- Complex Carbohydrate Research Center, University of Georgia Athens, Athens, GA, United States
The addition of N-acetyl glucosamine (GlcNAc) on the hydroxy group of serine/threonine residues is known as O-GlcNAcylation (OGN). The dynamic cycling of this monosaccharide on and off substrates occurs via O-linked β-N-acetylglucosamine transferase (OGT) and O-linked β-N-acetylglucosaminase (OGA) respectively. These enzymes are found ubiquitously in eukaryotes and genetic knock outs of the ogt gene has been found to be lethal in embryonic mice. The substrate scope of these enzymes is vast, over 15,000 proteins across 43 species have been identified with O-GlcNAc. OGN has been known to play a key role in several cellular processes such as: transcription, translation, cell signaling, nutrient sensing, immune cell development and various steps of the cell cycle. However, its dysregulation is present in various diseases: cancer, neurodegenerative diseases, diabetes. O-GlcNAc is heavily involved in cross talk with other post-translational modifications (PTM), such as phosphorylation, acetylation, and ubiquitination, by regulating each other’s cycling enzymes or directly competing addition on the same substrate. This crosstalk between PTMs can affect gene expression, protein localization, and protein stability; therefore, regulating a multitude of cell signaling pathways. In this review the roles of OGN will be discussed. The effect O-GlcNAc exerts over protein-protein interactions, the various forms of crosstalk with other PTMs, and its role as a nutrient sensor will be highlighted. A summary of how these O-GlcNAc driven processes effect the immune system will also be included.
Introduction
O-N-acetylglucosamine (O-GlcNAc) is a monosaccharide, post translational modification (PTM) covalently bound to serine and/or threonine residues. Unlike traditional forms of glycosylation, the addition of a O-GlcNAc moiety, O-GlcNAcylation, occurs within nucleocytoplasmic and mitochondrial compartments of the cell and remains as a monosaccharide opposed to further elaboration to a polysaccharide (1, 2). Similar to other PTMs like phosphorylation, O-GlcNAc is cycled on and off substrates, regulating their biological functions. However, unlike phosphorylation, the addition and removal of this modification is performed with a single set of enzymes, O-β-N-acetylglucosamine transferase (OGT) and O-β-N-acetylglucosamidase (OGA) respectively (3, 4).
These enzymes are found primarily in the nucleus, cytoplasm, and mitochondria in all metazoans, including plants viruses, and some bacteria (5, 6). Deletion of OGT or OGA is embryonically and perinatally lethal in mice, demonstrating its importance to survival and development (7, 8). The number of identified O-GlcNAcylated substrates is continuously growing at a rapid pace thanks to technological advances. Perhaps due to its ubiquitous nature and biological importance, various O-GlcNAc databases have been developed to mainstream pertinent information regarding O-GlcNAcylated substrates, such as corresponding literature references and, in some cases, the amino acid site of the modification (9, 10).
O-GlcNAcylation plays a role in a broad range of biological processes, such as transcription, translation, enzyme activity, cell division, protein localization and degradation. How these and other cellular operations are regulated is, in part, dependent on which substrates are O-GlcNAcylated and to what extent. As will be discussed later in more detail, OGT’s activity and substrate specificity vary with the concentration of UDP-GlcNAc within cells (11), the donor for O-GlcNAc, which is proportional to the flux of several metabolites (12–15). Thus, O-GlcNAc directly links the regulation of important biological processes with the cellular nutrient status to serve as a major nutrient sensor. This is further highlighted in metabolic diseases, such as diabetes and cancer, whose aberrant O-GlcNAc levels are correlated with pathologic phenotypes (16–20).
O-GlcNAc Cycling Enzymes
O-GlcNAc Transferase (OGT)
The ogt gene is highly conserved in numerous organisms from C. elegans to humans and is encoded on the X chromosome near the centromere, exhibiting greater than 60-80% amino acid identity between species (8). OGT was initially purified from rat liver and reticulocyte lysates (3, 21), sequenced and cloned (22, 23). The tertiary structure of the enzyme was determined in 2011 by overlapping two semi-complete crystal structures, the breakthrough of which contained a UDP and CKII peptide molecules bound to the active site (24). Belonging to the GT-B superfamily of glycosyltransferases, OGT is made up of four domains: 1) a N-terminal domain containing continuous helix-turn-helix tetratricopeptide repeats (TPRs), 2) C-terminal region bearing the GT41 catalytic domain containing two Rossmann-folded lobes, 3) an intervening region that bridges the two lobes and 4) a nuclear localization sequence between the TPR and catalytic domain (25). OGT has three different isoforms, all varying in the length of their respective TPR domains. ncOGT (nuclear cytoplasmic) and sOGT (short) have 13.5 and 2.5 TPRs, both of which are found in the cytoplasm and nucleus. mOGT (mitochondria) consists of 9 TPR domains and is localized in the mitochondrial inner membrane (26). ncOGT is the predominant isoform (27).
Much work has been done to elucidate OGT’s binding modes and substrate specificity, most of which is done through crystallization and related mutagenesis studies. One of the earliest was a crystal structure of OGT with the non-hydrolyzable GlcNAc derivative and OGT inhibiter UDP-5SGlcNAc, which identified an ordered bi-bi mechanism of glycosylation where UDP-GlcNAc initially binds to the active site and then is covered by the acceptor (28). Although there is one example of substrate binding to the catalytic domain alone (29), the majority of substrate binding is suggested to occur in the TPR domain of OGT. The removal of the TPR domain is known to abrogate OGT activity toward protein substrates but not to small peptide substrates (30, 31). Interestingly, unlike other small peptides, OGT does not modify the C-terminal domain (CTD) of RNA polymerase II, which consists of a degenerate seven amino acid repeats, if it has less than five repeats (35 amino acids). However, ten CTD repeats is an excellent substrate in vitro (32). Key structural features of the TPR domain have been identified thanks to crystal structures of various substrate-bound OGT complexes. The TPR domain forms a superhelix made up of two layers, the inner layer contains a highly conserved asparagine ladder which binds the amide backbone of the acceptor substrates (33). This ladder extends the length of the TPR region, the mutations of which were shown to impair protein O-GlcNAcylation (24, 34). Recent contributions in the literature have revealed the presence of two aspartate residues along the inner layer of the TPR domain proximal to the active site, the alanine mutation of which diminishes substrate binding (35). In this study and several earlier ones, an attempt to determine a consensus substrate sequence was attempted (3, 36, 37). From these investigations it is suggested that about half of the known OGT substrates contain acidic arginine or lysine residues within 7-11 amino acids of the functionalized threonine or serine.
Given these statistical findings, it has largely been agreed upon that there is no consensus sequence for OGT substrates. This may in part be due to OGT’s employment of adapter proteins. These are proteins that form complexes with OGT and subsequently direct substrate specificity by altering binding modes or localization. Various adapter proteins, such as mSin3a, PCG-1a and HDAC1 have been identified (38, 39), the most studied are OGT’s interactions and targeting by ten-eleven translocation (TET) family enzymes. OGT-TET complexes target OGT to chromatin or histones that are involved in chromatin remodeling (40). Although adapter protein identification and significance can be difficult to determine, recent advancements in labeling technology utilizing a biotin transferase-TPR fusion protein may provide additional examples of proteins acting in this manner (41). Expanding the list of known adapter proteins and their specific functions will help explain the promiscuity and specificity of OGT.
O-GlcNAcase (OGA)
The oga gene (mgea5), which was initially identified as a putative hyaluronidase and cloned from a meningioma, is present on the somatic chromosome 10. OGA protein was first purified from rat spleen (4) and rat brain (42). The rat brain OGA was sequenced and cloned and it was found to be identical to mgea5. OGA protein exists in two isoforms, the predominant 916 amino acid OGA-L and the less common short OGA-S truncated in the C-terminus. OGA consists of three domains: 1) a N-terminal catalytic domain, similar to glycoside hydrolase family 84 (GH84) enzymes with a [(β/α)8] triose-phosphate isomerase (TIM) barrel structure, 2) a stalk region or α-helical bundle, and 3) a C-terminal pseudo-histone acetyltransferase (HAT) domain which does not have any acetyl transferase activity (42).
Several crystallization studies in 2017 have elucidated various aspects of OGA’s structure (43, 44). These studies indicate that human OGA forms homo dimmers with a single stalk α-helix of the opposing monomer. Li and coworkers showed that removing one of the stalk helices exhibited 100-fold lower catalytic activity. OGA is cleaved in half by caspase 3 during apoptosis, but the enzyme remains active and the catalytic domain and HAT domains remain non-covalently associated (45). Two conserved aspartate residues were found in OGA’s active site and binding pocket flanking the O-GlcNAc glycosidic bond, potentially catalyzing its hydrolysis (43, 46). The OGA dimer forms a V-shaped cleft at the interface of the catalytic domain and stalk domain of the two monomers, providing a potential substrate binding pocket. For a more detailed discussion on OGT and OGA substrate specificity see (47).
