- 1Department: Apoptosis Research Centre, National University of Ireland Galway, Galway, Ireland
- 2ONK Therapeutics, Galway, Ireland
Genome engineered natural killer (NK) cell therapies are emerging as a promising cancer immunotherapy platform with potential advantages and remaining uncertainties. Feeder cells induce activation and proliferation of NK cells via cell surface receptor-ligand interactions, supported by cytokines. Feeder cell expanded NK cell products have supported several NK cell adoptive transfer clinical trials over the past decade. Genome engineered NK cell therapies, including CAR-NK cells, seek to combine innate and alloreactive NK cell anti-tumor activity with antigen specific targeting or additional modifications aimed at improving NK cell persistence, homing or effector function. The profound activating and expansion stimulus provided by feeder cells is integral to current applications of clinical-scale genome engineering approaches in donor-derived, primary NK cells. Herein we explore the complex interactions that exist between feeder cells and both viral and emerging non-viral genome editing technologies in NK cell engineering. We focus on two established clinical-grade feeder systems; Epstein-Barr virus transformed lymphoblastoid cell lines and genetically engineered K562.mbIL21.4-1BBL feeder cells.
Introduction
Natural killer (NK) cell-based adoptive cell transfer (ACT) is a promising experimental approach to cancer immunotherapy. The ability to administer NK cells across HLA barriers, without a risk of graft-versus-host disease has enabled NK cell products from a variety of cell sources to be evaluated in clinical trials (1–4). The therapeutic potential of unmodified NK ACT has been most apparent to date in the setting of hematological malignancy (1, 5, 6). The traditional paradigm that NK cells are more challenging to genetically engineer relative to T-cells is evolving (7, 8). Clinical scale gene editing of NK cells is established, most prominently through the addition of a chimeric antigen receptor (CAR) (3). CAR-NK cells add a potent layer of antigen specific activation to a complement of NK cell activating and inhibitory receptors which are the basis of innate and alloreactive NK cell recognition.
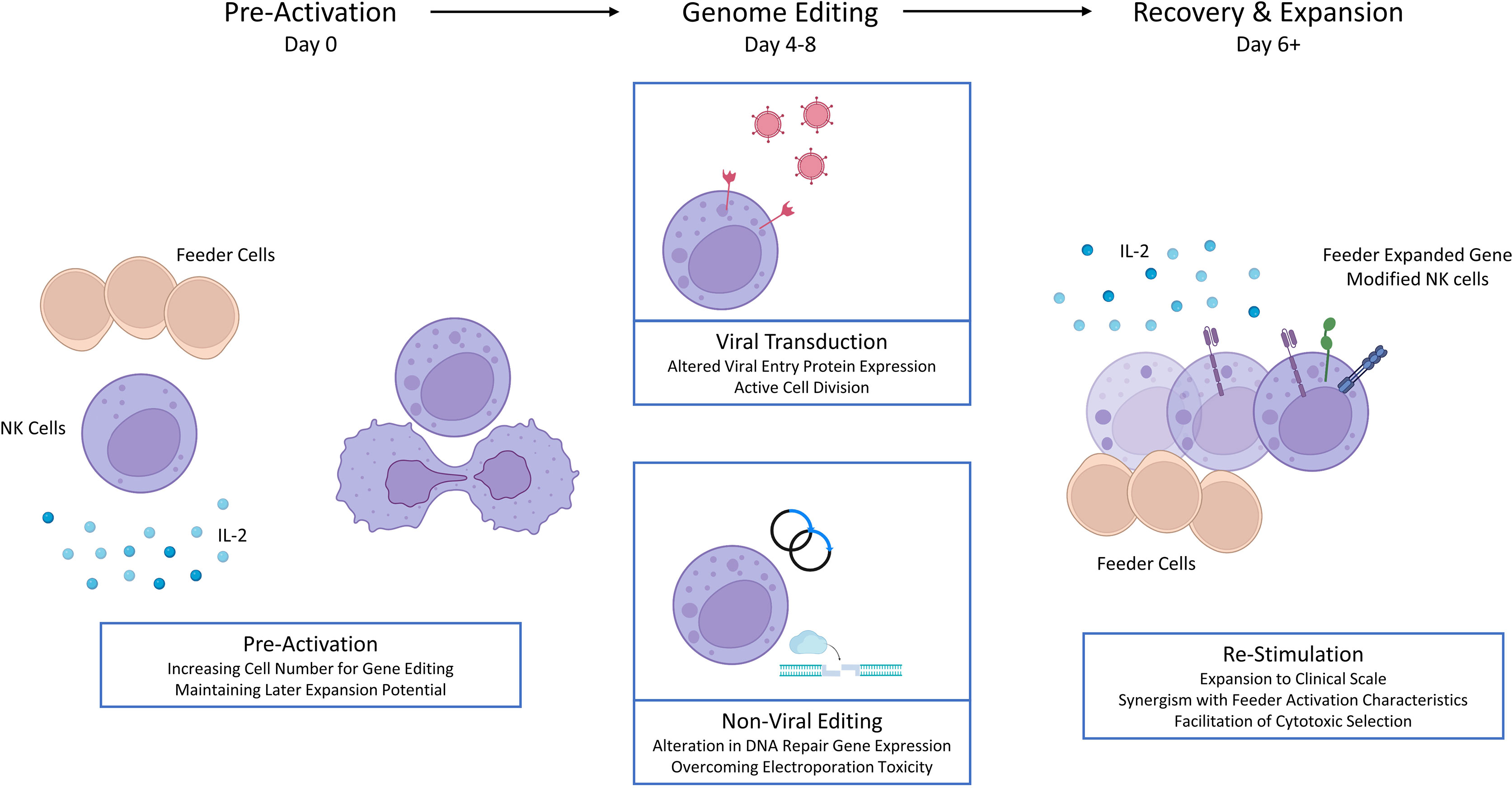
Primary NK cells are sourced from donor peripheral blood (PB) or umbilical cord blood (UCB). Although both sources are the basis of investigational therapies, UCB contains additional NK cell progenitors and is collected non-invasively or from existing banked material (9–11). Large scale NK cell production from small initial quantities relies upon feeder cells which naturally, or via further engineering, present ligands for NK cell receptors driving profound ex vivo NK cell expansion when combined with cytokines (Figure 1) (12, 13). To avoid a risk of proliferation and contamination, feeder cell lines are γ-irradiated prior to use. While the NK cell yield varies between donors, several approaches have been used to support clinical trials (6, 14, 15).
