- 1Institute of Hematology, School of Medicine, Jinan University, Guangzhou, China
- 2Department of Hematology, First Affiliated Hospital, Jinan University, Guangzhou, China
- 3Department of Pathology, School of Medicine, Jinan University, Guangzhou, China
Background: Heterogeneous T cells in acute myeloid leukemia (AML) have the combinatorial variety generated by different T cell receptors (TCRs). γδ T cells are a distinct subgroup of T cells containing TCRγ (TRGV) and TCRδ (TRDV) subfamilies with diverse structural and functional heterogeneity. Our previous study showed that clonally expanded TRDV T cells might benefit the immune response directed against AML. However, the features of the TRGV repertoire in AML remain unknown. To fully characterize the features of γδ T cells, we analyzed the distribution and clonality of TRGV I-III subfamilies (TRGV II is also termed TRVG 9), the proportions of γδ T cell subsets, and their effects on the overall survival (OS) of patients with AML.
Methods: In this study, the complementarity-determining region 3 (CDR3) size of TRGV subfamilies in γδ T cells of peripheral blood (PB) from de novo AML patients were analyzed by Genescan analysis. Expression levels of TRGV subfamilies were performed by real-time quantitative PCR. The proportions of total γδ T cells and their Vγ9+ Vδ2+ T cells subsets were detected by multicolor flow cytometry assay. We further compared the correlation among the TRGV gene expression levels, the proportion of Vγ9+ Vδ2+ T cells, and OS in AML.
Results: We first found that the distribution pattern and clonality of TRGV subfamilies were changed. The expression frequencies and gene expression levels of three TRGV subfamilies in AML samples were significantly lower than those in healthy individuals (HIs). Compared with HIs, the proportions of total γδ T cells and Vγ9+ Vδ2+ T cells were also significantly decreased in patients with AML. In addition, patients with AML who had higher expression levels of the TRGV gene and higher proportion of Vγ9+ Vδ2+ T cells showed better OS than their counterparts. Furthermore, high expression levels of TRGV 9 and proportion of Vγ9+ Vδ2+ T cells were identified as independent protective factors for complete remission in patients with AML.
Conclusions: The restriction of TRGV usage might be related to the preference of usage of γδ T cells. Higher expression of TRGV subfamilies might be associated with better OS in AML. Higher TRGV 9 expression and increased Vγ9+ Vδ2+ T cells subfamilies might indicate a better prognosis in patients with AML.
Introduction
Acute myeloid leukemia (AML) is a malignant clonal disease originating from hematopoietic stem cells and characterized by genetic and clinical heterogeneity and high mortality (1). Despite considerable progress in treating hematological malignancies, clinical outcomes of patients older than 60 years are unfavorable, and the overall long-term survival in patients with AML remains poor (2). Recent studies have revealed that T cell immunodeficiency is a common characteristic of patients with AML, mainly due to peripheral T cells that restricted oligoclonal T cell repertoires, reduced thymic output function, and lower activation and response to antigens (3, 4).
T cells recognize specific ligands by specific T cell receptors (TCRs), which are heterodimers consisting of either αβ and γδ chains. Genes encode for the variable domains of TRG (γ chain) and TRD (δ chain), which are assembled by somatic recombination from variable (V), diversity (D, only for TRD), and joining (J) segments and compose three hypervariable or complementarity-determining regions (CDR1, CDR2, and CDR3) that occur during T cell differentiation (5, 6). The TRG gene contains several different functional variable (TRGV) segments belonging to four subgroups (TRGV I–IV), and the TRD gene contains at least eight functional TRDV segments that are subdivided into eight TRDV subfamilies (TRDV 1–8) (5–9). Previous studies showed that TRGV IV was a pseudogene, which was a simple combination between TRGV IV and TRGC segment lacking TRGJ segment and there was no any rearrangement in CDR3 by sequencing (10, 11). Hence, the analysis of TRGV repertoire was acquired in three TRGV subfamilies in the present study. Nowadays, according to their TRD (TCRδ) chain usage, human γδ T cells are mainly divided into 2 major subsets including Vδ1 and Vδ2 in peripheral blood (PB). Several functional TRG (TCRγ) gene segments are generally divided into Vγ2, γ3, γ4, γ5, γ8, γ9, γ10 (also termed TRGV 2, TRGV 3, TRGV 4, TRGV 5, TRGV 8, TRGV 9 and TRGV10, respectively) (12, 13). The V-genes of TRGV 2-5 and TRGV 8 have a relatively high sequence similarity, which are different from TRGV 9 sequences. Different TCRγ chains and TCRδ chains can be combined to form different types of γδ T cells (14). Although Vδ1 T cells are predominantly associated with the Vδ1 comprising TRGV 2, TRGV 3, TRGV 4, TRGV 5, TRGV 8, which belonging to TRGV I subsets, the majority of Vδ2 T cells express an invariant TCR harboring TRGV 9, which belonging to TRGV II subsets (15). In addition, TRGV 10 belongings to TRGV III subsets (12). In the PB of healthy individuals (HIs), there is a predominant expression in the γδ T cell population, which is the cell expressing Vγ9 together with Vδ2, termed Vγ9+ Vδ2+ T cells (15, 16). The roles of some T cell subgroups in cancer are controversial because they have been suggested to play both an anti-tumor role and a pro-tumor role. The heterogeneous T cells in AML have the combinatorial variety generated by different TCRs, which might explain why some special T cell subsets have a controversial role in cancer immunity. Although PD-1+Vβ5.2+ and PD-1+Vβ12+ CD8+ T cells were thought to be related to poor prognosis in AML (17), our previous study found that clonally expanded TRDV T cells might benefit the immune response directed against AML (18). However, the features of the TRGV repertoire in AML remain unknown, and the cellular immunity characteristics of AML have yet to be fully elucidated. To further understand the heterogeneity of γδ T cells, in this study, we first analyzed the distribution pattern and clonality of TRGV subfamilies and further investigated correlation between expression levels of TRGV subfamilies and proportion of Vγ9+ Vδ2+ T cells and their clinical relevance in patients with AML.
Materials And Methods
Samples
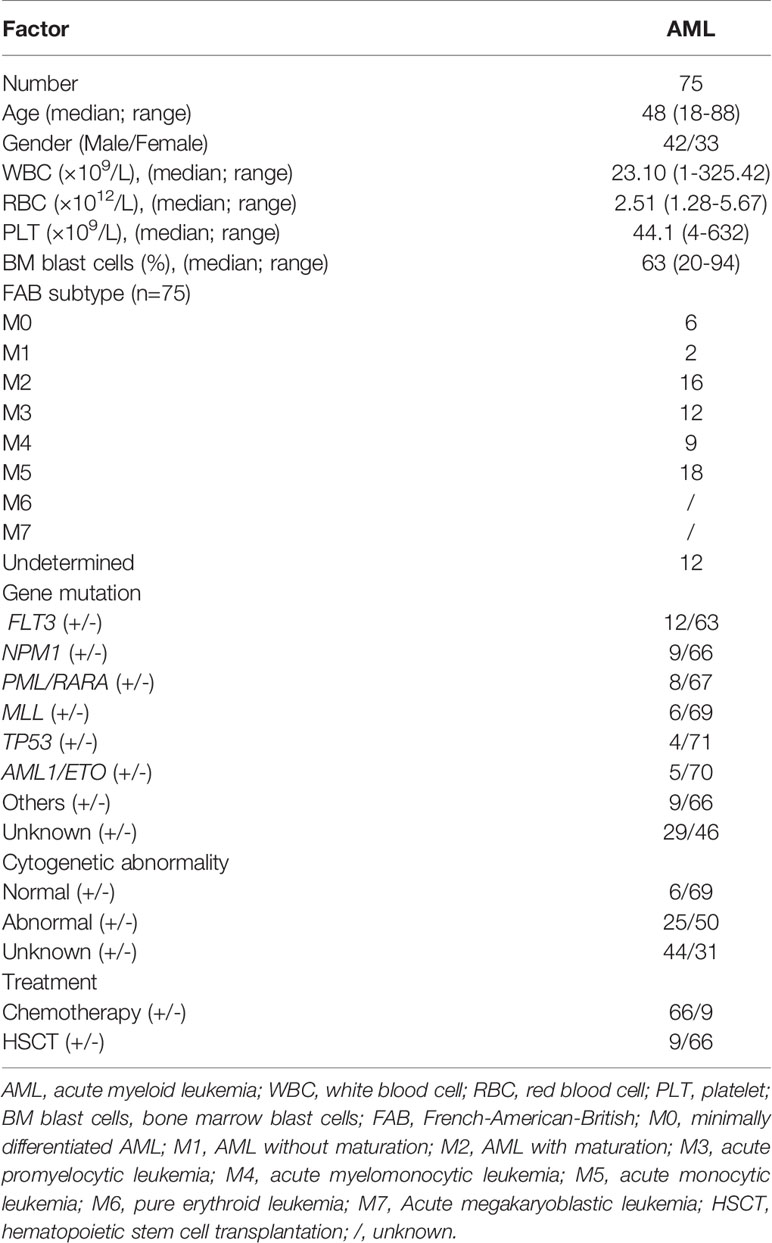
PB samples were collected from 75 patients with de novo AML (42 males and 33 females, median age 48 years, range 18–88 years) from January 2015 to December 2021. A total of 51 HIs (29 males and 22 females, median age 45 years, range 25–83 years) served as controls. Among the total samples, there were 56 patients with AML and 33 HIs were used to analyze the expression levels of TRGV subfamilies. Of the 56 patients identified, 50 patients with both available TRGV gene expression data and outcome information were eventually included in the survival analysis. In addition, the PB of extra 19 patients with AML and 18 HIs were analyzed by flow cytometry. Of the 19 patients, 18 patients with both available flow cytometry data and outcome information were also included in survival analysis. The clinical information was showed in Table 1 and Supplementary Table 1. Informed consent was obtained from all participants. The protocol of all experiments was approved by the Ethics Committee of First Affiliated Hospital, Medical School of Jinan University.
