
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 14 February 2022
Sec. Alloimmunity and Transplantation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.826074
This article is part of the Research Topic The Biological and Clinical Aspects of HLA-G, Volume II View all 16 articles
The human leukocyte antigen (HLA)-G is a non-classical HLA class I molecule, which has distinct features to classical HLA-A, -B, -C antigens, such as a low polymorphism, different splice variants, highly restricted, tightly regulated expression and immune modulatory properties. HLA-G expression in tumor cells and virus-infected cells, as well as the release of soluble HLA-G leads to escape from host immune surveillance. Increased knowledge of the link between HLA-G expression, viral infection and disease progression is urgently required, which highlights the possible use of HLA-G as novel diagnostic and prognostic biomarker for viral infections, but also as therapeutic target. Therefore, this review aims to summarize the expression, regulation, function and impact of HLA-G in the context of different viral infections including virus-associated cancers. The characterization of HLA-G-driven immune escape mechanisms involved in the interactions between host cells and viruses might result in the design of novel immunotherapeutic strategies targeting HLA-G and/or its interaction with its receptors on immune effector cells.
Accumulating evidence exists that immune suppressive mechanisms play a critical role in promoting viral infections by either suppressing the capacity of infected host cells to overcome viral infection or by preventing the elimination of virus-transformed cells by immune effector cells. A common mechanism to escape immune surveillance is a loss or downregulation of classical HLA class Ia antigens and the neoexpression of non-classical HLA class Ib antigens, such as HLA-E, -F and –G (1–3). While the expression of HLA class Ia antigens leads to a T cell-mediated control of host immune responses mediated by antigen presentation and recognition, neoexpression of HLA-G has immune modulatory properties by inhibiting both innate and adaptive immune responses thereby leading to an immune escape of virus-infected cells. Although diverse viruses exploit HLA-G to establish persistent infections, the underlying molecular mechanisms tremendously differ. In this review, the expression, regulation, and clinical impact of HLA-G neoexpression in the context of viral infections as well as the underlying molecular mechanisms will be summarized.
The non-classical HLA-G gene is located on the short arm of chromosome 6 and consists of 8 exons, 7 introns, a 5’ upstream regulatory region (URR) and a 3’ untranslated region (UTR). The primary HLA-G transcript is alternatively spliced into at least 7 mRNAs, which encode 4 membrane-bound (HLA-G1, -G2, -G3, -G4) and 3 soluble (sHLA-G; HLA-G5, -G6, -G7) protein isoforms (4). Each HLA-G isoform contains 1-3 extracellular domains (α1, α2, α3), which are encoded by exon 2, 3 and 4; exons 5 and 6 encode the transmembrane and cytoplasmic domain of the heavy chain (HC), while the presence of intronic sequences are highly variable (5). The HLA-G1 and sHLA-G5 isoforms have a comparable structure to classical HLA class Ia antigens (HLA-A, -B, -C) and contain the HLA class I HC, which is non-covalently linked to β2-microglobulin (β2-m) (6–9). Furthermore, a peptide is bound in the cleft of the α1 and α2 domains, while the α3 domain of both the membranous and the sHLA-G is bound by the CD8 co-receptor (10). Similar to the classical HLA class Ia molecules, HLA-G is capable of presenting a broad repertoire of peptides and requires peptide binding to be efficiently presented on the cell surface (11–13). The other HLA-G isoforms lack one or two extracellular domains, are smaller and not associated with β2-m. HLA-G1 to HLA-G4 are membrane-bound and have a transmembrane region. In contrast, HLA-G5 and HLA-G6 are soluble isoforms with the presence of intron 4, which contains a premature stop codon thereby preventing the translation of the transmembrane and cytoplasmic residue. HLA-G7 is also a soluble isoform due to the presence of intron 2 containing a pre-mature stop codon. Recently, a number of novel HLA-G isoforms have been identified, which could be generated by skipping exon 6 and by a deletion of the α1 domain (8, 14). It should be noted that sHLA-G1 could be generated by a metalloproteinase-dependent shedding (15). Furthermore, membrane-bound HLA-G can also circulate within the blood stream, in particular in form of extracellular vesicles (10).
HLA-G expression is generally restricted to immune privileged tissues, such as cytotrophoblasts, cornea and pancreatic islets (16–18). However, HLA-G neoexpression was detected under pathophysiological conditions, such as cancers, inflammatory diseases, auto-immune diseases and pathogen infections including viruses (19). It is known that HLA-G has immune suppressive properties by interacting with different inhibitory receptors, namely ILT2/LILRB1, ILT4/LILRB2 and KIR2DL4, which are expressed on various immune cell subpopulations (20–22). Other receptors have been recently discovered, which are able to bind HLA-G, in particular NKG2A (23), CD160 (24) and CD8 (25) thereby interacting with the α3 domain of the respective HLA-G isoforms. The structures of the HLA-G isoforms, their receptors including their HLA-G-binding domains and the cell types expressing these receptors are summarized in Figure 1. The interaction of these receptors with HLA-G leads to the inhibition of the cytolytic function of natural killer (NK) cells and CD8+ cytotoxic T lymphocytes (CTLs), macrophage-mediated cytotoxicity, allo-proliferative response of CD4+ T cells and of the maturation as well as function of dendritic cells (DCs).
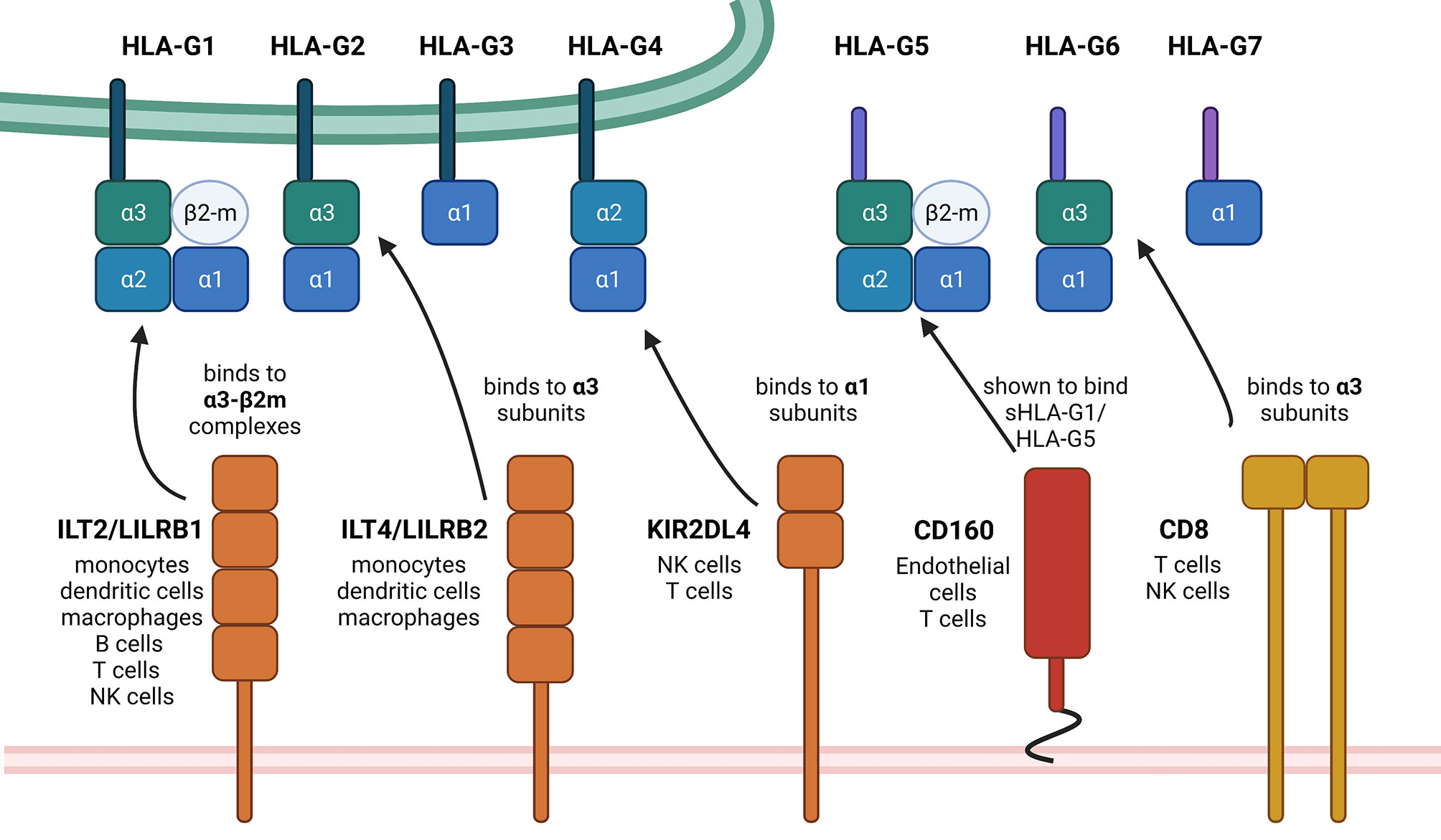
Figure 1 Summary of HLA-G isoforms with their binding positions for the so far known receptors. This schematic summary of the structural differences of HLA-G isoforms shows the so far known HLA-G receptors and also designates the exact binding position. The black arrows show exemplarily, but not exclusively possible interactions, depending on mentioned binding positions on the HLA-G ligands. There exist first evidence of a possible interaction between NKG2A with HLA-G, which still needs to be investigated in more detail. Therefore, NKG2A is not yet mentioned within this scheme. Created with BioRender.com.
