
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 03 February 2022
Sec. Alloimmunity and Transplantation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.791975
This article is part of the Research TopicThe Biological and Clinical Aspects of HLA-G, Volume IIView all 16 articles
Human leukocyte antigen (HLA)-G is a nonclassical MHC Class I molecule, which was initially reported as a mediator of immune tolerance when expressed in extravillous trophoblast cells at the maternal-fetal interface. HLA-G is the only known ligand of killer cell immunoglobulin-like receptor 2DL4 (KIR2DL4), an atypical family molecule that is widely expressed on the surface of NK cells. Unlike other KIR receptors, KIR2DL4 contains both an arginine–tyrosine activation motif in its transmembrane region and an immunoreceptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic tail, suggesting that KIR2DL4 may function as an activating or inhibitory receptor. The immunosuppressive microenvironment exemplified by a rewired cytokine network and upregulated immune checkpoint proteins is a hallmark of advanced and therapy-refractory tumors. Accumulating evidence has shown that HLA-G is an immune checkpoint molecule with specific relevance in cancer immune escape, although the role of HLA-G/KIR2DL4 in antitumor immunity is still uncharacterized. Our previous study had shown that HLA-G was a pivotal mediator of breast cancer resistance to trastuzumab, and blockade of the HLA-G/KIR2DL4 interaction can resensitize breast cancer to trastuzumab treatment. In this review, we aim to summarize and discuss the role of HLA-G/KIR2DL4 in the immune microenvironment of breast cancer. A better understanding of HLA-G is beneficial to identifying novel biomarker(s) for breast cancer, which is important for precision diagnosis and prognostic assessment. In addition, it is also necessary to unravel the mechanisms underlying HLA-G/KIR2DL4 regulation of the immune microenvironment in breast cancer, hopefully providing a rationale for combined HLA-G and immune checkpoints targeting for the effective treatment of breast cancer.
Breast cancer is the most common cancer diagnosed in women worldwide, with an estimated 2.3 million new cases and 0.7 million deaths in 2020 (1). Breast cancer is a very heterogeneous disease, both at molecular and histological levels. Five intrinsic subtypes of breast cancer were initially identified: Luminal-A, Luminal-B, HER2+, triple negative/basal like (TNBC) and normal like, which is based on the gene expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) (2). The character of each subtype was associated with incidence, treatment response, rate of disease progression and metastasis. The conventional treatment options for patients with breast cancer include surgery, chemotherapy, radiotherapy, hormonal therapy and targeted therapy (3). Increasing evidence has showed that breast cancer microenvironment is not only composed of tumor cells but also of other different cell types, including endothelial cells, several stromal cell types, and immune cells (4). Due to the potential of the immune system in various new treatment strategies, immunotherapy has attracted the attention of researchers, including adoptive cell therapy, oncolytic virus and the most noteworthy immune checkpoint blockade therapy (5). Monoclonal antibodies against PD-1/PD-L1 and CTLA-4 have now entered clinical trials for the clinical treatment of triple negative breast cancer (6). Our recent study has showed that human leukocyte antigen-G (HLA-G) was a pivotal mediator of HER2 positive breast cancer resistance to trastuzumab and blockade of HLA-G can improve the antitumor activity of NK cells significantly (7).
HLA-G is a non-classical HLA Class I molecule that is first found specifically expressed in extravillous trophoblasts (EVTs) and plays a major regulatory role in maternal-fetal immune tolerance (8). During pregnancy, the fetus expresses paternal HLA, which are foreign antigens for maternal tissue, yet the fetus is neither rejected nor attacked by the maternal immune system (9). This phenomenon is due to the immune tolerance elicited by interaction between EVTs expressing HLA-G and leukocytes expressing inhibitory receptors (8). In 1998, Paul and his colleagues first reported the abnormal HLA-G expression in melanoma lesions rather than adjacent normal tissues (10). Accumulated evidence have showed that HLA-G is abnormally highly expressed in a variety of tumor cells and is involved in immune escape of tumors (11). All these findings suggest that HLA-G might be an important immune checkpoint in breast cancer immunotherapy.
Unlike the classical HLA Class I molecule, such as HLA-A, HLA-B, and HLA-C, the gene of HLA-G displays limited polymorphism (12). Seven different transcripts of HLA-G gene, namely HLA-G1 to G7, have been identified. HLA-G1 can be translated into both membrane-binding isoform and soluble isoform; HLA-G2, G3, and G4 transcripts can be translated into membrane-binding proteins; HLA-G5, G6, and G7 transcripts are templates for soluble proteins (11). Different HLA-G isoforms might underlie the diverse functions in cancer immunotherapy.
According to the recent studies, there are multiple receptors of HLA-G, including immunoglobulin-like transcript (ILT)-2, ILT-4, and killer inhibitory receptor (KIR) 2DL4, which are differentially expressed on immune cells (11, 13). ILT-2 and ILT-4 are the type I transmembrane glycoproteins with four extracellular immunoglobulin like domains, a transmembrane region, and an intracellular tail, which have four or three immunoreceptor tyrosine-based inhibitory motifs (ITIMs). ILT-2 and ILT-4 are expressed on T cells, natural killer cells (NK), and dendritic cells (DC) (14). Based on the structure of the intracellular tail, ILT-2/ILT-4 can initiate inhibitory signaling after binding to HLA-G, leading to immune suppression. KIR2DL4 is a member of the killer cell immunoglobulin (Ig)-like receptor (KIR) family, with two atypical extracellular domains, a positively charged arginine residue in the transmembrane region and an ITIM domain in its intracellular tail. The charged arginine residue of KIR2DL4 can recruit activation adaptors that contain immunoreceptor tyrosine-based activation motifs (ITAMs) (14). KIR2DL4 has both the activation and inhibitory signaling domains, suggesting that it can function as both activating and inhibitory receptor.
