- 1School of Life Sciences, Shanghai University, Shanghai, China
- 2Shanghai Engineering Research Center of Organ Repair, Shanghai University, Shanghai, China
- 3State Key Laboratory of Virology, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
- 4University of Chinese Academy of Science, Beijing, China
- 5State Key Laboratory of Cell Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China
- 6Department of Head & Neck Surgery, Fudan University Shanghai Cancer Center/Cancer Institute, Shanghai, China
- 7Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 8Department of Respiration Medicine, Affiliated Nantong Hospital of Shanghai University, Nantong, China
- 9Cancer Center, Shanghai Tenth People’s Hospital, School of Medicine, Tongji University, Shanghai, China
Hepatocellular carcinoma (HCC) is one of the most common digestive system cancers (DSCs) with a poor prognosis. Zinc‐regulated transporter (ZRT)/iron‐regulated transporter (IRT) like protein transporters (ZIPs) encode membrane transport proteins, which are responsible for the absorption of zinc and play important roles in the pathogenesis of various human cancers. Tumor-associated macrophages (TAMs) are important participants in the regulation of tumor microenvironment and the development of HCC. Individual role of each ZIP involved in hepatocarcinogenesis remains elusive. In this study, the transcription patterns of ZIPs in the DSCs were screened firstly through GEPIA2 database. Interestingly, the analysis of the DSCs data showed the distinct mRNA levels of ZIPs between DSCs tissues and healthy controls. Notably, the transcription levels of ZIP2, ZIP5, ZIP8, ZIP9 and ZIP14 were decreased significantly in the tissues of human liver cancer compared to paracarcinoma liver tissues. To further confirm the mRNA transcriptional changes of Zips in HCC, N-Nitrosodiethylamine (DEN) combined with carbon tetrachloride (CCl4) inducing mouse model of HCC were established. Consistently, the mRNA levels of Zip2, Zip9, and Zip14 in liver tissues of HCC induced mice were also decreased compared with the healthy controls. In addition, mouse peritoneal elucidated macrophages (PEMs)-derived M1/M2 macrophages in vitro, as well as human patients of HCC-derived TAMs, were used to examine the transcription levels of ZIPs. Our results showed that both Zip2 and Zip9 were up-regulated in M2-polarized macrophages. Zip2 transcript was also up-regulated M1-polarized macrophages, but Zip9 was slightly down-regulated. TAMs generated from human liver cancer tissues also displayed a decrease in ZIP9 transcription compared to paracarcinoma tissues. To further explore the role of Zip9 in M1/M2 polarization, the siRNA knockdown results revealed that Zip9, but not Zip2, could promote M2 macrophage polarization and impair M1 macrophage polarization. Mechanistically, Zip9 enhances phosphorylated STAT6 to promote M2 macrophage polarization but suppresses the phosphorylation of IκBα/β to inhibit M1 macrophage polarization. Together, our results indicate that ZIP9 may involve in macrophages polarity in HCC development and may be a potent new biomarker for the diagnosis of HCC.
Introduction
Digestive system cancers (DSCs, including esophageal cancer, gastric cancer, colorectal cancer, cholangiocarcinoma, hepatocellular carcinoma, and pancreatic cancer) are common malignant cancers and the cause of death worldwide (1). Although the survival rates of these cancers have been greatly improved in recent years due to the advancement of early diagnosis and treatment strategies, the incidence and mortality of these cancers have increased (2, 3). Among these DSCs, hepatocellular carcinoma (HCC) is the sixth most common primary malignant tumor in the world, and it is the fourth leading cause of human death (4). It is well documented that the level of zinc was significantly lower in the human (hepatoma tissue) compared with surrounding “normal” or cirrhotic tissue (5). Zinc is the second essential trace element and plays an important role in all organisms, such as maintaining the stability and function of many metalloenzymes involved in protein synthesis, protein catabolism, energy metabolism, and RNA and DNA synthesis (6). An association between lower systemic zinc levels and DSCs has been demonstrated in various human disorders, such as hepatitis, colorectal cancer and gastric cancer (7–9).
The homeostasis of zinc in the body is mainly regulated by zinc transporters (10). These zinc transporters can be divided into two families, zinc transporters (CDF/ZnTs) and zinc‐regulated transporter (ZRT)/iron‐regulated transporter (IRT)‐like protein transporters (ZIPs). ZnTs are encoded by 10 genes and are responsible for zinc efflux, while ZIPs are encoded by 14 genes and are responsible for the influx of zinc into the cytoplasm (10). Multiple studies had highlighted the key functions of ZIP genes in DSCs, like colorectal cancer, esophageal carcinoma, pancreatic cancer, and hepatocellular carcinoma (11–15). Besides, emerging evidence indicated that ZIP genes were closely correlated with the prognosis of patients of DSCs. Some specific ZIP genes had been suggested to serve as cancer diagnostic or prognostic biomarkers. Lou et al. evaluated the prognostic values of ZIP family genes in gastric cancer (GC) patients by using a variety of online databases. Their results indicated that the high mRNA levels of ZIP7, ZIP11, ZIP14 were associated with better overall survival (OS), while ZIP1, ZIP2, ZIP3, ZIP4, ZIP5, ZIP6, ZIP8, ZIP9, ZIP12, ZIP13 were significantly related with worse OS in GC patients (16). In addition, ZIP3 has been reported to be downregulated in the early stage of pancreatic cancer (17). Xiao et al. reported that overexpression of ZIP4 increased cell migration and invasion in HepG2 cells (18). Zhu et al. found that ZIP4 was overexpressed in human pancreatic cancer tissues and cell lines (19). ZIP4 was also significantly enhanced in esophageal cancer (ESCC) specimens, which could serve as a novel prognostic to promote ESCC progression (20). Furthermore, ZIP5 was overexpressed in human esophageal cancer tissue in regions of high incidence, and downregulation of ZIP5 inhibited the proliferation, migration, and invasion of ESCC in vivo (21). Also, ZIP6 gene was up-regulated in ESCC tissue, high tumorous ZIP6 expression was significantly correlated with shorter OS (14). All these researches indicate that ZIPs function in the development of DSCs. However, to our knowledge, the relevant studies regarding the total transcription patterns of ZIP genes in the development of HCC are still limited.
Macrophages can be characterized by a high diversity of cell surface agents, transcriptional profiles, and different self-local environment-derived stimuli can induce the polarization of macrophage phenotypes (22). M1 macrophages are activated by interferon-γ (IFNγ) produced by CD8+ T cells and natural killer (NK) cells (23, 24). They are also activated by PAMPs, such as lipopolysaccharide (LPS), cytoplasmic receptors (25), the pro-inflammatory cytokines and chemokines (26). Those activated M1 macrophages can induce the synthesis of reactive oxygen species (ROS) and the release of nitric oxide (NO) (27). Macrophages polarized to M2 phenotype (induced by IL-4 and IL-13) can promote HCC progress (28). It has been shown that M2-polarized tumor associated macrophages (TAMs) influence HCC cells via the IL-6/STAT3 signaling pathway (29, 30). It also has been reported that HCC deteriorates on account of the upregulation of protein in M2 TAMs (31). More importantly, it has been elucidated that many transcription factors are involved in regulating M1/M2 polarization of macrophages, NF-κB promotes M1 polarization and STAT6 is essential to enhance the expression of M2 related genes (32, 33) Although some genes in Zip family are illustrated to be associated with the function of macrophages (34), the effect of Zips on the polarization of macrophages is poorly understood.
In this article, we focused on the transcriptome analysis of total ZIPs in human HCC patients and total Zips in DEN-induced HCC mice and their roles in polarized-macrophages in vitro and TAMs of HCC patients in vivo. Interestingly, our results showed that the mRNA level of ZIP9 (Zip9) was significantly down-regulated in tumors of both human and mouse HCC. Moreover, Zip9 knockdown specifically reduced M2 via the IL-4/STAT6 signaling pathways but enhanced M1 macrophage polarization via the NF-κB signaling pathways in response to inflammatory stimuli. Importantly, mRNA expression of ZIP9 was also down-regulated in human HCC-derived TAMs. This study may provide new insight into ZIP9 in HCC development and help to develop potent strategies for the diagnosis of patients with HCC.
Materials and Methods
Mice
C57BL6 background mice were purchased from the Model Animal Research Center (Nanjing University, Nanjing, China). All animals were housed in animal facilities at the Shanghai University under a standard 12‐h light/dark cycle with access to chow and water ad libitum. Experiments were conducted in accordance with the ethical statement.
DEN and CCl4- Induced Mouse Model of HCC
6-8 weeks of age male C57BL/6J mice were treated with DEN intraperitoneally (i.p., N0258‐1G; Sigma, St. Louis, MO, USA) at a dose of 100 mg·kg-1 or with vehicle alone (normal saline, i.p., referred as control). Four weeks later, mice were weekly i.p. injected with CCl4 (0.5 ml/kg, dissolved in olive oil) for 12 weeks. CCl4 was diluted 1: 9 in olive oil (A502795-0100, Sangon Biotech). The mice of the control group were treated with olive oil alone. Finally, mice were sacrificed for collection of liver tissues.
Collecting PEMs and Inducing M1/M2 Macrophages
To obtain peritoneal elucidated macrophages (PEMs), mice were injected i.p. with 3 ml sterile 3% Brewer thioglycollate medium; 3 days later, PEMs were harvested. PEMs were maintained in complete DMEM supplemented with 10% (vol/vol) FBS and penicillin/streptomycin (100 U/ml) (Sigma-Aldrich, Taufkirchen, Germany) at 37°C and 5% CO2. Post 24hrs of cell culture, media was replaced with fresh media containing either lipopolysaccharide (LPS; 1μg/ml; Sigma-Aldrich, L6529) and interferon γ (IFNγ; 100ng/ml; Peprotech, 315-05) to induce M1 macrophages or a combination of IL-4 (20ng/ml; Peprotech, 214-14) and IL-13 (20ng/ml; Peprotech, 210-13) to induce M2 macrophages.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from cells or tissues using Trizol (Takara). The RNA concentration and purity were determined spectrophotometrically. And cDNA was synthesized using approximately 1μg of total RNA and the reverse transcriptase M-MLV (Takara Bio, Shiga, Japan). Quantitative real-time PCR (RT-PCR) was carried out using 2× SYBR green (Takara Bio, Shiga, Japan) with specific primers for ZIPs (1–14), YM1, ARG1, IL6 and β-Actin (Table 1) in a C1000 thermal cycler (BIO-RAD CFX96, Hercules, CA, USA). β-Actin was used for normalization. The relative mRNA expression was calculated using the 2−ΔCT method or 2-ΔΔCT method.
