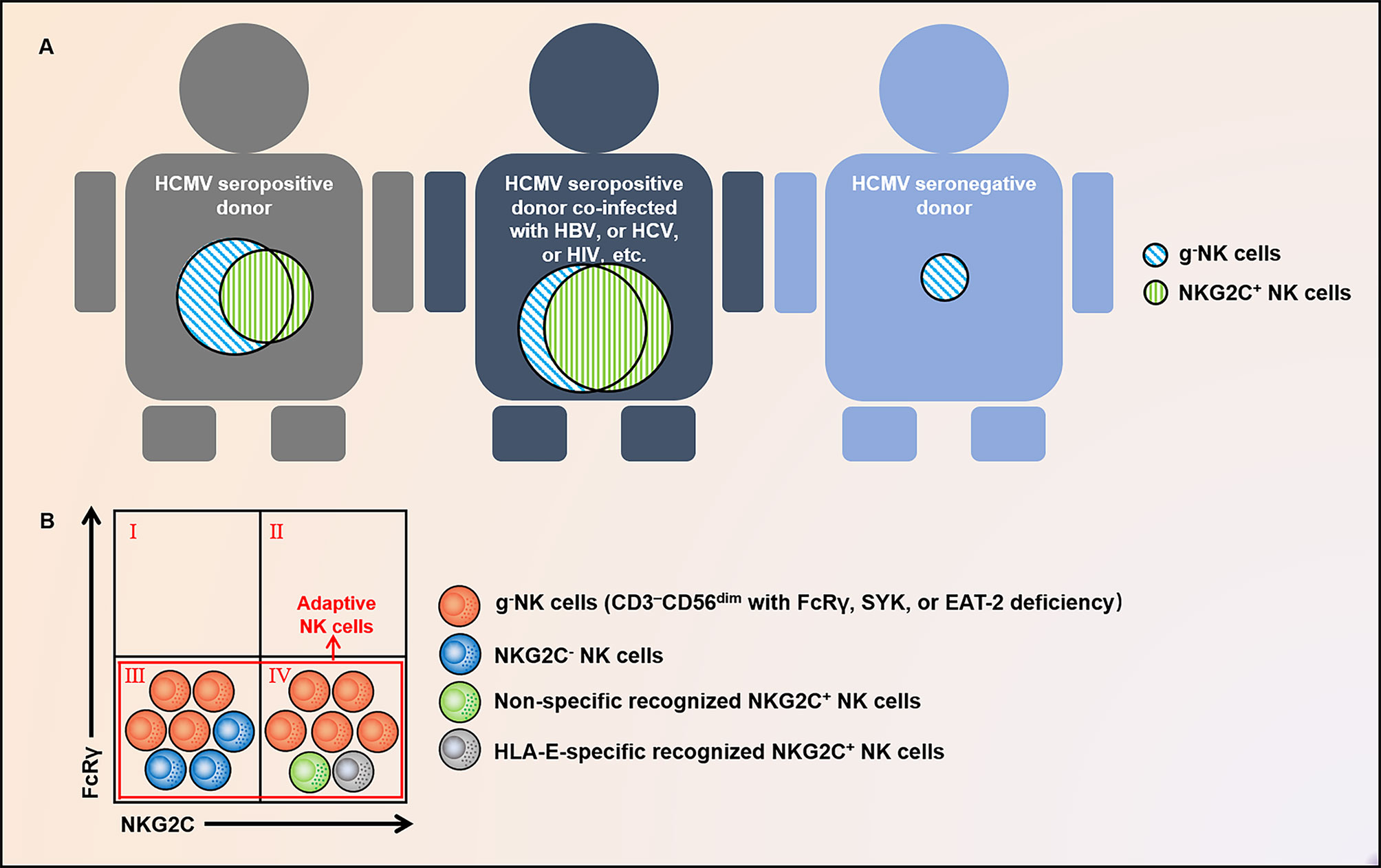
Figure 1 Distribution and relationship of g-NK and NKG2C+ NK cells. (A) g-NK cells persist and expand long-term in healthy individuals with prior HCMV infection, and their proportion will increase under the co-infection of HBV, HCV, HIV, and other chronic viruses. Exceptionally, a fraction of this subset has also been observed in HCMV- individuals. (B) g-NK cells characterized by FcRγ deficiency are different from NK cells marked by NKG2C and are composed of both NKG2C+ and NKG2C- subsets. In addition to this subset, adaptive NK cells comprise a subset of non-specifically recognized and HLA-E-specifically recognized NKG2C+ NK cells. These subgroups largely intersect. g-NK, γ chain-negative natural killer; HCMV, human cytomegalovirus; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; FcϵRIγ or FcRγ, IgE receptor subunit-γ; EAT2, EWS/FLI1-activated transcript 2; SYK, spleen tyrosine kinase; and HLA-E, HLA class I histocompatibility antigen; alpha chain E.
2 Discovery and Molecular Markers of Human Adaptive NK Cells
The discovery of human adaptive NK cells can be traced back to more than 1 decade ago (12, 15, 16, 18, 26–30). The expansion of a CD94/NKG2C+ NK subset in the PB of HCMV+ donors was first described by Guma et al. in 2004 (12). These NK cells express lower levels of NKp30 and NKp46 compared to CD94/NKG2A+ NK cells isolated from the same donors. The expansion of CD94/NKG2C+ NK cells is associated only with HCMV but not with HSV or EBV serology (12). In 2011, Lopez-Verges et al. described an expansion of CD57+NKG2Chi NK cells in bone marrow (BM) transplantation recipients with acute HCMV infection (26). In 2012, Foley et al. suggested that these HCMV-induced NKG2C+ NK cells were transplantable and expanded in response to recipient CMV Ag in vivo, thus demonstrating memory-like phenotype (27). Earlier, in the same year, Hwang et al. documented a group of FcRγ-deficient NK cells, g-NK, in the PB of one-third of healthy donors (15). These cells are considered likely to arise from clonal expansion, express low levels of NKp46 and NKp30, and respond poorly to K562 cells. However, g-NK cells express an almost normal level of CD16 and demonstrate an enhanced response to CD16 cross-linking. The association between the presence of g-NK cells and HCMV infection was described by Zhang, T. et al. in 2013 (16). In the same year, Beziat et al. studied the role of both self-specific inhibitory killer cell immunoglobulin-like receptors (KIRs) and activating KIRs during clonal expansion of NK cells in response to CMV infection (28). The association of FcRγ and NKG2C expression was finally described by Schlums et al. and Lee et al. in 2015 (18, 29) and further discussed by Muntasell et al. in 2016 (30). Additionally, HCMV-associated NKG2C+ NK cells were named adaptive NK cells by Muntasell et al. (30). As numerous researchers have considered NKG2C+ NK cells that have previously encountered HCMV as or even equal to adaptive NK cells, such generalization may ignore subsets of cells including NKG2C- g-NK cells (29); we hereby find a legible explanation. A study by Liu, L.L. et al. found that a characteristic footprint of adaptive NK cells existed in NKG2C-/- donors, including terminal differentiation phenotypes, functional reprogramming, and epigenetic remodeling of the interferon-gamma (IFN-γ) promoter (19).
Detailed studies have shown that HCMV-associated NK cells undergo extensive epigenetic modulation compared to c-NK cells. The defect in FcRγ is not due to gene mutation but due to hypermethylation of the FCER1G promoter. Epigenetic modification is also the underlying mechanism controlling a specified function. Compared with c-NK cells, adaptive NK cells have a superior ability to produce IFN-γ and tumor necrosis factor alpha (TNF-α) in response to stimuli through CD16. Consistently, IFNG and TNF, encoding IFN-γ and TNF-α, respectively, show hypomethylation at the gene loci, highlighting the mechanism of the elevated capacity of adaptive NK cells to release these cytokines following activation via CD16 and NKG2C (31, 32).
Though adaptive NK cells exhibit a stronger response to CD16 cross-linking, the expression of several signaling molecules of the CD16 pathway, including ZAP70, PLCγ1, PLCγ2, LCP2, and PIK3CA, is not significantly different between adaptive NK and c-NK cells. Of interest, SYK expression is absent in some adaptive NK cells due to promoter hypermethylation. The SYK-deficiency is closely related to HCMV infection, rather than HSV-1 or HSV-2, and can be passed along to daughter cells. The SYK-deficiency in adaptive NK cells does not impair their function but is associated with enhanced responsiveness to CD16 cross-linking. Notably, SYK-deficient adaptive NK cells have mainly been found among g-NK cells (18, 29). Another epigenetic signature of adaptive NK cells is the hypermethylation of the promoters of several important transcription factor (TF)-encoding genes, including ZBTB16 (encoding PLZF), SH2D1B (encoding EAT-2), and DAB2 (encoding DAB2). ZBTB16 silencing results in reduced expression of PLZF and deficiency of IL-12Rβ1 and IL-18Rα. Indeed, adaptive NK cells demonstrate a marginal response to IL-12 and IL-18 stimulation (18) (Figure 2).
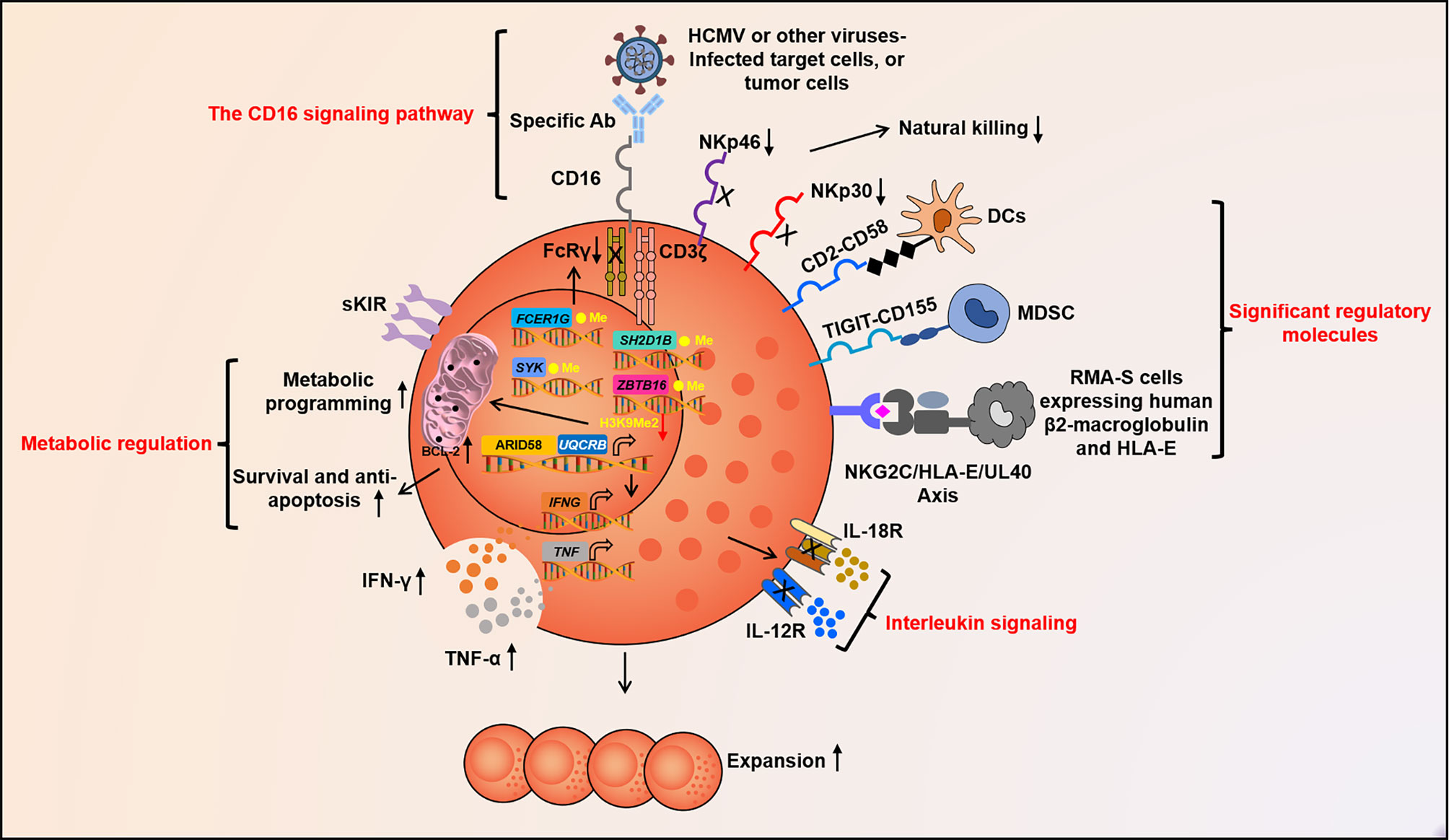
Figure 2 Molecular factors govern adaptive NK cell function. Fc segment of Abs can take shape specific signal of adaptive NK cells to recognize target cells by binding to CD16. NKG2C, CD2, TIGIT, and other unknown molecules may act as crucial signaling pathway for the activation or inhibition of adaptive NK cells. IL-12, IL-18, and other cytokines together constitute the pro-inflammatory cytokine signal, which guides adaptive NK cell differentiation, expansion, and activation. Epigenetic programming promotes stochastic DNA methylation-dependent allelic silencing of FCER1G, SYK, and SH2D1B. These ultimately contribute to IFNG promoter hypomethylation and format heterogeneous adaptive NK cell repertoire. The up-regulation of ARID5B and its inducing UQCRB with low H3K9Me2 in the promoter region alter metabolic reprogramming, prolong survival, and motivate abundant IFN-γ secretion of adaptive NK cells. Abs, antibodies; CD, cluster of differentiation; TIGIT, T-cell immunoglobulin and ITIM domain; interleukin, IL; IFN-γ, Interferon-gamma; TNF-α, Tumor necrosis factor alpha; and ARID5B, AT-rich interaction domain 5.
