- 1Infection and Inflammation Program, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 2Faculty of Medicine, The University of Queensland, Brisbane, QLD, Australia
- 3Genetics & Computational Biology Department, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 4Genecology Research Centre, University of the Sunshine Coast, Sunshine Coast, QLD, Australia
- 5School of Veterinary Science, The University of Queensland, Gatton, QLD, Australia
Employing the flatworm parasite Schistosoma mansoni as a model, we report the first application of CRISPR interference (CRISPRi) in parasitic helminths for loss-of-function studies targeting the SmfgfrA gene which encodes the stem cell marker, fibroblast growth factor receptor A (FGFRA). SmFGFRA is essential for maintaining schistosome stem cells and critical in the schistosome-host interplay. The SmfgfrA gene was targeted in S. mansoni adult worms, eggs and schistosomula using a catalytically dead Cas9 (dCas9) fused to a transcriptional repressor KRAB. We showed that SmfgfrA repression resulted in considerable phenotypic differences in the modulated parasites compared with controls, including reduced levels of SmfgfrA transcription and decreased protein expression of SmFGFRA, a decline in EdU (thymidine analog 5-ethynyl-2’-deoxyuridine, which specifically stains schistosome stem cells) signal, and an increase in cell apoptosis. Notably, reduced SmfgfrA transcription was evident in miracidia hatched from SmfgfrA-repressed eggs, and resulted in a significant change in miracidial behavior, indicative of a durable repression effect caused by CRISPRi. Intravenous injection of mice with SmfgfrA-repressed eggs resulted in granulomas that were markedly reduced in size and a decline in the level of serum IgE, emphasizing the importance of SmFGFRA in regulating the host immune response induced during schistosome infection. Our findings show the feasibility of applying CRISPRi for effective, targeted transcriptional repression in schistosomes, and provide the basis for employing CRISPRi to selectively perturb gene expression in parasitic helminths on a genome-wide scale.
Introduction
Schistosoma mansoni is a flatworm parasite that causes schistosomiasis, a disease which afflicts 250 million people in 74 countries (1–4). Currently, no anti-schistosome vaccines are available for human use and clinical treatment relies entirely on the single drug praziquantel (PZQ). The potential emergence of PZQ drug resistance is an ever-present concern (1). Effective vaccines and new treatments for reducing the global burden of schistosomiasis are thus needed urgently. The past few decades have witnessed new advances in schistosome developmental biology, genomics, proteomics and transcriptomics, and in our understanding of schistosome-induced pathogenesis, and the host-parasite interaction (1, 2, 5–10). Notably, complete genomic sequences of the three main schistosome species (S. mansoni (5, 6), S. japonicum (7, 8) and S. haematobium (9, 10)) have been released which provide valuable information to decipher the molecular biology of these blood flukes. However, progress in identifying and characterizing effective drug targets and vaccine candidates has been severely hampered by a general paucity of suitable molecular tools for modulation of critical genes in schistosomes. RNA interference (RNAi) has been developed as a post-transcriptional gene silencing tool over the past decade for loss-of-function studies in helminths (11–15), which generated variable levels of silencing efficiency and the outcomes were either transient or the inheritance of silencing effects was not fully penetrant (11–15).
The clustered regularly interspaced short palindromic repeat (CRISPR) approach has emerged as a novel genomic editing tool that has been broadly adapted in various organisms (16–26). CRISPR was first identified in bacteria as a defense mechanism against foreign genetic elements, utilizing RNA-guided CRISPR-associated protein (Cas) endonucleases to recognize and cleave invading viral DNA (27–32). The CRISPR system has been repurposed for transcription regulation using a catalytically dead version of Cas9 (dCas9) (with mutations at H840A and D10A) lacking endonucleolytic activity (33). The dCas9-sgRNA (single guide RNA) complex can specifically interfere with transcriptional initiation or transcriptional elongation, in a process which was termed CRISPR interference (CRISPRi) (33, 34). The dCas9 protein can also be fused to transcriptional repressor domains [eg. Krüppel-associated box (KRAB)] to achieve more effective transcriptional silencing (34–37). CRISPRi is a simple and cost-effective gene regulation tool with greater versatility, higher efficacy and specificity (38–40). To date, CRISPRi has been broadly employed for transcription repression, directed evolution, metabolic engineering and targeted genetic screening in mammalian cells, in the zebra fish, in Caenorhabditis elegans and a variety of unicellular organisms such as Synechococcus elongatus, Saccharomyces cerevisiae, Toxoplasma gondii and Plasmodium falciparum (33–45). To advance more efficient gene regulation in parasitic helminths, we applied CRISPRi in S. mansoni.
The life cycle and morphology of schistosomes are both complex. Adult S. mansoni pass eggs that escape the mammalian host in faeces. The egg gives rise to a ciliated larva, the miracidium that infects its specific Biomphalaria snail host, within which it undergoes a dramatic body conversion to produce an obligate asexually-reproducing adult, the mother sporocyst. Endogenous proliferation of stem cells (=germinal cells in the asexual stage) in the mother sporocyst leads to a new asexual stage - the daughter sporocyst (46). These ‘daughters’, in turn, can generate, by germinal cell proliferation, the next stage, the migratory cercariae, that escape from the snail into the aquatic environment (47). Cercariae seek and then penetrate the skin of a mammal, transform into schistosomula, which enters the vasculature to develop into dimorphic sexual adults. After paired with males, mature female worms lay eggs, which are crucial in host pathogenesis, immune modulation, and the transmission of schistosomiasis. Intra-mammalian development, driven by stem cells (defined as neoblast cells), gives rise to the remarkable reproductive capability of adults (48), over prolonged periods - indeed, schistosomes can live for over 30 years (49). To unravel the critical roles of stem cells in driving the schistosome life cycle (48, 50–53), characterization of the somatic stem cell marker SmFGFRA has drawn increased attention (50–55). Our previous study demonstrated SmFGFRA is abundantly expressed in different S. mansoni developmental stages (56). The distribution pattern of SmFGFRA in embryonic cells of immature eggs, in the neural mass of mature eggs and miracidia, and its co-location with EdU+ (thymidine analog 5-ethynyl-2’-deoxyuridine) cells in adult S. mansoni, strongly implicated its important roles in maintaining schistosome stem cells, in development of the nervous and reproductive systems, and in the host-parasite interplay (56).
Herein, for the first time, we report the application of CRISPRi in S. mansoni targeting the SmfgfrA gene. We firstly pre-screened eight sgRNAs, designed specifically targeting different loci of SmfgfrA, for effective inhibition of its transcription initiation and elongation. We determined the distinct phenotypic changes in SmfgfrA-repressed adult worms, schistosomula and eggs. Then, we injected the SmfgfrA-repressed eggs into the tail vein of mice and explored the pathogenicity induced by these SmfgfrA-repressed eggs in vivo.
Methods
Ethics
All experiments were approved by the Animal Ethics Committee (ethics number P242) of the QIMR Berghofer Medical Research Institute. The study was carried out based on the guidelines of the National Health and Medical Research Council of Australia, as published in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th edition, 2004 (www.nhmrc.gov.au). All work involving live S. mansoni parasites was conducted in quarantine-accredited premises.
Maintenance of parasites
Swiss mice (female, 6 weeks old) were infected with 100 S. mansoni cercariae subcutaneously. The infected mice were euthanized seven weeks post-infection and adult worms were harvested by portal perfusion using 37°C pre-warmed RPMI Medium 1640 (Gibco, Sydney, Australia). Adult worms were cultured overnight in RPMI complete medium [RPMI Medium 1640 (Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gibco) and 100 IU/ml penicillin and 100 μg/ml streptomycin (Gibco)] at 37°C in an atmosphere of 5% CO2. Mouse livers were removed at necropsy and liver eggs were isolated and purified as described (57). Liver eggs were cultured in RPMI complete medium at 37°C in 5% CO2. S. mansoni cercariae were obtained by shedding infected Biomphalaria glabrata snails under bright light. Schistosomula were obtained by mechanical transformation of cercariae in vitro and cultured in Basch’s medium as described (58).
Design of guide RNA targets and reconstruction of vectors
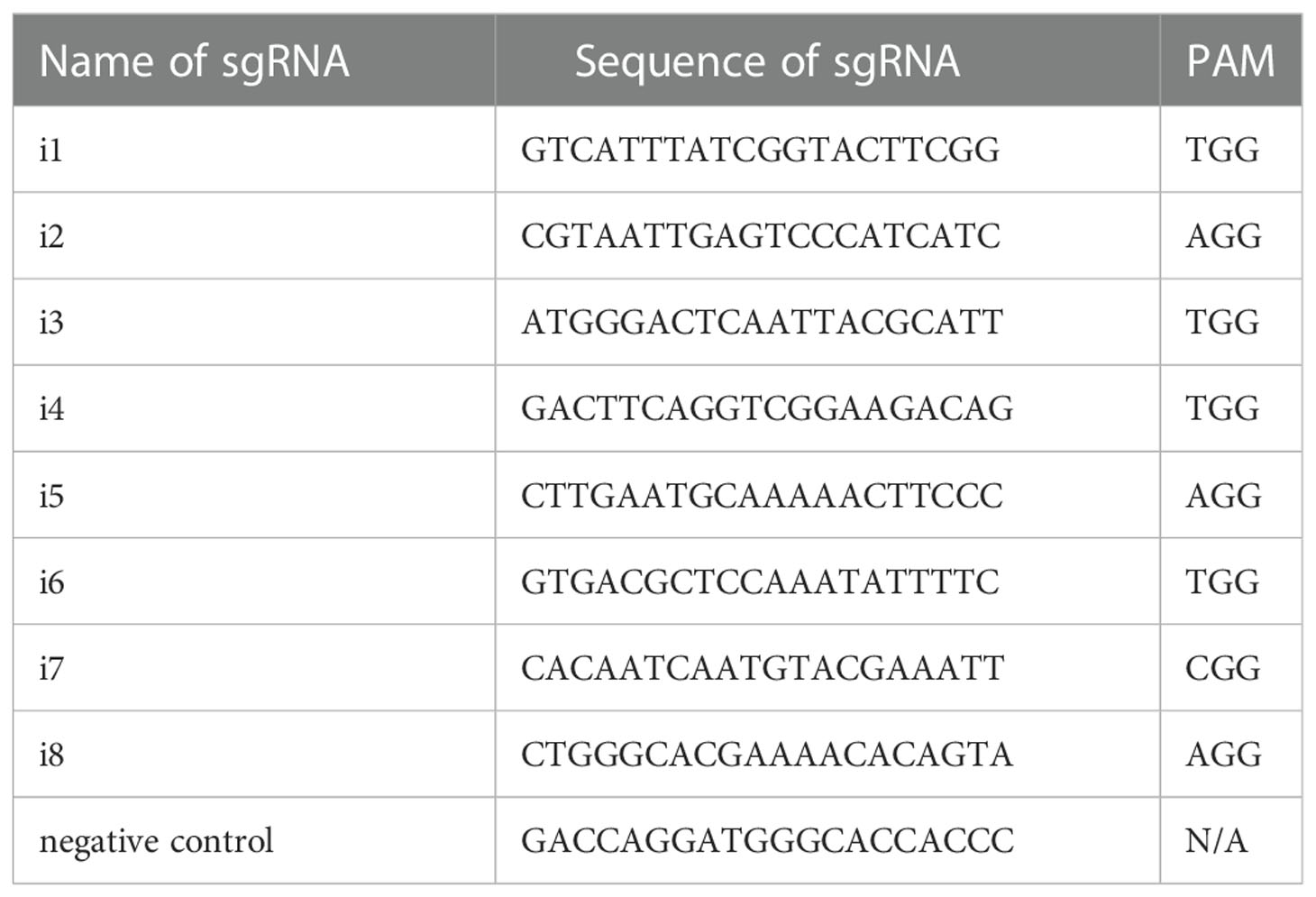
Single-guide RNAs (sgRNAs) were designed utilizing the web-based tools available at https://bioinfogp.cnb.csic.es/tools/breakingcas/ (59) and the Benchling application (https://benchling.com) to predict binding sites for the Streptococcus pyogenes dCas9 nuclease within the genome of S. mansoni. The location of the transcriptional start site (TSS) was determined using WormBase ParaSite (https://parasite.wormbase.org/index.html). SmfgfrA (Smp_175590) comprises thirteen exons separated by twelve introns spanning 51.95 kb on the reverse strand of S. mansoni chromosome 1, including a 64 bp 5’ untranslated region (5’ UTR) (Figure 1A). Eight sgRNAs (i1-i8) were designed uniquely targeting either DNA strand of SmfgfrA to determine whether CRISPRi could induce efficient repression of transcription initiation/elongation of this gene in S. mansoni. SgRNA i1, i2, i3, i7, and i6 align to nucleotides 667-686 (+67 bp to +86 bp relative to the predicted TSS), 703-722 (+103 bp to +122 bp), 707-726 (+107 bp to +126 bp), 744-763 (+144 bp to +163 bp), and 783-802 (+183 bp to +202 bp) in exon 1, respectively (Figure 1B); sgRNA i5 and i4 target nucleotides 871-890 (+271 bp to +290 bp) and 910-919 (+310 bp to +329 bp) in exon 2, respectively (Figure 1B). For the inhibition of transcription initiation, sgRNA i8 was designed to target residues 580-599 (-21 bp to -2 bp upstream of the TSS) of SmfgfrA (Figure 1B). All sgRNAs are adjacent to the protospacer adjacent motif (PAM), NGG. SgRNA oligonucleotides and PAM sequences are listed in Table 1. A non-targeting sgRNA (5’-GACCAGGATGGGCACCACCC-3’) was used as a negative control (NC). The sgRNAs were synthesized as double-stranded DNA fragments flanked by BstXI and XhoI restriction sites (Integrated DNA Technologies, Singapore) and inserted into the vector, pgRNA-humanized (a gift from Stanley Qi, Addgene plasmid #44248) (33). Expression of sgRNA is driven by the mouse U6 promoter. The CRISPRi vector PHR-SFFR-dCas9-BFP-KRAB (a gift from Stanley Qi & Jonathan Weissman, Addgene plasmid #46911) (34) contains a silencing-prone spleen focus forming virus (SFFV) promoter expressing dCas9 fused to the KRAB transcription repressor.