Patients and Tissue Samples
A total of 9 patients with histopathologically confirmed liver cancer were collected from Shanghai Zhongshan Hospital (Shanghai, China). All the samples were obtained via surgical resection or liver transplantation procedures. The diagnosis of liver cancer was performed following the WHO Classification of Cancers of the Digestive System (1). All the tissue samples were reviewed by 2 independent experienced pathologists. The study protocol was approved by the Ethical Committee of Shanghai Zhongshan Hospital. Written informed consent was obtained from all patients.
SiRNA Transfection
The synthesized siRNA was transfected into murine macrophages using Lipofectamine RNAiMAX (Invitrogen). siRNA sequences used for transfection was shown in Table 2.
Tissue Paraffin Section Preparation and HE Staining
The liver tissues were fixed in 4% paraformaldehyde or 10% neutral formaldehyde overnight then treated with different alcohol for dehydration, xylene solution for transparency, and finally embed the tissue in paraffin, used a microtome to cut the tissues into 3.5μm slices for HE staining and IHC staining. For HE staining, briefly, after deparaffinization and rehydration, sections were stained with hematoxylin solution for 5 min followed by dipping in 1% acid ethanol (1% HCl in 70% ethanol) and then rinsed with distilled water. Then the sections were stained with eosin solution for 3 min and followed by dehydration with graded alcohol and clearing in xylene. The slides were then examined and photographed under Olympus BX53 fluorescence microscope (Tokyo, Japan).
Immunohistochemistry
Paraffin-embedded liver tissue slides were kept at 60°C for 24 h, dewaxed with xylene and hydrated by gradient ethanol (100%-70%). The slides were incubated in a 1× antigen retrieval solution (YEASEN, 36310ES60) and an endogenous peroxidase blocker (ZSGB, SP-9000). Then the slices were incubated with goat serum working solution (ZSGB, SP-9000) for 10-15 min, and anti-CD68 primary antibody (CST, 76437T) at 4°C overnight, and finally washed with 1× PBS. The slides were rinsed with 1× PBS 3 times and incubated with the corresponding secondary antibody (ZSGB, SP-9000) for 30 min followed by 3, 3-diaminobenzidine (DAB) and hematoxylin staining, respectively. The slides were then examined and photographed using an Olympus BX53 fluorescence microscope (Tokyo, Japan).
Western Blot
Cells were lysed in RIPA buffer (50 mM Tris-HCl at pH 7.4,150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) with protease and phosphatase inhibitors, followed by denaturation at 99 °C for 10 min. Then the protein samples were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore). Next, the PVDF membranes were blocked with 5% bovine serum albumin for 1 h and incubated with the primary antibodies overnight, including rabbit anti-STAT6 (1:1000, CST, 5397), rabbit anti-p-STAT6 (1:1000, CST, 9361), rabbit anti-p-p65 (1: 1000, CST, 3033), rabbit anti-IKKα/β (1:1000, CST, 2697) and rabbit anti-tubulin (1:5000, ProteinTech, 11224-1-AP). After that, the PVDF membranes were disposed of with the relevant secondary antibodies (1:5000) for 1 h at room temperature and observed using the ECL kit chemiluminescence reagents (Millipore, Billerica, MA, USA). The signals of protein bands were detected with the Chemidoc detection system and quantified using Quantity One software (Bio-Rad).
Flow Cytometry and FACS
Livers of mice were harvested and pooled from 6-8 week C57BL/6J WT mice. Livers of human were collected from HCC patients in Shanghai Zhongshan Hospital (Shanghai, China). Red blood cells lysis was performed using ACK buffer. The panel of antibodies used in the experiments included APC-anti-F4/80 (Invitrogen, 17-4801-82), FITC-anti-CD11b (Invitrogen, 11-0112-82), APC-anti-CD11B (ThermoFisher, 11-0118-42), APC-anti-CD14 (ThermoFisher, 17-0149-42). Flow cytometry was performed using BD LSR II and BD FACS Aria III flow cytometers (BD Bioscience), and data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Statistics
Bioinformatics analysis of gene expression and Correlation analysis are available at the Gene Expression Profiling Interactive Analysis website (http://gepia2.cancer-pku.cn/#survival). Statistical data analysis was performed with Graph pad Prism 9.0 software. Data represent mean ± SD. Statistical significance was determined with 2-tailed unpaired Student’s t test between 2 groups. One-way or 2-way ANOVA with Holm-Sidak correction or with Newman-Keuls correction were used for multiple comparisons. Correlations were done using Pearson’s or Spearman’s tests. P< 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Results
ZIPs in DSCs Show Distinct Transcription Patterns in Samples From GEPIA2 Database
To explore the roles of ZIPs in DSCs, data were retrieved from patients with DSCs in the Gene Expression Profiling Interactive Analysis (GEPIA) and 72 normal samples from The Cancer Genome Atlas (TCGA) databases, we found except ZIP12, almost all ZIP genes had obviously different transcription patterns between tumor tissues and normal tissues (Figure 1). Our results showed that the transcription levels of five ZIPs were significantly elevated in cancer of esophageal carcinoma (ESCA). Seven ZIPs showed remarkable transcription increase in cancer of stomach adenocarcinoma (STAD). Seven ZIPs were significantly up-regulated in cancer tissues of cholangiocarcinoma (CHOL). Two ZIPs in rectum adenocarcinoma (READ) were notably down-regulated in cancers. Also, four ZIPs in liver hepatocellular carcinoma (LIHC) were substantially increased in cancers. Two ZIPs were significantly decreased in tumors of LIHC (Table 3). These data suggest that ZIPs have different transcription profiles in different DSCs, which implies their un-neglected physiological functions in the pathology of various DSCs.
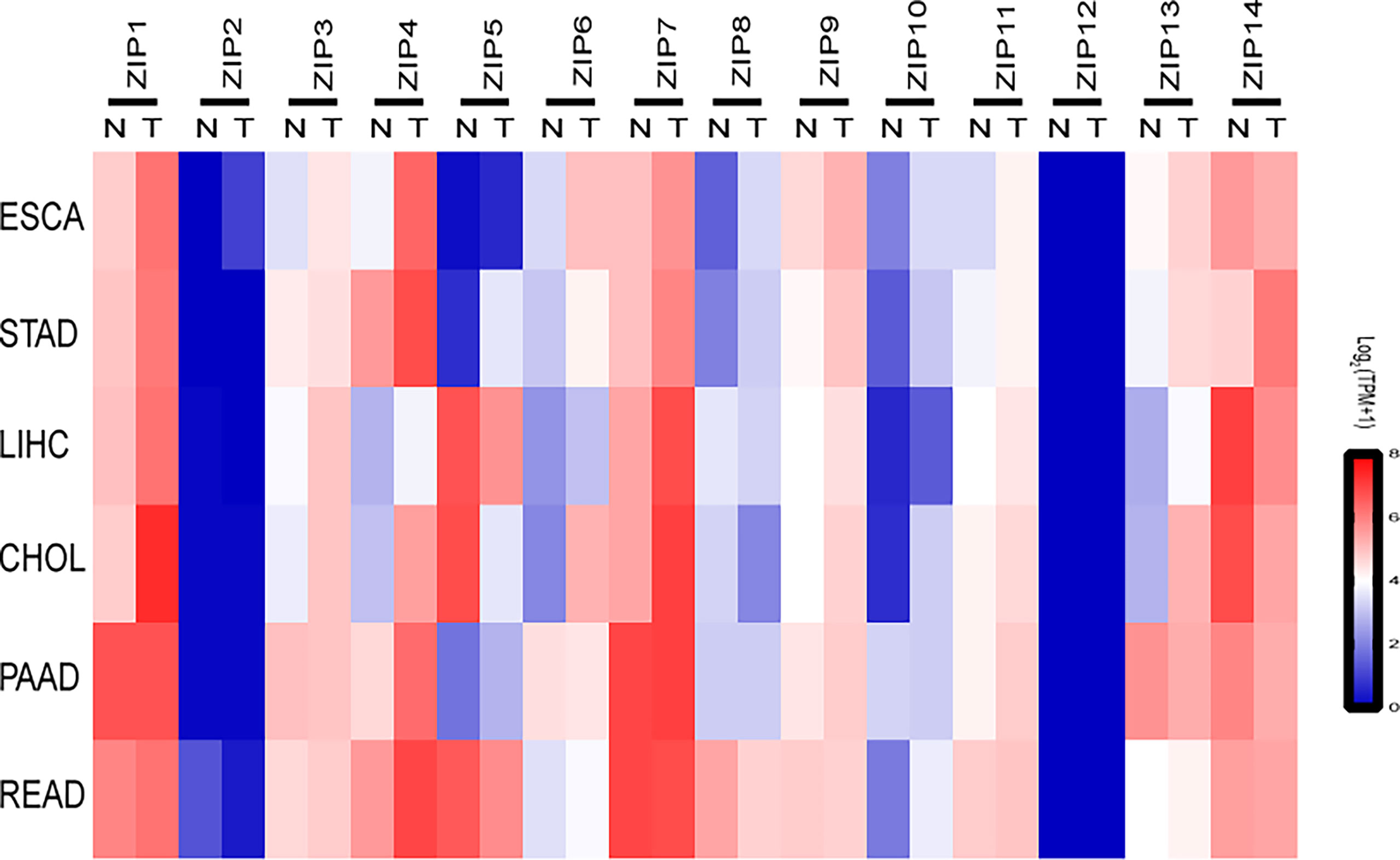
Figure 1 The transcription profiles of ZIPs in various cancers of the digestive system. Heat-map depicting the RNA expression profiles of ZIPs in a variety of human digestive system cancers based on RNA sequencing data from Gene Expression Profiling Interactive Analysis website (GEPIA2). Data were obtained from cancer patients (T) and healthy controls (N), and each sample was normalized by log2 (TPM+1). The red and blue regions represent the higher and the lower expression levels, respectively (TPM, transcripts per million; ESCA, Esophageal carcinoma; STAD, Stomach adenocarcinoma; LIHC, liver hepatocellular carcinoma; CHOL, Cholangiocarcinoma; PAAD, Pancreatic adenocarcinoma; READ, Rectum adenocarcinoma).
