- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Variable lymphocyte receptors (VLRs) play vital roles in adaptive immune system of agnathan vertebrate. In the present study, we first discover a novel VLR gene, VLR2, from an invertebrate, the Chinese mitten crab, Eriocheir sinensis. VLR2 has ten different isoforms formed via alternative splicing, which is different from that in agnathan vertebrate with the assembly of LRR modules. The longest isoform, VLR2-L, responds to Gram-positive bacteria Staphylococcus aureus challenge specifically, while shows no response to Gram-negative bacteria Vibrio parahaemolyticus challenge, confirmed by recombinant expression and bacterial binding experiments. Interestingly, VLR2s with short LRRs regions (VLR2-S8 and VLR2-S9) tend to bind to Gram-negative bacteria rather than Gram-positive bacteria. Antibacterial activity assay proves six isoforms of VLR2 have pluralistic antibacterial effects on bacteria which were never reported in invertebrate. These results suggest that the diversity and specificity of VLR2 resulted from alternative splicing and the length of the LRRs region. This pathogen-binding receptor diversity will lay the foundation for the study of immune priming. Furthermore, studying the immune function of VLR2 will provide a new insight into the disease control strategy of crustacean culture.
Introduction
Variable lymphocyte receptor (VLR) is a core immune factor in adaptive immune system of agnathan vertebrate. It is first discovered in Lampetra japonicum (1, 2). VLR is mainly composed of leucine-rich repeat (LRR) modules, and has two states: germline VLR (Germ variable lymphocyte receptor, gVLR) and mature VLR (1). The latter has diversity and can specifically bind to different antigens (1). The diversity of mature VLR genes is formed by the assembly of LRR modules, resulting in at least 1014 unique antigen-specific receptors, and the degree of mature VLR diversity is comparable to that of mammalian antibody diversity (3). A large number of different mature VLRs act as antigen receptors that specifically bind to various pathogens and enhance adaptive immune responses by activating the complement activation cascade (1, 4). In Eptatretus burgeri, the specific VLR against Nervous Necrosis Virus (NNV) reduces the infectivity effectively (5), another specific VLR against Viral Hemorrhagic Septicemia Virus (VHSV) is modified to work as a diagnostic tool for VHSV (6). Subsequently, VLR-like genes are also found in Branchiostoma floridae, while no LRR-box rearrangement assembly mechanism similar to that in agnathan vertebrate is detected (7).
In recent years, VLR genes or VLR-like genes have been found in invertebrate, such as Apostichopus japonicus, Litopenaeus vannamei, Eriocheir sinensis, etc. (8–10). In E. sinensis, the VLR gene exhibits strong bacteria-binding activity rather than antibacterial activity, and functions in crab immune priming through its immune memory-like characteristic (10). The above studies mainly focus on gene structure analysis and immune functions of VLR and VLR-like genes under pathogen challenge. However, it is still uncertain whether the VLR genes in invertebrate have diversity. If there is diversity of invertebrate VLRs, the formation mechanism and biological significances of this diversity also remain unknown.
As the model animal with easy culture and transportation of crustacean, the Chinese mitten crab, E. sinensis is an important freshwater cultured economic species. With the continuous expansion of breeding scale, large-scale diseases caused by bacteria, viruses and other pathogens also break out frequently causing huge losses to the industry of E. sinensis (11–14). Similar to other invertebrate sepcies, the E. sinensis is considered to own only the innate immune system, which can resist the invasion of foreign pathogens through humoral immunity and cellular immunity (15, 16). E. sinensis lacks an adaptive immune system and cannot produce diverse and specific antibodies against the invasion of foreign pathogens through Ig/TCR gene recombination (17). The recent studies show 30,600 Down syndrome cell adhesion molecule (Dscam) isoforms, which are generated via alternative splicing, can bind specifically with the original bacteria to facilitate efficient clearance in E. sinensis (18). Therefore, it is necessary to discover if more immune genes have the genetic diversity.
In this study, a novel VLR gene, VLR2, is discovered from E. sinensis. VLR2 has ten different isoforms, which are formed via alternative splicing. Recombinant expression, bacterial challenge and bacterial binding experiments are adopted to prove different isoforms own specific bacterial binding ability. This study should propose a new mechanism for the formation of VLR diversity and discover the biological significance of the diversity in the immune system of invertebrate. The diversity and specific bacterial binding ability of VLR2 provide insights for further studying the role of VLR in immune priming.
Materials and methods
Animal and sample preparation
Healthy male E. sinensis (100 ± 5 g) purchased from Sanshan market in Ningbo, China. All crabs were acclimated in tanks with filtered freshwater at room temperature (20 - 26°C) for one week before processing. Crab hemolymph was harvested from the last walking leg with an equal volume of ice-cold anticoagulant buffer (27 mM sodium citrate, 336 mM NaCl, 115 mM glucose, 9 mM EDTA, pH = 7.0), then immediately centrifuged at 800 g, 4°C for 5 min to isolate hemocytes. Crab tissues were collected after being euthanized in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines.
RNA isolation, cDNA synthesis and gene cloning of VLR2s
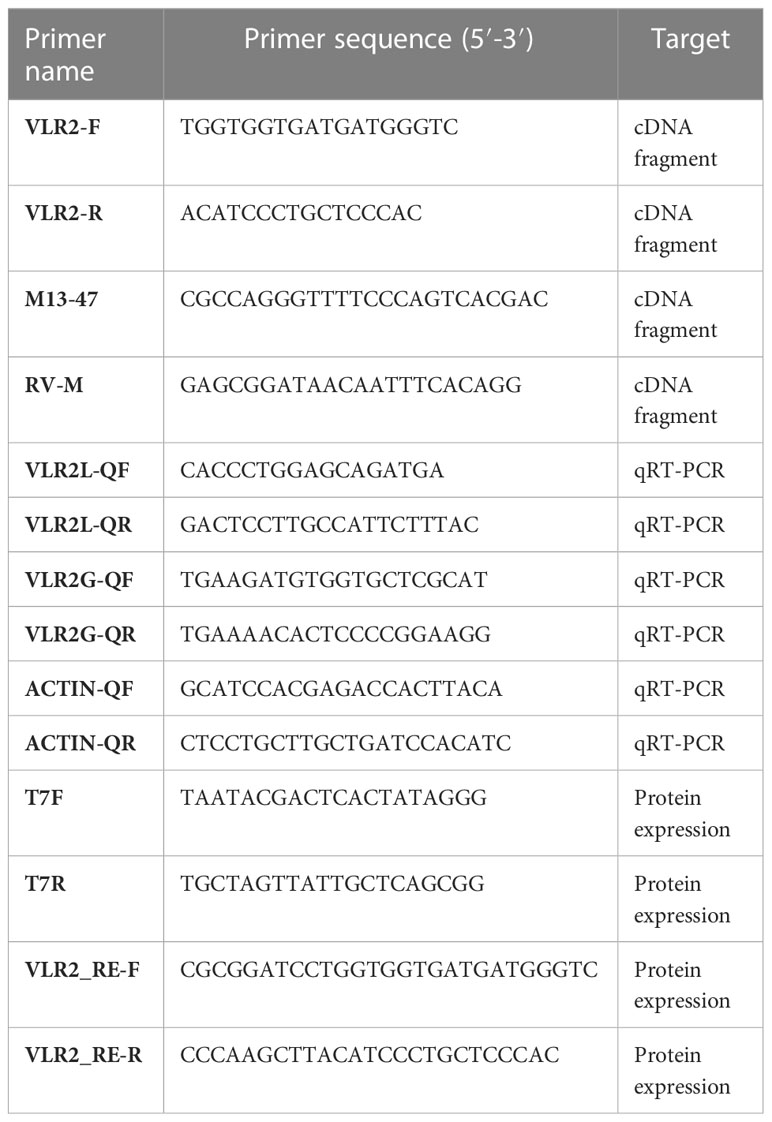
The total RNA was extracted using TRIzol reagent (Invitrogen) and the cDNA was synthesized using a PrimeScript™ RT Reagent Kit (Takara) according to the manufacture’s protocols. The raw data of the full-length transcriptome of the hepatopancreas of E. sinensis was downloaded from the NCBI database (accession number: SRR12632765). After the quality control and filter of raw reads, full-length reads were aligned to the E. sinensis genome (accession number: LQIF00000000) using the Hierarchical Indexing for Spliced Alignment of Transcripts (HISATS2) (19). Then, the aligned files was visualized using the Integrative Genomics Viewer (IGV) (20), and isomforms of VLR2 were observed in IGV. Based on these conserved regions in 5′-UTR and 3′-UTR of isomforms, a set of primers (VLR2-F and VLR2-R, Table 1) were designed to amplify the open reading frames (ORFs) of VLR2’s alternative splicing transcripts which span 5′-UTR to 3′-UTR. The PCR was performed in a 20 μL reaction volume consisting of 13.1 μL of sterile distilled H2O, 2.0 μL of 10 × PCR buffer,1.6 μL of dNTP (10 mM), 0.5 μL of each primer (10 mM), 0.3 μL (1 U) of Easy Taq polymerase (TransGen) and 2 μL of DNA template. The PCR programs were set as follows: 94°C for 2 min, followed by 32 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 1.5 min, and a final extension at 72°C for 5 min. The PCR products were analyzed by 1.2% agarose gel electrophoresis and purified by PCR purification kit (Axygen) as described in manufacture. The purified PCR products were cloned into the pMD19-T simple vector (TaKaRa). After being transformed into the competent cells of Escherichia coli DH5α, the positive recombinants were identified through the anti-Amp selection and PCR screening with M13-47 and RV-M primers (Table 1). Clones with different inserts were sequenced and the ORFs of VLR2′s alternative splicing transcripts were confirmed by aligning with VLR2 sequence among genome.
