- 1Department of Sport Rehabilitation, Shanghai University of Sport, Shanghai, China
- 2Department of Rehabilitation Medicine, Shanghai Shangti Orthopaedic Hospital, Shanghai, China
- 3College of Kinesiology, Shenyang Sport University, Shenyang, China
Rheumatoid arthritis (RA) is one of the most common autoimmune diseases that affect synovitis, bone, cartilage, and joint. RA leads to bone and cartilage damage and extra-articular disorders. However, the pathogenesis of RA is still unclear, and the lack of effective early diagnosis and treatment causes severe disability, and ultimately, early death. Accumulating evidence revealed that the regulatory network that includes long non-coding RNAs (lncRNAs)/circular RNAs (circRNAs), micro RNAs (miRNAs), and messenger RNAs (mRNA) plays important roles in regulating the pathological and physiological processes in RA. lncRNAs/circRNAs act as the miRNA sponge and competitively bind to miRNA to regulate the expression mRNA in synovial tissue, FLS, and PBMC, participate in the regulation of proliferation, apoptosis, invasion, and inflammatory response. Thereby providing new strategies for its diagnosis and treatment. In this review, we comprehensively summarized the regulatory mechanisms of lncRNA/circRNA-miRNA-mRNA network and the potential roles of non-coding RNAs as biomarkers and therapeutic targets for the diagnosis and treatment of RA.
Introduction
Rheumatoid arthritis (RA) is the most common autoimmune diseases with chronic, systemic inflammatory responses; it is characterized by persistent synovitis, bone, cartilage, and joint destruction (1, 2). Symmetrical pain, stiffness, and swelling of one or more joints are the main clinical symptoms of RA; and the joints involved are those in the hands, wrists, feet, and knees (3). The incidence of RA is very high affecting approximately 1% of the world population (4). As the disease progresses, it can lead to bone and cartilage damage and extra-articular disorders, such as cardiovascular disease (5) and organ damage (6); without active clinical treatment, RA can lead to severe disability, and ultimately, early death (2).
RA is a multifactorial and heterogeneous disease; accumulating evidence have documented that genetics is one of its key factors (7). Recently, the influences of environmental factors and gene-environment interactions have been revealed, providing new clues on disease pathogenesis (8). Despite the tremendous research efforts made in the past few years, the precise pathogenesis and etiology are not yet completely elucidated due to the complexity of the disease, resulting in the early diagnosis of RA remains difficult, and existing common serum biomarkers still lack specificity (9, 10). Approximately one third of patients with RA are serologically negative before the onset of severe clinical symptoms (2, 11); thus, many patients miss the best time for early treatment, leading to severe bone and cartilage damage, as well as permanent disability.
Non-coding RNA (ncRNA) is a class of RNA that is unable to encode proteins which mainly include microRNA (miRNA), long non-coding RNA (lncRNA), circular RNA (circRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA(snoRNA), small interfering RNA(siRNA), short hairpin RNA(shRNA) and Piwi-interactingRNA (piRNA) (12). In recent years, ncRNA has received much attention (13). ncRNA formerly known as transcriptional noise (14), however, accumulated evidence suggested that ncRNAs could serve as master regulators in a series of biological processes, such as transcription, splicing, and translation; they participate in the development and progression of many diseases, including RA (15, 16). The most commonly studied ncRNAs in RA are miRNAs (17), circRNAs (18), and lncRNAs (19). Numerous studies have shown that miRNAs, lncRNAs, and circRNAs are differentially expressed and participate in regulating the pathological and physiological processes in RA (20, 21). Significantly, new evidence indicated that lncRNAs and circRNAs compete to bind to miRNAs by competitive endogenous RNA (ceRNA) network, and they regulate their target mRNAs in the biological processes of many diseases (22, 23). This crosstalk includes lncRNA/miRNA and circRNA/miRNA, which are essential for the effective regulation of cellular signaling. In this review, we highlight the functional interactions between lncRNAs/circRNAs and miRNAs and describe the crosstalk in the lncRNA/circRNA-miRNA-mRNA axis of RA
miRNAs and RA
miRNAs belong to a class of short ncRNA molecules that are approximately 22–23 nucleotides in length and are generated endogenously. Mammalian miRNA genes are found in the introns; only approximately 20% miRNAs are found in exons or the combination of exons and introns (24). In general, most miRNAs are named by the combination of miR and a designated number and act as negative regulators; they silence their complementary mRNA expression by cleavage or translation suppression (25). The biosynthesis of miRNAs could be divided into two stages from nucleus to cytoplasm. In the nucleus, miRNA genes located in protein gene introns are transcribed to primary miRNA. Then, they are broken down and converted into pre-miRNA with approximately ~60 nucleotide by Drosha ribonuclease III and diGeorge syndrome critical region 8 (DGCR8); Drosha works by trimming 5’ and 3’ tails (18, 26). After the initial cut, pre-miRNA will translocate to the cytoplasm with the help of the membrane protein exportin 5. In the cytoplasm, miRNA-miRNA duplexes are formed by a Dicer endoribonuclease III known as an endonuclease, which dissociates the secondary structure. After the second initial cut, mature miRNA is formed. Subsequently, one of the miRNA duplexes combine with argonaute protein and transform into RNA-induced silencing complexes (RISCs), and the other is commonly wasted. Eventually, the mature RISC inhibits the translation and expression of target mRNA genes, resulting in the degradation of the message (24, 27).
The biological role of miRNA has been studied extensively for nearly 30 years. Research has shown that miRNAs target a third of all human genes that target mRNA genes and some genes that target DNA (25). In most instances, miRNAs act as inhibitory regulator at the post-transcriptional level by repressing the expression and translation of target mRNA genes, but there are instances when they accelerate the expression level of target genes (28). The miRNAs are highly tissue-specific and differentially expressed in different tissues, and these traits are related to the physiological development and pathological process of a variety of diseases, including cancer (29), stroke (30), heart disease (31), musculoskeletal disease (32), and autoimmune disease (33, 34).
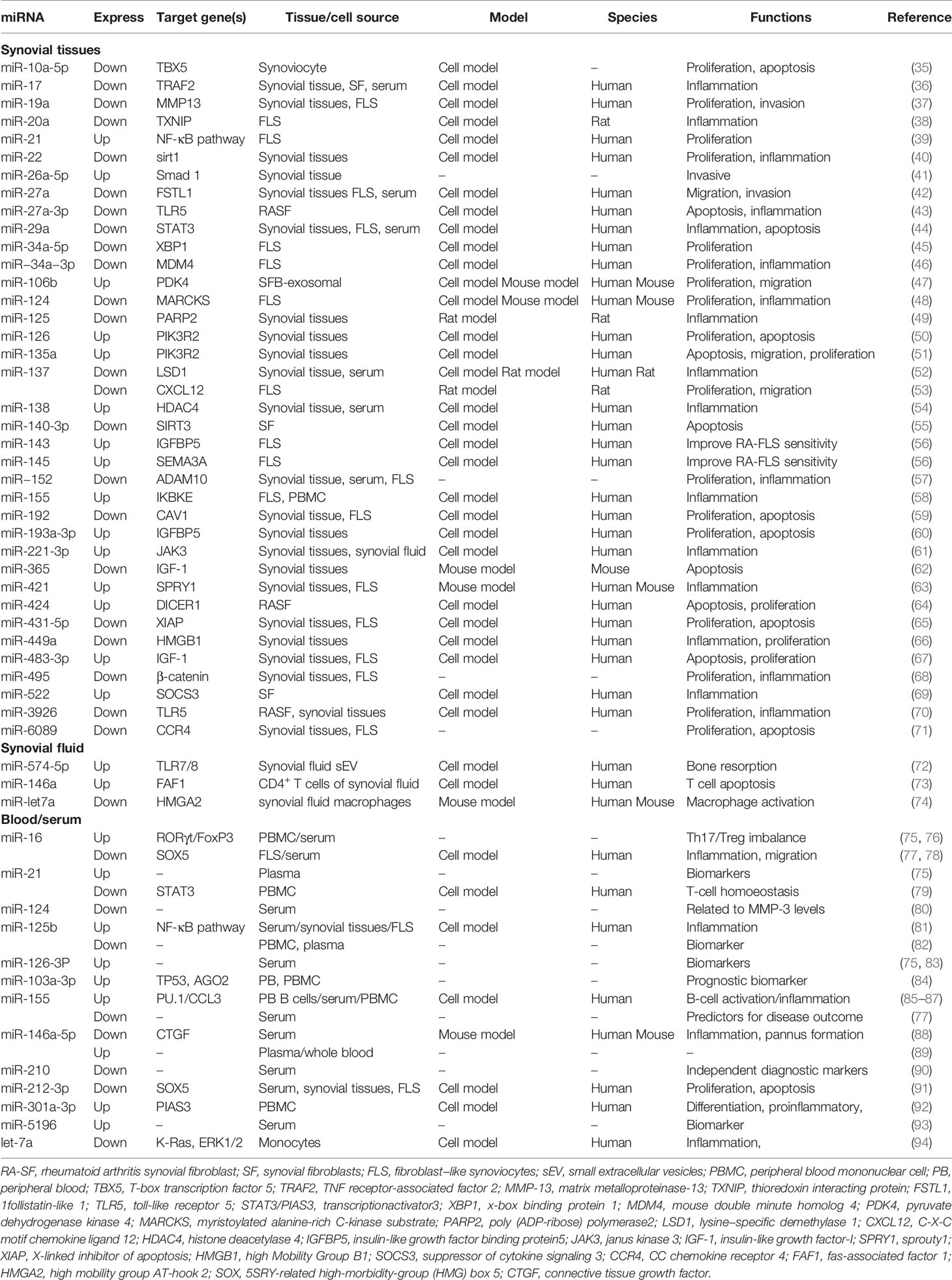
Studies confirmed that miRNAs in synovial tissue, synovial fluid, and blood of patients with RA showed significantly abnormal expression compared with those of healthy individuals (7) (Table 1). Synovial tissue is an important part of the knee joint, which mostly includes synovial macrophages and fibroblast-like synoviocytes (FLS) (95); synoviocyte proliferation, invasion, and migration are essential for the RA pathology (96). Among synovial and FLS miRNAs, miR-21 (39), miR-26a-5p (41), miR-126 (50), miR-135a (51), miR-138 (54), miR-143 (56), miR-145 (56), miR-155 (58), and miR-421 (63) are overexpressed, whereas miR-19a (37), miR-20a (38), miR-22 (40), miR-27a (42), miR-29a (44), miR-34a (45), miR-137 (53), miR-140-3p (55), miR−152 (57), and miR-495 (68) are down-regulated. The disturbed miRNAs enhance the expression level of proinflammatory cytokine (IL-6, IL-8, TNF-α, and IL-1β) and enzymes that erode the bone matrix (MMP-1 and MMP-3) by affecting Wnt (97, 98), NF-κB (81, 99), JAK/STAT (48, 100), and TLR (101, 102) pathways. Significantly, the disturbed synovial fibroblast-derived exosomal miRNAs were discovered in recent years. Liu et al. demonstrated that the expression level of miRNA miR-106b was significantly increased in synovial fluid-derived exosomes of RA, and it could target the pyruvate dehydrogenase kinase 4 (PDK4) gene; it could attenuate RA progression by regulating chondrocyte proliferation and migration (47). Furthermore, research found that RA synovial fibroblast (RASF)-exosomal miR-146a, miR-155, miR-323a, and miR-1307 are also involved in inducing local inflammation and attenuating octeoclastogenesis in RA (103).
As important diagnostic markers for RA, blood miRNAs have been studied extensively. The greatest number of studies focused on miR-146 (miR-146a and miR-146b) and miR-155 (104). However, their roles in RA are still controversial. miR-146a was described to be up-regulated in peripheral blood (PB) and peripheral blood mononuclear cells (PBMCs) from patients with RA (104–106); it is involved in the production of persistent proinflammatory cytokine and disturbance of the balance of Th17-Tregs (107). However, the expression level of miR-146a-5p in synovial fibroblast of RA patients is significantly down-regulated; miR-146a-5p could decrease inflammatory mediators, inhibit angiogenesis, and delay RA progression (88). Furthermore, the anti-rheumatic drugs, such as TNF inhibitors and methotrexate (MTX), could increase miR-146a-5p expression, suggesting that miR-146a-5p may be a potential novel biomarker for predicting and monitoring therapy outcome (87, 108). The roles of miR-155, miR-125b, miR-16, and miR-21 are also ambiguous. Some authors report that miR-155 and miR-16 are over-expressed in serum and PBMC (76, 87), and others report their down-regulation in serum (77, 78). miR-21 and miR-125b are over-expressed in plasma or serum (75, 81) but lowly expressed in PBMC (79, 82). These findings all suggest the tissue specificity of miRNA expression, and its biological function needs to be further studied. The dysregulation of miRNAs is also found in serum exosomes of patients with RA. Wang et al. found that the expression level of miRNA miR-17 was high in RA-exosomes by microarray analysis and real-time PCR; transforming growth factor beta receptor II (TGFBR II) was the direct target. miR-17 could dispute the homeostasis of Tregs by the participation of TGFBR II in the pathogenesis of RA (109). The abundant miRNAs in RA-exosomes provide a new idea and direction for the pathogenesis of RA, which is worthy of further study.
lncRNAs and RA
lncRNA is a newly identified RNA transcript with a length of more than 200 nucleotides; it has little or no protein-coding potential (15). There are approximately 92 343 lncRNA genes in humans, which are far more than the protein-coding genes (110), and the number is still increasing (111). There are five main categories classified by the localization between lncRNA and the closest protein-coding gene, as follows: intergenic, sense, antisense, intronic, and bidirectional (104, 112). lncRNA was a by-product of RNA polymerase II transcription and was originally called transcriptional noise of the genome; it was considered to have no biological function (113). However, subsequent studies found that lncRNA could regulate target gene expression at each stage from transcriptional and post-transcriptional to post-translational levels (114), and it is also a crucial regulator of a range of cellular transformation processes, such as apoptosis and intracellular transport (24). Furthermore, as a transcription inhibitor, lncRNA could influence the stability of miRNAs and RNA binding proteins as miRNA sponges, and it is involved in the epigenetic modification of DNA (115).
The functional role of dysregulation lncRNAs in the physiological development and pathological process of tumors (116) and cardiovascular disease (117) have been discovered, and the role of lncRNAs in autoimmune diseases was also revealed gradually, but its role is still unclear (118). Studies have discovered that lncRNAs are involved in regulating the development and differentiation of various immune cells, such as thymus T lymphocytes, macrophages, bone marrow B lymphocytes, and dendritic cells (119). lncRNAs are abnormally expressed in RA-associated immune cells and play a crucial role in the physiological and pathophysiological processes.
