- 1Department of Pathogen Biology, School of Basic Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department and Institute of Urology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 3Cytek Biosciences, R&D Clinical Reagents, Fremont, CA, United States
- 4Department of Paediatrics and Adolescent Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
Actin is an important cytoskeletal protein involved in signal transduction, cell structure and motility. Actin regulators include actin-monomer-binding proteins, Wiskott-Aldrich syndrome (WAS) family of proteins, nucleation proteins, actin filament polymerases and severing proteins. This group of proteins regulate the dynamic changes in actin assembly/disassembly, thus playing an important role in cell motility, intracellular transport, cell division and other basic cellular activities. Lymphocytes are important components of the human immune system, consisting of T-lymphocytes (T cells), B-lymphocytes (B cells) and natural killer cells (NK cells). Lymphocytes are indispensable for both innate and adaptive immunity and cannot function normally without various actin regulators. In this review, we first briefly introduce the structure and fundamental functions of a variety of well-known and newly discovered actin regulators, then we highlight the role of actin regulators in T cell, B cell and NK cell, and finally provide a landscape of various diseases associated with them. This review provides new directions in exploring actin regulators and promotes more precise and effective treatments for related diseases.
1 Introduction
Actin is a highly conserved protein that is abundantly expressed in most eukaryotic cells. Like intermediate filaments and microtubules, actin is a major component of the cytoskeleton and plays an extremely significant role in the structure, motility, activation and maintenance of cells.
Actin exists in the form of monomers (globular actin or G-actin) or filamentous polymers (filamentous actin or F-actin). G-actin is formed by a polypeptide chain containing 375 amino acid residues, which can be structurally divided into two parts: the endostructural domain and the exostructural domain (1). There are two clefts between these two domains, the nucleotide-binding cleft and the target-binding cleft, which binds to nucleotides and proteins, respectively, to regulate the activity of actin. F-actin is a right-handed helical structure consisting of two chains coiled around each other (2). G-actin and F-actin can interconvert through polymerization and depolymerization, and this process often requires the assistance of several regulatory proteins and the involvement of various small molecules. ATP-bound G-actin has greater affinity and nucleation when interacting with nucleation proteins, such as the Arp2/3 complex, which form stable oligomers consisting of three to four monomers (3) that are then extended and assembled into F-actin. Simultaneously, F-actin undergoes depolymerization via the hydrolysis of ATP. The rate of actin polymerization and depolymerization depends on the available concentration of G-actin and ATP. Due to the polarity of F-actin, in order to extend a filament in one direction, the rate of polymerization at the one end (barbed end), must be relatively faster than the depolymerization at the other end (pointed end). This phenomenon, known as “treadmilling” (4), maintains the dynamic equilibrium between G-actin and F-actin.
As an important component of microfilaments, actin plays a critical role in the maintenance of cell structure (5). Under the action of “treadmilling”, filaments extend at barbed ends, forming protrusions at the leading edge of the cell (6). This is the basis for the formation of lamellipodia, microvilli and filopodia (7), which are necessary for cell motility and food intake. Together with other molecules, actin is also involved in cellular endocytosis (8). In addition, actin also interacts with myosin. Myosin has ATPase activity and hydrolyzes ATP to generate energy, leading to the sliding of actin and myosin filaments against each other, thus generating the tension required for the basic mechanism of muscle contraction (9). Similar processes are also present in non-muscle cells, and the resulting contractile force is important for cell migration, cytoplasmic division, or other biological processes.
Actin regulators are a series of proteins that control the assembly/disassembly dynamics of actin for cell motility, intracellular transport, muscle contraction, cellular structure maintenance, cytokinesis and other fundamental cell activities. Well studied actin regulators include: 1) Actin Capping Proteins that interact with actin and regulate the capping of the ends of actin polymers, 2) Actin Depolymerizing Factors that severe and depolymerize actin filaments, 3) Actin-Related Protein 2/3 Complex (Arp2/3) that nucleates branched actin, and 4) Wiskott-Aldrich Syndrome Protein (WASP) Family that interacts with and activates Arp2/3 (Figure 1) (10).
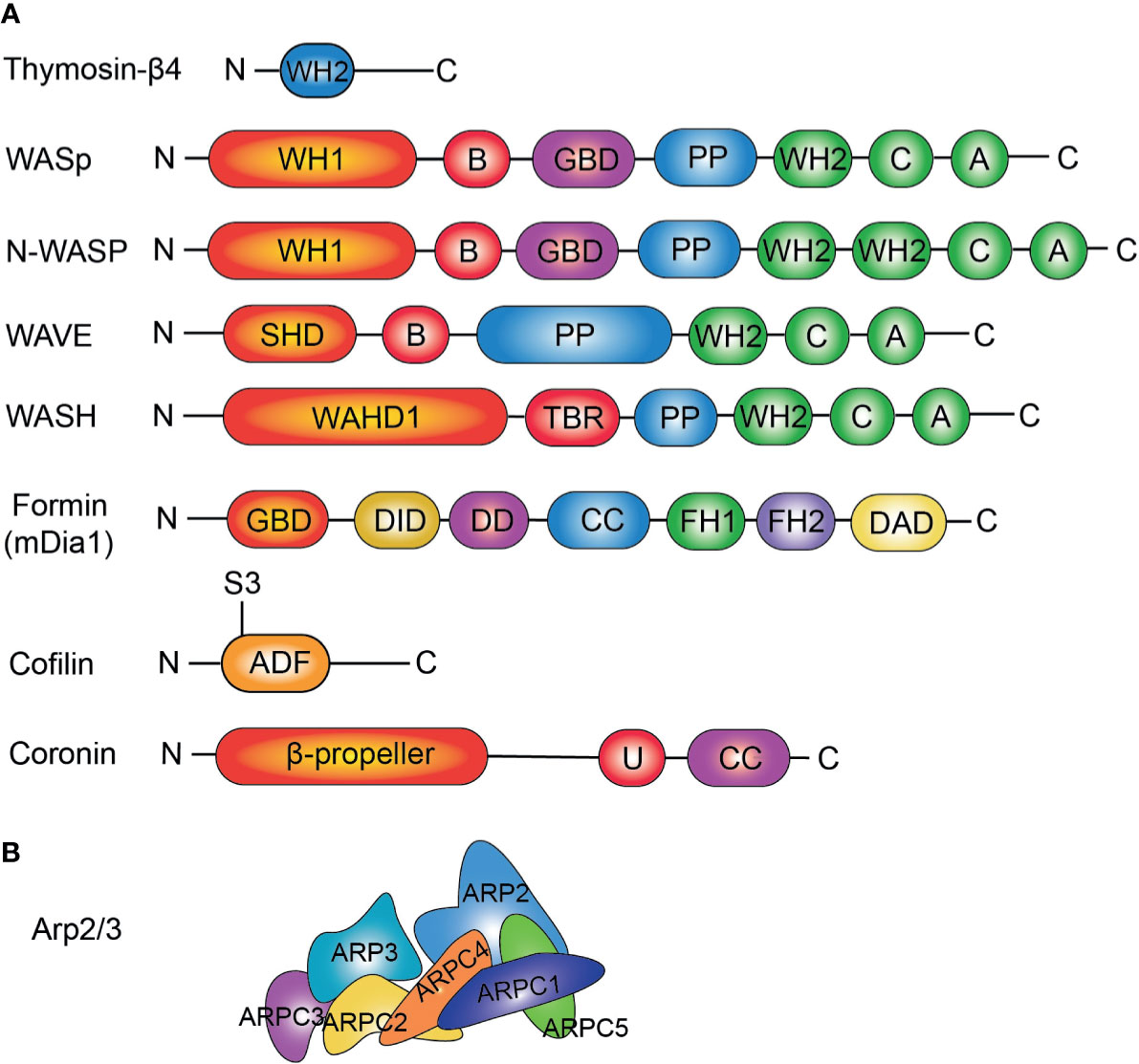
Figure 1 (A) Domain structures of actin regulators important for lymphocytes. THY, thymosin β actin-binding motif; WH1, WASP Homology domain-1; B, Basic domain; GBD, GTPase binding domain; PP, poly-proline; WH2, WASP Homology domain-2; C, Connecting sequence; A, Acidic sequence; SHD, SCAR-homology domain; WAHD1, WASH Homology domain; TBR, Tubulin binding region; HD, Helical domain; DID, diaphanous autoregulatory domain; DD, dimerization domain; CC, coiled coil region; FH1, formin homology domain 1; FH2, formin homology domain 2; DAD, diaphanous autoregulatory domain; ADF, actin depolymerizing factor; U, unique region; CC, coiled-coil. (B) Conformation of Arp2/3 complex. ARP2; ARP3; and ARP complex-1~5 (ARPC1~5) are shown.
Lymphocytes are an important constituent of leukocytes in humans, and include T lymphocytes (T cells), B lymphocytes (B cells) and natural killer cells (NK cells) that play an essential role in both innate and adaptive immunity (11). T lymphocytes are divided into T helper cells (Th cells), cytotoxic T cells (CTLs), regulatory T cells (Tregs) and other subsets according to biomarkers on the cell membrane and their functions, which are mainly involved in cell-mediated immunity. However, they are also indispensable in humoral immunity via their helper effector functions, such as cytokines release. Whereas B lymphocytes are the major players of humoral immunity and the exclusive source of antibodies (12, 13), NK cells are crucial to the innate immune system, which function in a cell-mediated and cytotoxic way that is essential in defending against tumors and viral infections (13). These three types of lymphocytes help form a sophisticated immune network to recognize and eliminate non-self antigens, thus maintaining the stability of the internal environment in humans.
In lymphocytes, actin is of great significance for cell activation, adhesion, and migration (14–18). For example, the actin cytoskeleton can mediate the formation of the supramolecular activation cluster (SMAG) or cap (15), which in the case of leukocytes, is the cellular immune synapse (IS) (19) that exerts regulatory effects on cellular signaling (15). In particular, actin plays an essential role in the developmental maturation of T cells. Studies have shown that Pak2-mediated actin cytoskeleton remodeling is important for T cell maturation in the thymus (20). In B cells, actin is involved in the regulation of B cell receptor (BCR) clustering, IS formation, antigen internalization and presentation (21). Concurrently, BCR-mediated antigen transport in B cells is also actin-dependent (22). In addition, differences in actin kinetics in various B cell subsets contribute to their specific regulation of activation (16). In NK cells, actin can be induced to reorganize by upstream signaling molecules, triggering downstream biological processes, such as granule polarization, synapse formation and target cell lysis (23), and the density of the actin cytoskeleton mediates the cytotoxic effects of NK cells (24). Thus, the regulation of the actin cytoskeleton is essential to the function and stability of the human immune system.
In this review, we will provide an overview of the structures and functions of several important actin regulators and how they play an important role in lymphocytes and regulating the human immune system (Figure 2). And we will also introduce some diseases related to the deficiency or dysfunction of these proteins and potential targets for treatment.
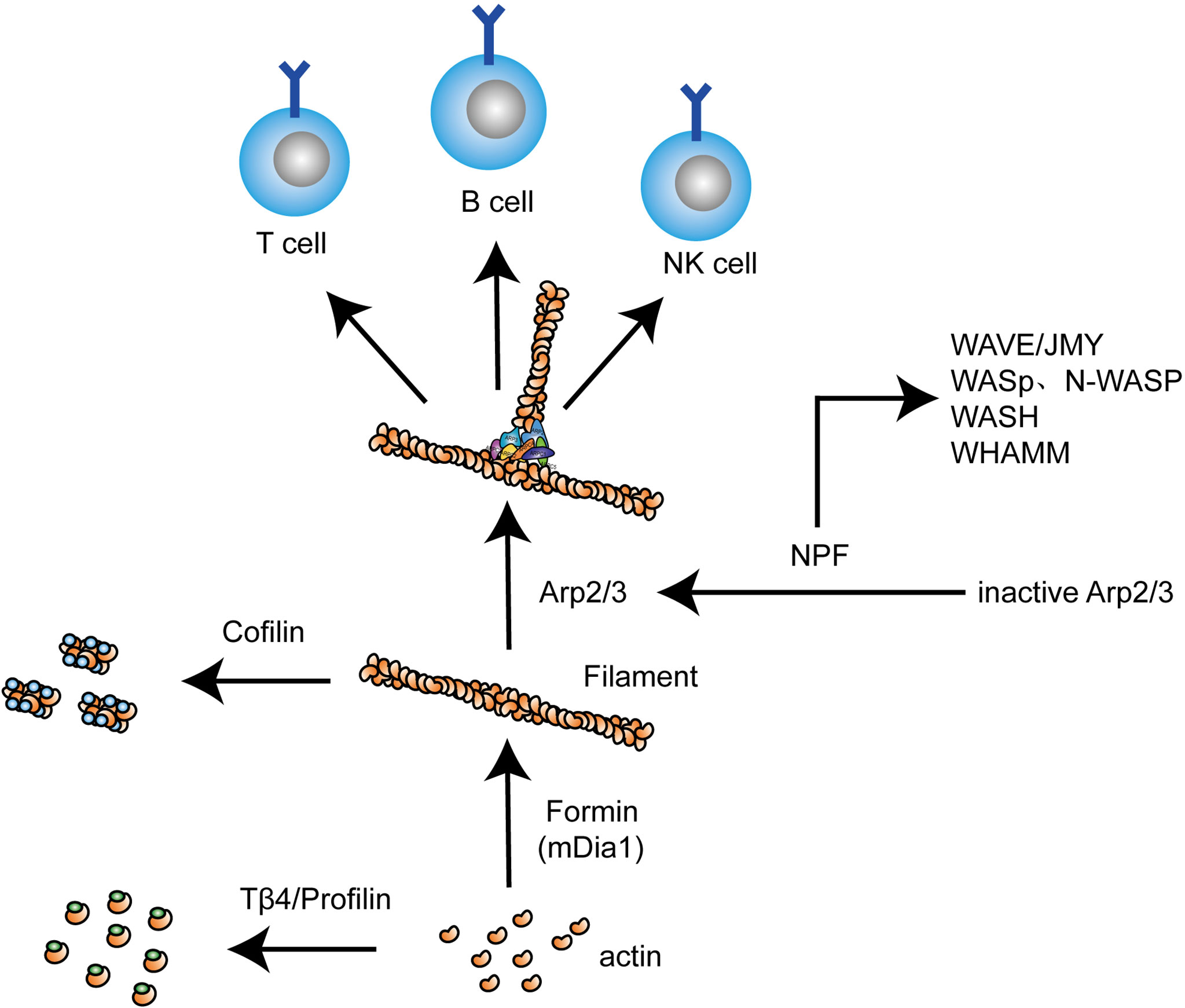
Figure 2 Different actin regulators play distinct roles in actin dynamics in lymphocytes. As actin-monomer-binding proteins, profilin and thymosin beta 4 (Tβ4) bind G-actin and prevent it from polymerizing, while formins act in an opposite manner, promoting actin filament polymerization. The activation of Arp2/3 complex requires NPFs including WAVE, JMY, WASp/N-WASp, WASH, and WAHMM, which is needed for branching F-actin. The severing of F-actin is mediated by cofilin. In the above process, actin undergoes dynamic changes in lymphocytes, thereby regulating cellular activity and function.
2 Structure and Basic Function
2.1 Actin-Monomer-Binding Proteins
2.1.1 Profilin
Found by Carlsson in 1976, profilin is a 12-15 kDa protein ubiquitously expressed in eukaryotic cells of a large variety of organisms and highly expressed under hyperoxia condition (25, 26). In different organisms, profilins have a highly conserved structure: 7 β-pleated-sheets and 4 helices (27), and have 4 isoforms containing 100-130 amino acids. Profilin I is expressed in many cell types while other isoforms have tissue specificity: profilin II is brain-specific and the expression of mouse profilin III and IV is in testis, while profilin III can also be found in rat kidney (28–31). At the cellular level, profilins localize in the ruffles of peripheral lamellae and the nascent stress fibers of spreading cells, but not in peripheral belts of stationary cells in epithelioid sheets, and it is likely that profilin levels are higher in areas of active actin dynamics (32, 33). The major ligands of profilin are actin monomers, N-WASp/WAVE, PI(4,5)-P2 and poly-L-proline (34). Phosphorylation at the residue threonine 89 of profilin by protein kinase A (PKA) enables enhanced binding to actin and poly-L-proline but has no effect on phosphatidylinositol-4,5-bisphosphate (PIP2) (35, 36). Besides PKA, profilin can also be phosphorylated by Rho-associated kinase (ROCK) and dephosphorylated at Ser-137 by Protein phosphatase 1 (PP1) (37).
Profilins play an important role in cell development, motility, membrane trafficking and signal transduction via binding to different ligands (38). Profilin’s main function is promoting actin polymerization (39). By binding to and changing the affinity of actin monomers, profilins slow down the elongation, thus optimizing the process of actin polymerization (40). This mechanism of action speeds up nucleotide exchange, which acts opposite of thymosin-β4 (41). In addition, profilins can also enhance microtubule growth directly and take part in single cell wound repair (42, 43). Different isoforms have their specific functions. Profilin I and II play opposite roles in cell motility, migration and membrane protrusion, whereby profilin I principally enhances these processes while profilin II suppresses them (44).
As profilin can regulate actin polymerization, it plays a crucial role in cell migration (45, 46). As for its interaction with WASp-interacting protein (WIP), proteins like Mena and vasodilator-stimulated phosphoprotein (VASP) can directly bind profilin through a proline-rich Actin Based Motility (ABM) motif, such as APPPPP (47–49). WASP-WIP’s function of regulating actin polymerization may be heightened by recruiting profilin to ABM-2 motifs on WIP (45). Also, in mammalian cell lines, profilin directly binds to actin monomers to sequester them when actin barbed ends are capped, this activity assures that the G-actin pool will not be monopolized by Arp2/3 and thereby formins have the chance to enter the G-actin pool and bind actin monomers, therefore profilins hinder actin assembly mediated by Arp2/3 to promote formin activity (50, 51). Thus, profilin selectively regulates actin monomers to flow from Arp2/3 to formins and Ena/VASP (51).
2.1.2 Thymosin-β4
Thymosin beta 4 (Tβ4) is a 43-amino-acid protein found to be widely expressed in thymocytes and hematopoietic cell lines, as well as in a variety of organs, such as brain, thymus, spleen, and lung (52–55). As one of the most abundant proteins in the highly conserved beta-thymosin family, Tβ4 is an important actin binding protein that binds to both the barbed and pointed ends of G-actin, which causes conformational changes in Tβ4 to spatially block actin polymerization (56, 57). This binding of Tβ4 to actin is influenced by nucleotides. It was demonstrated that the affinity of Tβ4 for ATP-actin is about 50-fold higher than that of ADP-actin (58). In addition to its role in the cytoplasm, Tβ4 also translocates to the nucleus and binds actin monomers to regulate actin dynamics (59). Overall, Tβ4’s effect on G-actin leads to an increased ratio of G-actin to F-actin in the cell, thereby regulating the cytoskeleton and affecting the biological activity of cells and tissues.
2.2 Wiskott-Aldrich Syndrome (WAS) Family of Proteins
Wiskott-Aldrich syndrome (WAS) is an X-linked primary immunodeficiency disease characterized by thrombocytopenia, eczema, episodes of fever, bloody diarrhea, recurrent bacterial infections, innate and adaptive immune deficiency, and a high rate of autoimmunity and malignancies (60, 61). The disease is due to mutations of the gene which encodes the WAS protein (WASp) (62). WASp was the first identified member of a family of proteins comprised of WASp/N-WASP, SCAR/WAVE, WHAMM/JMY/WHAMY, and WASH subfamilies (63). WAS proteins are nucleation-promoting factors (NPFs) that activate Arp2/3 to nucleate branched actin filaments in response to extracellular signals. This plays an important role in many cellular processes that happen at the cell surface, such as cellular motility, endocytosis, exocytosis and intracellular signal transduction (64). WAS family proteins are also present in the nucleus, where they regulate transcription and remodel chromatin (65). Distinct WAS family proteins participate in different cellular processes, therefore, they differ from each other in the structure of their N-terminal portions. However, contrary to their diverse N termini, all WAS family proteins possess an identical C-terminal structure, the verprolin homology (WH2)/connecting peptide/acidic domains (VCA) domain, which mediates interaction with the Arp2/3 complex and actin (Figure 1) (64). It was also reported that WAS family proteins function as polymerases, accelerating elongation of uncapped actin filaments (66). In summary, WAS family proteins have the following functions: nucleating branched actin filaments, linking actin networks to membranes, mediating transcription and chromatin remodeling, and accelerating filament elongation.
2.2.1 Wiskott-Aldrich Syndrome Protein (WASp)
WASp is a 502-amino-acid protein expressed exclusively in hematopoietic cells (62), which has several domains with different functions. The pleckstrin homology (PH) domain is close to the N-terminal region and is involved in the localization of WASp through interactions with other proteins or lipids, such as PIP2. The PH domain is very important since missense mutations in this region lead to severe diseases (67). The PH domain overlaps the Ena/VASP homology 1 (EVH1) domain (also known as WH1 domain), which constitutively interacts with the proline-rich region of the WIP in resting cells (68). The EVH1/WH1 domain is followed by a basic region (BR), a GTPase-binding domain (GBD), a poly-proline region (PP) and a VCA domain in the C-terminal (Figure 1) (60). In the inactive state, WASp is in an auto-inhibited conformation in which the GBD of WASp conceals the VCA domain. This prevents the interaction between WASp and Arp2/3 and inhibits actin polymerization. However, upon receptor activation mediated by extracellular stimulation, such as T cell receptor (TCR) and BCR signaling, WASp is recruited to the signaling site at the membrane and transformed to the active conformation (69). This process is induced by activated GTP-bound cell division control protein 42 homolog (Cdc42), which binds to the GBD and transforms WASp to an open-activated structure, allowing it to interact with Arp2/3 and promote actin polymerization. Additionally, proteins with Src homology 3 (SH3) domains, including Grb2, p47nck, Fyn, and Lck, combine with Cdc42 to enhance WASp conformational changes. Furthermore, Src-family kinases, such as Lyn and the Tec family of cytoplasmic tyrosine kinases, including Btk, Tec, and Itk, phosphorylate WASp at Tyr 291 (Y291) in a Cdc42-dependent fashion, which modulates the affinity of WASp for Cdc42 and later targets WASp for degradation via a ubiquitin-dependent proteasomal pathway (60, 68, 70). After activation of WASp, actin branching is mediated by the Arp2/3 complex, which is initiated by several steps. First, the V/WH2 segment of VCA binds to G-actin monomers, while the CA segment binds two sites on the Arp2/3 complex. These WASp-recruited actin monomers will be the first actin subunits added onto the newly branching filament. Second, the WASp-Arp2/3 complex undergoes a conformational change where two actin-related subunits in the complex, Arp2 and Arp3, are transformed into a short pitch conformation, which imitates an actin dimer within a filament. Finally, WASp interacts with an existing actin filament to initiate nucleation, which ensures only branched actin filaments are generated (64). After the filament branching process is finished, activated WASp is down-regulated by ubiquitination and proteasome degradation. In the resting state, WIP can inhibit WASp degradation by concealing its ubiquitination sites (69). Following activation, WIP undergoes phosphorylation and conformational changes, which exposes WASp’s ubiquitination sites in the WH1 domain. After ubiquitinated by E3 ligases, c‐Cbl and Cbl‐b, WASp is targeted for protease calpain degradation (68).
2.2.2 Neuronal Wiskott-Aldrich Syndrome Protein (N-WASP)
Another WASP family protein that plays a crucial role in cells is the Neuronal Wiskott-Aldrich Syndrome Protein (N-WASP). Unlike WASp, which is expressed exclusively in hematopoietic cells, the mRNA of N-WASP has been found to be widely localized in several organs including brain and colon (71). N-WASP has 50% homology with WASp at the protein level (71, 72), with the main structural difference being that N-WASP contains two verprolin homology sequences in the VCA structural domain (VVCA), which allows it to bind to Arp2/3 more efficiently than WASp (73). The structural similarity of N-WASP with WASp allows for their similar regulatory roles in cells. For example, expression of the N-WASP gene in WASp-deficient hematopoietic stem cells can partially rescue the signaling defect in T cells (74). However, mutations in the WASp gene predisposes someone to WAS syndrome, while N-WASP deletion is embryonic lethal (75). This suggests that the two proteins do not function in exactly the same way, which is supported by the experimental finding that N-WASP lacking the WASp-specific I30 region cannot rescue the chemotactic defects of WASP knockout Jurkat T-cells (76).
The activity of N-WASP is synergistically regulated by Cdc42 and PIP2. In the inactivated state, the VCA domain of N-WASP binds to the GBD and is in a self-inhibited state that cannot interact with Arp2/3 (77). N-WASP has binding sites for Cdc42 and PIP2. Binding of one of these molecules induces a conformational change in N-WASP that exposes the binding site for the other, which allows for both molecules to bind and activate N-WASP (78). N-WASP can also be activated by Src family kinases through phosphorylation (79) and proteins containing SH3 structural domains (such as cortactin) are also able to bind to N-WASP and activate its Arp2/3 binding capacity. Phosphorylation of cortactin by Erk facilitates this process, while phosphorylation by Src inhibits it (80). N-WASP and Cdc42 play a unique role in the formation of cellular filopodia (81) and the complex formed by N-WASP with WIP and Nck is necessary for the construction of dorsal ruffles (82).
2.2.3 Wiskott-Aldrich Syndrome Protein (WASp) and SCAR Homologue (WASH)
WASH is one of the novel members of the WASP protein family and has been found to be distributed in several human tissues including blood cells (83). The C-terminal structural domain (VCA domain) in WASH proteins, similar to WASP, enables it to function as a downstream effector molecule of Rho in Drosophila and as an Arp2/3 activator (84). Additionally, its unique N-terminal structural domains, including WASH homology domain 1 (WAHD1) and tubulin-binding region (TBR), give it the ability to interact directly with tubulin (85). Such structural properties allow WASH to localize to multiple types of endosomes and help maintain the stability of endosome morphology and the proper recycling pathway, especially for membrane surface receptors (86, 87). Also, WASH plays an important role in processes such as autophagy and cell differentiation (88, 89).
2.3 Nucleation Proteins
2.3.1 Arp2/3 Complex
The Arp2/3 complex is an important actin filament nucleation factor that was first purified from Acanthamoeba (90). It is evolutionarily conserved in most eukaryotic cells and consists of seven subunits (91), two of which, Arp2 and Arp3, are actin-related proteins that are structurally similar to actin (92). The other five subunits are ARPC1 through ARPC5, of which ARPC2 and ARPC4 form the center of the complex (93) called the clamp subunit. Recent studies have confirmed that the composition of the Arp2/3 complex was not constant (94), for instance, in humans, ARPC1 contained two isoforms, ARPC1A and ARPC1B, and different Arp2/3 complexes could play different biological roles.
