- Engineering Lab of Henan Province for Aquatic Animal Disease Control, College of Fisheries, Henan Normal University, Xinxiang, Henan, China
GSDME is the only direct executor of caspase-dependent pyroptosis in both canonical and non-canonical inflammasomes known to date in fish, and plays an important role in anti-bacterial infection and inflammatory response. In order to determine the regulation of GSDMEa on antibacterial infection in innate immune response, the CcGSDMEa gene in common carp (Cyprinus carpio haematopterus) was first identified and characterized, and then its function related to immune defense was investigated. Our results showed that the expressions of CcGSDMEa at the mRNA and protein levels were both significantly increased after Aeromonas hydrophila intraperitoneal infection at the early stage than that in the control group. We found that CcGSDMEa could be cleaved by inflammatory caspase (CcCaspase-1b) and apoptotic caspases (CcCaspase-3a/b and CcCaspase-7a/b). Interestingly, only the CcGSDMEa-NT (1-252 aa) displayed bactericidal activity to Escherichia coli and could punch holes in the membrane of HEK293T cells, whereas CcGSDMEa-FL (1-532 aa) and CcGSDMEa-CT (257-532 aa) showed no above activity and pore-forming ability. Overexpression of CcGSDMEa increased the secretion of CcIL-1β and the release of LDH, and could reduce the A. hydrophila burdens in fish. On the contrary, knockdown of CcGSDMEa reduced the secretion of CcIL-1β and the release of LDH, and could increase the A. hydrophila burdens in fish. Taken together, the elevated expression of CcGSDMEa was a positive immune response to A. hydrophila challenge in fish. CcGSDMEa could perform the pore-formation in cell membrane and the regulation on the secretion of IL-lβ, and further regulate the bacterial clearance in vivo. These results suggested that CcGSDMEa played an important role in immune defense against A. hydrophila and could provide a new insight into understanding the immune mechanism to resist pathogen invasion in teleost.
Introduction
The common carp (Cyprinus carpio haematopterus), as an important economic species, is famous for rich nutrition, fast growth rate and long-standing history of breeding, and contributes a lot to freshwater aquaculture (1). The expansion of farming scale and the emergence of intensive rearing bring economic benefits, but also lead to the outbreaks of various diseases at the meantime. Bacterial hemorrhagic septicemia caused by Aeromonas hydrophila is one of the main diseases, which is ferocious and causes fish massive die-offs in severe case. A. hydrophila is a Gram-negative short rod-shaped bacterium, which is widely distributed in various water and can cause severe diseases in fish, reptiles, amphibians and humans under certain conditions (2). In aquaculture, when environmental factors suddenly change and water quality deteriorates, A. hydrophila can infect various fish species, such as Indian major carp, mrigal Cirrhinus mrigala (3), rainbow trout Oncorhynchus mykiss (4), common carp C. carpio (5, 6), crucian carp Carassius auratus (7, 8), grass carp Ctenopharyngodon idellus (9), zebrafish Danio rerio (10), channel catfish Ictalurus punctatus (11–13), and tilapia Oreochromis niloticus (14), and lead to the outbreaks of bacterial hemorrhagic septicemia. This bacterial septicemia disease affected the quality and safety of the aquatic products, seriously restricted the sustainable development of healthy aquaculture, and resulted in considerable economic losses (2, 15). Hence, it is particularly important to determine the immune response of fish to the A. hydrophila infection, and to provide an effective strategy for controlling and preventing this bacterial septicemia.
In fish, innate immunity is considered as the major form of host defense, and it is also the efficient first line to resist invading pathogen (8). After the infection of A. hydrophila, the innate immune response is activated to improve the immune ability in teleost, and effectively eliminate pathogenic bacteria. Studies have shown that gasdermin E (GSDME) plays an indispensable role in combatting pathogen invasion through its NT-domain in teleost (16–20). The GSDME is composed of a conserved N-terminal pore-forming domain (N-PFD), a C-terminal auto-inhibitory domain (C-AID), and an intermediate linker region with a caspase hydrolysis site (18, 19, 21, 22). Under the normal condition, GSDME exists in inactive form with the full-length sequence in vivo, but under the stimulation of pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), Caspase-1/3/7 are activated and cleave GSDME in the linker region to release the NT domain, and then GSDME-NT oligomerization can penetrate cellular membrane, destroy cytomembrane integrity and lead to pyroptosis. Finally, the membrane rupture cells, as well as PAMPs and DAMPs, are phagocytosed by neutrophils to clear pathogenic bacteria and damaged cells (18, 19, 23–26).
Accompanying pyroptosis occurrence, massive cytoplasmic contents such as interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), lactate dehydrogenase (LDH) and ATP were released into the extracellular area at the same time (27, 28). As a key pro-inflammatory cytokine, IL-1β can trigger an inflammatory reaction at the site of pathogenic infection to clear pathogens (29–32). However, IL-1β, without signal peptide, could not be directly secreted to the outside of cell. Previous studies have shown that the predicted maximum diameter of IL-1β was 4.5 nm, and the intracellular IL-1β could be released to the extracellular through the pores formed by gasdermin (33, 34). However, LDH could not pass through the gasdermin pore because the predicted maximum diameter of LDH tetramer was 9.6 nm, and it could only be released upon membrane rupture (35). Hence, only IL-1β, not LDH, is released from intracellular area to extracellular area, which can be used as biological indicator of membrane pore formed by GSDME in fish (35). Both IL-1β and LDH are released into the extracellular simultaneously, and it is indicated that cell membrane rupture and pyroptosis appear.
Previous studies have shown that zebrafish DrGSDMEa and DrGSDMEb, turbot (Scophthalmus maximus) SmGSDMEb and tongue sole (Cynoglossus semilaevis) CsGSDME could be cleaved by caspases and formed the membrane pores to cause pyroptosis (16–19). However, up to now, the function of GSDMEa in common carp is still unclear. Herein, we investigated the roles of CcGSDMEa in the pore-formation of cellular membrane and regulation of IL-1β secretion after A. hydrophila infection. This study aims to clarify the roles of CcGSDMEa in immune response of common carp and provide a new insight into protecting fish against pathogen infection.
Materials and methods
Experimental animals and cell culture
Experiments involving live animals were conducted in accordance with the ‘‘Regulations for the Administration of Affairs Concerning Experimental Animals’’ promulgated by the State Science and Technology Commission of Henan Province. This study was approved by the Animal Care and Use Ethics Committee of the Henan Normal University.
The healthy common carp (23 ± 2 g, 11 ± 1 cm) were from the aquaculture base in Zhengzhou, Henan province. Before the experiment, the fish were randomly grouped with 50 fish in each group, and then acclimated in 200 L tanks for two weeks. During acclimation, the fish were maintained at 25 ± 2 °C, and pH 7.0 ± 0.2, with the dissolved oxygen concentration of 6.0 ± 0.2 mg/L and natural light/dark cycle, and 1/3 water in tank was exchanged with the fresh aerated water twice a week. The fish were fed with pellets at 5% body weight. The common carp (4% of stock) were randomly sampled for bacterial or virus detection, and no pathogen was detected in the sampled fish, the experimental fish were considered to be healthy (36).
Mycoplasma-free Human Embryonic Kidney (HEK) 293T cells were cultured in DMEM medium (Hyclone, USA) supplemented with 10% (v/v) Fetal bovine serum (FBS, Gibco, USA), 2 mM L-Glutamine, and 4.5 g Glucose, at 37 °C in a 5% CO2 incubator. For mycoplasma-free Epithelioma papulosum cyprini (EPC) cell, the M199 medium (Hyclone, USA) supplemented with 10% FBS, 1000 mg D-Glucose, 2200 mg NaHCO3, and 100 mg L-Glutamine was used, and cultured at 25˚C in an incubator with 5% CO2. The cells were passaged every 2-3 d at a split ratio of 1:3 when the cells reached 90% confluency.
Bacterial strain culture
A. hydrophila, which was stored in our laboratory and isolated from common carp (15), cultured on Luria-Bertani broth (LB) plates at 28°C overnight, and single colony of each strain was then inoculated into 5 mL LB medium at 28 °C with constant shaking (180 rpm), and then sub-cultured in 100 mL LB medium with the ratio of 1:100 at 28 °C at 180 rpm until the absorbance values (OD600) was 0.6 (37). The half lethal concentration (LC50) of bacteria was 7 × 107 CFU/mL. The bacteria were harvested by centrifugation at 8000 rpm at 4 °C for 10 min, washed three times with 0.75% physiological saline (0.75% NaCl), and then resuspended in 0.75% NaCl with the concentration of 7 × 106 CFU/mL.
RNA extraction and cDNA synthesis
Total RNA was extracted from the samples using RNA extraction reagent RNAiso (TaKaRa, Japan) on the basis of the protocol of manufacture. Total RNA was dissolved in the RNase free water (Sangon, China). The concentration was determined by NanoDrop 2000 spectrophotometer (Thermo company, USA), and the purity was detected through 1% agarose gel electrophoresis. First-strand cDNA used for gene cloning was synthesized using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Japan), and the PrimeScript™ RT Master Mix was used to synthesize cDNA template for quantitative Real Time PCR (qRT-PCR) (Vazyme, China). The reverse transcription products were stored at -20°C until used.
Gene cloning and bioinformatics analysis
The open reading frame (ORF) amplification of CcGSDMEa was performed in a 20 μL reaction system, which was made up of 1 μL cDNA template, 1 μL forward primer, 1 μL reverse primer, 10 μL 2 × Taq Master Mix, and 7 μL ddH2O. The PCR procedure was set as follows: 5 min at 94 °C for pre-denaturation, 34 cycles (30 s at 94°C for denaturation, 30 s at 58 °C for annealing, and 120 s at 72 °C for extension), and 10 min at 72°C for final extension. The products were analyzed on 1.5% agarose electrophoresis and purified by DNA gel extraction kit (Omega, USA). The purified DNA was ligated into the pMD-19T vector (Takara, Japan), and then transformed into Escherichia coli DH5α (Biomed, China). Positive clones were sequenced in Sangon Biotech Company (Shanghai, China).
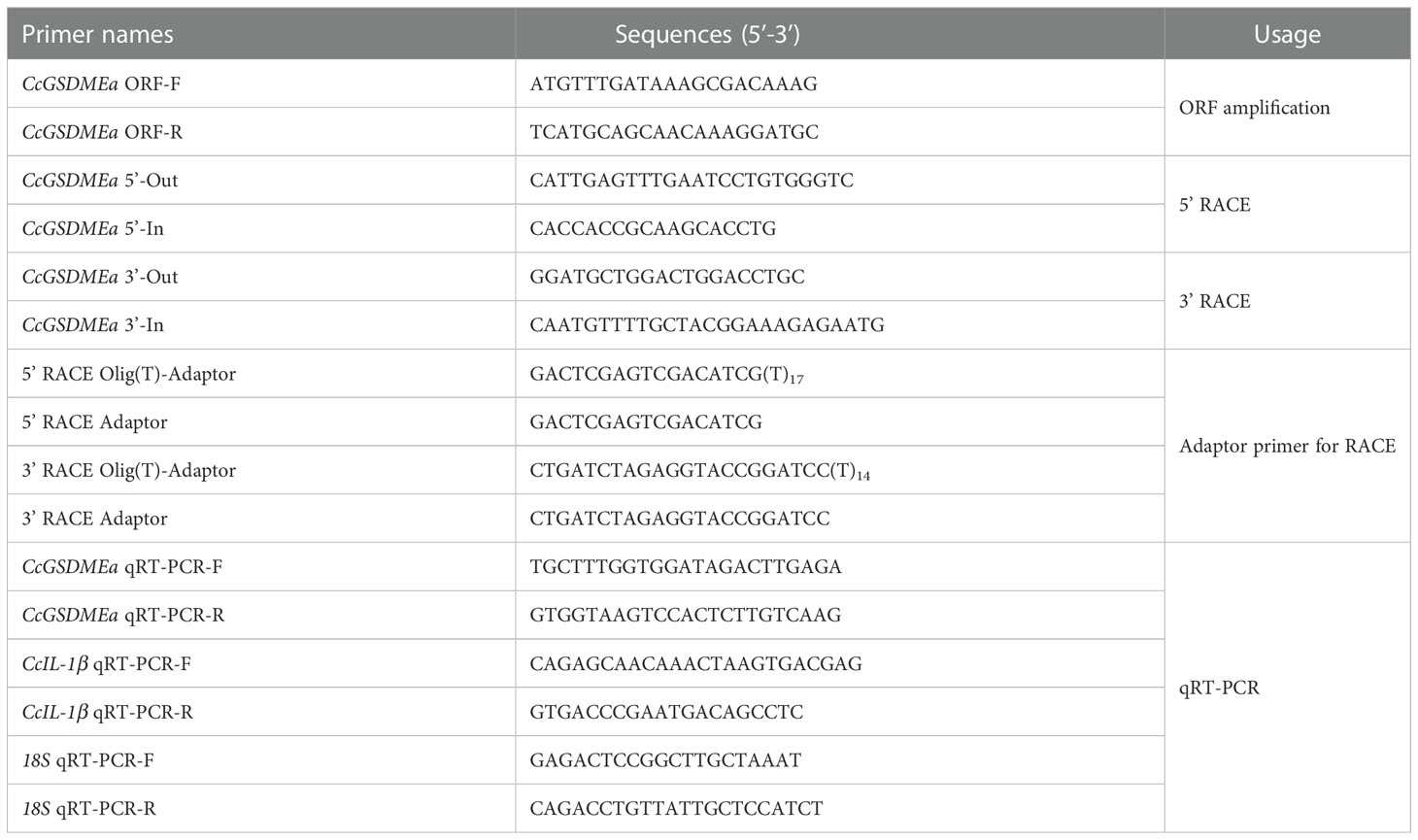
The rapid amplification of cDNA end (RACE) method was used to amplify the 3’ and 5’ end sequences of CcGSDMEa. According to the instruction of PrimeScript™ II 1st Strand cDNA Synthesis Kit, the Oligo dT Primer was replaced by the 3’ RACE Olig(T)-adaptor for reverse transcription to obtain the 3’ RACE template. With regard to 5’ RACE amplification template, the obtained cDNA product was purified with E.Z.N.A. Cycle-pure Kit (Omega, USA), and Poly (A) tail was added at the 3’ end after purification. A total of 50 μL reaction system was as follows: cDNA 10 μL, 5 × TdT buffer 10 μL, 1% BSA 5 μL, dATP (10 mmol/L) 2.5 μL, TdT enzyme 1 μL, and ddH2O 21.5 μL. Reaction condition was at 37 °C for 30 min and at 80 °C for 3 min. The obtained final product was 5’ RACE template. The PCR amplification was performed using the primers of 3’/5’ -Out and 3’ RACE Adaptor/5’ RACE Olig(T)-Adaptor for the first round PCR, and then followed by the nested PCR with the primers of 3’/5’ -In and 3’/5’ RACE Adaptor using the first round PCR products (diluted 50 times) as the template. The reaction system and PCR procedure were same as ORF amplification, except only 20 cycles for first round PCR. The primers used for gene cloning were shown in Table 1.