It has been shown that only NKG2Chi NK cells are expanded in HCMV seropositive individuals and experience epigenetic remodeling of the conserved non-coding sequence 1 (CNS1) of IFNG, similar to memory CD8+ T or Th1 cells. To cater to the engagement of NKG2C and 2B4, the accessibility of CNS1 is necessary to increase IFNG transcription. Indeed, a closed configuration of CNS1 has been detected in NKG2C- NK cells from HCMV+/- individuals and in NKG2C+ NK cells from HCMV- individuals. Remarkably, among HCMV+ individuals, the open configuration of CNS1 occurs exclusively in expanded NKG2Chi self-MHC specific KIRs (sKIRs)+ NK cells. Consistently, NKG2C+ NK cells from HCMV+ individuals do not show a closed configuration of CNS1, similar to NKG2C- NK cells. The CNS1 methylation that occurs in expanded NKG2Chi NK cells does not rely on the expression of sKIRs or CD57 and is stably retained in the daughter cells (31). Taken together, epigenetic modification is responsible for specific phenotypes of adaptive NK cells (Figure 2).
Collectively, adaptive NK cells have the following characteristics: 1) associated with prior HMCV infection; 2) lack expression of FcRγ, tyrosine kinase SYK, signaling molecules EAT-2 and DAB2, and TF PLZF; 3) do not necessarily express NKG2C or CD57; 4) express sKIRs but lack NKG2A; 5) down-regulation of natural cytotoxic receptors (NCRs) (i.e., NKp46 and NKp30), CD161, Siglec7, ITGA6, CD7, PECAM1, and TIM-3, and lack of IL-12R and IL-18R; 6) stable surface level of NKG2D and CD16, and up-regulation of ILT2, CD2, FAS, CD11a, CD44, CCR5, and Bcl-2; 7) mount long-term effective recall response directed by Fc receptor (FcR)-mediated Ab-dependent target recognition, possibly via specific recognition by NKG2C or other known or unknown receptor; 8) epigenetic imprinting; and 9) Ab-dependent enhanced functional capabilities, including Ab-dependent cell-mediated cytotoxicity (ADCC), in vitro expansion, and cytokine production (15, 16, 18, 29, 33, 34).
3 Functional Characteristics of Adaptive NK Cells
3.1 Expansion and Effector Function
The detailed mechanism mediating adaptive NK cell proliferation remains elusive, though NKG2C+ NK cell expansion can be observed both in vitro and in vivo (35). A similar response to different viral infections indicates that cellular factors mediate this expansion phenotype. Indeed, existing evidence suggests that IL-12, IL-15, and HLA-E are critical for the expansion of NKG2C+ NK cells (36–38). Consistently, HLA-E and inflammatory cytokines can be up-regulated during HCMV, hantavirus, or HIV infection. The involvement of cytokines in mediating NKG2C+ NK cell expansion is also supported by in vivo observations in humans infected with viruses. Up-regulated IL-12 and IL-15 are positively associated with HCMV serostatus in children (39). Moreover, IL-12Rβ1 deficiency impairs the generation of NK cells with adaptive features (40). However, the role of IL-12 in mediating the expansion of adaptive NK cells requires further study, given their low IL-12 receptor expression and marginal response to IL-12 plus IL-18 stimulation in vitro (18, 29). Of interest, adaptive NK cells show clonal expression of inhibitory KIRs specific for self-HLA class I molecules, suggesting that licensing is involved in adaptive NK cell generation or expansion (15, 18, 28). Notably, KIRs on adaptive NK cells tend to be ligands of HLA-C1/C2, which are also ligands for stimulating KIRs (such as 2DS4), which are correlated with resistance to viral infection (28, 41). Whether KIRs play a role in adaptive NK cell expansion requires further study.
CD16 is a low-affinity Fc receptor that is mainly expressed by NK cells but can also be detected on neutrophils and monocytes. The intracellular part of CD16 is short and, as a result, has a limited ability to transduce signals in isolation. CD16 couples with CD3ζ and FcRγ, to deliver an activating signal. Though many adaptive NK cells, mostly g-NK, do not express FcRγ, their response to CD16 cross-linking is even better than other NK cells that express both CD3ζ and FcRγ. While the underlying mechanism for this phenotype requires further study, it has been shown that knockout of FcRγ by CRISPR-Cas9-mediated gene editing can reconstitute this phenotype in c-NK cells (42). This finding is interesting because it indicates, for the first time, that FcRγ can inhibit the CD16 pathway of hNK cells. It also suggests that ADCC function can be enhanced by manipulating a single gene other than CD16 itself. FcRγ only contains one ITAM motif, while CD3ζ contains three. The simplest explanation is that the loss of FcRγ makes CD16 more available to CD3ζ, although the molecular mechanism of this finding warrants further study. Upon MCMV infection, Ly49H, through multiple surface expression, binds to m157 to drive the differential expansion of memory NK cell clones, such as TCR affinity. Consistent with experiments on mice that have reported that the expression of Ly49H has a fairly strong correlation with Ly49H+ NK cell expansion, a positive correlation between the expression of NKG2C and the expansion of NKG2C+ cells has also been observed in HCMV-seropositive donors (43, 44). However, such studies have not been reported for g-NK cells that are not specifically recognized via NKG2C, suggesting that the specifically activated signal CD16-Fc segment or some other unknown Ag recognition pathway is involved in avidity selection and expansion. Remarkably, the enhanced Ab-dependent response of g-NK cells is not limited to HCMV-infected target cells and also applies to HSV-1-infected target cells; this indicates that g-NK cells have considerable potential to mediate Ab-dependent cross-protection against broad-spectrum viral infections (16). In HCMV+ individuals, irrespective of NKG2C+ or NKG2C-, HCMV-specific Ab invariably induces an increase in IFN-γ and TNF-α production by g-NK cells, suggesting that the CD16 signaling pathway may be the crucial signal for adaptive NK cell responses (29). Notably, Lee et al. reported that g-NK cells can be significantly expanded in vitro following the application of autologous HCMV+ serum. It is unclear whether this expansion mainly relies on the CD16 pathway or whether other cytokines are also involved (29) (Figure 2).
NK cells are immune sentinels that eradicate target cells and release various cytokines and chemokines to tune the adaptive immune response (45). The activation of NK cells is determined by the balance between activating and inhibitory signals. NCRs, such as NKp30, NKp46, and NKG2D, are well-known to be important for natural killing activity, while CD16 mediates ADCC (46). A large portion of adaptive NK cells, mostly g-NK, have a limited number of membrane NCRs and therefore only show a marginal response when co-cultured with K562 cells (15).
3.2 Significant Regulatory Molecules
3.2.1 NKG2C
Human NKG2C, also known as KLRC2, is located at 12p13 in the NK complex (47). NKG2C binds to HLA-E and transmits an activating signal to NK cells. HCMV-encoded UL40 peptide can stabilize HLA-E on the surface of HCMV-infected cells and affect HLA-E presentation and binding to NKG2C (48). Given that NKG2C is a critical marker for adaptive NK cells, its role in regulating the function of adaptive NK cells has been well investigated. Interesting progress has been reported by Hammer et al., who demonstrated that UL40-encoded peptide-pulsed HLA-E-expressing cells can trigger an elevated frequency and number of NKG2C+ adaptive NK cells. Notably, due to the variability of the peptide, its ability to induce IFN-γ, TNF-α, and CCL3 secretion by NKG2C+ adaptive NK cells differs in the order of VMAPRTLFL > VMAPRTLIL > VMAPRTLVL, and the same phenomenon is also observed in the cytotoxicity assay (14). Merino et al. stimulated NKG2C chronically via anti-NKG2C and found a significant proliferation of adaptive NK cells with epigenetic remodeling similar to that of exhausted CD8+ T cells. Of note, this chronic stimulation also drives the expression of lymphocyte-activation gene 3 (LAG3), programmed death-1 (PD-1), and T-cell immunoglobulin and ITIM domain (TIGIT), which confer the phenotype of exhausted T cells (49).
In a healthy population, the NKG2C haplotype can be detected in approximately 20% of donors, while homozygous deletion is common (50). The NKG2C copy number not only determines the surface expression level of NKG2C receptor but also directly contributes to the abundance, differentiation, and distribution of adaptive NK cell subset in response to HCMV, as demonstrated by NKG2C+/del compared to NKG2C+/+ healthy individuals (30).
NK cells in NKG2C homozygous and hemizygous subjects show differences in response to NKG2C signals (i.e., iCa2+ influx), degranulation, and IL-15-dependent proliferation, further indicating that the receptor is involved in forming the HCMV-induced reconfiguration of the NK-cell compartment (51). Of note, the g-NK subset is composed of both NKG2C+ and NKG2C- NK cells. Lee et al. found that g-NK cells could respond to HCMV-infected target cells, regardless of NKG2C expression, in the presence of HCMV+ plasma (29). Liu, L.L. et al. made a similar observation in NKG2C-/- donors (19) (Figure 2). However, the frequency of NKG2C+ NK cells after g-NK expansion has not been well characterized. NKG2C- g-NK cells can be expanded in vitro (unpublished data), indicating that pathways other than NKG2C can mediate adaptive NK cell expansion.
3.2.2 CD2
Glycoprotein CD2 is a co-stimulatory receptor expressed mainly on T and NK cells, which binds to LFA3/CD58, a cell surface protein expressed on other cells, such as dendritic or endothelial cells. Previous studies have shown that CD2 is involved in the formation of immune synapses (ISs) between immune cells and Ag presenting cells and is up-regulated on memory and activated T cells. Similarly, the presence of CD2 has been observed in the ISs formed between NK and target cells. CD2 may interact with CD16 in cis to promote its reposition as a linkage between CD16 and the actin cytoskeleton (52). CD2 expression is significantly increased in adaptive NK cells of NKG2C+ and NKG2C-/- donors. Additionally, the interaction between CD2 and CD16 enhances the Ab-mediated response in adaptive NK cells. Co-ligation of CD2 and CD16 leads to significantly higher levels of phosphorylation of all signaling molecules, including CD3-ζ, ZAP70/SYK, SLP76, LAT, ERK1/2, and S6RP, suggesting that CD16 and CD2 synergistically activate the MAP kinase and mTOR pathways (19) (Figure 2).