Mononuclear Cell Isolation and γδ T Cell Sorting
The Ficoll–Hypaque gradient centrifugation method was used to isolate mononuclear cells from fresh PB. The γδ T cells were sorted by γδ monoclonal antibodies and MACS magnetic cell sorting technique (Miltenyi Biotec, Germany) (19). All samples were freshly obtained and subjected to immediate preparation.
RNA Isolation and cDNA Synthesis
According to the manufacturer’s recommendations, total RNA of γδ T cells was extracted by Trizol (Invitrogen, USA). Superscript II Kit (Gibco, USA) was used to synthesize the first single-strand complementary DNA (cDNA). Subsequently, the quality of cDNA was confirmed by RT-PCR for β2 microglubin (β2M) gene amplification (the primers of β2M gene for RT-PCR were list in Table 2) (20).
RT-PCR for TRGV Subfamily Amplification and Genescan Analysis for TRGV Subfamily Clonality Analysis
Three sense TRGV primers and a single TRGC reverse primer were used in unlabeled PCR for the amplification of the TRGV subfamilies. Runoff PCR was performed with fluorescent primers labeled at the 5’ end with the FAM fluorophore (Cγ-FAM) (TIB MOLBIOL GmbH, Germany). A DNA thermal cycler (BioMetra, Germany) was used to perform this reaction process. The primers are listed in Table 2. PCR was performed as described in our previous report (19–21). Aliquots of cDNA (1 μL) were amplified in 20 μL reactions with one of the three Vγ primers and a Cγ primer. The final reaction mixture contained 0.5 μM sense primer and antisense primer, 0.1 mM dNTPs, 1.5 mM MgCl2, 1× PCR buffer, and 1.25 U Taq polymerase (Promega, USA). After 3 min of denaturation at 94°C, 40 PCR cycles were carried out (94°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final elongation for 6 min at 72°C). All PCR products were stored at 4°C and ready for Genescan analysis (22).
Aliquots of the unlabeled PCR products (2 μL) were subjected to a cycle of runoff reaction with a fluorophore-labeled Cγ-FAM primer. The labeled runoff PCR products (2 μL) were heat-denatured at 94°C for 4 min with 9.5 μL of formamide (Hi-Di Formamide, ABI, USA) and 0.5 μL of size standards (GENESCAN™-500-LIZ™, Perkin Elmer, USA). The samples were then loaded on 3100 POP-4™ gel (Performance Optimized Polymer-4, ABI, USA) and resolved by electrophoresis in an ABI 3100 DNA sequencer for size and fluorescence intensity determination using Genescan software (23).
Real-Time Quantitative PCR (qPCR) for TRGV Gene
The gene expression levels of the TRGV subfamilies in cDNA of γδ T cells were determined by qPCR with SYBR Green I technique, and the β2-microglobulin (β2M) gene was used as an endogenous reference. The primers are listed in Table 2. qPCR was performed as described by Stams WAG et al. and our previous study (10, 24–26). In brief, qPCR was performed in a total volume of 20 μL with approximately 1 μL cDNA, 0.5 μM of each primer (one of the three TRGV sense primer and the antisense primer Cγ for TRGV amplification, β2M-for and β2M-back primers for β2M gene amplification), 2x RealMastrMix 10 μL (Tiangen, China). After 2 min of denaturation at 95°C, 40 PCR cycles were carried out (95°C for 15 s, 58°C for 20 s, and 72°C for 30 s). At the end of each run, melting curve analysis was performed starting at 65°C up to 95°C with an increase of 1°C per 2 s to verify primer specificities. Specific amplification of PCR products was analyzed by melting curve analysis. qPCR was repeated in at least three separate experiments. The following equation was used to calculate the relative expression level to the β2M gene for each target PCR. Relative mRNA expression = 2-ΔCt × 100% [ΔCt = Ct(TRGV subfamilies) – Ct(β2M)] (15).
Flow Cytometry
The following monoclonal antibodies APC/Cy7 anti-human CD3 (clone SK7), PE/Cy7 anti-human TCR γ/δ (clone B1), PerCP anti-human TCR Vδ2 (clone B6), and APC anti-human TCR Vγ9 (clone B3; Biolegend, USA) were used for cell surface staining following the manufacturer’s instructions (27). The stained cells were examined with BD FACS VERSE flow cytometer (BD, USA), and data were analyzed by Flowjo software (Flowjo LLC, USA).
Statistical Analysis
In this study, data were presented as median. Fisher’s exact test was used to compare expression frequencies of three TRGV subfamilies between AML patients and HIs. Kruskal–Wallis test was used for comparison of different gene expression levels from different TRGV subfamilies in AML and HIs. Differences in mRNA expression level of TRGV between two groups were analyzed using the Mann–Whitney U test. Pearson correlation analysis was used to analyze the correlation of mRNA expression levels of TRGV subfamilies between two groups. Binary logistic regression analysis was performed to determine associations between expression levels of three TRGV subfamilies and clinical outcome of the AML patients. Through Kaplan-Meier method and cox regression analysis the effect of TRGV expression and the proportion of Vγ9+ Vδ2+ T cells on prognosis of AML were analyzed. All analyses included the following variables: including gender, age, white blood cell (WBC), red blood cell (RBC), platelet (PLT), bone marrow (BM) blast cells, French-American-British (FAB) subtype, gene mutation and treatment in patients. Only values with P < 0.05 was regarded as statistically significant. All results were analyzed by SPSS 25.0 and GraphPad Prism 8.4.
Results
Expression Pattern and Clonality of the TRGV Repertoire in Patients With De Novo AML
In this study, the CDR3 region of three TRGV subfamily genes was analyzed by Genescan analysis in γδ T cells from 30 patients with de novo AML and 10 HIs to assess the spectral pattern visually. Diversity and clonality of TCR repertoire demonstrated the ability of specific amplifications to respond to antigen stimulation. Based on the CDR3 TCR rearrangement lengths, the clonality of γδ T cells was characterized as multipeaks and oligopeaks responding to polyclonality and oligoclonality. Polyclonality of the TRGV subfamily genes displayed a Gaussian distribution consisting of three or more peaks, and oligoclonality was a skewed spectral profile showing a single dominant peak. In this study, all patients with AML had a significantly skewed TCR repertoire with 16–21 of the three TRGV subfamilies (TRGV I, 9, and III) detected in each patient. Among AML samples, the most frequently expressed subfamily members were TRGV III (70%, 21/30) and TRGV 9 (66.67%, 20/30). TRGV I from patients with AML was detected only in 16 cases (53.33%, 16/30; Figures 1A–D). All of the three TRGV subfamilies could be detected in γδ T cells from HIs. The expression frequencies of the TRGV I and TRGV 9 subfamilies in patients with AML were lower than those in HIs (TRGV I: P = 0.007, TRGV 9: P = 0.043), whereas the TRGV III subfamily in AML was similar to that in HIs (P = 0.081; Figures 2A–C).