The amplified ORF was overlapped with the 3’/5’ terminal sequences to obtain the full length of CcGSDMEa cDNA sequence, implemented in software DNAMAN. CLC Main Workbench 6.8 was used to translate the amino acid sequences. Theoretical isoelectric point (pI) and molecular weight (MW) were respectively calculated on the basis of deduced amino acid sequences by the Compute pI/MW software (https://web.expasy.org/compute_pi/). The domains were predicted and annotated with Simple Modular Architecture Research Tool-SMART (http://smart.embl-heidelberg.de/), and the three-dimensional structure of protein was predicted by I-TASSER (https://zhanggroup.org/I-TASSER). Homologous sequences were searched and analyzed by the BLAST algorithm of NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were carried out using the DNAMAN 8.0, and phylogenetic tree was constructed using the neighbor-joining method, implemented in MEGA 7.0 program with a bootstrap test of 1000 replicates.
Spatial expression analysis by qRT-PCR
Various tissues including gill, liver, spleen, head kidney, trunk kidney, heart, intestine, brain, muscle, and skin were taken aseptically from six healthy fish (23 ± 2 g), respectively. Total RNA extraction and cDNA synthesis were performed as described above. The AceQ qPCR SYBR Green Master Mix (Vazyme, China) and LightCycler® 480 II instrument (Roche, Switzerland) were used for qRT-PCR to detect gene expression at mRNA level. The qRT-PCR was performed in a 20 μL reaction system (10 μL 2 × AceQ qPCR SYBR Green Master Mix, 1 μL specific primers, 3 μL cDNA template and 6 μL ddH2O), and cycling program (180 s at 95 °C for pre-incubation, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s for amplification). The amplification efficiency of qRT-PCR primers for CcGSDMEa and CcIL-1β gene was 97.34% and 99.07%, respectively. The 18S was used as the reference gene, and the amplification efficiency was 100.8% (Table 1). The results were analyzed by the 2-ΔΔCt method.
A. hydrophila challenge and sampling
The healthy common carp (23 ± 2 g) were randomly divided into two groups (the treated group and the control group), with 50 fish each group. Fish were injected intraperitoneally with 100 μL A. hydrophila suspension at a concentration of 7 × 106 CFU/mL in treated group. In the control group, the fish were injected with 0.75% NaCl in the same volume. The experiments were carried out in triplicate. The fish were not fed during the experiment period and maintained under the same conditions with those during the acclimation period. The mortality of fish was observed and recorded every day. After A. hydrophila challenge, six fish from each group were randomly selected at 0 h, 3 h, 6 h, 12 h, 24 h, 48 h and 96 h, respectively, which were anesthetized with ethyl 3-aminobenzoate (MS-222, Sigma, USA), and then sampled. Blood was collected from the caudal vein and centrifuged at 3000 rpm at 4 °C for 10 min to collect serum, which was used for Enzyme-Linked Immunosorbent Assay (ELISA). The tissues (gill, liver, spleen, head kidney, intestine and skin) were sampled immediately and stored in liquid nitrogen. Total RNA extraction, cDNA synthesis, and qRT-PCR were carried out as described above.
Plasmid construction
The different forms of CcGSDMEa were cloned into His-tagged pET-32a vector or GFP-tagged pEGFP-C1 vector through Hind III and KpnI sites to generate rCcGSDMEa-FL (1-532 aa) or GFP-CcGSDMEa-FL, rCcGSDMEa-NT (1-252 aa) or GFP-CcGSDMEa-NT and rCcGSDMEa-CT (257-532 aa) or GFP-CcGSDMEa-CT recombinant plasmids. CcCaspase-1a/1b/3a/3b/7a/7b sequences were inserted into N-DmrB-pcDNA3.1-3HA expression vector through EcoR I and KpnI sites, respectively. The CcGSDMEa, CcCaspase-1a/1b/3a/3b/7a/7b, pET-32a (+) vector, pEGFP-C1 vector and N-DmrB-pcDNA3.1-3HA vector were digested by restriction enzymes (New England Biolabs, USA), and then they were purified and ligated together by the T4 ligation enzyme (TakaRa, Japan). The recombinant plasmids were transferred into E. coli strain DH5α, and the positive clones were verified by sequencing. The primers used to construct plasmids were listed in Table 2. To remove the endotoxin, the constructed recombinant eukaryotic plasmids were extracted with an EndoFree Plasmid Kit following the instruction of manufacturer (Omega, USA).
CcGSDMEa cleavage assay
A total of 5 × 106 EPC cells were seeded per well in six-well plates for 16 h, then co-transfected with GFP-CcGSDMEa-FL (2.5 μg) and CcCaspase-1a/1b/3a/3b/7a/7b (2.5 μg) plasmids using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, USA) according to the instructions when the cells reached 90% confluency. 36 h later, the EPC cells were exposed to 1 μM AP20187 (a chemical inducer of dimerization, which could penetrate the cell membrane and induce the DmrB fusion protein to dimerize. Selleck, China) for 6 h to mediate CcCaspase-1a/1b/3a/3b/7a/7b dimerization (38, 39). Next, the cells were collected, and the total proteins were extracted using the protein extraction kit (Solarbio, China) according to the specification. In detail, the EPC cells were lysed in lysis buffer supplemented with 1% PMSF, protease inhibitor and phosphatase inhibitor on ice for 15 min, and then centrifuged at 12000 rpm at 4 °C for 10 min. Protein concentration was detected by BCA assay kit (Solarbio, China). Subsequently, the protein was mixed with 5 × protein loading buffer (Solarbio, China) and boiled for 10 min, and then used for western blotting.
Immunocytochemical staining
The HEK293T cells (5 × 105 cells/well) were seeded on coverslips into 6-well cell culture plates overnight, and the plasmids expressing GFP-CcGSDMEa-FL, GFP-CcGSDMEa-NT and GFP-CcGSDMEa-CT (1000 ng/well) were transfected with Lipofectamine 3000 Reagent when the cells reached 70% confluency, and the pEGFP-C1 vector was used as the control.
At 24 h after transfection, the cells were collected, and washed three times with sterile 1 × PBS buffer (Solarbio, China), and then fixed with 4% paraformaldehyde at room temperature for 1 h, washed three times. The cells were incubated with 0.1% Hoechst 33342 (Invitrogen, USA) for ten minutes for the sake of nucleus staining, or incubated with 0.1% DiI (Beyotime, China) for 10 min for the sake of cell plasma membrane staining, and then washed three times. The cell coverslips were mounted on glass slides with Antifade Mounting Medium (Beyotime, China) and air-dried at room temperature. The fluorescence images of cells were observed under a fluorescence microscope (Zeiss, Germany). In addition, the HEK293T cells were fixed with 2.5% glutaraldehyde at 4 °C overnight, and dehydrated using series gradient alcohol and tertiary butanol, and then the integrity of cytomembrane was observed under a biotypic scanning electron microscope (JEOL, Japan).
Assay for antibacterial effect and cytotoxicity
The different forms of rCcGSDMEa recombinant plasmids were transferred into E. coli strain BL21 (DE3), and the transformants were cultured in LB with 100 μg/mL Ampicillin (Amp), shaken at 200 rpm at 37 °C until OD600 to 0.6. The bacteria diluted at 104 times, were evenly spread with 50 μL on LB agar plates with or without 0.1 mM IPTG, and then cultured at 37 °C overnight. The CFU on the plates was counted and statistically calculated. At the same time, the growth curve of E. coli was tested by measuring the OD600 after induction by IPTG.
The cytotoxicity was monitored by evaluating the release of LDH from cells. The cell culture medium was centrifuged at 400 g for 5 min at 4°C, and the supernatant was collected. The LDH content released from the cells was detected according to the instruction of the Lactate Dehydrogenase Cytotoxicity Detection Kit (Beyotime, China). The untransfected cells were used as the blank background control, and the LDH released from the cells treated with LDH release agent was used as the positive control with maximum enzyme activity. Each sample was tested in triplicate. Cytotoxicity (%) = (treated sample A490 - blank background control A490)/(maximum enzyme activity A490 - blank background control A490) × 100.
Assay for bacterial clearance
The healthy fish (9 ± 0.5 g) were randomly divided into five groups (i.e., Control group: PBS; Knockdown groups: siControl group and siCcGSDMEa group; Overexpression groups: pEGFP-C1 group and GFP-CcGSDMEa-FL group). The primers of siRNAs (siCcGSDMEa-F/R, 5’-GGA UCU AUA GAG UUG GGA ATT-3’, 5’-UUC CCA ACU CUA UAG AUC CTT-3’; siControl-F/R, 5’-UUC UCC GAA CGU GUC ACG UTT-3’, 5’-ACG UGA CAC GUU CGG AGA ATT-3’) were designed and synthesized by Sangon Biotech Company (Shanghai, China). For the different knockdown groups, the fish were intramuscularly injected with 100 μL (10 μg) siControl or siCcGSDMEa, respectively. The fish were intramuscularly injected with 100 μL (10 μg) GFP-CcGSDMEa-FL or pEGFP-C1 plasmid in different overexpression groups, respectively (29, 40). In the control group, the fish were injected with 100 μL PBS by the same method. The experiments were carried out in triplicate. After 3 and 7 d, the tissues (gill, liver, spleen, trunk kidney and head kidney) were sampled in the aseptic condition and used for analyzing the efficiency of knockdown or overexpression by RT-qPCR.
In knockdown groups, overexpression groups and the control group, the fish were intraperitoneally injected with 100 μL A. hydrophila suspension (7× 106 CFU/mL) after knockdown or overexpression 3 d. The experiments were carried out in triplicate. After challenge for 24 h, the tissues were sampled respectively in the aseptic condition, and used to count the bacterial number in samples by plate count and detect the expression of CcIL-1β by RT-qPCR. Blood was collected from the caudal vein and centrifuged at 3000 rpm at 4 °C for 10 min to collect the serum, which was used for ELISA to measure the content of CcIL-1β.
Assay for CcIL-1β secretion
EPC cells (5 × 106 cells/well) were seeded into 6-well cell culture plates overnight, and then the siControl, siCcGSDMEa, pEGFP-C1 and GFP-CcGSDMEa-FL plasmids (1000 ng/well) were transfected when the cells reached 90% confluency, respectively. The untransfected cells were used as the control. The efficiency of knockdown or overexpression in vitro was analyzed after 24 h and 48 h, respectively. The untransfected cells and the cells transfected successfully with the above plasmids were divided into two subgroups, respectively. One group was stimulated by 10 μL A. hydrophila (1×104 CFU/well), and the other group was added with isometric PBS. After 3 h, the content of LDH and CcIL-1β in cell culture medium was assessed to determinate the role of CcGSDMEa in regulating IL-1β secretion.