Our recent study found that KIR2DL4 synergizes with FcRγ to enhance NK cell activation and degranulation, while HLA-G binding to KIR2DL4 impairs the cytotoxicity of NK cells in HER2 positive breast cancer microenvironment (7). These findings suggested that KIR2DL4 might provide a switch for NK cell activity via its association with HLA-G. Increasing evidence has showed that HLA-G is involved in both innate and adaptive immune responses required for immune escape. With the unique structure, whether KIR2DL4 participates in cancer immunotherapy remains to be explored. In this review, we aim to summarize the roles of HLA-G/KIR2DL4 in breast cancer immune microenvironment.
There are 8 exons and 7 introns in HLA-G gene. Most of the full-length HLA-G transcripts contains 7 exons, as exon 7 is usually removed by splicing. Compared with classical HLA Class I molecules, HLA-G is relatively short, the full length of which is only about 340 amino acids. The signal peptide was translated from exon 1; the extracellular α1, α2, and α3 domains were translated from exon 2-4, respectively; the transmembrane domain was generated by exon 5; and the intracellular cytoplasmic tail was generated by exon 6 (15). Accumulated evidences have shown that there are 7 isoforms of HLA-G. Each HLA-G isoform has its unique molecular structure due to different transcript splicing. HLA-G1 has both membrane-binding and soluble isoforms with extracellular α1, α2, and α3 domains, while HLA-G2/-G3/-G4 were membrane-binding isoforms. HLA-G2 has extracellular α1 and α3 domains; HLA-G3 has the only α1 domain; and HLA-G4 has α1 and α2 domains. HLA-G5/-G6/-G7 were soluble isoforms. HLA-G5 has extracellular α1, α2, and α3 domains; HLA-G6 has α1 and α3 domains; and HLA-G7 has the only α1 domains (11). The α1 and α2 domains contain the peptide-binding sites and the α3 domain can bind to β2-microglobulin (β2m) non-covalently. Recent studies have shown that a novel isoform of HLA-G without α1 domain can be detected in colorectal cancer patients and clear cell renal cell cancer patients, which might suggest distinct clinical significance (16, 17).
Killer cell immunoglobulin-like receptor 2DL4 (KIR2DL4) is the only KIR receptor recognized by HLA-G. KIR2DL4 belongs to the killer cell Ig-like receptors (KIR) family, which is expressed mainly on NK cells (7). KIRs can recognize both classical and nonclassical HLA Class I molecules. The KIR genes are located in chromosome 19q13.4, with each spanning 10,000-15,000 bp and separated by 1,000 bp. KIRs are encoded by up to nine exons: the leader peptide was encoded by the first two exons (exon 1 and exon 2), the two or three extracellular Ig-like domains (2D or 3D) by exons 3-5, the stem structure by exon 6, the transmembrane region by exon 7, and the cytoplasmic tail by exons 8-9 (18). Generally, KIRs have two or three extracellular immunoglobulin domains that were named KIR2D or KIR3D. Inhibitory KIRs including KIR2DL or KIR3DL contain a long cytoplasmic tail (L) with two immunoreceptor tyrosine-based inhibitory motifs (ITIMs). Activating KIRs like KIR2DS or KIR3DS contain a short cytoplasmic tail (S) and a positively charged residue in the transmembrane region, which can recruit adaptors that contain the immunoreceptor tyrosine-based activation motif (ITAM) (19). Unlike the conventional KIR2DL receptors, KIR2DL4 contains only one ITIM instead of two in its cytoplasmic tail and a positively charged arginine in its transmembrane region, which can recruit activation adaptors (19–21). Based on the unique structure, both activating and inhibitory functions have been described for KIR2DL4 on NK cells. Rajagopalan et al. found that KIR2DL4 is an activating receptor that can induce IFN-γ production by but not cytotoxicity of resting NK cells (22). Yusa and her colleagues showed that the single ITIM of KIR2DL4 can inhibit cytotoxic response of NK cells efficiently, which depends on SHP-2, but not SHP-1, and phosphorylated tyrosine (20). Our recent study has showed that HLA-G can desensitize breast cancer cells to trastuzumab by binding to the NK cell receptor KIR2DL4. Unless engaged by HLA-G, KIR2DL4 promotes ADCC function and forms a regulatory circuit with the IFN-γ production pathway, in which IFN-γ upregulates KIR2DL4 via JAK2/STAT1 signaling, and then KIR2DL4 synergizes with the CD16 to increase IFN-γ secretion by NK cells (7). Recent studies have showed that the IFN-γ production can impair the function of immune cells by upregulating PD-L1 on tumor cells (23–25). Consistent with previous reports, we observed that IFN-γ significantly increased the level of PD-L1 in breast cancer cells. Blockade of PD-L1 increased the cytotoxicity of NK cells against trastuzumab-treated HER2-overexpressing breast cancer cells (7). These findings suggested that since KIR2DL4 functions as either an activating receptor or an inhibitory receptor, it can affect the outcome of immunotherapy through the complicated cross-talk between different immune checkpoints and cytokines in the breast cancer immune microenvironment.
HLA-G expression in cancer microenvironment was regulated by several intracellular and extracellular mechanisms mediated by microRNAs, RNA-binding proteins, heat shock proteins, cytokines, et al. There are several microRNAs were correlated with the expression of HLA-G, such as miR-133a, miR-148a, miR-148b, miR-152, miR-199b-5p, miR-548q and miR-628-5p et.al, mainly investigated in vitro (26–28). Reches A et al. found that the RNA-binding protein, the heterogeneous nuclear ribonucleoprotein R (HNRNPR) can regulate the expression of HLA-G by binding to the 3’UTR of the HLA-G transcripts (29). Heat shock proteins was also reported to induce the HLA-G expression via the heat shock transcription factor 1 binding to the heat shock element of HLA-G promoter (30). The expression of HLA-G was regulated not only by intracellular post-transcriptional regulation, but also by extracellular mediators. HLA-G expression was found to be induced by progesterone via the interaction between progesterone receptor and progesterone response element (PRE) in the HLA-G promoter (31). Cytokines were also found to regulate the expression of HLA-G, such as GM-CSF, IL-10, TGF-β and IFN-β et al. (7, 32–34). All these findings indicated that the HLA-G expression in cancer microenvironment might be a key factor to investigate the progression of disease.