3.2.3 TIGIT
TIGIT is a momentous inhibitory molecule in the PVR/nectin family and is mainly expressed on T and NK cells, and its immune-modulatory function has been studied in the context of autoimmunity, viral immunity, and cancer (53). Ligating TIGIT with high-affinity CD155 and low-affinity CD112 can recruit SH2 domain-containing inositol-5-phosphatase (SHIP) to ITIM and inhibit the activation of T and NK cells. Indeed, tumor-infiltrating NK cells are found to express high levels of TIGIT. Blocking Abs that target TIGIT and CD155 interaction could up-regulate NK functions and inhibit tumor growth in a mouse model (54). PB adaptive NK cells express low levels of TIGIT and, therefore, in contrast to c-NK cells, are less sensitive to TIGIT pathway inhibition mediated by myeloid-derived inhibitory cells (MDSCs) (55) (Figure 2).
3.3 Metabolic Regulation
An isoform of AT-rich interaction domain 5 (ARID5B) is selectively up-regulated and involved in supporting mitochondrial membrane potential, the expression of electron transport chain (ETC) components, oxidative metabolism, survival, and IFN-γ production in adaptive NK cells. The increased metabolism observed in this subset appears to depend, at least in part, on the up-regulated expression of ARID5B and the induction of genes encoding components of ETC, including the ETC complex III gene UQCRB. Synchronously, a significantly inferior H3K9Me2 enrichment at the UQCRB promoter was observed. Beyond this, the decrease in ARID5B is associated with increased apoptosis and decreased expression of BCL-2, which suggests that ARID5B is essential for the survival of adaptive NK cells. As BCL-2 is located in mitochondria to counteract the production of ROS, a byproduct of ETC activity, increased BCL-2 in adaptive NK cells may be crucial to limit oxidative stress (56) (Figure 2). Taken together, enhancing ARID5B and mitochondrial function is a promising strategy to improve cell survival in NK cell-mediated immunotherapy.
3.4 Interleukin Signaling Regulation
The common gamma chain (γc) receptor family cytokines, including IL-2, IL-15, and IL-21, are required for the generation, persistence, and homeostasis of adaptive NK cells (57). IL-2 was first discovered as a T cell growth factor in 1976 and was later found to have various pleiotropic properties, including the ability to augment the cytolytic activities of NK cells and cytotoxic T cells. The IL-2 receptor α chain (IL-2Rα) binds IL-2 at intermediate-affinity (Kd ~ 10−9 M) but can combine with IL-2Rβ and IL-2Rγ to form a heterotrimer with a much higher affinity. Equally important, NK cells cannot survive for an extended time without IL-15 signaling, as IL-15 promotes the continuous expression of the key anti-apoptotic protein MCL-1, and adaptive NK cells are no exception. Moreover, IL-2 and IL-15 play a key role in NK cell function through the JAK1/3 and STAT5-dependent signaling pathway, which can bind to the IL-2R complex or IL-15Rβ/α on the surface of NK cells to induce the activation of β/γ-related JAK1 and JAK3 tyrosine kinases. This phosphorylation further leads to the recruitment and activation of STAT5 and the transcription of STAT5-target genes, such as Bcl2, GzmB, Idb2, Mcl1, Pim2, and Prf1, all of which are required for proliferation, survival, and cytotoxicity (58). Beyond the JAK/STAT pathway, γc family cytokines also function through other pathways, such as the MAP kinase- and phosphoinositol 3-kinase-dependent pathways (59).
Peptide recognition boosts adaptive NK cell expansion and differentiation under the assistance of short-term addition of IL-12 and IL-18, even though this subset does not express IL12/IL18R, especially g-NK, although the exact mechanism is unclear. The combined stimuli skew the adaptive NK phenotype and even contribute to hypomethylation of IFNG CNS1. Additionally, a short stimulation of IL-12 plus IL-18 induces down-regulation of FcRγ and CD7 in NKG2C+/- cells. Transcriptional levels of adaptive NK cell-related genes are also affected by the combination of pro-inflammatory signals plus peptide recognition, such as NCR3, SH2DB1, ZBTB32, ZBTB16, ZBTB20, ITGAL, and CRTAM. These signals drive sustained transcription of genes encoding activation and apoptosis markers, such as HLA-DR, TNFRSF9, LAG3, CTLA4, and PDCD1, as well as effector molecules, such as IL8, CSF2, IL10, GZMB, IFNG, TNF, CCL3, CCL4, and TNFSF10 (14).
4 Differentiation and Tissue Distribution of Adaptive NK Cells
Generally, hNK cells account for approximately 5%–15% of the total PB lymphocytes. The half-life of circulating NK cells is approximately 14 days, and the proliferation is 4%–5% per day. hNK cells stem from a common innate lymphoid progenitor (CILP) via an NK cell precursor (NKP), both of which are differentiated from a common lymphoid progenitor (CLP) (60). Subsequently, under the regulation of TFs, such as TOX, TBX21, ETS1, and E4BP4, NKP further differentiates into immature NK cells (iNK) in an Eomes-dependent manner to become highly proliferative CD56bright NK cells. Additionally, RUNX2, GATA3, and BACH2 constitute a regulatory network that dominates the formation of the mature CD56dim subgroup. Besides the down-regulation of CD56, terminal differentiation of NK cells involves the modulation of receptor profile, the acquisition of cytotoxic function, and even epigenetic modifications, which constitute the heterogeneity of NK cells (61). Although the differentiation of adaptive NK cells is poorly understood, it is feasible that adaptive NK cells may be derived from the CD56dimCD57+ subgroup. Holmes et al. considered that the TF regulation network shifts with the down-regulation of ZBTB16 and the up-regulation of BCL11B, which may be mutually antagonistic. Ultimately, BCL11B acts as the top of the pyramid to regulate YBX3, PBX4, SATB2, IRF4, and NFIC, in the formation of adaptive NK cells (62) (Figure 3).
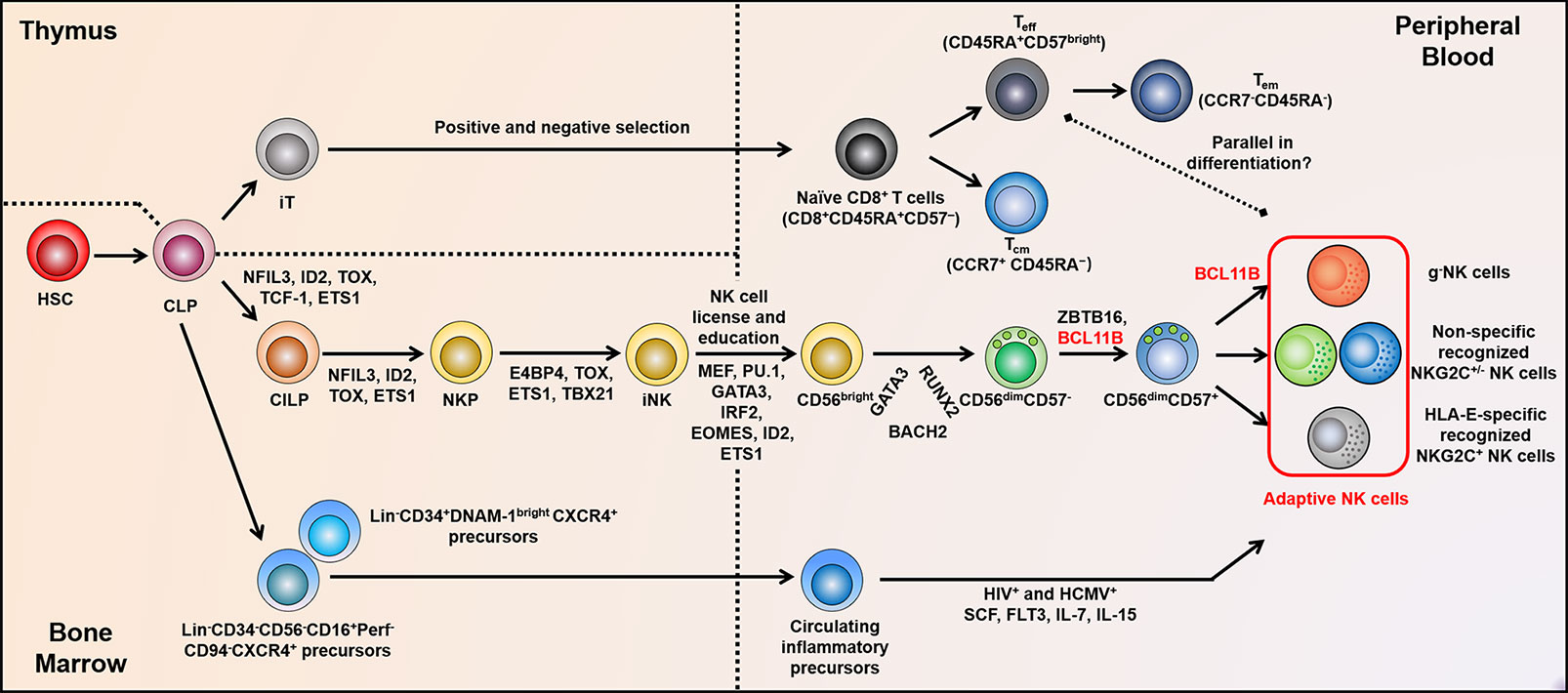
Figure 3 Adaptive NK cell development and differentiation. The positive and negative selection experienced by T cells in the thymus may be similar to the “license” and “education” of NK cells in BM. Subsequently, non-self KIRs expressed on NK cell surface facilitate adaptive NK cell diversity. TF networks show reciprocal regulation during distinct stages of NK cell differentiation, in particular, Bcl11b is fundamental for the differentiation of adaptive NK cells and c-NK cells. At the genome levels, adaptive NK cells accompanied by altered methylation properties are parallel to Teff cells. In addition, the circulating inflammatory progenitor cells in PB may as a source of adaptive NK cells during HCMV and HIV co-infection. BM, bone marrow; KIRs, killer cell immunoglobulin-like receptors; TF, transcription factor; HSC, hematopoietic stem cell; CILP, common innate lymphoid progenitor; NKP, NK cell precursor; CLP, common lymphoid progenitor; iT, immature T cell; iNK, immature NK cell; c-NK, conventional NK; and PB, peripheral blood.