Figure 1 Determinants and characteristics of feeder expanded NK cells. iKIR – inhibitory killer immunoglobulin receptor, EBV, Epstein-Barr Virus; mbIL21, membrane bound IL-21; TRAIL, TNF-related apoptosis-inducing ligand.
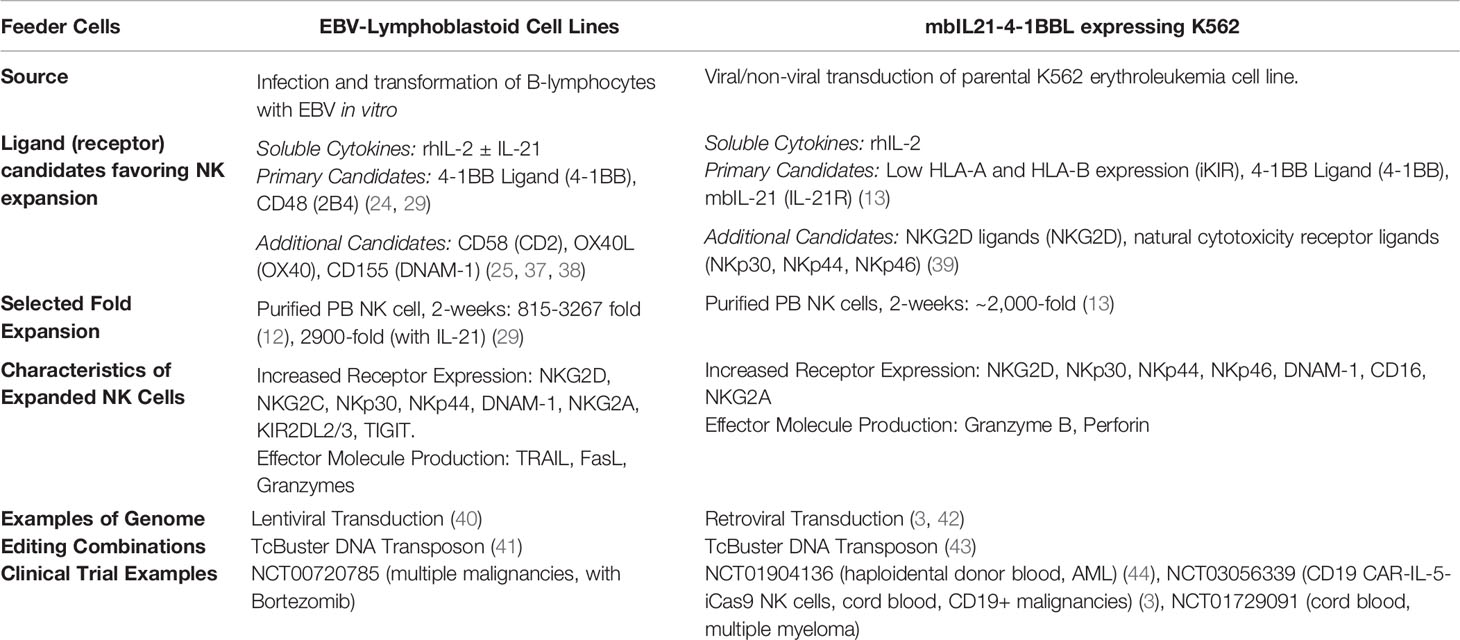
Feeder cells are integral to clinical scale, genetically modified primary NK cell production protocols (3, 16–18). This contrasts with genome engineered T-cells where antibody conjugated beads are the typical activating stimulus, induced pluripotent stem cell (iPSC) derived NK cells (a distinct and promising domain of investigational NK therapies) which are genetically engineered prior to differentiation and expansion, and NK cell lines (e.g. NK-92) with autonomous growth potential (2, 4, 17). The role of feeder cells in generating genetically modified primary NK cell products is multifaceted. Initial feeder cell stimulation may enhance the efficiency of gene editing through simple induction of cell division or more complex effects reflecting altered gene expression. Repeated feeder cell stimulation can support cellular recovery and expansion to clinical scale despite extensive ex vivo manipulation. This central role for feeder cells in the era of genome engineering compels us to examine the relationship between these vital tools. Herein we consider the interface between feeder cell-based primary NK cell activation and expansion with gene editing, focusing on prominent clinical-grade feeder cell approaches, Epstein-Barr Virus transformed lymphoblastoid cell lines (EBV-LCL) and genetically engineered K562.mbIL21.4-1BBL cells.
Prominent Feeder Cells for NK Expansion
In vitro infection of a mixed lymphocyte population with EBV creates an immortalized cell line with the characteristics of proliferating B-cells despite expression of relatively few viral genes (19). Early reports of preferential expansion of NK cells which occurs after co-culture of EBV-LCLs with PB mononuclear cells recognized the importance of both IL-2 and cell contact (20, 21). EBV-LCLs naturally express 4-1BBL (CD137L), the ligand for 4-1BB- an inducible stimulatory receptor on activated NK cells which is also upregulated by interactions with the Fc portion of antibodies (22, 23). Stimulation through 4-1BB is an important contributor to clinical scale NK cell expansion. EBV-LCLs express other ligands relevant to NK cell activation and expansion, including CD155, CD48 and CD58, through interactions with the NK cell activating receptors DNAM-1, 2B4 and CD2 respectively (24, 25). Dr. Richard Childs and colleagues at the National Institutes of Health (NIH) pioneered the use of EBV-LCL feeder cells as a clinical-grade NK cell expansion technique (26). Using enriched NK cell populations, mean 1,000-2,000-fold expansions are observed over 14 days and this process was subsequently automated using the GMP-compliant Miltenyi CliniMACS Prodigy (12, 27). Robust expansion of UCB NK cells has also been described from small amounts of starting material (28). The addition of a single IL-21 exposure at the outset enhances NK cell proliferation and enables prolonged expansion, overcoming senescence (29). The EBV-LCL expansion process was used to support a clinical trial at the NIH of expanded NK cells combined with Bortezomib, a sensitizing agent to the death ligand TRAIL, across a range of malignancies (NCT00720785).