ELISA
The levels of CcGSDMEa and CcIL-1β in serum were measured according to the method of ELISA (6, 40). Briefly, 96-well Immuno-Maxisorp plates were coated with the serum (1:1) in 2× coating buffer (0.05 M carbonate buffer, pH 9.6) overnight at 4 °C. Next, 10% skimmed milk powder was used as the blocking agent at 37 °C for 2 h, and then were subsequently washed three times (wash buffer: 20 mM PBST, 274 mM NaCl, 5.4 mM KCl, 20 mM Na2HPO4, 3.6 mM KH2PO4, 0.5 mL Tween-20, PH 7.4). Polyclonal antibodies against CcGSDMEa or CcIL-1β (1: 500, prepared in our laboratory) were added to the wells to incubation at 37 °C for 2 h and an addition wash. After incubation with horseradish peroxidase labeled Goat anti-Mouse IgG/HRP (Solarbio, China) at 37 °C for 1 h, the plates were washed again. 3,3’,5,5’ -tetramethylbenzidine (TMB, Sigma, USA) was used as the substrate for color development at 37 °C for 30 min. The reaction was terminated by adding 50 μL of termination solution (2M H2SO4). The OD450 absorbance value was detected in a microplate reader (PerkinElmer, USA) within 10 min, and the amount of excretive CcGSDMEa and CcIL-1β (pg/mL) was estimated by comparing to reference curves constructed using the corresponding standards.
Western blotting analysis
The protein samples (30 μg) were separated by SDS-PAGE before being transferred to a PVDF membrane (Millipore, USA) according to traditional protocol. The membranes were blocked in 5% w/v nonfat dry milk in TBST and then washed three times using TBST for 10 min every time. The mouse anti-GFP Ab (1:10,000, GenScript, China) or mouse anti-β-actin Ab (1:10,000, Beyotime, China) was incubated overnight at 4°C and washed another three times, then followed by incubation with the appropriate secondary goat anti-mouse IgG-HRP Ab (1:5000; Solarbio, China) and washed three times again, whereafter detected by super ECL detection reagent (Millipore, USA) using gel imaging system (Biorad, USA).
Statistical analysis
All values in this study were presented as means ± standard deviation (M ± SD) (n=3). The data statistical analysis was performed by SPSS 20.0 and GraphPad Prism 5.0. The statistical significance was determined using Student’s t test and one-way ANOVA followed by Tukey’s test. Significant difference was set as P <0.05 (*), P <0.01 (**), or P <0.001 (***).
Result
Structural analysis of CcGSDMEa
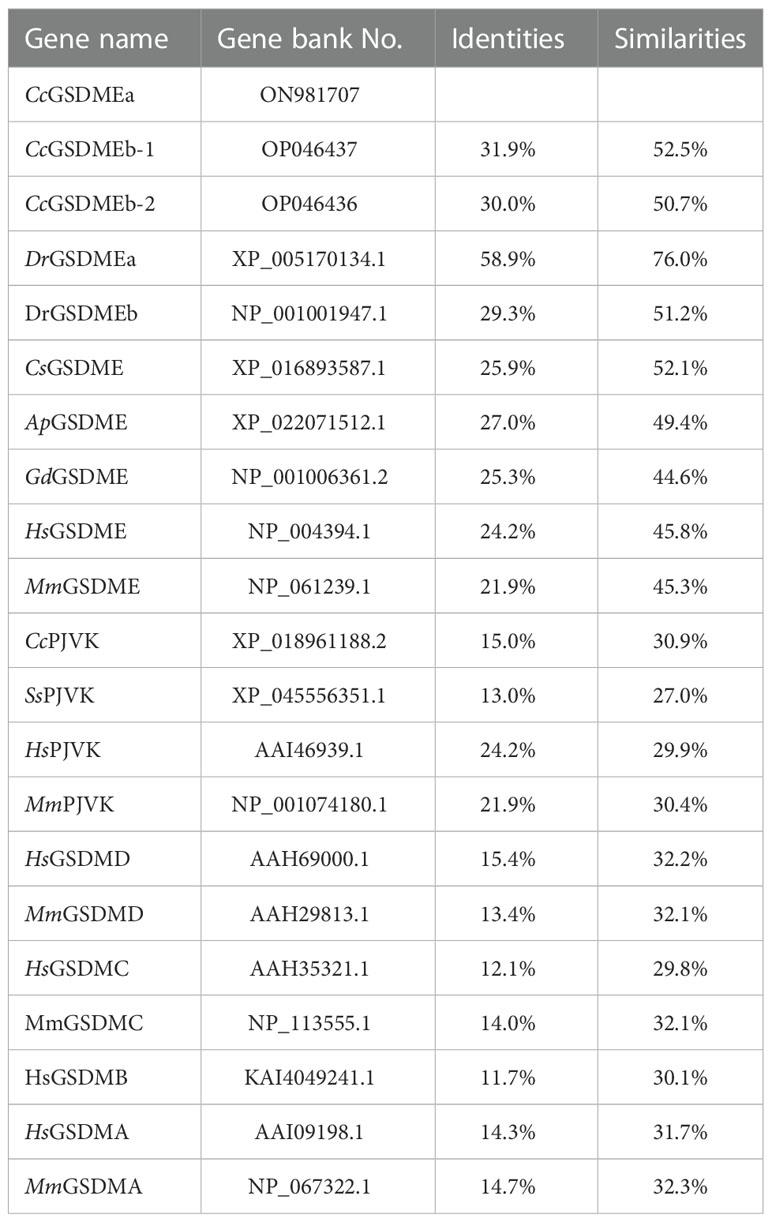
In this study, CcGSDMEa (GenBank accession number: ON981707), a homologous GSDME gene, was obtained from common carp, which is distributed within 7320 bp genomic fragment on chromosome A19 with 9 exons and 8 introns. The location and transcription direction of the adjacent genes to CcGSDMEa were consistent with those of DrGSDMEa and HsGSDME, and it was suggested that genes adjacent to CcGSDMEa loci shared an overall conserved chromosome synteny with those of humans and zebrafish (Figure 1A). The full length of CcGSDMEa cDNA sequence is 2736 bp with a 1599 bp ORF, a 177 bp 5’-untranslated region and a 960 bp 3’-untranslated region (Figure S1). The ORF encoded 532 amino acids was predicted with molecular weight of 58.57 kDa and theoretical isoelectric point of 4.92. It displayed high identities (58.9%) and similarities (76.0%) to zebrafish DrGSDMEa, but low identities (29.3% and 24.2%) and similarities (51.2% and 45.8%) to zebrafish DrGSDMEb and human HsGSDME, respectively (Table 3).
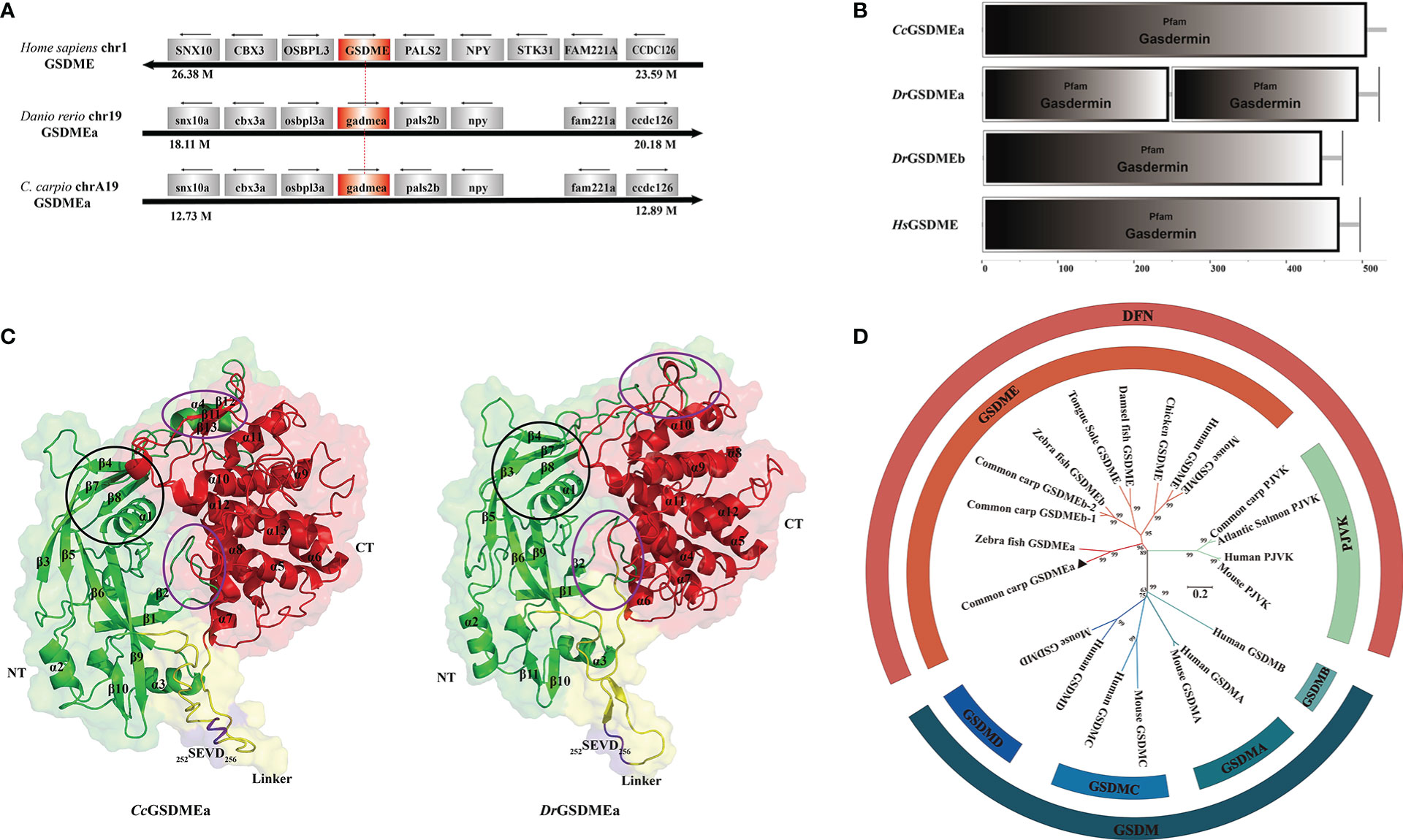
Figure 1 Structural feature and phylogenetic evolution of CcGSDMEa. (A) Gene synteny and chromosomal location analysis of CcGSDMEa. The genes adjacent to GSDME loci on human chromosome 1, zebrafish chromosome 19, common carp chromosome A19 are shown. Arrows indicate gene orientation. The small arrows above the gene indicate the direction in which the gene is transcribed. (B) Structure domains of CcGSDMEa, DrGSDMEa/b, and HsGSDME predicted by SMART. (C) The tertiary structures of CcGSDMEa and DrGSDMEa modeled by I-TASSER. The NT domains and CT domains are colored green and red, respectively. The motifs of tetrapeptide 252SEVD256 specifically recognized by caspases are marked in purple, and the linker regions are labeled in yellow. Secondary structure element α-helices and β-strands are indicated in images. The inter-domain interfaces are highlighted by purple ellipses. The α1, β3, β4, β7 and β8 marked by the black circle form the structure of the membrane inserted N-terminal domain. (D) Phylogenetic relationships of GSDM family members. The phylogenetic tree was constructed by MEGA (version 7.0) using the neighbor-joining method. The reliability of each node was estimated by bootstrap test with 1000 replications. The accession numbers of selected GSDM protein sequences from the National Center for Biotechnology Information Reference Sequence database (http://www.ncbi.nlm.nih.gov/RefSeq/) are shown in Table 3.
To better understand the function of CcGSDMEa, the features of domain and tertiary architecture were characterized. The CcGSDMEa contained one gasdermin domain, which is consistent with the other GSDM family members except for DrGSDMEa (Figure 1B). In addition, the CcGSDMEa shared highly conserved tertiary architectures with zebrafish DrGSDMEa, which had a conserved N-terminal pore-forming domain, C-terminal auto-inhibitory domain and a flexible and pliable hinge region (Figure 1C). In linker regions, the predicted tetrapeptide motif, which was specifically recognized by caspases, was 252SEVD256. This was the same as the cleavage site of DrGSDMEa. Phylogenetic tree revealed that CcGSDMEa was grouped with zebrafish DrGSDMEa with a high score of bootstrap tests. Interestingly, CcGSDMEa was not grouped with GSDMEb of common carp, but was clustered into a separate clade (Figure 1D), which provided us a better clue for future analysis of GSDME evolutionary function in teleost. In addition, both members of the deafness-associated gene (i.e., GSDME and PJVK) were clustered together, and they were distant from the other members (GSDMA, GSDMB, GSDMC and GSDMD).
Expression profile of CcGSDMEa after A. hydrophila challenge
CcGSDMEa was widely expressed in the tested tissues and exhibited a tissue-specific spatial expression pattern. CcGSDMEa was highly expressed in the skin and gill, followed by the head kidney and trunk kidney, and the lowest expression was in brain (Figure 2A). Moreover, the expression levels in immune-related tissues (head kidney, spleen, skin, intestine and gill) were markedly higher than those in heart, liver and brain.