Sequence and phylogenetic analysis
The deduced amino acid sequence was analyzed with Tbtools and visualized by DNASTAR. SMART (http://smart.embl-heidelberg.de/) and NCBI BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi/) were used to predict the structures and functional domains of VLR2 isoform proteins. The Protein Analysis Tools (http://www.bio-soft.net/sms/) was performed to predict the molecular weight (Mw) and theoretical isoelectric point (pI) of the protein. SignalP 5.0 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0) was used to predict signal peptides and TMHMM Server v.2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was used for transmembrane regions prediction. The three-dimensional structure models were built by SWISSMODEL (https://swissmodel.expasy.org/) and visualized by Swiss-PdbViewer.
Semi-quantitative PCR
Semi-quantitative PCR (Semi-qPCR) was applied to study the tissue expression pattern of VLR2 isoforms using two pairs of gene-specific primers (Table 1). VLR2L-QF and VLR2L-QR primers were specific to VLR2-L, VLR2G-QF and VLR2G-QR primers were suitable to all VLR2 isoforms including VLR2-L. The procedure of Semi-qPCR is the same as the PCR process as described above.
Challenge assays
The challenge assays were according to the previous method (21) with some modification. Vibrio parahaemolyticus and Staphylococcus aureus during logarithmic growth phase were centrifuged at 800 × g, 4°C for 10 min, and washed three times with sterile 0.1 M phosphate buffer saline (PBS, pH = 7.4). Finally, V. parahaemolyticus and S. aureus were diluted to 107 CFU/mL with PBS.
During the challenge experiment, 30 crabs were injected with 50 μL V. parahaemolyticus (1 × 107 CFU/mL, named as VP group), 30 crabs were injected with 50 μL S. aureus (1 × 107 CFU/mL, named as SA group), and the control group was injected with the equal volume of PBS (named as PBS group). At 3, 6, 12 and 24 h post-injection, six individuals from each group were randomly sampled to collect tissues. Then, tissues were used for RNA isolation and cDNA synthesis as described above.
Quantitative real-time PCR analysis of VLR2s
Quantitative real-time PCR (qRT-PCR) was applied to study the expression alterations of VLR2 isoforms in E. sinensis after challenge assays. Reactions were carried out on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems) using SYBR green II (TaKaRa) as the fluorescent dye. The β-actin was chosen as a reference gene for internal standardization. The PCR reaction was carried out in a total volume of 20 μL, containing 10 μL of TB Green Premix DimerEraser (TaKaRa), 0.4 μL of 50 ROX Reference Dye II (TaKaRa), 2 μL of the diluted cDNA, 0.6 μL of each primer (10 μM), and 6.4 μL of sterile distilled water. The PCR program was 95°C for 5 min, followed by 40 cycles of 95°C for 5 s and 55°C for 31 s. Dissociation curve analysis of the amplification products was performed at the end of each PCR reaction to confirm that only one PCR product was amplified. All samples were repeated in triplicates in the qRT-PCR analysis. Fold change for the gene expression relative to controls was determined by the 2-△△Ct method (22).
The construction of recombinant plasmids
The ORFs of the VLR2 variants (VLR2-L, VLR2-S2, VLR2-S3, VLR2-S5, VLR2-S8 and VLR2-S9) were amplified by PCR using a pair of primers containing BamH I and EcoR I sites (VLR2_RE-F and VLR2_RE-R, Table 1). The PCR products were analyzed by 1.2% agarose gel electrophoresis. Then the PCR products were purified by PCR purification kit (Axygen) and cloned into the pMD19-T simple vector (TaKaRa) as described in manufacture.
The recombinant plasmids (pMD19-T-VLR2s) and pET-32a (+) vector were digested with the restriction enzymes BamH I and EcoR I (Thermo), then the PCR products were inserted into pET-32a (+) vector. Afterward, the recombinant plasmids (pET-32a-VLR2s) were transformed into the Rosetta cells (ANG YU BIO) for vitro expression. The pET-32a (+) vector without insert fragment was selected as a negative control.
Expression and purification of recombinant VLR2s
Overnight culture of a single colony of Rosetta cells was diluted (1:100) with Luria-Bertani medium and incubated at 37°C with vigorous shaking to OD600 of 0.4–0.6. Isopropyl β-D-1-Thiogalactopyranoside (IPTG) was added at a final concentration of 0.2 mM to induce the protein expression for a period of 4 h at 25°C. The protein extract was analyzed by 15% (w/v) SDS-PAGE.
The cells were broken by supersonic wave at 0°C and centrifuged at 12,000 × g, 4°C for 30 min. The soluble fractions and insoluble fractions of broken cells were analyzed by 15% (w/v) SDS-PAGE. The supernatants containing recombinant proteins were collected for subsequent purification experiments and the precipitations containing recombinant proteins were denatured and renatured as previously reported (23) to obtain soluble proteins. The supernatant and soluble proteins were loaded on to a Ni-nitrilotriaceticacid-agarose affinity column (Novagen). The recombinant protein was eluted with four concentration of elution buffers (20 mM Tris pH 8.0, 500 mM NaCl, 50/100/200/500 mM imidazole) and dialyzed in PBS at 4°C overnight. The extracted protein was analyzed by 15% (w/v) SDS-PAGE and Western blot.
Assay of binding activity to bacteria
An equal volume of bacteria solution (0.5 mL, 3 × 108 CFU/mL) with PBS (control group) and purified proteins (final concentration, 1 mg/mL proteins) were mixed and incubated with gentle rocking at 4 °C for 30 min respectively. After centrifugation at 800 × g and 4 °C for 5 min, the supernatant was removed, and the cells were washed twice with PBS. The supernatant, washed and eluted fractions were analyzed by 15% (w/v) SDS-PAGE.