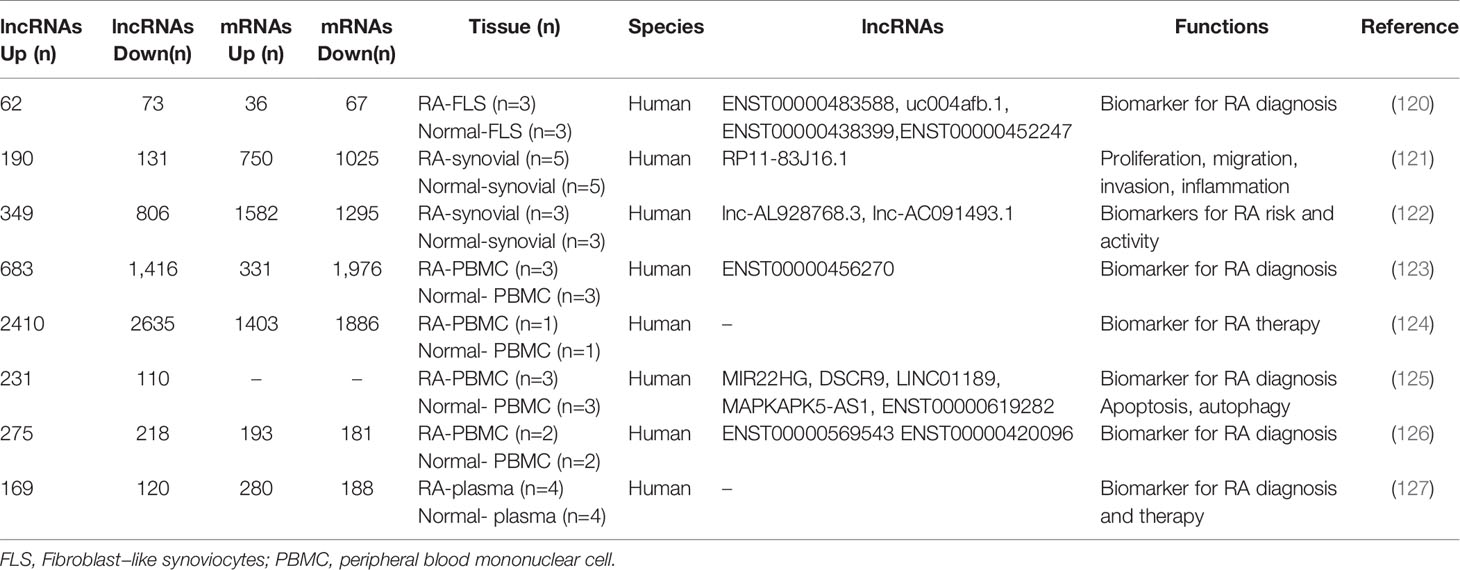
In recent years, microarray technology has been widely used in the study of RA regulatory network; increasing evidence shows the aberrant expression of lncRNAs in FLS, PBMS, plasma, and synovial tissues in RA patients (113) (Table 2). Zhang et al. found 135 differentially expressed lncRNAs (62 up-regulated and 73 down-regulated) and 103 differentially expressed mRNAs (36 up-regulated genes and 67 down-regulated genes) in three pairs of FLS samples through genome-wide analysis of the expression profiles (120). Luo et al. identified 2,410 up-regulated and 2,635 down-regulated lncRNAs and 1,403 up-regulated and 1,886 down-regulated mRNAs in PBMCs via microarrays. GO category and KEGG pathway analyses demonstrated that these differentially expressed transcripts are associated with multiple biological processes and signaling pathway, such as T cell receptor signaling pathway and TNF signaling pathway (124). Qin et al. also found that there are approximately 289 differentially expressed lncRNAs and 468 mRNAs in the plasma (127).
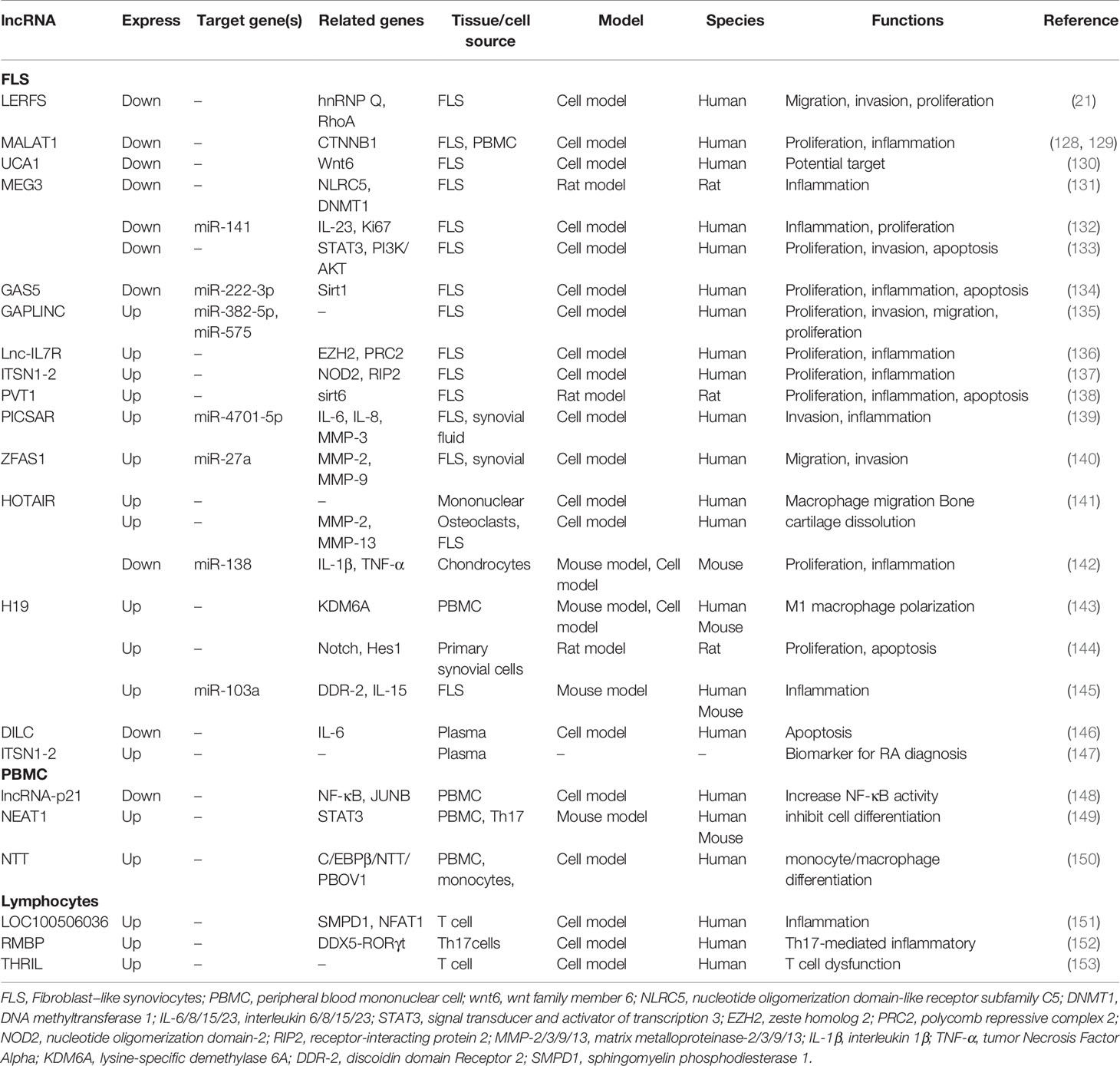
Many aberrantly expressed lncRNAs are reported by microarray analysis, but only a small number of differentially expressed genes participate in the development and progression of RA (Table 3). Research demonstrated that lncRNA LERFS (21), MALAT1 (128), UCA1 (130), GAS5 (154), and MEG3 (132) are down-regulated in FLS, whereas GAPLINC (135), Lnc-IL7R (136), ITSN1-2 (137), PVT1 (138), H19 (145), ZFAS1 (155), and PICSAR (139) are up-regulated. The dysregulation lncRNAs are involved in regulating synovial inflammation and cellular biological behavior of RA FLSs, including proliferation, migration, and invasion. lncRNAs may be promising therapeutic targets or biomarkers. Notably, lncRNA HOTAIR shows obvious tissue specificity in different RA tissues. Zhang et al. revealed that HOTAIR was lowly expressed in chondrocytes compared with normal individuals, and miR-138 was the direct target of HOTAIR. HOTAIR usually acts as protective regulator to delay the progression of RA by inhibiting inflammatory response and inactivating the NF-κB signaling pathway (142). Song and his colleagues found that in the differentiated osteoclasts and synoviocytes, HOTAIR was also down-regulated. It could promote the dissolution of bone and cartilage matrix by regulating MMP-2 and MMP-13 expressions. However, in blood mononuclear cells and serum exosomes, HOTAIR was overexpressed and participated in the migration of active macrophage (141). Furthermore, HOTAIR was also found to be up-regulated in serum. Based on these studies, we found that HOTAIR is easy to obtain and detect in blood and is expressed stably, making it a promising biomarker for RA diagnosis.
PBMC is a key component of host defense response and is readily available. However, there are few studies on lncRNA in PBMC at present. Existing studies demonstrated that NEAT1 (149), HIX003209 (156), and NTT (150) are up-regulated in PBMC, whereas H19 (143) and lincRNA-p21 (148) are down-regulated. Regulation of inflammation is an important mechanism of these aberrantly expressed lncRNAs. Yan et al. found that HIX003209 showed a significantly increased expression in PBMC from patients with RA; it could target miR-6089 directly and promote inflammation by regulating the TLR4/NF-κB pathway in macrophages. Furthermore, similar results were observed in lipopolysaccharide-mediated cell models; the overexpressed HIX003209 could function as a positive regulator of proliferation and activation (156). Spurlock et al. found that lincRNA-p21 was down-regulated and NF-κB activation marker phosphorylated p65 was up-regulated by analyzing blood samples and cell culture models from patients with RA; lincRNA-p21 could inhibit NF-κB activity directly. The regulation of lincRNA-p21 was one of the important mechanisms underlying the action of methotrexate against RA (148). Dysregulation lncRNAs, such as LOC100506036, THRIL, and RMBP, were also observed in lymphocytes and macrophages (157). However, their exact functions and mechanisms are still unclear and need further study.
circRNAs and RA
circRNAs are novel endogenous noncoding RNAs characterized by a closed circular structure and are approximately 500 ribonucleotides (nts) long (34); they include 1–5 exons without intervening introns (158). circRNAs are very stable because of their circular structure, which help them resist exonucleolytic decay through the cellular exosome ribonuclease complex. Studies have shown that circRNAs have a maximum half-life of 48 h, whereas linear mRNAs have only 4–9 h (159). Therefore, circRNAs are ideal biomarkers. The functions of circRNAs include the following: miRNA and RNA binding proteins (RBP) sponge; RNAP II elongation; and RNA maturation regulation (160). circRNAs are widely expressed in mammals and participate in the regulation of physiological and pathological processes for various diseases, such as cancer and osteoarticular and autoimmune diseases (161). Recently, many circRNA-related signaling pathways have been reported in autoimmune diseases, suggesting that circRNAs may serve as crucial immune regulators and potential biomarkers (118).
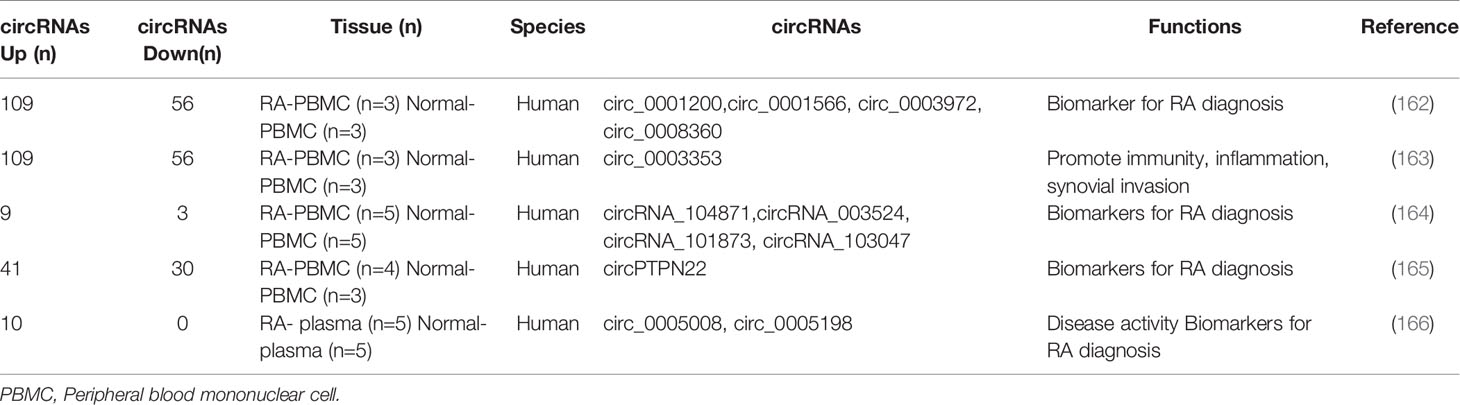
More aberrantly expressed circRNAs have been identified by gene microarray technology. A series of studies indicated that circRNAs were differentially expressed in PBMC and FLS (Table 4). Ouyang and his colleagues detected the expression of circRNA genes in PBMCs from 30 RA patients via quantitative real-time polymerase chain reaction (qRT-PCR). They confirmed that circRNAs circRNA_104871, circRNA_003524, circRNA_101873, and circRNA_103047 were up-regulated and may be promising biomarkers for diagnosis (164). Then, Ouyang et al. found that circRNAs were also differentially expressed in plasma. They clarified that circ_0005008 and circ_0005198 were overexpressed in the plasma of RA patients. Furthermore, circ_0005198 may target miR-4778-3p in RA-FLS (166). Wen et al. constructed a circRNA-miRNA network of differentially expressed genes in PBMC from patients with RA, and it contains 165 differentially expressed circRNAs and 63 differentially expressed miRNAs. After further RT-qPCR validation of the four significantly changed circRNAs (circRNA_0001200, circRNA_0001566, circRNA_0003972, and circRNA_0008360), they found that the expression was consistent with the results of sequencing, and these circRNAs may be promising biomarkers for diagnosis (162). After this study, Wen and his colleges further verified the presence of other circRNAs in PBMCs from patients with RA by high-throughput sequencing. The circ_0003353 and circ_0091685 were up-regulated, whereas circ_0005732, circ_0072428 were down-regulated. Then, the expression of circ_0003353 in fibroblast-like synoviocytes was further investigated for functional phenotype analysis; circ_0003353 was significantly highly expressed, it could promote of FLS inflammatory response and cell apoptosis, but inhibited cell proliferation (163).