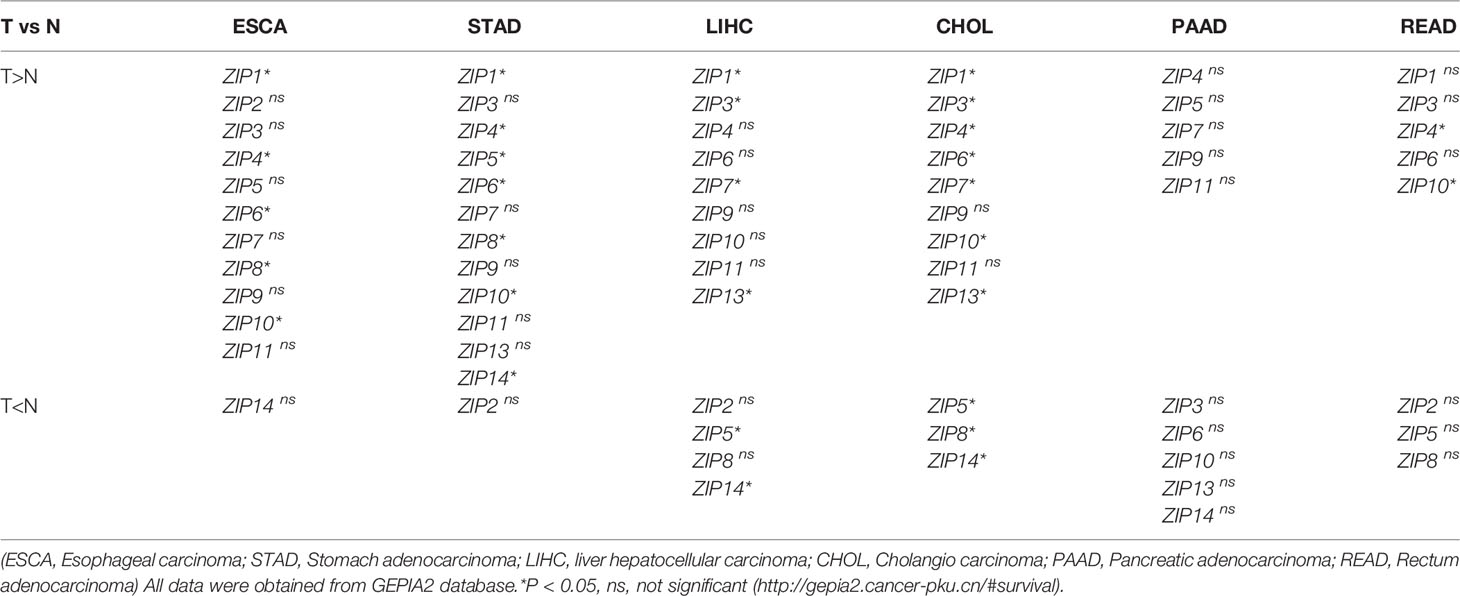
Table 3 Summary of ZIPs transcription levels in human digestive system cancers (T) and healthy controls (N).
The mRNA Levels of ZIP2/ZIP5/ZIP8/ZIP9/ZIP14 Are Decreased in the Liver Tissues of HCC Patients
Since the transcriptions of ZIPs in multiple DSCs showed distinct patterns between cancers and healthy controls, then we wondered the individual transcription profile of each ZIP in HCC. Next, we collected 9 pairs of cancer and paracarcinoma tissues from patients with HCC. We analyzed the mRNA levels of some HCC related genes, including AFP, GPC3, MST1 and some inflammation-related genes, such as TGFB, IL1B, IL6 and TNFA by RT-PCR firstly. Interestingly, TGF-B transcription was significantly increased in liver cancers compared with paracarcinoma tissues (Figures 2A, B). Additionally, liver cancer tissues of HCC patients showed increased numbers and size of tumors compared with paracarcinoma tissues by HE staining of tissue sections (Supplemental Figure 2). Then the transcription patterns of ZIPs in liver cancers and paracarcinoma tissues were analyzed. The results showed that the mRNA levels of ZIP2, ZIP5, ZIP8, ZIP9, and ZIP14 were significantly lower in liver tumor tissues than those in paracarcinoma tissues (Figure 2C). Particularly, the mRNA level of ZIP5 (approximately 4-fold, P=0.0005) and that of ZIP14 (approximately 3-fold, P=0.003) in the paracarcinoma tissues of HCC were significantly higher than those in tumor tissues. However, transcription of other ZIP genes, such as ZIP1, ZIP3, ZIP4, ZIP6, ZIP7, ZIP10, ZIP11, ZIP12, had no significant difference between human liver cancers and paracarcinoma tissues (Figure 2C). Collectively, all observations demonstrate that ZIP5, ZIP8 and ZIP9 show the decreased mRNA transcription profiles in cancer tissues of HCC patients.
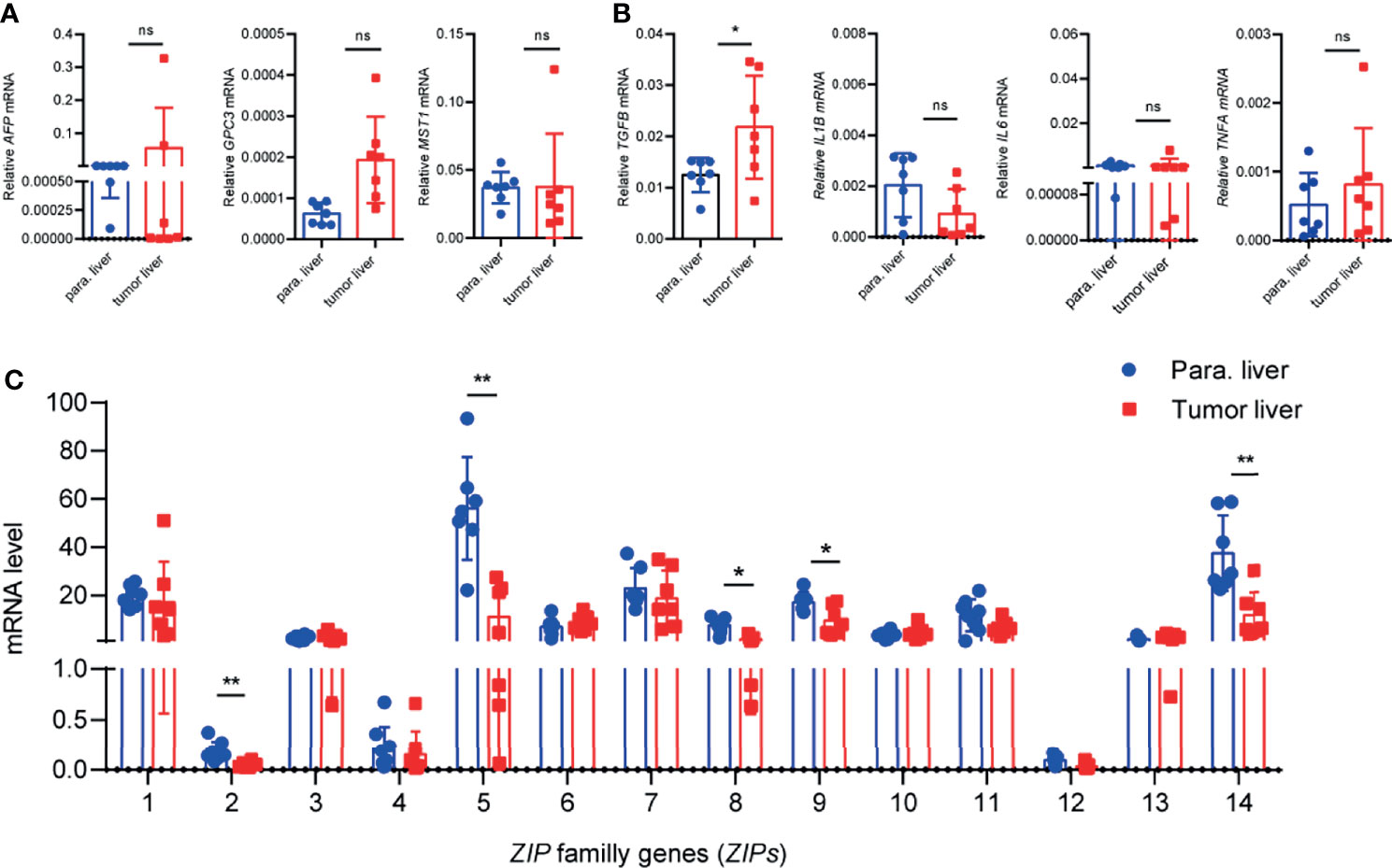
Figure 2 The transcription patterns of ZIPs in human liver cancer tissues and paracarcinoma liver tissues. (A) The mRNA levels of AFP, GPC3, MST1 were detected in human liver cancer tissues and paracarcinoma liver tissues by RT-PCR (n = 9). (B) Inflammation-related gene TGFB, IL1B, IL6, TNFA were measured in human liver cancer tissues and paracarcinoma liver tissues by RT-PCR (n = 9). (C) mRNA abundance profiles of ZIPs in human liver cancers and paracarcinoma liver tissues by RT-PCR (n = 9). The small circles represent the individual paracarcinoma liver tissues, and the squares represent the liver cancer tissues. The expressed values were calculated by the 2−ΔCT value. *P < 0.05, **P < 0.01, ns, not significant. The representative data from at least three independent experiments are shown. Data represent mean ± SD. By one-way ANOVA with Newman-Keuls or Holm-Sidak’s multiple comparisons test or Student’s t test.