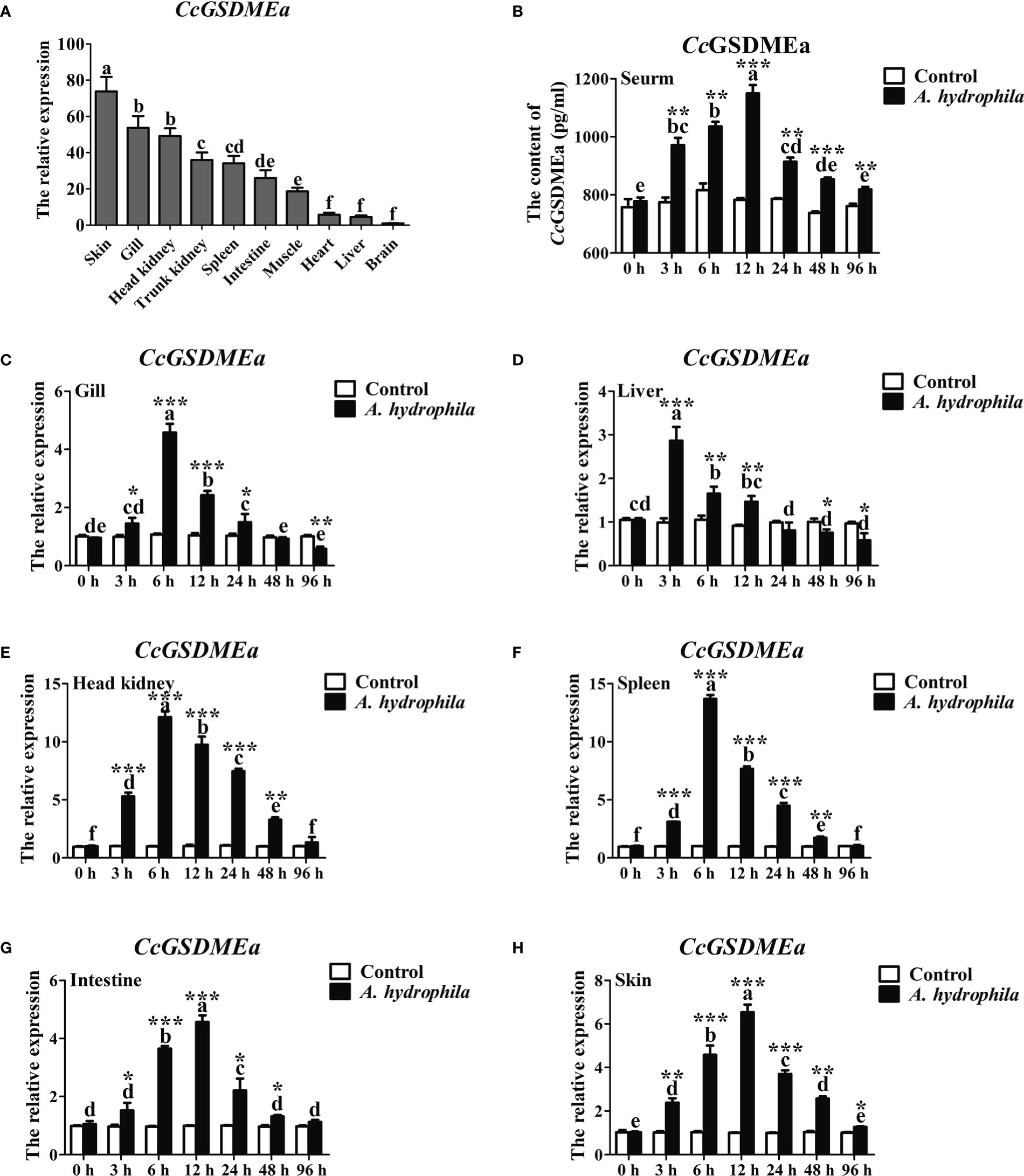
Figure 2 Expression profile of CcGSDMEa after A. hydrophila challenge. (A) Tissues distribution of CcGSDMEa in healthy C. carpio. The lowest expression level in brain was chosen as calibration (set as 1), and the relative expression of CcGSDMEa in other tissues was represented as fold-changes to the calibration. (B) The protein expression levels of CcGSDMEa in serum after A. hydrophila challenge. (C-H) The mRNA expression levels of CcGSDMEa in different tissues after A. hydrophila challenge. The control group was chosen as calibration (set as 1). The data were expressed as mean ± SD (n = 3). Significant difference between the different time points were analyzed using one-way ANOVA followed by Tukey’s test, and presented with the different lowercase letters in the group challenged by A. hydrophila (P<0.05), and the same letter was indicated no significant difference (P > 0.05). The significant differences between the A. hydrophila challenge group and the control group at the same time point were analyzed by student’s t test and indicated with asterisks (*, P < 0.05; **, P < 0.01, ***, P < 0.01).
To further analyze the expression profile of CcGSDMEa in response to bacterial infection, A. hydrophila was chosen to stimulate the juvenile common carp. The peaks of CcGSDMEa expression were at 3 h in liver, at 6 h in gill, head kidney and spleen, and at 12 h in intestine and skin (Figures 2C-H). Although the peaks of mRNA expression levels were different in various tissues and appeared at the different time points, the expression levels of CcGSDMEa were increased after A. hydrophila infection, compared with the control. The protein expression level of CcGSDMEa showed a trend of increasing at first and then decreasing with the time prolongation in serum, and the expression peak was observed at 12 h (Figure 2B). The expression levels among different time points showed significant differences. Meanwhile, the protein expression levels of CcGSDMEa were notably increased at the different time points compared with the control.
CcGSDMEa-NT was produced from CcGSDMEa cleavage by CcCaspase-1/3/7
To better understand the cleavage mechanism of CcGSDMEa, each of the CcCaspase-1a, CcCaspase-1b, CcCaspase-3a, CcCaspase-3b, CcCaspase-7a and CcCaspase-7b plasmids was co-transfected with CcGSDMEa-FL into EPC cells, and then treated with the dimerizing drug AP20187 for 6 h. The results showed that CcCaspase-1b, CcCaspase-3a, CcCaspase-3b, CcCaspase-7a and CcCaspase-7b could cleave CcGSDMEa to produce CcGSDMEa-NT, except for CcCaspase-1a (Figure 3A). The LDH released from cells co-transfected CcCaspase-1b/3a/3b and CcGSDMEa, were dramatically higher than that released from cells only transfected CcGSDMEa (Figure 3B). HEK293T cells were co-transfected with CcCaspase-1a/1b/3a/3b/7a/7b and CcGSDMEa, and the morphology of cell membrane was observed. The results showed that, except for the co-transfection of CcCaspase-1a and CcGSDMEa, membrane pores could be observed in the co-transfected cells (Figure 3C). It was indicated that any one of CcCaspase-1b/3a/3b/7a/7b could cleave CcGSDMEa to generate CcGSDMEa-NT, which could oligomerize on the cell membrane and penetrate the plasma membrane.
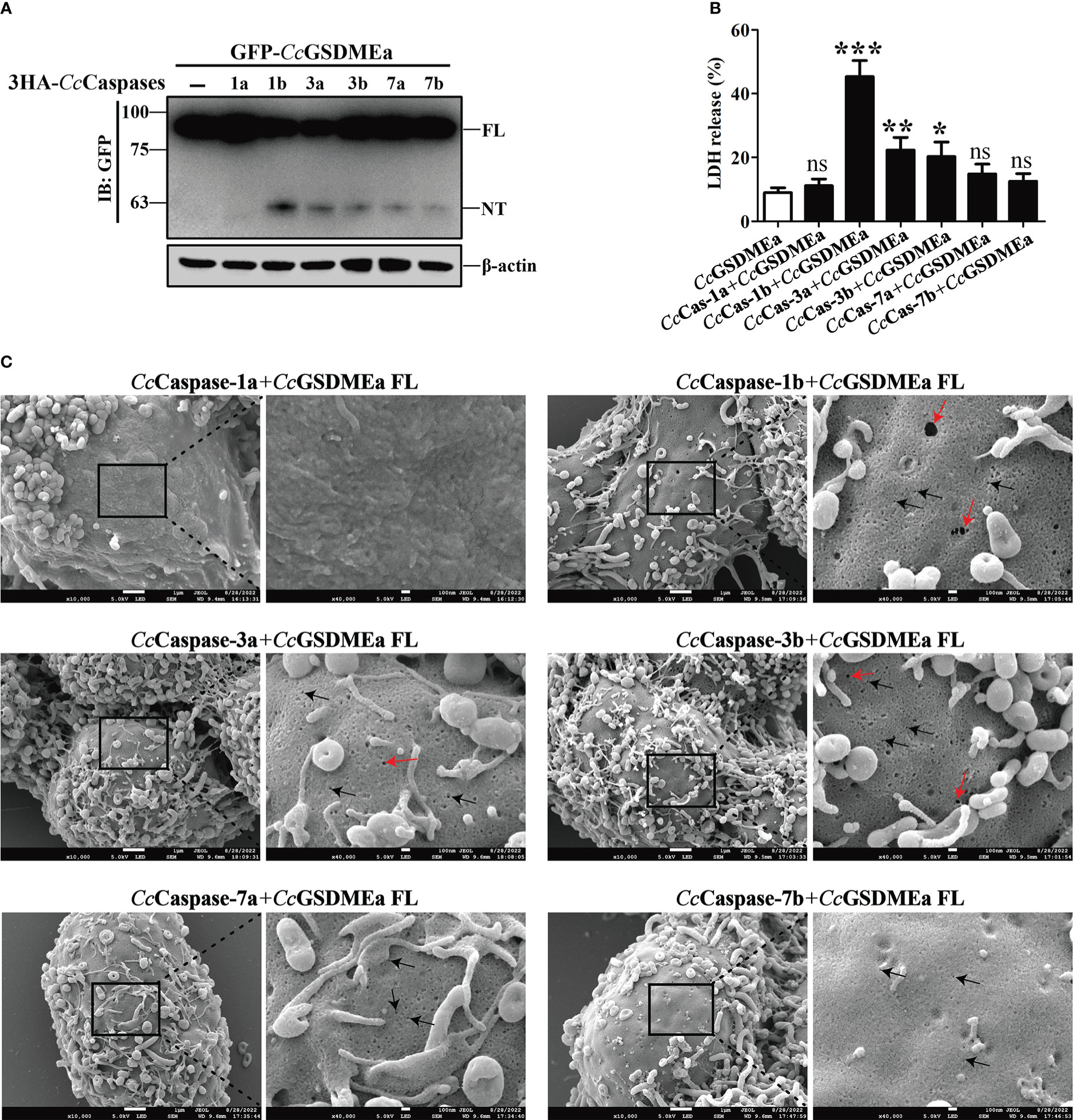
Figure 3 CcGSDMEa-NT was produced from CcGSDMEa cleavage by CcCaspase-1/3/7. (A) The western bolting result that CcGSDMEa was cleaved by CcCaspase-1b/3a/3b/7a/7b. (B) LDH release from the EPC cells co-transfected with CcGSDMEa and the CcCaspase-1a/1b/3a/3b/7a/7b plasmids. (C) Morphological observation of HEK293T cells co-transfected with CcGSDMEa and the CcCaspase-1a/1b/3a/3b/7a/7b plasmids. Red arrows display cell plasma membrane rupture, and dark arrows indicate the gasdermin membrane pores. Values are shown as mean ± SD (n = 3). The LDH release from the cells only transfected with CcGSDMEa was set as control. The significance difference of LDH release between the control cells and the co-transfected cells was analyzed by student’s t test and shown as: ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The membrane pore formation induced by CcGSDMEa-NT
To explore the pore forming function of CcGSDMEa, the CcGSDMEa-FL/NT/CT recombinant plasmids with GFP-tag were constructed (Figure 4A). The different recombinant plasmids of CcGSDMEa were transfected into HEK293T cells to observe the morphological changes and LDH release. The results showed that the release of LDH from cells expressing CcGSDMEa-NT was markedly higher than that of cells expressing CcGSDMEa-FL or CcGSDMEa-CT and pEGFP-C1 vector (Figure 4B). Cytomembrane pores and rupture phenomenon were also observed only in the transfected cells with CcGSDMEa-NT plasmids (Figure 4C). It was suggested that only CcGSDMEa could punch holes in the membrane and disrupt the integrity of cytomembrane through its NT domain.