Many studies have shown that HLA-G/KIR2DL4 expression in immune microenvironment were correlated with the prognosis and progression of breast cancer. In 2002, Lefebvre et al. found that HLA-G was up-regulated at high frequencies in human breast cancer and it may impair anti-tumor immunity by recognizing inhibitory receptors, ILT-2 (35). Meanwhile, Urosevic and colleagues made an editorial comment, proposing that HLA-G and related killer cell inhibitory receptor might represent another mechanism for tumor immune escape (36). However, a recent study has showed that HLA-G expression did not significantly correlate with poor clinical outcome of cancer patients, which indicated that HLA-G expression might not necessarily participate in immunosuppression in carcinogenesis (37). Palmisano et al. analyzed the HLA-G expression at both RNA and protein levels in 25 breast cancer patient tissues. They failed to detect HLA-G expression in breast cancer tissues and cell lines, which was later found attributed to stromal cell contamination in tissue samples (38). In 2003, a comparative study reported that soluble HLA-G can be detected in the malignant ascites of breast cancer patients (39). In the same year, Korkola JE et al. Found in microarray assays that HLA-G were differentially expressed in lobular versus ductal breast cancer (40). Later investigations have established that HLA-G is an important marker of tumor immune escape (41, 42). Several studies have shown that the expression of HLA-G is significantly associated with progression and poor prognosis in a variety of tumors including aggressive breast carcinoma (43–45). Based on previous studies, whether HLA-G can be used as a marker for clinical diagnosis and treatment of breast cancer patients has also been extensively investigated (46, 47). In 2010, a comparative study reported that estradiol/progesterone-induced HLA-G expression can inhibit allo-cytotoxic lymphocyte response to human breast cancer MCF-7 cells (48). Chen et al. found that high soluble HLA-G levels was significantly correlated with the increased infiltration of Treg in breast cancer patients, which indicated that HLA-G might play an important role in the immunosuppressive breast cancer microenvironment (49, 50). HLA-G expression was also associated with subtypes of breast cancer. Provatopoulou X et al. found that HLA-G expression was increased in co-existing ductal and lobular breast cancer patients, compared to those with pure ductal cancer or pure lobular cancer alone (51). Yang’s team reported that there were more cases with high expression of HLA-G in non-luminal than in luminal subtypes, and HLA-G expression was associated negatively with the density of tumor-infiltrating lymphocytes (52). The mRNA stability of HLA-G was found associated with a 14-bp insertion/deletion in exon 8 of the 3’ untranslated region. A series of studies have shown that HLA-G polymorphism could be a diagnostic and prognostic marker for the susceptibility and pathogenesis of breast cancer in populations from different regions (53–59). MicroRNAs were also found to be involved in the regulation of HLA-G expression in breast cancer. Tao et al. found that G protein-coupled estrogen receptor (GPER) mediated the regulation of HLA-G by miR-148a, which was induced by estrogen (E2) in 2 human breast cancer cell lines, MCF-7 and MDA-MB-231 (60). It has been shown that HLA-G is expressed on immune cells such as NK cells and T cells (61, 62).
KIR2DL4 is the only killer cell Ig-like family receptor recognized by HLA-G. In 1999, Long EO et al. first reported that KIR2DL4, which is expressed at the surface of all NK cells including activated NK cells and resting NK cells, can bind to cells expressing HLA-G (22, 63). Goodridge et al. found that there are two KIR2DL4 alleles with either 9 or 10 consecutive adenines (9A or 10A) in exon 6, which encodes the transmembrane domain (64). The 9A alleles can produce a secreted receptor due to the splicing out of the transmembrane region, which might cause a lack of KIR2DL4 expression (65). In contrast, the “10A” alleles encode a membrane-expressed receptor that is constitutively expressed on resting CD56bright and CD56dim NK cells (65). The It has been described that the interaction of the KIR2DL4 receptor with the HLA-G molecule is mediated by the α1 domain, which indicated that all the seven identified isoforms of HLA-G can be recognized by KIR2DL4 (66). In breast cancer immune microenvironment, HLA-G can bind to its receptor, such as KIR2DL4, to induce immunosuppression. Ueshima C et al. found that human mast cells expressing KIR2DL4 can promote invasion of HLA-G-expressing malignant cells and the subsequent metastasis of breast cancer and choriocarcinoma (67). Recently, we have found that HLA-G expression can predict a low trastuzumab response in HER2 positive breast cancer patients (7). We also detected abundant KIR2DL4 expression on infiltrating NK cells in HER2 positive breast cancer tissues. These findings indicated that HLA-G/KIR2DL4 in immune microenvironment might play an important role in the progression of breast cancer and become a new target for breast cancer treatment.