Although a series of dysregulation circRNAs were found in RA, their downstream pathways in regulating autoimmunity and inflammation are still poorly revealed. Existing studies have shown that the functions of dysregulation circRNAs are involved in regulating synovial inflammation response and cellular biological behavior of RA FLSs, including proliferation, migration, invasion, and apoptosis (Table 5). Cai et al. identified that circ_0088194 was up-regulated in RA FLS; it could act as miR-766-3p sponge and promote the expression of downstream target gene MMP2, thereby facilitating the fibroblast−like synoviocytes’ invasion and migration. It may be a novel and promising target for RA (167). Qu et al. suggested that circ-AFF2 was up-regulated in synovial tissues and FLS of RA; circ-AFF2 could bind to the miRNA miR-650; it enhances the expression level of downstream target 2’,3’-cyclic nucleotide phosphodiesterase (CNP) and promotes fibroblast−like synoviocyte proliferation, inflammatory response, migration, and invasion (168). Another study found that the up-regulated circ-AFF2 was also associate with FLS cell progression and inflammatory response via the miR-375/TAB2 axis (169). The circRNA circ-Sirt1 was up-regulated in FLS and MH7A cells; it participates in the inhibition of cell proliferation, promotion of apoptosis, and reduction of inflammation in MH7A by targeting the miR-132-mediated Sirt1 pathway (170). Many differentially expressed circRNAs genes should be further validated in vivo and in vitro to find possible targets and pathways and to provide a theoretical support for the development of novel RA biomarkers and molecularly targeted therapeutic drugs.
The Crosstalk of lncRNAs, miRNAs, and mRNAs in RA
Mounting evidence demonstrated that lncRNAs could interact with miRNAs in regulating mRNA expression via various post-transcriptional mechanisms (177). Four potential mechanisms were associated with the interactions of lncRNA, miRNA, and mRNA (23, 178), as follows: (1) lncRNAs sponge miRNAs as ceRNAs. ceRNA is a kind of RNAs acting as molecular sponges by competing for miRNA response elements (MREs), it could hinder the expression of other target genes, such as mRNAs, by contending with miRNA (179). In the lncRNA-miRNA-mRNA networks, lncRNAs competitively bind to miRNAs as miRNA sponges, inhibit miRNA expression, and enhance the translation of target mRNA. For example, lncRNA PRNCR1 directly binds to miR-326, thereby functioning as a miR-326 “sponge” to up-regulate the expression level of fascin actin-bundling protein 1(FSCN1) in oral squamous cell carcinoma (180). (2) miRNAs degrade lncRNAs. miRNAs directly target lncRNAs and regulate their stability and abundance, thereby affecting different cell functions in physiological and pathological processes. For example, miRNA miR-9 target lncRNA MALAT1, thereby silencing Ago2 and regulating the stability of MALAT1 in the nucleus of L428 (181). (3) lncRNAs bind to target mRNAs and directly compete with miRNAs. The overexpressed LncRNA BACE1AS combines with mRNA BACE1 to reverse the downward trend by miR-485-5p (182). (4) lncRNAs produce miRNAs. Some lncRNAs could generate miRNAs, thereby regulating the expression of downstream genes. For example, lncRNA H19 generates miR-675, thereby inhibiting the expression of insulin-like growth factor 1 receptor(IGF1R) (183). Recently, the lncRNA-miRNA-mRNA networks were gradually revealed to be involved in rheumatic diseases, such as RA (14) and systemic lupus erythematosus (SLE) (184); they participate in biological and pathological processes of diseases. This finding has become a hot topic, thereby attracting increasing attention especially in RA.
lncRNAs act as miRNA sponge, and this is the most studied mechanism in RA. lncRNAs competitively bind to miRNAs to regulate the expression level of downstream genes in synovial tissue, FLS, and PBMC. They participate in the regulation of the proliferation, apoptosis, invasion, and inflammatory response of RA-FLSs (Figure 1). Zhao et al. detected the expression of lncRNA in the serum and synovial tissues from patients with joint trauma or RA, and they found that the expression of FOXD2-AS1 was significantly increased. FOXD2-AS1 acts as miR-331-3p sponge modulator of downstream target gene STAT3 expression. The overexpressed FOXD2-AS1 increased the proliferation and invasion of fibroblast-like synoviocytes through the miR-331-3p/PIAS3 pathway, suggesting that FOXD2-AS1 may be a promising target for RA treatment (185). Tang et al. reported that in the synovial tissues of RA patients, the expressions of lncRNA PVT1 and miRNA miR-145-5p were negatively correlated. In addition, significantly up-regulated PVT1 and down-regulated miR-145-5p were found in the RA-FLS model (induced by TNF-α). The knockdown of PVT1 could directly target miR-145-5p to inhibit the over-proliferation of RA-FLS and the activation of NF-κB signaling pathway and to regulate the proliferation, apoptosis, and inflammatory response of RA-FLS (186). Wang and his colleagues revealed that the overexpressed PVT1 could directly target miR-543, enhance the expression level of SCUBE2, and promote proliferation and IL-1β secretion while inhibiting the apoptosis rate of FLSs (187). The two abovementioned studies showed that lncRNA PVT1 has two downstream miRNA targets, miR-145-5p and miR-543. Similar to circRNAs, lncRNAs could contain one or more binding sites to miRNAs, thereby serving a sponging function. Studies have clarified that lncRNA NEAT1 could bind to miR-129/miR-204 (188), miR-410-3p (189), and miR-23a (190), thereby regulating cell viability, migration, and inflammation in fibroblast−like synoviocytes from RA. Furthermore, the up-regulated lncRNA ZFAS1 (155, 191), HIX003209 (156), and XIST (192) and down-regulated lncRNA LINC01197 (193), GAS5 (154), and OIP5-AS1 (194) also sponged miRNAs, thereby participating in cell proliferation, differentiation, apoptosis, and inflammation in synovial tissue of RA via ZFAS1/miR-2682-5p/ADAMTS9 axis, ZFAS1/miR-296-5p/MMP-15 axis, HIX003209/miR-6089/TLR4 axis, lncRNA XIST/let-7c-5p/STAT3 axis, LINC01197/miR-150/THBS2 axis, GAS5/miR-128-3p/HDAC4 axis, and OIP5-AS1/miR-448/PON1, respectively. The details are presented in Table S1.
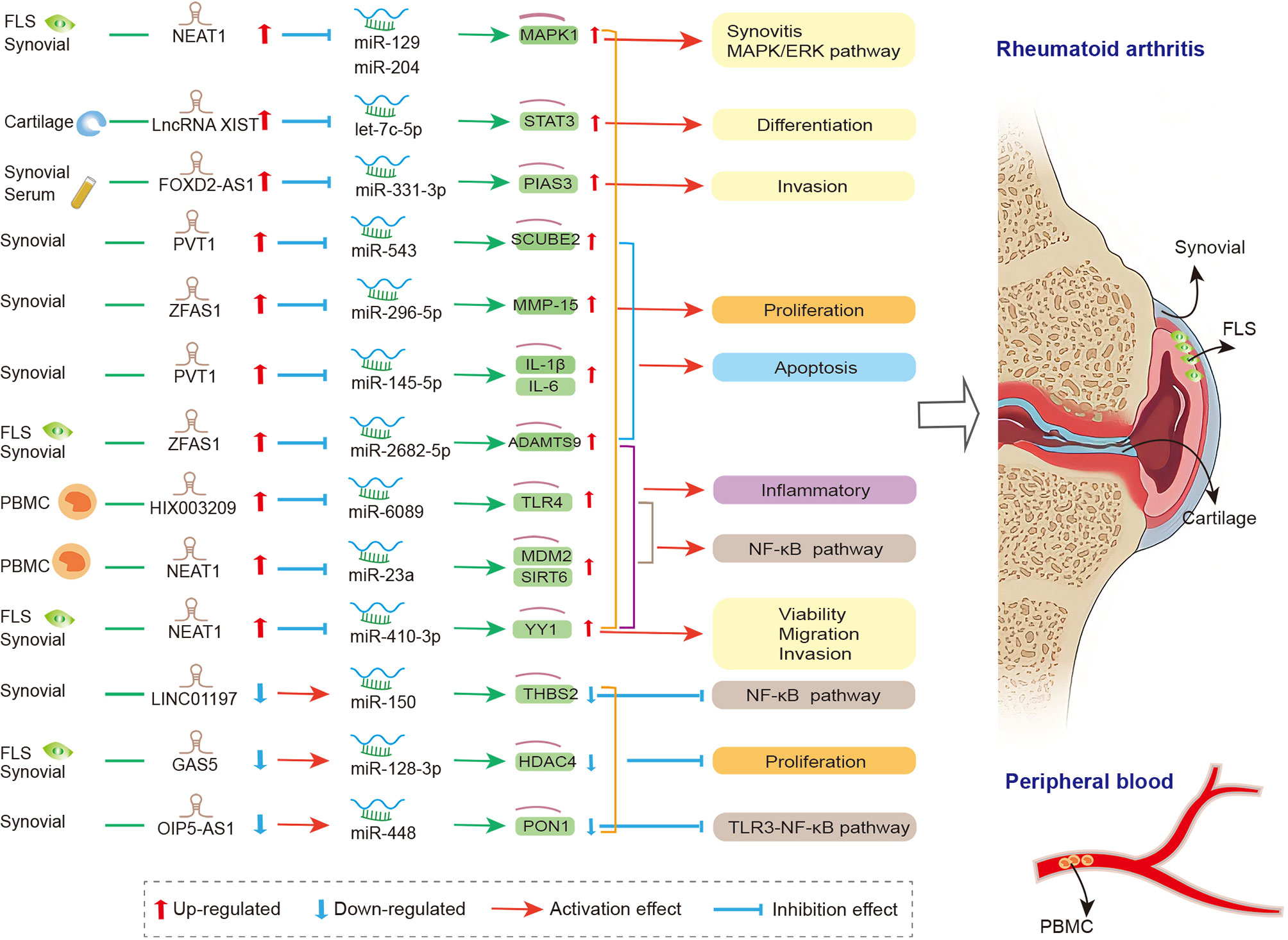
Figure 1 The crosstalk of lncRNA-miRNA-mRNA in RA. FLS, Fibroblast−like synoviocytes; PVT1, plasmacytoma variant translocation 1; SCUBE2, signal peptide-CUB-EGF-like containing protein 2; IL-1β, interleukin-1β; IL-6, interleukin-6; THBS2, thrombospondin-2; GAS5, growth arrest-specific transcript 5; HDAC4, histone deacetylase 4;PIAS3, protein inhibitor of activated STAT3; NEAT1, nuclear paraspeckle assembly transcript 1; MAPK1, mitogen-activated protein kinase 1; YY1, yin yang-1; mdm2, mouse double minute 2; Sirt6, sirtuin 6; PON1, paraoxonase 1; PBMC, peripheral blood mononuclear cell; NEAT1, nuclear paraspeckle assembly transcript 1; MDM2, murine double minute-2; SIRT6, sirtuin 6; MMP-15, matrix metalloproteinase-15; TLR4, toll-like-receptor 4; STAT3, signal transducer and activator of transcription 3.
Accumulating evidence has revealed the crucial role of lncRNA in modulating gene expression through the crosstalk of lncRNA-miRNA-mRNA in the immune and inflammatory pathways of RA. However, a new study found that lncRNA could be regulated by protein coding genes via the lncRNA-miRNA-mRNA axis, which was involved in the pathophysiologic process of RA (145). Mu et al. reported that lncRNA H19 was up-regulated, whereas miR-103a was down-regulated in RA-FLS. The expression of H19 could be greatly up-regulated when downstream target discoidin domain receptor 2(DDR-2) was activated, and miR-103a was the direct target of H19. Furthermore, miR-103a acts as a negative regulator that inhibits the expression of downstream genes interleukin 15 (IL-15) and dickkopf 1(DKK1). The study revealed that DDR-2 could exacerbate joint damage and inflammatory response via the H19-miR-103a network (145). Another study confirmed that protein coding gene forkhead box M1(FOXM1) was a new transcription regulator of lncRNA (195). Wang et al. indicated that FOXM1 and lncRNA LINC00152 were overexpressed in the FLS of patients with RA. FOXM1 overexpression could promote the expression of LINC00152, thereby acting as a transcription activator. LINC00152 could bind to miR-1270 and negatively regulate its expression. Intriguingly, the study found that the mRNA and protein levels of FOXM1 were positively regulated by LINC00152, and FOXM1 could also bind to LINC00152. Thus, LINC00152 and FOXM1 form a positive feedback loop in RA FLS. In summary, LINC00152 and FOXM1 could competitively bind with miR-1270; FOXM1/LINC00152/miR-1270 is a positive feedback loop involved in regulating the proliferation and apoptosis of RA-FLS (195). These studies revealed a novel molecular mechanism of pathophysiologic process in RA-FLS, thereby providing a new idea and direction for the future study of the pathological mechanism of RA.
The Crosstalk of circRNAs, miRNAs, and mRNAs in RA
For the past few years, the circRNA-miRNA-mRNA networks were gradually revealed. Studies have shown the presence of multiple miRNA complementary binding sites on circRNAs; circRNAs participate in the regulation of transcriptional and post-transcriptional levels by interacting with miRNA, thereby participating in the biological processes of many diseases (196, 197), such as central nervous system diseases (16), osteoarticular diseases (198), and cancer (199). Two main regulatory mechanisms of circRNA-miRNA-mRNA axis exist, as follows. 1) circRNAs sponge miRNAs. The “sponging” function reveals the regulatory mechanism, i.e., circRNAs may act as mRNA expression regulators by targeting seed sequences, thereby inhibiting the expression of miRNA. circRNA molecules usually contain one or more binding sites to which miRNA binds, thereby serving the sponging function (16). The circ_POLA2/miR-326/GNB1 axis could regulate lung cancer cell stemness and progression. Mechanistically, circ_POLA2 sponging miR-326 functioned as a ceRNA, thereby negatively regulating the expression of miR-326 target GNB1 (200). 2) miRNAs mediate circRNAs. miRNAs target circRNAs, thereby regulating the expression of downstream mRNA genes. miRNA miRNA-1224 could mediate the expression of circRNA circRNA-Filip1l by targeting downstream gene Ubr5, which is involved in the regulation of nociception (201).