Initial descriptions of the immunophenotype and in vitro function of EBV-LCL expanded NK cells, have been complemented by gene expression profiling (GEP) (12, 27, 30). Relative to freshly isolated NK cells, expanded populations adopt an activated phenotype with increased expression of activating receptors NKG2D, NKG2C, NKp30, NKp44, DNAM-1 and key effector molecules TRAIL, FasL and Granzymes. Described alterations in inhibitory receptors include increases in NKG2A positive cells and relative increases in both KIR2DL2/3 and TIGIT (12, 27). The diverse effects that this expansion process has on NK cells is highlighted by GEP with prominent upregulation of genes involved in metabolism, cytotoxicity and cellular growth (30). Many of these beneficial characteristics appear to be largely dependent on continued IL-2 exposure, and exogenous IL-2 administration alone may not overcome a loss of activation characteristics encountered in vivo (29).
An alternative, prominent feeder cell approach to NK cell expansion began with the recognition that K562, an erythroleukemia cell line deficient in HLA class I and a common target cell for in vitro NK cell cytotoxicity assays, provides a contact dependent expansion stimulus to NK cells (31). Expression of additional NK cell stimulatory molecules, primarily 4-1BBL and membrane bound (mb) cytokines (IL-15 or IL-21), introduced by viral transduction or DNA transposons, greatly enhances the expansion potential of K562 (32, 33). The impact of K562.4-1BBL feeder cell expression of mbIL15 or mbIL21 on the characteristics of expanded NK cells was reported by Denman et al. (13). Feeder cell expression of mbIL15 produced a mean 825-fold NK expansion (day 21), with telomere shortening relative to non-expanded NK cells and senescence characterized by loss of proliferation between weeks 4 and 6. Feeder cell expression of mbIL21 produced a mean 47,697-fold NK cell expansion (day 21), with an increased telomere length compared to baseline and sustained NK cell proliferation to week 6. Feeder cell expression of either mb cytokine was associated with a similar NK cell phenotype, including high expression of natural cytotoxicity receptors (NKp30, NKp44, NKp46) and KIR. Prominent activation and proliferation characteristics including high levels of granzyme B and perforin were observed by GEP. Relative to mbIL15, feeder cell expression of mbIL21 produced expanded NK cells with a similar in vitro cytotoxicity, but greater cytokine production. Feeder cell expression of mbIL21 has been shown to enhance NK cell metabolism through STAT3/cMyc pathway signaling (34). Several groups have reported on the use of K562.mbIL21.4-1BBL feeder cells to expand NK cells used in early phase clinical studies across PB and UCB NK cells, including the most prominent example of a CAR-NK cell therapy reported to date (3, 6, 14, 35). Membrane particles derived from K562.mbIL21.4-1BBL feeder cells have also been extracted and applied to NK expansion, overcoming the risk of viable feeder cell contamination, despite irradiation (36).
The characteristics of these prominent feeder cell approaches are summarized and compared in Table 1. Both are being exploited to facilitate clinical-scale primary NK cell genome editing, with important benefits of feeder cell stimulation highlighted in Figure 2. The reliance upon feeder cell expansion in current gene editing protocols has implications for the characteristics of these products. Some features may be synergistic with specific gene edits such as the generation of CAR-NK cells. For example, death ligands TRAIL and FasL are implicated in supporting NK cell serial killing, and subsequent secretion and/or shedding of TRAIL may trigger apoptosis of target antigen negative populations as described for CAR-T cells (45, 46). Alternatively, some characteristics are less desirable, such as the loss of activation features that may occur in vivo. Here, rational genome editing targets may overcome these limitations. Introduction of an IL-15 gene allows for autonomous production and enhances the functionality and persistence of CAR-NK cells, without inducing systemic alterations in IL-15 (42). Knockout of cytokine-inducible SH2-containing protein (CISH), a negative regulator of IL-15 signaling, has multiple effects relevant to NK cell therapies including enhanced cytotoxicity, metabolism and persistence which have been characterized in pre-clinical in vivo models (47–49). The induction of memory like characteristics, by transient exposure to IL-12, IL-15 and IL-18 in vitro, offers an alternative approach to enhancing in vivo NK cell performance. The beneficial characteristics of these cytokine induced memory like (CIML) NK cells (including enhanced in vivo persistence) and CAR triggered activation are synergistic, however the interaction between CIML-NK features and feeder mediated expansion is less clear (50).
Feeder Cells and Viral Transduction
Retroviral vectors are established gene delivery tools based upon the principle of reverse transcription of a viral RNA genome into double stranded DNA, which is then integrated into the host cell genome by the enzyme integrase, allowing for stable transgene expression (51). Gamma retroviruses efficiently deliver genes to replicating cells via nuclear membrane breakdown during mitosis, while lentiviral vectors can also transduce resting cells. Each of these classes are successfully used in manufacturing regulatory approved CAR-T products (52, 53). Several groups have described the role of K562.mbIL21.4-1BBL feeder cells in facilitating NK cell viral transduction. Using an RD114 pseudotyped retroviral vector Streltsova et al. described the transduction of feeder activated but not resting NK cells. Interestingly, the small, mature CD57+ NK cell subset detectable within the expanded population, failed to proliferate in response to the feeder stimulus and remained resistant to transduction, emphasizing the role of cellular division (54). In another example where feeder-free expansion and K562.mbIL21.4-1BBL feeder cells were compared, both facilitated transduction with a baboon envelope pseudotyped lentiviral vector, however subsequent re-expansion of sorted populations relied upon feeder cell stimulation (55). Interestingly, one proposed mechanism for the enhanced transduction achieved after activation relative to freshly isolated NK cells was upregulation of the viral entry proteins ASCT1 and ASCT2. Given the importance of cellular replication to successful retroviral engineering and the clinical-scale expansion required, the robust activating and expansion stimulus delivered by feeder cells has been central to the clinical translation of retrovirally engineered CAR-NK cells. In their phase I clinical trial report, Liu et al. describe an UCB derived, expanded NK cell product which expresses a CD19 CAR, IL-15, and an inducible caspase 9 suicide gene. This multi-cistronic gene was introduced on day 6 using a retroviral vector, after initial stimulation of purified UCB NK cells by K562.mbIL21.4-1BBL feeder cells. Later restimulation allowed for expansion sufficient to provide multiple clinical doses (3, 42). This groundbreaking study, which demonstrated clinical responses in the context of B-cell malignancies and long-term persistence of CAR-NK cells, established the promise of the CAR-NK cell platform.