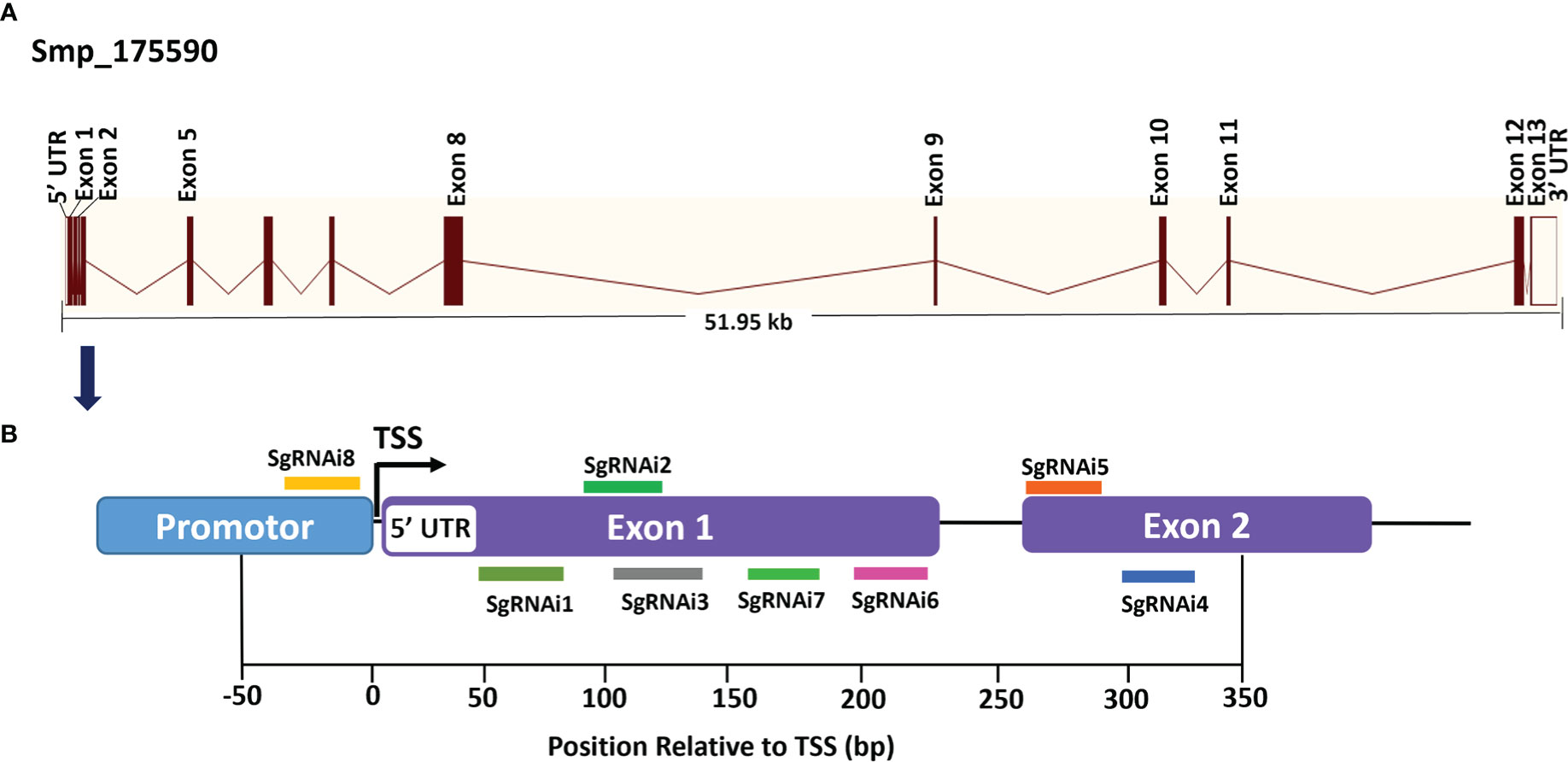
Figure 1 Gene structure of S. mansoni fibroblast growth factor receptor A (SmfgfrA) and locations of singe-guide RNAs (sgRNAs). (A) Schematic diagram of the SmfgfrA (Smp_175590) gene containing thirteen exons, twelve introns, 5’ untranslated region (5’ UTR) and 3’ UTR, spanning 51.95 kb on the reverse strand of S. mansoni chromosome 1. (B) Diagram of SmfgfrA indicating target sites of eight sgRNAs (i1-i8). SgRNAs (i1-i7) were designed for inhibition of transcription elongation within a window from +67 bp to +329 bp relative to the transcriptional start site (TSS) of SmfgfrA. SgRNA i8 aligns to the promotor region (-21 bp to -2 bp) of SmfgfrA.
Transfection of Schistosoma mansoni parasites
To perform CRISPRi repression targeting SmfgfrA in adult S. mansoni and select the most effective sgRNAs, 5 pairs of adults were separated into 5 males and 5 females and then subjected to electroporation in 200 μl Opti-MEM containing 3 μg PHR-SFFR-dCas9-BFP-KRAB vector and 3 μg of pgRNA vector reconstructed with: (1) negative control sgRNA (non-targeting RNA sequence) (NC); (2) i1 sgRNA (i1); (3) i2 sgRNA (i2); (4) i3 sgRNA (i3); (5) i4 sgRNA (i4); (6) i5 sgRNA (i5); (7) i6 sgRNA (i6); (8) i7 sgRNA (i7); (9) i8 sgRNA (i8); (10) i4 sgRNA and i5 sgRNA (i4+i5, 3 μg of each); (11) i4 sgRNA+i5 sgRNA+i6 sgRNA (i4+i5+i6, 3 μg of each); or (12) i5 sgRNA+i8 sgRNA (i5+i8, 3 μg of each). The mixture was transferred into a pre-chilled 4 mm electroporation cuvette (Bio-Rad, Sydney, Australia) and subjected to square wave electroporation using a single 20 millisecond pulse of 125 Volts (Gene Pulser Xcell Electroporator, Bio-Rad) (60). Thereafter, electroporated adult worms (5 males and 5 females per well) were cultured in RPMI complete medium at 37°C in 5% CO2 and collected three days post-electroporation. Wild type (WT) adult worms were not subjected to electroporation but were cultured under the same conditions as control. Eggs laid in vitro by adult female worms were collected three days post-electroporation and the collected eggs in each group were counted.
Based on the results in adult worms, we utilized two relatively more effective sgRNAs (sgRNA i4 and sgRNA i5) for repression of SmfgfrA in eggs and schistosomula. Briefly, 10,000 liver eggs or 2000 schistosomula were subjected to electroporation in 200 μl Opti-MEM containing 3 μg PHR-SFFR-dCas9-BFP-KRAB vector and 3 μg of pgRNA vector reconstructed with control sgRNA, sgRNA i4 and sgRNA i5, respectively. WT and NC-treated liver eggs or schistosomula were used as controls. After electroporation, eggs were maintained in RPMI complete medium and schistosomula were cultured in Basch’s medium (2) for three days. Hatched miracidia were collected from SmfgfrA-modulated eggs and control eggs cultured for seven days post-electroporation and the egg hatching efficiency (%) was determined by dividing the number of hatched eggs by the total number of eggs X 100.
Real-time PCR
Total RNAs were extracted from SmfgfrA-repressed and control (WT and NC-treated) S. mansoni eggs, schistosomula, adult worms and miracidia hatched from SmfgfrA-repressed and control eggs using RNeasy Mini Kits (Qiagen, Melbourne, Australia), followed by cDNA synthesis using QuantiTect Reverse Transcription Kits (Qiagen). For each extraction, 10, 000 eggs or 2000 schistosomula or 5 males and 5 females or 2000 miracidia were used. Three biological repeats and three technical repeats were undertaken. Real-time PCR was conducted using QuantiNova SYBR® Green PCR Kits (Qiagen) on a Mic qPCR Cycler (Bio Molecular Systems, Upper Coomera, QLD, Australia). Forward primer (5’-ATGGGACTCAATTACGCATT-3’) and reverse primer (5’-CACCACTGTCTTCCGACCTG-3’) for SmfgfrA were designed using the Primer 3 software (http://frodo.wi.mit.edu/), and the specificity of the primer sequences was confirmed by the Basic Local Alignment Search Tool (BLAST) at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). S. mansoni glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the house keeping reference gene (61). Real-time PCR reactions contained 10 µl 2xSYBR Green PCR Master Mix, 100 ng cDNA, and 0.7 µM of each primer. The cycling parameters were set as follows: 95°C for 5 min, 40 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s. Data analysis was performed using the Mic qPCR software (Bio Molecular Systems). Relative SmfgfrA transcription levels in each group were determined using the 2-ΔΔCt calculation (62) by normalizing to the control parasites (NC-treated parasites).
Caspase-3/-7 activity assay
S. mansoni soluble egg antigen (SEA), soluble worm antigen preparation (SWAP) and soluble schistosomula native antigens were prepared in PBST (PBS+0.1% v/v Tween-20) plus 10 mM HEPES and cOmplete™, Mini, EDTA-free Protease Inhibitor (Sigma-Aldrich, Sydney, Australia) as described (63, 64). Protein concentration was determined using the Bradford assay as described (65). The caspase-3/-7 activity of SWAP (0.4 mg/ml), SEA (0.2 mg/ml) and soluble schistosomula antigens (0.2 mg/ml) was determined using the Caspase-Glo® 3/7 Assay System (Promega, Sydney, Australia) according to the manufacturers’ instructions.