The most basic function of the Arp2/3 complex is nucleation and branching of the actin filament. Previous in vitro studies have shown that the Arp2/3 complex itself had a weak nucleation capacity (95) and required a class of proteins called nucleation-promoting factors (NPFs) for activation. NPFs can be divided into two types. Type I NPFs, including WAVE, WASp, N-WASP, WASH and junction-mediating and regulatory protein (JMY), contain a VCA structural domain (i.e., the verprolin-homology domain, the cofilin-homology domain and the acidic domain) that interacts with the Arp2/3 complex and binds to actin monomers. Type II NPFs, such as Cortactin, contain an acidic region that binds to Arp2/3 and weakly activates the Arp2/3 complex, as well as having effects such as stabilizing the filament branching structure (96). It was confirmed that the Arp2/3 complex contained two different activation sites for NPFs and functioned differently during activation (97), thus creating a prerequisite for the synergistic activation of the Arp2/3 complex by cortactin and WASP family proteins (98). During the activation of the Arp2/3 complex by NPFs, the complex binds to existing actin filaments (mother) and undergoes intra-subunit and inter-subunit changes (99). The one conformation is induced by clamp twisting within the subunit, which transforms the original twisted conformation of Arp into a flat conformation. The other conformation is created when the Arps form a short-pitch conformation, which mimics the end of the actin filament. As a result, the Arp2/3 complex initiates the formation of new filaments (daughter) on the mother chain and facilitates the formation of the actin branching network. Besides the activation of the Arp2/3 complex by proteins like NPFs, there are a variety of intracellular inhibitors that negatively regulate it. For instance, the binding of cofilin, an actin filament severing protein (discussed below), to the actin filament reduces the affinity of Arp2/3 for F-actin, thereby contributing to the detachment of Arp2/3 from the filament and debranching (100). This reduction in affinity may be achieved through direct competition of cofilin or conformational changes of the filament. Additionally, Glia Maturation Factor (GMF), which belongs to the actin depolymerizing factor homology (ADF-H) family with cofilin (101), was found to bind to Arp2/3, inhibiting its nucleation and inducing debranching (102, 103). Ydenberg et al. subsequently proposed that GMF mediated debranching via its two binding sites by binding the first actin subunit on the Arp2/3 and the filament branch (104). The process involved is similar to the severing mechanism of cofilin (104, 105), with GMF in coordination with cofilin regulates the debranching and severing of filaments. Coronins (discussed below) are also known to have an inhibitory effect on the branching of Arp2/3. Studies performed in different subgroups of coronins have confirmed their function in binding to Arp2/3 and inhibiting its actin nucleation activity (106–108), and they have synergistic effects with GMF in this regard (109). Similarly, the acidic motif of the newly discovered protein Arpin identified by Dang et al. can competitively inhibit the VCA domain of Arp2/3, thereby inhibiting the activity of the complex (110). Studies on the structure of Arp2/3 further confirmed that binding to Coronin, GMF and Arpin led to a shift of Arp2/3 into an open/inactive conformation, thus providing an explanation for their inhibitory effect (109). Moreover, the binding and hydrolysis of nucleotides affects the activity of the Arp2/3 complex (111, 112), and it was found that the hydrolysis of ATP played an important role in debranching (113). Furthermore, the effect of phosphorylation regulation on the Arp2/3 complex activity is of important interest (114–116), thus the regulation of Arp2/3 is a mutually coordinated and sophisticated process, which creates the prerequisite for its regulation of the actin cytoskeleton.
The nucleation effect of the Arp2/3 complex on actin makes it an important regulator of the cytoskeleton. Thus, the Arp2/3 complex plays a significant role in a variety of cellular activities. Experiments on Dictyostelium (117) and mice (118) suggest that defects in the Arp2/3 complex may be lethal. For cell migration, the Arp2/3 complex is localized in cell protrusions, including lamellipodia and pseudopodia (119, 120), which are critical for cell motility. Moreover, the Arp2/3 complex has been found to be essential for cell adhesion (121) and it also plays a key role in cellular endocytosis (122, 123), whereby disruption of Arp2/3 complex activators can lead to defects in endocytosis (124). In addition, the Arp2/3 complex is involved in Golgi-associated membrane transport processes (125, 126) as well as cellular phagocytosis (127), which is of great significance in the immune response. And recent studies have revealed that the WASH and Arp2/3 complexes were associated with centrosomes and that the actin nucleation mediated by them might function in cell mitosis (128, 129).
2.4 Actin Filament Polymerases
2.4.1 Formins
Formin homology proteins (Formins) are a family of highly conserved proteins that are widely found in eukaryotes and are involved in the regulation of the cytoskeleton (130). Members of the Formins family are morphologically diverse, with 15 different formin proteins present in humans (131), but relatively few formin proteins in yeast. Nevertheless, there are regions of similarity between most of these proteins, including formin homology 1 (FH1) and formin homology 2 (FH2) domains, which are key regions determining the regulation of actin polymerization by formin proteins. The FH1 domain is located in the N-terminal to the FH2 domain and contains multiple repeating units of polyproline that can act as a profilin ligand (132, 133). Polyproline segments are of variable length with non-proline residues inserted in between them, which are also important in the role of the FH1 structural domain (132, 134).
Profilin is an actin monomer-binding protein and its binding to the FH1 domain increases the profilin-actin affinity to the FH2-associated barbed end and helps in the rapid elongation of microfilaments (Figure 3) (135, 136). The crystal structure of the FH2 domain is in the form of a tethered dimer (137, 138), which is consistent with its ability to bind actin nuclei or filament barbed ends (139). The FH2 domain assists in nucleation by stabilizing actin oligomers (140) and can mediate the addition of G-actin to the barbed end of a filament while preventing the binding of capping proteins (141). In addition to the above two structural domains, some formin protein sequences contain some or all of the GBD, Diaphanous Inhibitory Domain (DID) and Diaphanous Autoregulatory Domain (DAD). The role of Formins in regulating the formation of unbranched actin filaments affects activities such as formation of cellular protrusions (7, 142, 143) and cytokinesis (144). In addition, Formins can regulate microtubule dynamics and link them to actin dynamics to coordinate cytoskeletal activities (145–147).
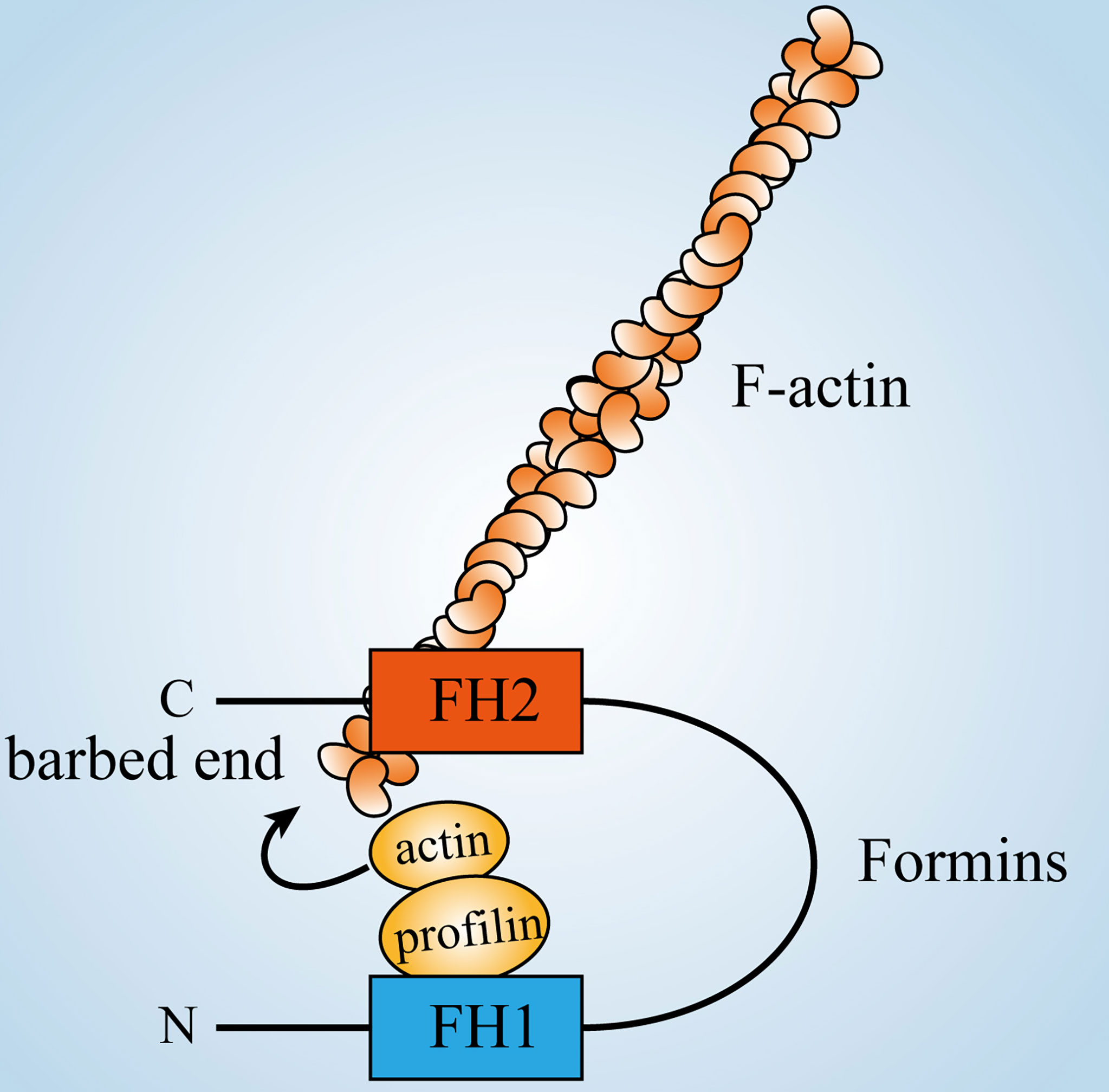
Figure 3 Formins’ role in regulating actin dynamics. The FH2 domain of formins close to the C-terminus can bind to the barbed end of filaments, while the FH1 domain near the N-terminus can bind to profilin-actin, thus increasing the concentration of actin monomers around filaments and positively promoting their polymerization.
2.5 Severing Proteins
2.5.1 Cofilin
Cofilin, a highly conserved protein with a molecular weight of 19 kDa, was first identified in chick brains and can bind to actin monomers on filaments in a 1:1 molar ratio (148, 149). As research progressed on cofilin, it was found to belong to the actin-depolymerizing factor (ADF)/cofilin family, which is a family of actin-binding proteins mainly containing three isoforms (Cofilin 1, Cofilin 2 and ADF). The structure of cofilin is relatively conserved consisting mainly of an ADF-H domain (UniProtP23528 and UniProt Q9Y281) which was found to interact with G-actin, filamentous actin or the Arp2/3 complex (150). And the tridimensional structure has two pairs of α-helices at the periphery, with a hybrid β-sheet sandwiched in between (151). It is currently believed that all eukaryotic cells contain at least one ADF/cofilin protein (152). In mammals, two cofilin isoforms are known. One is the NM-type (CFL1), which is widespread in non-muscle tissues, and the other is the M-type (CFL2), which is mainly expressed in muscle cells, but can also be seen in other tissues like testis (153, 154). For humans, the genes encoding CFL1 and CFL2 are localized on chromosomes 11 and 14, respectively (155).
As a member of the actin-binding protein family, the primary role of cofilin is to modulate actin dynamics. The cofilin protein contains a G/F site at the C-terminus and an F site at the N-terminal end (156), giving it the ability to bind to actin monomers (G-actin) or F-actin, in which it severs and depolymerizes actin filaments. Cofilin was initially thought to bind to some of the acidic residues at the N-terminal of actin by electrostatic interaction (157). Subsequent studies supported this by finding that cofilin binds to F-actin along the two initial helices between neighboring subunits (158), and that the ADF-H domain of cofilin binds between actin subdomains 1 and 3 to insert into its hydrophobic cleft (159). In vivo studies have shown that low concentrations of cofilin prefer to function in severing F-actin, while high concentrations tend to facilitate actin nucleation (160). A similar study also found that filaments are stabilized in the saturated cofilin state (160). However, this rule does not always hold. For example, in the thymus, it has been found that during cofilin saturation, cofilin was still able to depolymerize filaments in the presence of actin interacting protein 1 (Aip1), another important actin regulator (161). Many factors regulate the activity of cofilin. Phosphorylation or dephosphorylation of cofilin is a critical step in determining its activity (162), and phosphorylation at Ser-3 inactivates cofilin (163). pH level also affects the polymerization or depolymerization activity of cofilin on actin (164). In humans, depolymerization of actin mediated by cofilin increased when the pH was elevated from 6.5 to 8.0 (165). A similar study also found that cofilin favors binding to F-actin in a neutral or weakly acidic environment, while the extent of binding is greatly reduced in a weakly basic condition (166). Moreover, cofilin activity can also be influenced by phosphoinositide, where PIP2 is capable of inhibiting cofilin from binding to actin (167) and nucleotides, which cofilin has a higher affinity for ADP-actin compared to ATP-actin (168). It has also been proposed that the pH regulation of cofilin might be achieved through binding to phosphatidylinositol (169).
2.5.2 Coronin
Coronin is a 55 kDa protein that was first purified from growth-phase Dictyostelium Discoideum cells by E.L.deHostos and his co-workers. They named it “coronin” because of its interaction with the crown-shaped projections on the dorsal cortex of the cell. Through preliminary co-sedimentation experiments, they found coronin could colocalize with actin filaments (170). Different types and subtypes of coronins have been discovered and classified, however in this review we use the widely accepted classification method to divide them into 3 types: type I includes coronin 1A, coronin 1B, coronin 1C and the newly-found coronin 6. Type II includes coronin 2A and coronin 2B, and type III is coronin 7 in humans, or POD in nematode and Drosophila melanogaster (106). Up to now, coronin research has mainly focused on coronin 1A and less on the other types. Coronins are highly conserved proteins and the basic structure of the coronin family contains a 7-bladed β-propeller formed by 5 WD repeats on the N-terminal, a classical heptad coiled-coil domain on the C-terminal and an irregular secondary domain in the middle (171, 172). The N-terminal has phosphorylation sites, which mediate protein interactions and the coiled-coil domain regulates homo-oligomerization to help coronin 1A form a tripolymer structure (173, 174). Unlike other coronins, coronin 7 has two classical WD domains, which means two β propellers, but it has only one coiled-coil domain with an additional acidic domain, whose structure is similar to the acidic domain in the SCAR/WASP (172).
Encoded by the CORO gene, researches have shown that coronins are expressed in all eukaryotes, but as for the tissue specificity, coronin 1A is mainly expressed in hematopoietic tissues and immune cells (175). Coronin 1B and coronin 1C are ubiquitously expressed in different tissues and may be involved with cell migration (106). Type II coronins can be found only in vertebrates, they have different C-terminal structure from type I. Coronin 2A is expressed in testis, ovary, uterus and brain, while coronin 2B expression is mainly in the brain (176). Coronin 7 is found in mammals and POD in Caenorhabditis elegans, however being of similar genes, both localize and function within the Golgi complex in all kinds of cells (176).
Almost all the coronins belong to the actin filament-crosslinking and bundling protein family and they concentrate in the actin-rich areas on the cell membrane (177). The fundamental function of coronins is facilitating the actin depolymerization via interacting with Arp2/3 and actin filaments, and the binding site to actin is the KXRHXX-motif located near the N-terminal and the WD domain (178). Coronins, cofilins and WD repeat-containing protein 1 (WDR1) form an organized unit for the regulation of actin. The binding of coronin with actin enhances the binding of cofilin to actin as the filament twist is changed. Additionally, the cooperation between coronin and slingshot-1L increases the activity of cofilin (106, 179). In yeast, coronins bind Arp2/3 directly by a coiled-coil domain and suppress the nucleating activity of Arp2/3 (107). This mechanism is properly regulated by phosphorylation of protein kinase C (PKC) (173). However, recent studies demonstrated that while a high concentration of coronin restrains Arp2/3, a lower concentration of coronin could promote Arp2/3 nucleation (180).
2.5.3 WD Repeat-Containing Protein 1 (WDR1)
Aip1, also known as WDR1, is a conserved protein that belongs to the WD repeat domain-containing proteins, whose protein structure is mainly composed of two connected seven-bladed β-propellers (181). WDR1 alone has little effect on actin dynamics, but it assists cofilin in severing/depolymerizing of actin filaments (182, 183), a process that often also involves a third protein, coronin (discussed it above), which acts synergistically to promote the severing of filaments (184, 185). At the same time, studies in leukocytes have found that the actin-promoting effect of WDR1 is also influenced by caspase-11, the deletion of which leads to impaired motility of immune cells, including T cells (186).
3 Functions in Lymphocytes
3.1 T Lymphocytes
T cells are derived from the thymus. Mature T cells settle in thymus-dependent areas of peripheral immune organs, where they mediate adaptive cellular immune responses and also play an important secondary role in thymus-dependent antigen-induced humoral immune responses. The activity of actin regulators in T cells affects the development, motility, and functional effects of T cells.
3.1.1 Development
Bone marrow T cell progenitors locate to the thymus, where they develop into mature T cells via stages of differentiation, they then enter the peripheral lymphoid organs by the blood circulation. At this time, the naive mature T cells that come in contact with antigen will proliferate and differentiate into effector T cells, regulatory T cells or memory T cells that all have different functions (187). This is a very complex process in which various actin regulators are involved.
Earlier studies found that Tβ4 induces phenotypic changes in the human T cell line Molt-4 and may be involved in early cell differentiation (188). As an important member of NPFs, WASp also has an important regulatory role on T cell development. It has been shown that high expression of WASp inhibits the growth of T-cell lymphoma, while the loss of WASp inhibits T cell activation processes including decreasing T cell proliferation induced by TCR stimulation and preventing cytokine polarization and secretion (189). Specifically, CD8+ T cells from WASp-deficient mice are hyperactive with increased cytokine production, however these cells are also insufficient in CD8+ memory T cells differentiation and have increased apoptosis through upregulation of the Fas pathway. This further implicates a role for WASp in the survival and differentiation of CD8+ T cells (190). Additionally, double knockout (DKO) mice compared to single knockout of WASp or N-WASP mice, exhibit more pronounced abnormal thymic development and impaired T cell development, which is due to defects in cytoskeletal reorganization and migration (191). This indicates that N-WASP exhibits a synergistic effect with WASp during T cell development. WASH is required for the efficient proliferation of T cells after responding to CD3/CD28 signaling, which is related to its function in regulating the intracellular transport of T cell surface molecules, including TCR (192). Concurrently, the lack of coronin 1A leads to decreased naïve T cells and developments of severe combined immunodeficiency (SCID) (193). This is because coronin participates in the activation of calcium-calcineurin signaling that maintains the survival of naïve T cells (194), which was found have a similar effect on peripheral T cells (195). Also, abrupt reduction in the number of peripheral αβT cells and impaired late development of thymocytes caused by coronin1A defects indicate its significance in terminal T cell differentiation (196). For WDR1, it has been previously shown that defective expression of the WDR1 gene predisposes someone to autoinflammatory disease and thrombocytopenia, which are mainly manifested by abnormal neutrophil behavior (197, 198). A subsequent study reported a reduced rate of follicular helper T (Tfh) cells in patients with defective WDR1 gene expression and also showed a decreased Ca2+ response in TCR proximal signaling, suggesting that WDR1 may have an effect on T cell development and TCR signaling (199).
3.1.2 Cell Migration
Migration is one of the bases for accurate targeting of T lymphocytes to sites of infection (200). In response to chemokine stimulation, T cells can polarize, extend lamellipodium at the leading edge and produce a uropod at the rear end. Integrin alpha 4 beta 1 and alpha L beta 2 induce directional with T cell motility (201), which several actin regulators play a role in.
The migratory capacity of T cells partly depends on the production and location of actin and cofilin, which can be illustrated by the experimental fact that cofilin mRNA is distributed at the leading edge of migrating cells (202). Meanwhile, studies in Jurkat T cells revealed that cofilin is phosphorylated by the spatiotemporal regulation of LIM domain kinase (LIMK) and SSH1L, and this physiological process is crucial for SDF-1α-induced T cell migration (203). One possible regulatory mechanism is that following G-protein-coupled receptor stimulation, LIM-Kinase1 activity is inhibited in T cells via the Ras-MEK pathway to promote the dephosphorylation of cofilin (204). Activated cofilin may function at the leading edge of the cell to sever the F-actin, increasing the number of barbed ends to promote nucleation of actin polymerization there (152, 205). When the activity of MEK or cofilin is inhibited, lamellipodia extension is impaired, inhibiting the cell migration rate (206). This mechanism may play an important role in the localization of T cells during the immune response (204).
Another study on Jurkat T cells found that cell spreading on anti-CD3 sheet-like structures was inhibited by lack of Arp2 (207). Furthermore, preventing actin polymerization mediated by WASp/Arp2/3 inhibits T cell chemotaxis (208). This is due to the Arp2/3 complex being an important regulator of membrane protrusion formation, including lamellipodia (209), which is the main mode of T cell motility. Studies have found that in Arp3-impaired CTL, the cell motility pattern changed from lamellipodia-based to blebbing-like migration, a mode of movement resulting from the detachment of the plasma membrane from the actomyosin cortex (210), which decreases the migration speed (211). The different motility characteristics exhibited by T cells under different antigen affinities depends on the regulation of the Arp2/3 complex (212), which allows T cells to decelerate when encountering suitable antigens and is important to T cell bioactivity.
Unsurprisingly, N-WASP, the regulatory protein of the Arp2/3 complex, is also involved in T cell motility. An earlier study found that Cdc42 interacting protein 4 (CIP4), a protein that binds to Cdc42 and WASP/N-WASP and is involved in cytoskeletal regulation, is necessary for integrin-dependent T-cell migration, implying the participation of N-WASP in T cell motility (213). Specifically, in CD8+ T cells, N-WASP is a downstream effector molecule of NKG2D, which is a transmembrane receptor expressed mainly on CD8+ T cells and NK cells. And N-WASP is involved in the inhibitory effect of NKG2D on T cell chemotaxis under the regulation of activated Cdc42 upon CD3/NKG2D activation, and this pathway may also involve the downstream dephosphorylation of cofilin (Figure 4) (214).
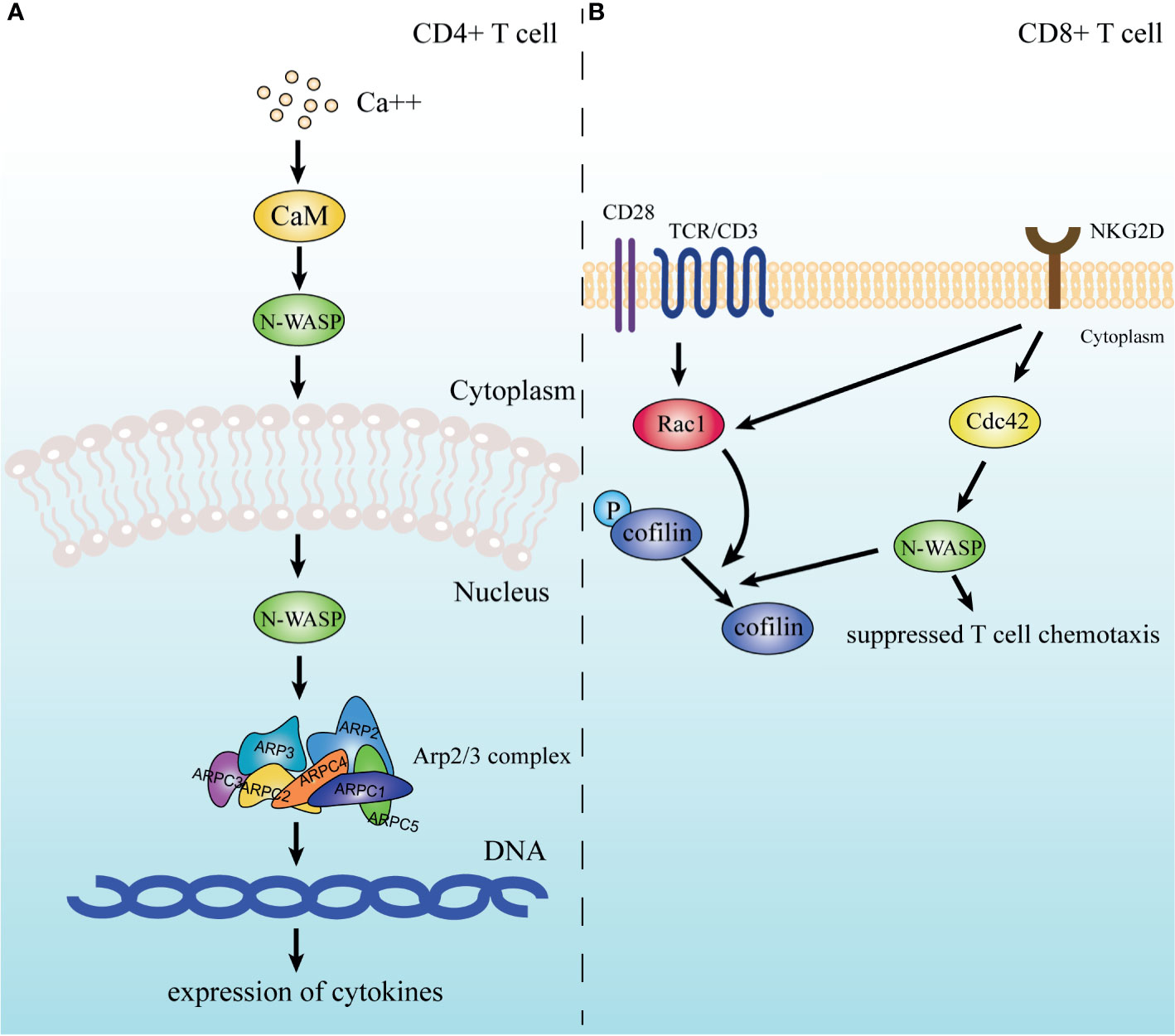
Figure 4 N-WASP is involved in signaling pathways of T cells. (A) In CD4+ T cells, N-WASP is activated by CaM and enters the nucleus to coordinate the Arp2/3 complex to participate in the expression of cytokines. (B) In CD8+ T cells, N-WASP receives signals from NKG2D and participates in downstream cofilin dephosphorylation while also mediating the inhibition of cellular chemotaxis.
Formin-like-1 (FMNL1) and mDia1 are known to be the predominantly expressed formin proteins in T cells (215). FMNL1-mediated cytoskeletal dynamics allows T cells to undergo shape changes for adapting to the environment, helping them to complete transendothelial migration (TEM) and translocation to sites of inflammation to exert immune effects (216). RhoA-regulated mDia is expressed in activated T cells and affects the functional exertion of Rac1 through the regulation of actin equilibrium, however overexpressed mDia negatively regulates the motility of T cells (46). Additionally, by studying T cells from p140mDia1-encoding knockout mice, Eisenmann et al. found that this protein is necessary for proper T cell development and chemotactic movement (217). Similarly, mDia1 deficiency inhibits T cell proliferation and chemotaxis induced migration, thus impairing the relocation of T cells to secondary lymphoid organs (218). Further studies revealed that mDia1 regulates microtubule dynamics and helps in T cell adhesion and translocation by inactivating glycogen synthase kinase (GSK) 3β and protecting adenomatous polyposis coli from phosphorylation (219).