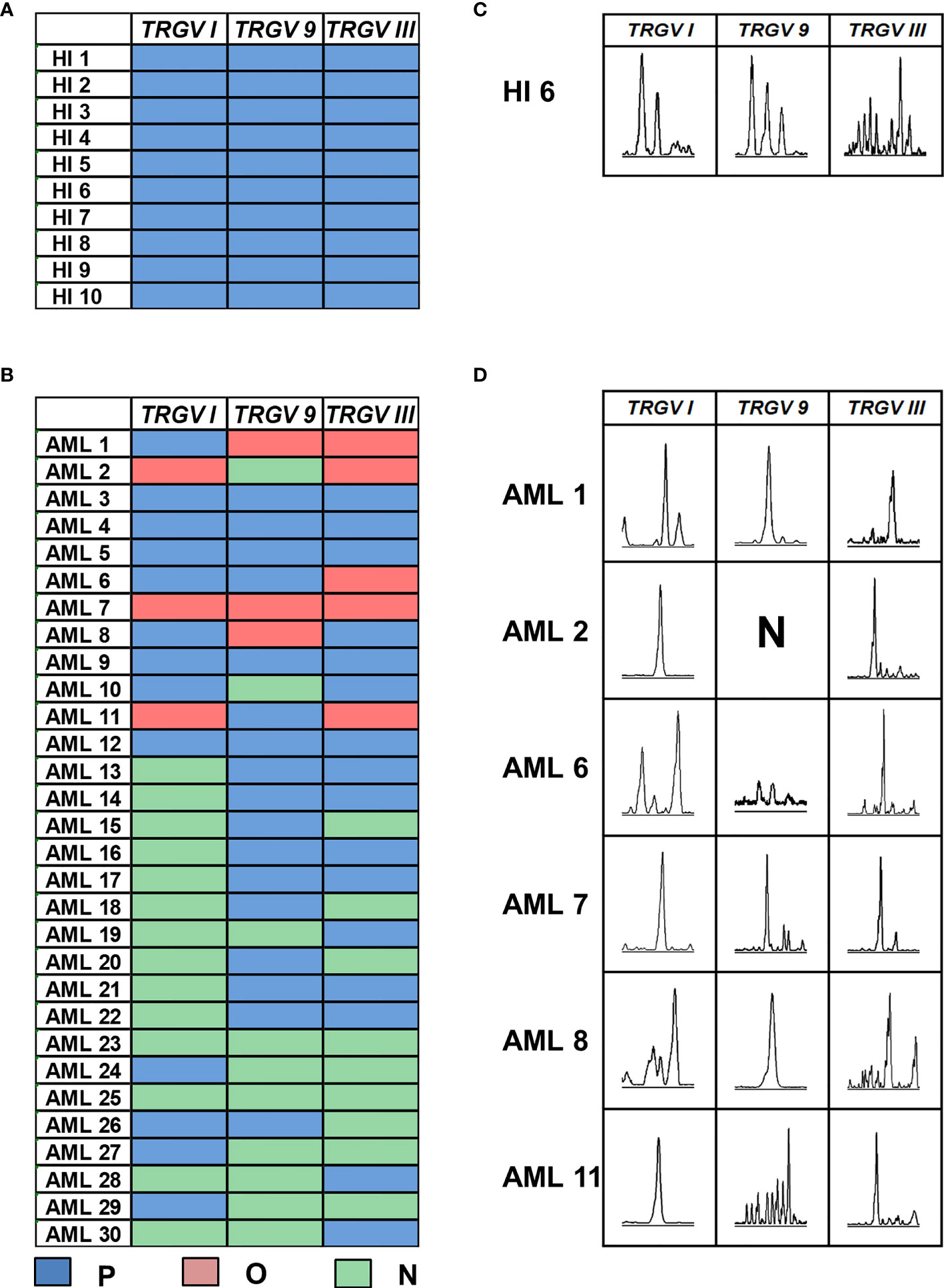
Figure 1 Distribution and clonality of TRGV subfamilies in γδ T cells from 10 healthy individuals and 30 patients with AML. The feature of distribution and clonality of TRGV subfamilies in healthy individuals and patients with AML (A, B). Expression pattern of TRGV subfamilies in one case of healthy individual and six AML patients (C, D). P, polyclonality; O, oligoclonality; N, negative.
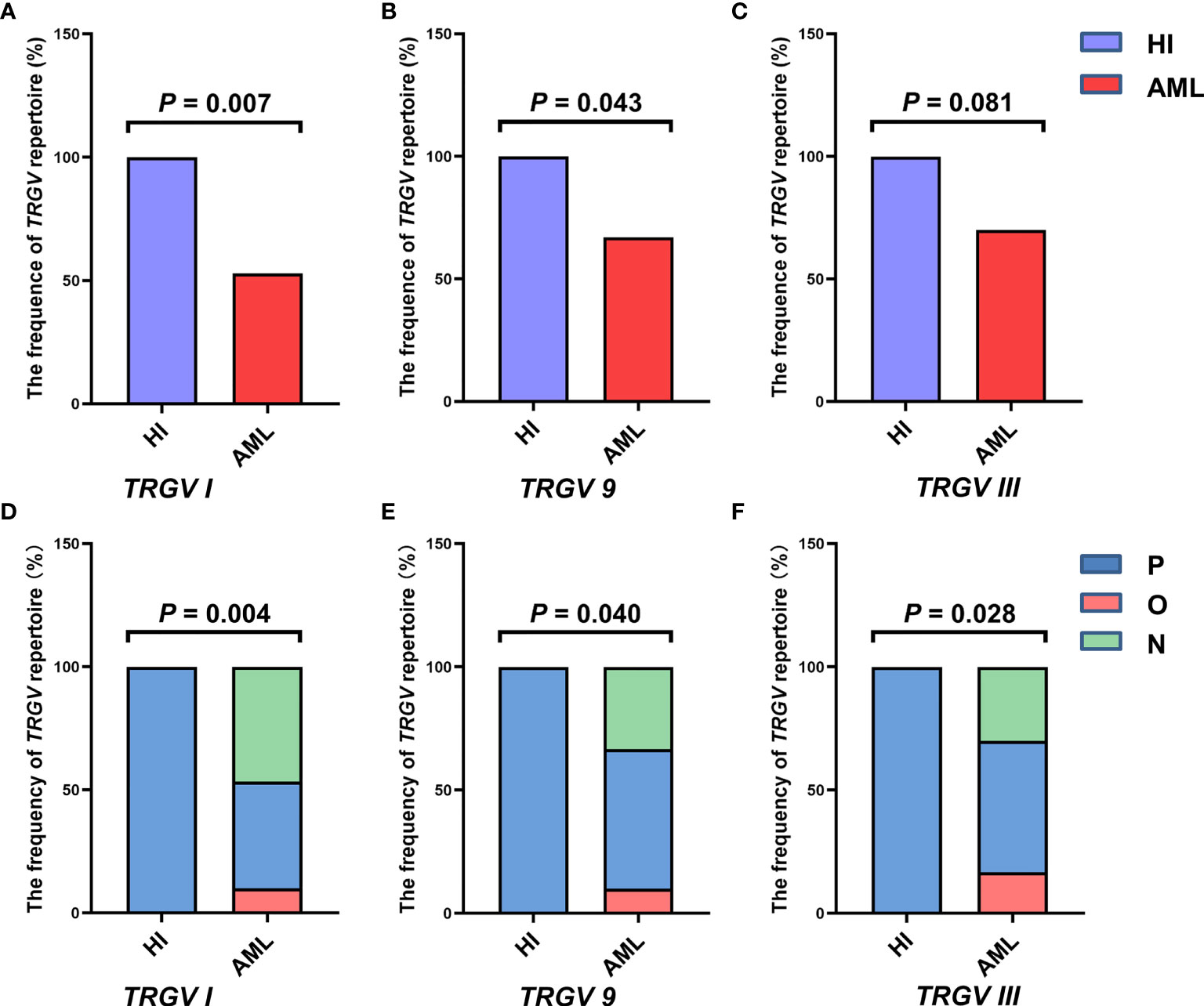
Figure 2 Frequencies of the TRGV subfamilies in γδ T cells from 10 healthy individuals and 30 patients with AML. The expression frequencies of three TRGV subfamilies in healthy individuals and patients with AML (using the Fisher’s exact test) (A–C). The clonal expansion frequency of the three TRGV subfamilies in healthy individuals and patients with AML (using the Fisher’s exact test) (D–F). P, polyclonality; O, oligoclonality; N, negative.
The deviation from the Gaussian profile could indicate a clonally expanded pattern. The PCR products produced only one peak, which represented that CDR3 lengths were identical, named oligoclonal pattern. We further analyzed the different clonotypic expansion patterns in HIs and patients with AML. Oligoclonal expansion was detected in the TRGV subfamily from six out of 30 cases in patients with AML (Figure 1D). The expression frequencies of clonally expanded TRGV subfamilies in the patients with AML were as follows: TRGV III (17%, 5/30), TRGV I (10%, 3/30), and TRGV 9 (10%, 3/30). However, there were no clonally expanded TRGV subfamilies that could be identified in HIs. Based on the clonally expanded pattern, we divided the clonal expansion frequency of the three TRGV subfamilies into three groups: polyclonality, oligoclonality and negative groups. The results showed a significant difference between patients with AML and HIs, and the clonal expansion frequencies of the TRGV subfamilies were statistically higher than those of HIs (TRGV I: P = 0.004; TRGV 9: P = 0.040; and TRGV III: P = 0.028; Figures 2D–F).