Antibacterial activity assay of the recombinant proteins
Bacteria suspended in PBS at a concentration of 2.6 × 107 CFU/mL was used for the antibacterial activity test. Fifty microliters of recombinant proteins (250 μg/mL) were mixed with the same volume of bacteria solution and incubated at 28°C for 2h respectively. Then, the mixtures were diluted (104 fold) and spread on Luria-Bertani or 2216E agar plates, and the CFU were calculated after culturing for 16 h at 28°C or 37°C. PBS and pET-32a were used as a blank control and negative control, respectively, and each group was repeated three times.
Statistical analysis
Statistical analyses were processed using SPSS 26.0 software. Data obtained from this study were presented as the mean ± standard deviation (S.D.). Data for challenge assays and antibacterial activity assay were first subjected to Levene’s test for testing the homogeneity of variances and then subjected to one-way analysis of variance (ANOVA) with Scheffé’s method post hoc analysis. Differences were considered statistically significant at P < 0.05.
Results
Sequence analysis of VLR2s
Ten ORF sequences of VLR2 isoforms were amplified and verified, which were classified into two types: long-type and short-type. The longest isoform, which was 1529 bp in ORF length, belongs to long-type and was termed as VLR2-L. While, other isoforms ranged from 309 to 801 bp in ORF length, were grouped into short-type and were termed as VLR2-S1 to VLR2-S9 according to ORF length. The VLR2-L encoded a 453-amino-acid-long protein with a predicted molecular weight of 50.81 kDa and a theoretical isoelectric point of 5.45 (Figure 1). VLR2-L contained LRR_NT, LRRs, LRR_CT and transmembrane domains, which were conserved among invertebrate VLRs. However, no transmembrane domain was found in VLR2-S1 to VLR2-S9, which only contained parts of LRR_NT, LRRs and LRR_CT domains. As shown in Figure 2A, all VLR2 isoforms were located in the same region of E. sinensis genome and were formed via alternative splicing. In addition, the crescent-shaped VLR solenoid structure, considered as a notable feature of VLR and resulting in highly specific binder, was detected in all VLR2 isoforms (Figure 2B). These ten ORF sequences were available at NCBI GenBank under accession numbers OP850588 - OP850597.
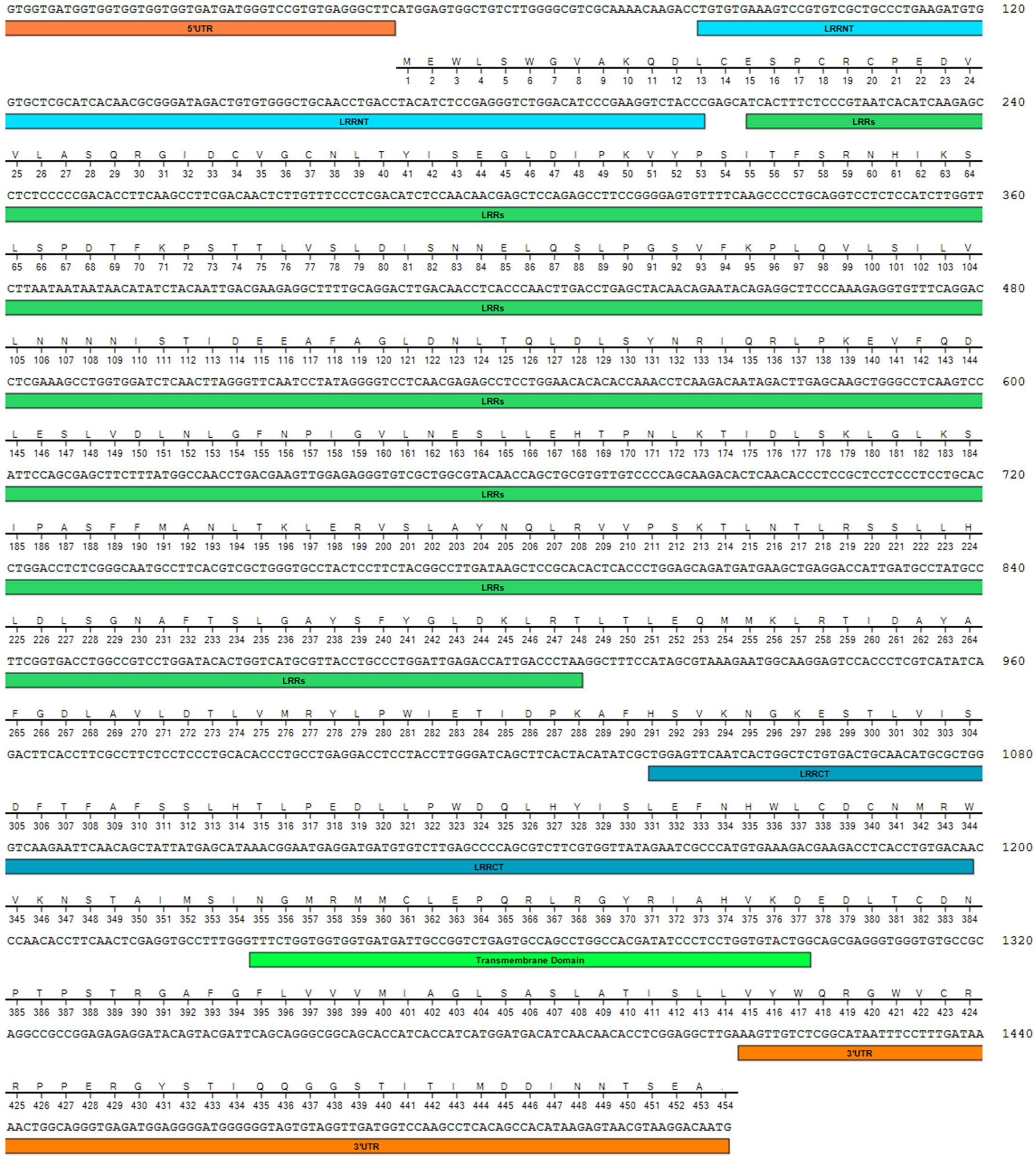
Figure 1 Nucleotide and deduced amino acid sequences of VLR2-L. The 5′-UTR and 3′-UTR were marked with orange, and the LRRNT, LRRs, LRRCT and transmembrane domain were marked with lightblue, green, navyblue and lightgreen respectively.