The Transcription Profiles of a Group of Zips in Liver Cancer Tissues of DEN- and CCl4- Induced Mouse Model of HCC Are Decreased
To further determine whether Zip genes were involved in hepatocarcinogenesis, a DEN and CCl4- induced mouse model of HCC was established as described by previous studies (35, 36) (Figure 3A). Then observation of the mRNA changes of Zips in the stage of HCC was performed. At the 32th weeks of DEN injection, which mimicked the occurrence stage of HCC (37, 38), tumors were clearly observed on the surface of the livers. HE staining of liver sections showed an increased nuclear-to-cytoplasmic ratio of cancer cells. The normal hepatic lobular structure was completely destroyed in liver sections of these mice (Figure 3B). By FACS analysis using anti-F4/80+ antibody, we confirmed the accumulation of intrahepatic macrophages in DEN-induced mice compared with vehicle-treated mice (Supplemental Figure 1). Furthermore, the mRNA levels of Zips, in the liver tissues of these mice were measured by RT-PCR, the results showed that Zip14 gene transcription was down-regulated, which was consistent with bioinformatics data (Figure 1). Besides, the mRNA levels of Zip2 and Zip9 were also down-regulated, which was consistent with the transcriptional pattern data of clinical HCC samples (Figures 2C, 3C). In summary, Zip2, Zip9, and Zip14 transcriptions were dramatically decreased in cancer tissues from DEN and CCl4- induced mice model of HCC.
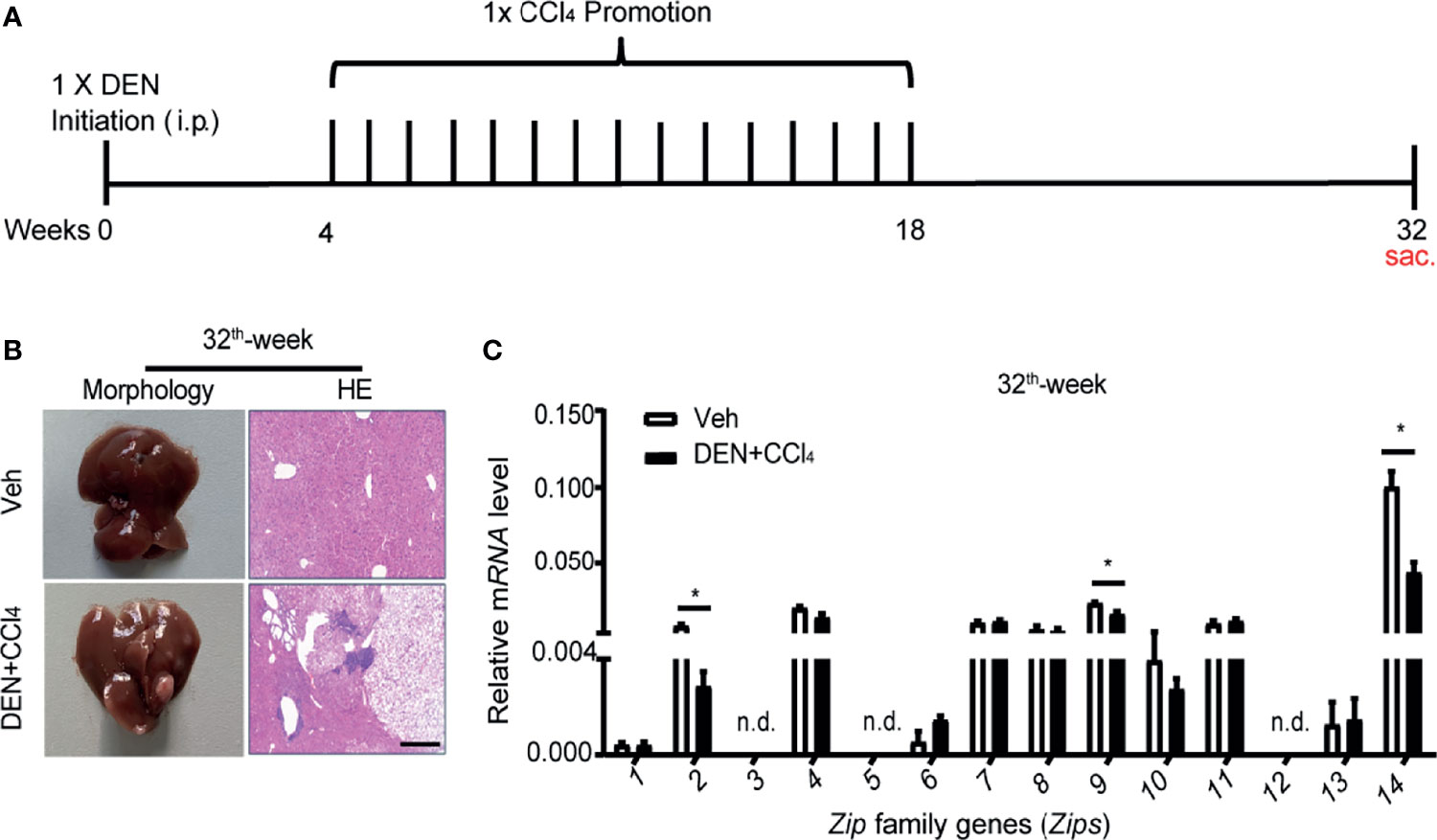
Figure 3 The transcription levels of Zips with DEN and CCl4- induced HCC model in mice. (A) Scheme of the DEN and CCl4- induced HCC model. WT male mice were treated with controls (Veh) or DEN (100 mg/kg) to induce HCC. Four-week later, control mice injected with olive oil (0.5 ml/kg; i.p.), whereas DEN treated mice were received CCl4 (0.5 ml/kg; i.p.) weekly. (B, C) After DEN treated 32 for weeks, the pathological characteristics were presented by morphology and HE staining of section of livers (B). The mRNA levels of Zips were detected by RT-PCR (C). Values are mean ± SD from 3 independent experiments. Using 2-tailed, unpaired Student’s t test (n = 7). P values are as followed: *P < 0.05, n.d., not detected.
The mRNA Levels of Zips Are Changed in M1 or M2 Polarized Macrophages
Since macrophages were suggested to play an important role in hepatocarcinogenesis previously (35), as reported before, we observed the infiltration of monocyte-derived macrophages in liver cancers by FACS analysis (Supplemental Figure 1). A large amount of CD68 positive macrophages were found in paracarcinoma tissue and liver cancers (Supplemental Figure 2). Given that macrophages could be typically categorized into M1 or M2 phenotypes by adapting to local microenvironment during the progress of HCC, we wondered whether Zips were involved in the polarization process of these macrophages. To address this, firstly, LPS plus IFNγ were added to the culture medium of the peritoneal elucidated macrophages (PEMs) to induce M1-polarized macrophages in vitro. Interestingly, the mRNA levels of M1 marker genes including Il6, Inos, Il1b and Tnfa were significantly augmented in PEMs (Figure 4A). These data strongly indicated that macrophages were polarized towards M1 phenotype. We then analyzed the transcription levels of Zips by RT-PCR in M1 phenotype macrophages. We observed that Zip2 and Zip7 mRNA were significantly increased in M1 macrophages (the mRNA of Zip2 was 7-fold higher than that in the untreated PEMs, P =0.032). Moreover, the mRNA levels of Zip6, Zip9, Zip10, Zip11, and Zip13 were comparable in M1 macrophages. But Zip5 and Zip12 were not detected in M1 macrophages (Figure 4B). We next detected the mRNA levels of Zips in M2-polarized macrophages. Under IL-4 and IL-13 stimulation, the mRNA levels of M2 marker genes including Ym1, Arg1, Fizz1, Il10, Il4 and Tgfb were detected. Ym1, Arg1, Fizz1 and Il10 mRNA expression were significantly increased in the M2- polarized macrophage cells compared with untreated cells (Figure 5A). Similarly, we also analyzed the transcription levels of Zips by RT-PCR in M2-polarized macrophages. The transcription levels of Zip2, Zip6, Zip7, Zip9, were elevated in M2 phenotype macrophages significantly. However, there were no significant differences in the mRNA levels of Zip1, Zip3, Zip4, Zip8, Zip10, Zip11, Zip13, and Zip14 (Figure 5B). Taken together, our data show that the mRNA levels of Zips are differently changed in M1 or M2 polarized macrophages.