The regulatory effects of Coronin and WDR1 on actin allow them to also influence T cell motility. Coronin1A helps T cell egress from the thymus and the coronin knock-out phenotype in mice shows deficiency in T cell migration, whereby there is abnormal accumulation of F-actin and failure of lamellipodia formation (106, 220). Furthermore, the enhanced effect of WDR1 on cofilin activity was shown to contribute to the chemotaxis of Jurkat cells, and also WDR1 is essential for their normal morphological maintenance and changes, which can be achieved by promoting the remodeling of the actin cytoskeleton (221).
3.1.3 Immune Synapse
T cells become activated when their T cell receptors (TCRs) bind specifically to antigen presented by major histocompatibility molecules (MHCs) on antigen-presenting cells (APCs). This binding to antigens induces signaling by the TCRs that lead to activation of signaling pathways involved in promoting T cell functions. This activation process requires signal transduction and amplification, which is supported by the stable platform created by the IS. The IS is a membrane structure formed on the surface of T cells at the contact site with APCs. The center of the IS is a cluster of TCRs with adhesion molecules surrounding the periphery (222, 223). This structure is also known as a SMAC, which includes the central supramolecular activation cluster (cSMAC), the peripheral SMAC (pSMAC) and the distal SMAC (dSMAC) (224, 225). IS formation is a dynamic process that depends on the activity of the actin cytoskeleton (223, 226), and therefore the contribution of actin regulators in this process is critical. Since IS formation is closely related to TCR signaling, which will be discussed below, we provide a brief overview of the actin regulators involved.
During IS formation, constant actin depolymerization and repolymerization contribute to the aggregation of related molecules, including TCR, LFA-1, and CD45. A number of experiments have shown that multiple actin regulators were important in IS formation. First, it was demonstrated that IS formation was halted with the inhibition of cofilin depolymerization activity (227). Second, the Arp2/3 complex was found to be important in the actin polymerization that is essential for IS formation. This was determined in patients with combined immunodeficiency (CID) that have a homozygotic mutation in the gene ARPC1B, which results in a defect in the development of the T cell IS (228–230). Additionally, ARPC3 is an essential protein involved in the TCR-related vesicular transport that enables TCR recycling between the intracellular and plasma membranes to continuously supply TCRs for activation and IS formation (231, 232). In this process, ARPC3 acts in concert with proteins such as IFT20 to regulate the assembly of a functional IS (233). Furthermore, the Arp2/3 complex is degraded intracellularly by GRAIL via Lys-48 and Lys-63 ubiquitination to regulate IS formation and keep T cells in a state of anergy (234), which is essential for immune tolerance of T cells. Along with Arp2/3, WASp also takes part in actin assembly and IS stabilization. It was found that downregulation of WASp causes actin foci to disappear and the symmetry of the IS to be destroyed (57). And WASp/N-WASp can function with Nck to regulate the interaction and movement between LAT and actin, which contributes to the formation of the IS (58). Similarly, Formins promote the formation of TCR microclusters through polymerizing F-actin during the early stages of IS formation in T cells (217, 235), and the aggregation of actin by Formins at the IS distal edge is needed to form the actin arc at the IS, which affects activities such as adhesion and signal transduction of T cells (236). A recent study also found that PKCδ-dependent phosphorylation of the formin protein, FMNL1β, negatively regulated F-actin polymerization at the IS and thus facilitated microtubule-organizing centers (MTOC) polarization (56). Furthermore, during IS formation in T cells, centrosome polarization requires the formation of stable and detyrosinated microtubules, which is a process mediated by the FH2 domain of INF2 (55). Thus, although the cytoskeletal components involved in IS formation is predominantly actin, other components may also be involved in this process and requires further research.
3.1.4 TCR Signaling
TCR signaling is initiated after T cells recognize a specific MHC-peptide complex (pMHC) on the APC. This process consists of interaction of receptors, followed by receptor activation and intracellular signal transduction. Coreceptors on T cells (CD4 or CD8), bind to the non-peptide-like region of MHCs, inducing recruitment and phosphorylation of tyrosines in the immunoreceptor tyrosine-based activation motif (ITAM) of the cytoplasmic segment of CD3 molecules. The process of phosphorylation is mainly mediated by Lck, a protein tyrosine kinase, which is an essential molecule for transmission of the external activity of the TCR into an internal signal. Phosphorylated ITAMs provide a binding site for ZAP-70, which is activated and continues the cascade of phosphorylation of downstream molecules and initiates several signaling pathways, including the PKC pathway, small G-protein pathway, and Ca2+ pathway, to regulate T cell activity (237–239). The understanding of the TCR signaling pathway involves millions of studies, and here we focus more on the role of actin regulators in it.
The role of the Arp2/3 complex in TCR signaling has long been of interest. Several studies have shown that the Arp2/3 complex maintains TCR downstream signaling (233, 240). A more definitive study suggests that deletion of APCR2 in T cells leads to reduced levels of cell surface TCRs, and that the Arp2/3 complex is also involved in the endosomal transport of TCRs, thus affecting the proximal TCR signaling (241). Indeed, Arp2/3 complex-mediated actin dynamics is a critical step in multiple pathways, and more often as an effector molecule of the distal TCR signaling, linking it to alterations in the actin cytoskeleton (Figure 5). Studies have indicated that after recognition of pMHC by TCR/CD3, stimulation is delivered to downstream tyrosine kinases like Fyn, LCK, and ZAP-70 (242), which then phosphorylate downstream molecules like LAT, SLP-76, Fyb, SLAP-130 (243), that ultimately activate the Arp2/3 complex. Multiple activators of the Arp2/3 complex, including NPFs, play significant roles in the signaling pathway. For instance, at the IS of T cells, the GTPase-binding domain of WASp binds to Cdc42-GTP, releasing the VAC domain and promoting the function of the WASp-Arp2/3 complex (70, 244). Also, PIP2 produced by TCR stimulation activates WASp to interact with the Arp2/3 complex (78, 245). However, the interaction between these two proteins can be inhibited by the dephosphorylation of protein tyrosine phosphatase PTP-PEST through proline, serine, threonine phosphatase interacting protein1 (PSTPIP1)-mediated binding to WASp (246). In CD4+ T lymphocytes, N-WASP interacts with CaM (71) to activate the Arp2/3 complex in the nucleus, which mediates the Ca2+ induced expression of cytokines (247). WAVE2 is another important activator of the Arp2/3 complex. WAVE2 receives signals from the Rac-GTPase as a complex, including Sra-1, Nap1, Abi-1/2, and WAVE2 (248, 249), and is recruited to the membrane (250) for activating the Arp2/3 complex and promoting lamellipodial spreading. Inhibition of WAVE2 expression can lead to impaired IS formation in T cells (251, 252). The acidic region of HS1, NTA, binds to the Arp2/3 complex and contributes to the stabilization of the F-actin branches (253, 254). Tyrosine phosphorylated HS1 was found to bind to the SH2 domain of Vav, stabilizing Vav at the IS (255), which in turn transmits the signal to the Arp2/3 complex.
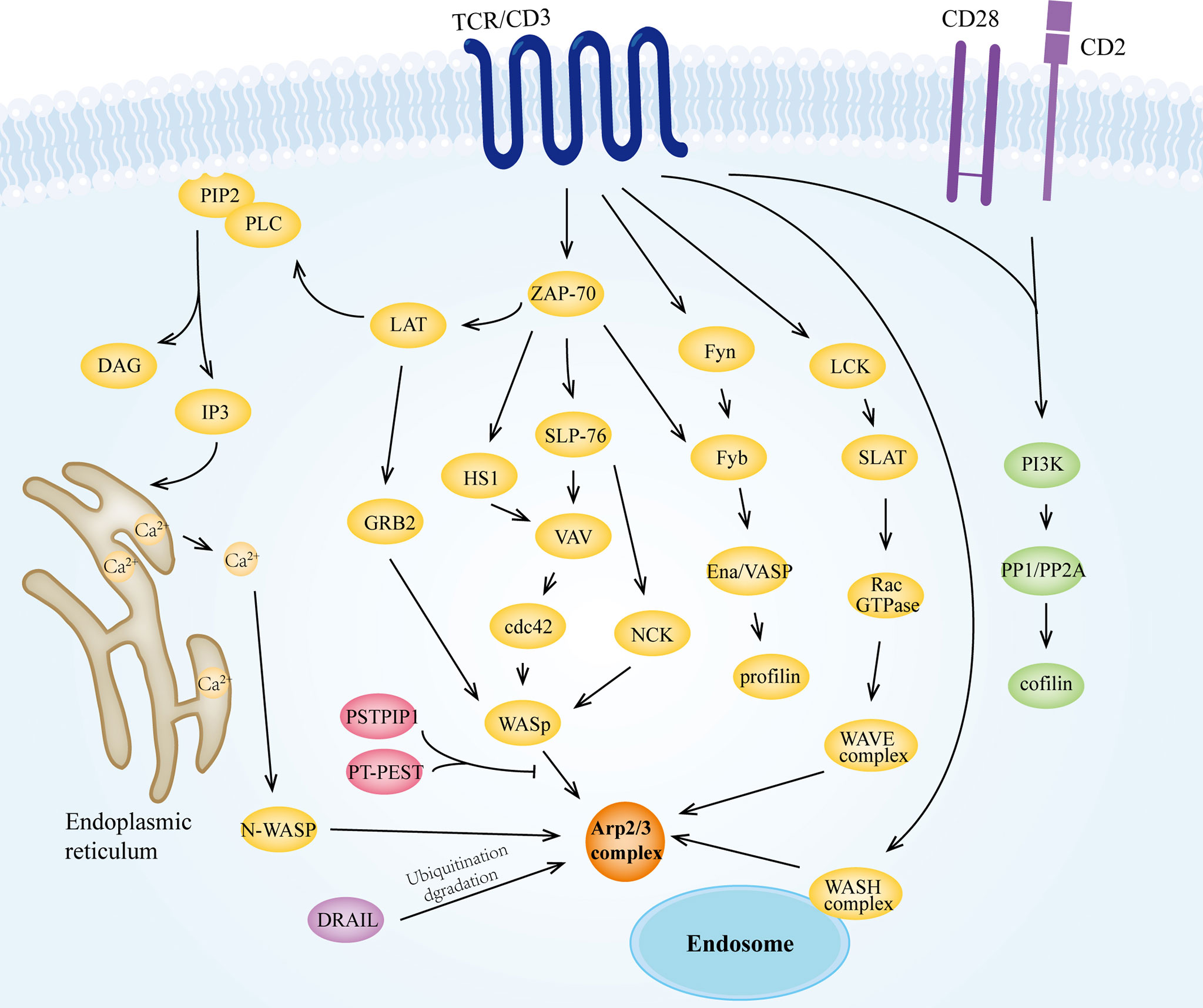
Figure 5 The Arp2/3 complex is involved in TCR signaling. Upon receiving signals from MHC molecules, TCR/CD3 can transmit stimuli to the Arp2/3 complex via tyrosine kinases such as Fyn, LCK, and ZAP-70. In this process, WASp family proteins play an important mediating role as direct activators of the Arp2/3 complex. The activated Arp2/3 complex has the ability to promote branched filament polymerization, which together with other T cell signaling-induced activation of actin regulators (e.g., cofilin and profilin) modulates the actin cytoskeleton, which is important for the biological activities of T cells, especially for IS formation.
Related to the above, Formins assist Arp2/3 in regulating the cytoskeleton during T cell activation (256). The Formin mDia 1/3 was shown to regulate the actin kinetics required for recruitment of activated ZAP70 to the IS to promote the phosphorylation of LAT. This suggests an essential role for Formins in early TCR signaling and therefore is important for positive selection of T cells. The downstream TCR signaling involves the interaction of FMNL1 with AHNAK1, which locates FMNL1 to the cell membrane and boosts calcium influx during cell activation (257). Also worth noting is that Inverted Formin2 (INF2), one of the Formins with both actin polymerization and depolymerization activities, is regulated in T cells by Cdc42 and Rac1, which helps MAL-mediated transport of Lck to the plasma membrane and participates in the signaling (258).
A recent study revealed the positive role of coronin in TCR signaling using coronin 1A-deficient T cells. These cells have elevated cAMP levels that activate PKA, leading to defective CREB phosphorylation and suppression of T cell immune responses by inhibiting CaMKK, a signaling molecule downstream of the TCR (259). However, this kind of functional disorder only causes autoimmunity suppression and has no influence on the response to foreign antigen (194).
Another actin regulator, cofilin, is involved in multiple T-cell signaling pathways, including TCR signaling (Figure 6). The cofilin protein sequence contains a phosphatidylinositol binding site which inhibits cofilin activity when bound to phosphatidylinositol (e.g. PIP2) on the plasma membrane, however this effect is independent of cofilin phosphorylation (167, 260). When Phospholipase C (PLC) is activated by TCR signaling as well as amplified signals from CD28 (261, 262), it triggers the decomposition of PIP2, allowing an increase in the amount of active cofilin (262), which may be an early event in T cell activation (263). Furthermore, co-stimulatory signals (e.g. CD40 and CD40L) are also required for T cell activation in addition to the stimulation of TCRs by pMHCs. In the absence of co-stimulatory signals, T cells are in a state of anergy. This is an important mechanism by which the body maintains immunosuppression, and it may also trigger certain pathological responses. It was confirmed that in resting human peripheral blood T lymphocytes (PB-T), dephosphorylation of cofilin occurs after co-stimulatory signals are received by co-receptors (e.g., CD2), rather than activation of TCR/CD3 receptors alone (264, 265). GTPase Ras is a key factor in this process, and dephosphorylation of cofilin requires the synergistic action of both Ras-MEK and Ras-PI3K pathways (266). Under the action of these two pathways, the protein serine/threonine phosphatases of type 1 (PP1) and type 2A (PP2A) bind to cofilin and mediate its dephosphorylation activation (267). It is worth noting that in studies on Jurkat T lymphocyte cell lines, Ras was unable to activate PI3K, reflecting the inconsistency in activation of cofilin by T lymphocytes of different origins (265, 268). Since cofilin contains nuclear localization sequences KKRKK (269), while actin does not, it can mediate actin nuclear ectopic processes upon dephosphorylation (263). This is necessary for the transcriptional activity of RNA polymerase II (270, 271). Additionally, in co-stimulated T cells, dephosphorylated cofilin can regulate the nuclear translocation of NF-κB and promote the production of anti-inflammatory factors, which is one of the mechanisms by which T helper 2 cells (Th2) exert anti-inflammatory effects (272). When cofilin is oxidized and inactivated, it affects costimulatory signaling and leads to T-cell anergy. Cofilin contains four cysteine residues, Cys39, Cys80, Cys140, and Cys148, all of which are potential sites for oxidation. Oxidants such as reactive oxygen species (ROS) may cause the formation of disulfide bonds between Cys39 and Cys80, resulting in the loss of cofilin’s depolymerization activity to F-actin although it can still bind to filaments (273). Moreover, oxidized cofilin tends to undergo mitochondrial translocation, leading to programmed T cell death (274). In contrast, the sensitivity of cofilin to PIP2 is decreased under reducing conditions, resulting in enhanced activity of cofilin (275).
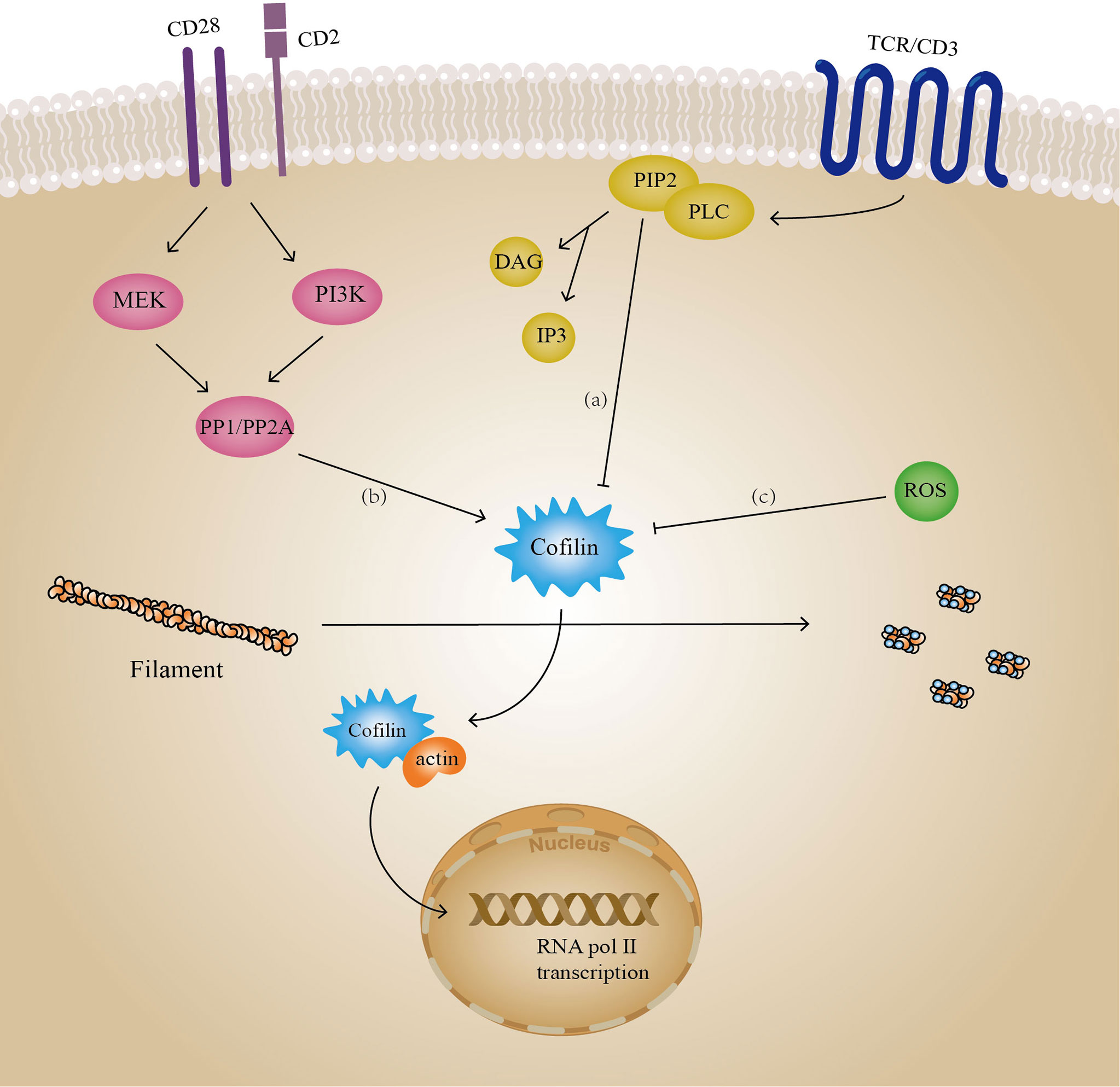
Figure 6 Cofilin involves in T cell signaling. Several signaling pathways in T cells contain cofilin, including: (A) PLC-PIP2 pathway belonging to the TCR signaling, (B) protein phosphorylation belonging to co-stimulation signaling, and (C) protein oxidation. Activated cofilin can mediate actin entry into the nucleus and play a regulatory role in gene transcription. PP1/PP2A: protein serine/threonine phosphatases of type 1 and type 2A, ROS, reactive oxygen species.
3.1.5 T Cell Cytotoxicity
Upon activation, naïve T cells proliferate and differentiate into different functional subpopulations in response to the local microenvironment and other factors. Some of these cells have killing activity and can differentiate into CTLs, predominantly CD8+ CTLs, but also CD4+ CTLs have been reported to exist in vivo (276). CTLs are able to recognize virally infected and tumorigenic cells and secrete perforin and granzyme to kill these target cells or mediate apoptosis through the death ligands pathway (277–279). Studies have revealed that the function of the Arp2/3 complex is critical for the formation of CTL synaptic protrusions and granule release (280), as well as the CTL’s cytolytic function (281). The importance of Arp2/3 in cytotoxicity is likely due to the actin cytoskeleton maintaining the IS of CTLs. In addition, Arp2/3 is involved in the cycling of TCR, CD8, and GLUT1, which enables the TCR signaling needed for inducing the cytotoxic effects of CTLs on target cells. Also, by binding to Arp2/3, WASp can promote the formation of protrusions and cause deformation of target cells, which was confirmed to enhance perforin and granzyme-mediated killing (280). Furthermore, Formins can also regulate TCR-mediated toxicity by affecting the centrosome polarity of T cells, which is vital for directed release of granules (215).
3.2 B Lymphocytes
B cells are important antibody-producing immune cells in humans, the homeostasis of B cells is significant to a well-balanced immunity. Stimulated by antigens, the activation of the BCR initiates the immunological response in B cells. The BCR signaling pathway is regulated by different molecules and their interaction maintains normal B cell function. In addition to BCRs, B cells also have Toll-like receptors (TLRs) that are involved in innate immunity and function to link specific immunity with non-specific immunity. As a component of B cell signal transduction, TLR signaling also contributes to regulation of B cell activity.
3.2.1 BCR/TLR Signaling
In BCR signaling, the unbranched filament formed by the Formin DIAPH1 assists Arp2/3 in the generation of actin foci, which is an important step in the antigen extraction by B cells (282). It was also found that the C-terminus of FHOS, another formin protein widely present in the human spleen, interacts with CD21, which is involved in the regulation of B cell signaling together with the subsequent actin kinetics (283). Upon B cell binding to APCs, the Arp2/3-mediated branched actin network promotes the aggregation of BCR-containing microclusters into a supramolecular activation cluster in the center of the IS, enhancing the BCR signaling (284). The experimental result that the inhibition of the Arp2/3 complex leads to reduced BCR motility also suggests a regulatory role of this complex on B cell signaling (285). N-WASP plays both similar and different roles compared with WASp. On the one hand, in WASp-deficient B cells, N-WASP plays a compensatory role to help BCR clustering and B cell spreading. Also, N-WASP can play a role in BCR microcluster aggregation into central clusters and promote B cell contraction. On the other hand, N-WASP is involved in the up- and down-regulation of BCR signaling. During upregulation, N-WASP and WASp synergistically promote microfilament formation, while N-WASP functions differently from WASP in removing F-actin during cell contraction and signal downregulation. A further study revealed that there is a mutual negative regulatory relationship between WASp and N-WASP. In the BCR signaling pathway, Btk activates WASp but inhibits N-WASP, whereas SHIP-1 can inhibit Btk and activate N-WASP (Figure 7) (286).
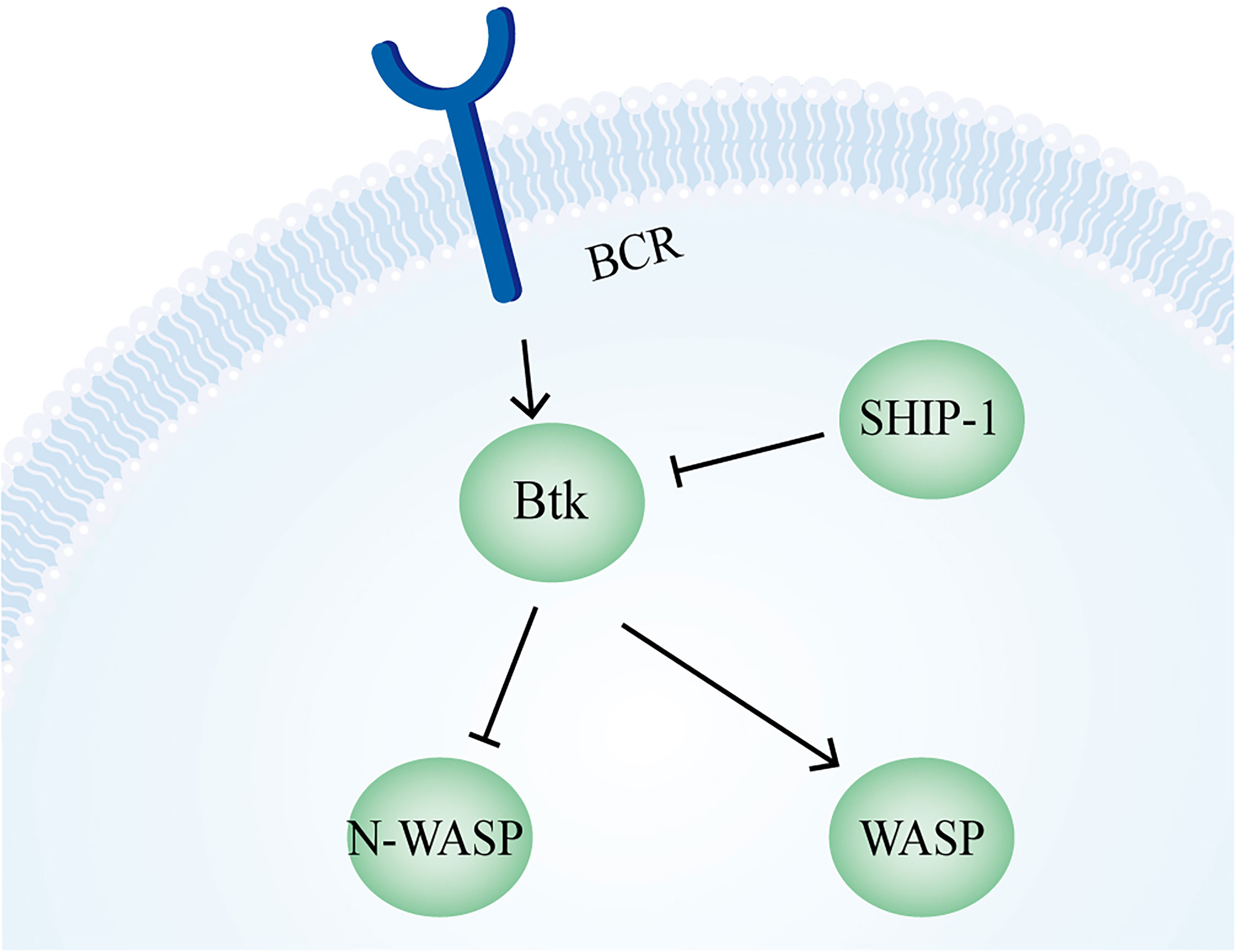
Figure 7 Regulation of N-WASP and WASP in B cells. In B cells, N-WASP is inhibited by Btk, which in turn is negatively regulated by SHIP-1. The regulation of WASP depends on Btk for activation.