O-GlcNAc Regulates Protein Function
In the first two and a half decades since the discovery of intracellular O-GlcNAc, around 500 O-GlcNAcylated proteins had been reported in the literature (48). Since then, advancements in analytical methods, specifically mass spectrometry and labeling techniques, have simplified and fast-tracked the elucidation of the O-GlcNAcome (49–52). Recently, multiple databases have been constructed mainstreaming the search for O-GlcNAcylated targets and even providing site-specific mapping when available (9, 10). As of November 2021, according to the https://www.oglcnac.mcw.edu/statistics/, there are over 15,000 GlcNAcylated proteins from 43 different species reported in the literature curated from around 2,300 articles. The role of these O-GlcNAcylated substrates extends to nearly every intracellular biological process imaginable: cell metabolism (53, 54), cell death (55), the circadian clock (56–58), cell cycle progression (13), various signaling pathways (59, 60), transcription (61), translation (62, 63), protein degradation (64), and cell development (65). Like other PTMs, such as phosphorylation, O-GlcNAc’s cycling conveys a bevy of biological outcomes by modifying the substrate’s function. A brief overview of these O-GlcNAc modified activities will be discussed below, however for a more complete discussion please see the references (53, 66).
A common result of O-GlcNAcylation is the relocalization of its substrate. This effect is most often observed with transcription factors, resulting in their activation or inhibition. NeuroD1 is a transcription factor in pancreatic β-cells which induces gene expression responsible for insulin production (67). Upon O-GlcNAcylation, this cytosolic protein is localized to the nucleus to initiate transcription (Figure 1). This was determined in the context of elevated glucose concentrations driving NeuroD1 nuclear translocation, which was ameliorated upon ogt silencing via siRNA (68). O-GlcNAcylation of β-catenin has the opposite effect. Modification at Ser23 was shown to induce plasma membrane localization, where it activates cell adhesion functionality, consequently blocking its nuclear localization and gene transcription (69).
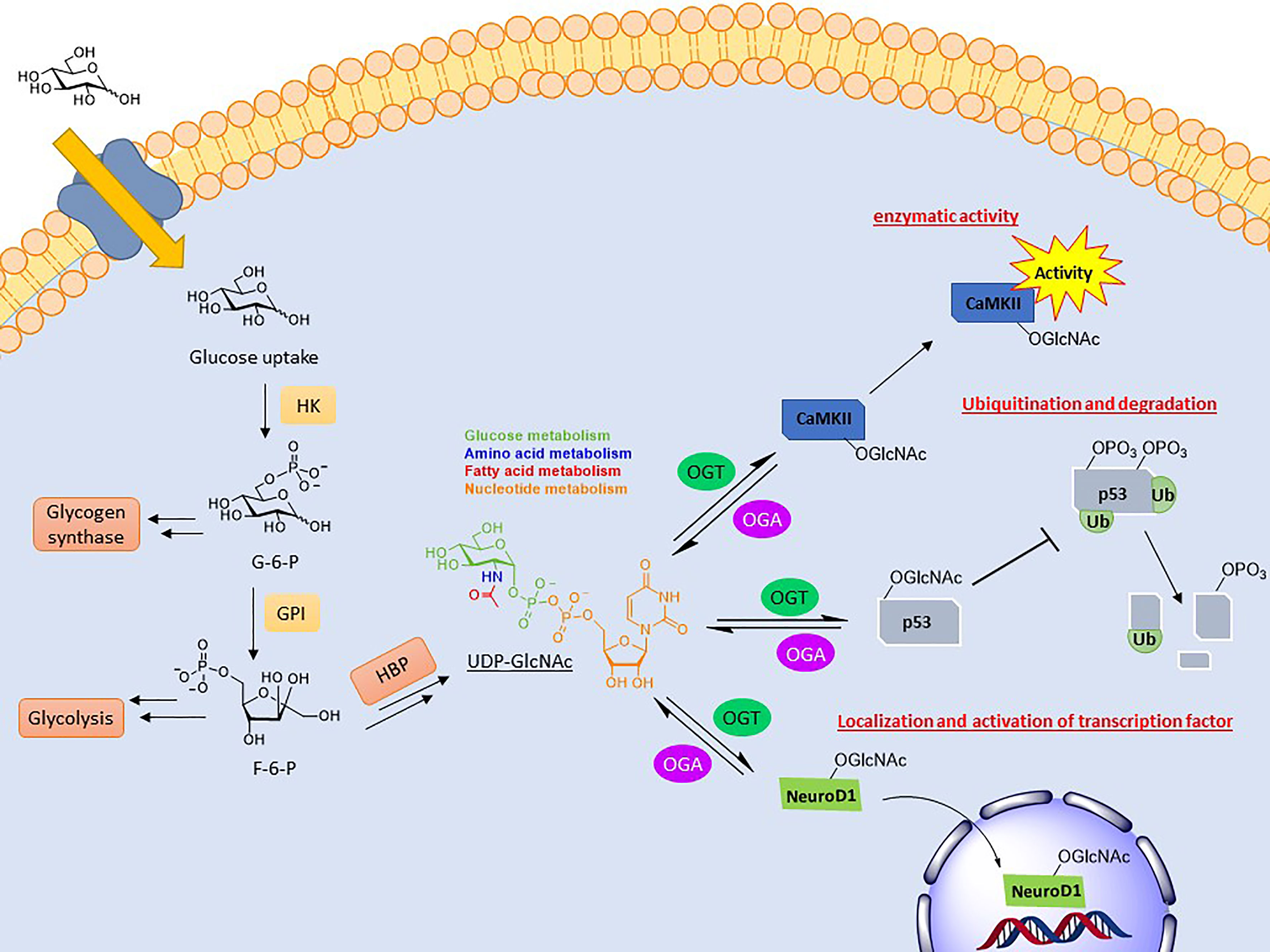
Figure 1 Depiction of the biosynthesis of UDP-GlcNAc and the effects conveyed on its substrates. UDP-GlcNAc is formed through the hexosamine biosynthetic pathway (HBP), combining several metabolites. O-GlcNAc cycling can modulate activity (CaMKII), stability (p53), and localization (NeuroD1) of its substrates.
Another key function of O-GlcNAc cycling is the inhibition or activation of enzymes. Calcium/calmodulin-dependent protein kinase II (CaMKII) is a regulatory kinase involved in Ca2+ release events important for heart and brain function. Hyperglycemia is correlated with the chronic activation of this protein, which has been shown to induce arrhythmias and other cardiomyopathic phenotypes (70). Elevated O-GlcNAcylation of this protein, specifically at S279, was shown to induce autonomous activation of CaMKII under hyperglycemic conditions even in the absence of Ca2+/calmodulin (Figure 1). FRET analysis demonstrated that this activation is perpetuated by inhibiting its reversion to an apo-conformation. Mutation of this site attenuated these effects (71).
The previous example also demonstrates how O-GlcNAcylation can alter substrate conformation and consequently its function. A more indirect example is the modification’s influence on heat shock proteins and other molecular chaperones. Elevated O-GlcNAc levels prior to heat stress causes increased HSP40 and HSP72 expression, which help to maintain protein folding and solubility during periods of stress (72). Additionally, NMR studies comparing O-GlcNAcylated and phosphorylated peptides found that the larger O-GlcNAc moiety destabilized α-helicies found in the phospho-peptides, and instead induces a bend (73).
O-GlcNAc Crosstalk With Other PTMs
The addition or removal of O-GlcNAc can alter a variety of biological activities. These changes often occur in concert with other PTMs in an inhibitory or promotive manner. This crosstalk phenomenon helps regulate the dynamic nature of cellular signaling in response to nutrient status. For a more detailed summary of O-GlcNAc cross talk with other PTMs, refer to the references (64, 74, 75).
The most direct form of crosstalk is O-GlcNAc’s relationship with phosphorylation. This is because both are serine and/or threonine modifications and, therefore, have the potential to compete for the same or proximal sites. This inhibitory form is referred to as reciprocal crosstalk. An example of this can be seen in the oncoprotein c-Myc at thr58, regulating its transactivation (76–78). Crosstalk PTMs more commonly occurs at a distance rather than at the same site. This can occur on near-by amino acids or at a distance, referred to as proximal or distal crosstalk, respectively. For these types, O-GlcNAcylation can either promote or inhibit the addition of subsequent PTMs. O-GlcNAcylation of p53, a tumor suppressor gene, at ser149 blocks its phosphorylation at thr155, which is known to induce its ubiquitin-proteasomal degradation. As a result, p53 accumulates in the cytoplasm, allowing for increased apoptotic activity (Figure 1) (79).
Furthermore, different PTMs often regulate each other’s cycling enzymes. Microarray studies have indicated that over 80% of the human kinome are substrates for OGT (53) and a search of the https://www.oglcnac.mcw.edu/statistics/ yields over 700 results for O-GlcNAcylated kinases as of November 2021. These modifications have been known to both inhibit and promote enzymatic activity and even change their substrate specificity, as is the case for Casein kinase 2α (CK2α). O-GlcNAcylation at ser347 blocks its phosphorylation at thr344. These two modified CK2α enzymes were screened against a protein microarray and found to have different substrate specificity (80).