However, it needs to be underlined that the interactions of HLA-G with immune effector cells are even much more diverse and complex as summarized so far, since also indirect immune modulatory effects of HLA-G on immune effector cells are known. Recently, it could be demonstrated that the non-classical HLA-E molecule can present peptides derived from HLA class Ia, but also from HLA-G leader sequences (26–30). HLA-E is a ligand for the inhibitory CD94/NKG2A,-B receptors and for the activating CD94/NKG2C receptor expressed on NK cells (31). This indirect mechanism modulates the immune effector mechanisms depending on the context of the engaged receptors. For example, the HLA-G-derived nonamer VMAPRTLFL presented on HLA-E molecules caused an enhanced NK cell-mediated lysis in an in vitro experiment of transfected 721.221 cells naturally lacking HLA class Ia/b molecules thereby representing valuable targets for NK cells. However, it needs to be addressed in much more detail whether this effect also plays a role in vivo, especially in the context of sHLA-G molecules within the TME of solid tumors inhibiting immune effector cells even prior to tumor infiltration.
In addition, immune suppressive cells are stimulated to secrete cytokines, like transforming growth factor (TGF)-β and interleukin (IL-10), which are able to increase HLA-G expression (32–35). Furthermore, the interaction of HLA-G with its receptors could lead to long-term immune modulatory effects by inducing and/or accumulating regulatory T cells (Tregs) (36), mesenchymal stem cells (MSCs) (37, 38) and myeloid-derived suppressor cells (MDSCs) (39, 40). In addition to the direct interaction of HLA-G with its appropriate receptors, the HLA-G-mediated immune suppression could be also caused by intercellular transfer mechanisms, such as trogocytosis, exosomes or tunneling nanotubes, which also leads to escape from the destruction by the host immune system.
So far, more than 88 different HLA-G alleles (41) have been discovered. Within its coding region, HLA-G shows a limited protein variability compared to classical HLA class Ia, but both the 5’ URR and the 3’ UTR contain a multitude of polymorphic sites affecting gene regulation (42). The major HLA-G polymorphisms associated with viral infections and their role in virus susceptibility are summarized in Table 1. Response elements for diverse transcriptional inducers have been identified, like the progesterone response elements (PRE), heat shock elements (HSE), interferon-stimulated response elements (ISRE) and elements responding to cyclic AMP (CRE) (50–54). In addition, negative regulators have been identified like the ras-responsive elements (RRE) (55). Many polymorphisms and single nucleotide variants within the regions of such response elements have been described (56, 57). In vitro evaluations of the effects of the most frequent 5’ URR haplotypes have indeed shown differential transcriptional regulation of HLA-G (58, 59). In addition, to the 5’ UTR, diverse polymorphic sites exist also in the 3’ UTR of the HLA-G transcript (60); many of them have been shown to change the affinity of gene-targeted sequencing for transcriptional or post-transcriptional factors, affecting splicing (61), microRNA (miRNA) binding (62) and RNA turnover (63). A variation of major significance for HLA-G expression is a 14 bp insertion/deletion (ins/del, rs66554220) in the 3’ UTR, which is associated with the alternative splicing of HLA-G and miRNA stability. Furthermore, the region downstream of the 14 bp ins/del polymorphic site has been suggested to be a target for HLA-G-specific miRNAs thereby leading to a direct downregulation of HLA-G. However, there exist controversial data regarding the role of the HLA-G 14 bp ins/del polymorphism and susceptibility to viral infections (64). Of note, population studies suggest that HLA-G haplotypes are strongly shaped by selective pressure throughout evolution thereby preserving protein-coding sequences and enabling divergence in regulatory sequences (42, 65).
There exist more than 220 viral species able to infect humans (66) and annually even more human pathogenic viruses are identified or even emerge including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (67). In 2008, before the SARS-CoV-2 pandemic started, Woolhouse and co-authors generated in silico prediction tools for estimating the emergence of novel human pathogenic viruses concluding that the formation/discovery of novel human viruses must be anticipated in public health planning and the future should prove this statement (68).
Infection with intracellular pathogens, such as viruses, could lead to the presentation of pathogen-relevant peptides, e.g. via HLA class I molecules to T cells resulting in T cell activation and elimination of the infected host cell, although numerous other molecular mechanisms also contribute to anti-viral immune surveillance. These include the induction of the MICA/MICB molecules as well as the UL16-binding proteins (ULBP)1-6, which are physiologically not expressed, but induced by cellular stress and viral infections and act as ligands for the activating NKG2D receptor present on NK cells and CTLs (69). In addition, intracellular, membranous and even secreted pattern recognition receptors exert a strong role in anti-viral immune surveillance. A prerequisite for appropriate immune effector cell functions is the recruitment of these immune effector cells to the virus-infected cells, a process, which involves a functional chemokine signaling. Indeed, the authors recently reviewed the distinct viral mechanisms targeting the above mentioned strategies involved in immune recognition and elimination of viral infections (70).
However, several human pathogenic viruses can successfully develop immune evasion by various molecular mechanisms, including the induced expression of immune checkpoints, like PD-L1 and HLA-G, in the virus-infected host cells (1, 71–73). In this review, we will summarize known examples of human viruses able to induce immunological tolerance towards host immune effector cells by HLA-G induction. Therefore, an overview of the tightly regulated HLA-G expression will be provided.
HLA-G is predominantly expressed in immune-privileged tissues, in particular placental and eye tissues (17, 74), but also insulin- and glucagon-positive cells within the endocrine islets of the pancreas (18) and the medullary thymic epithelial cells (75) and locally induce immunological tolerance thereby preventing tissue damages due to inflammatory reactions. The HLA-G gene expression itself is regulated at multiple levels, including the grade of promoter methylation (76), the acetylation of histones (77), the existence of transcriptional activators (CREB1, AIRE) (78, 79) as well as the lack of transcriptional repressors (RREB-1, LINE1) (55, 80). In addition to the transcriptional control, several factors contribute to a complex posttranscriptional gene regulation of HLA-G including many miRNAs, such as miR-148 family members with the highest affinity to the 3’ UTR of the HLA-G transcript and miRNA directed against the coding sequence (CDS) of HLA-G (81, 82) as well as long non-coding RNAs like HOTAIR (83). Furthermore, RNA-binding proteins (RBPs), like e.g. HNRNPR, have been reported with regulatory potential for the HLA-G mRNA (84).
Several cytokines, but also certain stress stimuli are known to enhance the HLA-G levels, like interferon (IFN)-γ, IL-10, TGF-β (76), hypoxia and heat stress (52, 85).
This complex regulatory network for the control of the HLA-G expression can be altered to induce or enhance the immune evasion of tumor cells, but also of infectious pathogens, e.g. viruses, bacteria and parasites (86). It is noteworthy that a number of viruses have an oncogenic potential and independently of viral infections a pathophysiological HLA-G neoexpression, which can be detected with high frequencies in virus-associated malignancies as well as independently of viral infections in solid and hematopoietic tumor diseases (87–90).
Human papillomavirus (HPV) infection is a common infection and has been linked to epithelial cancers including in particular cervical and head and neck cancers (91–93). HLA-G has an impact on the clinical course of persistent HPV infections, epithelial cell transformation, tumor growth, metastasis, formation and therapy resistance. In HPV-associated tumors like cervical cancer and head and neck squamous cell carcinoma (HNSCC), a role of HLA-G in HPV infections and in the initiation and progression has been described (94, 95) mediated by polymorphisms, methylation and deregulation of HLA-G (44).
HLA-G polymorphisms are genetic susceptibility and/or disease-relevant factors for cervical HPV infections and viral persistence of cervical cancers (Table 1). Most of the studies focused on the polymorphisms in the 3’ UTR of the HLA-G gene rather than on its promoter region or in its CDS. The HLA-G +14/G and +3142G alleles are risk factors for HPV infections and increase the risk of high-grade cervical lesions and are associated with cervical carcinogenesis (43). Interestingly, the HLA-G promoter methylation was not associated with the HPV infection status (96). Moreover, a codon 130C deletion was associated with an increased progression and reduced overall survival of patients with nasopharyngeal carcinoma (NPC) (97).
In comparison to HPV-negative HNSCC, higher HLA-G expression levels were found in HPV-positive HNSCC with HLA-G7 as the most frequent isoform (98). In addition to various HLA-G polymorphisms, which have been associated with the susceptibility of HPV infection, an influence of HLA-G on the immune modulation of HPV-positive HNSCC has been described. Furthermore, high levels of IFN-γ, but lower levels of IL-10, TGF-β, SOCS1/3 and programmed death receptor 1 (PD1) were found in HPV-positive HNSCC (98).
Infections with hepatitis B (HBV) and C virus (HCV) are major health threats worldwide (99, 100). Chronic HBV/HCV infection is followed by chronic hepatitis, which might lead to liver cirrhosis and hepatocellular carcinoma (HCC) (101). Despite the host immune response is crucial for the control of HBV and HCV, both viruses have developed different strategies to escape immune surveillance including HLA-G neoexpression (102). Indeed, a high frequency of HLA-G expression was found in tissues of HCV-infected patients. In addition, elevated sHLA-G serum levels were detected in patients with chronic HCV infection (103, 104). Furthermore, HLA-G expression was more prominent in high fibrosis specimens. In association with the increased HLA-G expression in liver, an enhanced inflammatory activity and liver fibrosis were demonstrated, which might have implications for progression and prognosis of liver diseases. HLA-G expression in the liver upon chronic HCV infection might be due to the composition of the inflammatory infiltrate, in particular of mast cells known to induce fibrosis by stimulating the proliferation of TGF-β-secreting hepatic stellate cells as demonstrated by Amiot and co-authors (105). In addition, hepatic stellate cells also secrete IL-10, which is able to upregulate HLA-G expression. Thus, the liver microenvironment including cytokines affecting the inflammatory response and fibrosis, such as IL-10 and TGF-β, are responsible for the induction of liver HLA-G expression in chronic hepatitis-diseased patients (104).