Recently, Allan et al. reported on the optimization of lentiviral transduction in primary NK cells which further explores the role that feeder cells play in the delicate balance of gene editing and expansion potential (40). Notably, while initial activation with IL-2 alone allowed for successful transduction relative to unstimulated NK cells, subsequent EBV-LCL feeder cell stimulated expansion was limited. Incorporation of EBV-LCL pre-stimulation modestly reduced integration rates but provided an overall higher yield of transduced cells at later timepoints reflecting a maintained expansion potential. The group went on to demonstrate that 5-7 days pre-activation prior to lentiviral transduction was optimal, balancing efficiency and fold-expansion, while the purity of transduced cells could be enhanced by immunomagnetic selection based on truncated protein selection markers. NK cells rapidly overwhelm dissipating feeder cells and were harvested for genetic engineering without repeated selection. In experiments where direct comparison was made between EBV-LCL and K562.mbIL21.4-1BBL feeder cells, similar results were obtained for transduction efficiency and expansion capacity. A distinct lymphoblastoid cell line, 721.221, engineered to express mb IL-21, has also been successfully combined with retroviral transduction to create CAR-NK cell products in a recent pre-clinical report (56).
Viral transduction approaches generally confer less toxicity to immune cells relative to electroporation with DNA based cargo. This improved viability, could be the basis of successful integration of feeder-free expansion approaches in the future. Feeder-free NK cell expansion relies upon cytokines and stimulating supplements, or antibodies, but in general results in a lower fold expansion. Feeder-free approaches offer logistical benefits and overcome the risk of viable feeder cell contamination, despite irradiation. Clinical scale examples of feeder-free expansion from primary NK cells have been reported, including using the supplement nicotinamide which confers a multitude of desirable characteristics to the expanded NK cell product beyond increasing cell number, including enhanced cytotoxicity and in vivo persistence (57, 58). Feeder-free approaches tend to rely upon larger volume apheresis product as a starting material, and to our knowledge have not been successfully combined with clinical-scale, stable gene editing of primary NK cells to date (59).
Feeder Cells and Non-Viral Gene Editing
Non-viral gene editing approaches are increasingly being applied to NK cells successfully and this field has been recently reviewed (8). Electroporation remains the most widely investigated non-viral approach to deliver diverse cargo including mRNA, DNA and protein to NK cells. This allows for transient gene expression from a non-integrated DNA plasmid or mRNA, or, genome level modification and stable expression when combined with transposon technology, or knock-in after delivery of endonucleases and a DNA template (8). Non-viral genome editing has potential safety advantages relative to viral transduction, but lacks the longer term safety data that is available for clinical products manufactured using viral approaches (60). DNA transposons are versatile gene vector systems, involving co-delivery of a transposon plasmid containing the genetic cargo, and a transposase enzyme which cuts and pastes the transposon into genomic DNA by recognition of flanking inverted terminal repeat (ITR) sequences (61). A variety of factors influence the efficiency of NK cell electroporation, including the type and concentration of cargo, pulse characteristics and electroporation buffer (62). The ultimate rate of stable integration achieved after DNA transposon electroporation may also be impacted by differences in the recovery and expansion rate of modified and non-modified populations, reflecting recovery conditions and the timing of restimulation.
Feeder cell-based activation and expansion is integral to early reports of the DNA transposon-based gene editing of primary NK cells. K562.mbIL21.4-1BBL feeder cell pre-activation supports enhanced electroporation efficiency and recovery when delivering CRISPR/Cas9 cargo (63). In a recent pre-print, Pomeroy et al. describe the application of the TcBuster™ transposon system to primary NK cells using the K562.mbIL21.4-1BBL pre-activation approach (43). Day 4 is identified as the optimal time for transposon delivery by electroporation, which may reflect the later upregulation of DNA sensors, and transposition efficiency was further enhanced by ribonuclease inhibitor pre-treatment of NK cells (transposase enzyme mRNA was simultaneously delivered with the DNA transposon). Their approach also incorporates Nanoplasmid™ to reduce the size of the plasmid selection cassette aiming to enhance DNA delivery, while others have applied minicircle technology with a similar goal (64, 65). Repeat stimulation 48h post electroporation allowed for clinically relevant expansion characteristics. Our group and collaborators have also recently applied TcBuster™ to primary NK cells using EBV-LCL pre-activation and expansion to successfully generate CAR-NK cells (41).
Viral RNA size may be a limiting factor for the efficiency of transduction in primary NK cells (40). DNA transposons allow for the delivery of larger transgenes and potentially can reduce the cost and variability of immune cell engineering. This ability has been leveraged to deliver multi-cistronic genes including a cytotoxic selection marker, allowing production of a homogenous gene modified product (66). Using these selection systems combined with robust feeder driven expansion would facilitate the outgrowth of gene modified cells from a lower percentage of transposition – an important consideration, as the ideal balance of efficiency, cellular toxicity, and subsequent expansion capability remains unclear. This was highlighted by the incidence of product derived lymphomas in two patients treated with a Piggybac transposon-based CAR-T product which were recently reported, although the transposon system was not directly implicated in this occurrence, despite extensive investigation (67).
The combination of endonucleases, with homologous DNA templates has been successfully applied to stable gene editing, including in NK cells. Cas9 mediated double strand breaks, repaired by homologous recombination or non-homologous end joining, can lead to integration of a tailored DNA template. This allows for targeted gene insertions to genetic safe-harbour sites, and simultaneous gene knock-out. The most effective examples of this approach to date, rely upon a non-integrating adenoviral vector to deliver the DNA template, shortly after electroporation delivery of gRNA/Cas9 complexes (68, 69). Considerably greater integration was achieved using a self-complementary adenovirus vector, providing for rapid availability of the dsDNA template for DNA repair. Notably, NK cell expansion with K562.mbIL21.4-1BBL feeder cells was implicated in the optimisation of this approach. Kararoudi et al. demonstrated upregulation in genes involved in homologous recombination (BRCA1 and BRCA2) in expanded NK cells, likely contributing to an improved efficiency of knock-in. Recently, efficient gene knock-out and knock-in of NK cells maintained in a feeder-free media was described using a highly active mutant of AsCas12a, a promising future approach to non-viral gene editing (70).