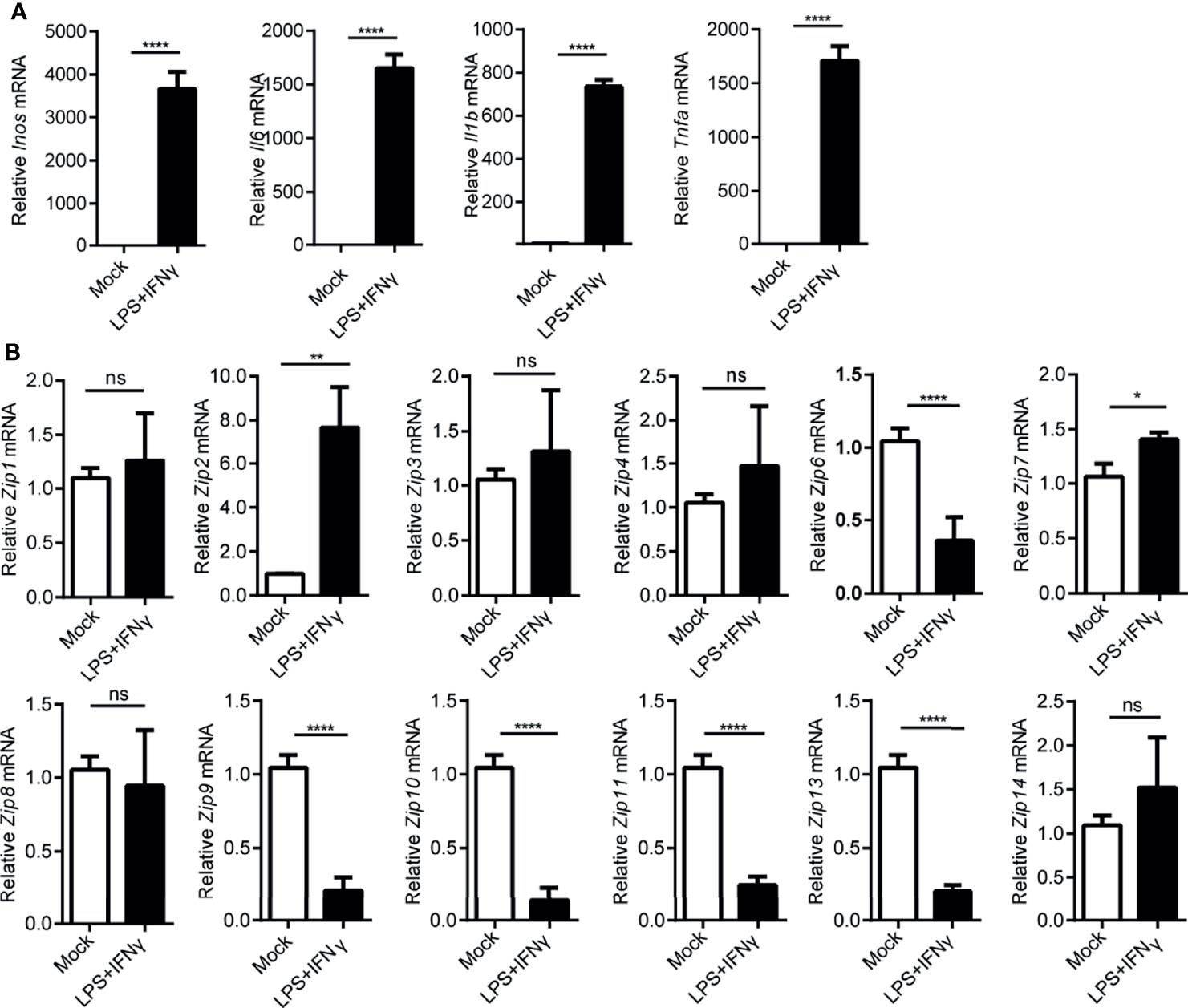
Figure 4 Transcription of Zip9 is downregulated during M1 macrophage polarity. (A) M1 macrophage characteristic genes were assessed by RT-PCR. M1 macrophages were generated by PEMs stimulating with LPS plus IFNγ. Data were normalized for β-actin mRNA levels. Results were presented as the average ± SD of three independent experiments. (B) PEMs were stimulated with LPS plus IFNγ for 6 h to measure Zips mRNA levels by RT-PCR. Results were presented as the average ± SD of three independent experiments. Data were normalized for β-actin mRNA levels. A, B was analyzed by Student’s t-test. The representative data from three independent experiments are shown. P values are as followed: *P < 0.05, **P < 0.01, and ****P < 0.0001, ns, not significant.
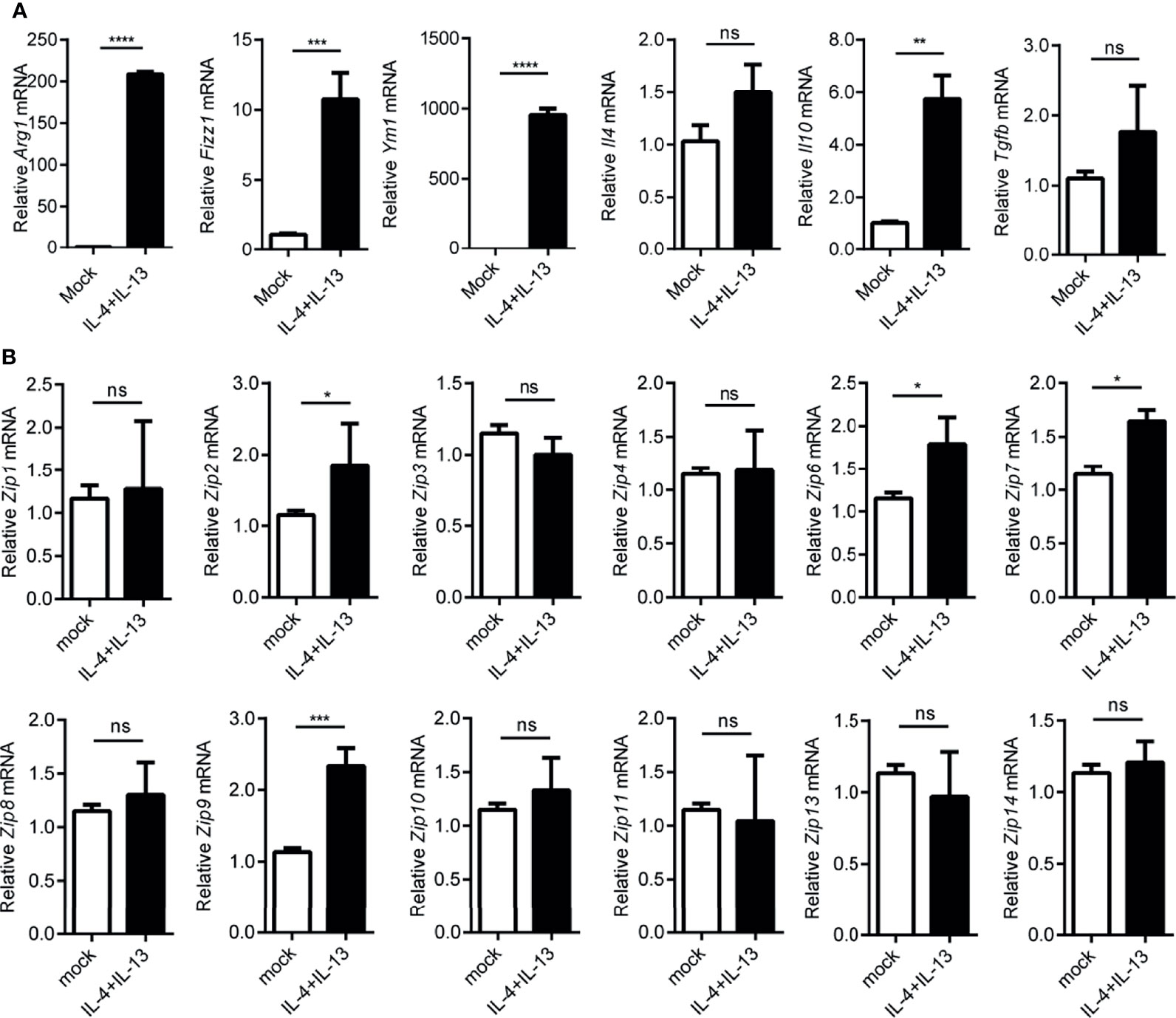
Figure 5 Transcription of Zip9 is upregulated during M2 macrophage polarity. (A) The mRNA levels of Arg1, Ym-1, Fizz1, and Il10, Il4, Tgfb were detected in IL-4 and IL-13 (20 ng/ml each) stimulated M2-polarized macrophages. (B) Zips transcription was detected by RT-PCR. PEMs were stimulated with IL-4 and IL-13 for 24 h. Data are shown as means ± S.D (n = 4). Data were normalized for β-actin mRNA levels. A, B was analyzed by two-tailed Student’s t-test. The representative data from three independent experiments are shown. P values are as followed: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, ns, not significant.
The Transcription of ZIP9 Is Decreased in Human TAMs From HCC Patients
To further confirm the transcriptional alternation of ZIP9, flow cytometry was used to sort tumor-associated macrophages (CD11B+ CD14high TAMs) generated from tumor liver tissues (tumor-TAMs) and paracarcinoma tissues (para-TAMs) of HCC patients. The process was shown in Figure 6A. The results showed that the percentage of tumor-TAMs was increased, as compared with para-TAMs (Figure 6B). The transcription levels of ZIPs were further verified by RT-PCR in these two patients-derived TAMs: tumor-TAMs and para-TAMs. Notably, a decrease in ZIP9 transcription was observed in tumor-TAMs. However, transcription of other ZIP genes, such as ZIP2, ZIP5, ZIP8, ZIP10, ZIP11, ZIP12, had no significant difference between tumor-TAMs and para-TAMs (Figure 6C). These findings indicate that ZIP9 is transcriptionally downregulated not only in HCC patient-derived TAMs but also in human liver cancer tissues, which suggests that ZIP9 may be involved in hepatocarcinogenesis by participating in TAMs differentiation.