Gene Expression Level of the TRGV Subfamily
Subsequently, we focused on detecting expression levels of TRGV subfamilies by qPCR, so we expanded the samples’ quantity, and further collected extra 26 AML samples on the basis of the original 30 samples. Therefore, three TRGV genes expression levels in a total of 56 patients with AML and 33 HIs as control were detected in our study. Results showed significant differences of expression levels in the TRGV subfamilies of HIs (χ2 = 9.998, P = 0.007) between TRGV I and TRGV 9 (P = 0.158), TRGV 9 and TRGV III (P = 0.002), and TRGV I and TRGV III (P = 0.082; Figure 3A). There were also significant differences in the TRGV subfamilies of AML (χ2 = 7.208, P = 0.027) between TRGV I and TRGV 9 (P = 0.679), TRGV 9 and TRGV III (P = 0.014), and TRGV I and TRGV III (P = 0.032; Figure 3B). We further compared the gene expression levels of the TRGV subfamilies in patients with AML and HIs. The gene expression levels of the three TRGV subfamilies in AML were lower than those in HIs (P < 0.001, P < 0.001, and P < 0.001; Figures 3C, G).
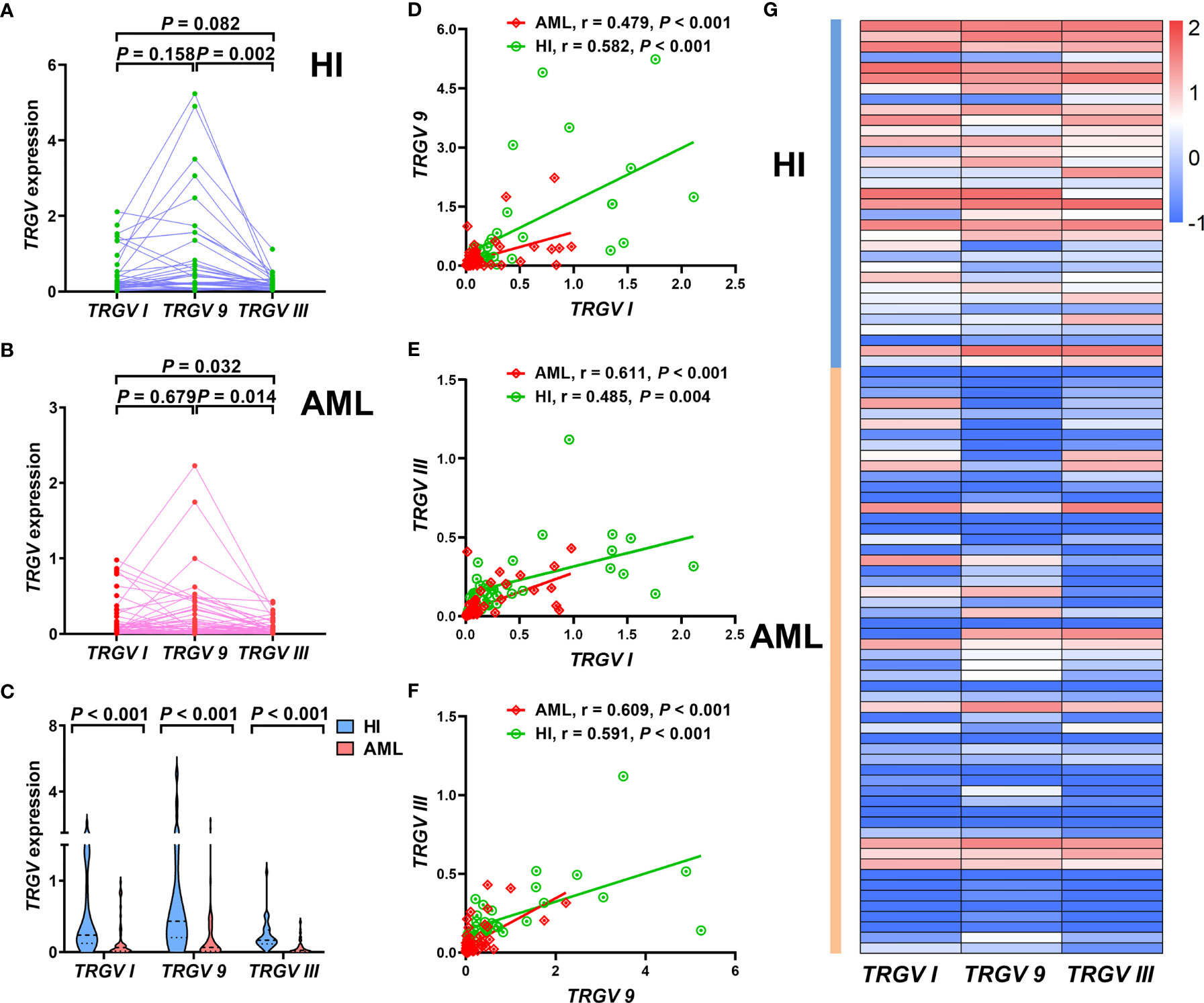
Figure 3 Pattern of expression levels of three TRGV subfamilies in γδ T cells from 33 cases with healthy individuals and 56 cases with AML (using the Mann Whitney test) (A–C). Correlations among three TRGV subfamilies in 33 healthy individuals and 56 patients with AML (using the Pearson correlation analysis) (D–F). Heatmap representing the expression levels of three TRGV subfamilies in 33 healthy individuals and 56 patients with AML (G).
We also obtained more insight to investigate the correlation of the gene expression levels of the three TRGV subfamilies in HIs and patients with AML. In HIs, a significant positive correlation was found in the expression levels of TRGV I and TRGV 9 (r = 0.582, P < 0.001), TRGV I and TRGV III (r = 0.485, P = 0.004), and TRGV 9 and TRGV III (r = 0.591, P < 0.001; Figures 3D–F). A positive correlation in the expression levels of TRGV I and TRGV 9 (r = 0.479, P < 0.001), TRGV I and TRGV III (r = 0.611, P < 0.001), and TRGV 9 and TRGV III (r = 0.609, P < 0.001) was also observed in patients with AML (Figures 3D–F).
Proportions of Total γδ T Cells and Vγ9+ Vδ2+ Subsets in patients With AML
Based on previous finding, we were more interested in proportions of total γδ T cells and Vγ9+ Vδ2+ T cell subsets from PB, so another 19 AML samples and 18 HIs were further collected and analyzed for FACS (Figures 4A–D). Compared with HIs, significantly lower proportions of total γδ T cells (median: 4.83% vs. 10.5%) and Vγ9+ Vδ2+ T cells (median: 57.9% vs. 84.25%) were found in patients with AML (P < 0.001 and P = 0.001, respectively; Figures 4E, F).
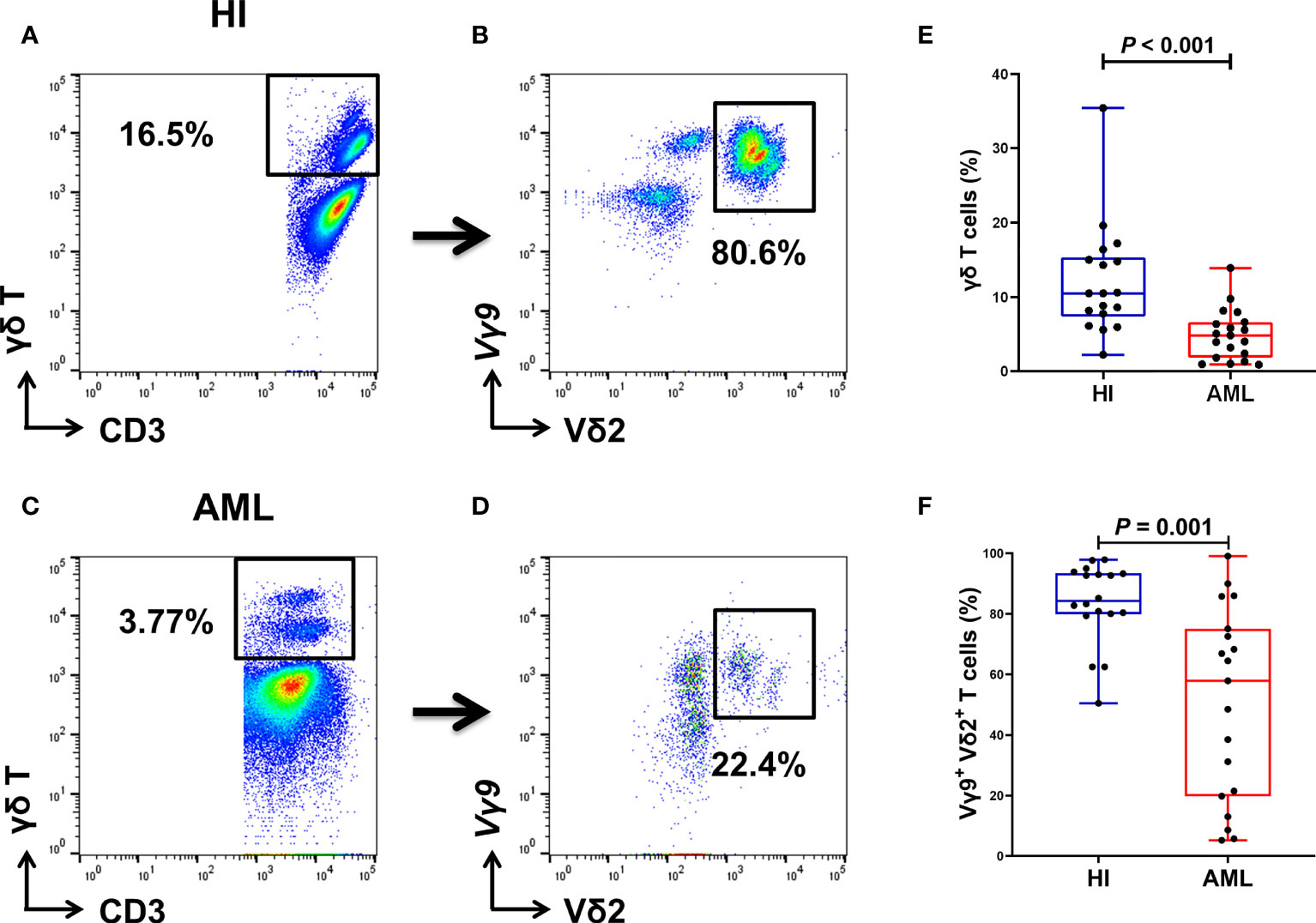
Figure 4 Gating strategy for identifying the percentage of γδ T cells from PB in 18 HIs and 19 patients with AML. Flow cytometry detection of the percentage of CD3+ γδ T cells and Vγ9+ Vδ2+ T cells in HIs (A, B) and patients with AML (C, D). The percentage of γδ T cells in HIs and patients with AML (using the Mann Whitney test) (E). Comparison of the percentages of Vγ9+ Vδ2+ T cells in HIs and patients with AML (using the Mann Whitney test) (F).