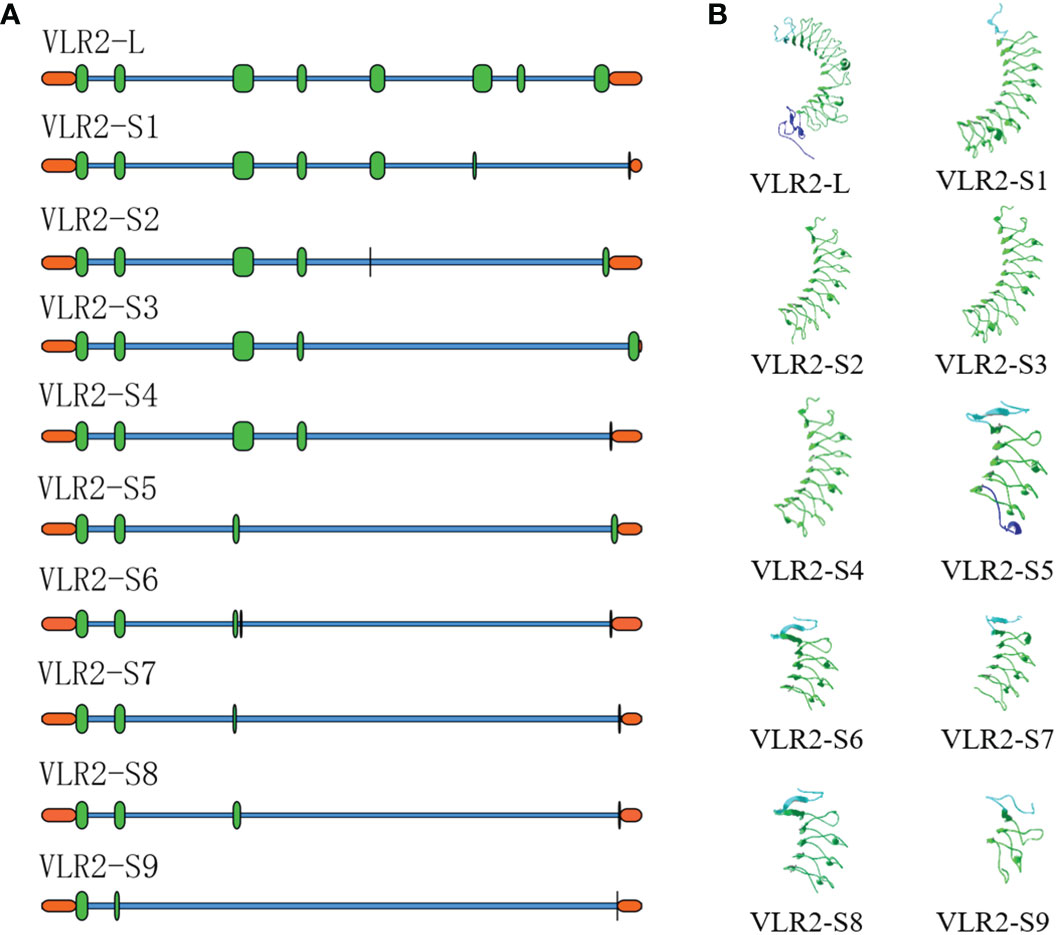
Figure 2 Sequence and structure analysis of VLR2 isoforms. (A). Multiple alignments of the isoforms to Eriocheir sinensis genome. The orange modules indicated UTRs, the green modules indicated exons and the blue lines indicated introns. (B). Three-dimensional structure prediction of VLR2 isoforms.
Expression pattern analysis
A broad spatial expression profile for VLR2-L and VLR2-G (all VLR2 isoforms including VLR2-L) was generated from tissues using Semi-qPCR (Figure 3). The hepatopancreas was considered as primary tissue responsible for both VLR2-L and VLR2-G expression. Interestingly, VLR2-L and VLR2-G performed consistent expression profile among most tissues except the stomach, where VLR2-G showed significantly higher expression than VLR2-L. This phenomenon indicated VLR2 was expressed as the long-type (VLR2-L) predominantly among most tissues except the stomach, while in the stomach, the predominantly-expressed VLR2 type was short-type (VLR2-S1 to VLR2-S9). The raw images of expression pattern analysis were available at Supplementary File 1 (Figures S1, S2).
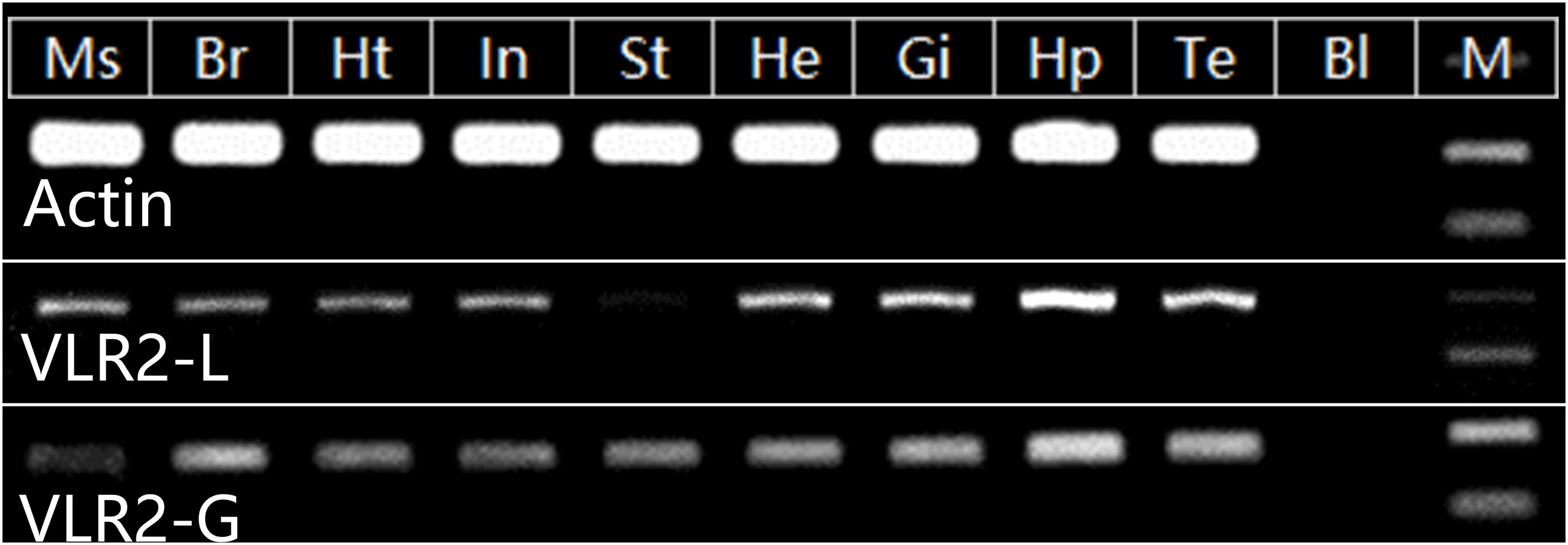
Figure 3 Tissue distribution of VLR2. Ms, muscle; Br, brain; Ht, heart; In, intestine; St, stomach; He, hemocytes; Gi, gill; Hp, hepatopancreas; Te, testis; Bl, blank; M, DL2000 Marker.
Expression patterns in response to bacterial challenge
To obtain preliminary knowledge on the immune function of VLR2s in response to pathogen challenge, the expression profiles of VLR2s in intestines, stomachs, gills and hepatopancreas of E. sinensis post-injection with V. parahaemolyticus and S. aureus were investigated (Figure 4; Supplementary File 2). The results showed that VLR2s responded differently to V. parahaemolyticus and S. aureus. During V. parahaemolyticus challenge, except in the early stage of injection (3 and 6 h post-injection), the transcription of VLR2s in intestines and stomachs decreased slightly, and no significant transcriptional change of VLR2s occurred in other cases, which indicated that VLR2s were insensitive to V. parahaemolyticus. During S. aureus challenge, the transcription of VLR2s tended to decrease at 3 and 6 h post-injection, then started to increase and exceeded the basal level at 12 and 24 h post-injection. Surprisingly, the transcription of VLR2s in stomachs dramatically increased at 24 h post-injection, and the increase of transcription in stomachs was much more than that in other tissues. Above results implied that VLR2s participated in the defense against S. aureus and the stomach might be the vital tissue where VLR2s played roles. In addition, the expression profiles of VLR2-L post-injection were consistent with that of VLR2-G basically, and transcript levels of VLR2-L were slightly lower than VLR2-G generally, suggesting VLR2-L was predominantly-expressed among VLR2s.