Western blotting
S. mansoni SWAP (40 µg) and SEA (20 µg) were separated on 12% (w/v) SDS-PAGE gels and transferred to an Immun-Blot low fluorescence-PVDF membrane (Bio-rad). The membrane was first blocked with Odyssey Blocking Buffer (TBS) (LI-COR Biosciences, Lincoln, Nebraska, USA) for 1 h at room temperature with shaking. Subsequently, the membrane was incubated with a polyclonal mouse anti-rSmFGFRA-L (anti-recombinant SmFGFRA extracellular ligand binding domain) antibody (previously generated in our laboratory (56)) (1:100 diluted in Odyssey buffer with 0.1% (v/v) Tween-20) for 1 h with shaking at room temperature. Following washes (4X) in Tris-Buffered Saline (TBS)/0.1% Tween-20 (TBST), the membrane was incubated with IRDye-labeled 680LT goat anti-mouse IgG antibody (Li-COR Biosciences) (1:15,000 diluted in Odyssey buffer with 0.1% Tween-20 and 0.01% SDS) for 1 h with shaking in a dark chamber. After four washes with TBST, the membrane was dried in the dark and visualized using the Odyssey CLx Infrared Imaging System (64). An anti-actin antibody (Sigma-Aldrich) was used to probe the expression of actin to ensure the equal loading of samples.
EdU staining
SmfgfrA-modulated parasites were cultured at 37°C under 5% CO2 in RPMI complete medium containing 10 µM EdU (Thermo Fisher Scientific, Brisbane, Australia), which specifically stains stem cells in S. mansoni (53). After 24 h, the stained worms were fixed in 10% formalin, paraffin embedded and sectioned. Three sections (4 µm/section, with 4 µm distance between each section) of EdU-labeled adult worms and schistosomula were subjected to EdU detection using a Click-iT™ EdU Cell Proliferation Kit (Alexa Fluor™ 488 dye) (Thermo Fisher Scientific) according to the manufacturers’ instructions. Nuclei in all tissue sections were also stained with Propidium iodide (PI) (Sigma-Aldrich) and visualized using a Zeiss 780 NLO confocal microscope (Zeiss, Oberkochen, Germany). Counting of EdU+ cell nuclei and PI+ cells was performed using QuPath software (https://qupath.github.io) (66). The percentage of EdU-positive cells was calculated by dividing the EdU+ cell number with the PI+ cell number X 100.
Miracidial behavioral assay
Miracidia were hatched in deionized water under light from SmfgfrA-repressed and control (WT, NC-treated) eggs. Miracidia were then harvested (58) and their behavior were monitored as described (67, 68). Briefly, approximately 30 S. mansoni miracidia in 100 μl deionized water were distributed evenly to a microscope slide. Miracidial movement (swimming) in the field of view (FOV) was detected utilizing an Olympus-CKX41 microscope equipped with an Olympus DPI Digital Microscope Camera DP22 (25 frames per second at 2.8-megapixel image quality). Miracidial movement was recorded for 1 minute by video and followed by analyzing with the FIJI software to calculate velocity (speed) of miracidial swimming, the tortuosity (the ratio of track length to maximum displacement) of miracidial swimming and the duration (time) of miracidia staying in the FOV (67). The miracidial movement velocity was determined in pixels employing the rolling mean subtraction approach (67, 68). The location of miracidial was monitored in each frame along an x-y axis and the trajectories were interpolated utilizing TrackMate (the plugin for FIJI software) (68, 69). Applying the MTrackJ (ImageJ plugin), the average velocity, duration and tortuosity of miracidia movement in each video were determined. The heatmaps were produced as mentioned (68, 70) for demonstrating the movement pattern of individual miracidia. Around 30 miracidia were used for each assay and 10 assays were performed for each group (totally 300 miracidia were examined in each group).
Intravenous injection of SmfgfrA-repressed eggs into mice
Swiss mice were injected intravenously (i.v.) with SmfgfrA-repressed or control (WT eggs and NC-treated eggs) eggs as described (60). Briefly, eggs were cultured for 48 h after electroporation, followed by three washes with chilled PBS. Then, 1000 eggs in 100 μl sterile PBS were injected into the lateral tail vein of female Swiss mice (8-9 weeks of age). Mice injected with sterile PBS and control eggs (WT eggs and NC-treated eggs) were served as negative control mice. Mice (5 mice/group) were euthanized two weeks post-injection.
Serum IgE level and lung granuloma size in mice exposed to SmfgfrA-repressed eggs
Two weeks post-injection, blood was obtained from each mouse and sera were prepared individually. The total immunoglobulin E (IgE) level in the serum samples was measured using an IgE mouse ELISA kit (Thermo Fisher Scientific, Waltham, Mass, USA) according to the manufacturer’s instructions.
To determine the size of granuloma that had formed in the lungs, the left lung of each mouse was fixed in 10% (v/v) formalin, paraffin embedded and sectioned. Sections (4 μm) of these paraffin blocks were stained with Hematoxylin and Eosin (H&E) to evaluate inflammatory infiltrates and the cellularity of granulomas. Slides were imaged using an Aperio Slide Scanner (Aperio Technologies, Vista, CA, USA) and analyzed using Aperio Image Scope v11.1.2.760 software (Leica Biosystems Imaging, Buffalo Grove, IL, USA). The degree of lung pathology was quantified by measurement of the area density of granulomatous lesions. The granuloma ratio in each lung sample was estimated from the total area of the granulomas in the lung sample divided by the total area of the lung tissue and X 100. For each group, 10 slides per lung (5 lungs/group), totaling 50 samples, were measured.
Statistical analysis
GraphPad Prism software (Version 8.2.1, La Jolla, CA, USA) was used for all statistical analyses. All data are shown as the mean ± SE. Differences between groups were analyzed for statistical significance by One-way ANOVA and, where appropriate, by two-tailed Student’s t-test. A statistically significant difference for a particular comparison was defined as a p value ≤ 0.05. * p value≤ 0.05, ** p value≤ 0.01, *** p value ≤ 0.001, **** p value ≤ 0.0001, not significant (ns).
Results
CRISPRi-mediated repression of SmfgfrA in Schistosoma mansoni adult worms
SmfgfrA transcription level in SmfgfrA-repressed adult worms
The transcription level of SmfgfrA was quantified in CRISPRi-modulated S. mansoni adult worms, liver eggs and schistosomula using real-time PCR assays. We found the most reduced level of transcription of SmfgfrA was induced by sgRNA i5 (51.6%, p<0.0001) in adult worms, followed by i4 (43.7%, p<0.0001), i6 (27.7%, p=0.0018), i7 (22.6%, p=0.0083), and i8 (15.4%, p=0.0184) (Figure 2A). No significant downregulation of SmfgfrA transcripts was observed in worms electroporated with i1, i2 or i3, compared with the control groups (WT and treated with NC) (Figure 2A). As a result, we selected i4 and i5 for subsequent study. To further investigate whether the gene repression efficiency was sex dependent, we determined SmfgfrA transcription levels in i4-treated and i5-treated male and female worms. Notably, both sgRNA i4 and sgRNA i5 induced similar transcriptional reductions in SmfgfrA in both male and female worms (Figure 2B); accordingly, we used mixed male and female adult worms (equal number of males and females) in all subsequent studies.
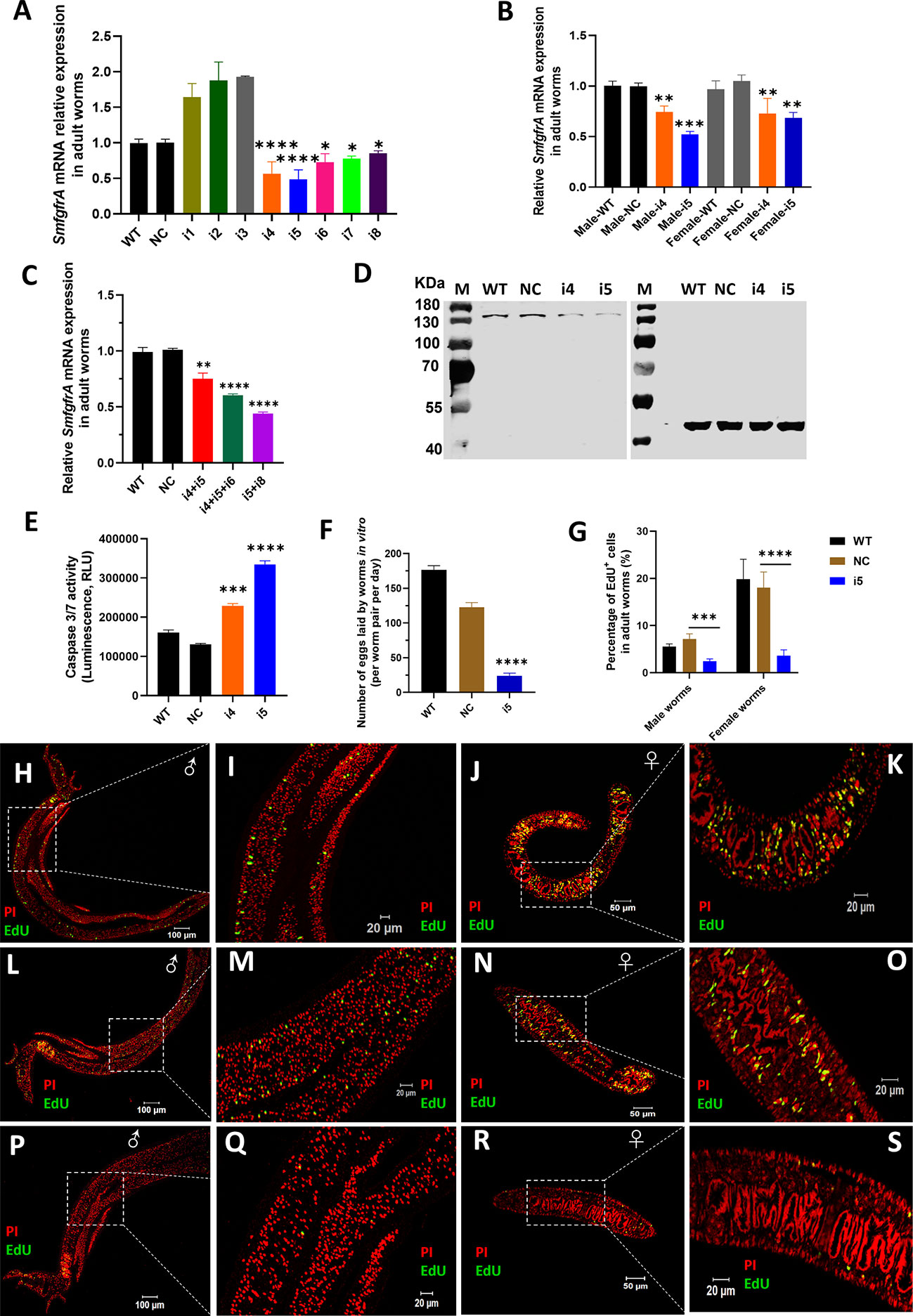
Figure 2 CRISPRi of SmfgfrA in S. mansoni adult worms. Transcription level of SmfgfrA was determined in (A) paired adult worms electroporated with a CRISPRi vector (PHR-SFFR-dCas9-BFP-KRAB) combined with reconstructed pg-RNA vector containing sgRNA (i1, i2, i3, i4, i5, i6, i7, i8), individually; (B) adult male and female worms treated with sgRNA i4 or i5; (C) paired worms treated with sgRNA i4+i5, i4+i5+i6, or i5+i8. Wild-type (WT) worms and worms treated with CRISPRi vector combined with pg-RNA vector containing a negative control (non-targeting) sgRNA (NC) were used as controls. Experiments in panels (A–C) were performed in triplicates and all data are shown as the mean ± SE. (D) Western blot demonstrating expression level of SmFGFRA protein in soluble worm antigen preparation (SWAP) of WT adult worms (Lane 1) and worms treated with NC (Lane 2), i4 (Lane 3) or i5 (Lane 4). An anti-actin antibody was employed to ensure equal protein loading. The electrophoresed SWAPs were transferred for the western blot analysis to two PVDF membranes which were probed simultaneously with the anti-rSmFGFRA-L antibody (left blot) and the anti-actin antibody (right blot). (E) Caspase-3/-7 activity measured in i4 and i5-treated adult worms. This assay was conducted in duplicate with all data presented as the mean ± SE. (F) The number of eggs laid by WT adult worms, NC-treated adult worms or i5-treated adult worms in vitro. This experiment was conducted in triplicates with all data shown as the mean ± SE. (G) The effect of SmfgfrA repression on the percentage of EdU+ cells in i5-treated male worms and female worms. WT and NC-treated male and female worms were used as controls. Cell nuclei in all samples were also stained with Propidium iodide (PI). Percentage of EdU+ cells in each sample was calculated by dividing the EdU+ cell number with PI+ cell number X 100. All data are shown as the mean ± SE (WT male worm: n=13, WT female worm: n=8, NC male worm: n=12, NC female worm: n=8, i5 male worm: n=10, i5 female worm: n=15). Confocal projections representative signals of EdU (green) and PI (red) in (H) WT male worm, (J) WT female worm, (L) NC-treated male worm, (N) NC-treated female worm, (P) i5-treated male worm and (R) i5-treated female worm. (I, K, M, O, Q, S) are magnified squared-region in (H, J, L, N, P, R), respectively. (Statistical significance was established employing One-way ANOVA by comparing with NC group: * p value≤ 0.05, *** p value ≤ 0.001). ** p value≤ 0.01, **** p value ≤ 0.0001.