So far, the role of HLA-G/KIR2DL4 in breast cancer immunotherapy has been progressively elucidated. Roberti et al. found that blockade of ILT-2 with antibodies can restore cetuximab-mediated ADCC in triple-negative breast cancer patients and revert immunosuppression mediated by HLA-G (68). In 2016, Ishibashi and colleagues found that an MHC Class II-binding peptide HLA-G26-40 can elicit tumor-reactive CD4+ T cell responses effectively (69). With the wide application of immunotherapy in cancer, the relationship between immunological signature and progression of breast cancer has also been intensively investigated. A study reported that the expression of HLA-G was associated with both improved relapse-free survival (RFS) and overall survival (OS) of basal-like breast cancer, which might indicate a better status of lymphocyte infiltrating (70). Zhang’s team reported that overexpression of HLA-G in breast cancer cells was induced by abnormal DNA methylation modification, which was mediated by DNA methyltransferase (DNMT) and ten-eleven translocation (TET) (71). Therefore, TET inhibitor can prevent aberrant HLA-G expression via maintenance of DNA methylation, which provides a novel potential target for cancer immunotherapy (71). Jørgensen et al. also found that HLA-G expression was regulated partially by DNA methylation since the DNA methyltransferase inhibitor, 5-aza-2’-deoxycytidine, induced HLA-G expression, suggesting the feasibility of manipulating HLA-G expression for immunotherapy in breast cancer (72). In 2020, Schwich et al. reported that soluble HLA-G can induce a an immunosuppressive/exhausted phenotype, and the purified soluble HLA-G1 protein or extracellular vesicles derived from an HLA-G1-positive human breast cancer cell line, SUM149, can affect the anti-tumor function of CD8+ T cells by binding to ILT-2 (73). Our recent study reported that KIR2DL4, an alternative receptor of HLA-G, might be a novel target in breast cancer immunotherapy (7). We found that HLA-G impaired trastuzumab-triggered ADCC by binding to KIR2DL4 on NK cells, and blockade of HLA-G/KIR2DL4 can enhance the antitumor activity of trastuzumab in vivo. However, unless engaged by HLA-G, KIR2DL4 can promote ADCC and form a regulatory circuit with the IFN-γ production pathway via JAK2/STAT1 signaling in NK cells. In addition, paracrine TGF-β and IFN-γ in breast cancer microenvironment can induce PD-1/PD-L1 expression on NK cells and tumor cells, which might further enhance intercellular signaling that leads to immunosuppression. These findings demonstrated the applicability of combined HLA-G/KIR2DL4 and PD-1/PD-L1 targeting in the treatment of trastuzumab-resistant breast cancer (Figure 1).
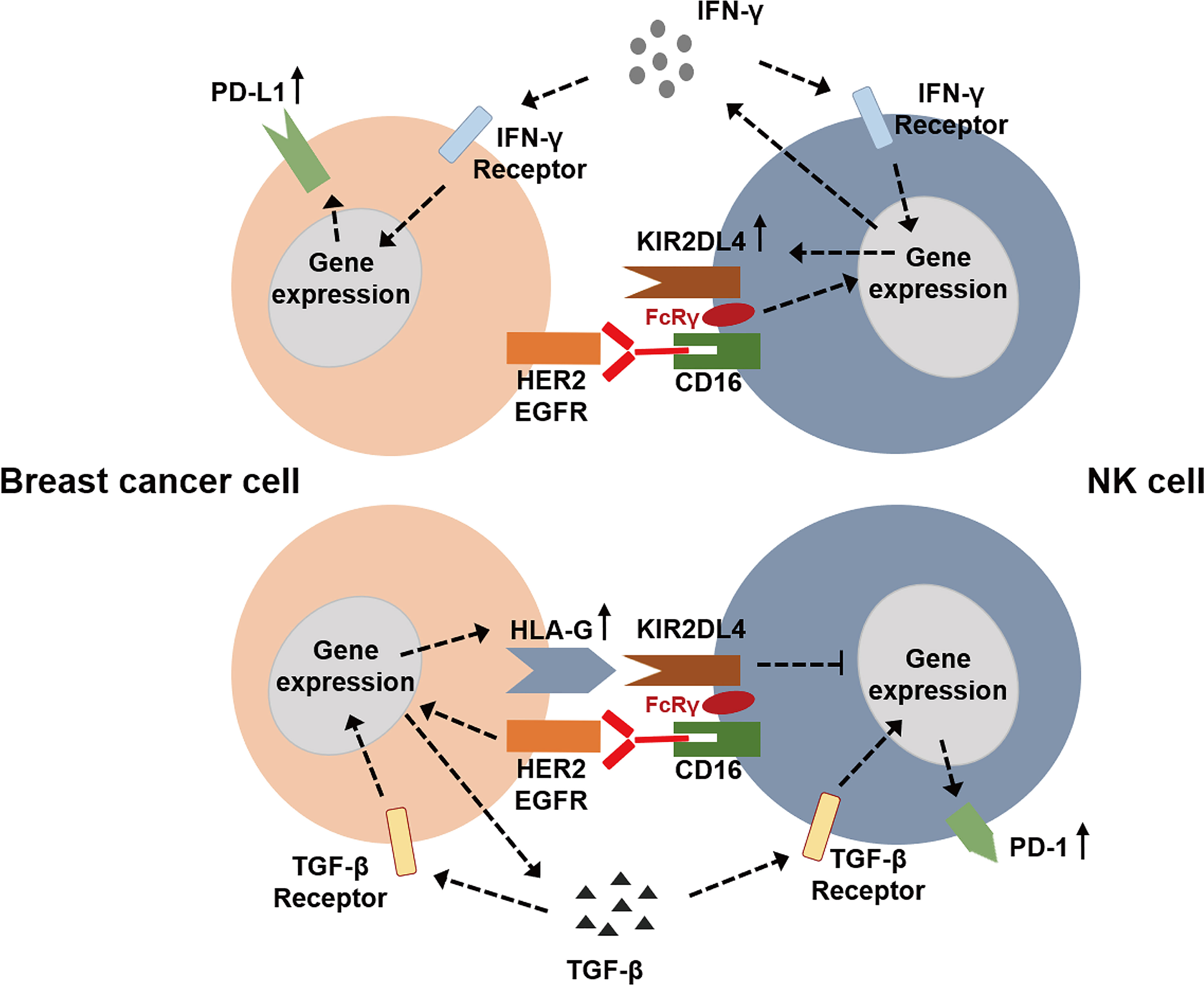
Figure 1 The potential roles of HLA-G/KIR2DL4 signaling in the NK cell mediated immunotherapy of breast cancer.