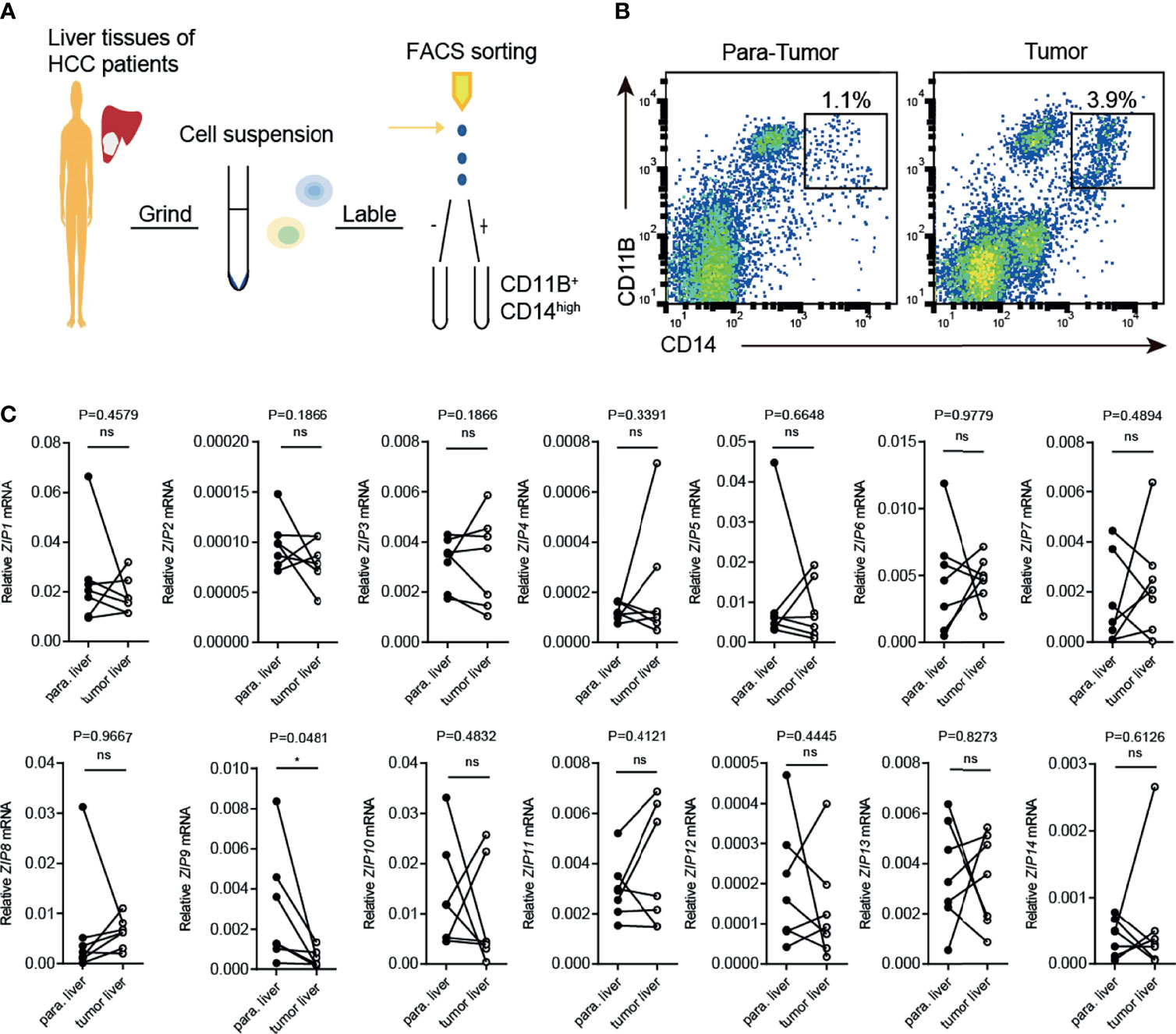
Figure 6 Transcription of Zip9 is downregulated in HCC patients-derived TAMs. (A) FACS protocol was used for isolating HCC patients-derived TAMs. (B) Human liver cancer tissues and paracarcinoma liver tissues were digested, stained for CD14, CD11B and then analyzed by flow cytometry. (C) The mRNA levels of ZIPs in TAMs isolated from human liver cancer tissues and paracarcinoma liver tissues were detected by RT-PCR (n = 7). The black points represent the individual paracarcinoma liver tissue, and the black circles represent the liver cancer tissue. The expressed values were calculated by the 2−ΔCT method. *P < 0.05, ns, not significant. The representative data from at least three independent experiments are shown. Data were determined with 2-tailed unpaired Student’s t test between 2 groups.
Zip9 Participates in Macrophage Polarization In Vitro
Above all, the results showed that ZIP2 (Zip2) and ZIP9 (Zip9) were significantly decreased in both cancer tissues of human HCC and mice HCC. In addition, the mRNA levels of Zip2 and Zip9 had significant changes in the polarized macrophages compared with native macrophages (Supplemental Figure 3A). These observations indicated that Zip2 and Zip9 might be involved in macrophages polarization. To investigate their roles in these processes, specific siRNA was used to knock down Zip2 or Zip9 in mouse PEMs, and the knockdown efficiency was confirmed by RT-PCR analysis (Figure 7A and Supplemental Figure 3B). The knockdown of Zip9 (SiZip9) significantly enhanced the transcription of M1 macrophage markers, such as Inos, Il1b and Tnfa (Figure 7B). However, reduced mRNA levels of M2 macrophage markers, such as Ym1, Arg1, Fizz1 were detected by RT-PCR in Zip9-specific knockdown macrophages (Figure 7C). Knockdown of Zip2 significantly promoted macrophages toward M1 polarized differentiation while knockdown of Zip2 had no changes in M2 polarization (Supplemental Figures 3C, D). Our data showed that Zip9, but not Zip2, could inhibit M1 macrophage polarization in response to LPS and IFNγ treatment in vitro and promote M2 macrophage polarization in response to IL-4 plus IL-13 stimulation in vitro. It had been reported that Zip8 could regulate the phosphorylation of IKK-β to attenuate the NF-κB signaling pathway (39), and increased cytosolic Zn2+ promotes the phosphorylation of STAT6 to enhance the anti-inflammation effects of IL-4 (40). To clarify the mechanism of how Zip9 is involved in regulating macrophage polarization, we determined the expression of signaling effectors of the NF-κB in M1 macrophages and IL4/STAT6 in M2 macrophages after Zip9 siRNA knockdown. As demonstrated, the levels of phosphorylation of IκBα/β (p-IκBα/β) and p65 (p-p65) were increased in the SiZip9 group following LPS plus IFNγ stimulation (Figure 7D) while phosphorylated STAT6 (p-STAT6) was significantly inhibited by Zip9 siRNA in IL-4 and IL-13 treated PEMs (Figure 7E). Taken together, our results indicate that Zip9 likely plays a role in classical M1/M2 polarization of macrophages. Zip9 inhibits M1 polarization by reducing phosphorylation of IκBα/β pathways while promoting M2 polarization by increasing phosphorylation of STAT6.
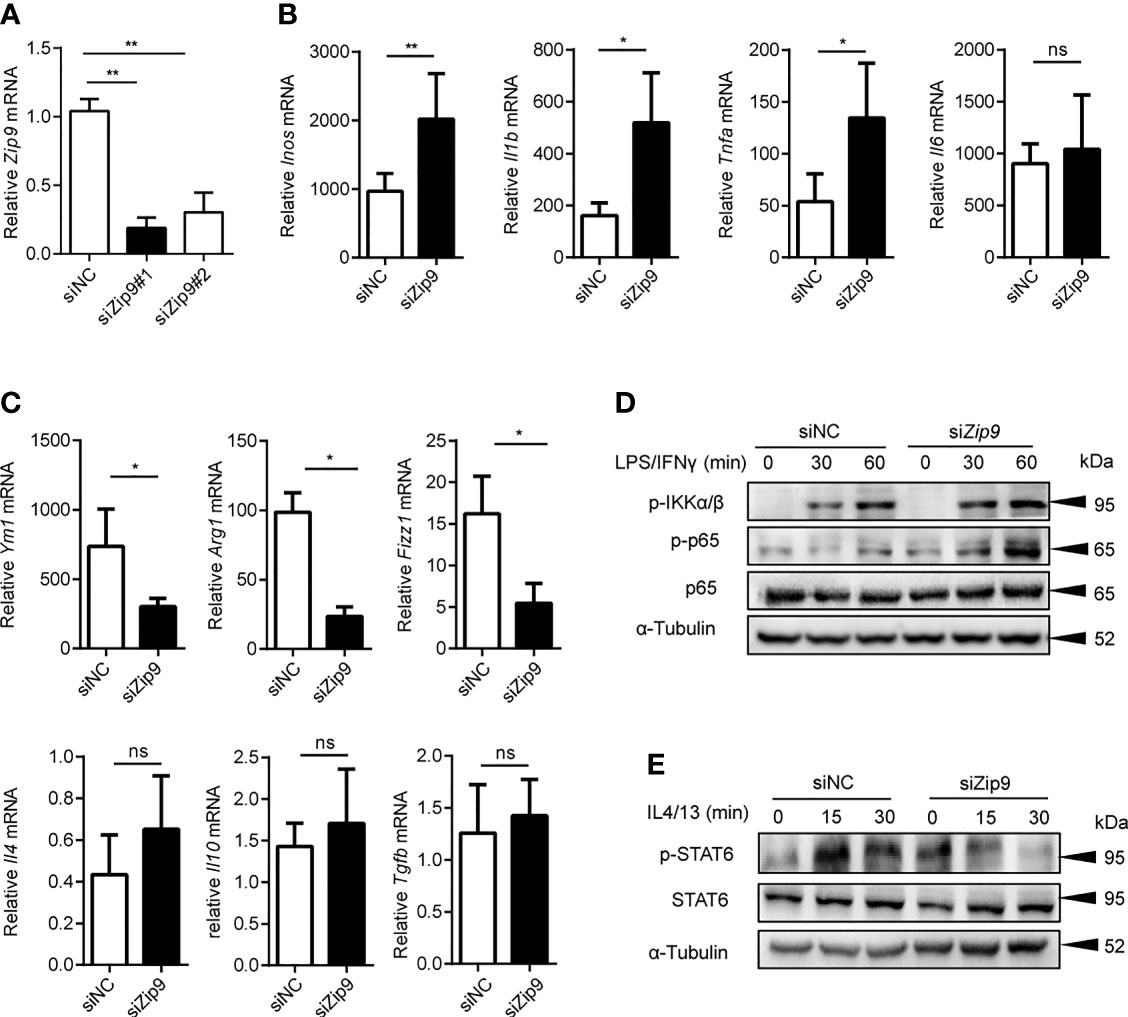
Figure 7 Zip9 modulates the macrophage polarity. (A) The siRNA knockdown efficiencies of Zip9. Zip9 was analyzed by RT-PCR following transfection with Zip9 siRNA 48 hours. (B) The analysis of M1-polarized macrophages characteristic genes (Inos, Il6, Il1b, Tnfa). Genes were detected by RT-PCR, following transfection with control small interfering RNA (siRNA) or Zip9 siRNA and stimulated with LPS plus IFNγ. (C) The analysis of M2-polarized macrophages characteristic genes (Ym1, Arg1, Fizz1, Il10, Il4, Tgfb). Genes were detected by RT-PCR, following transfection with control siRNA or Zip9 siRNA and stimulated with IL-4 and IL-13 (20 ng/ml each) *P < 0.05, **P < 0.01 and ns, not significant. Data were analyzed by Student’s t-test. Results were presented as the average ± SD of at least three independent experiments. (D) PEMs were pretreated with control small interfering RNA (siRNA) or Zip9 siRNA for 48h followed by 30/60 mins stimulation with IFNγ and LPS. Cell lysates were processed for immunoblotting with antibody against phospho-IκBα/β and phospho-p65 followed by stripping and reprobing with antibody against α-tubulin. (E) PEMs were pretreated with control small interfering RNA (siRNA) or Zip9 siRNA for 48h followed by 15/30 mins stimulation with IL-4 and IL-13. Cell lysates were processed for immunoblotting with antibody against phospho-STAT6 and total-STAT6 followed by stripping and reprobing with antibody against α-tubulin. Each panel is a representative experiment of at least three independent biological replicates.