It has been shown that CRISPRi conducted in Escherichia coli (E. coli) using multiplexed sgRNAs targeting the same gene can markedly increase gene repression efficiency compared with using single sgRNAs (33). To investigate this scenario in schistosomes, we tested the combination of sgRNA i4+i5, i4+i5+i6 and i5+i8, and this resulted in the downregulation of gene transcription by 25.6% (p=0.0017), 40.25% (p<0.0001) and 56.5% (p<0.0001) in the treated adult S. mansoni worms respectively (Figure 2C), compared with NC-treated parasites. Given the similar gene repression efficiency induced by the CRISPRi using single sgRNAs or multiplexed sgRNAs in silencing SmfgfrA, we decided to perform CRISPRi with individual sgRNA i5 and/or sgRNA i4 for subsequent phenotypic change studies.
Reduction of SmFGFRA protein expression in SmfgfrA-repressed adult worms
To determine whether the repression of SmfgfrA was reflected at the translational level, we performed western blot analysis using an anti-rSmFGFRA-L polyclonal antibody to probe SWAP extracted from CRISPRi- modulated adult worms. SEA generated from WT worms and NC-treated worms served as controls. A clearly decreased level of SmFGFRA protein expression was evident in SWAP extracted from i4-treated (Figure 2D, Lane 3) and i5-treated (Figure 2D, Lane 4) adult worms compared with that from WT adults (Figure 2D, Lane 1) and NC-treated adults (Figure 2D, Lane 2).
Increased caspase-3/-7 activity in SmfgfrA-repressed adult worms
Caspase-3 and caspase-7 are major executioner caspases that play critical roles in coordinating cell apoptosis, and thus determining their activities has been broadly used for monitoring apoptosis (71–73). Therefore, we determined the effect of SmfgfrA-repression on apoptosis in extracts of CRISPRi- modulated parasites by measuring the activity of caspase-3/-7. Remarkably, caspase-3/-7 activity in i4-treated and i5-treated adult worms was enhanced by 43% (p=0.0003) and 60.9% (p<0.0001), respectively (Figure 2E).
Since sgRNA i5 showed significantly higher efficiency than sgRNA i4 in adult S. mansoni, we selected sgRNA i5 for following studies.
Reduced adult worm egg production
To further investigate the effects of SmfgfrA-repression on egg production by adult females, the number of eggs laid in vitro by CRISPRi- modulated paired worms was determined. Notably, the egg production in i5-treated adult worms was dramatically depleted by 80.4% (p<0.0001) compared with NC-treated worms (Figure 2F).
Decline in EdU incorporation in SmfgfrA-repressed adult worms
EdU is able to be incorporated into newly synthesized cellular DNA, and stains only proliferating stem cells in schistosomes (53). To assess the effect of SmfgfrA-repression on schistosome stem cells, we monitored EdU incorporation in CRISPRi-modulated parasites. Representative confocal images of EdU signaling in sections of adult males and females are shown in Figures 2H–S. We found that ~7.17% and ~18% of cells in NC-treated male and NC-treated female worms, respectively, were EdU+, whereas the percentage of EdU+ cell nuclei was markedly reduced to ~2.53% (decreased by 64.7%, p=0.0007) in i5-treated male worms, and to ~3.8% (reduced by 78.9%, p<0.0001) in i5-treated female worms (Figure 2G).
CRISPRi-mediated silencing of SmfgfrA in Schistosoma mansoni schistosomula
In CRISPRi-modulated schistosomula, we found SmfgfrA-specific transcripts were clearly repressed by 17.9% (p=0.0006) in i5-treated parasites, while no significant change in the SmfgfrA mRNA level was observed in i4-treated schistosomula (Figure 3A). We then examined apoptosis in schistosomula following repression of SmfgfrA by measuring caspase-3/-7 activity. In i5-transfected schistosomula, the caspase-3/-7 activity was clearly increased by 21.7% (p=0.0057) but no significant difference was evident in i4-treated schistosomula, compared with the NC group (Figure 3B). Considering the clearly higher efficiency of sgRNA i5 than sgRNA i4 in schistosomula, we thus utilized sgRNA i5 for the following EdU staining analysis.
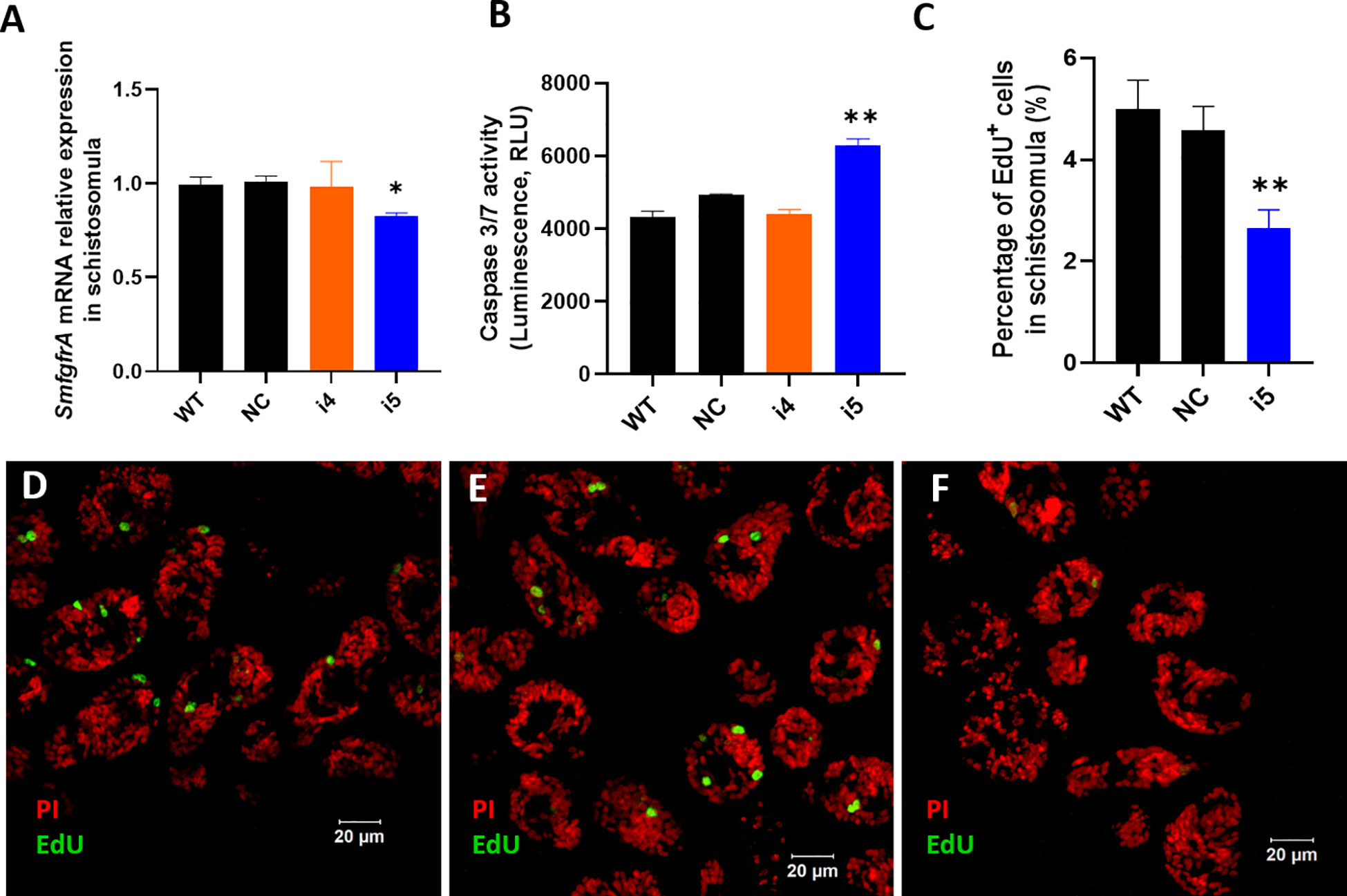
Figure 3 CRISPRi-mediated SmfgfrA repression in S. mansoni schistosomula. (A) SmfgfrA transcription level in untreated WT schistosomula and schistosomula treated with NC, i4 or i5. (B) Effect of SmfgfrA-repression on caspase-3/-7 activity in i4-treated or i5-treated schistosomula. WT schistosomula and schistosomula treated with NC were used as controls. Experiments in panels (A, B) was performed in duplicate with data presented as the mean ± SE. (C) The percentage of EdU+ cell nuclei in WT schistosomula and schistosomula treated with NC, i4 or i5. All data are demonstrated as the mean ± SE (WT: n=33; NC: n=42; i5: n=42). Confocal projections representing EdU (green) and PI (red) labeled (D) WT schistosomula, (E) NC-treated schistosomula and (F) i5-treated schistosomula. * p value≤ 0.05, ** p value≤ 0.01.
EdU labelling was undertaken to determine whether the silencing of SmfgfrA affected germinal cells in schistosomula. Figures 3D–F shows confocal images of EdU signal in sections of SmfgfrA-repressed schistosomula. We found the percentage of EdU+ cells in i5-treated schistosomula (~2.3% EdU+ cells) was considerably reduced by 45.6% (p=0.005) when compared with NC-treated schistosomula (~4.3% EdU+ cells) (Figure 3C).
CRISPRi-induced repression of SmfgfrA in Schistosoma mansoni eggs
Reduced SmfgfrA-specific transcripts
SmfgfrA transcription levels in CRISPRi-repressed eggs treated with i4 and i5 were substantially reduced by 46.9% (p=0.0329) and 67.3% (p=0.0056), respectively (Figure 4A). Notably, a marked decrease (25.3%, p=0.0025) of SmfgfrA-specific transcripts was also detected in miracidia hatched from i5-treated eggs whereas no significant change was evident in miracidia collected from i4-treated eggs (Figure 4B).