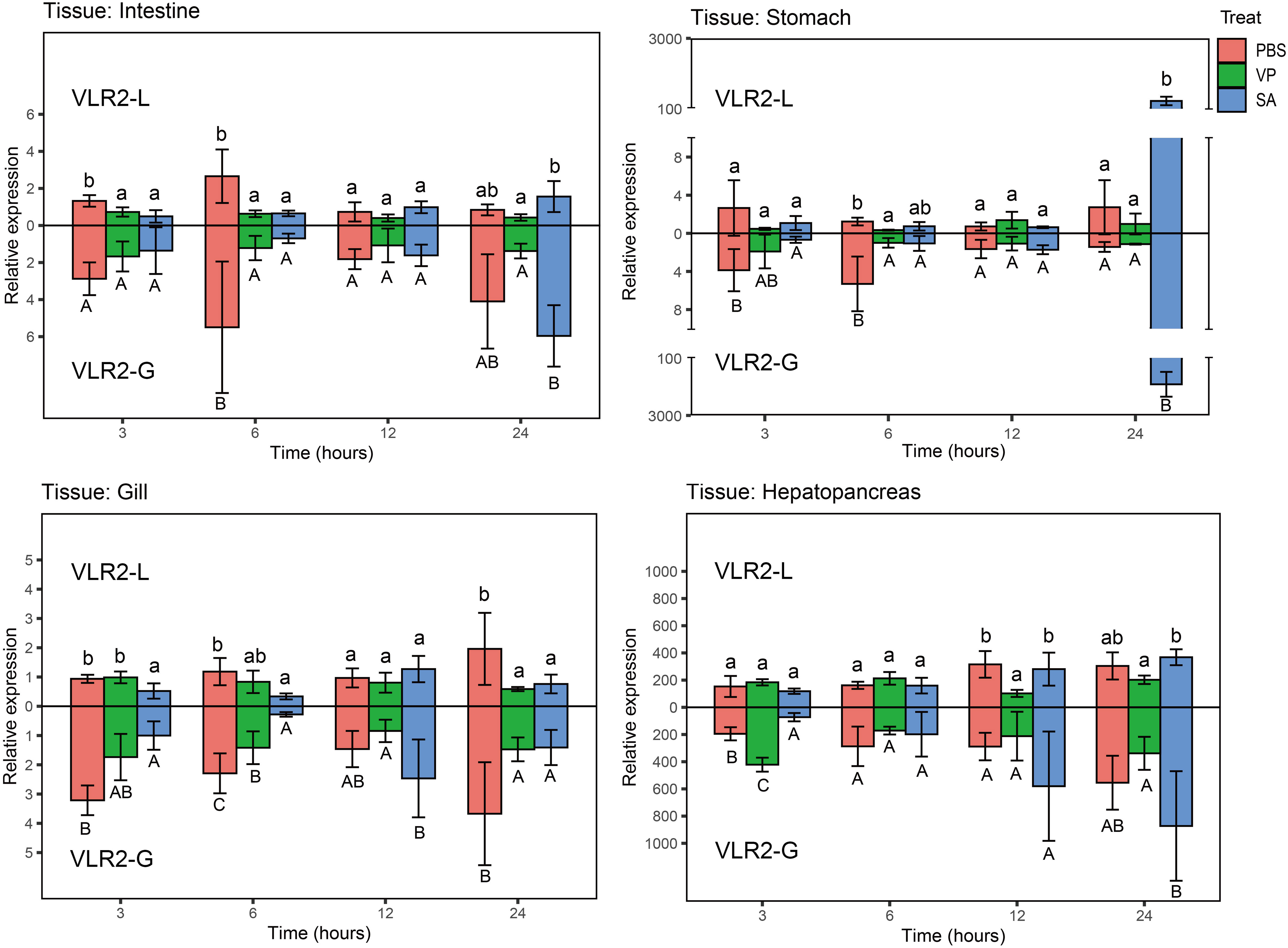
Figure 4 Expression profiles of VLR2 in immune-related tissues of E. sinensis challenged with Vibrio parahaemolyticus and Staphylococcus aureus. Each bar represented the mean ± SD (n = 4). Significant differences of the same group at p < 0.05 (n = 4, ANOVA) in different treatments were indicated by different letters while the same letters indicated no significant difference.
Bacteria-binding activity
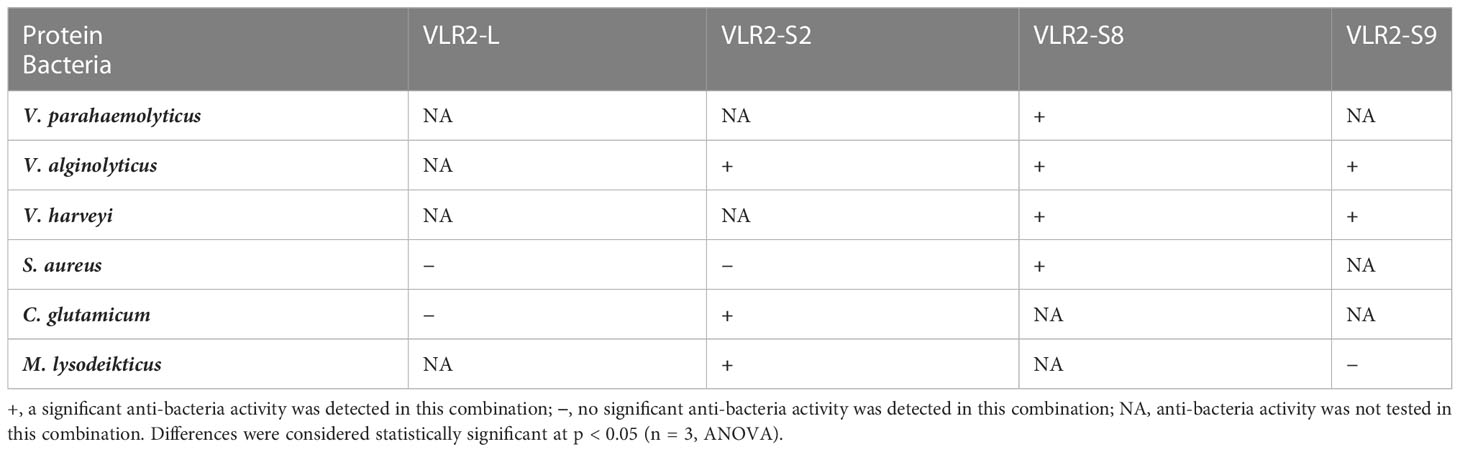
To investigate the specific bacteria-binding ability of VLR2 isoforms, six recombinant VLR2s (rVLR2-L, rVLR2-S2, rVLR2-S3, rVLR2-S5, rVLR2-S8 and rVLR2-S9) were adopted to bind three Gram-positive bacteria (S. aureus, Corynebacterium glutamicum and Micrococcus lysodeikticus) and three Gram-negative bacteria (V. parahaemolyticus, Vibrio alginolyticus and Vibrio harveyi) (Figure 5). The rVLR2-L bound with two Gram-positive bacteria (S. aureus and C. glutamicum) and no Gram-negative bacterium, and rVLR2-S2 bound with three Gram-positive bacteria (S. aureus, C. glutamicum and M. lysodeikticus) and only one Gram-negative bacterium (V. alginolyticus). The rVLR2-S8 bound with three Gram-negative bacteria (V. parahaemolyticus, V. alginolyticus and V. harveyi) and only one Gram-positive bacterium (S. aureus), and rVLR2-S9 bound with three Gram-negative bacteria (V. alginolyticus and V. harveyi) and only one Gram-positive bacterium (M. lysodeikticus). No bacterium was detected to bind with two medium-length rVLR2s, rVLR2-S3 and rVLR2-S5. These results suggested that, the longest rVLR2s (rVLR2-L and rVLR2-S2) and the shortest rVLR2s (rVLR2-S8 and rVLR2-S9) were biased toward binding Gram-negative bacteria and Gram-positive bacteria respectively. We speculated that VLR2 isoforms exhibited the specific bacteria-binding activity might be related with amino acid length. The raw images of bacteria-binding activity were available at Supplementary File 1 (Figures S3–S13).