As described for HCV-infected tissues, HLA-G is expressed at a high frequency in HBV-infected liver biopsies, but not in control hepatocytes (106). Furthermore, sHLA-G levels were also significantly higher in HBV-infected patients when compared to healthy controls (15, 107, 108). The HBV-mediated induction of HLA-G expression in hepatocytes might be caused by modulation of the hepatocyte expression of miRs, such as downregulation of the HLA-G-regulating miR-152 representing the most HLA-G mRNA affine member of the miR-148 family leading to neoexpression of HLA-G (109). These data further suggest that HLA-G and HLA-G-specific miRs are involved in HBV-induced HCC.
Human cytomegalovirus (HCMV) causes a life-long human infection, which may be life-threatening for immune-suppressed patients (110, 111). Like other viruses, HCMV has developed different strategies to escape immune surveillance including the modulation of HLA-G expression (112). Membrane-bound HLA-G was shown to be expressed in macrophages and monocytes undergoing lytic infection, while sHLA-G levels are increased in serum of patients with acute HCMV infection (113–115). Furthermore, an enhanced HLA-G expression has been associated with allograft tolerance after kidney transplantation (116). The single nucleotide polymorphism (SNP) (3142C>G) in the HLA-G gene of the recipient, but not in the transplant donor was associated with a higher susceptibility to HCMV infection after kidney transplantation. In addition, sHLA-G levels were associated with a higher susceptibility to HCMV infection (Table 1). These data suggested that both the recipient HLA-G+3142CC phenotype and sHLA-G levels represent predictive risk markers for HCMV infection (45). Recently, an association between other HLA-G 3’ UTR variants and kidney graft outcomes has been reported. In recipients with stable allograft function, significantly higher sHLA-G levels were found in patients who were +3010GG, +3142CC, +3187GG and +3196CC carriers in comparison to acute rejection recipients (117). Thus, there exists a direct association between this HLA-G 3’ UTR variants and sHLA-G levels in kidney recipients leading to graft acceptance. Therefore, monitoring of sHLA-G levels prior to transplantation might serve as suitable marker to predict kidney graft outcome. This was confirmed by a recent study demonstrating that the acute and chronic rejection rate of kidneys increased 1.06 times and 1.14 times, respectively, in kidney transplant recipients with low serum sHLA-G levels. The frequency of acute rejection was lower in patients with a 14 bp del/del polymorphism than that of ins/ins and ins/del polymorphisms. Based on these results, the HLA-G 3’ UTR polymorphism and the sHLA-G levels might represent useful markers for the prediction of rejection in kidney transplants (118).
Human herpesvirus (HHV)-6 is a β-herpesvirus comprising of the two viruses HHV-6A and HHV-6B (119–121) that cause both productive and life-long latent infections (122). HHV-6A/B induce HLA-G expression in mesothelial cells leading to impaired NK cell functions against virus-infected cells (123). HHV-6A/B express the viral protein U94, which has key functions in the viral life cycle and elicits immune responses. Furthermore, U94 has been shown to induce HLA-G expression by upregulating the expression of the transcription factor ATF3, which activates HLA-G expression and release (124). In line with these findings, patients suffering from systemic sclerosis showed elevated sHLA-G levels when being co-infected with HHV-6A. Furthermore, viral load correlated to NK cell dysfunction and disease severity (125).
Another member of the HHVs, the Epstein-Barr virus (EBV), has also been reported to induce the HLA-G expression by yet undefined molecular mechanisms (126). It could be speculated that the EBV-encoded viral IL-10 (vIL10), a known agonist of the human IL-10, might be an inducer of the HLA-G expression. Interestingly, EBV infections also affect the epigenetic control within the host cell genome, including altered DNA methylation patterns (127) as well as aberrant histone modifications (128), both mechanisms known as important regulators of HLA-G gene expression. It is noteworthy that EBV infections can be linked to various tumor entities, such as NPC, gastric adenocarcinoma (GC), classical Hodgkin lymphoma (cHL) and Burkitt lymphoma (BL) (129), which have been shown to exhibit pathophysiological HLA-G neoexpression.
The human immunodeficiency virus (HIV) is a RNA virus. Several studies suggested elevated sHLA-G in serum samples of untreated HIV patients, but membrane-bound HLA-G is also detected on immune cells of patients. Furthermore, HIV infection is not only inducing HLA-G on monocytes, macrophages and T lymphocytes (130), but also causes an increased HLA-E expression in infected CD4+ T helper cells (131), which exerts immune modulatory functions comparable to that of HLA-G thereby contributing to immune evasion of HIV-infected cells (131). HLA-E is the ligand for the inhibitory NK cell receptors CD94/NKG2A, -B and for the activating -C expressed on NK cells and CTL (132). HIV infection also induces the expression of the immune modulatory ligands PD-L1 and PD-L2 on macrophages (133), which interact with their receptor PD-1. The PD-L1/PD-1 interaction causes an inhibition of CTL activity, but also an increased IL-10 secretion, which contributes to immune evasion of HIV-infected CD4+ T helper cells (134).
The HIV-related induction of HLA-G expression has been currently well characterized. Certain HLA-G alleles increase the risk for heterosexual transmission of HIV in African women. Indeed, the HLA-G*010108 allele was associated with a 2.5-fold increased risk of HIV-1 infection and the two HLA-G*010108 alleles containing genotypes HLA-G*010108/010401 and G*010101/010108 were associated with an elevated risk of HIV-1 infection (Table 1) (46). Furthermore, children carrying the homozygous HLA-G genotype with the 14 bp deletion within the HLA-G 3’-UTR exhibited significantly reduced rates of perinatal HIV transmission (47, 135).
The overexpression of HLA-G in HIV-infected individuals may be secondary due to an increased release of IL-10. The HIV-encoded protein gp41 has been identified to induce IL-10 secretion in monocytes (136) thereby revealing a possible mechanism for the occurrence of HLA-G.
In addition, HIV infection causes a downregulation of the classical HLA class Ia molecules HLA-A and HLA-B (137), a known mechanism for (tumor) immune evasion, clearly demonstrating a deregulation of appropriate HLA class Ia/b signaling upon HIV infection.
The human T cell lymphotropic virus type 1 (HTLV-1) known to induce very aggressive adult T-cell lymphoma (ATL) also upregulates HLA-G expression, mainly HLA-G1 and HLA-G5 (138), and in analogy to HIV, the homozygous HLA-G genotype −14-bp/−14-bp genotype has a higher proviral load than the +14-bp/−14-bp and +14-bp/+14-bp genotypes (Table 1) (48). However, a detailed mechanism for the HTLV-1-mediated HLA-G increase has not yet been identified, but an IL-10-based mechanism analogous to the HIV-dependent HLA-G induction might be likely.
Influenza A virus (IAV) causes acute respiratory infections (139). Infections with IAV are known to induce HLA-G mRNA and protein expression in alveolar epithelial cells (140), which is a major inhibitory molecule of host immune responses to IAV infections. Although the detailed underlying mechanisms have not yet been determined, the study suggested an involvement of cytokines, in particular IFNs, which could cause the HLA-G neoexpression mediated by the viral encoded NS1 protein.
In 2019, a novel corona virus emerged that caused a worldwide pandemic and lead to the potentially life-threatening severe acute respiratory syndrome (SARS-CoV-2) (67). In patients with severe infection, high serum levels of sHLA-G were observed (141) and HLA-G was also found on the cell surface of diverse immune cells during the course of infection. HLA-G was detected on monocytes, T and B cells and showed a high-low-high pattern, probably correlating with infection, replication and clearance phase (142). However, these results were only compiled from a single patient and require further validation. Another clinical study found a correlation between high sHLA-G levels and improved disease outcome probably due to immune-dampening effects of HLA-G that suppress an excessive tissue-damage, possibly mediated by reduced neutrophil infiltration to the sites of infection (143). Disease outcome was not clearly linked to HLA-G variants despite a meta-analysis listed the HLA-G 3’UTR SNP rs9381042 as a candidate variant that was slightly overrepresented in a UK cohort of critically ill patients due to COVID-19 infection compared to the general population (Table 1) (49).
As summarized in this review, multiple viral infections are known to induce membranous HLA-G expression as well as its secretion in infected host cells. In some cases, the role of HLA-G in infections has been characterized to contribute to immune evasion of the infected host cells by inhibition of immune effector cells. So far, the detailed molecular mechanisms of this HLA-G neoexpression have not yet been identified. In many of these studies a correlation of the HLA-G neoexpression with elevated IL-10 levels is reported. Indeed, IL-10 is a known inducer of the HLA-G expression and has also been described in tumor cells independently of viral infections as inducer of HLA-G expression. Therefore, the very likely IL-10-based hypothesis of the virus-driven HLA-G induction is summarized in Figure 2.