Discussion
The landscape of investigational NK cell therapies has rapidly diversified. iPSC-NK, and donor-derived, primary NK cell products may become effective, off-the-shelf immunotherapies. Genome editing, supported by novel technologies and adaptation of existing techniques to NK cells appears fundamental to achieving this potential. Current protocols for clinical-scale genome editing of primary, donor-derived NK cells rely upon feeder cell stimulation. Initial stimulation increases the NK cell quantity for gene editing and improves the efficiency of retroviral transduction and transposon delivery. Repeat stimulation supports NK cell recovery and subsequent expansion, allowing for clinical scale and multi-dose manufacture. The characteristics of primary NK cells expanded with each of the feeder cell systems discussed herein apply to the final product. This may be beneficial by protecting against senescence during expansion or create a barrier through potential loss of activation characteristics occurring in vivo. Although few reports directly compare the two prominent feeder approaches discussed here, they share core activating pathways, and both are being successfully applied to support NK cell genome editing. Proposed feeder-free and novel approaches to NK expansion should also be evaluated for their performance in this critical supportive role to NK cell genome editing (36, 71, 72).
Author Contributions
MO’D conceived of the review and contributed to content and editing. MG wrote the manuscript and compiled figures. SK contributed to literature review and editing. SP contributed to literature review and editing. All authors contributed to the article and approved the submitted version.
Funding
Author MG is supported by Irish Clinical Academic Training (ICAT) Program fellowship funding. ICAT is supported by the Wellcome Trust and the Health Research Board (Grant Number 203930/B/16/Z), the Health Service Executive National Doctors Training and Planning and the Health and Social Care, Research and Development Division, Northern Ireland. Author SK is funded by the Irish Research Council- Employment Based Postgraduate Program under grant number EBPPG/2021/74.
Conflict of Interest
Author MO’D is chief scientific officer and equity holder in ONK therapeutics. SK and SP are employees of ONK therapeutics. MG has received research support from ONK therapeutics.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures 1 and 2 were created using biorender.com.
References
1. Miller J, Soignier Y, Panoskaltsis-Mortari A, McNearney S, Yun G, Fautsch S, et al. Successful Adoptive Transfer and In Vivo Expansion of Human Haploidentical NK Cells in Patients With Cancer. Blood (2005) 105:3051–7. doi: 10.1182/BLOOD-2004-07-2974
2. Tang X, Yang L, Li Z, Nalin AP, Dai H, Xu T, et al. First-in-Man Clinical Trial of CAR NK-92 Cells: Safety Test of CD33-CAR NK-92 Cells in Patients With Relapsed and Refractory Acute Myeloid Leukemia(2018) (Accessed October 18, 2021).
3. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N Engl J Med (2020) 382:545–53. doi: 10.1056/NEJMOA1910607
4. Bachanova V, Cayci Z, Lewis D, Maakaron J, Janakiram M, Bartz A, et al. Initial Clinical Activity of FT596, a First-in-Class, Multi-Antigen Targeted, Off-the-Shelf, Ipsc−Derived CD19 CAR NK Cell Therapy in Relapsed/Refractory B-Cell Lymphoma (Abstract). In: 62nd American Society of Hematology Meeting and Exposition (ASH) (2020) 136 (Supplement 1):8.
5. Cooley S, He F, Bachanova V, Vercellotti GM, DeFor TE, Curtsinger JM, et al. First-in-Human Trial of Rhil-15 and Haploidentical Natural Killer Cell Therapy for Advanced Acute Myeloid Leukemia. Blood Adv (2019) 3:1970. doi: 10.1182/BLOODADVANCES.2018028332
6. Ciurea SO, Schafer JR, Bassett R, Denman CJ, Cao K, Willis D, et al. Phase 1 Clinical Trial Using Mbil21 Ex Vivo–Expanded Donor-Derived NK Cells After Haploidentical Transplantation. Blood (2017) 130:1857. doi: 10.1182/BLOOD-2017-05-785659
7. Childs R, Carlsten M. Therapeutic Approaches to Enhance Natural Killer Cell Cytotoxicity Against Cancer: The Force Awakens. Nat Rev Drug Discov (2015) 14:487–98. doi: 10.1038/NRD4506
8. Robbins GM, Wang M, Pomeroy EJ, Moriarity BS. Nonviral Genome Engineering of Natural Killer Cells. Stem Cell Res Ther 2021 121 (2021) 12:1–9. doi: 10.1186/S13287-021-02406-6
9. Mehta RS, Shpall EJ, Rezvani K. Cord Blood as a Source of Natural Killer Cells. Front Med (2015) 2:93/BIBTEX. doi: 10.3389/FMED.2015.00093/BIBTEX
10. Perez SA, Sotiropoulou PA, Gkika DG, Mahaira LG, Niarchos DK, Gritzapis AD, et al. A Novel Myeloid-Like NK Cell Progenitor in Human Umbilical Cord Blood. Blood (2003) 101:3444–50. doi: 10.1182/BLOOD-2002-05-1501
11. Rutella S, Bonanno G, Marone M, de RD, Mariotti A, Voso MT, et al. Identification of a Novel Subpopulation of Human Cord Blood CD34–CD133–CD7–CD45+Lineage– Cells Capable of Lymphoid/NK Cell Differentiation After In Vitro Exposure to IL-15. J Immunol (2003) 171:2977–88. doi: 10.4049/JIMMUNOL.171.6.2977
12. Berg M, Lundqvist A, McCoy PJR, Samsel L, Fan Y, Tawab A, et al. Clinical Grade Ex Vivo-Expanded Human Natural Killer Cells Upregulate Activating Receptors and Death Receptor Ligands and Have Enhanced Cytolytic Activity Against Tumor Cells. Cytotherapy (2009) 11:341. doi: 10.1080/14653240902807034
13. Denman CJ, Senyukov VV, Somanchi SS, Phatarpekar PV, Kopp LM, Johnson JL, et al. Membrane-Bound IL-21 Promotes Sustained Ex Vivo Proliferation of Human Natural Killer Cells. PLoS One (2012) 7:30264. doi: 10.1371/JOURNAL.PONE.0030264
14. Shah N, Li L, McCarty J, Kaur I, Yvon E, Shaim H, et al. Phase I Study of Cord Blood-Derived Natural Killer Cells Combined With Autologous Stem Cell Transplantation in Multiple Myeloma. Br J Haematol (2017) 177:457. doi: 10.1111/BJH.14570
15. Kundu S, Gurney M, O’Dwyer M. Generating Natural Killer Cells for Adoptive Transfer: Expanding Horizons. Cytotherapy (2021) 23:559–66. doi: 10.1016/J.JCYT.2020.12.002
16. Xiao L, Cen D, Gan H, Sun Y, Huang N, Xiong H, et al. Adoptive Transfer of NKG2D CAR Mrna-Engineered Natural Killer Cells in Colorectal Cancer Patients. Mol Ther (2019) 27:1114. doi: 10.1016/J.YMTHE.2019.03.011
17. Vormittag P, Gunn R, Ghorashian S. Veraitch FS. A Guide to Manufacturing CAR T Cell Therapies. Curr Opin Biotechnol (2018) 53:164–81. doi: 10.1016/j.copbio.2018.01.025
18. Numbenjapon T, Serrano L, Chang W, Forman S, Jensen M, Cooper L. Antigen-Independent and Antigen-Dependent Methods to Numerically Expand CD19-Specific CD8+ T Cells. Exp Hematol (2007) 35:1083–90. doi: 10.1016/J.EXPHEM.2007.04.007
19. Neitzei H. A Routine Method for the Establishment of Permanent Growing Lymphoblastoid Cell Lines. Hum Genet (1986) 73:320–6. doi: 10.1007/BF00279094
20. Perussia B, Ramoni C, Anegon I, Cuturi M, Faust J, Trinchieri G. Preferential Proliferation of Natural Killer Cells Among Peripheral Blood Mononuclear Cells Cocultured With B Lymphoblastoid Cell Lines. Nat Immun Cell Growth Regul (1987) 6(4):171–88.