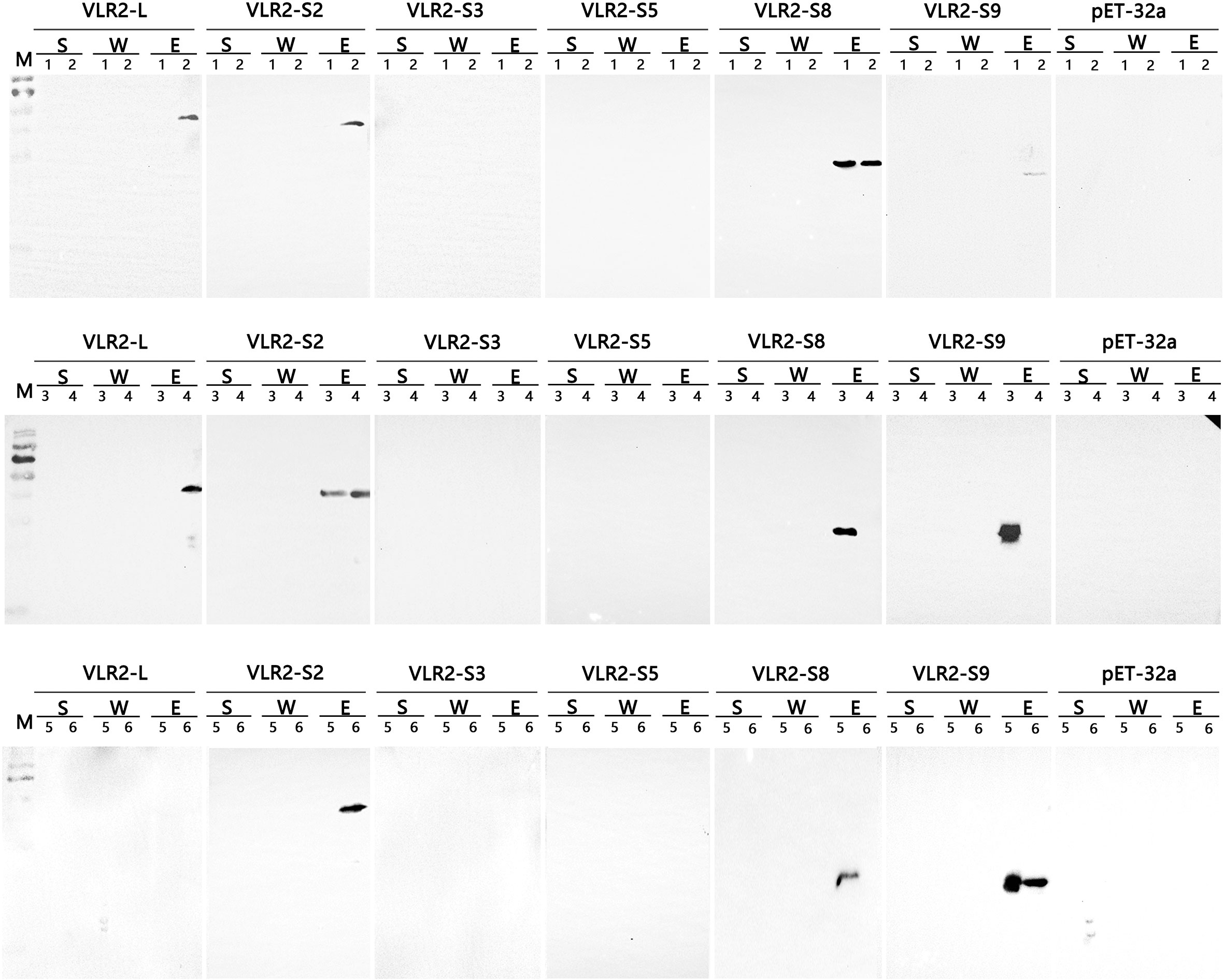
Figure 5 Binding activity of the recombinant VLR2 isoforms to Gram-negative bacteria V. parahaemolyticus (1), Vibrio alginolyticus (3) and Vibrio harveyi (5), and Gram-positive bacteria S. aureus (2), Corynebacterium glutamicum (4) and Micrococcus lysodeikticus (6). S, the supernatants separated by centrifugation after incubation; W, the waste liquid during washing cells; E, the precipitations eluted with PBS after centrifugation and washing.
Anti-bacteria activity
The rVLR2s with specific bacteria-binding activity (rVLR2-L, rVLR2-S2, rVLR2-S8 and rVLR2-S9) were employed to test anti-bacteria activity against bound bacteria (Figure 6; Table 2; Supplementary File 3). Significant anti-bacteria activity of rVLR-S8 against all four bound bacteria (V. parahaemolyticus, V. alginolyticus, V. harveyi and S. aureus) was observed. The rVLR2-S2 and rVLR2-S9 exhibited significant anti-bacteria activity against parts of bound bacteria, the rVLR2-S2 was against V. alginolyticus, C. glutamicum and M. lysodeikticus, and rVLR2-S9 was against V. alginolyticus and V. harveyi. However, no significant anti-bacteria activity of rVLR2-L against bound bacteria was detected.
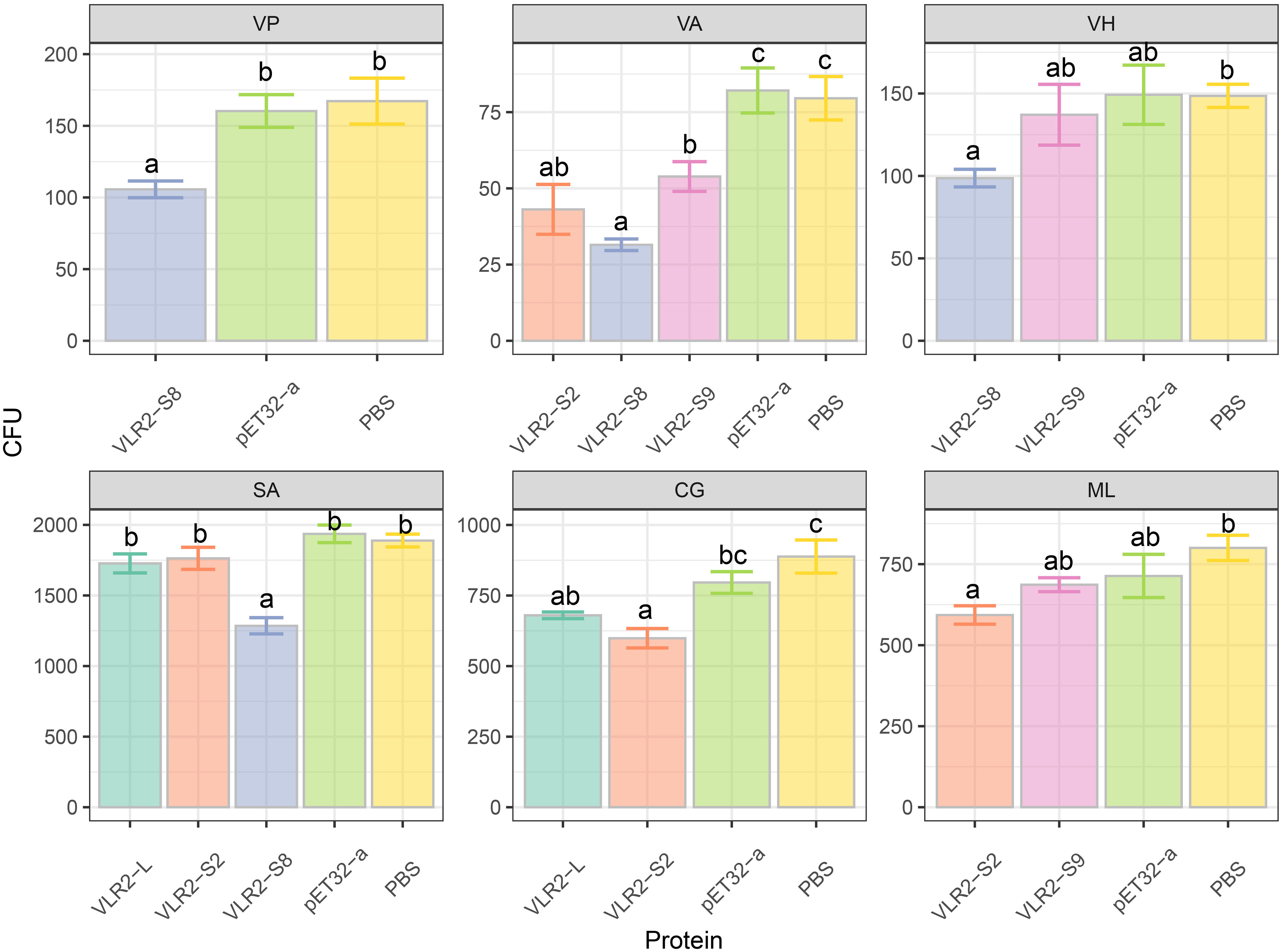
Figure 6 Anti-bacteria activity of the recombinant VLR2 isoforms to bound bacteria. Each bar represented the mean ± SD (n = 3). Significant differences of the same group at p < 0.05 (n = 3, ANOVA) in different treatments were indicated by different letters while the same letters indicated no significant difference.