The immune system played an important role in the occurrence and progression of breast cancer. Many immunotherapy approaches have been investigated for breast cancer, including antibodies against tumor antigens, immune checkpoint blockade and CAR-T cells (4). Recent studies have shown that the combination of conventional chemotherapy or radiotherapy with immunotherapy contributes to improved outcome of breast cancer patients (74, 75). The therapeutic strategies for breast cancer have shifted from cytotoxic therapy to priming anti-tumor immune responses. Nonetheless, only a small part of breast cancer patients can currently benefit from immunotherapy. The relatively low responsive rate and the immunosuppressive tumor microenvironment even in the responding populations have limited the benefit of immunotherapy for breast cancer patients. The abnormal expression of immune checkpoint molecules is the main cause of immune escape. Thus, immune checkpoint blockade represents an important approach to reversing the immunosuppressive status of tumor microenvironment.
In this review, we summarized the special structural features of HLA-G and KIR2DL4. Then, we reviewed the expression of HLA-G/KIR2DL4 in breast cancer immune microenvironment and the potential of HLA-G/KIR2DL4 application in breast cancer immunotherapy. HLA-G, a non-classical HLA Class I molecule, was highly expressed in breast cancer tissues and associated with tumor progression and poor prognosis of patients. HLA-G engagement of its cognate receptors, ILT-2, ILT-4 and KIR2DL4 expressed on immune cells, can significantly induce immunosuppression and result in tumor immune escape. KIR2DL4 is the only KIR receptor that binds to HLA-G. KIR2DL4 contains an ITIM domain in its cytoplasmic tail and a positively charged arginine in its transmembrane region, which can recruit activation adaptors containing a ITAM domain. Based on its special structure, KIR2DL4 can mediate a complicated cross-talk between immune checkpoint and cytokines in breast cancer microenvironment, and dictate distinct outcome of immunotherapy depending on whether or not HLA-G is engaged. A deep understanding of the regulatory role of HLA-G/KIR2DL4 in the immune microenvironment of breast cancer might provide new ideas for the treatment of breast cancer.
Conceptualization, funding acquisition: A-GY. Writing original draft, review and editing: GZ and LJ. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Sciences Foundation of China (No. 81630069 and 81872303).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
2. Barzaman K, Moradi-Kalbolandi S, Hosseinzadeh A, Kazemi MH, Khorramdelazad H, Safari E, et al. Breast Cancer Immunotherapy: Current and Novel Approaches. Int Immunopharmacol (2021) 98:107886. doi: 10.1016/j.intimp.2021.107886
4. Salemme V, Centonze G, Cavallo F, Defilippi P, Conti L. The Crosstalk Between Tumor Cells and the Immune Microenvironment in Breast Cancer: Implications for Immunotherapy. Front Oncol (2021) 11:610303. doi: 10.3389/fonc.2021.610303
5. Burguin A, Diorio C, Durocher F. Breast Cancer Treatments: Updates and New Challenges. J Pers Med (2021) 11(8):808. doi: 10.3390/jpm11080808
6. Farshbafnadi M, Pastaki Khoshbin A, Rezaei N. Immune Checkpoint Inhibitors for Triple-Negative Breast Cancer: From Immunological Mechanisms to Clinical Evidence. Int Immunopharmacol (2021) 98:107876. doi: 10.1016/j.intimp.2021.107876
7. Zheng G, Guo Z, Li W, Xi W, Zuo B, Zhang R, et al. Interaction Between HLA-G and NK Cell Receptor KIR2DL4 Orchestrates HER2-Positive Breast Cancer Resistance to Trastuzumab. Signal Transduct Target Ther (2021) 6(1):236. doi: 10.1038/s41392-021-00629-w
8. Xu X, Zhou Y, Wei H. Roles of HLA-G in the Maternal-Fetal Immune Microenvironment. Front Immunol (2020) 11:592010. doi: 10.3389/fimmu.2020.592010
9. Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The Role of Decidual Immune Cells on Human Pregnancy. J Reprod Immunol (2017) 124:44–53. doi: 10.1016/j.jri.2017.10.045
10. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G Expression in Melanoma: A Way for Tumor Cells to Escape From Immunosurveillance. Proc Natl Acad Sci USA (1998) 95(8):4510–5. doi: 10.1073/pnas.95.8.4510
11. Lin A, Yan WH. HLA-G/ILTs Targeted Solid Cancer Immunotherapy: Opportunities and Challenges. Front Immunol (2021) 12:698677. doi: 10.3389/fimmu.2021.698677
12. Zoehler B, Fracaro L, Senegaglia AC, Bicalho MDG. Infusion of Mesenchymal Stem Cells to Treat Graft Versus Host Disease: The Role of HLA-G and the Impact of its Polymorphisms. Stem Cell Rev Rep (2020) 16(3):459–71. doi: 10.1007/s12015-020-09960-1
13. Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent Advances in Our Understanding of HLA-G Biology: Lessons From a Wide Spectrum of Human Diseases. J Immunol Res (2016) 2016:4326495. doi: 10.1155/2016/4326495
14. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
15. Ishitani A, Geraghty DE. Alternative Splicing of HLA-G Transcripts Yields Proteins With Primary Structures Resembling Both Class I and Class II Antigens. Proc Natl Acad Sci USA (1992) 89(9):3947–51. doi: 10.1073/pnas.89.9.3947
16. Lin A, Zhang X, Zhang RL, Zhang JG, Zhou WJ, Yan WH. Clinical Significance of Potential Unidentified HLA-G Isoforms Without α1 Domain But Containing Intron 4 in Colorectal Cancer Patients. Front Oncol (2018) 8:361. doi: 10.3389/fonc.2018.00361