Additionally, tyrosine phosphorylation of OGT, resulting from stimulation of the insulin receptor, shows increased transferase activity (81). O-GlcNAcylation of other PTM cycling enzymes is also known to regulate activity. O-GlcNAc modification of the histone lysine methyltransferase MLL5 was demonstrated to promote methylation of H3K4, triggering cell lineage determination in HL60 lymphocytes (82).
Nutrient Sensing
While the majority of cellular glucose uptake is directed to glycolytic pathways primarily used for energy generation and storage, about 2-5% is processed through the hexosamine biosynthetic pathway (HBP) (15). The formation of the final product in this pathway, alpha uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc), combines to monitor the metabolism of glucose, amino acids, fatty acids, and nucleotides. Consequently, the cellular concentration of UDP-GlcNAc is responsive to nutrient levels and flux through the pathway (83, 84). This product is either transported to the ER and Golgi for constructing extracellular, endomembrane oligosaccharides or remains in the nucleus and cytoplasm as the donor for O-GlcNAcylation (1, 85, 86).
The intracellular UDP-GlcNAc levels are known to affect OGT’s activity and substrate specificity. This was demonstrated in 1999 by Hart and Kreppel using recombinant OGT in in vitro-based experiments comparing its activity and affinity for different peptide substrates under various UDP-GlcNAc concentrations. Remarkably multiple apparent Km values were found by varying UDP-GlcNAc concentrations for different substrates. Furthermore different substrates exhibited varying degrees of GlcNAcylation over a range of UDP-GlcNAc concentrations, indicating that O-GlcNAcylation is highly responsive to nutrient levels in a substrate specific manner (11). These early experiments have since been demonstrated in both cell and animal models, particularly in relationship to hyperglycemia or nutrient deprivation (87–89).
The nutrient status of a cell and the corresponding levels of UDP-GlcNAc and subsets of O-GlcNAcylated proteins act as a nutritional fingerprint imparting changes in biological processes, resulting in nutrient-based phenotypes. Due to this relationship O-GlcNAc is often referred to as a nutrient sensing rheostat (Figure 2). For example, O-GlcNAcylation of the proteasome subunit 19S is correlated with reduced activity (90), which has been demonstrated in the degradation of Sp1 transcription factor under nutrient deprivation (88). This increase in proteasome activity under reduced nutrient levels has been speculated to serve as a catabolic mechanism to regulate amino acid availability (54). Similarly O-GlcNAc has been shown to regulate cellular processes under increased nutrient levels, an example of which occurs in the insulin signaling pathway. Upon insulin stimulation, glucose uptake is increased and a signaling cascade initiates various processes (glycolysis, glycogen synthesis, lipogenesis). One of the early stages of this pathway is the phosphorylation of insulin receptor substrate 1 (IRS-1), which induces its association with PI3K and further propagates the signaling cascade towards Akt phosphorylation and activation (91). Various groups have shown that IRS-1 is O-GlcNAcylated upon insulin stimulation and that increasing this PTM, via OGA inhibition or OGT overexpression, reduces its association with PI3K, attenuating insulin signaling (92–94).
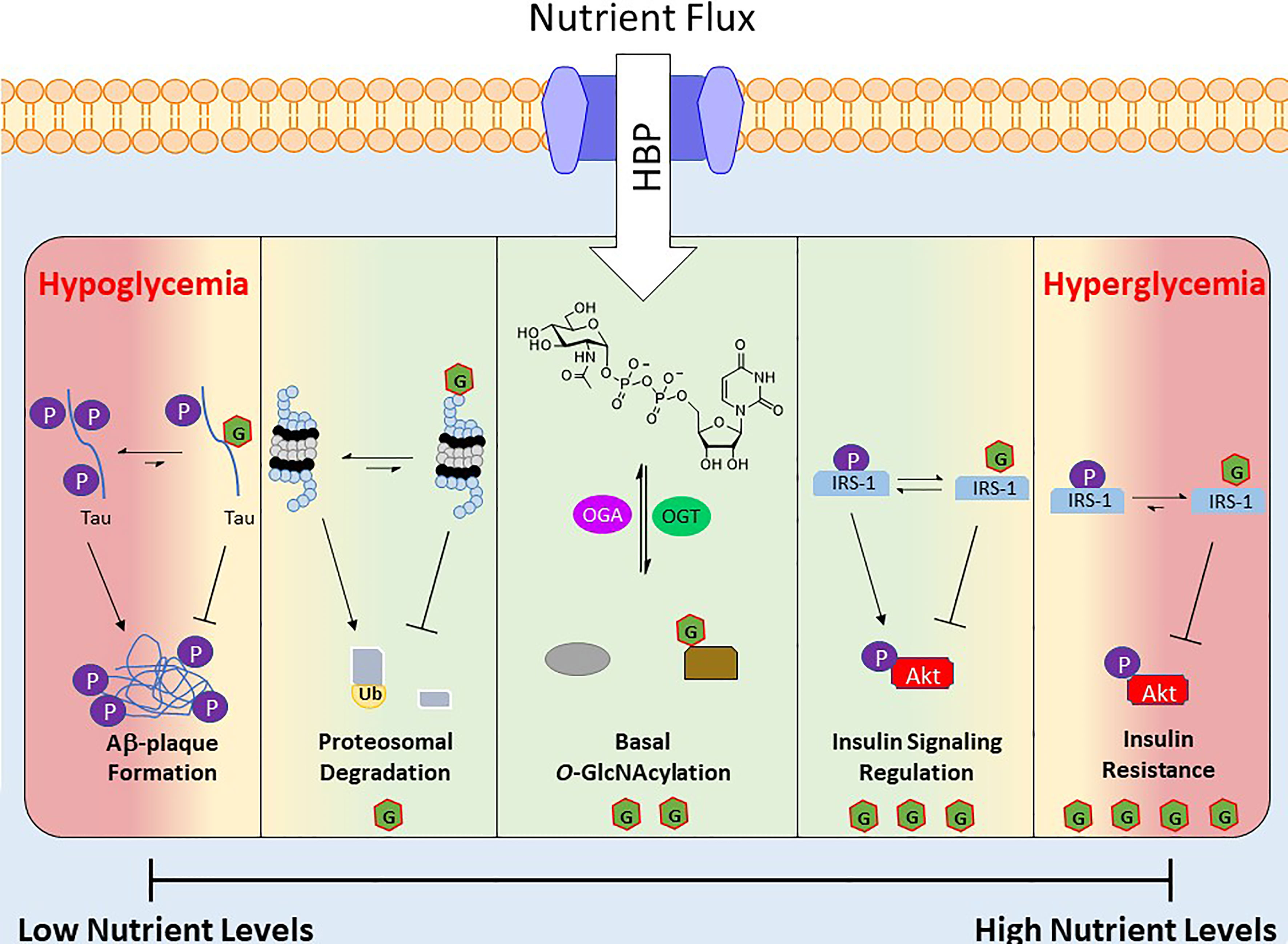
Figure 2 Depiction of O-GlcNAc’s role as a nutrient sensing rheostat. Nutrient flux directly impacts the levels of UDP-GlcNAc, which regulates the activity and substrate specificity of OGT. As a result the O-GlcNAcylated proteins and their corresponding biological functions are a response to the nutrient levels. Below are several examples of O-GlcNAc’s effect at various nutrient levels. Green area is “normal” effects of O-GlcNAcylation and the red areas are O-GlcNAcylation’s effects under extreme nutrient levels.
Although O-GlcNAc acts to regulate cellular functions during alterations in nutrient levels, prolonged extremes of nutritional flux are linked to disease states. For example, hyperglycemia in type 2 diabetes mellitus is associated with consistently elevated levels of O-GlcNAc (16). Similarly OGT overexpression or OGA inhibition has been shown to induce insulin resistance in various cell and animal models (95, 96), and insulin sensitivity is improved via OGT KO in mice (97). At the other end of the spectrum is hypoglycemia, which is consequently liked to decreased protein O-GlcNAcylation, is a common feature of many neurodegenerative diseases such as Alzheimer’s disease (AD) (98, 99). Studies in this area have found that hyperphosphorylation of β-amyloid precursor protein (APP) and Tau induces Aβ plaque formations, but O-GlcNAcylation of these proteins prevents this aggregation (Figure 2) (100).
O-GlcNAc’s Involvement in the Immune System
As described above, O-GlcNAc is extensively involved in a multitude of cellular processes, and the immune system is no exception. Many reviews have been written detailing O-GlcNAc’s role in various immune system aspects, such as: T-cell development (65), inflammation (101), infection (102), autoimmunity (103), lymphocytic cancers (104), immunometabolism (105), and broad overviews (106). In this section a brief overview of several aspects will be discussed highlighting O-GlcNAc’s function in the immune system, particularly as it relates to sensing nutritional cues that govern protein activities.