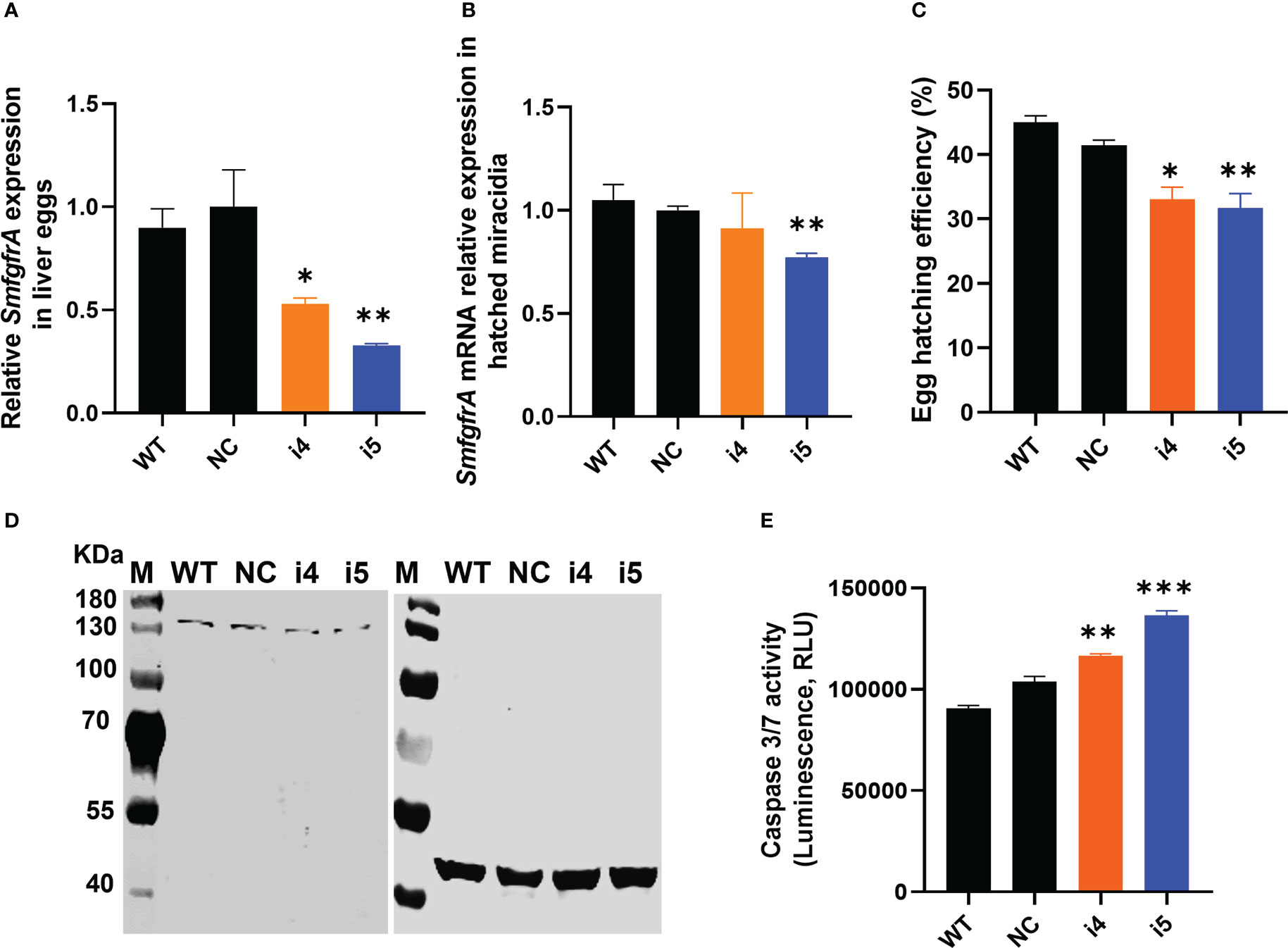
Figure 4 CRISPRi of SmfgfrA in S. mansoni eggs. (A) SmfgfrA mRNA level in eggs treated with i4 or i5. WT eggs and NC treated eggs were used as controls. (B) Transcription level of SmfgfrA in miracidia hatched from WT eggs and eggs treated with NC, i4 or i5. (C) Effects of SmfgfrA repression on the hatching of S. mansoni eggs. The hatching efficiency (%) of WT eggs and eggs treated with NC, i4 or i5 was calculated by dividing the number of hatched eggs with the total number of eggs (hatched and unhatched) X 100. Experiments in panels (A–C) was performed in triplicates with all data presented as the mean ± SE. (D) Western blot showing expression level of SmFGFRA protein in SEA of WT eggs (Lane 1) and eggs treated with NC (Lane 2), i4 (Lane 3) or i5 (Lane 4). An anti-actin antibody was utilized as control to ensure equal loading. The electrophoresed SEAs were transferred for the western blot analysis to two PVDF membranes which were probed simultaneously with the anti-rSmFGFRA-L antibody (left blot) and the anti-actin antibody (right blot). (E) Caspase-3/-7 activity in soluble egg antigen (SEA) of WT eggs and eggs treated with NC, i4 or i5. This assay was conducted in duplicate with all data presented as the mean ± SE. * p value≤ 0.05, ** p value≤ 0.01, *** p value ≤ 0.001.
Decreased hatching efficiency of SmfgfrA-repressed eggs
To determine whether SmfgfrA suppression had an effect on the hatching ability of modulated eggs, we assessed the hatching ability of WT eggs and eggs treated with NC, i4 or i5. Markedly reduced egg hatching efficiency was evident in i4-treated eggs (20.2%, p=0.0156) and i5-treated eggs (23.5%, p=0.0069) (Figure 4C), compared with eggs treated with NC.
Decreased SmFGFRA expression and increased apoptosis in modulated eggs
To determine the extent of gene regulation at the protein level, we extracted SEA from SmfgfrA-repressed eggs and performed western blotting and caspase-3/-7 activity assays. Consistent with the observations with adult worms, SmFGFRA expression in i4-treated (Figure 4D, Lane 3) and i5-treated (Figure 4D, Lane 4) eggs was clearly reduced, in comparison with that of NC-treated eggs (Figure 4D, Lane 2). Furthermore, caspase-3/-7 activity in i4-treated and i5-treated eggs was elevated by 11% (p=0.0097) and 24% (p=0.0002) compared to NC-treated eggs, respectively (Figure 4E).
In consistent with the results in adult worms and schistosomula, sgRNA i5 induced clearly higher repression efficacy than sgRNA i4 in eggs, we selected sgRNA i5 for the following CRISPRi studies in eggs.
Modified behavior of miracidia hatched from SmfgfrA-repressed eggs
Behavioral changes of miracidia hatched from SmfgfrA-repressed eggs were investigated by analyzing recordings of miracidial movement tracks. Heatmaps were created to illustrate the behavior of individual miracidia within a 1 minute recording. The heatmaps of control miracidia (WT miracidia and miracidia hatched from NC-treated eggs) depicted linear soft blue lines, indicating these miracidia had less circular and faster movement (Figures 5A, B). In contrast, there were more circular lines and more abundant red and yellow regions in heatmaps of miracidia hatched from the i5-treated eggs (Figure 5C), suggesting more turning and circling behavior and relatively slower swimming of these miracidia. Data analysis showed the average swimming velocity of miracidia hatched from i5-treated eggs was markedly decreased by 11.7% (p=0.0001) (Figure 5D and Supplementary Table 1). Notably, the average duration time and the movement tortuosity of miracidia hatched from i5-treated eggs were enhanced by 18.5% (p=0.0031) (Figure 5E and Supplementary Table 1) and 18.9% (p=0.003) (Figure 5F and Supplementary Table 1), respectively.
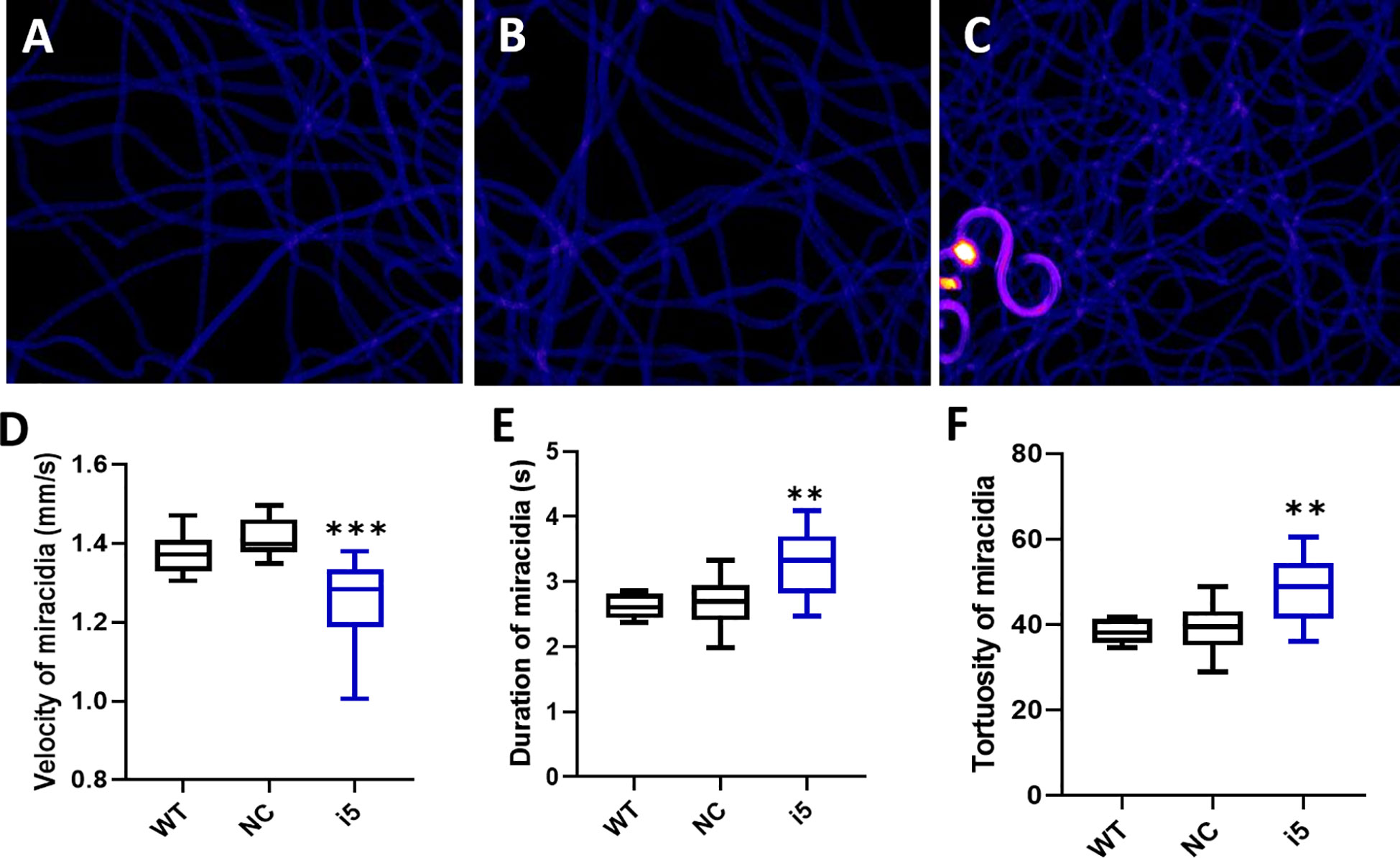
Figure 5 Behavioral changes in S. mansoni miracidia hatched from SmfgfrA-suppressed eggs. The behavior of miracidia hatched from WT eggs and eggs treated with NC or i5 was monitored. Heatmaps (A–C) represent the movement patterns of individual miracidia within a 1 min recording. Colors in the heatmaps show the time miracidia spent at a specific position. Black: absence; Blue: shorter time presence; Yellow and Red: longer time presence. Boxplots showing (D) velocity (E) duration, and (F) tortuosity of miracidial movement. Experiments were performed in biological duplicates and five technical repeats (n=10). ** p value≤ 0.01, *** p value ≤ 0.001.
Depleted granulomatous inflammation in lungs of mice injected with SmfgfrA-repressed eggs
To determine whether the repression of SmfgfrA in eggs could affect the formation of egg-induced granulomas in vivo, i5-treated eggs and controls (PBS, WT eggs and NC-treated eggs) were i.v. injected into the lateral vein of the tail of Swiss mice. In mice, schistosome eggs are transported to the lungs via the circulation leading to the development of lung granuloma (60, 74). Mice were euthanized two weeks post-injection and the left lung of each mouse was harvested for histological analysis. Representative digital microscopic images of mouse lung sections are shown in Figures 6A–D and indicate that more intense and severe granulomatous inflammation was evident in lungs of mice injected with WT eggs (Figure 6B) and NC-treated eggs (Figure 6C) compared with those of mice injected with i5-treated eggs (Figure 6D). Calculations of the granuloma ratio from individual lung tissue clearly showed that the sizes of lung granuloma in mice injected with i5-treated eggs were reduced by 60.8% (p=0.0003), compared with those in mice injected with NC-treated eggs (Figure 6E). Substantially smaller sized granulomas (p<0.0001) were also observed in mice injected with NC-treated eggs compared with those present in mice receiving WT eggs (Figure 6E), indicating that the electroporation of NC may have subsequently affected the granuloma formation occurred around eggs as previously reported (60).