The crosstalk of circRNAs, miRNAs, and mRNAs was also demonstrated in physiopathological process of RA (174), and the mechanism is that circRNAs act as the miRNA sponge and competitively bind to miRNA, thereby participating in the regulation of downstream genes in FLS, synovial tissue, and PBMC (Figure 2). In FLSs from patients with RA, Luo and his colleagues detected the expression levels of circRNA and miRNA via qRT-PCR and verified the interaction between them via dual-luciferase reporter assay. They indicated that the circMAPK9/miR-140-3p/PPM1A axis was involved in inhibiting inflammatory response, proliferation and migration and accelerating the apoptosis of fibroblast-like synoviocytes. The circRNA circMAPK9 was high expressed and targets miRNA miR-140-3p, and mRNA PPM1A was downstream target gene of miR-140-3p. The knocked down circMAPK9 sponged miR-140-3p and down-regulated PPM1A expression, thereby regulating the biological process of RA-FLSs (172). Hao et al. found that circRNA circ_0008360 was down-regulated in synovial tissue. And then, they used bioinformatics analysis to obtain a preliminary prediction. The results indicated that miR-135b-5p and histone deacetylase 4 (HDAC4) interacted with circ_0008360. They demonstrated that the circ_0008360 sponging miR-135b-5p positively regulated HDAC4 expression, thereby inhibiting the proliferation, migration, and inflammation and facilitating the apoptosis of RA-FLSs (174). Yang et al. found a high expression level of circRNA_09505 in PBMC from patients with RA. The in vitro macrophage cell model and in vivo collagen-induced arthritis (CIA) mice model indicated that circRNA_09505 could act as miR-6089 sponge through ceRNA mechanism, thereby activating IκBα/NF-κB signaling pathway, promoting miR-6089 direct target AKT1 expression, and exacerbating arthritis and inflammation (175).
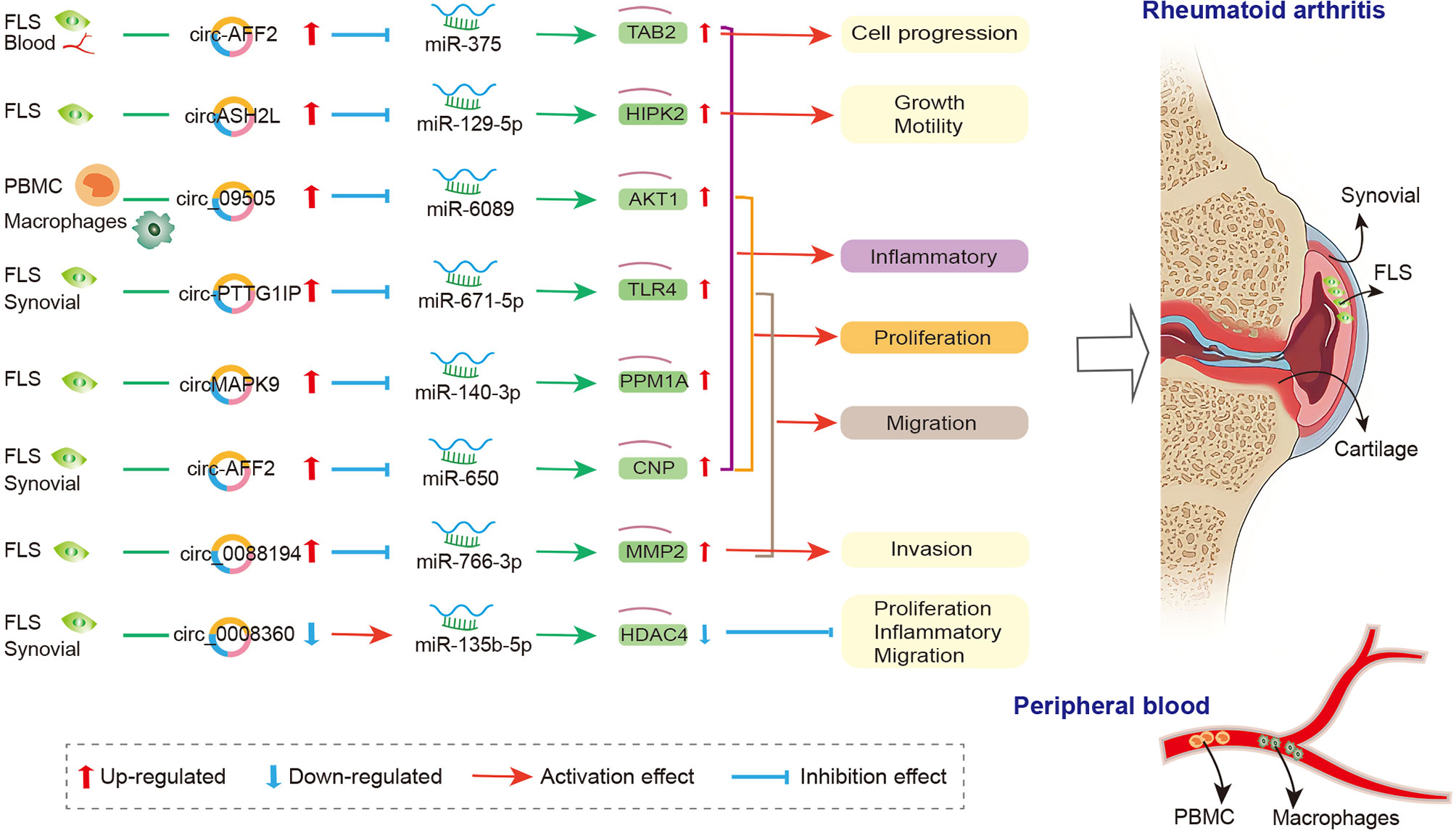
Figure 2 The crosstalk of circRNA-miRNA-mRNA in RA. FLS, Fibroblast−like synoviocytes; PBMC, peripheral blood mononuclear cell; MMP2, matrix metalloproteinase-2; CNP, 2’, 3’-cyclic nucleotide phosphodiesterase; TAB2, binding protein 2; TLR4, toll-like-receptor 4; PPM1A, protein phosphatase 1A; HIPK2, homeodomain-interacting protein kinase 2; HDAC4, histone deacetylase 4.
Other circRNAs, such as circ_0088194 (167), circ-AFF2 (168), circ-Sirt1 (170), circ-PTTG1IP (171), and circASH2L (173), also acted as miRNA sponge in RA; they regulated FLS proliferation, inflammation, and migration via circ_0088194/miR-766-3p/MMP2 axis, circ-AFF2/miR-650/CNP axis, circ-Sirt1/miR-132/Sirt1 pathway, circ-PTTG1IP/miR-671-5p/TLR4 axis, and circASH2L/miR-129-5p/HIPK2 axis, respectively. The details are presented in Table S2. The circRNA-miRNA-mRNA networks may have other biological functions in RA diseases besides proliferation, migration, invasion, and inflammatory response. To identify the function of specifically expressed circRNAs and miRNAs, Wen et al. first sequenced circRNAs and miRNAs in PBMCs from three pairs of RA patients and healthy controls. Then, the open source software platform cytoscape was used to build a circRNA-miRNA co-expression network that contained 228 circRNA–miRNA pairs. Further GO and KEGG analyses indicated that the significantly differentially expressed circRNAs were involved in apoptosis, inflammation, autophagy, and oxidative stress (162). This study presents the new idea that circRNAs might be related to the pathogenesis of RA worthy for further study.
Clinical Implications
RA is the most common autoimmune disease in the world (1). It leads to severe disability and early death (202). Therefore, early detection, diagnosis, and treatment are particularly important (2). Unfortunately, the markers used do not show a high degree of specificity and sensitivity (118). ncRNAs may serve as novel biomarkers because of their characteristics of stable expression in blood and body fluids. In recent years, a growing number of studies confirmed that ncRNAs (lncRNA, circRNA, and miRNA) play key roles in the physiopathological process of RA (34) and may become promising tools for disease diagnosis and prognosis and for prediction of treatment response. Some examples are the ncRNAs in blood (including plasma and serum) and synovial fluid, as follows: lncRNAs MEG3 (203), PICSAR (139); circRNAs circ_0044235 (204), Circ_AFF2 (169), circPTPN22 (165), circ_0005008 (166), and circ_0005198 (166); and miRNAs miR-23b (205), miR-103a-3p (84), and miR-125b (82, 206). Furthermore, several miRNAs could also function as potential biomarkers for RA complication, such as miR-146a-5p and miR-155-5p, which are reported to be possible biomarkers for the development of cardiovascular complications in RA (207). Moreover, the exosome-encapsulated miRNAs, such as miR-548a-3p (208) and miR-150-5p (209), may also be novel and promising targets for RA diagnosis and treatment.
Accumulated evidence demonstrated that the aberrantly expressed ncRNAs offer the opportunity to discover new targeted drugs for patients with RA (107, 161). ncRNA targeting treatment is more selective in RA treatment because of its low susceptibility to infection. Targeting lncRNAs LOC100652951and LOC10506036 modulates T cell inflammation in RA (151). miRNA miR-10a could also act as a regulator of inflammation in RA treatment (210). Furthermore, gene therapy in RA has received much attention in recent years, e.g., RNA interference (RNAi) (211) and antisense oligonucleotides(ASO) (161). RNAi is an intrinsic cellular mechanism that causes mRNA degradation through the interaction of miRNA and small interfering RNA(siRNA) molecules with complementary RNA molecules. Some biologicals of RNAi that target TNF and NF-κB have been used in RA animal models, but the results were not satisfactory (211). However, the treatment of STAT1 siRNA encapsulated by nanoparticles reduced joint deterioration in RA model mice; nanoparticles protected the siRNA from serum degradation (212). ASO is a promising nucleic acid therapy, and the ASO-based drug has been used in many diseases (213). Studies reported that the silencing of miR-223 using lentiviral vectors based on ASO could reduce disease severity of experimental arthritis (214). However, RA-related drugs are lacking. Few studies have investigated the clinical applicability of ncRNAs modulators in autoimmune diseases.
Conclusions
The studies on ncRNAs, especially the crosstalk of lncRNA/circRNA-miRNA-mRNA in autoimmune disorders, have received much attention. Although a series of published studies have revealed the role of lncRNA/circRNA-miRNA network in regulating inflammation and autoimmunity via Wnt3a/β-catenin and TLR/NF-κB signaling pathways in RA, the regulatory mechanism of ncRNAs is still unclear. More in-depth studies are needed to explore the interactions of lncRNA/circRNA-miRNA-mRNA. Elucidating the lncRNA/circRNA-miRNA-mRNA regulatory network and analyzing the interaction mechanism of these fundamental epigenetic regulators in the pathophysiology of RA are still a challenge. With the development of next-generation sequencing and other modern molecular biological techniques, more ncRNA molecular regulatory mechanisms and ncRNA targeted drugs will be uncovered. These would provide new strategies for the clinical diagnosis and targeted treatment for RA.
Author Contributions
X-AZ and X-QW: conceptualization, project administration, and funding acquisition. J-JH, X-AZ, and X-QW: writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
Supported by the Innovative Talents Support Program for Universities of Liaoning Province, No.WR2019024.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.810317/full#supplementary-material
References
1. Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ, Xu J. Rheumatoid Arthritis: Pathological Mechanisms and Modern Pharmacologic Therapies. Bone Res (2018) 6:15. doi: 10.1038/s41413-018-0016-9
2. Sparks JA. Rheumatoid Arthritis. Ann Intern Med (2019) 170(1):Itc1–itc16. doi: 10.7326/aitc201901010
3. Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Arthritis Rheum (2010) 62(9):2569–81. doi: 10.1002/art.27584
4. Myasoedova E, Crowson CS, Kremers HM, Therneau TM, Gabriel SE. Is the Incidence of Rheumatoid Arthritis Rising?: Results From Olmsted County, Minnesot-2007. Arthritis Rheum (2010) 62(6):1576–82. doi: 10.1002/art.27425
5. England BR, Thiele GM, Anderson DR, Mikuls TR. Increased Cardiovascular Risk in Rheumatoid Arthritis: Mechanisms and Implications. BMJ (2018) 361:k1036. doi: 10.1136/bmj.k1036
6. Conforti A, Di Cola I, Pavlych V, Ruscitti P, Berardicurti O, Ursini F, et al. Beyond the Joints, the Extra-Articular Manifestations in Rheumatoid Arthritis. Autoimmun Rev (2021) 20(2):102735. doi: 10.1016/j.autrev.2020.102735
7. Tsai CY, Hsieh SC, Liu CW, Lu CH, Liao HT, Chen MH, et al. The Expression of Non-Coding RNAs and Their Target Molecules in Rheumatoid Arthritis: A Molecular Basis for Rheumatoid Pathogenesis and Its Potential Clinical Applications. Int J Mol Sci (2021) 22(11):5689. doi: 10.3390/ijms22115689
8. Karlson EW, Deane K. Environmental and Gene-Environment Interactions and Risk of Rheumatoid Arthritis. Rheum Dis Clin North Am (2012) 38(2):405–26. doi: 10.1016/j.rdc.2012.04.002
9. Aletaha D, Smolen JS. Diagnosis and Management of Rheumatoid Arthritis: A Review. Jama (2018) 320(13):1360–72. doi: 10.1001/jama.2018.13103
10. Imas JJ, Ruiz Zamarreño C, Zubiate P, Sanchez-Martín L, Campión J, Matías IR. Optical Biosensors for the Detection of Rheumatoid Arthritis (RA) Biomarkers: A Comprehensive Review. Sensors (Basel) (2020) 20(21):6289. doi: 10.3390/s20216289
11. Pratt AG, Isaacs JD. Seronegative Rheumatoid Arthritis: Pathogenetic and Therapeutic Aspects. Best Pract Res Clin Rheumatol (2014) 28(4):651–9. doi: 10.1016/j.berh.2014.10.016
12. Chen X, Guo DY, Yin TL, Yang J. Non-Coding RNAs Regulate Placental Trophoblast Function and Participate in Recurrent Abortion. Front Pharmacol (2021) 12:646521. doi: 10.3389/fphar.2021.646521
13. Mazzone R, Zwergel C, Artico M, Taurone S, Ralli M, Greco A, et al. The Emerging Role of Epigenetics in Human Autoimmune Disorders. Clin Epigenet (2019) 11(1):34. doi: 10.1186/s13148-019-0632-2
14. Jiang H, Ma R, Zou S, Wang Y, Li Z, Li W. Reconstruction and Analysis of the lncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA Reveal Functional lncRNAs in Rheumatoid Arthritis. Mol Biosyst (2017) 13(6):1182–92. doi: 10.1039/c7mb00094d
15. Li Z, Li X, Jiang C, Qian W, Tse G, Chan MTV, et al. Long non-Coding RNAs in Rheumatoid Arthritis. Cell Prolif (2018) 51(1):e12404. doi: 10.1111/cpr.12404
16. Mehta SL, Chokkalla AK, Vemuganti R. Noncoding RNA Crosstalk in Brain Health and Diseases. Neurochem Int (2021) 149:105139. doi: 10.1016/j.neuint.2021.105139
17. Ospelt C, Gay S, Klein K. Epigenetics in the Pathogenesis of RA. Semin Immunopathol (2017) 39(4):409–19. doi: 10.1007/s00281-017-0621-5
18. Wang J, Yan S, Yang J, Lu H, Xu D, Wang Z. Non-Coding RNAs in Rheumatoid Arthritis: From Bench to Bedside. Front Immunol (2019) 10:3129. doi: 10.3389/fimmu.2019.03129
19. Taheri M, Eghtedarian R, Dinger ME, Ghafouri-Fard S. Dysregulation of Non-Coding RNAs in Rheumatoid Arthritis. BioMed Pharmacother (2020) 130:110617. doi: 10.1016/j.biopha.2020.110617
20. Zhou Q, Haupt S, Kreuzer JT, Hammitzsch A, Proft F, Neumann C, et al. Decreased Expression of miR-146a and miR-155 Contributes to an Abnormal Treg Phenotype in Patients With Rheumatoid Arthritis. Ann Rheum Dis (2015) 74(6):1265–74. doi: 10.1136/annrheumdis-2013-204377