Mounting evidence has shown that the development of adaptive NK cells and CD8+ T cells has an analogous process. Transcriptional profiling reveals that the differentiation axis of CD7loNKG2C+NKp30loCD57+ adaptive NK cells converges toward that of CD45RAhiCD28-CCR7-CD27-CD57+CD8+ T cells (62). Immature T cells complete positive and negative selection to obtain MHC restriction and central immune tolerance, respectively, in the thymus. Correspondingly, iNK cells undergo a similar process known as “missing self” and “education” or “license” by expressing self- and non-self KIRs to form a repertoire that binds to MHC-I ligands. Eventually, mature NK cells are formed with effector function and self-tolerance (63–65). Kim et al. and Bryceson et al. further suggested that HCMV-induced epigenetic modification and Ab-dependent expansion are mechanisms underlying the formation of this memory-like NK cell pool, paralleling with cytotoxic effector T cell (Teff) differentiation (18, 29). The genome-wide analysis illustrated that the methylation characteristic of early mature NK cells (CD56dimCD57brightEAT2+) was different from that of Teff cells. However, CD56dimCD57brightEAT2- adaptive NK cells are quite similar to those of Teff cells. Comparing adaptive NK cells with Teff cells, 61 differential methylation regions are found; nevertheless, there are up to 2372 differences between early mature NK (CD56dimCD57-) and adaptive NK cells (18). Heath et al. suggested that the extensive expansion of NKG2C+CD57+ NK cells after sensing stress served to keep pace with HCMV reactivation or other factors behind such expansion; this adaptive property is similar to that of virus-specific CD8+ T cells in that it does not involve replication. The accumulation of this NK subset may reflect the NK immune response, which is homologous with the expanded adaptive T cell response, marked by the accumulation of HCMV-specific effector memory T cells (Tem) (66). The integrative-omics analysis revealed that super-enhancers related to gene cohorts might coordinate NK cell function and localization. The TCF7-LEF1-MYC module in CD56bright NK cells may participate in preserving the progenitor cells, which can further replenish adaptive cells after BLIMP1 induction. TCF7-LEF1 is weakly expressed in adaptive NK cells that show a terminal differentiation state closer to that of Tem and Th1 cells (67). These results further illustrate the correlation between adaptive NK cells and Teff during differentiation and tissue colonization (Figure 3).
GATA-2 expression is confined to hematopoietic stem and progenitor cells (HSPC). Patients with heterozygous GATA2 mutation frequently exhibit immunodeficiency, such as no NK cell progenitors. However, in some patients with GATA-2 mutation, a durable survival or self-renewing PLZF- adaptive NK cell pool has been observed. These cells express low levels of perforin and GZMA but high levels of GZMB. Similar to adaptive NK cells in healthy individuals, these NK cells respond to FcR pathway-stimulation but not to IL-12 plus IL-18 (68). The expression of cytotoxic granule components is controlled by T-bet and Eomes (69). Consistently, T-bet expression in these adaptive NK cells is significantly lower than that in typical PLZF+ NK cells, but there is no clear difference in Eomes expression. PLZF- adaptive NK cells in patients with GATA2 mutation show functional properties related to those of adaptive NK cells from healthy individuals. Although they help to kill infected cells, the high-frequency adaptive NK cells in individuals with GATA2 mutation may cause inflammation-driven BM failure (68). Another important discovery suggested that the differentiation and function of adaptive NK cells can be independent of glycosylphosphatidylinositol (GPI) anchors in patients with paroxysmal nocturnal hemoglobinuria (PNH), which frequently undergoes somatic X-linked PIGA mutations that lead to a lack of GPI anchored membrane proteins on hematopoietic cells (70). In short, the homeostasis of adaptive NK cells in the periphery may not depend on HSPC or CD56bright precursor NK cells.
A recent study showed that in HIV+ anti-retroviral-treated and HCMV+ reactivated patients, HCMV-controlling NKG2C+ adaptive NK cells may be derived from novel circulating inflammatory precursors marked by Lin-CD34+ DNAM-1brightCXCR4+ and can rapidly differentiate into an NKG2C+KIR+CD57+ NK subset. Moreover, other Lin-CD34-CD56-CD16+Perf-CD94-CXCR4+ precursors from CLP also possess the potential to develop toward memory-like NKG2C+ NK cells (71) (Figure 3). It is inferred that the precursors of adaptive NK cells are released and circulate in PB during viral infection, where they exert powerful killing activity upon maturation. Overall, to date, it is unclear whether adaptive NK cells are specifically differentiated from a subgroup of CLP. No current studies have reported that c-NK cells from HCMV seronegative donors can be induced to differentiate into an adaptive NK cell state in vitro (72). Existing studies have shown that they are more likely to be NK cells with cytotoxic T-like specificity. Thus, the concrete mechanisms of their differentiation source and regulatory factors require further experimental data support.
After maturation, adaptive NK cells migrate to multiple peripheral organs but are preferentially present in non-lymphoid organs. In addition to PB, adaptive NK cells can colonize the tonsils, lymph nodes (LNs), liver, pleural fluid, and other sites. Interestingly, adaptive NK cells can also infiltrate into tumor tissues, such as in non-small cell lung cancer (NSCLC) (73) and colon cancer (unpublished data). Additionally, Shah et al. explored the presence of NK cell memory in primates and discovered a systematic distribution of Δg NK cells, referred to as g-NK, with adaptive features through a rhesus macaque model. They also found that in addition to PB, Δg NK cells were also distributed in the spleen, BM, multiple lymph nodes, and colon mucosa. Apart from CMV-induced expansion, rhesus cytomegalovirus (RhCMV)-primed Δg NK cells can also be affected by simian immunodeficiency virus (SIV) infection, following which they are recruited into the mucosa and effector tissues. Δg NK function is subverted by SIV infection through inhibition of the CD16-mediated CD3ζ-ZAP70 pathway. Most importantly, Δg NK cells chose CD3ζ-Zap70 signaling as an alternative pathway, which can modulate CD16 density, mucosal homing, and NK function while forgoing typical γ-chain/SYK signaling (74). This model provides an optimized experimental animal model for further exploration of the function of adaptive NK cells in the future.
5 Clinical Relevance of Adaptive NK Cells
The imprint mediated by HCMV infection on the adaptive NK cell repertoire is usually fixed at the initial encounter. The stability of the impact probably depends on the host and virus genetics, as well as environmental factors, such as age and viral load at initial infection. However, inefficient control of potential infections is associated with immunosenescence or dysregulated immunity, which may promote the expansion of pre-differentiated adaptive NK cells (75). In umbilical cord blood transplantation (UCBT) recipients, HCMV reactivation may induce rapid phenotypic reconfiguration, including the early and late acquisition of certain adaptive characteristics (76). Adaptive NK cells may play a part in in processes involving specific Abs, such as immune complex diseases or targeted Ab-based cancer therapy. As the definition of adaptive NK cells does not entirely overlap with that of the previously discovered NKG2C+ NK cells, it remains to be elucidated whether adaptive NK cells are also suitable for these models. Moreover, it is currently unclear whether adaptive NK cells may protect against or promote disease progression.
5.1 Viral Infection
HCMV impacts both innate and adaptive subsets and immune responses during its three-phase infection, that is, acute infection, persistence, and latency/reactivation, leading to immune system shaping (77). Early studies have shown that NK cells are critical for controlling herpesvirus infection (78). Not only are they involved in first-line innate defense, but the latest data based on mice show that memory-like NK cells with adaptive lymphocyte characteristics can be triggered in phase I of HCMV infection and can be preferentially maintained in the later phases. The presence of an abundance of adaptive NK cells may be associated with considerable clinical prognosis, especially for patients with cancer who use approved monoclonal antibodies (mAbs) that can effectively trigger ADCC. However, NKG2Chi NK cells account for 50% of the entire compartment in some HCMV-infected individuals, which will reduce the diversity of the overall NK pool and attenuate surveillance of heterologous infection or tumor progress (77).
In virologically suppressed patients with HIV, expanded adaptive NK cells express low levels of HLA-DR and CD38 and lack NKp30 and NKp46 expression. This may be damaging to NK-mediated immune surveillance in patients receiving combination antiretroviral therapy. The presence of this subgroup is related to HCMV serology and soluble CXCL10 in plasma (25). Likewise, in cHBV and HCMV co-infected individuals, adaptive NK cells show increased frequency and polarization compared to c-NK cells (79). Furthermore, adaptive NK cells are also present in individuals with HCV infection. Notably, direct antiviral therapy can alter the PD-1 expression and ADCC activity of adaptive NK cells, resulting in improved effector function (23). Another interesting finding indicates that in chronic HCV-infected HCMV+ subjects, adaptive NK cells account for most of the CD56negCD16+ population, in addition to constituting a fraction of CD56dimCD16+. Adaptive NK cell carriers have lower levels of liver enzymes and fibrosis, revealing that the involvement of adaptive NK cells in chronic HCV infection can effectively ease liver disease (80). Petitdemange et al. demonstrated that acute Chikungunya virus (CHIKV) infection promotes a transient change in the phenotype and function of NK cells, as evidenced by transient clonal expansion of NK cells co-expressing CD94/NKG2C and HLA-C1 alleles, decreased expression of NKp46, NKG2A, and CD161, and up-regulated expression of CD57, ILT2, and NKp44. Intriguingly, NKG2C can rapidly increase in response to acute CHIKV infection and enter the contraction phase following viral clearance. The clonal expansion of this subset is correlated with viral load, indicating that NK cells can sense CHIKV from the onset of infection, thereby helping to eliminate the virus (81). Hart et al. found that the frequency of adaptive NK cells is positively associated with a favorable prognosis of malaria after natural exposure to Plasmodium falciparum. Erythrocytes infected with P. falciparum can induce NK cell degranulation and be lysed via ADCC in the presence of plasma from patients infected with malaria. The significance of this research is that an imminent vaccine with IgG1 and IgG3 Abs against P. falciparum Ags expressed on the surface of infected erythrocytes could stimulate CD16 to ultimately achieve effective elimination of P. falciparum (82).
In the process of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), adaptive NK cells are enriched in patients compared to healthy donors. This difference is particularly obvious in patients with fatal outcomes. Moreover, the frequency of adaptive/memory NK cells in deceased patients increases statistically (83). Other work showed that adaptive NK cells had signs of expansion in patients with COVID-19, but this did not rely on the HCMV activation secondary to COVID-19. Compared to nonresponding adaptive NK cells, responding adaptive NK cells express HLA-DR, CD38, CD62L, and MIP-1, while the expression of NKG2A, NKG2D, TIGIT, and CD25 is marginal (84). Furthermore, NKG2C and HLA-E host genetic variation may determine the severity of COVID-19 symptoms. Indeed, the expression levels of KLRC2del and HLA-E*0101 are significantly higher in patients hospitalized with COVID19, especially in those requiring intensive care, compared to patients with mild symptoms (85).
5.2 Transplantation Immunity
As the first reconstituted lymphocyte after transplantation, NK cells may be strongly imprinted by HCMV, especially in a transplant setting where T-cell immune function is chronically impaired (86). Muccio et al. noticed that in UCBT recipients undergoing HCMV reactivation, the down-regulated expression of FcRγ was detected at a later time point (i.e., twelfth month), while other adaptive NK phenotypes were acquired early after UCBT (i.e., sixth month). Additionally, FcRγ was found to be more frequently down-regulated in NKG2C-CD57+ NK cells compared to the classic memory-like NKG2C+CD57+ subset. They further demonstrated that down-regulation of PLZF and FcRγ are independent events, as the down-regulation of FcRγ in NKG2C-CD57+ NK cells does not show a corresponding down-regulation of PLZF at 24 months post-transplantation. This may require HCMV reinfection/reactivation to achieve stable epigenetic reprogramming that allows for sustained FcRγ silencing. The expansion ability of adaptive NK cells is significantly improved in the presence of IgG Abs, suggesting that the low frequency of adaptive NK cells is related to the impaired humoral response in the UCBT recipient (76).
Whether the increased proportion of adaptive NK cells mentioned above (anti-COVID-19 immune response) is a double-edged sword for the body to defend against exogenous infection or tumor immunity remains a pressing research question. According to the existing results, the potential cross-reactivity of adaptive NK cells can be beneficial in the treatment of leukemia. Interestingly, patients with acute myeloid leukemia (AML) who undergo hematopoietic stem cell transplantation (HSCT) have a lower relapse rate and superior disease-free survival (DFS) when the donor and/or recipient is HCMV seropositive before transplantation; these individuals benefit from the expansion of CD56dimCD57+NKG2C+ adaptive NK cells in response to HCMV reactivation (87). Nguyen et al. considered that NKG2C+ adaptive NK cells can eradicate the minimal residual disease by cross-reactive recognition with HLA-E+ leukemic blasts (88). Ex vivo experiments have shown that adaptive NK cells enriched in HLA-E ligands have enhanced alloreactivity to HLA-mismatched targets and even serve as a specific and efficient killer of allogeneic pediatric T- and precursor B-cell acute lymphoblastic leukemia (ALL) blasts (89).