TRGV Repertoire and Its Clinical Relevance in AML
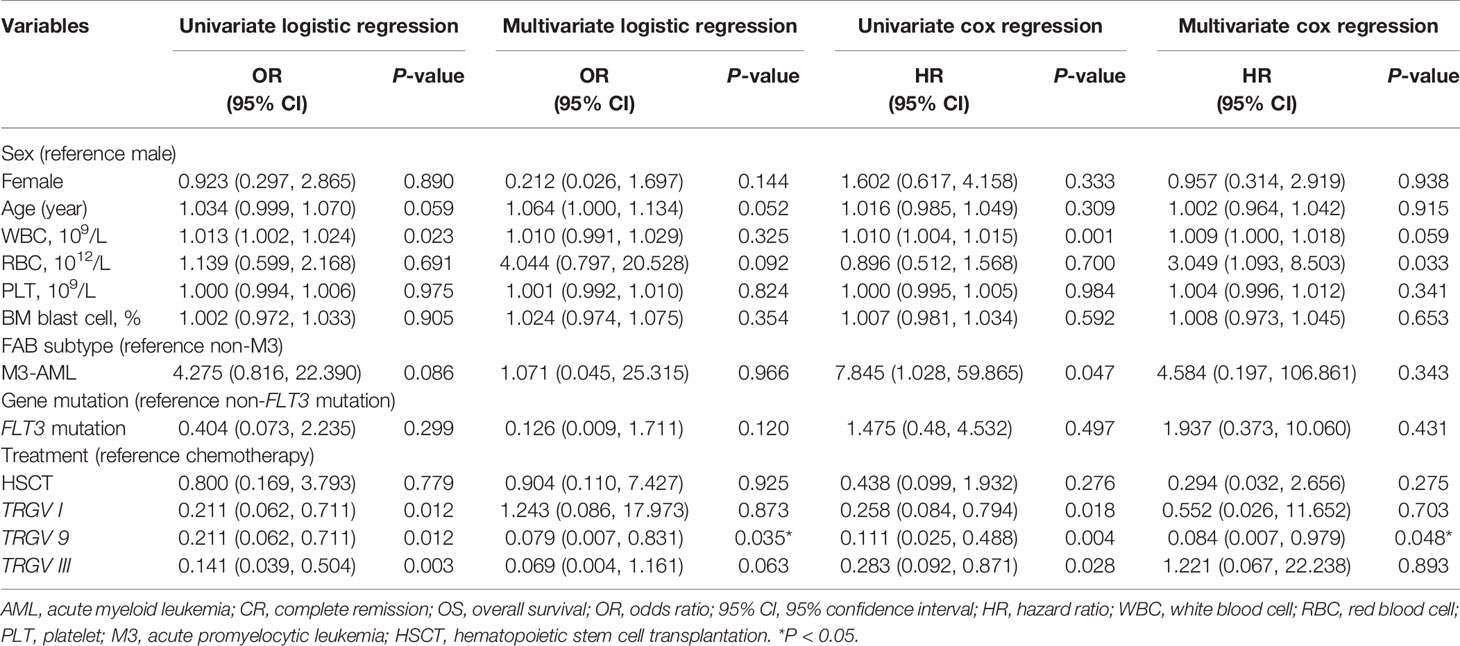
Despite the increased insight into the phenotype of γδ T cells, whether it correlates with clinical outcome remains poorly understood. To further understand the role of the TRGV subfamily and the prognosis of patients with AML, we analyzed correlation between the expression levels of TRGV subfamily genes and the frequency of Vγ9+ Vδ2+ T cells with the clinical prognosis of AML. We first focused on whether expression levels of TRGV subfamily genes affected AML clinical prognosis and assessed the clinical prognosis of the 56 AML patients. Due to 6 patients who refused therapy and voluntarily left the hospital, we finally analyzed the prognosis and outcome of 50 AML patients. Univariate and multivariate logistic regression analysis were used to analyze the expressive levels of three TRGV subfamilies and other impact factors, including gender, age, WBC, RBC, PLT, BM blast cells, AML subtype, gene mutation and treatment in patients with AML. The patients who followed up after first-cycle chemotherapy were divided into complete remission (CR) and non-CR groups based on BM smears and flow cytometry analysis. Univariate logistic regression analysis demonstrated high WBC counts was an independent risk factor for CR (P = 0.023, odds ratio (OR) = 1.013, 95% confidence interval (CI): 1.002-1.024), whereas high expression levels of TRGV I, TRGV 9, and TRGV III were the significant independent protective factors for CR (TRGV I: P = 0.012, OR = 0.211, 95% CI: 0.062-0.711; TRGV 9: P = 0.012, OR = 0.211, 95% CI: 0.062-0.711; TRGV III: P = 0.003, OR = 0.141, 95% CI: 0.039-0.504),. However, there was no significant difference in gender, age, RBC, PLT, BM blast cells, AML subtype, gene mutation and treatment (P > 0.05). Interestingly, multivariate logistic regression analysis showed that TRGV 9 expression was an independent protective factor for CR (TRGV 9: P = 0.035, OR = 0.079, 95% CI: 0.007-0.831). Besides, we further used univariate and multivariate cox regression analysis to further analyze the relationship between those factors and overall survival (OS) in AML patients. Univariate cox regression analysis showed that high counts of WBC (P = 0.001, hazard ratio (HR) = 1.010, 95% CI: 1.004-1.015) and the AML subtype (non-M3) (P = 0.047, HR = 7.845, 95% CI: 1.028-59.865) was associated with unfavorable OS in AML patients. Importantly, high expression levels of TRGV I, TRGV 9, and TRGV III were associated with favorable OS in AML patients (TRGV I: P = 0.018, HR = 0.258, 95% CI: 0.084-0.794; TRGV 9: P = 0.004, HR = 0.111, 95% CI: 0.025-0.488; TRGV III: P = 0.028, HR = 0.283, 95% CI: 0.092-0.871). Multivariate cox regression analysis also showed that high TRGV 9 expression could mark an improved OS in patients with AML (TRGV 9: P = 0.048, HR = 0.084, 95% CI: 0.007-0.979; Table 3).
Furthermore, we also used the same way to access the relationship between the proportion of γδ T cells, Vγ9+ Vδ2+ T cells and the prognosis of 19 AML patients. Univariate logistic regression analysis showed that the high proportion of Vγ9+ Vδ2+ T cells was an independent protected factor for CR (P = 0.044, OR = 0.963, 95% CI: 0.927-0.999), and age was an independent risk factor for AML-CR (P = 0.035, OR = 1.128, 95% CI: 1.009-1.261), but there was no significant difference in γδ T cells and other factors (gender, age, WBC, RBC, PLT, BM blast cells, AML subtype, gene mutation and treatment) (P > 0.05) (data were not showed). Due to insufficient numbers of AML samples, there was no significant difference in multivariate logistic regression analysis. There was one patient was voluntarily left the hospital because of impact of COVID-19 in total 19 AML patients, so we collected outcome of 18 AML patients. Univariate cox regression analysis showed that patients with high proportion of γδ T cells had low risk of death than those with low proportion (P = 0.008, hazard ratio (HR) = 0.109, 95% CI: 0.021-0.564), while multivariate cox regression analysis showed no significant difference (P > 0.05) (data were not showed).