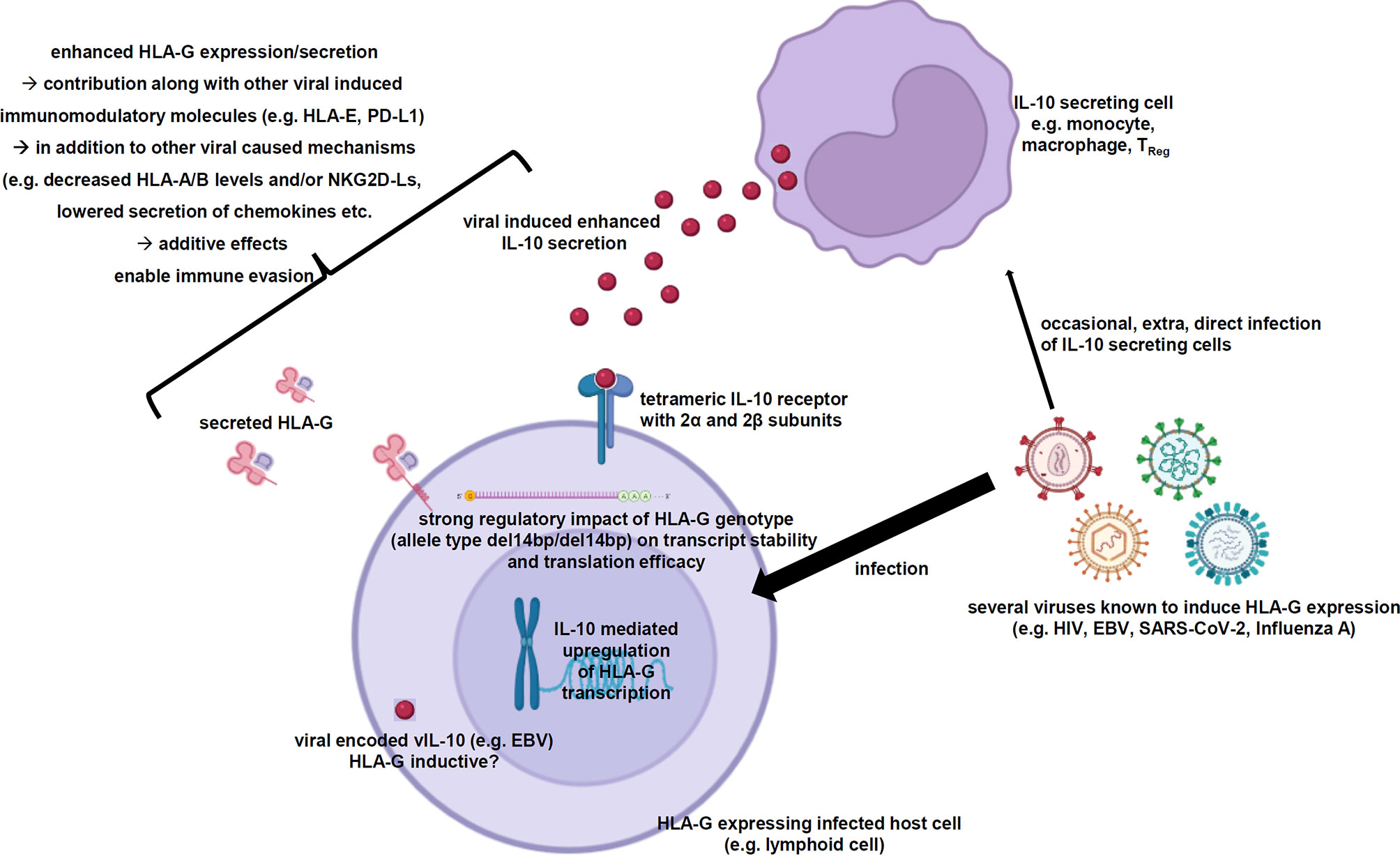
Figure 2 Scheme summarizing the virus-induced enhanced IL-10 secretion resulting in elevated HLA-G expression and secretion. The working hypothesis demonstrates the elevated IL-10 secretion of certain immune effector cells as consequence of viral infections, which is very likely the main driver of the increased HLA-G gene expression and secretion within the virally infected host cells strongly contributing to immune evasion. Created with BioRender.com.
Physiologically, IL-10 exerts its functions as an anti-inflammatory cytokine in preventing inflammatory and autoimmune pathologies by inhibition of certain immune effector cells thereby preventing inflammation-induced and autoimmune pathologies (144). Murine in vivo models inhibiting the IL-10R demonstrated that the viral infection with lymphocytic choriomeningitis virus resulted in a rapid resolution of the persistent infection. This blocked IL-10 signaling was also linked to an increased IFN-γ secretion by CD8+ T cells (145). Another murine study with IL-10-deficient mice could also demonstrate a better anti-viral T cell response against the lymphocytic choriomeningitis virus (146).
The predominant IL-10-expressing cells are Th2 cells and Tregs, but also macrophages, dendritic cells, eosinophils and neutrophils are known to secrete IL-10 (144). Some of these immune cell populations were directly infected by HLA-G-inducing viruses and subsequently shifted towards increased IL-10 secretion. In other cases, indirect effects, like e.g. an increased PD-L1/PD-1 signaling, caused an enhanced IL-10 secretion of these immune effector cells.
Once the HLA-G gene transcription has been induced upon IL-10 stimulation, the homozygous genotype del14bp/del14bp has additional beneficial effects due to an enhanced HLA-G mRNA stability by avoiding miRNA-based downregulation and thereby resulting in increased HLA-G protein levels.
It is also known that IL-10 stimulation affects the RNA expression profile in IL-10-stimulated cells (147), but whether HLA-G-regulating miRs are downregulated upon IL-10 signaling is so far unknown. In addition, further direct or indirect effects of IL-10 on other HLA-G-regulating factors are rather limited with the exception of the transcriptional activator CREB1, which induces the transcription of HLA-G and IL-10 (78, 148). Thus, the regulation of HLA-G expression upon viral infection is complex and further studies are urgently needed to gain deeper insights into the molecular mechanisms of the viral infection-driven HLA-G neoexpression.
SJ-B and BS designed the study. SJ-B, DS, OM, and BS wrote the manuscript. SJ-B and DS created the figures. All authors contributed to the article and approved the submitted version.
This work was funded by the German-Israeli Foundation (GIF; I-37-414.11-2016) and Dr. Werner Jackstädt Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
APC, antigen presenting cells; β2-m, β2-microglobulin; CDS, coding region, coding sequence; CTL, cytotoxic T lymphocyte; DC, dendritic cell; HC, heavy chain; HCMV, human cytomegalo virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; HHV, human herpesvirus; HLA, human leukocyte antigen; HNSCC, head and neck squamous cell carcinoma; HPV, human papilloma virus; HTLV-1, human I-lymphotropic virus type II; IAV, influenza A virus; IL, interleukin; MDSC, myeloid-derived suppressor cell; miRNA, microRNA; MSC, mesenchymal stem cells; NK, natural killer; NPC, nasopharyngeal carcinoma; PD1, programmed death receptor 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SNP, single nucleotide polymorphism; TGF-β, transforming growth factor β; Treg, regulatory T cell; ULBP, UL16-binding protein; URR, upstream regulatory region; UTR, untranslated region.
1. Rai KR, Shrestha P, Yang B, Chen Y, Liu S, Maarouf M, et al. Acute Infection of Viral Pathogens and Their Innate Immune Escape. Front Microbiol (2021) 12:672026. doi: 10.3389/fmicb.2021.672026
2. Monette A, Mouland AJ. T Lymphocytes as Measurable Targets of Protection and Vaccination Against Viral Disorders. Int Rev Cell Mol Biol (2019) 342:175–263. doi: 10.1016/bs.ircmb.2018.07.006
3. Finlay BB, McFadden G. Anti-Immunology: Evasion of the Host Immune System by Bacterial and Viral Pathogens. Cell (2006) 124(4):767–82. doi: 10.1016/j.cell.2006.01.034
4. Ishitani A, Geraghty DE. Alternative Splicing of HLA-G Transcripts Yields Proteins With Primary Structures Resembling Both Class I and Class II Antigens. Proc Natl Acad Sci USA (1992) 89(9):3947–51. doi: 10.1073/pnas.89.9.3947
5. Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: From Biology to Clinical Benefits. Trends Immunol (2008) 29(3):125–32. doi: 10.1016/j.it.2007.11.005
6. Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeu G, et al. Identification of HLA-G7 as a New Splice Variant of the HLA-G mRNA and Expression of Soluble HLA-G5, -G6, and -G7 Transcripts in Human Transfected Cells. Hum Immunol (2000) 61(11):1138–49. doi: 10.1016/S0198-8859(00)00197-X
7. da Silva Nardi F, König L, Wagner B, Giebel B, Santos Manvailer LF, Rebmann V, et al. Soluble Monomers, Dimers and HLA-G-Expressing Extracellular Vesicles: The Three Dimensions of Structural Complexity to Use HLA-G as a Clinical Biomarker. HLA (2016) 88(3):77–86. doi: 10.1111/tan.12844
8. Tronik-Le Roux D, Renard J, Vérine J, Renault V, Tubacher E, LeMaoult J, et al. Novel Landscape of HLA-G Isoforms Expressed in Clear Cell Renal Cell Carcinoma Patients. Mol Oncol (2017) 11(11):1561–78. doi: 10.1002/1878-0261.12119
9. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
10. Schwich E, Hò GGT, LeMaoult J, Bade-Döding C, Carosella ED, Horn PA, et al. Soluble HLA-G and HLA-G Bearing Extracellular Vesicles Affect ILT-2 Positive and ILT-2 Negative CD8 T Cells Complementary. Front Immunol (2020) 11:2046. doi: 10.3389/fimmu.2020.02046
11. Diehl M, Münz C, Keilholz W, Stevanović S, Holmes N, Loke YW, et al. Nonclassical HLA-G Molecules Are Classical Peptide Presenters. Curr Biol (1996) 6(3):305–14. doi: 10.1016/S0960-9822(02)00481-5
12. Munz C, Nickolaus P, Lammert E, Pascolo S, Stevanović S, Rammensee HG. The Role of Peptide Presentation in the Physiological Function of HLA-G. Semin Cancer Biol (1999) 9(1):47–54. doi: 10.1006/scbi.1998.0105
13. Di Marco M, Schuster H, Backert L, Ghosh M, Rammensee HG, Stevanović S. Unveiling the Peptide Motifs of HLA-C and HLA-G From Naturally Presented Peptides and Generation of Binding Prediction Matrices. J Immunol (2017) 199(8):2639–51. doi: 10.4049/jimmunol.1700938
14. Lin A, Zhang X, Zhang RL, Zhang JG, Zhou WJ, Yan WH. Clinical Significance of Potential Unidentified HLA-G Isoforms Without Alpha1 Domain But Containing Intron 4 in Colorectal Cancer Patients. Front Oncol (2018) 8:361. doi: 10.3389/fonc.2018.00361
15. Park GM, Lee S, Park B, Kim E, Shin J, Cho K, et al. Soluble HLA-G Generated by Proteolytic Shedding Inhibits NK-Mediated Cell Lysis. Biochem Biophys Res Commun (2004) 313(3):606–11. doi: 10.1016/j.bbrc.2003.11.153
16. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science (1990) 248(4952):220–3. doi: 10.1126/science.2326636
17. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in Human Cornea, an Immune-Privileged Tissue. Hum Immunol (2003) 64(11):1039–44. doi: 10.1016/j.humimm.2003.08.346