Immunosuppressed kidney transplant recipients (KTRs) induced by thymoglobulin have a poor immune response and may be infected after receiving transplants collected from HCMV seropositive donors. In KTRs receiving immunosuppressive therapy, primary HCMV infection, reactivation, or reinfection is associated with graft loss and reduced patient survival. The damaging effect of HCMV on KTRs can be prevented by post-transplant viremia screening, antiviral prophylaxis, and mTOR targeting drug treatment. However, the control of HCMV replication in KTRs depends on the recipients’ immune system to restrain the pathogen (90). Among KTRs that are clinically stable for more than 2 years after transplantation, HCMV seropositivity or HCMV DNA replication can alter the NK cell phenotype. In KTRs with active HCMV replication, an expanded FcRγ–LIR-1+NKG2C– NK subset exhibits vigorous ADCC function in the presence of immobilized HCMV glycoprotein B reactive Abs; however, low expression of perforin after co-culture with K562 suggests that the natural killing ability of this subset is weakened. These results demonstrate that HCMV can relapse in asymptomatic KTRs and that this recurrence leads to continuous exposure of NK cells to HCMV to promote the expansion and persistence of adaptive NK cells in vivo (91). This expansion also applies to the involvement of NKG2C+ adaptive NK cells in the control of HCMV in KTRs. Consistently, the incidence of viremia after transplantation is reduced in the case of transplantation of large numbers of NKG2C+ NK cells (92).
5.3 Tumor Immunity
Based on preclinical and clinical observations, as well as the aforementioned HSCT, adaptive NK cells promote the control of hematopoietic malignancies and prevent recurrence of disease. An analysis of 215 patients with hematological malignancies demonstrated that transplantation conditions significantly affect the functional NK cell pool. The levels of adaptive NK cells in recipients receiving non-myeloablative therapy (NMAC) are statistically higher than those in recipients receiving myeloablative therapy (MAC), and high HCMV neutralizing Ab titers have certain promotion significance for adaptive NK cell expansion. Multivariate analysis of DFS, relapse, and treatment-related mortality (TRM) suggests that NMAC recipients with a large number of adaptive NK cells have favorable clinical outcomes 6 months after HCT (93).
Similar effects have been observed for the treatment of multiple myeloma (MM) with CD38-specific Ab, daratumumab. CD38 is highly expressed in hematopoietic stem cells (HSCs) and MM and functions as a receptor-mediated adhesion to regulate the cyclase and hydrolase activities. Daratumumab targeting CD38 can lyse lymphoma cells or CD38+ immunosuppressive cells through Fc-mediated complement-dependent cytotoxicity (CDC), ADCC, and Ab-dependent cellular phagocytosis (ADCP) (94). Immunophenotypic characteristics and functional analysis of adaptive NK cells from newly diagnosed multiple myeloma (NDMM) patients showed that adaptive NK cells exhibit an observably lower level of CD38 expression compared to c-NK cells, suggesting that they can evade daratumumab-induced fratricide. A recent study has shown that knockdown of the ectoenzyme CD38 in high-affinity non-cleavable variant of CD16a (hnCD16a)-induced pluripotent stem cell (iPSC) NK cells enhances their metabolic capacity, particularly glycolysis and cysteine metabolism, and improves their relative resistance to oxidative stress. The metabolic profile of this subset is also observed in adaptive NK cells (95). Encouragingly, CD38low adaptive NK cells exert a powerful daratumumab-mediated ADCC in vitro, and the frequency of this subset is positively correlated with the effector function of daratumumab (96). In vivo or in vitro studies by Bigley et al. further support the effective anti-tumor effect of g-NK cells combined with therapeutic mAbs, daratumumab and elotuzumab, targeting signaling lymphocytic activation molecule F7 (SLAMF7), in MM. Consistent with the above, g-NK cells express minimal levels of CD38 and SLAMF7 on their surface. In NSG mice, following in vitro expansion, g-NK and c-NK cells were adoptively transferred and supplemented by daratumumab and rhIL-15 to achieve in vivo expansion. After 31 days, g-NK cells persisted in the blood and spleen, with more than 10-fold higher numbers than c-NK cells. In a disseminated orthotopic xenograft MM.1S model, compared to daratumumab plus c-NK cells, adoptive treatment with a combination of daratumumab and g-NK cells reduces average tumor burden by > 99.9%. Moreover, 5 of the 7 mice eliminated the burden of myeloma, with a survival rate of up to 100% after 60 days. Interestingly, the persistence of expanded g-NK cells is detected in the blood, spleen and, BM relative to that of c-NK cells in the MM model (57).
Contrary to the positive role played by adaptive NK cells in hematological tumors, HCMV infection and the existence of adaptive NK cells in solid tumors may be a negative factor. A solid tumor is a major adverse outcome of orthotopic liver transplantation (OLT), and 60%–90% of transplant recipients develop HCMV infection due to reactivation of the latent virus or new infection after long-term immunosuppressive treatment. HCMV can infect tumor cells, including medulloblastoma and colon carcinoma, and HCMV infection of tumor cells may disrupt the recognition by NK cells via down-regulating the expression of MHC class I molecules on tumor cells (97–99). Baryawno et al. discovered that HCMV infection of primary medulloblastomas and medulloblastoma cell lines further up-regulated COX-2 expression and PGE2 production in tumors, thereby stimulating tumor cell proliferation (100). The work by Achour et al. emphasized the complexity of the NK cell response and its clinical impact after OLT. Importantly, the new development of head and neck neoplasm or colon cancer was associated with the aberrant expansion of adaptive NK cells and robust production of TNF-α in HCMV+ patients. In contrast, NK cells from patients with genitourinary system tumors had classic iNK cell characteristics, including high expression of NKG2A and powerful IFN-γ production. TNF-α is closely related to the replication of CMV and can significantly increase malignant transformation by triggering the NF-κB transcription activator, eventually leading to immune failure in controlling various malignant tumors. Nevertheless, the level of IFN-γ can predict the long-term survival rate of patients with gastrointestinal stromal tumors after treatment with imatinib mesylate (101). In summary, in an immunosuppressive environment, the interaction between the NK repertoire and HCMV status may greatly hinder the spectrum of immune surveillance, which is preferential to the growth and development of specific neoplastic tumors after OLT.
6 Therapeutic Potential of Adaptive NK Cells
NK cell-based immunotherapy demonstrates promise. Currently, NK cell strategies for tumor immunotherapy include autologous or allogeneic NK cell therapy activated in vitro, a combination of NK cells and mAbs, such as immune checkpoint inhibitors (ICIs), rituximab, daratumumab, trastuzumab, and cetuximab, and construction of CAR-NK cells (102). Cytokine-induced human memory-like NK cells in NSG mice reveal an enhanced response to stimuli from several weeks to months after the initial adoptive transfer and a superior anti-tumor activity against AML. Similarly, in a first-in-human phase I clinical trial, adoptively transferred memory-like NK cells proliferate and expand in patients with AML and manifest a robust response to leukemia target (103). Chimeric Ag receptors (CARs) have been applied to improve the specific recognition of tumor cells by effector lymphocytes. CD19-CAR-memory-like NK cells targeting CD19 have shown a promising anti-tumor response to lymphoma in a preclinical study (104). For a long time, NK cells have been considered as short-lived innate effector cells (105). However, several lines of evidence indicate that adaptive NK cells have an unexpected long lifespan compared to c-NK cells, resembling adaptive memory cells, which can live for months to years (16). Based on the success of cytokine-induced memory-like NK and g-NK cells coupled with daratumumab in vivo and the durable longevity of adaptive NK cells, we hold the opinion that adaptive NK cells are suitable candidates for adoptive cell therapy.
Here, we propose several aspects to accelerate the clinical application of adaptive NK cells:
1) Developing Ag-specific Abs or NK cell engagers (NKCEs) that trigger the CD16 pathway to mimic TCR or BCR and significantly broadens the spectrum of Ag specificity of adaptive NK cells (106–108).
2) Stabilizing membrane CD16 and target Ag. Methods to significantly improve ADCC efficiency can be considered in terms of both adaptive NK and target cells, such as avoiding CD16 on the surface of adaptive NK cells being cleaved by A disintegrin and metalloproteinase-17 (ADAM17) and Matrix Metalloproteinase (MMP), enhancing its high-affinity binding to the Fc segment, or advancing of Ag presentation to the surface of tumor cells to reduce endocytosis (109, 110).
3) Manipulating co-regulatory pathways expressed on NK cells with either co-activated receptor agonist or co-inhibitory receptor blocking Abs, all of which are effective means to strengthen the activation and functions of adaptive NK cells, such as adding 4-1BB/CD28 and CD3ζ co-stimulatory signals, blocking the co-inhibitory pathway with EOS-448 against TIGIT, and stimulating the co-activation pathway of CD2, NKG2C, and 4-1BB (111).
4) Up-regulating ARID5B in adaptive NK cells to maintain their long-term survival. ARID5B is a transcriptional regulator that regulates anti-apoptosis, oxidative metabolism, and IFN-γ secretion in adaptive NK cells (56) (Figure 4).