17. Tronik-Le Roux D, Renard J, Vérine J, Renault V, Tubacher E, LeMaoult J, et al. Novel Landscape of HLA-G Isoforms Expressed in Clear Cell Renal Cell Carcinoma Patients. Mol Oncol (2017) 11(11):1561–78. doi: 10.1002/1878-0261.12119
18. Bruijnesteijn J, van der Wiel MKH, de Groot N, Otting N, de Vos-Rouweler AJM, Lardy NM, et al. Extensive Alternative Splicing of KIR Transcripts. Front Immunol (2018) 9:2846. doi: 10.3389/fimmu.2018.02846
19. Moradi S, Berry R, Pymm P, Hitchen C, Beckham SA, Wilce MC, et al. The Structure of the Atypical Killer Cell Immunoglobulin-Like Receptor, KIR2DL4. J Biol Chem (2015) 290(16):10460–71. doi: 10.1074/jbc.M114.612291
20. Yusa S, Catina TL, Campbell KS. SHP-1- and Phosphotyrosine-Independent Inhibitory Signaling by a Killer Cell Ig-Like Receptor Cytoplasmic Domain in Human NK Cells. J Immunol (2002) 168(10):5047–57. doi: 10.4049/jimmunol.168.10.5047
21. Faure M, Long EO. KIR2DL4 (CD158d), an NK Cell-Activating Receptor With Inhibitory Potential. J Immunol (2002) 168(12):6208–14. doi: 10.4049/jimmunol.168.12.6208
22. Rajagopalan S, Fu J, Long EO. Cutting Edge: Induction of IFN-Gamma Production But Not Cytotoxicity by the Killer Cell Ig-Like Receptor KIR2DL4 (CD158d) in Resting NK Cells. J Immunol (2001) 167(4):1877–81. doi: 10.4049/jimmunol.167.4.1877
23. Wudtiwai B, Makeudom A, Krisanaprakornkit S, Pothacharoen P, Kongtawelert P. Anticancer Activities of Hesperidin via Suppression of Up-Regulated Programmed Death-Ligand 1 Expression in Oral Cancer Cells. Molecules (2021) 26(17):5345. doi: 10.3390/molecules26175345
24. Xie C, Zhou X, Liang C, Li X, Ge M, Chen Y, et al. Apatinib Triggers Autophagic and Apoptotic Cell Death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 Signaling in Lung Cancer. J Exp Clin Cancer Res (2021) 40(1):266. doi: 10.1186/s13046-021-02069-4
25. An Z, Hu Y, Bai Y, Zhang C, Xu C, Kang X, et al. Antitumor Activity of the Third Generation EphA2 CAR-T Cells Against Glioblastoma is Associated With Interferon Gamma Induced PD-L1. Oncoimmunology (2021) 10(1):1960728. doi: 10.1080/2162402x.2021.1960728
26. Amodio G, Gregori S. HLA-G Genotype/Expression/Disease Association Studies: Success, Hurdles, and Perspectives. Front Immunol (2020) 11:1178. doi: 10.3389/fimmu.2020.01178
27. Tan B, Guo J, Wang L, Wang L, Gan X, Chen B. Expression and Change of miR-199b-5p, s HLA-G in Thyroid Carcinoma. Exp Mol Pathol (2021) 120:104643. doi: 10.1016/j.yexmp.2021.104643
28. Poras I, Yaghi L, Martelli-Palomino G, Mendes-Junior CT, Muniz YC, Cagnin NF, et al. Haplotypes of the HLA-G 3' Untranslated Region Respond to Endogenous Factors of HLA-G+ and HLA-G- Cell Lines Differentially. PloS One (2017) 12(1):e0169032. doi: 10.1371/journal.pone.0169032
29. Reches A, Nachmani D, Berhani O, Duev-Cohen A, Shreibman D, Ophir Y, et al. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J Immunol (2016) 196(12):4967–76. doi: 10.4049/jimmunol.1501550
30. Ibrahim EC, Morange M, Dausset J, Carosella ED, Paul P. Heat Shock and Arsenite Induce Expression of the Nonclassical Class I Histocompatibility HLA-G Gene in Tumor Cell Lines. Cell Stress Chaperones (2000) 5(3):207–18. doi: 10.1379/1466-1268(2000)005<0207:hsaaie>2.0.co;2
31. Castelli EC, Ramalho J, Porto IO, Lima TH, Felício LP, Sabbagh A, et al. Insights Into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front Immunol (2014) 5:476. doi: 10.3389/fimmu.2014.00476
32. Onno M, Le Friec G, Pangault C, Amiot L, Guilloux V, Drénou B, et al. Modulation of HLA-G Antigens Expression in Myelomonocytic Cells. Hum Immunol (2000) 61(11):1086–94. doi: 10.1016/s0198-8859(00)00191-9
33. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of Type 1 T Regulatory Cells (Tr1) by Tolerogenic DC-10 Requires the IL-10-Dependent ILT4/HLA-G Pathway. Blood (2010) 116(6):935–44. doi: 10.1182/blood-2009-07-234872
34. Boppana S, Mindur JE, Balashov KE, Dhib-Jalbut S, Ito K. Comparison of IFN-β Inducible Gene Expression in Primary-Progressive and Relapsing-Remitting Multiple Sclerosis. J Neuroimmunol (2013) 265(1-2):68–74. doi: 10.1016/j.jneuroim.2013.10.007
35. Lefebvre S, Antoine M, Uzan S, McMaster M, Dausset J, Carosella ED, et al. Specific Activation of the Non-Classical Class I Histocompatibility HLA-G Antigen and Expression of the ILT2 Inhibitory Receptor in Human Breast Cancer. J Pathol (2002) 196(3):266–74. doi: 10.1002/path.1039
36. Urosevic M, Trojan A, Dummer R. HLA-G and its KIR Ligands in Cancer–Another Enigma Yet to be Solved? J Pathol (2002) 196(3):252–3. doi: 10.1002/path.1057
37. van de Water RB, Krijgsman D, Houvast RD, Vahrmeijer AL. Kuppen PJK. A Critical Assessment of the Association Between HLA-G Expression by Carcinomas and Clinical Outcome. Int J Mol Sci (2021) 22(15):8265. doi: 10.3390/ijms22158265
38. Palmisano GL, Pistillo MP, Fardin P, Capanni P, Nicolò G, Salvi S, et al. Analysis of HLA-G Expression in Breast Cancer Tissues. Hum Immunol (2002) 63(11):969–76. doi: 10.1016/s0198-8859(02)00642-0
39. Singer G, Rebmann V, Chen YC, Liu HT, Ali SZ, Reinsberg J, et al. HLA-G is a Potential Tumor Marker in Malignant Ascites. Clin Cancer Res (2003) 9(12):4460–4.