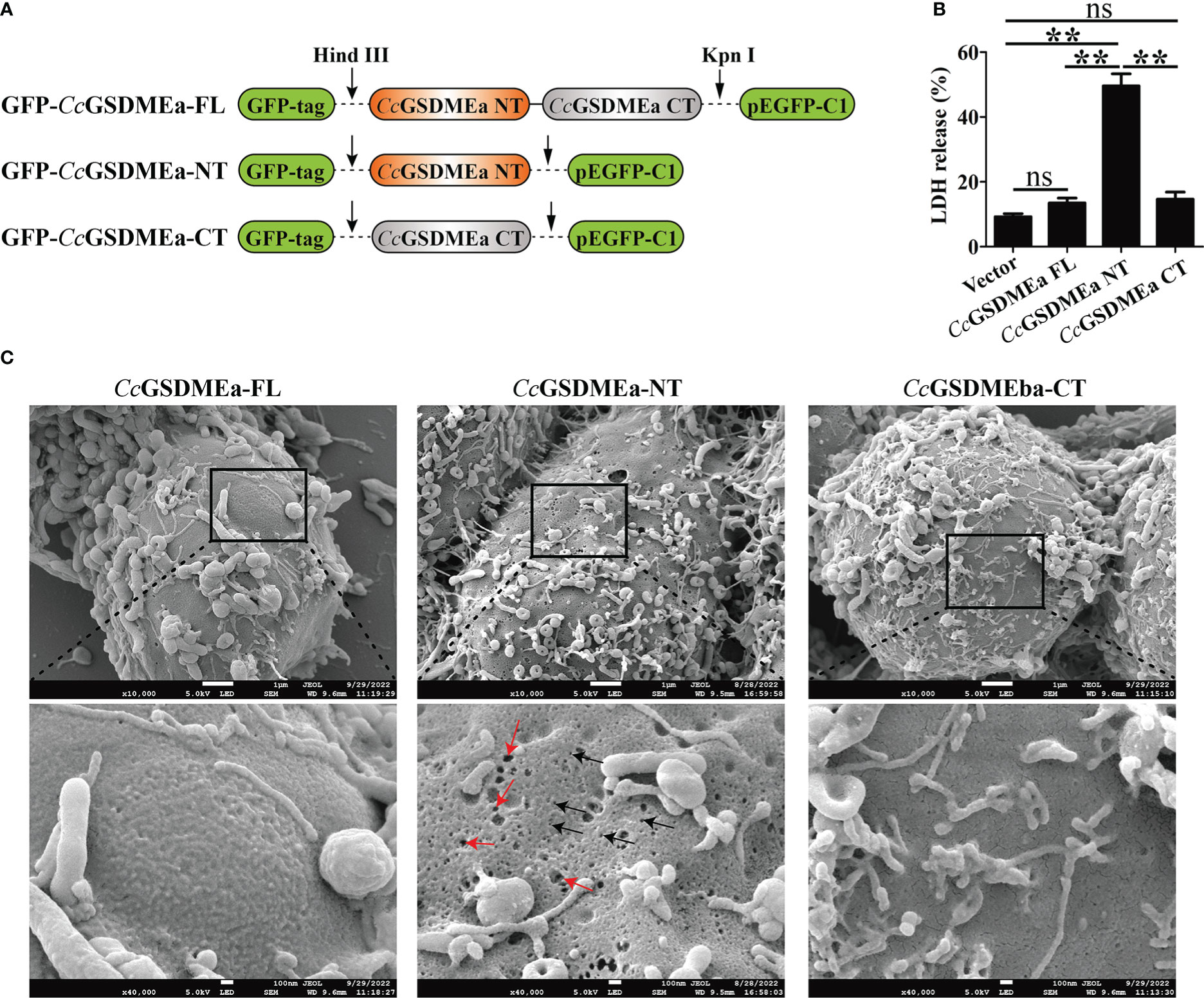
Figure 4 Membrane pore formation induced by CcGSDMEa-NT in HEK293T cells. (A) Schematic diagram of the recombinant plasmids of CcGSDMEa-FL (1-532 AA), CcGSDMEa-NT (1-250 AA), CcGSDMEa-CT (257-532 AA) and pEGFP-C1. (B) LDH release from cells transfected with 1000 ng of different forms of CcGSDMEa recombinant plasmids. (C) Morphological observation of HEK293T cells transfected with different forms of CcGSDMEa recombinant plasmids. Red arrows display cell plasma membrane rupture, and dark arrows indicate the membrane pores induced by CcGSDMEa-NT. The LDH release values are shown as M ± SD (n = 3). The significant differences were analyzed by student’s t test, “**” means P < 0.01, “ns” means no significant difference.
In addition, the subcellular localizations of CcGSDMEa-FL, NT and CT in HEK293T cells were varied (Figure 5A). The CcGSDMEa-FL was localized in the cytoplasm and cytomembrane, the CcGSDMEa-NT was only situated in the cytomembrane, and the CcGSDMEa-CT was distributed in the whole cells. We also found that CcGSDMEa-NT and DiI-labeled cytomembrane were completely colocalized (Figure 5B), and it was indicated that CcGSDMEa-NT could accumulate on the cellular membrane.
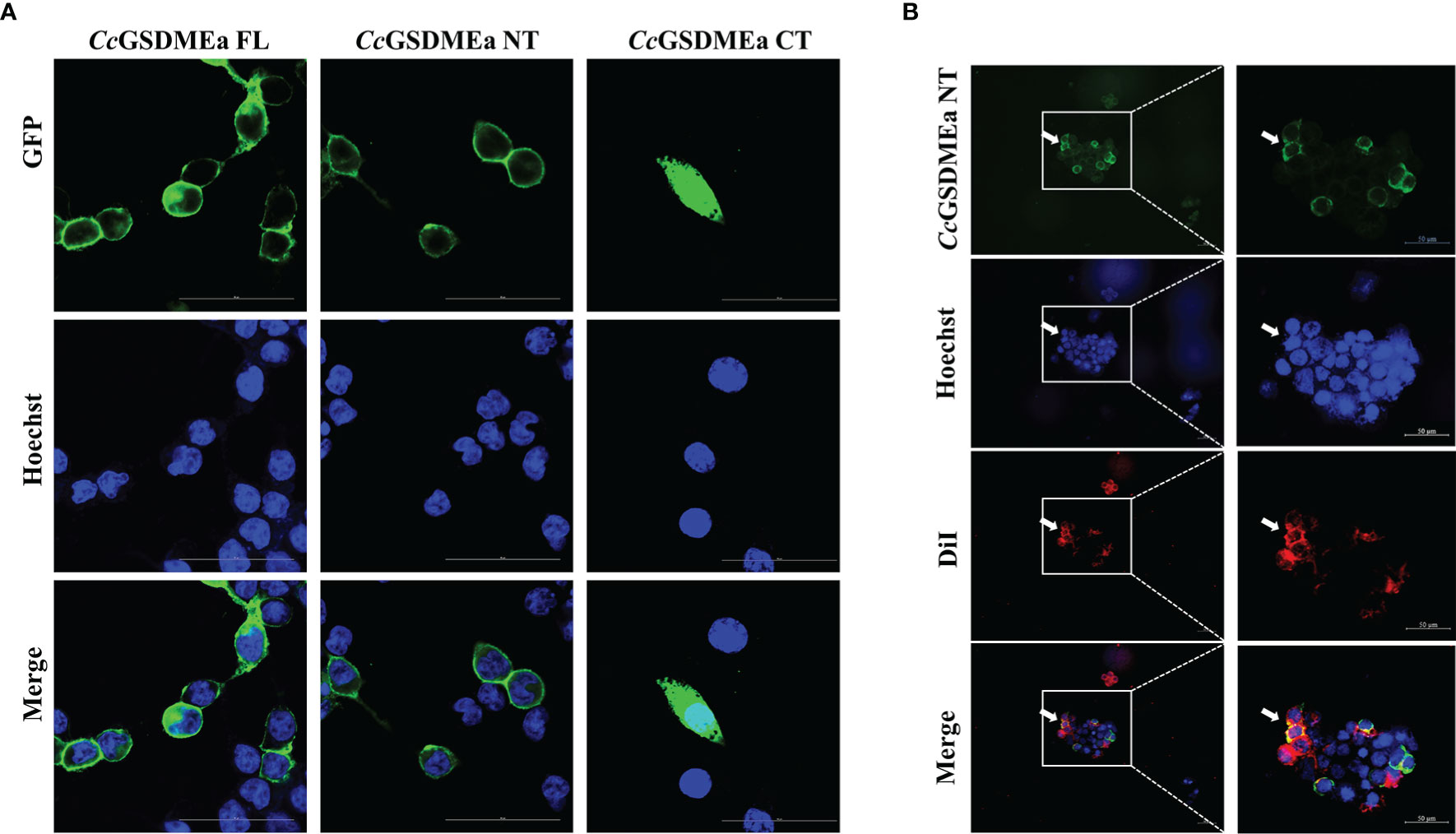
Figure 5 Subcellular localizations of CcGSDMEa different forms. (A) Subcellular localizations of CcGSDMEa-FL/NT/CT and Hoechst-stained nucleus in HEK293T cells. (B) Subcellular localization of CcGSDMEa-NT, Hoechst-stained nucleus and DiI-stained cytomembrane in HEK293T cells (white arrowheads). The original microscope images were on the left and the locally enlarged images were on the right in (B).
Bactericidal activity of CcGSDMEa to E. coli
In order to detect the bactericidal activity of CcGSDMEa, CcGSDMEa-FL, CcGSDMEa-NT and CcGSDMEa-CT were subcloned into pET-32a vector, respectively (Figure 6A). The recombinant plasmids rCcGSDMEa-FL/NT/CT were expressed in E. coli by IPTG-induced manner. Of note, the number of E. coli expressing rCcGSDMEa-NT was markedly reduced compared with that of expressing rCcGSDMEa-FL or rCcGSDMEa-CT after IPTG induction (Figures 6B, C). The growth curves of E. coli expressing different recombinant plasmids of CcGSDMEa were also been analyzed. The OD600 of E. coli expressing rCcGSDMEa-NT indicated little change at different time points after IPTG induction, but the OD600 values of E. coli expressing rCcGSDMEa-FL, rCcGSDMEa-CT and pET-32a vector showed an increasing trend with time prolongation after IPTG induction (Figure 6D). It was demonstrated that CcGSDMEa-NT could perform bactericidal function.
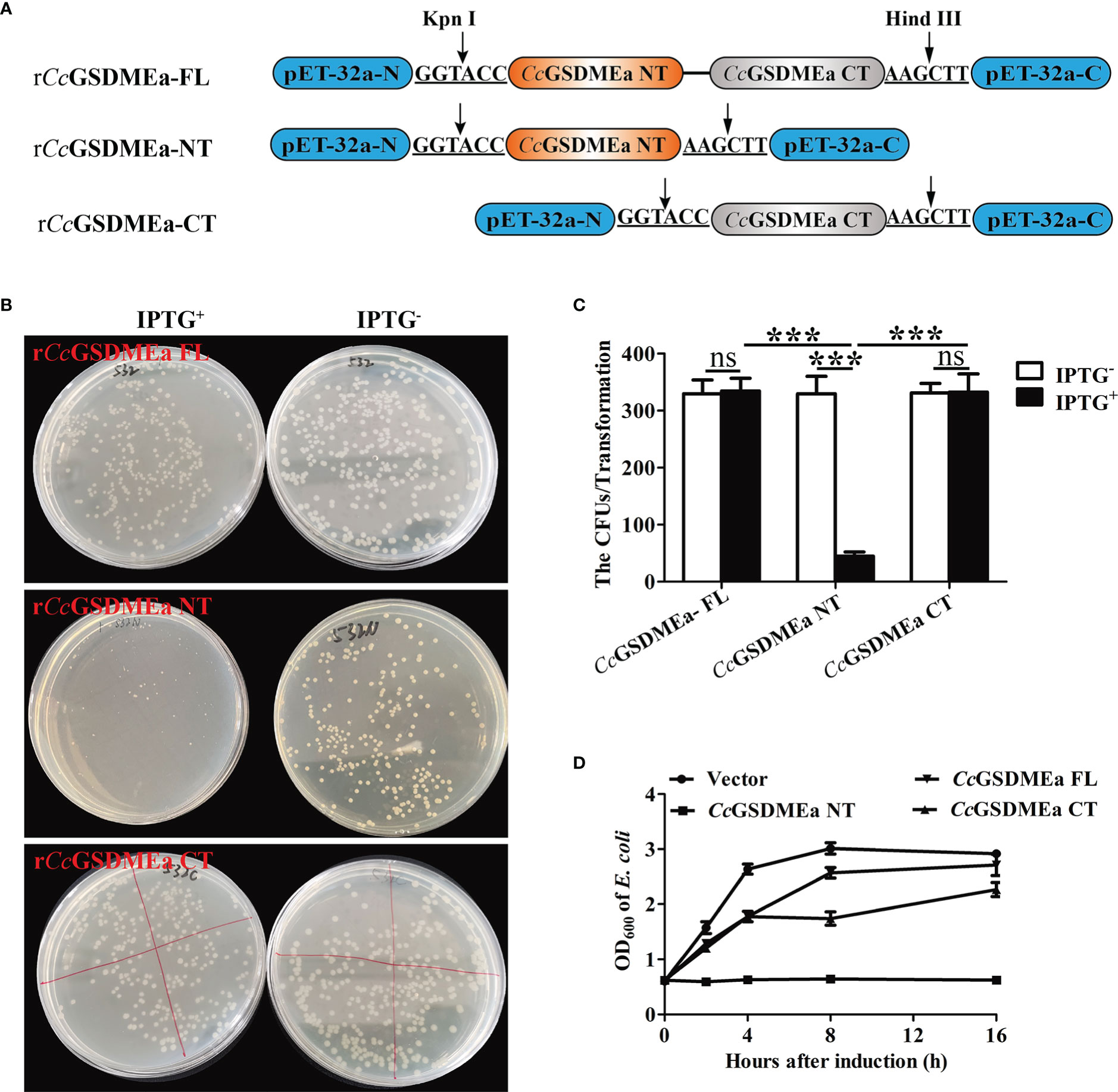
Figure 6 Bactericidal activity of CcGSDMEa to E. coli. (A) Schematic diagram of the recombinant plasmids of rCcGSDMEa-FL, rCcGSDMEa-NT and rCcGSDMEa-CT. (B) Colony growth of E. coli expressing rCcGSDMEa-FL/NT/CT. (C) Calculation of CFUs on the plates. (D) Growth curves of E. coli expressing rCcGSDMEa-FL/NT/CT after IPTG induction. Values are expressed as M ± SD (n = 3). The statistical difference was analyzed by student’s t test (***, P < 0.001; ns, no significant difference).