Discussion
VLR is a key immune factor in adaptive immune system of agnathan vertebrate and is first discovered in L. japonicum (1, 2). Recently, VLR genes or VLR-like genes are also identified in invertebrate and found probably to play a role in the invertebrate immune system (8–10). In this study, a novel VLR gene named VLR2 is identified in E. sinensis. VLR2 has multiple transcripts and performs a certain degree of diversity, which is similar to VLRs of agnathan vertebrate, rather than the reported invertebrate VLRs such as EsVLRA in E. sinensis (8), Aj-VLRA in A. japonicas (9) and LvLRRm in L. vannamei (10).
In agnathan vertebrate, there is a special formation mechanism of VLR diversity, when stimulated, the LRR modules on both sides of gVLR in lymphocytes will randomly insert into gVLR, and gradually form mature VLR with diversity (24). Although no similar assembly mechanism has been found in invertebrate, the alternative splicing can provide the genetic diversity (25), such as the Dscam in Drosophila melanogaster and E. sinensis (25, 26), the Toll-like receptor (TLR) genes TLR1, TLR2 and TLR3 in L. vannamei (27). In this study, the amplified ORFs of VLR2 are aligned with the E. sinensis genome and we find that ten isoforms are formed via alternative splicing. This is the first report of finding the diversity of VLRs in invertebrate, also suggests the mechanism of the diversity may be related to the alternative splicing.
LRR_NT, LRRs, LRR_CT, and transmembrane domains are the main features of VLRs, which present in both agnathan vertebrate and invertebrate (1, 8). Ten VLR2 isoforms in this study also have above structural characteristics, except that VLR2-S1 to VLR2-S9 do not contain transmembrane domains. The lengths of VLR2 isoforms (from 103 to 453 aa) are shorter than that of EsVLRA (799 aa) in E. sinensis (8) and Aj-VLRA (664 aa) in A. japonicus (9), which are closer to Petromyzon marinus VLRs (≤ 417 aa) (28). The three-dimensional structural model prediction shows that all ten VLR2 isoforms own crescent-shaped solenoid structures in LRRs regions (29) and these structures might perform microbial recognition functions (30), which has also been reported in EsVLRA (8).
VLR belongs to antigen receptors, which recognizes pathogens (8) and locates on the surface of cells in immune-related tissues (31). The hepatopancreas is considered as a critical immune tissue of crustaceans and the primary site for the production of immune recognition molecules (32). In this study, the hepatopancreas is the primary tissue responsible for VLR2 expression, which is consistent with the study of EsVLRA (8). Compared with other isoforms, almost no expression of VLR2-L is detected in the stomach. It suggests the immune function of VLR2-L may be different from short-type VLR2s (VLR2-S1 to S9) in E. sinensis.
Strangely, the experimental results of bacterial challenge show that, the transcription of VLR2-L in stomach increase sharply post S. aureus challenge, while remain steady post V. parahaemolyticus challenge. The dramatic expression change in stomach may be related to the function of the stomach as the first line against orally pathogenic infection (33). Furthermore, VLR2-L is detected to bind with S. aureus, rather than V. parahaemolyticus. Above findings imply that VLR2-L could defend against S. aureus specifically, which is comparable to VLRs with specific immune functions in agnathan vertebrate, including the Bacillus anthracis specific VLR in L. japonicum (3) and the NNV specific VLR in E. burgeri (5). In addition, we find that the isoforms with long LRRs regions (VLR2-L and VLR2-S2) tend to bind to Gram-positive bacteria, while the isoforms with short LRRs regions (VLR2-S8 and VLR2-S9) preferentially bind to Gram-positive bacteria. The specific binding mode of VLR2 isoforms is similar to that reported in Dscam, which has specific binding and scavenging ability for different pathogens (25). The relationship between binding mode and LRRs region length of VLR2s is similar to that reported in E. burgeri (5).
In agnathan vertebrate, VLRs bind to various pathogens specifically and exert the antibacterial effect by activating the complement activation cascade and regulating interleukins (1, 4). But, no VLR has been reported to perform anti-bacteria activity in invertebrate. In this study, we find that the VLR2 isoforms exhibit pluralistic anti-bacteria activity. The VLR2-S8 exert significant antibacterial effects on all bound bacteria, while VLR2-S2 and VLR2-S9 only perform significant antibacterial effects on parts of bound bacteria. Above results indicate that the anti-bacteria activities of VLR2 isoforms are pluralistic, which share similarities with VLRs of agnathan vertebrate to a certain extent. These VLR2s could be added to the feed to defend against bacteria of crabs (34).
Conclusions
In conclusion, we identify a novel VLR gene named VLR2 in E. sinensis and this gene can produce ten different isoforms via alternative splicing. The VLR2 isoforms play complex immune functions, which show a certain degree of specificity in the immune system of E. sinensis, and this specificity could be related to the LRRs region lengths of isoforms. This study shows that VLR2 isoforms exert vital immune functions in E. sinensis immune system, and the pathogen-binding receptor diversity based on VLR2 lay the foundation for the study of immune priming. In addition, studying the immune function of VLR2 also provides a new insight into the disease control strategy of crustacean culture.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number OP850588 - OP850597 (https://www.ncbi.nlm.nih.gov/popset/?term=OP850588%20-%20OP850597).
Author contributions
YX: Conceptualization, Methodology, Validation, Investigation, Writing–original draft. YY: Methodology, Validation, Investigation, Writing–review and editing. JZ: Methodology, Investigation. ZC: Resources, Funding acquisition, Writing–review and editing, Supervision. All authors contributed to the article and approved the submitted version.