21. Rabinowich H, Sedlmayr P, Herberman R, Whiteside T. Increased Proliferation, Lytic Activity, and Purity of Human Natural Killer Cells Cocultured With Mitogen-Activated Feeder Cells. Cell Immunol (1991) 135:454–70. doi: 10.1016/0008-8749(91)90290-R
22. Lin W, Voskens CJ, Zhang X, Schindler DG, Wood A, Burch E, et al. Fc-Dependent Expression of CD137 on Human NK Cells: Insights Into “Agonistic” Effects of Anti-CD137 Monoclonal Antibodies. Blood (2008) 112:699–707. doi: 10.1182/BLOOD-2007-11-122465
23. Alosaimi M, Hoenig M, Jaber F, Platt CD, Jones J, Wallace J, et al. Immunodeficiency and Epstein Barr Virus Induced Lymphoproliferationcaused by 4-1BB Deficiency. J Allergy Clin Immunol (2019) 144:574. doi: 10.1016/J.JACI.2019.03.002
24. Granzin M. Doctoral Thesis: Highly Efficenct Activation and Expansion of Natural Killer Cells for Clinical Use in Cancer Immunotherapy. Germany: Ruperto-Carola University of Heidelberg (2016).
25. Megyola C, Ye J, Bhaduri-McIntosh S. Identification of a Sub-Population of B Cells That Proliferates After Infection With Epstein-Barr Virus. Virol J 2011 81 (2011) 8:1–12. doi: 10.1186/1743-422X-8-84
26. Igarashi T, Wynberg J, Srinivasan R, Becknell B, McCoy JP, Takahashi Y, et al. Enhanced Cytotoxicity of Allogeneic NK Cells With Killer Immunoglobulin-Like Receptor Ligand Incompatibility Against Melanoma and Renal Cell Carcinoma Cells. Blood (2004) 104:170–7. doi: 10.1182/BLOOD-2003-12-4438
27. Granzin M, Soltenborn S, Müller S, Kollet J, Berg M, Cerwenka A, et al. Fully Automated Expansion and Activation of Clinical-Grade Natural Killer Cells for Adoptive Immunotherapy. Cytotherapy (2015) 17:621–32. doi: 10.1016/J.JCYT.2015.03.611
28. Vasu S, Berg M, Davidson-Moncada J, Tian X, Cullis H, Childs RW. A Novel Method to Expand Large Numbers of CD56+ Natural Killer Cells From a Minute Fraction of Selectively Accessed Cryopreserved Cord Blood for Immunotherapy After Transplantation. Cytotherapy (2015) 17:1582–93. doi: 10.1016/J.JCYT.2015.07.020
29. Granzin M, Stojanovic A, Miller M, Childs R, Huppert V, Cerwenka A. Highly Efficient IL-21 and Feeder Cell-Driven Ex Vivo Expansion of Human NK Cells With Therapeutic Activity in a Xenograft Mouse Model of Melanoma. Oncoimmunology (2016) 5(9):e1219007. doi: 10.1080/2162402X.2016.1219007
30. Park KU, Jin P, Sabatino M, Feng J, Civini S, Khuu H, et al. Gene Expression Analysis of Ex-Vivo Expanded and Freshly Isolated NK Cells From Cancer Patients. J Immunother (2010) 33:945. doi: 10.1097/CJI.0B013E3181F71B81
31. Robertson M, Cameron C, Lazo S, Cochran K, Voss S, Ritz J. Costimulation of Human Natural Killer Cell Proliferation: Role of Accessory Cytokines and Cell Contact-Dependent Signals(1996) (Accessed October 19, 2021).