CcGSDMEa promotes the expression of CcIL-1β in vivo
To better understand the role of CcGSDMEa in regulating CcIL-1β, the CcGSDMEa in fish was knocked down or overexpressed, and the expression of CcIL-1β at the mRNA and protein levels were detected. The knockdown and overexpression of CcGSDMEa were confirmed based on the results of CcGSDMEa mRNA expression levels in vivo. Figure 7 showed that the CcGSDMEa mRNA expression levels in GFP-CcGSDMEa group were markedly increased compared with the levels in pEGFP-C1 group at 3 d and 7 d (Figures 7A, B). However, the expression levels of CcGSDMEa in siCcGSDMEa group were lower than those in siControl group (Figures 7C, D).
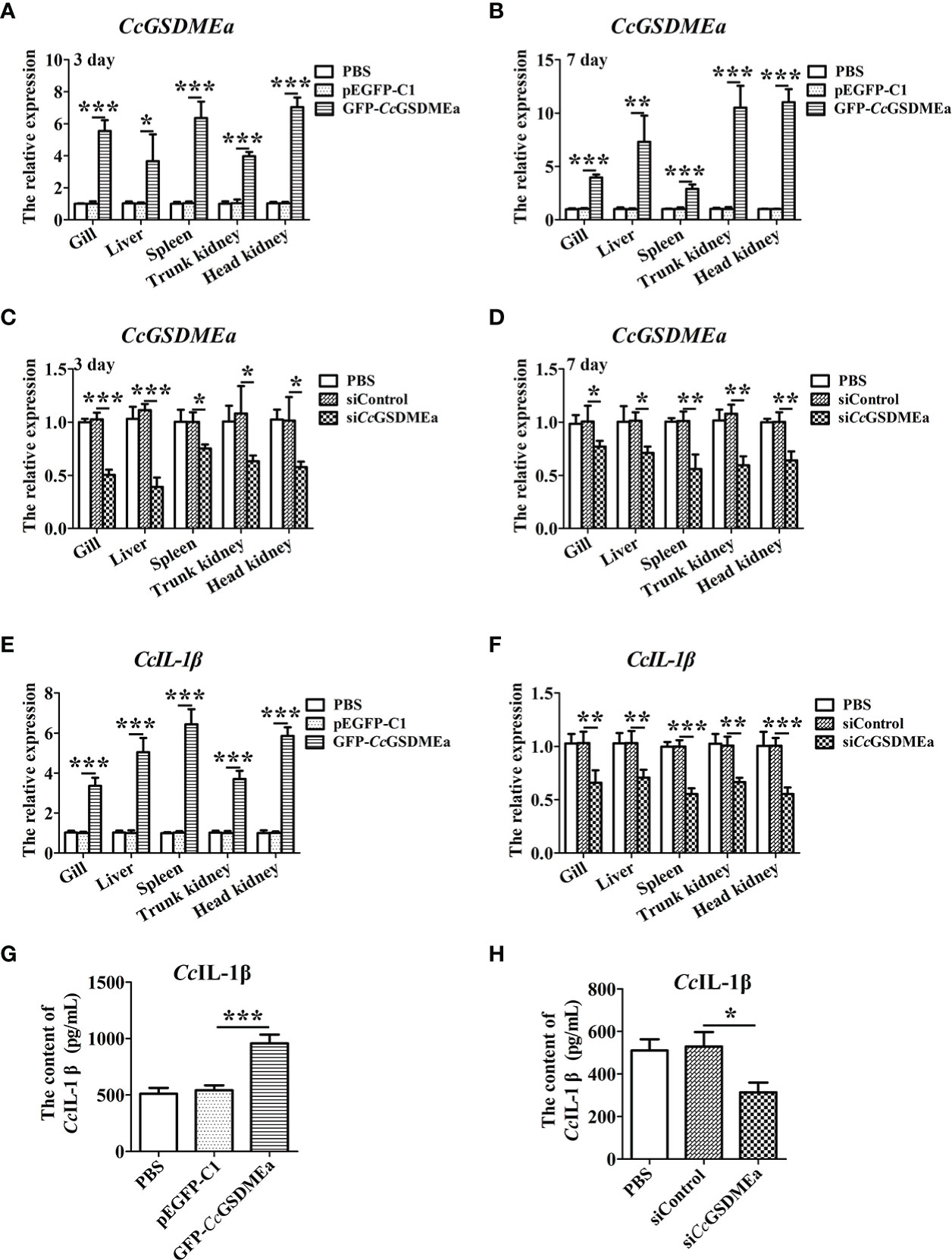
Figure 7 CcGSDMEa promotes the expression of CcIL-1β in vivo. (A-D) Overexpression efficiency and knockdown efficiency of CcGSDMEa in gill, liver, spleen, trunk kidney and head kidney of C. carpio. (E, F) The mRNA expression levels of CcIL-1β in the different groups after A. hydrophila challenge. (G, H) The protein expression levels of CcIL-1β in the different groups after A. hydrophila challenge. In each case, the mRNA expression level in the PBS group was chosen as calibration (set as 1). The statistical difference was analyzed by student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
In CcGSDMEa overexpression group, the CcIL-1β expressions at the mRNA and protein levels were notably higher compared with the expression levels in pEGFP-C1 group (Figures 7E, G). In CcGSDMEa knockdown group, the expression levels of CcIL-1β decreased compared with the siControl group (Figures 7F, H). These results suggested that CcGSDMEa could promote CcIL-1β expression.
CcGSDMEa prevents A. hydrophila colonization in vivo
As mentioned above, the expression of CcGSDMEa increased in fish when stimulated by A. hydrophila (Figure 2). After CcGSDMEa was overexpressed or knocked down, the bacterial burdens were examined in tissues of fish after A. hydrophila challenge for 24 h. As shown in Figure 8, the bacterial burdens were obviously lower in head kidney, spleen, gill, liver and trunk kidney in GFP-CcGSDMEa overexpression group than those in the pEGFP-C1 group. While, in siCcGSDMEa group, the bacterial burdens were observably increased in head kidney, gill, liver, spleen and trunk kidney, compared with those in siControl group. It was indicated that the CcGSDMEa could prevent the colonization of A. hydrophila and minish the bacterial burdens in fish.
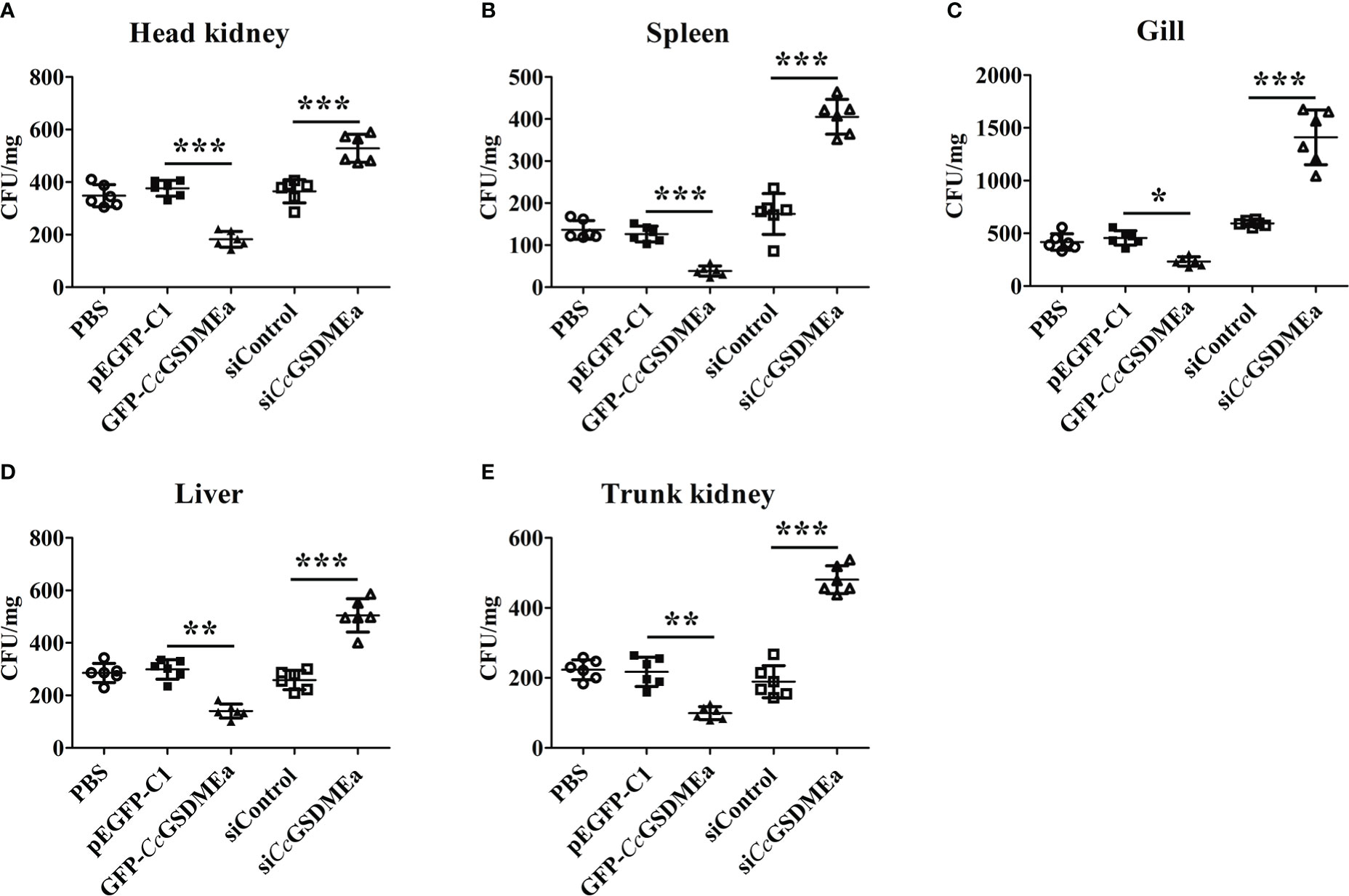
Figure 8 CcGSDMEa prevents A. hydrophila colonization in vivo. Bacterial burdens in head kidney (A), spleen (B), gill (C), liver (D) and trunk kidney (E) of different groups infected with A. hydrophila. The data are expressed as mean ± SD (n = 3) and analyzed statistically by student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
CcGSDMEa promotes the secretion of CcIL-1β in EPC cells
After EPC cells challenged with A. hydrophila, the expression levels of CcGSDMEa and CcIL-1β significantly increased, compared with the control group (Figure 9A), it was suggested that A. hydrophila could trigger the innate immune responses of EPC cells. Similarly, the contents of CcIL-1β protein in the cell culture medium were also increased after A. hydrophila challenge compared with the control group (Figure 9B), and it was manifested that A. hydrophila challenge could promote the secretion of CcIL-1β. Moreover, the LDH released from EPC cells treated by A. hydrophila were observably higher than that in the control group (Figure 9C), and indicted that A. hydrophila challenge might destroy cytomembrane integrity of EPC cells.