Discussion
In this study, we first analyzed the mRNA transcription profiles of ZIPs in digestive system cancers by bioinformatics, and revealed the significant changes of these gene transcripts between cancers and healthy controls (Figure 1). Previous reports showed that ZIPs were involved in the pathology of various digestive system cancers in clinical analysis and experimental investigations. Costello et al. performed immunological staining on normal pancreas and pancreatic sections and found that ZIP3 was significantly reduced in pancreatic cancer tissues (17). Li et al. observed the expression of ZIP4 in 17 clinical pancreatic cancer samples, and found that ZIP4 was significantly overexpressed in 16 samples compared with surrounding normal tissues, and the expression of ZIP4 mRNA in human pancreatic cancer cells was significantly higher than that in human pancreatic duct epithelium cells, indicating that up-regulation of ZIP4 may be involved in the pathogenesis and progression of pancreatic cancer (19). Jin et al. also showed that exogenous ZIP4 can promote the growth of pancreatic cancer through subcutaneous BALB/c nude mouse models in vivo and statistics of clinical data, which indicated ZIP4 can be a new diagnostic biomarker for pancreatic cancer (11). Sheng et al. tested the expression of ZIP7 in human colorectal cancer and five colorectal cancer cell lines, they showed that the expression level of ZIP7 in colorectal cancers was higher than that in normal colon tissues, and when the ZIP7 gene was knocked-down on colorectal cancer cells, the viability and proliferation ability of colorectal cancer cells were significantly decreased. Therefore, they speculated that ZIP7 may be a potential oncogene, which played an important role in the occurrence and development of colorectal cancer (12). These investigations were consistent with our observations (Figure 1). We hypothesize that ZIPs might play roles in the pathogenesis and progression of liver cancers. However, the relevant research about total transcription pattern of ZIPs in HCC is not clear.
In fact, it has been shown that the zinc element in the peripheral blood and liver cancer tissues of LIHC patients is significantly reduced (5). Small-scale clinical trials reported that long-term zinc supplementation can help reduce the occurrence of liver cancer (41). ZIPs are responsible for the absorption of zinc in the body (10). Therefore, we speculate that the abnormal absorption of zinc in LIHC due to the abnormal transcription of the ZIPs affects the body’s immune defensive response. In order to further test our hypothesis, nine matched samples of LIHC patients’ cancer and para-cancerous tissues were collected for detecting ZIPs mRNA levels. The results showed that the mRNA levels of ZIPs in cancerous and para-cancerous tissues were different. Besides previously reported ZIP14 (42), the transcription levels of ZIP2, ZIP5, ZIP8 and ZIP9 were also found a substantial decrease in liver cancer tissues (Figure 2C). By establishing DEN and CCl4- induced mouse model of HCC, we showed that Zip2, Zip9 and Zip14 among all the families were down-regulated in liver tissues of the mice with HCC induction (Figure 3C). The clues are worthy of further in-depth research on their roles in hepatocarcinogenesis through genetically engineered mouse models.
The involvement of macrophages in the development of HCC has been reported (43). The polarization of macrophages into type-1 or type-2 had different effects on the occurrence and development of HCC (22). Li et al. reported that the decrease of MST1 is beneficial to the occurrence and development of HCC (35). Furthermore, we are curious about whether Zips are involved in the regulation of macrophage polarity. Interestingly, our observations with both patient samples and mouse models demonstrated a large number of macrophages infiltrated in the liver cancer tissues (Supplemental Figures 1, 2). Notably, macrophages have been suggested to play an essential role in inflammatory microenvironment. The involvement of ZIP members in macrophage polarity was also shown before. THP-1 monocytes or macrophages showed zinc deficiency by adding zinc chelating agent TPEN in vitro, the expression of ZIP1 did not change, whereas the mRNA of ZIP2 gene was significantly increased (34). In addition, the phagocytic function of the THP1 cells with ZIP7 knockdown was severely inhibited, and further restored by supplementing exogenous Zn2+ (44). The expression of M2-macrophages polarization marker CD206 in zip7-deficient macrophages was increased, while the expression of M1 marker NOS2 was decreased (44). ZIP8 was also reported to be stimulated by LPS and TNFα, resulting in a rapid increase in intracellular zinc levels (39). Accordingly, increased zinc concentration negatively regulated NF-κB by inhibiting the IκB kinase β subunit (IKKβ) in the kinase domain, thereby inhibiting inflammation and increasing the survival rate of immune cells (39). Mechanistically, NF-κB was also reported to regulate the expression of ZIP8 in monocytes, macrophages, dendritic cells and lung epithelial cells. In innate immunity, the deficiency of ZIP10 could reduce the number of macrophages and monocytes in the inflammatory response induced by LPS, which is caused by an increase in P53-dependent mortality (45). Using LPS to stimulate human primary macrophages, it was found that the mRNA transcription level of ZIP14 was up-regulated (46). All these studies have shown that the involvement of ZIPs in the function of macrophages. More interestingly, in our study, we found the transcription levels of Zips were differentially regulated during the polarization of macrophages. Then the transcription patterns of ZIPs on human liver cancer-derived TAMs were detected. The results showed that ZIP9 was down transcription in tumor-TAMs compared with para-TAMs. Here, we demonstrated that Zip9 upregulation macrophage M2 polarization via IL4/STAT6 pathway and downregulation macrophage M1 polarization via IκBα/β-p65 pathway mechanistically. Further investigation on the crosstalk between these signaling events in the cytosol of macrophages is needed to uncover the detailed molecular mechanism for the purpose of discovering new biomarkers for the diagnosis of HCC in the future.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Shanghai Zhongshan Hospital ethics committee. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Ethics Committee of Shanghai University.
Author Contributions
BW conceptualized and designed the study. YG and DY contributed to the conceptual design, writing, editing, and generation of figures for this manuscript. TT, XZ, RZ, JR, DT, YM, HZ, YL and YW participated in the experimental work. YG, DY and BW wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Ministry of Science and Technology of China (2016YFD0500207), the National Natural Science Foundation of China (81961160738, 81825011, 81571617 and 81930038) and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19030200).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the core facility and technical support in Shanghai University. We are grateful to Weiyun Li and Fang Zhang for technical support for the establishment of HCC mice model.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.725595/full#supplementary-material
References
1. Zhou R, Zhang J, Cheng Y, Zeng D, Sun H, Rong X, et al. Immune Cell Infiltration as a Biomarker for the Diagnosis and Prognosis of Digestive System Cancer. Cancer Immunol Immunother (2019) 68(3):433–42. doi: 10.1007/s00262-018-2289-7
2. Fidler MM, Bray F, Soerjomataram I. The Global Cancer Burden and Human Development: A Review. Scand J Public Health (2018) 46(1):27–36. doi: 10.1177/1403494817715400
3. Wu C, Lin J, Weng Y, Zeng D, Xu J, Luo S, et al. Myeloid Signature Reveals Immune Contexture and Predicts the Prognosis of Hepatocellular Carcinoma. J Clin Invest (2020) 130(9):4679–93. doi: 10.1172/JCI135048
4. Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in Liver Cancer and Possible Treatment Approaches. Biochim Biophys Acta Rev Cancer (2020) 1873(1):188314. doi: 10.1016/j.bbcan.2019.188314
5. Liaw KY, Lee PH, Wu FC, Tsai JS, Lin-Shiau SY. Zinc, Copper, and Superoxide Dismutase in Hepatocellular Carcinoma. Am J Gastroenterol (1997) 92(12):2260–3.
7. Margalioth EJ, Schenker JG, Chevion M. Copper and Zinc Levels in Normal and Malignant Tissues. Cancer (1983) 52(5):868–72. doi: 10.1002/1097-0142(19830901)52:5<868::aid-cncr2820520521>3.0.co;2-k
8. Khayyatzadeh SS, Maghsoudi Z, Foroughi M, Askari G, Ghiasvand R. Dietary Intake of Zinc, Serum Levels of Zinc and Risk of Gastric Cancer: A Review of Studies. Adv BioMed Res (2015) 4:118. doi: 10.4103/2277-9175.157849
9. Khoshdel Z, Naghibalhossaini F, Abdollahi K, Shojaei S, Moradi M, Malekzadeh M. Serum Copper and Zinc Levels Among Iranian Colorectal Cancer Patients. Biol Trace Elem Res (2016) 170(2):294–9. doi: 10.1007/s12011-015-0483-4
10. Kambe T, Tsuji T, Hashimoto A, Itsumura N. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol Rev (2015) 95(3):749–84. doi: 10.1152/physrev.00035.2014
11. Jin H, Liu P, Wu Y, Meng X, Wu M, Han J, et al. Aberrant Expression of Zinc Transporter ZIP4 (SLC39A4) Significantly Contributes to Human Pancreatic Cancer Pathogenesis and Progression. Proc Natl Acad Sci USA (2007) 04(47):18636–41. doi: 10.1073/pnas.0709307104
12. Sheng N, Yan L, You W, Tan G, Gong J, Chen H, et al. Knockdown of SLC39A7 Inhibits Cell Growth and Induces Apoptosis in Human Colorectal Cancer Cells. Acta Biochim Biophys Sin (Shanghai) (2017) 9(10):926–34. doi: 10.1093/abbs/gmx094
13. Jin HY, Liu P, Wu YH, Meng XL, Wu MW, Han JH, et al. Exosomal Zinc Transporter ZIP4 Promotes Cancer Growth and Is a Novel Diagnostic Biomarker for Pancreatic Cancer. Cancer Sci (2018) 109(9):2946–56. doi: 10.1111/cas.13737
14. Cui XB, Shen YY, Jin TT, Li S, Li TT, Zhang SM, et al. SLC39A6: A Potential Target for Diagnosis and Therapy of Esophageal Carcinoma. J Transl Med (2015) 13(1):321. doi: 10.1186/s12967-015-0681-z
15. Zhang Y, Bharadwaj U, Logsdon CD, Chen C, Yao Q, Lin M. ZIP4 Regulates Pancreatic Cancer Cell Growth by Activating IL-6/STAT3 Pathway Through Zinc Finger Transcription Factor CREB. Clin Cancer Res (2010) 16(5):1423–30. doi: 10.1158/1078-0432.CCR-09-2405
16. Ding B, Lou W, Xu L, Li R, Fan W. Analysis the Prognostic Values of Solute Carrier (SLC) Family 39 Genes in Gastric Cancer. Am J Transl Res (2019) 11(1):486–98.