32. Imai C, Iwamoto S, Campana D. Genetic Modification of Primary Natural Killer Cells Overcomes Inhibitory Signals and Induces Specific Killing of Leukemic Cells. Blood (2005) 106:376–83. doi: 10.1182/BLOOD-2004-12-4797
33. Min B, Yang B, Kim Y-S, Park GM, Kim H, Kim H, et al. Harnessing Novel Engineered Feeder Cells Expressing Activating Molecules for Optimal Expansion of NK Cells With Potent Antitumor Activity. Cell Mol Immunol 2021 (2021) 19(2):296–298. doi: 10.1038/s41423-021-00759-9
34. Ojo EO, Sharma AA, Liu R, Moreton S, Checkley-Luttge M-A, Gupta K, et al. Membrane Bound IL-21 Based NK Cell Feeder Cells Drive Robust Expansion and Metabolic Activation of NK Cells. Sci Rep (2019) 9(14916). doi: 10.1038/S41598-019-51287-6
35. Zhao X-Y, Jiang Q, Jiang H, Hu L-J, Zhao T, Yu X-X, et al. Expanded Clinical-Grade Membrane-Bound IL-21/4-1BBL NK Cell Products Exhibit Activity Against Acute Myeloid Leukemia In Vivo. Eur J Immunol (2020) 50:1374–85. doi: 10.1002/EJI.201948375
36. Oyer L, Pandey V, Igarashi R, Somanchi S, Zakari A, Solh M, et al. Natural Killer Cells Stimulated With PM21 Particles Expand and Biodistribute In Vivo: Clinical Implications for Cancer Treatment. Cytotherapy (2016) 18:653–63. doi: 10.1016/J.JCYT.2016.02.006
37. Reuter D, Staege MS, Kühnöl CD, Föll J. Immunostimulation by OX40 Ligand Transgenic Ewing Sarcoma Cells. Front Oncol (2015) 5:242/BIBTEX. doi: 10.3389/FONC.2015.00242/BIBTEX
38. Thangaraj JL, Phan MTT, Kweon SH, Kim J, Lee JM, Hwang I, et al. Expansion of Cytotoxic Natural Killer Cells in Multiple Myeloma Patients Using K562 Cells Expressing OX40 Ligand and Membrane-Bound IL-18 and IL-21. Cancer Immunol Immunother (2021). doi: 10.1007/S00262-021-02982-9
39. Tremblay-Mclean A, Coenraads S, Kiani Z, Dupuy FP, Bernard NF. Expression of Ligands for Activating Natural Killer Cell Receptors on Cell Lines Commonly Used to Assess Natural Killer Cell Function 11 Medical and Health Sciences 1107 Immunology. BMC Immunol (2019) 20:1–13. doi: 10.1186/S12865-018-0272-X/FIGURES/3
40. Allan DSJ, Chakraborty M, Waller GC, Hochman MJ, Poolcharoen A, Reger RN, et al. Systematic Improvements in Lentiviral Transduction of Primary Human Natural Killer Cells Undergoing Ex Vivo Expansion. Mol Ther - Methods Clin Dev (2021) 20:559–71. doi: 10.1016/J.OMTM.2021.01.008
41. Gurney M, O’Reilly E, Corcoran S, Brophy S, Hardwicke D, Krawczyk J, et al. Abstract 1725: Tc Buster Transposon Engineered Cll-1 Car-Nk Cells Efficiently Target Acute Myeloid Leukemia. In: 63rd American Society of Haematology (ASH) Annual Meeting and Exposition (ASH) 138 (Supplement 1):1725
42. Liu E, Tong Y, Dotti G, Shaim H, Savoldo B, Mukherjee M, et al. Cord Blood NK Cells Engineered to Express IL-15 and a CD19-Targeted CAR Show Long-Term Persistence and Potent Anti-Tumor Activity. Leukemia (2018) 32:520. doi: 10.1038/LEU.2017.226
43. Pomeroy EJ, Lahr WS, Chang JW, Krueger J, Wick BJ, Slipek NJ, et al. Preprint: Non-Viral Engineering of CAR-NK and CAR-T Cells Using the Tc Buster Transposon System™. bioRxiv (2021) 2021.08.02.454772. doi: 10.1101/2021.08.02.454772
44. Ciurea SO, Kongtim P, Soebbing D, Trikha P, Behbehani G, Rondon G, et al. Decrease Post-Transplant Relapse Using Donor-Derived Expanded NK-Cells. Leuk 2021 (2021) 36:155–64. doi: 10.1038/s41375-021-01349-4
45. Prager I, Liesche C, van Ooijen H, Urlaub D, Verron Q, Sandström N, et al. NK Cells Switch From Granzyme B to Death Receptor–Mediated Cytotoxicity During Serial Killing. J Exp Med (2019) 216:2113–27. doi: 10.1084/JEM.20181454
46. DeSelm C, Palomba ML, Yahalom J, Hamieh M, Eyquem J, Rajasekhar VK, et al. Low-Dose Radiation Conditioning Enables CAR T Cells to Mitigate Antigen Escape. Mol Ther (2018) 26:2542. doi: 10.1016/J.YMTHE.2018.09.008
47. Putz EM, Guillerey C, Kos K, Stannard K, Miles K, Delconte RB, et al. Targeting Cytokine Signaling Checkpoint CIS Activates NK Cells to Protect From Tumor Initiation and Metastasis. Oncoimmunology (2017) 6:1–10. doi: 10.1080/2162402X.2016.1267892
48. Zhu H, Blum RH, Bernareggi D, Ask EH, Wu Z, Hoel HJ, et al. Metabolic Reprograming via Deletion of CISH in Human Ipsc-Derived NK Cells Promotes In Vivo Persistence and Enhances Anti-Tumor Activity. Cell Stem Cell (2020) 27:224–37.e6. doi: 10.1016/j.stem.2020.05.008
49. Daher M, Basar R, Gokdemir E, Baran N, Uprety N, Nunez Cortes AK, et al. Targeting a Cytokine Checkpoint Enhances the Fitness of Armored Cord Blood CAR-NK Cells. Blood (2021) 137:624. doi: 10.1182/BLOOD.2020007748
50. Gang M, Marin N, Wong P, Neal C, Marsala L, Foster M, et al. CAR-Modified Memory-Like NK Cells Exhibit Potent Responses to NK-Resistant Lymphomas. Blood (2020) 136:2308–18. doi: 10.1182/BLOOD.2020006619
51. Vargas JE, Chicaybam L, Stein RT, Tanuri A, Delgado-Cañedo A, Bonamino MH. Retroviral Vectors and Transposons for Stable Gene Therapy: Advances, Current Challenges and Perspectives. J Transl Med 2016 141 (2016) 14:1–15. doi: 10.1186/S12967-016-1047-X
52. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N Engl J Med (2017) 377:2531–44. doi: 10.1056/NEJMOA1707447
53. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N Engl J Med (2018) 380:45–56. doi: 10.1056/NEJMOA1804980