40. Korkola JE, DeVries S, Fridlyand J, Hwang ES, Estep AL, Chen YY, et al. Differentiation of Lobular Versus Ductal Breast Carcinomas by Expression Microarray Analysis. Cancer Res (2003) 63(21):7167–75.
41. Rebmann V, Regel J, Stolke D, Grosse-Wilde H. Secretion of sHLA-G Molecules in Malignancies. Semin Cancer Biol (2003) 13(5):371–7. doi: 10.1016/s1044-579x(03)00028-2
42. Rouas-Freiss N, Moreau P, Menier C, Carosella ED. HLA-G in Cancer: A Way to Turn Off the Immune System. Semin Cancer Biol (2003) 13(5):325–36. doi: 10.1016/s1044-579x(03)00023-3
43. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human Leukocyte Antigen G Expression: As a Significant Prognostic Indicator for Patients With Colorectal Cancer. Mod Pathol (2007) 20(3):375–83. doi: 10.1038/modpathol.3800751
44. Kleinberg L, Flørenes VA, Skrede M, Dong HP, Nielsen S, McMaster MT, et al. Expression of HLA-G in Malignant Mesothelioma and Clinically Aggressive Breast Carcinoma. Virchows Arch (2006) 449(1):31–9. doi: 10.1007/s00428-005-0144-7
45. Menier C, Prevot S, Carosella ED, Rouas-Freiss N. Human Leukocyte Antigen-G is Expressed in Advanced-Stage Ovarian Carcinoma of High-Grade Histology. Hum Immunol (2009) 70(12):1006–9. doi: 10.1016/j.humimm.2009.07.021
46. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G Expression in Classical HLA Class I-Negative Tumors is of Prognostic Value for Clinical Outcome of Early Breast Cancer Patients. J Immunol (2010) 185(12):7452–9. doi: 10.4049/jimmunol.1002629
47. Sayed D, Badr G, Maximous D, Mikhail NN, Abu-Tarboush F, Alhazza IM. HLA-G and its Relation to Proliferation Index in Detection and Monitoring Breast Cancer Patients. Tissue Antigens (2010) 75(1):40–7. doi: 10.1111/j.1399-0039.2009.01393.x
48. He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, et al. HLA-G Expression in Human Breast Cancer: Implications for Diagnosis and Prognosis, and Effect on Allocytotoxic Lymphocyte Response After Hormone Treatment In Vitro. Ann Surg Oncol (2010) 17(5):1459–69. doi: 10.1245/s10434-009-0891-9
49. Chen HX, Lin A, Shen CJ, Zhen R, Chen BG, Zhang X, et al. Upregulation of Human Leukocyte Antigen-G Expression and its Clinical Significance in Ductal Breast Cancer. Hum Immunol (2010) 71(9):892–8. doi: 10.1016/j.humimm.2010.06.009
50. Elliott RL, Jiang XP, Phillips JT, Barnett BG, Head JF. Human Leukocyte Antigen G Expression in Breast Cancer: Role in Immunosuppression. Cancer Biother Radiopharm (2011) 26(2):153–7. doi: 10.1089/cbr.2010.0924
51. Provatopoulou X, Kalogera E, Sagkriotis A, Zagouri F, Nonni A, Zografos GC, et al. Soluble Human Leukocyte Antigen-G Expression in Patients With Ductal and Lobular Breast Malignancy. Anticancer Res (2012) 32(3):1021–6.
52. Dong DD, Yie SM, Li K, Li F, Xu Y, Xu G, et al. Importance of HLA-G Expression and Tumor Infiltrating Lymphocytes in Molecular Subtypes of Breast Cancer. Hum Immunol (2012) 73(10):998–1004. doi: 10.1016/j.humimm.2012.07.321
53. Eskandari-Nasab E, Hashemi M, Hasani SS, Omrani M, Taheri M, Mashhadi MA. Association Between HLA-G 3'UTR 14-Bp Ins/Del Polymorphism and Susceptibility to Breast Cancer. Cancer Biomark (2013) 13(4):253–9. doi: 10.3233/cbm-130364
54. Ramos CS, Gonçalves AS, Marinho LC, Gomes Avelino MA, Saddi VA, Lopes AC, et al. Analysis of HLA-G Gene Polymorphism and Protein Expression in Invasive Breast Ductal Carcinoma. Hum Immunol (2014) 75(7):667–72. doi: 10.1016/j.humimm.2014.04.005
55. Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human Leukocyte Antigen-G (HLA-G) Polymorphism and Expression in Breast Cancer Patients. PloS One (2014) 9(5):e98284. doi: 10.1371/journal.pone.0098284
56. Zidi I, Dziri O, Zidi N, Sebai R, Boujelebene N, Ben Hassine A, et al. Association of HLA-G +3142 C>G Polymorphism and Breast Cancer in Tunisian Population. Immunol Res (2016) 64(4):961–8. doi: 10.1007/s12026-015-8782-6
57. Ouni N, Chaaben AB, Kablouti G, Ayari F, Douik H, Abaza H, et al. The Impact of HLA-G 3'utr Polymorphisms in Breast Cancer in a Tunisian Population. Immunol Invest (2019) 48(5):521–32. doi: 10.1080/08820139.2019.1569043
58. Kadiam S, Ramasamy T, Ramakrishnan R, Mariakuttikan J. Association of HLA-G 3'UTR 14-Bp Ins/Del Polymorphism With Breast Cancer Among South Indian Women. J Clin Pathol (2020) 73(8):456–62. doi: 10.1136/jclinpath-2019-205772