The Relationship Between TRGV Expression and Prognosis in AML Patients
The survival analysis demonstrated that the high expression levels of TRGV I, TRGV 9 and TRGV III were significant related to better OS (P = 0.011; P < 0.001; P = 0.019) (Figures 5A–C). To better understand the combination of three TRGV subfamilies in predicting the OS of AML patients, we divided patients into the following 3 groups: TRGV Ihigh TRGV 9high TRGV IIIhigh, TRGV I, TRGV 9, or TRGV IIIhigh and TRGV Ilow TRGV 9low TRGV IIIlow. Interestingly, the results suggested that the group of TRGV Ihigh TRGV 9high TRGV IIIhigh had longer survival time (P = 0.001) (Figure 5D). Next, we further access the proportion of Vγ9+ Vδ2+ T cells from PB with the clinical outcome of AML patients. The OS in high Vγ9+ Vδ2+ T cells were longer than those in low Vγ9+ Vδ2+ T cells group (P = 0.039) (Figure 5E).
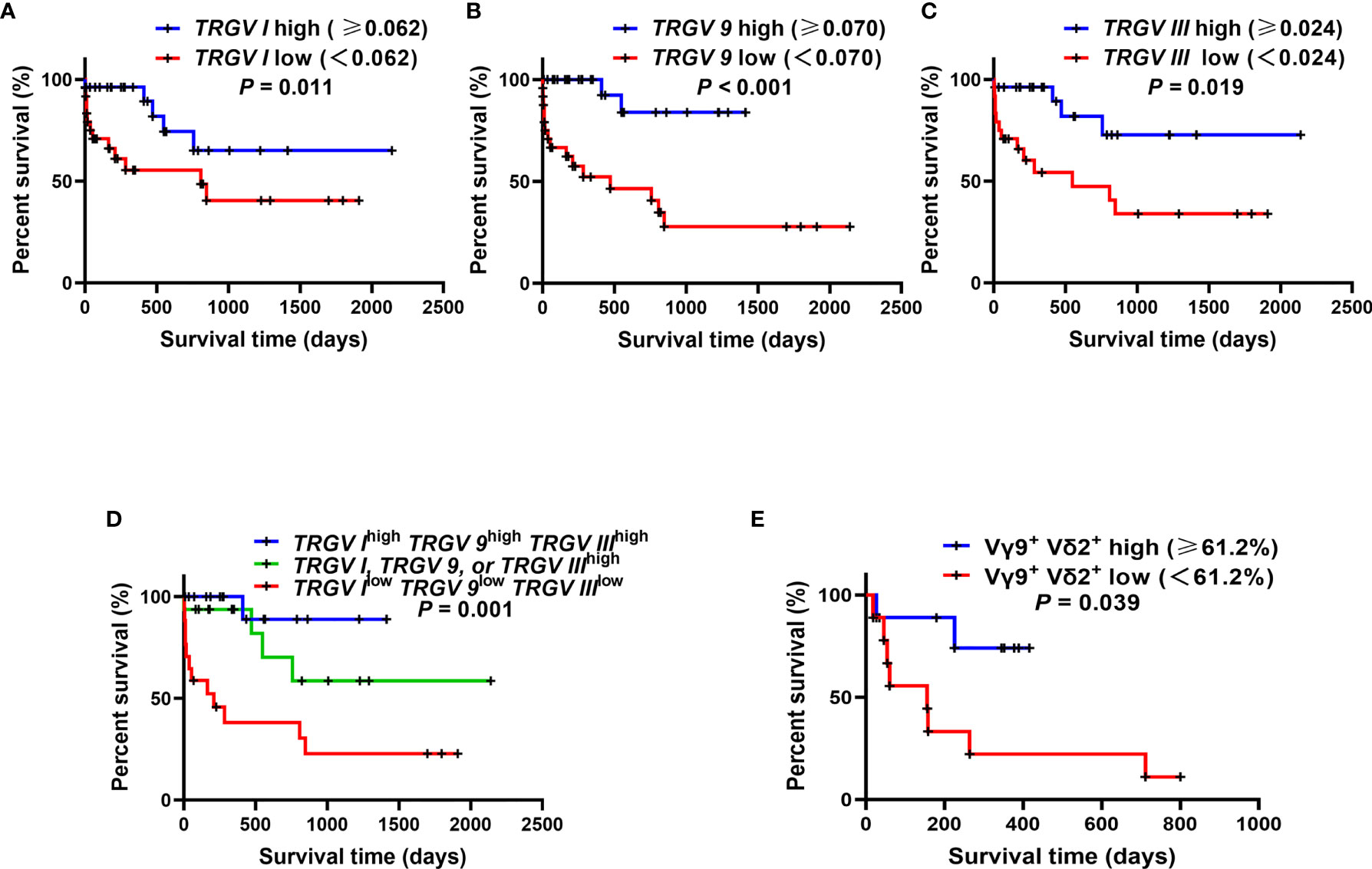
Figure 5 Overall survival (OS) analysis of the gene expression levels of three TRGV subfamilies in 50 AML patients and the percentages of Vγ9+ Vδ2+ T cells in 18 AML patients. Kaplan–Meier curves showed the OS for the high (blue line) and low (red line) TRGV expression groups (A–C). Kaplan–Meier curves showed the OS for the co-high expression (blue line), single high expression (green line), and co-low expression (red line) of TRGV I, TRGV 9 and TRGV III (D). Kaplan–Meier curves showed the OS for the high (blue line) and low (red line) percentages of Vγ9+ Vδ2+ T cells (E).
Discussion
The attractive features of γδ T cells include non-MHC-restricted antigen recognition and abundant cytokine secretion capacity, which have raised expectations for their application in cancer adoptive immunotherapy (28–30). The combinatorial variety generated by different TCRs might be the reason why γδ T cells can exert diverse actions in distinct pathological types of diseases (31). T cell immunodeficiency is a common feature in different hematological malignancies, including AML, immune thrombocytopenic purpura (ITP), B cell non-Hodgkin lymphoma, and graft-versus-host disease (GVHD) (21, 32–34). Analysis of alterations in the TCR repertoire is a practical approach that can help understand the involved immunological abnormalities and provide guidance for clinics in translational research (19). Analysis of the TRGV and TRDV repertoire provides a global picture of the distribution and clonal expansion of TCR γδ subfamilies in ITP, multiple myeloma (MM), and GVHD (21, 32, 35, 36). Our previous study also showed the clonally expanded TRDV T cells in AML (18). However, the features of the TRGV repertoire in AML remain unknown.
In this study, we investigated the expression pattern of TCR Vγ (TRGV) subfamilies and characterized the correlation between the expression of TRGV and clinical outcome in patients with AML. To further compare the difference in TCR repertoire diversity, three TRGV gene spectral profiles were examined by Genescan analysis. In HIs, polyclonal expanded T cells, which showed a small proportion of multiple peaks, were detected in the majority of the TRGV subfamily. By contrast, a clonotypic expansion pattern, which included a high peak together with one or a few lower peaks named oligoclonality, was a common pattern for each sample. Skewed expression of the TRGV repertoire was an obvious characteristic of patients with AML compared with HIs who expressed nearly all of the TRGV subfamilies, which indicated that patients with AML might have low diverse immune responses due to γδ T cell immunodeficiency. The T cell spectra are commonly characterized by a Gaussian distribution containing 6–8 peaks, which are named polyclonality in HIs, representing a repertoire that guarantees sufficiently diverse T cell clones (37). The clonally expanded T cell repertoire was also detected in all samples in this study. Multiple oligoclonal expanded TRGV subfamilies were demonstrated in patients with AML who were different from HIs. Thus, the oligoclonal TRGV repertoire might be associated with leukemia-associated antigen.