18. Cirulli V, Zalatan J, McMaster M, Prinsen R, Salomon DR, Ricordi C, et al. The Class I HLA Repertoire of Pancreatic Islets Comprises the Nonclassical Class Ib Antigen HLA-G. Diabetes (2006) 55(5):1214–22. doi: 10.2337/db05-0731
19. Gonzalez A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The Immunosuppressive Molecule HLA-G and Its Clinical Implications. Crit Rev Clin Lab Sci (2012) 49(3):63–84. doi: 10.3109/10408363.2012.677947
20. Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, et al. Human Inhibitory Receptors Ig-Like Transcript 2 (ILT2) and ILT4 Compete With CD8 for MHC Class I Binding and Bind Preferentially to HLA-G. Proc Natl Acad Sci USA (2003) 100(15):8856–61. doi: 10.1073/pnas.1431057100
21. Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G-Specific Receptor Expressed on All Natural Killer Cells. J Exp Med (1999) 189(7):1093–100. doi: 10.1084/jem.189.7.1093
22. Hofmeister V, Weiss EH. HLA-G Modulates Immune Responses by Diverse Receptor Interactions. Semin Cancer Biol (2003) 13(5):317–23. doi: 10.1016/S1044-579X(03)00022-1
23. Ho GT, Celik AA, Huyton T, Hiemisch W, Blasczyk R, Simper GS, et al. NKG2A/CD94 Is a New Immune Receptor for HLA-G and Distinguishes Amino Acid Differences in the HLA-G Heavy Chain. Int J Mol Sci (2020) 21(12):4362. doi: 10.3390/ijms21124362
24. Fons P, Chabot S, Cartwright JE, Lenfant F, L'Faqihi F, Giustiniani J, et al. Soluble HLA-G1 Inhibits Angiogenesis Through an Apoptotic Pathway and by Direct Binding to CD160 Receptor Expressed by Endothelial Cells. Blood (2006) 108(8):2608–15. doi: 10.1182/blood-2005-12-019919
25. Gao GF, Willcox BE, Wyer JR, Boulter JM, O'Callaghan CA, Maenaka K, et al. Classical and Nonclassical Class I Major Histocompatibility Complex Molecules Exhibit Subtle Conformational Differences That Affect Binding to CD8alphaalpha. J Biol Chem (2000) 275(20):15232–8. doi: 10.1074/jbc.275.20.15232
26. Rolle A, Meyer M, Calderazzo S, Jäger D, Momburg F. Distinct HLA-E Peptide Complexes Modify Antibody-Driven Effector Functions of Adaptive NK Cells. Cell Rep (2018) 24(8):1967–76.e4. doi: 10.1016/j.celrep.2018.07.069
27. Rolle A, Jager D, Momburg F. HLA-E Peptide Repertoire and Dimorphism-Centerpieces in the Adaptive NK Cell Puzzle? Front Immunol (2018) 9:2410. doi: 10.3389/fimmu.2018.02410
28. Llano M, Lee N, Navarro F, García P, Albar JP, Geraghty DE, et al. HLA-E-Bound Peptides Influence Recognition by Inhibitory and Triggering CD94/NKG2 Receptors: Preferential Response to an HLA-G-Derived Nonamer. Eur J Immunol (1998) 28(9):2854–63. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W
29. Vales-Gomez M, Reyburn HT, Erskine RA, López-Botet M, Strominger JL. Kinetics and Peptide Dependency of the Binding of the Inhibitory NK Receptor CD94/NKG2-A and the Activating Receptor CD94/NKG2-C to HLA-E. EMBO J (1999) 18(15):4250–60. doi: 10.1093/emboj/18.15.4250
30. Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of Human Histocompatibility Leukocyte Antigen (HLA)-E Complexed With HLA Class I Signal Sequence-Derived Peptides by CD94/NKG2 Confers Protection From Natural Killer Cell-Mediated Lysis. J Exp Med (1998) 187(5):813–8. doi: 10.1084/jem.187.5.813
31. Braud VM, Allan DS, O'Callaghan CA, Söderström K, D'Andrea A, Ogg GS, et al. HLA-E Binds to Natural Killer Cell Receptors CD94/NKG2A, B and C. Nature (1998) 391(6669):795–9. doi: 10.1038/35869
32. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and Posttranscriptional Regulations of the HLA-G Gene. J Immunol Res (2014) 2014:734068. doi: 10.1155/2014/734068
33. Dupin C, Lhuillier E, Létuvé S, Pretolani M, Thabut G, Mal H, et al. Inhibition of T Cell Alloreactivity by Bronchial Epithelium Is Impaired in Lung Transplant Recipients, Through Pathways Involving TGF-Beta, IL-10 and HLA-G. Transplantation (2017) 101(9):2192–9. doi: 10.1097/TP.0000000000001553
34. Gros F, Cabillic F, Toutirais O, Le Maux A, Sebti Y, Amiot L. Soluble HLA-G Molecules Impair Natural Killer/Dendritic Cell Crosstalk via Inhibition of Dendritic Cells. Eur J Immunol (2008) 38(3):742–9. doi: 10.1002/eji.200736918
35. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Modulation of Dendritic Cell Differentiation by HLA-G and ILT4 Requires the IL-6–STAT3 Signaling Pathway. Proc Natl Acad Sci USA (2008) 105(24):8357–62. doi: 10.1073/pnas.0803341105
36. Al-Khunaizi NR, Tabbara KS, Farid EM. Is There a Role for HLA-G in the Induction of Regulatory T Cells During the Maintenance of a Healthy Pregnancy? Am J Reprod Immunol (2020) 84(2):e13259. doi: 10.1111/aji.13259
37. Selmani Z, Naji A, Gaiffe E, Obert L, Tiberghien P, Rouas-Freiss N, et al. HLA-G is a Crucial Immunosuppressive Molecule Secreted by Adult Human Mesenchymal Stem Cells. Transplantation (2009) 87(9 Suppl):S62–6. doi: 10.1097/TP.0b013e3181a2a4b3
38. Nasef A, Mathieu N, Chapel A, Frick J, François S, Mazurier C, et al. Immunosuppressive Effects of Mesenchymal Stem Cells: Involvement of HLA-G. Transplantation (2007) 84(2):231–7. doi: 10.1097/01.tp.0000267918.07906.08
39. Kostlin N, Ostermeir A, Spring B, Schwarz J, Marmé A, Walter CB, et al. HLA-G Promotes Myeloid-Derived Suppressor Cell Accumulation and Suppressive Activity During Human Pregnancy Through Engagement of the Receptor ILT4. Eur J Immunol (2017) 47(2):374–84. doi: 10.1002/eji.201646564
40. Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in Tumor Escape Through Expansion of Myeloid-Derived Suppressor Cells and Cytokinic Balance in Favor of Th2 Versus Th1/Th17. Blood (2011) 117(26):7021–31. doi: 10.1182/blood-2010-07-294389
41. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA Database: Allele Variant Databases. Nucleic Acids Res (2015) 43(Database issue):D423–31. doi: 10.1093/nar/gku1161
42. Castelli EC, Ramalho J, Porto IOP, Lima THA, Felício LP, Sabbagh A, et al. Insights Into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476
43. Xu HH, Shi WW, Lin A, Yan WH. HLA-G 3' Untranslated Region Polymorphisms Influence the Susceptibility for Human Papillomavirus Infection. Tissue Antigens (2014) 84(2):216–22. doi: 10.1111/tan.12359
44. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the Polymorphism of HLA-G on its Function, Regulation, Evolution and Disease Association. Cell Mol Life Sci (2011) 68(3):369–95. doi: 10.1007/s00018-010-0580-7
45. Guberina H, Michita RT, Dolff S, Bienholz A, Trilling M, Heinemann FM, et al. Recipient HLA-G +3142 CC Genotype and Concentrations of Soluble HLA-G Impact on Occurrence of CMV Infection After Living-Donor Kidney Transplantation. Int J Mol Sci (2017) 18(11):2338. doi: 10.3390/ijms18112338
46. Matte C, Lajoie J, Lacaille J, Zijenah LS, Ward BJ, Roger M. Functionally Active HLA-G Polymorphisms are Associated With the Risk of Heterosexual HIV-1 Infection in African Women. AIDS (2004) 18(3):427–31. doi: 10.1097/00002030-200402200-00008
47. Fabris A, Catamo E, Segat L, Morgutti M, Arraes LC, de Lima-Filho JL, et al. Association Between HLA-G 3'UTR 14-Bp Polymorphism and HIV Vertical Transmission in Brazilian Children. AIDS (2009) 23(2):177–82. doi: 10.1097/QAD.0b013e32832027bf
48. Haddad R, Alves DCC, Rocha-Junior MC, Azevedo R, do Socorro Pombo-de-Oliveira M, Takayanagui OM, et al. HLA-G 14-Bp Insertion/Deletion Polymorphism is a Risk Factor for HTLV-1 Infection. AIDS Res Hum Retroviruses (2011) 27(3):283–8. doi: 10.1089/aid.2010.0165
49. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature (2021) 591(7848):92–8.