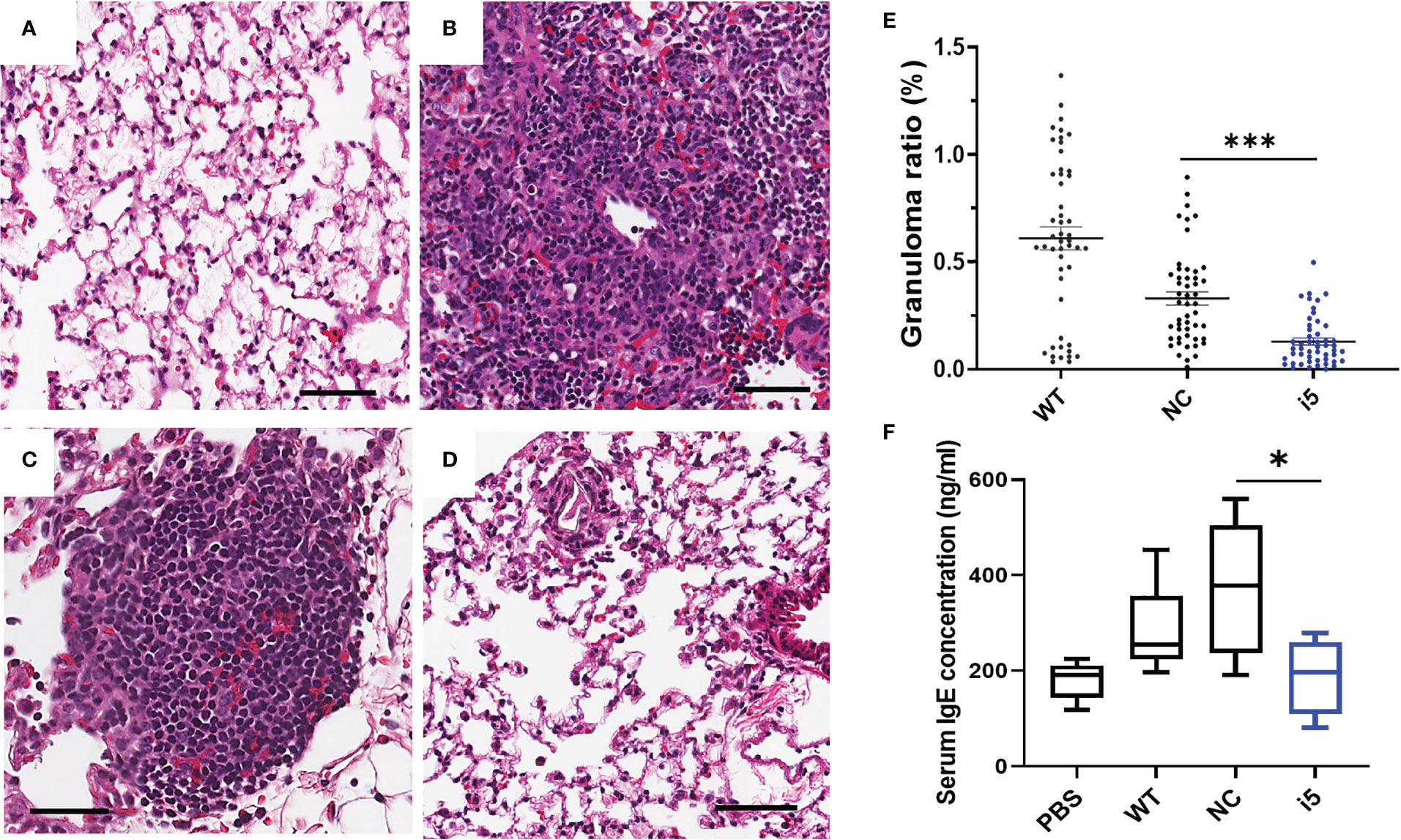
Figure 6 Reduced granulomatous inflammation and serum IgE levels in mice infected with SmfgfrA-repressed schistosome eggs. Representative digital microscopic images showed (A) lung from mouse injected with PBS and granulomas in lungs from mice injected with (B) WT eggs, (C) NC-treated or (D) i5-treated eggs. All sections were H&E stained. Scale bars in (A–D) = 50 µm. (E) Effect of SmfgfrA-repressed eggs on granuloma formation in the lungs of mice injected with i5-treated eggs. Mice in control groups were infected with untreated WT eggs or NC-treated eggs. Granuloma ratio in each lung section was calculated by dividing the total granuloma size with the lung size X100. Each group included 5 mice and 10 sections per lung were measured (n=50). All data are shown as the mean ± SE. (F) Total serum IgE level in mice 2 weeks after i.v. injection of i5-treated eggs. Mice injected with PBS, WT eggs or NC-treated eggs served as controls. * p value≤ 0.05, *** p value ≤ 0.001.
Reduced serum IgE level in mice exposed to SmfgfrA-repressed eggs
It is well recognized that during schistosome infection, the host immune response is highly Th2-polarized following egg laying (75). As a marker of the Th2-polarized response during infection, IgE is associated with the protective response against schistosomes by mediating macrophage toxicity (76). To determine whether SmfgfrA repression of eggs has any effect on the IgE response in infected mice, the levels of IgE were determined in sera collected from mice injected with PBS, WT eggs, NC-treated eggs and i5-treated eggs. Notably, the concentration of serum IgE in mice injected with i5-treated eggs was markedly reduced by 49.8% (p=0.00378) compared with that of mice injected with NC-treated eggs (Figure 6F).
Discussion
Since the award of the Nobel Prize in Chemistry (2020) for the discovery of the revolutionary CRISPR/Cas9 gene editing tool, advances in CRISPR technology have accelerated and the approach provides a powerful new avenue to undertake functional genomics studies. Recently, CRISPR/Cas9 has been repurposed for targeted gene regulation, providing a platform for RNA-guided transcription repression called CRISPR interference (CRISPRi). Using S. mansoni as a model, we show, for the first time, the feasibility and efficiency of applying CRISPRi to selectively tune genes in a parasitic worm. Efficient CRISPRi-mediated repression of SmfgfrA (encoding the stem cell marker SmFGFRA) was evidenced by the marked downregulation of SmfgfrA transcription and a decrease in the expression of SmFGFRA. These features were accompanied by distinct in vitro and in vivo phenotypic changes including a reduction in the number of stem cells and an elevated level of cell apoptosis in modulated parasites, and the decreased capacity of modulated eggs to induce granulomatous inflammation in infected mice together with a decline in serum IgE levels. This study points the way forward to undertake loss-of-function studies in schistosomes and other parasitic helminths.
To adapt CRISPRi for application in S. mansoni, we started with the screening of 8 sgRNAs (i1-i8) targeting -21 bp to +329 bp relative to the TSS of SmfgfrA, through identifying the efficiency of phenotypic changes induced by CRISPRi-SmfgfrA modulation in adult S. mansoni. Selection of the sgRNA targeting window for SmfgfrA was based on a previous study showing that the most effective sgRNAs for CRISPRi in mammalian cells target a region -50 bp to +300 bp relative to the TSS (77). Consistent with studies in mammalian cells and E. coli (33, 40, 78), we found that inhibition of both elongation and initiation of the SmfgfrA target gene via CRISPRi were achievable in schistosomes. Notably, the highest efficacy of repression (56.5%) was obtained when CRISPRi was performed by simultaneous recruiting sgRNA i5 and the sgRNA i8. This may indicate a relatively higher gene regulation efficiency occur when inhibiting the target gene elongation and initiation simultaneously in schistosomes, a feature which is worthy of further exploration. Considering the similar gene regulation efficiency of CRISPRi between using single sgRNA i5 and multiplexed sgRNAs i5+i8 in silencing SmfgfrA, we selected sgRNA i5 in subsequent phenotypic change studies.
The most reduced level of SmfgfrA transcription was observed in SmfgfrA-repressed eggs (67.3%), followed by adult worms (51.6%), and the lowest efficiency occurred in schistosomula (18.1%). A possible reason is that SmfgfrA is more highly expressed in eggs than in adult worms and schistosomula (56), resulting in the chromatin in eggs being more open (euchromatin) and providing greater access to dCas9 binding and functioning (79). Also, the morphology of eggs is relatively simple with a high stem-like cell content (80), compared with other developmental stages, which may indicate increased potential for manipulating the stem cells and stem cell marker genes (eg. SmfgfrA) in schistosomes. Furthermore, we found the CRISPRi-induced gene repression efficiency in adult worms is much higher than that observed in schistosomula, although adult worms and schistosomula present similar level of SmfgfrA transcripts (56). This might be explained by the more abundant distribution of this molecule in the tegument of adult worms than that in schistosomula (56), which may result in a relatively higher efficacy of SmfgfrA-repression in adult worms as genes with more tegumental location may provide more proximity to the external environment containing CRISPR components. In addition, the relatively larger surface area/volume ratio in adult worms than schistosomula (81) may also lead to a higher efficacy of delivering CRISPR components into adult flukes by electroporation.
It is noteworthy that the downregulated transcription of SmfgfrA was also detectable in miracidia hatched from SmfgfrA-repressed eggs, although the silencing effect declined in these hatched miracidia compared with that observed in modulated eggs. This might be because the eggs (obtained from the livers of mice infected with S. mansoni) used for CRISPRi are a mix of immature and mature eggs. The different morphology of these eggs (80, 82) may lead to different level of gene suppression efficiency (83). Therefore, it is possible that eggs with relatively higher gene silencing efficiency were not able to hatch, which was also evidenced by the decreased hatching efficiency of SmfgfrA-repressed eggs, resulting in the silencing effect being reduced in the hatched miracidia. The repression of SmfgfrA in miracidia hatched from SmfgfrA-modulated eggs was further confirmed by their significantly modified movement behavior in water, represented by enhanced turning and circling movement. This re-emphasizes the importance of SmfgfrA in the neuronal functioning and development of schistosomes, a characteristic explored in our previous study showing that SmFGFRA was abundant in the neural mass of mature eggs and miracidia (56). Together, these outcomes may indicate a long-term effect of CRISPRi-mediated gene repression in schistosomes, as described for Toxoplasma gondii (44).
Given the vital role of SmfgfrA in maintaining stem cells (49, 50, 53, 56, 84), we examined EdU incorporation in CRISPRi-SmfgfrA-modulated adult worms and schistosomula. Consistent with the decrease in SmfgfrA transcription levels in these two parasite stages, markedly reduced numbers of EdU+ stem cells were identified in both SmfgfrA-repressed adult worms and schistosomula, again highlighting the critical roles of SmfgfrA in maintaining schistosome stem cells. We also noted that suppression of SmfgfrA led to a greater reduction of EdU signal in female worms compared to male worms despite having similar levels of SmfgfrA-repressed transcripts. This implies that silencing of SmfgfrA may restrain the pairing of male and female worms as described previously (54), or negatively affect the development of the female reproductive system, leading to the pronounced reduction in the number of stem cells present in repressed females. Notably, a remarkably decreased egg production was observed in SmfgfrA-repressed worms, indicating the critical role of SmfgfrA in reproduction system of this parasite. This was also evidenced by the abundant expression of SmfgfrA in vitelline cells (56) which occupy the majority of the female worm and without them, the eggs will not form (1). Furthermore, the enhanced caspase-3/-7 activity, indicative of increased cell apoptosis, observed in SmfgfrA-repressed adult worms, schistosomula and eggs suggests that silencing of SmfgfrA may trigger apoptosis in the stem cells of schistosomes. This may be because schistosomes possess mechanisms to eliminate abnormal stem cells via apoptosis, as has been observed in mammalian stem cells (85, 86).