There are several instances during lymphocyte development and activation that requires a metabolic shift from relying on oxidative phosphorylation to glycolysis and glutaminolytic metabolism. In these situations, glucose and amino acids, particularly glutamine, uptake is dramatically increased, the latter of which leads to increased fatty acid and cholesterol synthesis (107–109). This influx in metabolites is necessary to fulfil the needs of increased cell growth and proliferation required in T-cell development (65) and lymphocyte activation (110). Accompanying this increase in nutrient flux is an increase in UDP-GlcNAc and O-GlcNAcylated protein levels (111), which have been found to impart signaling cues important to the required cellular process. One of the most studied of which is T-cell differentiation. Following TCR activation, T cells undergo several stages ultimately becoming various effector lineages based on its microenvironment and signaling. Increased O-GlcNAc levels were found during the transition from DN3 to DN4 stage and during positive selection, corresponding to increased glucose and glutamine uptake. In these studies, ogt knock out just before the DP stage in mice, diminished the population of mature T cells (112). O-GLcNAc has also been shown to be key in B cell survival and activity. Conditional ogt allele deletion at various stages of B cell development in mice showed increased apoptosis in mature B cells via defects in BAFFR mediated pathways, indicating OGT’s importance in maintaining homeostasis. The same study also showed that OGT KO had reduced B-cell activation corresponding to a decrease in NF-κB nuclear translocation (113).
While sufficient nutrient flux is important to survival, differentiation, and activation in immune cells, chronic aberration of nutrients levels are known to be detrimental. Hyperglycemic conditions, as in diabetes, increases the O-GlcNAcylation and activation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), similarly increasing the production of pro-inflammatory cytokines (114, 115). Hyperglycemia has also been shown to alter the polarization of macrophages to the less immunogenic M2 state, which correlated with tumorgenicity, suggesting a possible mechanism of immune system evasion (116).
One of the earliest indications of O-GlcNAc’s impact on the immune system came in 1991 from Kearse and Hart who demonstrated that, upon T-cell activation O-GlcNAcylated protein density rapidly change from the cytosol to the nucleus in a protein-specific manner (111). These results foreshadowed future research demonstrating O-GlcNAc’s regulation of key transcription factors, one of the most studied and significant of which being NF-κB (117). NF-κB is a dimeric protein that regulates cytokine production and cell proliferation, and is key in the activation and maturation of lymphocytes. Upon TCR or BCR-activation, the cytosolic dimer is phosphorylated resulting in the ubiquitination and degradation of its complexed inhibitor, inhibitor of κB protein (IκB). This dissociation event in turn induces nuclear localization of NF-κB and subsequent gene expression (118). O-GlcNAcylation of NF-κB’s subunits was found to be paramount for this translocalization process in B and T cells (117). Mutagenesis studies in T-cells revealed that O-GlcNAcylation of the subunit c-Rel was directly related to its nuclear localization, promoter binding and gene expression of IL2, IFNG, and CSF2 (119). Interestingly the location of this O-GlcNAcylation site is directly adjacent to the IκB binding domain of NF-κB and its phosphorylation was not found to be altered. Other studies performed in epithelial cell lines have demonstrated that O-GlcNAc functionalization of NF-κB is not only necessary for its translocation but also helps to mediate acetylation of p65 subunit which increases NF-κB gene transcription. This acetylation results from an interaction with the acetyltransferase CBP/p300, which is weakened by p65 phosphorylation (120). This inhibitory phosphorylation was shown to be blocked by O-GlcNAcylation of p65 on thr305 and ser319 (121). O-GlcNAc modification of NF-κB has been shown to inhibit activation in various cells. For example, In macrophages NF-κB activation promotes inducible nitric oxide synthase (iNOS) expression which is a key enzyme in the innate immune system for killing infectious bacteria (122). Overexpression of OGT in RAW264.7 macrophage cells and BV2 microglial cells resulted in suppressed iNOS expression corresponding to increased O-GlcNAcylation of p65 and c-Rel (123). Supporting this inhibitory effect, OGA inhibition in LPS treated RAW264.7 microglial cells increased iNOS expression (124). These results highlight the fact that O-GlcNAc’s regulatory roles are cell specific.
Discussion
Since its discovery in 1983 (1, 2), the O-linked-N-acetylglucosamine monosaccharide post-translational-modification has been demonstrated to be a key modification for regulating and maintaining a variety of biological functions in response to nutrient levels. These processes are controlled by changes in protein function driven by intricate crosstalk between O-GlcNAc and other PTMs. Aberrations in nutrient flux and O-GlcNAc levels has been demonstrated in numerous disease states. Disease phenotypes can be induced by directly altering O-GlcNAc cycling (enzyme inhibition or knockout) or via nutrient manipulation, indicating that changes in O-GlcNAcylation can have a direct effect on the progression or development of various pathologies.
Most work regarding O-GlcNAc in the immunology field has been focused around lymphocyte activation, survival, and development. The majority of these studies focus on macrophages, B cells and T cells. As a result, little is known about O-GlcNAc’s role in other immune cells. For example, changes in O-GlcNAcylation have been found in natural killer (NK) cells and neutrophils during cytotoxicity events and chemotactic stimulation respectively (125, 126). Furthermore glucosamine supplementation results in increased Rac activity in neutrophils, important for neutrophil mobilization, and decreased cytotoxic activity in NK cells (127, 128). These studies suggest that O-GlcNAc is involved in these processes, however specific mechanisms have yet to be determined. Even less work has been done with regards to dendritic cells, eosinophils, and basophils. Considering that O-GlcNAc’s effects may be cell specific, recall the effect on NF-κB and pro-/anti-inflammatory responses in various cells, these fundamental gaps in understanding may be significant.
Additionally, O-GlcNAc’s role in lymphoid cancers and autoimmune disease remains unexplored. Elevated glucose flux and metabolism is observed in nearly all cancers known as the Warburg effect. Consequently O-GlcNAcylated protein levels are also elevated (129). In Chronic Lymphoid Leukemia (CLL) O-GlcNAcylation levels of p53, Akt, c-Myc, and STAT5 are increased with respect to normal basal levels, promoting cell proliferation (130, 131). However, OGA inhibition with thiamet G increased cell sensitivity of chemotherapy for human leukemia cell lines (132). Although exciting, these contradictory effects highlight the need for further investigation before they can be applied in therapeutics.
Similar to cancer, increased glucose uptake in immune cells is a key feature of acute inflammation (133). Furthermore, prolonged hyperglycemia observed in type 2 diabetes mellitus is known to lead to chronic inflammation in patients that leads to an increased risk of autoimmunity development (134). A recent study performed by Liu et al. demonstrated enhanced pro-inflammatory cytokine production in virus-challenged primary mononuclear blood cells and pulmonary epithelial cells when treated with OGA inhibitor or glucosamine. These effects were extended to a mouse model and found to correlate with decreased survival rates. Further studies determined OGT’s interaction with interferon regulatory factor 5 (IRF5) as key regulator of the increased cytokine production (135). Conversely OGT was found to be paramount to the lineage stability of regulatory T cells (Treg cells). Ogt knockout in Treg cells in culture and mice exhibited a severe autoimmune phenotype resulting from attenuated IL-2/STAT5 signaling attributed to decreased glycosylation (136). This Ogt knockout-related Treg pathology was also found to exacerbate hepatitis in an autoimmune rat model (137). Taken together O-GlcNAc seems to play different roles in different aspects of autoimmunity. More studies are required to fully detail the relationship between aberrant nutrient flux, O-GlcNAcylation, and autoimmunity.