21. Zou Y, Xu S, Xiao Y, Qiu Q, Shi M, Wang J, et al. Long Noncoding RNA LERFS Negatively Regulates Rheumatoid Synovial Aggression and Proliferation. J Clin Invest (2018) 128(10):4510–24. doi: 10.1172/jci97965
22. Kleaveland B, Shi CY, Stefano J, Bartel DP. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell (2018) 174(2):350–362.e317. doi: 10.1016/j.cell.2018.05.022
23. Song G, Yang Z, Guo J, Zheng Y, Su X, Wang X. Interactions Among lncRNAs/circRNAs, miRNAs, and mRNAs in Neuropathic Pain. Neurotherapeutics (2020) 17(3):917–31. doi: 10.1007/s13311-020-00881-y
24. Mousavi MJ, Jamshidi A, Chopra A, Aslani S, Akhlaghi M, Mahmoudi M. Implications of the Noncoding RNAs in Rheumatoid Arthritis Pathogenesis. J Cell Physiol (2018) 234(1):335–47. doi: 10.1002/jcp.26911
25. Waller P, Blann AD. Non-Coding RNAs - A Primer for the Laboratory Scientist. Br J BioMed Sci (2019) 76(4):157–65. doi: 10.1080/09674845.2019.1675847
26. Zhao M, Hou Y, Du YE, Yang L, Qin Y, Peng M, et al. Drosha-Independent miR-6778-5p Strengthens Gastric Cancer Stem Cell Stemness via Regulation of Cytosolic One-Carbon Folate Metabolism. Cancer Lett (2020) 478:8–21. doi: 10.1016/j.canlet.2020.02.040
27. Ha M, Kim VN. Regulation of microRNA Biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
28. Wei F, Cao C, Xu X, Wang J. Diverse Functions of miR-373 in Cancer. J Transl Med (2015) 13:162. doi: 10.1186/s12967-015-0523-z
29. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci (2020) 21(5):1723. doi: 10.3390/ijms21051723
30. Ghoreishy A, Khosravi A, Ghaemmaghami A. Exosomal microRNA and Stroke: A Review. J Cell Biochem (2019) 120(10):16352–61. doi: 10.1002/jcb.29130
31. Kaur A, Mackin ST, Schlosser K, Wong FL, Elharram M, Delles C, et al. Systematic Review of microRNA Biomarkers in Acute Coronary Syndrome and Stable Coronary Artery Disease. Cardiovasc Res (2020) 116(6):1113–24. doi: 10.1093/cvr/cvz302
32. Vicente R, Noël D, Pers YM, Apparailly F, Jorgensen C. Deregulation and Therapeutic Potential of microRNAs in Arthritic Diseases. Nat Rev Rheumatol (2016) 12(4):211–20. doi: 10.1038/nrrheum.2015.162
33. Kabekkodu SP, Shukla V, Varghese VK, J DS, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and Their Role in Biological Functions and Diseases. Biol Rev Camb Philos Soc (2018) 93(4):1955–86. doi: 10.1111/brv.12428
34. Chen W, Liu D, Li QZ, Zhu H. The Function of ncRNAs in Rheumatic Diseases. Epigenomics (2019) 11(7):821–33. doi: 10.2217/epi-2018-0135
35. Hussain N, Zhu W, Jiang C, Xu J, Geng M, Wu X, et al. Down-Regulation of miR-10a-5p Promotes Proliferation and Restricts Apoptosis via Targeting T-Box Transcription Factor 5 in Inflamed Synoviocytes. Biosci Rep (2018) 38(2):BSR20180003. doi: 10.1042/bsr20180003
36. Akhtar N, Singh AK, Ahmed S. MicroRNA-17 Suppresses TNF-α Signaling by Interfering With TRAF2 and Ciap2 Association in Rheumatoid Arthritis Synovial Fibroblasts. J Immunol (2016) 197(6):2219–28. doi: 10.4049/jimmunol.1600360
37. Su J, Zhang J, Zhu J, Liu Y. The Promoting Effect of MMP13 on Mediating the Development of HFLS-RA by the Target of miR-19a Through IL-17 Signaling Pathway. J Cell Biochem (2020) 121(10):4282–94. doi: 10.1002/jcb.29609
38. Li XF, Shen WW, Sun YY, Li WX, Sun ZH, Liu YH, et al. MicroRNA-20a Negatively Regulates Expression of NLRP3-Inflammasome by Targeting TXNIP in Adjuvant-Induced Arthritis Fibroblast-Like Synoviocytes. Joint Bone Spine (2016) 83(6):695–700. doi: 10.1016/j.jbspin.2015.10.007
39. Chen Y, Xian PF, Yang L, Wang SX. MicroRNA-21 Promotes Proliferation of Fibroblast-Like Synoviocytes Through Mediation of NF-κb Nuclear Translocation in a Rat Model of Collagen-Induced Rheumatoid Arthritis. BioMed Res Int (2016) 2016:9279078. doi: 10.1155/2016/9279078
40. Zhang C, Fang L, Liu X, Nie T, Li R, Cui L, et al. miR-22 Inhibits Synovial Fibroblasts Proliferation and Proinflammatory Cytokine Production in RASF via Targeting SIRT1. Gene (2020) 724:144144. doi: 10.1016/j.gene.2019.144144
41. Zhang W, Chen L, Jiang Y, Shen Y. miR-26a-5p Regulates Synovial Fibroblast Invasion in Patients With Rheumatoid Arthritis by Targeting Smad 1. Med Sci Monit (2018) 24:5178–84. doi: 10.12659/msm.907816
42. Shi DL, Shi GR, Xie J, Du XZ, Yang H. MicroRNA-27a Inhibits Cell Migration and Invasion of Fibroblast-Like Synoviocytes by Targeting Follistatin-Like Protein 1 in Rheumatoid Arthritis. Mol Cells (2016) 39(8):611–8. doi: 10.14348/molcells.2016.0103
43. Chen L, Lu Q, Chen J, Feng R, Yang C. Upregulating miR-27a-3p Inhibits Cell Proliferation and Inflammation of Rheumatoid Arthritis Synovial Fibroblasts Through Targeting Toll-Like Receptor 5. Exp Ther Med (2021) 22(5):1227. doi: 10.3892/etm.2021.10661
44. Liu J, Fei D, Xing J, Du J. MicroRNA-29a Inhibits Proliferation and Induces Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Repressing STAT3. BioMed Pharmacother (2017) 96:173–81. doi: 10.1016/j.biopha.2017.09.120
45. Song AF, Kang L, Wang YF, Wang M. MiR-34a-5p Inhibits Fibroblast−Like Synoviocytes Proliferation via XBP1. Eur Rev Med Pharmacol Sci (2020) 24(22):11675–82. doi: 10.26355/eurrev_202011_23812
46. Hou C, Wang D, Zhang L. MicroRNA−34a−3p Inhibits Proliferation of Rheumatoid Arthritis Fibroblast−Like Synoviocytes. Mol Med Rep (2019) 20(3):2563–70. doi: 10.3892/mmr.2019.10516
47. Liu D, Fang Y, Rao Y, Tan W, Zhou W, Wu X, et al. Synovial Fibroblast-Derived Exosomal microRNA-106b Suppresses Chondrocyte Proliferation and Migration in Rheumatoid Arthritis via Down-Regulation of PDK4. J Mol Med (Berl) (2020) 98(3):409–23. doi: 10.1007/s00109-020-01882-2
48. Meng Q, Pan B, Sheng P. Histone Deacetylase 1 is Increased in Rheumatoid Arthritis Synovium and Promotes Synovial Cell Hyperplasia and Synovial Inflammation in the Collagen-Induced Arthritis Mouse Model via the microRNA-124-Dependent MARCKS-JAK/STAT Axis. Clin Exp Rheumatol (2021) 39(5):970–81.
49. Liu K, Zhang Y, Liu L, Yuan Q. miR-125 Regulates PI3K/Akt/mTOR Signaling Pathway in Rheumatoid Arthritis Rats via PARP2. Biosci Rep (2019) 39(1):BSR20180890. doi: 10.1042/bsr20180890
50. Gao J, Zhou XL, Kong RN, Ji LM, He LL, Zhao DB. microRNA-126 Targeting PIK3R2 Promotes Rheumatoid Arthritis Synovial Fibro-Blasts Proliferation and Resistance to Apoptosis by Regulating PI3K/AKT Pathway. Exp Mol Pathol (2016) 100(1):192–8. doi: 10.1016/j.yexmp.2015.12.015
51. Qu Y, Zhang YP, Wu J, Jie LG, Deng JX, Zhao DB, et al. Downregulated microRNA-135a Ameliorates Rheumatoid Arthritis by Inactivation of the Phosphatidylinositol 3-Kinase/AKT Signaling Pathway via Phosphatidylinositol 3-Kinase Regulatory Subunit 2. J Cell Physiol (2019) 234(10):17663–76. doi: 10.1002/jcp.28390
52. Sun W, Zhang Y, Wang G. MicroRNA-137-Mediated Inhibition of Lysine-Specific Demethylase-1 Prevents Against Rheumatoid Arthritis in an Association With the REST/mTOR Axis. Mol Pain (2021) 17:17448069211041847. doi: 10.1177/17448069211041847
53. Du J, Zhang F, Guo J. Mir−137 Decreases Proliferation, Migration and Invasion in Rheumatoid Arthritis Fibroblast−Like Synoviocytes. Mol Med Rep (2018) 17(2):3312–7. doi: 10.3892/mmr.2017.8225
54. Shao L, Hou C. miR-138 Activates NF-κb Signaling and PGRN to Promote Rheumatoid Arthritis via Regulating HDAC4. Biochem Biophys Res Commun (2019) 519(1):166–71. doi: 10.1016/j.bbrc.2019.08.092
55. Zu B, Liu L, Wang J, Li M, Yang J. MiR-140-3p Inhibits the Cell Viability and Promotes Apoptosis of Synovial Fibroblasts in Rheumatoid Arthritis Through Targeting Sirtuin 3. J Orthop Surg Res (2021) 16(1):105. doi: 10.1186/s13018-021-02236-5
56. Hong BK, You S, Yoo SA, Park D, Hwang D, Cho CS, et al. MicroRNA-143 and -145 Modulate the Phenotype of Synovial Fibroblasts in Rheumatoid Arthritis. Exp Mol Med (2017) 49(8):e363. doi: 10.1038/emm.2017.108
57. Guo J, Du J, Fei D, Xing J, Liu J, Lu H. Mir−152 Inhibits Rheumatoid Arthritis Synovial Fibroblast Proliferation and Induces Apoptosis by Targeting ADAM10. Int J Mol Med (2018) 42(1):643–50. doi: 10.3892/ijmm.2018.3636
58. Long L, Yu P, Liu Y, Wang S, Li R, Shi J, et al. Upregulated microRNA-155 Expression in Peripheral Blood Mononuclear Cells and Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. Clin Dev Immunol (2013) 2013:296139. doi: 10.1155/2013/296139
59. Li S, Jin Z, Lu X. MicroRNA-192 Suppresses Cell Proliferation and Induces Apoptosis in Human Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Downregulating Caveolin 1. Mol Cell Biochem (2017) 432(1-2):123–30. doi: 10.1007/s11010-017-3003-3
60. Qu SP, Li GW, Ma H, Xing Q. MicroRNA-193a-3p Participates in the Progression of Rheumatoid Arthritis by Regulating Proliferation and Apoptosis of MH7A Cells Through Targeting IGFBP5. Eur Rev Med Pharmacol Sci (2019) 23(11):4850–7. doi: 10.26355/eurrev_201906_18072
61. Quero L, Tiaden AN, Hanser E, Roux J, Laski A, Hall J, et al. miR-221-3p Drives the Shift of M2-Macrophages to a Pro-Inflammatory Function by Suppressing JAK3/STAT3 Activation. Front Immunol (2019) 10:3087. doi: 10.3389/fimmu.2019.03087
62. Wang X, Gong S, Pu D, Hu N, Wang Y, Fan P, et al. Up-Regulation of miR-365 Promotes the Apoptosis and Restrains Proliferation of Synoviocytes Through Downregulation of IGF1 and the Inactivation of the PI3K/AKT/mTOR Pathway in Mice With Rheumatoid Arthritis. Int Immunopharmacol (2020) 79:106067. doi: 10.1016/j.intimp.2019.106067
63. Jiang F, Zhou HY, Zhou LF, Wen YH, Gai HH, Wu GM. MicroRNA-421 Promotes Inflammatory Response of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis by Targeting SPRY1. Eur Rev Med Pharmacol Sci (2019) 23(19):8186–93. doi: 10.26355/eurrev_201910_19125
64. Wang S, Xu J, Guo Y, Cai Y, Ren X, Zhu W, et al. MicroRNA-497 Reduction and Increase of Its Family Member MicroRNA-424 Lead to Dysregulation of Multiple Inflammation Related Genes in Synovial Fibroblasts With Rheumatoid Arthritis. Front Immunol (2021) 12:619392. doi: 10.3389/fimmu.2021.619392
65. Wang Y, Zhang K, Yuan X, Xu N, Zhao S, Hou L, et al. miR-431-5p Regulates Cell Proliferation and Apoptosis in Fibroblast-Like Synoviocytes in Rheumatoid Arthritis by Targeting XIAP. Arthritis Res Ther (2020) 22(1):231. doi: 10.1186/s13075-020-02328-3
66. Cai Y, Jiang C, Zhu J, Xu K, Ren X, Xu L, et al. miR-449a Inhibits Cell Proliferation, Migration, and Inflammation by Regulating High-Mobility Group Box Protein 1 and Forms a Mutual Inhibition Loop With Yin Yang 1 in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Arthritis Res Ther (2019) 21(1):134. doi: 10.1186/s13075-019-1920-0
67. Wang Y, Hou L, Yuan X, Xu N, Zhao S, Yang L, et al. miR-483-3p Promotes Cell Proliferation and Suppresses Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Targeting IGF-1. BioMed Pharmacother (2020) 130:110519. doi: 10.1016/j.biopha.2020.110519
68. Fang L, Xu XF, Lu Y, Wu YY, Li JJ. MicroRNA-495 Attenuates Proliferation and Inflammatory Response in Rheumatoid Arthritis Fibroblast-Like Synoviocytes Through Attenuating β-Catenin Pathway. J Biol Regul Homeost Agents (2020) 34(3):837–44. doi: 10.23812/20-47-a-22
69. Wang X, Si X, Sun J, Yue L, Wang J, Yu Z. miR-522 Modulated the Expression of Proinflammatory Cytokines and Matrix Metalloproteinases Partly via Targeting Suppressor of Cytokine Signaling 3 in Rheumatoid Arthritis Synovial Fibroblasts. DNA Cell Biol (2018) 37(4):405–15. doi: 10.1089/dna.2017.4008
70. Fu D, Xiao C, Xie Y, Gao J, Ye S. MiR-3926 Inhibits Synovial Fibroblasts Proliferation and Inflammatory Cytokines Secretion Through Targeting Toll Like Receptor 5. Gene (2019) 687:200–6. doi: 10.1016/j.gene.2018.11.014