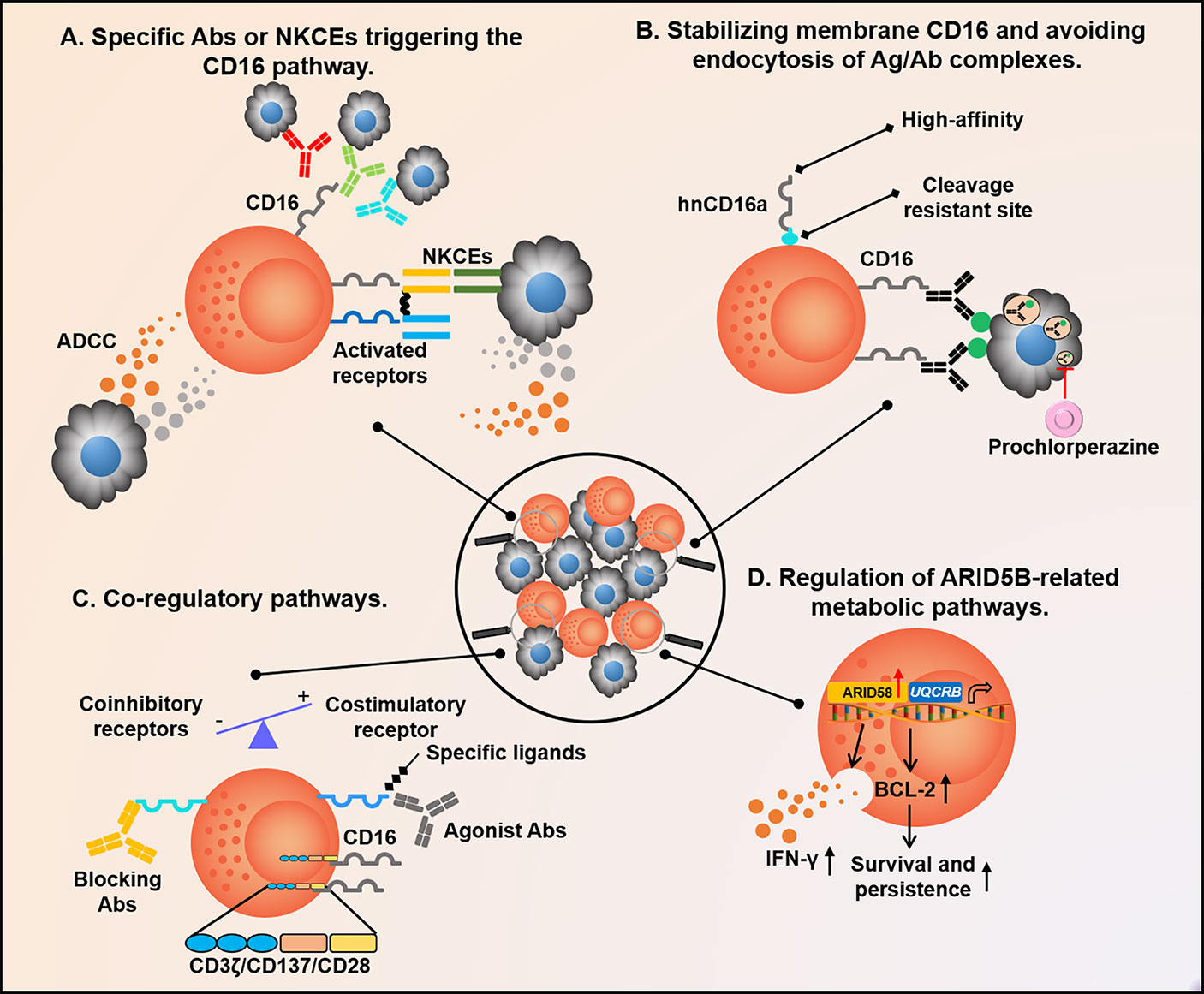
Figure 4 Future therapeutic strategies are based on adaptive NK cells. (A) When the CD16 pathway is activated by Abs or NKCEs binding to different targets, adaptive NK cells secrete numerous IFN-γ and TNF-α to regulate anti-tumor immunity; (B) Methods to significantly improve the efficiency of ADCC can be considered from two aspects of adaptive NK cells and target cells, such as avoiding CD16 on the surface of adaptive NK cells being cleaved by ADAM17 or MMP, enhancing its binding with Fc segment in a high-affinity manner, and advancing Ag presentation to the surface of tumor cells to reduce endocytosis; (C) Adding 4-1BB/CD28 and CD3ζ co-stimulatory signals to adaptive NK cells, or blocking the co-inhibitory pathway with the antagonistic Ab EOS-448 of TIGIT, and stimulating the co-activation pathway of CD2, NKG2C, and 4-1BB with agonist are all effective means to strengthen adaptive NK cell function; (D) Up-regulation of ARID5B expressed in adaptive NK cells can alter its metabolic characteristics, resulting in longer persistence and increased function. NKCEs, NK cell engagers; ADCC, Ab-dependent cell-mediated cytotoxicity; ADAM17, A disintegrin and metalloproteinase-17; and MMP, Matrix Metalloproteinase.
Our current results show that lentivirus infection in vitro can lead to down-regulation of FcRγ in c-NK cells, resulting in a prominent increase in the proportion of adaptive NK cells (unpublished data). Therefore, in the future, immunotherapy based on the above strategy will also apply to general NK cells or non-HCMV reactive memory-like NK cells. HCMV reactive adaptive NK cells have a unique advantage of being invoked as a cell therapy tool. They have a stronger Ab-dependent response-ability than c-NK cells and can maintain a longer survival time in vivo, which greatly exerts ADCC function and improves the curative effect.
7 Conclusion
A primary physiological role of NK cells is to provide a primary defense against pathogenic organisms during the initial response period when the adaptive immune system is activated. Although NK cells respond to various microorganisms, including bacteria and protozoa, they are particularly imperative in viral infections. Through Fc receptor-mediated recognition of target cells bound by Abs, adaptive NK cells can produce a robust response to infected cells, especially during chronic or recurrent infection, where reactive Abs are readily available. Considering that adaptive NK cells are present in a fraction of healthy individuals, the presence or absence and frequency of adaptive NK cells may accelerate immune heterogeneity among individuals against infection and cancer. Adaptive NK cells can serve as a novel tool for the clinical treatment of chronic diseases, such as malignancies and viral infections.
Author Contributions
TZ and FG took the lead in structuring and writing the manuscript, ZZ, YL, GS, and GY helped modify the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Key Project of Hunan Province (No. 2022WK2012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mujal AM, Delconte RB, Sun JC. Natural Killer Cells: From Innate to Adaptive Features. Annu Rev Immunol (2021) 39:417–47. doi: 10.1146/annurev-immunol-101819-074948
2. Vivier E, Ugolini S. Regulatory Natural Killer Cells: New Players in the IL-10 Anti-Inflammatory Response. Cell Host Microbe (2009) 6(6):493–5. doi: 10.1016/j.chom.2009.12.001
3. Correia AL, Guimaraes JC, Auf der Maur P, De Silva D, Trefny MP, Okamoto R, et al. Hepatic Stellate Cells Suppress NK Cell-Sustained Breast Cancer Dormancy. Nature (2021) 594(7864):566–71. doi: 10.1038/s41586-021-03614-z
4. Bjorkstrom NK, Strunz B, Ljunggren HG. Natural Killer Cells in Antiviral Immunity. Nat Rev Immunol (2021) 22:122–23. doi: 10.1038/s41577-021-00558-3
5. Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human NK Cells: Surface Receptors, Inhibitory Checkpoints, and Translational Applications. Cell Mol Immunol (2019) 16(5):430–41. doi: 10.1038/s41423-019-0206-4
6. Adams NM, Grassmann S, Sun JC. Clonal Expansion of Innate and Adaptive Lymphocytes. Nat Rev Immunol (2020) 20:694–707. doi: 10.1038/s41577-020-0307-4
7. Sheppard S, Sun JC. Virus-Specific NK Cell Memory. J Exp Med (2021) 218(4):e20201731. doi: 10.1084/jem.20201731
8. Sun JC, Beilke JN, Lanier LL. Adaptive Immune Features of Natural Killer Cells. Nature (2009) 457(7229):557–61. doi: 10.1038/nature07665
9. O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T Cell- and B Cell-Independent Adaptive Immunity Mediated by Natural Killer Cells. Nat Immunol (2006) 7(5):507–16. doi: 10.1038/ni1332
10. von Andrian UH. NK Cell Memory: Discovery of a Mystery. Nat Immunol (2021) 22(6):669–71. doi: 10.1038/s41590-021-00890-9
11. Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, et al. Critical Role for the Chemokine Receptor CXCR6 in NK Cell-Mediated Antigen-Specific Memory of Haptens and Viruses. Nat Immunol (2010) 11(12):1127–35. doi: 10.1038/ni.1953
12. Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of Human Cytomegalovirus Infection on the NK Cell Receptor Repertoire. Blood (2004) 104(12):3664–71. doi: 10.1182/blood-2004-05-2058
13. Guma M, Cabrera C, Erkizia I, Bofill M, Clotet B, Ruiz L, et al. Human Cytomegalovirus Infection Is Associated With Increased Proportions of NK Cells That Express the CD94/NKG2C Receptor in Aviremic HIV-1-Positive Patients. J Infect Dis (2006) 194(1):38–41. doi: 10.1086/504719
14. Hammer Q, Ruckert T, Borst EM, Dunst J, Haubner A, Durek P, et al. Peptide-Specific Recognition of Human Cytomegalovirus Strains Controls Adaptive Natural Killer Cells. Nat Immunol (2018) 19(5):453–63. doi: 10.1038/s41590-018-0082-6
15. Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T, et al. Identification of Human NK Cells That Are Deficient for Signaling Adaptor Fcrgamma and Specialized for Antibody-Dependent Immune Functions. Int Immunol (2012) 24(12):793–802. doi: 10.1093/intimm/dxs080
16. Zhang T, Scott JM, Hwang I, Kim S. Cutting Edge: Antibody-Dependent Memory-Like NK Cells Distinguished by Fcrgamma Deficiency. J Immunol (2013) 190(4):1402–6. doi: 10.4049/jimmunol.1203034
17. Wagner JA, Fehniger TA. Human Adaptive Natural Killer Cells: Beyond Nkg2c. Trends Immunol (2016) 37(6):351–3. doi: 10.1016/j.it.2016.05.001
18. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus Infection Drives Adaptive Epigenetic Diversification of NK Cells With Altered Signaling and Effector Function. Immunity (2015) 42(3):443–56. doi: 10.1016/j.immuni.2015.02.008
19. Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, et al. Critical Role of CD2 Co-Stimulation in Adaptive Natural Killer Cell Responses Revealed in NKG2C-Deficient Humans. Cell Rep (2016) 15(5):1088–99. doi: 10.1016/j.celrep.2016.04.005
20. Parga-Vidal L, van Gisbergen K. Area Under Immunosurveillance: Dedicated Roles of Memory CD8 T-Cell Subsets. Cold Spring Harb Perspect Biol (2020) 12(11):a037796. doi: 10.1101/cshperspect.a037796
21. Paust S, Blish CA, Reeves RK. Redefining Memory: Building the Case for Adaptive NK Cells. J Virol (2017) 91(20):e00169–17. doi: 10.1128/JVI.00169-17
22. Gill US, Golden-Mason L. HCMV Jogs the ‘Memory’ of NK Cells in HBV. J Hepatol (2019) 70(3):343–5. doi: 10.1016/j.jhep.2018.11.009
23. Mele D, Oliviero B, Mantovani S, Ludovisi S, Lombardi A, Genco F, et al. Adaptive Natural Killer Cell Functional Recovery in Hepatitis C Virus Cured Patients. Hepatology (2020) 73(1):79–90. doi: 10.1002/hep.31273
24. Peppa D, Pedroza-Pacheco I, Pellegrino P, Williams I, Maini MK, Borrow P. Adaptive Reconfiguration of Natural Killer Cells in HIV-1 Infection. Front Immunol (2018) 9:474. doi: 10.3389/fimmu.2018.00474
25. Zhou J, Amran FS, Kramski M, Angelovich TA, Elliott J, Hearps AC, et al. An NK Cell Population Lacking Fcrgamma Is Expanded in Chronically Infected HIV Patients. J Immunol (2015) 194(10):4688–97. doi: 10.4049/jimmunol.1402448
26. Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, et al. Expansion of a Unique CD57(+)NKG2Chi Natural Killer Cell Subset During Acute Human Cytomegalovirus Infection. Proc Natl Acad Sci USA (2011) 108(36):14725–32. doi: 10.1073/pnas.1110900108
27. Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. Human Cytomegalovirus (CMV)-Induced Memory-Like NKG2C(+) NK Cells Are Transplantable and Expand In Vivo in Response to Recipient CMV Antigen. J Immunol (2012) 189(10):5082–8. doi: 10.4049/jimmunol.1201964
28. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, et al. NK Cell Responses to Cytomegalovirus Infection Lead to Stable Imprints in the Human KIR Repertoire and Involve Activating Kirs. Blood (2013) 121(14):2678–88. doi: 10.1182/blood-2012-10-459545
29. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic Modification and Antibody-Dependent Expansion of Memory-Like NK Cells in Human Cytomegalovirus-Infected Individuals. Immunity (2015) 42(3):431–42. doi: 10.1016/j.immuni.2015.02.013
30. Muntasell A, Pupuleku A, Cisneros E, Vera A, Moraru M, Vilches C, et al. Relationship of NKG2C Copy Number With the Distribution of Distinct Cytomegalovirus-Induced Adaptive NK Cell Subsets. J Immunol (2016) 196(9):3818–27. doi: 10.4049/jimmunol.1502438
31. Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, et al. Human Cytomegalovirus Drives Epigenetic Imprinting of the IFNG Locus in NKG2Chi Natural Killer Cells. PloS Pathog (2014) 10(10):e1004441. doi: 10.1371/journal.ppat.1004441
32. Tesi B, Schlums H, Cichocki F, Bryceson YT. Epigenetic Regulation of Adaptive NK Cell Diversification. Trends Immunol (2016) 37(7):451–61. doi: 10.1016/j.it.2016.04.006
33. Capuano C, Battella S, Pighi C, Franchitti L, Turriziani O, Morrone S, et al. Tumor-Targeting Anti-Cd20 Antibodies Mediate In Vitro Expansion of Memory Natural Killer Cells: Impact of CD16 Affinity Ligation Conditions and In Vivo Priming. Front Immunol (2018) 9:1031. doi: 10.3389/fimmu.2018.01031
34. Rolle A, Brodin P. Immune Adaptation to Environmental Influence: The Case of NK Cells and HCMV. Trends Immunol (2016) 37(3):233–43. doi: 10.1016/j.it.2016.01.005
35. Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, et al. Expansion of CD94/NKG2C+ NK Cells in Response to Human Cytomegalovirus-Infected Fibroblasts. Blood (2006) 107(9):3624–31. doi: 10.1182/blood-2005-09-3682
36. Rolle A, Pollmann J, Ewen EM, Le VT, Halenius A, Hengel H, et al. IL-12-Producing Monocytes and HLA-E Control HCMV-Driven NKG2C+ NK Cell Expansion. J Clin Invest (2014) 124(12):5305–16. doi: 10.1172/JCI77440
37. Prod’homme V, Tomasec P, Cunningham C, Lemberg MK, Stanton RJ, McSharry BP, et al. Human Cytomegalovirus UL40 Signal Peptide Regulates Cell Surface Expression of the NK Cell Ligands HLA-E and Gpul18. J Immunol (2012) 188(6):2794–804. doi: 10.4049/jimmunol.1102068
38. Lauterbach N, Wieten L, Popeijus HE, Voorter CE, Tilanus MG. HLA-E Regulates NKG2C+ Natural Killer Cell Function Through Presentation of a Restricted Peptide Repertoire. Hum Immunol (2015) 76(8):578–86. doi: 10.1016/j.humimm.2015.09.003
39. Saghafian-Hedengren S, Sohlberg E, Theorell J, Carvalho-Queiroz C, Nagy N, Persson JO, et al. Epstein-Barr Virus Coinfection in Children Boosts Cytomegalovirus-Induced Differentiation of Natural Killer Cells. J Virol (2013) 87(24):13446–55. doi: 10.1128/JVI.02382-13
40. Mariano JL, Zenarruzabeitia O, Seroogy CM, Holland SM, Kuehn HS, Rosenzweig SD, et al. Intact IL-12 Signaling Is Necessary for the Generation of Human Natural Killer Cells With Enhanced Effector Function After Restimulation. J Allergy Clin Immunol (2014) 134(5):1190–3.e1. doi: 10.1016/j.jaci.2014.06.006
41. Djaoud Z, Parham P. Hlas, Tcrs, and Kirs, a Triumvirate of Human Cell-Mediated Immunity. Annu Rev Biochem (2020) 89:717–39. doi: 10.1146/annurev-biochem-011520-102754
42. Liu W, Scott JM, Langguth E, Chang H, Park PH, Kim S. Fcrgamma Gene Editing Reprograms Conventional NK Cells to Display Key Features of Adaptive Human NK Cells. iScience (2020) 23(11):101709. doi: 10.1016/j.isci.2020.101709
43. Adams NM, Geary CD, Santosa EK, Lumaquin D, Le Luduec JB, Sottile R, et al. Cytomegalovirus Infection Drives Avidity Selection of Natural Killer Cells. Immunity (2019) 50(6):1381–1390 e5. doi: 10.1016/j.immuni.2019.04.009
44. Grassmann S, Pachmayr LO, Leube J, Mihatsch L, Andrae I, Flommersfeld S, et al. Distinct Surface Expression of Activating Receptor Ly49H Drives Differential Expansion of NK Cell Clones Upon Murine Cytomegalovirus Infection. Immunity (2019) 50(6):1391–400.e4. doi: 10.1016/j.immuni.2019.04.015
45. Crinier A, Narni-Mancinelli E, Ugolini S, Vivier E. Snapshot: Natural Killer Cells. Cell (2020) 180(6):1280–1280 e1. doi: 10.1016/j.cell.2020.02.029
46. Chan CJ, Smyth MJ, Martinet L. Molecular Mechanisms of Natural Killer Cell Activation in Response to Cellular Stress. Cell Death Differ (2014) 21(1):5–14. doi: 10.1038/cdd.2013.26
47. Ryan JC, Naper C, Hayashi S, Daws MR. Physiologic Functions of Activating Natural Killer (NK) Complex-Encoded Receptors on NK Cells. Immunol Rev (2001) 181:126–37. doi: 10.1034/j.1600-065X.2001.1810110.x
48. Vietzen H, Ruckert T, Hartenberger S, Honsig C, Jaksch P, Geleff S, et al. Extent of Cytomegalovirus Replication in the Human Host Depends on Variations of the HLA-E/UL40 Axis. mBio (2021) 12(2):e02996–20. doi: 10.1128/mBio.02996-20
49. Merino A, Zhang B, Dougherty P, Luo XH, Wang JH, Blazar BR, et al. Chronic Stimulation Drives Human NK Cell Dysfunction and Epigenetic Reprograming. J Of Clin Invest (2019) 129(9):3770–85. doi: 10.1172/JCI125916
50. Miyashita R, Tsuchiya N, Hikami K, Kuroki K, Fukazawa T, Bijl M, et al. Molecular Genetic Analyses of Human NKG2C (KLRC2) Gene Deletion. Int Immunol (2004) 16(1):163–8. doi: 10.1093/intimm/dxh013
51. Muntasell A, Lopez-Montanes M, Vera A, Heredia G, Romo N, Penafiel J, et al. NKG2C Zygosity Influences CD94/NKG2C Receptor Function and the NK-Cell Compartment Redistribution in Response to Human Cytomegalovirus. Eur J Immunol (2013) 43(12):3268–78. doi: 10.1002/eji.201343773
52. Binder C, Cvetkovski F, Sellberg F, Berg S, Paternina Visbal H, Sachs DH, et al. Cd2 Immunobiology. Front Immunol (2020) 11:1090. doi: 10.3389/fimmu.2020.01090
53. Manieri NA, Chiang EY, Grogan JL. TIGIT: A Key Inhibitor of the Cancer Immunity Cycle. Trends Immunol (2017) 38(1):20–8. doi: 10.1016/j.it.2016.10.002
54. Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the Checkpoint Receptor TIGIT Prevents NK Cell Exhaustion and Elicits Potent Anti-Tumor Immunity. Nat Immunol (2018) 19(7):723–32. doi: 10.1038/s41590-018-0132-0
55. Sarhan D, Cichocki F, Zhang B, Yingst A, Spellman SR, Cooley S, et al. Adaptive NK Cells With Low TIGIT Expression Are Inherently Resistant to Myeloid-Derived Suppressor Cells. Cancer Res (2016) 76(19):5696–706. doi: 10.1158/0008-5472.CAN-16-0839
56. Cichocki F, Wu CY, Zhang B, Felices M, Tesi B, Tuininga K, et al. ARID5B Regulates Metabolic Programming in Human Adaptive NK Cells. J Exp Med (2018) 215(9):2379–95. doi: 10.1084/jem.20172168
57. Bigley AB, Spade S, Agha NH, Biswas S, Tang S, Malik MH, et al. Fcepsilonrigamma-Negative NK Cells Persist In Vivo and Enhance Efficacy of Therapeutic Monoclonal Antibodies in Multiple Myeloma. Blood Adv (2021) 5(15):3021–31. doi: 10.1182/bloodadvances.2020002440
58. Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The Emergence of Natural Killer Cells as a Major Target in Cancer Immunotherapy. Trends In Immunol (2019) 40(2):142–58. doi: 10.1016/j.it.2018.12.003
59. Leonard WJ, Lin JX, O’Shea JJ. The Gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity (2019) 50(4):832–50. doi: 10.1016/j.immuni.2019.03.028
60. Cherrier DE, Serafini N, Di Santo JP. Innate Lymphoid Cell Development: A T Cell Perspective. Immunity (2018) 48(6):1091–103. doi: 10.1016/j.immuni.2018.05.010
61. Cichocki F, Miller JS, Anderson SK, Bryceson YT. Epigenetic Regulation of NK Cell Differentiation and Effector Functions. Front Immunol (2013) 4:55. doi: 10.3389/fimmu.2013.00055
62. Holmes TD, Pandey RV, Helm EY, Schlums H, Han H, Campbell TM, et al. The Transcription Factor Bcl11b Promotes Both Canonical and Adaptive NK Cell Differentiation. Sci Immunol (2021) 6(57):eabc9801. doi: 10.1126/sciimmunol.abc9801
63. Sun JC, Lanier LL. NK Cell Development, Homeostasis and Function: Parallels With CD8(+) T Cells. Nat Rev Immunol (2011) 11(10):645–57. doi: 10.1038/nri3044
64. Yokoyama WM, Kim S. Licensing of Natural Killer Cells by Self-Major Histocompatibility Complex Class I. Immunol Rev (2006) 214:143–54. doi: 10.1111/j.1600-065X.2006.00458.x
65. Raulet DH, Vance RE. Self-Tolerance of Natural Killer Cells. Nat Rev Immunol (2006) 6(7):520–31. doi: 10.1038/nri1863
66. Heath J, Newhook N, Comeau E, Gallant M, Fudge N, Grant M. NKG2C(+)CD57(+) Natural Killer Cell Expansion Parallels Cytomegalovirus-Specific CD8(+) T Cell Evolution Towards Senescence. J Immunol Res (2016) 2016:7470124. doi: 10.1155/2016/7470124
67. Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, et al. Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell (2019) 176(1-2):348–60.e12. doi: 10.1016/j.cell.2018.11.045
68. Schlums H, Jung M, Han H, Theorell J, Bigley V, Chiang SC, et al. Adaptive NK Cells can Persist in Patients With GATA2 Mutation Depleted of Stem and Progenitor Cells. Blood (2017) 129(14):1927–39. doi: 10.1182/blood-2016-08-734236
69. Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, et al. Effector and Memory CD8+ T Cell Fate Coupled by T-Bet and Eomesodermin. Nat Immunol (2005) 6(12):1236–44. doi: 10.1038/ni1268
70. Corat MA, Schlums H, Wu C, Theorell J, Espinoza DA, Sellers SE, et al. Acquired Somatic Mutations in PNH Reveal Long-Term Maintenance of Adaptive NK Cells Independent of Hspcs. Blood (2017) 129(14):1940–6. doi: 10.1182/blood-2016-08-734285
71. Bozzano F, Della Chiesa M, Pelosi A, Antonini F, Ascierto ML, Del Zotto G, et al. HCMV-Controlling NKG2C+ NK Cells Originate From Novel Circulating Inflammatory Precursors. J Allergy Clin Immunol (2021) 147(6):2343–57. doi: 10.1016/j.jaci.2020.12.648
72. Cichocki F, Grzywacz B, Miller JS. Human NK Cell Development: One Road or Many? Front Immunol (2019) 10:2078. doi: 10.3389/fimmu.2019.02078
73. Kim KH, Yu HT, Hwang I, Park S, Park SH, Kim S, et al. Phenotypic and Functional Analysis of Human NK Cell Subpopulations According to the Expression of Fcepsilonrigamma and NKG2C. Front Immunol (2019) 10:2865. doi: 10.3389/fimmu.2019.02865