17. Costello LC, Levy BA, Desouki MM, Zou J, Bagasra O, Johnson LA, et al. Decreased Zinc and Downregulation of ZIP3 Zinc Uptake Transporter in the Development of Pancreatic Adenocarcinoma. Cancer Biol Ther (2011) 12(4):297–303. doi: 10.4161/cbt.12.4.16356
18. Xu X, Guo HJ, Xie HY, Li J, Zhuang RZ, Ling Q, et al. ZIP4, A Novel Determinant of Tumor Invasion in Hepatocellular Carcinoma, Contributes to Tumor Recurrence After Liver Transplantation. Int J Biol Sci (2014) 10(3):245–56. doi: 10.7150/ijbs.7401
19. Zhu B, Huo R, Zhi Q, Zhan M, Chen X, Hua ZC. Increased Expression of Zinc Transporter ZIP4, ZIP11, ZnT1, and ZnT6 Predicts Poor Prognosis in Pancreatic Cancer. J Trace Elem Med Biol (2021) 65:126734. doi: 10.1016/j.jtemb.2021.126734
20. Xia C, Chen X, Li J, Chen P. SLC39A4 as a Novel Prognosis Marker Promotes Tumor Progression in Esophageal Squamous Cell Carcinoma. Onco Targets Ther (2020) 13:3999–4008. doi: 10.2147/OTT.S245094
21. Jin J, Li Z, Liu J, Wu Y, Gao X, He Y. Knockdown of Zinc Transporter ZIP5 (SLC39A5) Expression Significantly Inhibits Human Esophageal Cancer Progression. Oncol Rep (2015) 34(3):1431–9. doi: 10.3892/or.2015.4097
22. Sica A, Invernizzi P, Mantovani A. Macrophage Plasticity and Polarization in Liver Homeostasis and Pathology. Hepatology (2014) 59(5):2034–42. doi: 10.1002/hep.26754
23. Montes VN, Turner MS, Subramanian S, Ding Y, Hayden-Ledbetter M, Slater S, et al. T Cell Activation Inhibitors Reduce CD8+ T Cell and Pro-Inflammatory Macrophage Accumulation in Adipose Tissue of Obese Mice. PloS One (2013) 8(7):e67709. doi: 10.1371/journal.pone.0067709
24. Xu Y, Romero R, Miller D, Kadam L, Mial TN, Plazyo O, et al. An M1-Like Macrophage Polarization in Decidual Tissue During Spontaneous Preterm Labor That Is Attenuated by Rosiglitazone Treatment. J Immunol (2016) 196(6):2476–91. doi: 10.4049/jimmunol.1502055
25. Murray PJ. Macrophage Polarization. Annu Rev Physiol (2017) 79(1):541–66. doi: 10.1146/annurev-physiol-022516-034339
26. Toh ML, Aeberli D, Lacey D, Yang Y, Santos LL, Clarkson M, et al. Regulation of IL-1 and TNF Receptor Expression and Function by Endogenous Macrophage Migration Inhibitory Factor. J Immunol (2006) 177(7):4818–25. doi: 10.4049/jimmunol.177.7.4818
27. Covarrubias A, Byles V, Horng T. ROS Sets the Stage for Macrophage Differentiation. Cell Res (2013) 23(8):984–5. doi: 10.1038/cr.2013.88
28. Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, et al. Alternatively Activated (M2) Macrophages Promote Tumour Growth and Invasiveness in Hepatocellular Carcinoma. J Hepatol (2015) 62(3):607–16. doi: 10.1016/j.jhep.2014.10.029
29. Yao RR, Li JH, Zhang R, Chen RX, Wang YH. M2-Polarized Tumor-Associated Macrophages Facilitated Migration and Epithelial-Mesenchymal Transition of HCC Cells via the TLR4/STAT3 Signaling Pathway. World J Surg Oncol (2018) 16(1):9. doi: 10.1186/s12957-018-1312-y
30. Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, et al. Tumor-Associated Macrophages Produce Interleukin 6 and Signal via STAT3 to Promote Expansion of Human Hepatocellular Carcinoma Stem Cells. Gastroenterology (2014) 147(6):1393–404. doi: 10.1053/j.gastro.2014.08.039
31. Li Z, Wu T, Zheng B, Chen L. Individualized Precision Treatment: Targeting TAM in HCC. Cancer Lett (2019) 458:86–91. doi: 10.1016/j.canlet.2019.05.019
32. Baker RG, Hayden MS, Ghosh S. NF-κB, Inflammation, and Metabolic Disease. Cell Metab (2011) 13(1):11–22. doi: 10.1016/j.cmet.2010.12.008
33. Yu T, Gan S, Zhu Q, Dai D, Xiao Y. Modulation of M2 Macrophage Polarization by the Crosstalk Between Stat6 and Trim24. Nat Commun (2019) 10(1):4353. doi: 10.1038/s41467-019-12384-2
34. Hamon R, Homan CC, Tran HB, Mukaro VR, Lester SE, Roscioli E, et al. Zinc and Zinc Transporters in Macrophages and Their Roles in Efferocytosis in COPD. PloS One (2014) 9(10):e110056. doi: 10.1371/journal.pone.0110056
35. Li W, Xiao J, Wang H, Wei B. STK4 Regulates TLR Pathways and Protects Against Chronic Inflammation-Related Hepatocellular Carcinoma. Eur J Immunol (2016) 125(11):4239–54. doi: 10.1172/JCI81203
36. Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKK Beta Couples Hepatocyte Death to Cytokine-Driven Compensatory Proliferation That Promotes Chemical Hepatocarcinogenesis. Cell (2005) 121(7):977–90. doi: 10.1016/j.cell.2005.04.014
37. Uehara T, Pogribny IP, Rusyn I, The DEN. And CCl4 -Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma. Curr Protoc Pharmacol (2014) 66:14.30.1–10. doi: 10.1002/0471141755
38. Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology (2016) 150(4):835–53. doi: 10.1053/j.gastro.2015.12.041
39. Liu MJ, Bao S, Gálvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, et al. ZIP8 Regulates Host Defense Through Zinc-Mediated Inhibition of NF-κB. Cell Rep (2013) 3(2):386–400. doi: 10.1016/j.celrep.2013.01.009
40. Dierichs L, Kloubert V, Rink L. Cellular Zinc Homeostasis Modulates Polarization of THP-1-Derived Macrophages. Eur J Nutr (2018) 57(6):2161–9. doi: 10.1007/s00394-017-1491-2
41. Hosui A, Kimura E, Abe S, Tanimoto T, Onishi K, Kusumoto Y, et al. Long-Term Zinc Supplementation Improves Liver Function and Decreases the Riskof Developing Hepatocellular Carcinoma. Nutrients (2018) 10(12):1955. doi: 10.3390/nu10121955
42. Franklin RB, Levy BA, Zou J, Hanna N, Desouki MM, Bagasra O, et al. ZIP14 Zinc Transporter Downregulation and Zinc Depletion in the Development and Progression of Hepatocellular Cancer. J Gastrointest Cancer (2012) 43(2):249–57. doi: 10.1007/s12029-011-9269-x
43. Ritz T, Krenkel O, Tacke F. Dynamic Plasticity of Macrophage Functions in Diseased Liver. Cell Immunol (2018) 330:175–82. doi: 10.1016/j.cellimm.2017.12.007
44. Xie W, Xue Q, Niu L, Wong KW. Zinc Transporter SLC39A7 Relieves Zinc Deficiency to Suppressalternative Macrophage Activation and Impairment of Phagocytosis. PloS One (2020) 15(7):e0235776. doi: 10.1371/journal.pone.0235776
45. Gao H, Zhao L, Wang H, Xie E, Wang X, Wu Q, et al. Metal Transporter Slc39a10 Regulates Susceptibility to Inflammatory Stimuli by Controlling Macrophage Survival. Proc Natl Acad Sci USA (2017) 114(49):12940–5. doi: 10.1073/pnas.1708018114
Keywords: Zip91, hepatocellular carcinoma, macrophages polarity, Zip2, ZIPs
Citation: Gou Y, Yang D, Tian T, Zhu X, Zhang R, Ren J, Tu D, Luo Y, Miao Y, Zhao H, Wang Y and Wei B (2022) The Transcription of ZIP9 Is Associated With the Macrophage Polarization and the Pathogenesis of Hepatocellular Carcinoma. Front. Immunol. 13:725595. doi: 10.3389/fimmu.2022.725595
Received: 15 June 2021; Accepted: 22 February 2022;
Published: 15 March 2022.
Edited by:
Peter Brossart, University of Bonn, GermanyReviewed by:
Limin Zheng, Sun Yat-sen University, ChinaSamah Mamdouh, Theodor Bilharz Research Institute, Egypt
Copyright © 2022 Gou, Yang, Tian, Zhu, Zhang, Ren, Tu, Luo, Miao, Zhao, Wang and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Wei, d2VpYmlud2h5QHNodS5lZHUuY24=; Yu Wang, bmVjazEzMEBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Yingying Gou1,2†
Yingying Gou1,2† Bin Wei
Bin Wei