Ca2+ is an important second messenger in BCR signaling, as BCR-induced Ca2+ release can act on cofilin via the PCLγ and CRAC pathways, and thus regulate B cell adhesion and lamellipodia formation (287). The Ca2+ mobilization decreases significantly in coronin1A deficient B cells after BCR stimulation, but the lack of coronin 1A makes no difference for B cell subset development and immune function (288).
Upon signal stimulation, cofilin is dephosphorylated by BCR via active Rap-GTP, severing actin filaments, and thereby increasing the mobility of the BCR and promoting B-cell spreading (289). Meanwhile, the Rap-cofilin pathway promotes the polarization of MTOC toward the APC contacting site, which is an important step in the process of B cell IS formation (290). In addition, cofilin is activated by TLR signaling stimulation, reducing the restriction of the BCR by the actin cytoskeleton and facilitating the signal transduction of the BCR (291) (Figure 8).
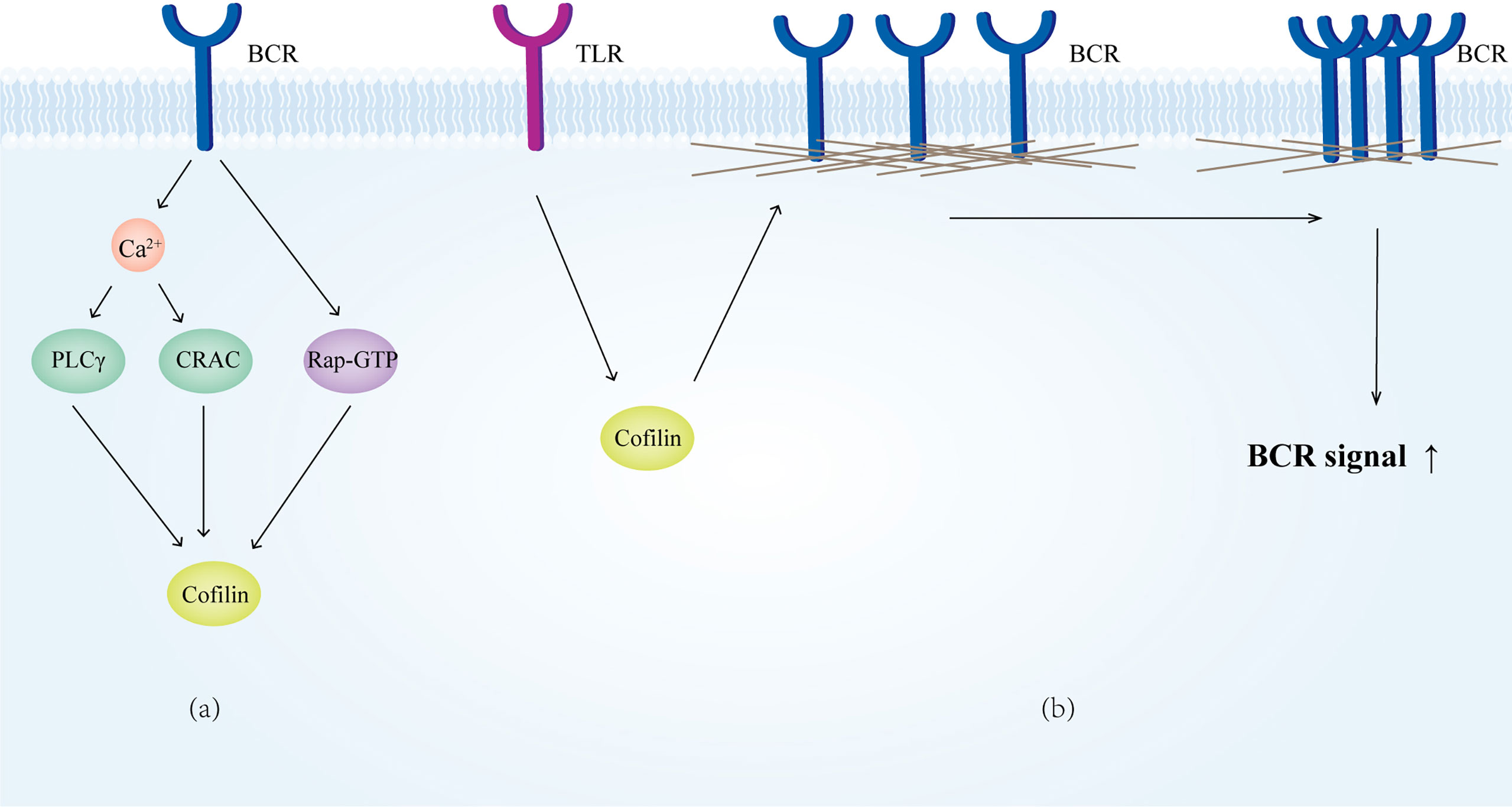
Figure 8 Cofilin in B cells. (A) Cofilin receives signals from the BCR. (B) Cofilin mediates the depolymerization of microfilaments on the membrane surface upon receiving TLR signaling, which increases BCR motility and facilitates the BCR signaling.
Recent studies have revealed that WDR1, together with LIMK, an enzyme that phosphorylates cofilin and negatively regulates it, constitute the WDR1-LIMK-cofilin axis, an important mechanism for regulating cSMAC formation and BCR signaling in B cells. This process involves WDR1 and LIMK working together to maintain cofilin at an equivalent activity (neither too high nor too low), thus achieving an optimal level of actin retrograde flow (292).
3.2.2 Others
Many other B cell activities are modified by actin regulators, such as anti-infection, cell development and antibody secretion.
For cell development, the latest studies revealed that the interaction between WASp and other signaling molecules facilitates B cell development and movement. Mst1 and WASp are significant for central and peripheral development of B cells and can adjust mutual localization and function (293). Deficiency of DOCK2 reduces the activation of WASp and accelerates its degradation, causing dysfunction of actin accumulation and affecting the early activation process of B cells (294). Similar to T cells, the reduction of marginal zone B cells in N-WASP conditional knockout mice indicates the promotional effect of this protein on peripheral B cell development. Further studies showed that conditional double knockout WASp and N-WASP exhibited more severe B cell developmental impairment and dysfunction than WASp KO, indicating the positive role of N-WASP in B cell development (72). More severe abnormalities were found in B-cells of WHR1-deficient patients than in T-cells. On the one hand, the number of B-cell precursors is reduced, and more peripheral blood B cells are in an immature stage; on the other hand, this defect leads to abnormal BCR/TLR signaling, generates elongated B-cell synapse, and affects B cell survival (199). Besides intrinsic accommodation of B cell growth, environmental factors such as the extracellular matrix components may influence the motility of B cells (295). The actin dynamics regulated by the Arp2/3 complex gives B cells this ability to exhibit adaptive mobility in different environments (296).
For the response to antigen stimulation, it has been demonstrated that in secondary lymphoid organs, cofilin mediates B cell responses to two common antigens, namely soluble and membrane-associated antigens, in the non-phosphorylated form (297). Differences in the distribution and kinetic characteristics of cofilin under the two antigen inductions explain the different reactivity of B cells to these two antigens (297). N-WASP in B cells is closely linked to autoimmune responses (298). It was found that specific knockdown of N-WASP in B cells resulted in elevated levels of autoantibodies, and mouse B cells lacking N-WASP exhibited higher levels and longer duration of activation. This suggests that N-WASP is a key inhibitor of B cell activation, and this inhibition is essential to terminate the immune response and prevent autoimmunity (286). Relatedly, simultaneous knockdown of WASp and N-WASP in B cells results in more severe proliferation defects in B cells and reduced levels of IgG autoantibodies compared to specific knockdown of the WASp gene, suggesting that N-WASP expression in B cells is required for the development of autoimmunity, which signifies N-WASP as a new therapeutic target to control autoimmunity in WAS patients (299). Analogously to that in T cells, the Arp2/3 complex is important to the formation and activity of the IS in B cells. It was demonstrated that this complex is involved in the formation of dynamic actin structures at the IS of B cells (282). Also, upon B lymphocyte activation, the HS1-dependent Arp2/3 complex is recruited at the IS through Syk-mediated tyrosine phosphorylation (300), which results in a reduction of Arp2/3 complex at the centrosome, leading to its nuclear separation and polarization to the IS (301), a process that maintains the immunomodulatory role of lymphocytes.
Additional benefits WASp provides for lymphocytes includes protection from DNA damage. WASp acts as a novel element to link irradiation-induced DNA damage signaling with the Golgi dispersal response (GDR), which has the ability of ensuring genome stability and contributes to cell survival under DNA damage. The loss of WASp triggers accumulated DNA damage and causes the failure of GDR, resulting in the dysfunction of human T and B lymphocytes (302).
3.3 NK Cells
As a vital member of lymphocytes, natural killer cells (NK cell) are not only involved in fighting tumors and infections, but also participate in hypersensitivity and autoimmune disease. Its natural killing activity is independent of antigen stimulation and is not restricted by MHC.
3.3.1 NK Cell Cytotoxicity
NK cells can kill target cells by forming IS and releasing lytic granules (303). The activity of NK cells is regulated by cytoskeletal proteins (304, 305), of which the Arp2/3 complex promotes the aggregation of NK receptors in the lytic synapse, allowing for stable binding between NK cells and target cells. This has been demonstrated in studies showing that the lack of Arp2/3 causes defective cell adhesion to target cells, which inhibits lytic synapse actin assembly, thus affecting the lysis activity of NK cells (306). Additionally, examination at the nanoscale level found that the Arp2/3 complex also plays an active role in lytic granule secretion (307).
Tβ4 enhances the cytotoxicity of NK cells by increasing the expression of intercellular adhesion molecule-1 (ICAM-1) and LFA-1, as well as cytolytic granule exocytosis (308). Furthermore, the expression of Tβ4 is upregulated by IL-18 and in turn, Tβ4 enhances IFN-γ secretion mediated by IL-18 in NK cells, which positively influences the immunomodulatory role of NK cells (309).
With respect to the WASp family, WASp is localized in the IS with F-actin and regulates cytotoxicity functions in NK cells (310, 311). This correlates with the interaction of DOCK8, which regulates the polarization of WASp and mediates the cytotoxicity of NK cells (312). The Y141 tyrosine of WASH can be phosphorylated by the Src family kinase Lck, thus exerting its role in promoting the movement and release of granules and assisting the cytotoxic effects of NK cells (313). Formins also help in NK cell activity. It has been demonstrated that the effect of hDia1 on NK cell toxic effects is actin-independent, instead it mediates the formation of microtubule networks through enrichment at the lytic synapse to facilitate the transport and secretion of lytic granules (306). In addition, the function of hDia1 in the formation of filopodial protrusions makes it a vital factor involved in the migration and adhesion of NK cells (306). What is more, cofilin is phosphorylated in response to LIMK stimulation and regulates the remodeling of the actin cytoskeleton in NK cells, thus affecting its cytotoxicity (314).
Finally, a recent mass spectrometry study of NK cell protein expression in severe aplastic anemia (SAA) patients found that the Arp2/3 complex is downregulated in NK cells (315), suggesting a role in the disease-protective function of NK cells in SAA.
3.3.2 Others
As for cell movement, the knockdown of N-WASP partially rescues the inhibition of NK cell migration towards CXCL12 gradient, suggesting its involvement in the positive regulation of NK cell motility (316). Furthermore, the results of in vitro experiments suggest that cofilin in NK-92 cells is stimulated by leptin under physiological conditions, which may promote cell migration and mediate the positive effects of leptin on NK cell morphology in a healthy environment (317).
It has been reported that WASp deficiency affects the activation of NK cells and DCs, as DCs in WAS KO mice have weaker induction of NK cells, thus influencing the immune response.
Up to now, it is believed that coronin has little function in NK cells, although further research is still needed (318).
4 Related Diseases
4.1 Cancer
Since actin filaments are important to cell motility, migration, adhesion, cell growth and cell division, aberrant functions of actin regulators could lead to abnormal cellular functions and cytokinesis, thus contributing to the development and migration of tumor cells (319). Moreover, aberrant actin regulators can also influence the cytotoxic functions of T cells and NK cells, resulting in tumorigenesis and progression.
Tβ4 is reported to have tumor-promoting functions that enhance the motility of cancerous cells. Myocardin-related transcription factors (MRTF), which are co-activators of serum response factor (SRF), are linked to epithelial-mesenchymal transition (EMT) and tumor metastasis. Studies have found Tβ4 can bind to actin competitively with MRTF, mediating the activation of the latter by TGFβ and regulating the cytoskeleton and the expression of tumor-associated proteins, thus enhancing the motility of tumor cells and leading to tumor progression and metastasis (320). Additionally, in glioma cells, Tβ4 inhibition reduces the migratory capacity and aggressiveness of tumor cells and is a potential target for the treatment of glioblastoma (321).
Many recent studies have focused on the relationship between WASp and cancer. WASp is important for tumor suppression and killing and considered a new possible therapeutic target in cancer. In T cells, constitutive activation of WASp improves cytotoxic clearing of tumor cells (322). Interestingly, studies have shown that WASp plays opposite roles in malignant and benign lymphocytes. In benign T and B cells, WASp is a tumor-suppressor protein, however it acts as a tumor activator in malignant lymphocytes, since the lack of WASp leads to an imbalance of CDC42/MAPK and NF-κB/AP-1 signaling pathways, which is important for the development of cancer (323). But a recent study reported that WASp and WIP are tumor suppressors in T cell lymphoma, because high expression of WASp prevents lymphoma growth (189). Further studies are needed to figure out the role of WASp in malignant lymphocytes. In WASp deficient mice, the antitumor functions of NK cells and DCs are weakened (324). WASp deficient patients have increased granzyme B and degranulation, along with enhanced production of IFN-γ. Additionally, the expression levels of DNAM-1, LAG-3, KLRG1 increase while CD56 expression decreases, indicating a lack of NK cell phenotype. This correlates with defective F-actin accumulation and disruption of lytic synapse formation. All these changes in NK cells lead to a weaker anti-tumor response (322, 325). In addition, inhibiting Cdc42, which activates NKG2D, WASp and N-WASp to regulate NK cell migration, may impact immunoreaction and evasion in tumors (316).
The Arp2/3 complex also shows strong relevance in cancer development. The contribution of the Arp2/3 complex in podosome formation (326, 327) makes it an important factor in enhancing the mobility of cancer cells. N-WASP can promote cancer progression by activating Arp2/3 and affecting the tumor killing function of NK cells. For example, it has been demonstrated that N-WASP exerts actin-regulatory activity in colorectal cancer metastasis (328), while in breast cancer cells, N-WASP enables rapid actin reconstitution, accumulating large amounts of F-actin at NK cell synapses, thus resisting NK cell attack and inducing immune escape (329). Furthermore, a clinical study on esophageal carcinoma showed that overexpressed WASH maintained the stem cell phenotype of cancer cells by promoting IL-8 production, thus promoting the progression of the carcinoma. Additionally, in vivo experiments found that the inhibition of WASH expression slowed the progression of tumors, which suggests WASH be a potential target to intervene in human esophageal carcinoma (330).
A study performed on an oral squamous cell carcinoma (SCC) cell line revealed that PI3K signaling-dependent formin, FHOD1, was specifically upregulated during EMT in tumor cells, which promoted morphological changes and enhanced the invasiveness of cancer cells (331). Similarly, an in vivo study found that mDia1 promoted the TEM capacity of leukemia cells and played an active role in the progression of leukemia, and conditional inhibition of mDia1 expression might be a latent way to treat leukemia (332). Studies in Molt-3 and Jurkat cells indicate that human formin-2 (FMN2) expression is inhibited by upregulated microRNA-144 (miR-144), affecting normal cellular activities and providing an explanation for the tumor suppression mechanism of miR-144 (333). Additionally, FMNL1 is expressed in a variety of malignant tissues, while an antigenic peptide (FMNL1-PP2) derived from FMNL1 was experimentally shown to induce specific T cells to exert killing effects on tumors, including lymphomas (334).
Cofilin is also closely associated with tumor development. The regulation of cofilin phosphorylation by Myeloid cell leukemia 1 (MCL-1) is associated with tumor cell activity in B-cell lymphoma 2, and the inhibition of this effect of MCL-1 has become important for tumor therapy (335). Moreover, in malignant T lymphoma cells, it was found that blocking the dephosphorylation of cofilin led to apoptosis, which might have a promising role in arresting tumor progression (336). Cofilin’s regulatory role on the cytoskeleton makes it an important molecule in regulating the migration of tumor cells (337). It has been demonstrated that chitinase 3-like 1 (Chi3l1) dysregulation, present in many solid tumors, inhibits the accumulation of phosphorylated cofilin, hence promoting tumor cell metastasis, however, activation of RIG-like helicase can rescue this process (338). Specifically, in breast cancer, various molecules, such as LMO2, regulate the actin cytoskeleton by interacting with cofilin and altering the dynamics of cell adhesion and lamellipodial, thereby promoting the metastasis of cancer cells and increasing their invasiveness (339–341). Additionally, in the tumor microenvironment, granulocytes or macrophages can mediate oxidative stress (342, 343), which may lead to a loss of functional activity of cofilin, and in turn affect the function of T cells. In tumor treatment, heat therapy can inhibit tumor cell migration by phosphorylating cofilin through high temperature, but it should be noted that the inhibitory effect of Hsp70 overexpression on this process can reduce the efficacy (344).
Analysis of the proteome of Pancreatic ductal adenocarcinoma (PDAC) tissues showed that protein level of WDR1 increased with the clinical progression of PDAC. Further studies revealed that WDR1 is associated with tumor cell growth and metastasis and is involved in the deubiquitination of β-Catenin in PDAC cells by interacting with USP7, thereby regulating Wnt/β-Catenin signaling in PDAC cells (345).
Overall, comprehensive understanding of the role of various actin regulators in tumorigenesis and metastasis can provide new ideas for precise therapy. Moreover, the critical role of actin regulators in immune cells can modulate tumor microenvironment and affect the outcome of immunotherapy. Therefore, further studies are required in this field to bring more benefit to patients with cancer.
4.2 HIV
Acquired immunodeficiency syndrome (AIDS) is a severe infectious disease caused by Human Immunodeficiency Virus (HIV) infection. Although researchers around the world have made great efforts, no effective drugs have been developed to cure AIDS, and there is also no effective vaccine that can be used for prevention. Several actin regulators are reported to be related to HIV infection and may provide latent therapeutic targets for AIDS.
The role of profilin in resisting HIV infection is controversial. In ADP-Heat Shock Protein (ADP-HSP) immunized macaques, profilin can up-regulate the expression of apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G), and increase the level of IgG against CD4+ T cells, which provides a new direction of therapy in HIV infection (346). However, another study has shown that through downregulating drebrin, another actin-binding protein, profilins accumulate and suppress the polymerization of actin in CD4+ T cells, which increases the entry of HIV (347).
HIV can cause the over-activation of cofilin (348), leading to a weakened T cell cycle and inhibiting T cell activation (349, 350). In vitro studies, however, revealed that the use of anti-human α4β7 integrin antibodies partially restored the motility of CD4+ T cells (348). Cofilin and the cytoskeleton have an important role in HIV infection (351). First, HIV can activate cofilin through CXCR4-mediated Gαi-dependent signaling pathways, promoting the depolymerization of cortical actin and facilitating its viral nuclear localization in resting T cells (352). Also, chemokines such as CCL19 and CCL21 can produce a similar reaction to promote the nuclear migration of HIV in memory T cells, which is one of the possible mechanisms for the establishment of HIV latency (353, 354). Furthermore, the migration of infected T cells can be restricted by HIV through the binding of Nef to the cellular kinase Pak2, which phosphorylates cofilin and inhibits actin dynamics (355).
In vitro and in vivo studies both showed that intracellular Tβ4 expression in T cells and macrophages was reduced after HIV-1 infection, indicating a regulatory effect of Tβ4 in T cell immunity (356).
N-WASP is one of the ways in which HIV-1 infects T cells. The Nef protein of HIV-1 inhibits both the activation of N-WASP and its recruitment in the T cell contact region, which also interferes with the role of its upstream regulatory molecules, Rac1 and Cdc42, in regulating actin remodeling, thus inhibiting the proper TCR signaling pathway (357).
Arp2/3 plays a critical role in HIV infection. Earlier studies have found that during pathogen infection the Arp2/3 complex of host cells can be activated by the vaccinia virus-encoded A36R protein, contributing to virus transmission (358). Also, baculovirus activates the Arp2/3 complex similarly, facilitating viral proliferation (359). The action of HIV on the Arp2/3 complex is an important component in the development of AIDs. Through the Rac1-IRSP53-WAVE2-Arp2/3 signaling pathway, HIV can invade host cells (360) and promote the assembly and release of virus-like particles (VLPs) (361). Lack of the Arp2/3 complex inhibits HIV infection of CD4 T cells, implying it could be a potent target for the treatment of AIDs (362, 363).
Furthermore, Dia1 and Dia2 also play an important role in early HIV-1 infection by helping to form a stable microtubule network and promoting viral uncoating (364). In HIV-1 infected DCs, Dia2 facilitates HIV-1 infection of T cells by regulating actin assembly to assist filopodia formation, while Slit2N was found to inhibit this effect (365, 366).
Determining the relationship between HIV infection and actin regulators will be helpful in developing more effective drugs in the prevention and treatment of AIDS in the future.
4.3 Others
Because of the vital role of the cytoskeleton in lymphocytes, actin regulators are also specifically related to other diseases including autoimmune diseases and infectious diseases.
High expression of Tβ4 was detected in patients with pulmonary tuberculosis, which is related to inflammation and angiogenesis mediated by HIF-1α and VEGF and may serve as a potential biomarker for diagnosis (367). Moreover, an analogue of Tβ4 was found to have a restorative effect on T-lymphocyte deficiency in uremic patients and used to rescue the immune function in uremic patients (368).
WAS directly influences the essential role of the functional activity of the Arp2/3 complex mediated by WASp in the human body (62). CID, a disease caused by a homozygous mutation in the gene encoding ARPC1B, is similar to WAS (230). And defects in ARPC4 in the epidermis may trigger Psoriasis-like skin complication epidermis (369), while pachygyria may be caused by cortical neuronal migration disorders due to overactivity of the Arp2/3 complex (370). Also, the decrease in cell migration caused by the inhibition of the Arp2/3 complex by factors such as prostaglandin E2 may be responsible for Hirschsprung disease (371, 372). Recent studies have also identified the significance of this complex in bone and neural tissues, suggesting that defects in Arp2/3 may be associated with the development of intervertebral disc defects (373) as well as Down syndrome and Alzheimer disease (374). In innate immune cells, defective WASp interrupts the autophagy-inflammasome axis (375). And further studies reveal that depending on the interaction with Arp2/3, WASp plays an essential role in the autophagy process, especially during the autophagosome formation and the delivery of autophagosomes to lysosomes (376), which contributes to the development of auto-inflammatory diseases. Additionally, the dysfunction of endogenous inhibitory proteins of Arp2/3 also leads to the development of several diseases. For example, the over-expression of coronin 1A causes the over activation of CD4+ T cells and CD8+ T cells in aplastic anemia and hemophagocytic syndrome (377).
Formins perform different roles in the development of several diseases. For example, the promotion of CD4+ T cell migration by FMNL1 may be one of the pathogenic mechanisms of equine recurrent uveitis (ERU) (378), suggesting its role in autoimmune diseases. In addition, it is noteworthy that in the chemical-induced asthma mouse model, the expression of cofilin in B cells differs from that in normal individuals (379), suggesting a link between cofilin and the progression of B cell-associated diseases.
Another point of interest is that mutations in the WDR1 gene have been found to cause lazy leukocyte syndrome, a disease characterized by impaired neutrophils, recurrent infections, and stomatitis (380–382). In combination with other findings (198, 383), this mutation impairs the regulation of actin by WDR1, leading to increased levels of F-actin, which induces an IL-18-mediated inflammatory response downstream. Thus, WDR1 is extremely important for neutrophil function and its deficiency can lead to auto-inflammation and immunodeficiency (179).
5 Conclusions
Here, we discussed a selection of the most representative actin regulators whose functions in universal cells have been studied extensively, therefore we focused on their roles in lymphocytes. Many of the identified actin regulators to date have important roles in the development and functional activity of lymphocytes. The majority of these roles are performed through cytoskeletal modulation by actin, leading to changes in cell structure or polarity that affect lymphocyte development and immune activity. It is also possible that actin regulators act as effector molecules involved in cellular signaling pathways, thereby regulating cellular activities such as gene expression, where actin may not be necessary. It is worth mentioning that these actin regulators do not exist independently. Actin regulators with similar/opposite functions coordinate/antagonize each other intracellularly to form a complete cytoskeletal regulatory network. Meanwhile, these actin regulators are not in complete parallel in terms of their functions, and multiple regulatory effects on lymphocytes need to be realized through interactions between upstream and downstream actin regulators. Therefore, damage or deletion of different actin regulators causes cellular developmental disorders and malfunctions to different degrees, leading to the development of various diseases. Particularly, in immune system diseases, abnormalities of these proteins often lead to immune overload or immunosuppression by affecting the normal signaling of intrinsic immune cells.
Unfortunately, not all of the actin regulators are fully discussed here since little is known about the functional role of some of the recently discovered actin regulators in lymphocytes. In particular, proteins with more specific functions, such as WASH, may have more scope for investigation in lymphocytes. In addition, we noticed that several actin regulators are closely related to AIDS and tumors, and previous studies have suggested the potential value of targeting these proteins for treatment, so a combine study of several related actin regulators could be followed as an entry point to find the most likely therapeutic modality.
Author Contributions
JS, XZ, and XF wrote the article and drew the figures. BY and CL organized the draft and revised the draft. HM and PL reviewed, proofread, and revised the draft. JS, XZ, and XF contribute equally to this work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (31970839), HUST Academic Frontier Youth Team (2018QYTD10), Independent Innovation Research Fund of Huazhong University of Science and Technology (2020kfyXGYJ017), and National Undergraduate Training Program for Innovation and Entrepreneurship (202110487122).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Elzinga M, Collins JH, Kuehl WM, Adelstein RS. Complete Amino-Acid Sequence of Actin of Rabbit Skeletal Muscle. Proc Natl Acad Sci USA (1973) 70(9):2687–91. doi: 10.1073/pnas.70.9.2687
2. Selby CC, Bear RS. The Structure of Actin-Rich Filaments of Muscles According to X-Ray Diffraction. J Biophys Biochem Cytol (1956) 2(1):71–85. doi: 10.1083/jcb.2.1.71
3. Goley ED, Welch MD. The ARP2/3 Complex: An Actin Nucleator Comes of Age. Nat Rev Mol Cell Biol (2006) 7(10):713–26. doi: 10.1038/nrm2026
4. Wegner A, Isenberg G. 12-Fold Difference Between the Critical Monomer Concentrations of the Two Ends of Actin Filaments in Physiological Salt Conditions. Proc Natl Acad Sci USA (1983) 80(16):4922–5. doi: 10.1073/pnas.80.16.4922
5. Heuser JE, Kirschner MW. Filament Organization Revealed in Platinum Replicas of Freeze-Dried Cytoskeletons. J Cell Biol (1980) 86(1):212–34. doi: 10.1083/jcb.86.1.212
6. Kovar DR, Pollard TD. Insertional Assembly of Actin Filament Barbed Ends in Association With Formins Produces Piconewton Forces. Proc Natl Acad Sci USA (2004) 101(41):14725–30. doi: 10.1073/pnas.0405902101
7. Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel Roles of Formin Mdia2 in Lamellipodia and Filopodia Formation in Motile Cells. PLoS Biol (2007) 5(11):e317. doi: 10.1371/journal.pbio.0050317
8. Doherty GJ, McMahon HT. Mechanisms of Endocytosis. Annu Rev Biochem (2009) 78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540
9. Huxley AF, Niedergerke R. Structural Changes in Muscle During Contraction; Interference Microscopy of Living Muscle Fibres. Nature (1954) 173(4412):971–3. doi: 10.1038/173971a0
10. Pollard TD. Actin and Actin-Binding Proteins. Cold Spring Harbor Perspect Biol (2016) 8(8):a018226. doi: 10.1101/cshperspect.a018226
11. Al-Shura AN. 7 - Lymphocytes. In: Al-Shura AN, editor. Advanced Hematology in Integrated Cardiovascular Chinese Medicine. London: Academic Press (2020). p. 41–6.