71. Lin S, Wang S, Zhang Z, Lu Y, Yang M, Chen P, et al. MiRNA-6089 Inhibits Rheumatoid Arthritis Fibroblast-Like Synoviocytes Proliferation and Induces Apoptosis by Targeting CCR4. Arch Physiol Biochem (2020), 1–8. doi: 10.1080/13813455.2020.1773862
72. Hegewald AB, Breitwieser K, Ottinger SM, Mobarrez F, Korotkova M, Rethi B, et al. Extracellular miR-574-5p Induces Osteoclast Differentiation via TLR 7/8 in Rheumatoid Arthritis. Front Immunol (2020) 11:585282. doi: 10.3389/fimmu.2020.585282
73. Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, et al. Altered microRNA Expression Profile With miR-146a Upregulation in CD4+ T Cells From Patients With Rheumatoid Arthritis. Arthritis Res Ther (2010) 12(3):R81. doi: 10.1186/ar3006
74. Zhu W, Yu J, Qiu S, Liu H, Wang Y, Xu X, et al. MiR-Let-7a Regulates Anti-Citrullinated Protein Antibody-Induced Macrophage Activation and Correlates With the Development of Experimental Rheumatoid Arthritis. Int Immunopharmacol (2017) 51:40–6. doi: 10.1016/j.intimp.2017.08.001
75. Churov AV, Oleinik EK, Knip M. MicroRNAs in Rheumatoid Arthritis: Altered Expression and Diagnostic Potential. Autoimmun Rev (2015) 14(11):1029–37. doi: 10.1016/j.autrev.2015.07.005
76. Wu YH, Liu W, Xue B, Zhang L, Liu XY, Liu B, et al. Upregulated Expression of microRNA-16 Correlates With Th17/Treg Cell Imbalance in Patients With Rheumatoid Arthritis. DNA Cell Biol (2016) 35(12):853–60. doi: 10.1089/dna.2016.3349
77. Filková M, Aradi B, Senolt L, Ospelt C, Vettori S, Mann H, et al. Association of Circulating miR-223 and miR-16 With Disease Activity in Patients With Early Rheumatoid Arthritis. Ann Rheum Dis (2014) 73(10):1898–904. doi: 10.1136/annrheumdis-2012-202815
78. Wei H, Wu Q, Shi Y, Luo A, Lin S, Feng X, et al. MicroRNA-15a/16/SOX5 Axis Promotes Migration, Invasion and Inflammatory Response in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Aging (Albany NY) (2020) 12(14):14376–90. doi: 10.18632/aging.103480
79. Dong L, Wang X, Tan J, Li H, Qian W, Chen J, et al. Decreased Expression of microRNA-21 Correlates With the Imbalance of Th17 and Treg Cells in Patients With Rheumatoid Arthritis. J Cell Mol Med (2014) 18(11):2213–24. doi: 10.1111/jcmm.12353
80. Pávková Goldbergová M, Lipková J, Fedorko J, Němec P, Gatterová J, Válková L, et al. Relationship of Epigenetic Variability of miR-124 to Extracellular Matrix Remodelling and Age-Related MMP-3 Expression in Rheumatoid Arthritis. Gen Physiol Biophys (2018) 37(6):703–10. doi: 10.4149/gpb_2018024
81. Zhang B, Wang LS, Zhou YH. Elevated microRNA-125b Promotes Inflammation in Rheumatoid Arthritis by Activation of NF-κb Pathway. BioMed Pharmacother (2017) 93:1151–7. doi: 10.1016/j.biopha.2017.07.042
82. Hruskova V, Jandova R, Vernerova L, Mann H, Pecha O, Prajzlerova K, et al. MicroRNA-125b: Association With Disease Activity and the Treatment Response of Patients With Early Rheumatoid Arthritis. Arthritis Res Ther (2016) 18(1):124. doi: 10.1186/s13075-016-1023-0
83. Cunningham CC, Wade S, Floudas A, Orr C, McGarry T, Wade S, et al. Serum miRNA Signature in Rheumatoid Arthritis and “At-Risk Individuals”. Front Immunol (2021) 12:633201. doi: 10.3389/fimmu.2021.633201
84. Anaparti V, Smolik I, Meng X, Spicer V, Mookherjee N, El-Gabalawy H. Whole Blood microRNA Expression Pattern Differentiates Patients With Rheumatoid Arthritis, Their Seropositive First-Degree Relatives, and Healthy Unrelated Control Subjects. Arthritis Res Ther (2017) 19(1):249. doi: 10.1186/s13075-017-1459-x
85. Alivernini S, Kurowska-Stolarska M, Tolusso B, Benvenuto R, Elmesmari A, Canestri S, et al. MicroRNA-155 Influences B-Cell Function Through PU.1 in Rheumatoid Arthritis. Nat Commun (2016) 7:12970. doi: 10.1038/ncomms12970
86. Elmesmari A, Fraser AR, Wood C, Gilchrist D, Vaughan D, Stewart L, et al. MicroRNA-155 Regulates Monocyte Chemokine and Chemokine Receptor Expression in Rheumatoid Arthritis. Rheumatol (Oxf) (2016) 55(11):2056–65. doi: 10.1093/rheumatology/kew272
87. Singh A, Patro PS, Aggarwal A. MicroRNA-132, miR-146a, and miR-155 as Potential Biomarkers of Methotrexate Response in Patients With Rheumatoid Arthritis. Clin Rheumatol (2019) 38(3):877–84. doi: 10.1007/s10067-018-4380-z
88. Sun W, Ma J, Zhao H, Xiao C, Zhong H, Ling H, et al. Resolvin D1 Suppresses Pannus Formation via Decreasing Connective Tissue Growth Factor Caused by Upregulation of miRNA-146a-5p in Rheumatoid Arthritis. Arthritis Res Ther (2020) 22(1):61. doi: 10.1186/s13075-020-2133-2
89. Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM. Utility of Select Plasma MicroRNA for Disease and Cardiovascular Risk Assessment in Patients With Rheumatoid Arthritis. J Rheumatol (2015) 42(10):1746–51. doi: 10.3899/jrheum.150232
90. Abdul-Maksoud RS, Sediq AM, Kattaia A, Elsayed W, Ezzeldin N, Abdel Galil SM, et al. Serum miR-210 and miR-155 Expression Levels as Novel Biomarkers for Rheumatoid Arthritis Diagnosis. Br J BioMed Sci (2017) 74(4):209–13. doi: 10.1080/09674845.2017.1343545
91. Liu Y, Zhang XL, Li XF, Tang YC, Zhao X. miR-212-3p Reduced Proliferation, and Promoted Apoptosis of Fibroblast-Like Synoviocytes via Down-Regulating SOX5 in Rheumatoid Arthritis. Eur Rev Med Pharmacol Sci (2018) 22(2):461–71. doi: 10.26355/eurrev_201801_14196
92. Tang X, Yin K, Zhu H, Tian J, Shen D, Yi L, et al. Correlation Between the Expression of MicroRNA-301a-3p and the Proportion of Th17 Cells in Patients With Rheumatoid Arthritis. Inflammation (2016) 39(2):759–67. doi: 10.1007/s10753-016-0304-8
93. Ciechomska M, Bonek K, Merdas M, Zarecki P, Swierkot J, Gluszko P, et al. Changes in MiRNA-5196 Expression as a Potential Biomarker of Anti-TNF-α Therapy in Rheumatoid Arthritis and Ankylosing Spondylitis Patients. Arch Immunol Ther Exp (Warsz) (2018) 66(5):389–97. doi: 10.1007/s00005-018-0513-y
94. Lai NS, Yu HC, Yu CL, Koo M, Huang HB, Lu MC. Anti-Citrullinated Protein Antibodies Suppress Let-7a Expression in Monocytes From Patients With Rheumatoid Arthritis and Facilitate the Inflammatory Responses in Rheumatoid Arthritis. Immunobiology (2015) 220(12):1351–8. doi: 10.1016/j.imbio.2015.07.007
95. Falconer J, Murphy AN, Young SP, Clark AR, Tiziani S, Guma M, et al. Review: Synovial Cell Metabolism and Chronic Inflammation in Rheumatoid Arthritis. Arthritis Rheumatol (2018) 70(7):984–99. doi: 10.1002/art.40504
96. Nygaard G, Firestein GS. Restoring Synovial Homeostasis in Rheumatoid Arthritis by Targeting Fibroblast-Like Synoviocytes. Nat Rev Rheumatol (2020) 16(6):316–33. doi: 10.1038/s41584-020-0413-5
97. Miao CG, Shi WJ, Xiong YY, Yu H, Zhang XL, Qin MS, et al. miR-375 Regulates the Canonical Wnt Pathway Through FZD8 Silencing in Arthritis Synovial Fibroblasts. Immunol Lett (2015) 164(1):1–10. doi: 10.1016/j.imlet.2015.01.003
98. Liu XG, Zhang Y, Ju WF, Li CY, Mu YC. MiR-21 Relieves Rheumatoid Arthritis in Rats via Targeting Wnt Signaling Pathway. Eur Rev Med Pharmacol Sci (2019) 23(3 Suppl):96–103. doi: 10.26355/eurrev_201908_18635
99. Yang B, Ge Y, Zhou Y, Wang J, Xie X, Li S, et al. miR-124a Inhibits the Proliferation and Inflammation in Rheumatoid Arthritis Fibroblast-Like Synoviocytes via Targeting PIK3/NF-κb Pathway. Cell Biochem Funct (2019) 37(4):208–15. doi: 10.1002/cbf.3386
100. Najm A, Masson FM, Preuss P, Georges S, Ory B, Quillard T, et al. MicroRNA-17-5p Reduces Inflammation and Bone Erosions in Mice With Collagen-Induced Arthritis and Directly Targets the JAK/STAT Pathway in Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Arthritis Rheumatol (2020) 72(12):2030–9. doi: 10.1002/art.41441
101. Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, et al. TLR2 Expression is Regulated by microRNA miR-19 in Rheumatoid Fibroblast-Like Synoviocytes. J Immunol (2012) 188(1):454–61. doi: 10.4049/jimmunol.1102348
102. Luo X, Cui J, Long X, Chen Z. TLRs Play Crucial Roles in Regulating RA Synoviocyte. Endocr Metab Immune Disord Drug Targets (2020) 20(8):1156–65. doi: 10.2174/1871530320666200427115225
103. Takamura Y, Aoki W, Satomura A, Shibasaki S, Ueda M. Small RNAs Detected in Exosomes Derived From the MH7A Synovial Fibroblast Cell Line With TNF-α Stimulation. PloS One (2018) 13(8):e0201851. doi: 10.1371/journal.pone.0201851
104. Kolarz B, Majdan M. Epigenetic Aspects of Rheumatoid Arthritis: Contribution of non-Coding RNAs. Semin Arthritis Rheum (2017) 46(6):724–31. doi: 10.1016/j.semarthrit.2017.01.003
105. Castro-Villegas C, Pérez-Sánchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limón P, et al. Circulating miRNAs as Potential Biomarkers of Therapy Effectiveness in Rheumatoid Arthritis Patients Treated With Anti-Tnfα. Arthritis Res Ther (2015) 17(1):49. doi: 10.1186/s13075-015-0555-z
106. Rezaeepoor M, Pourjafar M, Tahamoli-Roudsari A, Basiri Z, Hajilooi M, Solgi G. Altered Expression of microRNAs may Predict Therapeutic Response in Rheumatoid Arthritis Patients. Int Immunopharmacol (2020) 83:106404. doi: 10.1016/j.intimp.2020.106404
107. Evangelatos G, Fragoulis GE, Koulouri V, Lambrou GI. MicroRNAs in Rheumatoid Arthritis: From Pathogenesis to Clinical Impact. Autoimmun Rev (2019) 18(11):102391. doi: 10.1016/j.autrev.2019.102391
108. Liu Y, Han Y, Qu H, Fang J, Ye M, Yin W. Correlation of microRNA Expression Profile With Clinical Response to Tumor Necrosis Factor Inhibitor in Treating Rheumatoid Arthritis Patients: A Prospective Cohort Study. J Clin Lab Anal (2019) 33(7):e22953. doi: 10.1002/jcla.22953
109. Wang L, Wang C, Jia X, Yu J. Circulating Exosomal miR-17 Inhibits the Induction of Regulatory T Cells via Suppressing TGFBR II Expression in Rheumatoid Arthritis. Cell Physiol Biochem (2018) 50(5):1754–63. doi: 10.1159/000494793
110. Wu GC, Pan HF, Leng RX, Wang DG, Li XP, Li XM, et al. Emerging Role of Long Noncoding RNAs in Autoimmune Diseases. Autoimmun Rev (2015) 14(9):798–805. doi: 10.1016/j.autrev.2015.05.004
111. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: The Reference Human Genome Annotation for The ENCODE Project. Genome Res (2012) 22(9):1760–74. doi: 10.1101/gr.135350.111
112. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
113. Fang Y, Tu J, Han D, Guo Y, Hong W, Wei W. The Effects of Long non-Coding Ribonucleic Acids on Various Cellular Components in Rheumatoid Arthritis. Rheumatol (Oxf) (2020) 59(1):46–56. doi: 10.1093/rheumatology/kez472
114. Dykes IM, Emanueli C. Transcriptional and Post-Transcriptional Gene Regulation by Long Non-Coding RNA. Genomics Proteomics Bioinf (2017) 15(3):177–86. doi: 10.1016/j.gpb.2016.12.005
115. Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Mol Cell (2011) 43(6):904–14. doi: 10.1016/j.molcel.2011.08.018
116. Smolarz B, Zadrożna-Nowak A, Romanowicz H. The Role of lncRNA in the Development of Tumors, Including Breast Cancer. Int J Mol Sci (2021) 22(16):8427. doi: 10.3390/ijms22168427
117. Sallam T, Sandhu J, Tontonoz P. Long Noncoding RNA Discovery in Cardiovascular Disease: Decoding Form to Function. Circ Res (2018) 122(1):155–66. doi: 10.1161/circresaha.117.311802
118. Lodde V, Murgia G, Simula ER, Steri M, Floris M, Idda ML. Long Noncoding RNAs and Circular RNAs in Autoimmune Diseases. Biomolecules (2020) 10(7):1044. doi: 10.3390/biom10071044
119. Chen YG, Satpathy AT, Chang HY. Gene Regulation in the Immune System by Long Noncoding RNAs. Nat Immunol (2017) 18(9):962–72. doi: 10.1038/ni.3771
120. Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF, Guan XL, et al. Long Noncoding RNA Expression Profile in Fibroblast-Like Synoviocytes From Patients With Rheumatoid Arthritis. Arthritis Res Ther (2016) 18(1):227. doi: 10.1186/s13075-016-1129-4
121. Piao X, Zhou J, Hu J. Role of RP11-83J16.1, a Novel Long non-Coding RNA, in Rheumatoid Arthritis. Am J Transl Res (2020) 12(4):1397–414.