74. Shah SV, Manickam C, Ram DR, Kroll K, Itell H, Permar SR, et al. CMV Primes Functional Alternative Signaling in Adaptive Deltag NK Cells But Is Subverted by Lentivirus Infection in Rhesus Macaques. Cell Rep (2018) 25(10):2766–74.e3. doi: 10.1016/j.celrep.2018.11.020
75. Lopez-Botet M, Muntasell A, Martinez-Rodriguez JE, Lopez-Montanes M, Costa-Garcia M, Pupuleku A. Development of the Adaptive NK Cell Response to Human Cytomegalovirus in the Context of Aging. Mech Ageing Dev (2016) 158:23–6. doi: 10.1016/j.mad.2016.06.010
76. Muccio L, Falco M, Bertaina A, Locatelli F, Frassoni F, Sivori S, et al. Late Development of Fcepsilonrgamma(Neg) Adaptive Natural Killer Cells Upon Human Cytomegalovirus Reactivation in Umbilical Cord Blood Transplantation Recipients. Front Immunol (2018) 9:1050. doi: 10.3389/fimmu.2018.01050
77. Picarda G, Benedict CA. Cytomegalovirus: Shape-Shifting the Immune System. J Immunol (2018) 200(12):3881–9. doi: 10.4049/jimmunol.1800171
78. Biron CA, Byron KS, Sullivan JL. Severe Herpesvirus Infections in an Adolescent Without Natural Killer Cells. N Engl J Med (1989) 320(26):1731–5. doi: 10.1056/NEJM198906293202605
79. Schuch A, Zecher BF, Muller PA, Correia MP, Daul F, Rennert C, et al. NK-Cell Responses Are Biased Towards CD16-Mediated Effector Functions in Chronic Hepatitis B Virus Infection. J Hepatol (2019) 70(3):351–60. doi: 10.1016/j.jhep.2018.10.006
80. Oh JS, Ali AK, Kim S, Corsi DJ, Cooper CL, Lee SH. NK Cells Lacking Fcepsilonrigamma Are Associated With Reduced Liver Damage in Chronic Hepatitis C Virus Infection. Eur J Immunol (2016) 46(4):1020–9. doi: 10.1002/eji.201546009
81. Petitdemange C, Becquart P, Wauquier N, Beziat V, Debre P, Leroy EM, et al. Unconventional Repertoire Profile Is Imprinted During Acute Chikungunya Infection for Natural Killer Cells Polarization Toward Cytotoxicity. PloS Pathog (2011) 7(9):e1002268. doi: 10.1371/journal.ppat.1002268
82. Hart GT, Tran TM, Theorell J, Schlums H, Arora G, Rajagopalan S, et al. Adaptive NK Cells in People Exposed to Plasmodium Falciparum Correlate With Protection From Malaria. J Exp Med (2019) 216(6):1280–90. doi: 10.1084/jem.20181681
83. Varchetta S, Mele D, Oliviero B, Mantovani S, Ludovisi S, Cerino A, et al. Unique Immunological Profile in Patients With COVID-19. Cell Mol Immunol (2020) 18(3):604–12. doi: 10.21203/rs.3.rs-23953/v1
84. Maucourant C, Filipovic I, Ponzetta A, Aleman S, Cornillet M, Hertwig L, et al. Natural Killer Cell Immunotypes Related to COVID-19 Disease Severity. Sci Immunol (2020) 5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832
85. Vietzen H, Zoufaly A, Traugott M, Aberle J, Aberle SW, Puchhammer-Stockl E. Deletion of the NKG2C Receptor Encoding KLRC2 Gene and HLA-E Variants Are Risk Factors for Severe COVID-19. Genet Med (2021) 23(5):963–7. doi: 10.21203/rs.3.rs-34505/v1
86. Rocha V, Labopin M, Sanz G, Arcese W, Schwerdtfeger R, Bosi A, et al. Transplants of Umbilical-Cord Blood or Bone Marrow From Unrelated Donors in Adults With Acute Leukemia. N Engl J Med (2004) 351(22):2276–85. doi: 10.1056/NEJMoa041469
87. Cichocki F, Cooley S, Davis Z, DeFor TE, Schlums H, Zhang B, et al. CD56dimCD57+NKG2C+ NK Cell Expansion Is Associated With Reduced Leukemia Relapse After Reduced Intensity HCT. Leukemia (2016) 30(2):456–63. doi: 10.1038/leu.2015.260
88. Nguyen S, Beziat V, Dhedin N, Kuentz M, Vernant JP, Debre P, et al. HLA-E Upregulation on IFN-Gamma-Activated AML Blasts Impairs CD94/NKG2A-Dependent NK Cytolysis After Haplo-Mismatched Hematopoietic SCT. Bone Marrow Transplant (2009) 43(9):693–9. doi: 10.1038/bmt.2008.380
89. Liu LL, Beziat V, Oei VYS, Pfefferle A, Schaffer M, Lehmann S, et al. Ex Vivo Expanded Adaptive Nk Cells Effectively Kill Primary Acute Lymphoblastic Leukemia Cells. Cancer Immunol Res (2017) 5(8):654–65. doi: 10.1158/2326-6066.CIR-16-0296
90. Ataya M, Redondo-Pachon D, Llinas-Mallol L, Yelamos J, Heredia G, Perez-Saez MJ, et al. Pretransplant Adaptive NKG2C+ NK Cells Protect Against Cytomegalovirus Infection in Kidney Transplant Recipients. Am J Transplant (2020) 20(3):663–76. doi: 10.1111/ajt.15658
91. Makwana NB, Foley B, Lee S, Fernandez S, Irish AB, Price P. Asymptomatic CMV Infections in Long-Term Renal Transplant Recipients Are Associated With the Loss of Fcrgamma From LIR-1(+) NK Cells. Eur J Immunol (2016) 46(11):2597–608. doi: 10.1002/eji.201646422
92. Redondo-Pachon D, Crespo M, Yelamos J, Muntasell A, Perez-Saez MJ, Perez-Fernandez S, et al. Adaptive NKG2C+ NK Cell Response and the Risk of Cytomegalovirus Infection in Kidney Transplant Recipients. J Immunol (2017) 198(1):94–101. doi: 10.4049/jimmunol.1601236
93. Cichocki F, Taras E, Chiuppesi F, Wagner JE, Blazar BR, Brunstein C, et al. Adaptive NK Cell Reconstitution Is Associated With Better Clinical Outcomes. JCI Insight (2019) 4(2):e125553. doi: 10.1172/jci.insight.125553
94. van de Donk N, Richardson PG, Malavasi F. CD38 Antibodies in Multiple Myeloma: Back to the Future. Blood (2018) 131(1):13–29. doi: 10.1182/blood-2017-06-740944
95. Woan KV, Kim H, Bjordahl R, Davis ZB, Gaidarova S, Goulding J, et al. Harnessing Features of Adaptive NK Cells to Generate Ipsc-Derived NK Cells for Enhanced Immunotherapy. Cell Stem Cell (2021) 28(12):2062–75.e5. doi: 10.1016/j.stem.2021.08.013
96. Cho H, Kim KH, Lee H, Kim CG, Chung H, Choi YS, et al. Adaptive Natural Killer Cells Facilitate Effector Functions of Daratumumab in Multiple Myeloma. Clin Cancer Res (2021) 27(10):2947–58. doi: 10.1158/1078-0432.CCR-20-3418
97. Hawkins C, Croul S. Viruses and Human Brain Tumors: Cytomegalovirus Enters the Fray. J Clin Invest (2011) 121(10):3831–3. doi: 10.1172/JCI60005
98. Hartel H, Theiss J, Abdelaziz MO, Raftery MJ, Pecher G, Bogner E. HCMV-Mediated Interference of Bortezomib-Induced Apoptosis in Colon Carcinoma Cell Line Caco-2. Viruses (2021) 13(1):83. doi: 10.3390/v13010083
99. Lopez-Botet M, Angulo A, Guma M. Natural Killer Cell Receptors for Major Histocompatibility Complex Class I and Related Molecules in Cytomegalovirus Infection. Tissue Antigens (2004) 63(3):195–203. doi: 10.1111/j.1399-0039.2004.00210.x
100. Baryawno N, Rahbar A, Wolmer-Solberg N, Taher C, Odeberg J, Darabi A, et al. Detection of Human Cytomegalovirus in Medulloblastomas Reveals a Potential Therapeutic Target. J Clin Invest (2011) 121(10):4043–55. doi: 10.1172/JCI57147
101. Achour A, Baychelier F, Besson C, Arnoux A, Marty M, Hannoun L, et al. Expansion of CMV-Mediated NKG2C+ NK Cells Associates With the Development of Specific De Novo Malignancies in Liver-Transplanted Patients. J Immunol (2014) 192(1):503–11. doi: 10.4049/jimmunol.1301951
102. Guillerey C, Huntington ND, Smyth MJ. Targeting Natural Killer Cells in Cancer Immunotherapy. Nat Immunol (2016) 17(9):1025–36. doi: 10.1038/ni.3518
103. Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, et al. Cytokine-Induced Memory-Like Natural Killer Cells Exhibit Enhanced Responses Against Myeloid Leukemia. Sci Transl Med (2016) 8(357):357ra123. doi: 10.1126/scitranslmed.aaf2341
104. Gang M, Marin ND, Wong P, Neal CC, Marsala L, Foster M, et al. CAR-Modified Memory-Like NK Cells Exhibit Potent Responses to NK-Resistant Lymphomas. Blood (2020) 136(20):2308–18. doi: 10.1182/blood.2020006619
105. Peng H, Tian Z. Natural Killer Cell Memory: Progress and Implications. Front Immunol (2017) 8:1143. doi: 10.3389/fimmu.2017.01143
106. Morvan MG, Lanier LL. NK Cells and Cancer: You can Teach Innate Cells New Tricks. Nat Rev Cancer (2016) 16(1):7–19. doi: 10.1038/nrc.2015.5
107. Gauthier L, Morel A, Anceriz N, Rossi B, Blanchard-Alvarez A, Grondin G, et al. Multifunctional Natural Killer Cell Engagers Targeting Nkp46 Trigger Protective Tumor Immunity. Cell (2019) 177(7):1701–13.e16. doi: 10.1016/j.cell.2019.04.041
108. Capuano C, Pighi C, Battella S, De Federicis D, Galandrini R, Palmieri G. Harnessing CD16-Mediated NK Cell Functions to Enhance Therapeutic Efficacy of Tumor-Targeting Mabs. Cancers (Basel) (2021) 13(10):2500. doi: 10.3390/cancers13102500
109. Zhu H, Blum RH, Bjordahl R, Gaidarova S, Rogers P, Lee TT, et al. Pluripotent Stem Cell-Derived NK Cells With High-Affinity Noncleavable CD16a Mediate Improved Antitumor Activity. Blood (2020) 135(6):399–410. doi: 10.1182/blood.2019000621
110. Chew HY, De Lima PO, Gonzalez Cruz JL, Banushi B, Echejoh G, Hu L, et al. Endocytosis Inhibition in Humans to Improve Responses to ADCC-Mediating Antibodies. Cell (2020) 180(5):895–914.e27. doi: 10.1016/j.cell.2020.02.019
Keywords: HCMV, adaptive NK cells, ADCC, epigenetic remodeling, therapeutic potential
Citation: Gao F, Zhou Z, Lin Y, Shu G, Yin G and Zhang T (2022) Biology and Clinical Relevance of HCMV-Associated Adaptive NK Cells. Front. Immunol. 13:830396. doi: 10.3389/fimmu.2022.830396
Received: 07 December 2021; Accepted: 04 March 2022;
Published: 31 March 2022.
Edited by:
Nabila Jabrane-Ferrat, INSERM U1043 Centre de Physiopathologie de Toulouse Purpan, FranceReviewed by:
Subramaniam Malarkannan, Medical College of Wisconsin, United StatesMir Munir Rahim, University of Windsor, Canada
Copyright © 2022 Gao, Zhou, Lin, Shu, Yin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianxiang Zhang, emhhbmd0aTJAZ21haWwuY29t; Gang Yin, Z2FuZ3lpbkBjc3UuZWR1LmNu
 Fei Gao
Fei Gao Zhengwei Zhou1
Zhengwei Zhou1 Guang Shu
Guang Shu Gang Yin
Gang Yin Tianxiang Zhang
Tianxiang Zhang