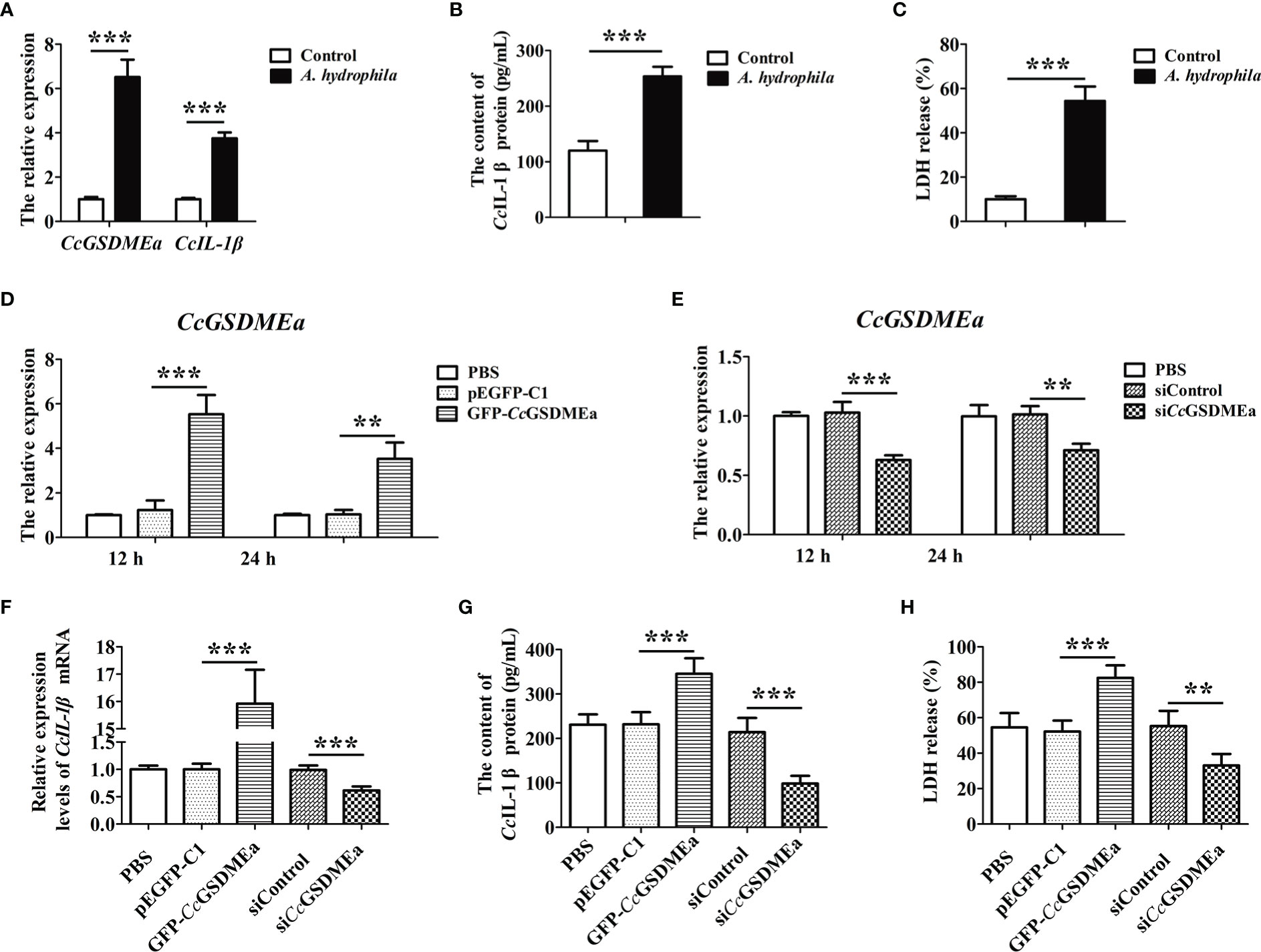
Figure 9 CcGSDMEa promotes the secretion of CcIL-1β in vitro. (A) The mRNA expression levels of CcGSDMEa and CcIL-1β in EPC cells after A. hydrophila challenge. (B) The content of CcIL-1β protein in the cell culture medium after A. hydrophila challenge. (C) The LDH release in EPC cells after A. hydrophila challenge. (D, E) The overexpression and knockdown efficiency of CcGSDMEa in EPC cells at 12 h and 24 h. (F-H) The mRNA expression levels of CcIL-1β, protein secretion contents of CcIL-1β and the LDH release from cells in overexpression or knockdown group after A. hydrophila challenge. The data were expressed as mean ± SD (n = 3). The significant differences were analyzed using student’s t test (**, P < 0.01; ***, P < 0.001).
To clarify the effect of CcGSDMEa on CcIL-1β secretion, the CcGSDMEa was overexpressed or knocked down in EPC cells, and then the contents of CcIL-1β protein in cell culture medium were examined after A. hydrophila stimulation. As shown in Figures 9D, E, the overexpression and knockdown of CcGSDMEa were both affirmed. After A. hydrophila challenge, in the CcGSDMEa overexpression group, expression level and secretion content of CcIL-1β and the release of LDH were significantly increased, compared with the pEGFP-C1 group (Figures 9F-H). It was illustrated that the overexpression of CcGSDMEa could intensify the effect of A. hydrophila challenge on CcIL-1β secretion and LDH release. However, in siCcGSDMEa group, after A. hydrophila challenge, the expression level and secretion content of CcIL-1β and the release of LDH were notably decreased compared with the siControl group. It was implied that the knockdown of CcGSDMEa could retard the effect of A. hydrophila on CcIL-1β secretion and LDH release. Taken together, these results revealed that CcGSDMEa played an important role in regulating the secretion of CcIL-1β.
Discussion
The different types of cell death, such as apoptosis, autophagy, necrosis or pyroptosis, mainly depend on the inducing factors and the different stages of the cell cycle. Among these types of programmed cell death, pyroptosis is a newly defined inflammatory programmed cell death that plays an important role in fighting bacterial infections and inflammatory diseases (41). Previous studies have shown that the gasdermin family members were the executors of pyroptosis and played key roles in the innate immunity (18, 42, 43). However, in teleost, only two members of gasdermin family (GSDME and PJVK) have been identified to date, and they were considered as the ancient members of the GSDM family (19, 21, 44). Since PJVK does not perform immune-related functions (35, 45), GSDME is considered to be the only executor of pyroptosis in teleost, and its role of innate immune to resist pathogen invasion has become a research hotspot.
In this study, the CcGSDMEa identified from common carp shares conserved chromosomal colinearity and tertiary structure with GSDME in other fish and human (17–19, 24). Similar to the structure of mouse GSDMA3, α1, α4 helix and β1-β2 hairpin in NT domain of CcGSDMEa provide the main surface for binding to the CT domain and then form the major autoinhibitory interactions (44). In addition, α1, β3, β4, β7 and β8 can form the N-terminal structure that inserts into cytomembrane. The α1 is the first α-helix that specifically interacts with phospholipids, and β3, β4, β7 and β8 are four parallel β-strands, which can insert into the membrane. These structural properties of CcGSDMEa might provide the basis for its functional role.
CcGSDMEa was ubiquitously expressed in all tested tissues, although the expression levels are different in the varied tissues. In zebrafish, DrGSDMEa was highly expressed in intestine, gill, head kidney and spleen, but lowly expressed in brain, heart and muscle (19). The same situation was found in common carp. The high expression of CcGSDMEa in immune-related tissues indicated that it played an important role in the immune surveillance system. Studying the expression changes of CcGSDMEa after A. hydrophila challenge is helpful to understand the immune response of fish to pathogen infection and provide new insights into the formulation of effective measures to prevent the occurrence of disease. The increased expression levels of CcGSDMEa (mRNA and protein) suggested that CcGSDMEa played an important role in immune response against the invasion of A. hydrophila. The peaks of CcGSDMEa expression levels appeared at the early stage of infection, indicated that fish immune response was more active and stronger at the early stage of pathogen infection, and it suggested that the early stage of infection was the best period to clear pathogens in fish.
GSDME could be cleaved by caspases to cause pyroptosis in teleost. For instance, DrGSDMEa could be cleaved by inflammatory caspases (caspase-B and caspase-19b) and apoptotic caspases (caspase-3a/b, caspase-7 and caspase-8a) in zebrafish (16), the SmGSDME could be cleaved by Smcaspase in turbot, and CsGSDME could be cleaved by caspase-1/3/7 in tongue sole (18). Similarly, CcGSDMEa was cleaved by inflammatory caspases (CcCaspase-1b) and apoptotic caspases (caspase-3a/b and caspase-7a/b) to release its N-terminal domain to form the pores on cell membrane. Moreover, CcGSDMEa cleaved by CcCaspase-1b/3a/3b also could disrupt the cell membrane. The different LDH release and the different degree of cell membrane destruction of the cells might be due to the different cleavage efficiency of CcGSDMEa by CcCaspase-1b/3a/3b/7a/7b. The tongue sole CsGSDME was cleaved with high efficiency by inflammatory Caspase-1, and with comparatively low efficiency by apoptotic Caspase-3/7 (18). To be different from this result, zebrafish DrGSDMEa was cleaved with high efficiency by Caspase-1/3b and with low efficiency by Caspase-3a/7 (17, 19). This suggested that the efficiency of GSDME being cleaved in fish was not only related to the different of Caspases, but also related to the specificity of species.
Studies have shown that the NT-domain of GSDME could punch holes in the cell membrane and induced pyroptotic bubble formation and membrane lysis (18, 19, 28, 33). The membrane pore formation, cytomembrane rupture and LDH release from cells were observed only in transfected cells with CcGSDMEa-NT plasmid, which was consistent with that found in zebrafish DrGSDMEa and turbot SmGSDMEb (16, 17). In addition, the number of E. coli expressing rCcGSDMEa-NT was markedly reduced after IPTG induction, this was consistent with the findings in tongue sole, turbot and zebrafish (16–18). Accumulating evidence suggested that CcGSDMEa-NT could not only punch holes in the membrane of HEK293T cells, but also directly destroy the integrity of bacterial membrane of E. coli, and showed that the CcGSDMEa exerted its bactericidal activity and cytotoxicity through its NT domain.
The antibacterial infection mechanism of GSDME remains to be poorly understood in fish at present. In this research, the increased bacterial burdens in various tissues in CcGSDMEa-knockdown group demonstrated that the ability to eliminate the invading bacteria was reduced in host. With respect to CcGSDMEa-overexpression group, it was shown that bacterial loads were obviously decreased in different tissues. These results mentioned above provided the direct evidence that CcGSDMEa played a key role in regulating bacterial clearance in fish. As a key node in the immune and inflammatory networks, IL-1β performs antibacterial defenses through involving in inflammation reaction in innate immunity (29, 30, 32, 46). Our results showed that CcGSDMEa could promote the expression and secretion of CcIL-1β, involve in inflammation reaction, and then help the host eliminate invading pathogens.
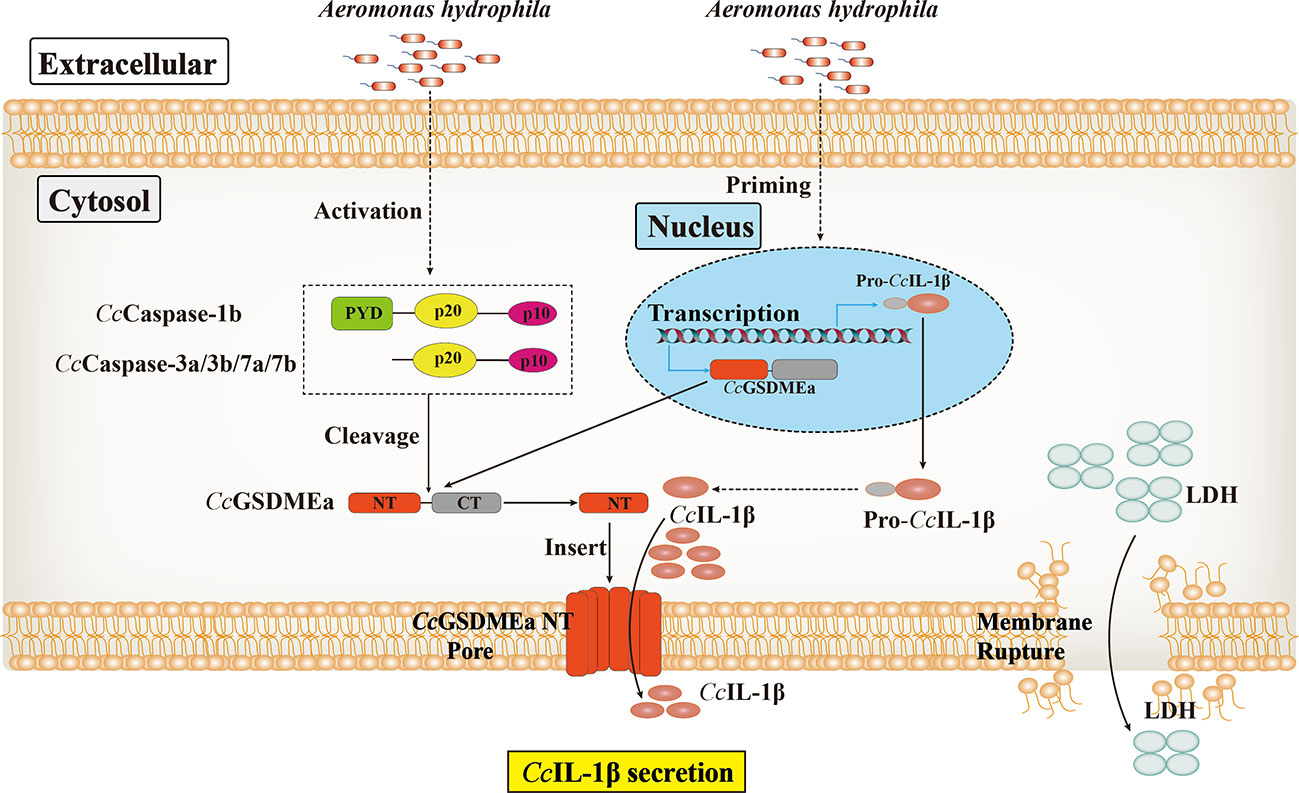
To sum up, CcGSDMEa could be cleaved by CcCaspase-1b/3a/3b/7a/7b to generate CcGSDMEa-NT domain, which could oligomerize and translocate to the plasma membrane, where it could form pores to promote pyroptosis. CcGSDMEa was involved in innate immune defense against A. hydrophila infection in common carp, through regulating the secretion of CcIL-1β, and it was indicated that CcGSDMEa played an important role in combating bacterial invasion (Figure 10). These results will provide a reference for us to further study the immune mechanism to defense against bacterial infection in fish, and also provide new strategies to prevent and control fish diseases for healthy and continuous aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Ethics Committee of the Henan Normal University.