54. Streltsova MA, Barsov E, Erokhina SA, Kovalenko EI. Retroviral Gene Transfer Into Primary Human NK Cells Activated by IL-2 and K562 Feeder Cells Expressing Membrane-Bound IL-21. J Immunol Methods (2017) 450:90–4. doi: 10.1016/J.JIM.2017.08.003
55. Colamartino ABL, Lemieux W, Bifsha P, Nicoletti S, Chakravarti N, Sanz J, et al. Efficient and Robust NK-Cell Transduction With Baboon Envelope Pseudotyped Lentivector. Front Immunol (2019) 10:2873. doi: 10.3389/fimmu.2019.02873
56. Yang Y, Badeti S, Tseng H, Ma MT, Liu T, Jiang J-G, et al. Superior Expansion and Cytotoxicity of Human Primary NK and CAR-NK Cells From Various Sources via Enriched Metabolic Pathways. Mol Ther - Methods Clin Dev (2020) 18:428–45. doi: 10.1016/J.OMTM.2020.06.014
57. Frei GM, Persi N, Lador C, Peled A, Cohen YC, Nagler A, et al. Nicotinamide, a Form of Vitamin B3, Promotes Expansion of Natural Killer Cells That Display Increased In Vivo Survival and Cytotoxic Activity. Blood (2011) 118:4035. doi: 10.1182/BLOOD.V118.21.4035.4035
58. Bachanova V, McKenna DH, Luo X, Defor TE, Cooley S, Warlick E, et al. First-in-Human Phase I Study of Nicotinamide-Expanded Related Donor Natural Killer Cells for the Treatment of Relapsed/Refractory Non-Hodgkin Lymphoma and Multiple Myeloma. Biol Blood Marrow Transplant (2019) 25:S175–6. doi: 10.1016/J.BBMT.2018.12.317
59. Li L, Li W, Wang C, Yan X, Wang Y, Niu C, et al. Adoptive Transfer of Natural Killer Cells in Combination With Chemotherapy Improves Outcomes of Patients With Locally Advanced Colon Carcinoma. Cytotherapy (2018) 20:134–48. doi: 10.1016/J.JCYT.2017.09.009
60. Irving M, Lanitis E, Migliorini D, Ivics Z, Guedan S. Choosing the Right Tool for Genetic Engineering: Clinical Lessons From Chimeric Antigen Receptor-T Cells. Hum Gene Ther (2021) 32:1044–58. doi: 10.1089/HUM.2021.173
61. Hudecek M, Ivics Z. Non-Viral Therapeutic Cell Engineering With the Sleeping Beauty Transposon System. Curr Opin Genet Dev (2018) 52:100–8. doi: 10.1016/J.GDE.2018.06.003
62. Ingegnere T, Mariotti FR, Pelosi A, Quintarelli C, De Angelis B, Tumino N, et al. Human CAR NK Cells: A New Non-Viral Method Allowing High Efficient Transfection and Strong Tumor Cell Killing. Front Immunol (2019) 0:957. doi: 10.3389/FIMMU.2019.00957
63. Pomeroy EJ, Hunzeker JT, Kluesner MG, Lahr WS, Smeester BA, Crosby MR, et al. A Genetically Engineered Primary Human Natural Killer Cell Platform for Cancer Immunotherapy. Mol Ther (2020) 28:52–63. doi: 10.1016/J.YMTHE.2019.10.009
64. Hudecek M, Gogishvili T, Monjezi R, Wegner J, Shankar R, Kruesemann C, et al. Minicircle-Based Engineering of Chimeric Antigen Receptor (CAR) T Cells. Recent Results Cancer Res (2016) 209:37–50. doi: 10.1007/978-3-319-42934-2_3
65. Bexte T, Botezatu L, Miskey C, Campe J, Reindl LM, Gebel V, et al. Non-Viral Sleeping Beauty Transposon Engineered CD19-CAR-NK Cells Show a Safe Genomic Integration Profile and High Antileukemic Efficiency. Blood (2021) 138:2797–7. doi: 10.1182/BLOOD-2021-153999
66. Hermanson DL, Barnett BE, Rengarajan S, Codde R, Wang X, Tan Y, et al. A Novel Bcma-Specific, Centyrin-Based CAR-T Product for the Treatment of Multiple Myeloma. Blood (2016) 128:2127. doi: 10.1182/BLOOD.V128.22.2127.2127
67. Micklethwaite K, Gowrishankar K, Gloss B, Li Z, Street J, Moezzi L, et al. Investigation of Product Derived Lymphoma Following Infusion of Piggybac Modified CD19 Chimeric Antigen Receptor T-Cells. Blood (2021) 138(16):1391–405. doi: 10.1182/BLOOD.2021010858
68. Kararoudi MN, Likhite S, Elmas E, Yamamoto K, Schwartz M, Sorathia K, et al. Preprint: CRISPR-Targeted CAR Gene Insertion Using Cas9/RNP and AAV6 Enhances Anti-AML Activity of Primary NK Cells. bioRxiv (2021):2021.03.17.435886. doi: 10.1101/2021.03.17.435886
69. Naeimi Kararoudi M, Likhite S, Elmas E, Schwartz M, Sorathia K, Yamamoto K, et al. Cd33 Targeting Primary Car-Nk Cells Generated by Crispr Mediated Gene Insertion Show Enhanced Anti-Aml Activity. Blood (2020) 136:3. doi: 10.1182/BLOOD-2020-142494
70. Zhang L, Zuris JA, Viswanathan R, Edelstein JN, Turk R, Thommandru B, et al. Ascas12a Ultra Nuclease Facilitates the Rapid Generation of Therapeutic Cell Medicines. Nat Commun 2021 121 (2021) 12:1–15. doi: 10.1038/s41467-021-24017-8
71. Tanaka Y, Nakazawa T, Nakamura M, Nishimura F, Matsuda R, Omoto K, et al. Ex Vivo-Expanded Highly Purified Natural Killer Cells in Combination With Temozolomide Induce Antitumor Effects in Human Glioblastoma Cells In Vitro. PLoS One (2019) 14:e0212455. doi: 10.1371/JOURNAL.PONE.0212455
Keywords: NK cells, expansion, transposon, genome editing, immunotherapy
Citation: Gurney M, Kundu S, Pandey S and O’Dwyer M (2022) Feeder Cells at the Interface of Natural Killer Cell Activation, Expansion and Gene Editing. Front. Immunol. 13:802906. doi: 10.3389/fimmu.2022.802906
Received: 27 October 2021; Accepted: 26 January 2022;
Published: 11 February 2022.
Edited by:
Evelyn Ullrich, Goethe University Frankfurt, GermanyReviewed by:
Hans Klingemann, NantKwest, Inc., United StatesAnna Dünkel, Fraunhofer Institute for Cell Therapy and Immunology (IZI), Germany
Copyright © 2022 Gurney, Kundu, Pandey and O’Dwyer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael O’Dwyer, michael.odwyer@nuigalway.ie
 Mark Gurney
Mark Gurney