Funding
The authors are very appreciative of the support by the National Key R&D Program of China (2018YFD0900303), the Ten Thousand Talents Program and the general scientific research project of department of education of Zhejiang Province (IF2022156)
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1105318/full#supplementary-material
References
1. Boehm T, Hirano M, Holland SJ, Das S, Schorpp M, Cooper MD. Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol (2018) 36:19–42. doi: 10.1146/annurev-immunol-042617-053028
2. Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature (2004) 430(6996):174–80. doi: 10.1038/nature02740
3. Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science (2005) 310(5756):1970–3. doi: 10.1126/science.1119420
4. Wu F, Chen L, Liu X, Wang H, Su P, Han Y, et al. Lamprey variable lymphocyte receptors mediate complement-dependent cytotoxicity. J Immunol (2013) 190(3):922–30. doi: 10.4049/jimmunol.1200876
5. Jung JW, JS L, Kim J, Im SP, Kim SW, Lazarte JMS, et al. Characterization of hagfish (Eptatretus burgeri) variable lymphocyte receptor-based antibody and its potential role in the neutralization of nervous necrosis virus. J Immunol (2020) 204(3):718–25. doi: 10.4049/jimmunol.1900675
6. Lee JS, Kim J, Im SP, Kim SW, Lazarte JMS, Jung JW, et al. Generation and characterization of hagfish variable lymphocyte receptor b against glycoprotein of viral hemorrhagic septicemia virus (VHSV). Mol Immunol (2018) 99:30–8. doi: 10.1016/j.molimm.2018.04.006
7. Cao DD, Liao X, Cheng W, Jiang YL, Wang WJ, Li Q, et al. Structure of a variable lymphocyte receptor-like protein from the amphioxus branchiostoma floridae. Sci Rep-Uk (2016) 6. doi: 10.1038/srep19951
8. Liu HR, Song CW, Ning JH, Liu Y, Cui ZX. Identification, functional characterization and the potential role of variable lymphocyte receptor EsVLRA from eriocheir sinensis in response to secondary challenge after vibrio parahaemolyticus vaccine. Fish Shellfish Immunol (2020) 98:201–9. doi: 10.1016/j.fsi.2020.01.011
9. Yang L, Yao F, Ba HZ, Qin T, Luan H, Li ZM, et al. Identification, expression pattern and potential role of variable lymphocyte receptor aj-VLRA from apostichopus japonicus in response to bacterial challenge. Fish Shellfish Immunol (2015) 45(2):221–30. doi: 10.1016/j.fsi.2015.04.010
10. Zhang HF, Li SH, Wang FX, Xiang JH, Li FH. Identification and functional study of an LRR domain containing membrane protein in litopenaeus vannamei. Dev Comp Immunol (2020) 109. doi: 10.1016/j.dci.2020.103713
11. Chen X, Wang J, Yue W, Liu J, Wang C. Hepatopancreas transcriptome analysis of Chinese mitten crab (Eriocheir sinensis) with white hepatopancreas syndrome. Fish Shellfish Immunol (2017) 70:302–7. doi: 10.1016/j.fsi.2017.08.031
12. Ding Z, Yao Y, Zhang F, Wan J, Sun M, Liu H, et al. The first detection of white spot syndrome virus in naturally infected cultured Chinese mitten crabs, eriocheir sinensis in China. J Virol Methods (2015) 220:49–54. doi: 10.1016/j.jviromet.2015.04.011
13. Ma Y, Dai T, Serwadda A, Shen H. Detecting a novel eriocheir sinensis reovirus by reverse transcription loop-mediated isothermal amplification assay. Lett Appl Microbiol (2016) 63(5):363–8. doi: 10.1111/lam.12630
14. Ning M, Yuan M, Liu M, Gao Q, Wei P, Gu W, et al. Characterization of cathepsin d from eriocheir sinensis involved in spiroplasma eriocheiris infection. Dev Comp Immunol (2018) 86:1–8. doi: 10.1016/j.dci.2018.04.018
15. Li C, Wang S, He J. The two NF-kappa b pathways regulating bacterial and WSSV infection of shrimp. Front Immunol (2019) 10:1785. doi: 10.3389/fimmu.2019.01785
16. Wang JJ, Yang B, Wang WL, Song XR, Jiang QF, Qiu LM, et al. The enhanced immune protection in Chinese mitten crab eriocheir sinensis against the second exposure to bacteria aeromonas hydrophila. Front Immunol (2019) 10:2041. doi: 10.3389/fimmu.2019.02041
17. Flajnik MF. A convergent immunological holy trinity of adaptive immunity in lampreys: Discovery of the variable lymphocyte receptors. J Immunol (2018) 201(5):1331–5. doi: 10.4049/jimmunol.1800965
18. Li XJ, Yang L, Li D, Zhu YT, Wang Q, Li WW. Pathogen-specific binding soluble down syndrome cell adhesion molecule (Dscam) regulates phagocytosis via membrane-bound dscam in crab. Front Immunol (2018) 9:801. doi: 10.3389/fimmu.2018.00801
19. Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods (2015) 12(4):357–60. doi: 10.1038/nmeth.3317
20. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform (2013) 14(2):178–92. doi: 10.1093/bib/bbs017
21. Pope EC, Powell A, Roberts EC, Shields RJ, Wardle R, Rowley AF. Enhanced cellular immunity in shrimp (Litopenaeus vannamei) after 'vaccination'. PloS One (2011) 6(6):e20960. doi: 10.1371/journal.pone.0020960
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
23. Liu A, Liu J, Liu F, Huang Y, Wang G, Ye H. Crustacean female sex hormone from the mud crab Scylla paramamosain is highly expressed in prepubertal males and inhibits the development of androgenic gland. Front Physiol (2018) 9:924. doi: 10.3389/fphys.2018.00924
24. Nagawa F, Kishishita N, Shimizu K, Hirose S, Miyoshi M, Nezu J, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol (2007) 8(2):206–13. doi: 10.1038/ni1419
25. Wan ZC, Li D, Li XJ, Zhu YT, Gao TH, Li WW, et al. B52 promotes alternative splicing of dscam in Chinese mitten crab, eriocheir sinensis. Fish Shellfish Immunol (2019) 87:460–9. doi: 10.1016/j.fsi.2019.01.027
26. Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell (2000) 101(6):671–84. doi: 10.1016/S0092-8674(00)80878-8
27. Zhang X, Li G, Jiang H, Li L, Ma J, Li H, et al. Full-length transcriptome analysis of litopenaeus vannamei reveals transcript variants involved in the innate immune system. Fish Shellfish Immunol (2019) 87:346–59. doi: 10.1016/j.fsi.2019.01.023
28. Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA (2005) 102(26):9224–9. doi: 10.1073/pnas.0503792102
29. Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. Slit: An extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes Dev (1990) 4(12A):2169. doi: 10.1101/gad.4.12a.2169
30. Rogozin IB, Iyer LM, Liang LZ, Glazko GV, Liston VG, Pavlov YI, et al. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol (2007) 8(6):647–56. doi: 10.1038/ni1463
31. Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol (2016) 16(9):537–52. doi: 10.1038/nri.2016.77
32. Gross PS, Bartlett TC, Browdy CL, Chapman RW, Warr GW. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the pacific white shrimp, litopenaeus vannamei, and the Atlantic white shrimp, l. setiferus. Dev Comp Immunol (2001) 25(7):565–77. doi: 10.1016/s0145-305x(01)00018-0
33. Soonthornchai W, Chaiyapechara S, Jarayabhand P, Soderhall K, Jiravanichpaisal P. Interaction of vibrio spp. with the inner surface of the digestive tract of penaeus monodon. PloS One (2015) 10(8):e0135783. doi: 10.1371/journal.pone.0135783
Keywords: variable lymphocyte receptor, alternative splicing, specific, diverse, immune system, Eriocheir sinensis
Citation: Xu Y, Yang Y, Zheng J and Cui Z (2023) Alternative splicing derived invertebrate variable lymphocyte receptor displays diversity and specificity in immune system of crab Eriocheir sinensis. Front. Immunol. 13:1105318. doi: 10.3389/fimmu.2022.1105318
Received: 22 November 2022; Accepted: 22 December 2022;
Published: 14 March 2023.
Edited by:
Mengqiang Wang, Ocean University of China, ChinaReviewed by:
Jianjian Lv, Yellow Sea Fisheries Research Institute (CAFS), ChinaYueling Zhang, Shantou University, China
Copyright © 2023 Xu, Yang, Zheng and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoxia Cui, Y3Vpemhhb3hpYUBuYnUuZWR1LmNu
 Yuanfeng Xu
Yuanfeng Xu Yanan Yang
Yanan Yang Jinbin Zheng1
Jinbin Zheng1 Zhaoxia Cui
Zhaoxia Cui