12. Cohn L, Hawrylowicz C, Ray A. 12 - Biology of Lymphocytes. In: Adkinson NF, Bochner BS, Burks AW, Busse WW, Holgate ST, Lemanske RF, editors. Middleton's Allergy (Eighth Edition). London: W.B. Saunders (2014). p. 203–14.
13. Omman RA, Kini AR. 9 - Leukocyte Development, Kinetics, and Functions. In: Keohane EM, Otto CN, Walenga JM, editors. Rodak's Hematology (Sixth Edition). St. Louis, MO: Elsevier (2020). p. 117–35.
14. Dupré L, Houmadi R, Tang C, Rey-Barroso J. T Lymphocyte Migration: An Action Movie Starring the Actin and Associated Actors. Front Immunol (2015) 6:586. doi: 10.3389/fimmu.2015.00586
15. Penninger JM, Crabtree GR. The Actin Cytoskeleton and Lymphocyte Activation. Cell (1999) 96(1):9–12. doi: 10.1016/s0092-8674(00)80954-x
16. Tolar P. Cytoskeletal Control of B Cell Responses to Antigens. Nat Rev Immunol (2017) 17(10):621–34. doi: 10.1038/nri.2017.67
17. Yan SLS, Hwang IY, Kamenyeva O, Kehrl JH. In Vivo F-Actin Filament Organization During Lymphocyte Transendothelial and Interstitial Migration Revealed by Intravital Microscopy. iScience (2019) 16:283–97. doi: 10.1016/j.isci.2019.05.040
18. Tang DD, Gerlach BD. The Roles and Regulation of the Actin Cytoskeleton, Intermediate Filaments and Microtubules in Smooth Muscle Cell Migration. Respir Res (2017) 18(1):54. doi: 10.1186/s12931-017-0544-7
19. Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T Cell Receptor-Proximal Signals Are Sustained in Peripheral Microclusters and Terminated in the Central Supramolecular Activation Cluster. Immunity (2006) 25(1):117–27. doi: 10.1016/j.immuni.2006.04.010
20. Phee H, Au-Yeung BB, Pryshchep O, O'Hagan KL, Fairbairn SG, Radu M, et al. Pak2 Is Required for Actin Cytoskeleton Remodeling, TCR Signaling, and Normal Thymocyte Development and Maturation. eLife (2014) 3:e02270. doi: 10.7554/eLife.02270
21. Fleire SJ, Goldman JP, Carrasco YR, Weber M, Bray D, Batista FD. B Cell Ligand Discrimination Through a Spreading and Contraction Response. Science (New York NY) (2006) 312(5774):738–41. doi: 10.1126/science.1123940
22. Brown BK, Song W. The Actin Cytoskeleton is Required for the Trafficking of the B Cell Antigen Receptor to the Late Endosomes. Traffic (Copenhagen Denmark) (2001) 2(6):414–27. doi: 10.1034/j.1600-0854.2001.002006414.x
23. Graham DB, Cella M, Giurisato E, Fujikawa K, Miletic AV, Kloeppel T, et al. Vav1 Controls DAP10-Mediated Natural Cytotoxicity by Regulating Actin and Microtubule Dynamics. J Immunol (Baltimore Md 1950) (2006) 177(4):2349–55. doi: 10.4049/jimmunol.177.4.2349
24. Gil-Krzewska A, Saeed MB, Oszmiana A, Fischer ER, Lagrue K, Gahl WA, et al. An Actin Cytoskeletal Barrier Inhibits Lytic Granule Release From Natural Killer Cells in Patients With Chediak-Higashi Syndrome. J Allergy Clin Immunol (2018) 142(3):914–27.e6. doi: 10.1016/j.jaci.2017.10.040
25. Vorum H, Ostergaard M, Hensechke P, Enghild JJ, Riazati M, Rice GE. Proteomic Analysis of Hyperoxia-Induced Responses in the Human Choriocarcinoma Cell Line JEG-3. Proteomics (2004) 4(3):861–7. doi: 10.1002/pmic.200300639
26. Carlsson L, Nystrom LE, Lindberg U, Kannan KK, Cid-Dresdner H, Lovgren S. Crystallization of a non-Muscle Actin. J Mol Biol (1976) 105(3):353–66. doi: 10.1016/0022-2836(76)90098-x
27. Krishnan K, Moens PDJ. Structure and Functions of Profilins. Biophys Rev (2009) 1(2):71–81. doi: 10.1007/s12551-009-0010-y
28. Witke W, Sutherland JD, Sharpe A, Arai M, Kwiatkowski DJ. Profilin I Is Essential for Cell Survival and Cell Division in Early Mouse Development. Proc Natl Acad Sci USA (2001) 98(7):3832–6. doi: 10.1073/pnas.051515498
29. Braun A, Aszodi A, Hellebrand H, Berna A, Fassler R, Brandau O. Genomic Organization of Profilin-III and Evidence for a Transcript Expressed Exclusively in Testis. Gene (2002) 283(1-2):219–25. doi: 10.1016/s0378-1119(01)00855-1
30. Obermann H, Raabe I, Balvers M, Brunswig B, Schulze W, Kirchhoff C. Novel Testis-Expressed Profilin IV Associated With Acrosome Biogenesis and Spermatid Elongation. Mol Hum Reprod (2005) 11(1):53–64. doi: 10.1093/molehr/gah132
31. Hu E, Chen Z, Fredrickson T, Zhu Y. Molecular Cloning and Characterization of Profilin-3: A Novel Cytoskeleton-Associated Gene Expressed in Rat Kidney and Testes. Exp Nephrol (2001) 9(4):265–74. doi: 10.1159/000052621
32. Wittenmayer N, Rothkegel M, Jockusch BM, Schluter K. Functional Characterization of Green Fluorescent Protein-Profilin Fusion Proteins. Eur J Biochem (2000) 267(16):5247–56. doi: 10.1046/j.1432-1327.2000.01600.x
33. Mayboroda O, Schluter K, Jockusch BM. Differential Colocalization of Profilin With Microfilaments in PtK2 Cells. Cell Motil Cytoskeleton (1997) 37(2):166–77. doi: 10.1002/(SICI)1097-0169(1997)37:2<166::AID-CM9>3.0.CO;2-6
34. Sohn RH, Goldschmidt-Clermont PJ. Profilin: At the Crossroads of Signal Transduction and the Actin Cytoskeleton. Bioessays (1994) 16(7):465–72. doi: 10.1002/bies.950160705
35. Sathish K, Padma B, Munugalavadla V, Bhargavi V, Radhika KV, Wasia R, et al. Phosphorylation of Profilin Regulates its Interaction With Actin and Poly (L-Proline). Cell Signal (2004) 16(5):589–96. doi: 10.1016/j.cellsig.2003.10.001
36. Gau D, Veon W, Zeng X, Yates N, Shroff SG, Koes DR, et al. Threonine 89 Is an Important Residue of Profilin-1 That Is Phosphorylatable by Protein Kinase A. PLoS One (2016) 11(5):e0156313. doi: 10.1371/journal.pone.0156313
37. Shao J, Diamond MI. Protein Phosphatase 1 Dephosphorylates Profilin-1 at Ser-137. PLoS One (2012) 7(3):e32802. doi: 10.1371/journal.pone.0032802
38. Yarmola EG, Bubb MR. Profilin: Emerging Concepts and Lingering Misconceptions. Trends Biochem Sci (2006) 31(4):197–205. doi: 10.1016/j.tibs.2006.02.006
39. Carlsson L, Nystrom LE, Sundkvist I, Markey F, Lindberg U. Actin Polymerizability Is Influenced by Profilin, a Low Molecular Weight Protein in Non-Muscle Cells. J Mol Biol (1977) 115(3):465–83. doi: 10.1016/0022-2836(77)90166-8
40. Zweifel ME, Courtemanche N. Profilin's Affinity for Formin Regulates the Availability of Filament Ends for Actin Monomer Binding. J Mol Biol (2020) 432(24):166688. doi: 10.1016/j.jmb.2020.10.022
41. Goldschmidt-Clermont PJ, Furman MI, Wachsstock D, Safer D, Nachmias VT, Pollard TD. The Control of Actin Nucleotide Exchange by Thymosin Beta 4 and Profilin. A Potential Regulatory Mechanism for Actin Polymerization in Cells. Mol Biol Cell (1992) 3(9):1015–24. doi: 10.1091/mbc.3.9.1015
42. Nakamura M, Verboon JM, Allen TE, Abreu-Blanco MT, Liu R, Dominguez ANM, et al. Autocrine Insulin Pathway Signaling Regulates Actin Dynamics in Cell Wound Repair. PLoS Genet (2020) 16(12):e1009186. doi: 10.1371/journal.pgen.1009186
43. Henty-Ridilla JL, Juanes MA, Goode BL. Profilin Directly Promotes Microtubule Growth Through Residues Mutated in Amyotrophic Lateral Sclerosis. Curr Biol (2017) 27(22):3535–43.e4. doi: 10.1016/j.cub.2017.10.002
44. Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, et al. Differential Remodeling of Actin Cytoskeleton Architecture by Profilin Isoforms Leads to Distinct Effects on Cell Migration and Invasion. Cancer Cell (2012) 22(5):615–30. doi: 10.1016/j.ccr.2012.09.027
45. Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a Protein Associated With Wiskott-Aldrich Syndrome Protein, Induces Actin Polymerization and Redistribution in Lymphoid Cells. Proc Natl Acad Sci USA (1997) 94(26):14671–6. doi: 10.1073/pnas.94.26.14671
46. Vicente-Manzanares M, Rey M, Pérez-Martínez M, Yáñez-Mó M, Sancho D, Cabrero JR, et al. The RhoA Effector Mdia Is Induced During T Cell Activation and Regulates Actin Polymerization and Cell Migration in T Lymphocytes. J Immunol (Baltimore Md 1950) (2003) 171(2):1023–34. doi: 10.4049/jimmunol.171.2.1023
47. Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, et al. The Proline-Rich Focal Adhesion and Microfilament Protein VASP Is a Ligand for Profilins. EMBO J (1995) 14(8):1583–9. doi: 10.1002/j.1460-2075.1995.tb07146.x
48. Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a Relative of VASP and Drosophila Enabled, Is Implicated in the Control of Microfilament Dynamics. Cell (1996) 87(2):227–39. doi: 10.1016/s0092-8674(00)81341-0
49. Purich DL, Southwick FS. ABM-1 and ABM-2 Homology Sequences: Consensus Docking Sites for Actin-Based Motility Defined by Oligoproline Regions in Listeria ActA Surface Protein and Human VASP. Biochem Biophys Res Commun (1997) 231(3):686–91. doi: 10.1006/bbrc.1997.6158
50. Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, et al. Profilin Regulates F-Actin Network Homeostasis by Favoring Formin Over Arp2/3 Complex. Dev Cell (2015) 32(1):43–53. doi: 10.1016/j.devcel.2014.10.027
51. Davidson AJ, Wood W. Unravelling the Actin Cytoskeleton: A New Competitive Edge? Trends Cell Biol (2016) 26(8):569–76. doi: 10.1016/j.tcb.2016.04.001
52. Low TL, Hu SK, Goldstein AL. Complete Amino Acid Sequence of Bovine Thymosin Beta 4: A Thymic Hormone That Induces Terminal Deoxynucleotidyl Transferase Activity in Thymocyte Populations. Proc Natl Acad Sci USA (1981) 78(2):1162–6. doi: 10.1073/pnas.78.2.1162
53. Gondo H, Kudo J, White JW, Barr C, Selvanayagam P, Saunders GF. Differential Expression of the Human Thymosin-Beta 4 Gene in Lymphocytes, Macrophages, and Granulocytes. J Immunol (Baltimore Md 1950) (1987) 139(11):3840–8.
54. Paulussen M, Landuyt B, Schoofs L, Luyten W, Arckens L. Thymosin Beta 4 mRNA and Peptide Expression in Phagocytic Cells of Different Mouse Tissues. Peptides (2009) 30(10):1822–32. doi: 10.1016/j.peptides.2009.07.010
55. Gómez-Márquez J, Dosil M, Segade F, Bustelo XR, Pichel JG, Dominguez F, et al. Thymosin-Beta 4 Gene. Preliminary Characterization and Expression in Tissues, Thymic Cells, and Lymphocytes. J Immunol (Baltimore Md 1950) (1989) 143(8):2740–4.
56. Safer D, Sosnick TR, Elzinga M. Thymosin Beta 4 Binds Actin in an Extended Conformation and Contacts Both the Barbed and Pointed Ends. Biochemistry (1997) 36(19):5806–16. doi: 10.1021/bi970185v
57. Dedova IV, Nikolaeva OP, Safer D, de la Cruz EM, dos Remedios CG. Thymosin Beta4 Induces a Conformational Change in Actin Monomers. Biophys J (2006) 90(3):985–92. doi: 10.1529/biophysj.105.063081
58. Carlier MF, Jean C, Rieger KJ, Lenfant M, Pantaloni D. Modulation of the Interaction Between G-Actin and Thymosin Beta 4 by the ATP/ADP Ratio: Possible Implication in the Regulation of Actin Dynamics. Proc Natl Acad Sci USA (1993) 90(11):5034–8. doi: 10.1073/pnas.90.11.5034
59. Huff T, Rosorius O, Otto AM, Müller CS, Ballweber E, Hannappel E, et al. Nuclear Localisation of the G-Actin Sequestering Peptide Thymosin Beta4. J Cell Sci (2004) 117(Pt 22):5333–41. doi: 10.1242/jcs.01404
60. Notarangelo LD, Miao CH, Ochs HD. Wiskott-Aldrich Syndrome. Curr Opin Hematol (2008) 15(1):30–6. doi: 10.1097/MOH.0b013e3282f30448
61. Halle-Bikovski A, Fried S, Rozentur-Shkop E, Biber G, Shaked H, Joseph N, et al. New Structural Insights Into Formation of the Key Actin Regulating WIP-WASp Complex Determined by NMR and Molecular Imaging. ACS Chem Biol (2018) 13(1):100–9. doi: 10.1021/acschembio.7b00486
62. Derry JM, Ochs HD, Francke U. Isolation of a Novel Gene Mutated in Wiskott-Aldrich Syndrome. Cell (1994) 78(4):635–44. doi: 10.1016/0092-8674(94)90528-2
63. Cianferoni A, Massaad M, Feske S, de la Fuente MA, Gallego L, Ramesh N, et al. Defective Nuclear Translocation of Nuclear Factor of Activated T Cells and Extracellular Signal-Regulated Kinase Underlies Deficient IL-2 Gene Expression in Wiskott-Aldrich Syndrome. J Allergy Clin Immunol (2005) 116(6):1364–71. doi: 10.1016/j.jaci.2005.09.006
64. Luan Q, Zelter A, MacCoss MJ, Davis TN, Nolen BJ. Identification of Wiskott-Aldrich Syndrome Protein (WASP) Binding Sites on the Branched Actin Filament Nucleator Arp2/3 Complex. Proc Natl Acad Sci USA (2018) 115(7):E1409–e18. doi: 10.1073/pnas.1716622115
65. Verboon JM, Sugumar B, Parkhurst SM. Wiskott-Aldrich Syndrome Proteins in the Nucleus: aWASH With Possibilities. Nucleus (Austin Tex) (2015) 6(5):349–59. doi: 10.1080/19491034.2015.1086051
66. Bieling P, Hansen SD, Akin O, Li TD, Hayden CC, Fletcher DA, et al. WH2 and Proline-Rich Domains of WASP-Family Proteins Collaborate to Accelerate Actin Filament Elongation. EMBO J (2018) 37(1):102–21. doi: 10.15252/embj.201797039
67. Imai K, Nonoyama S, Miki H, Morio T, Fukami K, Zhu Q, et al. The Pleckstrin Homology Domain of the Wiskott-Aldrich Syndrome Protein is Involved in the Organization of Actin Cytoskeleton. Clin Immunol (Orlando Fla) (1999) 92(2):128–37. doi: 10.1006/clim.1999.4746
68. Fried S, Matalon O, Noy E, Barda-Saad M. WIP: More Than a WASp-Interacting Protein. J Leukocyte Biol (2014) 96(5):713–27. doi: 10.1189/jlb.2RU0314-162R
69. Sun X, Wei Y, Lee PP, Ren B, Liu C. The Role of WASp in T Cells and B Cells. Cell Immunol (2019) 341:103919. doi: 10.1016/j.cellimm.2019.04.007
70. Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and Activation Mechanisms of the Wiskott-Aldrich Syndrome Protein. Nature (2000) 404(6774):151–8. doi: 10.1038/35004513
71. Miki H, Miura K, Takenawa T. N-WASP, a Novel Actin-Depolymerizing Protein, Regulates the Cortical Cytoskeletal Rearrangement in a PIP2-Dependent Manner Downstream of Tyrosine Kinases. EMBO J (1996) 15(19):5326–35. doi: 10.1002/j.1460-2075.1996.tb00917.x
72. Westerberg LS, Dahlberg C, Baptista M, Moran CJ, Detre C, Keszei M, et al. Wiskott-Aldrich Syndrome Protein (WASP) and N-WASP Are Critical for Peripheral B-Cell Development and Function. Blood (2012) 119(17):3966–74. doi: 10.1182/blood-2010-09-308197
73. Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two Tandem Verprolin Homology Domains Are Necessary for a Strong Activation of Arp2/3 Complex-Induced Actin Polymerization and Induction of Microspike Formation by N-WASP. Proc Natl Acad Sci USA (2000) 97(23):12631–6. doi: 10.1073/pnas.190351397
74. Klein C, Nguyen D, Liu CH, Mizoguchi A, Bhan AK, Miki H, et al. Gene Therapy for Wiskott-Aldrich Syndrome: Rescue of T-Cell Signaling and Amelioration of Colitis Upon Transplantation of Retrovirally Transduced Hematopoietic Stem Cells in Mice. Blood (2003) 101(6):2159–66. doi: 10.1182/blood-2002-05-1423
75. Snapper SB, Takeshima F, Antón I, Liu CH, Thomas SM, Nguyen D, et al. N-WASP Deficiency Reveals Distinct Pathways for Cell Surface Projections and Microbial Actin-Based Motility. Nat Cell Biol (2001) 3(10):897–904. doi: 10.1038/ncb1001-897
76. Jain N, Thanabalu T. Molecular Difference Between WASP and N-WASP Critical for Chemotaxis of T-Cells Towards SDF-1α. Sci Rep (2015) 5:15031. doi: 10.1038/srep15031
77. Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The Interaction Between N-WASP and the Arp2/3 Complex Links Cdc42-Dependent Signals to Actin Assembly. Cell (1999) 97(2):221–31. doi: 10.1016/s0092-8674(00)80732-1
78. Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of Multiple Signals Through Cooperative Regulation of the N-WASP-Arp2/3 Complex. Science (2000) 290(5492):801–6. doi: 10.1126/science.290.5492.801
79. Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, et al. Sustained Activation of N-WASP Through Phosphorylation Is Essential for Neurite Extension. Dev Cell (2002) 3(5):645–58. doi: 10.1016/s1534-5807(02)00324-6
80. Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src Phosphorylation of Cortactin Acts as a Switch On-Switch Off Mechanism That Controls its Ability to Activate N-WASP. Mol Cell Biol (2004) 24(12):5269–80. doi: 10.1128/mcb.24.12.5269-5280.2004
81. Miki H, Sasaki T, Takai Y, Takenawa T. Induction of Filopodium Formation by a WASP-Related Actin-Depolymerizing Protein N-WASP. Nature (1998) 391(6662):93–6. doi: 10.1038/34208
82. Banon-Rodriguez I, Saez de Guinoa J, Bernardini A, Ragazzini C, Fernandez E, Carrasco YR, et al. WIP Regulates Persistence of Cell Migration and Ruffle Formation in Both Mesenchymal and Amoeboid Modes of Motility. PLoS One (2013) 8(8):e70364. doi: 10.1371/journal.pone.0070364
83. Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human Subtelomeric WASH Genes Encode a New Subclass of the WASP Family. PLoS Genet (2007) 3(12):e237. doi: 10.1371/journal.pgen.0030237
84. Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash Functions Downstream of Rho and Links Linear and Branched Actin Nucleation Factors. Dev (Cambridge Engl) (2009) 136(16):2849–60. doi: 10.1242/dev.035246
85. Gomez TS, Billadeau DD. A FAM21-Containing WASH Complex Regulates Retromer-Dependent Sorting. Dev Cell (2009) 17(5):699–711. doi: 10.1016/j.devcel.2009.09.009
86. Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 Activator WASH Controls the Fission of Endosomes Through a Large Multiprotein Complex. Dev Cell (2009) 17(5):712–23. doi: 10.1016/j.devcel.2009.09.010
87. Zech T, Calaminus SD, Caswell P, Spence HJ, Carnell M, Insall RH, et al. The Arp2/3 Activator WASH Regulates α5β1-Integrin-Mediated Invasive Migration. J Cell Sci (2011) 124(Pt 22):3753–9. doi: 10.1242/jcs.080986
88. Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, et al. WASH Inhibits Autophagy Through Suppression of Beclin 1 Ubiquitination. EMBO J (2013) 32(20):2685–96. doi: 10.1038/emboj.2013.189
89. Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, et al. WASH is Required for the Differentiation Commitment of Hematopoietic Stem Cells in a C-Myc-Dependent Manner. J Exp Med (2014) 211(10):2119–34. doi: 10.1084/jem.20140169
90. Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a Cortical Complex Containing Two Unconventional Actins From Acanthamoeba by Affinity Chromatography on Profilin-Agarose. J Cell Biol (1994) 127(1):107–15. doi: 10.1083/jcb.127.1.107
91. Pollard TD, Beltzner CC. Structure and Function of the Arp2/3 Complex. Curr Opin Struct Biol (2002) 12(6):768–74. doi: 10.1016/s0959-440x(02)00396-2
92. Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, et al. Crystal Structure of Arp2/3 Complex. Science (2001) 294(5547):1679–84. doi: 10.1126/science.1066333
93. Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of Human Arp2/3 Complex Reveals Critical Roles of Individual Subunits in Complex Structure and Activity. Mol Cell (2001) 8(5):1041–52. doi: 10.1016/s1097-2765(01)00393-8
94. Abella JV, Galloni C, Pernier J, Barry DJ, Kjær S, Carlier MF, et al. Isoform Diversity in the Arp2/3 Complex Determines Actin Filament Dynamics. Nat Cell Biol (2016) 18(1):76–86. doi: 10.1038/ncb3286
95. Higgs HN, Pollard TD. Regulation of Actin Polymerization by Arp2/3 Complex and WASp/Scar Proteins. J Biol Chem (1999) 274(46):32531–4. doi: 10.1074/jbc.274.46.32531
96. Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, et al. Cortactin Promotes and Stabilizes Arp2/3-Induced Actin Filament Network Formation. Curr Biol (2001) 11(5):370–4. doi: 10.1016/s0960-9822(01)00098-7
97. Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 Complex is Bound and Activated by Two WASP Proteins. Proc Natl Acad Sci USA (2011) 108(33):E472–9. doi: 10.1073/pnas.1100236108
98. Helgeson LA, Nolen BJ. Mechanism of Synergistic Activation of Arp2/3 Complex by Cortactin and N-WASP. Elife (2013) 2:e00884. doi: 10.7554/eLife.00884
99. Shaaban M, Chowdhury S, Nolen BJ. Cryo-EM Reveals the Transition of Arp2/3 Complex From Inactive to Nucleation-Competent State. Nat Struct Mol Biol (2020) 27(11):1009–16. doi: 10.1038/s41594-020-0481-x
100. Chan C, Beltzner CC, Pollard TD. Cofilin Dissociates Arp2/3 Complex and Branches From Actin Filaments. Curr Biol CB (2009) 19(7):537–45. doi: 10.1016/j.cub.2009.02.060
101. Goroncy AK, Koshiba S, Tochio N, Tomizawa T, Sato M, Inoue M, et al. NMR Solution Structures of Actin Depolymerizing Factor Homology Domains. Protein Sci Publ Protein Soc (2009) 18(11):2384–92. doi: 10.1002/pro.248
102. Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, et al. GMF is a Cofilin Homolog That Binds Arp2/3 Complex to Stimulate Filament Debranching and Inhibit Actin Nucleation. Curr Biol CB (2010) 20(9):861–7. doi: 10.1016/j.cub.2010.03.026
103. Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, et al. Conformational Changes in the Arp2/3 Complex Leading to Actin Nucleation. Nat Struct Mol Biol (2005) 12(1):26–31. doi: 10.1038/nsmb870
104. Ydenberg CA, Padrick SB, Sweeney MO, Gandhi M, Sokolova O, Goode BL. GMF Severs Actin-Arp2/3 Complex Branch Junctions by a Cofilin-Like Mechanism. Curr Biol CB (2013) 23(12):1037–45. doi: 10.1016/j.cub.2013.04.058
105. Luan Q, Nolen BJ. Structural Basis for Regulation of Arp2/3 Complex by GMF. Nat Struct Mol Biol (2013) 20(9):1062–8. doi: 10.1038/nsmb.2628