We also found that the gene expression levels of the TRGV repertoire in γδ T cells between AML and HIs were different, and lower expression levels were found in TRGV genes in AML than in HIs. The change and pattern of TRGV subfamilies demonstrated that restrictive TRGV usage might be related to the preference of usage of γδ T cells. The biological significance of the difference observed remains unknown, so we attempted to characterize the association between the expression level of the TRGV repertoire and clinical patient characteristics. Our previous study showed that TIGIT+ Foxp3+ γδ T cells and TIGIT+ CD226- γδ T cells were related to the clinical outcome of patients with AML (38, 39). In the present study, we further analyzed the relationship between the expression of the TRGV repertoire and the OS of patients with AML. Our results showed that a higher expression level of TRGV subfamilies was associated with better OS in patients with AML, and patients with highly TRGV I, TRGV 9, and TRGV III genes co-expressed had better OS than their counterparts. Moreover, we found that TRGV 9 was an independent protective factor in AML-CR, thereby indicating that patients with high TRGV 9 expression may have the better prognosis than those with low expression. In addition, our data showed that increased Vγ9+ Vδ2+ T cells subfamilies in patients with AML might correlate with better therapeutic effects. Related research showed that γδ T cells played an essential role in cancer (40). Such cells have a long-term disease-free survival advantage to patients with AML and increased γδ T cells following hematopoietic stem cell transplantation (HSCT) (41, 42). The known pleiotropic effects of γδ T cells suggest multiple mechanisms by which γδ T cells might promote survival after HSCT, which were consistent with our findings in patients with AML. Understanding the characteristics of TRGV subsets in patients with AML may be helpful for clinical application and promote the treatment of patients. However, these γδ T cell subfamilies exerted certain anti-leukemia effects, so the anti-leukemia potency of γδ T cells could be exhausted due to prolonged antigenic stimulation. In the long run, we should choose a specific anti-tumor γδ T cell subgroup in γδ T cell immunotherapy and try to use a combination of γδ T cell adoptive immunotherapy and immune checkpoint inhibitors.
Conclusion
Taken together, in addition to the previously reported clonally expanded TRDV T cells in AML (18), our data further provide a detailed profile and feature of the TRGV repertoire in patients with AML. Importantly, the patients with AML who had high expression level of the TRGV gene or higher proportion of Vγ9+ Vδ2+ T cells were associated with favorable OS, which may be related to resorting anti-AML γδ T function. Further studies are required to confirm and dissect the detailed mechanisms. These findings could partially explain to promote our understanding of the cellular immune features of γδ T cells, which brings hope for immunotherapy to treat AML patients.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The protocol of all experiments was approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XW and ZJ were involved in experimental design and the concept development. XK and XL conducted the experiments. WW and XJ contributed to data analysis and figure preparation. JC and JL provided all samples and clinical data. ZJ, XK, and XW drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81800143, 81770150, 81200388, and 82170220), Natural Science Foundation of Guangdong Province (No. 2018A0303130220 and 2020A1515010817), the Science and Technology Planning Project of Guangzhou City of China (No. 201804010425), Medical Scientific Research Foundation of Guangdong Province (No. A2018565 and A2017198), Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (No. 202010559078), and Guangdong College Students’ Scientific and Technological Innovation (Nos. CX21283, CX21285, and CX20137).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We want to thank the Flow Facility of Biological Translational Research Institute of Jinan University, as well as the healthy volunteers who donated blood for this project. We also thank Professor Yangqiu Li from Jinan University, who gave valuable suggestions and guidance for the experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.823352/full#supplementary-material
Abbreviations
AML, acute myeloid leukemia; TCRs, T cell receptors; CDR, complementarity-determining region; PB, peripheral blood; HIs, healthy individuals; β2M, β2 microglubin; qPCR, real-time quantitative PCR; WBC, white blood cell; RBC, red blood cell; PLT, platelet; BM, bone marrow; FAB, French-American-British; CR, complete remission; OR, odds ratio; CI, confidence interval; OS, overall survival; HR, hazard ratio; ITP, immune thrombocytopenic purpura; GVHD, graft-versus-host disease; MM, multiple myeloma; HSCT, hematopoietic stem cell transplantation.
References
1. Vago L, Gojo I. Immune Escape and Immunotherapy of Acute Myeloid Leukemia. J Clin Invest (2020) 130(4):1552–64. doi: 10.1172/jci129204
2. Vasu S, Kohlschmidt J, Mrózek K, Eisfeld AK, Nicolet D, Sterling LJ, et al. Ten-Year Outcome of Patients With Acute Myeloid Leukemia Not Treated With Allogeneic Transplantation in First Complete Remission. Blood Adv (2018) 2(13):1645–50. doi: 10.1182/bloodadvances.2017015222
3. Murre C, Waldmann RA, Morton CC, Bongiovanni KF, Waldmann TA, Shows TB, et al. Human Gamma-Chain Genes are Rearranged in Leukaemic T Cells and Map to the Short Arm of Chromosome 7. Nature (1985) 316(6028):549–52. doi: 10.1038/316549a0
4. Shi L, Chen S, Zha X, Xu Y, Xu L, Yang L, et al. Enhancement of the Tcrζ Expression, Polyclonal Expansion, and Activation of T Cells From Patients With Acute Myeloid Leukemia After IL-2, IL-7, and IL-12 Induction. DNA Cell Biol (2015) 34(7):481–8. doi: 10.1089/dna.2015.2810
5. Forster A, Huck S, Ghanem N, Lefranc MP, Rabbitts TH. New Subgroups in the Human T Cell Rearranging V Gamma Gene Locus. EMBO J (1987) 6(7):1945–50. doi: 10.1002/j.1460-2075.1987.tb02456.x
6. Takihara Y, Tkachuk D, Michalopoulos E, Champagne E, Reimann J, Minden M, et al. Sequence and Organization of the Diversity, Joining, and Constant Region Genes of the Human T-Cell Delta-Chain Locus. Proc Natl Acad Sci USA (1988) 85(16):6097–101. doi: 10.1073/pnas.85.16.6097
7. Rabbitts TH, Lefranc MP, Stinson MA, Sims JE, Schroder J, Steinmetz M, et al. The Chromosomal Location of T-Cell Receptor Genes and a T Cell Rearranging Gene: Possible Correlation With Specific Translocations in Human T Cell Leukaemia. EMBO J (1985) 4(6):1461–5. doi: 10.1002/j.1460-2075.1985.tb03803.x
8. Lefranc MP, Rabbitts TH. The Human T-Cell Receptor Gamma (TRG) Genes. Trends Biochem Sci (1989) 14(6):214–8. doi: 10.1016/0968-0004(89)90029-7
9. Huck S, Dariavach P, Lefranc MP. Variable Region Genes in the Human T-Cell Rearranging Gamma (TRG) Locus: V-J Junction and Homology With the Mouse Genes. EMBO J (1988) 7(3):719–26. doi: 10.1002/j.1460-2075.1988.tb02868.x
10. Li Y, Chen S, Yang L, Li B, Chan JY, Cai D. TRGV and TRDV Repertoire Distribution and Clonality of T Cells From Umbilical Cord Blood. Transpl Immunol (2009) 20(3):155–62. doi: 10.1016/j.trim.2008.10.010
11. Zhang XM, Tonnelle C, Lefranc MP, Huck S. T Cell Receptor Gamma cDNA in Human Fetal Liver and Thymus: Variable Regions of Gamma Chains Are Restricted to V Gamma I or V9, Due to the Absence of Splicing of the V10 and V11 Leader Intron. Eur J Immunol (1994) 24(3):571–8. doi: 10.1002/eji.1830240312
12. Barros MS, de Araújo ND, Magalhães-Gama F, Pereira Ribeiro TL, Alves Hanna FS, Tarragô AM, et al. γδ T Cells for Leukemia Immunotherapy: New and Expanding Trends. Front Immunol (2021) 12:729085. doi: 10.3389/fimmu.2021.729085
13. Fichtner AS, Ravens S, Prinz I. Human γδ TCR Repertoires in Health and Disease. Cells (2020) 9(4):800. doi: 10.3390/cells9040800