Author Contributions
MM: Wrote the article and made the figures. GH: Edited the article and gave direction for sections. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Institutes of Health Grants R01DK61171 and R01GM116891.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Torres CR, Hart GW. Topography and Polypeptide Distribution of Terminal N-Acetylglucosamine Residues on the Surfaces of Intact Lymphocytes. Evidence for O-Linked GlcNAc. J Biol Chem (1984) 259(5):3308–17. doi: 10.1016/S0021-9258(17)43295-9
2. Holt GD, Haltiwanger RS, Torres CR, Hart GW. Erythrocytes Contain Cytoplasmic Glycoproteins. O-Linked GlcNAc on Band 4.1. J Biol Chem (1987) 262(31):14847–50. doi: 10.1016/S0021-9258(18)48100-8
3. Haltiwanger RS, Holt GD, Hart GW. Enzymatic Addition of O-GlcNAc to Nuclear and Cytoplasmic Proteins. Identification of a Uridine Diphospho-N-Acetylglucosamine:Peptide Beta-N-Acetylglucosaminyltransferase. J Biol Chem (1990) 265(5):2563–8. doi: 10.1016/S0021-9258(19)39838-2
4. Dong DL, Hart GW. Purification and Characterization of an O-GlcNAc Selective N-Acetyl-Beta-D-Glucosaminidase From Rat Spleen Cytosol. J Biol Chem (1994) 269(30):19321–30. doi: 10.1016/S0021-9258(17)32170-1
5. Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc Protein Modification in Plants: Evolution and Function. Biochim Biophys Acta (2010) 1800(2):49–56. doi: 10.1016/j.bbagen.2009.11.016
6. Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol (Lausanne) (2014) 5:183. doi: 10.3389/fendo.2014.00183
7. Keembiyehetty C, Love DC, Harwood KR, Gavrilova O, Comly ME, Hanover JA. Conditional Knock-Out Reveals a Requirement for O-Linked N-Acetylglucosaminase (O-GlcNAcase) in Metabolic Homeostasis. J Biol Chem (2015) 290(11):7097–113. doi: 10.1074/jbc.M114.617779
8. Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, et al. The O-GlcNAc Transferase Gene Resides on the X Chromosome and Is Essential for Embryonic Stem Cell Viability and Mouse Ontogeny. Proc Natl Acad Sci USA (2000) 97(11):5735–9. doi: 10.1073/pnas.100471497
9. Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, et al. The Human O-GlcNAcome Database and Meta-Analysis. Sci Data (2021) 8(1):25. doi: 10.1038/s41597-021-00810-4
10. Ma J, Li Y, Hou C, Wu C. O-GlcNAcAtlas: A Database of Experimentally Identified O-GlcNAc Sites and Proteins. Glycobiology (2021) 31(7):719–23. doi: 10.1093/glycob/cwab003
11. Kreppel LK, Hart GW. Regulation of a Cytosolic and Nuclear O-GlcNAc Transferase. Role of the Tetratricopeptide Repeats. J Biol Chem (1999) 274(45):32015–22. doi: 10.1074/jbc.274.45.32015
12. Wang J, Liu R, Hawkins M, Barzilai N, Rossetti L. A Nutrient-Sensing Pathway Regulates Leptin Gene Expression in Muscle and Fat. Nature (1998) 393(6686):684–8. doi: 10.1038/31474
13. Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-Linked Beta-N-Acetylglucosamine Protein Modification Cause Severe Defects in Mitotic Progression and Cytokinesis. J Biol Chem (2005) 280(38):32944–56. doi: 10.1074/jbc.M503396200
14. Zachara NE, Hart GW. Cell Signaling, the Essential Role of O-GlcNAc! Biochim Biophys Acta (2006) 1761(5-6):599–617. doi: 10.1016/j.bbalip.2006.04.007
15. Marshall S, Bacote V, Traxinger RR. Discovery of a Metabolic Pathway Mediating Glucose-Induced Desensitization of the Glucose Transport System. Role of Hexosamine Biosynthesis in the Induction of Insulin Resistance. J Biol Chem (1991) 266(8):4706–12. doi: 10.1016/S0021-9258(19)67706-9
16. Vaidyanathan K, Wells L. Multiple Tissue-Specific Roles for the O-GlcNAc Post-Translational Modification in the Induction of and Complications Arising From Type II Diabetes. J Biol Chem (2014) 289(50):34466–71. doi: 10.1074/jbc.R114.591560
17. Peterson SB, Hart GW. New Insights: A Role for O-GlcNAcylation in Diabetic Complications. Crit Rev Biochem Mol Biol (2016) 51(3):150–61. doi: 10.3109/10409238.2015.1135102
18. Hanover JA, Chen W, Bond MR. O-GlcNAc in Cancer: An Oncometabolism-Fueled Vicious Cycle. J Bioenerg Biomembr (2018) 50(3):155–73. doi: 10.1007/s10863-018-9751-2
19. Park J, Lai MKP, Arumugam TV, Jo D-G. O-GlcNAcylation as a Therapeutic Target for Alzheimer’s Disease. Neuromolecular Med (2020) 22(2):171–93. doi: 10.1007/s12017-019-08584-0
20. Zhu Y, Hart GW. Targeting O-GlcNAcylation to Develop Novel Therapeutics. Mol Aspects Med (2021) 79:100885. doi: 10.1016/j.mam.2020.100885
21. Starr CM, Hanover JA. Glycosylation of Nuclear Pore Protein P62. Reticulocyte Lysate Catalyzes O-Linked N-Acetylglucosamine Addition In Vitro. J Biol Chem (1990) 265(12):6868–73. doi: 10.1016/S0021-9258(19)39229-4
22. Lubas WA, Frank DW, Krause M, Hanover JA. O-Linked GlcNAc Transferase Is a Conserved Nucleocytoplasmic Protein Containing Tetratricopeptide Repeats. J Biol Chem (1997) 272(14):9316–24. doi: 10.1074/jbc.272.14.9316
23. Kreppel LK, Blomberg MA, Hart GW. Dynamic Glycosylation of Nuclear and Cytosolic Proteins. Cloning and Characterization of a Unique O-GlcNAc Transferase With Multiple Tetratricopeptide Repeats. J Biol Chem (1997) 272(14):9308–15. doi: 10.1074/jbc.272.14.9308
24. Joiner CM, Li H, Jiang J, Walker S. Structural Characterization of the O-GlcNAc Cycling Enzymes: Insights Into Substrate Recognition and Catalytic Mechanisms. Curr Opin Struct Biol (2019) 56:97–106. doi: 10.1016/j.sbi.2018.12.003
25. Seo HG, Kim HB, Kang MJ, Ryum JH, Yi EC, Cho JW. Identification of the Nuclear Localisation Signal of O-GlcNAc Transferase and Its Nuclear Import Regulation. Sci Rep (2016) 6:34614. doi: 10.1038/srep34614
26. Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and Nucleocytoplasmic Targeting of O-Linked GlcNAc Transferase. J Cell Sci (2003) 116(Pt 4):647–54. doi: 10.1242/jcs.00246
27. King DT, Males A, Davies GJ, Vocadlo DJ. Molecular Mechanisms Regulating O-Linked N-Acetylglucosamine (O-GlcNAc)-Processing Enzymes. Curr Opin Chem Biol (2019) 53:131–44. doi: 10.1016/j.cbpa.2019.09.001
28. Jínek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. The Superhelical TPR-Repeat Domain of O-Linked GlcNAc Transferase Exhibits Structural Similarities to Importin Alpha. Nat Struct Mol Biol (2004) 11(10):1001–7. doi: 10.1038/nsmb833
29. Cheung WD, Hart GW. AMP-Activated Protein Kinase and P38 MAPK Activate O-GlcNAcylation of Neuronal Proteins During Glucose Deprivation. J Biol Chem (2008) 283(19):13009–20. doi: 10.1074/jbc.M801222200
30. Iyer SP, Hart GW. Roles of the Tetratricopeptide Repeat Domain in O-GlcNAc Transferase Targeting and Protein Substrate Specificity. J Biol Chem (2003) 278(27):24608–16. doi: 10.1074/jbc.M300036200
31. Yang X, Qian K. Protein O-GlcNAcylation: Emerging Mechanisms and Functions. Nat Rev Mol Cell Biol (2017) 18(7):452–65. doi: 10.1038/nrm.2017.22
32. Comer FI, Hart GW. Reciprocity Between O-GlcNAc and O-Phosphate on the Carboxyl Terminal Domain of RNA Polymerase II. Biochemistry (2001) 40(26):7845–52. doi: 10.1021/bi0027480