106. Chan KT, Creed SJ, Bear JE. Unraveling the Enigma: Progress Towards Understanding the Coronin Family of Actin Regulators. Trends Cell Biol (2011) 21(8):481–8. doi: 10.1016/j.tcb.2011.04.004
107. Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, et al. Direct Regulation of Arp2/3 Complex Activity and Function by the Actin Binding Protein Coronin. J Cell Biol (2002) 159(6):993–1004. doi: 10.1083/jcb.200206113
108. Xie C, Jiang Y, Zhu Z, Huang S, Li W, Ou G. Actin Filament Debranching Regulates Cell Polarity During Cell Migration and Asymmetric Cell Division. Proc Natl Acad Sci USA (2021) 118(37):e2100805118. doi: 10.1073/pnas.2100805118
109. Sokolova OS, Chemeris A, Guo S, Alioto SL, Gandhi M, Padrick S, et al. Structural Basis of Arp2/3 Complex Inhibition by GMF, Coronin, and Arpin. J Mol Biol (2017) 429(2):237–48. doi: 10.1016/j.jmb.2016.11.030
110. Dang I, Gorelik R, Sousa-Blin C, Derivery E, Guérin C, Linkner J, et al. Inhibitory Signalling to the Arp2/3 Complex Steers Cell Migration. Nature (2013) 503(7475):281–4. doi: 10.1038/nature12611
111. Dayel MJ, Mullins RD. Activation of Arp2/3 Complex: Addition of the First Subunit of the New Filament by a WASP Protein Triggers Rapid ATP Hydrolysis on Arp2. PLoS Biol (2004) 2(4):E91. doi: 10.1371/journal.pbio.0020091
112. Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, et al. Effects of Arp2 and Arp3 Nucleotide-Binding Pocket Mutations on Arp2/3 Complex Function. J Cell Biol (2005) 168(2):315–28. doi: 10.1083/jcb.200408177
113. Martin AC, Welch MD, Drubin DG. Arp2/3 ATP Hydrolysis-Catalysed Branch Dissociation Is Critical for Endocytic Force Generation. Nat Cell Biol (2006) 8(8):826–33. doi: 10.1038/ncb1443
114. Michard C, Sperandio D, Baïlo N, Pizarro-Cerdá J, LeClaire L, Chadeau-Argaud E, et al. The Legionella Kinase LegK2 Targets the ARP2/3 Complex To Inhibit Actin Nucleation on Phagosomes and Allow Bacterial Evasion of the Late Endocytic Pathway. mBio (2015) 6(3):e00354–15. doi: 10.1128/mBio.00354-15
115. Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. P41-Arc Subunit of Human Arp2/3 Complex Is a P21-Activated Kinase-1-Interacting Substrate. EMBO Rep (2004) 5(2):154–60. doi: 10.1038/sj.embor.7400079
116. LeClaire LL 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 Complex is Necessary to Nucleate Actin Filaments. J Cell Biol (2008) 182(4):647–54. doi: 10.1083/jcb.200802145
117. Zaki M, King J, Fütterer K, Insall RH. Replacement of the Essential Dictyostelium Arp2 Gene by its Entamoeba Homologue Using Parasexual Genetics. BMC Genet (2007) 8:28. doi: 10.1186/1471-2156-8-28
118. Yae K, Keng VW, Koike M, Yusa K, Kouno M, Uno Y, et al. Sleeping Beauty Transposon-Based Phenotypic Analysis of Mice: Lack of Arpc3 Results in Defective Trophoblast Outgrowth. Mol Cell Biol (2006) 26(16):6185–96. doi: 10.1128/mcb.00018-06
119. Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, et al. Mammalian Actin-Related Protein 2/3 Complex Localizes to Regions of Lamellipodial Protrusion and Is Composed of Evolutionarily Conserved Proteins. Biochem J (1997) 328( Pt 1):105–12. doi: 10.1042/bj3280105
120. Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR. Spatial Control of Actin Polymerization During Neutrophil Chemotaxis. Nat Cell Biol (1999) 1(2):75–81. doi: 10.1038/10042
121. Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-Directed Actin Assembly: E-Cadherin Physically Associates With the Arp2/3 Complex to Direct Actin Assembly in Nascent Adhesive Contacts. Curr Biol (2002) 12(5):379–82. doi: 10.1016/s0960-9822(02)00661-9
122. Winter D, Podtelejnikov AV, Mann M, Li R. The Complex Containing Actin-Related Proteins Arp2 and Arp3 Is Required for the Motility and Integrity of Yeast Actin Patches. Curr Biol (1997) 7(7):519–29. doi: 10.1016/s0960-9822(06)00223-5
123. Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The Yeast Actin-Related Protein Arp2p Is Required for the Internalization Step of Endocytosis. Mol Biol Cell (1997) 8(7):1361–75. doi: 10.1091/mbc.8.7.1361
124. Benesch S, Polo S, Lai FP, Anderson KI, Stradal TE, Wehland J, et al. N-WASP Deficiency Impairs EGF Internalization and Actin Assembly at Clathrin-Coated Pits. J Cell Sci (2005) 118(Pt 14):3103–15. doi: 10.1242/jcs.02444
125. Matas OB, Martínez-Menárguez JA, Egea G. Association of Cdc42/N-WASP/Arp2/3 Signaling Pathway With Golgi Membranes. Traffic (2004) 5(11):838–46. doi: 10.1111/j.1600-0854.2004.00225.x
126. Chen JL, Lacomis L, Erdjument-Bromage H, Tempst P, Stamnes M. Cytosol-Derived Proteins are Sufficient for Arp2/3 Recruitment and ARF/coatomer-Dependent Actin Polymerization on Golgi Membranes. FEBS Lett (2004) 566(1-3):281–6. doi: 10.1016/j.febslet.2004.04.061
127. May RC, Caron E, Hall A, Machesky LM. Involvement of the Arp2/3 Complex in Phagocytosis Mediated by FcgammaR or CR3. Nat Cell Biol (2000) 2(4):246–8. doi: 10.1038/35008673
128. Farina F, Gaillard J, Guérin C, Couté Y, Sillibourne J, Blanchoin L, et al. The Centrosome is an Actin-Organizing Centre. Nat Cell Biol (2016) 18(1):65–75. doi: 10.1038/ncb3285
129. Farina F, Ramkumar N, Brown L, Samandar Eweis D, Anstatt J, Waring T, et al. Local Actin Nucleation Tunes Centrosomal Microtubule Nucleation During Passage Through Mitosis. EMBO J (2019) 38(11):e99843. doi: 10.15252/embj.201899843
130. Chalkia D, Nikolaidis N, Makalowski W, Klein J, Nei M. Origins and Evolution of the Formin Multigene Family That Is Involved in the Formation of Actin Filaments. Mol Biol Evol (2008) 25(12):2717–33. doi: 10.1093/molbev/msn215
131. Higgs HN, Peterson KJ. Phylogenetic Analysis of the Formin Homology 2 Domain. Mol Biol Cell (2005) 16(1):1–13. doi: 10.1091/mbc.e04-07-0565
132. Horan BG, Zerze GH, Kim YC, Vavylonis D, Mittal J. Computational Modeling Highlights the Role of the Disordered Formin Homology 1 Domain in Profilin-Actin Transfer. FEBS Lett (2018) 592(11):1804–16. doi: 10.1002/1873-3468.13088
133. Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. A Comparative Sequence Analysis Reveals a Common GBD/FH3-FH1-FH2-DAD Architecture in Formins From Dictyostelium, Fungi and Metazoa. BMC Genomics (2005) 6:28. doi: 10.1186/1471-2164-6-28
134. Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and Targeting of the Fission Yeast Formin Cdc12p in Cytokinesis. Mol Biol Cell (2008) 19(5):2208–19. doi: 10.1091/mbc.e07-07-0731
135. Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of Formin-Associated Actin Filament Elongation. Mol Cell (2006) 21(4):455–66. doi: 10.1016/j.molcel.2006.01.016
136. Paul AS, Pollard TD. The Role of the FH1 Domain and Profilin in Formin-Mediated Actin-Filament Elongation and Nucleation. Curr Biol CB (2008) 18(1):9–19. doi: 10.1016/j.cub.2007.11.062
137. Lu J, Meng W, Poy F, Maiti S, Goode BL, Eck MJ. Structure of the FH2 Domain of Daam1: Implications for Formin Regulation of Actin Assembly. J Mol Biol (2007) 369(5):1258–69. doi: 10.1016/j.jmb.2007.04.002
138. Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, et al. Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture. Cell (2004) 116(5):711–23. doi: 10.1016/s0092-8674(04)00210-7
139. Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, et al. Role of Formins in Actin Assembly: Nucleation and Barbed-End Association. Sci (New York NY) (2002) 297(5581):612–5. doi: 10.1126/science.1072309
140. Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of Formin-Induced Nucleation of Actin Filaments. Biochemistry (2003) 42(2):486–96. doi: 10.1021/bi026520j
141. Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a Processive Motor That Requires Profilin to Accelerate Actin Assembly and Associated ATP Hydrolysis. Cell (2004) 119(3):419–29. doi: 10.1016/j.cell.2004.09.039
142. Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-Related Formin Ddia2 Is Required for the Formation and Maintenance of Filopodia. Nat Cell Biol (2005) 7(6):619–25. doi: 10.1038/ncb1266
143. Block J, Breitsprecher D, Kühn S, Winterhoff M, Kage F, Geffers R, et al. FMNL2 Drives Actin-Based Protrusion and Migration Downstream of Cdc42. Curr Biol CB (2012) 22(11):1005–12. doi: 10.1016/j.cub.2012.03.064
144. Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, et al. Mdia2 Induces the Actin Scaffold for the Contractile Ring and Stabilizes its Position During Cytokinesis in NIH 3T3 Cells. Mol Biol Cell (2008) 19(5):2328–38. doi: 10.1091/mbc.e07-10-1086
145. Bartolini F, Gundersen GG. Formins and Microtubules. Biochim Biophys Acta (2010) 1803(2):164–73. doi: 10.1016/j.bbamcr.2009.07.006
146. Thurston SF, Kulacz WA, Shaikh S, Lee JM, Copeland JW. The Ability to Induce Microtubule Acetylation Is a General Feature of Formin Proteins. PLoS One (2012) 7(10):e48041. doi: 10.1371/journal.pone.0048041
147. Ishizaki T, Morishima Y, Okamoto M, Furuyashiki T, Kato T, Narumiya S. Coordination of Microtubules and the Actin Cytoskeleton by the Rho Effector Mdia1. Nat Cell Biol (2001) 3(1):8–14. doi: 10.1038/35050598
148. Nishida E, Maekawa S, Sakai H. Cofilin, a Protein in Porcine Brain That Binds to Actin Filaments and Inhibits Their Interactions With Myosin and Tropomyosin. Biochemistry (1984) 23(22):5307–13. doi: 10.1021/bi00317a032
149. Bamburg JR, Harris HE, Weeds AG. Partial Purification and Characterization of an Actin Depolymerizing Factor From Brain. FEBS Lett (1980) 121(1):178–82. doi: 10.1016/0014-5793(80)81292-0
150. Hild G, Kalmár L, Kardos R, Nyitrai M, Bugyi B. The Other Side of the Coin: Functional and Structural Versatility of ADF/Cofilins. Eur J Cell Biol (2014) 93(5-6):238–51. doi: 10.1016/j.ejcb.2013.12.001
151. Lappalainen P, Kessels MM, Cope MJT, Drubin DG. The ADF Homology (ADF-H) Domain: A Highly Exploited Actin-Binding Module. Mol Biol Cell (1998) 9(8):1951–9. doi: 10.1091/mbc.9.8.1951
152. Moon A, Drubin DG. The ADF/cofilin Proteins: Stimulus-Responsive Modulators of Actin Dynamics. Mol Biol Cell (1995) 6(11):1423–31. doi: 10.1091/mbc.6.11.1423
153. Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, et al. The Three Mouse Actin-Depolymerizing Factor/Cofilins Evolved to Fulfill Cell-Type–Specific Requirements for Actin Dynamics. Mol Biol Cell (2002) 13(1):183–94. doi: 10.1091/mbc.01-07-0331
154. Ono S, Minami N, Abe H, Obinata T. Characterization of a Novel Cofilin Isoform That Is Predominantly Expressed in Mammalian Skeletal Muscle. J Biol Chem (1994) 269(21):15280–6. doi: 10.1016/S0021-9258(17)36603-6
155. Gillett GT, Fox MF, Rowe PS, Casimir CM, Povey S. Mapping of Human non-Muscle Type Cofilin (CFL1) to Chromosome 11q13 and Muscle-Type Cofilin (CFL2) to Chromosome 14. Ann Hum Genet (1996) 60(3):201–11. doi: 10.1111/j.1469-1809.1996.tb00423.x
156. Shishkin S, Eremina L, Pashintseva N, Kovalev L, Kovaleva M. Cofilin-1 and Other ADF/Cofilin Superfamily Members in Human Malignant Cells. Int J Mol Sci (2016) 18(1):10. doi: 10.3390/ijms18010010
157. Muneyuki E, Nishida E, Sutoh K, Sakai H. Purification of Cofilin, a 21,000 Molecular Weight Actin-Binding Protein, From Porcine Kidney and Identification of the Cofilin-Binding Site in the Actin Sequence. J Biochem (1985) 97(2):563–8. doi: 10.1093/oxfordjournals.jbchem.a135091
158. McGough A, Pope B, Chiu W, Weeds A. Cofilin Changes the Twist of F-Actin: Implications for Actin Filament Dynamics and Cellular Function. J Cell Biol (1997) 138(4):771–81. doi: 10.1083/jcb.138.4.771
159. Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. Structure of the Actin-Depolymerizing Factor Homology Domain in Complex With Actin. J Cell Biol (2008) 182(1):51–9. doi: 10.1083/jcb.200803100
160. Andrianantoandro E, Pollard TD. Mechanism of Actin Filament Turnover by Severing and Nucleation at Different Concentrations of ADF/Cofilin. Mol Cell (2006) 24(1):13–23. doi: 10.1016/j.molcel.2006.08.006
161. Nadkarni AV, Brieher WM. Aip1 Destabilizes Cofilin-Saturated Actin Filaments by Severing and Accelerating Monomer Dissociation From Ends. Curr Biol (2014) 24(23):2749–57. doi: 10.1016/j.cub.2014.09.048
162. Wong DY, Sept D. The Interaction of Cofilin With the Actin Filament. J Mol Biol (2011) 413(1):97–105. doi: 10.1016/j.jmb.2011.08.039
163. Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, et al. Regulation of Actin Dynamics Through Phosphorylation of Cofilin by LIM-Kinase. Nature (1998) 393(6687):805–9. doi: 10.1038/31729
164. Yonezawa N, Nishida E, Sakai H. pH Control of Actin Polymerization by Cofilin. J Biol Chem (1985) 260(27):14410–2. doi: 10.1016/S0021-9258(17)38580-0
165. Hawkins M, Pope B, Maciver S, Weeds A. Human Actin Depolymerizing Factor Mediates a pH-Sensitive Destruction of Actin Filaments. Biochemistry (1993) 32(38):9985–93. doi: 10.1021/bi00089a014
166. Hayden S, Miller P, Brauweiler A, Bamburg J. Analysis of the Interactions of Actin Depolymerizing Factor With G-And F-Actin. Biochemistry (1993) 32(38):9994–10004. doi: 10.1021/bi00089a015
167. Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the Interactions of Cofilin, Destrin, and Deoxyribonuclease I With Actin by Phosphoinositides. J Biol Chem (1990) 265(15):8382–6. doi: 10.1016/S0021-9258(19)38897-0
168. Yeoh S, Pope B, Mannherz HG, Weeds A. Determining the Differences in Actin Binding by Human ADF and Cofilin. J Mol Biol (2002) 315(4):911–25. doi: 10.1006/jmbi.2001.5280
169. Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, et al. Cofilin Is a pH Sensor for Actin Free Barbed End Formation: Role of Phosphoinositide Binding. J Cell Biol (2008) 183(5):865–79. doi: 10.1083/jcb.200804161
170. de Hostos EL, Bradtke B, Lottspeich F, Guggenheim R, Gerisch G. Coronin, an Actin Binding Protein of Dictyostelium Discoideum Localized to Cell Surface Projections, has Sequence Similarities to G Protein Beta Subunits. EMBO J (1991) 10(13):4097–104. doi: 10.1002/j.1460-2075.1991.tb04986.x
171. Pieters J. Coronin 1 in Innate Immunity. Subcell Biochem (2008) 48:116–23. doi: 10.1007/978-0-387-09595-0_11
172. Uetrecht AC, Bear JE. Coronins: The Return of the Crown. Trends Cell Biol (2006) 16(8):421–6. doi: 10.1016/j.tcb.2006.06.002
173. Cai L, Holoweckyj N, Schaller MD, Bear JE. Phosphorylation of Coronin 1B by Protein Kinase C Regulates Interaction With Arp2/3 and Cell Motility. J Biol Chem (2005) 280(36):31913–23. doi: 10.1074/jbc.M504146200
174. Gatfield J, Albrecht I, Zanolari B, Steinmetz MO, Pieters J. Association of the Leukocyte Plasma Membrane With the Actin Cytoskeleton Through Coiled Coil-Mediated Trimeric Coronin 1 Molecules. Mol Biol Cell (2005) 16(6):2786–98. doi: 10.1091/mbc.e05-01-0042
175. Nal B, Carroll P, Mohr E, Verthuy C, Da Silva MI, Gayet O, et al. Coronin-1 Expression in T Lymphocytes: Insights Into Protein Function During T Cell Development and Activation. Int Immunol (2004) 16(2):231–40. doi: 10.1093/intimm/dxh022
176. Rybakin V, Stumpf M, Schulze A, Majoul IV, Noegel AA, Hasse A. Coronin 7, the Mammalian POD-1 Homologue, Localizes to the Golgi Apparatus. FEBS Lett (2004) 573(1-3):161–7. doi: 10.1016/j.febslet.2004.07.066
177. Rybakin V, Clemen CS. Coronin Proteins as Multifunctional Regulators of the Cytoskeleton and Membrane Trafficking. Bioessays (2005) 27(6):625–32. doi: 10.1002/bies.20235
178. Oku T, Itoh S, Okano M, Suzuki A, Suzuki K, Nakajin S, et al. Two Regions Responsible for the Actin Binding of P57, a Mammalian Coronin Family Actin-Binding Protein. Biol Pharm Bull (2003) 26(4):409–16. doi: 10.1248/bpb.26.409
179. Tur-Gracia S, Martinez-Quiles N. Emerging Functions of Cytoskeletal Proteins in Immune Diseases. J Cell Sci (2021) 134(3):jcs253534. doi: 10.1242/jcs.253534
180. Liu SL, Needham KM, May JR, Nolen BJ. Mechanism of a Concentration-Dependent Switch Between Activation and Inhibition of Arp2/3 Complex by Coronin. J Biol Chem (2011) 286(19):17039–46. doi: 10.1074/jbc.M111.219964
181. Voegtli WC, Madrona AY, Wilson DK. The Structure of Aip1p, a WD Repeat Protein That Regulates Cofilin-Mediated Actin Depolymerization. J Biol Chem (2003) 278(36):34373–9. doi: 10.1074/jbc.M302773200
182. Iida K, Yahara I. Cooperation of Two Actin-Binding Proteins, Cofilin and Aip1, in Saccharomyces Cerevisiae. Genes Cells Devoted Mol Cell Mech (1999) 4(1):21–32. doi: 10.1046/j.1365-2443.1999.00235.x
183. Okada K, Ravi H, Smith EM, Goode BL. Aip1 and Cofilin Promote Rapid Turnover of Yeast Actin Patches and Cables: A Coordinated Mechanism for Severing and Capping Filaments. Mol Biol Cell (2006) 17(7):2855–68. doi: 10.1091/mbc.e06-02-0135
184. Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin Disassembly by Cofilin, Coronin, and Aip1 Occurs in Bursts and is Inhibited by Barbed-End Cappers. J Cell Biol (2008) 182(2):341–53. doi: 10.1083/jcb.200801027
185. Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid Actin Monomer-Insensitive Depolymerization of Listeria Actin Comet Tails by Cofilin, Coronin, and Aip1. J Cell Biol (2006) 175(2):315–24. doi: 10.1083/jcb.200603149
186. Li J, Brieher WM, Scimone ML, Kang SJ, Zhu H, Yin H, et al. Caspase-11 Regulates Cell Migration by Promoting Aip1-Cofilin-Mediated Actin Depolymerization. Nat Cell Biol (2007) 9(3):276–86. doi: 10.1038/ncb1541
187. Kumar BV, Connors TJ, Farber DL. Human T Cell Development, Localization, and Function Throughout Life. Immunity (2018) 48(2):202–13. doi: 10.1016/j.immuni.2018.01.007
188. Kokkinopoulos D, Perez S, Papamichail M. Thymosin Beta 4 Induced Phenotypic Changes in Molt-4 Leukemic Cell Line. Blut (1985) 50(6):341–8. doi: 10.1007/bf00320927
189. Menotti M, Ambrogio C, Cheong TC, Pighi C, Mota I, Cassel SH, et al. Wiskott-Aldrich Syndrome Protein (WASP) is a Tumor Suppressor in T Cell Lymphoma. Nat Med (2019) 25(1):130–40. doi: 10.1038/s41591-018-0262-9
190. Liu Q, Zhang L, Shu Z, Yu T, Zhou L, Song W, et al. WASp Is Essential for Effector-To-Memory Conversion and for Maintenance of CD8(+)T Cell Memory. Front Immunol (2019) 10:2262. doi: 10.3389/fimmu.2019.02262
191. Cotta-de-Almeida V, Westerberg L, Maillard MH, Onaldi D, Wachtel H, Meelu P, et al. Wiskott Aldrich Syndrome Protein (WASP) and N-WASP Are Critical for T Cell Development. Proc Natl Acad Sci USA (2007) 104(39):15424–9. doi: 10.1073/pnas.0706881104
192. Piotrowski JT, Gomez TS, Schoon RA, Mangalam AK, Billadeau DD. WASH Knockout T Cells Demonstrate Defective Receptor Trafficking, Proliferation, and Effector Function. Mol Cell Biol (2013) 33(5):958–73. doi: 10.1128/mcb.01288-12
193. Jayachandran R, Pieters J. Regulation of Immune Cell Homeostasis and Function by Coronin 1. Int Immunopharmacol (2015) 28(2):825–8. doi: 10.1016/j.intimp.2015.03.045
194. Foger N, Rangell L, Danilenko DM, Chan AC. Requirement for Coronin 1 in T Lymphocyte Trafficking and Cellular Homeostasis. Science (2006) 313(5788):839–42. doi: 10.1126/science.1130563
195. Mueller P, Massner J, Jayachandran R, Combaluzier B, Albrecht I, Gatfield J, et al. Regulation of T Cell Survival Through Coronin-1-Mediated Generation of Inositol-1,4,5-Trisphosphate and Calcium Mobilization After T Cell Receptor Triggering. Nat Immunol (2008) 9(4):424–31. doi: 10.1038/ni1570
196. Mugnier B, Nal B, Verthuy C, Boyer C, Lam D, Chasson L, et al. Coronin-1A Links Cytoskeleton Dynamics to TCR Alpha Beta-Induced Cell Signaling. PLoS One (2008) 3(10):e3467. doi: 10.1371/journal.pone.0003467
197. Standing AS, Malinova D, Hong Y, Record J, Moulding D, Blundell MP, et al. Autoinflammatory Periodic Fever, Immunodeficiency, and Thrombocytopenia (PFIT) Caused by Mutation in Actin-Regulatory Gene WDR1. J Exp Med (2017) 214(1):59–71. doi: 10.1084/jem.20161228
198. Kile BT, Panopoulos AD, Stirzaker RA, Hacking DF, Tahtamouni LH, Willson TA, et al. Mutations in the Cofilin Partner Aip1/Wdr1 Cause Autoinflammatory Disease and Macrothrombocytopenia. Blood (2007) 110(7):2371–80. doi: 10.1182/blood-2006-10-055087
199. Pfajfer L, Mair NK, Jiménez-Heredia R, Genel F, Gulez N, Ardeniz Ö, et al. Mutations Affecting the Actin Regulator WD Repeat-Containing Protein 1 Lead to Aberrant Lymphoid Immunity. J Allergy Clin Immunol (2018) 142(5):1589–604.e11. doi: 10.1016/j.jaci.2018.04.023
200. Chimen M, Apta BH, McGettrick HM. Introduction: T Cell Trafficking in Inflammation and Immunity. Methods Mol Biol (Clifton NJ) (2017) 1591:73–84. doi: 10.1007/978-1-4939-6931-9_6
201. Hauzenberger D, Klominek J, Holgersson J, Bergström SE, Sundqvist KG. Triggering of Motile Behavior in T Lymphocytes via Cross-Linking of Alpha 4 Beta 1 and Alpha L Beta 2. J Immunol (Baltimore Md 1950) (1997) 158(1):76–84.