122. Sun L, Tu J, Liu C, Pan A, Xia X, Chen X. Analysis of lncRNA Expression Profiles by Sequencing Reveals That lnc-AL928768.3 and lnc-AC091493.1 are Novel Biomarkers for Disease Risk and Activity of Rheumatoid Arthritis. Inflammopharmacology (2020) 28(2):437–50. doi: 10.1007/s10787-019-00666-6
123. Yuan M, Wang S, Yu L, Qu B, Xu L, Liu L, et al. Long Noncoding RNA Profiling Revealed Differentially Expressed lncRNAs Associated With Disease Activity in PBMCs From Patients With Rheumatoid Arthritis. PloS One (2017) 12(11):e0186795. doi: 10.1371/journal.pone.0186795
124. Luo Q, Xu C, Li X, Zeng L, Ye J, Guo Y, et al. Comprehensive Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Rheumatoid Arthritis. Exp Ther Med (2017) 14(6):5965–73. doi: 10.3892/etm.2017.5284
125. Wen J, Liu J, Jiang H, Wan L, Xin L, Sun Y, et al. lncRNA Expression Profiles Related to Apoptosis and Autophagy in Peripheral Blood Mononuclear Cells of Patients With Rheumatoid Arthritis. FEBS Open Bio (2020) 10(8):1642–54. doi: 10.1002/2211-5463.12913
126. Li M, Ma K, Feng Z, Wang J, Zhou X, Zhou L. Differential Long non-Coding RNA Expression Profiles in the Peripheral Blood and CD4(+) T Cells of Patients With Active Rheumatoid Arthritis. Exp Ther Med (2020) 20(1):461–71. doi: 10.3892/etm.2020.8681
127. Qin W, Wang TH, Xie BH, Sun QQ, Huang H, Zhao BJ, et al. Plasma Long Non-Coding RNA Expression Profiles in Patients With Rheumatoid Arthritis. Clin Lab (2019) 65(8):1467–77. doi: 10.7754/Clin.Lab.2019.190144
128. Li GQ, Fang YX, Liu Y, Meng FR, Wu X, Zhang CW, et al. MALAT1-Driven Inhibition of Wnt Signal Impedes Proliferation and Inflammation in Fibroblast-Like Synoviocytes Through CTNNB1 Promoter Methylation in Rheumatoid Arthritis. Hum Gene Ther (2019) 30(8):1008–22. doi: 10.1089/hum.2018.212
129. Zhang TP, Zhu BQ, Tao SS, Fan YG, Li XM, Pan HF, et al. Long Non-Coding RNAs Genes Polymorphisms and Their Expression Levels in Patients With Rheumatoid Arthritis. Front Immunol (2019) 10:2529. doi: 10.3389/fimmu.2019.02529
130. Yan ZF, Zhao XY, Liu W, Liu XP. UCA1 Impacts Progress of Rheumatoid Arthritis by Inducing the Apoptosis of Fibroblast-Like Synoviocyte. Eur Rev Med Pharmacol Sci (2018) 22(4):914–20. doi: 10.26355/eurrev_201802_14370
131. Liu YR, Yang L, Xu QQ, Lu XY, Ma TT, Huang C, et al. Long Noncoding RNA MEG3 Regulates Rheumatoid Arthritis by Targeting NLRC5. J Cell Physiol (2019) 234(8):14270–84. doi: 10.1002/jcp.28126
132. Li G, Liu Y, Meng F, Xia Z, Wu X, Fang Y, et al. LncRNA MEG3 Inhibits Rheumatoid Arthritis Through miR-141 and Inactivation of AKT/mTOR Signalling Pathway. J Cell Mol Med (2019) 23(10):7116–20. doi: 10.1111/jcmm.14591
133. Lu X, Qian J. Downregulated MEG3 Participates in Rheumatoid Arthritis via Promoting Proliferation of Fibroblast-Like Synoviocytes. Exp Ther Med (2019) 17(3):1637–42. doi: 10.3892/etm.2018.7100
134. Yang Z, Lin SD, Zhan F, Liu Y, Zhan YW. LncRNA GAS5 Alleviates Rheumatoid Arthritis Through Regulating miR-222-3p/Sirt1 Signalling Axis. Autoimmunity (2021) 54(1):13–22. doi: 10.1080/08916934.2020.1846183
135. Mo BY, Guo XH, Yang MR, Liu F, Bi X, Liu Y, et al. Long Non-Coding RNA GAPLINC Promotes Tumor-Like Biologic Behaviors of Fibroblast-Like Synoviocytes as MicroRNA Sponging in Rheumatoid Arthritis Patients. Front Immunol (2018) 9:702. doi: 10.3389/fimmu.2018.00702
136. Ye Z, Xu J, Li S, Cai C, Li T, Sun L. Lnc−IL7R Promotes the Growth of Fibroblast−Like Synoviocytes Through Interaction With Enhancer of Zeste Homolog 2 in Rheumatoid Arthritis. Mol Med Rep (2017) 15(3):1412–8. doi: 10.3892/mmr.2017.6150
137. Yue T, Fan X, Zhang Z, Liu Z, Guo M, Bai F, et al. Downregulation of lncRNA ITSN1-2 Correlates With Decreased Disease Risk and Activity of Rheumatoid Arthritis (RA), and Reduces RA Fibroblast-Like Synoviocytes Proliferation and Inflammation via Inhibiting NOD2/RIP2 Signaling Pathway. Am J Transl Res (2019) 11(8):4650–66.
138. Zhang CW, Wu X, Liu D, Zhou W, Tan W, Fang YX, et al. Long non-Coding RNA PVT1 Knockdown Suppresses Fibroblast-Like Synoviocyte Inflammation and Induces Apoptosis in Rheumatoid Arthritis Through Demethylation of Sirt6. J Biol Eng (2019) 13:60. doi: 10.1186/s13036-019-0184-1
139. Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, et al. LncRNA PICSAR Promotes Cell Proliferation, Migration and Invasion of Fibroblast-Like Synoviocytes by Sponging miRNA-4701-5p in Rheumatoid Arthritis. EBioMedicine (2019) 50:408–20. doi: 10.1016/j.ebiom.2019.11.024
140. Ye Y, Gao X, Yang N. LncRNA ZFAS1 Promotes Cell Migration and Invasion of Fibroblast-Like Synoviocytes by Suppression of miR-27a in Rheumatoid Arthritis. Hum Cell (2018) 31(1):14–21. doi: 10.1007/s13577-017-0179-5
141. Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and Exosome-Derived Hotair Is a Critical Regulator and Potent Marker for Rheumatoid Arthritis. Clin Exp Med (2015) 15(1):121–6. doi: 10.1007/s10238-013-0271-4
142. Zhang HJ, Wei QF, Wang SJ, Zhang HJ, Zhang XY, Geng Q, et al. LncRNA HOTAIR Alleviates Rheumatoid Arthritis by Targeting miR-138 and Inactivating NF-κb Pathway. Int Immunopharmacol (2017) 50:283–90. doi: 10.1016/j.intimp.2017.06.021
143. Zhu X, Zhu Y, Ding C, Zhang W, Guan H, Li C, et al. LncRNA H19 Regulates Macrophage Polarization and Promotes Freund’s Complete Adjuvant-Induced Arthritis by Upregulating KDM6A. Int Immunopharmacol (2021) 93:107402. doi: 10.1016/j.intimp.2021.107402
144. Zhi LQ, Zhong Q, Ma JB, Xiao L, Yao SX, Wang X. LncRNA H19 Inhibitor Represses Synovial Cell Proliferation and Apoptosis in Rats With Rheumatoid Arthritis via Notch Signaling Pathway. Eur Rev Med Pharmacol Sci (2020) 24(15):7921. doi: 10.26355/eurrev_202008_22456
145. Mu N, Gu JT, Huang TL, Liu NN, Chen H, Bu X, et al. Blockade of Discoidin Domain Receptor 2 as a Strategy for Reducing Inflammation and Joint Destruction in Rheumatoid Arthritis Via Altered Interleukin-15 and Dkk-1 Signaling in Fibroblast-Like Synoviocytes. Arthritis Rheumatol (2020) 72(6):943–56. doi: 10.1002/art.41205
146. Wang G, Tang L, Zhang X, Li Y. LncRNA DILC Participates in Rheumatoid Arthritis by Inducing Apoptosis of Fibroblast-Like Synoviocytes and Down-Regulating IL-6. Biosci Rep (2019) 39(5):BSR20182374. doi: 10.1042/bsr20182374
147. Gong X, Fan X, Zhang Z, Cai Q, Guo M, Gao C, et al. Circulating lnc-ITSN1-2 Expression Presents a High Value in Diagnosis of Rheumatoid Arthritis and Correlates With Disease Activity. Int J Clin Exp Pathol (2017) 10(10):10451–8.
148. Spurlock CF, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate Inhibits NF-κb Activity via Long Intergenic (Noncoding) RNA-P21 Induction. Arthritis Rheumatol (2014) 66(11):2947–57. doi: 10.1002/art.38805
149. Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J. Knockdown of lncRNA NEAT1 Inhibits Th17/CD4(+) T Cell Differentiation Through Reducing the STAT3 Protein Level. J Cell Physiol (2019) 234(12):22477–84. doi: 10.1002/jcp.28811
150. Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, et al. lncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis. Int J Mol Sci (2018) 19(9):2806. doi: 10.3390/ijms19092806
151. Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung CH, et al. Increased Expression of Long Noncoding RNAs LOC100652951 and LOC100506036 in T Cells From Patients With Rheumatoid Arthritis Facilitates the Inflammatory Responses. Immunol Res (2016) 64(2):576–83. doi: 10.1007/s12026-015-8756-8
152. Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, et al. DDX5 and its Associated lncRNA Rmrp Modulate TH17 Cell Effector Functions. Nature (2015) 528(7583):517–22. doi: 10.1038/nature16193
153. Moharamoghli M, Hassan-Zadeh V, Dolatshahi E, Alizadeh Z, Farazmand A. The Expression of GAS5, THRIL, and RMRP lncRNAs Is Increased in T Cells of Patients With Rheumatoid Arthritis. Clin Rheumatol (2019) 38(11):3073–80. doi: 10.1007/s10067-019-04694-z
154. Peng T, Ji D, Jiang Y. Long non-Coding RNA GAS5 Suppresses Rheumatoid Arthritis Progression via miR-128-3p/HDAC4 Axis. Mol Cell Biochem (2021) 476(6):2491–501. doi: 10.1007/s11010-021-04098-1
155. Yang S, Yin W, Ding Y, Liu F. Lnc RNA ZFAS1 Regulates the Proliferation, Apoptosis, Inflammatory Response and Autophagy of Fibroblast-Like Synoviocytes via miR-2682-5p/ADAMTS9 Axis in Rheumatoid Arthritis. Biosci Rep (2020) 40(8):BSR20201273. doi: 10.1042/bsr20201273
156. Yan S, Wang P, Wang J, Yang J, Lu H, Jin C, et al. Long Non-Coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-κb Signaling Pathway in Rheumatoid Arthritis. Front Immunol (2019) 10:2218. doi: 10.3389/fimmu.2019.02218
157. Lao MX, Xu HS. Involvement of Long Non-Coding RNAs in the Pathogenesis of Rheumatoid Arthritis. Chin Med J (Engl) (2020) 133(8):941–50. doi: 10.1097/cm9.0000000000000755
158. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs Are Abundant, Conserved, and Associated With ALU Repeats. Rna (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
159. Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as Therapeutic Agents and Targets. Front Physiol (2018) 9:1262. doi: 10.3389/fphys.2018.01262
160. Xia X, Tang X, Wang S. Roles of CircRNAs in Autoimmune Diseases. Front Immunol (2019) 10:639. doi: 10.3389/fimmu.2019.00639
161. Deviatkin AA, Vakulenko YA, Akhmadishina LV, Tarasov VV, Beloukhova MI, Zamyatnin AA Jr, et al. Emerging Concepts and Challenges in Rheumatoid Arthritis Gene Therapy. Biomedicines (2020) 8(1):9. doi: 10.3390/biomedicines8010009
162. Wen J, Liu J, Zhang P, Jiang H, Xin L, Wan L, et al. RNA-Seq Reveals the Circular RNA and miRNA Expression Profile of Peripheral Blood Mononuclear Cells in Patients With Rheumatoid Arthritis. Biosci Rep (2020) 40(4). doi: 10.1042/bsr20193160
163. Wen J, Liu J, Wang X, Wang J. Expression and Clinical Significance of Circular RNAs Related to Immunity and Inflammation in Patients With Rheumatoid Arthritis. Int Immunopharmacol (2021) 92:107366. doi: 10.1016/j.intimp.2021.107366
164. Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, et al. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells From Rheumatoid Arthritis Patients. Cell Physiol Biochem (2017) 42(2):651–9. doi: 10.1159/000477883
165. Jiang Z, Zhong Z, Miao Q, Zhang Y, Ni B, Zhang M, et al. Circptpn22 as a Novel Biomarker and ceRNA in Peripheral Blood Mononuclear Cells of Rheumatoid Arthritis. Mol Med Rep (2021) 24(2):617. doi: 10.3892/mmr.2021.12256
166. Ouyang Q, Liu C, Lu X, Liang R, Zhao J, Yang M. Identification of Circular RNAs Circ_0005008 and Circ_0005198 in Plasma as Novel Biomarkers for New-Onset Rheumatoid Arthritis. Front Pharmacol (2021) 12:722017. doi: 10.3389/fphar.2021.722017
167. Cai Y, Liang R, Xiao S, Huang Q, Zhu D, Shi GP, et al. Circ_0088194 Promotes the Invasion and Migration of Rheumatoid Arthritis Fibroblast-Like Synoviocytes via the miR-766-3p/MMP2 Axis. Front Immunol (2021) 12:628654. doi: 10.3389/fimmu.2021.628654
168. Qu W, Jiang L, Hou G. Circ-AFF2/miR-650/CNP Axis Promotes Proliferation, Inflammatory Response, Migration, and Invasion of Rheumatoid Arthritis Synovial Fibroblasts. J Orthop Surg Res (2021) 16(1):165. doi: 10.1186/s13018-021-02306-8