59. Haghi M, Ranjbar M, Karari K, Samadi-Miandoab S, Eftekhari A, Hosseinpour-Feizi MA. Certain Haplotypes of the 3'-UTR Region of the HLA-G Gene are Linked to Breast Cancer. Br J BioMed Sci (2021) 78(2):87–91. doi: 10.1080/09674845.2020.1856495
60. Tao S, He H, Chen Q, Yue W. GPER Mediated Estradiol Reduces miR-148a to Promote HLA-G Expression in Breast Cancer. Biochem Biophys Res Commun (2014) 451(1):74–8. doi: 10.1016/j.bbrc.2014.07.073
61. Ostapchuk EO, Perfi L'eva YV, Talaeva S, Omarbaeva NA, Belyaev NN. Content of HLA-G(+) T Cells in the Peripheral Blood From Healthy Women and Breast Cancer Patients. Bull Exp Biol Med (2015) 159(5):649–51. doi: 10.1007/s10517-015-3038-y
62. Ostapchuk YO, Cetin EA, Perfilyeva YV, Yilmaz A, Skiba YA, Chirkin AP, et al. Peripheral Blood NK Cells Expressing HLA-G, IL-10 and TGF-β in Healthy Donors and Breast Cancer Patients. Cell Immunol (2015) 298(1-2):37–46. doi: 10.1016/j.cellimm.2015.09.002
63. Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G-Specific Receptor Expressed on All Natural Killer Cells. J Exp Med (1999) 189(7):1093–100. doi: 10.1084/jem.189.7.1093
64. Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three Common Alleles of KIR2DL4 (CD158d) Encode Constitutively Expressed, Inducible and Secreted Receptors in NK Cells. Eur J Immunol (2007) 37(1):199–211. doi: 10.1002/eji.200636316
65. Nowak I, Barcz E, Majorczyk E, Malinowski A, Wilczyński JR, Banasik M, et al. Genetic Polymorphism of KIR2DL4 in the Polish Population. Tissue Antigens (2015) 85(6):450–7. doi: 10.1111/tan.12544
66. Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G Alpha1 Domain Involve in KIR2DL4 Recognition. Cell Res (2005) 15(3):176–82. doi: 10.1038/sj.cr.7290283
67. Ueshima C, Kataoka TR, Hirata M, Furuhata A, Suzuki E, Toi M, et al. The Killer Cell Ig-Like Receptor 2dl4 Expression in Human Mast Cells and Its Potential Role in Breast Cancer Invasion. Cancer Immunol Res (2015) 3(8):871–80. doi: 10.1158/2326-6066.Cir-14-0199
68. Roberti MP, Juliá EP, Rocca YS, Amat M, Bravo AI, Loza J, et al. Overexpression of CD85j in TNBC Patients Inhibits Cetuximab-Mediated NK-Cell ADCC But can be Restored With CD85j Functional Blockade. Eur J Immunol (2015) 45(5):1560–9. doi: 10.1002/eji.201445353
69. Ishibashi K, Kumai T, Ohkuri T, Kosaka A, Nagato T, Hirata Y, et al. Epigenetic Modification Augments the Immunogenicity of Human Leukocyte Antigen G Serving as a Tumor Antigen for T Cell-Based Immunotherapy. Oncoimmunology (2016) 5(6):e1169356. doi: 10.1080/2162402x.2016.1169356
70. Martínez-Canales S, Cifuentes F, López De Rodas Gregorio M, Serrano-Oviedo L, Galán-Moya EM, Amir E, et al. Transcriptomic Immunologic Signature Associated With Favorable Clinical Outcome in Basal-Like Breast Tumors. PloS One (2017) 12(5):e0175128. doi: 10.1371/journal.pone.0175128
71. Zhang D, An X, Li Z, Zhang S. Role of Gene Promoter Methylation Regulated by TETs and DNMTs in the Overexpression of HLA-G in MCF-7 Cells. Exp Ther Med (2019) 17(6):4709–14. doi: 10.3892/etm.2019.7481
72. Jørgensen N, Sayed A, Jeppesen HB, Persson G, Weisdorf I, Funck T, et al. Characterization of HLA-G Regulation and HLA Expression in Breast Cancer and Malignant Melanoma Cell Lines Upon IFN-γ Stimulation and Inhibition of DNA Methylation. Int J Mol Sci (2020) 21(12):4307. doi: 10.3390/ijms21124307
73. Schwich E, Hò GT, LeMaoult J, Bade-Döding C, Carosella ED, Horn PA, et al. Soluble HLA-G and HLA-G Bearing Extracellular Vesicles Affect ILT-2 Positive and ILT-2 Negative CD8 T Cells Complementary. Front Immunol (2020) 11:2046. doi: 10.3389/fimmu.2020.02046
74. Bernal-Estévez DA, Ortíz Barbosa MA, Ortíz-Montero P, Cifuentes C, Sánchez R, Parra-López CA. Autologous Dendritic Cells in Combination With Chemotherapy Restore Responsiveness of T Cells in Breast Cancer Patients: A Single-Arm Phase I/II Trial. Front Immunol (2021) 12:669965. doi: 10.3389/fimmu.2021.669965
Keywords: HLA-G, KIR2DL4, breast cancer, immune microenvironment, immunotherapy
Citation: Zheng G, Jia L and Yang A-G (2022) Roles of HLA-G/KIR2DL4 in Breast Cancer Immune Microenvironment. Front. Immunol. 13:791975. doi: 10.3389/fimmu.2022.791975
Received: 09 October 2021; Accepted: 19 January 2022;
Published: 03 February 2022.
Edited by:
Wei-Hua Yan, Wenzhou Medical University, ChinaReviewed by:
Edgardo Carosella, Université Sorbonne Paris Cité, FranceCopyright © 2022 Zheng, Jia and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: An-Gang Yang, YWd5YW5nQGZtbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.