202. Maizels Y, Oberman F, Miloslavski R, Ginzach N, Berman M, Yisraeli JK. Localization of Cofilin mRNA to the Leading Edge of Migrating Cells Promotes Directed Cell Migration. J Cell Sci (2015) 128(10):1922–33. doi: 10.1242/jcs.163972
203. Nishita M, Tomizawa C, Yamamoto M, Horita Y, Ohashi K, Mizuno K. Spatial and Temporal Regulation of Cofilin Activity by LIM Kinase and Slingshot is Critical for Directional Cell Migration. J Cell Biol (2005) 171(2):349–59. doi: 10.1083/jcb.200504029
204. Klemke M, Kramer E, Konstandin MH, Wabnitz GH, Samstag Y. An MEK-Cofilin Signalling Module Controls Migration of Human T Cells in 3D But Not 2D Environments. EMBO J (2010) 29(17):2915–29. doi: 10.1038/emboj.2010.153
205. Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of Cofilin in Epidermal Growth Factor–Stimulated Actin Polymerization and Lamellipod Protrusion. J Cell Biol (2000) 148(3):531–42. doi: 10.1083/jcb.148.3.531
206. Zebda N, Bernard O, Bailly M, Welti S, Lawrence DS, Condeelis JS. Phosphorylation of ADF/cofilin Abolishes EGF-Induced Actin Nucleation at the Leading Edge and Subsequent Lamellipod Extension. J Cell Biol (2000) 151(5):1119–28. doi: 10.1083/jcb.151.5.1119
207. Nicholson-Dykstra SM, Higgs HN. Arp2 Depletion Inhibits Sheet-Like Protrusions But Not Linear Protrusions of Fibroblasts and Lymphocytes. Cell Motil Cytoskeleton (2008) 65(11):904–22. doi: 10.1002/cm.20312
208. Haddad E, Zugaza JL, Louache F, Debili N, Crouin C, Schwarz K, et al. The Interaction Between Cdc42 and WASP Is Required for SDF-1-Induced T-Lymphocyte Chemotaxis. Blood (2001) 97(1):33–8. doi: 10.1182/blood.v97.1.33
209. Samstag Y, Eibert SM, Klemke M, Wabnitz GH. Actin Cytoskeletal Dynamics in T Lymphocyte Activation and Migration. J Leukoc Biol (2003) 73(1):30–48. doi: 10.1189/jlb.0602272
210. Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, et al. Migration of Zebrafish Primordial Germ Cells: A Role for Myosin Contraction and Cytoplasmic Flow. Dev Cell (2006) 11(5):613–27. doi: 10.1016/j.devcel.2006.09.023
211. Obeidy P, Ju LA, Oehlers SH, Zulkhernain NS, Lee Q, Galeano Niño JL, et al. Partial Loss of Actin Nucleator Actin-Related Protein 2/3 Activity Triggers Blebbing in Primary T Lymphocytes. Immunol Cell Biol (2020) 98(2):93–113. doi: 10.1111/imcb.12304
212. Moreau HD, Lemaître F, Garrod KR, Garcia Z, Lennon-Duménil AM, Bousso P. Signal Strength Regulates Antigen-Mediated T-Cell Deceleration by Distinct Mechanisms to Promote Local Exploration or Arrest. Proc Natl Acad Sci USA (2015) 112(39):12151–6. doi: 10.1073/pnas.1506654112
213. Koduru S, Kumar L, Massaad MJ, Ramesh N, Le Bras S, Ozcan E, et al. Cdc42 Interacting Protein 4 (CIP4) Is Essential for Integrin-Dependent T-Cell Trafficking. Proc Natl Acad Sci USA (2010) 107(37):16252–6. doi: 10.1073/pnas.1002747107
214. Serrano-Pertierra E, Cernuda-Morollón E, López-Larrea C. NKG2D- and CD28-Mediated Costimulation Regulate CD8+ T Cell Chemotaxis Through Different Mechanisms: The Role of Cdc42/N-WASp. J Leukoc Biol (2014) 95(3):487–95. doi: 10.1189/jlb.0613316
215. Gomez TS, Kumar K, Medeiros RB, Shimizu Y, Leibson PJ, Billadeau DD. Formins Regulate the Actin-Related Protein 2/3 Complex-Independent Polarization of the Centrosome to the Immunological Synapse. Immunity (2007) 26(2):177–90. doi: 10.1016/j.immuni.2007.01.008
216. Thompson SB, Sandor AM, Lui V, Chung JW, Waldman MM, Long RA, et al. Formin-Like 1 Mediates Effector T Cell Trafficking to Inflammatory Sites to Enable T Cell-Mediated Autoimmunity. eLife (2020) 9:e58046. doi: 10.7554/eLife.58046
217. Eisenmann KM, West RA, Hildebrand D, Kitchen SM, Peng J, Sigler R, et al. T Cell Responses in Mammalian Diaphanous-Related Formin Mdia1 Knock-Out Mice. J Biol Chem (2007) 282(34):25152–8. doi: 10.1074/jbc.M703243200
218. Sakata D, Taniguchi H, Yasuda S, Adachi-Morishima A, Hamazaki Y, Nakayama R, et al. Impaired T Lymphocyte Trafficking in Mice Deficient in an Actin-Nucleating Protein, Mdia1. J Exp Med (2007) 204(9):2031–8. doi: 10.1084/jem.20062647
219. Dong B, Zhang SS, Gao W, Su H, Chen J, Jin F, et al. Mammalian Diaphanous-Related Formin 1 Regulates GSK3β-Dependent Microtubule Dynamics Required for T Cell Migratory Polarization. PLoS One (2013) 8(11):e80500. doi: 10.1371/journal.pone.0080500
220. Fulton LM, Taylor NA, Coghill JM, West ML, Foger N, Bear JE, et al. Altered T-Cell Entry and Egress in the Absence of Coronin 1A Attenuates Murine Acute Graft Versus Host Disease. Eur J Immunol (2014) 44(6):1662–71. doi: 10.1002/eji.201344155
221. Kato A, Kurita S, Hayashi A, Kaji N, Ohashi K, Mizuno K. Critical Roles of Actin-Interacting Protein 1 in Cytokinesis and Chemotactic Migration of Mammalian Cells. Biochem J (2008) 414(2):261–70. doi: 10.1042/bj20071655
222. Lee KH, Dinner AR, Tu C, Campi G, Raychaudhuri S, Varma R, et al. The Immunological Synapse Balances T Cell Receptor Signaling and Degradation. Sci (New York NY) (2003) 302(5648):1218–22. doi: 10.1126/science.1086507
223. Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, et al. The Immunological Synapse: A Molecular Machine Controlling T Cell Activation. Sci (New York NY) (1999) 285(5425):221–7. doi: 10.1126/science.285.5425.221
224. Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-Dimensional Segregation of Supramolecular Activation Clusters in T Cells. Nature (1998) 395(6697):82–6. doi: 10.1038/25764
225. Freiberg BA, Kupfer H, Maslanik W, Delli J, Kappler J, Zaller DM, et al. Staging and Resetting T Cell Activation in SMACs. Nat Immunol (2002) 3(10):911–7. doi: 10.1038/ni836
226. Hammer JA, Wang JC, Saeed M, Pedrosa AT. Origin, Organization, Dynamics, and Function of Actin and Actomyosin Networks at the T Cell Immunological Synapse. Annu Rev Immunol (2019) 37:201–24. doi: 10.1146/annurev-immunol-042718-041341
227. Eibert SM, Lee K-H, Pipkorn R, Sester U, Wabnitz GH, Giese T, et al. Cofilin Peptide Homologs Interfere With Immunological Synapse Formation and T Cell Activation. Proc Natl Acad Sci (2004) 101(7):1957–62. doi: 10.1073/pnas.0308282100
228. Kuijpers TW, Tool ATJ, van der Bijl I, de Boer M, van Houdt M, de Cuyper IM, et al. Combined Immunodeficiency With Severe Inflammation and Allergy Caused by ARPC1B Deficiency. J Allergy Clin Immunol (2017) 140(1):273–7.e10. doi: 10.1016/j.jaci.2016.09.061
229. Somech R, Lev A, Lee YN, Simon AJ, Barel O, Schiby G, et al. Disruption of Thrombocyte and T Lymphocyte Development by a Mutation in ARPC1B. J Immunol (2017) 199(12):4036–45. doi: 10.4049/jimmunol.1700460
230. Brigida I, Zoccolillo M, Cicalese MP, Pfajfer L, Barzaghi F, Scala S, et al. T-Cell Defects in Patients With ARPC1B Germline Mutations Account for Combined Immunodeficiency. Blood (2018) 132(22):2362–74. doi: 10.1182/blood-2018-07-863431
231. Finetti F, Baldari CT. Compartmentalization of Signaling by Vesicular Trafficking: A Shared Building Design for the Immune Synapse and the Primary Cilium. Immunol Rev (2013) 251(1):97–112. doi: 10.1111/imr.12018
232. Soares H, Lasserre R, Alcover A. Orchestrating Cytoskeleton and Intracellular Vesicle Traffic to Build Functional Immunological Synapses. Immunol Rev (2013) 256(1):118–32. doi: 10.1111/imr.12110
233. Galgano D, Onnis A, Pappalardo E, Galvagni F, Acuto O, Baldari CT. The T Cell IFT20 Interactome Reveals New Players in Immune Synapse Assembly. J Cell Sci (2017) 130(6):1110–21. doi: 10.1242/jcs.200006
234. Ichikawa D, Mizuno M, Yamamura T, Miyake S. GRAIL (Gene Related to Anergy in Lymphocytes) Regulates Cytoskeletal Reorganization Through Ubiquitination and Degradation of Arp2/3 Subunit 5 and Coronin 1A. J Biol Chem (2011) 286(50):43465–74. doi: 10.1074/jbc.M111.222711
235. Thumkeo D, Katsura Y, Nishimura Y, Kanchanawong P, Tohyama K, Ishizaki T, et al. Mdia1/3-Dependent Actin Polymerization Spatiotemporally Controls LAT Phosphorylation by Zap70 at the Immune Synapse. Sci Adv (2020) 6(1):eaay2432. doi: 10.1126/sciadv.aay2432
236. Murugesan S, Hong J, Yi J, Li D, Beach JR, Shao L, et al. Formin-Generated Actomyosin Arcs Propel T Cell Receptor Microcluster Movement at the Immune Synapse. J Cell Biol (2016) 215(3):383–99. doi: 10.1083/jcb.201603080
237. Gaud G, Lesourne R, Love PE. Regulatory Mechanisms in T Cell Receptor Signalling. Nat Rev Immunol (2018) 18(8):485–97. doi: 10.1038/s41577-018-0020-8
238. Courtney AH, Lo WL, Weiss A. TCR Signaling: Mechanisms of Initiation and Propagation. Trends Biochem Sci (2018) 43(2):108–23. doi: 10.1016/j.tibs.2017.11.008
239. Smith-Garvin JE, Koretzky GA, Jordan MS. T Cell Activation. Annu Rev Immunol (2009) 27:591–619. doi: 10.1146/annurev.immunol.021908.132706
240. Kumari S, Depoil D, Martinelli R, Judokusumo E, Carmona G, Gertler FB, et al. Actin Foci Facilitate Activation of the Phospholipase C-γ in Primary T Lymphocytes via the WASP Pathway. Elife (2015) 4:e58046. doi: 10.7554/eLife.04953
241. Zhang Y, Shen H, Liu H, Feng H, Liu Y, Zhu X, et al. Arp2/3 Complex Controls T Cell Homeostasis by Maintaining Surface TCR Levels via Regulating TCR(+) Endosome Trafficking. Sci Rep (2017) 7(1):8952. doi: 10.1038/s41598-017-08357-4
242. Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, et al. ZAP-70: An Essential Kinase in T-Cell Signaling. Cold Spring Harb Perspect Biol (2010) 2(5):a002279. doi: 10.1101/cshperspect.a002279
243. Krause M, Sechi AS, Konradt M, Monner D, Gertler FB, Wehland J. Fyn-Binding Protein (Fyb)/SLP-76-Associated Protein (SLAP), Ena/vasodilator-Stimulated Phosphoprotein (VASP) Proteins and the Arp2/3 Complex Link T Cell Receptor (TCR) Signaling to the Actin Cytoskeleton. J Cell Biol (2000) 149(1):181–94. doi: 10.1083/jcb.149.1.181
244. Zeng R, Cannon JL, Abraham RT, Way M, Billadeau DD, Bubeck-Wardenberg J, et al. SLP-76 Coordinates Nck-Dependent Wiskott-Aldrich Syndrome Protein Recruitment With Vav-1/Cdc42-Dependent Wiskott-Aldrich Syndrome Protein Activation at the T Cell-APC Contact Site. J Immunol (2003) 171(3):1360–8. doi: 10.4049/jimmunol.171.3.1360
245. Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP Activation by CDC42 and Phosphatidylinositol 4, 5-Bisphosphate. J Cell Biol (2000) 150(6):1299–310. doi: 10.1083/jcb.150.6.1299
246. Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. Fyn and PTP-PEST-Mediated Regulation of Wiskott-Aldrich Syndrome Protein (WASp) Tyrosine Phosphorylation is Required for Coupling T Cell Antigen Receptor Engagement to WASp Effector Function and T Cell Activation. J Exp Med (2004) 199(1):99–112. doi: 10.1084/jem.20030976
247. Tsopoulidis N, Kaw S, Laketa V, Kutscheidt S, Baarlink C, Stolp B, et al. T Cell Receptor-Triggered Nuclear Actin Network Formation Drives CD4(+) T Cell Effector Functions. Sci Immunol (2019) 4(31):eaav1987. doi: 10.1126/sciimmunol.aav1987
248. Billadeau DD, Nolz JC, Gomez TS. Regulation of T-Cell Activation by the Cytoskeleton. Nat Rev Immunol (2007) 7(2):131–43. doi: 10.1038/nri2021
249. Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, et al. Sra-1 and Nap1 Link Rac to Actin Assembly Driving Lamellipodia Formation. EMBO J (2004) 23(4):749–59. doi: 10.1038/sj.emboj.7600084
250. Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an Essential Intermediate Between Rac and WAVE in the Regulation of Membrane Ruffling. Nature (2000) 408(6813):732–5. doi: 10.1038/35047107
251. Nolz JC, Gomez TS, Zhu P, Li S, Medeiros RB, Shimizu Y, et al. The WAVE2 Complex Regulates Actin Cytoskeletal Reorganization and CRAC-Mediated Calcium Entry During T Cell Activation. Curr Biol (2006) 16(1):24–34. doi: 10.1016/j.cub.2005.11.036
252. Zipfel PA, Bunnell SC, Witherow DS, Gu JJ, Chislock EM, Ring C, et al. Role for the Abi/wave Protein Complex in T Cell Receptor-Mediated Proliferation and Cytoskeletal Remodeling. Curr Biol (2006) 16(1):35–46. doi: 10.1016/j.cub.2005.12.024
253. Uruno T, Zhang P, Liu J, Hao JJ, Zhan X. Haematopoietic Lineage Cell-Specific Protein 1 (HS1) Promotes Actin-Related Protein (Arp) 2/3 Complex-Mediated Actin Polymerization. Biochem J (2003) 371(Pt 2):485–93. doi: 10.1042/bj20021791
254. Uruno T, Liu J, Zhang P, Fan Y, Egile C, Li R, et al. Activation of Arp2/3 Complex-Mediated Actin Polymerization by Cortactin. Nat Cell Biol (2001) 3(3):259–66. doi: 10.1038/35060051
255. Gomez TS, McCarney SD, Carrizosa E, Labno CM, Comiskey EO, Nolz JC, et al. HS1 Functions as an Essential Actin-Regulatory Adaptor Protein at the Immune Synapse. Immunity (2006) 24(6):741–52. doi: 10.1016/j.immuni.2006.03.022
256. Tabdanov E, Gondarenko S, Kumari S, Liapis A, Dustin ML, Sheetz MP, et al. Micropatterning of TCR and LFA-1 Ligands Reveals Complementary Effects on Cytoskeleton Mechanics in T Cells. Integr Biol Quantitative Biosci Nano Macro (2015) 7(10):1272–84. doi: 10.1039/c5ib00032g
257. Han Y, Yu G, Sarioglu H, Caballero-Martinez A, Schlott F, Ueffing M, et al. Proteomic Investigation of the Interactome of FMNL1 in Hematopoietic Cells Unveils a Role in Calcium-Dependent Membrane Plasticity. J Proteomics (2013) 78:72–82. doi: 10.1016/j.jprot.2012.11.015
258. Andrés-Delgado L, Antón OM, Madrid R, Byrne JA, Alonso MA. Formin INF2 Regulates MAL-Mediated Transport of Lck to the Plasma Membrane of Human T Lymphocytes. Blood (2010) 116(26):5919–29. doi: 10.1182/blood-2010-08-300665
259. Jayachandran R, Pieters J. Induction of Allograft Tolerance While Maintaining Immunity Against Microbial Pathogens: Does Coronin 1 Hold a Key? Transplantation (2020) 104(7):1350–7. doi: 10.1097/TP.0000000000003101
260. Gorbatyuk VY, Nosworthy NJ, Robson SA, Bains NP, Maciejewski MW, Dos Remedios CG, et al. Mapping the Phosphoinositide-Binding Site on Chick Cofilin Explains How PIP2 Regulates the Cofilin-Actin Interaction. Mol Cell (2006) 24(4):511–22. doi: 10.1016/j.molcel.2006.10.007
261. Michel F, Attal-Bonnefoy G, Mangino G, Mise-Omata S, Acuto O. CD28 as a Molecular Amplifier Extending TCR Ligation and Signaling Capabilities. Immunity (2001) 15(6):935–45. doi: 10.1016/S1074-7613(01)00244-8
262. Van Rheenen J, Song X, Van Roosmalen W, Cammer M, Chen X, DesMarais V, et al. EGF-Induced PIP2 Hydrolysis Releases and Activates Cofilin Locally in Carcinoma Cells. J Cell Biol (2007) 179(6):1247–59. doi: 10.1083/jcb.200706206
263. Samstag Y, Eckerskorn C, Wesselborg S, Henning S, Wallich R, Meuer SC. Costimulatory Signals for Human T-Cell Activation Induce Nuclear Translocation of Pp19/Cofilin. Proc Natl Acad Sci (1994) 91(10):4494–8. doi: 10.1073/pnas.91.10.4494
264. Samstag Y, Henning SW, Bader A, Meuer SC. Dephosphorylation of Pp19: A Common Second Signal for Human T Cell Activation Mediated Through Different Accessory Molecules. Int Immunol (1992) 4(11):1255–62. doi: 10.1093/intimm/4.11.1255
265. Samstag Y, Nebl G. Ras Initiates Phosphatidyl-Inositol-3-Kinase (PI3K)/PKB Mediated Signalling Pathways in Untransformed Human Peripheral Blood T Lymphocytes. Adv Enzyme Regul (2005) 45:52–62. doi: 10.1016/j.advenzreg.2005.02.005
266. Nebl G, Fischer S, Penzel R, Samstag Y. Dephosphorylation of Cofilin is Regulated Through Ras and Requires the Combined Activities of the Ras-Effectors MEK and PI3K. Cell Signal (2004) 16(2):235–43. doi: 10.1016/S0898-6568(03)00133-5
267. Ambach A, Saunus J, Konstandin M, Wesselborg S, Meuer SC, Samstag Y. The Serine Phosphatases PP1 and PP2A Associate With and Activate the Actin-Binding Protein Cofilin in Human T Lymphocytes. Eur J Immunol (2000) 30(12):3422–31. doi: 10.1002/1521-4141(2000012)30:12<3422::AID-IMMU3422>3.0.CO;2-J
268. Wabnitz GH, Nebl G, Klemke M, Schröder AJ, Samstag Y. Phosphatidylinositol 3-Kinase Functions as a Ras Effector in the Signaling Cascade That Regulates Dephosphorylation of the Actin-Remodeling Protein Cofilin After Costimulation of Untransformed Human T Lymphocytes. J Immunol (2006) 176(3):1668–74. doi: 10.4049/jimmunol.176.3.1668
269. Iida K, Matsumoto S, Yahara I. The KKRKK Sequence is Involved in Heat Shock-Induced Nuclear Translocation of the 18-kDa Actin-Binding Protein, Cofilin. Cell Struct Funct (1992) 17(1):39–46. doi: 10.1247/csf.17.39
270. Rungger D, Rungger-Brändle E, Chaponnier C, Gabbiani G. Intranuclear Injection of Anti-Actin Antibodies Into Xenopus Oocytes Blocks Chromosome Condensation. Nature (1979) 282(5736):320–1. doi: 10.1038/282320a0
271. Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of Actin-Binding Proteins and Actin Antibodies Demonstrates Involvement of Nuclear Actin in Transcription of Lampbrush Chromosomes. Cell (1984) 39(1):111–22. doi: 10.1016/0092-8674(84)90196-X
272. Wabnitz GH, Kirchgessner H, Jahraus B, Umansky L, Shenolikar S, Samstag Y. Protein Phosphatase 1α and Cofilin Regulate Nuclear Translocation of NF-κb and Promote Expression of the Anti-Inflammatory Cytokine Interleukin-10 by T Cells. Mol Cell Biol (2018) 38(22):e00041-18. doi: 10.1128/MCB.00041-18
273. Klemke M, Wabnitz GH, Funke F, Funk B, Kirchgessner H, Samstag Y. Oxidation of Cofilin Mediates T Cell Hyporesponsiveness Under Oxidative Stress Conditions. Immunity (2008) 29(3):404–13. doi: 10.1016/j.immuni.2008.06.016
274. Wabnitz G, Goursot C, Jahraus B, Kirchgessner H, Hellwig A, Klemke M, et al. Mitochondrial Translocation of Oxidized Cofilin Induces Caspase-Independent Necrotic-Like Programmed Cell Death of T Cells. Cell Death Dis (2010) 1(7):e58–e. doi: 10.1038/cddis.2010.36
275. Schulte B, John I, Simon B, Brockmann C, Oelmeier SA, Jahraus B, et al. A Reducing Milieu Renders Cofilin Insensitive to Phosphatidylinositol 4, 5-Bisphosphate (PIP2) Inhibition. J Biol Chem (2013) 288(41):29430–9. doi: 10.1074/jbc.M113.479766
276. Takeuchi A, Saito T. CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Front Immunol (2017) 8:194. doi: 10.3389/fimmu.2017.00194
277. Thiery J, Lieberman J. Perforin: A Key Pore-Forming Protein for Immune Control of Viruses and Cancer. Sub-Cell Biochem (2014) 80:197–220. doi: 10.1007/978-94-017-8881-6_10
278. Stinchcombe JC, Griffiths GM. Secretory Mechanisms in Cell-Mediated Cytotoxicity. Annu Rev Cell Dev Biol (2007) 23:495–517. doi: 10.1146/annurev.cellbio.23.090506.123521
279. Martínez-Lostao L, Anel A, Pardo J. How Do Cytotoxic Lymphocytes Kill Cancer Cells? Clin Cancer Res Off J Am Assoc Cancer Res (2015) 21(22):5047–56. doi: 10.1158/1078-0432.Ccr-15-0685
280. Tamzalit F, Wang MS, Jin W, Tello-Lafoz M, Boyko V, Heddleston JM, et al. Interfacial Actin Protrusions Mechanically Enhance Killing by Cytotoxic T Cells. Sci Immunol (2019) 4(33):eaav5445. doi: 10.1126/sciimmunol.aav5445
281. Randzavola LO, Strege K, Juzans M, Asano Y, Stinchcombe JC, Gawden-Bone CM, et al. Loss of ARPC1B Impairs Cytotoxic T Lymphocyte Maintenance and Cytolytic Activity. J Clin Invest (2019) 129(12):5600–14. doi: 10.1172/jci129388
282. Roper SI, Wasim L, Malinova D, Way M, Cox S, Tolar P. B Cells Extract Antigens at Arp2/3-Generated Actin Foci Interspersed With Linear Filaments. eLife (2019) 8:e48093. doi: 10.7554/eLife.48093
283. Gill MB, Roecklein-Canfield J, Sage DR, Zambela-Soediono M, Longtine N, Uknis M, et al. EBV Attachment Stimulates FHOS/FHOD1 Redistribution and Co-Aggregation With CD21: Formin Interactions With the Cytoplasmic Domain of Human CD21. J Cell Sci (2004) 117(Pt 13):2709–20. doi: 10.1242/jcs.01113
284. Bolger-Munro M, Choi K, Scurll JM, Abraham L, Chappell RS, Sheen D, et al. Arp2/3 Complex-Driven Spatial Patterning of the BCR Enhances Immune Synapse Formation, BCR Signaling and B Cell Activation. Elife (2019) 8:e44574. doi: 10.7554/eLife.44574
285. Rey-Suarez I, Wheatley BA, Koo P, Bhanja A, Shu Z, Mochrie S, et al. WASP Family Proteins Regulate the Mobility of the B Cell Receptor During Signaling Activation. Nat Commun (2020) 11(1):439. doi: 10.1038/s41467-020-14335-8
286. Liu C, Bai X, Wu J, Sharma S, Upadhyaya A, Dahlberg CI, et al. N-Wasp is Essential for the Negative Regulation of B Cell Receptor Signaling. PLoS Biol (2013) 11(11):e1001704. doi: 10.1371/journal.pbio.1001704
287. Maus M, Medgyesi D, Kiss E, Schneider AE, Enyedi Á, Szilágyi N, et al. B Cell Receptor-Induced Ca2+ Mobilization Mediates F-Actin Rearrangements and is Indispensable for Adhesion and Spreading of B Lymphocytes. J Leukocyte Biol (2013) 93(4):537–47. doi: 10.1189/jlb.0312169
288. Combaluzier B, Mueller P, Massner J, Finke D, Pieters J. Coronin 1 is Essential for IgM-Mediated Ca2+ Mobilization in B Cells But Dispensable for the Generation of Immune Responses In Vivo. J Immunol (2009) 182(4):1954–61. doi: 10.4049/jimmunol.0801811
289. Freeman SA, Lei V, Dang-Lawson M, Mizuno K, Roskelley CD, Gold MR. Cofilin-Mediated F-Actin Severing is Regulated by the Rap GTPase and Controls the Cytoskeletal Dynamics That Drive Lymphocyte Spreading and BCR Microcluster Formation. J Immunol (2011) 187(11):5887–900. doi: 10.4049/jimmunol.1102233
290. Wang JC, Lee JY-J, Christian S, Dang-Lawson M, Pritchard C, Freeman SA, et al. The Rap1–Cofilin-1 Pathway Coordinates Actin Reorganization and MTOC Polarization at the B Cell Immune Synapse. J Cell Sci (2017) 130(6):1094–109. doi: 10.1242/jcs.191858
291. Freeman SA, Jaumouillé V, Choi K, Hsu BE, Wong HS, Abraham L, et al. Toll-Like Receptor Ligands Sensitize B-Cell Receptor Signalling by Reducing Actin-Dependent Spatial Confinement of the Receptor. Nat Commun (2015) 6(1):1–17. doi: 10.1038/ncomms8015
292. Bolger-Munro M, Choi K, Cheung F, Liu YT, Dang-Lawson M, Deretic N, et al. The Wdr1-LIMK-Cofilin Axis Controls B Cell Antigen Receptor-Induced Actin Remodeling and Signaling at the Immune Synapse. Front Cell Dev Biol (2021) 9:649433. doi: 10.3389/fcell.2021.649433
293. Huang L, Sun X, Yang D, Dai X, Jiang P, Bai X, et al. WASP and Mst1 Coregulate B-Cell Development and B-Cell Receptor Signaling. Blood Adv (2020) 4(3):573–85. doi: 10.1182/bloodadvances.2018027870
294. Jing Y, Kang D, Liu L, Huang H, Chen A, Yang L, et al. Dedicator of Cytokinesis Protein 2 Couples With Lymphoid Enhancer-Binding Factor 1 to Regulate Expression of CD21 and B-Cell Differentiation. J Allergy Clin Immunol (2019) 144(5):1377–90.e4. doi: 10.1016/j.jaci.2019.05.041
295. Smaniotto S, Mendes-da-Cruz DA, Carvalho-Pinto CE, Araujo LM, Dardenne M, Savino W. Combined Role of Extracellular Matrix and Chemokines on Peripheral Lymphocyte Migration in Growth Hormone Transgenic Mice. Brain Behav Immun (2010) 24(3):451–61. doi: 10.1016/j.bbi.2009.11.014
296. Rey-Barroso J, Calovi DS, Combe M, German Y, Moreau M, Canivet A, et al. Switching Between Individual and Collective Motility in B Lymphocytes is Controlled by Cell-Matrix Adhesion and Inter-Cellular Interactions. Sci Rep (2018) 8(1):5800. doi: 10.1038/s41598-018-24222-4
297. Liu C, Miller H, Orlowski G, Hang H, Upadhyaya A, Song W. Actin Reorganization is Required for the Formation of Polarized B Cell Receptor Signalosomes in Response to Both Soluble and Membrane-Associated Antigens. J Immunol (2012) 188(7):3237–46. doi: 10.4049/jimmunol.1103065
298. Dahlberg CI, Torres ML, Petersen SH, Baptista MA, Keszei M, Volpi S, et al. Deletion of WASp and N-WASp in B Cells Cripples the Germinal Center Response and Results in Production of IgM Autoantibodies. J Autoimmun (2015) 62:81–92. doi: 10.1016/j.jaut.2015.06.003
299. Volpi S, Santori E, Abernethy K, Mizui M, Dahlberg CI, Recher M, et al. N-WASP is Required for B-Cell-Mediated Autoimmunity in Wiskott-Aldrich Syndrome. Blood (2016) 127(2):216–20. doi: 10.1182/blood-2015-05-643817
300. Hao JJ, Carey GB, Zhan X. Syk-Mediated Tyrosine Phosphorylation is Required for the Association of Hematopoietic Lineage Cell-Specific Protein 1 With Lipid Rafts and B Cell Antigen Receptor Signalosome Complex. J Biol Chem (2004) 279(32):33413–20. doi: 10.1074/jbc.M313564200
301. Obino D, Farina F, Malbec O, Sáez PJ, Maurin M, Gaillard J, et al. Actin Nucleation at the Centrosome Controls Lymphocyte Polarity. Nat Commun (2016) 7:10969. doi: 10.1038/ncomms10969
302. Wen KK, Han SS, Vyas YM. Wiskott-Aldrich Syndrome Protein Senses Irradiation-Induced DNA Damage to Coordinate the Cell-Protective Golgi Dispersal Response in Human T and B Lymphocytes. J Allergy Clin Immunol (2020) 145(1):324–34. doi: 10.1016/j.jaci.2019.09.026
303. Iizuka Y, Cichocki F, Sieben A, Sforza F, Karim R, Coughlin K, et al. UNC-45A Is a Nonmuscle Myosin IIA Chaperone Required for NK Cell Cytotoxicity via Control of Lytic Granule Secretion. J Immunol (2015) 195(10):4760–70. doi: 10.4049/jimmunol.1500979
304. Mace EM, Orange JS. Lytic Immune Synapse Function Requires Filamentous Actin Deconstruction by Coronin 1a. Proc Natl Acad Sci (2014) 111(18):6708–13. doi: 10.1073/pnas.1314975111
305. Orange JS, Harris KE, Andzelm MM, Valter MM, Geha RS, Strominger JL. The Mature Activating Natural Killer Cell Immunologic Synapse is Formed in Distinct Stages. Proc Natl Acad Sci (2003) 100(24):14151–6. doi: 10.1073/pnas.1835830100
306. Butler B, Cooper JA. Distinct Roles for the Actin Nucleators Arp2/3 and Hdia1 During NK-Mediated Cytotoxicity. Curr Biol CB (2009) 19(22):1886–96. doi: 10.1016/j.cub.2009.10.029
307. Carisey AF, Mace EM, Saeed MB, Davis DM, Orange JS. Nanoscale Dynamism of Actin Enables Secretory Function in Cytolytic Cells. Curr Biol (2018) 28(4):489–502.e9. doi: 10.1016/j.cub.2017.12.044
308. Lee HR, Yoon SY, Kang HB, Park S, Kim KE, Cho YH, et al. Thymosin Beta 4 Enhances NK Cell Cytotoxicity Mediated by ICAM-1. Immunol Lett (2009) 123(1):72–6. doi: 10.1016/j.imlet.2009.02.008
309. Lee HR, Yoon SY, Song SB, Park Y, Kim TS, Kim S, et al. Interleukin-18-Mediated Interferon-Gamma Secretion Is Regulated by Thymosin Beta 4 in Human NK Cells. Immunobiology (2011) 216(10):1155–62. doi: 10.1016/j.imbio.2011.04.002
310. Gismondi A, Cifaldi L, Mazza C, Giliani S, Parolini S, Morrone S, et al. Impaired Natural and CD16-Mediated NK Cell Cytotoxicity in Patients With WAS and XLT: Ability of IL-2 to Correct NK Cell Functional Defect. Blood (2004) 104(2):436–43. doi: 10.1182/blood-2003-07-2621
311. Orange JS, Ramesh N, Remold-O'Donnell E, Sasahara Y, Koopman L, Byrne M, et al. Wiskott-Aldrich Syndrome Protein is Required for NK Cell Cytotoxicity and Colocalizes With Actin to NK Cell-Activating Immunologic Synapses. Proc Natl Acad Sci USA (2002) 99(17):11351–6. doi: 10.1073/pnas.162376099
312. Ham H, Guerrier S, Kim J, Schoon RA, Anderson EL, Hamann MJ, et al. Dedicator of Cytokinesis 8 Interacts With Talin and Wiskott-Aldrich Syndrome Protein to Regulate NK Cell Cytotoxicity. J Immunol (2013) 190(7):3661–9. doi: 10.4049/jimmunol.1202792
313. Huang L, Zhu P, Xia P, Fan Z. WASH Has a Critical Role in NK Cell Cytotoxicity Through Lck-Mediated Phosphorylation. Cell Death Dis (2016) 7(7):e2301. doi: 10.1038/cddis.2016.212
314. Duvall MG, Fuhlbrigge ME, Reilly RB, Walker KH, Kılıç A, Levy BD. Human NK Cell Cytoskeletal Dynamics and Cytotoxicity Are Regulated by LIM Kinase. J Immunol (2020) 205(3):801–10. doi: 10.4049/jimmunol.2000186
315. Liu H, Zhang T, Chen Y, Liu C, Qi W, Shao Y, et al. Proteomics Analysis Reveals Alterations of NK Cells in Patients With Severe Aplastic Anemia. Int J Lab Hematol (2020) 42(3):308–15. doi: 10.1111/ijlh.13175
316. Serrano-Pertierra E, Cernuda-Morollón E, López-Larrea C. Wiskott-Aldrich Syndrome Protein (WASp) and N-WASp Are Involved in the Regulation of NK-Cell Migration Upon NKG2D Activation. Eur J Immunol (2012) 42(8):2142–51. doi: 10.1002/eji.201142070
317. Oswald J, Büttner M, Jasinski-Bergner S, Jacobs R, Rosenstock P, Kielstein H. Leptin Affects Filopodia and Cofilin in NK-92 Cells in a Dose-and Time-Dependent Manner. Eur J Histochem: EJH (2018) 62(1):2848. doi: 10.4081/ejh.2018.2848
318. Tchang VSY, Stiess M, Siegmund K, Karrer U, Pieters J. Role for Coronin 1 in Mouse NK Cell Function. Immunobiology (2017) 222(2):291–300. doi: 10.1016/j.imbio.2016.09.011
319. Gibieža P, Petrikaitė V. The Regulation of Actin Dynamics During Cell Division and Malignancy. Am J Cancer Res (2021) 11(9):4050–69.