50. Yie SM, Li LH, Li GM, Xiao R, Librach CL. Progesterone Enhances HLA-G Gene Expression in JEG-3 Choriocarcinoma Cells and Human Cytotrophoblasts In Vitro. Hum Reprod (2006) 21(1):46–51. doi: 10.1093/humrep/dei305
51. Yie SM, Xiao R, Librach CL. Progesterone Regulates HLA-G Gene Expression Through a Novel Progesterone Response Element. Hum Reprod (2006) 21(10):2538–44. doi: 10.1093/humrep/del126
52. Ibrahim EC, Morange M, Dausset J, Carosella ED, Paul P. Heat Shock and Arsenite Induce Expression of the Nonclassical Class I Histocompatibility HLA-G Gene in Tumor Cell Lines. Cell Stress Chaperones (2000) 5(3):207–18. doi: 10.1379/1466-1268(2000)005<0207:HSAAIE>2.0.CO;2
53. Chu W, Gao J, Murphy WJ, Hunt JS. A Candidate Interferon-Gamma Activated Site (GAS Element) in the HLA-G Promoter Does Not Bind Nuclear Proteins. Hum Immunol (1999) 60(11):1113–8. doi: 10.1016/S0198-8859(99)00091-9
54. Gobin SJ, Biesta P, de Steenwinkel JEM, Datema G, van den Elsen PJ. HLA-G Transactivation by cAMP-Response Element-Binding Protein (CREB). An Alternative Transactivation Pathway to the Conserved Major Histocompatibility Complex (MHC) Class I Regulatory Routes. J Biol Chem (2002) 277(42):39525–31. doi: 10.1074/jbc.M112273200
55. Flajollet S, Poras I, Carosella ED, Moreau P. RREB-1 is a Transcriptional Repressor of HLA-G. J Immunol (2009) 183(11):6948–59. doi: 10.4049/jimmunol.0902053
56. Tan Z, Shon AM, Ober C. Evidence of Balancing Selection at the HLA-G Promoter Region. Hum Mol Genet (2005) 14(23):3619–28. doi: 10.1093/hmg/ddi389
57. Hviid TV, Rizzo R, Melchiorri L, Stignani M, Baricordi OR. Polymorphism in the 5' Upstream Regulatory and 3' Untranslated Regions of the HLA-G Gene in Relation to Soluble HLA-G and IL-10 Expression. Hum Immunol (2006) 67(1-2):53–62. doi: 10.1016/j.humimm.2005.12.003
58. Dias FC, Bertol BC, Poras I, Souto BM, Mendes-Junior CT, Castelli EC. The Genetic Diversity Within the 1.4 Kb HLA-G 5' Upstream Regulatory Region Moderately Impacts on Cellular Microenvironment Responses. Sci Rep (2018) 8(1):5652. doi: 10.1038/s41598-018-24009-7
59. Hviid TV, Sorensen S, Morling N. Polymorphism in the Regulatory Region Located More Than 1.1 Kilobases 5' to the Start Site of Transcription, the Promoter Region, and Exon 1 of the HLA-G Gene. Hum Immunol (1999) 60(12):1237–44. doi: 10.1016/S0198-8859(99)00130-5
60. Castelli EC, Mendes-Junior CT, Deghaide NHS, de Albuquerque RS, Muniz YCN, Simões RT, et al. The Genetic Structure of 3'untranslated Region of the HLA-G Gene: Polymorphisms and Haplotypes. Genes Immun (2010) 11(2):134–41. doi: 10.1038/gene.2009.74
61. Hviid TV, Hylenius S, Rørbye C, Nielsen LG. HLA-G Allelic Variants are Associated With Differences in the HLA-G mRNA Isoform Profile and HLA-G mRNA Levels. Immunogenetics (2003) 55(2):63–79. doi: 10.1007/s00251-003-0547-z
62. Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-Specific Targeting of microRNAs to HLA-G and Risk of Asthma. Am J Hum Genet (2007) 81(4):829–34. doi: 10.1086/521200
63. Yie SM, Li LH, Xiao R, Librach CL. A Single Base-Pair Mutation in the 3'-Untranslated Region of HLA-G mRNA is Associated With Pre-Eclampsia. Mol Hum Reprod (2008) 14(11):649–53. doi: 10.1093/molehr/gan059
64. de Almeida BS, Muniz YCN, Prompt AH, Castelli EC, Mendes-Junior CT, Donadi EA. Genetic Association Between HLA-G 14-Bp Polymorphism and Diseases: A Systematic Review and Meta-Analysis. Hum Immunol (2018) 79(10):724–35. doi: 10.1016/j.humimm.2018.08.003
65. Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, et al. Worldwide Genetic Variation at the 3' Untranslated Region of the HLA-G Gene: Balancing Selection Influencing Genetic Diversity. Genes Immun (2014) 15(2):95–106. doi: 10.1038/gene.2013.67
66. Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. Human Viruses: Discovery and Emergence. Philos Trans R Soc Lond B Biol Sci (2012) 367(1604):2864–71. doi: 10.1098/rstb.2011.0354
67. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus From Patients With Pneumonia in China, 2019. N Engl J Med (2020) 382(8):727–33. doi: 10.1056/NEJMoa2001017
68. Woolhouse ME, Howey R, Gaunt E, Reilly L, Chase-Topping M, Savill N. Temporal Trends in the Discovery of Human Viruses. Proc Biol Sci (2008) 275(1647):2111–5. doi: 10.1098/rspb.2008.0294
69. Schmiedel D, Mandelboim O. Disarming Cellular Alarm Systems-Manipulation of Stress-Induced NKG2D Ligands by Human Herpesviruses. Front Immunol (2017) 8:390. doi: 10.3389/fimmu.2017.00390
70. Jasinski-Bergner S, Mandelboim O, Seliger B. Molecular Mechanisms of Human Herpes Viruses Inferring With Host Immune Surveillance. J Immunother Cancer (2020) 8(2):e000841. doi: 10.1136/jitc-2020-000841
71. Wang L, Ning S. TRIMming Type I Interferon-Mediated Innate Immune Response in Antiviral and Antitumor Defense. Viruses (2021) 13(2):279. doi: 10.3390/v13020279
72. El Baba R, Herbein G. Immune Landscape of CMV Infection in Cancer Patients: From "Canonical" Diseases Toward Virus-Elicited Oncomodulation. Front Immunol (2021) 12:730765. doi: 10.3389/fimmu.2021.730765
73. Naz SS, Aslam A, Malik T. An Overview of Immune Evasion Strategies of DNA and RNA Viruses. Infect Disord Drug Targets (2021) 21(7):e300821192322. doi: 10.2174/1871526521666210317161329
74. McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, et al. Human Placental HLA-G Expression is Restricted to Differentiated Cytotrophoblasts. J Immunol (1995) 154(8):3771–8.
75. Mallet V, Blaschitz A, Crisa L, Schmitt C, Fournel S, King A, et al. HLA-G in the Human Thymus: A Subpopulation of Medullary Epithelial But Not CD83(+) Dendritic Cells Expresses HLA-G as a Membrane-Bound and Soluble Protein. Int Immunol (1999) 11(6):889–98. doi: 10.1093/intimm/11.6.889
76. Dunker K, Schlaf G, Bukur J, Altermann WW, Handke D, Seliger B. Expression and Regulation of non-Classical HLA-G in Renal Cell Carcinoma. Tissue Antigens (2008) 72(2):137–48. doi: 10.1111/j.1399-0039.2008.01090.x
77. Mouillot G, Marcou C, Rousseau P, Rouas-Freiss N, Carosella ED, Moreau P. HLA-G Gene Activation in Tumor Cells Involves Cis-Acting Epigenetic Changes. Int J Cancer (2005) 113(6):928–36. doi: 10.1002/ijc.20682
78. Friedrich M, Stoehr C, Jasinski-Bergner S, Hartmann A, Wach S, Wullich B, et al. Characterization of the Expression and Immunological Impact of the Transcriptional Activator CREB in Renal Cell Carcinoma. J Transl Med (2020) 18(1):371. doi: 10.1186/s12967-020-02544-0
79. Melo-Lima BL, Poras I, Passos GA, Carosella ED, Donadi EA, Moreau P. The Autoimmune Regulator (Aire) Transactivates HLA-G Gene Expression in Thymic Epithelial Cells. Immunology (2019) 158(2):121–35. doi: 10.1111/imm.13099
80. Ikeno M, Suzuki N, Kamiya M, Takahashi Y, Kudoh J, Okazaki T. LINE1 Family Member is Negative Regulator of HLA-G Expression. Nucleic Acids Res (2012) 40(21):10742–52. doi: 10.1093/nar/gks874
81. Friedrich M, Vaxevanis CK, Biehl K, Mueller A, Seliger B. Targeting the Coding Sequence: Opposing Roles in Regulating Classical and non-Classical MHC Class I Molecules by miR-16 and miR-744. J Immunother Cancer (2020) 8(1):e000396. doi: 10.1136/jitc-2019-000396
82. Jasinski-Bergner S, Reches A, Stoehr C, Massa C, Gonschorek E, Huettelmaier S, et al. Identification of Novel microRNAs Regulating HLA-G Expression and Investigating Their Clinical Relevance in Renal Cell Carcinoma. Oncotarget (2016) 7(18):26866–78. doi: 10.18632/oncotarget.8567
83. Sun J, Chu H, Ji J, Huo G, Song Q, Zhang X. Long non-Coding RNA HOTAIR Modulates HLA-G Expression by Absorbing miR-148a in Human Cervical Cancer. Int J Oncol (2016) 49(3):943–52. doi: 10.3892/ijo.2016.3589
84. Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, et al. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J Immunol (2016) 196(12):4967–76. doi: 10.4049/jimmunol.1501550
85. Garziera M, Scarabel L, Toffoli G. Hypoxic Modulation of HLA-G Expression Through the Metabolic Sensor HIF-1 in Human Cancer Cells. J Immunol Res (2017) 2017:4587520. doi: 10.1155/2017/4587520
86. Amiot L, Vu N, Samson M. Immunomodulatory Properties of HLA-G in Infectious Diseases. J Immunol Res (2014) 2014:298569. doi: 10.1155/2014/298569
87. Amiot L, Vu N, Samson M. Biology of the Immunomodulatory Molecule HLA-G in Human Liver Diseases. J Hepatol (2015) 62(6):1430–7. doi: 10.1016/j.jhep.2015.03.007
88. Silva TG, Crispim JCO, Miranda FA, Hassumi MK, de Mello JMY, Simões RT, et al. Expression of the Nonclassical HLA-G and HLA-E Molecules in Laryngeal Lesions as Biomarkers of Tumor Invasiveness. Histol Histopathol (2011) 26(12):1487–97.