169. Zhi L, Liang J, Huang W, Ma J, Qing Z, Wang X. Circ_AFF2 Facilitates Proliferation and Inflammatory Response of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via the miR-375/TAB2 Axis. Exp Mol Pathol (2021) 119:104617. doi: 10.1016/j.yexmp.2021.104617
170. Zhang S, Zhao J, Ma W. Circ-Sirt1 Inhibits Proliferation, Induces Apoptosis, and Ameliorates Inflammation in Human Rheumatoid Arthritis Fibroblast-Like Synoviocytes. Autoimmunity (2021) 54(8):514–25. doi: 10.1080/08916934.2021.1969550
171. Chen L, Huang H, Chen L, Xu L, Chen J, Lu Q. Circ-PTTG1IP/miR-671-5p/TLR4 Axis Regulates Proliferation, Migration, Invasion and Inflammatory Response of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. Gen Physiol Biophys (2021) 40(3):207–19. doi: 10.4149/gpb_2021014
172. Luo Z, Chen S, Chen X. CircMAPK9 Promotes the Progression of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via the miR-140-3p/PPM1A Axis. J Orthop Surg Res (2021) 16(1):395. doi: 10.1186/s13018-021-02550-y
173. Li X, Qu M, Zhang J, Chen K, Ma X. CircASH2L Facilitates Tumor-Like Biologic Behaviours and Inflammation of Fibroblast-Like Synoviocytes via miR-129-5p/HIPK2 Axis in Rheumatoid Arthritis. J Orthop Surg Res (2021) 16(1):302. doi: 10.1186/s13018-021-02432-3
174. Hao J, Chen Y, Yu Y. Circular RNA Circ_0008360 Inhibits the Proliferation, Migration, and Inflammation and Promotes Apoptosis of Fibroblast-Like Synoviocytes by Regulating miR-135b-5p/HDAC4 Axis in Rheumatoid Arthritis. Inflammation (2021). doi: 10.1007/s10753-021-01538-4
175. Yang J, Cheng M, Gu B, Wang J, Yan S, Xu D. CircRNA_09505 Aggravates Inflammation and Joint Damage in Collagen-Induced Arthritis Mice via miR-6089/AKT1/NF-κb Axis. Cell Death Dis (2020) 11(10):833. doi: 10.1038/s41419-020-03038-z
176. Tang X, Wang J, Xia X, Tian J, Rui K, Xu H, et al. Elevated Expression of ciRS-7 in Peripheral Blood Mononuclear Cells From Rheumatoid Arthritis Patients. Diagn Pathol (2019) 14(1):11. doi: 10.1186/s13000-019-0783-7
177. Liao J, Wang J, Liu Y, Li J, Duan L. Transcriptome Sequencing of lncRNA, miRNA, mRNA and Interaction Network Constructing in Coronary Heart Disease. BMC Med Genomics (2019) 12(1):124. doi: 10.1186/s12920-019-0570-z
178. Yoon JH, Abdelmohsen K, Gorospe M. Functional Interactions Among microRNAs and Long Noncoding RNAs. Semin Cell Dev Biol (2014) 34:9–14. doi: 10.1016/j.semcdb.2014.05.015
179. Li Y, Huo C, Lin X, Xu J. Computational Identification of Cross-Talking ceRNAs. Adv Exp Med Biol (2018) 1094:97–108. doi: 10.1007/978-981-13-0719-5_10
180. Liu DK, Li YJ, Tian B, Sun HM, Li QY, Ren BF. LncRNA PRNCR1 Aggravates the Malignancy of Oral Squamous Cell Carcinoma by Regulating miR-326/FSCN1 Axis. Eur Rev Med Pharmacol Sci (2021) 25(8):3226–34. doi: 10.26355/eurrev_202104_25731
181. Leucci E, Patella F, Waage J, Holmstrøm K, Lindow M, Porse B, et al. microRNA-9 Targets the Long non-Coding RNA MALAT1 for Degradation in the Nucleus. Sci Rep (2013) 3:2535. doi: 10.1038/srep02535
182. Faghihi MA, Zhang M, Huang J, Modarresi F, van der Brug MP, Nalls MA, et al. Evidence for Natural Antisense Transcript-Mediated Inhibition of microRNA Function. Genome Biol (2010) 11(5):R56. doi: 10.1186/gb-2010-11-5-r56
183. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a Developmental Reservoir of miR-675 That Suppresses Growth and Igf1r. Nat Cell Biol (2012) 14(7):659–65. doi: 10.1038/ncb2521
184. Xu H, Chen W, Zheng F, Tang D, Liu D, Wang G, et al. Reconstruction and Analysis of the Aberrant lncRNA-miRNA-mRNA Network in Systemic Lupus Erythematosus. Lupus (2020) 29(4):398–406. doi: 10.1177/0961203320908927
185. Zhao Q, Zhao F, Liu C, Xu T, Song K. LncRNA FOXD2-AS1 Promotes Cell Proliferation and Invasion of Fibroblast-Like Synoviocytes by Regulation of miR-331-3p/PIAS3 Pathway in Rheumatoid Arthritis. Autoimmunity (2021) 54(5):254–63. doi: 10.1080/08916934.2021.1919879
186. Tang J, Yi S, Liu Y. Long non-Coding RNA PVT1 can Regulate the Proliferation and Inflammatory Responses of Rheumatoid Arthritis Fibroblast-Like Synoviocytes by Targeting microRNA-145-5p. Hum Cell (2020) 33(4):1081–90. doi: 10.1007/s13577-020-00419-6
187. Wang J, Kong X, Hu H, Shi S. Knockdown of Long Non-Coding RNA PVT1 Induces Apoptosis of Fibroblast-Like Synoviocytes Through Modulating miR-543-Dependent SCUBE2 in Rheumatoid Arthritis. J Orthop Surg Res (2020) 15(1):142. doi: 10.1186/s13018-020-01641-6
188. Chen J, Luo X, Liu M, Peng L, Zhao Z, He C, et al. Silencing Long non-Coding RNA NEAT1 Attenuates Rheumatoid Arthritis via the MAPK/ERK Signalling Pathway by Downregulating microRNA-129 and microRNA-204. RNA Biol (2021) 18(5):657–68. doi: 10.1080/15476286.2020.1857941
189. Wang Y, Hou L, Yuan X, Xu N, Zhao S, Yang L, et al. LncRNA NEAT1 Targets Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via the miR-410-3p/YY1 Axis. Front Immunol (2020) 11:1975. doi: 10.3389/fimmu.2020.01975
190. Rao Y, Fang Y, Tan W, Liu D, Pang Y, Wu X, et al. Delivery of Long Non-Coding RNA NEAT1 by Peripheral Blood Monouclear Cells-Derived Exosomes Promotes the Occurrence of Rheumatoid Arthritis via the MicroRNA-23a/MDM2/SIRT6 Axis. Front Cell Dev Biol (2020) 8:551681. doi: 10.3389/fcell.2020.551681
191. Zheng J, Zeng P, Zhang H, Zhou Y, Liao J, Zhu W, et al. Long Noncoding RNA ZFAS1 Silencing Alleviates Rheumatoid Arthritis via Blocking miR-296-5p-Mediated Down-Regulation of MMP-15. Int Immunopharmacol (2021) 90:107061. doi: 10.1016/j.intimp.2020.107061
192. Wang ZQ, Xiu DH, Jiang JL, Liu GF. Long non-Coding RNA XIST Binding to Let-7c-5p Contributes to Rheumatoid Arthritis Through its Effects on Proliferation and Differentiation of Osteoblasts via Regulation of STAT3. J Clin Lab Anal (2020) 34(11):e23496. doi: 10.1002/jcla.23496
193. Zhao F, Dong J, Guo J, Bi L. Inhibiting Role of Long non-Coding RNA LINC01197 in Inflammation in Rheumatoid Arthritis Through the microRNA-150/THBS2 Axis. Exp Cell Res (2020) 394(2):112136. doi: 10.1016/j.yexcr.2020.112136
194. Qing P, Liu Y. Inhibitory Role of Long non-Coding RNA OIP5-AS1 in Rheumatoid Arthritis Progression Through the microRNA-448-Paraoxonase 1-Toll-Like Receptor 3-Nuclear Factor κb Axis. Exp Physiol (2020) 105(10):1708–19. doi: 10.1113/ep088608
195. Wang W, Guo P, Chen M, Chen D, Cheng Y, He L. FOXM1/LINC00152 Feedback Loop Regulates Proliferation and Apoptosis in Rheumatoid Arthritis Fibroblast-Like Synoviocytes via Wnt/β-Catenin Signaling Pathway. Biosci Rep (2020) 40(1):BSR20191900. doi: 10.1042/bsr20191900
196. Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, et al. An Emerging Function of circRNA-miRNAs-mRNA Axis in Human Diseases. Oncotarget (2017) 8(42):73271–81. doi: 10.18632/oncotarget.19154
197. Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA Code: Emerging Implications for Cancer Diagnosis and Treatment. Mol Oncol (2019) 13(4):669–80. doi: 10.1002/1878-0261.12468
198. Chen X, Ouyang Z, Shen Y, Liu B, Zhang Q, Wan L, et al. CircRNA_28313/miR-195a/CSF1 Axis Modulates Osteoclast Differentiation to Affect OVX-Induced Bone Absorption in Mice. RNA Biol (2019) 16(9):1249–62. doi: 10.1080/15476286.2019.1624470
199. Zhang M, Bai X, Zeng X, Liu J, Liu F, Zhang Z. circRNA-miRNA-mRNA in Breast Cancer. Clin Chim Acta (2021) 523:120–30. doi: 10.1016/j.cca.2021.09.013
200. Fan Z, Bai Y, Zhang Q, Qian P. CircRNA Circ_POLA2 Promotes Lung Cancer Cell Stemness via Regulating the miR-326/GNB1 Axis. Environ Toxicol (2020) 35(10):1146–56. doi: 10.1002/tox.22980
201. Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang Q, et al. MicroRNA-1224 Splicing CircularRNA-Filip1l in an Ago2-Dependent Manner Regulates Chronic Inflammatory Pain via Targeting Ubr5. J Neurosci (2019) 39(11):2125–43. doi: 10.1523/jneurosci.1631-18.2018
202. Lacaille D, Avina-Zubieta JA, Sayre EC, Abrahamowicz M. Improvement in 5-Year Mortality in Incident Rheumatoid Arthritis Compared With the General Population-Closing the Mortality Gap. Ann Rheum Dis (2017) 76(6):1057–63. doi: 10.1136/annrheumdis-2016-209562
203. Wang Z, Zhang W, Li S, Chen D, Wang L, Xu B, et al. Multiple Time-Point Assessment of lncRNA MEG3 Shows Potential to Monitor Treatment Efficacy in Rheumatoid Arthritis Patients. Biomark Med (2021) 15(14):1261–70. doi: 10.2217/bmm-2021-0070
204. Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, et al. Identification of Circular RNAs Hsa_Circ_0044235 in Peripheral Blood as Novel Biomarkers for Rheumatoid Arthritis. Clin Exp Immunol (2018) 194(1):118–24. doi: 10.1111/cei.13181
205. Liu X, Ni S, Li C, Xu N, Chen W, Wu M, et al. Circulating microRNA-23b as a New Biomarker for Rheumatoid Arthritis. Gene (2019) 712:143911. doi: 10.1016/j.gene.2019.06.001
206. Duroux-Richard I, Pers YM, Fabre S, Ammari M, Baeten D, Cartron G, et al. Circulating miRNA-125b Is a Potential Biomarker Predicting Response to Rituximab in Rheumatoid Arthritis. Mediators Inflamm (2014) 2014:342524. doi: 10.1155/2014/342524
207. López-Pedrera C, Pérez-Sánchez C, Ramos-Casals M, Santos-Gonzalez M, Rodriguez-Ariza A, Cuadrado MJ. Cardiovascular Risk in Systemic Autoimmune Diseases: Epigenetic Mechanisms of Immune Regulatory Functions. Clin Dev Immunol (2012) 2012:974648. doi: 10.1155/2012/974648
208. Wang Y, Zheng F, Gao G, Yan S, Zhang L, Wang L, et al. MiR-548a-3p Regulates Inflammatory Response via TLR4/NF-κb Signaling Pathway in Rheumatoid Arthritis. J Cell Biochem (2018). doi: 10.1002/jcb.26659
209. Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic Potential of Mesenchymal Cell-Derived miRNA-150-5p-Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF. J Immunol (2018) 201(8):2472–82. doi: 10.4049/jimmunol.1800304
210. Mu N, Gu J, Huang T, Zhang C, Shu Z, Li M, et al. A Novel NF-κb/YY1/microRNA-10a Regulatory Circuit in Fibroblast-Like Synoviocytes Regulates Inflammation in Rheumatoid Arthritis. Sci Rep (2016) 6:20059. doi: 10.1038/srep20059
211. Rai MF, Pan H, Yan H, Sandell LJ, Pham CTN, Wickline SA. Applications of RNA Interference in the Treatment of Arthritis. Transl Res (2019) 214:1–16. doi: 10.1016/j.trsl.2019.07.002
212. Scheinman RI, Trivedi R, Vermillion S, Kompella UB. Functionalized STAT1 siRNA Nanoparticles Regress Rheumatoid Arthritis in a Mouse Model. Nanomed (Lond) (2011) 6(10):1669–82. doi: 10.2217/nnm.11.90
213. Banerjee D, Roeker LE, Grogan M, Swiecicki P, Poterucha J, Heimbach J, et al. Outcomes of Patients With Familial Transthyretin Amyloidosis After Liver Transplantation. Prog Transplant (2017) 27(3):246–50. doi: 10.1177/1526924817715463
Keywords: noncoding RNA, rheumatoid arthritis, miRNA, circRNA, lncRNA, review
Citation: Han J-J, Wang X-Q and Zhang X-A (2022) Functional Interactions Between lncRNAs/circRNAs and miRNAs: Insights Into Rheumatoid Arthritis. Front. Immunol. 13:810317. doi: 10.3389/fimmu.2022.810317
Received: 19 November 2021; Accepted: 06 January 2022;
Published: 07 February 2022.
Edited by:
Kunihiro Ichinose, Nagasaki University, JapanReviewed by:
Consuelo Romero-Sanchez, El Bosque University, ColombiaJin Hou, Second Military Medical University, China
Copyright © 2022 Han, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-An Zhang, emhhbmd4YTI3MjVAMTYzLmNvbQ==; Xue-Qiang Wang, cWlhbmc4OTdAMTYzLmNvbQ==
 Juan-Juan Han
Juan-Juan Han Xue-Qiang Wang
Xue-Qiang Wang Xin-An Zhang
Xin-An Zhang