14. Li Y, Li G, Zhang J, Wu X, Chen X. The Dual Roles of Human γδ T Cells: Anti-Tumor or Tumor-Promoting. Front Immunol (2020) 11:619954. doi: 10.3389/fimmu.2020.619954
15. Zheng R, Wu X, Huang X, Chen Y, Yang Q, Li Y, et al. Gene Expression Pattern of Treg and TCR Vγ Subfamily T Cells Before and After Specific Immunotherapy in Allergic Rhinitis. J Transl Med (2014) 12:24. doi: 10.1186/1479-5876-12-24
16. Beetz S, Wesch D, Marischen L, Welte S, Oberg HH, Kabelitz D. Innate Immune Functions of Human Gammadelta T Cells. Immunobiology (2008) 213(3-4):173–82. doi: 10.1016/j.imbio.2007.10.006
17. Huang J, Tan J, Chen Y, Huang S, Xu L, Zhang Y, et al. A Skewed Distribution and Increased PD-1+Vβ+CD4+/CD8+ T cells in Patients With Acute Myeloid Leukemia. J Leukoc Biol (2019) 106(3):725–32. doi: 10.1002/jlb.Ma0119-021r
18. Jin Z, Luo Q, Lu S, Wang X, He Z, Lai J, et al. Oligoclonal Expansion of TCR Vδ T Cells may be a Potential Immune Biomarker for Clinical Outcome of Acute Myeloid Leukemia. J Hematol Oncol (2016) 9(1):126. doi: 10.1186/s13045-016-0353-3
19. Chen XM, Zhang T, Qiu D, Feng JY, Jin ZY, Luo Q, et al. Gene Expression Pattern of TCR Repertoire and Alteration Expression of IL-17A Gene of γδ T Cells in Patients With Acute Myocardial Infarction. J Transl Med (2018) 16(1):189. doi: 10.1186/s12967-018-1567-7
20. Geng S, Weng J, Du X, Lai P, Huang X, Chen S, et al. Comparison of the Distribution and Clonal Expansion Features of the T-Cell γδ Repertoire in Myelodysplastic Syndrome-RAEB and RAEB With Progression to AML. DNA Cell Biol (2012) 31(10):1563–70. doi: 10.1089/dna.2012.1769
21. Zhang X, Chen S, Yang L, Li B, Zhu K, Li Y. The Feature of TRGV and TRDV Repertoire Distribution and Clonality in Patients With Immune Thrombocytopenic Purpura. Hematology (2009) 14(4):237–44. doi: 10.1179/102453309x439755
22. Xuan L, Wu X, Wu M, Zhang Y, Liu H, Fan Z, et al. Effect of Granulocyte Colony-Stimulating Factor Mobilization on the Expression Patterns, Clonality and Signal Transduction of TRAV and TRBV Repertoire. Immunobiology (2012) 217(8):816–22. doi: 10.1016/j.imbio.2012.05.004
23. Jin Z, Wu X, Chen S, Yang L, Liu Q, Li Y. Distribution and Clonality of the Vα and Vβ T-Cell Receptor Repertoire of Regulatory T Cells in Leukemia Patients With and Without Graft Versus Host Disease. DNA Cell Biol (2014) 33(3):182–8. doi: 10.1089/dna.2013.2277
24. Stams WA, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, van Wering ER, et al. Sensitivity to L-Asparaginase Is Not Associated With Expression Levels of Asparagine Synthetase in T(12;21)+ Pediatric ALL. Blood (2003) 101(7):2743–7. doi: 10.1182/blood-2002-08-2446
25. Xu L, Wu H, Wu X, Li Y, He D. The Expression Pattern of Bcl11a, Mdm2 and Pten Genes in B-Cell Acute Lymphoblastic Leukemia. Asia Pac J Clin Oncol (2018) 14(2):e124–e8. doi: 10.1111/ajco.12690
26. Liao Z, Lv X, Liu S, He Z, Chen S, Wang L, et al. Different Aberrant Expression Pattern of Immune Checkpoint Receptors in Patients With PTCL and NK/T-CL. Asia Pac J Clin Oncol (2018) 14(5):e252–e8. doi: 10.1111/ajco.12850
27. Tan J, Chen S, Lu Y, Yao D, Xu L, Zhang Y, et al. Higher PD-1 Expression Concurrent With Exhausted CD8+ T Cells in Patients With De Novo Acute Myeloid Leukemia. Chin J Cancer Res (2017) 29(5):463–70. doi: 10.21147/j.issn.1000-9604.2017.05.11
28. Dar AA, Patil RS, Chiplunkar SV. Insights Into the Relationship Between Toll Like Receptors and Gamma Delta T Cell Responses. Front Immunol (2014) 5:366. doi: 10.3389/fimmu.2014.00366
29. Alnaggar M, Xu Y, Li J, He J, Chen J, Li M, et al. Allogenic Vγ9vδ2 T Cell as New Potential Immunotherapy Drug for Solid Tumor: A Case Study for Cholangiocarcinoma. J Immunother Cancer (2019) 7(1):36. doi: 10.1186/s40425-019-0501-8
30. Xu Y, Xiang Z, Alnaggar M, Kouakanou L, Li J, He J, et al. Allogeneic Vγ9vδ2 T-Cell Immunotherapy Exhibits Promising Clinical Safety and Prolongs the Survival of Patients With Late-Stage Lung or Liver Cancer. Cell Mol Immunol (2021) 18(2):427–39. doi: 10.1038/s41423-020-0515-7
31. Zhao Y, Niu C, Cui J. Gamma-Delta (γδ) T Cells: Friend or Foe in Cancer Development? J Transl Med (2018) 16(1):3. doi: 10.1186/s12967-017-1378-2
32. Minculescu L, Marquart HV, Ryder LP, Andersen NS, Schjoedt I, Friis LS, et al. Improved Overall Survival, Relapse-Free-Survival, and Less Graft-Vs.-Host-Disease in Patients With High Immune Reconstitution of TCR Gamma Delta Cells 2 Months After Allogeneic Stem Cell Transplantation. Front Immunol (2019) 10:1997. doi: 10.3389/fimmu.2019.01997
33. Kasakovski D, Xu L, Li Y. T Cell Senescence and CAR-T Cell Exhaustion in Hematological Malignancies. J Hematol Oncol (2018) 11(1):91. doi: 10.1186/s13045-018-0629-x
34. Chen C, Liu S, Jiang X, Huang L, Chen F, Wei X, et al. Tumor Mutation Burden Estimated by a 69-Gene-Panel is Associated With Overall Survival in Patients With Diffuse Large B-Cell Lymphoma. Exp Hematol Oncol (2021) 10(1):20. doi: 10.1186/s40164-021-00215-4
35. Ma Y, Lei H, Tan J, Xuan L, Wu X, Liu Q. Characterization of γδ Regulatory T Cells From Peripheral Blood in Patients With Multiple Myeloma. Biochem Biophys Res Commun (2016) 480(4):594–601. doi: 10.1016/j.bbrc.2016.10.098
36. Xuan L, Wu X, Zhang Y, Fan Z, Ling Y, Huang F, et al. Granulocyte Colony-Stimulating Factor Affects the Distribution and Clonality of TRGV and TRDV Repertoire of T Cells and Graft-Versus-Host Disease. J Transl Med (2011) 9:215. doi: 10.1186/1479-5876-9-215
37. Xu Y, Xu L, Chen C, Zhang Y, Zeng C, Jin Z, et al. Age-Related Immune Profile of the T Cell Receptor Repertoire, Thymic Recent Output Function, and miRNAs. BioMed Res Int (2020) 2020:5910823. doi: 10.1155/2020/5910823
38. Jin Z, Ye W, Lan T, Zhao Y, Liu X, Chen J, et al. Characteristic of TIGIT and DNAM-1 Expression on Foxp3+ γδ T Cells in AML Patients. BioMed Res Int (2020) 2020:4612952. doi: 10.1155/2020/4612952
39. Jin Z, Lan T, Zhao Y, Du J, Chen J, Lai J, et al. Higher TIGIT(+)CD226(-) γδ T Cells in Patients With Acute Myeloid Leukemia. Immunol Invest (2020) 51(1):40–50. doi: 10.1080/08820139.2020.1806868
40. Lo Presti E, Di Mitri R, Pizzolato G, Mocciaro F, Dieli F, Meraviglia S. γδ Cells and Tumor Microenvironment: A Helpful or a Dangerous Liason? J Leukoc Biol (2018) 103(3):485–92. doi: 10.1002/jlb.5mr0717-275rr
41. Godder KT, Henslee-Downey PJ, Mehta J, Park BS, Chiang KY, Abhyankar S, et al. Long Term Disease-Free Survival in Acute Leukemia Patients Recovering With Increased Gammadelta T Cells After Partially Mismatched Related Donor Bone Marrow Transplantation. Bone Marrow Transplant (2007) 39(12):751–7. doi: 10.1038/sj.bmt.1705650
42. Perko R, Kang G, Sunkara A, Leung W, Thomas PG, Dallas MH. Gamma Delta T Cell Reconstitution Is Associated With Fewer Infections and Improved Event-Free Survival After Hematopoietic Stem Cell Transplantation for Pediatric Leukemia. Biol Blood Marrow Transplant (2015) 21(1):130–6. doi: 10.1016/j.bbmt.2014.09.027
Keywords: acute myeloid leukemia, γδ T cells, TRGV repertoire, clonality, prognosis
Citation: Kong X, Zheng J, Liu X, Wang W, Jiang X, Chen J, Lai J, Jin Z and Wu X (2022) High TRGV 9 Subfamily Expression Marks an Improved Overall Survival in Patients With Acute Myeloid Leukemia. Front. Immunol. 13:823352. doi: 10.3389/fimmu.2022.823352
Received: 27 November 2021; Accepted: 24 January 2022;
Published: 10 February 2022.
Edited by:
Xiaoli Wu, Tianjin University, ChinaReviewed by:
Ahmed Gaballa, Karolinska Institutet (KI), SwedenWinfried F. Pickl, Medical University of Vienna, Austria
Copyright © 2022 Kong, Zheng, Liu, Wang, Jiang, Chen, Lai, Jin and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenyi Jin, amluemhlbnlpam51QDE2My5jb20=; Xiuli Wu, c2l1bGllckAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Xueting Kong1†
Xueting Kong1† Xiaxin Liu
Xiaxin Liu Zhenyi Jin
Zhenyi Jin Xiuli Wu
Xiuli Wu