89. Diepstra A, Poppema S, Boot M, Visser L, Nolte IM, Niens M, et al. HLA-G Protein Expression as a Potential Immune Escape Mechanism in Classical Hodgkin's Lymphoma. Tissue Antigens (2008) 71(3):219–26. doi: 10.1111/j.1399-0039.2008.01005.x
90. Loustau M, Anna F, Dréan R, Lecomte M, Langlade-Demoyen P, Caumartin J. HLA-G Neo-Expression on Tumors. Front Immunol (2020) 11:1685. doi: 10.3389/fimmu.2020.01685
91. Brianti P, De Flammineis E, Mercuri SR. Review of HPV-Related Diseases and Cancers. New Microbiol (2017) 40(2):80–5.
92. Kombe Kombe AJ, Li B, Zahid A, Mengist HM, Bounda GA, Zhou Y, et al. Epidemiology and Burden of Human Papillomavirus and Related Diseases, Molecular Pathogenesis, and Vaccine Evaluation. Front Public Health (2020) 8:552028. doi: 10.3389/fpubh.2020.552028
93. Scott-Wittenborn N, Fakhry C. Epidemiology of HPV Related Malignancies. Semin Radiat Oncol (2021) 31(4):286–96. doi: 10.1016/j.semradonc.2021.04.001
94. Aggarwal R, Sharma M, Mangat N, Suri V, Bhatia T, Kumar P, et al. Understanding HLA-G Driven Journey From HPV Infection to Cancer Cervix: Adding Missing Pieces to the Jigsaw Puzzle. J Reprod Immunol (2020) 142:103205. doi: 10.1016/j.jri.2020.103205
95. Dong DD, Yang H, Li K, Xu G, Song LH, Fan XL, et al. Human Leukocyte Antigen-G (HLA-G) Expression in Cervical Lesions: Association With Cancer Progression, HPV 16/18 Infection, and Host Immune Response. Reprod Sci (2010) 17(8):718–23. doi: 10.1177/1933719110369183
96. Gillio-Tos A, da Graça Bicalho M, Fiano V, Grasso C, Tarallo V, De Marco L, et al. Case-Control Study of HLA-G Promoter Methylation Status, HPV Infection and Cervical Neoplasia in Curitiba, Brazil: A Pilot Analysis. BMC Cancer (2012) 12:618. doi: 10.1186/1471-2407-12-618
97. Ghandri N, Gabbouj S, Farhat K, Bouaouina N, Abdelaziz H, Nouri A, et al. Association of HLA-G Polymorphisms With Nasopharyngeal Carcinoma Risk and Clinical Outcome. Hum Immunol (2011) 72(2):150–8. doi: 10.1016/j.humimm.2010.10.006
98. Sarmah N, Baruah MN, Baruah S. Immune Modulation in HLA-G Expressing Head and Neck Squamous Cell Carcinoma in Relation to Human Papilloma Virus Positivity: A Study From Northeast India. Front Oncol (2019) 9:58. doi: 10.3389/fonc.2019.00058
99. Yuen MF, Chen DS, Dusheiko GM, Janssen HLA, Lau DTY, Locarnini SA, et al. Hepatitis B Virus Infection. Nat Rev Dis Primers (2018) 4:18035. doi: 10.1038/nrdp.2018.35
100. Kaplan DE. Hepatitis C Virus. Ann Intern Med (2020) 173(5):ITC33–48. doi: 10.7326/AITC202009010
101. Yoneda S, Umemura T, Katsuyama Y, Kamijo A, Joshita S, Komatsu M, et al. Association of Serum Cytokine Levels With Treatment Response to Pegylated Interferon and Ribavirin Therapy in Genotype 1 Chronic Hepatitis C Patients. J Infect Dis (2011) 203(8):1087–95. doi: 10.1093/infdis/jiq165
102. Rehermann B. Pathogenesis of Chronic Viral Hepatitis: Differential Roles of T Cells and NK Cells. Nat Med (2013) 19(7):859–68. doi: 10.1038/nm.3251
103. Bertol BC, Dias FC, Debortoli G, Souto BM, Mendonça PB, Araújo RC, et al. HLA-G Liver Expression and HLA-G Extended Haplotypes are Associated With Chronic Hepatitis C in HIV-Negative and HIV-Coinfected Patients. Clin Immunol (2020) 217:108482. doi: 10.1016/j.clim.2020.108482
104. Weng PJ, Fu YM, Ding SX, Xu DP, Lin A, Yan WH. Elevation of Plasma Soluble Human Leukocyte Antigen-G in Patients With Chronic Hepatitis C Virus Infection. Hum Immunol (2011) 72(5):406–11. doi: 10.1016/j.humimm.2011.02.008
105. Amiot L, Vu N, Rauch M, L'Helgoualc'h A, Chalmel F, Gascan H, et al. Expression of HLA-G by Mast Cells is Associated With Hepatitis C Virus-Induced Liver Fibrosis. J Hepatol (2014) 60(2):245–52. doi: 10.1016/j.jhep.2013.09.006
106. Souto FJ, Crispim JCO, Ferreira SC, da Silva ASM, Bassi CL, Soares CP, et al. Liver HLA-G Expression is Associated With Multiple Clinical and Histopathological Forms of Chronic Hepatitis B Virus Infection. J Viral Hepat (2011) 18(2):102–5. doi: 10.1111/j.1365-2893.2010.01286.x
107. Shi WW, Lin A, Xu DP, Bao WG, Zhang JG, Chen SY, et al. Plasma Soluble Human Leukocyte Antigen-G Expression is a Potential Clinical Biomarker in Patients With Hepatitis B Virus Infection. Hum Immunol (2011) 72(11):1068–73. doi: 10.1016/j.humimm.2011.06.012
108. Laaribi AB, Bortolotti D, Hannachi N, Mehri A, Hazgui O, Yahia HB, et al. Increased Levels of Soluble HLA-G Molecules in Tunisian Patients With Chronic Hepatitis B Infection. J Viral Hepat (2017) 24(11):1016–22. doi: 10.1111/jvh.12718
109. Bian X, Si Y, Zhang M, Wei R, Yang X, Ren H, et al. Down-Expression of miR-152 Lead to Impaired Anti-Tumor Effect of NK via Upregulation of HLA-G. Tumour Biol (2016) 37(3):3749–56. doi: 10.1007/s13277-015-3669-7
110. Asberg A, Jardine AG, Bignamini AA, Rollag H, Pescovitz MD, Gahlemann CC, et al. Effects of the Intensity of Immunosuppressive Therapy on Outcome of Treatment for CMV Disease in Organ Transplant Recipients. Am J Transplant (2010) 10(8):1881–8. doi: 10.1111/j.1600-6143.2010.03114.x
111. Fishman JA. Infection in Organ Transplantation. Am J Transplant (2017) 17(4):856–79. doi: 10.1111/ajt.14208
112. Park B, Spooner E, Houser BL, Strominger JL, Ploegh HL. The HCMV Membrane Glycoprotein US10 Selectively Targets HLA-G for Degradation. J Exp Med (2010) 207(9):2033–41. doi: 10.1084/jem.20091793
113. Onno M, Pangault C, Le Friec G, Guilloux V, André P, Fauchet R. Modulation of HLA-G Antigens Expression by Human Cytomegalovirus: Specific Induction in Activated Macrophages Harboring Human Cytomegalovirus Infection. J Immunol (2000) 164(12):6426–34. doi: 10.4049/jimmunol.164.12.6426
114. Yan WH, Lin A, Chen BG, Chen SY. Induction of Both Membrane-Bound and Soluble HLA-G Expression in Active Human Cytomegalovirus Infection. J Infect Dis (2009) 200(5):820–6. doi: 10.1086/604733
115. Rizzo R, Gabrielli L, Bortolotti D, Gentili V, Piccirilli G, Chiereghin A, et al. Study of Soluble HLA-G in Congenital Human Cytomegalovirus Infection. J Immunol Res (2016) 2016:3890306. doi: 10.1155/2016/3890306
116. Rebmann V, da Silva Nardi F, Wagner B, Horn PA. HLA-G as a Tolerogenic Molecule in Transplantation and Pregnancy. J Immunol Res (2014) 2014:297073. doi: 10.1155/2014/297073
117. Durmanova V, Bandzuchova H, Zilinska Z, Tirpakova J, Kuba D, Buc M, et al. Association of HLA-G Polymorphisms in the 3'UTR Region and Soluble HLA-G With Kidney Graft Outcome. Immunol Invest (2019) 48(6):644–58. doi: 10.1080/08820139.2019.1610888
118. Darbas S, Yilmaz VT, Kocak H, Kisaoglu A, Demiryilmaz I, Aydinli B, et al. New Markers for Predictions of Acute and Chronic Rejection and Graft Outcomes in Kidney Transplant Recipients; HLA-G Gene 3'UTR 14 Bp Polymorphism and sHLA-G. Gene (2021) 790:145712. doi: 10.1016/j.gene.2021.145712
119. Salahuddin SZ, Ablashi DV, Markham PD, Josephs SF, Sturzenegger S, Kaplan M, et al. Isolation of a New Virus, HBLV, in Patients With Lymphoproliferative Disorders. Science (1986) 234(4776):596–601. doi: 10.1126/science.2876520
120. Ablashi DV, Josephs SF, Buchbinder A, Hellman K, Nakamura S, Llana T, et al. Human B-Lymphotropic Virus (Human Herpesvirus-6). J Virol Methods (1988) 21(1-4):29–48. doi: 10.1016/0166-0934(88)90050-X
121. Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, et al. Classification of HHV-6A and HHV-6B as Distinct Viruses. Arch Virol (2014) 159(5):863–70.