33. Lazarus MB, Jiang J, Kapuria V, Bhuiyan T, Janetzko J, Zandberg WF, et al. HCF-1 Is Cleaved in the Active Site of O-GlcNAc Transferase. Science (2013) 342(6163):1235–9. doi: 10.1126/science.1243990
34. Levine ZG, Fan C, Melicher MS, Orman M, Benjamin T, Walker S. O-GlcNAc Transferase Recognizes Protein Substrates Using an Asparagine Ladder in the Tetratricopeptide Repeat (TPR) Superhelix. J Am Chem Soc (2018) 140(10):3510–3. doi: 10.1021/jacs.7b13546
35. Joiner CM, Levine ZG, Aonbangkhen C, Woo CM, Walker S. Aspartate Residues Far From the Active Site Drive O-GlcNAc Transferase Substrate Selection. J Am Chem Soc (2019) 141(33):12974–8. doi: 10.1021/jacs.9b06061
36. Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, et al. The Active Site of O-GlcNAc Transferase Imposes Constraints on Substrate Sequence. Nat Struct Mol Biol (2015) 22(9):744–50. doi: 10.1038/nsmb.3063
37. Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, et al. O-Linked N-Acetylglucosamine Proteomics of Postsynaptic Density Preparations Using Lectin Weak Affinity Chromatography and Mass Spectrometry. Mol Cell Proteomics (2006) 5(5):923–34. doi: 10.1074/mcp.T500040-MCP200
38. Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc Transferase to Promoters by Corepressor Msin3a: Coupling Protein O-GlcNAcylation to Transcriptional Repression. Cell (2002) 110(1):69–80. doi: 10.1016/S0092-8674(02)00810-3
39. Correia JC, Ferreira DMS, Ruas JL. Intercellular: Local and Systemic Actions of Skeletal Muscle PGC-1s. Trends Endocrinol Metab (2015) 26(6):305–14. doi: 10.1016/j.tem.2015.03.010
40. Ito R, Katsura S, Shimada H, Tsuchiya H, Hada M, Okumura T, et al. TET3-OGT Interaction Increases the Stability and the Presence of OGT in Chromatin. Genes Cells (2014) 19(1):52–65. doi: 10.1111/gtc.12107
41. Stephen HM, Praissman JL, Wells L. Generation of an Interactome for the Tetratricopeptide Repeat Domain of O-GlcNAc Transferase Indicates a Role for the Enzyme in Intellectual Disability. J Proteome Res (2021) 20(2):1229–42. doi: 10.1021/acs.jproteome.0c00604
42. Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-Glycosylation of Nuclear and Cytosolic Proteins: Cloning and Characterization of a Neutral, Cytosolic Beta-N-Acetylglucosaminidase From Human Brain. J Biol Chem (2001) 276(13):9838–45. doi: 10.1074/jbc.M010420200
43. Li B, Li H, Lu L, Jiang J. Structures of Human O-GlcNAcase and Its Complexes Reveal a New Substrate Recognition Mode. Nat Struct Mol Biol (2017) 24(4):362–9. doi: 10.1038/nsmb.3390
44. Roth C, Chan S, Offen WA, Hemsworth GR, Willems LI, King DT, et al. Structural and Functional Insight Into Human O-GlcNAcase. Nat Chem Biol (2017) 13(6):610–2. doi: 10.1038/nchembio.2358
45. Butkinaree C, Cheung WD, Park S, Park K, Barber M, Hart GW. Characterization of Beta-N-Acetylglucosaminidase Cleavage by Caspase-3 During Apoptosis. J Biol Chem (2008) 283(35):23557–66. doi: 10.1074/jbc.M804116200
46. Çetinbaş N, Macauley MS, Stubbs KA, Drapala R, Vocadlo DJ. Identification of Asp174 and Asp175 as the Key Catalytic Residues of Human O-GlcNAcase by Functional Analysis of Site-Directed Mutants. Biochemistry (2006) 45(11):3835–44. doi: 10.1021/bi052370b
47. Stephen HM, Adams TM, Wells L. Regulating the Regulators: Mechanisms of Substrate Selection of the O-GlcNAc Cycling Enzymes OGT and OGA. Glycobiology (2021) 31(7):724–33. doi: 10.1093/glycob/cwab005
48. Hart GW, Housley MP, Slawson C. Cycling of O-Linked Beta-N-Acetylglucosamine on Nucleocytoplasmic Proteins. Nature (2007) 446(7139):1017–22. doi: 10.1038/nature05815
49. Ma J, Hart GW. O-GlcNAc Profiling: From Proteins to Proteomes. Clin Proteomics (2014) 11(1):8. doi: 10.1186/1559-0275-11-8
50. Wu ZL, Tatge TJ, Grill AE, Zou Y. Detecting and Imaging O-GlcNAc Sites Using Glycosyltransferases: A Systematic Approach to Study O-GlcNAc. Cell Chem Biol (2018) 25(11):1428–35.e3. doi: 10.1016/j.chembiol.2018.07.007
51. Worth M, Li H, Jiang J. Deciphering the Functions of Protein O-GlcNAcylation With Chemistry. ACS Chem Biol (2017) 12(2):326–35. doi: 10.1021/acschembio.6b01065
52. Hu CW, Worth M, Li H, Jiang J. Chemical and Biochemical Strategies To Explore the Substrate Recognition of O-GlcNAc-Cycling Enzymes. Chembiochem (2019) 20(3):312–8. doi: 10.1002/cbic.201800481
53. Hart GW. Nutrient Regulation of Signaling and Transcription. FASEB J (2018) 32(S1):98.1–.1. doi: 10.1096/fasebj.2018.32.1_supplement.98.1
54. Bond MR, Hanover JA. O-GlcNAc Cycling: A Link Between Metabolism and Chronic Disease. Annu Rev Nutr (2013) 33:205–29. doi: 10.1146/annurev-nutr-071812-161240
55. Banerjee S, Sangwan V, McGinn O, Chugh R, Dudeja V, Vickers SM, et al. Triptolide-Induced Cell Death in Pancreatic Cancer Is Mediated by O-GlcNAc Modification of Transcription Factor Sp1 *. J Biol Chem (2013) 288(47):33927–38. doi: 10.1074/jbc.M113.500983
56. Kaasik K, Kivimäe S, Allen JJ, Chalkley RJ, Huang Y, Baer K, et al. Glucose Sensor O-GlcNAcylation Coordinates With Phosphorylation to Regulate Circadian Clock. Cell Metab (2013) 17(2):291–302. doi: 10.1016/j.cmet.2012.12.017
57. Hart GW. How Sugar Tunes Your Clock. Cell Metab (2013) 17(2):155–6. doi: 10.1016/j.cmet.2013.01.008
58. Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, et al. O-GlcNAc Signaling Entrains the Circadian Clock by Inhibiting BMAL1/CLOCK Ubiquitination. Cell Metab (2013) 17(2):303–10. doi: 10.1016/j.cmet.2012.12.015
59. Ong Q, Han W, Yang X. O-GlcNAc as an Integrator of Signaling Pathways. Front Endocrinol (Lausanne) (2018) 9(599). doi: 10.3389/fendo.2018.00599
60. Naseem S, Parrino SM, Buenten DM, Konopka JB. Novel Roles for GlcNAc in Cell Signaling. Commun Integr Biol (2012) 5(2):156–9. doi: 10.4161/cib.19034
61. Hanover JA, Krause MW, Love DC. Bittersweet Memories: Linking Metabolism to Epigenetics Through O-GlcNAcylation. Nat Rev Mol Cell Biol (2012) 13(5):312–21. doi: 10.1038/nrm3334
62. Datta B, Ray MK, Chakrabarti D, Wylie DE, Gupta NK. Glycosylation of Eukaryotic Peptide Chain Initiation Factor 2 (eIF-2)-Associated 67-kDa Polypeptide (P67) and Its Possible Role in the Inhibition of eIF-2 Kinase-Catalyzed Phosphorylation of the eIF-2 Alpha-Subunit. J Biol Chem (1989) 264(34):20620–4. doi: 10.1016/S0021-9258(19)47108-1
63. Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A Functional RNAi Screen Links O-GlcNAc Modification of Ribosomal Proteins to Stress Granule and Processing Body Assembly. Nat Cell Biol (2008) 10(10):1224–31. doi: 10.1038/ncb1783
64. Ruan HB, Nie Y, Yang X. Regulation of Protein Degradation by O-GlcNAcylation: Crosstalk With Ubiquitination. Mol Cell Proteomics (2013) 12(12):3489–97. doi: 10.1074/mcp.R113.029751
65. Abramowitz LK, Hanover JA. T Cell Development and the Physiological Role of O-GlcNAc. FEBS Lett (2018) 592(23):3943–9. doi: 10.1002/1873-3468.13159
66. Tarbet HJ, Toleman CA, Boyce M. A Sweet Embrace: Control of Protein-Protein Interactions by O-Linked β-N-Acetylglucosamine. Biochemistry (2018) 57(1):13–21. doi: 10.1021/acs.biochem.7b00871