Author contributions
YZ and JZ: conceived, designed and performed the experiments, and analyzed data, as well as wrote the draft manuscript and submitted the manuscript. DQ, FG and YG: carried out the experiments and analyzed data. XJ and LZ: performed data interpretation and acquired finding. XK: designed this study and provided a critical editing of the manuscript and was responsible for forming the finalizing manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the National Natural Science Foundation of China (U2004154).
Acknowledgments
We thank the Engineering Lab of Henan Province for Aquatic Animal Disease Control and Engineering Technology Research Center of Henan Province for Aquatic Animal Cultivation (Henan Normal University, Xinxiang, China) for providing instruments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1110322/full#supplementary-material
References
1. Nakajima T, Hudson MJ, Uchiyama J, Makibayashi K, Zhang J. Common carp aquaculture in neolithic China dates back 8,000 years. Nat Ecol Evol (2019) 3(10):1415–8. doi: 10.1038/s41559-019-0974-3
2. Lafferty KD, Harvell CD, Conrad JM, Friedman CS, Kent ML, Kuris AM, et al. Infectious diseases affect marine fisheries and aquaculture economics. Ann Rev Mar Sci (2015) 7:471–96. doi: 10.1146/annurev-marine-010814-015646
3. Basu M, Swain B, Maiti NK, Routray P, Samanta M. Inductive expression of toll-like receptor 5 (TLR5) and associated downstream signaling molecules following ligand exposure and bacterial infection in the Indian major carp, mrigal (Cirrhinus mrigala). Fish Shellfish Immunol (2012) 32(1):121–31. doi: 10.1016/j.fsi.2011.10.031
4. Orozova P, Sirakov I, Austin DA, Austin B. Recovery of bacillus mycoides, b. pseudomycoides and Aeromonas hydrophila from common carp (Cyprinus carpio) and rainbow trout (Oncorhynchus mykiss) with gill disease. J Fish Dis (2018) 41(1):125–9. doi: 10.1111/jfd.12686
5. Di G, Li H, Zhang C, Zhao Y, Zhou C, Naeem S, et al. Label-free proteomic analysis of intestinal mucosa proteins in common carp (Cyprinus carpio) infected with aeromonas hydrophila. Fish Shellfish Immunol (2017) 66:11–25. doi: 10.1016/j.fsi.2017.04.025
6. Feng JC, Cai ZL, Zhang XP, Chen YY, Chang XL, Wang XF, et al. The effects of oral rehmannia glutinosa polysaccharide administration on immune responses, antioxidant activity and resistance against Aeromonas hydrophila in the common carp, cyprinus carpio l. Front Immunol (2020) 11:904. doi: 10.3389/fimmu.2020.00904
7. Podok P, Xu L, Xu D, Lu L. Different expression profiles of interleukin 11 (IL-11), intelectin (ITLN) and purine nucleoside phosphorylase 5a (PNP 5a) in crucian carp (Carassius auratus gibelio) in response to cyprinid herpesvirus 2 and aeromonas hydrophila. Fish Shellfish Immunol (2014) 38(1):65–73. doi: 10.1016/j.fsi.2014.03.001
8. Zhang J, Wang L, Zhao Y, Kong X, Wu F, Zhao X. Molecular characterization and expression analysis of toll-like receptors 5 and 22 from natural triploid carassius auratus. Fish Shellfish Immunol (2017) 64:1–13. doi: 10.1016/j.fsi.2017.03.004
9. Weifeng M, Yaping W, Wenbo W, Bo W, Jianxin F, Zuoyan Z. Enhanced resistance to Aeromonas hydrophila infection and enhanced phagocytic activities in human lactoferrin-transgenic grass carp (Ctenopharyngodon idellus). Aquaculture (2004) 242(1-4):93–103. doi: 10.1016/j.aquaculture.2004.07.020
10. Lu AJ, Hu XC, Wang Y, Zhu AH, Shen LL, Tian J, et al. Skin immune response in the zebrafish, Danio rerio (Hamilton), to Aeromonas hydrophila infection: a transcriptional profiling approach. J Fish Dis (2015) 38(2):137–50. doi: 10.1111/jfd.12214
11. Bebak J, Wagner B, Burnes B, Hanson T. Farm size, seining practices, and salt use: risk factors for aeromonas hydrophila outbreaks in farm-raised catfish, Alabama, USA. Prev Vet Med (2015) 118(1):161–8. doi: 10.1016/j.prevetmed.2014.11.001
12. Mu X, Pridgeon JW, Klesius PH. Comparative transcriptional analysis reveals distinct expression patterns of channel catfish genes after the first infection and re-infection with aeromonas hydrophila. Fish Shellfish Immunol (2013) 35(5):1566–76. doi: 10.1016/j.fsi.2013.08.027
13. Peatman E, Mohammed H, Kirby A, Shoemaker CA, Yildirim-Aksoy M, Beck BH. Mechanisms of pathogen virulence and host susceptibility in virulent Aeromonas hydrophila infections of channel catfish (Ictalurus punctatus). Aquaculture (2018) 482:1–8. doi: 10.1016/j.aquaculture.2017.09.019
14. Pachanawan A, Phumkhachorn P, Rattanachaikunsopon P. Potential of psidium guajava supplemented fish diets in controlling Aeromonas hydrophila infection in tilapia (Oreochromis niloticus). J Biosci Bioeng (2008) 106(5):419–24. doi: 10.1263/jbb.106.419
15. Zhao XL, Jin ZH, Di GL, Li L, Kong XH. Molecular characteristics, pathogenicity and medication regimen of Aeromonas hydrophila isolated from common carp (Cyprinus carpio l. ). J Vet Med Sci (2019) 81(12):1769–75. doi: 10.1292/jvms.19-0025
16. Chen H, Wu X, Gu Z, Chen S, Zhou X, Zhang Y, et al. Zebrafish gasdermin e cleavage-engaged pyroptosis by inflammatory and apoptotic caspases. Dev Comp Immunol (2021) 124:104203. doi: 10.1016/j.dci.2021.104203
17. Chen S, Jin P, Chen H, Wu D, Li S, Zhang Y, et al. Dual function of a turbot inflammatory caspase in mediating both canonical and non-canonical inflammasome activation. Dev Comp Immunol (2021) 121:104078. doi: 10.1016/j.dci.2021.104078
18. Jiang S, Gu H, Zhao Y, Sun L. Teleost gasdermin e is cleaved by caspase 1, 3, and 7 and induces pyroptosis. J Immunol (2019) 203(5):1369–82. doi: 10.4049/jimmunol.1900383
19. Li JY, Wang YY, Shao T, Fan DD, Lin AF, Xiang LX, et al. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1beta maturation and gasdermin e-mediated pyroptosis. J Biol Chem (2020) 295(4):1120–41. doi: 10.1074/jbc.RA119.011751
20. Song Z, Zou J, Wang M, Chen Z, Wang Q. A comparative review of pyroptosis in mammals and fish. J Inflammation Res (2022) 15:2323–31. doi: 10.2147/JIR.S361266
21. Daskalov A, Glass NL. Gasdermin and gasdermin-like pore-forming proteins in invertebrates, fungi and bacteria. J Mol Biol (2022) 434(4):167273. doi: 10.1016/j.jmb.2021.167273
22. Feng S, Fox D, Man SM. Mechanisms of gasdermin family members in inflammasome signaling and cell death. J Mol Biol (2018) 430(18 Pt B):3068–80. doi: 10.1016/j.jmb.2018.07.002
23. Chen J, Ge L, Shi X, Liu J, Ruan H, Heng D, et al. Lobaplatin induces pyroptosis in cervical cancer cells via the caspase-3/GSDME pathway. Anticancer Agents Med Chem (2022) 22(11):2091–7. doi: 10.2174/1871520621666211018100532
24. Dong S, Shi Y, Dong X, Xiao X, Qi J, Ren L, et al. Gasdermin e is required for induction of pyroptosis and severe disease during enterovirus 71 infection. J Biol Chem (2022) 298(5):101850. doi: 10.1016/j.jbc.2022.101850
25. Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun (2017) 8:14128. doi: 10.1038/ncomms14128
26. Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature (2017) 547(7661):99–103. doi: 10.1038/nature22393
27. Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U.S.A. (2016) 113(28):7858–63. doi: 10.1073/pnas.1607769113
28. Sborgi L, Ruhl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, et al. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J (2016) 35(16):1766–78. doi: 10.15252/embj.201694696
29. Chen S, Ma X, Wu D, Yang D, Zhang Y, Liu Q. Scophthalmus maximus interleukin-1beta limits Edwardsiella piscicida colonization in vivo. Fish Shellfish Immunol (2019) 95:277–86. doi: 10.1016/j.fsi.2019.10.050
30. Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol (2009) 27:519–50. doi: 10.1146/annurev.immunol.021908.132612
31. Fabiani C, Sota J, Tosi GM, Franceschini R, Frediani B, Galeazzi M, et al. The emerging role of interleukin (IL)-1 in the pathogenesis and treatment of inflammatory and degenerative eye diseases. Clin Rheumatol (2017) 36(10):2307–18. doi: 10.1007/s10067-016-3527-z
32. Munoz-Wolf N, Lavelle EC. A guide to IL-1 family cytokines in adjuvanticity. FEBS J (2018) 285(13):2377–401. doi: 10.1111/febs.14467
33. Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature (2016) 535(7610):111–6. doi: 10.1038/nature18590
34. Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin d causes pyroptosis by forming membrane pores. Nature (2016) 535(7610):153–8. doi: 10.1038/nature18629
35. Kovacs SB, Miao EA. Gasdermins: Effectors of pyroptosis. Trends Cell Biol (2017) 27(9):673–84. doi: 10.1016/j.tcb.2017.05.005
36. Zhou ZX, Zhang BC, Sun L. Poly(I:C) induces antiviral immune responses in Japanese flounder (Paralichthys olivaceus) that require TLR3 and MDA5 and is negatively regulated by Myd88. PloS One (2014) 9(11):e112918. doi: 10.1371/journal.pone.0112918
37. Zhang J, Li L, Kong X, Wu F, Zhou C, Nie G, et al. Expression patterns of toll-like receptors in natural triploid Carassius auratus after infection with aeromonas hydrophila. Vet Immunol Immunopathol (2015) 168(1-2):77–82. doi: 10.1016/j.vetimm.2015.08.009
38. Wang Z, Gu Z, Hou Q, Chen W, Mu D, Zhang Y, et al. Zebrafish GSDMEb cleavage-gated pyroptosis drives septic acute kidney injury in vivo. J Immunol (2020) 204(7):1929–42. doi: 10.4049/jimmunol.1901456
39. Boucher D, Monteleone M, Coll RC, Chen KW, Ross CM, Teo JL, et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J Exp Med (2018) 215(3):827–40. doi: 10.1084/jem.20172222
40. Chen H, Ding S, Tan J, Yang D, Zhang Y, Liu Q. Characterization of the Japanese flounder NLRP3 inflammasome in restricting Edwardsiella piscicida colonization in vivo. Fish Shellfish Immunol (2020) 103:169–80. doi: 10.1016/j.fsi.2020.04.063
41. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol (2009) 7(2):99–109. doi: 10.1038/nrmicro2070
42. He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin d is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res (2015) 25(12):1285–98. doi: 10.1038/cr.2015.139
43. Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature (2015) 526(7575):660–5. doi: 10.1038/nature15514
44. Broz P, Pelegrin P, Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol (2020) 20(3):143–57. doi: 10.1038/s41577-019-0228-2
45. Xia S. Biological mechanisms and therapeutic relevance of the gasdermin family. Mol Aspects Med (2020) 76:100890. doi: 10.1016/j.mam.2020.100890
Keywords: common carp, Aeromonas hydrophila, GSDMEa, caspases, gasdermin pore, IL-lβ secretion
Citation: Zhao Y, Zhang J, Qiao D, Gao F, Gu Y, Jiang X, Zhu L and Kong X (2023) CcGSDMEa functions the pore-formation in cytomembrane and the regulation on the secretion of IL-lβ in common carp (Cyprinus carpio haematopterus). Front. Immunol. 13:1110322. doi: 10.3389/fimmu.2022.1110322
Received: 28 November 2022; Accepted: 19 December 2022;
Published: 06 January 2023.
Edited by:
Heng Chi, Ocean University of China, ChinaReviewed by:
Jingguang Wei, South China Agricultural University, ChinaXue Qiao, Dalian Ocean University, China
Copyright © 2023 Zhao, Zhang, Qiao, Gao, Gu, Jiang, Zhu and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianghui Kong, eGhrb25nQGh0dS5jbg==
†These authors have contributed equally to this work and share first authorship
 Yanjing Zhao
Yanjing Zhao Jie Zhang†
Jie Zhang† Xinyu Jiang
Xinyu Jiang Lei Zhu
Lei Zhu Xianghui Kong
Xianghui Kong