122. Scotet E, Peyrat MA, Saulquin X, Retiere C, Couedel C, Davodeau F, et al. Frequent Enrichment for CD8 T Cells Reactive Against Common Herpes Viruses in Chronic Inflammatory Lesions: Towards a Reassessment of the Physiopathological Significance of T Cell Clonal Expansions Found in Autoimmune Inflammatory Processes. Eur J Immunol (1999) 29(3):973–85.
123. Caselli E, Campioni D, Cavazzini F, Gentili V, Bortolotti D, Cuneo A, et al. Acute Human Herpesvirus-6A Infection of Human Mesothelial Cells Modulates HLA Molecules. Arch Virol (2015) 160(9):2141–9.
124. Rizzo R, D'Accolti M, Bortolotti D, Caccuri F, Caruso A, Di Luca D, et al. Human Herpesvirus 6A and 6B Inhibit In Vitro Angiogenesis by Induction of Human Leukocyte Antigen G. Sci Rep (2018) 8(1):17683.
125. Caselli E, Soffritti I, D'Accolti M, Bortolotti D, Rizzo R, Sighinolfi G, et al. HHV-6a Infection and Systemic Sclerosis: Clues of a Possible Association. Microorganisms (2019) 8(1):39. doi: 10.3390/microorganisms8010039
126. Gazit E, Sherf M, Balbin E, Muratov A, Goldstein I, Loewenthal R. HLA-G Expression is Induced in Epstein-Barr Virus-Transformed B-Cell Lines by Culture Conditions. Hum Immunol (2007) 68(6):463–8. doi: 10.1016/j.humimm.2007.02.009
127. Birdwell CE, Queen KJ, Kilgore PCSR, Rollyson P, Trutschl M, Cvek U, et al. Genome-Wide DNA Methylation as an Epigenetic Consequence of Epstein-Barr Virus Infection of Immortalized Keratinocytes. J Virol (2014) 88(19):11442–58. doi: 10.1128/JVI.00972-14
128. Leong MML, Cheung AKL, Dai W, Tsao SW, Tsang CM, Dawson CW, et al. EBV Infection is Associated With Histone Bivalent Switch Modifications in Squamous Epithelial Cells. Proc Natl Acad Sci USA (2019) 116(28):14144–53. doi: 10.1073/pnas.1821752116
129. Bauer M, Jasinski-Bergner S, Mandelboim O, Wickenhauser C, Seliger B. Epstein-Barr Virus-Associated Malignancies and Immune Escape: The Role of the Tumor Microenvironment and Tumor Cell Evasion Strategies. Cancers (Basel) (2021) 13(20):5189. doi: 10.3390/cancers13205189
130. Lozano JM, González R, Kindelán JM, Rouas-Freiss N, Caballos R, Dausset J, et al. Monocytes and T Lymphocytes in HIV-1-Positive Patients Express HLA-G Molecule. AIDS (2002) 16(3):347–51. doi: 10.1097/00002030-200202150-00005
131. Martini F, Agrati C, D'Offizi G, Poccia F. HLA-E Up-Regulation Induced by HIV Infection may Directly Contribute to CD94-Mediated Impairment of NK Cells. Int J Immunopathol Pharmacol (2005) 18(2):269–76. doi: 10.1177/039463200501800209
132. Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, et al. HLA-E Expression and its Clinical Relevance in Human Renal Cell Carcinoma. Oncotarget (2016) 7(41):67360–72. doi: 10.18632/oncotarget.11744
133. Rodriguez-Garcia M, Porichis F, de Jong OG, Levi K, Diefenbach TJ, et al. Expression of PD-L1 and PD-L2 on Human Macrophages is Up-Regulated by HIV-1 and Differentially Modulated by IL-10. J Leukoc Biol (2011) 89(4):507–15. doi: 10.1189/jlb.0610327
134. Said EA, Dupuy FP, Trautmann L, Zhang Y, Shi Y, El-Far M, et al. Programmed Death-1-Induced Interleukin-10 Production by Monocytes Impairs CD4+ T Cell Activation During HIV Infection. Nat Med (2010) 16(4):452–9. doi: 10.1038/nm.2106
135. Fernandes A, Sanches R, Matos F, Massaro J, Donadi E. HLA-G 14bp Polymorphism on HIV-1 Perinatal Transmission (P3041). J Immunol (2013) 190(1 Supplement):55.21.
136. Barcova M, Kacani L, Speth C, Dierich MP. Gp41 Envelope Protein of Human Immunodeficiency Virus Induces Interleukin (IL)-10 in Monocytes, But Not in B, T, or NK Cells, Leading to Reduced IL-2 and Interferon-Gamma Production. J Infect Dis (1998) 177(4):905–13. doi: 10.1086/515230
137. Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, et al. The Selective Downregulation of Class I Major Histocompatibility Complex Proteins by HIV-1 Protects HIV-Infected Cells From NK Cells. Immunity (1999) 10(6):661–71. doi: 10.1016/S1074-7613(00)80065-5
138. Zhao G, Zhang L, Peng H, Zhang S, Zhang C. Effects of HTLV-1 Protein Tax on HLA-G Expression. Chin J Microbiol Immunol (2020) (12):290–4.
139. Krammer F, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, et al. Influenza. Nat Rev Dis Primers (2018) 4(1):3. doi: 10.1038/s41572-018-0002-y
140. LeBouder F, Khoufache K, Menier C, Mandouri Y, Keffous M, Lejal N, et al. Immunosuppressive HLA-G Molecule is Upregulated in Alveolar Epithelial Cells After Influenza A Virus Infection. Hum Immunol (2009) 70(12):1016–9. doi: 10.1016/j.humimm.2009.07.026
141. Al-Bayatee NT, Ad'hiah AH. Soluble HLA-G is Upregulated in Serum of Patients With Severe COVID-19. Hum Immunol (2021) 82(10):726–32. doi: 10.1016/j.humimm.2021.07.007
142. Zhang S, Gan J, Chen BG, Zheng D, Zhang JG, Lin RH, et al. Dynamics of Peripheral Immune Cells and Their HLA-G and Receptor Expressions in a Patient Suffering From Critical COVID-19 Pneumonia to Convalescence. Clin Transl Immunol (2020) 9(5):e1128. doi: 10.1002/cti2.1128
143. Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Spadaro S, et al. Increased sHLA-G Is Associated With Improved COVID-19 Outcome and Reduced Neutrophil Adhesion. Viruses (2021) 13(9):1855. doi: 10.3390/v13091855
144. Saraiva M, O'Garra A. The Regulation of IL-10 Production by Immune Cells. Nat Rev Immunol (2010) 10(3):170–81. doi: 10.1038/nri2711
145. Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, Crotty S, et al. Resolution of a Chronic Viral Infection After Interleukin-10 Receptor Blockade. J Exp Med (2006) 203(11):2461–72. doi: 10.1084/jem.20061462
146. Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MBA. Interleukin-10 Determines Viral Clearance or Persistence In Vivo. Nat Med (2006) 12(11):1301–9. doi: 10.1038/nm1492
147. Rossato M, Curtale G, Tamassia N, Castellucci M, Mori L, Gasperini S, et al. IL-10-Induced microRNA-187 Negatively Regulates TNF-Alpha, IL-6, and IL-12p40 Production in TLR4-Stimulated Monocytes. Proc Natl Acad Sci USA (2012) 109(45):E3101–10. doi: 10.1073/pnas.1209100109
Keywords: human leukocyte antigen G, viral infection, immune escape, virus-induced tumors, interleukin 10
Citation: Jasinski-Bergner S, Schmiedel D, Mandelboim O and Seliger B (2022) Role of HLA-G in Viral Infections. Front. Immunol. 13:826074. doi: 10.3389/fimmu.2022.826074
Received: 30 November 2021; Accepted: 20 January 2022;
Published: 14 February 2022.
Edited by:
Wei-Hua Yan, Wenzhou Medical University, ChinaReviewed by:
Mar Vales-Gomez, Spanish National Research Council (CSIC), SpainCopyright © 2022 Jasinski-Bergner, Schmiedel, Mandelboim and Seliger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Seliger, YmFyYmFyYS5zZWxpZ2VyQHVrLWhhbGxlLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.