67. Chae JH, Stein GH, Lee JE. NeuroD: The Predicted and the Surprising. Mol Cells (2004) 18(3):271–88.
68. Andrali SS, Qian Q, Ozcan S. Glucose Mediates the Translocation of NeuroD1 by O-Linked Glycosylation. J Biol Chem (2007) 282(21):15589–96. doi: 10.1074/jbc.M701762200
69. Ha JR, Hao L, Venkateswaran G, Huang YH, Garcia E, Persad S. β-Catenin is O-GlcNAc Glycosylated at Serine 23: Implications for β-Catenin’s Subcellular Localization and Transactivator Function. Exp Cell Res (2014) 321(2):153–66. doi: 10.1016/j.yexcr.2013.11.021
70. Anderson ME, Brown JH, Bers DM. CaMKII in Myocardial Hypertrophy and Heart Failure. J Mol Cell Cardiol (2011) 51(4):468–73. doi: 10.1016/j.yjmcc.2011.01.012
71. Erickson JR, Pereira L, Wang L, Han G, Ferguson A, Dao K, et al. Diabetic Hyperglycaemia Activates CaMKII and Arrhythmias by O-Linked Glycosylation. Nature (2013) 502(7471):372–6. doi: 10.1038/nature12537
72. Zachara NE, O’Donnell N, Cheung WD, Mercer JJ, Marth JD, Hart GW. Dynamic O-GlcNAc Modification of Nucleocytoplasmic Proteins in Response to Stress. A Survival Response of Mammalian Cells. J Biol Chem (2004) 279(29):30133–42. doi: 10.1074/jbc.M403773200
73. Elbaum MB, Zondlo NJ. OGlcNAcylation and Phosphorylation Have Similar Structural Effects in α-Helices: Post-Translational Modifications as Inducible Start and Stop Signals in α-Helices, With Greater Structural Effects on Threonine Modification. Biochemistry (2014) 53(14):2242–60. doi: 10.1021/bi500117c
74. van der Laarse SAM, Leney AC, Heck AJR. Crosstalk Between Phosphorylation and O-GlcNAcylation: Friend or Foe. FEBS J (2018) 285(17):3152–67. doi: 10.1111/febs.14491
75. Zeidan Q, Hart GW. The Intersections Between O-GlcNAcylation and Phosphorylation: Implications for Multiple Signaling Pathways. J Cell Sci (2010) 123(Pt 1):13–22. doi: 10.1242/jcs.053678
76. Kamemura K, Hayes BK, Comer FI, Hart GW. Dynamic Interplay Between O-Glycosylation and O-Phosphorylation of Nucleocytoplasmic Proteins: Alternative Glycosylation/Phosphorylation of THR-58, a Known Mutational Hot Spot of C-Myc in Lymphomas, is Regulated by Mitogens. J Biol Chem (2002) 277(21):19229–35. doi: 10.1074/jbc.M201729200
77. Chou TY, Hart GW, Dang CV. C-Myc is Glycosylated at Threonine 58, a Known Phosphorylation Site and a Mutational Hot Spot in Lymphomas. J Biol Chem (1995) 270(32):18961–5. doi: 10.1074/jbc.270.32.18961
78. Chou TY, Dang CV, Hart GW. Glycosylation of the C-Myc Transactivation Domain. Proc Natl Acad Sci USA (1995) 92(10):4417–21. doi: 10.1073/pnas.92.10.4417
79. Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, et al. Modification of P53 With O-Linked N-Acetylglucosamine Regulates P53 Activity and Stability. Nat Cell Biol (2006) 8(10):1074–83. doi: 10.1038/ncb1470
80. Tarrant MK, Rho HS, Xie Z, Jiang YL, Gross C, Culhane JC, et al. Regulation of CK2 by Phosphorylation and O-GlcNAcylation Revealed by Semisynthesis. Nat Chem Biol (2012) 8(3):262–9. doi: 10.1038/nchembio.771
81. Whelan SA, Lane MD, Hart GW. Regulation of the O-Linked Beta-N-Acetylglucosamine Transferase by Insulin Signaling. J Biol Chem (2008) 283(31):21411–7. doi: 10.1074/jbc.M800677200
82. Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, et al. GlcNAcylation of a Histone Methyltransferase in Retinoic-Acid-Induced Granulopoiesis. Nature (2009) 459(7245):455–9. doi: 10.1038/nature07954
83. Marshall S, Nadeau O, Yamasaki K. Dynamic Actions of Glucose and Glucosamine on Hexosamine Biosynthesis in Isolated Adipocytes: Differential Effects on Glucosamine 6-Phosphate, UDP-N-Acetylglucosamine, and ATP Levels. J Biol Chem (2004) 279(34):35313–9. doi: 10.1074/jbc.M404133200
84. Zachara NE, Hart GW. O-GlcNAc a Sensor of Cellular State: The Role of Nucleocytoplasmic Glycosylation in Modulating Cellular Function in Response to Nutrition and Stress. Biochim Biophys Acta (2004) 1673(1-2):13–28. doi: 10.1016/j.bbagen.2004.03.016
85. Perez M, Hirschberg CB. Translocation of UDP-N-Acetylglucosamine Into Vesicles Derived From Rat Liver Rough Endoplasmic Reticulum and Golgi Apparatus. J Biol Chem (1985) 260(8):4671–8. doi: 10.1016/S0021-9258(18)89122-0
86. Bond MR, Hanover JA. A Little Sugar Goes a Long Way: The Cell Biology of O-GlcNAc. J Cell Biol (2015) 208(7):869–80. doi: 10.1083/jcb.201501101
87. Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, et al. O-GlcNAc Regulates FoxO Activation in Response to Glucose. J Biol Chem (2008) 283(24):16283–92. doi: 10.1074/jbc.M802240200
88. Han I, Kudlow JE. Reduced O Glycosylation of Sp1 Is Associated With Increased Proteasome Susceptibility. Mol Cell Biol (1997) 17(5):2550–8. doi: 10.1128/MCB.17.5.2550
89. Yki-Järvinen H, Virkamäki A, Daniels MC, McClain D, Gottschalk WK. Insulin and Glucosamine Infusions Increase O-Linked N-Acetyl-Glucosamine in Skeletal Muscle Proteins In Vivo. Metabolism (1998) 47(4):449–55. doi: 10.1016/S0026-0495(98)90058-0
90. Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc Modification Is an Endogenous Inhibitor of the Proteasome. Cell (2003) 115(6):715–25. doi: 10.1016/S0092-8674(03)00974-7
91. White MF. IRS Proteins and the Common Path to Diabetes. Am J Physiol Endocrinol Metab (2002) 283(3):E413–22. doi: 10.1152/ajpendo.00514.2001
92. Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW. Regulation of Insulin Receptor Substrate 1 (IRS-1)/AKT Kinase-Mediated Insulin Signaling by O-Linked Beta-N-Acetylglucosamine in 3T3-L1 Adipocytes. J Biol Chem (2010) 285(8):5204–11. doi: 10.1074/jbc.M109.077818
93. Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, et al. Phosphoinositide Signalling Links O-GlcNAc Transferase to Insulin Resistance. Nature (2008) 451(7181):964–9. doi: 10.1038/nature06668
94. Klein AL, Berkaw MN, Buse MG, Ball LE. O-Linked N-Acetylglucosamine Modification of Insulin Receptor Substrate-1 Occurs in Close Proximity to Multiple SH2 Domain Binding Motifs. Mol Cell Proteomics (2009) 8(12):2733–45. doi: 10.1074/mcp.M900207-MCP200
95. Vosseller K, Wells L, Lane MD, Hart GW. Elevated Nucleocytoplasmic Glycosylation by O-GlcNAc Results in Insulin Resistance Associated With Defects in Akt Activation in 3T3-L1 Adipocytes. Proc Natl Acad Sci USA (2002) 99(8):5313–8. doi: 10.1073/pnas.072072399
96. McClain DA, Lubas WA, Cooksey RC, Hazel M, Parker GJ, Love DC, et al. Altered Glycan-Dependent Signaling Induces Insulin Resistance and Hyperleptinemia. Proc Natl Acad Sci USA (2002) 99(16):10695–9. doi: 10.1073/pnas.152346899
97. Shi H, Munk A, Nielsen TS, Daughtry MR, Larsson L, Li S, et al. Skeletal Muscle O-GlcNAc Transferase is Important for Muscle Energy Homeostasis and Whole-Body Insulin Sensitivity. Mol Metab (2018) 11:160–77. doi: 10.1016/j.molmet.2018.02.010
98. Kuljiš RO, Salković-Petrišić M. Dementia, Diabetes, Alzheimer’s Disease, and Insulin Resistance in the Brain: Progress, Dilemmas, New Opportunities, and a Hypothesis to Tackle Intersecting Epidemics. J Alzheimers Dis (2011) 25(1):29–41. doi: 10.3233/JAD-2011-101392
99. Liu F, Shi J, Tanimukai H, Gu J, Gu J, Grundke-Iqbal I, et al. Reduced O-GlcNAcylation Links Lower Brain Glucose Metabolism and Tau Pathology in Alzheimer’s Disease. Brain (2009) 132(Pt 7):1820–32. doi: 10.1093/brain/awp099
100. Dias WB, Hart GW. O-GlcNAc Modification in Diabetes and Alzheimer’s Disease. Mol Biosyst (2007) 3(11):766–72. doi: 10.1039/b704905f
101. Li Y, Xie M, Men L, Du J. O-GlcNAcylation in Immunity and Inflammation: An Intricate System (Review). Int J Mol Med (2019) 44(2):363–74. doi: 10.3892/ijmm.2019.4238
102. Quik M, Hokke CH, Everts B. The Role of O-GlcNAcylation in Immunity Against Infections. Immunology (2020) 161(3):175–85. doi: 10.1111/imm.13245
103. Qiang A, Slawson C, Fields PE. The Role of O-GlcNAcylation in Immune Cell Activation. Front Endocrinol (Lausanne) (2021) 12:596617. doi: 10.3389/fendo.2021.596617