The potentially life-threatening pathology of schistosomiasis is evoked by schistosome eggs that are lodged in mammalian host tissue which induce granulomatous inflammation around the transiting eggs (87). During a mature schistosome infection when eggs are produced, the immune response of the host is polarized to a Th2-type immunity. Together with this enhancement of the Th2 response, serum IgE levels and the number of circulating eosinophils are also considerably elevated (76, 88). To determine whether CRISPRi-mediated suppression of SmfgfrA affected the formation of granulomas around the genetically modified eggs in vivo, we injected SmfgfrA-repressed eggs and control eggs into tail veins of mice. We found remarkably decreased sizes of pulmonary circumoval granulomas around SmfgfrA-repressed eggs in the lungs of the mice, a feature characteristic of fewer host immune cells (eosinophils, macrophages and neutrophils) accumulating around the modulated eggs (1). In addition, a considerably reduced level of serum IgE (a Th2 response marker (76, 88)) was observed in mice injected with SmfgfrA-repressed eggs compared with those given non-edited eggs. Collectively, these outcomes emphasize the critical role played by SmFGFRA in the formation of schistosome egg-induced lung granulomatous inflammation. This concept is also supported by the abundant distribution of SmFGFRA in the von Lichtenberg’s layer of S. mansoni eggs (56), a layer heavily involved in the release of immunogenic secretions through eggshell pores into the host circulatory system which regulate the host immune response and subsequent granuloma formation (89, 90).
These outcomes indicate the feasibility of applying CRISPRi for targeted transcriptional regulation in schistosomes, but challenges remain and further additional improvement and optimization of the approach will need to be considered before this revolutionary technique can be applied on a larger scale in parasitic helminths. Strategies developed for other organisms, including utilizing different dCas9 orthologues such as CRISPR1 dCas9 derived from Streptococcus thermophilus (Sth. dCas9), may improve gene regulation efficiency, a strategy that has already been undertaken in bacteria (44, 45). Another tactic could be the employment of an alternative transcription repressor (eg. KRAB-MeCP2), which has been shown to improve gene suppression outcomes in mammalian cells (91), may also improve CRISPRi performance in schistosomes.
In conclusion, these findings provide a blueprint for selectively regulating gene expression by employing CRISPRi in schistosomes, with the potential of the approach to be extended to the study of other parasitic helminths. With its adaptability and scalability to target diverse gene loci and with appropriate refinements and modifications of the methodology to ensure appropriate levels of gene regulation efficacy, CRISPRi can provide the basis to markedly accelerate the functional characterization of not only the genomes of schistosomes, but also a range of other parasitic helminths as well.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Conceived and designed the experiments: HY, XD, DM, JF and HS performed the experiments: XD, NC, and SM analyzed the data: XD, HY, DM, CF, and MJ contributed reagents/materials/analysis tools: XD and HY wrote the paper: XD, HY, DM, JF and MJ. All authors contributed to the article and approved the submitted version.
Funding
DM is a National Health and Medical Research Council (NHMRC) of Australia Leadership Fellow and receives Program (APP1132975), Project (APP1098244) and Investigator Grant (APP1194462) support from the NHMRC for his research on schistosomes and schistosomiasis. XD holds a Research Training Program (RTP) Scholarship and Graduate School Scholarship from the University of Queensland, Australia. HY holds QIMR Berghofer Medical Research Institute Seed Funding Grants.
Acknowledgments
We thank Mary Duke from QIMR Berghofer Medical Research Institute for the maintaining of the S. mansoni life cycle and the provision of parasite materials for this study. B. glabrata snails were provided by the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) through NIH-NIAID Contract HHSN272201700014I for distribution through BEI Resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1105719/full#supplementary-material
References
1. McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou X-N. Schistosomiasis (Primer). Nat Rev Dis Primers (2018) 4:13. doi: 10.1038/s41572-018-0013-8
2. Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet (2006) 368(9541):1106–18. doi: 10.1016/S0140-6736(06)69440-3
3. McManus DP, Bergquist R, Cai P, Ranasinghe S, Tebeje BM, You H. Schistosomiasis–from immunopathology to vaccines. Semin Immunopathol (2020) 42:355–71. doi: 10.1007/s00281-020-00789-x
4. Deol AK, Fleming FM, Calvo-Urbano B, Walker M, Bucumi V, Gnandou I, et al. Schistosomiasis–assessing progress toward the 2020 and 2025 global goals. N Engl J Med (2019) 381(26):2519–28. doi: 10.1056/NEJMoa1812165
5. Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, et al. The genome of the blood fluke schistosoma mansoni. Nature (2009) 460(7253):352. doi: 10.1038/nature08160
6. Protasio AV, Tsai IJ, Babbage A, Nichol S, Hunt M, Aslett MA, et al. A systematically improved high quality genome and transcriptome of the human blood fluke schistosoma mansoni. PloS Negl Trop Dis (2012) 6(1):e1455. doi: 10.1371/journal.pntd.0001455
7. Zhou Y, Zheng H, Chen X, Zhang L, Wang K, Guo J, et al. The schistosoma japonicum genome reveals features of host-parasite interplay. Nature (2009) 460(7253):345. doi: 10.1038/nature08140
8. Luo F, Yin M, Mo X, Sun C, Wu Q, Zhu B, et al. An improved genome assembly of the fluke schistosoma japonicum. PloS Negl Trop Dis (2019) 13(8):e0007612. doi: 10.1371/journal.pntd.0007612
9. Young ND, Jex AR, Li B, Liu S, Yang L, Xiong Z, et al. Whole-genome sequence of schistosoma haematobium. Nat Genet (2012) 44(2):221–5. doi: 10.1038/ng.1065
10. Stroehlein AJ, Korhonen PK, Chong TM, Lim YL, Chan KG, Webster B, et al. High-quality schistosoma haematobium genome achieved by single-molecule and long-range sequencing. GigaScience (2019) 8(9):giz108. doi: 10.1093/gigascience/giz108
11. Dalzell JJ, Warnock ND, Mcveigh P, Marks NJ, Mousley A, Atkinson L, et al. Considering rnai experimental design in parasitic helminths. Parasitology (2012) 139(5):589–604. doi: 10.1017/S0031182011001946
12. Correnti JM, Brindley PJ, Pearce EJ. Long-term suppression of cathepsin b levels by rna interference retards schistosome growth. Mol Biochem Parasitol (2005) 143(2):209–15. doi: 10.1016/j.molbiopara.2005.06.007
13. Fanelli E, Di Vito M, Jones JT, De Giorgi C. Analysis of chitin synthase function in a plant parasitic nematode, meloidogyne artiellia, using rnai. Gene (2005) 349:87–95. doi: 10.1016/j.gene.2004.11.045
14. Rinaldi G, Morales ME, Cancela M, Castillo E, Brindley PJ, Tort JF. Development of functional genomic tools in trematodes: Rna interference and luciferase reporter gene activity in fasciola hepatica. PloS Negl Trop Dis (2008) 2(7):e260. doi: 10.1371/journal.pntd.0000260
15. Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M, Plasterk RH. Long-term gene silencing by rnai. Nature (2006) 442(7105):882. doi: 10.1038/442882a
16. Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol (2013) 31(3):230. doi: 10.1038/nbt.2507
17. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (2013) 339(6121):819–23. doi: 10.1126/science.1231143
18. Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, et al. Genome engineering of drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics (2013) 194(4):1029–35. doi: 10.1534/genetics.113.152710
19. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. Rna-guided human genome engineering Via Cas9. Science (2013) 339(6121):823–6. doi: 10.1126/science.1232033
20. Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a crispr-cas system. Nat Biotechnol (2013) 31(3):227. doi: 10.1038/nbt.2501
21. Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in c. elegans Via a crispr-Cas9 system. Nat Methods (2013) 10(8):741. doi: 10.1038/nmeth.2532
22. Bryant JM, Baumgarten S, Glover L, Hutchinson S, Rachidi N. Crispr in parasitology: Not exactly cut and dried! Trends Parasitol (2019) 35(6):409–22. doi: 10.1016/j.pt.2019.03.004
23. Castelletto ML, Gang SS, Hallem EA. Recent advances in functional genomics for parasitic nematodes of mammals. J Exp Biol (2020) 223(Pt Suppl 1):jeb206482. doi: 10.1242/jeb.206482
24. Gang SS, Castelletto ML, Bryant AS, Yang E, Mancuso N, Lopez JB, et al. Targeted mutagenesis in a human-parasitic nematode. PloS Pathog (2017) 13(10):e1006675. doi: 10.1371/journal.ppat.1006675
25. Nakayama K-i, Ishita Y, Chihara T, Okumura M. Screening for Crispr/Cas9-induced mutations using a Co-injection marker in the nematode pristionchus pacificus. Dev Genes Evol (2020) 230(3):257–64. doi: 10.1007/s00427-020-00651-y
26. Ittiprasert W, Mann VH, Karinshak SE, Coghlan A, Rinaldi G, Sankaranarayanan G, et al. Programmed genome editing of the omega-1 ribonuclease of the blood fluke, schistosoma mansoni. Elife (2019) 8:e41337. doi: 10.7554/eLife.41337
27. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in escherichia coli, and identification of the gene product. J Bacteriol (1987) 169(12):5429–33. doi: 10.1128/jb.169.12.5429-5433.1987
28. Jansen R, Embden J, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol (2002) 43(6):1565–75. doi: 10.1046/j.1365-2958.2002.02839.x
30. Ledford H. Crispr: Gene editing is just the beginning. Nat News (2016) 531(7593):156. doi: 10.1038/531156a
31. Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, et al. Direct observation of r-loop formation by single rna-guided Cas9 and cascade effector complexes. Proc Natl Acad Sci U S A (2014) 111(27):9798–803. doi: 10.1073/pnas.1402597111
32. Dickinson DJ, Goldstein B. Crispr-based methods for caenorhabditis elegans genome engineering. Genetics (2016) 202(3):885–901. doi: 10.1534/genetics.115.182162
33. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing crispr as an rna-guided platform for sequence-specific control of gene expression. Cell (2013) 152(5):1173–83. doi: 10.1016/j.cell.2013.02.022
34. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. Crispr-mediated modular rna-guided regulation of transcription in eukaryotes. Cell (2013) 154(2):442–51. doi: 10.1016/j.cell.2013.06.044
35. Xiao B, Yin S, Hu Y, Sun M, Wei J, Huang Z, et al. Epigenetic editing by Crispr/Dcas9 in plasmodium falciparum. Proc Natl Acad Sci U S A (2019) 116(1):255–60. doi: 10.1073/pnas.1813542116
36. Walker MP, Lindner SE. Ribozyme-mediated, multiplex crispr gene editing and crispr interference (Crispri) in rodent-infectious plasmodium yoelii. J Biol Chem (2019) 294(24):9555–66. doi: 10.1074/jbc.RA118.007121
37. Cámara E, Lenitz I, Nygård Y. A crispr activation and interference toolkit for industrial saccharomyces cerevisiae strain Ke6-12. Sci Rep (2020) 10(1):1–13. doi: 10.1038/s41598-020-71648-w
38. Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, et al. Crispr interference efficiently induces specific and reversible gene silencing in human ipscs. Cell Stem Cell (2016) 18(4):541–53. doi: 10.1016/j.stem.2016.01.022
39. Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by crispr interference in mycobacteria. Nat Commun (2015) 6(1):1–11. doi: 10.1038/ncomms7267