320. Morita T, Hayashi K. Tumor Progression Is Mediated by Thymosin-β4 Through a Tgfβ/MRTF Signaling Axis. Mol Cancer Res MCR (2018) 16(5):880–93. doi: 10.1158/1541-7786.Mcr-17-0715
321. Wirsching HG, Krishnan S, Florea AM, Frei K, Krayenbühl N, Hasenbach K, et al. Thymosin β 4 Gene Silencing Decreases Stemness and Invasiveness in Glioblastoma. Brain J Neurol (2014) 137(Pt 2):433–48. doi: 10.1093/brain/awt333
322. Kritikou JS, Oliveira MM, Record J, Saeed MB, Nigam SM, He M, et al. Constitutive Activation of WASp Leads to Abnormal Cytotoxic Cells With Increased Granzyme B and Degranulation Response to Target Cells. JCI Insight (2021) 6(6):e140273. doi: 10.1172/jci.insight.140273
323. Han SS, Wen KK, Vyas YM. Deficiency of Wiskott-Aldrich Syndrome Protein has Opposing Effect on the Pro-Oncogenic Pathway Activation in Nonmalignant Versus Malignant Lymphocytes. Oncogene (2021) 40(2):345–54. doi: 10.1038/s41388-020-01533-3
324. Catucci M, Zanoni I, Draghici E, Bosticardo M, Castiello MC, Venturini M, et al. Wiskott-Aldrich Syndrome Protein Deficiency in Natural Killer and Dendritic Cells Affects Antitumor Immunity. Eur J Immunol (2014) 44(4):1039–45. doi: 10.1002/eji.201343935
325. Kritikou JS, Dahlberg CI, Baptista MA, Wagner AK, Banerjee PP, Gwalani LA, et al. IL-2 in the Tumor Microenvironment is Necessary for Wiskott-Aldrich Syndrome Protein Deficient NK Cells to Respond to Tumors in vivo. Sci Rep (2016) 6:30636. doi: 10.1038/srep30636
326. Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich Syndrome Protein Regulates Podosomes in Primary Human Macrophages. Proc Natl Acad Sci USA (1999) 96(17):9648–53. doi: 10.1073/pnas.96.17.9648
327. Calle Y, Jones GE, Jagger C, Fuller K, Blundell MP, Chow J, et al. WASp Deficiency in Mice Results in Failure to Form Osteoclast Sealing Zones and Defects in Bone Resorption. Blood (2004) 103(9):3552–61. doi: 10.1182/blood-2003-04-1259
328. Yan P, Liu J, Zhou R, Lin C, Wu K, Yang S, et al. LASP1 Interacts With N-WASP to Activate the Arp2/3 Complex and Facilitate Colorectal Cancer Metastasis by Increasing Tumour Budding and Worsening the Pattern of Invasion. Oncogene (2020) 39(35):5743–55. doi: 10.1038/s41388-020-01397-7
329. Al Absi A, Wurzer H, Guerin C, Hoffmann C, Moreau F, Mao X, et al. Actin Cytoskeleton Remodeling Drives Breast Cancer Cell Escape From Natural Killer-Mediated Cytotoxicity. Cancer Res (2018) 78(19):5631–43. doi: 10.1158/0008-5472.Can-18-0441
330. Huang L, Lian J, Chen X, Qin G, Zheng Y, Zhang Y. WASH Overexpression Enhances Cancer Stem Cell Properties and Correlates With Poor Prognosis of Esophageal Carcinoma. Cancer Sci (2017) 108(12):2358–65. doi: 10.1111/cas.13400
331. Gardberg M, Kaipio K, Lehtinen L, Mikkonen P, Heuser VD, Talvinen K, et al. FHOD1, a Formin Upregulated in Epithelial-Mesenchymal Transition, Participates in Cancer Cell Migration and Invasion. PLoS One (2013) 8(9):e74923. doi: 10.1371/journal.pone.0074923
332. Thompson SB, Wigton EJ, Krovi SH, Chung JW, Long RA, Jacobelli J. The Formin Mdia1 Regulates Acute Lymphoblastic Leukemia Engraftment, Migration, and Progression In Vivo. Front Oncol (2018) 8:389:. doi: 10.3389/fonc.2018.00389
333. Jin J, Wang Y, Xu Y, Zhou X, Liu Y, Li X, et al. MicroRNA-144 Regulates Cancer Cell Proliferation and Cell-Cycle Transition in Acute Lymphoblastic Leukemia Through the Interaction of FMN2. J Gene Med (2017) 19(6-7):e2898. doi: 10.1002/jgm.2898
334. Schuster IG, Busch DH, Eppinger E, Kremmer E, Milosevic S, Hennard C, et al. Allorestricted T Cells With Specificity for the FMNL1-Derived Peptide PP2 Have Potent Antitumor Activity Against Hematologic and Other Malignancies. Blood (2007) 110(8):2931–9. doi: 10.1182/blood-2006-11-058750
335. Young AI, Timpson P, Gallego-Ortega D, Ormandy CJ, Oakes SR. Myeloid Cell Leukemia 1 (MCL-1), an Unexpected Modulator of Protein Kinase Signaling During Invasion. Cell Adh Migr (2018) 12(6):513–23. doi: 10.1080/19336918.2017.1393591
336. Samstag Y, Dreizler E-M, Ambach A, Sczakiel G, Meuer SC. Inhibition of Constitutive Serine Phosphatase Activity in T Lymphoma Cells Results in Phosphorylation of Pp19/Cofilin and Induces Apoptosis. J Immunol (1996) 156(11):4167–73.
337. Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, et al. Cofilin Determines the Migration Behavior and Turning Frequency of Metastatic Cancer Cells. J Cell Biol (2007) 179(4):777–91. doi: 10.1083/jcb.200707009
338. Ma B, Herzog EL, Moore M, Lee C-M, Na SH, Lee CG, et al. RIG-Like Helicase Regulation of Chitinase 3-Like 1 Axis and Pulmonary Metastasis. Sci Rep (2016) 6(1):1–13. doi: 10.1038/srep26299
339. Liu Y, Wang Z, Huang D, Wu C, Li H, Zhang X, et al. LMO2 Promotes Tumor Cell Invasion and Metastasis in Basal-Type Breast Cancer by Altering Actin Cytoskeleton Remodeling. Oncotarget (2017) 8(6):9513. doi: 10.18632/oncotarget.13434
340. Tahtamouni LH, Shaw AE, Hasan MH, Yasin SR, Bamburg JR. Non-Overlapping Activities of ADF and Cofilin-1 During the Migration of Metastatic Breast Tumor Cells. BMC Cell Biol (2013) 14(1):1–16. doi: 10.1186/1471-2121-14-45
341. Wang L-H, Xiang J, Yan M, Zhang Y, Zhao Y, Yue C-F, et al. The Mitotic Kinase Aurora-A Induces Mammary Cell Migration and Breast Cancer Metastasis by Activating the Cofilin-F-Actin Pathway. Cancer Res (2010) 70(22):9118–28. doi: 10.1158/0008-5472.CAN-10-1246
342. Otsuji M, Kimura Y, Aoe T, Okamoto Y, Saito T. Oxidative Stress by Tumor-Derived Macrophages Suppresses the Expression of CD3 ζ Chain of T-Cell Receptor Complex and Antigen-Specific T-Cell Responses. Proc Natl Acad Sci (1996) 93(23):13119–24. doi: 10.1073/pnas.93.23.13119
343. Schmielau J, Finn OJ. Activated Granulocytes and Granulocyte-Derived Hydrogen Peroxide are the Underlying Mechanism of Suppression of T-Cell Function in Advanced Cancer Patients. Cancer Res (2001) 61(12):4756–60.
344. Simard JP, Reynolds DN, Kraguljac AP, Smith GS, Mosser DD. Overexpression of HSP70 Inhibits Cofilin Phosphorylation and Promotes Lymphocyte Migration in Heat-Stressed Cells. J Cell Sci (2011) 124(14):2367–74. doi: 10.1242/jcs.081745
345. Li H, Liu X, Jiang S, Zhou X, Yao L, Di Y, et al. WD Repeat-Containing Protein 1 Maintains β-Catenin Activity to Promote Pancreatic Cancer Aggressiveness. Br J Cancer (2020) 123(6):1012–23. doi: 10.1038/s41416-020-0929-0
346. Bergmeier LA, Babaahmady K, Pido-Lopez J, Heesom KJ, Kelly CG, Lehner T. Cytoskeletal Proteins Bound to Heat-Shock Protein 70 may Elicit Resistance to Simian Immunodeficiency Virus Infection of CD4(+) T Cells. Immunology (2010) 129(4):506–15. doi: 10.1111/j.1365-2567.2009.03202.x
347. Gordon-Alonso M, Rocha-Perugini V, Alvarez S, Ursa A, Izquierdo-Useros N, Martinez-Picado J, et al. Actin-Binding Protein Drebrin Regulates HIV-1-Triggered Actin Polymerization and Viral Infection. J Biol Chem (2013) 288(39):28382–97. doi: 10.1074/jbc.M113.494906
348. He S, Fu Y, Guo J, Spear M, Yang J, Trinité B, et al. Cofilin Hyperactivation in HIV Infection and Targeting the Cofilin Pathway Using an Anti-α4β7 Integrin Antibody. Sci Adv (2019) 5(1):eaat7911. doi: 10.1126/sciadv.aat7911
349. Wu Y, Yoder A, Yu D, Wang W, Liu J, Barrett T, et al. Cofilin Activation in Peripheral CD4 T Cells of HIV-1 Infected Patients: A Pilot Study. Retrovirology (2008) 5(1):1–6. doi: 10.1186/1742-4690-5-95
350. He S, Wu Y. Relationships Between HIV-Mediated Chemokine Coreceptor Signaling, Cofilin Hyperactivation, Viral Tropism Switch and HIV-Mediated CD4 Depletion. Curr HIV Res (2019) 17(6):388–96. doi: 10.2174/1570162X17666191106112018
351. Guo J, Wang W, Yu D, Wu Y. Spinoculation Triggers Dynamic Actin and Cofilin Activity That Facilitates HIV-1 Infection of Transformed and Resting CD4 T Cells. J Virol (2011) 85(19):9824–33. doi: 10.1128/JVI.05170-11
352. Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, et al. HIV Envelope-CXCR4 Signaling Activates Cofilin to Overcome Cortical Actin Restriction in Resting CD4 T Cells. Cell (2008) 134(5):782–92. doi: 10.1016/j.cell.2008.06.036
353. Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, et al. Establishment of HIV-1 Latency in Resting CD4+ T Cells Depends on Chemokine-Induced Changes in the Actin Cytoskeleton. Proc Natl Acad Sci (2010) 107(39):16934–9. doi: 10.1073/pnas.1002894107
354. Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, Lewin SR. CCR7 Ligands CCL19 and CCL21 Increase Permissiveness of Resting Memory CD4+ T Cells to HIV-1 Infection: A Novel Model of HIV-1 Latency. Blood J Am Soc Hematol (2007) 110(13):4161–4. doi: 10.1182/blood-2007-06-097907
355. Stolp B, Reichman-Fried M, Abraham L, Pan X, Giese SI, Hannemann S, et al. HIV-1 Nef Interferes With Host Cell Motility by Deregulation of Cofilin. Cell Host Microbe (2009) 6(2):174–86. doi: 10.1016/j.chom.2009.06.004
356. Shahabuddin M, McKinley G, Potash MJ, Volsky DJ. Modulation of Cellular Gene Expression of HIV Type 1 Infection as Determined by Subtractive Hybridization Cloning: Downregulation of Thymosin Beta 4 In Vitro and In Vivo. AIDS Res Hum Retroviruses (1994) 10(11):1525–9. doi: 10.1089/aid.1994.10.1525
357. Haller C, Rauch S, Michel N, Hannemann S, Lehmann MJ, Keppler OT, et al. The HIV-1 Pathogenicity Factor Nef Interferes With Maturation of Stimulatory T-Lymphocyte Contacts by Modulation of N-Wasp Activity. J Biol Chem (2006) 281(28):19618–30. doi: 10.1074/jbc.M513802200
358. Frischknecht F, Moreau V, Röttger S, Gonfloni S, Reckmann I, Superti-Furga G, et al. Actin-Based Motility of Vaccinia Virus Mimics Receptor Tyrosine Kinase Signalling. Nature (1999) 401(6756):926–9. doi: 10.1038/44860
359. Ohkawa T, Volkman LE, Welch MD. Actin-Based Motility Drives Baculovirus Transit to the Nucleus and Cell Surface. J Cell Biol (2010) 190(2):187–95. doi: 10.1083/jcb.201001162
360. Harmon B, Campbell N, Ratner L. Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step. PLoS Pathog (2010) 6(6):e1000956. doi: 10.1371/journal.ppat.1000956
361. Thomas A, Mariani-Floderer C, López-Huertas MR, Gros N, Hamard-Péron E, Favard C, et al. Involvement of the Rac1-IRSp53-Wave2-Arp2/3 Signaling Pathway in HIV-1 Gag Particle Release in CD4 T Cells. J Virol (2015) 89(16):8162–81. doi: 10.1128/jvi.00469-15
362. Spear M, Guo J, Turner A, Yu D, Wang W, Meltzer B, et al. HIV-1 Triggers WAVE2 Phosphorylation in Primary CD4 T Cells and Macrophages, Mediating Arp2/3-Dependent Nuclear Migration. J Biol Chem (2014) 289(10):6949–59. doi: 10.1074/jbc.M113.492132
363. Komano J, Miyauchi K, Matsuda Z, Yamamoto N. Inhibiting the Arp2/3 Complex Limits Infection of Both Intracellular Mature Vaccinia Virus and Primate Lentiviruses. Mol Biol Cell (2004) 15(12):5197–207. doi: 10.1091/mbc.e04-04-0279
364. Delaney MK, Malikov V, Chai Q, Zhao G, Naghavi MH. Distinct Functions of Diaphanous-Related Formins Regulate HIV-1 Uncoating and Transport. Proc Natl Acad Sci USA (2017) 114(33):E6932–e41. doi: 10.1073/pnas.1700247114
365. Shrivastava A, Prasad A, Kuzontkoski PM, Yu J, Groopman JE. Slit2N Inhibits Transmission of HIV-1 From Dendritic Cells to T-Cells by Modulating Novel Cytoskeletal Elements. Sci Rep (2015) 5:16833. doi: 10.1038/srep16833
366. Aggarwal A, Iemma TL, Shih I, Newsome TP, McAllery S, Cunningham AL, et al. Mobilization of HIV Spread by Diaphanous 2 Dependent Filopodia in Infected Dendritic Cells. PLoS Pathog (2012) 8(6):e1002762. doi: 10.1371/journal.ppat.1002762
367. Kang YJ, Jo JO, Ock MS, Yoo YB, Chun BK, Oak CH, et al. Over-Expression of Thymosin β4 in Granulomatous Lung Tissue With Active Pulmonary Tuberculosis. Tuberculosis (Edinburgh Scotland) (2014) 94(3):323–31. doi: 10.1016/j.tube.2014.01.003
368. Abiko T, Sekino H. Synthesis of a New Biological Response Modifier Thyrnosin Beta(4) Analogue and its Restorative Effect on Depressed. Mediators Inflamm (1992) 1(2):113–9. doi: 10.1155/s096293519200019x
369. van der Kammen R, Song JY, de Rink I, Janssen H, Madonna S, Scarponi C, et al. Knockout of the Arp2/3 Complex in Epidermis Causes a Psoriasis-Like Disease Hallmarked by Hyperactivation of Transcription Factor Nrf2. Development (2017) 144(24):4588–603. doi: 10.1242/dev.156323
370. Schaffer AE, Breuss MW, Caglayan AO, Al-Sanaa N, Al-Abdulwahed HY, Kaymakçalan H, et al. Biallelic Loss of Human CTNNA2, Encoding αn-Catenin, Leads to ARP2/3 Complex Overactivity and Disordered Cortical Neuronal Migration. Nat Genet (2018) 50(8):1093–101. doi: 10.1038/s41588-018-0166-0
371. Wu F, Wen Z, Zhi Z, Li Y, Zhou L, Li H, et al. MPGES-1 Derived PGE2 Inhibits Cell Migration by Regulating ARP2/3 in the Pathogenesis of Hirschsprung Disease. J Pediatr Surg (2019) 54(10):2032–7. doi: 10.1016/j.jpedsurg.2019.01.001
372. Tang W, Cai P, Huo W, Li H, Tang J, Zhu D, et al. Suppressive Action of miRNAs to ARP2/3 Complex Reduces Cell Migration and Proliferation via RAC Isoforms in Hirschsprung Disease. J Cell Mol Med (2016) 20(7):1266–75. doi: 10.1111/jcmm.12799
373. Tessier S, Doolittle AC, Sao K, Rotty JD, Bear JE, Ulici V, et al. Arp2/3 Inactivation Causes Intervertebral Disc and Cartilage Degeneration With Dysregulated TonEBP-Mediated Osmoadaptation. JCI Insight (2020) 5(4):e131382. doi: 10.1172/jci.insight.131382
374. Lauterborn JC, Cox CD, Chan SW, Vanderklish PW, Lynch G, Gall CM. Synaptic Actin Stabilization Protein Loss in Down Syndrome and Alzheimer Disease. Brain Pathol (2020) 30(2):319–31. doi: 10.1111/bpa.12779
375. Lee PP, Lobato-Marquez D, Pramanik N, Sirianni A, Daza-Cajigal V, Rivers E, et al. Wiskott-Aldrich Syndrome Protein Regulates Autophagy and Inflammasome Activity in Innate Immune Cells. Nat Commun (2017) 8(1):1576. doi: 10.1038/s41467-017-01676-0
376. Rivers E, Rai R, Lotscher J, Hollinshead M, Markelj G, Thaventhiran J, et al. Wiskott Aldrich Syndrome Protein Regulates non-Selective Autophagy and Mitochondrial Homeostasis in Human Myeloid Cells. Elife (2020) 9:e55547. doi: 10.7554/eLife.55547
377. Chanez-Paredes S, Montoya-Garcia A, Schnoor M. Cellular and Pathophysiological Consequences of Arp2/3 Complex Inhibition: Role of Inhibitory Proteins and Pharmacological Compounds. Cell Mol Life Sci (2019) 76(17):3349–61. doi: 10.1007/s00018-019-03128-y
378. Degroote RL, Uhl PB, Amann B, Krackhardt AM, Ueffing M, Hauck SM, et al. Formin Like 1 Expression is Increased on CD4+ T Lymphocytes in Spontaneous Autoimmune Uveitis. J Proteomics (2017) 154:102–8. doi: 10.1016/j.jprot.2016.12.015
379. Haenen S, Vanoirbeek JA, De Vooght V, Schoofs L, Nemery B, Clynen E, et al. Proteomic Alterations in B Lymphocytes of Sensitized Mice in a Model of Chemical-Induced Asthma. PLoS One (2015) 10(9):e0138791. doi: 10.1371/journal.pone.0138791
380. Kuhns DB, Fink DL, Choi U, Sweeney C, Lau K, Priel DL, et al. Cytoskeletal Abnormalities and Neutrophil Dysfunction in WDR1 Deficiency. Blood (2016) 128(17):2135–43. doi: 10.1182/blood-2016-03-706028
381. Pinkerton PH, Robinson JB, Senn JS. Lazy Leucocyte Syndrome–Disorder of the Granulocyte Membrane? J Clin Pathol (1978) 31(4):300–8. doi: 10.1136/jcp.31.4.300
382. Miller ME, Oski FA, Harris MB. Lazy-Leucocyte Syndrome. A New Disorder of Neutrophil Function. Lancet (Lond Engl) (1971) 1(7701):665–9. doi: 10.1016/s0140-6736(71)92679-1
Keywords: actin regulators, T cell, B cell, NK cell, WAS
Citation: Sun J, Zhong X, Fu X, Miller H, Lee P, Yu B and Liu C (2022) The Actin Regulators Involved in the Function and Related Diseases of Lymphocytes. Front. Immunol. 13:799309. doi: 10.3389/fimmu.2022.799309
Received: 21 October 2021; Accepted: 01 February 2022;
Published: 16 March 2022.
Edited by:
Frédéric Rieux-Laucat, INSERM U1163 Institut Imagine, FranceReviewed by:
Narcisa Martinez Quiles, Complutense University of Madrid, SpainShoichiro Ono, Emory University, United States
Copyright © 2022 Sun, Zhong, Fu, Miller, Lee, Yu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Yu, eXViaW5nQGh1c3QuZWR1LmNu; Chaohong Liu, Y2hhb2hvbmdsaXU4MEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
 Jianxuan Sun
Jianxuan Sun Xingyu Zhong
Xingyu Zhong Xiaoyu Fu
Xiaoyu Fu Heather Miller3
Heather Miller3 Bing Yu
Bing Yu Chaohong Liu
Chaohong Liu