104. de Jesus T, Shukla S, Ramakrishnan P. Too Sweet to Resist: Control of Immune Cell Function by O-GlcNAcylation. Cell Immunol (2018) 333:85–92. doi: 10.1016/j.cellimm.2018.05.010
105. Machacek M, Slawson C, Fields PE. O-GlcNAc: A Novel Regulator of Immunometabolism. J Bioenerg Biomembr (2018) 50(3):223–9. doi: 10.1007/s10863-018-9744-1
106. Chang Y-H, Weng C-L, Lin K-I. O-GlcNAcylation and Its Role in the Immune System. J BioMed Sci (2020) 27(1):57–. doi: 10.1186/s12929-020-00648-9
107. Loos JA, Roos D. Changes in the Carbohydrate Metabolism of Mitogenically Stimulated Human Peripheral Lymphocytes. 3. Stimulation by Tuberculin and Allogenic Cells. Exp Cell Res (1973) 79(1):136–42. doi: 10.1016/0014-4827(73)90498-9
108. Jones RG, Thompson CB. Revving the Engine: Signal Transduction Fuels T Cell Activation. Immunity (2007) 27(2):173–8. doi: 10.1016/j.immuni.2007.07.008
109. Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T Cell Responses Rely on Amino Acid Transporter ASCT2 Facilitation of Glutamine Uptake and Mtorc1 Kinase Activation. Immunity (2014) 40(5):692–705. doi: 10.1016/j.immuni.2014.04.007
110. Doughty CA, Bleiman BF, Wagner DJ, Dufort FJ, Mataraza JM, Roberts MF, et al. Antigen Receptor-Mediated Changes in Glucose Metabolism in B Lymphocytes: Role of Phosphatidylinositol 3-Kinase Signaling in the Glycolytic Control of Growth. Blood (2006) 107(11):4458–65. doi: 10.1182/blood-2005-12-4788
111. Kearse KP, Hart GW. Lymphocyte Activation Induces Rapid Changes in Nuclear and Cytoplasmic Glycoproteins. Proc Natl Acad Sci USA (1991) 88(5):1701–5. doi: 10.1073/pnas.88.5.1701
112. Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, et al. Glucose and Glutamine Fuel Protein O-GlcNAcylation to Control T Cell Self-Renewal and Malignancy. Nat Immunol (2016) 17(6):712–20. doi: 10.1038/ni.3439
113. Wu JL, Chiang MF, Hsu PH, Tsai DY, Hung KH, Wang YH, et al. O-GlcNAcylation Is Required for B Cell Homeostasis and Antibody Responses. Nat Commun (2017) 8(1):1854. doi: 10.1038/s41467-017-01677-z
114. Oh H, Ghosh S. NF-κb: Roles and Regulation in Different CD4(+) T-Cell Subsets. Immunol Rev (2013) 252(1):41–51. doi: 10.1111/imr.12033
115. Yang WH, Park SY, Nam HW, Kim DH, Kang JG, Kang ES, et al. NFkappaB Activation Is Associated With Its O-GlcNAcylation State Under Hyperglycemic Conditions. Proc Natl Acad Sci USA (2008) 105(45):17345–50. doi: 10.1073/pnas.0806198105
116. Rodrigues Mantuano N, Stanczak MA, Oliveira IA, Kirchhammer N, Filardy AA, Monaco G, et al. Hyperglycemia Enhances Cancer Immune Evasion by Inducing Alternative Macrophage Polarization Through Increased O-GlcNAcylation. Cancer Immunol Res (2020) 8(10):1262–72. doi: 10.1158/2326-6066.CIR-19-0904
117. Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-Linked N-Acetylglucosaminyltransferase in Lymphocytes Activation. EMBO J (2007) 26(20):4368–79. doi: 10.1038/sj.emboj.7601845
118. Hayden MS, Ghosh S. Signaling to NF-Kappab. Genes Dev (2004) 18(18):2195–224. doi: 10.1101/gad.1228704
119. Ramakrishnan P, Clark PM, Mason DE, Peters EC, Hsieh-Wilson LC, Baltimore D. Activation of the Transcriptional Function of the NF-κb Protein C-Rel by O-GlcNAc Glycosylation. Sci Signal (2013) 6(290):ra75. doi: 10.1126/scisignal.2004097
120. Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-Kappa B P65 by PKA Stimulates Transcriptional Activity by Promoting a Novel Bivalent Interaction With the Coactivator CBP/P300. Mol Cell (1998) 1(5):661–71. doi: 10.1016/S1097-2765(00)80066-0
121. Ma Z, Chalkley RJ, Vosseller K. Hyper-O-GlcNAcylation Activates Nuclear Factor κ-Light-Chain-Enhancer of Activated B Cells (NF-κb) Signaling Through Interplay With Phosphorylation and Acetylation. J Biol Chem (2017) 292(22):9150–63. doi: 10.1074/jbc.M116.766568
122. Nathan C. Role of iNOS in Human Host Defense. Science (2006) 312(5782):1874–5. doi: 10.1126/science.312.5782.1874b
123. Hwang SY, Hwang JS, Kim SY, Han IO. O-GlcNAc Transferase Inhibits LPS-Mediated Expression of Inducible Nitric Oxide Synthase Through an Increased Interaction With Msin3a in RAW264.7 Cells. Am J Physiol Cell Physiol (2013) 305(6):C601–8. doi: 10.1152/ajpcell.00042.2013
124. Hwang JS, Kim KH, Park J, Kim SM, Cho H, Lee Y, et al. Glucosamine Improves Survival in a Mouse Model of Sepsis and Attenuates Sepsis-Induced Lung Injury and Inflammation. J Biol Chem (2019) 294(2):608–22. doi: 10.1074/jbc.RA118.004638
125. Yao AY, Tang HY, Wang Y, Feng MF, Zhou RL. Inhibition of the Activating Signals in NK92 Cells by Recombinant GST-sHLA-G1a Chain. Cell Res (2004) 14(2):155–60. doi: 10.1038/sj.cr.7290215
126. Madsen-Bouterse SA, Xu Y, Petty HR, Romero R. Quantification of O-GlcNAc Protein Modification in Neutrophils by Flow Cytometry. Cytometry A (2008) 73(7):667–72. doi: 10.1002/cyto.a.20569
127. Cicchetti G, Allen PG, Glogauer M. Chemotactic Signaling Pathways in Neutrophils: From Receptor to Actin Assembly. Crit Rev Oral Biol Med (2002) 13(3):220–8. doi: 10.1177/154411130201300302
128. Božič J, Stoka V, Dolenc I. Glucosamine Prevents Polarization of Cytotoxic Granules in NK-92 Cells by Disturbing FOXO1/ERK/paxillin Phosphorylation. PloS One (2018) 13(7):e0200757. doi: 10.1371/journal.pone.0200757
129. Ma Z, Vosseller K. Cancer Metabolism and Elevated O-GlcNAc in Oncogenic Signaling. J Biol Chem (2014) 289(50):34457–65. doi: 10.1074/jbc.R114.577718
130. Shi Y, Tomic J, Wen F, Shaha S, Bahlo A, Harrison R, et al. Aberrant O-GlcNAcylation Characterizes Chronic Lymphocytic Leukemia. Leukemia (2010) 24(9):1588–98. doi: 10.1038/leu.2010.152
131. Freund P, Kerenyi MA, Hager M, Wagner T, Wingelhofer B, Pham HTT, et al. O-GlcNAcylation of STAT5 Controls Tyrosine Phosphorylation and Oncogenic Transcription in STAT5-Dependent Malignancies. Leukemia (2017) 31(10):2132–42. doi: 10.1038/leu.2017.4
132. Ding N, Ping L, Shi Y, Feng L, Zheng X, Song Y, et al. Thiamet-G-Mediated Inhibition of O-GlcNAcase Sensitizes Human Leukemia Cells to Microtubule-Stabilizing Agent Paclitaxel. Biochem Biophys Res Commun (2014) 453(3):392–7. doi: 10.1016/j.bbrc.2014.09.097
133. Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E, et al. Hexosamine Pathway Metabolites Enhance Protein Quality Control and Prolong Life. Cell (2014) 156(6):1167–78. doi: 10.1016/j.cell.2014.01.061
134. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory Therapy With Canakinumab for Atherosclerotic Disease. N Engl J Med (2017) 377(12):1119–31. doi: 10.1056/NEJMoa1707914
135. Wang Q, Fang P, He R, Li M, Yu H, Zhou L, et al. O-GlcNAc Transferase Promotes Influenza A Virus-Induced Cytokine Storm by Targeting Interferon Regulatory Factor-5. Sci Adv (2020) 6(16):eaaz7086. doi: 10.1126/sciadv.aaz7086
136. Liu B, Salgado OC, Singh S, Hippen KL, Maynard JC, Burlingame AL, et al. The Lineage Stability and Suppressive Program of Regulatory T Cells Require Protein O-GlcNAcylation. Nat Commun (2019) 10(1):354. doi: 10.1038/s41467-019-08300-3
Keywords: GlcNAc, immune system, post translational modification, protein-protein interactions, glycobiology, nutrient sensing, cell signaling, lymphocyte activation
Citation: Mannino MP and Hart GW (2022) The Beginner’s Guide to O-GlcNAc: From Nutrient Sensitive Pathway Regulation to Its Impact on the Immune System. Front. Immunol. 13:828648. doi: 10.3389/fimmu.2022.828648
Received: 03 December 2021; Accepted: 05 January 2022;
Published: 31 January 2022.
Edited by:
Junji Xing, Houston Methodist Research Institute, United StatesReviewed by:
Willayat Yousuf Wani, Northwestern University, United StatesChad Slawson, University of Kansas Medical Center Research Institute, United States
Copyright © 2022 Mannino and Hart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerald W. Hart, R2VyYWxkLkhhcnRAdWdhLmVkdQ==
 Michael P. Mannino
Michael P. Mannino Gerald W. Hart
Gerald W. Hart