40. Long L, Guo H, Yao D, Xiong K, Li Y, Liu P, et al. Regulation of transcriptionally active genes Via the catalytically inactive Cas9 in c. elegans and d. rerio. Cell Res (2015) 25(5):638–41. doi: 10.1038/cr.2015.35
41. Li S, Jendresen CB, Grünberger A, Ronda C, Jensen SI, Noack S, et al. Enhanced protein and biochemical production using crispri-based growth switches. Metab Eng (2016) 38:274–84. doi: 10.1016/j.ymben.2016.09.003
42. Peters JM, Koo B-M, Patino R, Heussler GE, Hearne CC, Qu J, et al. Enabling genetic analysis of diverse bacteria with mobile-crispri. Nat Microbiol (2019) 4(2):244–50. doi: 10.1038/s41564-018-0327-z
43. Huang C-H, Shen CR, Li H, Sung L-Y, Wu M-Y, Hu Y-C. Crispr interference (Crispri) for gene regulation and succinate production in cyanobacterium s. elongatus pcc 7942. Microb Cell Fact (2016) 15(1):196. doi: 10.1186/s12934-016-0595-3
44. Markus BM, Boydston EA, Lourido S. Crispr-mediated transcriptional repression in toxoplasma gondii. Msphere (2021) 6(5):e00474–21. doi: 10.1128/mSphere.00474-21
45. Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, et al. Programmable transcriptional repression in mycobacteria using an orthogonal crispr interference platform. Nat Microbiol (2017) 2(4):1–9. doi: 10.1038/nmicrobiol.2016.274
46. Whitfield PJ, Evans NA. Parthenogenesis and asexual multiplication among parasitic platyhelminths. Parasitology (1983) 86(Pt 4):121–60. doi: 10.1017/s0031182000050873
47. Beaumier CM, Gillespie PM, Hotez PJ, Bottazzi ME. New vaccines for neglected parasitic diseases and dengue. Transl Res (2013) 162(3):144–55. doi: 10.1016/j.trsl.2013.03.006
48. Lu Z, Sessler F, Holroyd N, Hahnel S, Quack T, Berriman M, et al. Schistosome sex matters: A deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci Rep (2016) 6:31150. doi: 10.1038/srep31150
49. Wendt GR, Collins JJ 3rd. Schistosomiasis as a disease of stem cells. Curr Opin Genet Dev (2016) 40:95–102. doi: 10.1016/j.gde.2016.06.010
50. Wang B, Collins JJ III, Newmark PA. Functional genomic characterization of neoblast-like stem cells in larval schistosoma mansoni. Elife (2013) 2:e00768. doi: 10.7554/eLife.00768
51. Wang B, Lee J, Li P, Saberi A, Yang H, Liu C, et al. Stem cell heterogeneity drives the parasitic life cycle of schistosoma mansoni. Elife (2018) 7:e35449. doi: 10.7554/eLife.35449
52. You H, Jones MK, Whitworth DJ, McManus DP. Innovations and advances in schistosome stem cell research. Front Immunol (2021) 12:599014. doi: 10.3389/fimmu.2021.599014
53. Collins JJ III, Wang B, Lambrus BG, Tharp ME, Iyer H, Newmark PA. Adult somatic stem cells in the human parasite schistosoma mansoni. Nature (2013) 494(7438):476–9. doi: 10.1038/nature11924
54. Hahnel S, Quack T, Parker-Manuel SJ, Lu Z, Vanderstraete M, Morel M, et al. Gonad rna-specific qrt-pcr analyses identify genes with potential functions in schistosome reproduction such as Smfz1 and smfgfrs. Front Genet (2014) 5:170. doi: 10.3389/fgene.2014.00170
55. Sarfati DN, Li P, Tarashansky AJ, Wang B. Single-cell deconstruction of stem-Cell-Driven schistosome development. Trends Parasitol (2021) . 37(9):790–802. doi: 10.1016/j.pt.2021.03.005
56. Du X, McManus DP, Fogarty CE, Jones MK, You H. Schistosoma mansoni fibroblast growth factor receptor a orchestrates multiple functions in schistosome biology and in the host-parasite interplay. Front Immunol (2022) 13:868077. doi: 10.3389/fimmu.2022.868077
57. Dalton J, Day S, Drew A, Brindley P. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology (1997) 115(1):29–32. doi: 10.1017/s0031182097001091
58. Du X, Jones MK, Nawaratna SS, Ranasinghe S, Xiong C, Cai P, et al. Gene expression in developmental stages of schistosoma japonicum provides further insight into the importance of the schistosome insulin-like peptide. Int J Mol Sci (2019) 20(7):1565. doi: 10.3390/ijms20071565
59. Oliveros JC, Franch M, Tabas-Madrid D, San-León D, Montoliu L, Cubas P, et al. Breaking-Cas–interactive design of guide rnas for crispr-cas experiments for ensembl genomes. Nucleic Acids Res (2016) 44(W1):W267–W71. doi: 10.1093/nar/gkw407
60. You H, Mayer JU, Johnston RL, Sivakumaran H, Ranasinghe S, Rivera V, et al. Crispr/Cas9-mediated genome editing of schistosoma mansoni acetylcholinesterase. FASEB J (2021) 35(1):e21205. doi: 10.1096/fj.202001745RR
61. Barber RD, Harmer DW, Coleman RA, Clark BJ. Gapdh as a housekeeping gene: Analysis of gapdh mrna expression in a panel of 72 human tissues. Physiol Genomics (2005) 21(3):389–95. doi: 10.1152/physiolgenomics.00025.2005
62. Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ (–delta delta ct) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath (2013) 3(3):71.
63. Zheng X, Hu X, Zhou G, Lu Z, Qiu W, Bao J, et al. Soluble egg antigen from schistosoma japonicum modulates the progression of chronic progressive experimental autoimmune encephalomyelitis Via Th2-shift response. J Neuroimmunol (2008) 194(1-2):107–14. doi: 10.1016/j.jneuroim.2007.12.001
64. Du X, McManus DP, Cai P, Hu W, You H. Identification and functional characterisation of a schistosoma japonicum insulin-like peptide. Parasitol Vectors (2017) 10(1):1–12. doi: 10.1186/s13071-017-2095-7
65. Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: Theoretical and experimental studies. Anal Biochem (1996) 236(2):302–8. doi: 10.1006/abio.1996.0171
66. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. Qupath: Open source software for digital pathology image analysis. Sci Rep (2017) 7(1):1–7. doi: 10.1038/s41598-017-17204-5
67. Wang T, Wyeth RC, Liang D, Bose U, Ni G, McManus DP, et al. A biomphalaria glabrata peptide that stimulates significant behaviour modifications in aquatic free-living schistosoma mansoni miracidia. PloS Negl Trop Dis (2019) 13(1):e0006948. doi: 10.1371/journal.pntd.0006948
68. Fogarty CE, Zhao M, McManus DP, Duke MG, Cummins SF, Wang T. Comparative study of excretory–secretory proteins released by schistosoma mansoni-resistant, susceptible and naïve biomphalaria glabrata. Parasitol Vectors (2019) 12(1):1–17. doi: 10.1186/s13071-019-3708-0
69. Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, et al. Trackmate: An open and extensible platform for single-particle tracking. Methods (2017) 115:80–90. doi: 10.1016/j.ymeth.2016.09.016
70. Wyeth RC, Braubach OR, Fine A, Croll RP. Videograms: A method for repeatable unbiased quantitative behavioral analysis without scoring or tracking. In: Zebrafish neurobehavioral protocols. Totowa, New Jersey, United States: Humana Press (2011). p. 15–33.
71. Shim MK, Yoon HY, Lee S, Jo MK, Park J, Kim J-H, et al. Caspase-3/-7-Specific metabolic precursor for bioorthogonal tracking of tumor apoptosis. Sci Rep (2017) 7(1):1–15. doi: 10.1038/s41598-017-16653-2
72. Zhu M, Chen M, Zuscik M, Wu Q, Wang YJ, Rosier RN, et al. Inhibition of β-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheumatol (2008) 58(7):2053–64. doi: 10.1002/art.23614
73. Walsh JG, Cullen SP, Sheridan C, Lüthi AU, Gerner C, Martin SJ. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci U S A (2008) 105(35):12815–9. doi: 10.1073/pnas.0707715105
74. Eltoum IA, Wynn TA, Poindexter RW, Finkelman FD, Lewis FA, Sher A, et al. Suppressive effect of interleukin-4 neutralization differs for granulomas around schistosoma mansoni eggs injected into mice compared with those around eggs laid in infected mice. Infect Immun (1995) 63(7):2532–6. doi: 10.1128/iai.63.7.2532-2536.1995
75. Pearce EJ M, Kane C, Sun J, Taylor J J, McKee AS, Cervi L. Th2 response polarization during infection with the helminth parasite schistosoma mansoni. Immunol Rev (2004) 201(1):117–26. doi: 10.1111/j.0105-2896.2004.00187.x
76. Zhang P, Mutapi F. Ige: A key antibody in schistosoma infection. Electron J Biol (2006) 2:11–4. doi: 10.1186/1475-4924-2-11
77. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale crispr-mediated control of gene repression and activation. Cell (2014) 159(3):647–61. doi: 10.1016/j.cell.2014.09.029
78. Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered crispr-cas system. Nucleic Acids Res (2013) 41(15):7429–37. doi: 10.1093/nar/gkt520
79. Verkuijl SA, Rots MG. The influence of eukaryotic chromatin state on crispr–Cas9 editing efficiencies. Curr Opin Biotechnol (2019) 55:68–73. doi: 10.1016/j.copbio.2018.07.005
80. Jurberg AD, Gonçalves T, Costa TA, de Mattos ACA, Pascarelli BM, de Manso PPA, et al. The embryonic development of schistosoma mansoni eggs: Proposal for a new staging system. Dev Genes Evol (2009) 219(5):219. doi: 10.1007/s00427-009-0285-9
81. Sankaranarayanan G, Coghlan A, Driguez P, Lotkowska ME, Sanders M, Holroyd N, et al. Large Crispr-Cas-Induced deletions in the oxamniquine resistance locus of the human parasite schistosoma mansoni. Wellcome Open Res (2020) 5:178. doi: 10.12688/wellcomeopenres.16031.2
82. Jamieson BG. Schistosoma: Biology, pathology and control. (Boca Raton, Florida, United States:CRC Press) (2017).
83. Rinaldi G, Eckert SE, Tsai IJ, Suttiprapa S, Kines KJ, Tort JF, et al. Germline transgenesis and insertional mutagenesis in schistosoma mansoni mediated by murine leukemia virus. PloS Pathog (2012) 8(7):e1002820. doi: 10.1371/journal.ppat.1002820
84. Itoh N, Ornitz DM. Evolution of the fgf and fgfr gene families. Trends Genet (2004) 20(11):563–9. doi: 10.1016/j.tig.2004.08.007
85. Cheung H-H, Liu X, Rennert OM. Apoptosis: Reprogramming and the fate of mature cells. Int Sch Res (2012) 2012:e685852. doi: 10.5402/2012/685852
86. Sarig R, Rotter V. Can an ips cell secure its genomic fidelity? Cell Death Differ (2011) 18(5):743–4. doi: 10.1038/cdd.2011.14
87. Costain AH, MacDonald AS, Smits HH. Schistosome egg migration: Mechanisms, pathogenesis and host immune responses. Front Immunol (2018), 9:3042. doi: 10.3389/fimmu.2018.03042
88. Dutra WO, Correa-Oliveira R, Dunne D, Cecchini LF, Fraga L, Roberts M, et al. Polarized Th2 like cells, in the absence of Th0 cells, are responsible for lymphocyte produced il-4 in high ige-producer schistosomiasis patients. BMC Immunol (2002) 3(1):1–8. doi: 10.1186/1471-2172-3-8
89. Ashton P, Harrop R, Shah B, Wilson R. The schistosome egg: Development and secretions. Parasitology (2001) 122(3):329–38. doi: 10.1017/s0031182001007351
90. Takaki KK, Rinaldi G, Berriman M, Pagán AJ, Ramakrishnan L. Schistosoma mansoni eggs modulate the timing of granuloma formation to promote transmission. Cell Host Microbe (2021) 29(1):58–67.e5. doi: 10.1016/j.chom.2020.10.002
Keywords: Schistosoma mansoni, CRISPR interference, fibroblast growth factor receptor A, stem cells, transcription regulation
Citation: Du X, McManus DP, French JD, Collinson N, Sivakumaran H, MacGregor SR, Fogarty CE, Jones MK and You H (2023) CRISPR interference for sequence-specific regulation of fibroblast growth factor receptor A in Schistosoma mansoni. Front. Immunol. 13:1105719. doi: 10.3389/fimmu.2022.1105719
Received: 23 November 2022; Accepted: 28 December 2022;
Published: 13 January 2023.
Edited by:
Thiago Almeida Pereira, Stanford University, United StatesReviewed by:
David Roquis, Technical University of Munich, GermanyMin Hu, Huazhong Agricultural University, China
Copyright © 2023 Du, McManus, French, Collinson, Sivakumaran, MacGregor, Fogarty, Jones and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong You, SG9uZy5Zb3VAcWltcmJlcmdob2Zlci5lZHUuYXU=
†Deceased
 Xiaofeng Du
Xiaofeng Du Donald P. McManus
Donald P. McManus Juliet D. French
Juliet D. French Natasha Collinson1
Natasha Collinson1 Conor E. Fogarty
Conor E. Fogarty Malcolm K. Jones
Malcolm K. Jones Hong You
Hong You